Abstract
Trikatu Churna (TC) comprising Zingiber officinale rhizome, Piper longum, and Piper nigrum fruit, is effective in treating liver diseases and has high nutraceutical values. However, the efficacy of TC in treating alcoholic liver disease (ALD) and its mechanism remain largely unknown. This study evaluated the hepatoprotective effects of different doses of TC as well as to identify the bioactive components and determine their mechanism of action against ethanol-induced ALD. A compound-target network analysis model of TC was established to identify its potential bioactive compounds and pathways that might regulate its hepatoprotective effects. Further, in-vivo studies were performed to validate the potential of TC (200 mg/kg and 400 mg/kg b.w.) in the treatment and management of ALD. The study revealed that both the dosages of TC demonstrate significant (p > 0.0001) hepatoprotective effects by improving body weight, total bilirubin, serum glutamic oxaloacetic transaminase (SGOT), serum glutamic pyruvic transaminase (SGPT), serum alkaline phosphate (ALP), total cholesterol, total protein, globulin, albumin, and liver morphology. The High-performance thin-layer chromatography (HPTLC) fingerprinting of TC showed the presence of piperine. Network pharmacology identifies the role of TC in regulating various signaling processes including Advanced glycation end products-receptor for advanced glycation end products (AGE-RAGE), Hypoxia-inducible factors (HIF-1), Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-Kappa B), and Phosphatidylinositol 3-kinase/protein kinase B (PI3K/Akt) signaling to exert its anti-inflammatory, antioxidant and anti-apoptotic role in managing ALD. Based on the bioinformatics analysis, some of the key targets of TC were found to be Prostaglandin-Endoperoxide Synthase 2 (PTGS2) or Cyclooxygenase-2 (COX-2), Sirtuin 1 (SRT1), and caspase-3. These effects may serve as a novel therapeutic option for the treatment of ALD. These preclinical validation studies for the ethnopharmacological potential of TC in ALD treatment further paved the way for researchers to perform next-level translational and clinical studies. Further, in-depth experimental studies for the validation of these bioinformatics-based results will give a clearer picture of mechanisms.
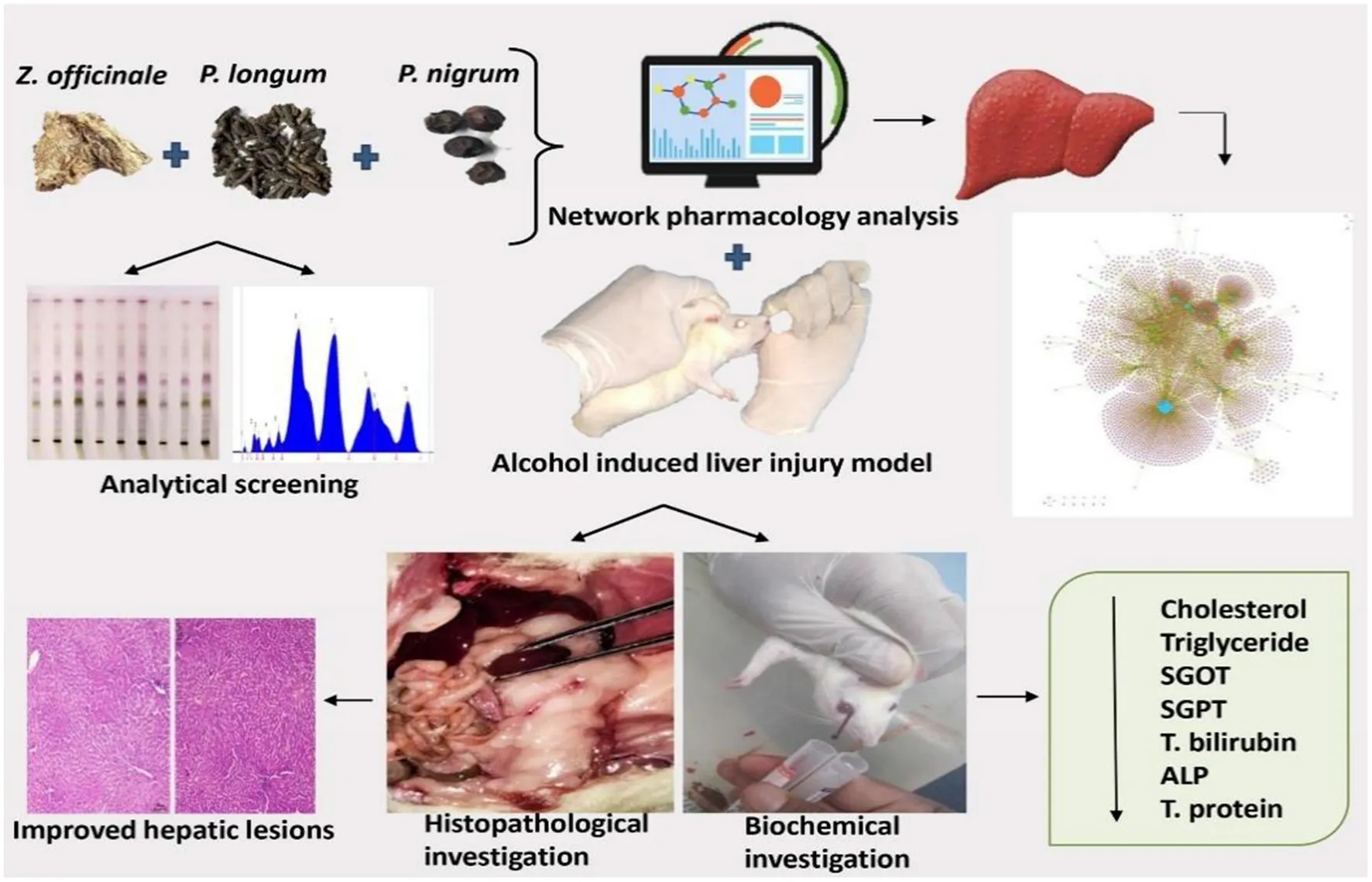
Introduction
Globally, liver diseases and cirrhosis account for 23.59% of deaths, placing them 27th among all of the leading causes of death (1). In India alone, liver disease and cirrhosis make up to 2.74% of all causes of death (1). Globally, two main types of liver disease exist; alcoholic liver disease (ALD) and non-alcoholic fatty liver disease (NAFLD) (2). In India, alcoholism is the leading cause of chronic liver disease (3). According to WHO estimates, 140 million people worldwide suffer from alcohol dependence, and the disease is responsible for 3.8% of global mortality and 4.6% of disability-adjusted life years (DALYs) (4). ALD is a consequence of liver damage caused by acetaldehyde build-up and oxidative stress produced by alcohol consumption (5, 6). It includes manifestations of liver damage, such as fatty liver, alcoholism, or chronic hepatitis with cirrhosis or fibrosis (5, 6).
Further, no reliable hepatoprotective agent has been found to stimulate the liver's functions efficiently and protect against liver injuries with less toxicity (7). Plant-based hepatoprotective remedies to treat liver disorders have therefore received adequate attention on a global scale (8). A herbal formula like Trikatu Churna (TC), aka Triushna and Vayosh in Ayurveda, is derived from fine (#80) powder of three popular herbs, namely Zingiber officinale (Z. officinale) rhizome, Piper longum (P. longum) and Piper nigrum (P. nigrum) fruit (7). These three herbs are in use for thousands of years in the Indian traditional medical system to manage liver diseases (7).
These herbs have a similar bitter taste, but they are used differently. TC herbs are used in folk medicine for treating fever, asthma, diabetes, nasal diseases, obesity, anorexia, respiratory diseases, skin diseases, jaundice, and alcohol indigestion (9, 10). Several scientific reports claim that TC has anti-helminthic, anti-bacterial, hypo-lipidemic, anti-inflammatory, bioenhancing, anti-arthritic, immunomodulatory, adaptogenic, anti-anorectic, antioxidant, and antitumor properties (11–21). As part of describing the treatment of liver disease, Ayurvedic texts mention numerous formulations, 50% of which contain TC or its constituents (22). Of all the three ingredients, P. longum and P. nigrum contain an alkaloid piperine as the chief constituent (22), which has shown significant hepatoprotective effects against tert-butyl hydroperoxide and carbon tetrachloride (CCl4)-induced hepatotoxicity in the intoxicated mice (23). In another study, ethanol extract of TC exhibits hepatoprotective activity against CCl4-induced hepatotoxicity (24). Therefore, all these aforesaid reports suggest promising hepatoprotective properties of TC.
Despite the fact that TC is used in traditional medicine to treat several chronic liver diseases, no scientific evidence has yet been established that how it will alleviate ALD. As part of our study, we applied network pharmacology approaches to further understand the TC mechanism of action and active potential components. The emerging field of network pharmacology integrates concepts from system biology and computational biology to study, predict and illustrate the interactions between numerous components and targets in herbs at a systemic and molecular level (25). The potential of this approach to explore the multi-targeting and synergistic rationale of Ayurvedic herbs is well established (26, 27). The networks show the interactions between TC components and their molecular targets with liver diseases. This pioneering research might provide new insights into the pharmacodynamics of Ayurvedic drugs like TC and help to identify new leads and targets related to various diseases. It is an effective tool for analyzing in silico Ayurvedic medicine systems to determine the scientific rationale, active ingredients, and reaction mechanisms, but also requires further experimental exploration (28).
Therefore, for the first time, Ayurvedic claims of TC hepatoprotective effects on chronic liver injury caused by ethanol have been validated experimentally by using two doses of TC in Wistar rats and network pharmacology.
Materials and methods
Network pharmacological analysis
Phytochemical dataset and active compound screening
The phytochemicals present in each constituent herb were compiled from six different database sources: CMAUP (29), Duke (30), IMPPAT (31), NPASS (32), PCIDB and TCM-mesh (33). The phytochemicals were checked for their chemical information from the PubChem database (http://www.ncbi.nlm.nih.gov/), and duplicate entries were removed to obtain a unique set of phytochemicals. Secondly, ADME parameters of the phytochemicals were acquired using FAFdrugs4 (http://fafdrugs4.mti.univ-paris-diderot.fr) (34). FAFdrugs4 is a free ADME-Tox filtering tool that is conductive in screening our compounds. The phytochemicals were screened on the basis of a “drug-like soft” filter and referred to as potentially-active compounds (PACs) of TC (Figure 1).
Figure 1
Candidate protein targets of TC
The potential human protein targets of phytochemicals were retrieved from the STITCH database (35). For the selection, only those compound protein targets within the score ≥700 were compiled and used for constructing the interaction network to study their relationship. To facilitate subsequent analysis, the target names were converted to UniProt IDs using the UniProt database (https://www.uniprot.org), a universal protein database.
Liver disease-associated genes
Liver disease-associated genes were collected from DisGeNET database (36). Herein, 38 liver disease terms were used and annotated using Medical Subject Headings (MeSH) or UMLS vocabularies (Supplementary Table S1). Only datasets corresponding to the curated gene-disease pairs were used in the present study. The target genes were labeled by using their Uniprot IDs.
Network construction and analysis
The network construction, visualization, and investigation were performed using an open-source software of Cytoscape (version 3.0) (37). For the network analysis, the Network analyzer utility of the Cytoscape was used.
Protein-protein interactions (PPIs) are central to the proper functioning of the most basic molecular mechanisms underlying cellular life and are often disturbed in disease states. To elucidate the regulatory effect of TC on PPIs associated with ALD, a protein interaction network (PIN) was derived from STRING, within the confidence, score range ≥900 (38). The Molecular Complex Detection (MCODE) algorithm was used to cluster the PIN and organize proteins into various modules to examine module-regulation of TC as per the mentioned approach (39). The MCODE algorithm detects densely connected regions among the protein interaction networks and offers a directed mode for fine-tuning of clusters among the PINs. It also provides a controlled mode for fine-tuning of the clusters to examine their interconnectivity in detail (40). Gene ontology (GO) based annotation as well as protein-class annotation of the protein targets among the networks were carried out using PANTHER (41). KEGG-pathway enrichment analysis was done using Database for Annotation, Visualization, and Integrated Discovery (DAVID) (42).
Experimental analysis
The plan of the experimental study is demonstrated in Figure 2.
Figure 2
Chemicals
We used silymarin, ethanol, saturated picric acid, and staining reagent hematoxylin and eosin (H&E) that were of the standard protocol and analytical grade (Sigma, USA). Biochemical parameters were analyzed by using an automated analyzer and diagnostic kits (ERBA, Czech Republic).
Collection and authentication of herbs
The crude herbs such as raw tuber/rhizome of Z. officinale and fruits of P. longum and P. nigrum were procured from the local market of Jammu, Jammu, and Kashmir, India and authenticated from the botanical institute of Savitribai Phule Pune University, Pune, India with specimen voucher number DNM/19. The samples were identified and authenticated using floras, taxonomic literature, macroscopic (external structure of the plant)/microscopic (palisade ratio, stomatal index, cell structure, epidermis, vascular bundle arrangement) characters. Their nomenclature is done as per IPNI (theplantlist.com).
Preparation and physicochemical analysis of testing formulation
The herbs were ground separately (#80) and blended in equal proportions to produce a homogeneous mixture. The prepared formulation was weighed and packed in an air-tight container under aseptic conditions (43). Physicochemical analysis of each herb and TC was done according to the Ayurvedic Pharmacopeia of India (API) guidelines. Physicochemical tests were performed to determine the foreign matter, moisture content, total ash value, acid insoluble ash, alcohol soluble extractive, and water-soluble extractive value (44). High-performance thin-layer chromatography (HPTLC) was performed to standardize and fingerprint TC. Based on the existing knowledge, chromatographic separation was carried out on HPTLC plates precoated with silica gel 60 F254 (E. Merck) of 0.2 mm thickness supported with aluminum sheet support. Plates were developed in a glass twin trough chamber (CAMAG automatic TLC sampler 4) that was pre-saturated with the mobile phase. The experimental condition was maintained at 20 ± 2°C. After derivatizing plates with anisaldehyde sulphuric acid reagent and photo documentation with CAMAG Reprostar 3 photo documentation platform at 550 nm, piperine could be detected. The solvent system consisting of toluene-ethyl acetate-glacial acetic acid (8:2:0.1, v/v/v) was used to resolve piperine from the matrix of TC (45–47). Densitometric determinations were performed in fluorescence mode by CAMAG TLC scanner.
Dosage fixation
For the present study, the doses of TC were calculated by extrapolating the maximum tolerated dose from existing scientific evidence on the toxicity of TC. TC at 2,000 mg/kg body weight once orally and 5, 50, and 300 mg/kg body weight once daily for 28 days were well tolerated by the Charles Foster rats (both male and female) (48). The safety of consuming the herb was also a great concern despite its potential, so all these evidence are crucial in justifying its safety. Also, a study on an ethanol extract of TC at an oral dose of 150 mg/kg exhibited a significant protective effect by lowering serum levels of glutamic oxaloacetic transaminase, glutamic pyruvic transaminase, alkaline phosphatase, and total bilirubin (24). By considering all these data from the previous studies, the two different dose groups of 200 and 400 mg/kg orally were considered for evaluation of the hepatoprotective activity.
Experimental design
An animal study was conducted at Agharkar research institute, Pune, India, according to the recommendations and approval of the Institutional Animal Ethical Committee with approval number ARI/IAEC/2019/11. A total of 24 male Wistar rats 6–8 weeks old (180–200 g) were procured and housed under standard environmental conditions (normal daily light cycle, 40–60% relative humidity and 25°C temperature). Animals were fed on a standard chow diet and provided with purified water ad libitum. In addition, animals were marked with a saturated picric acid solution in water for proper identification.
Ethanol-induced hepatotoxicity model in rats
Hepatotoxicity was induced by oral administration of ethanol once daily for 28 days. As tolerance developed, ethanol dosage was gradually increased in order to maintain blood alcohol levels between 150 and 300 mg/dl. The starting dose was 8 g/kg per day for 15 days and then increased gradually by 1 g/kg per day for the next 7 days and then final dose for the next 7 days and the rest of the study was 16 g/kg per day. Following 28 days, six rats in total were randomly selected and 2 ml of blood was collected under light anesthesia from the retro-orbital space of the eye for biochemical analysis: serum glutamic oxaloacetic transaminase (SGOT), serum glutamic pyruvic transaminase (SGPT), serum alkaline phosphate (ALP), total bilirubin, cholesterol, triglyceride, total protein, albumin, and globulin levels to confirm hepatotoxicity. After confirmation of toxicity, animals were randomly divided into four groups (each group containing 6 rats): control group (no treatment, only ethanol), Test drug 1 group (TC 200 mg/kg + ethanol), Test drug 2 group (TC 400 mg/kg + ethanol), Standard group (Silymarin 100 mg/kg + ethanol). Rats were weighed and given following treatment along with a standard diet for 28 days. A suspension of TC was made with distilled water for oral administration to the animals. Even after induction of hepatotoxicity, in the dosing schedule, ethanol is not stopped and is given to the control group and treatment groups along with the treatment to maintain the state of hepatotoxicity as it is known that liver has tendency to regenerate by itself also. Total duration of study is for 56 days (28 days for induction of hepatotoxicity and 28 days for treatment schedule).
Biochemical investigation and histopathology
On the 29th day of treatment, under light anesthesia, 2 ml of the blood was conveniently collected from the retro-orbital space of each rat and sent for biochemical investigations. The serum from the blood was separated by centrifuging it at 3,000 rpm, 4°C for 10 min and used for measurement of the various biochemical markers SGOT, SGPT, ALP, total bilirubin, cholesterol, triglycerides, total protein, albumin, and globulin. Furthermore, rats of each group were sacrificed by giving a 50 mg/kg dose of pentobarbital intraperitoneally, and then the dissection was done. The slices of the left liver lobe and left kidney (from six animals of each group) were removed and fixed in 10% formalin for 24 h. After 24 h, tissues were embedded in paraffin and 5–6 μm sections were routinely stained with hematoxylin and eosin (H&E) and assessed in a light microscope. All alterations from the normal structure were registered.
Statistical analysis
Results were presented as mean ± SEM, the difference between the groups was statistically determined by one-way analysis of variance (ANOVA) followed by the Tukey-Kramer Multiple Comparisons Test. The comparison of weight before treatment but after induction of disease (BT) and after treatment at end of the study (AT) was statistically determined by paired t-test with a level of significance at P < 0.05. The level of significance was noted and interpreted accordingly.
Results
Network pharmacology-based analysis
For further exploring the global effects of TC and its potential role in managing ALD, a network pharmacology study was performed.
Phytochemical dataset of TC
A phytochemical search among the databases (mentioned in the Material and methods section) resulted in the compilation of 1,127 PCs in TC (Figure 3A).
Figure 3
The information of each herb of TC and their corresponding PCs was used to construct herb-phytochemical (H-PC) network (Figure 3B). The herb-phytochemical association along with the chemical information of each PC is given in Supplementary Table S1.
ADME analysis of the PCs using FAFdrugs4 leads to the selection of 418 PCs (referred to as PACs) when screened on the basis of a “drug-like soft” filter (Supplementary Table S1).
TC-protein target network
Human target proteins of TC were compiled from the STITCH database, which were further used to construct TC-protein target (T-PT) network. For the STITCH confidence score ≥700, 2,765 protein targets were obtained for 198 PCs, resulting in the T-PT network of size 2,963 nodes and 7,055 edges (Supplementary Figure S1; Supplementary Table S2).
A sub-network specific to PACs was derived from the T-PT network, to highlight the regulatory PACs and their specific human protein targets. This resulted in a sub-network of size of 1,100 nodes and 1,967 edges containing 95 PACs and their 1,005 protein targets (Figure 4A; Supplementary Table S3).
Figure 4
The protein targets of PACs majorly belonged to the class of metabolite interconversion enzyme, transmembrane signal receptor, and protein modifying enzymes (Figure 4B).
To decipher the biological classification of protein targets of PACs, gene ontology analysis was also carried out. As shown in Figures 4C,D, GO analysis indicated that the enriched molecular function corresponds to binding (GO:0005488) and catalytic activity (GO:0003824). For the biological process, the proteins were involved in the cellular process (GO:0009987), biological regulation (GO:0065007), response to stimulus (GO:0050896), metabolic process (GO:0008152), signaling (GO:0023052), and immune system processes (GO:0002376). GO-based annotation and the protein class association are detailed in Supplementary Table S3.
Liver disease-gene association network
Based on 38 liver disease terms and their associated curated gene-disease DisGeNET data, 516 unique proteins were harvested. The proteins and their associated liver disease term were used for obtaining the liver-specific disease gene (LD-G) network. In curated gene-disease pairs, only 24 of 38 liver disease terms could be obtained, which resulted in an LD-G network of size 539 nodes (515 proteins and 24 disease terms) and 1,308 edges (Supplementary Figure S2; Supplementary Table S4).
Relationship between TC targets and ALD genes
To explore ALD in detail, all the protein targets linked with ALD were derived from the LD-G network. The proteins were checked for their regulatory phytochemicals in the T-PT network and a sub-network specific to ALD was derived, which is referred to as ALD-PT network. The network consisted of 51 nodes and 63 edges, demonstrating the information of 11 ALD proteins (Figure 5A) and 40 PCs of TC (Figure 5B; Supplementary Table S5).
Figure 5
In addition to managing ALD, these 40 PCs also have the ability to regulate other liver diseases. This can be seen in Figure 5C, where 453 protein targets of these 40 PCs are linked with 24 liver disease terms. When checked for the pathway association of these proteins, the proteins were playing a major role in the pathways associated with cancer and signaling processes like advanced glycation end products (AGE-RAGE) signaling, Hypoxia-inducible factors (HIF-1) signaling pathway, interlukin-17 (IL-17) signaling, chemokine, and cytokine signaling, apoptosis signaling, Phosphatidylinositol 3-kinase—protein kinase B (PI3k-Akt) signaling pathway, etc (Figure 5D; Supplementary Table S6). Pathway analysis also highlighted the direct role of 26 proteins in the ALD disease pathway. The mapping of these proteins onto the ALD-disease pathway is given in Figure 6.
Figure 6
Among the disease association network, liver cirrhosis and liver carcinoma were identified as the major disease targets. It is interesting to note that 15 of these 40 PCs belong to PACs, further highlighting their role as lead molecules for future drug-discovery procedures against liver complications. These 15 PACs and their regulators can be checked in the ALD-PT network. The network topology attributes of the ALD-PT network were analyzed for their degree centrality measure, betweenness and closeness centrality. Among the network P01375 and P04040 were highlighted as nodes with high-degree centrality value of 19 each, reflecting high neighborhood connectivity of these nodes as degree centrality is a measure of number of connections a node of interest possesses. P01375 is a tumor necrosis factor encoded by TNFA gene, where TNF shows multifunctional properties for the liver heath including liver regeneration and inflammation, hepatocyte apoptosis and necroptosis, etc. (49). The other node among the network with highest degree value is P04040, a catalase protein encoded by CAT gene which is commonly expressed in liver. When checked for the betweenness centrality, a measure that quantifies the role of node to control the flow of information among the network, Q03181 and P01375 were showing high betweenness of 1 and 0.49 respectively (50, 51). Q03181 is a Peroxisome proliferator-activated receptor delta and play a significant role in liver health by acting as a major class of metabolic regulators (52).
Closeness centrality takes into account the average distance of all shortest path between a node to all other nodes among the network, where high closeness indicates high proximity to all other nodes. Among the ALD-PT network, Q03181 and C_0013 (1-butanol) were showing high closeness centrality values of 1.0 and 0.66, respectively (53). The degree, betweenness and closeness centrality values of each node among the ALD-PT network is provided in Supplementary Table S5. The information on multi-targeting PCs, including PACs in managing ALD among the ALD-PT network, obtained from network analysis has been shown in Table 1.
Table 1
| Serial no. | Phytochemical ID | Name of the compound | PAC (Yes or No) | No. of ALD targets | Associated plant | Part of the plant |
|---|---|---|---|---|---|---|
| C_0241 | Zinc | N | 8 | Z. officinale | Rhizome | |
| P. nigrum, P. longum | Fruit | |||||
| C_0032 | 3-(1-Methylpyrrolidin-2-yl)pyridine | Y | 3 | P. nigrum | Fruit | |
| C_0237 | Iron | N | 3 | Z. officinale | Rhizome | |
| P. nigrum, P. longum | Fruit | |||||
| C_0005 | Acetaldehyde | N | 3 | Z. officinale | Rhizome | |
| C_0598 | Quercetin | Y | 3 | P. nigrum | Fruit | |
| Z. officinale | Rhizome | |||||
| C_0324 | Nicotine | N | 3 | P. nigrum | Fruit | |
| C_0600 | Linoleic acid | N | 2 | P. nigrum, P. longum | Fruit | |
| Z. officinale | Rhizome | |||||
| C_0242 | Silicon dioxide | N | 2 | P. nigrum | Fruit | |
| C_0065 | Cysteine | N | 2 | Z. officinale | Rhizome | |
| P. nigrum | Fruit | |||||
| C_0474 | Oleic acid | N | 2 | P. nigrum, P. longum | Fruit | |
| Z. officinale | Rhizome | |||||
| C_0028 | Melatonin | Y | 2 | P. nigrum | Fruit | |
| C_0059 | 9,12-Octadecadienoic acid | N | 2 | P. longum, P. nigrum | Fruit |
The information of multi-targeting PCs, including PACs in managing ALD among the ALD-PT network.
TC regulated human PIN (TRH-PIN) of ALD
Toward elucidating the global effects of the TC in managing ALD, a PIN network of ALD-specific protein targets of TC was constructed and analyzed in detail. For this, high-confidence protein-protein interaction data of 11 proteins among the ALD-PT network was obtained from STRING via restricting the confidence score ≥900. This resulted in the compilation of 569 protein interactors of these 11 proteins leading to the network of size 580 nodes and 7,947 edges (Figure 7, TRH-PIN). The protein-protein interaction data of TRH-PIN along with the confidence score can be checked in Supplementary Table S7.
Figure 7
The information of topologically important nodes was also inferred for the TRH-PIN. Network analysis highlighted the role of P0CG48 and Q04206 as hub nodes among the PIN, owing to their high connectivity (or high degree value) with ability to regulate 207 and 194 other proteins, respectively. P0CG48 is a human Polyubiquitin-C protein encoded by UBC gene which is highly expressed in the liver and dysregulation of this gene is linked to defective proliferation of hepatocytes (54). The other hub-protein, Q04206 is a Transcription factor p65 encoded by RELA (NFKB3) gene responsible for master regulation of inflammatory and cell death processes with central link to the hepatocellular injury and liver fibrosis (51).
As already discussed, betweenness centrality measures the information-flow among the network. The nodes with high betweenness represent critical points of the network and are referred to as bottlenecks (55). Among the TRH-PIN, P0CG48 appeared as bottleneck with high betweenness centrality value of 0.10. Thus suggests the high relevance of this node in terms of its connectivity and control over the communication among the TRH-PIN. The degree and betweenness centrality values of each node among the TRH-PIN can be checked in Supplementary Table S7.
To understand the modular architecture of the TRH-PIN of ALD, the network was subjected to module identification using MCODE. MCODE returned six highly organized and connected topological clusters (Figure 8; Table 2). To access the biological role of clusters, GO-based annotation of each cluster was carried out. The GO-based analysis highlighted the major association of proteins in molecular processes like binding (GO:0005488) and transcription regulator activity (GO:0140110). While the major biological processes were cellular processes (GO:0009987) and cellular anatomical entities (GO:0110165). The cluster proteins were showing their involvement among the pathways like PI3K-Akt signaling (hsa04151), Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kappa B) signaling (hsa04064), and pathways in cancer (hsa05200) especially hepatocellular carcinoma (hsa05225). A detailed description of the protein composition of each cluster and their associated GO annotation is given in Supplementary Table S8.
Figure 8
Table 2
| MCODE score | Nodes | Edges | Protein targets of TC | Regulatory phytochemicals | Regulatory PACs | |
|---|---|---|---|---|---|---|
| Cluster-1 | 33.412 | 69 | 1136 | 20 | 31 | 11 |
| Cluster-2 | 25.935 | 32 | 402 | 16 | 38 | 11 |
| Cluster-3 | 11.677 | 94 | 543 | 41 | 75 | 36 |
| Cluster-4 | 10.298 | 48 | 242 | 25 | 36 | 15 |
| Cluster-5 | 7.6 | 76 | 285 | 37 | 60 | 23 |
| Cluster-6 | 4.16 | 51 | 104 | 26 | 40 | 14 |
MCODE cluster analysis of TRH-PIN of ALD with the information of regulatory phytochemicals of each cluster.
The regulatory phytochemicals of each cluster were also checked via searching for cluster proteins among the T-PT network. As a result, the regulatory phytochemicals in each cluster can be checked in Table 2. The multi-module regulatory potential of the PACs was also identified by searching the participation of protein targets of PACs among the clusters. This led to the identification of PACs like C_0032, C_0598 and C_0028 toward directly regulating the proteins of all the identified six-clusters, thus highlighting their high multi-module regulatory potential.
Physicochemical analysis
During powder preparation, the average mass yield% of Z. officinale, P. longum, and P. nigrum was 64.50, 63.20, and 88.57%, respectively. Organoleptic and physicochemical analyses of Z. officinale, P. longum, P. nigrum powder, and TC are recorded in Tables 3, 4, respectively. Results of physicochemical analysis of TC come under permissible values of pharmacopeial standards. Fingerprinting and monographs of HPTLC data of TC in triplicates were stipulated in Figure 9 and Table 5. Results indicate the presence of complex compounds. Piperine was found in TC.
Table 3
| Organoleptic tests | Z. officinale powder | P. longum powder | P. nigrum powder | TC |
|---|---|---|---|---|
| Sound | Not applicable | Not applicable | Not applicable | Not applicable |
| Touch | Rough | Rough | Rough | Rough |
| Color | Light yellow | Greenish black | Blackish brown | Light yellowish-brown |
| Taste | Pungent | Strongly pungent | Pungent | Astringent |
| Smell | Aromatic | Aromatic | Characteristic faint odor | Characteristic |
Organoleptic analysis of prepared herbal powder.
Table 4
| Name of test | Z. officinale powder | API standards | P. longum powder | API standards | P. nigrum powder | API standards | TC |
|---|---|---|---|---|---|---|---|
| Moisture content | 4% | NMT 5% | 3.8% | NMT 5% | 3.8% | NMT 5% | 4% |
| Foreign matter | NIL | NMT 2% | NIL | NMT 2% | NIL | NMT 2% | – |
| Total ash | 4.46% | NMT 6% | 6.32% | NMT 7% | 4.10% | NMT 5% | 2.47% |
| Acid insoluble ash | 0.83% | NMT 1.5% | 0.21% | NMT 0.5% | 0.30% | NMT 0.5% | 0.61% |
| Alcohol soluble extractive | 6.21% | NLT 3% | 8.63% | NLT 5% | 10.25% | NLT 6% | 15.70% |
| Water soluble extractive | 14.17% | NLT 10% | 10.48% | NLT 7% | 9.36% | NLT 6% | 35.29% |
Physicochemical analysis of prepared herbal powder.
NMT, not more than; NLT, not less than; API, Ayurvedic Pharmacopeia of India.
Figure 9
Table 5
| Batch | Peak | Start position (Rf) | Start height (AU) | MAX position (Rf) | Max height (AU) | Max% | End position (Rf) | End height (AU) | Area (AU) | Area% |
|---|---|---|---|---|---|---|---|---|---|---|
| A | 1 | 0.03 | 0.8 | 0.04 | 19.5 | 1.34 | 0.06 | 0 | 144 | 0.22 |
| 2 | 0.08 | 0.7 | 0.1 | 56 | 3.84 | 0.12 | 35.5 | 781.7 | 1.17 | |
| 3 | 0.12 | 36.5 | 0.13 | 45.8 | 3.15 | 0.15 | 3.6 | 527 | 0.79 | |
| 4 | 0.15 | 3.9 | 0.19 | 44.6 | 3.07 | 0.22 | 13.4 | 924.7 | 1.38 | |
| 5 | 0.22 | 13.8 | 0.25 | 63.9 | 4.39 | 0.27 | 27.5 | 1,142.6 | 1.71 | |
| 6 | 0.27 | 27.6 | 0.37 | 376.7 | 25.87 | 0.49 | 33 | 22,934.2 | 34.31 | |
| 7 | 0.49 | 33.3 | 0.59 | 360.8 | 24.78 | 0.67 | 0.1 | 18,912.9 | 28.29 | |
| 8 | 0.68 | 0.1 | 0.8 | 198.5 | 13.63 | 0.83 | 23.8 | 8,580 | 12.84 | |
| 9 | 0.83 | 123.9 | 0.85 | 134.8 | 9.26 | 0.97 | 26.4 | 6,659 | 9.96 | |
| 10 | 0.97 | 26.6 | 1.04 | 155.4 | 10.67 | 1.12 | 0.9 | 6,239.2 | 9.33 | |
| B | 1 | 0.08 | 0.4 | 0.11 | 37.8 | 3.51 | 0.14 | 4.5 | 651.2 | 1.6 |
| 2 | 0.14 | 5.5 | 0.16 | 32.1 | 2.99 | 0.18 | 27.6 | 541 | 1.33 | |
| 3 | 0.18 | 27.8 | 0.19 | 35 | 3.26 | 0.22 | 17.3 | 654.8 | 1.61 | |
| 4 | 0.22 | 17.6 | 0.25 | 50 | 4.65 | 0.27 | 35.7 | 1,001.7 | 2.47 | |
| 5 | 0.27 | 35.9 | 0.35 | 326.7 | 30.39 | 0.48 | 20.5 | 16,837.8 | 41.47 | |
| 6 | 0.48 | 20.7 | 0.55 | 274.8 | 25.57 | 0.59 | 45.7 | 9,577.1 | 23.59 | |
| 7 | 0.59 | 46.5 | 0.61 | 77.8 | 7.24 | 0.66 | 1 | 1,744.2 | 4.3 | |
| 8 | 0.67 | 0.1 | 0.8 | 87.5 | 8.14 | 0.83 | 60.4 | 3,677.9 | 9.06 | |
| 9 | 0.83 | 60.8 | 0.85 | 72.4 | 6.74 | 0.97 | 8.3 | 2,996.8 | 7.38 | |
| 10 | 0.97 | 8.2 | 1.04 | 80.7 | 7.51 | 1.12 | 0.8 | 2,915.4 | 7.18 | |
| C | 1 | 0.09 | 0.1 | 0.11 | 37.1 | 3.84 | 0.14 | 1.8 | 590 | 1.65 |
| 2 | 0.14 | 2.6 | 0.17 | 3.12 | 3.12 | 0.18 | 25.7 | 429.7 | 1.2 | |
| 3 | 0.15 | 25.8 | 0.2 | 3.64 | 3.64 | 0.22 | 18.7 | 720.3 | 2.01 | |
| 4 | 0.23 | 18.8 | 0.26 | 4.87 | 4.87 | 0.27 | 36.9 | 1,012.1 | 2.83 | |
| 5 | 0.28 | 37.1 | 0.35 | 30.2 | 30.2 | 0.49 | 21.3 | 15,008.7 | 41.89 | |
| 6 | 0.49 | 21.4 | 0.56 | 26.86 | 26.86 | 0.6 | 40.6 | 8,833.6 | 24.66 | |
| 7 | 0.6 | 41.6 | 0.62 | 7.32 | 7.32 | 0.67 | 0.1 | 1,557.4 | 4.35 | |
| 8 | 0.68 | 1.2 | 0.8 | 7.16 | 7.16 | 0.83 | 49.9 | 2,989.5 | 8.34 | |
| 9 | 0.83 | 49.9 | 0.86 | 6.21 | 6.21 | 0.96 | 3.4 | 2,281.7 | 6.37 | |
| 10 | 0.97 | 3.3 | 1.04 | 6.78 | 6.78 | 1.11 | 1 | 2,402.2 | 6.71 |
HPTLC data—Batch A, B, and C of TC.
Experimental analysis
Hepatotoxicity induced model
Induction of hepatotoxicity was confirmed, as all the biochemical parameters were raised from their normal range (Table 6).
Table 6
| Animal no. | SGOT | SGPT | Total bilirubin | ALP | Cholesterol | Triglyceride | T. protein | Albumin | Globulin |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 85.6 | 65 | 0.55 | 198 | 164 | 123 | 6.1 | 3.1 | 4 |
| 2 | 120 | 72 | 0.6 | 181 | 213 | 153 | 6.6 | 2.87 | 3.6 |
| 3 | 90.2 | 49 | 0.76 | 156 | 157 | 122 | 5.9 | 2.9 | 4.1 |
| 4 | 110 | 65 | 0.82 | 210 | 232 | 120 | 5.4 | 3.2 | 3.9 |
| 5 | 126 | 77 | 0.76 | 169 | 311 | 116 | 6.9 | 3.6 | 3.2 |
| 6 | 98.12 | 51 | 0.52 | 198 | 298 | 145 | 8 | 2.8 | 3.2 |
| Mean of the observation | 104.9 | 63.1 | 0.67 | 185.3 | 229.1 | 129.8 | 6.48 | 3.08 | 3.6 |
| Normal value of parameters | 45–80 | 17–35 | 0.3–0.7 | 120–180 | 120–160 | 60–120 | 4–8 | 2–5 | 2–4 |
Results of biochemical parameters of six rats after induction of hepatotoxicity.
Body weight
Changes that took place in the weights of rats BT and AT from each group were statistically determined by Paired t-test, which are presented in Table 7. Mean and SEM represents the mean and standard error of the mean difference in animals of a group. It was found that treatment with TC and silymarin resulted in a significant weight increase in rats. However, on applying ANOVA (Table 8), no significant difference in the body weight of rats was found.
Table 7
| Group | Mean differences ±S. E. M of differences | P value | Result |
|---|---|---|---|
| Control | 13.83 ± 17.75 | >0.05 | NS |
| TD1 | 47.33 ± 8.54 | < 0.01 | ** |
| TD2 | 47.5 ± 13.69 | < 0.05 | * |
| Standard | 62.5 ± 6.5 | < 0.001 | *** |
Comparative data of before and after treatment for control, TD1, TD2, and standard group by applying paired t-test-body weight.
TD1, test drug 1; TD2, test drug 2; NS, not significant; * significant; ** very significant; *** highly significant.
Table 8
| Groups | Control | TD1 | TD2 | Standard | Results of ANOVA |
|---|---|---|---|---|---|
| Body weight | 277.67 ± 12.32 | 311.83 ± 7.56 | 304.50 ± 10.31 | 312.50 ± 11.901 | NS |
| SGOT | 119.67 ± 7.96 | 78.92 ± 3.43 | 53.62 ± 4.0 | 63.95 ± 13.57 | *** |
| SGPT | 62.17 ± 8.03 | 44.83 ± 5.71 | 29.22 ± 3.99 | 31.12 ± 2.70 | ** |
| Total bilirubin | 0.76 ± 0.04 | 0.58 ± 0.08 | 0.38 ± 0.05 | 0.33 ± 0.03 | *** |
| Sr. ALP | 225.33 ± 12.06 | 171 ± 9.76 | 163.83 ± 8.58 | 144.83 ± 7.86 | *** |
| Cholesterol | 263.17 ± 13.22 | 196 ± 10.24 | 153.33 ± 11.74 | 161.67 ± 9.47 | *** |
| Triglyceride | 165.83 ± 6.90 | 102 ± 9.92 | 94 ± 11.39 | 111.83 ± 6.02 | *** |
| T. Protein | 6.90 ± 0.17 | 5 ± 0.46 | 4.87 ± 0.59 | 4.68 ± 0.43 | ** |
| Albumin | 4.32 ± 0.24 | 3.23 ± 0.28 | 3.03 ± 0.31 | 2.81 ± 0.48 | * |
| Globulin | 3.32 ± 0.27 | 2.45 ± 0.17 | 2.24 ± 0.21 | 2.53 ± 0.13 | ** |
Comparative biochemical data of all groups (ANOVA test).
TD1, Test drug 1; TD2, Test drug 2; NS, not significant; Sr. ALP, serum alkaline phosphatase, T. Protein, total protein.
* Significant; ** very significant; *** highly significant.
Biochemical parameters
The biochemical parameters of the different groups are expressed as mean ± SEM in Table 8 along with the statistically determined significance level of one-way analysis of variance (ANOVA). Further, the Tukey Krammer Multiple Comparisons test to compare treatment efficacy between the groups' results is performed and recorded in Table 9. TC at both doses shows highly significant potential in reducing SGOT, SGPT, ALP, total bilirubin, cholesterol, triglycerides, total protein, albumin, and globulin compared to the control group. But only, the higher dose of 400 mg/kg has shown a significant difference in reducing SGOT, SGPT, total bilirubin, cholesterol, total protein, albumin, and globulin parameters compared to the standard drug, which was less shown by TC (200 mg/kg) dose.
Table 9
| Biochemical parameter | Control vs. TD1 | Control vs. TD2 | Control vs. standard | TD1 vs. TD2 | TD1 vs. standard | TD2 vs. standard |
|---|---|---|---|---|---|---|
| SGOT | < 0.00001 | < 0.00001 | < 0.00001 | < 0.001 | < 0.05 | >0.05 |
| *** | *** | *** | *** | * | NS | |
| SGPT | < 0.001 | < 0.00001 | < 0.01 | < 0.001 | < 0.01 | >0.05 |
| *** | *** | *** | *** | ** | NS | |
| Total bilirubin | < 0.0001 | < 0.00001 | < 0.00001 | < 0.0001 | < 0.00001 | >0.05 |
| *** | *** | *** | *** | *** | NS | |
| Sr. ALP | < 0.00001 | < 0.00001 | < 0.00001 | >0.05 | < 0.001 | < 0.05 |
| *** | *** | *** | NS | *** | * | |
| Cholesterol | < 0.00001 | < 0.00001 | < 0.00001 | < 0.0001 | < 0.001 | >0.05 |
| *** | *** | *** | *** | *** | NS | |
| Triglyceride | < 0.00001 | < 0.00001 | < 0.00001 | >0.05 | >0.05 | < 0.05 |
| *** | *** | *** | NS | NS | * | |
| Total protein | < 0.00001 | < 0.00001 | < 0.00001 | >0.05 | >0.05 | >0.05 |
| *** | *** | *** | NS | NS | NS | |
| Albumin | < 0.00001 | < 0.001 | < 0.0001 | >0.05 | >0.05 | >0.05 |
| *** | *** | *** | NS | NS | NS | |
| Globulin | < 0.00001 | < 0.00001 | < 0.0001 | >0.05 | >0.05 | >0.05 |
| *** | *** | *** | NS | NS | NS |
Comparative results after applying Tukey-Krammer test between groups.
TD1, Test drug 1; TD2, Test drug 2; NS, not significant; Sr. ALP, serum alkaline phosphatase; T. Protein, Total Protein.
* Significant; ** very significant; *** highly significant.
Histopathology
Histopathology changes in the liver and kidney of all groups are illustrated in Figure 10, and data of different groups are summarized in Table 10. Fatty accumulation, inflammatory cellular infiltration, degeneration changes, and necrosis of hepatic tissues caused due to alcohol intoxication were improved in the treatment group of both doses of TC. But higher dose, 400 mg/kg of TC, has shown tremendous potential in reversing hepatic lesions. There is no fatty cell accumulation, inflammatory cells, and only minimal degenerative changes comparable to that of Silymarin.
Figure 10
Table 10
| Group | Vascular tissue | Hepatocytes | Hepatic parenchyma | Inflammatory cellular changes | Overall pathological lesion score | |
|---|---|---|---|---|---|---|
| Histopathological analysis—liver | ||||||
| Control | Focal congestion of central vein | Cellular swelling, granular cytoplasm, and presence of fatty accumulation. Mild degenerative changes and focal necrotic changes with loss of cellular border | Focal necrotic changes with loss of nucleus | Focal inflammatory cellular infiltration of mononuclear cells | Moderate (+3) | |
| TD1 | Focal congestion of central vein | Mild degenerative changes with granular cytoplasm and presence of fatty accumulation | Focal necrotic changes with loss of nucleus | Focal inflammatory cellular infiltration of mononuclear cells | Mild (+2) | |
| TD2 | No changes | Minimal degenerative changes with granular cytoplasmic changes with the occasional presence of fatty accumulation | Focal changes | Absent | Minimal (+1) | |
| Standard | No changes | Minimal degenerative changes with granular cytoplasmic changes with the occasional presence of fatty accumulation | Focal changes | Absent | Minimal (+1) | |
| Histopathological analysis—kidney | ||||||
| Groups | Vascular tissue | Renal parnchyma | Tubuarepitelium | Renal tubules | Glomeruli | Overall Pathological lesion score |
| Control | Focal congestion in medulla and cortex | Focal interstitial hemorrhages | Cellular swelling and tubular degeneration with focal necrosis in the medullary region | Mild to moderate degenerative changes with granular cytoplasm and enlarged tubules | Focal hypertrophic features of glomeruli in the cortical region | Mild (+1) |
| TD1 | Focal congestion in medulla and cortex | Focal interstitial hemorrhages | Focal area of cellular swelling and tubular degeneration with focal necrosis in the medullary region | Focal degenerative changes with granular cytoplasm and enlarged tubules | Focal hypertrophic features of glomeruli in the cortical region | Minimal (+1) to Mild (+2) |
| TD2 | No changes | No changes | Focal areas of cellular swelling and tubular degeneration with focal necrosis and presence of an eosinophilic mass in the tubular lumen | No changes | No changes | Minimal (+1) |
| Standard | No changes | No changes | Focal and minimal changes with cellular swelling | No changes | No changes | Minimal (+1) |
Results of histopathology of liver.
Overall Grade score as—NAD, no abnormality detected, minimal changes (+1), mild changes (+2), moderate changes (+3), severe changes (+4).
Discussion
Network pharmacology-based analysis
In the present study, we analyzed the candidate components and targets associated with TC with the aid of network pharmacology analysis and discovered the association between 1,127 PCs and their 2,765 protein targets. ADME screening has resulted in the identification of 418 active compounds referred to as PACs with the ability to regulate 1,005 protein targets. Metabolite inter-conversion enzymes are an enriched class of protein targets, where P35354 and P42574 emerged as major targets, being regulated by 13 PACs of TC. P34354 is a Prostaglandin G/H synthase 2 encoded by Prostaglandin-Endoperoxide Synthase 2 (PTGS2) and also known as cyclooxygenase-2 (COX-2). COX-2 is a crucial enzyme in the formation of prostaglandins and thus plays a significant role in the inflammatory responses. COX-2 has been shown to be involved in carcinogenesis via apoptosis inhibition, where selective inhibitors of COX-2 can provide chemoprevention in the case of liver tumors (56). In such a case, it may be assumed that these 13 regulators of PTGS2 could contribute to providing an anti-inflammatory effect of TC via reducing the production of prostaglandins, which promote inflammation. The other major protein target of TC, P42574 is caspase-3 that is involved in apoptosis execution. The association of caspases in liver disease and their inhibitors are well studied in relation to liver diseases (57). Thus, suggesting the high therapeutic relevance of TC in managing liver diseases. GO-based annotation of the protein targets revealed the synergistic role of TC in regulating a multitude of cellular functions to elicit its therapeutic effect.
The construction and analysis of ALD-specific networks highlight the therapeutic efficacy of the formulation in managing ALD via its 40 PCs pathway association of the protein targets of ALD-specific 40 PCs revealed that TC mainly regulates pathways associated with cancer and signaling processes like HIF-1, chemokine signaling, PI3K-Akt, etc. The role of these signaling pathways in the progression of ALD is well-known, for example, interactions among cytokines and chemokines play a significant role in disease development (58). Also, HIF-1α plays a crucial role in the development and prognosis of ALD, and targeting the same offers an attractive strategy for the treatment of ALD (59). This suggests that the enriched pathways might be the crucial pharmacological mechanism of TC for ALD management. Also, mapping of protein targets on the ALD-disease pathway reflects the direct regulation of TC. For example, Sirtuin 1 encoded by the SRT1 gene, is a target of TC whose reduced levels have been linked to the inflammatory processes observed in ALD patients. This study also highlighted the multi-targeting roles of PACs of TC, which offer an additional advantage to these PCs in future drug-discovery procedures for their multi-targeting ability toward ALD proteins. Phytochemical-specific regulatory potential of TC upon the ALD-specific Human-PIN was also identified, where both direct and indirect control of TC over the clusters were identified. It was found that direct control takes place through their protein targets and indirect via proteins interaction of the TC protein targets. The regulation over clusters was mainly through PI3K-Akt signaling, NF-kappa B signaling, and pathways involved in cancer, especially hepatocellular carcinoma. Inflammation plays a vital role in disease progression, leading to fibrosis, which can later develop into cirrhosis. As PI3K-Akt activates immune cells via regulating inflammatory cytokines, the regulation of this pathway may contribute to achieving therapeutic effects for ALD (55). Also, NF-kappa B performs a wide range of cellular functions, influencing inflammation in Kupffer cells and survival of hepatocytes, thus linking injury, fibrosis, and hepatocellular carcinoma (60). These data support the therapeutic efficacy of TC toward ALD.
Physicochemical analysis
Foreign matter, moisture content, total ash value, acid insoluble ash, water-soluble ash, water-soluble extract, and alcohol soluble extract of Z. officinale, P. longum, and P. nigrum were found under the permissible limits of pharmacopeia standards. In crude drugs, a number of constituents are present, and they have the selective solubility rate in different solvents such as water and alcohol. Water can extract ether polar substances such as alkaloids, tannins, and flavonoids, but alcohol extracts the compounds that seem insoluble in water, such as sterols and phenols. Water and alcohol soluble extract content in TC is found to be more than that of their individual powder, indicating a higher concentration of substances in TC. As TC has higher concentration of substances, so it may show more activity than individual herbs. Combination of herbs are increasing its potency.
If maintained under the same conditions, HPTLC analysis shows that all the three batches of TC have an Rf value of 0.43, indicating the presence of Piperine (45–47). Piperine is an essential and major component of TC formulation and its presence in the sample suggests its standard quality. The fingerprinting of the TC presents that at Rf 0.80, all the three batches show a maximum peak, in the first batch maximum% age is demonstrated by 0.37 Rf i.e., to be 25.87%, in the second batch maximum% age is shown by 0.35 Rf i.e., to be 30.39%, in third batch maximum% age is shown by 0.35 Rf i.e., 30.20%. Second and third, both the batches show maximum% age of amount at Rf 0.35, and that is almost equal to 30%.
Experimental analysis
Body weight
Liver diseases cause insufficient absorption and metabolism of nutrients resulting in malnutrition and decreased body weight. This study evidenced a significant increase in rats' weights among TD2, TD1, and standard groups which is not evident in the control group directed toward the reversal of liver injury by TC (61).
Biochemical parameters
Alcohol consumption causes substantial diseases, and the liver is the most adversely affected organ. Liver function test (LFT), is a well-known biochemical test that is used to determine the health and function of the liver through the measurement of liver enzymes (SGOT, SGPT, alkaline phosphate), bilirubin, and protein in the blood (62). During intoxication of alcohol, there are structural changes, leading to an increase in the permeability of the cell membrane. As there is an increase in permeability of the membrane, it further produces SGOT and SGPT leakage into blood circulation as shown by abnormally high levels of serum hepatic markers. Elevated LFT from the normal values is the indication of liver injury (62). TC shows a reduction in SGOT and SGPT levels in these respective studies of alcohol-mediated hepatotoxicity which may be attributed to its cell membrane stabilizing ability, hepatic enzymes leakage prevention, and translocation into the serum (63). Liver steatosis is the earliest stage of liver disease in chronic ethanol consumption, with the characteristic of fat accumulation (64). In this study, alcohol-induced hepatic steatosis was demonstrated by increased cholesterol, triglycerides values, and fatty accumulation in liver histology (64). Data from the present study the first show that TC can reverse the ethanol-induced hepatotoxicity. Oral treatment of TC by both doses significantly reduced the elevated biochemical markers compared with the control group. The highest suppressive effect on decreasing SGOT, SGPT, total bilirubin, and cholesterol was induced by the high dose of TC 400 mg, respectively, comparable to that of silymarin, and decreased triglyceride level more than that of silymarin.
The total protein content consists of globulin and albumin protein. Albumin is a very common protein found in the blood with various functions, produced by the liver only. The blood levels of these plasma proteins are decreased when there is extensive liver damage. Hypoalbuminaemia may occur in liver disease, with significant destruction of hepatocytes and hyperglobulinaemia in chronic inflammatory diseases such as cirrhosis and chronic hepatitis (64, 65). Usually, in liver diseases, synthesis of this protein decreases, but in this study after administration of ethanol, it is noted that the values of total protein, globulin, and albumin neither increased nor decreased, the values just are in the normal range. But after treatment, it was noticed that there is a difference in the means of each treatment. In all the three treatment groups-total protein, globulin, and albumin values are found significantly less compared to that of the control group also reflecting toward the improved functional capacity of the liver by TC (64).
Histopathology
Pretreatment with ethanol resulted in damage to the liver as infiltration of mononuclear cells, distortion of the normal structure of the hepatocytes cells, loss of nucleus, presence of necrotic cells, and accumulation of fatty cells were detected in many regions of the liver sections (66). Treatment with TC at both doses and silymarin has helped in modulating degenerative changes, hepatic lesions, inflammatory cellular infiltration, and fatty accumulation in hepatocytes which is evident in the control group. The higher dose of 400 mg/kg of TC showed a remarkable liver regeneration by improving hepatic lesions, inflammation, and the only occasional presence of fatty changes (66).
Histopathology of the kidney shows mild pathological changes in the control group, demonstrating harmful effects of chronic consumption of ethanol on the kidney. TC, however, also demonstrates improvement in pathological lesions of the kidney compared to that of silymarin indicating its renal protective ability (67).
Possible scenario of hepatoprotection mechanism-TC
The exact pathogenesis of alcoholic liver injury is yet not known that why only some chronic alcoholics develop the complete sequence of changes in the liver while the others don't. Ethanol and its metabolites are responsible for ill effects on the liver. Biomedical and cellular events such as—changes in fat contents in the liver, cytoskeletal and membrane damage of a hepatic cell, hepatic fibrogenesis, oxidative damage to membrane and proteins, inflammatory reaction, activation of stellate cell and kupffer, lactic acidosis and collagen formation, which occurs due to chronic alcohol consumption culminate in morphologic lesions of alcoholic steatosis (fatty liver), alcoholic hepatitis and alcoholic cirrhosis, which is evidenced by elevated biochemical parameter SGOT, SGPT, ALP, total bilirubin, cholesterol, triglyceride level, and histopathology. TC helps in reducing the elevated biochemical parameters. Also, TC reduces oxidative stress, fatty changes, inflammatory reactions, and damage to hepatocytes; thus, acclimating to hepatoprotection. Previous studies also report their antioxidant and anti-inflammatory activity in numerous in vivo and in vitro models. The ability of TC to cope with the oxidative stress and inflammation induced by alcohol could be accountable to the antioxidant and anti-inflammatory capacity of the herb (14, 16, 20, 68). The association of protein targets of TC in metabolic pathways, signaling processes like HIF1α, PI3k-Akt, NF-kappa B, chemokine/cytokine, and apoptotic pathways as observed in network pharmacological analysis confirms its role in reducing inflammation, oxidative stress, and fibrosis. Especially the molecular targets of TC like PTGS2, SRT1, and caspases are of direct relevance in achieving its desired hepatoprotective effects. Most importantly, TC help to lower the level of liver enzymes in relieving the symptoms of ALD, and metabolic enzymes were recognized as the major target class of phytochemicals considering network analyses. The possible scenario of TC action in interfering with hepatoprotective activity is depicted in Figure 11.
Figure 11
Limitations and future perspective
As many Rfs were found in fingerprinting, TC can be evaluated for these compounds by running with multiple standard markers in HPTLC or by other chromatographic tests. The process will also enable the quantification and standardized analysis of other compounds, allowing for a complete physicochemical profile to be generated. The quantification of each compound also plays a major role in its therapeutic effect. So, all the compounds present in the TC should be quantified in future studies to find out in which quantity these effects can be produced.
TC, its metabolites, or different extracts can be evaluated for its hepatoprotective effect by using different in vivo and in vitro hepatotoxicity models, multiple doses, chronic toxicity, and multi-centric clinical trials. Furthermore, the study indicates that TC and silymarin can improve kidney damage due to alcohol consumption. Therefore, both these drugs can be evaluated for the kidney protective effect in different experimental models and clinical trials, and therefore results can be validated. Also, through network pharmacology techniques, the possible involved mechanisms in the execution of this effect can also be ruled out.
The pathways detected in executing hepatoprotective effects should also be verified in more experimental studies. Pathway specific animal should be conducted to understand the clear mechanism and developing the novel treatment for ALD.
Conclusion
Our research presents a novel and feasible approach for screening active ingredients of TC herb responsible for ALD treatment along with the mechanism involved. In this study, we adopted a systematic network pharmacology approach to investigate the hepatoprotective effects of TC in ALD and it indicates that the hepatoprotective effects of TC are manifested by the multitude of mechanisms involving regulating cancer and signaling processes like AGE-RAGE signaling, HIF-1 signaling, NF-Kappa B signaling, PI3K/Akt signaling responsible for anti-inflammatory, anti-oxidant and anti-apoptotic role. Furthermore, in vivo experiments validate the therapeutic effects of TC at doses of 200 and 400 mg/kg. TC improved ALD symptoms by inhibiting inflammation, inhibiting cellular evasion, reducing fatty cell accumulation, and lowering liver enzymes. TC high dose of 400 mg/kg showed comparable effects to silymarin, but less at a dose of 200 mg/kg. In addition, TC also showed renal protective action by ameliorating kidney lesions caused due to ethanol.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 32070671 and 32270690) and Regional Innovation Cooperation between Sichuan and Guangxi provinces (Project No. 2020YFQ0019).
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
An animal study was conducted at Agharkar Research Institute, Pune, India, according to the recommendations and approval of the Institutional Animal Ethical Committee with approval number ARI/IAEC/2019/11.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Acknowledgments
The authors kindly acknowledge Banaras Hindu University for providing their valuable support and facilities. Authors are thankful to the Researchers Supporting Project Number (RSP-2021/335), King Saud University, Riyadh, Saudi Arabia.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2022.1063118/full#supplementary-material
Supplementary Figure S1Trikatu-protein target (T-PT) Network: Network of size 2,963 nodes 7,055 edges, containing the information of 198 phytochemicals (cyan colored diamond nodes) and their 2,765 protein targets (pink colored circular nodes). The size of the nodes varies according to their degree value in the net-work, where node with the highest degree is bigger in size corresponding to the other nodes. The protein targets were compiled from STITCH having interaction score ≥700.
Supplementary Figure S2Liver specific disease gene (LD-G) Network. The proteins and their associated liver Disease term was used for obtaining liver specific disease gene (LD-G) Network, as per the curated gene-disease pair data available in DisGeNET. LD-G network comprise of 539 nodes (515 proteins and 24 disease terms) and 1,308 edges, where pink nodes are specific to proteins and green denote disease class.
Supplementary Table S138 liver disease terms considered in the study (as per DisGeNET data).
Supplementary Table S2Trikatu-protein target (T-PT) network specific to PACs.
Supplementary Table S3Protein class association using PANTHER.
Supplementary Table S4Disgnet disease-protein association data corresponding to Liver diseases (LD-G network).
Supplementary Table S5Protein targeting ability of PCs in the ALD-PT network.
Supplementary Table S6KEGG pathway enrichment.
Supplementary Table S7Network analysis of TRH-PIN.
Supplementary Table S8MCODE cluster information along with the GO-based annotation and protein-class enrichment.
References
1.
SharmaRJadhavM. Hepatoprotective activity of Shrikhand Asava—a preclinical study. Int J Ayu Pharm Chem. (2021) 15:2.
2.
WangJLiPJiangZYangQMiYLiuYet al. Diagnostic value of alcoholic liver disease (ALD)/non-alcoholic fatty liver disease (NAFLD) index combined with gamma-glutamyltransferase in differentiating ALD and NAFLD. Korean J Intern Med. (2016) 31:479–87. 10.3904/kjim.2015.253
3.
BlackJMHawksJH. Medical-Surgical Nursing: Clinical Management for Positive OuTComes, 8th edition, volume 1. St Louis, MO: Saunders (2005), p. 1336–52.
4.
GargVGargHKhanATrehanpatiNKumarASharmaBCet al. Granulocyte colony-stimulating factor mobilizes CD34+ cells and improves survival of patients with acute-on-chronic liver failure. Gastroenterology. (2012) 142:505–12. 10.1053/j.gastro.2011.11.027
5.
RonisMJHuangJICrouchJAMercadoCYIrbyDAValentineCRet al. Cytochrome P450 CYP 2E1 induction during chronic alcohol exposure occurs by a two-step mechanism associated with blood alcohol concentrations in rats. J Pharmacol Exp Therap. (1993) 264:944–50.
6.
DasSKVasudevanDM. Alcohol-induced oxidative stress. Life Sci. (2007) 81:177–87. 10.1016/j.lfs.2007.05.005
7.
MaZZhangBFanYWangMKebebeDLiJet al. Traditional Chinese medicine combined with hepatic targeted drug delivery systems: a new strategy for the treatment of liver diseases. Biomed Pharmacother. (2019) 117:109128. 10.1016/j.biopha.2019.109128
8.
KhudhairDHAl-GareebAIAl-KuraishyHMEl-KademAHElekhnawyENegmWAet al. Combination of Vitamin C and curcumin safeguards against methotrexate-induced acute liver injury in mice by synergistic antioxidant effects. Front Med. (2022) 9:866343. 10.3389/fmed.2022.866343
9.
The Ayurvedic Formulary of India. Part II. Depart. Ind. System Med. Homeo.New Delhi: The Ayurvedic Formulary of India (2002). p. 322–3.
10.
KaushikR. Trikatu—a combination of three bioavailability enhancers. Int J Green Pharmacy. (2018) 12:S437–41.
11.
MalvankarPR. Anthelmintic activity of water extracts of Trikatuchurna and its individual ingredients on Indian earthworms. Int J Pharma Biol Sci. (2012) 3:374–8.
12.
DahikarSBBhutadaSAVibhuteSKSonvaleVCTambekarDHKastureSBet al. Evaluation of antibacterial potential of Trikatuchurna and its ingredients: an in-vitro study. Int J Phytomed. (2010) 2:412–7. 10.5138/ijpm.2010.0975.0185.02058
13.
Siva kumarVSiva kumarS. Effect of an indigenous herbal compound preparation “Trikatu” on the lipid profiles of atherogenic diet and standard diet fed Rattusnorvegicus. Phytother Res. (2004) 18:976–81. 10.1002/ptr.1586
14.
MurunikkaraVRasoolM. Trikatu, an herbal compound as immunomodulatory and anti-inflammatory agent in the treatment of rheumatoid arthritis—an experimental study. Cell Immunol. (2014) 287:62–8. 10.1016/j.cellimm.2013.12.002
15.
AtalCKZutshiURaoPG. Scientific evidence on the role of Ayurvedic herbals on bioavailability of drugs. J Ethnopharmacol. (1981) 4:229–32. 10.1016/0378-8741(81)90037-4
16.
SabinaEPNagarSRasoolM. A role of piperine on monosodium urate crystal-induced inflammation-an experimental model of gouty arthritis. Inflammation. (2011) 34:184–92. 10.1007/s10753-010-9222-3
17.
NehaJMishraRN. Immunomodulator activity of Trikatu mega Ext. Int J Res Pharma Biomed Sci. (2011) 2:160–64.
18.
DossHMGanesanRRasoolM. Trikatu, an herbal compound ameliorates rheumatoid arthritis by the suppression of inflammatory immune responses in rats with adjuvant-induced arthritis and on cultured fibroblast like synoviocytes via the inhibition of the NFκB signaling pathway. Chem Biol Interact. (2016) 258:175–86. 10.1016/j.cbi.2016.09.003
19.
KulkarniVSSuranaSJ. Reversal of CRF and stress-induced anorexia by an ayurvedic formulation. Brazil J Pharmacog. (2011) 22:404–11. 10.1590/S0102-695X2011005000166
20.
NehaJMishraRN. Antioxidant activity of Trikatu mega Ext. Int J Res Pharma Biomed Sci. (2011) 2:624–28.
21.
D'souzaPFAshokaSRanjanMSShabarayaAR. Anti tumor activity of mercaptopurine in combination with trikatu and gomutra on 20-Methylcholantrene induced Carcinogenesis. J App Pharm Sci. (2013) 3:20–4. 10.7324/JAPS.2013.3804
22.
HarwanshRKMukherjeeKBhadraSKarABahadurSMitraAet al. Cytochrome P450 inhibitory potential and RP-HPLC standardization of Trikatu—a Rasayana from Indian Ayurveda. J Ethno Pharmacol. (2014) 153:674–81. 10.1016/j.jep.2014.03.023
23.
PatelJAShahUS. Hepatoprotective activity of Piper longum traditional milk extract on carbon tetrachloride induced liver toxicity in Wistar rats. Boletín Latinoam Caribe Plantas Med Arom. (2009) 8:121–9.
24.
KumarSVSMishraSH. Hepatoprotective activity of Trikatu churna-an ayurvedic formulation. Indian J Pharm Sci. (2004) 66:365–7.
25.
ChandranUMehendaleNTilluGPatwardhanB. Network pharmacology of ayurveda formulation Triphala with special reference to anti-cancer property. Combinatorial Chem High Throughput Screen. (2015) 18:846–54. 10.2174/1386207318666151019093606
26.
ChoudharyNSinghV. Insights about multi-targeting and synergistic neuromodulators in Ayurvedic herbs against epilepsy: integrated computational studies on drug-target and protein-protein inter-action networks. Sci Rep. (2019) 9:10565. 10.1038/s41598-019-46715-6
27.
ChoudharyNSinghV. Multi-scale mechanism of antiviral drug-alike phytoligands from Ayurveda in managing COVID-19 and associated metabolic comorbidities: insights from network pharmacology. Mol Divers. (2022) 7:1–20. 10.1007/s11030-021-10352-x
28.
LiSWangNHongMTanHYPanGFengYet al. Hepatoprotective effects of a functional formula of three Chinese medicinal herbs: experimental evidence and network pharmacology-based identification of mechanism of action and potential bioactive components. Molecules. (2018) 23:352. 10.3390/molecules23020352
29.
ZengXZhangPWangYQinCChenSHeWet al. CMAUP: a database of collective molecular activities of useful plants. Nucleic Acids Res. (2019) 47:D1118–27. 10.1093/nar/gky965
30.
Duke. Handbook of Phytochemical Constituent Grass, Herbs and Other Economic Plants.Milton Park: Routledge (2017). 10.1201/9780203752623
31.
MohanrajKKarthikeyanBSVivek-AnanthRPChandRPAparnaSRMangalapandiPet al. IMPPAT: a curated database of Indian Medicinal Plants, Phytochemistry and Therapeutics. Sci Rep. (2018) 8:4329. 10.1038/s41598-018-22631-z
32.
ChoudharyNChoudharySKumarASinghV. Deciphering the multi-scale mechanisms of Tephrosia purpurea against polycystic ovarian syndrome (PCOS) and its major psychiatric comorbidities: Studies from network pharmacological perspective. Gene. (2021) 773:145385. 10.1016/j.gene.2020.145385
33.
ZengXZhangPHeWQinCChenSTaoLet al. NPASS: natural product activity and species source database for natural product research, discovery and tool development. Nucleic Acids Res. (2018) 46:D1217–22. 10.1093/nar/gkx1026
34.
ZhangRZYuSJBaiHNingK. TCM-Mesh: the database and analytical system for network pharmacology analysis for TCM preparations. Sci Rep. (2017) 7:2821. 10.1038/s41598-017-03039-7
35.
LagorceDBouslamaLBecotJMitevaMAVilloutreixBO. FAF-Drugs4: free ADME-tox filtering computations for chemical biology and early stages drug discovery. Bioinformatics. (2017) 33:3658–60. 10.1093/bioinformatics/btx491
36.
SzklarczykDSantosAVon MeringCJensenLJBorkPKuhnMet al. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. (2016) 44:D380–4. 10.1093/nar/gkv1277
37.
PiñeroJRamírez-AnguitaJMSaüch-PitarchJRonzanoFCentenoESanzFet al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. (2019) 48:D845–55. 10.1093/nar/gkz1021
38.
ShannonPMarkielAOzierOBaligaNSWangJTRamageDet al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) 13:2498–504. 10.1101/gr.1239303
39.
SzklarczykDGableALLyonDJungeAWyderSHuerta-CepasJet al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. (2019) 47:D607–13. 10.1093/nar/gky1131
40.
BroheeSVan HeldenJ. Evaluation of clustering algorithms for protein-protein interaction networks. BMC Bioinform. (2006) 7:488. 10.1186/1471-2105-7-488
41.
MiHMuruganujanAHuangXEbertDMillsCGuoXet al. (2019). Protocol update for large-scale genome and gene function analysis with the PAN-THER classification system (v.14.0). Nat Protoc. 14:703–21. 10.1038/s41596-019-0128-8
42.
DennisGShermanBTHosackDAYangJGaoWLaneHCet al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. (2003) 4:3. 10.1186/gb-2003-4-5-p3
43.
JohriRKZutshiU. An Ayurvedic formulation “Trikatu” and its constituents. J Ethnopharmacol. (1992) 37:85–91. 10.1016/0378-8741(92)90067-2
44.
The Ayurvedic Pharmacopoeia of India (API). Part 1, Vol 1, 3, 4, New Delhi, The Controller of Publication, Civil Lines, 1st Edition. New Delhi: API.
45.
ShailajanSSayedNJoshiHTiwariB. Standardization of an ayurvedic formulation: Trikatu Churna using Bioanalytical tools. IJRAP (2011), 2:1676–8.
46.
GargSMishraAGuptaR. Fingerprint profile of selected ayurvedic churnas/preparations: an overview. Altern Integ Med. (2013) 2:6. 10.4172/2327-5162.1000125
47.
SinghASharmaHNarshimhajiCVSinghRSrikanthN. HPTLC detection of Carica papaya dried seed (adulterant) in Trikatu based on piperine. J Biol Active Prod Nat. (2019) 9:141–61. 10.1080/22311866.2019.1605933
48.
ChandaDShankerKPalALuqmanSBawankuleDUManiDet al. Safety evaluation of Trikatu, a generic ayurvedic medicine in Charles Foster rats. J Toxicol Sci. (2009) 34:99–108. 10.2131/jts.34.99
49.
TiegsGHorstAK. TNF in the liver: targeting a central player in inflammation. Semin Immunopathol. (2022) 44:445–59. 10.1007/s00281-022-00910-2
50.
KoschützkiDSchreiberF. Centrality analysis methods for biological networks and their application to gene regulatory networks. Gene Regul Syst Bio. (2008) 2:193–201. 10.4137/GRSB.S702
51.
YuHKimPMSprecherETrifonovVGersteinM. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol. (2007) 3:e59. 10.1371/journal.pcbi.0030059
52.
WangYNakajimaTGonzalezFJTanakaN. PPARs as metabolic regulators in the liver: lessons from liver-specific PPAR-null mice. Int J Mol Sci. (2020) 21: 2061. 10.3390/ijms21062061
53.
SoofiATaghizadehMTabatabaeiSMRezaei TaviraniMShakibHNamakiSet al. Centrality analysis of protein-protein interaction networks and molecular docking prioritize potential drug-targets in type 1 diabetes. Iran J Pharm Res. (2020) 19:121–34. 10.22037/ijpr.2020.113342.14242
54.
YeRXuHWanCPengSWangLXuHet al. Antibacterial activity and mechanism of action of ε-poly-L-lysine. Biochem Biophys Res Commun. (2013) 439:148–53. 10.1016/j.bbrc.2013.08.001
55.
LueddeTSchwabeRF. NF-κB in the liver—linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. (2011) 8:108–18. 10.1038/nrgastro.2010.213
56.
RicoteC. Role of cyclooxygenase-2 in the pathogenesis of chronic liver diseases. Med Clin (Barc). (2003) 121:743–8. 10.1016/s0025-7753(03)74082-2
57.
WoolbrightBLDingWXJaeschkeH. Caspase inhibitors for the treatment of liver disease: friend or foe?Expert Rev Gastroenterol Hepatol. (2017) 11:397–9. 10.1080/17474124.2017.1300060
58.
KawarataniHTsujimotoTDouharaATakayaHMoriyaKNamisakiTet al. The effect of inflammatory cytokines in alcoholic liver disease. Mediators Inflamm. (2013) 495156. 10.1155/2013/495156
59.
MatsudaSKobayashiMKitagishiY. Roles for PI3K/AKT/PTEN pathway in cell signaling of nonalcoholic fatty liver disease. ISRN Endocrinol. (2013) 2013:472432. 10.1155/2013/472432
60.
ChuQGuXZhengQZhuH. Regulatory mechanism of HIF-1α and its role in liver diseases: a narrative review. Ann Transl Med. (2022) 10:109. 10.21037/atm-21-4222
61.
SaaluLCOgunladeBAjayiGOOyewopoAOAkunnaGGOgunmodedeOSet al. The hepato-protective potentials of Moringa oleifera leaf extract on alcohol—induced hepatotoxicity in Wistar rats. Am J Biotechnol Mol Sci. (2012) 2:6–14. 10.5251/ajbms.2012.2.1.6.14
62.
PariLKarthikesanK. Protective role of caffeic acid against alcohol-induced biochemical changes in rats. Fundam Clin Pharmacol. (2007) 21:355–61. 10.1111/j.1472-8206.2007.00505.x
63.
VermeulenNPBessemsJGVan de StraatR. Molecular aspects of paracetamol-induced hepatotoxicity and its mechanism-based prevention. Drug Metals Dev. (1992) 24:367–407. 10.3109/03602539208996298
64.
VijayNPadmaaM. Hepatoprotective activity of Chenopodium album Linn. against alcohol induced liver damage. Pharmacologyonline. (2012) 3:e460. 10.5138/ijpm.v3i4.460
65.
VicasSILasloVTimarAVBaltaCHermanHCiceuAet al. Nano selenium-enriched probiotics as functional food products against cadmium liver toxicity. Materials. (2021) 14:2257. 10.3390/ma14092257
66.
HoWYYeapSKHoCLAbdul RahimRAlitheenNB. Hepatoprotective activity of Elephantopus scaber on alcohol-induced liver damage in mice: evidence-based complement. Altern Med. (2012) 417953:8. 10.1155/2012/417953
67.
VicasSILasloVTimarAVBaltaCHermanHCiceuAet al. Functional food product based on nanoselenium-enriched Lactobacillus casei against cadmium kidney toxicity. Appl Sci. (2021) 11:4220. 10.3390/app11094220
68.
MarshallWBangertS. Plasma proteins and enzymes. Clin Chem. (2007) 6:253.
Summary
Keywords
ethanol-induced hepatotoxicity, hepatoprotective, herbal formulation, network pharmacology, Piper longum, Piper nigrum, Trikatu Churna, Zingiber officinale
Citation
Sharma R, Jadhav M, Choudhary N, Kumar A, Rauf A, Gundamaraju R, AlAsmari AF, Ali N, Singla RK, Sharma R and Shen B (2022) Deciphering the impact and mechanism of Trikatu, a spices-based formulation on alcoholic liver disease employing network pharmacology analysis and in vivo validation. Front. Nutr. 9:1063118. doi: 10.3389/fnut.2022.1063118
Received
06 October 2022
Accepted
28 October 2022
Published
16 November 2022
Volume
9 - 2022
Edited by
Monika Thakur, Amity University, India
Reviewed by
Harshit Agarwal, University of Bayreuth, Germany; Prabhat Suman, Central University of Punjab, India
Updates
Copyright
© 2022 Sharma, Jadhav, Choudhary, Kumar, Rauf, Gundamaraju, AlAsmari, Ali, Singla, Sharma and Shen.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rohit Sharma rohitsharma@bhu.ac.inBairong Shen bairong.shen@scu.edu.cn
This article was submitted to Nutrition and Food Science Technology, a section of the journal Frontiers in Nutrition
†ORCID: Ruchi Sharma orcid.org/0000-0002-4931-3396
Rajeev K. Singla orcid.org/0000-0002-3353-7897
Rohit Sharma orcid.org/0000-0002-3682-3573
Bairong Shen orcid.org/0000-0003-2899-1531
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.