Abstract
Reputed as a significant metabolic disorder, non-alcoholic fatty liver disease (NAFLD) is characterized by high-fat deposits in the liver and causes substantial economic challenges to any country's workforce. Previous studies have indicated that some lactic acid bacteria may effectively prevent or treat NAFLD. Overall, L. acidophilus KLDS1.0901 protected against HFD-induced NAFLD by improving liver characteristics and modulating microbiota composition, and thus could be a candidate for improving NAFLD. This study aimed to assess the protective effects of L. acidophilus KLDS1.0901 on a high-fat diet(HFD)-induced NAFLD. First, hepatic lipid profile and histological alterations were determined to study whether L. acidophilus KLDS1.0901 could ameliorate NAFLD. Then, the intestinal permeability and gut barrier were explored. Finally, gut microbiota was analyzed to elucidate the mechanism from the insights of the gut–liver axis. The results showed that Lactobacillus KLDS1.0901 administration significantly decreased body weight, Lee's index body, fat rate, and liver index. L. acidophilus KLDS1.0901 administration significantly improved lipid profiles by decreasing the hepatic levels of total cholesterol (TC), triglyceride (TG), and low-density lipoprotein cholesterol (LDL-C) and by increasing the high-density lipoprotein cholesterol (HDL-C) levels. A conspicuous decrease of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) in serum was observed after L. acidophilus KLDS1.0901 administration. Meanwhile, the H&E and Oil Red O-stained staining showed that L. acidophilus KLDS1.0901 significantly reduced liver lipid accumulation of HFD-fed mice by decreasing the NAS score and lipid area per total area. Our results showed that L. acidophilus KLDS1.0901 administration decreased the interleukin-6 (IL-6), interleukin-1β (IL-1β), and tumor necrosis factor-alpha (TNF-α) concentrations accompanied by the increase of interleukin-10 (IL-10). L. acidophilus KLDS1.0901 administration could improve the intestinal barrier function by upregulating the mRNA levels of occludin, claudin-1, ZO-1, and Muc-2, which were coupled to the decreases of the concentration of LPS and D-lactic acid. Notably, L. acidophilus KLDS1.0901 administration modulated the gut microbiota to a near-normal pattern. Hence, our results suggested that L. acidophilus KLDS1.0901 can be used as a candidate to ameliorate NAFLD.
Introduction
Soaring obesity rates have been closely linked to non-communicable metabolic diseases like chronic liver and cardiovascular diseases as approximately 90% of patients are diagnosed with severe obesity and non-alcoholic fatty liver disease (NAFLD) (1, 2). NAFLD is a highly prevalent liver disease characterized by non-frequent alcohol consumption (3, 4) and could progress to serious conditions such as liver fibrosis, cancer, and cirrhosis (52). Because the pathogenesis of NAFLD is still unclear, studies are ongoing to develop an effective treatment protocol for NAFLD (3). Natural products incorporated into diets (ginkgolide C, polyphenols, yeast-fermented wall-broken bee pollen, among others) could be effective against NAFLD (5–7). Furthermore, next-generation sequencing techniques have revealed a possible correlation between NAFLD pathogenesis and changes in the intestinal microbiome (8), thus offering a potential strategy for diet-induced NAFLD (9).
As a widely used probiotic in foods, Lactobacillus acidophilus exerts several benefits after proliferating in the gastrointestinal tract, including ameliorating type 2 diabetes, enteric infections, allergic dermatitis, and renal failure and hepatic failure, various forms of inflammatory bowel disease, lactose intolerance, and possible anticancer activity. Furthermore, other studies have reported its potential to control serum cholesterol concentrations, reduce tumor development risks, and ensure better digestion to boost host immunity (10–12). Researchers have been paying more attention to L. acidophilus to improve abnormal glucose and lipid metabolism, especially NAFLD. Andreasen et al. reported that L. acidophilus NCFM could improve insulin sensitivity and the systemic inflammatory response in human subjects (52). It has been found that L. acidophilus LA5 could improve the saturated fat-induced obesity mouse model through the enhanced intestinal Akkermansia muciniphila (5). L. acidophilus NX2-6 showed the potential against oleic acid-induced steatosis, mitochondrial dysfunction, endoplasmic reticulum stress, and inflammatory responses (6). L. acidophilus SNZ 86 could alleviate Western diet-induced non-alcoholic fatty liver disease in rats via modulation of autophagy through the AMPK/SIRT-1 pathway (7). An Egyptian study by Abdel Monem with Zigazag University randomized patients with NASH to probiotic Acidophilus capsule (2 billion Lactobacillus acidophilus) or placebo for 17 months and measured improved AST and ALT in treated patients (8). L. acidophilus showed a significant reduction in the liver/body weight ratio and a significant improvement in steatosis compared to the patients with NAFLD (9). Yogurt fermented with L. acidophilus improves body mass index and fasting insulin levels without affecting serum leptin and adiponectin levels in NAFLD (10).
Lactobacillus acidophilus KLDS1.0901 was isolated from traditional fermented dairy products in Sinkiang Province, China, and preserved in our laboratory. Our previous studies showed L. acidophilus KLDS1.0901 with antioxidative activity had good tolerance to acid and bile salt and strong adhesion ability (11). Furthermore, L. acidophilus KLDS1.0901 could alleviate type 2 diabetes by regulating hepatic glucose, lipid metabolism, and gut microbiota in mice (12). L. acidophilus KLDS1.0901 also could prevent chronic alcoholic liver injury in mice by protecting the intestinal barrier and regulating gut microbiota and liver-related pathways (13). Thus, we hypothesized that L. acidophilus KLDS1.0901 would possess the ability to alleviate NAFLD. The aim of this study was to assess the protective effects of L. acidophilus KLDS1.0901 on HFD-induced NAFLD. First, hepatic lipid profile and histological alterations were determined to study whether L. acidophilus KLDS1.0901 could ameliorate NAFLD. However, the intestinal permeability and gut barrier were explored. Finally, gut microbiota was analyzed to elucidate the mechanism from the insights of the gut–liver axis.
Materials and methods
Bacterial strain and culture
Traditional fermented dairy products from Xinjiang Province, China were used to obtain Lactobacillus acidophilus KLDS 1.0901 and stored in 20% (v/v) glycerol at −20°C. The bacteria was incubated for 18 h in de Man Rogosa and Sharpe (MRS) broth (2% v/v) at 37°C and subcultured two times. Bacterial cultures were then centrifuged (6,000 rpm for 10 min at 4°C), washed three times with PBS solution, and the supernatant discarded. Cells were then resuspended in PBS at 5 × 109 CFU/mL.
Animals and experiment design
Following the method of Nguyen et al. (13), 6-week-old male C57BL/6J mice (n = 24) were obtained from the Vital River Laboratory Animal Technology Company (Beijing, China). Study animals were housed in a sterile animal room at 22 ±0.5°C, 55±5% humidity with 12 h light/12 h dark cycles. They had ad libitum access to chow and water ad libitum throughout the study. Following a 7-day acclimatization phase, mice were randomly divided into three groups (n = 8 mice per group). Control group (NC) mice were fed a D12450B diet, while others were fed a D12492 high-fat diet for 8 weeks. Feed formulas are reported in Supplementary Table S1. Both groups were gavaged with 0.2 mL of sterile PBS solution, and for the L. acidophilus KLDS1.0901 group (KLDS1.0901), the mice were administered with 0.2 mL of the L. acidophilus KLDS1.0901 (109 CFU/d). Study animals were humanely sacrificed after 12 h of fasting and blood samples were obtained. Colon and liver samples as well as colon and cecum content were collected and stored at−80°C for further analysis. The Northeast Agricultural University Guide for the Care and Use of Laboratory Animals was followed to ensure strict ethical animal procedures. In addition, our study was approved by the Northeast Agricultural University Animal Ethics Committee.
Histopathological analysis of liver
In reference to the previous method with a slight modification (14), we placed all liver tissues in paraffin, in thin slices of 5 μm thick, and stained with hematoxylin and eosin (H&E) and oil O red after deparaffinization. The sections were observed under a light microscope (Nikon E100, 200 × magnification) for lesions and other histological features. The lipid droplet area (percentage of total area) of each group was analyzed using the software Image J. NAFLD activity integral was calculated based on Supplementary Table S2.
Determination of TC, TG, LDL-C, and HDL-C in liver
The liver tissues were homogenized in aseptic PBS (1:9, w/v), succeeded by centrifugation at 10,000 rpm for 10 min at 4°C. The concentration of TC, TG, HDL-C, and LDL-C was detected using mouse ELISA kits (Conodi creatures, Fujian, China) based on the instructions of the manufacturer.
Determination of ALT and AST in Serum
Each mice group's serum AST and ALT levels were obtained using mouse kits (Conodi Creatures, Fujian, China) by following the manufacturer's directives.
ELISA measurement of inflammatory cytokines
Aseptic PBS (1:9, w/v) was used to homogenize mice liver tissues, followed by centrifugation (10,000 rpm, 10 min, 4°C). Based on the manufacturer's directives, we identified the concentration of TNF-α, IL-6, IL-10, and IL-1β with mouse ELISA kits (Conodi creatures, Fujian, China).
Measurement of the intestinal permeability
Following the manufacturer's instructions, the Enzyme-Linked Immunosorbent Assay (ELISA) kits (Conodi creatures, Fujian, China) were used to evaluate the concentrations of D-lactic acid (D-LA) and lipopolysaccharide (LPS) in mice serum samples. The serum samples were acquired before the mice were sacrificed.
Real-time quantitative polymerase chain reaction (RT-qPCR) analysis
Real-time quantitative polymerase chain reaction was used to determine relative mRNA expression levels of tight junction protein ZO-1, claudin-1, occludin, and Muc-2. Colon tissue RNA levels were obtained and quantified using the Total RNA kit (Vazyme, Nanjing, China), and 2000C Ultra-micro UV spectrophotometer (Thermo Fisher Scientific Inc., USA), respectively. We used the Transcriptor First Strand cDNA Synthesis kit (Promega, Madison, USA) to synthesize cDNA for this study. The GoTaq°R SYBR-Green qPCR Master Mix (Promega, Madison, USA) was used to perform RT-qPCR corrections. The relative mRNA expressions of specific genes were calculated by the 2−ΔΔCT method. GAPDH genes are used as internal reference genes. Supplementary Table S3 displays the specific gene primers as designed by Sangon Biotech Co., Ltd (Shanghai, China).
DNA extraction and 16S rRNA gene sequencing
Fecal samples served as the substrate for the total bacterial genomic DNA using the Fast DNA SPIN extraction kits (MP Biomedicals, Santa Ana, CA, USA). DNA molecular size and quantification were performed using a 0.8% agarose gel electrophoresis and the NanoDrop NC-2000 spectrophotometer, respectively. The V3–V4 region of bacterial 16S rRNA genes was then amplified using PCR with the forward primer 338F (5'-ACTCCTACGGGAGGCAGCA-3') and the reverse primer 806R (5'-GGACTACHVGGGTWTCTAAT-3'). PCR amplicons were purified with Agencourt AMPure Beads (Beckman Coulter, Indianapolis, IN) and quantified using the PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). Finally, the MiSeq Reagent kit v3 (Shanghai Personal Biotechnology Co., Ltd, Shanghai, China) was used to carry out the sequencing on the Illumina MiSeq platform.
Statistical analysis
All data were analyzed using SPSS 22.0 software (SPSS Inc., Chicago, IL, USA). Statistical analysis of Duncan's multiple range tests after one-way analysis of variance (ANOVA). For all analyses, at p < 0.05, the differences were considered significant.
Results
Effect of L. acidophilus KLDS1.0901 administration on body weight, body fat rate, Lee's index, and liver index of HFD-fed mice
High-fat diet significantly increased (p < 0.01) the final body weight, whereas the L. acidophilus KLDS1.0901 administration reduced (p < 0.01) it when compared with the HFD group (Figure 1A). The body fat rate of the mice in the HFD group was sharply elevated when compared to that of the NC group (p < 0.01), however, L. acidophilus KLDS1.0901 administration inhibited the increase (p < 0.01) (Figure 1B). Lee's index in the HFD group was higher (p < 0.01) than that in the control group, but Lee's index was remarkably decreased (p < 0.01) after L. acidophilus KLDS1.0901 administration (Figure 1C). In the analysis of the liver index, compared with the NC group, mice in the HFD group showed a marked increase (p < 0.01), which was reversed (p < 0.05) by the L. acidophilus KLDS1.0901 administration (Figure 1D).
Figure 1
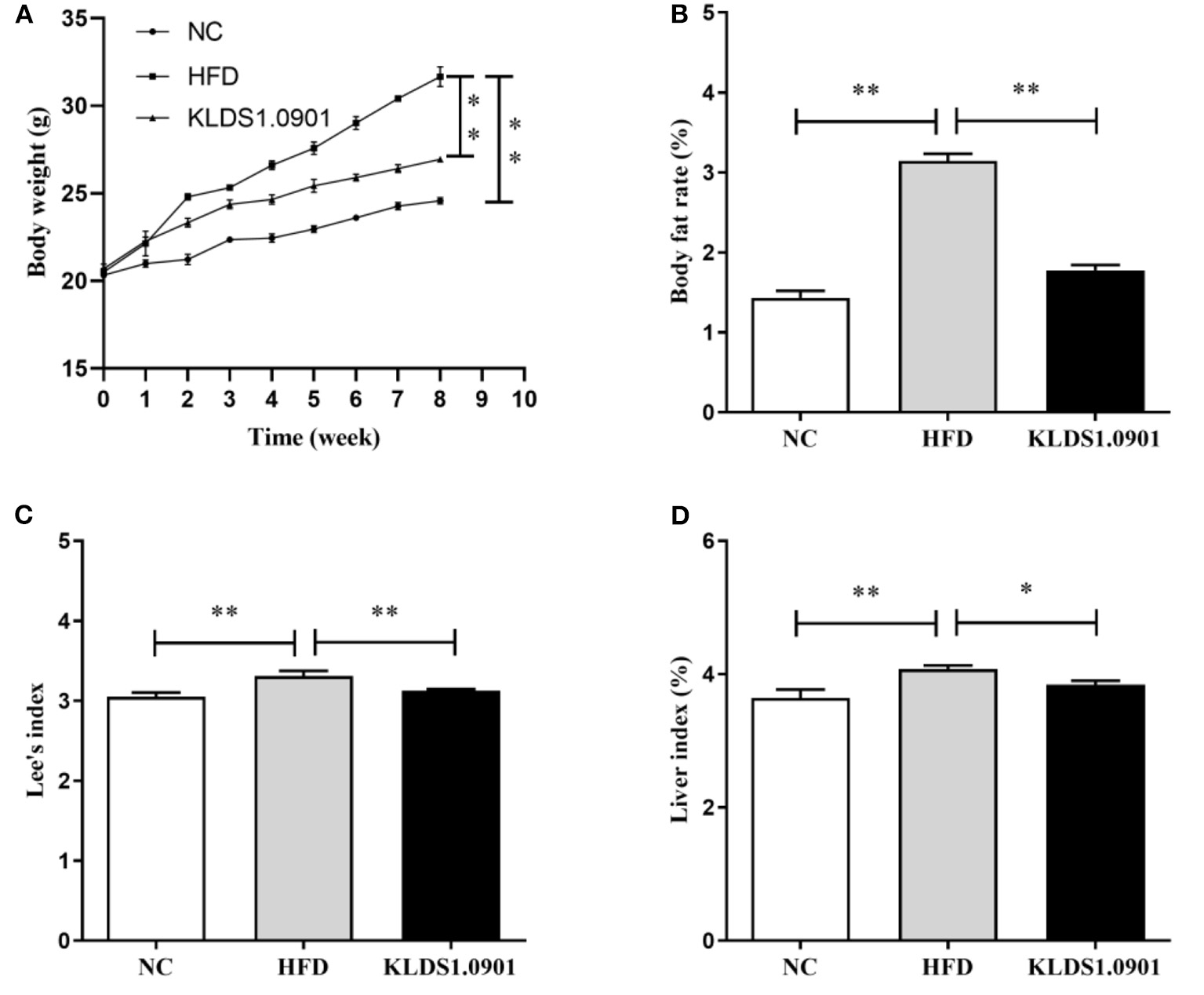
Effect of L. acidophilus KLDS1.0901 administration on body weight (A), body fat rate (B), Lee's index (C), and liver index (D) of HFD-fed mice. NC, normal control group; HFD, high-fat diet group; and KLDS1.0901, L. acidophilus KLDS1.0901 group. *p < 0.05 and **p < 0.01 indicated that there was a significant difference when compared with the HFD group.
Effect of L. acidophilus KLDS1.0901 administration on lipid accumulation and liver function in HFD-fed mice
To analyze hepatic lipid accumulation, the levels of TG, TC, LDL-C, and HDL-C in the liver of mice were determined by ELISA. As shown in Figures 2A–D. The lipid profiles including TG, TC, and LDL-C were pronouncedly (p < 0.01) enhanced in the HFD group when compared with the control group. However, L. acidophilus KLDS1.0901 administration suppressed the increases in these lipid parameters (p < 0.01). On the contrary, the HDL-C level was significantly lowered (p < 0.01) in the HFD group when compared with that in the NC group, L. acidophilus KLDS1.0901 administration reversed this trend (p < 0.01). As shown in Figures 3A, B. In order to study liver function, the serum ALT and AST levels of mice were examined by ELISA. A significant increase (p < 0.01) of ALT and AST in serum was observed in the HFD group. Notably, L. acidophilus KLDS1.0901 administration conspicuously inhibited this effect (p < 0.01).
Figure 2
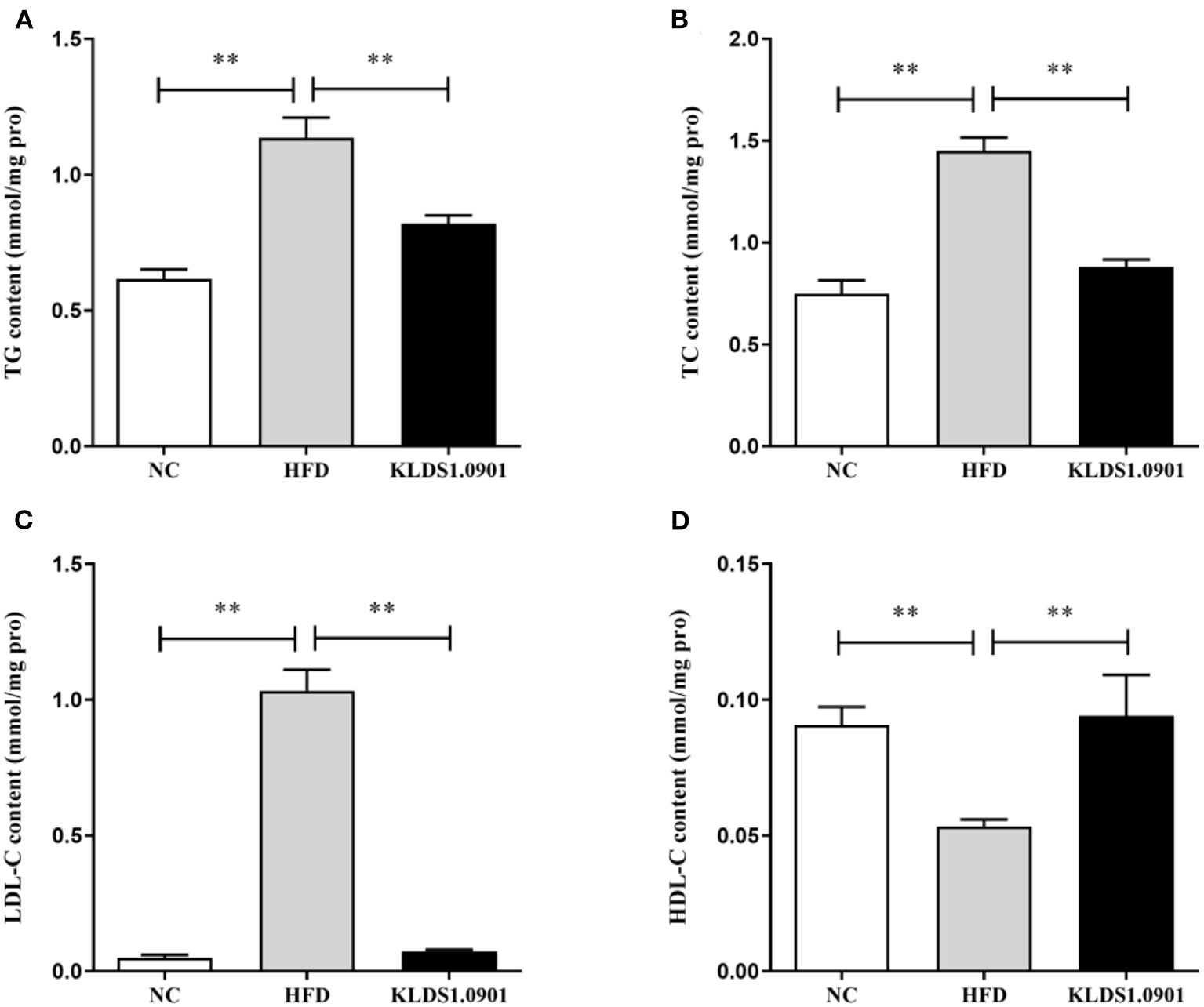
Effects of L. acidophilus KLDS1.0901 administration on lipid accumulation in HFD-fed mice. NC, normal control group; HFD, high-fat diet group; and KLDS1.0901, L. acidophilus KLDS1.0901 group. (A) Hepatic triglyceride (TG) level; (B) hepatic total cholesterol (TC) level; (C) hepatic low-density lipoprotein cholesterol (HDL-C) level; and (D) hepatic high-density lipoprotein cholesterol (LDL-C) level. Values are expressed as mean ± SD (n = 8). **p < 0.01 indicated that there was a significant difference when compared with the HFD group.
Figure 3
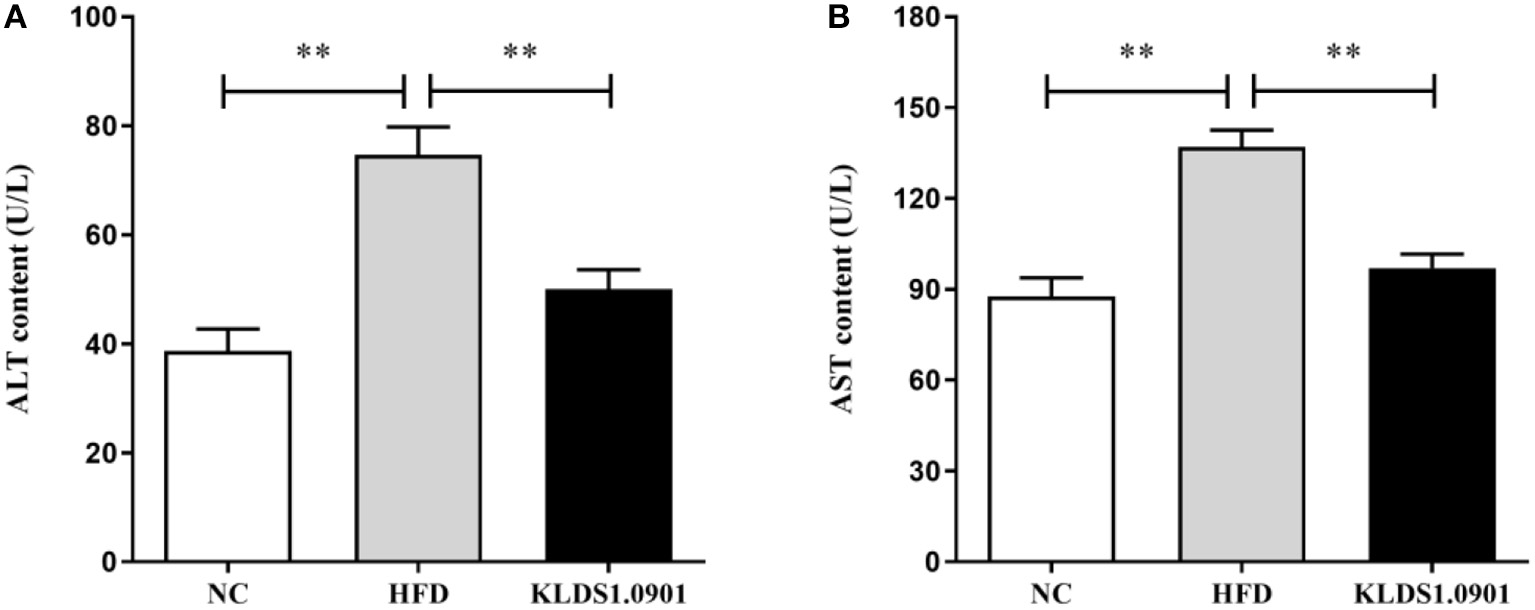
Effects of L. acidophilus KLDS1.0901 administration on liver function. NC, normal control group; HFD, high-fat diet group; and KLDS1.0901, L. acidophilus KLDS1.0901 group. (A) Serum alanine aminotransferase (ALT) level; and (B) serum aspartate aminotransferase (AST) level. Values are expressed as mean ± SD(n = 8). **p < 0.01 indicated that there was a significant difference when compared with the HFD group.
Effects of L. acidophilus KLDS1.0901 administration on histological alterations of liver
In the NC group, there was no steatosis, and the tissue structure was clear and complete. The H&E staining showed that HFD blurred the boundary and induced the regular round fat hole. In particular, HFD detrimentally caused substantial fat accumulation in the liver during 8 weeks of feeding, whereas L. acidophilus KLDS1.0901 administration effectively restored the trend caused by HFD (Figure 4A). As shown in Figure 4B, in view of the NAFLD activity score (NAS), the HFD-induced increase was significantly (p < 0.01) reduced by L. acidophilus KLDS1.0901 administration. Quantification of lipid area per total area also suggested that L. acidophilus KLDS1.0901 administration significantly reduced (p < 0.01) liver lipid accumulation of HFD-fed mice (Figure 4C).
Figure 4
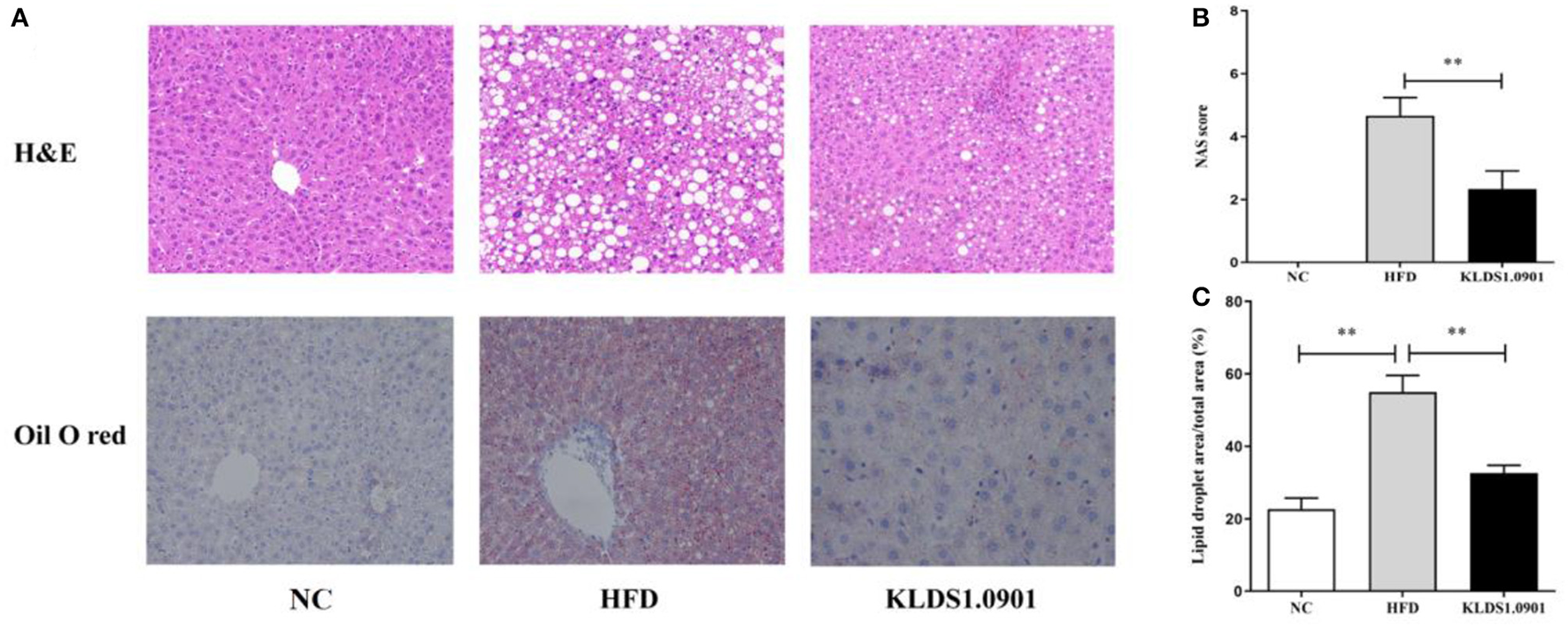
Effects of L. acidophilus KLDS1.0901 administration on histological alterations of the liver. NC, normal control group; HFD, high-fat diet group; and KLDS1.0901, L. acidophilus KLDS1.0901 group. (A) The representative histological changes of liver sections; (B) NAFLD Activity Score (NAS); and (C) quantification of Oil Red O-stained hepatic lipid droplets. Values are expressed as mean ± SD (n = 8). **p < 0.01 indicated that there was a significant difference when compared with the HFD group.
Effect of L. acidophilus KLDS1.0901 administration on hepatic inflammation
As shown in Figure 5, our cytokine analyses showed that the concentrations of IL-6, IL-1β, and TNF-α increased considerably in the HFD mice compared to the NC group (p < 0.01); however, L. acidophilus KLDS1.0901 administration significantly decreased them (p < 0.01). While the concentration of IL-10 was significantly decreased (p < 0.01), L. acidophilus KLDS1.0901 administration significantly increased it (p < 0.05).
Figure 5
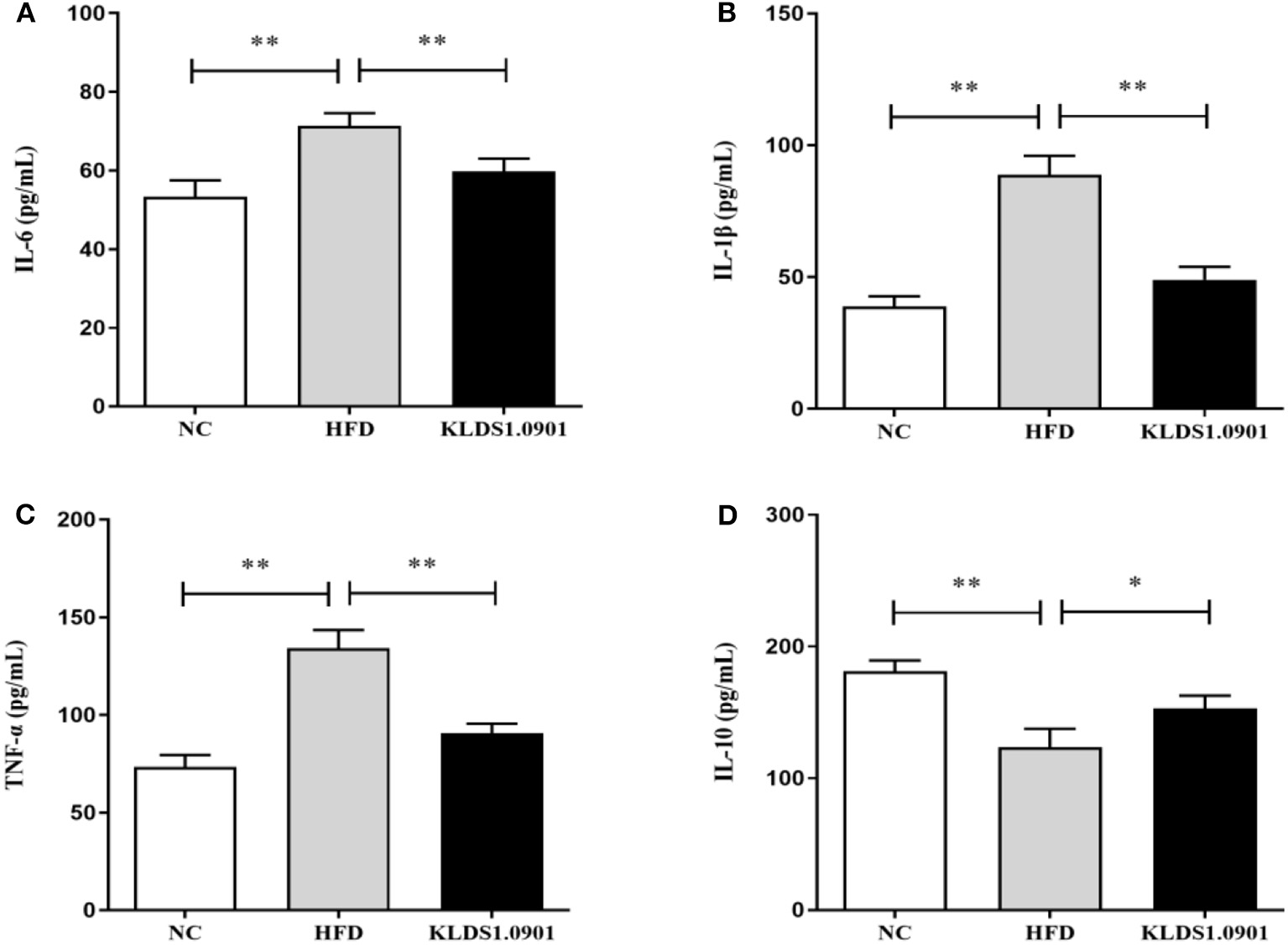
Effects of L. acidophilus KLDS1.0901 administration on hepatic inflammation. NC, normal control group; HFD, high-fat diet group; and KLDS1.0901, L. acidophilus KLDS1.0901 group. (A) IL-6; (B) IL-1β; (C) TNF-α; and (D) IL-10. Values are expressed as mean ± SD (n = 8). *p < 0.05 and **p < 0.01 indicated that there was a significant difference when compared with the HFD group.
Effect of L. acidophilus KLDS1.0901 administration on the intestinal permeability and gut barrier
Lipopolysaccharides and D-lactic acid are standard indicators of intestinal barrier damage, the concentration of LPS and D-lactic acid was examined by ELISA. As shown in Figures 6A, B, the concentration of LPS and D-lactic acid increased substantially (p < 0.01) in the HFD group, resulting in the anticipated mucosal damage, but L. acidophilus KLDS1.0901 administration drastically reduced (p < 0.01) the concentration of LPS and D-lactic acid, thus improving intestinal permeability. To study the effect of L. acidophilus KLDS1.0901 administration on the gut barrier, we determined the mRNA levels of occludin, claudin-1, ZO-1, and Muc-2. The results showed that the mRNA levels of these four genes were strikingly downregulated (p < 0.01) in the HFD group, but L. acidophilus KLDS1.0901 administration significantly upregulated (p < 0.01) the mRNA levels of these four genes (Figures 6C–F).
Figure 6
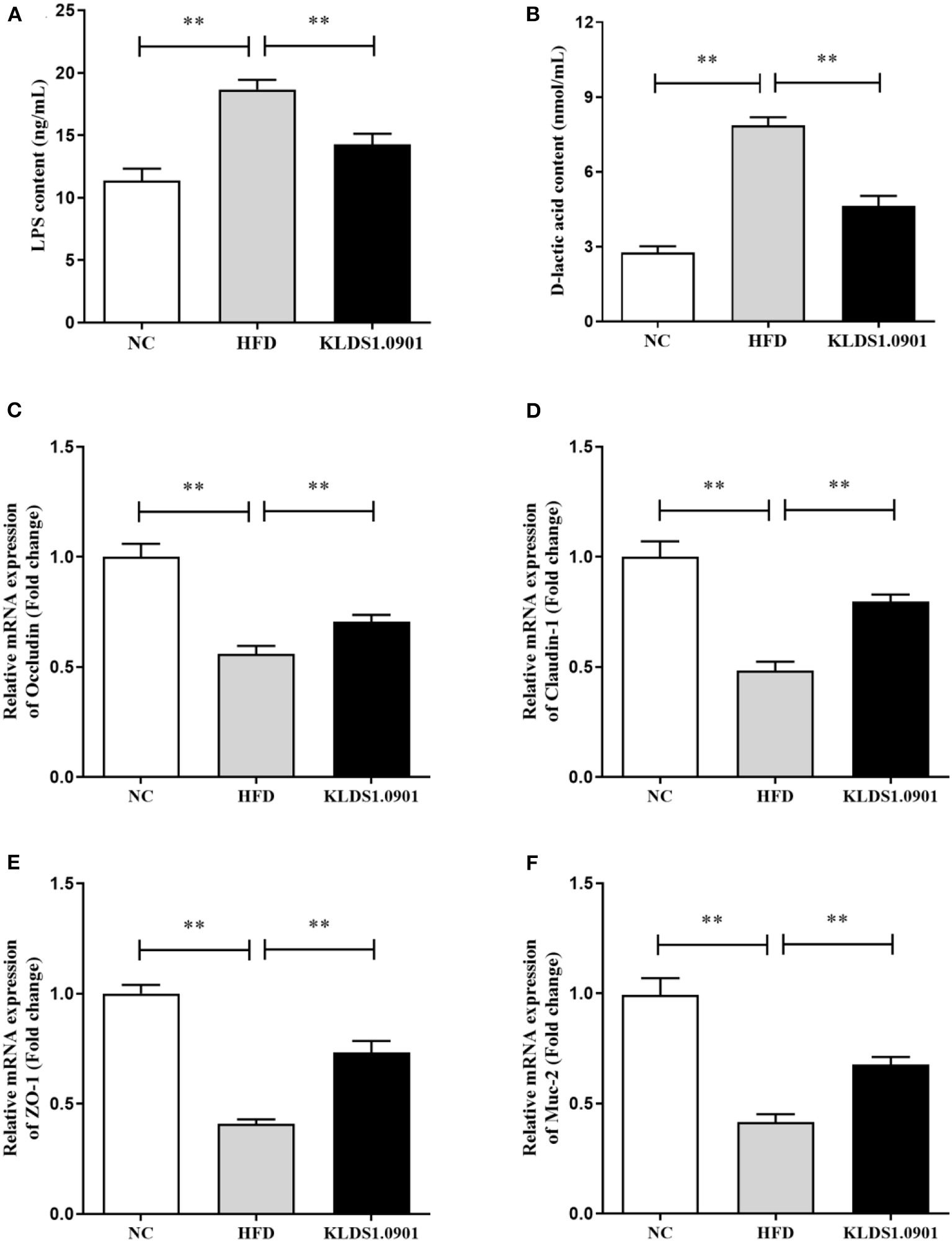
Effects of L. acidophilus KLDS1.0901 administration on the intestinal permeability and gut barrier. NC, normal control group; HFD, high-fat diet group; and KLDS1.0901, L. acidophilus KLDS1.0901 group. (A) Serum LPS concentration; (B) serum D-lactic acid concentration; (C) occludin; (D) claudin-1; (E) ZO-1; and (F) Muc-2. Values are expressed as mean ± SD (n = 8). **p < 0.01 indicated that there was a significant difference when compared with the HFD group.
Effect of L. acidophilus KLDS1.0901 administration on gut microbiota
We sequenced the 16S rDNA V3–V4 variable region to analyze the cecal gut microbiota. This enabled us to gain insights as to whether L. acidophilus KLDS1.0901 administration modulated the bacterial communities of NAFLD-induced mice. The results showed that the OTU determined the gut microbiota diversity of each study group, with the common abundance shown with a Venn diagram (177 OTU in all groups). Our results showed 207, 211, and 216 different microorganisms in the NC, HFD, and MC groups, respectively (Figure 7A). The α-diversity reflecting the microbial community diversity was assessed by the Chao 1 and Shannon indexes (Figures 7B, C). There were no significant changes in Chao 1 index among the three groups. However, the Shannon index in the HFD mice was lower than that in the NC group (p < 0.05), which was significantly elevated by L. acidophilus KLDS1.0901 administration (p < 0.05). The hierarchical clustering tree results of the current study grouped the NC and KLDS 1.0901 cohorts together and then clustered with the HFD communities (Figure 7D).
Figure 7
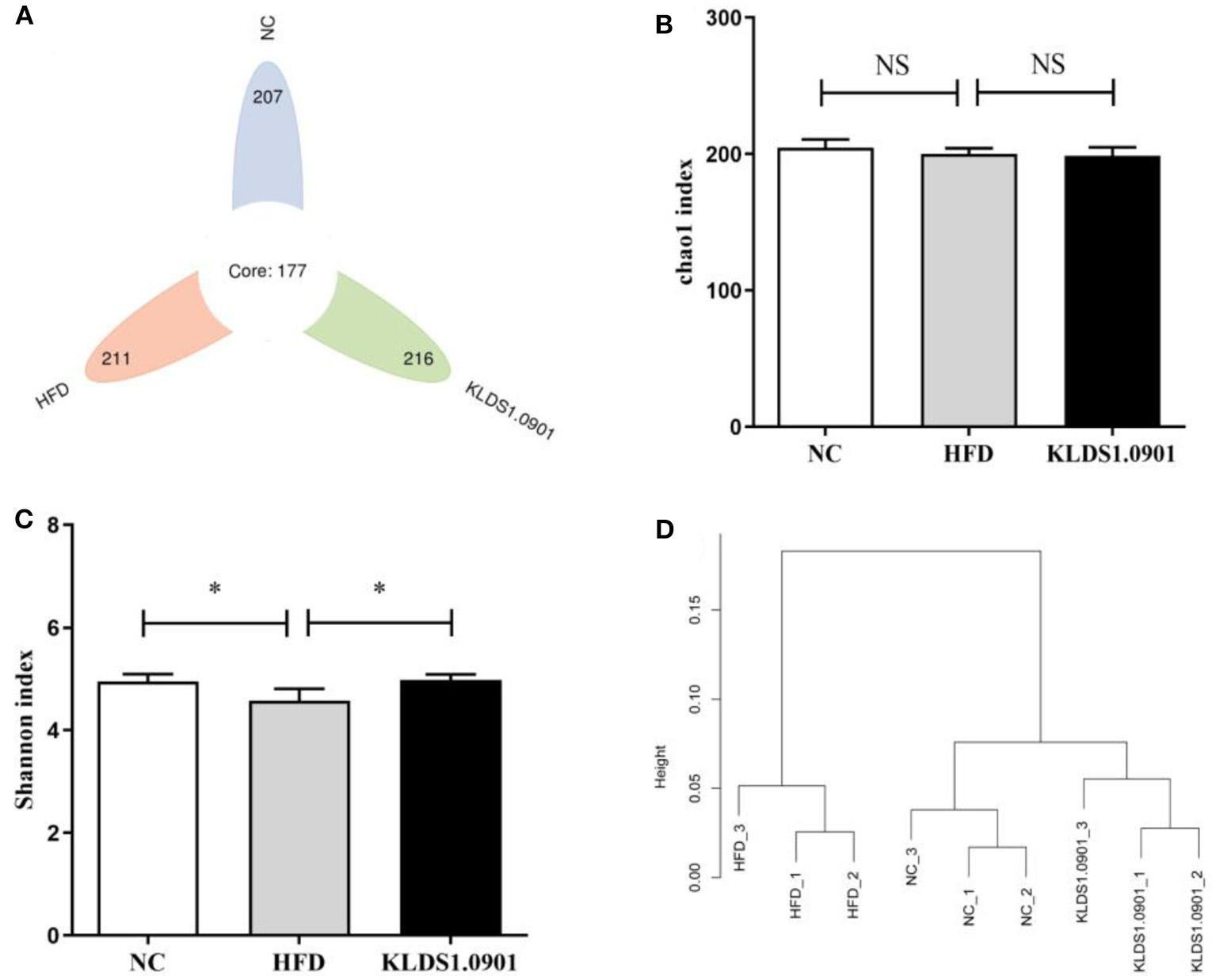
L. acidophilus KLDS1.0901 administration regulates (A) the number of OTUs, (B) Chao 1 index, (C) Shannon index, and (D) the hierarchical clustering tree of weighted UniFrac distances. NC, normal control group; HFD, high-fat diet group; and KLDS1.0901, L. acidophilus KLDS1.0901 group. Values are expressed as mean ± SD (n = 8). *p < 0.05 indicated that there was a significant difference when compared with the HFD group. NS indicated that there was no significant difference when compared with the HFD group.
Our phylum-level results indicated that Firmicutes and Bacteriodetes make up at least 80% of all groups (Figure 8A). The HFD group had a high abundance of Firmicutes and depleted levels of Bacteriodetes. This group also had a significantly high ratio of Firmicutes to Bacteriodetes. We also observed that L. acidophilus KLDS1.0901 administration reversed this trend by lowering Firmicutes levels, increasing Bacteroidetes abundance, and reducing the HFD-induced Firmicutes-to-Bacteroidetes ratio.
Figure 8
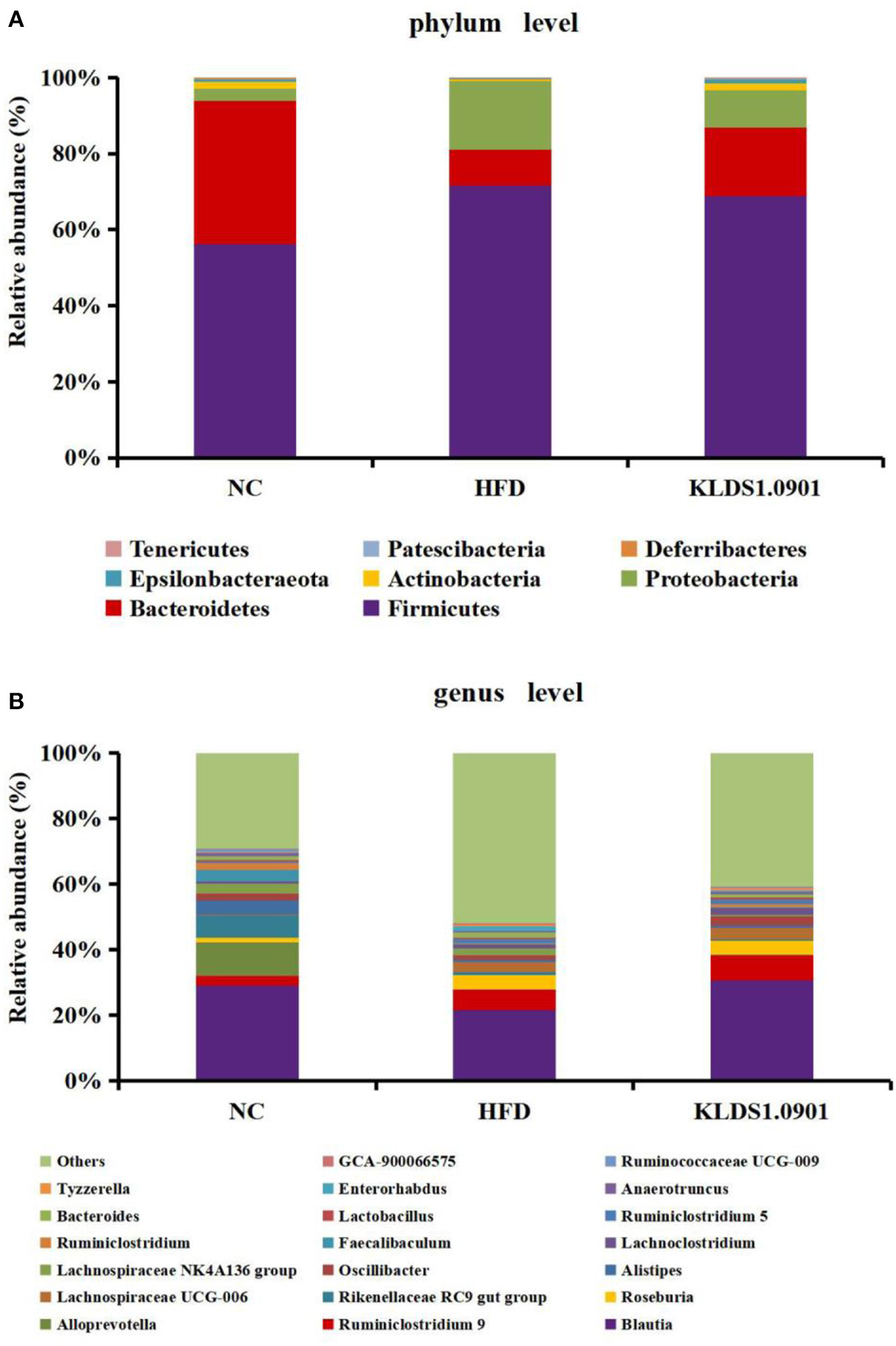
Changes of the gut microbiota at the phylum (A) and genus (B) level after L. acidophilus KLDS1.0901 administration. NC, normal control group; HFD, high-fat diet group; and KLDS1.0901, L. acidophilus KLDS1.0901 group.
The genus-level gut microbiota results for the different groups are shown in Figure 8B. We observed that in the HFD group, the relative abundances of Roseburia, Lachnospiraceae UCG-006, Bacteroides, and Enterorhabdus were much higher than that in the NC group, which were suppressed by L. acidophilus KLDS1.0901 administration. However, the HFD group had low levels of Blautia, Alistipes, Oscillibacter, Faecalibaculum, Ruminiclostridium, Lactobacillus, and Ruminococcaceae UCG-009, which were reversed by L. acidophilus KLDS1.0901 administration. Our observations indicated that L. acidophilus KLDS1.0901 administration modulated the composition of the intestinal microorganisms of HFD-fed mice.
Discussion
The prevalence of NAFLD has been linked to the onset and exacerbation of other metabolic disorders like type 2 diabetes (T2D) and obesity, thus posing a significant public health concern. It has been posited previously that the progression of NAFLD could be driven by lipid metabolism disorders (2). Accumulating evidence suggests that some lactic acid bacteria have the potential to relieve NAFLD (13, 15). Our previous studies showed L. acidophilus KLDS1.0901 with antioxidative activity had good tolerance to acid and bile salt and strong adhesion ability (11). Furthermore, L. acidophilus KLDS1.0901 could alleviate type 2 diabetes by regulating hepatic glucose, lipid metabolism, and gut microbiota in mice (12). L. acidophilus KLDS1.0901 also could prevent chronic alcoholic liver injury in mice by protecting the intestinal barrier and regulating gut microbiota and liver-related pathways (13). However, the underlying mechanisms were unclear. Thus, in this study, the possible mechanisms by which the same lactobacilli strains could prevent obesity were mined and increased the final body weight, body fat rate, Lee's index, and liver index.
In this study, HFD significantly increased the final body weight, body fat rate, Lee's index, and liver index, which was reversed by the L. acidophilus KLDS1.0901 administration, which was in line with the results of Naudin et al. (16), expressing L. acidophilus KLDS1.0901 had the protective effects of HFD-induced NAFLD. HFD (manifested in hyperlipidemia) is known to trigger NAFLD by elevating TC, TG, and LDL-C levels, with a corresponding drop in host HDL-C levels (17). Interestingly, some prior studies have confirmed that reducing TC, TG, and LDL-C levels can alleviate NAFLD (18, 19, 51). In this study, the HFD significantly elevated the hepatic levels of TC, TG, and LDL-C and lowered the HDL-C levels, resulting in lipid metabolism disorder. However, L. acidophilus KLDS1.0901 administration reversed this trend and agrees with the effects of L. plantarum NA136 and L. johnsonii BS15 as reported earlier (20, 21). Meanwhile, the H&E and Oil Red O-stained staining showed that HFD detrimentally caused substantial fat accumulation in the liver during 8 weeks of feeding and increased the NAS score and lipid area per total area. These results indicated that L. acidophilus KLDS1.0901 administration could have protective effects from hepatic steatosis due to lowered lipid content in NAFLD mice.
Furthermore, elevated lipid accumulation can be toxic and trigger liver injury via hepatic parenchymal cell inflammation. These liver functions are mostly measured by serum AST and ALT levels (22). In this study, a significant increase of ALT and AST in serum was observed in the HFD group. Notably, L. acidophilus KLDS1.0901 administration conspicuously inhibited this effect. These results demonstrated that L. acidophilus KLDS1.0901 administration protects liver functions and mitigates HFD-induced liver injury. Host tissue damage and repair, immune modulation, and inflammation activities are generally regulated by cytokines. Similarly, cytokines such as IL-6, IL-1β, and TNF-α can be mechanistically stimulated by intestinal bacterial communities (23). It has been reported that Lactobacillus and Pediococcus ameliorate the progression of NAFLD through the modulation of cytokines (24). Our results showed that HFD induction increased the IL-6, IL-1β, and TNF-α concentrations accompanied by the decrease of IL-10. However, L. acidophilus KLDS1.0901 administration effectively restored the trend, indicating that L. acidophilus KLDS1.0901 administration could alleviate the NAFLD by modulating the concentrations of cytokines.
The gut and the liver communicate through the gut–liver axis, which consists of the gut, the liver, and the intestinal barrier (25). Tight junctions, composed of ZO-1, occludin, and claudin, link the intestinal epithelial cells and maintain intestinal barrier integrity (26). Both intestinal barrier malfunction and dysbiosis of the gut microbiota play important roles in the pathophysiology of liver diseases (27). NAFDL-associated gut microbiota dysbiosis induced by long-term consumption of a high-fat and high-fructose diet may disrupt intestinal barrier function by reducing the expression of intestinal tight junction proteins (occludin, claudin-1, and ZO-1) (28).
Homeostasis in intestinal epithelial cell permeability and regulating barrier functions are modulated by ZO-1 (29), with occludin serving as a tight junction protein molecule (30). Claudins and mucins (Muc1–Muc6), on the contrary, regulate inflammation, intestinal epithelial homeostasis, and the colon mucus layer (31, 32). Intestinal inflammation typically results when the absence of tight junctions weakens the intestinal barriers and allows bacterial invasion (33). Our results showed that the mRNA levels of these four genes were strikingly downregulated in the HFD group, but L. acidophilus KLDS1.0901 administration upregulated the mRNA levels of these four genes. Intestinal inflammation results in “leaky gut,” a condition where bacteria and their metabolites trigger the release of cellular inflammatory factors with deleterious effects on the host (34). Several previous investigations have demonstrated that LPS regulates intestinal flora and inflammation responses involved in the onset of metabolic diseases (35). In particular, NAFLD onset has been correlated with the presence of bacterial LPS from enteric gram-negative flora (53). In this study, the concentration of LPS and D-lactic acid was significantly increased (p < 0.05) in the HFD group, but L. acidophilus KLDS1.0901 administration could drastically reduce the concentration of LPS and D-lactic acid. These findings indicated that L. acidophilus KLDS1.0901 administration could improve the intestinal barrier function induced by HFD.
The gut microbiome is a key environmental factor in the onset of NAFLD (36). Firmicutes and Bacteroidetes are essential participants in host energy metabolism (37). The phylum-level study of a mixed lactobacilli treatment administered to HFD-fed mice indicated that the Firmicutes bacterial group increased with a corresponding drop in the relative abundance of Bacteriodetes (38–40). These researchers discovered that an increased Firmicutes to Bacteriodetes ratio increased calories absorption, harvestable energy levels, and obesity biomarkers (41, 42), which was in line with the results of Yu et al. (15). The results showed that the F/B ratio was significantly lower in the L. acidophilus KLDS1.0901 group, suggesting that 1.0901 may regulate energy metabolism in mice by modulating intestinal flora, which, in turn, affects the degree of fat accumulation.
The genus-level gut microbiota results for the different groups are shown in Figure 8B. We observed that in the HFD group, the relative abundances of Roseburia, Lachnospiraceae UCG-006, Bacteroides, and Enterorhabdus were much higher when compared to the NC group, which were suppressed by L. acidophilus KLDS1.0901 administration. However, the HFD group had low levels of Blautia, Alistipes, Oscillibacter, Faecalibaculum, Ruminiclostridium, Lactobacillus, and Ruminococcaceae UCG-009, which were reversed by L. acidophilus KLDS1.0901 administration. Our observations indicated that L. acidophilus KLDS1.0901 administration modulated the composition of the intestinal microbiome of HFD-fed mice.
Genus-level results of the current study indicate that the relative abundances of Roseburia, Lachnospiraceae UCG-006, Bacteroides, and Enterorhabdus elevated while that of Blautia, Alistipes, Oscillibacter, Faecalibaculum, Ruminiclostridium, Lactobacillus, and Ruminococcaceae UCG-009 were significantly decreased in the HFD group when compared to the NC group. It has been reported that Roseburia, which is known for suppressing SCFAs-producing bacteria, was enriched in HFD-induced NAFLD mice (43, 44). Enterorhabdus secretes LPS and is thus implicated in obesity, insulin resistance, and pro-inflammatory factors proliferation (21). Moreover, a substantial relative abundance of Bacteroides has been observed in individuals with NAFLD (45). Conversely, SCFAs secreted by, Blautia, is known to be pathogen-inhibiting and promotes healthy intestinal microbiota (46). Alistipes was negatively correlated with obesity (47). Lactobacillus is known to be pathogen-inhibiting and promotes healthy intestinal microbiota (48). Previous observations note that HFD reduced the relative abundance of Faecalibaculum (49). In addition, Ruminiclostridium were linked to weight reduction or a lean phenotype (50). In summary, L. acidophilus KLDS1.0901 further alleviates NAFLD through the intestinal-liver axis by regulating the ecological imbalance of intestinal microbiota caused by a high-fat diet and improving intestinal barrier function.
Conclusion
Lactobacillus KLDS1.0901 administration could significantly decrease body weight gain, Lee's index body, fat rate, and liver index. Lactobacillus KLDS1.0901 administration could significantly improve lipid profiles by decreasing the hepatic levels of TC, TG, and LDL-C and ALT and AST in serum and increasing the HDL-C levels. L. acidophilus KLDS1.0901 administration could decrease the IL-6, IL-1β, and TNF-α concentrations accompanied by the decrease of IL-10. L. acidophilus KLDS1.0901 administration could improve the intestinal barrier function by upregulating the mRNA levels of occludin, claudin-1, ZO-1, and Muc-2, which were coupled to the decreases of the concentration of LPS and D-lactic acid. Notably, L. acidophilus KLDS1.0901 administration modulated the gut microbiota to the normal pattern. Hence, our study provides guidance for the selection and application of presumed probiotics in the treatment of NAFLD.
Statements
Data availability statement
The data presented in the study are deposited in the NCBI repository (https://www.ncbi.nlm.nih.gov/sra/), accession number PRJNA925467.
Ethics statement
The animal study was reviewed and approved by Northeast Agricultural University Animal Ethics Committee.
Author contributions
BL and BT designed the study. YWan and FJ performed the experiments. YWang and ZW wrote the manuscript. XS and ZX analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
The present research study was financially supported by the National Natural Science Foundation of China (32101919), the Young Elite Scientist Sponsorship Program by CAST (YESS20200271), and the China Postdoctoral Science Foundation (2022M721071).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2023.1147423/full#supplementary-material
References
1.
Samuel VT Shulman GI . Nonalcoholic fatty liver disease as a nexus of metabolic and hepatic diseases. Cell Metab. (2018) 27:22–41. 10.1016/j.cmet.2017.08.002
2.
Machado M Marques-Vidal P Cortez-Pinto H . Hepatic histology in obese patients undergoing bariatric surgery. J Hepatol. (2006) 45:600–6. 10.1016/j.jhep.2006.06.013
3.
Cobbina E Akhlaghi F . Non-alcoholic fatty liver disease (NAFLD)–pathogenesis, classification, and effect on drug metabolizing enzymes and transporters. Drug Metab Rev. (2017) 49:197–211. 10.1080/03602532.2017.1293683
4.
Bedossa P . Pathology of non-alcoholic fatty liver disease. Liver International. (2017) 37:85–9. 10.1111/liv.13301
5.
Sun L Bao L Phurbu D Qiao S Sun S Perma Y et al . Amelioration of metabolic disorders by a mushroom-derived polyphenols correlates with the reduction of Ruminococcaceae in gut of DIO mice. Food Sci Hum Wellness. (2021) 10:442–51. 10.1016/j.fshw.2021.04.006
6.
Yan S Kai W Xiaoying W Aiqun O Feiran W Liming W et al . Effect of fermented bee pollen on metabolic syndrome in high-fat diet-induced mice. Food Sci Hum Wellness. (2021) 10:345–55. 10.1016/j.fshw.2021.02.026
7.
Tang C Zhou W Shan M Lu Z Lu Y . Yogurt-derived Lactobacillus plantarum Q16 alleviated high-fat diet-induced non-alcoholic fatty liver disease in mice. Food Sci Hum Wellness. (2022) 11:1428–39. 10.1016/j.fshw.2022.04.034
8.
Guss DA Mohanty SR . Gut microbia dysbiosis in non-alcoholic fatty liver disease. Transl Cancer Res. (2016) 5:S152–5. 10.21037/tcr.2016.07.07
9.
Cui Y Wang Q Chang R Zhou X Xu C . Intestinal barrier function–non-alcoholic fatty liver disease interactions and possible role of gut microbiota. J Agric Food Chem. (2019) 67:2754–62. 10.1021/acs.jafc.9b00080
10.
Altamirano-Ríos AV Guadarrama-Lezama A Arroyo-Maya IJ Hernández-Álvarez AJ Orozco-Villafuerte J . Effect of encapsulation methods and materials on the survival and viability of Lactobacillus acidophilus: a review. Int J Food Sci Technol. (2022) 57:4027–40. 10.1111/ijfs.15779
11.
Evivie SE Huo G Igene J Bian X . Some current applications, limitations and future perspectives of lactic acid bacteria as probiotics. Food Nutr Res. (2017) 61:1318034. 10.1080/16546628.2017.1318034
12.
Parra Huertas RA . Review lactic acid bacteria: functional role in the foods. Biotecnología en el Sector Agropecuario y Agroindustrial. (2010) 8:93–105.
13.
Nguyen HT Gu M Werlinger P Cho JH Cheng J Suh J . Lactobacillus sakei MJM60958 as a potential probiotic alleviated non-alcoholic fatty liver disease in mice fed a high-fat diet by modulating lipid metabolism, inflammation, Gut Microbiota Int J Mol Sci. (2022) 23:13436. 10.3390/ijms232113436
14.
Wang G Jiao T Xu Y Li D Si Q Hao J et al . Bifidobacterium adolescentis and Lactobacillus rhamnosus alleviate non-alcoholic fatty liver disease induced by a high-fat, high-cholesterol diet through modulation of different gut microbiota-dependent pathways. Food Funct. (2020) 11:6115–27. 10.1039/C9FO02905B
15.
Yu YS Gi Soo Y Jieun C Chang-Ho K Byung Yong K Seung-Jo Y et al . Lactobacillus lactis and Pediococcus pentosaceus-driven reprogramming of gut microbiome and metabolome ameliorates the progression of non-alcoholic fatty liver disease. Clin Transl Med. (2021) 11:e634. 10.1002/ctm2.634
16.
Naudin CR Maner-Smith K Owens JA Wynn GM Robinson BS Matthews JD et al . Lactococcus lactis subspecies cremoris elicits protection against metabolic changes induced by a western-style diet. Gastroenterology. (2020) 159:639–51. e5. 10.1053/j.gastro.2020.03.010
17.
Nabavi S Rafraf M Somi MH Homayouni-Rad A Asghari-Jafarabadi M . Effects of probiotic yogurt consumption on metabolic factors in individuals with nonalcoholic fatty liver disease. J Dairy Sci. (2014) 97:7386–93. 10.3168/jds.2014-8500
18.
Ye H Qian L Zhengzhe Z Maocheng S Changhui Z Tiehua Z . Effect of a novel potential probiotic Lactobacillus paracasei Jlus66 isolated from fermented milk on nonalcoholic fatty liver in rats. Food Funct. (2017) 8:4539–46. 10.1039/C7FO01108C
19.
Zhao Z Wang C Zhang L Zhao Y Duan C Zhang X et al . Lactobacillus plantarum NA136 improves the non-alcoholic fatty liver disease by modulating the AMPK/Nrf2 pathway. Appl Microbiol Biotechnol. (2019) 103:5843–50. 10.1007/s00253-019-09703-4
20.
Xin J Dong Z Hesong W Xueqin N Dan Y Kangcheng P et al . Preventing non-alcoholic fatty liver disease through Lactobacillus johnsonii BS15 by attenuating inflammation and mitochondrial injury and improving gut environment in obese mice. Appl Microbiol Biotechnol. (2014) 98:6817–29. 10.1007/s00253-014-5752-1
21.
Zhao Z Long C Yujuan Z Chao W Cuicui D Ge Y et al . Lactobacillus plantarum NA136 ameliorates nonalcoholic fatty liver disease by modulating gut microbiota, improving intestinal barrier integrity, attenuating inflammation. Appl Microbiol Biotechnol. (2020) 104:5273–82. 10.1007/s00253-020-10633-9
22.
Zhao L Yu J Yuxin N Tianzhu Z Cuicui D Cheng H et al . Protective effects of Lactobacillus plantarum C88 on chronic ethanol-induced liver injury in mice. J Funct Foods. (2017) 35:97–104. 10.1016/j.jff.2017.05.017
23.
DeCicco LA Rikans LE Tutor CG Hornbrook KR . Serum liver concentrations of tumor necrosis factor α and interleukin-1β following administration of carbon tetrachloride to male rats. Toxicol Lett. (1998) 98:115–21. 10.1016/S0378-4274(98)00110-6
24.
Lee NY Yoon SJ Han DH Gupta H Youn G Shin M et al . Lactobacillus and Pediococcus ameliorate progression of non-alcoholic fatty liver disease through modulation of the gut microbiome. Gut Microbes. (2020) 11:882–99. 10.1080/19490976.2020.1712984
25.
Yao M Lingling Q Yanmeng L Baohong W Lanjuan L . An update on the efficacy and functionality of probiotics for the treatment of non-alcoholic fatty liver disease. Engineering. (2021) 7:679–86. 10.1016/j.eng.2020.01.017
26.
Artis D . Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat Rev Immunol. (2008) 8:411–20. 10.1038/nri2316
27.
Paolella G Mandato C Pierri L Poeta M Stasi MD Vajro P . Gut-liver axis and probiotics: their role in non-alcoholic fatty liver disease. WJG. (2014) 20:15518. 10.3748/wjg.v20.i42.15518
28.
Gangarapu V Yildiz K Baysal B . Role of gut microbiota: obesity and NAFLD. Turkish J Gastroenterol. (2014) 25:2. 10.5152/tjg.2014.7886
29.
Dan Y Marchiando AM Weber CR Raleigh DR Wang Y Le S et al . MLCK-dependent exchange and actin binding region-dependent anchoring of ZO-1 regulate tight junction barrier function. Proc Natl Acad Sci U S A. (2010) 107:8237–41. 10.1073/pnas.0908869107
30.
Buschmann MM Shen L Rajapakse H Raleigh DR Wang Y Wang Y et al . Occludin OCEL-domain interactions are required for maintenance and regulation of the tight junction barrier to macromolecular flux. Mol Biol Cell. (2013) 24:3056–68. 10.1091/mbc.e12-09-0688
31.
Barmeyer C Fromm M Schulzke JD . Active passive involvement of claudins in the pathophysiology of intestinal inflammatory diseases. Pflügers Archiv - Eur J Physiol. (2017) 469:15–26. 10.1007/s00424-016-1914-6
32.
Dawson PA Huxley S Gardiner B Tran T McAuley JL Grimmond S et al . Reduced mucin sulfonation and impaired intestinal barrier function in the hyposulfataemic NaS1 null mouse. Gut. (2009) 58:910–9. 10.1136/gut.2007.147595
33.
Liping S Shen L Clayburgh DR . Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology. (2009) 136:551–63. 10.1053/j.gastro.2008.10.081
34.
Wang W Zhao J Gui W Sun D Dai H Xiao L et al . Tauroursodeoxycholic acid inhibits intestinal inflammation and barrier disruption in mice with non-alcoholic fatty liver disease. Br J Pharmacol. (2018) 175:469–84. 10.1111/bph.14095
35.
Ritze Y Bárdos G Claus A Ehrmann V Bergheim I Schwiertz A et al . Lactobacillus rhamnosus GG protects against non-alcoholic fatty liver disease in mice. PLoS ONE. (2014) 9:e80169. 10.1371/journal.pone.0080169
36.
Walker AW Julian P . Microbiology. Fighting obesity with bacteria. Science. (2013) 341:1069–70. 10.1126/science.1243787
37.
Indiani C Rizzardi K Castelo P Ferraz L Darrieux M Manzano Parisotto T . Childhood obesity and firmicutes/bacteroidetes ratio in the gut microbiota: a systematic review. Childhood Obesity. (2018) 14:501–9. 10.1089/chi.2018.0040
38.
Chang C-J Lin C-S Lu C-C Martel J Ko Y-F Ojcius DM et al . Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nat Commun. (2015) 6:7489. 10.1038/ncomms8489
39.
Ilseung C Shingo Y Laura C Methé BA Jiri Z Kelvin L et al . Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. (2012) 488:621–6. 10.1038/nature11400
40.
Zhang X Man Z Chi-Tang H Xiaojing G Zufang W Peifang W et al . Metagenomics analysis of gut microbiota modulatory effect of green tea polyphenols by high fat diet-induced obesity mice model. J Funct Foods. (2018) 46:268–77. 10.1016/j.jff.2018.05.003
41.
Turnbaugh PJ Ley RE Mahowald MA Vincent M Mardis ER Gordon JI . An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. (2006) 444:1027–31. 10.1038/nature05414
42.
Komaroff AL . The microbiome and risk for obesity and diabetes. JAMA. (2017) 317:355–6. 10.1001/jama.2016.20099
43.
Liu JP Wen-Li Z Shui-Jiao C Hong-Yun W Ya-Ni Y Yi-You Z et al . Effects of different diets on intestinal microbiota and nonalcoholic fatty liver disease development. World J Gastroenterol. (2016) 22:7353. 10.3748/wjg.v22.i32.7353
44.
Hu W Gao W Liu Z Fang Z Wang H Zhao J et al . Specific strains of Faecalibacterium prausnitzii ameliorate nonalcoholic fatty liver disease in mice in association with gut microbiota regulation. Nutrients. (2022) 14:2945. 10.3390/nu14142945
45.
Boursier J Mueller O Barret M Machado M Fizanne L Araujo-Perez F et al . The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. (2016) 63:764–75. 10.1002/hep.28356
46.
Li X Wang Y Xing Y Xing R Liu Y Xu Y . Changes of gut microbiota during silybin-mediated treatment of high-fat diet-induced non-alcoholic fatty liver disease in mice. Hepatol Res. (2020) 50:5–14. 10.1111/hepr.13444
47.
Zhu Z Zhu B Sun Y Ai C Wang L Wen C et al . Sulfated polysaccharide from sea cucumber and its depolymerized derivative prevent obesity in association with modification of gut microbiota in high-fat diet-fed mice. Mol Nutr Food Res. (2018) 62:1800446. 10.1002/mnfr.201800446
48.
Shi J Xie Q Yue Y Chen Q Zhao L Evivie SE et al . Gut microbiota modulation and anti-inflammatory properties of mixed lactobacilli in dextran sodium sulfate-induced colitis in mice. Food Funct. (2021) 12:5130–43. 10.1039/D1FO00317H
49.
Hu M Zhang L Ruan Z Han P Yu Y . The regulatory effects of citrus peel powder on liver metabolites and gut flora in mice with non-alcoholic fatty liver disease (NAFLD). Foods. (2021) 10:3022. 10.3390/foods10123022
50.
Wang X Linlin S Xinping W Yi F Yuan W . MDG-1, an Ophiopogon polysaccharide, restrains process of non-alcoholic fatty liver disease via modulating the gut-liver axis. Int J Biol Macromol. (2019) 141:1013–21. 10.1016/j.ijbiomac.2019.09.007
51.
Cao F Ding Q Zhuge H Shanglei L Chang K Le C et al . Lactobacillus plantarum ZJUIDS14 alleviates non-alcoholic fatty liver disease in mice in association with modulation in the gut microbiota. Front Nutr. (2023) 9:3290. 10.3389/fnut.2022.1071284
52.
Adams LA Lymp JF St Sauver J Sanderson SO Lindor KD Feldstein A et al . The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. (2005) 129:113–21. 10.1053/j.gastro.2005.04.014
53.
Peverill W Powell LW Skoien R . Evolving concepts in the pathogenesis of NASH: beyond steatosis and inflammation. Int J Mol Sci. (2014) 15:8591–638. 10.3390/ijms15058591
Summary
Keywords
Lactobacillus acidophilus , high-fat diet, non-alcoholic fatty liver disease, lipid accumulation, gut microbiota
Citation
Wang Y, Wang Z, Wan Y, Jin F, Shi X, Xing Z, Tian B and Li B (2023) Assessing the in vivo ameliorative effects of Lactobacillus acidophilus KLDS1.0901 for induced non-alcoholic fatty liver disease treatment. Front. Nutr. 10:1147423. doi: 10.3389/fnut.2023.1147423
Received
18 January 2023
Accepted
20 February 2023
Published
20 March 2023
Volume
10 - 2023
Edited by
Daxi Ren, Zhejiang University, China
Reviewed by
Yuxing Guo, Nanjing Normal University, China; Guangtian Cao, China Jiliang University, China; Zhen Wu, Ningbo University, China
Updates
Copyright
© 2023 Wang, Wang, Wan, Jin, Shi, Xing, Tian and Li.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Tian tianbo@neau.edu.cnBailiang Li 15846092362@163.com
This article was submitted to Nutrition and Metabolism, a section of the journal Frontiers in Nutrition
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.