Abstract
Our dietary choices not only affect our body but also shape the microbial community inhabiting our large intestine. The colonic microbiota strongly influences our physiology, playing a crucial role in both disease prevention and development. Hence, dietary strategies to modulate colonic microbes have gained notable attention. However, most diet-colonic microbiota research has focused on adults, often neglecting other key life stages, such as infancy and older adulthood. In this narrative review, we explore the impact of various dietary patterns on the colonic microbiota from early infancy to centenarian age, aiming to identify age-specific diets promoting health and well-being by nourishing the microbiota. Diversified diets rich in fruits, vegetables, and whole grains, along with daily consumption of fermented foods, and moderate amounts of fish and lean meats (two to four times a week), increase colonic microbial diversity, the abundance of saccharolytic taxa, and the production of beneficial microbial metabolites. Most of the current knowledge of diet-microbiota interactions is limited to studies using fecal samples as a proxy. Future directions in colonic microbiota research include personalized in silico simulations to predict the impact of diets on colonic microbes. Complementary to traditional methodologies, modeling has the potential to reduce the costs of colonic microbiota investigations, accelerate our understanding of diet-microbiota interactions, and contribute to the advancement of personalized nutrition across various life stages.
1 Introduction
The human gastrointestinal tract hosts a diverse and dynamic microbial community, including bacteria, archaea, fungi, and viruses, which play key roles in host health and wellbeing. Most microbes are found in the large intestine or colon, with an estimated concentration of 1011 cells/mL (1), although colonic microbial abundance can vary depending on the sample site, analytical method used, and host physiology. Fecal samples are non-invasive proxies to study the relationship between colonic microbes and human health, particularly microbial composition and function. These analyses have revealed the impact of colonic commensals on host nutrition, metabolism, and the immune and neurological systems (2–8). However, many crucial aspects of this relationship remain unknown. Defining what constitutes a healthy colonic microbiota composition and function is an ongoing challenge (9, 10).
One of the major challenges in investigating the colonic microbiota is its substantial compositional variability, which occurs both within the same individual over time and between individuals (11, 12). This variation limits predicting how each individual’s microbiota may respond to interventions and their subsequent impact on host health. Colonic microbes interact with one another and the host, forming a dynamic network influenced by various individual and environmental factors. These factors mainly include dietary habits, host health status, genetics, age, gender, geographical location, lifestyle behaviors, and antibiotic use (13).
Among the factors under host control, diet is key. Dietary compounds not absorbed by the small intestine reach the colon, are fermented by colonic microbes, and produce metabolites that influence host physiology. Dietary interventions can rapidly alter the composition and function of the colonic microbiota (14). For instance, non-digestible carbohydrates have prebiotic properties. Their consumption promotes the growth of saccharolytic microorganisms and increases the production of beneficial short-chain fatty acids (SCFAs) (15, 16). In contrast, a diet lacking non-digestible carbohydrates and with excessive intake of protein and fat from animal origin increases the abundance of pathogens and the production of potentially deleterious molecules (17–19). Recently, phytochemicals, bioactive compounds found in plants, have attracted attention for their potential prebiotic effect, as well as antioxidant and anti-inflammatory properties (20).
However, the long-term effect of dietary patterns on colonic microbes remains unclear, including the persistence of diet-induced alterations in the colonic microbiota when changing dietary exposures. Furthermore, diet-colonic microbiota research has predominantly focused on adults (ages 18-65), limiting our understanding of how diet influences colonic microbes in other life stages. Notably, dietary patterns during infancy (under 3 years) and older adulthood (over 65 years) differ from those in adulthood, likely influencing the composition and function of the colonic microbiota and highlighting the need for more investigations specific to these age groups (21, 22). Literature reviews on the influence of diet on colonic microbiota across the human lifespan are scarce and often do not fully cover all life stages (23, 24). Considering these knowledge gaps, this narrative review examines how different dietary patterns influence the human colonic microbiota across early infancy, weaning, adulthood, older adulthood, and centenarian age. This review aims to identify age-appropriate diets for microbiota modulation, providing insights into promoting health and wellbeing.
2 Search strategy
A narrative review was conducted to evaluate the impact of dietary patterns on the human colonic microbiota across infancy (under 3 years), adulthood (between 18 and 65 years), older adulthood (between 65 and 100 years), and centenarian age (over 100 years). Articles were primarily identified through searches in the PubMed and Google Scholar databases using the terms “gut microbiota”, “diet”, and “infant OR adult OR older adult OR centenarian”. Additional records evaluating the effect of dietary patterns on the colonic microbiota were identified by replacing “diet” with terms referring to specific dietary patterns (e.g., “gut microbiota” AND “western diet” AND “infant OR adult OR older adult OR centenarian”). Only studies involving humans and published in English were identified. No specific inclusion and exclusion criteria were applied. Articles were prioritized based on their publication date, giving preference to the most recent studies. When available, meta-analyses, systematic reviews, and randomized controlled trials were prioritized.
3 Principal colonic microbes and produced microbial metabolites
The colonic microbiota is dynamic, and its composition varies throughout life. Its development is proposed to have an initial phase until the first 14 months of postnatal life, followed by a transitional period between 15 and 30 months, reaching stability after 31 months (25). Multiple factors influence the composition and function of colonic commensals over life, resulting in a unique profile of colonic microbes for each individual (13, 25). Despite this variability, certain taxa and gene functions prevail in healthy adults based on fecal data, suggesting the existence of core microbial functional groups (26, 27). The principal microbial taxa in the colonic microbiota and their associated microbial metabolites are summarized in Supplementary Tables 1, 2, respectively.
To date, the mechanisms by which colonic microbes influence host health remain unclear. Under favorable conditions, colonic microbes may support host homeostasis by competing with pathogens for resources, producing anti-microbial metabolites, and modulating host immune responses (28, 29). Conversely, disruptions in the colonic microbiota can favor the growth of pathogens and the production of pro-inflammatory metabolites, compromising the integrity of the colonic epithelial barrier. This allows luminal molecules and microbes to enter the bloodstream, potentially triggering excessive host immune responses. Over time, these responses may lead to a chronic inflammatory state and increased disease risk. Additionally, it is plausible that certain diseases may alter the colonic environment, disrupting the colonic microbiota and creating a vicious cycle that perpetuates disease.
4 Influence of dietary patterns on colonic microbes
4.1 Infancy
During the first months of postnatal life, the colonic microbiota composition is mainly affected by the gestational age, mode of delivery, and type of feeding (human milk versus infant formula) (25, 30, 31). Systematic literature reviews suggested that full-term pregnancy, natural birth, and breastfeeding provide greater opportunities for the infant colonic microbiota to thrive (32, 33). In contrast, pre-term pregnancy, C-section, and formula-feeding are associated with disruptions of the microbial community (25, 31). In regards to feeding in early infancy, breast milk contains oligosaccharides that promote the growth of the genus Bifidobacterium and support beneficial microbial cross-feeding interactions (34, 35). In addition, breast milk is not sterile and contains microbes that can colonize the infant’s colon, as well as bioactive compounds like antibodies and lactoferrin, which reduce pathogen colonization (36, 37).
In contrast, infant formulas are predominantly made with bovine milk, have higher protein content, and are often supplemented with fructooligosaccharides and galactooligosaccharides to provide prebiotic effects. While infant formulas meet the nutritional requirements for infant development, they fail to mimic the bifidogenic effect of human milk. Systematic reviews have found that, compared to breastfed infants, formula-fed infants exhibit a lower fecal abundance of the genus Bifidobacterium, an increased abundance of pathogens, and a higher expression of microbial genes associated with amino acid metabolism (38, 39).
Exclusive breastfeeding is recommended for the first 6 months of life (40). Nutrients from complementary foods reaching the colon unabsorbed mature the infant’s colonic microbiota toward a more adult-like configuration (Table 1). Non-digestible carbohydrates are preferentially fermented by colonic microbes, producing SCFAs and gases (Supplementary Table 2), and their availability in the colon limits the fermentation of other dietary compounds (41, 42). In vitro evidence suggests that carbohydrate fermentation primarily occurs in the proximal colon, while other macronutrients are metabolized in the distal colon (43, 44) (Figure 1). Notably, SCFAs are molecules that exert well-documented health benefits, and their reduced fecal levels are frequently observed in preterm infants, adults with autoimmune, metabolic, and gastrointestinal diseases, and older adults with neurological disorders (15, 45–48). However, excessive fermentation of rapidly fermented fibers can increase gas production in individuals with functional gastrointestinal disorders, leading to bloating, pain, and discomfort (49).
TABLE 1
| Food category | Food intervention trials | Observational studies | In vitro fecal fermentations | References |
| Meats (e.g., beef, pork) | ↑Clostridiales, Clostridium XIVa ↓Enterobacteriaceae ↑alpha diversity (Chao1 and Shannon indexes) | ↓Bacteroides ↑alpha diversity (Shannon index) | Not evaluated | (72, 82, 84, 85, 94, 95) |
| Dairy (e.g., yogurt, cheese) | Not evaluated | ↓Bacteroides, Clostridiaceae ↑alpha diversity (Shannon index) | ↑Bifidobacterium, Lactobacillus, Enterococcaceae ↓Enterobacteriaceae ↑acetate, propionate, butyrate | (72, 84, 88, 96) |
| Infant cereals (e.g., whole-grain and refined cereals) | ↑Bacteroidales, Bacteroides ↓Enterobacteriaceae, Escherichia-Shigella | ↑alpha diversity (richness and Shannon index) | ↑Bacteroidaceae, Prevotellaceae, Ruminococcaceae ↑acetate | (81–83, 85, 86) |
| Fruits and vegetables (e.g., apple, berries, carrot) | Not evaluated | ↑alpha diversity (Shannon index) | ↑Bifidobacterium, Lactobacillus, Streptococcus, Ruminococcus, Faecalibacterium ↓Clostridium, Enterobacteriaceae ↑acetate, propionate | (83, 87, 88) |
| Sweets (e.g., cakes, desserts, chocolates) | Not evaluated | ↓Bifidobacterium, Clostridium cluster IV | Not evaluated | (50) |
Impact of complementary foods on the colonic microbiota of weaning infants.
FIGURE 1

Fermentation of dietary compounds by the colonic microbiota. Metabolites in blue benefit the host, while those in red are potentially deleterious. A decrease in carbohydrate fermentation is observed from the proximal to the distal colon, mainly producing short-chain fatty acids (SCFAs) and gases. On the other hand, amino acid, fat, and phytochemical fermentation increase from the proximal to the distal colon. Microbial fermentation of amino acids produces SCFAs, branched-chain fatty acids (BCFAs), biogenic amines, hydrogen sulfide (H2S), and ammonia. Fermentation of dietary fats is involved in trimethylamine production, whereas the fermentation of polyphenols leads to the generation of multiple bioactive molecules.
Longitudinal evidence demonstrated that colonic microbial diversity and stability increase during weaning, and genes associated with carbohydrate degradation, vitamin biosynthesis, and production of SCFAs are enriched (50). The abundances of the genera Bifidobacterium and Lactobacillus decrease, while the phyla Bacillota and Bacteroidota and the genera Clostridium and Bacteroides are enriched (50–52). Furthermore, the dominant fungal species Debaryomyces hansenii is replaced by Saccharomyces cerevisiae and the viral richness and diversity decrease (53, 54).
Diet-induced microbiota changes during infancy may have long-lasting effects, influencing susceptibility to diseases later in life (55–57). Weaning is crucial for the colonic microbiota development, influencing health and wellbeing later in life. Although many studies have characterized the influence of feeding type (breastmilk versus infant formula) on the development of the infant gut microbiota (25, 31, 38, 39), the impact of complementary foods on colonic microbes of weaning infants remains underexplored. A recent systematic review of interventional trials assessing the effects of complementary foods on fecal microbiota composition in weaning infants found that whole-grain cereals and meats increased the fecal abundance of SCFA-producing bacterial taxa and increased microbial richness (58). However, the review was limited to only seven clinical trials characterizing the microbiota using 16S rRNA gene sequencing, highlighting the scarcity of food interventions in this field.
Another notable knowledge gap is the limited understanding of how maternal diet influences the infant’s colonic microbiota. Maternal intake of plant-based dietary compounds during pregnancy has been reported to influence the neonatal fecal microbiota (59), while increased maternal dietary diversity and consumption of fermented foods during pregnancy were associated with lower fecal alpha diversity in infants (60). Additionally, the mother’s diet affects the microbial and nutritional composition of breastmilk, which may impact colonic microbes of breastfed infants (61). A scoping review identified associations between maternal dietary patterns and infant fecal microbiota composition. Maternal consumption of seafood, fermented dairy, fruits and veggies, and nuts was linked to an increased abundance of beneficial taxa in the infant fecal microbiota (33). Conversely, maternal intake of artificial sweeteners and high-fat diets were associated with negative alterations in the infant microbiota (33).
Artificial sweeteners, along with emulsifiers, thickeners, stabilizers, preservatives, and colorants, are chemicals added during food production to extend shelf-life or modify sensory properties. Although little is known about the effect of food additives on colonic microbes, a systematic review of randomized controlled trials, encompassing participants from infants to older adults, found that maltodextrin impacts the composition of the fecal microbiota, notably the abundance of the genera Lactobacillus and Bifidobacterium (62). Furthermore, another systematic review, including in vivo and in vitro studies, reported that exposure to colorants, sweeteners, emulsifiers, and preservatives is linked with perturbations in the colonic microbiota and adverse health effects (63). These findings highlight the need for further research and support recommendations to reduce the consumption of ultra-processed foods (64).
Another topic that requires further research is the interplay between micronutrients and the infant colonic microbiota. Vitamins are mostly absorbed in the upper small intestine, while minerals typically have lower bioavailability and reach the colon in greater amounts (65). Colonic microbes influence micronutrient levels by synthesizing vitamins and modulating mineral absorption (66, 67). In turn, micronutrients affect their composition and function. For instance, a systematic review of observational human studies reported that vitamin B12 supplementation may increase the alpha diversity of the fecal microbiota in adults and older adults but not in infants (68). In contrast, iron intervention supplementation in infants, commonly used to combat malnutrition, may increase the fecal abundance of pathogens, as observed in randomized controlled trials profiling the microbiota using 16S rRNA gene sequencing (69, 70).
Current nutritional guidelines recommend introducing infants to a diverse range of complementary foods (64, 71). Consistently, fecal microbiota observational studies have shown that greater dietary diversity during weaning is associated with increased microbial diversity and richness (72, 73). Importantly, dietary diversity in infancy is essential to prevent nutrient deficiencies often linked with monotonous diets, which can contribute to perturbations in the colonic microbiota and subsequent elevated risk of disease (74). In line with these recommendations, various fruits, vegetables, and whole grains are recommended for weaning infants (64, 71, 75). These foods are sources of vitamins, minerals, complex carbohydrates and phytochemicals, leading to beneficial alterations in colonic microbes (Table 1). Phytochemicals have recently attracted attention in colonic microbiota research for their ability to support the growth of beneficial taxa and inhibit pathogens (76), in addition to having anti-inflammatory and antioxidant properties. Polyphenols are the most prominent phytochemicals, with less than 10% absorbed in the small intestine and the majority reaching the distal colon, where they may be converted into more bioactive and bioavailable molecules by colonic microbes (77, 78). This microbial conversion is associated with various health benefits to the host, such as cardioprotective and antimetabolic syndrome effects, as observed in adults (79, 80).
Whole-grain infant cereal interventions in weaning infants increased the fecal abundance of SCFAs-producing bacteria, such as the genus Bacteroides, while reducing the abundance of pathogens belonging to the family Enterobacteriaceae, as determined using 16S rRNA gene sequencing (81, 82). Consistently, observational trials profiling the fecal microbiota of weaning infants by 16S rRNA sequencing, positively correlated the intake of complex carbohydrates with increased fecal alpha diversity and abundance of Lachnospiraceae and Ruminococcaceae families (83, 84). These findings were further confirmed by a longitudinal trial that used shotgun metagenomic sequencing to evaluate the composition of the infant fecal microbiota during the first year of life. The study highlighted the key role of complex carbohydrates in supporting the maturation of the colonic microbiota in weaning infants, as evidenced by an increased fecal alpha diversity and abundance of SCFA-producing bacterial genera (85). In vitro fecal fermentations using inoculum from weaning infants reported that whole grain cereals and numerous fruits and vegetables promoted the growth of the Ruminococcaceae and Prevotellaceae families to the detriment of the Enterobacteriaceae family, leading to acetate production after 24 h of fermentation (86–88). An in silico investigation recently identified berries as promising candidates for increasing the production of acetate and propionate by the infant’s fecal microbiota when consumed with breastmilk (89). Similarly, a 24-h in vitro fermentation study using weaning infant inoculum found that berries increased acetate and propionate production (90).
Nutritional guidelines also recommend moderate consumption (one serving per day) of lean meats and fermented dairy for infants to meet protein and micronutrient requirements (64, 71, 75). Introducing protein-rich foods to infants increased their colonic availability of amino acids, as their protein digestion and absorption are not yet fully developed (91). In the colon, the fermentation of unabsorbed amino acids and small peptides produces SCFAs (30% of protein mass), branched-chain fatty acids (BCFAs), tryptophan-derivatives, and biogenic amines, but also pro-inflammatory metabolites, such as hydrogen sulfide (H2S) and nitrogen-derivatives (92, 93). Consistently, clinical trials demonstrated that introducing protein-rich foods to weaning infants increases their fecal abundance of SCFA-producing taxa. For instance, pureed beef interventions in infants increased fecal abundance of Clostridium XIVa members, reduced the abundance of the Enterobacteriaceae family, and increased alpha diversity, as determined using 16S rRNA sequencing (82, 94, 95). Similarly, an observational study profiling the infant fecal microbiota using 16S rRNA amplicon sequencing reported a positive association between meat intake and increased fecal alpha diversity (72). For dairy products, their consumption by weaning infants was negatively associated with the abundance of the Bacteroides genus and the Clostridiaceae family, according to an observational study that profiled the fecal microbiota of infants aged 6-24 months using 16S rRNA gene sequencing (96). In turn, the fermentation of bovine milk for 10 h using feces from infants at weaning age increased the abundance of taxa from the Bifidobacterium genus in vitro (88).
An observational trial profiling the microbiota of weaning infants by 16S rRNA amplicon sequencing reported that dietary patterns characterized by the consumption of foods rich in protein and in fiber-rich foods, such as meats, cheese, and wholegrain bread, have been positively correlated with the increased fecal abundance of taxa from the Lachnospiraceae family, decreased abundance of taxa from the Bifidobacteriaceae family, and increased microbial alpha diversity (72). On the other hand, the consumption of diets rich in fat but low in carbohydrates has been associated with reduced fecal levels of SCFAs and increased abundance of pathogens in post-weaning infants (97). Dietary fats are normally well-digested and absorbed, with less than 5% of ingested fat reaching the colon (98). Importantly, their influence on colonic microbes depends on their type: as concluded by a systematic review of adult studies, saturated fatty acids are associated with reduced colonic microbiota richness and diversity, whereas polyunsaturated fatty acids do not exhibit this effect (99). Furthermore, certain fatty acids have anti-microbial activity and can reduce the abundance of pathogenic taxa (100). In this context, the Mediterranean diet, rich in polyunsaturated fatty acids from olive oil, was recently adapted to children (aged 3 and older) to help prevent obesity and cardiometabolic diseases in infancy and later life. The adapted diet emphasizes the daily consumption of fruits, vegetables, legumes, nuts, whole grains, olive oil, and dairy, along with weekly consumption of fish, eggs, and meats (101).
4.2 Adulthood
The complete maturation of the colonic microbiota is believed to occur around 3 years old (102). Nevertheless, evidence suggests its continued functional and compositional development during childhood and adolescence (103, 104). Compared to other life stages, more is known about the influence of dietary patterns on the colonic microbiota of adults. This topic has been extensively reviewed in recent publications (105–107). Therefore, only the impact of common diets on the colonic microbes of adults is briefly discussed here (Table 2).
TABLE 2
| Diet | Definition | Microbial composition | Microbial function | Observed effect of diet on host health | References |
| Western diet | High intake of protein, fat, and sugars. Low consumption of complex carbohydrates | ↑Bacteroides, Alistipes, Penicillium ↓Methanobrevibacter | ↑Amino acid and bile acid metabolism and production of nitrogen derivatives and BCFAs | Adherence to a Western diet increased risk factors for metabolic and cardiovascular diseases compared to a prudent dietary pattern | (108–111) |
| Plant-based diet | Exclusive or predominant consumption of plant-based foods (e.g., vegan or vegetarian diets) | ↑Prevotella, Faecalibacterium, Ruminococcus, Candida, Methanobrevibacter | ↑Degradation of complex carbohydrates and SCFA production | A meta-analysis of observational trials found that adherence to vegetarian or vegan diets was linked with lower risk of cardiovascular diseases and cancer | (108–110, 274, 275) |
| Mediterranean diet | High consumption of fruits, vegetables, and olive oil. Moderate of fish, dairy, and lean meats | ↑Bacteroides, Prevotella, Faecalibacterium | ↑Acetate and propionate | A meta-analysis linked adherence to the Mediterranean diet with a reduction of all-cause mortality | (125, 126) |
| Fermented foods diet | High consumption of fermented foods (e.g., yogurt, kimchi, cheese, bread) | ↑Lactobacillus ↑alpha diversity | ↑conjugated linoleic acid | Decreased biomarkers of inflammation in a 10-week intervention with a fermented foods diet compared to baseline values | (127, 128) |
| Low FODMAP diet | Low consumption of rapidly fermentable carbohydrates and polyols | ↓Bifidobacterium No changes in microbial diversity | No changes in SCFA and BCFA production | Alleviates pain and discomfort in patients suffering from irritable bowel syndrome. May have negative health outcomes | (130, 131) |
| Low gluten diet | Low consumption of gluten-containing foods | ↓Bifidobacterium, Dorea, Veillonellaceae No changes in microbial diversity | ↓Degradation of carbohydrates | Alleviates symptoms in patients with celiac disease or gluten sensitivity. May have negative health outcomes | (132, 133, 276) |
| Ketogenic diet | High consumption of fat and protein, and restricted consumption of carbohydrates | ↓Bifidobacterium, Eubacterium, Faecalibacterium, Roseburia | ↓total SCFAs, acetate, butyrate | Alleviates symptoms in patients with epilepsy and induces fat loss. A systematic review suggested that adherence to Ketogenic diets may increase risk for obesity, type-2 diabetes, and depression | (135) |
Impact of various dietary patterns on the adult colonic microbiota.
Observational trials reported that Western diets (rich in fat, protein of animal origin, and simple sugars, and low in complex carbohydrates) were associated with increased fecal abundance of the genera Alistipes, Bacteroides, and Penicillium, and less methanogenic archaea, resulting in higher expression of microbial genes related to bile acid metabolism, amino acid fermentation, and production of BCFAs (108–110). Adherence to this dietary pattern is linked with increased susceptibility to chronic inflammatory, and intestinal, metabolic, neurological, and cardiovascular diseases (111). Consistently, diets high in protein but low in carbohydrates (30% protein, 35% carbohydrate, and 35% fat as calories) and high-fat diets (40% fat as calories) have been linked with perturbations in the colonic microbiota and adverse health outcomes in adults (18, 112, 113). These findings are supported by a systematic review concluding that increased intake of saturated fatty acids, predominantly found in animal-based foods, is associated with reduced colonic microbiota richness and diversity in adults (99). Recently, diets rich in high industrially processed food and low in fiber from plants have been associated with colonic microbiota perturbations, altered profile of plasma metabolites, and an increased risk of disease development, including metabolic disorders and colorectal cancer (19, 114).
On the other hand, diets predominantly based on plants, like vegan and vegetarian diets, are associated with a higher fecal abundance of saccharolytic taxa and enrichment of microbial genes involved in complex carbohydrates degradation and production of SCFAs (108, 110). Notably, colonic microbiota alterations in adults due to adherence to plant-based diets have been linked to protective effects against metabolic and cardiovascular diseases (115). Recently, adherence to a plant-based African heritage diet and consumption of a traditional fermented beverage were associated with lower levels of circulating inflammatory biomarkers in healthy adults (116). These effects may be explained by the high content of non-digestible carbohydrates and polyunsaturated fatty acids in these diets. A meta-analysis concluded that non-digestible carbohydrates support the growth of saccharolytic commensals and stimulate beneficial microbial cross-feeding interactions in adults, producing SCFAs (117). Additionally, the consumption of polyunsaturated fatty acids has been linked with beneficial alterations in colonic microbiota composition in both observational and interventional trials (118, 119).
However, a cross-sectional study found no differences in the fecal concentration of SCFAs and BCFAs between vegans and omnivores (who consumed at least three servings of meat per week) (120). Furthermore, omnivores had higher fecal alpha diversity measured by the Shannon index (120). Consistent with these findings, flexitarian diets (primarily plant-based but including occasional meat, fish, and dairy) have been associated with increased fecal alpha diversity and abundance of beneficial taxa in adults (121–123). These findings suggest that moderate consumption of animal products (e.g., three times per week) alongside a plant-rich diet may offer additional benefits to the colonic microbiota.
In this context, the Mediterranean diet is a prime example of a healthy dietary pattern. This diet is typical of countries around the Mediterranean basin, varying according to the region. It traditionally consists of a high consumption of fruits, vegetables, and olive oil (at least two servings per day), along with a moderate intake of seafood, fish, and dairy (two or more servings per week), and occasional consumption of lean meats (no more than two servings per week) (124). A recent systematic review of observational and interventional trials found that following the Mediterranean diet increased the alpha diversity of the colonic microbiota in adults (125). It also increased the abundance of the genera Faecalibacterium, Prevotella, and Bacteroides and the production of SCFAs, particularly acetate and propionate (125). Additionally, adherence to the Mediterranean diet has been associated with lower mortality rates (126). Similarly, consuming fermented foods, including yogurt, kefir, kimchi, and sourdough bread is encouraged to provide health benefits. Adults consuming diets enriched in fermented foods (at least three servings per week) showed an increased fecal abundance of various Lactobacillus species, as well as increased production of conjugated linoleic acid (127). Furthermore, the intake of fermented foods has been associated with increased fecal alpha diversity and lower levels of circulating cytokines (128).
These observations highlight the importance of a balanced and diversified diet to nourish the adult colonic microbiota, aligning with current nutritional recommendations (64). Conversely, restrictions on macronutrients, notably carbohydrates, are associated with perturbations in the colonic microbiota, raising concerns about the potential long-term deleterious effects of such restrictions (129). Restricted diets are typically recommended for managing pre-existing medical conditions. For instance, the low FODMAP diet, characterized by low consumption of rapidly fermentable carbohydrates, is recommended for individuals with disorders of gut-brain interaction (also known as functional gastrointestinal disorders) to alleviate discomfort (130). However, a systematic review and a meta-analysis of interventional studies concluded that this diet reduces the fecal abundance of the genus Bifidobacterium in adults with irritable bowel syndrome (131). Similarly, gluten-free or low-gluten diets are recommended for individuals with Celiac Disease or gluten sensitivity. In healthy adults, adherence to these diets has been associated with the decreased fecal abundance of the genera Bifidobacterium and Dorea and the family Veillonellaceae, and decreased expression of microbial genes involved in carbohydrate degradation (132, 133). The ketogenic diet is characterized by high fat intake (70-80% of total energy), moderate protein consumption (10-20% of total energy), and restriction of carbohydrates (less than 10% of total energy). Variants of this diet are recommended for individuals with epilepsy (for example, the medium-chain triglyceride diet) or following a rapid weight-loss strategy (134). A systematic review concluded that this diet decreases the fecal abundance of the Bifidobacterium genus and potentially reduces the abundance of butyrate-producing genera, leading to reduced levels of acetate and butyrate (135). These alterations in the colonic microbiota may contribute to an increased risk of obesity, type-2 diabetes, and depression (135). Further evidence supports these adverse outcomes, as adherence to the ketogenic diet reduced glucose tolerance in healthy adults (136).
4.3 Older adulthood
The colonic microbiota remains stable during adulthood until it undergoes modifications in its composition and function at around 65 years old, when changes in lifestyle and physiology occur with aging. The metabolism and physical activity levels decrease, antibiotics are used more frequently, the diet becomes less diversified, colon motility decreases, and fecal retention time increases (137). Observational evidence suggests that older adults have high inter-individual variation in the colonic microbiota, which is strongly influenced by their health status (138).
Healthy older adults have fecal microbiota similar to that of healthy young adults, although observational studies suggest increased diversity of methanogenic archaea and lower viral richness (139–141). On the other hand, unhealthy aging (e.g., frailty or chronic diseases) is associated with a reduced abundance of the genera Bifidobacterium, Faecalibacterium, and Eubacterium, and an increased abundance of the family Enterobacteriaceae and genera Streptococcus, Clostridium, Penicillium, Candida, and Aspergillus (142–144). In addition, the expression of genes synthesizing vitamins and fatty acids is reduced (143). These alterations in the colonic microbiota are associated with chronic inflammation and an increased risk of morbidity and mortality (145).
A recent systematic review evaluated the effect of different dietary patterns on the fecal microbiota of older adults, including a total of 38 intervention trials, most of which profiled the microbiota using 16S rRNA sequencing (146). Diets rich in plant foods and including animal products in moderation (such as daily consumption of dairy and lean meats or fish two to four times per week) increased the fecal abundance of saccharolytic taxa and the production of SCFAs (146). Similarly, an observational study that evaluated the fecal microbiota of older adults using shotgun metagenomic sequencing found that adherence to the healthy plant-based diet index was associated with a greater fecal abundance of bacterial saccharolytic species, along with enriched pathways for the biosynthesis of branched-chain amino acids (147). These results may be explained by the high content of non-digestible carbohydrates and phytochemicals in these diets. For instance, greater consumption of dietary fiber among older adults was associated with an increased fecal abundance of the order Clostridiales, which includes butyrate-producing bacteria, and increased expression of pathways involved in polysaccharide degradation, according to an observational study employing shotgun metagenomic sequencing (148). Furthermore, an intervention trial reported that adherence to a polyphenol-rich diet (total polyphenols around 1,300 mg/day) increased the fecal abundance of butyrate-producing bacteria, as profiled by 16S rRNA sequencing, and reduced blood pressure in older adults (149).
In contrast, the systematic review of intervention trials found that high intakes of fat, protein, and simple sugars but low consumptions of complex carbohydrates are associated with the growth of opportunistic pathogens, production of pro-inflammatory toxins, and frailty (146). Furthermore, an intervention study that profiled the fecal microbiota of older adults using 16S rRNA sequencing found that diets rich in fat but low in carbohydrates have been associated with decreased abundance of the genus Bifidobacterium (150). Consistently, a longitudinal study in older adults linked higher red meat intake to increased plasma concentration of trimethylamine N-oxide, a microbial metabolite associated with cardiovascular risk (151). These observations are consistent with results observed for young adults, highlighting the importance of a diversified diet primarily composed of fruits, vegetables, and whole cereals to nourish beneficial colonic microbes (Table 3). In line with these findings, a longitudinal study of over 100,000 participants followed for 30 years assessed the impact of long-term dietary patterns on aging. Diets rich in plant-based foods, such as fruits, vegetables, whole grains, and nuts, and including animal-based foods in moderation, like low-fat dairy, were associated with healthy aging, defined as survival to the age of 70 years with intact cognitive, physical, and mental functions, and without chronic diseases (152).
TABLE 3
| Diet | Definition | Microbial composition | Microbial function | Observed effect of diet on host health | Reference |
| Western diet | High consumption of processed meats and refined grains | ↑Alistipes, Desulfovibrio, Ruminococcus ↓Faecalibacterium, Prevotella | ↑Amino acid metabolism ↓SCFA production | Adherence to a Western diet was associated with increased body mass index compared to a Prudent diet | (17) |
| High-protein diet | Consumption of protein higher than the recommended dietary intake (> 0.8 g protein/kg bodyweight/day) | No changes in taxa abundance or microbial diversity | No changes in the production of organic acids | No changes in appetite after 6-moth intervention compared to control (habitual diet) | (158, 159) |
| Prudent diet | High consumption of fruits, vegetables, nuts, fish, and chicken | ↑Clostridium, Faecalibacterium, Lachnospira ↓Desulfovibrio, Ruminococcus | ↑Complex carbohydrate degradation and SCFA production | Adherence to a Prudent diet was associated with reduced body mass index compared to a Western diet | (17) |
| Mediterranean diet | High consumption of fruits, vegetables, and olive oil. Moderate of fish, dairy, and lean meats | ↑Roseburia, Eubacterium, Faecalibacterium ↓Ruminococcus torques | ↑SCFA and BCFA production ↓Secondary bile acids, p-cresol, ethanol | Reduced frailty and chronic inflammation, and improved cognitive function after 1-year intervention compared to control (habitual diet) | (277) |
| Polyphenol-rich diet | High consumption of polyphenol-rich foods (e.g., berries, pomegranate, green tea, dark chocolate) | ↑Faecalibacterium, Butyricicoccus, Ruminococcaceae ↓Streptococcus, Enterobacteriaceae | Not evaluated | Increased serum concentration of indole 3-propionic acid and decreased of zonulin after 8-week intervention compared to the control diet (low-polyphenol diet) | (149, 278) |
Impact of various dietary patterns on the colonic microbiota of older adults.
However, increased protein consumption appears to be beneficial in older adults, who typically have slower protein digestion and absorption compared to younger individuals (153). For instance, a cross-sectional analysis of the fecal microbiota of older men, characterized using 16S rRNA gene sequencing, revealed that higher protein intake was associated with increased fecal alpha diversity (154). Additionally, a meta-analysis of observational trials concluded that protein intake was negatively associated with frailty in older individuals (155). Therefore, a daily intake of 1.2 g protein/kg bodyweight, compared to the standard recommendations of 0.66-0.8, has been proposed to support good health and maintain functionality in older populations (156). In line with this recommendation, a narrative review suggested that high-fiber diets enriched with protein from legumes, dairy, and lean meats could promote a balanced colonic microbiota (eubiosis), also contributing to muscle synthesis and overall metabolic health in older adults (157). In this context, complex carbohydrates are important to mitigate excessive protein fermentation in the colon and the consequent production of deleterious metabolites, such as hydrogen sulfide and nitrogen-derivatives (92, 93). As demonstrated by intervention trials, protein intakes exceeding the recommended dietary allowance did not alter fecal microbiota composition, characterized using 16S rRNA sequencing, or function in older adults when accompanied by a prudent diet (158, 159).
4.4 Centenarian age
Recently, colonic microbiota investigations have focused on centenarians (individuals aged 100 or older) as models for healthy aging. Compared to younger controls, observational studies demonstrated that centenarians exhibit higher fecal bacterial and viral diversity (160–162). A systematic review of 27 observational studies suggests that their high fecal microbial diversity and abundance of health-promoting taxa contribute to healthy aging and longevity (163).
In terms of microbial composition, the genera Alistipes, Parabacteroides, Clostridium, and Methanobrevibacter are enriched in the feces of healthy centenarians, while the butyrate-producing species Faecalibacterium prausnitzii and Eubacterium rectale are depleted (160, 162, 164). However, no age-related changes have been observed in the fecal fungal microbiota (165). Concerning functionality, centenarians have lower fecal butyrate concentrations but higher levels of BCFAs, ammonium, and secondary bile acids compared to younger controls (160). Furthermore, their fecal microbiome is enriched in genes associated with SCFA production from amino acids, secondary bile acid metabolism, and the degradation of xenobiotics, plant-based fats, and tryptophan (166–168). In contrast, they have fewer genes involved in carbohydrate and animal fat metabolism (166–168).
Few investigations have assessed the interaction between diet and colonic microbes in centenarians, with current knowledge primarily derived from longitudinal and cross-over studies. Overall, adherence to a diverse, plant-rich diet has been associated with a higher abundance of microbial taxa linked to longevity, as observed in Italian, Chinese, and South Korean centenarians (166, 169–172). Additionally, the consumption of fermented soybean paste was positively associated with the distinct fecal microbiota composition of South Korean centenarians, according to an observational study that profiled the microbiota using 16S rRNA gene sequencing (172). A cross-sectional study of Estonian centenarians found that cereal consumption and lower adherence to Western dietary patterns were linked to longevity (162). These observations highlight the importance of a prudent, plant-rich diet for healthy aging, aligning with dietary patterns that support a balanced colonic microbiota across life stages.
Interestingly, Estonian centenarians were also more exposed to animals and experienced lower sanitary conditions during their childhood (162). Similarly, early-life exposure to animals has been shown to contribute to the development of the fecal microbiota in infancy (25). Taken together, these findings suggest that early exposure to environmental microbes may play a crucial role in supporting the long-term balance of colonic microbiota throughout life.
5 Dietary recommendations for nourishing a balanced colonic microbiota
The evidence gathered in this review suggests that diets promoting a balanced colonic microbiota in infants post-weaning, adults, older adults, and centenarians share similar compositions (Figure 2). These diets are diverse and primarily based on plant foods, such as fruits, vegetables, and whole grains. These foods are rich in complex carbohydrates, polyunsaturated fatty acids, and polyphenols, which support the growth of saccharolytic microbes, for example, taxa from the genera Bifidobacterium, Faecalibacterium, Prevotella, Eubacterium, and Ruminococcus, and the production of beneficial metabolites, such as SCFAs and conjugated fatty acids (76, 117, 119). Additionally, polyunsaturated fatty acids and polyphenols have anti-inflammatory and antioxidant properties, and their metabolism by colonic microbes may confer further benefits to the host (76, 173). For instance, a 2-month intervention in adults consuming cereals enriched with polyphenols, dietary fiber, and omega-3 fatty acids increased the fecal abundance of Bacteroides species while reducing fecal calprotectin levels, a biomarker for intestinal inflammation (174).
FIGURE 2
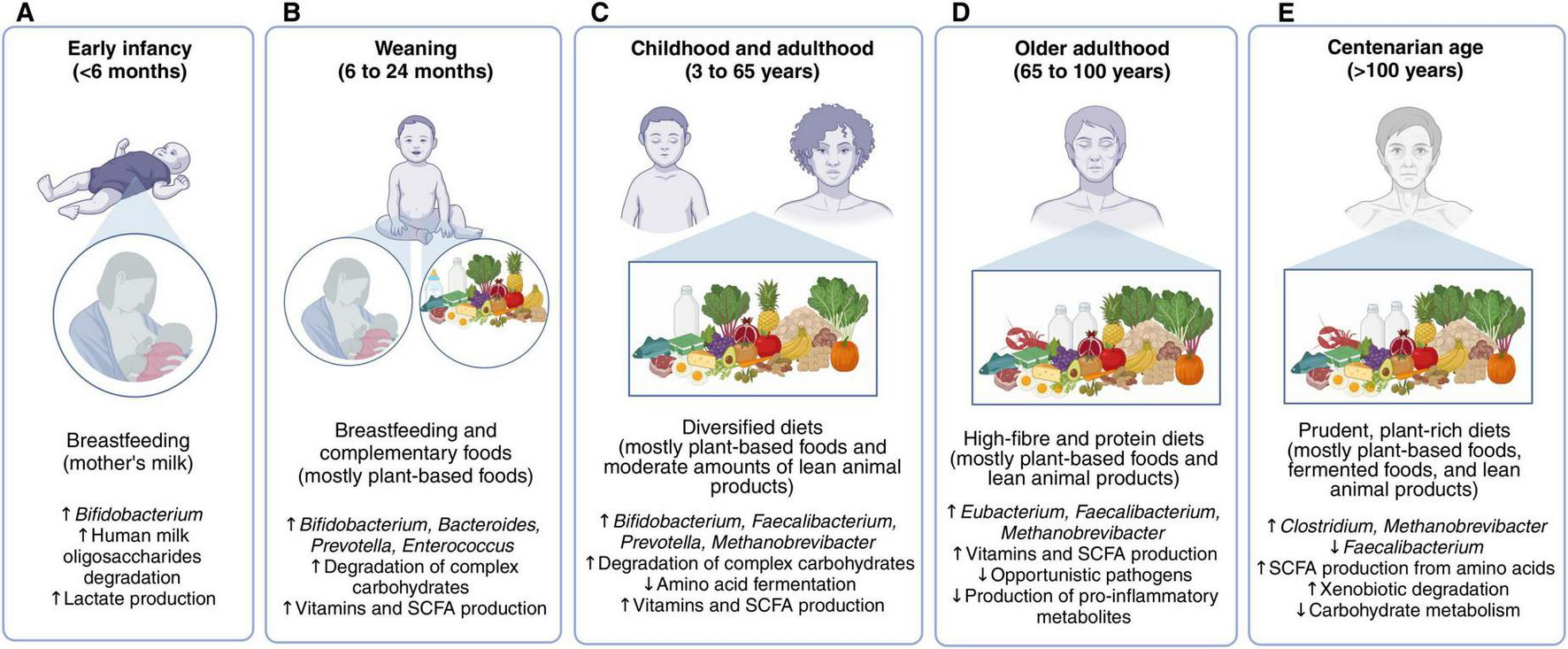
Favorable diets for nourishing the colonic microbiota in different life stages. In early infancy (A), breastfeeding is the optimal dietary strategy, but mother’s milk does not meet all the necessary nutritional requirements once the infant reaches the weaning period. In this stage (B), diets should combine breastfeeding with complementary foods, mainly fruits, vegetables, whole cereals, and animal products in moderation. The colonic microbiota matures as the introduction of solid foods increases and breastmilk or infant-formula decreases, achieving an adult state in which breast milk is no longer consumed. Children, adolescents, and adults have similar diets for promoting a balanced microbiota (C). These are rich in non-digestible carbohydrates, phytochemicals, and polyunsaturated fatty acids found in fruits, vegetables, nuts, pulses and whole grains, also containing moderate amounts of fish, fermented dairy products, and lean meats. A higher intake of protein is recommended for older adults, particularly from dairy and lean meats (D). Centenarians have a similar dietary pattern to that of older adults (E).
In addition to plant-based foods, favorable diets also include moderate consumption of animal-based foods, such as fish and lean meats (two to four servings per week). These foods are rich sources of essential amino acids and micronutrients, supporting colonic microbial diversity and contributing to meeting nutritional needs (120). Their consumption is particularly important for infants and older adults to promote physiological development and reduce disease risk (155). Daily consumption of fermented foods, such as fermented dairy, is also encouraged, as they contain lactic acid bacteria, supporting colonic eubiosis (127). Importantly, the intake of animal foods should be paired with the consumption of complex carbohydrates to mitigate excessive microbial fermentation of animal protein and fat. Notably, these recommendations for nourishing the colonic microbiota align with current dietary guidelines (64).
6 Limitations and future perspectives in diet-colonic microbiota research
Most of the knowledge on diet-colonic microbiota interaction in humans comes from studies using fecal samples as proxies. Compared to biopsies, fecal sampling is a non-invasive and cost-effective approach (175). However, they predominantly represent microbial communities from the distal colon, providing limited information about microbes attached to the colon’s mucosa or inhabiting other parts of the gastrointestinal tract (176). As a result, diet-induced changes observed in fecal microbial function and composition may not accurately reflect microbial alterations throughout the entire large intestine. Furthermore, current culture and sequencing techniques are unable to fully characterize the human colonic microbiota (177). Ultimately, due to technical limitations, our current understanding of how dietary patterns influence colonic commensals remains largely inferred and may not fully capture the complexity of host-microbe interactions.
Diet-colonic microbiota research has traditionally focused on adults. Hence, there is an opportunity for further research to expand our knowledge on diet-microbiota interactions in other life stages, such as infancy and older adulthood. Notably, weaning plays a critical role in colonic microbiota development (72). Investigating the impact of complementary feeding on host-microbiota associations early in life is a promising field to promote health and wellbeing and prevent diseases. Similarly, the continuous rise in human life expectancy urges more comprehension of the relationship between colonic microbiota and aging. The confounding effects of diseases on colonic microbes often challenge research on older populations. Therefore, studies involving both healthy and unhealthy older adults are necessary to deepen our understanding of how diet-microbiota modulations can support functionality during aging.
Among dietary compounds, the effects of complex carbohydrates on colonic microbes are well-characterized, whereas less is known about protein, fatty acids, polyphenols, micronutrients, and food additives. Notably, polyphenols and polyunsaturated fatty acids have been associated with health benefits, in which colonic microbes seem to play a critical role (76, 119). A better understanding of the impact of these nutrients on the colonic microbiota is needed, including in the long term. Future research should also investigate how dietary compounds interact with each other and their combined effects on the microbiota. Increased knowledge of the role of various nutrients in shaping colonic microbes, alongside individual factors (e.g., age, health status, activity level), will help researchers and medical professionals to tailor nutritional recommendations based on individual needs.
These challenges highlight the need for complementary methods to study the influence of dietary patterns on the colonic microbiota. Randomized clinical trials are the gold standard approach to measure the effect of food interventions on colonic microbes and resulting host health outcomes. They are also useful for validating findings from animal models and in vitro studies. However, clinical trials are time and resource-consuming, are prone to confounding factors, and rely on participant compliance and the accuracy of food questionnaires, limiting their feasibility (178). Furthermore, dietary assessment tools vary across microbiome studies, highlighting the need for standardized methods to improve comparability between studies targeting different age groups.
A promising strategy to address these limitations is to combine traditional methodologies with mathematical modeling. Mathematical models can investigate hypotheses that cannot be efficiently evaluated in vitro or in vivo, using a fraction of the time and cost of traditional approaches. In silico pipelines for predicting the effect of diets on personalized microbial communities have already been proposed (179, 180). Ongoing development and validation of these models could expand colonic microbiota research to traditionally underrepresented populations. For instance, in silico approaches were used to predict compositional and functional changes in the colonic microbiota of infants, children with different clinical conditions, healthy adults, and adults with Crohn’s disease according to the diet (89, 181–184). Moreover, models are flexible and can create personalized simulations based on input data, contributing toward personalized nutrition. Ultimately, integrating mathematical models with traditional approaches can reduce the costs of colonic microbiota research and accelerate our understanding of the relationship between diet, colonic microbes, and host health.
7 Conclusion
Colonic microbes ferment non-absorbed dietary compounds producing bioactive metabolites that influence host physiology. Therefore, identifying dietary patterns that support colonic eubiosis across different life stages is crucial for promoting host health and well-being. The evidence gathered in this review suggests that diets nourishing the colonic microbiota throughout human life are primarily composed of plant-based foods and include daily consumption of fermented foods, such as dairy products, and moderate amounts of fish and lean meats (two to four times a week). However, most diet-colonic microbiota investigations have focused on adults, neglecting weaning infants and older adults. Notably, weaning is a critical period for colonic microbiota development, setting the foundation for later life. In older adulthood, colonic microbes have a crucial role in maintaining functionality and promoting healthy aging. The limited understanding of how diets influence colonic microbes of infants and older adults is a significant barrier to using colonic microbiota modulation strategies to promote health. Further investigation of the long-term effects of dietary patterns on colonic microbes across different life stages is necessary to overcome some of the current limitations in diet-colonic microbiota research.
Statements
Author contributions
VG: Conceptualization, Formal Analysis, Visualization, Writing – original draft. NR: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. NS: Supervision, Writing – review & editing. CW: Funding acquisition, Supervision, Writing – review & editing. JM: Supervision, Writing – review & editing. WM: Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the New Zealand Ministry for Business, Innovation and Employment (MBIE, grant no. 3710040) through the High-Value Nutrition National Science Challenge.
Acknowledgments
VG was supported by a Riddet Institute PhD Scholarship. The PhD stipend and the project are funded by the High-Value Nutrition National Science Challenge.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2025.1591341/full#supplementary-material
References
1.
SenderRFuchsSMiloR. Revised estimates for the number of human and bacteria cells in the body.PLoS Biol. (2016) 14:e1002533. 10.1371/journal.pbio.1002533
2.
GaundalLMyhrstadMRudIGjøvaagTByfuglienMRetterstølKet alGut microbiota is associated with dietary intake and metabolic markers in healthy individuals.Food Nutr Res. (2022) 66:8580. 10.29219/fnr.v66.8580
3.
BojovićKIgnjatovićÐSoković BajićSVojnović MilutinovićDTomićMGolićNet alGut microbiota dysbiosis associated with altered production of short chain fatty acids in children with neurodevelopmental disorders.Front Cell Infect Microbiol. (2020) 10:223. 10.3389/fcimb.2020.00223
4.
FranzosaESirota-MadiAAvila-PachecoJFornelosNHaiserHReinkerSet alGut microbiome structure and metabolic activity in inflammatory bowel disease.Nat Microbiol. (2019) 4:293–305. 10.1038/s41564-018-0306-4
5.
KangJKhatibLDilmoreAHestonMUllandTJohnsonSet alGut microbiome features associate with cognitive scores in individuals at risk for Alzheimer’s disease.Alzheimers Dement. (2024) 20:e085801. 10.1002/alz.085801
6.
Valles-ColomerMFalonyGDarziYTigchelaarEWangJTitoRet alThe neuroactive potential of the human gut microbiota in quality of life and depression.Nat Microbiol. (2019) 4:623–32. 10.1038/s41564-018-0337-x
7.
Lloyd-PriceJArzeCAnanthakrishnanASchirmerMAvila-PachecoJPoonTet alMulti-omics of the gut microbial ecosystem in inflammatory bowel diseases.Nature. (2019) 569:655–62. 10.1038/s41586-019-1237-9
8.
MeiZWangFBhosleADongDMehtaRGhaziAet alStrain-specific gut microbial signatures in type 2 diabetes identified in a cross-cohort analysis of 8,117 metagenomes.Nat Med. (2024) 30:2265–76. 10.1038/s41591-024-03067-7
9.
JoosRBoucherKLavelleAArumugamMBlaserMClaessonMet alExamining the healthy human microbiome concept.Nat Rev Microbiol. (2025) 23:192–205. 10.1038/s41579-024-01107-0
10.
RinninellaERaoulPCintoniMFranceschiFMiggianoGGasbarriniAet alWhat is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases.Microorganisms. (2019) 7:14. 10.3390/microorganisms7010014
11.
OlssonLBoulundFNilssonSKhanMGummessonAFagerbergLet alDynamics of the normal gut microbiota: A longitudinal one-year population study in Sweden.Cell Host Microbe. (2022) 30:726–39.e3. 10.1016/j.chom.2022.03.002
12.
ZhernakovaAKurilshikovABonderMTigchelaarESchirmerMVatanenTet alPopulation-based metagenomics analysis reveals markers for gut microbiome composition and diversity.Science. (2016) 352:565–9. 10.1126/science.aad3369
13.
ManorODaiCKornilovSSmithBPriceNLovejoyJet alHealth and disease markers correlate with gut microbiome composition across thousands of people.Nat Commun. (2020) 11:5206. 10.1038/s41467-020-18871-1
14.
DavidLMaternaAFriedmanJCampos-BaptistaMBlackburnMPerrottaAet alHost lifestyle affects human microbiota on daily timescales.Genome Biol. (2014) 15:R89. 10.1186/gb-2014-15-7-r89
15.
DürholzKHofmannJIljazovicAHägerJLucasSSarterKet alDietary short-term fiber interventions in arthritis patients increase systemic SCFA levels and regulate inflammation.Nutrients. (2020) 12:3207. 10.3390/nu12103207
16.
OjoOOjoOZandNWangX. The effect of dietary fibre on gut microbiota, lipid profile, and inflammatory markers in patients with type 2 diabetes: A systematic review and meta-analysis of randomised controlled trials.Nutrients. (2021) 13:1805. 10.3390/nu13061805
17.
ShikanyJDemmerRJohnsonAFinoNMeyerKEnsrudKet alAssociation of dietary patterns with the gut microbiota in older, community-dwelling men.Am J Clin Nutr. (2019) 110:1003–14. 10.1093/ajcn/nqz174
18.
WanYWangFYuanJLiJJiangDZhangJet alEffects of dietary fat on gut microbiota and faecal metabolites, and their relationship with cardiometabolic risk factors: A 6-month randomised controlled-feeding trial.Gut. (2019) 68:1417–29. 10.1136/gutjnl-2018-317609
19.
WangKLoCMehtaRNguyenLWangYMaWet alAn empirical dietary pattern associated with the gut microbial features in relation to colorectal cancer risk.Gastroenterology. (2024) 167:1371–1383.e4. 10.1053/j.gastro.2024.07.040
20.
SweeneyMBurnsGSturgeonNMearsKStoteKBlantonC. The effects of berry polyphenols on the gut microbiota and blood pressure: A systematic review of randomized clinical trials in humans.Nutrients. (2022) 14:2263. 10.3390/nu14112263
21.
GuQSableCBrooks-WilsonAMurphyR. Dietary patterns in the healthy oldest old in the healthy aging study and the Canadian longitudinal study of aging: A cohort study.BMC Geriatr. (2020) 20:106. 10.1186/s12877-020-01507-w
22.
RobinsonSMarriottLPooleJCrozierSBorlandSLawrenceWet alDietary patterns in infancy: The importance of maternal and family influences on feeding practice.Br J Nutr. (2007) 98:1029–37. 10.1017/S0007114507750936
23.
GolshanyHHelmySMorsyNKamalAYuQFanL. The gut microbiome across the lifespan: How diet modulates our microbial ecosystem from infancy to the elderly.Int J Food Sci Nutr. (2024) 0:1–27. 10.1080/09637486.2024.2437472
24.
BradleyEHaranJ. The human gut microbiome and aging.Gut Microbes. (2024) 16:2359677. 10.1080/19490976.2024.2359677
25.
StewartCAjamiNO’BrienJHutchinsonDSmithDWongMet alTemporal development of the gut microbiome in early childhood from the TEDDY study.Nature. (2018) 562:583–8. 10.1038/s41586-018-0617-x
26.
ZhangJGuoZXueZSunZZhangMWangLet alA phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities.ISME J. (2015) 9:1979–90. 10.1038/ismej.2015.11
27.
HuttenhowerCGeversDKnightRAbubuckerSBadgerJChinwallaAet alStructure, function and diversity of the healthy human microbiome.Nature. (2012) 486:207–14. 10.1038/nature11234
28.
CashHWhithamCBehrendtCHooperL. Symbiotic bacteria direct expression of an intestinal bactericidal lectin.Science. (2006) 313:1126–30. 10.1126/science.1127119
29.
MacphersonAGattoDSainsburyEHarrimanGHengartnerHZinkernagelRMA. Primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria.Science. (2000) 288:2222–6. 10.1126/science.288.5474.2222
30.
HillCLynchDMurphyKUlaszewskaMJefferyIO’SheaCet alEvolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort.Microbiome. (2017) 5:4. 10.1186/s40168-016-0213-y
31.
BäckhedFRoswallJPengYFengQJiaHKovatcheva-DatcharyPet alDynamics and stabilization of the human gut microbiome during the first year of life.Cell Host Microbe. (2015) 17:690–703. 10.1016/j.chom.2015.04.004
32.
RutayisireEHuangKLiuYTaoF. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants’ life: A systematic review.BMC Gastroenterol. (2016) 16:86. 10.1186/s12876-016-0498-0
33.
TaylorRKeaneDBorregoPArcaroK. Effect of maternal diet on maternal milk and breastfed infant gut microbiomes: A scoping review.Nutrients. (2023) 15:1420. 10.3390/nu15061420
34.
VatanenTPlichtaDSomaniJMünchPArthurTHallAet alGenomic variation and strain-specific functional adaptation in the human gut microbiome during early life.Nat Microbiol. (2019) 4:470–9. 10.1038/s41564-018-0321-5
35.
DurantiSLugliGMilaniCJamesKMancabelliLTurroniFet alBifidobacterium bifidum and the infant gut microbiota: An intriguing case of microbe-host co-evolution.Environ Microbiol. (2019) 21:3683–95. 10.1111/1462-2920.14705
36.
MoossaviSSepehriSRobertsonBBodeLGorukSFieldCet alComposition and variation of the human milk microbiota are influenced by maternal and early-life factors.Cell Host Microbe. (2019) 25:324–35.e4. 10.1016/j.chom.2019.01.011
37.
DolanSBoesman-FinkelsteinMFinkelsteinR. Antimicrobial activity of human milk against pediatric pathogens.J Infect Dis. (1986) 154:722–5. 10.1093/infdis/154.4.722
38.
MancabelliLTarracchiniCMilaniCLugliGFontanaFTurroniFet alMulti-population cohort meta-analysis of human intestinal microbiota in early life reveals the existence of infant community state types (ICSTs).Comput Struct Biotechnol J. (2020) 18:2480–93. 10.1016/j.csbj.2020.08.028
39.
InchingoloFInchingoloALatiniGFerranteLde RuvoECampanelliMet alDifference in the intestinal microbiota between breastfeed infants and infants fed with artificial milk: A systematic review.Pathogens. (2024) 13:533. 10.3390/pathogens13070533
40.
World Health Organization [WHO]. Breastfeeding Recommendations. (2023). Available online at: https://www.who.int/health-topics/breastfeeding(accessed February 13, 2023).
41.
GratzSHazimSRichardsonAScobbieLJohnstoneAFyfeCet alDietary carbohydrate rather than protein intake drives colonic microbial fermentation during weight loss.Eur J Nutr. (2019) 58:1147–58. 10.1007/s00394-018-1629-x
42.
ZengXXingXGuptaMKeberFLopezJLeeYet alGut bacterial nutrient preferences quantified in vivo.Cell. (2022) 185:3441–56.e19. 10.1016/j.cell.2022.07.020
43.
MacfarlaneGMacfarlaneSGibsonG. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon.Microb Ecol. (1998) 35:180–7. 10.1007/s002489900072
44.
SmithEMacfarlaneG. Enumeration of amino acid fermenting bacteria in the human large intestine: Effects of pH and starch on peptide metabolism and dissimilation of amino acids.FEMS Microbiol Ecol. (1998) 25:355–68. 10.1111/j.1574-6941.1998.tb00487.x
45.
FrankDSt AmandALFeldmanRABoedekerECHarpazNPaceNR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases.Proc Natl Acad Sci. (2007) 104:13780–5. 10.1073/pnas.0706625104
46.
UngerMSpiegelJDillmannKGrundmannDPhilippeitHBürmannJet alShort chain fatty acids and gut microbiota differ between patients with Parkinson’s disease and age-matched controls.Park Relat Disord. (2016) 32:66–72. 10.1016/j.parkreldis.2016.08.019
47.
ZhaoLLouHPengYChenSZhangYLiX. Comprehensive relationships between gut microbiome and faecal metabolome in individuals with type 2 diabetes and its complications.Endocrine. (2019) 66:526–37. 10.1007/s12020-019-02103-8
48.
ArboleyaSBinettiASalazarNFernándezNSolísGHernández-BarrancoAet alEstablishment and development of intestinal microbiota in preterm neonates.FEMS Microbiol Ecol. (2012) 79:763–72. 10.1111/j.1574-6941.2011.01261.x
49.
StaudacherHLomerMAndersonJBarrettJMuirJIrvingPet alFermentable carbohydrate restriction reduces luminal bifidobacteria and gastrointestinal symptoms in patients with irritable bowel syndrome.J Nutr. (2012) 142:1510–8. 10.3945/jn.112.159285
50.
Gómez-MartínMSaturioSArboleyaSHerrero-MorínDCalzónMLópezTet alAssociation between diet and fecal microbiota along the first year of life.Food Res Int. (2022) 162:111994. 10.1016/j.foodres.2022.111994
51.
FallaniMAmarriSUusijarviAAdamRKhannaSAguileraMet alDeterminants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres.Microbiology. (2011) 157:1385–92. 10.1099/mic.0.042143-0
52.
Fahur BottinoGBonhamKPatelFMcCannSZieffMNaspoliniNet alEarly life microbial succession in the gut follows common patterns in humans across the globe.Nat Commun. (2025) 16:660. 10.1038/s41467-025-56072-w
53.
LimEZhouYZhaoGBauerIDroitLNdaoIet alEarly life dynamics of the human gut virome and bacterial microbiome in infants.Nat Med. (2015) 21:1228–34. 10.1038/nm.3950
54.
ScheiKAvershinaEØienTRudiKFollestadTSalamatiSet alEarly gut mycobiota and mother-offspring transfer.Microbiome. (2017) 5:107. 10.1186/s40168-017-0319-x
55.
ArrietaMStiemsmaLDimitriuPThorsonLRussellSYurist-DoutschSet alEarly infancy microbial and metabolic alterations affect risk of childhood asthma.Sci Transl Med. (2015) 7:307ra152. 10.1126/scitranslmed.aab2271
56.
HeiskanenMAatsinkiAHakonenPKartiosuoNMunukkaELahtiLet alAssociation of long-term habitual dietary fiber intake since infancy with gut microbiota composition in young adulthood.J Nutr. (2024) 154:744–54. 10.1016/j.tjnut.2024.01.008
57.
OluwagbemigunKO’DonovanABerdingKLyonsKAlexyUSchmidMet alLong-term dietary intake from infancy to late adolescence is associated with gut microbiota composition in young adulthood.Am J Clin Nutr. (2021) 113:647–56. 10.1093/ajcn/nqaa340
58.
Geniselli da SilvaVTonkieJRoyNSmithNWallCKrugerMet alThe effect of complementary foods on the colonic microbiota of weaning infants: A systematic review.Crit Rev Food Sci Nutr. (2024) [Online ahead of print]. 10.1080/10408398.2024.2439036
59.
García-MantranaISelma-RoyoMGonzálezSParra-LlorcaAMartínez-CostaCColladoM. Distinct maternal microbiota clusters are associated with diet during pregnancy: Impact on neonatal microbiota and infant growth during the first 18 months of life.Gut Microbes. (2020) 11:962–78. 10.1080/19490976.2020.1730294
60.
DawsonSClarkeGPonsonbyALoughmanAMohebbiMBorgeTet alA gut-focused perinatal dietary intervention is associated with lower alpha diversity of the infant gut microbiota: Results from a randomised controlled trial.Nutr Neurosci. (2024) [Online ahead of print]. 10.1080/1028415X.2024.2413233
61.
BabakobiMReshefLGihazSBelgorodskyBFishmanABujanoverYet alEffect of maternal diet and milk lipid composition on the infant gut and maternal milk microbiomes.Nutrients. (2020) 12:2539. 10.3390/nu12092539
62.
AlmutairiRBassonAWearshPCominelliFRodriguez-PalaciosA. Validity of food additive maltodextrin as placebo and effects on human gut physiology: Systematic review of placebo-controlled clinical trials.Eur J Nutr. (2022) 61:2853–71. 10.1007/s00394-022-02802-5
63.
Abiega-FranyuttiPFreyre-FonsecaV. Chronic consumption of food-additives lead to changes via microbiota gut-brain axis.Toxicology. (2021) 464:153001. 10.1016/j.tox.2021.153001
64.
U.S. Department of Agriculture, U.S. Department of Health and Human Services. Dietary Guidelines for Americans, 2020-2025.Washington, DC: U.S. Department of Agriculture (2020).
65.
TondeurMSchauerCChristofidesAAsanteKNewtonSSerfassRet alDetermination of iron absorption from intrinsically labeled microencapsulated ferrous fumarate (sprinkles) in infants with different iron and hematologic status by using a dual-stable-isotope method.Am J Clin Nutr. (2004) 80:1436–44. 10.1093/ajcn/80.5.1436
66.
RodionovDArzamasovAKhoroshkinMIablokovSLeynSPetersonSet alMicronutrient requirements and sharing capabilities of the human gut microbiome.Front Microbiol. (2019) 10:1316. 10.3389/fmicb.2019.01316
67.
GonzálezAGálvezNMartínJReyesFPérez-VictoriaIDominguez-VeraJ. Identification of the key excreted molecule by Lactobacillus fermentum related to host iron absorption.Food Chem. (2017) 228:374–80. 10.1016/j.foodchem.2017.02.008
68.
GuettermanHHueySKnightRFoxAMehtaSFinkelsteinJ. Vitamin B-12 and the gastrointestinal microbiome: A systematic review.Adv Nutr. (2022) 13:530–58. 10.1093/advances/nmab123
69.
PopovicABourdonCWangPGuttmanDSoofiSBhuttaZet alMicronutrient supplements can promote disruptive protozoan and fungal communities in the developing infant gut.Nat Commun. (2021) 12:6729. 10.1038/s41467-021-27010-3
70.
JaeggiTKortmanGMorettiDChassardCHoldingPDostalAet alIron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants.Gut. (2015) 64:731–42. 10.1136/gutjnl-2014-307720
71.
World Health Organization [WHO]. WHO Guideline for Complementary Feeding of Infants and Young Children 6–23 Months of Age. (2023). Available online at: https://www.who.int/publications/i/item/9789240081864(accessed July 15, 2024).
72.
LaursenMAndersenLMichaelsenKMølgaardCTrolleEBahlMet alInfant gut microbiota development is driven by transition to family foods independent of maternal obesity.mSphere. (2016) 1:e0069-15. 10.1128/mSphere.00069-15
73.
HomannCRosselCDizzellSBervoetsLSimioniJLiJet alInfants’ First Solid Foods: Impact on gut microbiota development in two intercontinental cohorts.Nutrients. (2021) 13:2639. 10.3390/nu13082639
74.
FontaineFTurjemanSCallensKKorenO. The intersection of undernutrition, microbiome, and child development in the first years of life.Nat Commun. (2023) 14:3554. 10.1038/s41467-023-39285-9
75.
New Zealand Ministry of Health. Healthy Eating Guidelines for New Zealand Babies and Toddlers (0–2 years old). (2021). Available online at: https://www.health.govt.nz/system/files/documents/publications/healthy-eatingguidelines-for-new-zealand-babies-and-toddlers-nov21-v2.pdf(accessed July 1, 2023).
76.
MaGChenY. Polyphenol supplementation benefits human health via gut microbiota: A systematic review via meta-analysis.J Funct Foods. (2020) 66:103829. 10.1016/j.jff.2020.103829
77.
ManachCWilliamsonGMorandCScalbertARémésyC. Bioavailability and bioefficacy of polyphenols in humans. I. Review of 97 bioavailability studies.Am J Clin Nutr. (2005) 81:230S–42S. 10.1093/ajcn/81.1.230S
78.
SelmaMBeltránDLunaMRomo-VaqueroMGarcía-VillalbaRMiraAet alIsolation of human intestinal bacteria capable of producing the bioactive metabolite isourolithin A from ellagic acid.Front Microbiol. (2017) 8:1521. 10.3389/fmicb.2017.01521
79.
Moreno-IndiasISánchez-AlcoholadoLPérez-MartínezPAndrés-LacuevaCCardonaFTinahonesFet alRed wine polyphenols modulate fecal microbiota and reduce markers of the metabolic syndrome in obese patients.Food Funct. (2016) 7:1775–87. 10.1039/C5FO00886G
80.
Calderón-PérezLLlauradóECompanysJPla-PagàLPedretARubióLet alInterplay between dietary phenolic compound intake and the human gut microbiome in hypertension: A cross-sectional study.Food Chem. (2021) 344:128567. 10.1016/j.foodchem.2020.128567
81.
Plaza-DiazJBernalMSchutteSChenollEGenovésSCodoñerFet alEffects of whole-grain and sugar content in infant cereals on gut microbiota at weaning: A randomized trial.Nutrients. (2021) 13:1496. 10.3390/nu13051496
82.
KrebsNSherlockLWestcottJCulbertsonDHambidgeKFeazelLet alEffects of different complementary feeding regimens on iron status and enteric microbiota in breastfed infants.J Pediatr. (2013) 163:416–23.e4. 10.1016/j.jpeds.2013.01.024
83.
LeongCHaszardJLawleyBOtalATaylorRSzymlek-GayEet alMediation analysis as a means of identifying dietary components that differentially affect the fecal microbiota of infants weaned by modified baby-led and traditional approaches.Appl Environ Microbiol. (2018) 84:e914–8. 10.1128/AEM.00914-18
84.
KhineWRahayuESeeTKuahSSalminenSNakayamaJet alIndonesian children fecal microbiome from birth until weaning was different from microbiomes of their mothers.Gut Microbes. (2020) 12:1761240. 10.1080/19490976.2020.1761240
85.
LalliMSaloTHakolaLKnipMVirtanenSVatanenT. Associations between dietary fibers and gut microbiome composition in the EDIA longitudinal infant cohort.Am J Clin Nutr. (2025) 121:83–99. 10.1016/j.ajcnut.2024.11.011
86.
GamageHTetuSChongRAshtonJPackerNPaulsenI. Cereal products derived from wheat, sorghum, rice and oats alter the infant gut microbiota in vitro.Sci Rep. (2017) 7:14312. 10.1038/s41598-017-14707-z
87.
ParkarSFrostJRosendaleDStoklosinskiHJobsisCHedderleyDet alThe sugar composition of the fibre in selected plant foods modulates weaning infants’ gut microbiome composition and fermentation metabolites in vitro.Sci Rep. (2021) 11:9292. 10.1038/s41598-021-88445-8
88.
ParkarSRosendaleDStoklosinskiHJobsisCHedderleyDGopalP. Complementary food ingredients alter infant gut microbiome composition and metabolism in vitro.Microorganisms. (2021) 9:2089. 10.3390/microorganisms9102089
89.
da SilvaVSmithNMullaneyJWallCRoyNMcNabbW. Food-breastmilk combinations alter the colonic microbiome of weaning infants: An in silico study.mSystems. (2024) 9:e00577-24. 10.1128/msystems.00577-24
90.
da SilvaVMullaneyJRoyNSmithNWallCTattonCet alComplementary foods in infants: An in vitro study of the faecal microbial composition and organic acid production.Food Funct. (2025) 16:3465–81. 10.1039/D5FO00414D
91.
SchaartMde BruijnATibboelDRenesIvan GoudoeverJ. Dietary protein absorption of the small intestine in human neonates.J Parenter Enter Nutr. (2007) 31:482–6. 10.1177/0148607107031006482
92.
MacfarlaneGGibsonGBeattyECummingsJ. Estimation of short-chain fatty acid production from protein by human intestinal bacteria based on branched-chain fatty acid measurements.FEMS Microbiol Ecol. (1992) 10:81–8. 10.1111/j.1574-6941.1992.tb00002.x
93.
WangSvan GeffenMVenemaKMommersAJonkersDvan SchootenFet alEffect of protein fermentation products on gut health assessed in an in vitro model of human colon (TIM-2).Mol Nutr Food Res. (2023) 67:2200574. 10.1002/mnfr.202200574
94.
QasemWAzadMHossainZAzadEJorgensenSCastillo San JuanSet alAssessment of complementary feeding of Canadian infants: Effects on microbiome & oxidative stress, a randomized controlled trial.BMC Pediatr. (2017) 17:54. 10.1186/s12887-017-0805-0
95.
TangMMaCWeinheimer-HausERobertsonCKofonowJBermanLet alDifferent gut microbiota in U.S. formula-fed infants consuming a meat vs. dairy-based complementary foods: A randomized controlled trial.Front Nutr. (2023) 9:1063518. 10.3389/fnut.2022.1063518
96.
Smith-BrownPMorrisonMKrauseLDaviesP. Microbiota and body composition during the period of complementary feeding.J Pediatr Gastroenterol Nutr. (2019) 69:726–32. 10.1097/MPG.0000000000002490
97.
De FilippoCCavalieriDDi PaolaMRamazzottiMPoulletJMassartSet alImpact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa.Proc Natl Acad Sci. (2010) 107:14691–6. 10.1073/pnas.1005963107
98.
MoralesPFujioSNavarretePUgaldeJMagneFCarrasco-PozoCet alImpact of dietary lipids on colonic function and microbiota: An experimental approach involving orlistat-induced fat malabsorption in human volunteers.Clin Transl Gastroenterol. (2016) 7:e161. 10.1038/ctg.2016.20
99.
WoltersMAhrensJRomaní-PérezMWatkinsCSanzYBenítez-PáezAet alDietary fat, the gut microbiota, and metabolic health – A systematic review conducted within the MyNewGut project.Clin Nutr. (2019) 38:2504–20. 10.1016/j.clnu.2018.12.024
100.
MatsueMMoriYNagaseSSugiyamaYHiranoROgaiKet alMeasuring the antimicrobial activity of lauric acid against various bacteria in human gut microbiota using a new method.Cell Transplant. (2019) 28:1528–41. 10.1177/0963689719881366
101.
CasasRRuiz-LeónAArgenteJAlasalvarCBajoubABertomeuIet alA new mediterranean lifestyle pyramid for children and youth: A critical lifestyle tool for preventing obesity and associated cardiometabolic diseases in a sustainable context.Adv Nutr. (2025) 16:100381. 10.1016/j.advnut.2025.100381
102.
YatsunenkoTReyFManaryMTrehanIDominguez-BelloMContrerasMet alHuman gut microbiome viewed across age and geography.Nature. (2012) 486:222–7. 10.1038/nature11053
103.
ZengSPatangiaDAlmeidaAZhouZMuDPaul RossRet alA compendium of 32,277 metagenome-assembled genomes and over 80 million genes from the early-life human gut microbiome.Nat Commun. (2022) 13:5139. 10.1038/s41467-022-32805-z
104.
RoswallJOlssonLKovatcheva-DatcharyPNilssonSTremaroliVSimonMet alDevelopmental trajectory of the healthy human gut microbiota during the first 5 years of life.Cell Host Microbe. (2021) 29:765–76.e3. 10.1016/j.chom.2021.02.021
105.
RinninellaETohumcuERaoulPFioraniMCintoniMMeleMet alThe role of diet in shaping human gut microbiota.Best Pract Res Clin Gastroenterol. (2023) 6:101828. 10.1016/j.bpg.2023.101828
106.
DuncansonKWilliamsGHoedtECollinsCKeelySTalleyN. Diet-microbiota associations in gastrointestinal research: A systematic review.Gut Microbes. (2024) 16:2350785. 10.1080/19490976.2024.2350785
107.
RossFPatangiaDGrimaudGLavelleADempseyERossRet alThe interplay between diet and the gut microbiome: Implications for health and disease.Nat Rev Microbiol. (2024) 22:671–86. 10.1038/s41579-024-01068-4
108.
DavidLMauriceCCarmodyRGootenbergDButtonJWolfeBet alDiet rapidly and reproducibly alters the human gut microbiome.Nature. (2014) 505:559–63. 10.1038/nature12820
109.
WuGChenJHoffmannCBittingerKChenYKeilbaughSet alLinking long-term dietary patterns with gut microbial enterotypes.Science. (2011) 334:105–8. 10.1126/science.1208344
110.
HoffmannCDolliveSGrunbergSChenJLiHWuGet alArchaea and fungi of the human gut microbiome: Correlations with diet and bacterial residents.PLoS One. (2013) 8:e66019. 10.1371/journal.pone.0066019
111.
WellingtonNShanmuganathanMde SouzaRZulyniakMAzabSBloomfieldJet alMetabolic trajectories following contrasting prudent and western diets from food provisions: Identifying robust biomarkers of short-term changes in habitual diet.Nutrients. (2019) 11:2407. 10.3390/nu11102407
112.
BeaumontMPortuneKSteuerNLanACerrudoVAudebertMet alQuantity and source of dietary protein influence metabolite production by gut microbiota and rectal mucosa gene expression: A randomized, parallel, double-blind trial in overweight humans.Am J Clin Nutr. (2017) 106:1005–19. 10.3945/ajcn.117.158816
113.
RussellWGratzSDuncanSHoltropGInceJScobbieLet alHigh-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health.Am J Clin Nutr. (2011) 93:1062–72. 10.3945/ajcn.110.002188
114.
LiYPetersBYuBPerreiraKDaviglusMChanQet alBlood metabolomic shift links diet and gut microbiota to multiple health outcomes among Hispanic/Latino immigrants in the U.S.medRxiv [Preprint]. (2024): 10.1101/2024.07.19.24310722
115.
SidhuSKokCKunasegaranTRamadasA. Effect of plant-based diets on gut microbiota: A systematic review of interventional studies.Nutrients. (2023) 15:1510. 10.3390/nu15061510
116.
TembaGPechtTKullayaVVadaqNMoshaMUlasTet alImmune and metabolic effects of African heritage diets versus Western diets in men: A randomized controlled trial.Nat Med. (2025) [Online ahead of print]. 10.1038/s41591-025-03602-0
117.
SoDWhelanKRossiMMorrisonMHoltmannGKellyJet alDietary fiber intervention on gut microbiota composition in healthy adults: A systematic review and meta-analysis.Am J Clin Nutr. (2018) 107:965–83. 10.1093/ajcn/nqy041
118.
SchoelerMEllero-SimatosSBirknerTMayneris-PerxachsJOlssonLBrolinHet alThe interplay between dietary fatty acids and gut microbiota influences host metabolism and hepatic steatosis.Nat Commun. (2023) 14:5329. 10.1038/s41467-023-41074-3
119.
LiuHLiXZhuYHuangYZhangQLinSet alEffect of plant-derived n-3 polyunsaturated fatty acids on blood lipids and gut microbiota: A double-blind randomized controlled trial.Front Nutr. (2022) 9:830960. 10.3389/fnut.2022.830960
120.
TrefflichIDietrichSBrauneAAbrahamKWeikertC. Short- and branched-chain fatty acids as fecal markers for microbiota activity in vegans and omnivores.Nutrients. (2021) 13:1808. 10.3390/nu13061808
121.
CotillardACartier-MeheustALitwinNChaumontSSaccareauMLejzerowiczFet alA posteriori dietary patterns better explain variations of the gut microbiome than individual markers in the American Gut Project.Am J Clin Nutr. (2022) 115:432–43. 10.1093/ajcn/nqab332
122.
MaukonenMKoponenKHavulinnaAKaartinenNNiiranenTMéricGet alAssociations of plant-based foods, red and processed meat, and dairy with gut microbiome in Finnish adults.Eur J Nutr. (2024) 63:2247–60. 10.1007/s00394-024-03406-x
123.
BaldeonAMcDonaldDGonzalezAKnightRHolscherH. Diet quality and the fecal microbiota in adults in the American gut project.J Nutr. (2023) 153:2004–15. 10.1016/j.tjnut.2023.02.018
124.
Bach-FaigABerryELaironDReguantJTrichopoulouADerniniSet alMediterranean diet pyramid today. Science and cultural updates.Public Health Nutr. (2011) 14:2274–84. 10.1017/S1368980011002515
125.
KhavandegarAHeidarzadehAAngooraniPHasani-RanjbarSEjtahedHLarijaniBet alAdherence to the Mediterranean diet can beneficially affect the gut microbiota composition: A systematic review.BMC Med Genomics. (2024) 17:91. 10.1186/s12920-024-01861-3
126.
EleftheriouDBenetouVTrichopoulouAVecchiaCBamiaC. Mediterranean diet and its components in relation to all-cause mortality: Meta-analysis.Br J Nutr. (2018) 120:1081–97. 10.1017/S0007114518002593
127.
TaylorBLejzerowiczFPoirelMShafferJJiangLAksenovAet alConsumption of fermented foods is associated with systematic differences in the gut microbiome and metabolome.mSystems. (2020) 5:e00901–19. 10.1128/msystems.00901-19
128.
WastykHFragiadakisGPerelmanDDahanDMerrillBYuFet alGut-microbiota-targeted diets modulate human immune status.Cell. (2021) 184:4137–53.e14. 10.1016/j.cell.2021.06.019
129.
SchoonakkerMvan PeetPvan den BurgENumansMDucarmonQPijlHet alImpact of dietary carbohydrate, fat or protein restriction on the human gut microbiome: A systematic review.Nutr Res Rev. (2024) 38:238–55. 10.1017/S0954422424000131
130.
HalmosEPowerVShepherdSGibsonPMuirJ. A Diet Low in FODMAPs reduces symptoms of irritable bowel syndrome.Gastroenterology. (2014) 146:67–75.e5. 10.1053/j.gastro.2013.09.046
131.
SoDLoughmanAStaudacherH. Effects of a low FODMAP diet on the colonic microbiome in irritable bowel syndrome: A systematic review with meta-analysis.Am J Clin Nutr. (2022) 116:943–52. 10.1093/ajcn/nqac176
132.
HansenLRoagerHSøndertoftNGøbelRKristensenMVallès-ColomerMet alA low-gluten diet induces changes in the intestinal microbiome of healthy Danish adults.Nat Commun. (2018) 9:1–13. 10.1038/s41467-018-07019-x
133.
BonderMTigchelaarECaiXTrynkaGCenitMHrdlickovaBet alThe influence of a short-term gluten-free diet on the human gut microbiome.Genome Med. (2016) 8:45. 10.1186/s13073-016-0295-y
134.
Borowicz-ReuttKKrawczykMCzerniaJ. Ketogenic diet in the treatment of epilepsy.Nutrients. (2024) 16:1258. 10.3390/nu16091258
135.
RewLHarrisMGoldieJ. The ketogenic diet: Its impact on human gut microbiota and potential consequent health outcomes: A systematic literature review.Gastroenterol Hepatol Bed Bench. (2022) 15:326–42. 10.22037/ghfbb.v15i4.2600
136.
HengistADaviesRWalhinJBuniamJMerrellLRogersLet alKetogenic diet but not free-sugar restriction alters glucose tolerance, lipid metabolism, peripheral tissue phenotype, and gut microbiome: RCT.Cell Rep Med. (2024) 5:101667. 10.1016/j.xcrm.2024.101667
137.
PrestonJBiddellB. The physiology of ageing and how these changes affect older people.Medicine. (2021) 49:1–5. 10.1016/j.mpmed.2020.10.011
138.
WilmanskiTDienerCRappaportNPatwardhanSWiedrickJLapidusJet alGut microbiome pattern reflects healthy ageing and predicts survival in humans.Nat Metab. (2021) 3:274–86. 10.1038/s42255-021-00348-0
139.
BianGGloorGGongAJiaCZhangWHuJet alThe gut microbiota of healthy aged chinese is similar to that of the healthy young.mSphere. (2017) 2:e00327-17. 10.1128/mSphere.00327-17
140.
GregoryAZablockiOZayedAHowellABolducBSullivanM. The gut virome database reveals age-dependent patterns of virome diversity in the human gut.Cell Host Microbe. (2020) 28:724–40.e8. 10.1016/j.chom.2020.08.003
141.
MihajlovskiADoréJLevenezFAlricMBrugèreJ. Molecular evaluation of the human gut methanogenic archaeal microbiota reveals an age-associated increase of the diversity.Environ Microbiol Rep. (2010) 2:272–80. 10.1111/j.1758-2229.2009.00116.x
142.
AhmadHMejiaJKrychLKhakimovBKotWBechshøftRet alGut mycobiome dysbiosis is linked to hypertriglyceridemia among home dwelling elderly danes.bioRxiv [Preprint]. (2020): 10.1101/2020.04.16.044693
143.
ZhangSZengBChenYYangMKongFWeiLet alGut microbiota in healthy and unhealthy long-living people.Gene. (2021) 779:145510. 10.1016/j.gene.2021.145510
144.
GhoshTShanahanFO’TooleP. Toward an improved definition of a healthy microbiome for healthy aging.Nat Aging. (2022) 2:1054–69. 10.1038/s43587-022-00306-9
145.
ShintouoCMetsTBeckweeDBautmansIGhogomuSSouopguiJet alIs inflammageing influenced by the microbiota in the aged gut? A systematic review.Exp Gerontol. (2020) 141:111079. 10.1016/j.exger.2020.111079
146.
Hairul HishamHLimSNeohCAbdul MajeedAShaharSRamasamyK. Effects of non-pharmacological interventions on gut microbiota and intestinal permeability in older adults: A systematic review: Non-pharmacological interventions on gut microbiota/barrier.Arch Gerontol Geriatr. (2025) 128:105640. 10.1016/j.archger.2024.105640
147.
LiYWangDSatijaAIveyKLiJWilkinsonJet alPlant-based diet index and metabolic risk in men: Exploring the role of the gut microbiome.J Nutr. (2021) 151:2780–9. 10.1093/jn/nxab175
148.
MaWNguyenLSongMWangDFranzosaECaoYet alDietary fiber intake, the gut microbiome, and chronic systemic inflammation in a cohort of adult men.Genome Med. (2021) 13:102. 10.1186/s13073-021-00921-y
149.
Del Bo’CBernardiSCherubiniAPorriniMGargariGHidalgo-LiberonaNet alA polyphenol-rich dietary pattern improves intestinal permeability, evaluated as serum zonulin levels, in older subjects: The MaPLE randomised controlled trial.Clin Nutr. (2021) 40:3006–18. 10.1016/j.clnu.2020.12.014
150.
NagpalRNethBWangSCraftSYadavH. Modified Mediterranean-ketogenic diet modulates gut microbiome and short-chain fatty acids in association with Alzheimer’s disease markers in subjects with mild cognitive impairment.EBioMedicine. (2019) 47:529–42. 10.1016/j.ebiom.2019.08.032
151.
LiJLiYIveyKWangDWilkinsonJFrankeAet alInterplay between diet and gut microbiome, and circulating concentrations of trimethylamine N-oxide: Findings from a longitudinal cohort of US men.Gut. (2022) 71:724–33. 10.1136/gutjnl-2020-322473
152.
TessierAWangFKoratAEliassenAChavarroJGrodsteinFet alOptimal dietary patterns for healthy aging.Nat Med. (2025) [Online ahead of print]. 10.1038/s41591-025-03570-5
153.
MilanAD’SouzaRPundirSPileggiCBarnettMMarkworthJet alOlder adults have delayed amino acid absorption after a high protein mixed breakfast meal.J Nutr Health Aging. (2015) 19:839–45. 10.1007/s12603-015-0500-5
154.
FarsijaniSCauleyJPeddadaSLangsetmoLShikanyJOrwollEet alRelation between dietary protein intake and gut microbiome composition in community-dwelling older men: Findings from the osteoporotic fractures in men study (MrOS).J Nutr. (2022) 152:2877–87. 10.1093/jn/nxac231
155.
Coelho-JúniorHRodriguesBUchidaMMarzettiE. Low protein intake is associated with frailty in older adults: A systematic review and meta-analysis of observational studies.Nutrients. (2018) 10:1334. 10.3390/nu10091334
156.
BauerJBioloGCederholmTCesariMCruz-JentoftAMorleyJet alEvidence-based recommendations for optimal dietary protein intake in older people: A position paper from the PROT-AGE study group.J Am Med Dir Assoc. (2013) 14:542–59. 10.1016/j.jamda.2013.05.021
157.
ProkopidisKCervoMGandhamAScottD. Impact of protein intake in older adults with sarcopenia and obesity: A gut microbiota perspective.Nutrients. (2020) 12:2285. 10.3390/nu12082285
158.
MitchellSMcKenzieEMitchellCMilanAZengND’SouzaRet alA period of 10 weeks of increased protein consumption does not alter faecal microbiota or volatile metabolites in healthy older men: A randomised controlled trial.J Nutr Sci. (2020) 9:e25. 10.1017/jns.2020.15
159.
FluitmanKWijdeveldMDavidsMvan RuitenCReindersIWijnhovenHet alPersonalized dietary advice to increase protein intake in older adults does not affect the gut microbiota, appetite or central processing of food stimuli in community-dwelling older adults: A six-month randomized controlled trial.Nutrients. (2023) 15:332. 10.3390/nu15020332
160.
SatoYAtarashiKPlichtaDAraiYSasajimaSKearneySet alNovel bile acid biosynthetic pathways are enriched in the microbiome of centenarians.Nature. (2021) 599:458–64. 10.1038/s41586-021-03832-5
161.
JohansenJAtarashiKAraiYHiroseNSørensenSVatanenTet alCentenarians have a diverse gut virome with the potential to modulate metabolism and promote healthy lifespan.Nat Microbiol. (2023) 8:1064–78. 10.1038/s41564-023-01370-6
162.
SeppESmidtIRööpTŠtšepetovaJKõljalgSMikelsaarMet alComparative analysis of gut microbiota in centenarians and young people: Impact of eating habits and childhood living environment.Front Cell Infect Microbiol. (2022) 12:851404. 10.3389/fcimb.2022.851404
163.
BadalVVaccarielloEMurrayEYuKKnightRJesteDet alThe gut microbiome, aging, and longevity: A systematic review.Nutrients. (2020) 12:3759. 10.3390/nu12123759
164.
WangNLiRLinHFuCWangXZhangYet alEnriched taxa were found among the gut microbiota of centenarians in East China.PLoS One. (2019) 14:e0222763. 10.1371/journal.pone.0222763
165.
WuLZengTDeligiosMMilanesiLLangilleMZinelluAet alAge-related variation of bacterial and fungal communities in different body habitats across the young, elderly, and centenarians in Sardinia.mSphere. (2020) 5:e00558-19. 10.1128/msphere.00558-19
166.
RampelliSSoveriniMD’AmicoFBaroneMTavellaTMontiDet alShotgun meta genomics of gut microbiota in humans with up to extreme longevity and the increasing role of xenobiotic degradation.mSystems. (2020) 5:e00124-20. 10.1128/msystems.00124-20
167.
WuLZengTZinelluARubinoSKelvinDCarruC. A cross-sectional study of compositional and functional profiles of gut microbiota in Sardinian Centenarians.mSystems. (2019) 4:e00325-19. 10.1128/msystems.00325-19
168.
WuLXieXLiYLiangTZhongHYangLet alGut microbiota as an antioxidant system in centenarians associated with high antioxidant activities of gut-resident Lactobacillus.Npj Biofilms Microbiomes. (2022) 8:1–17. 10.1038/s41522-022-00366-0
169.
PalmasVPisanuSMadauVCasulaEDeleddaACusanoRet alGut microbiota markers and dietary habits associated with extreme longevity in healthy Sardinian Centenarians.Nutrients. (2022) 14:2436. 10.3390/nu14122436
170.
LuanZSunGHuangYYangYYangRLiCet alMetagenomics study reveals changes in gut microbiota in centenarians: A cohort study of Hainan centenarians.Front Microbiol. (2020) 11:1474. 10.3389/fmicb.2020.01474
171.
WangFYuTHuangGCaiDLiangXSuHet alGut microbiota community and its assembly associated with age and diet in Chinese centenarians.J Microbiol Biotechnol. (2015) 25:1195–204. 10.4014/jmb.1410.10014
172.
KimBChoiCShinHJinSBaeJHanMet alComparison of the gut microbiota of centenarians in longevity villages of South Korea with those of other age groups.J Microbiol Biotechnol. (2019) 29:429–40. 10.4014/jmb.1811.11023
173.
MiyamotoJIgarashiMWatanabeKKarakiSMukouyamaHKishinoSet alGut microbiota confers host resistance to obesity by metabolizing dietary polyunsaturated fatty acids.Nat Commun. (2019) 10:4007. 10.1038/s41467-019-11978-0
174.
Hornero-RamirezHMorissetteAMarcotteBPenhoatALecomteBPanthuBet alMultifunctional dietary approach reduces intestinal inflammation in relation with changes in gut microbiota composition in subjects at cardiometabolic risk: The SINFONI project.Gut Microbes. (2025) 17:2438823. 10.1080/19490976.2024.2438823
175.
TangQJinGWangGLiuTLiuXWangBet alCurrent sampling methods for gut microbiota: A call for more precise devices.Front Cell Infect Microbiol. (2020) 10:151. 10.3389/fcimb.2020.00151
176.
FlynnKRuffinMITurgeonDSchlossP. Spatial variation of the native colon microbiota in healthy adults.Cancer Prev Res. (2018) 11:393–402. 10.1158/1940-6207.CAPR-17-0370
177.
AlmeidaAMitchellABolandMForsterSGloorGTarkowskaAet alA new genomic blueprint of the human gut microbiota.Nature. (2019) 568:499–504. 10.1038/s41586-019-0965-1
178.
KostisJDobrzynskiJ. Limitations of randomized clinical trials.Am J Cardiol. (2020) 129:109–15. 10.1016/j.amjcard.2020.05.011
179.
DienerCGibbonsSResendis-AntonioO. MICOM: Metagenome-scale modeling to infer metabolic interactions in the gut microbiota.mSystems. (2020) 5:e606–19. 10.1128/mSystems.00606-19
180.
BaldiniFHeinkenAHeirendtLMagnusdottirSFlemingRThieleI. The Microbiome Modeling Toolbox: From microbial interactions to personalized microbial communities.Bioinformatics. (2019) 35:2332–4. 10.1093/bioinformatics/bty941
181.
ShaabanRBusiSWilmesPGuéantJHeinkenA. Personalized modeling of gut microbiome metabolism throughout the first year of life.Commun Med. (2024) 4:1–12. 10.1038/s43856-024-00715-4
182.
BauerEThieleI. From metagenomic data to personalized in silico microbiotas: Predicting dietary supplements for Crohn’s disease.Npj Syst Biol Appl. (2018) 4:1–9. 10.1038/s41540-018-0063-2
183.
BlascoTPérez-BurilloSBalzeraniFHinojosa-NogueiraDLerma-AguileraAPastorizaSet alAn extended reconstruction of human gut microbiota metabolism of dietary compounds.Nat Commun. (2021) 12:4728. 10.1038/s41467-021-25056-x
184.
HaghebaertMLarocheBSalaLMondotSDoréJ. A mechanistic modelling approach of the host–microbiota interactions to investigate beneficial symbiotic resilience in the human gut.J R Soc Interface. (2024) 21:20230756. 10.1098/rsif.2023.0756
185.
SinghHTorralbaMMonceraKDiLelloLPetriniJNelsonKet alGastro-intestinal and oral microbiome signatures associated with healthy aging.GeroScience. (2019) 41:907–21. 10.1007/s11357-019-00098-8
186.
Sayols-BaixerasSDekkersKBaldanziGJönssonDHammarULinYet alStreptococcus species abundance in the gut is linked to subclinical coronary atherosclerosis in 8973 participants from the SCAPIS Cohort.Circulation. (2023) 148:459–72. 10.1161/CIRCULATIONAHA.123.063914
187.
MaJLiZZhangWZhangCZhangYMeiHet alComparison of the gut microbiota in healthy infants with different delivery modes and feeding types: A cohort study.Front Microbiol. (2022) 13:868227. 10.3389/fmicb.2022.868227
188.
ArmougomFHenryMVialettesBRaccahDRaoultD. Monitoring bacterial community of human gut microbiota reveals an increase in Lactobacillus in obese patients and methanogens in anorexic patients.PLoS One. (2009) 4:e7125. 10.1371/journal.pone.0007125
189.
LarsenNVogensenFvan den BergFNielsenDAndreasenAPedersenBet alGut microbiota in human adults with type 2 diabetes differs from non-diabetic adults.PLoS One. (2010) 5:e9085. 10.1371/journal.pone.0009085
190.
MitsouEKirtzalidouEOikonomouILiosisGKyriacouA. Fecal microflora of Greek healthy neonates.Anaerobe. (2008) 14:94–101. 10.1016/j.anaerobe.2007.11.002
191.
KashtanovaDKlimenkoNStrazheskoIStarikovaEGlushchenkoOGudkovDet alCross-sectional study of the gut microbiota composition in moscow long-livers.Microorganisms. (2020) 8:1162. 10.3390/microorganisms8081162
192.
ZengQLiDHeYLiYYangZZhaoXet alDiscrepant gut microbiota markers for the classification of obesity-related metabolic abnormalities.Sci Rep. (2019) 9:13424. 10.1038/s41598-019-49462-w
193.
HasegawaSGotoSTsujiHOkunoTAsaharaTNomotoKet alIntestinal dysbiosis and lowered serum lipopolysaccharide-binding protein in Parkinson’s Disease.PLoS One. (2015) 10:e0142164. 10.1371/journal.pone.0142164
194.
LingZLiZLiuXChengYLuoYTongXet alAltered fecal microbiota composition associated with food allergy in infants.Appl Environ Microbiol. (2014) 80:2546–54. 10.1128/AEM.00003-14
195.
Kowalska-DuplagaKGosiewskiTKapustaPSroka-OleksiakAWędrychowiczAPieczarkowskiSet alDifferences in the intestinal microbiome of healthy children and patients with newly diagnosed Crohn’s disease.Sci Rep. (2019) 9:18880. 10.1038/s41598-019-55290-9
196.
TakahashiKNishidaAFujimotoTFujiiMShioyaMImaedaHet alReduced abundance of butyrate-producing bacteria species in the fecal microbial community in Crohn’s Disease.Digestion. (2016) 93:59–65. 10.1159/000441768
197.
PiccaAPonzianiFCalvaniRMariniFBiancolilloACoelho-JúniorHet alGut microbial, inflammatory and metabolic signatures in older people with physical frailty and Sarcopenia: Results from the BIOSPHERE Study.Nutrients. (2020) 12:65. 10.3390/nu12010065
198.
JoossensMHuysGCnockaertMPreterVVerbekeKRutgeertsPet alDysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives.Gut. (2011) 60:631–7. 10.1136/gut.2010.223263
199.
UedaAShinkaiSShiromaHTaniguchiYTsuchidaSKariyaTet alIdentification of Faecalibacterium prausnitzii strains for gut microbiome-based intervention in Alzheimer’s-type dementia.Cell Rep Med. (2021) 2:100398. 10.1016/j.xcrm.2021.100398
200.
DemirciMTokmanHUysalHDemiryasSKarakullukcuASaribasSet alReduced Akkermansia muciniphila and Faecalibacterium prausnitzii levels in the gut microbiota of children with allergic asthma.Allergol Immunopathol. (2019) 47:365–71. 10.1016/j.aller.2018.12.009
201.
GeravandMFallahPYaghoobiMSoleimanifarFFaridMZinatizadehNet alInvestigation of Enterococcus faecalis population in patients with polyp and colorectal cancer in comparison of healthy individuals.Arq Gastroenterol. (2019) 56:141–5. 10.1590/S0004-2803.201900000-28
202.
LiWWuXHuXWangTLiangSDuanYet alStructural changes of gut microbiota in Parkinson’s disease and its correlation with clinical features.Sci China Life Sci. (2017) 60:1223–33. 10.1007/s11427-016-9001-4
203.
BojovićKSokovićBSVojnović MilutinovićDTomićMGolićNTolinačkiM. Gut microbiota dysbiosis associated with altered production of short chain fatty acids in children with neurodevelopmental disorders.Front Cell Infect Microbiol. (2020) 10:223. 10.3389/fcimb.2020.00223
204.
KnollRForslundKKultimaJMeyerCKullmerUSunagawaSet alGut microbiota differs between children with Inflammatory Bowel Disease and healthy siblings in taxonomic and functional composition: A metagenomic analysis.Am J Physiol-Gastrointest Liver Physiol. (2017) 312:G327–39. 10.1152/ajpgi.00293.2016
205.
ZhouYZhiF. Lower Level of Bacteroides in the gut microbiota is associated with inflammatory bowel disease: A meta-analysis.BioMed Res Int. (2016) 2016:e5828959. 10.1155/2016/5828959
206.
JakobssonHAbrahamssonTJenmalmMHarrisKQuinceCJernbergCet alDecreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by Caesarean section.Gut. (2014) 63:559–66. 10.1136/gutjnl-2012-303249
207.
Kovatcheva-DatcharyPNilssonAAkramiRLeeYDe VadderFAroraTet alDietary fiber-induced improvement in glucose metabolism is associated with increased abundance of prevotella.Cell Metab. (2015) 22:971–82. 10.1016/j.cmet.2015.10.001
208.
LiJZhaoFWangYChenJTaoJTianGet alGut microbiota dysbiosis contributes to the development of hypertension.Microbiome. (2017) 5:14. 10.1186/s40168-016-0222-x
209.
WenNSunLGengQZhengG. Gut microbiota changes associated with frailty in older adults: A systematic review of observational studies.World J Clin Cases. (2024) 12:6815–25. 10.12998/wjcc.v12.i35.6815
210.
FrémontMCoomansDMassartSDe MeirleirK. High-throughput 16S rRNA gene sequencing reveals alterations of intestinal microbiota in myalgic encephalomyelitis/chronic fatigue syndrome patients.Anaerobe. (2013) 22:50–6. 10.1016/j.anaerobe.2013.06.002
211.
ZuoKLiJLiKHuCGaoYChenMet alDisordered gut microbiota and alterations in metabolic patterns are associated with atrial fibrillation.GigaScience. (2019) 8:giz058. 10.1093/gigascience/giz058
212.
XuYWangYLiHDaiYChenDWangMet alAltered fecal microbiota composition in older adults with frailty.Front Cell Infect Microbiol. (2021) 11:696186. 10.3389/fcimb.2021.696186
213.
AhrensAHyötyläinenTPetroneJIgelströmKGeorgeCGarrettTet alInfant microbes and metabolites point to childhood neurodevelopmental disorders.Cell. (2024) 187:1853–73.e15. 10.1016/j.cell.2024.02.035
214.
TeixeiraFSGrześkowiakŁSalminenSLaitinenKBressanJGouveia PeluzioM. Faecal levels of Bifidobacterium and Clostridium coccoides but not plasma lipopolysaccharide are inversely related to insulin and HOMA index in women.Clin Nutr. (2013) 32:1017–22. 10.1016/j.clnu.2013.02.008
215.
ZhuLBakerSGillCLiuWAlkhouriRBakerRet alCharacterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH.Hepatology. (2013) 57:601–9. 10.1002/hep.26093
216.
VictoriaMElenaVAmparoGMaríaJAdrianaGIreneAet alGut microbiota alterations in critically ill older patients: A multicenter study.BMC Geriatr. (2022) 22:373. 10.1186/s12877-022-02981-0
217.
WangLAlammarNSinghRNanavatiJSongYChaudharyRet alGut microbial dysbiosis in the irritable bowel syndrome: A systematic review and meta-analysis of case-control studies.J Acad Nutr Diet. (2020) 120:565–86. 10.1016/j.jand.2019.05.015
218.
BellocchiCFernández-OchoaÁMontanelliGVigoneBSantanielloAMilaniCet alMicrobial and metabolic multi-omic correlations in systemic sclerosis patients.Ann N Y Acad Sci. (2018) 1421:97–109. 10.1111/nyas.13736
219.
MurrosKHuynhVTakalaTSarisP. Desulfovibrio bacteria are associated with Parkinson’s disease.Front Cell Infect Microbiol. (2021) 11:652617. 10.3389/fcimb.2021.652617
220.
DinhDRamadassBKattulaDSarkarRBraunsteinPTaiAet alLongitudinal analysis of the intestinal microbiota in persistently stunted young children in South India.PLoS One. (2016) 11:e0155405. 10.1371/journal.pone.0155405
221.
KarlssonCÖnnerfältJXuJMolinGAhrnéSThorngren-JerneckK. The microbiota of the gut in preschool children with normal and excessive body weight.Obesity. (2012) 20:2257–61. 10.1038/oby.2012.110
222.
PngCLindénSGilshenanKZoetendalEMcSweeneyCSlyLet alMucolytic bacteria with increased prevalence in IBD mucosa augmentin vitroutilization of mucin by other bacteria.Off J Am Coll Gastroenterol ACG. (2010) 105:2420–8. 10.1038/ajg.2010.281
223.
SantacruzAColladoMGarcía-ValdésLSeguraMMartín-LagosJAnjosTet alGut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women.Br J Nutr. (2010) 104:83–92. 10.1017/S0007114510000176
224.
CamaraAKonateSTidjani AlouMKodioATogoACortaredonaSet alClinical evidence of the role of Methanobrevibacter smithii in severe acute malnutrition.Sci Rep. (2021) 11:5426. 10.1038/s41598-021-84641-8
225.
ShoubridgeACarpenterLFlynnEPapanicolasLCollinsJGordonDet alSevere cognitive impairment is linked to a reduced gut microbiome capacity to synthesise immunomodulators, neurotransmitters, and amino acids required for autophagy in residents of long-term aged care.mdRxiv [Preprint]. (2023). 10.1101/2023.03.06.23286878
226.
SokolHLeducqVAschardHPhamHJegouSLandmanCet alFungal microbiota dysbiosis in IBD.Gut. (2017) 66:1039–48. 10.1136/gutjnl-2015-310746
227.
StratiFCavalieriDAlbaneseDDe FeliceCDonatiCHayekJet alNew evidences on the altered gut microbiota in autism spectrum disorders.Microbiome. (2017) 5:24. 10.1186/s40168-017-0242-1
228.
LingZZhuMLiuXShaoLChengYYanXet alFecal fungal dysbiosis in Chinese patients with Alzheimer’s Disease.Front Cell Dev Biol. (2021) 8:631460. 10.3389/fcell.2020.631460
229.
GaoRXiaKWuMZhongHSunJZhuYet alAlterations of gut mycobiota profiles in adenoma and colorectal cancer.Front Cell Infect Microbiol. (2022) 12:839435. 10.3389/fcimb.2022.839435
230.
ZouRWangYDuanMGuoMZhangQZhengH. Dysbiosis of gut fungal microbiota in children with autism spectrum disorders.J Autism Dev Disord. (2021) 51:267–75. 10.1007/s10803-020-04543-y
231.
BridgmanSAzadMFieldCHaqqABeckerAMandhanePet alFecal short-chain fatty acid variations by breastfeeding status in infants at 4 months: Differences in relative versus absolute concentrations.Front Nutr. (2017) 4:11. 10.3389/fnut.2017.00011
232.
KimKYaoYJuS. Short chain fatty acids and fecal microbiota abundance in humans with obesity: A systematic review and meta-analysis.Nutrients. (2019) 11:2512. 10.3390/nu11102512
233.
WuLHanYZhengZPengGLiuPYueSet alAltered gut microbial metabolites in amnestic mild cognitive impairment and Alzheimer’s Disease: Signals in host–microbe interplay.Nutrients. (2021) 13:228. 10.3390/nu13010228
234.
Da SilvaHTeterinaAComelliETaibiAArendtBFischerSet alNonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance.Sci Rep. (2018) 8:1466. 10.1038/s41598-018-19753-9
235.
SzczesniakOHestadKHanssenJRudiK. Isovaleric acid in stool correlates with human depression.Nutr Neurosci. (2016) 19:279–83. 10.1179/1476830515Y.0000000007
236.
GallGGuttulaKKellingrayLTettAten HoopenRKemsleyEet alMetabolite quantification of faecal extracts from colorectal cancer patients and healthy controls.Oncotarget. (2018) 9:33278–89. 10.18632/oncotarget.26022
237.
KokCBrabecBChichlowskiMHarrisCMooreNWamplerJet alStool microbiome, pH and short/branched chain fatty acids in infants receiving extensively hydrolyzed formula, amino acid formula, or human milk through two months of age.BMC Microbiol. (2020) 20:337. 10.1186/s12866-020-01991-5
238.
TriconSBurdgeGKewSBanerjeeTRussellJGrimbleRet alEffects of cis-9,trans-11 and trans-10,cis-12 conjugated linoleic acid on immune cell function in healthy humans.Am J Clin Nutr. (2004) 80:1626–33. 10.1093/ajcn/80.6.1626
239.
RacineNWatrasACarrelAAllenDMcVeanJClarkRet alEffect of conjugated linoleic acid on body fat accretion in overweight or obese children123.Am J Clin Nutr. (2010) 91:1157–64. 10.3945/ajcn.2009.28404
240.
MoloneyFYeowTMullenANolanJRocheH. Conjugated linoleic acid supplementation, insulin sensitivity, and lipoprotein metabolism in patients with type 2 diabetes mellitus123.Am J Clin Nutr. (2004) 80:887–95. 10.1093/ajcn/80.4.887
241.
GrahamSArvelaOWiseG. Long-term neurologic consequences of nutritional vitamin B12 deficiency in infants.J Pediatr. (1992) 121:710–4. 10.1016/S0022-3476(05)81897-9
242.
KivipeltoMAnnerboSHultdinJBäckmanLViitanenMFratiglioniLet alHomocysteine and holo-transcobalamin and the risk of dementia and Alzheimers disease: A prospective study.Eur J Neurol. (2009) 16:808–13. 10.1111/j.1468-1331.2009.02590.x
243.
JärvinenEIsmailKMuniandyMBoglLHeinonenSTummersMet alBiotin-dependent functions in adiposity: A study of monozygotic twin pairs.Int J Obes. (2016) 40:788–95. 10.1038/ijo.2015.237
244.
SheaMBoothSMassaroJJacquesPD’AgostinoRSr.Dawson-HughesBet alVitamin K and Vitamin D Status: Associations with inflammatory markers in the framingham offspring study.Am J Epidemiol. (2008) 167:313–20. 10.1093/aje/kwm306
245.
SheaKCushmanMBoothSBurkeGChenHKritchevskyS. Associations between vitamin K status and haemostatic and inflammatory biomarkers in community-dwelling adults.Thromb Haemost. (2014) 112:438–44. 10.1160/TH13-12-1003
246.
YilmazCYucaSYilmazNBektaşMÇaksenH. Intracranial hemorrhage due to vitamin K deficiency in infants: A clinical study.Int J Neurosci. (2009) 119:2250–6. 10.3109/00207450903170437
247.
PhamVLacroixCBraeggerCChassardC. Lactate-utilizing community is associated with gut microbiota dysbiosis in colicky infants.Sci Rep. (2017) 7:11176. 10.1038/s41598-017-11509-1
248.
ChassardCDapoignyMScottKCrouzetLDel’hommeCMarquetPet alFunctional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome.Aliment Pharmacol Ther. (2012) 35:828–38. 10.1111/j.1365-2036.2012.05007.x
249.
AnRWilmsELogtenbergMvan TrijpMScholsHMascleeAet alIn vitro metabolic capacity of carbohydrate degradation by intestinal microbiota of adults and pre-frail elderly.ISME Commun. (2021) 1:1–12. 10.1038/s43705-021-00065-5
250.
JiangTSuarezFLevittMNelsonSZieglerE. Gas production by feces of infants.J Pediatr Gastroenterol Nutr. (2001) 32:534–41. 10.1097/00005176-200105000-00009
251.
YangXXiuWWangJLiLHeCGaoC. CO2 Is beneficial to gut microbiota homeostasis during colonoscopy: Randomized controlled trial.J Clin Med. (2022) 11:5281. 10.3390/jcm11185281
252.
YamanoHYoshikawaKKimuraTYamamotoEHaradaEKudouTet alCarbon dioxide insufflation for colonoscopy: Evaluation of gas volume, abdominal pain, examination time and transcutaneous partial CO2 pressure.J Gastroenterol. (2010) 45:1235–40. 10.1007/s00535-010-0286-5
253.
JangiSGandhiRCoxLLiNvon GlehnFYanRet alAlterations of the human gut microbiome in multiple sclerosis.Nat Commun. (2016) 7:12015. 10.1038/ncomms12015
254.
SoaresATahanSFagundes-NetoUde MoraisM. [Breath methane in children with chronic constipation].Arq Gastroenterol. (2002) 39:66–72. 10.1590/s0004-28032002000100012
255.
PiragineEMalanimaMLucenteforteEMartelliACalderoneV. Circulating Levels of Hydrogen Sulfide (H2S) in patients with age-related diseases: A systematic review and meta-analysis.Biomolecules. (2023) 13:1023. 10.3390/biom13071023
256.
LuQChenJJiangLGengTTianSLiaoYet alGut microbiota-derived secondary bile acids, bile acids receptor polymorphisms, and risk of cardiovascular disease in individuals with newly diagnosed type 2 diabetes: A cohort study.Am J Clin Nutr. (2024) 119:324–32. 10.1016/j.ajcnut.2023.08.023
257.
LiYZhangDHeYChenCSongCZhaoYet alInvestigation of novel metabolites potentially involved in the pathogenesis of coronary heart disease using a UHPLC-QTOF/MS-based metabolomics approach.Sci Rep. (2017) 7:15357. 10.1038/s41598-017-15737-3
258.
HuangWHsuHLiuJYangTChenJChangJet alThe association of ursodeoxycholic acid use with colorectal cancer risk: A nationwide cohort study.Medicine. (2016) 95:e2980. 10.1097/MD.0000000000002980
259.
HillDBuckR. Infants fed breastmilk or 2′-FL supplemented formula have similar systemic levels of microbiota-derived secondary bile acids.Nutrients. (2023) 15:2339. 10.3390/nu15102339
260.
KeanIWagnerJWijeyesekeraADe GoffauMThurstonSClarkJet alProfiling gut microbiota and bile acid metabolism in critically ill children.Sci Rep. (2022) 12:10432. 10.1038/s41598-022-13640-0
261.
PanJXiaJDengFLiangWWuJYinBet alDiagnosis of major depressive disorder based on changes in multiple plasma neurotransmitters: A targeted metabolomics study.Transl Psychiatry. (2018) 8:1–10. 10.1038/s41398-018-0183-x
262.
MancaRDe MarcoMSoininenHRuffiniLVenneriA. Changes in neurotransmitter-related functional connectivity along the Alzheimer’s disease continuum.Brain Commun. (2025) 7:fcaf008. 10.1093/braincomms/fcaf008
263.
AbdulamirHAbdul-RasheedOAbdulghaniE. Serotonin and serotonin transporter levels in autistic children.Saudi Med J. (2018) 39:487–94. 10.15537/smj.2018.5.21751
264.
SongQHwangCLiYWangJParkJLeeSet alGut-derived ammonia contributes to alcohol-related fatty liver development via facilitating ethanol metabolism and provoking ATF4-dependent de novo lipogenesis activation.Metabolism. (2024) 151:155740. 10.1016/j.metabol.2023.155740
265.
FismanMBallMBlumeW. Hyperammonemia and Alzheimer’s Disease.J Am Geriatr Soc. (1989) 37:1102–1102. 10.1111/j.1532-5415.1989.tb06935.x
266.
OzanneBNelsonJCousineauJLambertMPhanVMitchellGet alThreshold for toxicity from hyperammonemia in critically ill children.J Hepatol. (2012) 56:123–8. 10.1016/j.jhep.2011.03.021
267.
IkematsuNKashiwagiMHaraKWatersBMatsusueATakayamaMet alOrgan distribution of endogenous p-cresol in hemodialysis patients.J Med Invest. (2019) 66:81–5. 10.2152/jmi.66.81
268.
GabrieleSSaccoRCerulloSNeriCUrbaniATripiGet alUrinary p-cresol is elevated in young French children with autism spectrum disorder: A replication study.Biomarkers. (2014) 19:463–70. 10.3109/1354750X.2014.936911
269.
BanerjeeTMeyerTShafiTHostetterTMelamedMZhuYet alFree and total p-cresol sulfate levels and infectious hospitalizations in hemodialysis patients in CHOICE and HEMO.Medicine. (2017) 96:e5799. 10.1097/MD.0000000000005799
270.
AlexeevELanisJKaoDCampbellEKellyCBattistaKet alMicrobiota-derived indole metabolites promote human and murine intestinal homeostasis through regulation of interleukin-10 receptor.Am J Pathol. (2018) 188:1183–94. 10.1016/j.ajpath.2018.01.011
271.
EhrlichAPachecoAHenrickBTaftDXuGHudaMet alIndole-3-lactic acid associated with Bifidobacterium-dominated microbiota significantly decreases inflammation in intestinal epithelial cells.BMC Microbiol. (2020) 20:357. 10.1186/s12866-020-02023-y
272.
HietbrinkFBesselinkMRenooijWde SmetMDraismaAvan der HoevenHet alSystemic inflammation increases intestinal permeability during experimental human endotoxemia.Shock. (2009) 32:374–8. 10.1097/SHK.0b013e3181a2bcd6
273.
VatanenTKosticAd’HennezelESiljanderHFranzosaEYassourMet alVariation in microbiome LPS immunogenicity contributes to autoimmunity in humans.Cell. (2016) 165:842–53. 10.1016/j.cell.2016.04.007
274.
TrefflichIJabakhanjiAMenzelJBlautMMichalsenALampenAet alIs a vegan or a vegetarian diet associated with the microbiota composition in the gut? Results of a new cross-sectional study and systematic review.Crit Rev Food Sci Nutr. (2020) 60:2990–3004. 10.1080/10408398.2019.1676697
275.
DinuMAbbateRGensiniGCasiniASofiF. Vegetarian, vegan diets and multiple health outcomes: A systematic review with meta-analysis of observational studies.Crit Rev Food Sci Nutr. (2017) 57:3640–9. 10.1080/10408398.2016.1138447
276.
Rubio-TapiaARahimMSeeJLahrBWuTMurrayJ. Mucosal recovery and mortality in adults with celiac disease after treatment with a gluten-free diet.Off J Am Coll Gastroenterol ACG. (2010) 105:1412. 10.1038/ajg.2010.10
277.
GhoshTRampelliSJefferyISantoroANetoMCapriMet alMediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: The NU-AGE 1-year dietary intervention across five European countries.Gut. (2020) 69:1218–28. 10.1136/gutjnl-2019-319654
278.
PeronGMeroñoTGargariGHidalgo-LiberonaNMiñarroALozanoEet alA polyphenol-rich diet increases the gut microbiota metabolite indole 3-propionic acid in older adults with preserved kidney function.Mol Nutr Food Res. (2022) 66:2100349. 10.1002/mnfr.202100349
Summary
Keywords
gut microbiota, diet, infant, adult, older adult, centenarian
Citation
Geniselli da Silva V, Roy NC, Smith NW, Wall C, Mullaney JA and McNabb WC (2025) Dietary patterns influencing the human colonic microbiota from infancy to centenarian age: a narrative review. Front. Nutr. 12:1591341. doi: 10.3389/fnut.2025.1591341
Received
11 March 2025
Accepted
12 May 2025
Published
04 June 2025
Volume
12 - 2025
Edited by
Alfonso Benítez-Páez, Spanish National Research Council (CSIC), Spain
Reviewed by
Chiara Devirgiliis, CREA (Consiglio per la ricerca in agricoltura e l’analisi dell’economia agraria), Italy
Kotryna Simonyté Sjödin, Umeå University, Sweden
Updates
Copyright
© 2025 Geniselli da Silva, Roy, Smith, Wall, Mullaney and McNabb.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vitor Geniselli da Silva, vgenisel@massey.ac.nz
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.