- Department of Physics, Randall Centre for Cell and Molecular Biophysics, King's College London, London, United Kingdom
Bridging the gap between traditional immunology and nanoscale biophysics has proved more difficult than originally thought. For cell biology applications however, super-resolution microscopy has already facilitated considerable advances. From neuronal segmentation to nuclear pores and 3D focal adhesion structure—nanoscopy has begun to illuminate links between nanoscale organization and function. With immunology, the explanation must go further, relating nanoscale biophysical phenomena to the manifestation of specific diseases, or the altered activity of specific immune cell types in a bodily compartment. What follows is a summary of how nanoscopy has elucidated single cell immunological function, and what might be achieved in the future to link quantifiable, nanoscale, biophysical phenomena with cell and whole tissue functionality. We explore where the gaps in our understanding occur, and how they might be addressed.
The so called immunological synapse has proved fertile ground for the intersection of nanoscopy with immunology [1]. The word synapse originates from Greek sun–together and hapsis–joining. Like a neuronal synapse, the immune synapse (IS) describes a communication zone at the interface between two cells. In the case of the T cell synapse, a T lymphocyte and an antigen presenting cell (APC) communicate through an intricately arranged array of receptors. Archetypal immune synapses can be recapitulated through use of supported lipid bilayers (SLBs) loaded with peptide bound major histocompatibility complex (p-MHC) [2], or more simply through the use of activating antibodies bound to glass, directed at the T cell receptor [3, 4]. In the past few years, several proteins in the T cell membrane have been found to be organized on nano-length scales, and links to cell function have begun to emerge.
In the fields of super-resolution and biophysics, the IS has had a dual function–as an area for cell biological insight in itself, and as a proving ground for super-resolution technique development. Cell biology has benefitted from the system as headway has been made in understanding the role of nanoscale molecular organization of the TCR [5–7], LFA-1 [8, 9], LAT [10, 11] and the nanoscale meshworks formed from fibers like actin [12]. Elsewhere, super-resolution imaging has been used to study nanoscale structures such as the nuclear pore complex, mechanisms of segmentation within organelles such as the mitochondria, or the axon and dendrites of neurons [13, 14]. In these cases, the use of diverse nanoscopic methodologies helped independent researchers report and verify the same structures.
The organization of the TCR has been subject to rather intense investigation over the last few years, in part because of controversy over its nanoscale organization, and in part due to artifacts emanating from the imaging. It therefore provides a useful case study of many researchers being involved in the elucidation of a phenomenon using new technology.
Bjorn Lillemeier's group were among the first to show that T cell receptor forms nanoclusters in activated cells, which group together into islands to amplify a signal, allowing T cells to surpass a threshold for activation [6]. In a noisy, live environment, resolution was reduced, but new information afforded by this technique showed that the TCR and LAT indeed organize into nanoscale islands upon activation by a single pMHC-TCR interaction in vivo.This, the authors posit, is a nanoscale spatial mechanism for signal amplification—one that negates the need for 10 or more antigen peptides to be presented for full T cell activation [15].
However, the existence of TCR clusters in antigen experienced “resting” cells is less clear, and new analysis suggests that pre-clustered TCR may be an artifactual symptom of molecular overcounting [8, 16]. Alternatively, such clusters may represent ligand-independent triggering of the TCR that happens when T cells come into contact with various surfaces commonly used for in vitro assays, including poly-l-lysine [17]. Molecular overcounting occurs when multi-blinking fluorescent emitters are detected more than once, in slightly different places, resulting in a single molecule looking like a cluster in the output data. Varying the label density is a possible way to rule out false clusters in super resolution localization data [8]. When this technique was used, no TCR clusters were found in resting T cells. Biologically, new data suggests that the T cell receptor would seem to benefit from being a single entity in that this would speed the scanning phase in resting T cells: if TCRs are non-clustering, they have a higher probability of coming into contact with pMHC [18]. Recent work by Cai et al. supports this. Here, high resolution lattice lightsheet microscopy, along with quantification using the displacement of quantum dots showed how microvilli are used by T cells to speed the scanning process [19]. Jung et al. used variable angle TIRF and localization of fluorescently labeled TCR and Zap70 (Syk family kinase involved in TCR activation) to observe nano-clustering at the tips of the microvilli [20]. The precise characteristics of TCR and Zap70 clusters, and the function of the clustering observed at the dwell sites of microvilli requires further investigation. The Zap70 catch and release model [21] appears to match well with microvillar scanning, which might provide the amplification needed for further signaling and full activation in the presence of a strong, albeit rare signal. Together, these works provide evidence that the nanoscale spatial localization of multiple signaling molecules like this are likely to be involved in the regulation and amplification of an initial TCR-pMHC signal.
Nanoscale T Cell Migration
Intrinsically coupled to their function of antigen recognition, T cells are necessarily highly migratory. The precisely orchestrated timing of targeted T lymphocyte migration is also called “homing,” and describes a voyage through peripheral tissues, lymphoid vessels and organs, and along vasculature, during which a process of differentiation into many different T cell subtypes occurs—themselves with intrinsic migratory capacities [22]. In the lymph node, naïve T cells undertake the scanning of dendritic cells—interactions which can total 500 to 5,000 cell contacts per hour per T cell [23]. Integrin based adhesions have long been known to be important for these events, but only recently have we been able to observe their machinations on the nanoscale. Molecular clustering is necessary for the structured formation of adhesions in T cells [24], and new nanoscopy techniques have allowed us to show that there are levels of organization beyond microscale adhesion clusters. The dynamics of such clusters likely occur across many timescales, and while live cell PALM in the T cell synapse has provided important insights into nanocluster dynamics with temporal resolution of 1–2 s [10, 11, 25–27], fast dynamics will rely on the development of much faster localization imaging.
Nanoscale T cell adhesions are short lived, small and distinct from conventional focal adhesions observed in other cell types. Recent super-resolution microscopy work has shown that T cell adhesions based around LFA-1 integrin are very small throughout the cell—on the length scale of nascent adhesions at around 100 nm in diameter [9]. Those adhesions that are anchored to ligand on the outside of the cell, and to actin on the inside remain engaged from the leading edge through the lamella, but have a total lifespan of < 1 min. As the field progresses, further nanoscopic investigation may help us to answer questions about the precise composition and compartmentalization of nanoscale T cell adhesions. One possibility is that actin and the membrane work together to compartmentalize nano-adhesions, a concept pioneered by Kusumi [28, 29] and suggested for T cells by Lillemeier [6].
Recent work has linked the modulation of such nanoscale adhesions with altered migration due to the loss of function mutation of a phosphatase associated with autoimmune disease: PTPN22 [9]. Upon loss of this phosphatase, effector CD8+ and CD4+ cells migrate considerably faster and alter their nano-adhesion structure as well as their link to the actin cytoskeleton. In humans suffering from rheumatoid arthritis, PTPN22 mutation or deficiency is linked to an accumulation of such effector T cells in the joints [30]. Therefore, it is possible that phosphatases like PTPN22 act as nanoscale control switches for integrin based adhesions, and that the lack of this control causes a default fast migration phenotype, contributing to the mis-localization of cells and development of autoimmune disease conditions. The effects on the base migration phenotype and accompanying nano-adhesion architecture caused by this mutation warrant more investigation. This might be done in other fast moving cell types affected by phosphatase mutation [31], as well as in specific T cell subtypes such as T effector memory cells that migrate in the periphery or central memory T cells that migrate mainly in the secondary lymph nodes [32].
Migration Plasticity and Nano-Adhesion Memory
The base structure of nanoscale adhesions may be immunologically important in the case of memory T cells which commit themselves to special patterns of migration. Individual T cells must dynamically alter their style of migration as they move through different environments in the periphery and in the lymph nodes [33]. When inside the lymph nodes, cells use actin to push and squeeze through their surroundings [34], and LFA-1 integrin to enhance DC scanning and migration speed [35]. Along blood vessels, migration and diapedesis relies on actin “treadmilling” and integrin based adhesions to anchor themselves and translate actin flow into cell movement [35]. In the diverse environment of the periphery, it is likely that these modes of migration are mixed and complimentary and are probably true for other exploratory immune cells. Interestingly, it is also clear that subtypes of T cells develop and maintain set migratory patterns as they differentiate. Some predominantly move around the peripheral tissues (e.g., stem memory T cells), or remain resident there (T resident memory), while others reside largely in the secondary lymph nodes, such as the central memory T cells [32, 36].
Insight into the nanoscale behavior of adhesion receptors and effectors may provide immunological understanding as to why these cells maintain such behaviors. One question might be whether cells coded for certain kinds of migration also maintain “cluster memory,” where adhesion clusters adopt specific clustering formations in the membrane based on preformed intracellular clusters. Such preformed clusters of integrins and associated intermediates/effectors have been observed in vesicles in other cells [37]. If such vesicles are preformed and stored in pools, they could be used to quickly enact specific kinds of prepackaged migration, by using spatial platforms for fast signaling in a migratory niche. The precise arrangement of molecules within vesicles, and their mechanism of formation is unknown, but may soon be accessible with the combination of new optical and analytical tools. This hypothesis of nanoscale spatial memory might extend to vesicular adhesion receptors and effectors—importantly, integrins can be pre-activated within such vesicles [37].
The optical tools to analyze such phenomena exist. Lattice light sheet microscopy (LLSM) already allows for high spatiotemporal resolution at low laser power. Whole 3D volumes can be acquired very quickly, enabling imaging of single cell dynamics with high temporal resolution. Low phototoxicity and fast 3D imaging with high signal to noise has allowed the confirmation and measurement of dynamics of the well-studied immunological synapse in a 3D matrix between a T cell and a dendritic cell [38], as well as microvilli [19]. Recent advances in adaptive optics for microscopes has increased the reach of the lattice light-sheet technique, achieving sub 100 nm lateral and axial resolution even in highly scattering tissues [39]. The technique has also been used to get single molecule localization information, and has provided impressive PAINT images in fixed cells, where the narrow and homogenous Bessel beam lightsheet allowed for superior optical sectioning [38].
While LLSM has the advantage of high speed and low dose, allowing access to fast processes that occur in immune cells, it has not yet been used for quantification of single molecules in live cells. Extending this technique to identify single molecule involved in nanoclusters in live cells might be achievable through its combination with new live cell cluster quantification techniques [40]. Instead of focusing on optics or fluorescent protein development, the algorithm relies on statistics to increase the temporal resolution, by calculating the fewest points required to reliably state the relative size and density of an under-sampled nanocluster. The algorithm works with low molecule numbers, such that enough information is collected in only a few raw frames to extract statistically relevant nanoscale information about cluster dynamics. Through advances in such quantitation techniques, live cell microscopy techniques and reversibly switchable or replenishable fluorophore engineering [41–43], it is likely that the dynamics of individual protein clusters of signaling molecules will be investigable in live T cells. Point pattern analysis at physiologically relevant spatio-temporal resolution will provide highly quantifiable data, potentially allowing sensitive processes to be statistically analyzed.
Increasing the Super-Resolution Sample Size
One concern within circles of immunologists is the relevance of tiny changes in the properties of nanoscale signaling domains to immune system function. That changes within nanoscale signaling domains are very small might seem like truism, but there is a valid point here. While researchers might use T cells sourced from several different people and observe high replicability across many samples, the nanoscale change may be tiny: cluster size changes of a few nanometers for example. Several functional changes have been linked to nanoscale alterations: the increased clustering of LAT derived from sub-cellular vesicles that correlates with T cell receptor activation and synapse formation [10], the co-clustering of Zap70 and TCR with LAT [25]as well as Lck [7, 8, 27] upon T cell activation. Recently, links to mechanism have been posited during the activation of B cells, where actin and tetraspanin reorganizes B cell receptors [44], in macrophages, where Src family kinases cause actin to rearrange FcγR1 nanoclusters [45], and in the regulation and formation of the natural killer cell lytic synapse [46].
Such studies provide important insight into phenomena in single cells. However, the low n-numbers lend themselves to study of the average changes within a heterogeneous dataset composed of diverse clusters, and do not allow access to population data when looking at multiple cell types in the same experiment as is common in immunology labs. This denies researchers access to possible minority phenomena–nanoscale machinations that start small, residing at the tail ends of data distributions, and result in whole cell change, as well as the effect of the change on the rest of the immune cell population in a given niche.
High Throughput Trans-Scale Microscopy
While the average super-resolution experiment relies on data from a few tens of cells, the jewel in the crown of immunology, flow cytometry, is able to analyze tens of thousands of cells in a matter of minutes. This allows for imaging and analysis of a diverse population, often giving a snapshot of immune cell populations in a given bodily niche. If super-resolution microscopy is to be used for population based imaging, the number of cells imaged, and their diversity must be increased. To achieve this, researchers have been making efforts toward increasing the throughput of an average super-resolution experiment by automating the process [47, 48], and by using flat field illumination to increase the number of cells collectable in each image [49, 50].
Flat field super-resolution microscopy attempts to increase the illumination area to match the capabilities of scientific complementary metal oxide semiconductor (sCMOS) cameras, which offer a huge field of view [49]. sCMOS cameras used for localization microscopy also have lower readout noise and rapid readout rates, but are not as sensitive as EMCCD cameras. The latter have been the equipment of choice for most single molecule localization microscopy applications. However, while EM amplifies the signal it also amplifies the camera noise. sCMOS cameras do not use electron multiplication but have extremely low camera noise. Together, this means that contrast is higher and therefore single molecules can be localized more precisely by the latest sCMOS cameras than by EMCCD [38]. Combined with the faster readout rate and large field of view, as well as potential for better quantum yields in the future, sCMOS will likely supersede EMCCD cameras for localization microscopy.
Most super-resolution microscopes illuminate the sample such that there is a relatively narrow Gaussian distribution of intensity from the center of the field of view, which is made narrower in some cases by the use of a condenser lens to increase excitation intensity. Fluorophores near the edge of this illumination zone are less strongly excited than those in the center, and as a result are less precisely localized. To address this, Suliana Manley's group set up a flat field microscope, which relies on an inexpensive microlens array to image 100 μm2 regions with super-resolution, more than quadrupling the current limits [49]. This could well be applied to immunology where samples of a mixed population of T cells must be imaged all at once, identified separately and characterized on the nanoscale by super-resolution.
Automated super-resolution imaging and single particle tracking has been performed by Masato Yasui of the Ueda lab in Osaka [47]. His system uses fast focusing based on pixel intensity at the iris to select the correct focal plane, then machine learning to find and image cells with a given morphology and fluorescent signal. Combined with automated addition of chemicals or desired treatments using robotics, the system has the capacity to image several 100 cells per day. Single particle tracking or super-resolution localization microscopy can then be carried out in multiple conditions, resulting in hundreds of cells per condition, and millions of analyzable molecules. In conjunction are servers that can handle the localization of huge numbers of molecules, and localization and analysis algorithms that make most efficient use of the CPU or GPU of the systems involved. As an automated process, new ways to validate the quality of super-resolution data [51, 52], could be built into the process of screening before image processing.
Minority Biology
Increasing the n number from an average of 10 cells to 1,000 cells may also allow us access to new biological phenomena, undetectable when comparing the means of two conditions. Heterogeneity of clustering in cells is clear—usually the distributions display a wide range of cluster shapes, densities and colocalizations with other clustered/non-clustered molecules. Data comprised of many cells and many thousands more single molecule detections has the potential to be mined in various ways to reveal minority or rare nanoscale phenomena.
Minority phenomena represent putative events that have a lot of power for change, but occur at the fringes of distributions. One might imagine a molecular cluster exhibiting divergent behavior. Such a cluster might be made of precisely organized layers of kinases, phosphatases and mechanical intermediates [such as a focal adhesion [53]] which at a given quorum of molecular participants [molecular conformations and phosphorylation states considered] initiates a signal cascade affecting neighboring clusters. This seed cluster could build up a level of signaling that then reorganizes actin [through the local action of Rho-GTPases for example [54]] or the strength of an adhesion [through modulation of vinculin binding proteins [55]] to change the direction of cell migration or to initiate the formation of a full synapse after initial microvillus based scanning.
Such a cluster would certainly be overlooked by conventional comparison of mean values between two separate distributions representing the characteristics of thousands of clusters. The kind of N-numbers available by increasing the throughput of super-resolution microscopy may allow for statistical analysis of segregated minority events at the tail ends of the population distribution on the nanoscale but also at single cell or cell population level. The transition from “scanning” to “swarming” mode of neutrophils in the lymph node is a good example of a single cell instigating a change in the behavior of the population [56]. High throughput, trans-scale type experiments may aid us to investigate such phenomena, allowing elucidation of the link between initial nanoscale behavior and cell to population wide functional outcomes.
Functional Super-Resolution Imaging
Nanoscale colocalization of transmembrane proteins with intracellular signaling intermediates with well-characterized functions can provide some level of insight into functional output. Such “proxy” measures are likely to lead to important discoveries, and with sub 10 nm resolution can complement FRET techniques. However, clear links between nanoscale molecular arrangements and their mechanisms will require the combination of super-resolution microscopy with direct functional readouts, which together might provide clearer answers about the importance of nanoscale events and how they relate to cell function, cell population function and immune system function (Figure 1).
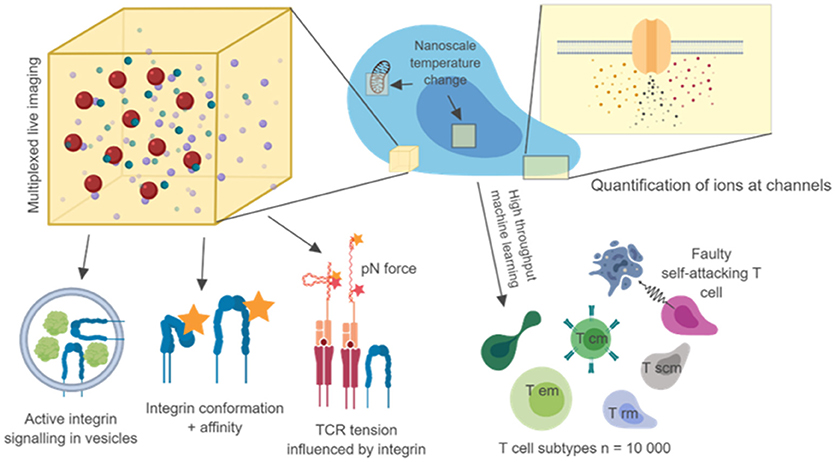
Figure 1. Nanoscale imaging with multiple metrics and high throughput microscopy. Nanoscale readouts, combined with high throughput analysis may link single cell, multi cell and in vivo behavior to separate T cell subtypes and disease phenotypes. Combining 3D nanoscale live-cell imaging of many targets with other nanoscale measures such as tension, temperature, conformation will help us to make causal links to single cell behavior. The use of machine learning and automation to image 1,000 s of cells and billions of molecules will help us to create true trans-scale experiments. Diagram created using biorender.io.
One example of linking function to super resolved coordinate data is Travis Moore's work linking LFA-1 integrin conformation with its affinity for ligand [57]. By labeling the head group of the integrin and the inner leaflet of the plasma membrane, Moore and the AIC at Janelia research institute used interferometric PALM to directly detect conformational change in the protein by measuring the distance between the two [57]. The structure of LFA-1 integrin relates to its affinity, therefore it is important that the researchers confirmed that the integrin structures predicted from x ray crystallography matched those in cells.
DNA tension sensors represent another way single molecule data could be enriched, and have successfully been used to investigate the force applied by TCR when it binds to pMHC [58]. Here, the authors describe a positive relationship between the pulling force of TCR upon peptide-MHC and full T cell activation. LFA-1 binding to its ligand ICAM-1 augmented TCR/pMHC tension, implying crosstalk between the two pathways. Such an interaction represents a catch bond—one that becomes stronger as the force across it is increased. Other molecules that undergo catch bond behavior have been investigated by optical traps, such as vinculin [59]: an intracellular actin/integrin linker that reveals cryptic binding sites as it is stretched. Many techniques can now be utilized on a single cell or single molecule level to map forces in cells [59]. If such techniques can be multiplexed with super resolution microscopy, this may allow for matching tension with the spatio-temporal arrangement of molecules on the nanoscale. Adding layers of information to localization data could provide a route to discover whether specific structures or cluster types (with regards molecular content, density, size, and location) link to the mechanical tension they exert on their surroundings. This might also be used to find out whether clusters of molecules such as integrins are homogenous in their mechanical tension within single clusters. By imaging cells live, we may be able to discern whether these characteristics play a role in the basis of many immune cell functions, such as the formation of microvilli, of migration in response to chemokine, or of immune synapse formation. Temperature sensors represent another avenue of development which may be combined with super-resolution imaging. Recently, it was reported that mitochondria are maintained at a temperature of 50 degrees celcius, far above that of the cytoplasm [60] using a mito-targeted organic probe [61]. ERthermAC, a thermosensitive organic dye, has been used to detect large temperature changes in the endoplasmic reticulum of adipocytes [62]. It should not be discounted therefore that these temperature changes and others might occur in immune cells.
Finally, the imaging of ions at super-resolution would be useful for many areas related to immune cell behavior. Changes in ion flow through single channels are good candidates for minority events that may cause cell wide nanoscale changes that translate to function. Calcium ions are highly relevant to T cell activation, delivered through store operated calcium channels (SOC) [63] but also T-type channels [64] upon activation of the T cell receptor. Potassium ions are shown to be inhibitory to T cells, particular in the tumor microenvironment [65]. In addition, potassium efflux may be involved in chronic maintenance of TEM cells, but several different potassium channels have compensatory roles, and the dynamics of ion flow at the level of single receptors is unknown [65]. The investigation of ion flux at super-resolution is therefore an area ripe for study in the context of peripheral T cell migration, interaction with target cells and during a failed response to cancer. Imaging ions, their respective channels/pumps and intermediates on the nanoscale has the potential to reveal more about how T cells respond quickly to different environments, and may be achievable soon through the development of a raft of ion sensing probes [61].
Conclusion
Through the combination of cell friendly single molecule imaging, artifact controls, high throughput microscopy and multiple metrics on single molecules, trans-scale experiments can be undertaken. Such techniques may hold the key to many immunological phenomena that start on the nanoscale, representing the natural next step for immune function basic research. They may help us to understand how immune cells can be used for cancer immunotherapy [66], how the immune system co-develops with the body's bacterial population [67], and how immune cells malfunction and attack our own cells, causing autoimmune disease. The nano-scale is now truly accessible to immunologists, and the right combinations of techniques will soon help us link that nanoscale world to holistic, causal outcomes.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by ERC Starter Grant #337187. MS was supported by the King's Bioscience Institute and the Guy's and St Thomas' Charity Prize Ph.D. Programme in Biomedical and Translational Science.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Dustin ML. The immunological synapse. Cancer Immunol Res. (2014) 2:1023–33. doi: 10.1158/2326-6066.CIR-14-0161
2. Valvo S, Mayya V, Seraia E, Afrose J, Novak-Kotzer H, Ebner D, et al. Comprehensive analysis of immunological synapse phenotypes using supported lipid bilayers. Methods Mol Biol. (2017) 1584:423–41. doi: 10.1007/978-1-4939-6881-7_26
3. Smoligovets AA, Smith AW, Wu HJ, Petit RS, Groves JT. Characterization of dynamic actin associations with T-cell receptor microclusters in primary T cells. J. Cell Sci. (2012) 125:735–42. doi: 10.1242/jcs.092825
4. Kumari S, Curado S, Mayya V, Dustin ML. T cell antigen receptor activation and actin cytoskeleton remodeling. Biochim Biophys Acta Biomembr. (2014) 1838:546–56. doi: 10.1016/j.bbamem.2013.05.004
5. Sherman E, Barr V, Samelson LE. Super-resolution characterization of TCR-dependent signaling clusters. Immunol Rev. (2013) 251:21–35. doi: 10.1111/imr.12010
6. Hu YS, Cang H, Lillemeier BF. Superresolution imaging reveals nanometer- and micrometer-scale spatial distributions of T-cell receptors in lymph nodes. Proc Natl Acad Sci USA. (2016) 113:7201–6. doi: 10.1073/pnas.1512331113
7. Owen DM, Rentero C, Rossy J, Magenau A, Williamson D, Rodriguez M, et al. PALM imaging and cluster analysis of protein heterogeneity at the cell surface. J Biophotonics (2010) 3:446–54. doi: 10.1002/jbio.200900089
8. Baumgart F, Arnold AM, Leskovar K, Staszek K, Fölser M, Weghuber J, et al. Varying label density allows artifact-free analysis of membrane-protein nanoclusters. Nat Methods (2016) 13:661–4. doi: 10.1038/nmeth.3897
9. Burn GL, Cornish GH, Potrzebowska K, Samuelsson M, Griffié J, Minoughan S, et al. Superresolution imaging of the cytoplasmic phosphatase PTPN22 links integrin-mediated T cell adhesion with autoimmunity. Sci Signal. (2016) 9:ra99. doi: 10.1126/scisignal.aaf2195
10. Williamson DJ, Owen DM, Rossy J, Magenau A, Wehrmann M, Gooding JJ, et al. Pre-existing clusters of the adaptor Lat do not participate in early T cell signaling events. Nat Immunol. (2011) 12:655–62. doi: 10.1038/ni.2049
11. Griffié J, Shlomovich L, Williamson DJ, Shannon M, Aaron J, Khuon S, et al. 3D Bayesian cluster analysis of super-resolution data reveals LAT recruitment to the T cell synapse. Sci Rep. (2017) 7:4077. doi: 10.1038/s41598-017-04450-w
12. Ashdown GW, Cope A, Wiseman PW, Owen DM. Molecular flow quantified beyond the diffraction limit by spatiotemporal image correlation of structured illumination microscopy data. Biophys J. (2014) 107:L21–3. doi: 10.1016/j.bpj.2014.09.018
13. Sahl SJ, Hell SW, Jakobs S. Fluorescence nanoscopy in cell biology. Nat Rev Mol Cell Biol. (2017) 18:685–701. doi: 10.1038/nrm.2017.71
14. Baddeley D, Bewersdorf J. Biological insight from super-resolution microscopy: what we can learn from localization-based images. Annu. Rev. Biochem. (2018) 87:965–89. doi: 10.1146/annurev-biochem-060815-014801
15. Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature (2002) 419:845–9. doi: 10.1038/nature01076
16. Baumgart F, Arnold AM, Rossboth BK, Brameshuber M, Schütz GJ. What we talk about when we talk about nanoclusters. Methods Appl. Fluoresc. (2018) 7:013001. doi: 10.1088/2050-6120/aaed0f
17. Santos AM, Ponjavic A, Fritzsche M, Fernandes RA, de la Serna JB, Wilcock MJ, et al. Capturing resting T cells: the perils of PLL. Nat Immunol. (2018) 19:203–5. doi: 10.1038/s41590-018-0048-8
18. Rossboth B, Arnold AM, Ta H, Platzer R, Kellner F, Huppa JB, et al. TCRs are randomly distributed on the plasma membrane of resting antigen-experienced T cells. Nat Immunol. (2018) 19:821–7. doi: 10.1038/s41590-018-0162-7
19. Cai E, Marchuk K, Beemiller P, Beppler C, Rubashkin MG, Weaver VM, et al. Visualizing dynamic microvillar search and stabilization during ligand detection by T cells. Science (2017) 356:eaal3118. doi: 10.1126/science.aal3118
20. Jung Y, Riven I, Feigelson SW, Kartvelishvily E, Tohya K, Miyasaka M, et al. Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proc Natl Acad Sci USA. (2016) 113:E5916–24. doi: 10.1073/pnas.1605399113
21. Katz ZB, Novotná L, Blount A, Lillemeier BF. A cycle of Zap70 kinase activation and release from the TCR amplifies and disperses antigenic stimuli. Nat Immunol. (2017) 18:86–95. doi: 10.1038/ni.3631
22. Chang JT, Wherry EJ, Goldrath AW. Molecular regulation of effector and memory T cell differentiation. Nat Immunol. (2014) 15:1104–15. doi: 10.1038/ni.3031
23. Miller MJ, Hejazi AS, Wei SH, Cahalan MD. Parker I. T cell repertoire scanning is promoted by dynamic dendritic cell behavior and random T cell motility in the lymph node. Proc Natl Acad Sci USA. (2004) 101:998–1003. doi: 10.1073/pnas.0306407101
24. Hogg N, Laschinger M, Giles K, McDowall A. T-cell integrins: more than just sticking points. J Cell Sci. (2003) 116:4695–705. doi: 10.1242/jcs.00876
25. Sherman E, Barr V, Manley S, Patterson G, Balagopalan L, Akpan I, et al. Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor. Immunity (2011) 35:705–20. doi: 10.1016/j.immuni.2011.10.004
26. Lillemeier BF, Mörtelmaier MA, Forstner MB, Huppa JB, Groves JT, et al. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. (2010) 11:90–6. doi: 10.1038/ni.1832
27. Rossy J, Owen DM, Williamson DJ, Yang Z, Gaus K. Conformational states of the kinase Lck regulate clustering in early T cell signaling. Nat Immunol. (2013) 14:82–9. doi: 10.1038/ni.2488
28. Shibata AC, Fujiwara TK, Chen L, Suzuki KG, Ishikawa Y, Nemoto YL, et al. Archipelago architecture of the focal adhesion: membrane molecules freely enter and exit from the focal adhesion zone. Cytoskeleton (2012) 69:380–92. doi: 10.1002/cm.21032
29. Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J Cell Biol. (2002) 157:1071–82. doi: 10.1083/jcb.200202050
30. Burn GL, Svensson L, Sanchez-Blanco C, Saini M, Cope AP. Why is PTPN22 a good candidate susceptibility gene for autoimmune disease? FEBS Lett. (2011) 585:3689–98. doi: 10.1016/j.febslet.2011.04.032
31. Bottini N, Peterson EJ, Bottini N, Peterson EJ. Tyrosine phosphatase PTPN22: multifunctional regulator of immune signaling, development, and disease. Annu Rev Immunol. (2014) 32:83–119. doi: 10.1146/annurev-immunol-032713-120249
32. Mueller SN, Gebhardt T, Carbone FR, Heath WR. Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol. (2013) 31:137–61. doi: 10.1146/annurev-immunol-032712-095954
33. Krummel MF, Friedman RS, Jacobelli J. Modes and mechanisms of T cell motility: roles for confinement and Myosin-IIA. Curr Opin Cell Biol. (2013) 30:9–16. doi: 10.1016/j.ceb.2014.05.003
34. Hons M, Kopf A, Hauschild R, Leithner A, Gaertner F, Abe J, et al. Chemokines and integrins independently tune actin flow and substrate friction during intranodal migration of T cells. Nat Immunol. (2018) 19:606–16. doi: 10.1038/s41590-018-0109-z
35. Teijeira A, Hunter MC, Russo E, Proulx ST, Frei T, Debes GF, et al. T cell migration from inflamed skin to draining lymph nodes requires intralymphatic crawling supported by ICAM-1/LFA-1 Interactions. Cell Rep. (2017) 18:857–65. doi: 10.1016/j.celrep.2016.12.078
36. Buchholz VR, Schumacher TN, Busch DH. T cell fate at the single-cell level. Annu Rev Immunol. (2016) 34:65–92. doi: 10.1146/annurev-immunol-032414-112014
37. Alanko J, Ivaska J. Endosomes: emerging platforms for integrin-mediated FAK signalling. Trends Cell Biol. (2016) 26:391–8. doi: 10.1016/j.tcb.2016.02.001
38. Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science (2014) 346:1257998. doi: 10.1126/science.1257998
39. Liu TL, Upadhyayula S, Milkie DE, Singh V, Wang K, Swinburne IA, et al. Observing the cell in its native state: imaging subcellular dynamics in multicellular organisms. Science (2018) 360:eaaq1392. doi: 10.1126/science.aaq1392
40. Griffie J, Owen D, Rubin-Delanchy P, Burn G. Quantitative analysis of membrane protein clustering from live-cell, single-molecule super-resolution microscopy data. Biophys J. (2017) 112:144a−5a. doi: 10.1016/j.bpj.2016.11.794
41. Kotera I, Iwasaki T, Imamura H, Noji H, Nagai T. Reversible dimerization of Aequorea victoria fluorescent proteins increases the dynamic range of FRET-based indicators. ACS Chem Biol. (2010) 5:215–22. doi: 10.1021/cb900263z
42. Arai Y, Takauchi H, Ogami Y, Fujiwara S, Nakano M, Matsuda T, et al. A spontaneously blinking fluorescent protein for simple single laser super-resolution live cell imaging. bioRxiv (2017) 200014. doi: 10.1101/200014
43. Tiwari DK, Arai Y, Yamanaka M, Matsuda T, Agetsuma M, Nakano M, et al. A fast- and positively photoswitchable fluorescent protein for ultralow-laser-power RESOLFT nanoscopy. Nat Methods (2015) 12:515–8. doi: 10.1038/nmeth.3362
44. Mattila PK, Feest C, Depoil D, Treanor B, Montaner B, Otipoby KL, et al. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity (2013) 38:461–74. doi: 10.1016/j.immuni.2012.11.019
45. Lopes FB, Bálint Š, Valvo S, Felce JH, Hessel EM, Dustin ML, et al. Membrane nanoclusters of FcγRI segregate from inhibitory SIRPα upon activation of human macrophages. J Cell Biol. (2017) 216:1123–41. doi: 10.1083/jcb.201608094
46. Mace EM, Orange JS. High- and super-resolution microscopy imaging of the NK cell immunological synapse. Methods Mol Biol. (2016) 1441:141–50. doi: 10.1007/978-1-4939-3684-7_12
47. Yasui M, Hiroshima M, Kozuka J, Sako Y, Ueda M. Automated single-molecule imaging in living cells. Nat Commun. (2018) 9:3061. doi: 10.1038/s41467-018-05524-7
48. Holden SJ, Pengo T, Meibom KL, Fernandez Fernandez C, Collier, J, Manley S. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc Natl Acad Sci USA. (2014) 111:4566–71. doi: 10.1073/pnas.1313368111
49. Douglass KM, Sieben C, Archetti A, Lambert A, Manley S. Super-resolution imaging of multiple cells by optimized flat-field epi-illumination. Nat Photonics (2016) 10:705–8. doi: 10.1038/nphoton.2016.200
50. Rowlands CJ, Ströhl F, Ramirez PPV, Scherer KM, Kaminski CF. Flat-field super-resolution localization microscopy with a low-cost refractive beam-shaping element. Sci Rep. (2018) 8:5630. doi: 10.1038/s41598-018-24052-4
51. Culley S, Albrecht D, Jacobs C, Pereira PM, Leterrier C, Mercer J, et al. NanoJ-SQUIRREL: quantitative mapping and minimisation of super-resolution optical imaging artefacts. bioRxiv (2017) 158279. doi: 10.1101/158279
52. Fox-Roberts P, Marsh R, Pfisterer K, Jayo A, Parsons M, Cox S. Local dimensionality determines imaging speed in localization microscopy. Nat Commun. (2017) 8:13558. doi: 10.1038/ncomms13558
53. Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, et al. Nanoscale architecture of integrin-based cell adhesions. Nature (2010) 468:580–4. doi: 10.1038/nature09621
54. Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, et al. Cell migration: integrating signals from front to back. Science (2003) 302:1704–9. doi: 10.1126/science.1092053
55. Goult BT, Zacharchenko T, Bate N, Tsang R, Hey F, Gingras AR, et al. RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover. J Biol Chem. (2013) 288:8238–49. doi: 10.1074/jbc.M112.438119
56. Lämmermann T. In the eye of the neutrophil swarm-navigation signals that bring neutrophils together in inflamed and infected tissues. J Leukoc Biol. (2016) 100:55–63. doi: 10.1189/jlb.1MR0915-403
57. Moore TI, Aaron J, Chew TL, Springer TA. Measuring integrin conformational change on the cell surface with super-resolution microscopy. Cell Rep. (2018) 22:1903–2. doi: 10.1016/j.celrep.2018.01.062
58. Liu Y, Blanchfield L, Ma VP, Andargachew R, Galior K, Liu Z, et al. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc Natl Acad Sci USA. (2016) 113:5610–5. doi: 10.1073/pnas.1600163113
59. Huang DL, Bax NA, Buckley CD, Weis WI, Dunn AR. Vinculin forms a directionally asymmetric catch bond with F-actin. Science (2017) 357:703–6. doi: 10.1126/science.aan2556
60. Chrétien D, Bénit P, Ha HH, Keipert S, El-Khoury R, Chang YT, et al. Mitochondria are physiologically maintained at close to 50°C. PLOS Biol. (2018) 16:e2003992. doi: 10.1371/journal.pbio.2003992
61. Arai S, Suzuki M, Park SJ, Yoo JS, Wang L, Kang NY, et al. Mitochondria-targeted fluorescent thermometer monitors intracellular temperature gradient. Chem Commun. (2015) 51:8044–7. doi: 10.1039/C5CC01088H
62. Arai S, Lee SC, Zhai D, Suzuki M, Chang YT. A molecular fluorescent probe for targeted visualization of temperature at the endoplasmic reticulum. Sci Rep. (2015) 4:6701. doi: 10.1038/srep06701
63. Zhang Y, Ge C, Zhu C, Salaita K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nat Commun. (2014) 5:5167. doi: 10.1038/ncomms6167
64. Lacinova L, Weiss N. It takes two T to shape immunity: emerging role for T-type calcium channels in immune cells. Gen Physiol Biophys (2016) 35:393–6. doi: 10.4149/gpb_2016034
65. Eil R, Vodnala SK, Clever D, Klebanoff CA, Sukumar M, Pan JH, et al. Ionic immune suppression within the tumour microenvironment limits T cell effector function. Nature (2016) 537:539–43. doi: 10.1038/nature19364
66. Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, et al. Systemic immunity is required for effective cancer immunotherapy. Cell (2017) 168:487–502.e15. doi: 10.1016/j.cell.2016.12.022
Keywords: T cells, immunology, SMLM, nanoscopy, super-resolution
Citation: Shannon M and Owen DM (2019) Bridging the Nanoscopy-Immunology Gap. Front. Phys. 6:157. doi: 10.3389/fphy.2018.00157
Received: 24 September 2018; Accepted: 18 December 2018;
Published: 24 January 2019.
Edited by:
Jorge Bernardino de La Serna, United Kingdom Research and Innovation, United KingdomReviewed by:
Florian Baumgart, Technische Universität Wien, AustriaSusanne Fenz, Universität Würzburg, Germany
Ana Mafalda Santos, University of Oxford, United Kingdom
Copyright © 2019 Shannon and Owen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dylan M. Owen, ZHlsYW4ub3dlbkBrY2wuYWMudWs=
 Michael Shannon
Michael Shannon Dylan M. Owen
Dylan M. Owen