Abstract
Introduction:
The combination of BRAF and tyrosine kinase (TK) inhibitors has been demonstrated to be highly effective in inhibiting tumor development and is an approach for overcoming resistance in clinical trials. Accordingly, a novel series of 1,2,4-oxadiazole/quinazoline-4-one hybrids was developed as antiproliferative multitargeted inhibitors.
Methods:
The structures of the newly synthesized compounds 9a-o were validated using IR, NMR, MS, and elemental techniques. 9a–o were tested as antiproliferative agents.
Results and Discussion:
The results showed that the majority of the tested compounds showed significant antiproliferative action with 9b, 9c, 9h, 9k, and 9l being the most potent. Compounds 9b, 9c, 9h, 9k, and 9l were tested as EGFR and BRAFV600E inhibitors. These in vitro tests revealed that compounds 9b, 9c, and 9h are strong antiproliferative agents that may act as dual EGFR/BRAFV600E inhibitors. 9b, 9c, and 9h were further investigated for their inhibitory effect on mutant EGFR (EGFRT790M), and the results showed that the tested compounds had considerable inhibitory action. Cell cycle study and apoptosis detection demonstrated that compound 9b exhibits cell cycle arrest at the G2/M transition. Molecular docking simulations reveal the binding mechanism of the most active antiproliferative agents.
1 Introduction
Drug developers have spent decades generating selective medicines for specific targets (Medina-Franco et al., 2013; Zhou et al., 2019). Despite the effectiveness of many single-target selective medications, the advancement of multifactorial disorders such as cancer and neurological diseases included many signaling pathways (Fu et al., 2017; Raghavendra et al., 2018). As a result, there is a growing interest in developing treatments that address many targets at the same time.
There are currently two opposing methodologies for designing multi-targeting medicines. The first technique involves establishing an additive or synergistic effect of various medications operating on separate targets through combination drug therapy. Preclinical evidence of enhanced apoptosis and delayed resistance to BRAF (Rapidly Accelerated Fibrosarcoma, B-family) inhibitors (Paraiso et al., 2010; Flaherty et al., 2012), for example, prompted the FDA to approve a combination of dabrafenib (BRAF inhibitor) and trametinib (MEK inhibitor) for the treatment of metastatic melanoma with BRAF mutations (Robert et al., 2015; Wahid et al., 2018). The use of palbociclib and letrozole in the treatment of advanced breast cancer is another example of successful combination therapy (Finn et al., 2016).
The second approach is designing and generating multiple-targeting medicines that synergistically block numerous carcinogenic pathways (Keith et al., 2005; Boran and Iyengar, 2010). The method of multi-targeting therapies is finding a single agent that can operate on two or more targets simultaneously. Cabozantinib, also known as cabometyx, was approved by the FDA as a small molecule dual-targeting inhibitor of the tyrosine kinases c-Met (mesenchymal-epithelial transition factor) and VEGFR-2 (Vascular Endothelial Growth Factor Receptor) and has been demonstrated to suppress tumor growth, metastasis, and angiogenesis (Food and Administration, 1997).
On the other hand, the combination of BRAF and tyrosine kinase (TK) inhibitors has been demonstrated to be highly effective in inhibiting tumor development and is an approach for overcoming resistance in clinical trials. Vemurafenib (BRAFV600E inhibitor) resistance in thyroid cancer can be addressed by combining it with EGFR (Epidermal Growth Factor Receptor) inhibitors (Notarangelo et al., 2017). This combination has also shown promising results in BRAFV600E colorectal cancer (Mondaca et al., 2018). In addition, various compounds have been discovered in vitro that include the key pharmacophoric groups required to inhibit tyrosine kinases, such as EGFR/VEGFR-2 and BRAF (Okaniwa et al., 2012; Zhang et al., 2013). Compound I (Figure 1) inhibited wild-type BRAF and EGFR with IC50 values in the nanomolar range. Additionally, imidazo [1,2-b]pyridazine II inhibited BRAF and VEGFR-2.
FIGURE 1

Structures of compounds I and II.
Heterocyclic moieties form the largest and most varied class of organic molecules. In medicinal chemistry, compounds containing heterocyclic nuclei have gained great interest because of their diverse therapeutic actions (Padwa and Bur, 2007). Heterocyclics play a crucial role in the breakdown of all living things and participate in various biochemical processes necessary for life (Kitadai and Maruyama, 2018). The heteroaromatic framework resembles biologically active compounds such as nucleic acids, hormones, and neurotransmitters (Meanwell, 2017). As a result, these moieties could be used to design safer medications. Heterocycles are often found in nature and have been exploited to develop anti-cancer drugs that target many sites and disrupt cancer growth pathways (Sharma et al., 2017). Heterocyclic rings can be modified with various substituents to cover various chemicals, making them ideal for designing anti-cancer drugs.
Nitrogen-containing heterocyclic chemicals significantly affect about 75% of FDA-approved anti-cancer drugs (Kerru et al., 2020). Quinazolinone, a bicyclic system composed of benzene and pyrimidinone rings, is one of the most common nitrogen-containing heterocycles in medicinal chemistry, found in various compounds with diverse biological activity. Idelalisib III (Do et al., 2016), Ispinesib IV (Purcell et al., 2010), and Halofuginone V (Figure 2) (Derbyshire et al., 2012; McLaughlin et al., 2014) are examples of recently approved or marketed medications with anti-cancer properties.
FIGURE 2

Examples of approved or commercialized anti-cancer medicines with the quinazoline-4-one scaffold.
Depending on the position of the keto or oxo group, three distinct forms are possible: quinazolin-2(1H)-one VI, quinazoline-2,4-(1H,3H)-di-one VII, and quinazolin-4(3H)-one VIII (Figure 3). Among these, quinazolin-4-one VIII is the most commonly used scaffold in synthetic processes or as a structural component of natural compounds (Sharma et al., 2017). This last scaffold is adaptable, allowing up to six potential substitutes in positions 2, 3, 5, 6, 7, and 8.
FIGURE 3

Different forms of quinazolinones.
In recent publications (Hisham et al., 2022; Hisham et al., 2023), we present the design and synthesis of a new series of quinazoline-4-one/chalcone hybrids that function as dual inhibitors of EGFR and BRAFV600E with antiproliferative activity. The target compounds were tested in vitro against various cancer cell lines and the EGFR and BRAFV600E enzymes. Compound IX (Figure 4) was the most potent derivative, with a GI50 of 1.16 µM, compared to the reference drug Doxorubicin (GI50 = 1.14 µM). Compound IX showed significant inhibitory activity against EGFR and BRAFV600E, with IC50 values of 0.11 µM and 0.65 µM, respectively. Moreover, apoptosis assay results revealed that compound IX enhanced the level of active caspase-3, 8, and 9 with significant induction of cytochrome c and Bax levels and downregulation of the anti-apoptotic Bcl-2 levels.
FIGURE 4

Structures of quinazoline-4-one and 1,2,4-oxadiazole-based derivatives as anticancer agents.
On the other hand, literature reviews reveal that 1,2,4-oxadiazoles have statistical significance in bioorganic and medicinal chemistry. They are recognized for their diverse pharmacological characteristics (Benassi et al., 2020; El Mansouri et al., 2020; Loboda et al., 2020). The 1,2,4-oxadiazole exhibits bioisosteric equivalence with ester and amide moieties. When unstable conditions (e.g., hydrolysis) are identified, 1,2,4-oxadiazole is a highly effective alternative (Hendawy, 2022). The substantial biological impact of 1,2,4-oxadiazole derivatives on cancer cells can be attributed to various mechanisms of action. For example, we developed and synthesized novel 1,2,4-oxadiazole-based derivatives linked to a triaryl-imidazole moiety, with compound X (Figure 4) being the most potent (Youssif et al., 2022). In vitro studies assessed the antiproliferative effects of recently identified compounds inhibiting p38α and BRAFV600E. These compounds showed effective antiproliferative and kinase inhibition.
Another set of 1,2,4-oxadiazole-based compounds (compound XI, Figure 4) were designed, synthesized, and tested for antiproliferative properties against EGFR-TK. The experiment showed promising antiproliferative effects against cancer cell lines, with low micromolar IC50 values against EGFR, compared to the reference doxorubicin (Unadkat et al., 2021).
1.1 Rational design
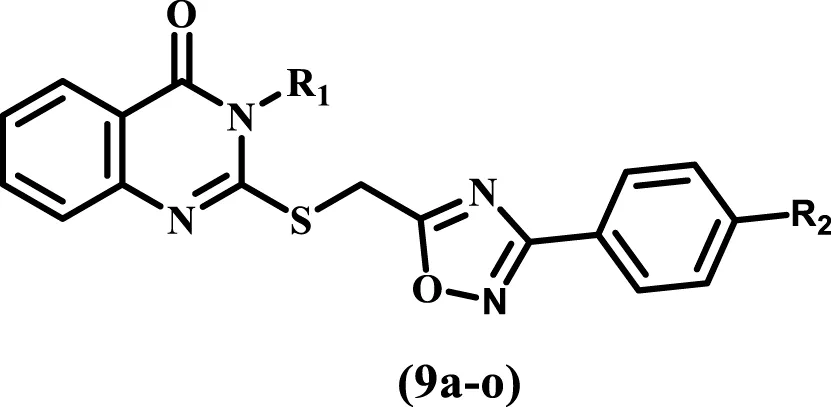
Consistent with prior findings and continuing our efforts to develop dual or multitargeted antiproliferative agents (Al-Wahaibi et al., 2020; Alshammari et al., 2022; Al-Wahaibi et al., 2023a; Abdel-Aziz et al., 2023; Al-Wahaibi et al., 2023b; Al-Wahaibi et al., 2023c; Al-Wahaibi et al., 2023d; Al-Wahaibi et al., 2023e), this study’s strategy was to design and synthesize new antiproliferative agents based on quinazoline-4-one/1,2,4-oxadiazole hybrids (Figure 5) to obtain new anti-tumor agents with synergistic activity.
FIGURE 5

Structures of new targets 9a-o.
The substitutions on the nitrogen atom of the quinazoline moiety were changed between alkyl (methyl, ethyl, and allyl) and aryl (phenyl or tolyl) moieties to examine the impact of rigidity on the antiproliferative activity of these compounds. In addition, different substituents, such as a chlorine atom (electron withdrawing group) or a methoxy group (electron donating group), were used to investigate the different electronic impacts of these substituents on 9a-o’s antiproliferative activity.
All of the novel analogs were examined for cell viability effect against a normal human mammary gland epithelial (MCF-10A) cell line as well as for antiproliferative activity against four human cancer cell lines: colon (HT-29), pancreatic (Panc-1), lung (A-549), and breast (MCF-7). Furthermore, the compounds with the highest antiproliferative activity were investigated in vitro as multi-targeting inhibitors of EGFR, EGFRT790M, and BRAFV600E enzymes. The study was expanded to include one of the most active derivatives, 9b, as a representative agent to evaluate its mechanistic effects on the cell cycle and induction of apoptosis. Finally, docking studies were performed on the most active compounds against the selected enzymes to explain their in vitro results. Furthermore, the ADME analyses were performed to investigate their pharmacokinetic features.
2 Results and discussion
2.1 Chemistry
Scheme 1 summarizes the synthetic pathways of the new target compounds 9a-o. Anthranilic acid (1) was refluxed in ethanol with isothiocyanate derivatives 2a-e for 8 h. After the reaction was completed (as determined by TLC), the resulting white precipitate was collected by filtration and recrystallized from an ethanol/dioxane mixture (1:1) to give the corresponding quinazoline derivatives 3a-e in 90%–95% yields (Moussa et al., 2018). On the other hand, compounds 6a-c, amidoxime derivatives, were synthesized in 50%–60% yields over two steps, Scheme 2. The first step involved reacting the corresponding aldehydes 4a-c with 28% aqueous ammonia and iodine in THF for 2–3 h to yield the corresponding aryl nitrile derivatives 5a-c in 76%–80% (Yan et al., 2017). The second step was a 12- to 18-h methanol reflux of compounds 5a-c with hydroxylamine chloride and sodium carbonate (Youssif et al., 2019). Compounds 6a-c were reacted with chloroacetyl chloride in dry acetone to yield benzimidamides (7a-c), which were cyclized by refluxing in toluene to the corresponding 3-aryl-5-(chloromethyl)-1,2,4-oxadiazole derivatives 8a-c as a yellow oil. Compounds 8a-c were purified using column chromatography with hexane: ethyl acetate (9:1) as an eluent (Minin et al., 2023). For example, the 1H NMR spectrum of compound 8b confirmed the disappearance of two protons from the NH2 group of the corresponding amidoxime 7b. Moreover, the spectrum displayed a singlet signal corresponding to the methylene protons (Cl-CH2) at δ 4.74. The spectra also revealed a characteristic pair of doublets in the aromatic region for 4-ClC6H4 at δ 8.01 and 7.46.
SCHEME 1

Synthesis of the new target compounds 9a-o
SCHEME 2

Structure Activity Relationship (SAR) analysis of new targets 9a-o.
Reagents and Conditions: a) Triethylamine, ethanol, Reflux 8 h; b) ammonia (28%), I2, THF, Stirring 1h; c) NH2OHHCl, Na2CO3, THF, Reflux 18 h; d) Chloroacetyl Chloride, K2CO3, Dry acetone, Stirring 24hrs; e) Toluene, Reflux 10 h; f) K2CO3, KI, DMF, Stirring 24 h.
Finally, the target novel compounds, 9a-o, were synthesized in high yields by coupling compounds 3a-e with the corresponding 1,2,4-oxadiazoles 8a-c in DMF using anhydrous K2CO3 and KI and stirring for 24 h at room temperature. 9a-o were purified via ethanol recrystallization. The structures of 9a-o were elucidated using 1H NMR, 13C NMR, and elemental microanalyses. The 1H NMR spectra of compound 9l, as an example, confirmed the presence of ethyl group characteristic signals in the form of triplet at δ 1.31 (t, J = 7.1 Hz, 3H, N-CH2CH3) and quartet at δ 4.11 (q, J = 7.1 Hz, 2H, N-CH2). The spectrum also revealed two distinct singlet signals: at δ 3.81 (s, 3H, OCH3) and at δ 4.91 (s, 2H, S-CH2). Additionally, the spectrum revealed a pair of doublets of the aromatic ring’s para di substitution pattern and extra signals for the aromatic protons in the quinazoline moiety. The 13C NMR spectrum of 9l indicated the presence of ethyl group characteristic signals at δ 39.56 and δ 13.01, methylene group at δ 26.62, and methoxy group at δ 55. Elemental microanalysis of 9l confirmed that the calculated data (%) were C, 60.90; H, 4.60; N, 14.20; S, 8.13, while the found data (%) were C, 61.13; H, 4.74; N, 14.37; S, 8.20.
2.2 Biology
2.2.1 Assay of cell viability effect
The human mammary gland epithelial (MCF-10A) normal cell line was used to test the viability of novel compounds 9a-o (Mahmoud et al., 2022; Mekheimer et al., 2022). After 4 days of incubation on MCF-10A cells, the cell viability of compounds 9a-o was determined using the MTT test. Table 1 demonstrates that none of the compounds examined were cytotoxic, and all hybrids showed more than 89% cell viability at 50 µM.
TABLE 1
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Comp | Cell viability % | R1 | R2 | Antiproliferative activity IC50 ± SEM (nM) | ||||
| A-549 | MCF-7 | Panc-1 | HT-29 | Average (GI50) | ||||
| 9a | 90 | Phenyl | H | 46 ± 4 | 50 ± 4 | 48 ± 4 | 48 ± 4 | 48 |
| 9b | 89 | Phenyl | Cl | 22 ± 2 | 26 ± 2 | 24 ± 2 | 24 ± 2 | 24 |
| 9c | 91 | Phenyl | OMe | 24 ± 2 | 28 ± 3 | 26 ± 2 | 25 ± 2 | 26 |
| 9d | 90 | p-Tolyl | H | 54 ± 4 | 58 ± 5 | 54 ± 5 | 55 ± 5 | 55 |
| 9e | 91 | p-Tolyl | Cl | 40 ± 3 | 44 ± 4 | 42 ± 4 | 42 ± 4 | 42 |
| 9f | 92 | p-Tolyl | OMe | 50 ± 4 | 53 ± 5 | 52 ± 5 | 52 ± 5 | 52 |
| 9g | 90 | m-Tolyl | H | 49 ± 4 | 52 ± 5 | 50 ± 5 | 50 ± 5 | 50 |
| 9h | 89 | m-Tolyl | Cl | 28 ± 2 | 31 ± 3 | 29 ± 2 | 30 ± 3 | 30 |
| 9i | 91 | m-Tolyl | OMe | 65 ± 6 | 69 ± 6 | 66 ± 6 | 68 ± 6 | 67 |
| 9j | 90 | Ethyl | H | 69 ± 6 | 76 ± 7 | 68 ± 6 | 68 ± 6 | 70 |
| 9k | 93 | Ethyl | Cl | 32 ± 3 | 35 ± 3 | 34 ± 3 | 34 ± 3 | 34 |
| 9l | 90 | Ethyl | OMe | 36 ± 3 | 40 ± 4 | 38 ± 3 | 37 ± 3 | 38 |
| 9m | 92 | Allyl | H | 62 ± 6 | 65 ± 6 | 64 ± 6 | 62 ± 6 | 63 |
| 9n | 91 | Allyl | Cl | 43 ± 4 | 47 ± 4 | 45 ± 4 | 44 ± 4 | 45 |
| 9o | 89 | Allyl | OMe | 56 ± 4 | 59 ± 5 | 57 ± 5 | 57 ± 5 | 57 |
| Erlotinib | ND | — | — | 30 ± 3 | 40 ± 3 | 30 ± 3 | 30 ± 3 | 33 |
Cell viability percent and antiproliferative activity (IC50 values) of compounds 9a-o.
ND: not determined.
2.2.2 Assay of antiproliferative action
The MTT assay was used to investigate the antiproliferative activity of hybrids 9a-o versus four human cancer cell lines using Erlotinib as a control: colon cancer (HT-29) cell line, pancreatic cancer (Panc-1) cell line, lung cancer (A-549) cell line, and breast cancer (MCF-7) cell line (El-Sherief et al., 2019; Al-Wahaibi et al., 2022). Table 1 displays the median inhibitory concentration (IC50) and GI50 (average IC50) against the four cancer cell lines.
In general, the hybrids 9a-o had significant antiproliferative action with GI50 values ranging from 24 nM to 70 nM versus the four cancer cell lines evaluated, compared to Erlotinib, which had a GI50 value of 33 nM. Compounds 9b, 9c, 9h, 9k, and 9l were the most potent five derivatives, with GI50 values of 24, 26, 30, 34, and 38 nM, making 9b, 9c, and 9h more potent than Erlotinib (GI50 = 33 nM). Out of all the newly synthesized hybrids 9a-o, compound 9b (R1 = phenyl, R2 = Cl) had the highest potency, with a GI50 value of 24 nM, which was 1.4 times more potent than the reference Erlotinib (GI50 = 33 nM).
The type of the aryl/alkyl moieties at position 3 of the quinazoline moiety appears to be critical for 9a-o hybrids antiproliferative activity. The GI50 value of compound 9h (R1 = m-tolyl, R2 = Cl) was 30 nM, less potent than compound 9b but still more potent than the reference erlotinib (GI50 = 33 nM).
Moreover, Compounds 9e (R1 = p-tolyl, R2 = Cl), 9k (R1 = ethyl, R2 = Cl), and 9n (R1 = allyl, R2 = Cl) demonstrated GI50 values of 42, 34, and 45 nM, respectively, being less potent than compounds 9b, 9h, and even Erlotinib. These results indicated the importance of the quinazoline moiety position three substitution pattern on antiproliferative activity, with activity rising in the following order: phenyl > m-tolyl > p-tolyl > ethyl > allyl.
Compound 9c (R1 = phenyl, R2 = OMe) rated second in activity against the four cancer cell lines, with a GI50 value of 26 nM, slightly less effective than 9b but still more potent than Erlotinib (GI50 = 33 nM). The unsubstituted phenyl derivative, 9a (R1 = phenyl, R2 = H), was less potent than 9b and 9c, indicating that the substitution pattern at the fourth position of the phenyl group in the 1,2,4-oxadiazole moiety affects the antiproliferative activity of these hybrids, with activity increasing in the order Cl > OMe > H. Regardless of the nature of the substitution pattern at position 3 of the quinazoline moiety, the same rule (Cl > OMe > H in activity) applies to other derivatives.
2.2.3 EGFR inhibitory assay
The EGFR-TK test (Abdel-Aziz et al., 2023) was used to assess the inhibitory potency of the most effective antiproliferative derivatives 9b, 9c, 9h, 9k, and 9l against EGFR, and the results are shown in Table 2. This assay’s results are in line with the antiproliferative assay’s, which found that compounds 9b (R1 = phenyl, R2 = Cl), 9c (R1 = phenyl, R2 = OMe), and 9h (R = m-tolyl, R2 = Cl), the most effective antiproliferative hybrids, were the most potent derivatives of EGFR inhibitors, with IC50 values of 57 ± 4 nM, 64 ± 5 nM, and 72 ± 5 nM, respectively, surpassing the reference drug Erlotinib (IC50 = 80 ± 5). Compounds 9k (R1 = ethyl, R2 = Cl) and 9l (R1 = ethyl, R2 = OMe) demonstrated significant anti-EGFR activity, with IC50 values of 84 ± 6 nM and 91 ± 07 nM, respectively, which were less potent than Erlotinib. These findings show that compounds 9b, 9c, and 9h have significant EGFR inhibitory action and could be used as antiproliferative agents.
TABLE 2
| Compound | EGFR inhibition IC50 ± SEM (nM) | BRAFV600E inhibition IC50 ± SEM (nM) | EGFRT790M inhibition IC50 ± SEM (nM) |
|---|---|---|---|
| 9b | 57 ± 4 | 48 ± 4 | 10 ± 1 |
| 9c | 64 ± 5 | 51 ± 5 | 11 ± 2 |
| 9h | 72 ± 5 | 57 ± 5 | 15 ± 2 |
| 9k | 84 ± 6 | 62 ± 5 | ND |
| 9l | 91 ± 7 | 70 ± 5 | ND |
| Erlotinib | 80 ± 5 | 60 ± 5 | ND |
| Vemurafenib | ND | 30 ± 3 | ND |
| Osimertinib | ND | ND | 8 ± 1 |
IC50 values of compounds 9b, 9c, 9h, 9k, and 9l against EGFR, BRAFV600E and EGFRT790M.
ND: not determined.
2.2.4 BRAFV600E inhibitory assay
An in vitro investigation assessed the anti-BRAFV600E activity of 9b, 9c, 9h, 9k, and 9l (Youssif et al., 2022). The enzyme assay demonstrated that the five hybrids examined substantially inhibited BRAFV600E, with IC50 values ranging from 48 to 70 nM, Table 2. In all cases, the IC50 of the examined compounds is greater than that of the reference Vemurafenib (IC50 = 30). Compounds 9b, 9c, and 9h demonstrated the most effective inhibitory activity against BRAFV600E (IC50 = 48, 51, and 57 nM, respectively) and were discovered to be potent inhibitors of cancer cell growth (GI50 = 24, 26, and 30 nM, respectively). As a result, compounds 9b, 9c, and 9h are effective antiproliferative agents that function as dual EGFR/BRAFV600E inhibitors.
2.2.5 EGFRT790M inhibitory assay
The HTRF KinEASE-TK assay (
Miles et al., 2020) was used to evaluate the inhibitory action of the most potent hybrids, 9b, 9c, and 9h, against mutant-type EGFR (EGFR
T790M). As demonstrated in
Table 2, compounds 9b, 9c, and 9h displayed excellent inhibitory effect against EGFR
T790M, with IC
50values of 10 ± 1, 11 ± 1, and 15 ± 1 nM, respectively, being equivalent to the reference Osimertinib (IC
50= 8 ± 1 nM), which may explain their robust antiproliferative activity. These findings suggested that phenyl and
m-tolyl substitutions in the quinazoline moiety’s third position, as well as chlorine atom and methoxy substitutions in the
para-position of the phenyl group in the 1,2,4-oxadiazole moiety, are required for the inhibitory impact on EGFR, BRAF
V600E, and EGFR
T790M.
1. The type of the aryl/alkyl moieties at position 3 of the quinazoline moiety appears to be critical for 9a-o hybrids antiproliferative activity, with activity increasing in the following order: phenyl > m-tolyl > p-tolyl > ethyl > allyl.
2. The substitution pattern at the fourth position of the phenyl group in the 1,2,4-oxadiazole moiety affects the antiproliferative activity of these hybrids as well as EGFR, BRAFV600E, and EGFRT790M inhibition, with activity increasing in the order Cl > OMe > H.
3. Regardless of the nature of the substitution pattern at position 3 of the quinazoline moiety, the same rule (Cl > OMe > H in activity) applies to other derivatives.
2.2.6 Cell cycle analysis and apoptosis assays
2.2.6.1 Cell cycle analysis
Compound 9b was investigated for its effects on cell cycle progression and apoptosis induction in A-549 cells. A lung cancer (A-549) cell line was treated for 24 h with an IC50 concentration of 9b. The cell line was labeled with PI/Annexin V, and flow cytometry was done with a BD FASC Caliber (El-Sherief et al., 2018). The results (Figure 6) showed that A-549 treated with compound 9b had a significant percentage of cell accumulation (29%) in the G2/M phase after 24 h of incubation, indicating cell cycle arrest at the G2/M transition.
FIGURE 6

Cell cycle analysis results for compound 9b.
2.2.6.2 Apoptosis induction assay
To assess 9b′s potential to induce apoptosis, A-549 cells were labeled with Annexin V/PI, grown for 24 h, and evaluated. Examining early and late apoptosis demonstrated that compound 9b could produce high levels of apoptosis, with a necrosis percentage 6.43 (Figures 7, 8).
FIGURE 7

Apoptosis induction results of compound 9b.
FIGURE 8

Cell cycle analysis and apoptosis induction results of compound 9b.
2.3 Docking simulations
Starting with RSCB deposited crystal structure of EGFR protein having Erlotinib as a co-crystallized ligand (PDB: 1M17) (Bhat et al., 2022), and re-docking of Erlotinib revealed a docking score (S) of −7.30 kcal/mol and an RMSD of 1.28 Å, in addition to formation of the two characteristic H-bond interactions with two of key amino acid residues, Gln767 and Met769, indicating validity of docking study parameters. While running docking simulations within EGFR active site (PDB ID: 1M17) showed that most of the test derivatives (9a-o) showed moderate to strong binding interactions (S = −5.93 to −7.52; c.f. Erlotinib: 7.30 kcal/mol) as listed in Table 3. These interactions were variable between H-bond and/or H-pi, with key amino acid residues (Met 769, Lys 721, Gly 772, and Leu 694) lining the active site, as shown in Supplementary Figure S1 (Supplementary Material).
TABLE 3
| Comp | R1 | R2 | Docking score (S; kcal/mol) | ||
|---|---|---|---|---|---|
| EGFR (1M17) | EGFRT790M (2JIU) | BRAFV600E (5JRQ) | |||
| 9a | Phenyl | H | −6.0118 | −7.1900 | −7.0458 |
| 9b | Phenyl | Cl | −6.5073 | −7.4268 | −7.0985 |
| 9c | Phenyl | OMe | −6.0388 | −7.2876 | −7.2278 |
| 9d | p-Tolyl | H | −6.0314 | −6.7971 | −6.9232 |
| 9e | p-Tolyl | Cl | −6.4310 | −5.6713 | −7.3078 |
| 9f | p-Tolyl | OMe | −6.8251 | −6.7228 | −6.9971 |
| 9g | m-Tolyl | H | −7.5281 | −6.8538 | −6.8646 |
| 9h | m-Tolyl | Cl | −6.2685 | −6.4894 | −6.8470 |
| 9i | m-Tolyl | OMe | −6.8622 | −6.7373 | −7.6516 |
| 9j | Ethyl | H | −6.6348 | −6.9894 | −6.2829 |
| 9k | Ethyl | Cl | −5.7431 | −6.3041 | −6.5824 |
| 9l | Ethyl | OMe | −5.9343 | −6.1808 | −6.7029 |
| 9m | Allyl | H | −6.1516 | −7.2311 | −6.8390 |
| 9n | Allyl | Cl | −6.2599 | −6.3498 | −6.8475 |
| 9o | Allyl | OMe | −6.7193 | −6.7397 | −6.8732 |
| Erlotinib | −7.3034 | — | — | ||
| Osimertinib | — | −8.5638 | — | ||
| Vemurafenib | — | --- | −9.3319 | ||
Oxadiazoles docking scores in EGFR, EGFRT790M, and BRAFV600E active sites.
Remarkably, derivative 9b (R1 = Ph. and R2 = Cl) showed a higher docking score (S = −6.51 kcal/mol) over its methoxy congener (S = −6.04 kcal/mol) and has the best docking score over other p-chloro derivatives (9e, 9h, and 9k). Visual inspection of the docking pose with the lowest RMSD value and highest docking score of compound 9b, we observed a stabilizing H-bonding and H-pi binding interactions through N-phenyl and amidic carbonyl group of quinazoline ring with Lys721 amino acid residue, Figure 9. Such interactions were not found in other p-chloro derivatives, 9e and 9k (except with derivative 9h), because of the hydrophilic tale of Gln738 amino acid residue, which repels closely found non-hydrophilic groups as methyl group of 9e and ethyl group of 9k, as shown in Supplementary Figure S1.
FIGURE 9

Binding Interactions of 9b within EGFR (PDB ID: 1M17) active site showing both H-bonds and H-Pi interactions as green-dotted arrows and lines, respectively.
Whereas upon working on binding interactions within mutant EGFR (EGFRT790M; PDB ID: 2JIU) active site, the p-Cl derivative (9b) gave the highest docking score (S = −7.43 kcal/mol) among all 15 derivatives tested, as shown in Table 3. Most test derivatives commonly interacted through H-bonds and/or H-pi interactions with Lys 745 and Leu 718, as seen with docking poses of derivatives 9b and 9h, Figure 10.
FIGURE 10

Docking poses of compound 9b (top figure) and 9h (bottom figure) within the active site of EGFRT790M (PDB ID: 2JIU) showing H-Pi interactions with key amino acid residues.
Finally, docking scores of derivatives (9a-o) interactions within BRAF (PDB ID: 5JRQ) (Umar et al., 2020) active site were high and so close to each other (S = −6.24 to −7.65 kcal/mol). Additionally, multiple interactions varying from the weak Pi-Pi interactions, through H-Pi to the strong H-bonds with either Phe 583, Val 471, Asp 594, or Lys 483, as shown in Figure 11 of compound 9l.
FIGURE 11

2D-Binding interactions of compound 9l within BRAFV600E (PDB ID: 5JRQ) active site showing H-bonds (as blue-arrows) and pi-pi interactions (as green-dotted lines) with Asp 594 and Phe 583, respectively.
To summarize, all the 15 derivatives showed good binding profiles within target proteins EGFR, mutant EGFR (EGFRT790M), and BRAFV600E, as seen from their docking scores and interactions with amino acid residues lining their active sites, and this could be used to explain the possible mechanism by which such class of compounds inhibit these proteins activity.
2.4 Calculation of ADME properties
The drug-likeness of new compounds 9a-o was calculated using the SwissADME website (Daina et al., 2017) to predict their transport properties through membranes like GIT and/or BBB. All the test compounds obey Lipinski’s rule of five (RO5) with MLogP below 5, in addition to having both a topological polar surface area below 140 Å2 and molar refractivity below 130, indicating their facile transport through cell membranes and hence better oral bioavailability (F), as shown in Table 4.
TABLE 4
| Cpd | MW | Lipinski Parametersa | F | MR | Water Solubility Silicos-IT class |
GI absorption | BBB | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HBA | HBD | nrotb | TPSA | MLogP | |||||||
| 9a | 412.46 | 5 | 0 | 5 | 99.11 | 3.6 | 0.55 | 116.84 | Poor | High | No |
| 9b | 446.91 | 5 | 0 | 5 | 99.11 | 4.08 | 0.55 | 121.85 | Poor | High | No |
| 9c | 442.49 | 6 | 0 | 6 | 108.34 | 3.28 | 0.55 | 123.33 | Poor | High | No |
| 9d | 426.49 | 5 | 0 | 5 | 99.11 | 3.81 | 0.55 | 121.81 | Poor | High | No |
| 9e | 460.94 | 5 | 0 | 5 | 99.11 | 4.29 | 0.55 | 126.82 | Poor | High | No |
| 9f | 456.52 | 6 | 0 | 6 | 108.34 | 3.49 | 0.55 | 128.3 | Poor | High | No |
| 9g | 426.49 | 5 | 0 | 5 | 99.11 | 3.81 | 0.55 | 121.81 | Poor | High | No |
| 9h | 460.94 | 5 | 0 | 5 | 99.11 | 4.29 | 0.55 | 126.82 | Poor | High | No |
| 9i | 456.52 | 6 | 0 | 6 | 108.34 | 3.49 | 0.55 | 128.3 | Poor | High | No |
| 9j | 364.42 | 5 | 0 | 5 | 99.11 | 2.92 | 0.55 | 101.57 | Poor | High | No |
| 9k | 398.87 | 5 | 0 | 5 | 99.11 | 3.15 | 0.55 | 106.58 | Poor | High | No |
| 9l | 394.45 | 6 | 0 | 6 | 108.34 | 2.35 | 0.55 | 108.06 | Poor | High | No |
| 9m | 376.43 | 5 | 0 | 6 | 99.11 | 3.08 | 0.55 | 105.9 | Poor | High | No |
| 9n | 410.88 | 5 | 0 | 6 | 99.11 | 3.3 | 0.55 | 110.91 | Poor | High | No |
| 9o | 406.46 | 6 | 0 | 7 | 108.34 | 2.5 | 0.55 | 112.4 | Poor | High | No |
Theoretical calculations of ADME properties of compounds (9a-o) using swissADME software.
MW, molecular weight; HBA, H-bond acceptor; HBD, H-bond donor; nrotb, no. of rotatable bonds; TPSA, topological polar surface area (Å2); MLogP, n-octanol/water distribution coefficient; F, Abbott bioavailability scores (0–1).
Drug lead-like character: MW ≤ 500, HBA ≤10, HBD ≤5, nrotb ≤10, TPSA ≤140, lipophilicity parameter MLogP ≤5; MR, 40–130; BBB, Blood-Brain Barrier.
3 Conclusion
A novel set of quinazoline-4-one/1,2,4-oxadiazole hybrids (9a-o) was designed and synthesized as EGFR, EGFRT790M, and BRAFV600E inhibitors in the search for multitargeted antiproliferative scaffold. The novel hybrids showed encouraging antiproliferative actions. Compounds 9b, 9c, 9h, 9k, and 9l were evaluated as EGFR and BRAFV600E inhibitors. These in vitro experiments demonstrated that compounds 9b, 9c, and 9h are potent antiproliferative agents capable of acting as dual EGFR/BRAFV600E inhibitors. 9b, 9c, and 9h were further studied for their inhibitory effect on mutant EGFR (EGFRT790M), with the results indicating that the evaluated compounds had a significant inhibitory effect. Cell cycle analysis and apoptosis induction assay of 9b revealed cell cycle arrest at the G2/M phase, which can induce apoptosis. EGFR and BRAFV600E docking simulations inside their active regions shed light on these compounds’ possible modes of inhibition. ADME calculations revealed that all test compounds satisfy Lipinski’s rule of five (RO5) with MLogP <5, with easy transport through cell membranes and higher oral bioavailability. These new hybrids may have potential as anti-cancer drugs after optimization.
4 Experimental
4.1 Chemistry
4.1.1 General details
The starting materials, quinazolines 3a-e (Moussa et al., 2018) and 3-aryl-5-(chloromethyl)-1,2,4-oxadiazole derivatives, compounds 8a-c (Minin et al., 2023), were prepared according to literature methods.
4.1.2 General procedures for the synthesis of compounds (9a-o)
To a stirred solution of quinazoline derivatives (0.60 mmol, 1 eq), compounds 3a-e, in DMF (5 mL), anhydrous K2CO3 (0.72 mmol, 1.20 eq, 0.10 g) was added and stirred for 1h at room temperature. Then, 3-aryl-5-(chloromethyl)-1,2,4-oxadiazole derivatives, compounds 8a-c, (0.60 mmol, 1 eq.) was added, and KI (0.60 mmol, 1 eq, 0.10 g) was also added to the reaction mixture and stirring was continued for 24 h. After completion of the reaction (checked by TLC using Hexane: Ethyl acetate 2:1), the reaction mixture was poured into crushed ice while stirring. The obtained precipitate was filtered off, washed several times with water, dried at 60°C, and crystallized from ethanol.
4.1.3 2-((3-Phenyl-1,2,4-oxadiazol-5-yl)methylthio)-3-phenylquinazolin-4(3H)-one (9a)
Yield: 0.21 g (85%), White solid, m.p: 162°C–164°C, Rf. 0.66 (Hexane: Ethyl acetate, 2:1, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.07 (d, J = 7.7 Hz, 1H, Ar-H), 7.99 (d, J = 6.4 Hz, 2H, Ar-H), 7.81 (t, J = 7.4 Hz, 1H, Ar-H), 7.60 (d, J = 11.0 Hz, 4H, Ar-H), 7.57–7.52 (m, 4H, Ar-H), 7.47 (t, J = 8.0 Hz, 2H, Ar-H), 4.79 (s, 2H, S-CH2); 13C NMR (100 MHz, δ ppm DMSO-d6): 177.09, 167.84, 160.56, 155.46, 146.85, 135.49, 135.01, 131.59, 130.22, 129.67, 129.46, 129.25, 126.98, 126.61, 126.32, 126.03, 125.90, 119.60, 26.97; Anal. Calc. (%) For C23H16N4O2S: C, 66.97; H, 3.91; N, 13.58; S, 7.77. Found: C, 66.81; H, 3.85; N, 13.82; S, 7.85.
4.1.4 2-((3-(4-Chlorophenyl)-1,2,4-oxadiazol-5-yl)methylthio)-3 phenylquinazolin-4(3H)-one (9b)
Yield: 0.24 g (89%), White solid, m.p: 172°C–174°C, Rf. 0.67 (Hexane: Ethyl acetate, 2:1, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.07 (dd, J = 8.2, 1.5 Hz, 1H, Ar-H), 8.00 (d, J = 8.7 Hz, 2H, Ar-H p-Cl C6H4), 7.84–7.79 (m, 1H, Ar-H), 7.63 (d, J = 8.7 Hz, 2H, Ar-H p-Cl C6H4), 7.62–7.60 (m, 3H, Ar-H), 7.57–7.52 (m, 2H, Ar-H), 7.51–7.44 (m, 2H, Ar-H), 4.78 (s, 2H, S-CH2); 13C NMR (100 MHz, δ ppm DMSO-d6): 177.84, 167.52, 161.02, 155.92, 147.31, 136.82, 135.95, 135.47, 130.70, 130.14, 129.93, 129.89, 129.24, 127.08, 126.79, 126.36, 125.33, 120.08, 27.44; Anal. Calc. (%) For C23H15ClN4O2S: C, 61.81; H, 3.38; N, 12.54; S, 7.17. Found: C, 61.97; H, 3.50; N, 12.71; S, 7.28.
4.1.5 2-((3-(4-Methoxyphenyl)-1,2,4-oxadiazol-5-yl)methylthio)-3-phenylquinazolin-4(3H)-one (9c)
Yield: 0.23 g (88%), White solid, m.p: 186°C–188°C, Rf. 0.65 Hexane: Ethyl acetate, 2:1, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.07 (d, J = 7.7 Hz, 1H, Ar-H), 7.92 (d, J = 8.7 Hz, 2H, Ar-H p-OCH3 C6H4), 7.81 (t, J = 7.3 Hz, 1H, Ar-H), 7.62 (d, J = 3.9 Hz, 3H, Ar-H), 7.59–7.35 (m, 4H, Ar-H), 7.09 (d, J = 8.7 Hz, 2H, Ar-H p-OCH3 C6H4), 4.76 (s, 2H, S-CH2), 3.82 (s, 3H, OCH3); 13C NMR (100 MHz, δ ppm DMSO-d6): 176.64, 167.52, 161.71, 160.54, 155.46, 146.85, 135.48, 134.99, 130.20, 129.64, 129.45, 128.67, 126.60, 126.30, 125.89, 119.60, 118.28, 114.63, 55.36, 26.89; Anal. Calc. (%) For C24H18N4O3S: C, 65.14; H, 4.10; N, 12.66; S, 7.25. Found: C, 64.91; H, 4.27; N, 12.89; S, 7.23.
4.1.6 2-((3-Phenyl-1,2,4-oxadiazol-5-yl)methylthio)-3-p-tolylquinazolin-4(3H)-one (9d)
Yield: 0.22 g (86%), White solid, m.p: 168°C–170°C, Rf. 0.69 (Hexane: Ethyl acetate, 2:1, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.06 (d, J = 7.5 Hz, 1H, Ar-H), 7.98 (d, J = 6.5 Hz, 2H, Ar-H), 7.81 (t, J = 7.4 Hz, 3H, Ar-H), 7.59–7.52 (m, 4H, Ar-H), 7.47 (d, J = 6.1 Hz, 1H, Ar-H), 7.39 (q, J = 8.2 Hz, 2H, Ar-H), 4.76 (s, 2H, S-CH2), 2.42 (s, 3H, CH3); 13C NMR (100 MHz, δ ppm DMSO-d6): 177.12, 167.83, 160.62, 155.78, 146.87, 139.99, 135.01, 132.86, 131.63, 130.16, 129.29, 129.18, 126.99, 126.63, 126.32, 126.03, 125.89, 119.60, 26.96, 20.89; Anal. Calc. (%) For C24H18N4O2S: C, 67.59; H, 4.25; N, 13.14; S, 7.52. Found: C, 67.34; H, 4.43; N, 13.40; S, 7.68.
4.1.7 2-((3-(4-Chlorophenyl)-1,2,4-oxadiazol-5-yl)methylthio)-3-p-tolylquinazolin-4(3H)-one (9e)
Yield: 0.25 g (90%), White solid, m.p: 156°C–158°C, Rf. 0.70 (Hexane: Ethyl acetate, 2:1, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.05 (d, J = 6.9 Hz, 1H, Ar-H), 7.98 (d, J = 7.5 Hz, 2H, Ar-H p-Cl C6H4), 7.80 (t, J = 6.4 Hz, 1H, Ar-H), 7.62 (d, J = 7.5 Hz, 2H, Ar-H p-Cl C6H4), 7.45 (d, J = 7.5 Hz, 2H, Ar-H), 7.42–7.31 (m, 4H, Ar-H), 4.76 (s, 2H, S-CH2), 2.42 (s, 3H, CH3); 13C NMR (100 MHz, δ ppm DMSO-d6): 177.42, 167.06, 160.62, 155.76, 146.86, 140.00, 136.36, 135.02, 132.84, 130.17, 129.47, 129.17, 128.80, 126.62, 126.34, 125.89, 124.87, 119.59, 26.99, 20.90; Anal. Calc. (%) For C24H17ClN4O2S: C, 62.54; H, 3.72; N, 12.16; S, 6.96. Found: C, 62.37; H, 3.80; N, 12.42; S, 6.89.
4.1.8 2-((3-(4-Methoxyphenyl)-1,2,4-oxadiazol-5-yl)methylthio)-3-p-tolylquinazolin-4(3H)-one (9f)
Yield: 0.24 g (89%), White solid, m.p: 165°C–167°C, Rf. 0.68 (Hexane: Ethyl acetate, 2:1, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.06 (d, J = 7.6 Hz, 1H, Ar-H), 7.91 (d, J = 8.5 Hz, 2H, Ar-H p-OCH3 C6H4), 7.80 (t, J = 7.4 Hz, 1H, Ar-H), 7.48 (d, J = 7.8 Hz, 2H, Ar-H), 7.39 (q, J = 7.7 Hz, 4H, Ar-H), 7.09 (d, J = 8.5 Hz, 2H, Ar-H p-OCH3 C6H4), 4.74 (s, 2H, S-CH2), 3.82 (s, 3H, OCH3), 2.42 (s, 3H, CH3); 13C NMR (100 MHz, δ ppm DMSO-d6): 176.65, 167.54, 161.73, 160.62, 155.74, 146.87, 139.98, 134.97, 132.86, 130.15, 129.17, 128.68, 126.62, 126.29, 125.90, 119.59, 118.30, 114.65, 55.38, 26.90, 20.90; Anal. Calc. (%) For C25H20N4O3S: C, 65.77; H, 4.42; N, 12.27; S, 7.02. Found: C, 65.62; H, 4.61; N, 12.41; S, 6.98.
4.1.9 2-((3-Phenyl-1,2,4-oxadiazol-5-yl)methylthio)-3-m-tolylquinazolin-4(3H)-one (9g)
Yield: 0.22 g (86%), White solid, m.p: 170°C–172°C, Rf. 0.69 (Hexane: Ethyl acetate, 2:1, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.06 (dd, J = 8.2, 1.4 Hz, 1H, Ar-H), 7.98 (dd, J = 7.9, 1.7 Hz, 2H, Ar-H), 7.82–7.78 (m, 1H, Ar-H), 7.59–7.54 (m, 3H, Ar-H), 7.49–7.45 (m, 3H, Ar-H), 7.41 (d, J = 7.7 Hz, 1H, Ar-H), 7.31 (d, J = 8.7 Hz, 2H, Ar-H), 4.76 (s, 2H, S-CH2), 2.40 (s, 3H, CH3); 13C NMR (100 MHz, δ ppm DMSO-d6): 177.15, 167.86, 160.57, 155.56, 146.87, 139.34, 135.40, 135.07, 131.65, 130.91, 129.65, 129.49, 129.31, 127.01, 126.63, 126.44, 126.38, 126.04, 125.92, 119.60, 26.99, 20.79; Anal. Calc. (%) For C24H18N4O2S: C, 67.59; H, 4.25; N, 13.14; S, 7.52. Found: C, 67.36; H, 4.09; N, 13.41; S, 7.60.
4.1.10 2-((3-(4-Chlorophenyl)-1,2,4-oxadiazol-5-yl)methylthio)-3-m-tolylquinazolin-4(3H)-one (9h)
Yield: 0.25 g (90%), White solid, m.p: 178°C–180°C, Rf. 0.70 (Hexane: Ethyl acetate, 2:1, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.09 (d, J = 7.7 Hz, 1H, Ar-H), 8.02 (d, J = 8.6 Hz, 2H, Ar-H p-Cl C6H4), 7.84 (t, J = 8.4 Hz, 1H, Ar-H), 7.66 (d, J = 8.6 Hz, 2H, Ar-H p-Cl C6H4), 7.60–7.41 (m, 4H, Ar-H), 7.34 (d, J = 10.4 Hz, 2H, Ar-H), 4.79 (s, 2H, S-CH2), 2.43 (s, 3H, CH3); 13C NMR (100 MHz, δ ppm DMSO-d6): 177.37, 167.03, 160.50, 155.49, 146.81, 139.28, 136.33, 135.35, 134.99, 130.86, 129.61, 129.43, 129.23, 128.76, 126.58, 126.40, 126.31, 125.86, 124.85, 119.57, 26.95, 20.75; Anal. Calc. (%) For C24H17ClN4O2S: C, 62. 54; H, 3.72; N, 12.16; S, 6.96. Found: C, 62.39; H, 3.85; N, 12.40; S, 6.89.
4.1.11 2-((3-(4-Methoxyphenyl)-1,2,4-oxadiazol-5-yl)methylthio)-3-m-tolylquinazolin-4(3H)-one (9i)
Yield: 0.24 g (89%), White solid, m.p: 194°C–196°C, Rf .0.68 (Hexane: Ethyl acetate, 2:1, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.09 (d, J = 7.6 Hz, 1H, Ar-H), 7.95 (d, J = 8.7 Hz, 2H, Ar-H p-OCH3 C6H4), 7.84 (t, J = 7.3 Hz, 1H, Ar-H), 7.59–7.47 (m, 3H, Ar-H), 7.44 (d, J = 7.6 Hz, 1H, Ar-H), 7.34 (d, J = 10.8 Hz, 2H, Ar-H), 7.12 (d, J = 8.7 Hz, 2H, Ar-H p-OCH3 C6H4), 4.77 (s, 2H, S-CH2), 3.85 (s, 3H, OCH3), 2.43 (s, 3H, CH3); 13C NMR (100 MHz, δ ppm DMSO-d6): 176.66, 167.51, 161.72, 160.52, 155.52, 146.83, 139.28, 135.37, 135.00, 130.85, 129.62, 129.43, 128.67, 126.58, 126.41, 126.31, 125.89, 119.57, 118.28, 114.65, 55.37, 26.90, 20.75; Anal. Calc. (%) For C25H20N4O3S: C, 65.77; H, 4.42; N, 12.27; S, 7.02. Found: C, 65.62; H, 4.61; N, 12.49; S, 7.14.
4.1.12 2-((3-Phenyl-1,2,4-oxadiazol-5-yl)methylthio)-3-ethylquinazolin-4(3H)-one (9j)
Yield: 0.17 g (79%), White solid, m.p: 114°C–116°C, Rf. 0.60 (Hexane: Ethyl acetate, 2:1, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.04 (d, J = 8.0 Hz, 1H, Ar-H), 7.97 (d, J = 6.4 Hz, 2H, Ar-H), 7.74 (t, J = 7.4 Hz, 1H, Ar-H), 7.57–7.54 (m, 3H, Ar-H), 7.42 (t, J = 7.4 Hz, 1H, Ar-H), 7.35 (d, J = 8.4 Hz, 1H, Ar-H), 4.94 (s, 2H, S-CH2), 4.11 (q, J = 7.0 Hz, 2H, N-CH2), 1.31 (t, J = 7.0 Hz, 3H, CH3); 13C NMR (100 MHz, δ ppm DMSO-d6): 177.21, 167.84, 160.13, 154.45, 146.41, 134.73, 131.60, 129.27, 126.95, 126.38, 126.22, 126.00, 125.66, 118.84, 26.70, 13.04; Anal. Calc. (%) For C19H16N4O2S: C, 62.62; H, 4.43; N, 15.37; S, 8.80. Found: C, 62.89; H, 4.51; N, 15.62; S, 8.71.
4.1.13 2-((3-(4-Chlorophenyl)-1,2,4-oxadiazol-5-yl)methylthio)-3-ethylquinazolin-4(3H)-one (9k)
Yield: 0.20 g (84%), White solid, m.p: 118°C–120°C, Rf. 0.62 (Hexane: Ethyl acetate, 2:1, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.04 (dd, J = 7.9, 1.0 Hz, 1H, Ar-H), 7.98 (d, J = 8.6 Hz, 2H, Ar-H p-Cl C6H4), 7.75–7.72 (m, 1H, Ar-H), 7.60 (d, J = 8.6 Hz, 2H, Ar-H p-Cl C6H4), 7.44–7.41 (m, 1H, Ar-H), 7.35 (d, J = 8.1 Hz, 1H, Ar-H), 4.94 (s, 2H, S-CH2), 4.11 (q, J = 7.0 Hz, 2H, N-CH2), 1.31 (t, J = 7.0 Hz, 3H, CH3); 13C NMR (100 MHz, δ ppm DMSO-d6): 177.49, 167.05, 160.11, 154.42, 146.39, 136.35, 134.72, 129.43, 128.73, 126.37, 126.22, 125.64, 124.82, 118.83, 39.58, 26.67, 13.03; Anal. Calc. (%) For C19H15ClN4O2S: C, 57.21; H, 3.79; N, 14.05; S, 8.04. Found: C, 57.49; H, 3.86; N, 14. 27; S, 8.12.
4.1.14 2-((3-(4-Methoxyphenyl)-1,2,4-oxadiazol-5-yl)methylthio)-3-ethylquinazolin-4(3H)-one (9l)
Yield: 0.20 g (84%), White solid, m.p: 134°C–136°C, Rf. 0.57 (Hexane: Ethyl acetate, 2:1, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.04 (dd, J = 8.0, 1.3 Hz, 1H, Ar-H), 7.91 (d, J = 8.9 Hz, 2H, Ar-H p-OCH3 C6H4), 7.76–7.71 (m, 1H, Ar-H), 7.44–7.40 (m, 1H, Ar-H), 7.37 (d, J = 7.6 Hz, 1H, Ar-H), 7.07 (d, J = 8.9 Hz, 2H, Ar-H p-OCH3 C6H4), 4.91 (s, 2H, S-CH2), 4.11 (q, J = 7.1 Hz, 2H, N-CH2), 3.81 (s, 3H, OCH3), 1.31 (t, J = 7.1 Hz, 3H, CH3); 13C NMR (100 MHz, δ ppm DMSO-d6): 176.74, 167.52, 161.71, 160.11, 154.42, 146.40, 134.69, 128.63, 126.35, 126.9, 125.65, 118.83, 118.25, 114.63, 55.35, 39.56, 26.62, 13.01; Anal. Calc. (%) For C20H18N4O3S: C, 60.90; H, 4.60; N, 14.20; S, 8.13. Found: C, 61.13; H, 4.74; N, 14.37; S, 8.20.
4.1.15 2-((3-Phenyl-1,2,4-oxadiazol-5-yl)methylthio)-3-allylquinazolin-4(3H)-one (9m)
Yield: 0.18 g (80%), White solid, m.p: 110°C–112°C, Rf. 0.61(Hexane: Ethyl acetate, 2:1, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.04 (d, J = 7.8 Hz, 1H, Ar-H), 7.97 (d, J = 6.8 Hz, 2H, Ar-H), 7.75 (t, J = 7.5 Hz, 1H, Ar-H), 7.62–7.49 (m, 3H, Ar-H), 7.43 (t, J = 7.5 Hz, 1H, Ar-H), 7.37 (d, J = 8.1 Hz, 1H, Ar-H), 6.11–5.83 (m, 1H, =CH), 5.24 (d, J = 10.4 Hz, 1H, =CH2), 5.15 (d, J = 17.3 Hz, 1H, =CH2), 4.92 (s, 2H, S-CH2), 4.72 (d, J = 4.0 Hz, 2H, N-CH2); 13C NMR (100 MHz, δ ppm DMSO-d6): 177.20, 167.84, 160.19, 154.93, 146.44, 134.94, 131.64, 131.25, 129.30, 126.96, 126.53, 126.37, 126.01, 125.74, 118.73, 117.60, 45.97, 26.80; Anal. Calc. (%) For C20H16N4O2S: C, 63.81; H, 4.28; N, 14.88; S, 8.52. Found: C, 64.05; H, 4.42; N, 15.17; S, 8.64.
4.1.16 2-((3-(4-Chlorophenyl)-1,2,4-oxadiazol-5-yl)methylthio)-3-allylquinazolin-4(3H)-one (9n)
Yield: 0.21 g (85%), White solid, m.p: 115°C–117°C, Rf. 0.63 (Hexane: Ethyl acetate, 2:1, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.07 (dd, J = 8.0, 1.2 Hz, 1H, Ar-H), 7.99 (d, J = 8.7 Hz, 2H, Ar-H p-Cl C6H4), 7.80–7.74 (m, 1H, Ar-H), 7.63 (d, J = 8.7 Hz, 2H, Ar-H p-Cl C6H4), 7.49–7.43 (m, 1H, Ar-H), 7.39 (d, J = 8.2, 1H, Ar-H), 6.04–5.90 (m, 1H, =CH), 5.26 (dd, J = 10.4, 1.2 Hz, 1H, =CH2), 5.17 (dd, J = 17.2, 1.2 Hz, 1H, =CH2), 4.93 (s, 2H, S-CH2), 4.74 (d, J = 5.1 Hz, 2H, N-CH2); 13C NMR (100 MHz, δ ppm DMSO-d6): 177.39, 167.02, 160.10, 154.80, 146.37, 136.32, 134.81, 131.19, 129.38, 128.68, 126.47, 126.27, 125.68, 124.80, 118.69, 117.61, 45.92, 26.72; Anal. Calc. (%) For C20H15ClN4O2S: C, 58.46; H, 3.68; N, 13.64; S, 7.80. Found: C, 58.70; H, 3.73; N, 13. 91; S, 7. 94.
4.1.17 2-((3-(4-Methoxyphenyl)-1,2,4-oxadiazol-5-yl)methylthio)-3-allylquinazolin 4(3H)-one (9o)
Yield: 0.20 g (83%), White solid, m.p: 122°C–124°C, Rf. 0.59 (Hexane: Ethyl acetate, 2:1, v/v). 1H NMR (400 MHz, δ ppm DMSO-d6): 8.07 (d, J = 7.6 Hz, 1H, Ar-H), 7.92 (d, J = 8.7 Hz, 2H, Ar-H p-OCH3 C6H4), 7.78 (t, J = 7.2 Hz, 1H, Ar-H), 7.46 (t, J = 7.4 Hz, 1H, Ar-H), 7.40 (d, J = 8.4 Hz, 1H, Ar-H), 7.09 (d, J = 8.7 Hz, 2H, Ar-H p-OCH3 C6H4), 6.01–5.91 (m, 1H, =CH), 5.26 (d, J = 10.4 Hz, 1H, =CH2), 5.17 (d, J = 17.3 Hz, 1H, =CH2), 4.91 (s, 2H, S-CH2), 4.74 (d, J = 4.7 Hz, 2H, 2H, N-CH2), 3.82 (s, 3H, OCH3); 13C NMR (100 MHz, δ ppm DMSO-d6): 176.73, 167.53, 161.73, 160.16, 154.89, 146.43, 134.88, 131.24, 128.64, 126.51, 126.33, 125.74, 118.72, 118.28, 117.58, 114.66, 55.37, 45.93, 26.70; Anal. Calc. (%) For C21H18N4O3S: C, 62.05; H, 4.46; N, 13.78; S, 7.89. Found: C, 62.29; H, 4.53; N, 14.02; S, 7.81.
4.2 Biology
4.2.1 Assay of cell viability of 9a-o
The human mammary gland epithelial (MCF-10A) normal cell line was used to test the viability of compounds 9a-o (Mahmoud et al., 2022; Mekheimer et al., 2022). Refer to Supplementary Appendix A (Supplementary Material) for more details.
4.2.2 Assay of antiproliferative action
The MTT assay was used to investigate the antiproliferative activity of hybrids 9a-o versus four human cancer cell lines using Erlotinib as a control: colon cancer (HT-29) cell line, pancreatic cancer (Panc-1) cell line, lung cancer (A-549) cell line, and breast cancer (MCF-7) cell line (El-Sherief et al., 2019; Al-Wahaibi et al., 2022). See Supplementary Appendix A for more information.
4.2.3 EGFR inhibitory assay
The EGFR-TK test (Abdel-Aziz et al., 2023) assessed the inhibitory potency of the most effective antiproliferative derivatives 9b, 9c, 9h, 9k, and 9l against EGFR. For more details, see Supplementary Appendix A.
4.2.4 BRAFV600E inhibitory assay
An in vitro investigation assessed the anti-BRAFV600E activity of 9b, 9c, 9h, 9k, and 9l (Youssif et al., 2022). Refer to Supplementary Appendix A for more details.
4.2.5 EGFRT790M inhibitory assay
The HTRF KinEASE-TK assay (Miles et al., 2020) was used to evaluate the inhibitory action of the most potent hybrids, 9b, 9c, and 9h, against mutant-type EGFR (EGFRT790M). For more details, see Supplementary Appendix A.
4.2.6 Cell cycle analysis and apoptosis detection
Compound 9b was investigated for its effects on cell cycle progression and apoptosis induction in A-549 cells. A lung cancer (A-549) cell line was treated for 24 h with an IC50 concentration of 9b. The cell line was labeled with PI/Annexin V, and flow cytometry was done with a BD FASC Caliber (El-Sherief et al., 2018). See Supplementary Appendix A for more details.
4.3 Docking study
Molecular docking simulations of 15 derivatives (9a-o) were performed via Molecular Operating Environment (MOE®) software according to reported protocols (Abdel-Aziz et al., 2023) within the active site of EGFR tyrosine kinase domain (PDB ID: 1M17), mutant EGFR kinase domain T790M (EGFRT790M; PDB ID: 2JIU), and mutant BRAF kinase domain (BRAFV600E; PDB ID: 5JRQ) crystals structures downloaded from RSCB protein data bank (https://www.rcsb.org/). For more details, see Supplementary Appendix A.
4.4 Calculations of SwissADME
Pharmacokinetics and drug-likeness prediction for all the newly synthesized compounds was performed using the online tool SwissADME predictor software (http://www.swissadme.ch/) made by the Swiss Institute of Bioinformatics.
Statements
Data availability statement
Data will be available upon request from the authors.
Author contributions
AM: Formal Analysis, Methodology, Software, Writing–original draft. OA-G: Investigation, Supervision, Validation, Visualization, Writing–original draft. YM: Methodology, Supervision, Writing–original draft. KD: Funding acquisition, Writing–original draft. SB: Investigation, Validation, Visualization, Writing–review and editing. BY: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Researchers Supporting Project Number (RSP2024R388) King Saud University, Riyadh, Saudi Arabia. The authors also acknowledge support from the KIT-Publication Fund of the Karlsruhe Institute of Technology.
Acknowledgments
This work was funded by the Researchers Supporting Project Number (RSP2024R388) King Saud University, Riyadh, Saudi Arabia. The authors also acknowledge support from the KIT-Publication Fund of the Karlsruhe Institute of Technology.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2024.1447618/full#supplementary-material
References
1
Abdel-Aziz M. Beshr E. A. Gomaa H. A. Mostafa Y. A. Youssif B. G. Hayallah A. M. et al (2023). Discovery of new cyanopyridine/chalcone hybrids as dual inhibitors of EGFR/BRAFV600E with promising antiproliferative properties. Arch. Pharm.356 (4), 2200464. 10.1002/ardp.202200464
2
Alshammari M. B. Aly A. A. Youssif B. G. Bräse S. Ahmad A. Brown A. B. et al (2022). Design and synthesis of new thiazolidinone/uracil derivatives as antiproliferative agents targeting EGFR and/or BRAFV600E. Front. Chem.10, 1076383. 10.3389/fchem.2022.1076383
3
Al-Wahaibi L. H. Abou-Zied H. A. Beshr E. A. Youssif B. G. Hayallah A. M. Abdel-Aziz M. (2023a). Design, synthesis, antiproliferative actions, and DFT studies of new bis–pyrazoline derivatives as dual EGFR/BRAFV600E inhibitors. Int. J. Mol. Sci.24 (10), 9104. 10.3390/ijms24109104
4
Al-Wahaibi L. H. Abou-Zied H. A. Hisham M. Beshr E. A. Youssif B. G. Bräse S. et al (2023b). Design, synthesis, and biological evaluation of novel 3-cyanopyridone/pyrazoline hybrids as potential apoptotic antiproliferative agents targeting EGFR/BRAFV600E inhibitory pathways. Molecules28 (18), 6586. 10.3390/molecules28186586
5
Al-Wahaibi L. H. El-Sheref E. M. Hammouda M. M. Youssif B. G. (2023c). One-pot synthesis of 1-Thia-4-azaspiro [4.4/5] alkan-3-ones via schiff base: design, synthesis, and apoptotic antiproliferative properties of dual EGFR/BRAFV600E inhibitors. Pharmaceuticals16 (3), 467. 10.3390/ph16030467
6
Al-Wahaibi L. H. El-Sheref E. M. Hassan A. A. Bräse S. Nieger M. Youssif B. G. et al (2023d). Synthesis and structure determination of substituted thiazole derivatives as EGFR/BRAFV600E dual inhibitors endowed with antiproliferative activity. Pharmaceuticals16 (7), 1014. 10.3390/ph16071014
7
Al-Wahaibi L. H. Gouda A. M. Abou-Ghadir O. F. Salem O. I. Ali A. T. Farghaly H. S. et al (2020). Design and synthesis of novel 2, 3-dihydropyrazino [1, 2-a] indole-1, 4-dione derivatives as antiproliferative EGFR and BRAFV600E dual inhibitors. Bioorg. Chem.104, 104260. 10.1016/j.bioorg.2020.104260
8
Al-Wahaibi L. H. Mahmoud M. A. Mostafa Y. A. Raslan A. E. Youssif B. G. (2023e). Novel piperine-carboximidamide hybrids: design, synthesis, and antiproliferative activity via a multi-targeted inhibitory pathway. J. Enzyme Inhibition Med. Chem.38 (1), 376–386. 10.1080/14756366.2022.2151593
9
Al-Wahaibi L. H. Mostafa Y. A. Abdelrahman M. H. El-Bahrawy A. H. Trembleau L. Youssif B. G. (2022). Synthesis and biological evaluation of indole-2-carboxamides with potent apoptotic antiproliferative activity as EGFR/CDK2 dual inhibitors. Pharmaceuticals15 (8), 1006. 10.3390/ph15081006
10
Benassi A. Doria F. Pirota V. (2020). Groundbreaking anticancer activity of highly diversified oxadiazole scaffolds. Int. J. Mol. Sci.21 (22), 8692. 10.3390/ijms21228692
11
Bhat M. A. Tüzün B. Alsaif N. A. Khan A. A. Naglah A. M. (2022). Synthesis, characterization, molecular modeling against EGFR target and ADME/T analysis of novel purine derivatives of sulfonamides. J. Mol. Struct.1257, 132600. 10.1016/j.molstruc.2022.132600
12
Boran A. D. Iyengar R. (2010). Systems approaches to polypharmacology and drug discovery. Curr. Opin. drug Discov. & Dev.13 (3), 297–309.
13
Daina A. Michielin O. Zoete V. (2017). SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep.7 (1), 42717. 10.1038/srep42717
14
Derbyshire E. R. Mazitschek R. Clardy J. (2012). Characterization of Plasmodium liver stage inhibition by halofuginone. ChemMedChem7 (5), 844–849. 10.1002/cmdc.201200045
15
Do B. Mace M. Rexwinkle A. (2016). Idelalisib for treatment of B-cell malignancies. Am. J. health-system Pharm.73 (8), 547–555. 10.2146/ajhp150281
16
El Mansouri A.-E. Oubella A. Maatallah M. AitItto M. Y. Zahouily M. Morjani H. et al (2020). Design, synthesis, biological evaluation and molecular docking of new uracil analogs-1, 2, 4-oxadiazole hybrids as potential anticancer agents. Bioorg. & Med. Chem. Lett.30 (19), 127438. 10.1016/j.bmcl.2020.127438
17
El-Sherief H. A. Youssif B. G. Abdelazeem A. H. Abdel-Aziz M. Abdel-Rahman H. M. (2019). Design, synthesis and antiproliferative evaluation of novel 1, 2, 4-triazole/schiff base hybrids with EGFR and B-RAF inhibitory activities. Anti-Cancer Agents Med. Chem. Former. Curr. Med. Chemistry-Anti-Cancer Agents.19 (5), 697–706. 10.2174/1871520619666181224115346
18
El-Sherief H. A. Youssif B. G. Bukhari S. N. A. Abdelazeem A. H. Abdel-Aziz M. Abdel-Rahman H. M. (2018). Synthesis, anticancer activity and molecular modeling studies of 1, 2, 4-triazole derivatives as EGFR inhibitors. Eur. J. Med. Chem.156, 774–789. 10.1016/j.ejmech.2018.07.024
19
Finn R. S. Martin M. Rugo H. S. Jones S. Im S.-A. Gelmon K. et al (2016). Palbociclib and letrozole in advanced breast cancer. N. Engl. J. Med.375 (20), 1925–1936. 10.1056/nejmoa1607303
20
Flaherty K. T. Infante J. R. Daud A. Gonzalez R. Kefford R. F. Sosman J. et al (2012). Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N. Engl. J. Med.367 (18), 1694–1703. 10.1056/nejmoa1210093
21
Food U. Administration D. (1997). Center for drug evaluation and research, guidance for industry: dissolution testing of immediate release solid oral dosage forms.
22
Fu R.-g. Sun Y. Sheng W.-b. Liao D.-f. (2017). Designing multi-targeted agents: an emerging anticancer drug discovery paradigm. Eur. J. Med. Chem.136, 195–211. 10.1016/j.ejmech.2017.05.016
23
Hendawy O. M. (2022). A comprehensive review of recent advances in the biological activities of 1, 2, 4‐oxadiazoles. Arch. Pharm.355 (7), 2200045. 10.1002/ardp.202200045
24
Hisham M. Hassan H. A. Gomaa H. A. Youssif B. G. Hayalah A. M. Abdel-Aziz M. (2023). Design, synthesis, and antiproliferative activity of quinazolin-4-one/chalcone hybrids via the egfr inhibition pathway. Anti-Cancer Agents Med. Chem. Former. Curr. Med. Chemistry-Anti-Cancer Agents.23 (17), 1932–1943. 10.2174/1871520623666230727104933
25
Hisham M. Hassan H. A. Gomaa H. A. Youssif B. G. Hayallah A. M. Abdel-Aziz M. (2022). Structure-based design, synthesis and antiproliferative action of new quinazoline-4-one/chalcone hybrids as EGFR inhibitors. J. Mol. Struct.1254, 132422. 10.1016/j.molstruc.2022.132422
26
Keith C. T. Borisy A. A. Stockwell B. R. (2005). Multicomponent therapeutics for networked systems. Nat. Rev. Drug Discov.4 (1), 71–78. 10.1038/nrd1609
27
Kerru N. Gummidi L. Maddila S. Gangu K. K. Jonnalagadda S. B. (2020). A review on recent advances in nitrogen-containing molecules and their biological applications. Molecules25 (8), 1909. 10.3390/molecules25081909
28
Kitadai N. Maruyama S. (2018). Origins of building blocks of life: a review. Geosci. Front.9 (4), 1117–1153. 10.1016/j.gsf.2017.07.007
29
Loboda K. B. Valjavec K. Štampar M. Wolber G. Žegura B. Filipič M. et al (2020). Design and synthesis of 3, 5-substituted 1, 2, 4-oxadiazoles as catalytic inhibitors of human DNA topoisomerase IIα. Bioorg. Chem.99, 103828. 10.1016/j.bioorg.2020.103828
30
Mahmoud M. A. Mohammed A. F. Salem O. I. Gomaa H. A. Youssif B. G. (2022). New 1, 3, 4‐oxadiazoles linked with the 1, 2, 3‐triazole moiety as antiproliferative agents targeting the EGFR tyrosine kinase. Arch. Pharm.355 (6), 2200009. 10.1002/ardp.202200009
31
McLaughlin N. P. Evans P. Pines M. (2014). The chemistry and biology of febrifugine and halofuginone. Bioorg. & Med. Chem.22 (7), 1993–2004. 10.1016/j.bmc.2014.02.040
32
Meanwell N. (2017). A synopsis of the properties and applications of heteroaromatic rings in medicinal chemistry. Adv. Heterocycl. Chem., 245–361. 10.1016/bs.aihch.2016.11.002
33
Medina-Franco J. L. Giulianotti M. A. Welmaker G. S. Houghten R. A. (2013). Shifting from the single to the multitarget paradigm in drug discovery. Drug Discov. today18 (9-10), 495–501. 10.1016/j.drudis.2013.01.008
34
Mekheimer R. A. Allam S. M. Al-Sheikh M. A. Moustafa M. S. Al-Mousawi S. M. Mostafa Y. A. et al (2022). Discovery of new pyrimido [5, 4-c] quinolines as potential antiproliferative agents with multitarget actions: rapid synthesis, docking, and ADME studies. Bioorg. Chem.121, 105693. 10.1016/j.bioorg.2022.105693
35
Miles D. Chen Y. Paprcka S. L. Foley C. N. Grange R. Leleti M. R. et al (2020). Discovery and characterization of potent and selective AXL receptor tyrosine kinase inhibitors for cancer therapy. Eur. J. Cancer138, S38–S39. 10.1016/s0959-8049(20)31175-8
36
Minin D. Popkov S. Pesochinskaya K. Aleksanov D. (2023). Synthesis of [4-(3-aryl-1, 2, 4-oxadiazol-5-ylmethylthio)-2-methylphenoxy] acetic acids as new potential PPAR agonists. Russ. Chem. Bull.72 (7), 1648–1653. 10.1007/s11172-023-3944-5
37
Mondaca S. Lacouture M. Hersch J. Yaeger R. (2018). Balancing RAF, MEK, and EGFR inhibitor doses to achieve clinical responses and modulate toxicity in BRAF V600E colorectal cancer, 2. JCO precision oncol. 10.1200/PO.18.00088
38
Moussa G. Alaaeddine R. Alaeddine L. M. Nassra R. Belal A. S. Ismail A. et al (2018). Novel click modifiable thioquinazolinones as anti-inflammatory agents: design, synthesis, biological evaluation and docking study. Eur. J. Med. Chem.144, 635–650. 10.1016/j.ejmech.2017.12.065
39
Notarangelo T. Sisinni L. Condelli V. Landriscina M. (2017). Dual EGFR and BRAF blockade overcomes resistance to vemurafenib in BRAF mutated thyroid carcinoma cells. Cancer cell Int.17 (1), 86–89. 10.1186/s12935-017-0457-z
40
Okaniwa M. Hirose M. Imada T. Ohashi T. Hayashi Y. Miyazaki T. et al (2012). Design and synthesis of novel DFG-out RAF/vascular endothelial growth factor receptor 2 (VEGFR2) inhibitors. 1. Exploration of [5, 6]-fused bicyclic scaffolds. J. Med. Chem.55 (7), 3452–3478. 10.1021/jm300126x
41
Padwa A. Bur S. K. (2007). The domino way to heterocycles. Tetrahedron63 (25), 5341–5378. 10.1016/j.tet.2007.03.158
42
Paraiso K. Fedorenko I. Cantini L. Munko A. Hall M. Sondak V. et al (2010). Recovery of phospho-ERK activity allows melanoma cells to escape from BRAF inhibitor therapy. Br. J. cancer102 (12), 1724–1730. 10.1038/sj.bjc.6605714
43
Purcell J. W. Davis J. Reddy M. Martin S. Samayoa K. Vo H. et al (2010). Activity of the kinesin spindle protein inhibitor ispinesib (SB-715992) in models of breast cancer. Clin. Cancer Res.16 (2), 566–576. 10.1158/1078-0432.ccr-09-1498
44
Raghavendra N. M. Pingili D. Kadasi S. Mettu A. Prasad S. (2018). Dual or multi-targeting inhibitors: the next generation anticancer agents. Eur. J. Med. Chem.143, 1277–1300. 10.1016/j.ejmech.2017.10.021
45
Robert C. Karaszewska B. Schachter J. Rutkowski P. Mackiewicz A. Stroiakovski D. et al (2015). Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med.372 (1), 30–39. 10.1056/nejmoa1412690
46
Sharma V. Kamal R. Kumar V. (2017). Heterocyclic analogues as kinase inhibitors: a focus review. Curr. Top. Med. Chem.17 (22), 2482–2494. 10.2174/1568026617666170307113744
47
Umar A. B. Uzairu A. Shallangwa G. A. Uba S. (2020). QSAR modelling and molecular docking studies for anti-cancer compounds against melanoma cell line SK-MEL-2. Heliyon6 (3), e03640. 10.1016/j.heliyon.2020.e03640
48
Unadkat V. Rohit S. Parikh P. Sanna V. Singh S. (2021). Rational design-aided discovery of novel 1, 2, 4-oxadiazole derivatives as potential EGFR inhibitors. Bioorg. Chem.114, 105124. 10.1016/j.bioorg.2021.105124
49
Wahid M. Jawed A. Mandal R. K. Dar S. A. Akhter N. Somvanshi P. et al (2018). Recent developments and obstacles in the treatment of melanoma with BRAF and MEK inhibitors. Crit. Rev. oncology/hematology125, 84–88. 10.1016/j.critrevonc.2018.03.005
50
Yan G. Zhang Y. Wang J. (2017). Recent advances in the synthesis of aryl nitrile compounds. Adv. Synthesis & Catal.359 (23), 4068–4105. 10.1002/adsc.201700875
51
Youssif B. G. Gouda A. M. Moustafa A. H. Abdelhamid A. A. Gomaa H. A. Kamal I. et al (2022). Design and synthesis of new triarylimidazole derivatives as dual inhibitors of BRAFV600E/p38α with potential antiproliferative activity. J. Mol. Struct.1253, 132218. 10.1016/j.molstruc.2021.132218
52
Youssif B. G. Mohamed M. F. Al-Sanea M. M. Moustafa A. H. Abdelhamid A. A. Gomaa H. A. (2019). Novel aryl carboximidamide and 3-aryl-1, 2, 4-oxadiazole analogues of naproxen as dual selective COX-2/15-LOX inhibitors: design, synthesis and docking studies. Bioorg. Chem.85, 577–584. 10.1016/j.bioorg.2019.02.043
53
Zhang Q. Diao Y. Wang F. Fu Y. Tang F. You Q. et al (2013). Design and discovery of 4-anilinoquinazoline ureas as multikinase inhibitors targeting BRAF, VEGFR-2 and EGFR. MedChemComm4 (6), 979–986. 10.1039/c3md00096f
54
Zhou J. Jiang X. He S. Jiang H. Feng F. Liu W. et al (2019). Rational design of multitarget-directed ligands: strategies and emerging paradigms. J. Med. Chem.62 (20), 8881–8914. 10.1021/acs.jmedchem.9b00017
Summary
Keywords
quinazolinone, oxadiazole, kinases, apoptosis, antiproliferative, EGFR, BRAF
Citation
Mohamed AM, Abou-Ghadir OMF, Mostafa YA, Dahlous KA, Bräse S and Youssif BGM (2024) Design and synthesis of new 1,2,4-oxadiazole/quinazoline-4-one hybrids with antiproliferative activity as multitargeted inhibitors. Front. Chem. 12:1447618. doi: 10.3389/fchem.2024.1447618
Received
11 June 2024
Accepted
05 August 2024
Published
30 August 2024
Volume
12 - 2024
Edited by
Xuekui Xia, Biology Institute of Shandong Academy of Sciences, China
Reviewed by
Chao Liu, Shandong University, China
Ravikumar Jimmidi, Baylor College of Medicine, United States
Updates
Copyright
© 2024 Mohamed, Abou-Ghadir, Mostafa, Dahlous, Bräse and Youssif.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bahaa G. M. Youssif, bgyoussif2@gmail.com, bahaa.youssif@pharm.aun.edu.eg; Stefan Bräse, braese@kit.edu
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.