Abstract
Volatile esters are key flavor components in most plants, including Lavandula x intermedia (lavandin). The final step in ester biosynthesis is catalyzed by Lavandula x intermedia alcohol acyltransferases (LiAATases), which attach alcohols to acyl groups. However, the functional role and mechanism of LiAATases remain poorly understood. Here, we predicted their structural models using AlphaFold2 and identified potential active site residues through the GalaxyWEB program. Catalytic assays were optimized at pH 7.5 and 30 °C. Substrate specificity for alcohols was assessed for both enzymes. Gene expression analysis revealed that LiAATase1 and LiAATase2 were most highly expressed in the petals and pistils, respectively, with peak expression occurring at stage 4 for LiAATase1 and stage 1 for LiAATase2. Our study aims to elucidate the functional properties of alcohol acyltransferases in Lavandula x intermedia, contributing to an understanding of ester biosynthesis and specificity in this species.
Introduction
Esters are a key class of volatile compounds that play a significant role in the formation of plant aromas (Liu et al., 2023; Aschariyaphotha et al., 2024). Alcohol acyltransferase (AATase) catalyzes the transfer of an acyl group from acyl-CoA to an alcohol, resulting in ester production (Aharoni et al., 2004; Defilippi et al., 2005; Seixas et al., 2023). This enzyme plays a critical role in the biosynthesis of volatile esters in various plant species, including climacteric fruits such as melons, bananas, and apples, as well as non-climacteric plants like strawberries, passionfruit, pineapple, and certain flowers like Gypsophila, Clarkia breweri, and Karawek (Beekwilder et al., 2004; Canessa et al., 2004; Defilippi et al., 2005; Guterman et al., 2006; Khanom and Ueda, 2008; Mascarell-Creus et al., 2009; de Jong et al., 2010; Shah et al., 2012; Ghezzi et al., 2017; Moon et al., 2021; Wang et al., 2021; Liu et al., 2023; Saez et al., 2024).
Plant lavender essential oils (EOs) primarily consist of volatile monoterpenes and sesquiterpenes, which may include alcohols (Patrignani et al., 2021; Wani et al., 2021; Bavarsad et al., 2023; Liu et al., 2024a; Liu et al., 2025c). These oils serve various physiological and ecological functions, such as allelopathy, plant defense, and pollinator attraction (Benabdelkader et al., 2011; Ebadollahi et al., 2020; Karpiński, 2020). Additionally, lavender EOs possess considerable economic importance due to their extensive applications in cosmetics, personal care products and alternative medicine (Lesage-Meessen et al., 2015; de Groot and Schmidt, 2016a; de Groot and Schmidt, 2016b). The enzyme Lavandula x intermedia alcohol acetyltransferase (LiAATase) is essential for the biosynthesis of volatile esters by catalyzing the transfer of acyl groups from acyl-CoA to alcohols. LiAATase is part of a larger enzyme family responsible for alcohol acetylation. Despite its importance in aroma production, the function and mechanism of LiAATases in lavender remain poorly understood.
Herein, we used the NPS@ server to predict the secondary structures of LiAATase1 and LiAATase2, and structural models were generated using AlphaFold2. The GalaxyWEB program was employed to identify potential active site residues. Optimal catalytic conditions for both LiAATase1 and LiAATase2 were determined to be pH 7.5 at 30 °C. Alcohol substrate specificities for both enzymes were assessed. Gene expression analysis revealed that LiAATase1 and LiAATase2 exhibited the highest expression in the petals and pistils, respectively. Furthermore, peak expression of LiAATase1 occurred at stage 4, while LiAATase2 showed highest expression at stage 1, among the five developmental stages analyzed. This study aimed to characterize alcohol acetyltransferases in Lavandula x intermedia, contributing to the understanding of their role in ester biosynthesis and specificity.
Results
Secondary structure prediction of LiAATase1 and LiAATase2
Using the amino acid sequences of LiAATase1 (UniProt code A0A0K0LCG5) and LiAATase2 (UniProt code A0A0K0LBP0) (Figure 1), we predicted their secondary structures via the NPS@ server (Combet et al., 2000). The predicted secondary structures of LiAATase1 and LiAATase2 consist of alpha helices (37.17% and 33.78% of residues, respectively), along with multiple strands and coils (Figure 2). LiAATase1 contains 155 residues in helices, while LiAATase2 has 151.
FIGURE 1

Sequence alignment of alcohol acetyltransferase family. The sequence alignment employs the ClustalW default color scheme, where conserved amino acids are highlighted with more intense colors than non-conserved residues. The reference proteins included in the alignment are as follows: A0A0K0LBP0, Lavandula x intermedia (Lavandin, Lavandula angustifolia x Lavandula latifolia); D0QJ94, Vasconcellea cundinamarcensis (Mountain papaya, Carica candamarcensis); Q64FJ6, Malus domestica (Apple, Pyrus malus); Q6QLX4, Solanum lycopersicum (Tomato, Lycopersicon esculentum); P0DO25, Actinidia deliciosa (Kiwi); A0A0K0LCG5, Lavandula x intermedia (Lavandin, Lavandula angustifolia x Lavandula latifolia); Q9FVF1, Fragaria ananassa (Strawberry, Fragaria chiloensis x Fragaria virginiana).
FIGURE 2

Prediction of secondary structure models of LiAATases (Lavandula x intermedia alcohol acyltransferases). The secondary structure predictions for LiAATase1 and LiAATase2 are shown in (a,b) respectively.
Prediction and quality assessment of LiAATase1 and LiAATase2 structures
The three-dimensional (3D) structures of LiAATase1 and LiAATase2 were predicted using AlphaFold2 (Jumper et al., 2021; Wayment-Steele et al., 2023), a deep learning-based tool that provides highly accurate and reliable protein structure predictions, surpassing traditional homology modeling methods.
To evaluate the quality of the predicted structures (Figures 3a,d), we employed the Ramachandran plot to analyze the dihedral angles of the protein backbones, ensuring they fell within acceptable regions, which indicates a valid protein conformation. For LiAATase1, 88.7% of the residues were in the most favored region, 10.8% in the additionally allowed region, 0.5% in the generously allowed region, and none in the disallowed region (Figure 3b; Table 1). For LiAATase2, 90.5% of residues were in the most favored region, 9.2% in the additionally allowed region, 0.3% in the generously allowed region, and none in the disallowed region (Figure 3e; Table 1).
FIGURE 3

Structural prediction and quality assessment of LiAATase1 and LiAATase2. The three-dimensional (3D) structures of (a) LiAATase1 and (d) LiAATase2 were predicted using AlphaFold2. Both models are depicted as cyan ribbon diagrams from two distinct orientations, with α-helices in pink and β-sheets in cyan. Structural validation was performed using Ramachandran plot analysis [(b) for LiAATase1, (e) for LiAATase2], where the most favorable residue conformations are highlighted in red, and less favorable regions are shown in progressively lighter shades. Additionally, (c, f) ProSA analysis yielded Z-scores of −10.90 (LiAATase1) and −5.68 (LiAATase2), confirming the high quality of the predicted models.
TABLE 1
| Constructs | Residues in most favored regions | Residues in additional allowed regions | Residues in generously allowed regions | Residues in disallowed regions | ||||
|---|---|---|---|---|---|---|---|---|
| Residues | Number of residues | % of residues | Number of residues | % of residues | Number of residues | % of residues | Number of residues | % of residues |
| LiAATase1a | 338 | 88.7 | 41 | 10.8 | 2 | 0.5 | 0 | 0 |
| LiAATase2b | 353 | 90.5 | 36 | 9.2 | 1 | 0.3 | 0 | 0 |
Ramchandran plot analysis of structural models of the two alcohol acetyltransferases using PDBsum.
Number of end-residues (excl. Gly and Pro): 2; Number of glycine residues (shown as triangles): 13; Number of proline residues: 21.
Number of end-residues (excl. Gly and Pro): 2; Number of glycine residues (shown as triangles): 28; Number of proline residues: 27.
ProSA analysis of the models showed Z-scores of −10.90 for LiAATase1 and -10.79 for LiAATase2 (Figures 3c,f). Both Z-scores were well below −10.00, demonstrating excellent model quality. However, the slight difference between the two Z-scores does not suggest a significant structural discrepancy in the model.
Although the overall fold of LiAATase1 closely resembles that of LiAATase2 (Figure 4), the root mean square deviation (RMSD) for all atoms was 2.25 Å, and the sequence identity was 27.47% (Figure 4).
FIGURE 4

Comparative structural analysis of LiAATase1 (in magenta) and LiAATase2 (in cyan). LiAATase1 adopted an overall fold similar to that of LiAATase2, but the root mean square deviation (RMSD) value for all atoms was large (2.25 Å), and the amino acid sequence identity was only 27.47%.
Predicting LiAATase1 and LiAATase2 active sites
Using the predicted models (Figures 3a,d, 4, 5a,b), we employed the GalaxyWEB program (Ko et al., 2012; Heo et al., 2013; Heo et al., 2016; Seok et al., 2021) to identify the active sites of LiAATase1 and LiAATase2. The analysis revealed that the active site residues of LiAATase1 include I156, R225, P238, S239, R240, V241, H273, A274, V275, N276 (Figures 5a,c). For LiAATase2, the active site residues are T254, S256, K257, F258, N285, T286, V287, N288, K334, and D368 (Figures 5b,c). These residues are likely involved in interactions with the alcohol substrate, potentially forming bonds with the substrate’s side chain atoms.
FIGURE 5
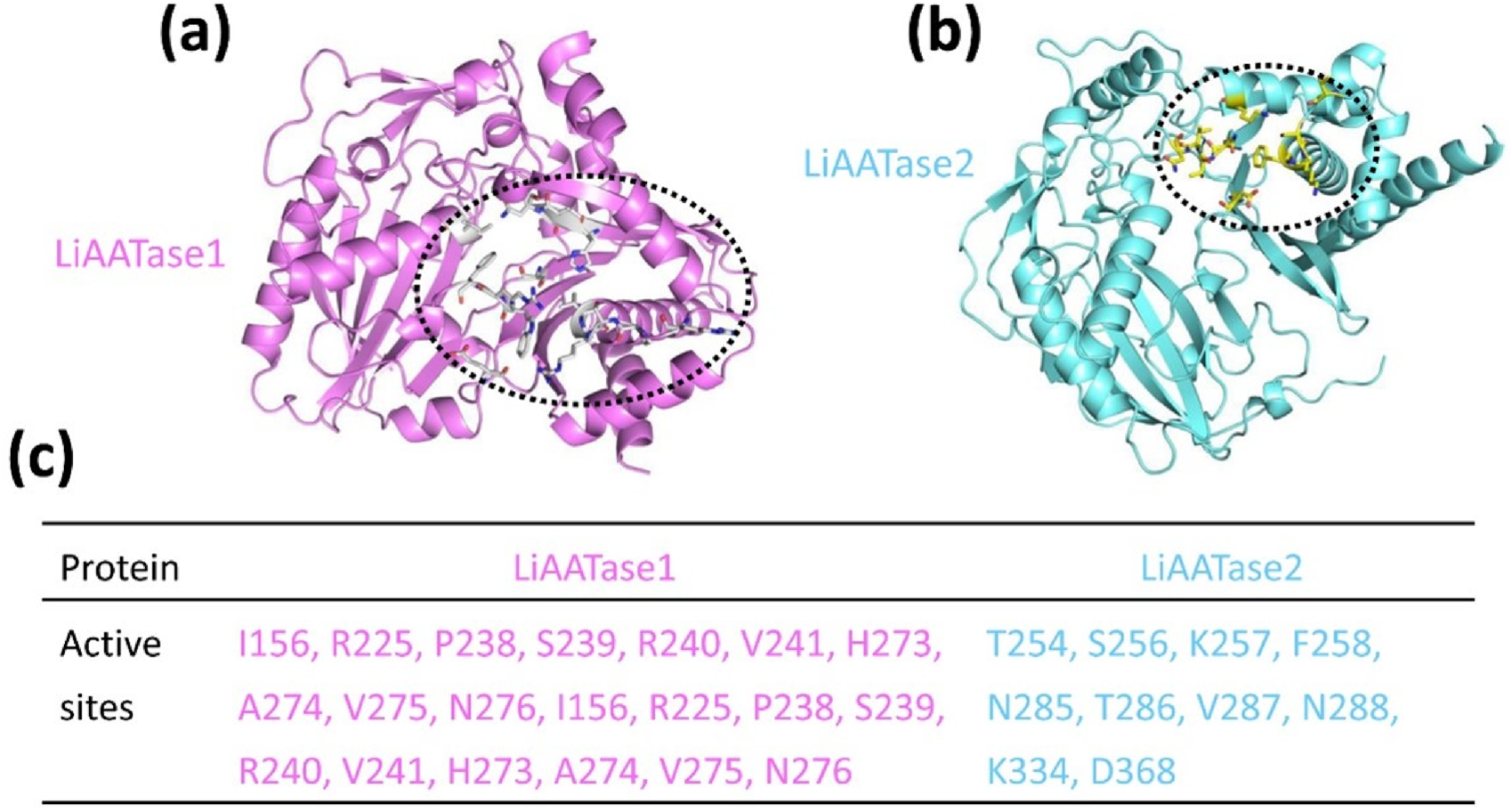
Predicting (a) LiAATase1 and (b) LiAATase2 active site residues using the GalaxyWEB program. LiAATase1 and LiAATase2 were colored in magenta and cyan, respectively. (c) The residues in the active site of LiAATase1 and LiAATase2.
Effects of temperature and pH on the activity of LiAATase1 and LiAATase2
To determine the optimal conditions for alcohol acetyltransferase activity of LiAATase1 and LiAATase2, we examined the effects of temperature and pH on enzyme activity (Figure 6). Activity was tested across a temperature range of 21–39°C, revealing an increase in activity up to 30°C, after which it declined (Figures 6a,b). The enzymes showed optimal activity within a pH range of 5.5–9.5, with peak activity occurring at pH 7.5 before decreasing (Figures 6c,d). Based on these results, we selected pH 7.5 and 30 °C as the optimal conditions for further activity assessments.
FIGURE 6

The effects of temperature and pH on the relative activity (%) of LiATTase1 and LiATTase2. Both enzymes exhibited maximal activity at 30 °C (a,b). Similarly, their optimal pH was 7.5 (c,d).
Alcohol substrate specificities of LiAATase1 and LiAATase2
Alcohol acetyltransferases are well-known for their role in producing acetate esters and are often associated with ethylene-dependent or ripening-specific processes in many plants (Beekwilder et al., 2004; Defilippi et al., 2005; Aschariyaphotha et al., 2024). The activity of LiAATases was determined under the optimal conditions (pH 7.5 at 30 °C). We found that both LiAATase1 and LiAATase2 demonstrated high activity with medium-chain alcohols, particularly 1-hexanol, while showing minimal activity with short-chain alcohols like methanol and ethanol. The enzyme activity increased with the length of the alcohol chain, with 1-hexanol exhibiting 4.1 to 5.5 times higher activity than methanol (Figure 7). This trend may be attributed to alcohol acyltransferase’s preference for utilizing acyl-CoA residues, as the enzyme tends to react more readily with acyl-CoA substitutes other than acetyl-CoA.
FIGURE 7

Specific activity and relative activity of LiAATase1 and LiATTase2 using different alcohols. The catalytic activity of both enzymes was determined under pH 7.5 and 30 °C.
Spatial and temporal analyses of genes LiAATase1 and LiAATase2
To establish a detailed spatial and temporal expression profile, RT-qPCR (real-time quantitative polymerase chain reaction) was used to quantify the expression levels of the target genes LiAATase1 and LiAATase2 across different flower organs and developmental stages. The results showed that LiAATase1 expression was highest in the petals (Figure 8a), while LiAATase2 expression peaked in the pistils (Figure 8b). Regarding developmental stages, LiAATase1 expression was greatest at stage 4 (Figure 8c), whereas LiAATase2 expression was highest at stage 1 (Figure 8d). This tissue- and stage-specific expression pattern highlights the significant role of LiAATase1 and LiAATase2 in lavender essential oil biosynthesis, with flower tissues being the primary site of expression.
FIGURE 8

Gene expression profiles of LiAATase1 and LiAATase2. (a,b) Expression levels of genes LiAATase1 and LiAATase2 in different flower organs (petals, stamens, pistils and sepals). (c,d) Expression levels of LiAATase1 and LiAATase2 genes during different flowering stages. Flower development was classified into five stages, i.e., flowers with tightly closed buds (stage 1), flowers with slightly open buds (stage 2), flowers beginning to open (stage 3), flowers in full bloom (stage 4), and flowers completely wilted (stage 5). The relative expression levels of genes LiAATase1 and LiAATase2 were quantified using RT-qPCR (real-time quantitative polymerase chain reaction). Expression ratios are shown as log2 values, and values above zero indicate upregulation of gene expression.
Discussion
In this work, the secondary structures of LiAATase1 and LiAATase2 were predicted using the NPS@ server, and their structural models were generated with AlphaFold2. Active site residues were identified through the GalaxyWEB program. Optimal catalytic conditions for both enzymes were determined to be pH 7.5 and 30 °C. Substrate specificity assays revealed the alcohol preferences of LiAATase1 and LiAATase2. Gene expression analysis indicated that LiAATase1 was most highly expressed in petals, while LiAATase2 showed peak expression in pistils. Additionally, LiAATase1 had its highest expression at stage 4, whereas LiAATase2 peaked at stage 1 among the five developmental stages examined. This study highlights key characteristics of alcohol acetyltransferases in lavender, offering insights into their roles in ester biosynthesis and specificity in Lavandula x intermedia.
Alcohol acetyltransferase (EC 2.3.1.84) plays a crucial role in aroma biosynthesis by catalyzing the formation of esters from acyl-CoA and alcohols (Beekwilder et al., 2004; Defilippi et al., 2005; Khanom and Ueda, 2008). Despite their low sequence identity, alcohol acetyltransferases share highly conserved three-dimensional structures, with partially conserved active sites. These active sites bind acyl-CoA and alcohol, generally featuring an HXXXXD motif that forms a substrate reaction channel (Bayer et al., 2004; Galaz et al., 2013; Zheng et al., 2016). These differ from LiAATases, suggesting that alcohol acetyltransferases may employ distinct catalytic mechanisms. To further investigate these, we are exploring the structural and mechanistic features of LiAATase-catalyzed reactions using experimental approaches such as crystallography. This structural arrangement allows independent binding of the substrate and co-substrate, catalyzing C-O bond formation and promoting the synthesis of corresponding ester compounds (Bayer et al., 2004; Ma et al., 2005; Morales-Quintana et al., 2011; Cumplido-Laso et al., 2012; Morales-Quintana et al., 2012; Galaz et al., 2013; Morales-Quintana et al., 2013; Morales-Quintana et al., 2015; Zheng et al., 2016; Liu et al., 2023; Saez et al., 2024). These enzymes are capable of pairing various alcohols with acyl-CoA, resulting in the production of a diverse array of esters, which contributes to the complexity of ester profiles (Horton et al., 2003; Guterman et al., 2006; Lilly et al., 2006). The alcohol component of an ester corresponds to the alcohols primarily synthesized in the plant, while the acyl component reflects the specificity of the alcohol acetyltransferases for different acyl-CoA molecules. For instance, strawberry alcohol acetyltransferase shows strong activity with hexanol and both acetyl- and butyl-CoA, while banana alcohol acetyltransferase is highly active with butanol and acetyl-CoA but less so with butyl-CoA (Zhang et al., 2014; Hu et al., 2016). Therefore, alcohol acetyltransferases from different plants exhibit unique substrate specificities for both alcohols and acyl-CoA.
Additionally, alcohol acetyltransferase has also been identified in the native Californian flower C. breweri, where it plays a key role in the esterification of benzyl alcohols within the flower (Defilippi et al., 2005; Wang et al., 2021; Liu et al., 2023). A strong affinity for aromatic alcohols, including benzyl alcohol and cinnamyl alcohol. Benzyl acetate, a significant compound in the fragrance industry, is a prominent aroma in this flower. It is the primary component of jasmine and gardenia essential oils and is commonly used as a minor constituent in various other oils.
The engineering of volatile emissions in plants has primarily focused on terpenoids. For example, a tomato variety with low endogenous linalool levels in the fruit was transformed with C. breweri linalool synthase under the control of the fruit-specific E8 promoter (Beekwilder et al., 2004). This led to significantly elevated levels of S-linalool in the fruit. The same gene was introduced into carnation flowers to induce linalool emission. However, overexpression of C. breweri linalool synthase in carnations resulted in the accumulation of linalool as a nonvolatile glucopyranoside conjugate, suggesting that endogenous enzymes in petunia sequester volatile linalool as a nonvolatile form (Moon et al., 2021; Lee and Trinh, 2022). In Arabidopsis leaves constitutively expressing a strawberry linalool synthase, linalool production occurred alongside its glycosylated and hydroxylated derivatives. Furthermore, overexpression of alcohol dehydrogenase in tomato fruit increased the levels of hexanol and cis-3-hexenol, at the expense of aldehyde production (Beekwilder et al., 2004).
In conclusion, our study presents a novel approach to comprehensively explore the functional mechanisms of alcohol acetyltransferases in Lavandula x intermedia, aiming to improve the quality of lavender essential oils.
Materials and methods
Bioinformatics analysis
The amino acid sequences of LiAATase1 (UniProt code A0A0K0LCG5) and LiAATase2 (UniProt code A0A0K0LBP0) were analyzed using the ProtParam online server (https://web.expasy.org/protparam/) to predict their chemical properties and physicochemical parameters.
Prediction of structural models
Structural predictions for LiAATase1 and LiAATase2 were performed using the AlphaFold2 program (Jumper et al., 2021; Wayment-Steele et al., 2023). Secondary structures were predicted with the NPS@ server (Combet et al., 2000), while active site residues were identified using the GalaxyWEB program (Ko et al., 2012; Heo et al., 2013; Heo et al., 2016; Seok et al., 2021). Multiple sequence alignment data were obtained from the LSQKAB program within the CCP4 suite (Collaborative Computational Project, 1994), and the root mean square deviation (RMSD) for Cα atoms was calculated. Structural images were generated using PyMOL 2.3.4 (https://www.pymol.org/2/).
Quality assessment of structural models
To validate the tertiary structures, Ramachandran plots for LiAATase1 and LiAATase2 were generated using the PDBsum database (Laskowski, 2004; de Beer et al., 2014; Laskowski et al., 2017; Laskowski, 2022). This tool assesses protein structure quality by detecting geometric errors, thereby improving the accuracy of the models. The Ramachandran plot specifically evaluates the stereochemical properties of the structures by displaying the dihedral angles of amino acid residues, highlighting the allowed conformational regions and identifying disallowed orientations.
Additionally, ProSA (Protein Structure Analysis) is a widely used tool for analyzing and validating predicted protein models (Wiederstein and Sippl, 2007). The z-score reflects overall model quality and is plotted against the z-scores of all experimentally determined protein chains in the current PDB. The plot distinguishes between structure groups (e.g., X-ray, NMR) using color-coding. This allows assessment of whether the input structure’s z-score falls within the expected range for native proteins of comparable size.
Protein isolation and purification
LiAATase1 and LiAATase2 were isolated and purified with slight modifications to previously described methods (Egea-Cortines et al., 2019; Nazeer et al., 2019). Fresh flowers were first washed with tap water, then with distilled water, and dried in the shade for 5–6 days at room temperature. The dried samples were powdered in liquid nitrogen using a mortar and pestle. Total protein was extracted using a Tris-buffer saline solution (150 mM NaCl, 20 mM Tris-HCl, PVP, pH 7.4), with a slight adjustment to the extraction buffer ratio (1:7, w/v). The sample was filtered through three layers of muslin cloth, and the filtrate was stirred magnetically at 4 °C overnight. The sample was then centrifuged at 18,000 rpm for 30 min at 4°C, and the supernatant was collected while discarding the pellet.
For protein purification, the crude extract was precipitated using varying ammonium sulfate saturation levels (20%, 35%, 55%, 75%, and 90%). A 75% saturated ammonium sulfate solution yielded the best quality pellet after centrifugation at 18,000 rpm for 30 min at 4 °C. The supernatant was then precipitated with 75% ammonium sulfate saturation and stored at −20 °C overnight to ensure complete protein precipitation. The following morning, the sample was centrifuged again at 18,000 rpm for 30 min at 4°C, with the supernatant discarded. The resulting pellet was washed six times with acetone and then dried. The protein was dialyzed against distilled water for 12 h at 4°C, and purified using size exclusion chromatography (SEC).
Enzymatic activity assays
Enzyme assays were conducted in 25-mL glass syringes with Luer lock caps, following a previously described method with minor modifications (Hu et al., 2016; Stribny et al., 2016; Egea-Cortines et al., 2019). The reaction mixtures contained a glycerol buffer (50 mM potassium phosphate, pH 7.5, 10% (w/v) glycerol), which included higher alcohol as a co-substrate, acetyl-CoA as the second co-substrate, and a cell extract. The volume ratio of higher alcohol, acetyl-CoA, and cell extract was 10:1:4, with a final reaction volume of 1.5 mL. Isoamyl alcohol (0.01–100 mM final concentration), isobutanol (60 mM), or 2-phenylethanol (30 mM) were used as substrates, along with 0.8 mM acetyl-CoA. Substrate concentrations were carefully measured. After adding all components, entrapped air was removed using the plunger, and the syringe was attached to an orbital shaker. Following a 30-min incubation, 1.5-mL samples were transferred to 15-mL vials containing 0.35 g NaCl for ester quantification via gas chromatography. Enzyme activity was terminated by adding 60 mL of a saturated KSCN solution.
RT-qPCR analysis of gene expression levels
Gene expression of LiAATase1 and LiAATase2 was quantified using real-time quantitative polymerase chain reaction (RT-qPCR) with PowerUp SYBR Green Master Mix (Applied Biosystems). Total RNA was extracted from various developmental stages and tissues using the Universal Plant Total RNA Extraction Kit (Bioteke, Beijing, China) according to the manufacturer’s instructions. cDNA was synthesized from the RNA samples using the PrimeScript first Strand cDNA Synthesis Kit (Takara, Kyoto, Japan). The primers used in the experiments are listed in Table 2. RT-qPCR analysis was performed on an Applied Biosystems QuantStudio five instrument. Data were analyzed using the 2−ΔΔCT method (Livak and Schmittgen, 2001; Schmittgen and Livak, 2008; Hawkins and Guest, 2017; Green and Sambrook, 2018; Liu et al., 2024a; Liu et al., 2024b; Liu et al., 2025a; Liu et al., 2025b; Liu et al., 2025c), and relative expression ratios were presented as log2 values in histograms. A ratio greater than zero indicated upregulation, while a ratio less than zero indicated downregulation. Beta-actin was used as a reference gene for normalization, and a positive control with beta-actin was included in the analysis.
TABLE 2
| Constructs | Primers | Primer sequence (5′-3′) |
|---|---|---|
| Beta-actin | Forward primer | ggcagtttggacaagagaagacacagtcacc |
| Reverse primer | ttttttgattaaaaaaaaaagctgaaatcataatatttttaat | |
| LiAATase1 | Forward primer | atggcgatgattattacaaaacaaattttgaggccatcatctc |
| Reverse primer | tcaagtatccaatttattgtaattggcttggagcacttggaag | |
| LiAATase2 | Forward primer | atggcatccaccaaaaccctgaccttc |
| Reverse primer | tcacaatgctgaaagattgagagtcctggcag |
Primers used for RT-qPCR in this study.
Statistical analysis
All experiments were conducted at least in triplicate. The data were expressed as mean ± SD. Statistical analysis was conducted using Origin 8.5, Microsoft Excel 2013 and SPSS 19.0. In the all statistical evaluations, p < 0.05 was considered statistically significant, and p < 0.01 was considered high statistically significant.
Statements
Data availability statement
The datasets generated in this study are available in online repositories. Details regarding the repository names and accession numbers are provided in the article and Supplementary Material.
Author contributions
DL: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review and editing. YD: Investigation, Writing – original draft. AA: Investigation, Writing – original draft. LZ: Investigation, Writing – original draft. DS: Investigation, Writing – original draft. HD: Investigation, Writing – original draft. XW: Investigation, Writing – original draft. YZ: Investigation, Writing – original draft. BS: Investigation, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Our research work is financially supported by grants from Xinjiang Key Laboratory of Lavender Conservation and Utilization (LCUZ2405), and Start-up Fund for Doctoral Research Established by Yili Normal University (2024RCYJ08).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2025.1627286/full#supplementary-material
References
1
Aharoni A. Giri A. P. Verstappen F. W. A. Bertea C. M. Sevenier R. Sun Z. et al (2004). Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell16, 3110–3131. 10.1105/tpc.104.023895
2
Aschariyaphotha W. Noichinda S. Bodhipadma K. Wongs-Aree C. (2024). Characterization of alcohol acetyltransferases in the ripe flesh of ‘Monthong’ and ‘Chanthaburi 1′ durians. Plant Physiology Biochem.206, 108241. 10.1016/j.plaphy.2023.108241
3
Bavarsad N. H. Bagheri S. Kourosh-Arami M. Komaki A. (2023). Aromatherapy for the brain: lavender's healing effect on epilepsy, depression, anxiety, migraine, and Alzheimer's disease: a review article. Heliyon9, e18492. 10.1016/j.heliyon.2023.e18492
4
Bayer A. Ma X. Stöckigt J. (2004). Acetyltransfer in natural product biosynthesis––functional cloning and molecular analysis of vinorine synthase. Bioorg. and Med. Chem.12, 2787–2795. 10.1016/j.bmc.2004.02.029
5
Beekwilder J. Alvarez-Huerta M. Neef E. Verstappen F. W. A. Bouwmeester H. J. Aharoni A. (2004). Functional characterization of enzymes forming volatile esters from strawberry and banana. Plant Physiol.135, 1865–1878. 10.1104/pp.104.042580
6
Benabdelkader T. Zitouni A. Guitton Y. Jullien F. Maitre D. Casabianca H. et al (2011). Essential oils from wild populations of Algerian Lavandula stoechas L.: composition, chemical variability, and in vitro biological properties. Chem. and Biodivers.8, 937–953. 10.1002/cbdv.201000301
7
Canessa J. A. Larach J. A. Massardo T. Parra J. Jofré J. González P. et al (2004). Fluorodeoxiglucose F18 positron emission tomography imaging (F18FDG) for the assessment of rising levels of serum CA 19-9 in pancreatic mucinous cystadenocarcinoma. Report of one case. Rev. Médica Chile132, 347–352. 10.4067/s0034-98872004000300010
8
Collaborative Computational Project, N (1994). The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D. Biol. Crystallogr.50, 760–763. 10.1107/s0907444994003112
9
Combet C. Blanchet C. Geourjon C. Deléage G. (2000). NPS@: network protein sequence analysis. Trends Biochem. Sci.25, 147–150. 10.1016/s0968-0004(99)01540-6
10
Cumplido-Laso G. Medina-Puche L. Moyano E. Hoffmann T. Sinz Q. Ring L. et al (2012). The fruit ripening-related gene FaAAT2 encodes an acyl transferase involved in strawberry aroma biogenesis. J. Exp. Bot.63, 4275–4290. 10.1093/jxb/ers120
11
De Beer T. a.P. Berka K. Thornton J. M. Laskowski R. A. (2014). PDBsum additions. Nucleic Acids Res.42, D292–D296. 10.1093/nar/gkt940
12
Defilippi B. G. Kader A. A. Dandekar A. M. (2005). Apple aroma: alcohol acyltransferase, a rate limiting step for ester biosynthesis, is regulated by ethylene. Plant Sci.168, 1199–1210. 10.1016/j.plantsci.2004.12.018
13
De Groot A. Schmidt E. (2016a). Essential oils, Part V: peppermint oil, lavender oil, and lemongrass oil. Dermatitis27, 325–332. 10.1097/der.0000000000000218
14
De Groot A. C. Schmidt E. (2016b). Essential oils, Part III: chemical composition. Dermatitis27, 161–169. 10.1097/der.0000000000000193
15
De Jong B. Anaya C. Masera O. Olguín M. Paz F. Etchevers J. et al (2010). Greenhouse gas emissions between 1993 and 2002 from land-use change and forestry in Mexico. For. Ecol. Manag.260, 1689–1701. 10.1016/j.foreco.2010.08.011
16
Ebadollahi A. Ziaee M. Palla F. (2020). Essential oils extracted from different species of the lamiaceae plant family as prospective bioagents against several detrimental pests. Molecules25, 1556. 10.3390/molecules25071556
17
Egea-Cortines M. Zhang T. Huo T. Ding A. Hao R. Wang J. et al (2019). Genome-wide identification, characterization, expression and enzyme activity analysis of coniferyl alcohol acetyltransferase genes involved in eugenol biosynthesis in Prunus mume. Plos One14, e0223974. 10.1371/journal.pone.0223974
18
Galaz S. Morales‐Quintana L. Moya‐León M. A. Herrera R. (2013). Structural analysis of the alcohol acyltransferase protein family from Cucumis melo shows that enzyme activity depends on an essential solvent channel. FEBS J.280, 1344–1357. 10.1111/febs.12127
19
Ghezzi A. Li X. Lew L. K. Wijesekera T. P. Atkinson N. S. (2017). Alcohol-induced neuroadaptation is orchestrated by the histone acetyltransferase CBP. Front. Mol. Neurosci.10, 103. 10.3389/fnmol.2017.00103
20
Green M. R. Sambrook J. (2018). Analysis and normalization of real-time polymerase chain reaction (PCR) experimental data. Cold Spring Harb. Protoc.2018, pdb.top095000. 10.1101/pdb.top095000
21
Guterman I. Masci T. Chen X. Negre F. Pichersky E. Dudareva N. et al (2006). Generation of phenylpropanoid pathway-derived volatiles in transgenic plants: rose alcohol acetyltransferase produces phenylethyl acetate and benzyl acetate in petunia flowers. Plant Mol. Biol.60, 555–563. 10.1007/s11103-005-4924-x
22
Hawkins S. F. C. Guest P. C. (2017). Multiplex analyses using real-time quantitative PCR, Methods Mol. Biol., 1546,125–133. 10.1007/978-1-4939-6730-8_8
23
Heo L. Lee H. Seok C. (2016). GalaxyRefineComplex: refinement of protein-protein complex model structures driven by interface repacking. Sci. Rep.6, 32153. 10.1038/srep32153
24
Heo L. Park H. Seok C. (2013). GalaxyRefine: protein structure refinement driven by side-chain repacking. Nucleic Acids Res.41, W384–W388. 10.1093/nar/gkt458
25
Horton C. E. Huang K.-X. Bennett G. N. Rudolph F. B. (2003). Heterologous expression of the Saccharomyces cerevisiae alcohol acetyltransferase genes in Clostridium acetobutylicum and Escherichia coli for the production of isoamyl acetate. J. Industrial Microbiol. Biotechnol.30, 427–432. 10.1007/s10295-003-0070-0
26
Hu Z. Li X. Wang H. Niu C. Yuan Y. Yue T. (2016). A novel method to quantify the activity of alcohol acetyltransferase Using a SnO 2 -based sensor of electronic nose. Food Chem.203, 498–504. 10.1016/j.foodchem.2016.02.087
27
Jumper J. Evans R. Pritzel A. Green T. Figurnov M. Ronneberger O. et al (2021). Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589. 10.1038/s41586-021-03819-2
28
Karpiński T. M. (2020). Essential oils of lamiaceae family plants as antifungals. Biomolecules10, 103. 10.3390/biom10010103
29
Khanom M. M. Ueda Y. (2008). Bioconversion of aliphatic and aromatic alcohols to their corresponding esters in melons (Cucumis melo L. cv. Prince melon and cv. Earl's favorite melon). Postharvest Biol. Technol.50, 18–24. 10.1016/j.postharvbio.2008.02.015
30
Ko J. Park H. Heo L. Seok C. (2012). GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res.40, W294–W297. 10.1093/nar/gks493
31
Laskowski R. A. (2004). PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res.33, D266–D268. 10.1093/nar/gki001
32
Laskowski R. A. (2022). 1: a standalone program for generating PDBsum analyses. Protein Sci.31, e4473. 10.1002/pro.4473
33
Laskowski R. A. Jabłońska J. Pravda L. Vařeková R. S. Thornton J. M. (2017). PDBsum: structural summaries of PDB entries. Protein Sci.27, 129–134. 10.1002/pro.3289
34
Lee J.-W. Trinh C. T. (2022). Controlling selectivity of modular microbial biosynthesis of butyryl-CoA-derived designer esters. Metab. Eng.69, 262–274. 10.1016/j.ymben.2021.12.001
35
Lesage-Meessen L. Bou M. Sigoillot J.-C. Faulds C. B. Lomascolo A. (2015). Essential oils and distilled straws of lavender and lavandin: a review of current use and potential application in white biotechnology. Appl. Microbiol. Biotechnol.99, 3375–3385. 10.1007/s00253-015-6511-7
36
Lilly M. Bauer F. F. Lambrechts M. G. Swiegers J. H. Cozzolino D. Pretorius I. S. (2006). The effect of increased yeast alcohol acetyltransferase and esterase activity on the flavour profiles of wine and distillates. Yeast23, 641–659. 10.1002/yea.1382
37
Liu D. Abdiriyim A. Zhang L. Ruzitohti B. (2025a). Functional and mechanistic insights into the stealth protein full-length CpsY is conducive to understanding immune evasion mechanisms by Mycobacterium tuberculosis. Tuberculosis152, 102616. 10.1016/j.tube.2025.102616
38
Liu D. Abdiriyim A. Zhang L. Yu F. (2025b). Functional and mechanistic insights into the fatty-acid CoA ligase FadK in Escherichia coli. Front. Bioscience-Landmark36701, 36701. 10.31083/fbl36701
39
Liu D. Deng H. Song H. (2025c). Insights into the functional mechanisms of the sesquiterpene synthase GEAS and GERDS in lavender. Int. J. Biol. Macromol.299, 140195. 10.1016/j.ijbiomac.2025.140195
40
Liu D. Song H. Deng H. Abdiriyim A. Zhang L. Jiao Z. et al (2024a). Insights into the functional mechanisms of three terpene synthases from Lavandula angustifolia (Lavender). Front. Plant Sci.15, 1497345. 10.3389/fpls.2024.1497345
41
Liu D. Tian Z. Tusong K. Mamat H. Luo Y. (2024b). Expression, purification and characterization of CTP synthase PyrG in Staphylococcus aureus. Protein Expr. Purif.221, 106520. 10.1016/j.pep.2024.106520
42
Liu G. Huang L. Lian J. (2023). Alcohol acyltransferases for the biosynthesis of esters. Biotechnol. Biofuels Bioprod.16, 93. 10.1186/s13068-023-02343-x
43
Livak K. J. Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods25, 402–408. 10.1006/meth.2001.1262
44
Ma X. Koepke J. Panjikar S. Fritzsch G. Stöckigt J. (2005). Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. J. Biol. Chem.280, 13576–13583. 10.1074/jbc.m414508200
45
Mascarell-Creus A. Cañizares J. Vilarrasa-Blasi J. Mora-García S. Blanca J. Gonzalez-Ibeas D. et al (2009). An oligo-based microarray offers novel transcriptomic approaches for the analysis of pathogen resistance and fruit quality traits in melon (Cucumis melo L.). BMC Genomics10, 467. 10.1186/1471-2164-10-467
46
Moon H. Y. Kim H. J. Kim K. S. Yoo S. J. Lee D. W. Shin H. J. et al (2021). Molecular characterization of the Saccharomycopsis fibuligera ATF genes, encoding alcohol acetyltransferase for volatile acetate ester formation. J. Microbiol.59, 598–608. 10.1007/s12275-021-1159-8
47
Morales-Quintana L. Fuentes L. Gaete-Eastman C. Herrera R. Moya-León M. A. (2011). Structural characterization and substrate specificity of VpAAT1 protein related to ester biosynthesis in mountain papaya fruit. J. Mol. Graph. Model.29, 635–642. 10.1016/j.jmgm.2010.11.011
48
Morales-Quintana L. Moya-León M. A. Herrera R. (2012). Molecular docking simulation analysis of alcohol acyltransferases from two related fruit species explains their different substrate selectivities. Mol. Simul.38, 912–921. 10.1080/08927022.2012.672738
49
Morales-Quintana L. Moya-León M. A. Herrera R. (2015). Computational study enlightens the structural role of the alcohol acyltransferase DFGWG motif. J. Mol. Model.21, 216. 10.1007/s00894-015-2762-6
50
Morales-Quintana L. Nuñez-Tobar M. X. Moya-León M. A. Herrera R. (2013). Molecular dynamics simulation and site-directed mutagenesis of alcohol acyltransferase: a proposed mechanism of catalysis. J. Chem. Inf. Model.53, 2689–2700. 10.1021/ci400409s
51
Nazeer M. Waheed H. Saeed M. Ali S. Y. Choudhary M. I. Ul-Haq Z. et al (2019). Purification and characterization of a nonspecific lipid transfer protein 1 (nsLTP1) from ajwain (trachyspermum ammi) seeds. Sci. Rep.9, 4148. 10.1038/s41598-019-40574-x
52
Patrignani F. Prasad S. Novakovic M. Marin P. D. Bukvicki D. (2021). Lamiaceae in the treatment of cardiovascular diseases. Front. Biosci.26, 612–643. 10.2741/4909
53
Saez D. Rodríguez-Arriaza F. Urra G. Fabi J. P. Hormazábal-Abarza F. Méndez-Yáñez A. et al (2024). Unraveling the key step in the aroma puzzle: insights into alcohol acyltransferases in strawberries. Plant Physiology Biochem.212, 108668. 10.1016/j.plaphy.2024.108668
54
Schmittgen T. D. Livak K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc.3, 1101–1108. 10.1038/nprot.2008.73
55
Seixas I. Santos D. Vasconcelos I. Mira N. P. Mendes-Ferreira A. (2023). Insights into the transcriptional regulation of poorly characterized alcohol acetyltransferase-encoding genes (HgAATs) shed light into the production of acetate esters in the wine yeast Hanseniaspora guilliermondii. FEMS Yeast Res.23, foad021. 10.1093/femsyr/foad021
56
Seok C. Baek M. Steinegger M. Park H. Lee G. R. Won J. (2021). Accurate protein structure prediction: what comes next?Biodesign9, 47–50. 10.34184/kssb.2021.9.3.47
57
Shah V. Sáez C. A. Lobos M. G. Macaya E. C. Oliva D. Quiroz W. et al (2012). Variation in patterns of metal accumulation in thallus parts of lessonia trabeculata (laminariales; phaeophyceae): implications for biomonitoring. PLoS ONE7, e50170. 10.1371/journal.pone.0050170
58
Stribny J. Querol A. Pérez-Torrado R. (2016). Differences in enzymatic properties of the Saccharomyces kudriavzevii and Saccharomyces uvarum alcohol acetyltransferases and their impact on aroma-active compounds production. Front. Microbiol.7, 897. 10.3389/fmicb.2016.00897
59
Wang Q. Al Makishah N. H. Li Q. Li Y. Liu W. Sun X. et al (2021). Developing clostridia as cell factories for short- and medium-chain ester production. Front. Bioeng. Biotechnol.9, 661694. 10.3389/fbioe.2021.661694
60
Wani A. R. Yadav K. Khursheed A. Rather M. A. (2021). An updated and comprehensive review of the antiviral potential of essential oils and their chemical constituents with special focus on their mechanism of action against various influenza and coronaviruses. Microb. Pathog.152, 104620. 10.1016/j.micpath.2020.104620
61
Wayment-Steele H. K. Ojoawo A. Otten R. Apitz J. M. Pitsawong W. Hömberger M. et al (2023). Predicting multiple conformations via sequence clustering and AlphaFold2. Nature625, 832–839. 10.1038/s41586-023-06832-9
62
Wiederstein M. Sippl M. J. (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res.35, W407–W410. 10.1093/nar/gkm290
63
Zhang J. Zhang C. Qi Y. Dai L. Ma H. Guo X. et al (2014). Acetate ester production by Chinese yellow rice wine yeast overexpressing the alcohol acetyltransferase-encoding gene ATF2. Genet. Mol. Res.13, 9735–9746. 10.4238/2014.november.27.1
64
Zheng J. Navarro-Retamal C. Gaete-Eastman C. Herrera R. Caballero J. Alzate-Morales J. H. (2016). Structural and affinity determinants in the interaction between alcohol acyltransferase from F. x ananassa and several alcohol substrates: a computational study. Plos One11, e0153057. 10.1371/journal.pone.0153057
Summary
Keywords
Lavandula x intermedia (lavandin), alcohol acetyltransferase, structural prediction, enzyme activity assay, gene expression levels
Citation
Liu D, Du Y, Abdiriyim A, Zhang L, Song D, Deng H, Wen X, Zhang Y and Sun B (2025) Molecular functional mechanisms of two alcohol acetyltransferases in Lavandula x intermedia (lavandin). Front. Chem. 13:1627286. doi: 10.3389/fchem.2025.1627286
Received
12 May 2025
Accepted
27 May 2025
Published
11 June 2025
Volume
13 - 2025
Edited by
Yusuke Kato, National Agriculture and Food Research Organization, Japan
Reviewed by
Luis Morales-Quintana, Autonomous University of Chile, Chile
Ramesh Maruthi Chingle, National Institutes of Health (NIH), United States
Updates
Copyright
© 2025 Liu, Du, Abdiriyim, Zhang, Song, Deng, Wen, Zhang and Sun.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dafeng Liu, dafeli@sina.cn, dafeli-dafeli@hotmail.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.