- 1Agro-Biotechnology Research Institute, Jilin Academy of Agricultural Sciences, Changchun, China
- 2Crop Germplasm Institute, Jilin Academy of Agricultural Sciences, Gongzhuling, China
- 3Department of Biology, Beijing Normal University, Beijing, China
- 4Institute of Plant Protection, Jilin Academy of Agricultural Sciences, Gongzhuling, China
Heat shock proteins (HSPs) perform a fundamental role in protecting plants against abiotic stresses. Previous studies have made great efforts in the functional analysis of individual family members, but there has not yet been an overall analysis or expression profiling of the HSP70 gene family in soybeans (Glycine max L.). In this study, an investigation of the soybean genome revealed 61 putative HSP70 genes, which were evaluated. These genes were classified into eight sub-families, denoted I–VIII, based on a phylogenetic analysis. In each sub-family, the constituent parts of the gene structure and motif were relatively conserved. These GmHSP70 genes were distributed unequally on 17 of the 20 chromosomes. The analysis of the expression profiles showed that 53 of the 61 GmHSP70 genes were differentially expressed across the 14 tissues. However, most of the GmHSP70s were differentially expressed in a tissue-specific expression pattern. Furthermore, the expression of some of the duplicate genes was partially redundant, while others showed functional diversity. The quantitative real-time PCR (qRT-PCR) analysis of the 61 soybean HSP70 genes confirmed their stress-inducible expression patterns under both drought and heat stress. These findings provide a thorough overview of the evolution and modification of the GmHSP70 gene family, which will help to determine the functional characteristics of the HSP70 genes in soybean growth and development.
Introduction
Heat shock proteins (HSPs) are a group of proteins induced by heat shock, and they are found in virtually all living organisms, from bacteria to humans (De Maio, 1999). In plants, the HSP gene family members play important roles in developmental processes, as well as different kinds of environmental stress conditions, such as heat and drought (Swindell et al., 2007; Cho and Choi, 2009), low and high temperatures (Krishna et al., 1995; Sabehat et al., 1998; Lopez-Matas et al., 2004), salinity (Zou et al., 2012), and heavy metals (Kim et al., 2014). HSPs have recently been discovered to be associated with plant responses to infection by pathogens such as nematodes (Maimbo et al., 2007). Based on their molecular weight, HSPs are classified into five major families: sHSP, small heat shock protein; HSP60, chaperonin family; HSP70, 70-kDa-heat shock protein; HSP90 and HSP100 family (Wang et al., 2004). Of these, the HSP70 family is of greatest interest, as it is fundamental to plant developmental processes and functions during heat stress (Sung et al., 2001).
HSP70 proteins are central components of the cellular network of molecular chaperones and folding catalysts (Mayer and Bukau, 2005), and were first identified and characterized in the early 1960s (Ritossa, 1996). HSP70s are comprised of two major functional domains. One is a conserved ~44-kD N-terminal ATPase domain (NBD), which is also called the nucleotide binding domain and the other is a ~18-kD substrate binding domain (SBD) with a ~10-kD variable C-terminal “lid” (Dragovic et al., 2006). The diverse biological function of the HSP70 gene family has been well characterized in many plants. For example, in Arabidopsis, the cpHsc70-1 mutant plants exhibited defective phenotypes after germinated seeds were treated with heat stress (Su and Li, 2008), and AtHSP70-15 was shown to play an essential role during heat response (Jungkunz et al., 2011). HSP70s have also been reported to play roles in response to heat stress in rubber trees (Hevea brasiliensis) (Zhang et al., 2009), wheat (Triticum aestivum L) (Francki et al., 2002; Duan et al., 2011), pepper (Capsicum annuum L) (Guo et al., 2014), and cucumber (Cucumis sativus) (Li et al., 2014). HSP70 also shows complex developmental regulation during the vegetative and reproductive phases of growth. In tomato, for example, HSP70 transcripts were detected in mature anthers (Duck and Folk, 1994). In alfalfa (Medicago sativa), the MsHSP70-1 plays an important role in the development of alfalfa nodules (He et al., 2008). Recently, some studies have suggested that the HSP70 family members may play various roles in plant RNA virus multiplication, such as viral protein folding, virion assembly and protein expression (Nagy et al., 2011). Among the plant (+) RNA viruses, it has been reported that HSP70 participates in the assembly of the replicase complexes of the TBSV (Hafrén et al., 2010; Mathioudakis et al., 2012), TuMV (Dufresne et al., 2009; Wang et al., 2009a), and rice stripe virus (Wang et al., 2009b). These results indicate that the HSP70 family members are multi-functional in the life cycle of plants (Friedman and Brandon, 2001; Jiang et al., 2014).
Soybean (Glycine max L.) is one of the most economically and nutritionally crucial crops in the world. It provides not only vegetable protein and edible oil, but also essential amino acids for humans and animals. However, soybean production is threatened by drought, as well as other environmental stresses. With the sequencing of the soybean genome (Schmutz et al., 2010), a few members of the HSP family were characterized in this species (Jinn et al., 1995; Fuganti et al., 2010; Mohammadi et al., 2012; Liu and Whitham, 2013; Lopes-Caitar et al., 2013; Wang et al., 2014). However, most of their functions remain to be determined. There remains no genome-wide characterization of the HSP70 family in soybean to date.
In our study, all of the non-redundant sets of the soybean HSP70 genes were scanned and integrated, and we also determined their chromosomal locations, predicted their protein structures with available software and network stations, and analyzed the expression levels of the soybean HSP70 genes using qRT-PCR in order to determine their tolerance to drought and heat stresses. Our findings will be useful resources for future studies to unravel the functions of the GmHSP70 genes and will contribute to our understanding of the evolutionary history of the HSP70 genes in different species.
Materials and Methods
Genome-wide Identification of HSP70 Proteins in Soybean
The whole soybean genome sequence was downloaded from the annotation database (DB) Phytozome v9.1 (Joint Genome Institute, JGI). To gather the probable candidate soybean HSP70 amino acid sequences, the HMM profile of the HSP70 domain was first downloaded from the Pfam (http://www.sanger.ac.uk/Software/Pfam/) database (Pfam: PF00012) and then submitted as a query in a BLASTP (P = 0.001) search of the soybean genome database (Finn et al., 2014). All obtained protein sequences were examined for the presence of the HSP70 domain by SMART (http://smart.embl-heidelberg.de/) tools (Letunic et al., 2004).
Soybean HSP70 gene information, including the locations on the chromosomes, genomic sequences, full CDS sequences, protein sequences, and 2000 bp of the nucleotide sequences upstream of the translation initiation codon were downloaded from the website Phytozome (www.phytozome.net). The molecular weight (Da) and isoelectric point (pI) of each gene were calculated using compute pI/Mw tool from ExPASy (http://www.expasy.org/tools/) (Gasteiger et al., 2003).
Subcellular Localization and Promoter Analysis of the GMHSP70 Family
Subcellular localization was predicted using the WoLF PSORT (http://www.genscript.com/psort/wolf_psort.html) (Horton et al., 2007) and TargetP 1.1 (http://www.cbs.dtu.dk/services/TargetP/) (Emanuelsson et al., 2007). The 5′ upstream region, including a 2000 bp DNA sequence of each gene of the GmHSP70 family, was subjected to the plantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) for a cis-element scan.
Multiple Alignment and Phylogenetic Analysis
We constructed two phylogenetic trees to classify the soybean HSP70 genes in this paper. The first one contained only soybean HSP70 protein sequences and the second included also Arabidopsis thaliana and rice HSP70 sequences. The gene sequences and protein sequences of Arabidopsis and rice HSPs were acquired from TAIR (Lamesch et al., 2012), and TIGR (Yuan et al., 2003), respectively. Multiple sequence alignment of all predicted HSP70 protein sequences were performed with Clustal X2.0 software using default parameters. The phylogenetic trees were constructed using MEGA 5.0 with the Neighbor-Joining (NJ) method, and a bootstrap analysis was conducted using 1000 replicates with a pairwise gap deletion mode (Tamura et al., 2011).
Gene Structure Analysis and Identification of Conserved Motifs
To investigate the diversity and structure of members of the GmHSP70 gene family, we compared the exon/intron organization between the cDNA sequences with the corresponding genomic DNA sequences of HSP70 by using Gene Structure Display Server (GSDS; http://gsds.cbi.pku.edu.cn/) (Guo et al., 2007). In addition, their amino acid sequences were subjected to “predict the domain and motif analyses” online with MEME (http://meme.sdsc.edu/meme/website/intro.html) (Bailey et al., 2006). The parameters were as follows: number of repetitions: any; maximum number of motifs: 10; and optimum motif widths: 6 to 200 amino acid residues.
Chromosomal Location and Gene Duplication
The chromosomal location image of the GmHSP70 genes was generated by MapChart software, according to the chromosomal position information provided in the Phytozome database. In order to identify the tandem and segmental duplications, two genes in the same species, located in the same clade of the phylogenetic tree, were defined as being coparalogs. The SoyBase browser (http://soybase.org/gb2/gbrowse/gmax1.01/) was queried in order to detect the segmental duplication coordinates of the target genes (Grant et al., 2010). The coparalogs were deemed be the results of segmental duplication if they were located on duplicated chromosomal blocks (Wei et al., 2007). The paralogs were deemed to be tandem duplicated genes if two genes were separated by five or fewer genes in a 100 kb region (Wang et al., 2010). The local alignment of two protein sequences was calculated using the Smith-Waterman algorithm (http://www.ebi.ac.uk/Tools/psa/).
RNA-Seq Atlas Analysis
To acquire the tissue-specific transcript data, the 61 GmHSP70 genes were investigated based on the RNA Seq-Atlas from fourteen tissues (http://soybase.org/soyseq/), including underground tissues (root and nodule), seed development (seed 10-DAF, seed 14-DAF, seed 21-DAF, seed 25-DAF, seed 28-DAF, seed 35-DAF, and seed 42-DAF) and aerial tissues (young leaf, flower, pod-shell 10-DAF, pod-shell 14-DAF, and 1 cm pod). The expression data were gene-wise normalized and the heat map was drawn using MeV v4.8 software (http://www.tm4.org/) (Mar et al., 2011).
Plant Growth and Stress Treatments
Soybean Williams 82 seeds were germinated in vermiculite in a light chamber at 25 ± 2°C for about 2 weeks. For heat stress, the seedlings were transferred to a growth chamber at 42 ± 1°C, while for drought stress, 15% (w/v) PEG3000 was used. The leaves of heat-stressed and drought-stressed plants were collected at 0, 3, 6, 12, and 24-h intervals. After collection, the samples were immediately frozen in liquid N2, and stored at −80°C for RNA extraction. Three biological replicates were obtained per sample.
RNA Extraction and qRT-PCR Analysis
The total RNA from all of the samples was isolated using Trizol reagent (Invitrogen, USA), followed by DNaseI (Promega, USA) treatment for the purpose of removing any traces of genomic DNA. The first-strand cDNA synthesis was performed using the TransScript First-Strand cDNA Synthesis SuperMix Kit (TransGen, China). Gene-specific primers for the 61 GmHSP70 genes were designed using Primer 5.0 (Additional File 13). A housekeeping gene constitutively expressed in soybean, Actin (LOC100792119), was used as a reference for normalization. The real-time quantitative reverse transcription-polymerase chain reaction (PCR) was conducted using an Applied Biosystems StepOne Real-Time PCR System (Applied Biosystems, USA). The reaction volume consisted of 10 μL SYBR premix Ex Taq™ (2×) mixture, 1 μL cDNA (diluted 10 times), 0.4 μL upstream primer (10 pM), 0.4 μL downstream primer (10 pM) and 8.2 μL ddH2O (20 μL in total). The reaction was performed with the following cycling profile: 95°C for 30s, 40 cycles at a denaturation of 95°C for 5s, and 60°C for 30s. Three technical replicates were performed for each sample. The calculation of the gene expression levels followed the 2−ΔΔCT method described by Livak and Schmittgen (2001).
Results
Identification of the HSP70 Gene Family in Soybean
A soybean genome blast, as well as online software identification, were utilized to identify a total of 61 soybean HSP70 genes based on their nomenclature (Table 1). All of the 61 members contained the domains PF00012, based on Pfam and SMART tests. The proteins that had only one of these domains, or that did not have an integral open reading frame, were excluded. Then, the protein sequences (Additional File 1), coding sequences (CDS) (Additional File 2), genomic sequences (Additional File 3), and 2000 bp of the nucleotide sequences upstream from the translation initiation codon (Additional File 4) were downloaded from the Phytozome database (www.phytozome.net). The basic information of all of the soybean HSP70 genes (including gene name, chromosome location, ORF length, exon and intron number, protein length, molecular weight and pI value) is provided in Table 1. The GmHSP70s encoded proteins varied from 134 to 924 amino acids (aa) in length. Among these proteins the Glyma18g13077 protein sequence was the shortest, with 134 amino acids. The Glyma20g16070 was the longest protein sequence, with 927 amino acids. The predicted molecular weights of the GmHSP70 candidates were distributed in a range from 15188 Da (Glyma18g13077, 134aa) to 102932 Da (Glyma20g16070, 927aa). The predicted isoelectric points (pI) of the GmHSP70 candidates were between 4.89 (Glyma05g03770) and 9.94 (Glyma18g13077). The predicted protein instability indices showed that only 19 of the 61 GmHSP70 candidates could be considered stable proteins (cutoff < 40), according to the ExPASy analysis.
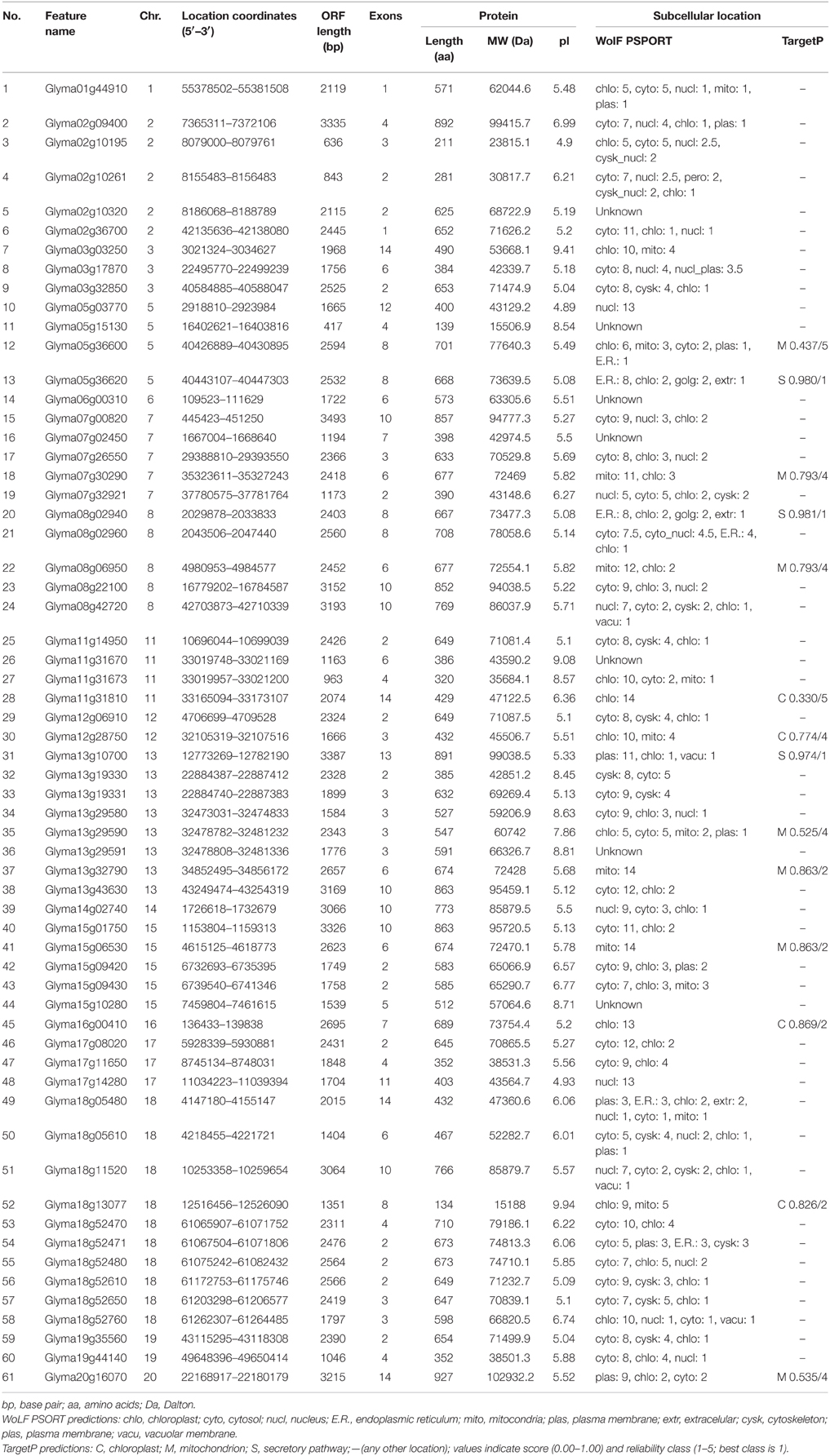
Table 1. List of 61 HSP70 genes identified in soybean, their sequence characteristics and subcellular localization.
Subcellular Localization and cis-acting Elements of GmHSP70 Family
The Wolf PSORT assessment revealed that within the 61 GmHSP70 proteins, 27 soybean HSP70 proteins were localized to the cytosol, 11 to the chloroplast, six to the nucleus, four to the mitochondria, three to the plasma membrane, two to the endoplasmic reticulum, one to the cytoskeleton, and seven were not identified by WoLF PSORT. The TargetP analysis revealed that seven soybean HSP70 proteins were located in the mitochondria, four in the chloroplast, and three in the secretory pathway and 47 in other compartments. These detailed parameters are provided in Table 1.
Plant gene promoter, an important cis-acting element that is located upstream of the start codon region, is the control center of gene transcription. Our cis-acting element analysis identified a total of 115 types of cis-regulatory elements in the promoter regions of GmHSP70s (see Additional Files 5, 6). These cis-regulatory elements could be broadly divided into eight categories: Light responsive elements, Promoter related elements, Hormone responsive elements, Environmental stress-related elements, Development related elements, Site-binding related elements and other elements). The light responsive category contains 42 kinds of cis-regulatory elements and accounts for the largest proportion in the total. Twelve types of cis-regulatory elements were found to be involved in plant hormone responsiveness, which cover most types of hormone, including ABRE, abscisic acid responsive element; ERE, ethylene responsive element; GARE-motif, p-box, gibberellins responsive element; TCA-element, salicylic acid responsiveness; TGA-element, auxin-responsive element; CGTCA-motif, TATC-box, TGACG-motif, the MeJA-responsiveness; and AuxRR-core, auxin responsiveness element. The third category contained 11 types of cis-regulatory elements that might respond to environmental stress, such as heat stress, Box-W1 and W3, fungal elicitor responsive element; MBS, MYB binding site involved in drought-responsiveness; LTR, low-temperature responsiveness; and W-box, wound-responsive element. The flowing category is related to plant growth and development, and contains 15 types of cis-regulatory elements: AACA-motif, AAGAA-motif, AC-I, AC-II, CAT-box, CCAAT-motif, O2-site, as1, circadian, GCN4-motif, RY-element, Skn-1_motif, HD-Zip1, HD-Zip2, and MSA-like. Two categories were promoter related elements and site-binding related elements; they contained eight and seven types of cis-regulatory elements, respectively. Finally, the functions of the remaining 20 types of cis-regulatory elements are unclear (Additional File 6).
Phylogenetic, Gene Structure, and Motif Analysis of the HSP70 Proteins in Soybean
Based on the unrooted phylogenetic tree of the 61 GmHSP70 protein sequences, the soybean HSP70 gene family was categorized into eight major sub-families (sub-family I, II, III, IV, V, VI, VII, and VIII). Sub-family I (containing 29 members) was the largest group, followed by sub-family VIII (12), sub-family VII (7), sub-family III (6), and sub-family II (4). The sub-families IV, V, VI were the smallest, with only one GmHSP70 gene member each (Figure 1A). Correspondingly, Figure 1B provides a detailed illustration of the relative lengths of the introns, and the conservation of the corresponding exon sequences within each HSP70 gene within the soybean. The number of introns in all of these genes ranged from 0 to 13. However, no intron was found in the Glyma13g19330, Glyma02g36700, Glyma17g08020, Glyma18g52470, Glyma18g52471, Glyma11g31670, Glyma07g32921, or Glyma01g44910, whereas 13 introns were found in the Glyma20g16070, Glyma03g03250, Glyma11g31810, and Glyma18g05480. The MEME of the total 61 GmHSP70 genes were also analyzed (Figure 2). The results revealed the conserved domains or motifs shared among related proteins, and identified 15 conserved motifs (Additional Files 7 and 8). The type, order, and number of motifs were similar in proteins with the same sub-family, but differed from the proteins in other sub-families.
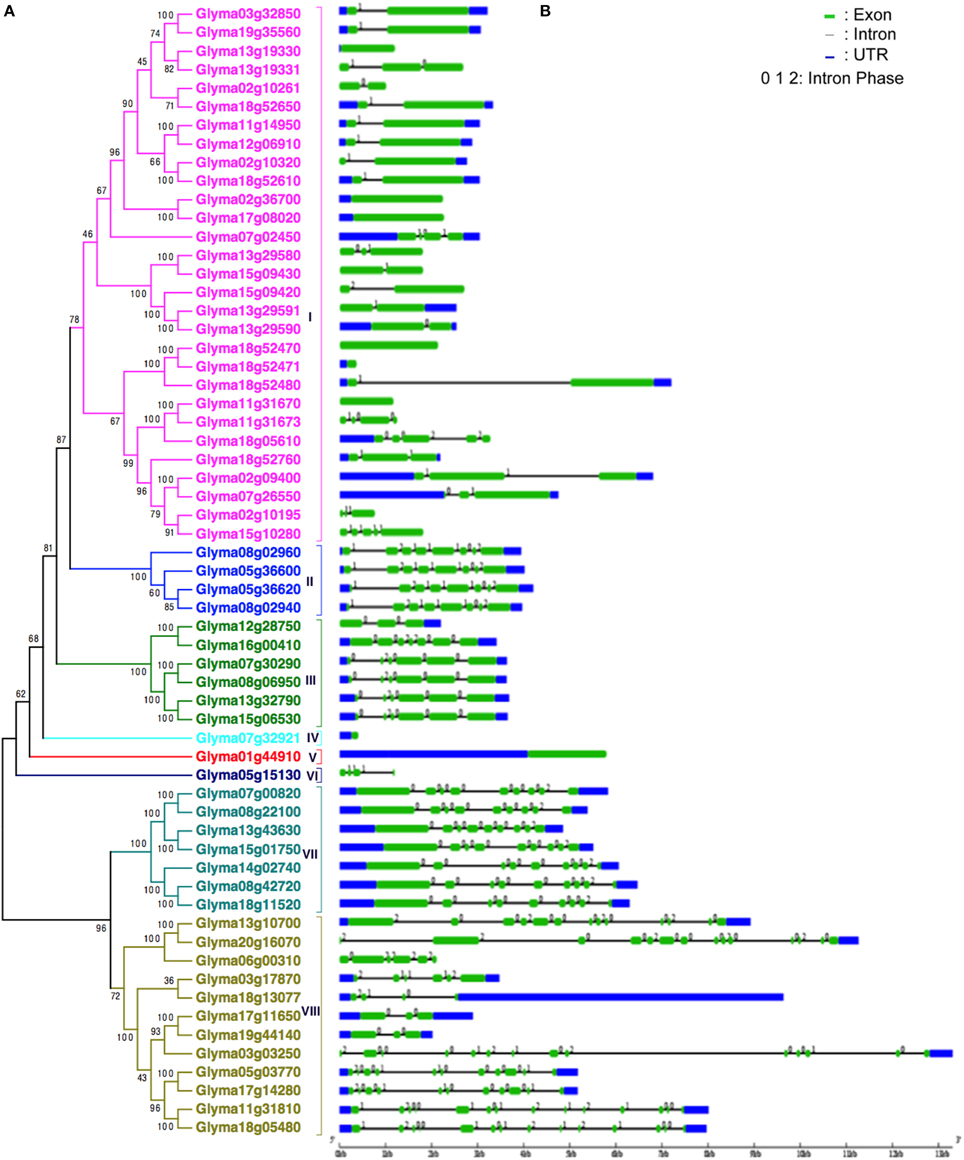
Figure 1. Phylogenetic relationships and gene structure of soybean HSP70 genes. (A) The unrooted tree was generated with the MEGA5.0 program using the full-length amino acid sequences of the 61 soybean HSP70 proteins by the Neighbor-Joining (NJ) method, with 1000 bootstrap replicates. Subfamilies of HSP70 genes (I–VIII) are highlighted with different colored backgrounds and vertical bars next to the gene names of the tree. (B) Exon/intron organization of soybean HSP70 genes. Green boxes represent exons and black lines represent introns. The untranslated regions (UTRs) are indicated by blue boxes. The numbers 0, 1, and 2 represents the splicing phases. The sizes of exons and introns can be estimated using the scale at the bottom.
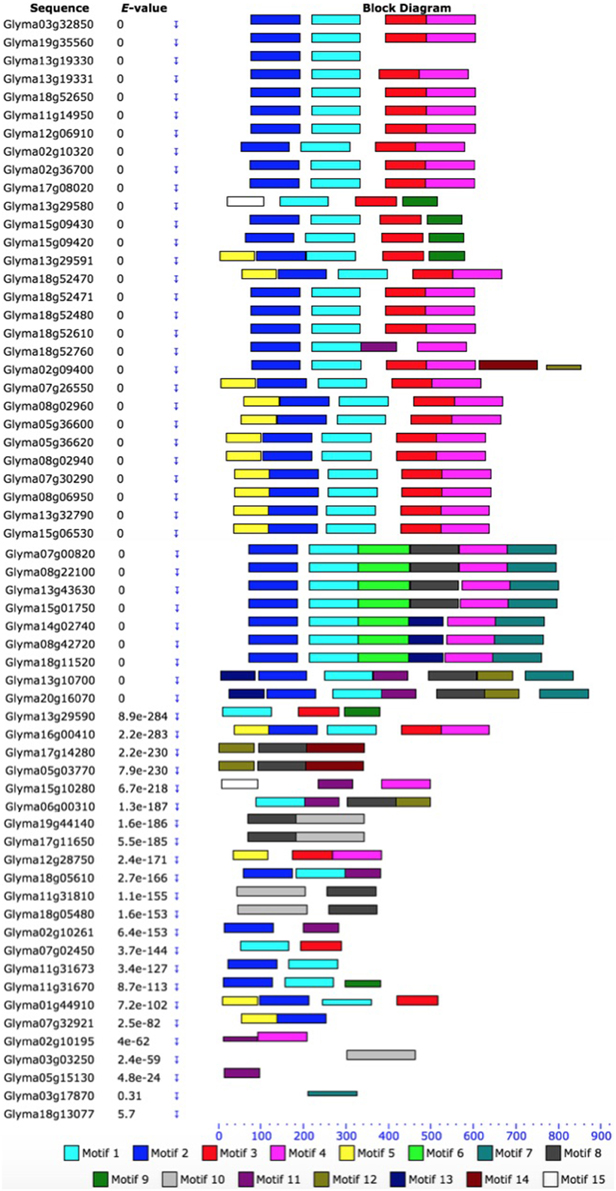
Figure 2. Schematic representation of the conserved motifs in soybean HSP70 proteins elucidated from publicly available data. Each colored box represents a motif in the protein, with the motif name indicated in the box on the right. The length of the protein and motif can be estimated using the scale at the bottom. The scale at the top of the image may be used to estimate motif length. aa, amino acids. A detailed motif introduction is shown in Additional File 7 and 8.
Chromosomal Location and Gene Duplication
The 61 putative GmHSP70 gene candidates were distributed across 17 of the 20 chromosomes in the soybean genome. Among them, chromosome 18 had the highest density of GmHSP70 genes, with 10 members; chromosome 13 had the next highest density, with eight genes; five GmHSP70 genes were located in each of chromosomes 2, 7, 8, and 15; four GmHSP70 genes were situated in chromosomes 5 and 11; three GmHSP70 genes were positioned in chromosomes 3 and 17; two genes each were found in chromosomes 12 and 19; only one gene was found in each of chromosomes 1, 6, 14, 16, and 20; and no GmHSP70 genes were detected in chromosome 3, 9, or 10 (Figure 3). We also investigated the gene duplication events of the GmHSP70 family, and the results showed that there were 24 sister pairs in the 61 GmHSP70 genes (Additional File 10). Only one pair of paralogous genes (Glyma03g17870 and Glyma18g13077) did not reach this standard. The other 23 paralog pairs were all fitted for duplications of the HSP70 genes. Four of the 23 GmHSP70 paralog gene pairs were detected within a distance of less than 5 kb (>100 kb) on chromosomes 11, 13, and 18, which were identified as tandem duplications. The other 19-paralog gene pairs were segmental duplications. These results suggest that segmental duplication played a crucial role in the expansion of the HSP70 gene family in soybeans.
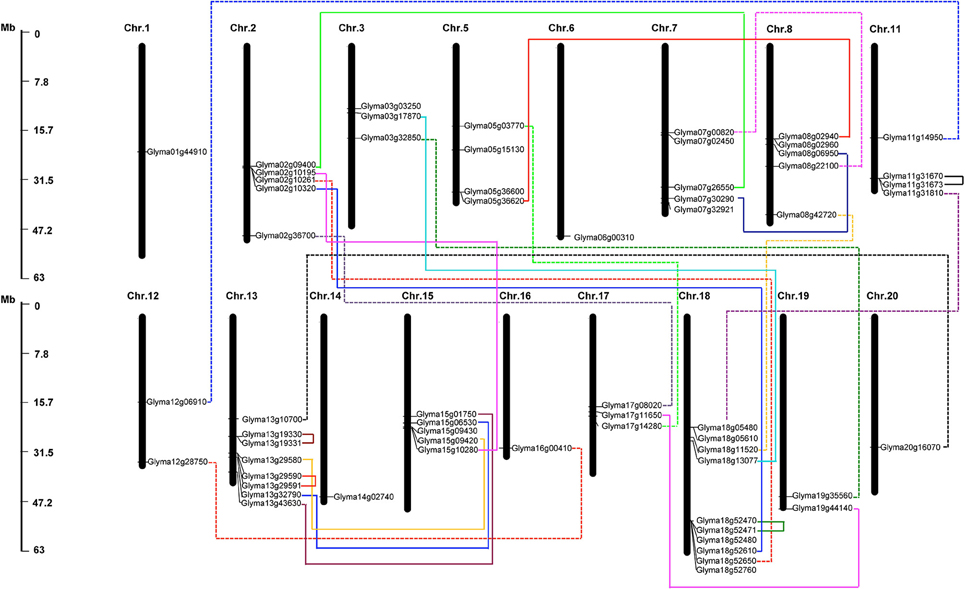
Figure 3. Chromosomal map and duplication event coordinates of paralogous GmHSP70 gene candidates. The identity of each linkage group is indicated at the top of each bar. Only the chromosomes where GmHSP70 genes were mapped are shown. Possible duplicated genes are connected by different color lines. The bar located on the left side indicates chromosome sizes in megabases. The scale represents the length of the chromosome.
Comparative Analysis of the GmHSP70 Genes in Soybean, Arabidopsis, and Rice
The development of comparative genomics has enabled the analysis of the same protein families among different species. The NJ phylogenetic tree was constructed using 111 full-length protein sequences in order to reveal the evolutionary relationships among soybean (61), Arabidopsis (18), and rice (32) HSP70 proteins (Figure 4 and Additional File 9) in our study. The 18 Arabidopsis HSP70 proteins were identified from the TAIR database, and, according to previous studies, the AtHSP70 gene family is divided into five sub-families (Lin et al., 2001). There were 32 OsHSP70 family members identified from the TIGR database, while rice contained six sub-families (Jung et al., 2013; Sarkar et al., 2013). Therefore, based on their phylogenetic relationships, the combined soybean, rice, and Arabidopsis phylogenetic trees can be divided into eight distinct sub-families (class I–VIII; Figure 4). Among the eight clusters, class I was the largest, containing 48 members, and composed of 29 members from soybean, 13 from rice, and 6 from Arabidopsis. Class VII was the second largest, containing 10 members from soybean, four from rice, and four from Arabidopsis. Class II contained 13 members (four soybean, six rice, and three Arabidopsis members). The HSP70 members in Class III were six soybean, five rice, and four in Arabidopsis. Class IV was a small sub-family, which only included one soybean HSP70 gene (Glyma07g32921), and no rice or Arabidopsis. Class V contained three members: one soybean, one rice, and one Arabidopsis. Class VI contained three rice and one soybean member. Class VIII only contained 9 soybean members. With the exceptions of classes IV and VIII, all of the other sub-families contained rice, Arabidopsis, and soybean HSP70 genes. These results suggest that the main characteristics of this family of plants were generated before the dicot/monocot split.
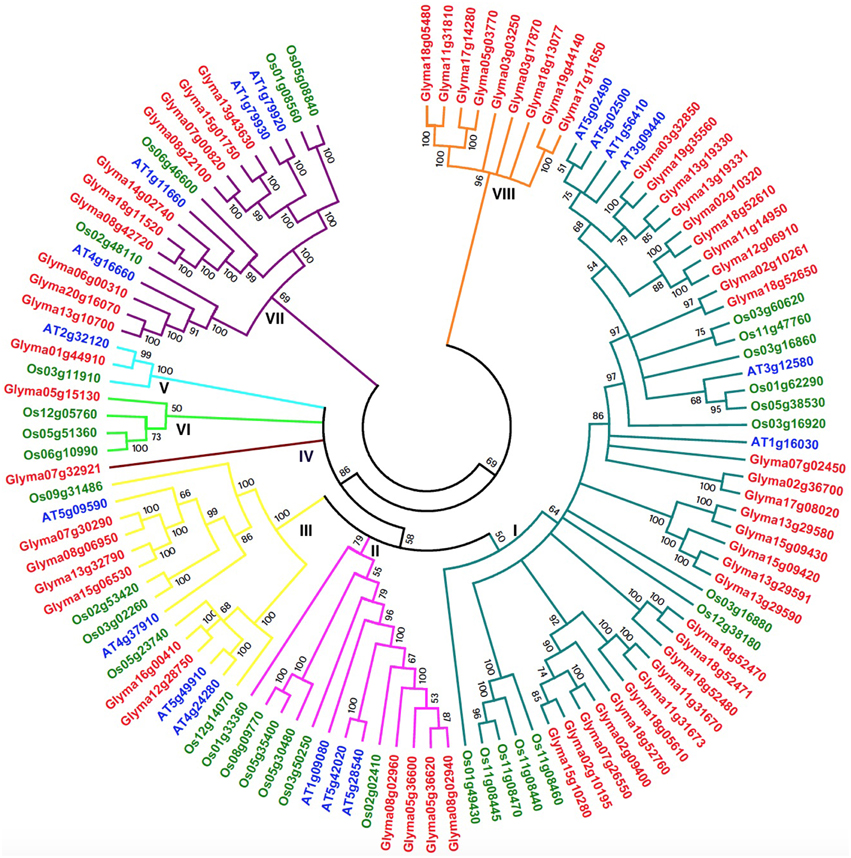
Figure 4. Phylogenetic tree of full-length HSP70 proteins from soybean, Arabidopsis and rice. The 61 soybean, 18 Arabidopsis and 32 rice HSP70 protein sequences were aligned by Clustal X 1.83 and the phylogenetic tree was constructed using MEGA5.0 by the Neighbor-Joining (NJ) method. The Bootstrap value was 1000 replicates. Each HSP70 subfamily is indicated by a specific color.
The combined phylogenetic tree also revealed that the paralogous and orthologous relationships among the HSP70 family members, in particular the duplicated genes, are contained in the paralogous pairs of each species, which supports the occurrence of species-specific HSP70 gene duplication events. In contrast, 24 pairs of paralogous genes from soybean, eight pairs from rice, and five pairs from Arabidopsis were identified (Additional Files 10, 11). Two pairs of orthologous genes from soybean and Arabidopsis were identified, where one was the pair of Glyma01g44910 and At2g32120 in sub-family V, and the other pair was Glyma07g02450 and At1g16030. These results suggest that the ortholog pair originated from the common ancestral genes that existed before the divergence of the monocots and dicots. The fact that the genes in the paralogs accounted for most of the family members confirmed the fact that the soybean had undergone two duplication events after the monocot/dicot split, and most of the HSP70 genes in the soybean had expanded in a species-specific manner.
Expression Patterns of Soybean GmHSP70 Genes in Various Tissues
In order to obtain more insight into the temporal and spatial expression patterns of the soybean HSP70 genes during soybean development, the RNA-Seq Atlas of the Glycine max was searched, and the RNA-Seq atlas data of the soybean HSP70 genes (Additional File 12) were downloaded from the Soybase (http://soybase.org/soyseq/). Due to the fact that the expression profiles of eight GmHSP70 genes (Glyma02g10195, Glyma02g10261, Glyma07g32921, Glyma11g31673, Glyma13g19331, Glyma13g29591, Glyma18g13077, and Glyma18g52471) were not obtained in the soybean database, only the expression patterns of 53/61 GmHSP70 genes were examined. The data analysis revealed that most soybean HSP70 genes exhibit broad expression patterns (Figure 5). With the exception of the six GmHSP70 genes (Glyma06g00310, Glyma07g02450, Glyma11g31670, Glyma15g10280, Glyma18g05610, and Glyma18g52760), which were not expressed in any tissues, the other 47 GmHSP70 genes were expressed in at least one of the seven tissues (young leaves, flowers, 1cm pod, pod-shell, roots, nodules, and seed). This particularly applied to five genes, Glyma05g36620, Glyma13g19330, Glyma03g32850, and Glyma19g35560, which were highly expressed in all types of tissues. These findings indicate that the GmHSP70 genes were involved in multiple processes during the development of the soybean. The heat map also revealed that the majority of the GmHSP70s showed preferential expressions. Based on a hierarchical clustering analysis, the 53 HSP70 genes were mainly clustered into three groups (A, B, and C), as shown in Figure 5. In Group A, with the exception of the six un-expressed genes (Glyma06g00310, Glyma07g02450, Glyma11g31670, Glyma15g10280, Glyma18g05610, and Glyma18g52760), the genes showed partial expressions in the seed (from 10 to 42 DAF). The other two groups (B and C) were the opposite, with most of the genes showing down-expression in the seeds compared with the other tissues. Four GmHSP70 genes (Glyma15g09420, Glyma18g52470, Glyma18g52650, and Glyma11g14960) showed markedly high transcript abundance profiles in only a single tissue type.
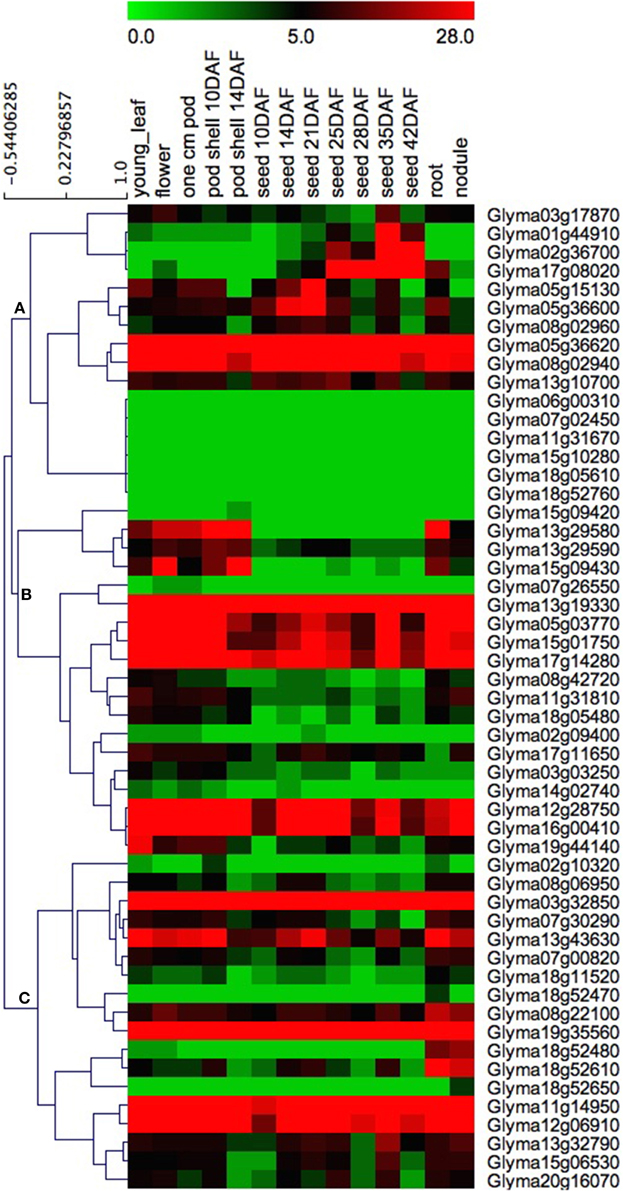
Figure 5. Heat map of the expression profiles of GmHSP70 candidate genes in 14 tissues. RNA-seq relative expression data from 14 tissues were used to reconstruct the expression patterns of soybean genes. Genes were clustered into three groups (A–C). The raw data was normalized and retrieved from the online database http://soybase.org/soyseq/. The normal relative expression levels of 53 GmHSP70 genes are shown in Additional File 12.
qRT-PCR Analysis of Soybean HSP70 Gene Expression under Heat and Drought Stress
In order to gain more insight into the roles of soybean HSP70 genes in heat and drought tolerance, the expression profiles of 61 soybean HSP70 genes in response to drought (0, 3, 6, 12, 24 h) and heat (0, 3, 6, 12, 24 h) stresses were reanalyzed by qRT-PCR. These genes expressed diversely under both stresses (Figures 6A,B). For the drought treatment (15% PEG), these genes could be divided into four clusters. Cluster 1 contains 31 (50.8%) members of detectable GmHSP70 genes, which were widely upregulated after 6 h by drought treatment, and up to the highest expression level at 12 h, then downregulated at 24 h after drought treatment. Cluster 2 mainly consists of 13 (21.3%) genes, which were widely downregulated by drought treatment. Cluster 3 has 6 (9.8%) members, which showed a weakly upregulated at 6 or 12 h, but downregulated at 24 h after drought treatment. Cluster 4 contains 11 (18%) genes, which were mainly upregulated with the increased level of drought treatment after 24 h, and the Glyma02g36700 in particular was highly induced at 3 h after drought treatment. Overall, 30 GmHSP70 genes were up-regulated from two-fold to 222-fold after drought treatment relative to the control, and the other GmHSP70 genes were down-regulated (<0.5-fold) at some stage or showed no changes during the drought stress treatment (Additional File 14).
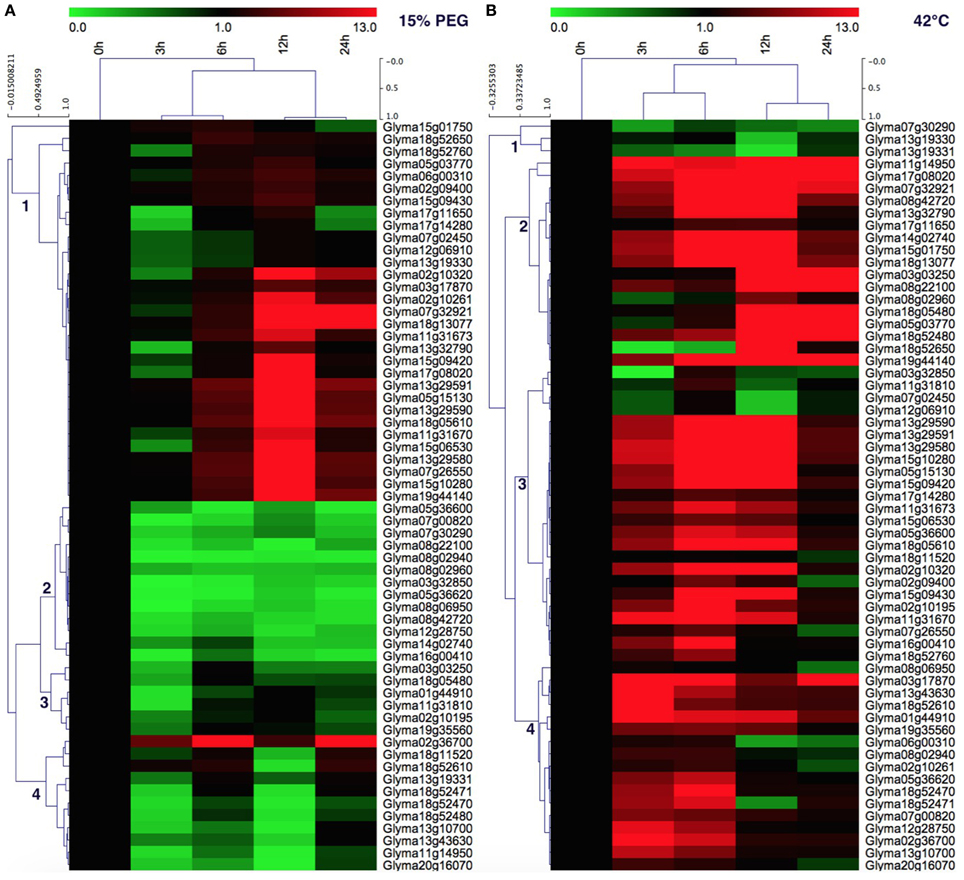
Figure 6. Quantitative RT-PCR analysis of the GmHSP70 gene expression in soybean leaves in response to drought stress and 42°C heat stress. (A) Expression levels of 61 GmHSP70 family genes under drought stress. (B) Expression levels of 61 GmHSP70 family genes under heat stress. The normalized relative expression level of the 0 h treatment time point was set up as 1. Leaves collected at 0, 3, 6, 12, and 24 h post-drought or at 0, 3, 6, 12, and 24 h post heat stress. Means are calculated from technical triplicate qRT-PCR measurements within three biological replicates. The qRT-PCR data was shown in the Additional File 14.
For the heat stress treatment, the heatmap also showed the genes clustered in four groups. Cluster 1 has 3 (4.91%) members of 61 detectable GmHSP70 genes, which were widely downregulated after 42°C treatment. The 17 members of cluster 2 were mainly upregulated after the heat treatment, and exhibited the highest expression after 12 h. Most members (24) of cluster 3 were mainly upregulated after 3 h under the 42°C treatment, but were downregulated after 24 h. And the cluster 4 members (17) had a similar expression with cluster 3, but the difference was most of genes downregulated after 12 h. Overall, 55 of GmHSP70 genes were upregulated from two-fold to 241-fold after 42°C treatment; only 6 GmHSP70 genes were downregulated or nearly unchanged under the heat treatment.
Notably, 29 GmHSP70 genes showed upregulation under both drought and heat stress conditions. Five soybean HSP70 genes were downregulated both under the drought and heat stress. 26 upregulated genes typically induced by heat were downregulated during drought stress. In contrast, only one gene, Glyma18g11520, was downregulated by the heat treatment and upregulated by the drought stress (Figure 6 and Additional File 14).
Discussion
Preliminary analysis of the HSP70 gene family has been performed in the model plants Arabidopsis and rice (Lin et al., 2001; Jung et al., 2013; Sarkar et al., 2013). However, this family has not previously been studied in soybean. Therefore, we performed an overall analysis of the HSP70 gene family in soybean, including analysis of their phylogeny, chromosomal location, gene structure, conserved motifs and expression profiles. The Arabidopsis genome is 125 Mbp in size, with 26,500 predicted coding genes, while the soybean is 1150 Mbp, with 46,400 predicted coding genes. Therefore, the soybean possesses a 9.2-fold larger genome size and a 1.75-fold higher gene count than the Arabidopsis (Sterck et al., 2007; Cannon and Shoemaker, 2012). The HSP70 gene family in soybean is by far the largest, compared with that in other plant species (18 in Arabidopsis, 32 in rice, and 30 in cotton) (Lin et al., 2001; Jung et al., 2013; Sarkar et al., 2013; Zhang et al., 2014).
HSP70 is a multi-genic family. Various members constituting this family were present in different cellular compartments (Cho and Choi, 2009; Kose et al., 2012). In our study, with the analysis of subcellular localization of GmHSP70 proteins, we found that multiple HSP70 members were identified in various cellular compartments in soybean. For instance, we found 34 putative nucleo/cytosolic members in soybean, compared with the OsHSP70 proteins, which contained 11 putative nucleo/cytosolic members and five cytosolic HSP70 in Arabidopsis (Lin et al., 2001; Jung et al., 2013; Sarkar et al., 2013). The GmHSP70 proteins encoded for 11 chloroplast, four mitochondria, three plasma membrane, and two endoplasmic reticulum.
Analyses of cis-elements in the promoters suggest that GmHSP70s might respond to regulate plant growth and development, as well as different environmental stresses and stimuli (Additional File 6). In a previous study, the promoters for 11 Arabidopsis HSP70 genes were examined for the presence of two major temperature responsive cis-elements, heat shock element (HSE), and C-repeat or dehydration responsive element (CRT/DRE) (Sung et al., 2001). HSE has been linked with the heat inducible expression of many heat shock genes (Czarnecka et al., 1989). CRT/DRE is known to be associated with drought- and cold-inducible expression of many genes (Yamaguchi-Shinozaki and Shinozaki, 1994). In our cis-acting elements analysis of GmHSP70s, we found that 45 of 61 GmHSP70 genes contain the HSE element, and one gene (Glyma03g17870) contains the DRE element. These results suggest that most of the GmHSP70s might be significantly related to heat-stress response.
Based on the phylogenetic, gene structure, and motif analysis of the HSP70 proteins in soybean, we discovered that the most closely related members in the same sub-families share similar exon/intron structures and intron numbers, and these results were also consistent with the characteristics defined in the above phylogenetic analysis (Figure 1B). For example, in sub-family I, most members contain 1 to 5 exons, while those in sub-family II contain 8 exons. In the terminal branch of the phylogenetic tree, the number of exons/introns were very similar in some of the sister pairs (Figure 1 and Additional File 10). However, there were still some sister pairs that showed changes in their intron/exon structure and numbers, for instance, Glyma13g19330 and Glyma13g19331. These findings indicated that some intron loss, along with intron gain events, might have occurred during the structural evolution between the two families of soybean HSP70 encoding genes. The same situation was also revealed by motif analysis; the type, order, and number of motifs were similar in proteins with the same sub-family, but differed from the proteins in other sub-families.
We also investigated gene duplication events in order to further understand the expansion mechanism of the soybean HSP70 gene family. The duplications of individual genes, chromosomal segments, or entire genomes, have been major forces in the evolution of plant genome structure and content during the process of genome evolution (Sémon and Wolfe, 2007). The soybean genome is speculated to undergo at least two rounds of genome-wide duplication, followed by multiple segmental duplication, tandem duplication, and transposition events. A tandem duplication event is confirmed by the presence of two or more genes on the same chromosome, while a segmental duplication event is defined as gene duplication on different chromosomes (Schlueter et al., 2007).
Our heat map data showed that most GmHSP70 genes were expressed in different tissues and organs of soybean. These results indicate that they may participate in growth and development. The expression patterns of the paralogous pairs in the different sub-families were revealed by heat maps, and it was determined that most of the paralogous pairs with high sequence similarity had similar expression patterns in different tissues. For instance, the Glyma03g32850 and Glyma19g35560 pair showed a strong expression in all of the tissues types. However, the Glyma13g29580 and Glyma15g09430 had a low expression in all the seed stages. An expression divergence was also found in the paralogous pairs. For example, in the Glyma11g31810 and Glyma18g05480 pair, Glyma18g05480 was only highly expressed in the roots and nodules, with little or no expression in the other tissue types. Meanwhile, its paralog, Glyma11g31810, was not only expressed in the roots and nodules, but also had an obvious expression in the young leaves, flowers, and pods. The paralogous pair of Glyma02g10320 and Glyma18g52610 showed under-expression in all of the tissue types, with the exception of Glyma18g52610 exhibiting an obvious expression in the roots and nodules. When the Glyma07g00820 and Glyma08g22100 pair was compared with Glyma07g00820, it was found that Glyma08g22100 also displayed an obvious expression in the pods and seeds. Similar cases have also been observed in the Glyma13g10700 and Glyma20g16070 pair (Figure 5 and Additional File 10).
Previous studies have shown that HSP70 genes play an important role in plants in response to various stresses, such as drought, salt, freezing temperatures, and heat (Kurepa et al., 2009). In this study, the expression profiles of the GmHSP70 genes under drought and heat stresses (Figure 6 and Additional File 14) revealed that the soybean HSP70 genes were widely involved in responding to drought and heat stresses. It is interesting to note that most of GmHSP70 genes (55) were strongly upregulated during heat stress, which indicates that those genes might be mainly involved in the heat stress biological pathway. Some GmHSP70 genes showed the same expression profile under the heat or drought treatment, being either upregulated or downregulated. This result suggests that these GmHSP70 genes were co-expressed in responses to drought and heat stresses, and those genes might play shared roles in drought and heat stresses. However, the gene expressions profiles which responded to the different stress conditions usually tended to be different; some heat-regulated genes were downregulated in drought stress, which indicated that two sets of GmHSP70 genes were involved in the response to the drought and heat stress, respectively. These results also imply that the signaling pathways in the plants' abiotic stress responses are very complicated systems. From an applied perspective, the identification of the HSP70s with potential value in the improvement of stress resistance of the soybean would likely benefit from targeting such genes which are part of the abiotic and biotic stress response networks.
Conclusions
The soybean (Glycine max L.) genome contains 61 members of the HSP70 gene family, and these genes are located within 17 chromosomes. The genome distribution, gene organizations, and gene structures suggest a complex evolutionary history of this family in soybeans. The soybean HSP70 family is greatly contributed to by segment/chromosomal duplications, which may be associated with the soybean genome fusion events. Their expression profiles in various tissues and responses to drought and heat stress conditions demonstrate that this gene family is widely involved in soybean organ development, as well as drought and heat stress responses. The overall picture of this gene family and their potential involvement in growth, development, and stress responses, will facilitate further research on the HSP70 gene family, particularly in regards to their evolutionary history and biological functions.
Author Contributions
LZ, HZ, and YD planned and designed the study. LZ performed the computational analysis, executed the experiments, generated the figures, and drafted the manuscript. YZ and QD contributed to the execution of the abiotic and biotic experiments. HL and GX also contributed to the sample preparations. YW provided the nematode material, along with contributing by leading the biotic experiment. LZ and YZ performed the qRT-PCR data analysis. QL and YD contributed to the discussion portion of the manuscript. All of the authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the National Natural Science Foundation of China (31300282, 31501327), the National High Technology Research and Development Program of China (2012AA101106-3) and the Ministry of Agriculture of China for Transgenic Research (2014ZX08004-003). The Youth Scientific Research Foundation of Jilin (20150520123JH). The authors report no conflicts of interest. The authors alone are responsible for the content and composition of this paper.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00773
References
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucl. Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Cannon, S. B., and Shoemaker, R. C. (2012). Evolutionary and comparative analyses of the soybean genome. Breed. Sci. 61, 437–444. doi: 10.1270/jsbbs.61.437
Cho, E. K., and Choi, Y. J. (2009). A nuclear-localized HSP70 confers thermoprotective activity and drought-stress tolerance on plants. Biotechnol. Lett. 31, 597–606. doi: 10.1007/s10529-008-9880-5
Czarnecka, E., Key, J. L., and Gurley, W. B. (1989). Regulatory domains of the Gmhsp17.5 heat shock promoter soybean. Mol. Cell. Biol. 9, 3457–3463.
De Maio, A. (1999). Heat shock proteins: facts, thoughts, and dreams. Shock 11, 1–12. doi: 10.1097/00024382-199901000-00001
Dragovic, Z., Broadley, S. A., Shomura, Y., Bracher, A., and Hartl, F. U. (2006). Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of HSP70s. EMBO J. 25, 2519–2528. doi: 10.1038/sj.emboj.7601138
Duan, Y. H., Guo, J., Ding, K., Wang, S. J., Zhang, H., Dai, X. W., et al. (2011). Characterization of a wheat HSP70 gene and its expression in response to stripe rust infection and abiotic stresses. Mol. Biol. Rep. 38, 301–307. doi: 10.1007/s11033-010-0108-0
Duck, N. B., and Folk, W. R. (1994). HSP70 heat shock protein cognate is expressed and stored in developing tomato pollen. Plant Mol. Biol. 26, 1031–1039. doi: 10.1007/BF00040686
Dufresne, P. J., Thivierge, K., Cotton, S., Beauchemin, C., Ide, C., Ubalijoro, E., et al. (2009). Heat shock 70-protein interaction with Turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology 374, 217–227. doi: 10.1016/j.virol.2007.12.014
Emanuelsson, O., Brunak, S., von Heijne, G., and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971. doi: 10.1038/nprot.2007.131
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Francki, M. G., Berzonsky, W. A., Ohm, H. W., and Anderson, J. M. (2002). Physical location of a HSP70 gene homologue on the centromere of chromosome 1B of wheat (Triticum aestivum L.). Theor. Appl. Genet. 104, 184–191. doi: 10.1007/s00122-001-0781-4
Friedman, M., and Brandon, D. L. (2001). Nutritional and health benefits of soy proteins. J. Agric. Food Chem. 49, 1069–1086. doi: 10.1021/jf0009246
Fuganti, R., Machado Mde, F., Lopes, V. S., Silva, J. F., Arias, C. A., Marin, S. R., et al. (2010). Size of AT (n) insertions in promoter region modulates Gmhsp17.6-L mRNA transcript levels. J. Biomed. Biotechnol. 2010:847673. doi: 10.1155/2010/847673
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucl. Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Grant, D., Nelson, R. T., Cannon, S. B., and Shoemaker, R. C. (2010). SoyBase, the USDA ARS soybean genetics and genomics database. Nucleic Acids Res. 38, D843–D846. doi: 10.1093/nar/gkp798
Guo, A. Y., Zhu, Q. H., Chen, X., and Luo, J. C. (2007). GSDS: a gene structure display server. Yi Chuan 29, 1023–1026. doi: 10.1360/yc-007-1023
Guo, M., Zhai, Y. F., Lu, J. P., Chai, L., Chai, W. G., Gong, Z. H., et al. (2014). Characterization of CaHSP70-1, a pepper heat-shock protein gene in response to heat stress and some regulation exogenous substances in Capsicum annuum L. J. Mol. Sci. 15, 19741–19759. doi: 10.3390/ijms151119741
Hafrén, A., Hofius, D., Rönnholm, G., Sonnewald, U., and Mäkinen, K. (2010). HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell 22, 523–535. doi: 10.1105/tpc.109.072413
He, Z., Xie, R., Wang, Y., Zou, H., Zhu, J., and Yu, G. (2008). Cloning and characterization of a heat shock protein 70 gene, MsHSP70-1, in Medicago sativa. Acta Biochim. Biophys. Sin. 40, 209–216. doi: 10.1111/j.1745-7270.2008.00394.x
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Jiang, S., Lu, Y., Li, K., Lin, L., Zheng, H., Yan, F., et al. (2014). Heat shock protein 70 is necessary for Rice stripe virus infection in plants. Mol. Plant Pathol. 15, 907–917. doi: 10.1111/mpp.12153
Jinn, T. L., Chen, Y. M., and Lin, C. Y. (1995). Characterization and physiological function of Class I low-molecular-mass, heat-shock protein complex in soybean. Plant Physiol. 108, 693–701.
Jung, K. H., Gho, H. J., Nguyen, M. X., Kim, S. R., and An, G. (2013). Genome-wide expression analysis of HSP70 family genes in rice and identification of a cytosolic HSP70 gene highly induced under heat stress. Funct. Integr. Genomics 13, 391–402. doi: 10.1007/s10142-013-0331-6
Jungkunz, I., Link, K., Vogel, F., Voll, L. M., Sonnewald, S., and Sonnewald, U. (2011). AtHSP70-15-deficient Arabidopsis plants are characterized by reduced growth, a constitutive cytosolic protein response and enhanced resistance to TuMV. Plant J. 66, 983–995. doi: 10.1111/j.1365-313X.2011.04558.x
Kim, B. M., Rhee, J. S., Jeong, C. B., Seo, J. S., Park, G. S., Lee, Y. M., et al. (2014). Heavy metals induce oxidative stress and trigger oxidative stress-mediated heat shock protein (hsp) modulation in the intertidal copepod Tigriopus japonicus. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 166, 65–74. doi: 10.1016/j.cbpc.2014.07.005
Kose, S., Furuta, M., and Imamoto, N. (2012). Hikeshi, a nuclear import carrier for HSP70s, protects cells from heat shock-induced nuclear damage. Cell 149, 578–589. doi: 10.1016/j.cell.2012.02.058
Krishna, P., Sacco, M., Cherutti, J. F., and Hill, S. (1995). Cold-induced accumulation of hsp90 transcripts in Brassica napus. Plant Physiol. 107, 915–923.
Kurepa, J., Wang, S., Li, Y., and Smalle, J. (2009). Proteasome regulation, plant growth and stress tolerance. Plant Signal Behav. 4, 924–927. doi: 10.4161/psb.4.10.9469
Lamesch, P., Berardini, T. Z., Li, D., Swarbreck, D., Wilks, C., Sasidharan, R., et al. (2012). The arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40, D1202–D1210. doi: 10.1093/nar/gkr1090
Letunic, I., Copley, R. R., Schmidt, S., Ciccarelli, F. D., Doerks, T., Schultz, J., et al. (2004). SMART 4.0: towards genomic data integration. Nucleic Acids Res. 32, D142–D144. doi: 10.1093/nar/gkh088
Li, H., Liu, S. S., Yi, C. Y., Wang, F., Zhou, J., Xia, X. J., et al. (2014). Hydrogen peroxide mediates abscisic acid-induced HSP70 accumulation and heat tolerance in grafted cucumber plants. Plant Cell Environ. 37, 2768–2780. doi: 10.1111/pce.12360
Lin, B. L., Wang, J. S., Liu, H. C., Chen, R. W., Meyer, Y., Barakat, A., et al. (2001). Genomic analysis of the HSP70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 6, 201–208. doi: 10.1379/1466-1268(2001)006<0201:GAOTHS>2.0.CO;2
Liu, J. Z., and Whitham, S. A. (2013). Overexpression of a soybean nuclear localized type-III DnaJ domain-containing HSP40 reveals its roles in cell death and disease resistance. Plant J. 74, 110–221. doi: 10.1111/tpj.12108
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lopes-Caitar, V. S., de Carvalho, M. C., Darben, L. M., Kuwahara, M. K., Nepomuceno, A. L., Dias, W. P., et al. (2013). Genome-wide analysis of the Hsp20 gene family in soybean: comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genomics 14:577. doi: 10.1186/1471-2164-14-577
Lopez-Matas, M. A., Nuñez, P., Soto, A., Allona, I., Casado, R., Collada, C., et al. (2004). Protein cryoprotective activity of a cytosolic small heat shock protein that accumulates constitutively in chestnut stems and is up-regulated by low and high temperatures. Plant Physiol. 134, 1708–1717. doi: 10.1104/pp.103.035857
Maimbo, M., Ohnishi, K., Hikichi, Y., Yoshioka, H., and Kiba, A. (2007). Induction of a small heat shock protein and its functional roles in Nicotiana plants in the defense response against Ralstonia solanacearum. Plant Physiol. 145, 1588–1599. doi: 10.1104/pp.107.105353
Mar, J. C., Wells, C. A., and Quackenbush, J. (2011). Defining an informativeness metric for clustering gene expression data. Bioinformatics 27, 1094–1100. doi: 10.1093/bioinformatics/btr074
Mathioudakis, M. M., Veiga, R., Ghita, M., Tsikou, D., Medina, V., Canto, T., et al. (2012). Pepino mosaic virus capsid protein interacts with a tomato heat shock protein cognate 70. Virus Res. 163, 28–39. doi: 10.1016/j.virusres.2011.08.007
Mayer, M. P., and Bukau, B. (2005). HSP70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684. doi: 10.1007/s00018-004-4464-6
Mohammadi, P. P., Moieni, A., Hiraga, S., and Komatsu, S. (2012). Organ-specific proteomic analysis of drought-stressed soybean seedlings. J. Proteomics 75, 1906–1923. doi: 10.1016/j.jprot.2011.12.041
Nagy, P. D., Wang, R. Y., Pogany, J., Hafrén, A., and Makinen, K. (2011). Emerging picture of host chaperone and cyclophilin roles in RNA virus replication. Virology 411, 374–382. doi: 10.1016/j.virol.2010.12.061
Sabehat, A., Lurie, S., and Weiss, D. (1998). Expression of small heat-shock proteins at low temperatures. A possible role in protecting against chilling injuries. Plant Physiol. 117, 651–658. doi: 10.1104/pp.117.2.651
Sarkar, N. K., Kundnani, P., and Grover, A. (2013). Functional analysis of HSP70 superfamily proteins of rice (Oryza sativa). Cell Stress Chaperones 18, 427–437. doi: 10.1007/s12192-012-0395-6
Schlueter, J. A., Lin, J. Y., Schlueter, S. D., Vasylenko-Sanders, I. F., Deshpande, S., Yi, J., et al. (2007). Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genomics. 8:330. doi: 10.1186/1471-2164-8-330
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Sémon, M., and Wolfe, K. H. (2007). Consequences of genome duplication. Curr. Opin. Genet. Dev. 17, 505–512. doi: 10.1016/j.gde.2007.09.007
Sterck, L., Rombauts, S., Vandepoele, K., Rouze, P., and Van de Peer, Y. (2007). How many genes are there in plants (…and why are they there)? Curr. Opin. Plant Biol. 10, 199–203. doi: 10.1016/j.pbi.2007.01.004
Su, P. H., and Li, H. M. (2008). Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiol. 146, 1231–1241. doi: 10.1104/pp.107.114496
Sung, D. Y., Vierling, E., and Guy, C. L. (2001). Comprehensive expression profile analysis of the Arabidopsis HSP70 gene family. Plant Physiol. 126, 789–800. doi: 10.1104/pp.126.2.789
Swindell, W. R., Huebner, M., and Weber, A. P. (2007). Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 8:125. doi: 10.1186/1471-2164-8-125
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Wang, L., Guo, K., Li, Y., Tu, Y., Hu, H., Wang, B. R., et al. (2010). Expression profiling and integrative analysis of the CESA/CSL superfamily in rice. BMC Plant Biol. 10:282. doi: 10.1186/1471-2229-10-282
Wang, L., Yang, S., Han, L., Fan, D., and Zhao, K. (2014). Phenotypic plasticity of HSP70s gene expression during diapause: signs of evolutionary responses to cold stress among Soybean Pod Borer populations (Leguminivora glycinivorella) in Northeast of China. PLoS ONE 9:e109465. doi: 10.1371/journal.pone.0109465
Wang, R. Y., Stork, J., and Nagy, P. D. (2009a). A key role for heat shock protein 70 in the localization and insertion of tombusvirus replication proteins to intracellular membranes. J. Virol. 83, 3276–3287. doi: 10.1128/JVI.02313-08
Wang, R. Y., Stork, J., Pogany, J., and Nagy, P. D. (2009b). A temperature sensitive mutant of heat shock protein 70 reveals an essential role during the early steps of tombusvirus replication. Virology 394, 28–38. doi: 10.1016/j.virol.2009.08.003
Wang, W., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Wei, F., Coe, E., Nelson, W., Bharti, A. K., Engler, F., Butler, E., et al. (2007). Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 3:e123. doi: 10.1371/journal.pgen.0030123
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. doi: 10.1105/tpc.6.2.251
Yuan, Q., Ouyang, S., Liu, J., Suh, B., Cheung, F., and Sultana, R. (2003). The TIGR rice genome annotation resource: annotating the rice genome and creating resources for plant biologists. Nucleic Acids Res. 31, 229–233. doi: 10.1093/nar/gkg059
Zhang, Y., Wang, M., Chen, J., Rong, J., and Ding, M. (2014). Genome-wide analysis of HSP70 superfamily in Gossypium raimondii and the expression of orthologs in Gossypium hirsutum. Yi Chuan 36, 921–933. doi: 10.3724/SP.J.1005.2014.0921
Zhang, Z. L., Zhu, J. H., Zhang, Q. Q., and Cai, Y. B. (2009). Molecular characterization of an ethephon-induced HSP70 involved in high and low-temperature responses in Hevea brasiliensis. Plant Physiol. Biochem. 47, 954–959. doi: 10.1016/j.plaphy.2009.06.003
Keywords: soybean (Glycine max L.), HSP70 gene family, genome-wide analysis, phylogeny, gene structure, expression pattern
Citation: Zhang L, Zhao H-K, Dong Q-L, Zhang Y-Y, Wang Y-M, Li H-Y, Xing G-J, Li Q-Y and Dong Y-S (2015) Genome-wide analysis and expression profiling under heat and drought treatments of HSP70 gene family in soybean (Glycine max L.). Front. Plant Sci. 6:773. doi: 10.3389/fpls.2015.00773
Received: 29 June 2015; Accepted: 09 September 2015;
Published: 25 September 2015.
Edited by:
Janila Pasupuleti, International Crops Research Institute for the Semi Arid Tropics, IndiaReviewed by:
Fan Chen, Institute of Genetics and Developmental Biology, ChinaXingliang Hou, South China Botanical Garden, China
Copyright © 2015 Zhang, Zhao, Dong, Zhang, Wang, Li, Xing, Li and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qi-Yun Li, Institute of Plant Protection, Jilin Academy of Agricultural Sciences, No. 303 Kemaoxi Street, Gongzhuling, JiLin 136100, China, qyli1225@126.com;
Ying-Shan Dong, Agro-Biotechnology Research Institute, Jilin Academy of Agricultural Sciences, No. 1363 Shengtai Street, Jing Yue District, ChangChun, JiLin 130033, China, ysdong@cjaas.com
 Ling Zhang
Ling Zhang Hong-Kun Zhao
Hong-Kun Zhao Qian-Li Dong
Qian-Li Dong Yuan-Yu Zhang
Yuan-Yu Zhang Yu-Min Wang
Yu-Min Wang Hai-Yun Li
Hai-Yun Li Guo-Jie Xing
Guo-Jie Xing Qi-Yun Li
Qi-Yun Li Ying-Shan Dong
Ying-Shan Dong