- 1State Key Laboratory of Crop Stress Biology for Arid Areas, College of Agronomy, Northwest A&F University, Yangling, China
- 2Guizhou Rapeseed Institute, Guizhou Academy of Agricultural Sciences, Guiyang, China
The diploid Aegilops tauschii is the D-genome donor to hexaploid wheat (Triticum aestivum) and represents a potential source for genetic study in common wheat. The ubiquitous wax covering the aerial parts of plants plays an important role in protecting plants against non-stomatal water loss. Cuticular waxes are complex mixtures of very-long-chain fatty acids, alkanes, primary and/or secondary alcohols, aldehydes, ketones, esters, triterpenes, sterols, and flavonoids. In the present work, primary alcohols were identified as the major components of leaf cuticular wax in Ae. tauschii, with C26:0-OH being the dominant primary alcohol. Analysis by scanning electron microscope revealed that dense platelet-shaped wax crystals were deposited on leaf surfaces of Ae. tauschii. Ten putative wax biosynthetic genes encoding fatty acyl-coenzyme A reductase (FAR) were identified in the genome of Ae. tauschii. Five of these genes, Ae.tFAR1, Ae.tFAR2, Ae.tFAR3, Ae.tFAR4, and Ae.tFAR6, were found expressed in the leaf blades. Heterologous expression of the five Ae.tFARs in yeast (Saccharomyces cerevisiae) showed that Ae.tFAR1, Ae.tFAR2, Ae.tFAR3, Ae.tFAR4, and Ae.tFAR6 were predominantly responsible for the accumulation of C16:0, C18:0, C26:0, C24:0, and C28:0 primary alcohols, respectively. In addition, nine Ae.tFAR paralogous genes were located on D chromosome of wheat and the wheat nullisomic–tetrasomic lines with the loss of Ae.tFAR3 and Ae.tFAR4 paralogous genes had significantly reduced levels of primary alcohols in the leaf blades. Collectively, these data suggest that Ae.tFAR1, Ae.tFAR2, Ae.tFAR3, Ae.tFAR4, and Ae.tFAR6 encode alcohol-forming FARs involved in the biosynthesis of primary alcohols in the leaf blades of Ae. tauschii. The information obtained in Ae. tauschii enables us to better understand wax biosynthesis in common wheat.
Introduction
The outermost surfaces of land plants are covered by the cuticle, which is composed of a cutin polymer matrix and cuticular waxes. Cutin is a three dimensional polymer of mainly C16 and C18 hydroxy fatty acids, which are cross-linked in ester bonds directly or through a glycerol backbone (Pollard et al., 2008; Li and Beisson, 2009). Cuticular wax is a mixture of lipids, consisting of, for example, very-long-chain fatty acids (VLCFAs, C20 to C34) and their derivatives, including alcohols, aldehydes, alkanes, ketones, and wax esters (Kunst and Samuels, 2003; Samuels et al., 2008; Lee and Suh, 2013). Cutin and waxes are integrated into the outer portion of the polysaccharide cell wall, forming a lipid-impregnated zone known as the cuticular layer, and are also layered on top of the cuticular layer to create a hydrophobic surface devoid of polysaccharides called the cuticle proper (Jeffree, 2006). Cuticular wax has multiple functions in plants, with the primary one being as a water permeability barrier that restricts non-stomatal water loss from epidermal surfaces (Kerstiens, 2006; Domínguez et al., 2011). Cuticular wax also protects plants against ultraviolet radiation, pathogen attack, and insect destruction (Eigenbrode and Espelie, 1995; Jenks et al., 2002; Solovchenko and Merzlyak, 2003; Pfündel et al., 2006).
Wax composition can be quite distinct depending on species, organs, and developmental stage. The composition also tends to be influenced by a variety of environmental factors, such as light, temperature and humidity (Kolattukudy, 1996; Samuels et al., 2008). The biosynthesis of wax has been thoroughly characterized in Arabidopsis. It starts with the de novo synthesis of C16 or C18 fatty acid in the plastids of epidermal cells. Then, elongation of C16 and C18 fatty acyl-CoAs via fatty acid elongase (FAE) complex in the endoplasmic reticulum (ER) leads to the production of VLCFA wax precursors (Samuels et al., 2008; Kunst and Samuels, 2009). Following elongation is the production of wax components by converting long-chain fatty acyl-CoAs via two different pathways: the acyl-reduction pathway, producing primary alcohols and wax esters (Samuels et al., 2008), and the decarbonylation pathway, generating aldehydes, alkanes, secondary alcohols, and ketones (Schneider-Belhaddad and Kolattukudy, 2000). Fatty acyl reductase catalyzes the conversion of fatty acyl to primary alcohols and different fatty acyl-CoA reductases (FARs) generally show distinct specificities for acyl chain lengths, leading to the production of fatty alcohols with various chain lengths. In Arabidopsis, FAR2/MS2 and FAR6 preferentially produce sporopollenin-related long-chain fatty alcohols (Doan et al., 2009; Chen et al., 2011). FAR1, FAR4, and FAR5 are primarily responsible for generating suberin-associated C22:0-OH, C20:0-OH, and C18:0-OH, respectively (Domergue et al., 2010), while FAR3/CER4 in Arabidopsis is associated with the production of C24:0, C26:0, C28:0, and C30:0 primary fatty alcohols (Rowland et al., 2006). Some FAR genes related to wax biosynthesis have subsequently been identified and characterized in bread wheat. Expression of these TaFARs in various heterologous systems leads to the accumulation of C18:0-OH to C30:0-OH. TaFAR3 catalyzes the biosynthesis of C28 primary alcohol, which is the major wax component in wheat leaf, whereas TaFAR1 and TaFAR4 generate C22 and C24 primary alcohols, respectively (Wang et al., 2015b, 2016).
In wheat, one of the key staple crops worldwide, primary alcohols of C20 to C32 are important cuticular wax components (Wang et al., 2015a). In a previous work, 32 genes were identified encoding FARs associated with wax biosynthesis in the leaf blade of wheat and 12 of them are located on D genome (Wang et al., 2016). Of 12 genes, TaFAR2 and TaFAR5 are responsible for the biosynthesis of C18 and C22 primary alcohols, respectively (Wang et al., 2015c, 2016). However, information regarding the rest of genes encoding alcohol-forming enzymes on D genome is still lacking. Common wheat is an allohexaploid species (Triticum aestivum, 2n = 42, AABBDD), deriving from the hybridization between tetraploid wheat Triticum turgidum (2n = 28, AABB) and diploid grass Aegilops tauschii (2n = 14, DD) (Pestsova et al., 2000; Jia et al., 2013). Being the D-genome donor to hexaploid wheat, Ae. tauschii serves as an important resource for wheat breeding and provides ample information for genomic study in wheat. The availability of complete genome data makes the utility of this species even further (Jia et al., 2013). Thus, taking advantage of Ae. tauschii, we may have a better understanding of mechanisms of primary alcohol biosynthesis in wheat. The objective of our present work was to identify FARs involved in the biosynthesis of wax primary alcohols in the leaf blades of Ae. tauschii and through analysis of these FARs to acquire more information on alcohol-forming FARs in hexaploid wheat.
Materials and Methods
Plant Materials and Growth Conditions
The wild diploid grass Ae. tauschii, hexaploid wheat (T. aestivum L.) cv. Chinese Spring (CS) and wheat nullisomic–tetrasomic lines were grown at the experimental farm of Northwest A&F University, Yangling, China. Eight-week old leaves at the seedling stage and flag leaves, leaf sheaths, peduncles, glumes, and anthers (160 days old) at the flowering stage were used for analysis of wax composition and micromorphology. Simultaneously, materials were collected for analysis of tissue-specific expression patterns of Ae.tFARs. The seedling leaves and flag leaves were used for quantitative real-time PCR (qRT-PCR) and cuticular wax analysis in wild-type wheat and nullisomic–tetrasomic lines.
Wax Extraction and Chemical Characterization
Wax load and chemical composition were determined in Ae. tauschii and hexaploid wheat. Various organs including seedling leaves, flag leaves, leaf sheaths, peduncles, glumes, and anthers from Ae. tauschii and leaves from wheat were harvested and immersed in chloroform for 60 s to extract waxes. For analysis by GC-MS and GC-FID, dried wax samples were derivatized with equal amounts of pyridine (Alfa Aesar) and BSTFA [bis-N,O-(trimethylsilyl)trifluoroacetamide] (Fluka) for 40 min at 70°C in a small GC vials. Then, samples were dried under nitrogen gas before being re-dissolved in chloroform (Wang et al., 2015c). n-tetracosane (C24) was added as an internal standard. Wax compounds were quantified by gas chromatography equipped with mass spectrometric detector (GC-MS) (GCMS-QP2010, SHIMADZU, Japan). Each wax component load was calculated based on peak area of each compound and that of the C24 internal standard. The sample surface areas were determined using ImageJ software.
Scanning Electron Microscopy (SEM) Analysis
To observe the micromorphology of wax crystals, various organs including seedling leaves, flag leaves, leaf sheaths, peduncles, glumes, and anthers were gently flattened and dried in oven. Sample segments were carefully mounted onto scanning electron microscopy (SEM) stubs, and then coated with superfine platinum powder in an ion sputter coater (Hitachi E-1045, Japan). A Hitachi S-4800 field emission SEM was used to view the surfaces of coated samples at an accelerating voltage of 5 kV (Wang et al., 2016).
Isolation of Ae.tFAR Genes from Ae. tauschii Leaf
Total RNA was extracted from seedling leaves of wild Ae. tauschii using Trizol extraction reagent (TaKaRa, Japan). Then, 1 μg RNA was used for first-strand cDNA synthesis by PrimeScript® reverse transcriptase (TaKaRa) with primer oligo (dT)18. To amplify the putative Ae.tFARs, PCR was performed with HS PrimeSTAR® DNA polymerase (TaKaRa) and the following cycling conditions: initial denaturation at 94°C for 1 min, followed by 35 cycles, 98°C for 10 s, 58°C for 30 s, 72°C for 2 min. Gene-specific primers Ae.tFARx were used to amplify the full-length coding sequences (Supplementary Table 1). The amplified DNA fragments were separated and purified using the DNA purification kit (TIANGEN, China). These corresponding fragments were cloned into pMD-18T vector (TaKaRa) and sequenced. Subsequent construction of expression vectors was made using this clone as template.
Functional Expression of Ae.tFARs in Yeast
The entire coding sequences of Ae.tFAR1, Ae.tFAR2, Ae.tFAR3, Ae.tFAR4, and Ae.tFAR6 were amplified using primers shown in Supplementary Table 1. The amplified DNA fragments were double digested and cloned into the corresponding sites of the yeast (Saccharomyces cerevisiae) expression vector pYES3 (Invitrogen) under the control of GAL1 promoter to generate pYES3-Ae.tFARx. A vector of p416 MET25-FLAG3:Sur4-F262A/K266L was used to obtain the yeast mutant INVSur4# (a mutation in ketoacyl-CoA synthase, KCS), which allows for the production of fatty acids with longer chain lengths in yeast (Denic and Weissman, 2007). The p416 MET25-FLAG3:Sur4-F262A/K266L was co-transformed with pYES3-Ae.tFARx into wild-type INVSc1 strain (MATa his3-D1 leu2 trp1-289 ura3-52). The corresponding empty vectors were used as positive control as detailed in Supplementary Table 2.
The yeast transformation was carried out as described by Gietz and Woods (2002). Yeast cells transformed with different vector combinations were cultured on synthetic complete (SC) selection medium lacking tryptophan and uracil with 2% (w/v) glucose for 48 h, induced by 2% (w/v) galactose for 12 h and followed by incubation in 0.1 M potassium phosphate and 2% (w/v) glucose for 24 h at 28°C. For extraction of lipids from yeast cells, 40 mL of liquid cultures were collected, re-suspended in 20% KOH/50% ethanol, and extracted three times with hexane at 70°C for 5 min as described by Wang et al. (2011).
For analysis of fatty alcohols, lipid mixtures from yeast were separated and purified by thin layer chromatography (TLC) plate (5 cm × 10 cm, silica gel, HSGF254, 0.15–0.20 mm) using chloroform as the mobile phase. Different lipids on the plates were then stained with primuline and observed by UV light. The fatty alcohol bands were scratched off and were further analyzed by GC-MS and GC-FID.
Quantitative Real-Time PCR Analysis
Transcript levels were determined by qRT-PCR. Various tissues including seedling leaves, flag leaves, leaf sheaths, peduncles, glumes, and anthers from Ae. tauschii and seedling leaves and flag leaves from wheat were used for total RNA extractions. The RNA samples were then used for cDNA synthesis by PrimeScript® reverse transcriptase following standard protocols. The qRT-PCR was performed using SYBR® Premix Ex TaqTM (TaKaRa) with gene-specific primers (Supplementary Table 1). Ae. tauschii GAPDH gene (GenBank accession no. KD518701) and wheat Actin gene (GenBank accession no. DQ115883) were used as internal expressed controls. Primers of GAPDH and Actin were designed to amplify a 177- and 118-bp fragment, respectively. Real-time PCR was carried out in 25 μl reaction volume using a CFX96 real-time PCR detection system (BIO-RAD, United States) with the following program: 1 cycle of 30 s at 95°C, followed by 40 cycles of 5 s at 95°C and 30 s at 60°C.
Phylogenetic Analysis
Multiple sequence alignments were performed with DNAMAN program using amino acid sequences of FARs from different species. A phylogenetic tree was created with the MEGA software (version 5.1) based on a Maximum Likelihood method (1,000 replicates). TMHMM Server (version 2.0) was used to predict potential transmembrane domains.
Results
Chemical Composition and Micromorphology of Wax in the Leaf Blades of Ae. tauschii
To investigate genes involved in wax biosynthesis in Ae. tauschii, we first analyzed the wax load and composition from various organs, including seedling leaves, flag leaves, leaf sheaths, peduncles, glumes, and anthers by GC-MS and GC-FID. The results showed that the waxes from different organs consisted of the same chemical classes, including fatty acids, primary alcohols, esters, aldehydes, alkanes, hentriacontane-14, 16-dione (β-diketone), and OH-β-diketone (Table 1), however, the relative amount of each chemical class was quite different depending on organs. Alkanes and β-diketones were the dominant chemical species in panicle and glume, respectively, while fatty acids and aldehydes were major chemical classes in anthers. Primary alcohol was found to be the richest chemical class in leaves, accounting for 84% of total wax load in seedling leaves and 76% in flag leaves, whereas only small amounts of alkanes and aldehydes were detected in leaves (Figure 1 and Table 1). The high level of primary alcohol was also observed in leaf sheaths, being 57% of total wax load (Table 1). The chain lengths of primary alcohols ranged from C20 to C32, among which C26 was the most prevalent one in seedling leaf, flag leaf, and leaf sheath (Figures 1A–C). Intermediate levels of primary alcohols, β-diketone and alkanes were observed. Chain length distributions of primary alcohols in peduncles and glumes varied from C20 to C32 and the dominant chain length was C28, which is different from that in leaves (Figures 1D,E). These results suggest that primary alcohols are the key wax components and vary considerably depending on organs in leaves of Ae. tauschii.
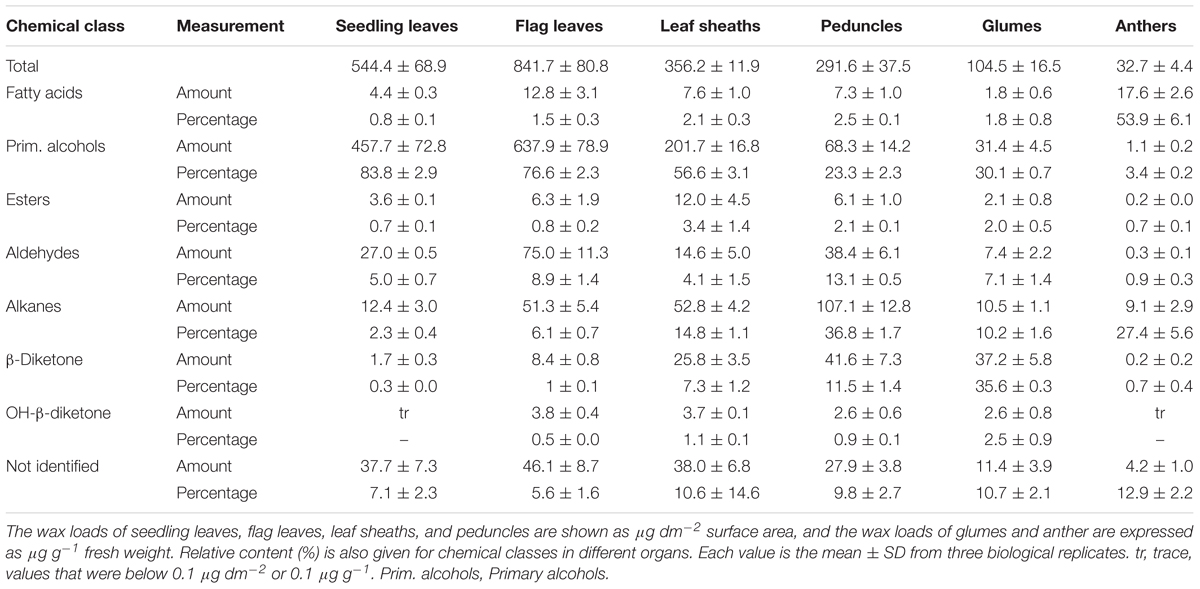
TABLE 1. Wax composition of seedling leaves, flag leaves, leaf sheaths, peduncles, glumes and anthers of in Aegilops tauschii.
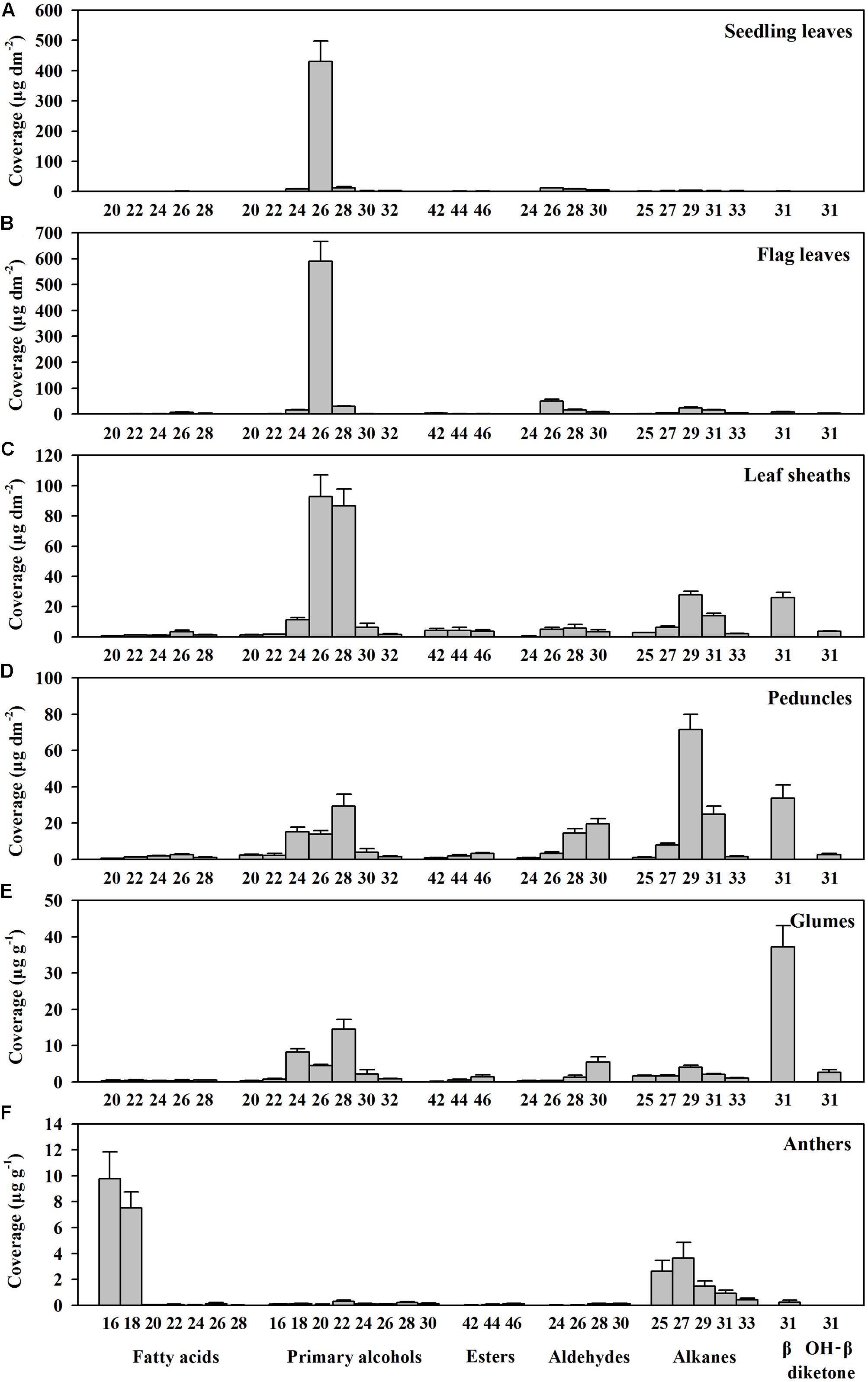
FIGURE 1. Chain length distributions within compound classes of cuticular wax from various organs of Aegilops tauschii. Cuticular wax mixtures were extracted from (A) seedling leaves, (B) flag leaves, (C) leaf sheaths, (D) peduncles, (E) glumes, and (F) anthers of Ae. tauschii. The coverage of individual compound is expressed as μg per unit dm2 (seedling leaves, flag leaves, leaf sheaths, and peduncles) and μg per unit g fresh weight (glumes and anthers). Values are means of three biological replicates. Error bars = SD.
Since wax morphology is influenced by wax composition (Zhang et al., 2013), we investigated the morphology of wax crystals deposited in various organs by SEM. In seedling leaves, a dense array of wax platelets was found covering both adaxial and abaxial surfaces (Figures 2A–D), while in flag leaves, wax crystal density of both sides was different, with thicker platelet-shaped wax crystals being on adaxial surface (Figures 2E,F) and scattered platelet-shaped crystals being on abaxial surface (Figures 2G,H). Similarly, platelet-shaped crystals were found in leaf sheaths, peduncles, and glumes, but distributed sparsely (Figures 2I–N). In anthers, wave-shaped wax crystals were found (Figures 2O,P). These observations are consistent with studies showing that platelet-shaped wax crystals are associated with the deposition of primary alcohols in wheat leaf blades (Hallam and Chambers, 1970; Gülz, 1994).
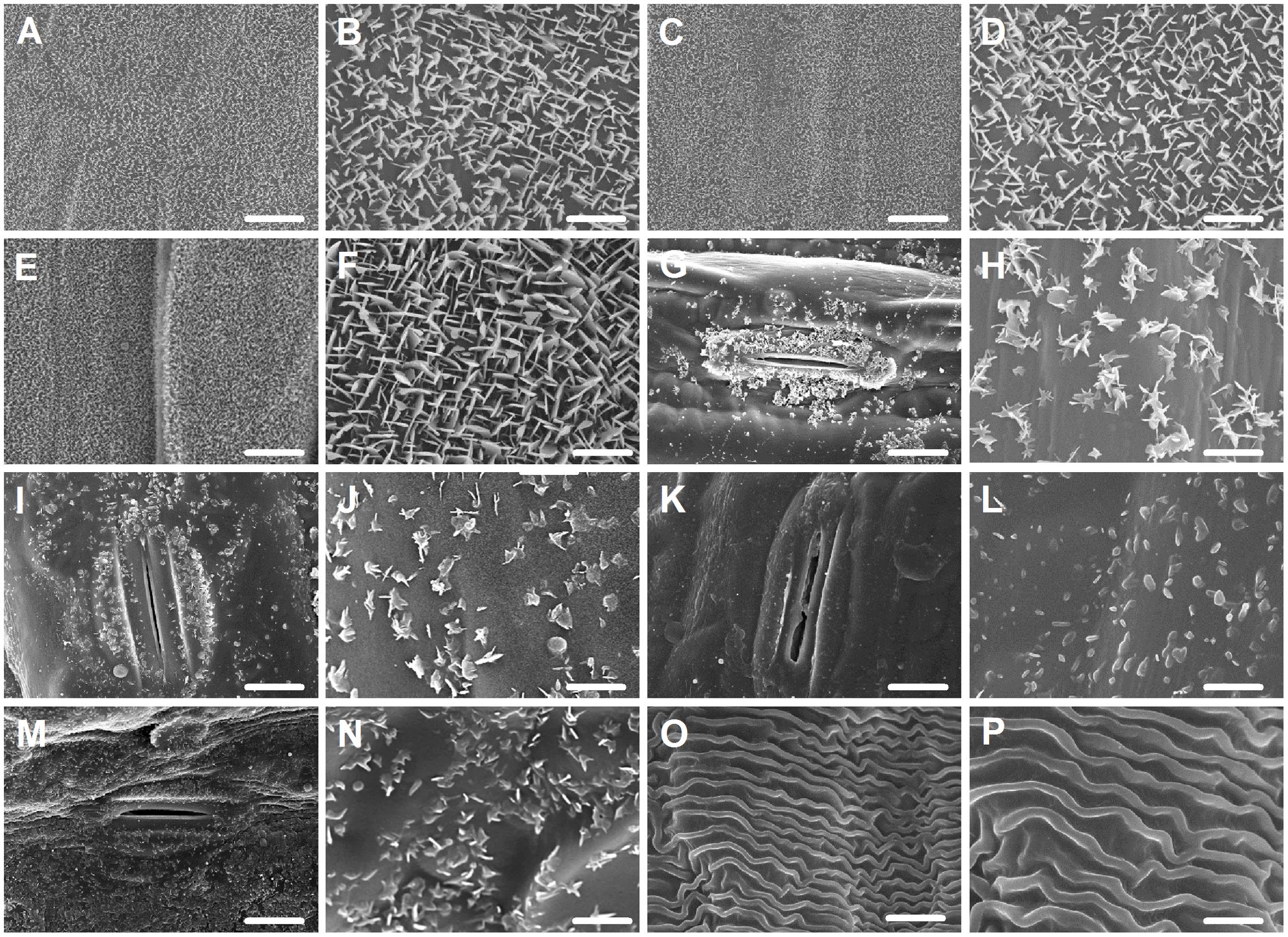
FIGURE 2. Epicuticular wax crystals on the surfaces of various organs in Ae. tauschii detected by scanning electron microscope (SEM). (A,B) Adaxial surface of seedling leaf. (C,D) Abaxial surface of seedling leaf. (E,F) Adaxial surface of flag leaf. (G,H) Abaxial surface of flag leaf. (I,J) Leaf sheaths. (K,L) Peduncles. (M,N) Glumes. (O,P) Anthers. (A,C,E,G,I,K,M,O) Were detected at 2,000× magnification, Scale bars = 10 μm. (B,D,F,H,J,L,N,P) Were detected at 10,000× magnification, Scale bars = 2 μm.
Previous studies have shown that formation of primary alcohols was catalyzed by FAR in the acyl-reduction pathway (Kunst and Samuels, 2003; Samuels et al., 2008). Since high level of primary alcohols are present in Ae. tauschii leaves, FAR may play a significant role in the accumulation of leaf cuticular wax in Ae. tauschii. We thus continued to search for the genes encoding FARs in Ae. tauschii.
Molecular Identification of Ae.tFARs from Ae. tauschii Leaf
Using the amino acid sequence of Arabidopsis CER4/FAR3 (GenBank accession no. NP567936) as a query, a BLAST search was performed in the Ae. tauschii genome database (NCBI BioProject ID: PRJNA182898). In total, 10 FAR-like genes (designated as Ae.tFAR1 to Ae.tFAR10) were obtained, with amino acid sequences being 24–47% and 40–100% identical with Arabidopsis CER4 and wheat FARs over the entire length, respectively (Supplementary Tables 3, 5).
Tissue-specific expression patterns of these putative Ae.tFAR genes were determined by qRT-PCR using RNA derived from various organs, including seedling leaves, flag leaves, leaf sheaths, peduncles, glumes, and anthers (Figure 3A). The results showed that Ae.tFAR3 was the most expressed gene in seedling leaves and flag leaves, while Ae.tFAR5 was largely restricted to seedling leaves and anthers. Ae.tFAR4 was found to be widely expressed in all aerial organs, whereas Ae.tFAR7 to Ae.tFAR10 genes were expressed only at later developmental stages of Ae. tauschii. Ae.tFAR1, Ae.tFAR2, and Ae.tFAR6 transcripts were detected in all organs, but at low levels.
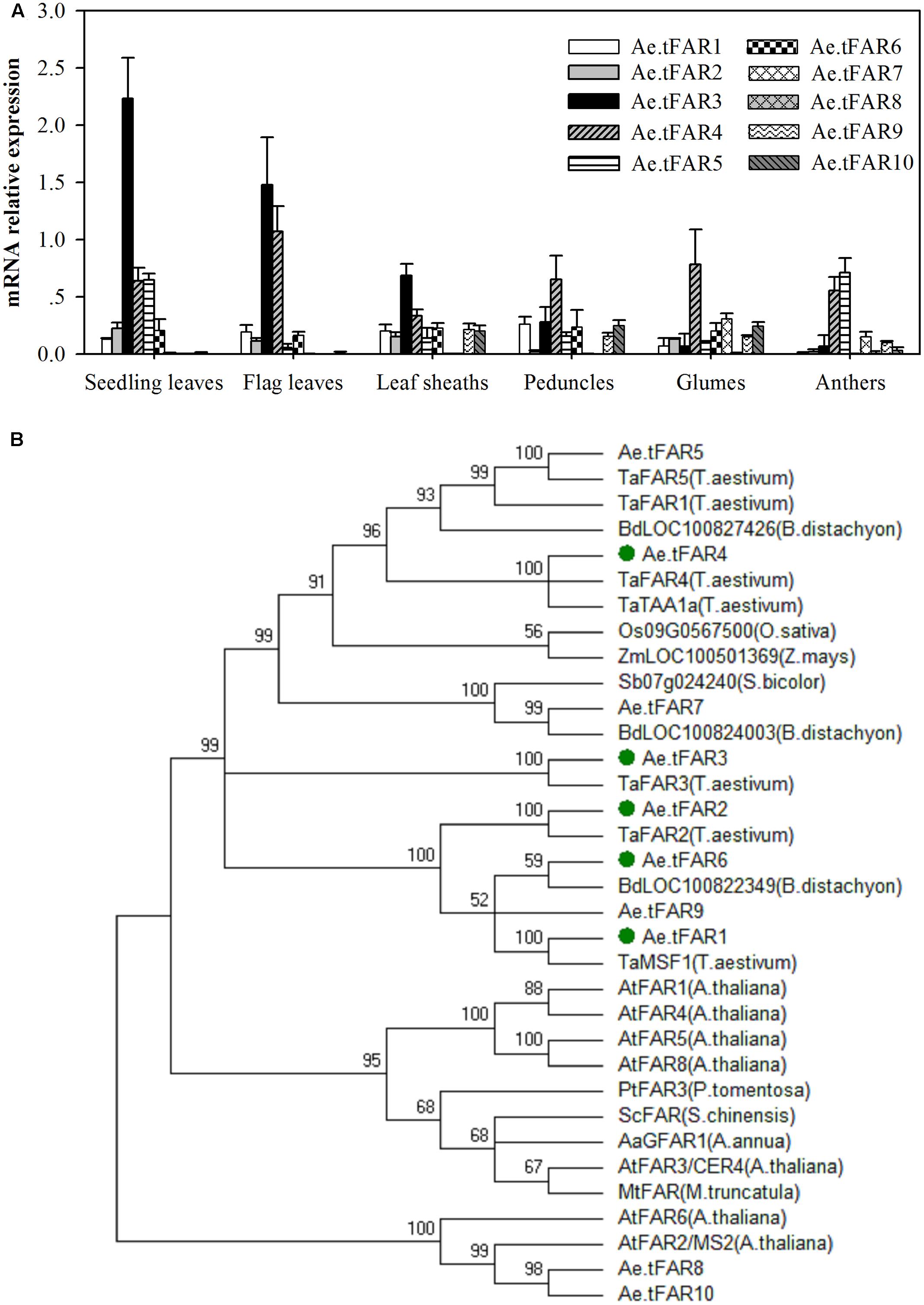
FIGURE 3. Gene expression and phylogenetic analysis of Ae.tFAR. (A) Tissue-specific gene expression patterns of Ae.tFAR by quantitative real-time PCR (qRT-PCR). Each value is the mean of three biological replicates. Error bars = SD. (B) Phylogenetic tree of the alcohol-forming fatty acyl-CoA reductases (FARs) from different species. The GenBank accession numbers of the FAR sequences from different plant species are summarized in Supplementary Table 4.
In order to identify FARs associated with biosynthesis of leaf cuticular wax, FAR genes expressed in leaves of Ae. tauschii were selected for further analysis. Of six genes (Ae.tFAR1–6) expressed in leaves, Ae.tFAR5 was 100% identical with a known alcohol-forming FAR (TaFAR5) in hexaploid wheat (Wang et al., 2015c). Thus, we subsequently focused on the other five FAR-like genes (Ae.tFAR1–4 and Ae.tFAR6) in Ae. tauschii. A phylogenetic analysis using neighbor-joining methods showed that the five Ae.tFARs were highly related to FARs from other species and belonged to the same monocot clade as wheat FARs (Figure 3B). These five Ae.tFARs were further analyzed to determine whether they are involved in the biosynthesis of primary alcohols in the leaf blades of Ae. tauschii.
Analysis of Ae.tFARs
To isolate the five FAR candidates, reverse transcription-PCR (RT-PCR) was performed with sets of specific primers and templates that were derived from leaf blades of Ae. tauschii. Sequence analysis revealed that Ae.tFAR1, Ae.tFAR2, Ae.tFAR3, Ae.tFAR4, and Ae.tFAR6 cDNA contained open reading frames of 1,497, 1,470, 1,497, 1,524, and 1500bp, encoding a polypeptide of 498, 489, 498, 507, and 499 amino acids, respectively. The molecular masses of Ae.tFAR1, Ae.tFAR2, Ae.tFAR3, Ae.tFAR4, and Ae.tFAR6 protein were predicted to be 57.0, 55.8, 55.7, 57.3, and 56.7 kDa, respectively.
A predictive protein analysis indicated that the five Ae.tFARs contain two distinct FAR protein domains: a Rossmann-fold NADB binding domain (FAR-N_SDR_e) linked with a FAR domain (FAR_C) (Figure 4A), in accordance with FAR proteins from other plant species, such as TaFAR1, TaFAR2, TaFAR3, and TaFAR4 in wheat (Wang et al., 2015b, 2016), and AtFAR5 and AtFAR8 in Arabidopsis (Chacón et al., 2013). Proteins with these two domains are believed to function as FARs, acting on medium- and long-chain fatty acids. Furthermore, the Rossmann-fold domain had two classical conserved motifs, a NAD(P)H binding site motif (TGXXGXXG) and an active site motif (YXXXK) (Figure 4B). Analysis by TMHMM program revealed that Ae.tFAR3 and Ae.tFAR4 contain one transmembrane helix between residues 391 to 413 and 411 to 433, respectively. This feature was found to be similar with that in jojoba FAR, which also contains membrane-spanning domains (Metz et al., 2000). However, no transmembrane domain was found in Ae.tFAR1, Ae.tFAR2, and Ae.tFAR6, which is similar to the alcohol-forming Arabidopsis FAR3/CER4 (Rowland et al., 2006, Figure 4B). Based on these analyses, we hypothesized that these five Ae.tFAR proteins might have the reductive activity of FAR and be involved in primary alcohol biosynthesis in leaf blades of Ae. tauschii.
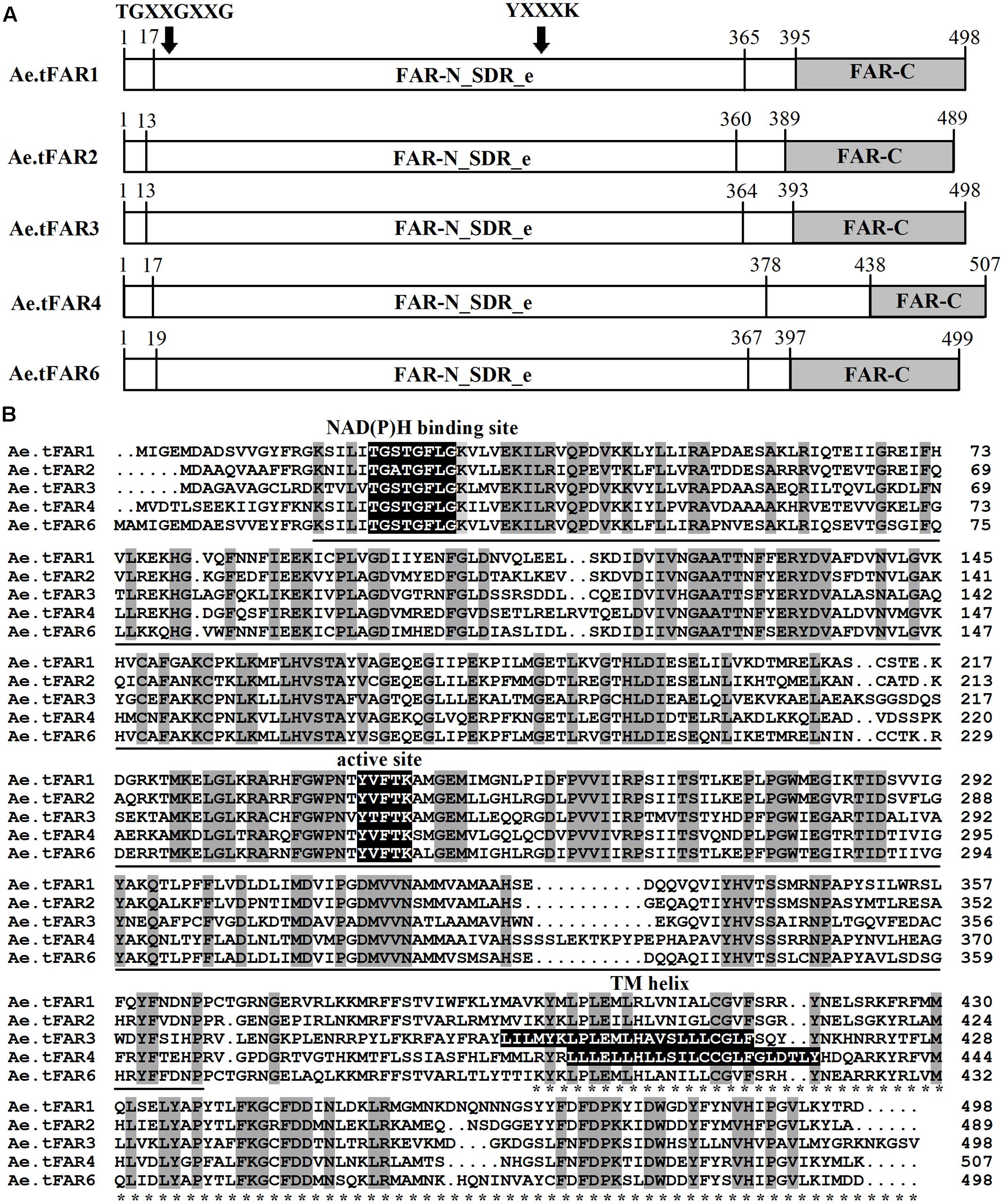
FIGURE 4. The structure and functional domains of the five Ae.tFAR proteins and amino acid sequence alignment. (A) Predicted functional domains of Ae. tauschii FARs. The conserved FAR-N_SDR_e and FAR_C domains are marked by underline and asterisks, respectively. The numbers indicate amino acid positions. (B) Amino acid sequences of the five Ae.tFARs. Identical amino acids are highlighted in dark gray. Three conserved motifs, NAD(P)H binding site motif (TGXXGXXG), active site motif (YXXXK), and TM (transmembrane) helix motif are highlighted in black (where X represents any amino acid).
Heterologous Expression of Putative Ae.tFARs in Yeast
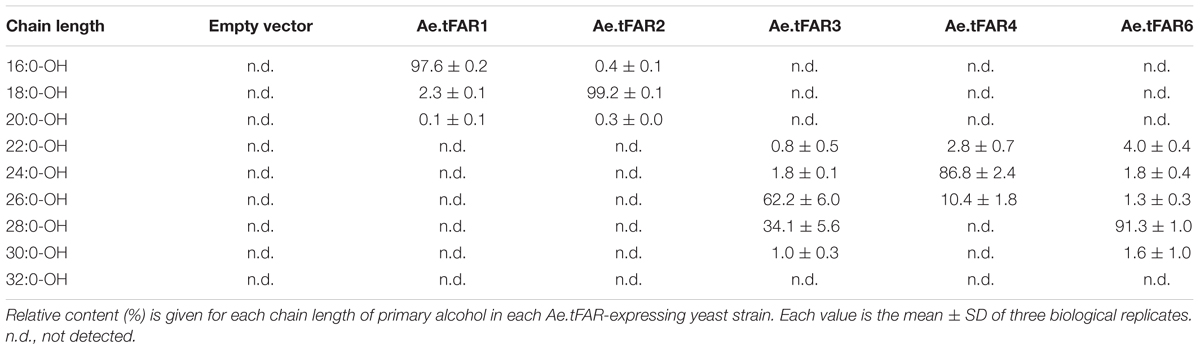
To prove that Ae.tFAR1, Ae.tFAR2, Ae.tFAR3, Ae.tFAR4, and Ae.tFAR6 are FARs responsible for the production of primary alcohols, we expressed the five Ae.tFAR genes in a yeast system. The open reading frames of the five Ae.tFAR genes were cloned into the yeast expression vector pYES3 downstream of the GAL1 promoter. Since C20 to C32 primary alcohols are components of leaf cuticular wax in Ae. tauschii, the presence of same chain lengths of VLCFA substrates in yeast is required. However, wild-type yeast can not produce VLCFAs of longer than C28, which may hamper us from correctly assessing the catalytic capacities of Ae.tFARs (Oh et al., 1997; Rowland et al., 2006; Domergue et al., 2010). To circumvent this problem, we used a yeast mutant INVSur#4 by introducing the vector p416 MET25-FLAG3:Sur4-F262A/K266L into wild-type yeast. This yeast mutant was capable of producing ≥C28 fatty acids (Denic and Weissman, 2007; Bernard et al., 2012, Supplementary Table 2). When Ae.tFARs were expressed in INVSur4#, all transgenic yeast cells produced novel chemical compounds (Figure 5), which were identified as primary alcohols ranging from C16 to C20 in yeast cells expressing Ae.tFAR1 and Ae.tFAR2 (Figures 5B,C), from C22 to C30 in yeast cells expressing Ae.tFAR3 and Ae.tFAR6 (Figures 5D,F), and from C22 to C26 in yeast cells expressing Ae.tFAR4 (Figure 5E). In contrast, yeast cells hosting empty vectors accumulated no primary alcohols (Figure 5A). It is noted that all these primary alcohols produced by Ae.tFARs in yeast were consistent with those detected in the leaf blades of Ae. tauschii in our study (Figure 5G).
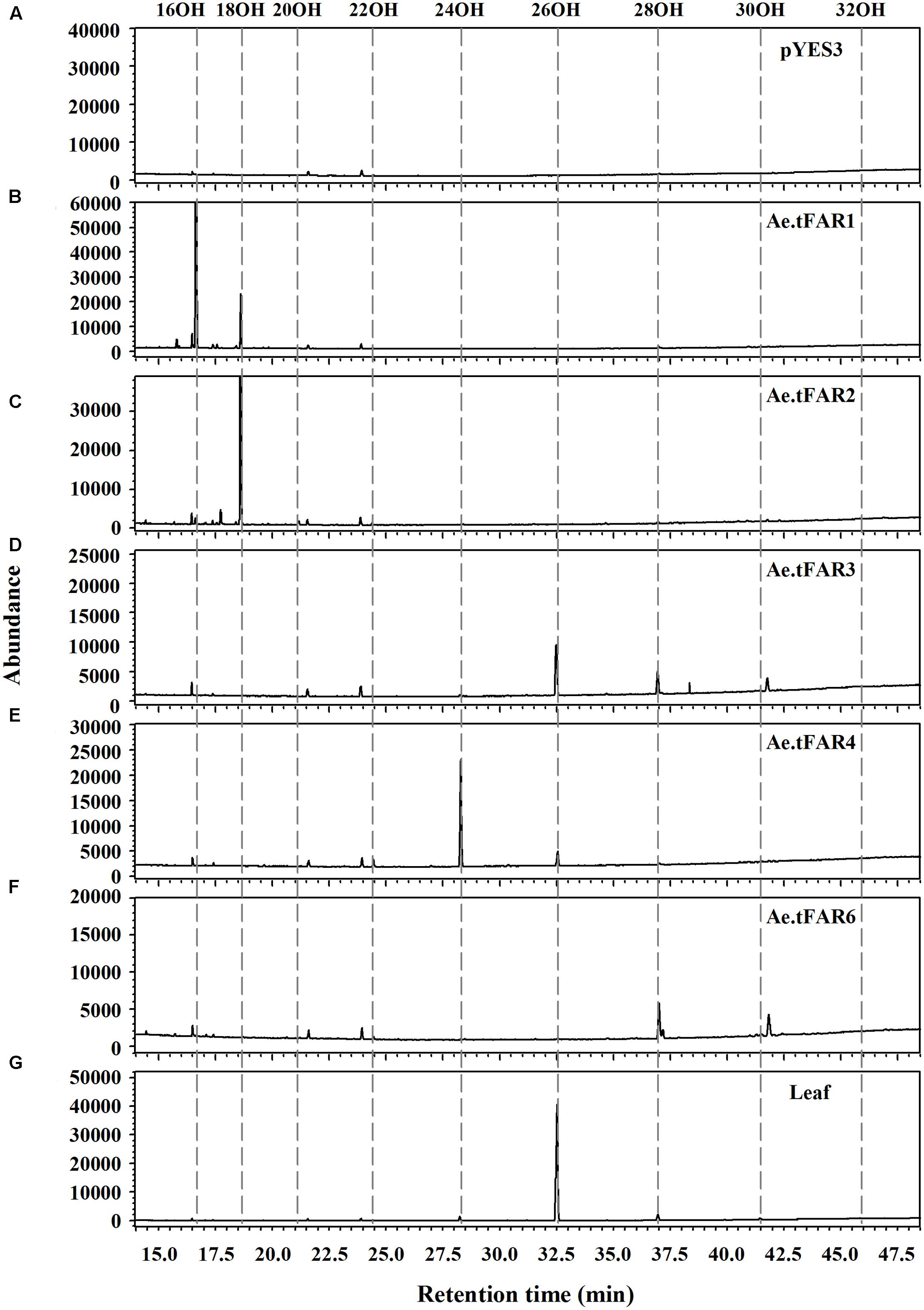
FIGURE 5. Heterologous expression of the five Ae.tFARs in yeast mutant INVSur4#. The yeast expressing (A) empty vector control pYES3 or vector harboring (B) Ae.tFAR1 (C) Ae.tFAR2, (D) Ae.tFAR3, (E) Ae.tFAR4 or (F) Ae.tFAR6. Transgenic yeast cells were grown in stringent medium lacking tryptophan and uracil. Primary alcohols were separated from other neutral lipids by thin layer chromatography (TLC). In empty vector control, no primary alcohols were detected. (G) All primary alcohols in transgenic yeast were also observed in the leaf blades of Ae. tauschii.
Further analysis revealed that each Ae.tFAR showed distinct chain length preferences. Expression of Ae.tFAR1 resulted in large quantities of long-chain C16 alcohol (97.6%), and very small amounts of C18 (2.3%) and C20 (0.1%) alcohols. In contrast, expression of Ae.tFAR2 mainly produced C18:0-OH (99.2%), with traces of C16:0-OH (0.4%) and C20:0-OH (0.3%) being detected. It appeared that Ae.tFAR4 preferred producing C24:0-OH (86.8%) to C26:0-OH (10.4%) and C22:0-OH (2.8%). Ae.tFAR6 was primarily responsible for generating C28:0-OH (91.3%). For Ae.tFAR3, the major product was C26:0-OH (62.2%), followed by C28 (34.1%), C24 (1.8%), C30 (1.0%), and C22 (0.8%) alcohols (Table 2). These results confirmed that the five Ae.tFARs from Ae. tauschii are FAR, being able to produce long and very-long-chain primary alcohols. Notably, Ae.tFAR3 mainly produced C26 primary alcohol, which is the richest alcohol species in the leaf blades of Ae. tauschii.
Expression Levels and Chromosomal Localization of Ae.tFAR Paralogous Genes in Wheat
In order to further confirm that the five Ae.tFARs are able to generate primary alcohols, we investigated the functions of Ae.tFAR paralogous genes in wheat. Ae.tFAR amino acid sequences were used as queries to search the wheat genome database (Ensembl Plants). Nine sequences in wheat D genome were found to be identical with Ae.tFAR amino sequences (Supplementary Table 3), while no sequence was found to match with Ae.tFAR6 amino acid sequence. The paralogous genes of Ae.tFAR1, Ae.tFAR2, Ae.tFAR3, Ae.tFAR4, Ae.tFAR5, Ae.tFAR7, Ae.tFAR8, Ae.tFAR9, and Ae.tFAR10 are located on wheat chromosome 3D, 7D, 5D, 4D, 4D, 7D, 1D, 3D, and 4D, respectively. We then examined the relative expression levels of these Ae.tFAR paralogous genes in seedling and flag leaves of wheat. The results showed that the Ae.tFAR3 and Ae.tFAR4 paralogs, Traes_5DL_5597A11EC.1 and Traes_4DL_9480F40CF.1, were highly expressed in wheat seedling leaf and flag leaves, respectively (Figure 6), highlighting the importance of Ae.tFAR3 and Ae.tFAR4 in the primary alcohol biosynthesis in the leaf blades.
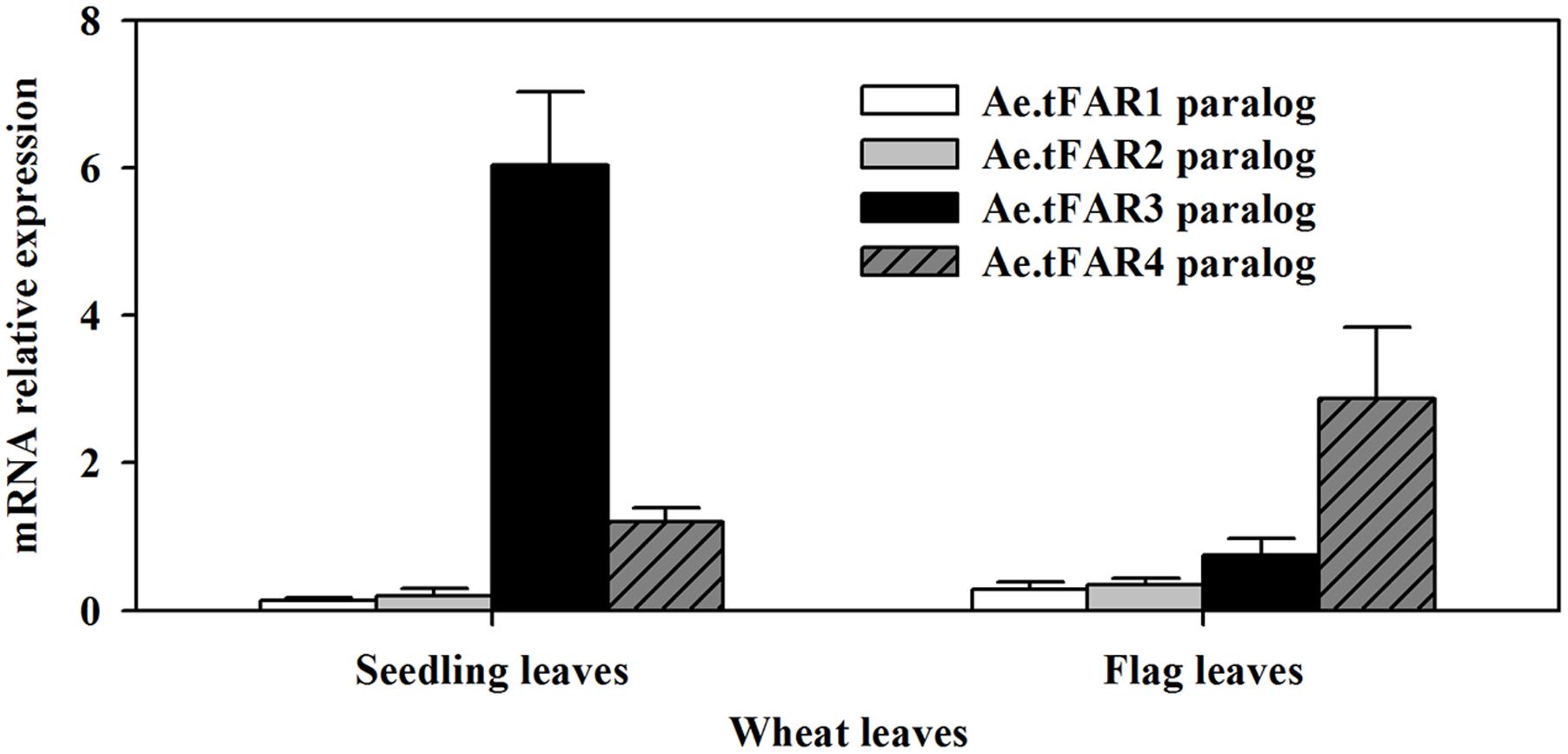
FIGURE 6. Expression analyses of Ae.tFAR paralogous genes in hexaploid wheat. The relative expression levels of Ae.tFAR paralogous genes were determined in seedling leaves and flag leaves of wheat (cv. Chinese Spring, CS) by qRT-PCR analysis. Each value is the mean of three independent parallel experiments. Error bars represent the SD.
The wheat nullisomic–tetrasomic substitution lines were used to further investigate Ae.tFAR3 and Ae.tFAR4 paralogs in hexaploid wheat. cDNA was isolated from a set of seven nullisomic–tetrasomic substitution lines of wheat cv. CS with the loss of one D-chromosome and then was used as template for the amplification of Ae.tFAR3 and Ae.tFAR4 paralogs. As shown in Figure 7A, no amplification product was detected in N5DT5B and N4DT4A, which lacked the whole of chromosome 5D and 4D, respectively. In contrast, PCR products were obtained from all the other six nullisomic–tetrasomic substitution lines. Our results further confirm that Ae.tFAR3 and Ae.tFAR4 paralogs (Traes_5DL_5597A11EC.1 and Traes_4DL_9480F40CF.1) are located on wheat chromosome 5D and 4D. The substitution lines N5DT5B and N4DT4A lacking Ae.tFAR3 and Ae.tFAR4 paralogs, respectively, were then further analyzed. The wax mixtures from seedling and flag leaves of these substitution lines lacking of corresponding Ae.tFAR3 and Ae.tFAR4 paralogs showed a significant reduction in primary alcohols. Compared with wild-type wheat, the N5DT5B substitution line lacking of Ae.tFAR3 paralog (Traes_5DL_5597A11EC.1) exhibited a large decrease in the amount of C26:0-OH and C28:0-OH primary alcohols in seedling leaf (Figure 7B), while N4DT4A with the loss of Ae.tFAR4 paralog (Traes_4DL_9480F40CF.1) showed a notable decrease in C24:0-OH and C26:0-OH primary alcohols in seedling and flag leaves (Figures 7B,C). Altogether, these results might hint that Ae.tFAR3 and Ae.tFAR4 encode active FARs, catalyzing the biosynthesis of primary alcohols in the leaf blades of Ae. tauschii.
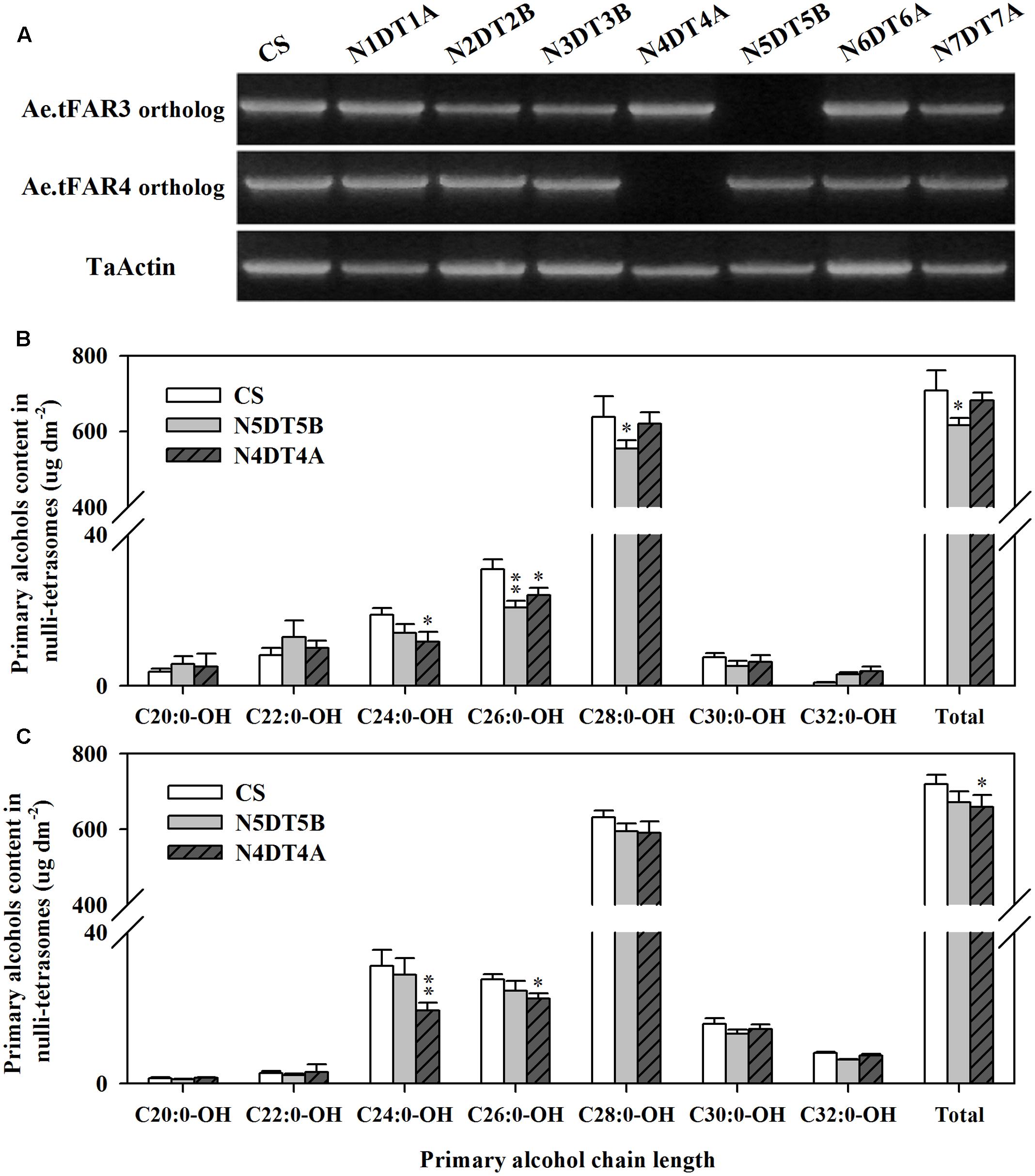
FIGURE 7. Chromosomal localization of Ae.tFAR paralogs and primary alcohol profiles of wheat nullisomic lines. (A) Chromosomal localization of Ae.tFAR paralogs in nullisomic–tetrasomic lines of wheat (cv. CS). N5DT5B and N4BT4A are abbreviations for nullisomic–tetrasomics of common wheat, namely nulli5D–tetra5B and nulli4B–tetra4A, respectively. The coverage and chain length distributions of primary alcohols from (B) seedling leaves and (C) flag leaves in CS and the nullisomic–tetrasomic lines. Each value is the mean of three independent measurements. Error bars indicate SD, and significant differences were assessed according to Student’s t-test (∗ for P < 0.05; ∗∗ for P < 0.01).
Discussion
Fatty acyl-CoA reductase catalyzes the production of primary alcohols in the biosynthesis of cuticular wax in plants. In a previous work, we have identified 12 genes, which are located on D genome in wheat, are associated with wax biosynthesis (Wang et al., 2016). Of 12 genes, TaFAR2 and TaFAR5 are responsible for the biosynthesis of C18 and C22 primary alcohols, respectively (Wang et al., 2015c, 2016). However, knowledge on the rest of genes on D genome encoding alcohol-forming enzymes is still lacking. As Ae. tauschii contributes to the D-genome of bread wheat, study on wax biosynthesis in Ae. tauschii may provide insights on wax biosynthesis in bread wheat. In the present work, we identified and characterized five Ae.tFARs in leaves of Ae. tauschii, including Ae.tFAR1, Ae.tFAR2, Ae.tFAR3, Ae.tFAR4, and Ae.tFAR6. We have concluded that the five Ae.tFARs are involved in the biosynthesis of primary fatty alcohols in the leaf blades of Ae. tauschii. The evidence presented includes: (1) Gene expression patterns of Ae.tFARs are associated with deposition sites of primary alcohols. (2) Heterologous expression of Ae.tFAR1, Ae.tFAR2, Ae.tFAR3, Ae.tFAR4, and Ae.tFAR6 in yeast primarily produces C16:0-OH, C18:0-OH, C26:0-OH, C24:0-OH, and C28:0-OH, respectively. (3) The nullisomic–tetrasomic lines of hexaploid wheat lacking corresponding Ae.tFAR paralogous genes have significantly reduced levels of primary alcohols in the leaf blades.
Investigation of wax profiles from different organs of Ae. tauschii confirmed that primary alcohols are the dominant wax components in the leaf blades. Studies have shown that micromorphology and density of wax vary depending on wax composition and content of each component. It is proposed that platelet-like wax crystals are often associated with primary alcohols (Baker, 1982; Barthlott et al., 1998). In this study, the appearance of dense platelet-like crystals coincided with the high level of primary alcohols in the leaf blades of Ae. tauschii. When primary alcohols were much reduced in peduncle and glumes, the platelet-like crystals became scarce, further supporting the correlation between primary alcohol content and density of platelet-like crystals.
Like other FARs, the five Ae.tFARs possess Rossmann-fold NADB binding domain and FAR_C domain, and an active site YXXXK motif, indicating a catalytic role of these Ae.tFARs. Expression in yeast confirmed that the five Ae.tFARs produce primary alcohols ranging from C16:0 to C30:0. Ae.tFAR1, Ae.tFAR2, Ae.tFAR4, and Ae.tFAR6 are mainly responsible for generating C16:0-OH, C18:0-OH, C24:0-OH, and C28:0-OH, respectively. Interestingly, Ae.tFAR3 was most important for producing C26 primary alcohol and in the meanwhile, C26 primary alcohol was identified as the dominant wax component in the leaf blades of Ae. tauschii, suggesting the essential role of Ae.tFAR3 in wax accumulation of Ae. tauschii. To our best knowledge, the functions of these fatty alcohols-forming FARs have not yet been verified in Ae. tauschii. Arabidopsis CER4/FAR3 and wheat TaFAR1 and TaFAR5 are capable of producing C28 primary alcohol in plants, however, they are not able to generate primary alcohols beyond C26, at least, in yeast strains W3031A and INVSc1 (Rowland et al., 2006; Wang et al., 2015c, 2011). We speculate that the difference is due to the unavailability of substrates past C26 and/or the lack of substrate specificities of these FARs in yeast. In our experiment, we used INVSur4# to produce substrates with a wider range of chain lengths. Expression of Ae.tFAR3 and Ae.tFAR6 in INVSur4# resulted in the production of primary alcohols, with chain lengths being C26 or longer than C26. In contrast, three other Ae.tFARs (Ae.tFAR1, Ae.tFAR2, and Ae.tFAR4) were still unable to make primary alcohols longer than C26 even when substrates of chain length long enough were available, indicating that Ae.tFAR1, Ae.tFAR2, and Ae.tFAR4 are incapable of catalyzing the synthesis of >C26 primary alcohols in yeast. It appears that FARs can catalyze the production of a few chain lengths of primary alcohols in Ae. tauschii, however, each one shows strong preference for one major chain length of substrate and thus produces one major product. For example, Ae.tFAR3 primarily produces C26 primary alcohol, while Ae.tFAR4 principally generates C24 primary alcohol, reflecting the distinct substrate specificities of Ae.tFARs with respect to acyl chain length. However, it is unknown so far what determines chain length specificities of Ae.tFARs in Ae. tauschii. A prior analysis of FAR8 in Arabidopsis has shown that chain length specificity is associated with several amino acids at the junction of the Rossmann-fold NADB binding domain and the FAR_C domain (Chacón et al., 2013). Whether this is also the case for Ae.tFARs merits further investigation.
It is recognized that Ae. tauschii is the D-genome donor to hexaploid wheat. Nine Ae.tFAR paralogous genes were found locating on D genome of hexaploid wheat. The nullisomic–tetrasomic substitution lines of wheat were used in this study to further validate the role of Ae.tFARs in the biosynthesis of primary alcohols. One wheat gene (Traes_5DL_5597A11EC.1) identical to Ae.tFAR3 is located on 5D genome and the loss of 5D in the nullisomic–tetrasomic substitution line N5DT5B led to an obvious reduction in the amounts of primary alcohols in wheat leaves. Another wheat gene (Traes_4DL_9480F40CF.1) sharing 99% identity with Ae.tFAR4 is located on 4D genome in wheat. Examination on the nullisomic–tetrasomic substitution line N4DT4A lacking of 4D genome revealed a substantial decrease in the contents of primary alcohols in flag leaves. The analysis of these wheat genes, which share 40–100% identity to Ae.tFARs, further supports that Ae.tFAR3 and Ae.tFAR4 are involved in the biosynthesis of primary alcohols.
The expression patterns of Ae.tFARs are consistent with their roles in the biosynthesis of primary alcohols in Ae. tauschii. The transcripts of Ae.tFAR3, Ae.tFAR4, and Ae.tFAR6 were all found in leaves, in keeping with their functions in the production of C20 to C30 primary alcohols, which are components of leaf cuticular wax in Ae. tauschii. The highest level of expression was observed for Ae.tFAR3 in the leaf blades, agreeing with the highest accumulation of C26 primary alcohol. In addition to wax biosynthesis, FAR genes have been reported contributing to the biosynthesis of suberin, cutin, and sporopollenin in plants (Metz et al., 2000; Wang et al., 2002; Pollard et al., 2008; Domergue et al., 2010; Beisson et al., 2012). The expression levels of Ae.tFAR1 and Ae.tFAR2 were low in leaves, and Ae.tFAR1 and Ae.tFAR2 were mainly involved in the generation of C16 and C18 primary alcohols, respectively. It is thus possible that these two genes function in the biosynthesis of suberin- and sporopollenin-associated primary alcohols in Ae. Tauschii. However, further evidence is required to draw a conclusion regarding the role of Ae.tFAR1 and Ae.tFAR2 in Ae. tauschii.
In summary, we have cloned Ae.tFAR1, Ae.tFAR2, Ae.tFAR3, Ae.tFAR4, and Ae.tFAR6 from Ae. tauschii and provided evidence that Ae.tFAR3, Ae.tFAR4, and Ae.tFAR6 encode FARs, which are involved in the production of wax-related primary alcohols. Particularly, Ae.tFAR3 is mainly responsible for generating C26:0-OH in the leaf blades of Ae. tauschii where C26 primary alcohol is also the dominant wax component.
Author Contributions
MW, YW, and ZW contributed to the experimental design. MW, HW, JX, CL, and YW performed the experiments and analyzed the data. MW wrote the manuscript and ZW revised it.
Funding
This research was supported by grants from the National Natural Science Foundation of China (31271794, 31471568) and Science and Technology Innovation Team Project of Shaanxi Province, China (2014KCT-25).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We are grateful to Dr. Jérôme Joubès at Université Bordeaux Ségalen, France, for providing the vector p416 MET25-FLAG3:Sur4-F262A/K266L and yeast strain.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01012/full#supplementary-material
References
Baker, E. A. (1982). “Chemistry and morphology of plant epicuticular waxes,” in The Plant Cuticle, eds D. Cutler, K. L. Alvin, and C. E. Price (London: Academic Press), 139–165.
Barthlott, W., Neinhuis, C., Cutler, D., Ditsch, F., Meusel, I., Theisen, I., et al. (1998). Classification and terminology of plant epicuticular waxes. Bot. J. Linn. Soc. 126, 237–260. doi: 10.1111/j.1095-8339.1998.tb02529.x
Beisson, F., Li-Beisson, Y. H., and Pollard, M. (2012). Solving the puzzles of cutin and suberin polymer biosynthesis. Curr. Opin. Plant Biol. 15, 329–337. doi: 10.1016/j.pbi.2012.03.003
Bernard, A., Domerguem, F., Pascal, S., Jetter, R., Renne, C., Faure, J. D., et al. (2012). Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERSUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24, 3106–3118. doi: 10.1105/tpc.112.099796
Chacón, M. G., Fournier, A. E., Tran, F., Dittrich-Domergue, F., Pulsifer, I. P., Domergue, F., et al. (2013). Identification of amino acids conferring chain length substrate specificities on fatty alcohol-forming reductases FAR5 and FAR8 from Arabidopsis thaliana. J. Biol. Chem. 288, 30345–30355. doi: 10.1074/jbc.M113.499715
Chen, W., Yu, X. H., Zhang, K., Shi, J., Oliveira, S. D., Schreiber, L., et al. (2011). Male sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol. 157, 842–853. doi: 10.1104/pp.111.181693
Denic, V., and Weissman, J. S. (2007). A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 130, 663–677. doi: 10.1016/j.cell.2007.06.031
Doan, T. T., Carlsson, A. S., Hamberg, M., Bülow, L., Stymne, S., and Olsson, P. (2009). Functional expression of five arabidopsis fatty acyl-coA reductase genes in Escherichia coli. J. Plant. Physiol. 166, 787–796. doi: 10.1016/j.jplph.2008.10.003
Domergue, F., Shrestha, J., Morant, M., Pinot, F., Matsuno, M., Swanson, R., et al. (2010). Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol. 153, 1539–1554. doi: 10.1104/pp.110.158238
Domínguez, E., Heredia-Guerrero, J. A., and Heredia, A. (2011). The biophysical design of plant cuticles: an overview. New Phytol. 189, 938–949. doi: 10.1111/j.1469-8137.2010.03553.x
Eigenbrode, S. D., and Espelie, K. E. (1995). Effects of plant epicuticular lipids on insect herbivores. Annu. Rev. Entomol. 40, 171–194. doi: 10.1146/annurev.en.40.010195.001131
Gietz, R. D., and Woods, R. A. (2002). Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96. doi: 10.1016/S0076-6879(02)50957-5
Gülz, P. G. (1994). Epicuticular leaf waxes in the evolution of the plant kingdom. J. Plant Physiol. 143, 453–464. doi: 10.1016/S0176-1617(11)81807-9
Hallam, N. D., and Chambers, T. C. (1970). The leaf waxes of the genus Eucalyptus ‘L’Hérit. Aust. J. Bot. 18, 335–389. doi: 10.1071/BT9700335
Jeffree, C. E. (2006). “The fine structure of the plant cuticle,” in Annual Plant Reviews 23: Biology of the Plant Cuticle, eds M. Riederer and C. Müller (Oxford: Blackwell), 11–125.
Jenks, M. A., Eigenbrode, S. D., and Lemieux, B. (2002). “Cuticular waxes of Arabidopsis,” in The Arabidopsis Book, eds C. R. Somerville and E. M. Meyerowitz (Rockville, MD: American Society of Plant Biologists Press).
Jia, J. Z., Zhao, S. C., Kong, X. Y., Li, Y. R., Zhao, G. Y., He, W. M., et al. (2013). Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496, 91–95. doi: 10.1038/nature12028
Kerstiens, G. (2006). Water transport in plant cuticles: an update. J. Exp. Bot. 57, 2493–2499. doi: 10.1093/jxb/erl017
Kolattukudy, P. E. (1996). “Biosynthetic pathways of cutin and waxes, their sensitivity to environmental stresses,” in Plant Cuticles: An Integrated Functional Approach, ed. G. Kersteins (Oxford: BIOS Scientific Publishers Press), 83–108.
Kunst, L., and Samuels, A. L. (2003). Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 42, 51–80. doi: 10.1016/S0163-7827(02)00045-0
Kunst, L., and Samuels, L. (2009). Plant cuticles shine: advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 12, 721–727. doi: 10.1016/j.pbi.2009.09.009
Lee, S. B., and Suh, M. C. (2013). Recent advances in cuticular wax biosynthesis and its regulation in Arabidopsis. Mol. Plant 6, 246–249. doi: 10.1093/mp/sss159
Li, Y., and Beisson, F. (2009). The biosynthesis of cutin and suberin as an alternative source of enzymes for the production of bio-based chemicals and materials. Biochimie 91, 685–691. doi: 10.1016/j.biochi.2009.03.016
Metz, J. G., Pollard, M. R., Anderson, L., Hayes, T. R., and Lassner, M. W. (2000). Purification of a jojoba embryo fatty acyl-coenzyme A reductase and expression of its cDNA in high erucic acid rapeseed. Plant Physiol. 122, 635–644. doi: 10.1104/pp.122.3.635
Oh, C.-S., Toke, D. A., Mandala, S., and Martin, C. E. (1997). ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272, 17376–17384. doi: 10.1074/jbc.272.28.17376
Pestsova, E., Korzun, V., Goncharov, N. P., Hammer, K., Ganal, M. W., and Röder, M. S. (2000). Microsatellite analysis of Aegilops tauschii germplasm. Theor. Appl. Genet. 101, 100–106. doi: 10.1007/s001220051456
Pfündel, E. E., Agati, G., and Cerovic, Z. G. (2006). “Optical properties of plant surfaces,” in Biology of the Plant Cuticle, eds M. Riederer and C. Müller (Oxford: Blackwell), 216–249.
Pollard, M., Beisson, F., Li, Y. H., and Ohlrogge, J. B. (2008). Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci. 13, 236–246. doi: 10.1016/j.tplants.2008.03.003
Rowland, O., Zheng, H., Hepworth, S. R., Lam, P., Jetter, R., Kunst, L., et al. (2006). CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol. 14, 866–877. doi: 10.1104/pp.106.086785
Samuels, L., Kunst, L., and Jetter, R. (2008). Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59, 683–707. doi: 10.1146/annurev.arplant.59.103006.093219
Schneider-Belhaddad, F., and Kolattukudy, P. E. (2000). Solubilization, partial purification, and characterization of a fatty aldehyde decarbonylase from a higher plant, Pisum sativum. Arch. Biochem. Biophys. 377, 341–349. doi: 10.1006/abbi.2000.1798
Solovchenko, A., and Merzlyak, M. (2003). Optical properties and contribution of cuticle to UV protection in plants: experiments with apple fruit. Photochem. Photobiol. Sci. 2, 861–866. doi: 10.1039/b302478d
Wang, A., Xia, Q., Xie, W., Dumonceaux, T., Zou, J., Datla, R., et al. (2002). Male gametophyte development in bread wheat (Triticum aestivum L.): molecular, cellular, and biochemical analyses of a sporophytic contribution to pollen wall ontogeny. Plant J. 30, 613–623. doi: 10.1046/j.1365-313X.2002.01313.x
Wang, M. L., Wang, Y., Wu, H. Q., Xu, J., Li, T. T., Hegebarth, D., et al. (2016). Three TaFAR genes function in the biosynthesis of primary alcohols and the response to abiotic stresses in Triticum aestivum. Sci. Rep. 6:25008. doi: 10.1038/srep25008
Wang, Y., Wang, J. H., Chai, G. Q., Li, C. L., Hu, Y. G., Chen, X. H., et al. (2015a). Developmental changes in composition and morphology of cuticular waxes on leaves and spikes of glossy and glaucous wheat (Triticum aestivum L.). PLoS ONE 10:e0141239. doi: 10.1371/journal.pone.0141239
Wang, Y., Wang, M. L., Sun, Y. L., Hegebarth, D., Li, T. T., Jetter, R., et al. (2015b). Molecular characterization of TaFAR1 involved in primary alcohol biosynthesis of cuticular wax in hexaploid wheat. Plant Cell Physiol. 56, 1944–1961. doi: 10.1093/pcp/pcv112
Wang, Y., Wang, M. L., Sun, Y. L., Wang, Y. T., Li, T. T., Chai, G. Q., et al. (2015c). FAR5, a fatty acyl-coenzyme A reductase, is involved in primary alcohol biosynthesis of the leaf blade cuticular wax in wheat (Triticum aestivum L.). J. Exp. Bot. 66, 1165–1178. doi: 10.1093/jxb/eru457
Wang, Z. H., Guhling, O., Yao, R. N., Li, F., Yeats, T. H., Rose, J. K. C., et al. (2011). Two oxidosqualene cyclases responsible for biosynthesis of tomato fruit cuticular triterpenoids. Plant Physiol. 155, 540–552. doi: 10.1104/pp.110.162883
Keywords: fatty acyl-coenzyme A reductase, primary alcohol, cuticular wax, Saccharomyces cerevisiae, Aegilops tauschii, Triticum aestivum
Citation: Wang M, Wu H, Xu J, Li C, Wang Y and Wang Z (2017) Five Fatty Acyl-Coenzyme A Reductases Are Involved in the Biosynthesis of Primary Alcohols in Aegilops tauschii Leaves. Front. Plant Sci. 8:1012. doi: 10.3389/fpls.2017.01012
Received: 11 February 2017; Accepted: 26 May 2017;
Published: 12 June 2017.
Edited by:
Basil J. Nikolau, Iowa State University, United StatesReviewed by:
Owen Rowland, Carleton University, CanadaFrederic Domergue, Centre National de la Recherche Scientifique, France
Copyright © 2017 Wang, Wu, Xu, Li, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhonghua Wang, zhwangnew@126.com Yong Wang, wangyong2114@163.com
†These authors have contributed equally to this work.
 Meiling Wang
Meiling Wang Hongqi Wu1†
Hongqi Wu1†