- 1Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing, China
- 2College of Agronomy and Biotechnology, Southwest University, Chongqing, China
- 3Genomics Lab, Department of Plant Breeding and Genetics, Bahauddin Zakariya University, Multan, Pakistan
The sensitivity to abscisic acid (ABA) by its receptors, pyrabactin resistance-like proteins (PYLs), is considered a most important factor in activating the ABA signal pathway in response to abiotic stress. However, it is still unknown which PYL is the crucial ABA receptor mediating response to drought stress in cotton (Gossypium hirsutum L.). Here, we reported the identification and characterization of highly induced ABA receptor GhPYL9-11A in response to drought in cotton. It is observed that GhPYL9-11A was highly induced by ABA treatment. GhPYL9-11A binds to protein phosphatase 2Cs (PP2Cs) in an ABA-independent manner. Moreover, the GhPYL-11A-PP2C interactions are partially disrupted by mutations, proline (P84) and histidine (H111), in the gate-latch region. Transgenic Arabidopsis overexpressing GhPYL9-11A plants were hypersensitive to ABA during seed germination and early seedling stage. Further, the increased in root growth and up regulation of drought stress-related genes in transgenic Arabidopsis as compared to wild type confirmed the potential role of GhPYL9-11A in abiotic stress tolerance. Consistently, the expression level of GhPYL9-11A is on average higher in drought-tolerant cotton cultivars than in drought-sensitive cottons under drought treatment. In conclusion, the manipulation of GhPYL9-11A expression could be a useful strategy for developing drought-tolerant cotton cultivars.
Introduction
Abscisic acid (ABA) is a multifunctional phytohormone that regulates multiple aspects of plant growth and development, including seed dormancy and germination (Fang et al., 2008), vegetative development (Finkelstein et al., 2002), root elongation (Xing et al., 2016), leaf senescence (Liang et al., 2014), and fruit ripening (Zhang et al., 2009). ABA has also long been known to be a key endogenous messenger in the response to abiotic and biotic stresses, such as drought, salinity, cold, and pathogen infection (Choi et al., 2000; Xiong et al., 2002; Zhu, 2002; Sheard and Zheng, 2009; Yoshida et al., 2014). ABA-regulated stress responses include changes in gene expression, increased stomatal closure, reduced transpiration rate, protection of photosynthesis, and regulation of plant growth.
Previous studies have demonstrated that pyrabactin resistance-like (PYL), clade A protein phosphatase 2C (PP2C), and sucrose non-fermenting 1- related protein kinase 2 (SnRK2) proteins are the three core components of the ABA signaling cascade (Park et al., 2009; Sheard and Zheng, 2009; Zhu, 2016). The primary step in initiating the ABA signaling pathway is triggered by the PYL ABA receptors (Sheard and Zheng, 2009), which contain a central hydrophobic ligand-binding pocket (Iyer et al., 2001). In the absence of ABA, two β-loops, termed the gate and latch, together with several nearby structural elements form a large open pocket for ligand binding. When ABA is present, ABA binds to this pocket, leading to a series of conformational rearrangements, including gate and latch closure, which sequester ABA within the pocket. The ABA-bound PYL receptor physically interacts with and sequesters the constitutive SnRK2 repressor, PP2C, allowing SnRK2 kinases autophosphorylation. ABA-activated SnRK2 kinases subsequently phosphorylate and activate downstream ABA-responsive element-binding transcription factors that induce the expression of ABA-responsive genes (Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009).
The PYL proteins belong to the START/Bet V1 superfamily (Iyer et al., 2001). There are fourteen PYL members in Arabidopsis, and this family is divided into three subfamilies based on amino acid sequences similarity (Zhao et al., 2013, 2016). All PYL proteins have highly conserved amino-acid residues in key interaction domains, including ABA-binding pocket, and the two entrance loops. Despite these similar structural features, PYLs have distinct properties and different expression patterns that explain the versatility of ABA signaling in plant growth and development. For example, Arabidopsis PYR1, PYL1, and PYL2 are able to form dimers in solution, whereas PYL proteins 4–10 exist as monomers (Hao et al., 2011). PYR1, PYL1, PYL2, and PYL3 interact with several clade A PP2Cs in an ABA-dependent manner. In contrast, most other PYL proteins except for PYL7 inhibit PP2Cs independently of ABA (Bai et al., 2013; Li et al., 2013; Zhao et al., 2013). Recently, several genetic studies have elucidated the significance of these receptors in ABA response; pyr1 pyl1 pyl2 pyl4 quadruple mutants show ABA insensitivity (Park et al., 2009; Nishimura et al., 2010; Gonzalez-Guzman et al., 2012), and a pyl8 single mutant has altered ABA responses during seed germination and seedling growth, as well as decreased drought resistance (Antoni et al., 2013; Zhao et al., 2014). PYL9 enhances drought resistance by limiting transpiration water loss and regulating senescence in old leaves and growth in young tissues (Zhao et al., 2016). Moreover, both PYL9 and PYL8 act as critical regulators of lateral root formation in response to ABA (Xing et al., 2016).
Upland cotton (Gossypium hirsutum L.) is an important economic crop and is an important source of fiber and oil (Wang et al., 2012, 2017; Shan et al., 2014). Compared with the model plants Arabidopsis and rice, cotton has higher tolerance to drought and salt stress (Liang et al., 2016). However, its growth and development, as well as fiber yield and quality are also significantly affected by severe environmental conditions, especially drought stress (He et al., 2016). To date, a number of candidate genes for drought tolerance in cotton have been identified. Several stress-related genes, including GhABF2 (Liang et al., 2016), GhNAC2 (Gunapati et al., 2016), GhATAF1 (He et al., 2016), GhbHLH130 (Guang et al., 2014), GhSARP1 (Liu et al., 2016), GhMKK3 (Wang C. et al., 2016), Di19-1 and Di19-2 (Qin et al., 2016), have been shown to regulate stress response in an ABA-dependent manner. Therefore, understanding ABA signal transduction in cotton will accelerate the molecular breeding of stress-tolerant cotton cultivars.
Because ABA has an essential role in plant growth and development, understanding how each PYL protein affects plant physiology has both basic and applied agricultural significance. Although there have been a number of studies of ABA receptors in the model plant Arabidopsis and other crops, such as rice and soybean, functional analysis of PYLs in upland cotton has not been reported. Upland cotton is a typical allotetraploid crop that was formed about 1–2 MYA by hybridization between an A-genome ancestor and a D-genome ancestor (Chen et al., 2007; Lin et al., 2010; Paterson et al., 2012; Li et al., 2014; Cao, 2015; Yuan et al., 2015), and the PYL gene family expanded in this process. In this study, we carried out genome-wide analysis to identify all PYL genes in the two diploid progenitor species, G. raimondii and G. arboretum, and two tetraploids G. hirsutum and G. barbadense. Genome-wide expression profiling analysis revealed that GhPYL9-11A was the most highly expressed PYL gene during drought stress in upland cotton. Consistent with a function in drought stress response, overexpression of GhPYL9-11A in Arabidopsis remarkably increases ABA sensitivity and enhances drought tolerance. The future characterization of GhPYL9-11A will further our understanding of ABA-mediated stress response in cotton, and facilitate its application in breeding drought tolerant cotton cultivars.
Materials and Methods
Plant Materials and Growth Conditions
Upland cotton (G. hirsutum) seeds of cultivar Y18R, a restores variety developed by our lab (Shi et al., 2012), were planted during the regular cotton cultivation season in 2015 and 2016 in the experimental farm at the Biotechnology Research Institute, Chinese Academy of Agricultural Sciences (Beijing, China). The recommended cultural and plant measures were adopted during the growth of cotton crop. For expression analysis, various organs including leaf, root, stem, and seed, were collected after 1 week of the onset of flowering stage. For analysis of ABA-induced GhPYL9-11A expression, cotton plants were grown in the Greenhouse with a 12-h-light (30°C)/12-h-dark (26°C) photoperiod with about 300 μM m-2s-1 photon density and 45% humidity, and the leaves of 3-weeks old plants were sprayed with 100 μM ABA (Sigma–Aldrich, United States). Ten plants were collected at 0, 1, 3, and 6 h after treatment and stored at -70°C for RNA isolation.
Arabidopsis thaliana Col-0 seed and GhPYL9-11A transgenic plants were sterilized for approximately 10 min in 10% bleach and then rinsed in sterile deionized water three times. Sterilized seeds were sown on petri dishes containing 0.8% agar media with aaa12 MS nutrients and 1% sucrose. After stratification at 4°C for 2 days, the dishes were moved to a growth chamber at 22°C with a 16-h light/8-h dark cycle and 70% humidity. All seedlings were grown vertically before transplanting to media supplemented different concentrations of ABA and mannitol.
Gene Cloning and Expression Analysis
The full-length open reading frame of GhPYL9-11A was amplified from G. hirsutum L. using the Invitrogen RACE system (Invitrogen, United States). Total RNA was extracted using the EASYspin reagent (YPHBio, China). After DNase treatment, approximately 2 μg total RNA was reverse-transcribed using the ReverTra Ace qPCR RT Master Kit (Toyobo, Japan). Quantitative RT-PCR (qRT-PCR) analysis was performed using SYBR Green I PCR mix (SSoFast EvaGreen Supermix, Bio-Rad, United States) and the Bio-Rad CFX96 real time PCR system. Data were analyzed with Opticon monitor software (Bio-Rad). The cotton GhHISTON3 (GhHIS3) and Arabidopsis Actin 8 genes, both with stability expression in the different tissues, developmental stages and environmental conditions (Zhu et al., 2017), were used as internal controls. Primers used are listed in Table S1. Values are means ± SD of three biological repeats. Student’s t-test was used for statistical analysis.
Vector Construction and Arabidopsis Transformation
The GhPYL9-11A full-length coding sequence was cloned into pBI121-35S to generate the pBI121-35S:GhPYL9-11A overexpression construct. The constructs were transfected into Agrobacterium tumefaciens GV3101 by electroporation, and then transformed into Arabidopsis Col-0 pants using the floral dipping method (Liu L. et al., 2015). Homozygous transgenic Arabidopsis lines were obtained, and the lines GO16, GO21, and GO22, which have high levels of GhPYL9-11A expression, were selected for further analysis. The primers used for vector construction are listed in Table S1.
Yeast Two-Hybrid Assay
The Y2H assay was performed using the MatchmakerTM Gold Yeast Two-hybrid System (Clontech, United States). Full length cDNA of GhPYL9-11A was amplified and cloned into the pGBKT7 vector, and AtABI1, AtABI2, GhPP2C1, and GhPP2C2 full length cDNAs were amplified and cloned into the pGADT7 vector. GhPYL9-11A point mutations were introduced by site-directed mutagenesis using pBI121-35S:GhPYL9-11A as a template, and GhPYL9-11AP84S and GhPYL9-11AH111A were cloned into the pGBKT7 vector. Each pair of AD and BD constructs was co-transformed according to the manufacturer’s protocol. The positive yeast liquid culture was serially diluted to OD600 = 0.6 and 3 μL of dilutions 1:10, 1:100, 1:1,000, and 1:10,000 were inoculated onto various plates [Synthetic Dropout Medium (SD) -leucine (Leu)/tryptophan (Trp), SD-Leu/Trp/histidine (His), and SD-Leu/Trp/His supplemented with 10 μM ABA]. All plates were incubated at 28°C for 2 days and then photographed.
Germination Assay
Surface-sterilized seeds were sown on aaa12 MS without or with ABA (0.3 and 0.5 μM) or mannitol (150 and 300 μM). After 5 days seeds with green shoots were scored as germinated.
Drought Stress Treatment
To test the drought tolerance of 35S:GhPYL9-11A transgenic Arabidopsis, the homozygous F2 generation was subjected to drought stress in soil in a growth chamber. Seed from GO1, GO2, GO3, and wild type lines were imbibed at 4°C for 2 days and directly planted in soil in the same pots (30 cm in diameter). All plants were grown at room temperature (22–25°C) under long-day conditions (16 h light/8 h dark) in the culture room. After 2 weeks of seedling emergence, drought treatment was established by withholding the water for 4 weeks. The experiment was comprised of five replications. Watering was resumed 1 day after the 4 weeks of drought treatment, and the survival rate (SR) was calculated 3 days later.
Chlorophyll Measurement
After measuring the fresh weight of the samples, five seedlings for each sample were used to calculated the total chlorophyll contents as the absorbance at 652 nm following (Liang et al., 2014).
Photosynthesis Parameters and Water Loss Measurement
For measurement of photosynthesis parameters, water was withheld for 7 days beginning when plants were 14-days old. Ten independent plants were used for each GhPYL9-11A overexpression line. Photosynthesis parameters were measured as previously described by (Liang et al., 2015). For determination of water loss, the whole aboveground rosettes of 21-day-old plants were cut from the base and weighed at different time points.
Electrolyte Leakage Measurement
The measurement of electrolyte leakage was performed according to a previously described method (Lin A. et al., 2012). Ten different leaf disks from wild-type and GhPYL9-11A overexpression plants were placed into a flask containing 10 mL distilled deionized water. After shaking at room temperature for 6 h at approximately 120 rpm, the conductivity (Ci) was measured with a conductivity meter (Leici-DDS-307A). Then, the disks were boiled for 20 min to kill the leaf tissues and shaken for 1 h to completely release the electrolytes into the solution. The conductivity (Cm) was measured again. The electrolyte leakage was calculated according to the equation (Ci/Cm) × 100%.
Hydrogen Peroxide Production and Measurement of Antioxidant Enzyme Activities
For hydrogen peroxide production and measurement of antioxidant enzyme activities, Arabidopsis plants were grown on soil for 14 days and leaves were collected from the plants after water was withheld for 7 days. Measurement of H2O2 production was performed according to a previously described method (Liang et al., 2015), and quantified using the Hydrogen peroxidase assay kit (Beyotime, China) according to the manufacturer’s instructions. The quantification of CAT, SOD, and POD activity was performed according to a previously described method (Liang et al., 2016). Total protein was measured using Bradford protein assay kit (Sangon Biotech, China).
Protein Sequence and Phylogenetic Analysis
For phylogenetic analysis of GhPYL9-11A homologs in plants, 20 protein sequences were obtained through National Center for Biotechnology Information database using BLASTp searches with the GhPYL9-11A protein as a query. All sequences were aligned using Clustal W. Phylogenetic trees were constructed using MEGA 6.0 based on the neighbor-joining method. Topological robustness was assessed by bootstrap analysis with 1,000 replicates (Liu L. et al., 2015).
Results
Transcriptome Analysis of Cotton PYL ABA Receptor
Pyrabactin resistance-like protein coding sequences were identified in upland cotton species, G. hirsute, G. barbadense, G. raimondii, and G. arboretum, with complete genomes or genome assemblies. We then compared the expression profiles of the PYL genes under drought stress. Twenty-two PYL proteins were identified from each of the two diploid progenitor species G. raimondii and G. arboretum, and 44 and 36 members were identified in the tetraploids G. hirsutum and G. barbadense, respectively (Table S2). High-resolution temporal expression profiling revealed that two GhPYL genes, Gh_A11G08070 and Gh_D11G1013, were most highly expressed during drought stress in upland cotton (G. hirsutum) (Figure 1 and Table S2). The proteins encoded by Gh_A11G0870 and Gh_D11G1013 share approximately 80 and 79% amino acid identity with AtPYL9, an ABA receptor, respectively.
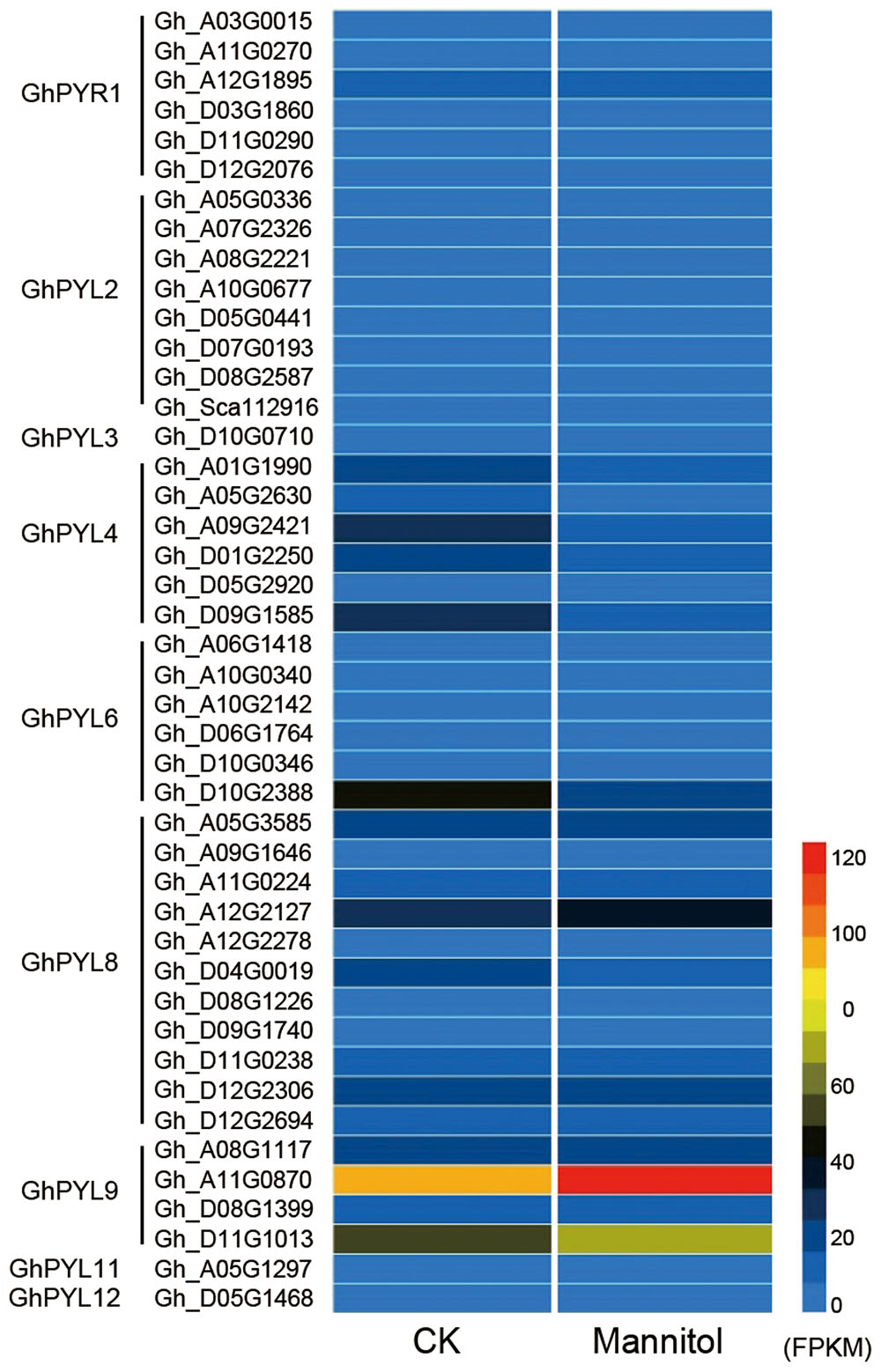
FIGURE 1. Heat map of pyrabactin resistance-like (PYL) family genes in upland cotton. Detailed information about gene annotations can be found in Table S1. Gene annotation data were obtained from the Cotton Functional Genomics Database (https://cottonfgd.org/).
Both Gh_A11G0870 and Gh_D11G1013 belong to the PYL9 subfamily of PYL proteins. There are two members each in G. raimondii and G. arboretum, and four members each in G. hirsutum and G. barbadense. Based on phylogenetic analysis, the PYL proteins could be further divided into two distinct subgroups, PYL9-I and PLY9-II (Figure S1). Interestingly, all members of PYL9-I in both diploid and tetraploid cotton were highly expressed during seed germination and drought stress, whereas PLY9-II members were with low expression level (Table S2). These results suggest that PYL9-I proteins may play important roles in the regulation of seed germination and drought responses in cotton. Because Gh_A11G08070 (hereafter referred to as GhPYL9-11A) belongs to the drought stress-responsive PYL9-I subfamily, its function was investigated in detail.
Structural Features of GhPYL9-11A
Using protein database searches, we identified a number of putative GhPYL9-11A orthologs in various higher plants. GhPYL9-11A homologs were only found in the plant kingdom, indicating that PYL9 proteins are plant-specific. The identification of PYL proteins in both monocots and dicots suggests that PYL proteins are evolutionally conserved across plant species. The existence of PYL9 in different plant species might also suggest that its function in drought stress response is also conserved. Phylogenetic analysis of the cotton GhPYL9-11A protein and its 20 homologs showed that GyhPYL9-11A is most closely related to LOC18613997 of Theobroma cacao, and shares approximately 80% amino acid sequence identity with related proteins in Figure 2A.
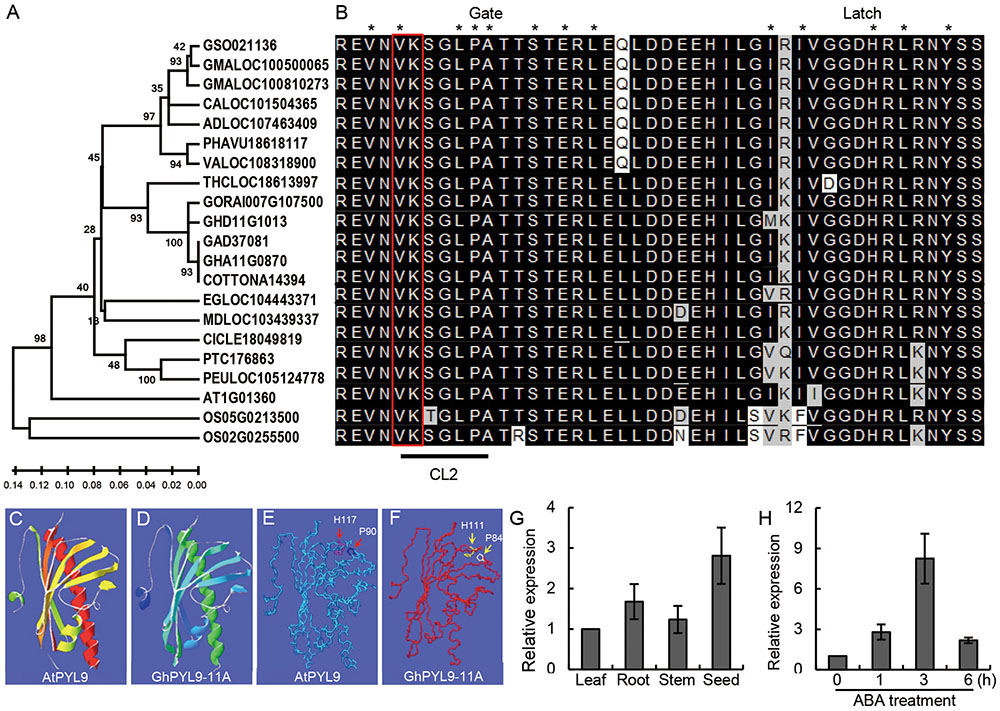
FIGURE 2. Phylogenetic analyses of GhPYL9-11A homologs and organic-specific expression of GhPYL9-11A. (A) Phylogenetic relationship of GhPYL9-11A homologs in plants. AT (Arabidopsis thaliana), OS (Oryza sativa), GSO (Glycine soja), GMA (Glycine max), CAL (Citrus clementina), AD (Arachis duranensis), PHAVU (Phaseolus vulgaris), VA (Vigna angularis), THC (Theobroma cacao), GORA (Gossypium raimondii), GH (Gossypium hirsutum), GA (Gossypium arboreum), EG (Eucalyptus grandis), MD (Malus domestica), CICLE (Citrus clementine), PT (Populus trichocarpa), PEU (Populus euphratica). (B) Amino acid alignment of the PYL9 CL2/gate-latch region for the proteins shown in (A). The black line indicates the CL2 domain. Two amino acid residues ‘VK’ are marked with a red open rectangle. The residues in contact with the ABA molecule are designated by asterisk. (C–F) Comparison of the predicted 3-D structures of AtPYL9 and GhPYL9-11A. The conserved proline and histidine residues in the gate and latch region are marked with red arrows in AtPYL9 (E) and yellow arrows in GhPYL9-11A (F). (G) Expression of GhPYL9-11A in various organs determined by quantitative real time (qRT)-PCR analysis. Roots, leaves, and stems were harvested from 3-week-old plants. (H) Time-course of the response of GhPYL-11A to 100 μM ABA treatment.
PYL9 family proteins have a pyrabactin resistance1/pyr1-like/regulatory component of ABA receptor (PYR/PYL/RCAC)-like structure (amino acids 28–172) typical of ABA receptors (Figure 2B). We analyzed the 3D structure of this conserved motif in GhPYL9-11A using SWISSMODEL. The predicted structure is a common helix-grip structural feature consisting of four α-helixes and seven antiparallel β-sheets and is highly similar to the Arabidopsis AtPYL9 PYR/PYL/RCAC motif (Figures 2C,D).
Expression Pattern of GhPYL9-11A
Quantitative RT-PCR analysis showed that GhPYL9-11A is expressed preferentially in seeds and roots, with a low level of expression in the leaf and stem (Figure 2E). Moreover, time-course analysis revealed that GhPYL9-11A was rapidly induced by ABA, with expression levels increasing approximately 2.8-fold after 1 h of ABA application (Figure 2F). Levels of GhPYL9-11A transcription reached a maximum after 4 h of treatment, and were approximately eightfold higher than without hormone treatment. These results clearly demonstrate that this gene is transcribed in response to ABA and thus is potentially involved in ABA response.
GhPYL9-11A Interacts with Clade A PP2Cs in an ABA-Independent Manner
It has been shown that PYL proteins can interact with PP2Cs in yeast two-hybrid (Y2H) assays in an ABA-dependent or ABA-independent manner (Bai et al., 2013; Zhao et al., 2013). To investigate whether GhPYL9-11A binds clade A PP2Cs, we tested the interaction of GhPYL9-11A with two Arabidopsis PP2C proteins, A. thaliana ABSCISIC ACID INSENSITIVE1 (AtABI1) and AtABI2, in Y2H assays in the presence or absence of ABA. As shown in Figure 3, GhPYL9-11A interacted with both AtABI1 and AtABI2 in an ABA-independent manner. We further tested the interaction between GhPYL9-11A and two cotton PP2Cproteins, GhPP2C1 (Gh_A05G0308) and GhPP2C2 (Gh_A07G0123), that share high similarity with AtABI1 (63% for GhPP2C1 and 71% for GhPP2C2, Figure S2). GhPYL9-11A also binds to both GhPP2C1 and GhPP2C2 in an ABA-independent manner (Figure 3).
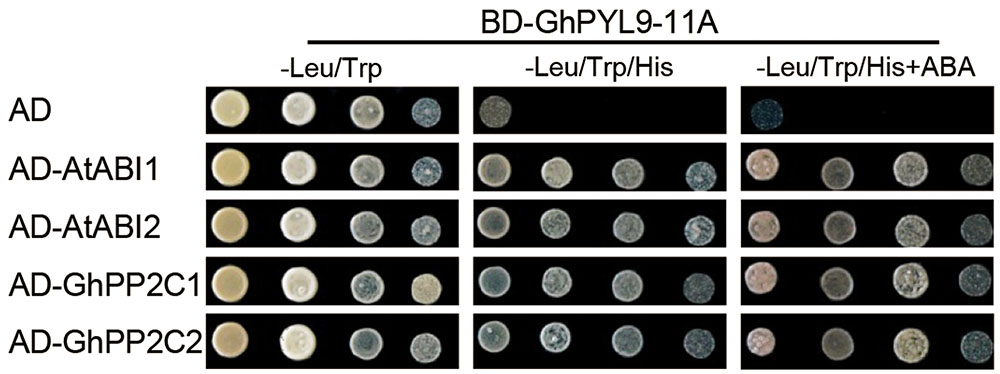
FIGURE 3. Interactions between GhPYL9-11A and PP2C proteins in yeast. The BD-GhPYL9-11A fusion was co-expressed with AD-AtABI1, AD-AtABI2, AD-GhPP2C1, or AD-GhPP2C2 in yeast. Interactions were determined by growth assays on media lacking leucine (Leu), tryptophan (Trp), and histidine (His) in the presence and absence of 10 μM ABA.
The mutated conserved proline (P88S) in the gate region and histidine (H111A) in the latch region of GhPYL9-11A, and tested the ability of the mutated proteins, GhPYL9-11AP84S and GhPYL9-11AH111A, to interact with AtABI1, AtABI2, GhPP2C1, and GhPP2C2 in Y2H assays. Both the GhPYL9-11AP84S and GhPYL9-11AH111A mutations reduced the level of interaction with AtABI1, AtABI2, GhPP2C1, and GhPP2C2 both in the presence and absence of ABA (Figure 4). These data suggest that GhPYL-11A-PP2C interactions are partially mediated by P84 and H111 in the gate-latch region of GhPYL9-11A.
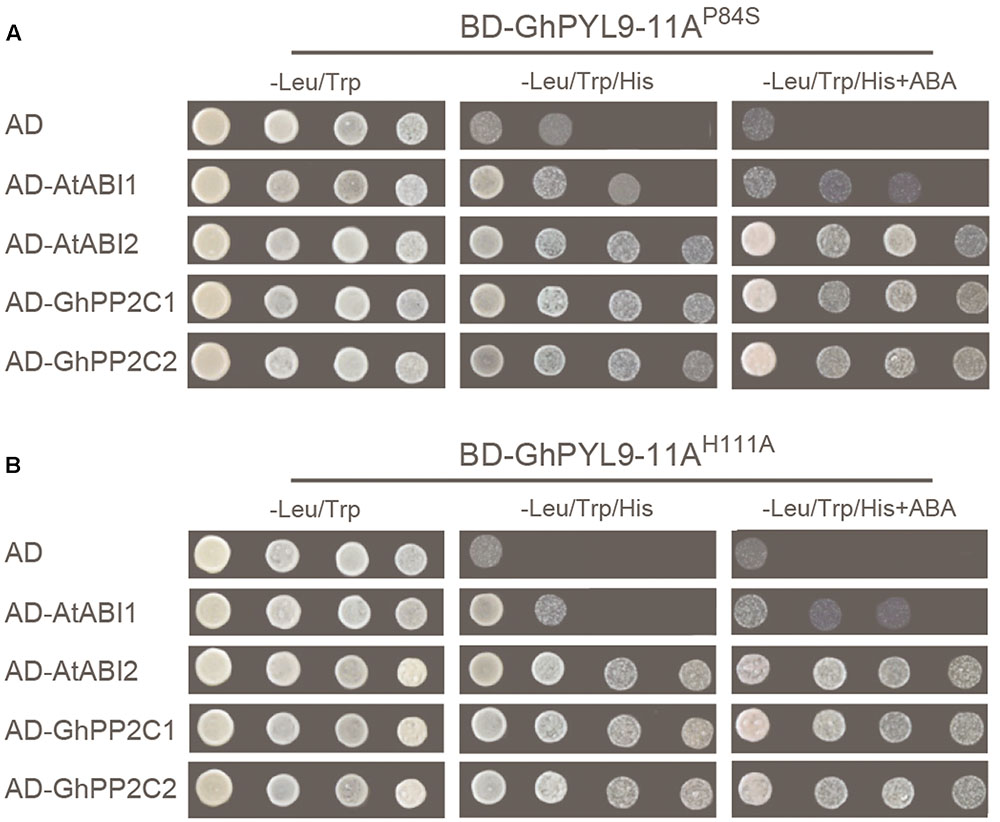
FIGURE 4. GhPYL-11A-PP2C interactions are partially mediated by proline (P84) and histidine (H111) in the gate-latch region of GhPYL9-11A. Replacement of P84 by serine (GhPYL9-11AP84S) (A) and H111 by alanine (GhPYL9-11AH111A) (B) abolished binding to AtABI1 both in the absence and presence of 10 μM ABA. However, these mutations partially affected binding to AtABI2, GhPP2C1, and GhPP2C2 both in the presence and absence of 10 μM ABA.
Overexpression of GhPYL9-11A in Arabidopsis Leads to an ABA-Hypersensitive Phenotype during Germination
To investigate the biological function of GhPYL9-11A in plants, the GhPYL9-11A coding sequence was cloned into the plant binary vector pBI121 in front of the 35S promoter and transformed into Arabidopsis though Agrobacterium tumefaciens-mediated transformation. A set of 27 positive independent transgenic lines had up-regulated expression of GhPYL9-11A. Among them, three independent homozygous T3 GhPYL9-11A-overexpressing (GO) lines, GO16, GO21, and GO22, with high levels of GhPYL9-11A transcript, were selected for further analysis (Figure 5A).
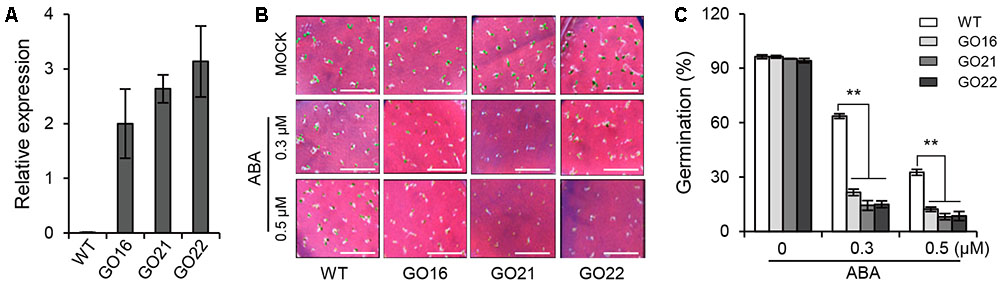
FIGURE 5. The effect of GhPYL9-11A overexpression on Arabidopsis seed germination. (A) The expression levels of GhPYL9-11A in transgenic Arabidopsis plants determined by qRT-PCR. GO, GhPYL9-11A overexpression lines. Values are means ± SD of three biological repeats. (B) Germinated seeds from wild-type plants and GO lines grown for 5 days on half-strength Murashige and Skoog (aaa12 MS) in the presence or absence of ABA. (C) Seed germination rate under the different growth conditions in (B). Values are means ± SD of five measurements. ∗∗P ≤ 0.01; Student’s t-test.
To determine whether GhPYL9-11A is a functional ABA receptor, we treated three GO plants and wild type with 0.3 and 0.5 μM ABA and then compared their seed germination phenotypes. Under control conditions, there was no significant difference in seed germination rate between GO plants and wild type. However, the seed germination rates of the three GO transgenic Arabidopsis lines were less than 20 and 10% in the presence of 0.3 μM ABA and 0.5 μ M ABA, respectively, whereas the germination rates of wild type were 63.5 and 32.6%, respectively (Figures 5B,C). These observations suggest that overexpression of GhPYL9-11A represses seed germination in the presence of ABA in Arabidopsis.
GhPYL9-11A Overexpression in Arabidopsis Results in ABA-Hypersensitive Phenotypes during Seedling Growth
We further examined the effects of GhPYL9-11A-overexpression on Arabidopsis seedling root growth. We found no obvious difference in root development between GO transgenic plants and wild type at different stages of seedling growth. However, ABA treatment repressed root growth to a larger extent in the GO lines compared with wild type; in the presence of 0.3 M ABA the root lengths of in the three GO lines were reduced by 37.5–47.9% compared with control conditions, but in wild type root lengths were only reduced by 28.8%. In the absence of ABA, and in the presence of 0.5 μM ABA, root lengths were 44.8–52.6% of GO plants while 34.8% of wild type (Figures 6A–D). These results suggest that overexpression of GhPLY9-11A in Arabidopsis significantly increases sensitivity to ABA. Thus, we deduced that GhPYL9-11A may act as an ABA receptor in cotton.
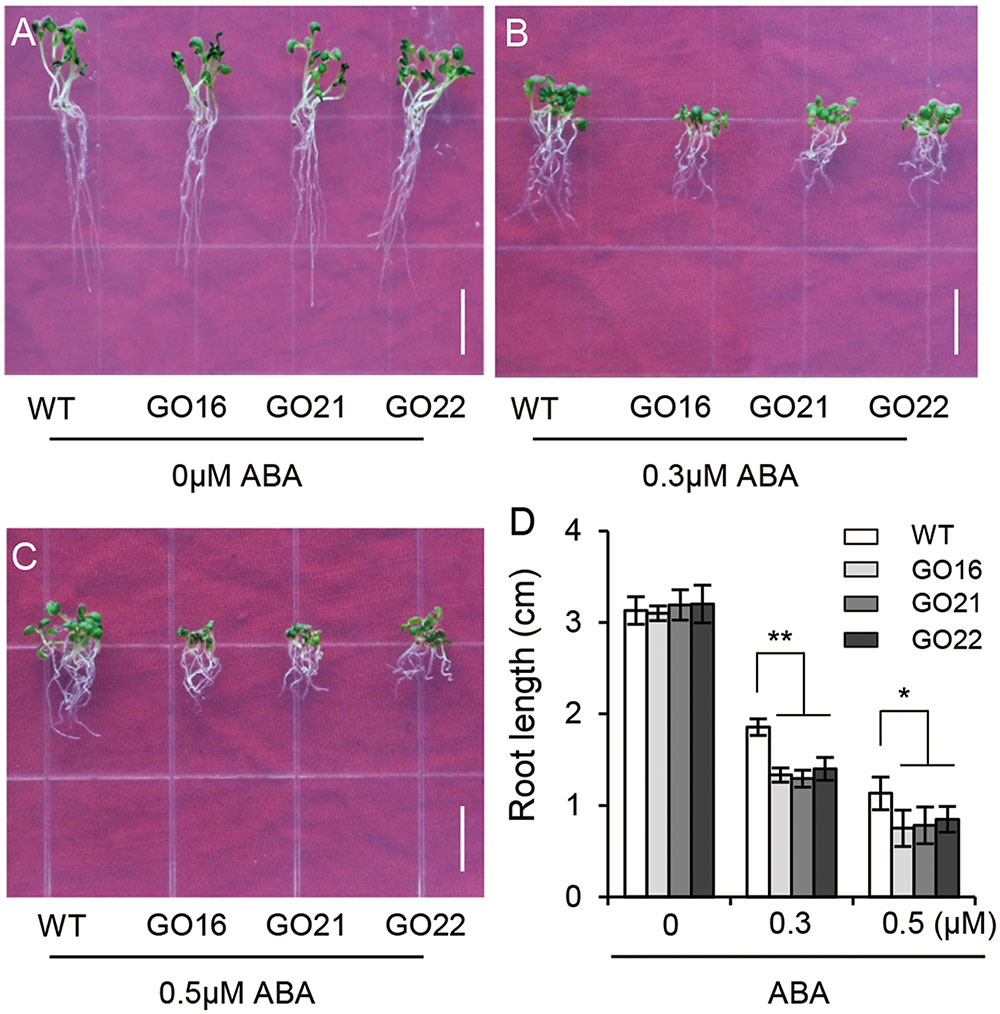
FIGURE 6. Differential ABA responsiveness of GhPYL9-11A overexpression and wild-type Arabidopsis seedlings. (A–C) Phenotypes of wild type and three independent 35S:GhPYL9-11A transgenic Arabidopsis lines, GO16, GO21, and GO22, grown in aaa12 MS medium supplemented with 0 μM (A), 0.3 μM (B), and 0.5 μM (C) ABA. Bar = 1 cm. (D) Seedling primary roots lengths under the different growth conditions in (A–C). Measurements were performed for least 30 independent seedlings. Results are shown as mean ± SD of three replicates; Student’s t-test. ∗P ≤ 0.05, ∗∗P ≤ 0.01.
The Seeds of Arabidopsis GO Lines Are Hypersensitive to Osmotic Stress
Keeping in view the ability of ABA to regulate seed germination, we examined the effects of GhPYL9-11A overexpression on seed germination under osmotic stress imposed by adding 150 and 300 mM mannitol to aaa12 MS media. Compared with wild-type Arabidopsis, GO plants had a significantly decreased ratio of seed germination under osmotic stress (Figure 7), but no difference in seed germination rate between GO plants and wild type was observed under control conditions. The repression of seed germination under osmotic stress in the GhPYL9-11A overexpression lines is consistent with hypersensitivity to ABA.
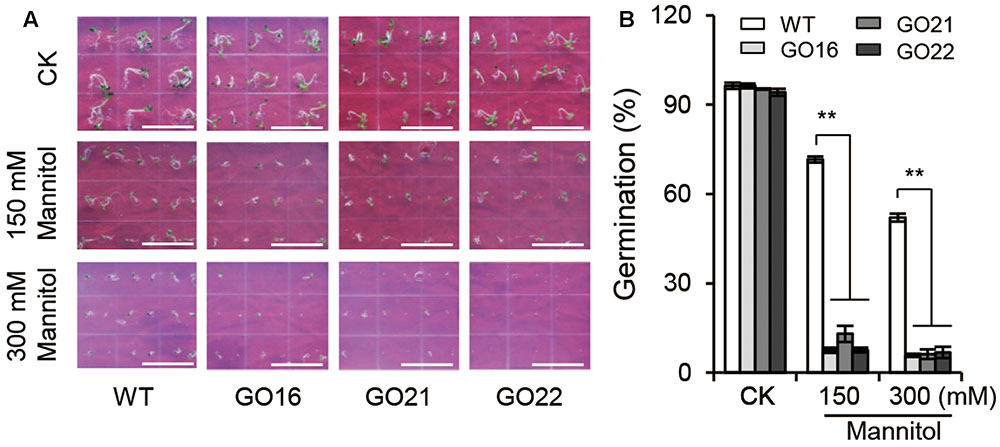
FIGURE 7. Effect of GhPYL9-11A overexpression on Arabidopsis seed germination under mannitol treatment. (A) Germinated seeds from wild-type and GhPYL9-11A overexpression plants grown for 5 days on 1/2 MS with or without mannitol. (B) Seed germination rate under the different growth conditions in (A). Values are means ± SD of five measurements. ∗∗P ≤ 0.01; Student’s t-test.
GhPYL9-11A Overexpression in Arabidopsis Enhances Tolerance to Drought Stress
The ABA-dependent signaling pathway has an essential role in response to drought stress therefore, we examined whether overexpression of GhPYL9-11A was sufficient to confer drought stress tolerance to Arabidopsis. The Arabidopsis 35S:GhPYL9-11A plants, GO16, GO21, and GO22, exhibited significantly increased drought tolerance during the vegetative stage (Figures 8A–F). GO plants also had reduced chlorophyll degradation (Figure 8G), reduced water loss (Figure 8H), enhanced photosynthetic rate (Figure 8I), reduced electrolyte leakage (Figure 8J), reduced accumulation of toxic hydrogen peroxide (Figure 8K), and enhanced activities of antioxidant enzymes including catalase (CAT, Figure 8L), superoxide dismutase (SOD, Figure 8M) and peroxidase (POD, Figure 8N). Consistent with these observations, the SR and total biomass were significantly increased after drought treatment in GO lines compared with the wild type (Figures 8O,P). Moreover, four ABA-dependent drought stress-associated genes, ABF2, RAB18, RD29A, and RD29B, were more strongly induced after drought treatment in the rosette leaves of Arabidopsis GO lines than in wild type (Figures 9A–D). Taken together, these data provide strong evidence that GhPYL9-11A overexpression confers drought tolerance in plants by activating the ABA-dependent signaling pathway.
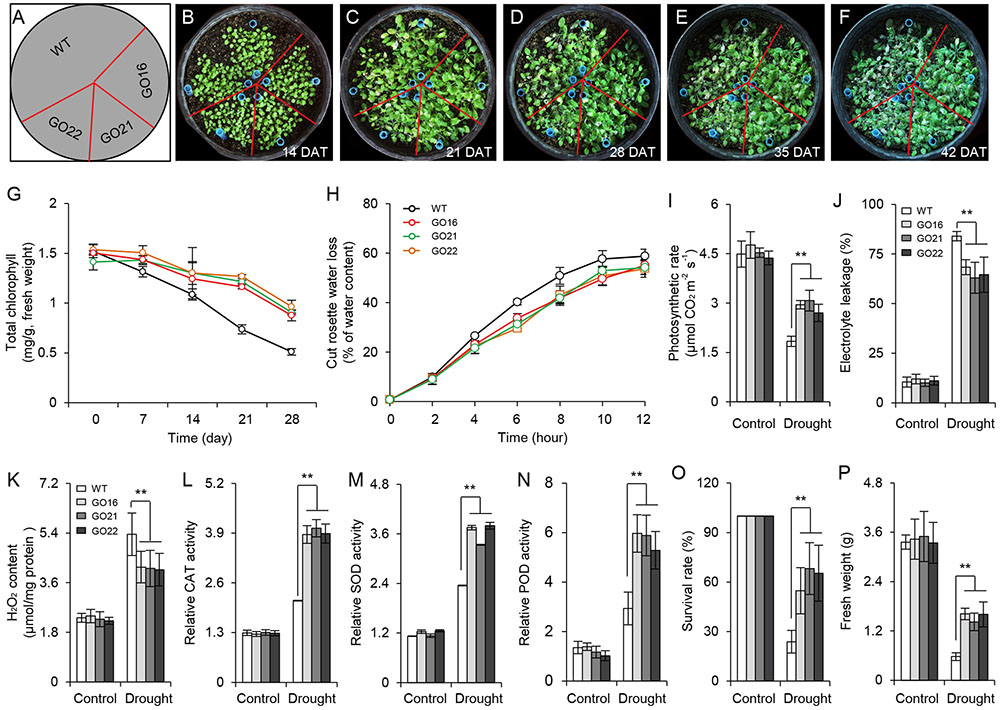
FIGURE 8. Transgenic Arabidopsis harboring 35S:GhPYL9-11A exhibit increased drought tolerance. (A–F) 35S:GhPYL9-11A confers drought tolerance to Arabidopsis. After 2 weeks of growth, water was withheld for 28 days under short-day conditions before watering was resumed. Representative images of plants taken 7, 14, 21, and 28 days after watering was withheld. DAT, day after transplanting. (G) Chlorophyll content of mature leaves in wild type and GO plants at the indicated times after watering were withheld. Values are means ± SD of 20 measurements; Student’s t-test. (H) Cumulative transpirational water loss from the rosettes of wild-type and GO Arabidopsis at the indicated times after detachment. Values are means ± SD of 20 measurements; Student’s t-test. (I–N) Physiological parameters of GO plants under drought stress treatment. Parameters were measured after water was withheld for 7 days. Overexpression of GhPYL9-11A in Arabidopsis increased the photosynthetic rate (I), reduced electrolyte leakage (J), reduced H2O2 content (K), and increased the activities of antioxidant enzymes, including CAT (L), SOD (M), and POD (N). Error bars indicate SD (n = 5). ∗∗P ≤ 0.01; Student’s t-test. (O) Survival rate (SR) of the wild-type and GO Arabidopsis. After 2 weeks of growth, plants were subjected to drought stress by withholding water for 4 weeks. Survival was scored 3 days after watering was resumed. Error bars indicate SD (n = 3); Student’s t-test. (P) Relative fresh weights of wild type and GO lines. After watering was withheld for 28 days, the rosettes were collected and weighted. Plants grown under well-watered conditions served as the control. Error bars indicate SD (n = 3). ∗∗P ≤ 0.01; Student’s t-test.
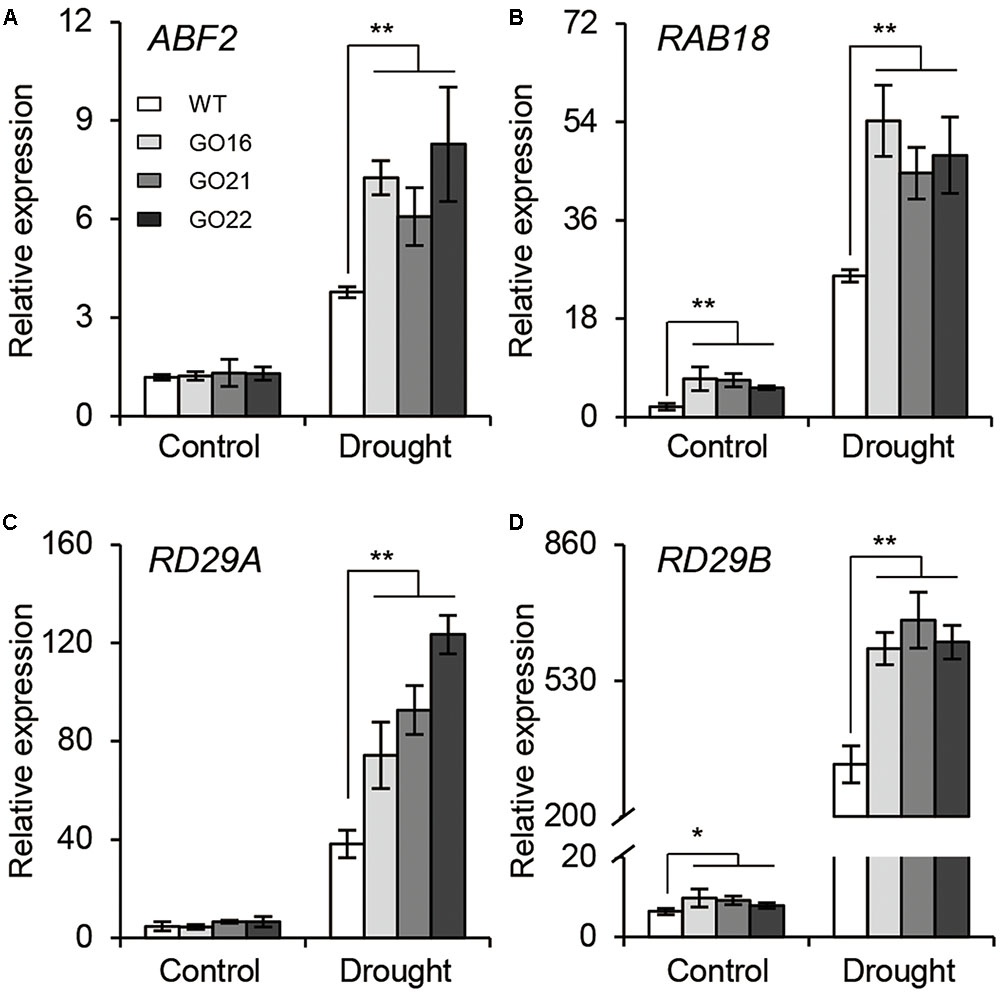
FIGURE 9. Expression of ABA-responsive genes in GhPYL9-11A overexpression plants under control and drought conditions. QRT-PCR analysis of ABA-responsive genes, including ABF2 (A), RAB18 (B), DR29A (C), and RD29B (D), in wild type and Arabidopsis GhPYL9-11A overexpression lines treated with 400 μM mannitol for 24 h. Values are means ± SD of three biological repeats. ∗P ≤ 0.05, ∗∗P ≤ 0.01; Student’s t-test.
GhPYL9-11A Expression Level Associates with Drought Stress Tolerance in Different Cultivated Cotton Cultivars
Our results clearly demonstrate that overexpression of GhPYL9-11A in Arabidopsis enhances drought stress tolerance. Thus, we speculated that the expression level of GhPLY-11A may be correlated with drought tolerance in cultivated cotton varieties. To test this hypothesis, we phenotyped the drought tolerance of 226 cotton varieties. These varieties were indexed by plant SR under severe drought stress, and 20 drought-tolerant (with SR ranging from 17.0 to 49.3%) and 20 drought-sensitive (with SR ranging from 65.2 to 95.9%) cotton varieties were selected for further analysis (Table S3). We next compared the seedling-stage expression levels of GhPYL9-11A in drought-tolerant and drought-sensitive cultivated cotton accessions under drought treatment. The average expression levels of GhPYL9-11A in the 20 drought-tolerant cotton varieties were significantly higher than in the drought-sensitive varieties under drought treatment (Figure 10B). However, the expression level did not differ under control conditions (Figure 10A). We further examined the relationship between the GhPYL9-11A expression levels and SR in 20 drought-tolerant and 20 drought-sensitive cultivated cotton varieties under drought stress and found that GhPYL9-11A expression is positively correlated with drought tolerance (R = 0.483; Figure 10C). These results further suggest that GhPYL9-11A plays an important role in drought tolerance in cultivated cotton.
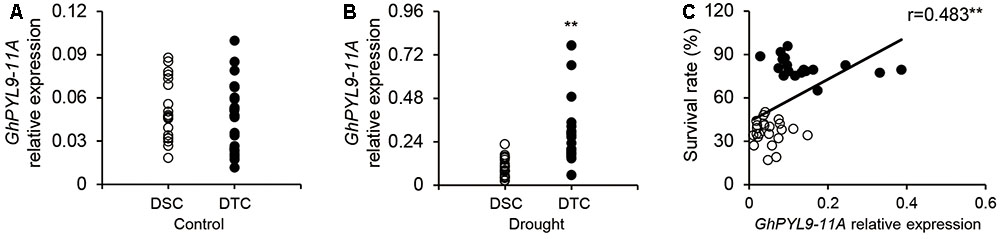
FIGURE 10. Expression level of GhPYL-11A in drought-tolerant and drought-sensitive cultivated cotton varieties is correlated with SR under severe drought stress. (A,B) The expression level of GhPYL9-11A in drought-tolerant and drought-sensitive cultivated cotton varieties treated with H2O (A) and 400 mM mannitol (B) for 24 h was determined by qRT-PCR. DSC, drought-sensitive cultivated cotton. DTC, drought-tolerant cultivated cotton. In (A,B), asterisks indicate significant differences (∗∗P < 0.01). (C) Correlation between the SR of cotton seedlings grown under severe drought conditions and the expression level of GhPYL9-11A.
Discussion
Recently, significant progress has been made in the identification and characterization of the PYL ABA receptors in several plants (Sheard and Zheng, 2009; Bai et al., 2013). However, to our knowledge, up to now, no ABA receptors from upland cotton, which is widely grown in over 80 countries and provides the material for about 90% of the world’s cotton lint production (Wang et al., 2012; Wang Y. et al., 2016; Paterson and Wendel, 2015; Liang et al., 2016), have been reported. Owing to allopolyploidization, the number of PYL genes has doubled in upland cotton compared with diploid cotton. However, to date, it remains largely unknown which PYL members is the crucial ABA receptor mediating response to drought stress in tetraploid cotton. Therefore, it is interesting and important to determine the functions and regulatory mechanisms of the ABA receptors in tetraploid cotton. In this study, we identified 22 ABA receptors orthologous to Arabidopsis PYLs in both the A- and D-genomes of the tetraploid cotton G. hirsutum using the recently released cotton genome sequence (Wang et al., 2012; Li et al., 2014, 2015; Liu X. et al., 2015). We further demonstrated that GhPYL9-11A is a functional ABA receptor, and similar to AtPYL9, interacts with PP2Cs in ABA-independent manner and regulates drought resistance and leaf senescence in Arabidopsis (Zhao et al., 2016). We found that GhPYL9-11A overexpression in Arabidopsis results in ABA-hypersensitive seed germination and root development phenotypes. Moreover, overexpression of GhPYL9-11A also significantly enhances drought and osmotic stress tolerance.
Abscisic acid was discovered in the 1960s, and to date, numerous biochemical and genetic studies have revealed that ABA is a vital regulator of multiple stress responses (Hurst, 1995; Finkelstein et al., 2002; Zhu, 2002; Sheard and Zheng, 2009; Zhao et al., 2014). ABA enhances plant survival under stress conditions by inducing seed and bud dormancy, inhibiting germination, closing stomata, and accelerating old leaf senescence and abscission. A variety of abiotic stresses sharply increase endogenous ABA levels and activates ABA signaling pathways leading to elevated plant survival. Furthermore, exogenously applied ABA has been shown to induce the expression of genes known to enhance stress tolerance. Our previous studies have demonstrated that several ABA-responsive genes, such as GhABF2 (Liang et al., 2016), GhDr1 (Ding et al., 2011), and GhSRK2D (Lin F. et al., 2012), play important roles in ABA signal transduction in response to drought stress in cotton. In this study, through comparative analysis of the transcript levels of 44 PYL genes, we found that GhPYL9-11A (Gh_A11G08070) is the most highly expressed during both seed germination and drought stress (Figure 1 and Table S2). Consistently, the homologs of GhPYL9-11A, Gorai_007G107500 and Cotton_A14394, in diploid cotton G. raimondii and G. arboreum, respectively, also show highly expressed pattern during seed germination and drought stress (Table S2). Moreover, the expression level of GhPYL9-11A is associated with drought stress in different cultivated cotton cultivars (Figure 10). We also confirmed that expression of GhPYL9-11A is indeed significantly induced by exogenous ABA treatment (Figure 2). These findings suggest that among the cotton PYLs, GhPYL9-11A may be the most important regulator of ABA signaling during seed germination and under drought stress.
Pyrabactin resistance-likes are ABA receptors that act at the apex of a negative regulatory pathway and regulate ABA signaling by binding and inhibiting PP2Cs (Melcher et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Park et al., 2009; Santiago et al., 2009; Sheard and Zheng, 2009). Similar to AtPYL-PP2C interactions in Arabidopsis, GhPYL9-11A interacts with AtABI1 and AtABI2, as well as with GhPP2C1 and GhPP2C2 in Y2H assays (Figure 3). Moreover, the interaction between GhPL9-11A and PP2C proteins does not depend on ABA. Previous studies have revealed that PYL-PP2C interactions are partially associated with two conserved amino acid residues preceding the CL2/gate-latch domain (Figure 4), and there are several polymorphisms in these residues, including VI, VV, VK, VQ, VT, VR, VN, IT, LV, and LK, in Arabidopsis, rice, soybean, poplar, and maize (Bai et al., 2013; Zhao et al., 2013). These polymorphisms are closely correlated with ABA-dependent or -independent interactions with PP2C. GhPYL9-11A belong to subgroup II of the GhPYL family, and have the typical VK combination in the CL2/gate-latch domain (Figure 2B). Consistent with the ABA-independent interaction between GhPYL0-11A and PP2Cs, VK is correlated with ABA-independent PYL-PP2C interactions. Intriguingly, GhPYL9-11A and AtPYL9 share high amino acid sequence identity and 3D structures, and both proteins contain the conserved proline (P84) and histidine (H111) residues in the gate-latch domain (Figure 2C). However, the site-directed mutants GhPYL9-11AP84S and GhPYL9-11AH111A are able to interact with AtABI2, GhPP2C1, and GhPP2C2, but not AtABI1 in an ABA-independent manner (Figures 3, 4), suggesting that residues in other regions of GhPYL9-11A also regulate the ABA-independent interactions with PP2Cs in cotton.
To escape unfavorable environmental conditions at unusual times or of atypical severity, plants increase seed dormancy and inhibit growth by elevating ABA content and activating core ABA signaling pathways (Zhu, 2002; Zhao et al., 2016). Although recent studies have greatly improved our knowledge about the PYL family in several plants, the basic biological functions of PYLs in tetraploid cotton remain largely unknown. Here, we clearly demonstrate that GhPYL9-11A overexpression in Arabidopsis has multiple physiological effects, including enhanced ABA sensitivity and inhibition of seed germination under mannitol treatment (Figures 3, 4). GhPYL9-11A overexpression also significantly increased seedling drought and osmotic tolerance, enhanced antioxidant enzyme activities, reduced the change in photosynthetic rate, reduced chlorophyll degradation, water loss, and electrolyte leakage, and induced the expression of a subset of ABA-regulated genes (Figures 8, 9). The combination of inhibition of seed germination and enhanced seedling abiotic stress tolerance in GhPYL9-11A overexpression lines is consistent with plant survival. These results and the observation that drought tolerant cotton varieties have higher GhPYL9-11A expression levels suggests that GhPYL9-11A may have played a role in the evolution of drought and osmotic tolerance in cotton.
Conclusion
GhPYL9-11A acts as a positive regulator of the ABA signaling pathway by binding to type PP2C protein phosphatases in an ABA-independent manner. Our study provides valuable insight into the function of GhPYL9-11A in the regulation of ABA signaling. In addition, the improved drought and osmotic tolerance of both seeds and seedlings of Arabidopsis GhPYL9-11A overexpression lines is encouraging for future efforts to genetically engineer plant drought and osmotic tolerance. Further manipulation of GhPYL-11A expression in cotton could aid the development of drought and osmotic tolerant cultivars.
Author Contributions
CL, SG, and RZ designed research. CL, YLiu, YLi, ZM, RY, YW, SK, MA, and GS performed research. CL, TZ, and RZ analyzed data. CL, WM, SG, and RZ wrote the paper.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31601349 to CL) and the Ministry of Agriculture (Grant No. 2016ZX08005004 to RZ, Grant No. 2016ZX08009003-003-004 to CL), and the Innovation Program of Chinese Academy of Agricultural Sciences.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01453/full#supplementary-material
FIGURE S1 | Phylogenetic analysis of PYL9 proteins in two diploid progenitor species G. raimondii and G. arboretum, and 4 two tetraploids G. hirsutum and G. barbadense. The scale bar indicates 0.1 amino acid substitution per site.
FIGURE S2 | Protein sequence alignment of PP2C protein, AtABI1, AtABI2 GhPP2C1, and GhPP2C2 in Arabidopsis and cotton using Cluster X. Conserved amino acid are highlighted in shades of black and gray. White letter with black background (100% identity), white letter with gray background (60%).
TABLE S1 | Primers used in this study.
TABLE S2 | Expression pattern of PYL genes in cotton.
TABLE S3 | Drought-tolerance and drought-sensitive cotton varieties used for correlation analysis between survival rate and GhPYL9-11A expression level.
References
Antoni, R., Gonzalez-Guzman, M., Rodriguez, L., Peirats-Llobet, M., Pizzio, G. A., Fernandez, M. A., et al. (2013). PYRABACTIN RESISTANCE1-LIKE8 plays an important role for the regulation of abscisic acid signaling in root. Plant Physiol. 161, 931–941. doi: 10.1104/pp.112.208678
Bai, G., Yang, D. H., Zhao, Y., Ha, S., Yang, F., Ma, J., et al. (2013). Interactions between soybean ABA receptors and type 2C protein phosphatases. Plant Mol. Biol. 83, 651–664. doi: 10.1007/s11103-013-0114-4
Cao, X. (2015). Whole genome sequencing of cotton–a new chapter in cotton genomics. Sci. China Life Sci. 58, 515–516. doi: 10.1007/s11427-015-4862-z
Chen, Z. J., Scheffler, B. E., Dennis, E., Triplett, B. A., Zhang, T., Guo, W., et al. (2007). Toward sequencing cotton (Gossypium) genomes. Plant Physiol. 145, 1303–1310. doi: 10.1104/pp.107.107672
Choi, H., Hong, J., Ha, J., Kang, J., and Kim, S. Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. doi: 10.1074/jbc.275.3.1723
Ding, Z., Zhang, R., and Guo, S. (2011). Cloning and bioinformatics analysis of cotton gene GhDr1 related to resistance to abiotic stress. Biotechnol. Bull. 1, 99–106. doi: 10.1093/pcp/pcw090
Fang, J., Chai, C., Qian, Q., Li, C., Tang, J., Sun, L., et al. (2008). Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J. 54, 177–189. doi: 10.1111/j.1365-313X.2008.03411.x
Finkelstein, R. R., Gampala, S. S., and Rock, C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl.), S15–S45. doi: 10.1105/tpc.010441
Gonzalez-Guzman, M., Pizzio, G. A., Antoni, R., Vera-Sirera, F., Merilo, E., Bassel, G. W., et al. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24, 2483–2496. doi: 10.1105/tpc.112.098574
Guang, Y., Song, G., Zhang, J., Wang, X., and Tan, C. (2014). Molecular cloning and expression analysis of GhbHLH130, encoding a novel bHLH transcription factor in uplant cotton (Gossypium hirsutum L.). Cotton Sci. 26, 363–370. doi: 10.1126/scisignal.2005051
Gunapati, S., Naresh, R., Ranjan, S., Nigam, D., Hans, A., Verma, P. C., et al. (2016). Expression of GhNAC2 from G. herbaceum, improves root growth and imparts tolerance to drought in transgenic cotton and Arabidopsis. Sci. Rep. 6:24978. doi: 10.1038/srep24978
Hao, Q., Yin, P., Li, W., Wang, L., Yan, C., Lin, Z., et al. (2011). The molecular basis of ABA-independent inhibition of PP2Cs by a subclass of PYL proteins. Mol. Cell 42, 662–672. doi: 10.1016/j.molcel.2011.05.011
He, X., Zhu, L., Xu, L., Guo, W., and Zhang, X. (2016). GhATAF1, a NAC transcription factor, confers abiotic and biotic stress responses by regulating phytohormonal signaling networks. Plant Cell Rep. 35, 2167–2179. doi: 10.1007/s00299-016-20276
Iyer, L. M., Koonin, E. V., and Aravind, L. (2001). Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins 43, 134–144. doi: 10.1002/1097-0134(20010501)43:2<134::AID-PROT1025>3.0.CO;2-I
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530. doi: 10.1038/nbt.3208
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572. doi: 10.1038/ng.2987
Li, W., Wang, L., Sheng, X., Yan, C., Zhou, R., Hang, J., et al. (2013). Molecular basis for the selective and ABA-independent inhibition of PP2CA by PYL13. Cell Res. 23, 1369–1379. doi: 10.1038/cr.2013.143
Liang, C., Meng, Z., Meng, Z., Malik, W., Yan, R., Lwin, K. M., et al. (2016). GhABF2, a bZIP transcription factor, confers drought and salinity tolerance in cotton (Gossypium hirsutum L.). Sci. Rep. 6:35040. doi: 10.1038/srep35040
Liang, C., Wang, Y., Zhu, Y., Tang, J., Hu, B., Liu, L., et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. U.S.A. 111, 10013–10018. doi: 10.1073/pnas.1321568111
Liang, C., Zheng, G., Li, W., Wang, Y., Hu, B., Wang, H., et al. (2015). Melatonin delays leaf senescence and enhances salt stress tolerance in rice. J. Pineal Res. 59, 91–101. doi: 10.1111/jpi.12243
Lin, A., Wang, Y., Tang, J., Xue, P., Li, C., Liu, L., et al. (2012). Nitric oxide and protein S-nitrosylation are integral to hydrogen peroxide-induced leaf cell death in rice. Plant Physiol. 158, 451–464. doi: 10.1104/pp.111.184531
Lin, F., Zhang, R., Sun, G., Sun, B., and Guo, S. (2012). Molecular cloning and its functional analysis of GhSRK20 gene from cotton. J. Agric. Sci. Technol. 14, 36–42.
Lin, L., Pierce, G. J., Bowers, J. E., Estill, J. C., Compton, R. O., Rainville, L. K., et al. (2010). A draft physical map of a D-genome cotton species (Gossypium raimondii). BMC Genomics 11:395. doi: 10.1186/1471-2164-11-395
Liu, L., Tong, H., Xiao, Y., Che, R., Xu, F., Hu, B., et al. (2015). Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. U.S.A. 112, 11102–11107. doi: 10.1073/pnas.1512748112
Liu, X., Zhao, B., Zheng, H. J., Hu, Y., Lu, G., Yang, C. Q., et al. (2015). Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci. Rep. 5:14139. doi: 10.1038/srep14139
Liu, Y., Zhang, X., Zhu, S., Zhang, H., Li, Y., Zhang, T., et al. (2016). Overexpression of GhSARP1 encoding a E3 ligase from cotton reduce the tolerance to salt in transgenic Arabidopsis. Biochem. Biophys. Res. Commun. 478, 1491–1496. doi: 10.1016/j.bbrc.2016.07.033
Melcher, K., Ng, L. M., Zhou, X. E., Soon, F. F., Xu, Y., Suino-Powell, K. M., et al. (2009). A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462, 602–608. doi: 10.1038/nature08613
Miyazono, K., Miyakawa, T., Sawano, Y., Kubota, K., Kang, H. J., Asano, A., et al. (2009). Structural basis of abscisic acid signalling. Nature 462, 609–614. doi: 10.1038/nature08583
Nishimura, N., Hitomi, K., Arvai, A. S., Rambo, R. P., Hitomi, C., Cutler, S. R., et al. (2009). Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science 326, 1373–1379. doi: 10.1126/science.1181829
Nishimura, N., Sarkeshik, A., Nito, K., Park, S. Y., Wang, A., Carvalho, P. C., et al. (2010). PYR/PYL/RCAR family members are major in-vivo ABI1 protein phosphatase 2C-interacting proteins in Arabidopsis. Plant J. 61, 290–299. doi: 10.1111/j.1365-313X.2009.04054.x
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Paterson, A. H., and Wendel, J. F. (2015). Unraveling the fabric of polyploidy. Nat. Biotechnol. 33, 491–493. doi: 10.1038/nbt.3217
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Qin, L. X., Nie, X. Y., Hu, R., Li, G., Xu, W. L., and Li, X. B. (2016). Phosphorylation of serine residue modulates cotton Di19-1 and Di19-2 activities for responding to high salinity stress and abscisic acid signaling. Sci. Rep. 6:20371. doi: 10.1038/srep20371
Santiago, J., Dupeux, F., Round, A., Antoni, R., Park, S. Y., Jamin, M., et al. (2009). The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462, 665–668. doi: 10.1038/nature08591
Shan, C. M., Shangguan, X. X., Zhao, B., Zhang, X. F., Chao, L. M., Yang, C. Q., et al. (2014). Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat. Commun. 5:5519. doi: 10.1038/ncomms6519
Sheard, L. B., and Zheng, N. (2009). Signal advance for abscisic acid. Nature 462, 575–576. doi: 10.1038/462575a
Shi, Y., Zhang, R., Wu, X. P., Meng, Z., and Guo, S. (2012). Cloning and characterization of a somatic embryogenesis receptor-like kinase gene in cotton (Gossypium hirsutum). J. Integr. Agric. 11, 898–909. doi: 10.1016/S2095-3119(12)60080-X
Wang, C., Lu, W., He, X., Wang, F., Zhou, Y., Guo, X., et al. (2016). The cotton mitogen-activated protein kinase kinase 3 functions in drought tolerance by regulating stomatal responses and root growth. Plant Cell Physiol. 57, 1629–1642. doi: 10.1093/pcp/pcw090
Wang, M., Tu, L., Lin, M., Lin, Z., Wang, P., Yang, Q., et al. (2017). Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat. Genet. 49, 579–587. doi: 10.1038/ng.3807
Wang, K., Wang, Z., Li, F., Ye, W., Wang, J., Song, G., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103. doi: 10.1038/ng.2371
Wang, Y., Liang, C., Wu, S., Zhang, X., Tang, J., Jian, G., et al. (2016). Significant improvement of cotton Verticillium wilt resistance by manipulating the expression of Gastrodia antifungal proteins. Mol. Plant 9, 1436–1439. doi: 10.1016/j.molp.2016.06.013
Xing, L., Zhao, Y., Gao, J., Xiang, C., and Zhu, J. K. (2016). The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci. Rep. 6:27177. doi: 10.1038/srep27177
Xiong, L., Schumaker, K. S., and Zhu, J. K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14(Suppl.), S165–S183.
Yoshida, T., Mogami, J., and Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Yuan, D., Tang, Z., Wang, M., Gao, W., Tu, L., Jin, X., et al. (2015). The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 5:17662. doi: 10.1038/srep17662
Zhang, M., Yuan, B., and Leng, P. (2009). The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J. Exp. Bot. 60, 1579–1588. doi: 10.1093/jxb/erp026
Zhao, Y., Chan, Z., Gao, J., Xing, L., Cao, M., Yu, C., et al. (2016). ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. U.S.A. 113, 1949–1954. doi: 10.1073/pnas.1522840113
Zhao, Y., Chan, Z., Xing, L., Liu, X., Hou, Y. J., Chinnusamy, V., et al. (2013). The unique mode of action of a divergent member of the ABA-receptor protein family in ABA and stress signaling. Cell Res. 23, 1380–1395. doi: 10.1038/cr.2013.149
Zhao, Y., Xing, L., Wang, X., Hou, Y. J., Gao, J., Wang, P., et al. (2014). The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 7, ra53. doi: 10.1126/scisignal.2005051
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Keywords: ABA, ABA receptor, GhPYL9, PP2C, seed germination, drought stress, cotton
Citation: Liang C, Liu Y, Li Y, Meng Z, Yan R, Zhu T, Wang Y, Kang S, Ali Abid M, Malik W, Sun G, Guo S and Zhang R (2017) Activation of ABA Receptors Gene GhPYL9-11A Is Positively Correlated with Cotton Drought Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 8:1453. doi: 10.3389/fpls.2017.01453
Received: 11 May 2017; Accepted: 04 August 2017;
Published: 23 August 2017.
Edited by:
Hui Xia, Shanghai Agrobiological Gene Center, ChinaReviewed by:
Abid Ali, University of Agriculture, Faisalabad, PakistanAndrew Wood, Southern Illinois University Carbondale, United States
Copyright © 2017 Liang, Liu, Li, Meng, Yan, Zhu, Wang, Kang, Ali Abid, Malik, Sun, Guo and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Zhang, zhangrui@caas.cn Sandui Guo, guosandui@caas.cn
†These authors have contributed equally to this work.
 Chengzhen Liang
Chengzhen Liang Yan Liu1†
Yan Liu1† Yanyan Li
Yanyan Li Zhigang Meng
Zhigang Meng Rong Yan
Rong Yan Tao Zhu
Tao Zhu Muhammad Ali Abid
Muhammad Ali Abid Waqas Malik
Waqas Malik Sandui Guo
Sandui Guo Rui Zhang
Rui Zhang