- 1The State Key Laboratory of Plant Genomics, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
- 2Institute of Environment and Sustainable Development in Agriculture, Chinese Academy of Agricultural Sciences, Beijing, China
Alternanthera philoxeroides is a notoriously invasive weed that can readily adapt to different environmental conditions. Control of this weed is difficult, and it spreads easily and causes damage to native habitats and agriculture. In this study, our goal was to investigate the molecular mechanisms that lead to the ability of A. philoxeroides to invade new habitats, to adapt to environmental stresses, and to cause damage. We developed a simple and highly effective potato virus X-based virus-induced gene silencing (VIGS) approach. The VIGS approach was first used to silence the phytoene desaturase gene, which resulted in the expected photo-bleaching phenotype. Next, the VIGS approach was used to silence two additional genes, drought-induced protein gene 15 (ApDRI15) and salinity-induced protein gene 1 (ApSI1). When ApDRI15 was knocked down, the plants were more sensitive to drought stress than the control plants, with smaller leaves, shorter internodes, and lower biomass. The ApDRI15-silenced plants had lower relative water content, lower free proline levels, and higher water loss rates than the control. Silencing of ApSI1 significantly decreased tolerance to salinity, and the ApSI1-silenced plants were withered and smaller. These results indicate that the pgR107 VIGS approach is a simple and highly effective tool for dissecting gene function in A. philoxeroides. Further experiments with the VIGS approach will enhance our understanding of the molecular mechanisms of the adaptability and plasticity of A. philoxeroides and improve our ability to combat the damage caused by this weed.
Introduction
Alternanthera philoxeroides (Max.), also called alligator weed, is a notoriously invasive weed. It originated in the Parana river region in southern America, but now has invaded the United States, Australia, New Zealand, China, and India (Sainty et al., 1998). This stoloniferous and amphibious weed grows in both terrestrial and aquatic conditions. A. philoxeroides can adapt to different habitats and to fluctuating environments (Wang et al., 2008, 2009; Dong et al., 2012; Fan et al., 2013). For instance, in arid land, its leaves are small, with short internodes and thin cavities, but in water the leaves are large, with long internodes and wide cavities (Wang et al., 2008; Dong et al., 2010). The plasticity and adaptability of A. philoxeroides facilitates invasion into new habitats, which damages agricultural production and ecological balance (Ye et al., 2003; You et al., 2016). A. philoxeroides has been become one of the most notoriously destructive weeds worldwide, but there are no effective measures to control this weed (Schooler et al., 2007). Therefore, there is a critical need for exploration of the underlying mechanisms involved in the plasticity and adaptability of A. philoxeroides.
Most of the major studies on A. philoxeroides have focused on plant morphology, ecology, taxonomy, and weed management (Sainty et al., 1998; Ye et al., 2003; Wang et al., 2005, 2008; Dong et al., 2010; Wu et al., 2016a,b). However, the underlying molecular mechanisms of invasion of this weed remain unclear, primarily because of a shortage of analytical techniques. Virus-induced gene silencing (VIGS) is a powerful approach in molecular biology and genetics, widely used to dissect gene function in plants. The modified VIGS vectors come from many plant RNA and DNA viruses, which are used in diverse dicot and monocot plants. For instance, there are VIGS vectors widely used in dicot plants, including tobacco rattle virus-derived vector (Ratcliff et al., 2001; Liu et al., 2002), bean pot mottle virus-derived vector (Zhang and Ghabrial, 2006), cabbage leaf curl virus-derived vector (Turnage et al., 2002), potato virus X vector (later modified pgR106/107 VIGS vector) (Faivre-Rampant et al., 2004), and tobacco mosaic rattle virus (George et al., 2010). In monocot plants, there are also several VIGS systems for analyzing the function of genes, such as barley stripe mosaic virus (Holzberg et al., 2002), brome mosaic virus (Ding et al., 2006), and bamboo mosaic virus (Liou et al., 2014). Very recently, cucumber mosaic virus-based VIGS in maize (Wang et al., 2016) and foxtail mosaic virus-based VIGS have been reported in barley, wheat, and foxtail millet (Setaria italica) (Liu et al., 2016). Thus, our goal was to develop a VIGS system for assessing gene function and for controlling A. philoxeroides invasion and damage.
In this study, we firstly explored several types of VIGS systems and successfully developed a pgR107 (PVX-based vector) VIGS approach in A. philoxeroides. First, we isolated a phytoene desaturase (PDS) gene from A. philoxeroides to use as a marker gene in the development of the VIGS system. Then, we employed this VIGS system to examine the role of the A. philoxeroides drought-induced protein gene 15 (DRI15, GenBank DQ985704.1) and the salinity-induced protein gene 1 (SI1, GenBank DQ489701) in response to stress from drought and high salinity. The pgR107 VIGS system is a simple and effective tool for analyzing A. philoxeroides gene function that will facilitate control of weed invasion and damage.
Materials and Methods
Plant Materials, Infiltration and Growth Conditions
Alternanthera philoxeroides was collected from field in Sichuan province in China, and cultured it in water supplemented with hoagland’s solution in growth chamber for asexual propagation. Then newly germinating internodes of branches were transferred to a pot for a single clone culture with many branches. The 3–4 internodes cut from different branches were inserted into cultural soil (two internodes above the ground) for propagation. When the two new internodes grew, the two opposite leaves at top internodes were used for VIGS infiltration. The new expanding leaves of inoculation plants were used for RNA and proteins extraction. All plants were grown in a growth chamber at 25°C with a 12-h light/12-h dark photoperiod cycle. The intensity of light is 2000 lum/sqf.
PDS, DRI15, and SI1 cDNA Isolation and Plasmid Construction
pgR107 vector (Jones et al., 1999; Lu et al., 2003) was kindly given by Professor Zhendong Tian in Huazhong Agricultural University of China, was firstly applied to the asexual propagation plant of A. philoxeroides in this study. pgR107 vector is binary vector based on pGreen0000 backbone, in which CP promoter contains CalI-SmalI-SalI cloning sites.
The A. philoxeroides PDS gene was amplified by PCR using DNA from A. philoxeroides leaves. The PCR primers were designed to amplify sequences conserved in other species of plants (Supplementary Table S1). The 936 base pair PDS fragment was amplified by PCR (Supplementary Data Sheet S1), and a 300 base pair specific fragment (Figure 1A in box) was inserted in the Sal I/Cla I sites of the pgR107 vector. The resulting vector was named pgRPDS (Figure 1A).
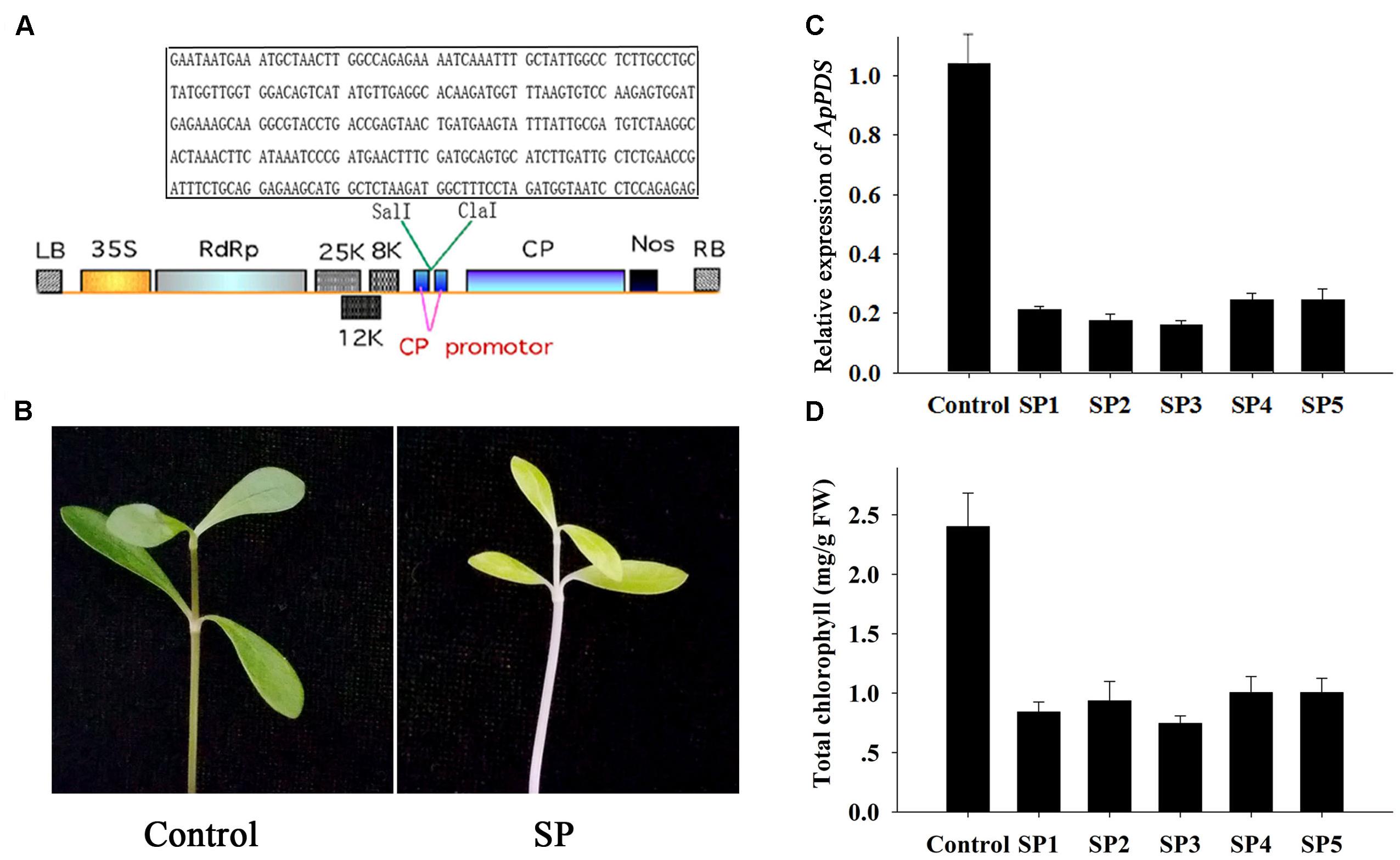
FIGURE 1. Development of ApPDS-silenced plants. (A) T-DNA schematic structure of the pgRPDS virus vector. LB and RB indicate the left and right T-DNA border sequences. 35S indicates the 35S promoter of cauliflower mosaic virus. RdRp indicates the potato virus X (PVX) 165K RNA-dependent RNA polymerase. 8K, 12K, and 25K indicate the PVX triple gene block movement proteins. CP indicates the viral coat protein gene. Nos indicates the nopaline synthase transcriptional terminator. The specific target sequence of the ApPDS gene is contained in the box. (B) The photo-bleaching phenotype. The control was plants agro-infiltrated with the empty vector. SP indicates the ApPDS-silenced plants, exhibiting photo-bleaching. (C) The expression level of ApPDS. SP1 through SP5 are five replicate ApPDS-silenced plants. (D) The total chlorophyll levels of new leaves from PDS-silenced plants. SP1 through SP5 are five ApPDS-silenced plants.
The ApDRI15 and ApSI1 genes were first identified from a database of sequenced cDNAs prepared from A. philoxeroides leaves. After we blast two gene nucleotide sequences in NCBI, the specific sequence fragments in A. philoxeroides were selected for analyzing their functions. Then a special sequence fragment of ApDRI15 (260 bp) or ApSI1 (290 bp) was PCR-amplified with its specific primers (Supplementary Table S1). The ApDRI15 and ApSI1 PCR fragments were inserted in the sense orientation into the Sal I/Cla I sites of the pgR107 virus vector to create the pgRDRI15 and pgRDSI1 vectors, respectively (Figures 2A, 4A in box).
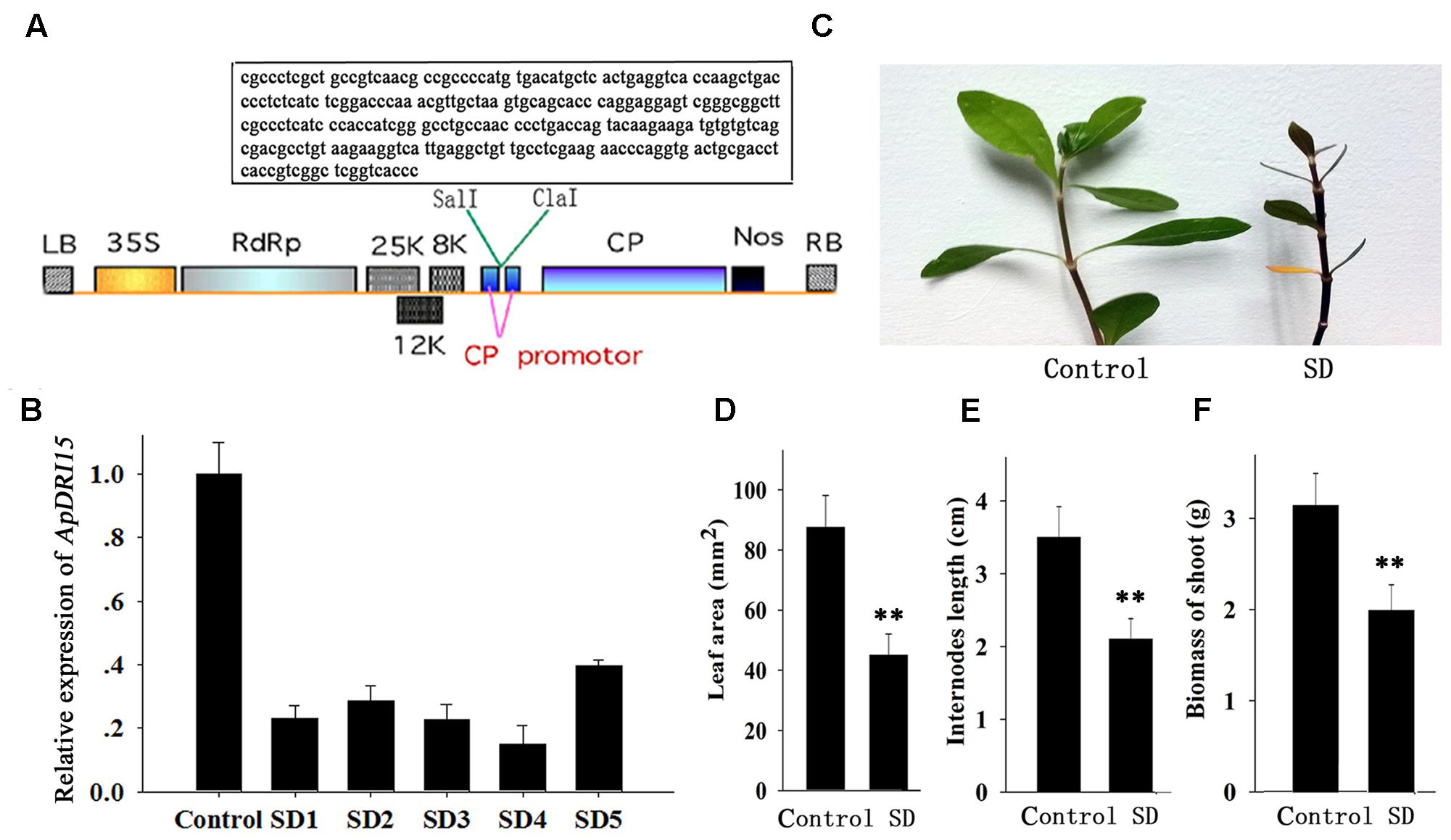
FIGURE 2. Sensitivity of ApDRI15-silenced plants to drought stress. (A) T-DNA schematic structure of pgRDRI15 virus vector. LB and RB indicate the left and right T-DNA border sequences. 35S indicates the 35S promoter of cauliflower mosaic virus. RdRp indicates the PVX 165K RNA-dependent RNA polymerase. 8K, 12K, and 25K indicate the PVX triple gene block movement proteins. CP indicates the viral coat protein gene. Nos indicates the nopaline synthase transcriptional terminator. The specific target sequence of the ApDRI15 gene is displayed in the box. (B) The expression level of ApDRI15. The control is the plants agro-infiltrated with the empty vector. SD1 through SD5 are five replicate ApDRI15-silenced plants. Expression in the silenced plants was normalized to the expression in the control plant. (C) Drought-sensitive phenotype of the ApDRI15-silenced plants. SD is the ApDRI15-silenced plant. (D) The leaf area of the ApDRI15-silenced plants and the control. (E) The internode length of the ApDRI15-silenced plants and the control. (F) The shoot biomass of the ApDRI15-silenced plants and the control. Error bars represent the standard derivation of three biological replicated [n = 36 in (D,E), n = 24 in (F)]. Double asterisks indicate statistically significant differences compared to the control, as determined using the Student’s t-test (p < 0.01).
RNA Extraction and Analysis
Total RNA was extracted and purified from A. philoxeroides leaves using Plant Total RNA Isolation Kits (Sangon, Shanghai, China). One microgram of RNA was reverse-transcribed using the BluePrint 1st strand cDNA Synthesis Kit (Takara, Dalian, China) according to the manufacturer’s protocol.
The quantitative real-time PCR (qPCR) assay was conducted using the SYBR Premix Ex Taq kit (Takara) according to the manufacturer’s guidelines. All PCRs were run on an IQ5 multicolor detection system (Bio-Rad, Hercules, CA, United States). Gene expression was quantified using the ΔΔCt algorithm. To normalize gene expression, the universal actin gene was used as an internal standard. The gene-specific primers are listed in Supplementary Table S1. The experiment was repeated using three biological replicates.
Agrobacterium Infiltration
The pgRPDS, pgRDRI15, and pgRSI vectors were introduced into Agrobacterium tumefaciens strain GV3101 with helper plasmid pJICSa_Rep (carries tetracycline as selection mark (5 μg/ml) was needed for replicating) by electroporation. Agrobacterial cells were grown, collected, and resuspended in MMA solution [10 mM MES (2-(N-Morpholino) ethanesulfonic acid), 10 mM MgCl2, 200 μM acetosyringone] to a final OD600 of 1.2. The A. philoxeroides shoots with 3–4 internodes (6–8 leaves) were used. The undersides of two fully expanded leaves were inoculated with Agrobacterium cells using a 1-mL needleless syringe. When another new 3–4 internodes grew after inoculation, they were cut and inserted a new pot with two internodes underground, which was regarded as silenced plants for salinity and drought treatment. Forty-five plants for each gene were repeated at least. Infiltrated plants were grown in a growth chamber at 25°C with a 12-h light/12-h dark photoperiod cycle.
Transpirational Water Loss Assay
Transpirational water loss assay of detached leaves was conducted according to previous reports (Duan et al., 2012). Leaves of ApDRI15-silenced and control plants were detached and placed on an electronic balance for continuously weighting under room temperature condition. The reduction in fresh weight of the samples was presumed to be the result of water loss. This experiment was repeated three times.
Relative Water Content
The relative water content of leaves was measured using the method of Parida et al. (2007). Fully expanded leaves were cut from the plants, and the fresh weight was recorded immediately. Then, the fresh portions were immersed in distilled water for 4 h and the turgid weight was recorded. Finally, the dry weight was recorded after drying for 48 h at 80°C in an oven. The relative water content was calculated according to the following formula:
Relative water content (%) = (Fresh weight - Dry weight)/(Turgid weight - Dry weight) × 100.
Chlorophyll Content Assay
Chlorophyll content was measured using the method of Arnon (1949). Extracts were obtained from 0.1 g (fresh weight) leaf samples and were homogenized in 1 mL of 80% acetone to quantify the chlorophyll content via spectrophotometric analysis.
Biomass Accumulation and Determination of Proline Content
Similar size of plants was used for each treatment. The total above-ground fresh weight biomass (including shoot and leaves) was measured immediately after harvesting after 15 days stress treatment and then the average biomass plant-1. The free proline content was measured using the method described by Bates et al. (1973). Leaf segments were homogenized in 3% sulfosalicylic acid, and the homogenates were centrifuged at 3000 × g for 20 min. Mixtures containing 2 mL of sample supernatant, 2 mL of acetic acid, and 2 mL of 2.5% acid ninhydrin solution were boiled for 30 min, and the absorbance at 520 nm (A520) was measured.
Results
Photo-Bleaching Phenotype in ApPDS-Silenced Plants
The PDS gene is widely used as a marker gene in many plant VIGS assays because the photo-bleaching phenotype is easy to visualize (Liu et al., 2002). Therefore, our first step was to isolate the A. philoxeroides PDS gene. Several nucleotide sequences of the PDS gene from diverse plants in the NCBI database were aligned (Supplementary Figure S1). We then designed PCR primers in highly conserved regions, as shown in Supplementary Table S1 and Figure S1. A 936 base-pair PDS fragment was obtained (Supplementary Data Sheet S1). A 300 base-pair specific fragment of the PDS gene (Figure 1A in box) was selected for gene silencing. The new leaves of A. philoxeroides plants were infiltrated with Agrobacterium cells containing pgRPDS or empty vector with a 1-mL syringe. Fourteen days after injection, the emerging leaves and internodes were photo-bleached in PDS-silenced plants presenting lighter green, while no changes were observed in plants infiltrated with empty vector (Figure 1B). The other pictures showing the similar lighter green phenotype were shown as Supplementary Figure S2. Additionally, the virus did not damage either these plants or the untreated plants. These data indicate that the pgR107 VIGS system can be used effectively and safely to dissect gene function in A. philoxeroides.
To analyze the effect of PDS silencing, qPCR was used to monitor the expression level of the gene in the agro-infiltrated plants. As shown Figure 1C, the PDS expression level was reduced in the new emerging leaves from five PDS-silenced plants. PDS was expressed at only 15–25% of the level expressed in plants injected with empty vector (the control). The chlorophyll contents of these treated plant leaves were tested to further confirm effective PDS knockdown. The total chlorophyll levels of new leaves from PDS-silenced plants were significantly lower than in leaves from the control (Figure 1D). The mean concentration of chlorophyll in PDS-silenced plants was only 35% of the control concentrations. The successful knockdown of the PDS gene indicates that the pgR107 VIGS system works well for dissection of A. philoxeroides gene function.
VIGS Knockdown of ApDRI15 Increased A. philoxeroides Susceptibility to Drought Stress
Alternanthera philoxeroides adapts readily to various abiotic stresses including drought and salinity. We isolated a drought response-related gene, named ApDRI15, which was submitted to the NCBI database. A sequence fragment of ApDRI15 was inserted into the pgR107 virus vector to create the pgRDRI15 vector (Figure 2A). The ApDRI15-silenced plants were created by agro-infiltration with GV3101 containing pgRDRI15. The normal plant inoculating with pgR107 VIGS vector (empty vector) served as control. As shown in Figure 2B, the ApDRI15 expression level in the five ApDRI15-silenced plants was only 15–40% of the expression level in the control leaves. The silenced plants with <25% of the control expression level were selected for further analysis with the drought-response assay.
The ApDRI15-silenced plants and the control plants were grown in a greenhouse without watering. After 15 days (Figure 2C), the silenced plants were noticeably different from the controls. The silenced plants had significantly smaller leaves, shorter internodes, and less biomass than the controls (Figures 2D–F). After 21 days of drought, the silenced plants had wilted shoots and withered old leaves, evidence of greater drought sensitivity compared to the controls. Ten days after re-watering, 92% of control plants had recovered but only 42% of ApDRI15-silenced plants had recovered (Figures 3A,B).
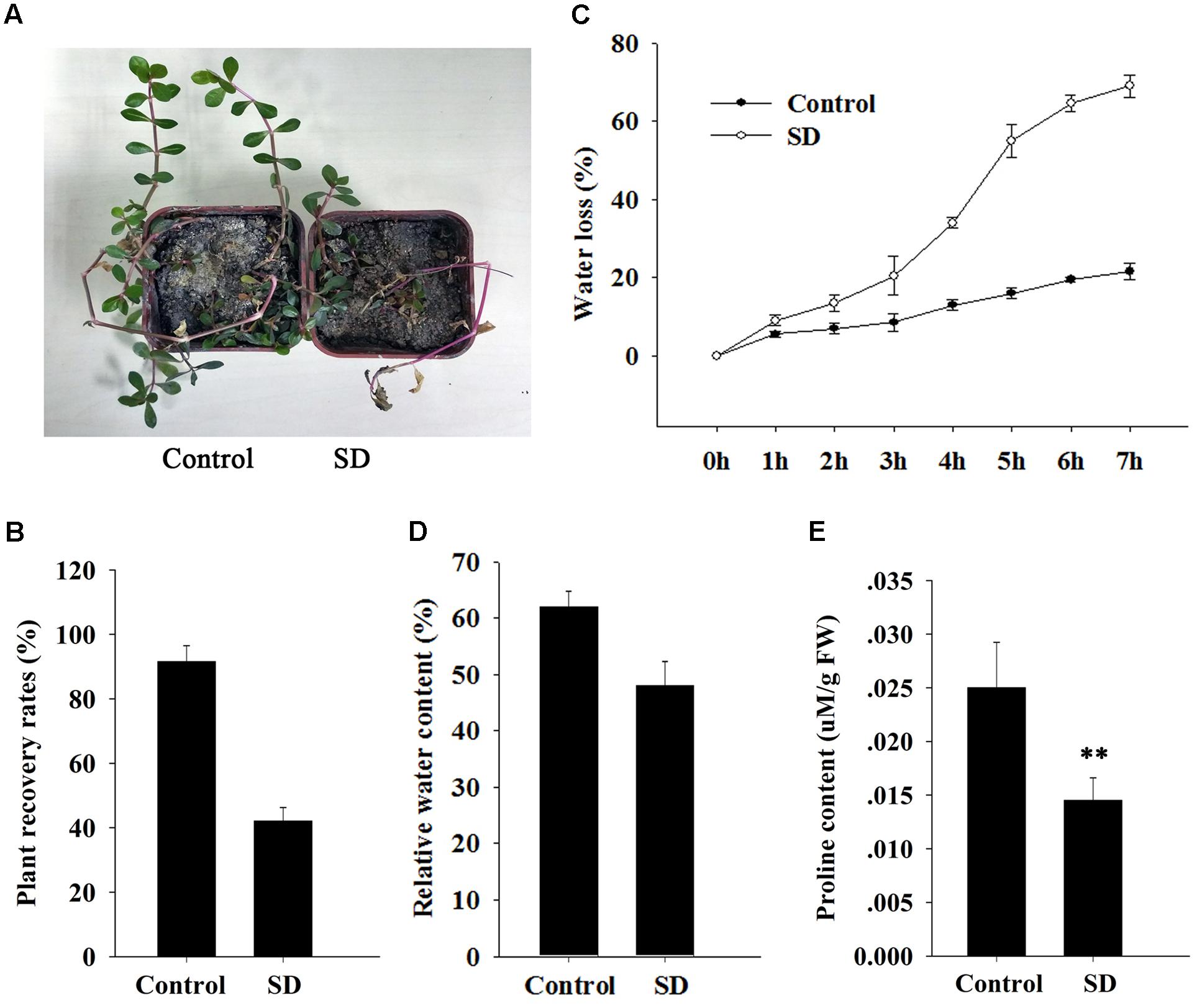
FIGURE 3. Effect of ApDRI15 knockdown on survival and physiology of plants under drought stress. (A) Recovery of ApDRI15-silenced plants and control plants after re-watering. Control plants are plants agro-infiltrated with the empty vector. SD indicates ApDRI15-silenced plants. (B) Recovery rates of ApDRI15 and control plants. (C) Water loss rates of detached leaves from ApDRI15-silenced plants and control plants. (D) Relative water contents of leaves of ApDRI15-silenced plants and control plants. (E) Free proline concentrations of ApDRI15-silenced plants and control plants. Error bars represent the standard derivation of three biological replicates [n = 92 in (B), n = 30 in (C,D), n = 15 in (E)]. Double asterisks indicate statistically significant differences compared to the control, as determined using the Student’s t-test (p < 0.01).
We measured physiological parameters in order to evaluate the function of ApDRI15 in the drought stress response. The detached leaf transpiration rate in the silenced plants was significantly higher than that in the controls (Figure 3C). The average leaf weight in the ApDRI15-silenced plants decreased by 70% after 8 h, whereas in the control plants the decrease was only 25%. The relative water content in leaves of silenced plants (48%) was lower than in leaves of control plants (62%) (Figure 3D). In addition, we monitored the level of free proline, an osmoprotective molecule that accumulates under drought stress, which has been reported as standard maker for plant drought resistance (Li et al., 2017). The average proline content of the ApDRI15-silenced plants was significantly lower than in the control plants (Figure 3E). These results suggested that the role of ApDRI15 in drought stress resistance can be dissected using a pgR107 VIGS approach in A. philoxeroides.
Knockdown of ApSI1 Attenuated the Resistance of Plants to Salinity Stress
We isolated another salinity inducible gene, ApSI1, that was deposited in NCBI database, and characterized its function with the pgR107 VIGS approach. As described above, the special fragment of ApSI1 was cloned, and the resulting vector pgRSI was generated (Figure 4A). ApSI1-silenced plants and control plants were generated following agro-infiltration. The ApSI1 expression levels in the silenced plants were significantly reduced, averaging only 22% of the control levels (Figure 4B). The ApSI1-silenced plants with low ApSI1 expression levels (less than 22% of the control levels) were used to perform a salinity stress analysis. When silenced plants and control plants were treated with 300 μM sodium chloride, the ApSI1-silenced plant leaves were a paler shade of green than the control leaves. After 20 days of sodium chloride treatment, the ApSI1-silenced plants were withering, yet the control plants grew a bit yellowish under sodium chloride (Figure 4C). To evaluate the role of ApSI1 in growth under salinity stress, we measured the total chlorophyll and the biomass of the silenced plants and the control plants 15 days after sodium chloride treatment. The average total chlorophyll level of ApSI1-silenced plant leaves was significantly lower than that of control leaves (Figure 4D). The fresh biomass of the silenced plants averaged 3.9 g per silenced plant shoot, but for the controls the value was 5.5 g (Figure 4E). The dissection of the role of ApSI1 in salinity tolerance further demonstrates that the pgR107 VIGS approach is an effective tool for analysis of gene function in A. philoxeroides.
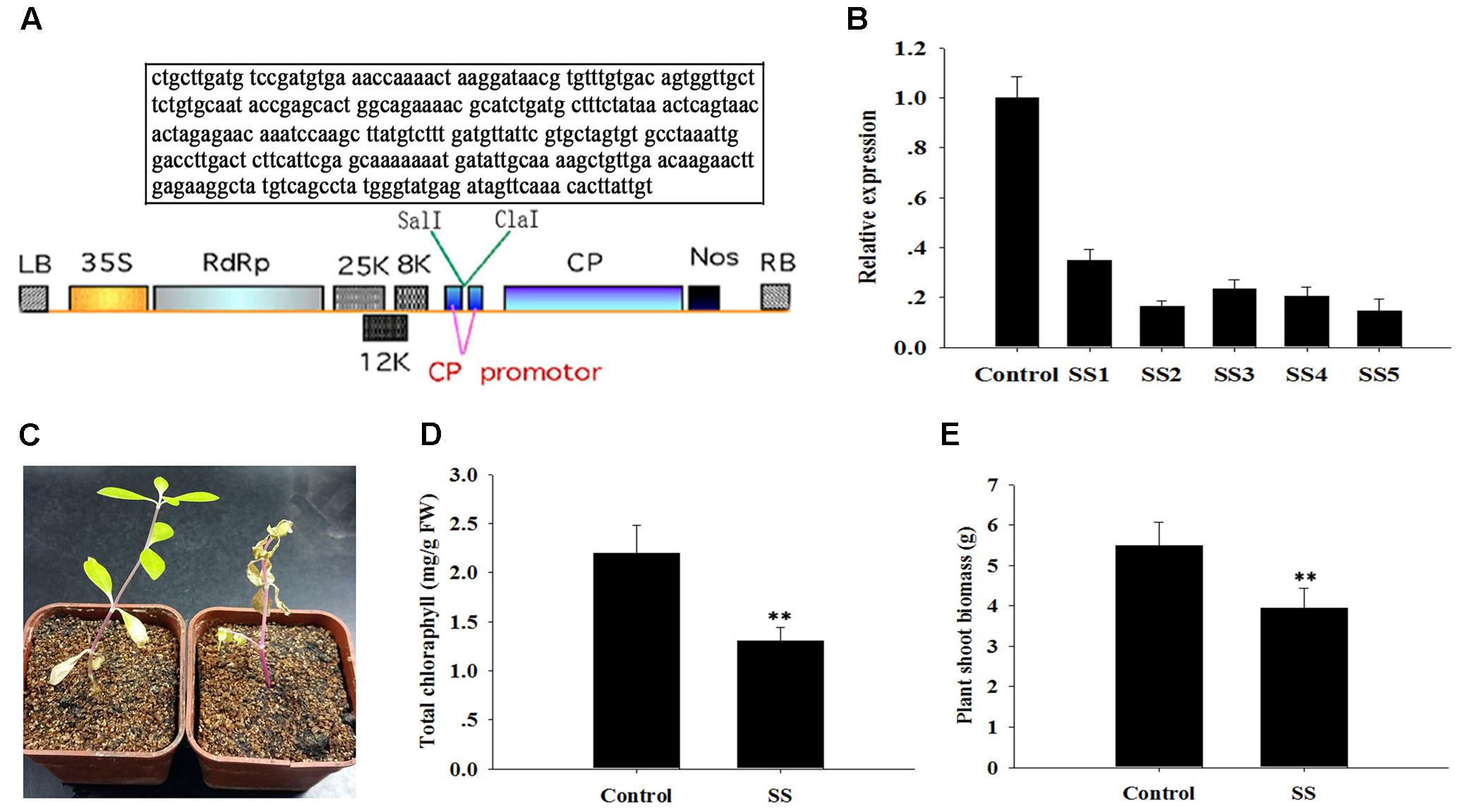
FIGURE 4. Sensitivity of ApSI1-silenced plants to salinity stress. (A) T-DNA schematic structure of pgRDSI1 virus vector. LB and RB indicate the left and right T-DNA border sequences. 35S indicates the 35S promoter of cauliflower mosaic virus. RdRp indicates the PVX 165K RNA-dependent RNA polymerase. 8K, 12K, and 25K indicate the PVX triple gene block movement proteins. CP indicates the viral coat protein gene. Nos indicates the nopaline synthase transcriptional terminator. The specific target sequence of ApSI1 gene is displayed in the box. (B) The expression level of ApsI1. The control was plants agro-infiltrated with the empty vector. SS1 through SS5 are five replicate ApSI1-silenced plants. Expression in the ApSI1-silenced plants was normalized to the control. (C) Salinity-sensitive phenotype of ApSI1-silenced plants. SS indicates ApSI1-silenced plants. (D) Total chlorophyll contents of the ApSI1-silenced plants and control plants. (E) Shoot biomass of ApSI1-silenced plants and control plants. Error bars represent the standard derivation of three biological replicates (n = 24). Double asterisks indicate statistically significant differences between ApSI1-silenced and control plants, as determined using the Student’s t-test (p < 0.01).
Discussion
Alternanthera philoxeroides is a notoriously invasive weed, and is difficult to prevent from its damages (Sainty et al., 1998; Pan et al., 2006; Fan et al., 2013). To date, the researches in the underlying molecular mechanisms of A. philoxeroides, exclude taxonomy and ecology and the integrated management is only limited to analyses of genome-wide DNA methylation and gene expression (Gao et al., 2010, 2015), due to insufficient molecular approaches. In this study, we successfully developed a simple and highly effective method in A. philoxeroides, the pgR107 VIGS system, using the ApPDS marker gene. We also used this VIGS approach to determine the roles of the ApDRI15 and ApSI1 genes in the A. philoxeroides response to drought and salinity stresses, respectively.
The disease symptoms of plants injected with the PVX vector are mild, which aids in the analysis of gene function (Faivre-Rampant et al., 2004; Jada et al., 2013; Ramanna et al., 2013). We used the pgR107 virus vector to silence A. philoxeroides genes, without obvious disease symptoms. Our results demonstrate that the PVX VIGS system (pgR107 vector) is extremely useful for dissecting gene function in A. philoxeroides, as demonstrated with the ApPDS, ApDRI15, and ApSI1 genes. Additionally, because the A. philoxeroides line used in this study was developed through internode reproduction in a greenhouse for 2 years, further analysis of gene function in A. philoxeroides from other habitats is warranted because of the strong phenotypic plasticity of the species.
Virus-induced gene silencing is a valuable tool that can effectively silence individual genes or families of genes critical for plant development or resistance to biotic and abiotic stresses (George et al., 2015). This tool is a simple and rapid method for assessing gene function (Burch-Smith et al., 2004; Robertson, 2004; Becker and Lange, 2010; Cakir et al., 2010). A. philoxeroides can readily adapt to terrestrial and aquatic habitats. Moreover, the vegetative regeneration clones can exploit extremely diverse habitats, including dry lands, lakes, and high-salt areas, exhibiting notable morphological differences (Huai et al., 2003; Pan et al., 2006; Geng et al., 2007; Gao et al., 2010). Thus, this weed is able to colonize a wide range of habitats (Geng et al., 2006, 2007; Li and Ye, 2006; Pan et al., 2006; Wu et al., 2016a,b). In this study, we dissected the function of the drought-related gene ApDRI15 and the salinity-related gene ApSI1 using the VIGS method. ApDRI15 knockdown significantly increased susceptibility of A. philoxeroides plants to drought stresses, resulting in lower biomass, smaller leaves, shorter internodes, higher water loss rates, lower relative water content, and lower proline level compared to the control plants. When the ApSI1 gene was silenced in plants, the silenced plants withered, grew pale, and had lower biomass compared to the control plants. Therefore, the PVX VIGS system can be used to silence A. philoxeroides genes to determine their function, thereby elucidating the molecular mechanisms of invasion and colonization.
In this study, A. philoxeroides DRI15 were isolated for analyzing its function against drought stress. In NCBI database, A. philoxeroides DRI15 gene had been previously submitted, and annotated as drought-induced gene. We also further confirmed that the DRI15 expression was response to drought stress by qPCR analysis (Supplementary Figure S3). However, DRI15 has not been identified in other plants. The structural ortholog of A. philoxeroides DRI15 putative protein is highly similar with ribosomal protein s3a and s1-a (identification of 93.1 and 91.8% at protein level, respectively, Supplementary Data Sheet S2). The ribosomal protein served as a multi-functional protein interacted with p53, which was associated with cellular stress in human (Goodin and Rutherford, 2002; Zhan, 2005; Yadavilli et al., 2009). Thereby, the function of DRI15 in plants should be extensively explored in drought-tolerance.
We selected a salt-induced gene of A. philoxeroides, SI1, for evaluating its function by VIGS method. Up-to-now, there were many researches involved in salt-induced genes. For instance, Salicornia brachiata SI-1 and SI-2 genes were overexpressed in tobacco, conferring salinity tolerance (Yadav et al., 2014; Kumari et al., 2017). A salt-responsive gene in wheat, TaDi19A, was identified to play a vital role in the plant salt-tolerance (Li et al., 2010). In the present study, ApSI1 played important roles in A. philoxeroides plant salt-tolerance according to the results of silenced plants under salt treatment.
Conclusion
We have successfully developed a pgR107 VIGS approach in A. philoxeroides. By dissecting the role of the ApDRI15 and ApSI1 genes in the response of plants to the stress of drought and salinity, we have demonstrated that the PVX VIGS system is a simple and rapid method for assessing the role of individual genes and gene families in A. philoxeroides. This tool will be invaluable for revealing the potential molecular mechanisms of A. philoxeroides invasion and colonization and for developing measures to check weed invasion, control damage to agriculture, and protect ecological balance in invaded habits.
Author Contributions
J-HW and G-LZ designed the experimental strategy, analyzed the data, and drafted the manuscript. CB and W-DF edited the manuscript. CB and PW cloned the A. philoxeroides PDS gene, created the ApDRI15 and ApSI1 constructs, and set up the pgR107 VIGS system. QF measured the water content. LW conducted the transpirational water loss assay. Z-NZ carried out the chlorophyll content assay. ZS measured and calculated the biomass. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant from National Key Research and Development Project (2016YFSF03) and the State Key Laboratory of Cotton Biology Open Fund (CB2017B04).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01579/full#supplementary-material
FIGURE S1 | Alignment of several nucleotide sequences of the PDS gene from diverse plants in the NCBI database.
FIGURE S2 | The photo-bleaching phenotype. The control was plants agro-infiltrated with the empty vector in (A–E) indicate the ApPDS-silenced plants, exhibiting photo-bleaching.
FIGURE S3 | The relative expression level of ApDRI15 in normal plants under the control (normal-watering) and drought treatment. Error bars represent the standard derivation of three biological replicates (n = 45).
DATA SHEET S1 | The partial sequence of ApPDS including 936bp.
DATA SHEET S2 | The alignment of structural ortholog of Alternanthera philoxeroides DRI15 putative protein in Phyre2 database.
References
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts, polyphenoxidase in beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Bates, L., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water stress studies. Plant Soil 39, 205–207. doi: 10.1016/j.dental.2010.07.006
Becker, A., and Lange, M. (2010). VIGS-genomics goes functional. Trends Plant Sci. 15, 1–4. doi: 10.1016/j.tplants.2009.09.002
Burch-Smith, T. M., Anderson, J. C., Martin, G. B., and Dinesh-Kumar, S. P. (2004). Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 39, 734–746. doi: 10.1111/j.1365-313X.2004.02158.x
Cakir, C., Gillespie, M. E., and Scofield, S. R. (2010). Rapid determination of gene function by virus-induced gene silencing in wheat and barley. Crop Sci. 50, 77–84. doi: 10.2135/cropsci2009.10.0567
Ding, X. S., Schneider, W. L., Chaluvadi, S. R., Mian, M. R., and Nelson, R. S. (2006). Characterization of a Brome mosaic virus strain and its use as a vector for gene silencing in monocotyledonous hosts. Mol. Plant Microbe Interact. 19, 1229–1239. doi: 10.1094/MPMI-19-1229
Dong, B. C., Alpert, P., Guo, W., and Yu, F. H. (2012). Effects of fragmentation on the survival and growth of the invasive, clonal plant Alternanthera philoxeroides. Biol Invasions 14, 1101–1110. doi: 10.1007/s10530-011-0141-5
Dong, B. C., Yu, G. L., Guo, W., Zhang, M. X., Dong, M., and Yu, F. H. (2010). How internode length, position and presence of leaves affect survival and growth of Alternanthera philoxeroides after fragmentation? Evol. Ecol. 24, 1447–1461. doi: 10.1007/s10682-010-9390-5
Duan, J., Zhang, M., Zhang, H., Xiong, H., Liu, P., Ali, J., et al. (2012). OsMIOX, a myoinositoloxygenase gene, improves drought tolerance through scavenging of reactive oxygen species in rice (Oryza Sativa L.). Plant Sci. 196, 143–151. doi: 10.1016/j.plantsci.2012.08.003
Faivre-Rampant, O., Gilroy, E. M., Hrubikova, K., Hein, I., Millam, S., Loake, G. J., et al. (2004). Potato virus X-induced gene silencing in leaves and tubers of potato. Plant Physiol. 134, 1308–1316. doi: 10.1104/pp.103.037507
Fan, S., Yu, D., and Liu, C. (2013). The invasive plant Alternanthera philoxeroides was suppressed more intensively than its native congener by a native generalist: implications for the biotic resistance hypothesis. PLOS ONE 8:e83619. doi: 10.1371/journal.pone.0083619
Gao, L., Geng, Y., Li, B., Chen, J., and Yang, J. (2010). Genome-wide DNA methylation alterations of Alternanthera philoxeroides in natural and manipulated habitats: implications for epigenetic regulation of rapid responses to environmental fluctuation and phenotypic variation. Plant Cell Environ. 33, 1820–1827. doi: 10.1111/j.1365-3040.2010.02186.x
Gao, L., Geng, Y., Yang, H., Hu, Y., and Yang, J. (2015). Gene expression reaction norms unravel the molecular and cellular processes underpinning the plastic phenotypes of Alternanthera Philoxeroides in contrasting hydrological conditions. Front. Plant Sci. 6:991. doi: 10.3389/fpls.2015.00991
Geng, Y. P., Pan, X. Y., Xu, C. Y., Zhang, W. J., Li, B., and Chen, J. K. (2006). Phenotypic plasticity of invasive Alternanthera philoxeroides in relation to different water availability, compared to its native congener. Acta Oecol. 30, 380–385. doi: 10.1016/j.actao.2006.07.002
Geng, Y. P., Pan, X. Y., Xu, C. Y., Zhang, W. J., Li, B., Chen, J.-K., et al. (2007). Phenotypic plasticity rather than locally adapted ecotypes allows the invasive alligator weed to colonize a wide range of habitats. Biol. Invasions 9, 245–256. doi: 10.1007/s10530-006-9029-1
George, G. M., Ruckle, M. E., and Lloyd, J. R. (2015). Virus-induced gene silencing as a scalable tool to study drought tolerance in Plants. Methods Mol. Biol. 1287, 243–253. doi: 10.1007/978-1-4939-2453-0_18
George, G. M., van der Merwe, M. J., Nunes-Nesi, A., Bauer, R., Fernie, A. R., Kossmann, J., et al. (2010). Virus-induced gene silencing of plastidial soluble inorganic pyrophosphatase impairs essential leaf anabolic pathways and reduces drought stress tolerance in Nicotiana benthamiana. Plant Physiol. 154, 55–66. doi: 10.1104/pp.110.157776
Goodin, J. L., and Rutherford, C. L. (2002). Characterization of human ribosomal S3a, gene expression during adenosine 3’:5’ cyclic monophosphate induced neuroendocrine differentiation of LNCaP cells – Regulation of S3a gene expression in LNCaP. Mol. Biol. Rep. 29, 301–316. doi: 10.1023/A:1020457400377
Holzberg, S., Brosio, P., Gross, C., and Pogue, G. P. (2002). Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 30, 315–327. doi: 10.1046/j.1365-313X.2002.01291.x
Huai, H., Jin, Y., Zhang, B., Cui, Y., Gao, H., and Wu, X. (2003). The diversity of habitats occupied by alien Alternanthera philoxeroides. J. Weed Sci. 1, 18–20.
Jada, B., Soitamo, A. J., and Lehto, K. (2013). Organ-specific alterations in tobacco transcriptome caused by the PVX-derived P25 silencing suppressor transgene. BMC Plant Biol. 13:8. doi: 10.1186/1471-2229-13-8
Jones, L., Hamilton, A. J., Voinnet, O., Thomas, C. L., Maule, A. J., and Baulcombe, D. C. (1999). RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11, 2291–2301. doi: 10.1105/tpc.11.12.2291
Kumari, J., Udawat, P., Dubey, A. K., Haque, M. I., Rathore, M. S., and Jha, B. (2017). Overexpression of SbSI-1, a nuclear protein from Salicornia brachiata confers drought, and salt stress tolerance, and maintains photosynthetic efficiency in transgenic tobacco. Front. Plant Sci. 8:1215. doi: 10.3389/fpls.2017.01215
Li, J., and Ye, W. H. (2006). Genetic diversity of alligator weed ecotypes is not the reason for their different responses to biological control. Aquat. Bot. 85, 155–158. doi: 10.1016/j.aquabot.2006.02.006
Li, Q., Zhang, X., Lv, Q., Zhu, D., Qiu, T., Xu, Y., et al. (2017). Physcomitrella patens dehydrins (PpDHNA and PpDHNC) confer salinity and drought tolerance to transgenic Arabidopsis plants. Front. Plant Sci. 8:1316. doi: 10.3389/fpls.2017.01316
Li, S., Xu, C., Yang, Y., and Xia, G. (2010). Functional analysis of TaDi19A, a salt-responsive gene in wheat. Plant Cell Environ. 33, 117–129. doi: 10.1111/j.1365-3040.2009.02063.x
Liou, M. R., Huang, Y. W., Hu, C. C., Lin, N. S., and Hsu, Y. H. (2014). A dual gene-silencing vector system for monocot and dicot plants. Plant Biotechnol. J. 12, 330–343. doi: 10.1111/pbi.12140
Liu, N., Xie, K., Jia, Q., Zhao, J., Chen, T., Li, H., et al. (2016). Foxtail mosaic virus-induced gene silencing in monocot plants. Plant Physiol. 171, 1801–1807. doi: 10.1104/pp.16.00010
Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. doi: 10.1046/j.1365-313X.2002.01297.x
Lu, R., Malcuit, I., Moffett, P., Ruiz, M. T., Peart, J., Wu, A. J., et al. (2003). High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 22, 5690–5699. doi: 10.1093/emboj/cdg546
Pan, X., Geng, Y., Zhang, W., Li, B., and Chen, J. (2006). The influence of abiotic stress and phenotypic plasticity on the distribution of invasive Alternanthera philoxeroides along a riparian zone. Acta Oecol. 30, 333–341. doi: 10.1016/j.actao.2006.03.003
Parida, A. K., Dasgaonkar, V. S., Phalak, M. S., Umalkar, G. V., and Aurangabadkar, L. P. (2007). Alterations in photosynthetic pigments, protein, and osmotic components in cotton genotypes subjected to short-term drought stress followed by recovery. Plant Biotechnol. Rep. 1, 37–48. doi: 10.1007/s11816-006-0004-1
Ramanna, H., Ding, X. S., and Nelson, R. S. (2013). Rationale for developing new virus vectors to analyze gene function in grasses through virus-induced gene silencing. Methods Mol. Biol. 975, 15–32. doi: 10.1007/978-1-62703-278-0_2
Ratcliff, F., Martin-Hernandez, A. M., and Baulcombe, D. C. (2001). Technical advance: tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245. doi: 10.1046/j.0960-7412.2000.00942.x
Robertson, D. (2004). VIGS vectors for gene silencing: many targets, many tools. Annu. Rev. Plant Biol. 55, 495–519. doi: 10.1146/annurev.arplant.55.031903.141803
Sainty, G., McCorkelle, G., and Julien, M. (1998). Control and spread of alligator weed Alternanthera philoxeroides (Mart.) Griseb., in Australia: lessons for other regions. Wetl. Ecol. Manag. 5, 195–201. doi: 10.1023/A:1008248921849
Schooler, S. S., Yeates, A. G., Wilson, J. R., and Julien, M. H. (2007). Herbivory, mowing, and herbicides differently affect production and nutrient allocation of Alternanthera philoxeroides. Aquat. Bot. 86, 62–68. doi: 10.1016/j.aquabot.2006.09.004
Turnage, M. A., Muangsan, N., Peele, C. G., and Robertson, D. (2002). Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J. 30, 107–114. doi: 10.1046/j.1365-313X.2002.01261.x
Wang, B., Li, W., and Wang, J. (2005). Genetic diversity of Alternanthera philoxeroides in China. Aquat. Bot. 81, 277–283. doi: 10.1016/j.aquabot.2005.01.004
Wang, N., Yu, F. H., Li, P. X., He, W. M., Liu, F. H., Liu, J. M., et al. (2008). Clonal integration affects growth, photosynthetic efficiency and biomass allocation, but not the competitive ability, of the alien invasive Alternanthera philoxeroides under severe stress. Ann. Bot. 101, 671–678. doi: 10.1093/aob/mcn005
Wang, N., Yu, F. H., Li, P. X., He, W. M., Liu, J., Yu, G. L., et al. (2009). Clonal integration supports the expansion from terrestrial to aquatic environments of the amphibious stoloniferous herb Alternanthera philoxeroides. Plant Biol. 11, 483–489. doi: 10.1111/j.1438-8677.2008.00133.x
Wang, R., Yang, X., Wang, N., Liu, X., Nelson, R. S., Li, W., et al. (2016). An efficient virus-induced gene silencing vector for maize functional genomics research. Plant J. 86, 102–115. doi: 10.1111/tpj.13142
Wu, H., Carrillo, J., and Ding, J. (2016a). Invasion by alligator weed, Alternanthera philoxeroides, is associated with decreased species diversity across the latitudinal gradient in China. J. Plant Ecol. 9, 311–319. doi: 10.1093/jpe/rtv060
Wu, H., Ismail, M., and Ding, J. (2016b). Global warming increases the interspecific competitiveness of the invasive plant alligator weed, Alternanthera philoxeroides. Sci. Total Environ. 575, 1415–1422. doi: 10.1016/j.scitotenv.2016.09.226
Yadav, N. S., Singh, V. K., Singh, D., and Jha, B. (2014). A novel gene SbSI-2 encoding nuclear protein from a halophyte confers abiotic stress tolerance in E. coli and tobacco. PLOS ONE 9:e101926. doi: 10.1371/journal.pone.0101926
Yadavilli, S., Mayo, L. D., Higgins, M., Lain, S., Hegde, V., and Deutsch, W. A. (2009). Ribosomal protein S3: a multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA Repair 8, 1215–1224. doi: 10.1016/j.dnarep.2009.07.003
Ye, W., Li, J., Cao, H., and Ge, X. (2003). Genetic uniformity of Alternanthera philoxeroides in South China. Weed Res. 43, 297–302. doi: 10.1046/j.1365-3180.2003.00346.x
You, W. H., Han, C. M., Fang, L. X., and Du, D. L. (2016). Propagule pressure, habitat conditions and clonal integration influence the establishment and growth of an invasive clonal plant, Alternanthera philoxeroides. Front. Plant Sci. 7:568. doi: 10.3389/fpls.2016.00568
Zhan, Q. (2005). Gadd45a, a p53- and BRCA1-regulated stress protein, in cellular response to DNA damage. Mutat. Res. 569, 133–143. doi: 10.1016/j.mrfmmm.2004.06.055
Keywords: Alternanthera philoxeroides, virus induced gene silencing (VIGS), drought stress, salinity stress, pgR107 VIGS vector
Citation: Bai C, Wang P, Fan Q, Fu W-D, Wang L, Zhang Z-N, Song Z, Zhang G-L and Wu J-H (2017) Analysis of the Role of the Drought-Induced Gene DRI15 and Salinity-Induced Gene SI1 in Alternanthera philoxeroides Plasticity Using a Virus-Based Gene Silencing Tool. Front. Plant Sci. 8:1579. doi: 10.3389/fpls.2017.01579
Received: 29 March 2017; Accepted: 29 August 2017;
Published: 12 September 2017.
Edited by:
Roger Deal, Emory University, United StatesReviewed by:
Gavin Mager George, Stellenbosch University, South AfricaYuhui Chen, Noble Research Institute, LLC, United States
Copyright © 2017 Bai, Wang, Fan, Fu, Wang, Zhang, Song, Zhang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia-He Wu, wujiahe@im.ac.cn Guo-Liang Zhang, zhangguoliang@caas.cn
†Co-first authors and have contributed equally to this work.
 Chao Bai
Chao Bai Peng Wang
Peng Wang Qiang Fan1
Qiang Fan1 Wei-Dong Fu
Wei-Dong Fu Le Wang
Le Wang Zhen Song
Zhen Song Guo-Liang Zhang
Guo-Liang Zhang Jia-He Wu
Jia-He Wu