- 1Future Food Laboratory, Innovation Center of Yangtze River Delta, Zhejiang University, Jiaxing, Zhejiang, China
- 2Department of Food Science and Nutrition, Zhejiang University, Hangzhou, Zhejiang, China
- 3Zhejiang Key Laboratory of Agri-food Resources and High-value Utilization, Zhejiang University, Hangzhou, Zhejiang, China
- 4College of Food Science and Technology, Shanghai Ocean University, Shanghai, China
Aquatic products are a crucial source of dietary protein, especially in regions with abundant marine resources. However, with the expansion of global trade, the risk of microbial contamination in these products has increased, leading to serious public health concerns due to extended transportation and varying regulatory standards. Foodborne illnesses associated with aquatic products not only impact consumer health but also result in significant economic losses due to reduced market confidence, brand damage, and costly recalls. This review systematically examines the role of traceability technologies in enhancing microbial safety in aquatic products. Emphasis is placed on the integration of genome sequencing, artificial intelligence, and digital monitoring systems within the traceability framework. The evaluation considers specific performance indicators, including detection sensitivity (for example, the minimum limit of detection for target pathogens), source attribution resolution (for example, ≤20 core-genome SNP differences or unique wgMLST allelic profiles), and time-to-result in outbreak scenarios, as well as accessibility for small-scale producers and scalability across diverse aquaculture environments. In particular, we outline how artificial intelligence can be integrated with genome sequencing. For instance, WGS-derived genomic fingerprints can be transformed into machine learning models for rapid and highly sensitive microbial source prediction, thereby enhancing real-time decision-making capability along the aquatic product supply chain. Traceability systems have proven effective in enabling real-time monitoring and rapid response to contamination events. Technologies such as genome sequencing and AI significantly enhance detection speed and accuracy, contributing to improved food safety management. Nonetheless, challenges remain, including technological barriers for small-scale producers, fragmented international standards, and low public awareness. To overcome these limitations, future efforts should focus on developing cost-effective and user-friendly traceability tools, promoting global standardization, strengthening regulatory frameworks, and increasing public engagement. Furthermore, innovative approaches involving big data analytics, and AI hold great promise for advancing microbial safety and ensuring the integrity of aquatic product supply chains.
1 Introduction
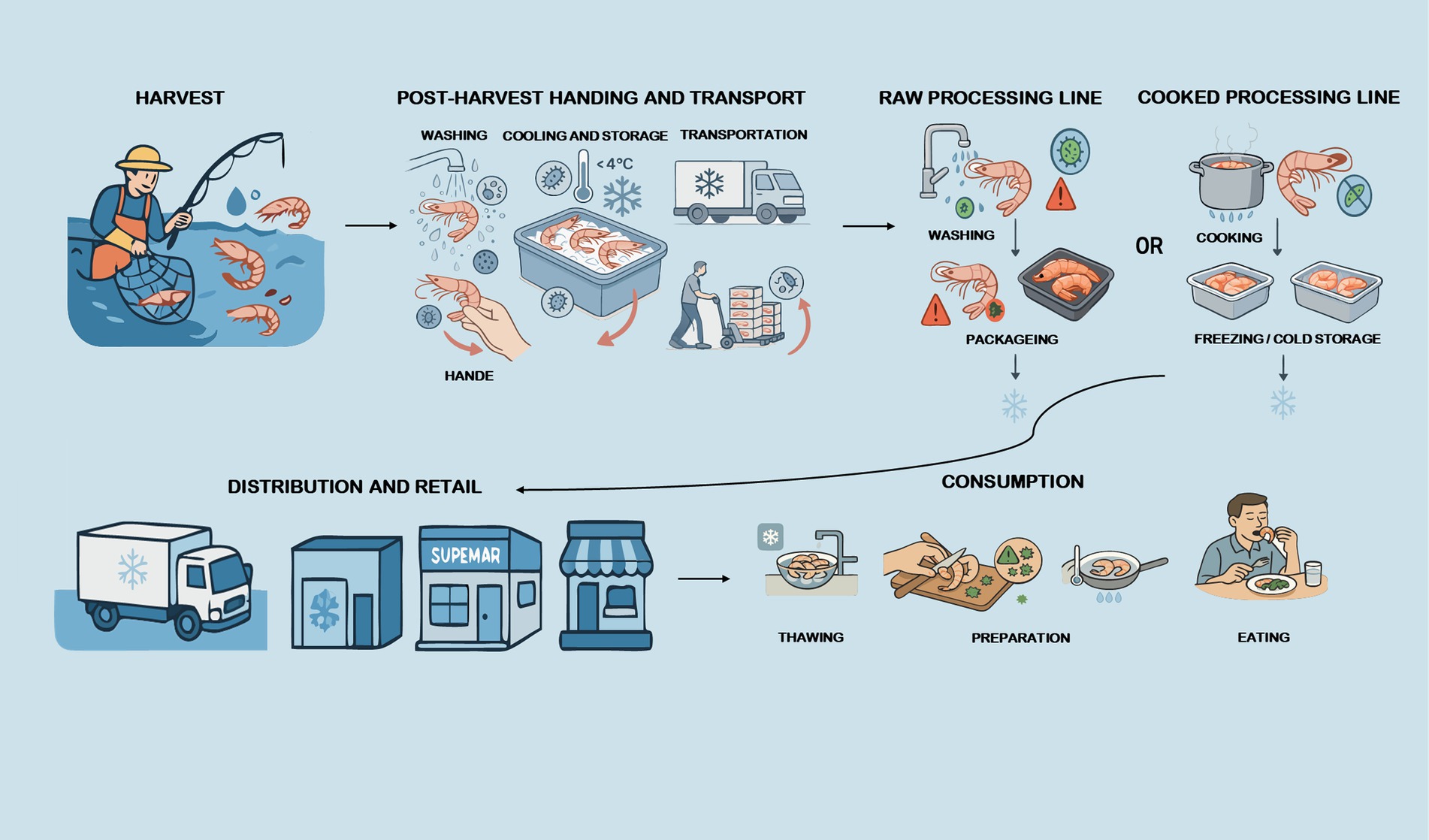
Aquatic products are rich in proteins, omega-3 fatty acids, vitamins and minerals, with good nutritional value, and they have become an important part of the global food market (1). According to the Food and Agriculture Organization of the United Nations, global per capita aquatic product consumption was 20.6 kg in 2021, more than double the average of 9.9 kg in the 1960s (2). Despite their nutritional benefits, aquatic products are uniquely susceptible to contamination by foodborne microorganisms along the supply chain (e.g., aquaculture, processing, distribution, retail, consumption), posing significant public health risks (Figure 1) (3). Common foodborne microorganisms in aquatic products include Salmonella, Vibrio parahaemolyticus, Vibrio vulnificus, norovirus, hepatitis A virus, etc. (4). These microorganisms pose significant public health risks due to their potential transmission from aquatic products to humans. For example, whole genome sequencing analyses have revealed high genomic similarity between isolates from aquatic foods and those from clinical cases, indicating possible direct transmission routes (5).
To mitigate these public health risks and interrupt the transmission chain, effective traceability systems are imperative for assuring the microbial safety of aquatic food chains (6). However, aquatic supply chains face numerous challenges, including fragmented production sites, variable environmental conditions (e.g., water temperature, salinity), and high perishability (7). Traditional approaches, which rely heavily on laboratory culture plate counts for microorganism detection, suffer from slow identification and response times, leading to delayed responses during outbreaks (8). Instead, proactive end-to-end traceability enables real-time monitoring of critical control points (e.g., harvest zones, processing facilities, and logistics), empowering stakeholders to identify, isolate, and mitigate contamination risks before products reach consumers (9).
In this review, we emphasize advancements and gaps in addressing microbial hazards across aquatic supply chains. We evaluated the emerging tools for their efficacy in microorganism tracking and supply chain digitization, and discussed barriers to implementation. By bridging interdisciplinary insights from microbiology, data science, and supply chain governance, this work aims to inform policymakers, industry leaders, and researchers in advancing safer and more transparent aquatic food systems.
2 Microbial hazards in aquatic products
2.1 Vibrio spp.
Vibrio spp. are a group of Gram-negative halophilic bacteria, widely distributed in the global ocean, estuarine and coastal waters (10, 11). Vibrio spp. exhibit the preference for salty environments and demonstrate facultative anaerobic capacities, enabling survival with or without oxygen (12). In an investigation by Ma et al. (13), it is reported that global prevalence of V. cholerae, V. parahaemolyticus and V. vulnificus in fish was 9.56, 24.77, and 5.29%, respectively. Aquatic products (e.g., shrimps, mussels, oysters) are the potential carriers of Vibrio spp. (14). Common Vibrio species detected in the aquatic foods include Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus (15). Epidemiological data from Australia indicates that a level of 97% of V. parahaemolyticus-associated enteric infection outbreaks nationwide were attributed by the consumption of oysters (16).
Cultural methods (e.g., ISO 21872-1:2017) with the use of Thiosulfate citrate bile salts sucrose (TCBS) medium remain as the gold standard for the detection of Vibrio spp. in the aquatic foods (Supplementary Table 1). Additionally, the molecular approaches (e.g., PCR, qPCR) are also widely employed for detecting the specific genes: ctxA gene for V. cholerae, ldh, tlh, tdh and trh gene for V. parahaemolyticus, vvh gene for V. vulnificus (17). However, PCR is unable to discriminate between non-viable and viable cells, potentially leading to overestimate of contamination loads. To overcome this constraint, qPCR approaches incorporating the viability-selective dyes, such as propidium monoazide (PMA) and ethidium monoazide (EMA), have been applied for the detection of total viable microorganisms (18).
2.2 Salmonella spp.
Salmonella spp., a facultative anaerobic gram-negative foodborne microorganism, often causes human infections through contaminated food (19). This bacterium can survive under low temperature and high salty condition, rendering the possibility of their persistence in aquatic foods (Supplementary Table 2) (20). According to the White-Kauffmann classification scheme, Salmonella spp. are classified into two species, S. bongori and S. enterica. This classification is based on the antigenic variations in the surface structures: lipopolysaccharide (LPS), flagella, and capsular polysaccharide. S. enterica includes six subspecies, enterica, salamae, arizonae, diarizonae, houtenae, and indica, with over 2,600 serovars (21). In aquatic products (such as fish, shellfish, shrimp, mollusks, etc.), Salmonella contamination can result from aquaculture waters (such as water contaminated with faeces or sewage), contaminated feed, cross-contamination during processing, or improper transportation and storage conditions (22). Salmonellosis is prone to lead to fever, diarrhea, vomiting and other gastroenteritis symptoms, especially in children, the elderly and immunocompromised people (23). In the study by Ferrari et al. (24), it is revealed that global Salmonella-related foodborne outbreaks related to fish consumption reach up to 12%. The study further demonstrated substantial geographic variation in Salmonella serovars distribution across aquatic food products. For instance, in Africa, S. enteritidis, S. Hadar, S. Kentucky and S. Blockley were found to be the most abundant serovars in aquatic food, whereas S. Newport emerged as the primary serovar in North America.
2.3 Listeria monocytogenes
Listeria monocytogenes, a resilient foodborne microorganism, poses a significant microbial hazard in aquatic food products (e.g., shellfish, fish) (Supplementary Table 3) (25). The consumption of food contaminated by L. monocytogenes could cause listeriosis with a range of symptoms including fever, nausea, vomiting, diarrhea, headache, etc. (25). The disease is especially dangerous for vulnerable populations including pregnant women, newborns, the elderly, and immunocompromised individuals, potentially causing septicemia, meningitis, or fetal loss (26). L. monocytogenes is highly tolerant to cold, high salt concentrations, and acidic environments, allowing it to survive and even multiply under refrigeration, making it particularly difficult to eliminate in aquatic food chains (27). Studies suggest that contamination sources for L. monocytogenes in aquatic foods may include cross-contamination during processing, environmental exposure, or contact with contaminated ice used for refrigeration (28). It is estimated that the global incidence of Listeria monocytogenes in aquatic foods reached 11% (29).
2.4 Pseudomonas spp.
Pseudomonas spp. are gram-negative, psychrophilic, facultative anaerobic bacteria, one of the most common spoiling microorganisms in aquatic foods, such as fish, shellfish, crustaceans and refrigerated processed products (Supplementary Table 4) (30). Although Pseudomonas spp. do not often directly cause foodborne diseases, as a typical spoilage bacterium, it can lead to rapid spoilage of aquatic products by decomposing proteins and lipids, significantly shortening the shelf life, and may indirectly affect food safety (31).
2.5 Norovirus
Norovirus is a common cause of gastroenteritis and is widely found in the natural environment (Supplementary Table 5) (32). Noroviruses are non-enveloped viruses with a single-stranded positive-stranded RNA genome, which belongs to the family Caliciviridae (33). HuNoVs can be divided into the genogroups of GI-GX. They are spread through contaminated food, water or contact. Aquatic products are the potential carriers of norovirus (34). Particularly during aquaculture and harvesting, viruses can enter aquatic products through contaminated water sources or unsanitary handling, increasing the risk of human infection. In the study by Li et al. (35), the prevalence of human noroviruses in shellfish was detected as 29% around the world. Symptoms of norovirus infection include nausea, vomiting, diarrhea and abdominal pain, and while usually self-limiting, the health risks are greater for the elderly, the immunocompromised and children (36).
2.6 Hepatitis viruses
Hepatitis viruses, particularly hepatitis A (HAV) and hepatitis E (HEV), are significant microorganisms transmitted through the fecal-oral route (37). Both HAV (the Picornaviridae family) and HEV (the Hepeviridae family) are small, non-enveloped RNA viruses (38). HAV is classified into six genotypes (I, II, III, IV, V, VI) and HEV mainly contains eight genotypes (39). Aquatic products, especially shellfish such as oysters, clams, and mussels, may accumulate these viruses if harvested from contaminated waters or cross-contamination during distribution, processing or retail (Supplementary Table 6). According to a comprehensive literature review, foodborne transmission of HAV is frequently associated with contaminated shellfish and fresh produce, while HEV is increasingly linked to undercooked pork products (40). Acute hepatitis caused by HAV or HEV infection typically presents with symptoms such as fatigue, loss of appetite, nausea, vomiting, and jaundice (38). It is important to note that conventional cooking methods may not always completely inactivate these viruses, particularly HEV (41). It was found that it was only 50% inactivated at 56 °C for 1 h and 96% at 60 °C for 1 h (41).
3 Contamination points along the supply chain of aquatic products
3.1 Aquaculture
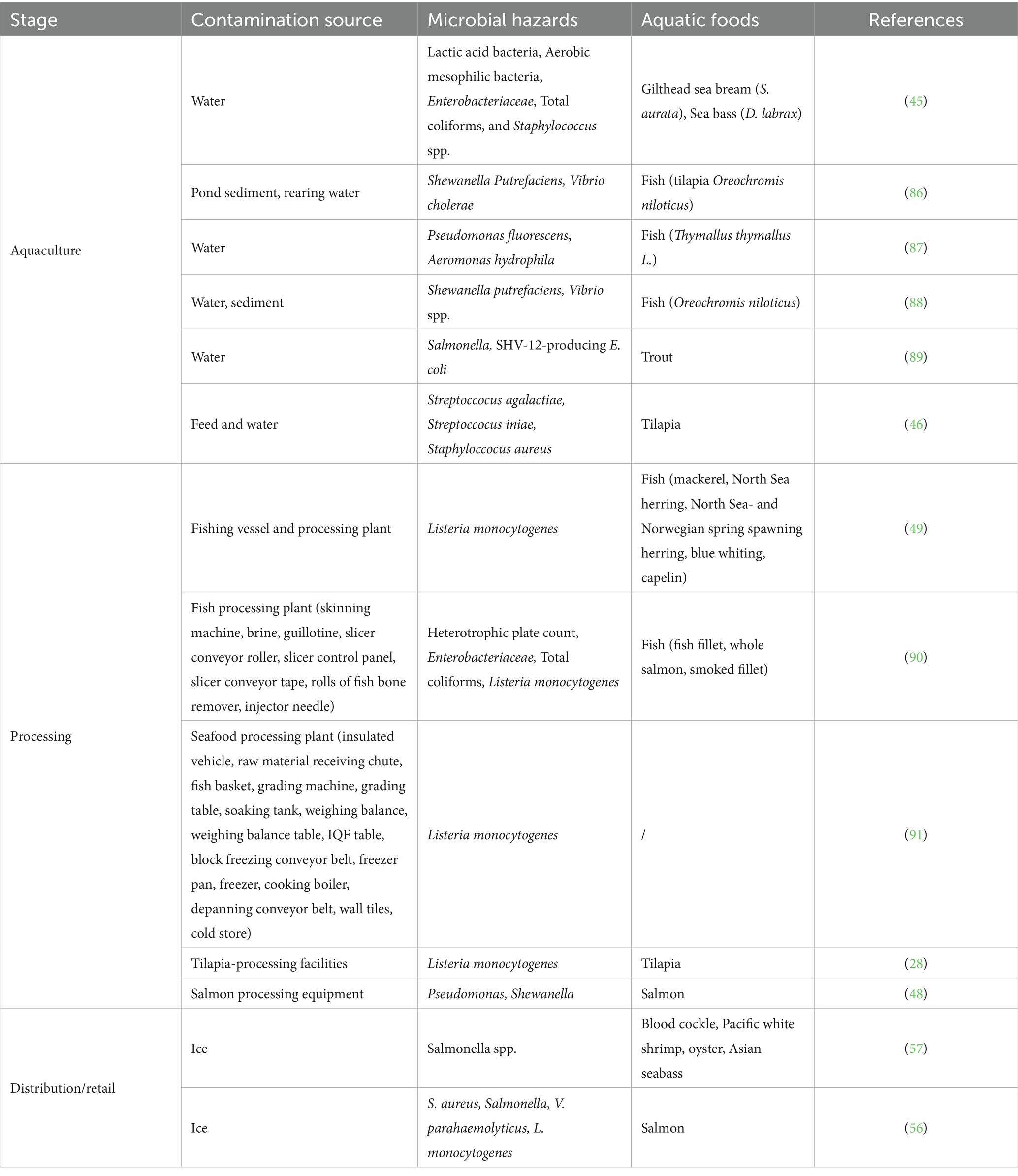
Water is one of the potential microbial contamination sources in aquaculture (42). The quality of aquaculture water (e.g., microbial contamination levels, water temperature, salinity) contributes to the microbial contamination of aquatic products. For instance, Flannery et al. (43) investigated the norovirus contamination of oysters cultivated at a wastewater treatment plant (WWTP) outfall, where oysters were initially free from microbial contamination. Their findings revealed a significant correlation between norovirus levels in oysters and concentrations in effluent wastewater, highlighting water as a critical vector for contamination. Similarly, a study in the coastal oyster breeding farms and fishing ports detected adenovirus (AdVs) and norovirus (NoVs) in water samples at rates up to 40.6 and 11.1%, respectively (44). These viruses, linked to direct discharges of domestic sewage, livestock wastewater, and fishing market effluents into coastal waters, were bioaccumulated by shellfish, posing significant food safety risks. Notably, lower water temperatures were associated with higher detection rates of AdVs and NoVs, particularly during winter months, likely due to enhanced viral stability in colder conditions. Additionally, in the study by Correia Peres Costa et al. (45), it was found that the contamination of lactic acid bacteria, aerobic mesophilic bacteria, Enterobacteriaceae, and total coliforms in fish was highly associated with the microbial levels of harvesting water.
Besides aquaculture water, feed is also an important source of contamination, especially when it is improperly stored or comes from unqualified suppliers and can carry microorganisms (46). Furthermore, a study conducted in Ghana indicates that Escherichia coli, Acinetobacter spp., and Citrobacter spp. are frequently detected in fish feed, which can cause diseases in aquatic animals and affect human food safety through the food chain (47). Furthermore, a study conducted in Ghana found a significant positive correlation between the prevalence of bacterial pathogens in the feed and their detection rate in diseased fish tissues (46). Therefore, in the full-chain traceability of aquatic products, tracking and controlling microorganisms in feed are crucial steps to ensure aquatic product safety.
3.2 Processing
Cross-contamination is a common risk in the processing of aquatic products. Common sources of contamination during processing include direct contact surfaces for aquatic products, non-direct contact surfaces, processing personnel, and physical/chemical treatment stages. When raw food and cooked food processing facilities are not strictly separated, or the tools and equipment used are not cleaned and disinfected in a timely manner, microorganisms can easily spread from raw water products to cooked food or other foods (48). According to Svanevik et al. (49), pre-capture analysis of fishing vessels revealed contamination of sift boxes, sorting chambers, and pipelines, likely due to exposure to seawater or seabird droppings, with Escherichia coli detected in the water samples of refrigerated seawater tank before and after capture, causing fish cross-contamination from poor-quality seawater near sewage outlets. It is also found that over 68% of factory water samples tested positive for enterococci (0.6–2.2 log CFU/100 mL), with landing tanks, washing tanks, and sorting machines exceeding 1.8 log CFU/100 mL. Listeria monocytogenes was found in fish and contact points (fishing gear, RSW tanks, conveyor belts, and water samples), indicating potential contamination transfer from vessels to factories. In addition, Møretrø et al. (48) found that industrially processed salmon fillets exhibited higher levels of Pseudomonas compared to manually gutted/sliced fish. The dominant Pseudomonas sequence types (based on partial 16S rRNA gene) in fish fillets were also prevalent in isolates from equipment post-cleaning, indicating a net transfer of Pseudomonas from processing equipment to fillets. These strains colonizing factories likely originate from live salmon or seawater. Pseudomonas species are well-adapted to food processing environments, dominating in factories even after cleaning, due to their ability to grow at low temperatures, minimal growth requirements, resistance to bactericidal agents, and propensity to form biofilms, enhancing their survival and colonization in processing plants. Therefore, maintaining good hygiene in processing plants is crucial to ensure the microbiological safety of aquatic food.
In addition to food contact surfaces, non-food-contact areas such as walls, floors, drainage systems, and sewers in the factories can also serve as sources of microbial contamination for aquatic foods during processing (50). Berrang and Frank (51) demonstrated that when a high-pressure water jet strikes the drain wall, residual Listeria innocua cells are instantaneously aerosolized into the facility’s air, where those bacteria can disperse horizontally up to 4 meters and vertically up to 2.4 meters. These microscopic droplets disperse through turbulent mixing in the air and then, under the combined effects of gravity and aerodynamic forces, rapidly settle onto equipment, worktables, and exposed seafood, driving cross-contamination. Aerosol generation is driven by the shear forces from the water impact dislodging droplets from the biofilm layer on the drain surface; notably, even in the absence of standing water, a residual biofilm alone can produce substantial aerosols upon impact. While most droplets settle within minutes, this brief period is sufficient for extensive dispersion and deposition. Therefore, during cleaning operations, direct high-pressure spraying of drains or outlets should be avoided, and integrated measures such as optimized drain design, controlled ventilation and routine monitoring and sanitation should be implemented to minimize the risk of aerosol-mediated cross-contamination. Furthermore, temperature control during processing is also crucial. Improper temperature treatment (such as cold chain breaks or substandard processing temperatures) can cause microorganisms to survive and grow on aquatic products. A study on the impact of changes in cold chain conditions for fish pointed out that improper temperature control significantly affects the microbial composition and greatly reduces the quality and shelf life of the fish meat (52).
3.3 Distribution/retail
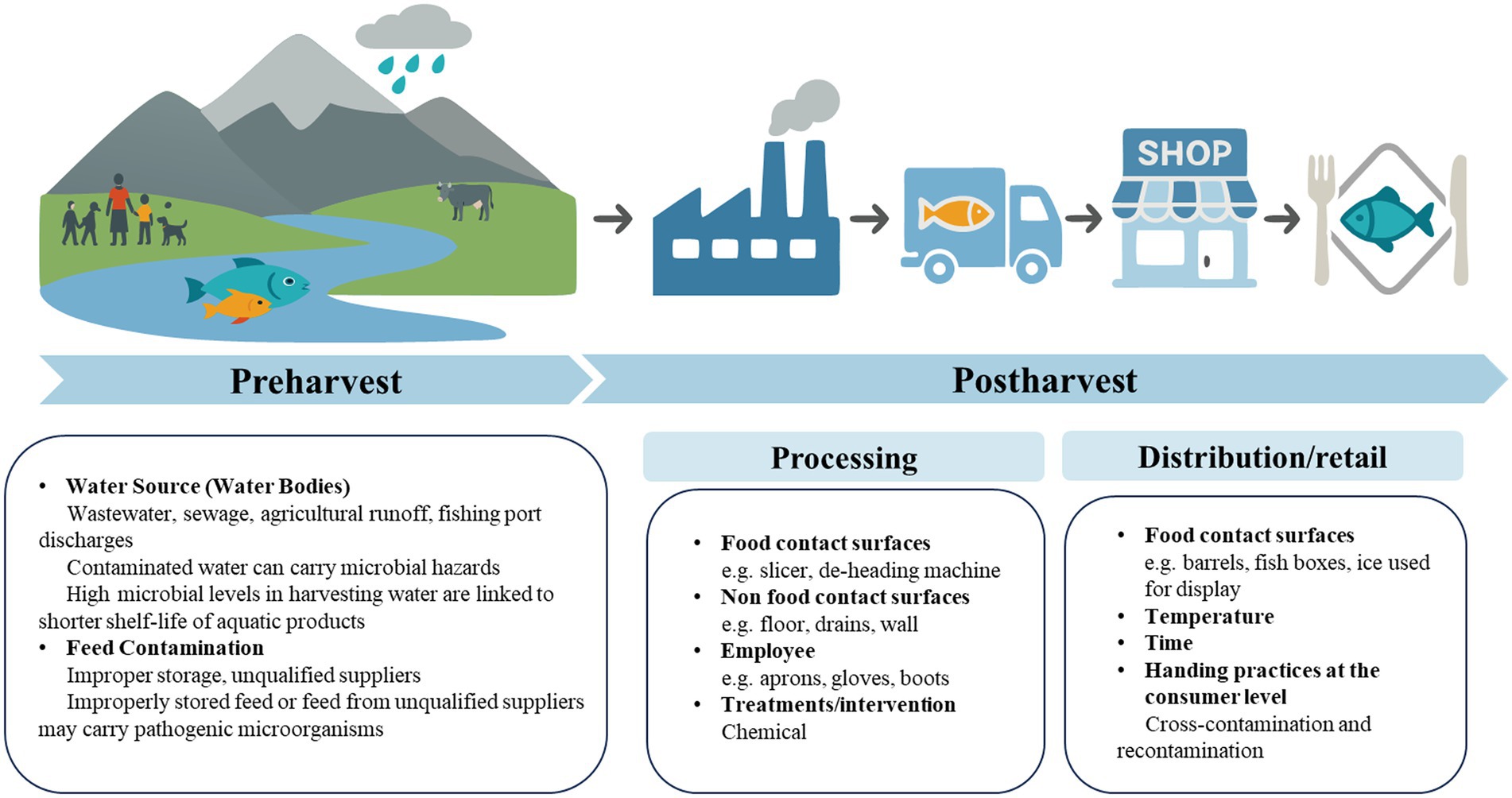
Distribution and retail are also one of the major points highly associated with the microbial contamination along the aquatic supply chain (53) (Figure 2; Table 1). Cold chain transportation is crucial for maintaining the freshness of aquatic products, and if the cold chain is disrupted during transportation or storage, it can cause temperature rises, promoting microbial growth (54). European Food Safety Authority highlighted that transport conditions critically influenced microbial contamination risk in aquatic products. Under abuse scenarios (e.g., temperature fluctuations or delayed icing), fish tubs (triple-layer polyethylene containers with freshwater and ice) significantly reduced bacterial growth during extended storage (3–5 days) by minimizing fish temperature increases compared to fish boxes (high-density polyethylene/HDPE containers with layered fish and ice), widely used by the industry (55). To mitigate contamination during the transportation and storage, the study mandated using pre-cooled, intact containers with lids, maintaining near-0 °C water fully covering aquatic products, ensuring complete ice coverage over the water surface with timely replenishment, limiting storage to ≤5 days, and minimizing physical stress. Consistent temperature control via water circulation and cool environments is also essential for the vessels of storage and transportation. Besides the vessels, ice used in distribution or retail of aquatic product may sever as a source of microbial contamination, potentially transfer microbial cells to the products through cross-contamination (56). Once contaminated, the ice can act as both a reservoir and a vector, enabling microbial transfer during the storage, transportation, or retail display of aquatic products. Inadequate ice hygiene or repeated use of recycled ice further amplifies the risk. The study conducted by Atwill and Jeamsripong (57) reported that aquatic foods displayed on ice exhibited 1.7-fold higher prevalence of Salmonella contamination compared to those not stored on ice (p < 0.0001).
Additionally, microbial contamination during the retail stage of the aquatic food chain is influenced by personal hygiene of the handlers, packaging materials, and environmental conditions (58). For example, a study on edible bivalve shellfish found that the contamination rate of hepatitis A virus (HAV) was higher in samples from retail markets (6.1%) compared to those from aquaculture farms (1.8%) (59), highlighting the retail stage as a critical point for microbial hazards. In open-air wet markets, inadequate temperature control and minimal packaging easily lead to microbial growth. For instance, one study reported a 97% detection rate of Escherichia coli (E. coli) in live fish displayed at open-air wet markets, with an average concentration of 3.0 log CFU/g. And 58% of samples contained extended-spectrum beta-lactamase (ESBL)-producing E. coli, with an average concentration of 2.3 log CFU/g, and 28% tested positive for Salmonella. In contrast, modern supermarket, which employ cold chain systems, pre-packaging, and standardized protocols, exhibit lower contaminationlevels: 71% for E. coli (1.6 log CFU/g), 8% for ESBL-producing E. coli (1.6 log CFU/g), and 8% for Salmonella (60). These data indicated retail markets as critical high-risk points for microbial contamination, highlighting the need for improved hygiene practices to reduce microbial hazards along the aquatic food chain (61).
4 Traceability technologies in the aquatic supply chain
4.1 Conventional methods
Culture-dependent methods, as the gold standards, are widely used in the field of food safety, for the identification or counting of microorganisms along the aquatic supply chain (Supplementary Table 6) (62). By using selective media, specific bacteria (such as Salmonella spp., Vibrio spp., etc.) in aquatic products can be effectively isolated and identified (17). Generally, culture-dependent methods include the steps of sample preparation, enrichment, dilution, plating, enumeration, and isolation of single species colonies for further characterization (63). However, traditional culture methods often take a longer time to obtain results and do not provide real-time data.
4.2 Emerging technologies
With the advancement of technology, the emerging traceability technology provides a more efficient and accurate solution for the traceability of aquatic microorganisms, which can monitor and analyze the transmission path of microorganisms in real time.
4.2.1 Molecular methods
Polymerase Chain Reaction (PCR) technology has been widely applied in the microbial traceability of aquatic products (Table 2) (64). Among its various forms, multiplex PCR enables the simultaneous amplification of multiple DNA fragments, thereby facilitating the detection of diverse microorganisms in aquaculture environments. Real-time PCR, when combined with high-resolution melting (HRM) curve analysis, supports high-throughput source tracking and can be integrated with production data to accurately identify contamination sources. This technique can also be integrated with production data to accurately pinpoint sources of contamination. To further enhance sensitivity and specificity, many studies have employed a two-round nested RT-PCR approach, which involves an initial RT-PCR followed by a second PCR using internal primers. For example, nested RT-PCR has been used to screen for hepatitis A virus (HAV) in mussels from Southern Italy, with the presence of infectious particles later confirmed through cell culture and RT-PCR (Supplementary Table 5) (65). In Galicia, a broad-spectrum nested RT-PCR based on Erker’s method and targeting the HEV ORF2 region was used to genotype HEV-positive samples (66). In addition, RT-booster-PCR has been developed to enhance detection sensitivity. This method involves a second amplification round using the same primers as those used in conventional RT-PCR. RT-booster-PCR has been successfully applied in the detection of norovirus, proving especially valuable as a complementary tool to RT-PCR when dealing with samples containing low viral loads or complex matrices (67). Due to its rapidity, sensitivity, and specificity, PCR technology holds significant promise for microorganism identification and contamination source tracing in the aquaculture industry.
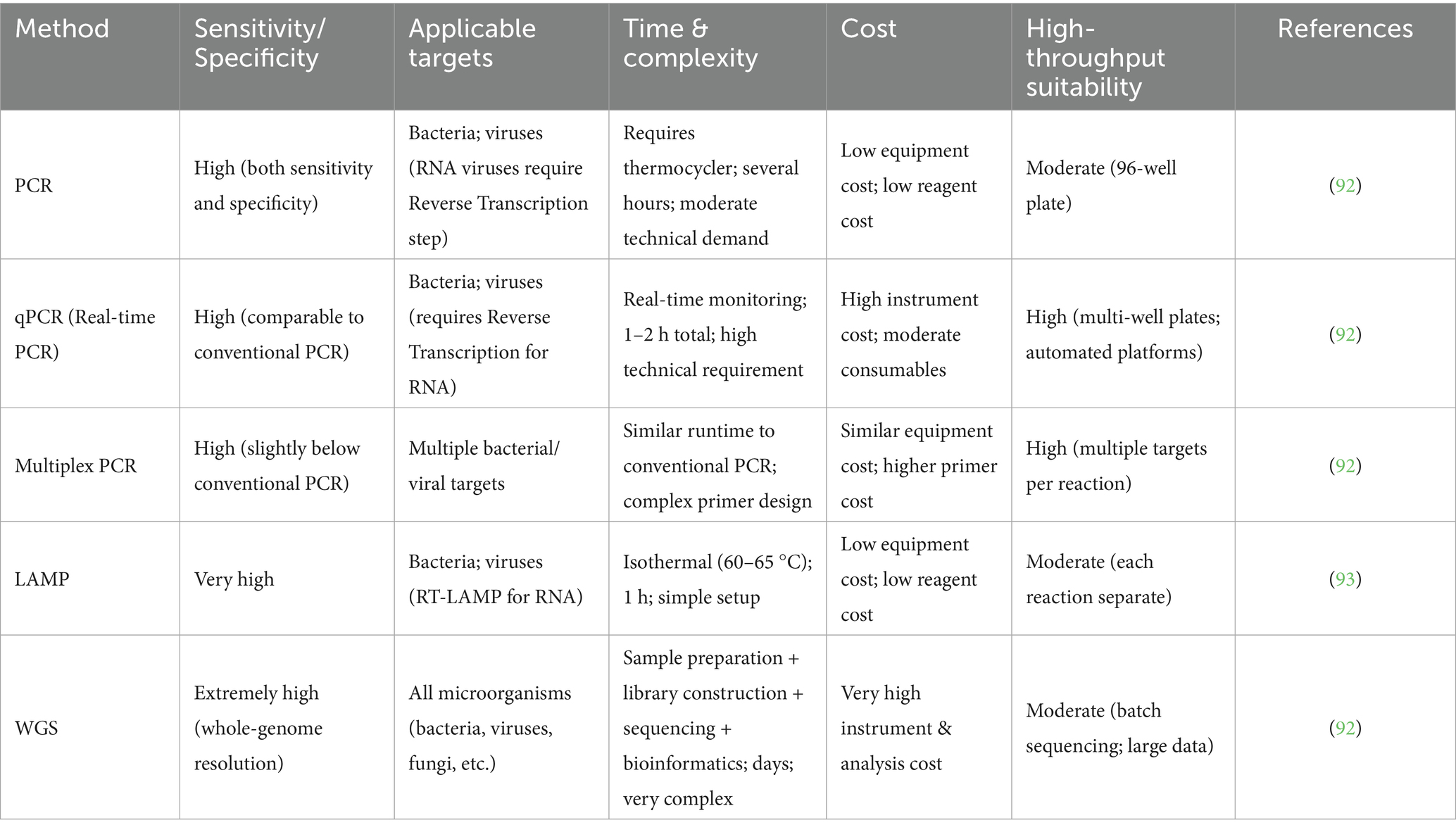
Table 2. Comparisons of molecular detection methods for microorganisms along the aquatic supply chain.
Loop-mediated isothermal amplification (LAMP) represents a highly sensitive and specific approach for the detection of microorganisms in aquatic products (68). Through the utilization of a meticulously designed set of primers, LAMP enables the amplification of target microorganism nucleic acid sequences under a constant temperature condition. This allows for the accurate identification of multiple food - borne microorganisms present in fish, shellfish, and other aquatic product items. Compared with conventional techniques, LAMP stands out for its rapid turnaround, streamlined workflow and true on-site applicability, offering great potential to become an indispensable tool for microbial surveillance and traceability in the aquaculture industry. While LAMP is rapid, highly sensitive, and suitable for on-site detection, it requires complex primer design and may be prone to non-specific amplification. Future development could focus on improving multiplexing capability and integration with portable devices to enhance its applicability in diverse aquatic supply chain settings.
Whole genome sequencing (WGS) is increasingly employed as a culture-independent method for microbial traceability in aquatic products (69). Unlike traditional approaches such as 16S rRNA gene sequencing, which are limited in resolution due to targeting a single gene, WGS enables comprehensive analysis by examining the entire genome of microbial hazards. A typical WGS workflow integrates several bioinformatics tools: Fastp for read trimming and quality filtering; FastQC and MultiQC for quality assessment; Kraken2 for contamination detection; and SPAdes or Shovill for de novo genome assembly. QUAST is used to evaluate assembly quality, while Prokka performs genome annotation. Downstream comparative genomics analyses often involve tools such as MLST and cgMLST for typing, Snippy for single nucleotide polymorphism (SNP) detection, Roary for pan-genome analysis, and Mash for estimating genomic distances. These analyses facilitate the assessment of genetic relatedness among isolates for source attribution along the supply chain of aquatic products (70).
To infer evolutionary relationships, phylogenetic trees are constructed based on aligned core genome sequences. Maximum likelihood methods-implemented in tools like IQ-TREE, RAxML, and PhyML-provide robust phylogenetic inference under explicit evolutionary models. IQ-TREE, in particular, offers efficient and accurate tree estimation with integrated model selection and ultrafast bootstrap (UFBoot) support (71). For large datasets, approximate Maximum likelihood algorithms like FastTree or distance-based methods such as RapidNJ offer rapid generation of initial tree topologies. The reliability of phylogenetic inference critically depends on the appropriate choice of substitution models (e.g., GTR) and accounting for rate heterogeneity (e.g., using a gamma distribution). Source attribution and transmission pathway analyses combine genomic data with epidemiological metadata, including sampling time, geographic location, and exposure history (70). Isolates with high genomic similarity, typically clustering in the same monophyletic group and differing by no more than 20 core genome SNPs with strong bootstrap support, are considered to originate from a common source (70). In contrast, differences exceeding 100 SNPs generally indicated unrelated sources. For example, incorporating temporal and spatial metadata allows for the reconstruction of transmission chains, especially when early food or environmental isolates closely match clinical cases that emerge later (72). Based on WGS, some countries and regions have established public health networks for enhanced the surveillance of foodborne microorganism along the supply chain. Programs such as the FDA’s GenomeTrakr network highlight the power of WGS-based surveillance to rapidly identify contamination sources and support timely public health responses (72). Similarly, in the United States, PulseNet, led by the Centers for Disease Control and Prevention (CDC), is a molecular subtyping network for foodborne pathogens. Initially based on pulsed-field gel electrophoresis (PFGE), PulseNet has evolved to incorporate WGS, enabling standardized data sharing and real-time collaboration across laboratories to facilitate rapid outbreak detection and source tracing (73). In China, a national foodborne disease molecular tracing network (TraNet) has been established to address foodborne diseases (74). The establishment of this network provides strong technical support for identifying the causes and accurately tracing the source of foodborne disease outbreaks across regions and even internationally. However, WGS, while offering unmatched resolution for microbial source attribution, remains limited in routine aquatic product monitoring by high costs, data processing requirements, and the need for specialized bioinformatics expertise. Advances in portable sequencing platforms and automated analysis pipelines may broaden its accessibility and facilitate near real-time source tracking.
4.2.2 Biomarkers for aquatic product microbiology traceability
Microbial DNA profiling serves as a direct biological fingerprint for tracing the origin of microbiological communities on aquatic products. By characterizing the unique composition of a microbial community, a field known as microbial biogeography, this method can link a sample to a specific geographic region based on its distinctive microbial signature (75). Concurrently, stable isotope analysis of the host organism acts as a robust geochemical tracer, determining the geographic origin of the aquatic product itself (76). By revealing where the host fish lived and foraged, this analysis provides a powerful environmental context, or a geographical proxy, for the microbial community it harbors.
The advancement presented herein lies in the synergistic combination of microbial DNA profiling and host isotopic geolocation, which collectively enhance the precision of origin assignment within a unified analytical framework. By employing an advanced analytical framework, such as a Bayesian model, the probabilistic origin assignment from the microbial DNA (the direct biological evidence) can be powerfully refined using the geographical origin data from the host’s isotopes (the environmental context) (Figure 3). This dual-layered approach transforms two separate analyses into a single, high-confidence verification system. The result is a multidimensional and exceptionally robust traceability framework, capable of pinpointing the geographic source of microbial communities on aquatic products with unprecedented precision.
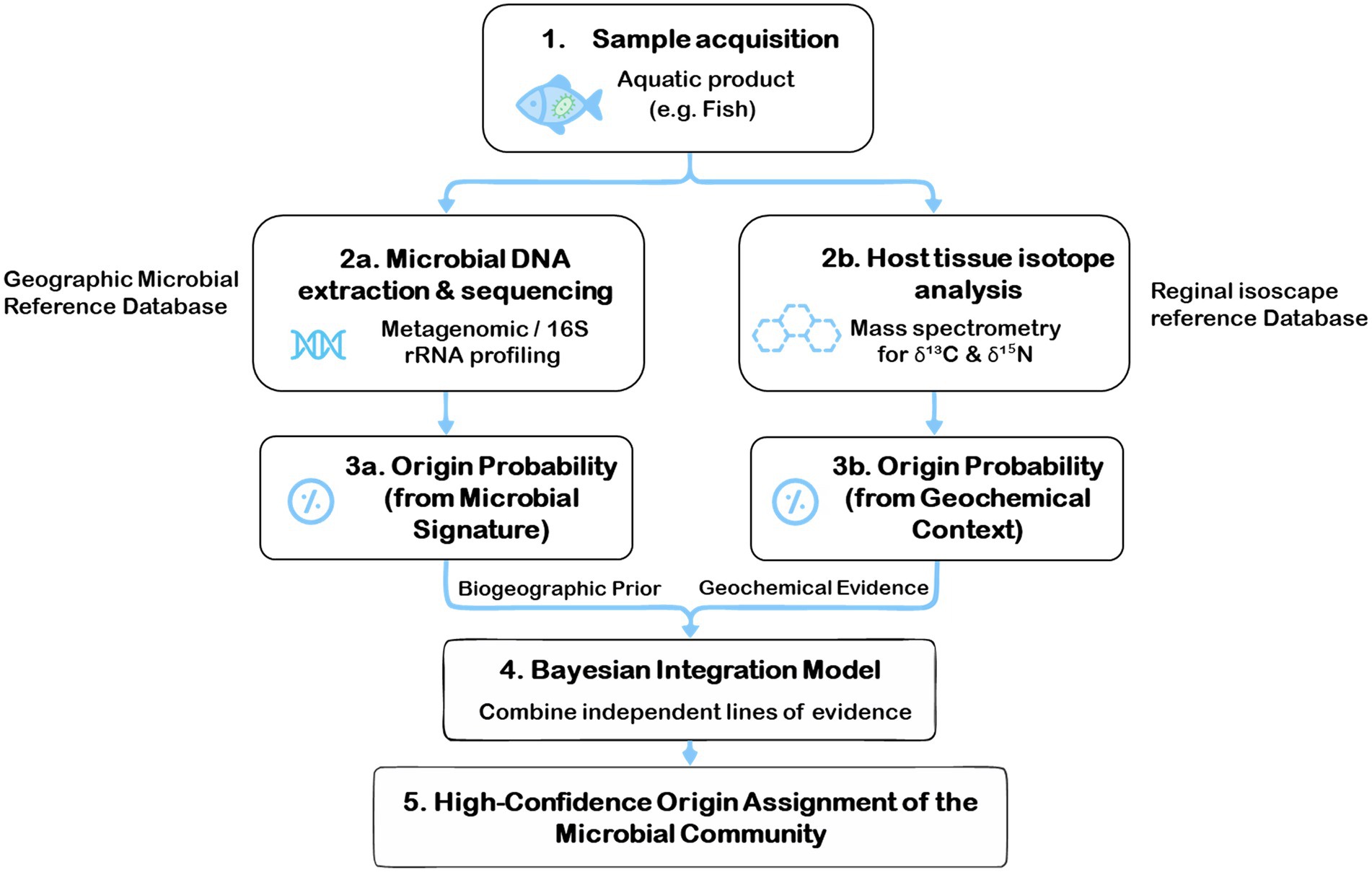
Figure 3. A framework for tracing the origin of microbial communities in the aquatic supply chain (75, 76).
4.2.3 Applications of AI in microbial source tracking of aquatic products
In recent years, the integration of artificial intelligence (AI) and big data technologies into microbial source tracking has shown immense promise, particularly for ensuring the safety of aquatic products. AI methods, especially machine learning (ML), can simulate human intelligence to analyze large-scale genomic and microbiome datasets, thereby significantly enhancing the speed and accuracy of contamination source identification.
The primary application of AI in the source tracking of aquatic products involves direct tracing based on the inherent genetic information or community structures of microorganisms, which act as unique “identity fingerprints.” When tracing specific microbial contamination events along the supply chain, WGS provides forensic-level precision. For instance, U.S. agencies including the Centers for Disease Control and Prevention (CDC), the Food and Drug Administration (FDA), and the Department of Agriculture (USDA) have co-developed machine learning models trained on WGS data to accurately estimate the sources of Salmonella infections, achieving remarkable accuracy (77). The core workflow of this approach involves creating standardized whole-genome multilocus sequence typing (wgMLST) allelic profiles from bacterial WGS data to serve as ‘genomic fingerprints’. A Random Forest machine learning model is then trained on a reference database of these fingerprints from known origins. Finally, the fingerprint of an unknown isolate is fed into this trained model to generate a probability report pinpointing its most likely source.
Beyond tracking a single microorganism, AI can leverage an entire microbial community as a “natural barcode” to verify the geographic origin of aquatic products. This capability is crucial for ensuring supply chain integrity and safety, as it helps confirm that a product is genuinely from its stated source. In one study, researchers combined 16S rRNA gene metabarcoding with supervised machine learning algorithms to successfully trace the geographic origin of aquatic products (78). This approach begins by sequencing the V3-V4 hypervariable region of the bacterial 16S rRNA gene from two clam tissues, the gills and digestive glands, to construct Amplicon Sequence Variant (ASV) profiles that represent the microbial community composition. As a key step, these ASV profiles are converted into binary “presence/absence” matrices to serve as input features for machine learning. Specific models were trained for different traceability objectives: a Bagging-enhanced multinomial logistic regression for provenance tracking (a multi-class task) and a Random Forest to distinguish between legal and illegal harvesting (a binary-class task). To ensure reliability, all models were trained using sample data from the second year and validated on a temporally independent dataset from the first year. Finally, a “consensus model” was created by fusing the prediction probabilities from the separate models for gills and digestive glands to further improve the final traceability accuracy.
As microbiome datasets grow in scale and dimensionality, the performance of traditional machine learning methods can face bottlenecks. To address this challenge, researchers have introduced advanced deep learning frameworks like the Ontology-aware Neural Network for Microbiome Source Tracking (ONN4MST) (79). The method’s workflow begins by preparing the input data: microbial community abundance profiles paired with a predefined hierarchical map of sample environments, known as a biome ontology. A crucial subsequent step employs a random forest algorithm for intelligent feature selection, identifying a core set of approximately 1,462 key indicators from the vast number of microbial species. This streamlined data is then used to train the central neural network, which learns to associate specific microbial ‘fingerprints’ with their precise, multi-layered location within the biome ontology. Once trained, the model can make predictions with just a single forward pass. This efficient architecture allows ONN4MST to achieve millisecond-level response times (about 20 s for 100 samples) on datasets with nearly one million samples, all while maintaining a low memory footprint (around 22 GB). Critically, compared to traditional tools like FEAST and SourceTracker, its accuracy improves from 0.89 to 0.98 in benchmark tests against tools like FEAST and SourceTracker, representing a comprehensive optimization of speed, memory, and precision.
In summary, artificial intelligence and big data are fundamentally reshaping the landscape of microbial source tracking for aquatic products. From high-precision, genome-based pathogen tracing and microbial community-based origin verification to the development of advanced deep learning frameworks for large-scale data analysis, machine learning provides scalable, high-throughput solutions for accurately identifying and verifying microbial sources in aquatic products. As computational algorithms and microbiome databases continue to evolve, AI is poised to play an increasingly vital role in safeguarding the microbial safety and traceability of aquatic food products. AI enables high-accuracy microbial source tracking by integrating complex multi-dimensional datasets, but its performance depends heavily on the availability of large, diverse, and high-quality training data. Combining AI with complementary approaches such as WGS or stable isotope analysis could improve predictive robustness and practical adoption in regulatory frameworks (see Figure 4).
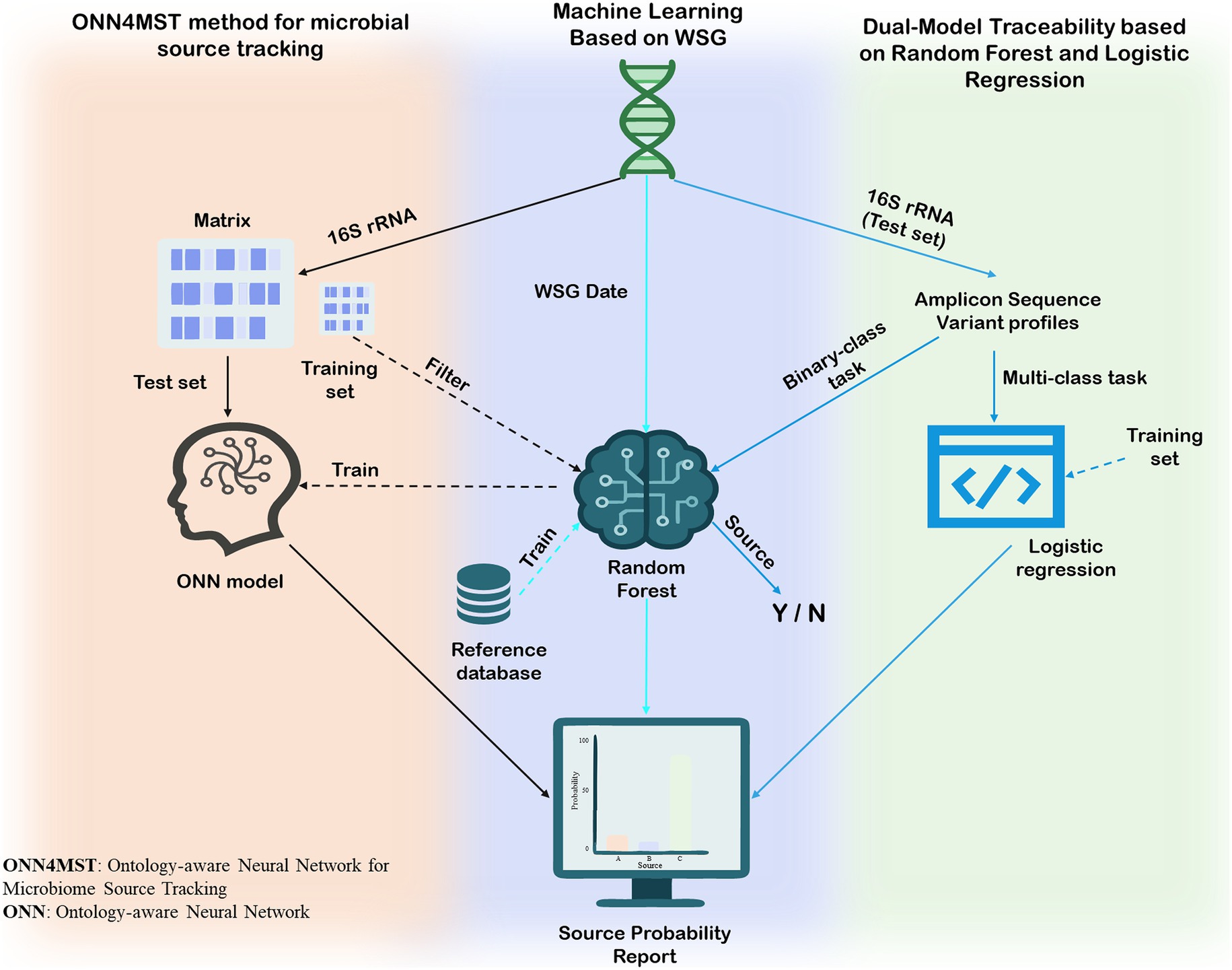
Figure 4. Application of three artificial intelligence methods in microbial source tracking in the aquatic supply chain.
5 Challenges in aquatic product traceability
5.1 Technical and data barriers
One of the primary technical hurdles in the microbial traceability of aquatic products is the lack of uniformity in technology adoption among different countries and regions (80). Developed nations generally have the requisite infrastructure, technological capabilities, and regulatory frameworks to implement efficient traceability systems. Conversely, numerous developing countries, which are significant sources of aquatic products, frequently lack the essential technology, equipment, and skilled workforce to establish comparable systems.
Furthermore, the internal systems utilized by enterprises, such as Enterprise Resource Planning (ERP) and Laboratory Information Management Systems (LIMS), often diverge from government regulatory platforms in terms of data formats and transmission protocols. In practice, mechanisms for verifying data authenticity and preventing tampering prior to uploading, such as blockchain technology, are under - exploited. This makes it arduous to guarantee the auditability of the traceability chain. All these factors have a detrimental impact on the microbial traceability of aquatic products.
5.2 Policy and standardization gaps
Currently, there is no dedicated international standard specifically addressing the traceability of microbial hazards in aquatic products. Most countries base their systems on the general principles of the Codex Alimentarius and their respective national frameworks such as Regulation (EC) No 178/2002 in the European Union and China’s Food Safety Law. However, the depth and specificity of these standards vary widely (81, 82). Existing traceability systems typically emphasize batch origin and distribution pathways, but often lack harmonized provisions on microbial threshold limits, quantitative risk assessment methodologies, and laboratory capability verification mechanisms.
Furthermore, significant regional disparities exist in the enforcement and implementation of these standards. Regulatory bodies in developed countries, such as the U.S. FDA and Japan’s Ministry of Health, Labour and Welfare, have established mature practices regarding regulatory oversight, enterprise audits, sample testing, and legal enforcement. In contrast, many developing countries face limitations in regulatory infrastructure and enforcement capacity, which undermine their ability to establish effective deterrents against microbial hazards in aquatic supply chains.
This disparity underscores the urgent need for globally harmonized traceability standards and capacity-building efforts to ensure consistent food safety governance across regions.
5.3 Challenges of traceability in complex supply chains
The aquatic product supply chain is intricate and globally dispersed. Traceability can be easily disrupted when responsibilities are fragmented among numerous stakeholders. Aquaculture and capture fisheries involve harvesters, processors, distributors, and retailers, who may be located in different countries and subject to different authorities (e.g., fisheries management agencies and food safety regulators). Insufficient coordination often gives rise to gaps. A report on Southeast Asian fisheries noted that government oversight is plagued by fragmented responsibilities across agencies and regional authorities, and enforcement loopholes impede effective monitoring (83).
Likewise, the existence of multiple private and public traceability systems, such as eco-labels, catch certificates, and import controls, can lead to overlaps without integration, resulting in confusion regarding responsibilities. These disconnects delay recall actions. Outbreak investigations may require piecing together invoices, Hazard Analysis and Critical Control Points (HACCP) records, and catch reports from various sources (84). Even when traceability data is available, complex processing procedures (e.g., the mixing of frozen fish batches) make it difficult to correlate microorganism test results with final products. In essence, multi-layered supply chains, cross-border trade, and siloed institutions reduce transparency.
6 Future perspectives
The development of aquatic microorganism traceability will greatly benefit from innovations in microbial detection methods. Highly accurate and rapid detection technologies will help identify contamination promptly, enabling timely responses and corrective measures. Developing mature on-site detection technologies, especially those that do not destroy the samples, can combine both cost-effectiveness and practical monitoring results, thus lowering the detection threshold and promoting widespread adoption. Additionally, continuous monitoring of environmental parameters such as water temperature, pH, dissolved oxygen, and turbidity in aquaculture systems can also serve as early warning indicators for microbial risk, further enhancing prevention and control capabilities.
However, building a full-chain traceability system for microorganisms faces significant challenges in handling vast amounts of data. Due to difficulties in data sharing and lack of interoperability between different stages of the supply chain, more intelligent tools, such as artificial intelligence, big data analysis, and blockchain technology, are urgently needed to improve data integration and risk identification. Therefore, developing and integrating intelligent systems to support aquatic microorganism traceability is of paramount importance.
The development of such technologies also requires guidance and support from policies and regulations. A review concluded that although many countries have implemented HACCP systems and corresponding inspection regulations, coordination between regulations remains necessary in managing hazards within increasingly globalized supply chains (85). Governments can promote the establishment of mandatory microorganism genome data upload and sharing mechanisms through legislation, building a global foodborne microorganism database, which is crucial for responding swiftly to international food safety incidents.
7 Conclusion
Aquatic products play a vital role in global food security, especially in regions abundant in marine resources. However, with the expansion of international trade, ensuring the microbial safety of these products has become an increasingly complex challenge. Foodborne microorganisms associated with aquatic products continue to pose serious public health risks and economic burdens.
Traceability technologies have emerged as essential tools for addressing these challenges. By enabling real-time monitoring and precise tracking of each stage in the aquatic product supply chain, these systems significantly enhance the ability to prevent, detect, and respond to microbial contamination. The integration of advanced technologies such as Internet of Things (IoT) sensors, genome sequencing, and big data analytics further strengthens the effectiveness and responsiveness of traceability frameworks.
Despite these advancements, significant obstacles remain. The uneven access to technological infrastructure, regulatory fragmentation across borders, and limited consumer awareness hinder the full potential of traceability systems. To move forward, efforts should focus on developing cost-effective and scalable traceability solutions, harmonizing international standards, enhancing policy frameworks, and promoting public engagement.
Prospectively, the integration of advanced technologies, including synthetic biology, artificial intelligence, and real-time data platforms, holds great promise for developing more robust and intelligent traceability systems. These innovations will contribute to achieving a safer, more transparent, and sustainable global aquatic product supply chain.
Author contributions
JZ: Conceptualization, Writing – original draft, Writing – review & editing. TD: Writing – review & editing. JA: Writing – review & editing. ZZ: Conceptualization, Project administration, Supervision, Writing – review & editing. XL: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Key R&D Program of China (2024YFD2402201).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer QJ declared a shared affiliation with the author ZZ to the handling editor at the time of review.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2025.1673037/full#supplementary-material
References
1. Gephart, JA, Agrawal Bejarano, R, Gorospe, K, Godwin, A, Golden, CD, Naylor, RL, et al. Globalization of wild capture and farmed aquatic foods. Nat Commun. (2024) 15:8026. doi: 10.1038/s41467-024-51965-8
2. FAO (2025). Fishery and Aquaculture Statistics – Yearbook 2022. Rome: Food and Agriculture Organization of the United Nations (FAO).
3. Stentiford, GD, Peeler, EJ, Tyler, CR, Bickley, LK, Holt, CC, Bass, D, et al. A seafood risk tool for assessing and mitigating chemical and pathogen hazards in the aquaculture supply chain. Nat Food. (2022) 3:169–78. doi: 10.1038/s43016-022-00465-3
4. Bondad-Reantaso, M, Arthur, JR, and Subasinghe, R. Understanding and applying risk analysis in aquaculture Food and Agriculture Organization of the United Nations (2008) (Rome: FAO Fisheries and Aquaculture Technical Paper No. 519).
5. Li, M, Xu, H, Tian, Y, Zhang, Y, Jiao, X, and Gu, D. Comparative genomic analysis reveals the potential transmission of Vibrio parahaemolyticus from freshwater food to humans. Food Microbiol. (2023) 113:104277. doi: 10.1016/j.fm.2023.104277
6. El Sheikha, AF, and Xu, J. Traceability as a key of seafood safety: reassessment and possible applications. Rev Fish Sci Aquac. (2017) 25:158–70. doi: 10.1080/23308249.2016.1254158
7. Ohore, OE, Wei, Y, Wang, Y, Nwankwegu, AS, and Wang, Z. Tracking the influence of antibiotics, antibiotic resistomes, and salinity gradient in modulating microbial community assemblage of surface water and the ecological consequences. Chemosphere. (2022) 305:135428. doi: 10.1016/j.chemosphere.2022.135428
8. Kabiraz, MP, Majumdar, PR, Mahmud, MMC, Bhowmik, S, and Ali, A. Conventional and advanced detection techniques of foodborne pathogens: A comprehensive review. Heliyon. (2023) 9:e15482. doi: 10.1016/j.heliyon.2023.e15482
9. Taylor, B, Ofori, KF, Parsaeimehr, A, Akdemir Evrendilek, G, Attarwala, T, and Ozbay, G. Exploring the complexities of seafood: from benefits to contaminants. Foods. (2025) 14:1461. doi: 10.3390/foods14091461
10. Roy, PK, Roy, A, Jeon, EB, DeWitt, CAM, Park, JW, and Park, SY. Comprehensive analysis of predominant pathogenic bacteria and viruses in seafood products. Compr Rev Food Sci Food Saf. (2024) 23:e13410. doi: 10.1111/1541-4337.13410
11. Vu, TTT, Alter, T, and Huehn, S. Prevalence of Vibrio spp. in retail seafood in Berlin, Germany. J Food Prot. (2018) 81:593–7. doi: 10.4315/0362-028X.JFP-17-366
12. Diner Rachel, E, Kaul, D, Rabines, A, Zheng, H, Steele Joshua, A, Griffith John, F, et al. Pathogenic Vibrio species are associated with distinct environmental niches and planktonic taxa in Southern California (USA) aquatic microbiomes. mSystems. (2021) 6:10–1128. doi: 10.1128/msystems.00571-21
13. Ma, J, Zhu, X, Hu, R, Qi, Z, Sun, W, Hao, Z, et al. A systematic review, meta-analysis and meta-regression of the global prevalence of foodborne Vibrio spp. infection in fishes: a persistent public health concern. Mar Pollut Bull. (2023) 187:114521. doi: 10.1016/j.marpolbul.2022.114521
14. FAO. (2002). Risk assessment of Campylobacter spp. in broiler chickens and Vibrio spp. in seafood. Available online at: https://www.fao.org/4/y8145e/y8145e00.htm#Contents (Accessed June 10, 2025).
15. Jiang, D, Han, H, Guo, Y, Zhang, R, Zhan, L, Zhou, Y, et al. Epidemiological characteristics of sporadic foodborne diseases caused by Vibrio parahaemolyticus - China, 2013-2022. China CDC Wkly. (2024) 6:1354–9. doi: 10.46234/ccdcw2024.269
16. Fearnley, E, Leong, LEX, Centofanti, A, Dowsett, P, Combs, BG, Draper, ADK, et al. Vibrio parahaemolyticus foodborne illness associated with oysters, Australia, 2021-2022. Emerg Infect Dis. (2024) 30:2271–8. doi: 10.3201/eid3011.240172
17. Zeidler, C, Szott, V, Alter, T, Huehn-Lindenbein, S, and Fleischmann, S. Prevalence of Vibrio spp. in seafood from German supermarkets and fish markets. Foods. (2024) 13:3987. doi: 10.3390/foods13243987
18. Di Salvo, E, Panebianco, F, Panebianco, A, and Ziino, G. Quantitative detection of viable but Nonculturable Vibrio parahaemolyticus in frozen bivalve Molluscs. Foods. (2023) 12:2373. doi: 10.3390/foods12122373
19. Zhang, Y, Liao, X, Ding, T, and Feng, J. Tolerance variations and mechanisms of Salmonella enterica serovar Newport in response to long-term hypertonic stress. Food Qual Saf. (2024) 8:fyad068. doi: 10.1093/fqsafe/fyad068
20. Hocking, AD. Foodborne microorganisms of public health significance. Austr Vet J. (2003) 77:54. doi: 10.1111/j.1751-0813.1999.tb12437.x
21. Popoff, MY, Bockemühl, J, and Gheesling, LL. Supplement 2002 (no. 46) to the Kauffmann-White scheme. Res Microbiol. (2004) 155:568–70. doi: 10.1016/j.resmic.2004.04.005
22. FAO. Report of the FAO expert workshop on application of biosecurity measures to control Salmonella contamination in sustainable aquaculture. Rome: Food and Agriculture Organization of the United Nations (FAO) (2010).
23. Amagliani, G, Brandi, G, and Schiavano, GF. Incidence and role of Salmonella in seafood safety. Food Res Int. (2012) 45:780–8. doi: 10.1016/j.foodres.2011.06.022
24. Ferrari, RG, Rosario, DKA, Cunha-Neto, A, Mano, SB, Figueiredo, EES, and Conte-Junior, CA. Worldwide epidemiology of Salmonella serovars in animal-based foods: a meta-analysis. Appl Environ Microbiol. (2019) 85:e519–91. doi: 10.1128/AEM.00591-19
25. Abdollahzadeh, E, Ojagh, SM, Hosseini, H, Irajian, G, and Ghaemi, EA. Prevalence and molecular characterization of Listeria spp. and Listeria monocytogenes isolated from fish, shrimp, and cooked ready-to-eat (RTE) aquatic products in Iran. LWT. (2016) 73:205–11. doi: 10.1016/j.lwt.2016.06.020
26. Jackson, KA, Iwamoto, M, and Swerdlow, D. Pregnancy-associated listeriosis. Epidemiol Infect. (2010) 138:1503–9. doi: 10.1017/S0950268810000294
27. Chintagari, S, Hazard, N, Edwards, G, Jadeja, R, and Janes, M. Risks associated with fish and seafood. Microbiol Spectr. (2017) 5:10–1128. doi: 10.1128/microbiolspec.pfs-0013-2016
28. Vázquez-Sánchez, D, Galvão, JA, and Oetterer, M. Contamination sources, serogroups, biofilm-forming ability and biocide resistance of Listeria monocytogenes persistent in tilapia-processing facilities. J Food Sci Technol Mysore. (2017) 54:3867–79. doi: 10.1007/s13197-017-2843-x
29. Costa, JCCP, Bolívar, A, Alberte, TM, Zurera, G, and Pérez-Rodríguez, F. Listeria monocytogenes in aquatic food products: spotlight on epidemiological information, bio-based mitigation strategies and predictive approaches. Microb Pathog. (2024) 197:106981. doi: 10.1016/j.micpath.2024.106981
30. Bloomfield, SJ, Palau, R, Holden, ER, Webber, MA, and Mather, AE. Genomic characterization of Pseudomonas spp. on food: implications for spoilage, antimicrobial resistance and human infection. BMC Microbiol. (2024) 24:20. doi: 10.1186/s12866-023-03153-9
31. Shahrokhi, GR, Rahimi, E, and Shakerian, A. The prevalence rate, pattern of antibiotic resistance, and frequency of virulence factors of Pseudomonas aeruginosa strains isolated from fish in Iran. J Food Qual. (2022) 2022:1–8. doi: 10.1155/2022/8990912
32. Belliot, G, Lopman, BA, Ambert-Balay, K, and Pothier, P. The burden of norovirus gastroenteritis: an important foodborne and healthcare-related infection. Clin Microbiol Infect. (2014) 20:724–30. doi: 10.1111/1469-0691.12722
33. Chhabra, P, de Graaf, M, Parra, GI, Chan, MC, Green, K, Martella, V, et al. Updated classification of norovirus genogroups and genotypes. J Gen Virol. (2019) 100:1393–406. doi: 10.1099/jgv.0.001318
34. Fouillet, A, Fournet, N, Forgeot, C, Jones, G, Septfons, A, Franconeri, L, et al. Large concomitant outbreaks of acute gastroenteritis emergency visits in adults and food-borne events suspected to be linked to raw shellfish, France, December 2019 to January 2020. Eurosurveillance. (2020) 25:2000060. doi: 10.2807/1560-7917.ES.2020.25.7.2000060
35. Li, Y, Xue, L, Gao, J, Cai, W, Zhang, Z, Meng, L, et al. A systematic review and meta-analysis indicates a substantial burden of human noroviruses in shellfish worldwide, with GII.4 and GII.2 being the predominant genotypes. Food Microbiol. (2023) 109:104140. doi: 10.1016/j.fm.2022.104140
36. Barrabeig, I, Rovira, A, Buesa, J, Bartolomé, R, Pintó, R, Prellezo, H, et al. Foodborne norovirus outbreak: the role of an asymptomatic food handler. BMC Infect Dis. (2010) 10:269. doi: 10.1186/1471-2334-10-269
37. Nemes, K, Persson, S, and Simonsson, M. Hepatitis a virus and hepatitis e virus as food- and waterborne pathogens—transmission routes and methods for detection in food. Viruses. (2023) 15:1725. doi: 10.3390/v15081725
38. Sander, A, Corman, VM, Lukashev, AN, and Drexler, JF. Evolutionary origins of enteric hepatitis viruses. Cold Spring Harb Perspect Med. (2018) 8:a031690. doi: 10.1101/cshperspect.a031690
39. Vaughan, G, Goncalves Rossi, LM, Forbi, JC, de Paula, VS, Purdy, MA, Xia, G, et al. Hepatitis A virus: host interactions, molecular epidemiology and evolution. Infect Genet Evol. (2014) 21:227–43. doi: 10.1016/j.meegid.2013.10.023
40. Di Cola, G, Fantilli, AC, Pisano, MB, and Ré, VE. Foodborne transmission of hepatitis A and hepatitis E viruses: A literature review. Int J Food Microbiol. (2021) 338:108986. doi: 10.1016/j.ijfoodmicro.2020.108986
41. Crossan, C, Baker, PJ, Craft, J, Takeuchi, Y, Dalton, HR, and Scobie, L. Hepatitis E virus genotype 3 in shellfish, United Kingdom. Emerg Infect Dis. (2012) 18:2085–7. doi: 10.3201/eid1812.120924
42. Su, X, Sutarlie, L, and Loh, XJ. Sensors, biosensors, and analytical Technologies for Aquaculture Water Quality. Research. (2020) 2020:8272705. doi: 10.34133/2020/8272705
43. Flannery, J, Keaveney, S, Rajko-Nenow, P, O'Flaherty, V, and Doré, W. Concentration of norovirus during wastewater treatment and its impact on oyster contamination. Appl Environ Microbiol. (2012) 78:3400–6. doi: 10.1128/AEM.07569-11
44. Nagarajan, V, Chen, J, Hsu, G, Chen, H, Chao, H, Huang, S, et al. Surveillance of adenovirus and norovirus contaminants in the water and shellfish of major oyster breeding farms and fishing ports in Taiwan. Pathogens. (2022) 11:316. doi: 10.3390/pathogens11030316
45. Correia Peres Costa, JC, Floriano, B, Bascón Villegas, IM, Rodríguez-Ruiz, JP, Posada-Izquierdo, GD, Zurera, G, et al. Study of the microbiological quality, prevalence of foodborne pathogens and product shelf-life of Gilthead Sea bream (Sparus aurata) and sea bass (Dicentrarchus labrax) from aquaculture in estuarine ecosystems of Andalusia (Spain). Food Microbiol. (2020) 90:103498. doi: 10.1016/j.fm.2020.103498
46. Diyie, RL, Osei-Atweneboana, MY, Armah, E, Yankson, K, and Aheto, DW. Contamination of fish feed with pathogenic organisms: implications on fish diseases in aquaculture systems. Ghana J Sci Technol Dev. (2024) 2:2343–6727. doi: 10.47881/283.967x
47. Al-Musawi, ML, Hussein, SB, Hassan, ZAA, Hamd, MT, and Jasim, RI. Microbial contamination in imported fish feed to Iraq. J Phys Conf Ser. (2021) 1999:012026. doi: 10.1088/1742-6596/1999/1/012026
48. Møretrø, T, Moen, B, Heir, E, Hansen, AÅ, and Langsrud, S. Contamination of salmon fillets and processing plants with spoilage bacteria. Int J Food Microbiol. (2016) 237:98–108. doi: 10.1016/j.ijfoodmicro.2016.08.016
49. Svanevik, CS, Roiha, IS, Levsen, A, and Lunestad, BT. Microbiological assessment along the fish production chain of the Norwegian pelagic fisheries sector – results from a spot sampling programme. Food Microbiol. (2015) 51:144–53. doi: 10.1016/j.fm.2015.05.016
50. Belias, A, Sullivan, G, Wiedmann, M, and Ivanek, R. Factors that contribute to persistent Listeria in food processing facilities and relevant interventions: a rapid review. Food Control. (2022) 133:108579. doi: 10.1016/j.foodcont.2021.108579
51. Berrang, ME, and Frank, JF. Generation of airborne Listeria innocua from model floor drains. J Food Prot. (2012) 75:1328–31. doi: 10.4315/0362-028X.JFP-12-021
52. Chen, B, Xu, T, Yan, Q, Karsli, B, Li, D, and Xie, J. Effect of temperature fluctuations on large yellow croaker fillets (Larimichthys crocea) in cold chain logistics: a microbiological and metabolomic analysis. J Food Eng. (2025) 386:112290. doi: 10.1016/j.jfoodeng.2024.112290
53. Trafiałek, J, Zwolinski, M, and Kolanowski, W. Assessing hygiene practices during fish selling in retail stores. Br Food J. (2016) 118:2053–67. doi: 10.1108/BFJ-12-2015-0477
54. Kai, L., Gao, W., and Cong, C. (2023) Oyster cold chain logistics monitoring system based on RFID and cloud platforms. Fourth International Conference on Signal Processing and Computer Science. SPIE.
55. EFSA Panel on Biological Hazards (BIOHAZ). The use of the so-called ‘tubs’ for transporting and storing fresh fishery products. EFSA J. (2020) 18:e6091. doi: 10.2903/j.efsa.2020.6091
56. Liao, X, Shen, W, Wang, Y, Bai, L, and Ding, T. Microbial contamination, community diversity and cross-contamination risk of food-contact ice. Food Res Int. (2023) 164:112335. doi: 10.1016/j.foodres.2022.112335
57. Atwill, ER, and Jeamsripong, S. Bacterial diversity and potential risk factors associated with Salmonella contamination of seafood products sold in retail markets in Bangkok, Thailand. PeerJ. (2021) 9:e12694. doi: 10.7717/peerj.12694
58. Prabhakar, P, Lekshmi, M, Ammini, P, Nayak, BB, and Kumar, S. Salmonella contamination of seafood in landing centers and retail Markets of Mumbai, India. J AOAC Int. (2020) 103:1361–5. doi: 10.1093/jaoacint/qsaa042
59. Namsai, A, Louisirirotchanakul, S, Wongchinda, N, Siripanyaphinyo, U, Virulhakul, P, Puthavathana, P, et al. Surveillance of hepatitis A and E viruses contamination in shellfish in Thailand. Lett Appl Microbiol. (2011) 53:608–13. doi: 10.1111/j.1472-765X.2011.03152.x
60. Amin, MB, Talukdar, PK, Sraboni, AS, Islam, MR, Mahmud, ZH, Berendes, D, et al. Prevalence and antimicrobial resistance of major foodborne pathogens isolated from pangas and tilapia fish sold in retail markets of Dhaka city, Bangladesh. Int J Food Microbiol. (2024) 418:110717. doi: 10.1016/j.ijfoodmicro.2024.110717
61. Wong, H, Jiang, H, Lin, H, and Wang, Y. Microbiological quality of seafood marketed in Taiwan. J Food Prot. (2015) 78:1973–9. doi: 10.4315/0362-028X.JFP-15-152
62. Jasson, V, Jacxsens, L, Luning, P, Rajkovic, A, and Uyttendaele, M. Alternative microbial methods: an overview and selection criteria. Food Microbiol. (2010) 27:710–30. doi: 10.1016/j.fm.2010.04.008
63. Hameed, S, Xie, L, and Ying, Y. Conventional and emerging detection techniques for pathogenic bacteria in food science: a review. Trends Food Sci Technol. (2018) 81:61–73. doi: 10.1016/j.tifs.2018.05.020
64. Boldura, O, and Popescu, S. PCR: A powerful method in food safety field In: A Samadikuchaksaraei, editor. Polymerase chain reaction for biomedical applications. London: IntechOpen (2016).
65. Croci, L, De Medici, D, Ciccozzi, M, Di Pasquale, S, Suffredini, E, and Toti, L. Contamination of mussels by hepatitis A virus: a public-health problem in southern Italy. Food Control. (2003) 14:559–63. doi: 10.1016/S0956-7135(02)00133-0
66. Rivadulla, E, Varela, MF, Mesquita, JR, Nascimento, MSJ, and Romalde, JL. Detection of hepatitis E virus in shellfish harvesting areas from Galicia (northwestern Spain). Viruses. (2019) 11:618. doi: 10.3390/v11070618
67. Croci, L, Losio, MN, Suffredini, E, Pavoni, E, Di Pasquale, S, Fallacara, F, et al. Assessment of human enteric viruses in shellfish from the northern Adriatic Sea. Int J Food Microbiol. (2007) 114:252–7. doi: 10.1016/j.ijfoodmicro.2006.09.015
68. Chen, A, Ma, B, Zhang, Y, Huang, W, Xiao, Y, Li, J, et al. Probe-based dual-chip digital loop-mediated isothermal amplification for the simultaneous detection of Staphylococcus aureus and Salmonella enteritidis in livestock and aquatic products. Food Control. (2025) 168:110988. doi: 10.1016/j.foodcont.2024.110988
69. Rheman, S, Hossain, S, Sarker, MS, Akter, F, Khor, L, Gan, HM, et al. Nanopore sequencing for identification and characterization of antimicrobial-resistant Escherichia coli and Salmonella spp. from tilapia and shrimp sold at wet markets in Dhaka, Bangladesh. Front Microbiol. (2024) 7:1329620. doi: 10.3389/fmicb.2024.1329620
70. Atxaerandio-Landa, A, Arrieta-Gisasola, A, Laorden, L, Bikandi, J, Garaizar, J, Martinez-Malaxetxebarria, I, et al. A practical bioinformatics workflow for routine analysis of bacterial WGS data. Microorganisms. (2022) 10:2364. doi: 10.3390/microorganisms10122364
71. Nguyen, L, Schmidt, HA, von Haeseler, A, and Minh, BQ. Iq-tree: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. (2015) 32:268–74. doi: 10.1093/molbev/msu300
72. Allard, MW, Strain, E, Melka, D, Bunning, K, Musser, SM, Brown, EW, et al. Practical value of food pathogen traceability through building a whole-genome sequencing network and database. J Clin Microbiol. (2016) 54:1975–83. doi: 10.1128/JCM.00081-16
73. Nadon, C, Van Walle, I, Gerner-Smidt, P, Campos, J, Chinen, I, Concepcion-Acevedo, J, et al. PulseNet international: vision for the implementation of whole genome sequencing (WGS) for global food-borne disease surveillance. Eurosurveillance. (2017) 22:30544. doi: 10.2807/1560-7917.ES.2017.22.23.30544
74. Li, W, Wu, S, Fu, P, Liu, J, Han, H, Bai, L, et al. National molecular tracing network for foodborne disease surveillance in China. Food Control. (2018) 88:28–32. doi: 10.1016/j.foodcont.2017.12.032
75. Meriggi, N, Russo, A, Renzi, S, Cerasuolo, B, Nerini, M, Ugolini, A, et al. Enhancing seafood traceability: tracking the origin of seabass and seabream from the tuscan coast area by the analysis of the gill bacterial communities. Anim Microbiome. (2024) 6:13. doi: 10.1186/s42523-024-00300-z
76. Cusa, M, St John Glew, K, Trueman, C, Mariani, S, Buckley, L, Neat, F, et al. A future for seafood point-of-origin testing using DNA and stable isotope signatures. Rev Fish Biol Fish. (2022) 32:597–621. doi: 10.1007/s11160-021-09680-w
77. Rose, EB, Author, MKSC, Tolar, B, Pettengill, J, Batz, M, Bazaco, M, et al. Attribution of Salmonella enterica to food sources by using whole-genome sequencing data. Emerg Infect Dis. (2025) 4:783–90. doi: 10.3201/eid3104.241172
78. Peruzza, L, Cicala, F, Milan, M, Rovere, GD, Patarnello, T, Boffo, L, et al. Preventing illegal seafood trade using machine-learning assisted microbiome analysis. BMC Biol. (2024) 22:202. doi: 10.1186/s12915-024-02005-w
79. Zha, Y, Chong, H, Qiu, H, Kang, K, Dun, Y, Chen, Z, et al. Ontology-aware deep learning enables ultrafast and interpretable source tracking among sub-million microbial community samples from hundreds of niches. Genome Med. (2022) 14:43. doi: 10.1186/s13073-022-01047-5
80. FAO. (2025). The State of World Fisheries and Aquaculture. Available online at: https://www.fao.org/publications/fao-flagship-publications/the-state-of-world-fisheries-and-aquaculture/en (Accessed June 10, 2025).
81. CJO. (2021). Food Safety Law of China. Available online at: https://www.chinajusticeobserver.com/law/x/food-safety-law-of-the-peoples-republic-of-china20210429/chn (Accessed June 10, 2025).
82. EU. (2002). Regulation (EC) No 178/2002 of the European Parliament and of the Council of 28 January 2002. Available online at: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:32002R0178. (Accessed June 10, 2025).
83. GSMA. (2025). Advancing Sustainable Fisheries with Digital Technologies: The Gulf of Thailand | Mobile for Development. Available online at: https://www.gsma.com/solutions-and-impact/connectivity-for-good/mobile-for-development/gsma_resources/advancing-sustainable-fisheries-with-digital-technologies-the-gulf-of-thailand/ (Accessed June 6, 2025).
84. CDC. (2024). Colorado Stops a Multistate Salmonella Outbreak Linked to Seafood. Available online at: https://www.cdc.gov/foodcore/php/success-stories/colorado-stops-outbreak.html (Accessed June 10, 2025).
85. Sheng, L, and Wang, L. The microbial safety of fish and fish products: recent advances in understanding its significance, contamination sources, and control strategies. Compr Rev Food Sci Food Saf. (2021) 20:738–86. doi: 10.1111/1541-4337.12671
86. Pakingking, R, Palma, P, and Usero, R. Quantitative and qualitative analyses of the bacterial microbiota of tilapia (Oreochromis niloticus) cultured in earthen ponds in the Philippines. World J Microbiol Biotechnol. (2015) 31:265–75. doi: 10.1007/s11274-014-1758-1
87. Gołaś, I, Szmyt, M, Potorski, J, Łopata, M, Gotkowska-Płachta, A, and Glińska-Lewczuk, K. Distribution of Pseudomonas fluorescens and Aeromonas hydrophila bacteria in a recirculating aquaculture system during farming of European grayling (Thymallus thymallus L.) broodstock. Water. (2019) 11:376. doi: 10.3390/w11020376
88. Al-Harbi, AH, and Uddin, N. Bacterial diversity of tilapia (Oreochromis niloticus) cultured in brackish water in Saudi Arabia. Aquaculture. (2005) 250:566–72. doi: 10.1016/j.aquaculture.2005.01.026
89. Antunes, P, Campos, J, Mourao, J, Pereira, J, Novais, C, and Peixe, L. Inflow water is a major source of trout farming contamination with Salmonella and multidrug resistant bacteria. Sci Total Environ. (2018) 642:1163–71. doi: 10.1016/j.scitotenv.2018.06.143
90. Skowron, K, Wiktorczyk, N, Grudlewska, K, Wałecka-Zacharska, E, Paluszak, Z, Kruszewski, S, et al. Phenotypic and genotypic evaluation of Listeria monocytogenes strains isolated from fish and fish processing plants. Ann Microbiol. (2019) 69:469–82. doi: 10.1007/s13213-018-1432-1
91. Selvaganapathi, R, Jeyasekaran, G, Shakila, RJ, Sukumar, D, Kumar, MP, and Sivaraman, B. Occurrence of Listeria monocytogenes on the seafood contact surfaces of Tuticorin coast of India. J Food Sci Technol. (2018) 55:2808–12. doi: 10.1007/s13197-018-3230-y
92. Aladhadh, M. A review of modern methods for the detection of foodborne pathogens. Microorganisms. (2023) 11:1111. doi: 10.3390/microorganisms11051111
93. Sadeghi, Y, Kananizadeh, P, Moghadam, SO, Alizadeh, A, Pourmand, MR, Mohammadi, N, et al. The sensitivity and specificity of loop-mediated isothermal amplification and PCR methods in detection of foodborne microorganisms: a systematic review and Meta-analysis. Iran J Public Health. (2021) 50:2172–82. doi: 10.18502/ijph.v50i11.7571
Keywords: aquatic supply chain, food safety, microbial contamination, traceability technology, artificial intelligence
Citation: Zhang J, Ding T, Ahn J, Zhang Z and Liao X (2025) Tracing microbial hazards in the aquatic supply chain: challenges, technologies, and future directions. Front. Nutr. 12:1673037. doi: 10.3389/fnut.2025.1673037
Edited by:
Wei Huamao, Ningbo University, ChinaReviewed by:
Zhaohui Qiao, Ningbo University, ChinaQingqing Jiang, Shanghai Ocean University, China
Gaoshang Li, Ningbo University, China
Copyright © 2025 Zhang, Ding, Ahn, Zhang and Liao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinyu Liao, eGlueXVfbGlhb0B6anUuZWR1LmNu; Zhaohuan Zhang, emgtemhhbmdAc2hvdS5lZHUuY24=
 Jiayi Zhang
Jiayi Zhang Tian Ding
Tian Ding Juhee Ahn
Juhee Ahn Zhaohuan Zhang
Zhaohuan Zhang Xinyu Liao
Xinyu Liao