Abstract
Mitochondria are dynamic organelles that integrate bioenergetics, biosynthesis, and signaling in cells and regulate redox homeostasis, apoptotic pathways, and cell proliferation and differentiation. Depending on the environmental conditions, the mitochondrial morphology dynamically changes to match the energy demands. The mitochondrial dynamics is related to the initiation, migration, and invasion of diverse human cancers and thus affects cancer metastasis, metabolism, drug resistance, and cancer stem cell survival. We reviewed the current image-based analytical tools and machine-learning techniques for phenotyping mitochondrial morphology in different cancer cell lines from confocal microscopy images. We listed and applied pipelines and packages available in ImageJ/Fiji, CellProfiler, MATLAB, Java, and Python for the analysis of fluorescently labeled mitochondria in microscopy images and compared their performance, usability and applications. Furthermore, we discussed the potential of automatic mitochondrial segmentation, classification and prediction of mitochondrial abnormalities using machine learning techniques. Quantification of the mitochondrial morphology provides potential indicators for identifying metabolic changes and drug responses in cancer cells.
Introduction
Cancer cells have different metabolic profiles from healthy cells to ensure their unregulated proliferation and survival in tumor microenvironments. Aerobic glycolysis, which is known as the Warburg effect, is a hallmark of cancer cell metabolism [1]. In addition to ATP generation via mitochondrial oxidative phosphorylation (OXPHOS), glycolytic flux is often enhanced despite the presence of oxygen. Aerobic glycolysis provides metabolic intermediates for tumor biomass expansion, is an alternative pathway for generating energy (ATP), reduces the oxygen requirement of the hypoxic tumor interior, evades mitochondria-mediated apoptosis, and exports lactate to create an acidic extracellular environment that favors cancer cell survival. However, cancer cells exhibit metabolic plasticity to survive and thrive in various tumor microenvironments [2]. In addition to aerobic glycolysis, previous studies have revealed that cancer cells could adopt a variety of metabolic phenotypes [3, 4], such as high OXPHOS, intermediate type (high glycolysis and OXPHOS), and idling type (low glycolysis and low OXPHOS), and gene expression profiles predicted via systems biology simulations. Cancer cells might reconfigure their metabolic pathways to facilitate epithelial-mesenchymal transition and metastasis [5].
As the hub for bioenergetics, metabolism, signal transduction, and cellular survival, mitochondria play crucial roles in cancer cell proliferation, progression, and response to treatment [6–9]. Mitochondria are also motile organelles that constantly undergo fission, fusion, mitophagy, and biogenesis cycles. Processes called mitochondrial dynamics are crucial for the mitochondrial network, DNA (mtDNA) integrity, ATP synthesis, ROS generation, and cell cycle progression [6, 10, 11]. Mitochondrial fusion is associated with a well-developed mitochondrial network and increased OXPHOS capacity. In contrast, mitochondrial fission is related to fragmented mitochondria, enhanced mitophagy (elimination of damaged mitochondria), increased uncoupling, and increased ROS production [12]. The cellular mitochondrial phenotype correlates with the cellular metabolic state.
At the molecular level, mitochondrial fission/fusion events are driven by dynamin-related GTPases [10]. Mitochondrial fusion is mediated by mitofusins (Mfn-1 and Mfn-2) [13] located on the outer mitochondrial membrane (OMM) and optical atrophy-1 (Opa-1) [14, 15] proteins located on the inner mitochondrial membrane (IMM), and these proteins drive OMM and IMM fusion via sequential steps [16], respectively. Mitochondrial fission is mediated by dynamin-related protein 1 (Drp-1) [17] recruited to the OMM at endoplasmic reticulum (ER)-mitochondria contacts [18] and by other cofactors, such as mitochondrial fission factor (Mff), mitochondrial fission 1 protein (Fis1), MiD49, and MiD51 [19]. The activities of fusion/fission proteins are not only regulated by gene expression levels but also controlled by posttranslational modifications [10, 20], metabolic cues [12], cellular signaling, and crosstalk between fusion and fission proteins. For instance, Fis1 exhibits dual functions of blocking fusion proteins (Opa-1, Mfn-1, and Mfn-2) and enhancing the recruitment of the fission protein Drp-1, which results in the fragmentation of mitochondria [21].
Cancer cell progression and metastasis involve alterations in the mitochondrial network morphology. In general, a fragmented mitochondrial network facilitates cancer cell proliferation and metastasis [22]. The downregulation of fusion proteins (e.g., Opa1 and mitofusins) and the upregulation of fission proteins (e.g., Drp-1) have been observed in various cancer cells, with some exceptions [10]. In addition to genomic and proteomic profiles, the mitochondrial network morphology might serve as biomarkers for the cancer cell metabolic status and response to antitumor treatments [23]. A previous study revealed that an increased fraction of punctate mitochondria is associated with increased glycolysis relative to OXPHOS and the response to antitumor drugs in cancer cell lines and cancer cells harvested from patients.
Alterations in the mitochondrial morphology and structure may indicate early events of carcinogenesis or tumor progression [24], which suggests that the mitochondrial phenotype might provide a biomarker for cancer diagnosis and treatment targets [25, 26]. However, the identification of reliable and quantitative tools for evaluating the mitochondrial morphology and network parameters has been challenging [27]. Imaging techniques provide potential solutions. Electron microscopy has been used to investigate the morphologies of the mitochondrial network in human cancer, particularly in pathological tissues [28, 29]. However, electron microscopy requires special preparation procedures and is not applicable to live specimens. Consequently, imaging using confocal microscopy is another solution for measuring mitochondrial morphological features. Nonetheless, manually analyzing and tracking microscopy images of mitochondrial structures can be labor intensive, and image analysis techniques are thus crucial for obtaining objective and quantifiable results. Hence, in this review, we focus on automatic (or semiautomatic) pipelines established for confocal microscopy image analysis, specifically for analyzing the mitochondrial morphology and dynamics in live cells.
Image Processing Pipelines
Fluorescence microscopy images intuitively show the shapes, intensities, positions, and distributions of labeled organelles to study their morphologies. Mitochondria have been labeled with mitochondria-targeting fluorescent proteins or organic dyes, such as MitoTracker Red, tetramethylrhodamine methyl ester perchlorate (TMRM), and JC1, for imaging [30, 31]. The acquisition of images using microscopy is often classified spatially (2D images vs. 3D images) and temporally (snapshot images vs. time-lapse images). Flat, two-dimensional (2D) images only capture a single slice of the xy-plane. The information along the z-axis is limited. However, with the pinhole nature of confocal microscopes blocking out-of-focus light, a three-dimensional (3D) image is obtained by taking multiple z-slices (known as a z-stack). Although 3D images reveal more of the cells’ internal structure, the imaging of multiple slices is more time consuming, requires more computational power to process the resulting data, and risks an overdose of incoming photons, which leads to phototoxicity and photobleaching. A similar rule applies to taking multiple snapshots for a time-series image. Cells and mitochondria are prone to damage from the intense laser beams under the confocal microscope, and the fluorescent signals decrease as more images are taken over time.
After obtaining fluorescent images, the next step is to determine a proper method for analyzing the images and revealing the morphological information. Although manual classification is straightforward and intuitive, it is laborious and lacks precise and quantitative criteria. Therefore, researchers have developed software and pipelines to automatically process and analyze images in a high-throughput manner. Several image analysis tools or software packages are currently available with detailed pipelines and procedures or are released as open-source software (Table 1). The selection of tools from all of these sources according to the image features and functional needs can make image analysis more efficient and effective. Within this scope, mitochondrial fluorescent image analysis tools in different platforms and applications are reviewed and compared, including traditional image processing pipelines as well as emerging machine learning and deep learning methods.
TABLE 1
| Software/Algorithm | Image dimension | Input images | Output analysis | Features | Tested cells | Reference | Link |
| CellProfiler | |||||||
| Rees et al. | 2D | TIF images with a mitochondrial channel and nucleal channel | Count, area and intensity of nuclei, mitochondrial puncta, and cytoplasm | Step-by-step construction and detailed description of the execution and parameters of automated segmentation to allow user implementation | SN4741 mouse midbrain-derived neuronal cell line (a rotenone-based in vitro Parkinson’s disease model) | [35] | — |
| Reis et al. | 2D | Fluorescent images of mitochondria. (softWoRx is used for deconvolution) | Parameters provided by CellProfiler measure functions such as area and shape, Zernike, and texture-related parameters | Classifies mitochondria into networked, fragmented, and swollen morphologies using a Random Forest algorithm | Human MCF-7 breast cancer cells | [36] | — |
| ImageJ | |||||||
| MiNA | 2D/3D (limited options) | Single-channel image with 8-bit or 16-bit color | Skeleton parameters | Focuses on a skeleton analysis of mitochondrial networks and classifies mitochondria into two types: individuals and networks | C2C12 cells, SH-SY5Y cells | [45] | https://github.com/StuartLab/MiNA |
| 1) Parameters related to branch lengths: mean branch length, median branch length, standard deviation of branch lengths | |||||||
| 2) Parameters related to the number of branches: individuals, networks, mean branches/network, median branches/network, standard deviation, branches/network | |||||||
| Mitochondria Analyzer | 2D/3D | Fluorescent images of mitochondria | Particle analysis such as “Mito Count”, “Area”, “Perimeter”, “Form Factor” and “Aspect Ratio” for 2D images and “Volume”, “Surface Area”, and “Sphericity” for 3D images | Performs particle analysis and skeleton analysis and provides various parameters for both 2D and 3D images | MIN6 cells, pancreatic β-cells | [46] | https://github.com/AhsenChaudhry/Mitochondria-Analyzer |
| Skeleton analysis such as “Branches”, “Branch Junctions”, “Branch Length” and “Mean Branch Length” | |||||||
| Chustecki et al. | 2D | Time-series images of mitochondria | Cell areas and dynamic and network parameters, such as the diffusion rate, moving speed, connectivity, and average degree of the social network | Dynamic analysis focusing on the colocalization events, which is the most suitable approach for analyzing the morphology of discrete and separated mitochondria | Wild-type (WT) and mutant A. thaliana | [47] | — |
| MATLAB | |||||||
| Mytoe | 2D | TIF/most common image file types | Mitochondrial length, tortuosity, intensity, number of branches and motion parameters (speed, direction, etc.) | Applying optical flow estimation to time-series images for motion analysis | U2OS cells | [52] | https://sites.google.com/view/andreribeirolab/home/software |
| Multipage and multichannel images are supported | |||||||
| MitoSPT | 2D | Time series images | Net distances traveled by the mitochondria and individual mitochondria location information | Track whole-cell mitochondrial movements | Adult human dermal fibroblasts | [53] | https://github.com/kandelj/MitoSPT |
| Preprocessing using an ImageJ macro | |||||||
| MitoWave | 2D | Time-series fluorescent images of mitochondrial labeled with TMRM (membrane potential-related dye) | Transition time points of the inner mitochondrial membrane potentials during ischemia, five different membrane potential oscillating patterns (scalograms) during I/R, predominant frequencies of mitochondrial clusters, and mitochondrial cluster size | Analysis of time-series data focusing on the mitochondrial membrane potential | Neonatal mouse cardiac myocytes | [51] | https://github.com/dashok1/MitoWave |
| Zamponi et al. | 2D | Fluorescent images of mitochondria | Cluster mass (i.e., number of pixels) and pixel degree; c1, and c2 are obtained using computational models | Easy to use and can be combined with computational model simulations | MEFs | [49] | https://github.com/nahuelzamponi/MitoCluster |
| Mitometer | 2D/3D | TIF image stacks with time series | Morphological parameters, such as area, solidity, and perimeter, along with dynamic parameters, such as fusion rate, fission rate, distance, and speed | Dynamic analysis including fusion/fission rates and speeds from both 2D and 3D images | Neoblastic pheochromocytoma 12 cells | [50] | https://github.com/aelefebv/Mitometer |
| Non-small-cell lung carcinoma H1299 cells | |||||||
| Human foreskin fibroblast (HFF) cells | |||||||
| Primary normal breast epithelial cell lines | |||||||
| Two ER/PR + breast cancer cell lines (MCF-7 and T-47D), three TNBC cell lines (MDA-MB-231, MDA-MB-468, and BT-549) | |||||||
| Two TNBC PDX cell lines (HCI-010 and HCI-002) | |||||||
| Others | |||||||
| Momito (Java) | 2D | Confocal images/binary images (JPEG) | Mitochondrial length, connectivity (ECs and J/E), cristae structureetc. | Probabilistic approach that considers multiple possible connection patterns of skeleton configurations | WT MEFs and human U2OS cells | [48] | www.uqtr.ca/LaboMarcGermain |
| MitoGraph (C++) | 2D/3D | Fluorescent images and binary images of mitochondria as well as VTK data | Morphological parameters, such as volume, average width, and total length | 3D skeleton reconstruction with high performance | Saccharomyces cerevisiae, HMEC-1 cells, C2C12 cells, HEK293 and 293T cells, H1299 cells | [39] | https://github.com/vianamp/MitoGraph |
| Network parameters, such as number of nodes with different degrees, locations of nodes and average edge length | |||||||
List of publicly available tools for analyzing the mitochondrial morphology and dynamics.
CellProfiler
CellProfiler is a multifunctional software developed by the Broad Institute that allows researchers to perform automatic analyses in several biological areas, such as genomics, organelle morphologies, and protein staining [32]. CellProfiler has a friendly graphical user interface (GUI) and is open-source software written in MATLAB [32] and Python [33, 34]. CellProfiler is flexible in terms of adjusting its parameters to suit different image sets. Due to its extensibility, several image processing pipelines have been developed to analyze and process mitochondrial images in an automated, high-throughput manner [35, 36]. CellProfiler pipelines need both nuclear and mitochondrial information to determine mutual relationships. Thus, two dyes (one for nuclei and the other for mitochondria) are needed for bioimaging. CellProfiler pipelines are optimized for small and round mitochondria and less effective for extensive mitochondrial networks.
Reis et al. [36] developed a fluorescent image analysis pipeline based on CellProfiler that extracts mitochondrial morphological features from microscopic 3D images of human MCF-7 breast cancer cells. This pipeline classifies mitochondria into networked, fragmented, and swollen types using the random forest method, and Zernike [37] binary information and the mitochondrial area and shape are the most relevant features. The detailed steps are described in Table 2 and the Supplementary Material.
TABLE 2
| Stage | Name(s) | Descriptions |
|---|---|---|
| Before the pipelines | softWoRxTM | An external tool to deconvolute 3D images |
| Preprocessing | – Identify prim automatic | Finding nuclei and cell locations |
| – Identify secondary | ||
| – Rescale intensity | Adjusting pixel intensities for mitochondria to accommodate uneven illumination in the images | |
| – Apply threshold | ||
| – Correct illumination calculate | ||
| – Correct illumination apply | ||
| – Smooth or enhance | ||
| Mitochondrial segmentation | – Identify prim automatic with otsu adaptive threshold | Finding mitochondrial locations |
| Mitochondrial profiling | – Measure object area shape | Calculating mitochondrial morphologies; relating mitochondria and their owner cells |
| – Measure object neighbors | ||
| – Relate objects |
List of tools and functions in the CellProfiler pipeline developed by Reis et al. [36].
Rees et al. [35] studied the SN4741 mouse midbrain neuronal cell line as an in vitro model of Parkinson’s disease by adding the mitochondrial complex I inhibitor rotenone to cause mitochondrial fragmentation. These researchers used CellProfiler to analyze microscopic images and compare the number of mitochondrial puncta and small and round mitochondria. They also provide a comprehensive description of the steps, settings, and parameters for implementing and reproducing the protocols. The detailed steps are described in Table 3 and the Supplementary Material.
TABLE 3
| Stage | Name(s) | Descriptions |
|---|---|---|
| Preprocessing | – Correct illumination calculate | Adjusting pixel intensities to accommodate uneven illumination in the images |
| – Correct illumination apply | ||
| Segmentation | – Identify primary/secondary/tertiary objects | Finding nuclei, cell bodies, and cytoplasm locations |
| Image enhancement | – Enhance or suppress feature | Enhancing mitochondrial speckle signals |
| Mitochondrial segmentation | – Identify primary objects with Otsu thresholding “Per Object” | Marking mitochondrial areas |
| Mitochondrial morphology | – Measure object size and shape | Finding mitochondrial morphologies; relating mitochondria and their owner cells |
| – Measure object intensity | ||
| – Relate objects |
List of tools and functions in the CellProfiler pipeline developed by Rees et al. [35].
MitoGraph
MitoGraph [38, 39] is an open-source automated platform for analyzing 3D mitochondrial morphologies from fluorescent images. It is written in C++ and includes comprehensive documentation and examples. MitoGraph processes 3D bioimages and turns mitochondrial networks into surfaces and node-and-edge structures (skeletons). The outputs could be further analyzed by other programs, such as R scripts, and visualized using ParaView and MayaVi. Although MitoGraph was initially designed for mitochondria in budding yeast [39, 40], MitoGraph has been used for images of mitochondria in various mammalian cell lines, including HMEC-1 cells (human microvascular endothelial cells), C2C12 cells (mouse myoblasts), HEK29d and 293T cells (human embryonic kidney cells and their derived mutant cells containing the SV40 T-antigen), C6 (rat glial) cells, and H1299 (human non-small-cell lung carcinoma) cells. The detailed steps are described in Table 4 and the Supplementary Material.
TABLE 4
| Stage | Name(s) | Descriptions |
|---|---|---|
| Preprocessing | – GenerateFramesMaxProj.ijm | Marking regions of interest (ROIs) using maximum intensity projections as boundary references |
| – CropCells.ijm | Cropping image according to ROIs | |
| Mitochondrial morphology | Skeletonization by MitoGraph | Converting mitochondrial images to graph-based networks |
Another study [41] also used MitoGraph to analyze the mitochondrial network structures (such as loops), complexity, and connectivity from wild-type and Δdnm1Δfzo1 (both fission- and fusion-inhibited) budding yeasts. The authors concluded that the wild-type mitochondrial networks were more evenly distributed than their Δdnm1Δfzo1 counterparts for better communication efficiency.
ImageJ/Fiji and Java-Based Software
ImageJ [42, 43] and its battery-included Fiji distribution [44] are the software programs of choice for microscopy image processing and analysis. Written in Java, ImageJ is a cross-platform tool available for Linux, Mac OS, and Windows. Due to its open-source nature, many plugins, macros, and pipelines have been developed for ImageJ. MiNA (Mitochondrial Network Analysis) [45] and Mitochondria Analyzer [46] are two prime examples of ImageJ-based image analysis pipelines for studying the mitochondrial morphology.
Mitochondrial Network Analysis
MiNA [45] provides simplified, free, and user-friendly analysis methods for 2D mitochondrial morphologies, including puncta, rods, and branched networks. MiNA focuses on 2D image processing and is suitable for images of flat mammalian cells such as C2C12 cells, SH-SY5Y cells, and mouse embryonic fibroblasts (MEFs). MiNA may produce skewed results for thicker specimens. After analysis, MiNA yields mitochondrial network parameters such as the mean branch length, number of individuals and networks, and mitochondrial footprint (i.e., total area). The detailed steps of MiNA are described in Table 5 and the Supplementary Material.
TABLE 5
| Stage | Name(s) | Descriptions |
|---|---|---|
| Preprocessing | – Unsharp mask | Enhancing the edge and sharpening the images without changing contrast |
| – Enhance local contrast (CLAHE) | Equalizing histograms locally with limited ranges of changes to avoid overamplifying noise | |
| – Median filters | Eliminating salt-and-pepper noise | |
| Thresholding | – IsoData algorithm | Converting to black-and-white (binary) images |
| Mitochondrial morphology | – Skeletonization | Categorizing mitochondrial morphologies into networks and individuals |
| – Categorization |
List of tools and functions in the MiNA [45] pipeline.
Mitochondria Analyzer
Mitochondria Analyzer is another ImageJ pipeline and plugin for analyzing fluorescent 2D, 3D, and 4D (3D time-series) images of mitochondria [46]. Mitochondria Analyzer has been used to demonstrate mitochondrial fragmentation in both primary mouse β-cells under glucose stimulation and the pancreatic beta-cell line MIN6 under palmitate treatment. The mitochondria analyzer revealed that the experimental groups exhibit a higher mitochondrial count, a lower average area, and a shorter branch length. The detailed steps of Mitochondria Analyzer are described in Table 6 and the Supplementary Material.
TABLE 6
| Stage | Name(s) | Descriptions |
|---|---|---|
| Preprocessing | – DcceonvolutionLab2 | For deconvolution of z-stack (3D) images |
| – Subtract background | Subtracting the background pixel intensity with the Rolling Ball Algorithm [88] | |
| – Sigma filter plus | Enhancing local contrast | |
| – Gamma correction | Enhancing faint objects | |
| Thresholding | – Weighted mean (adaptive), mean, median, or MidGray methods | Converting to black-and-white (binary) images |
| Weighted mean (adaptive) produces the best results | ||
| Noise reduction | – Despeckle | Removing residual noises |
| – Remove outliers | ||
| Mitochondrial morphology | – Skeletonize (2D/3D) | Converting mitochondrial images to graph-based networks |
| Analysis | – Analyze particles | Mitochondrial count, area, perimeter, form factor, and aspect ratio from 2D images |
| – 3D object counter | Mitochondrial count, volume, surface area, and sphericity from 3D images | |
| – 3D Particle analyzer (from MorphoLibJ package) | ||
| – Analyze skeleton (2D/3D) | Branches, branch junctions, branch length, and mean branch length from 2D/3D images | |
| Visualization | – 3D Viewer | Visualization of 3D images |
| – Volume Viewer |
List of tools and functions in the Mitochondria Analyzer [46] pipeline.
ImageJ Pipeline Reported by Chustecki’s ImageJ Pipeline
The ImageJ pipeline developed by Chustecki et al. [47] is a novel approach for analyzing mitochondrial dynamics using time-series bioimages. This pipeline has been used to reveal the mitochondrial spatial distribution, moving speed, connectivity, and interactions between mitochondria in the plant Arabidopsis thaliana. The pipeline is suitable for separated and punctate-like mitochondrial images such as those in A. thaliana. The detailed steps are described in Table 7 and the Supplementary Material.
TABLE 7
| Stage | Name(s) | Descriptions |
|---|---|---|
| Analysis | – TrackMate | To detect and track the dynamic behaviors in time-series fluorescent images |
| – Mosaic ParticleTracker |
List of tools and functions in the ImageJ pipeline by Chustecki et al. [47].
Momito
Momito [48] is a Java-based tool for automatic 2D mitochondrial analysis from confocal microscopy images. Momito uses a probabilistic approach to compare two mitochondrial morphologies under different conditions and thus more accurately determine their differences. Momito can manage undesired image artifacts such as overlapping mitochondria by considering possible connection patterns to produce desirable mitochondrial network skeleton configurations. Momito produces comprehensive results for morphological analysis, such as the numbers of ends, tubules, junctions, and clusters, to quantify the mitochondrial network connectivity and distributions. The detailed steps of the Momito pipeline are described in Table 8 and the Supplementary Material.
TABLE 8
| Stage | Name(s) | Descriptions |
|---|---|---|
| Thresholding | – Median smoothing | Converting to black-and-white (binary) images |
| – Contrast enhancement | ||
| – Auto local threshold | ||
| Noise reduction | – Manual global threshold | Removing low-intensity noises |
| – Multiplying pixels from two images | ||
| – Erosion | ||
| Mitochondrial morphology | – Skeletonize | Converting mitochondrial images to graph-based networks and quantifying the mitochondrial network connectivity |
| – MitoMo algorithm | ||
| Analysis | – MitoMo algorithm | Mitochondrial area, count, length, and connectivity |
| – Data consolidation | Compiling data obtained from the analysis and comparing the distributions | |
| – EMD comparison |
List of tools and functions in the Momito [48] pipeline.
MATLAB-Based Software
As a programming platform used in matrix operations, graphs and visualization, MATLAB is a common analysis tool for images of mitochondria. Here, we review several methods that utilizing MATLAB to study mitochondrial morphologies.
Mitocluster
Zamponi et al. [49] published Mitocluster, a MATLAB-based image analysis pipeline to analyze mitochondrial networks as skeletons and elucidate the mitochondrial cluster mass and pixel connectivity. Sukhorukov et al. [51] used Mitocluster with bioimage data to estimate fission and fusion frequencies and developed a computational model to simulate mitochondrial dynamics events in a graph-based manner. The detailed steps of the Mitocluster pipeline are described in Table 9 and the Supplementary Material.
TABLE 9
| Stage | Name(s) | Descriptions |
|---|---|---|
| Thresholding | – im2bw | Conversion to black-and-white (binary) images |
| Mitochondrial morphology | – bimorph | Converting binary images to graph-based networks and finding mitochondrial network clusters |
| – bwlabel | ||
| Analysis | – Graph-based analysis of the mitochondrial network | Degree of each connecting point |
List of tools and functions in the Mitocluster [49] pipeline.
Mitometer
Mitometer [50] analyzes the mitochondrial morphology and fission-fusion dynamics from time-series 2D and 3D microscopic TIFF images. Mitometer not only measures morphological features such as the mitochondrial sizes and shapes but also dynamic parameters such as movement speeds and fission/fusion rates. The detailed steps of the Mitometer pipeline are described in Table 10 and the Supplementary Material.
TABLE 10
| Stage | Name(s) | Descriptions |
|---|---|---|
| Preprocessing | – Diffuse background removal algorithm | Removing background noise and smoothing the image |
| – Gaussian filter | ||
| Thresholding | – Otsu’s threshold | Making a binary mask for mitochondrial segmentation |
| Mitochondrial morphology | – Frame-by-frame global assignment tracking algorithm | Tracking mitochondrial movements and fission/fusion events |
| Analysis | – Morphological parameters | Mitochondrial areas (volumes), solidity, and perimeters |
| – Dynamics parameters | Mitochondrial fission/fusion rates, movement speed, and distance between mitochondria |
List of tools and functions in the Mitometer [50] pipeline.
MitoWave
MitoWave [51], which is based on the MATLAB programming language and Fiji image analysis platform, can be used to study mitochondrial inner membrane potential (ΔΨm) fluctuations from 2D time-series microscopic images. MitoWave was developed to measure the ΔΨm oscillation patterns in neonatal mouse ventricular myocytes (NMVMs) undergoing ischemia/reperfusion (I/R) episodes. The developers classified the reperfusion ΔΨm patterns into five distinct types and compared the ΔΨm transition time points during ischemia, the predominant frequencies of mitochondrial clusters, and the size of mitochondrial clusters. These researchers found that the oscillation frequency of the membrane potential is inversely correlated with the area of mitochondrial clusters. The detailed steps of the MitoWave pipeline are described in Table 11 and the Supplementary Material.
TABLE 11
| Stage | Name(s) | Descriptions |
|---|---|---|
| Preprocessing | – StackReg | Aligning time-series images |
| Thresholding | – Median filter | Making masks of mitochondria |
| – Enhance contrast | ||
| – Auto local threshold (Niblack) | ||
| Mitochondrial morphology | – Analyze Particles | Finding mitochondrial particles; those with a radius less than 60 pixels are omitted |
| Analysis | – Intensity and oscillation analysis using wavelet transform algorithms | Calculating membrane potential oscillations using wavelet transform |
List of tools and functions in the MitoWave [51] pipeline.
Mytoe
Mytoe [52] is a MATLAB-based tool for the analysis of the mitochondrial morphology, dynamics, and motion from 2D time-series images. This tool uses optical flow (OF) estimation to obtain mitochondrial dynamics parameters, including the speed, moving direction, and wiggling ratio of each branch. Another study [29] combined Mytoe and fractal analysis algorithms to study the drug response of the mitochondrial network in the DMS273 small cell lung carcinoma (SCLC) cell line compared with that of a nontumorigenic control lung cell line (BEAS-2B). The detailed steps of the MitoWave pipeline are described in Table 12 and the Supplementary Material.
TABLE 12
| Stage | Name(s) | Descriptions |
|---|---|---|
| Preprocessing | – Median filter | Removing background noises |
| – Top-hat filter | ||
| Thresholding | – Otsu’s algorithm | Obtaining binary masks |
| Mitochondrial morphology | – Thinning procedure with two iterations | Converting to skeletal images |
| Analysis | – Morphological parameters | Cell area, mitochondrial area, and mitochondrial count |
| – Optical flow (OF) estimation | Motion estimation: speed, moving direction, and wiggle ratio of each branch | |
| – MitoMorF and FracLac | Fractal analysis of the mitochondrial morphology and dynamics [29] |
List of tools and functions in the Mytoe [52] pipeline.
MitoSPT
MitoSPT [53] is an easy-to-use, robust, and computationally inexpensive MATLAB package that quantifies the distribution of mitochondrial motility in time-series 2D images. MitoSPT was used by its developers to study adult human dermal fibroblasts [53], and the results showed that the net distances traveled by individual mitochondria followed a lognormal distribution and were not directly correlated with their spatial locations in the cell. The detailed steps of the MitoWave pipeline are described in Table 13 and the Supplementary Material.
TABLE 13
| Stage | Name(s) | Descriptions |
|---|---|---|
| Preprocessing | – Convolution | Provided by the MitoSPT pipeline |
| – Fast fourier transform (FFT) | ||
| – Filtering | ||
| Thresholding | – Manually | Obtaining binary (black-and-white) images |
| Mitochondrial morphology and tracking | – bwconncomp | Tracking centroid locations of all identified mitochondrial objects in time-series images |
| – regionprops |
List of tools and functions in the MitoSPT [53] pipeline.
Machine Learning and Deep Learning for Mitochondrial Bioimaging Analysis
Over the past few years, machine learning (ML) and deep learning (DL) have achieved tremendous success in computer vision and have outperformed other conventional image processing methods. Modern microscopes are often connected to digital systems, and the acquired microscopy images must be processed using computational methods. As a result, scientists have developed novel image analysis methods, and one of the primary approaches involves the application of DL to microscopy data. Since then, several DL methods have shown strong potential for the analysis of microscopy images, including U-Net [54] and UNet++ [55] for image segmentation. DeepImageJ provides access to pretrained deep learning models to perform common bioimage processing tasks, including classification, segmentation, denoising, and virtual staining [56].
The analysis task most commonly applied to microscopy images is the detection and segmentation of objects such as nuclei, mitochondria, and other organelles. After segmentation, scientists can further measure the morphology and properties of segmented organelles. The traditional segmentation method is based on human visual inspection, which requires professional knowledge and is time-consuming. Moreover, manual completion of the segmentation task is rarely possible for large datasets. Therefore, the use of deep learning technology to aid the completion of these tasks appears to be a reasonable approach. For example, Ounkomol et al. [57] trained a DL model using transmitted light images as the input and successfully predicted fluorescence images. Falk et al. [58] adopted U-Net for microscopy image analysis to count and detect cells. Another primary microscopy image analysis using deep learning was designed to improve the quality of images or to reconstruct the images. Several DL algorithms, such as CARE [59] and Noise2Void [60], were developed to recover high-resolution images from low-intensity, noisy, and low-quality images. In addition to restoration, other DL algorithms assist with the postprocessing steps used in super-resolution microscopy techniques (e.g., DeepSTORM [61]).
As mentioned above, the segmentation of organelles and the analysis of mitochondrial morphology have become essential tools for studying mitochondrial function. Traditionally, mitochondria exhibit complex movements and morphology, which impedes the construction of a robust system for analyzing mitochondrial images. Several DL-based tools have been developed to solve problems related to mitochondrial segmentation. For instance, Fischer et al. [62] developed MitoSegNet, a toolbox that quantifies the mitochondrial morphology using the deep learning segmentation tool and the morphological analysis tool. Chai et al. [63] implemented mitochondrial segmentation with fluorescence microscopy images using a fully convolutional network (FCN) and U-Net. U-Net has also been utilized to segment mitochondria from label-free phase-contrast microscopy images [64] or brightfield images [65]. Classification is another crucial application in the analysis of bioimages of mitochondria. Recently, Collier and Taylor [66] trained a supervised machine learning model to classify the mitochondrial morphologies using known mitochondrial phenotypes, including fragmented, normal, and hypertubular phenotypes. MitoMo [67] with a MATLAB GUI automatically segments and classifies the mitochondrial morphology using the K-nearest neighbor (KNN) and naïve Bayesian methods. MitoMo also provides pixel-based methods for analyzing the motion of molecules, individual mitochondria, and classes of mitochondria. Fogo et al. [68] implemented decision tree algorithms in R to classify mitochondria in primary neurons according to their size and geometric descriptors. Leonard et al. [69] used the conditional inference algorithm in R to categorize individual mitochondrial subtypes into four morphological bins (puncta, rod, network, and large and round). MitoHacker [70] provides a set of tools with Jupyter notebook semigraphical user interfaces, including Cell Catcher, Mito Catcher, and MiA, to process cellular images, extract mitochondrial networks, and perform morphometric measurements. Iqbal et al. [71] built a convolutional neural network-based framework called mitochondrial organelle movement classification (MOMC) to classify mitochondrial movements according to the mitochondrial shape, positions, and fission/fusion events. GoogLeNet-22, ResNet-50, and an inception model extract low-level, middle-level, and high-level features to differentiate mitochondrial organelle positions.
To summarize, the image analysis tasks are classified into four categories with the available ML/DL tools based on their specific applications: 1) Mitochondria Segmentation: U-Net is the most commonly used DL architecture that can be implemented for mitochondria segmentation. MitoSegNet [63] and MitoHacker [71] also provide toolboxes for mitochondria segmentation tasks. 2) Mitochondria Classification: MitoMo [68] provides the toolbox that uses ML methods to classify mitochondria. Fogo et al. [69] and Leonard et al. [70] also provide guidelines for mitochondrial classification using ML algorithms. 3) Motion analysis: MOMC [72] is a DL-based algorithm used for mitochondrial movement classification. MitoMo [68] can also be used for movement detection. 4) Image Restoration: CARE [60] and Noise2Void [61] are algorithms developed for recovering high-resolution images from low-quality microscopy images.
Discussion
Mitochondria adjust their morphologies and functions in response to biological or metabolic signals in the cellular environment, including changes in tumor microenvironments. Close relationships between mitochondrial dynamics in cancer cells and diseases have been revealed. Mitochondrial dynamics also play crucial roles in cancer cell metastasis, metabolic rewiring, drug resistance, and cancer stem cell survival. The structural alterations in human tumor mitochondria are heterogeneous and not specifically correlated with any neoplasms.
Fluorescence microscopy provides direct observations of mitochondrial morphologies in a snapshot and fission-fusion dynamics in a time series. Image analysis pipelines are studied procedures that facilitate automation to reduce menial work and increase throughput. After analyses using the image analysis pipelines, quantitative morphological indicators can be extracted and provide clues for understanding mitochondrial profiles and network morphology, such as whether the mitochondrial network is network-like or fragmented under certain conditions. These indicators quantify the morphological features of mitochondria and describe the relationship between the value of each indicator and the morphology indicator in detail.
Several mitochondria analysis tools mentioned above have been tested using confocal microscopy images of mitochondria (Figure 1). Each method has its advantages and disadvantages. For example, CellProfiler provides flexible and versatile methods and algorithms with a user-friendly interface but lacks 3D pipeline references for mitochondrial image analysis. MitoGraph provides precise 3D skeleton results and visualization with ParaView but is not available for Windows systems and has limited thresholding methods for different cell lines. MiNA uses fast and adjustable ImageJ pipelines but works only for 2D images focusing on skeleton network analysis and provides fewer mitochondrial indicators than other software packages. We summarize a flowchart to help select the appropriate tool for analyzing mitochondrial images (Figure 2). Notably, preprocessing and thresholding parameters should be optimized in each software program based on the specific cell type to obtain more accurate results.
FIGURE 1
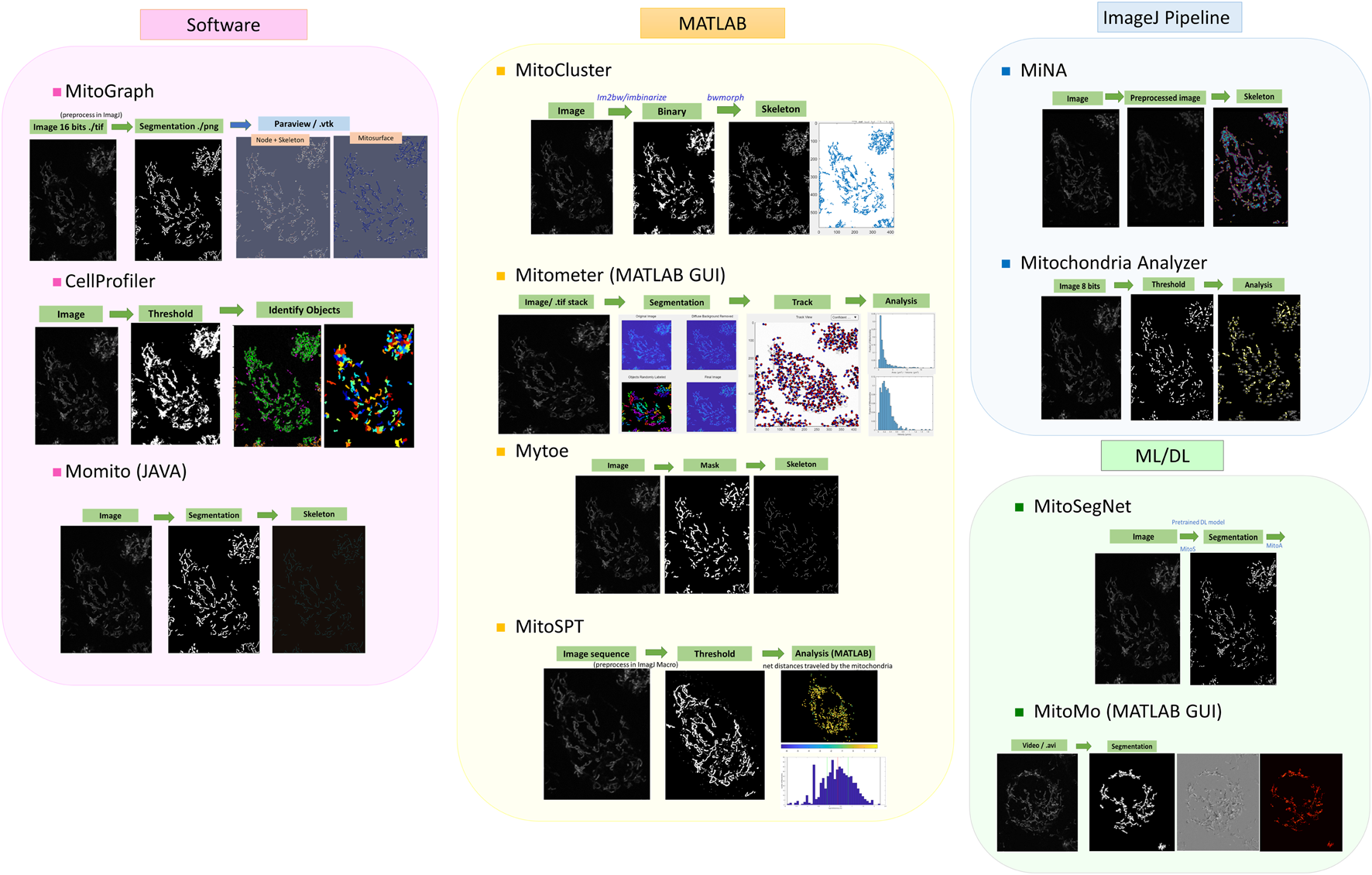
Examples of published tools for mitochondrial image analysis. The image analysis pipelines are roughly classified according to the programming languages, although some software programs were developed with mixtures of programming languages and tools. The images are TMRM-fluorescent-labeled mitochondria obtained from PANC-1 cell lines.
FIGURE 2
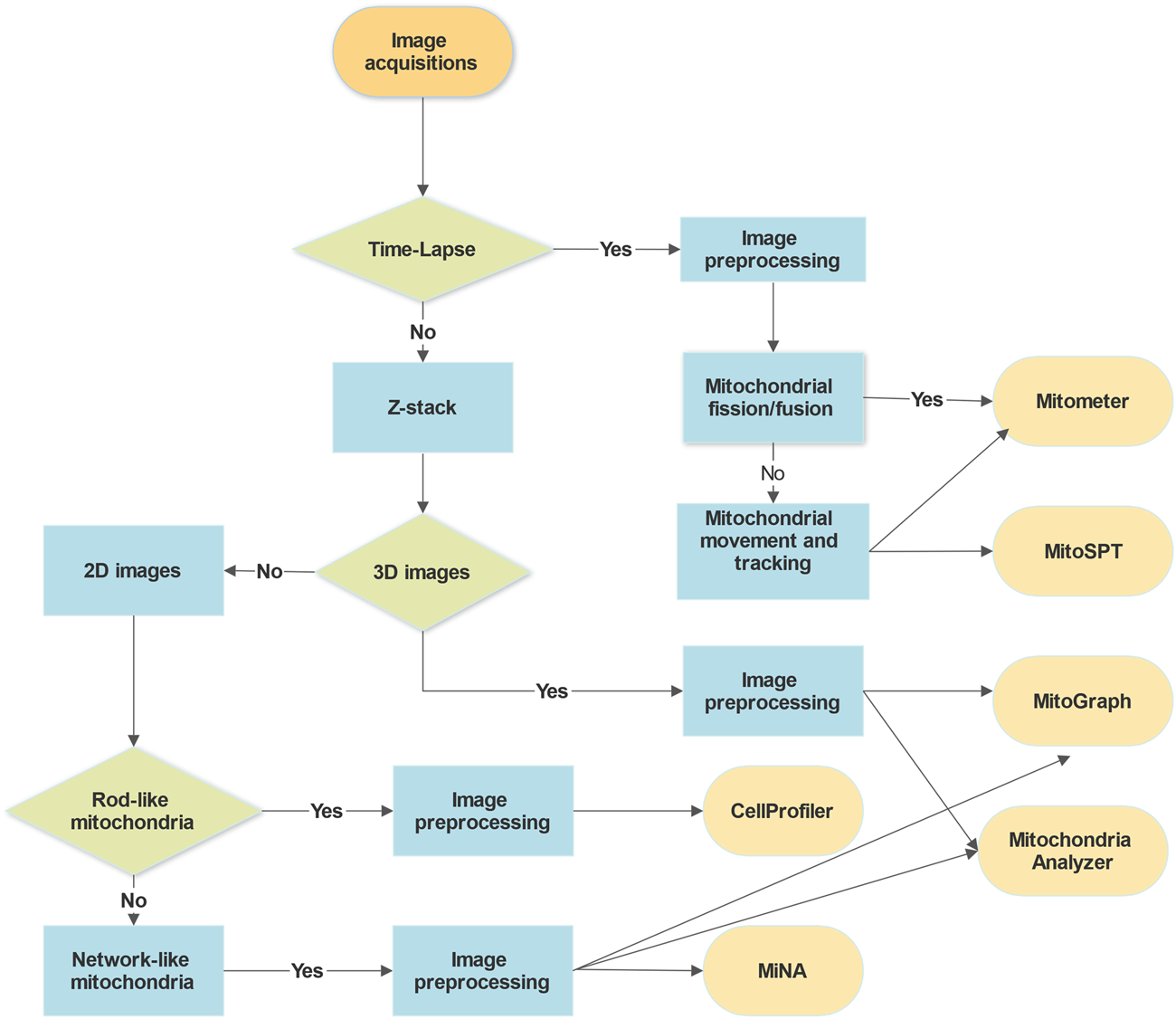
Decision flow chart for the selection of image analysis tools. The choice of analysis tools for mitochondrial microscopic images depends mainly on the imaging acquisition methods and types, the mitochondrial morphologies and cell types, and the purposes of the analysis. The requirements for image preprocessing, adjusting parameters, and programming languages of the software might be other things to consider.
Cancer cells are diverse and display heterogeneity in different types of tissue, cancer stages, and tumor positions. To illustrate, we used our own mitochondrial image analysis pipeline [72] adapted from Mitochondria Analyzer to test cancerous and noncancerous cell lines and compare differences in the mitochondrial size, amount, shape, and network structures in 2D (Figure 3). The mitochondrial morphologies and network structures vary among different types of cells and tissues depending on the cellular functions.
FIGURE 3
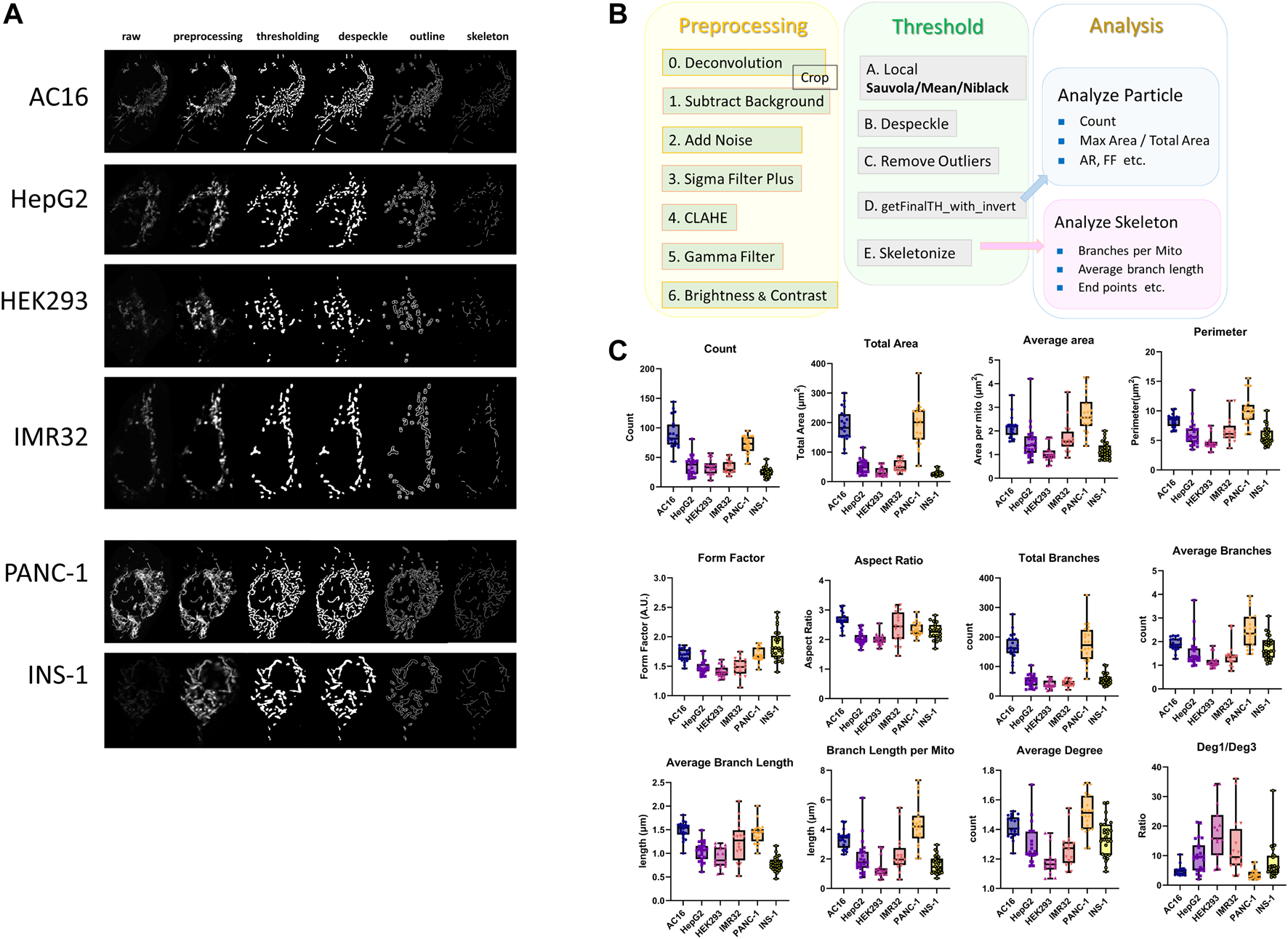
Comparison of mitochondrial morphologies between noncancerous and cancerous cell lines. (A) Fluorescence microscopy images of the AC16 human cardiomyocyte, HepG2 human liver hepatocellular carcinoma, HEK293 human embryonic kidney, IMR32 neuroblast, PANC-1 human pancreatic carcinoma, and INS-1 rat insulinoma cell lines. Mitochondria were labeled with TMRM and imaged with a Zeiss LSM800 laser-scanning confocal microscope and a 1.40-NA 63× objective. (B) Workflow of the 2D image analysis pipeline for confocal images of mitochondria [74], which was adapted from the Mitochondria Analyzer [35] ImageJ pipeline. The image analysis pipeline is divided into three steps: preprocessing, thresholding, and analysis. In the preprocessing step, images are first deconvoluted using ZEISS ZEN 2 (Blue Edition) software, and the ImageJ commands “Subtract Background”, “Add Noise” (optional), “Sigma Filter Plus”, “CLAHE”, “Gamma” and “Brightness and Contrast” are then applied to reduce the noise, increase the contrast, and make the images appropriate and ready for thresholding. In the thresholding step, the local adaptive threshold algorithm “Sauvola”, “Mean” or “Niblack” is applied to the preprocessed images obtained from the previous step in most cases. After thresholding, the ImageJ commands “Despeckle” and “Remove Outliers” are applied to remove the salt-and-pepper noise, and the ImageJ macro “getFinalTH_with_invert”, which fills the tiny holes regarded as artifactual holes due to noise in the images, is then applied. After thresholding and noise removal, the binary images are converted into skeleton images with one-pixel-width lines representing the network frame of mitochondria using the “Skeletonize” ImageJ command. Branches with lengths less than the threshold are removed from the skeleton images using the image macro “Prune” prior to the skeleton analysis if serious overskeletonization occurs. For skeleton images, “Analyze Skeleton” is used to extract and calculate network information such as “Total Branches”, “Average Branch Length”, “Total Deg 1 Points” and “Average Degree.” After completing the binarization and skeletonization steps, “Analyze Particles” is applied to the binary images to perform particle analysis and obtain information on indicators such as “Count”, “Total Area”, “Perimeter” and “Form Factor” that describe the features related to the number and profile of mitochondria. (C) Boxplots of the 2D particle analysis and skeleton analysis of mitochondrial networks in six different cell lines. The analysis feature “Count” is the number of total mitochondria in one cell. “Total area” measures the total area of the mitochondria in one cell, and “Average area” denotes the average mitochondrial area in a cell. “Perimeter” measures the average length of the mitochondrial boundary in a cell. “Form factor” is defined as the perimeter 2/(4π*area), and “Aspect ratio” is the length of the major axis divided by the length of the minor axis. “Total branches” represents the number of total edges of the mitochondrial network in one cell, and “Average branches” is defined as the average number of branches per mitochondrion in a particular cell. “Average branch length” is the mean value of the average length of branches of each mitochondrion in a cell. “Branch length per mito” is the mean value of the total length of branches of each mitochondrion in a cell. “Average degree” is defined as the mean value of the average degree of each separated mitochondrion as connected components in cells. “Deg. 1/Deg. 3” is the ratio of total degree 1 nodes to total degree three nodes in the mitochondrial network.
Determining the most appropriate tools to utilize among all image analysis sources according to the image features and functional needs may make image analysis more efficient and effective. Some image analysis tools are open-source and freely available, whereas others, such as MATLAB, Image-Pro Plus, and Imaris, are proprietary image analysis programs that may require a paid license. The next steps are to encourage further development and ensure that studies are more reproducible, although several challenges remain. First, the mitochondrial sizes and morphologies are heterogeneous [73, 74] due to the use of diverse cell types, different cultures and various imaging parameters. Variations even exist within the population from the same tumor [23]. The processing pipeline might need to be constantly adjusted to accommodate these variations. Second, mitochondria are small (0.5–3 µm) [73, 75, 76], and super-resolution microscopy (higher resolution than traditional confocal microscopy) may be needed to observe their fine structures (e.g., cristae) on a submicron scale. Third, the intense laser beams used to illuminate chromophores might result in phototoxicity and photobleaching [77, 78], which affects the image quality and limits how many slices or snapshots can be obtained from live-cell imaging.
Regarding hardware, novel microscopy techniques with high resolution and a limited laser dosage will be suitable for mitochondrial imaging to address these challenges. For example, light-sheet microscopy illuminates the sample from its side in a sheet-like manner and provides high resolution, fast acquisition, and low photodamage [79, 80]. Super-resolution radial fluctuations (SRRFs) [81–84] enable the super-resolution imaging of live cells without complex hardware by measuring the radial symmetry in each pixel and calculating the location of the fluorophores.
Regarding software, kinetic modeling methods [49, 85–87] have been used to estimate mitochondrial dynamics parameters from limited data. Additionally, machine learning can train models to gather quantitative information and recognize specific patterns, shapes and patterns in mitochondrial images to optimize and accelerate image analysis. Machine learning can also help 1) reconstruct high-resolution data from low-resolution data in both time and space, 2) increase the throughput of image processing pipelines, and 3) classify mitochondria in different cell types or pathological conditions based on features extracted from the mitochondrial image analysis. Bioimage analysis using modern DL methods is faster when the models are trained and ready compared with the analyses performed using the above-mentioned conventional methods. In addition, with a considerable amount of data, the DL methods will exhibit better performance in certain analysis tasks, such as segmentation and classification. Several studies have focused on cell classification methods based on images of mitochondria to detect whether a cell is healthy. For example, USK-net models have been applied to measure the closeness of cells to the healthy or tumorous classes based on the morphology of healthy mitochondria or cancerous mitochondria in electron micrographs [88]. However, DL methods may face serious difficulties in analyzing bioimages without sufficient data and proper data preprocessing, which rarely occurs when using conventional algorithms. Therefore, the selection of using either traditional or DL methods deeply depends on the quality and quantity of mitochondrial bioimages. Another option is combining the strength of traditional image analysis for preprocessing images and ML/DL algorithms for downstream segmentation and classification tasks. Ultimately, image analysis can be accelerated to obtain unbiased and reliable results.
Scientists have been researching the relationship between mitochondria and cancer cells to further explore the potential application in mitochondrial morphology analysis. Cancer cells reprogram metabolism to lead to mitochondrial dysfunction and changes in the mitochondrial morphology. In addition to molecular methods, image-based features are also important in determining a clinical cancer diagnosis. Morphological abnormalities in mitochondria are related to cancer function and the metabolic state; therefore, analyzing descriptors of the mitochondrial morphology, shape, and movement will identify potential indicators for cancerous cells. Overall, image acquisition, software selection, methods for preprocessing, parameters of algorithms, and additional adjustments differ according to the cell lines, cellular conditions, microscope settings, image quality, and the purposes of the analysis, which are all critical factors to consider when developing mitochondrial image analysis tools.
Statements
Author contributions
A-CW, W-WT and C-HC conceived the research. C-HC and A-CW performed the analyses. C-HC, W-WT, C-MH and A-CW wrote the manuscript. A-CW secured funding.
Funding
The work was supported by the Ministry of Science and Technology in Taiwan (Grant MOST110-2636-B-002-017 and MOST 111-2636-B-002-018).
Acknowledgments
We thank Yi-Ju Lee for performing experiments and acquiring confocal images. We thank the National Center for High-performance Computing (NCHC) in Taiwan for providing computational and storage resources.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphy.2022.855775/full#supplementary-material
Abbreviations
DL, Deep learning; IMM, Inner mitochondrial membrane; Mfn, Mitofusin (including Mfn-1 and Mfn-2); Drp-1, Dynamin-related protein 1; ML, Machine learning; Mff, Mitochondrial fission factor; OMM, Outer mitochondrial membrane; Opa-1, Optical atrophy-1 protein; OXPHOS, Oxidative phosphorylation; MiD49 and MiD51, Mitochondrial dynamics proteins of 49 and 51 kDa; TMRM, Tetramethylrhodamine methyl ester perchlorate.
References
1.
MudassarFShenHO’NeillGHauE. Targeting Tumor Hypoxia and Mitochondrial Metabolism with Anti-parasitic Drugs to Improve Radiation Response in High-Grade Gliomas. J Exp Clin Cancer Res (2020) 39:208. 10.1186/s13046-020-01724-6
2.
ShanMDaiDVudemAVarnerJDStroockAD. Multi-scale Computational Study of the Warburg Effect, Reverse Warburg Effect and Glutamine Addiction in Solid Tumors. Plos Comput Biol (2018) 14:e1006584. 10.1371/journal.pcbi.1006584
3.
JiaDLuMJungKHParkJHYuLOnuchicJNet alElucidating Cancer Metabolic Plasticity by Coupling Gene Regulation with Metabolic Pathways. Proc Natl Acad Sci U.S.A (2019) 116:3909–18. 10.1073/pnas.1816391116
4.
JiaDPaudelBBHayfordCEHardemanKNLevineHOnuchicJNet alDrug-Tolerant Idling Melanoma Cells Exhibit Theory-Predicted Metabolic Low-Low Phenotype. Front Oncol (2020) 10:1426. 10.3389/fonc.2020.01426
5.
KangXWangJLiC. Exposing the Underlying Relationship of Cancer Metastasis to Metabolism and Epithelial-Mesenchymal Transitions. iScience (2019) 21:754–72. 10.1016/j.isci.2019.10.060
6.
BolandMLChourasiaAHMacleodKF. Mitochondrial Dysfunction in Cancer. Front Oncol (2013) 3:292. 10.3389/fonc.2013.00292
7.
PorporatoPEFilighedduNPedroJMB-SKroemerGGalluzziL. Mitochondrial Metabolism and Cancer. Cell Res (2018) 28:265–80. 10.1038/cr.2017.155
8.
GaudeEFrezzaC. Defects in Mitochondrial Metabolism and Cancer. Cancer Metab (2014) 2:10. 10.1186/2049-3002-2-10
9.
AshtonTMMcKennaWGKunz-SchughartLAHigginsGS. Oxidative Phosphorylation as an Emerging Target in Cancer Therapy. Clin Cancer Res (2018) 24:2482–90. 10.1158/1078-0432.CCR-17-3070
10.
MaYWangLJiaR. The Role of Mitochondrial Dynamics in Human Cancers. Am J Cancer Res (2020) 10:1278–93.
11.
ChanDC. Mitochondrial Dynamics and its Involvement in Disease. Annu Rev Pathol Mech Dis (2020) 15:235–59. 10.1146/annurev-pathmechdis-012419-032711
12.
LiesaMShirihaiOS. Mitochondrial Dynamics in the Regulation of Nutrient Utilization and Energy Expenditure. Cel Metab (2013) 17:491–506. 10.1016/j.cmet.2013.03.002
13.
ChenHDetmerSAEwaldAJGriffinEEFraserSEChanDC. Mitofusins Mfn1 and Mfn2 Coordinately Regulate Mitochondrial Fusion and Are Essential for Embryonic Development. J Cel Biol (2003) 160:189–200. 10.1083/jcb.200211046
14.
AlaviMVFuhrmannN. Dominant Optic Atrophy, OPA1, and Mitochondrial Quality Control: Understanding Mitochondrial Network Dynamics. Mol Neurodegeneration (2013) 8:32. 10.1186/1750-1326-8-32
15.
SongZChenHFiketMAlexanderCChanDC. OPA1 Processing Controls Mitochondrial Fusion and Is Regulated by mRNA Splicing, Membrane Potential, and Yme1L. J Cel Biol (2007) 178:749–55. 10.1083/jcb.200704110
16.
SongZGhochaniMMcCafferyJMFreyTGChanDC. Mitofusins and OPA1 Mediate Sequential Steps in Mitochondrial Membrane Fusion. MBoC (2009) 20:3525–32. 10.1091/mbc.E09-03-0252
17.
SmirnovaEGriparicLShurlandD-Lvan der BliekAM. Dynamin-related Protein Drp1 Is Required for Mitochondrial Division in Mammalian Cells. MBoC (2001) 12:2245–56. 10.1091/mbc.12.8.2245
18.
SteffenJKoehlerCM. ER-mitochondria Contacts: Actin Dynamics at the ER Control Mitochondrial Fission via Calcium Release. J Cel Biol (2018) 217:15–7. 10.1083/jcb.201711075
19.
LosónOCSongZChenHChanDC. Fis1, Mff, MiD49, and MiD51 Mediate Drp1 Recruitment in Mitochondrial Fission. MBoC (2013) 24:659–67. 10.1091/mbc.E12-10-0721
20.
AdaniyaSMO-UchiJCypressMWKusakariYJhunBS. Posttranslational Modifications of Mitochondrial Fission and Fusion Proteins in Cardiac Physiology and Pathophysiology. Am J Physiology-Cell Physiol (2019) 316:C583–C604. 10.1152/ajpcell.00523.2018
21.
YuRJinSBLendahlUNistérMZhaoJ. Human Fis1 Regulates Mitochondrial Dynamics through Inhibition of the Fusion Machinery. EMBO J (2019) 38. 10.15252/embj.201899748
22.
ChenHChanDC. Mitochondrial Dynamics in Regulating the Unique Phenotypes of Cancer and Stem Cells. Cel Metab (2017) 26:39–48. 10.1016/j.cmet.2017.05.016
23.
GiedtRJFumene FeruglioPPathaniaDYangKSKilcoyneAVinegoniCet alComputational Imaging Reveals Mitochondrial Morphology as a Biomarker of Cancer Phenotype and Drug Response. Sci Rep (2016) 6:32985. 10.1038/srep32985
24.
PendinDFiladiRPizzoP. The Concerted Action of Mitochondrial Dynamics and Positioning: New Characters in Cancer Onset and Progression. Front Oncol (2017) 7:102. 10.3389/fonc.2017.00102
25.
LiLQiRZhangLYuYHouJGuYet alPotential Biomarkers and Targets of Mitochondrial Dynamics. Clin Translational Med (2021) 11:e529. 10.1002/ctm2.529
26.
RothKGMambetsarievIKulkarniPSalgiaR. The Mitochondrion as an Emerging Therapeutic Target in Cancer. Trends Mol Med (2020) 26:119–34. 10.1016/j.molmed.2019.06.009
27.
RafelskiSM. Mitochondrial Network Morphology: Building an Integrative, Geometrical View. BMC Biol (2013) 11:71. 10.1186/1741-7007-11-71
28.
Arismendi-MorilloG. Electron Microscopy Morphology of the Mitochondrial Network in Human Cancer. Int J Biochem Cel Biol (2009) 41:2062–8. 10.1016/j.biocel.2009.02.002
29.
MirzapoiazovaTLiHNathanASrivstavaSNasserMWLennonFet alMonitoring and Determining Mitochondrial Network Parameters in Live Lung Cancer Cells. Jcm (2019) 8:1723. 10.3390/jcm8101723
30.
SamantaSHeYSharmaAKimJPanWYangZet alFluorescent Probes for Nanoscopic Imaging of Mitochondria. Chem (2019) 5:1697–726. 10.1016/j.chempr.2019.03.011
31.
Cottet-RousselleCRonotXLeverveXMayolJ-F. Cytometric Assessment of Mitochondria Using Fluorescent Probes. Cytometry (2011) 79A:405–25. 10.1002/cyto.a.21061
32.
CarpenterAEJonesTRLamprechtMRClarkeCKangIFrimanOet alCellProfiler: Image Analysis Software for Identifying and Quantifying Cell Phenotypes. Genome Biol (2006) 7:R100. 10.1186/gb-2006-7-10-r100
33.
KamentskyLJonesTRFraserABrayM-ALoganDJMaddenKLet alImproved Structure, Function and Compatibility for CellProfiler: Modular High-Throughput Image Analysis Software. Bioinformatics (2011) 27:1179–80. 10.1093/bioinformatics/btr095
34.
McQuinCGoodmanAChernyshevVKamentskyLCiminiBAKarhohsKWet alCellProfiler 3.0: Next-Generation Image Processing for Biology. Plos Biol (2018) 16:e2005970. 10.1371/journal.pbio.2005970
35.
ReesDJRobertsLCarla CarisiMMorganAHBrownMRDaviesJS. Automated Quantification of Mitochondrial Fragmentation in an In Vitro Parkinson's Disease Model. Curr Protoc Neurosci (2020) 94:e105. 10.1002/cpns.105
36.
ReisYBernardo-FauraMRichterDWolfTBrorsBHamacher-BradyAet alMulti-parametric Analysis and Modeling of Relationships between Mitochondrial Morphology and Apoptosis. PLoS One (2012) 7:e28694. 10.1371/journal.pone.0028694
37.
BhatiaABWolfE. On the circle Polynomials of Zernike and Related Orthogonal Sets. Math Proc Camb Phil Soc (1954) 50:40–8. 10.1017/S0305004100029066
38.
HarwigMCVianaMPEgnerJMHarwigJJWidlanskyMERafelskiSMet alMethods for Imaging Mammalian Mitochondrial Morphology: A Prospective on MitoGraph. Anal Biochem (2018) 552:81–99. 10.1016/j.ab.2018.02.022
39.
VianaMPLimSRafelskiSM. Quantifying Mitochondrial Content in Living Cells. Methods Cel Biol (2015) 125:77–93. 10.1016/bs.mcb.2014.10.003
40.
RafelskiSMVianaMPZhangYChanY-HMThornKSYamPet alMitochondrial Network Size Scaling in Budding Yeast. Science (2012) 338:822–4. 10.1126/science.1225720
41.
VianaMPBrownAIMuellerIAGoulCKosloverEFRafelskiSM. Mitochondrial Fission and Fusion Dynamics Generate Efficient, Robust, and Evenly Distributed Network Topologies in Budding Yeast Cells. Cel Syst (2020) 10:287–97. e5. 10.1016/j.cels.2020.02.002
42.
SchneiderCARasbandWSEliceiriKW. NIH Image to ImageJ: 25 Years of Image Analysis. Nat Methods (2012) 9:671–5. 10.1038/nmeth.2089
43.
RuedenCTSchindelinJHinerMCDeZoniaBEWalterAEArenaETet alImageJ2: ImageJ for the Next Generation of Scientific Image Data. BMC Bioinformatics (2017) 18:529. 10.1186/s12859-017-1934-z
44.
SchindelinJArganda-CarrerasIFriseEKaynigVLongairMPietzschTet alFiji: an Open-Source Platform for Biological-Image Analysis. Nat Methods (2012) 9:676–82. 10.1038/nmeth.2019
45.
ValenteAJMaddalenaLARobbELMoradiFStuartJA. A Simple ImageJ Macro Tool for Analyzing Mitochondrial Network Morphology in Mammalian Cell Culture. Acta Histochem (2017) 119:315–26. 10.1016/j.acthis.2017.03.001
46.
ChaudhryAShiRLucianiDS. A Pipeline for Multidimensional Confocal Analysis of Mitochondrial Morphology, Function, and Dynamics in Pancreatic β-cells. Am J Physiology-Endocrinology Metab (2020) 318:E87–E101. 10.1152/ajpendo.00457.2019
47.
ChusteckiJMGibbsDJBasselGWJohnstonIG. Network Analysis of Arabidopsis Mitochondrial Dynamics Reveals a Resolved Tradeoff between Physical Distribution and Social Connectivity. Cel Syst (2021) 12:419–31. e4. 10.1016/j.cels.2021.04.006
48.
OuelletMGuillebaudGGervaisVLupien St-PierreDGermainM. A Novel Algorithm Identifies Stress-Induced Alterations in Mitochondrial Connectivity and Inner Membrane Structure from Confocal Images. Plos Comput Biol (2017) 13:e1005612. 10.1371/journal.pcbi.1005612
49.
ZamponiNZamponiECannasSABilloniOVHelgueraPRChialvoDR. Mitochondrial Network Complexity Emerges from Fission/fusion Dynamics. Sci Rep (2018) 8:363. 10.1038/s41598-017-18351-5
50.
LefebvreAEYTMaDKessenbrockKLawsonDADigmanMA. Automated Segmentation and Tracking of Mitochondria in Live-Cell Time-Lapse Images. Nat Methods (2021) 18:1091–102. 10.1038/s41592-021-01234-z
51.
AshokDO’RourkeB. MitoWave: Spatiotemporal Analysis of Mitochondrial Membrane Potential Fluctuations during I/R. Biophysical J (2021) 120:3261–71. 10.1016/j.bpj.2021.05.033
52.
LihavainenEMäkeläJSpelbrinkJNRibeiroAS. Mytoe: Automatic Analysis of Mitochondrial Dynamics. Bioinformatics (2012) 28:1050–1. 10.1093/bioinformatics/bts073
53.
KandelJChouPEckmannDM. Automated Detection of Whole-Cell Mitochondrial Motility and its Dependence on Cytoarchitectural Integrity. Biotechnol Bioeng (2015) 112:1395–405. 10.1002/bit.25563
54.
RonnebergerOFischerPBroxT. U-net: Convolutional Networks for Biomedical Image Segmentation. In: NavabNHorneggerJWellsWMFrangiAF, editors. Medical Image Computing and Computer-Assisted Intervention (MICCAI) Lecture Notes in Computer Science. Cham: Springer International Publishing (2015). p. 234–41. 10.1007/978-3-319-24574-4_28
55.
ZhouZRahman SiddiqueeMMTajbakhshNLiangJ. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support (2018) 11045:3–11. 10.1007/978-3-030-00889-5_1
56.
Gómez-de-MariscalEGarcía-López-de-HaroCOuyangWDonatiLLundbergEUnserMet alDeepImageJ: A User-Friendly Environment to Run Deep Learning Models in ImageJ. Nat Methods (2021) 18:1192–5. 10.1038/s41592-021-01262-9
57.
OunkomolCSeshamaniSMaleckarMMCollmanFJohnsonGR. Label-free Prediction of Three-Dimensional Fluorescence Images from Transmitted-Light Microscopy. Nat Methods (2018) 15:917–20. 10.1038/s41592-018-0111-2
58.
FalkTMaiDBenschRÇiçekÖAbdulkadirAMarrakchiYet alU-net: Deep Learning for Cell Counting, Detection, and Morphometry. Nat Methods (2019) 16:67–70. 10.1038/s41592-018-0261-2
59.
WeigertMSchmidtUBootheTMüllerADibrovAJainAet alContent-aware Image Restoration: Pushing the Limits of Fluorescence Microscopy. Nat Methods (2018) 15:1090–7. 10.1038/s41592-018-0216-7
60.
KrullABuchholzT-OJugF. Noise2Void - Learning Denoising from Single Noisy Images. In: 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). IEEE (2019). 2132–24. 10.1109/CVPR.2019.00223
61.
NehmeEWeissLEMichaeliTShechtmanY. Deep-STORM: Super-resolution Single-Molecule Microscopy by Deep Learning. Optica (2018) 5:458. 10.1364/OPTICA.5.000458
62.
FischerCABesora-CasalsLRollandSGHaeusslerSSinghKDuchenMet alMitoSegNet: Easy-To-Use Deep Learning Segmentation for Analyzing Mitochondrial Morphology. iScience (2020) 23:101601. 10.1016/j.isci.2020.101601
63.
ChaiXBaQYangG. Characterizing Robustness and Sensitivity of Convolutional Neural Networks for Quantitative Analysis of Mitochondrial Morphology. Quant Biol (2018) 6:344–58. 10.1007/s40484-018-0156-3
64.
GuoSMaYPanYSmithZJChuK. Organelle-specific Phase Contrast Microscopy Enables Gentle Monitoring and Analysis of Mitochondrial Network Dynamics. Biomed Opt Express (2021) 12:4363–79. 10.1364/BOE.425848
65.
HsuC-MLeeY-JWeiA-C. Convolutional Neural Networks Predict Mitochondrial Structures from Label-free Microscopy Images. In: ChangR-F, editor. International Forum on Medical Imaging in Asia 2021. Bellingham, United States: SPIE (2021). p. 39. 10.1117/12.2591089
66.
CollierJJTaylorRW. Machine Learning Algorithms Reveal the Secrets of Mitochondrial Dynamics. EMBO Mol Med (2021) 13:e14316. 10.15252/emmm.202114316
67.
ZahediAOnVPhandthongRChailiARemarkGBhanuBet alDeep Analysis of Mitochondria and Cell Health Using Machine Learning. Sci Rep (2018) 8:16354. 10.1038/s41598-018-34455-y
68.
FogoGMAnzellARMaherasKJRaghunayakulaSWiderJMEmausKJet alMachine Learning-Based Classification of Mitochondrial Morphology in Primary Neurons and Brain. Sci Rep (2021) 11:5133. 10.1038/s41598-021-84528-8
69.
LeonardAPCameronRBSpeiserJLWolfBJPetersonYKSchnellmannRGet alQuantitative Analysis of Mitochondrial Morphology and Membrane Potential in Living Cells Using High-Content Imaging, Machine Learning, and Morphological Binning. Biochim Biophys Acta (Bba) - Mol Cel Res (2015) 1853:348–60. 10.1016/j.bbamcr.2014.11.002
70.
RohaniAKashatusJASessionsDTSharminSKashatusDF. Mito Hacker: a Set of Tools to Enable High-Throughput Analysis of Mitochondrial Network Morphology. Sci Rep (2020) 10:18941. 10.1038/s41598-020-75899-5
71.
IqbalMSEl-AshramSHussainSKhanTHuangSMehmoodRet alEfficient Cell Classification of Mitochondrial Images by Using Deep Learning. J Opt (2019) 48:113–22. 10.1007/s12596-018-0508-4
72.
TsengW-WChuC-HChangCLeeY-JZhaoSHoY-Pet alMetabolic Regulation of Mitochondrial Morphologies in Pancreatic Beta Cells: Bioenergetics-Mitochondrial Dynamics Coupling (2021). BioRxiv. 10.1101/2021.10.04.462897
73.
WiemerslageLLeeD. Quantification of Mitochondrial Morphology in Neurites of Dopaminergic Neurons Using Multiple Parameters. J Neurosci Methods (2016) 262:56–65. 10.1016/j.jneumeth.2016.01.008
74.
Das NevesRPJonesNSAndreuLGuptaREnverTIborraFJ. Connecting Variability in Global Transcription Rate to Mitochondrial Variability. Plos Biol (2010) 8:e1000560. 10.1371/journal.pbio.1000560
75.
PaladeGE. The fine Structure of Mitochondria. Anat Rec (1952) 114:427–51. 10.1002/ar.1091140304
76.
ShamiGJChengDVerhaeghPKoekGWisseEBraetF. Three-dimensional Ultrastructure of Giant Mitochondria in Human Non-alcoholic Fatty Liver Disease. Sci Rep (2021) 11:3319. 10.1038/s41598-021-82884-z
77.
IchaJWeberMWatersJCNordenC. Phototoxicity in Live Fluorescence Microscopy, and How to Avoid it. Bioessays (2017) 39:1700003. 10.1002/bies.201700003
78.
LaissuePPAlghamdiRATomancakPReynaudEGShroffH. Assessing Phototoxicity in Live Fluorescence Imaging. Nat Methods (2017) 14:657–61. 10.1038/nmeth.4344
79.
FaderoTCGerbichTMRanaKSuzukiADiSalvoMSchaeferKNet alLITE Microscopy: Tilted Light-Sheet Excitation of Model Organisms Offers High Resolution and Low Photobleaching. J Cel Biol (2018) 217:1869–82. 10.1083/jcb.201710087
80.
HuiskenJSwogerJDel BeneFWittbrodtJStelzerEHK. Optical Sectioning Deep inside Live Embryos by Selective Plane Illumination Microscopy. Science (2004) 305:1007–9. 10.1126/science.1100035
81.
CulleySToshevaKLMatos PereiraPHenriquesR. SRRF: Universal Live-Cell Super-resolution Microscopy. Int J Biochem Cel Biol (2018) 101:74–9. 10.1016/j.biocel.2018.05.014
82.
HanYLuXZhangZLiuWChenYLiuXet alUltra-fast, Universal Super-resolution Radial Fluctuations (SRRF) Algorithm for Live-Cell Super-resolution Microscopy. Opt Express (2019) 27:38337–48. 10.1364/OE.27.038337
83.
GustafssonNCulleySAshdownGOwenDMPereiraPMHenriquesR. Fast Live-Cell Conventional Fluorophore Nanoscopy with ImageJ through Super-resolution Radial Fluctuations. Nat Commun (2016) 7:12471. 10.1038/ncomms12471
84.
VenkatachalapathyMBelapurkarVJoseMGautierANairD. Live Cell Super Resolution Imaging by Radial Fluctuations Using Fluorogen Binding Tags. Nanoscale (2019) 11:3626–32. 10.1039/c8nr07809b
85.
PatelPKShirihaiOHuangKC. Optimal Dynamics for Quality Control in Spatially Distributed Mitochondrial Networks. Plos Comput Biol (2013) 9:e1003108. 10.1371/journal.pcbi.1003108
86.
ShahSIPaineJGPerezCUllahG. Mitochondrial Fragmentation and Network Architecture in Degenerative Diseases. PLoS One (2019) 14:e0223014. 10.1371/journal.pone.0223014
87.
SukhorukovVMDikovDReichertASMeyer-HermannM. Emergence of the Mitochondrial Reticulum from Fission and Fusion Dynamics. Plos Comput Biol (2012) 8:e1002745. 10.1371/journal.pcbi.1002745
88.
MishraMSchmittSZischkaHStrasserMNavabNMarrCet alQuantifying Structural Heterogeneity of Healthy and Cancerous Mitochondria Using a Combined Segmentation and Classification USK-Net. In: TetkoIVKůrkováVKarpovPTheisF, editors. Artificial neural networks and machine learning – ICANN 2019: workshop and special sessions: 28th international conference on artificial neural networks, munich, germany, september 17–19, 2019, proceedings Lecture notes in computer science; Cham. Springer International Publishing (2019). p. 289–98. 10.1007/978-3-030-30493-5_30
Summary
Keywords
cancer, mitochondrial dynamics, confocal microscopic images, bioimage analysis, machine learning, mitochondrial morphology
Citation
Chu C-H, Tseng W-W, Hsu C-M and Wei A-C (2022) Image Analysis of the Mitochondrial Network Morphology With Applications in Cancer Research. Front. Phys. 10:855775. doi: 10.3389/fphy.2022.855775
Received
15 January 2022
Accepted
21 March 2022
Published
13 April 2022
Volume
10 - 2022
Edited by
Yen-Liang Liu, China Medical University,Taiwan
Reviewed by
Leonel Malacrida, Universidad de la República,Uruguay
Tai-Yen Chen, University of Houston,United States
Updates
Copyright
© 2022 Chu, Tseng, Hsu and Wei.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: An-Chi Wei, acwei86@ntu.edu.tw
†These authors have contributed equally to this work and share first authorship
This article was submitted to Optics and Photonics, a section of the journal Frontiers in Physics
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.