- Department of Biological Sciences, University of Lethbridge, Lethbridge, AB, Canada
Angiosperms do not contain a distinct germline, but rather develop gametes from gametophyte initials that undergo cell division. These gametes contain cells that give rise to an endosperm and the embryo. DNA methylation is decreased in the vegetative nucleus (VN) and central cell nuclei (CCN) resulting in expression of transposable elements (TEs). It is thought that the siRNAs produced in response to TE expression are able to travel to the sperm cells and egg cells (EC) from VN and CCN, respectively, in order to enforce silencing there. Demethylation during gametogenesis helps ensure that even newly integrated TEs are expressed and therefore silenced by the resulting siRNA production. A final form of epigenetic control is modification of histones, which includes accumulation of the H3 variant HTR10 in mature sperm that is then completely replaced following fertilization. In females, the histone isoforms present in the EC and CCN differ, potentially helping to differentiate the two components during gametogenesis.
Introduction
Flowering plants do not contain a distinct germline and instead maintain undifferentiated cells in the sporophyte, which continually produces vegetative tissues and organs. These stem cells serve as the gametophyte initials, which undergo meiosis and give rise to microspores and megaspores. The spores develop into male and female gametophytes that are then responsible for the production of gametes (Yadegari and Drews, 2004; Twell, 2011).
The male gametophyte in Arabidopsis thaliana undergoes only two divisions to generate gametes, the first of which produces a vegetative cell as well as a germ cell. The germ cell then divides a second time. The result is a male gamete that contains two sperm cells (SC) and a vegetative nuclei (VN; Twell, 2011). The female gametophyte consists of four different cell types for a total of seven cells. Critically, it contains the egg cell (EC) that gives rise to the embryo, as well as the two central cell nuclei (CCN) from which the endosperm develops. In addition the female gametophyte contains two accessory cells, synergid cells, which are crucial for pollen tube attraction and three antipodal cells of unknown function (Sundaresan and Alandete-Saez, 2010).
The SCs present in pollen are responsible for a double fertilization event, in which one fuses with the two nuclei of central cell, resulting in a triploid (3n) endosperm nucleus which is a terminally differentiated tissue analogous to the placenta in mammals. The second SC fuses with the EC, producing an embryo that develops into a mature plant (Law and Jacobsen, 2010). We will discuss the epigenetic processes operating in plant gametes including DNA methylation, small RNAs, and histones modifications.
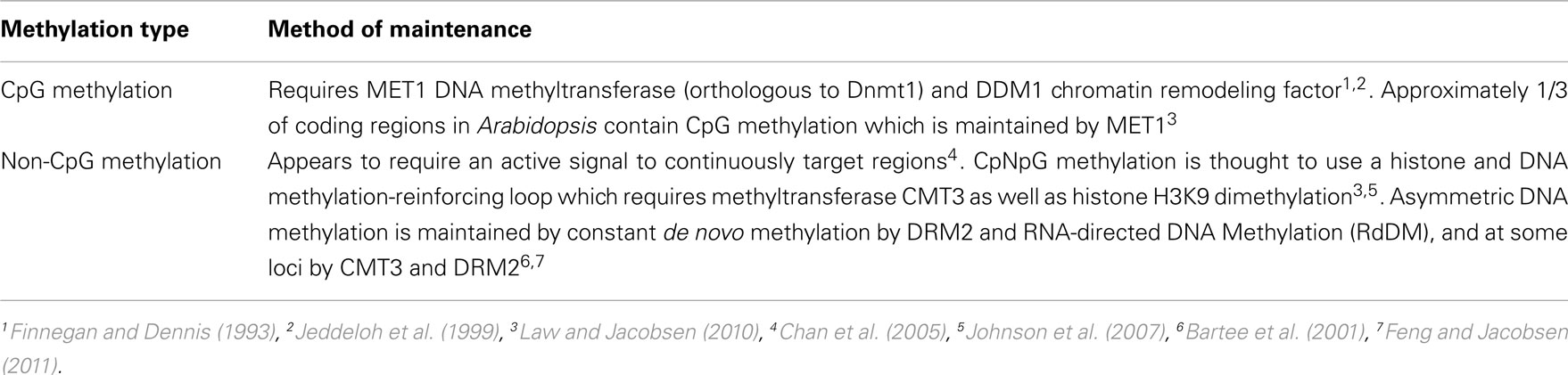
DNA Methylation
De novo methylation of DNA is catalyzed predominately by DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2), and maintained via three different pathways. These pathways are DNA METHYLTRANSFERASE 1 (MET1)-, CHROMOMETHYLASE (CMT3)-, and DRM2-dependent (Chan et al., 2005; Law and Jacobsen, 2010). For details on how maintenance methylation occurs in plants see Table 1.
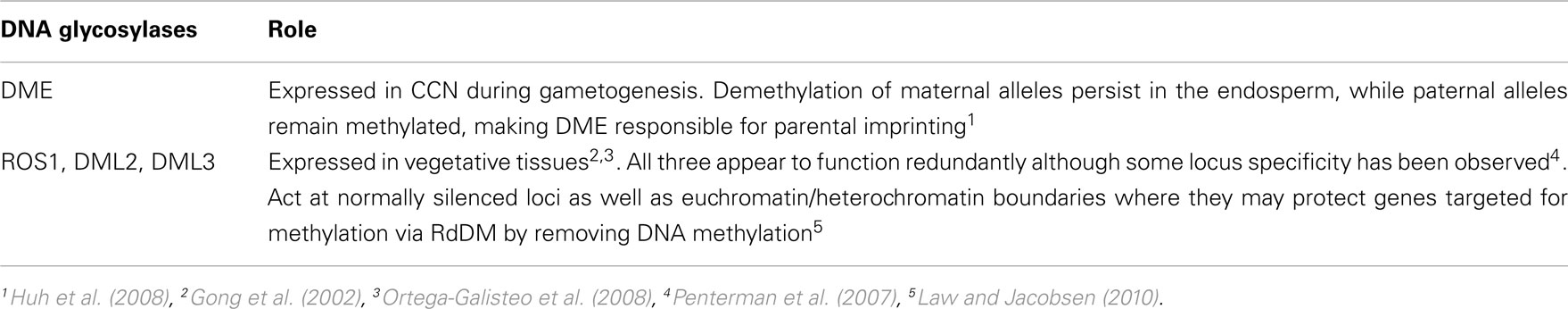
In Arabidopsis, the protein DEMETER (DME) is responsible for active demethylation in reproductive organs. DME is a bifunctional DNA glycosylase/lyase that works with the base excision repair pathway to demethylate DNA by removing 5-methylcytosine and replacing it with cytosine (Choi et al., 2002; Gehring et al., 2006). In addition to active demethylation, as described in Table 2, passive demethylation may also occur.
MULTICOPY SUPPRESSOR of IRA1 (MSI1) transcriptionally represses MET1 during female gametogenesis, a process that also requires RETINOBLASTOMA RELATED 1 (RBR1). MSI1/RBR1-dependent passive demethylation compliments DME-dependent active demethylation (DME) in the activation of imprinted genes (Jullien et al., 2008).
Gene imprinting occurs when the parent of origin impacts the expression level of a particular allele, resulting in differential expression of maternal and paternal (Gehring et al., 2004). The exact number of imprinted genes in plants is unknown, but recent analysis has discovered that more than 200 genes that appear to show imprinting (Gehring et al., 2011; Hsieh et al., 2011; Wolff et al., 2011).
While it is accepted that the majority of loci are only imprinted in the endosperm, there are conflicting viewpoints on the number of genes with parentally biased expression in the embryo. Jahnke and Scholten (2009) found the first reported incident of imprinting in a plant embryo when they described maternal expressed in embryo 1 (mee1) in maize. mee1 is only expressed following fertilization and is unique to the embryo and endosperm. In situ hybridization found mee1 expression throughout the embryo (Jahnke and Scholten, 2009). Recent work by Hsieh et al. (2011) saw no examples of imprinting in the Arabidopsis embryo and while a study by Gehring et al. (2011) found 18 imprinted genes in the embryo, it was concluded that all 18 might have been false positives.
In mammals, imprints are established following reprogramming of the germline during which the methylation status of the genome is reset (Migicovsky and Kovalchuk, 2011). Plants behave very differently, and one possible explanation for this is that control of imprinting in angiosperms such as Arabidopsis is maintained via the endosperm, a tissue that does not contribute to the germline, meaning that there is no need to erase and then apply imprints each generation (Xiao et al., 2003; Scott and Spielman, 2004).
DNA Methylation: Male Gametogenesis
Male gametogenesis in Arabidopsis results in the production of pollen grains containing a VN as well as two SCs (Twell, 2011). Transposons, which are methylated and therefore silent in most tissues, are demethylated and expressed in the VN of the pollen but not in SCs (Table 3; Slotkin et al., 2009). Decreased methylation in pollen is supported by the down regulation of RdDM components as well as the absence of DDM1 in the VN. Reduction of DNA methylation in the VN is likely an active process, as it is separated from SCs by only two cell divisions (Slotkin et al., 2009).
Slotkin et al. (2009) suspected the existence of a DNA demethylase active only in the VN, where it is responsible for removing DNA methylation from some transposable elements (TEs). It is crucial to protect the genome of SCs from transposons but not the genome of the VN, as it does not contribute genetic material to the progeny (Law and Jacobsen, 2010). In addition, an increase of 21 nt siRNAs that do not correspond to transposons expressed in SCs has been found in VN. It is thought that siRNAs produced in the VN may have the ability to travel to SCs, reinforcing silencing (Slotkin et al., 2009). The impact of methylation on TEs is discussed further in Section “Regulation of Transposable Elements Via Small RNAs.”
In addition, the DNA methyltransferase MET1 plays an important role during male gametogenesis in maintaining silencing. This has been demonstrated on genes such as FWA and FIS2, which are only expressed in the maternal genome of the endosperm. The use of maintenance methylation as opposed to de novo makes the establishment of imprints distinct in plants versus animals (Jullien et al., 2006).
DNA Methylation: Female Gametogenesis
In Arabidopsis, paternal imprinting results from demethylation in the CCN prior to fertilization, which combines with a SC to form the endosperm, resulting in maternal-only expression in the endosperm (Hsieh et al., 2009; Law and Jacobsen, 2010). Cases of paternal-only expression may also occur, although there are fewer examples of this (Hsieh et al., 2011). DME is essential in the CCN and seeds with a mutant maternal dme have CpG methylation levels similar to other tissues and do not complete development (Choi et al., 2002; Hsieh et al., 2009, 2011). However, the enzyme disappears following fertilization, resulting in an almost entirely demethylated maternal genome in the endosperm (Hsieh et al., 2009, 2011; Table 3). The end result is that the endosperm inherits two genomes with differing epigenetic states, a necessary requirement for imprinting (Bauer and Fischer, 2011).
So far only one imprinted gene has been found in the embryo, emphasizing that the majority of plant imprinting occurs in the endosperm. In addition, the majority of imprinting results in maternal-only expression, as was seen with 52 of the 65 imprinted genes identified in a recent study by Wolff et al. (2011). Another study by Gehring et al. (2011) found maternal-only expression for 165 of the 208 genes uncovered. Finally, work by Hsieh et al. (2011) found that among 43 imprinted genes, 34 of them were maternally expressed. Even among those that are paternally expressed, maternal demethylation may occur; for example demethylation at the tandem repeats downstream of PHE1 gene allows for PcG-mediated silencing of the maternal allele (Villar et al., 2009; Hsieh et al., 2011).
This maternal demethylation is likely the cause of the hypomethylation seen in endosperm tissue in comparison to the embryo. These findings correspond to previous observations that chromatin in the endosperm is less tightly bound, a characteristic often caused by hypomethylated DNA (Baroux et al., 2007; Gehring et al., 2009). In fact, studying differentially methylated regions (DMRs) in the endosperm and embryo has shown that approximately 90% are more methylated in the embryo. Many of these imprinted regions are those enriched for TEs, which are also the best candidates for imprinting as they show increased expression in the endosperm and low levels of transcription in other parts of the plants (Gehring et al., 2009; Kohler and Weinhofer-Molisch, 2010).
Recent work by Luo et al. (2011) in rice found that only one of 56 imprinted loci occurred in the embryo, and it also showed maternal-only expression in the endosperm. The locus identified was Os10g0750, which is a homolog of Ole e 1, the major allergen in olive and thought to control pollen tube emergence and production (de Dios Alche et al., 2004; Luo et al., 2011). Zemach et al. (2010) showed many genes that are hypomethylated and preferentially expressed in the endosperm are involved in rice endosperm biogenesis, such as starch synthesizing enzymes, indicating the crucial role of DNA methylation on gene expression. In addition, short TEs lost the most CHH methylation in the rice endosperm, while showing hypermethylation in the embryo, further suggesting the enhancement of transposon silencing via the endosperm, similar to what occurs in Arabidopsis (Zemach et al., 2010).
Despite a decrease of methylation in the endosperm, findings by Hsieh et al. (2009) actually indicated increased levels of CpNpN methylation in both endosperm and embryo tissues in comparison to adult plant tissue. They suggested that this might be the result of RdDM activity. In fact, endosperm hypermethylation has been shown to be a highly specific process performed by targeted siRNA (Hsieh et al., 2009). It was suggested that the high levels of maternally derived siRNAs that accumulate in the endosperm as a result of hypomethylation may be generated in the CCN, and possibly responsible for local hypermethylation and silencing in the EC as well as the developing embryo, a process similar to the one described in pollen (Hsieh et al., 2009).
Regulation of Transposable Elements via Small RNAs
Transposable elements are usually heterochromatic and transcriptionally silent, however they play a key role in the structure of the eukaryotic genome. If a transposon is activated the result may be extremely negative for the host, as it could be inserted into important regions of the genome, or support rearrangements causing genomic instability. In response to this possibility, many eukaryotes have developed the ability to target transposons and ensure they are inactivated (Biemont, 2009; Calarco and Martienssen, 2011).
However, TEs do have several important roles in the genome including chromatin formation and centromere function (Chueh et al., 2009; van der Heijen and Bortvin, 2009). This means that although TEs are maintained because of their positive contributions, the fact that they continue to be present in the genome makes further invasion possible, even though genomes may attempt to silence them using epigenetic processes such as small RNAs (Biemont, 2009).
Demethylation during gametogenesis, as described above, may help reveal TEs within the genome that could be expressed and ensure they are silenced. The benefit of this system is that even newly integrated transposons would be expressed, resulting in siRNA production and therefore silencing (Law and Jacobsen, 2010). Work by Teixeira et al. (2009), demonstrated that unlike other regions of the Arabidopsis genome, loci responsible for the production of siRNAs can be re-methylated even when methylation is lost in previous generations (Teixeira et al., 2009). It is possible that reactivation of TEs in surrounding cells which are not a part of the germline allows for improved siRNA silencing in the plant germline, a role suggestive of PIWI proteins in animals. In the animal germ line, piRNAs are responsible for silencing TEs through a mechanism in which active TEs result in significant production of siRNAs that are then responsible for TE inactivation. It is possible siRNAs function similarly in plants (Aravin et al., 2008; Armenta-Medina et al., 2011). The role of small RNAs in silencing TEs clearly indicates their importance for maintaining genome integrity in angiosperm gametes (Mosher and Melnyk, 2010; Le Trionnaire et al., 2011).
Regulation of Transposable Elements via Small RNAs: Male Gametogenesis
Two distinct nuclei exist in pollen, and DNA hypomethylation is seen in the pollen VN but not the SC. SCs maintain high levels of methylation, which prevent TEs from being activated, a mechanism that is reinforced by siRNAs from the VN, which is a terminal tissue (Slotkin et al., 2009). As DDM1 (Table 1) is downregulated in the VN of Arabidopsis, reactivation of TEs occurs. The VN degenerates in the pollen tube prior to the SCs entering the ovule and so the cells responsible for fertilization lack active TEs, resulting in paternal imprinting (Slotkin et al., 2009).
Small RNA pathway components that show a strong increase in SCs include AGO9, DDM1, DRB4, MET1, and SUVH5, and their presence in pollen is likely a result of enrichment in SCs (Borges et al., 2008). Many of the enriched transcripts are involved in RdDM including DDM1, as previously described, and SUVH5, a histone methyltransferase that helps maintain non-CG methylation (Ebbs and Bender, 2006; Borges et al., 2008; Table 3). Although DCL3 was not found to be expressed in SCs, DCL1 was, as well as the AGO1-homolog AGO5. This suggests that although RdDM and maintenance of DNA methylation are important in SCs, they likely occur as a result of a novel small RNA pathway due to the absence of certain important transcripts such as DCL3 (Borges et al., 2008).
Regulation of Transposable Elements via Small RNAs: Female Gametogenesis
Demethylation of the maternal genome in the endosperm supports transposon reactivation while TEs remain silenced in the embryo (Hsieh et al., 2009). However, studying the transcriptome of female gametophytes is much more difficult than it is for males, due to its location and the relative rarity of female gametophytic cells (Le Trionnaire et al., 2011). Still, there is early evidence that similar to males, components of small RNA pathways may be present in female gametophytes. It has been speculated that down regulation of MET1 early during female gametogenesis results in the production of Pol IV-dependent siRNAs, also known as p4 siRNAs (Bourc’his and Voinnet, 2010). However, it is also currently thought that DME-directed genome demethylation is what allows the transcription and production of p4 siRNAs (Mosher and Melnyk, 2010). A recent study by Mosher et al. (2011) indicated that locus 08002 in Arabidopsis does not require differential DNA methylation for maternal-specific expression of p4 siRNAs. More work is still needed to investigate this potential link.
Work in Arabidopsis revealed several different AGO transcripts (AGO1, AGO2, and AGO5), which bind to siRNAs and direct chromatin remodeling, as well as DCL1 transcripts indicating that small RNA pathways are definitely present in the female gametophyte (Wuest et al., 2010; Le Trionnaire et al., 2011). AGO9 plays a crucial role in silencing of transposons in the ovule; ago9 exhibits reactivation of transposons in the ovule (Feng et al., 2010; Olmedo-Monfil et al., 2010). While ago9 mutants can initiate gametophyte development in somatic tissue, their ortholog in maize, ago104 results in the megaspore mother cell producing unreduced gametophytes (Grossniklaus, 2011; Singh et al., 2011). For this reason, AGO104 is required for the production of male and female spores in maize. In fact, ago104 showed reduced methylation at non-CG sites as well as increased transcription of repeats (Singh et al., 2011). Finally, in rice, MEL1, another AGO protein, has a crucial role in sporogenesis that is likely achieved via small RNA-mediated gene silencing (Nonomura et al., 2007).
Two functionally uncharacterized paralogs of PAZ/Piwi-domain encoding genes, AT5G21150 and AT5G21030 have also been uncovered, providing further evidence that small RNAs play an important role in female gametogenesis, potentially protecting against TEs in a manner similar to that which occurs in males (Wuest et al., 2010; Le Trionnaire et al., 2011).
Histone Modifications and Histone Replacement
Histones help control essential processes in the genome such as transcription, replication, chromosome condensation, and segregation, as well as DNA repair (Hauser et al., 2011). Among the modifications of histones, in particular H3 and H4, which help to control gene expression, are acetylation and methylation of histone lysine residues (Hauser et al., 2011). In addition to changes in DNA methylation, imprinted genes in Arabidopsis may be regulated via histone 3 lysine 27 methylation (H3K27me) by Polycomb Group Activity (Jullien and Berger, 2009). Histones can also be replaced by histone variants that may cause changes in expression.
Histone Modifications and Histone Replacement: Male Gametogenesis
Histones are an extremely important form of epigenetic control, and specific histone H3 variants characterize the angiosperm male germline. Despite a single cell division separating the VN from the GN, they show significant differences in chromatin structure, a differentiation that depends on histones (Ueda and Tanaka, 1995).
For example, Lilium longiflorum has been shown to possess three novel histone genes; gH2A, gH2B, and gH3; that only recognize the nuclei of the male gametic (both generative and sperm) cells and appear to be correlated to chromatin structure due to their association with the nucleosome (Ueda and Tanaka, 1995; Ueda et al., 2000). In addition, a gradual decrease in histone H1 levels has been shown to exist in only the VN of Lilium resulting in mature pollen where the VN contains very little histone H1. In comparison, histone H2B is maintained at a similar level in both the VN and GN (Tanaka et al., 1998). It is thought that histone H1 is involved not only in chromatin structure, but also the condensation of chromosomes during the mitotic phase (Woodcock et al., 2006). Therefore it is possible that the decrease of H1 in the VN helps to specify the developmental fate of a cell that, unlike the GN, does not divide again (Tanaka, 1997; Tanaka et al., 1998).
In Arabidopsis, the H3 variant HTR10 is expressed in the germline and mature sperm, but is completely replaced by other H3.3 variants in the zygote upon fertilization (Ingouff et al., 2007; Table 3). The impact of this replacement is not yet fully understood, however it is hypothesized that it is a part of global reprogramming events, similar to the H3.3 replacement that occurs in the mammalian germline. It is unique from DNA methylation reprogramming because it occurs in the zygote, as opposed to being limited to accessory cells in plants (Hajkova et al., 2008; Feng et al., 2010; Twell, 2011). This is likely an active process as paternal HTR10 signals are completely removed from the SC that fertilizes the egg only hours before S phase of the first zygote division, suggesting it occurs in a replication-independent manner (Ingouff et al., 2007; Feng et al., 2010).
Histone Modifications and Histone Replacement: Female Gametogenesis
Histone modifications in female gametes remain significantly less well understood than in males. However, an epigenetic dimorphism between the EC and central cell is known to exist, including the level of specific H3.3 isoforms found in each cell.
Only one H3.3 isoform (HTR5) present in SCs has also been found in ECs, while two – HTR8 and HTR14 – have been found in central cells. This dimorphism appears to be eliminated following fertilization, potentially indicating that it is established in order to distinguish the EC from the CCN during gametogenesis. The resetting of H3 variants in zygote chromatin suggests that these epigenetic marks are not transmitted to progeny. However, it is still possible that some loci may escape remodeling following fertilization, resulting in a maternal imprint, and further work is needed to determine whether or not this occurs (Ingouff et al., 2010; Baroux et al., 2011).
Recent work has provided further evidence of histone modifications resulting in epigenetic dimorphism between EC and CCN late during female gametogenesis in Arabidopsis. EC accumulates high levels of histone H3 lysine 9 dimethylation (H3K9me2) and LIKE HETEROCHROMATIN PROTEIN1 (LHP1) localization in comparison to the CCN. In addition, there is selective depletion of histone H2B in the EC and synergids of mature gametophytes (Pillot et al., 2010). These changes result in differences in transcriptional activity, due to the association of LHP1 with loci enriched in H3K27me3 and of a transcriptionally repressed state (Exner et al., 2009; Pillot et al., 2010).
In maize, variation in histone marks including H3K9ac and H3K4me3, both of which are associated with active transcription states, showed a much higher level of repressed chromatin in the EC versus the CCN, consistent with the pattern of the repressive H3K9me (Garcia-Aguilar et al., 2010). In fact, the ability of the RdDM pathway to silence gene expression is reinforced by methylation of H3K9me in Arabidopsis, indicating a link between changes in histones and the DNA methylation pathways previously discussed (Jackson et al., 2002).
Concluding Remarks
DNA methylation, regulation of transposons via small RNAs, and histone modifications all represent effective ways of controlling the epigenetic status of plant gametes, efficiently altering the way plants develop. Although knowledge of the role that these mechanisms play in developing male and female plant gametes remains limited, it is already recognized how essential they are for successful plant reproduction. In particular, there is a strong need for more research with regards to the role that epigenetics plays during female gametogenesis, an area which remains difficult to study due to the deep embedding of the female gametophyte within maternal tissues. Ultimately, comprehending the ability of epigenetic mechanisms to modify inheritance and gene expression during gametogenesis is an important step in understanding the way plant reproduction works.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aravin, A. A., Sachidanandam, R., Bourc’his, D., Schaefer, C., Pezic, D., Toth, K. F., Bestor, T., and Hannon, G. J. (2008). A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799.
Armenta-Medina, A., Demesa-Arevalo, E., and Vielle-Calzada, J. P. (2011). Epigenetic control of cell speciation during female gametogenesis. Sex. Plant Reprod. 24, 137–147.
Baroux, C., Pecinka, A., Fuchs, J., Schubert, I., and Grossniklaus, U. (2007). The triploid endosperm genome of Arabidopsis adopts a peculiar, parental-dosage-dependent chromatin organization. Plant Cell 19, 1782–1794.
Baroux, C., Raissig, M., and Grossniklaus, U. (2011). Epigenetic regulation and reprogramming during gamete formation in plants. Curr. Opin. Genet. Dev. 21, 124–133.
Bartee, L., Malagnac, F., and Bender, J. (2001). Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 15, 1753–1758.
Bauer, M., and Fischer, R. (2011). Genome demethylation and imprinting in the endosperm. Curr. Opin. Plant Biol. 14, 162–167.
Biemont, C. (2009). Are transposable elements simply silenced or are they under house arrest? Trends Genet. 25, 333–334.
Borges, F., Gomes, G., Gardner, R., Moreno, N., McCormick, S., Feijo, J., and Becker, J. (2008). Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol. 148, 1168–1181.
Bourc’his, D., and Voinnet, O. (2010). A small-RNA perspective on gametogenesis, fertilization, and early zygotic development. Science 330 617–622.
Calarco, J. P., and Martienssen, R. A. (2011). Genome reprogramming and small interfering RNA in the Arabidopsis germline. Curr. Opin. Genet. Dev. 21, 134–139.
Chan, S. W., Henderson, I. R., and Jacobsen, S. E. (2005). Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6, 351–360.
Choi, Y., Gehring, M., Johnson, L., Hannon, M., Harada, J. J., Goldberg, R. B., Jacobsen, S. E., and Fischer, R. L. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110, 33–42.
Chueh, A. C., Northrop, E. L., Brettingham-Moore, K. H., Choo, K. H., and Wong, L. H. (2009). LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 5, e1000354. doi: 10.1371/journal.pgen.1000354
de Dios Alche, J., M’rani-Alaoui, M., Castro, A. J., and Rodriguez-Garcia, M. (2004). Ole e 1, the major allergen from olive (Olea europea L.) pollen, increases its expression and is released to the culture medium during in vitro germination. Plant Cell Physiol. 45, 1149–1157.
Ebbs, M. L., and Bender, J. (2006). Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18, 1166–1176.
Exner, V., Aichinger, E., Shu, H., Wildhaber, T., Alfarano, P., Caflisch, A., Gruissem, W., Kohler, C., and Hennig, L. (2009). The chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 is essential for H3K27me3 binding and function during Arabidopsis development. PLoS ONE 4, e5335. doi: 10.1371/journal.pone.0005335
Feng, S., and Jacobsen, S. (2011). Epigenetic modifications in plants: an evolutionary perspective. Curr. Opin. Plant Biol. 14, 179–186.
Feng, S., Jacobsen, S., and Reik, W. (2010). Epigenetic reprogramming in plant and animal development. Science 330, 622–627.
Finnegan, E. J., and Dennis, E. S. (1993). Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 21, 2383–2388.
Garcia-Aguilar, M., Michaud, C., Leblanc, O., and Grimanelli, D. (2010). Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22, 3249–3267.
Gehring, M., Bubb, K. L., and Henikoff, S. (2009). Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324, 1447–1451.
Gehring, M., Choi, Y., and Fischer, R. L. (2004). Imprinting and seed development. Plant Cell 16(Suppl.), S203–S213.
Gehring, M., Huh, J. H., Hsieh, T. F., Penterman, J., Choi, Y., Harada, J. J., Goldberg, R. B., and Fischer, R. L. (2006). DETEMER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124, 495–506.
Gehring, M., Missirian, V., and Henikoff, S. (2011). Genomic analysis of parent-of-origin allelic expression in Arabidopsis thaliana seeds. PLoS ONE 6, e23687. doi: 10.1371/journal.pone.0023687
Gong, Z., Morales-Ruiz, T., Ariza, R., Roldan-Arjona, T., David, L., and Zhu, J. K. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111, 803–814.
Grossniklaus, U. (2011). Plant germline development: a tale of cross-talk, signaling, and cellular interactions. Sex. Plant Reprod. 24, 91–95.
Hajkova, P., Ancelin, K., Waldmann, T., Lacoste, N., Lange, U. C., Cesari, F., Lee, C., Almouzni, G., Schneider, R., and Surani, M. A. (2008). Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature 452, 877–881.
Hauser, M. T., Aufsatz, W., Jonak, C., and Lusching, C. (2011). Transgenerational epigenetic inheritance in plants. Biochim. Biophys. Acta. 1809, 459–468.
Hsieh, T. F., Ibarra, C. A., Silva, P., Zemach, A., Eshed-Williams, L., Fischer, R. L., and Zilberman, D. (2009). Genome-wide demethylation of Arabidopsis endosperm. Science 324, 1451–1454.
Hsieh, T. F., Shin, J., Uzawa, R., Silva, P., Cohen, S., Bauer, M., Hashimoto, M., Kirkbride, R., Harada, J., Zilberman, D., and Fischer, R. (2011). Regulation of imprinting gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. U.S.A. 108, 1755–1762.
Huh, J. H., Bauer, M. J., Hsieh, T. F., and Fischer, R. L. (2008). Cellular programming of plant gene imprinting. Cell 132, 735–744.
Ingouff, M., Rademacher, S., Holec, S., Solijc, L., Xin, N., Readshaw, A., Foo, S. H., Lahouze, B., Sprunk, S., and Berger, F. (2010). Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr. Biol. 20, 2137–2143.
Ingouff, M., Hamaura, Y., Gourgues, M., Higashiyama, T., and Berger, F. (2007). Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr. Biol. 17, 1032–1037.
Jackson, J. P., Lindroth, A. M., Cao, X., and Jacobsen, S. E. (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 415, 556–560.
Jahnke, S., and Scholten, S. (2009). Epigenetic resetting of a gene imprinted in plant embryos. Curr. Biol. 19, 1677–1681.
Jeddeloh, J. A., Stokes, T. L., and Richards, E. J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22, 94–97.
Johnson, L. M., Bostick, M., Zhang, X., Kraft, E., Henderson, I., Callis, J., and Jacobsen, S. E. (2007). The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr. Biol. 17, 379–384.
Jullien, P. E., and Berger, F. (2009). Gamete-specific epigenetic mechanisms shape genomic imprinting. Curr. Opin. Plant Biol. 12, 637–642.
Jullien, P. E., Kinoshita, T., Ohad, N., and Berger, F. (2006). Maintenance of DNA methylation during Arabidopsis life cycle is essential for parental imprinting. Plant Cell 18, 1360–1372.
Jullien, P. E., Mosquna, A., Ingouff, M., Sakata, T., Ohad, N., and Berger, F. (2008). Retinoblastoma and its binding partner MSII control imprinting in Arabidopsis. PLoS Biol. 6, e194. doi: 10.1371/journal.pbio.0060194
Kohler, C., and Weinhofer-Molisch, I. (2010). Mechanisms and evolution of genomic imprinting in plants. Heredity 105, 57–63.
Law, J., and Jacobsen, S. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220.
Le Trionnaire, G., Grant-Downton, R. T., Kourmpetli, S., Dickinson, H. G., and Twell, D. (2011). Small RNA activity and function in angiosperm gametophytes. J. Exp. Bot. 62, 1601–1610.
Luo, M., Taylor, J. M., Spriggs, A., Zhang, H., Wu, X., Russell, S., Singh, M., and Koltunow, A. (2011). A genome-wide survey of imprinted genes in rice seeds reveals imprinting primarily occurs in the endosperm. PLoS Genet. 7, e1002125. doi: 10.1371/journal.pgen.1002125
Migicovsky, Z., and Kovalchuk, I. (2011). Epigenetic memory in mammals. Front. Gene 2:28. doi: 10.3389/fgene.2011.00028
Mosher, R. A., and Melnyk, C. W. (2010). SiRNAs and DNA methylation: seed epigenetics. Trends Plant Sci. 15, 204–210.
Mosher, R. A., Melnyk, C. W., Kelly, K. A., Dunn, R. M, Studholme, D. J., and Baulcombe, D. C. (2009). Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460, 283–286.
Mosher, R. A., Tan, E. H., Shin, J., Rischer, R. L., Pikaard, C. S., and Baulcombe, D. C. (2011). An atypical epigenetic mechanism affects uniparental expression of Pol IV-Dependent siRNAs. PLoS ONE 6, e25756. doi: 10.1371/journal.pone.0025756
Nonomura, K.-I., Morohoshi, A., Nakano, M., Eiguchi, M., Miyao, A., Hirochika, H., and Kurata, N. (2007). A germ cell-specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19, 2583–2594.
Okada, T., Bhalla, P. L., and Singh, M. B. (2006). Expressed sequence tag analysis of Lilium longiflorum generative cells. Plant Cell Physiol. 47, 698–705.
Olmedo-Monfil, V., Duran-Figueroa, N., Arteaga-Vazquez, M., Demesa-Arevalo, E., Autran, D., Grimanelli, D., Slotkin, R. K., Martienssen, R. A., and Vielle-Calzada, J.-P. (2010). Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464, 628–632.
Ortega-Galisteo, P., Morales-Ruiz, T., Ariza, R. R., and Roldan-Arjona, T. (2008). Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 67, 671–681.
Penterman, J., Zilberman, D., Huh, J. H., Ballinger, T., Henikoff, S., and Fischer, R. L. (2007). DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. U.S.A. 104, 6752–6757.
Pillot, M., Baroux, C., Vazquez, M. A., Autran, D., Leblanc, O., Vielle-Calzada, J. P., Grossniklaus, U., and Grimanelli, D. (2010). Embryo and endosperm inherit distinct chromatin and transcriptional states from the female gametes in Arabidopsis. Plant Cell 22, 307–320.
Scott, R. J., and Spielman, M. (2004). Epigenetics: imprinting in plants and mammals – the same but different? Curr. Biol. 14, R201–R203.
Singh, M., Goel, S., Melley, R. B., Dantec, C., Parrinello, H., Michaud, C., Leblanc, O., and Grimanelli, D. (2011). Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23, 443–458.
Slotkin, R., Vaugh, M., Borges, F., Tanurdzic, M., Becker, J., Feijo, J., and Martienssen, R. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–472.
Sundaresan, V., and Alandete-Saez, M. (2010). Pattern formation in miniature: the female gametophyte of flowering plants. Development 137, 179–189.
Tanaka, I. (1997). Differentiation of generative and vegetative cells in angiosperm pollen. Sex. Plant Reprod. 10, 1–7.
Tanaka, I., Ono, K., and Fukuda, T. (1998). The developmental fate of angiosperm pollen is associated with a preferential decrease in the level of histone H1 in the vegetative nucleus. Planta 206, 561–569.
Teixeira, F. K., Heredia, F., Sarazin, A., Roudier, F., Boccara, M., Ciaudo, C., Cruaud, C., Poulain, J., Berdasco, M., Fraga, M. F., Voinnet, O., Wincker, P., Esteller, M., and Colot, V. (2009). A role for RNAi in the selective correction of DNA methylation defects. Science 323, 1600–1604.
Twell, D. (2011). Male gametogenesis and germline specification in flowering plants. Sex Plant Reprod. 24, 149–160.
Ueda, K., Kinoshita, Y., Xu, Z.-J., Ide, N., Ono Michiyuki, O., Akahori, Y., Tanaka, I., and Inoue, M. (2000). Unusual core histones specifically expressed in male gametic cells of Lilium longiflorum. Chromosoma 108, 491–500.
Ueda, K., and Tanaka, I. (1995). Male gametic nucleus-specific H2B and H3 histones, designated gH2B and gH3, in Lilium longiflorum. Planta 197, 289–295.
van der Heijen, G. W., and Bortvin, A. (2009). Transient relaxation of transposon silencing at the onset of mammalian meiosis. Epigenetics 4, 76–79.
Villar, C. B., Erilova, A., Makarevich, G., Trosch, R., and Kohler, C. (2009). Control of PHERES1 imprinting in Arabidopsis by direct tandem repeats. Mol. Plant 2, 654–660.
Wolff, P., Weinhofer, I., Seguin, J., Roszak, P., Beisel, C., Donoghue, M., Spillane, C., Nordborg, M., Rehmseier, M., and Kohler, C. (2011). High-resolution analysis of parent-of-origin allelic expression in the Arabidopsis endosperm. PLoS Genet. 7, e1002126. doi: 10.1371/journal.pgen.1002126
Woodcock, C., Skoultchi, A., and Yuhong, F. (2006). Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 14, 17–25.
Wuest, S, Vijverbeg, K., Schmidt, A., Weiss, M., Gheyselinck, J., Lohr, M., Wellmer, F., Rahnenfuher, J., von Mering, C., and Grossniklaus, U. (2010). Arabidopsis female gametophyte gene expression maps reveals similarities between plant and animal gametes. Curr. Biol. 20, 506–512.
Xiao, W., Gehring, M., Choi, Y., Margossian, L., Pu, H., Harada, J. J., Goldberg, R. B., Pennell, R. I., and Fischer, R. L. (2003). Imprinting of the MEA polycomb gene is controlled by antagonism between MET1 methyltransferase and DME glycosylase. Dev. Cell 5, 891–901.
Keywords: epigenetic modifications, genomic imprinting, transposon reactivation, plant gametogenesis, DNA methylation, histone modifications
Citation: Migicovsky Z and Kovalchuk I (2012) Epigenetic modifications during angiosperm gametogenesis. Front. Plant Sci. 3:20. doi: 10.3389/fpls.2012.00020
Received: 08 October 2011; Accepted: 19 January 2012;
Published online: 06 February 2012.
Edited by:
Shawn Kaeppler, University of Wisconsin-Madison, USAReviewed by:
Nathan M. Springer, University of Minnesota, USAJoseph F. Petolino, Dow AgroSciences, USA
Daphné Autran, Institut de Recherche pour le Développement, France
Copyright: © 2012 Migicovsky and Kovalchuk. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Igor Kovalchuk, University of Lethbridge, 4401 University Drive, Lethbridge, AB, Canada e-mail:aWdvci5rb3ZhbGNodWtAdWxldGguY2E=