- 1Key Laboratory of Biogeography and Bioresource in Arid Land, Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences, Urumqi, China
- 2USDA Forest Service, Rocky Mountain Research Station, Provo, UT, USA
- 3Library, Xinjiang Normal University, Urumqi, China
- 4Shanghai Chenshan Plant Science Research Center, Shanghai Chenshan Botanical Garden, Chinese Academy of Sciences, Shanghai, China
Population genetic studies provide a foundation for conservation planning, especially for endangered species. Three chloroplast SSRs (mtrnSf-trnGr, mtrnL2-trnF, and mtrnL5-trnL3) and the internal transcribed spacer were used to examine the population structure of Helianthemum in northwestern China. A total of 15 populations of the genus were collected. Nine chloroplast haplotypes and two nuclear genotypes were detected. Both the nuclear and chloroplast data showed two lineages in Helianthemum songaricum, respectively, distributed in Yili Valley and western Ordos Plateau. A total of 66.81% (p < 0.001) of the genetic variation was supported by this lineage split. A Mantel test showed a significant correlation between genetic distance and geographical distance (r = 0.937, p < 0.001). Based on genetic analyses, cpSSRs data support strong genetic divergence between regions. We speculate that the climate change during the late Tertiary and early Quaternary isolated H. songaricum into their current distribution, resulting in interruption of gene flow, leading to isolation and genetic divergence between the two regions. Meanwhile, possible selfing would increase genetic drift in small fragmented populations, that might account for the observed genetic divergence in both regions. Given the loss of genetic diversity and genetic divergence in small populations of Helianthemum in northwestern China immediate conservation management steps should be taken on the species.
Introduction
Helianthemum is a shrub or subshrub mostly distributed in the Mediterranean, extending to Central Asia (Yang and Michael, 2007). Helianthemum songaricum and H. ordosicum in northwestern China is disjunctively distributed in Yili Valley of Xinjiang and western Ordos Plateau of Inner Mongolia, growing in rocky hills and slopes in steppe-desert regions between 1000 and 1400 m. It has spine-tipped branches, stipulate leaves, yellow flowers, and insect pollinated. Because of anthropogenic activities, such as grazing, mining, and the heavy harvest of firewood over the last few decades, H. songaricum and H. ordosicum has been in decline and become highly fragmented. As a result, it was listed as endangered in the China Species Red List (Fu, 1992).
There is disagreement with the taxonomic status of species in Helianthemum in northwestern China. Initially, taxonomists considered only one species of Helianthemum in northwestern China, Helianthemum songaricum Schrenk (Li, 1990). However, later studies found that there was a significant difference of pollen morphology and chromosome number between populations in Yili Valley and those in western Ordos Plateau. In the Yili Valley pollen was striate with a cytotype of 2n = 20, and in the western Ordos Plateau the pollen was perforate with 2n = 40 (Mo et al., 1997; Cao et al., 2000). Based on these results, a new taxon, H. ordosicum, was proposed in western Ordos (Zhao et al., 2000). More recently, Yang and Michael (2007) recognized only one species, H. songaricum. In our previous phylogeographic study, two chloroplast intergenic spacers data supported two species: H. songaricum and H. ordosicum. (Su et al., 2011).
Correctly defining the taxonomy and populations (i.e., intraspecific manage units, MU) is essential to make effective conservation strategies for endangered species (Frankham et al., 2002). Incorrect taxonomy would lead to ill-conceived management strategies. For example, outcrossing of different taxa can create inviable or infertile offspring (Barton and Hewitt, 1981; Coyne and Orr, 1989) and dismantle of coadapted complexes (Mayr, 1963; Shields, 1982; Templeton, 1987), leading to outbreeding depression. Outcrossing depression can also be observed in crosses at the intraspecific level (Geiger, 1988; Waser and Price, 1989). Populations that have adapted to different habitats could also suffer from outbreeding depression and should treated as a distinct manage unit to avoid outcrossing (Frankham et al., 2002).
Information on genetic structure can resolve ambiguous classifications and establish a foundation for conservation genetic planning. Chloroplast simple sequence repeats (cpSSRs) are a highly polymorphic molecular tool in the population genetic analysis (Vendramin et al., 1996; Morgante et al., 1997; Ebert and Peakall, 2009). Besides their high mutation rates, they have other specific features. Because of their uniparental inheritance, they could show pronounced level of population differentiation (Ennos, 1994; Vendramin et al., 1999; Flannery et al., 2006). In addition, in monoecious species, uniparentally inherited genomes have only half the effective population size (Birky, 1988), therefore, these genomes are sensitive to historical bottlenecks (Morgante et al., 1997). Though the apparent advantages, chloroplast DNA markers might only provide partly genetic information of a species (Mäder et al., 2010), and combination with biparentally inherited nuclear DNA markers would present a more integral view of population structure and demography history (Burban and Petit, 2003; Petit et al., 2005).
The primary conclusion in our previous study, that was significant genetic divergence existed between Yili Valley and western Ordos Plateau, was only based on two chloroplast spacers (trnD-trnT and rps16-trnK) and need to be further improved with nuclear genome data. In addition, population structure in the two regions are still unclear because of the limited polymorphism in the two chloroplast spacers (Su et al., 2011). Here, we use three highly polymorphic cpSSRs and nuclear Internal Transcribed Spacer (ITS) sequence to investigate the full genetic structure of Helianthemum in northwestern China to address the following questions: (1) Whether populations of Helianthemum in western Ordos Plateau represent a distinct taxa, H. ordosicum? (2) if so, what is the genetic structure within the two species? (3) What are the conservation implications from the genetic structure analysis?
Materials and Methods
Sampling
In 2010 and 2014, H. songaricum and H. ordosicum was sampled throughout its distribution in northwestern China. A total of 15 populations of the species were collected: nine from Yili Valley and six from western Ordos Plateau (Figure 1A). The geographical locations of the collection sites are presented in Table 1. Six to twelve individuals were sampled in each population. Fresh leaves were dried in silica gel and stored at 4°C until DNA extraction.
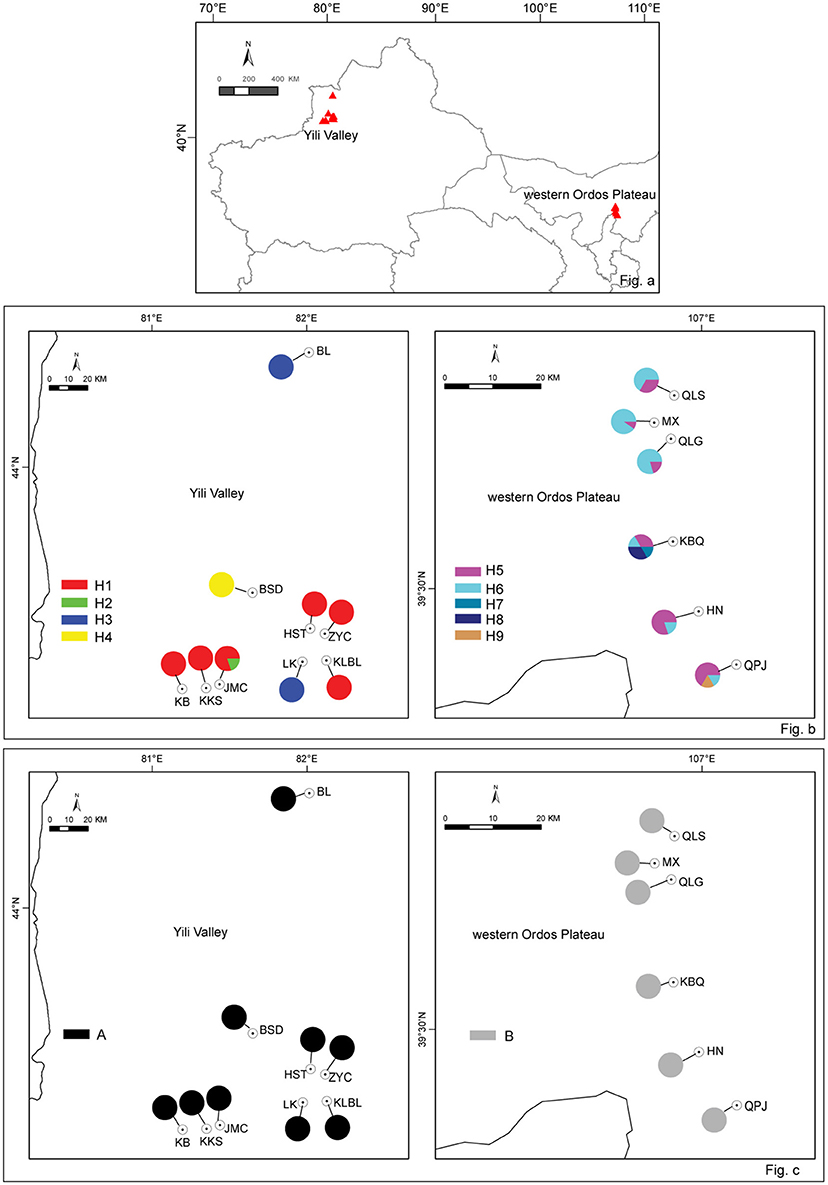
Figure 1. Sampling distribution of Helianthemum in China (a), the cp haplotype distribution (b), and ITS genotypes distribution in Helianthemum (c). Population numbers correspond to those in Table 1; cp haplotypes to those in Table 2, pie-charts represent haplotype frequency.
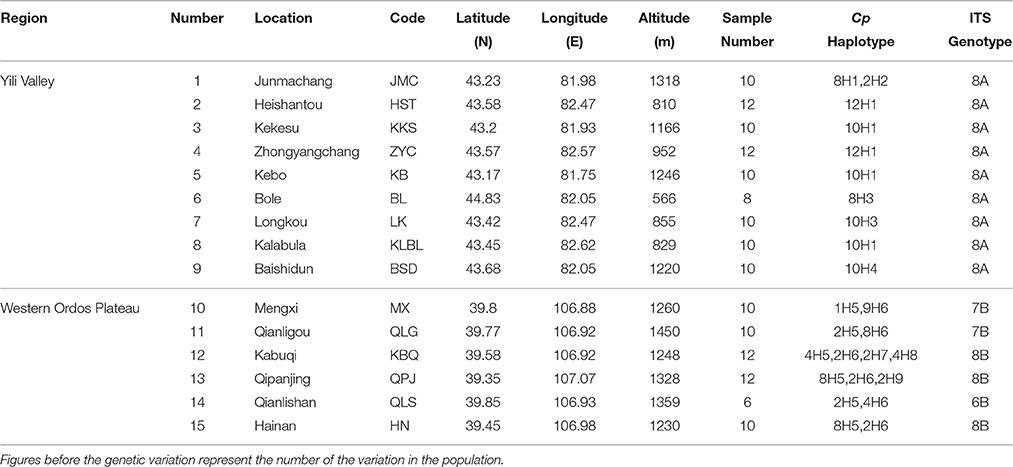
Table 1. Details of sample locations, sample size, and genetic variation for 15 populations of Helianthemum.
DNA Extraction, CpSSRS, and ITS Sequencing
Total genomic DNA was extracted from dried leaves by the CTAB method (Doyle and Doyle, 1987). Polymerase chain reactions (PCR) were carried out in a volume of 25 μL reaction mixtures containing 4 mM MgCl2, 0.2 mM dNTP, 0.5 μmol primer, and 1 U Taq polymerase (Applied Biosystems, Foster City, Calif.), implemented in a Biorad T100 thermocycler (Biorad). The cpSSRs were amplified using three Helianthemum-specific polymorphic loci detected in regions trnL-trnF, trnL5-trnL3, and trnS-trnG: mtrnSf-trnGr, mtrnL2-trnF, and mtrnL5-trnL3 (see Soubani et al., 2014), and following the temperature profile: 95°C for 4 min; 30 cycles of 92°C for 45 s; 57°C for 45 s; and 72°C for 1 min; linked a extension at 72°C for 10 min; ITS2 region was amplified using primers of Sun et al. (1994), and following the temperature profile: 94°C for 5 min; 35 cycles of 94°C for 30 s; 52°C for 45 s; and 72°C for 1 min; linked a extension at 72°C for 8 min. The cpSSRs products were separated by capillary electrophoresis, with an ABI 3730xl (Applied Biosystems) automated sequencer. CpSSRs fragment sizes were determined in Geneious version 7.0 using the package Plugin (Kearse et al., 2012), using Gene-flo 625 (Chimerx) as the internal lane standard. ITS2 amplified primers were used in sequencing reactions conducting in the DYEnamic ET Terminator Kit (Amersham Pharmacia Biotech). Sequencing were carried out in ABI 3730xl. ITS2 electropherograms were edited and assembled in SEQUENCHER 4.8 (Gene Codes, Ann Arbor, MI, USA), then the sequences were aligned in CLUSTALW (Thompson et al., 1994), and refined by visual inspection.
Population Genetic Analysis
For ease of presentation in this study, the terms “locus” refers to a cpSSR site, and “allele” refers to a length-variant at a cpSSR site. Alleles of the three plastid loci were scored, respectively, treated as ordered characters and then combined together as multilocus haplotypes, assuming a stepwise pattern in mutation (Ohta and Kimura, 1973). Using stirling probability distribution and Bayes's theorem, the completeness of haplotype sampling in this study was estimated (Dixon, 2006).
The number of different alleles (Na), the effective number of alleles (Ne), and Nei's genetic distence (Nei, 1978), were caculated in GenAlEx 6.5 software (Peakall and Smouse, 2006). Within-population diversity (hS), total gene diversity (hT), genetic differentiation index (GST, leaves out mutation steps between haplotypes; NST, includes mutation steps between haplotypes) were calculated in the program HAPLONST, using U-test to determine whether NST is significantly larger than GST.
Using pairwise population differentiation measures (FST) as the variance components (Wright, 1965), analysis of molecular variance (AMOVA) was performed to study the partition of total genetic variation within and among populations, conducted in ARLEQUIN v.3.01 (Excoffier et al., 1992). The significance test used 10,000 permutations. To evaluate the population genetic structure, a Neighbor-Joining network (NJ) of the 15 populations was constructed in MEGA 6.0 (Tamura et al., 2013), using Nei's genetic distance matrix. This genetic distance matrix was also used to perform principal coordinate (PCO) analysis in GenAlEx 6.5 (Peakall and Smouse, 2006). To reveal the genetic divergence between the two regions, a Mantel test was performed in ARLEQUIN v.3.01, with 10,000 permutations significance test. Geographical distance was calculated in GEODIS 2.5 (Posada et al., 2000), natural-log transformed in Excel 2000, and then correlated with the Nei's genetic distances.
Phylogenetic Analysis
To analyse the genealogical relationships among all the chloroplast haplotypes, a network was constructed using median-joining method conducted in NETWORK v. 4.600 (Bandelt et al., 1999).
Results
Allele and Sequence Analysis
A total of 13 alleles were detected in the three cpSSRs: three alleles in mtrnSf-trnGr, four alleles in mtrnL2-trnF, and six alleles in mtrnL5-trnL3. The 154 individuals sampled from 15 populations yielded 9 haplotypes (Table 2). Using the method described in Dixon (2006), the estimated probability of haplotype completeness was 1.0, suggesting that we have sampled almost all potential haplotypes in this study. For the ITS2 region, the aligned sequence length was 449 bp, and one informative nucleotide substitution (G/T) was found in position 173. Two nuclear genotypes (A and B) were identified in 116 individuals from 15 populations. GenBank accession numbers of the ITS2 sequences are KY314618-KY314619.
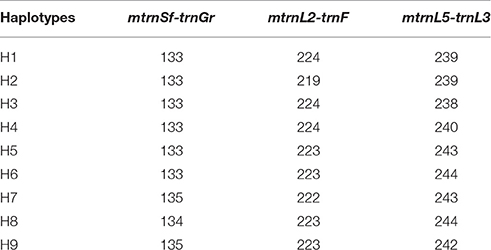
Table 2. Nine haplotypes of Helianthemum recognized on basis of three chloroplast SSRs, mtrnSf-trnGr, mtrnL2-trnF, and mtrnL5-trnL3.
Haplotype Patterns
The cpSSR haplotypes were partitioned among the two regions: Yili Valley and western Ordos Plateau. Haplotypes H1-H4 were distributed in Yili Valley, and haplotypes H5–H9 were distributed in western Ordos Plateau. Between the two regions, there was no shared haplotypes (Figure 1B). In Yili Valley, haplotype H1 was widespread in six populations of the total nine populations; rare haplotype H2 was found in population JMC; haplotype H3 was isolated in populations BL and LK, and haplotype H4 was isolated in population BSD. In western Ordos Plateau, haplotypes H5 and H6 were found in each population; rare haplotypes, H7 and H8 were found in population KBQ, and H9 was found in population QPJ (Figure 1B). For ITS genotype, all the individuals in Yili Valley contained one genotype (A), and all the individuals in western Ordos Plateau contained the other genotype (B) (Figure 1C).
The genetic relationships among the nine haplotypes also supports the disjunct geography of the Yili Valley and Ordos Plateau. The haplotypes found in the two geographic regions are also distinct based on the haplotype network (Figure 2). These two regional lineages, haplotypes H1-H4 corresponding to Yili Valley and H5-H9 corresponding to western Ordos Plateau, were connected by at least two hypothetical haplotypes (mv3 and mv5).
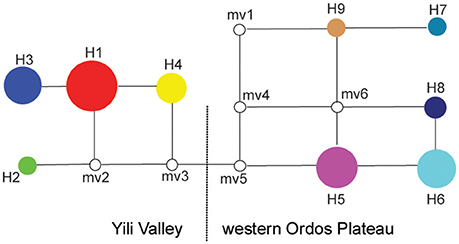
Figure 2. Median-joining network of Helianthemum haplotypes. The blank circles indicate missing or inferred haplotypes; the circle size is proportional to haplotype frequency; haplotypes in the network showed in the same colors correspond to those in the geographical distribution, Figure 1B.
Genetic Diversity and Genetic Structure
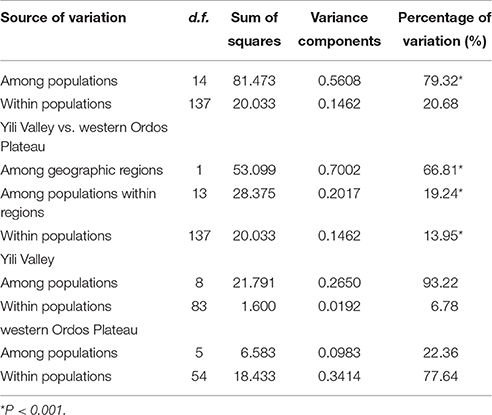
The mean different alleles number (Na) was 1.267, and effective alleles number (Ne) was 1.150. Across the entire study area, total gene diversity (hT) was 0.805 (SE 0.0632), and within-population gene diversity (hS) was 0.209 (SE 0.0679). Genetic differentiation index GST was 0.740 (SE 0.0784), and NST was 0.803 (SE 0.0617). As shown by the results of a U-test (U = 0.99, p < 0.01), NST was significantly higher than GST, suggesting a significant phylogeographical structure within Helianthemum. In Yili Valley, hT was 0.583 (SE 0.1501), hS was 0.04 (SE 0.0395), and genetic differentiation index was (GST = 0.932, NST = 0.934); in western Ordos Plateau, hT was 0.634 (SE 0.0748), hS was 0.463 (SE 0.0836), and genetic differentiation index was (GST = 0.270, NST = 0.226). AMOVA analysis showed that 79.32% (p < 0.001) of the total variation occurred among populations. When populations were grouped by geographical region, 66.81% (p < 0.001) of the total variation occurred among the regions (Table 3). In Yili Valley, 93.22% (p < 0.001) of the total variation occurred among populations; in western Ordos Plateau, 22.36% (p < 0.001) of the total variation occurred among populations. Mantel's test showed a significant correlation between genetic distance and geographical distance (r = 0.937, p < 0.001, Figure 3).
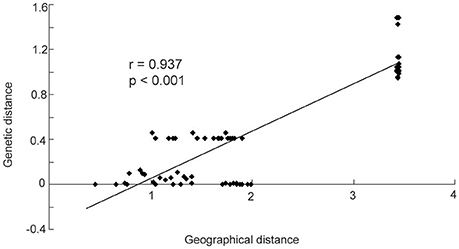
Figure 3. The figure shows a significant relationship between geographic and genetic distance (r = 0.937, p < 0.001).
The PCO plot illustrates the distinct differences between regions and differences within regions. The first two axis accounted for 76.14 and 13.66% of the total variation, respectively (Figure 4). The first axis separated all the Helianthemum populations into two groups, one including populations in Yili Valley and the other including populations in western Ordos Plateau. The second axis separated all the populations in Yili Valley into three groups, one including population BSD, one including populations BL and LK, and another included the remaining populations. The PCO plots suggested a high genetic divergence between Yili Valley and western Ordos Plateau population, and also a high genetic divergence among populations within Yili Valley. The PCO plot was consistent with the structure of the NJ network (Figure 5). In the NJ network, all the populations from Yili Valley clustered into a clade (Yili Valley clade), sister to the other clade containing all the populations from western Ordos Plateau (western Ordos clade). Yili Valley clade consists of two inner clade: the first inner clade contains populations BSD, BL, and LK, and the second clade contains the remaining populations. In the first inner clade, population BSD are separate from populations BL and LK. In western Ordos clade, populations MX, QLG, and QLS from the north of the plateau cluster together, drifting apart from populations HN, KBQ, and QPJ, from the south of the plateau.
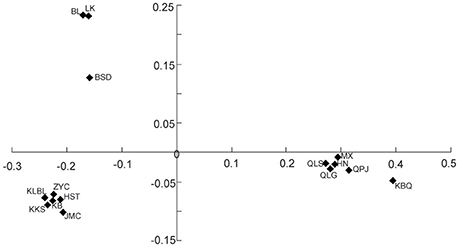
Figure 4. Plots of the first two coordinates based on pairwise population differentiation (Nei' s) matrix of Helianthemum.
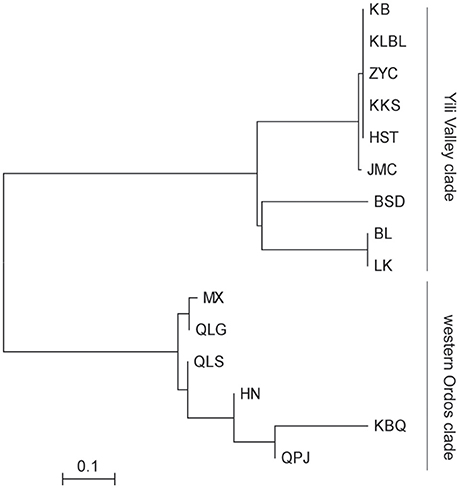
Figure 5. Neighbor-Joining tree of the 15 Helianthemum populations constructed using Nei's genetic distance matrix.
Discussion
Genetic Divergence Between Yili Valley and Western Ordos Plateau
Both the nuclear and chloroplast phylogenetic analyses showed two distinct lineages in Helianthemum, distributed in Yili Valley and western Ordos Plateau (Figures 1, 2). NJ network and PCO plots also indicated the similar result (Figures 4, 5). AMOVA analysis and Mantel test both showed a high level of genetic divergence between the two regions. In addition, ploidy in H. songaricum is 2X while in H. ordosicum is 4X. Wiley (1978) stated “A single lineage of ancestral descendant populations of organisms which maintains its identity from other such lineages and which has its own evolutionary tendencies and historical fates, should be defined as species status.” The two single lineages in Helianthemum in northwestern China clarified the taxonomic confusion of the genus, supporting that populations in western Ordos Plateau should be given species rank, H. ordosicum, supposed by Zhao et al. (2000).
In early Tertiary, the terrain and climate of northwestern China were very different from the arid and mountainous conditions of today. Some species of ancient Mediterranea flora, such as Helianthemum, spread along the relic of ancient Mediterranea distribution, across the Hexi Corridor, arrived to Alxa desert (Czenda, 1977; Liu, 1995). In the late Tertiary, uplifting of the northern Tibetan Plateau caused extensive aridification in northwestern China (Zheng et al., 2003; Sun et al., 2008). During the Quaternary, glaciation began to developed in the Northern Hemisphere, and the colder climate reached its maximum at about 0.8–0.6 Ma (Williams et al., 1993). Due to the dramatic climate change, many plant species of deserts in northwestern China gradually became extinct (Liu, 1995). This climatic history suggests that ancestral Helianthemum distributed in Hexi Corridor, a passage connecting Yili Valley with western Ordos Plateau, became extinct leaving those distributed in Yili Valley and western Ordos Plateau as relics. This hypothesis is supported by the haplotype relationships shown in the median-joining network (Figure 2). The putative extinction of Helianthemum along this corridor limited gene flow between the two regions. In addition to restricted gene flow, the climate in Yili Valley likely differentiated from that of the western Ordos Plateau. Because of the Tianshan Mountains, the Yili Valley has a Central Asia climate (Liu, 1995) with hot- dry summers, and mild-humid springs and winters (Shi et al., 2005). However, climate in western Ordos Plateau is typically drier throughout the year (Walker, 1974). Helianthemum in these two regions have inhabited distinct habitats for several millennia, harboring unique populations.
Genetic Diversity and Genetic Structure
Total genetic diversity of the two species are both moderate (H. songaricum: hT = 0.583; H. ordosicum: hT = 0.634), compared with other desert plants, such as Ammopiptanthus mongolicus (hT = 0.434), A. nunas (hT = 0.041) (Su et al., 2016), Reaumuria soongorica (hT = 0.312) (Qian et al., 2008), and are both higher than that in the previous phylogeography study of Helianthemum (H. songaricum, hT = 0.162; H. ordosicum, hT = 0.566), using two chloroplast intergenic spacers (Su et al., 2011). The inconsistency is due to higher polymorphism in the three cpSSRs than the two chloroplast spacers.
The cpSSRs data showed signs for genetic divergence in both H. songaricum and H. ordosicum. AMOVA analysis demonstrated significant genetic divergence among populations in both H. songaricum and H. ordosicum. The genetic divergence in the two species were also supported by NJ network and PCO analysis. As shown by the NJ network and PCO plots (Figures 4, 5), populations BL, LK, and BSD clustered together, apparently separated from the other populations in Yili Valley; populations HN, KBQ, and QPJ clustered together, apparently separated from the other populations in western Ordos Plateau. The significant genetic divergence among populations in the two species might be attributed to several factors. First, the seed viability is poor. In a Helianthemum flower, most ovules are unfertilized, or fertilized but with abnormal development, usually leaving only 1-3 well-developed seeds. Thus, seed production is very low (Ma et al., 2007). In addition, the seed requires a dormancy period, and with the poor water translocation due to the hard testa (Cao et al., 2000), the germination is very low (Ma et al., 2007). Second, habitats of the two species are both highly fragmented. In Yili Valley, there are several mountain ranges that subdivide Helianthemum habitat into five valleys (Zhang, 2006). The collected sites of H. songaricum are located in different secondary valleys. Similar in western Ordos Plateau, collection sites of H. ordosicum located in different valleys, along the Table Mountains. Within each species, the numerous geographic barriers isolate the populations, obstructing gene flow among them, and consequently likely decreasing genetic diversity and increasing the genetic divergence. Third, a reduced population size in the two species might also affect the population structure by increased selfing, mating among related individuals, and genetic drift. Based on congeners (Rodríguez-pérez, 2005; Aragón and Escudero, 2008), H. songaricum is likely an outcrosser but also self-compatible. Increased selfing or mating among related individuals in small population would reduce heterozygosity (Schaal and Leverich, 1996), and increased genetic drift would fix alleles randomly (Lynch et al., 1995), resulting in an alteration of population allele composition, inducing the population as an unique genetic sector (Gaudeul et al., 2000). The pattern of single haplotypes found in nearly all the populations in the Yili Valley suggests selfing is a fixture of this region. Compared to H. songaricum, H. ordosicum populations have greater population diversity (Table 3), and typically multiple haplotypes per population (Figure 1). These contrasting patterns suggest a greater degree of selfing or inbreeding in H. songaricum that could be caused by barriers to gene flow, differences in the abundance of pollinators or adaptation to a selfing life history.
Implications for Conservation in Helianthemum
Habitat fragmentation is a significant threat to the survival of plant species in many terrestrial ecosystems (Young et al., 1996). Our data observed low genetic diversity in isolated small populations in Helianthemum in northwestern China. Low genetic diversity can reduce population fitness and viability, weakening the population's ability to respond to changing selection pressures, increasing the extinction vulnerability (Young et al., 1996). We suggest an effective conservation management program incorporated our genetic analysis in following manners: (i) Significant genetic divergence showed by both nuclear and chloroplast data indicates two single evolutionary lineages in Helianthemum in northwestern China. The two species should be treated, respectively, when performed a management strategy. (ii) For in situ conservation, all the natural habitat of Helianthemum populaton should be preserved by local governments. Nature reserves for H. ordosicum have been set up in western Ordos Plateau. However, in Yili Valley, conservation of H. songaricum has not been given enough attention, and we propose nature reserves for H. songaricum should be set up at once. In addition, for serious fragmentation in both species, extinct populations should be reestablished to connect remnant populations in each species, using progenies from populations with nearest geographic distance. Also, population sizes should be augmented by transplanting progenies propagated from original populations. (iii) For both species, ex situ conservation site should be established first. Seeds collection should capture all detected genetic variations to represent the genetic diversity of each species in maximum, avoiding artificially induced bottlenecks (Maunder et al., 2001). Because of genetic uniqueness of populations BSD, BL, and LK in H. songaricum, seeds collections in these populations should be deposited as separate stocks. Meanwhile, seedlings of each species should be cultured for their future restoration. In H. songaricum, crossing individuals from unique populations (BSD, BL, and LK) and the rest of the populations should be tested ex situ to prevent potential outcrossing depression.
Author Contributions
Conceived and designed the experiments: ZS, BR. Performed the experiments: ZS. Analyzed the data: ZS, BR, LZ, and XJ. Contributed reagents/materials/analysis tools: ZS. Wrote the paper: ZS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by grants from Natural Science Foundation of Xinjiang (2014211A073). We thank USDA Forest Service, Rocky Mountain Research Station, for the helps in the experiment.
References
Aragón, C., and Escudero, A. (2008). Mating system of Helianthemum squamatum (Cistaceae), a gypsophile specialist of semi-arid Mediterranean environments. Bot. Helv. 118, 129–137. doi: 10.1007/s00035-008-0855-x
Bandelt, H. J., Forster, P., and Röhl, A. (1999). Median joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 14, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Barton, N. H., and Hewitt, G. M. (1981). “Hybrid zones and speciation,” in Evolution and Speciation. Essays in Honor of M. J. D. White, eds W. R. Atchley and D. S. Woodruff (Cambridge, UK: Cambridge University Press), 109.
Birky, C. W. (1988). “Evolution and variation in plant chloroplast and mitochondrial genomes,” in Plant Evolutionary Biology, eds L. Gottlieb and S. Jain (London: Chapman and Hall), 23–53.
Burban, C., and Petit, R. J. (2003). Phylogeography of maritime pine inferred with organelle markers having contrasted inheritance. Mol. Ecol. 12, 1487–1495. doi: 10.1046/j.1365-294X.2003.01817.x
Cao, R., Duan, F. Z., Ma, H., and Wang, L. Q. (2000). The biodiversity and population biology of a relic species - Helianthemum songaricum. Chinese Sci. Abstr. 6, 220–222. (in Chinese with English abstract).
Coyne, J. A., and Orr, H. A. (1989). Patterns of speciation in Drosophila. Evolution 43, 362–381. doi: 10.2307/2409213
Dixon, C. J. (2006). A means of estimating the completeness of haplotype sampling using the Stirling probability distribution. Mol. Ecol. Notes 6, 650–652. doi: 10.1111/j.1471-8286.2006.01411.x
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem. Bull. 19, 11–15.
Ebert, D., and Peakall, R. (2009). Chloroplast simple sequence repeats (cpSSRs): technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Mol. Ecol. 9, 673–690. doi: 10.1111/j.1755-0998.2008.02319.x
Ennos, R. (1994). Estimating the relative rates of pollen and seed migration among plant populations. Heredity (Edinb). 72, 250–259. doi: 10.1038/hdy.1994.35
Excoffier, L., Smouse, P. E., and Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes - application to human mitochondrial DNA restriction data. Genetics 131, 479–491.
Flannery, M. L., Mitchell, F. J., Coyne, S., Kavanagh, T. A., Burke, J. I., Salamin, N., et al. (2006). Plastid genome characterisation in Brassica and Brassicaceae using a new set of nine SSRs. Theor. Appl. Genet. 113, 1221–1231. doi: 10.1007/s00122-006-0377-0
Frankham, R., Ballou, J. D., and Briscoe, D. A. (2002). Introduction to Conservation Genetics. Cambridge, UK: Cambridge University Press.
Gaudeul, M., Taberlet, P., and Till-Bottraud, I. (2000). Genetic diversity in an endangered alpine plant, Eryngium alpinum L. (Apiaceae), inferred from amplified fragment length polymorphism markers. Mol. Ecol. 9, 1625–1637. doi: 10.1046/j.1365-294x.2000.01063.x
Geiger, H. H. (1988). “Epistasis and heterosis,” in Proceedings of the Second International Conference on Quantitative Genetics, eds B. S. Weir, E. J. Eisen, M. M. Goodman, and G. Namkoong (Sunderland, MA: Sinauer Associates), 395–399.
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Liu, Y. X. (1995). A study on origin and formation of the Chinese desert flora. J. Syst. Evol. 33, 131–143.
Lynch, M., Conery, J., and Burger, R. (1995). Mutational meltdowns in sexual populations. Evolution 49, 1067–1080. doi: 10.2307/2410432
Ma, X. P., Zhao, C. L., and Song, Y. X. (2007). The present situation and conservation countermeasures of threatened plant Helianthemum songaricum Schrenk. J. Agr. Sci. 28, 72–75. (in Chinese with English abstract).
Mäder, G., Zamberlan, P. M., Fagundes, N. J., Magnus, T., Salzano, F. M., Bonatto, S. L., et al. (2010). The use and limits of ITS data in the analysis of intraspecific variation in Passiflora L. (Passifloraceae). Genet. Mol. Biol. 33, 99–108. doi: 10.1590/S1415-47572009005000101
Maunder, M., Cowan, R. S., Stranc, P., and Fay, M. F. (2001). The genetic status and conservation management of two cultivated bulb species extinct in the wild: Tecophilaea cyanocrocus (Chile) and Tulipa sprengeri (Turkey). Conserv. Genet. 2, 193–201. doi: 10.1023/A:1012281827757
Mayr, E. (1963). Animal Species and Evolution. Cambridge, MA: Harvard University Press. doi: 10.4159/harvard.9780674865327
Mo, R. G., Bai, X. L., Ma, Y. Q., and Cao, R. (1997). On the intraspecific variations of pollon morphology and pollen geography of a relic species - Helianthemum songaricum Schrenk. Acta Bot. Bor. Occidental. Sin. 17, 528–532.
Morgante, M., Felice, N., and Vendramin, G. G. (1997). “Analysis of hyper-variable chloroplast microsatellites in Pinus halepensis reveals a dramatic genetic bottleneck,” in Molecular Tools for Screening Biodiversity: Plants and Animals, eds A. Karp, P. G. Isaac, and D. S. Ingram (London: Chapman and Hall), 407–412.
Nei, M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89, 583–590.
Ohta, T., and Kimura, M. (1973). A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population. Genet. Res. 22, 201–204. doi: 10.1017/S0016672300012994
Peakall, R., and Smouse, P. E. (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295. doi: 10.1111/j.1471-8286.2005.01155.x
Petit, R. J., Duminll, J., Fineschi, S., Hampe, A., Salvini, D., and Vendramin, G. G. (2005). Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 14, 689–701. doi: 10.1111/j.1365-294x.2004.02410.x
Posada, D., Crandall, K. A., and Templeton, A. R. (2000). GeoDis: a program for the cladistic nested analysis of the geographical distribution of genetic haplotypes. Mol. Ecol. 9, 487–488. doi: 10.1046/j.1365-294x.2000.00887.x
Qian, Z. Q., Xu, L., Wang, Y. L., Yang, J., and Zhao, G. F. (2008). Ecological genetics of Reaumuria soongorica (Pall.) Maxim. population in the oasis-desert ecotone in Fukang, Xinjiang, and its implications for molecular evolution. Biochem. Syst. Ecol. 36, 593–601. doi: 10.1016/j.bse.2008.01.008
Rodríguez-pérez, J. (2005). Breeding system, flower visitors and seedling survival of two endangered species of Helianthemum (Cistaceae). Ann. Bot. Lond. 95, 1229–1236. doi: 10.1093/aob/mci137
Schaal, B. A., and Leverich, W. J. (1996). Molecular variation in isolated plant populations. Plant Spec. Biol. 11, 33–40. doi: 10.1111/j.1442-1984.1996.tb00106.x
Shi, Y. F., Cui, Z. J., and Su, Z. (2005). The Quaternary Glaciations and Environmental Variations in China. Hebei: Hebei Science and Technology Publishing House.
Shields, W. M. (1982). Philopatry, Inbreeding, and the Evolution of Sex. Albany, NY: State University of New York Press.
Soubani, E., Hedrén, M., and Widén, B. (2014). Phylogeography of the European rock rose Helianthemum nummularium (Cistaceae): incongruent patterns of differentiation in plastid DNA and morphology. Bot. J. Linn. Soc. 176, 311–331. doi: 10.1111/boj.12209
Su, Z. H., Pan, B. R., Zhang, M. L., and Shi, W. (2016). Conservation genetics and geographic patterns of genetic variation of endangered shrub Ammopiptanthus (Fabaceae) in northwestern China. Conserv. Genet. 17, 485–496. doi: 10.1007/s10592-015-0798-x
Su, Z. H., Zhang, M. L., and Sanderson, S. C. (2011). Chloroplast phylogeography of Helianthemum songaricum (Cistaceae) from Northwestern China: implications for preservation of genetic diversity. Conserv. Genet. 12, 1525–1537. doi: 10.1007/s10592-011-0250-9
Sun, J. M., Zhang, L. Y., Deng, C. L., and Zhu, R. X. (2008). Evidence for enhanced aridity in the Tarim Basin of China since 5.3 Ma. Q. Sci. Rev. 27, 1012–1023. doi: 10.1016/j.quascirev.2008.01.011
Sun, Y., Skinner, D. Z., Liang, G. H., and Hulbert, S. H. (1994). Phylogenetic analysis of Sorghum and related taxa using Internal Transcribed Spacer of nuclear ribosomal DNA. Theor. Appl. Genet. 89, 26–32. doi: 10.1007/BF00226978
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Templeton, A. R. (1987). “Inferences on natural population structure from genetic studies on captive mammalian populations,” in Mammalian Dispersal Patterns, eds B. D. Chepko-Sade and Z. T. Halpin (Chicago, IL: University of Chicago Press), 257–272.
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). Clustal-W—improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Vendramin, G. G., Degen, B., Petit, R. J., Anzidei, M., Madaghiele, A., and Ziegenhagen, B. (1999). High level of variation at Abies alba chloroplast microsatellite loci in Europe. Mol. Ecol. 8, 1117–1126. doi: 10.1046/j.1365-294x.1999.00666.x
Vendramin, G. G., Lelli, L., Rossi, P., and Morgante, M. (1996). A set of primers for the amplification of 20 chloroplast microsatellites in Pinaceae. Mol. Ecol. 5, 595–598. doi: 10.1111/j.1365-294X.1996.tb00353.x
Walker, J. W. (1974). Evolution of exine structure in pollen of primitive angiosperms. Am. J. Bot. 61, 891–902. doi: 10.2307/2441626
Waser, N. M., and Price, M. V. (1989). Optimal outcrossing in Ipomopsis aggregata: seed set and offspring fitness. Evolution 43, 1097–1109. doi: 10.2307/2409589
Wiley, E. O. (1978). The evolutionary species concept reconsidered. Syst. Biol. 27, 17–26. doi: 10.2307/2412809
Williams, M. A. J., Dunkerley, D. L., De Dekker, P., Kershaw, A. P., and Stokes, T. (1993). Quaternary Environments. London: Edward Arnold.
Wright, S. (1965). The interpretation of population structure by fstatistics with special regards to systems of mating. Evolution 19, 395–420. doi: 10.2307/2406450
Young, A., Boyle, T., and Brown, T. (1996). The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol. 11, 413–418. doi: 10.1016/0169-5347(96)10045-8
Zhang, J. M. (2006). Studies on the geological structures and characteristic of terrain and landform in Yili river basin. J. Shihezi Univ. 24, 442–445. (in Chinese with English abstract).
Zhao, Y.-Z., Cao, R., and Zhu, Z.-Y. (2000). A new species of Helianthemum mill. (Cistaceae). J. Syst. Evol. Acta Phytotaxonom. Sinica 38, 294–296. (in Chinese with English abstract).
Keywords: Helianthemum, Yili Valley, western Ordos Plateau, genetic diversity, genetic structure, conservation implication
Citation: Su Z, Richardson BA, Zhuo L and Jiang X (2017) Divergent Population Genetic Structure of the Endangered Helianthemum (Cistaceae) and Its Implication to Conservation in Northwestern China. Front. Plant Sci. 7:2010. doi: 10.3389/fpls.2016.02010
Received: 27 September 2016; Accepted: 19 December 2016;
Published: 05 January 2017.
Edited by:
Badri Padhukasahasram, Illumina, USAReviewed by:
Robert Henry, University of Queensland, AustraliaJinfeng Chen, University of California, Riverside, USA
Hovirag Lancioni, University of Perugia, Italy
Copyright © 2017 Su, Richardson, Zhuo and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhihao Su, c3V6aEBtcy54amIuYWMuY24=
 Zhihao Su
Zhihao Su Bryce A. Richardson2
Bryce A. Richardson2 Xiaolong Jiang
Xiaolong Jiang