- National Key Lab of Crop Genetic Improvement, National Center of Oil Crop Improvement (Wuhan), College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
A novel cytoplasmic male sterility (CMS) in Brassica napus (inap CMS) was selected from the somatic hybrid with Isatis indigotica (Chinese woad) by recurrent backcrossing. The male sterility was caused by the conversion of tetradynamous stamens into carpelloid structures with stigmatoid tissues at their tips and ovule-like tissues in the margins, and the two shorter stamens into filaments without anthers. The feminized development of the stamens resulted in the complete lack of pollen grains, which was stable in different years and environments. The pistils of inap CMS displayed normal morphology and good seed-set after pollinated by B. napus. Histological sections showed that the developmental alteration of the stamens initiated at the stage of stamen primordium differentiation. AFLP analysis of the nuclear genomic composition with 23 pairs of selective primers detected no woad DNA bands in inap CMS. Twenty out of 25 mitochondrial genes originated from I. indigotica, except for cox2-2 which was the recombinant between cox2 from woad and cox2-2 from rapeseed. The novel cox2-2 was transcribed in flower buds of inap CMS weakly and comparatively with the fertile B. napus addition line Me harboring one particular woad chromosome. The restorers of other autoplasmic and alloplasmic CMS systems in rapeseed failed to restore the fertility of inap CMS and the screening of B. napus wide resources found no fertility restoration variety, showing its distinct origin and the related mechanism of sterility. The reasons for the mitochondrial rearrangements and the breeding of the restorer for the novel CMS system were discussed.
Introduction
Cytoplasmic male sterility (CMS) is a maternally inherited trait that fails to produce viable pollens, and has been reported in a large number of plant species. CMS is encoded in the mitochondrial genome and can arise spontaneously due to mutation in the genome (autoplasmy) or can be expressed following cytoplasmic substitutions due to nuclear-mitochondrial incompatibility (alloplasmy) (Prakash et al., 2009). A CMS fertility restoration system has been used as a suitable pollination control system to produce commercial hybrid seed for many crops, because it is easy to maintain.
Cytoplasmic male sterility phenotypes encompass a wide range of reproductive abnormalities, including degenerate anthers, aborted pollen, carpelloid and petaloid stamens (for review see Chase, 2007). Many CMS genes result from mtDNA rearrangements, as at least 10 essential mitochondrial genes are involved in the origination of CMS genes among 28 types of CMS from 13 crop species (Chen and Liu, 2014). Intriguingly, sequences of the known functional mitochondrial genes are not necessarily included in some CMS genes, for several rice CMS-associated ORFs consist of sequences of putative mitochondrial ORFs (Chen and Liu, 2014). Additionally, some CMS genes (such as orf125 in radish CMS-Kos and its variant orf138 in Brassica CMS-Ogu) are non-chimeric genes with the sequences from single source (Bonhomme et al., 1992; Iwabuchi et al., 1999). CMS genes have been shown to cause mitochondrial dysfunction by disrupting the mitochondrial membranes, reducing the mitochondrial ATP level and increasing the reactive oxygen species content (Hanson and Bentolila, 2004; Chen and Liu, 2014; Horn et al., 2014). Up to now, nine Rf (restorer of fertility) genes have been isolated in seven plant species, such as Rfo (Rfk1) in radish and Brassica (Chen and Liu, 2014). Most of the identified Rf genes encode PPR (pentatricopeptide repeat) proteins, but Rf genes are also highly multifarious and the respective restoration of fertility in CMS/Rf systems may be realized by various mechanisms at genomic, post-transcriptional, translational, or post-translational, and metabolic levels (Chen and Liu, 2014).
Stable CMS lines of crops could be produced by introducing the alien cytoplasm from the relatives through interspecific-/intergeneric hybridizations, or by mediating the mitochondrial rearrangement via somatic fusions (Prakash et al., 2009). For Brassica crops, since a sterility inducing cytoplasm identified in a wild population of Raphanus sativus (Ogura CMS) was introduced into B. napus and B. oleracea (Bannerot et al., 1974), a spectrum of alloplasmic CMS lines of diverse origins have been obtained by combining the cytoplasm of Brassica coenospecies with crop nuclei, particularly in B. juncea (see reviews by Delourme and Budar, 1999; Budar et al., 2004; Prakash et al., 2009; Yamagishi and Bhat, 2014). More recently, a novel CMS system in B. juncea incorporating the cytoplasm of B. fruticulosa was developed (Atri et al., 2016).
Somatic hybridization via protoplast fusion is a possible alternative for gene transfer from wild relatives to crops, by combining both nuclear and cytoplasmic genomes of two distantly related species, genera or even tribes. In somatic hybrids from protoplast fusion, the chloroplasts are usually inherited from one of the parents, whereas the mitochondrial DNA (mtDNA) is rearranged and may include DNA from both parents (Prakash et al., 1999). Intergenomic mitochondrial recombination with high frequency was found to occur in various somatic hybrids from the combinations of different species within Brassicaceae (Landgren and Glimelius, 1994; Dieterich et al., 2003; Leino et al., 2003; Oshima et al., 2010; Liu et al., 2015), leading to novel CMS genes. In the B. napus CMS Tournefortii-Stiewe produced by protoplast fusion with B. tournefortii (Dieterich et al., 2003), the mitochondrial rearrangement at upstream of the gene atp9 generated a chimeric orf193 that was co-transcribed with atp9. But another B. napus alloplasmic CMS Tournefortii by sexual hybridization with the same species had a chimeric orf263 at the vicinity of the atp6 (Landgren et al., 1996). Furthermore, recombinant mitochondrial genomes were also generated when protoplasts of fertile and sterile cytoplasm were fused (Sakai and Imamura, 1990; Wang et al., 1995; Meur et al., 2006).
From the mitochondrial genomes of nap, pol, ogu, and hau CMS sequenced (Handa, 2003; Chen et al., 2011; Tanaka et al., 2012; Heng et al., 2014), comparative analysis revealed that the CMS associated genes were localized close to the edge of syntenic sequence blocks, together with short repeats and overlapped repeats. These repeats were responsible for reorganization of mitochondrial genomes via homologous recombination. Overlapping homologous sequences from the fusion parents were produced when the parental mitochondria fused and the recombination occurred. In the cybrid derived from the somatic hybrid between Nicotiana tabacum and Hyoscyamus niger after being backcrossed with N. tabacum, the mitochondrial genome was highly recombinant with at least 35 intergenomic rearrangement events detected between two parents (Sanchez-Puerta et al., 2015). Recombination occurred via homologous mechanisms involving the double-strand break repair and/or break-induced replication pathways.
Currently, the autoplasmic pol CMS system from natural mutations in B. napus is still predominant for rapeseed hybrid production in China, but the alloplasmic Ogura CMS-fertility restoration system is widely used in Europe and Canada. Although several CMS systems have been reported, they are not used commercially due to undesirable CMS phenotype and lack of fertility restoration genes. In our previous study, the intertribal somatic hybrids between B. napus and Isatis indigotica Fort. (Chinese woad) of the Isatideae tribe within the Brassicaceae family were obtained and backcrossed successively to B. napus (Du et al., 2009; Kang et al., 2014), resulting in the development of one novel B. napus CMS line with carpelloid stamens (named as inap CMS). Herein, the histological and genetical characterizations of inap CMS were described, together with the mitochondrial rearrangements.
Materials and Methods
Plant Materials
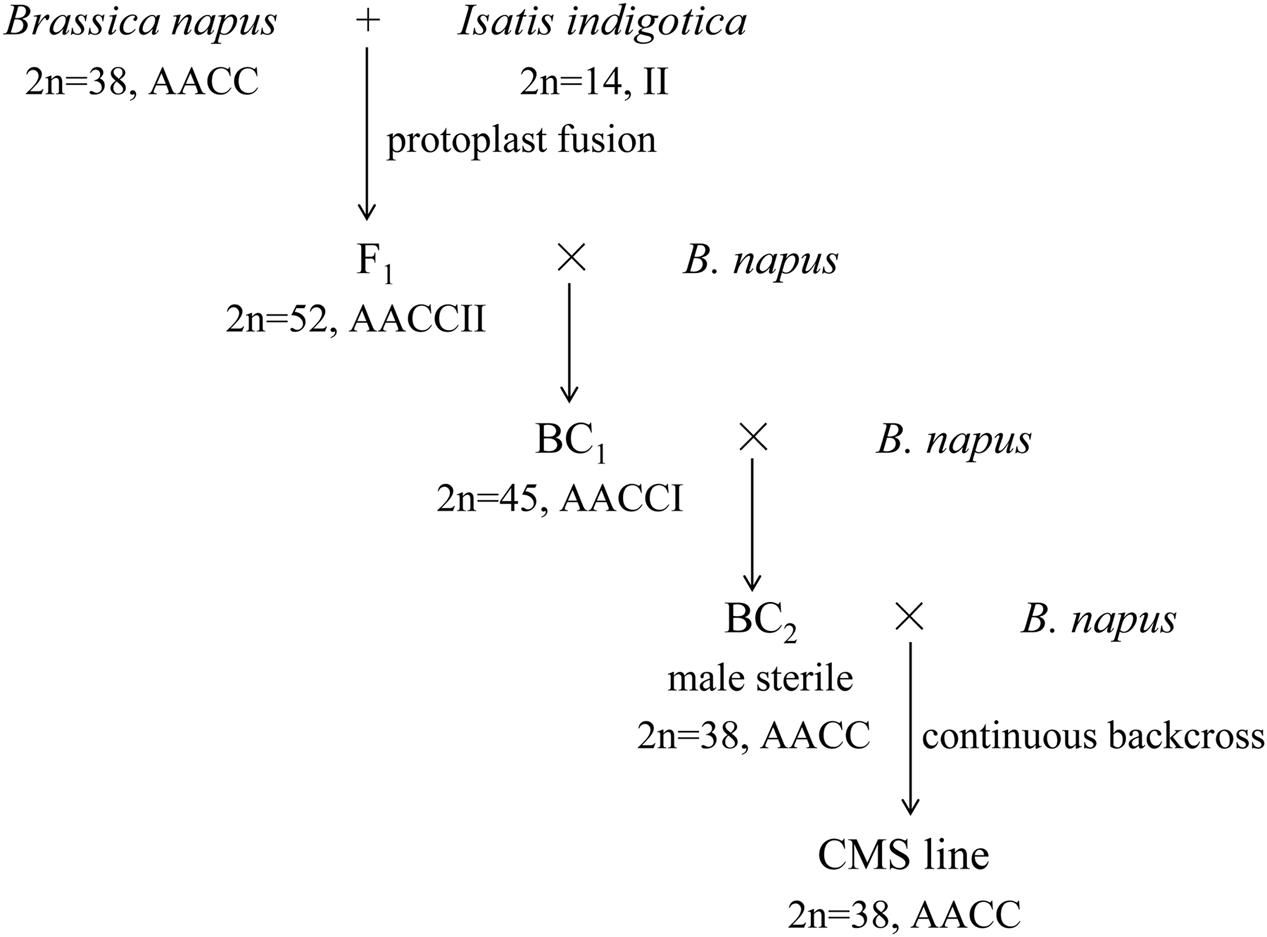
The intertribal somatic hybrid (As1, 2n = 52, AACCII) between B. napus L. cv. Huashuang 3 (2n = 38, AACC) and I. indigotica Fort. (Chinese woad, 2n = 14, II) was previously produced by protoplast fusion (Du et al., 2009). The backcrossed progenies with the same chromosome number and complement as B. napus were male sterile due to the development of carpelloid stamens (Kang et al., 2014). The phenotype of the carpelloid stamens was stably maintained after backcrosses with Huashuang 3 (Figure 1), which showed the characteristic of the cytoplasmic inheritance. Then, the novel CMS line of B. napus was developed and named as inap CMS. Its stability of male sterility was observed for 3 years in two locations and two seasons: Qinghai University experimental field in summer (Xining, altitude 2261 m, 101°77′E, 36°62′N, average temperature 19°C) and Huazhong Agricultural University in spring.
Nuclear and Organelle DNA Analysis
For nuclear analysis, total DNA was extracted and purified from young leaves of the parents and CMS line according to the method by Dellaporta et al. (1983). AFLP analysis was carried out according to the procedures of Vos et al. (1995) with some modifications. Genomic DNA (75 ng) of each sample was digested by using the restriction endonucleases EcoR I and Mse I for 6 h at 37°C and denaturalized for 1 h at 65°C. Then two adapters were ligated to the sticky ends of the digested DNA at 22°C overnight, and the resulting ligation products were amplified by PCR with primers matching the adapters. After being diluted to 15 times, resultant PCR products were amplified using 23 pairs of randomly selected primers. Finally, the PCR products were separated on 6% polyacrylamide gels and DNA bands were visualized by silver staining.
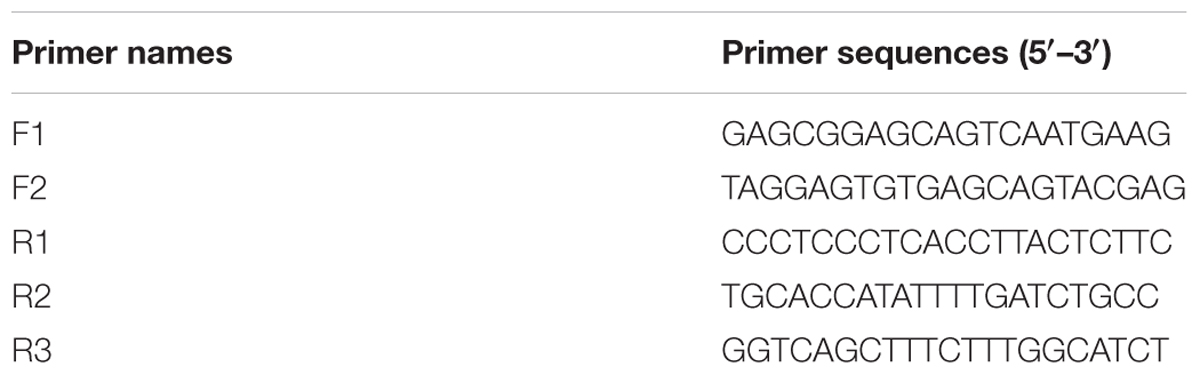
For mtDNA analysis, 24 mitochondrial genes were examined in parents and CMS line (Supplementary Table S1). A total of 20 μl reaction contained 4 μl 5Phusion HF Buffer, 0.4 μl 10 mM dNTPs, 0.5 μl forward and reverse primer (10 μM), 0.6 μl 100% DMSO, 1 μl template DNA (50 ng/μl) and 0.2 μl Phusion Hot Start II High-Fidelity DNA Polymerase (2 U/μl). PCR amplification was carried out with an initial denaturation step at 98°C for 30 s followed by 35 cycles of 94°C for 5 s, 65°C for 20 s (optimal annealing temperature was determined by Tm calculator on website: www.thermoscientific.com/pcrwebtools), 72°C for 45 s, and a final10-min extension at 72°C. PCR products were separated on 0.8% agarose gels and purified using the TIANgel Midi Purification Kit (TIANGEN BIOTECH (BEIJING) CO., LTD., Beijing, China). The 3′-A overhang of blunt-end DNA fragments was added using DNA A-Tailing Kit (TaKaRa BIO INC, Dalian, China). Then the products were cloned into the pMD18-T vector and sequenced. The primers for amplifying complete sequence of cox2-1 and cox2-2 were designed based on the rapeseed mtDNA sequence (accession number: AP006444, Table 1).
To distinguish inap CMS from Ogura and pol CMS, the primers of orf138 (GenBank: AB055443) and orf224 (GenBank: EU254235) were designed (Supplementary Table S1). Reactions (10 μl) contained 1 × Taq buffer, 2 mM MgCl2, 5 mM dNTPs, 5 μM forward and reverse primer, 1 U Taq DNA polymerase and 50 ng total DNA. DNA fragments were amplified after a 5-min denaturation at 94 for 30 cycles (94°C for 45 s, 57°C for 30 s, 72°C for 90 s), and a 10-min extension step at 72°C.
Reverse Transcription-PCR
Total RNA was extracted from flower buds of inap CMS line, B. napus cv. Huashuang 3, I. indigotica and the male fertile B. napus monosomic addition line Me (Kang et al., 2014) using the Eastep® Super Total RNA Extraction Kit (Shanghai Promega Biological Products, Ltd, Shanghai, China). cDNA was synthesized from total RNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc., America). RT-PCR amplification was carried out with an initial denaturation step at 94°C for 5 min followed by 35 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 90 s, and a final 10-min extension at 72°C.
Histological Studies
The inflorescences of B. napus and CMS line were first fixed by 50% FAA solution and stained with Ehrlich’s haematoxylin solution. After rinsed with distilled water, the samples were dehydrated with a graded ethanol series (15, 30, 50, 70, 85, 95, and 100%) for 4 h in each. Then samples were gradually cleared in 20, 40, 60, 80, and 100% chloroform for 2 h, and added paraffin fragments into 100% chloroform twice and kept in an incubator at 37°C for 2 days. The samples were embedded in Paraffin with ceresin (Sinopharm Chemical Reagent Co., Ltd, Shanghai, China). The tissues were sectioned into 8 μm sections, floated on drops of distilled water and dried overnight onto Poly-L-Lysine-coated slides at 37°C. After deparaffinized with xylene, the tissues sections were mounted by neutral balsam. All micrographs were taken with a Nikon Digital DS-Ri1.
For scanning electron microscopy, the young flower buds were fixed with 2.5% glutaraldehyde in 0.05 M phosphate buffer (pH 6.8) at room temperature for 2 h, then post-fixed in 1% osmium tetroxide in the same buffer at 4°C for 1 h. The samples were dehydrated with a graded ethanol series, then critical point dried using liquid CO2. After mounting, the samples were shadowed with gold and observed with a JSM-6390LV scanning microscope (NTC, Japan).
Restorer Search for inap CMS
The restorer lines of different CMS systems were used to test their ability to restore the fertility of inap CMS (Table 2). The seeds of Ogura and pol CMS lines and restorer lines were kindly provided by Prof. Jiangsheng Wu from our University. B. napus cv. Yudal was reported to restore Tournefortii-Stiewe CMS with recombinant mtDNA of both B. tournefortii and B. napus (Dieterich et al., 2003). The disomic rape-radish addition line (DAL-f) (2n = 40, AACC+1IIR) was selected, for it acted as the restorer for a novel CMS of B. napus with Raphanus cytoplasm and also carpel-like stamens (Budahn et al., 2008). However, the addition with the chromosome F did not restore Ogura CMS of B. napus. One B. napus monosomic addition line Me carrying one chromosome of Chinese woad and the same cytoplasm of inap CMS was used, as it produced anthers with viable pollen grains (Kang et al., 2014). After inap CMS line was pollinated by these different lines by hand, the seeds were harvested and F1 plants were assessed for the degree of male sterility.

TABLE 2. The restorers of other cytoplasmic male sterility (CMS) lines and the male fertility of their progenies with inap CMS.
Results
Phenotype of inap CMS in B. napus
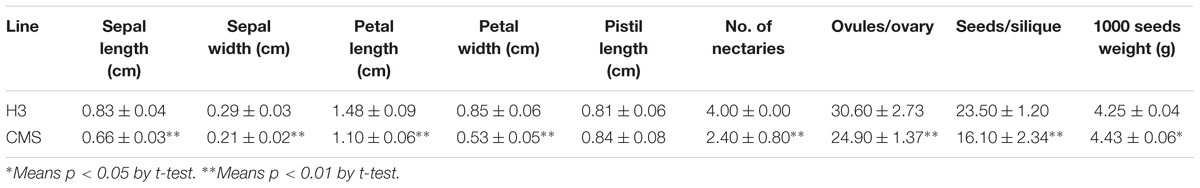
The inap CMS of B. napus was selected from the somatic hybrids with Chinese woad by successive backcrossing to recover the B. napus chromosome complement (Figure 1). The male sterility was attributed to the developmental conversion of its tetradynamous stamens into the carpelloid structures with stigmatoid tissues at the tips and ovule-like tissues in the margins, and two shorter stamens into filaments (Figures 2A–E). The two carpelloid structures at the same side fused together in some flowers, and still remained after the flower faded and as the pods developed (Figure 2F). The size and developmental complex of carpelloid structures diminished after successive backcrosses. The petals and sepals of its flowers were significantly smaller than the donor B. napus cv. Huashuang 3 (Table 3), and the number of nectaries reduced to 2.4 from 4 of Huashuang 3 due to the fused carpelloid stamens. Although the pistil length of the flowers was close to Huashuang 3, the seeds per pod (16.1) of inap CMS after pollination by Huashuang 3 were fewer than those in Huashuang 3 (23.5) by selfing (Table 3), likely some ovules were inviable (24.9/30.9). The negative effect of the cytoplasm from inap CMS on the defect of female development might cause the yield penalty at certain degrees, and optimal combinations needed to be screened for its correction. The male sterility phenotype was expressed consistently during recurrent backcrosses, and in Wuhan and Xining for 3 years, indicating its genetic and environmental stability. Then the male sterility was also easily maintained by Huashuang 3 and all other B. napus genotypes used (see below), as inap CMS had good seed-sets and still produced the male sterile progenies after pollinated by them.

FIGURE 2. Flower phenotype of inap CMS and Brassica napus donor. (A,B) The opened flower and the one with sepals and petals removed of Huashuang 3. (C,D) The whole flower and that with sepals and petals removed of inap CMS. (E) The carpelloid stamen with stigmatoid structures at its tips and ovules in the margins. (F) The inflorescences of Huashang 3 (left) and the CMS (right). Bars: (A–C) 5 mm; (D,E) 2 mm; (F) 10 cm.
The plants of inap CMS grew more slowly and flowered later almost 2 weeks than Huashuang 3 in Wuhan, but they show no leaf chlorosis, likely for they maintained the chloroplast genetic component from Huashuang 3 (Du et al., 2009; Kang et al., 2014). But death of flower buds occurred at the initial stage of flowering, and later the flower buds developed and opened normally.
Stamen Development of inap CMS
The stages of floral development in Arabidopsis thaliana were defined by Smyth et al. (1990) using scanning electron microscopy. With this standard, the early flower developmental processes of inap CMS line and its maintainer line Huanshuang 3 were observed and compared. They showed no differences until the stamen primordium appeared (Figures 3A,B,E,F). Subsequently, the stamen primordium developed into stamen in the maintainer (Figures 3C,D), while the stamen converted into the carpelloid structure in the CMS (Figures 3G,H). The ovule-like structure appeared on the carpelloid tissues in the CMS line at the stage 9 (data not shown). By histological studies, disordered cell division occurred at the sites of the stamen primordium in the flower buds at stage 4, and petal primordium would arise (Figures 3I–P). The stamen primordium developed into the carpelloid structure as a result of disorganized cell proliferation. So the male sterility of inap CMS initiated at the stage of stamen primordium differentiation.
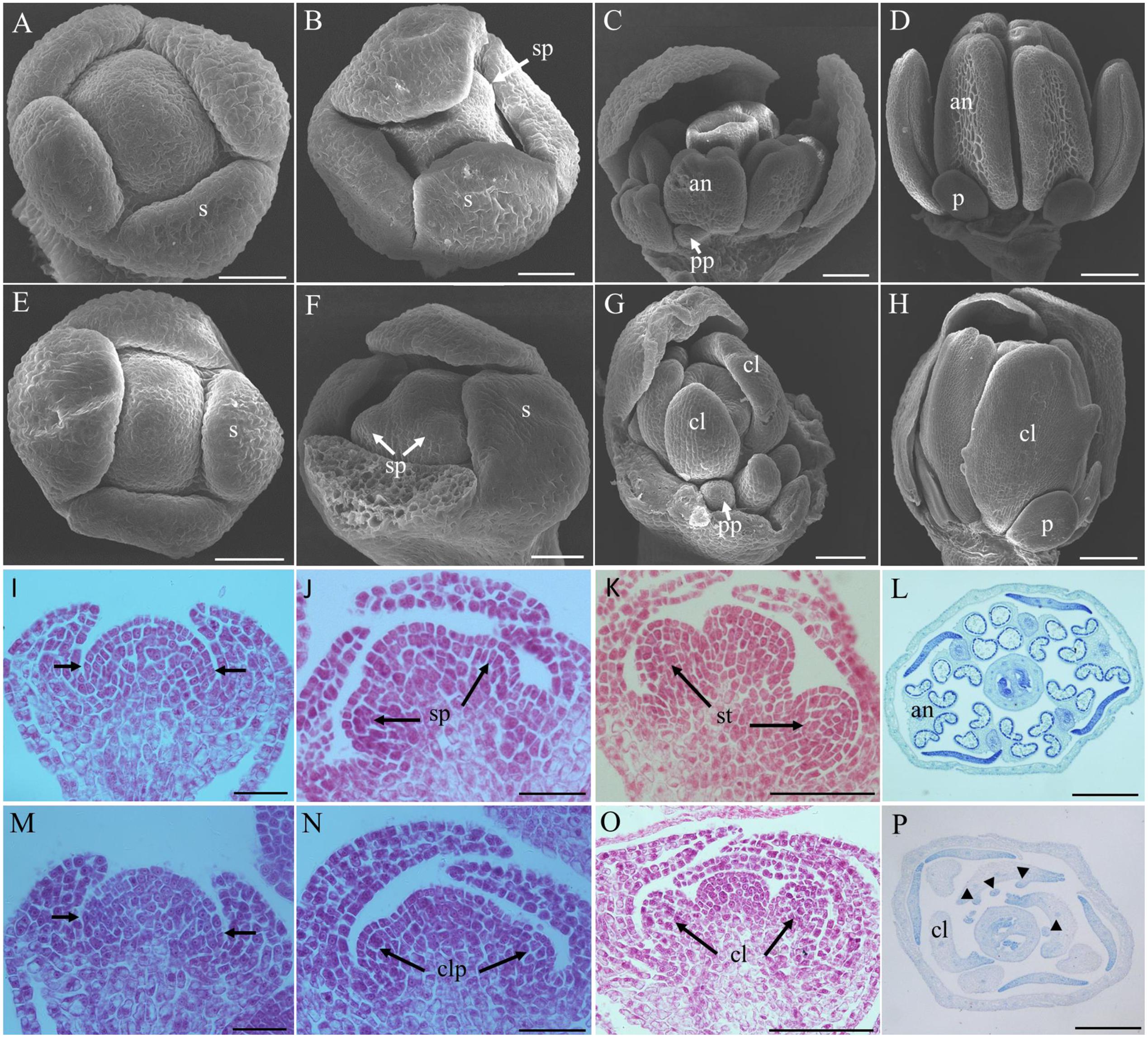
FIGURE 3. Comparative histology of flower development in B. napus and inap CMS line. (A,E) Flower buds at stage 4 of B. napus and inap CMS. (B,F) The stamen primordia are visible (arrows) at stage 5. (C,G) At stage 7, the anthers are visible in B. napus, while the stamen are converted to the carpelloid structure in inap CMS line. (D,H) Flower buds of B. napus and inap CMS at stage 9. (I,M) Flower buds at stage 4. Arrows indicate the sites where the stamen primordium and petal primordium will arise. (J,N) Flower buds at stage 5. (K,O) Flower buds at stage 6. (L,P) Cross sections of flower buds of B. napus and inap CMS. Triangles indicate ovule-like structures on the carpelloid organs in inap CMS. an, anther; cl, carpelloid structure; clp, carpelloid primordium; pp, petal primordium; p, petal; s, sepal; sp, stamen primordium; st, stamen. Bars: (A,B,E,F,K,O) 50 μm; (C,G,L,P) 100 μm; (D,H) 200 μm; (I,J,M,N) 20 μm.
Nuclear and Cytoplasmic Composition of inap CMS
With randomly selected 23 pairs of AFLP selective primers, no I. indigotica genomic bands were found in inap CMS line, showing no woad chromosomal segments introgressed after successive backcrosses with B. napus.
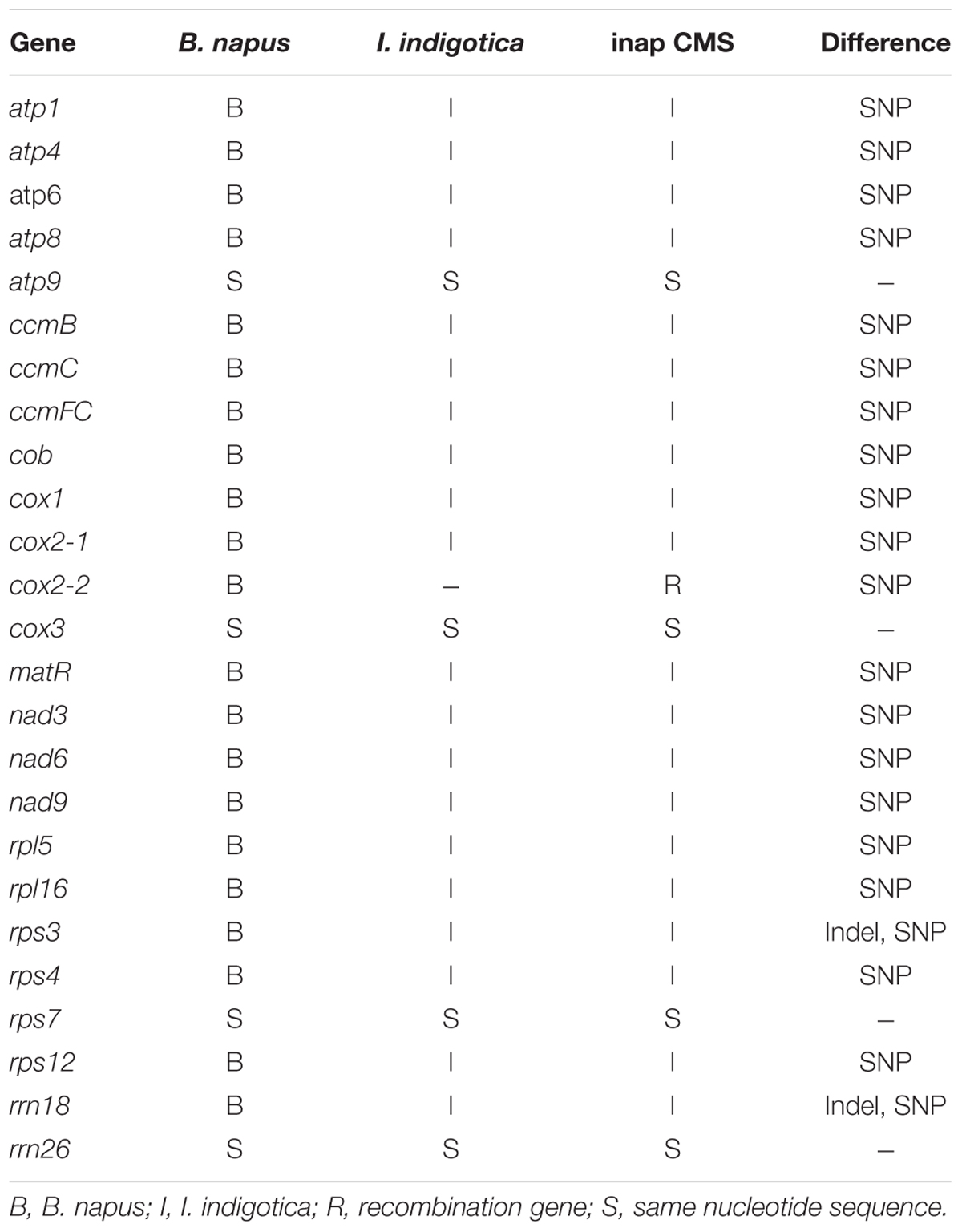
It has been shown that the somatic hybrid contained the recombinant mitochondrial genome from B. napus and I. indigotica (Kang et al., 2014). As most of mitochondrial genes had no fragment length differences between parents and the hybrid from PCR amplification, 25 mitochondrial genes in the CMS line were sequenced and found that 20 of them originated from I. indigotica (Table 4). Only the genes atp9, cox3, rps7, and rrn26 showed no sequence differences between parents and CMS line. Several SNPs were identified at the first exon and the intron of cox2-2 between CMS and B. napus, while one same band was detected by PCR amplification using its primers. Two copies of cox2 genes were reported to exist in Brassica species, except for B. nigra and B. carinata (Handa, 2003; Yamagishi et al., 2014), which showed a consistent sequence of the first exon and intron and diverged from each other at the second exon. The cox2-1 was homologous to the mitochondrial cox2 genes of other plants, but cox2-2 had an extension with no homology to any other sequences. To obtain the full sequence of cox2-1 and cox2-2 of inap CMS and parents, the primers were redesigned (Table 1 and Figure 4A). Only one copy of the cox2 (GenBank: KY656165) was obtained from I. indigotica, having 99.77% sequence identity with cox2-1 of B. napus (Figure 4B and Supplementary Figure S1). Multiple sequence alignment indicated that the first exon and intron of the cox2-2 of the CMS was from the cox2 of I. indigotica, but the second exon was derived from the cox2-2 of B. napus (Figure 4 and Supplementary Figure S1). RT-PCR analysis showed that this cox2-2 was transcribed in inap CMS flower buds, but the expression level was very low (Supplementary Figure S2). To examine the effect of nuclear restorer gene on the expression of the cox2-2, the expression level was compared between the fertile B. napus addition line Me and inap CMS, but showed no significant difference (Supplementary Figure S2).
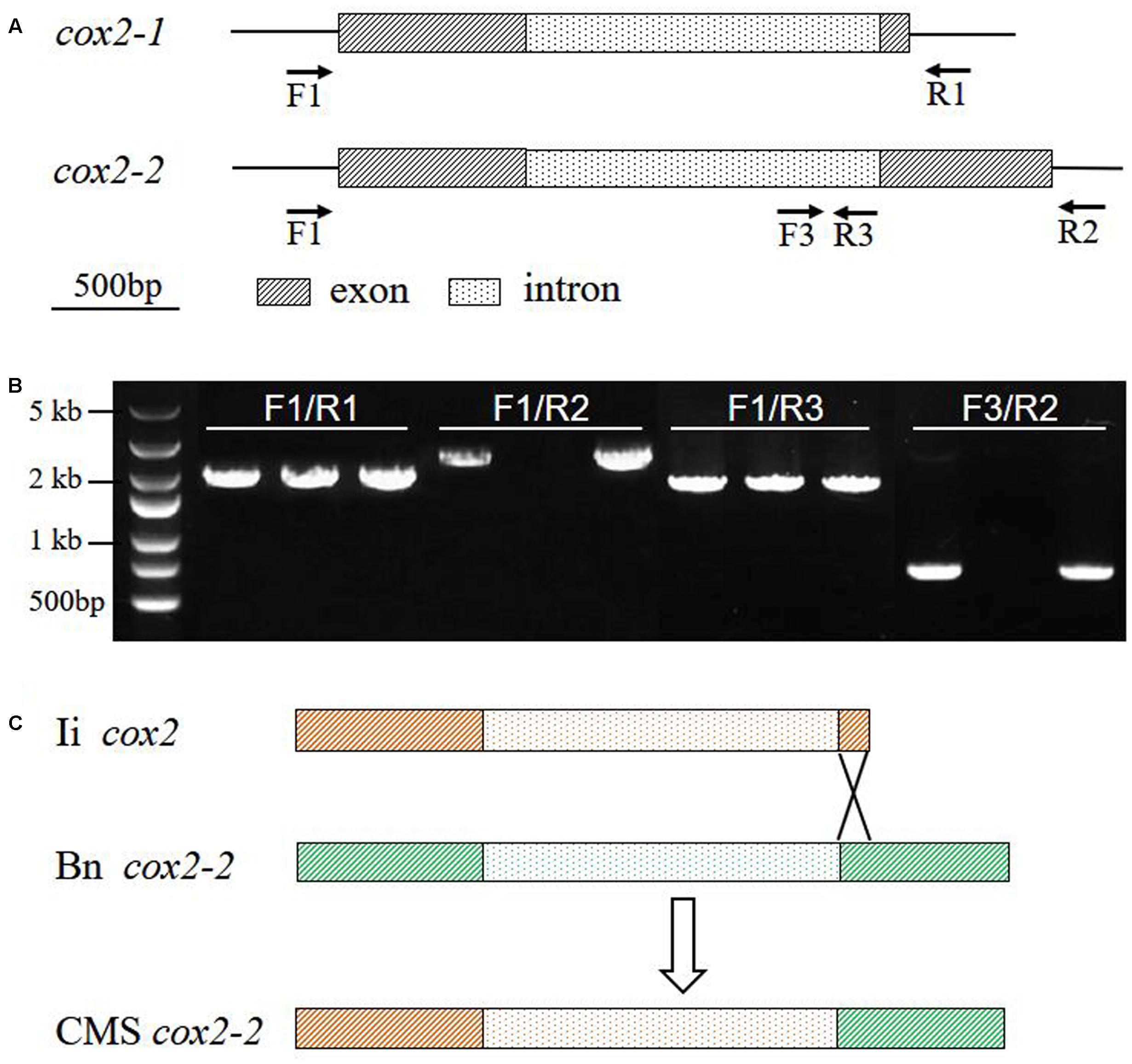
FIGURE 4. The origin of cox2-2 in inap CMS. (A) The binding sites of primers used for specific amplification of mtDNA fragments. (B) PCR amplification products of mtDNA fragments from B. napus, I. indigotica and inap CMS. (C) Gene structures, showing that the gene cox2-2 of inap CMS was the recombinant from cox2 of I. indigotica and cox2-2 of B. napus via homologous recombination. Bn, B. napus; Ii, I. indigotica.
To determine whether inap CMS was different from Ogura and pol CMS in sterility genes, the primers of their respective CMS-associated genes orf138 and orf224 were used for PCR amplification, but did not produce the products, showing that inap CMS did not share these genes (Supplementary Figure S3).
Different Fertility Restoration Relationship of inap CMS from Other CMS Systems
As shown in Table 2, all the F1 hybrids between inap CMS and these restorers of various CMS systems were male sterile and still produced the carpelloid stamens, except for those with the B. napus addition line harboring the woad chromosome E. Although the two alloplamic Ogura CMS lines had the similar phenotype of inap CMS, their restorers with the introgression of restoration gene from one specific radish chromosome F failed to restore our inap CMS. Furthermore, the hybrids between inap CMS and the disomic rape-radish chromosome addition line f (DAL-f) (2n = 39, AACC+1R) with the radish chromosome carrying the restoration gene still displayed male sterility. To identify restorers of inap CMS in B. napus, 112 cultivars of wide origins were used as pollinators (data not shown) and the pods with good seed-sets were produced, but none was found to function as the restorer.
Discussion
The novel inap CMS in B. napus selected from the intertribal somatic hybrids contained the recombinant mtDNA of both parents but biased to woad, and had the stamens converted into carpelloid structures. The sterile phenotype was stable during years and under different ecological environment. The developmental conversion of stamens occurred at the stage of stamen primordium differentiation. It was different from other CMS lines in phenotype, CMS associated genes, and restorer and maintainer relationships.
Homeotic conversion of stamens into carpelloid structures were described in several CMS lines of wheat (Ogihara et al., 1997), tobacco (Farbos et al., 2001), carrot (Linke et al., 2003), rapeseed (Leino et al., 2003), cauliflower (Kamiñski et al., 2012), and broccoli (Shu et al., 2015). Stamens of the CMS lines were replaced by carpelloid organs, thus resembled the B-class genes APETALA3 (AP3) and PISTILLATA (PI) mutants. In B. napus CMS line with the rearranged Arabidopsis mitochondria, downregulation of AP3 in the stamen primordia and subsequent repression of PI resulted in the conversion of stamens into carpelloid organ (Carlsson et al., 2007a). Similarly, the B-class genes were transcriptionally downregulated in carpelloid CMS flowers of tobacco, carrot and wheat (Farbos et al., 2001; Linke et al., 2003; Hama et al., 2004). The mitochondrial background had a distinct influence on nuclear gene expression (Carlsson et al., 2007b). This indicated that CMS-associated mitochondria genes regulated the expression of these nuclear MADS-box genes by mitochondrial retrograde signaling (MRS). Wheat Calmodulin-Binding Protein 1 (WCBP1) significantly upregulated in young spikes of the pistillody line (Yamamoto et al., 2013), indicating that CMS- associated genes played a role in development of pistil-like stamens by MRS involved Ca2+ signaling pathway in wheat CMS.
The most mitochondrial genes of inap CMS were found to originate from I. indigotica (Table 4), which was likely caused by the asymmetric fusion to produce the hybrids (Du et al., 2009). The protoplasts of B. napus were treated by 3 mM iodoacetate (IOA), and those of I. indigotica were irradiated with UV (Du et al., 2009). IOA could inactivate the glyceraldehyde-3-phosphate dehydrogenase, and resulted in the prevention of the glycolytic pathway (Zhao et al., 2008). Only when B. napus protoplasts for fusion contained the normal cytoplasm, the fused cells could divide, formed calli and regenerated plants. Thus, the mitochondria from I. indigotica were retained, and those from B. napus eliminated but several segments were introgressed into the mitochondria of the hybrid by recombination (Table 4). Such situation was also reported in other asymmetric somatic hybrids (Morgan and Maliga, 1987; Akagi et al., 1995). MtDNA sequencing of an Ogura-CMS cybrid derived from somatic fusions between B. napus and sterile radish showed that the mtDNA component mainly inherited from radish (Wang et al., 2012). The B. napus origin of the chloroplast DNA of inap CMS was in accord with the conclusion that the chloroplasts were generally from the iodoacetate-treated parent (Morgan and Maliga, 1987; Leino et al., 2003). This might be due to the better compatibility of recipient chloroplasts with the recipient nucleus, or the loss of the irradiated chloroplasts.
Two copies of cox2 gene in Brassica species (Handa, 2003; Yamagishi et al., 2014) arose from duplication, for it was involved in a large repeat sequence. Via this large repeat, the Brassica mitochondrial genome could be recombined into two subgenomic circles. I. indigotica contained only one copy of this gene. Of two copies of cox2 gene in inap CMS, cox2-1 was from I. indigotica, but novel cox2-2 was the recombinant between cox2 of woad and cox2-2 of rapeseed (Figure 4 and Supplementary Figure S1). Rearrangements in the cox2 region were also reported in somatic hybrids between B. napus and A. thaliana (Landgren and Glimelius, 1994; Leino et al., 2003), B. napus and R. sativus Kosena CMS (Sakai and Imamura, 1990). CMS-associated genes identified in many CMS systems (Wang et al., 1995; Chen et al., 2011; Tanaka et al., 2012; Heng et al., 2014) were located in flanking sequence of the known mitochondrial genes, and co-transcribed with it. Protoplast fusion offered an opportunity for mitochondrial genome recombination, leading to appearance of novel CMS genes. Three atp9 genes were present in the alloplasmic Tournefortii-Stiewe CMS of B. napus, which was generated by protoplast fusion with B. tournefortii (Dieterich et al., 2003). MtDNA rearrangements happened in the upstream of one of these genes and generated a chimeric gene orf193 that was co-transcribed with atp9. An alloplasmic B. juncea CMS line derived from somatic hybridization with Diplotaxis catholica had two copies of coxI gene, and the recombinant coxI-2 gene was suggested to cause CMS (Pathania et al., 2007). For the novel cox2-2 gene was expressed in the inap CMS flowers weakly and insignificantly from restorer plant, it was likely not responsible for male sterility.
The identification of the restorer and maintainer relationship is one of the most classical methods to distinguish different types of sterile cytoplasm (Wan et al., 2008). The test results showed that the fertility of inap CMS failed to be restored by restorer lines from Ogura, pol, nap and Tournefortii-Stiewe CMS systems. Budahn et al. (2008) found that rapeseed plants with the alien radish cytoplasm showed pistilloid stamens with ovules and stigmatoid tips, and the radish chromosome F could eliminate the disadvantageous effect of alien cytoplasm in B. napus. Additionally, the chromosome F did not restore Ogura CMS. In spite of the similar phenotype with inap CMS, the radish chromosome F was unable to restore its fertility, indicating that these two CMS lines had different CMS-associated genes and mechanisms of male sterility. No restorers were screened among 112 B. napus cultivars. This further testified that the fertility restoration gene(s) of alloplasmic CMS lines generally existed in nuclear genome of cytoplasm donor species. The restoration gene of a B. napus CMS line with rearranged A. thaliana mtDNA was located on chromosome III of A. thaliana (Leino et al., 2004). The restorer line for Nsa CMS of B. napus developed by the somatic hybridization with Sinapis arvensis was one disomic addition line with the restoration gene on the alien chromosome (Wei et al., 2010). Similarly, the restoration gene(s) for our inap CMS also existed on one specific woad chromosome, because only the monosomic alien addition line of B. napus with woad chromosome e among the complete set of such lines and also the same cytoplasm as inap CMS produced the normal flowers with fertile pollen grains (Kang et al., 2014). To breed the restorer line for inap CMS, the chromosomal segment carrying the fertility restoration gene(s) should be introgressed into one of rapeseed chromosome and the lines (2n = 38) with high restoration capability selected, as excellently exemplified by the long-term development of the radish introgression carrying the Rfo restorer gene for the Ogu-INRA CMS in B. napus (Delourme et al., 1991, 1998, 1999; Primard-Brisset et al., 2005). The identification of some progeny plants (2n = 38) with the restored flower development and pollen fertility from the addition line gave the expectation that the new CMS and fertility restoration system should be developed for rapeseed hybrid production in near future.
Author Contributions
ZL and LK conceived and designed the experiments, analyzed the data and wrote the paper. LK performed the experiments. PL participated in mtDNA analysis. AW performed the histological section experiments. XG provided technical expertise for molecular analysis and scientific discussions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This study was supported by National Natural Science Foundation of China project (No. 31601331) and China Postdoctoral Science Foundation (Grant No. 2015M582238).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00529/full#supplementary-material
References
Akagi, H., Shimada, H., and Fujimura, T. (1995). High-frequency inter-parental recombination between mitochondrial genomes of rice cybrids. Curr. Genet. 29, 58–65. doi: 10.1007/BF00313194
Atri, C., Kaur, B., Sharma, S., Gandhi, N., Verma, H., Goyal, A., et al. (2016). Substituting nuclear genome of Brassica juncea (L.) Czern & Coss. In cytoplasmic background of Brassica fruticulosa results in cytoplasmic male sterility. Euphytica 209, 31–40. doi: 10.1007/s10681-015-1628-4
Bannerot, H., Boulidard, L., Cauderon, Y., and Tempé, J. (1974). “Transfer of cytoplasmic male sterility from Raphanus sativus to Brassica oleracea,” in Proceedings of the Eucarpia Meeting on Cruciferae, Dundee, 52–54.
Bonhomme, S., Budar, F., Lancelin, D., Small, I., Defrance, M., and Pelletier, G. (1992). Sequence and transcript analysis of the Nco2.5 Ogura-specific fragment correlated with cytoplasmic male sterility in Brassica cybrids. Mol. Gen. Genet. 235, 340–348. doi: 10.1007/BF00279379
Budahn, H., Schrader, O., and Peterka, H. (2008). Development of a complete set of disomic rape-radish chromosome-addition lines. Euphytica 162, 117–128. doi: 10.1007/s10681-007-9609-x
Budar, F., Delourme, R., and Pelletier, G. (2004). “Male sterility,” in Biotechnology in Agriculture and Forestry, eds E. C. Pua, and C. J. Douglas (NewYork, NY: Springer), 43–64.
Carlsson, J., Lagercrantz, U., Sundstrom, J., Teixeira, R., Wellmer, F., Meyerowitz, E. M., et al. (2007a). Microarray analysis reveals altered expression of a large number of nuclear genes in developing cytoplasmic male sterile Brassica napus flowers. Plant J. 49, 452–462. doi: 10.1111/j.1365-313X.2006.02975.x
Carlsson, J., Leino, M., and Glimelius, K. (2007b). Mitochondrial genotypes with variable parts of Arabidopsis thaliana DNA affect development in Brassica napus lines. Theor. Appl. Genet. 115, 627–641. doi: 10.1007/s00122-007-0593-2
Chase, C. D. (2007). Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 23, 81–90. doi: 10.1016/j.tig.2006.12.004
Chen, J. M., Guan, R. Z., Chang, S. X., Du, T. Q., Zhang, H. S., and Xing, H. (2011). Substoichiometrically different mitotypes coexist in mitochondrial genomes of Brassica napus L. PLoS ONE 6:e17662. doi: 10.1371/journal.pone.0017662
Chen, L. T., and Liu, Y. G. (2014). Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 65, 579–606. doi: 10.1146/annurev-arplant-050213-040119
Dellaporta, S. L., Wood, J., and Hicks, J. B. (1983). A plant DNA mini preparation: version II. Plant Mol. Biol. Rep. 1, 19–21.
Delourme, R., and Budar, F. (1999). “Male sterility,” in Biology of Brassica coenospecies, ed. C. Gómez-Campo (Amsterdam: Elsevier Science), 185–216.
Delourme, R., Eber, F., and Renard, M. (1991). “Radish cytoplasmic male sterility in rapeseed: breeding restorer lines with a good female fertility,” in Proceedings of the 8th International Rapeseed Conference, Saskatoon, SK,1506–1510.
Delourme, R., Foisset, N., Horvais, R., Barret, P., Champagne, G., Cheung, W. Y., et al. (1998). Characterization of the radish introgression carrying the Rfo restorer gene for the Ogu-INRA cytoplasmic male sterility in rapeseed (Brassica napus L.). Theor. Appl. Genet. 97, 129–134. doi: 10.1007/s001220050876
Delourme, R., Horvais, R., Vallée, P., and Renard, M. (1999). “Double low restored F1 hybrids can be produced with the Ogu-INRA CMS in rapeseed,” in Proceedings of the 10th International Rapeseed Conference, Canberra, ACT, 26–29.
Dieterich, J. H., Braun, H. P., and Schmitz, U. K. (2003). Alloplasmic male sterility in Brassica napus (CMS ‘Tournefortii-Stiewe’) is associated with a special gene arrangement around a novel atp9 gene. Mol. Genet. Genomics 269, 723–731. doi: 10.1007/s00438-003-0886-3
Du, X. Z., Ge, X. H., Yao, X. C., Zhao, Z. G., and Li, Z. Y. (2009). Production and cytogenetic characterization of intertribal somatic hybrids between Brassica napus and Isatis indigotica and backcross progenies. Plant Cell Rep. 28, 1105–1113. doi: 10.1007/s00299-009-0712-4
Farbos, I., Mouras, A., Bereterbide, A., and Glimelius, K. (2001). Defective cell proliferation in the floral meristem of alloplasmic plants of Nicotiana tabacum leads to abnormal floral organ development and male sterility. Plant J. 26, 131–142. doi: 10.1046/j.1365-313x.2001.01011.x
Hama, E., Takumi, S., Ogihara, Y., and Murai, K. (2004). Pistillody is caused by alterations to the class-B MADS-box gene expression pattern in alloplasmic wheats. Planta 218, 712–720. doi: 10.1007/s00425-003-1157-6
Handa, H. (2003). The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 31, 5907–5916. doi: 10.1093/nar/gkg795
Hanson, M. R., and Bentolila, S. (2004). Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16, S154–S169. doi: 10.1105/tpc.015966
Heng, S. P., Wei, C., Jing, B., Wan, Z. J., Wen, J., Yi, B., et al. (2014). Comparative analysis of mitochondrial genomes between the hau cytoplasmic male sterility (CMS) line and its iso-nuclear maintainer line in Brassica juncea to reveal the origin of the CMS-associated gene orf288. BMC Genomics 15:322. doi: 10.1186/1471-2164-15-322
Horn, R., Gupta, K. J., and Colombo, N. (2014). Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion 19, 198–205. doi: 10.1016/j.mito.2014.04.004
Iwabuchi, M., Koizuka, N., Fujimoto, H., Sakai, T., and Imamura, J. (1999). Identification and expression of the kosena radish (Raphanus sativus cv. Kosena) homologue of the ogura radish CMS-associated gene, orf138. Plant Mol. Biol. 39, 183–188. doi: 10.1023/A:1006198611371
Kamiñski, P., Dyki, B., and Stêpowska, A. A. (2012). Improvement of cauliflower male sterile lines with Brassica nigra cytoplasm, phenotypic expression and possibility of practical application. J. Agric. Sci. 4, 190–200. doi: 10.5539/jas.v4n4p190
Kang, L., Du, X. Z., Zhou, Y. Y., Zhu, B., Ge, X. H., and Li, Z. Y. (2014). Development of a complete set of monosomic alien addition lines between Brassica napus and Isatis indigotica (Chinese woad). Plant Cell Rep. 33, 1355–1364. doi: 10.1007/s00299-014-1621-8
Landgren, M., and Glimelius, K. (1994). A high frequency of intergenomic mitochondrial recombination and an overall biased segregation of B. campestris or recombined B. campestris mitochondria were found in somatic hybrids made within Brassicaceae. Theor. Appl. Genet. 87, 854–862. doi: 10.1007/bf00221138
Landgren, M., Zetterstrand, M., Sundberg, E., and Glimelius, K. (1996). Alloplasmic male-sterile Brassica lines containing B. tournefortii mitochondria express an ORF 3’ of the atp6 gene and a 32 kDa protein. Plant Mol. Biol. 32, 879–890. doi: 10.1007/BF00020485
Leino, M., Teixeira, R., Landgren, M., and Glimelius, K. (2003). Brassica napus lines with rearranged Arabidopsis mitochondria display CMS and a range of developmental aberrations. Theor. Appl. Genet. 106, 1156–1163. doi: 10.1007/s00122-002-1167-y
Leino, M., Thyselius, S., Landgren, M., and Glimelius, K. (2004). Arabidopsis thaliana chromosome III restores fertility in a cytoplasmic male-sterile Brassica napus line with A. thaliana mitochondrial DNA. Theor. Appl. Genet. 109, 272–279. doi: 10.1007/s00122-004-1644-6
Linke, B., Nothnagel, T., and Borner, T. (2003). Flower development in carrot CMS plants: mitochondria affect the expression of MADS box genes homologous to GLOBOSA and DEFICIENS. Plant J. 34, 27–37. doi: 10.1046/j.1365-313X.2003.01703.x
Liu, J., Xiang, R. Y., Wang, W. M., Mei, D. S., Li, Y. C., Mason, A. S., et al. (2015). Cytological and molecular analysis of Nsa CMS in Brassica napus L. Euphytica 206, 279–286. doi: 10.1007/s10681-015-1443-y
Meur, G., Gaikwad, K., Bhat, S. R., Prakash, S., and Kirti, P. B. (2006). Homeotic-like modification of stamens to petals is associated with aberrant mitochondrial gene expression in cytoplasmic male sterile Ogura Brassica juncea. J. Genet. 85, 133–139. doi: 10.1007/BF02729019
Morgan, A., and Maliga, P. (1987). Rapid chloroplast segregation and recombination of mitochondrial DNA in Brassica cybrids. Mol. Gen. Genet. 209, 240–246. doi: 10.1007/BF00329649
Ogihara, Y., Futami, K., Tsuji, K., and Murai, K. (1997). Alloplasmic wheats with Aegilops crassa cytoplasm which express photoperiod-sensitive homeotic transformations of anthers, show alterations in mitochondrial DNA structure and transcription. Mol. Gen. Genet. 255, 45–53. doi: 10.1007/s004380050473
Oshima, M., Kikichi, R., Imamura, J., and Handa, H. (2010). Origin of the CMS gene locus in rapeseed cybrid mitochondria: active and inactive recombination produces the complex CMS gene region in the mitochondrial genomes of Brassicaceae. Genes Genet. Syst. 85, 311–318. doi: 10.1266/ggs.85.311
Pathania, A., Kumar, R., Kumar, V. D., Ashutosh, Dwivedi, K. K., Kirti, P. B., et al. (2007). A duplicated coxI gene is associated with cytoplasmic male sterility in an alloplasmic Brassica juncea line derived from somatic hybridization with Diplotaxis catholica. J. Genet. 86, 93–101. doi: 10.1007/s12041-007-0014-y
Prakash, S., Bhat, S. R., Quiros, C. F., Kirti, P. B., and Chopra, V. L. (2009). Brassica and its close allies: cytogenetics and evolution. Plant Breed. Rev. 31, 21–187. doi: 10.1002/9780470593783.ch2
Prakash, S., Takahata, Y., Kirti, P. B., and Chopra, V. L. (1999). “Cytogenetics,” in Biology of Brassica Coenospecies, ed. C. Gómez-Campo (Amsterdam: Elsevier Science), 59–106.
Primard-Brisset, C., Poupard, J. P., Horvais, R., Eber, F., Pelletier, G., Renard, M., et al. (2005). A new recombined double low restorer line for the Ogu-INRA cms in rapeseed (Brassica napus L.). Theor. Appl. Genet. 111, 736–746. doi: 10.1007/s00122-005-2059-8
Sakai, T., and Imamura, J. (1990). Intergeneric transfer of cytoplasmic male sterility between Raphanus sativus (cms line) and Brassica napus through cytoplast-protoplast fusion. Theor. Appl. Genet. 80, 421–427. doi: 10.1007/BF00210083
Sanchez-Puerta, M. V., Zubko, M. K., and Palmer, J. D. (2015). Homologous recombination and retention of a single form of most genes shape the highly chimeric mitochondrial genome of a cybrid plant. New Phytol. 206, 381–396. doi: 10.1111/nph.13188
Shu, J. S., Liu, Y. M., Li, Z. S., Zhang, L. L., Fang, Z. Y., Yang, L. M., et al. (2015). Organelle simple sequence repeat markers help to distinguish carpelloid stamen and normal cytoplasmic male sterile sources in broccoli. PLoS ONE 10:e0138750. doi: 10.1371/journal.pone.0138750
Smyth, D. R., Bowman, J. L., and Meyerowitz, E. M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. doi: 10.1105/tpc.2.8.755
Tanaka, Y., Tsuda, M., Yasumoto, K., Yamagishi, H., and Terachi, T. (2012). A complete mitochondrial genome sequence of Ogura-type male-sterile cytoplasm and its comparative analysis with that of normal cytoplasm in radish (Raphanus sativus L.). BMC Genomics 13:352. doi: 10.1186/1471-2164-13-352
Vos, P., Hogers, R., Bleeker, M., Reijans, M., van de Lee, T., Hornes, M., et al. (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23, 4407–4414. doi: 10.1093/nar/23.21.4407
Wan, Z. J., Jing, B., Tu, J. X., Ma, C. Z., Shen, J. X., Yi, B., et al. (2008). Genetic characterization of a new cytoplasmic male sterility system (hau) in Brassica juncea and its transfer to B. napus. Theor. Appl. Genet. 116, 355–362. doi: 10.1007/s00122-007-0673-3
Wang, H. M., Ketela, T., Keller, W. A., Gleddie, S. C., and Brown, G. G. (1995). Genetic correlation of the orf224/atp6 gene region with Polima CMS in Brassica somatic hybrids. Plant Mol. Biol. 27, 801–807. doi: 10.1007/BF00020232
Wang, J., Jiang, J. J., Li, X. M., Li, A. M., Zhang, Y. T., Guan, R. Z., et al. (2012). Complete sequence of heterogenous-composition mitochondrial genome (Brassica napus) and its exogenous source. BMC Genomics 13:675. doi: 10.1186/1471-2164-13-675
Wei, W. H., Li, Y. C., Wang, L. J., Liu, S. Y., Yan, X. H., Mei, D. S., et al. (2010). Development of a novel Sinapis arvensis disomic addition line in Brassica napus containing the restorer gene for Nsa CMS and improved resistance to Sclerotinia sclerotiorum and pod shattering. Theor. Appl. Genet. 120, 1089–1097. doi: 10.1007/s00122-009-1236-6
Yamagishi, H., and Bhat, S. R. (2014). Cytoplasmic male sterility in Brassicaceae crops. Breed. Sci. 64, 38–47. doi: 10.1270/jsbbs.64.38
Yamagishi, H., Tanaka, Y., and Terachi, T. (2014). Complete mitochondrial genome sequence of black mustard (Brassica nigra; BB) and comparison with Brassica oleracea (CC) and Brassica carinata (BBCC). Genome 57, 577–582. doi: 10.1139/gen-2014-0165
Yamamoto, M., Shitsukawa, N., Yamada, M., Kato, K., Takumi, S., Kawaura, K., et al. (2013). Identification of a novel homolog for a calmodulin-binding protein that is upregulated in alloplasmic wheat showing pistillody. Planta 237, 1001–1013. doi: 10.1007/s00425-012-1812-x
Keywords: Brassica napus, Isatis indigotica, cytoplasmic male sterility, mitochondrial rearrangement, carpelloid stamen
Citation: Kang L, Li P, Wang A, Ge X and Li Z (2017) A Novel Cytoplasmic Male Sterility in Brassica napus (inap CMS) with Carpelloid Stamens via Protoplast Fusion with Chinese Woad. Front. Plant Sci. 8:529. doi: 10.3389/fpls.2017.00529
Received: 10 January 2017; Accepted: 24 March 2017;
Published: 06 April 2017.
Edited by:
Maoteng Li, Huazhong University of Science and Technology, ChinaReviewed by:
Shengwu Hu, Northwest A&F University, ChinaLiezhao Liu, Southwest University, China
Jacob A. Tennessen, Oregon State University, USA
Copyright © 2017 Kang, Li, Wang, Ge and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zaiyun Li, lizaiyun@mail.hzau.edu.cn.
 Lei Kang
Lei Kang Pengfei Li
Pengfei Li Aifan Wang
Aifan Wang Xianhong Ge
Xianhong Ge Zaiyun Li
Zaiyun Li