- 1Key Laboratory of Bio-resource and Eco-environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu, China
- 2School of Life Sciences, Capital Normal University, Beijing, China
Synopsis
This work demonstrates that PpABI3 contributes to freezing tolerance regulation in Physcomitrella patens.
Transcription factor ABSCISIC ACID INSENSITIVE3 (ABI3) is known to play a major role in regulating seed dormancy, germination, seedling development as well as stress responses. ABI3 is conserved among land plants; however, its roles in non-seed plants under stress conditions have not been well characterized. In this study, we report that ABI3 is involved in freezing tolerance regulation during cold acclimation at least in part through ABA signaling pathway in moss Physcomitrella patens (P. patens). Deletion of PpABI3 (Δabi3-1) compromises the induction of genes related to cold response and antioxidative protection, resulting in reduced accumulation of cryoprotectants and antioxidants. In addition, photosystem II (PSII) activity is repressed in Δabi3-1 during cold acclimation partially due to alternations of photosynthetic protein complexes compositions. The gametophyte of Δabi3-1 displays severe growth inhibition and developmental deficiency under low temperature condition, while two independent complementary lines display phenotypes similar to that of wild-type P. patens (WT). Furthermore, the freezing tolerance of Δabi3-1 was significantly affected by deletion of PpABI3. These data revealed that PpABI3 plays an important role in low temperature response and freezing tolerance in P. patens.
Introduction
Nearly two-thirds of the world's plants grow in regions below 0°C (Beck et al., 2004; Beike et al., 2015). Freezing stress adversely affects plant growth and crop productivity globally while plants have evolved mechanisms to tolerate freezing stress through cold acclimation. In this way, plants can efficiently activate their cellular signal transduction and metabolisms strategies to acquire enhanced freezing tolerance (Miura and Furumoto, 2013). Previous studies have documented that cold acclimation is a complex process involving many biochemical and physiological changes in plant cells, including alterations of lipid compositions, accumulation of antioxidants and reduction of photoinhibition through non-photochemical quenching (NPQ) (Thomashow, 1999; Beike et al., 2015). During cold acclimation, expression profile of cold-responsive genes (COR) is reprogrammed to activate cold responsive signaling and these CORs are coordinated with multiple signaling pathways to enhance freezing tolerance in plants (Chinnusamy et al., 2007; Jeon and Kim, 2013; Miura and Furumoto, 2013).
ABSCISIC ACID INSENSITIVE3 (ABI3) is a plant-specific B3 domain-containing transcription factor (TF), which is conserved in higher plant (Romanel et al., 2009). In angiosperm plants, ABI3 participates in seed development and maturation (Monke et al., 2012; Delmas et al., 2013). In addition, ABI3 also plays essential regulatory roles in plastids development, vegetative growth, flowering time regulation and abiotic stress responses (Parcy et al., 1997; Rohde et al., 2000; Frank et al., 2005; Khandelwal et al., 2010). Transcriptomic analysis revealed that ABI3 appears to affect overlapping sets of stress-responsive genes mediated by CBF1/DREB1A, a key TF regulating freezing tolerance in Arabidopsis (Tamminen et al., 2001). Over-expression of ABI3 gene enhances freezing tolerance in response to low temperature in Arabidopsis (Tamminen et al., 2001).
P. patens is a representative ancestor of land plant which highly tolerates drought, salt, osmotic and cold stress (Frank et al., 2005; Takezawa et al., 2015). Data showed that PpABI3A enhances ABA-induced Em-GUS expression in P. patens in a manner similar to that in angiosperms (Marella et al., 2006). PpABI3s are required for P. patens vegetative tissue to tolerate desiccation via regulating multiple ABA-responsive genes, which might participate in water deficit responsive modulation (Khandelwal et al., 2010; Yotsui et al., 2016). PpABI3 regulates ABA-responsive genes through ABRE element in their promoter. Specifically, RY motif in Em genes is also involved in PpABI3-mediated transcription regulation (Sakata et al., 2010). Furthermore, PpABI3 might regulate set of ABA-responsive gene transcription via the ACTT-core element in coordination with nuclear factor Y (NF-Y) complex (Yotsui et al., 2013). In fact, one of the three homologs of PpABI3 could partially rescue the phenotype of abi3-6 mutant of Arabidopsis (Marella et al., 2006). These results indicate that ABA signaling and ABI3 are conserved in angiosperm plants and bryophytes. The whole-genome transcriptomic and proteomic data suggested that cold acclimation in P. patens might be similar to that in higher plants (e.g., Arabidopsis). However, whether PpABI3 plays a regulatory role in cold acclimation and freezing tolerance of P. patens is not clear.
In this study, we report that PpABI3 is involved in regulating cold response and freezing tolerance in P. patens. PpABI3 deletion mutant Δabi3-1, in which all the three PpABI3 homologs were deleted, displays severe growth inhibition and developmental deficiency of gametophytes under low temperature condition, while complementary expression three gene of PpABI3 could rescue these phenotype and display similar to that of WT. In addition, deletion of PpABI3 compromises the induction of genes related to cold response and reduce the accumulation of antioxidants and cryoprotectants. Furthermore, photosystem II activity is significantly repressed in Δabi3-1 during cold acclimation and the mutant exhibits lower freezing tolerance despite after cold acclimation. Together, all the results in this study demonstrated that PpABI3 is involved in regulating cold acclimation and freezing tolerance in P. patens.
Results
Disruption of PpABI3 Inhibits Gametophyte Growth of P. patens under Cold Condition
P. patens genome encodes three ABI3 genes, PpABI3A, PpABI3B, and PpABI3C, which share higher identity in the conserved basic domains (Marella et al., 2006; Figures S1A,B; Table S2). To investigate the possible regulatory role of PpABI3 in cold response, expression of PpABI3 genes was first determined in WT with or without cold acclimation. We found that PpABI3A and PpABI3B were significantly induced in protonema incubated under 10°C for 2 weeks, while expression of PpABI3C was only slightly induced by low temperature (Figure S1C). These data indicated that all the PpABI3 genes might positively regulate cold response in P. patens. The effect of PpABI3 disruption on gametophyte growth and development under low temperature condition was then investigated. PpABI3 deletion mutant line Δabi3-1, in which all three PpABI3 genes were deleted by sequential gene targeting, was generated (Khandelwal et al., 2010; Figures S2A,B). Seven-day-old protonematal tissues of WT and mutant strains were then cultured at growth temperature (25°C) or low temperature (10°C) for 4 weeks. Compared to the growth condition, the growth of WT and Δabi3-1 were both repressed under 10°C. The biomass of Δabi3-1 was significantly lower than that of wild type during cold stress (Figures 1A,C). Meanwhile, Δabi3-1 generated less gametophore during cold incubation than that of WT, although this activity was even higher in the mutant when grown at 25°C (Figures 1A,D). Interestingly, we found that transcription of two genes, PpSHI1 (Pp1s373_11V6) and PpSHI2 (Pp1s19_109V6) which function in auxin biosynthesis (Eklund et al., 2010), was higher in Δabi3-1 compared to that in WT when grown at 25°C, which might explain why the number of gametophore per colony in the mutant is higher than in WT, and suggest a possible role of PpABI3 in auxin biosynthesis (Figure S3). In addition, the cormus of Δabi3-1 was shorter and possessed fewer leaves after low temperature incubation (Figures 1B,E,F; Figure S4).
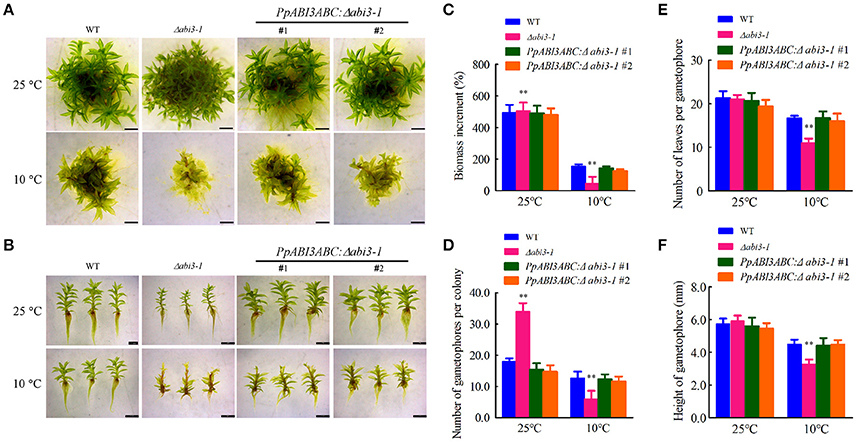
Figure 1. Deletion of PpABI3 inhibits growth of P. patens under cold condition. (A) Analysis of gametophyte growth. Seven-day-old tissues of WT, Δabi3-1 and two complementary lines were inoculated on fresh BCD medium, and then grown under growth temperature (25°C) or lower temperature (10°C) for 4 weeks before photographs were taken. (B) Representative images of gametophores of each strains. Gametophores of wild type, Δabi3-1and two complementary lines incubated under 25°C or 10°C were collected to show their different phenotypes. Bar = 2 mm for all panels. (C) Analysis of biomass increment in (A). Fresh weight of each strain was determined for biomass increment analysis in relation to the starting inoculated biomass. Error bars represent SD (n = 3) and Two-way ANOVA was used to determine the statistical significance (**P < 0.01). (D) Number of gametophores that emerged on the colonies of each strain. The colonies were incubated under 25°C or 10°C for 4 weeks and gametophores that emerged were counted. Error bars indicate mean ± SD (n = 6) and Two-way ANOVA was used to determine the statistical significance (**P < 0.01). (E) Number of leaves that generated per gametophore of each strain. The colonies were incubated under 25°C or 10°C for 4 weeks and representative gametophores were collected for estimation of the leaves amount. Error bars indicate mean ± SD (n = 10) and Two-way ANOVA was used to determine the statistical significance (**P < 0.01). (F) Mean height of gametophores analysis for the wild type, Δabi3-1 and PpABI3ABC:Δabi3-1 lines. Error bars indicate mean ± SD (n > 60) and Two-way ANOVA was used to determine the statistical significance (**P < 0.01).
We have also generated the complementary lines (PpABI3ABC:Δabi3-1) by introducing all the three PpABI3 genes into Δabi3-1 background and confirmed the complementary lines by RT-PCR assay (Figure S2C). Data showed that the growth inhibition in Δabi3-1 was rescued by PpABI3A, PpABI3B and PpABI3C in the complementary lines (Figures 1A–F). The elevated electrolyte leakage (EL) in Δabi3-1 under low temperature condition was also decreased to a level similar to WT in both complementary lines (Figure S2D). These results together demonstrated that deletion of PpABI3 represses the gametophyte growth of P. patens under low temperature condition and PpABI3 might be involved in regulating growth and development in P. patens.
PpABI3 Participates in Regulating Cryoprotectants and Antioxidants Accumulation during Cold Acclimation
Since PpABI3 plays a role in cold response, we then hypothesized that they might contribute to cold acclimation regulation. Cold stress imposes cell injury and oxidative damages to plants (Mahajan and Tuteja, 2005; Demidchik et al., 2014; Hossain et al., 2015). To determine whether PpABI3 is involved in regulating cell injury and redox homeostasis during cold acclimation, WT and Δabi3-1 mutant were subjected to 25°C or 10°C for 2 weeks, then EL and ROS accumulation were determined. Data showed that both EL and ROS were induced in WT and Δabi3-1 after incubation under 10°C (Figures 2A,B). However, the accumulation of EL and H2O2 in Δabi3-1 were significantly higher than that in WT (Figures 2A,B), indicating that cold stress disrupted cellular redox homeostasis and caused severely cell injury in Δabi3-1. We further detected the accumulation of MDA, a marker of lipid oxidation in response to environmental stresses (Davey et al., 2005). The content of MDA was also higher in Δabi3-1 compared to that of WT under low temperature condition (Figure 2C). These results indicated that PpABI3 is involved in low temperature response in P. patens. In addition, accumulations of cryoprotectants including proline, soluble sugar and soluble protein, all of which could contribute to cold acclimation, were significantly lower in Δabi3-1 than that in WT (Figures 2D–F). Consistently, we found that the expression of LEA-like protein and sucrose synthase which function in plant cold response (Beike et al., 2015) were also much lower in Δabi3-1 under low temperature condition (Figures 2G,H). These results demonstrated that PpABI3 plays a regulatory role in redox homeostasis and cryoprotectants accumulation during cold acclimation.
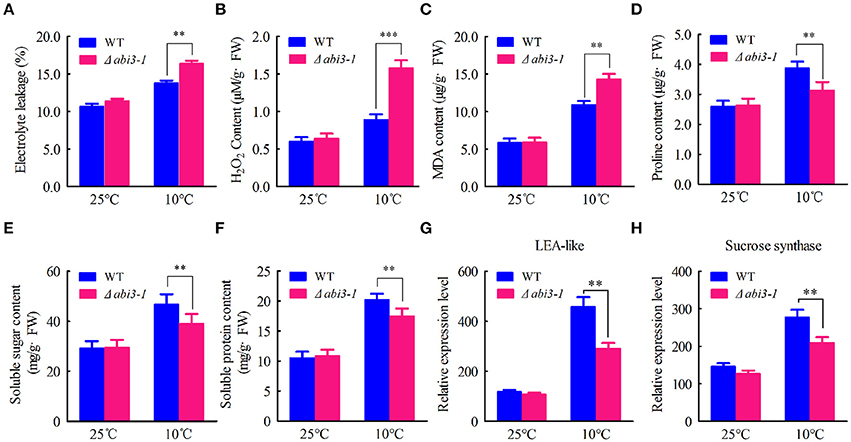
Figure 2. PpABI3 participates in low temperature response of P. patens. (A–C) Analysis of cell injury and oxidative damage. Tissues of WT and Δabi3-1 with or without cold acclimation under 10°C were collected to determine electrolyte leakage (EL) rate (A), hydrogen peroxide (H2O2) content (B) and malonyldialdehyde (MDA) content (C). (D–F) Analysis the accumulation of cryoprotectants. Tissues of WT and Δabi3-1 with or without cold acclimation were collected to estimate proline content (D), total soluble sugar content (E) and total soluble protein content (F). Error bars represent SD (n = 3) and Two-way ANOVA was used to determine the statistical significance (**P < 0.01). (G,H) Quantitative RT-PCR to analyse the expression of a late embryogenesis abundant (LEA)-like protein-coding gene (Pp1s267_21V6.1) and a sucrose synthase-coding gene (Pp1s93_98V6.1). PpACTIN5 (Pp1S381_21V6) was used as internal control. Error bars represent SD (n = 3) and Two-way ANOVA was used to determine the statistical significance (**P < 0.01; ***P < 0.001).
Cellular antioxidative signaling will be induced to protect cells from oxidative damage by scavenging ROS in plants during cold acclimation (Blokhina et al., 2003; Gill and Tuteja, 2010). We then determined the enzymatic activity of ascorbate peroxidase (APX), catalase (CAT), peroxidase (POD) as well as superoxide dismutase (SOD) in WT and Δabi3-1 under control and cold conditions. Data showed that the activity of APX, CAT, POD and SOD were relatively increased in both WT and Δabi3-1 during cold acclimation (Figures 3A–D). However, the increase amplitudes of the four enzymes in Δabi3-1 were significantly lower than in WT (Figures 3A–D). These results suggest that PpABI3 contributes to redox homeostasis regulation during cold acclimation in P. paten. We further found that transcription level of genes encoding antioxidant enzymes, PpAPX (Pp1s277_34V6), PpCAT (Pp1s422_8V6), PpPOD (Pp1s98_2V6), and PpSOD (Pp1s22_320V6), in Δabi3-1 were much lower than that in WT during cold acclimation (Figures 4A–D). These results were consistent with the lower antioxidant enzymatic activity in Δabi3-1 during cold acclimation (Figures 3A–D).
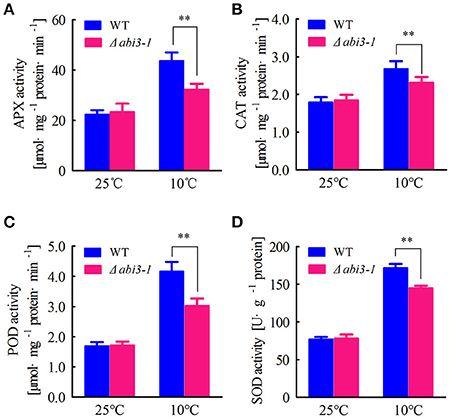
Figure 3. PpABI3 is involved in regulating enzymatic antioxidative signaling. (A–D) Antioxidant enzymatic activities analysis. Protonema after incubation under standard growth temperature (25°C) or low temperature (10°C) for 2 weeks were harvested to analyze ascorbate peroxidase (APX) activities (A), catalase (CAT) activities (B), glutathione peroxidase (POD) activities (C) and superoxide dismutase (SOD) (D). Error bars represent SD (n = 3) and two-way ANOVA was used to determine the statistical significance (**P < 0.01). Experimental details are found in Materials and Methods.
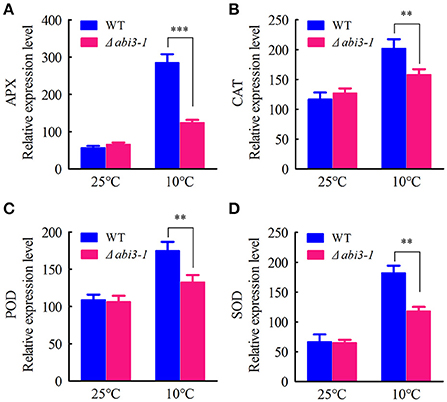
Figure 4. Expression of antioxidant enzymes mediated by PpABI3 during cold acclimation. (A–D) The transcriptional levels of genes encoding ascorbate peroxidase (APX: Pp1S277_34V6), catalase (CAT: Pp1S422_8V6), glutathione peroxidase (POD: Pp1S98_2V6) and superoxide dismutase (SOD: Pp1S22_320V6) were analyzed by quantitative RT-PCR in wild-type and Δabi3-1 with or without cold acclimation. Error bars represent SD (n = 3), and two-way ANOVA was used to determine the statistical significance (**P < 0.01; ***P < 0.001).
PpABI3 Participates in Regulating Expression of Multiple Genes during Cold Acclimation
To investigate the mechanism by which PpABI3 regulates cold acclimation, we next analyzed the contributions of PpABI3 in regulating cold-responsive genes (COR) during cold acclimation. We found that the cold-induction target genes such as PpCOR47 (Pp1s442_22V6.2), PpRD29A (Pp1s203_40V6.1), and PpCSP3 (PP1S103_65V6.1) were drastically repressed in Δabi3-1 compared to WT (Figure 5A), which correlates with the growth inhibition of Δabi3-1 under low temperature. Interestingly, two transcription factors, PpDREB1/CBF (Pp1s60_228V6.1) and PpAP2/EREBP (Pp1s373_18V6.1), which also contributed to cold acclimation regulation (Beike et al., 2015), were induced significantly in WT during cold acclimation, while their accumulation in Δabi3-1 was also partially repressed (Figure 5B), suggesting that PpABI3 might regulate cold acclimation in part through PpDREB1/CBF and PpAP2/EREBP. We also determined the transcription of PpABI5 (Pp1s49_161V6) in WT and Δabi3-1 P. patens. Although ABI3 and ABI5 are intensely related in higher plants (Lim et al., 2013), we did not observe any detectable difference of PpABI5 transcription between WT and Δabi3-1 under both control and cold conditions (Figure 5B). Together, these results suggested that PpABI3 is involved in cold acclimation by regulating COR genes expression in P. patens.
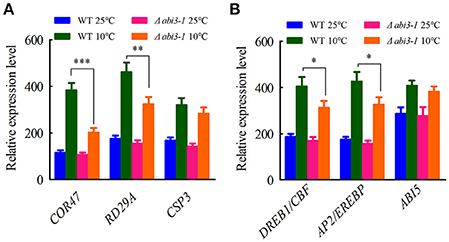
Figure 5. Cold-responsive genes are regulated by PpABI3 during cold acclimation. (A) Relative transcription of three cold responsive genes. The transcriptional levels of three genes: PpCOR47 (Dehydrin-coding gene: Pp1s442_22V6.2), PpRD29A (Desiccation-responsive protein 29A: PP1S203_40V6.1) and PpCSP3 (Cold shock protein: Pp1s103_65V6.1) were analyzed by quantitative RT-PCR. (B) Relative transcription of representative transcription factors. The transcriptional levels of three cold responsive transcription factors: DREBP1/CBF (DRE/CRT binding factor: Pp1s60_228V6.1), AP2/EREBP (APETALA2/ethylene-responsive element binding protein: Pp1s373_18V6.1) and ABI5 (ABSCISIC ACID INSENSITIVE5: Pp1s49_161V6) were analyzed by quantitative RT-PCR. Error bars represent SD (n = 3), and two-way ANOVA was used to determine the statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001).
Interestingly, expression of photosynthetic genes including PpPsaA (PhpapaCp039), PpPsaB (PhpapaCp040), PpPsbA (PhpapaCp046), PpPsbD (PhpapaCp044), PpPsbO (Pp1s60_65V6.1) as well as PpPsbP (Pp1s63_71V6.1) were also determined, aiming to analyze the possible effect of PpABI3 disruption on photosynthesis related transcriptome regulation. The results showed that all these candidate genes expressed at similar level under control condition in wild-type and mutant lines (Figures 6A–F). However, expression of these genes decreased during cold acclimation in both WT and Δabi3-1, indicating that low temperature might repress photosynthetic activity through restricting expression of these core components of photosynthetic apparatus (Figures 6A–F). Interestingly, we found that expression of PpPsbA and PpPsbD was relatively higher in Δabi3-1 than that in WT although other genes showed similar expression level in both lines (Figures 6C,D), indicating that transcription of several photosynthetic proteins might be affected due to PpABI3 deletion during cold acclimation. Together, we proposed that PpABI3 might participate in photosynthesis regulation at least in part by modulating expression of photosynthetic genes during cold acclimation. On the other hand, we still need to determine whether photosynthesis and photosynthetic apparatus were affected in Δabi3-1.
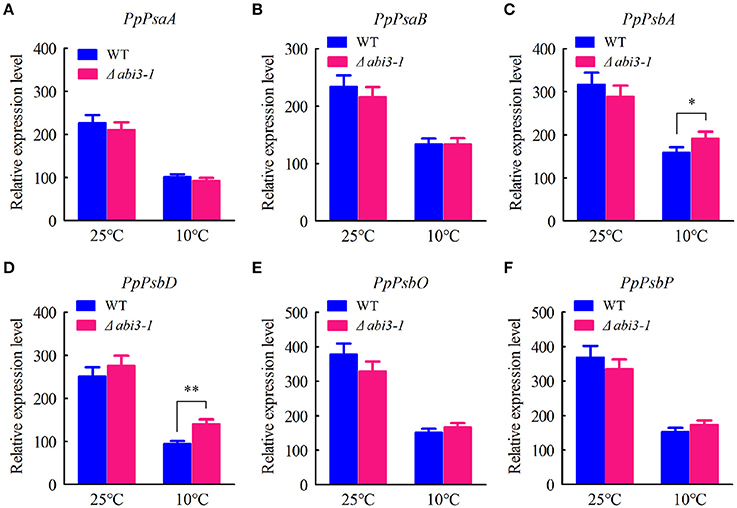
Figure 6. Expression pattern of photosynthetic genes. (A–F) The transcriptional levels of genes related to photosystem manganese-stabilizing including PpPsaA (PhpapaCp039), PpPsaB (PhpapaCp040), PpPsbA (PhpapaCp046), PpPsbD (PhpapaCp044), PpPsbO (Pp1s60_65V6.1) and PpPsbP (Pp1s63_71V6.1) were determined by quantitative RT-PCR assay. Error bars represent SD (n = 3), and two-way ANOVA was used to determine the statistical significance (*P < 0.05; **P < 0.01).
Disruption of PpABI3 Reduces PSII Activity in P. patens during Cold Acclimation
Low temperature significantly represses photosynthetic capacity in plants (Hurry et al., 2000; Paul and Foyer, 2001; Ensminger et al., 2006). We then analyzed the putative function of PpABI3 in regulating photosynthesis during cold acclimation. The maximum photochemical efficiency of PSII [variable fluorescence (Fv)/maximun fluorescence (Fm)] were determined. Data showed that, Fv/Fm reduced in both WT and Δabi3-1 after 2-week cold acclimation, and it was significantly lower in Δabi3-1 compared to WT (Figure 7A; Figure S5). In contrast, the non-photochemical quenching (NPQ) were induced during cold incubation, whereas this upregulation was dramatically higher in Δabi3-1 (Figures 7B–D). These results indicate that Δabi3-1 might suffer severer photoinhibition during cold acclimation. We further determined the light-responsive efficiency of PSII quantum yield (ΦPSII) in WT and Δabi3-1. Under growth condition, increase of light intensity represses the efficiency of ΦPSII, and the light curve of ΦPSII in Δabi3-1 was similar to that in WT (Figure 7E). After incubation in 10°C for 2 weeks, ΦPSII decreased rapidly along with light intensity increasing in both WT and Δabi3-1, however, the decline amplitude of ΦPSII in Δabi3-1 was more remarkable (Figure 7F). Furthermore, there exists no obvious difference of the maximum value of ETR between WT and Δabi3-1 under control condition. However, The maximum value of ETR in Δabi3-1 was significantly lower than that in WT, and the decrease of ETR in Δabi3-1 was more rapidly than that in WT under low temperature condition (Figures 7G,H), indicating a restricted electron transport, which might result from photoinhibition. In fact, long term low temperature incubation caused photoinhibition was consistent to growth inhibition of Δabi3-1. These results suggest that PpABI3 contributes to maintaining the PSII activity in response to cold stress.
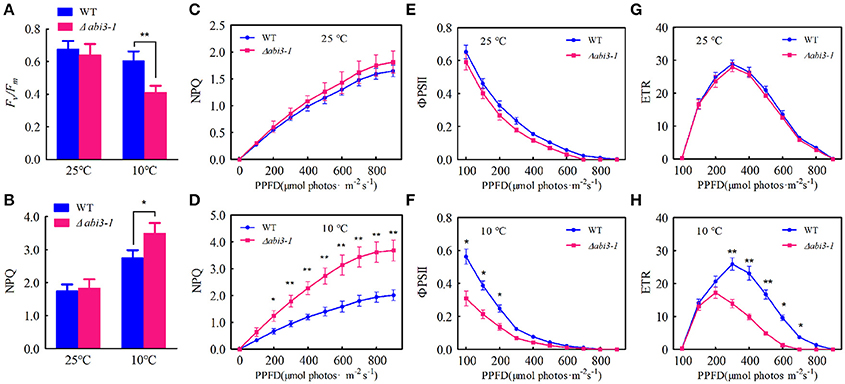
Figure 7. Deletion of PpABI3 reduces PSII activity during cold acclimation. (A–H) Chlorophyll fluorescence analysis. Two-week-old protonema of WT and Δabi3-1 with or with cold acclimation were collected to determine the maximum photochemical efficiency of PSII (Fv/Fm) (A), steady-state non-photochemical quenching (NPQ) (B), light-response curves of non-photochemical quenching (NPQ) (C,D), the efficiency of PSII quantum yield (ΦPSII) (E,F) and PSII electron transport rate (ETR) (G,H). Error bars represent SD (n = 3), two-way ANOVA was used to determine the statistical significance (*P < 0.05; **P < 0.01). Experimental details are found in the Materials and Methods.
Assembly of PSII is Perturbed in Δabi3-1 during Cold Acclimation
PSII activity defects might be caused by alternation of the compositions of photosynthetic apparatus (Baker, 1991; Sato et al., 1995; Lokstein et al., 2002). To analyze the possible role of PpABI3 in regulating compositions of photosynthetic protein complexes during cold acclimation, thylakoid membranes of both WT and Δabi3-1 were solubilized with n-dodecyl β-D-maltoside (DM) and subjected to Blue Native (BN)-gel electrophoretic assay. Data revealed that during cold acclimation, there exist significant differences in compositions of PSII-LHCII supercomplexes and PSI/PSII dimers between WT and Δabi3-1 (Figure 8A). Compared to growth condition, PSII-LHCII supercomplexes were reduced in both WT and Δabi3-1. However, accumulation of this supercomplexe in Δabi3-1 was significantly lower than that in WT (Figure 8A). In addition, the accumulation of PSI/PSII dimers in Δabi3-1 was slightly higher than that in WT during cold acclimation, which might result from reduction of PSII supercomplexes (Figure 8A). Interestingly, protein level of ATP synthase was elevated in both wild-type and mutant lines during cold acclimation (Figure 8A), indicating a regulatory role of photosynthetic phosphorylation during cold acclimation. Two-dimesional analysis (SDS-PAGE follows BN gel electrophoretic assay) showed that the distribution of subunits in PSII core proteins such as D1 and D2 in Δabi3-1 were obviously different from that in WT, although the accumulation of other protein subunits was more or less similar between control and cold conditions (Figure 8B). Together, these results suggested that disruption of PpABI3 significantly perturbs assembly of PSII during cold acclimation and results in decreased photosynthetic capacity.
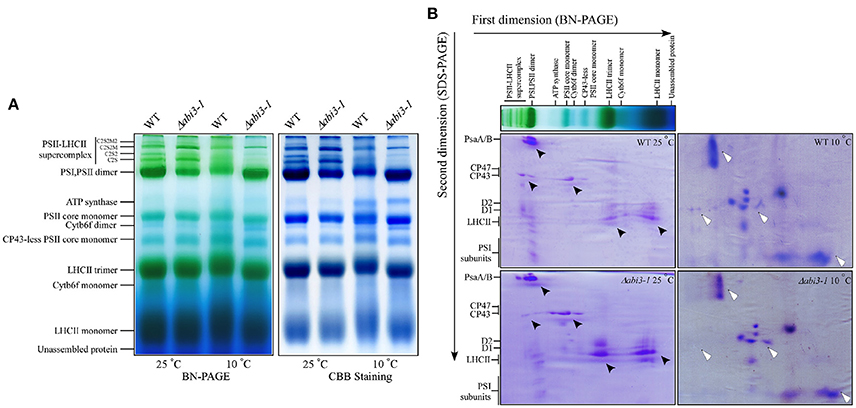
Figure 8. Deletion of PpABI3 affects assembly of PSII during cold acclimation. (A) Blue Native (BN)-gel electrophoresis analysis of thylakoid membrane protein complexes. Thylakoid membranes were isolated from WT and Δabi3-1 with or without cold acclimation, then solubilized with 2% DM and separated by BN-PAGE. A sample with an equal amount of chlorophyll (25 μg) was loaded in each lane. The BN gel (left) was further stained with Coomassie Brilliant blue (CBB; right). (B) Two-dimensional (2D) separation [BN-gel follows SDS-PAGE] of thylakoid protein complexes. Complexes in BN-gel were subsequently separated by SDS-urea-PAGE and stained with CBB to show their constituent subunits. Protein distribution differences between WT and Δabi3-1 were indicated by triangle symbol (black, standard condition; white, after cold acclimation).
PpABI3 is Involved in ABA-Dependent Freezing Tolerance in P. patens
Cold acclimation is essential for freezing tolerance in plants (Guy, 1990; Thomashow, 1999). Plants acquire enhanced freezing tolerance after cold acclimation (Shinozaki and Yamaguchi-Shinozaki, 2000; Kaplan et al., 2004; Chinnusamy et al., 2007). Since cold acclimation related cellular signaling and metabolism were significantly disturbed in Δabi3-1, which accumulated less cryoprotectants and antioxidants with disrupted redox homeostasis during cold acclimation, we proposed that PpABI3 disruption might impair freezing tolerance in P. patens. To confirm this hypothesis, both WT and Δabi3-1 were left untreated or treated under 10°C for 2 weeks (NA or CA) and then frozen to −4°C. After thawing, freezing tolerance was determined by quantifying EL from injured cells. As expected, the freezing tolerance was enhanced both in WT and Δabi3-1 after cold acclimation, respectively (Figure 9A). However, EL in Δabi3-1 was significantly higher than that in WT (Figure 9A), indicating a lower freezing tolerance of Δabi3-1. These data suggested that PpABI3 positively modulates freezing tolerance via regulating cold acclimation in P. patens.
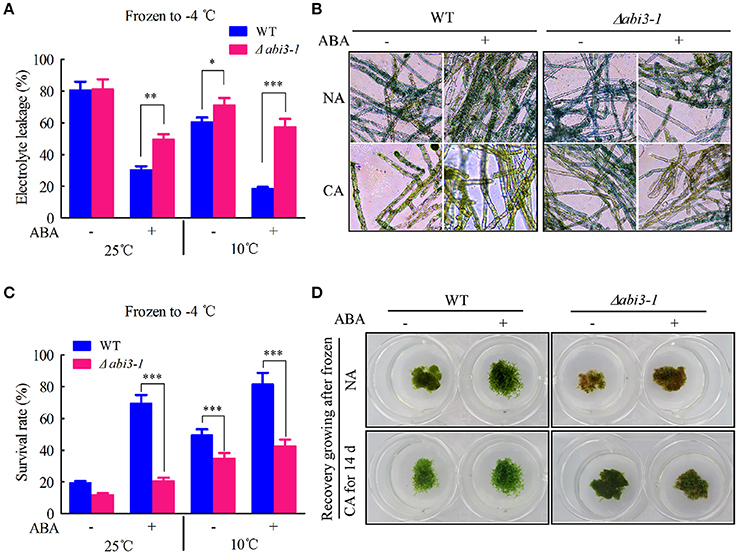
Figure 9. PpABI3 is involved in ABA-dependent freezing tolerance regulation. (A) Electrolyte leakage rate analysis. Tissues of WT and Δabi3-1 with or without cold acclimation were treated or not with 1 μM ABA, then frozen to −4°C. After thawing, electrolyte leakage was measured to estimate cell injury. Error bars represent SD (n = 3), and two-way ANOVA was used to determine the statistical significance (*P < 0.05; **P < 0.01; ***P < 0.001). (B) Cell death rate estimation. Frozen-thawed cells were stained with 0.5% Evans Blue to analysis the cell death of WT and Δabi3-1. (C) Survival rate analysis. Tissues of WT and Δabi3-1 with or without cold acclimation were treated or not with 1 μM ABA, then frozen to −4°C, after thawing the cells survival rate were estimated according to Evans Blue staining in (B). Error bars represent SD (n = 3), and two-way ANOVA was used to determine the statistical significance (***P < 0.001). (D) Recovery growing after frozen to −4°C. Frozen-thawed protonematal tissues of WT and Δabi3-1 were transferred onto fresh BCD medium and regrown under standard condition for 7 days to estimate the recovery growth capacity.
Previous study has revealed that cold acclimation contributes to freezing tolerance in P. patens at least in part depends on ABA signaling (Bhyan et al., 2012). We further investigated the regulatory role of PpABI3 in ABA signaling associated cold acclimation and subsequent freezing tolerance in P. patens. Results showed that exogenous ABA treatment significantly enhanced freezing tolerance in WT, while Δabi3-1 failed to acquire additional freezing tolerance after ABA treatment with or without cold acclimation (Figure 9A). To confirm that the EL is associated with actual cell death resulted from freezing, cell survival and cell death were analyzed in WT and Δabi3-1 by cells staining with Evans Blue. Data revealed that cold acclimation repressed freezing induced cell death in both WT and Δabi3-1, however, disruption of PpABI3 led to higher level of cell death and which is correlated with the EL during freezing stress (Figures 9B,C). Consistently, exogenous ABA cannot enhance freezing tolerance due to PpABI3 deletion, since Δabi3-1 displayed similar cell death rate whether with or without ABA treatment (Figures 9B,C). Subsequently, the frozen-thawed P. patens of WT and Δabi3-1 were left for further recovery analysis. We found that the recovery growth was consistent with the survival rate (Figures 9C,D), indicating that PpABI3 and ABA signaling also contribute to P. patens recovery growth after freezing stress is released. Together, these data suggested that PpABI3 is involved in ABA-dependent freezing tolerance regulation in P. patens during cold acclimation.
Discussion
To elucidate the involvement of PpABI3 in the cold acclimation and freezing tolerance in P. patens, PpABI3 deletion mutant Δabi3-1 was used for investigation. To avoid artificial results, we also generated complementary lines by introducing all the three PpABI3 genes into Δabi3-1 background. We found that the growth inhibition of Δabi3-1 under low temperature condition was remarkable, and due to PpABI3 loss-of-function, Δabi3-1 exhibited less freezing tolerance than WT. Disruption of PpABI3 resulted in lower survival rate and repressed recovery growing after frozen to −4°C. H2O2 and MDA contents in Δabi3-1 were higher than that in WT. In addition, the accumulation of cryoprotectants such as proline in Δabi3-1 was lower than that in WT during cold acclimation. Due to disruption of PpABI3, activity of ROS scavenging enzymes was significantly lower during cold acclimation. Cold-induction of cold-responsive (COR) genes transcription in Δabi3-1 was repressed during cold acclimation compared to WT. Photosynthetic genes expression and photosynthetic apparatus compositions were significantly altered in Δabi3-1. These results were consistent to the freezing-sensitive phenotype of Δabi3-1 and demonstrated that PpABI3 is involved in cold acclimation and freezing tolerance regulation in P. patens.
ABA contributes to freezing tolerance in P. patens during cold acclimation (Bhyan et al., 2012). In this study, we found that exogenous ABA treatment enhanced the freezing tolerance in WT, however, Δabi3-1 exhibited no significant difference in freezing tolerance with or without ABA treatment. These results were similar to the previous work, which showed that ABA-insensitive lines have barely increased freezing tolerance after ABA incubation (Bhyan et al., 2012). Together these results, we propose that ABA signaling contributes to cold acclimation and freezing tolerance at least in part through PpABI3. In fact, the ACTT-core element (5′-TCCACTTGTC-3′) in the promoter of several ABA-responsive genes is required for PpABI3 transcription regulation (Yotsui et al., 2013). It would be interesting to determine whether those genes targeted by PpABI3 during cold response also contain this element. On the other hand, Δabi3-1 also acquired partial freezing tolerance after cold acclimation, because the acclimated Δabi3-1 mutant can partially withstand freezing temperatures than the non-acclimated ones, supporting that PpABI3-independent pathways exist to regulate cold acclimation. In fact, C-repeat binding factors (CBFs), including CBF1, CBF2, and CBF3, contribute to cold acclimation in Arabidopsis (Medina et al., 1999; Gilmour et al., 2004). In addition, we found that induction of TFs such as PpDREB1/CBF and PpAP2/EREBP was partially inhibited in the Δabi3-1 during cold acclimation, suggesting that there might exists a crosstalk between PpABI3 and PpCBFs in cold acclimation. To further determine the function of PpABI3, the transcriptomic profile of wild type and Δabi3-1 during cold acclimation needs to be investigated in the future studies.
During cold acclimation, plants need to adapt to a long-term of environmental changes (Huner et al., 1998) and overcome energy imbalance caused by adverse environment (Miura and Furumoto, 2013). Energy imbalance might cause excess PSII excitation pressure which leads to photoinhibition or photodamage (Gray et al., 1997; Huner et al., 1998; Miura and Furumoto, 2013; Pinnola et al., 2013). Photoinhibiton reduces photosynthetic activity, resulting in plant growth repression (Takahashi and Badger, 2011). Results in this study showed that PSII activity in Δabi3-1 was much lower than that in WT, especially during cold acclimation, which might explain the repressed growth due to PpABI3 disruption. In addition, the NPQ was much higher under low temperature condition in Δabi3-1, suggesting the Δabi3-1 processed higher excess PSII excitation energy which needs to be dissipated (Pinnola et al., 2013). Low temperature-caused excess PSII excitation would lead to down-regulation of PSII activity through dissipation of excess energy (which resulted in increase of NPQ and decrease of ETR) or inactivation of PSII irreversibly and subsequently inhibit photosynthetic activity (which resulted in decrease of ΦPSII) (Oquist and Huner, 2003). Our data showed that both ETR and ΦPSII were decreased in Δabi3-1 compared to WT during cold acclimation, suggesting that disruption of PpABI3 decreased photosynthetic capacity might due to photoinhibition. Moreover, compositions of photosynthetic protein complexes in Δabi3-1 were clearly different from that in WT, suggesting that the steady state levels of photosynthetic proteins were altered because of PpABI3 disruption, which would cause severely impacts on plant photosynthesis during cold acclimation.
Taken together, we identified that PpABI3 was involved in cold acclimation induced freezing tolerance in P. patens. However, we still need to investigate how PpABI3 modulates cellular ROS homeostasis and photosynthetic protein complexes compositions during cold acclimation. Previous studies have revealed that cold stress causes photoinhibition and negatively regulates plant photosynthesis by affecting the repair process of PSII (Lei et al., 2014; Gururani et al., 2015; Zhang et al., 2016). Since PSII repair needs efficient and appropriate synthesis of a variety of proteins de novo, we proposed that TF PpABI3 might function in such process and contributes to freezing tolerance in P. patens. In addition, we should further determine whether redox homeostasis is involved in PpABI3-mediated proteins synthesis during cold acclimation.
Materials and Methods
Plant Materials and Cold Acclimation
Protonema tissues of P. patens (Gransden, wild-type), and Δabi3-1 were grown axenically on BCD medium containing 0.5% (w/v) glucose, 0.75% (w/v) agar, supplemented with 5 mM ammonium tartrate and cultured in greenhouse at 25°C under 16 h light/8 h darkness with light intensity of 50 μmol m−2 s−1 (Wang et al., 2014). After 1 week, protonematal tissues were transferred onto ammonium tartrate-free BCD medium for gametophyte growth (Cove et al., 2009). Two-week-old protonema was transferred to 25°C or 10°C under standard growth light condition for low temperature incubation.
Plasmid Construction and Transformation
The full coding sequences of PpABI3A, PpABI3B, and PpABI3C were amplified with the PpABI3A-CF/CR, PpABI3B-CF/CR, and PpABI3C-CF/CR primers, respectively. The resulting fragments were then cloned into the modified pTFH15.3 vector between the EcoRV and ApaI sites. Primer sequences are found in Table S1. Transformation of P. patens protoplasts and isolation of transgenic lines was performed as described (Khandelwal et al., 2010).
Growth Analysis
One-week-old wild-type and Δabi3-1 P. patens generated from 1.5 mL protonema suspension were transferred to 25°C and 10°C under standard growth light. Biomass formation starting at the day of inoculation and ending at week four was estimated by measuring fresh weight of per petri dish tissues. The measured raw data were transformed to a percentage value in relation to the inoculated biomass at the starting point which was defined as 0% and growth rate was calculated.
ABA Treatment and Freezing Tolerance Analyzes
Freezing tolerance was determined by measuring electrolyte leakage (EL) after the protonematal tissues were thawed from equilibrium freezing (−4°C) as previously described (Minami et al., 2003) with slight modifications. Protonema under growth condition or incubated under low temperature (10°C) for 2 weeks were transferred to fresh BCD medium added with 1 μM ABA (Sigma) and incubated at 25°C for 24 h. Tissues were collected in test tubes containing 0.5 mL of sterile distilled water. Put the tubes in a liquid bath of 50% ethylene glycol and kept at −1°C for 10 min. Adding pieces of ice into the liquid bath to initiate freezing, and incubating at −1°C for 1 h. The tubes were then cooled to −4°C at a rate of 2°C per hour. Then, remove liquid bath and keep the test tubes at 4°C in darkness until completely thawed. Add 2 mL distilled water to each thawed sample, and incubate them at room temperature for 2 h with gentle shaking in the dark, and then EL of the frozen-thawed tissues was measured. For recovery growing, small pieces of the protonematal tissues after frozen-thawing were inoculated onto a fresh BCD medium and cultured at standard growth condition.
Electrolyte Leakage Measurements and Evans Blue Staining
Electrolyte leakage (EL) was measured as previous description (Komatsu et al., 2013). The protonematal tissues thawed after frozen to −4°C were stained for 1 h in 0.5% (w/v) solution of Evans Blue (Sigma, E2129) and observed using microscopy as described to determine survival rate during freezing stress (Wertman et al., 2012).
Hydrogen Peroxide Content Assay
The H2O2 content was analyzed as previously described (Xu et al., 2012). Briefly, approximately 0.5 g protonematal tissues were homogenized in an ice bath with 5 mL 0.1% (w/v) trichloroacetic acid (TCA). The homogenate was centrifuged at 12,000 g for 20 min at 4°C. Then 0.5 mL of the supernatant was added to 0.5 mL 10 mM potassium phosphate buffer (pH 7.0) and 1 mL 1 M KI. The absorbance of the supernatant was read at 390 nm.
Malonyldialdehyde Content Assay
Malonyldialdehyde (MDA) content was measured as the method described by Lei et al. (2010) with slight modifications. Approximately 0.2 g of protonematal tissues were homogenized with 5 mL of 5% TCA in an ice bath. The homogenate was centrifuged at 1,000 g for 10 min at 4°C. Aliquots of the supernatant and 0.5% thiobarbituric acid (TBA) in 20% TCA solution were transferred into a new tube. The mixture was incubated in water bath at 100°C for 30 min, then cooled to room temperature and centrifuged at 8,000 g for 5 min. The supernatant was subjected for absorbance reading at A535/A600 using a spectrophotometer. The MDA content was calculated from the subtracted absorbance using the Extinction Coefficient of 155 mM−1 cm−1.
Proline Content Determination
Proline content was estimated using the acid-ninhydrin method according to Bates et al. (1973) and Wang et al. (2008) with modifications. 0.5 g protonematal tissues were homogenized with 5 mL of 3% (w/v) sulphosalicylic acid and filtered to obtain the clear filtrate. Glacial acetic acid and ninhydrin reagent (1 mL each) were added to 1 mL of the filtrate. The mixture was heated in an oven at 100°C for 1 h and the reaction was terminated in an ice bath. The reaction mixture was extracted with 4 mL toluene and absorption of the chromophore was read at 520 nm. Proline content was calculated using a standard curve constructed with L-proline standards.
Antioxidant Enzymatic Activity Analysis
For the enzymatic activity assays, 0.3 g protonema were ground with 3 mL ice-cold 25 mM Hepes buffer (pH 7.8) containing 0.2 mM EDTA, 2 mM ascorbate and 2% polyvinylpyrrolidone (PVP). The homogenates were centrifuged at 12,000 g for 20 min at 4°C and the supernatants were used for the enzymatic activity determination (Zhang et al., 2015). The superoxide dismutase (SOD, EC 1.15.1.1) activity was assayed with NBT following the method of Stewart and Bewley (1980). The catalase (CAT, EC 1.11.1.6) activity was measured as the decline in the absorbance at 240 nm with the method of Patra and Mishra (1979). Peroxidase (POD, EC 1.11.1.7) activity was determined using guaiacol as substrate according to Hammerschmidt et al. (1982). The ascorbate peroxidase (APX, EC 1.11.1.11) activity was measured at 290 nm (Nakano and Asada, 1981).
Quantitative RT-PCR Assays
Total RNA was extracted as previous description (Xi et al., 2010). DNaseI (Sigma) was added to remove DNA in extracted RNA. Two micrograms of RNA was reverse transcribed by ThermoScript TM RT-PCR System (Invitrogen). The cDNA was amplified by using SYBR Premix Ex Taq (TaKaRa). PpACTIN5 was used as internal control. Genomic DNA was extracted from 1 week-old protonema as previously described (Allen et al., 2006). The absence of PpABI3A, PpABI3B, and PpABI3C were confirmed in Δabi3-1 by PCR using DNA as template. RT-PCR assay was performed with cDNA synthesized from total RNA from wild-type and Δabi3-1 to further confirm the expression of PpABI3. All primers are described in Table S1.
Chlorophyll Fluorescence Analysis
Chlorophyll fluorescence was analyzed using chlorophyll fluorometer (IMAG-MINI PAM-2000; Heinz Walz, Effeltrich) with red (630 nm) pulse modulated measuring light at room temperature. P. patens were dark-adapted for 20 min before measurements. Values of Fv/Fm (maximum PSII photochemical efficiency) and nonphotochemical quenching (NPQ) were averaged from an approximately rectangular interested area (Gould et al., 2010). Color-indexed images of the samples showing chlorophyll fluorescence parameters were captured using the Imaging PAM software. To analyze the PSII quantum yield (ΦPSII) and the electron transport rate (ETR), P. patens were illuminated at the following light intensities: 0, 84, 155, 187, 307, 369, 461, 789, and 909 μmol photons m−2 s−1 (Lu et al., 2011).
Thylakoid Membranes Isolation and BN-PAGE Assays
Thylakoid membranes were isolated as described by Lu et al. (2011) with minor modifications. Samples of P. patens were crushed and homogenized with ice-cold extraction solution I [50 mM HEPES-KOH, pH 7.5, 330 mM sorbitol, 2 mM EDTA, 1 mM MgCl2, 5 mM ascorbate, 0.05% bovine serum albumin and 10 mM NaF]. Ascorbic acid was added to the buffer immediately before homogenization. The homogenates was filtered through four layers of cheesecloth and centrifuged at 2,000 g for 4 min at 4°C. The precipitation was resuspended in solution II [50 mM HEPES-KOH, pH 7.5, 5 mM sorbitol, 10 mM NaF], then centrifuged at 2,000 g for 4 min at 4°C. The thylakoid pellet was resuspended and centrifuged twice in solution III [50 mM HEPES-KOH, pH 7.5, 100 mM sorbitol, 10 mM MgCl2, 10 mM NaF]. The final pellet was resuspended in a small volume of solution III. After chlorophylls were extracted with 80% (v/v) aqueous acetone and quantified (Wellburn, 1994), thylakoids were rapidly frozen in liquid nitrogen and stored at −80°C for further assays.
BN-PAGE assay was performed as described (Malnoe et al., 2014). Thylakoids were solubilized with 2% (w/v) dodecyl β-D-maltoside (Sigma) on ice for 30 min. After centrifugation at 13,000 g for 10 min at 4°C, the supernatant was supplemented with 0.1 vol sample buffer containing 100 mM Bis Tris-HCl, pH 7.0, 500 mM 6-amino-caproicacid, 30% (w/v) glycerol, 5% (w/v) Serva blue G, and subjected to BN-PAGE with a gradient of 5–13.5% Bis-Tris mini separation gel. Electrophoresis was performed at 4°C with cathode buffer (50 mM Tricine, 15 mM Bis-Tris, pH 7.0, and 0.01% Coomassie Brilliant Blue G 250) and anode buffer (50 mM Bis-Tris, pH 7.0) for 25 min at 50 V and then 2 h at 150 V.
Statistical Analysis
Means of at least three biological replicates were measured for each assay. Two-way ANOVA was used for comparison between different treatments. A difference was considered to be statistically significant when P < 0.05, P < 0.01, or P < 0.001, respectively.
Author Contributions
TT, HZ, and HL designed the experiments. TT performed major of the study. YS, XP, GW, and FB provided assistance. TT, HZ, YH, and HL analyzed the data. HZ and TT wrote the manuscript and contributed to discussion.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Professor R. S. Quatrano (Washington University, USA) for providing the wild-type and Δabi3-1 of P. patens. This work was supported by the National Natural Science Foundation of China (31470342 and 31670235 to HL; 31600201 to HZ), the National Basic Research Program of China (2015CB150100 to HL) and the National Transgenic Major Project of China (2016ZX08009-003-002 to HL).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01599/full#supplementary-material
Abbreviations
ABA, abscisic acid; ABI3, ABSCISIC ACID INSENSITIVE3; APX, ascorbate peroxidase; BN-PAGE, blue-native polyacrylamide gel electrophoresis; CAT, catalase; CBB, Coomassie Brilliant Blue; Chl, chlorophyll; COR, cold-responsive genes; EL, electrolyte leakage; ETR, electron transport rate; MDA, malondialdehyde; NF-Y, nuclear factor Y; NPQ, non-photochemical quenching; PAM, pulse-amplitude modulated; POD, peroxidase; PPFD, photosynthetically active photon flux density; ROS, reactive oxygen species; RWC, relative water content; SOD, superoxide dismutase; ΦPSII, PSII quantum yield.
References
Allen, G. C., Flores-Vergara, M. A., Krasynanski, S., Kumar, S., and Thompson, W. F. (2006). A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 1, 2320–2325. doi: 10.1038/nprot.2006.384
Baker, N. R. (1991). A possible role for photosystem II in environmental perturbations of photosynthesis. Physiol. Plant 81, 563–570. doi: 10.1111/j.1399-3054.1991.tb05101.x
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Beck, E. H., Heim, R., and Hansen, J. (2004). Plant resistance to cold stress: mechanisms and environmental signals triggering frost hardening and dehardening. J. Biosci. 29, 449–459. doi: 10.1007/BF02712118
Beike, A. K., Lang, D., Zimmer, A. D., Wust, F., Trautmann, D., Wiedemann, G., et al. (2015). Insights from the cold transcriptome of Physcomitrella patens: global specialization pattern of conserved transcriptional regulators and identification of orphan genes involved in cold acclimation. New Phytol. 205, 869–881. doi: 10.1111/nph.13004
Bhyan, S. B., Minami, A., Kaneko, Y., Suzuki, S., Arakawa, K., Sakata, Y., et al. (2012). Cold acclimation in the moss Physcomitrella patens involves abscisic acid-dependent signaling. J. Plant Physiol. 169, 137–145. doi: 10.1016/j.jplph.2011.08.004
Blokhina, O., Virolainen, E., and Fagerstedt, K. V. (2003). Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann. Bot. 91, 179–194. doi: 10.1093/aob/mcf118
Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. doi: 10.1016/j.tplants.2007.07.002
Cove, D. J., Perroud, P. F., Charron, A. J., McDaniel, S. F., Khandelwal, A., and Quatrano, R. S. (2009). Culturing the moss Physcomitrella patens. Cold Spring Harb. Protoc. 2009:pdbprot5136. doi: 10.1101/pdb.prot5136
Davey, M. W., Stals, E., Panis, B., Keulemans, J., and Swennen, R. L. (2005). High-throughput determination of malondialdehyde in plant tissues. Anal. Biochem. 347, 201–207. doi: 10.1016/j.ab.2005.09.041
Delmas, F., Sankaranarayanan, S., Deb, S., Widdup, E., Bournonville, C., Bollier, N., et al. (2013). ABI3 controls embryo degreening through Mendel's I locus. Proc. Natl. Acad. Sci. U.S.A. 110, E3888–E3894. doi: 10.1073/pnas.1308114110
Demidchik, V. I., Straltsova, D., Medvedev, S. S., Pozhvanov, G. A., Sokolik, A., and Yurin, V. (2014). Stress-induced electrolyte leakage: the role of K+-permeable channels and involvement in programmed cell death and metabolic adjustment. J. Exp. Bot. 65, 1259–1270. doi: 10.1093/jxb/eru004
Eklund, D. M., Thelander, M., Landberg, K., Staldal, V., Nilsson, A., Johansson, M., et al. (2010). Homologues of the Arabidopsis thaliana SHI/STY/LRP1 genes control auxin biosynthesis and affect growth and development in the moss Physcomitrella patens. Development 137, 1275–1284. doi: 10.1242/dev.039594
Ensminger, I., Busch, F., and Huner, N. (2006). Photostasis and cold acclimation: sensing low temperature through photosynthesis. Physiol. Plant 126, 28–44. doi: 10.1111/j.1399-3054.2006.00627.x
Frank, W., Ratnadewi, D., and Reski, R. (2005). Physcomitrella patens is highly tolerant against drought, salt and osmotic stress. Planta 220, 384–394. doi: 10.1007/s00425-004-1351-1
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Gilmour, S. J., Fowler, S. G., and Thomashow, M. F. (2004). Arabidopsis transcriptional activators CBF1, CBF2, CBF3 have matching functional activities. Plant Mol. Biol. 54, 767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4
Gould, K. S., Dudle, D. A., and Neufeld, H. S. (2010). Why some stems are red: cauline anthocyanins shield photosystem II against high light stress. J. Exp. Bot. 61, 2707–2717. doi: 10.1093/jxb/erq106
Gray, G. R., Chauvin, L. P., Sarhan, F., and Huner, N. P. A. (1997). Cold acclimation and freezing tolerance (A complex interaction of light and temperature). Plant Physiol. 114, 467–474. doi: 10.1104/pp.114.2.467
Gururani, M. A., Venkatesh, J., and Tran, L. S. P. (2015). Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol. Plant 8, 1304–1320. doi: 10.1016/j.molp.2015.05.005
Guy, C. L. (1990). Cold acclimation and freezing stress tolerance: role of protein metabolism. Annu. Rev. Plant Biol. 41, 187–223. doi: 10.1146/annurev.pp.41.060190.001155
Hammerschmidt, R., Nuckles, E. M., and Kuć, J. (1982). Association of enhanced peroxidase activity with induced systemic resistance of cucumber to Colletotrichum lagenarium. Physiol. Plant Pathol. 20, 73–82. doi: 10.1016/0048-4059(82)90025-X
Hossain, M. A., Bhattacharjee, S., Armin, S. M., Qian, P., Xin, W., Li, H. Y., et al. (2015). Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: insights from ROS detoxification and scavenging. Front. Plant Sci. 6:420. doi: 10.3389/fpls.2015.00420
Huner, N. P. A., Oquist, G., and Sarhan, F. (1998). Energy balance and acclimation to light and cold. Trends Plant Sci. 3, 224–230. doi: 10.1016/S1360-1385(98)01248-5
Hurry, V., Strand, Å., Furbank, R., and Stitt, M. (2000). The role of inorganic phosphate in the development of freezing tolerance and the acclimatization of photosynthesis to low temperature is revealed by the pho mutants of Arabidopsis thaliana. Plant J. 24, 383–396. doi: 10.1046/j.1365-313x.2000.00888.x
Jeon, J., and Kim, J. (2013). Cold stress signaling networks in Arabidopsis. J. Plant Biol. 56, 69–76. doi: 10.1007/s12374-013-0903-y
Kaplan, F., Kopka, J., Haskell, D. W., Zhao, W., Schiller, K. C., Gatzke, N., et al. (2004). Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 136, 4159–4168. doi: 10.1104/pp.104.052142
Khandelwal, A., Cho, S. H., Marella, H., Sakata, Y., Perroud, P. F., Pan, A., et al. (2010). Role of ABA and ABI3 in desiccation tolerance. Science 327:546. doi: 10.1126/science.1183672
Komatsu, K., Suzuki, N., Kuwamura, M., Nishikawa, Y., Nakatani, M., Ohtawa, H., et al. (2013). Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat. Commun. 4:2219. doi: 10.1038/ncomms3219
Lei, T., Feng, H., Sun, X., Dai, Q. L., Zhang, F., Liang, H. G., et al. (2010). The alternative pathway in cucumber seedlings under low temperature stress was enhanced by salicylic acid. Plant Growth Regul. 60, 35–42. doi: 10.1007/s10725-009-9416-6
Lei, Y., Zheng, Y., Dai, K., Duan, B., and Cai, Z. (2014). Different responses of photosystem I and photosystem II in three tropical oilseed crops exposed to chilling stress and subsequent recovery. Trees 28, 923–933. doi: 10.1007/s00468-014-1007-0
Lim, S., Park, J., Lee, N., Jeong, J., Toh, S., Watanabe, A., et al. (2013). ABA-INSENSITIVE3, ABA-INSENSITIVE5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25, 4863–4878. doi: 10.1105/tpc.113.118604
Lokstein, H., Tian, L., Polle, J. E., and DellaPenna, D. (2002). Xanthophyll biosynthetic mutants of Arabidopsis thaliana: altered nonphotochemical quenching of chlorophyll fluorescence is due to changes in photosystem II antenna size and stability. Biochim. Biophys. Acta 1553, 309–319. doi: 10.1016/S0005-2728(02)00184-6
Lu, Y., Hall, D. A., and Last, R. L. (2011). A small zinc finger thylakoid protein plays a role in maintenance of photosystem II in Arabidopsis thaliana. Plant Cell 23, 1861–1875. doi: 10.1105/tpc.111.085456
Mahajan, S., and Tuteja, N. (2005). Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophys. 444, 139–158. doi: 10.1016/j.abb.2005.10.018
Malnoe, A., Wang, F., Girardbascou, J., Wollman, F., and De Vitry, C. (2014). Thylakoid FtsH protease contributes to photosystem II and cytochrome b6f remodeling in Chlamydomonas reinhardtii under stress conditions. Plant Cell 26, 373–390. doi: 10.1105/tpc.113.120113
Marella, H. H., Sakata, Y., and Quatrano, R. S. (2006). Characterization and functional analysis of ABSCISIC ACID INSENSITIVE3-like genes from Physcomitrella patens. Plant J. 46, 1032–1044. doi: 10.1111/j.1365-313X.2006.02764.x
Medina, J., Bargues, M., Terol, J., Pérez-Alonso, M., and Salinas, J. (1999). The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol. 119, 463–470.
Minami, A., Nagao, M., Arakawa, K., Fujikawa, S., and Takezawa, D. (2003). Abscisic acid-induced freezing tolerance in the moss Physcomitrella patens is accompanied by increased expression of stress-related genes. J. Plant Physiol. 160, 475–483. doi: 10.1078/0176-1617-00888
Miura, K., and Furumoto, T. (2013). Cold signaling and cold response in plants. Int. J. Mol. Sci. 14, 5312–5337. doi: 10.3390/ijms14035312
Monke, G., Seifert, M., Keilwagen, J., Mohr, M., Grosse, I., Hahnel, U., et al. (2012). Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res. 40, 8240–8254. doi: 10.1093/nar/gks594
Nakano, Y., and Asada, K. (1981). Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 22, 867–880.
Oquist, G., and Huner, N. P. (2003). Photosynthesis of overwintering evergreen plants. Annu. Rev. Plant Biol. 54, 329–355. doi: 10.1146/annurev.arplant.54.072402.115741
Parcy, F., Valon, C., Kohara, A., Misera, S., and Giraudat, J. (1997). The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell 9, 1265–1277. doi: 10.1105/tpc.9.8.1265
Patra, H. K., and Mishra, D. (1979). Pyrophosphatase, peroxidase and polyphenoloxidase activities during leaf development and senescence. Plant Physiol. 63, 318–323. doi: 10.1104/pp.63.2.318
Paul, M. J., and Foyer, C. H. (2001). Sink regulation of photosynthesis. J. Exp. Bot. 52, 1383–1400. doi: 10.1093/jexbot/52.360.1383
Pinnola, A., Dall'Osto, L., Gerotto, C., Morosinotto, T., Bassi, R., and Alboresi, A. (2013). Zeaxanthin binds to light-harvesting complex stress-related protein to enhance nonphotochemical quenching in Physcomitrella patens. Plant Cell 25, 3519–3534. doi: 10.1105/tpc.113.114538
Rohde, A., Kurup, S., and Holdsworth, M. (2000). ABI3 emerges from the seed. Trends Plant Sci. 5, 418–419. doi: 10.1016/S1360-1385(00)01736-2
Romanel, E. A., Schrago, C. G., Counago, R. M., Russo, C. A., and Alves-Ferreira, M. (2009). Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS ONE 4:e5791. doi: 10.1371/journal.pone.0005791
Sakata, Y., Nakamura, I., Taji, T., Tanaka, S., and Quatrano, R. S. (2010). Regulation of the ABA-responsive Em promoter by ABI3 in the moss Physcomitrella patens: Role of the ABA response element and the RY element. Plant Signal. Behav. 5, 1061–1066. doi: 10.4161/psb.5.9.11774
Sato, N., Sonoike, K., Tsuzuk, M., and Kawaguchi, A. (1995). Impaired photosystem II in a mutant of Chlamydomonas reinhardtii defective in sulfoquinovosyl diacylglycerol. Eur. J. Biochem. 234, 16–23. doi: 10.1111/j.1432-1033.1995.016_c.x
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. doi: 10.1016/S1369-5266(00)00067-4
Stewart, R. R. C., and Bewley, J. D. (1980). Lipid peroxidation associated with accelerated aging of soybean axes. Plant Physiol. 65, 245–248. doi: 10.1104/pp.65.2.245
Takahashi, S., and Badger, M. R. (2011). Photoprotection in plants: a new light on photosystem II damage. Trends Plant Sci. 16, 53–60. doi: 10.1016/j.tplants.2010.10.001
Takezawa, D., Watanabe, N., Ghosh, T. K., Saruhashi, M., Suzuki, A., Ishiyama, K., et al. (2015). Epoxycarotenoid-mediated synthesis of abscisic acid in Physcomitrella patens implicating conserved mechanisms for acclimation to hyperosmosis in embryophytes. New Phytol. 206, 209–219. doi: 10.1111/nph.13231
Tamminen, I., Makela, P., Heino, P., and Palva, E. T. (2001). Ectopic expression of ABI3 gene enhances freezing tolerance in response to abscisic acid and low temperature in Arabidopsis thaliana. Plant J. 25, 1–8. doi: 10.1046/j.1365-313x.2001.00927.x
Thomashow, M. F. (1999). Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Wang, X., Yang, P., Gao, Q., Liu, X., Kuang, T., Shen, S., et al. (2008). Proteomic analysis of the response to high-salinity stress in Physcomitrella patens. Planta 228, 167–177. doi: 10.1007/s00425-008-0727-z
Wang, X., Zhou, S., Chen, L., Quatrano, R. S., and He, Y. (2014). Phospho-proteomic analysis of developmental reprogramming in the moss Physcomitrella patens. J. Proteomics 108, 284–294. doi: 10.1016/j.jprot.2014.05.012
Wellburn, A. R. (1994). The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 144, 307–313. doi: 10.1016/S0176-1617(11)81192-2
Wertman, J., Lord, C. E., Dauphinee, A. N., and Gunawardena, A. H. (2012). The pathway of cell dismantling during programmed cell death in lace plant (Aponogeton madagascariensis) leaves. BMC Plant Biol. 12:115. doi: 10.1186/1471-2229-12-115
Xi, D., Yang, H., Jiang, Y., Xu, M., Shang, J., Zhang, Z., et al. (2010). Interference between tobacco necrosis virus and turnip crinkle virus in Nicotiana benthamiana. J. Phytopathol. 158, 263–269. doi: 10.1111/j.1439-0434.2009.01607.x
Xu, F., Zhang, D. W., Zhu, F., Tang, H., Lv, X., Cheng, J., et al. (2012). A novel role for cyanide in the control of cucumber (Cucumis sativus L.) seedlings response to environmental stress. Plant Cell Environ. 35, 1983–1997. doi: 10.1111/j.1365-3040.2012.02531.x
Yotsui, I., Saruhashi, M., Kawato, T., Taji, T., Hayashi, T., Quatrano, R. S., et al. (2013). ABSCISIC ACID INSENSITIVE3 regulates abscisic acid-responsive gene expression with the nuclear factor Y complex through the ACTT-core element in Physcomitrella patens. New Phytol. 199, 101–109. doi: 10.1111/nph.12251
Yotsui, I., Serada, S., Naka, T., Saruhashi, M., Taji, T., Hayashi, T., et al. (2016). Large-scale proteome analysis of abscisic acid and ABSCISIC ACID INSENSITIVE3-dependent proteins related to desiccation tolerance in Physcomitrella patens. Biochem. Biophys. Res. Commun. 471, 589–595. doi: 10.1016/j.bbrc.2016.02.024
Zhang, D. W., Deng, X. G., Fu, F. Q., and Lin, H. H. (2015). Induction of plant virus defense response by brassinosteroids and brassinosteroid signaling in Arabidopsis thaliana. Planta 241, 875–885. doi: 10.1007/s00425-014-2218-8
Zhang, Z. S., Jin, L. Q., Li, Y. T., Tikkanen, M., Li, Q. M., Ai, X. Z., et al. (2016). Ultraviolet-B radiation (UV-B) relieves chilling-light-induced PSI photoinhibition and accelerates the recovery of CO2 assimilation in Cucumber (Cucumis sativus L.) leaves. Sci. Rep. 6, 344–355. doi: 10.1038/srep34455
Keywords: Physcomitrella patens, cold acclimation, freezing tolerance, ABI3, ABA, photosynthetic protein complexes
Citation: Tan T, Sun Y, Peng X, Wu G, Bao F, He Y, Zhou H and Lin H (2017) ABSCISIC ACID INSENSITIVE3 Is Involved in Cold Response and Freezing Tolerance Regulation in Physcomitrella patens. Front. Plant Sci. 8:1599. doi: 10.3389/fpls.2017.01599
Received: 29 April 2017; Accepted: 31 August 2017;
Published: 12 September 2017.
Edited by:
Mukesh Jain, Jawaharlal Nehru University, IndiaReviewed by:
Kazuo Nakashima, Japan International Research Center for Agricultural Sciences, JapanAxel De Zelicourt, Université Paris-Sud, France
Copyright © 2017 Tan, Sun, Peng, Wu, Bao, He, Zhou and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huapeng Zhou, zhouhuapeng@scu.edu.cn
Honghui Lin, hhlin@scu.edu.cn
 Tinghong Tan
Tinghong Tan Yanni Sun1
Yanni Sun1 Fang Bao
Fang Bao Yikun He
Yikun He Huapeng Zhou
Huapeng Zhou Honghui Lin
Honghui Lin