- 1Key Laboratory of Plant Resources Conservation and Germplasm Innovation in Mountainous Region (Guizhou University), Ministry of Education, Institute of Agro-Bioengineering, College of Life Sciences, Guizhou University, Guiyang, China
- 2Centre for Bioinformatics Research, SRKR Engineering College, Bhimavaram, India
- 3Finnish Museum of Natural History, University of Helsinki, Helsinki, Finland
- 4Organismal and Evolutionary Biology (OEB) Research Programme, Department of Biological and Environmental Sciences, University of Helsinki, Helsinki, Finland
microRNAs (miRNAs) play an important role as key regulators controlling the post-transcriptional events in plants across development, abiotic and biotic stress, tissue polarity and also in defining the evolutionary basis of the origin of the post-transcriptional machinery. Identifying patterns of regulated and co-regulated small RNAs, in particular miRNAs and their sequence variants with the availability of next generation sequencing approaches has widely demonstrated the role of miRNAs and their temporal regulation in maintaining plant development and their response to stress conditions. Although the role of canonical miRNAs has been widely explored and functional diversity is revealed, those works for isomiRs are still limited and urgent to be carried out across plants. This relative lack of information with respect to isomiRs might be attributed to the non-availability of large-scale detection of isomiRs across wide plant species. In the present research, we addressed this by developing Plant isomiR Atlas, which provides large-scale detection of isomiRs across 23 plant species utilizing 677 smallRNAs datasets and reveals a total of 98,374 templated and non-templated isomiRs from 6,167 precursors. Plant isomiR Atlas provides several visualization features such as species specific isomiRs, isomiRs and canonical miRNAs overlap, terminal modification classifications, target identification using psRNATarget and TargetFinder and also canonical miRNAs:target interactions. Plant isomiR Atlas will play a key role in understanding the regulatory nature of miRNAome and will accelerate to understand the functional role of isomiRs. Plant isomiR Atlas is available at www.mcr.org.in/isomir.
One Sentence Summary
Plant isomiR Atlas will play a key role in understanding the regulatory nature of miRNAome and will accelerate the understanding and diversity of functional targets of plants isomiRs.
Introduction
Mechanistic understanding of plant RNA biology and identifying classes and patterns of smallRNAs, a class of non-coding RNAs has evolved particularly after the advent of the next-generation sequencing. Non-coding RNAs such as microRNAs, long non-coding RNAs (lncRNAs), miRtrons (miRNAs located in introns), artificial miRNAs, siRNAs, phasiRNAs, and trans-acting siRNAs play an important role in regulating the post-transcriptional responses at various levels, such as at different stages of development in response to abiotic and biotic stress (Lin et al., 2018) or by acting as targets for endogenous silencing mechanism (Neilsen et al., 2012; Sablok et al., 2015). Accelerated application of next generation sequencing techniques has revealed not only the concordance and dis-concordant patterns of miRNA regulation but has also enabled the identification and discovery of the miRNA variants, binding site efficiency, phase shifting and phasiRNAs (Yang et al., 2018). Fundamental application of miRNAs either from the sequence conservation or from the binding site conservation viewpoint has unfolded conserved regulatory relation accross plants, e.g. members from MIR156 target SQUAMOSA promoter binding protein-like (SPL) genes in plants which involved in development (Stief et al., 2014). Nonetheless, recently explored miRNA derived siRNAs piggybacking for virus resistance (Incarbone et al., 2018) can be widely exploited for developing miRNA-based approaches for plant domestication.
After the first report of the microRNAs canonical variants (isomiRs) in Oryza sativa (Morin et al., 2008a), these canonical variants (isomiRs) have been shown to play an important role in regulating the post-transcriptional responses in response to several abiotic and biotic responses (Gozmanova et al., 2017) and recently much emphasis has been laid on the discovery of these variants to unravel the biogenesis pathway and target potential of these isomiRs. Relative features such as length variations have classified these isomiRs into several classes, which includes 5′- isomiRs, 3′- isomiRs, polymorphic isomiRs, template, and non-templated isomiRs (Neilsen et al., 2012; Rogans and Rey, 2016). Plant isomiR biogenesis pathway has not been widely studied, however, several reports hypothesize the ribonuclease DICER-LIKE 1 (DCL) cleavage variations as the prominent cause of isomiR biogenesis (Ruby et al., 2006; Landgraf et al., 2007; Kuchenbauer et al., 2008; Morin et al., 2008a,b). For non-templated isomiRs, their origin is thought as coming from nucleotidyltransferase like PAPD4 (Burroughs et al., 2010) describing the addition of templated or non-templated nucleotides to isomiRs terminus. It has been previously shown that some of the observed substitutions, with few relative occurrences within the internal sites may be an effect of post-transcriptional RNA editing (Neilsen et al., 2012; Rogans and Rey, 2016). Although uridylation events have been primarily linked to the terminal modification or additions leading to the biogenesis of isomiRs, concluding evidences reveal that the complete biogenesis pathway remains a challenging question.
Irrespective of its biogenesis pathway, target site accessibility and whether or not these isomiRs play a role in expanding the target predictions has been recently addressed on a large scale in model plant species (Ahmed et al., 2014). Ahmed et al. (2014) performed comprehensive analysis of isomiR terminal heterogeneity in demonstrating the effect on the accessibility of target binding demonstrating that combination of miRNAs and isomiRs increased the accuracy of target prediction in A. thaliana (Ahmed et al., 2014). Additionally, several targets were shared by members of isomiRs and they were enriched for certain GO terms (Ahmed et al., 2014). This is further evident from the recent reports in Phaseoleae, where target binding conversation and enhancement was seen in canonical miRNA and terminal isoforms of the miR1510 family (Fei et al., 2018). Furthermore, many targets of isomiRs were also targets of canonical miRNAs indicating that miRNA-mediated gene regulation were also assisted by isomiRs (Ahmed et al., 2014).
Terminal modifications, which can be addition or substitution events have been primarily defined as an essential criteria for the classification of isomiRs and have been widely used as a. golden rule to identify and classify isomiRs by several tools such as isomiRID (de Oliveira et al., 2013), isomiRage (Muller et al., 2014), DeAnnoIso (Zhang et al., 2016), miR-isomiRExp (Guo et al., 2016), and isomiR2Function (Yang et al., 2017). Although the above-mentioned tools present a classification approach for the identification of isomiRs, large-scale identification and functional repertoire of these isomiRs across plant species and their cross-conservation of isomiRs and their corresponding targets present a knowledge gap that is yet to be addressed. In plants, recently developed isomiRBank (Zhang et al., 2016) represents one such repository, which displays isomiRs detection across 4 plant species and lacks the large-scale detection of isomiRs and their corresponding classification and target diversity taking into account the terminal modifications as described previously (Neilsen et al., 2012; Rogans and Rey, 2016). In plants, preferential association of the miRNAs with the AGO has been widely demonstrated and well described (Fátyol et al., 2015; Incarbone et al., 2018). miR390 presents an classical example of AGO divergence, which has preference for loading on AGO7 for the production of the TAS3 ta-siRNAs (Axtell et al., 2006; Incarbone et al., 2018) but has also shown to be loading on AGO2 due to its 5′A, which assists in the AGO2 loading (Mi et al., 2008) thus demonstrating that the terminal nucleotides at the 5′ or 3′ can influence the preferential AGO selection. With the limited knowledge about the diversity and distribution of the terminal nucleotides in isomiRs with respect to the canonical miRNA, it is still a challenging task to describe the preferential association of the isomiRs to AGO and the preferential terminal nucleotides which affect the miRNA+isomiR target abundance.
Additionally, the role of the isomiRs generated from the canonical miRNAs and the target mimics has not been defined. Moreover, it is worth highlighting that previously developed visualization approaches such as isomiRBank have largely focused on the detection of templated isomiRs, which presents a gap in understanding of non-templated isomiRs in plants. To address this knowledge gap, such as (1) low number of species present in isomiRBank (Zhang et al., 2016), (2) absence of the non-templated isomiR repository, (3) targets of the detected isomiRs, (4) consensus target calling through the implementation of several target prediction approaches and (5) isomiRs and target interactions including target mimics, we present Plant isomiR Atlas (www.mcr.org.in/isomir), which is a large scale repository of plant specific isomiRs across 23 pecies representing a total of 677 datasets and presents both templated and non-templated isomiRs profiling with support for two target prediction algorithms and target mimics. We believe that the profiled isomiRs and the ease of visualization of the isomiRs along with the target classification and miRNA mimic under one common framework will accelerate the functional discovery of isomiRs and will increase the targeted miRNA'ome in plants.
Materials and Methods
Data Resources
Genome datasets with corresponding features in GFF3 format, annotations, and transcripts were downloaded for 23 plant species: Amborella trichopoda, Arabidopsis lyrata, Arabidopsis thaliana, Brachypodium distachyon, Brassica rapa, Carica papaya, Citrus clementina, Citrus sinensis, Glycine max, Gossypium raimondii, Malus domestica, Manihot esculenta, Medicago truncatula, Nelumbo nucifera, Oryza sativa, Populus trichocarpa, Setaria italic, Solanum lycopersicum, Solanum tuberosum, Sorghum bicolor, Triticum aestivum, Vitis vinifera, and Zea mays from Phytozome version 11 accessible through www.phytozome.net/ (Goodstein et al., 2011). smallRNA datasets relevant to these species were identified through curated literature searches and their information can be found in Table S1. For the aforementioned species, pre-miRNAs and mature miRNAs corresponding to these species were downloaded from miRBase release 21 accessible through http://www.mirbase.org (Kozomara and Griffiths-Jones, 2013) and plant microRNA database (PMRD) accessible through: bioinformatics.cau.edu.cn/PMRD (Zhang et al., 2009). tRNAs and rRNAs were downloaded from RFAM (http://rfam.xfam.org/), plant snoRNAs are available from (http://bioinf.scri.sari.ac.uk/cgi-bin/plant_snorna/home).
Large Scale Detection of Plant IsomiRs, Targets, and Endogenous miRNAs
For the identification of plant isomiRs, quality control, and adaptor trimming were done by a perl script which calls cutadapt (Martin, 2011) and indicates counts of high-quality reads. All the clean reads were further mapped to RFAM (http://rfam.xfam.org/), tRNAs (http://rfam.xfam.org/), and plant snoRNAs database (http://bioinf.scri.sari.ac.uk/cgi-bin/plant_snorna/home) to remove the background noise coming from other RNA classes. Reads with read count lower than 10 were also removed from further analysis to avoid reads generated by any sequencing error or representing the low coverage reads. The entire bioinformatic workflow for Plant isomiR Atlas is described in Figure 1, which outlines the detection of templated and non-templated isomiRs. Briefly, to identify templated isomiRs, remain reads were mapped on miRNA hairpins of corresponding species allowing no mismatch. Reads successfully mapped to hairpins were defined as templated isomiRs. For reads failed mapping to any miRNA hairpin, they were mapped to genome allowing no mismatch to make sure they were not generated from other place in genome. Among them, reads failed mapping to the genome were then mapped to miRNA hairpins again allowing two mismatches. Reads successfully mapped to hairpins were defined as non-templated isomiRs. After templated and non-templated isomiRs had been identified, they were classified in three aspects: location, substitution and seed corruptness. More details on isomiR identification can be found at the Method page of the database (http://www.mcr.org.in/isomir/method.php?pg=2). We classified isomiRs in three aspects: location, substitution and seed corruptness. Substitution patterns have been defined as 5 V for substitutions happened at 5′ terminus, 3 V for substitution happened at 3′ terminus, TS for substitutions happened internally and tandemly, MS for substitutions happened internally and randomly, SS for single substitutions happened internally, CV for substitutions happened terminally and internally. Location patterns have been defined as non-drift for isomiRs generated from the same loci that generates canonical miRNAs. Additionally, [5′/3′][+/–] have been defined for those isomiRs whose 5′ or 3′ terminal position is upstream or downstream of the corresponding canonical miRNAs, e.g., 5′+3 indicates that isomiR has three additional bases at its 5′ terminus compared to canonical miRNAs. Seed corruptness patterns have been defined as [F/f] for whether the similar seed sequence was between 2 and 8 bases and [C/c] for whether the seed sequence was the same with overlapped canonical miRNA's. Since isomiRs shows sequence difference to the canonical miRNA, all the identified non-drift isomiRs are non-templated, which are defined on the basis of the internal SNPs counted as mismatches against miRNA hairpin.
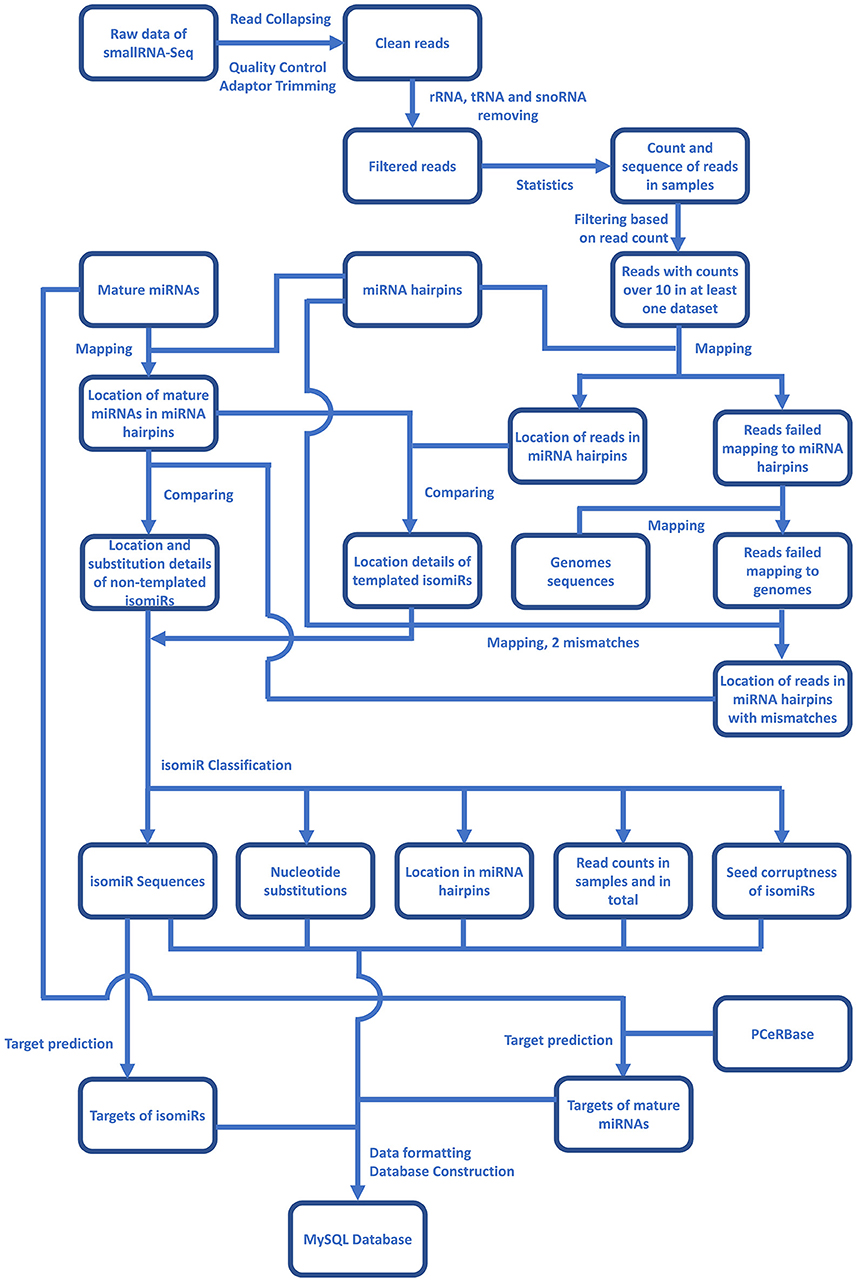
Figure 1. An overlay of the bioinformatics workflow in Plant isomiR Atlas. Mapping reads on genome and miRNA precursors were carried out by Bowtie1. isomiRs of both types were identified and classified in details by algorithm of isomiR2Function (Yang et al., 2017).
Taking into account the differences in the target prediction algorithms, we implemented two target prediction tools, psRNATarget (Dai and Zhao, 2011) and TargetFinder (Fahlgren et al., 2007) with default parameters. For the identification of the miRNA-target interactions, all the miRNAs-target interactions were downloaded from PCeRBase (Yuan et al., 2017) including the target mimics interactions. Since we aimed toward the identification of the isomiRs spanning across the datasets, the read normalization was therefore not done as the proposed goal of the Plant isomiR Atlas is large scale detection of isomiRs and read normalization can bias the detection of the isomiRs, where each of the datasets embedded may have different read counts in the respective experiments. Plant isomiR Atlas has been developed using the MySQL as a backend database and uses PHP as a front end. Plant isomiR Atlas is hosted on a 16 core Intel Xeon machine with Ubuntu as an operating system and allows for the rapid searches and easy to browse interface is equipped with several search options.
Results and Discussion
High Throughput IsomiR Profiling Across Plant Clade
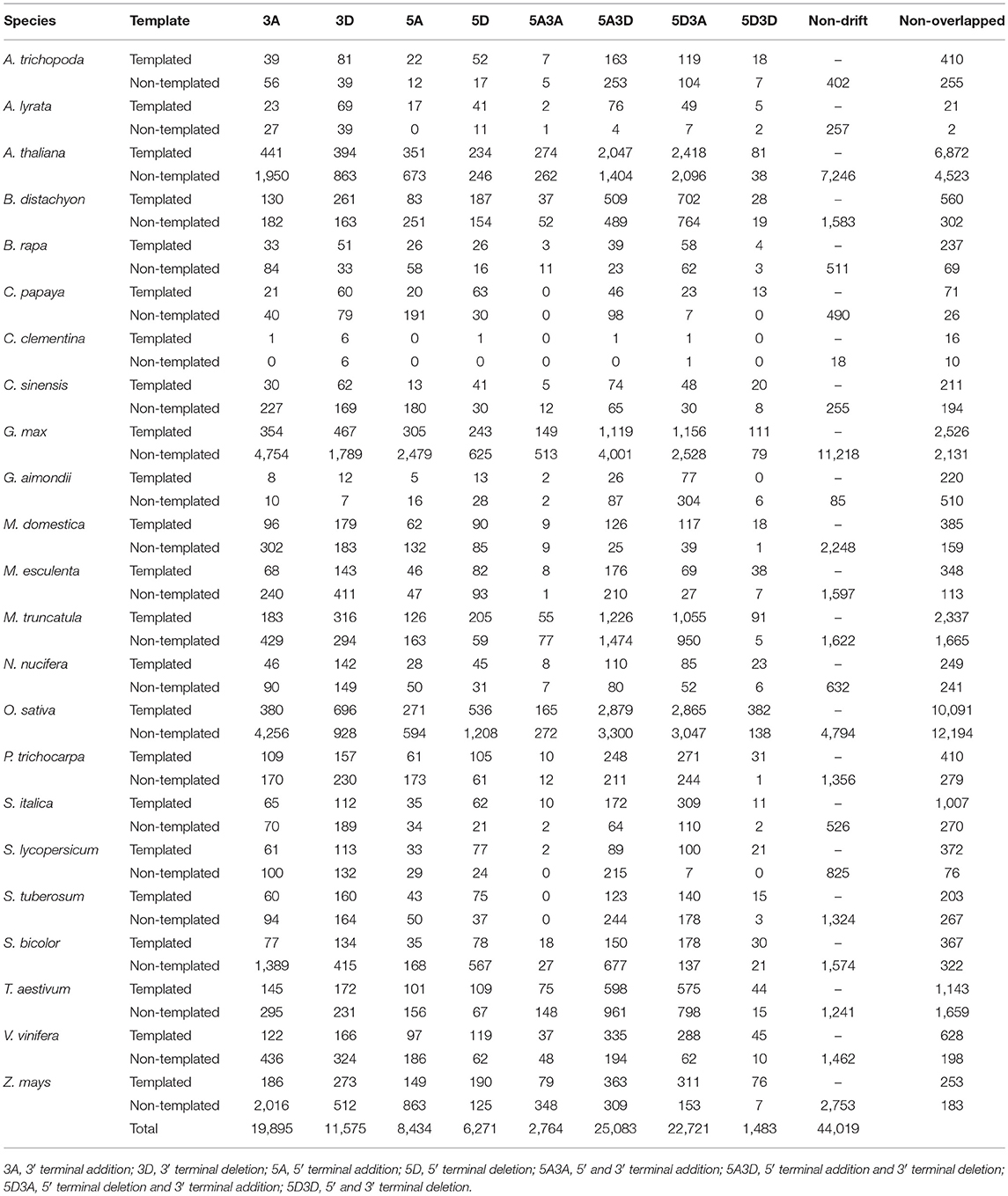
Identification and profiling of isomiRs has recently gained importance taking into account the increasing reports of isomiRs present in plants genomes and their consecutive involvement in response to abiotic stresses (Ahmed et al., 2014; Sablok et al., 2015). Several genomics and bioinformatics approaches have been developed, however large-scale mining of isomiRs and their relationship with canonical miRNAs and corresponding target divergence and conversation are some of the questions which demand attention to understand the origin and diversity of isomiRs. Plant isomiR Atlas fills this knowledge gap by providing large-scale isomiRs across 23 species. High throughput profiling of isomiRs across plant genomes using the criteria (see Material and Methods) allowed the detection of unique 98,374 templated and non-templated isomiRs from 6,167 precursors, with species G. max (13,478), O. sativa (24,503), A. thaliana (17,516), and M. truncatula (7,584) showing the maximum number of unique isomiRs across the studied plant species (Table 1). It is noteworthy to mention that number of unique isomiR may be influenced by available sequencing data for the respective species. A total of 44,019 non-drift events were observed in non-templated isomiRs (Table 2). Among the drift events, 5A3D and 5D3A showed maximum abundance with 25,083 and 22,721, respectively (Table 2).
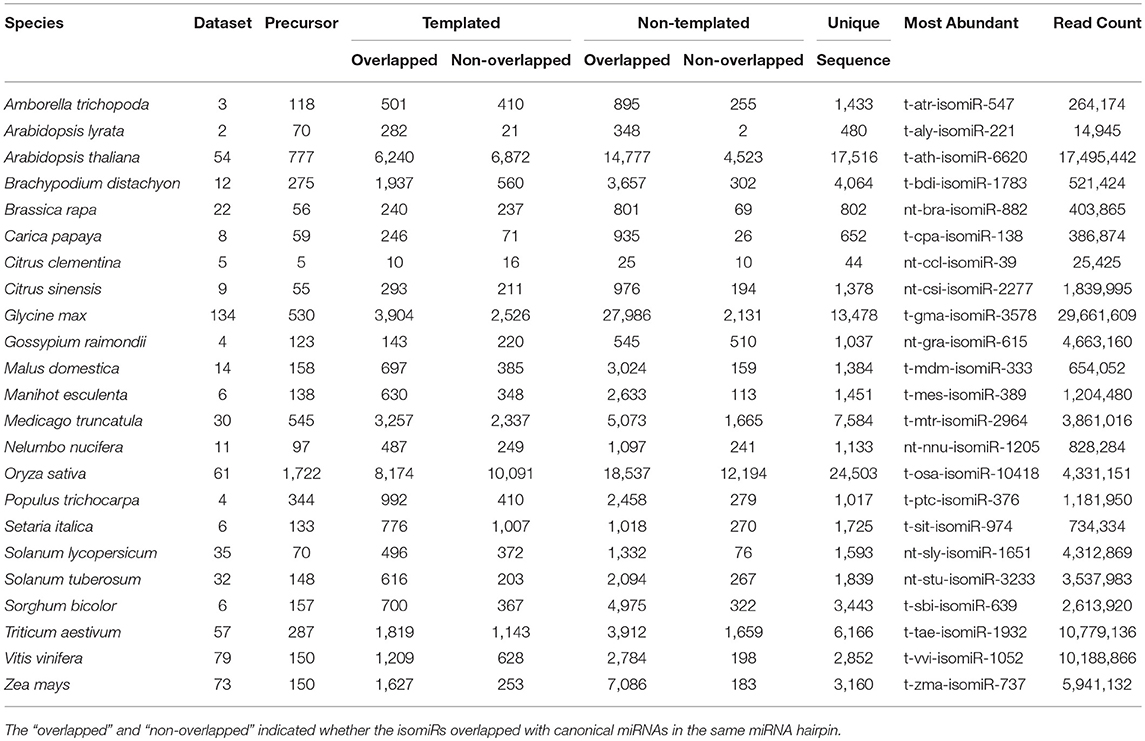
Table 1. Table showing the species enlisted in the Plant isomiR Atlas along with information on number of datasets, precursor, template, and non-templated isomiRs identified for each species and the most abundant isomiR in each species with the read count corresponding for that isomiR.
Terminal modifications play an important role in defining the isomiRs and have been primarily linked with post-transcriptional uridylation events, cytidine additions or nucleotide addition that might regulate the 3′-5′ miRNA degradation as previously observed in Larix leptolepis, A. thaliana and other model plants (Zhang et al., 2013). Previously, template based terminal modification have been described as a measure of the base-line events that might indicate the isomiR biogenesis. We leveraged and focused on both the templated and non-templated isomiRs and interestingly observed that the non-templated isomiRs (Table S2) share the same pattern of terminal modification as templated isomiRs (Table S3). Table S3 details the observed terminal modification with detailed information on types of substitution such as internal single SNP are classified under SS, internal random SNPs are classified under MS, internal SNPs and terminal substitution are classified under CV, TS for having internal tandem SNPs, and 5 V/3 V for 5′- or 3′- end substitutions. Among the terminal modification cytidine addition was one the most dominant event in species such as A. thaliana, B. distachyon, G. max, S. bicolor, and Z. mays supporting the previous observations (Kulcheski et al., 2011; Zhang et al., 2013; Jacobs et al., 2016). Interestingly, we observed that the cytidine addition was also one of most prominent in the A. trichopoda suggesting that the cytidine addition for isomiR biogenesis might be an evolutionary conserved phenomenon (Table S3).
Plant isomiR Atlas Visualization
Plant isomiR Atlas is an easy to browse web-repository of plant specific isomiRs and allows for the selection and visualization of profiled isomiRs along with the targets and canonical miRNAs information according to the species of interest. Among the features offered, users are allowed to browse the dataset information on the basis of species (Figure 2a). In the result page, tabulated dataset information, which is hyperlinked to the respective SRA accession used for the identification of isomiRs (Figure 2b) was shown. The parameter selection page offers several features such as overlap of the isomiR to the canonical miRNA, seed corruptness, mismatch type in case of non-templated isomiRs. Additionally, isomiRs can be further filtered based on the depth of the isomiRs and the types of the isomiRs classified on the basis of 5′- or 3′- end substitutions events (Figure 2c). Circular graphs and tabulated information for the drift events embedded within the Plant isomiR Atlas allows for the visualization of the associated drift events, nucleotide substitutions, and substitution patterns on the basis of species or on the basis of the selected miRNA precursors (Figures 2d–f).

Figure 2. Schematic representation of browsing of Plant isomiR Atlas: (a) sub section shows the species selection according to species name or present an option to select all the species; (b) tabulated list of the datasets present for each species and also hyperlinked pages for the embedded datasets. Additionally, a final panel to allow user to search for particular accession; (c) the parameter page allows the user to define several parameters such as define species, mismatch, seed corruptness, overlap and minimum, and maximum length of isomiR with additional selection based on the depth of isomiR and the type of the isomiR; (d) Tabulated list of the isomiRs for the selected species with hyperlinks to see the isomiR associated information and does the isomiRs generated drift with the observed variation; (e) circular graphs showing the proportion of the variations observed in the species defined isomiRs; (f) circular graphs showing the proportion of the variations observed in the particular isomiR defined in the species; (g) isomiR classified page displaying multitude of information such as isomiR sequence, length, alignment of the isomiR with mature canonical miRNAs, potential origin of the isomiRs, isomiR overlap with other species, sequencing depth for the selected isomiR in the SRA datasets spanning for the selected species and across the other species in which the isomiRs shared the sequence conservation, target prediction for the selected isomiR using the psRNATarget and Targetfinder and a consensus overlap between the two target prediction algorithms.
Plant isomiR Atlas provides isomiR specific pages for each identified isomiRs (Figure 2g) with pre-defined features such as isomiR sequence, overlap with the canonical miRNAs, type of modifications and target predictions coming from the psRNATarget and TargetFinder. Diverse roles and functions of miRNAs have been well described previously and miRNAs interactions with the canonical target site in the mRNAs have been a subject of wide explorations. Recently, it has been shown that the miRNAs not only show lineage specific conserved functions but also diverge functions also, which increase the target repertoire of the plant miRNAs (Baldrich et al., 2018) It is noteworthy to highlight that these interactions have not only shown at the miRNAs level but also the level of siRNAs, which have alternative biogenesis pathway through the RdDM pathway. Irrespective of the biogenesis pathway, post-transcriptional regulatory role of miRNAs has been widely demonstrated ranging from the developmental to the functional landscape of plant abiotic and biotic stress. From the development point of view, regulatory role of the miR156 and their canonical post-transcriptional control of the SPL- family of genes have been widely illustrated. More details on capability of Plant isomiR Atlas can be found at the Help page of the database (http://www.mcr.org.in/isomir/help.php?pg=5).
In confluence with the recent reports, it is becoming increasingly evident that although the miRNA targets are evolutionary conserved for some families of miRNAs, they might however have additional lineage specific targets. From the recent miRNAs and siRNAs including the phasiRNAs studies, Ma et al. (2017) highlighted the functional divergence of the miR482/2118 family, which has in addition to the evolutionary conserved targets such as NB-LRR genes by showing additional functional targets such as protein kinases and zinc finger nucleases in Litchi chinensis Sonn. Taking into account these reports, it becomes imperative to explore the target site feasibility and also miRNA-target interactions. Ahmed et al. (2014) directed the conclusion that isomiRs coupled with canonical miRNAs can increase the functional target predictions. Taking this into account, we implement two target prediction algorithms in Plant isomiR Atlas viz. psRNATarget (Dai and Zhao, 2011) and TargetFinder (Fahlgren et al., 2007) independently and also consensus target predicted by both target prediction algorithms. Availability of these two target predictions allows the robust consensus selection of the predicted targets and allow for the identification of the functionally and evolutionarily conserved isomiRs with respect to the isomiR targets and will increase the target repertoire of the classified isomiRs for each species. For each isomiR, a cross-species conservation based on sequence conservation and also the concurrent read depth are provided. Recent studies have shown that genic miRNAs have different rates of evolutions such as the family targeting the chalcone 4 -O-glucosyltransferase (4 CGT) (Baldrich et al., 2018) and also the family specific miRNAs such as os-miR7695 in O. sativa (Baldrich et al., 2018) and miR-1510 in legumes (Baldrich et al., 2018). As the relationship between the canonical miRNAs and potential targets unravel with more diverse species, it is understandable that the corresponding isomiR diversity and their targets will eventually lead to better understanding of miRNAome. Taking this into account, Plant isomiR Atlas provides both the read based as well as the cross-species conservation of the identified targets for the isomiRs, which will accelerate the identification and development of the specific species isomiRs having shared or novel targets.
Conclusion
To conclude, we have developed a large-scale plant specific isomiR repository, which features 23 plants species reresenting a total of 677 datasets and addresses both the templated and non-templated isomiRs profiling. Easy to browse plant isomiR atlas will accelerate the functional profiling of the templated and non-templated isomiRs and to understand the isomiR biogenesis and its conclusive regulatory roles. Understanding of the concurrent role of the isomiRs and the canonical miRNAs will contribute to increased understanding of non-coding miRNAome diversity and regulation genomics for strengthening the plant functional genomics.
Author Contributions
GS conceived and designed the research. KY and GS analyzed the data. KY coded the Plant isomiR Atlas. SM hosted the Plant isomiR Atlas. XW took part in discussions. GS and KY wrote the manuscript.
Funding
This project is supported by grants from the National Natural Science Foundation of China (31560549, 31760566) and The Provincial Nation-class Discipline of Biology Foundation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01881/full#supplementary-material
References
Ahmed, F., Senthil-Kumar, M., Lee, S., Dai, X., Mysore, K. S., and Zhao, P. X. (2014). Comprehensive analysis of small RNA-seq data reveals that combination of miRNA with its isomiRs increase the accuracy of target prediction in Arabidopsis thaliana. RNA Biol. 11, 1414–1429. doi: 10.1080/15476286.2014.996474
Axtell, M. J., Jan, C., Rajagopalan, R., and Bartel, D. P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127, 565–577. doi: 10.1016/j.cell.2006.09.032
Baldrich, P., Beric, A., and Meyers, B. C. (2018). Despacito: the slow evolutionary changes in plant microRNAs. Curr. Opin. Plant Biol. 42, 16–22. doi: 10.1016/j.pbi.2018.01.007
Burroughs, A. M., Ando, Y., de Hoon, M. J., Tomaru, Y., Nishibu, T., Ukekawa, R., et al. (2010). A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 20, 1398–1410. doi: 10.1101/gr.106054.110
Dai, X., and Zhao, P. X. (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39, W155–W159. doi: 10.1093/nar/gkr319
de Oliveira, L. F., Christoff, A. P., and Margis, R. (2013). isomiRID: a framework to identify microRNA isoforms. Bioinformatics 29, 2521–2523. doi: 10.1093/bioinformatics/btt424
Fahlgren, N., Howell, M. D., Kasschau, K. D., Chapman, E. J., Sullivan, C. M., Cumbie, J. S., et al. (2007). High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2:e219. doi: 10.1371/journal.pone.0000219
Fátyol, K., Ludman, M., and Burgyán, J. (2015). Functional dissection of a plant Argonaute. Nucleic Acids Res. 44, 1384–1397. doi: 10.1093/nar/gkv1371
Fei, Q., Yu, Y., Liu, L., Zhang, Y., Baldrich, P., Dai, Q., et al. (2018). Biogenesis of a young, 22-nt microRNA in Phaseoleae species by precursor-programmed uridylation. bioRxiv. p.310920. doi: 10.1101/310920
Goodstein, D. M., Shu, S., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2011). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Gozmanova, M., Baev, V., Apostolova, E., Sablok, G., and Yahubyan, G. (2017). Growing diversity of plant MicroRNAs and MIR-derived small RNAs. Plant Epigenet. 49–67. doi: 10.1007/978-3-319-55520-1_3
Guo, L., Yu, J., Liang, T., and Zou, Q. (2016). miR-isomiRExp: a web-server for the analysis of expression of miRNA at the miRNA/isomiR levels. Sci. Rep. 6:23700. doi: 10.1038/srep23700
Incarbone, M., Ritzenthaler, C., and Dunoyer, P. (2018). Peroxisomal targeting as a sensitive tool to detect protein-small RNA interactions through in vivo Piggybacking. Front. Plant Sci. 9:135.
Jacobs, T. B., Lawler, N. J., LaFayette, P. R., Vodkin, L. O., and Parrott, W. A. (2016). Simple gene silencing using the trans-acting siRNA pathway. Plant Biotechnol. J. 14, 117–127. doi: 10.1111/pbi.12362
Kozomara, A., and Griffiths-Jones, S. (2013). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73. doi: 10.3389/fpls.2018.00135.
Kuchenbauer, F., Morin, R. D., Argiropoulos, B., Petriv, O. I., Griffith, M., Heuser, M., et al. (2008). In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res. 18, 1787–1797. doi: 10.1101/gr.077578.108
Kulcheski, F. R., de Oliveira, L. F., Molina, L. G., Almerão, M. P., Rodrigues, F. A., Marcolino, J., et al. (2011). Identification of novel soybean microRNAs involved in abiotic and biotic stresses. BMC Genomics 12:307. doi: 10.1186/1471-2164-12-307
Landgraf, P., Rusu, M., Sheridan, R., Sewer, A., Iovino, N., Aravin, A., et al. (2007). A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414. doi: 10.1016/j.cell.2007.04.040
Lin, J. S., Kuo, C. C., Yang, I. C., Tsai, W. A., Shen, Y. H., Lin, C. C., et al. (2018). MicroRNA160 modulates plant development and heat shock protein gene expression to mediate heat tolerance in Arabidopsis. Front. Plant Sci. 9:68. doi: 10.3389/fpls.2018.00068
Ma, W., Chen, C., Liu, Y., Zeng, M., Meyers, B. C., Li, J., et al. (2017). Coupling of microRNA-directed phased small interfering RNA generation from long noncoding genes with alternative splicing and alternative polyadenylation in small RNA-mediated gene silencing. New Phytol. 217, 1535–1550. doi: 10.1111/nph.14934
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17:10. doi: 10.14806/ej.17.1.200
Mi, S., Cai, T., Hu, Y., Chen, Y., Hodges, E., Ni, F., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5' terminal nucleotide. Cell 133, 116–127. doi: 10.1016/j.cell.2008.02.034
Morin, R. D., Aksay, G., Dolgosheina, E., Ebhardt, H. A., Magrini, V., Mardis, E. R., et al. (2008a). Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 18, 571–584. doi: 10.1101/gr.6897308
Morin, R. D., O'Connor, M. D., Griffith, M., Kuchenbauer, F., Delaney, A., Prabhu, A. L., et al. (2008b). Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 18, 610–621. doi: 10.1101/gr.7179508
Muller, H., Marzi, M. J., and Nicassio, F. (2014). IsomiRage: from functional classification to differential expression of miRNA isoforms. Front. Bioeng. Biotechnol. 2:38. doi: 10.3389/fbioe.2014.00038
Neilsen, C. T., Goodall, G. J., and Bracken, C. P. (2012). IsomiRs–the overlooked repertoire in the dynamic microRNAome. Trends Genet. 28, 544–549. doi: 10.1016/j.tig.2012.07.005
Rogans, S. J., and Rey, C. (2016). Unveiling the micronome of Cassava (Manihot esculenta Crantz). PLoS ONE 11:e0147251. doi: 10.1371/journal.pone.0147251
Ruby, J. G., Jan, C., Player, C., Axtell, M. J., Lee, W., Nusbaum, C., et al. (2006). Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127, 1193–1207. doi: 10.1016/j.cell.2006.10.040
Sablok, G., Srivastva, A. K., Suprasanna, P., Baev, V., and Ralph, P. J. (2015). isomiRs: increasing evidences of isomirs complexity in plant stress functional biology. Front. Plant Sci. 6:949. doi: 10.3389/fpls.2015.00949
Stief, A., Altmann, S., Hoffmann, K., Pant, B. D., Scheible, W. R., and Bäurle, I. (2014). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26, 1792–1807. doi: 10.1105/tpc.114.123851
Yang, K., Sablok, G., Qiao, G., Nie, Q., and Wen, X. (2017). isomiR2Function: an integrated workflow for identifying microRNA variants in plants. Front. Plant Sci. 8:322. doi: 10.3389/fpls.2017.00322
Yang, K., Wen, X., and Sablok, G. (2018). Method for the Large-Scale Identification of phasiRNAs in Brachypodium distachyon. Brachypodium Genomics 187–194. doi: 10.1007/978-1-4939-7278-4_14
Yuan, C., Meng, X., Li, X., Illing, N., Ingle, R. A., Wang, J., et al. (2017). PceRBase: a database of plant competing endogenous RNA. Nucleic Acids Res. 45, D1009–D1014. doi: 10.1093/nar/gkw916
Zhang, J., Zhang, S., Li, S., Han, S., Wu, T., Li, X., et al. (2013). A genome-wide survey of microRNA truncation and 3' nucleotide addition events in larch (Larix leptolepis). Planta 237, 1047–1056. doi: 10.1007/s00425-012-1823-7
Zhang, Y., Zang, Q., Zhang, H., Ban, R., Yang, Y., Iqbal, F., et al. (2016). DeAnnIso: a tool for online detection and annotation of isomiRs from small RNA sequencing data. Nucleic Acids Res. 44, W166–W175. doi: 10.1093/nar/gkw427
Keywords: isomiRs, microRNAs, plants, post-transcriptional machinery, functional targets
Citation: Yang K, Wen X, Mudunuri SB and Sablok G (2019) Plant IsomiR Atlas: Large Scale Detection, Profiling, and Target Repertoire of IsomiRs in Plants. Front. Plant Sci. 9:1881. doi: 10.3389/fpls.2018.01881
Received: 09 March 2018; Accepted: 05 December 2018;
Published: 22 January 2019.
Edited by:
Rosalba Giugno, Università degli Studi di Verona, ItalyReviewed by:
Firoz Ahmed, Jeddah University, Saudi ArabiaQuan Zou, University of Electronic Science and Technology of China, China
Copyright © 2019 Yang, Wen, Mudunuri and Sablok. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaopeng Wen, xpwensc@hotmail.com
Gaurav Sablok, sablokg@gmail.com
†These authors have contributed equally to this work
 Kun Yang
Kun Yang Xiaopeng Wen1*
Xiaopeng Wen1* Suresh B. Mudunuri
Suresh B. Mudunuri Gaurav Sablok
Gaurav Sablok