- State Key Laboratory of Grassland Agro-Ecosystem, School of Life Sciences, Lanzhou University, Lanzhou, China
Secondary metabolites of the flavonoid pathway participate in plant defense, and bHLH and MYB transcription factors regulate the synthesis of these metabolites. Here, we define the regulatory mechanisms in response to pathogens. Two transcription factors from Populus alba var. pyramidalis, PalbHLH1 and PalMYB90, were overexpressed together in poplar, and transcriptome analysis revealed differences in response to pathogen infection. The transgenic plants showed elevated levels of several key flavonoid pathway components: total phenols, proanthocyanidins (PAs), and anthocyanins and intermediates quercetin and kaempferol. Furthermore, PalbHLH1 and PalMYB90 overexpression in poplar enhanced antioxidase activities and H2O2 release and also increased resistance to Botrytis cinerea and Dothiorella gregaria infection. Gene expression profile analysis showed most genes involved in the flavonoid biosynthesis pathway or antioxidant response to be upregulated in MYB90/bHLH1-OE poplar, but significant differential expression occurred in response to pathogen infection. Specifically, expression of PalF3H (flavanone 3-hydroxylase), PalDFR (dihydroflavonol 4-seductase), PalANS (anthocyanin synthase), and PalANR (anthocyanin reductase), which function in initial, middle, and final steps of anthocyanin and PA biosynthesis, respectively, was significantly upregulated in D. gregaria-infected MYB90/bHLH1-OE poplar. Our results highlight that PalbHLH1 and PalMYB90 function as transcriptional activators of flavonoid pathway secondary-metabolite synthesis genes, with differential mechanisms in response to bacterial or fungal infection.
Introduction
The Populus species comprises some of the most valuable woody plants in the world due to their rapid growth, easy propagation, adaptation, and wide application in forestation (Polle et al., 2013). However, disease outbreaks in poplar are widespread and threaten poplar plantations and production. Poplar diseases on plantations are mainly induced by fungal or bacterial pathogens that can spread widely in leaves, stems, and even roots, causing widespread poplar death (Ostry and McNabb Jr, 1985; Newcombe and Bradshaw Jr, 1996; Newcombe and Ostry, 2001). Thus, the development of disease-resistant poplar cultivars is becoming necessary and urgent. In general, a successful disease resistance breeding program relies on an understanding of the genetic basis of resistance and interactions between hosts and pathogens, yet genetic information on disease resistance in Populus is scarce.
Secondary metabolic products play significant roles in plant resistance to pathogens. Among these metabolites, flavonoids, such as anthocyanins and condensed tannins (also called proanthocyanidins, PAs), accumulate in the seed coat, leaf, stem, bark, and root in response to biotic (Lorenc-Kukuła et al., 2005; Zhang et al., 2013) and abiotic (Dixon et al., 2005; Paolocci et al., 2007) stresses. These molecules also serve as adjuncts in the defense mechanisms of plants (Winkel-Shirley, 2002; Dixon et al., 2005; Koes et al., 2005; Lepiniec et al., 2006) and are involved in the antioxidant response to increase plant resistance to various stresses (Winkel-Shirley, 2002; Dixon et al., 2005). In poplar, PA accumulation can prevent pathogen development by either reducing mycelium growth of infectious fungi (Yuan et al., 2012; Wang et al., 2017) or inhibiting extracellular hydrolases of invading pathogens (Scalbert, 1991). Moreover, a high anthocyanin content can enhance the ROS (reactive oxygen species) scavenging induced by pathogen attack or abiotic stress (Yamasaki et al., 1996). For example, when plants are exposed to UV radiation, flavonols facilitate high antioxidant reactions, and their biosynthesis is upregulated to increase protection against this stress in Ligustrum vulgare plants (Agati et al., 2011). Therefore, to uncover the resistance mechanism of flavonoids, their biosynthetic pathways and regulation should be clarified.
The biosynthetic pathways of flavonoids are regulated by many transcription factors, such as R2R3-MYB (Cho et al., 2016; Wang et al., 2017), WD-repeat (WDR), and basic helix-loop-helix (bHLH) transcription factors (Hichri et al., 2011; Huang et al., 2013). In particular, MYB family transcription factors are emerging as central players in the coordinated activation of sets of genes specific to the anthocyanin and tannin pathways (Wang et al., 2017). In poplar, overexpression of MYB19 from P. trichocarpa activates expression of CHS1 (chalcone synthase 1) and ANS2 (anthocyanins synthase 2), which promotes the accumulation of anthocyanins in hybrid poplar (Cho et al., 2016). In tomato, coexpression of two transcription factors, Delila (Del, bHLH) and Rosea1 (Ros1, MYB), from snapdragon (Antirrhinum majus) (Butelli et al., 2008; Maligeppagol et al., 2013) results in accumulation of anthocyanins, which not only extends the shelf life of the transgenic tomatoes but also increase their resistance to Botrytis cinerea (Zhang et al., 2013). Anthocyanin production is also commonly induced under stress conditions (Gould, 2004). In tomato, overexpression of AtMYB12, Del, and Ros1 increases anthocyanin biosynthetic gene expression and activates primary metabolism (Zhang et al., 2015). In addition, expression of Del from snapdragon was shown to enhance leaf and flower anthocyanin production and antioxidant activities in tobacco by regulating the transcript levels of NtCHS, NtCHI, NtF3H, NtDFR, and NtANS (Naing et al., 2017). Altogether, evidence thus far indicates that overexpression of the Del and Ros1 transcription factors can enhance flavonoid biosynthesis and antioxidant activities and improve resistance to biotic and abiotic stress in transgenic plants. However, it remains unclear whether similar flavonoid biosynthesis response mechanisms to fungal and bacterial infection occurs in poplar.
Populus alba var. pyramidalis has been widely cultivated in northern China due to its advantageous properties of rapid growth, lack of seed catkins, erect stems, and high biomass production (Xu, 1988; Zhang et al., 2008; Xu et al., 2011). However, wood diseases and insect herbivory occur extensively in P. alba, resulting in massive production loss worldwide. Therefore, improving the disease resistance of P. alba var. pyramidalis using molecular genetic breeding technology is imperative. With the publication of P. alba var. pyramidalis (Ma et al., 2018) genomic data, screening for candidate genes thought to be involved in the formation of specific poplar traits, such as resistance to pathogens and other stresses, is possible through genome comparison.
In this study, the PalbHLH1 and PalMYB90 transcription factors were overexpressed together in P. alba var. pyramidalis, and accumulation of secondary metabolites from the flavonoid pathway was examined. We also investigated the abilities of the transgenic plants to resist pathogens and analyzed their gene expression profiles. Our findings may be utilized to alter flavonoid biosynthesis via metabolic engineering methods to improve poplar resistance to fungal and bacterial pathogens.
Materials and Methods
Plant Materials
Two-year-old Populus euphratica seedlings and P. alba var. pyramidalis saplings were collected from Xinjiang Province, China, planted in pots with loam soil, and grown in a semi-greenhouse under a constant temperature of 25°C, a 16/8-h (light/dark) photoperiod and 60% relative air humidity. Saplings and seedlings at similar growth stages were used for pathogen inoculations. P. alba var. Pyramidalis tissue culture saplings were cultured in WPM, which is a type of woody plant medium (0.1 mg L−1 NAA, 400 mg L−1 cefotaxime, 9.0 mg L−1 hygromycin, 20 g L−1 sucrose, and 8.3–8.4 g L−1 agar) (Lloyd and McCown, 1980), and plantlets were grown under a 14/10-h (light/dark) cycle with supplemental light (56.25 µmol m−2 s−1) in a light growth chamber at 25°C.
PalbHLH1 and PalMYB90 Gene Cloning
Total RNA was isolated from the leaves of P. alba var. pyramidalis saplings using Plant Mini Kit (Qiagen, Germany). First-strand cDNA was synthesized from 2 μg of total RNA in a 20-μl reaction mixture using the RT-AMV transcriptase kit (TaKaRa, Dalian, China). The coding sequences of PalbHLH1 and PalMYB90, were amplified using gene-specific primers (Table S1 in Datasheet 1) designed based on the PalbHLH1/PalMYB90 gene sequences respectively, in the P. alba var. pyramidalis genome (Ma et al., 2018). PCR was carried out with Pfu DNA polymerase (TaKaRa) in a total volume of 50 μl with a thermal cycler program of 98°C for 2 s, 38 cycles of 98°C for 10 s, 55°C for 5 s, and 72°C for 2 min, and a final extension step at 72°C for 10 min. The amplification products were inserted into the plant binary vector pCAMBIA1305 through intermediate vectors pMD19 and pCXSN using the enzyme digestion–linked cloning system to produce 35S::PalbHLH1 and 35S::PalMYB90 constructs (Figure 2A). Positive clones were verified by DNA sequencing and aligned with sequences from the P. alba var. pyramidalis genome (Ma et al., 2018).
Phylogenetic Analysis
The homologs of PalMYB90 (PalRos1, PAYT030711.1) from other species were retrieved by BLAST searches (http://www.phytozome.com) and aligned using MAGE 5.0 (Tamura et al., 2011) and Genedoc (Lynnon Corporation, USA).The accession numbers are shown in Table S6 in Datasheet 1. The full-length coding sequence of the PalbHLH1 gene (PalDel1, PAYT035597.1) and homologs from other plants (Heim et al., 2003) were downloaded from National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov) and aligned using MAGE 5.0 (Tamura et al., 2011). The accession numbers are listed in Table S5 in Datasheet 1. Phylogenetic analyses based on amino acid sequences were performed using the neighbor-joining (NJ) and Maximum likelihood (ML) method via MAGE 5.0. Phylogenetic analyses were performed using bootstrapping with 1,000 replicates.
Quantitative Real-Time PCR
Quantitative real-time PCR (qRT-PCR) analysis was performed using a Thermal Cycler Dice Real-Time System TP800 (TaKaRa, Dalian, China) and the specific primers for PalbHLH1 and PalMYB90 shown in Table S3 in Datasheet 1. qRT-PCR was performed in a 25 µL reaction volume containing 12.5 µL of SYBR Premix ExTaq™ (Takara, Dalian, China), and the data were analyzed as described by Tsai et al. (2006). CYC063 was used as the internal reference gene for qRT-PCR (Qu et al., 2016). Differences in gene expression are expressed as the fold change relative to the control, which was calculated using the 2−△△Ct method (Livak and Schmittgen, 2001). Each measurement was carried out in triplicate, and error bars represent the SE of the mean of fold changes for three biological replicates (Table S1 in Datasheet 1). A standard curve was generated using an accurately quantified plasmid containing the target gene and diluted into a series of concentration gradients for the PCR reaction.
Plant Transformation
Agrobacterium-mediated methods (Jia et al., 2010) were used for poplar transformation with some modifications (Ma et al., 2018). One-year-old P. alba var. pyramidalis clones propagated from cuttings grown in a greenhouse at 25°C under a 16-h light/8-h dark cycle (100 µmol/m2/s) were used for transformation. After disinfection with 12% sodium hypochlorite, leaves were cut into pieces and placed on WPM (with 2 mg/L zeatin, 1 mg/L naphthalene acetic acid, 100 µmol/L acetosyringone, 20 g L−1sucrose, and 8.3–8.4 g L−1 agar) for induction. The poplar leaf discs were infected with recombinant Agrobacterium tumefaciens strain GV3101, and putative transgenic plants were selected on WPM supplemented with 9 mg/L hygromycin. The explants were induced to produce new plants under aseptic conditions, which were used for the transformation process performed according to the P. alba var. pyramidalis transformation protocol (Ma et al., 2018).
DMACA Staining
To determine PA accumulation, leaves of WT and transgenic lines at the same growth stage were stained for about 20 min with 0.3% (w/v) 4-dimethylaminocinnamaldehyd (DMACA) in a cold mixture of methanol and 6 M HCl (1:1, v/v), rinsed with several changes of 70% (v/v) ethanol and observed under a dissecting microscope. PA-containing cells stained blue, and the stained leaves were preserved in 70% (v/v) ethanol (Li et al., 1996). In brief, after decolorization with 30% acetic acid/methanol for 6–10 h, the leaves were washed two to three times with 75% ethanol and stained with DMACA dye solution for 5–10 min, and the staining results were compared.
Quantification of Anthocyanin, Quercetin, Kaempferol, Total Phenol, and Tannin Contents
For analysis of anthocyanin levels, 0.5 g of poplar leaves at the same growth stage was ground rapidly in liquid nitrogen; 5 ml of methanol:0.1% ascorbic acid was quickly added to the ground material, and the mixture was sonicated for 1 h. The material was then wrapped in aluminum foil to protect the reaction from light, followed by shaking for 24 h at 120 rpm. After centrifugation at 2,500×g for 10 min, 1 ml of the supernatant was mixed with 1 ml ultrapure water, and 1 ml chloroform was added to remove chlorophyll. After centrifugation, the absorbance of the clear supernatant at 530 nm was determined using a spectrophotometer, and the total anthocyanin concentration was calculated using the molar absorbance of cyanidin-3-O-glucoside (Wang et al., 2013): y = 0.3305x − 0.0024, R² = 0.9956 (Figure S5A).
Quercetin and kaempferol contents were determined by chromatographic separation using a DIKMA Diamonsil (250 × 4.6 mm, 5 μm) chromatographic column. Fresh leaves (0.1 g) were collected and ground quickly in liquid nitrogen; 1 ml of cold acetone was added, and the mixture was homogenized. After centrifugation at 12,000 rpm for 10 min, the samples were subjected to disintegration via ultrasonication for 2 min. The supernatant was mixed with an equal volume of hydrochloric acid and incubated in a water bath at 70°C for 40 min. An equal volume of ethyl acetate was added to the sample, and the mixture was centrifuged at 12,000 rpm for 10 min. The mobile phase was composed of acetonitrile (A) and 0.1% phosphoric acid solution (B) and applied at a flow rate of 1.0 ml/min. Detection was carried out at 360 nm. Finally, the contents of quercetin and kaempferol were determined according to standard curves: y = 0.1543x + 0.0155 (R² = 0.9973) and y = 0.1062x + 0.006 (R² = 0.9981), respectively (Figures S5B, C).
The total phenol content was assessed using the folinephenolcolorimetric method (Singleton and Rossi, 1965). Briefly, 100 mg of fresh leaves was ground quickly in liquid nitrogen. After the addition of 1 ml of 80% methanol, the mixture was shaken in the dark for 2 h at 4°C. After centrifugation, the supernatant was diluted with 3 ml of distilled water, after which 0.5 ml of forinol (50% water solution) and 2 ml of a 20% Na2CO3 solution were added, followed by incubation in a water bath at 45°C for 15 min. After cooling for 10 min, absorbance at 650 nm was measured using a spectrophotometer, and the total phenol content was calculated according to the standard curve of gallic acid: y = 0.2791x + 0.0086, R² = 0.9964 (Figure S5D).
For extraction of soluble tannin, 0.5 g fresh leaf tissue was ground in liquid nitrogen and extracted with 5 ml pre-prepared extraction solution (70% acetone and 0.5% ascorbic acid). The samples were ultrasonicated for 1 h and then shaken 24 h in darkness at 20°C. The sample was then centrifuged at 10,000 r/min for 10 min. The supernatant was transferred to another clean centrifuge tube, and excess NaCl was added. After shaking for 5 min, the acetone phase was collected, and the residue was re-extracted twice. The acetone phase was freeze-dried and dissolved in 2 ml methanol containing 0.5% ascorbic acid; 1.5 ml concentrated hydrochloric acid, 3% methanol-soluble vanillin and 0.5 ml final extract were added to a clean test tube for reaction in the dark for 15 min. The absorbance at 550 nm of the final reaction solution was assessed by spectrophotometry to calculate the final content of soluble condensed tannin using a standard curve.
The insoluble tannin content was also measured by spectrophotometry. After extracting soluble tannins, the residue was collected and thoroughly air-dried. Formic acid/butanol (5 ml) was added, and after ultrasonic treatment for 1 h, the absorbance of the reaction solution at 500 nm was measured. After treatment in boiling water for 1 h, absorbance at 500 nm was measured again, and the difference between the two measurements was considered the insoluble tannin content, as calculated using a standard curve. For the tannin standard curve, the preparation method was to absorb 0, 0.1, 0.25, 0.5, 1, and 2 mg/ml tannin standard solution, 1 ml each and placed in the cuvette. Different volumes (0, 0.1, 0.25, 0.5, 1, 2 ml) of the 1 mg/ml standard tannin solution was absorbed to a colorimetric plate. Absorbance was measured at 550 nm, and the standard working curve was drawn. The following regression equation was obtained: y = 0.4463x + 0.0456, R2 = 0.9972 (Figure S5E). Each experiment was performed three times with at least three replicate plants per treatment and included controls.
Examination of Antioxidant Enzyme Activities
To investigate the tolerance of MYB90/bHLH1-OE poplar plants to various stresses, we evaluated the activities of peroxidase (POD) and superoxide dismutase (SOD) and release of hydrogen peroxide (H2O2). Wild type (WT), MYB90/bHLH1-2, and MYB90/bHLH1-5 saplings were grown in soil for 2 months. The eighth leaf from the top was harvested and used to measure SOD and POD activities and the H2O2 content using a kit (Comin Biotechnology, China) according to the manufacturer’s protocol. Specific operating instructions can be obtained from the company’s website (www.cominbio.com). Each sample included at least three individual plants, and all experiments were conducted in triplicate.
Transgenic Plant Pathogen Challenge
To evaluate resistance to pathogens, mature leaves of 3-month-old poplar saplings were incubated with D. gregaria or B. cinerea. D. gregaria was grown on potato dextrose agar medium for 3 days at 28°C; B. cinerea was grown on malt extract peptone agar medium for 3–5 days at 25°C (Zheng et al., 2006; Huang et al., 2012; Karim et al., 2015). The pathogen-inoculated leaves were covered with a transparent plastic dome to maintain high humidity, and disease development under a constant temperature of 25°C and a 16/8-h (light/dark) photoperiod was evaluated after 5 days. The relative lesion areas of the poplar leaves were evaluated using ImageJ software. Each sample included at least three individual plants, and all experiments were conducted in triplicate.
Transcriptome Analysis of Transgenic Poplar After Pathogen Infection
One-year-old P. alba var. pyramidalis saplings were cultivated and infected by the two pathogens under the conditions described above. Three individuals of sapling (WT and MYB90/bHLH1-OE5) incubated with a pathogen were selected as biological replicates. Total RNA was extracted for transcriptome sequencing using the cetyltrimethyl ammonium bromide (CTAB) procedure (Chang et al., 1993). cDNA libraries were constructed and then sequenced at ANOROAD Genome (Beijing, China) using the Illumina HiSeq 2500 sequencing platform (Illumina Inc., CA, USA) with a 150-bp read length. Image output data from the sequencer were transformed into raw sequence data by base calling; raw data were stored in FASTQ format. Sequence data from this study can be found at Genome Sequence Archive (http://bigd.big.ac.cn/) under accession number CRA001044. Sequenced reads were mapped to the P. alba var. pyramidalis genome using Tophat2 software, and expression levels were calculated using cufflinks software (Trapnell et al., 2012). Differentially expressed genes (DEGs) were identified by cuffdiff based on the parameters q < 0.05 and log2 (fold change) ≥1 or ≤ –1. We plotted heatmap based on FPKM through the ggplot2 package (Ginestet, 2011) in R (https://cran.r-project.org/mirrors.html).
GO and KEGG Enrichment Analyses
Gene Ontology (GO) analysis was classified using Web Gene Ontology Annotation Plot Software (WEGO; http://wego.genomocs.org.cn). The classifications were calculated using custom-designed Perl scripts. The enrichment of the Kyoto Encyclopedia of Genes and genes (KEGG) was carried out using KOBAS 3.0 software (http://kobas.cbi.pku.edu.cn/anno_iden.php), based on homologous genes identified from P. trichocarpa using a reciprocity Basic Local Alignment Search Tool (BLAST) analysis (Tuskan et al., 2006; Marchant et al., 2015). In order to prove that the software combination of “Tophat2 and cufflink” is accurate for mapping, gene expression qualification and DEGs screening for the RNA-seq, we repeated the results of D.gregaria group by using Hisat2 for mapping and feature_count for gene expression qualification, and finally Deseq2 and edgeR for DEGs screening. The repeated results were showed in Supplementary Datasheet 2.
Statistical Analysis
Data are presented as the mean ± standard error for each group from three independent experiments. Statistical analysis of the qRT-PCR and secondary metabolite content results was performed using one-way analysis of variance in SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). The LSD and Duncan tests were applied to compare means and determine significant differences for multiple samples with a significance level P = 0.05. We used Origin software (version 8.0) and AI (Adobe Illustrator CC 2017) to plot SPSS statistical data.
Results
Isolation and Characterization of the PalbHLH1 and PalMYB90 Genes of P. alba var. pyramidalis
To investigate the potential mechanisms of the transcription factors PalbHLH1 and PalMYB90 involved in the poplar response to pathogens, we cloned the PalbHLH1 and PalMYB90 genes from the P. alba var. pyramidalis genome (Ma et al., 2018) (Table S1 in Datasheet 1) and aligned them with homologous genes from P. euphratica (Ma et al., 2013), P. trichocarpa (Tuskan et al., 2006), and A. majus. The PalbHLH1 and PalMYB90 sequences were identified via phylogeny analysis (Figures 1A, B) and sequencing with specific primers (Table S2 in Datasheet 1). PalbHLH1 contains a bHLH-MYC-N domain and an HLH domain (Figure 1C); PalMYB90 contains two tandem R2R3-Myb DNA-binding domains (Figure 1D). All of these protein domains are conserved and similar to their homologs from A. majus.
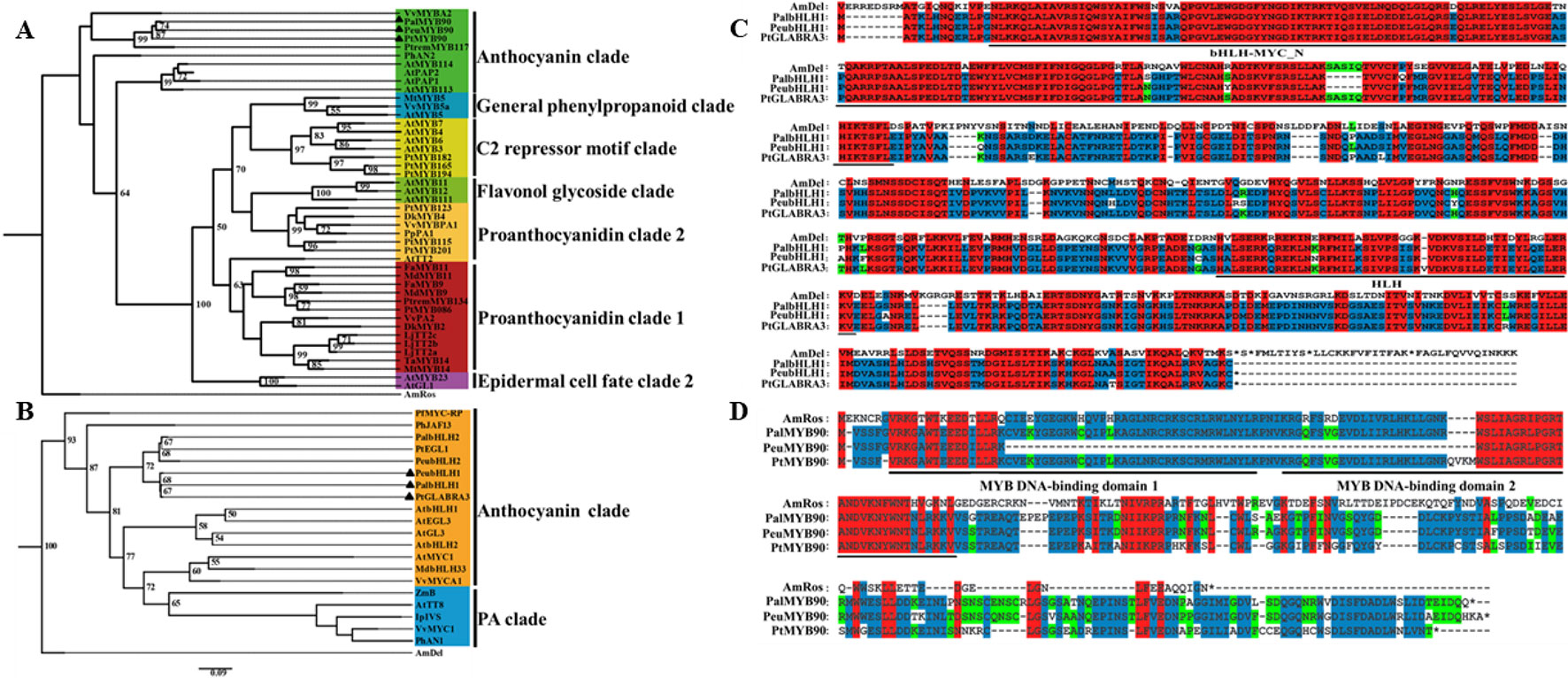
Figure 1 Phylogenetic analyses and multiple alignments of PalbHLH1 and PalMYB90. (A) Phylogenetic tree of MYB90 and selected R2R3-Myb proteins from other plant species obtained using the Maximum Likelihood (ML) method in MEGA v.5.1 software. (B) Phylogenetic tree of bHLH1 with other bHLH sequences. The scale bar represents 0.05 substitutions per site. (C, D) The bHLH domains of bHLH1 and R2R3-Myb of MYB90 are both labeled with black lines. Identical amino acids are shaded in red, and similar amino acids are shaded in blue and green.
Construction of Transgenic Poplar Coexpressing PalbHLH1 and PalMYB90
To explore the functions of PalbHLH1 and PalMYB90 coexpressed in poplar, the two genes were cloned and ligated into the intermediate vector pCXSN and pMD19, respectively, and the two genes carrying the 35S promoter and NOS terminator were sequentially ligated into the pCAMBIA1305 vector by restriction enzyme ligation, and positive clones were confirmed through sequencing (Figure 2A). The expression plasmids were transformed into P. alba var. pyramidalis leaf discs using the Agrobacterium-mediated method as described by Jia et al. (2010), with some modifications (Ma et al., 2018), and two strongly expressing lines, MYB90/bHLH1-OE2 and MYB90/bHLH1-OE5, were obtained (Figure 2B; Figures S1, S2A, S2B and Table S3 in Datasheet 1). Phenotypic analysis showed no significant differences in the height, weight, or length-to-width ratio of MYB90/bHLH1-OE2 or MYB90/bHLH1-OE5 leaves compared to those of WT poplar (Figures S2C–E). However, compared with WT leaves, the anthocyanin content of MYB90/bHLH1-OE was 1.9- to 2.8-fold higher (P < 0.05, Figures 2C, D) and the quercetin and kaempferol contents of MYB90/bHLH1-OE were1- to 1.1-fold higher (P < 0.01, Figures 2F, G). The total amount of flavonoids containing phenol rings (i.e. the total phenol content) in MYB90/bHLH1-OE2 and MYB90/bHLH1OE5 leaves was 1.6- to 2-fold higher than that in WT leaves (P < 0.05, Figure 2E). Interestingly, PalbHLH1 expression in the MYB90/bHLH1-OE5 line was up to 1400-fold higher than that in the WT plant (Figure S2B), and its expression in the MYB90/bHLH1-OE2 line was 130 times higher than that in the WT plant (Figure S2B). This increase affected the accumulation of anthocyanids, tannins, and other secondary metabolic products in the leaves of the two overexpressing lines. Moreover, the flavonoid content in MYB90/bHLH1-OE5 was 0.2- to 0.5-fold higher than that in MYB90/bHLH1-OE2. These results indicate that overexpression of PalbHLH1 and PalMYB90 enhances the accumulation of secondary metabolites in the flavonoid pathway.
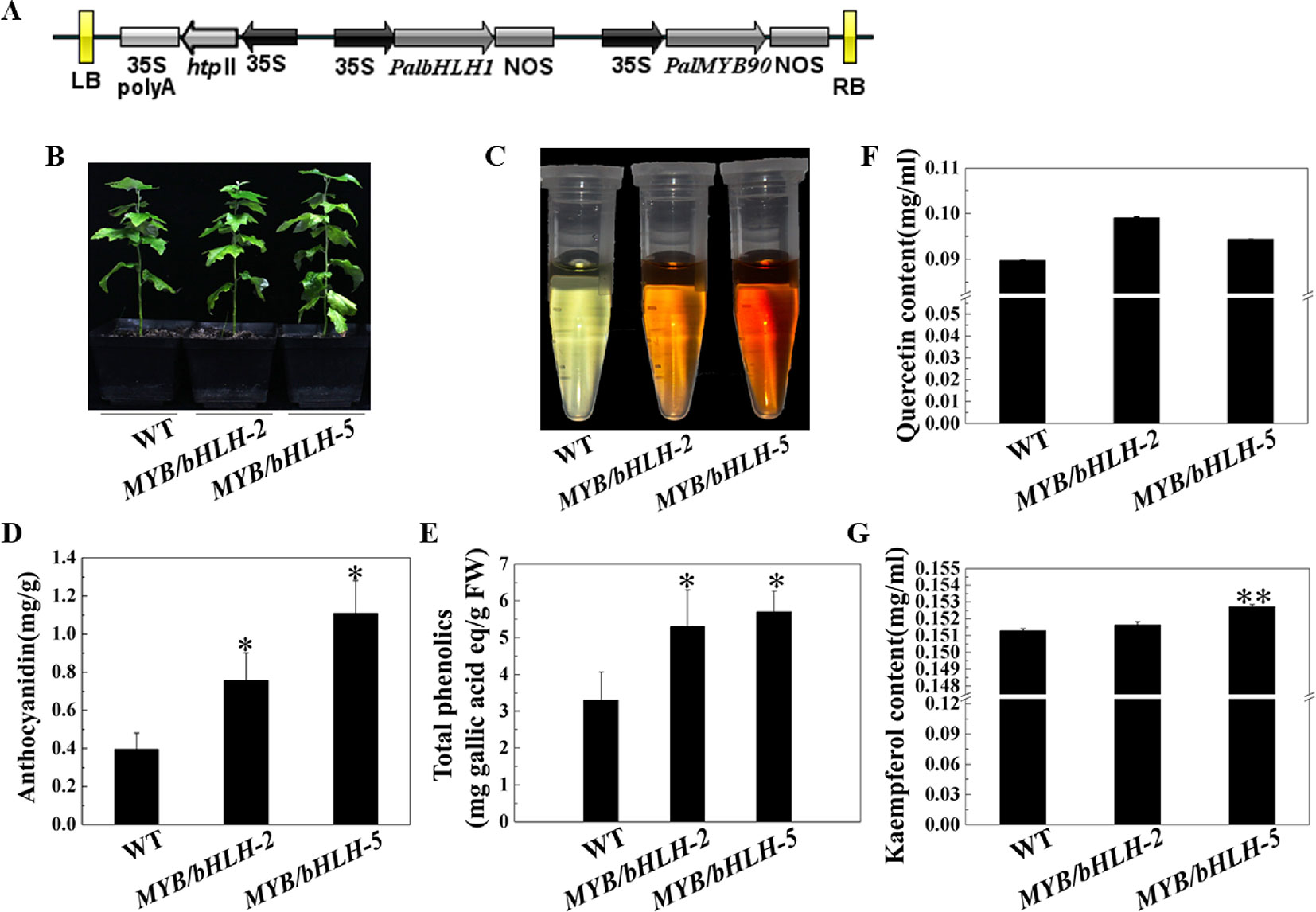
Figure 2 Characterization of transgenic poplar plants. (A) Construction of the MYB90/bHLH1-OE plasmid. (B) Growth of transgenic (MYB90/bHLH1-OE) and wild-type (WT) poplar plants. (C) Color of anthocyanin extraction. (D) Accumulation of total anthocyanins in the leaves of MYB90/bHLH1-OE and WT plants. (E) Accumulation of total phenolics in the leaves of MYB90/bHLH1-OE and WT plants. (F, G) Contents of quercetin and kaempferol in the leaves of MYB90/bHLH1-OE and WT poplar plants, respectively. The leaves of 2-month-old poplar grown in pots were used in these experiments. Error bars indicate ± SE. Asterisks (*) indicate significant differences, P < 0.05. Asterisks (**) indicate significant differences, P ≤ 0.01.
Coexpression of PalbHLH1 and PalMYB90 Genes Increases the Antioxidant Capacities of Transgenic Poplar
To study antioxidant capacities, we first measured the content of tannins, which has been confirmed to enhance the antioxidant capacity of plants (Li et al., 2007). Leaves of transgenic and WT plants were stained with DMACA, which reacts specifically with tannins and flavan-3-ols to form a blue chromophore (Figure 3A). As expected, the intensity of DMACA staining of WT leaves was reduced compared to that of the overexpressing lines, indicating a higher concentration of PAs in the leaves of the latter. These results were confirmed by comparing tannin levels in leaves of control and OE lines, and compared to WT, two transgenic lines (OE2 and OE5) showed significantly increased contents (P < 0.05) of both soluble and insoluble PA in their leaves (Figures 3B, C).
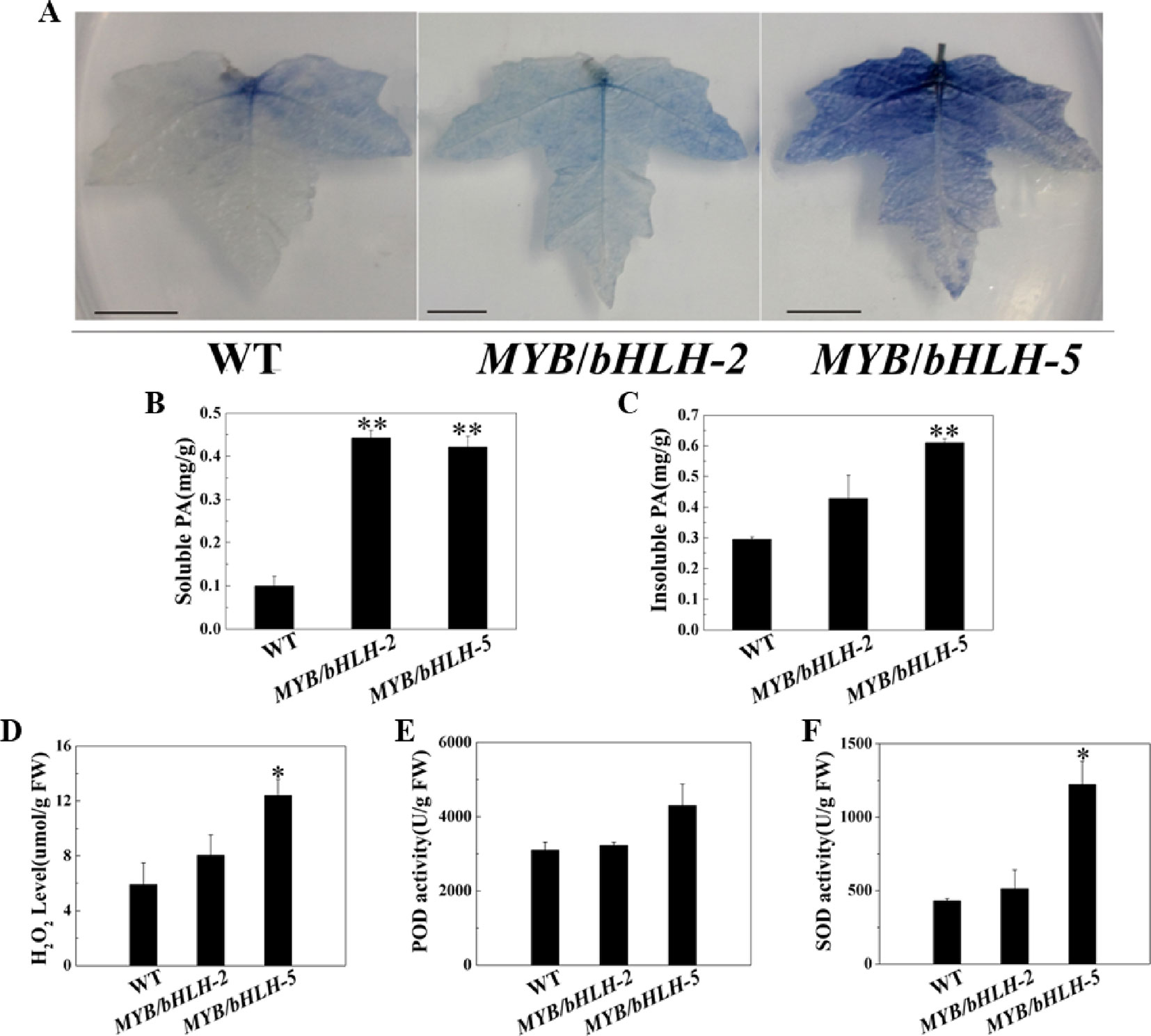
Figure 3 Quantification of insoluble and soluble tannin (PA) and reactive oxygen species in wild-type (WT) and transgenic lines (MYB90/bHLH1-OE 2 and MYB90/bHLH1-OE 5). (A) PA accumulation detected by DMACA staining. (B) Quantification of soluble PA. (C) Quantification of insoluble PA. (D) Changes in H2O2 content and (E, F) activities of two antioxidases, POD and SOD, in MYB90/bHLH1-OE poplar leaves. Values represent the means of three replications. Error bars indicate ± SE. Asterisks indicate significant differences: *P < 0.05. Asterisks (**) indicate significant differences, P ≤ 0.01.
To further prove that the transgenic plants have stronger antioxidant capacity, we determined POD/SOD activities and the H2O2 content. H2O2 levels were increased according to PalbHLH1 and PalMYB90 expression levels (Figures 3D–F). In addition, the activity of SOD was markedly increased in MYB90/bHLH1-OE5 compared to that in WT and MYB90/bHLH1-OE2 (P < 0.05). Interestingly, POD activity did not differ among WT, MYB90/bHLH1-OE2, and MYB90/bHLH1-OE5 plants (P > 0.05). A high anthocyanin content can enhance the ROS scavenging induced by pathogen attack or abiotic stress (Yamasaki et al., 1996), and the two transgenic lines (MYB90/bHLH1-OE2 and MYB90/bHLH1-OE5) exhibited significantly increased anthocyanin contents, as described above. Therefore, we suspect that the content of ROS was not only affected by the content of anthocyanids in the overexpressing plants; that is, the accumulation of H2O2 was not regulated solely by anthocyanins. Overexpression of PalbHLH1 and PalMYB90 itself may also affect ROS accumulation in MYB90/bHLH1-OE lines to aid in plant disease defense (Cui et al., 2018). In fact, accumulation of H2O2 and other ROS is one of the earliest events in host-pathogen recognition, and it has been postulated to play an important role in plant defense (Baker and Orlandi, 1995). Accordingly, elevated SOD and POD help to coordinate H2O2 accumulation to control the effects of excessive ROS on plants. Together, the elevated levels of antioxidants in the transgenic plants reduced the tissue-damaging effect of ROS and thus likely increased resistance in the MYB90/bHLH1-OE2 and MYB90/bHLH1-OE5 lines. These results indicate that overexpression of PalbHLH1 and PalMYB90 can enhance the PA concentrations and antioxidant capacity in transgenic poplar.
PalbHLH1/PalMYB90 Overexpression Enhances Resistance to D. gregaria and B. cinerea in Transgenic Poplar
Poplar plants serve as hosts for fungal pathogens, such as Aspergillus niger and D. gregaria (Scalbert, 1991; Yuan et al., 2012), and bacterial pathogens. To determine the initial effects of PalbHLH1/PalMYB90 overexpression with regard to pathogen resistance, we compared the relative lesion areas of infected leaves. The leaves of transgenic and WT plants were inoculated with D. gregaria and B. cinerea hyphal suspensions for 5–8 days. Regardless of whether D. gregaria or B. cinerea was applied, the lesion area of the WT lines was two times larger than those of the overexpression lines (Figures 4A, B), indicating that MYB90/bHLH1-OE2 and MYB90/bHLH1-OE5 had significantly stronger ability to resist both pathogens than did WT (P < 0.01). To further visualize the degree of leaf infection by pathogens, we quantitatively analyzed the size of the lesion areas using ImageJ software, with the MYB90/bHLH1-OE lines only 25–30% of the WT- D. gregaria lesion areas and 35–45% of the WT- B. cinerea lesion areas (Figure 4C). However, no significant differences between MYB90/bHLH1-OE2 and MYB90/bHLH1-OE5 lesion areas were observed (Figures 4A–C). We speculate that although the expression levels of PalbHLH1 were significantly different between the two transgenic lines and the flavonoid contents also differed, their abilities to resist disease did not differ after a certain point.
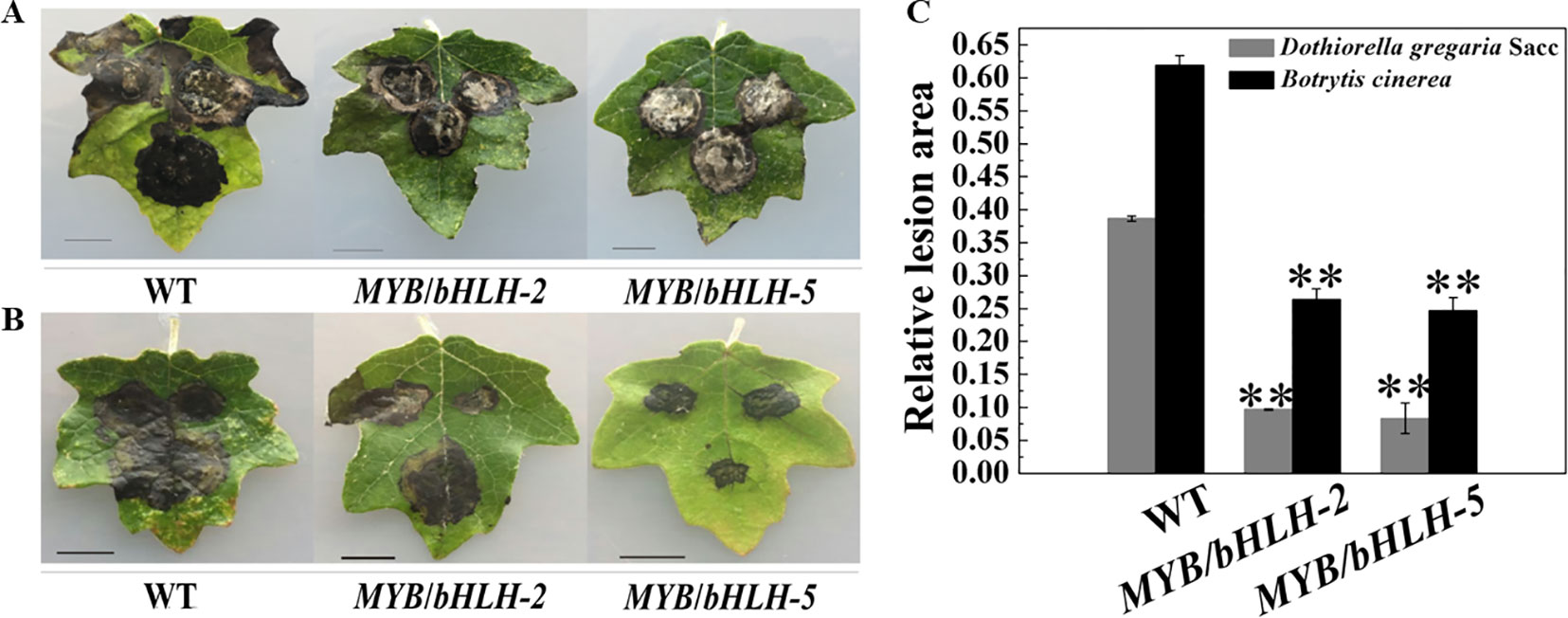
Figure 4 Resistance of transgenic poplar to pathogen infection. Disease symptoms of three leaves of wild-type (WT) and MYB90/bHLH1-OE plants after B. cinerea infection for 4 days (A) or D. gregaria infection for 7 days (B). (C) Ratios of the lesion areas of infected leaves from WT and MYB90/bHLH1-OE plants (OE-2 and OE-5 lines). Values represent the means of three replications. Error bars indicate ± SE. Asterisks indicate significant differences: **P ≤ 0.01.
Determination of Gene Expression Induced by PalbHLH1/PalMYB90 and Pathogen Infection
To investigate genes related to the improved pathogen resistance of the transgenic poplar, we analyzed gene expression profiles in transgenic lines and after infection by two pathogens. The results showed most of the genes involved in the biosynthesis of anthocyanins and flavonoids were upregulated in the transgenic plants, especially after infection. The results showed most of the genes involved in the biosynthesis of anthocyanins and flavonoids were upregulated in the transgenic plants, especially after infection (Supplementary Datasheet 3). In brief, PalbHLH1 and PalMYB90 expression levels were upregulated in the transgenic lines, and both genes were expressed at particularly higher levels after infection by the two pathogens (Figures S3A, B). Transcriptome analysis showed that overexpression of PalbHLH1/PalMYB90 promoted expression of genes involved in flavonoid biosynthesis, such as those encoding fasciclin-like arabinogalactan 6 (FLA), MYB 46, POD, laccase/diphenol oxidase family protein, WRKY 70, NAC 12, ERF 1 (ethylene responsive factor), and basic chitinase (Figures S3C, D).
In total, 56 genes, including those encoding the transcription factors MYB113, ERF4 and ERF5, and bHLH factors, were differentially expressed after infection by D. gregaria or B. cinerea (Figures S3E, F), though responses to D. gregaria and B. cinerea differed. After infection by D. gregaria, genes coding for several key enzymes involved in the regulation of secondary metabolite processes and biosynthesis, such as CHS (chalcone synthase), chalcone and stilbene synthase (PAYT009441.1), F3H (flavanone 3-hydroxylase), and DFR (dihydroflavonol 4-reductase), were significantly upregulated in MYB90/bHLH1-OE lines compared with in control plants (Figures S4A–C). Conversely, no differences among these genes were found after infection by B. cinerea. Pathogen infection might have induced differential expression of only 17 genes, including PalbHLH1, two late embryogenesis abundant (LEA) genes and the gene encoding glucosyl transferase 8 (Figures S4D–F). Furthermore, with reference to the latest research (Yu et al., 2019), we focus on the expression levels of genes coding for PalF3’H, PalDFR, PalANS, and PalANR, which catalyze the initial, middle, and final steps of anthocyanin and PA biosynthesis, respectively, were significantly upregulated in MYB90/bHLH1-OE poplar after infection by D. gregaria but not after infection by B. cinerea (Figure 5).
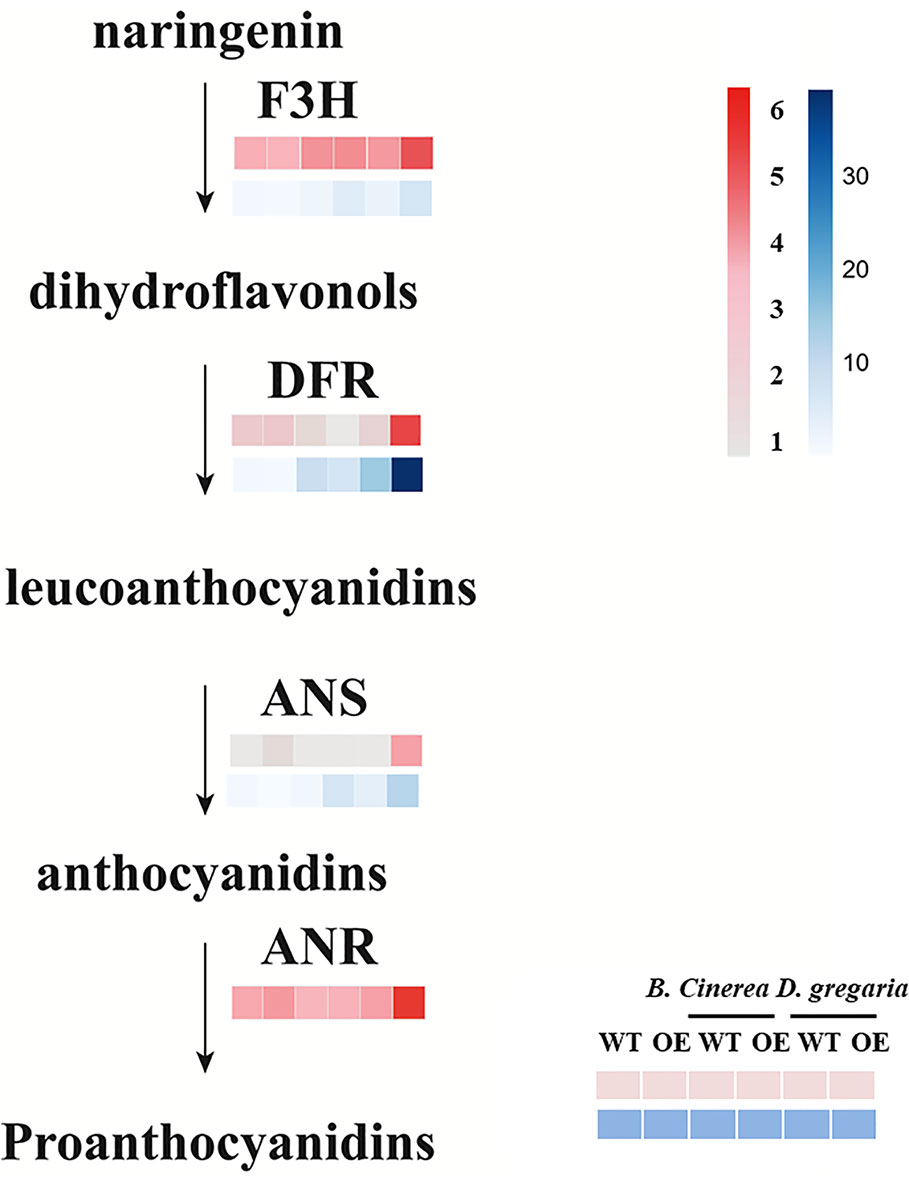
Figure 5 Expression patterns of flavonoid pathway genes in transgenic and control plants infected by two pathogens. Among them, pink blocks are gene expression patterns based on transcriptome data analysis, while blue blocks are qRT-PCR validation results for these genes. Error bars indicate ± SE.
To further validate the transcriptome results, we selected 12 genes related to flavonoid synthesis for qRT-PCR analyses in both MYB90/bHLH1-OE poplar lines (OE-2 and OE-5): F3H, DFR, ANS1, FLA, WRKY70, NAC12, ERF1, ERF4, ERF5, MYB113, LEA, and Prx1 (encoding class III peroxidase). The results showed that the expression levels of the PalF3’H, PalDFR, and PalANS genes in two transgenic lines were generally higher than those in wild type after infection by the two pathogens. Most of the 12 genes of the OE-5 line examined here, showed basically similar expression patterns with that of in RNA-seqs (Figure 6, Table S4 in Datasheet 1).
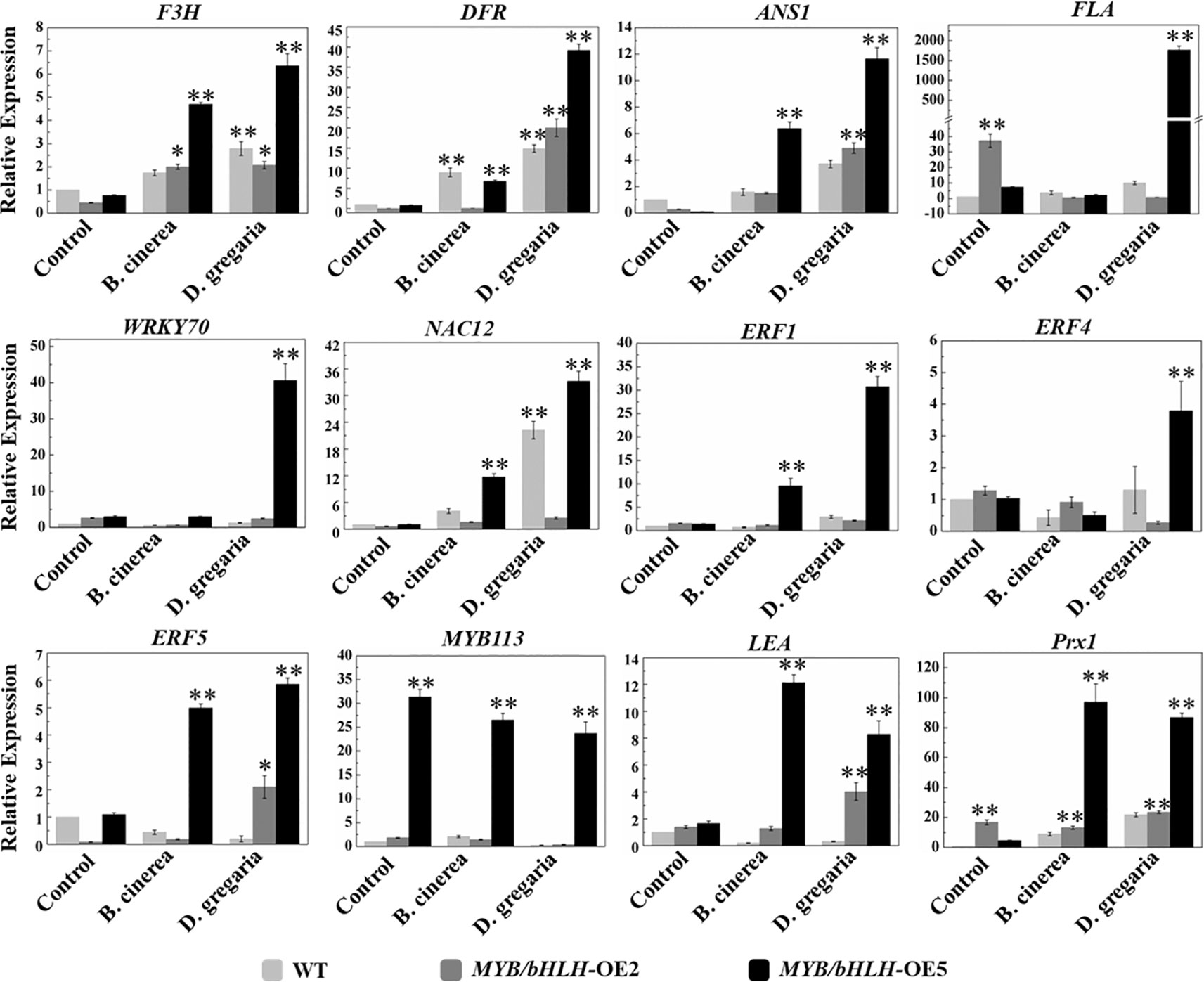
Figure 6 Quantitative real-time PCR analysis of the transcript levels in WT and both independent MYB90/bHLH1-OE lines before and after being infected by two types of pathogens. The CYC063 expression was set as the internal control. In the figure, light gray bar represents WT, dark gray and black bars represent MYB90/bHLH1-OE2 and MYB90/bHLH1-OE5 lines, respectively. Each measurement was carried out in triplicate, and error bars represent the SE of the mean of fold changes for three biological replicates. Asterisks indicate significant differences: *P < 0.05, **P ≤.01.
Anthocyanins Enhance the Resistance of P. euphratica to D. gregaria Infection
In addition to studying the mechanism of enhanced resistance to fungal pathogens in P. alba var. pyramidalis, we were also interested in the disease resistance and mechanism of different poplar varieties. Therefore, we compared the pathogen resistance abilities of P. euphratica and P. alba var. pyramidalis by infecting the leaves of saplings of both with D. gregaria. Both poplar species are closely related species but show significant difference either in resistance to pathogens and abiotic stresses or morphology (Ma et al., 2013; Ma et al., 2018). The relative lesion area of P. euphratica leaves was significantly smaller than that of P. alba var. pyramidalis leaves (P < 0.05) (Figures 7A, D), indicating that the former had a greater ability to resist D. gregaria infection. The anthocyanin content in P. euphratica leaves was also significantly higher than that in P. alba var. pyramidalis leaves (P < 0.05) (Figures 7B, C), and the expression level of PalbHLH1 in P. euphratica (PeuGLABRA3-1) was higher than that in P. alba var. pyramidalis (P < 0.05) (Figure 7E). We also tested the expression level of the structural genes of anthocyanin pathway for P. euphratica and P. alba var. pyramidalis, such as F3H and DFR. The results showed that the expression level of these genes in P. euphratica was higher than that in P. alba var. pyramidalis (Figure 7E and Table S7 in Datasheet 1). However, the expression levels of PalMYB90 in the two species were lower than the limit of detection. In general, compared to P. alba var. pyramidalis, P. euphratica displayed higher resistance to D. gregaria.
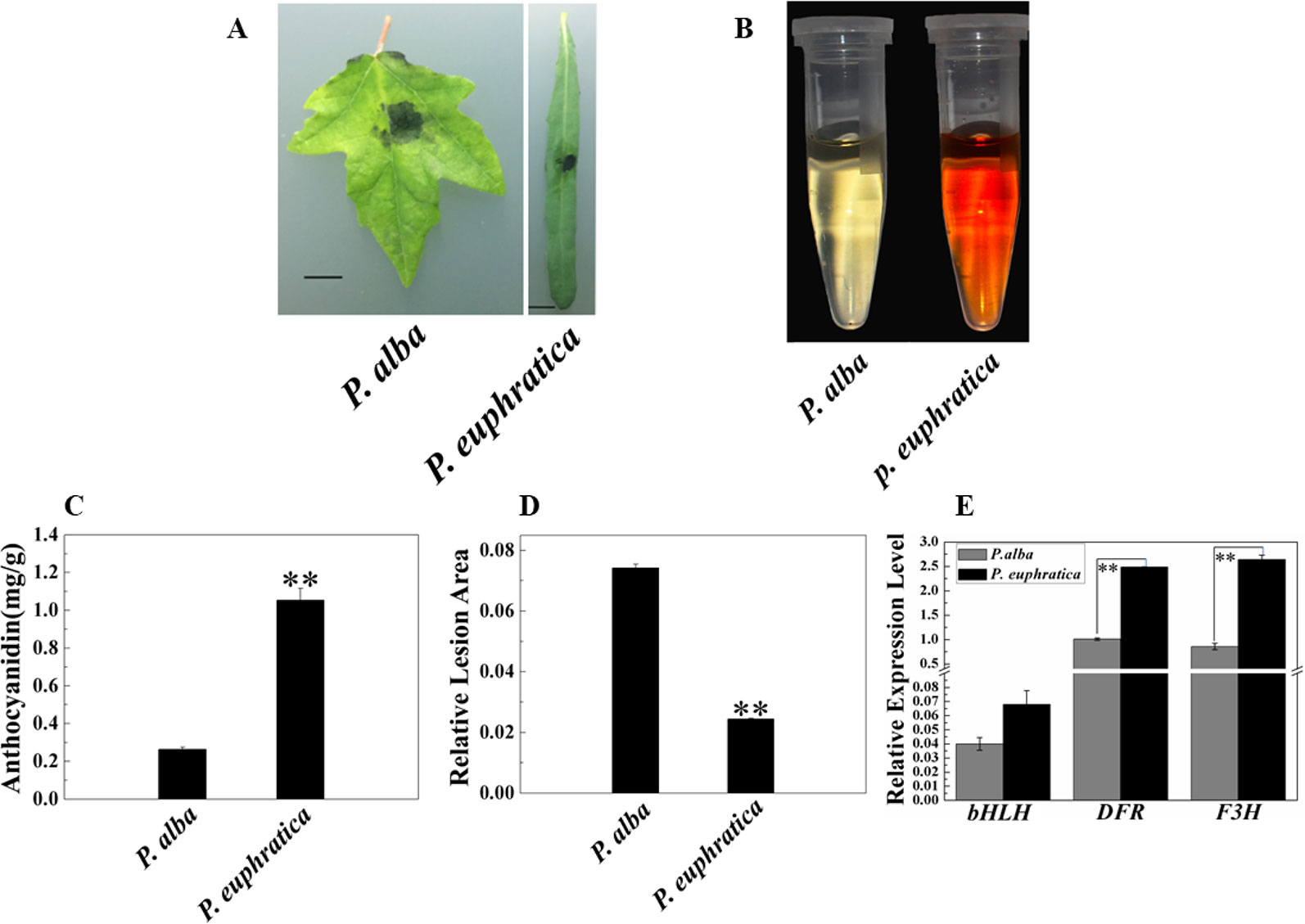
Figure 7 Relationships of anthocyanin content and responses to pathogen infection in P. alba var. pyramidalis and P. euphratica leaves. (A) Disease symptoms induced by D. gregaria on the leaves of two sister poplars, P. alba var. pyramidalis and P. euphratica. (B) Colors of anthocyanin extracted from P. alba var. pyramidalis and P. euphratica leaves. (C) Total anthocyanin contents in P. alba var. pyramidalis and P. euphratica leaves. (D) Relative lesion areas of the third leaves of P. alba var. pyramidalis and P. euphratica plants after D. gregaria infection for 7 days. (E) Relative expression levels of bHLH1, DFR, and F3H in P. alba var. pyramidalis and P. euphratica leaves. Error bars indicate the standard error (SE) of the average anthocyanin content, **P < 0.01.
Discussion
PalbHLH1 and PalMYB90 Overexpression Enhances Accumulation of Secondary Metabolites in the Flavonoid Pathway
Secondary metabolites in the flavonoid pathway are involved in responses to various stresses (Lepiniec et al., 2006), and their production is regulated by a series of transcription factors, including MYB, basic bHLH, and WD-repeat proteins (Baudry et al., 2004; Chezem and Clay, 2015), acting on related genes. Transgenic expression of anthocyanin-regulating transcription factors in ornamental plants leads to accumulation of anthocyanin products in other plants (Nishihara and Nakatsuka, 2011). For example, anthocyanin production was enhanced in tomato by coexpression of the transcription factors Del and Ros1 derived from snapdragon (A. majus) (Butelli et al., 2008; Maligeppagol et al., 2013). Del and Ros1 overexpression also increases expression of genes related to anthocyanin biosynthesis (Zhang et al., 2015; Ullah et al., 2017; Wang et al., 2017). These findings provide a plausible strategy and model for improving the disease resistance of plants to different pathogenic microorganisms. In this study, we would like to investigate whether over-expression of orthologous can improve disease resistance in poplar, and whether the resistant mechanisms are consistent with in that of in tomato. We overexpressed the two endogenous poplar genes involved in flavonoid metabolism and assessed disease resistance of the poplar species with the weakest resistance. Coexpression of PalbHLH1 and PalMYB90 from P. alba var. pyramidalis was driven by two 35S promoters in transgenic P. alba var. pyramidalis. Overexpression of PalbHLH1 and PalMYB90 not only promoted accumulation of secondary metabolites, such as anthocyanins and PAs, but also enhanced the contents of their intermediate products quercetin and kaempferol. PAs are effective molecules in chemical defense against foliar rust infection in poplar (Ullah et al., 2017). Depending on the level of anthocyanin accumulation, the total polyphenol, soluble PA, and insoluble PA contents increased with PalbHLH1 and PalMYB90 expression levels across transgenic poplar lines. From this perspective, the regulation of anthocyanin accumulation affects not only the colors of plant flowers and fruits but also the biosynthesis of secondary metabolic products in other tissues, such as leaves, roots, and stems, all contributing to improving plant resistance to pathogens (Zhang et al., 2013). This study has important implications for environmental restoration.
PalbHLH1 and PalMYB90 Overexpression Induces Activation of Regulatory Pathways Involved in Pathogen Resistance
Secondary metabolite accumulation in the flavonoid pathway contributes to improving plant pathogen resistance. For example, PA accumulation can reduce the growth of mycelia of fungi infecting poplar (Yuan et al., 2012; Ullah et al., 2017; Wang et al., 2017) or inhibit extracellular hydrolases of invading pathogens (Scalbert, 1991); when accumulated at high levels, anthocyanins act as ROS scavengers to reduce the oxidative bursts induced by pathogen infection (Yamasaki et al., 1996). Together, these factors decrease the damage caused by pathogen infection in plants. The synthesis of these metabolites is regulated by a series of complex pathways that are involved in plant growth, development, and responses to environmental stimuli (Butelli et al., 2008; Maligeppagol et al., 2013). Compared with tomato, Del and Ros (PalbHLH1 and PalMYB90) of poplar exhibit functional novelty. Transcriptome analysis showed that overexpression of PalbHLH1 and PalMYB90 promoted expression of many transcription factors associated with biosynthesis in the flavonoid pathway, which is involved in pathogen resistance. For example, MYB113, a homologous gene of MYB115, which encodes a member of the MYB transcription factor family, affects the expression of enzymes involved in later steps of anthocyanin biosynthesis, such as CHI1, CHS4, and DFR1, and leads to anthocyanin accumulation in potato (Liu et al., 2016) and PA accumulation in poplar (Wang et al., 2017). These findings suggest that MYB113 directly or indirectly regulates these genes.
Furthermore, pathogen infection induced expression of key genes in the flavonoid pathway. For example, after D. gregaria infection, Gene Ontology (GO) annotations of DEGs included mainly oxidoreductase activity, secondary metabolite processes, and intracellular parts. DFR is an important enzyme that catalyzes the conversion of dihydroflavonol into leucoanthocyanidin, which can be further transformed into anthocyanin and PA, respectively (Dixon et al., 2013). F3H and FLS, two important enzymes controlling the biosynthesis of the key intermediate products quercetin and kaempferol, were also induced by pathogen infection (Lepiniec et al., 2006), which led to more quercetin and kaempferol accumulation in transgenic lines than in WT plants. The genes encoding other enzymes involved in the anthocyanin and flavonoid pathways, such as leucoanthocyanidin dioxygenase, chalcone and stilbene synthase (PAYT005204.1), WRKY75, WD40 domain protein, glutathione S-transferase 19 (GST19), and flavin-dependent monooxygenase 1, were also upregulated. Among them, the WD40 protein activates early biosynthetic genes of the flavonoid pathway (e.g. F3H, DFR, ANS, ANR) as well as genes that are specific to the anthocyanins pathway (e.g. ANS and UFGT) (reviewed by Dixon et al., 2013). The expression levels of PalF3H, PalDFR, PalANS, PalANR, which function in the initial, middle, and final steps of anthocyanin and PA biosynthesis, respectively, were significantly upregulated in MYB90/bHLH1-OE poplar after D. gregaria infection.
After B. cinerea infection, only 17 genes were differentially expressed. One was the gene encoding glucosyl transferase 8, which is responsible for the biosynthesis of flavone O-glucosides in the flavonoid pathway and improves tolerance to UV radiation and drought (Peng et al., 2017). Compared with the DEGs from MYB90/bHLH1-OE poplar infected by D. gregaria, the related genes after infection by B. cinerea expressed no significantly difference in transcriptome analysis. Thus, we performed qRT-PCR to verify expression levels of the DEGs regulated by two pathogen infection (Figure 6). The results showed that expression of most of the DEGs were different significantly and positively correlated with the results of the phenotype (Figure 4). These results suggest that the MYB90/bHLH1-OE lines responded differently to B. cinerea and D. gregaria infection. Moreover, expression of genes encoding some antioxidants or antioxidases, such as SOD, POD, and laccase/diphenol oxidase, was enhanced by PalbHLH1/PalMYB90 overexpression. These results indicate that the accumulation of secondary metabolites, especially anthocyanins, is likely associated with higher antioxidant activities (Fini et al., 2011; Qiu et al., 2014). Interestingly, the content of H2O2 also increased significantly in the transgenic lines; however, because the content of H2O2 itself was low, the final H2O2 content was low even though the content increased significantly. Therefore, we speculate that the content of H2O2 is not only affected by flavonoids and the activities of antioxidant enzymes but may also be related to overexpression of PalMYB90 and PalbHLH1. Indeed, overexpression of MYB90 and bHLH1 genes is a type of stress itself when compared with WT. It has been shown that lower levels of H2O2 play an important role in plant defense, and we suggest that lower levels of H2O2 in transgenic plants also contribute to plant defense against pathogens.
PalbHLH1 Expression Improves Poplar Resistance to Pathogen Infection
Metabolic responses to specific stressors are conserved among flowering plants. For example, the pathways of antimicrobial compound biosynthesis in response to pathogen infection are conserved (Mansfield, 2000; Dixon, 2001). Thus, comparing stress responses among closely related species may help in the elucidation of candidate genes involved in specific stress responses and the formation of adaptive traits. In this study, we compared the expression levels of bHLH1, F3H, and DFR and the anthocyanin content in leaves infected with D. gregaria between P. euphratica and P. alba var. pyramidalis. We found that both bHLH1, F3H, and DFR expression and anthocyanin accumulation were induced more rapidly in P. euphratica than in P. alba var. pyramidalis. In addition, the relative lesion area in P. euphratica leaves was smaller than that in P. alba var. pyramidalis leaves, indicating that the higher anthocyanin accumulation in P. euphratica leaves, positively regulated by bHLH1 might contribute to improve its resistance to D. gregaria. P. euphratica prefers to grow in arid or semiarid environments and has high resistance to drought and salinity (Zhang et al., 2008; Ma et al., 2013). Therefore, high PalbHLH1 expression levels in combination with anthocyanin accumulation in P. euphratica should enhance its tolerance to severe conditions (Fini et al., 2011; Nakabayashi et al., 2014; Naing et al., 2017).
Application of PalbHLH1/PalMYB90 Overexpression to Improve Pathogen Resistance in Poplar
P. alba var. pyramidalis has been widely cultivated for urban afforestation and ecological restoration from northwest to northern China because of its advantageous traits of rapid growth, lack of seed catkins, erect stems, and high biomass production (Xu, 1988; Zhang et al., 2008; Xu et al., 2011; Ma et al., 2018). Nonetheless, poplar plants, particularly P. alba var. pyramidalis, are easily infected by numerous pathogens, such as B. cinerea, D. gregaria, and Melampsora larici-populina (Miranda et al., 2007; Yuan et al., 2012). Thus, breeding poplars for disease resistance is becoming increasingly urgent and necessary. In this study, we found that overexpression of PalbHLH1 and PalMYB90 enhanced the accumulation of secondary metabolites, resulting in improved resistance to bacterial/fungal pathogen infection. Our study provides an efficient genetic engineering strategy for increasing the accumulation of secondary metabolic products in transgenic poplar plants to enhance their capacity of pathogen resistance.
Conclusion
In summary, Overexpression of PalbHLH1/PalMYB90 led to enhanced secondary metabolic accumulation and antioxidase activities, which all contribute to enhance the resistance of poplars to pathogen infection.
Data Availability Statement
The datasets generated for this study can be found in the Genome Sequence Archive website (http://bigd.big.ac.cn/) under accession number CRA001044.
Author Contributions
DW supervised the project. QB and BD performed the experiments. QB, JM and YF participated in analyzing the transcript data. SS, YF, QL and JL provided assistance in the experiments and transcript analysis. All authors read and approved the final manuscript.
Funding
The research was supported by the National Science Foundation of China (No. 31870580) and the Fundamental Research Funds for the Central Universities (lzujbky-2017-k14).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the Core Facility of School of Life Sciences, Lanzhou University, for providing us with qRT-PCR facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01772/full#supplementary-material
Figure S1 | Validation of agarose gel electrophoresis analysis of positive plants using the marker gene Hyg.
Figure S2 | Verification of PalbHLH1 and PalMYB90 gene expression in positive plants. (A) Expression level of the PalMYB90 gene. (B) Expression level of the PalbHLH1 gene, E=1.07. (C) Heights of poplar plants. (D) Length-to-width ratios of leaves. (E) Weights of poplar plants. Error bars indicate the standard deviations of five independent experiments.
Figure S3 | Gene expression profiles before and after infection by two pathogens. (A) Expression profiles of PalbHLH1 before and after infection by two pathogens. (B) Expression profiles of PalMYB90 before and after infection by two pathogens. (C) and (D) Transcriptome analysis of WT and transgenic poplar. (E) Expression profiles of differentially expressed genes (DEGs) of WT and transgenic poplar. (F) DEG analysis of plants infected by pathogen compared with WT and transgenic poplar.
Figure S4 | Overexpression of PalbHLH1 and PalMYB90 causes global transcriptional reprogramming in transgenic poplar. (A) Differential expression between MYB90/bHLH1-OE and WT plants after D. gregaria infection. (B) and (C) Transcriptome analysis of DEGs after D. gregaria infection. (D) Differential expression between MYB90/bHLH1-OE and WT plants after B. cinerea infection. (E) and (F) Transcriptome analysis of DEGs after B. cinerea infection.
Figure S5 | Graphs displaying five standard curves. (A) Standard curve for determination of anthocyanin content. (B) Standard curve for determination of quercetin content. (C) Standard curve for determination of kaempferol content. (D) Standard curve for determination of total phenol content. (E) Standard curve for determination of tanin content.
Datasheets 1 | Related gene accession numbers and primers for certain PCR and qPCR.
Datasheets 2 | The repeated results using Deseq2 and edegR for down-regulated DEGs screening. In the data, WT group gene expression was down-regulated compared to OE group.
Datasheets 3 | Read mapping results of WT, MYB90/bHLH1-OE line, WT infested by B. cinerea (WT-B. cinerea), WT infested by D. gregaria (WT-D. gregaria), MYB90/bHLH1-OE line infested by B. cinerea (OE-B. cinerea) and MYB90/bHLH1-OE line infested by D. gregaria (OE-D. gregaria).
References
Agati, G., Biricolti, S., Guidi, L., Ferrini, F., Fini, A., Tattini, M. (2011). The biosynthesis of flavonoids is enhanced similarly by UV radiation and root zone salinity in L. vulgare leaves. J. Plant Physiol. 168, 204–212. doi: 10.1016/j.jplph.2010.07.016
Baker, C. J., Orlandi, E. W. (1995). Active oxygen in plant pathogen. Annu. Rev. Phytopathol. 33 (1), 299–321. doi: 10.1146/annurev.py.33.090195.001503
Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B., Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39 (3), 366–380. doi: 10.1111/j.1365-313X.2004.02138.x
Butelli, E., Titta, L., Giorgio, M., Mock, H. P., Matros, A., Peterek, S., et al. (2008). Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308. doi: 10.1038/nbt.1506
Chang, S., Puryear, J., Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11 (2), 113–116. doi: 10.1007/BF02670468
Chezem, W. R., Clay, N. K. (2015). Regulatory and Biosynthetic Mechanisms Underlying Plant Chemical Defense Responses to Biotic Stresses. Mol. Mech. Plant Adaptation 5, 117–146. doi: 10.1002/9781118860526
Cho, J. S., Nguyen, V. P., Jeon, H. W., Kim, M. H., Eom, S. H., Lim, Y. J., et al. (2016). Overexpression of PtrMYB119, a R2R3-MYB transcription factor from Populus trichocarpa, promotes anthocyanin production in hybrid poplar. Tree Physiol. 36, 1162–1176. doi: 10.1093/treephys/tpw046
Cui, B., Pan, Q., Clarke, D., Villarreal, M. O., Umbreen, S., Yuan, B., et al. (2018). S-nitrosylation of the zinc finger protein SRG1 regulates plant immunity. Nat. Commun. 9 (1), 4226. doi: 10.1038/s41467-018-06578-3
Dixon, R. A., Xie, D. Y., Sharma, S. B. (2005). Proanthocyanidins: a final frontier in flavonoid research? New Phytol. 165, 9–28. doi: 10.1111/j.1469-8137.2004.01217.x
Dixon, R. A., Liu, C., Jun, J. H. (2013). Metabolic engineering of anthocyanins and condensed tannins in plants. Curr. Opin. Biotechnol. 24, 329–335. doi: 10.1016/j.copbio.2012.07.004
Dixon, R. A. (2001). Natural products and plant disease resistance. Nature 411 (6839), 843. doi: 10.1038/35081178
Fini, A., Brunetti, C., Di Ferdinando, M., Ferrini, F., Tattini, M. (2011). Stress-induced flavonoid biosynthesis and the antioxidant machinery of plants. Plant Signal. Behav. 6, 709–711. doi: 10.4161/psb.6.5.15069
Ginestet, C. (2011). ggplot2: elegant graphics for data analysis. J. Royal Stat. Soc.: Series A (Statistics Soc.) 174 (1), 245–246. doi: 10.1111/j.1467-985X.2010.00676_9.x
Gould, K. S. (2004). Nature’s Swiss Army Knife: The Diverse Protective Roles of Anthocyanins in Leaves. Biomed Res. Int. 2004 (5), 314–320. doi: 10.1155/S1110724304406147
Heim, M. A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., Bailey, P. C. (2003). The basic helix–loop–helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20 (5), 735–747. doi: 10.1093/molbev/msg088
Hichri, I., Barrieu, F., Bogs, J., Kappel, C., Delrot, S., Lauvergeat, V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62 (8), 2465–2483. doi: 10.1093/jxb/erq442
Huang, Y., Liu, H., Jia, Z., Fang, Q., Luo, K. (2012). Combined expression of antimicrobial genes (Bbchit1 and LJAMP2) in transgenic poplar enhances resistance to fungal pathogens. Tree physiol. 32 (10), 1313–1320. doi: 10.1093/treephys/tps079
Huang, W., Sun, W., Lv, H., Luo, M., Zeng, S., Pattanaik, S., et al. (2013). A R2R3-MYB transcription factor from Epimedium sagittatum regulates the flavonoid biosynthetic pathway. PloS one 8, e70778. doi: 10.1371/journal.pone.0070778
Jia, Z., Gou, J., Sun, Y., Yuan, L., Tang, Q., Yang, X., et al. (2010). Enhanced resistance to fungal pathogens in transgenic Populus tomentosa Carr. by overexpression of an nsLTP-like antimicrobial protein gene from motherwort (Leonurus japonicus). Tree Physiol 30 (12), 1599–1605. doi: 10.1093/treephys/tpq093
Karim, A., Jiang, Y., Guo, L., Ling, Z., Ye, S., Duan, Y., et al. (2015). Isolation and characterization of a subgroup IIa WRKY transcription factor PtrWRKY40 from Populus trichocarpa. Tree Physiol. 35, 1129–1139. doi: 10.1093/treephys/tpv084
Koes, R., Verweij, W., Quattrocchio, F. (2005). Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10 (5), 236–242. doi: 10.1016/j.tplants.2005.03.002
Lepiniec, L., Debeaujon, I., Routaboul, J. M., Baudry, A., Pourcel, L., Nesi, N., et al. (2006). Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57, 405–430. doi: 10.1146/annurev.arplant.57.032905.105252
Li, Y. G., Tanner, G., Larkin, P. (1996). The DMACA-HCl protocol and the threshold proanthocyanidin content for bloat safety in forage legumes. J. Sci. Food Agr. 70, 89–101. doi: 10.1002/(SICI)1097-0010
Li, H. B., Cheng, K. W., Wong, C. C., Fan, K. W., Chen, F., Jiang, Y. (2007). Evaluation of antioxidant capacity and total phenolic content of different fractions of selected microalgae. Food chem. 102 (3), 771–776. doi: 10.1016/j.foodchem.2006.06.022
Liu, Y., Lin-Wang, K., Espley, R. V., Wang, L., Yang, H., Yu, B., et al. (2016). Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 67, 8) 2159–2176. doi: 10.1093/jxb/erw014
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression datausing real-time quantitative PCR and the 2(-[△] [△] C(T)) method. Methods 25 (4), 402–408. doi: 10.1006/meth.2001.1262
Lloyd, G., McCown, B. (1980). Commercially feasible micropropagation of mountain laural (Kalmla latlfolia) by use of shoot tip cultures. Comb. Proc. Int. Plant. Prop. Soc. 30, 421c. 3.
Lorenc-Kukuła, K., Jafra, S., Oszmiański, J., Szopa, J. (2005). Ectopic expression of anthocyanin 5-o-glucosyltransferase in potato tuber causes increased resistance to bacteria. J. Agr. Food Chem. 53, 272–281. doi: 10.1021/jf048449p
Ma, T., Wang, J., Zhou, G., Yue, Z., Hu, Q., Chen, Y., et al. (2013). Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 4, 2797. doi: 10.1038/ncomms3797
Ma, J., Wan, D., Duan, B., Bai, X., Bai, Q., Chen, N., et al. (2018). Genome sequence and genetic transformation of a widely distributed and cultivated poplar. Plant Biotechnol. J. 17 (2), 451–460. doi: 10.1111/pbi.12989
Maligeppagol, M., Chandra, G. S., Navale, P. M., Deepa, H., Rajeev, P. R., Asokan, R., et al. (2013). Anthocyanin enrichment of tomato (Solanumly copersicum L.) fruit by metabolic engineering. Curr. Sci. 105, 72–80.
Mansfield, J. W. (2000). “Antimicrobial compounds and resistance,” in Mechanisms of resistance to plant diseases (Dordrecht: Springer), 325–370. doi: 10.1007/978-94-011-3937-3_10
Marchant, A., Mougel, F., Almeida, C., Jacquin-Joly, E., Costa, J., Harry, M. (2015). De novo transcriptome assembly for a non-model species, the blood-sucking bug Triatoma brasiliensis, a vector of Chagas disease. Genetica 143 (2), 225–239. doi: 10.1007/s10709-014-9790-5
Miranda, M., Ralph, S. G., Mellway, R., White, R., Heath, M. C., Bohlmann, J., et al. (2007). The transcriptional response of hybrid poplar (Populus trichocarpa × P. deltoides) to infection by Melampsora medusae leaf rust involves induction of flavonoid pathway genes leading to the accumulation of proanthocyanidins. Mol. Plant. Microbe In. 20 (7), 816–831. doi: 10.1094/MPMI-20-7-0816
Naing, A. H., Park, K. I., Ai, T. N., Chung, M. Y., Han, J. S., Kang, Y. W., et al. (2017). Overexpression of snapdragon Delila (Del) gene in tobacco enhances anthocyanin accumulation and abiotic stress tolerance. BMC Plant Biol. 17, 65. doi: 10.1186/s12870-017-1015-5
Nakabayashi, R., Yonekura-Sakakibara, K., Urano, K., Suzuki, M., Yamada, Y., Nishizawa, T., et al. (2014). Enhancement of oxidative and drought tolerance in Arabidopsis by over accumulation of antioxidant flavonoids. Plant J. 77 (3), 367–379. doi: 10.1111/tpj.12388
Newcombe, G., Bradshaw, H. D., Jr. (1996). Quantitative trait loci conferring resistance in hybrid poplar to Septoria populicola, the cause of leaf spot. Can. J. Forest Res. 26 (11), 1943–1950. doi: 10.1139/x26-219
Newcombe, G., Ostry, M. (2001). Recessive resistance to Septoria stem canker of hybrid poplar. Phytopathology 91, 1081–1084. doi: 10.1094/PHYTO.2001.91.11.1081
Nishihara, M., Nakatsuka, T. (2011). Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol. Lett. 33 (3), 433–441. doi: 10.1007/s10529-010-0461-z
Ostry, M. E., McNabb, H. S., Jr. (1985). Susceptibility of Populus species and hybrids to disease in the north central United States. Plant Dis. 69, 755–757. doi: 10.1094/PD-69-755
Paolocci, F., Robbins, M. P., Madeo, L., Arcioni, S., Martens, S., Damiani, F. (2007). Ectopic expression of a basic helix-loop-helix gene transactivates parallel pathways of proanthocyanidin biosynthesis. Structure, expression analysis, and genetic control of leucoanthocyanidin 4-reductase and anthocyanidin reductase genes in Lotus corniculatus. Plant Physiol. 143 (1), 504–516. doi: 10.1104/pp.106.090886
Peng, M., Shahzad, R., Gul, A., Subthain, H., Shen, S., Lei, L., et al. (2017). Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. Nat. Commun. 8, 1975. doi: 10.1038/s41467-017-02168-x
Polle, A., Janz, D., Teichmann, T., Lipka, V. (2013). Poplar genetic engineering: promoting desirable wood characteristics and pest resistance. Appl. Microbiol. Biot. 97 (13), 5669–5679. doi: 10.1007/s00253-013-4940-8
Qiu, Z. B., Wang, Y. F., Zhu, A. J., Peng, F. L., Wang, L. S. (2014). Exogenous sucrose can enhance tolerance of Arabidopsis thaliana seedlings to salt stress. Bio. Plant 58, 611–617. doi: 10.1007/s10535-014-0444-3
Qu, C. P., Xu, Z. R., Hu, Y. B., Lu, Y., Yang, C. J., Sun, G. Y., et al. (2016). RNA-SEQ reveals transcriptional level changes of poplar roots in different forms of nitrogen treatments. Front. Plant Sci. 7, 51. doi: 10.3389/fpls.2016.00051
Scalbert, A. (1991). Antimicrobial properties of tannins. Phytochemistry 30, 3875–3883. doi: 10.1016/0031-9422(91)83426-L
Singleton, V. L., Rossi, J. A. (1965). Colorimetry of total phenolics with phosphomolybdic- phosphotungstic acid reagents. Am. J. Enol. Viticult. 16 (3), 144–158.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011). MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 28 (10), 2731–2739. doi: 10.1093/molbev/msr121
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7 (3), 562–578. doi: 10.1038/nprot.2012.016
Tsai, C. J., Harding, S. A., Tschaplinski, T. J., Lindroth, R. L., Yuan, Y. (2006). Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol. 172, 47–62. doi: 10.1111/j.1469-8137.2006.01798.x
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Sci. 313 (5793), 1596–1604. doi: 10.1126/science.1128691
Ullah, C., Unsicker, S. B., Fellenberg, C., Constabel, C. P., Schmidt, A., Gershenzon, J., et al. (2017). Flavan-3-ols are an Effective Chemical Defense against Rust infection. Plant Physiol. 175 (4), 1560–1578. doi: 10.1104/pp.17.00842
Wang, L., Jiang, Y., Yuan, L., Lu, W., Yang, L., Karim, A., et al. (2013). Isolation and characterization of cDNAs encoding leucoanthocyanidin reductase and anthocyanidin reductase from Populus trichocarpa. PloS One 8 (5), e64664. doi: 10.1371/journal.pone.0064664
Wang, N., Xu, H., Jiang, S., Zhang, Z., Lu, N., Qiu, H., et al. (2017). MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90 (2), 276–292. doi: 10.1111/tpj.13487
Winkel-Shirley, B. (2002). Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 5, 218–223. doi: 10.1016/S1369-5266(02)00256-X
Xu, X., Tong, L., Li, F., Kang, S., Qu, Y. (2011). Sap flow of irrigated Populus alba var. pyramidalis and its relationship with environmental factors and leaf area index in an arid region of Northwest China. J. Forest Res. 16, 144–152. doi: 10.1007/s10310-010-0220-y
Yamasaki, H., Uefuji, H., Sakihama, Y. (1996). Bleaching of the red anthocyanin induced by superoxide radical. Arch. Biochem. Biophys. 332, 183–186. doi: 10.1006/abbi.1996.0331
Yu, K., Jun, J. H., Duan, C., Dixon, R. A. (2019). VvLAR1 and VvLAR2 Are Bifunctional Enzymes for Proanthocyanidin Biosynthesis in Grapevine. Plant physiol. 180 (3), 1362–1374. doi: 10.1104/pp.19.00447
Yuan, L., Wang, L., Han, Z., Jiang, Y., Zhao, L., Liu, H., et al. (2012). Molecular cloning and characterization of PtrLAR3, a gene encoding leucoanthocyanidin reductase from Populus trichocarpa, and its constitutive expression enhances fungal resistance in transgenic plants. J. Exp. Bot. 63 (7), 2513–2524. doi: 10.1093/jxb/err425
Zhang, Z. H., Kang, X. Y., Li, D. L., Chen, H. W. (2008). Pollen development and multi-nucleate microspores of Populus bolleana Lauche. Forest. Stud. China 10 (2), 107–111. doi: 10.1007/s11632-008-0027-5
Zhang, Y., Butelli, E., De Stefano, R., Schoonbeek, H. J., Magusin, A., Pagliarani, C., et al. (2013). Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr. Biol. 23, 1094–1100. doi: 10.1016/j.cub.2013.04.072
Zhang, Y., Butelli, E., Alseekh, S., Tohge, T., Rallapalli, G., Luo, J., et al. (2015). Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 6, 8635. doi: 10.1038/ncomms9635
Keywords: PalbHLH1 and PalMYB90, Populus alba var. pyramidalis, flavonoid, Dothiorella gregaria Sacc, Botrytis cinerea
Citation: Bai Q, Duan B, Ma J, Fen Y, Sun S, Long Q, Lv J and Wan D (2020) Coexpression of PalbHLH1 and PalMYB90 Genes From Populus alba Enhances Pathogen Resistance in Poplar by Increasing the Flavonoid Content. Front. Plant Sci. 10:1772. doi: 10.3389/fpls.2019.01772
Received: 11 June 2019; Accepted: 18 December 2019;
Published: 26 February 2020.
Edited by:
Jens Staal, Ghent University, BelgiumReviewed by:
Chaofeng Li, The University of Tokyo, JapanHui Zhou, Anhui Academy of Agricultural Sciences, China
Copyright © 2020 Bai, Duan, Ma, Fen, Sun, Long, Lv and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongshi Wan, wandsh@lzu.edu.cn
†These authors have contributed equally to this work
 Qiuxian Bai
Qiuxian Bai Bingbing Duan†
Bingbing Duan† Qiming Long
Qiming Long Dongshi Wan
Dongshi Wan