- 1Beijing Advanced Innovation Center for Tree Breeding by Molecular Design, Beijing Forestry University, Beijing, China
- 2College of Biological Sciences and Biotechnology, Beijing Forestry University, Beijing, China
- 3College of Horticulture and Plant Protection, Yangzhou University, Yangzhou, China
Extensive studies have been carried out on chloroplast gene expression and chloroplast development; however, the regulatory mechanism is still largely unknown. Here, we characterized Pigment-Defective Mutant4 (PDM4), a P-type PPR protein localized in chloroplast. The pdm4 mutant showed seedling-lethal and albino phenotype under heterotrophic growth conditions. Transmission electron microscopic analysis revealed that thylakoid structure was totally disrupted in pdm4 mutant and eventually led to the breakdown of chloroplasts. The levels of several chloroplast- and nuclear-encoded proteins are strongly reduced in pdm4 mutant. Besides, transcript profile analysis detected that, in pdm4 mutant, the expression of plastid-encoded RNA polymerase-dependent genes was markedly affected, and deviant chloroplast rRNA processing was also observed. In addition, we found that PDM4 functions in the splicing of group II introns and may also be involved in the assembly of the 50S ribosomal particle. Our results demonstrate that PDM4 plays an important role in chloroplast gene expression and chloroplast development in Arabidopsis.
Introduction
Chloroplasts are known for providing energy and carbon resource to the plant cell and are also indispensable for plant development and growth (Bryant et al., 2011). Derived from cyanobacterial ancestors, the chloroplasts belong to semi-autonomous organelles which possess their own genomes. Over the last billion years, the chloroplast genome has lost numerous genes in higher plants and generally remains about 120 genes which encode primary components of translation, transcription, and photosynthesis apparatus, as well as contains some critical biogenesis-related genes such as accD, clpP1, matK, ycf1, and ycf2 (Sato et al., 1999; Leister, 2003; Ouyang et al., 2017). Although the chloroplast genome is small and with limited coding information, the transcriptional process is much more complex than that of prokaryotes which are usually organized in polycistronic transcriptional units. In particular, RNA processing from polycistronic precursors and editing are strikingly different between chloroplast and prokaryotes (Sugita and Sugiura, 1996; Sato et al., 1999).
Generally, in higher plant, two RNA polymerases with different origins participate in the transcription of plastid genes, including a plastid-encoded RNA polymerase (PEP) and nuclear-encoded RNA polymerases (Hajdukiewicz et al., 1997; Liere and Börner, 2007; NEP). During chloroplast development, the plastidic genetic system is first established within the proplastids. During this stage, the genes of PEP components and related ribosomal proteins were transcribed by NEP that is critical for the nascent construction of the plastid-genetic background. As a result, the activity of the transcriptional apparatus in the proplastid is remarkably raised. At the second stage, the well-assembled and functional PEP starts to transcribe plastid-encoded genes. Meanwhile, the photosynthetic proteins which are encoded by nuclear genes exhibit a high expression level and eventually lead to the establishment of photosynthesis systems (Mullet, 1993; Majeran et al., 2010; Tiller and Bock, 2014). PEP activity is also essential for the fully active chloroplasts formation because it promotes the expression of photosynthesis-related genes (Pfalz and Pfannschmidt, 2013). Subunits of the PEP core are present in two plastid protein preparations; one is associated with thylakoid and envelope membranes, and these are protein:DNA-complexes termed transcriptionally active chromosomes (TACs) (Krause and Krupinska, 2000; Pfalz et al., 2006; Krupinska et al., 2012). So far, the richest protein data set resulted from protein mass spectrometry analysis of isolated pTACs from Arabidopsis (Arabidopsis thaliana) and mustard (Sinapis alba), in which 35 proteins were identified (Pfalz et al., 2006). Eighteen of these proteins were denoted pTAC proteins, and three of them (pTAC2, -6, and -12) were shown to be required for plastid gene expression (Pfalz et al., 2006). Besides, PEP forms a complex with PEP-associated proteins (PAPs), and the Arabidopsis thaliana nuclear genome contains at least 12 PAP genes (Yu et al., 2014), and all PAPs have also been identified in the nucleoid or TAC proteomes (Pfalz et al., 2006; Majeran et al., 2012; Melonek et al., 2016).
The complexity of the chloroplast gene expression system is also highly regulated at the post-transcriptional which mainly reflects the extensive modifications exerted on transcripts during RNA processing (Chi et al., 2015). For instance, defects in endonucleolytic cleavage polycistronic transcripts would result in a blocking translation in chloroplast mRNAs (Sugiura et al., 1998; Walter et al., 2010). And critical sites of chloroplast RNAs that are essential for chloroplasts development in higher plant can be correctly spliced or edited (Bobik et al., 2017; Du et al., 2017; Zhang J. et al., 2017).
Chloroplast RNA metabolism refers to a substantial number of RNA-binding proteins (Stern et al., 2010). Due to the limited coding capacity of the chloroplast genome, the chloroplast gene expression is controlled both by plastid-encoded and nucleus-encoded proteins (Germain et al., 2013; Stoppel and Meurer, 2013; Belcher et al., 2015). As nucleus-encoded factors, it has been demonstrated that the pentatricopeptide repeat (PPR) proteins participate in chloroplast gene expression and function (Barkan and Small, 2014). Members of the PPR protein family are considerably numerous in land plants with up to 450 representatives in Arabidopsis (Lurin et al., 2004), and this family is characterized by PPR repeat with highly degenerate unit of 35 amino acids (Lurin et al., 2004). In addition, according to the variation in length and amino acid composition of PPR repeats, the PPR proteins have been used to define two categories; P-class PPR proteins are mainly composed of typical 35-amino acid sequence repeats, and the PLS-class members contain triplets of motifs by varied amino acid lengths and sometimes with an additional C-terminal domain (Lurin et al., 2004; Cheng et al., 2016).
The PPR proteins can directly bind to chloroplast RNAs and prevent targeted RNAs from RNase degradation and/or facilitate or directly participate in related processing (Small and Peeters, 2000; Pogson and Albrecht, 2011). It is widely accepted that the PLS subgroup proteins are mostly involved in RNA editing, whereas the P subgroup proteins play crucial roles in intron splicing, RNA stabilization, and translation process (Barkan and Small, 2014). For example, either PPR protein DYW2 or NUWA can be interacted with CLB19, function in editing of organelle RNA (Guillaumot et al., 2017). Seedling Lethal1 (SEL1/PDM1), a PPR protein was proved to participate in the plastid gene expression and chloroplast development at an early stage (Pyo et al., 2013). Through specifically recognizing RNA sequence of 23S–4.5S rRNA precursor, PPR protein SOT1 performs the endonucleolytic activity during the maturation of 23S and 4.5S rRNA in chloroplast (Zhou et al., 2017). SOT5/EMB2279 is involved in intron splicing of plastid rpl2, and PpPPR_66 acts as a processing factor to assist ndhA pre-mRNA splicing by bounding preferentially to the specific region (Huang et al., 2018; Ito et al., 2018). Besides, some PPR proteins are proved to be required for the accumulation/assembly of plastid ribosomes (Williams and Barkan, 2004). Synthesis of the rRNAs and proteins, with correct folding, maturation/modification, and further assembly into functional particles, are highly coordinated. The 30S subunit includes 16S rRNA and about 20 ribosomal proteins, while the 50S subunit consists of 23S, 5S, and 4.5S rRNAs and about 30 ribosomal proteins (Yamaguchi et al., 2003).
In this study, we report the pdm4 mutant of Arabidopsis, which was generated by T-DNA insertion and screened as defective pigment and seedling-lethal phenotype. The corresponding gene, PDM4, encodes a putative chloroplast PPR protein. Subsequent genetic and molecular analyses suggest that PDM4 is involved in the regulation of plastid gene expression and chloroplast development.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana Columbia (Col-0) ecotype and the mutant pdm4 (SALK_034168, obtained from the Arabidopsis Biological Resource Center; ecotype Columbia) were used in all experiments. The seeds were sterilized by 0.8% NaClO for 10 min, followed by 5 times rinsing with sterile double-distilled water. Then they were plated on 1/2 MS medium with 1% sucrose and 0.8% agar (pH 6.0) at 4°C in the dark for 48 h after sterilizing. Next, they were transferred to long-day conditions (16 h light, 8 h dark) at 22°C. The T-DNA insertion was confirmed by PCR analysis and subsequent sequencing with the primers LBb1.3 (5′-ATTTTGCCGATTTCGGAAC-3′) and pdm4-RP. The homozygous pdm4 mutant line was verified by PCR using specific primers pdm4-LP 5′-TCACTAACCAATAACACCACC-3′ and pdm4-RP 5′-ATTGCTTGTGAGCCTTGGT-3′.
Total RNA Isolation and Reverse Transcription (RT)-PCR; Quantitative Real-Time PCR (qRT-PCR) Analysis
Three total RNA samples were extracted from light grown 3-week-old mutant and wild-type plants with RNeasy Mini Kit (Tiangen Biotech Company, Beijing, China). For RT-PCR analysis, first strand cDNA was synthesized by the one-step reverse-transcription system (TransGen Biotech, Beijing, China), and the operation was carried out based on previous protocol (Zhang J. et al., 2017). The qRT-PCR was performed, and the primers used in this analysis were according to Du et al. (2017). All the measurement for each sample was repeated three times.
Measurement of Chlorophyll
For measuring the chlorophyll content, leaves from 3-week-old Arabidopsis seedlings were collected. One hundred milligram of leaves was ground in liquid nitrogen into fine powder and mixed thoroughly with 250 μl 80% acetone and quantified on a UV2800 spectrophotometer (Unico Instruments Co., Ltd, USA). We calculated the chlorophyll content from the absorbance following the method of Arnon (1949). Three biological replicates, each with three repeats, were analyzed for each sample.
Optical and Transmission Electron Microscopy
To observe the development of the embryos between wild-type and the mutant plants, the seeds of heterozygous pdm4 mutant line were removed from siliques and totally cleared in Hoyer’s buffer (chloral hydrate, 50 g; gum Arabic, 3.75 g; and glycerol, 2.5 ml were dissolved in 15 ml of water) according to Meinke et al. (1994). The individual embryo in the cleared seed was examined by light microscopy (Zeiss, Axioskop, Germany) using differential interference contrast (DIC; Du et al., 2017). For transmission electron microscopy (TEM) analysis, the samples were first cut into ultra-thin pieces (LKB-8800, LKB, Broma, Sweden) and stained with alkaline lead citrate and uranyl acetate and then examined with a transmission electron microscope (JEM 1200EX, JEOL, Japan).
Northern-Blot Analysis
For northern-blot analysis, total RNA from wild-type and pdm4 seedlings was extracted and determined by using thermo NanoDrop 2000 (Thermo, USA). Three equal content RNA samples of the wild type and pdm4 mutant were separated on 1.3% (w/v) agarose-formaldehyde gels and subsequently blotted to a nylon membrane. Next, the membrane was hybridized with a specific probe labeled with 32P. The labeled probes were obtained by using the Prime-a-Gene Labeling Kit (SGMB01-Promega-U1100, USA). The sequences of the primers were according to Du et al. (2017). All the analysis was performed at least in three independent repeats.
Subcellular Localization of Green Fluorescent Protein
In order to study the subcellular localization of PDM4, two-week-old complemented Arabidopsis seedlings (COM) were cut into small pieces and incubated in a solution (10 mM MES, 20 mM CaCl2, 0.5 M mannitol, pH 5.8, containing 0.1 g ml−1 macerozyme and 0.1 g ml−1 cellulase) for 4 h at room temperature in the dark. Protoplasts were then isolated according to Dovzhenko et al. (2003) and assessed for fusion gene expression with a confocal microscope (TCS SP5 CLSM; Leica). The signal of green fluorescence was detected, and red fluorescence represents the auto-fluorescence of chlorophyll.
Protein Extraction and Western Blot Analyses
The total proteins were extracted from 3-week-old wild type and pdm4 mutant with NB1 buffer (pH 8.0), including 1 mM MgCl2, 5 mM DTT, 0.5 M sucrose, 50 mM Tris MES, 10 mM EDTA, and protease inhibitor cocktail. The different protein samples were separated by 15% SDS-page and then transferred onto PVDF membranes. And then, membranes were incubated with specific primary antibodies. The antibodies were used in this study according to Xiao et al. (2012). Signals were detected using enhanced chemiluminescence method (Du et al., 2017), and signal intensity of protein band was analyzed by “ImageJ” software. These experiments were repeated at least three times independently.
Chloroplast Isolation and Fractionation
Chloroplast isolation method was performed as described previously with minor modifications (Aronsson and Jarvis, 2002). Exactly, 21-day-old Arabidopsis plants were ground in extraction buffer (5 mM EGTA, 50 mM HEPES-KOH pH 8.05, 0.33 M sorbitol, 5 mM EDTA, 10 mM NaHCO3, and 5 mM MgCl2). After filtering through Miracloth, the sample was centrifuged for 1 min at 1,000 g. The supernatants were removed, and pellets were re-suspended and then loaded onto Percoll gradients (70 and 40% in isolated buffer respectively); then intact chloroplasts were collected and washed three times with washing buffer (3 mM MgSO4, 0.33 M sorbitol, and 50 mM HEPES-KOH, pH 8.0). Chloroplasts were fractionated into the thylakoid membrane, stromal and envelope fractions as described by Du et al. (2017).
RNA Immunoprecipitation Assays
The procedures used for coimmunoprecipitation and immunoblot assays were described previously (Terzi and Simpson, 2009) using 3-week-old 35S:PDM4-GFP complemented seedlings. Anti-GFP magnetic beads were obtained from Abcam company (ab290, http://www.abcam.com/). The sequences of primers used to detect RNA content that coimmunoprecipitated with PDM4-GFP are listed in Supplemental Table 1.
Results
Characterization of the pdm4 Mutant
To identify PPR genes involved in chloroplast development, we screened a collection of T-DNA inserted mutation lines localized in PPR genes (Du et al., 2017; Zhang J. et al., 2017). In this study, we obtained a new mutant, designated as pigment-defective mutant4 (pdm4). The position of T-DNA insertion in pdm4 was confirmed by PCR and subsequent sequencing, and the result exhibited T-DNA inserted in 165 base pairs downstream of putative start codon (Figure 1A).
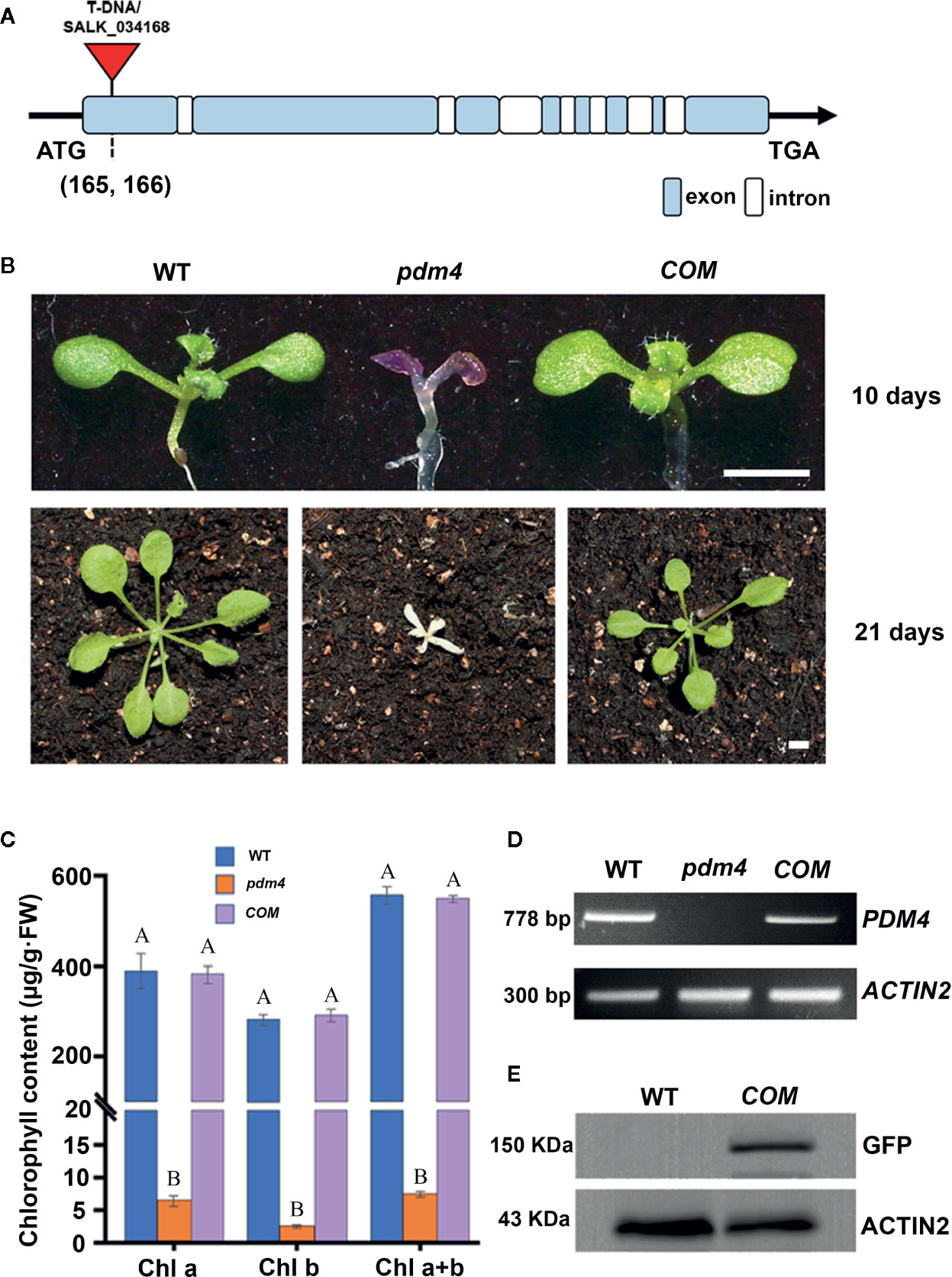
Figure 1 Identification and characterization of the pdm4 mutant. (A) Gene structure of the PDM4 (AT5G27270). Exons indicated by the wathet boxes, introns by the white boxes, and the T-DNA insertion indicated by the red triangle; ATG represents the initiation codon, and TGA represents the stop codon. (B) Pigment-defective phenotypes and complementation of the pdm4 mutant. The cDNA of the PDM4 was cloned into a binary expression vector with the GFP tag and complementation of the pdm4 mutant (COM). WT, wild type. 10-day-old plants were grown on sucrose-supplemented medium (up lane), and 21-day-old plants were grown in soil (down lane). Scale bar: 3 mm. (C) The chlorophyll content of wild-type, pdm4 and COM. Chlorophyll was extracted from 14-day-old seedlings and quantified. Values given are μg/g fresh weight ± SD (n = 3). Values not connected by the same letters are significantly different (Student’s t test, p < 0.05). The average of three replicates is shown. (D) Reverse transcription PCR analysis. RT-PCR was performed using specific primers for AT5G27270 or ACTIN2. (E) Total proteins from wild-type and COM (15 μg) were separated by SDS-PAGE, followed by immunoblot analysis with the anti-GFP. The experiments of (D, E) were repeated three times independently.
When cultivated on 1/2 MS medium supplemented with 1% sucrose, the pdm4 mutant had developed purple cotyledons that were gradually bleached to white with extended growth period (Figure 1B). After being transferred into the soil, the pdm4 mutant was unable to grow and died shortly (Figure 1B). Homozygous pdm4 plants are kept by segregating from a self-pollinated heterozygous plant with a ratio 3:1 in green and albino plants (data not shown). Thus, the albino phenotype is inherited as a recessive mutation. To confirm that the disruption of PDM4 results in the lethal phenotype, we performed the functional complementation analysis. The result indicated that full-length coding region of PDM4 gene fused a GFP tag at its C terminus successfully rescued the pdm4 phenotypes. Among the 56 T1 transgenic lines analyzed, 16 lines were homozygous pdm4 plants and showed a wild-type looking phenotype (Figure 1B).
The concentration of chlorophyll (μg/g FW) in the pdm4 was significantly reduced compared with the wild type (Figure 1C). As expected, chlorophyll accumulated in COM plants was equivalent to the level of the wild-type plants. By reverse transcription-PCR analysis, obvious signals were obtained from COM and wild-type plants but not observed in homozygous pdm4 mutant; this result demonstrated the expression of PDM4 was completely suppressed (Figure 1D). Eventually, western blot result showed that PDM4-GFP proteins were located at about 150 kilodaltons (kDa) in complemented lines by using a GFP antibody, which is in accordance with the predicted protein molecular weight of GFP-tagged PDM4 (Figure 1E). The complementation analysis of the pdm4 phenotype indicated that the PDM4-GFP is a functional protein, and PDM4 gene was responsible for the phenotype of the pdm4.
Chloroplast Development and Accumulation of Photosynthetic Proteins in pdm4
Considering that most photosynthetic pigment defects may result in a retarded chloroplast development, we assessed the possibility that the pdm4 mutation causes ultrastructural changes in the chloroplasts, and plastids from 3-week-old seedling mesophyll cells were examined by transmission electron microscopy (Figures 2A–D). The chloroplasts from wild type contained well-developed membrane systems featured with typical grana structure connected by the stroma lamellae, and the stroma thylakoid and grana thylakoid were easily distinguished (Figures 2A, B). Relative to wild-type chloroplasts, the pdm4 plastids are smaller, deformed, and devoid of thylakoid membrane and granal stacks, and meanwhile, the membrane spacing was not clear (Figures 2C, D).
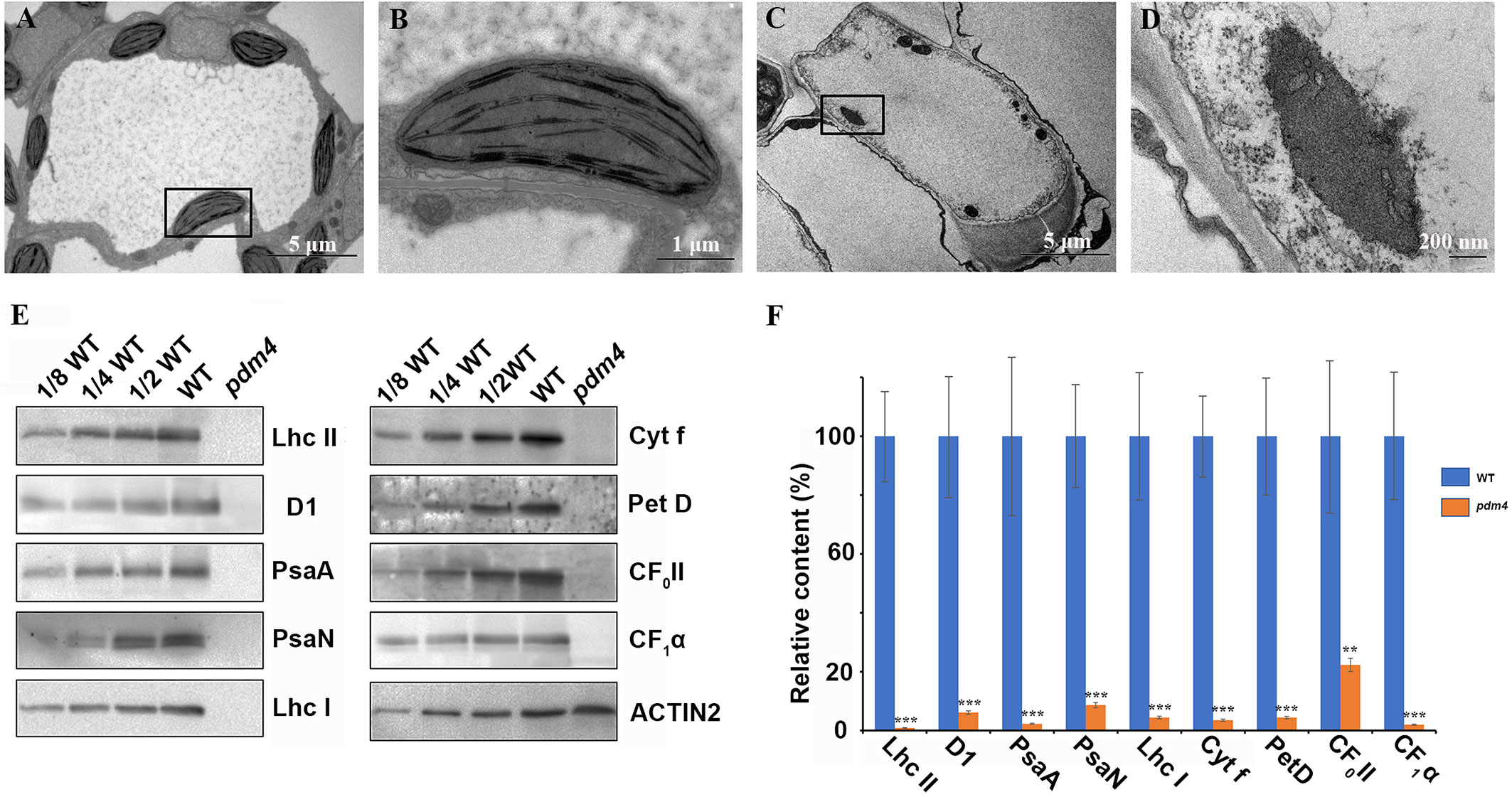
Figure 2 Ultrastructure of plastids and chloroplast proteins in pdm4. (A–D) The thylakoid membrane organization in the chloroplasts of wild type (A, B) and pdm4 (C, D). Scale bars: 5 μm in (A) and (C); 1 μm in (B), and 500 nm in (D). (E) Accumulation of representative subunits of photosynthetic protein complexes determined by western blot analysis with specific antibodies. Total proteins from wild-type and pdm4 seedlings were extracted and separated by 15% SDS-PAGE. Probes used specific anti-Lhc II, anti-D1, anti-PsaA, anti-PsaN, anti-Lhc I, anti-Cyt f, anti-Pet D, anti-CF0 II, anti-CF1α, and anti-ACTIN antibodies. (F) Semi-quantitative analysis of chloroplast proteins. After immunoblot analysis, the average signal intensities for each protein were quantified by the ImageJ software for three independent times. The protein relative contents (per unit of total protein) were determined and compared. Error bars represent standard errors. The relative protein level of pdm4 mutants was obtained when protein level of wild type was set to 100. Similar result to that presented in (E) was obtained from three independent experiments. Results from a representative experiment are shown. The asterisks indicate significant differences between WT and pdm4 (Student’s t test; **p < 0.01; ***p < 0.001).
To obtain more information concerning the molecular lesion in pdm4 to further explain the defects of chloroplasts development in corresponding mutant, total proteins were isolated, and equal sample volumes were loaded in denaturing polyacrylamide gel electrophoresis. Then we performed western blot analysis to detect accumulation of core subunits of photosynthetic complexes, including D1 and LHC II of PSII (encoded by psbA and lhcb2), PsaA, PsaN, and LHC I of PSI (encoded by psaA, psaN, and Lhca1), Cyt f and petD, the subunits of Cyt b6/f, and the CF1α, CF0 II of the ATP synthase. All these proteins were markedly reduced, even barely detected in pdm4 mutant (Figures 2E, F). So, due to the dramatically decreased of some representative subunits of photosynthetic complexes in pdm4 mutant, we got a conclusion that PDM4 is essential for the normal accumulation of thylakoid proteins.
Homozygous pdm4 Embryos Show Delay in Embryogenesis
To reveal the defects in pdm4 development and embryos, we examined developing seeds at various developmental stages by using the differential interference contrast (DIC) optics. Within the immature siliques of heterozygous pdm4, the segregation ratio of the green and white seeds was close to 3:1 (data not shown) and consistent with the segregation in albino phenotype (Figures 3A, B). Moreover, assessment of cleared seeds from the same heterozygous silique indicated that the wild-type and heterozygous seeds underwent normal developmental stages (Figures 3C–G), whereas embryo development of homozygous seeds was seriously disrupted (Figures 3H–L). Different developmental stages of the seeds in wild-type were well-defined and in normal condition, and no difference was visible in pdm4 compared with wild-type embryos before or at the early globular stage (Figure S1). But from the late globular stage to the early heart stage, developmental deviation of mutant embryos became apparent (Figures 3C–L).
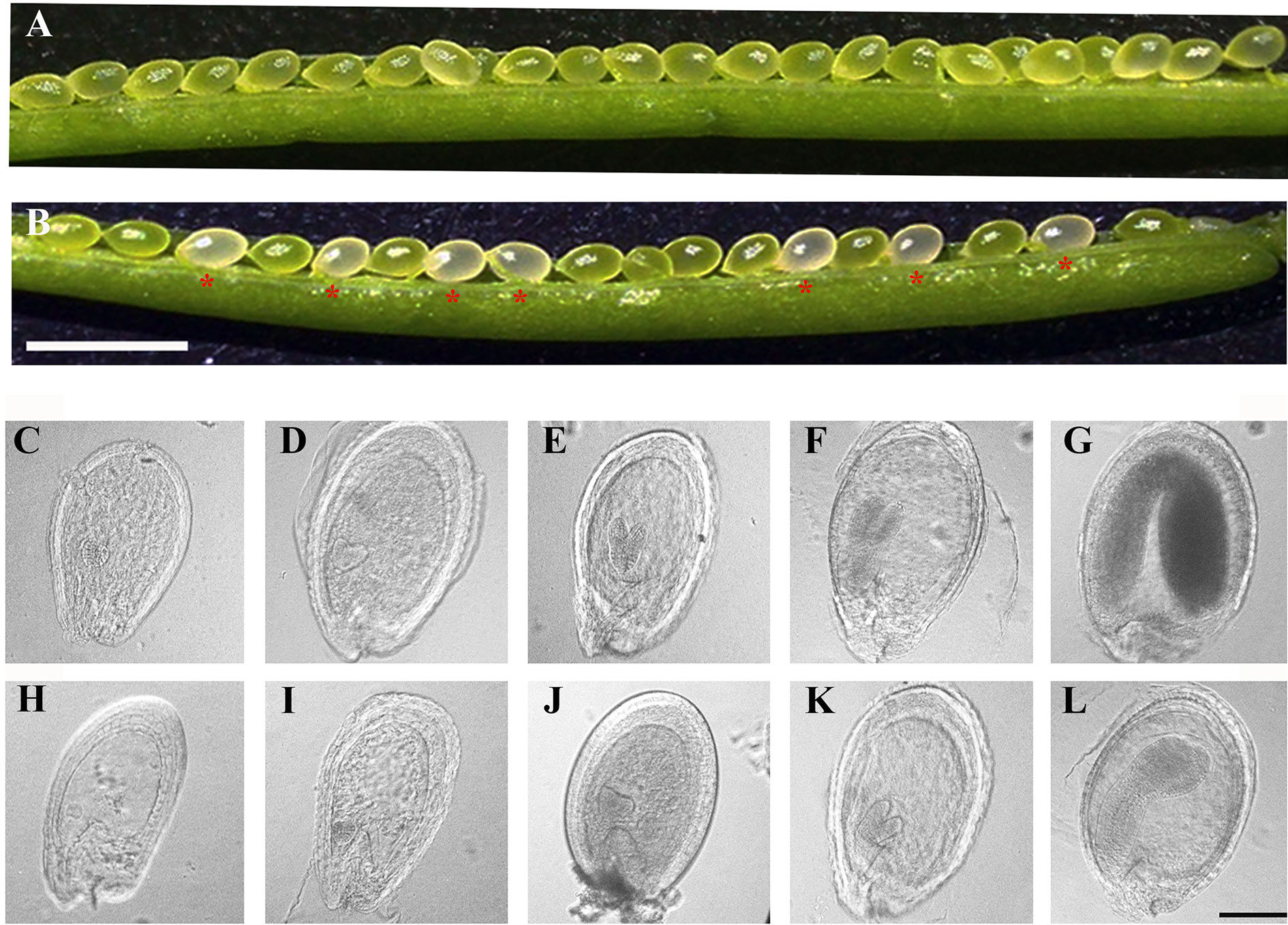
Figure 3 Phenotypic analysis in siliques and embryogenesis in pdm4 mutant. (A) Seeds in wild-type plants. (B) Seed segregation in siliques from a pdm4/+ heterozygous plants. (C–L) Cleared seeds observed under differential interference contrast (DIC). Embryo development of wild type (C–G) and pdm4 (H–L) at globular, heart, torpedo, walking-stick, and cotyledon stages. Scale bars: (A, B), 2 mm; (C–L), 1 mm.
PDM4 Encodes a Novel P-Type PPR Protein Localized in the Chloroplast
Based on the redefined PPR motif, PDM4 is classified P-type PPR protein and possesses 16 PPR repeat domains (Figure S2). Sequence analysis of the PDM4 gene revealed that it contains eight exons with a 3,114 base pair open reading frame, which encodes a polypeptide of 1,038 amino acids. By Chlorop1.1 software prediction, PDM4 possesses a putative 31 amino acid transit peptide at the N terminus. To investigate the subcellular distribution of PDM4, we extracted and observed the protoplasts from COM plants, and the results showed that the fusion proteins were exclusive to chloroplasts and colocalized with chlorophyll (Figure 4A). To further confirm the sublocalization of PDM4 within chloroplasts, intact chloroplasts from COM plants were isolated and further fractionated into stroma, envelop, and thylakoid membrane. Figure 4B shows that the GFP-tagged PDM4 protein is mainly located in the chloroplast stroma, but not in the thylakoid or envelope membrane fraction. Furthermore, qRT-PCR assay demonstrated that PDM4 is universally transcribed throughout the various developmental stages; especially, a high expression level in the seedling and leaf and relatively low expression in the flower, stem, and root are observed (Figure 4C).
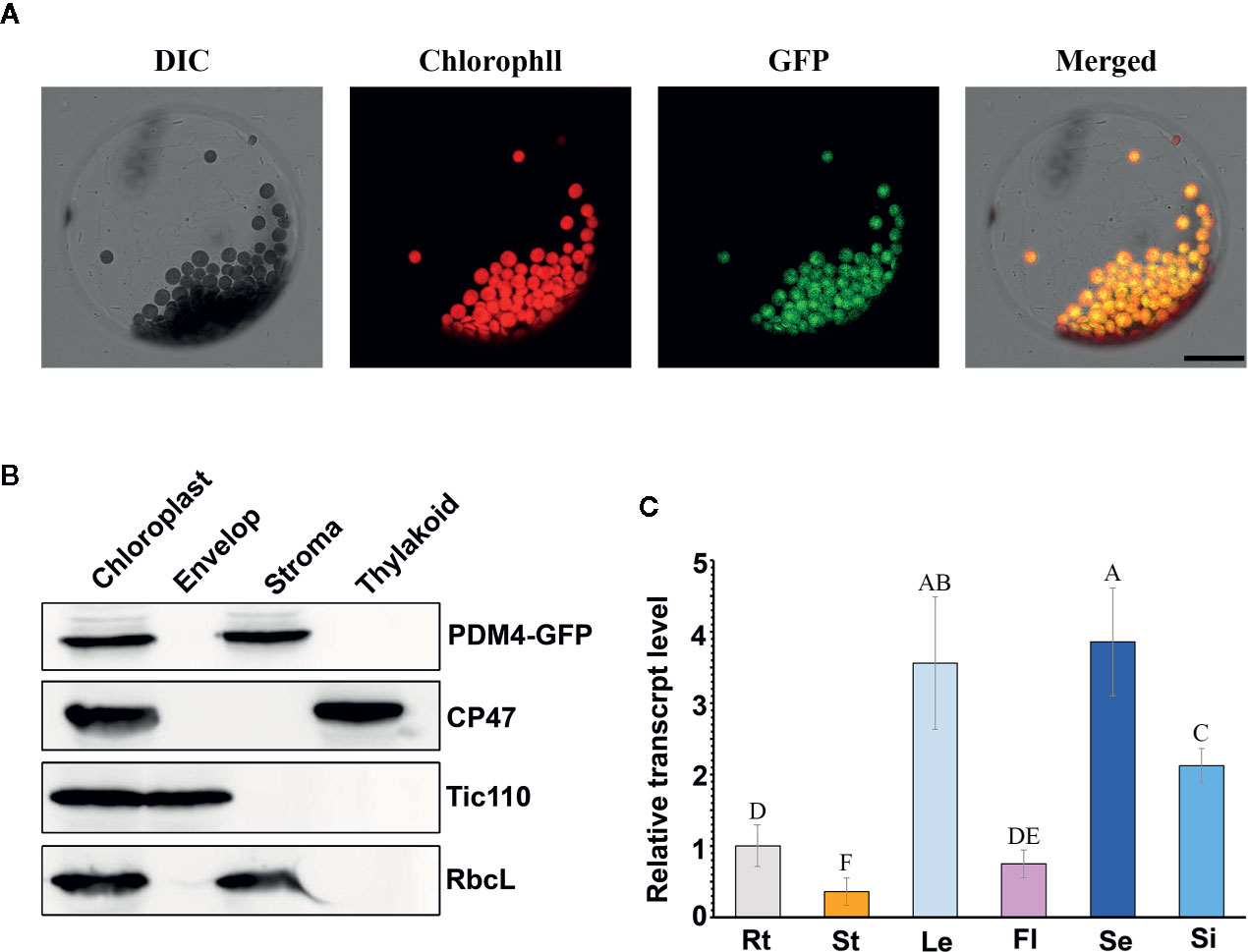
Figure 4 PDM4 protein location and gene expression pattern. (A) Localization of the PDM4 protein within the chloroplast using the GFP assay by protoplasts of complement line. Scale bars: 25 nm. (B) Immunolocalization of PDM4. Intact chloroplasts were isolated from the leaves of GFP-tagged complemented Arabidopsis, and then separated into envelop, stroma, and thylakoid membrane fractions. Antisera were used against GFP, the integral thylakoid membrane protein CP47, the integral inner envelop membrane protein Tic110, and abundant stroma protein RbcL. (C) Quantitative real-time (qRT)-PCR analysis of the PDM4 gene in the root (Rt), stem (St), leaf (Le), flower (Fl), seedling (Se), and silique (Si). Values not connected by the same letters are significantly different (Student’s t test, p < 0.05). All the experiments were repeated three times independently.
To identify PDM4 homologs in several species, a search was carried out using NCBI protein database and the protein sequence of PDM4 as the template. High similarity of protein sequences was selected for bioinformatics analysis. The PDM4 homologous proteins exhibited a relatively high level of similarity in Populus trichocarpa (Potri.T071500, 58.0%) and Glycine max (Glyma.08G106500, 54.8%), while they showed a low level in Volvox carteri (Vocar.0001s1331, 23.4%) (Figure S3). To further reveal the relationship between the PDM4 and other family member proteins in different species, we constructed a phylogenetic tree (www.phylogeny.fr) of the closest amino acid sequences from Figure S3, and analysis revealed that PDM4-like proteins are present in most chloroplastida including monocots, dicots, ferns, mosses, and algae (Figure S4).
The PDM4 Mutation Affects Plastid Gene Expression
Growing mass of information supports the idea that the PPR proteins are always involved in the regulation of plastid gene expression (Barkan and Small, 2014). Depending on the different RNA polymerases required, plastid genes can be roughly divided into three categories: genes of class I are mainly synthesized by PEP, genes of class II are synthesized by both PEP and NEP, and genes of class III are synthesized by NEP. To assess the possibility that PDM4 functions in gene expression in plastid, we examined the transcript levels of three types of genes both in the pdm4 mutant and wild-type plant by qRT-PCR analysis.
We chose the six genes as representation of PEP-dependent (class I) type, psaA, psbA, psbB, petB, ndhA, and rbcL. The results indicated that the transcription level of PEP-dependent genes in pdm4 was dramatically decreased from about 40 to 90% (Figure 5A). In contrast, the transcript levels of the plastid genes, which were tested as the representatives of class III (NEP-dependent) type, including rpoA, accD, rpoB, ycf2, rpoC1, and rpoC2, were all increased by varying degrees in pdm4 (Figure 5B). As for the class II genes, synthesized by both NEP and PEP, ndhB, atpB, and ndhF expression levels showed a reverse trend. The transcript levels of ndhB and atpB decreased by nearly 20%, while the transcripts of ndhF were obviously upregulated (Figure 5C). The contents of representative genes were also confirmed by northern-blot analysis and the result is comparable to the qRT-PCR analysis (Figure 5D). To further study whether the increased transcription level of rpoB resulted in an enhancement at protein level, we detected the RpoB protein content in the pdm4 mutant by western blot analysis. Compared with wild type, the result showed an obviously reduced level of RpoB in the pdm4 mutant (Figure 5E).

Figure 5 Chloroplast gene expression in pdm4 relative to the wild type. (A) The expression levels of plastid-encoded polymerase (PEP)-dependent chloroplast genes. (B) The expression levels of nuclear-encoded polymerase (NEP)-dependent chloroplast genes. (C) The expression levels of both PEP- and NEP-dependent chloroplast genes. The asterisks indicate significant differences between WT and pdm4 (Student’s t test; *p < 0.05; **p < 0.01; ***p < 0.001). (D) RNA gel-blot hybridization analysis of transcript levels for the different chloroplast gene classes. 5 μg of total RNA from 3-week old plants was size fractionated by agarose gel electrophoresis, transferred to a nylon membrane, and probed with 32P-labeled cDNA probes for psaA, psbA, psbB, rbcL, rpoA, rpoB, and 18S rRNA. (E) RpoB protein content was indicated by western blot. Total protein (50 μg or the indicated dilution of the wild-type sample) from 3-week-old seedlings was loaded per lane. Actin was used as a loading control. These experiments obtained similar results each time. Results from a representative experiment of three times are shown.
PDM4 Involved in RNA Splicing of Multiple Chloroplast Group II Introns
Dozens of P-type PPR proteins have been reported to function in RNA splicing in chloroplast (Beick et al., 2008; Barkan and Small, 2014). To determine whether PDM4 influences the splicing of the group II introns, representative splicing event was assayed in the pdm4 mutant by performing RT-PCR analysis (Figure 6A). Compared with the wild-type plants, the unspliced precursors of ndhA, petB, ycf3-int-1, petD, clpp1-int-1 accumulated to an increased level in the pdm4 mutant (Figure 6A). The observation of altered intron processing was also confirmed by northern-blot analysis. The result indicated that unspliced precursors of ycf3, petB, petD, and ndhA were present and accumulated in a high level in pdm4 and absent in the wild type. By contrast, the transcripts of rps14 showed a higher efficiency of accumulation than the wild type (Figure 6B). Besides, RNA immunoprecipitation, followed by a quantitative PCR assay using the GFP antibody and COM plants, indicated that PDM4 was specifically associated with these target sequences in the ndhA, petB, ycf3-1, and petD transcripts (Figure 6C). The RNA immunoprecipitation efficiency was supported by western blot analysis (Figure S5), and the transcripts containing 18S rRNA were used as control.
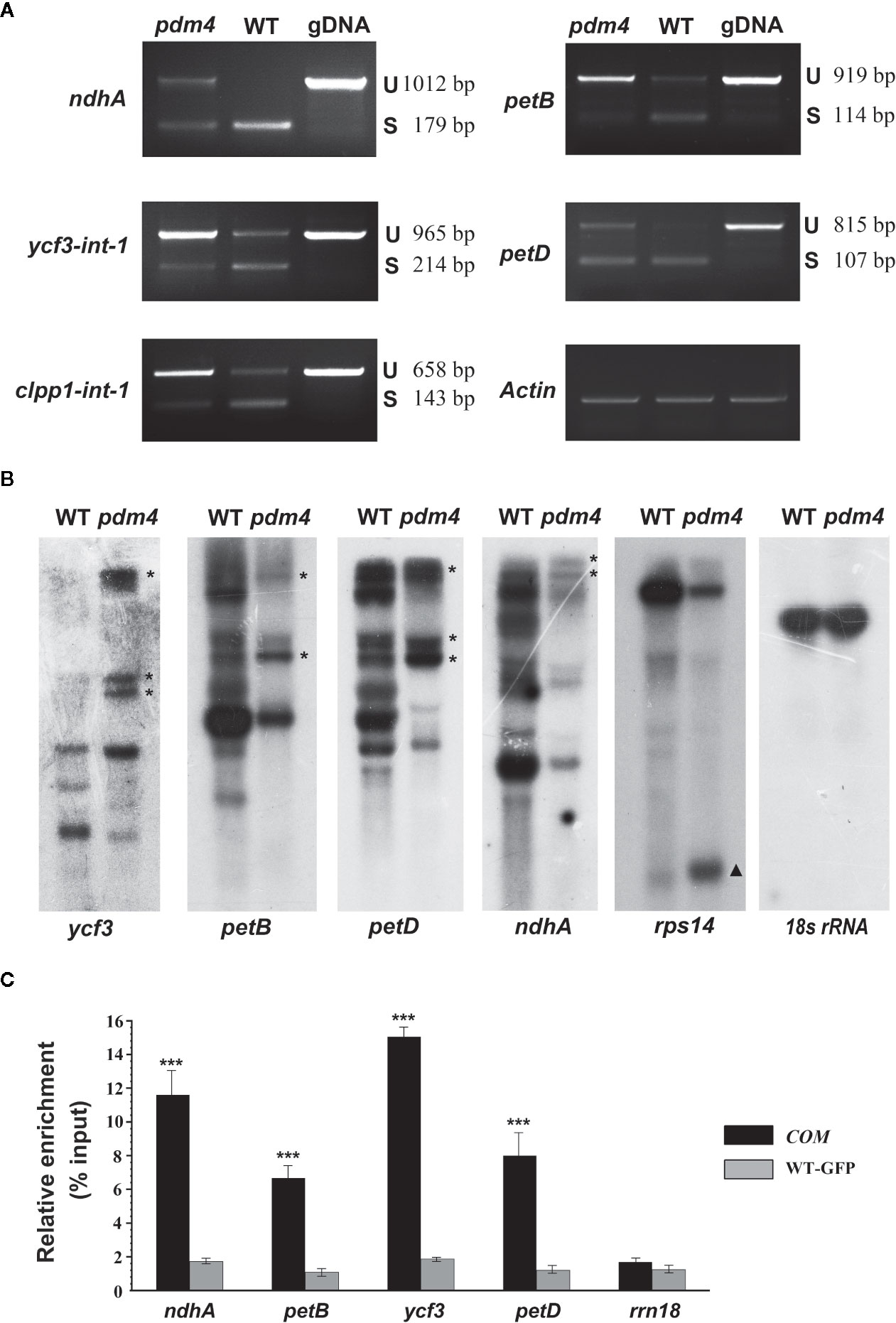
Figure 6 PDM4 is involved in RNA splicing of Arabidopsis group II introns. (A) Inefficient splicing of group II introns of ndhA, petB, ycf3-int-1, petD, and clpp1-int-1 in the mutant as determined by reverse-transcription (RT)-PCR. S, introns are spliced; U, introns are retained. (B) Northern-blot analysis for the transcript accumulation and pattern of ycf3, petB, petD, ndhA, rps14, and 18S rRNA. 5 μg of total RNA from 3-week-old seedlings was analyzed by hybridization to specific probes for each gene. Gene precursor indicated by the asterisk. The results shown are representatives of three independent biological replicates. (C) RNA immunoprecipitation analysis followed by a quantitative PCR assay. 3-week-old 35S:PDM4-GFP complemented seedlings were used. IP+, anti-GFP immunoprecipitation; IP−, mock immunoprecipitation. The asterisks indicate significant differences between WT-GFP and COM (Student’s t test; ***p < 0.001). Data are means (± SE) obtained from three replicates.
pdm4 Mutant Is Defective in Chloroplast rRNA Accumulation
The chloroplast rRNAs, as well as two tRNAs, are arranged in one operon, and transcription depends on both PEP and NEP (Tiller and Bock, 2014). When denatured rRNA samples were separated on denaturing agarose gels, it was shown that the rRNA fragmentation pattern in the wild type was obviously different from pdm4 mutant by using the ethidium bromide staining method (Figure 7A). The signal intensities of the 1.5 and 1.1-kb RNA corresponding to chloroplast 16S rRNA and a breakdown product of the chloroplast 23S rRNA were dramatically reduced in the pdm4 mutant (Figure 7A).
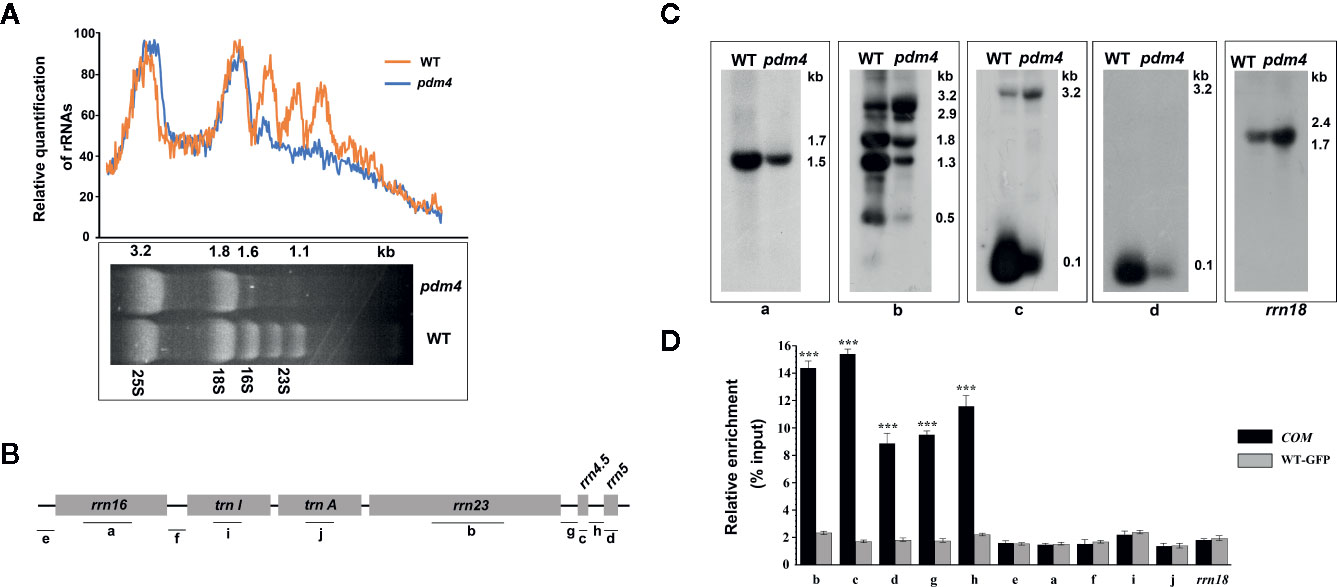
Figure 7 The chloroplast rRNA processing in pdm4 mutant. (A) The contents of rRNAs in wild type and pdm4. 5 μg of total RNA from 3-week-old wild type and pdm4 seedlings was separated by denaturing gel. 23S* is the breakdown product of the chloroplast 23S rRNA. rRNA was quantified using “ImageJ” software. (B) Diagram of the chloroplast rRNA operon and the locations of the probes (a–d) used for the RNA gel-blot analysis and used for RNA immunoprecipitation analysis (a–j). (C) RNA gel analysis of 16S rRNA (probe a), 23S rRNA (probes b), 4.5S rRNA (probe c), and 5S rRNA (probe d). The sizes of the transcripts (in kb) are shown. The 18S rRNA is shown as a loading control. (D) RNA immunoprecipitation analysis followed by a quantitative PCR assay. Probes b, d, c, a, i, f, and rrn18 are fragments from the 23S rRNA, 5S rRNA, 4.5S rRNA, 16S rRNA, trnI, trnA, and 18S rRNA, respectively and probes e, f, g, h for the intergenic region. The asterisks indicate significant differences between WT-GFP and COM (Student’s t test; ***p < 0.001). Data are means (± SE) obtained from three replicates.
To study the impaired accumulation of chloroplast rRNAs in detail, we performed northern-blot analyses to detect the 16S, 4.5S, 5S, and 23S rRNA by using four probes with an internal region (probes a, b, c, and d, respectively, as shown in Figure 7B). Upregulated levels of the 3.2-kb 23S–4.5S rRNA precursor were detected in the pdm4 mutant, whereas the levels of the 0.12-kb 5S, 0.1-kb 4.5S, 0.5-kb 23S, and 1.5-kb 16S mature rRNAs were drastically downregulated (Figure 7C). We also have tested the interaction and/or association between PDM4 and its targets by RNA immunoprecipitation and qRT-PCR methods and using specific primers for transcripts containing the 23S, 5S, 4.5S, 16S rRNA, and two tRNAs, as well as the intergenic region (Figure 7B, probes e, f, g, h). We detected enrichment fragments of 23S, 5S, 4.5S rRNA, and the intergenic region in the anti-GFP immunoprecipitated, but not of 16S, trnI, trnA, and 18S rRNA (Figure 7D). Results from northern blots and RNA-Co-IPs support the conclusion that PDM4 associates with rRNA and affects rRNA processing. The RNA immunoprecipitation efficiency was also supported by western blot analysis (Figure S5).
Discussion
Contrary to the increasing information about the subfamily classification and organellar targeting of PPR proteins in plants, the cellular role and function of most PPR proteins are still so far from fully being apprehended (Lee et al., 2019). Among these PPR proteins, several mutants show pigment defective or lethal phenotypes, such as sel1 (Pyo et al., 2013), ecd1 (Jiang et al., 2018), ppr4, emb2654 (Lee et al., 2019), and sot5/emb2279 (Huang et al., 2018) in addition to pdm2 and pdm3 mutants (Du et al., 2017; Zhang J. et al., 2017) which have been reported in our lab before. These works suggest that most functional PPR proteins located in the chloroplast mainly play a critical role in accumulation of chlorophyll and are essential for plant survival. In this study, we identified and characterized a novel PPR protein PDM4; the pigment defective phenotype of the mutant and complementation analysis with the PDM4 gene demonstrated that PDM4 is indispensable for plant survival and growth.
Chlorophyll accumulation is a prerequisite for the maintenance of functional photosynthetic reaction centers and light-harvesting complexes (Pan et al., 2013). In the pdm4, the albino phenotype with a decrease of chlorophyll contents was observed (Figure 1C), indicating the photosystem complexes might be impaired, and in accordance with this hypothesis, the result of western blot analysis confirmed this possibility (Figure 2E). The chloroplast ultrastructure of pdm4 exhibited an abnormal morphology, and the structure of the thylakoid membrane was seriously disturbed, suggesting that the lethal phenotype in these plants was probably attributed to developmental defects in chloroplasts. Furthermore, proper development and biogenesis of chloroplast play an essential role in the vast majority of important biological processes, including cell proliferation, secondary metabolite synthesis, and embryogenesis (Yin et al., 2012). Compared with our previous studies in pdm2 and pdm3 mutants, the development of homozygous embryo was also retarded especially after globular-to-heart stage (Figures 3H–L), suggesting that disruption of PDM4 is the primary cause for embryogenesis defection in the pdm4 mutant. Several PPR mutants have similar developmental defects during the embryogenesis process, e.g., Atppr2 (Lu et al., 2011), fac19 (Yu et al., 2012), At_dek36 (Wang et al., 2017), blx (Sun et al., 2018), and ecd1 (Jiang et al., 2018). Borisjuk et al. (2005) suggested that the probable reason for these developmenal defects is energy source transformation from the endosperm to the chloroplast in seed development because chloroplast formed transiently and transformed into storage organelles during embryo development. This view also coincided with Barkan and Small (2014). Thus, blocking embryo development in pdm4 mutant may result from the defective chloroplast by some indirect effects.
The expression of plastid-encoded genes of photosynthesis was closely linked with the developmental state of the chloroplast (Chi et al., 2008; Zhang Z. et al., 2017). qRT-PCR and northern-blot analysis showed that the levels of PEP-dependent transcripts were dramatically reduced; on the contrary, levels of NEP-dependent transcripts were obviously increased, suggesting that NEP functions correctly or efficiently in pdm4 (Figures 5A–D). Compared with upregulated transcriptional level, the protein level of RpoB is significantly decreased in pdm4 mutant (Figure 5E), and we obtained a similar result in the pdm3 mutant (Zhang J. et al., 2017). It suggests that RpoB protein biosynthesis or stability is affected in the pdm4, leading to a dysfunctional PEP complex, just like the case that happened in the pdm3 (Zhang J. et al., 2017). Thus, PDM4 is necessary for an efficient and functional PEP transcription machinery. As to the reduced PEP activity, one possible reason is that PDM4 acts as a participant of nucleoid proteins, like PAPs or pTACs, or merely associates with them to participate in the regulation of PEP activity because pdm4 mutant shares some similarities in molecular phenotypes with other PEP-related mutants (Figures 5 and 6; Zhang J. et al., 2017; He et al., 2019). As we know, pTACs always interacted with thylakoid membrane; we had not detected any signals with PDM4 in the thylakoid fragments (Figure 4B); this may indicate that there exists a weakly interaction between PDM4 and pTACs although none of the presented data show PEP/TAC-association or that PEP-activity is actually PDM4 dependent. Another explanation is that the stability of the transcripts may be decreased, or the rate of mRNA turnover is enhanced in those mutants. In fact, dozens of PPR proteins that affect the stability of chloroplast gene transcripts have been identified (Barkan and Small, 2014).
In higher plants, dozens of P-type PPR proteins are targeted to the chloroplasts and have been proved to be necessary in removing some distinct introns (Barkan and Small, 2014). For example, PPR5 was confirmed to promote the splicing of the group II intron within rpl16 in Arabidopsis (Rojas et al., 2018); and in rice, AL2 collaborates a subset of chloroplast associated proteins to regulate splicing of both chloroplast group II and I introns (Liu et al., 2016). In this investigation, the disruption of PDM4 affected the splicing of ndhA, petB, clpP1-1, ycf3-1, and petD transcripts, and to our surprise, rps14 transcript accumulation was improved in this study (Figure 6B). This result does not fully coincide with pdm3 in the respective transcript splicing events, e.g. the affected splicing sites are ndhB, clpP1-1, and trnA. Alternately, this probably suggests partially distinctive and redundant functions refer to PDM3 and PDM4 in the regulation of chloroplast gene expression. A recent work has shown that BFA2, a P-type PPR protein in the chloroplast, affects atpF-atpA transcript splicing by combining to the intergenic region of atpF-atpA and acts as a specific barrier to prevent atpH/F mRNA from exoribonuclease degradation (Zhang et al., 2019). Thus, we could not rule out the possibility that PDM4 acted as a barrier and resulted in a high efficiency in transcript accumulation of the rps14 in pdm4 mutant.
The result of agarose gel electrophoresis analysis validated that chloroplast rRNA was dramatically reduced in the pdm4 mutant (Figure 7A). It is reported that PPR proteins are also involved in pre-rRNA processing and lead to reduced rRNA levels (Barkan and Small, 2014). To further assess this possibility, chloroplast rRNA was analyzed in detail by northern-blot analysis (Figure 7C). Compared with the wild type, 3.1-kb RNA representing the precursor of 23S and 4.5S rRNA was accumulated more in pdm4 (Figure 7C). As a consequence of the increased precursor of rRNA in pdm4, we deduced that protein PDM4 was involved in the cleavage precursor of rRNA in the chloroplast, especially during the maturation of 50S rRNA. As pre-rRNA processing and ribosome assembly are intimately linked in the chloroplast, therefore, lesions in the ribosome assembly are frequently found in mutants with rRNA processing defects (Prikryl et al., 2008; Chi et al., 2011; Asakura et al., 2012). Coimmunoprecipitation assays produced evidence that PDM4 associates with rRNA and affects rRNA processing (Figure 7D), and we can also draw the conclusion that the PDM4 protein was specifically associated with these target sequences in the ndhA, petB, ycf3-1, and petD transcripts (Figure 6C). Thus, in pdm4 mutant, the aberrant maturation and accumulation of chloroplast rRNA mutant may be due to a defect in ribosomal biogenesis/assembly. The pmd4 mutant also showed an obvious downregulated level in the mature form of 16S rRNA, whereas an accumulation of precursor rRNA was not detected (Figure 7C). The decrease in mature 16S rRNA in pdm4 appeared to be regarded as an indirect consequence of the defects in the 50S subunit biogenesis/assembly because PDM4 associated with the 30S particle was not found (Figure 7). This conclusion is further sustained by the identification of the RH22 in Arabidopsis (Chi et al., 2012).
PDM4 is required for PEP activity, polycistronic accumulation, and rRNA maturation. But it is hard to rule out which process is a dominant factor; because of the pleiotropic nature of knockout plants, it is particularly true when general plastid translation can be affected by the lack of ribosome processing and/or assembly, which indirectly results in dysfunctional transcription machine as the results of disrupted expression of the plastid encoded polymerase (PEP) and in turn affects RNA processing patterns and levels (Legen et al., 2002; Stoppel and Meurer, 2011). Furthermore, in pdm4, chloroplast development is severely damaged. We can find that PDM4 affecting the transcription level of PEP-dependent genes, RNA splicing of multiple chloroplast group II introns, and chloroplast rRNA accumulation. These processes influence and restrict each other, which together leads to the loss of chloroplast development. And based on our research, the key or the direct reason of this phenomenon is still not very clear. So, further study of PDM4 function should facilitates the general understanding the mechanism of plastid gene expression and chloroplast development.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
JX designed the study. XW, LZ, YM, XL, LW, and JX performed the research. XW, XL, and JX analyzed the data. JX and XW wrote the paper. All authors discussed the results and made comments on the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the National Natural Science Foundation of China (grants. 31970653, 91954202 and 31670181).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Arabidopsis Biological Resource Center for the pdm4 mutant line. And we thank professor Jinxing Lin (Beijing Forestry University, Beijing 10083, China) for language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01198/full#supplementary-material
References
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts: polyphenoloxidase in beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Aronsson, H., Jarvis, P. (2002). A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 529, 215–220. doi: 10.1016/s0014-5793(02)03342-2
Asakura, Y., Galarneau, E., Watkins, K. P., Barkan, A., van-Wijk, K. J. (2012). Chloroplast RH3 DEAD box RNA helicases in maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis. Plant Physiol. 159, 961–974. doi: 10.1104/pp.112.197525
Barkan, A., Small, I. (2014). Pentatricopeptide Repeat Proteins in Plants. Annu. Rev. Plant Biol. 65, 415–442. doi: 10.1146/annurev-arplant-050213-040159
Beick, S., Schmitz-Linneweber, C., Williams-Carrier, R., Jensen, B., Barkan, A. (2008). The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 28, 5337–5347. doi: 10.1128/MCB.00563-08
Belcher, S., Williams-Carrier, R., Stiffler, N., Barkan, A. (2015). Large-scale genetic analysis of chloroplast biogenesis in maize. Biochim. Biophys. Acta 1847, 1004–1016. doi: 10.1016/j.bbabio.2015.02.014
Bobik, K., McCray, T. N., Ernest, B., Fernandez, J. C., Howell, K. A., Lane, T., et al. (2017). The chloroplast RNA helicase ISE2 is required for multiple chloroplast RNA processing steps in Arabidopsis thaliana. Plant J. 91, 114–131. doi: 10.1111/tpj.13550
Borisjuk, L., Nguyen, T. H., Neuberger, T., Rutten, T., Tschiersch, H., Claus, B., et al. (2005). Gradients of lipid storage, photosynthesis and plastid differentiation in developing soybean seeds. New Phytol. 167, 761–776. doi: 10.1111/j.1469-8137.2005.01474.x
Bryant, N., Lloyd, J., Sweeney, C., Myouga, F., Meinke, D. (2011). Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol. 155, 1678–1689. doi: 10.1104/pp.110.168120
Cheng, S., Gutmann, B., Zhong, X., Ye, Y., Fisher, M. F., Bai, F., et al. (2016). Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85, 532–547. doi: 10.1111/tpj.13121
Chi, W., Ma, J., Zhang, D., Guo, J., Chen, F., Lu, C., et al. (2008). The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol. 147, 573–584. doi: 10.1104/pp.108.116194
Chi, W., He, B., Mao, J., Li, Q., Ma, J., Ji, D., et al. (2012). The function of RH22, a DEAD RNA helicase, in the biogenesis of the 50S ribosomal subunits of Arabidopsis chloroplasts. Plant Physiol. 158, 693–707. doi: 10.1104/pp.111.186775
Chi, W., Feng, P., Ma, J., Zhang, L. (2015). Metabolites and chloroplast retrograde signaling. Curr. Opin. Plant Biol. 25, 32–38. doi: 10.1016/j.pbi.2015.04.006
Chi, Y. H., Semmes, O. J., Jeang, K. T. (2011). A proteomic study of TAR-RNA binding protein (TRBP)-associated factors. Cell Biosci. 1, 9. doi: 10.1186/2045-3701-1-9
Dovzhenko, A., Bosco, C. D., Meurer, J., Koop, H. U. (2003). Efficient regeneration from cotyledon protoplasts in Arabidopsis thaliana. Protoplasma 222 (1-2), 107–111. doi: 10.1007/s00709-003-0011-9
Du, L., Zhang, J., Qu, S., Zhao, Y., Su, B., Lv, X., et al. (2017). The pentratricopeptide repeat protein pigment-defective mutant2 is involved in the regulation of chloroplast development and chloroplast gene expression in Arabidopsis. Plant Cell Physiol. 58, 747–759. doi: 10.1093/pcp/pcx004
Germain, A., Hotto, A. M., Barkan, A., Stern, D. B. (2013). RNA processing and decay in plastids. WIREs RNA 4, 295–316. doi: 10.1002/wrna.1161
Guillaumot, D., Lopez-Obando, M., Baudry, K., Avon, A., Rigaill, G., Falcon de Longevialle, A., et al. (2017). Two interacting PPR proteins are major Arabidopsis editing factors in plastid and mitochondria. Proc. Natl. Acad. Sci. U. S. A. 114, 8877–8882. doi: 10.1073/pnas.1705780114
Hajdukiewicz, P. T., Allison, L. A., Maliga, P. (1997). The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16, 4041–4048. doi: 10.1093/emboj/16.13.4041
He, P., Wu, S., Jiang, Y., Zhang, L., Tang, M., Xiao, G., et al. (2019). GhYGL1d, a pentatricopeptide repeat protein, is required for chloroplast development in cotton. BMC Plant Biol. 19, 350–350. doi: 10.1093/emboj/16.13.4041
Huang, W., Zhu, Y., Wu, W., Li, X., Zhang, D., Yin, P., et al. (2018). The pentatricopeptide repeat protein SOT5/EMB2279 is required for plastid rpl2 and trnK intron splicing. Plant Physiol. 177, 684–697. doi: 10.1104/pp.18.00406
Ito, A., Sugita, C., Ichinose, M., Kato, Y., Yamamoto, H., Shikanai, T., et al. (2018). An evolutionarily conserved P-subfamily pentatricopeptide repeat protein is required to splice the plastid ndhA transcript in the moss Physcomitrella patens and Arabidopsis thaliana. Plant J. 94, 638–648. doi: 10.1111/tpj.13884
Jiang, T., Zhang, J., Rong, L., Feng, Y., Wang, Q., Song, Q., et al. (2018). ECD1 functions as an RNA-editing trans-factor of rps14-149 in plastids and is required for early chloroplast development in seedlings. J. Exp. Bot. 69, 3037–3051. doi: 10.1093/jxb/ery139
Krause, K., Krupinska, K. (2000). Molecular and functional properties of highly purified transcriptionally active chromosomes from spinach chloroplasts. Physiol. Plant. 109, 188–195. doi: 10.1034/j.1399-3054.2000.100211.x
Krupinska, K., Melonek, J., Krause, K. (2012). New insights into plastid nucleoid structure and functionality. Planta 237, 653–664. doi: 10.1007/s00425-012-1817-5
Lee, K., Park, S. J., Han, J. H., Jeon, Y., Pai, H.-S., Kang, H. (2019). A chloroplast-targeted pentatricopeptide repeat protein PPR287 is crucial for chloroplast function and Arabidopsis development. BMC Plant Biol. 19, 244–244. doi: 10.1186/s12870-019-1857-0
Legen, J., Kemp, S., Krause, K., Profanter, B., Herrmann, R. G., Maier, R. M. (2002). Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 31, 171–188. doi: 10.1186/s12870-019-1857-0
Liere, K., Börner, T. (2007). Transcription and transcriptional regulation in plastids. Cell and Molecular Biology of Plastids 19, 121–174. doi: 10.1007/4735_2007_0232
Leister, D. (2003). Chloroplast research in the genomic age. Trends Genet. 19, 47–56. doi: 10.1016/s0168-9525(02)00003-3
Liu, C., Zhu, H., Xing, Y., Tan, J., Chen, X., Zhang, J., et al. (2016). Albino Leaf 2 is involved in the splicing of chloroplast group I and II introns in rice. J. Exp. Bot. 67, 5339–5347. doi: 10.1093/jxb/erw296
Lu, Y., Li, C., Wang, H., Chen, H., Berg, H., Xia, Y. (2011). AtPPR2, an Arabidopsis pentatricopeptide repeat protein, binds to plastid 23S rRNA and plays an important role in the first mitotic division during gametogenesis and in cell proliferation during embryogenesis. Plant J. 67, 13–25. doi: 10.1111/j.1365-313X.2011.04569.x
Lurin, C., Andrés, C., Aubourg, S., Bellaoui, M., Bitton, F., Bruyère, C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 16, 2089–2103. doi: 10.1105/tpc.104.022236
Majeran, W., Friso, G., Ponnala, L., Connolly, B., Huang, M., Reidel, E., et al. (2010). Structural and metabolic transitions of C4 leaf development and differentiation defined by microscopy and quantitative proteomics in maize. Plant Cell. 22, 3509–3542. doi: 10.1105/tpc.110.079764
Majeran, W., Friso, G., Asakura, Y., Qu, X., Huang, M., Ponnala, L., et al. (2012). Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves; a new conceptual framework for nucleoid functions. Plant Physiol. 158, 156–189. doi: 10.1104/pp.111.188474
Meinke, D. W., Franzmann, L. H., Nickle, T. C., Yeung, E. C. (1994). Leafy cotyledon mutants of Arabidopsis. Plant Cell. 6, 1049–1064. doi: 10.1105/tpc.6.8.1049
Melonek, J., Oetke, S., Krupinska, K. (2016). Multifunctionality of plastid nucleoids as revealed by proteome analyses. Biochim. Biophys. Acta 1864, 1016–1038. doi: 10.1016/j.bbapap.2016.03.009
Mullet, J. E. (1993). Dynamic regulation of chloroplast transcription. Plant Physiol. 103, 309–313. doi: 10.1104/pp.103.2.309
Ouyang, M., Li, X., Zhao, S., Pu, H., Shen, J., Adam, Z., et al. (2017). The crystal structure of Deg9 reveals a novel octameric-type HtrA protease. Nat. Plants 12, 973–982. doi: 10.1038/s41477-017-0060-2
Pan, X., Liu, Z., Li, M., Chang, W. (2013). Architecture and function of plant light-harvesting complexes II. Curr. Opin. Struct. Biol. 23, 515–525. doi: 10.1016/j.sbi.2013.04.004
Pfalz, J., Pfannschmidt, T. (2013). Essential nucleoid proteins in early chloroplast development. Trends Plant Sci. 18, 186–194. doi: 10.1016/j.tplants.2012.11.003
Pfalz, J., Liere, K., Kandlbinder, A., Dietz, K. J., Oelmüller, R. (2006). pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 18, 176–197. doi: 10.1105/tpc.105.036392
Pogson, B. J., Albrecht, V. (2011). Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol. 155, 1545–1551. doi: 10.1104/pp.110.170365
Prikryl, J., Watkins, K. P., Friso, G., van Wijk, K. J., Barkan, A. (2008). A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 36, 5152–5165. doi: 10.1093/nar/gkn492
Pyo, Y. J., Kwon, K.-C., Kim, A., Cho, M. H. (2013). Seedling Lethal1, a pentatricopeptide repeat protein lacking an E/E+ or DYW domain in Arabidopsis, is involved in plastid gene expression and early chloroplast development. Plant Physiol. 163, 1844–1858. doi: 10.1104/pp.113.227199
Rojas, M., Ruwe, H., Miranda, R. G., Zoschke, R., Hase, N., Schmitz-Linneweber, C., et al. (2018). Unexpected functional versatility of the pentatricopeptide repeat proteins PGR3, PPR5 and PPR10. Nucleic Acids Res. 46, 10448–10459. doi: 10.1093/nar/gky737
Sato, S., Nakamura, Y., Kaneko, T., Asamizu, E., Tabata, S. (1999). Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 6, 283–290. doi: 10.1093/dnares/6.5.283
Small, I. D., Peeters, N. (2000). The PPR motif - a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 45–47. doi: 10.1016/s0968-0004(99)01520-0
Stern, D. B., Goldschmidt-Clermont, M., Hanson, M. R. (2010). Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61, 125–155. doi: 10.1146/annurev-arplant-042809-112242
Stoppel, R., Meurer, J. (2011). The cutting crew - ribonucleases are key players in the control of plastid gene expression. J. Exp. Bot. 63, 1663–1673. doi: 10.1093/jxb/err401
Stoppel, R., Meurer, J. (2013). Complex RNA metabolism in the chloroplast: an update on the psbB operon. Planta 237, 441–449. doi: 10.1007/s00425-012-1782-z
Sugita, M., Sugiura, M. (1996). Regulation of gene expression in chloroplasts of higher plants. Plant Mol. Biol. 32, 315–326. doi: 10.1007/bf00039388
Sugiura, M., Hirose, T., Sugita, M. (1998). Evolution and mechanism of translation in chloroplasts. Annu. Rev. Genet. 32, 437–459. doi: 10.1146/annurev.genet.32.1.437
Sun, Y., Huang, J., Zhong, S., Gu, H., He, S., Qu, L. (2018). Novel DYW-type pentatricopeptide repeat (PPR) protein BLX controls mitochondrial RNA editing and splicing essential for early seed development of Arabidopsis. J. Genet. Genomics 45, 155–168. doi: 10.1016/j.jgg.2018.01.006
Terzi, L. C., Simpson, G. G. (2009). Arabidopsis RNA immunoprecipitation. Plant J. 59, 163–168. doi: 10.1111/j.1365-313X.2009.03859.x
Tiller, N., Bock, R. (2014). The translational apparatus of plastids and its role in plant development. Mol. Plant 7, 1105–1120. doi: 10.1093/mp/ssu022
Walter, M., Piepenburg, K., Schöttler, M. A., Petersen, K., Kahlau, S., Tiller, N., et al. (2010). Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. Plant J. 64, 851–863. doi: 10.1111/j.1365-313X.2010.04377.x
Wang, G., Zhong, M., Shuai, B., Song, J., Zhang, J., Han, L., et al. (2017). E+ subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and Arabidopsis. New Phytol. 214, 1563–1578. doi: 10.1111/nph.14507
Williams, P. M., Barkan, A. (2004). A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 36, 675–686. doi: 10.1046/j.1365-313x.2003.01915.x
Yamaguchi, K., Beligni, M. V., Prieto, S., Haynes, P. A., McDonald, W. H., Yates, J. R., et al. (2003). Proteomic characterization of the Chlamydomonas reinhardtii chloroplast ribosome. Identification of proteins unique to the 70S ribosome. J. Biol. Chem. 278, 33774–33785. doi: 10.1074/jbc.M301934200
Xiao, J., Li, J., Ouyang, M., Yun, T., He, B., Ji, D., et al. (2012). DAC is involved in the accumulation of the cytochrome b6/f complex in Arabidopsis thaliana. Plant Physiol. 160, 1911–1922. doi: 10.1104/pp.112.204891
Yin, T., Pan, G., Liu, H., Wu, J., Li, Y., Zhao, Z., et al. (2012). The chloroplast ribosomal protein L21 gene is essential for plastid development and embryogenesis in Arabidopsis. Planta 235, 907–921. doi: 10.1007/s00425-011-1547-0
Yu, D., Jiang, L., Gong, H., Liu, C.-M. (2012). EMBRYONIC FACTOR 19 encodes a pentatricopeptide repeat protein that is essential for the initiation of zygotic embryogenesis in Arabidopsis. J. Integr. Plant Biol. 54, 55–64. doi: 10.1111/j.1744-7909.2011.01089.x
Yu, Q. B., Huang, C., Yang, Z. N. (2014). Nuclear-encoded factors associated with the chloroplast transcription machinery of higher plants. Front. Plant Sci. 5, 316. doi: 10.3389/fpls.2014.00316
Zhang, J., Xiao, J., Li, Y., Su, B., Xu, H., Shan, X., et al. (2017). PDM3, a pentatricopeptide repeat-containing protein, affects chloroplast development. J. Exp. Bot. 68, 5615–5627. doi: 10.1093/jxb/erx360
Zhang, Z., Cui, X., Wang, Y., Wu, J., Gu, X., Lu, T. (2017). The RNA editing factor WSP1 is essential for chloroplast development in rice. Mol. Plant 10, 86–98. doi: 10.1016/j.molp.2016.08.009
Zhang, L., Zhou, W., Che, L., Rochaix, J.-D., Lu, C., Li, W., et al. (2019). PPR protein BFA2 is essential for the accumulation of the atpH/F transcript in chloroplasts. Front. Plant Sci. 10, 446. doi: 10.3389/fpls.2019.00446
Keywords: pigment-defective mutant4, chloroplast, pentatricopeptide repeat protein, development, gene expression
Citation: Wang X, Zhao L, Man Y, Li X, Wang L and Xiao J (2020) PDM4, a Pentatricopeptide Repeat Protein, Affects Chloroplast Gene Expression and Chloroplast Development in Arabidopsis thaliana. Front. Plant Sci. 11:1198. doi: 10.3389/fpls.2020.01198
Received: 11 January 2020; Accepted: 23 July 2020;
Published: 11 August 2020.
Edited by:
Yan Lu, Western Michigan University, United StatesReviewed by:
Zhong-Nan Yang, Shanghai Normal University, ChinaSheng Teng, Shanghai Institutes for Biological Sciences (CAS), China
Jeannette Pfalz, Friedrich Schiller University Jena, Germany
Copyright © 2020 Wang, Zhao, Man, Li, Wang and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianwei Xiao, xiaojianwei@bjfu.edu.cn
 Xinwei Wang1,2
Xinwei Wang1,2 Jianwei Xiao
Jianwei Xiao