- 1Molecular Crop Research Unit, Department of Biochemistry, Chulalongkorn University, Bangkok, Thailand
- 2Amity Institute of Biotechnology, Amity University, Uttar Pradesh, Lucknow Campus, Lucknow, India
- 3Plant Molecular Biology Laboratory, Department of Botany, Dayanand Anglo-Vedic (PG) College, Chhatrapati Shahu Ji Maharaj University, Kanpur, India
- 4Omics Sciences and Bioinformatics Center, Faculty of Science, Chulalongkorn University, Bangkok, Thailand
Calcium (Ca2+) ion is a critical ubiquitous intracellular second messenger, acting as a lead currency for several distinct signal transduction pathways. Transient perturbations in free cytosolic Ca2+ ([Ca2+]cyt) concentrations are indispensable for the translation of signals into adaptive biological responses. The transient increase in [Ca2+]cyt levels is sensed by an array of Ca2+ sensor relay proteins such as calmodulin (CaM), eventually leading to conformational changes and activation of CaM. CaM, in a Ca2+-dependent manner, regulates several transcription factors (TFs) that are implicated in various molecular, physiological, and biochemical functions in cells. CAMTA (calmodulin-binding transcription activator) is one such member of the Ca2+-loaded CaM-dependent family of TFs. The present review focuses on Ca2+ as a second messenger, its interaction with CaM, and Ca2+/CaM-mediated CAMTA transcriptional regulation in plants. The review recapitulates the molecular and physiological functions of CAMTA in model plants and various crops, confirming its probable involvement in stress signaling pathways and overall plant development. Studying Ca2+/CaM-mediated CAMTA TF will help in answering key questions concerning signaling cascades and molecular regulation under stress conditions and plant growth, thus improving our knowledge for crop improvement.
Introduction
Plants have evolved several strategies, majorly through molecular mechanisms, to optimize growth and ameliorate tolerance toward environmental constraints (Tripathi et al., 2020). One of the basic mechanisms is an increase in the concentration of [Ca2+]cyt in response to various external stimulus (Hepler, 2005). Ca2+ signatures are considered as core regulators of many adaptive and developmental processes. They are characterized by stimulus-driven signals resulting from the cumulative action of carriers, pumps, and channels. These transporters drive Ca2+ signals spatially and temporally to act inside a cell. Spatial and temporal Ca2+ signals are transmitted and decoded by a bunch of Ca2+-binding proteins such as calmodulin (CaM) and calcineurin. These proteins further spread relayed information to generate specific downstream response by regulating transcription factors (TFs). TFs are DNA-binding regulatory proteins involved in distinctive expression of genes that regulate developmental processes and environmental stress responses (Hong, 2016; Mitsis et al., 2020). CaM, upon interaction with Ca2+, undergoes conformational changes to modulate several TFs (Reddy, 2001). TFs contribute to various facets of cellular processes and act as a toolkit to signals perceived from within and outside an organism (Mitsis et al., 2020). Intriguingly, TFs constitute a major portion of eukaryotic genome, accounting for approximately 5% of the total genome (approximately 2,000 genes) in humans (Tupler et al., 2001). In Arabidopsis thaliana, 11.8% of their genome encodes for TFs (approximately 3,000 genes) (Arabidopsis Genome Initiative, 2000). TFs such as DREB (Agarwal et al., 2006; Chen et al., 2008; Lata and Prasad, 2011; Jan et al., 2017), NAC (Tran et al., 2004; Nakashima et al., 2007; Puranik et al., 2012; Marques et al., 2017), bHLH (Sun et al., 2018; Wang et al., 2020), WRKY (Park et al., 2005; Phukan et al., 2016), and MYB (Levy et al., 2005; Roy, 2016; Zhang et al., 2018) had been previously reported to play crucial roles in stress biology and plant growth. In a similar vein, CAMTA (calmodulin-binding transcription activator) in a Ca2+/CaM-driven modus had been involved in carrying out important functions by modulating plant stress responses and overall development (Bouche et al., 2002, 2005; Galon et al., 2010a; Liu et al., 2015; Shkolnik et al., 2019). The present review recapitulates the progress made in comprehending the involvement of Ca2+/CaM-mediated CAMTA regulation in stress adaptation and plant development.
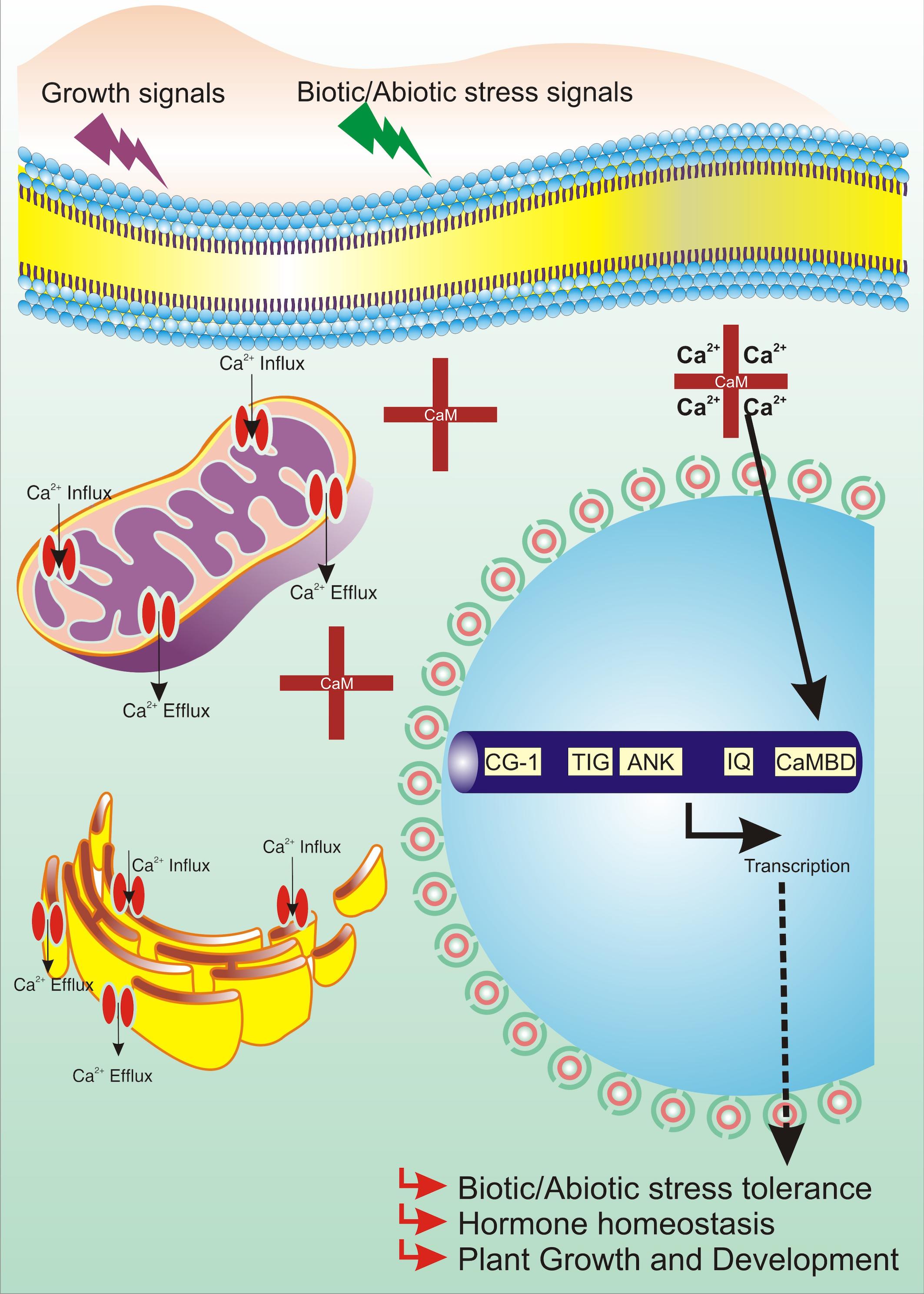
Calcium: A Second Messenger and Its Interaction With Calmodulin
A second messenger is a molecule that acts to transmit signals from a receptor to a target (Bush, 1995). Ca2+ is one of the best characterized second messengers. It is hypothesized that Ca2+, as a messenger ion, emerged early during cell evolution (Edel et al., 2017). The role of Ca2+ as a primary nutrient in sustaining the structural integrity of cell walls and modulating diverse physiological processes such as biotic and abiotic stresses had been elucidated well by extensive biochemical and genetic studies (Poovaiah et al., 1993; Zielinski, 1998; Reddy, 2001; Snedden and Fromm, 2001; Sanders et al., 2002; Harper et al., 2004; Reddy et al., 2004; Bouche et al., 2005; Hepler, 2005; Aghdam et al., 2012; Thor, 2019; Tian et al., 2020). Ca2+ spikes are usually the outcome of two opposing reactions occurring in cells: Ca2+ influx (entry) via dedicated channels or Ca2+ efflux (exit) via specific pumps (Xiong et al., 2006; Tuteja and Mahajan, 2007). The regulation of nanomolar concentration of Ca2+ is achieved by two mechanisms. First, Ca2+-ATPases pump [Ca2+]cyt to the exterior or into organelles such as vacuoles and endoplasmic reticulum. Second, the opening of Ca2+-permeable ion channels results in the Ca2+ influx to the cytosol (Medvedev, 2005; Tuteja and Mahajan, 2007; Swarbreck et al., 2013; Demidchik and Shabala, 2018). Transmembrane Ca2+ gradients maintain sufficient energy to drive Ca2+ flux passively into the cytosol from the apoplast or the organelles (Medvedev, 2005; Swarbreck et al., 2013; Demidchik and Shabala, 2018).
Much evidence indicates that Ca2+-mediated signaling is implicated in the relay of stress signals such as light (Kim et al., 2003; Liang et al., 2009; Hochmal et al., 2015; Hou et al., 2019), temperature (Teige, 2019), salt (Rahman A. et al., 2016; Manishankar et al., 2018; Seifikalhor et al., 2019), cold (Shi et al., 2014; Yuan et al., 2018b), and gravity (Kordyum, 2003; Toyota et al., 2008; Salmi et al., 2011); oxidative signals such as pathogen attack (Kiep et al., 2015; Aldon et al., 2018; Tian et al., 2019) and reactive oxygen species (Mori and Schroeder, 2004; Monshausen et al., 2009; Kurusu et al., 2015); and hormone signals such as ethylene (Li et al., 2018; Zhu et al., 2018), abscisic acid (ABA) (Edel and Kudla, 2016; Chen et al., 2017; Yuenyong et al., 2018), gibberellins (Abbasi et al., 2004; Nakata et al., 2009; Li et al., 2013), and auxins (Vanneste and Friml, 2013; Hazak et al., 2019). In plants, oodles of Ca2+-binding proteins function as Ca2+ sensors decoding complex Ca2+ signatures (Kudla et al., 2018). These binding proteins sense changes in [Ca2+]cyt and/or [Ca2+]nuc, regulating downstream signaling processes and hence drawing physiological response against them (Day et al., 2002; Boonburapong and Buaboocha, 2007). Ca2+ sensors are basically proteins with highly conserved one or multiple helix-turn-helix structures-EF-hand (Nakayama et al., 2000; Day et al., 2002). Approximately, 250 EF-hand comprising putative Ca2+ sensors are known in A. thaliana, which accounts for about 1% of the reported proteome (Day et al., 2002). Ca2+ sensors are divided into three categories, namely, (i) CaM and CMLs (calmodulin-like proteins) (Bender and Snedden, 2013), (ii) CBLs (calcineurin-B-like proteins) (Luan, 2009), and (iii) CPKs (Ca2+-dependent protein kinases) and CCaMK (calcium and calcium/calmodulin-dependent protein kinase) (Cheng et al., 2002).
Calmodulins are a class of extensively studied Ca2+ sensors and are known to regulate diverse cellular processes in plants, including stress responses and plant development (Zielinski, 1998; Bouche et al., 2002; Bergey et al., 2014; Zeng et al., 2015; Dubrovina et al., 2019). CaM is a 17-kDa dumbbell-shaped cytosolic acidic protein, with a supple joint in the middle (Meador et al., 1992). CaMs contain four EF-hand motifs and are highly conserved among eukaryotes (Zielinski, 1998; Reddy et al., 2002). It has been ascertained that around 300 proteins interact with CaMs in plants (Reddy et al., 2002; Bouche et al., 2005; Popescu et al., 2007). Among the known protein–protein interactions, CaMs are implicated to have maximum interacting partners (Lee et al., 2010). Conformational changes upon Ca2+/CaM association lead to contact of hydrophobic surface within each domain (Liu et al., 2017). This ultimately elevates the CaM’s Ca2+ sensor bustle, thus resulting in an interaction with its target proteins (Snedden and Fromm, 2001). CaM can act either directly by interacting with key target enzymes or indirectly with the help of specific kinases (Zielinski, 1998; Mt and Mn, 2008; Villalobo et al., 2019). Interestingly, Ca2+ ions interact with CaM in a cooperative manner, such that even small changes in the level of [Ca2+]cyt lead to huge alteration in the levels of active CaM (Beccia et al., 2015; Fernandes and Oliveira-Brett, 2017). Various CaM proteins display differential expression and possess variable affinity to Ca2+, including their downstream target proteins (McCormack et al., 2005; Popescu et al., 2007; Chinpongpanich et al., 2015). Many Ca2+- and Ca2+/CaM-associated TFs, implicated in stress signaling, are found in plants (Singh and Virdi, 2013; Shen et al., 2020). However, few TFs are specifically transcribed to distinct Ca2+ signal durations and amplitude. For example, the binding of Ca2+ can directly regulate the activity of certain TFs such as the DREAM (downstream regulatory element antagonist modulator) protein (Carrion et al., 1999). Similarly, AtNIG1 (A. thaliana NaCl-inducible gene 1) is also classified as a Ca2+-dependent TF, which is targeted in the nucleus, indicating that it is a nuclear Ca2+-binding protein (Kim and Kim, 2006). Taking Ca2+/CaM-associated TFs into account, AtGT2L, a member of GT-2 subfamily, upon interaction with Ca2+/CaM, is responsive to freezing and salinity stresses in plants (Xi et al., 2012). WRKY is also implicated to be a Ca2+/CaM-dependent gene (Park et al., 2005; Yan et al., 2018). AtWRKY7 is well reported to be involved in Ca2+/CaM-dependent gene regulation and has a role in pathogen incursion (Park et al., 2005). Experimental results have revealed that CaM binds exclusively to the Ca2+-regulated CaM-binding domain (CaMBD) of AtWRKY7 (Park et al., 2005). In addition, WRKY45, WRKY43, WRKY53, and WRKY50 bind to various isoforms of CaM in a Ca2+-driven behavior (Popescu et al., 2007). Myb is another TF that is well characterized as a Ca2+/CaM-dependent protein. It acts upstream to a number of defense-responsive, salinity, and drought-receptive genes (Do Heo et al., 1999; Stracke et al., 2001; Li et al., 2019). CaM also supports transcriptional repression interceded by CBNAC/NTL9 (Kim et al., 2007). The TF ZmNAC84 has been reported to interact physically with ZmCCaMK both in vivo and in vitro. ZmNAC84, after ABA induction, has a partially overlapping expression profile with ZmCCaMK. Its overexpression renders drought tolerance and resistance to oxidative stress induction owing to drought (Zhu et al., 2016). Moreover, Cam7, also known as ZBF3, is a CaM isoform in A. thaliana that works as a transcriptional regulator. The upstream elements to TSS of light-inducible genes are directly targeted by ZBF3 to promote photomorphogenesis (Yadav et al., 2005; Kushwaha et al., 2008). In a similar context, recently, CaM proteins have also been implicated in salt stress. “Khao Dawk Mali 105” rice variety, which overexpressed OsCam1-1, shows differential expression of several genes implicated in salt stress signaling, hormonal regulation, lipid/carbohydrate/secondary metabolism, photosynthesis, etc. Owing to OsCam1-1 overexpression, the rate of photosynthesis declines in transgenic rice, whereas content of sucrose and starch increases under salt stress. This study further revealed that CaM under salt stress condition boosts several metabolic enzymes implicated in energy pathways of plant cells, which either conserve or generate energy under limited photosynthesis (Yuenyong et al., 2018). Thus, Ca2+/CaM acts as an intermediate complex between stimulus perception and gene expression (Figure 1).
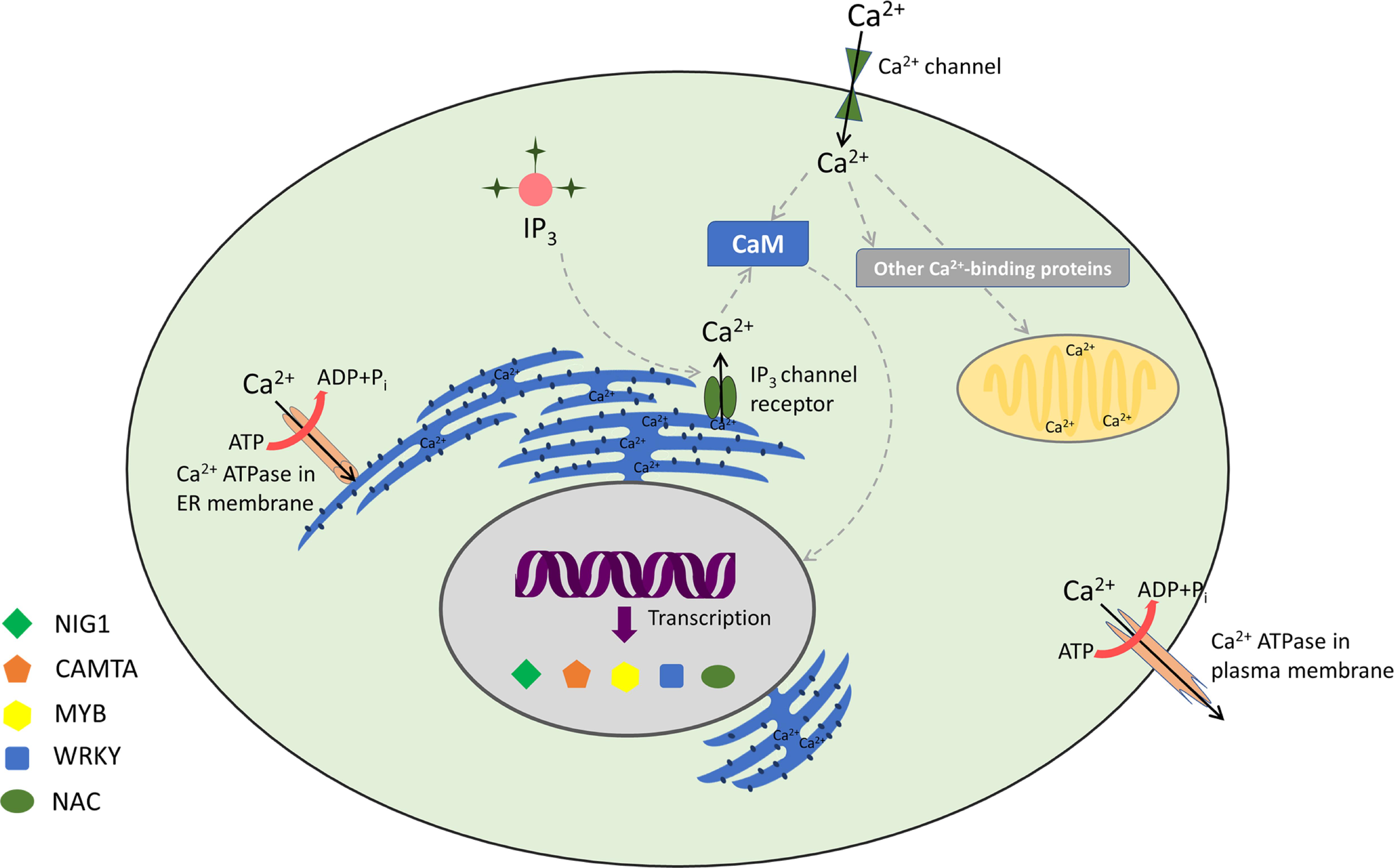
Figure 1. Decoding of calcium signatures: calcium signaling and its sensors with associated transcription factors.
Calmodulin-Binding Transcription Activator
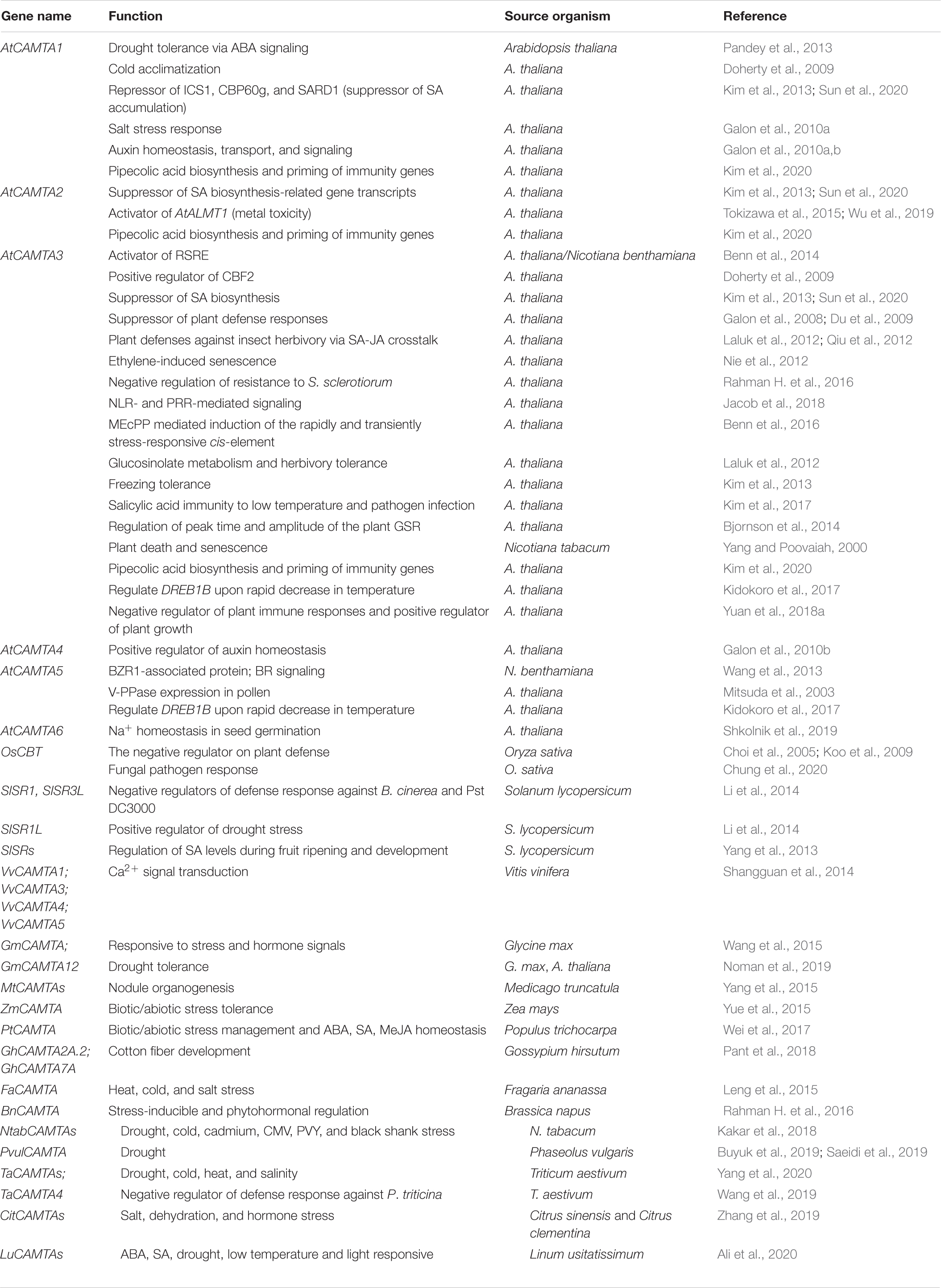
Calmodulin-binding transcription activator also known as SR (signal responsive) protein (Yang and Poovaiah, 2000) or EICBP (ethylene-induced CaM-binding proteins) (Reddy et al., 2000) is considered as the largest and best characterized family of CaM-binding TFs (Finkler et al., 2007; Kim et al., 2009). The presence of this novel protein that binds to DNA was for the very first time identified during the process of isolation of partial cDNA clone (CG-1) from parsley (Petroselinum crispum) (e Silva, 1994). It was shown that CGCG was the signature motif required for the binding of this protein to DNA. Upon exposure to UV-containing white light, the CG-1 coding mRNA was rapidly and transiently accumulated in parsley cultured cells; however, until then, CG-1 was not designated as a Ca2+/CaM-binding protein (e Silva, 1994). Later, Yang and Poovaiah (2000) identified NtER1 as a CaM-binding protein engaged in ethylene-regulated plant death and senescence, indicating its potential application in prolonged storage life of horticultural crops. They also showed that CaM binds with a very high affinity (Kd∼12 nM) in a Ca2+-dependent manner to NtER1 (Yang and Poovaiah, 2000). Yang and Poovaiah (2002) further reported a homolog of NtER1 and five related genes in Arabidopsis (AtSRs). Their results evidently suggested AtSRs as a CaM-binding gene family with CGCG as a core motif in their promoters. The study also indicated probable implication of CAMTAs in numerous signal transduction pathways in plants (Yang and Poovaiah, 2002). Since then, CAMTA TF had been reported in various multicellular organisms. With the advancement in computational techniques, proteins similar to CAMTA had been reported in unicellular eukaryotes such as the ciliates, Paramecium tetraurelia and Tetrahymena thermophila (Finkler et al., 2007). The orthologs of CAMTA had also been reported in Arabidopsis (Yang and Poovaiah, 2002; Bouche et al., 2005); humans (Song et al., 2006); flies (Han et al., 2006); and in various crop plants such as tobacco (Yang and Poovaiah, 2000), rice (Choi et al., 2005), tomato (Yang et al., 2012), grapevine (Shangguan et al., 2014), corn (Yue et al., 2015), strawberry (Leng et al., 2015), barrelclover (Yang et al., 2015), soybean (Wang et al., 2015), poplar (Wei et al., 2017), and cotton (Pant et al., 2018). Deciphering the CAMTA gene expression patterns and functional redundancies in model plants and various crops laid a foundation for further investigation of its role, which can be beneficially applied to agricultural sector. CAMTA as a TF has been extensively studied for its involvement in stress and developmental biology (Galon et al., 2008; Doherty et al., 2009; Pandey et al., 2013; Sun et al., 2020) (discussed in CAMTA Transcriptional Response Under Abiotic Stress Conditions and CAMTA Transcriptional Response Under Biotic Stress Conditions).
The most important characteristic of this TF is its affinity for CaM, indicating its participation in Ca2+ sensing. Ca2+ signatures are effectively decoded by CAMTAs to generate explicit gene expression patterns (Liu et al., 2015). Liu et al. deciphered how different Ca2+ signatures are decoded by CAMTAs for specific gene expression patterns. The group devised a dynamic model based on thermodynamic and kinetic principles for understanding the Ca2+–CaM–CAMTA binding and the subsequent gene expression patterns. The modeling analysis unraveled that Ca2+ signals in the form of elevated [Ca2+]cyt are non-linearly amplified upon binding of Ca2+, CaM, and CAMTAs. They further combined experimental data with mathematical modeling to comprehend the information flow from Ca2+ signatures to CAMTA−regulated gene expression patterns (Liu et al., 2015). This study provided great insight about the involvement of Ca2+/CaM in regulation of CAMTAs. The succeeding section focuses on the transcriptional machinery of CAMTA with respect to its molecular and physiological functions in regulating stress responses and overall plant development.
CAMTA Domain Organization
Calmodulin-binding transcription activators were reported to consist of multiple functional domains (Finkler et al., 2007). These domains have evolutionary conserved amino acid sequence, organized into a specific conserved order. The predicted functional domains of CAMTA include (a) NLS (nuclear localization signals) for targeting protein into nucleus. These signals have been reported in almost all the CAMTAs, but their localization varies in different organisms; (b) CG-1 domain, which is a unique domain implicated in DNA binding; for any protein to be characterized as CAMTA, the presence of CG-1 domain is obligatory; (c) TIG, which is involved in non-specific DNA interactions in TFs (Aravind and Koonin, 1999) and also implicated in protein dimerization (Muller et al., 1995); (d) ankyrin (ANK) repeats, which are tandem repeats of about 33 amino acids in a variety of eukaryotic and viral proteins and contribute in protein–protein interactions (Sedgwick and Smerdon, 1999; Rubtsov and Lopina, 2000); (e) CaMBD, which is implicated in the association of Ca2+-loaded CaM to CAMTAs; (f) IQ motifs, which are implicated in the binding of CaM and CaM-like proteins and are regions of low complexity with a consensus motif IQXXXRGXXX (Bahler and Rhoads, 2002). In some cases, the presence of transcription activation domains (TADs) has also been mapped. The presence of TAD was reported in AtCAMTA1 (Bouche et al., 2002), HsCAMTA2 (Song et al., 2006), and DmCAMTA (Han et al., 2006). However, owing to lack of sequence homology in this domain, it cannot be inferred that all CAMTAs possess TAD. All these domains are pivotal for the transcription of CAMTA. Nonetheless, certain variations exist; for example, the mapping of Ca2+-loaded CaM domain on AtCAMTA1 reveals a binding site adjacent to IQ motif, indicating multiple CaM binding sites (Bouche et al., 2002). Similarly, in OsCAMTA, the Ca2+-dependent CaMBD and Ca2+-independent CaM dissociation domains were mapped on C-termini (Choi et al., 2005). Recently, in Gossypium species, GrCAMTA5.2, GrCAMTA5.3, and GhCAMTA3D.1, GhCAMTA5D.1 were depicted as TIG lacking CAMTAs in Gossypium raimondii and Gossypium hirsutum, respectively (Pant et al., 2018). Similar to Gossypium study, five TaCAMTAs (Triticum aestivum CAMTA), namely, TaCAMTA1-A/B/D and TaCAMTA5-A/D, also lacked TIG domain (Yang et al., 2020). The domain organization of six Arabidopsis CAMTAs, analyzed bioinformatically, is shown in Figure 2.
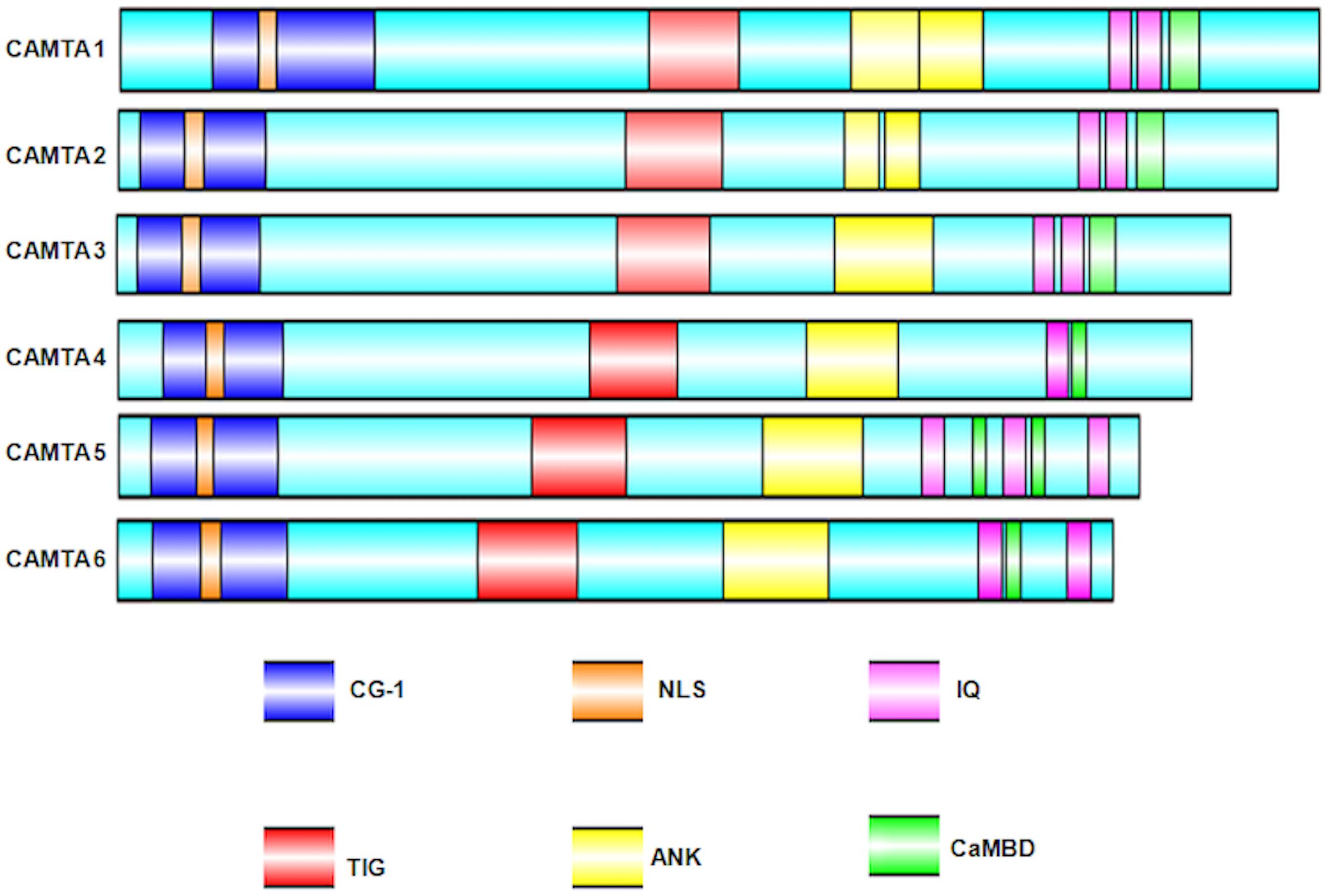
Figure 2. Schematic representation of the Arabidopsis thaliana CAMTA domain organization. Domain organization was obtained using NCBI/BLAST/CD-SEARCH (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). NLS was specifically searched in Motif scan (http://myhits.isb-sib.ch/cgi-bin/motif_ scan). CaMBD were searched using Calmodulin Target Database (http://calcium.uhnres.utoronto.ca/ctdb/ctdb/). The domain structures were drawn using Illustrator for Biological sequences software (http://ibs.biocuckoo.org/).
CAMTA Transcriptional Response Under Abiotic Stress Conditions
The underpinning mechanisms of CAMTA under abiotic stress conditions had been reasonably elucidated (Pandey et al., 2013; Noman et al., 2019; Shkolnik et al., 2019). Under normal conditions, where plants are not exposed to any stress, there is minimal induction of CAMTA genes. This could be either because the genes in this family of TFs share functional redundancy or the genes in this family are expressed under certain specific environmental conditions. Taking the model plant A. thaliana into account, six CAMTAs were reported: CAMTA1, CAMTA2, CAMTA3, CAMTA4, CAMTA5, and CAMTA6. It was shown that all these genes were rapidly and differentially triggered by environmental cues such as high temperatures, salinity, H2O2, physical wounds; and hormones such as ethylene, ABA, methyl jasmonate (MeJA), and salicylic acid (SA) (Yang and Poovaiah, 2002; Kim et al., 2013, 2017; Pandey et al., 2013; Shkolnik et al., 2019). Considering the first member of Arabidopsis CAMTA gene family, AtCAMTA1, its role has been reported in auxin signaling (increased expression of AUX/IAA-IAA29), transport, and homeostasis. For transgenic plants possessing AtCAMTA1 promoter, GUS construct displayed cell-specific expression profiles of auxin. The involvement of CAMTA1 in auxin signaling is established by chemical impairment of polar auxin transport. Gene expression profiling of auxin transport inhibitor 1-N-naphthylphthalamic acid to plants also supports the responsiveness of CAMTA1 in auxin pathways. Genome-wide transcriptome of camta1 mutant mapped 63 upregulated genes. Further analysis revealed that 17 genes were involved in auxin signaling. Moreover, on analysis of hypocotyl elongation, the camta1 mutants were hyperresponsive to auxin exposure as compared to wild type (Galon et al., 2010a). It was further reported that CAMTA1, CAMTA2, and CAMTA3 are negative regulators of auxin and are associated with genes responsible for red light and high light responses, whereas CAMTA4, CAMTA5, and CAMTA6 are positive regulators of auxins and are associated with genes responsible for blue light and darkness responses (Galon et al., 2010b). In a similar vein, phytohormonal regulation of CAMTAs is also extended to brassinosteroid (BR) signaling in which BZR1 (involved in BR signaling cascade) has CAMTA5 as an associated protein (Wang et al., 2013).
Additionally, camta1 responded severely to cold stress, indicating its role in cold stress management. CAMTA protein provides a much-needed connecting link between Ca2+ signatures and cold acclimatization. CBF TF plays a pivotal role in cold acclimation of plants. In A. thaliana, CBF genes induce approximately 100 more genes (termed as CBF regulon) upon exposure to low temperatures (Maruyama et al., 2004; Vogel et al., 2005). CAMTA binds to consensus sequence located in the promoter of the CBF2 gene. The 1,000-bp upstream region of CBF2 gene comprises seven conserved consensus motifs: CM1 to CM7. The CAMTA signature domain CG-1 matches the CM2 motif of CBF2. CBF2 expression is impaired in camta3 mutants and impaired to a much higher extent in camta1 and camta3 double mutant. These are indications of the intersecting functions of different CAMTA proteins. Therefore, CAMTA controls CBF regulon, which confers freezing tolerance to plants (Doherty et al., 2009). Moreover, CAMTA1, CAMTA2, and CAMTA3 work in recital to induce CBF1, CBF2, and CBF3 and confer freezing tolerance to plants at low temperatures. Furthermore, at warmer temperatures, these three CAMTAs collectively inhibit SA biosynthesis. It had been previously shown that exposure to low temperatures increases SA levels (Scott et al., 2004). SA biosynthesis at low temperatures involves ICS (isochorismate synthase) pathway, encircling ICS1, CBP60g, and SARD1 genes. At warm temperatures, CAMTA1, CAMTA2, and CAMTA3 suppress the accretion of ICS1, CBP60g, and SARD1. Such suppression by CAMTA proteins was not observed at low temperatures (Kim et al., 2013). More neoteric inventions have elucidated that the suppression of genes involved in SA biosynthesis by CAMTA at high temperatures (22°C) is overcome when plants are infected by biotrophic or hemibiotrophic pathogens and grown at low temperatures (4°C) for more than a week. Suppression of SA pathway by CAMTA3 under normal condition involves an N-terminal repression module (NRM). NRM works autonomously of CaM binding to CaMBD or the IQ. This finding is different owing to the fact that CAMTA3 repressional assertion involves the binding of CaM to CaMBD. To sum up, the repression activity of CAMTA3 at low temperatures and pathogen infection is governed by related mechanisms (Kim et al., 2017). To further elaborate, three dehydration-responsive element (DRE) binding protein 1/C-repeat binding factors (DREB1/CBFs) are master regulators of cold-responsive gene expression. DREB1 stimulates cold-responsive transcriptional cascade, eventually inducing several genes implicated in cold stress response. Rapid and gradual temperature decreases are sensed as cold stress by plants, which results in the activation of DREB1 genes. CAMTA3 and CAMTA5 are responsive to rapid decline in temperatures by inducing the expression of DREB1s. However, CAMTA3 and CAMTA5 do not respond to gradual declines in temperatures. In contrast to circadian clock associated 1 and late elongated hypocotyl (acts as transcriptional activators to regulate DREB1 expression only during the day), AtCAMTA3 and AtCAMTA5 function both during the day and night (Kidokoro et al., 2017).
Drought and salinity are considered as few of the worst conditions in agriculture and horticulture. The role of CAMTA1 has been extended to drought response (Pandey et al., 2013). The mutants had impaired photosystem II efficiency and water use efficiency, hence making plants susceptible to drought. The mutants had relatively low water content and retarded growth. Microarray analysis revealed drought revival, osmotic balance, apoptosis, DNA methylation, and photosynthesis as investigative pathways in drought-treated camta1 mutants. It has also been shown that CAMTA1 regulates a broad spectrum of stress-inducible genes such as RD26, ERD7, RAB18, LTPs, COR78, CBF1, and HSPs. To ascertain the claims, CAMTA recognition cis-element was found to be enriched in the 1,000-bp upstream regions of the listed genes (Pandey et al., 2013). Very recently, CAMTA6 has been shown to be associated with sodium (Na+) homeostasis during early seed germination. The camta6 mutants accumulate less NaCl and display tolerance to salinity and ABA. AtHKT1 (high affinity K+ transporter 1 encoding a Na+/K+ transporter) expression was limited to radicles and did not increase upon subjecting camta6 to salt stress or ABA treatment. In addition, even the transcriptome of CAMTA6 under control and salt stress condition revealed 1,020 upregulated and 1,467 downregulated salt-responsive genes in the wild type (Shkolnik et al., 2019). With regard to context, although CAMTA2 in plants has been less explored, studies highlight the regulation of AtALMT1 (aluminum-activated malate transporter 1) by CAMTA2 in association with WRKY46 (Tokizawa et al., 2015; Wu et al., 2019). Next, considering the involvement of CAMTAs in plant development, the role of CAMTA1 and CAMTA5 is also extended in pollen development. The AVP1 promoter activity has been reported to be activated by AtCAMTA1 in cultured cells. AtCAMTA1 triggers the GUS reporter expression driven by P281 promoters of AVP1 but does not stimulate the GUS reporter expression in mutants with disrupted despaired CGCG-box, indicating that AtCAMTA1 attaches to CGCG-box and elevates AVP1 expression. AtCAMTA1 in collaboration with AtCAMTA5, cooperatively elevates their gene function and enhance AVP1 expression during pollen development (Mitsuda et al., 2003). Apart from A. thaliana, CAMTAs from other plant species have also been implicated very well in abiotic stress responses. Various in silico and quantitative reverse transcription–polymerase chain reaction (qRT-PCR) approaches were deployed to assess the involvement of CAMTAs under different abiotic stress conditions. For example, Zea mays (corn) ZmCAMTAs in drought, cold, salt, and hormonal signaling (Yue et al., 2015); Fragaria ananassa (Strawberry) FaCAMTAs in heat, cold, salt, and ethylene stress (Leng et al., 2015); T. aestivum (wheat) TaCAMTAs in cold, salt, heat, and drought stress (Yang et al., 2020); Citrus sinensis (sweet orange) CitCAMTAs in salinity, dehydration, and hormonal response (Zhang et al., 2019); and Linum usitatissimum (Flax) LuCAMTAs in drought, low temperature, and light responses (Ali et al., 2020) (discussed in detail in Recent Discoveries of CAMTAs in Various Crop Plants).
CAMTA Transcriptional Response Under Biotic Stress Conditions
Transient Ca2+ levels are known to be elevated during pathogen attack (Ma and Berkowitz, 2007), which eventually activates CaM. Similar to the role of CAMTA1 in abiotic stress tolerance, the role of CAMTA3 in response to biotic stress has been elucidated to a great extent (Galon et al., 2008; Laluk et al., 2012; Benn et al., 2016; Kim et al., 2017, 2020; Jacob et al., 2018). Ca2+-loaded CaM regulates the activity of CAMTA3 either by negative regulation of CAMTA3 or by positive regulation of a negative repressor TF (Galon et al., 2008). The bacterial pathogen Pseudomonas syringae and the fungal pathogen Botrytis cinerea did not heavily affect camta3 in comparison to their effect in control plants. This is suggestive of the fact that CAMTA3 is responsible for suppression of biotic defense responses, which may be primarily achieved either by binding of CAMTA3 with the promoters of suppressed genes or by expressing a TF that is repressed. Transcriptomics approach on camta3 revealed an attenuated expression of six genes and an enhanced expression of 99 genes out of which 32 genes are related to defense response such as WRKY33, PR1, chitinase, etc. Apart from model plants, the role of CAMTA3 in biotic response was also extended to crop plants. OsCBT encoding CaM-binding protein was functionally characterized in plant defense. Oscbt-1 mutants were significantly tolerant to rice blast fungus Magnaporthe grisea and the bacterial pathogen Xanthomonas oryzae (Choi et al., 2005; Koo et al., 2009). Furthermore, for the identification of transcriptional network governed by OsCBT, transcriptome analysis between wild-type (WT) and oscbt-1 rice grown in pathogen-free conditions was performed. The analysis revealed 81 up-regulated genes and 200 down-regulated genes. Majority of these genes were implicated in biotic stress responses. The gene expression patterns of fungal pathogen response were also found to be significantly affected in oscbt-1 mutant, suggesting the probability of OsCBT in regulating rice defense response (Chung et al., 2020).
Pathogen attack on plants causes intracellular Ca2+ spikes; however, how Ca2+ mediates this interaction-based SA level fluctuation remains unclear. CAMTA3 has been reported to link Ca2+ transients to SA-mediated immune response. CAMTA3 also interacts with EDS1 promoter to impair its expression (Du et al., 2009). Further research has led to the establishment of the role of CAMTA3 in plant defense and ethylene-induced senescence (Nie et al., 2012). Edr2 (enhanced disease resistance 2) suppressors were screened to establish the components of EDR2 pathways. This led to the identification of gain-of-function mutation in signal responsive1 (SR1) or CAMTA3. Edr2 mutants have enhanced tolerance for Golovinomyces cichoracearum (powdery mildew), indicating that EDR2 is a negative regulator for powdery mildew. The sr1-4D mutation restrains edr2-mediated powdery mildew tolerance. Post mildew infection, as compared to wild type, the edr2 plants displayed necrotic lesions with little powder. However, the edr2 sr1-4D plants phenotypically resemble that of the control plants with oodles of conidia on leaves. The sr1-4D gain-of-function also represses elevated ethylene induced senescence. Thus, CAMTA3 negatively impacts plant immune responses. It also directly interacts with the promoter of NDR1 (non-race specific disease resistance1), a key module in resistance to P. syringae–mediated plant immunity. Moreover, CAMTA3 also regulates EIN3 (ethylene insensitive3), which contributes to ethylene-induced senescence (Nie et al., 2012).
As discussed previously, CAMTA1, 2, and 3 play pivotal roles in SA-mediated plant immunity. Pip (pipecolic acid) regulates the process of systemic acquired resistance (SAR) in plants (Bernsdorff et al., 2016). CAMTA1, 2, and 3 suppress the synthesis of Pip by inducing ALD1 (AGD2-like defense response protein 1). ALD1 is known to encode an enzyme required for Pip biosynthesis. The induction of ALD1 results in an accumulation of Pip, which in turn increases the concentration of NPR1 (SA receptor protein). Thus, CAMTA123 triple mutation induces plant defense machinery and instigates SAR (Kim et al., 2020). Moreover, CBP60g and SARD1 TFs are regulators of SA and NHP (N-hydroxypipecolic acid). Both SA and NHP have been extensively linked to plant immunity. CBP60g has been identified as a direct target of CAMTA3 through chromatin immuno-precipitation (ChIP) and electrophoretic mobility shift assay (EMSA) assays. Thus, CAMTAs suppress SA and NHP biosynthesis by regulating SARD1 and CBP60g expression (Lenzoni et al., 2018; Sun et al., 2020).
CAMTA3, as defense against insect herbivory, has also been studied in plants (Qiu et al., 2012). Loss-of-function mutation of AtSR1 suppresses resistance to feeding by Trichoplusia ni (generalist herbivore), sustaining radically elevated larval weight gains and jasmonate (JA) accumulation owing to wound. The responsiveness of AtSR1 mutant to T. ni is attributed to low glucosinolate (GS) levels with remarkably decreased levels in indol-3-ylmethyl (I3M) and 4-methylsulfinylbutyl (4MSOB), the two well-characterized herbivory deterrents. Induction of the various genes implicated in GS metabolism such as IQD1, MYB51, and AtST5a also changes with alterations in AtSR1 transcript levels. Thus, AtSR1 through Ca2+ signaling cascade acts as a key module of plant resistance to insect herbivory. Additionally, Ca2+/CaM-dependent signaling plays a major role in GS metabolism through CAMTA3 involvement (Laluk et al., 2012). Furthermore, upon pathogen exposure, plants allocate their energy resources (at the expense of growth) to defend themselves against invading pathogens. Yuan et al. (2018a) well-documented AtSR1 as a negative regulator of plant immune responses ( PTI-, ETI-, SA-, and JA-mediated signaling pathways) and a positive regulator of plant growth (auxin and BR signaling pathways). EMSA and ChIP assays demonstrated that AtSR1 maintains a strong balance between plant immunity and growth by interacting with the CGCG motif present in the upstream sequences of its potential target genes (Yuan et al., 2018a). The same has also been previously established by Li et al. (2014), who identified seven SISR/CAMTA genes in tomato (Solanum lycopersicum). Functional analysis using VIGS indicated that both SlSR1 and SlSR3L negatively regulate plant defense responses. SISRs were explicitly induced upon P. syringae pv. tomato (Pst) DC3000 and B. cinerea infection. The knockouts of SlSR1 or SlSR3L displayed higher tolerance to Pst DC3000 and B. cinerea with subsequent accumulation of H2O2. Moreover, the genes implicated in pattern-triggered immunity, defense responses, and ethylene and SA pathways were significantly elevated (Li et al., 2014).
To cope with environmental constraints, plants have synchronized mechanisms underlying GSR (general stress response) networks and stress-specific networks (Benn et al., 2014). RSRE (rapid stress response element), the functional motif of GSR, has been extensively studied to explore the new horizons of stress signaling (Walley and Dehesh, 2010). CAMTA3 has been very well studied as a pivotal modulator of RSRE-mediated GSR (Bjornson et al., 2014). The function of CAMTA in RSRE response was hypothesized on the basis that Ca2+ ion inducers, flagellin22 (flg22) and oligogalacturonic acid, enhance the RSRE activity, and Ca2+ ion chelator, EGTA, reduces the RSRE activity upon wounding. CAMTA3 lies downstream of MEKK1, facilitating the regulation of peak time and amplitude of plant GSR and, in conjugation with CAMTA2 and CAMTA4, stimulates RSRE (Benn et al., 2014; Bjornson et al., 2014). Moreover, the methylerythritol cyclodiphosphate (MEcPP)–triggered induction of GSR by transduction of CAMTA3 has also been reported (Benn et al., 2016). CCaMKS and CDPKs all together act to relay the Ca2+ signals essential for RSRE activation. Induction of RSRE by Ca2+ outburst is in accordance with the defined and established role of this transducer in stress signaling (Dodd et al., 2010). CAMTA3 conceivably has also been hypothesized to negatively regulate PTI (PAMP-triggered immunity) and defense by direct targeting of BAK1 and JIN1 through suppression of JA signaling pathway (Rahman H. et al., 2016). CAMTA3 also controls the transcriptional regulation of early convergence of leucine-rich repeat–containing protein (NLR) and pattern recognition receptor (PRR) signaling. CAMTAs, being a probable target of pathogen effectors, promote pathogen virulence. Furthermore, camta3 mutants activate NLRs, resulting in host cell death. Deciphering the NLR-driven CAMTA activity underlines the connection of CAMTA in plant innate immunity (Lolle et al., 2017; Jacob et al., 2018). Hence, ever since its discovery, CAMTAs have been implicated in various developmental and cellular processes either themselves acting as a key TF or targeting another TF through direct or indirect association. The overall involvement of CAMTAs in diverse molecular processes is shown diagrammatically in Figure 3.
Recent Discoveries of CAMTAs in Various Crop Plants
The function of CAMTAs has been established in crop plants like corn (Z. mays), strawberry (F. ananassa), barrelclover (Medicago truncatula), poplar (Populus trichocarpa), soybean (Glycine max), grapevine (Vitis vinifera), tomato (S. lycopersicum), and cotton (Gossypium spp.), where they were previously and largely unknown (Yang et al., 2013, 2015; Shangguan et al., 2014; Leng et al., 2015; Wang et al., 2015; Yue et al., 2015; Wei et al., 2017; Pant et al., 2018). Nine ZmCAMTAs have been reported, upon scanning the entire genome of maize. These CAMTAs were shown to share homology with rice CAMTA. Moreover, an exceedingly high degree of sequence and structural pattern existed among the ZmCAMTAs, indicating that their origin is from a single ancestral sequence. All these CAMTAs possess highly tissue-specific expression and display slight diversity in their gene organization. The qRT-PCR of ZmCAMTAs under various abiotic stress (drought, salt, and cold), biotic stress (RBSDV-rice black-streaked dwarf virus infection), and stress hormones (ABA, auxin, SA, and MeJA) reveals its role as a stress-responsive gene. The occurrence of many stress-related cis-regulatory element in the 1.5-kb promoter region of ZmCAMTAs further confirmed its role in stress regulation (Yue et al., 2015). Taking into account tomato (S. lycopersicum), a horticulturally important crop, signal-responsive (SR)/CAMTA TF is well implicated in postharvest biology. SlSRs, in response to stress conditions, also coordinate Ca2+ signaling with signal transduction pathways implicated in stress during fruit ripening and storage, thus indicating its significant role in horticultural sector (Yang et al., 2013). As previously discussed, SlSRs are involved in plant defense responses and drought management. In S. lycopersicum, SlSR1 and SlSR3L negatively regulate plant defenses against B. cinerea and Pst DC3000. Conversely, SlSR1L is involved in the positive regulation of drought stress responses (Li et al., 2014).
In F. ananassa, 4 CAMTAs, namely, FaCAMTA1, FaCAMTA3, FaCAMTA4, and FaCAMTA5, have been reported, upon the exploitation of BLAST and HMMER tool bioinformatically, using the CAMTA family domain file. All these CAMTAs possessed known domain organization comprising CG1, TIG, ANK repeats, and IQ motifs. Under heat, cold, salt, and ethylene stresses, the FaCAMTAs exhibit distinct expression patterns (Leng et al., 2015). CAMTA genes have also been identified in M. truncatula genome, specifying its role in nodule organogenesis. The MtCAMTA genes display tissue-specific expression and respond to various stress-related hormones. Upon Sinorhizobium meliloti infection, the expression profiling of the MtCAMTA genes revealed that there is an alleviation in the suppression of most MtCAMTA genes expression, indicating the CAMTA involvement in early nodulation signaling pathway. Moreover, the promoters of early rhizobial infection response genes display the presence of CAMTA binding motifs (Yang et al., 2015). Similarly, seven CAMTAs were identified in the genome of P. trichocarpa, and their expression patterns were analyzed in roots and leaves. The qRT-PCR analysis showed that their expression was induced by pathogenic infection with Alternaria alternata; mannitol, NaCl, and cold stress; and phytohormones including SA, ABA, and MeJA (Wei et al., 2017). This was further substantiated as their promoter analysis revealed that most PtCAMTAs contain phytohormone or stress-related cis-regulatory elements. In G. max, 15 CAMTA genes were identified, sharing sequence homology with their Arabidopsis counterparts. All GmCAMTAs were profoundly expressed in root and leaf tissue. Their transcript abundance was induced significantly upon dehydration, cold, salinity, and hormonal treatments such as ABA, SA, and MeJA. In concurrence to their receptiveness to these signals, the promoter regions of these genes have been reported to be enriched with stress-related cis-regulatory elements (Wang et al., 2015). Complete functional analysis of GmCAMTA12, in relation to drought stress, has been performed. The promoter region of GmCAMTAs contained ABRE, SARE, G-box, and W-box cis-regulatory elements. Arabidopsis T3 overexpression lines of GmCAMTA12 display enhanced drought tolerance and increased survival and germination rate under drought. GmCAMTA12 overexpression lines performed better in terms of physiological parameters such as proline and malondialdehyde contents, catalase activity, and electrolyte leakage (Noman et al., 2019). In V. vinifera, 10 SR/CAMTAs were identified bioinformatically as Ca2+/CaM-binding protein, belonging to four gene groups: VvCAMTA1, VvCAMTA3, VvCAMTA4, and VvCAMTA5 and localized on 5, 7, 1, and 5 chromosomes, respectively. Expression analysis of VvCAMTAs depicted its role mainly in Ca2+ signal relay, with significantly high expression in bud, fruit, and inflorescence (Shangguan et al., 2014). Additionally, in four different Nicotiana species (Nicotiana tabacum, Nicotiana sylvestris, Nicotiana tomentosiformis, and Nicotiana benthamiana), 29 CAMTAs (13 in N. tabacum, 6 in N. sylvestris, and 5 each in N. tomentosiformis and N. benthamiana) were identified by evolutionary and expression analyses, which provided great insights into CAMTAs origin, expansion, and response toward stress conditions and plant development (Kakar et al., 2018). NtabCAMTAs are among the early responsive genes to biotic stress (cucumber mosaic virus-M -M strain of CMV, potato virus Y -Mn strain of PVY, and fungal pathogen black shank or Phytophthora nicotianae) and played pivotal roles in plant defense. NtabCAMTAs were also significantly responsive to abiotic conditions such as cold, drought, and cadmium stress (Kakar et al., 2018).
The identification and functional characterization of CAMTA gene family were also extended to cotton. Six CAMTA members were identified in Gossypium arboreum, 7 in G. raimondii, and 9 in G. hirsutum. Segmental duplication and purifying selection play key roles in the expansion and evolution of CAMTAs in cotton genome. Expression profiling indicated that cotton CAMTAs were predominantly expressed in various stages of fiber development (0, 6, 9 12, and 25 days after anthesis). Precisely, the specific involvement of GhCAMTA2A.2 and GhCAMTA7A in maintaining fiber strength has been reported (Pant et al., 2018). CAMTA gene family has also been classified in Phaseolus vulgaris (common bean). P. vulgaris contained 11 chromosomes, of which five chromosomes harbored eight CAMTA genes with 11 to 12 introns per gene. PvulCAMTAs shared similarity with GmCAMTAs owing to similar genome organization. In silico analysis revealed that PvulCAMTAs are involved in salt stress signaling and were further substantiated through RNA-seq and qRT-PCR analyses (Buyuk et al., 2019). In concurrence to the above study, PvulCAMTA1 has also been characterized to impart drought tolerance (Saeidi et al., 2019). CAMTA gene family had also been well reported in citrus (Zhang et al., 2019). Nine CAMTA genes were found in the citrus (C. sinensis and Citrus clementina) genomes. Most of the CitCAMTAs were found to be highly conserved during evolution. All CitCAMTA genes (except CitCAMTA4) were expressed in minimum one plant tissue corroborating their involvement in leaf, root, stem, seed, cotyledon, and fruit development. Hormone and stress experiments revealed varying expression profiles of CitCAMTAs, indicating stress adaptation. Upon salt stress (0- to 24-h treatment), CitCAMTA1, CitCAMTA5, and CitCAMTA9 were significantly expressed. The expression of CitCAMTA7 first increased and then decreased as the time of salt treatment progressed. The expression of CitCAMTA3 was suppressed at all time points. For the remaining CitCAMTAs, i.e., CitCAMTA2, CitCAMTA6, and CitCAMTA8, there were no obvious changes observed. Upon dehydration stress (0- to 12-h treatment), CitCAMTA5, CitCAMTA6, and CitCAMTA8 levels slightly declined, whereas for CitCAMTA3 and CitCAMTA9, the expression levels were up-regulated reaching a maximum at 12 h of treatment. CitCAMTA1 and CitCAMTA2 showed moderate level of expression under dehydration stress. While most of the CitCAMTAs were responsive to salt and dehydration stress, CitCAMTA4 remain unaltered. Lastly, the authors investigated the involvement of CitCAMTAs in hormonal signaling (SA, ETH, IAA, 6- BA, ABA, MeJA, and GA3). All the CitCAMTAs (except CitCAMTA4) were responsive to at least one phytohormone. CitCAMTA1, CitCAMTA5, CitCAMTA7, and CitCAMTA9 positively correlated to all the hormones, whereas CitCAMTA3 showed a negative correlation. These observations led the authors to hypothesize the involvement of CitCAMTAs in abiotic stress pathways and hormonal regulation (Zhang et al., 2019). Genome-wide identification also led to the identification of 15 wheat CAMTA genes, which were classified into three groups A, B, and C containing seven, six, and two TaCAMTAs, respectively (Yang et al., 2020). Most of the TaCAMTAs contained stress-responsive cis-regulatory elements. All the 15 TaCAMTAs expressed differentially in various tissues and under different abiotic stresses. The expression analysis of TaCAMTA1-A and 1-D revealed that they were significantly expressed under drought, cold, heat, and salt stress. TaCAMTA genes belonging to the same homologous group had similar expression profiles under particular stress condition. For example, TaCAMTA1-A/B/D had almost similar expression profile under drought stress, TaCAMTA5-A/D under salt stress, and TaCAMTA1-A/B/D under heat stress. This observation implies that homologous TaCAMTA genes from the same group might share same functionality. However, few homologous TaCAMTA genes belonging to the same group displayed varying expression profiles under drought, cold, heat, and salinity stress. For instance, TaCAMTA3-A/D was significantly expressed upon cold stress, but the expression profiles of TaCAMTA1-B and TaCAMTA3-B were comparatively stable. This might be due to the functional differentiation in some homologous TaCAMTA genes. Additionally, in the study, it was depicted that 584 genes in the wheat genome can be potential targets of TaCAMTA, indicating the probable role of CAMTA in stress biology and plant development (Yang et al., 2020). Furthermore, TaCAMTA4 had been demonstrated to function as negative regulator of defense response against Puccinia triticina (Wang et al., 2019). TaCAMTA4 was found to be homologous to AtCAMTA4 and has been very well implicated as a CaM-binding protein in the wheat–P. triticina interaction system. This was proved by cloning and functionally characterizing TaCAMTA4 by using the CaM-encoding gene (TaCaM4-1) as a bait, for subsequent screening of cDNA library from P. triticina–infected Triticum leaves. The EMSA results revealed that TaCaM4-1 binds to TaCAMTA4 by the C-terminal CaMBD in Ca2+-dependent manner. Bimolecular fluorescence complementation (BiFC) analysis showed cytoplasm and nucleus as the probable interaction site of TaCAMTA4 and TaCaM4-1 (Wang et al., 2019). In a similar vein, nine CAMTA genes were identified in Flax (L. usitatissimum) upon genome-wide identification study. These nine CAMTA proteins were classified into three groups, depending on the phylogenetic analysis. Various hormonal (ABA and SA) and stress-related (drought, low temperature, and light) cis-regulatory elements were found to be enriched in promoter region of LuCAMTAs genes. In addition, the miRNA target analysis showed different miRNA families (miRN30, miRN9, miR2275, miR164, miR159, miR164, miRN15, miR395, miR156, miRN28, and miR164) as probable targets of the LuCAMTAs genes (except LuCAMTA9) (Ali et al., 2020).
Conclusion and Future Perspective
Improving crop yield and productivity for the ever-growing population has been a major challenge all over the world. The demand for ever-increasing food supply has put forth a tremendous pressure on the agriculture sector. The advent of new biotechnological tools and techniques has robustly contributed to crop improvement and horticultural science (Limera et al., 2017; Adlak et al., 2019). In this regard, several TFs, including CAMTA, had been identified and functionally characterized. Ever since the discovery of CAMTAs, its role had been comprehensively reported in developmental and stress biology in various model and crop plants (Table 1). The objective ahead is to understand the fundamental functionality and molecular mechanism behind CAMTA protein expression and downstream processing. A deeper understanding of the mechanisms underlying the involvement of Ca2+ and CaM in CAMTA transcript expression would provide great insights into its probable function in various tissues and response to environmental cues. In addition, deciphering the probable functions and mechanisms underlying the complex regulatory property of CAMTA proteins through its ability to transduce Ca2+ signatures via CaM can improve our knowledge of phenotypic plasticity in plants. However, because of the possible functional redundancy or at least overlapping functions, molecular characterization of individual CAMTA genes still poses a great challenge ahead for researchers. Rigorously developed CRISPR/Cas9-mediated genome editing techniques that rapidly generate mutations at multiple loci using one single-guide RNA will effectively help characterizing each of the gene family members. Nonetheless, the involvement of CAMTA as stress-related gene and its subsequent study in this direction would widen and provide vast knowledge for improvement in crop yield, productivity, and strategies to cope with adverse conditions in arid or semiarid regions of the world.
Author Contributions
ZI drafted the manuscript. MSI helped in drawing the figures. SPS edited the manuscript. TB reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded using research grants from the Second Century Fund (C2F Post-Doc fellowship). The fellowship for ZI was supported by C2F, Chulalongkorn University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
ZI is thankful to TB for providing the facilities and infrastructure.
References
Abbasi, F., Onodera, H., Toki, S., Tanaka, H., and Komatsu, S. (2004). OsCDPK13, a calcium-dependent protein kinase gene from rice, is induced by cold and gibberellin in rice leaf sheath. Plant Mol. Biol. 55, 541–552. doi: 10.1007/s11103-004-1178-y
Adlak, T., Tiwari, S., Tripathi, M., Gupta, N., Sahu, V. K., Bhawar, P., et al. (2019). Biotechnology: an advanced tool for crop improvement. Curr. J. Appl. Sci. Technol. 33, 1–11. doi: 10.9734/cjast/2019/v33i130081
Agarwal, P. K., Agarwal, P., Reddy, M. K., and Sopory, S. K. (2006). Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 25, 1263–1274. doi: 10.1007/s00299-006-0204-8
Aghdam, M. S., Hassanpouraghdam, M. B., Paliyath, G., and Farmani, B. (2012). The language of calcium in postharvest life of fruits, vegetables and flowers. Sci. Hortic. 144, 102–115. doi: 10.1016/j.scienta.2012.07.007
Aldon, D., Mbengue, M., Mazars, C., and Galaud, J.-P. (2018). Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 19:665. doi: 10.3390/ijms19030665
Ali, E., Raza, M. A., Cai, M., Hussain, N., Shahzad, A. N., Hussain, M., et al. (2020). Calmodulin-binding transcription activator (CAMTA) genes family: genome-wide survey and phylogenetic analysis in flax (Linum usitatissimum). PLoS One 15:e0236454. doi: 10.1371/journal.pone.0236454
Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. doi: 10.1038/35048692
Aravind, L., and Koonin, E. V. (1999). Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches1. J. Mol. Biol. 287, 1023–1040. doi: 10.1006/jmbi.1999.2653
Bahler, M., and Rhoads, A. (2002). Calmodulin signaling via the IQ motif. FEBS Lett. 513, 107–113. doi: 10.1016/s0014-5793(01)03239-2
Beccia, M. R., Sauge-Merle, S., Lemaire, D., Brémond, N., Pardoux, R., Blangy, S., et al. (2015). Thermodynamics of calcium binding to the calmodulin N-terminal domain to evaluate site-specific affinity constants and cooperativity. JBIC J. Biol. Inorg. Chem. 20, 905–919. doi: 10.1007/s00775-015-1275-1
Bender, K. W., and Snedden, W. A. (2013). Calmodulin-related proteins step out from the shadow of their namesake. Plant Physiol. 163, 486–495. doi: 10.1104/pp.113.221069
Benn, G., Bjornson, M., Ke, H., De Souza, A., Balmond, E. I., Shaw, J. T., et al. (2016). Plastidial metabolite MEcPP induces a transcriptionally centered stress-response hub via the transcription factor CAMTA3. Proc. Natl. Acad. Sci. U.S.A. 113, 8855–8860. doi: 10.1073/pnas.1602582113
Benn, G., Wang, C. Q., Hicks, D. R., Stein, J., Guthrie, C., and Dehesh, K. (2014). A key general stress response motif is regulated non-uniformly by CAMTA transcription factors. Plant J. 80, 82–92. doi: 10.1111/tpj.12620
Bergey, D., Kandel, R., Tyree, B., Dutt, M., and Dhekney, S. (2014). The role of calmodulin and related proteins in plant cell function: an ever-thickening plot. Springer Sci. Rev. 2, 145–159.
Bernsdorff, F., Döring, A.-C., Gruner, K., Schuck, S., Bräutigam, A., and Zeier, J. (2016). Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and-independent pathways. Plant Cell 28, 102–129. doi: 10.1105/tpc.15.00496
Bjornson, M., Benn, G., Song, X., Comai, L., Franz, A. K., Dandekar, A. M., et al. (2014). Distinct roles for mitogen-activated protein kinase signaling and CALMODULIN-BINDING TRANSCRIPTIONAL ACTIVATOR3 in regulating the peak time and amplitude of the plant general stress response. Plant Physiol. 166, 988–996. doi: 10.1104/pp.114.245944
Boonburapong, B., and Buaboocha, T. (2007). Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biol. 7:4. doi: 10.1186/1471-2229-7-4
Bouche, N., Scharlat, A., Snedden, W., Bouchez, D., and Fromm, H. (2002). A novel family of calmodulin-binding transcription activators in multicellular organisms. J. Biol. Chem. 277, 21851–21861. doi: 10.1074/jbc.m200268200
Bouche, N., Yellin, A., Snedden, W. A., and Fromm, H. (2005). Plant-specific calmodulin-binding proteins. Annu. Rev. Plant Biol. 56, 435–466. doi: 10.1146/annurev.arplant.56.032604.144224
Bush, D. S. (1995). Calcium regulation in plant cells and its role in signaling. Annu. Rev. Plant Biol. 46, 95–122. doi: 10.1146/annurev.pp.46.060195.000523
Buyuk, l., l̇lha, E., Şener, D., Özsoy, A. U., and Aras, S. (2019). Genome-wide identification of CAMTA gene family members in Phaseolus vulgaris L. and their expression profiling during salt stress. Mol. Biol. Rep. 46, 2721–2732. doi: 10.1007/s11033-019-04716-8
Carrion, A. M., Link, W. A., Ledo, F., Mellstram, B., and Naranjo, J. R. (1999). DREAM is a Ca 2+-regulated transcriptional repressor. Nature 398, 80. doi: 10.1038/18044
Chen, J.-Q., Meng, X.-P., Zhang, Y., Xia, M., and Wang, X.-P. (2008). Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol. Lett. 30, 2191–2198. doi: 10.1007/s10529-008-9811-5
Chen, Y., Zhou, X., Chang, S., Chu, Z., Wang, H., Han, S., et al. (2017). Calcium-dependent protein kinase 21 phosphorylates 14-3-3 proteins in response to ABA signaling and salt stress in rice. Biochem. Biophys. Res. Commun. 493, 1450–1456. doi: 10.1016/j.bbrc.2017.09.166
Cheng, S.-H., Willmann, M. R., Chen, H.-C., and Sheen, J. (2002). Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 129, 469–485. doi: 10.1104/pp.005645
Chinpongpanich, A., Phean-O-Pas, S., Thongchuang, M., Qu, L.-J., and Buaboocha, T. (2015). C-terminal extension of calmodulin-like 3 protein from Oryza sativa L.: interaction with a high mobility group target protein. Acta Biochim. Biophys. Sin. 47, 880–889. doi: 10.1093/abbs/gmv097
Choi, M. S., Kim, M. C., Yoo, J. H., Moon, B. C., Koo, S. C., Park, B. O., et al. (2005). Isolation of a calmodulin-binding transcription factor from rice (Oryza sativa L.). J. Biol. Chem. 280, 40820–40831. doi: 10.1074/jbc.m504616200
Chung, J.-S., Koo, S. C., Jin, B. J., Baek, D., Yeom, S.-I., Chun, H. J., et al. (2020). Rice CaM-binding transcription factor (OsCBT) mediates defense signaling via transcriptional reprogramming. Plant Biotechnol. Rep. 14, 309–321. doi: 10.1007/s11816-020-00603-y
Day, I. S., Reddy, V. S., Ali, G. S., and Reddy, A. S. N. (2002). Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol. 3:research0056.1.
Demidchik, V., and Shabala, S. (2018). Mechanisms of cytosolic calcium elevation in plants: the role of ion channels, calcium extrusion systems and NADPH oxidase-mediated ‘ROS-Ca2+ Hub’. Funct. Plant Biol. 45, 9–27. doi: 10.1071/fp16420
Do Heo, W., Lee, S. H., Kim, M. C., Kim, J. C., Chung, W. S., Chun, H. J., et al. (1999). Involvement of specific calmodulin isoforms in salicylic acid-independent activation of plant disease resistance responses. Proc. Natl. Acad. Sci. U.S.A. 96, 766–771. doi: 10.1073/pnas.96.2.766
Dodd, A. N., Kudla, J. R., and Sanders, D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61, 593–620.
Doherty, C. J., Van Buskirk, H. A., Myers, S. J., and Thomashow, M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984. doi: 10.1105/tpc.108.063958
Du, L., Ali, G. S., Simons, K. A., Hou, J., Yang, T., Reddy, A. S. N., et al. (2009). Ca 2+/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158. doi: 10.1038/nature07612
Dubrovina, A. S., Aleynova, O. A., Ogneva, Z. V., Suprun, A. R., Ananev, A. A., and Kiselev, K. V. (2019). The Effect of Abiotic Stress Conditions on Expression of Calmodulin (CaM) and Calmodulin-Like (CML) Genes in Wild-Growing Grapevine Vitis amurensis. Plants 8:602. doi: 10.3390/plants8120602
e Silva, O. D. C. (1994). CG-1, a parsley light-induced DNA-binding protein. Plant Mol. Biol. 25, 921–924. doi: 10.1007/bf00028887
Edel, K. H., and Kudla, J. (2016). Integration of calcium and ABA signaling. Curr. Opin. Plant Biol. 33, 83–91. doi: 10.1016/j.pbi.2016.06.010
Edel, K. H., Marchadier, E., Brownlee, C., Kudla, J., and Hetherington, A. M. (2017). The evolution of calcium-based signalling in plants. Curr. Biol. 27, R667–R679.
Fernandes, I. P., and Oliveira-Brett, A. M. (2017). Calcium-induced calmodulin conformational change. Electrochemical evaluation. Bioelectrochemistry 113, 69–78. doi: 10.1016/j.bioelechem.2016.10.002
Finkler, A., Ashery-Padan, R., and Fromm, H. (2007). CAMTAs: calmodulin- binding transcription activators from plants to human. FEBS Lett. 581, 3893–3898. doi: 10.1016/j.febslet.2007.07.051
Galon, Y., Aloni, R., Nachmias, D., Snir, O., Feldmesser, E., Scrase-Field, S., et al. (2010a). Calmodulin-binding transcription activator 1 mediates auxin signaling and responds to stresses in Arabidopsis. Planta 232, 165–178. doi: 10.1007/s00425-010-1153-6
Galon, Y., Snir, O., and Fromm, H. (2010b). How calmodulin binding transcription activators (CAMTAs) mediate auxin responses. Plant Signal. Behav. 5, 1311–1314. doi: 10.4161/psb.5.10.13158
Galon, Y., Nave, R., Boyce, J. M., Nachmias, D., Knight, M. R., and Fromm, H. (2008). Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 582, 943–948. doi: 10.1016/j.febslet.2008.02.037
Han, J., Gong, P., Reddig, K., Mitra, M., Guo, P., and Li, H.-S. (2006). The fly CAMTA transcription factor potentiates deactivation of rhodopsin, a G protein-coupled light receptor. Cell 127, 847–858. doi: 10.1016/j.cell.2006.09.030
Harper, J. F., Breton, G., and Harmon, A. (2004). Decoding Ca2+ signals through plant protein kinases. Annu. Rev. Plant Biol. 55, 263–288. doi: 10.1146/annurev.arplant.55.031903.141627
Hazak, O., Mamon, E., Lavy, M., Sternberg, H., Behera, S., Schmitz-Thom, I., et al. (2019). A novel Ca2+-binding protein that can rapidly transduce auxin responses during root growth. PLoS Biol. 17:e3000085. doi: 10.1371/journal.pbio.3000085
Hepler, P. K. (2005). Calcium: a central regulator of plant growth and development. Plant Cell 17, 2142–2155. doi: 10.1105/tpc.105.032508
Hochmal, A. K., Schulze, S., Trompelt, K., and Hippler, M. (2015). Calcium-dependent regulation of photosynthesis. Biochim. Biophys. Acta 1847, 993–1003. doi: 10.1016/j.bbabio.2015.02.010
Hong, J. C. (2016). “General aspects of plant transcription factor families,” in Plant Transcription Factors (Amsterdam: Elsevier), 35–56. doi: 10.1016/b978-0-12-800854-6.00003-8
Hou, J., Li, J., Yang, Y., Wang, Z., Chang, B., Yu, X., et al. (2019). Physiological and transcriptomic analyses elucidate that exogenous calcium can relieve injuries to potato plants (Solanum tuberosum L.) under weak light. Int. J. Mol. Sci. 20:5133. doi: 10.3390/ijms20205133
Jacob, F., Kracher, B., Mine, A., Seyfferth, C., Blanvillain-Baufumé, S., Parker, J. E., et al. (2018). A dominant-interfering camta3 mutation compromises primary transcriptional outputs mediated by both cell surface and intracellular immune receptors in Arabidopsis thaliana. New Phytol. 217, 1667–1680. doi: 10.1111/nph.14943
Jan, A. U., Hadi, F., Midrarullah, A. A., and Rahman, K. (2017). Role of CBF/DREB gene expression in abiotic stress tolerance. A Review. Int. J. Hortic. Agric. 2, 1–12. doi: 10.1007/978-0-387-79143-2_2
Kakar, K. U., Nawaz, Z., Cui, Z., Cao, P., Jin, J., Shu, Q., et al. (2018). Evolutionary and expression analysis of CAMTA gene family in Nicotiana tabacum yielded insights into their origin, expansion and stress responses. Sci. Rep. 8:10322.
Kidokoro, S., Yoneda, K., Takasaki, H., Takahashi, F., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2017). Different cold-signaling pathways function in the responses to rapid and gradual decreases in temperature. Plant Cell 29, 760–774. doi: 10.1105/tpc.16.00669
Kiep, V., Vadassery, J., Lattke, J., Maaß, J. P., Boland, W., Peiter, E., et al. (2015). Systemic cytosolic Ca2+ elevation is activated upon wounding and herbivory in Arabidopsis. New Phytol. 207, 996–1004. doi: 10.1111/nph.13493
Kim, H. S., Park, B. O., Yoo, J. H., Jung, M. S., Lee, S. M., Han, H. J., et al. (2007). Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. J. Biol. Chem. 282, 36292–36302. doi: 10.1074/jbc.m705217200
Kim, J., and Kim, H.-Y. (2006). Functional analysis of a calcium-binding transcription factor involved in plant salt stress signaling. FEBS Lett. 580, 5251–5256. doi: 10.1016/j.febslet.2006.08.050
Kim, K.-N., Lee, J.-S., Han, H., Choi, S. A., Go, S. J., and Yoon, I. S. (2003). Isolation and characterization of a novel rice Ca 2+-regulated protein kinase gene involved in responses to diverse signals including cold, light, cytokinins, sugars and salts. Plant Mol. Biol. 52, 1191–1202. doi: 10.1023/b:plan.0000004330.62660.a2
Kim, M. C., Chung, W. S., Yun, D.-J., and Cho, M. J. (2009). Calcium and calmodulin-mediated regulation of gene expression in plants. Mol. Plant 2, 13–21. doi: 10.1093/mp/ssn091
Kim, Y., Gilmour, S. J., Chao, L., Park, S., and Thomashow, M. F. (2020). Arabidopsis CAMTA transcription factors regulate pipecolic acid biosynthesis and priming of immunity genes. Mol. Plant 13, 157–168. doi: 10.1016/j.molp.2019.11.001
Kim, Y., Park, S., Gilmour, S. J., and Thomashow, M. F. (2013). Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 75, 364–376. doi: 10.1111/tpj.12205
Kim, Y. S., An, C., Park, S., Gilmour, S. J., Wang, L., Renna, L., et al. (2017). CAMTA-mediated regulation of salicylic acid immunity pathway genes in Arabidopsis exposed to low temperature and pathogen infection. Plant Cell 29, 2465–2477. doi: 10.1105/tpc.16.00865
Koo, S. C., Choi, M. S., Chun, H. J., Shin, D. B., Park, B. S., Kim, Y. H., et al. (2009). The calmodulin-binding transcription factor OsCBT suppresses defense responses to pathogens in rice. Mol. Cells 27, 563–570. doi: 10.1007/s10059-009-0081-4
Kordyum, E. (2003). Calcium signaling in plant cells in altered gravity. Adv. Space Res. 32, 1621–1630. doi: 10.1016/s0273-1177(03)90403-0
Kudla, J., Becker, D., Grill, E., Hedrich, R., Hippler, M., Kummer, U., et al. (2018). Advances and current challenges in calcium signaling. New Phytol. 218, 414–431. doi: 10.1111/nph.14966
Kurusu, T., Kuchitsu, K., and Tada, Y. (2015). Plant signaling networks involving Ca2+ and Rboh/Nox-mediated ROS production under salinity stress. Front. Plant Sci. 6:427. doi: 10.3389/fpls.2015.00427
Kushwaha, R., Singh, A., and Chattopadhyay, S. (2008). Calmodulin7 plays an important role as transcriptional regulator in Arabidopsis seedling development. Plant Cell 20, 1747–1759. doi: 10.1105/tpc.107.057612
Laluk, K., Prasad, K., Savchenko, T., Celesnik, H., Dehesh, K., Levy, M., et al. (2012). The calmodulin-binding transcription factor SIGNAL RESPONSIVE1 is a novel regulator of glucosinolate metabolism and herbivory tolerance in Arabidopsis. Plant Cell Physiol. 53, 2008–2015. doi: 10.1093/pcp/pcs143
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Lee, K., Thorneycroft, D., Achuthan, P., Hermjakob, H., and Ideker, T. (2010). Mapping plant interactomes using literature curated and predicted protein protein interaction data sets. Plant Cell 22, 997–1005. doi: 10.1105/tpc.109.072736
Leng, X., Han, J., Wang, X., Zhao, M., Sun, X., Wang, C., et al. (2015). Characterization of a calmodulin-binding transcription factor from strawberry (Fragaria ananassa). Plant Genome 8, 1–12.
Lenzoni, G., Liu, J., and Knight, M. R. (2018). Predicting plant immunity gene expression by identifying the decoding mechanism of calcium signatures. New Phytol. 217, 1598–1609. doi: 10.1111/nph.14924
Levy, M., Wang, Q., Kaspi, R., Parrella, M. P., and Abel, S. (2005). Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 43, 79–96. doi: 10.1111/j.1365-313x.2005.02435.x
Li, J., Han, G., Sun, C., and Sui, N. (2019). Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 14, 1613131. doi: 10.1080/15592324.2019.1613131
Li, S., Han, X., Yang, L., Deng, X., Wu, H., Zhang, M., et al. (2018). Mitogen-activated protein kinases and calcium-dependent protein kinases are involved in wounding-induced ethylene biosynthesis in Arabidopsis thaliana. Plant Cell Environ. 41, 134–147. doi: 10.1111/pce.12984
Li, X., Huang, L., Zhang, Y., Ouyang, Z., Hong, Y., Zhang, H., et al. (2014). Tomato SR/CAMTA transcription factors SlSR1 and SlSR3L negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance. BMC Plant Biol. 14:286. doi: 10.1186/s12870-014-0286-3
Li, Z.-Y., Xu, Z.-S., Chen, Y., He, G.-Y., Yang, G.-X., Chen, M., et al. (2013). A novel role for Arabidopsis CBL1 in affecting plant responses to glucose and gibberellin during germination and seedling development. PLoS One 8:e56412. doi: 10.1371/journal.pone.0056412
Liang, W., Wang, M., and Ai, X. (2009). The role of calcium in regulating photosynthesis and related physiological indexes of cucumber seedlings under low light intensity and suboptimal temperature stress. Sci. Hortic. 123, 34–38. doi: 10.1016/j.scienta.2009.07.015
Limera, C., Sabbadini, S., Sweet, J. B., and Mezzetti, B. (2017). New biotechnological tools for the genetic improvement of major woody fruit species. Front. Plant Sci. 8:1418. doi: 10.3389/fpls.2017.01418
Liu, F., Chu, X., Lu, H. P., and Wang, J. (2017). Molecular mechanism of multispecific recognition of Calmodulin through conformational changes. Proc. Natl. Acad. Sci. U.S.A. 114, E3927–E3934.
Liu, J., Whalley, H. J., and Knight, M. R. (2015). Combining modelling and experimental approaches to explain how calcium signatures are decoded by calmodulin†binding transcription activators (CAMTAs) to produce specific gene expression responses. New Phytol. 208, 174–187.
Lolle, S., Greeff, C., Petersen, K., Roux, M., Jensen, M. K., Bressendorff, S., et al. (2017). Matching NLR immune receptors to autoimmunity in camta3 mutants using antimorphic NLR alleles. Cell Host Microbe 21, 518–529.e4.
Luan, S. (2009). The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 14, 37–42. doi: 10.1016/j.tplants.2008.10.005
Ma, W., and Berkowitz, G. A. (2007). The grateful dead: calcium and cell death in plant innate immunity. Cell. Microbiol. 9, 2571–2585. doi: 10.1111/j.1462-5822.2007.01031.x
Manishankar, P., Wang, N., Köster, P., Alatar, A. A., and Kudla, J. (2018). Calcium signaling during salt stress and in the regulation of ion homeostasis. J. Exp. Bot. 69, 4215–4226. doi: 10.1093/jxb/ery201
Marques, D. N., Dos Reis, S. P., and De Souza, C. R. (2017). Plant NAC transcription factors responsive to abiotic stresses. Plant Gene 11, 170–179. doi: 10.1016/j.plgene.2017.06.003
Maruyama, K., Sakuma, Y., Kasuga, M., Ito, Y., Seki, M., Goda, H., et al. (2004). Identification of cold- inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 38, 982–993. doi: 10.1111/j.1365-313x.2004.02100.x
McCormack, E., Tsai, Y.-C., and Braam, J. (2005). Handling calcium signaling: arabidopsis CaMs and CMLs. Trends Plant Sci. 10, 383–389. doi: 10.1016/j.tplants.2005.07.001
Meador, W. E., Means, A. R., and Quiocho, F. A. (1992). Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science 257, 1251–1255. doi: 10.1126/science.1519061
Medvedev, S. (2005). Calcium signaling system in plants. Russ. J. Plant Physiol. 52, 249–270. doi: 10.1007/s11183-005-0038-1
Mitsis, T., Efthimiadou, A., Bacopoulou, F., Vlachakis, D., Chrousos, G. P., and Eliopoulos, E. (2020). Transcription factors and evolution: an integral part of gene expression. World Acad. Sci. J. 2, 3–8.
Mitsuda, N., Isono, T., and Sato, M. H. (2003). Arabidopsis CAMTA family proteins enhance V-PPase expression in pollen. Plant Cell Physiol. 44, 975–981. doi: 10.1093/pcp/pcg137
Monshausen, G. B., Bibikova, T. N., Weisenseel, M. H., and Gilroy, S. (2009). Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 21, 2341–2356. doi: 10.1105/tpc.109.068395
Mori, I. C., and Schroeder, J. I. (2004). Reactive oxygen species activation of plant Ca2+ channels. A signaling mechanism in polar growth, hormone transduction, stress signaling, and hypothetically mechanotransduction. Plant Physiol. 135, 702–708. doi: 10.1104/pp.104.042069
Mt, S., and Mn, W. (2008). Ca2+/calmodulin-dependent protein kinases. Cell. Mol. Life. Sci. 65, 2637–2657.
Muller, C. W., Rey, F. A., Sodeoka, M., Verdine, G. L., and Harrison, S. C. (1995). Structure of the NF- ÎoB p50 homodimer bound to DNA. Nature 373, 311–317.
Nakashima, K., Tran, L. S. P., Van Nguyen, D., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51, 617–630. doi: 10.1111/j.1365-313x.2007.03168.x
Nakata, M., Yuasa, T., Takahashi, Y., and Ishida, S. (2009). CDPK1, a calcium-dependent protein kinase, regulates transcriptional activator RSG in response to gibberellins. Plant Signal. Behav. 4, 372–374. doi: 10.4161/psb.4.5.8229
Nakayama, S., Kawasaki, H., and Kretsinger, R. (2000). “Evolution of EF-hand proteins,” in Calcium Homeostasis, eds E. Carafoli and J. Krebs (Berlin: Springer), 29–58. doi: 10.1007/978-3-642-58306-3_2
Nie, H., Zhao, C., Wu, G., Wu, Y., Chen, Y., and Tang, D. (2012). SR1, a calmodulin-binding transcription factor, modulates plant defense and ethylene-induced senescence by directly regulating NDR1 and EIN3. Plant Physiol. 158, 1847–1859. doi: 10.1104/pp.111.192310
Noman, M., Jameel, A., Qiang, W.-D., Ahmad, N., Liu, W.-C., Wang, F.-W., et al. (2019). Overexpression of GmCAMTA12 enhanced drought tolerance in arabidopsis and soybean. Int. J. Mol. Sci. 20:4849. doi: 10.3390/ijms20194849
Pandey, N., Ranjan, A., Pant, P., Tripathi, R. K., Ateek, F., Pandey, H. P., et al. (2013). CAMTA 1 regulates drought responses in Arabidopsis thaliana. BMC Genomics 14:216. doi: 10.1186/1471-2164-14-216
Pant, P., Iqbal, Z., Pandey, B. K., and Sawant, S. V. (2018). Genome-wide comparative and evolutionary analysis of Calmodulin-binding Transcription Activator (CAMTA) family in Gossypium species. Sci. Rep. 8:5573.
Park, C. Y., Lee, J. H., Yoo, J. H., Moon, B. C., Choi, M. S., Kang, Y. H., et al. (2005). WRKY group IId transcription factors interact with calmodulin. FEBS Lett. 579, 1545–1550. doi: 10.1016/j.febslet.2005.01.057
Phukan, U. J., Jeena, G. S., and Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7:760. doi: 10.3389/fpls.2016.00760
Poovaiah, B. W., Reddy, A. S. N., and Feldman, L. (1993). Calcium and signal transduction in plants. Crit. Rev. Plant Sci. 12, 185–211. doi: 10.1007/978-3-642-83974-0_14
Popescu, S. C., Popescu, G. V., Bachan, S., Zhang, Z., Seay, M., Gerstein, M., et al. (2007). Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. U.S.A. 104, 4730–4735. doi: 10.1073/pnas.0611615104
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Qiu, Y., Xi, J., Du, L., Suttle, J. C., and Poovaiah, B. W. (2012). Coupling calcium/calmodulin-mediated signaling and herbivore-induced plant response through calmodulin-binding transcription factor AtSR1/CAMTA3. Plant Mol. Biol. 79, 89–99. doi: 10.1007/s11103-012-9896-z
Rahman, A., Nahar, K., Hasanuzzaman, M., and Fujita, M. (2016). Calcium supplementation improves Na+/K+ ratio, antioxidant defense and glyoxalase systems in salt-stressed rice seedlings. Front. Plant Sci. 7:609. doi: 10.3389/fpls.2016.00609
Rahman, H., Xu, Y.-P., Zhang, X.-R., and Cai, X.-Z. (2016). Brassica napus Genome Possesses Extraordinary High Number of CAMTA Genes and CAMTA3 Contributes to PAMP Triggered Immunity and Resistance to Sclerotinia sclerotiorum. Front. Plant Sci. 7:581. doi: 10.3389/fpls.2016.00581
Reddy, A. S. N. (2001). Calcium: silver bullet in signaling. Plant Sci. 160, 381–404. doi: 10.1016/s0168-9452(00)00386-1
Reddy, A. S. N., Reddy, V. S., and Golovkin, M. (2000). A calmodulin binding protein from Arabidopsis is induced by ethylene and contains a DNA-binding motif. Biochem. Biophys. Res. Commun. 279, 762–769. doi: 10.1006/bbrc.2000.4032
Reddy, V. S., Ali, G. S., and Reddy, A. S. N. (2002). Genes encoding calmodulin-binding proteins in the Arabidopsis genome. J. Biol. Chem. 277, 9840–9852. doi: 10.1074/jbc.m111626200
Reddy, V. S., Day, I. S., Thomas, T., and Reddy, A. S. N. (2004). KIC, a novel Ca2+ binding protein with one EF-hand motif, interacts with a microtubule motor protein and regulates trichome morphogenesis. Plant Cell 16, 185–200. doi: 10.1105/tpc.016600
Roy, S. (2016). Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 11:e1117723. doi: 10.1080/15592324.2015.1117723
Rubtsov, A. M., and Lopina, O. D. (2000). Ankyrins. FEBS Lett. 482, 1–5. doi: 10.1016/s0014-5793(00)01924-4
Saeidi, K., Zare, N., Baghizadeh, A., and Asghari-Zakaria, R. (2019). Phaseolus vulgaris genome possesses CAMTA genes, and phavuCAMTA1 contributes to the drought tolerance. J. Genet. 98:31. doi: 10.1007/s11118-011-9244-y
Salmi, M. L., Ul Haque, A., Bushart, T. J., Stout, S. C., Roux, S. J., and Porterfield, D. M. (2011). Changes in gravity rapidly alter the magnitude and direction of a cellular calcium current. Planta 233, 911–920. doi: 10.1007/s00425-010-1343-2
Sanders, D., Pelloux, J. R. M., Brownlee, C., and Harper, J. F. (2002). Calcium at the crossroads of signaling. Plant Cell 14, S401–S417.
Scott, I. M., Clarke, S. M., Wood, J. E., and Mur, L. A. (2004). Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis. Plant Physiol. 135, 1040–1049. doi: 10.1104/pp.104.041293
Sedgwick, S. G., and Smerdon, S. J. (1999). The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24, 311–316. doi: 10.1016/s0968-0004(99)01426-7
Seifikalhor, M., Aliniaeifard, S., Shomali, A., Azad, N., Hassani, B., Lastochkina, O., et al. (2019). Calcium signaling and salt tolerance are diversely entwined in plants. Plant Signal. Behav. 14:1665455. doi: 10.1080/15592324.2019.1665455
Shangguan, L., Wang, X., Leng, X., Liu, D., Ren, G., Tao, R., et al. (2014). Identification and bioinformatic analysis of signal responsive/calmodulin-binding transcription activators gene models in Vitis vinifera. Mol. Biol. Rep. 41, 2937–2949. doi: 10.1007/s11033-014-3150-5
Shen, Q., Fu, L., Su, T., Ye, L., Huang, L., Kuang, L., et al. (2020). Calmodulin HvCaM1 negatively regulates salt tolerance via modulation of HvHKT1s and HvCAMTA4. Plant Physiol. 183, 1650–1662. doi: 10.1104/pp.20.00196
Shi, H., Ye, T., Zhong, B., Liu, X., and Chan, Z. (2014). Comparative proteomic and metabolomic analyses reveal mechanisms of improved cold stress tolerance in bermudagrass (Cynodon dactylon (L.) Pers.) by exogenous calcium. J. Integr. Plant Biol. 56, 1064–1079. doi: 10.1111/jipb.12167
Shkolnik, D., Finkler, A., Pasmanik-Chor, M., and Fromm, H. (2019). CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR 6: a key regulator of Na+ homeostasis during germination. Plant Physiol. 180, 1101–1118. doi: 10.1104/pp.19.00119
Singh, P., and Virdi, A. S. (eds) (2013). “Ca 2+, calmodulin and plant-specific calmodulin-binding proteins: implications in abiotic stress adaptation,” in Stress Signaling in Plants: Genomics and Proteomics Perspective, Vol. Volume 1, (Berlin: Springer), 1–23. doi: 10.1007/978-1-4614-6372-6_1
Snedden, W. A., and Fromm, H. (2001). Calmodulin as a versatile calcium signal transducer in plants. New Phytol. 151, 35–66. doi: 10.1046/j.1469-8137.2001.00154.x
Song, K., Backs, J., Mcanally, J., Qi, X., Gerard, R. D., Richardson, J. A., et al. (2006). The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell 125, 453–466. doi: 10.1016/j.cell.2006.02.048
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/s1369-5266(00)00199-0
Sun, T., Huang, J., Xu, Y., Verma, V., Jing, B., Sun, Y., et al. (2020). Redundant CAMTA transcription factors negatively regulate the biosynthesis of salicylic acid and N-hydroxypipecolic acid by modulating the expression of SARD1 and CBP60g. Mol. Plant 13, 144–156. doi: 10.1016/j.molp.2019.10.016
Sun, X., Wang, Y., and Sui, N. (2018). Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 503, 397–401. doi: 10.1016/j.bbrc.2018.07.123
Swarbreck, S. M., Colaço, R., and Davies, J. M. (2013). Plant calcium-permeable channels. Plant Physiol. 163, 514–522. doi: 10.1104/pp.113.220855
Teige, M. (2019). Chloroplasts use calcium signals to call for help under heat stress. Plant Cell Physiol. 60, 492–493. doi: 10.1093/pcp/pcz039
Thor, K. (2019). Calcium—nutrient and messenger. Front. Plant Sci. 10:440. doi: 10.3389/fpls.2019.00440
Tian, W., Hou, C., Ren, Z., Wang, C., Zhao, F., Dahlbeck, D., et al. (2019). A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572, 131–135. doi: 10.1038/s41586-019-1413-y
Tian, W., Wang, C., Gao, Q., Li, L., and Luan, S. (2020). Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat. Plants 6, 1–10. doi: 10.1079/9780851995120.0001
Tokizawa, M., Kobayashi, Y., Saito, T., Kobayashi, M., Iuchi, S., Nomoto, M., et al. (2015). Sensitive to proton rhizotoxicity1, calmodulin binding transcription activator2, and other transcription factors are involved in aluminum-activated malate transporter1 expression. Plant Physiol. 167, 991–1003. doi: 10.1104/pp.114.256552
Toyota, M., Furuichi, T., Tatsumi, H., and Sokabe, M. (2008). Critical consideration on the relationship between auxin transport and calcium transients in gravity perception of Arabidopsis seedlings. Plant Signal. Behav. 3, 521–524. doi: 10.4161/psb.3.8.6339
Tran, L.-S. P., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Tripathi, D. K., Singh, V. P., Chauhan, D. K., Sharma, S., Prasad, S. M., Dubey, N. K., et al. (2020). Plant Life Under Changing Environment: Responses and Management. Cambridge, MA: Academic Press.
Tupler, R., Perini, G., and Green, M. R. (2001). Expressing the human genome. Nature 409, 832–835. doi: 10.1038/35057011
Tuteja, N., and Mahajan, S. (2007). Calcium signaling network in plants: an overview. Plant Signal. Behav. 2, 79–85. doi: 10.4161/psb.2.2.4176
Vanneste, S., and Friml, J. (2013). Calcium: the missing link in auxin action. Plants 2, 650–675. doi: 10.3390/plants2040650
Villalobo, A., González-Muñoz, M., and Berchtold, M. W. (2019). Proteins with calmodulin-like domains: structures and functional roles. Cell. Mol. Life Sci. 76, 2299–2328. doi: 10.1007/s00018-019-03062-z
Vogel, J. T., Zarka, D. G., Van Buskirk, H. A., Fowler, S. G., and Thomashow, M. F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41, 195–211. doi: 10.1111/j.1365-313x.2004.02288.x
Walley, J. W., and Dehesh, K. (2010). Molecular mechanisms regulating rapid stress signaling networks in Arabidopsis. J. Integr. Plant Biol. 52, 354–359. doi: 10.1111/j.1744-7909.2010.00940.x
Wang, C., Shang, J.-X., Chen, Q.-X., Oses-Prieto, J. A., Bai, M.-Y., Yang, Y., et al. (2013). Identification of BZR1-interacting proteins as potential components of the brassinosteroid signaling pathway in Arabidopsis through tandem affinity purification. Mol. Cell. Proteomics 12, 3653–3665. doi: 10.1074/mcp.m113.029256
Wang, G., Zeng, H., Hu, X., Zhu, Y., Chen, Y., Shen, C., et al. (2015). Identification and expression analyses of calmodulin-binding transcription activator genes in soybean. Plant Soil 386, 205–221. doi: 10.1007/s11104-014-2267-6
Wang, Y., Wei, F., Zhou, H., Liu, N., Niu, X., Yan, C., et al. (2019). TaCAMTA4, a calmodulin-interacting protein, involved in defense response of wheat to Puccinia triticina. Sci. Rep. 9:641.
Wang, Z., Jia, C., Wang, J.-Y., Miao, H.-X., Liu, J.-H., Chen, C., et al. (2020). Genome-wide analysis of basic helix-loop-helix transcription factors to elucidate candidate genes related to fruit ripening and stress in Banana (Musa acuminata L. AAA Group, cv. Cavendish). Front. Plant Sci. 11:650. doi: 10.3389/fpls.2020.00650
Wei, M., Xu, X., and Li, C. (2017). Identification and expression of CAMTA genes in Populus trichocarpa under biotic and abiotic stress. Sci. Rep. 7:17910.
Wu, L., Sadhukhan, A., Kobayashi, Y., Ogo, N., Tokizawa, M., Agrahari, R. K., et al. (2019). Involvement of phosphatidylinositol metabolism in aluminum-induced malate secretion in Arabidopsis. J. Exp. Bot. 70, 3329–3342. doi: 10.1093/jxb/erz179
Xi, J., Qiu, Y., Du, L., and Poovaiah, B. W. (2012). Plant-specific trihelix transcription factor AtGT2L interacts with calcium/calmodulin and responds to cold and salt stresses. Plant Sci. 185, 274–280. doi: 10.1016/j.plantsci.2011.11.013
Xiong, T.-C., Bourque, S., Lecourieux, D., Amelot, N., Grat, S., Brière, C., et al. (2006). Calcium signaling in plant cell organelles delimited by a double membrane. Biochim. Biophys. Acta 1763, 1209–1215. doi: 10.1016/j.bbamcr.2006.09.024
Yadav, V., Mallappa, C., Gangappa, S. N., Bhatia, S., and Chattopadhyay, S. (2005). A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17, 1953–1966. doi: 10.1105/tpc.105.032060
Yan, C., Fan, M., Yang, M., Zhao, J., Zhang, W., Su, Y., et al. (2018). Injury activates Ca2+/calmodulin-dependent phosphorylation of JAV1-JAZ8-WRKY51 complex for jasmonate biosynthesis. Mol. Cell 70, 136–149.e7.
Yang, F., Dong, F., Liu, Y., Chai, J., Zhao, H., Lv, M., et al. (2020). Genome-wide identification and expression analysis of the calmodulin-binding transcription activator (CAMTA) gene family in wheat (Triticum aestivum L.). BMC Genetics 21:105. doi: 10.1186/s12863-020-00916-5
Yang, T., Peng, H., Whitaker, B. D., and Conway, W. S. (2012). Characterization of a calcium/calmodulin-regulated SR/CAMTA gene family during tomato fruit development and ripening. BMC Plant Biol. 12:19. doi: 10.1186/1471-2229-12-19
Yang, T., Peng, H., Whitaker, B. D., and Jurick, W. M. (2013). Differential expression of calcium/calmodulin- regulated SlSRs in response to abiotic and biotic stresses in tomato fruit. Physiol. Plant. 148, 445–455. doi: 10.1111/ppl.12027
Yang, T., and Poovaiah, B. W. (2000). An early ethylene up-regulated gene encoding a calmodulin-binding protein involved in plant senescence and death. J. Biol. Chem. 275, 38467–38473. doi: 10.1074/jbc.m003566200
Yang, T., and Poovaiah, B. W. (2002). A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 277, 45049–45058. doi: 10.1074/jbc.m207941200
Yang, Y., Sun, T., Xu, L., Pi, E., Wang, S., Wang, H., et al. (2015). Genome-wide identification of CAMTA gene family members in Medicago truncatula and their expression during root nodule symbiosis and hormone treatments. Front. Plant Sci. 6:459. doi: 10.3389/fpls.2015.00459
Yuan, P., Du, L., and Poovaiah, B. (2018a). Ca2+/Calmodulin-Dependent AtSR1/CAMTA3 plays critical roles in balancing plant growth and immunity. Int. J. Mol. Sci. 19:1764. doi: 10.3390/ijms19061764
Yuan, P., Yang, T., and Poovaiah, B. (2018b). Calcium signaling-mediated plant response to cold stress. Int. J. Mol. Sci. 19:3896. doi: 10.3390/ijms19123896
Yue, R., Lu, C., Sun, T., Peng, T., Han, X., Qi, J., et al. (2015). Identification and expression profiling analysis of calmodulin-binding transcription activator genes in maize (Zea mays L.) under abiotic and biotic stresses. Front. Plant Sci. 6:576. doi: 10.3389/fpls.2015.00576
Yuenyong, W., Chinpongpanich, A., Comai, L., Chadchawan, S., and Buaboocha, T. (2018). Downstream components of the calmodulin signaling pathway in the rice salt stress response revealed by transcriptome profiling and target identification. BMC Plant Biol. 18:335. doi: 10.1186/s12870-018-1538-4
Zeng, H., Xu, L., Singh, A., Wang, H., Du, L., and Poovaiah, B. (2015). Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 6:600. doi: 10.3389/fpls.2015.00600
Zhang, J., Pan, X., Ge, T., Yi, S., Lv, Q., Zheng, Y., et al. (2019). Genome-wide identification of citrus CAMTA genes and their expression analysis under stress and hormone treatments. J. Hortic. Sci. Biotechnol. 94, 331–340. doi: 10.1080/14620316.2018.1504631
Zhang, T., Zhao, Y., Wang, Y., Liu, Z., and Gao, C. (2018). Comprehensive analysis of MYB gene family and their expressions under abiotic stresses and hormone treatments in Tamarix hispida. Front. Plant Sci. 9:1303. doi: 10.3389/fpls.2018.01303
Zhu, L., Jin, X., Xie, Q., Yao, Q., Wang, X., and Li, H. (2018). Calcium-dependent protein kinase family genes involved in ethylene-induced natural rubber production in different Hevea brasiliensis cultivars. Int. J. Mol. Sci. 19:947. doi: 10.3390/ijms19040947
Zhu, Y., Yan, J., Liu, W., Liu, L., Sheng, Y., Sun, Y., et al. (2016). Phosphorylation of a NAC transcription factor by ZmCCaMK regulates abscisic acid-induced antioxidant defense in maize. Plant Physiol. 171, 1651–1664. doi: 10.1104/pp.16.00168
Keywords: calcium, calmodulin, development, transcription factor, CAMTA, stress
Citation: Iqbal Z, Iqbal MS, Singh SP and Buaboocha T (2020) Ca2+/Calmodulin Complex Triggers CAMTA Transcriptional Machinery Under Stress in Plants: Signaling Cascade and Molecular Regulation. Front. Plant Sci. 11:598327. doi: 10.3389/fpls.2020.598327
Received: 14 September 2020; Accepted: 30 October 2020;
Published: 03 December 2020.
Edited by:
Jean Molinier, UPR2357 Institut de Biologie Moléculaire des Plantes (IBMP), FranceReviewed by:
Gang Ma, University of Connecticut, United StatesShiu Cheung Lung, The University of Hong Kong, Hong Kong
Copyright © 2020 Iqbal, Shariq Iqbal, Singh and Buaboocha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teerapong Buaboocha, teerapong.b@chula.ac.th
 Zahra Iqbal
Zahra Iqbal Mohammed Shariq Iqbal
Mohammed Shariq Iqbal Surendra Pratap Singh
Surendra Pratap Singh Teerapong Buaboocha
Teerapong Buaboocha