- Commonwealth Scientific and Industrial Research Organisation (CSIRO) Agriculture and Food, Canberra, ACT, Australia
The reproductive success of many plants depends on their capacity to respond appropriately to their environment. One environmental cue that triggers flowering is the extended cold of winter, which promotes the transition from vegetative to reproductive growth in a response known as vernalization. In annual plants of the Brassicaceae, the floral repressor, FLOWERING LOCUS C (FLC), is downregulated by exposure to low temperatures. Repression is initiated during winter cold and then maintained as the temperature rises, allowing plants to complete their life cycle during spring and summer. The two stages of FLC repression, initiation and maintenance, are distinguished by different chromatin states at the FLC locus. Initiation involves the removal of active chromatin marks and the deposition of the repressive mark H3K27me3 over a few nucleosomes in the initiation zone, also known as the nucleation region. H3K27me3 then spreads to cover the entire locus, in a replication dependent manner, to maintain FLC repression. FLC is released from repression in the next generation, allowing progeny of a vernalized plant to respond to winter. Activation of FLC in this generation has been termed resetting to denote the restoration of the pre-vernalized state in the progeny of a vernalized plant. It has been assumed that resetting must differ from the activation of FLC expression in progeny of plants that have not experienced winter cold. Considering that there is now strong evidence indicating that chromatin undergoes major modifications during both male and female gametogenesis, it is time to challenge this assumption.
Introduction
Plants must respond to environmental challenges to ensure their survival or reproductive success by changing patterns of gene expression. Gene expression is modulated not only by transcription factors but also by the associated chromatin environment, which can influence expression long after the event that triggered changes in local chromatin. Vernalization, the promotion of flowering in response to the prolonged cold of winter, is a well-studied example of the long-term memory provided by the epigenome (reviewed in Berry and Dean, 2015). In annual plants, the vernalized state is established during winter, maintained through the life of the vernalized plant but is reset in the progeny so that each generation can respond appropriately to the seasonal cycles (Lang, 1965). The timing of resetting differs in perennials in which vegetative growth is restored in some shoots on return to warmer conditions (reviewed in Turck and Coupland, 2014).
In Arabidopsis thaliana, the key gene in the vernalization response is the repressor of flowering, FLOWERING LOCUS C (FLC), which is downregulated by low temperatures. Initially repression is transient, but FLC repression is stabilized as the duration of the exposure to low temperature increases (Angel et al., 2011). As ambient temperatures rise at the end of winter, FLC repression is maintained allowing the induction of two promoters of flowering, FLOWERING LOCUS T (FT) and SUPPRESSOR OF CONSTANS 1 (SOC1), and the subsequent transition to flowering (Helliwell et al., 2006; Searle et al., 2006). FLC is released from repression in the next generation allowing the progeny of a vernalized plant to respond to winter (Sheldon et al., 2000). Activation of FLC in the next generation has been termed resetting to denote restoration of the non-vernalized state in the progeny of a vernalized plant. The process of resetting the vernalized state has long intrigued biologists because tissues giving rise to male and female gametes are not set aside early in embryo development as they are in mammals, but rather develop from somatic tissue that has accumulated changes to the epigenome during vegetative growth (reviewed in Gehring, 2019).
The molecular events associated with the long-term repression of FLC have been elucidated. In contrast to the gradual decline in the abundance of FLC mRNA that occurs during an extended period at low temperatures, the downregulation of transcription occurs more rapidly, and this may be initiated by a cold-induced physical change in FLC chromatin (Csorba et al., 2014; Finnegan, 2015; Helliwell et al., 2015; Rosa et al., 2016). Cold exposure also results in the induction of antisense transcripts, collectively known as COOLAIR, as well as VERNALIZATION INSENSITIVE 3 (VIN3), a PHD domain protein that associates with a vernalization-specific polycomb repression complex, Polycomb Repressive Complex 2 (PRC2; Sung and Amasino, 2004; de Lucia et al., 2008; Swiezewski et al., 2009). Initially repression of FLC is transient but becomes stabilized in a time dependent manner that is cell (and locus) autonomous (Angel et al., 2011; Berry et al., 2015; Yang et al., 2017). This switch is associated with stabilization of the nucleosome in the +1 position relative to transcription, loss of active chromatin marks, and gain of H3K27me3 within the nucleation region of FLC chromatin that encompasses the +1 nucleosome (Finnegan and Dennis, 2007; Angel et al., 2011; Finnegan, 2015). After winter ends, H3K27me3 then spreads across the entire locus in a DNA replication-dependent process (Finnegan and Dennis, 2007; Angel et al., 2011; Hyun et al., 2013; Finnegan, 2015; Yang et al., 2017). PRC2 activity is essential for the switch between transient and stable repression, suggesting that H3K27me3 accumulation is important for stable repression (Gendall et al., 2001; Helliwell et al., 2011; Yang et al., 2017). Recruitment of PRC2 to FLC chromatin is facilitated by the binding of the B3 transcriptional repressors, VAL1 and/or VAL2, to RY-1 and RY-2 motifs (TGCATG; R, purine, Y, pyrimidine; Swaminathan et al., 2008) that constitute a cold memory element (CME) within the nucleation region of FLC, during cold exposure (Questa et al., 2016; Yuan et al., 2016). VAL1 interacts directly with components of PRC1 and PRC2 (AtBMI1 and LHP1, respectively), and SAP18, part of the SIN3-histone deactylase complex that in turn binds the histone deactylase HDA19, to shut down transcription and indirectly recruit PRC2 (Questa et al., 2016; Yuan et al., 2016). It has long been thought that resetting reverses these changes by the removal of H3K27me3 and activation of FLC transcription. This could occur prior to, or during, gamete formation in the vernalized plant or post-fertilization, in the developing embryo. To determine whether resetting is a process unique to vernalized plants, we must first consider the timing and genetic requirements for expression of FLC in each generation of non-vernalized plants.
FLC Expression Must be Activated in the Embryos of Non-Vernalized Plants
An FLC::GUS reporter construct, which mirrors the endogenous FLC gene, is expressed throughout the somatic tissues of non-vernalized plants and in both the carpel and stamens, but there is no expression in either the developing gametophytic embryo sac or in mature pollen (Figure 1A; Sheldon et al., 2008; Choi et al., 2009). Thus, even in the absence of vernalization, FLC is repressed during gametogenesis and must, therefore, be activated in the next generation. Activation of FLC::GUS is initiated at the earliest stage of embryo development although some β-glucuronidase (GUS) activity may result from maternal mRNA inherited via the egg cell (Sheldon et al., 2008; Tao et al., 2017; Luo et al., 2020). FLC::GUS expression continues to increase until early heart stage and is then maintained for the duration of embryo development (Figure 1A; Sheldon et al., 2008; Tao et al., 2017; Luo et al., 2020). In somatic tissue, a complex of proteins, the FRI complex (FRIc), which includes FRI, FRL1, FES, FLX, and SUF4, is required for expression of FLC (Choi et al., 2011). However, FLC::GUS is activated in young embryos in both fri and suf4 mutants, suggesting that FRIc is not essential for activation of FLC in the early stages, although it promotes FLC expression in globular embryos and throughout the later stages of embryo development (Choi et al., 2009; Tao et al., 2019). This raises the question of what initiates FLC expression in the zygote.
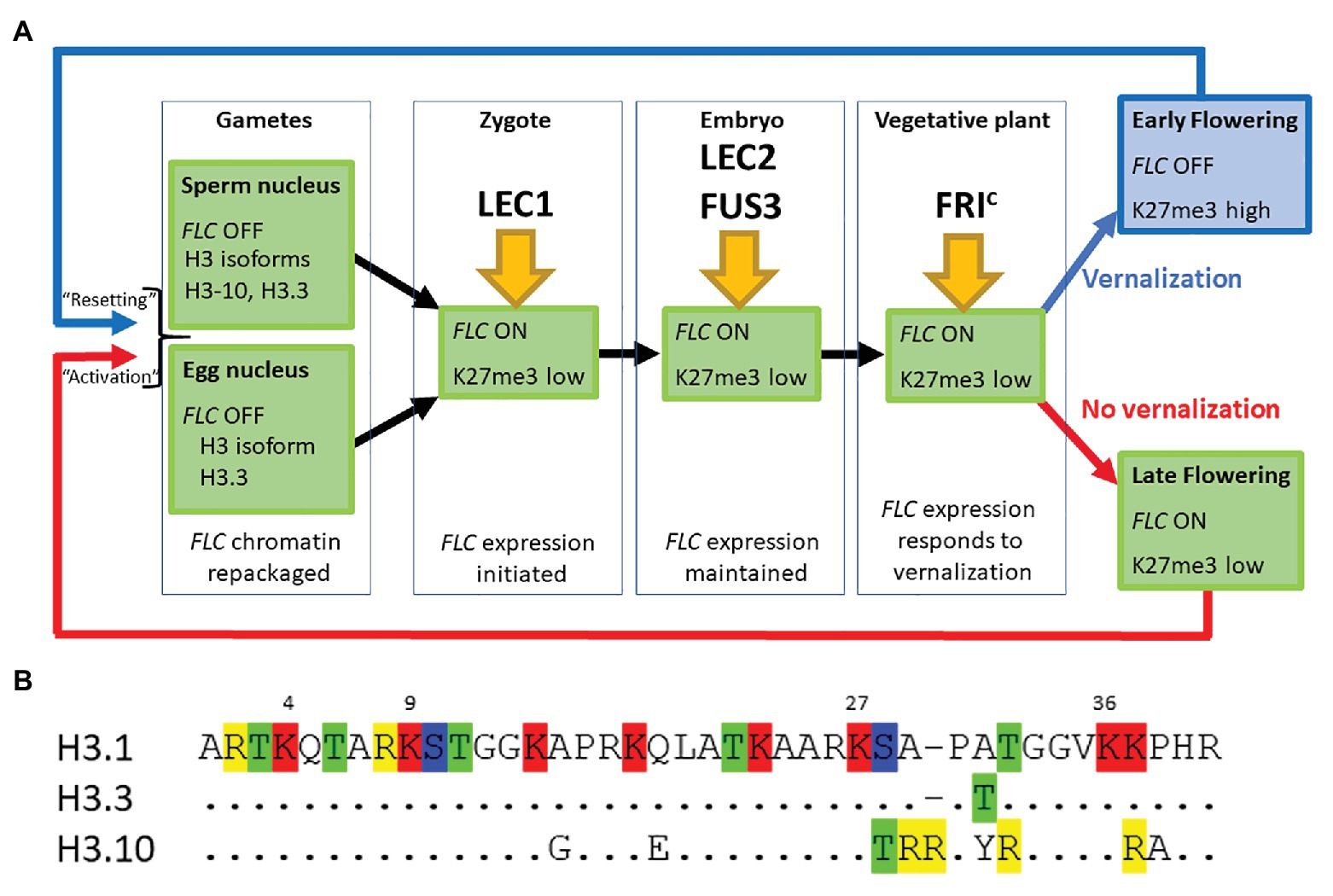
Figure 1. (A) FLOWERING LOCUS C (FLC) expression is established in the zygote and maintained through the life of the plant unless the plant becomes vernalized by exposure to prolonged cold during winter. FLC is inactive in mature gametes of both vernalized and non-vernalized plants where the chromatin is repackaged and epigenetic marks are lost. The major transcription factors determining FLC expression at each developmental stage are indicated. (B) The first 40 amino acids of three different isoforms of Histone H3 are shown. Single letter nomenclature is used and the dots shown in H3.3 and H3.10 indicate that the sequence is conserved with H3.1. The commonly methylated lysines are numbered above the sequence. Amino acids that can be modified are highlighted; R, me; T, ph; K, ac/me; S, ph. H3.10 and H3.3 each have the potential for novel modifications not seen in H3.1 and the substitution S-T28 (H3.10) may disrupt the activity of the kinase that normally phosphorylates that position. The changes adjacent to K27 may also prevent methylation of that residue. H3.1 denoted the protein encoded by HTR3, H3.10 by HTR10, and H3.3 by HTR5.
As FLC expression is observed as early as 1 day after pollination (DAP), it was proposed that activation of FLC would most likely involve the action of an embryo transcription factor (Tao et al., 2017). LEAFY COTYLEDON 1 (LEC1), which is expressed in the zygote and throughout embryogenesis, is a component of a seed specific NF-Y (nuclear factor of the Y box) transcription factor that is a master regulator of embryo development (Lotan et al., 1998; Le et al., 2010). NF-Y transcription factors are conserved across kingdoms and have been shown to act as pioneer transcription factors that can bind their target recognition site in the context of nonmodified chromatin or even closed chromatin marked with the repressive modification, H3K27me3 (Fleming et al., 2013). They are comprised of three subunits, NF-YA, a site-specific DNA binding protein that targets CCAAT motifs and two proteins with a histone-domain fold, NF-YB and NF-YC that are structurally related to Histone H2B and H2A, respectively (reviewed in Mantovani, 1999).
To investigate the role of NF-Y on FLC activation, a null mutant of lec1, one of 10 NF-YB subunits encoded by the Arabidopsis genome, was introduced into ColFRISf2. Loss of LEC1 activity partially suppresses the late flowering phenotype in non-vernalized plants, and the expression of both FLC::GUS and endogenous FLC is reduced throughout embryo development. Once established, the level of FLC expression appears to be fixed as the proportion of embryos showing weak, intermediate, or strong expression remains consistent across the first 3 DAP (Table 1A; Tao et al., 2017). FLC expression is even lower in embryos where there was decreased activity of all five members of the LEC1 clade, indicating that NF-YB subunits from this clade act redundantly to activate the level of FLC in the zygote (Tao et al., 2017). Several lines of evidence indicate that LEC1 directly activates FLC. Mutation of the four putative NF-Y binding sites in the FLC promoter leads to early flowering; LEC1::FLAG is enriched at this region of the FLC promoter; FLC expression is induced in response to ectopic induction of a LEC1 transgene and this is associated with the establishment of active chromatin at FLC (Tao et al., 2017).
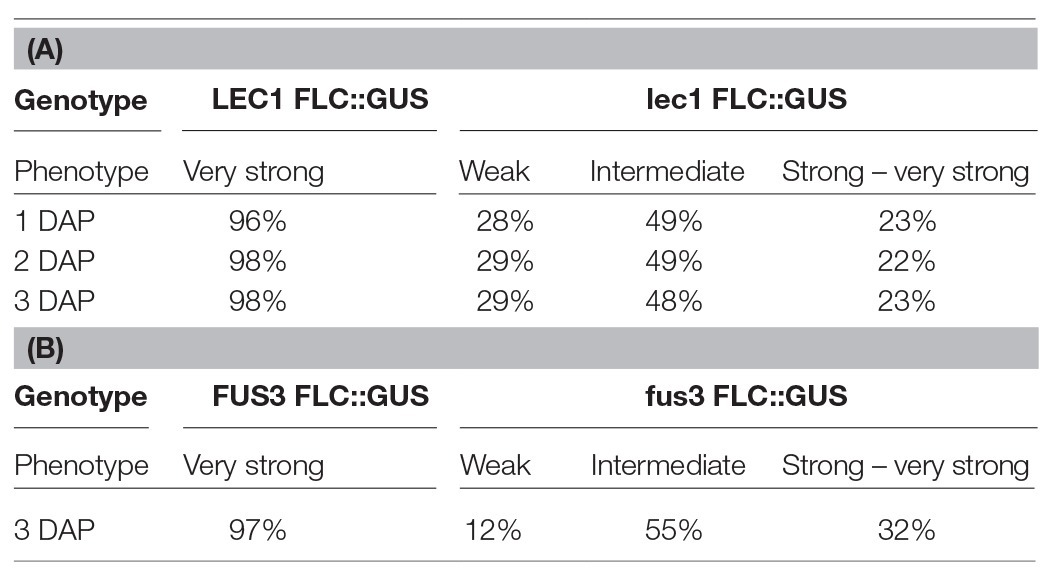
Table 1. GUS expression during embryo development is determined by the activity of LEC1 in the zygote (A) or FUS3 activity in the embryo (B; adapted from Tao et al., 2017, 2019).
While LEC1 is key to the activation of FLC in the zygote, two other embryo-specific transcription factors, LEC2 and FUSCA 3 (FUS3), are needed to maintain FLC expression from 2 DAP (Figure 1A; Tao et al., 2019). LEC2 and FUS3 are B3 domain transcription factors that are in the same subfamily as VAL1 and VAL2, proteins that recruit PRC2 to FLC chromatin during vernalization (Yuan et al., 2016). In lec2 or fus3 single mutants, FLC::GUS is re-activated in the zygote, just as it is in wild type plants, but FLC::GUS expression in some embryos declined from 2 DAP (lec2) and 3 DAP (fus3; Table 1B). Binding of LEC2 and FUS3 to the CME within FLC chromatin is facilitated by LEC1. As enrichment of first LEC2 and then FUS3 increases from 2 to 6 DAP, there is a corresponding decrease in enrichment of VAL1 at the CME, suggesting that LEC2 and FUS3 antagonize VAL1 binding at the CME during embryogenesis to ensure that FLC transcription is maintained following activation by LEC1 (Tao et al., 2019). LEC2 and FUS3 interact with FRI and are required for the recruitment of FRI to the CME and adjacent regions of the FLC locus, consistent with the finding that FRI promotes FLC expression from 3 DAP. While LEC2 and FUS3 play an important role in maintaining FLC expression during early embryogenesis, these proteins are not expressed in post-embryonic stages of Arabidopsis allowing VAL1/2 to bind the CME and mediate FLC repression during vernalization.
What Do We Know About Resetting of the Vernalized State?
The resetting of epigenetic regulation in plants varies with the epigenetic modifier involved (Gehring, 2019); for example, there is little evidence to support widespread erasure and replacement of DNA methylation in the developing plant embryo, as occurs in mammalian embryos, because epialleles can be stably inherited between plant generations (Becker et al., 2011; Schmitz et al., 2011). In contrast, the vernalized state, which is mediated by changes in histone modifications at FLC chromatin, is reset in each generation (Sheldon et al., 2000). Resetting of FLC expression in male and female gametes, the zygote, and developing embryo was examined using an FLC::GUS reporter construct (Sheldon et al., 2008; Choi et al., 2009).
Although some GUS activity can be detected in the developing anther, there is no GUS activity in mature pollen or the female gametophyte of vernalized plants (Sheldon et al., 2008; Choi et al., 2009). The timing of FLC::GUS reactivation differs between the paternally and maternally inherited transgene. The paternally derived FLC::GUS is expressed in up to 50% single-celled zygotes and expression continues throughout embryo development. When inherited from the vernalized maternal parent, expression of FLC::GUS is not detected until about 3 DAP (Sheldon et al., 2008). Neither maternally nor paternally inherited FLC:GUS are expressed in the fertilized central cell or the developing endosperm (Sheldon et al., 2008).
A mutagenesis screen for resetting mutants identified a hypomorphic mutation in EARLY FLOWERING 6 (ELF6) that impairs resetting of the vernalized state and FLC expression in the progeny of vernalized elf6-5 plants (Crevillen et al., 2014). ELF6 is a jumonji-domain-containing protein that demethylates di- and tri-methylated H3K27, and consistent with this, there is a small increase in H3K27me3 in some regions of FLC chromatin associated with somewhat lower FLC expression in non-vernalized elf6-5 plants compared to wild type. Curiously, a null mutant, elf6-3, which also has reduced FLC expression prior to vernalization, shows no effect on resetting (Crevillen et al., 2014; Tao et al., 2017). A second weak mutant of ELF6, elf6-4, also has no effect on the resetting of FLC in progeny of a vernalized plant (Tao et al., 2017). Taken together, these observations suggest that ELF6 plays at most a minor role in reactivating FLC expression to pre-vernalized levels.
LEC1, LEC2, and FUS3 are essential for resetting expression of FLC in the progeny of vernalized plants just as they are for activation of FLC in the embryos of non-vernalized plants (Figure 1A; Tao et al., 2017, 2019). Consistent with this, the timing of FLC::GUS expression is similar in embryos of both vernalized and non-vernalized plants (Tao et al., 2017).
Discussion
Our comparison of the genetic requirements for resetting of FLC in the progeny of a vernalized plant and those associated with the activation of FLC in the young embryo of non-vernalized parents indicates that there is essentially no difference between these processes (Figure 1A). This may seem surprising given that repression of FLC during vernalization causes the depletion of active chromatin marks followed by an enrichment with the repressive chromatin modification H3K27me3 across the entire FLC locus. We suggest that changes in the nucleosome composition of chromatin that occur during gametogenesis could account in part for these findings (Figures 1A,B).
Firstly, the chromatin of mature sperm cells differs from that in somatic tissue as sperm chromatin lacks histone H3.1, the H3 isoform associated with H3K27me3 (Figure 1A). Instead, sperm chromatin is enriched in H3.10, encoded by the sperm-specific gene HTR10, and an H3.3 isoform encoded by HTR5 (Okada et al., 2005; Rotman et al., 2005; Brownfield et al., 2009; Ingouff et al., 2010; Borg et al., 2011). It has recently been shown that amino acid substitutions around the critical K27 residue in H3.10 (Figure 1B) prevent trimethylation of this residue by PRC2 (Borg et al., 2020). Indeed, H3K27me3 is barely detectable in sperm chromatin and, consistent with this, components of PRC2 are not expressed in Arabidopsis sperm cells (Borg et al., 2020). Similarly in monocots, H3K27me3 is observed only in the chromatin of the vegetative cell but not of sperm cells (Sano and Tanaka, 2010; Houben et al., 2011; Pandey et al., 2013). This suggests that the chromatin associated with FLC loci, inherited through the paternal gamete of a vernalized plant, would have been stripped of the repressive H3K27me3 mark prior to fertilization. It is hardly surprising then that an ELF6 null mutant has little effect on resetting (Crevillen et al., 2014; Tao et al., 2017).
Secondly, chromatin dynamics during the development of the mature egg cell are extremely complex and include waves of depletion and presumptive restoration of H3K27m3 (as judged by chromatin localization of LHP1; Baroux and Autran, 2015). Like the sperm cell nuclei, the egg nucleus has a novel complement of histone variants, expressing only HTR5, an isoform of H3.3 (Figures 1A,B). Recent data suggest that the vernalized state is transmitted through egg cell chromatin and that the vernalized state is not erased immediately following fertilization (Luo et al., 2020); this is consistent with the observation that FLC::GUS inherited from a vernalized maternal parent is not detected until about 3 DAP (Sheldon et al., 2008).
Finally, after fertilization, the paternally inherited H3.10 and the maternally inherited H3.3 are actively removed from chromatin in the zygote but the somatic complement of histone variants is not restored until after de novo synthesis of H3.1 and H3.3 during the first zygotic mitosis (Ingouff et al., 2007, 2010). It seems likely then that resetting is merely a consequence of the normal processes by which chromatin is remodeled during gametogenesis and following fertilization.
In conclusion, we suggest that the steps required to activate FLC expression in the young embryo are nearly identical regardless of whether the embryo is the progeny of a vernalized or non-vernalized plant. In each case, the chromatin associated with FLC loci undergoes major reprogramming during male and female gametogenesis. A shared route to FLC activation (Figure 1A) is supported by genetic data showing that the pioneer transcription factor NF-Y and the SWIR chromatin remodeling complex that deposits H2A.Z into chromatin are essential for activating FLC expression (Choi et al., 2011; Tao et al., 2017), with LEC2, FUS3, and FRIc being required to fully activate FLC expression beyond the day after fertilization (Choi et al., 2009; Tao et al., 2019).
Author Contributions
EF developed the concept and wrote the manuscript. MR and CH edited the manuscript and designed the figures. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Angel, A., Song, J., Dean, C., and Howard, M. (2011). A Polycomb-based switch underlying quantitative epigenetic memory. Nature 476, 106–108. doi: 10.1038/nature10241
Baroux, C., and Autran, D. (2015). Chromatin dynamics during cellular differentiation in the female reproductive lineage of flowering plants. Plant J. 83, 160–176. doi: 10.1111/tpj.12890
Becker, C., Hagmann, J., Muller, J., Koenig, D., Stegle, O., Borgwardt, K., et al. (2011). Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 480, 245–249. doi: 10.1038/nature10555
Berry, S., and Dean, C. (2015). Environmental perception and epigenetic memory: mechanistic insight through FLC. Plant J. 83, 133–148. doi: 10.1111/tpj.12869
Berry, S., Hartley, M., Olsson, T. S. G., Dean, C., and Howard, M. (2015). Local chromatin environment of a Polycomb target gene instructs its own epigenetic inheritance. eLife 4:e07205. doi: 10.7554/eLife.07205
Borg, M., Brownfield, L., Khatab, H., Sidorova, A., Lingaya, M., and Twell, D. (2011). The R2R3 MYB transcription factor DUO1 activates a male germline-specific regulon essential for sperm cell differentiation in Arabidopsis. Plant Cell 23, 534–549. doi: 10.1105/tpc.110.081059
Borg, M., Jacob, Y., Susaki, D., LeBlanc, C., Buendia, D., Axelsson, E., et al. (2020). Targeted reprogramming of H3K27me3 resets epigenetic memory in plant paternal chromatin. Nat. Cell Biol. 22, 621–629. doi: 10.1038/s41556-020-0515-y
Brownfield, L., Hafidh, S., Borg, M., Sidorova, A., and Twell, D. (2009). A plant germline-specific integrator of sperm specification and cell cycle progression. PLoS Genet. 5:e1000430. doi: 10.1371/journal.pgen.1000430
Choi, J., Hyun, Y., Kang, M. -J., Yun, H. I., Yun, J. -Y., Lister, C., et al. (2009). Resetting and regulation of FLOWERING LOCUS C expression during Arabidopsis reproductive development. Plant J. 57:918–931. doi: 10.1111/j.1365-313X.2008.03776.x
Choi, K., Kim, J., Hwang, H. -J., Kim, S., Park, C., Kim, S. Y., et al. (2011). The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23, 289–303. doi: 10.1105/tpc.110.075911
Crevillen, P., Yang, H., Cui, X., Greeff, C., Trick, M., Qiu, Q., et al. (2014). Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 515, 587–590. doi: 10.1038/nature13722
Csorba, T., Questa, J. I., Sun, Q., and Dean, C. (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. U. S. A. 111, 16160–16165. doi: 10.1073/pnas.1419030111
de Lucia, F., Crevillen, P., Jones, A. M. E., Greb, T., and Dean, C. (2008). A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. U. S. A. 105, 16831–16836. doi: 10.1073/pnas.0808687105
Finnegan, E. J. (2015). Time-dependent stabilization of the +1 nucleosome is an early step in the transition to stable cold-induced repression of FLC. Plant J. 84, 875–885. doi: 10.1111/tpj.13044
Finnegan, E. J., and Dennis, E. S. (2007). Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr. Biol. 17, 1978–1983. doi: 10.1016/j.cub.2007.10.026
Fleming, J. D., Pavesi, G., Benatti, P., Imbriano, C., Mantovani, R., and Struhl, K. (2013). NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res. 23, 1195–1209. doi: 10.1101/gr.148080.112
Gehring, M. (2019). Epigenetic dynamics during flowering plant reproduction: evidence for reprogramming? New Phytol. 224, 91–96. doi: 10.1111/nph.15856
Gendall, A. R., Levy, Y. Y., Wilson, A., and Dean, C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107, 525–535. doi: 10.1016/s0092-8674(01)00573-6
Helliwell, C. A., Anderssen, R. A., Robertson, M., and Finnegan, E. J. (2015). How is FLC repressed by cold? Trends Plant Sci. 20, 76–82. doi: 10.1016/j.tplants.2014.12.004
Helliwell, C. A., Robertson, M., Finnegan, E. J., Buzas, D. M., and Dennis, E. S. (2011). Vernalization-repression of Arabidopsis FLC requires promoter sequences but not antisense transcripts. PLoS One 6:e21513. doi: 10.1371/journal.pone.0021513
Helliwell, C. A., Wood, C. C., Robertson, M., Peacock, W. J., and Dennis, E. S. (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight complex. Plant J. 46, 183–192. doi: 10.1111/j.1365-313X.2006.02686.x
Houben, A., Kumke, K., Nagaki, K., and Hause, G. (2011). CENH3 distribution and differential chromatin modifications during pollen development in rye (Secale cereale L.). Chromosom. Res. 19, 471–480. doi: 10.1007/s10577-011-9207-6
Hyun, Y., Yun, H., Park, K., Ohr, H., Lee, O., Kim, D. -H., et al. (2013). The catalytic subunit of Arabidopsis DNA polymerase α ensures stable maintenance of histone modification. Development 140, 156–166. doi: 10.1242/dev.084624
Ingouff, M., Hamamura, Y., Gourgues, M., Higashiyama, T., and Berger, F. (2007). Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr. Biol. 17, 1032–1037. doi: 10.1016/j.cub.2007.05.019
Ingouff, M., Rademacher, S., Holec, S., Soljic, L., Xin, N., Readshaw, A., et al. (2010). Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in Arabidopsis. Curr. Biol. 20, 2137–2143. doi: 10.1016/j.cub.2010.11.012
Lang, A. (1965). “Physiology of flower initiation” in Encyclopedia of plant physiology. ed. W. Ruhland (Berlin: Springer Verlag), 1489–1536.
Le, B. H., Cheng, C., Bui, A. Q., Wagmaister, J. A., Henrya, K. F., Pelletier, J., et al. (2010). Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc. Natl. Acad. Sci. U. S. A. 107, 8063–8070. doi: 10.1073/pnas.1003530107
Lotan, T., Ohto, M., Yee, K. M., West, M. A. L., Lo, R., Kwong, R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. doi: 10.1016/s0092-8674(00)81463-4
Luo, X., Ou, Y., Li, R., and He, Y. (2020). Maternal transmittion of the epigenetic ‘memory of winter cold’ in Arabidopsis. Nat. Plants 6, 1211–1218. doi: 10.1038/s41477-020-00774-0
Mantovani, R. (1999). The molecular biology of the CCAAT-binding factor NF-Y. Gene 239, 15–27. doi: 10.1016/s0378-1119(99)00368-6
Okada, T., Endo, M., Singh, M. B., and Bhalla, P. L. (2005). Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J. 44, 557–568. doi: 10.1111/j.1365-313X.2005.02554.x
Pandey, P., Houben, A., Kumlehn, J., Melzer, M., and Rutten, T. (2013). Chromatin alterations during pollen development in Hordeum vulgare. Genome Res. 141, 50–57. doi: 10.1159/000351211
Questa, J. I., Song, J., Geraldo, N., An, A., and Dean, C. (2016). Arabidopsis transcriptional repressor VAL1 triggers Polycomb silencing at FLC during vernalization. Science 353, 485–488. doi: 10.1126/science.aaf7354
Rosa, S., Duncan, S., and Dean, C. (2016). Mutually exclusive sense–antisense transcription at FLC facilitates environmentally induced gene repression. Nat. Commun. 7:13031. doi: 10.1038/ncomms13031
Rotman, N., Durbarry, N., Wardle, A., Yang, W. C., Chabourd, A., Faure, J. E., et al. (2005). A novel class of MYB factors controls sperm-cell formation in plants. Curr. Biol. 15, 244–248. doi: 10.1016/j.cub.2005.01.013
Sano, Y., and Tanaka, I. (2010). Distinct localization of histone H3 methylation in the vegetative nucleus of lily pollen. Cell Biol. Int. 34, 253–259. doi: 10.1042/CBI20090124
Schmitz, R. J., Schultz, M. D., Lewsey, M. G., O’Malley, R. C., Urich, M. A., Libiger, O., et al. (2011). Transgenerational epigenetic instability is a source of novel methylation variants. Science 334, 369–373. doi: 10.1126/science.1212959
Searle, I., He, Y., Turck, F., Vincent, C., Fornara, F., Krober, S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912. doi: 10.1101/gad.373506
Sheldon, C. C., Hills, M. J., Lister, C., Dean, C., Dennis, E. S., and Peacock, W. J. (2008). Resetting of FLOWERING LOCUS C expression after epigenetic repression by vernalization. Proc. Natl. Acad. Sci. U. S. A. 105, 2214–2219. doi: 10.1073/pnas.0711453105
Sheldon, C. C., Rouse, D. T., Finnegan, E. J., Peacock, W. J., and Dennis, E. S. (2000). The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. U. S. A. 97, 3753–3758. doi: 10.1073/pnas.060023597
Sung, S., and Amasino, R. M. (2004). Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427, 159–164. doi: 10.1038/nature02195
Swaminathan, K., Peterson, K., and Jack, T. (2008). The plant B3 superfamily. Trends Plant Sci. 13, 647–655. doi: 10.1016/j.tplants.2008.09.006
Swiezewski, S., Liu, F., Magusin, A., and Dean, C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462, 799–802. doi: 10.1038/nature08618
Tao, Z., Hu, H., Luo, X., Jia, B., Du, J., and He, Y. (2019). Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis. Nat. Plants 5, 424–435. doi: 10.1038/s41477-019-0402-3
Tao, Z., Shen, L., Gu, X., Wang, Y., Yu, H., and He, Y. (2017). Embryonic epigenetic reprogramming by a pioneer transcription factor in plants. Nature 551, 124–128. doi: 10.1038/nature24300
Turck, F., and Coupland, G. (2014). Natural variation in epigenetic gene regulation and its effects on plant developmental traits. Evolution 68, 620–631. doi: 10.1111/evo.12286
Yang, H., Berry, S., Olsson, T. S. G., Hartley, M., Howard, M., and Dean, C. (2017). Distinct phases of Polycomb silencing to hold epigenetic memory of cold in Arabidopsis. Science 357, 1142–1145. doi: 10.1126/science.aan1121
Keywords: vernalized state, epigenetic memory, histone H3, chromatin, gametogenesis
Citation: Finnegan EJ, Robertson M and Helliwell CA (2021) Resetting FLOWERING LOCUS C Expression After Vernalization Is Just Activation in the Early Embryo by a Different Name. Front. Plant Sci. 11:620155. doi: 10.3389/fpls.2020.620155
Edited by:
Richard Amasino, University of Wisconsin-Madison, United StatesReviewed by:
Yuehui He, Chinese Academy of Sciences (CAS), ChinaMartin Howard, John Innes Centre, United Kingdom
Copyright © 2021 Finnegan, Robertson and Helliwell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: E. Jean Finnegan, jean.finnegan@csiro.au
 E. Jean Finnegan*
E. Jean Finnegan* Masumi Robertson
Masumi Robertson Chris A. Helliwell
Chris A. Helliwell