- 1Institute of Cytology and Genetics (ICG), Siberian Branch of Russian Academy of Sciences (SB RAS), Novosibirsk, Russia
- 2N.I. Vavilov All-Russian Research Institute of Plant Genetic Resources (VIR), Saint Petersburg, Russia
Polyphenol oxidase (PPO) is an oxidoreductase. In damaged plant tissues, it catalyzes enzymatic browning by oxidizing o-diphenols to highly reactive o-quinones, which polymerize producing heterogeneous dark polymer melanin. In intact tissues, functions of PPO are not well understood. The aim of the study was to investigate the barley PPO gene family and to reveal the possible involvement of Ppo genes in melanization of barley grain, which is controlled by the Blp1 gene. Based on known barley Ppo genes on chromosome 2H (Ppo1 and Ppo2), two additional genes—Ppo3 and Ppo4—were found on chromosomes 3H and 4H, respectively. These genes have one and two exons, respectively, contain a conserved tyrosinase domain and are thought to be functional. Comparative transcriptional analyzes of the genes in samples of developing grains (combined hulls and pericarp tissues) were conducted in two barley lines differing by melanin pigmentation. The genes were found to be transcribed with increasing intensity (while grains mature) independently from the grain color, except for Ppo2, which is transcribed only in black-grained line i:BwBlp1 accumulating melanin in grains. Analysis of this gene’s expression in detached hulls and pericarps showed its elevated transcription in both tissues in comparison with yellow ones, while it was significantly higher in hulls than in pericarp. Segregation analysis in two F2 populations obtained based on barley genotypes carrying dominant Blp1 and recessive ppo1 (I) and dominant Blp1 and recessive ppo1 and ppo2 (II) was carried out. In population I, only two phenotypic classes corresponding to parental black and white ones were observed; the segregation ratio was 3 black to 1 white, corresponding to monogenic. In population II, aside from descendants with black and white grains, hybrids with a gray phenotype — light hulls and dark pericarp — were observed; the segregation ratio was 9 black to 3 gray to 4 white, corresponding to the epistatic interaction of two genes. Most hybrids with the gray phenotype carry dominant Blp1 and a homozygous recessive allele of Ppo2. Based on transcription and segregation assays one may conclude involvement of Ppo2 but not Ppo1 in melanin formation in barley hulls.
Introduction
Polyphenol oxidases (PPO) are oxidoreductase enzymes widespread among plant species. Enzymes of this class share a tyrosinase domain that contains two conserved copper-binding motifs: CuA and CuB (Marusek et al., 2006). PPOs can exert two types of activity: monophenolase (E.C. 1.14.18.1), which catalyzes the o-hydroxylation of monophenols forming o-diphenols, and diphenolase (E.C. 1.10.3.), which oxidizes o-diphenols to o-quinones. The highly reactive o-quinones are then polymerized nonenzymatically giving rise to dark-colored compounds: melanins (Nicolas et al., 1994). PPOs located in plastids are thought to be spatially separated from their phenolic substrates, which are found in vacuoles of plant cells (Vaughn and Duke, 1984). Therefore, oxidation of phenolic substrates to melanins usually occurs after a mechanical impact damaging cell compartments. This process is called the enzymatic browning reaction and considered to be an important part of the plant immune response to stress (Zhang, 2023). However, the undesirable darkening of plants caused by the enzymatic browning reaction during the postharvest process has substantial negative effects on commercial value of agricultural products (Taranto et al., 2017; Moon et al., 2020). One way to avoid this is to breed varieties with inactive PPO; this approach requires knowledge about functions of Ppo genes in different plant tissues and about allelic diversity of these genes.
Besides enzymatic browning, additional functions in plant physiology are attributed to PPO and should be taken into consideration when Ppo-deficient varieties are bred. For example, it was shown that a knockout of the single gene encoding PPO results in the appearance of dark necrotic spots on leaves of walnut (Juglans regia), and this phenomenon was associated with disruption of tyrosine metabolism and accumulation of a toxic compound called tyramine in cells (Araji et al., 2014). On the other hand, a knockout of Ppo genes in potato (Solanum tuberosum) also has a significant effect on metabolism of phenolic compounds, thereby increasing accumulation of end products of the phenylpropanoid biosynthetic pathway (in the mutant lines compared to the wild type) and enhancing resistance to late blight (Llorente et al., 2011; Llorente et al., 2014). Given that PPO is localized to chloroplasts, possible participation of PPOs in photosynthesis and in protection of the photosynthetic apparatus from damage under abiotic stress conditions has been discussed (Boeckx et al., 2015; Boeckx et al., 2017). Additionally, PPO activity can be associated with genetically determined synthesis of constitutive melanin pigments in plant seed coats. Black pigmentation of plant seeds is a common trait, and for some plant species, melanin presence has been confirmed by chemical extraction and identification methods (Glagoleva et al., 2020). For example, melanin has been isolated from hulls of sunflower seeds (Nicolaus et al., 1964; Gracheva and Zheltobryukhov, 2016), watermelon seeds (Nicolaus et al., 1964), and chestnuts (Yao et al., 2012), sesame (Dossou et al., 2022) as well as from seeds of cereals like oat (Varga et al., 2016), barley (Shoeva et al., 2020), and wheat (Bazhenov et al., 2023).
Melanin is the least investigated plant pigment owing to its nonobvious functions and laborious extraction. Nevertheless, it is believed that the dark color of seeds provides some advantages to plants during adaptation to unfavorable environmental conditions (Glagoleva et al., 2020). For example, the barley landraces with black seeds are grown in the most arid regions of Syria, whereas white-grained landraces are adapted to milder growing conditions (Ceccarelli et al., 1987). Specimens with black grains have turned out to be more cold- and drought-tolerant than specimens with white grains (Ceccarelli et al., 1987; Weltzien, 1988). Besides abiotic stress, melanin is believed to participate in protection of plants from biotic stress. Varieties of oat with a dark spike color caused by melanin are less affected by Fusarium infection than are varieties without dark husk pigments (Loskutov et al., 2016). Barley recombinant inbred lines with black grains demonstrated lower incidence of Fusarium head blight and lower accumulation of the mycotoxin deoxynivalenol than did lines with yellow grains (Choo et al., 2015). Although melanin is a widespread pigment in the plant kingdom, there is little information on how it is synthesized. In some plant species, a possible relation between the activity of Ppo genes and the biosynthesis of melanin pigments in intact tissues has been reported. For example, in watermelon, the gene encoding PPO was identified as a candidate gene responsible for melanin synthesis (Li et al., 2020). Comparative transcriptome studies on peanut (Wan et al., 2016) and sesame (Wang et al., 2020) have revealed increased expression of genes encoding PPO in melanin-accumulating seeds. In addition, an involvement of PPO in melanin synthesis in persimmon peel was suggested (Qi et al., 2020).
Barley (Hordeum vulgare L.) is one of the most widely cultivated crops in the world. It is believed that the presence of melanin in the hulls of barley seeds is a common characteristic of barley populations (Glagoleva et al., 2022). The black color of the barley grain is under monogenic control by the Blp1 (Black lemma and pericarp 1) locus on chromosome 1H (Costa et al., 2001; Long et al., 2019). Based on existing knowledge about the locus, it can be argued that the gene determining melanin synthesis in the barley grain does not encode PPO (Jia et al., 2017; Long et al., 2019), but in a comparative RNA-seq analysis of white- and black-grained near-isogenic lines (NILs), the Blp1 locus was shown to affect the expression of the Ppo genes that have been proposed as components of a gene network controlling melanin formation in the barley grain (Glagoleva et al., 2017). Two genes encoding PPO have already been found in the barley genome, which are referred to as Ppo1 and Ppo2 and are localized to the long arm of chromosome 2H (Taketa et al., 2010). Besides them, two additional Ppo genes are thought to exist in the barley genome because of the presence of two additional Ppo genes on chromosomes 3H and 4H in the related wild barley species Hordeum chilense (Rodríguez-Suárez and Atienza, 2014). However, functions of the Ppo genes with respect to melanin formation in the barley grain have not been studied. Thus, the aim of the study was to investigate the barley polyphenol oxidase gene family and to reveal the possible involvement of Ppo genes in melanin synthesis in the barley grain.
Materials and methods
Plant material
NIL i:BwBlp1 (also known as BW062; accession number in NordGen collection: NGB20470; www.nordgen.org) carrying the Blp1.b allele of the Blp1 (Black lemma and pericarp 1) locus mapped to chromosome 1HL (Druka et al., 2011) and characterized by melanin accumulation in the husk and grain pericarp was used for the study. The NIL was developed in the genetic background of spring cultivar Bowman (“Bowman From Fargo,” NGB22812), which features the absence of melanin pigmentation in the grain. Two NILs with naked caryopsis that were designated as i:Bwnud1 and i:BwBlp1nud1 were also employed in the study, which are characterized by unpigmented and melanin-containing grains, respectively. These lines were developed based on Intense blue aleurone (NGB20651, characterized by naked caryopsis and the absence of melanin in the grain) and i:BwBlp1 lines. The scheme of obtaining of lines is presented in Supplementary Figure 1. Barley specimens carrying mutant alleles of genes Ppo1 and Ppo2 served as parental ones for segregation analysis. Variety 4970 (U004, Institute of Plant Science and Resources, Okayama University, Japan, BarleyDB, http://earth.nig.ac.jp/~dclust/cgi-bin/index.cgi?lang=en) has a recessive allele of the Ppo1 gene, and variety Shogor 3 (C 97-3) (I677) carries both recessive Ppo1 and Ppo2 genes (Taketa et al., 2010). Plants were grown in a greenhouse at the Institute of ICG SB RAS (Novosibirsk, Russia) under a 12-h photoperiod in a temperature range of 20–25°C.
Identification of Ppo genes and in silico sequence analysis
The BLAST algorithm run against databases NCBI (https://www.ncbi.nlm.nih.gov/) and Ensembl Plants (https://plants.ensembl.org/index.html) was carried out to identify sequences of Ppo genes in genomes of barley and other cereals with 80% homology as a selection criterion. Tyrosinase domain sequences of genes Ppo1 (AB549330) and Ppo2 (AB549331) from ref (Taketa et al., 2010) were used as a query. Multiple sequence alignment was performed in the MUSCLE software (Madeira et al., 2019). Gene structure was predicted using FGENESH+ (Solovyev, 2004). Prediction of genes’ functional domains was performed by means of the InterPro database (Mitchell et al., 2019). A phylogenetic tree was constructed in the MEGA X software (Kumar et al., 2018) by the neighbor-joining method with a bootstrap support of 1000. The IDT PrimerQuest tool (http://eu.idtdna.com/PrimerQuest/Home/) was used to design PCR primers. The full list of PCR primers utilized in this work can be found in Supplementary Table 1.
Total-RNA extraction and gene expression analysis by quantitative reverse-transcription PCR (qRT-PCR)
Total RNA from i:BwBlp1 and cv. Bowman was isolated at three stages of spike development: from developing spikelets at the booting stage (46 days after sowing, BBCH code 49), from combined tissues of the grain pericarp and husk at the early dough stage (67 days after sowing, BBCH code 77), and at the soft-dough stage (71 days after sowing, BBCH code 83). Furthermore, total RNA from the hulls and pericarp (separated from each other by peeling with tweezers) of lines i:BwBlp1, i:Bwnud1, i:BwBlp1nud1, and cv. Bowman was isolated at the early dough stage (67 days after sowing, BBCH code 77). RNA was extracted from the samples as three biological replicates, each obtained by pooling material from three plants. The RNA isolation was performed using the RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) including DNase I treatment (QIAGEN RNase-Free DNase Set). RNA concentration in the final solution was determined on a NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). cDNA was synthesized from 1 μg of total RNA by reverse transcription with the (dT)15 primer and RevertAid™ Kit (Thermo Fisher Scientific Inc.). qRT-PCR was conducted by means of the SYNTOL SYBR Green I Kit (Synthol, Moscow, Russia) according to the manufacturer’s protocol. The amplification was performed using a QuantStudio 5 RealTime PCR System (Thermo Fisher Scientific Inc.). Each reaction was carried out in three technical replicates. Relative expression levels were calculated by the standard-curve method and normalized to Actin gene expression (von Zitzewitz et al., 2005). The Mann–Whitney U test was performed to evaluate the significance of differences in expression levels between samples.
PPO activity assay
The total PPO activity in mature seeds was evaluated with L-tyrosine according to a method described by (Taranto et al., 2012) with modifications. Seeds of cv. Bowman and i:BwBlp1 line were ground and incubated in a 0.01 M L-tyrosine solution (1 g per 4 mL of the solution) at 37°C for 19 h; after centrifugation at 12000 rpm, absorbance of the collected supernatant was measured at 405 nm on a UV/visible spectrophotometer (Bio-Rad, USA). The reported data are presented as the average of three biological replicates per sample. The Mann–Whitney U test was performed to evaluate the significance of differences between samples.
Segregation analysis
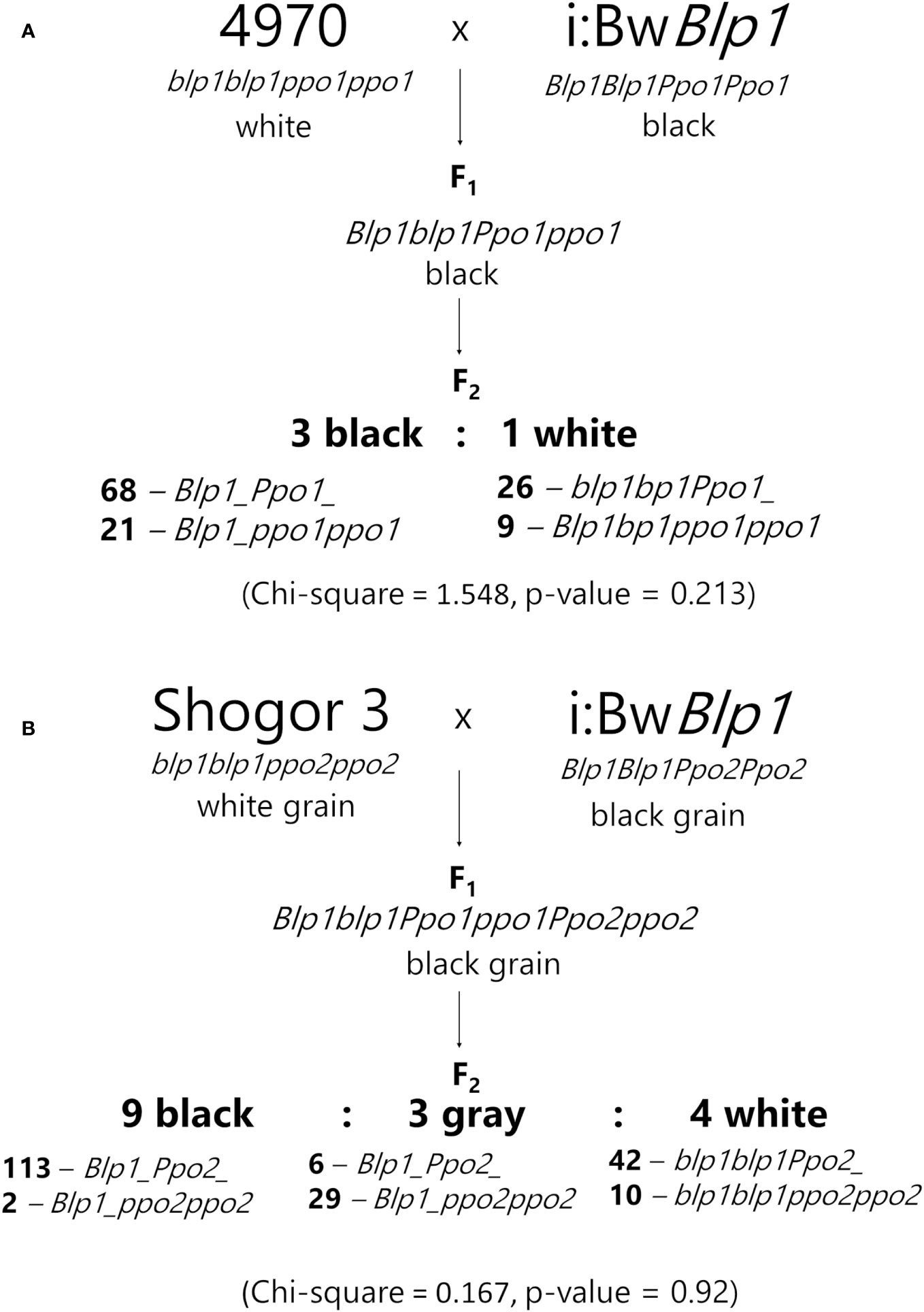
Specimens of barley 4970 and Shogor 3 were crossed with NIL i:BwBlp1 to obtain two F2 populations: 4970 × i:BwBlp1 (124 plants) and Shogor 3 × i:BwBlp1 (202 plants). The color of grains was determined visually after maturity, and the mode of inheritance of the trait was evaluated by the Chi-square test. DNA was extracted from leaves of each F2 plant according to the procedure described by (Plaschke et al., 1995) and was genotyped by PCR with primers that allow to distinguish alleles of genes Ppo1, Ppo2 (Taketa et al., 2010), and Blp1 followed by electrophoresis of amplicons in a 3% agarose gel (Supplementary Table 1).
Results
Identification of new members of the polyphenol oxidase gene family in barley
Based on tyrosinase domain sequences of Ppo1 (AB549330) and Ppo2 (AB549331), two additional Ppo genes were identified in the barley genome via a homology search by the BLAST algorithm: HORVU.MOREX.r3.3HG0294590, localized to the distal region of the long arm of chromosome 3H, and HORVU.MOREX.r3.4HG0418160, situated in the distal region of the long arm of chromosome 4H. These genes were designated as Ppo3 and Ppo4, respectively.
The length of nucleotide sequence of the Ppo3 gene amounted to 2084 bp, including a 1743 bp protein-coding region and 5′ and 3′ untranslated regions (115 and 226 bp, respectively). No introns were found in the Ppo3 gene (Figure 1). The length of nucleotide sequence of the Ppo4 gene is 2851 bp, including 5′ and 3′ untranslated regions (53 and 621 bp, respectively). The protein-coding part of the Ppo4 gene consists of one intron (491 bp) and two exons (807 and 879 bp) (Figure 1). A comparison of structural organization of the barley polyphenol oxidase family genes indicated that genes Ppo3 and Ppo4 differ in their exon-intron structure from previously described genes Ppo1 and Ppo2, which contain three exons and two introns each (Figure 1). Despite the differences in the structure of the identified Ppo genes, as well as their high variation within the family, all four genes have a functional tyrosinase domain containing two conserved Cu-binding motifs (Supplementary Figure 2).
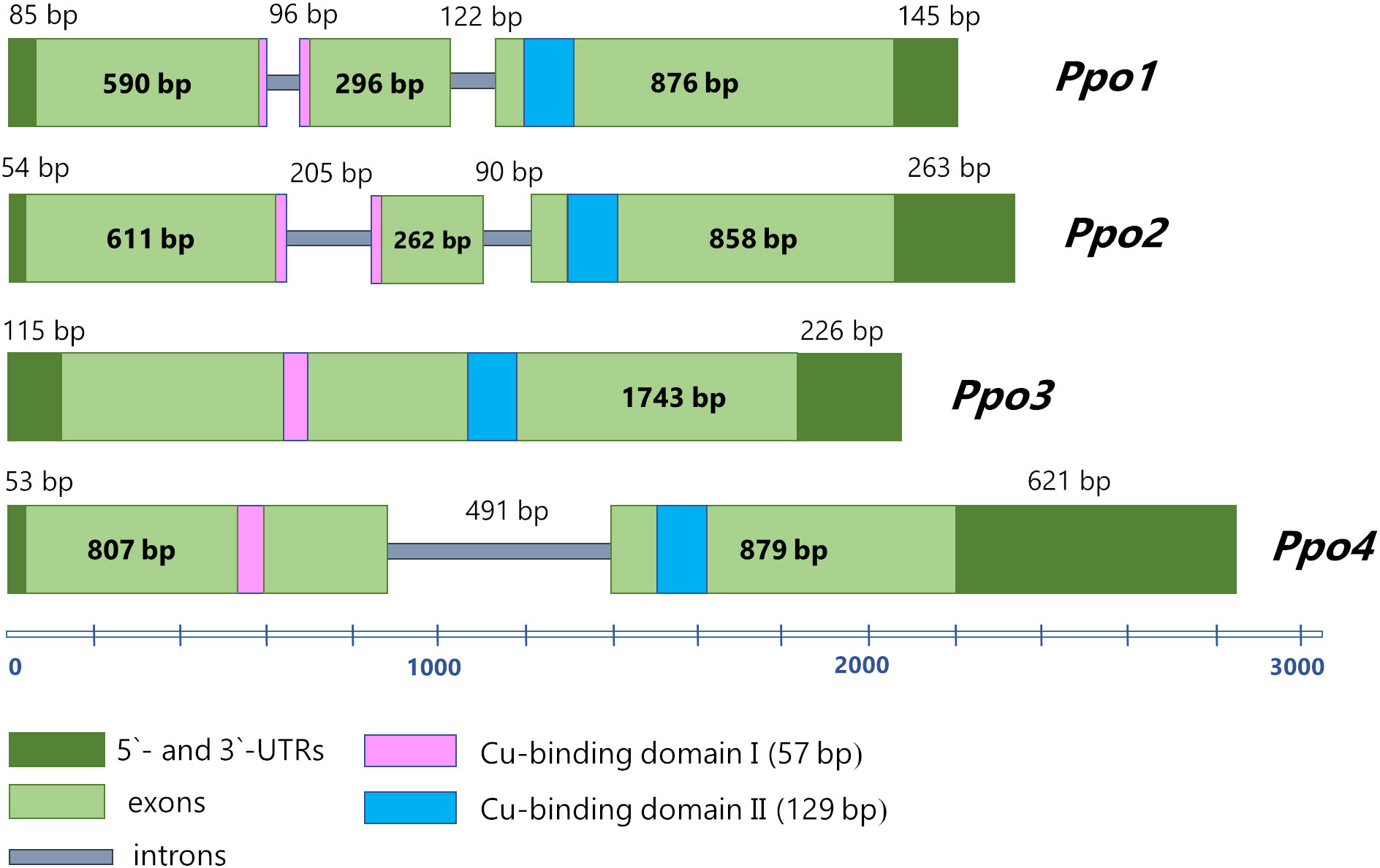
Figure 1 The structure of Ppo genes in barley. 3` and 5`-UTRs are designated by dark green, exons are light green, introns are grey. The Cu-binding domains I and II are highlighted by pink and blue, respectively.
Phylogenetic analysis of Ppo genes in barley
Tyrosinase domain sequences of Ppo genes from the following species were identified and used to construct a phylogenetic tree: Triticum aestivum (11 genes), Triticum urartu (two genes), Aegilops tauschii (three genes), Hordeum chilense (two genes), Oryza sativa (two genes), Zea mays (two genes), Brachypodium distachyon (two genes), and Sorghum bicolor (two genes). The PPO sequence of guinea yam (Dioscorea rotundata) served as an outgroup. The phylogenetic tree constructed from these sequences is displayed in Figure 2. The complete list of gene sequences with identification numbers from the Ensemble Plants database is provided in Supplementary Table 2.
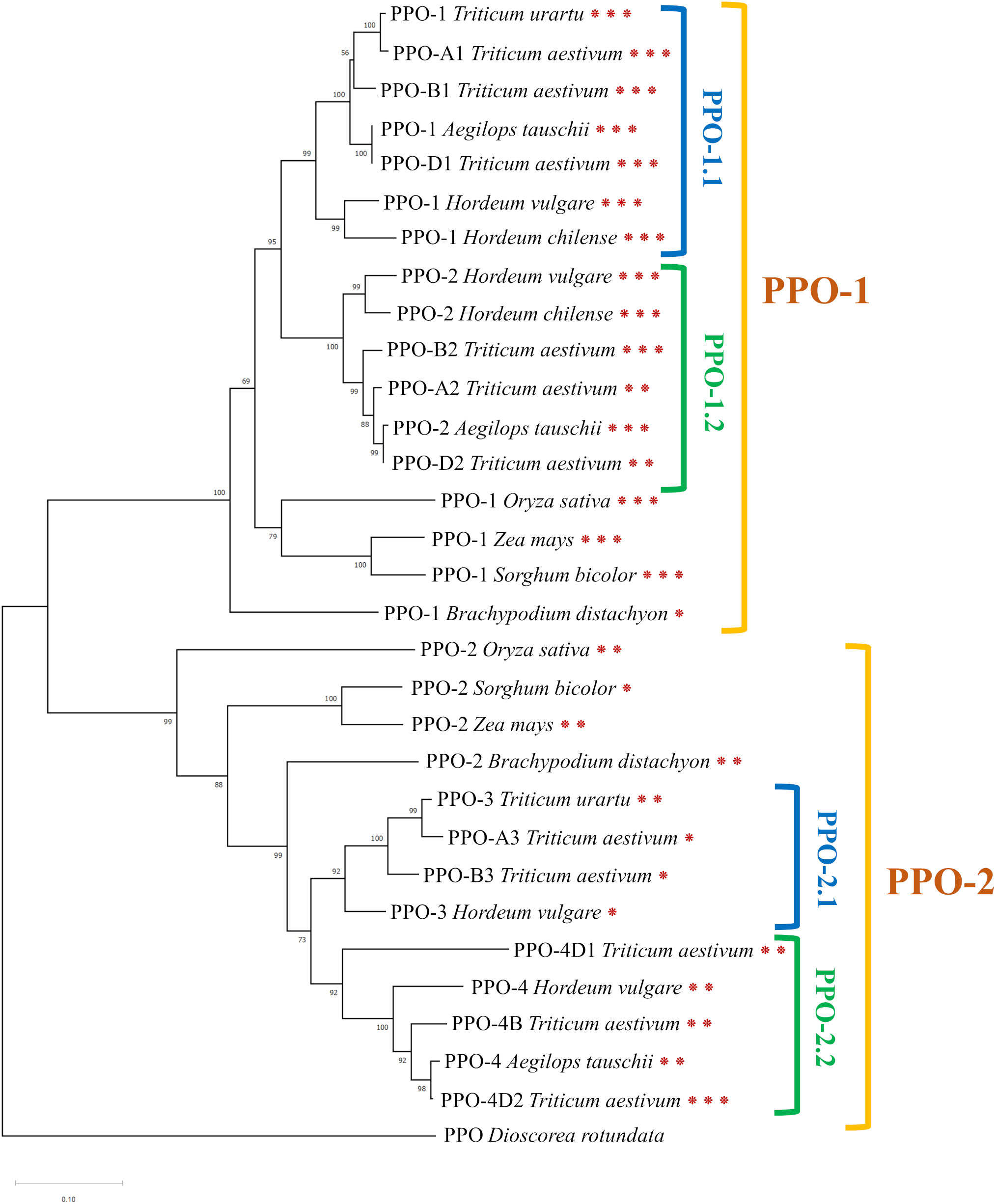
Figure 2 The phylogenetic tree constructed based on amino acid sequences of the tyrosinase domain of PPO (neighbor-joining method with a bootstrap support of 1000). Asterisks indicate the number of exons.
Ppo genes of the cereal species under study are divided into two main clusters, between which two subclusters can be distinguished (Figure 2). The first cluster, designated as PPO-1, includes the Ppo1 and Ppo2 genes of barley species (H. vulgare and H. chilense) and highly homologous sequences of Triticeae species: T. aestivum, A. tauschii, and T. urartu. Besides, the PPO-1 cluster contains Ppo genes from rice (O. sativa), maize (Z. mays), Brachypodium (B. distachyon), and sorghum (S. bicolor). Within this cluster, the Ppo sequences of Triticeae species are divided into two subclusters, PPO-1.1 and PPO-1.2, represented by two groups of duplicated genes on chromosome 2.
In the second cluster, referred to as PPO-2, two subclusters can be distinguished too: PPO-2.1 and PPO-2.2 (Figure 2). The PPO-2.1 cluster includes the barley Ppo3 gene and Ppo genes of Triticeae species located in the third group of chromosomes: PPO-3 of T. urartu, PPO-A3 of T. aestivum, and PPO-B3 of T. aestivum. The PPO-2.2 cluster includes the Ppo4 gene of barley and homologous genes of T. aestivum and A. tauschii on chromosome 4. Just as the PPO-1 cluster, the PPO-2 cluster contains such genes of rice, maize, Brachypodium, and sorghum.
It was noted that genes in clusters PPO-1 and PPO-2 have different exon-intron structures (Figure 2). Most of the genes in the PPO-1 group have three exons, except for PPO-A2 and PPO-D2 genes of T. aestivum, which have two exons, and the PPO-1 gene of B. distachyon, whose sequence contains one exon. Genes in the PPO-2 group are generally characterized by the presence of two exons; however, Ppo-2 of S. bicolor and genes from the PPO-2.1 subgroup have only one exon, with the exception of the PPO-3 gene of T. urartu. In the PPO-2.2 subgroup, the PPO-4D2 gene of T. aestivum has three exons.
These data suggested that the Ppo3 and Ppo4 genes identified in the barley genome are not unique and are represented by orthologous genes in other cereals.
Expression analysis of Ppo genes and PPO activity assay
Comparative expression analysis of Ppo genes in the barley spike during maturation was performed next. The barley NIL i:BwBlp1, which accumulates melanin pigments in the hulls and grain pericarp, and its melaninless parental cv. Bowman were used. The analysis was performed on tissues of the hulls and pericarp combined at each of three time points: the booting stage, early dough stage, and soft-dough stage, (Figure 3). None of the Ppo genes were found to be expressed at the booting stage regardless of the genotype, but expression increased as the plants matured. At the early dough and soft-dough stages, it was possible to detect differences in expression between Ppo genes depending on the presence of melanin. The transcriptional activity of genes Ppo1 and Ppo4 increases during spike development but does not differ significantly between i:BwBlp1 and cv. Bowman. The Ppo2 gene is not transcriptionally active in Bowman at any of the stages examined, whereas elevated expression of this gene was registered in the melanin-accumulating line (Figure 3). Increased expression of the Ppo3 gene in the i:BwBlp1 line in comparison with Bowman was observed, but the differences between the lines were not statistically significant.
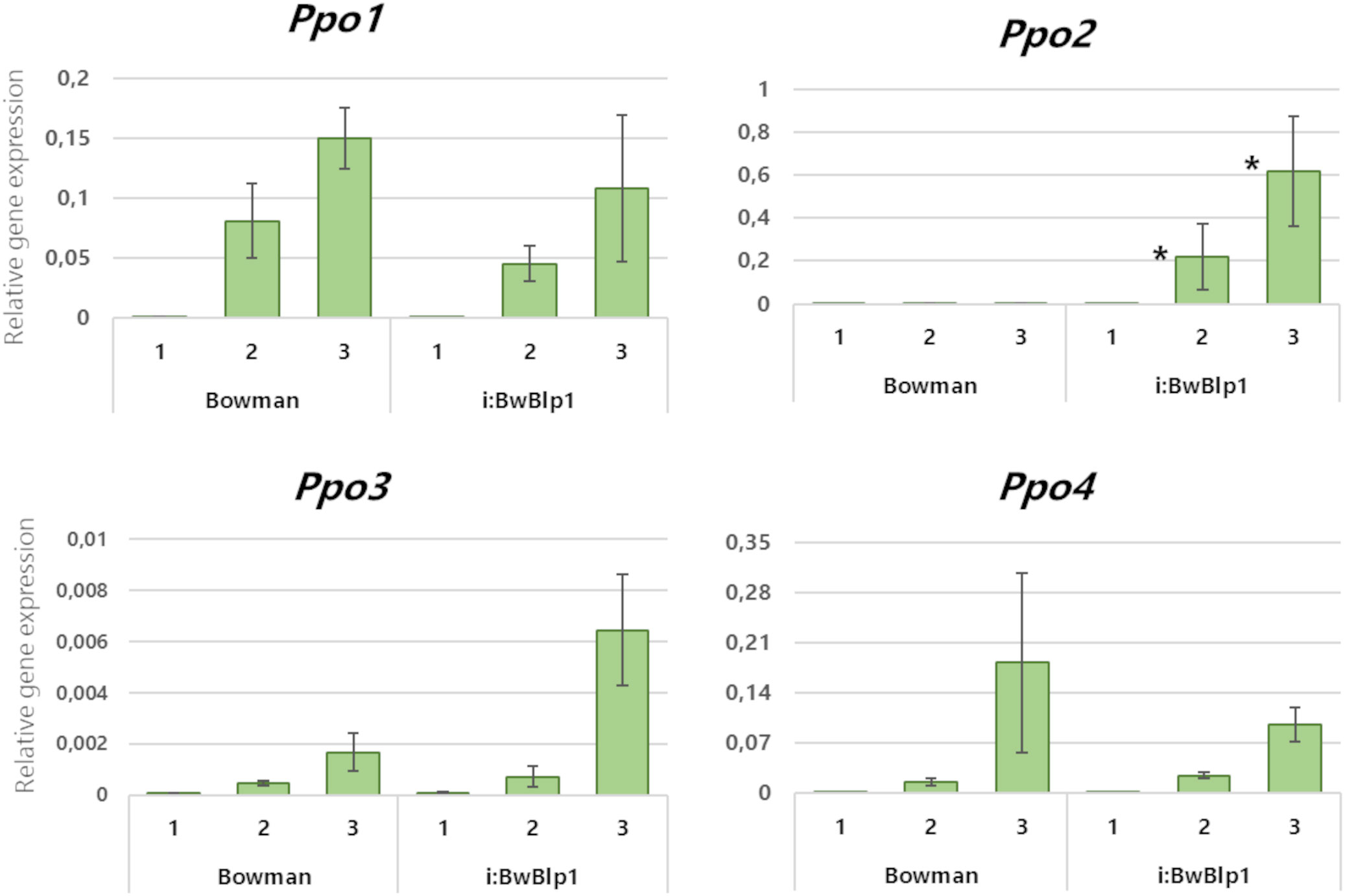
Figure 3 Relative expression levels of Ppo genes in combined tissues of the pericarp and lemma from hulled barley lines in the course of spike maturation. 1: Booting stage, 2: early dough stage, 3: soft-dough stage. *A difference is statistically significant (U test, p < 0.05) between i:BwBlp1 and cv. Bowman at the same developmental stage.
Expression analysis of the Ppo2 gene was performed on detached tissues of the hulls and pericarp at the early dough stage in hulled i:BwBlp1 and cv. Bowman and its naked derivatives: lines i:BwBlp1nud1 and i:Bwnud1 (Figure 4). In hulled lines, elevated transcription of Ppo2 was detected in both melanin-containing tissues—hulls and pericarp— in comparison with noncolored ones, while in naked lines, transcription of Ppo2 was increased in melanin-containing hulls, but not in pericarp, in comparison with tissues of noncolored line. However, in both sets of lines (hulled and naked) Ppo2 expression was significantly higher in melanin-containing hulls in comparison with the pericarp. It can be hypothesized that, of the four genes encoding PPO in barley, Ppo2 may participate in melanin biosynthesis in barley grain.
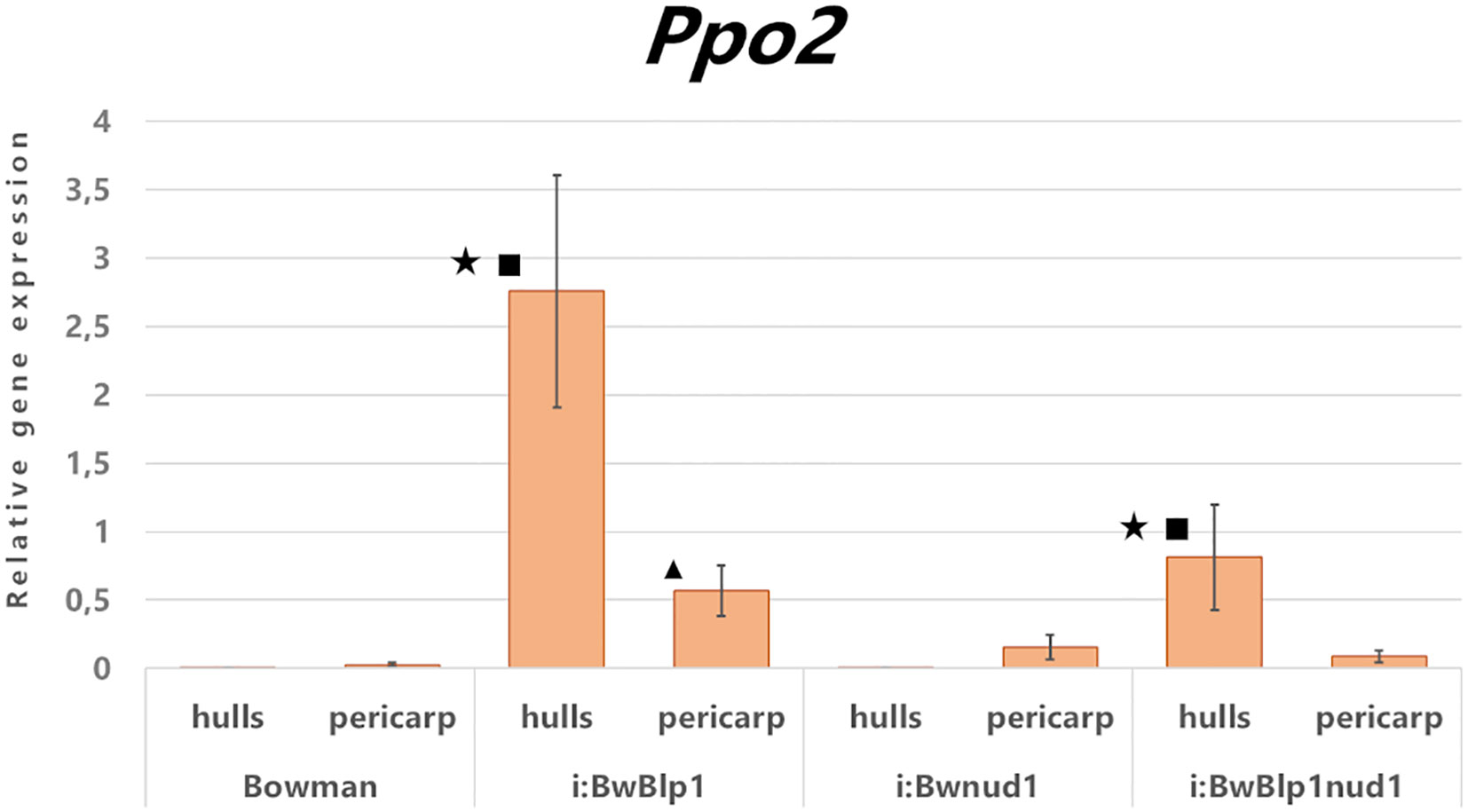
Figure 4 Relative expression levels of the Ppo2 gene in detached tissues of the pericarp and hulls from hulled (Bowman, i:BwBlp1) and naked (i:Bwnud1, i:BwBlp1nud1) lines at the early dough stage. Star – a difference is statistically significant (U test, p < 0.05) between the hulls and pericarp within a line, square – a difference is statistically significant between lines with black and white hulls, triangle – a difference is statistically significant between lines with black and white pericarps.
To confirm the obtained comparative transcriptional data, the PPO activity assay was performed on mature grain of cv. Bowman and of i:BwBlp1 line. The observed activity of PPO in the i:BwBlp1 line was significantly higher in comparison with cv. Bowman: about two times higher (Supplementary Figure 3). Overall the observed increased expression of the Ppo genes in envelopes of black grains coincides with enhanced PPO activity in mature grains.
Testing of Ppo involvement in melanin synthesis in the barley grain
To evaluate the involvement of the Ppo2 gene in melanin formation in the barley grain, two F2 populations were generated: 4970 × i:BwBlp1 (124 plants) and Shogor 3 × i:BwBlp1 (202 plants). Variety 4970 carries a recessive Ppo1 gene resulting from a mutation leading to a stop codon in the first exon (Taketa et al., 2010). The Shogor 3 variety is characterized by both recessive Ppo1 and Ppo2 genes: it has a single-nucleotide insertion in the second exon of the Ppo1 gene and a 8 bp insertion in the first exon of the Ppo2 gene (Taketa et al., 2010). Both lines do not accumulate melanin in grain envelopes (Figure 5A).

Figure 5 (A) Phenotypes of parental lines used in crosses, (B) the gray phenotype revealed among F2 plants in the Shogor 3 × i:BwBlp1 cross.
In F1, the black color of the hulls and grain pericarp was observed in all plants resulting from the crosses, just as in the parental i:BwBlp1 line. In the 4970 × i:BwBlp1 cross, F2 plants could be divided into two groups according to the grain color. The segregation ratio between melanin-pigmented and unpigmented plants was 3:1 (Chi-square = 1.548, p-value = 0.213). The F2 plants were genotyped with the primers for the Ppo1 and Blp1 genes, and the presence of melanin in the barley grain was found to depend only on the allelic state of the Blp1 gene (not on alleles of the Ppo1 gene; Figure 6A).
In the F2 population resulting from the Shogor 3 × i:BwBlp1 cross, three phenotypic classes were identified: the black color of the grain: corresponding to the i:BwBlp1 phenotype, gray: with a lighter gray color of the grain, and white: melanin was not observed in the grain, corresponding to the phenotype of Shogor 3 (Figure 5B). The black, gray, and white phenotypic classes were present in a ratio of 9:3:4 (Chi-square = 0.167, p-value = 0.92), which may indicate an epistatic interaction between genes Ppo2 and Blp1. It turned out that 29 of the 35 plants with the gray phenotype had the Ppo2 gene in the recessive homozygous state and a dominant allele of the Blp1 gene in a homozygous or heterozygous state (Figure 6B, Supplementary Figure 4). All plants with melanin in the grain carried a dominant Blp1 allele, and none of the plants characterized by the simultaneous presence of the dominant Blp1 gene allele and the recessive Ppo2 gene allele showed complete absence of melanin. Visual examination of hybrids carrying dominant Blp1 and recessive Ppo2 revealed that they had noncolored hulls and awns and dark pigments in underlying tissues. From these data, we can theorize that Ppo2 is responsible for tissue-specific melanin accumulation in hulls.
Discussion
PPO are well-researched plant enzymes. They carry out a protective enzymatic browning reaction that proceeds when the integrity of plant cell compartments is disrupted (Vaughn and Duke, 1984). PPO-induced browning of produce is undesirable, and therefore ways to prevent it are being actively sought, including the development of plant varieties with inactive PPO (Taranto et al., 2012; Hamdan et al., 2022). In barley, the strategies to avoid discoloration of barley-based food products include selecting genotypes with low discoloration such as proanthocyanidin-free genotypes, lowering the total polyphenol content or PPO activity through abrasion, heat treatment, exclusion of oxygen, and the use of enzyme inhibitors (Quinde-Axtell et al., 2006); breeding of PPO-deficient barley varieties is also possible because alleles of Ppo genes encoding inactive enzymes have been revealed in barley collections (Taketa et al., 2010).
Two Ppo genes mapped to the long arm of chromosome 2H have been found previously in the barley genome (Taketa et al., 2010). Here, we discovered two additional Ppo genes: Ppo3 and Ppo4. They were shown to encode intact functional domains and to be expressed in the barley spike. During our phylogenetic analysis, orthologs of these genes were identified in other cereal species. It is known that the size of gene families encoding PPO can vary widely among plant species (Tran et al., 2012; Jukanti, 2017). Such differences in the number of PPO genes are explained by the acquisition of new genes as a result of segmental duplications in plant genomes and by the loss of previously duplicated genes (Tran et al., 2012). Segmental duplications of Ppo genes are thought to have occurred in a common ancestor of monocotyledon species. Some of these ancestral genes have been lost during evolution; for example, in rice (O. sativa), only two Ppo genes have been identified (Yu et al., 2008). Alternatively, additional duplicated genes have been acquired; for instance, eight Ppo genes have been found in sorghum (Cai et al., 2013). Barley genes Ppo1 and Ppo2 are known to have arisen via segmental duplication of the long arm of chromosome 2 in a common ancestor of the Triticeae tribe (Taketa et al., 2010; Taranto et al., 2017). These genes—together with orthologous genes identified in maize, rice, sorghum, and Brachypodium—constitute the PPO-1 cluster (Figure 2). The other two barley genes, Ppo3 and Ppo4, together with their orthologous genes form another cluster: PPO-2 (Figure 2). Because the PPO-2 cluster contains one Ppo gene each from maize and sorghum, and because separate orthologous genes for Ppo3 and Ppo4 have been identified only in common wheat (T. aestivum) and related species, we can hypothesize that these genes have arisen through a duplication that occurred in an ancestor of the Triticeae tribe.
Within the same plant species, Ppo family members are reportedly often differ in their spatial and temporal expression profiles among different vegetative and reproductive tissues and between baseline and a response to various environmental cues. Looking at the clusterization of Ppo genes in Figure 2, it is worth noting that barley genes Ppo1 and Ppo2 cluster together with the S. bicolor Ppo-1 gene [SbPpo3 according to the nomenclature of (Cai et al., 2013)], for which a thylakoid transfer domain was predicted, while barley genes Ppo3 and Ppo4 cluster with another sorghum gene: Ppo-2 (SbPpo8), whose protein contains a predicted N-terminal signal peptide for targeting to the secretory pathway. From these data, one may conclude that the physiological functions of revealed Ppo3 and Ppo4 can differ from Ppo1 and Ppo2 belonging to different clades. Among the genes from the PPO-1 clade, wheat Ppo1 and Ppo2 genes have been reported to determine PPO activity in the grain and are considered targets for the breeding of PPO-deficient varieties (Beecher and Skinner, 2011). The rice Phr1 gene (PPO-1 of O. sativa in Figure 2), which belongs to clade PPO-1 too, participates together with Bh4 in the control of black pigments’ synthesis in rice hulls (Fukuda et al., 2012) and determines phenol reaction phenotype (which could be revealed by immersing the grain in a phenol solution) (Yu et al., 2008). In barley, Ppo1 and Ppo2 control the phenol reaction phenotype of awns and grain, respectively (Taketa et al., 2010), while roles of these genes in melanin synthesis within the barley grain have not been studied. Because barley genes Ppo1 and Ppo2 cluster together with the Phr1 gene, which controls the black hull trait in rice, one could say the functions of these genes are the same. Previously, enhanced total expression of both Ppo genes in grain envelopes combining hulls and pericarp tissues of the black-grained i:BwBlp1 line has been observed in comparison with white-grained Bowman (Glagoleva et al., 2017). Here, we distinguished and analyzed expression of each identified Ppo gene in combined tissues of hulls and pericarp (Figure 3) and in separated ones (Figure 4). It was found that Ppo2 makes a major contribution to total Ppo expression in black grain envelopes; moreover, the expression of only this gene was higher in the presence of the Blp1 locus. The enhanced transcriptional activity of the Ppo2 gene in seed envelopes coincides with increased total PPO activity observed in whole mature grain of melanin-accumulating line (Supplementary Figure 3). The subsequent transcriptional assay of the Ppo2 gene in detached hulls and pericarp tissues of hulled lines with melanin pigmentation and without it revealed enhanced transcription of Ppo2 in black hulls and pericarp in comparison with the yellow ones. In naked sister lines, elevated transcription was observed in melanin-accumulating hulls, but not in pericarp in comparison with line without melanin. However, in both sets of lines (hulled and naked) transcription of Ppo2 was significantly higher in hulls than in pericarp (Figure 4). Such differential expression of different members of one gene family in distinct tissues is a well-known phenomenon in plants. For example, among eight S. bicolor Ppo genes, only SbPpo5 and SbPpo7 were detected in the ovary or panicle tissues that developed under normal conditions, while the expression of the other genes (SbPPO1–SbPPO3, SbPPO6, and SbPPO8) was induced by stressful environments (involving a phytohormone or acid, alkaline, wounding, or heat shock treatments) (Cai et al., 2013). Among 11 Ppo genes of artichoke, three (Ppo6, Ppo7, and Ppo11) showed significant upregulation of transcripts in capitula tissues after cutting in comparison with the other genes; these genes were chosen as targets for CRISPR/Cas9-based inhibition of the enzymatic browning to maintain high quality of capitula after processing and during marketing (Pompili et al., 2023).
Although Ppo2 demonstrated elevated transcription in both melanin-containing tissues—hulls and pericarp—in comparison with noncolored Bowman’s tissues, segregation analysis of two F2 populations derived by crossing the black-grained i:BwBlp1 line with barley specimens carrying mutations in coding part of Ppo1 (variety 4970) or both Ppo1 and Ppo2 (Shogor 3) inferred the involvement of Ppo2 in melanin formation in the hulls but not in the pericarp. In the first population (based on 4970), the segregation was not different from monogenic: 3:1, for which the Blp1 locus is responsible (Woodward, 1941; Costa et al., 2001; Jia et al., 2017; Long et al., 2019). By contrast, in our second population (based on Shogor 3), the segregation ratio was 9:3:4, which corresponds to digenic mode of inheritance with the epistatic interaction between the two genes. Plants with the dominant Blp1 gene and the recessive Ppo2 gene were found to have an intermediate phenotype with light hulls and awns, which was designated by us as the gray phenotype. This observation allows us to propose the involvement of Ppo2 in melanogenesis in hulls. In pericarp tissue that possesses dark pigmentation in the presence of recessive Ppo1 as well as of Ppo2, it is possible that other oxidases participate in melanin formation under the control of the same Blp1 gene. In this regard, multicopper oxidases called laccases should be considered too; they can oxidize a variety of phenolic substrates, and for these enzymes, functions beyond lignification have been proposed (Cai et al., 2006). In general, Ppo2 is thought to be regulated by major gene Blp1, whose molecular functions have not been determined yet. Inactive PPO is not common among barley taxa. So far, only 51 phenol-negative barley accessions have been identified among 8894 samples analyzed, i.e., the grains do not discolor when immersed in a phenol solution (Takeda and Lin Chang, 1996). On the other hand, the loss of PPO activity and of dark pigmentation in seeds could have occurred during domestication of plants (Inoue et al., 2015; Balarynová et al., 2022). In rice, the allelic state of the Phr1 gene is a taxonomic trait that separates two subspecies, indica and japonica, with recessive Phr1 widespread within the japonica subspecies, and recessive Bh4 within the indica subspecies (Yu et al., 2008). Crossing of uncolored specimens from japonica and indica leads to progenies with dark pigments of hulls (Fukuda et al., 2012). It is likely that in barley, inactive forms of PPO (in contrast to recessive Blp1) did not spread within a region where selection for white grains took place. The mutations in the Ppo genes may have occurred later in yellow-grained barleys and have not been under domestication-related selection in barley.
Conclusions
Despite widespread occurrence of melanin pigments in nature, their synthesis in intact tissues of plants is not well understood. PPOs are considered the main participants of melanin production in damaged plant tissues when such an enzyme located in chloroplasts and its substrates located in vacuoles are released and interact giving rise to melanin. In undamaged tissues, such a mechanism of melanogenesis is not obvious, although there is evidence of the presence of substrates of PPOs in chloroplasts, and melanin granules in chloroplasts have been detected in some species. Here, the focus of the study is to test the hypothesis that Ppo genes are responsible for melanin formation in the barley grain, which accumulates melanin in the hulls and pericarp. The Ppo gene family was investigated, and two new genes coding for PPOs were found in barley and characterized for the first time. These genes proved to be functionally active and expressed in the barley spike. Among the four Ppo genes of barley, Ppo2 was shown to be upregulated in melanin-accumulating hulls and pericarp in comparison with the tissues of the line, not accumulating melanin, but transcription in hulls was significantly higher than in the pericarp. Moreover, recessive Ppo2 in combination with dominant Blp1 resulted in hybrids with discolored hulls and a dark pericarp. Although our results imply the involvement of Ppo2 in melanization of barley hulls, definitive proof can be obtained only by knocking out Ppo2 in black-grained barely genotypes by genetic engineering techniques.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author/s.
Author contributions
AG: Formal Analysis, Investigation, Methodology, Validation, Writing – original draft. TK: Investigation, Methodology, Resources, Writing – review & editing. EK: Conceptualization, Writing – review & editing. OS: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study was funded by the Russian Science Foundation, grant number 21-76-10024. The cultivation of the barley plants at the Greenhouse Facility was supported by ICG project FWNR-2022-0017.
Acknowledgments
The English language was corrected and certified by shevchuk-editing.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1320770/full#supplementary-material
Supplementary Table 1 | The list of primers used in the study.
Supplementary Table 2 | Identification numbers of Ppo genes from the Ensembl Plants and NCBI databases used for the phylogenetic analysis.
Supplementary Figure 1 | The scheme of obtaining of NILs i:Bwnud1 and i:BwBlp1nud1. Lines with a dark-blue aleurone and i:BwBlp1 lines based on the cv. Bowman genetic background were used. PCR primers specific to genes Blp1 and Nud1 were employed for marker-assisted selection.
Supplementary Figure 2 | The alignment of amino acid sequences of PPO.
Supplementary Figure 3 | PPO activity assay in cv. Bowman and i:BwBlp1 line. A405 means absorbance at 405 nm wavelength after incubation of grinded seeds in L-tyrosine solution. * - significant differences between samples (U test, p < 0.05).
Supplementary Figure 4 | Electropherograms of the Ppo2 gene alleles screening in F2 population of Shogor 3 × i:BwBlp1 cross. The amplicons lengths: 88 bp for dominant allele of the Ppo2 gene, 96 bp for the recessive one.
References
Araji, S., Grammer, T. A., Gertzen, R., Anderson, S. D., Mikulic-Petkovsek, M., Veberic, R., et al. (2014). Novel roles for the polyphenol oxidase enzyme in secondary metabolism and the regulation of cell death in walnut. Plant Physiol. 164, 1191–1203. doi: 10.1104/pp.113.228593
Balarynová, J., Klčová, B., Sekaninová, J., Kobrlová, L., Cechová, M. Z., Krejčí, P., et al. (2022). The loss of polyphenol oxidase function is associated with hilum pigmentation and has been selected during pea domestication. New Phytol. 235, 1807–1821. doi: 10.1111/nph.18256
Bazhenov, M. S., Litvinov, D. Y., Divashuk, M. G. (2023). Melanin found in wheat spike husks 542842. doi: 10.1101/2023.05.30.542842
Beecher, B., Skinner, D. Z. (2011). Molecular cloning and expression analysis of multiple polyphenol oxidase genes in developing wheat (Triticum aestivum) kernels. J. Cereal Sci. 53, 371–378. doi: 10.1016/j.jcs.2011.01.015
Boeckx, T., Winters, A., Webb, K. J., Kingston-Smith, A. H. (2017). Detection of potential chloroplastic substrates for polyphenol oxidase suggests a role in undamaged leaves. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00237
Boeckx, T., Winters, A. L., Webb, K. J., Kingston-Smith, A. H. (2015). Polyphenol oxidase in leaves: Is there any significance to the chloroplastic localization? J. Exp. Bot. 66, 3571–3579. doi: 10.1093/jxb/erv141
Cai, X., Davis, E. J., Ballif, J., Liang, M., Bushman, E., Haroldsen, V., et al. (2006). Mutant identification and characterization of the laccase gene family in Arabidopsis. J. Exp. Bot. 57, 2563–2569. doi: 10.1093/jxb/erl022
Cai, Y., Dong, Z., Zhao, S., Han, Y., Shao, Y., Lu, M., et al. (2013). Genome-wide analysis of polyphenol oxidase genes and their transcriptional patterns during grain development in Sorghum. Int. J. Plant Sci. 174, 710–721. doi: 10.1086/669909
Ceccarelli, S., Grando, S., Leur, J. A. G. V. (1987). Genetic diversity in barley landraces from Syria and Jordan. Euphytica 36, 389–405. doi: 10.1007/BF00041482
Choo, T. M., Vigier, B., Savard, M. E., Blackwell, B., Martin, R., Wang, J., et al. (2015). Black barley as a means of mitigating deoxynivalenol contamination. Crop Sci. 55, 1096. doi: 10.2135/cropsci2014.05.0405
Costa, J. M., Corey, A., Hayes, P. M., Jobet, C., Kleinhofs, A., Kopisch-Obusch, A., et al. (2001). Molecular mapping of the Oregon Wolfe Barleys: a phenotypically polymorphic doubled-haploid population. Theor. Appl. Genet. 103, 415–424. doi: 10.1007/s001220100622
Dossou, S. S. K., Luo, Z., Wang, Z., Zhou, W., Zhou, R., Zhang, Y., et al. (2022). The dark pigment in the sesame (Sesamum indicum L.) seed coat: isolation, characterization, and its potential precursors. Front. Nutr. 9. doi: 10.3389/fnut.2022.858673
Druka, A., Franckowiak, J., Lundqvist, U., Bonar, N., Alexander, J., Houston, K., et al. (2011). Genetic dissection of barley morphology and development. Plant Physiol. 155, 617–627. doi: 10.1104/pp.110.166249
Fukuda, A., Shimizu, H., Shiratsuchi, H., Yamaguchi, H., Ohdaira, Y., Mochida, H. (2012). Complementary genes that cause black ripening hulls in F 1 plants of crosses between indica and japonica rice cultivars. Plant Production Sci. 15, 270–273. doi: 10.1626/pps.15.270
Glagoleva, A. Y., Novokreschenov, L. A., Shoeva, O. Y., Kovaleva, O. N., Khlestkina, E. K. (2022). Studying grain color diversity in the barley collection of VIR. Trudy po prikladnoy botanike genetike i selektsii 183, 76–84. doi: 10.30901/2227-8834-2022-3-76-84
Glagoleva, A. Y., Shmakov, N. A., Shoeva, O. Y., Vasiliev, G. V., Shatskaya, N. V., Börner, A., et al. (2017). Metabolic pathways and genes identified by RNA-seq analysis of barley near-isogenic lines differing by allelic state of the Black lemma and pericarp (Blp) gene. BMC Plant Biol. 17, 182. doi: 10.1186/s12870-017-1124-1
Glagoleva, A. Y., Shoeva, O. Y., Khlestkina, E. K. (2020). Melanin pigment in plants: current knowledge and future perspectives. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00770
Gracheva, N. V., Zheltobryukhov, V. F. (2016). A method for obtaining melanins from sunflower husk and studying its antioxidant activity. News Kazan Technological Univ. 19 (15), 154–157.
Hamdan, N., Lee, C. H., Wong, S. L., Fauzi, C. E. N. C. A., Zamri, N. M. A., Lee, T. H. (2022). Prevention of enzymatic browning by natural extracts and genome-editing: A review on recent progress. Molecules 27, 1101. doi: 10.3390/molecules27031101
Inoue, T., Yuo, T., Ohta, T., Hitomi, E., Ichitani, K., Kawase, M., et al. (2015). Multiple origins of the phenol reaction negative phenotype in foxtail millet, Setaria italica (L.) P. Beauv., were caused by independent loss-of-function mutations of the polyphenol oxidase (Si7PPO) gene during domestication. Mol. Genet. Genomics 290, 1563–1574. doi: 10.1007/s00438-015-1022-x
Jia, Q., Wang, J., Zhu, J., Hua, W., Shang, Y., Yang, J., et al. (2017). Toward identification of black lemma and pericarp gene blp1 in barley combining bulked segregant analysis and specific-locus amplified fragment sequencing. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01414
Jukanti, A. (2017). Polyphenol oxidases (PPOs) in plants. Springer Singapore. doi: 10.1007/978-981-10-5747-2
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Li, B., Lu, X., Gebremeskel, H., Zhao, S., He, N., Yuan, P., et al. (2020). Genetic mapping and discovery of the candidate gene for black seed coat color in watermelon (Citrullus lanatus). Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01689
Llorente, B., Alonso, G. D., Bravo-Almonacid, F., Rodríguez, V., López, M. G., Carrari, F., et al. (2011). Safety assessment of nonbrowning potatoes: opening the discussion about the relevance of substantial equivalence on next generation biotech crops. Plant Biotechnol. J. 9, 136–150. doi: 10.1111/J.1467-7652.2010.00534.X
Llorente, B., López, M. G., Carrari, F., Asís, R., Naranjo, R. D. D. P., Flawiá, M. M., et al. (2014). Downregulation of polyphenol oxidase in potato tubers redirects phenylpropanoid metabolism enhancing chlorogenate content and late blight resistance. Mol. Breed. 34, 2049–2063. doi: 10.1007/S11032-014-0162-8
Long, Z., Jia, Y., Tan, C., Zhang, X.-Q., Angessa, T., Broughton, S., et al. (2019). Genetic mapping and evolutionary analyses of the black grain trait in barley. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01921
Loskutov, I. G., Blinova, E. V., Gavrilova, O. P., Gagkaeva, T. Y. (2016). The valuable characteristics of oats genotypes and resistance to Fusarium disease. Vavilov J. Genet. Breed. 20, 286–294. doi: 10.18699/VJ16.151
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Marusek, C. M., Trobaugh, N. M., Flurkey, W. H., Inlow, J. K. (2006). Comparative analysis of polyphenol oxidase from plant and fungal species. J. inorganic Biochem. 100, 108–123. doi: 10.1016/J.JINORGBIO.2005.10.008
Mitchell, A. L., Attwood, T. K., Babbitt, P. C., Blum, M., Bork, P., Bridge, A., et al. (2019). InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 47, D351–D360. doi: 10.1093/nar/gky1100
Moon, K. M., Kwon, E. B., Lee, B., Kim, C. Y. (2020). Recent trends in controlling the enzymatic browning of fruit and vegetable products. Molecules 25, 2754. doi: 10.3390/molecules25122754
Nicolas, J. J., Richard-Forget, F. C., Goupy, P. M., Amiot, M.-J., Aubert, S. Y. (1994). Enzymatic browning reactions in apple and apple products. Crit. Rev. Food Sci. Nutr. 34, 109–157. doi: 10.1080/10408399409527653
Nicolaus, R. A., Piattelli, M., Fattorusso, E. (1964). The structure of melanins and melanogenesis—IV. Tetrahedron 20, 1163–1172. doi: 10.1016/s0040-4020(01)98983-5
Plaschke, J., Ganal, M. W., Roder, M. S. (1995). Detection of genetic diversity in closely related bread wheat using microsatellite markers. Theor. Appl. Genet. 91–91, 1001–1007. doi: 10.1007/BF00223912
Pompili, V., Mazzocchi, E., Moglia, A., Acquadro, A., Comino, C., Rotino, G. L., et al. (2023). Structural and expression analysis of polyphenol oxidases potentially involved in globe artichoke (C. cardunculus var. scolymus L.) tissue browning. Sci. Rep. 13, 12288. doi: 10.1038/s41598-023-38874-4
Qi, Y., Liu, J., Liu, Y., Yan, D., Wu, H., Li, R., et al. (2020). Polyphenol oxidase plays a critical role in melanin formation in the fruit skin of persimmon (Diospyros kaki cv. ‘Heishi’). Food Chem. 330, 127253. doi: 10.1016/j.foodchem.2020.127253
Quinde-Axtell, Z., Powers, J., Baik, B.-K. (2006). Retardation of discoloration in barley flour gel and dough. Cereal Chem. 83, 385–390. doi: 10.1094/CC-83-0385
Rodríguez-Suárez, C., Atienza, S. G. (2014). Polyphenol oxidase genes in Hordeum Chilense and implications in tritordeum breeding. Mol. Breed. 34, 1867–1877. doi: 10.1007/s11032-014-0145-9
Shoeva, O., Mursalimov, S. R., Gracheva, N. V., Glagoleva, A.Yu., Börner, A., Khlestkina, E. K. (2020). Melanin formation in barley grain occurs within plastids of pericarp and husk cells. Sci. Rep. 10, 1–9. doi: 10.1038/s41598-019-56982-y
Solovyev, V. (2004). “Statistical approaches in eukaryotic gene prediction,” in Handbook of Statistical Genetics (Hoboken, New Jersey, U.S.: John Wiley & Sons, Ltd). doi: 10.1002/0470022620.bbc06
Takeda, K., Lin Chang, C. (1996). Inheritance and geographical distribution of phenol reaction-less varieties of barley. Euphytica 90, 217–221. doi: 10.1007/BF00023861
Taketa, S., Matsuki, K., Amano, S., Saisho, D., Himi, E., Shitsukawa, N., et al. (2010). Duplicate polyphenol oxidase genes on barley chromosome 2H and their functional differentiation in the phenol reaction of spikes and grains. J. Exp. Bot. 61, 3983–3993. doi: 10.1093/jxb/erq211
Taranto, F., Delvecchio, L. N., Mangini, G., Faro, L. D., Blanco, A., Pasqualone, A. (2012). Molecular and physico-chemical evaluation of enzymatic browning of whole meal and dough in a collection of tetraploid wheats. J. Cereal Sci. 55, 405–414. doi: 10.1016/j.jcs.2012.02.005
Taranto, F., Pasqualone, A., Mangini, G., Tripodi, P., Miazzi, M., Pavan, S., et al. (2017). Polyphenol oxidases in crops: biochemical, physiological and genetic aspects. Int. J. Mol. Sci. 18, 377. doi: 10.3390/ijms18020377
Tran, L. T., Taylor, J. S., Constabel, C. P. (2012). The polyphenol oxidase gene family in land plants: Lineage-specific duplication and expansion. BMC Genomics 13, 1. doi: 10.1186/1471-2164-13-395
Varga, M., Berkesi, O., Darula, Z., May, N. V., Palágyi, A. (2016). Structural characterization of allomelanin from black oat. Phytochemistry 130, 313–320. doi: 10.1016/j.phytochem.2016.07.002
Vaughn, K. C., Duke, S. O. (1984). Function of polyphenol oxidase in higher plants. Physiologia Plantarum 60, 106–112. doi: 10.1111/j.1399-3054.1984.tb04258.x
von Zitzewitz, J., Szűcs, P., Dubcovsky, J., Yan, L., Francia, E., Pecchioni, N., et al. (2005). Molecular and structural characterization of barley vernalization genes. Plant Mol. Biol. 59, 449–467. doi: 10.1007/s11103-005-0351-2
Wan, L., Li, B., Pandey, M. K., Wu, Y., Lei, Y., Yan, L., et al. (2016). Transcriptome analysis of a new peanut seed coat mutant for the physiological regulatory mechanism involved in seed coat cracking and pigmentation. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01491
Wang, L., Dossou, S. S. K., Wei, X., Zhang, Y., Li, D., Yu, J., et al. (2020). Transcriptome dynamics during black and white sesame (Sesamum indicum L.) seed development and identification of candidate genes associated with black pigmentation. Genes 11, 1–14. doi: 10.3390/genes11121399
Weltzien, E. (1988). Evaluation of barley (Hordeum vulgare L.) landrace populations originating from different growing regions in the near East*. Plant Breed. 101, 95–106. doi: 10.1111/j.1439-0523.1988.tb00273.x
Woodward, R. W. (1941). INHERITANCE OF A MELANINLIKE PIGMENT IN THE GLUMES AND CARYOPSES OF BARLEY ‘. J. Agric. Res. 63, 21–28.
Yao, Z., Qi, J., Wang, L. (2012). Isolation, fractionation and characterization of melanin-like pigments from chestnut (Castanea mollissima) shells. J. Food Sci. 77, 671–676. doi: 10.1111/j.1750-3841.2012.02714.x
Yu, Y., Tang, T., Qian, Q., Wang, Y., Yan, M., Zeng, D., et al. (2008). Independent losses of function in a polyphenol oxidase in rice: differentiation in grain discoloration between subspecies and the role of positive selection under domestication. Plant Cell 20, 2946–2959. doi: 10.1105/tpc.108.060426
Keywords: Black lemma and pericarp 1, enzymatic browning, epistasis, genetic segregation, melanin
Citation: Glagoleva AY, Kukoeva TV, Khlestkina EK and Shoeva OY (2024) Polyphenol oxidase genes in barley (Hordeum vulgare L.): functional activity with respect to black grain pigmentation. Front. Plant Sci. 14:1320770. doi: 10.3389/fpls.2023.1320770
Received: 12 October 2023; Accepted: 18 December 2023;
Published: 08 January 2024.
Edited by:
Jianping Wang, University of Florida, United StatesReviewed by:
Francesca Taranto, CNR-IBBR Institute of Bioscience and BioResources, ItalyYanhao Xu, Hubei Academy of Agricultural Sciences, China
Maria Dellino, University of Bari Aldo Moro, Italy
Copyright © 2024 Glagoleva, Kukoeva, Khlestkina and Shoeva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anastasiia Y. Glagoleva, glagoleva@bionet.nsc.ru
 Anastasiia Y. Glagoleva
Anastasiia Y. Glagoleva Tat’jana V. Kukoeva1
Tat’jana V. Kukoeva1 Elena K. Khlestkina
Elena K. Khlestkina Olesya Y. Shoeva
Olesya Y. Shoeva