- 1Plant Cell Technology Laboratory, Graduate School of Horticulture, Chiba University, Matsudo, Japan
- 2Institute for Advanced Academic Research (IAAR), Chiba University, Chiba, Japan
- 3RIKEN Center for Sustainable Resource Science, Yokohama, Japan
- 4Graduate School of Bioagricultural Sciences, Nagoya University, Nagoya, Japan
- 5Plant Molecular Science Center, Chiba University, Chiba, Japan
- 6Research Center for Space Agriculture and Horticulture, Chiba University, Matsudo, Japan
The ectopic overexpression of developmental regulator (DR) genes has been reported to improve the transformation in recalcitrant plant species because of the promotion of cellular differentiation during cell culture processes. In other words, the external plant growth regulator (PGR) application during the tissue and cell culture process is still required in cases utilizing DR genes for plant regeneration. Here, the effect of Arabidopsis BABY BOOM (BBM) and WUSCHEL (WUS) on the differentiation of tobacco transgenic cells was examined. We found that the SRDX fusion to WUS, when co-expressed with the BBM-VP16 fusion gene, significantly influenced the induction of autonomous differentiation under PGR-free culture conditions, with similar effects in some other plant species. Furthermore, to understand the endogenous background underlying cell differentiation toward regeneration, phytohormone and RNA-seq analyses were performed using tobacco leaf explants in which transgenic cells were autonomously differentiating. The levels of active auxins, cytokinins, abscisic acid, and inactive gibberellins increased as cell differentiation proceeded toward organogenesis. Gene Ontology terms related to phytohormones and organogenesis were identified as differentially expressed genes, in addition to those related to polysaccharide and nitrate metabolism. The qRT-PCR four selected genes as DEGs supported the RNA-seq data. This differentiation induction system and the reported phytohormone and transcript profiles provide a foundation for the development of PGR-free tissue cultures of various plant species, facilitating future biotechnological breeding.
1 Introduction
Plants can regenerate a whole new plant from a single cell through dedifferentiation and redifferentiation, which is an ability named totipotency. Artificial control of totipotency in cell and tissue culture has been used for plant conservation, breeding, and scientific research because it accelerates not only plant propagation, but also the production of genetically modified plants. Cell differentiation is controlled by the external application of plant growth regulators (PGRs), such as auxins and cytokinins, to the culture medium; however, the optimal hormonal conditions for differentiation vary depending on plant species, genotypes, and tissue types. Therefore, establishment of optimal hormonal conditions to induce differentiation is required for every explant type. To date, several genes have been found to affect cell differentiation, such as ISOPENTENYLTRANSFERASE (IPT) (Akiyoshi et al., 1984; Barry et al., 1984), SHOOT MERISTEMLESS (STM) (Barton and Poethig, 1993), LEAFY COTYLEDON1 (LEC1) (Lotan et al., 1998), LEAFY COTYLEDON2 (LEC2) (Stone et al., 2001), GROWTH-REGULATING FACTOR-GRF-INTERACTING FACTOR (GRF-GIF) (Debernardi et al., 2020), BABY BOOM (BBM) (Boutilier et al., 2002), and WUSCHEL (WUS) (Laux et al., 1996). Therefore, these genes are also called developmental regulators (DRs).
BBM encodes an AP2/ERF transcription factor (Boutilier et al., 2002), which functions as a key regulator of zygotic embryogenesis in Arabidopsis (Chen et al., 2022a). Somatic embryo formation from the tissues of regenerated transgenic plants where Brassica BBM was overexpressed has been reported in Arabidopsis, rapeseed, tobacco, and sweet pepper (Boutilier et al., 2002; Heidmann et al., 2011). Also, during cell culture, the introduction and overexpression of Brassica BBM in Chinese white poplar calli induced somatic embryos (Deng et al., 2009).
WUS encodes a homeobox transcription factor that mainly localizes to the shoot apical meristem (SAM) to maintain stem cell identity (Laux et al., 1996). WUS is also involved in embryogenesis, and its overexpression induces somatic embryogenesis in Arabidopsis (Zuo et al., 2002; Ikeda et al., 2009). In addition to its homeodomain, the C-terminal region of the WUS protein contains three transcriptional regulatory domains: an acidic domain, a WUS-box, and an ethylene-responsive element binding factor-associated amphiphilic repression (EAR-like) motif (Ikeda et al., 2009).
Methodology using ectopic expression of these DRs induces somatic embryogenesis and shoot formation in transgenic cells, consequently improving transformation efficiency (Gordon-Kamm et al., 2019; Duan et al., 2022; Lian et al., 2022; Cody et al., 2023). In particular, a combination of BBM and WUS has been frequently used in biotechnological studies to improve the transformation efficiency of recalcitrant monocot cultivars (Lowe et al., 2016; Mookkan et al., 2017; Lowe et al., 2018; Gordon-Kamm et al., 2019; Suo et al., 2021; Johnson et al., 2023).
However, these previous studies using DRs have not completely omitted the application of PGRs to the tissue or cell culture media for plant regeneration. The main bottleneck in artificial plant cellular differentiation is the difficulty in establishing the appropriate culture conditions, especially PGR application recipe, for each explant material. To overcome the obstacles, a versatile methodology to regulate cellular differentiation using DRs would be useful for biotechnological breedings. In addition, detailed information on the gene expression profiles and physiology regulated by DRs during cell differentiation will be informative for further improvements in plant molecular science and biotechnological breeding. Several studies have reported that phytohormone signaling in regenerated transgenic plants is altered by the ectopic expression of DRs (Li et al., 2019; Chen et al., 2022b; Zhou et al., 2023). However, how DR expression affects endogenous gene expression and physiology in cells undergoing differentiation has not been understood completely.
In this study, we utilized Arabidopsis BBM and WUS and modified the expressions and proteins to achieve autonomous differentiation of the transgenic cells. Here, we report that Brassicaceae Arabidopsis BBM and WUS successfully induced autonomous dedifferentiation and redifferentiation of transgenic cells without the application of PGRs during the culture process in Solanaceae tobacco, petunia, and Asteraceae lettuce. Using the differentiating transgenic tobacco cells obtained with the above system, we performed phytohormone measurements and RNA-seq analyses. We herein provide information on the physiological and gene expression backgrounds that enable autonomous cell dedifferentiation.
2 Materials and methods
2.1 Plant materials and growth conditions
Arabidopsis thaliana (Col-0) seeds were surface-sterilized in sodium hypochlorite solution (1% effective chlorine concentration) for 7–10 minutes, and then germinated on Murashige and Skoog (MS) medium containing 1% sucrose, 0.5 g l-1 2-Morpholinoethanesulfonic acid, monohydrate (MES), and 0.8% agar. Two-week-old seedlings were acclimatized and grown at 22°C under an 8 h light/16 h dark cycle. In vitro-grown tobacco (Nicotiana tabacum ‘Petit Havana’ SR-1) (Maliga et al., 1973) and petunia (Petunia x hybrida ‘White Creepia’) plants were maintained with half-strength MS medium containing 3% sucrose and 0.8% agar at 25°C under continuous fluorescent light. Lettuce (Lactuca sativa ‘Cisco’, ‘Watson’, and ‘Berkeley’) (TAKII & Co.,Ltd, Kyoto, Japan) seeds were surface-sterilized in sodium hypochlorite solution (1% effective chlorine concentration) for 20 min, and then grown on half-strength MS medium containing 1% sucrose and 0.8% agar at 22°C under an 8 h light/16 h dark cycle.
2.2 Vector construction
The primers used for vector construction are listed in Supplementary Table 1. BBM (AT5G17430) coding region (CDS) was PCR-amplified using cDNA from A. thaliana pistils at 7 day-after flowering and was then cloned into pCR® 8/GW/TOPO (Thermo Fisher Scientific, Inc., Waltham, M.A., USA). The BBM CDS was inserted into pGWB2 (Nakagawa et al., 2007) using the LR reaction (Thermo Fisher Scientific, Inc.). The PCR-amplified fragment of Ipomoea batatas Myb (IbMyb) expression cassette (35Sp:IbMyb:NosT), which induces anthocyanin biosynthesis in the transgenic tobacco cells for visible marker selection (Sato et al., 2023), was further inserted into PmeI-SbfI site using the In-Fusion® HD cloning kit (Takara Bio Inc., Shiga, Japan), producing a pGWB_BBM binary vector (Supplementary Figure 1B).The 35Sp:ΩBBMVP16:HSPT region was cloned into an entry vector (pUC_35Sp:ΩBBMVP16) and transferred to pGWB1 (Nakagawa et al., 2007) via the LR reaction. Then an IbMyb expression cassette was further inserted into PmeI-SbfI site by In-Fusion®, producing a pGWB_ΩBBMVP16 binary vector (Supplementary Figure 1C). The CDS of WUS (AT2G17950) was amplified using cDNA from A. thaliana flower buds by PCR. The amplicons of A. thaliana ribosomal protein subunit 5A (RPS5A) promoter (AT3G11940) and the WUS CDS were inserted into a NotI site on pUC_35Sp:ΩBBMVP16 entry vector through In-Fusion®. The constructed 35Sp:ΩBBMVP16:NosT_RPS5Ap:WUS:NosT region was amplified, then inserted into the BamHI-SalI site on pKI1.1R vector (Tsutsui and Higashiyama, 2017) through In-Fusion®. Further, the IbMyb expression cassette was inserted into a PmeI site by In-Fusion®. The final binary vector was named pKI_ΩBBMVP16&WUS (Supplementary Figure 1G). PCR-amplified fragments of RPS5A promoter, SRDXWUSm1:NosT, and 35S promoter were inserted into the HpaI-NruI site on pKI_ΩBBMVP16&WUS vector through In-Fusion®, producing a binary vector pKI_ΩBBMVP16&SRDXWUSm1 (Supplementary Figure 1J). pGWB_GFP (Supplementary Figure 1A) was obtained by LR reaction of pCR® 8/GW/TOPO_eGFP and pGWB2 (Nakagawa et al., 2007). The PCR-amplified 35Sp:BBM:NosT region of pGWB_BBM (Supplementary Figure 1B) was inserted into the XmaI-PsiI sites of pKI_ΩBBMVP16&WUS (Supplementary Figure 1G) through In-Fusion®, producing a binary vector pKI_BBM&WUS (Supplementary Figure 1F). The SmaI-PsiI region was removed from pKI_ΩBBMVP16&WUS (Supplementary Figure 1G), and then self-ligated using Ligation high Ver. 2 (TOYOBO Co. Ltd., Osaka, Japan) to produce the binary vector pKI_WUS (Supplementary Figure 1D). The SmaI-PsiI regions of pKI_ΩBBMVP16&SRDXWUSm1 (Supplementary Figure 1J) were removed and self-ligated to produce the binary vector pKI_SRDXWUSm1 (Supplementary Figure 1E). The WUS:NosT:35Sp region was inserted into the PmlI-NruI site on pKI_ΩBBMVP16&SRDXWUSm1 (Supplementary Figure 1J) through In-Fusion®, producing a binary vector pKI_ΩBBMVP16&SRDXWUS (Supplementary Figure 1I). The ApaI-PmlI sites of pKI_ΩBBMVP16&SRDXWUSm1 (Supplementary Figure 1J) was replaced with the HSPT:RPS5Ap:WUS (partial) fragment, producing pKI_ΩBBMVP16&WUSm1 (Supplementary Figure 1H). All the series of T-DNA schemes constructed for Agrobacterium-mediated gene transfer are shown in Supplementary Figure 1. BBM and WUS expression was controlled by the cauliflower mosaic virus 35S and A. thaliana RPS5A promoters, respectively. The expression and functions of BBM and WUS were examined. For BBM, the 5’-leader sequence of tobacco mosaic virus (translational enhancer Ω) (Gallie, 2002) was inserted upstream of the CDS, and a translational activator domain of VP16 from herpes simplex virus (Triezenberg et al., 1988) was fused to the C-terminal (ΩBBMVP16, Supplementary Figure 1C). For WUS, two amino acid mutations were inserted into the WUS-box (WUSm1) and an artificial strong repression domain, Superman Repression Domain X (SRDX) (Hiratsu et al., 2003), was fused to the N-terminal end of WUSm1 (Ikeda et al., 2009) (SRDXWUSm1, Supplementary Figure 1E).
2.3 Agrobacterium-mediated gene transfer
The constructed binary vectors were transferred into Agrobacterium tumefaciens strain EHA105. Agrobacterium tumefaciens containing each binary vector was grown in Lysogeny Broth (LB) liquid medium containing 20 mg l-1 rifampicin, 30 mg l-1 chloramphenicol, and an appropriate antibiotic for each binary vector (50 mg l-1 kanamycin for pGWB, or 100 mg l-1 spectinomycin for pKI) at 28°C overnight in a shaking incubator (120 rpm). The Agrobacterium solution was diluted with MS liquid medium (OD600 = 0.1) containing 3% sucrose and 100 μM acetosyringone (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan). In vitro-grown tobacco leaves were cut into leaf explants (5.5-mm diameter) using a cork borer (NONAKARIKAKI Co., Ltd, Tokyo, Japan). For infection, 10–20 leaf explants were soaked in Agrobacterium solution for 1 min, then co-cultured on MS medium containing 3% sucrose and 0.8% agar for 3 days in the dark. Afterward, the leaf explants were transferred to an MS selection medium containing 3% sucrose, 0.8% agar, 40 mg l-1 hygromycin B, and 25 mg l-1 meropenem (Sumitomo Dainippon Pharma Co., Ltd., Tokyo, Japan), and subcultured to a new MS selection medium every two weeks. Two and four weeks after Agrobacterium infection (2WAI and 4WAI), the cell differentiation phenotypes were evaluated by counting the number of calli and shoots originating from each leaf explant culture. Three independent experiments were conducted to evaluate each binary vector.
Agrobacterium-mediated transformations for lettuce and petunia were carried out similarly to tobacco with minor modifications. Leaf discs (5.0-mm diameter) were infected, and a half-strength MS medium was used for Agrobacterium inoculation and co-culture. After co-culture, the leaf discs were cultured without selection pressure of hygromycin for a few weeks, followed by subculturing to a new MS selection medium containing 10−40 mg l-1 hygromycin B, 25 mg l-1 meropenem, 3% sucrose, and 0.8% agar. Shoots that emerged from the leaf explants were detached and transferred to the MS selection medium. Each grown and rooted plant were maintained in the culture vessels with MS selection medium.
2.4 Genomic PCR analysis
Genomic DNA was isolated from tobacco calli and shoots originating from infected leaf explants. For petunia and lettuce, small leaves derived from the shoots showing hygromycin resistance were used for genomic PCR. The calli and shoots were homogenized in DNA extraction buffer (containing 20 mM Tris-HCl (pH 8.0), 10 mM EDTA (pH 8.0), and 10% SDS) and then centrifuged. After a phenol/chloroform treatment and ethanol precipitation, the obtained pellets were dissolved in water. Primers for IbMyb, Arabidopsis BBM and WUS, and Virulence D (VirD, for pTiBo542) detection (Supplementary Table 1) were used for PCR.
2.5 Real-time quantitative RT-PCR (qRT-PCR)
Total RNA was isolated using ISOSPIN Plant RNA (NIPPON GENE Co., Ltd., Tokyo, Japan). The cDNA was synthesized from 200 ng of total RNA using ReverTra Ace® (TOYOBO Co., Ltd). qRT-PCR was performed using KOD SYBR qPCR Mix (TOYOBO Co., Ltd) and Applied Biosystems StepOnePlus Real Time PCR System (Thermo Fisher Scientific, Inc.), using Comparative CT (ΔΔCT) method. For all qRT-PCR analyses, N. tabacum Elongation Factor 1-alpha (EF1α, AF120093) was used as the reference gene (Schmidt and Delaney, 2010). The primers used in each qRT-PCR analysis were listed in Supplementary Table 1. All primers that amplify WUS specifically do not distinguish WUS, SRDXWUS, and SRDXWUSm1.
2.6 RT-PCR analysis
Independent tobacco calli and shoots originating from the infected leaf explants were sampled as putative transgenic individuals. Total RNA was isolated using ISOSPIN Plant RNA. The cDNA was synthesized from 200 ng of total RNA using ReverTra Ace®. RT-PCR was performed using the primers listed in Supplementary Table 1.
2.7 Sample preparation for phytohormone-quantitative analysis and RNA-seq
Five groups were used for phytohormone-quantitative analysis and RNA-seq: non-infected leaf explants (control), leaf cell cultures at 2 and 4WAI with ΩBBMVP16&WUS (2w:ΩBBMVP16&WUS and 4w:ΩBBMVP16&WUS), and leaf cell cultures at 2 and 4WAI with ΩBBMVP16&SRDXWUSm1 (2w:ΩBBMVP16&SRDXWUSm1 and 4w:ΩBBMVP16&SRDXWUSm1) (Figure 1A). Each Agrobacterium infection was carried out three or four times as a biological replicate. Five to ten leaf explants were collected.
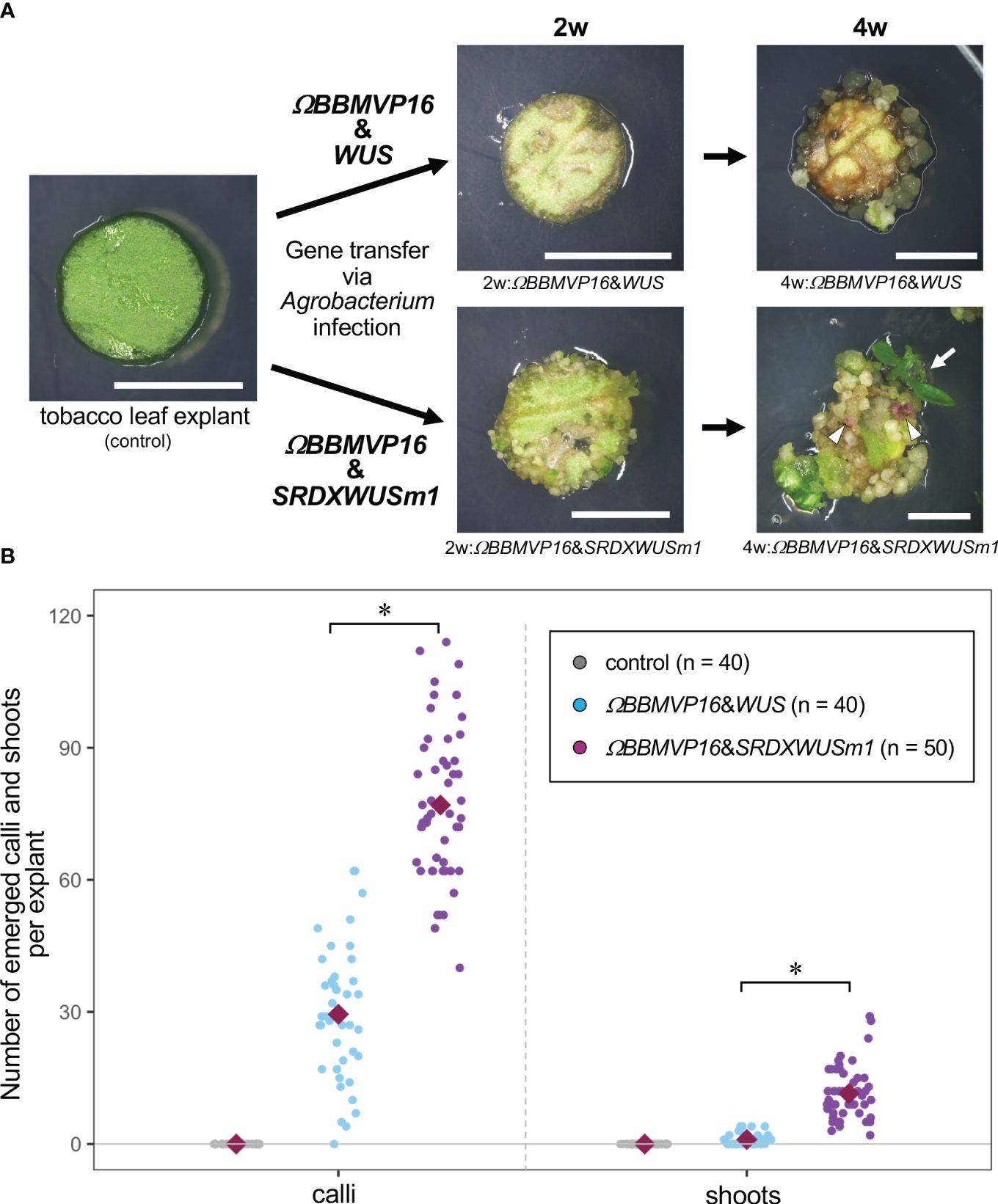
Figure 1 Autonomous differentiation of tobacco leaf cells with introduced Arabidopsis BABY BOOM (BBM) and WUSCHEL (WUS) genes. (A) The phenotypes of tobacco leaf explant cultures with introduced ΩBBMVP16&WUS (upper photos) and ΩBBMVP16&SRDXWUSm1 (lower photos) via Agrobacterium infection. Photos at two weeks (2w) and four weeks (4w) after Agrobacterium infection are shown, respectively. The left image is an explant used for infection (Φ 5.5 mm). A differentiated shoot from the leaf explant cells with introduced ΩBBMVP16&SRDXWUSm1 is marked with a white arrow. White triangles represent anthocyanin pigmentation rarely observed in calli. Bars = 5 mm. (B) The number of calli and shoots that emerged from a leaf explant 4 weeks after the infection. Three biological replicates of Agrobacterium infection were carried out, and the data is indicated together. Red, green, and blue dot-plots represent the number of calli (left plots) and shoots (right plots) that originated from a non-infected (control) leaf explant or one with introduced ΩBBMVP16&WUS and ΩBBMVP16&SRDXWUSm1, respectively. Each violet diamond represents the mean. Asterisks indicate significant differences (p < 0.01) between ΩBBMVP16&WUS and ΩBBMVP16&SRDXWUSm1, detected by Welch’s t-test.
For phytohormone-quantitative analysis, the collected samples were ground in liquid nitrogen with mortar and pestle and kept at −80°C until the analysis (following section 3.8). For total RNA extraction, the samples were soaked in RNA Save (Cosmo Bio Co., Ltd., Tokyo, Japan) and kept at −80°C until the experiments (following section 3.9).
2.8 Phytohormone-quantitative analysis
Phytohormone profiles in the differentiating leaf cultures were analyzed by a highly sensitive and high-through method that allows a single run using ‘MS-probe’ (chemical derivatization) and liquid chromatography-tandem mass spectrometry (Kojima et al., 2009). Phytohormone extraction and semi-purification from ground and frozen tissues (100 mg) were performed as described previously (Kojima et al., 2009; Kojima and Sakakibara, 2012). Contents of cytokinins were quantified with ultra-performance liquid chromatography (UPLC)-electrospray interface (ESI) tandem quadrupole mass spectrometer (qMS/MS) (ACQUITY UPLC™ System/Xevo-TQS) (Waters Corp., Milford, M.A., USA). Abscisic acid (ABA), indole-3-acetic acid (IAA), indole-3-acetyl-L-aspartic acid (IAAsp), gibberellins (GAs), jasmonic acid (JA), jasmonyl-isoleucine (JA-Ile) and salicylic acid (SA) were quantified with ultra-high performance liquid chromatography (UHPLC) electrospray interface (ESI) quadrupole-orbitrap mass spectrometer (UHPLC/Q-Exactive™) (Thermo Fisher Scientific, Inc.) (Shinozaki et al., 2015). The procedures were performed by RIKEN CSRS (Kanagawa, Japan).
2.9 RNA-seq analysis
Total RNA was isolated from each sample using ISOSPIN Plant RNA. An RNA-seq library was prepared using the Lasy-Seq ver. 1.1 protocol (Kamitani et al., 2019) using 120 ng of total RNA. Each library was sequenced in triplicates using the single-end mode of the Illumina HiSeqX platform (Illumina Inc., San Diego, C.A., USA). Library preparation and sequencing were performed by Clockmics Inc. (Osaka, Japan).
The raw RNA-seq reads were pre-processed in Trimmomatic ver. 0.39 (Bolger et al., 2014) using default parameters, and the trimmed reads were aligned to the reference tobacco TN90 genome (Sierro et al., 2014) using HISAT2 ver. 2.2.1 (Kim et al., 2015). Gene expression levels were quantified using StringTie ver. 2.2.1 (Pertea et al., 2015). Principal component analysis (PCA) was performed using the graphical user interface for the R package TCC (TCC-GUI ver. 1.0 pipeline) (Su et al., 2019) using the expression data. Differentially expressed genes (DEGs) between the two groups were detected using the TCC-GUI pipeline with a false discovery rate (FDR) of 0.05, and between the three groups using the method of combining the R packages baySeq ver. 2.24.0 and TCC ver. 1.30.0 (Osabe et al., 2019) with FDR of 0.05. The expression levels of the DEGs were visualized using the R package heatmap3 ver. 1.1.9 (Zhao et al., 2014) using read count data normalized by the R package TCC. Gene Ontology (GO) enrichment analysis of the DEGs was performed using the R package clusterProfiler ver. 3.18.1 (Yu et al., 2012). GO term networks were also visualized using the same R package. DEGs were annotated from the NCBI database (https://www.ncbi.nlm.nih.gov/). Orthologous genes of the DEGs in A. thaliana were identified using OrthoDB v11 (https://www.orthodb.org/), and annotations of the orthologous genes were obtained from the TAIR database (https://www.arabidopsis.org/). Commonly upregulated DEGs in two–and three–group comparisons were visualized using the R package VennDiagram ver. 1.7.1 (Chen and Boutros, 2011). The raw RNA-seq data was deposited at the NCBI Sequence Read Archive (https://www.ncbi.nlm.nih.gov) with an accession number PRJNA1049661.
3 Results
3.1 Effects of Arabidopsis BBM and WUS genes introduced into tobacco, petunia, and lettuce leaf cells
First, we tested the gene expression scheme using Arabidopsis BBM and WUS as DRs, which enabled autonomous cell differentiation of transgenic tobacco cells in Agrobacterium-infected leaf explants during the culture process.
Leaf explants from the experimental controls, non-infected explants, and explants infected with pGWB_35Sp:GFP, did not show any reaction to cell differentiation, and were entirely browned by 4WAI (Supplementary Figure 1A). The results indicated that tobacco leaf cells could not differentiate or survive on a medium without PGRs, and the stress of Agrobacterium infection and gene transfer did not have different effects.
The BBM and ΩBBMVP16-introduced leaf cells also died by 4WAI (Supplementary Figures 1B, C). A previous study reported that overexpression of SRDXWUSm1 enhanced adventitious shoot formation in Arabidopsis (Ikeda et al., 2020). However, WUS and SRDXWUSm1-introduced leaf cells did not survive (Supplementary Figures 1D, E), indicating that the introduction of Arabidopsis BBM or WUS alone was insufficient to induce tobacco cell differentiation. Next, the combination of BBM and WUS was examined. Although the introduction of BBM&WUS was insufficient (Supplementary Figure 1F), ΩBBMVP16&WUS-introduced leaf cells differentiated into calli after 4WAI (Figure 1A; Supplementary Figure 1G). Furthermore, ΩBBMVP16&SRDXWUSm1-introduced leaf cells showed a more accelerated differentiation phenotype after 4WAI (Figure 1A; Supplementary Figure 1J). Unlike the ΩBBMVP16&WUS-introduced leaf cells, ΩBBMVP16&SRDXWUSm1-introduced leaf cells differentiated into greenish, organ-like structures, in addition to small calli at 2WAI (Figure 1A; Supplementary Figure 1J). As of 4WAI, the calli proliferated vigorously, and a few adventitious shoots emerged from ΩBBMVP16&SRDXWUSm1-introduced leaf cells. Significantly higher numbers of calli and shoots were obtained from leaf explant cultures introduced with ΩBBMVP16&SRDXWUSm1 (Figure 1B; Supplementary Figure 2). To investigate whether amino acid mutations in WUS-box (WUSm1) or SRDX fusion to the N-terminal of WUS (SRDXWUS) was involved in the accelerated differentiation phenotype of ΩBBMVP16&SRDXWUSm1-introduced leaf cells, ΩBBMVP16&WUSm1 (Supplementary Figure 1H) and ΩBBMVP16&SRDXWUS (Supplementary Figure 1I) were evaluated. While ΩBBMVP16&WUSm1-introduced leaf cells did not induce any cell differentiation and died by 4WAI (Supplementary Figure 1H), ΩBBMVP16&SRDXWUS-introduced leaf cells differentiated calli and shoots vigorously (Supplementary Figure 1I). Compared to ΩBBMVP16&SRDXWUS, the number of calli that emerged from a leaf explant culture was higher in ΩBBMVP16&SRDXWUSm1, whereas the number of shoots did not show significant differences between the two constructs (Supplementary Figure 2). In the present study, introducing Arabidopsis ΩBBMVP16&WUS, ΩBBMVP16&SRDXWUS, and ΩBBMVP16&SRDXWUSm1 succeeded in the autonomous differentiation of tobacco leaf cells without applying PGRs during the tissue culture process.
To confirm the observed phenotypes (Figure 1; Supplementary Figure 1) were linked to expression of the introduced genes, real-time qRT-PCR was performed for tobacco leaf explants introduced the ΩBBMVP16&WUS and ΩBBMVP16&SRDXWUSm1 (Supplementary Figure 3). As a result, the transcription of Arabidopsis BBM and WUS was confirmed in the infected leaf explants during the culture by 4WAI. Compared to WUS driven by RPS5A promoter, BBM driven by 35S promoter showed significantly higher expression levels in all the 2 and 4WAI leaf explants.
We further evaluated whether the autonomously differentiated calli and shoots were the transgenics expressing BBM and WUS. Genomic PCR analysis was performed using calli and shoots originated from ΩBBMVP16&WUS- and ΩBBMVP16&SRDXWUSm1-introduced leaf cells. As a result, the Arabidopsis BBM and WUS were detected in the callus and shoot lines obtained with ΩBBMVP16&WUS and ΩBBMVP16&SRDXWUSm1, but the VirD gene from Agrobacterium was not, showing that these were transgenic (Supplementary Figure 4). IbMyb, which has been introduced as a visible marker of anthocyanin pigmentation (Kim et al., 2010; Sato et al., 2023), was also detected; however, anthocyanin pigmentation was hardly observed in the differentiated cells (Figure 1A; Supplementary Figures 1G, I, J). To further verify that each transgene was transcribed in transgenic calli and shoots, RT-PCR was performed. As a result, all the IbMyb, Arabidopsis BBM, and WUS expressions were detected (Supplementary Figure 5). Thus, the downstream anthocyanin biosynthesis pathway induced by IbMyb may have been hampered in cells expressing BBM and WUS.
Furthermore, we tested the effect of these expression constructs with different plant species, petunia and lettuce, which are receptive to Agrobacterium-mediated gene transfer. As a result, autonomous differentiation of calli and shoots from leaf explants cultured on the PGR-free medium was observed at 4WAI (Supplementary Figure 6). The presence of transgenes in the obtained hygromycin-resistant shoots was confirmed (Supplementary Figure 7).
The ectopic overexpression of DRs has been reported to cause morphogenic abnormalities in transgenic plants (Boutilier et al., 2002; Srinivasan et al., 2007; Chen et al., 2022b), and the higher expression level linked to severe phenotype in tobacco (Srinivasan et al., 2007). The obtained tobacco transgenic plants in the present study showed malformation with various intensities. The qRT-PCR of BBM and WUS in the plant lines showing ‘nearly normal’, ‘moderate,’ and ‘severe’ malformations showed similar co-relation between DRs levels and morphologies (Supplementary Figure 8).
3.2 Phytohormone contents in differentiating tobacco leaf cells
Cultured cells differentiating without external PGR treatment are suitable for analyses to obtain pure endogenous cellular behaviors. Phytohormones are important regulators of cell division and differentiation. Because the differentiation of leaf cells caused by the introduction of ΩBBMVP16&WUS and ΩBBMVP16&SRDXWUSm1 showed dramatically different patterns (Figure 1A; Supplementary Figures 1G, J), the underlying hormonal behaviors were analyzed. Six phytohormones (auxins, cytokinins, GAs, ABA, SA, and JA) and their metabolites in the leaf explant cultures at 2 and 4WAI were measured (Figure 2; Supplementary Table 2).
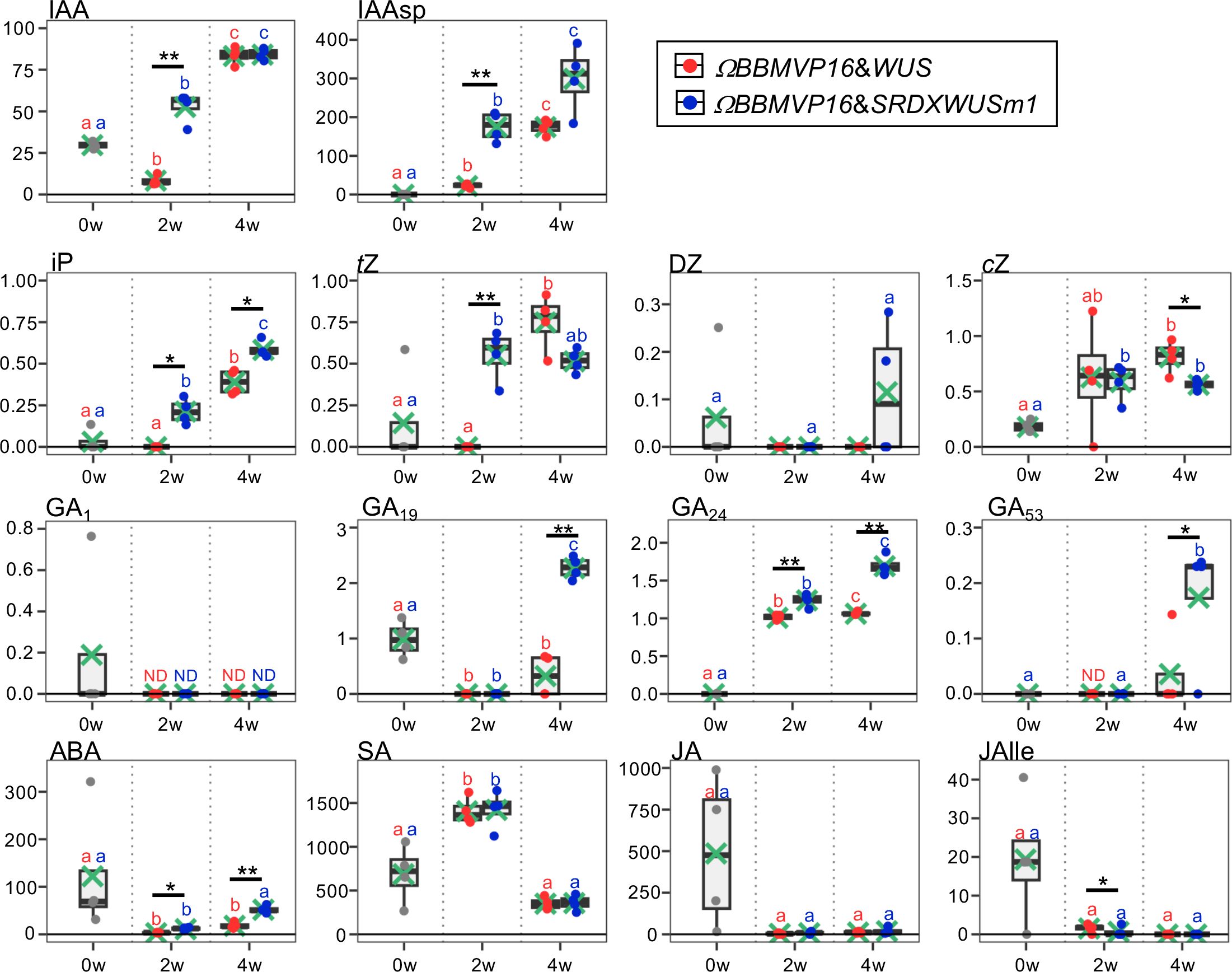
Figure 2 Phytohormone content in differentiating leaf explant cells with introduced ΩBBMVP16&WUS and ΩBBMVP16&SRDXWUSm1. The phytohormone content in the leaf explant cultures was measured at two and four weeks after Agrobacterium infection. The y-axis represents the concentration (pmol/g fresh weight (FW)) and the x-axis represents the culture period (0, 2, and 4 weeks (w)). Orange and blue dots represent the values of ΩBBMVP16&WUS- and ΩBBMVP16&SRDXWUSm1-introduced leaf explant cultures, respectively. Non-infected leaf explants were analyzed as 0-week cultured samples (0w), and the results are indicated by gray dots. Light green crosses and vertical black bars associated with boxplots represent the mean and standard deviation, respectively (n = 4). The horizontal black bar in each boxplot represents the median value. IAA, Indole-3-acetic acid; IAAsp, indole-3-acetyl-L-aspartic acid; iP, N6-(Δ2-isopentenyl) adenine; tZ, trans-Zeatin; DZ, dihydrozeatin; cZ, cis-Zeatin; GA, Gibberellin; ABA, Abscisic acid; SA, Salicylic acid; JA, Jasmonic acid; JAIle, Jasmonic acid with isoleucine. Asterisks indicate significant differences (*p < 0.05, **p < 0.01) between ΩBBMVP16&WUS- and ΩBBMVP16&SRDXWUSm1-introduced leaf explant cultures during the same culture period, as detected by Welch’s t-test. Different lowercase letters in the same color indicate significant differences (p < 0.05) among the three time points (0, 2, and 4w), as detected using the Tukey-Kramer’s test. ND, not detected (below the quantification limit). All measurement data are shown in Supplementary Table 2.
The active auxin indole-3-acetic acid (IAA) and the irreversible catabolite indole-3-acetyl-L-aspartic acid (IAAsp) showed similar behaviors in the ΩBBMVP16&WUS- and ΩBBMVP16&SRDXWUSm1-introduced leaf cultures by 4WAI. At 2WAI, compared with non-infected (0-week cultured) leaf explants, the IAA content in ΩBBMVP16&SRDXWUSm1 was significantly higher, whereas it was significantly lower in ΩBBMVP16&WUS. The IAA content was further elevated to a similar level by 4WAI in both ΩBBMVP16&WUS- and ΩBBMVP16&SRDXWUSm1-introduced leaf cultures. The IAAsp content in ΩBBMVP16&SRDXWUSm1-introduced leaf cultures was also higher than that in ΩBBMVP16&WUS-introduced leaf cultures, especially at 2WAI.
In comparison with the non-infected leaf explants, similar cytokinin content, except for dihydrozeatin (DZ), was observed in both ΩBBMVP16&WUS and ΩBBMVP16&SRDXWUSm1-introduced leaf cultures. At 2WAI, N6-(Δ2-isopentenyl) adenine (iP) content was yet negligible in ΩBBMVP16&WUS, whereas it was significantly higher in ΩBBMVP16&SRDXWUSm1. The iP elevations in both ΩBBMVP16&WUS- and ΩBBMVP6&SRDXWUSm1-introduced leaf cultures continued for 4WAI, and the overall contents in the ΩBBMVP16&SRDXWUSm1-introduced leaf cultures were significantly higher than those in ΩBBMVP16&WUS. An earlier elevation of trans-zeatin (tZ) was observed in ΩBBMVP16&SRDXWUSm1 than in ΩBBMVP16&WUS, and the content level was maintained by 4WAI. Although tZ was negligible in the ΩBBMVP16&WUS-introduced leaf cultures at 2WAI, it reached a level similar to that of ΩBBMVP16&SRDXWUSm1 at 4WAI. Elevated cis-zeatin (cZ) level was detected in ΩBBMVP16&SRDXWUSm1 compared to the non-infected leaf cultures at 2WAI, and each level was maintained until 4WAI. cZ content at 4WAI was significantly higher in ΩBBMVP16&WUS than in ΩBBMVP16&SRDXWUSm1.
The content behaviors were various depending on the GA type. One of the active GAs, GA1, was maintained at negligible levels by 4WAI in both ΩBBMVP16&WUS- and ΩBBMVP16&SRDXWUSm1-introduced leaf cultures. Significantly higher levels of GA19, GA24, and GA53 were detected in the ΩBBMVP16&SRDXWUSm1-introduced leaf cultures at 4WAI than in those of ΩBBMVP16&WUS, whereas continuous elevation of GA24 was also observed in the ΩBBMVP16&WUS-introduced leaf cultures. The content of GA4, an active GA, was below the detection level in all samples analyzed (Supplementary Table 2).
Compared to the non-infected leaf explants, the ABA content was lower in the ΩBBMVP16&WUS-introduced leaf cultures throughout the four weeks of culture from infection. In contrast, the ABA level was elevated in the ΩBBMVP16&SRDXWUSm1-introduced leaf cultures at the 4WAI, showing overall higher levels than that of ΩBBMVP16&WUS.
No significant differences in SA, JA, and JA-Ile content were detected throughout the four weeks of culture in the comparison of ΩBBMVP16&WUS- and ΩBBMVP16&SRDXWUSm1-introduced leaf cultures. Statistically, the JA-Ile content at 2WAI was significantly lower in the ΩBBMVP16&SRDXWUSm1-introduced leaf cultures. Focusing on active auxin (IAA) and cytokinins (iP, tZ), each phytohormone level tended to increase earlier and was higher in the leaf explant cultures at 2WAI with ΩBBMVP16&SRDXWUSm1-introduced leaf cultures than in those with ΩBBMVP16&WUS.
3.3 Gene expression profiles in differentiating tobacco leaf cells
To analyze transcriptional behavior during cell differentiation, we performed RNA-seq of tobacco leaf cells cultured for 2 or 4WAI with ΩBBMVP16&WUS and ΩBBMVP16&SRDXWUSm1, and 0-week cultured leaf explants without Agrobacterium infection (control). PCA showed that the three biological replicates of each sample represented by the same color clustered together (Figure 3). Notably, the groups of 2w:ΩBBMVP16&SRDXWUSm1 and 4w:ΩBBMVP16&WUS were located close together.
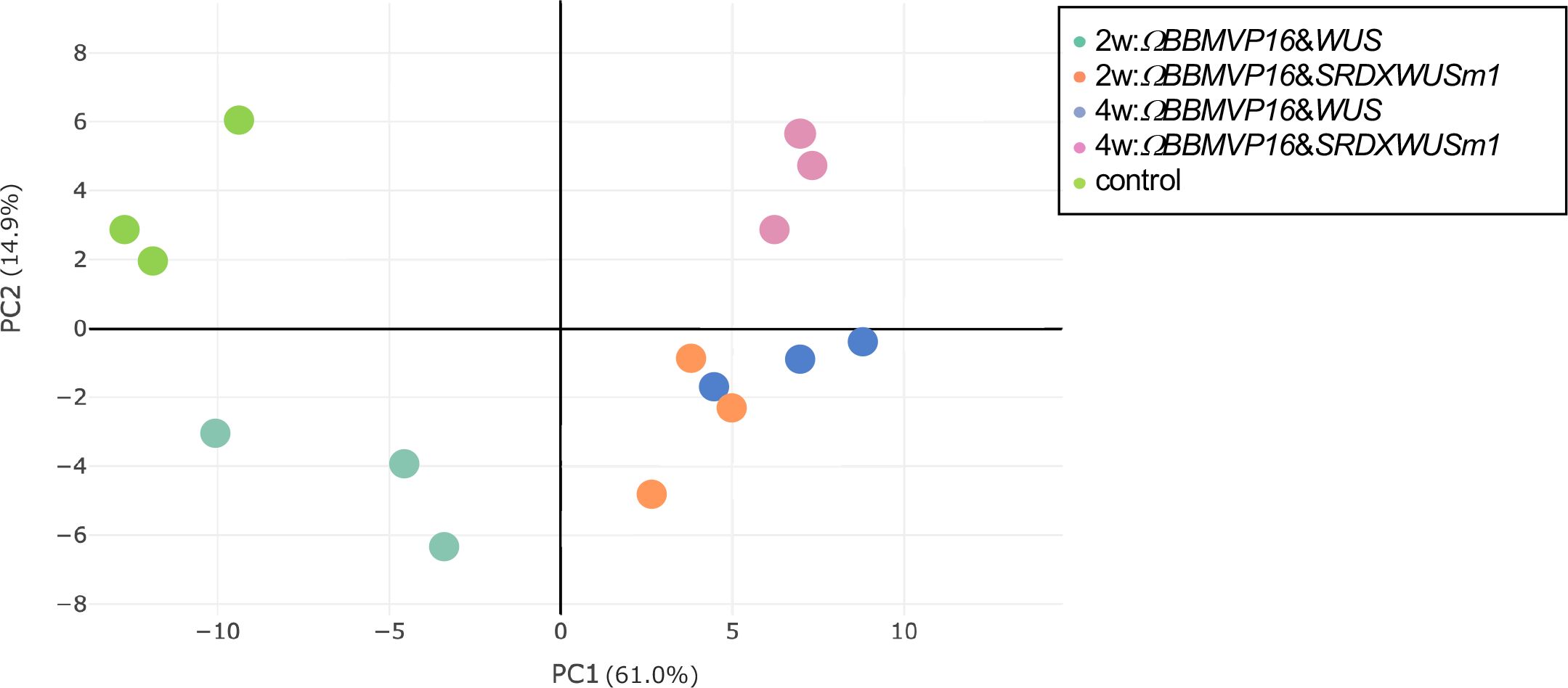
Figure 3 Principal component analysis of gene expression levels in tobacco leaf explant cultures. The ‘control’ is the experimental control (non-infected leaf explant). Plots with the same color indicate biological replicates. 2w and 4w, 2 (or 4) weeks after Agrobacterium infection.
First, we compared the gene expression of 2w:ΩBBMVP16&WUS (G1) and 2w:ΩBBMVP16&SRDXWUSm1 (G2) (Figure 4A), where differences in the swiftness of differentiation and phytohormone levels were observed (Figures 1A, 2; Supplementary Figures 1G, J). A total of 179 genes were identified as DEGs (FDR < 0.05). Among the DEGs, 128 genes were significantly upregulated in 2w:ΩBBMVP16&SRDXWUSm1 (G1 < G2), whereas the expression of 51 genes was significantly upregulated in 2w:ΩBBMVP16&WUS (G1 > G2) (Figure 4A). GO analysis of the 128 DEGs (G1 < G2) detected 16 GO terms related to cell wall, auxin, cytokinin, and water (Figure 4B; Supplementary Figure 9). The putative A. thaliana orthologs for the detected genes are listed in Supplementary Table 3. Focusing on phytohormones, the genes categorized into auxin-related GO terms included orthologs of AUXIN-RESPONSIBLE PROTEIN (AUX/IAAs) (two DEGs) and PIN-FORMED (PINs) (one DEG) (Figure 4C; Supplementary Table 3). Genes categorized into the cytokinin-related GO were the orthologs of HISTIDINE PHOSPHOTRANSFER PROTEIN 6 (HP6) (two DEGs) (Figure 4C; Supplementary Table 3). No significant GO terms were detected when using the 51 genes that were upregulated in 2w:ΩBBMVP16&WUS.
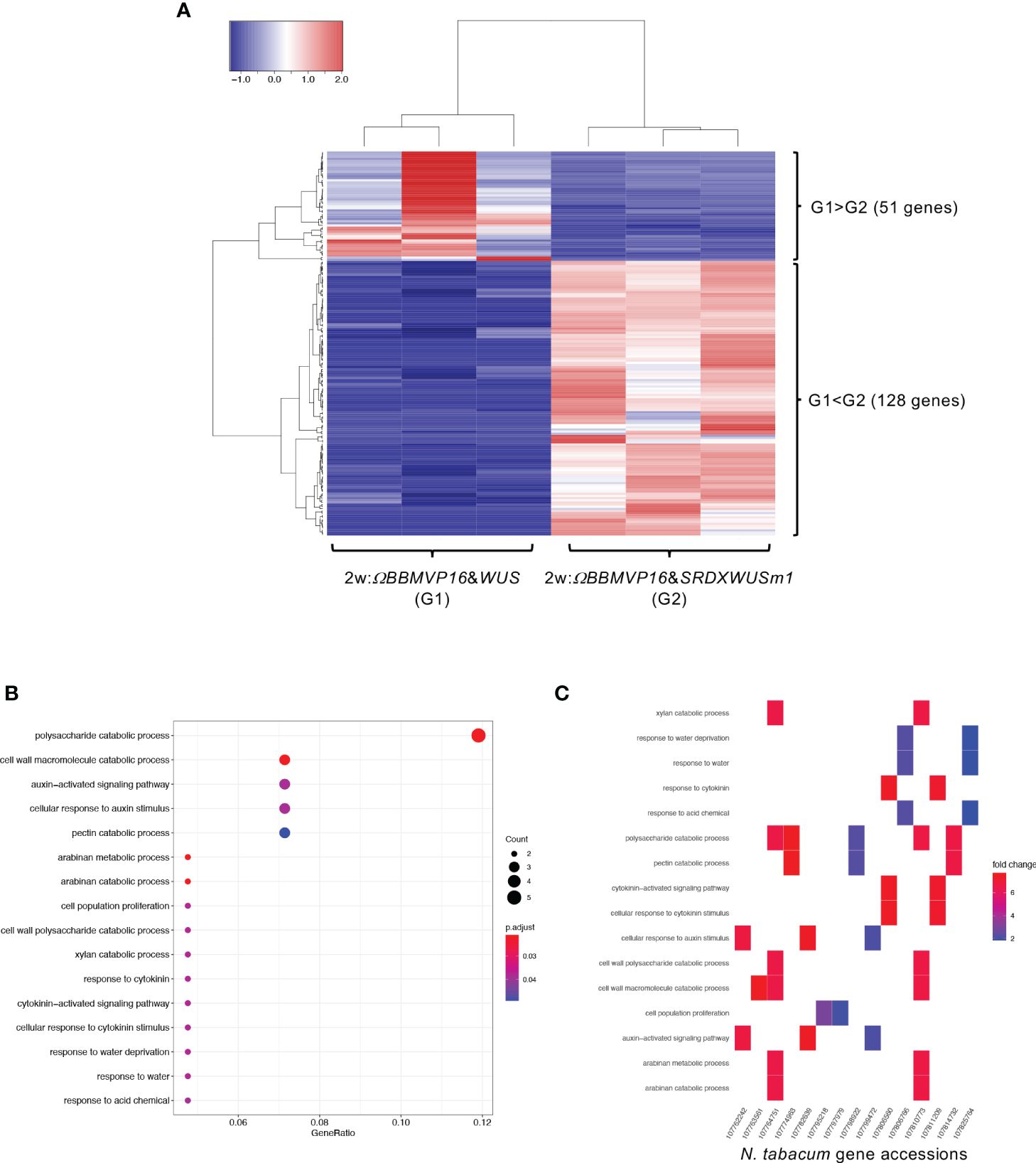
Figure 4 Differential expression analysis for two-group RNA-seq count data (2w:ΩBBMVP16&WUS vs 2w:ΩBBMVP16&SRDXWUSm1). Group1 (G1) is 2w:ΩBBMVP16&WUS, Group2 (G2) is 2w:ΩBBMVP16&SRDXWUSm1. (A) Hierarchical clustering heatmap of 179 DEGs (FDR < 0.05) generated using TCC-GUI. Red and blue indicate high and low gene expressions, respectively. (B) Visualization of GO enrichment analysis using 128 DEGs (G1 < G2). All detected 16 GO terms are shown. (C) N. tabacum gene accessions included 16 GO terms (B). The color of the square represents the Log2 fold-change (M value of the MA plot).
We then compared gene expression between 4w:ΩBBMVP16&WUS (G1) and 4w:ΩBBMVP16&SRDXWUSm1 (G2) (Figure 5A), where differences in the type of differentiation were observed (Figure 1A; Supplementary Figures 1G, J). Among the 4,358 genes detected as significant DEGs (FDR < 0.05), 2,380 genes were expressed at significantly higher levels in 4w:ΩBBMVP16&SRDXWUSm1 (G1 < G2), whereas 1,978 genes were expressed at significantly lower levels (Figure 5A). With the 2,380 DEGs upregulated in 4w:ΩBBMVP16&SRDXWUSm1, GO terms related to photosynthesis were the most enriched, and GO terms related to auxin and ABA were also enriched (Figure 5B; Supplementary Figures 10A, 11). In contrast, GO terms related to ribosomes were highly enriched in the downregulated 1,978 genes (Figure 5C; Supplementary Figure 10B). Focusing on the phytohormones, GO terms ‘auxin-activated signaling pathway’ and ‘cellular response to auxin stimulus’ included the orthologs of AUX/IAAs (10 DEGs), AUXIN RESPONSES FACTORS (ARFs) (6 DEGs), PIN-likes (PILSs) (3 DEGs), and PIN6 (1 DEG) (Supplementary Table 4; Supplementary Figure 12). The GO term ‘response to ABA’ (Figure 5B) included orthologs of LATE EMBRYOGENESIS ABUNDANT (LEAs/EMs) (seven DEGs), PYRABACTIN RESISTANCE-LIKE (PYLs/PYRs/RCARs) (four DEGs), and ABSCISIC ACID-INSENSITIVE 5 (ABI5) (two DEGs) (Supplementary Table 4; Supplementary Figure 12).
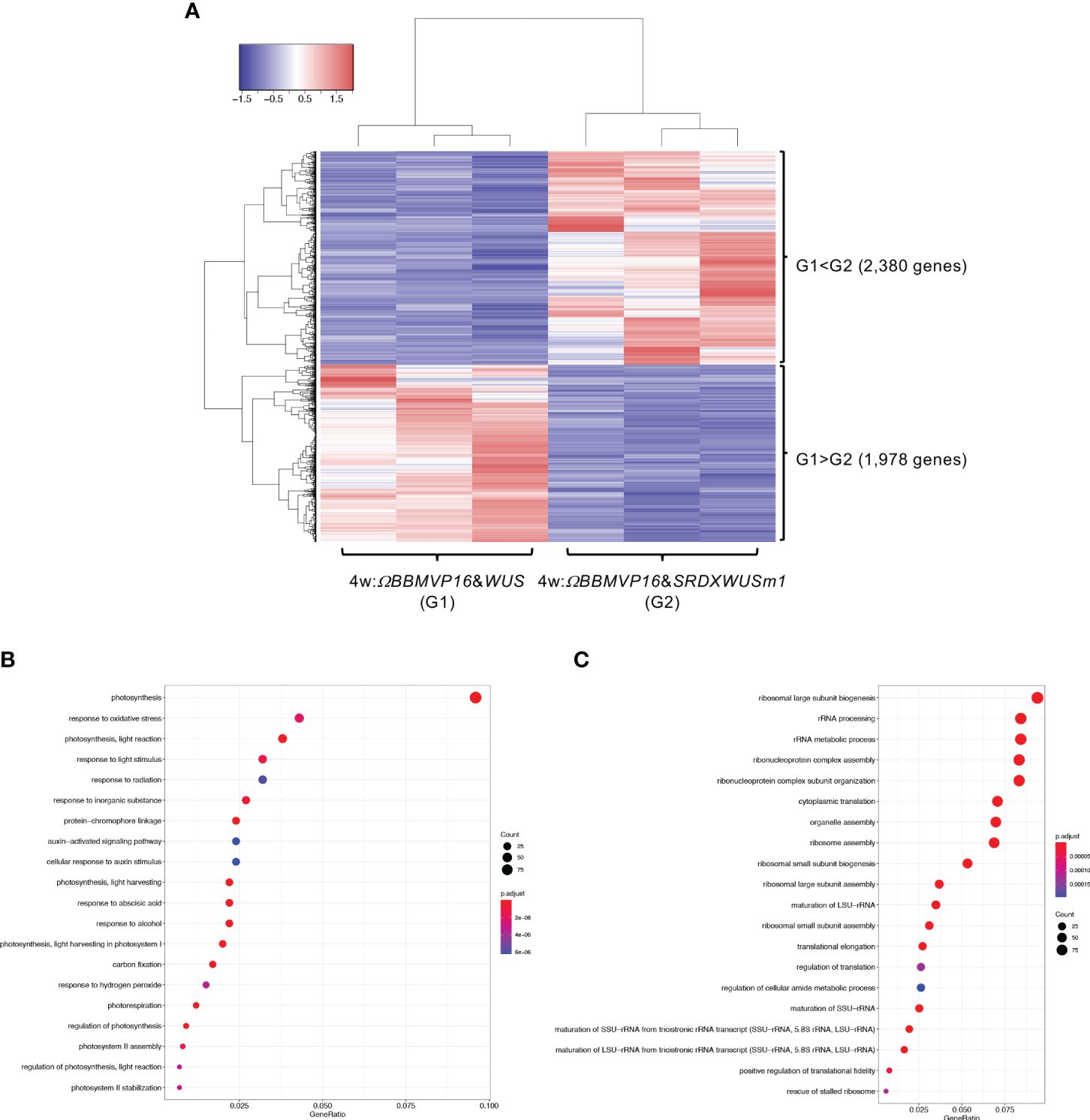
Figure 5 Differential expression analysis for two-group RNA-seq count data (4w:ΩBBMVP16&WUS vs 4w:ΩBBMVP16&SRDXWUSm1). Group1 (G1) is 4w:ΩBBMVP16&WUS, Group2 (G2) is 4w:ΩBBMVP16&SRDXWUSm1. (A) Hierarchical clustering heatmap of 4,358 DEGs (FDR < 0.05) generated using TCC-GUI. Red and blue indicate high and low gene expressions, respectively. (B) Visualization of GO enrichment analysis using 2,380 DEGs (G1 < G2). The top 20 GO terms are listed. (C) Visualization of GO enrichment analysis using 1,978 DEGs (G1 > G2). The top 20 GO terms are listed. All GO terms are shown in Supplementary Figure 6. The top 20 GO terms were selected based on FDR values for each comparison (G1 < G2 and G1 > G2).
In summary, GO terms involved in phytohormones were enriched with genes upregulated by introducing ΩBBMVP16&SRDXWUSm1 at 2 and 4WAI, compared to ΩBBMVP16&WUS. The GO terms related to phytohormones differed at each time point; GO terms related to auxin and cytokinin were enriched at 2WAI, whereas those related to auxin and ABA were enriched at 4WAI. These GO terms were associated with signaling or transport but not with biosynthesis and metabolism.
Next, the RNA-seq data obtained from ΩBBMVP16&SRDXWUSm1-introduced leaf cells, which showed remarkable differentiation patterns for up to 4 weeks of culture, were analyzed by comparing the three groups: non-infected leaf explants (control: G1), 2w:ΩBBMVP16&SRDXWUSm1 (G2), and 4w:ΩBBMVP16&SRDXWUSm1 (G3) (Figure 6A). Among the 15,100 significant DEGs (FDR < 0.05), 117 genes were gradually upregulated (G1 < G2 < G3), whereas 100 genes were gradually downregulated (G1 > G2 > G3) during the culture process after Agrobacterium infection (Figure 6A). Although no significant GO terms (FDR < 0.05) were detected for the 117 upregulated genes, GO analysis of the 100 downregulated genes revealed 13 significant GO terms (FDR < 0.05) (Figure 6B; Supplementary Figure 13). Among the 100 downregulated genes, GO terms related to stimuli (five DEGs) included the ortholog genes of OSMOTIN-LIKE34 (OSM34) and MILDEW RESISTANCE LOCUS O (MLOs). GO terms related to glutamine and asparagine (four DEGs) included GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1 (ASN1), GLUTAMINE SYNTHETASE 2 (GS2), and GLUTAMATE DEHYDROGENASE (GDH) (Figure 6C; Supplementary Table 5). In addition, eight GO terms (FDR < 0.05) related to shoot, morphogenesis, and organ formation were detected with two DEGs (LOC107773826 and LOC107815131) (Figure 6C), including the orthologs of TEOSINTE BRANCHED1/CYCLOIDEA/PCF3/4/10 (TCP3/4/10) (Supplementary Table 5).
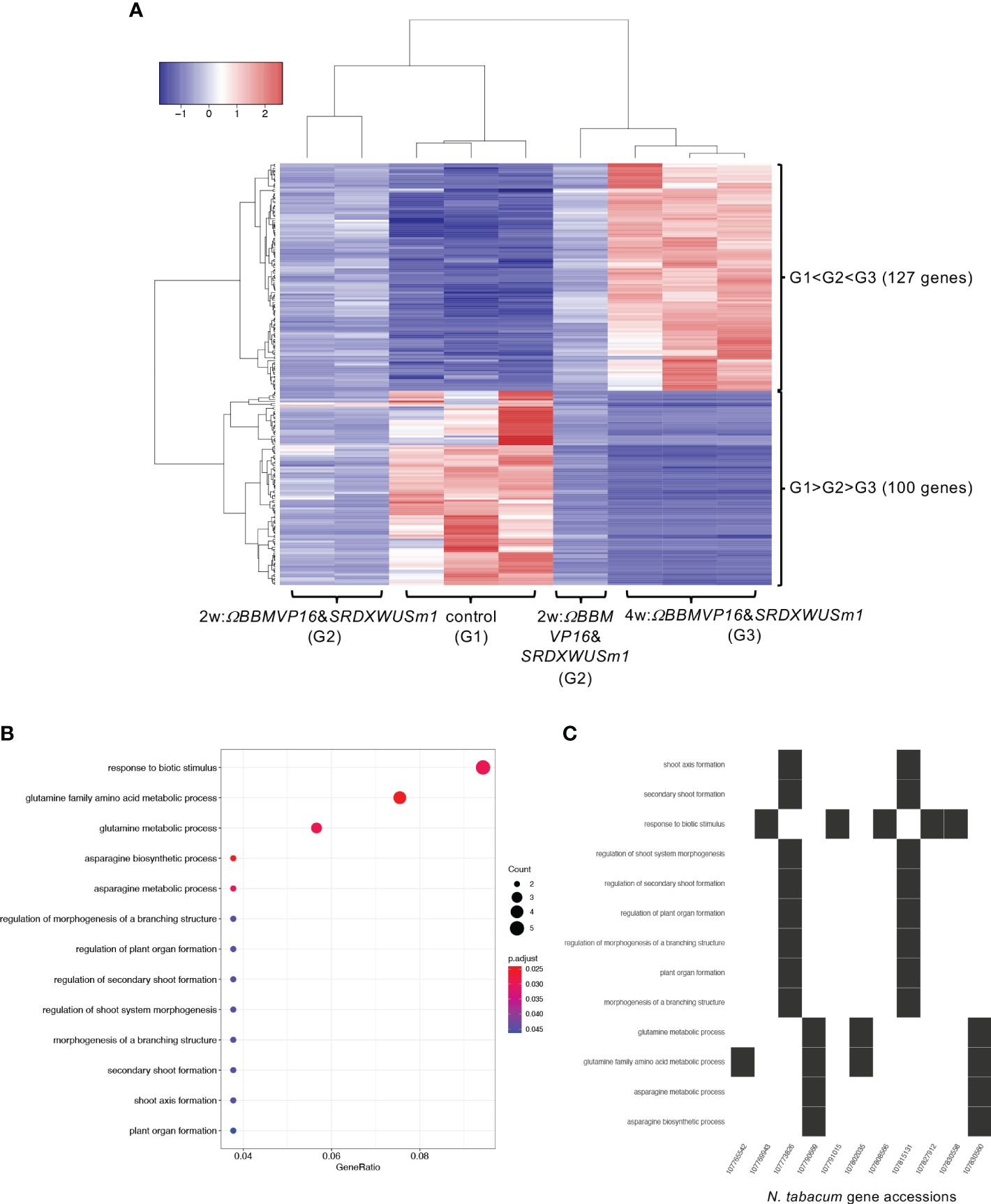
Figure 6 Differential expression analysis for three-group RNA-seq count data (control vs 2w:ΩBBMVP16&SRDXWUSm1 vs 4w:ΩBBMVP16&SRDXWUSm1). Group1 (G1) is control (non-infected leaf explants), Group2 (G2) is 2w:ΩBBMVP16&SRDXWUSm1, and Group3 (G3) is 4w:ΩBBMVP16&SRDXWUSm1. (A) Hierarchical clustering heatmap of 117 genes identified as DEGs (FDR < 0.05) based on baySeq with TCC. Red and blue indicate high and low gene expressions, respectively. (B) Visualization of the GO enrichment analysis using 100 DEGs (G1 > G2 > G3). (C) N. tabacum gene accessions included 13 GO terms (G1 > G2 > G3).
In a three-group comparison using ΩBBMVP16&WUS-introduced leaf cultures at 2WAI (G2) and 4WAI (G3) and non-infected leaf explants (control: G1) (Supplementary Figure 14A), 9,161 genes in total were identified as DEGs (FDR < 0.05). Among the 9,161 significant DEGs (FDR < 0.05), only two genes were upregulated (G1 < G2 < G3), whereas 74 genes were downregulated (G1 > G2 > G3) during the culture process after Agrobacterium infection (Supplementary Figure 14A). The 28 GO terms related to stimuli, metabolism, and catabolism were significantly (FDR < 0.05) enriched in the 74 downregulated genes (Supplementary Figure 14B). Because GO enrichment analysis could not be applied to only the two upregulated genes, we obtained each gene description from NCBI; LOC107761077 was annotated as the ortholog of AT-HOOK MOTIF NUCLEAR-LOCALIZED PROTEIN 15 (AHL15/AGF2), and LOC107809734 was an uncharacterized gene (Supplementary Table 6).
The DEGs detected in this study were summarized as Venn diagram (Supplementary Figure 15); significantly upregulated in ΩBBMVP16&SRDXWUSm1-introduced leaf cells at both 2 (Figure 4) and 4WAI (Figure 5), and gradually upregulated during the culture process in ΩBBMVP16&SRDXWUSm1-introduced leaf cells (Figure 6). Among 19 genes commonly detected as significantly upregulated in ΩBBMVP16&SRDXWUSm1-introduced leaf cells at both 2 (blue) and 4WAI (green in Supplementary Figure 15), cell proliferation and auxin-related GO terms were included (FDR < 0.05) (Supplementary Table 7). In this analysis, one common gene LOC107801243 was detected in the three-group comparison (purple in Supplementary Figure 15; Supplementary Table 7). On the other hand, no significant GO terms were detected for the intersect DEGs that were downregulated (data not shown).
3.4 qRT-PCR of the genes detected as DEGs in ΩBBMVP16&SRDXWUSm1-introduced leaf cell cultures
To evaluate the actual expression behaviors of DEGs affected by ΩBBMVP16&SRDXWUSm1 introduction, qRT-PCR was performed for four genes: LOC107773826 down-regulated over time (Supplementary Table 5), LOC107782639 and LOC107795218 showed higher levels compared to ΩBBMVP16&WUS (Supplementary Table 7), and LOC107801243 upregulated over time and higher compared to ΩBBMVP16&WUS (Supplementary Table 7). LOC107773826 and LOC107782639 are the putative orthologs of Arabidopsis TCP3/4/10 and IAA26, respectively (Supplementary Tables 3-5). The LOC107795218 is annotated as a putative phytosulfokine-3 gene (XM_016617810). The LOC107801243 is identified as a putative paralogue gene of NAD+-dependent protein deacetylase HST1 (XP_016480848), as detected by tBLASTn search (NCBI). Expression analysis revealed that LOC107773826 (TCP3/4/10) was consistently downregulated in all infected leaf cell cultures at 2 and 4WAI compared to the control (non-infected leaf explant). Notably, expression levels were lower in leaf cells introduced with the ΩBBMVP16&SRDXWUSm1 than in those with ΩBBMVP16&WUS (Figure 7A). Both LOC107782639 (IAA26) and LOC107795218 (Phytosulfokine-3) showed higher expression levels in ΩBBMVP16&SRDXWUSm1-introduced leaf cells than in ΩBBMVP16&WUS-introduced cells at 2 and 4 WAI (Figure 7A). LOC107801243 (HST1) showed high expression in the ΩBBMVP16&SRDXWUSm1-introduced leaf cell cultures, further upregulated at 4WAI (Figure 7A). These qRT-PCR results corresponded with DEGs identified from RNA-seq analysis.
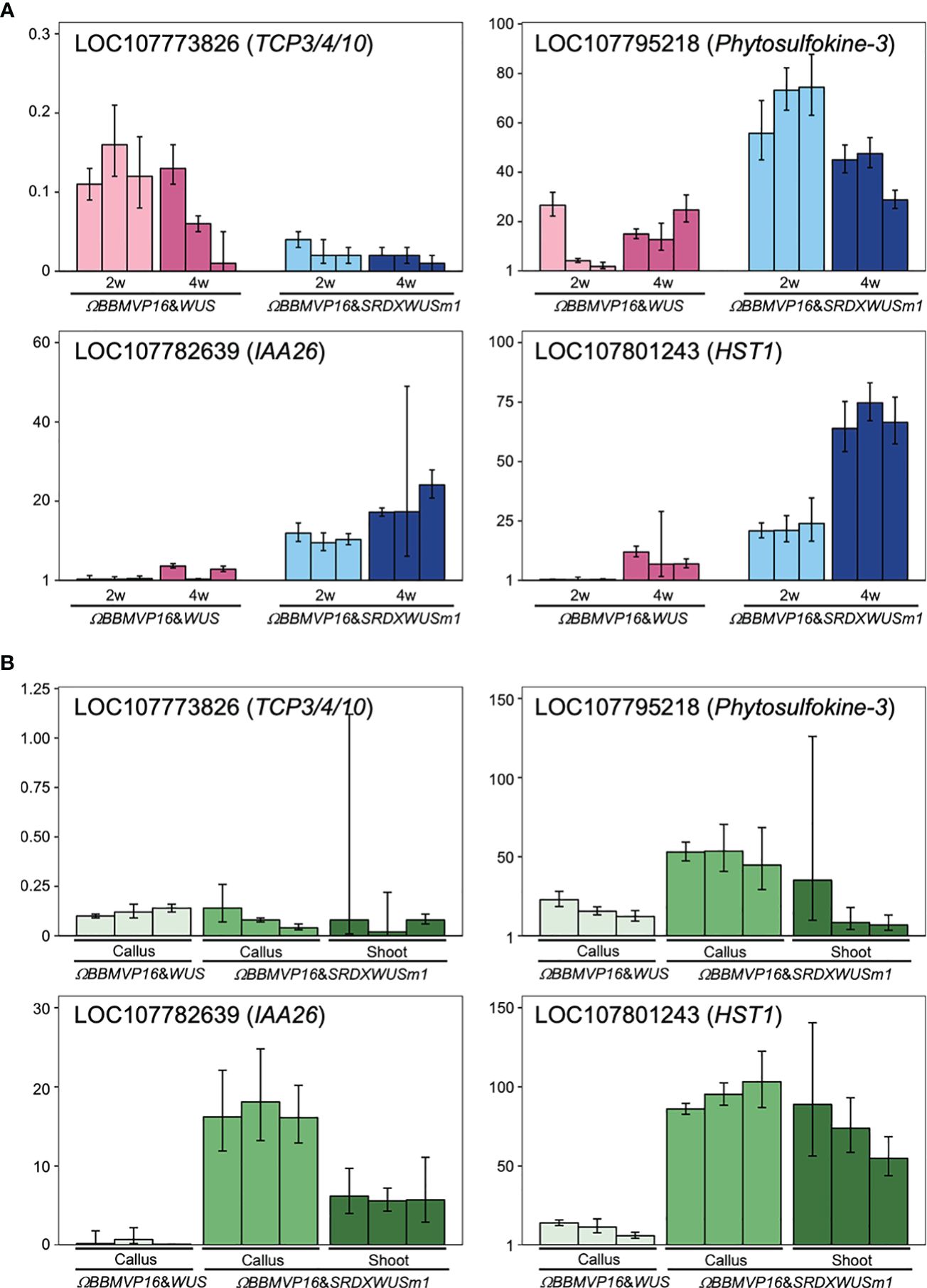
Figure 7 Relative expression levels of four genes detected as DEGs by ΩBBMVP16&SRDXWUSm1 introduction. (A) q-RT PCR analysis with samples derived from three biological replicates of infection with ΩBBMVP16&WUS and ΩBBMVP16&SRDXWUSm1. The x-axis represents the 2 or 4-week-cultured leaf explant cultures (2w and 4w) introduced with each construct. (B) qRT-PCR with transgenic calli and shoots samples. The shoots introduced with ΩBBMVP16&WUS were not analyzed because of the scarce formation (Figure 1B). Three independent samples detached from the differentiating leaf explant cell culture were analyzed. The y-axis in all graphs represents the relative quantification (RQ; 2-ΔΔCt) compared to the control (non-infected leaf explant). The error bars represent the maximum and minimum RQ values of three reaction replicates (95.0% confidence level).
Furthermore, qRT-PCR of the same four genes was performed for transgenic calli and shoots derived from leaf explant cultures. Compared to the non-infected leaf explants (control), LOC107773826 (TCP3/4/10) expression was downregulated in all tested calli and shoots (Figure 7B). In general, expression levels of LOC107782639 (IAA26), LOC107795218 (Phytosulfokine-3), and LOC107801243 (HST1) were higher inΩBBMVP16&SRDXWUSm1 samples compared to ΩBBMVP16&WUS, particularly in callus (Figure 7B).
4 Discussion
4.1 Modification of Arabidopsis BBM and WUS enabled autonomous cell differentiation of the transgenic cells without external PGR treatment
In the present study, we demonstrated that the combined expression of Arabidopsis BBM and WUS in transgenic tobacco, petunia, and lettuce leaf cells resulted in autonomous cell differentiation without the application of PGRs during culture process (Figure 1A; Supplementary Figures 1-7). Single and combined introduction of BBM and WUS, which are regulated by the 35S and RPS5A promoters, respectively, was insufficient to induce cell differentiation (Supplementary Figures 1B, D, F). A single introduction of ΩBBMVP16, in which the translation enhancer omega sequence and the VP16 transcriptional activator were applied, was still insufficient (Supplementary Figure 1C). In addition, a single introduction of SRDXWUSm1, in where two amino acid mutations were inserted into the WUS-box and an artificial strong repression domain, SRDX, was fused to the N-terminal end of WUSm1, did not induce cell differentiation (Supplementary Figure 1E). In contrast, the combined expression of ΩBBMVP16 and WUS (ΩBBMVP16&WUS) successfully induced transgenic calli (Figure 1A; Supplementary Figure 1G). In a previous study, Arabidopsis and Brassica napus BBM were evaluated in tobacco, and inducible differentiation from leaf explant cells was achieved by the supportive application of cytokinins, and leaves from the 35S::BBM transgenic tobacco plants did not cause cellular differentiation in PGR-free culture conditions (Srinivasan et al., 2007). Taken together with the previous insights and our results, the single expression of translationally and functionally enhanced BBM could alter the cell physiology involved in endogenous phytohormone for differentiation; however, it might still be insufficient to cause morphological alterations as apparent differentiation. As for WUS, the expression of Arabidopsis WUS induces adventitious shoot formation from the root tissues in Arabidopsis and tobacco (Rashid et al., 2007; Ikeda et al., 2009, 2020). Therefore, to induce adventitious shoot differentiation from somatic cells by the ectopic expression of WUS, it may be critical that somatic cells have a root-pertaining genetic background. Thus, the differentiation achieved by the combined expression of ΩBBMVP16 and WUS (or SRDXWUS, SRDXWUSm1) suggests that these DRs synergistically influence gene expression and physiology, resulting in cellular differentiation. In leaf cell cultures exhibiting autonomous differentiation, relatively lower levels of WUS expression were observed compared to BBM expression levels (Supplementary Figure 3). This observation supports the speculation that WUS triggers cell proliferation and organogenesis with substantial physiological modifications by BBM. The malformation patterns observed in the regenerated transgenic plants were associated with higher DR expression levels, particularly a significant BBM level (Supplementary Figure 8). To regulate the morphology of regenerated plants, employing inducible promoters would improve the current system.
The combined expression of ΩBBMVP16 and SRDXWUSm1 (or SRDXWUS) showed a different differentiation pattern compared to ΩBBMVP16&WUS, showing faster and more accelerated shoot differentiation in tobacco transgenic cells (Figure 1A; Supplementary Figures 1I, J). A similar tendency was observed in the case of petunia and lettuce (Supplementary Figure 6). The ectopic expression of SRDXWUSm1 is more effective than WUS in inducing adventitious shoot formation from somatic cells in Arabidopsis (Ikeda et al., 2020). Similarly, our study demonstrated a significant effect on autonomous shoot organogenesis in tobacco cells through the expression of SRDXWUS or SRDXWUSm1 in combination with ΩBBMVP16, indicating that SRDX fusion to the N-terminal of WUS was effective for the cellular differentiation also in tobacco cell (Supplementary Figures 1, 2). Regarding the WUS-box mutation, WUSm1 showed a reduced ability for autonomous callus formation compared to WUS (Supplementary Figures 1G, H, 2). In contrast, SRDXWUSm1 induced higher callus formation than SRDXWUS (Supplementary Figure 2). WUS function as a repressor in maintaining pluripotent stem cells, and WUS-box is crucial for this role (Ikeda et al., 2009). It has been suggested that the fusion of the artificial strong repression domain SRDX to the WUSm1 generates a protein with enhanced repression activity, thereby promoting organogenesis through cytokinin signaling (Ikeda et al., 2020). Alternatively, it could be postulated that the acceleration of stem cell maintenance repression disruption by the WUS-box mutation was facilitated by SRDX fusion, thereby stimulating cell division in tobacco cells.
4.2 Phytohormone behaviors underlining dedifferentiation and redifferentiation
Because the present BBM and WUS expression system enabled autonomous cell differentiation on a PGR-free medium, pure endogenous phytohormone behaviors underlying cellular differentiation could be analyzed. To understand the swiftness and differences in cell differentiation observed in this study, phytohormone-quantitative analysis was performed (Figure 2; Supplementary Table 2). Active auxin (IAA) and cytokinin (iP and tZ) levels were higher in 2w:ΩBBMVP16&SRDXWUSm1 and 4w:ΩBBMVP16&WUS, in which callus induction was observed. The timing of the increase in active auxin/cytokinin content coincided with callus induction in tobacco leaf explants (Figure 1A). This result is consistent with the acknowledged theory that increased auxin and cytokinin levels in tissue culture medium promote dedifferentiation (Skoog and Miller, 1957; Phillips and Garda, 2019).
Focusing on 4WAI, the calli that emerged from ΩBBMVP16&SRDXWUSm1 induced a more shoot-like structure, indicating that redifferentiation was promoted compared to ΩBBMVP16&WUS. The contents of GA19, GA24, and GA53, which are precursors of active GA1 or GA4 (He et al., 2020), were significantly higher in ΩBBMVP16&SRDXWUSm1 than in ΩBBMVP16&WUS, whereas all the active GAs (GA1, GA3, GA4, and GA7) were not present at the quantitative level in any of the analyzed samples (Figure 2; Supplementary Table 2). Active GAs negatively regulate shoot regeneration in Arabidopsis (Ezura and Harberd, 1995). Active GAs and cytokinins act antagonistically during shoot regeneration in tobacco (Engelke et al., 1973). In our study, the accumulation of active GA precursors (Figure 2) suggests that active GAs synthesis was suppressed and endogenous cytokinins were not antagonized for shoot regeneration in ΩBBMVP16&SRDXWUSm1-introduced leaf cells.
In the present study, a significant increase in ABA was observed in 4w:ΩBBMVP16&SRDXWUSm1. Although the substantial contribution of ABA to cellular differentiation has not yet been elucidated (Shin et al., 2020), ABA is known to crosstalk with other phytohormones, and is involved in plant growth regulation and stress (Skubacz et al., 2016). Thus, the observed elevation suggests the involvement of ABA in organogenesis in the coordinated balance between auxin and cytokinin that maintained this level.
4.3 Gene expression differences between the differentiated cells caused by ΩBBMVP16&WUS and ΩBBMVP16&SRDXWUSm1
In RNA-seq analysis, GO terms such as ‘auxin-activated signaling pathways’ and ‘response to cytokinin’ were enriched with the DEGs upregulated in 2w:ΩBBMVP16&SRDXWUSm1 (Figures 4B, C; Supplementary Figure 9). These GO terms included the putative orthologs of AUX/IAAs and PIN1/3/4/7 (Supplementary Table 3), which coincided with increased levels of active auxin (Figure 2). The other detected ortholog in the GO terms, Arabidopsis HP6, has been known to act as an inhibitor of cytokinin signaling (Mähönen et al., 2006; Müller and Sheen, 2007; Besnard et al., 2014). Moreover, it has also been reported that overexpression of HP6 in SAM suppresses type-A ARRs, which repress cytokinin signaling (Besnard et al., 2014). In our study, the tobacco orthologous gene of Arabidopsis HP6 was upregulated in 2w:ΩBBMVP16&SRDXWUSm1 more than in 2w:ΩBBMVP16&WUS. The calli induced by ΩBBMVP16&SRDXWUSm1 had greenish organ-like structures (Figure 1A; Supplementary Figure 1J). Therefore, it was suggested that upregulation of HP6 promoted the suppression of type-A ARRs, resulting in the activation of cytokinin signaling, cell division, and near-organogenic differentiation. Active cytokinin content was also higher in 2w:ΩBBMVP16&SRDXWUSm1 than in 2w:ΩBBMVP16&WUS (Figure 2), which is consistent with this hypothesis. Previous studies have reported that HP6 is directly activated by auxin (Bishopp et al., 2011; Besnard et al., 2014) and affects the localization of PIN1 in Arabidopsis (Moreira et al., 2013). Such interactions of HP6 with auxin and PINs may be involved in the differences in the differentiation phenotypes caused by ΩBBMVP16&WUS and ΩBBMVP16&SRDXWUSm1. In addition, GO terms related to the cell wall, especially the ‘polysaccharide catabolic process’ were significantly upregulated in 2w:ΩBBMVP16&SRDXWUSm1 compared to 2w:ΩBBMVP16&WUS (Figures 4B, C; Supplementary Table 3). A previous study reported that WUS homeobox-containing 13 (WOX13) is a key regulator to promote callus formation by modifying the cell wall properties in Arabidopsis (Ikeuchi et al., 2022). WOX13 directly upregulates cell wall-related genes involved in the polysaccharide catabolic process (Ikeuchi et al., 2022). WOX13 conserves the WUS-homeodomain and an acidic domain but lacks the WUS-box (Dolzblasz et al., 2016) and targets sequences that also target WUS (Ma et al., 2019; Ikeuchi et al., 2022). The promoted callus formation by SRDXWUSm1 observed in our study (Supplementary Figure 2) could reflect a similar activation to WOX13.
The 4w:ΩBBMVP16&SRDXWUSm1 leaf cells differentiated into calli and shoots, in contrast to 4w:ΩBBMVP16&WUS, in which only callus formation was observed (Figure 1A; Supplementary Figures 1G, J). The result that the active cytokinin iP content was higher (Figure 2) was reminiscent of the differentiation tendency of N. tabacum in tissue culture, where a higher ratio of cytokinin to auxin induces organogenesis (Skoog and Miller, 1957; Phillips and Garda, 2019). Although the GO terms related to cytokinin were not enriched, GO terms related to auxin and ABA were detected in the DEG upregulated in 4w:ΩBBMVP16&SRDXWUSm1 (Figure 5B; Supplementary Figures 10A, 11; Supplementary Table 4). In addition to auxin transport, response, and signaling, GO terms related to auxins included accessions involved in shoot development (Figure 5B; Supplementary Figures 10A, 11; Supplementary Table 4). This result may reflect the adventitious shoot formation observed in the present study (Figure 1A; Supplementary Figure 1J). Although the role of ABA in plant regeneration has not been extensively studied, several studies have suggested its effects on shoot regeneration (Shin et al., 2020). For example, the leucine-rich repeat receptor-like kinase RECEPTOR-LIKE PROTEIN KINASE1 (RPK1), which is involved in ABA signaling, promotes shoot regeneration in Arabidopsis (Motte et al., 2014). As mentioned above, ABA is also known to crosstalk with other phytohormones (Skubacz et al., 2016), as reported that ABA-induced ABI5 inhibited PINs accumulation and auxin activity, for instance (Yuan et al., 2014). The fact that many GO terms related to stress were detected (Figure 5B; Supplementary Figure 10A) implies the involvement of ABA. In our study, the putative orthologs of ABI5 and PINs were detected as DEG that were upregulated in 4w:ΩBBMVP16&SRDXWUSm1 (Supplementary Table 4), suggesting that phytohormone crosstalk, especially by auxin and ABA, was influenced by organogenesis, such as shoot formation.
The primary GO terms detected in DEGs downregulated in 4w:ΩBBMVP16&SRDXWUSm1 were related to metabolism, which is likely involved in transcription and translation (Figure 5C; Supplementary Figure 10B). This result reflects the dominance of the above metabolic pathways for active cell division in callus formation (4w:ΩBBMVP16&WUS) rather than for organogenesis (4w:ΩBBMVP16&SRDXWUSm1).
4.4 Changes in gene expression over time in ΩBBMVP16&SRDXWUSm1-introduced leaf cells
No significant changes in the expression of genes related to phytohormones or regeneration were observed over time in ΩBBMVP16&WUS-introduced leaf cells, whereas GO terms related to the regulation of plant organogenesis were significantly downregulated over time in ΩBBMVP16&SRDXWUSm1-introduced leaf cells. Genes related to the regulation of plant organogenesis were identified as the TCP3/4/10 orthologs (Supplementary Table 5), and qRT-PCR also showed the significant downregulation in ΩBBMVP16&SRDXWUSm1-introduced leaf cells and differentiated calli and shoots (Figure 7). TCP4 is involved in multiple plant developmental processes, such as leaf and flower morphogenesis, secondary cell wall biosynthesis, senescence, and hormone signaling in Arabidopsis (Palatnik et al., 2003; Schommer et al., 2008; Nag et al., 2009; Sarvepalli and Nath, 2011; Li et al., 2012; Sun et al., 2017). A previous study reported promotion of shoot regeneration in tcp3/4/10 mutant in Arabidopsis, indicating that TCPs are strong negative regulators of de novo shoot regeneration (Yang et al., 2020). The downregulation of TCP3/4/10 over time in ΩBBMVP16&SRDXWUSm1-introduced leaf cell cultures might account for the enhanced differentiation, while the significant difference in the expression level between differentiated calli and shoots was not observed in our study (Figure 7B). In addition, GO terms related to glutamine and asparagine were gradually downregulated in ΩBBMVP16&SRDXWUSm1-introduced leaf cells (Figure 6B; Supplementary Figure 13), and the ortholog genes of GDHs, GS2, and ASN1 were annotated (Supplementary Table 5). These genes are upregulated in the dark to activate nitrogen assimilation (Yoneyama and Suzuki, 2020; Liu et al., 2022). In the present study, chloroplast differentiation was observed in ΩBBMVP16&SRDXWUSm1-introduced leaf cells (Figure 1A; Supplementary Figure 1J), and all culture processes were performed under continuous fluorescent light. These results and the environmental conditions may account for the downregulation of these genes. In addition, GO terms related to stimuli were detected from five DEGs, including OSM34 and MLOs orthologs, which have been reported to be involved in the ABA response (Lim and Lee, 2014; Park and Kim, 2021). The relationship between the elevated ABA level at 4WAI compared to that at 2WAI (Figure 2) and the downregulation of these ortholog genes cannot be explained at present, although it could imply that altered ABA signaling influenced cellular differentiation.
The present study identified 19 genes as the common intersections among DEGs upregulated during culture over time by ΩBBMVP16&SRDXWUSm1 compared to ΩBBMVP16&WUS. Among these genes, LOC107801243 was upregulated compared to both the control and ΩBBMVP16&WUS (Supplementary Figure 15; Supplementary Table 7). The qRT-PCR analysis of selected genes LOC107782639 (IAA26), LOC107795218 (Phytosulfokine-3), and LOC107801243 (NAD+-dependent protein deacetylase HST1) showed similar behavior to those detected by RNA-seq, showing upregulation in both differentiated calli and shoots (Figure 7). The IAA26 belongs to the Aux/IAA transcription factor family and is known to respond to auxin. Therefore, the increased levels of auxins (Figure 2) may have contributed to the upregulation of LOC107782639. Phytosulfokine is a plant peptide growth factor that affects plant growth and differentiation (Ding et al., 2022). A study has shown that altered expression of an Arabidopsis phytosulfokine receptor influenced the callus formation capacity in response to PGRs (Matsubayashi et al., 2006). Considering the higher expression of LOC107795218 (Phytosulfokine-3) was observed in calli induced with ΩBBMVP16&SRDXWUSm1 (Figure 7B), enhanced LOC107795218 expression may be associated with callus induction. In the yeast Saccharomyces cerevisiae, it has been reported that NAD+-dependent protein deacetylase HST1 is a sensor of cellular NAD+ levels involved in homeostasis (Bedalov et al., 2003). NAD+ is converted from nicotinic acid imported from the culture medium. Additionally, yeast HST1 represses the expression of thiamin biosynthesis genes by binding to their promoters, thereby regulating thiamin homeostasis (Li et al., 2010). Nicotinic acid and thiamin are components of the MS medium. These findings and our results (Figure 7; Supplementary Figure 15; Supplementary Table 7) suggest that LOC107801243 was highly upregulated in actively differentiating cells importing extracellular vitamins. In our study, no orthologs of LEC1, LEC2, ABSCISIC ACIDINSENSITIVE3 (ABI3), and FUSCA3 (FUS3), which are known to be activated by BBM (Horstman et al., 2017), were detected. The present study examined DEGs resulting from the combined expression of SRDXWUS or SRDXWUSm1 with ΩBBMVP16. It is possible that these combinations could have influenced gene networks differently compared to the single expression of BBM alone. Future functional analysis of found DEGs and investigation of gene networks related to known DRs would provide further insights into cellular differentiation and organogenesis.
5 Conclusion
In this study, we demonstrated the functional efficacy of co-expressing Brassicaceae Arabidopsis BBM and WUS genes in inducing autonomous cellular differentiation with some plant cultivars from the Solanaceae and Asteraceae families. The functional enhancement by fusing VP16 to BBM and the functional modification of the WUS by the addition of SRDX showed drastic effect on accelerating the differentiation of transgenic cells toward organogenesis. The ectopic expressions of these DRs consequently influenced phytohormone levels and gene expressions involved in auxin response, metabolism, and organogenesis, resulting in fine-tuned physiology for differentiation. A noteworthy aspect of this study is the complete omission of external PGR application during tissue and cell culture for plant regeneration, a typically prerequisite process for transformation and genome-editing studies. Further investigations on the functional conservation of BBM and WUS in different plant species would improve the versatility of this system and enable its simplification. Challenges to develop an accurate regulation system for gene expression and the levels are also ongoing.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Bioproject accession number: PRJNA1049661, https://dataview.ncbi.nlm.nih.gov/object/PRJNA1049661?reviewer=r5siqhguq12lgug1tqlrkmfmk3.
Author contributions
YS: Investigation, Methodology, Writing – original draft. MM: Data curation, Writing – review & editing. SK: Investigation, Writing – original draft. BP: Investigation, Writing – original draft. MK: Investigation, Writing – review & editing. YT: Investigation, Writing – review & editing. HS: Investigation, Writing – review & editing. TI: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Japan Society for the Promotion of Science KAKENHI Grants (JP17K07600 to TI, JP19H04852 to TI, JP23H04732 to TI) and JST SPRING grants (JPMJSP2109 to YS).
Acknowledgments
We thank Dr. Nakagawa (Shimane University) for providing pGWB vectors. pUC vectors containing either the attL1_35Sp:ΩBBM&VP16:HSPT_attL2 or the 35Sp:SRDXWUSm1 region were provided by Dr. Ikeda (Fukui Prefectural University). The cDNA library construction and sequencing were conducted by ClockMics, Inc. (Osaka, Japan). Phytohormone-quantitative analysis was conducted by RIKEN CSRS (Kanagawa, Japan).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1308417/full#supplementary-material
References
Akiyoshi, D. E., Klee, H., Amasino, R. M., Nester, E. W., Gordon, M. P. (1984). T-DNA of Agrobacterium tumefaciens encodes an enzyme of cytokinin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 81, 5994–5998. doi: 10.1073/pnas.81.19.5994
Barry, G. F., Rogers, S. G., Fraley, R. T., Brand, L. (1984). Identification of a cloned cytokinin biosynthetic gene. Proc. Natl. Acad. Sci. U.S.A. 81, 4776–4780. doi: 10.1073/pnas.81.15.4776
Barton, M. K., Poethig, R. S. (1993). Formation of the shoot apical meristem in Arabidopsis thaliana: an analysis of development in the wild type and in the shoot meristemless mutant. Dev. 119, 823–831. doi: 10.1242/dev.119.3.823
Bedalov, A., Hirao, M., Posakony, J., Nelson, M., Simon, J. A. (2003). NAD+-dependent deacetylase Hst1p controls biosynthesis and cellular NAD+ levels in Saccharomyces cerevisiae. Mol. Cell Biol. 23, 7044–7054. doi: 10.1128/MCB.23.19.7044-7054.2003
Besnard, F., Rozier, F., Vernoux, T. (2014). The AHP6 cytokinin signaling inhibitor mediates an auxin-cytokinin crosstalk that regulates the timing of organ initiation at the shoot apical meristem. Plant Signal. Behav. 9, e28788. doi: 10.4161/psb.28788
Bishopp, A., Help, H., El-Showk, S., Weijers, D., Scheres, B., Friml, J., et al. (2011). A mutually inhibitory interaction between auxin and cytokinin specifies vascular pattern in roots. Curr. Biol. 21, 917–926. doi: 10.1016/j.cub.2011.04.017
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boutilier, K., Offringa, R., Sharma, V. K., Kieft, H., Ouellet, T., Zhang, L., et al. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell. 14, 1737–1749. doi: 10.1105/tpc.001941
Chen, B., Maas, L., Figueiredo, D., Zhong, Y., Reis, R., Li, M., et al. (2022a). BABY BOOM regulates early embryo and endosperm development. Proc. Natl. Acad. Sci. U.S.A. 119, e2201761119. doi: 10.1073/pnas.2201761119
Chen, H., Boutros, P. C. (2011). VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 12, 35. doi: 10.1186/1471-2105-12-35
Chen, J., Tomes, S., Gleave, A. P., Hall, W., Luo, Z., Xu, J., et al. (2022b). Significant improvement of apple (Malus domestica Borkh.) transgenic plant production by pre-transformation with a Baby boom transcription factor. Hortic. Res. 9, uhab014. doi: 10.1093/hr/uhab014
Cody, J. P., Maher, M. F., Nasti, R. A., Starker, C. G., Chamness, J. C., Voytas, D. F. (2023). Direct delivery and fast-treated Agrobacterium co-culture (Fast-TrACC) plant transformation methods for Nicotiana benthamiana. Nat. Protoc. 18, 81–107. doi: 10.1038/s41596-022-00749-9
Debernardi, J. M., Tricoli, D. M., Ercoli, M. F., Hayta, S., Ronald, P., Palatnik, J. F., et al. (2020). A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 38, 1274–1279. doi: 10.1038/s41587-020-0703-0
Deng, W., Luo, K., Li, Z., Yang, Y. (2009). A novel method for induction of plant regeneration via somatic embryogenesis. Plant Sci. 177, 43–48. doi: 10.1016/j.plantsci.2009.03.009
Ding, S., Lv, J., Hu, Z., Wang, J., Wang, P., Yu, J., et al. (2022). Phytosulfokine peptide optimizes plant growth and defense via glutamine synthetase GS2 phosphorylation in tomato. EMBO J. 42, e111858. doi: 10.15252/embj.2022111858
Dolzblasz, A., Nardmann, J., Clerici, E., Causier, B., van der Graaff, E., Chen, J., et al. (2016). Stem cell regulation by Arabidopsis WOX Genes. Mol. Plant 9, 1028–1039. doi: 10.1016/j.molp.2016.04.007
Duan, H., Maren, N. A., Ranney, T. G., Liu, W. (2022). New opportunities for using WUS/BBM and GRF-GIF genes to enhance genetic transformation of ornamental plants. Ornam. Plant Res. 2, 1–7. doi: 10.48130/OPR-2022-0004
Engelke, A. L., Hamzi, H. Q., Skoog, F. (1973). CytokininOPR-2022-000 regulation of shoot development and leaf form in tobacco plantlets. Am. J. Bot. 60, 491–495. doi: 10.1002/j.1537-2197.1973.tb05949.x
Ezura, H., Harberd, N. P. (1995). Endogenous gibberellin levels influence in-vitro shoot regeneration in Arabidopsis thaliana (L.) Heynh. Planta. 197, 301–305. doi: 10.1007/BF00202651
Gallie, D. R. (2002). The 5’-leader of tobacco mosaic virus promotes translation through enhanced recruitment of eIF4F. Nucleic Acids Res. 30, 3401–3411. doi: 10.1093/nar/gkf457
Gordon-Kamm, B., Sardesai, N., Arling, M., Lowe, K., Hoerster, G., Betts, S., et al. (2019). Using Morphogenic Genes to Improve recovery and regeneration of transgenic plants. Plants (Basel). 8, 38. doi: 10.3390/plants8020038
He, J., Xin, P., Ma, X., Chu, J., Wang, G. (2020). Gibberellin metabolism in flowering plants: An Update and Perspectives. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00532
Heidmann, I., De Lange, B., Lambalk, J., Angenent, G. C., Boutilier, K. (2011). Efficient sweet pepper transformation mediated by the BABY BOOM transcription factor. Plant Cell Rep. 30, 1107–1115. doi: 10.1007/s00299-011-1018-x
Hiratsu, K., Matsui, K., Koyama, T., Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739. doi: 10.1046/j.1365-313X.2003.01759.x
Horstman, A., Li, M., Heidmann, I., Weemen, M., Chen, B., Muino, J. M., et al. (2017). The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. Plant Physiol. 175, 848–857. doi: 10.1104/pp.17.00232
Ikeda, M., Mitsuda, N., Ohme-Takagi, M. (2009). Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell. 21, 3493–3505. doi: 10.1105/tpc.109.069997
Ikeda, M., Takahashi, M., Fujiwara, S., Mitsuda, N., Ohme-Takagi, M. (2020). Improving the efficiency of adventitious shoot induction and somatic embryogenesis via modification of WUSCHEL and LEAFY COTYLEDON 1. Plants (Basel). 9, 1434. doi: 10.3390/plants9111434
Ikeuchi, M., Iwase, A., Ito, T., Tanaka, H., Favero, D. S., Kawamura, A., et al. (2022). Wound-inducible WUSCHEL-RELATED HOMEOBOX 13 is required for callus growth and organ reconnection. Plant Physiol. 188, 425–441. doi: 10.1093/plphys/kiab510
Johnson, K., Cao Chu, U., Anthony, G., Wu, E., Che, P., Jones, T. J. (2023). Rapid and highly efficient morphogenic gene-mediated hexaploid wheat transformation. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1151762
Kamitani, M., Kashima, M., Tezuka, A., Nagano, A. J. (2019). Lasy-Seq: a high-throughput library preparation method for RNA-Seq and its application in the analysis of plant responses to fluctuating temperatures. Sci. Rep. 9, 7091. doi: 10.1038/s41598-019-43600-0
Kim, C. Y., Ahn, Y. O., Kim, S. H., Kim, Y. H., Lee, H. S., Catanach, A. S., et al. (2010). The sweet potato IbMYB1 gene as a potential visible marker for sweet potato intragenic vector system. Physiol. Plant 139, 229–240. doi: 10.1111/j.1399-3054.2010.01365.x
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kojima, M., Kamada-Nobusada, T., Komatsu, H., Takei, K., Kuroha, T., Mizutani, M., et al. (2009). Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 50, 1201–1214. doi: 10.1093/pcp/pcp057
Kojima, M., Sakakibara, H. (2012). Highly sensitive high-throughput profiling of six phytohormones using MS-probe modification and liquid chromatography-tandem mass spectrometry. Methods Mol. Biol. 918, 151–164. doi: 10.1007/978-1-61779-995-2_11
Laux, T., Mayer, K. F., Berger, J., Jürgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Dev. 122, 87–96. doi: 10.1242/dev.122.1.87
Li, Z., Li, B., Shen, W. H., Huang, H., Dong, A. (2012). TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J. 71, 99–107. doi: 10.1111/j.1365-313X.2012.04973.x
Li, M., Petteys, B. J., McClure, J. M., Valsakumar, V., Bekiranov, S., Frank, E. L., et al. (2010). Thiamine biosynthesis in Saccharomyces cerevisiae is regulated by the NAD+-dependent histone deacetylase Hst1. Mol. Cell Biol. 30, 3329–3341. doi: 10.1128/MCB.01590-09
Li, K., Wang, J., Liu, C., Li, C., Qiu, J., Zhao, C., et al. (2019). Expression of AtLEC2 and AtIPTs promotes embryogenic callus formation and shoot regeneration in tobacco. BMC Plant Biol. 19, 314. doi: 10.1186/s12870-019-1907-7
Lian, Z., Nguyen, C. D., Liu, L., Wang, G., Chen, J., Wang, S., et al. (2022). Application of developmental regulators to improve in planta or in vitro transformation in plants. Plant Biotechnol. J. 20, 1622–1635. doi: 10.1111/pbi.13837
Lim, C. W., Lee, S. C. (2014). Functional roles of the pepper MLO protein gene, CaMLO2, in abscisic acid signaling and drought sensitivity. Plant Mol. Biol. 85, 1–10. doi: 10.1007/s11103-013-0155-8
Liu, X., Hu, B., Chu, C. (2022). Nitrogen assimilation in plants: current status and future prospects. J. Genet. Genomics 49, 394–404. doi: 10.1016/j.jgg.2021.12.006
Lotan, T., Ohto, M., Yee, K. M., West, M. A., Lo, R., Kwong, R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 93, 1195–1205. doi: 10.1016/S0092-8674(00)81463-4
Lowe, K., La Rota, M., Hoerster, G., Hastings, C., Wang, N., Chamberlin, M., et al. (2018). Rapid genotype "independent" Zea mays L. (maize) transformation via direct somatic embryogenesis. In Vitro Cell. Dev. Biol. Plant 54, 240–252. doi: 10.1007/s11627-018-9905-2
Lowe, K., Wu, E., Wang, N., Hoerster, G., Hastings, C., Cho, M. J., et al. (2016). Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell. 28, 1998–2015. doi: 10.1105/tpc.16.00124
Ma, Y., Miotk, A., Šutiković, Z., Ermakova, O., Wenzl, C., Medzihradszky, A., et al. (2019). WUSCHEL acts as an auxin response rheostat to maintain apical stem cells in Arabidopsis. Nat. Commun. 10, 5093. doi: 10.1038/s41467-019-13074-9
Mähönen, A. P., Bishopp, A., Higuchi, M., Nieminen, K. M., Kinoshita, K., Törmäkangas, K., et al. (2006). Cytokinin signaling and its inhibitor AHP6 regulate cell fate during vascular development. Science. 311, 94–98. doi: 10.1126/science.1118875
Maliga, P., Sz-Breznovits, A., Márton, L. (1973). Streptomycin-resistant plants from callus culture of haploid tobacco. Nat. New Biol. 244, 29–30. doi: 10.1038/newbio244029a0
Matsubayashi, Y., Ogawa, M., Kihara, H., Niwa, M., Sakagami, Y. (2006). Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiol. 142, 45–53. doi: 10.1104/pp.106.081109
Mookkan, M., Nelson-Vasilchik, K., Hague, J., Zhang, Z. J., Kausch, A. P. (2017). Selectable marker independent transformation of recalcitrant maize inbred B73 and sorghum P898012 mediated by morphogenic regulators BABY BOOM and WUSCHEL2. Plant Cell Rep. 36, 1477–1491. doi: 10.1007/s00299-017-2169-1
Moreira, S., Bishopp, A., Carvalho, H., Campilho, A. (2013). AHP6 inhibits cytokinin signaling to regulate the orientation of pericycle cell division during lateral root initiation. PloS One 8, e56370. doi: 10.1371/journal.pone.0056370
Motte, H., Vercauteren, A., Depuydt, S., Landschoot, S., Geelen, D., Werbrouck, S., et al. (2014). Combining linkage and association mapping identifies RECEPTOR-LIKE PROTEIN KINASE1 as an essential Arabidopsis shoot regeneration gene. Proc. Natl. Acad. Sci. U.S.A. 111, 8305–8310. doi: 10.1073/pnas.1404978111
Müller, B., Sheen, J. (2007). Advances in cytokinin signaling. Science. 318, 68–69. doi: 10.1126/science.1145461
Nag, A., King, S., Jack, T. (2009). miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 22534–22539. doi: 10.1073/pnas.0908718106
Nakagawa, T., Kurose, T., Hino, T., Tanaka, K., Kawamukai, M., Niwa, Y., et al. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41. doi: 10.1263/jbb.104.34
Osabe, T., Shimizu, K., Kadota, K. (2019). Accurate classification of differential expression patterns in a bayesian framework with robust normalization for multi-group RNA-Seq count data. Bioinform. Biol. Insights 13, 1177932219860817. doi: 10.1177/1177932219860817
Palatnik, J. F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J. C., et al. (2003). Control of leaf morphogenesis by microRNAs. Nature. 425, 257–263. doi: 10.1038/nature01958
Park, E. J., Kim, T. H. (2021). Arabidopsis OSMOTIN 34 functions in the ABA signaling pathway and is regulated by proteolysis. Int. J. Mol. Sci. 22, (15). doi: 10.3390/ijms22157915
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Phillips, G. C., Garda, M. (2019). Plant tissue culture media and practices: an overview. In Vitro Cell. Dev. Biol. Plant 55, 242–257. doi: 10.1007/s11627-019-09983-5
Rashid, S. Z., Yamaji, N., Kyo, M. (2007). Shoot formation from root tip region: a developmental alteration by WUS in transgenic tobacco. Plant Cell Rep. 26, 1449–1455. doi: 10.1007/s00299-007-0342-7
Sarvepalli, K., Nath, U. (2011). Hyper-activation of the TCP4 transcription factor in Arabidopsis thaliana accelerates multiple aspects of plant maturation. Plant J. 67, 595–607. doi: 10.1111/j.1365-313X.2011.04616.x
Sato, Y., Fukuda, M., Chukwurah, P. N., Igawa, T. (2023). Development of an inducible excision system of a visual marker Ipomoea batatas Myb gene from the genome of transgenic cells. Plant Biotechnol. (Tokyo). 40, 175–179. doi: 10.5511/plantbiotechnology.23.0309a
Schmidt, G. W., Delaney, S. K. (2010). Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genomics 283, 233–241. doi: 10.1007/s00438-010-0511-1
Schommer, C., Palatnik, J. F., Aggarwal, P., Chételat, A., Cubas, P., Farmer, E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PloS Biol. 6, e230. doi: 10.1371/journal.pbio.0060230
Shin, J., Bae, S., Seo, P. J. (2020). De novo shoot organogenesis during plant regeneration. J. Exp. Bot. 71, 63–72. doi: 10.1093/jxb/erz395
Shinozaki, Y., Hao, S., Kojima, M., Sakakibara, H., Ozeki-Iida, Y., Zheng, Y., et al. (2015). Ethylene suppresses tomato (Solanum lycopersicum) fruit set through modification of gibberellin metabolism. Plant J. 83, 237–251. doi: 10.1111/tpj.12882
Sierro, N., Battey, J. N., Ouadi, S., Bakaher, N., Bovet, L., Willig, A., et al. (2014). The tobacco genome sequence and its comparison with those of tomato and potato. Nat. Commun. 5, 3833. doi: 10.1038/ncomms4833
Skoog, F., Miller, C. O. (1957). Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc Exp. Biol. 11, 118–130.
Skubacz, A., Daszkowska-Golec, A., Szarejko, I. (2016). The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01884
Srinivasan, C., Liu, Z., Heidmann, I., Supena, E. D., Fukuoka, H., Joosen, R., et al. (2007). Heterologous expression of the BABY BOOM AP2/ERF transcription factor enhances the regeneration capacity of tobacco (Nicotiana tabacum L.). Planta. 225, 341–351. doi: 10.1007/s00425-006-0358-1
Stone, S. L., Kwong, L. W., Yee, K. M., Pelletier, J., Lepiniec, L., Fischer, R. L., et al. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. U.S.A. 98, 11806–11811. doi: 10.1073/pnas.201413498
Su, W., Sun, J., Shimizu, K., Kadota, K. (2019). TCC-GUI: a Shiny-based application for differential expression analysis of RNA-Seq count data. BMC Res. Notes. 12, 133. doi: 10.1186/s13104-019-4179-2
Sun, X., Wang, C., Xiang, N., Li, X., Yang, S., Du, J., et al. (2017). Activation of secondary cell wall biosynthesis by miR319-targeted TCP4 transcription factor. Plant Biotechnol. J. 15, 1284–1294. doi: 10.1111/pbi.12715
Suo, J., Zhou, C., Zeng, Z., Li, X., Bian, H., Wang, J., et al. (2021). Identification of regulatory factors promoting embryogenic callus formation in barley through transcriptome analysis. BMC Plant Biol. 21, 145. doi: 10.1186/s12870-021-02922-w
Triezenberg, S. J., Kingsbury, R. C., Mcknight, S. L. (1988). Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev. 2, 718–729. doi: 10.1101/gad.2.6.718
Tsutsui, H., Higashiyama, T. (2017). pKAMA-ITACHI vectors for highly efficient CRISPR/Cas9-mediated gene knockout in Arabidopsis thaliana. Plant Cell Physiol. 58, 46–56. doi: 10.1093/pcp/pcw191
Yang, W., Choi, M. H., Noh, B., Noh, Y. S. (2020). De novo shoot regeneration controlled by HEN1 and TCP3/4 in Arabidopsis. Plant Cell Physiol. 61, 1600–1613. doi: 10.1093/pcp/pcaa083
Yoneyama, T., Suzuki, A. (2020). Light-independent nitrogen assimilation in plant leaves: Nitrate incorporation into glutamine, glutamate, aspartate, and asparagine traced by 15N. Plants (Basel). 9, (10). doi: 10.3390/plants9101303
Yu, G., Wang, L. G., Han, Y., He, Q. Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 16, 284–287. doi: 10.1089/omi.2011.0118
Yuan, T. T., Xu, H. H., Zhang, K. X., Guo, T. T., Lu, Y. T. (2014). Glucose inhibits root meristem growth via ABA INSENSITIVE 5, which represses PIN1 accumulation and auxin activity in Arabidopsis. Plant Cell Environ. 37, 1338–1350. doi: 10.1111/pce.12233
Zhao, S., Guo, Y., Sheng, Q., Shyr, Y. (2014). Heatmap3: an improved heatmap package with more powerful and convenient features. BMC Bioinform. 15, 1–2. doi: 10.1186/1471-2105-15-S10-P16
Zhou, R., Zhao, Y., Cheng, P., Zhang, B., Liu, Z., Wang, S., et al. (2023). GmBBM7 promotes callus and root growth during somatic embryogenesis of soybean (Glycine max). Biotechnol. Biotechnol. Equip. 37, 2238833. doi: 10.1080/13102818.2023.2238833
Keywords: autonomous cell differentiation, BABY BOOM, WUSCHEL, transgenic cells, PGR-free, phytohormone analysis, RNA-seq
Citation: Sato Y, Minamikawa MF, Pratama BB, Koyama S, Kojima M, Takebayashi Y, Sakakibara H and Igawa T (2024) Autonomous differentiation of transgenic cells requiring no external hormone application: the endogenous gene expression and phytohormone behaviors. Front. Plant Sci. 15:1308417. doi: 10.3389/fpls.2024.1308417
Received: 06 October 2023; Accepted: 18 March 2024;
Published: 03 April 2024.
Edited by:
Pejman Azadi, Agricultural Biotechnology Research Institute of Iran, IranReviewed by:
Alfred (heqiang) Huo, University of Florida, United StatesSiddharth Tiwari, National Agri-Food Biotechnology Institute, India
Copyright © 2024 Sato, Minamikawa, Pratama, Koyama, Kojima, Takebayashi, Sakakibara and Igawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomoko Igawa, tigawa@chiba-u.jp
 Yuka Sato
Yuka Sato Mai F. Minamikawa
Mai F. Minamikawa Berbudi Bintang Pratama
Berbudi Bintang Pratama Shohei Koyama
Shohei Koyama Mikiko Kojima3
Mikiko Kojima3 Hitoshi Sakakibara
Hitoshi Sakakibara Tomoko Igawa
Tomoko Igawa