- Department of Plant Protection, Faculty of Agriculture, University of Kurdistan, Sanandaj, Iran
Oak decline is a complex disorder that seriously threatens the survival of Zagros forests. In an extensive study on taxonomy and pathology of fungi associated with oak decline in the central and northern part of Zagros forests, 462 fungal isolates were obtained from oak trees showing canker, gummosis, dieback, defoliation, and partial or total death symptoms. Based on inter-simple sequence repeat (ISSR) fingerprinting patterns, morphological characteristics, and sequences of ribosomal DNA (28S rDNA and ITS) and protein coding loci (acl1, act1, caM, tef-1α, rpb1, rpb2, and tub2), 24 fungal species corresponding to 19 genera were characterized. Forty percent of the isolates were placed in eight coelomycetous species from seven genera, namely, Alloeutypa, Botryosphaeria, Cytospora, Didymella, Gnomoniopsis, Kalmusia, and Neoscytalidium. Of these, four species are new to science, which are introduced here as taxonomic novelties: Alloeutypa iranensis sp. nov., Cytospora hedjaroudei sp. nov., Cytospora zagrosensis sp. nov., and Gnomoniopsis quercicola sp. nov. According to pathogenicity trials on leaves and stems of 2-year-old Persian oak (Quercus brantii) seedlings, Alternaria spp. (A. alternata, A. atra, and A. contlous), Chaetomium globosum, and Parachaetomium perlucidum were recognized as nonpathogenic. All coelomycetous species were determined as pathogenic in both pathogenicity trials on leaves and seedling stems, of which Gnomoniopsis quercicola sp. nov., Botryosphaeria dothidea, and Neoscytalidium dimidiatum were recognized as the most virulent species followed by Biscogniauxia rosacearum.
1 Introduction
Oaks (Quercus L.) are the dominant vegetation of Zagros forests in western Iran with significant economic and environmental values (Heydari et al., 2013; Shiravand and Hosseini, 2020). Over the last two decades, oak decline as a serious ecological damage is spreading in this part of the world, which is caused by various biotic and abiotic factors affected by climate change, drought, wildfire, air pollution, irresponsible exploitation, and mismanagement (Ahmadi et al., 2014; Hosseini et al., 2018; Ahmadi et al., 2019; Shiravand and Hosseini, 2020; Mirhashemi et al., 2023). Fungi are well-known biotic agents, which affect trees under stress and cause canker, gummosis, vascular tissues necrosis, dieback, wilting, and partial and total death symptoms (Mirabolfathy et al., 2013; Ghasemi-Esfahlan et al., 2016, 2017; Safaee et al., 2017; Alidadi et al., 2019; Hanifeh et al., 2019; Moradi-Amirabad et al., 2019; Sabernasab et al., 2019; Di Lecce et al., 2020; Bashiri et al., 2020a, b, 2022). A wide variety of fungal species are associated with declined woody plants including coelomycetous fungi, a well-known morphological group belonging to the class Dothideomycetes (Wijayawardene et al., 2016). The most common pathogenic coelomycetous fungal species in association with declined trees showing canker, gummosis, dieback, wilting, and wood discoloration and necrosis are members of the Botryosphaeriaceae (e.g., Botryosphaeria, Diplodia, Lasiodiplodia, and Neofusicoccum spp.) (Alves et al., 2004; Turco et al., 2006; Linaldeddu et al., 2013; Dreaden et al., 2014; Smahi et al., 2017; Mahamedi et al., 2020), Diatrypaceae (e.g., Diatrype, Diatrypella, Eutypa, and Eutypella spp.) (Luque et al., 2000; Acero et al., 2004; Vasilyeva and Stephenson, 2004; Trouillas and Gubler, 2010; Lynch et al., 2013, 2014), Cytosporaceae (e.g., Cytospora spp.) (Preston, 1945; Spaulding, 1961; Senanayake et al., 2017; Shang et al., 2020, 2020; Pan et al., 2021), Gnomoniaceae (e.g., Discula and Gnomoniopsis spp.) (Moricca and Ragazzi, 2008; Sogonov et al., 2008; Walker et al., 2010; Jiang et al., 2021), and Graphostromataceae (e.g., Biscogniauxia spp.) (Raimondo et al., 2016; Safaee et al., 2017; Bahmani et al., 2021; Bashiri et al., 2022). Several studies on fungi associated with oak decline, mainly in the central part of Zagros forests, have introduced different fungal species most commonly belonging to the Cytosporaceae, Diatrypaceae, Biscogniauxia, and phoma-like genera (Mirabolfathy et al., 2013; Mehrabi et al., 2015; Ghobad-Nejhad et al., 2017, 2016, 2019; Alidadi et al., 2018, 2019; Ghasemi-Esfahlan et al., 2019; Sabernasab et al., 2019; Bashiri et al., 2022). In an extensive research on taxonomy and pathogenicity of fungal species associated with oak trees showing canker, gummosis, defoliation, wilting, dieback, and partial or total death symptoms in central and northern parts of Zagros forests, we identified 24 fungal species including 8 coelomycetous species. In this paper, we focus on the taxonomy and pathology of these species and introduce four new species as taxonomic novelties.
2 Materials and methods
2.1 Sampling, disease symptoms, and isolation of fungi
During a survey from July to November 2017, twigs and branches of oak trees showing external disease symptoms, dieback, wilting, canker, gummosis and twig/branch discoloration, and bark cracking were collected from Zagros forests of five provinces: Ilam, Kermanshah, Kurdistan, Lorestan, and West Azarbaijan. Cross-sections of the samples were examined to classify internal disease symptoms. To isolate fungi, small pieces of disinfested woody tissues (with 70% ethanol for 3 min) were transferred on potato dextrose agar (PDA) supplemented with 100 mg/L streptomycin sulfate and ampicillin and incubated at 20–25°C. Pure cultures were obtained using single spore or hyphal tip methods on tap water agar (2% WA). The isolates were stored on PDA at 4°C. Representative isolates were deposited in the culture collection of the Iranian Research Institute of Plant Protection (IRAN, Tehran, Iran) and the CBS collection of the Westerdijk Institute, Utrecht, The Netherlands. Isolates that were sequenced and studied morphologically are available in Table 1.
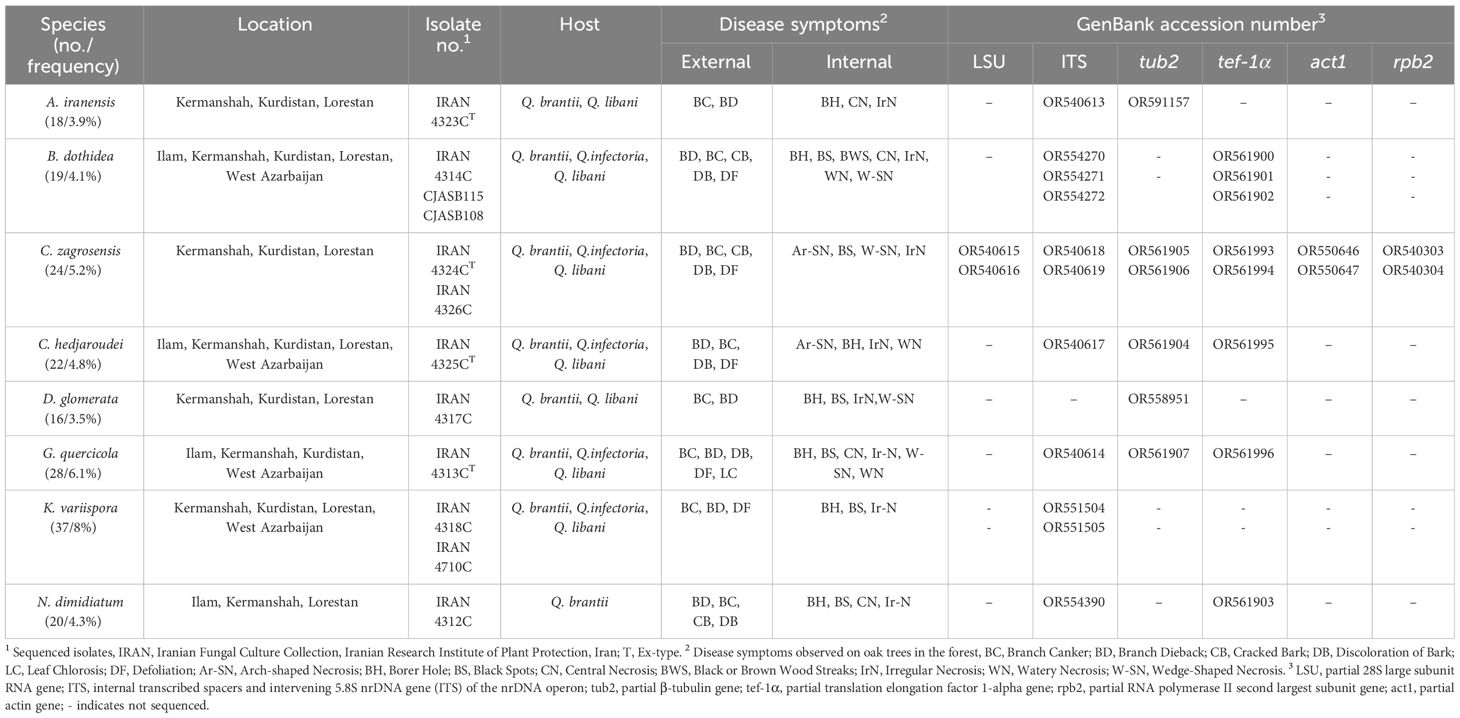
Table 1 Isolates, frequency, location, host, and collection codes of identified fungal species, disease symptoms observed and sequences generated in this study.
2.2 Molecular experiments
Total DNA was extracted from isolates grown on potato dextrose broth (PDB) after 7–10 d at 25°C using the modified Raeder and Broda (1985) method as described by Abdollahzadeh et al. (2009). To reduce the number of isolates for sequencing and microscopy, fungal isolates were grouped at the species level using the inter-simple sequence repeat (ISSR) technique based on DNA profiles generated with M13 primer (data not shown), and representative isolates of each group were sequenced. According to fungal taxonomic groups or genera deduced from morphology, different gene regions were amplified and sequenced using the following primer pairs: LROR/LR5 for a part of 28S nrDNA (LSU) (Vilgalys and Hester, 1990; Cubeta et al., 1991), ITS5/ITS4 for the ITS1-5.8S nrDNA-ITS2 region (ITS) (White et al., 1990), ACT512F/ACT783R for a part of actin (act1) (Carbone and Kohn, 1999), 5F (or 5F2)/7cR for a part of RNA polymerase II second largest subunit (rpb2) (Woudenberg et al., 2013), EF1-728F/EF1-986R (Vilgalys and Hester, 1990) for a part of translation elongation factor 1-alpha (tef-1α), and Bt2a (or T1)/Bt2b for a part of β-tubulin (tub2) (O’Donnell and Cigelnik, 1997; Glass and Donaldson, 1999). The PCR mixtures (25 µL) consisted of 1×PCR buffer, 3 mM MgCl2, 200 µM of dNTPs, 5 pmol of each primer, 1 U of Taq DNA polymerase, and 1 µL of template DNA (50–100 ng/µL). For amplification of ITS and tub2 regions, we followed the PCR conditions as described by Bashiri et al. (2022). The PCR conditions for tef-1α and rpb2 were as follows: an initial denaturation step of 5 min at 95°C followed by 35 cycles of 30 s at 95°C, 30 s at 52°C (tef-1α)/45 s at 54°C (rpb2)/1 min at 55°C (LSU)/30 s at 58°C (act1), and 90 s at 72°C, with a final extension of 7 min at 72°C. PCR products were purified and sequenced by Elim Biopharm (USA) via FAZABiotec Co. (Tehran, Iran) and BGI (China) via BMG (Bio Magic Gene) Co. (Karaj, Iran). Consensus sequences were prepared with BioEdit v. 7.0.0 (Hall, 2004). All new sequences generated in this study were submitted to GenBank (Table 1).
2.3 Phylogeny
The generated sequences together with the sequences retrieved from GenBank were aligned using Clustal X v. 1.83 (Thompson et al., 1997) or online MAFFT v. 7 and edited manually in BioEdit v. 7.0.0, where necessary. Each locus was aligned separately and the alignments were concatenated with Mesquite 2.75 (Maddison and Maddison, 2023). Both single and combined loci were analyzed by Maximum Parsimony (MP), Maximum Likelihood (ML), and Bayesian Inference (BI). MP was performed using PAUP v. 4.0b10 (Swofford, 2003). ML and BI were carried out through the online CIPRES Science Gateway (Miller et al., 2012) using RAxML-HPC BlackBox v. 8.2.10 (Stamatakis, 2014) and MrBayes v. 3.2.6 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003), respectively. MP analysis was executed according to Abdollahzadeh et al. (2013). Optimal nucleotide substitution models were detected for each locus using MrModelTest v. 2.3 (Nylander, 2004). The ML and Bayesian analyses were executed as described by Abdollahzadeh et al. (2020). Phylograms were plotted using FigTree v. 1.4.3 and edited in Adobe Illustrator CS2 v. 12.0.0. Alignments and trees were deposited in TreeBASE (www.treebase.org; S30794, S30795, and S30796) and taxonomic novelties registered in MycoBank (Crous et al., 2004).
2.4 Phenotypic and microscopic studies
Depending on the fungal morphological group, culture characteristics and microscopic fungal structures were examined and recorded from cultures grown on PDA, oat meal agar (OA), malt extract agar (MEA), and yeast malt agar (YMA) at room temperature. Sporulation and fruiting body production were induced under a combination of near-UV and cool-white fluorescent lights under 12-h light/12-h dark conditions. The structure and dimensions of microscopic features were determined and measured in 100% lactic acid or distilled water using an Olympus DP72 camera on an Olympus BX51 microscope and a Cell Sense Entry measurement module. To compute the dimensions of each fungal structure, mean, standard deviation, and 95% confidence intervals were estimated based on at least 30 microscopic measurements. Dimensions are presented as the range of measurements with extremes in brackets followed by mean ± standard deviation.
2.5 Pathogenicity tests
Pathogenicity tests of recognized species were performed on leaves and stems of 2-year-old oak (Q. brantii) seedlings in Petri plates and under greenhouse conditions, respectively. The surface of leaves and stems was disinfested with 70% ethanol. Small pieces of bark (≈ 3 × 3 mm) were cut from the stems of potted seedlings and small scratches (≈3 mm) were made on the leaves. Mycelial plugs of 7-d-old colonies on PDA were inoculated on wounded leaves and stems. Controls were inoculated with sterile PDA plugs. To evaluate the phytotoxic activity of fungal isolates, inoculated leaves were incubated at 25°C for a period of 7 days. The inoculated seedling wounds were wrapped with Parafilm and placed under greenhouse conditions (22–28°C) and watered as needed. As the seedlings declined, the external symptoms were recorded and the extent of vascular discoloration (lesion length) was measured. To confirm Koch’s postulates, fungal isolates were re-isolated from inoculated leaves and stems on PDA at 25°C and morphologically re-examined. The greenhouse trials were performed as a completely randomized nested design (CRND) with three replications per treatment. To determine differences in lesion lengths caused by inoculated fungi, one-way ANOVA was executed. Homogeneity of variance and normality assumption were examined using Bartlett and Shapiro–Wilk’s tests, respectively. Least significant difference (LSD) values were calculated (p = 0.05) using SAS v. 9.1.3.
3 Results
3.1 Disease symptoms, fungal isolates, and species identification
A total of 462 fungal isolates were collected from oak trees (Q. brantii, Q. infectoria, and Q. libani), of which 184 isolates (40%) were accommodated in coelomycetous Dothideomycetes. Various external and internal symptoms (Figures 1, 2) were observed on trees’ and twigs’ cross-sections listed in Table 1. Primarily, borer feeding sites (e.g., Buprestidae) were observed in association with irregular and central wood necrosis and no correlation was found between fungal species and type of disease symptoms.

Figure 1 External symptoms on oak trees. (A) Defoliation, (B, C) dieback, and (D–F) cankers and discoloration on twigs and branches.
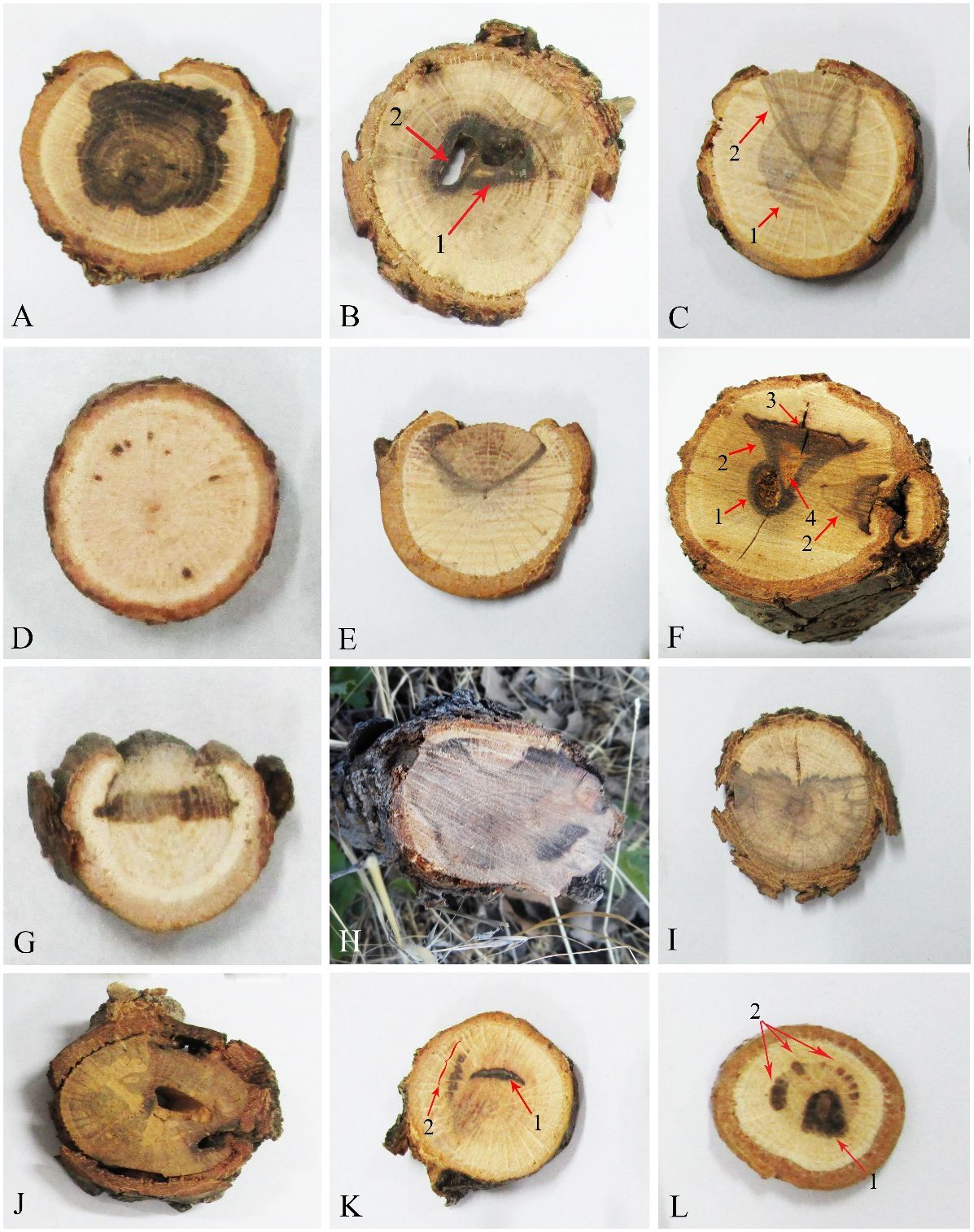
Figure 2 Internal wood symptoms in cross-sections of trunks and branches of oak trees. (A, H, I) Extended irregular necrosis. (B) Co-occurrence of central necrosis (1) and borer holes (2). (C) Co-occurrence of central necrosis (1) and wedge-shaped necrosis. (D) Black spots. (E) Wedge-shaped necrosis. (F) Co-occurrence of borer hole (1), wedge-shaped necrosis (2), black wood streaking (3), and wood decay (4). (G) Irregular necrosis. (J) Discolored wood areas surrounding borer holes. (K) Co-occurrence of arch-shaped necrosis (1) and black spots (2). (L) Co-occurrence of central necrosis (1) and black spots (2).
According to DNA profiles generated with M13 primer (data not shown), 36 isolates as representatives of recognized DNA banding patterns were selected for morphological and phylogenetic analyses.
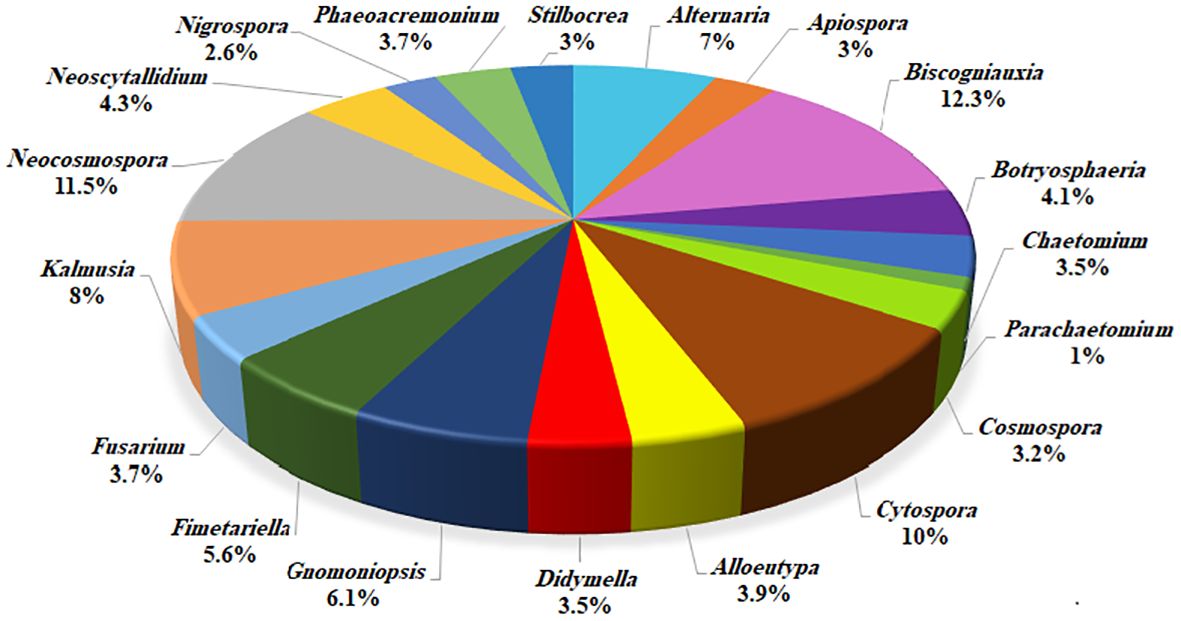
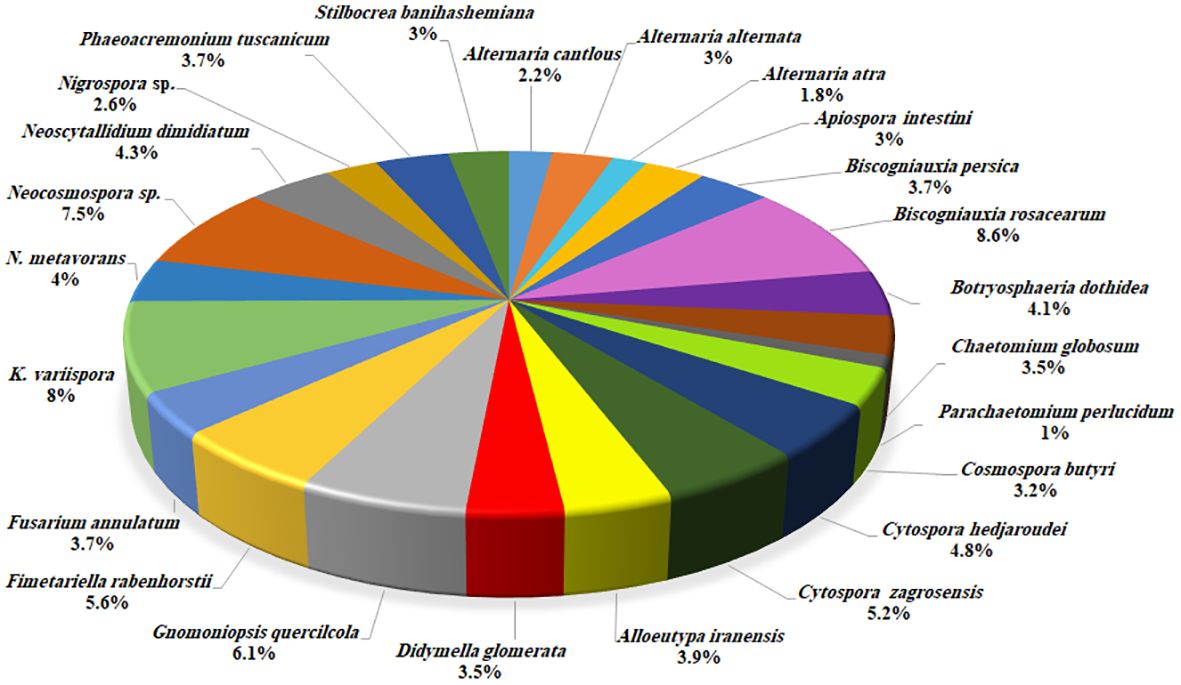
Based on morphology and DNA sequence data (LSU, ITS, rpb1, rpb2, tef-1α, tub2, acl1, act1, and caM), 24 fungal species corresponding to 19 genera were recognized (Figures 3, 4). Of these, eight species with 184 isolates (40%) were placed in coelomycetous Dothideomycetes, namely, four known species—Botryosphaeria dothidea (19 isolates/4.1%), Didymella glomerata (16 isolates/3.5%), Kalmusia variispora (37 isolates/8.1%), and Neoscytalidium dimidiatum (20 isolates/4.3%)—and four new species that are described and named Alloeutypa iranensis sp. nov. (18 isolates/3.9%), Cytospora hedjaroudei sp. nov. (22 isolates/4.8%), Cytospora zagrosensis sp. nov. (24 isolates/5.2%), and Gnomoniopsis quercicola sp. nov. (28 isolates/6.1%) (Table 1, Figure 4). Biscogniauxia, Neocosmospora, and Cytospora were the most prevalent fungi with frequencies of 12.3%, 11.5%, and 10%, respectively (Figure 3). Among the coelomycetous fungi, Cytospora and Kalmusia were the most common fungi associated with the decline of oak trees followed by Gnomoniopsis, Neoscytalidium, Botryosphaeria, Alloeutypa, and Didymella (Figure 3). Moreover, Biscogniauxia rosacearum, K. variispora, and Neocosmospora sp. were the most prevalent fungi at the species level followed by G. quercicola, Fimetariella rabenhorstii, C. zagrosensis, and C. hedjaroudei (Figure 4).
3.2 Phylogenetic analyses
Megablast search of the GenBank nucleotide database with ITS sequences revealed that our isolates are close to the members of Diatrypaceae (Eutypa/Eutypella), Cytosporaceae (Cytospora), and Gnomoniaceae (Discula/Gnomoniopsis). Thus, based on blast searches, preliminary phylogenetic analyses, literature, and fungal databases (indexfun-gorum/speciesfungorum/mycobank), three datasets were provided and analyzed.
The first DNA sequence dataset (ITS: 71, tub2: 47 sequences) consisted of our selected isolate IRAN 4323C, 68 isolates belonging to 66 species of the Diatrypaceae family, and two outgroups Xylaria hypoxylon CBS 122620/Kretzschmaria deusta CBS 826.72. The aligned datasets of ITS (691) and tub2 (454) were combined and subjected to MP, ML, and BI. After alignment, the combined dataset consisted of 1,145 characters including alignment gaps. Of these, 490 were constant, 192 were variable and parsimony-uninformative, and 463 were parsimony-informative. MP analysis of the remaining 463 parsimony-informative characters resulted in 729 most parsimonious trees (TL = 2,925, CI = 0.41, RI = 0.6, HI = 0.59). MrModelTest revealed that the general time-reversible model of evolution (Rodríguez et al., 1990), including estimation of invariable sites and assuming a discrete gamma distribution (GTR+I+G) with six rate categories (lsetnst = 6, rates = invgamma) and dirichlet (1,1,1,1) base frequencies, is the best nucleotide substitution model for both loci (ITS, tub2). The Bayesian analyses of the concatenated alignments of two loci generated 3,682 trees from which 920 trees were discarded as burn-in. The consensus tree and posterior probability values (PP) were calculated from the remaining 2,762 trees. The average standard deviation of split frequencies was 0.009962 at the end of the run. The RAxML search of the dataset with 760 distinct alignment patterns produced a best-scoring ML tree (lnL = −14,040.126555). The ML and Bayesian phylogenetic trees were mapped on the MP tree shown in Figure 5 with MP/ML/BI bootstrap support and posterior probability values at the nodes. In these analyses, our isolate IRAN 4323C was placed in a distinct clade in Alloeutypa, a new genus recently introduced in Diatrypaceae, which is recognized here as a new species named Alloeutypa iranensis sp. nov. close to A. flavovirens CBS 272.87 and isolate MFLU 19-0911 (Figure 5). A. iranensis showed remarkable differences in nucleotide sequences with A. flavovirens CBS 272.87 (ITS: 7 substitutions; tub2: 7 substitutions, 21 deletions/insertions) and MFLU 19-0911 (ITS: 5 substitutions; tub2: 6 substitutions, 25 deletions/insertions). It is also significantly distinct from the type species A. milinensis FSATAS 4309 based on ITS (12 substitutions, 1 deletion/insertion) and tub2 (16 substitutions, 25 deletions/insertions) sequence data. Isolate MFLU 19-0911 is placed in a distinct clade and is apparently a representative of a new Alloeutypa species.

Figure 5 One of the 729 most parsimonious trees of Diatrypaceae family obtained from analyses of combined ITS and tub2 sequence data. MP/ML/BI bootstrap support values and posterior probabilities are shown at the nodes. The phylogenetic tree was rooted with Kretzschmaria deusta CBS 826.72/Xylaria hypoxylon CBS 122620. The novel taxon is in boldface. T Ex-type.
The second dataset consisting of DNA sequences of six loci (LSU, ITS, rpb2, act1, tef-1α, and tub2) was analyzed to examine the taxonomic position of our Cytospora isolates. New generated sequences were aligned with available authentic sequences of Cytospora species and Diaporthe vaccinii CBS 160.32 as an outgroup (LSU: 43, ITS: 47, rpb2: 41, act1: 44, tef-1α: 39, tub2: 29 sequences). The concatenated alignment of LSU (803), ITS (593), rpb2 (664), act1 (311), tef-1α (420), and tub2 (503) was subjected to MP, ML, and BI. After alignment, the dataset consisted of 3,295 characters including alignment gaps. Of these, 2,283 were constant, 222 were variable and parsimony-uninformative, and 790 were parsimony-informative. MP analysis of the remaining 790 parsimony-informative characters resulted in 162 most parsimonious trees (TL = 3,345, CI = 0.48, RI = 0.67, HI = 0.52). MrModelTest revealed that the general time-reversible model of evolution (Rodríguez et al., 1990), including estimation of invariable sites and assuming a discrete gamma distribution (GTR+I+G) with six rate categories (lsetnst = 6, rates = invgamma) and dirichlet (1,1,1,1) base frequencies, is the best nucleotide substitution model for all loci (LSU, ITS, rpb2, act1, tef-1α, and tub2). The Bayesian analyses of the concatenated alignments of six loci generated 1,172 trees from which 292 trees were discarded as burn-in. The consensus tree and posterior probability values (PP) were calculated from the remaining 880 trees. The average standard deviation of split frequencies was 0.009890 at the end of the run. The RAxML search of the dataset with 1,272 distinct alignment patterns produced a best-scoring ML tree (lnL = −20,279.905622). The MP and ML trees were mapped on the Bayesian phylogenetic tree as shown in Figure 6 with MP/ML/BI bootstrap support and posterior probability values at the nodes.
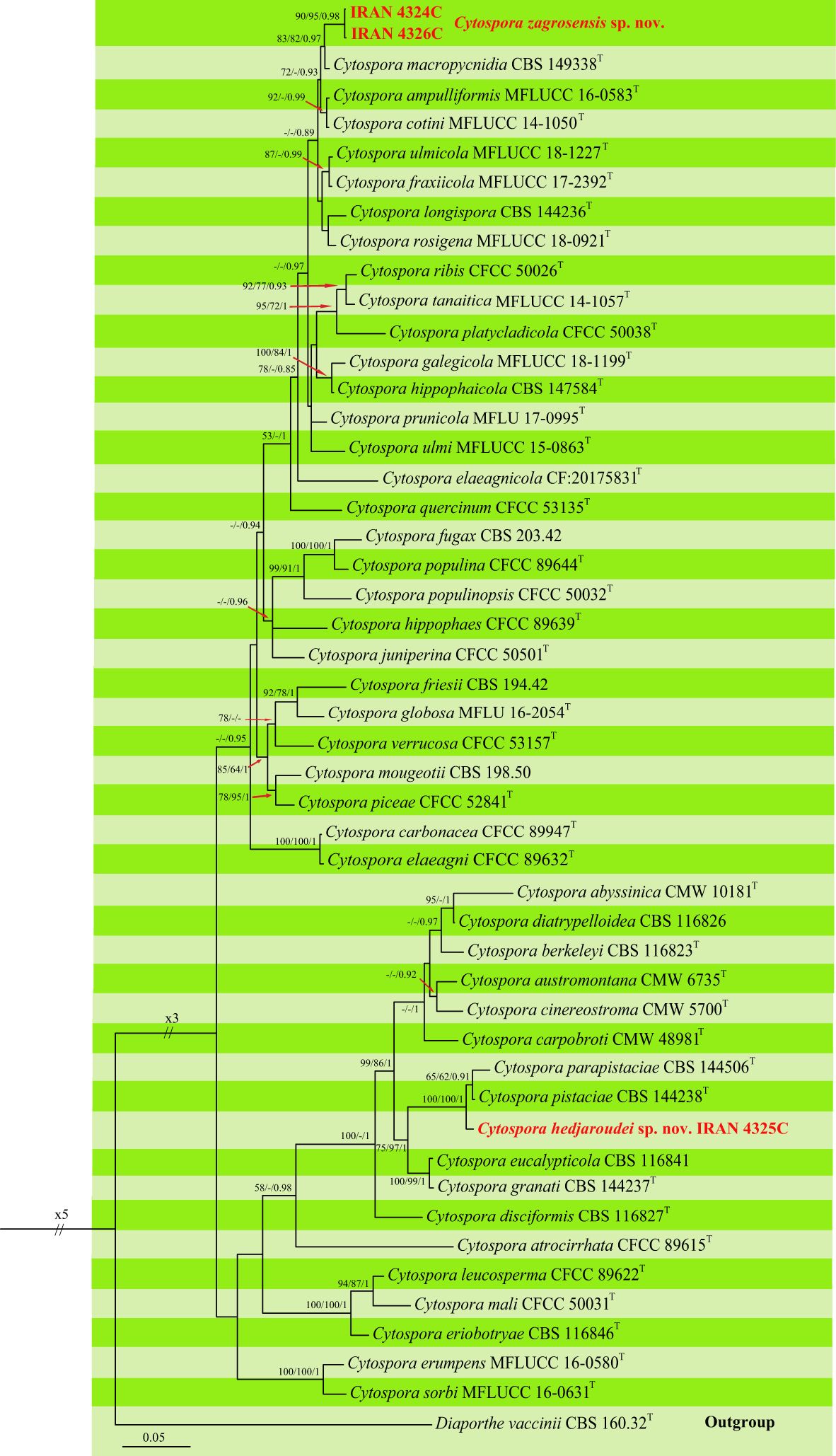
Figure 6 Phylogenetic tree of Cytospora spp. inferred from Bayesian analysis of combined LSU, ITS, rpb2, ef1-a, act1, and tub2 sequence data. MP/ML/BI bootstrap support values and posterior probabilities are shown at the nodes. The phylogenetic tree was rooted with Diaporthe vaccinii CBS 160.32. The new species are in boldface. T Ex-type.
Our isolates were placed in two distinct clades differentiated from all other Cytospora species that are recognized here as two new species. Two isolates, IRAN 4324C and IRAN 4326C, formed a well-supported clade as a sister group with C. macropycnidia CBS 149338 and named Cytospora zagrosensis sp. nov. This species is differentiated from C. macropycnidia in ITS (2 substitutions), rpb2 (2 substitutions), tef-1α (6 substitutions, 2 deletions/insertions), and tub2 (11 substitutions, 3 deletions/insertions) nucleotide sequences. The third isolate IRAN 4325C represents the second new species close to C. pistaciae CBS 144238 and C. parapistaciae CBS 144506, which is named Cytospora hedjaroudei sp. nov. (Figure 6). Based on nucleotide sequences, C. hedjaroudei differs from two closely related species C. pistaciae (ITS: 5 substitutions, 1 deletion/insertion; tef-1α 1 substitution) and C. parapistaciae (ITS: 8 substitutions, 28 deletions/insertions; tef-1α: 4 substitutions, 1 deletion/insertion). It is noticeable that ITS sequence data are enough to separate C. hedjaroudei from both C. pistaciae and C. parapistaciae.
The third dataset contained three loci DNA sequences (ITS: 31, tef-1α: 23, and tub2: 26 sequences) of our selected isolate IRAN 4313C, 29 Gnomoniopsis species, and Melanconis stilbostoma CBS 109778 as an outgroup. The aligned single gene sequences were concatenated and subjected to MP, ML, and BI. The combined dataset consisted of 1,918 characters (ITS: 606, tef-1α: 370, and tub2: 942) including alignment gaps. Of these, 1,099 were constant, 278 were variable and parsimony-uninformative, and 541 were parsimony-informative. MP analysis of the remaining 541 parsimony-informative characters resulted in three most parsimonious trees (TL = 2,367, CI = 0.55, RI = 0.6, HI = 0.45). MrModelTest revealed that the general time-reversible model of evolution (Rodríguez et al., 1990), including estimation of invariable sites and assuming a discrete gamma distribution (GTR+I+G) with six rate categories (lsetnst = 6, rates = invgamma) and dirichlet (1,1,1,1) base frequencies, is the best nucleotide substitution model for all loci (ITS, tef-1α, and tub2). The Bayesian analyses of the concatenated alignments of three loci generated 4,892 trees from which 1,222 trees were discarded as burn-in. The consensus tree and posterior probability values (PP) were calculated from the remaining 3,670 trees. The average standard deviation of split frequencies was 0.009837 at the end of the run. The RAxML search of the dataset with 976 distinct alignment patterns produced a best-scoring ML tree (lnL = −13,465.491131). The ML and Bayesian phylogenetic trees were mapped on the MP tree shown in Figure 7. Our isolate IRAN 4313C was clustered in a strongly supported clade close to G. paraclavulata CBS 123202 with significant nucleotide differences in all three loci ITS (5 substitutions, 2 deletions/insertions), tef-1α (35 substitutions, 8 deletions/insertions), and tub2 (58 substitutions, 9 deletions/insertions). Thus, it is recognized as a new species and named Gnomoniopsis quercicola sp. nov. (Figure 7).
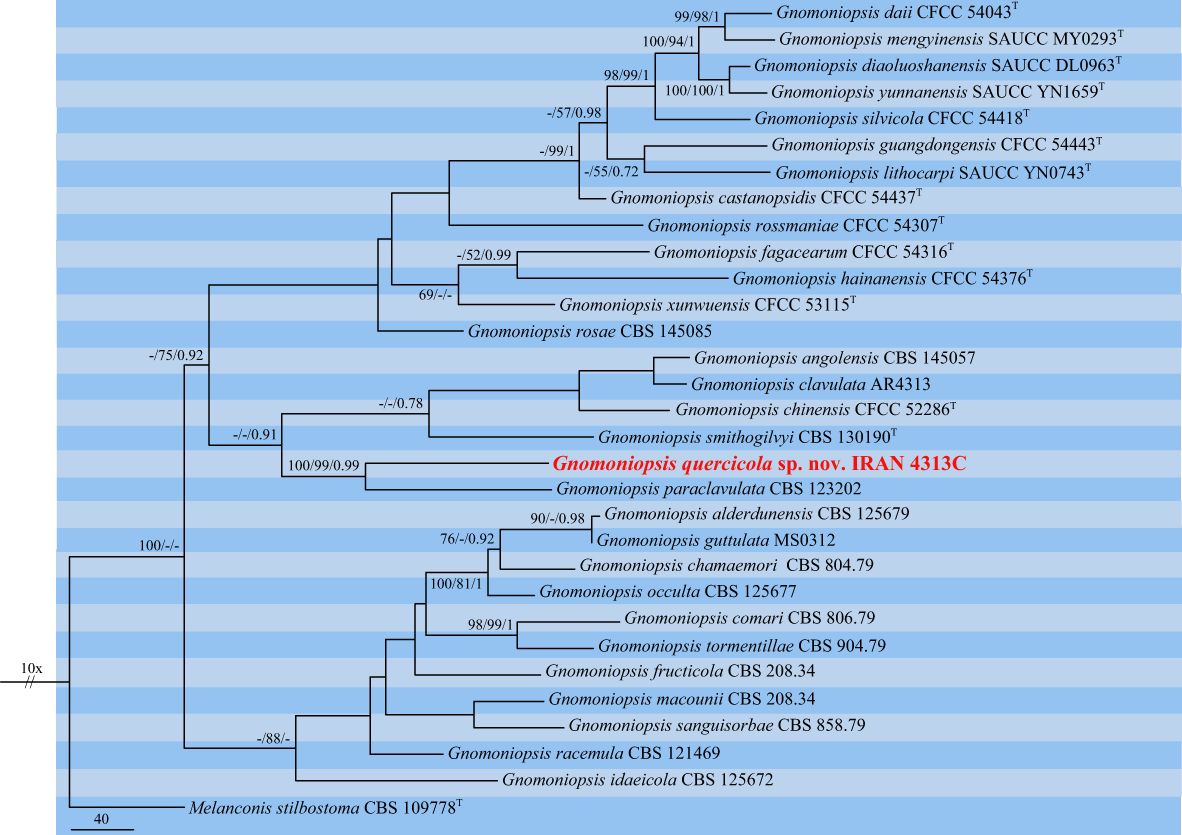
Figure 7 One of the three most parsimonious trees of Gnomoniopsis spp. obtained from combined ITS, tef-1α, and tub2 sequence data. MP/ML/BI bootstrap support values and posterior probabilities are shown at the nodes. The phylogenetic tree was rooted with Melanconis stilbostoma CBS 109778. The novel species is in boldface. T Ex-type.
3.3 Taxonomy
Based on phylogenetic analyses and morphology, eight coelomycetous fungal species were characterized and associated with declined oak trees in the central and northern part of Zagros forests in Iran. Of these, four species were recognized as new fungal species for science, which are described here as follows:
Alloeutypa iranensis S. Bashiri & Abdollahz., sp. nov. (Figure 8)
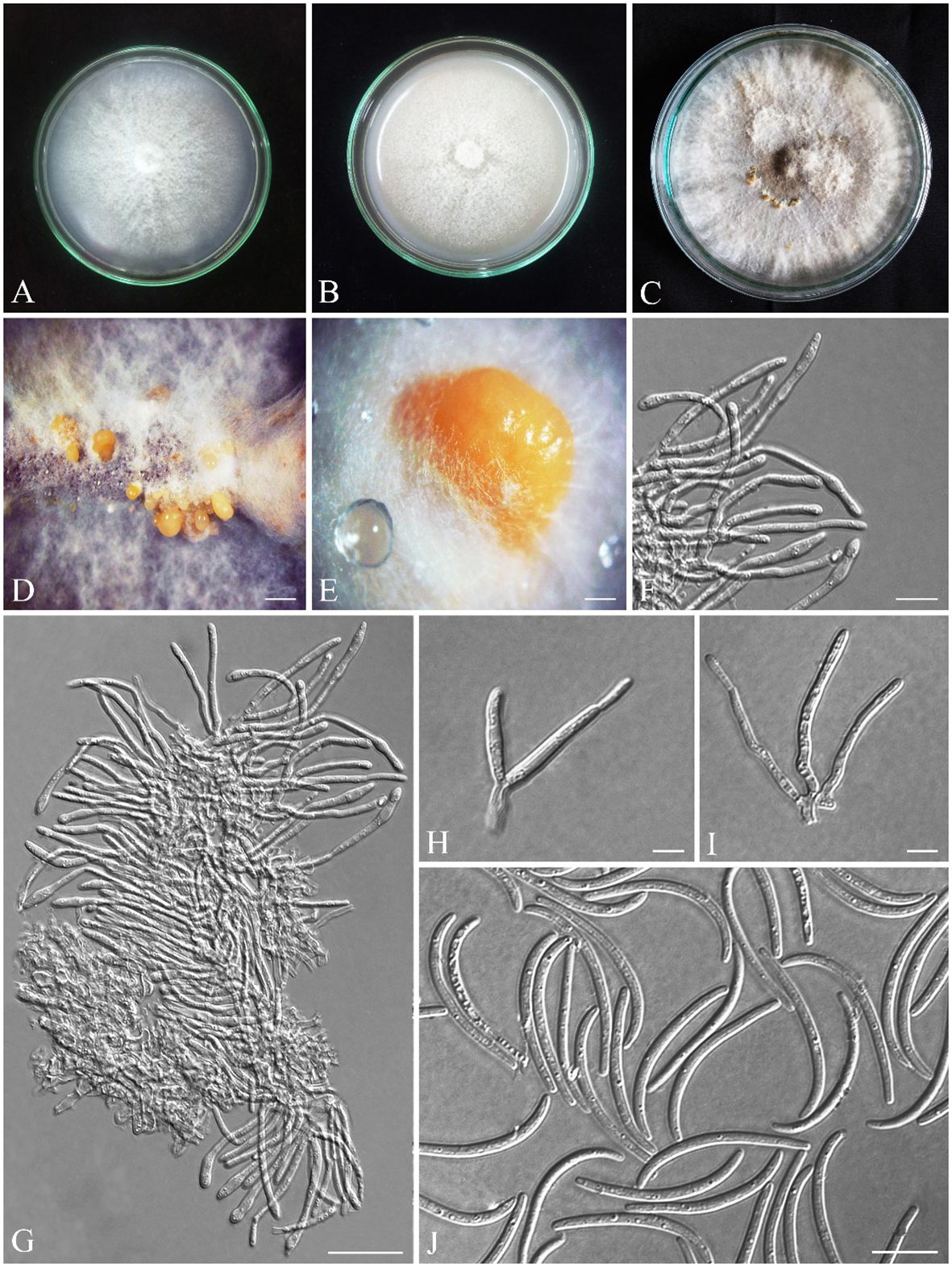
Figure 8 Seven-day-old colonies of Alloeutypa iranensis (IRAN 18255F, Holotype). (A) On PDA. (B) On OA. (C) Three-month-old colonies on MEA at room temperature. (D, E) Conidiomata. (F–I) Conidiophores and conidiogenous cells. (J) Conidia. Scale bars: (D) 900 μm; (E) 400 μm; (F, H, I) 5 μm; (G, J) 10 μm.
MycoBank: MB850401.
Diagnosis: A. iranensis is the third species to be described in Alloeutypa. Since the type species A. milinensis was described based on the sexual morph and we have only observed the asexual morph, it is only possible to morphologically distinguish A. iranensis from A. flavovirens based on the larger dimensions of conidiomata (0.9–3.9 mm vs. 0.7–1.3 mm), conidiophores (12.6 × 3 μm vs. 4.9 × 2.4 μm), and conidiogenous cells (32.4 × 2.6 μm vs. 11.5× 2.4 μm) and shorter conidia (28.5 μm vs. 34 μm).
3.3.1 Etymology
iranensis refers to the country where the fungus was first found.
3.3.2 Type
IRAN, Lorestan Province, Khorramabad, from Q. brantii branches, 33°18′50.2″ N/48°43′12.0″ E, 27 September 2016, S. Bashiri, IRAN 18255F (Holotype) (GenBank ITS: OR540613; tub2: OR591157), ex-type IRAN 4323C = CBS 149771.
3.3.3 Description
3.3.4 Sexual morph
not observed.
3.3.5 Asexual morph
Pycnidia produced on MEA, superficial, solitary or aggregated, sub-conical, globose to sub-globose, shiny, with smooth surface, cream to pale yellow (Figures 8D, E). Peridium thick, comprising dark cream, thick-walled, cells of textura angularis. Conidiophores branched, arising from pseudoparenchymatous cells or interwoven hyphae, (5.6–) 11–16 (–17.1) × (2–) 2.4–4 (–5.4) μm (av. ± SD = 12.6 ± 0.5 × 3 ± 0.1 µm). Conidiogenous cells cylindrical, in dense palisades, straight or curved, apex mostly inflated, (18–) 24–43 (–51.7) × (1.8–) 2–3.5 (–3.9) μm (av. ± SD = 32.4 ± 1.3 × 2.6 ± 0.1 µm). Conidia filiform, curved or rarely straight, with blunt apex and flattened base, hyaline, (23.3–) 25–30 (–33.3) × (1.6–) 1.7–2 (–2.5) μm (av. ± SD = 28.5 ± 0.3 × 2 ± 0.02 µm) (Figures 8F–J).
3.3.6 Culture characteristics
Colonies on PDA cottony, dense, white, margin smooth, reverse yellowish-white, reaching 75 mm diameter after 7 days at room temperature in the dark; on MEA flat, white and becoming grayish white with age, margin smooth, reverse grayish, reaching 60 mm diameter after 7 days at room temperature in the dark; on OA cottony, white and becoming grayish white with age, margin smooth, reverse grayish, reaching 90 mm diameter after 7 days at room temperature in the dark (Figures 8A–C). Mycelial clumps produced on MEA, erect, white, and floccose on culture.
3.3.7 Additional specimens examined
Iran, Kurdistan Province, Baneh, from Q. libani branches, 22 August 2016, S. Bashiri, IRAN 4837C = CBS 149772 (36°05′01.8″ N/45°40′32.8″ E), CJASB222/CJASB223 (36°03′46.7″ N/45°38′32.3″ E); Iran, Kurdistan Province, Sarvabad, from Q. libani branches, 15 August 2016, S. Bashiri, CJASB225 (35°22′95.7″ N/46°12′89.5″ E), CJASB226 (35°09′87.1″ N/46°32′42.4″ E); Iran, Kurdistan Province, Marivan, from Q. libani branches, 15 August 2016, S. Bashiri, IRAN 4895C (35°24′78.4″ N/46°10′73.4″ E), CJASB228 (35°28′83.4″ N/46°06′22.8″ E); Iran, Kermanshah Province, Eslamabad-e-Gharb, from Q. brantii branches 11 October 2016, S. Bashiri, IRAN 4897 (33°58′68.9″ N/46°29′92.0″ E); Iran, Kermanshah Prov-ince, Paveh, from Q. brantii branches, 21 October 2016, S. Bashiri, CJASB230/CJASB231(34°57′06.9″ N/46°27′80.5″ E); Iran, Lorestan Province, Chegini, from Q. brantii branches, 27 September 2016, S. Bashiri, CJASB232 (35°32′49.7″ N/48°05′23.8″ E), IRAN 4896C/CJASB235 (35°33′76.3″ N/46°06′18.8″ E); Iran, Lorestan Province, Khorramabad, from Q. brantii branches, 25 September 2016, S. Bashiri, CJASB234 (33°39′40.4″ N/46°16′85.5″ E); Iran, Lorestan Province, Kuhdasht, from Q. brantii branches, 27 Sep-tember 2016, S. Bashiri, CJASB236 (33°31′28.4″ N/47°47′24.6″ E); Iran, Lorestan Province, Poldokhtar, from Q. brantii branches, 27 September 2016, S. Bashiri, CJASB237/CJASB238 (33°18′12.2″ N/47°48′95.0″ E).
Cytospora hedjaroudei S. Bashiri & Abdollahz., sp. nov. (Figure 9).
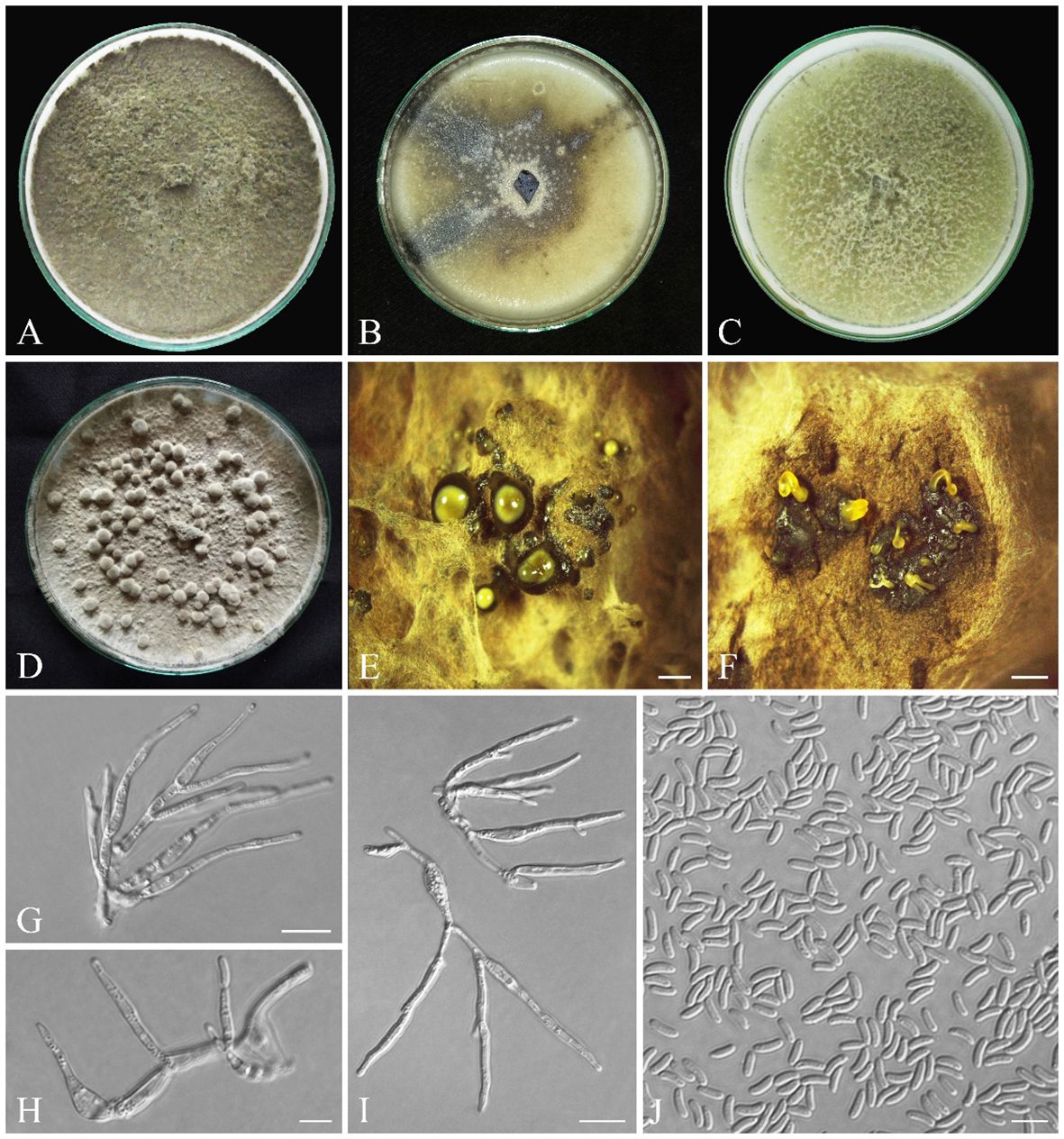
Figure 9 Seven-day-old colonies of Cytospora hedjaroudei (IRAN 18257F, Holotype). (A) On PDA. (B) On OA. (C) On MEA. (D) Fourteen-day-old colonies on PDA at room temperature. (E) Conidiomata. (F) Cross-section of conidiomata. (G–I) Conidiogenous cells. (J) Conidia. Scale bars: (E, F) 900 μm; (G, I) 10 μm; (H, J) 5 μm.
MycoBank: MB850403.
3.3.8 Diagnosis
C. hedjaroudei is phylogenetically close to C. pistaciae and C. parapistaciae but is morphologically differentiated by having distinct conidiophores that are absent and reduced to conidiogenous cells in both C. pistaciae and C. parapistaciae. Conidiomata with multiple internal locules are seen in C. hedjaroudei, while in both C. pistaciae and C. parapistaciae, conidiomata have a single internal locule. It is also differentiated from C. pistaciae by having significantly larger conidiogenous cells (12–25 × 2–5 µm vs. 7.1–8.9 × 1.1–1.5 µm) and slightly larger conidia (3.2–) 4–5 (–9.1) × (1–) 1.3–1.7 (–1.9) µm vs. (3.5–) 4–4.8 (–5.5) × (1.0–) 1.1–1.3 (–1.5) µm and from C. parapistaciae by having significantly larger conidiomata (800–2,800 µm vs. 335–590 µm), conidiogenous cells (12–25 × 2–5 µm vs. 7.6–9.6 × 1.2–1.6) and conidia (4–5 × 1.3–1.7 (av. = 4.5 × 1.5) µm vs. 3.5–4.3 × 0.9–1.1 µm).
3.3.9 Etymology
Named after Dr. Ghorbanali Hedjaroude, emeritus professor of Tehran University, who significantly contributed to the knowledge of mycology in Iran.
3.3.10 Type
IRAN, West Azarbaijan Province, Sardasht, from Q. infectoria branches, 36°18′15.8″ N/45°28′36.6″ E, 7 August 2016, S. Bashiri, IRAN 18257F (Holotype) (GenBank ITS: OR540617; tef-1α: OR561995; tub2: OR561904), ex-type IRAN 4325C = CBS 149769.
3.3.11 Description
3.3.12 Sexual morph
not observed.
3.3.13 Asexual morph
Conidiomata on PDA pycnidial, globose, black-gray, covered by smoke gray mycelium, scattered, aggregated or solitary, with multiple internal locules, erumpent at maturity, exuding pale yellow or black conidial masses, 0.8–2.8 mm diameter (Figures 9D–F). Conidiophores hyaline, filamentous, branched or unbranched, thin walled, (4.9–) 7–22 (–25.5) µm (av. 14.2 ± 1.1 µm). Conidiogenous cells hyaline, smooth, cylindrical to subcylindrical, tapering to the apices, enteroblastic, phialidic with periclinal thickening, (8.5–) 12–25 (–33.8) × (1.5–) 2–5 (–2.8) µm (av. ± SD = 17.8 ± 1.3 × 2.2 ± 0.1 µm). Conidia hyaline, thin walled, smooth, eguttulate, allantoid, aseptate, (3.2–) 4–5 (–9.1) × (1–) 1.3–1.7 (–1.9) µm (av. ± SD = 4.5 ± 0.2 × 1.5 ± 0.1 µm) (Figures 9G–J).
3.3.14 Culture characteristics
Colonies on PDA fast-growing, cottony, with dense aerial mycelium, smoke gray (21′′′′f) to olivaceous gray (21′′′′′i), margin smooth, reverse smoke gray (21′′′′f), reaching 90 mm diameter after 7 days at room temperature in the dark; on MEA cottony, with aerial mycelium, olivaceous buff (21′′′d) to greenish olivaceous (23′′′i), margin smooth, reverse olivaceous buff (21′′′d), reaching 90 mm diameter after 7 days at room temperature in the dark; on OA appressed, olivaceous buff (21′′′d) at the margin to greenish olivaceous (23′′′i), with some greenish olivaceous (23′′′i) to dull green (27′′m) sectors, reverse olivaceous buff (21′′′d), reaching 90 mm diameter after 7 days at room temperature in the dark (Figures 9A–C).
3.3.15 Additional specimens examined
Iran, Kurdistan Province, Marivan, from Q. libani branches, 16 August 2016, S. Bashiri, IRAN 4842C = CJASB196 (35°35′57.4″ N/46°06′52.4″ E); Iran, Kurdistan Province, Marivan, from Q. brantii branches, 16 August 2016, S. Bashiri, CJASB197 (35°09′53.7″ N/46°30′69.9″ E); Iran, Kurdistan Province, Sarvabad, from Q. brantii branches, 16 August 2016, S. Bashiri, CJASB194 (35°09′87.1″ N/46°32′42.4″ E), CJASB195 (35°22′95.7″ N/46°12′89.4″ E); Iran, Kurdistan Province, Baneh, from Q. libani branches, 21 August 2016, S. Bashiri, CJASB190 (35°44′91.5″ N/45°49′74.0″ E), CJASB191 (22 August 2016, 36°08′19.9″ N/45°42′31.8″ E), CJASB193 (23 August 2016, 35°57′18.3″ N/46°00′58.5″ E); Iran, Kurdistan Province, Baneh, from Q. brantii branches, 23 August 2016, S. Bashiri, IRAN 4903C (35°50′76.5″ N/45°56′67.1″ E); Iran, West Azarbaijan Province, Sardasht, from Q. infectoria branches, 8 August 2016, S. Bashiri, CJASB184 (36°10′36.0″ N/45°30′15.3″ E); Iran, West Azarbaijan Province, Sardasht, from Q. libani branches, 8 August 2016, S. Bashiri, IRAN 4904C (36°07′23.4″ N/45°28′08.5″ E); Iran, Ilam Province, Eyvan, from Q. brantii branches, 15 September 2016, S. Bashiri, CJASB186/CJASB187 (33°50′84.8″ N/46°12′21.0″ E); Iran, Ilam Province, Sarableh, from Q. brantii branches, 15 September 2016, S. Bashiri, IRAN 4902C/CJASB189 (33°47′40.9″ N/46°30′93.1″ E); Iran, Kermanshah Province, Gilan-e-Gharb, from Q. brantii branches, 9 October 2016, S. Bashiri, CJASB199 (34°54′36.6″ N/46°32′65.6″ E), CJASB200 (15 September 2016, 34°05′02.3″ N/46°02′15.6″ E); Iran, Kermanshah Province, Paveh, from Q. brantii branches, 21 October 2016, S. Bashiri, CJASB201 (34°57′06.9″ N/46°27′80.5″ E); Iran, Kermanshah Province, Ravansar, from Q. brantii branches, 24 September 2016, S. Bashiri, CJASB198 (34°03′12.5″ N/46°27′19.3″ E); Iran, Lorestan Province, Chegini, from Q. brantii branches, 24 September 2016, S. Bashiri, CJASB202/CJASB203 (35°33′76.3″ N/46°06′18.8″ E); Iran, Lorestan Province, Poldokhtar, from Q. brantii branches, 27 September 2016, S. Bashiri, CJASB204 (33°18′12.2″ N/47°48′95.0″ E).
Cytospora zagrosensis S. Bashiri & Abdollahz., sp. nov. (Figure 10).
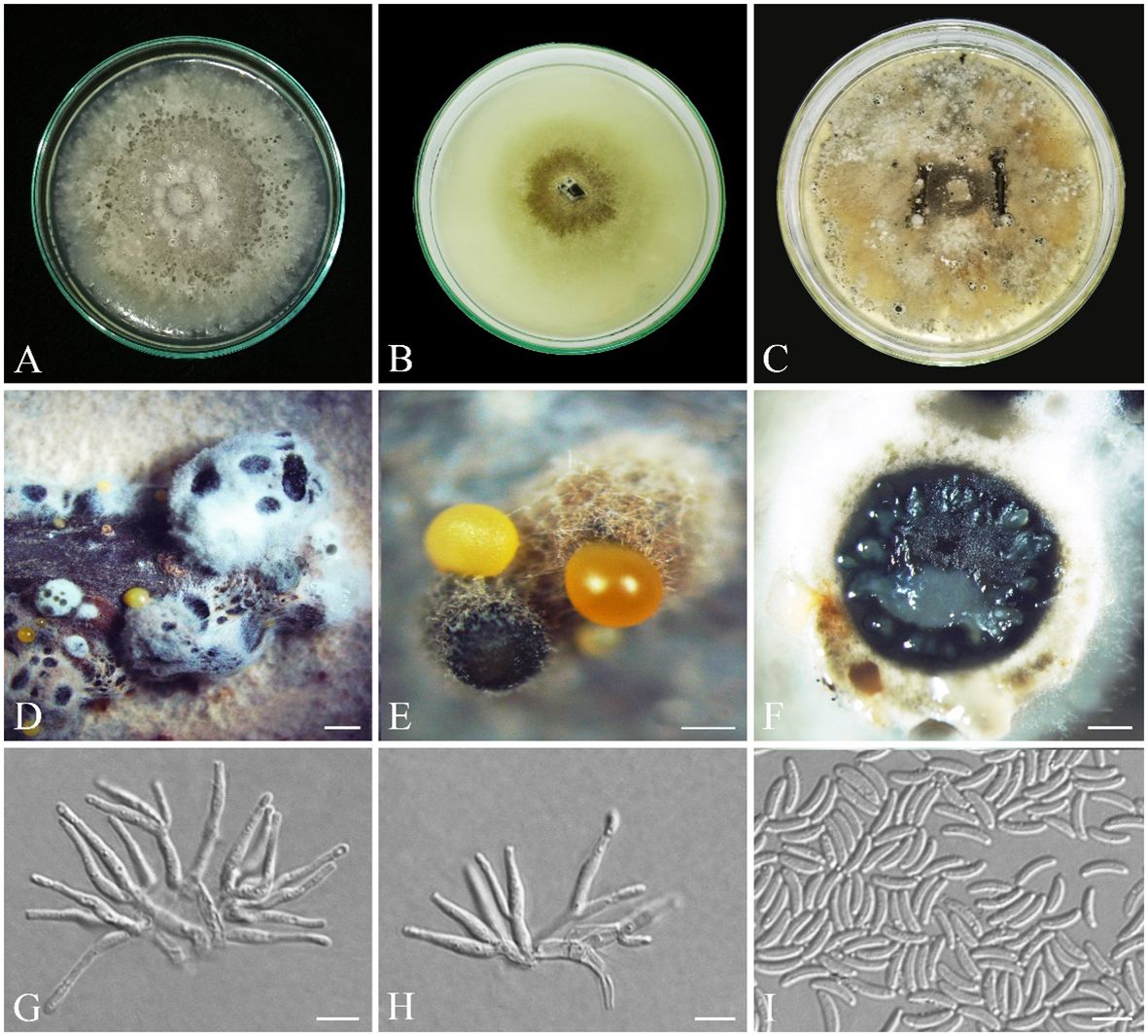
Figure 10 Seven-day-old colonies of Cytospora zagrosensis (IRAN 18256F, Holotype). (A) On PDA. (B) On OA. (C) Fourteen-day-old colonies on MEA at room temperature. (D, E) Conidiomata. (F) Cross-section of conidiomata. (G, H). Conidiogenous cells. (I) Conidia. Scale bars: (D) 100 μm; (E) 600 μm; (F) 400 μm; (G–I) 5 μm.
MycoBank: MB850404.
3.3.16 Diagnosis
Cytospora zagrosensis differs from closely related species C. macropycnidia by having smaller conidiomata (1.1–3.6 mm vs. 2.3–4.3 mm) and conidiogenous cells (av. = 12.8 × 2 µm vs. 17 × 2.5 μm) and long-narrow conidia (av. = 6.4 × 1.7 µm vs. 6 × 2.1 µm).
3.3.17 Etymology
zagrosensis refers to the Zagros forests located at the western part of Iran, from which the fungus was first collected.
3.3.18 Type
IRAN, Kurdistan Province, Baneh, from Q. libani branches, 35°18′81.1″ N/46°21′89.2″ E, 22 August 2016, S. Bashiri, IRAN 18256F (Holotype) (GenBank LSU: OR540615; ITS: OR540618; rpb2: OR540303; tef-1α: OR5611993; tub2: OR561905; act1: OR550646), ex-type IRAN 4324C = CBS 149765.
3.3.19 Description
3.3.20 Sexual morph
not observed.
3.3.21 Asexual morph
Conidiomata on PDA pycnidial, brown to black, covered by buff to white mycelium, aggregated or solitary, with multiple internal locules, erumpent at maturity, exuding yellow or pale orange conidial masses, 1.1–3.6 mm diameter (Figures 10D–F). Conidiophores hyaline, thin walled, reduced to short discrete basal cells with whorls of two to three concentric conidiogenous cells on the tips. Conidiogenous cells hyaline, smooth, subcylindrical, tapering to the apices, enteroblastic, phialidic with periclinal thickening, (6.1–) 10–16 (–19.5) × (1.4–) 1.7–2.3 (–2.5) μm (av. ± SD = 12.8 ± 0.4 × 2 ± 0.1 µm). Conidia hyaline, thin walled, smooth, eguttulate, allantoid, aseptate, (5–) 5.5–7 (–8) × (1.3–) 1.4–1.9 (–2) μm (av. ± SD = 6.4 ± 0.1 × 1.7 ± 0.1 µm) (Figures 10G–I).
3.3.22 Culture characteristics
Colonies on PDA cottony, olivaceous buff (21′′′d) to smoke gray (21′′′′f), margin slightly irregular or lobate, reverse pale luteous (17d) to luteous (17i), reaching 80 mm diameter after 7 days at room temperature in the dark, on MEA cottony, olivaceous buff (21′′′d) to smoke gray (21′′′′f), reverse buff (21′′′d) to whitish, margin smooth, reaching 50 mm diameter after 7 days at room temperature in the dark; on OA appressed, olivaceous buff (21′′′d) to greenish olivaceous (23′′′i), margin smooth, reverse olivaceous buff (21′′′d), reaching 60 mm diameter after 7 days at room temperature in the dark (Figures 10A–C).
3.3.23 Additional specimens examined
Iran, Kermanshah Province, Paveh, from Q. brantii branches, 21 October 2016, S. Bashiri, IRAN 4326C = CBS 149767 (GenBank LSU: OR540616; ITS: OR540619; rpb2: OR540304; tef-1α: OR561994; tub2: OR561906; act1: OR550647) (34°57′06.9″ N/46°27′80.5″ E), CJASB165/CJASB166/CJASB177/CJASB178 (34°57′06.9″ N/46°27′80.5″ E); Iran, Kermanshah Province, Javanrud, from Q. brantii branches, 21 October 2016, S. Bashiri, CJASB167 (34°49′26.8″ N/46°30′15.1″ E); Iran, Kermanshah Province, Gilan-e-Gharb, from Q. brantii branches, 9 October 2016, S. Bashiri, IRAN 4838C = CJASB179/CJASB169 (34°54′36.6″ N/46°32′65.6″ E), CJASB168/CJASB180 (15 September 2016, 34°05′02.3″ N/46°02′15.6″ E); Iran, Kermanshah Province, Eslamabad-e-Gharb, from Q. brantii branches, 10 October 2016, S. Bashiri, CJASB163 (33°47′87.6″N/46°52′11.4″ E), CJASB164 (11 October 2016, 33°58′68.9″ N/46°29′92.0″ E); Iran, Kurdistan Province, Marivan, from Q. libani branches, 16 August 2016, S. Bashiri, IRAN 4839C = CJASB161 (35°24′97.6″ N/46°16′08.5″ E), CJASB176 (35°38′57.4″ N/46°06′52.4″ E); Iran, Kurdistan Province, Marivan, from Q. brantii branches, 15 August 2016, S. Bashiri, CJASB162 (35°24′78.4″ N/46°10′73.4″ E); Iran, Kurdistan Province, Marivan, from Q. infectoria branches, 28 July 2016, S. Bashiri, CJASB175 (35°28′83.4″ N/46°06′22.8″ E); Iran, Kurdistan Province, Sarvabad, from Q. libani branches, 15 August 2016, S. Bashiri, CJASB160 (35°22′90.1″ N/46°09′28.3″ E), CJASB174 (35°23′86.3″ N/46°12′49.4″ E); Iran, Lorestan Province, Chegini, from Q. brantii branches, 26 September 2016, S. Bashiri, CJASB170 (33°28′01.4″ N/48°00′43.3″ E), CJASB182 (27 September 2016, 33°32′49.7″ N/48°05′23.8″ E); Iran, Lorestan Province, Poldokhtar, from Q. brantii branches, 27 September 2016, S. Bashiri, IRAN 4901C (33°18′12.2″ N/47°48′95.0″ E), CJASB181 (33°14′33.1″ N/47°39′64.6″ E); Iran, Lorestan Province, Kuhdasht, from Q. brantii branches, 27 September 2016, S. Bashiri, CJASB172 (33°31′28.4″ N/47°47′.246″ E).
Gnomoniopsis quercicola S. Bashiri & Abdollahz., sp. nov. (Figure 11).
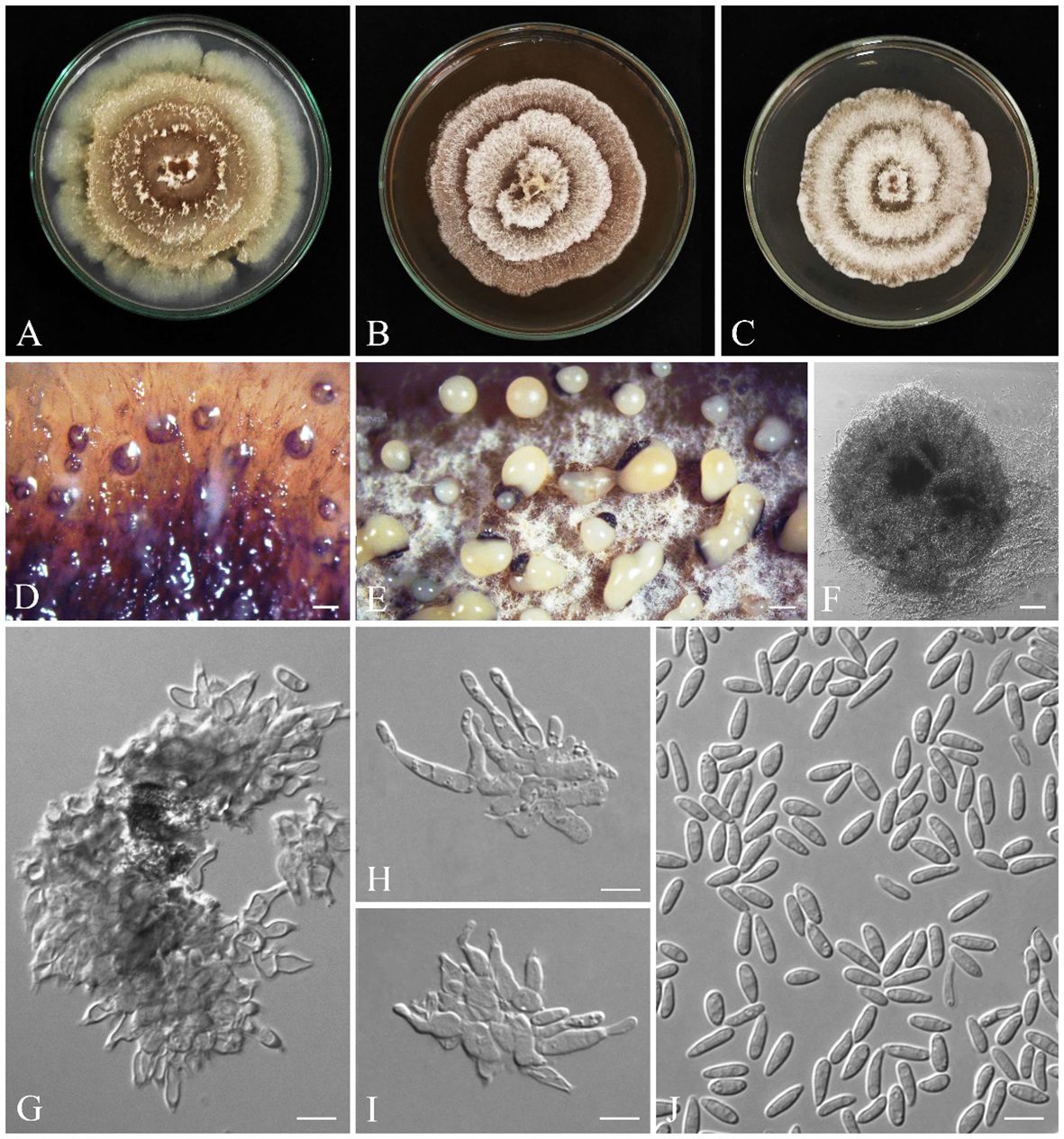
Figure 11 Seven-day-old colonies of Gnomoniopsis quercicola (IRAN 18252F, Holotype). (A) On PDA. (B) On MEA. (C) On YMA at room temperature. (D–F) Conidiomata. (G–I) Conidiogenous cells. (J) Conidia. Scale bars: (D, E) 200; (F) 25 μm; (G–J) 5 μm.
MycoBank: MB850402.
3.3.24 Diagnosis
Generally, it is difficult to discriminate all Gnomoniopsis species based on morphology. Gnomoniopsis quercicola clusters with G. paraclavulata in a distinct clade. It is discriminated from G. paraclavulata by having smaller conidia (6.2 ± 0.1 × 2.2 ± 0.04 µm vs. 7.5 ± 0.5 × 3 ± 0.3 μm) and a faster growing rate (on PDA: 90 mm/9 d vs. 90 mm/40 d; on MEA and MYA: 90/14 d vs. 90 mm/40 d).
3.3.25 Etymology
quercicola refers to Quercus, the host genus from which the fungus was first isolated.
3.3.26 Type
IRAN, Kurdistan Province, Baneh, from Q. brantii branches, 36°07′17.7″ N/45°41′37.0″ E, 22 August 2016, S. Bashiri, IRAN 18252F (Holotype) (GenBank ITS: OR540614; tef-1α: OR561996 tub2: OR561907), ex-type IRAN 4313C = CBS 149773.
3.3.27 Description
3.3.28 Sexual morph
not observed.
3.3.29 Asexual morph
Conidiomata on PDA aggregated or solitary, globose, without neck, semi-immersed, dark yellow to brown, erumpent at maturity, exuding pale yellow slimy conidial mass after 14 days at room temperature (Figures 11D–F). Conidiophores are indistinct, often reduced to conidiogenous cells. Conidiogenous cells cylindrical to lageniform, tapering toward apex, hyaline, phialidic with periclinal thickening, (4.3–) 5–12 (–13.9) × (1.6–) 2–3 (–4.5) µm (av. ± SD = 8.5 ± 0.4 × 2.5 ± 0.1 µm). Conidia oval to oblong, hyaline, smooth, often straight, rarely slightly curved, base subtruncate to bluntly rounded, (4.5–) 5–7 (–8.3) × (1.3–) 1.9–2.5 (–2.8) µm (av. ± SD = 6.2 ± 0.1 × 2.2 ± 0.04 µm) (Figures 11G–J).
3.3.30 Culture characteristics
Colonies on PDA cottony, with aerial mycelium, colony margin forming a concentric ring with sparse aerial mycelium, followed by additional rings, creating a lobed rosette-like appearance, buff (21′′′d) to olivaceous (19′′k) at the center, olivaceous buff (21′′′d) to greenish olivaceous (23′′′i), reaching 90 mm after 9 days at room temperature in the dark; on MEA cottony, with aerial mycelium, spreading out in concentric rings, creating a lobed rosette-like appearance, buff (21′′′d) to whitish, reaching 90 mm after 14 days at room temperature in the dark; on MYA cottony, with aerial mycelium, spreading out in concentric rings, creating a lobed rosette-like appearance, buff (21′′′d) to whitish, reaching 90 mm after 14 days at room temperature in the dark (Figures 11A–C).
3.3.31 Additional specimens examined
Iran, Kurdistan Province, Baneh, from Q. libani branches, 22 August 2016, S. Bashiri, IRAN 4875 = CBS 149774/CJASB296 (36°05′01.8″ N/45°40′32.8″ E), CJASB298 (23 August 2016, 35°52′30.8″ N/45°59′19.6″ E); Iran, Kurdistan Province, Baneh, from Q. brantii branches, 23 August 2016, S. Bashiri, CJASB299 (35°52′.308″ N/45°59′.196″ E), CJASB294 (23 August 2016, 36°07′17.7″ N/45°41′37.0″ E); Iran, Kurdistan Province, Sarvabad, from Q. libani branches, 15 August 2016, S. Bashiri, CJASB300 (35°22′90.1″ N/46°09′28.3″ E), CJASB302(16 August 2016, 35°23′86.3″ N/46°12′49.4″ E)/CJASB303 (16 August 2016, 35°05′64.3″ N/46°35′30.9″ E); Iran, Kurdistan Province, Sarvabad, from Q. brantii branches, 16 August 2016, S. Bashiri, CJASB304 (35°06′79.9″ N/46°32′58.1″ E); Iran, Kurdistan Province, Sarvabad, from Q. infectoria branches, 15 August 2016, S. Bashiri, CJASB301 35°22′95.7″ N/46°12′89.5″ E; Iran, West Azarbaijan Province, Sardasht, from Q. libani branches, 8 August 2016, S. Bashiri, CJASB283/IRAN 4905C (36°06′07.2″ N/45°29′64.5″ E), CJASB286 (36°10′20.1″ N/45°29′12.3″ E); Iran, West Azarbaijan Province, Sardasht, from Q. infectoria branches, 8 August 2016, S. Bashiri, CJASB285 (36°07′23.4″ N/45°28′08.5″ E), CJASB286 (36°10′20.1″ N/45°29′12.3″ E); Iran, West Azarbaijan Province, Piranshahr, from Q. infectoria branches, 7 August 2016, S. Bashiri, CJASB287/CJASB288 (36°23′12.2″ N/45°23′33.9″ E), CJASB286 (36°10′20.1″ N/45°29′12.3″ E); Iran, Kermanshah Province, Ravansar, from Q. brantii branches, 21 October 2016, S. Bashiri, CJASB305/CJASB306 (34°03′12.5″ N/46°27′19.3″ E); Iran, Kermanshah Province, Paveh, from Q. brantii branches, 21 October 2016, S. Bashiri, IRAN 4907C/CJASB308/CJASB309 (34°57′06.9″ N/46°27′80.5″ E); Iran, Ilam Province, Ilam, from Q. brantii branches, 13 September 2016, S. Bashiri, CJASB289/CJASB290 (33°42′15.6″ N/46°22′63.5″ E), CJASB291 (33°35′67.3″ N/46°26′50.6″ E); Iran, Ilam Province, Eyvan, from Q. brantii branches, 13 September 2016, S. Bashiri, IRAN 4906C/CJASB293 (33°50′84.8″ N/46°12′21.0″ E).
3.4 Pathogenicity tests
In this survey, pathogenicity of representative isolates was examined under in vitro (on leaves) and greenhouse (on seedling stems) conditions (Figures 12–15, Supplementary Figures 1–4). In both experiments, 19 fungal species were determined as pathogenic, and Alternaria spp. (A. alternata, A. atra, and A. contlous), Chaetomium globosum, and Parachaetomium perlucidum were nonpathogenic.

Figure 12 Pathogenicity tests and disease symptoms caused by A. iranensis on oak seedlings in greenhouse (A–C, G) and leaves under in vitro (D, E) conditions. (A) Inoculated plants after 2 months. (B) Control. (C) Necrotic lesion on stems. (D, E) Necrotic spot on leaves and control. (F) A. iranensis colony re-isolated from inoculated seedlings. (G) Stem cross-sections showing wood necrosis and discoloration.

Figure 13 Pathogenicity tests and disease symptoms caused by C. hedjaroudei on oak seedlings in greenhouse (A–C, G) and leaves under in vitro (D, E) conditions. (A) Inoculated plants after 2 months. (B) Control. (C) Necrotic lesion on stems. (D, E) Necrotic spot on leaves and control. (F) C. hedjaroudei colony re-isolated from inoculated seedlings. (G) Stem cross-sections showing wood necrosis and discoloration.
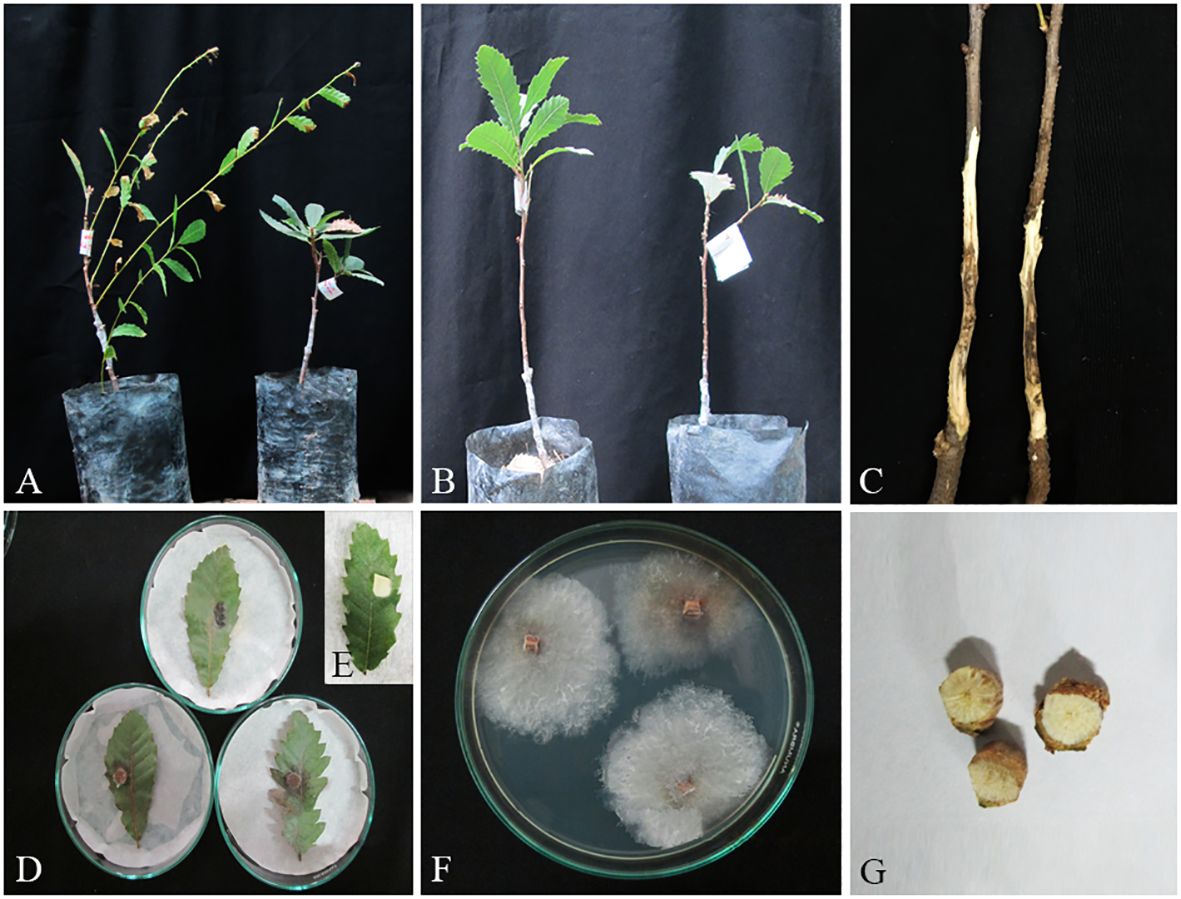
Figure 14 Pathogenicity tests and disease symptoms caused by C. zagrosensis on oak seedlings in greenhouse (A–C, G) and leaves under in vitro (D,E) conditions. (A) Inoculated plants after 2 months. (B) Control. (C) Necrotic lesion on stems. (D, E) Necrotic spot on leaves and control. (F) C. zagrosensis colony re-isolated from inoculated seedlings. (G) Stem cross-sections showing wood necrosis and discoloration.

Figure 15 Pathogenicity tests and disease symptoms caused by G. quercicola on oak seedlings in greenhouse (A–C, G) and leaves under in vitro (D,E) conditions. (A) Inoculated plants after 1 month. (B) Control. (C) Necrotic lesion on stems. (D, E) Necrotic spot on leaves and control. (F) G. quercicola colony re-isolated from inoculated seedlings. (G) Stem cross-sections showing wood necrosis and discoloration.
In the phytotoxicity assay under in vitro conditions on oak leaves, we qualitatively evaluated and grouped pathogenic species in four classes as highly virulent (Biscogniauxia persica, B. rosacearum, Botryosphaeria dothidea, Gnomoniopsis quercicola, and Neoscytalidium dimidiatum), moderately virulent (Cosmospora butyri, Didymella glomerata, Fusarium anulatum, Kalmusia variispora, and Neocosmospora sp.), weakly virulent (Alloeutypa iranensis, Cytospora hedjaroudei, C. zagrosensis, Fimetariella rabenhorstii, Neocosmospora metavorans, Phaeoacremonium tuscanicum, and Stilbocrea anihashemiana), and very weak virulent (Apiospora intestini and Nigrospora sp.).
In pathogenicity tests under greenhouse conditions, Biscogniauxia rosacearum, B. persica, Botryosphaeria dothidea, G. quercicola, and N. dimidiatum were the most virulent species, which caused a quick decline and death of inoculated seedlings after 28–30 days. Thus, to evaluate the pathogenicity of these species in a distinct analysis, we recorded the external and internal symptoms and analyzed them after 30 days (Table 2). Pathogenicity of the other examined species were recorded after 60 days and analyzed separately (Table 3). In the first statistical analyses, significant differences were determined in lesion lengths between species (F = 54.22, p < 0.001) and B. rosacearum was the most virulent species and G. quercicola was the most virulent coelomycetous species. Gnomoniopsis quercicola showed high phytotoxic activity on leaves examined under in vitro conditions (Figure 15D). Severe leaf yellowing and defoliation on inoculated seedlings and extended brown wood necrosis in cross-sections of inoculated stems were observed 28–30 days after inoculation under greenhouse conditions (Figure 15). Botryosphaeria dothidea and N. dimidiatum showed high phytotoxic activity on leaves (Supplementary Figures 1–4D) and inoculated seedlings showed severe leaf blight and defoliation together with wedge-shaped and irregular wood necrosis in cross-sections of inoculated stems 28–30 days after inoculation (Supplementary Figures 1–4). The second statistical analysis also indicated significant differences in lesion lengths between species (F = 318.33, p < 0.001) and all coelomycetous species; A. iranensis, C. hedjaroudei, C. zagrosensis, D. glomerata, and K. variispora were pathogenic and caused leaf necrosis, blight, and defoliation on oak seedlings under greenhouse conditions 60 days after inoculation. Moreover, cross-sections of inoculated stems showed an intermediate vascular discoloration and irregular wood necrosis (Table 3, Figures 12–14, Supplementary Figures 2, 3). All inoculated pathogens were re-isolated from inoculated leaves and seedling stems, confirming Koch’s postulates (Figures 12–15, Supplementary Figures 1–4).
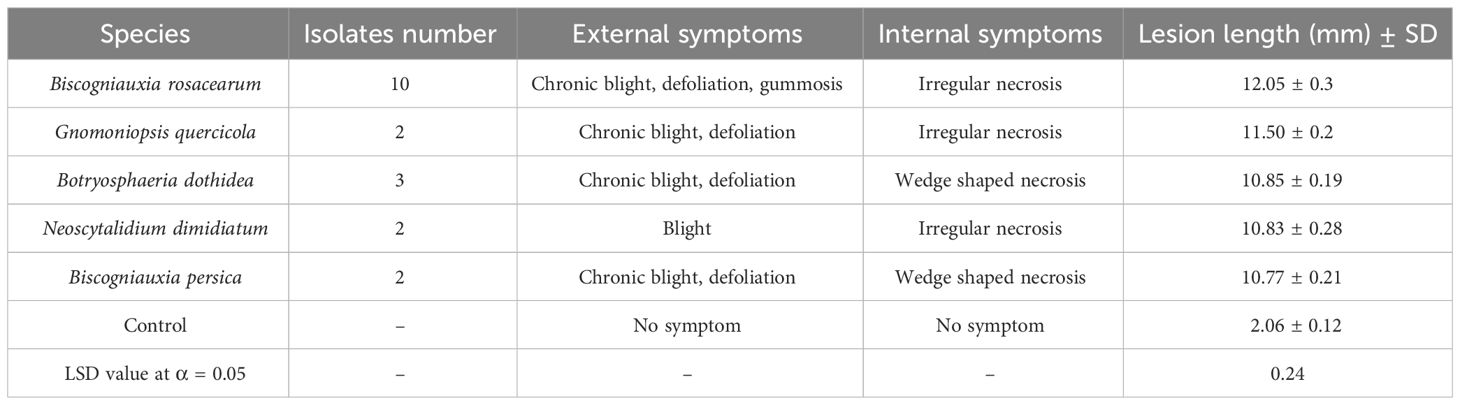
Table 2 Number of isolates, observed symptoms, and mean lesion length measured in pathogenicity studies of five highly virulent species recorded and analyzed after 30 days.
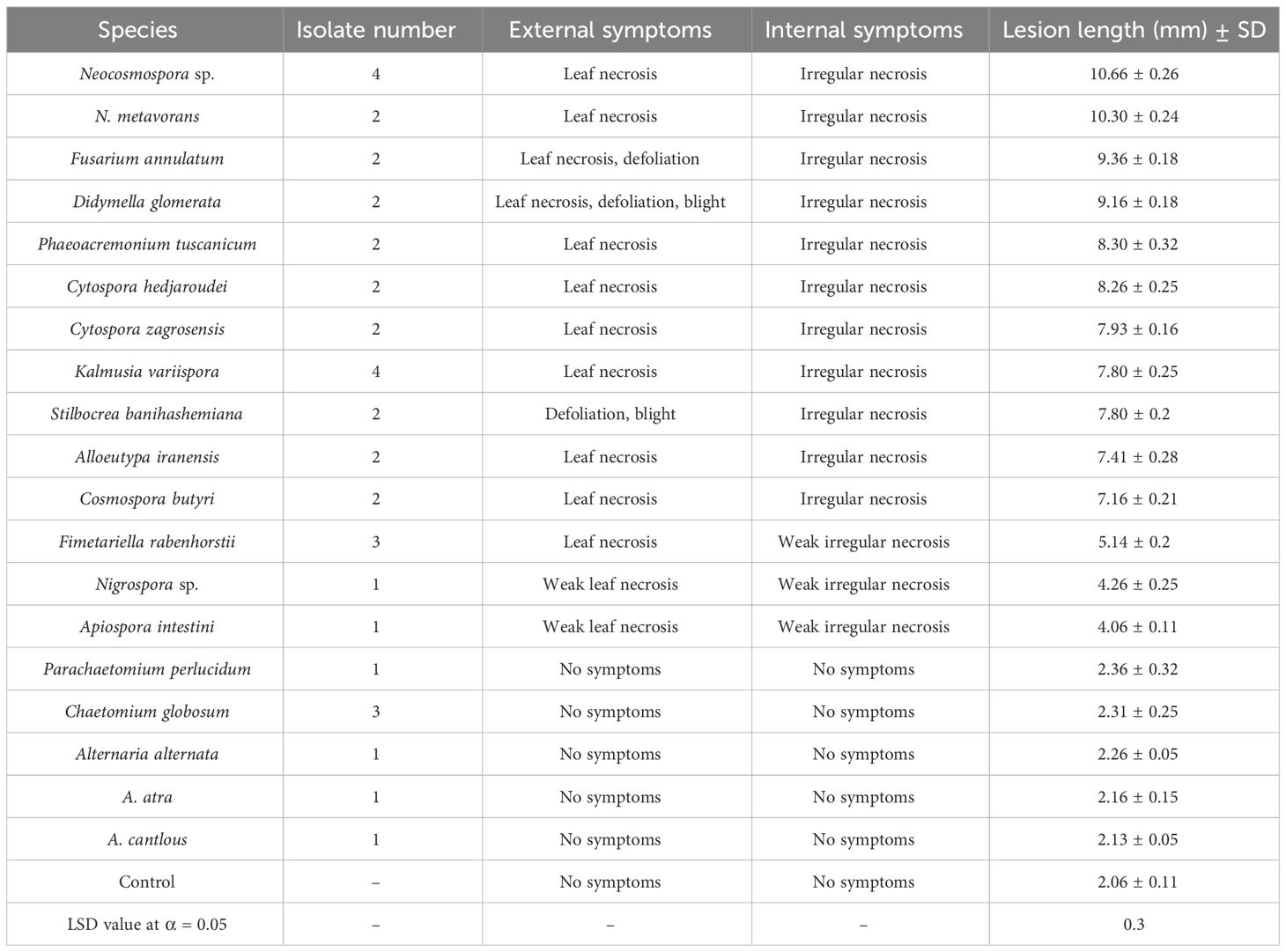
Table 3 Number of isolates, observed symptoms, and mean lesion length measured in pathogenicity studies of 19 species recorded and analyzed after 60 days.
4 Discussion
In this study, we focused on oak decline, a growing threat to Zagros forests, and collected a large fungal collection from oak trees showing various types of external and internal disease symptoms in Zagros forests located in five western provinces: Ilam, Kermanshah, Kurdistan, Lorestan, and West Azarbaijan. Generally, depending on a variety of biotic and abiotic factors (e.g., host plant, microbial communities, and climate factors), management and exploitation systems, method and time of sampling, and isolation procedure in a study composition, the frequency and diversity of fungal species associated with declined woody plants vary in different geographic regions around the world (Lynch et al., 2013; Linaldeddu et al., 2016; Hashemi et al., 2017; Alidadi et al., 2018; Ghobad-Nejhad et al., 2018; Alidadi et al., 2019; Pinna et al., 2019; Mahamedi et al., 2020; Bahmani et al., 2021; Nahvi Moghadam et al., 2022). Based on DNA sequence data and morphological features, we characterized 24 fungal species belonging to 19 genera from 10 different families, including Apiosporaceae (Apiospora and Nigrospora), Bionecteriaceae (Stilbocrea), Bombardiaceae (Fimetariella), Botryosphaeriaceae (Botryosphaeria and Neoscytalidium), Chaetomiaceae (Chaetomium, Parachaetomium), Cytosporaceae (Cytospora), Diatrypaceae (Alloeutypa), Didymellaceae (Didymella), Didymosphaeriaceae (Kalmusia), Gnomoniaceae (Gnomoniopsis), Graphostromataceae (Biscogniauxia), Nectriaceae (Cosmospora, Fusarium, and Neocosmospora), Pleosporaceae (Alternaria), and Togninaceae (Phaeoacremonium). In this paper, we have listed and provided frequency and distribution of the identified fungi at the genus and species levels and focused on phylogeny and pathology of coelomycetous fungi, a well-known morphological group in fungal pathogens.
Biscogniauxia (B. rosacearum and B. persica), Neocosmospora (N. metavorans and Neocosmospora sp.), and Cytospora (C. hedjaroudei and C. zagrosensis) were the most prevalent identified fungi associated with oak decline. Among the coelomycetous fungi, members of two well-known families Botryosphaeriaceae (Botryosphaeria and Neoscytalidium) and Cytosporaceae (Cytospora) in association with decline of woody plants were isolated from declined oak trees in the central and northern part of Zagros forests.
Botryosphaeriaceae members are common fungal pathogens, endophytes, or saprobes on woody plants (Phillips et al., 2013; Moricca et al., 2016; Kazemzadeh Chakusary et al., 2019; Mahamedi et al., 2020; Brazee et al., 2023). Botryosphaeria and Neoscytalidium species are among the most virulent members of Botryosphaeriaceae associated with canker, dieback, fruit rot, and decline symptoms in a broad spectrum of woody plants (including Quercus spp.) (Luque et al., 2000; Sánchez et al., 2003; Linaldeddu et al., 2009; Marsberg et al., 2017; Kazemzadeh Chakusary et al., 2019; Mahamedi et al., 2020; Derviş and Özer, 2023). A search of Index Fungorum (November 2023; www.indexfungorum.org) lists more than 200 Botryosphaeria species in which DNA sequence data are available for some 20 species. Thus far, five Neoscytalidium species, namely, N. dimidiatum, N. hylocereum, N. novaehollandiae, N. oculi, and N. orchidacearum, have been described (November 2023; www.indexfungorum.org). Of these, N. novaehollandiae and N. orchidacearum have been reduced to synonymy with N. dimidiatum (Zhang et al., 2021). Botryosphaeria dothidea has previously been isolated as saprophyte from pycnidia on dead twigs of an unknown Quercus species in Gardane-Heyran, north of the country (Abdollahzadeh, 2009; Abdollahzadeh et al., 2013), but it is reported here for the first time as a pathogenic species isolated from necrotic wood tissues of oak trees in Iran. We isolated this species from all three oak species in Ilam, Kermanshah, Kurdistan, Lorestan, and West Azarbaijan provinces, while N. dimidiatum was found only from Q. brantii in Kermanshah, Ilam, and Lorestan provinces with a higher mean annual temperature. Neoscytalidium dimidiatum has already been reported as an endophytic (Ghasemi-Esfahlan et al., 2019) or pathogenic fungal species (Alidadi et al., 2019; Sabernasab et al., 2019) associated with Persian oak (Q. brantii) in Zagros forests located in Kermanshah, Ilam, and Lorestan provinces, but it is reported for the first time from Kurdistan and West Azarbaijan provinces. In this study, B. dothidea and N. dimidiatum were determined as highly virulent in both pathogenicity experiments on leaves and seedling stems of Q. brantii. Pathogenicity of both species on oak has previously been confirmed (Sánchez et al., 2003; Turco et al., 2006; Alidadi et al., 2019; Sabernasab et al., 2019).
Members of diaporthalean fungi are associated with several diseases including canker and dieback in economically and ecologically important woody plants such as Quercus species (Luque et al., 2000; Lynch et al., 2014; Fan et al., 2018; Jiang et al., 2018, 2019; Zhu et al., 2019, 2021). Among the diaporthalean fungi, Cytospora with approximately 700 species listed in Index Fungorum (November 2023; www.indexfungorum.org) is the most common and widespread genus associated with a wide variety of woody plants around the world, which causes various disease symptoms such as canker and dieback or found as endophyte and saprobe (Adams et al., 2005; Lawrence et al., 2018; Azizi et al., 2020; Fan et al., 2020; Hanifeh et al., 2022; Ilyukhin et al., 2023). Cytospora species are found in association with canker and dieback diseases on various Quercus species (Kowalski, 1991; Adams et al., 2005; Lynch et al., 2014; Fan et al., 2018; Lawrence et al., 2018; Pinna et al., 2019; Pan et al., 2021). Very few Cytospora species have been reported from oak trees (Q. brantii) in Iran including C. intermedia from Fars Province (Fotouhifar et al., 2008), Cystospora sp.1 and Cytospora sp.2 from Kohgiluyeh and Boyer-Ahmad (Ghobad-Nejhad et al., 2017, 2018), and C. ribis from Ilam (Alidadi et al., 2019) provinces. In this study, based on morphology and phylogenetic analyses of a dataset combining sequences of five loci LSU, ITS, rpb2, ef-1α, tub2, and act1, we characterized two new species named C. hedjaroudei sp. nov. and C. zagrozensis sp. nov. isolated from all three Quercus species. Cytospora zagrosensis isolates were collected from Kermanshah, Kurdistan, and Lorestan provinces while C. hedjaroudei was also collected from West Azarbaijan and Ilam provinces. Thus far, some 15 Cytospora species have been reported from oak trees including the most known or recently described species: Cytospora chrysosperma, C. quercicola, C. quercinum, C. prunicola, C. pubescentis, and C. vinacea (Preston, 1945; Spaulding, 1961; Senanayake et al., 2017; Shang et al., 2020, 2020; Pan et al., 2021). Cytospora hedjaroudei and C. zagrosensis can be distinguished from closely related species-based morphology and DNA sequenced data. In pathogenicity experiments on oak leaves and seedling stems, both Cytospora species were recognized as weakly virulent.
Gnomoniaceae, as the second largest family of Diaporthales, is found on leaves and twigs of hardwood trees, shrubs, and herbaceous plants (Walker et al., 2010). Gnomoniopsis Berl., as a distinct genus based on the type species Gnomoniopsis chamaemori (Fr.) Berl., is isolated only from three plant families Fagaceae, Onagraceae, and Rosaceae (Wang et al., 2022). Thus far, several species, namely, G. clavulata, G. paraclavulata, G. daii, G. fagacearum, and G. silvicola, have been recorded on various Quercus species (Sogonov et al., 2008; Walker et al., 2010; Jiang et al., 2021; Jankowiak et al., 2022). To date, Discula quercina is the only Gnomoniaceae species reported from Q. infectoria, Q. macranthera, and Q. rubra in Iran (Khabiri, 1958; Ghasemi-Esfahlan et al., 2019; Hanifeh et al., 2019). In this study, we characterized a new Gnomoniopsis species named Gnomoniopsis quercicola sp. nov. from all three Quercus species in Zagros forests of West Azarbaijan, Kurdistan, Kermanshah, and Ilam provinces. Gnomoniopsis quercicola was placed in a distinct clade close to G. paraclavulata with significant differences in DNA sequence data and growth rate of colony on culture media. This species was the most pathogenic coelomycetous species and the second highly virulent species after B. rosacearum in both pathogenicity experiments on oak leaves and seedling stems.
Diatrypaceae family members are other fungal pathogens that have been isolated in association with canker and dieback diseases of a broad spectrum of woody hosts (Acero et al., 2004; Trouillas and Gubler, 2010; Trouillas et al., 2011, 2011; Mehrabi et al., 2015; Mehrabi et al., 2016; Mehrabi et al., 2019; Zhu et al., 2021). Several Diatrypaceae members belonging to various genera (e.g., Alloeutypa, Cryptovalsa, Diatrype, Diatrypella, Eutypa, Eutypella, and Libertella) have been isolated from oak trees (Acero et al., 2004; Mehrabi and Hemmati, 2013; Mehrabi et al., 2015; Mehrabi et al., 2016; Mehrabi et al., 2019; Zhu et al., 2021). Among these genera, Alloeutypa has recently been introduced to encompass a new species, A. milinensis (type species), and a new combination, Alloeutypa flavovirens (Ma et al., 2023). In this study, we recognized a new species in Alloeutypa close to A. flavovirens named Alloeutypa iranensis sp. nov. Based on morphology and DNA sequence data, this species is obviously different from A. flavovirens and A. milinensis. In pathogenicity experiments, A. iraniensis was weakly virulent on oak leaves and seedling stems.
Didymella glomerata (Didymellaceae) and K. variispora (Didymosphaeriaceae = Montagnulaceae), two phoma-like fungal pathogens, are seen among the pathogenic fungi associated with woody plants showing canker, dieback, and decline symptoms or as endophyte and saprobe (Holz et al., 1989; Thomidis et al., 2011; Verkley et al., 2014; Alidadi et al., 2018, 2019; Ghobad-Nejhad et al., 2017; Bahmani et al., 2021; Nahvi Moghadam et al., 2022). Recently, D. glomerata was isolated from Q. brantii in Zagros forests of Ilam as endophyte and pathogen (Alidadi et al., 2019). Here, we have isolated this species from Q. brantii and Q. libani for the first time from Kurdistan, Kermanshah, and Lorestan provinces. Didymella glomerata in both pathogenicity experiments was determined as moderately virulent. Recently, pathogenicity of D. glomerata on Q. brantii seedlings was confirmed (Alidadi et al., 2019). Additionally, K. variispora has been isolated from Q. brantii as an endophyte in Kohgiluyeh and Boyer-Ahmad (Ghobad-Nejhad et al., 2017) and associated with oak decline in Ilam provinces (Alidadi et al., 2018), while we have isolated this species from all three Quercus species (Q. brantii, Q. infectoria, and Q. libani) growing in Zagros forests in Ilam, Kermanshah, Kurdistan, Lorestan, and West Azarbaijan provinces. In pathogenicity experiments, K. variispora was moderately and weakly virulent on oak leaves and seedling stems, respectively. Based on our knowledge, the pathogenicity of K. variispora is confirmed on oak trees for the first time.
The majority of identified fungi (19 out of 24, 79%), including all coelomycetous species, were determined as pathogenic; thus, it is important to consider them in various aspects of pathogenicity, host range, geographic distribution, genetic diversity, and management. Since phytotoxic secondary metabolites are major biochemical weapons, as pathogenicity or virulence factors in fungal pathogens causing canker, dieback, and decline diseases in woody plants (Evidente et al., 2019), we have studied and characterized phytotoxic compounds of some species such as B. rosacearum (Masi et al., 2021), F. rabenhorstii (Bashiri et al., 2020a), and S. banihashemiana, previously identified as S. macrostoma (Di Lecce et al., 2020). At the time being, phytotoxic metabolites of six species, A. iranensis, G. quercicola, N. dimidiatum, Ph. tuscanicum, Cosmospora butyri, and Neocosmospora sp., are being investigated.
In this study, branch canker, dieback, and defoliation were obviously dominant external symptoms of declined oak trees accompanied by borer hole and central and irregular necrosis as common internal symptoms on twigs’ cross-sections. We found no correlation between fungal species isolated and type of internal and external symptoms as we can infer from several studies (Mohammadi et al., 2014; Hashemi and Mohammadi, 2016; Esparham et al., 2020; Soltaninejad et al., 2017; Bashiri et al., 2022). Most of the identified species were isolated in association with borer hole (feeding site of insects) on infected trees; thus, it is necessary to consider the role of insects in transmission, penetration, and emergence of opportunistic fungal pathogens, weakness, and decline of oak trees in combination with fungal species. In the last decades, many fungi have been isolated from insects or feeding sites of larvae including Biscogniauxia rosacearum, B. persica, B. dryophila, Neocosmospora ewallaceae, N. ambrosia, N. metavorans, Fusarium spp., Diplodia corticola, Dothiorella iberica, and Cryphonectria naterciae (Pazoutova et al., 2010; O’Donnell et al., 2012; Freeman et al., 2013; Bateman et al., 2016; Herr et al., 2016; Bashiri et al., 2022).
According to the composition and diversity of the identified fungal species, the pathogenicity of the majority of species, and the presence of some well-known and new highly virulent species (e.g., Biscogniauxia rosacearum, Botryosphaeria dothidea, N. dimidiatum, and G. quercicola sp. nov.) together with their association with insect borer hole, we strongly recommend to investigate in more detail all identified fungi specifically the most virulent ones and new fungal species to discover their interaction with growing oak species, as well as their relationships together and with insects and abiotic stress. Concurrently, regarding the threats facing Zagros oak forests including drought, heat and dust stresses due to climate change, wildfire, plant diseases (charcoal canker), and pests (borer beetles and green oak tortrix), it is crucial to prevent inappropriate human exploitation and forest management through training and improving welfare of local communities and following environmental friendly approaches dealing with plant diseases and pests.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
SB: Formal analysis, Investigation, Software, Writing – original draft. JA: Conceptualization, Funding acquisition, Methodology, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. SB (under PostDoc research project number S01/9/3891) and JA were supported by the University of Kurdistan.
Acknowledgments
The authors thank Dr. B. Bahramnejad, Department of Plant Production and Genetics Engineering, University of Kurdistan for advice on statistical analyses.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1377441/full#supplementary-material
References
Abdollahzadeh, J. (2009). Taxonomy and phylogeny of the Botryosphaeriaceae in Iran. Tarbiat Modares University, Tehran, Iran. 200 pp.
Abdollahzadeh, J., Groenewald, J. Z., Coetzee, M. P. A., Wingfield, M. J., Crous, P. W. (2020). Evolution of lifestyles in Capnodiales. Stud. Mycol. 95, 381–414. doi: 10.1016/j.simyco.2020.02.004
Abdollahzadeh, J., Mohammadi Goltapeh, E., Javadi, A., Shams-bakhsh, M., Zare, R., Phillips, A. J. L. (2009). Barriopsis Iraniana and Phaeobotryon cupressi: two new species of the Botryosphaeriaceae from trees in Iran. Persoonia 23, 1–8. doi: 10.3767/003158509X467552
Abdollahzadeh, J., Zare, R., Phillips, A. J. L. (2013). Phylogeny and taxonomy of Botryosphaeria and Neofusicoccum species in Iran, with description of Botryosphaeria scharifii sp. nov. Mycologia 105, 210–220. doi: 10.3852/12-107
Acero, F. J., González, V., Sánchez-Ballesteros, J., Rubio, V., Checa, J., Bills, G. F., et al. (2004). Molecular phylogenetic studies on the Diatrypaceae based on rDNA-ITS sequences. Mycologia 96, 249–259. doi: 10.1080/15572536.2005.11832975
Adams, G. C., Wingfeld, M. J., Common, R., Roux, J. (2005). Phylogenetic relationships and morphology of Cytospora species and related teleomorphs (Ascomycota, Diaporthales, Valsaceae) from Eucalyptus. Stud. Mycol. 52, 1–144.
Ahmadi, E., Kowsari, M., Azadfar, D., Salehi-Jouzani, G. (2019). Bacillus pumilus and Stenotrophomonas maltophilia as two potentially causative agents involved in Persian oak decline in Zagros forests (Iran). For. Pathol. 49, e12541. doi: 10.1111/efp.12541
Ahmadi, R., Kiadaliri, H., Mataji, A., Kafaki, S. (2014). Oak forest decline zonation using AHP model and GIS technique in Zagros forests of Ilam province. J. Biodivers. Environ. Sci. 4, 141–150.
Alidadi, A., Javan-Nikkhah, M., Kowsari, M., Karami, S., Ebrahimi Rastaghi, M. (2018). Some species of fungi associated with declined Persian oak trees in Ilam province with emphasis on new records to mycobiota of Iran. Rostaniha 19, 75–91. doi: 10.22092/botany.2019.122177.1105
Alidadi, A., Kowsari, M., Javan-Nikkhah, M., Jouzani, G. S., Rastaghi, M. E. (2019). New pathogenic and endophytic fungal species associated with Persian oak in Iran. Eur. J. Plant Pathol. 155, 1017–1032. doi: 10.1007/s10658-019-01830-y
Alves, A., Correia, A., Luque, J., Phillips, A. (2004). Botryosphaeria corticola, sp. nov. on Quercus species, with notes and description of Botryosphaeria stevensii and its anamorph, Diplodia mutila. Mycologia 96, 598–613. doi: 10.1080/15572536.2005.11832956
Azizi, R., Ghosta, Y., Ahmadpour, A. (2020). Morphological and molecular characterization of Cytospora species involved in apple decline in Iran. Mycol. Iran. 7, 205–218. doi: 10.22043/MI.2021.123907
Bahmani, Z., Abdollahzadeh, J., Amini, J., Evidente, A. (2021). Biscogniauxia rosacearum the charcoal canker agent as a pathogen associated with grapevine trunk diseases in Zagros region of Iran. Sci. Rep. 11, 14098. doi: 10.1038/s41598-021-93630-w
Bashiri, S., Abdollahzadeh, J., Evidente, A. (2020a). Phaeoacremonium tuscanicum, a new fungal pathogen associated with oak decline in Zagros forests, Iran. Mycol. Iran. 7, 247–252. doi: 10.22043/MI.2021.124424
Bashiri, S., Abdollahzadeh, J., Evidente, A. (2022). Diagnosing and pathogenicity of Biscogniauxia species, the causal agents of oak charcoal canker and decline in Zagros forests of Iran. J. Plant Pathol. 104, 1011–1025. doi: 10.1007/s42161-022-01124-z
Bashiri, S., Abdollahzadeh, J., Lecce, R. D., Alioto, D., Gorecki, M., Pescitelli, G., et al. (2020b). Rabenchromenone and rabenzophenone, phytotoxic tetrasubstituted chromenone and hexasubstituted benzophenone constituents produced by the oak-decline-associated fungus Fimetariella rabenhorstii. J. Nat. Prod. 83, 447–452. doi: 10.1021/acs.jnatprod.9b01017
Bateman, C., Šigut, M., Skelton, J., Smith, K. E., Hulcr, J. (2016). Fungal associates of the Xylosandrus compactus (Coleoptera: Curculionidae, Scolytinae) are spatially segregated on the insect body. Environ. Entomol. 45, 883–890. doi: 10.1093/ee/nvw070
Brazee, N. J., Munck, I. A., McLaughlin, K., Ferreira, S., Keleher, N. (2023). Diplodia twig canker (Diplodia gallae) of northern red oak (Quercus rubra) in the northeastern United States. For. Pathol. 53, 12822. doi: 10.1111/efp.12822
Carbone, I., Kohn, L. M. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91, 553–556. doi: 10.1080/00275514.1999.12061051
Crous, P. W., Gams, W., Stalpers, J. A., Robert, V., Stegehuis, G. (2004). MycoBank: an online initiative to launch mycology into the 21st century. Stud. Mycol. 50, 19–22.
Cubeta, M. A., Echandi, E., Abenerthy, T., Vilgalys, R. (1991). Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of ribosomal RNA gene. Phytopathology 81, 1395–1400. doi: 10.1094/Phyto-81-1395
Derviş, S., Özer, G. (2023). Plant-associated neoscytalidium dimidiatum—Taxonomy, host range, epidemiology, virulence, and management strategies: A comprehensive review. J. Fungi. 9, 1048. doi: 10.3390/jof9111048
Di Lecce, R., Bashiri, S., Masi, M., Alioto, D., Tuzi, A., Abdollahzadeh, J., et al. (2020). Phytotoxic metabolites from Stilbocrea macrostoma, a fungal pathogen of Quercus brantii in Iran. Nat. Prod. Res. 35, 5857–5861. doi: 10.1080/14786419.2020.1797731
Dreaden, T. J., Black, A. W., Mullerin, S., Smith, J. A. (2014). First report of Diplodia quercivora causing shoot dieback and branch cankers on Live oak (Quercus virginiana) in the United States. Plant Dis. 98, 282. doi: 10.1094/PDIS-07-13-0736-PDN
Esparham, N., Mohammadi, H., Gramaje, D. (2020). A survey of trunk disease pathogens within citrus trees in Iran. Plants 9, 1–20. doi: 10.3390/plants9060754
Evidente, A., Cimmino, A., Masi, M. (2019). Phytotoxins produced by pathogenic fungi of agrarian plants. Phytochem. Rev. 18, 843–870. doi: 10.1007/s11101-019-09624-0
Fan, X. L., Bezerra, J. D., Tian, C. M., Crous, P. W. (2018). Families and genera of diaporthalean fungi associated with canker and dieback of tree hosts. Pers.: Mol. Phyl. Evol. Fungi. 40, 119–134. doi: 10.3767/persoonia.2018.40.05
Fan, X. L., Bezerra, J. D. P., Tian, C. M., Crous, P. W. (2020). Cytospora (Diaporthales) in China. Pers.: Mol. Phyl. Evol. Fungi. 45, 1–45. doi: 10.3767/persoonia.2020.45.01
Fotouhifar, K. B., Hedjaroude, G. A., Okhovvat, S., Javan-nikkhah, M., Ershad, D., Moussavi, S. (2008). New records of form-genus Cytospora in Iran (II). Rostaniha 9, 49–66.
Freeman, S., Sharon, M., Maymon, M., Mendel, Z., Protasov, A., Aoki, T., et al. (2013). Fusarium euwallaceae sp. nov. a symbiotic fungus of Euwallacea sp., an invasive ambrosia beetle in Israel and California. Mycologia 105, 1595–1606. doi: 10.3852/13-066
Ghasemi-Esfahlan, S., Arzanlou, M., Babai-Ahari, A. (2016). Detection of Biscogniauxia mediterranea, the causal agent of charcoal rot disease on oak trees in Arasbaran forests and evaluation of its pathogenicity on oak under glasshouse conditions. Iran. J. Plant Pathol. 52, 217–230.
Ghasemi-Esfahlan, S., Arzanlou, M., Tavakoli, M. (2017). Detection of Biscogniauxia mediterranea, one of the causal agents of oak decline in Sardasht (West Azarbaijan Province). Iran. J. Plant Pathol. 53, 349–351.
Ghasemi-Esfahlan, S., Khodaei, S., Karimi, K., Tavakoli, M., Pertot, I., Arzanlou, M. (2019). Biodiversity study of endophytic fungi associated with two Quercus species in Iran. For. Syst. 28, e003–e0015. doi: 10.5424/fs/2019281-14528
Ghobad-Nejhad, M., Asgari, B., Chaharmiri Dokhaharani, S. (2017). Notes on some endophytic fungi isolated from Quercus brantii in Dena region of Kohgiluyeh and Boyer-Ahmad province Iran. Mycol. Iran. 4, 1–12. doi: 10.22043/MI.2018.115893
Ghobad-Nejhad, M., Meyn, R., Langer, E. (2018). Endophytic fungi isolated from healthy and declining Persian oak (Quercus brantii) in western Iran. Nova Hedwigia 107, 273–290. doi: 10.1127/nova_hedwigia/2018/0470
Glass, N. L., Donaldson, G. C. (1999). Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 61, 1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995
Hall, T. A. (2004). BioEdit. Version 7.0.0. Department of Microbiology (United States: North Carolina State University).
Hanifeh, S., Zafari, D., Soleimani, M. J., Arzanlou, M. (2022). Multigene phylogeny, morphology, and pathogenicity trials reveal novel Cytospora species involved in perennial canker disease of apple trees in Iran. Fungal Biol. 126, 707–726. doi: 10.1016/j.funbio.2022.08.009
Hanifeh, S., Zafari, D., Soleimani, M., Ravanlou, A. (2019). Discula quercina as a possible causal agent of dieback on oak trees in Iran. For. Pathol. 49, 12468–12475. doi: 10.1111/efp.12468
Hashemi, H., Mohammadi, H. (2016). Identification and characterization of fungi associated with internal wood lesions and decline disease of willow and poplar trees in Iran. For. Pathol. 46, 341–352. doi: 10.1111/efp.12269
Hashemi, H., Mohammadi, H., Abdollahzadeh, J. (2017). Symptoms and fungi associated with elm trees decline in Iran. Eur. J. For. Res. 136, 857–879. doi: 10.1007/s10342-017-1075-y
Herr, J. R., Scully, E. D., Geib, S. M., Hoover, K., Carlson, J. E., Geiser, D. M. (2016). Genome sequence of Fusarium isolate MYA-4552 from the midgut of Anoplophora glabripennis, an invasive, wood-boring beetle. Genome Announc. 4, e00544–e00560. doi: 10.1128/genomeA.00544-16
Heydari, M., Poorbabaei, H., Rostami, T., Begim-Faghir, M., Salehi, A., Ostad Hashmei, R. (2013). Plant species in oak (Quercus brantii Lindl.) understory and their relationship with physical and chemical properties of soil in different altitude classes in the Arghavan valley protected area, Iran. Casp. J. Environ. Sci. 11, 97–110.
Holz, G., Morley, M., Schreuder, W. (1989). Pathogenicity of Phoma glomerata, P. cava, P. eupyrena and Cytospora chrysosperma on blackthorn (Acacia mellifera subsp. detinens). Agricola 7, 37–42.
Hosseini, A., Hosseini, S. M., Linares, J. C. (2018). Site factors and stand conditions associated with Persian oak decline in Zagros mountain forests. For. Syst. 26, e014. doi: 10.5424/fs/2017263-11298
Huelsenbeck, J. P., Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Ilyukhin, E., Nguyen, H. D., Castle, A. J., Ellouze, W. (2023). Cytospora paraplurivora sp. nov. isolated from orchards with fruit tree decline syndrome in Ontario, Canada. PloS One 18, 279490. doi: 10.1371/journal.pone.0279490
Jankowiak, R., Stępniewska, H., Bilański, P., Taerum, S. J. (2022). Fungi as potential factors limiting natural regeneration of pedunculate oak (Quercus robur L.) in mixed-species forest stands in Poland. Plant Pathol. 71, 805–817. doi: 10.1111/ppa.13529
Jiang, N., Fan, X. L., Crous, P. W., Tian, C. M. (2019). Species of Dendrostoma (Erythrogloeaceae, Diaporthales) associated with chestnut and oak canker diseases in China. MycoKeys 48, 67–96. doi: 10.3897/mycokeys.48.31715
Jiang, N., Voglmayr, H., Bian, D. R., Piao, C. G., Wang, S. K., Li, Y. (2021). Morphology and phylogeny of gnomoniopsis (Gnomoniaceae, diaporthales) from fagaceae leaves in China. J. Fungi. 7, 792–810. doi: 10.3390/jof7100792
Jiang, N., Voglmayr, H., Tian, C. (2018). New species and records of Coryneum from China. Mycologia 110, 1172–1188. doi: 10.1080/00275514.2018.1516969
Kazemzadeh Chakusary, M., Mohammadi, H., Khodaparast, S. A. (2019). Diversity and pathogenicity of Botryosphaeriaceae species on forest trees in the north of Iran. Eur. J. For. Res. 138, 685–704. doi: 10.1007/s10342-019-01200-7
Khabiri, E. (1958). Contribution à la mycoflore de l Iran. Troisième liste. Rev. Mycologique 23, 408–412.
Kowalski, T. (1991). Oak decline: I. Fungi associated with various disease symptoms on overground portions of middle-aged and old oak (Quercus robur L.). Eur. J. For. Res. 21, 136–151. doi: 10.1111/j.1439-0329.1991.tb01418.x
Lawrence, D. P., Holland, L. A., Nouri, M. T., Travadon, R., Abramians, A., Michailides, T. J., et al. (2018). Molecular phylogeny of Cytospora species associated with canker diseases of fruit and nut crops in California, with the descriptions of ten new species and one new combination. IMA fungus 9, 333–369. doi: 10.5598/imafungus.2018.09.02.07
Linaldeddu, B. T., Deidda, A., Scanu, B., Franceschini, A., Alves, A., Abdollahzadeh, J., et al. (2016). Phylogeny, morphology and pathogenicity of Botryosphaeriaceae, Diatrypaceae and Gnomoniaceae associated with branch diseases of hazelnut in Sardinia (Italy). Eur. J. Plant Pathol. 146, 259–279. doi: 10.1007/s10658-016-0912-z
Linaldeddu, B. T., Franceschini, A., Alves, A., Phillips, A. J. L. (2013). Diplodia quercivora sp. nov.: a new species of Diplodia found on declining Quercus canariensis trees in Tunisia. Mycologia 105, 1266–1274. doi: 10.3852/12-370
Linaldeddu, B. T., Scanu, B., Schiaffino, A., Zanda, A., Franceschini, A. (2009). First report of Botryosphaeria dothidea causing canker and branch dieback on Quercus suber in Italy. J. Plant Pathol. 91, 97.
Luque, J., Parlade, J., Pera, J. (2000). Pathogenicity of fungi isolated from Quercus suber in Catalonia (NE Spain). For. Syst. 30, 247–263. doi: 10.1046/j.1439-0329.2000.00208.x
Lynch, S. C., Zambino, P. J., Mayorquin, J. S., Wang, D. H., Eskalen, A. (2013). Identification of new fungal pathogens of coast live oak in California. Plant Dis. 97, 1025–1036. doi: 10.1094/PDIS-11-12-1055-RE
Lynch, S. C., Zambino, P. J., Scott, T. A., Eskalen, A. (2014). Occurence, incidence and associations among fungal pathogens and Agrilus auroguttatus and their roles in Quercus agrifolia decline in California. For. Syst. 44, 62–74. doi: 10.1111/efp.12070
Ma, H. X., Yang, Z. E., Song, Z. K., Qu, Z., Li, Y., Zhu, A. H. (2023). Taxonomic and phylogenetic contributions to Diatrypaceae from southeastern Tibet in China. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1073548
Maddison, W. P., Maddison, D. R. (2023) Mesquite: a modular system for evolutionary analysis. Available online at: http://www.mesquiteproject.org.
Mahamedi, A. E., Phillips, A. J., Lopes, A., Djellid, Y., Arkam, M., Eichmeier, A., et al. (2020). Diversity, distribution and host association of Botryosphaeriaceae species causing oak decline across different forest ecosystems in Algeria. Eur. J. Plant Pathol. 158, 745–765. doi: 10.1007/s10658-020-02116-4
Marsberg, A., Kemler, M., Jami, F., Nagel, J. H., Postma-Smidt, A., Naidoo, S., et al. (2017). Botryosphaeria dothidea: a latent pathogen of global importance to woody plant health. Mol. Plant Pathol. 18, 477–488. doi: 10.1111/mpp.12495
Masi, M., Bashiri, S., Cimmino, A., Bahmani, Z., Abdollahzadeh, J., Evidente, A. (2021). Phytotoxins produced by two Biscogniauxia rosacearum strains, causal agents of grapevine trunk diseases, and charcoal canker of oak trees in Iran. Toxins 13, 812. doi: 10.3390/toxins13110812
Mehrabi, M., Asgari, B., Hemmati, R. (2019). Two new species of Eutypella and a new combination in the genus Peroneutypa (Diatrypaceae). Mycol. Prog. 18, 1057–1069. doi: 10.1007/s11557-019-01503-4
Mehrabi, M., Hemmati, R. (2013). First report of the genus Libertella in Iran. Iran. J. Plant Pathol. 49, 357–358.
Mehrabi, M., Hemmati, R., Vasilyeva, L. N., Trouillas, F. P. (2015). A new species and a new record of Diatrypaceae from Iran. Mycosphere 6, 60–68. doi: 10.5943/mycosphere/6/1/7
Mehrabi, M., Hemmati, R., Vasilyeva, L. N., Trouillas, F. P. (2016). Diatrypella macrospora sp. nov. and new records of diatrypaceous fungi from Iran. Phytotaxa 252, 43–55. doi: 10.11646/phytotaxa.252.1.4
Miller, M. A., Pfeiffer, W., Schwartz, T. (2012) The CIPRES science gateway. Available online at: http://www.phylo.org/index.php/portal (Accessed August 2023).
Mirabolfathy, M., Ju, Y. M., Hsieh, H. M., Rogers, J. D. (2013). Obolarina persica sp. nov., associated with dying Quercus in Iran. Mycoscience 54, 315–320. doi: 10.1016/j.myc.2012.11.003
Mirhashemi, H., Heydari, M., Karami, O., Ahmadi, K., Mosavi, A. (2023). Modeling climate change effects on the distribution of oak forests with nachine learning. Forests 14, 469. doi: 10.3390/f14030469
Mohammadi, H., Kazemi, S., Frahmand, H. (2014). Phaeoacremonium and Botryosphaeriaceae species associated with cypress (Cupressus sempervirens L.) decline in Kerman province (Iran). Phytopathol. Mediterr. 53, 27–39. doi: 10.14601/Phytopathol_Mediterr-12717
Moradi-Amirabad, Y., Rahimian, H., Babaeizad, V., Denman, S. (2019). Brenneria spp. and Rahnella victoriana associated with acute oak decline symptoms on oak and hornbeam in Iran. For. Syst. 49, e12535. doi: 10.1111/efp.12535
Moricca, S., Linaldeddu, B. T., Ginetti, B., Scanu, B., Franceschini, A., Ragazzi, A. (2016). Endemic and emerging pathogens threatening cork oak trees: Management options for conserving a unique forest ecosystem. Plant Dis. 100, 21842193. doi: 10.1094/PDIS-03-16-0408-FE
Moricca, S., Ragazzi, A. (2008). Fungal endophytes in mediterranean oak forests: A lesson from discula quercina. Phytopathology 98, 380–386. doi: 10.1094/PHYTO-98-4-0380
Nahvi Moghadam, J., Khaledi, E., Abdollahzadeh, J., Amini, J. (2022). Seimatosporium marivanicum, Sporocadus kurdistanicus, and Xenoseimatosporium kurdistanicum: Three new pestalotioid species associated with grapevine trunk diseases from the Kurdistan Province, Iran. Mycol. Prog. 21, 427–446. doi: 10.1007/s11557-021-01764-y
Nylander, J. A. A. (2004). MrModeltest. Version 2. Evolutionary Biology Centre (Uppsala, Sweden: Uppsala University). Program distributed by the author.
O’Donnell, K., Cigelnik, E. (1997). Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 7, 103–116. doi: 10.1006/mpev.1996.0376
O’Donnell, K., Humber, R. A., Geiser, D. M., Kang, S., Park, B., Robert, V. A. R. G., et al. (2012). Phylogenetic diversity of insecticolous fusaria inferred from multilocus DNA sequence data and their molecular identification via FUSARIUM-ID and Fusarium MLST. Mycologia 104, 427–445. doi: 10.3852/11-179
Pan, M., Zhu, H., Liang, L., Tian, C., Fan, X. (2021). Studies of canker and dieback of oak tree in China, with two Cytospora species described. Plant Pathol. 70, 2005–2015. doi: 10.1111/ppa.13435
Pazoutova, S., Srutka, P., Holusa, J., Chudickova, M., Kolarik, M. (2010). The phylogenetic position of Obolarina dryophila (Xylariales). Mycol. Prog. 9, 501–507. doi: 10.1007/s11557-010-0658-5
Phillips, A. J. L., Alves, A., Abdollahzadeh, J., Slippers, B., Wingfield, M. J., Groenewald, J. Z., et al. (2013). The Botryosphaeriaceae: genera and species known from culture. Stud. Mycol. 76, 51–167. doi: 10.3114/sim0021
Pinna, C., Linaldeddu, B. T., Deiana, V., Maddau, L., Montecchio, L., Lentini, A. (2019). Plant pathogenic fungi associated with Coraebus florentinus (Coleoptera: Buprestidae) attacks in declining oak forests. Forests 10, 488. doi: 10.3390/f10060488
Raeder, U., Broda, P. (1985). Rapid preparation of DNA from filamentous fungi. J. Appl. Microbiol. 1, 17–20. doi: 10.1111/j.1472-765X.1985.tb01479.x
Raimondo, M. L., Lops, F., Carlucci, A. (2016). Charcoal canker of pear, plum and quince trees caused by Biscogniauxia rosacearum sp. nov. in Southern Italy. Plant Dis. 100, 1813–1822. doi: 10.1094/PDIS-09-15-1037-RE
Rodríguez, F., Oliver, J. F., Martín, A., Medina, J. R. (1990). The general stochastic model of nucleotide substitution. J. Theor. Biol. 142, 485–501. doi: 10.1016/S0022-5193(05)80104-3
Ronquist, F., Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Sabernasab, M., Jamali, S., Marefat, A., Abbasi, S. (2019). Morphological and molecular characterization of Neoscytalidium novaehollandiae, the cause of Quercus brantii dieback in Iran. Phytopathol. Mediterr. 58, 347–357. doi: 10.14601/Phytopathol_Mediter-10621
Safaee, D., Khodaparast, S. A., Mirabolfathy, M., Mousanejad, S. (2017). A multiplex PCR-based technique for identification of Biscogniauxia mediterranea and Obolarina persica causing charcoal disease of oak trees in Zagros forests. For. Pathol. 47, 12330–12338. doi: 10.1111/efp.12330
Sánchez, M. E., Venegas, J., Romero, M. A., Phillips, A. J. L., Trapero, A. (2003). Botryosphaeria and related taxa causing oak canker in southwestern Spain. Plant Dis. 87, 1515–1521. doi: 10.1094/PDIS.2003.87.12.1515
Senanayake, I. C., Crous, P. W., Groenewald, J. Z., Maharachchikumbura, S. S. N., Jeewon, R., Phillips, A. J. L., et al. (2017). Families of Diaporthales based on morphological and phylogenetic evidence. Stud. Mycol. 86, 217–296. doi: 10.1016/j.simyco.2017.07.003
Shang, Q. J., Hyde, K. D., Camporesi, E., Maharachchikumbura, S. S. N., Norphanphoun, C., Brooks, S., et al. (2020). Additions to the genus Cytospora with sexual morphs in Cytosporaceae. Mycosphere 11, 189–224. doi: 10.5943/mycosphere/11/1/2
Shiravand, H., Hosseini, S. A. (2020). A new evaluation of the influence of climate change on Zagros oak forest dieback in Iran. Theor. Appl. Climatol. 141, 685–697. doi: 10.1007/s00704-020-03226-z
Smahi, H., Belhoucine-Guezouli, L., Berraf-Tebbal, A., Chouih, S., Arkam, M., Franceschini, A., et al. (2017). Molecular characterization and pathogenicity of Diplodia corticola and other Botryosphaeriaceae species associated with canker and dieback of Quercus suber in Algeria. Mycosphere 8, 1261–1272. doi: 10.5943/mycosphere/8/2/10
Sogonov, M. V., Castlebury, L. A., Rossman, A. Y., Mejía, L. C., White, J. F. (2008). Leaf-inhabiting genera of the gnomoniaceae, diaporthales. Stud. Mycol. 62, 1–79. doi: 10.3114/sim.2008.62.01
Soltaninejad, N., Mohammadi, H., Massumi, H. (2017). Isolation, identification and pathogenicity of Botryosphaeriaceae and Phaeoacremonium species associated with decline of Prunus species in Iran. J. Plant Pathol. 99, 571–581. doi: 10.4454/jpp.v99i3.3937
Spaulding, P. (1961). Foreign diseases of forest trees of the world: an annotated list. US Depart. Agric. 197, 1–357.
Stamatakis, A. (2014). RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1844–1849. doi: 10.1093/bioinformatics/btu033
Swofford, D. L. (2003). PAUP*: phylogenetic analysis using parsimony (*and other methods) (Sunderland, MA, USA: Sinauer Associates).
Thomidis, T., Michailides, T. J., Exadaktylou, E. (2011). Phoma glomerata (Corda) Wollenw. & Hochapfel a new threat causing cankers on shoots of peach trees in Greece. Eur. J. Plant Pathol. 131, 171. doi: 10.1007/s10658-011-9796-0
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., Higgins, D. G. (1997). The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Trouillas, F. P., Gubler, W. D. (2010). Pathogenicity of Diatrypaceae species in grapevines in California. Plant Dis. 94, 867–872. doi: 10.1094/PDIS-94-7-0867
Trouillas, F. P., Pitt, W. M., Sosnowski, M. R., Huang, R., Peduto, F., Loschiavo, A., et al. (2011). Taxonomy and DNA phylogeny of Diatrypaceae associated with Vitis vinifera and other woody plants in Australia. Fungal Divers. 49, 203–223. doi: 10.1007/s13225-011-0094-0
Turco, E., Marianelli, L., Vizzuso, C., Ragazzi, A., Gini, R., Selleri, B., et al. (2006). First report of botryosphaeria dothidea on sycamore, red oak and english oak in Northwestern Italy. Plant Dis. 90, 1106. doi: 10.1094/PD-90-1106B
Vasilyeva, L. N., Stephenson, S. L. (2004). Pyrenomycetes of the great smoky mountains national park. I. Diatrype fr. (Diatrypaceae). Fungal Divers. 17, 191–201.
Verkley, G. J. M., Dukik, K., Renfurm, R., Göker, M., Stielow, J. B. (2014). Novel genera and species of Coniothyrium-like fungi in Montagnulaceae (Ascomycota). Persoonia 32, 25–51. doi: 10.3767/003158514X679191
Vilgalys, R., Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4239–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Walker, D. M., Castlebury, L. A., Rossman, A. Y., Sogonov, M. V., White, J. F. (2010). Systematics of genus Gnomoniopsis (Gnomoniaceae, Diaporthales) based on a three gene phylogeny, host associations and morphology. Mycologia 102, 1479–1496. doi: 10.3852/10-002
Wang, S., Zhang, Z., Liu, R., Liu, S., Liu, X., Zhang, X. (2022). Morphological and phylogenetic analyses reveal four new species of gnomoniopsis (Gnomoniaceae, diaporthales) from China. J. Fungi. 8, 770. doi: 10.3390/jof8080770
White, T. J., Bruns, T., Lee, S. J. W. T., Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols: A guide to methods and applications, vol. 18 . Eds. Innis, M. A., Gelfand, D. H., Sninsky, J. J., White, T. J. (Academic Press, New York, NY), 315–322.
Wijayawardene, N. N., Hyde, K. D., Wanasinghe, D. N., Papizadeh, M., Goonasekara, I. D., Camporesi, E., et al. (2016). Taxonomy and phylogeny of dematiaceous coelomycetes. Fungal Divers. 77, 1–316. doi: 10.1007/s13225-016-0360-2
Woudenberg, J. H. C., Groenewald, J. Z., Binder, M., Crous, P. W. (2013). Alternaria redefined. Stud. Mycol. 75, 171–212. doi: 10.3114/sim0015
Zhang, W., Groenewald, J. Z., Lombard, L., Schumacher, R. K., Phillips, A. J. L., Crous, P. W. (2021). Evaluating species in botryosphaeriales. Pers.: Mol. Phyl. Evol. Fungi. 46, 63–115. doi: 10.3767/persoonia.2021.46.03
Zhu, H., Pan, M., Bonthond, G., Tian, C., Fan, X. (2019). Diaporthalean fungi associated with canker and dieback of trees from Mount Dongling in Beijing, China. MycoKeys 59, 67. doi: 10.3897/mycokeys.59.38055
Keywords: oak decline, pathogenicity, phylogeny, systematics, Zagros
Citation: Bashiri S and Abdollahzadeh J (2024) Taxonomy and pathogenicity of fungi associated with oak decline in northern and central Zagros forests of Iran with emphasis on coelomycetous species. Front. Plant Sci. 15:1377441. doi: 10.3389/fpls.2024.1377441
Received: 27 January 2024; Accepted: 18 March 2024;
Published: 18 April 2024.
Edited by:
Franco Nigro, University of Bari Aldo Moro, ItalyReviewed by:
Alan John Lander Phillips, Universidade de Lisboa, PortugalAbhijeet Shankar Kashyap, National Bureau of Agriculturally Important Microorganisms (ICAR), India
Copyright © 2024 Bashiri and Abdollahzadeh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jafar Abdollahzadeh, j.abdollahzadeh@uok.ac.ir
 Samaneh Bashiri
Samaneh Bashiri Jafar Abdollahzadeh
Jafar Abdollahzadeh