- 1Citrus Research and Education Center, Department of Microbiology and Cell Science, Institute of Food and Agricultural Sciences (IFAS), University of Florida, Lake Alfred, FL, United States
- 2Citrus Research and Education Center, Horticultural Sciences Department, Institute of Food and Agricultural Sciences (IFAS), University of Florida, Lake Alfred, FL, United States
- 3Biochemistry Department, Faculty of Agriculture, Zagazig University, Zagazig, Egypt
Citrus canker disease affects citrus production. This disease is caused by Xanthomonas citri subsp. citri (Xcc). Previous studies confirmed that during Xcc infection, PthA4, a transcriptional activator like effector (TALE), is translocated from the pathogen to host plant cells. PthA4 binds to the effector binding elements (EBEs) in the promoter region of canker susceptibility gene LOB1 (EBEPthA4-LOBP) to activate its expression and subsequently cause canker symptoms. Previously, the Cas12a/CBE co-editing method was employed to disrupt EBEPthA4-LOBP of pummelo, which is highly homozygous. However, most commercial citrus cultivars are heterozygous hybrids and more difficult to generate homozygous/biallelic mutants. Here, we employed Cas12a/CBE co-editing method to edit EBEPthA4-LOBP of Hamlin (Citrus sinensis), a commercial heterozygous hybrid citrus cultivar grown worldwide. Binary vector GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1 was constructed and shown to be functional via Xcc-facilitated agroinfiltration in Hamlin leaves. This construct allows the selection of transgene-free regenerants via GFP, edits ALS to generate chlorsulfuron-resistant regenerants as a selection marker for genome editing resulting from transient expression of the T-DNA via nCas9-mPBE:ALS2:ALS1, and edits gene(s) of interest (i.e., EBEPthA4-LOBP in this study) through ttLbCas12a, thus creating transgene-free citrus. Totally, 77 plantlets were produced. Among them, 8 plantlets were transgenic plants (#HamGFP1 - #HamGFP8), 4 plantlets were transgene-free (#HamNoGFP1 - #HamNoGFP4), and the rest were wild type. Among 4 transgene-free plantlets, three lines (#HamNoGFP1, #HamNoGFP2 and #HamNoGFP3) contained biallelic mutations in EBEpthA4, and one line (#HamNoGFP4) had homozygous mutations in EBEpthA4. We achieved 5.2% transgene-free homozygous/biallelic mutation efficiency for EBEPthA4–LOBP in C. sinensis cv. Hamlin, compared to 1.9% mutation efficiency for pummelo in a previous study. Importantly, the four transgene-free plantlets and 3 transgenic plantlets that survived were resistant against citrus canker. Taken together, Cas12a/CBE co-editing method has been successfully used to generate transgene-free canker‐resistant C. sinensis cv. Hamlin in the T0 generation via biallelic/homozygous editing of EBEpthA4 of the canker susceptibility gene LOB1.
Introduction
Citrus is grown worldwide as one of the most popular fruits, which can be eaten fresh or consumed as juice. However, global citrus production faces many biotic and abiotic challenges, including citrus bacterial canker and Huanglongbing, droughts, flooding, and freezes (Hu et al., 2014; Cimen and Yesiloglu, 2016; Dala-Paula et al., 2019; Wang, 2019). New citrus cultivars are urgently needed to undertake these challenges. CRISPR/Cas mediated genome editing is deemed the most promising approach to breed new citrus cultivars, owing to its short time requirement, precise genetic improvement and predictable results (Chen et al., 2019; Gao, 2021; Cao et al., 2022; Huang et al., 2022a). To date, SpCas9/gRNA from Streptococcus pyogenes, SaCas9/gRNA from Staphylococcus aureus, LbCas12a/crRNA from Lachnospiraceae bacterium and base editor derived from SpCas9/gRNA have been successfully adapted to modify citrus genomes for gene function study and new citrus cultivar breeding (Jia and Wang, 2014a; Jia et al., 2016; Peng et al., 2017; Zhang et al., 2017; Jia et al., 2017a, Jia et al., 2017b; LeBlanc et al., 2018; Zhu et al., 2019; Jia et al., 2019a; Dutt et al., 2020; Huang et al., 2020; Jia and Wang, 2020; Huang and Wang, 2021; Jia et al., 2021; Alquezar et al., 2022; Jia et al., 2022; Mahmoud et al., 2022; Parajuli et al., 2022; Yang et al., 2022; Huang et al., 2022b, 2023; Su et al., 2023).
CRISPR genome editing has been used to breed disease-resistant varieties for many crops (Caserta et al., 2020; Bowen et al., 2022; Liu et al., 2022). For citrus, a lot of work has been done for generating canker-resistant citrus cultivars with most focused on editing Citrus sinensis lateral organ boundary 1 (CsLOB1), the citrus canker susceptibility gene (Hu et al., 2014). Interestingly, editing the DOWNY MILDEW RESISTANCE 6 (DMR6) gene, which encodes 2-oxoglutarate Fe(II)-dependent dioxygenases, in citrus also results in improved resistance to Xcc (Parajuli et al., 2022). Citrus canker is one of the most economically important citrus diseases worldwide and is caused by Xanthomonas citri subsp. citri (Ference et al., 2018). Previous studies demonstrate that during Xcc infection, PthA4, a transcriptional activator like effector (TALE), is transported from Xcc cells to host plant cells. Once inside the cell nucleus, PthA4 binds to the effector binding elements (EBEs) in the promoter region of CsLOB1 (CsLOBP) to activate its expression and expression of downstream genes, which consequently leads to canker symptom formation (Hu et al., 2014; Zhang et al., 2016; Duan et al., 2018; Zou et al., 2021; de Souza-Neto et al., 2023). Therefore, editing either CsLOB1 coding region or the EBE recognized by PthA4 (EBEpthA4) has been adopted to create canker-resistant citrus. For instance, canker-resistant Duncan grapefruit was developed through editing CsLOB1 coding region using SpCas9/gRNA (Jia et al., 2017b). Moreover, canker-resistant Duncan grapefruit, Hamlin, Pummelo and Wanjincheng orange were created by disrupting EBEpthA4 or the TATA box of CsLOB1 using spCas9/gRNA, LbCas12a/crRNA and base editor (Peng et al., 2017; Jia and Wang, 2020; Jia et al., 2022; Huang et al., 2022b). However, it must be kept in mind that all of the aforementioned canker-resistant genome-edited citrus plants are transgenic, which have not been commercialized owing to regulations and public perception concerns (Jia et al., 2006; Gong et al., 2021; Turnbull et al., 2021).
Transgene-free genome editing, on the other hand, is promising to overcome such issues. Multiple strategies have been developed to produce transgene-free plants after target-gene editing with CRISPR/Cas (Kocsisova and Coneva, 2023). Some examples include delivering DNA-free gene editing reagents such as ribonucleoproteins (Woo et al., 2015; Malnoy et al., 2016; Subburaj et al., 2016; Svitashev et al., 2016; Liang et al., 2017; Andersson et al., 2018; Su et al., 2023), novel delivery vectors such as viruses (Mei et al., 2019; Ma et al., 2020; Liu et al., 2023), unconventional selection methods to bypass integration of transgenes (Veillet et al., 2019; Alquezar et al., 2022; Huang et al., 2022b, 2023; Wei et al., 2023), graft-mobile editing systems (Yang et al., 2023), and so on. Initially, Alquezar et al. and Huang et al. independently developed transgene-free citrus using CBE-mediated editing (Alquezar et al., 2022; Huang et al., 2022b). Recently, two studies have been reported to generate transgene-free genome-edited citrus resisting against canker in the T0 generation. Su et al. took advantage of Cas12a/crRNA ribonucleoprotein to disrupt the coding region of LOB1 (Su et al., 2023), and another study employed Cas12/CBE co-editing method to edit EBEPthA4-LOBP of pummelo, which is highly homozygous and relatively easy to work with (Huang et al., 2023). Notably, the latter took advantage of green fluorescent protein for selecting transgene-free transformants, Agrobacterium-mediated transient expression of cytosine base editor (CBE) to edit ALS encoding acetolactate synthase to confer herbicide chlorsulfuron resistance as a selection marker, and Cas12a/CRISPR RNA for editing gene(s) of interest (Huang et al., 2023). Intriguingly, Cas12/CBE co-editing method has been successfully employed for transgene-free genome editing of multiple plant species, including potato, tomato, and tobacco in addition to citrus (Huang et al., 2023).
In this study, we successfully used Cas12/CBE co-editing strategy to modify EBEPthA4-LOBP of sweet orange cv. Hamlin, a heterozygous hybrid between pummelo (C. maxima) and mandarin (C. reticulata) and an important citrus cultivar grown worldwide. Using this co-editing strategy, we have generated multiple transgene-free biallelic/homozygous EBEPthA4-LOBP Hamlin mutants, which are canker resistant and demonstrated its usefulness in genetic improvements of heterozygous commercial citrus varieties.
Materials and methods
Plasmid construction
SpCas9p is one version of codon-optimized SpCas9 (Ma et al., 2015b), which was successfully employed to edit citrus genome (Jia et al., 2021). From 35S-SpCas9p:DunLOBP (Jia et al., 2021), codon-optimized SpCas9p was amplified using primer Cas9p-5-AflII (5′-AGGTCTTAAGGACAAGAAGTACTCGATCGGCCTCGCCATCG GCACCAACAGCGTCGGCTGGGCGGTGATCAC-3′) and Cas9p-3-MluI (5′-AGTCACGCGT CTTCTTTTTCTTAGCCTGTCCGGCCTT-3′). After digestion with AflII and MluI, part of SpCas9p was cloned into nCas9-PBE vector from Addgene (Addgene plasmid #98164) to form nCas9-mPBE vector. nCas9-PBE (plant base editor) was used to perform base editing in rice, wheat and maize (Zong et al., 2017). To construct GFP-p1380N-CmYLCV-nCas9-mPBE, the BamHI- EcoRI-cut nCas9-mPBE fragment was ligated with BamHI- EcoRI-cut GFP-p1380N-CmYLCV-nCas9-PBE (Huang et al., 2023).
From 35S-SpCas9p:DunLOBP (Jia et al., 2021), the AtU6-26 promoter was amplified using AtU6-26-5-XhoI (5′-AGGTCTCGAGTCGTTGAACAACGGAAACTCGACTTGCCTT-3′) and AtU6-26-3-phos (5′-phosphorylated-aatcactacttcgactctagctgt-3′), and the sgRNA-ALSBE-NosT fragment was PCR-amplified using sgRNA-ALSBE1-P1 (5′-phosphorylated-GcaggtcccTcggaggatgatGTTTTAGAGCTAGAAATAGCAAGT-3′) and NosT-3-SpeI (5′-aggtactagTCCGATCTAGTAACATAGATGACA-3′). Through three-way ligation, XhoI-cut AtU6-26 and SpeI-digested sgRNA-ALSBE1-NosT were inserted into XhoI-XbaI-treated pUC-NosT-MCS to construct pUC-NosT-AtU6-26-sgRNA-ALSBE1. pUC-NosT-MCS was constructed previously, which harbors the XhoI-AscI-XbaI-PmeI multiple enzyme sites (Jia et al., 2019a). From pUC-NosT-AtU6-26-sgRNA-ALSBE (Huang et al., 2023), the AtU6-26-sgRNA-ALSBE2 fragment was amplified using AtU6-26-5-XhoI and NosT-3-BsaI (5′-ATTCGGTCTCCCATGTATGAT AATCATCGCAAGACCGGC-3′), and the AtU6-26-sgRNA-ALSBE1 was PCR-amplified using AtU6-26-5-BsaI (5′-TCGAGGTCTCCCATGTCGTTGAACAACGGAAACTCGACTTGCCTT-3′) and NosT-3-SpeI from pUC-NosT-AtU6-26-sgRNA-ALSBE1. Through three-way ligation, XhoI-BsaI-cut XhoI-AtU6-26-sgRNA-ALSBE2-BsaI and BsaI-SpeI-digested BsaI-AtU6-26-sgRNA-ALSBE1-SpeI were inserted into XhoI-XbaI-treated pUC-NosT-MCS to construct pUC-NosT-AtU6-26-sgRNA-ALSBE2-ALSBE1. Subsequently, the EcoRI-NosT-AtU6-26-sgRNA-ALSBE2-AtU6-26-sgRNA-ALSBE1-PmeI fragment from pUC-NosT-AtU6-26-sgRNA-ALSBE2-ALSBE1 were cloned into EcoRI-PmeI-cut GFP-p1380N-CmYLCV-nCas9-mPBE to generate GFP-p1380N-CmYLCV-nCas9-mPBE:ALS2:ALS1.
Finally, the AscI-PmeI-cut CmYLCV-nCas9-mPBE:ALS2:ALS1 fragment GFP-p1380N-CmYLCV-nCas9-mPBE:ALS2:ALS1 was clone into AscI-PmeI-cut vector GFP-p1380N-ttLbCas12a:LOBP1-AscI-XbaI-PmeI, which was constructed previously (Huang et al., 2023), to form GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1 (Figure 1, Supplementary Figure 1). Notably, GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1 contained both GFP and nptII selectable genes that can be used for selection of putative non-transgenic transformants (Figure 1, Supplementary Figure 1) with GFP being an easier option for visual selection in our experience (Huang et al., 2023).
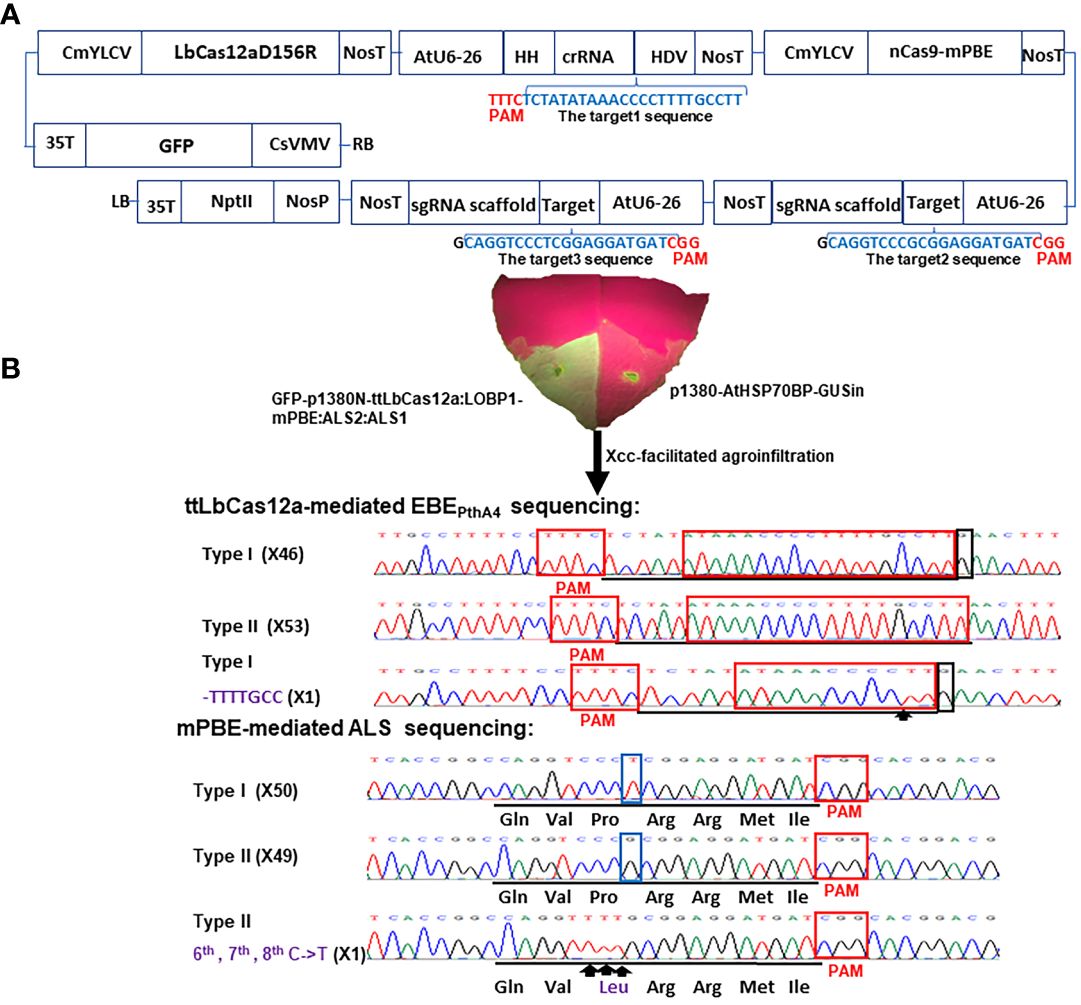
Figure 1 Schematic representation of the binary vector GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1 and its functional test. (A) Schematic diagram of GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1. LOBP: the promoter region of LOB1. LB and RB, the left and right borders of the T-DNA region; CsVMV, the cassava vein mosaic virus promoter; GFP, green fluorescent protein; 35T, the cauliflower mosaic virus 35S terminator; CmYLCV, the cestrum yellow leaf curling virus promoter; NosP and NosT, the nopaline synthase gene promoter and its terminator; ttLbCas12a, temperature-tolerant LbCas12a containing the single mutation D156R; AtU6-26, Arabidopsis U6-26 promoter; target1, the 23 nucleotides of Type II LOBP highlighted by blue, was located downstream of protospacer-adjacent motif (PAM); HH, the coding sequence of hammerhead ribozyme; HDV, the coding sequence of hepatitis delta virus ribozyme; nCas9-mPBE, a plant codon-optimized base editor composed of rat cytidine deaminase APOBEC1, Cas9-D10A nickase (nCas9) and uracil glycosylase inhibitor (UGI); AtU6-26, Arabidopsis U6-26 promoter; target2 and target3, the 20 nucleotides of two CsALS alleles highlighted by blue, were located upstream of protospacer-adjacent motif (PAM); NptII, the coding sequence of neomycin phosphotransferase II. (B) Xcc-facilitated agroinfiltration of the binary vector GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1. Xcc-pre-treated Hamlin leaf was agroinfiltrated with Agrobacterium cells harboring vector GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1. After four days, GFP fluorescence was observed and photographed. A negative control was Agrobacterium cells harboring p1380-AtHSP70BP-GUSin. ttLbCas12a-directed LOBP indels and mPBE-mediated CsALS base editing were analyzed through Sanger sequencing. Among 200 colonies sequenced, there were expected mutations for LOBP and CsALS. x inside the parentheses indicates number of Sanger sequencing. The targeted sequence within LOBP and CsALS was underlined by black lines, and the mutant site was pointed out with arrows and highlighted by purple. Type I LOBP has one more G than Type II LOBP downstream of EBEPthA4, and the G was highlighted by a black rectangle. The nucleotides different between two alleles of CsALS were highlighted by blue rectangles.
The binary vector GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1 was transformed into A. tumefaciens strain EHA105 via electroporation. Recombinant Agrobacterium cells were employed for Xcc-facilitated agroinfiltration or citrus epicotyl transformation.
Xanthomonas citri subsp. citri-facilitated agroinfiltration in Hamlin
Grown in a greenhouse at 28°C, C. sinensis cv. ‘Hamlin’ was pruned to generate uniform shooting before Xcc-facilitated agroinfiltration. It should be pointed out that ttLbCas12a performed better at 28°C than at 22°C in a previous study (Schindele and Puchta, 2020).
Xcc-facilitated agroinfiltration was performed as described previously (Jia and Wang, 2014b). Briefly, the fully-expanded young Hamlin leaves were first treated with Xcc, which was re-suspended in sterile tap water at a concentration of 5 × 108 CFU/mL. Twenty-four hours later, the Xcc-treated leaf areas were inoculated with Agrobacterium cells harboring vectors GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1, or p1380-AtHSP70BP-GUSin. p1380-AtHSP70BP-GUSin was constructed before (Jia and Wang, 2014b), and used as a no-GFP-fluorescence control. Four days after agroinfiltration, GFP was observed and photographed, and genomic DNA was extracted from the Hamlin leaves treated by agroinfiltration.
Agrobacterium-mediated Hamlin transformation
As described previously (Jia et al., 2019b), Hamlin epicotyl transformation was conducted with minor modifications. Briefly, Hamlin epicotyl explants were co-incubated with Agrobacterium cells harboring the binary vector GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1. After cocultivation in darkness for 3 days at 25°C, the epicotyl explants were placed on regeneration medium containing 50 mg/L kanamycin for one week at 28°C, and then the epicotyl explants were placed on regeneration medium containing 40 μg/L chlorsulfuron at 28°C (Veillet et al., 2019). All regenerated plants were subjected to genome editing analysis and GFP inspection. It is noteworthy that we initially placed Hamlin epicotyls on medium with 100 mg/L kanamycin for two weeks; three rounds of selection with 40 ng/mL chlorsulfuron, each lasting two weeks as reported previously (Veillet et al., 2019). However, very few shoots were regenerated. Based on our observations, we adapted the protocol by reducing the kanamycin concentration to 50 mg/L for one week, and conducting two rounds of selection with 40 ng/mL chlorsulfuron, each extending to three weeks.The GFP-positive and transgene-free regenerated shoots were selected and micro-grafted on ‘Carrizo’ citrange rootstock plants (C. sinensis (L.) Osbeck x Poncirus trifoliata (L.) Raf.) for further analysis. Six months later, they were used for PCR analysis with the primers Npt-Seq-5 (5′-TGTGCTCGACGTTGTCACTGAAGC-3′) and 35T-3 (5´-TTCGGGGGATCTGGATTTT AGTAC-3′).
PCR amplification of mutagenized CsALS and LOBP
Genomic DNA was extracted from the Xcc-facilitated-agroinfiltrated Hamlin leaves or each regenerated Hamlin line. To analyze GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1-mediate CsALS and LOBP mutations, PCR was carried out using either primers CsALSP1 (5′- Atctgtatcgccacctcggggcccggc-3′) and CsALSP2 (5′- TGGCCGCCCAGATGTTGCTAAAAGG-3′), or primers LOB21 (5′- ACACCTTGGTAATTTTGACATTAGGTA-3′) and LOB22 (5′- TGAGAGAAGAAAACTGTTGGGTTGTAG-3′). The PCR products were sequenced through cloning and colony sequencing. Primers CsALSP1 and CsALSP2 were designed to amplify the sgRNA-target CsALS region for the mutation analysis via Sanger sequencing, and primers LOB21 and LOB2 were employed to analyze LOBP. For PCR product direct sequencing, the CsALSP1/CsALSP2-amplified and LOB21/LOBP22-amplified PCR products were purified and subjected to sequencing using CsALSP3 (5′- tggtcagcgggctcgccgacgcgct-3′) as to CsALS, and primer LOB4 (5′-CGTCATTCAATTAAAATTAATGAC-3′) as to LOBP. Direct sequencing of PCR products was employed to genotype CRISPR/Cas-mediated indels (Ma et al., 2015a; Jia et al., 2016). Ten random colonies for each transgenic and transgene-free Hamin line were selected for sequencing. Mutation rates were calculated by the mutant colonies/10 colonies. Chromas Lite program was used to analyze the sequencing results.
GFP detection
An Omax camera was installed onto a Zeiss Stemi SV11 dissecting microscope for photographing GFP fluorescence. Under illumination of the Stereo Microscope Fluorescence Adapter (NIGHTSEA), GFP fluorescence of the Hamlin leaves treated by Xcc-facilitated agroinfiltration and GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1-transformed Hamlin was observed. Subsequently, the Hamlin leaves were photographed with the Omax Toupview software connected to the Omax camera.
Canker symptom assay in citrus
Wild type, transgenic and transgene-free Hamlin plants were grown in a greenhouse at the Citrus Research and Education Center, University of Florida. Prior to Xcc inoculation, all plants were trimmed to generate new shoots. Leaves of similar age were infiltrated with either Xcc or XccΔpthA4:dLOB1.5 (5 × 108 CFU/mL) using needleless syringes. At three, six and nine days post inoculation (DPI), canker symptoms were observed and photographed.
Whole genome sequencing analysis of transgene-free Hamlin plant #HamNoGFP4
Genomic DNA of transgene-free Hamlin plant #HamNoGFP4 was subjected to whole genome sequencing at Novogene (Sacramento, CA, USA). The whole genome data of #HamNoGFP4 were released at NCBI (NCBI Bio-project ID: PRJNA1073671). DNA Library construction, sequencing, and data analysis were performed as follows. Following the manufacturer’s protocol of short read DNA sequencing from Illumina (Kozarewa et al., 2009), the library was prepared. After quality control, quantification, and normalization of the DNA libraries, 150-bp paired-end reads were generated using the Illumina NovaSeq 6000 platform according to the manufacturer’s instructions at Novogene. The raw paired-end reads were filtered to remove the low-quality reads using fastp program version 0.22.0 (Chen et al., 2018). To assess the target site mutations of mutated plants, the high quality paired-end short genomic reads were mapped to sweet orange (C. sinensis) (Wang et al., 2021) reference genome using Bowtie2 software version 2.2.6 (Langmead and Salzberg, 2012). The mutations (single nucleotide polymorphisms, deletions and insertions) for the mutated plant genomes were identified using the SAMtools package version 1.2 (Li et al., 2009) and deepvariant program version 1.4.0 (Poplin et al., 2018). The identified mutations were filtered by quality and sequence depth (mapping quality > 10 and mapping depth > 10). The mutations of target sites were visualized using IGV software version 2.15.4 (Robinson et al., 2011). The off-target sites were predicted using CRISPR-P 2.0 (Liu et al., 2017) and the Cas-OFFinder (Bae et al., 2014) program and aligning target sequence with whole genome using blast program. Based on the mapping results, mutations of off-target sites were detected using the SAMtools package version 1.2 and deepvariant program version 1.4.0. To detect the potential foreign DNA contamination, the high quality paired-end short genomic reads were mapped to the plasmid sequences using Bowtie2 software version 2.2.6.
Results
Construction of the binary vector GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1
In a previous study, GFP-p1380N-ttLbCas12a:LOBP1-PBE:ALS was constructed to produce transgene-free pummelo, which is a highly homozygous diploid (Wu et al., 2018). One sgRNA was designed to target both alleles of acetolactate synthase (CsALS) and another sgRNA for both alleles of EBEPthA4-LOBP in pummelo (Huang et al., 2023). C. sinensis cv. Hamlin is a heterozygous hybrid between pummelo (C. maxima) and mandarin (C. reticulata). In this study, the promoter region of CsLOB1 (CsLOBP) from mandarin was designated as Type I LOBP, whereas CsLOBP from pummelo was named as Type II LOBP. As a result, Hamlin has Type I LOBP and Type II LOBP. Notably, Type I LOBP has one more G than Type II LOBP downstream of EBEPthA4, whose detailed sequence is ATAAACCCCTTTTGCCTT (Figure 1). Similarly, two alleles of CsALS from mandarin and from pummelo were named as Type I CsALS and Type II CsALS, respectively. As a result, Hamlin also has Type I CsALS and Type II CsALS. Notably, Type I CsALS has T at the position of 9, and Type II CsALS has G (the PAM labeled as positions from 21 to 23) (Figure 1). Consequently, two different sgRNAs were designed to target the ALS gene of Hamlin in vector GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1 (Figure 1). On the other hand, one crRNA was designed to target the conserved sequence of both alleles of EBEPthA4-LOBP of Hamlin. Based on the reference genome of sweet orange genome (V3) from HZAU (http://citrus.hzau.edu.cn/index.php) (Liu et al., 2017), the genomic position for target 1 (TCTATATAAACCCCTTTTGCCTT) is from 3970724-3970746 on chromosome 7. The genomic positions for target 2 (caggtcccGcggaggatgat) and targets 3 (caggtcccTcggaggatgat) are from 15794167-15794186 on chromosome 7, since target 2 and targets 3 are two alleles of CsALS (Figure 1A). To enhance editing efficiency, base editor nCas9-PBE of GFP-p1380N-ttLbCas12a:LOBP1-PBE:ALS was optimized to form nCas9-mPBE in GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1, which harbors the codon-optimized SpCas9p backbone for plants (Figure 1A). SpCas9p has been used to produce homozygous/biallelic mutations for multiple plant species in the T0 generation, such as Arabidopsis, rice, and citrus (Ma et al., 2015b; Jia and Wang, 2020).
Evaluation of GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1 efficacy via Xcc-facilitated agroinfiltration
Xcc-facilitated agroinfiltration of Hamlin leaf was used to test whether GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1 could be employed to edit citrus EBEPthA4-LOBP by ttLbCas12a and edit CsALS by nCas9-mPBE. It must be pointed out that cestrum yellow leaf curling virus (CmYLCV) promoter was used to drive ttLbCas12a and nCas9-mPBE expression in GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1 (Figure 1, Supplementary Figure 1), since CmYLCV outperformed CaMV 35S for citrus genome editing (Huang et al., 2022a). In addition, in order to promote editing, the coding sequence of hammerhead ribozyme (HH) at the 5’ end of crRNA, and the coding sequence of hepatitis delta virus ribozyme (HDV) were placed at the 3’ end of crRNA (Figure 1, Supplementary Figure 1) (Tang et al., 2017). Genomic DNA was extracted from GFP-expressing Hamlin leaf and subjected to PCR, ligation and E. coli transformation. Sanger sequencing results showed that one colony contained ttLbCas12a-directed indels in EBEPthA4-LOBP among one batch of 100 colonies sequenced, and one colony contained the nCas9-PBE-mediated cytosine-to-thymine base conversion in CsALS among another batch of 100 colonies sequenced (Figure 1B). Therefore, GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1 was functional to edit both EBEPthA4-LOBP and CsALS in Hamlin. Though the same genomic DNA was used as PCR template, two pairs of primers, CsALSP1/CsALSP2 and LOB21/LOBP22, were separately used to analyze mutant CsALS and LOBP. It is likely that ttLbCas12a-directed indels in LOBP and the nCas9-PBE-mediated cytosine-to-thymine base conversion in CsALS took place in the same cells even though we could not totally exclude other possibilities.
Generation of transgene-free EBEPthA4-LOBP edited Hamlin via the co-editing method
Hamlin epicotyls were transformed with recombinant Agrobacterium cells harboring GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1 (Figure 1) (Jia et al., 2019b). Three-day after co-cultivation, the epicotyls were first selected on 50 mg/L kanamycin for one week, then transferred to selective medium containing 40 μg/L chlorsulfuron for six weeks, during which new chlorsulfuron-containing medium was used after three-week cultivation. After six week herbicide selection, Hamlin epicotyls were cultivated on chlorsulfuron-free medium. In addition, the shoot generation was done at 28°C to facilitate LbCas12aD156-mediated EBEPthA4-LOBP editing (Schindele and Puchta, 2020; Jia et al., 2022).
In the presence of chlorsulfuron, 77 shoots were established. Among them, 8 shoots were GFP-positive, named as #HamGFP1 to #HamGFP8. Unexpectedly, five lines (#HamGFP4, #HamGFP5, #HamGFP6, #HamGFP7 and #HamGFP8) died after grafting, however, three GFP-positive shoots (#HamGFP1, #HamGFP2 and #HamGFP3) survived (Figure 2A). On the other hand, 69 shoots had no GFP expression, designated as #HamNoGFP1 to #HamNoGFP69 (Figure 2A).

Figure 2 GFP detection and PCR verification of GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1-transformed and transgene-free genome-edited Hamlin plants. (A) GFP fluorescence was observed in transgenic Hamlin plants, whereas wild type and transgene-free genome-edited plants did not show GFP. (B) Using a pair of primers Npt-Seq-5 and 35T-3PCR, wild type, transgenic and transgene-free Hamlin plants were analyzed. The wild type Hamlin and plasmid GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1 were used as controls. M, 1kb DNA ladder.
Based on the results of direct sequencing of PCR products from transgenic Hamlin, ttLbCas12a-directed indels and mPBE-mediated C-to-T conversions were observed in #HamGFP1, #HamGFP2 and #HamGFP3 (Figure 3). Among the 69 no-GFP-expressing shoots, four shoots, #HamNoGFP1 to #HamNoGFP4, had mutations in CsALS and EBEPthA4-LOBP (Figure 4), whereas the rest, from #HamNoGFP5 to #HamNoGFP69, had wild type CsALS and EBEPthA4-LOBP (Figure 4). Therefore, the four no-GFP-expressing lines (#HamNoGFP1, #HamNoGFP2, #HamNoGFP3, #HamNoGFP4) are likely transgene-free EBEPthA4-LOBP-edited lines.
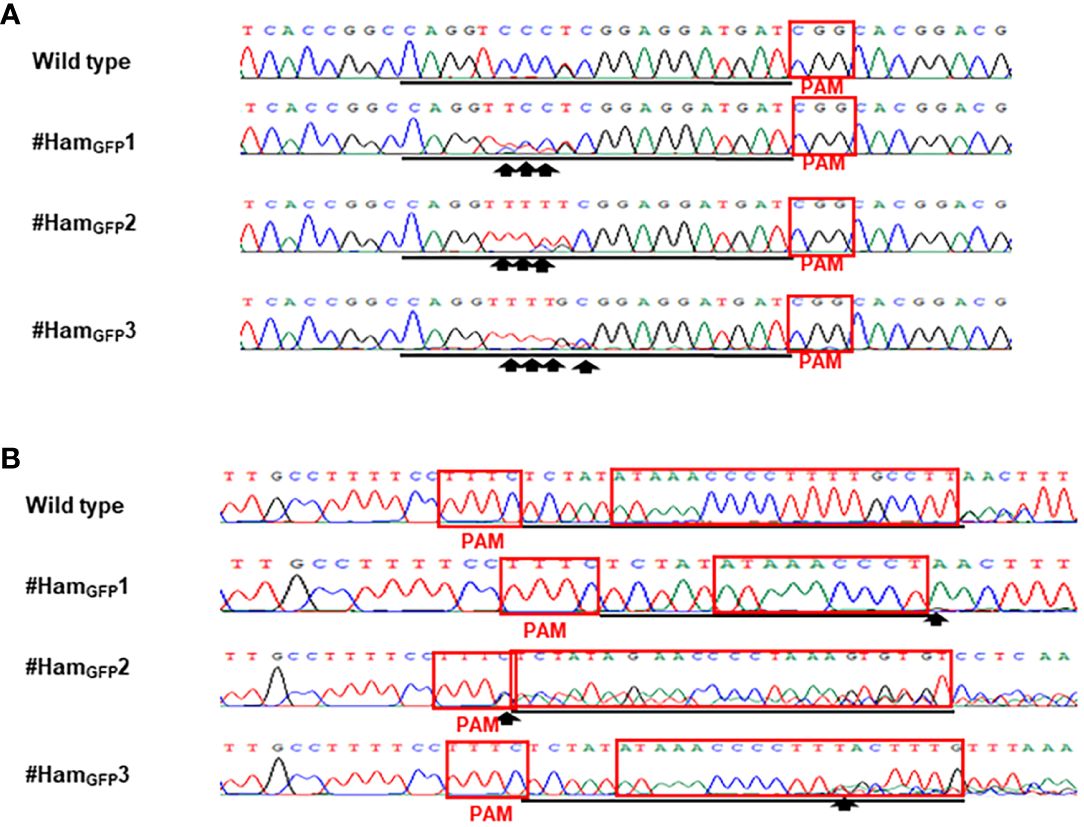
Figure 3 Detection of genome editing of GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1-transformed Hamlin by direct sequencing of CsALS PCR products (A) and LOBP PCR products (B). (A) The chromatograms of direct sequencing of CsALS PCR products. Primers CsALSP1 and CsALSP2 were used to amplify CsALS from wild type and transgenic Hamlin. Direct sequencing primer was CsALSP3. The base editing sites were shown by arrows. (B) The chromatograms of direct sequencing of LOBP PCR products. Primers LOB21 and LOB22 were used to amplify LOBP from wild type and transgenic Hamlin. Direct sequencing primer was LOB4. The mutation site or the beginning sites of multiple peaks were shown by arrows. The targeted sequence was underlined by black lines and EBEPthA4-LOBP was highlighted by red rectangles.
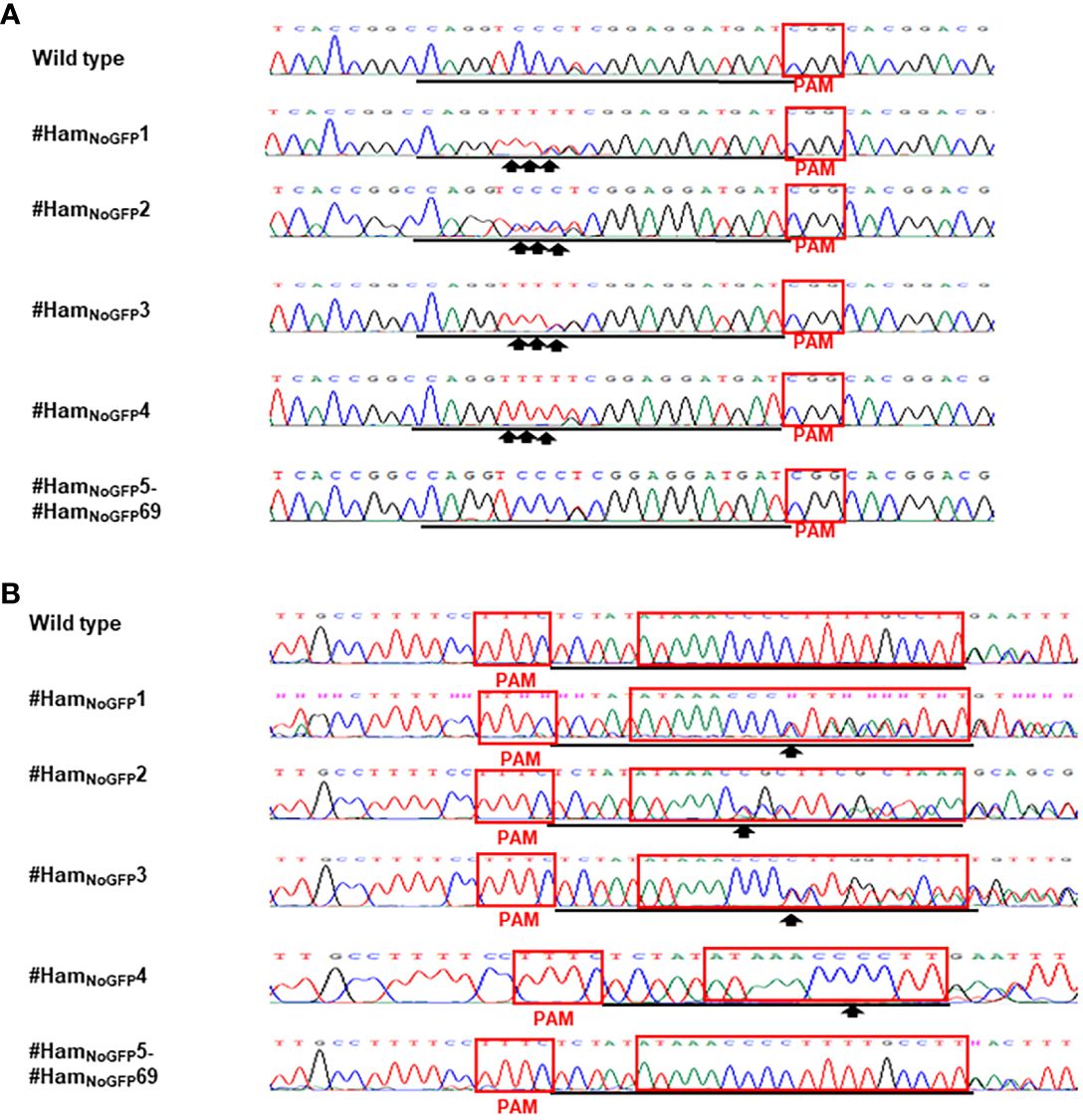
Figure 4 Detection of genome editing of no-GFP-expressing Hamlin by direct sequencing of CsALS PCR products (A) and LOBP PCR products (B). (A) The chromatograms of direct sequencing of CsALS PCR products. Primers CsALSP1 and CsALSP2 were used to amplify CsALS from wild type and transgenic Hamlin. Direct sequencing primer was CsALSP3. The base editing sites were shown by arrows. (B) The chromatograms of direct sequencing of LOBP PCR products. Primers LOB21 and LOB22 were used to amplify LOBP from wild type and transgenic Hamlin. Direct sequencing primer was LOB4. The mutation site or the beginning sites of double peaks were shown by arrows. The targeted sequence was underlined by black lines and EBEPthA4-LOBP was highlighted by red rectangles.
To further verify whether the four no-GFP-expressing lines are transgene-free, we analyzed nptII gene in GFP-positive shoots (#HamGFP1 to #HamGFP3) and no-GFP-expressing shoots (#HamNoGFP1 to #HamNoGFP4). Using a pair of primers Npt-Seq-5 and 35T-3, nptII gene was subjected to PCR analysis. Three GFP-positive Hamlin had the expected nptII PCR products (Figure 2B), which verified that #HamGFP1 to #HamGFP3 were transgenic. No nptII PCR products were observed in no-GFP-expressing Hamlin lines from #HamNoGFP1 to #HamNoGFP4. The results confirmed that #HamNoGFP1, #HamNoGFP2, #HamNoGFP3, and #HamNoGFP4 were transgene-free plants (Figure 2B).
Sanger sequencing was employed to analyze mutation genotypes of the edited lines through cloning. As for EBEPthA4-LOBP, the results demonstrated that #HamGFP1, #HamGFP2 and #HamGFP3 contained biallelic, chimeric and chimeric mutations, respectively (Supplementary Figures 2, 3). As for ALS, #HamGFP1, #HamGFP2 and #HamGFP3 contained chimeric, biallelic and biallelic mutations, respectively (Supplementary Figures 2, 3). Intriguingly, #HamGFP3 harbored C to T substitutions at C10 of CsALS sgRNA (Supplementary Figure 3A), which is somehow out of nCas9-PBE-mediated C-to-T conversion ranges from position 3 to 9 within the protospacer, counting the PAM as positions 21-23 (Zong et al., 2017). Previous study already showed that CBE could convert C to T outside the editing window (Lv et al., 2020). In addition, 7 colonies had the C to T substitution at C10 among 10 colonies sequenced. Thus, the C to T substitutions at C10 of CsALS in #HamGFP3 could attribute to nCas9-PBE activity rather than sequencing errors. #HamGFP2 contained 24 bps deletions from EBEPthA4-LOBP, including part of PAM (Supplementary Figure 2D).
For the transgene-free edited lines, Sanger sequencing results showed that #HamNoGFP1, #HamNoGFP2, #HamNoGFP3 and #HamNoGFP4 contained biallelic, biallelic, biallelic and homozygous mutations, respectively, in EBEPthA4-LOBP (Figures 5B, D, 6, B). As for ALS, Sanger sequencing results indicated that #HamNoGFP1, #HamNoGFP2, #HamNoGFP3 and #HamNoGFP4 harbored biallelic, chimeric, biallelic and homozygous mutations, respectively (Figures 5A, C, 6A, C). Notably, #HamNoGFP4 had homozygous mutations in both EBEPthA4-LOBP and CsALS (Figures 6C, D).
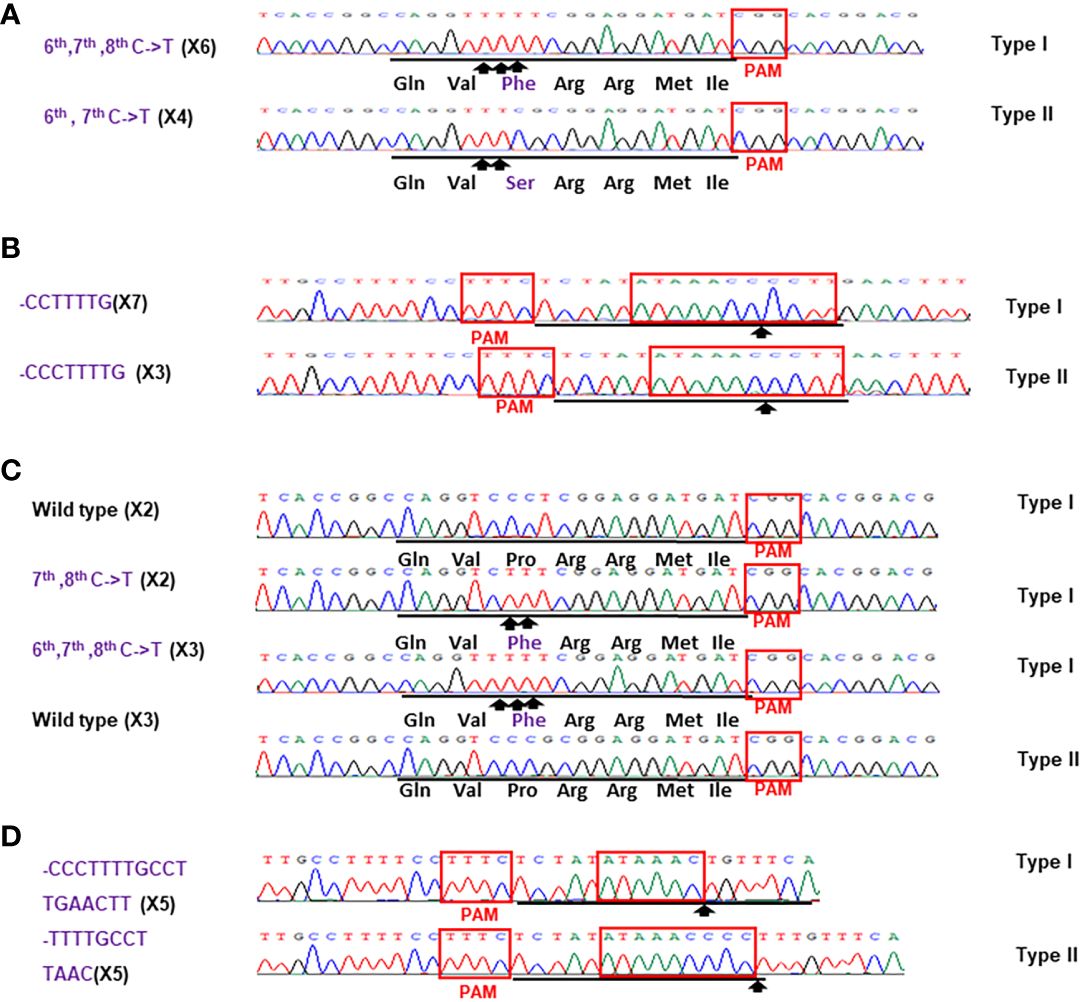
Figure 5 Sanger analysis of #HamNoGFP1 (A, B) and #HamNoGFP2 (c, d). Sanger sequencing results of #HamNoGFP1 and #HamNoGFP2. (A) As for CsALS of #HamNoGFP1, Type I allele contained 6th, 7th, 8th C->T changes, and Type II allele was 6th, 7th C->T mutant among 10 colonies sequenced. (B) As for EBEPthA4-LOBP of #HamNoGFP2, Type I allele had CCTTTTG deletion, and Type II allele had CCCTTTTG deletion. (C) As for CsALS of #HamNoGFP2, wild type and mutants were present among 10 colonies sequenced. (D) As for EBEPthA4-LOBP of #HamNoGFP2, five of them are CCCTTTTGCCTTGAACTT deletion from Type I allele, and five of them are TTTTGCCTTAAC deletion from Type II allele.
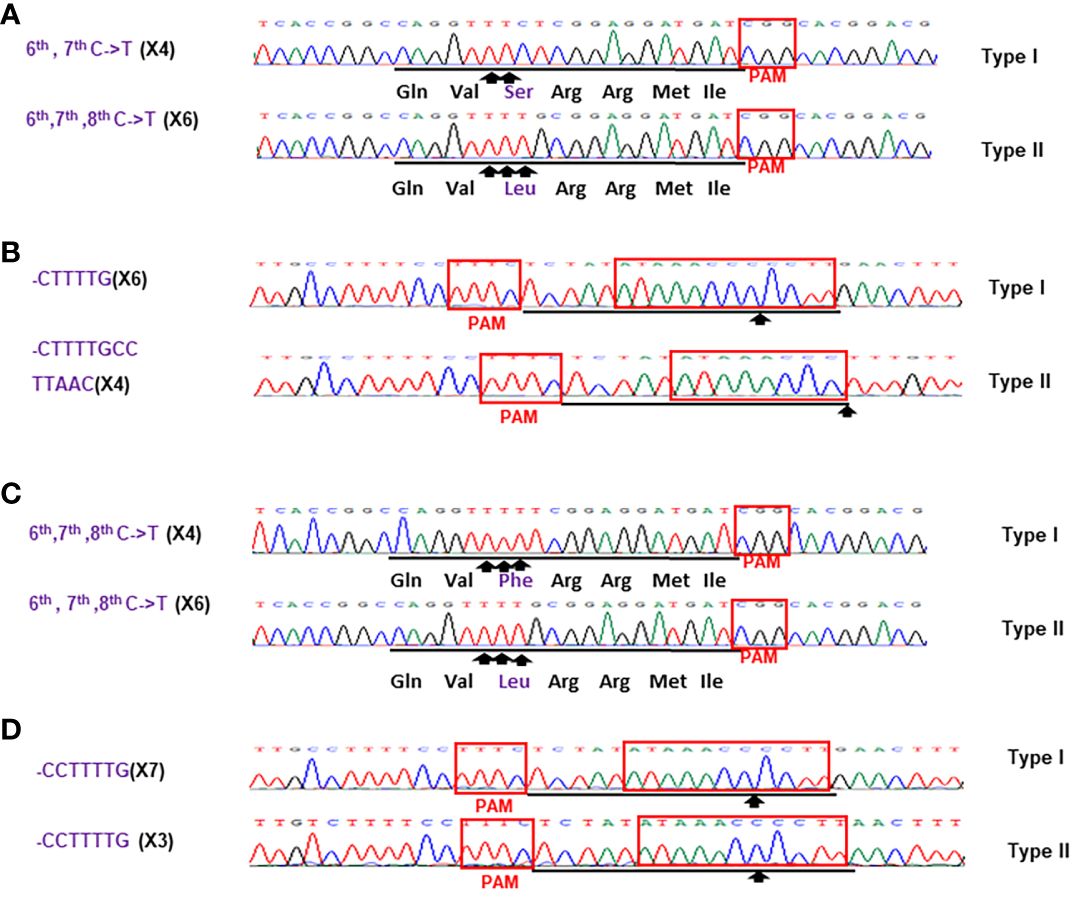
Figure 6 Sanger analysis of #HamNoGFP3 (A, B) and #HamNoGFP4 (C, D). Sanger sequencing results of #HamNoGFP3 and #HamNoGFP4. (A) As for CsALS of #HamNoGFP3, Type I allele was 6th, 7th C->T mutant, and Type II allele had 6th, 7th, 8th C->T mutation among 10 colonies sequenced. (B) As for EBEPthA4-LOBP of #HamNoGFP3, Type I allele harbored CTTTTG deletion, and Type II allele contained CTTTtGCcttAAC deletion. (C) As for CsALS of #HamNoGFP4, Type I and Type II allele were 6th, 7th, 8th C->T mutant among 10 colonies sequenced. (D) As for EBEPthA4-LOBP of #HamNoGFP4, Type I and Type II allele had CCTTTTG deletion.
Sanger sequencing results of transgenic and transgene-free Hamlin demonstrated that EBEPthA4-LOBP mutations were independent of those of ALS, which is consistent with the previous study in pummelo (Huang et al., 2023). ttLbCas12a deleted several bps from the target site, which occurred ≥10th bp distal to the PAM site except that of #HamGFP2 (Figure 5, 6, Supplementary Figures 2, 3), which is consistent with previous work in citrus (Jia et al., 2022; Huang et al., 2023). The #HamGFP2 harbored 24 bps deletions overlapping with PAM in EBEPthA4-LOBP (Supplementary Figure 2D). nCas9-mPBE catalyzed the targeted conversion of cytosine to thymine from position 6 to 8 within the protospacer except that of #HamGFP3, which is consistent with previous work in rice, wheat and maize (Zong et al., 2017). Notably, the EBEPthA4-LOBP mutation rates were 100% in #HamGFP1, #HamGFP2, #HamNoGFP1, #HamNoGFP2, #HamNoGFP3 and #HamNoGFP4 (Supplementary Figure 2, Figures 5, 6).
Canker resistance of transgenic and transgene-free Hamlin plants
Since Hamlin plants from #HamNoGFP5 to #HamNoGFP69 had no mutations in EBEPthA4-LOBP (Figure 4), they were not tested with Xcc inoculation. Three transgenic plants (#HamGFP1-3) and four transgene-free plants (#HamNoGFP1-4) were evaluated for canker resistance. Wild type Hamlin was used as a control. All plants were inoculated with Xcc and XccΔpthA4:dLOB1.5 at a concentration of 5 × 108 CFU/mL. XccΔpthA4:dLOB1.5 was included as a positive control because dLOB1.5 is a designed TALE, which binds to the sequence 5´- TAAAGCAGCTCCTCCTCATCCCTT- 3′ in the promoter region of LOB1, away from the EBE region, to activate LOB1 expression (Supplementary Figure 4A) (Jia and Wang, 2020). Therefore, dLOB1.5 can mimic PthA4 function, and XccΔpthA4:dLOB1.5 can cause citrus canker, giving that its recognizing sequence is intact (Jia and Wang, 2020) whereasXccΔpthA4 cannot cause canker (Hu et al., 2014). Sanger sequencing results indicated that the dLOB1.5 binding sites were intact among all Hamlin plants (Supplementary Figure 4B).
At three, six and nine days post inoculation (DPI) with Xcc, canker symptoms were observed on wild type Hamlin plants, whereas no canker symptoms were observed on transgene-free #HamNoGFP1, #HamNoGFP2; #HamNoGFP3, #HamNoGFP4 and three transgenic Hamlin plants (Figure 7, Supplementary Figure 5). The results indicated that all plants, transgenic or not, harboring 100% mutations in EBEPthA4-LOBP (Figures 5, 6, Supplementary Figure 2), resisted against Xcc infection as expected. Intriguingly, #HamGFP3 containing 90% mutation in EBEPthA4-LOBP also resisted against Xcc (Figure 7, Supplementary Figure 3). Consistently, Cas9/sgRNA-transformed DLOB9 harboring 89.36% mutation rate in CsLOB1 also showed Xcc resistance in a previous study (Jia et al., 2017a). At three, six and nine DPI with XccΔpthA4:dLOB1.5, canker symptoms developed on all treated plants (Figure 7, Supplementary Figure 5), since there was no editing in the dLOB1.5 binding sites (Supplementary Figure 4B). Therefore, it was the EBEPthA4-LOBP disruption that conferred Hamlin resistance against Xcc.
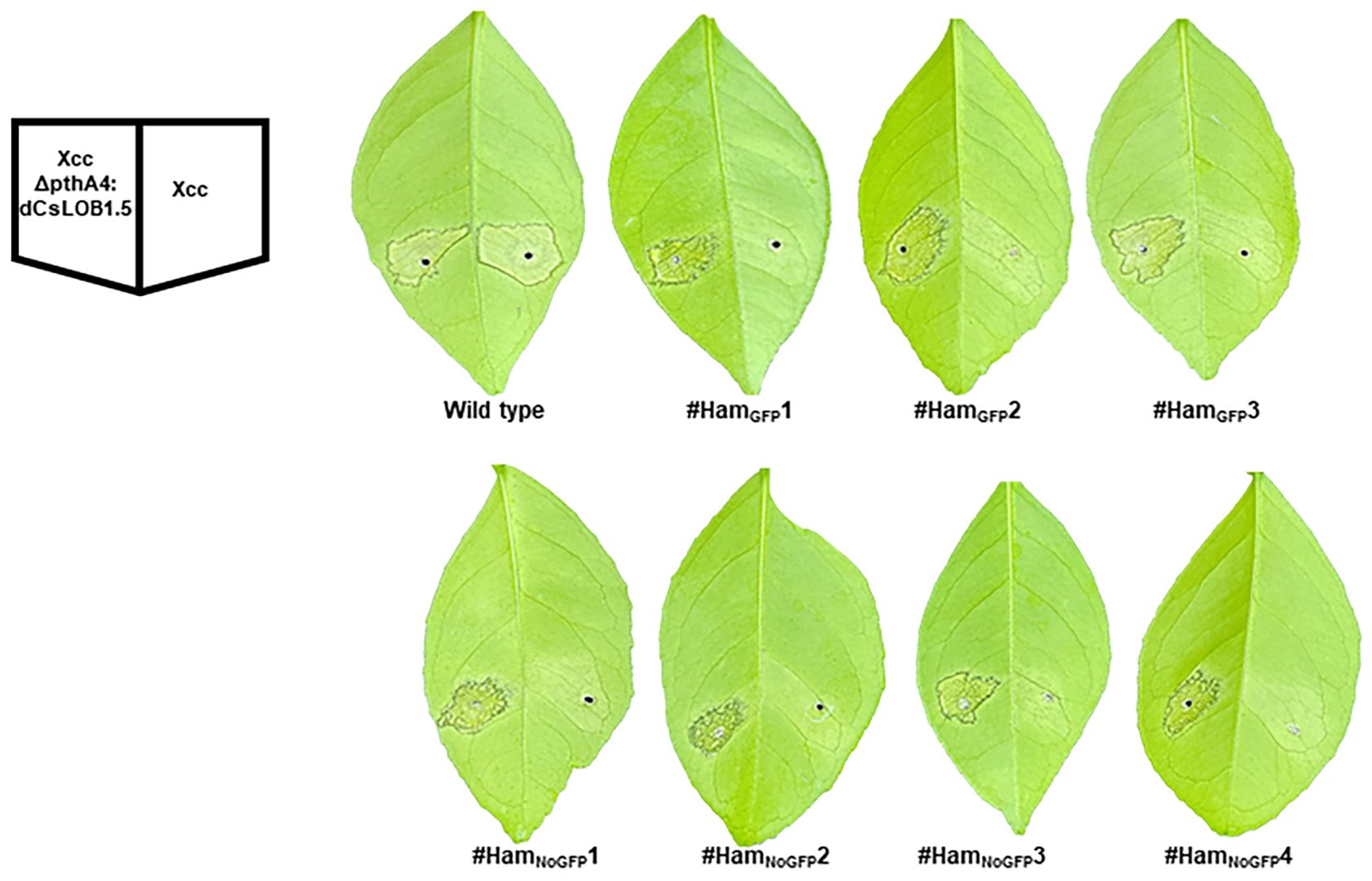
Figure 7 Canker-resistance in the transgenic and transgene-free EBEPthA4-LOBP-edited Hamlin plants. Six days post Xcc inoculation, citrus canker symptoms were observed on wild type Hamlin, whereas no canker symptoms were observed on LOBP-edited Hamlin plants. As expected, XccpthA4:Tn5 (dCsLOB1.5) caused canker symptoms on all plants. dCsLOB1.5 induces LOB1 to cause canker symptoms by recognizing a different region from EBEPthA4-LOBP.
Whole genome sequencing analysis of #HamNoGFP4
To further confirm whether the transgene-free genome-edited lines are indeed free of plasmid sequences, we conducted a whole genome sequencing analysis of #HamNoGFP4. #HamNoGFP4 was selected because it contains homozygous EBEPthA4-LOBP and CsALS mutations (Figures 6C, D). We obtained high quality paired-end short genomic reads for #HamNoGFP4 which were mapped to the T-DNA sequence corresponding to GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1 using Bowtie2 software version 2.2.6. No T-DNA sequences were mapped to the genomic DNA of #HamNoGFP4, confirming it was transgene-free. Furthermore, whole genome sequencing analysis indicated that #HamNoGFP4 harbored homozygous mutations in both EBEPthA4-LOBP and ALS (Supplementary Figure 6), which was consistent with Sanger sequencing results (Figure 6).
To analyze off-target mutations, CRISPR-P 2.0 (Liu et al., 2017) and the Cas-OFFinder (Bae et al., 2014) program were used to find the potential off-targets of crRNAs and sgRNAs of GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1. No potential off-target was identified for EBEPthA4-LOBP and CsALS (mismatch number ≤3). Since no potential off-target was identified for EBEPthA4-LOBP and CsALS (mismatch number ≤3), we changed the mismatch number ≤4. Consequently, 1 potential off-target was identified for EBEPthA4-LOBP, whereas 1 and 4 potential off-targets were identified for type I and type II CsALS alleles, respectively. However, analyses of the whole genome sequencing for the potential off-targets (mismatch number ≤4) did not identify any off-target mutations for both EBEPthA4-LOBP and CsALS.
Discussion
In this study, we successfully employed the co-editing strategy, which couples ttLbCas12a with base editor nCas9-mPBE and GFP selection, to produce transgene-free canker-resistant Hamlin in the T0 generation via transient expression. For this purpose, the binary vector GFP-p1380N-ttLbCas12a:LOBP1-mPBE:ALS2:ALS1 was constructed (Figure 1, Supplementary Figure 1). This plasmid contains GFP, which facilitates the selection of transgene-free regenerants, nCas9-mPBE:ALS2:ALS1 which edits ALS to generate chlorsulfuron-resistant regenerants as a selection marker for genome editing resulting from transient expression of the T-DNA, and ttLbCas12a which edits gene(s) of interest (i.e., EBEPthA4-LOBP in this study). In addition, this plasmid contains nptII gene, which can also be used for selection of non-transgenic transformants. In a previous study, the co-editing method was used to generate transgene-free tobacco, tomato, potato, and pummelo (Huang et al., 2023). This study further demonstrates that the co-editing strategy can be used for genetic improvements of elite citrus varieties that are heterozygous hybrids via transgene-free genome editing in the T0 generation. In the previous study, two transgene-free homozygous/biallelic EBEPthA4–LOBP pummelo mutants were identified from 107 generated shoots, representing 1.9% transgene-free homozygous/biallelic mutation efficiency. Here, four transgene-free homozygous/biallelic EBEPthA4–LOBP C. sinensis cv. Hamlin mutants were identified from 77 generated shoots, representing 5.2% transgene-free homozygous/biallelic mutation efficiency. The improvement in transgene-free homozygous/biallelic mutation efficiency might result from the optimization of the base editor. The Cas9 nickase (nCas9) in the base editor construct of the previous study (Huang et al., 2023) was codon optimized for cereal crops which are monocots (Zong et al., 2017). Here, we have replaced the nCas9 with SpCas9p backbone which was codon optimized for both monocots and dicots, including citrus (Ma et al., 2015b; Jia and Wang, 2020).
To date, two methods have been employed to develop transgene-free citrus in the T0 generation: 1) PEG-mediated embryogenic protoplast infection with LbCas12aU/crRNA RNP (Su et al., 2023); 2) Agrobacterium-mediated epicotyl transformation with Cas12a/CBE co-editing (Huang et al., 2023). Both methods have their pros and cons. As for LbCas12aU/crRNA RNP method, all regenerants containing the target editing should be regarded as transgene-free, owing to no foreign DNA involved during PEG infection. Most importantly, RNP method had a very high biallelic/homozygous mutation rate, which is up to 97.4%. However, the reagents for LbCas12aU/crRNA RNP method were expensive, and 10 months were needed to establish canker-resistant Hamlin (Su et al., 2023). As for Cas12a/CBE co-editing method, the reagents related to Agrobacterium transformation were cheap, and 6 months were needed to establish canker-resistant citrus in this study. Since binary vector was used for epicotyl transformation, whole genome sequencing must be carried out to exclude potential T-DNA insertion in the chromosome (Huang et al., 2023). In addition, homozygous/biallelic mutation efficiency, which is 5.2% in this study, is much lower than that of RNP method. It is worthy to test whether PEG-mediated protoplast infection with Cas12a/CBE can produce transgene-free citrus with higher homozygous/biallelic efficiency.
In this study, one transgene-free line was chimeric in the ALS gene and 3 transgenic lines were chimeric in either the CsALS gene or the EBEPthA4–LOBP among the 77 regenerated shoots. Epicotyls were used as explants for Cas12a/CBE co-editing here, previous study indicated that a high frequency of chimeric shoots were observed when citrus epicotyl was the target explants for Agrobacterium-mediated transformation (Domínguez et al., 2004). Consistently, the chimeric/mosaic shoots were commonly developed during Agrobacterium-mediated citrus epicotyl transformation with CRISPR/Cas (Peng et al., 2017; Zhang et al., 2017). Remarkably, there were no regenerants containing chimeric CsLOB1 when LbCas12aU/crRNA RNP was employed to infect protoplasts to develop transgene-free Hamlin (Su et al., 2023). The underneath mechanisms for the chimeric mutations in the two methods remain to be explored.
Transgene-free genome editing of plants in the T0 generation is especially useful for vegetatively propagated and perennial plant species. Compared to transgenic plants, transgene-free genome edited plants have multiple promising properties: 1) Easier path for deregulation and commercialization. Both USDA Animal & Plant Health Inspection Service (APHIS) and US Environmental Protection Agency exempt transgene-free genome-edited plants (Turnbull et al., 2021; Su et al., 2023), 2) Alleviate the potentially deleterious effects from the T-DNA integrated into the host genome stably (O’Malley and Ecker, 2010), 3) Reducing off-target mutations by eliminating the constitutive expression of genome editing systems. Off-target mutations are another critical factor for consideration during genetic improvement by genome editing. Transient expression of Cas/gRNA DNA, mRNA, and RNP in embryogenic protoplasts, calli, or immature embryo cells has been reported to generate transgene-free plants without causing off-target mutations (Woo et al., 2015; Zhang et al., 2016; Liang et al., 2017). This probably results from the short functional time of Cas/gRNA during transient expression, as suggested by previous studies (Huang et al., 2022a; Randall et al., 2021). In addition to the co-editing method, transformation of citrus embryogenic protoplast cells has also been successfully used to generate transgene-free canker-resist Hamlin by editing CsLOB1 coding region (Su et al., 2023). It is noteworthy that our work here targeted CsLOB1 promoter elements, EBEPthA4-LOBP. Even though editing either the coding region or promoter region of LOB1 generates canker-resistant citrus varieties, it remains to be determined whether there are any phenotypical changes between the two different editions.
In summary, our improved co-editing approach provides a cost-effective, time-saving and one-step method to produce transgene-free genome-edited citrus in the T0 generation. This strategy has the potential to be expanded to other plant species, especially those that have long juvenility or/and must be produced through vegetative propagation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA1073671.
Author contributions
HJ: Investigation, Methodology, Writing – original draft, Writing – review & editing. AO: Methodology, Resources, Writing – review & editing. JX: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. JD: Investigation, Writing – review & editing. YW: Investigation, Writing – review & editing. YF: Investigation, Writing – review & editing. WW: Investigation, Writing – review & editing. ZH: Investigation, Writing – review & editing. JG: Resources, Writing – review & editing. NW: Conceptualization, Funding acquisition, Investigation, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This project was supported by funding from Florida Citrus Initiative Program, Citrus Research and Development Foundation 18-025, U.S. Department of Agriculture National Institute of Food and Agriculture grants 2023-70029-41280, 2022-70029-38471, 2021-67013-34588, and 2018-70016-27412, FDACS Specialty Crop Block Grant Program AM22SCBPFL1125, and Hatch project [FLA-CRC-005979] to NW.
Acknowledgments
We thank Wang lab members for constructive suggestions and insightful discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1385768/full#supplementary-material
Supplementary Figure 6 | Whole genome sequencing analysis of #HamNoGFP4. (A) Based on whole genome sequencing, two alleles of CsALS of #HamNoGFP4 contained the identical 6th, 7th, 8th C->T mutations. (B) As for EBEPthA4-LOBP of #HamNoGFP4, there was 7 bp deletion of CCTTTTG from EBE region of Type I and Type II allele. The mutations were showed by horizontal bar chart. The vertical bar chart showed the sequence depth for each base.
References
Alquezar, B., Bennici, S., Carmona, L., Gentile, A., Pena, L. (2022). Generation of transfer-DNA-Free base-Edited citrus plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.835282
Andersson, M., Turesson, H., Olsson, N., Falt, A. S., Ohlsson, P., Gonzalez, M. N., et al. (2018). Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant 164, 378–384. doi: 10.1111/ppl.12731
Bae, S., Park, J., Kim, J.-S. (2014). Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475. doi: 10.1093/bioinformatics/btu048
Bowen, J., Brummell, D., Gapper, N. (2022). Biotechnological approaches for reducing fruit losses caused by pathogenic infection. Curr. Opin. Biotechnol. 78, 102795. doi: 10.1016/j.copbio.2022.102795
Cao, H., Vu, G., Gailing, O. (2022). From genome sequencing to CRISPR-based genome editing for climate-resilient forest trees. Int. J. Mol. Sci. 23(2), 966. doi: 10.3390/ijms23020966
Caserta, R., Teixeira-Silva, N. S., Granato, L. M., Dorta, S. O., Rodrigues, C. M., Mitre, L. K., et al. (2020). Citrus biotechnology: What has been done to improve disease resistance in such an important crop? Biotechnol. Res. Innov. 3, 95–109. doi: 10.1016/j.biori.2019.12.004
Chen, K., Wang, Y., Zhang, R., Zhang, H., Gao, C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. doi: 10.1146/annurev-arplant-050718-100049
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Cimen, B., Yesiloglu, T. (2016). “Rootstock breeding for abiotic stress tolerance in citrus,” in Abiotic and Biotic Stress in Plants - Recent Advances and Future Perspectives. Eds. Shanker, A. K., Shanker, C. (InTech, Rijeka), 23. doi: 10.5772/62047
Dala-Paula, B. M., Plotto, A., Bai, J., Manthey, J. A., Baldwin, E. A., Ferrarezi, R. S., et al. (2019). Effect of Huanglongbing or greening disease on orange juice quality, a review. Front. Plant Sci. 9, 1976. doi: 10.3389/fpls.2018.01976
Domínguez, A, Cervera, M, Pérez, RM, Romero, J, Fagoaga, C, Cubero, J, López, MM, Juárez, JA, Navarro, L, Peña, L (2004). Characterisation of regenerants obtained under selective conditions after agrobacterium-mediated transformation of citrus explants reveals production of silenced and chimeric plants at unexpected high frequencies Mol Breed 14, 171–183. doi: 10.1023/B:MOLB.0000038005.73265.61
de Souza-Neto, R. R., Vasconcelos, F. N. D. C., Teper, D., Carvalho, I. G. B., Takita, M. A., Benedetti, C. E., et al. (2023). The expansin gene csLIEXP1 is a direct target of csLOB1 in citrus. Phytopathology 113, 1266–1277. doi: 10.1023/B:MOLB.0000038005.73265.61
Duan, S., Jia, H., Pang, Z., Teper, D., White, F., Jones, J., et al. (2018). Functional characterization of the citrus canker susceptibility gene CsLOB1. Mol. Plant Pathol. 19, 1908–1916. doi: 10.1111/mpp.12667
Dutt, M., Mou, Z., Zhang, X., Tanwir, S. E., Grosser, J. W. (2020). Efficient CRISPR/Cas9 genome editing with Citrus embryogenic cell cultures. BMC Biotechnol. 20, 58. doi: 10.1186/s12896-020-00652-9
Ference, C. M., Gochez, A. M., Behlau, F., Wang, N., Graham, J. H., Jones, J. B. (2018). Recent advances in the understanding of Xanthomonas citri ssp. citri pathogenesis and citrus canker disease management. Mol. Plant Pathol. 19, 1302–1318. doi: 10.1111/mpp.12638
Gao, C. (2021). Genome engineering for crop improvement and future agriculture. Cell 184, 1621–1635. doi: 10.1016/j.cell.2021.01.005
Gong, Z., Cheng, M., Botella, J. R. (2021). Non-GM genome editing approaches in crops. Front. Genome Ed. 3, 817279. doi: 10.3389/fgeed.2021.817279
Hu, Y., Zhang, J., Jia, H., Sosso, D., Li, T., Frommer, W. B., et al. (2014). Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. U.S.A. 111, E521–E529. doi: 10.1073/pnas.1313271111
Huang, X., Huang, S., Han, B., Li, J. (2022a). The integrated genomics of crop domestication and breeding. Cell 185, 2828–2839. doi: 10.1016/j.cell.2022.04.036
Huang, X., Jia, H., Xu, J., Wang, Y., Wen, J., Wang, N. (2023). Transgene-free genome editing of vegetatively propagated and perennial plant species in the T0 generation via a co-editing strategy. Nat. Plants 9, 1591–1597. doi: 10.1038/s41477-023-01520-y
Huang, X., Wang, N. (2021). Highly efficient generation of canker resistant sweet orange enabled by an improved CRISPR/Cas9 system. bioRxiv. doi: 10.1101/2021.10.20.465103
Huang, X., Wang, Y., Wang, N. (2022b). Base editors for citrus gene editing. Front. Genome Ed. 4, 852867. doi: 10.3389/fgeed.2022.852867
Huang, X., Wang, Y., Xu, J., Wang, N. (2020). Development of multiplex genome editing toolkits for citrus with high efficacy in biallelic and homozygous mutations. Plant Mol. Biol. 104, 297–307. doi: 10.1007/s11103-020-01043-6
Jia, H., Omar, A., Orbović, V., Wang, N. (2021). Biallelic editing of the LOB1 promoter via CRISPR/Cas9 creates canker-resistant 'Duncan' grapefruit. Phytopathology 112, 308–314. doi: 10.1094/PHYTO-04-21-0144-R
Jia, H., Orbovic, V., Jones, J. B., Wang, N. (2016). Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection. Plant Biotechnol. J. 14, 1291–1301. doi: 10.1111/pbi.12495
Jia, H., Orbović, V., Wang, N. (2019a). CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol. J. 17, 1928–1937. doi: 10.1111/pbi.13109
Jia, H., Pang, Y., Chen, X., Fang, R. (2006). Removal of the selectable marker gene from transgenic tobacco plants by expression of Cre recombinase from a tobacco mosaic virus vector through agroinfection. Transgenic Res. 15, 375–384. doi: 10.1007/s11248-006-0011-6
Jia, H., Wang, N. (2014a). Targeted genome editing of sweet orange using cas9/sgRNA. PloS One 9, e93806. doi: 10.1371/journal.pone.0093806
Jia, H., Wang, N. (2014b). Xcc-facilitated agroinfiltration of citrus leaves: a tool for rapid functional analysis of transgenes in citrus leaves. Plant Cell Rep. 33, 1993–2001. doi: 10.1007/s00299-014-1673-9
Jia, H., Wang, N. (2020). Generation of homozygous canker-resistant citrus in the T0 generation using CRISPR-SpCas9p. Plant Biotechnol. J. 18, 1190–1192. doi: 10.1111/pbi.13375
Jia, H., Wang, Y., Su, H., Huang, X., Wang, N. (2022). LbCas12a-D156R efficiently edits LOB1 effector binding elements to generate canker-resistant citrus plants. Cells 11, 315. doi: 10.3390/cells11030315
Jia, H., Xu, J., Orbović, V., Zhang, Y., Wang, N. (2017a). Editing citrus gneome via SaCas9/sgRNA system. Front. Plant Sci. 8, 2135. doi: 10.3389/fpls.2017.02135
Jia, H., Zhang, Y., Orbović, V., Xu, J., White, F. F., Jones, J. B., et al. (2017b). Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 15, 817–823. doi: 10.1111/pbi.12677
Jia, H., Zou, X., Orbović, V., Wang, N. (2019b). Genome editing in citrus tree with CRISPR/cas9. Plant Genome Editing CRISPR Systems. 1917, 235–241. doi: 10.1007/978-1-4939-8991-1_17
Kocsisova, Z., Coneva, V. (2023). Strategies for delivery of CRISPR/Cas-mediated genome editing to obtain edited plants directly without transgene integration. Front. Genome Ed 5. doi: 10.3389/fgeed.2023.1209586
Kozarewa, I., Ning, Z., Quail, M. A., Sanders, M. J., Berriman, M., Turner, D. J. (2009). Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat. Methods 6, 291–295. doi: 10.1038/nmeth.1311
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
LeBlanc, C., Zhang, F., Mendez, J., Lozano, Y., Chatpar, K., Irish, V. F., et al. (2018). Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J. 93, 377–386. doi: 10.1111/tpj.13782
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liang, Z., Chen, K., Li, T., Zhang, Y., Wang, Y., Zhao, Q., et al. (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8, 14261. doi: 10.1038/ncomms14261
Liu, H., Ding, Y., Zhou, Y., Jin, W., Xie, K., Chen, L. L. (2017). CRISPR-P 2.0: an improved CRISPR-cas9 tool for genome editing in plants. Mol. Plant 10, 530–532. doi: 10.1016/j.molp.2017.01.003
Liu, Y., Zhang, C., Wang, X., Li, X., You, C. (2022). CRISPR/Cas9 technology and its application in horticultural crops. Hortic. Plant J. 8, 395–407. doi: 10.1016/j.hpj.2022.04.007
Liu, Q., Zhao, C., Sun, K., Deng, Y., Li, Z. (2023). Engineered biocontainable RNA virus vectors for non-transgenic genome editing across crop species and genotypes. Mol. Plant 16, 616–631. doi: 10.1016/j.molp.2023.02.003
Lv, X., Qiu, K., Tu, T., He, X., Peng, Y., Ye, J., et al. (2020). Development of a simple and quick method to assess base editing in human cells. Mol. Ther. - Nucleic Acids 20, 580–588. doi: 10.1016/j.omtn.2020.03.004
Ma, X., Chen, L., Zhu, Q., Chen, Y., Liu, Y. G. (2015a). Rapid decoding of sequence-specific nuclease-induced hetero-zygous and biallelic mutations by direct sequencing of PCR products. Mol. Plant 8, 1285–1287. doi: 10.1016/j.molp.2015.02.012
Ma, X., Zhang, X., Liu, H., Li, Z. (2020). Highly efficient DNA-free plant genome editing using virally delivered CRISPR-Cas9. Nat. Plants 6, 773–779. doi: 10.1038/s41477-020-0704-5
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015b). A robust CRISPR/cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Mahmoud, L. M., Kaur, P., Stanton, D., Grosser, J. W., Dutt, M. (2022). A cationic lipid mediated CRISPR/Cas9 technique for the production of stable genome edited citrus plants. Plant Methods 18, 1–14. doi: 10.1186/s13007-022-00870-6
Malnoy, M., Viola, R., Jung, M.-H., Koo, O-J., Kim, S., Kim, J-S., et al. (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7, 1904. doi: 10.3389/fpls.2016.01904
Mei, Y., Beernink, B. M., Ellison, E. E., Konečná, E., Neelakandan, A. K., Voytas, D. F., et al. (2019). Protein expression and gene editing in monocots using foxtail mosaic virus vectors. Plant Direct 3, 1–16. doi: 10.1002/pld3.181
O’Malley, R. C., Ecker, J. R. (2010). Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J. 61, 928–940. doi: 10.1111/j.1365-313X.2010.04119.x
Parajuli, S., Huo, H., Gmitter, F. G., Duan, Y., Luo, F., Deng, Z. (2022). Editing the CsDMR6 gene in citrus results in resistance to the bacterial disease citrus canker. Hortic. Res. 9 (2022), uhac082 doi: 10.1093/hr/uhac082
Peng, A., Chen, S., Lei, T., Xu, L., He, Y., Wu, L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. doi: 10.1111/pbi.12733
Poplin, R., Chang, P. C., Alexander, D., Schwartz, S., Colthurst, T., Ku, A., et al. (2018). A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 36, 983–987. doi: 10.1038/nbt.4235
Randall, LB, Sretenovic, S, Wu, Y, Yin, D, Zhang, T, Eck, JV, Qi, Y. (2021). Genome- and transcriptome-wide off-target analyses of an improved cytosine base editor. Plant Physiol 187, 73–87. doi: 10.1093/plphys/kiab264
Robinson, J., Thorvaldsdóttir, H., Winckler, W., Guttman, M., Lander, E., Getz, G., et al. (2011). Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. doi: 10.1038/nbt.1754
Schindele, P., Puchta, H. (2020). Engineering CRISPR/LbCas12a for highly efficient, temperature-tolerant plant gene editing. Plant Biotechnol. J. 18, 1118–1120. doi: 10.1111/pbi.13275
Su, H., Wang, Y., Xu, J., Omar, A. A., Grosser, J. W., Calovic, M., et al. (2023). Generation of the transgene-free canker-resistant Citrus sinensis using Cas12a/crRNA ribonucleoprotein in the T0 generation. Nat. Commun. 14, 3957. doi: 10.1038/s41467-023-39714-9
Subburaj, S., Chung, S. J., Lee, C., Ryu, S. -M., Kim, D. H., Kim, J.-S., et al. (2016). Site-directed mutagenesis in Petunia × hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 35, 1535–1544. doi: 10.1007/s00299-016-1937-7
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K., Cigan, A. M. (2016). Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat. Commun. 7, 13274. doi: 10.1038/ncomms13274
Tang, X., Lowder, L. G., Zhang, T., Malzahn, A. A., Zheng, X., Voytas, D. F., et al. (2017). A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 3, 17018. doi: 10.1038/nplants.2017.18
Turnbull, C., Lillemo, M., Hvoslef-Eide, T. A. K. (2021). Global regulation of genetically modified crops amid the gene edited crop boom – a review. Front. Plant Sci. 12, 630396. doi: 10.3389/fpls.2021.630396
Veillet, F., Perrot, L., Chauvin, L., Kermarrec, M. P., Guyon-Debast, A., Chauvin, J. E., et al. (2019). Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci. 20, 402. doi: 10.3390/ijms20020402
Wang, N. (2019). The citrus huanglongbing crisis and potential solutions. Mol. Plant 12, 607–609. doi: 10.1016/j.molp.2019.03.008
Wang, L., Huang, Y., Liu, Z., He, J., Jiang, X., He, F., et al. (2021). Somatic variations led to the selection of acidic and acidless orange cultivars. Nat. Plants 7, 954–965. doi: 10.1038/s41477-021-00941-x
Wei, T., Jiang, L., You, X., Ma, P., Xi, Z., Wang, N. N. (2023). Generation of herbicide-resistant soybean by base editing. Biology 12, 741. doi: 10.3390/biology12050741
Woo, J. W., Kim, J., Kwon, S. I., Corvalan, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi: 10.1038/nbt.3389
Wu, G. A., Terol, J., Ibanez, V., López-García, A., Pérez-Román, E., Borredá, C., et al. (2018). Genomics of the origin and evolution of Citrus. Nature 554, 311–316. doi: 10.1038/nature25447
Yang, L., Machin, F., Wang, S., Saplaoura, E., Kragler, F. (2023). Heritable transgene-free genome editing in plants by grafting of wild-type shoots to transgenic donor rootstocks. Nat. Biotechnol. 41, 958–967. doi: 10.1038/s41587-022-01585-8
Yang, W., Ren, J., Liu, W., Liu, D., Xie, K., Zhang, F., et al. (2022). An efficient transient gene expression system for protein subcellular localization assay and genome editing in citrus protoplasts. Hortic. Plant J. 9, 425–436. doi: 10.1016/j.hpj.2022.06.006
Zhang, J., Carlos Huguet Tapia, J., Hu, Y., Jones, J., Wang, N., Liu, S., et al. (2016). Homologs of CsLOB1 in citrus function as disease susceptibility genes in citrus canker. Mol. Plant Pathol. 18, 798–810. doi: 10.1111/mpp.12441
Zhang, F., LeBlanc, C., Irish, V. F., Jacob, Y. (2017). Rapid and efficient CRISPR/Cas9 gene editing in Citrus using the YAO promoter. Plant Cell Rep. 36, 1883–1887. doi: 10.1007/s00299-017-2202-4
Zhu, C., Zheng, X., Huang, Y., Ye, J., Chen, P., Zhang, C., et al. (2019). Genome sequencing and CRISPR/Cas9 gene editing of an early flowering mini-Citrus (Fortunella hindsii). Plant Biotechnol. J. 17, 2199–2210. doi: 10.1111/pbi.13132
Zong, Y., Wang, Y., Li, C., Zhang, R., Chen, K., Ran, Y., et al. (2017). Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminasefusion. Nat. Biotechnol. 35, 438–440. doi: 10.1038/nbt.3811
Keywords: transgene-free genome editing, CRISPR, Cas12a, Citrus, Xanthomonas, citrus canker
Citation: Jia H, Omar AA, Xu J, Dalmendray J, Wang Y, Feng Y, Wang W, Hu Z, Grosser JW and Wang N (2024) Generation of transgene-free canker-resistant Citrus sinensis cv. Hamlin in the T0 generation through Cas12a/CBE co-editing. Front. Plant Sci. 15:1385768. doi: 10.3389/fpls.2024.1385768
Received: 13 February 2024; Accepted: 15 March 2024;
Published: 26 March 2024.
Edited by:
Elena Palomo Ríos, Universidad de Málaga, SpainReviewed by:
Bhabesh Borphukan, Washington State University, United StatesAlessandra Alves De Souza, Secretariat of Agriculture and Food Supply of São Paulo State, Brazil
Copyright © 2024 Jia, Omar, Xu, Dalmendray, Wang, Feng, Wang, Hu, Grosser and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nian Wang, nianwang@ufl.edu
 Hongge Jia
Hongge Jia Ahmad A. Omar
Ahmad A. Omar Jin Xu
Jin Xu Javier Dalmendray
Javier Dalmendray Yuanchun Wang
Yuanchun Wang Yu Feng1
Yu Feng1 Jude W. Grosser
Jude W. Grosser Nian Wang
Nian Wang