- 1State Key Laboratory of Crop Biology, College of Life Sciences, Shandong Agricultural University, Taian, Shandong, China
- 2Donald Danforth Plant Science Center, St. Louis, MO, United States
Plants have evolved interconnected regulatory pathways which enable them to respond and adapt to their environments. In plants, stress memory enhances stress tolerance through the molecular retention of prior stressful experiences, fostering rapid and robust responses to subsequent challenges. Mounting evidence suggests a close link between the formation of stress memories and effective future stress responses. However, the mechanism by which environmental stressors trigger stress memory formation is poorly understood. Here, we review the current state of knowledge regarding the RNA-based regulation on stress memory formation in plants and discuss research challenges and future directions. Specifically, we focus on the involvement of microRNAs (miRNAs), small interfering RNAs (siRNAs), long non-coding RNAs (lncRNAs), and alternative splicing (AS) in stress memory formation. miRNAs regulate target genes via post-transcriptional silencing, while siRNAs trigger stress memory formation through RNA-directed DNA methylation (RdDM). lncRNAs guide protein complexes for epigenetic regulation, and AS of pre-mRNAs is crucial to plant stress memory. Unraveling the mechanisms underpinning RNA-mediated stress memory formation not only advances our knowledge of plant biology but also aids in the development of improved stress tolerance in crops, enhancing crop performance and global food security.
Introduction
As sessile organisms, plants must respond and adapt to fluctuating, and sometimes near-lethal, environmental conditions over their entire lifespan (Trivedi et al., 2020; Zhang et al., 2022). To endure adverse environmental conditions, plants have evolved stress tolerance mechanisms, including the establishment of a “stress memory” (Crisp et al., 2016; Hilker and Schmülling, 2019; Liu et al., 2022). Stress memory is formed through transient exposure to mild or severe stress and allows primed plants to respond robustly and swiftly to subsequent stressors, facilitating their recovery (Crisp et al., 2016; Liu et al., 2022). Understanding how plants stress memory works is crucial for improving crop resilience and productivity, which can help ensure food security in the face of changing environmental conditions and growing global food demands.
Following the initial stress exposure, stress memory modulates the expression of key genes through epigenetic modifications such as chromatin re-modelling, DNA methylation, nucleosome positioning, histone modification, and non-coding RNA-mediated regulation (Liu et al., 2022). In addition, a growing body of evidence suggests that stress memory involves RNA-mediated regulation via gene silencing and/or activation (Crisp et al., 2016). Here, we review the roles played by diverse RNAs in plant stress memory by summarizing recent research advances and providing generalized examples. MicroRNAs (miRNAs) and small interfering RNAs (siRNAs) are involved in plant stress memory via post-transcriptional gene silencing and RNA-directed DNA methylation, respectively (Song et al., 2019; Li et al., 2020). Long non-coding RNAs (lncRNAs) and alternative splicing (AS) also play crucial roles in stress memory (Ohama et al., 2017; Chaudhary et al., 2019; Yu et al., 2019; Ling et al., 2021). Finally, we investigate the mechanisms of RNA-mediated stress memory in plants and suggest possible future research directions (Table 1).
The role of miRNAs in plant stress memory: post-transcriptional silencing
In plants, miRNAs regulate gene expression and/or silencing by binding to complementary sequences within target messenger RNAs (mRNAs), resulting in translational repression and/or transcript degradation (Sunkar et al., 2012; Ha and Kim, 2014). miRNAs are critical for regulating plant growth and reproduction, as well as biotic and abiotic stress responses (Wang et al., 2009; Zhao et al., 2019; Liebsch and Palatnik, 2020; Li et al., 2023; Xu et al., 2023). Recent research suggests that miRNAs may also be key regulators of plant stress memory. Specifically, miRNAs can respond quickly to environmental and developmental cues via the post-transcriptional silencing of stress-responsive target genes (Figure 1A).
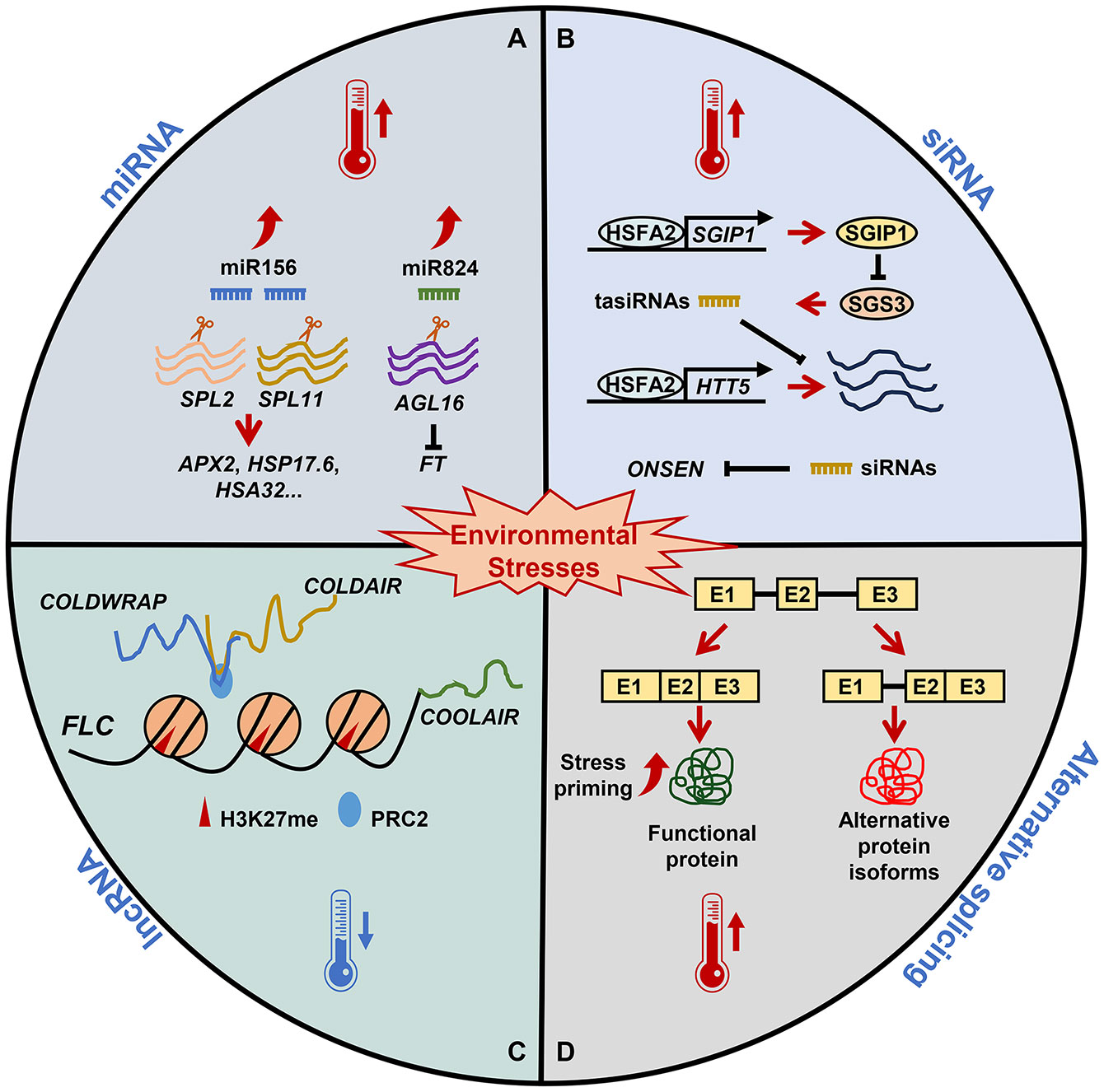
Figure 1 The role of RNA-based regulation in plant stress memory. Under nonstress conditions, miRNAs, siRNAs, lncRNAs, and alternative splicing are essential for plant growth and development. (A) Under heat stress, miR156 expression is induced and SPL genes are post-transcriptionally downregulated, affecting the expression of heat stress memory-related genes. Parallel to miR824 induction, its target AGL16 is decreased. AGL16 downregulation in response to heat leads to a fine-tuning of FT. (B) Heat stress upregulates HSFA2, which activates SGIP1 to trigger the transgenerational degradation of SGS3, leading to the suppression of tasiRNA biosynthesis. The reduced tasiRNA levels converge to activate HTT5. In addition, siRNA-mediated regulation and the initiation of transgenerational transposition of ONSEN are involved in heat stress memory. (C) Winter cold triggers high levels of H3K27me3 at FLC mediated by the PRC2 complex. lncRNAs, including COOLAIR, COLDAIR, and COLDWRAP, are important for FLC repression. COOLAIR decreases H3K36me3 at FLC; COLDAIR recruits PRC2 and promotes FLC repression; and COLDWRAP may help PRC2 spread to the FLC promoter and stabilize H3K27me3. (D) Exposure to sub-lethal heat stress results in the priming of plants, which establishes splicing-linked heat-stress memory. This heat priming executes correct splicing and ensures the functional RNA/proteins resulting in an effective adaptive response that ensures the survival of plants.
Most recently, improved high-throughput sequencing techniques have revealed the close relationship between miRNAs and stress memory. With drought pretreatment, 195 miRNAs, including 186 drought memory-specific and nine significantly differentially expressed shared miRNAs, were identified as candidate drought memory-related miRNAs in wheat (Triticum aestivum) (Yue et al., 2022). In Arabidopsis thaliana, overexpression of the wheat drought memory-related miRNA tae-miR531_L-2 significantly improves drought tolerance in transgenic plants (Yue et al., 2022). A recent study of the miRNAome of wheat seedlings subjected to water deficit and heat stress revealed the long-term impact of stress on plant physiology and gene regulation and suggested that miRNAs and their target genes play important roles in transgenerational stress adaptation (Liu et al., 2020). In coffee (Coffea arabica) subjected to repeated cycles of drought, the transcriptional levels of miRNA-guide stress-related genes are different, which exhibit distinct transcriptional memory behavior (Guedes et al., 2018). Specifically, their complex regulation of target V-myb myeloblastosis viral oncogene (MYB) homologs highlights the crucial role of MYB at the crossroads of plant miRNA-mediated stress memory (Guedes et al., 2018).
Recent work has also revealed that multiple miRNAs participate in heat stress memory (Stief et al., 2014b). Mutant plants with impaired small RNA (sRNA) biogenesis, specifically ago1 (Argonaute1) and dcl1 (Dicer-like1) mutants, are unable to acquire thermotolerance, indicating that heat stress memory requires miRNA intermediation (Stief et al., 2014a). Isoforms of miR156 are highly-induced following exposure to heat stress, and repression of their target genes SQUAMOSA promoter binding protein-like 2 (SPL2) and SPL11, which is required for the transcriptional induction of heat stress memory genes, including ASCORBATE PEROXIDASE 2 (APX2), HEAT STRESS ASSOCIATED 32 (HSA32), HEAT SHOCK TRANSCRIPTION FACTOR A2 (HSFA2), HEAT SHOCK PROTEIN 17.6A (HSP17.6A) and HEAT SHOCK PROTEIN 22 (HSP22) (Stief et al., 2014a). Moreover, miR156h overexpression-induced acquired thermotolerance results from the increased expression of heat stress memory-related genes (Stief et al., 2014a). Notably, heat stress-induced miR156 induction has been observed in alfalfa (Medicago sativa), field mustard (Brassica rapa), ginkgo (Ginkgo biloba), banana (Musa acuminata), safflower (Carthamus tinctorius), and wheat (T. aestivum), suggesting that this miRNA may have conserved functions in both development and heat stress memory (Wang et al., 2014; Ragupathy et al., 2016; Matthews et al., 2019; Zhu et al., 2019; Chang et al., 2020; Kouhi et al., 2020). As miR156 is an important regulator of developmental transitions, this signaling module may be used to integrate stress memory and plant development (Cho et al., 2012; Cheng et al., 2021).
Similarly, heat-responsive induction of miR824 appears to be dependent on HEAT SHOCK TRANSCRIPTION FACTOR A1 (HSFA1) in A. thaliana (Szaker et al., 2019). HSFA1a directly regulating miR824 promoter to activate the transcription under heat stress, and AGAMOUS LIKE 16 (AGL16) is a target gene of miR824. miR824-dependent AGL16 downregulation is primarily manifested post stress exposure, suggesting that miR824 may post-transcriptionally modulate FLOWERING LOCUS T (FT)-driven development in response to environmental signals (Hu et al., 2014; Szaker et al., 2019). Notably, heat-mediated regulation of the miR824/AGL16 module is conserved in multiple Brassicaceae species (Kutter et al., 2007). This module integrates multiple abiotic stimuli under complex climatic conditions, and therefore may hold considerable potential for enhancing plant stress resistance (Kutter et al., 2007). In B. rapa, the differential expression of bra-miR168 following heat stress is correlated with braAGO1 transcription, suggestive of their potential roles as key regulators of transgenerational stress memory (Bilichak et al., 2015).
Overall, the involvement of post-transcriptional regulatory miRNA modules in plant stress memory highlights the flexibility of plants to effectively respond and adapt to fluctuating and stressful environmental conditions. Additionally, as an indicator of their long-term adaptability, stress memory allows plants to respond rapidly and robustly to future stressors, effectively acting as an inoculation.
The role of siRNAs in stress memory: RNA-directed DNA methylation
siRNAs are a class of non-coding RNA molecules produced through the processing of long double-stranded RNA (dsRNA) precursors (Lee et al., 2023). Similarly to miRNAs, siRNAs actively participate in RNA interference (RNAi) by binding to complementary mRNA sequences, resulting in mRNA cleavage and subsequent degradation (Liu et al., 2023). Notably, siRNAs can also induce silencing and RNA-directed DNA methylation (RdDM) to enforce epigenetic states, which may be involved in plant stress memory (Liu et al., 2019).
In A. thaliana, heat stress-induced HSFA2 expression results in the suppression of the SUPPRESSOR OF GENE SILENCING 3 (SGS3) protein, which is involved in siRNA production (Liu et al., 2019) (Figure 1B). This pathway leads to inhibited siRNA biosynthesis, a decrease in methylation, and the suppression of transposons and stress-responsive loci, thus allowing stress memory to persist (Liu et al., 2019). Similarly, a retrotransposon known as ONSEN (Japanese for “hot spring”) is significantly activated in response to heat stress because it is targeted by HSFA1 and HSFA2 (Ito et al., 2011; Matsunaga et al., 2012). Heat-induced ONSEN transcription and transposition are promoted in mutant plants with impaired siRNA biogenesis (Matsunaga et al., 2012). Although both the ONSEN transcripts and extrachromosomal DNA decayed over time, new ONSEN insertions were observed in the progeny of stressed siRNA-deficient plants (Matsunaga et al., 2015). An siRNA-mediated mechanism is involved in the new insertions happen in the progeny. Heat activated ONSEN was transposed to the next generation and increased in copy number in the host genome (Matsunaga et al., 2015). Together, these studies highlight the involvement of siRNA-guide epigenetic mechanisms in the formation of transgenerational stress memory.
Emerging roles of long non-coding RNAs in stress memory
lncRNAs, which are typically longer than 200 nucleotides (nt), are a large and diverse class of eukaryotic genes which contribute to an array of regulatory processes (Palos et al., 2023). For example, lncRNAs play an important role in coping with environmental stress during plant growth and development (Song et al., 2021). Specifically, lncRNAs mediate epigenetic modifications, and by studying their participation in stress memory we may begin to unravel the intricate interplay between non-coding RNAs and epigenetic regulation (Figure 1C).
Strand-specific whole-transcriptome RNA sequencing of repeatedly drought-stressed rice (Oryza sativa) revealed that the lncRNA TCONS_00028567, a predicted precursor of the miRNA are strongly upregulated at the second drought treatment stage, but downregulated at the three re-water treatment stage (Li et al., 2019). Such expression variability of TCONS_00028567 is related to the repeatedly short-term drought treatment, suggesting that TCONS_00028567 may regulate short-term drought memory in rice. In switchgrass (Panicum virgatum), lncRNAs targeting the biosynthesis of ABA and trehalose were upregulated during the first and second drought cycles, but lncRNAs regulating ethylene signaling were suppressed in the second drought cycle, thereby preventing leaf senescence and supporting plant development under stressful conditions (Zhang et al., 2018).
In A. thaliana, low-temperature-responsive epigenetic modifications are induced during vernalization, resulting in the repression of the FLOWERING LOCUS C (FLC) gene until flowering commences (Zhu et al., 2021). The FLC locus includes three different cold-induced lncRNAs: COOLAIR, COLDAIR, and COLDWARP (Hung et al., 2022). COOLAIR is highly induced when plants experience a dip below freezing temperature, likely analogous to the first frost in autumn. Then, upregulation of COOLAIR leads to decreased FLC expression and disruption of the synchronized replacement of H3K36 with H3K27me3 methylation at the FLC nucleation site (Csorba et al., 2014; Nielsen et al., 2024). Both COLDAIR and COLDWRAP bind to FLC chromatin, and thus stably silence FLC by recruiting PHD-PRC2 to its specific chromatin location in response to cold temperatures (Heo and Sung, 2011; Kim and Sung, 2017). Thus, the cold-induced Polycomb nucleation mechanism locks in the FLC silenced transcriptional state to maintain the epigenetic memory of cold exposure (Zhao et al., 2021). It appears that lncRNAs act as guides for protein complexes mediating epigenetic regulation, facilitating the formation of cold stress memory in plants.
Overall, these findings highlight the importance of lncRNAs to the formation of stress memory in plants. We anticipate that an increasing number of lncRNAs will be discovered across a diverse range of plant species, and that many of these will be found to be involved in plant stress memory formation and enhanced stress tolerance.
The relationship between AS and stress memory
AS results in the generation of multiple transcripts from the same gene, thereby increasing proteomic diversity and regulating gene expression and mRNA levels (Chaudhary et al., 2019). To maximize metabolic efficiency under stressful conditions, plants may make more proteins with disordered domains via AS in order to diversify substrate specificity and maintain sufficient regulatory capacity (Ling et al., 2021). According to recent research, splicing-linked memory formation during the priming phase is crucial for guaranteeing the availability of correctly-spliced transcripts or proteins which are essential for increased stress tolerance (Figure 1D).
Recent studies in A. thaliana suggest that AS may be a novel component of heat shock memory formation (Ling et al., 2021). For example, priming plants with non-lethal heat stress results in the de-repression of splicing following a second exposure to heat stress, while non-primed plants exhibit significant splicing repression. An array of heat shock protein (HSP) genes, such as HSP21, HSP101, HSP70.10, HSP70.6, HSP90.5, and HSP100.3, have been found to undergo AS in response to heat stress/priming, primarily through intron retention (Ling et al., 2018). Specifically, the levels of intron-retained isoforms were found to be higher during heat shock, with the exception of HSP70.17. The constitutively-spliced isoform of HSP70.10 was mainly expressed during heat priming, although multiple isoforms were observed during other phases (Ling et al., 2018). In contrast, several isoforms of HSP90.6 were observed during the priming phase, but not during other phases (Ling et al., 2018).
Such observations link “splicing memory” to the ability of plants to survive subsequent, and perhaps otherwise lethal, heat stress events. Therefore, priming-induced splicing memory may represent a general feature of the plant heat stress response (Sanyal et al., 2018). AS-mediated stress memory formation may itself be mediated by epigenetic coding. Integrated studies in pine (Pinus spp.) implicate AS as an important mechanism mediating stress response and memory. Specifically, changes in spliceosome-related proteins were observed during heat stress and recovery, as in the cases of SERINE/ARGININE-RICH SPLICING FACTOR RSZ22 (RSZ22), GLYCINE-RICH RNA-BINDING PROTEIN RZ1A (RZ1A), and UNCHARACTERIZED PROTEIN DUF4050 (DUF4050) (Roces et al., 2022). Moreover, exon-skipping events may be induced during drought memory formation in rice (O. sativa), with 920 drought memory-associated genes exhibiting differential AS patterns (Yang et al., 2022).
Plants undergo changes in their gene expression patterns in response to stress exposure. AS contributes to both protein diversity and functional plasticity, allowing plants to adapt to adverse conditions (Chaudhary et al., 2019). For example, certain splice variants may be preferentially produced in response to specific stressors, leading to the activation of tailored defense mechanisms (Kutter et al., 2007). This process allows plants to retain a “memory” of previous stress events, enabling them to mount faster and more effective responses upon subsequent exposure to similar stressors. By deciphering the intricate connections between AS and stress response, researchers may be able to develop crop varieties with enhanced resilience to environmental stressors.
Conclusion and future prospects
In this review, we summarized the role of non-coding RNAs such as miRNAs, siRNAs, and lncRNAs, as well as AS, in the regulation of plant stress memory formation. We highlighted the interconnected regulatory pathways which enable plants to remember past stress events and to use those stored responses to better adapt to new challenges. The involvement of non-coding RNAs in stress memory is demonstrated through their ability to quickly respond to environmental and developmental cues, enhancing stress tolerance and contributing to epigenetic regulation.
However, many questions remain regarding RNA-mediated stress memory formation. First, how do different stressors differentially mediate the formation of stress memories in plants? Second, how can acquired stress memories be efficiently transferred to offspring to enhance the stress resistance of subsequent generations? Third, are there alternative mechanisms (i.e., other than epigenetic) of stress memory transmission between generations? Answering these questions will significantly improve our understand of RNA-mediated stress memory formation, thereby allowing the development of improved crop varieties with enhanced stress tolerance. This research will be crucial in addressing global food security challenges posed by climate change, population growth, and other factors. By unraveling the mysteries of RNA-mediated stress memory formation, we can not only develop more resilient crop varieties but also lay the foundation for sustainable agriculture practices that address the complex challenges of climate change on a global scale.
Author contributions
W-BX: Writing – original draft, Writing – review & editing. FC: Writing – review & editing. PL: Writing – review & editing. KY: Writing – original draft, Writing – review & editing. Q-HG: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Natural Science Foundation of Shandong Province (Grant ZR2022MC051 and ZR2021QC126) in China is gratefully acknowledged.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bilichak, A., Ilnytskyy, Y., Wóycicki, R., Kepeshchuk, N., Fogen, D., Kovalchuk, I. (2015). The elucidation of stress memory inheritance in Brassica rapa plants. Front. Plant Sci. 6, 5. doi: 10.3389/fpls.2015.00005
Chang, B., Ma, K., Lu, Z., Lu, J., Cui, J., Wang, L., et al. (2020). Physiological, transcriptomic, and metabolic responses of ginkgo biloba L. @ to drought, salt, and heat stresses. Biomolecules 10 (12), 1635. doi: 10.3390/biom10121635
Chaudhary, S., Jabre, I., Reddy, A. S. N., Staiger, D., Syed, N. H. (2019). Perspective on alternative splicing and proteome complexity in plants. Trends Plant Sci. 24, 496–506. doi: 10.1016/j.tplants.2019.02.006
Cheng, Y. J., Shang, G. D., Xu, Z. G., Yu, S., Wu, L. Y., Zhai, D., et al. (2021). Cell division in the shoot apical meristem is a trigger for miR156 decline and vegetative phase transition in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 118 (46), e2115667118. doi: 10.1073/pnas.2115667118
Cho, S. H., Coruh, C., Axtell, M. J. (2012). miR156 and miR390 regulate tasiRNA accumulation and developmental timing in Physcomitrella patens. Plant Cell 24, 4837–4849. doi: 10.1105/tpc.112.103176
Crisp, P. A., Ganguly, D., Eichten, S. R., Borevitz, J. O., Pogson, B. J. (2016). Reconsidering plant memory: Intersections between stress recovery, RNA turnover, and epigenetics. Sci. Adv. 2, e1501340. doi: 10.1126/sciadv.1501340
Csorba, T., Questa, J. I., Sun, Q., Dean, C. (2014). Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. U.S.A. 111, 16160–16165. doi: 10.1073/pnas.1419030111
Guedes, F., Nobres, P., Rodrigues Ferreira, D. C., Menezes-Silva, P. E., Ribeiro-Alves, M., Correa, R. L., et al. (2018). Transcriptional memory contributes to drought tolerance in coffee (Coffea canephora) plants. Environ. Exp. Bot. 147, 220–233. doi: 10.1016/j.envexpbot.2017.12.004
Ha, M., Kim, V. N. (2014). Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524. doi: 10.1038/nrm3838
Heo, J. B., Sung, S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79. doi: 10.1126/science.1197349
Hilker, M., Schmülling, T. (2019). Stress priming, memory, and signalling in plants. Plant Cell Environ. 42, 753–761. doi: 10.1111/pce.13526
Hu, J. Y., Zhou, Y., He, F., Dong, X., Liu, L. Y., Coupland, G., et al. (2014). miR824-regulated AGAMOUS-LIKE16 contributes to flowering time repression in arabidopsis. Plant Cell 26, 2024–2037. doi: 10.1105/tpc.114.124685
Hung, F. Y., Shih, Y. H., Lin, P. Y., Feng, Y. R., Li, C., Wu, K. (2022). WRKY63 transcriptional activation of COOLAIR and COLDAIR regulates vernalization-induced flowering. Plant Physiol. 190, 532–547. doi: 10.1093/plphys/kiac295
Ito, H., Gaubert, H., Bucher, E., Mirouze, M., Vaillant, I., Paszkowski, J. (2011). An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472, 115–119. doi: 10.1038/nature09861
Kim, D. H., Sung, S. (2017). Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs. Dev. Cell 40, 302–312.e304. doi: 10.1016/j.devcel.2016.12.021
Kouhi, F., Sorkheh, K., Ercisli, S. (2020). MicroRNA expression patterns unveil differential expression of conserved miRNAs and target genes against abiotic stress in safflower. PLoS One 15, e0228850. doi: 10.1371/journal.pone.0228850
Kutter, C., Schöb, H., Stadler, M., Meins, F., Jr., Si-Ammour, A. (2007). MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell 19, 2417–2429. doi: 10.1105/tpc.107.050377
Lee, Y. Y., Kim, H., Kim, V. N. (2023). Sequence determinant of small RNA production by DICER. Nature 615, 323–330. doi: 10.1038/s41586-023-05722-4
Li, J., Cao, Y., Zhang, J., Zhu, C., Tang, G., Yan, J. (2023). The miR165/166-PHABULOSA module promotes thermotolerance by transcriptionally and posttranslationally regulating HSFA1. Plant Cell 35, 2952–2971. doi: 10.1093/plcell/koad121
Li, P., Yang, H., Wang, L., Liu, H., Huo, H., Zhang, C., et al. (2019). Physiological and transcriptome analyses reveal short-term responses and formation of memory under drought stress in rice. Front. Genet. 10, 55. doi: 10.3389/fgene.2019.00055
Li, T., Gonzalez, N., Inzé, D., Dubois, M. (2020). Emerging connections between small RNAs and phytohormones. Trends Plant Sci. 25, 912–929. doi: 10.1016/j.tplants.2020.04.004
Liebsch, D., Palatnik, J. F. (2020). MicroRNA miR396, GRF transcription factors and GIF co-regulators: a conserved plant growth regulatory module with potential for breeding and biotechnology. Curr. Opin. Plant Biol. 53, 31–42. doi: 10.1016/j.pbi.2019.09.008
Ling, Y., Mahfouz, M. M., Zhou, S. (2021). Pre-mRNA alternative splicing as a modulator for heat stress response in plants. Trends Plant Sci. 26, 1153–1170. doi: 10.1016/j.tplants.2021.07.008
Ling, Y., Serrano, N., Gao, G., Atia, M., Mokhtar, M., Woo, Y. H., et al. (2018). Thermopriming triggers splicing memory in Arabidopsis. J. Exp. Bot. 69, 2659–2675. doi: 10.1093/jxb/ery062
Liu, H., Able, A. J., Able, J. A. (2020). Transgenerational effects of water-deficit and heat stress on germination and seedling vigour-new insights from durum wheat microRNAs. Plants (Basel) 9 (2), 189. doi: 10.3390/plants9020189
Liu, H., Able, A. J., Able, J. A. (2021). Small RNAs and their targets are associated with the transgenerational effects of water-deficit stress in durum wheat. Sci. Rep. 11(11), 3613. doi: 10.1038/s41598-021-83074-7
Liu, H., Able, A. J., Able, J. A. (2022). Priming crops for the future: rewiring stress memory. Trends Plant Sci. 27, 699–716. doi: 10.1016/j.tplants.2021.11.015
Liu, J., Feng, L., Gu, X., Deng, X., Qiu, Q., Li, Q., et al. (2019). An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 29, 379–390. doi: 10.1038/s41422-019-0145-8
Liu, S., Han, Y., Li, W. X., Ding, S. W. (2023). Infection defects of RNA and DNA viruses induced by antiviral RNA interference. Microbiol. Mol. Biol. Rev. 87, e0003522. doi: 10.1128/mmbr.00035-22
Matsunaga, W., Kobayashi, A., Kato, A., Ito, H. (2012). The effects of heat induction and the siRNA biogenesis pathway on the transgenerational transposition of ONSEN, a copia-like retrotransposon in Arabidopsis thaliana. Plant Cell Physiol. 53, 824–833. doi: 10.1093/pcp/pcr179
Matsunaga, W., Ohama, N., Tanabe, N., Masuta, Y., Masuda, S., Mitani, N., et al. (2015). A small RNA mediated regulation of a stress-activated retrotransposon and the tissue specific transposition during the reproductive period in Arabidopsis. Front. Plant Sci. 6, 48. doi: 10.3389/fpls.2015.00048
Matthews, C., Arshad, M., Hannoufa, A. (2019). Alfalfa response to heat stress is modulated by microRNA156. Physiol. Plant 165, 830–842. doi: 10.1111/ppl.12787
Nielsen, M., Menon, G., Zhao, Y., Mateo-Bonmati, E., Wolff, P., Zhou, S., et al. (2024). COOLAIR and PRC2 function in parallel to silence FLC during vernalization. Proc. Natl. Acad. Sci. U.S.A. 121, e2311474121. doi: 10.1073/pnas.2311474121
Ohama, N., Sato, H., Shinozaki, K., Yamaguchi-Shinozaki, K. (2017). Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 22, 53–65. doi: 10.1016/j.tplants.2016.08.015
Palos, K., Yu, L., Railey, C. E., Nelson Dittrich, A. C., Nelson, A. D. L. (2023). Linking discoveries, mechanisms, and technologies to develop a clearer perspective on plant long noncoding RNAs. Plant Cell 35, 1762–1786. doi: 10.1093/plcell/koad027
Ragupathy, R., Ravichandran, S., Mahdi, M. S., Huang, D., Reimer, E., Domaratzki, M., et al. (2016). Deep sequencing of wheat sRNA transcriptome reveals distinct temporal expression pattern of miRNAs in response to heat, light and UV. Sci. Rep. 6, 39373. doi: 10.1038/srep39373
Roces, V., Lamelas, L., Valledor, L., Carbó, M., Cañal, M. J., Meijón, M. (2022). Integrative analysis in Pinus revealed long-term heat stress splicing memory. Plant J. 112, 998–1013. doi: 10.1111/tpj.15990
Sanyal, R. P., Misra, H. S., Saini, A. (2018). Heat-stress priming and alternative splicing-linked memory. J. Exp. Bot. 69, 2431–2434. doi: 10.1093/jxb/ery111
Song, L., Fang, Y., Chen, L., Wang, J., Chen, X. (2021). Role of non-coding RNAs in plant immunity. Plant Commun. 2, 100180. doi: 10.1016/j.xplc.2021.100180
Song, X., Li, Y., Cao, X., Qi, Y. (2019). MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 70, 489–525. doi: 10.1146/annurev-arplant-050718-100334
Stief, A., Altmann, S., Hoffmann, K., Pant, B. D., Scheible, W. R., Bäurle, I. (2014a). Arabidopsis miR156 Regulates Tolerance to Recurring Environmental Stress through SPL Transcription Factors. Plant Cell 26, 1792–1807. doi: 10.1105/tpc.114.123851
Stief, A., Brzezinka, K., Lämke, J., Bäurle, I. (2014b). Epigenetic responses to heat stress at different time scales and the involvement of small RNAs. Plant Signal Behav. 9, e970430. doi: 10.4161/15592316.2014.970430
Sunkar, R., Li, Y. F., Jagadeeswaran, G. (2012). Functions of microRNAs in plant stress responses. Trends Plant Sci. 17, 196–203. doi: 10.1016/j.tplants.2012.01.010
Szaker, H. M., Darkó, É., Medzihradszky, A., Janda, T., Liu, H. C., Charng, Y. Y., et al. (2019). miR824/AGAMOUS-LIKE16 module integrates recurring environmental heat stress changes to fine-tune poststress development. Front. Plant Sci. 10, 1454. doi: 10.3389/fpls.2019.01454
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., Singh, B. K. (2020). Plant-microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
Wang, J. W., Czech, B., Weigel, D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138, 738–749. doi: 10.1016/j.cell.2009.06.014
Wang, Y., Wu, F., Bai, J., He, Y. (2014). BrpSPL9 (Brassica rapa ssp. pekinensis SPL9) controls the earliness of heading time in Chinese cabbage. Plant Biotechnol. J. 12, 312–321. doi: 10.1111/pbi.12138
Xu, W. B., Zhao, L., Liu, P., Guo, Q. H., Wu, C. A., Yang, G. D., et al. (2023). Intronic microRNA-directed regulation of mitochondrial reactive oxygen species enhances plant stress tolerance in Arabidopsis. New Phytol. 240, 710–726. doi: 10.1111/nph.19168
Yang, H., Li, P., Jin, G., Gui, D., Liu, L., Zhang, C. (2022). Temporal regulation of alternative splicing events in rice memory under drought stress. Plant Divers. 44, 116–125. doi: 10.1016/j.pld.2020.11.004
Yu, Y., Zhang, Y., Chen, X., Chen, Y. (2019). Plant noncoding RNAs: hidden players in development and stress responses. Annu. Rev. Cell Dev. Biol. 35, 407–431. doi: 10.1146/annurev-cellbio-100818-125218
Yue, H., Zhang, H., Su, N., Sun, X., Zhao, Q., Weining, S., et al. (2022). Integrate small RNA and degradome sequencing to reveal drought memory response in wheat (Triticum aestivum L.). Int. J. Mol. Sci. 23 (11), 5917. doi: 10.3390/ijms23115917
Zhang, C., Tang, G., Peng, X., Sun, F., Liu, S., Xi, Y. (2018). Long non-coding RNAs of switchgrass (Panicum virgatum L.) in multiple dehydration stresses. BMC Plant Biol. 18, 79. doi: 10.1186/s12870-018-1288-3
Zhang, H., Zhu, J., Gong, Z., Zhu, J. K. (2022). Abiotic stress responses in plants. Nat. Rev. Genet. 23, 104–119. doi: 10.1038/s41576-021-00413-0
Zhao, J., Yuan, S., Zhou, M., Yuan, N., Li, Z., Hu, Q., et al. (2019). Transgenic creeping bentgrass overexpressing Osa-miR393a exhibits altered plant development and improved multiple stress tolerance. Plant Biotechnol. J. 17, 233–251. doi: 10.1111/pbi.12960
Zhao, Y., Zhu, P., Hepworth, J., Bloomer, R., Antoniou-Kourounioti, R. L., Doughty, J., et al. (2021). Natural temperature fluctuations promote COOLAIR regulation of FLC. Genes Dev. 35, 888–898. doi: 10.1101/gad.348362.121
Zhu, P., Lister, C., Dean, C. (2021). Cold-induced Arabidopsis FRIGIDA nuclear condensates for FLC repression. Nature 599, 657–661. doi: 10.1038/s41586-021-04062-5
Keywords: stress memory, environmental stresses, microRNA, small interfering RNA, long noncoding RNA, alternative splicing
Citation: Xu W-B, Cao F, Liu P, Yan K and Guo Q-H (2024) The multifaceted role of RNA-based regulation in plant stress memory. Front. Plant Sci. 15:1387575. doi: 10.3389/fpls.2024.1387575
Received: 18 February 2024; Accepted: 15 April 2024;
Published: 26 April 2024.
Edited by:
Ze-Ting Song, Yunnan University, ChinaReviewed by:
Zhihua Hua, Ohio University, United StatesCopyright © 2024 Xu, Cao, Liu, Yan and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian-Huan Guo, qhguo@sdau.edu.cn; Kang Yan, kangyan@sdau.edu.cn
 Wei-Bo Xu1
Wei-Bo Xu1 Peng Liu
Peng Liu Kang Yan
Kang Yan