Abstract
Monkeypox virus (MPV) infection has developed into a re-emerging disease, and despite the potential of tecovirimat and cidofovir drugs, there is currently no conclusive treatment. The treatment’s effectiveness and cost challenges motivate us to use In Silico approaches to seek natural compounds as candidate antiviral inhibitors. Using Maestro 11.5 in Schrodinger suite 2018, available natural molecules with validated chemical structures collected from Eximed Laboratory were subjected to molecular docking and ADMET analysis against the highly conserved A42R Profilin-like Protein of Monkeypox Virus Zaire-96-I-16 (PDB: 4QWO) with resolution of 1.52 Å solved 3D structure. Compared to the FDA-approved Tecovirimat, molecular docking revealed that Salsoline derivatives, Genistein, Semisynthetic derivative of kojic acid, and Naringenin had strengthened affinity (−8.9 to −10 kcal/mol) to 4QWO, and the molecular dynamic’s simulation confirmed their high binding stability. In support of these results, the hydrogen bond analysis indicated that the Salsoline derivative had the most robust interaction with the binding pockets of 4QWO among the four molecules. Moreover, the comparative free energy analyses using MM-PBSA revealed an average binding free energy of the complexes of Salsoline derivative, Genistein, Semisynthetic derivative of kojic acid, Naringenin, of −106.418, −46.808, −50.770, and −63.319 kJ/mol, respectively which are lower than −33.855 kJ/mol of the Tecovirimat complex. Interestingly, these results and the ADMET predictions suggest that the four compounds are promising inhibitors of 4QWO, which agrees with previous results showing their antiviral activities against other viruses.
Introduction
Monkeypox virus (MPV) infection has become persistent (Marennikova et al., 1972). MPV was initially identified in human populations in 1970 in the Democratic Republic of the Congo, and subsequently, the disease’s occurrence remained limited to this region (Marennikova et al., 1972). Later, MPV extended its presence to several African countries (Bunge et al., 2022; Meyer et al., 2002). In 1996 and 1997, the Democratic Republic of the Congo experienced outbreaks with low case fatality rates and attack rates higher than the norm. Since 2017, Nigeria has recorded over 500 suspected cases, with more than 200 confirmed cases and a case-fatality ratio exceeding 3% (Heymann et al., 1998). The virus’s first appearance outside Africa was recorded in 2003 in the USA (Reynolds et al., 2006). From 2018 to 2022, more than 40 countries, including the United Kingdom, Spain, Portugal, and Germany, identified a hundred infections (Do; Kumar et al., 2022). Historically, the case fatality rate for monkeypox in the general population varied from 0% to 11% and was more significant in young children (Nuzzo et al., 2022).
MPV is a double-enveloped DNA virus classified within the Poxviruses genus, part of the Poxvirus family (Kugelman et al., 2014). Two distinct groups of MPV have been identified: the Congo Basin group (Central African group) and the West African group. The Congo Basin group is thought to be more transmissible and has a history of causing more severe illnesses. (Likos et al., 2005). Based on the recent genomic sequencing data, the DNA sequences of the MPV strains found in Europe, specifically in Portugal, align with the West African clade, which implies a potentially less severe manifestation of the disease (Velavan and Meyer, 2022). In 2022, potential viral mutations are suggested to be responsible for the rapid transmission of monkeypox outbreak (Kumar et al., 2022) through direct contact with the blood, bodily fluids, skin lesions, or mucous secretions of sick animals, and animal-to-human infection (Angelo et al., 2019). Although several animals, such as rope squirrels and tree squirrels, have displayed indications of MPV infection (Tesh et al., 2004), the natural reservoir for MPV has not yet been found (Arita and Henderson, 1976) (World health organization, 2022a). MPV incubation (World health organization, 2022b; Kabuga and El Zowalaty, 2019) covers two stages: i) the invasion period (0–5 days long), which is characterized by fever, severe headache, enlarged lymph nodes, painful back and muscle pain, and significant weakness, and ii) the rash phase which frequently begins 1–3 days following the fever symptoms (McCollum and Damon, 2014). Several thousand lesions or sores merge in severe cases, leading to skin shedding (World health organization, 2022a; Kabuga and El Zowalaty, 2019; McCollum and Damon, 2014). A range of complications are known to be associated with MPV, including secondary infections, pneumonia, sepsis, encephalitis, and corneal infections leading to vision loss.
Several studies have revealed that smallpox immunization prevents 85% of the MPV infections. Therefore, a novel two-dose vaccine based on the attenuated modified vaccinia virus (Ankara strain) was authorized in 2019; however, this treatment option still confronts limited supplies and cost challenges (World health organization, 2022b; Gruber, 2022). The European Medicines Agency 2022 authorized an antiviral agent known as Tecovirimat (TPOXX®) to treat monkeypox, initially developed to treat smallpox but is not yet widely available (Sherwat et al., 2022; Rizk et al., 2022). Tecovirimat, an antiviral agent thwarting orthopoxvirus activity and preventing the production of infective virions, gained FDA approval on 13 July 2018. It has also secured European marketing authorization for treating Smallpox, Monkeypox, and Cowpox virus infections. Tecovirimat targets the viral protein p37, an essential component of the orthopoxvirus replication complex responsible for the maturation of the virus particles and their release from infected cells. By inhibiting the activity of p37, Tecovirimat effectively interferes with the viral replication cycle, preventing the spread of the virus within the host (Shiryaev et al., 2021; Sen Gupta et al., 2023).
Despite the genomics analysis of MPV, the proteome study is still challenging, and there is only a minimal solved protein structure, such as the highly conserved A42R protein, the first protein structure (resolution: 1.52 Å) deposited in the Protein Data Bank (PDB: 4QWO). Encoded by gp153 locus, the A42R protein role is still discussed despite the biochemical data suggesting its implication in replicating an assembly of orthopoxviruses. The current study’s findings and knowledge of its structure indicate that A42R could be a therapeutic target for structure-based drug design (Minasov et al., 2022; Bajrai et al., 2022).
The current study collected available natural molecules with validated chemical structures from the Eximed Laboratory. Using in silico approaches, they were evaluated as candidate inhibitors of the A42R Profilin-like Protein of Monkeypox Virus. The ADMET predictions for the selected Salsoline Derivatives, Genistein, a Semisynthetic Derivative of Kojic Acid, and Naringenin show promising results, justifying their potential for experimental validation, which represents the weakness of the present study.
Materials end methods
Ligand preparation
Available Naturel molecules with validated chemical structures were collected from Eximed Laboratory (https://eximedlab.com/) on 30 August 2022 (Alesawy et al., 2021) and subsequently prepared for docking calculations using the LigPrep utility available in the Maestro 11.5 edition of the Schrödinger Software program. This process utilized the OPLS3 force field and involved selecting ionization states for pH values 7.0 and 2.0. Furthermore, the ligands were allowed to generate a maximum of 32 stereoisomers (Mssillou et al., 2024).
Protein preparation
The three-dimensional crystal structure of the A42R Profilin-like Protein from the Monkeypox Virus (PDB: 4QWO), with a resolution of 1.52 Å, was retrieved from the Protein Data Bank in PDB format (Preet et al., 2022). The structure was subjected to the preparation procedure using the Protein Preparation Wizard in Schrödinger-Maestro v11.5, which includes the removal of all water molecules, the conversion of selenomethionine residues to methionine, and the addition of hydrogen atoms to heavy atoms. Minimization was carried out employing the OPLS3 force field, with a maximum allowable heavy atom RMSD (root-mean-square-deviation) set at 0.30.
The grid box coordinates were defined as x: 1.26, y: 6.4, and z: 25.9. The box had a volumetric spacing of 20 × 20 × 20. To establish the connection between the ligand and the protein-based grid box, the ‘Extra Precision’ (XP) mode was utilized, and the results were evaluated using the XP Gscore (Mssillou et al., 2024).
Three distinct modes were used to evaluate the prospective ligands, including high throughput virtual screening (HTVS), standard precision (SP), and extra precision (XP). This screening protocol was structured to iteratively improve ligand placements, beginning with HTVS, followed by SP mode, and culminating in XP mode (Veeramachaneni et al., 2015). Furthermore, ligand refinement involved applying the properties of absorption, metabolism, distribution, and excretion (ADMET) parameters and adhering to Lipinski’s Rule of Five using the QikProp tool within the Schrödinger Software, specifically version 11.5 of Maestro (Chebaibi et al., 2024). This evaluation was conducted based on the physicochemical and pharmacokinetic characteristics of various molecules analyzed in our study. Key factors considered included molecular weight, the number of hydrogen bond donors and acceptors, total solvent-accessible surface area, the blood-brain partition coefficient, the octanol/water partition coefficient, and aqueous solubility (Kumar et al., 2023; Sharma et al., 2022).
A redocking method was implemented to assess the molecular docking protocol’s reliability. This entailed docking the co-crystallized ligand into its binding site. The precision and validity of the docking protocol were determined by achieving a root mean square deviation (RMSD) value below 2 Å between the initial and docked ligand poses (Beniwal et al., 2022).
Molecular dynamics (MD) simulations
MD simulations were conducted utilizing GROMACS software (version 2019.3) to investigate the conformational dynamics of the most favorable docking complexes using the CHARMM 27 force field (Al-Khafaji and Taskin Tok, 2021). The protein topology was constructed using the GROMACS pdb2gmx modules with Chemistry at Harvard Macromolecular Mechanics force-field (CHARMm ff) (Chen et al., 2014), and the ligand topology was generated via the SwissParam server (Zoete et al., 2011). The docked structures were immersed in a simulation box with dimensions of 9.6 nm on each side and solvated using the three-point transferable intermolecular potential (TIP3P) solvent (Mark and Nilsson, 2001). To ensure system neutrality, ten chloride (Cl were introduced when needed. The system was then subjected to energy minimization using the steepest descent algorithm, with a maximum force threshold of 1,000 kJ/mol/nm. The pressure and temperature were then set to 1 bar and 300 K using the Nose-Hoover thermostat and isotropic Parrinello-Rahman barostat (Mahmoudi Gomari et al., 2022). Finally, a 100 ns simulation was carried out for each docked complex. We employed custom scripts derived from the results of molecular dynamics simulations to compute several metrics, such as root mean square deviation (RMSD) (utilizing ‘gmx rms’), root mean square fluctuation of residues (with ‘gmx rmsf’), Solvent Accessible Surface Area (SASA), the radius of gyration (Rg), and the count of hydrogen bonds (utilizing ‘gmx hbond').
MM-PBSA calculation
The binding of free energies for the ligands (ΔEbind) was calculated using the Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) method (Kumari et al., 2014). ΔEbind is calculated using the following equations:
Equation 1: Ecomplex is the total MMPBSA energy of the protein-ligand complex. Eprotein and Eligand are the total solution-free energies of the isolated protein and ligand, respectively. Equation 2: MM-G/PBSA is the sum of electrostatic (Eele), van der Waals (EvdW), polar (Gpolar), and nonpolar (Gnonpolar) energies.
Results and discussion
Virtual screening results
Collected compounds were initially assessed for their inhibitory effects against MPV. Based on a combination of pharmacokinetic and pharmacological criteria, a subset of 6,360 molecules were selected for further analysis through High Throughput Virtual Screening (HTVS). Among these, a refined selection of 900 natural products underwent evaluation using Standard Precision (SP) and Extra Precision (XP), identifying 61 promising candidates (Supplemental Files). Based on the docking score, glide emodel score, and glide energy score, the Salsoline derivative, Genistein, Semisynthetic derivative of kojic acid, and Naringenin have been selected as candidate ligands to the active site of the A42R Profilin-like Protein from the MPV (PDB: 4QWO) (Figure 1).
FIGURE 1

Selected natural products as potential MPV inhibitors.
Salsoline, a tetrahydroisoquinoline alkaloid extract from Salsola plants, is known for its diverse biological activities, including antibacterial effectiveness against Staphylococcus aureus, Streptococcus mutans, Bacillus subtilis, Streptococcus pneumoniae (Ou and Kwok, 2004; Wang et al., 2004; Wang et al., 2020) and antiviral activity against influenza A and B viruses (Wang et al., 2004). In the present in silico investigation, the Salsoline derivative exhibited the highest docking score, glide emodel score, and glide energy score, registering at −5.679, −49.575, and −36.546 kcal/mol, respectively (Table 1).
TABLE 1
| Compound | Glide gscore (Kcal/mol) | Glide emodel (Kcal/mol) | Glide energy (Kcal/mol |
|---|---|---|---|
| Salsoline derivative | −5.679 | −49.575 | −36.546 |
| Genistein | −5.617 | −47.838 | −33.885 |
| Semisynthetic derivative of kojic acid | −5.585 | −46.373 | −33.428 |
| Naringenin | −5.278 | −40.661 | −38.038 |
| Tecovirimat | −3.477 | −30.647 | −25.482 |
Docking results with ligands in A42R Profilin-like Protein from MPV (PDB:4QWO).
Genistein, identified as a potential A42R inhibitor, displayed a docking score, glide emodel score, and glide energy score of −5.617, −47.838, and −33.885 kcal/mol, respectively (Table 1). This phytoestrogen isoflavone is abundantly found in sources like soy and dairy products. It manifests various biological actions such as antioxidant activity, vermifuge activity, DNA topoisomerase inhibition, and tyrosine-protein kinase inhibition (Dixon and Ferreira, 2002; Polkowski and Mazurek, 2000). Notably, it has demonstrated antiviral efficacy against several viruses, including HSV-1, cytomegalovirus, and human immunodeficiency virus (LeCher et al., 2019).
Naringenin, the fourth selected molecule, displayed a docking score, glide emodel score, and glide energy score of −5.278, −40.661, and −38.038 kcal/mol, respectively. As a flavonoid in the flavanone class, it is predominantly found in citrus fruits, grapefruit leaves, and celery seeds. Naringenin’s diverse activities encompass anti-atherogenic, anti-inflammatory, anti-mutagenic, anticancer, and antiviral effects (Yin et al., 2018; Karim et al., 2018; Ke et al., 2017; Pinho-Ribeiro et al., 2016; Salehi et al., 2019).
Genistein and naringenin, isomers of the well-known antimicrobial and antiviral isocoumarins, demonstrated significant inhibitory effects on the profilin-like protein A42R. The structural similarity between these flavonoids and isocoumarins may underlie their observed bioactivity, reinforcing the potential of these compounds as antiviral agents. Previous studies on isocoumarins have shown their broad-spectrum bioactivity, which could inform further exploration of genistein and naringenin against MPV proteins (Ramanan et al., 2016; Sudarshan et al., 2015; Sudarshan et al., 2019).
Various microorganisms form kojic acid from the aerobic degradation of carbohydrates. Kojic acid and its derivatives are known for their biological activities, including antimicrobial and antiviral (Aytemir and Özçelik, 2010), antitumor (Nawarak et al., 2008), antidiabetic (Xiong and Pirrung, 2008), and anticancer (Yoo et al., 2010) activities. Through our virtual screening, a semisynthetic derivative of kojic acid emerged as a potential MPV inhibitor, garnering a docking score, glide emodel score, and glide energy score of −5.585, −46.373, and −33.428 kcal/mol, respectively (Table 1).
In the present study, as a reference, the Tecovirimat was docked within the active site of the A42R and displayed a docking score, glide model score, and glide energy score of −3.477, −30.647, and −25.482 kcal/mol, respectively (Table 1). Compared to Tecovirimat, this result underscores the heightened effectiveness of the four selected natural molecules as a candidate inhibitor of A42R.
Figures 2, 3 illustrate the molecular docking interactions, revealing the nature and quantity of potential bonds formed between the ligands and the active site of the A42R. The Salsoline derivative exhibited notable interactions, establishing three hydrogen bonds with ARG 115, ARG 122, and ASP 123 residues and a Pi cation bond with residue ARG 122. Genistein established two hydrogen bonds with residues ARG 122 and ASP 123, intriguingly, 2 Pi-Pi stacking bonds with TYR A 118 and TYR B 118. The semisynthetic kojic acid derivative displayed two hydrogen bonds with ARG 122 and ASP 123 residues and a Pi-Pi stacking bond with TYR A 118. Naringenin established a single hydrogen bond with residue ASP 123 in its interactions. These interactions highlight the diverse bonding patterns contributing to the potential ligand binding and inhibitory effects against the A42R Profilin-like Protein.
FIGURE 2

2D diagrams of candidate ligand’s interactions with the active sites. (A) Salsoline derivative; (B) Genistein; (C) Semisynthetic derivative of kojic acid; (D) Naringenin.
FIGURE 3

3D diagrams of candidate ligands interactions with the active sites. (A) Salsoline derivative; (B) Genistein; (C) Semisynthetic derivative of kojic acid; (D) Naringenin.
Bioavailability hinges on the interplay between the active compound’s absorption, distribution, metabolism, and excretion (ADME) rates, with its physicochemical attributes playing a crucial role (El-Seedi et al., 2012). ADME predictions offer insights into the natural product characteristics and pharmacokinetic attributes of compounds identified as potential inhibitors for A42R. Interestingly, the selected four compounds exhibit a molecular mass below 500 Mol and possess favorable values (≤5 and ≤10, respectively) for hydrogen bond donors and acceptors (Table 2). Moreover, the oral bioavailability of drug compounds is significantly impacted by the magnitude of the solvent-exposed surface area (Khan et al., 2019). Interestingly, the Salsoline derivative, Genistein, the Semisynthetic derivative of kojic acid, and Naringenin demonstrate satisfactory values falling within the 300 to 1,000 range. Our study’s results agree with the predicted apparent Caco-2 cell permeability (QPP Caco), representing the gut-blood barrier (with values below 25 indicating poor permeability and those above 500 signifying excellent permeability). Each selected candidate inhibitor of A42R Profilin-like Protein had an acceptable QPP Caco value, ranging from 56.587 to 186.558 nm/s.
TABLE 2
|
Compound |
MMa | Donors HBb | Acceptors HBc | SASAd | QPP cacoe | QP logPo/wf | QPlogBBg | QP logSh | % human oral Absorptioni |
|---|---|---|---|---|---|---|---|---|---|
| Salsoline derivative | 380.443 | 2 | 7 | 673.712 | 186.558 | 2.309 | −1.386 | −4.443 | 81.11 |
| Genistein | 270.241 | 2 | 3.75 | 485.362 | 163.953 | 1.717 | −1.335 | −3.089 | 76.639 |
| Semisynthetic derivative of kojic acid | 302.24 | 3 | 8.45 | 505.571 | 56.587 | −0.235 | −1.797 | −2.411 | 56.937 |
| Naringenin | 272.257 | 2 | 4 | 499.696 | 130.037 | 1.656 | −1.398 | −3.432 | 74.482 |
ADME properties of natural products selected as potential MPV inhibitors.
MM: mass of molecules (acceptable range: 500 mol).
Donors HB: donor of hydrogen bonds (acceptable range: ≤5).
Acceptors HB: acceptors of hydrogen bonds (acceptable range: ≤10).
SASA: Total solvent accessible surface area using a probe with a 1.4 radius (acceptable range: 300–1,000 radius).
QPP, Caco: Predicted apparent Caco-2, cell permeability in nm/s. Caco-2, cells is a model for the gut-blood barrier (˂25-poor, ˃500-great).
QP, logPo/w: Predicted octanol/water partition coefficient (acceptable range: −2–6.5).
QPlogBB: Predicted blood-brain partition coefficient (acceptable range: −3–1.2).
QP, logS: predicted aqueous solubility, S in mol/dm−3 (acceptable range: −6.5–0.5).
Predicted human oral absorption on 0%–100% scale (<25% is poor, and >80% is high).
Furthermore, these compounds exhibit advantageous blood-brain partition coefficient values, falling within the −3 to −1.2 range (as reported in reference (Pinho-Ribeiro et al., 2016)), suggesting their potential ability to cross the blood-brain barrier. As shown in Table 2, the anticipated oral absorption rates for these molecules range from 56% to 81%. Based on these promising results, the subsequent exploration aimed to delve into the molecular dynamics aspects of the four selected compounds.
Molecular dynamics (MD) simulation
The application of MD simulation revealed new insights into the stability and dynamic nature of protein-ligand interactions, configured space of sampling, inter-atomic forces calculation, generation of the trajectory that is then followed by the dynamics of a protein’s interaction with its ligand during the molecular motions and effect at a specific time (Shukla and Tripathi, 2020). In this study, MD simulations have been applied to assess the structural stability of A42R (PDB: 4QWO), lasting 100 ns, both in its unbound (apo) and ligand-bound states. In addition, Root-Mean-Square-Deviation (RMSD) analysis (Belhassan et al., 2021; Ouassaf et al., 2021) has been used to assess the stability of these systems. The RMSD analysis revealed the following average values for the 4QWO protein in various states: 0.23 nm for the unbound form, 0.15 nm for the Salsoline derivative complex, 0.21 nm for the Genistein complex, 0.17 nm for the Semisynthetic derivative of kojic acid complex, 0.15 nm for the Naringenin complex, and 0.15 nm for the Tecovirimat complex (Table 3). The values of all three systems consistently established that the simulations remained stable throughout the 100 nanoseconds, providing a reliable measure of conformational changes over time. The backbone RMSD plot analysis revealed the complexes’ stability during the entire MD simulation despite a minor deviation from their initial conformations (Figure 4). The MD simulation results indicate that the RMSD of the protein backbone in the APO protein (without any compound) was higher than that of the Salsoline derivative, Genistein, Semisynthetic derivative of kojic acid, Naringenin, and Tecovirimat-protein complex structures which suggests less stability of the APO protein under physiological conditions. For the 4QWO/Genistein and 4QWO/Semisynthetic derivative of kojic acid complexes, the RMSD graphs exhibited an upward trend, with RMSD values increasing from 0.10 to 0.40 nm between 0 and 70 ns, indicating that the compounds were adjusting to a new conformation within the binding pocket. Subsequently, the RMSD values plateaued and stabilized at around 0.20 nm, not exceeding the 0.3 nm threshold. In contrast, the 4QWO/Salsoline derivative, 4QWO/Naringenin, and 4QWO/Tecovirimat complexes achieved stability at approximately 20 ns, and there was no significant deviation in the protein backbone atoms for the remainder of the simulation, resulting in a final RMSD values ranging from 0.20 to 0.18 nm.
TABLE 3
|
Complex |
Average RMSD (nm) | Average RMSF (nm) | Radius of gyration (nm) | Average SASA (backbone, nm2) |
|---|---|---|---|---|
| Apo 4QWO | 0.23 ± 0.04 | 0.11 ± 0.04 | 2.04 ± 0.01 | 142.73 ± 1.36 |
| 4QWO - Salsoline derivative | 0.15 ± 0.02 | 0.09 ± 0.04 | 2.01 ± 0.01 | 143.33 ± 1.28 |
| 4QWO - Genistein | 0.22 ± 0.04 | 0.10 ± 0.03 | 2.00 ± 0.01 | 143.69 ± 1.25 |
| 4QWO - Semisynthetic derivative of kojic acid | 0.21 ± 0.04 | 0.10 ± 0.04 | 2.00 ± 0.01 | 143.26 ± 1.26 |
| 4QWO - Naringenin | 0.17 ± 0.03 | 0.10 ± 0.03 | 2.02 ± 0.01 | 143.10 ± 1.24 |
| 4QWO - Tecovirimat | 0.15 ± 0.02 | 0.09 ± 0.03 | 2.01 ± 0.01 | 142.64 ± 1.26 |
The calculated parameters for all the systems were obtained after 100 ns MD simulations.
Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) analysis.
FIGURE 4

Root mean square deviation (RMSD) of protein backbone vs simulation time for solvated 4QWO protein free and complexed with (A) Salsoline derivative, (B) Genistein, (C) Semisynthetic derivative of kojic acid, (D) Naringenin, and (E) Tecovirimat during 100 ns of molecular dynamics simulations.
Root mean square fluctuation (RMSF)
RMSF analysis was employed to explore the impact of ligand binding on the structural flexibility of the protein and the behavior of critical amino acids (Shao and Hall, 2017); a lower RMSF value suggests increased rigidity, while a higher value indicates greater flexibility (Khan et al., 2021). Figure 5 presents the RMSF profiles of Apo 4QWO, 4QWO - Salsoline derivative, Genistein, Semisynthetic derivative of kojic acid, Naringenin, and Tecovirimat complexes. A comparison to the Apo form of 4QWO reveals that the fluctuations in the residues of the ligand-bound complexes are stable, mainly in the ligand-binding regions. Additionally, the average RMSF values of 4QWO free, 4QWO - Salsoline derivative, Genistein, Semisynthetic derivative of kojic acid, Naringenin, and Tecovirimat complexes were 0.10, 0.09, 0.11, 0.10, and 0.09, respectively (Table 3), indicating that ligand binding has contributed to maintaining the structural stability of 4QWO.
FIGURE 5
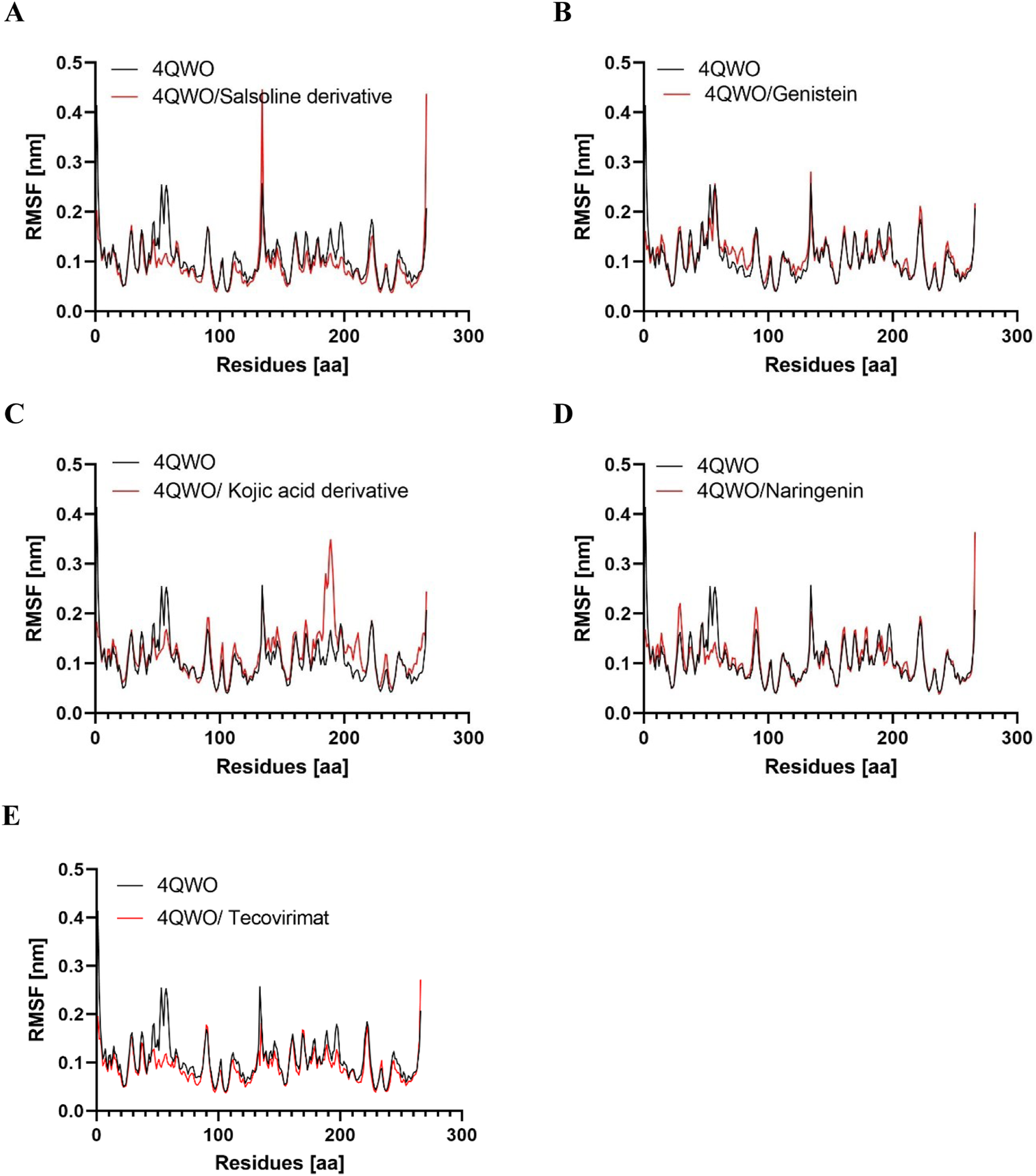
Root mean square deviation (RMSF) values of 4QWO alone and in complex with (A) Salsoline derivative, (B) Genistein, (C) Semisynthetic derivative of kojic acid, (D) Naringenin, and (E) Tecovirimat vs. the number of residues.
Radius of gyration (Rg)
To investigate the impact of various ligands on the overall compactness of the protein’s structure (El-Seedi et al., 2012), we examined the Rg over time; a ligand exhibiting a higher Rg value is more likely to display flexibility, indicating instability. Conversely, a lower Rg value suggests a denser and tightly packed conformation (Sharif et al., 2021). The average Rg values for Apo 4QWO, 4QWO-Salsoline derivative, Genistein, Semisynthetic derivative of kojic acid, Naringenin, and Tecovirimat complexes were determined to be 2.04, 2.01, 2.00, 2.00, 2.02, and 2.01, respectively (Table 3) which indicate that the binding to 4QWO did not induce a substantial alteration in compactness. Additionally, the graphical representation (Figure 6) illustrates that Rg values for the 4QWO - Tecovirimat, 4QWO - Salsoline derivative, and 4QWO - Semisynthetic derivative of kojic acid complexes achieved a stable equilibrium during the 100 ns simulation, in contrast to the 4QWO - Genistein and 4QWO - Naringenin complexes.
FIGURE 6

The radius of gyration (Rg) for backbone atoms of 4QWO alone and in complex with (A) Salsoline derivative, (B) Genistein, (C) Semisynthetic derivative of kojic acid, (D) Naringenin, and (E) Tecovirimat throughout the simulation.
Solvent accessible surface area (SASA)
SASA is defined as the area of a protein exposed to the surrounding solvent (Durham et al., 2009). The interpretation relies on assessing the connection between the surface of the macromolecule-ligand complex and the water molecules that are surrounded by (Priya et al., 2014). The variation in SASA for Apo 4QWO, 4QWO with a Salsoline derivative, Genistein, a Semisynthetic derivative of kojic acid, Naringenin, and Tecovirimat over a 100 ns period (Figure 7) showed an average value of 142.73 nm2, 143.33 nm2, 143.69 nm2, 143.26 nm2, 143.10 nm2, and 142.64 nm2, respectively (Table 3), which indicates that the ligands binding has no significant difference in the SASA values.
FIGURE 7

The comparative SASA values for backbone atoms of 4QWO alone and in complex with (A) Salsoline derivative, (B) Genistein, (C) Semisynthetic derivative of kojic acid, (D) Naringenin, and (E) Tecovirimat throughout the simulation.
Hydrogen bonds analysis
Hydrogen bond analysis is used to comprehend how the examined molecules are recognized at the molecular level, their interactions, and selectivity within the receptors. (Bissantz et al., 2010). This analysis helped monitor the protein-ligand interactions that emerged during MD simulation based on secondary structural elements. During the MD trajectories of the complexes, we quantified the number of hydrogen bonds formed. Figure 8 illustrates the counts of hydrogen bonds and pairs formed within a 0.35 nm distance for the 4QWO-Salsoline derivative, 4QWO-Genistein, 4QWO-Semisynthetic derivative of kojic acid, 4QWO-Naringenin, and 4QWO-Tecovirimat complexes throughout the MD simulation. The results revealed that the Salsoline derivative established an average of 2.67 hydrogen bonds with the active pocket of 4QWO. Similarly, Genistein and the Semisynthetic derivative of kojic acid acted as ligands, interacting with 4QWO within the binding site with an average of 2.45 and 2.96 hydrogen bonds, respectively. However, the average number of hydrogen bonds for the 4QWO-Naringenin and 4QWO-Tecovirimat complexes was 1.47 and 1.70, respectively. The hydrogen bond analysis plot indicated that the Salsoline derivative maintained a more robust interaction with the binding pockets of 4QWO throughout the simulation compared to the Genistein, the Semisynthetic derivative of kojic acid, and the Naringenin (Figure 5).
FIGURE 8

Hydrogen bond numbers made between (A) Salsoline derivative, (B) Genistein, (C) Semisynthetic derivative of kojic acid, (D) Naringenin, and (E) Tecovirimat in the4QWO protein active site residues during MD simulations.
Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) analysis
The MMPBSA analysis (Rostami et al., 2022) was used to evaluate the complexes’ binding affinities. All the trajectory snapshots were employed to calculate the primary forces governing the interactions between the protein and ligand. The total binding free energies (ΔEbind) for these complexes were computed in kJ/mol, and the results are presented in Table 4. The average binding free energy of the complexes, which include the Salsoline derivative, Genistein, Semisynthetic derivative of kojic acid, Naringenin, and Tecovirimat, were found to be −106.418, −46.808, −50.770, and −63.319 kJ/mol, respectively which are lower than −33.855 kJ/mol of the Tecovirimat complex. As indicated in Table 4, the Salsoline derivative exhibited the lowest binding energy, suggesting a weaker binding strength in this complex. This observation is consistent with the contributions of Van der Waals (VdW), electrostatic (Elec), and polar solvation energies to the binding energy, which align with the docking results.
TABLE 4
|
Complex |
Binding energy (kJ/mol) | SASA energy (kJ/mol) | Polar solvation energy (kJ/mol) | Electrostatic energy (kJ/mol) | Van der waals energy (kJ/mol) |
|---|---|---|---|---|---|
| 4QWO - Salsoline derivative | −106.418 + /- 58.464 | −15.086 + /- 4.008 | 108.736 + /- 26.747 | −67.391 + /- 25.660 | −132.676 + /- 43.347 |
| 4QWO - Genistein | −46.808 + /- 35.016 | −10.932 +/- 2.747 | 100.615 +/- 31.227 | −51.243 +/- 38.730 | −85.248 + /- 30.935 |
| 4QWO - Semisynthetic derivative of kojic acid | −50.770 + /- 24.495 | −10.521 +/- 2.052 | 114.180 + /- 32.332 | −71.260 + /- 31.135 | −83.169 + /- 19.987 |
| 4QWO - Naringenin | −63.319 + /- 22.461 | −12.709 + /- 1.899 | 95.126 + /- 28.289 | −44.171 + /- 33.057 | −101.565 + /- 19.561 |
| 4QWO - Tecovirimat | −33.855 + /- 28.604 | −6.387 + /- 3.545 | 32.325 + /- 34.411 | −7.179 + /- 11.858 | −52.614 + /-31.400 |
MM-PBSA calculations of binding free energy for All the complexes.
Conclusion
The recurrent spread of the Monkeypox virus has emerged as a global threat, posing significant health risks. Based on our virtual screening and molecular dynamics simulation, the Salsoline derivative, Genistein, the semi-synthetic derivative of kojic acid, and Naringenin have shown significant potential as inhibitors of the profilin-like protein A42R from the Monkeypox Zaire-96-I-16 virus (PDB: 4QWO).
Given that the absorption, distribution, metabolism, excretion, and toxicity (ADMET) predictions yielded promising results, we strongly recommend experimental validation to confirm the binding affinities of these compounds against A42R. This represents a limitation of the present study but also highlights an essential avenue for future research.
Looking forward, clinical trials to evaluate the anti-MPV activities of these natural compounds in patients infected with MPV are not only appropriate but also highly justified to further validate their therapeutic potential. Such studies could pave the way for developing new antiviral treatments, contributing significantly to public health efforts in managing and potentially curbing future outbreaks of the Monkeypox virus.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
MC: Conceptualization, Data curation, Formal Analysis, Methodology, Project administration, Resources, Supervision, Visualization, Writing–review and editing, Investigation, Software, Validation, Writing–original draft. MB: Data curation, Formal Analysis, Investigation, Methodology, Validation, Visualization, Writing–review and editing. FA: Investigation, Methodology, Validation, Visualization, Writing–review and editing. MS: Validation, Writing–review and editing. IM: Writing–review and editing, Investigation. MA-S: Investigation, Writing–review and editing, Methodology, Supervision, Validation. AK: Investigation, Methodology, Supervision, Writing–review and editing, Data curation, Formal Analysis, Funding acquisition, Resources, Software. RH: Investigation, Writing–review and editing. DB: Investigation, Writing–review and editing, Data curation, Formal Analysis, Methodology, Supervision. SA: Formal Analysis, Investigation, Methodology, Writing–review and editing, Data curation. RB: Formal Analysis, Investigation, Writing–review and editing, Resources, Supervision, Validation, Visualization. RD: Formal Analysis, Resources, Supervision, Visualization, Writing–review and editing, Conceptualization, Funding acquisition, Methodology, Project administration.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The authors extend their appreciation to the Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number: ISP23-81.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1
Alesawy M. S. Elkaeed E. B. Alsfouk A. A. Metwaly A. M. Eissa I. H. (2021). In silico screening of semi-synthesized compounds as potential inhibitors for SARS-CoV-2 papain-like protease: pharmacophoric features, molecular docking, ADMET, toxicity and DFT studies. Molecules26 (21), 6593. 10.3390/molecules26216593
2
Al-Khafaji K. Taskin Tok T. (2021). Understanding the mechanism of amygdalin’s multifunctional anti-cancer action using computational approach. J. Biomol. Struct. Dyn.39 (5), 1600–1610. 10.1080/07391102.2020.1736159
3
Angelo K. M. Petersen B. W. Hamer D. H. Schwartz E. Brunette G. (2019). Monkeypox transmission among international travellers—serious monkey business?. J. Travel Med.26 (5), taz002. 10.1093/jtm/taz002
4
Arita I. Henderson D. A. (1976). Monkeypox and whitepox viruses in west and central Africa. Bull. World Health Organ.53 (4), 347–353.
5
Aytemir M. D. Özçelik B. (2010). A study of cytotoxicity of novel chlorokojic acid derivatives with their antimicrobial and antiviral activities. Eur. J. Med. Chem.45 (9), 4089–4095. 10.1016/j.ejmech.2010.05.069
6
Bajrai L. H. Alharbi A. S. El-Day M. M. Bafaraj A. G. Dwivedi V. D. Azhar E. I. (2022). Identification of antiviral compounds against monkeypox virus profilin-like protein A42R from plantago lanceolata. Molecules27 (22), 7718. 10.3390/molecules27227718
7
Belhassan A. Zaki H. Chtita S. Alaqarbeh M. Alsakhen N. Benlyas M. et al (2021). Camphor, artemisinin and sumac phytochemicals as inhibitors against COVID-19: computational approach. Comput. Biol. Med.136, 104758. 10.1016/j.compbiomed.2021.104758
8
Beniwal M. Jain N. Jain S. Aggarwal N. (2022). Design, synthesis, anticancer evaluation and docking studies of novel 2-(1-isonicotinoyl-3-phenyl-1 H-pyrazol-4-yl)-3-phenylthiazolidin-4-one derivatives as Aurora-A kinase inhibitors. BMC Chem.16 (1), 61. 10.1186/s13065-022-00852-8
9
Bissantz C. Kuhn B. Stahl M. (2010). A medicinal chemist’s guide to molecular interactions. J. Med. Chem.53 (14), 5061–5084. 10.1021/jm100112j
10
Bunge E. M. Hoet B. Chen L. Lienert F. Weidenthaler H. Baer L. R. et al (2022). The changing epidemiology of human monkeypox—a potential threat? A systematic review. PLoS neglected Trop. Dis.16 (2), e0010141. 10.1371/journal.pntd.0010141
11
Chebaibi M. Mssillou I. Allali A. Bourhia M. Bousta D. Boschi Gonçalves R. F. et al (2024). Antiviral activities of compounds derived from medicinal plants against SARS-CoV-2 based on molecular docking of proteases. J. Biol. Biomed. Res.1 (1), 10–30. 10.69998/j2br1
12
Chen K.-C. Sun M. F. Chen H. Y. Lee C. C. Chen C. Y. C. (2014). Potential smoothened inhibitor from traditional Chinese medicine against the disease of diabetes, obesity, and cancer. BioMed Res. Int.2014, 1–12. 10.1155/2014/873010
13
Dixon R. A. Ferreira D. (2002). Genistein. Phytochemistry60 (3), 205–211. 10.1016/s0031-9422(02)00116-4
14
World Health Organization (2022a). US monkeypox outbreak: situation summary.
15
Durham E. Dorr B. Woetzel N. Staritzbichler R. Meiler J. (2009). Solvent accessible surface area approximations for rapid and accurate protein structure prediction. J. Mol. Model.15 (9), 1093–1108. 10.1007/s00894-009-0454-9
16
El-Seedi H. R. El-Said A. M. A. Khalifa S. A. M. Göransson U. Bohlin L. Borg-Karlson A. K. et al (2012). Biosynthesis, natural sources, dietary intake, pharmacokinetic properties, and biological activities of hydroxycinnamic acids. J. Agric. food Chem.60 (44), 10877–10895. 10.1021/jf301807g
17
Gruber M. F. (2022). Current status of monkeypox vaccines. npj Vaccines7 (1), 1–3. 10.1038/s41541-022-00527-4
18
Heymann D. L. Szczeniowski M. Esteves K. (1998). Re-emergence of monkeypox in Africa: a review of the past six years. Br. Med. Bull.54 (3), 693–702. 10.1093/oxfordjournals.bmb.a011720
19
Kabuga A. I. El Zowalaty M. E. (2019). A review of the monkeypox virus and a recent outbreak of skin rash disease in Nigeria. J. Med. Virology91 (4), 533–540. 10.1002/jmv.25348
20
Karim N. Jia Z. Zheng X. Cui S. Chen W. (2018). A recent review of citrus flavanone naringenin on metabolic diseases and its potential sources for high yield-production. Trends Food Sci. and Technol.79, 35–54. 10.1016/j.tifs.2018.06.012
21
Ke J. Y. Banh T. Hsiao Y. H. Cole R. M. Straka S. R. Yee L. D. et al (2017). Citrus flavonoid naringenin reduces mammary tumor cell viability, adipose mass, and adipose inflammation in obese ovariectomized mice. Mol. Nutr. and food Res.61 (9), 1600934. 10.1002/mnfr.201600934
22
Khan M. F. Verma G. Akhtar W. Shaquiquzzaman M. Akhter M. Rizvi M. A. et al (2019). Pharmacophore modeling, 3D-QSAR, docking study and ADME prediction of acyl 1, 3, 4-thiadiazole amides and sulfonamides as antitubulin agents. Arabian J. Chem.12 (8), 5000–5018. 10.1016/j.arabjc.2016.11.004
23
Khan S. A. Zia K. Ashraf S. Uddin R. Ul-Haq Z. (2021). Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 via integrated computational approach. J. Biomol. Struct. Dyn.39 (7), 2607–2616. 10.1080/07391102.2020.1751298
24
Kugelman J. R. Johnston S. C. Mulembakani P. M. Kisalu N. Lee M. S. Koroleva G. et al (2014). Genomic variability of monkeypox virus among humans, Democratic Republic of the Congo. Emerg. Infect. Dis.20 (2), 232–239. 10.3201/eid2002.130118
25
Kumar H. Sharma A. Kumar D. Marwaha M. G. Dhanawat M. Aggarwal N. et al (2023). Synthesis, biological evaluation and in silico studies of some new analogues of 3, 5-disubstituted thiazolidin-2, 4-dione. Future Med. Chem.15 (24), 2257–2268. 10.4155/fmc-2023-0237
26
Kumar N. Acharya A. Gendelman H. E. Byrareddy S. N. (2022). The 2022 outbreak and the pathobiology of the monkeypox virus. J. Autoimmun.131, 102855. 10.1016/j.jaut.2022.102855
27
Kumari R. Kumar R. Lynn A. (2014). g_mmpbsa⋅ A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model.54 (7), 1951–1962. 10.1021/ci500020m
28
LeCher J. C. Diep N. Krug P. W. Hilliard J. K. (2019). Genistein has antiviral activity against herpes B virus and acts synergistically with antiviral treatments to reduce effective dose. Viruses11 (6), 499. 10.3390/v11060499
29
Likos A. M. Sammons S. A. Olson V. A. Frace A. M. Li Y. Olsen-Rasmussen M. et al (2005). A tale of two clades: monkeypox viruses. J. General Virology86 (10), 2661–2672. 10.1099/vir.0.81215-0
30
Mahmoudi Gomari M. Rostami N. Omidi-Ardali H. Arab S. S. (2022). Insight into molecular characteristics of SARS-CoV-2 spike protein following D614G point mutation, a molecular dynamics study. J. Biomol. Struct. Dyn.40 (12), 5634–5642. 10.1080/07391102.2021.1872418
31
Marennikova S. Seluhina E. M. Mal'ceva N. N. Cimiskjan K. L. Macevic G. R. (1972). Isolation and properties of the causal agent of a new variola-like disease (monkeypox) in man. Bull. World Health Organ.46 (5), 599–611.
32
Mark P. Nilsson L. (2001). Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A105 (43), 9954–9960. 10.1021/jp003020w
33
McCollum A. M. Damon I. K. (2014). Human monkeypox. Clin. Infect. Dis.58 (2), 260–267. 10.1093/cid/cit703
34
Meyer H. Perrichot M. Stemmler M. Emmerich P. Schmitz H. Varaine F. et al (2002). Outbreaks of disease suspected of being due to human monkeypox virus infection in the Democratic Republic of Congo in 2001. J. Clin. Microbiol.40 (8), 2919–2921. 10.1128/jcm.40.8.2919-2921.2002
35
Minasov G. Inniss N. L. Shuvalova L. Anderson W. F. Satchell K. J. (2022). Structure of the Monkeypox profilin-like protein A42Rreveals potential function differences from cellular profilins. bioRxiv, 2022. 10.1107/S2053230X22009128
36
Mssillou I. Agour A. Lefrioui Y. Chebaibi M. (2024). LC-TOFMS analysis, in vitro and in silico antioxidant activity on NADPH oxidase, and toxicity assessment of an extract mixture based on Marrubium vulgare L. and Dittrichia viscosa L. J. Biol. Biomed. Res.1 (1), 31–45. 10.69998/j2br2
37
Nawarak J. Huang-Liu R. Kao S. H. Liao H. H. Sinchaikul S. Chen S. T. et al (2008). Proteomics analysis of kojic acid treated A375 human malignant melanoma cells. J. Proteome Res.7 (9), 3737–3746. 10.1021/pr7008737
38
Nuzzo J. B. Borio L. L. Gostin L. O. (2022). The WHO declaration of monkeypox as a global public health emergency. Jama328, 615. 10.1001/jama.2022.12513
39
Ou S. Kwok K. C. (2004). Ferulic acid: pharmaceutical functions, preparation and applications in foods. J. Sci. Food Agric.84 (11), 1261–1269. 10.1002/jsfa.1873
40
Ouassaf M. Belaidi S. Chtita S. Lanez T. Abul Qais F. Md Amiruddin H. (2021). Combined molecular docking and dynamics simulations studies of natural compounds as potent inhibitors against SARS-CoV-2 main protease. J. Biomol. Struct. Dyn.40, 11264–11273. 10.1080/07391102.2021.1957712
41
Pinho-Ribeiro F. A. Zarpelon A. C. Fattori V. Manchope M. F. Mizokami S. S. Casagrande R. et al (2016). Naringenin reduces inflammatory pain in mice. Neuropharmacology105, 508–519. 10.1016/j.neuropharm.2016.02.019
42
Polkowski K. Mazurek A. P. (2000). Biological properties of genistein. A review of in vitro and in vivo data. Acta Pol. Pharm.57 (2), 135–155.
43
Preet G. Oluwabusola E. T. Milne B. F. Ebel R. Jaspars M. (2022). Computational repurposing of mitoxantrone-related structures against monkeypox virus: a molecular docking and 3D pharmacophore study. Int. J. Mol. Sci.23 (22), 14287. 10.3390/ijms232214287
44
Priya G. Doss C. Nagasundaram N. (2014). Molecular docking and molecular dynamics study on the effect of ERCC1 deleterious polymorphisms in ERCC1-XPF heterodimer. Appl. Biochem. Biotechnol.172 (3), 1265–1281. 10.1007/s12010-013-0592-5
45
Ramanan M. Sinha S. Sudarshan K. Aidhen I. S. Doble M. (2016). Inhibition of the enzymes in the leukotriene and prostaglandin pathways in inflammation by 3-aryl isocoumarins. Eur. J. Med. Chem.124, 428–434. 10.1016/j.ejmech.2016.08.066
46
Reynolds M. G. Yorita K. Kuehnert M. Davidson W. Huhn G. Holman R. et al (2006). Clinical manifestations of human monkeypox influenced by route of infection. J. Infect. Dis.194 (6), 773–780. 10.1086/505880
47
Rizk J. G. Lippi G. Henry B. M. Forthal D. N. Rizk Y. (2022). Prevention and treatment of monkeypox. Drugs82, 957–963. 10.1007/s40265-022-01742-y
48
Rostami N. Choupani E. Hernandez Y. Arab S. S. Jazayeri S. M. Gomari M. M. (2022). SARS‐CoV‐2 spike evolutionary behaviors; simulation of N501Y mutation outcomes in terms of immunogenicity and structural characteristic. J. Cell. Biochem.123 (2), 417–430. 10.1002/jcb.30181
49
Salehi B. Fokou P. V. T. Sharifi-Rad M. Zucca P. Pezzani R. Martins N. et al (2019). The therapeutic potential of naringenin: a review of clinical trials. Pharmaceuticals12 (1), 11. 10.3390/ph12010011
50
Sen Gupta P. S. Panda S. K. Nayak A. K. Rana M. K. (2023). Identification and investigation of a cryptic binding pocket of the P37 envelope protein of monkeypox virus by molecular dynamics simulations. J. Phys. Chem. Lett.14 (13), 3230–3235. 10.1021/acs.jpclett.3c00087
51
Shao Q. Hall C. K. (2017). Allosteric effects of gold nanoparticles on human serum albumin. Nanoscale9 (1), 380–390. 10.1039/c6nr07665c
52
Sharif M. Hossen N. L. Shaikat M. M. Haidary F. Ema T. I. Dey D. et al (2021). Molecular optimization, docking and dynamic simulationstudy of selective natural aromatic components to block E2-CD81 complex formation inpredating protease inhibitor resistant HCV influx. Int. J. Pharm. Res.13 (2), 3511–3525. 10.31838/ijpr/2021.13.02.408
53
Sharma R. K. Bibi S. Chopra H. Khan M. S. Aggarwal N. Singh I. et al (2022). In silico and in vitro screening constituents of eclipta alba leaf extract to reveal antimicrobial potential. Evid. Based Complementary Altern. Med.2022 (1), 1–14. 10.1155/2022/3290790
54
Sherwat A. Brooks J. T. Birnkrant D. Kim P. (2022). Tecovirimat and the treatment of monkeypox—past, present, and future considerations. N. Engl. J. Med.387, 579–581. 10.1056/nejmp2210125
55
Shiryaev V. A. Skomorohov M. Y. Leonova M. V. Bormotov N. I. Serova O. A. Shishkina L. N. et al (2021). Adamantane derivatives as potential inhibitors of p37 major envelope protein and poxvirus reproduction. Design, synthesis and antiviral activity. Eur. J. Med. Chem.221, 113485. 10.1016/j.ejmech.2021.113485
56
Shukla R. Tripathi T. (2020). “Molecular dynamics simulation of protein and protein–ligand complexes,” in Computer-aided drug design (Springer), 133–161.
57
Sudarshan K. Boda A. k. Dogra S. Bose I. Yadav P. N. Aidhen I. S. (2019). Discovery of an isocoumarin analogue that modulates neuronal functions via neurotrophin receptor TrkB. Bioorg. and Med. Chem. Lett.29 (4), 585–590. 10.1016/j.bmcl.2018.12.057
58
Sudarshan K. Manna M. K. Aidhen I. S. (2015). Synthesis of 3‐arylisocoumarins by using acyl anion Chemistry and synthesis of thunberginol A and cajanolactone A. Eur. J. Org. Chem.2015 (8), 1797–1803. 10.1002/ejoc.201403524
59
Tesh R. B. Watts D. M. Sbrana E. Siirin M. Popov V. L. Xiao S. Y. (2004). Experimental infection of ground squirrels (Spermophilus tridecemlineatus) with monkeypox virus. Emerg. Infect. Dis.10 (9), 1563–1567. 10.3201/eid1009.040310
60
Veeramachaneni G. K. kodamala K. R. Chalasani L. M. J S B. Talluri V. R. (2015). High-throughput virtual screening with e-pharmacophore and molecular simulations study in the designing of pancreatic lipase inhibitors. Drug Des. Dev. Ther.9, 4397–4412. 10.2147/dddt.s84052
61
Velavan T. P. Meyer C. G. (2022). Monkeypox 2022 outbreak: an update. Trop. Med. and Int. Health27, 604–605. 10.1111/tmi.13785
62
Wang R. F. Feng Wang R. Wei Yang X. Mei Ma C. Nong Li J. Shoyama Y. (2004). A bioactive alkaloid from the flowers of Trollius chinensis. Heterocycles-Sendai Inst. Heterocycl. Chem.63 (6), 1443–1448. 10.3987/com-04-10062
63
Wang S. Yan M. Guo Y. Sun R. Jin H. Gong Y. (2020). In vivo and in vitro effects of Salsola collina on gastrointestinal motility in rats. Iran. J. Basic Med. Sci.23 (3), 383–389. 10.22038/IJBMS.2019.40613.9605
64
World health organization . MonkeyPox, 2022b.
65
Xiong X. Pirrung M. C. (2008). Modular synthesis of candidate indole-based insulin mimics by Claisen rearrangement. Org. Lett.10 (6), 1151–1154. 10.1021/ol800058d
66
Yin J. Liang Y. Wang D. Yan Z. Yin H. Wu D. et al (2018). Naringenin induces laxative effects by upregulating the expression levels of c-Kit and SCF, as well as those of aquaporin 3 in mice with loperamide-induced constipation. Int. J. Mol. Med.41 (2), 649–658. 10.3892/ijmm.2017.3301
67
Yoo D. S. Lee J. Choi S. S. Rho H. S. Cho D. H. Shin W. C. et al (2010). A modulatory effect of novel kojic acid derivatives on cancer cell proliferation and macrophage activation. Die Pharmazie-An Int. J. Pharm. Sci.65 (4), 261–266. 10.1691/ph.2010.9764
68
Zoete V. Cuendet M. A. Grosdidier A. Michielin O. (2011). SwissParam: a fast force field generation tool for small organic molecules. J. Comput. Chem.32 (11), 2359–2368. 10.1002/jcc.21816
Summary
Keywords
monkeypox virus, natural compounds, virtual screening, molecular dynamics simulation, tecovirimat
Citation
Chebaibi M, Bourhia M, Amrati Fe-z, Slighoua M, Mssillou I, Aboul-Soud MAM, Khalid A, Hassani R, Bousta D, Achour S, Benhida R and Daoud R (2024) Salsoline derivatives, genistein, semisynthetic derivative of kojic acid, and naringenin as inhibitors of A42R profilin-like protein of monkeypox virus: in silico studies. Front. Chem. 12:1445606. doi: 10.3389/fchem.2024.1445606
Received
10 June 2024
Accepted
26 August 2024
Published
10 September 2024
Volume
12 - 2024
Edited by
Belgin Sever, Anadolu University, Türkiye
Reviewed by
Kasireddy Sudarshan, Purdue University, United States
Navidha Aggarwal, Maharishi Markandeshwar University, India
Updates
Copyright
© 2024 Chebaibi, Bourhia, Amrati, Slighoua, Mssillou, Aboul-Soud, Khalid, Hassani, Bousta, Achour, Benhida and Daoud.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rachid Daoud, rachid.daoud@um6p.ma; Asaad Khalid, akahmed@jazanu.edu.sa; Mohamed Chebaibi, mohamed.chebaibi@usmba.ac.ma
† Present address: Fatima ez-zahra Amrati, Laboratory of Cell Biology and Molecular Genetics (LBCGM), Department of Biology, Faculty of Sciences, Ibn Zohr University, Agadir, Souss Massa, Morocco Meryem Slighoua, Ministry of Health and Social Protection, Higher Institute of Nursing Professions and Health Techniques, Marrakech, Morocco
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.