Abstract
Mycobacterium tuberculosis (Mtb) is a significant and highly pathogenic intracellular microorganism responsible for tuberculosis (TB). The global TB crisis has been exacerbated by the emergence and spread of multidrug-resistant Mtb strains, resulting in elevated mortality rates. Mtb MmcO exhibits scavenging activity against reactive oxygen species (ROS), thereby supporting Mtb survival. However, the molecular mechanism underlying MmcO function remains poorly understood. Herein, the hydrodynamic radius of MmcO was determined to be 5.9 ± 0.3 nm. A structural model of MmcO was predicted using AlphaFold2 and subsequently evaluated for accuracy using a Ramachandran plot and ProSA analysis. Site-directed mutagenesis revealed that substitutions H120A, H122A, H161A, or H163A nearly abolished the activity, while mutations H120R, H122R, H161R, or H163R led to minor alterations in the activity. The addition of Triton X-100 or Ca2+ significantly enhanced MmcO activity, whereas EDTA or other metal ions markedly inhibited its activity to varying extents. MmcO, a multicopper oxidase, plays a role in maintaining redox homeostasis in Mtb, a function critical for bacterial survival in host macrophages. Our study reveals that Cu2+ is essential for enzymatic activity, while Ni2+, Mn2+, and Zn2+ inhibit function, likely due to improper metal coordination. Given its importance in oxidative stress resistance, MmcO presents a promising drug target for Mtb therapy. Therefore, this study offers valuable insights for developing novel therapeutic agents targeting Mtb.
Introduction
Tuberculosis (TB) is a persistent infectious disease caused by Mycobacterium tuberculosis (Mtb) and remains a significant global health challenge (Getahun et al., 2015; Reid et al., 2019; Pai et al., 2016; Dowdy and Behr, 2022; Furin et al., 2019). According to the World Health Organization, despite the extensive implementation of intensive chemotherapy over the past several decades, approximately one-quarter of the global population remains infected with Mtb (https://www.who.int/news-room/fact-sheets/detail/tuberculosis). TB severity is exacerbated by co-morbid conditions, such as infections with HIV or SARS-CoV-2 (Dheda et al., 2022; Flores-Lovon et al., 2022; Udoakang et al., 2023; Dhana et al., 2022; Rule et al., 2021). The emergence of drug-resistant Mtb strains poses a major challenge to TB treatment, which exhibit resistance to key anti-TB drugs, including isoniazid, pyrazinamide, and rifampicin (Dominguez et al., 2023; Naidoo and Dookie, 2022; Dean et al., 2022). These drug-resistant strains significantly compromise the efficacy of TB treatment, resulting in elevated mortality rates (Tiberi et al., 2018; Micoli et al., 2021; Lange et al., 2019; Jenkins et al., 2014; Getahun et al., 2015; Dheda et al., 2017; Furin et al., 2019; Reid et al., 2019). Therefore, a deeper understanding of Mtb pathogenesis is urgently required to facilitate the development of more effective therapeutic strategies against TB.
Multicopper oxidases play key roles in bacterial pathogenesis, with homologs in Pseudomonas aeruginosa and Salmonella enterica contributing to copper detoxification and oxidative stress resistance. In Mtb, MmcO may function similarly to SodA and KatG in mitigating reactive oxygen and nitrogen species, highlighting its potential as a novel drug target.
Mtb has the ability to evade host immune surveillance and clearance, allowing it to persist within host macrophages for extended periods (O’Garra et al., 2013; Getahun et al., 2015; Tiberi et al., 2018). Within macrophages, Mtb is exposed to diverse oxidative stress conditions, including fluctuations in pH, reactive oxygen species (ROS), enzymatic degradation, nutrient deprivation, and nitrogen intermediates (Aderem and Underhill, 1999; Agarwal et al., 2021; Kinkar et al., 2019; Sharma et al., 2023; Raya et al., 2022). Mtb MmcO has been shown to exhibit ROS scavenging activity, effectively neutralizing ROS produced in the xanthine-xanthine oxidase enzyme system and, more critically, ROS generated in activated THP-1 cells (Kinkar et al., 2019). The cysteine residue in the N-terminus of Mtb MmcO aligns with a putative signal peptidase cleavage site and likely serves as the site for lipid modification that anchors the protein to the membrane. MmcO represents a promising target for the development of novel therapeutic agents against Mtb. However, the functional mechanisms and detailed molecular roles of Mtb MmcO are not well known.
Here, the structural model of MmcO was predicted using AlphaFold2 and subsequently evaluated using a Ramachandran plot and ProSA. The results of site-directed mutagenesis experiments demonstrated that substitutions H120A, H122A, H161A, or H163A nearly abolished enzymatic activity, while mutations H120R, H122R, H161R, or H163R caused minor alterations in the activity. The addition of Triton X-100 or Ca2+ significantly enhanced MmcO activity, whereas EDTA or other metal ions markedly reduced activity to varying extents. These findings highlight two critical sites on MmcO essential for its activity and provide intriguing insights into the metal ion preferences of Mtb MmcO. These offers a potential foundation for the development of novel antitubercular therapies.
Results
Bioinformatics analysis
The multicopper oxidase MmcO of Mtb contains a single conserved domain, the multicopper oxidase domain, spanning amino acids 45–504 (Figure 1a; Supplementary Figure S1). The sequence of MmcO was obtained from the UniProt database (accession number I6WZK7). The estimated molecular weight of MmcO is approximately 53.8 kDa. Its molecular formula was determined to be C2386H3717N667O715S19, and its isoelectric point (pI) was calculated to be 5.9.
FIGURE 1

The cluster organization and structural model of MmcO. (a) Schematic diagram illustrating the signal (orange) and multicopper oxidase (cyan) domains. (b) The MmcO structure, predicted using AlphaFold2, is shown in ribbon representation from two orientations, with the conserved domain colored as depicted in Figure 1A. (c) Structural validation of MmcO was carried out using Ramachandran Plot analysis, where the most favored regions are highlighted in red, with progressively lighter shades indicating less favored regions. (d) ProSA analysis indicated that the Z-score of the MmcO structure was −11.27.
The efficacy of codon optimization was evaluated by analyzing the codon adaptation index (CAI) and GC content. The optimized CAI for MmcO was determined to be 84.6% (Table 1). The GC content, which is considered optimal within the range of 30%–70%, was optimized to 58.3% for MmcO, falling well within the acceptable range (Table 1).
TABLE 1
| Optimization for codon | Codon adaptation index (CAI) value | GC content value |
|---|---|---|
| Before codon optimization | 67.2% | 64.0% |
| After codon optimization | 84.6% | 58.3% |
Codon optimization of MmcO.
Prediction and quality assessment of MmcO structure
The three-dimensional (3D) structure of MmcO was predicted using the AlphaFold2 (Jumper et al., 2021; Wayment-Steele et al., 2023; Tunyasuvunakool et al., 2021) (Figure 1b). In contrast to traditional homology modeling approaches utilized in earlier studies, AlphaFold2 leverages deep learning techniques to achieve improved accuracy and reliability in protein structure prediction (Supplementary Figure S2).
The quality of the predicted MmcO structural model was evaluated using a Ramachandran plot, which assessed whether the dihedral angles of the protein backbone were within acceptable regions, thereby validating the structural integrity of the model (Figure 1c; Supplementary Figure S2). The analysis revealed that 86.9% of the residues were located in the most favored regions, 12.6% in the additionally allowed regions, 0.5% in the generously allowed regions, and none in the disallowed regions (Figure 1c; Table 2). Furthermore, the structure included 2 terminal residues, 55 glycine residues, and 41 proline residues (Figure 1c; Table 2). The quality of structural model was further confirmed using ProSA, an interactive web tool for detecting errors in 3D protein structures, which provided a Z-score of −11.27, indicating the high quality of the predicted structure (Figure 1d).
TABLE 2
| Constructs | Residues in most favored regions | Residues in additional allowed regions | Residues in generously allowed regions | Residues in disallowed regions | ||||
|---|---|---|---|---|---|---|---|---|
| Residues | Number of residues | % of residuesa | Number of residues | % of residues | Number of residues | % of residues | Number of residues | % of residues |
| Statistics | 353 | 86.9 | 51 | 12.6 | 2 | 0.5 | 0 | 0 |
Ramchandran plot analysis of MmcO structural model using PDBsum.
Note: Number of end-residues (excl. Gly and Pro): 2; Number of glycine residues (shown as triangles): 55; Number of proline residues: 41.
Overall, the structural model of MmcO is of good quality and is suitable for further following analysis.
Characterization of MmcO by dynamic light scattering
Dynamic light scattering (DLS) experiments were conducted to further examine the oligomeric state of MmcO by determining its diameter following centrifugation. The regularized DLS histograms were analyzed, showing that the hydrodynamic radius of MmcO was 5.9 ± 0.3 nm (Figure 2). This result indicated that MmcO existed in a monomeric form.
FIGURE 2

Dynamic light scattering (DLS) spectrum of MmcO. The hydrodynamic radius of MmcO was measured as 5.9 ± 0.3 nm, showing the existence of monomeric MmcO.
MmcO crystals resisted optimization
The primary objective of this study was to elucidate the structure of full-length MmcO in order to investigate the relationship between its functional and structural characteristics. However, despite considerable effort, optimization of MmcO crystallization proved unsuccessful (Figure 3a).
FIGURE 3

The crystal showed limitations in obtaining optimized diffracting MmcO crystals. (a) MmcO crystals were of poor quality. (b) Despite extensive optimization efforts, attempts to improve the crystallization of MmcO83-504 (in red frame) were unsuccessful in producing suitable diffraction patterns. Consequently, (c) the diffraction image obtained for MmcO83-504 was of low quality, with few clearly resolvable diffraction spots.
Following initial crystal screening of three recombinant MmcO proteins (MmcO45-504 and MmcO83-504), crystals of MmcO83-504 were successfully obtained in 8% (w/v) polyethylene glycol (PEG) 3,350 with 0.1 M sodium formate at pH 4.5. However, despite extensive efforts to optimize the crystallization conditions (Figure 3b), the MmcO83-504 crystals showed poor diffraction quality and were resistant to further optimization (Figure 3c). Consequently, crystal optimization remains ongoing.
MmcO83-504 accounts for the full catalytic activity of full-length MmcO
Full-length MmcO was truncated based on the structural model predicted using AlphaFold2 (Tunyasuvunakool et al., 2021; Jumper et al., 2021; Tunyasuvunakool et al., 2021; Wayment-Steele et al., 2023; Abramson et al., 2024). However, the specific role of MmcO45-504 or MmcO83-504 in regulating the multicopper oxidase activity of the full-length MmcO remained unclear. Both MmcO45-504 and MmcO83-504 were expressed and purified using the same protocols as full-length MmcO. Our results showed that the copper oxidase activity of MmcO45-504 and MmcO83-504 was identical to that of the full-length MmcO (Figure 4), demonstrating that MmcO83-504 plays a crucial role in maintaining the activity of the full-length MmcO.
FIGURE 4

Comparative analysis of the multicopper oxidase activity between MmcO45-504, MmcO83-504 and MmcO. The multicopper oxidase activity of MmcO45-504 or MmcO83-504 was identical to that of full-length MmcO, showing that MmcO45-504 or MmcO83-504 was responsible for all full-length MmcO activity. The activity of wild-type (WT) MmcO was set to 100%.
Enzymatic activity for MmcO site-directed mutation
Mutagenesis experiments were performed based on the sequence alignment results (Figure 5; Supplementary Figure S1). The mutations H120A, H122A, H161A, or H163A resulted in the complete loss of copper oxidase activity when compared to the wild-type (WT) protein (Figure 6a). In contrast, mutations H120R, H122R, H161R, or H163R caused only minor changes in activity (Figure 6b). This result can be attributed to the stabilizing effects of His and Arg residues, which possess positive charges. Consequently, residues H120, H122, H161, and H163 are crucial for MmcO activity, and their proximity to the substrate underscores the importance of these mutations.
FIGURE 5
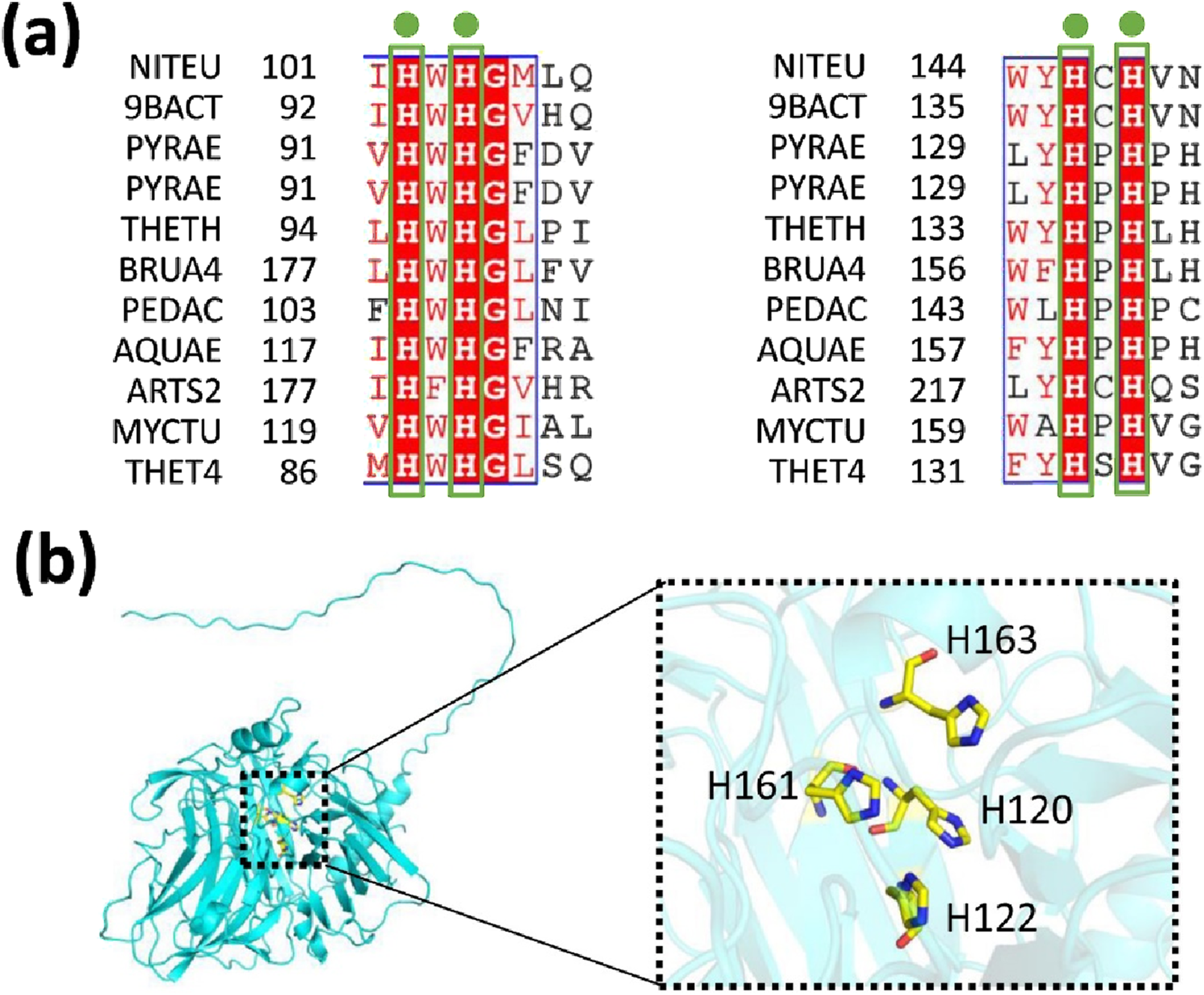
Relative multicopper oxidase activities of wild-type (WT) MmcO and its indicated mutants. (a) Sequence alignment of conserved residues from various species. NITEU, Nitrosomonas europaea; 9BACT, Escherichia coli; PYRAE, hyperthermophilic archaeon Pyrobaculum aerophilum; PYRAE, Pyrobaculum aerophilum str. IM2; THETH, Thermus thermophilus; BRUA4, Ochrobactrum; PEDAC, Pediococcus acidilactici; AQUAE, hyperthermophile aquifex aeolicus; ARTS2, Arthrobacter sp. FB24; MYCTU, Mycobacterium tuberculosis; THET4, Thermothelomyces thermophilus. (b) The conserved amino acids are identified as: H120, H122, H161 and H163. These conserved residues are highlighted with sticks and colored yellow. The structural model of MmcO is displayed in a cyan cartoon format.
FIGURE 6

Relative multicopper oxidase activities of wild-type (WT) MmcO and its indicated mutants. (a) H120A, H122A, H161A or H163A almost completely abolished the activity, whereas, (b) H120R, H122R, H161R or H163R caused slight change in the activity compared to WT protein. The OD (optical density) value at 412 nm was measured using ABTS (2,2′-azino-bis-3-ethylbenzthiazoline-6-sulphonic acid). The activity of wild-type (WT) MmcO was set to 100%.
Effects of metal ions, EDTA and TritonX-100 on MmcO activity
The influence of metal ions on MmcO activity was investigated (Figure 7). MmcO activity was significantly inhibited by Ni+ (Nickel), Mn2+ (Manganese), and Zn2+ (Zinc), with inhibition exceeding 80% (Figure 7). Co2+ (Cobalt), Cu2+ (Copper), Fe2+ (Ferrous), K+ (Potassium), Na+ (Sodium), and Ag+ (Silver) ions also inhibited MmcO activity to varying degrees (40%–70% inhibition) (Figure 7). In contrast to these inhibitory effects, Ca2+ (Calcium) ions enhanced MmcO activity by 130% (Figure 7). The inhibition of MmcO by Ni2+, Mn2+, and Zn2+ is likely due to their competitive displacement of Cu2+ at the active site, thereby impairing electron transfer efficiency. Furthermore, pre-treatment with EDTA (Ethylene diamine tetraacetic acid) before dialysis resulted in a 90% reduction in MmcO activity (Figure 7), implying the essential role of metal ions in MmcO function. Treatment with Triton X-100 led to a 172% increase in activity (Figure 7), suggesting that MmcO may be membrane-associated.
FIGURE 7

Effects of TritonX-100, EDTA and metal ions on MmcO activity. Triton X-100 or Ca2+ dramatically increased the activity of MmcO, whereas EDTA or other metal ions significantly decreased the activity to varying degrees. The OD value was measured at 412 nm. The activity of WT (wild-type) MmcO without addition of metal ions or chelators was set to 100%.
Discussion
In this study, we determined the hydrodynamic radius of MmcO to be 5.9 ± 0.3 nm. The structural model of MmcO was predicted using AlphaFold2 and subsequently evaluated with the Ramachandran plot and ProSA. Based on sequence alignment results, we performed mutagenesis experiments and found that mutations H120A, H122A, H161A, or H163A almost completely abolished MmcO activity, while mutations H120R, H122R, H161R, or H163R caused only minor alterations in activity. Treatment with Triton X-100 or Ca2+ significantly enhanced MmcO activity, whereas EDTA and other metal ions inhibited activity to varying extents. Our findings identified key residues involved in catalytic activity, providing valuable insights for the development of new therapeutic agents targeting Mycobacterium tuberculosis (Mtb).
Due to the failure to obtain MmcO crystals, we conducted a more comprehensive investigation into its functional mechanism. To assist in this analysis, we employed SWISS-MODEL (https://Swissmodel.expasy.org/) to identify structural homologs of MmcO (Supplementary Table S1). Our search revealed that MmcO exhibited amino acid sequence identities of 36.13%, 33.41%, 33.16%, 32.95%, and 32.91% with terpene synthases from uncultured bacterium, Canariomyces arenarius, Aspergillus niger, Thermus thermophilus, and Pseudomonas thermotolerans, respectively (Supplementary Table S1). These findings offer valuable insights into the structural and functional mechanisms of MmcO in Mtb.
The preservation of residual activity in histidine-to-arginine mutants suggests that MmcO’s active site has compensatory electron transfer pathways. Further structural studies, including molecular dynamics simulations, are needed to explore these adaptive mechanisms.
Given its role in oxidative stress defense, MmcO is a promising therapeutic target. Inhibitors designed to disrupt Cu2+ binding, similar to existing copper-chelating antibiotics, could enhance Mtb susceptibility to host immune responses. To conclude, our study offers a new perspective for exploring the complex functional mechanisms of MmcO in Mtb.
Materials and methods
Bioinformatics analysis
The predicted amino acid sequence of MmcO (UniProt accession number I6WZK7) was analyzed using the ProtParam online server (https://web.expasy.org/protparam/) to assess its chemical properties and physicochemical parameters. All gene sequences were optimized according to a previously described method (Liu et al., 2025b) using the online tool (https://www.novopro.cn/tools/codon-optimization.html).
Protein construction, expression and purification
MmcO constructs, expression and purification were as described previously (Kinkar et al., 2019).
Structure prediction and quality assessment of MmcO
The three-dimensional (3D) structure of MmcO was predicted using the AlphaFold2 program (Jumper et al., 2021; Tunyasuvunakool et al., 2021; Wayment-Steele et al., 2023). The MmcO sequences were obtained from the UniProt database under entry ID I6WZK7. Structural visualizations were created using PyMOL 2.3.4 (https://www.pymol.org/2/).
To validate the tertiary structures, PDBsum (https://www.ebi.ac.uk/thornton-srv/databases/pdbsum/Generate.html) was employed to generate the Ramachandran plot for MmcO. This tool is instrumental in the quality assessment and validation of protein structures, as it identifies geometric errors and helps ensure the accuracy of the models. Additionally, the Ramachandran plot examines the stereochemical properties of the structures by evaluating the dihedral angles of the constituent amino acid residues. The plot identifies the allowed regions for residue positioning and highlights disallowed orientations.
In parallel, ProSA (Protein Structure Analysis) is a widely used tool for protein structure validation, primarily employed to analyze and verify predicted protein models (Wiederstein and Sippl, 2007). The Z-score value reflects the overall quality of the model. Its value is presented in a plot that includes the Z-scores of all experimentally determined protein chains in the current PDB. In this plot, structures from different sources (e.g., X-ray, NMR) are differentiated by color. The Z-score can be used to determine whether the score of the input structure falls within the typical range observed for native proteins of comparable size.
Dynamic light scattering experiments
To investigate the oligomeric state, MmcO diameter was measured using dynamic light scattering (DLS) with a Dynapro DLS instrument (Malvern Zetasizer, Malvern, United Kingdom), following previously established protocols (Liu et al., 2024a; Liu et al. 2024b; Liu et al. 2025a). The protein was concentrated to approximately 2.7 mg/mL and then subjected to centrifugation at 12,000 rpm for 5 min. MmcO was subsequently placed into a 1-cm path length cuvette, and data acquisition was carried out over 30 runs, with an equilibration period of 120 s. The resulting DLS data were analyzed using the Zetasizer software (Ver. 6.20), and regularized DLS histograms were generated. The diameter was continuously monitored throughout the analysis.
Enzymatic activity assays of wild-type MmcO
The MmcO activity assays were performed based on the oxidation of ABTS (2,2′-azino-bis-3-ethylbenzthiazoline-6-sulphonic acid), following previously described methods with some modifications (Kinkar et al., 2019; Park et al., 1999). The assay was conducted in a 50 mM sodium acetate buffer (pH 4.0) (Sigma-Aldrich, City of Saint Louis, State of Missouri, United States) containing 2 mM ABTS (Sigma-Aldrich, City of Saint Louis, State of Missouri, United States) and 2 mM CuSO4 (Sigma-Aldrich, City of Saint Louis, State of Missouri, United States). The reaction mixture was incubated with shaking for 2 h. The optical density (OD) at 412 nm was measured, and the specific activity was calculated using the molar absorptivity coefficient ε = 18,400 M-1cm-1.
Enzymatic activity assays for site-directed mutagenesis MmcO
Primers for site-directed mutagenesis of MmcO were designed in our lab and synthesized by Shanghai Sangon Biotechnology (Shanghai, China) (Tables 3, 4; Supplementary Figures S3–S4). The plasmid pET21a containing the gene encoding MmcO was served as the template, and PCR was carried out using Q5 polymerase (New England Biolabs, Ipswich, Massachusetts, United States) for the site-directed mutagenesis (Tables 3, 4; Supplementary Figures S3–S4). The success of the mutagenesis was confirmed through nucleotide sequencing by Shanghai Sangon Biotechnology (Shanghai, China). The mutated MmcO genes were then inserted into the pET21a vector. Expression and purification were conducted using the same protocols as for the wild-type (WT) MmcO. Additionally, enzymatic assays of the site-directed mutagenesis proteins were performed under conditions identical to those used for WT MmcO.
TABLE 3
| Constructs | Primers | Primer sequence (5′-3′) |
|---|---|---|
| pET21a-MmcO | Forward primer | TCCGTCGACAAGCTTATGCCGGAACTGGCGACTTCTGG |
| Reverse primer | CTCGAGTGCGGCCGCTCACAGGATATAGTCCAGACGGGTAGCCATGC | |
| pET21a-MmcO45-504 | Forward primer | TCCGTCGACAAGCTTGCGGGCATGACCGCGGC |
| Reverse primer | CTCGAGTGCGGCCGCTCACAGGATATAGTCCAGACGGGTAGCCATGC | |
| pET21a-MmcO83-504 | Forward primer | TCCGTCGACAAGCTTGTGTCTACCCTGACCTACGGTAATACCATTCCG |
| Reverse primer | CTCGAGTGCGGCCGCTCACAGGATATAGTCCAGACGGGTAGCCATGC |
Primers used for constructions of MmcO plasmids.
Note: The endonuclease restriction sites were underlined and displayed in bold.
TABLE 4
| Primers | Primer sequence (5′-3′) |
|---|---|
| H120A (F) | CTAGCGTAgcaTGGCACGGTATTGCTCTGCG |
| H120A (R) | CGTGCCAtgcTACGCTAGTTGGATCACCCAG |
| H122A (F) | GTACACTGGgcaGGTATTGCTCTGCGTAACGATATGGATG |
| H122A (R) | GCAATACCtgcCCAGTGTACGCTAGTTGGATCACC |
| H161A (F) | CTGGGCTgcaCCTCATGTAGGCCTGCAAG |
| H161A (R) | CATGAGGtgcAGCCCAGTAAGTACCCGGATCC |
| H163A (F) | CTCACCCTgcaGTAGGCCTGCAAGGCGAC |
| H163A (R) | CAGGCCTACtgcAGGGTGAGCCCAGTAAGTACCC |
| H120R (F) | CTAGCGTAcgtTGGCACGGTATTGCTCTGCG |
| H120R (R) | CGTGCCAacgTACGCTAGTTGGATCACCCAG |
| H122R (F) | GTACACTGGcgtGGTATTGCTCTGCGTAACGATATGGATG |
| H122R (R) | GCAATACCacgCCAGTGTACGCTAGTTGGATCACC |
| H161R (F) | CTGGGCTcgtCCTCATGTAGGCCTGCAAG |
| H161R (R) | CATGAGGacgAGCCCAGTAAGTACCCGGATCC |
| H163R (F) | CTCACCCTcgtGTAGGCCTGCAAGGCGAC |
| H163R (R) | CAGGCCTACacgAGGGTGAGCCCAGTAAGTACCC |
Primers used for generating site-directed mutants of MmcO.
Note: Mutated regions in the sequence are underlined and bold.
Statistical analysis
Experiments were performed at least three times, and the results are presented as mean ± SD. Statistical analysis was performed using Origin 8.5, Microsoft Excel 2013, and SPSS 19.0. Statistical significance was determined by the p-value; p < 0.05 and p < 0.01 were considered to be significant and highly significant, respectively.
Statements
Data availability statement
The datasets generated for this study can be found in online repositories. The names of the repositories and accession numbers can be found in the article and its Supplementary Material.
Author contributions
DL: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review and editing. FY: Investigation, Writing – original draft. YL: Investigation, Writing – original draft. AH: Investigation, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Our research work is financially supported by grants from Xinjiang Key Laboratory of Lavender Conservation and Utilization (LCUZ2405), and Start-up Fund for Doctoral Research Established by Yili Normal University (2024RCYJ08).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2025.1565715/full#supplementary-material
References
1
Abramson J. Adler J. Dunger J. Evans R. Green T. Pritzel A. et al (2024). Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature630, 493–500. 10.1038/s41586-024-07487-w
2
Aderem A. Underhill D. M. (1999). Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol.17 (1), 593–623. 10.1146/annurev.immunol.17.1.593
3
Agarwal P. Gordon S. Martinez F. O. (2021). Foam cell macrophages in tuberculosis. Front. Immunol.12, 775326. 10.3389/fimmu.2021.775326
4
Dean A. S. Tosas Auguet O. Glaziou P. Zignol M. Ismail N. Kasaeva T. et al (2022). 25 years of surveillance of drug-resistant tuberculosis: achievements, challenges, and way forward. Lancet Infect. Dis.22 (7), e191–e196. 10.1016/S1473-3099(21)00808-2
5
Dhana A. Hamada Y. Kengne A. P. Kerkhoff A. D. Rangaka M. X. Kredo T. et al (2022). Tuberculosis screening among ambulatory people living with HIV: a systematic review and individual participant data meta-analysis. Lancet Infect. Dis.22 (4), 507–518. 10.1016/S1473-3099(21)00387-X
6
Dheda K. Gumbo T. Maartens G. Dooley K. E. McNerney R. Murray M. et al (2017). The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respir. Med.5, 291–360. 10.1016/S2213-2600(17)30079-6
7
Dheda K. Perumal T. Moultrie H. Perumal R. Esmail A. Scott A. J. et al (2022). The intersecting pandemics of tuberculosis and COVID-19: population-level and patient-level impact, clinical presentation, and corrective interventions. Lancet Respir. Med.10 (6), 603–622. 10.1016/S2213-2600(22)00092-3
8
Dominguez J. Boeree M. J. Cambau E. Chesov D. Conradie F. Cox V. et al (2023). Clinical implications of molecular drug resistance testing for Mycobacterium tuberculosis: a 2023 TBnet/RESIST-TB consensus statement. Lancet Infect. Dis.23 (4), e122–e137. 10.1016/S1473-3099(22)00875-1
9
Dowdy D. W. Behr M. A. (2022). Are we underestimating the annual risk of infection with Mycobacterium tuberculosis in high-burden settings?Lancet Infect. Dis.22 (9), e271–e278. 10.1016/S1473-3099(22)00153-0
10
Flores-Lovon K. Ortiz-Saavedra B. Cueva-Chicana L. A. Aperrigue-Lira S. Montes-Madariaga E. S. Soriano-Moreno D. R. et al (2022). Immune responses in COVID-19 and tuberculosis coinfection: a scoping review. Front. Immunol.13, 992743. 10.3389/fimmu.2022.992743
11
Furin J. Cox H. Pai M. (2019). Tuberculosis. Lancet393 (10181), 1642–1656. 10.1016/S0140-6736(19)30308-3
12
Getahun H. Matteelli A. Chaisson R. Raviglione M. (2015). Latent Mycobacterium tuberculosis infection. N. Engl. J. Med.373 (12), 1179–1180. 10.1056/NEJMc1508223
13
Jenkins H. E. Tolman A. W. Yuen C. M. Parr J. B. Keshavjee S. Perez-Velez C. M. et al (2014). Incidence of multidrug-resistant tuberculosis disease in children: systematic review and global estimates. Lancet383 (9928), 1572–1579. 10.1016/S0140-6736(14)60195-1
14
Jumper J. Evans R. Pritzel A. Green T. Figurnov M. Ronneberger O. et al (2021). Highly accurate protein structure prediction with AlphaFold. Nature596 (7873), 583–589. 10.1038/s41586-021-03819-2
15
Kinkar E. Kinkar A. Saleh M. (2019). The multicopper oxidase of Mycobacterium tuberculosis (MmcO) exhibits ferroxidase activity and scavenges reactive oxygen species in activated THP-1 cells. Int. J. Med. Microbiol.309 (7), 151324. 10.1016/j.ijmm.2019.06.004
16
Lange C. Dheda K. Chesov D. Mandalakas A. M. Udwadia Z. Horsburgh C. R. Jr (2019). Management of drug-resistant tuberculosis. Lancet394 (10202), 953–966. 10.1016/S0140-6736(19)31882-3
17
Liu D. Deng H. Song H. (2025a). Insights into the functional mechanisms of the sesquiterpene synthase GEAS and GERDS in lavender. Int. J. Biol. Macromol.299, 140195. 10.1016/j.ijbiomac.2025.140195
18
Liu D. Dou W. Song H. Deng H. Tian Z. Chen R. et al (2025b). Insights into the functional mechanism of the non-specific lipid transfer protein nsLTP in Kalanchoe fedtschenkoi (Lavender scallops). Protein Expr. Purif.226, 106607. 10.1016/j.pep.2024.106607
19
Liu D. Song H. Deng H. Abdiriyim A. Zhang L. Jiao Z. et al (2024a). Insights into the functional mechanisms of three terpene synthases from Lavandula angustifolia (Lavender). Front. Plant Sci.15, 1497345. 10.3389/fpls.2024.1497345
20
Liu D. Tian Z. Tusong K. Mamat H. Luo Y. (2024b). Expression, purification and characterization of CTP synthase PyrG in Staphylococcus aureus. Protein Expr. Purif.221, 106520. 10.1016/j.pep.2024.106520
21
Micoli F. Bagnoli F. Rappuoli R. Serruto D. (2021). The role of vaccines in combatting antimicrobial resistance. Nat. Rev. Microbiol.19 (5), 287–302. 10.1038/s41579-020-00506-3
22
Naidoo K. Dookie N. (2022). Can the GeneXpert MTB/XDR deliver on the promise of expanded, near-patient tuberculosis drug-susceptibility testing?Lancet Infect. Dis.22 (4), e121–e127. 10.1016/S1473-3099(21)00613-7
23
O'Garra A. Redford P. S. McNab F. W. Bloom C. I. Wilkinson R. J. Berry M. P. (2013). The immune response in tuberculosis. Annu. Rev. Immunol.31, 475–527. 10.1146/annurev-immunol-032712-095939
24
Pai M. Behr M. A. Dowdy D. Dheda K. Divangahi M. Boehme C. C. et al (2016). Tuberculosis. Nat. Rev. Dis. Prim.2, 16076. 10.1038/nrdp.2016.76
25
Park Y. S. Suzuki K. Taniguchi N. Gutteridge J. M. C. (1999). Glutathione peroxidase‐like activity of caeruloplasmin as an important lung antioxidant. FEBS Lett.458 (2), 133–136. 10.1016/s0014-5793(99)01142-4
26
Raya D. Shreya A. Kumar A. Giri S. K. Salem D. R. Gnimpieba E. Z. et al (2022). Molecular regulation of conditioning film formation and quorum quenching in sulfate reducing bacteria. Front. Microbiol.13, 1008536. 10.3389/fmicb.2022.1008536
27
Reid M. J. A. Arinaminpathy N. Bloom A. Bloom B. R. Boehme C. Chaisson R. et al (2019). Building a tuberculosis-free world: the Lancet Commission on tuberculosis. Lancet393 (10178), 1331–1384. 10.1016/S0140-6736(19)30024-8
28
Rule R. Mitton B. Govender N. P. Hoffmann D. Said M. (2021). Spinal epidural abscess caused by Aspergillus spp masquerading as spinal tuberculosis in a person with HIV. Lancet Infect. Dis.21 (11), e356–e362. 10.1016/S1473-3099(20)30979-8
29
Sharma K. K. Singh D. Mohite S. V. Williamson P. R. Kennedy J. F. (2023). Metal manipulators and regulators in human pathogens: a comprehensive review on microbial redox copper metalloenzymes “multicopper oxidases and superoxide dismutases”. Int. J. Biol. Macromol.233, 123534. 10.1016/j.ijbiomac.2023.123534
30
Tiberi S. du Plessis N. Walzl G. Vjecha M. J. Rao M. Ntoumi F. et al (2018). Tuberculosis: progress and advances in development of new drugs, treatment regimens, and host-directed therapies. Lancet Infect. Dis.18 (7), e183–e198. 10.1016/S1473-3099(18)30110-5
31
Tunyasuvunakool K. Adler J. Wu Z. Green T. Zielinski M. Zidek A. et al (2021). Highly accurate protein structure prediction for the human proteome. Nature596 (7873), 590–596. 10.1038/s41586-021-03828-1
32
Udoakang A. J. Djomkam Zune A. L. Tapela K. Nganyewo N. N. Olisaka F. N. Anyigba C. A. et al (2023). The COVID-19, tuberculosis and HIV/AIDS: Ménage à Trois. Front. Immunol.14, 1104828. 10.3389/fimmu.2023.1104828
33
Wayment-Steele H. K. Ojoawo A. Otten R. Apitz J. M. Pitsawong W. Hömberger M. et al (2023). Predicting multiple conformations via sequence clustering and AlphaFold2. Nature625 (7996), 832–839. 10.1038/s41586-023-06832-9
34
Wiederstein M. Sippl M. J. (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res.35 (Web Server), W407–W410. 10.1093/nar/gkm290
Summary
Keywords
tuberculosis (TB), Mycobacterium tuberculosis (MTB), multicopper oxidase MmcO, site-specific mutation, enzymatic activity assays
Citation
Liu D, Yu F, Luo Y and Hanate A (2025) Functional and structural insights into the multicopper oxidase MmcO from Mycobacterium tuberculosis: implications for drug targeting. Front. Chem. 13:1565715. doi: 10.3389/fchem.2025.1565715
Received
23 January 2025
Accepted
12 May 2025
Published
27 May 2025
Volume
13 - 2025
Edited by
Gucheng Zeng, Sun Yat-sen University, China
Reviewed by
Ramesh Maruthi Chingle, National Institutes of Health (NIH), United States
Wadee Abdullah Al-shehari, Harbin Medical University, China
Updates
Copyright
© 2025 Liu, Yu, Luo and Hanate.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dafeng Liu, dafeli@sina.com, dafeli@hotmail.com
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.