Abstract
Introduction:
Azadirachta indica (neem) shows medicinal potential against chronic diseases, but clinical translation is challenging. This study aimed to analyze neem compounds using topological indices (TIs) to predict physicochemical properties.
Methods:
Valency-based indices, including Zagreb and atom bond connectivity indices, were used to characterize boiling point, vaporization, enthalpy, mass, and refractivity. Regression analysis and multi-criteria decision-making methods were employed for predictive modeling and compound ranking.
Results:
Statistical metrics demonstrated the predictive power of the models. Ranking methods provided a hierarchical ordering of compounds based on therapeutic potential.
Discussion:
This study contributes to analogous prediction, optimization, and virtual screening of neem compounds using a cost-effective approach. The findings offer insight into neem compound properties, potentially accelerating drug discovery and development.
1 Introduction
Azadirachta indica, informally known as neem, is a tree belonging to the family Meliaceae. It is used in pharmaceuticals, animal nutrition, agriculture, cosmetics, dental hygiene, personal care, and fuel production (Nwanekezie et al., 2023). Neem extracts (NEs) have been patented for their antibacterial properties, targeting drug-resistant bacteria, and treating breast carcinomas (Nandi and Ghosh, 2023). NEs have anticancer effects on the liver and lung carcinomas, and clinical trials have shown their safe modulation of biological systems in cancer treatment. Studies have revealed that nimbins, a bioactive neem compound, act against four dengue virus strains (Khan et al., 2024). Continued neem consumption can reduce dengue-related morbidity and human pathogens mortality (Wylie and Merrell, 2022). Research suggests that incorporating neem into anti-diabetic agents enhances their effectiveness (Abdullah et al., 2023). NEs inhibit SARS-CoV-2 3-Chymotrypsin (3C) and impede their adhesion to the vascular epithelium. These studies highlight potential of NE in treating COVID-19 (Johnson et al., 2021). Neem leaf extracts show promise against HIV, malaria, and cancer cell proliferation (Eze et al., 2022). Nimbolide, azadirachtin, and gedunin are bioactive neem compounds that influence biological processes in animals (Sarkar et al., 2021). NEs may be suitable for managing HIV, cancer, skin diseases like psoriasis and obesity (Maji and Modak, 2021; Mohan et al., 2023; Tiwari, 2023). NE’s bactericidal, fungicidal, and insecticidal traits are vital for ecological cultivation with arthropod control (Modi and Soni, 2023). Recent research has focused on exploring the potential of neem as a novel therapeutic agent, particularly for its antimicrobial, anti-inflammatory, antioxidant, and anticancer properties (Puttongsiri et al., 2025). There is growing interest in neem compounds for the treatment of drug-resistant pathogens and biofilm-forming organisms (Kumar Singh et al., 2025).
Although the benefits of Azadirachta indica are vast and hold significant pharmacological promise, several challenges hinder its wider clinical application. Various obstacles that impede the advancement of neem-based pharmaceuticals include a) biodiversity, b) toxicity, c) regulatory inconsistencies, d) inefficient and labor-intensive extraction processes, and e) the requirement for precise elucidation of the molecular mechanism (Tembe-Fokunang et al., 2019). A bibliometric analysis of reputable publications and biodiversity studies from various countries concluded that sufficient funding is required for neem research to understand neem biodiversity and toxicity for its safe application in the treatment of diseases (Onasanya et al., 2022). A brief description of these obstacles is as follows:
Being a native to the Indian subcontinent, the neem tree has been successfully introduced to African regions, but its global distribution remains limited as shown in Supplementary Material (Bakewell-Stone, 2024). Research on the toxicity of neem extracts has revealed their effectiveness when administered orally over short periods of time. Nevertheless, prolonged use may result in adverse toxic effects including renal dysfunction, substantial decreases in arterial blood pressure, and hypoglycaemic responses (Braga et al., 2021; Saha et al., 2024; Stinguel et al., 2024; Tumanjong et al., 2024; Ogundipe et al., 2025). Addressing toxicity involves elucidating the precise mechanisms of action, establishing clinical efficacy, and assessing the safety profile of neem-based therapeutics (Modi and Soni, 2023). Furthermore, the development of standardized protocols for extract preparation is essential, because suboptimal processing techniques may lead to detrimental health consequences (Islas et al., 2020).
Internationally recognized databases reveal a significant gap in the information pertaining to the physicochemical characteristics of neem phytochemicals. The phytochemicals in the neem are likely to be composed of numerous flexible molecules. Traditional methodologies struggle to capture the conformation-dependent properties of these molecules and their interactions with biological targets, necessitating interdisciplinary approaches (Ramadhan et al., 2023). The main challenges include: 1) collecting comprehensive, high-quality data on a diverse array of neem compounds, 2) choosing appropriate molecular descriptors, and 3) ensuring rigorous validation and defining the applicability domain of the models. Addressing these challenges is crucial to improve the efficiency and sustainability of neem-derived drug development for chronic diseases (Parmar et al., 2025).
Developing precise QSPR models and prioritizing lead compounds can help to overcome these challenges. These processes facilitate the prediction of properties and hierarchical arrangements that are crucial for efficient virtual screening (Noviandy et al., 2025). QSPR is a theoretical framework used in medical research to enhance the scrutiny of pharmaceutical agents intended to treat specific ailments with desired characteristics derived from their physicochemical characteristics and biological efficacy (Sorgun and Birgin, 2025). This vital methodology supplements experimental research with computational analyses and plays a crucial role in toxicity analysis and virtual screening of pharmaceutical compounds (Hasani et al., 2025).
Molecular descriptors/topological indices are indispensable tools for QSPR studies, enabling the theoretical prediction of physicochemical properties (Huang et al., 2024; Ozge, 2024; Yu et al., 2024). These descriptors accurately identify the information embedded in molecular structures, which is governed by the interconnectivity and spatial configuration of atoms (Sahu and Ojha, 2023). The importance of topological indices (TIs) stems from their ability to facilitate the prediction of their theoretical chemical properties, thereby bolstering QSPR methodologies (Ullah et al., 2023). These numerical descriptors assign a polynomial or number to the correlation between the positions of the atoms in a chemical graph structure and their innate physical properties (Hakeem et al., 2023; Tharmalingam et al., 2023; Yang et al., 2023).
In the literature, the first and second Zagreb indices have applications in complexity and molecular chirality studies of chemical compounds (Hakami et al., 2025). When studying the heat of formation of octanes and heptane, the augmented Zagreb index was found to be a suitable predictive index (Ali et al., 2021). The heat of formation, stability, and strain energy of alkanes and cycloalkanes are highly correlated with the atom bond connectivity (ABC) index (Rahul et al., 2022). Vukičević and Gašperov defined the Adriatic indices. There are three types of indices: variable, discrete, and extended. Discrete Adriatic descriptors are among the closest groups of these descriptors and contain 148 descriptors (Kulli et al., 2021). These indices predict the enthalpy of vaporization, heat capacity, Log P, relative retention time, and biological activity, which are necessary for virtual screening of drug compounds (Anuradha et al., 2024).
TIs have been used in QSPR studies to investigate medications used for diverse medical conditions. The physical attributes of biochemical networks (Ullah et al., 2024), antiviral drugs used for treating headaches (Sardar et al., 2023), and nonsteroidal anti-inflammatory drugs, including opiates and antidepressants, (Gnanaraj et al., 2023), were analyzed by TIs. Drugs used for cardiac dysfunction (Kuriachan and Parthiban, 2025), schizophrenia, tuberculosis, malignancies, viral infections, cancer (Arockiaraj et al., 2025; Kuriachan and Angamuthu, 2025; Nasir, 2025; Qin et al., 2025; Sorgun and Birgin, 2025), and respiratory disorders, such as asthma, were analysed by TIs (Adnan et al., 2022; Balasubramaniyan and Chidambaram, 2023; Gnanaraj et al., 2023; Hakeem, 2023; Huang et al., 2023; Pattabiraman and Cancan, 2023; Zaman et al., 2023). The properties crucial to COVID-19 medicines and anti-hepatitis drugs were investigated using M-polynomial and NM-polynomial (Asghar, 2025), geometric-quadratic index, quadratic-geometric indices, and the first and second inverse Nirmala indices (Das et al., 2023; Nagarajan et al., 2023). Adriatic indices have been used to analyze curcumin- and benzophenone-conjugated PAMAM dendrimers, boron triangular nanosheets, and some nanostructures (Anuradha and Jaganthan, 2023; Anuradha and Jaganathan, 2023).
Multiple-criteria decision-making (MCDM) methodologies, such as VIseKriterijumska Optimizacija I Kompromisno Raspoređivanje (VIKOR) and Simple Additive Weighting (SAW), enable hierarchical ordering of phytochemicals based on various parameters (Zuo et al., 2023). Hierarchical ordering in drug design streamlines discovery by prioritizing candidates based on criteria. This method optimizes resources, provides structure-activity relationship insights, facilitates data-driven decisions, and optimizes combinatorial chemistry. By ranking compounds, researchers identify promising candidates, allocate resources effectively, navigate large libraries and increase success in developing new therapeutics. Various drugs used for treating lung disorders (Ashraf and Idrees, 2024), cancer (Li et al., 2022; Farooq, 2024), eye disorders, kidney cancer (Husin et al., 2024), anti-psychotic drugs (Saeed and Idrees, 2024),and multiple sclerosis (Farooq et al., 2025) have used MCDM to rank drug compounds.
QSAR/QSPR analysis of neem compounds has traditionally been conducted using experimental methods. Recent investigations have utilized laboratory techniques, such as gas chromatography-mass spectrometry and liquid chromatography coupled with quadrupole time-of-flight mass spectrometry, to identify and quantify the bioactive constituents in neem extracts. Density Functional Theory analysis remains the sole computational method employed to evaluate the reactivity and stability of neem phytochemicals (Costa et al., 2021; Ojha et al., 2021). Neem compounds present substantial opportunities for drug discovery and optimization; however, many of their critical properties have not been sufficiently investigated.
In literature, phytochemical compounds like curcumin, resveratrol were examined widely using TIs (Çolakoǧlu, 2022; Preetha et al., 2024; Zaman et al., 2024). However, neem compounds’ pharmacological activities through molecular descriptors remains unexplored. Hence this article attempts to explore them through QSPR modelling and ranking techniques. Linear and quadratic regression methods were applied to create QSPR models. The model was developed using a set of descriptor formulations linked to specific physicochemical properties of a range of chemicals that exhibited biological activity. The results were used to evaluate and rank the neem chemicals. The implications of these findings can enhance pharmacist capabilities across various stages of drug development, namely, analogs evaluation, predicting drug compositions, and virtual screening.
2 Materials and methods
2.1 Neem phytochemicals
Neem, the botanical entity known as a “Pharmaceutical Wonder,” is a repository for an extensive array of medicinal properties. This plant contains approximately 300 distinct phytochemicals, each characterized by its unique chemical composition and structural complexity. The therapeutic efficacy of neem is attributed to its complex phytochemical profile, which includes gallic acid, limonoids, saponins, nimbins, catechins, glycoproteins, and flavonoids, which contribute to its diverse medicinal properties (Sandhir et al., 2021). This study focused on a subset of 11 phytochemicals selected for their extensive research history and frequent utilization across various scientific disciplines, namely, azadirone, nimbin, nimbolide (Sarkar et al., 2021), azadirachtin (Nagini et al., 2024), stigmasterol (Bakrim et al., 2022), tiglic acid (Khanpara and Jadeja, 2022), catechin (Monika et al., 2023), scopoletin (Antika et al., 2022), odoratone, tirucallol (Fernandes et al., 2019), and sugiol (Bajpai et al., 2021). The structures of the phytochemicals (as per PubChem database) are shown in Table 1.
TABLE 1
| Structure of potent Azadrichita indica bio active compounds | |
|---|---|

|

|
| (a) Azadirone | (b) Azadirachtin |

|

|
| (c) Stigmasterol | (d) Tiglic Acid |

|

|
| (e) Catechin | (f) Scopoletin |

|

|
| (g) Odoratone | (h) Tirucallol |

|

|
| (i) Nimbin | (j) Nimbolide |

|
|
| (k) Sugiol | |
Therapeutic neem phytochemicals - molecular structure.
2.2 Valency based indices and drug likeness prediction
Numerous physicochemical characteristics are crucial for predicting drug-likeness, particularly Absorption, Digestion, Metabolism, Excretion, and Toxicity (ADMET) properties, which are governed by Lipinski’s rule of five. However, these properties of neem phytochemicals have not been extensively studied, leading to a scarcity of data in internationally recognized databases. TI is a crucial alternative method for analyzing and predicting the properties of these compounds. This study employed valency-based topological indices (TIs) to describe a range of chemical, physical, and biological activities using edge partition technique. The following section offers a succinct overview of the properties and indices expected to predict the properties examined in this study (Abdullah et al., 2023).
The efficacy of pharmaceutical compounds is significantly influenced by their solubility, with more soluble compounds generally demonstrating a higher potency. The solubility is determined by the octanol-water partition coefficient, expressed as Log P, which can be derived from the boiling point of a molecule (Karami et al., 2022). The dissolution process is affected by polarizability and vaporization enthalpy, which involves heat absorption. These characteristics are linked to enhanced drug effects and mitigated impact reduction due to evaporation. In pharmaceutical research, mass determination is critical because it influences whether substances remain suspended or sink in the liquid media. For pharmacological molecules, a lower mass is typically preferred because of its association with crystallization processes. Drug behavior is also influenced by molar refraction, a property that is related to both refractive index and polarizability. Increased refractive index and polarizability can enhance the interaction of a drug with light and other molecules, making these properties particularly relevant for phototherapeutic applications (Idrees et al., 2025). These attributes play a significant role in drug absorption, distribution, and formulation. This study aims to classify and rank NEs based on their physicochemical properties by deriving the QSPR for vaporization enthalpy, boiling point, polarizability, molar refractivity, and monoisotopic mass (Nagar et al., 2020; Khalikova et al., 2023). TIs, hypothesized to be the best predictors of the aforementioned properties, were used in this study. The following section offers a concise description of the definitions, notations, terminology, and formulations of the TIs utilized in this study.
Throughout this article, denotes any chemical graph, the set of all vertices represents the collection of all edges . Two vertices ,and , are said to be neighbors if there exists an edge to link them. Let represent an edge in the graph that ends at and . The number of edges incident on any vertex is the degree of the vertex and is denoted by . Degree corresponds to the valency of atom. TIs are based on the edge-partition technique, whose summation runs over all partitions. The indices utilized in this study are formulated in Equations 1–10.
The atom bond connectivity (ABC) index was formulated based on the connectivity between the atoms proposed by Estrada et al., as follows:
The first Zagreb index (M1) is one of the elementary indices established by Trinajstic and Gutman and is defined as
An augmented Zagreb index (AUZ) was developed based on the ABC index.
The randic type lodeg index (Heat Capacity predictor) is denoted by RLI and is defined by
The sum lordeg index (a good predictor of Log P Value) is denoted by SLI and is defined by
The inverse sum indeg index (a good predictor of TSA) is denoted by ISI and is defined as:
The misbalance lodeg index (a good predictor of Enthalpy of Vaporization) is denoted by MLI and is defined by
The misbalance index (a good predictor of the standard enthalpy of vaporization) is denoted by MDI and is formulated as
The max-min rodeg index denoted as MMRDI (a good predictor of density), is defined by
The inverse sum lordeg index (ISLI), a good predictor of total surface area, is defined by
The physicochemical properties taken from Chemspider and calculated indices numerical values are shown in Tables 2, 3 respectively.
TABLE 2
| Property | Enthalpy of vaporization | Boiling point | Polarizability | Molar refractivity | Monoisotopic Mass |
|---|---|---|---|---|---|
| NEs | |||||
| Azadirone | 77.6 | 506 | 49 | 123.7 | 436.26 |
| Azadirachtin | 131.3 | 792.4 | 66.6 | 168 | 720.26 |
| Stigmasterol | 88.7 | 501.1 | 51.2 | 129.1 | 412.37 |
| Tiglic Acid | 47.9 | 198.5 | 10.6 | 26.7 | 100.05 |
| Catechin | — | 630.4 | 29.2 | 73.6 | 290.08 |
| Scopoletin | 69.2 | 413.5 | 19.2 | 48.3 | 192.04 |
| Odoratone | 98.4 | 571.2 | 53.6 | 135.2 | 472.36 |
| Tirucalol | 88.3 | 498.9 | 52.9 | 133.4 | 426.39 |
| Nimbin | 90.1 | 606.1 | 54.8 | 138.1 | 540.24 |
| Nimbolide | 90.4 | 608.6 | 47.7 | 120.4 | 466.2 |
| Sugiol | 72.1 | 437.2 | 35.5 | 89.7 | 300.21 |
Physicochemical characteristics of bioactive compounds from Azadirachta indica.
TABLE 3
| TI | ISI | MDI | MMRDI | ISLI | ABC | M1 | RLI | SLI | AUZ | MLI |
|---|---|---|---|---|---|---|---|---|---|---|
| NEs | ||||||||||
| Azadirone | 43.845 | 44 | 47.664 | 21.547 | 25.983 | 198 | 27.3274 | 64.193 | 301.78 | 18.6562 |
| Azadirachtin | 70.2026 | 68 | 76.437 | 35.887 | 40.875 | 306 | 44.4979 | 99.675 | 542.53 | 29.7 |
| Stigmasterol | 38.99 | 40 | 42.897 | 20.273 | 23.67 | 168 | 25.6985 | 50.483 | 271.2 | 14.8535 |
| Tiglic Acid | 5.7 | 8 | 9.4282 | 4.8263 | 4.53 | 26 | 1.6684 | 7.9502 | 37.516 | 4.7998 |
| Catechin | 26.35 | 23 | 29.582 | 11.686 | 16.647 | 114 | 20.7007 | 39.726 | 174.44 | 10.7632 |
| Scopoletin | 16.9667 | 13 | 16.901 | 10.646 | 10.744 | 72 | 9.7478 | 25.705 | 97.656 | 6.5392 |
| Odoratone | 53.8 | 54 | 45.083 | 24.777 | 27.692 | 208 | 31.1197 | 58.012 | 381.57 | 20.977 |
| Tirucalol | 40.6857 | 42 | 53.359 | 17.359 | 30.342 | 180 | 24.5695 | 59.16 | 278.09 | 17.96915 |
| Nimbin | 50.547 | 52 | 53.283 | 24.929 | 29.388 | 209 | 32.2078 | 73.497 | 350.3 | 19.0154 |
| Nimbolide | 48.5143 | 40 | 49.691 | 22.897 | 27.275 | 237 | 31.4463 | 70.153 | 270 | 17.1059 |
| Sugiol | 28.4089 | 30 | 33.205 | 15.01 | 17.538 | 118 | 16.8485 | 41.18 | 187.65 | 13.2349 |
Topological indices for analysis of Azadirachta indica phytochemicals.
To aid a better understanding graphical representation is provided in Figure 1.
FIGURE 1

TIs and Properties of Neem compounds - Graphical Representation.
The correlation coefficient is a crucial metric for evaluating the efficacy of any QSPR model. The computed correlation coefficients are graphically shown in Figure 2.
FIGURE 2

Correlation coefficient between Properties and TIs of Azadrichta Indica.
2.3 Regression model for phytochemicals of neem
Linear regression is a widely employed algorithm with a high level of predictability and adaptability, and is easily interpretable for QSPR analysis. The physicochemical properties were viewed as the dependent variables in this model, and the TIs of neem chemicals were considered as the independent variables. The following equations provide linear and quadratic regression models: where (dependent variable) denotes the physico chemical property of phytochemical, is the intercept (constant), and denote the regression coefficient (constant), and TI is the topological index (independent variable). The constants A (intercept) and (regression coefficient) were computed for the five physicochemical properties and ten TIs considered for the study.
2.4 Ranking of compounds
The hierarchical arrangement of drug compounds is a crucial component of the virtual screening methodology. Machine learning derived MCDM techniques are pivotal in compound ranking. This study used the VIKOR and SAW rankings to classify neem compounds and evaluate their comparative accuracy. These techiques facilitates drug design by prioritizing candidates, assessing drug-likeness, balancing multiple parameters, and supporting decision making (Magaji Yuguda et al., 2023). This process helps researchers to focus on the most promising compounds with desired properties, such as potency, selectivity, and safety. Ranking allows the rapid screening of large compound libraries by reducing the number of experimental assays required (Gates and Hamed, 2020). The following sections provide a detailed explanation of this process.
2.4.1 VIKOR ranking
VIKOR, an acronym for VIseKriterijumska Optimizacija I Kompromisno Raspoređivanje, represents a sophisticated MCDM approach. This methodology facilitates the selection of an optimal solution by concurrently optimizing multiple parameters. In the pharmaceutical industry, VIKOR is used to coordinate drug candidates and identify a compromise solution that accurately approximates the ideal. This technique is particularly valuable in contexts where a balanced solution is more appropriate than an absolute optimal choice. The emphasis on compromise ranking in VIKOR is especially beneficial when no single pharmaceutical agent satisfies all ideal criteria. It considers both group utility () and individual regret () to avoid bias. This methodology provides a clear hierarchical structure and decision-making stability (Gerdes et al., 2021; Feng et al., 2022). The processes involved in this ranking system is described as follows.
2.4.1.1 Alternatives, criterions, and decision matrix
The compounds that need ranking are termed a set of alternatives (). Indices that influence the drug likeness of compounds are criteria. The indices providing the best-fit regression models derived from QSPR analysis were considered as criteria ). Construct a decision matrix γ , where represents the performance of alternatives under criterion
2.4.1.2 Criteria weights
To reflect the relative importance of each criterion in the decision process, weights were assigned through a systematic approach, satisfying
2.4.1.3 Best (maximum) and worst (minimum) alternatives
Identify the maximum and minimum of the alternatives.
The beneficial criteria are calculated using
The non-beneficial criteria are calculated using
Utility and Regret Measures: Compute the Utility Measure and the Regret Measure
Utility Measure: , Regret Measure:
2.4.1.4 VIKOR index
Parameter helps balance the majority rule and individual dominance. Compute the VIKOR Ranking with the VIKOR index where is the weight of the strategy, typically , where , , ,
2.4.1.5 VIKOR ranking
Arrange alternatives in ascending order. The alternative possessing the minimal value is ranked as one (best alternative), and the rest follow in order.
2.4.2 SAW ranking
The Simple Additive Weighting (SAW) method is a widely used multicriteria decision-making approach for ranking alternatives based on multiple criteria and their associated weights. SAW is characterized by their straightforward nature and ease of implementation, rendering them a prevalent choice for decision-making across various domains, including business, finance, and project management (
Peng et al., 2021;
Feng et al., 2022;
Idrees et al., 2025). This process is delineated into distinct phases.
1. Define decision matrix using the criterions used to evaluate the alternatives
2. Normalize the decision matrix to ensure that all criteria are on the same scale and allows comparison using . This process ensures that all alternatives are normalized such that one criterion does not dominate because of a larger magnitude.
3. Weights and Weighted Scores: The Weights are assigned to each criterion to reflect their relative importance. In this study, they were determined based on their performance in producing the best-fit regression models satisfying the equation . The weighted scores are calculated for each alternative on each criterion by multiplying its performance value by the corresponding weight: Weighted Score
4. Aggregation: This total score for each alternative on each criterion is termed as the total score and calculated by: Total Score
5. The evaluation of alternatives was conducted through a hierarchical ranking system, where higher numerical values corresponded to enhanced effectiveness.
3 Results
A regression-based QSPR model was developed using degree-based TIs derived from the molecular structures. The statistical parameters were characterized as follows. The correlation coefficient (R) was used to quantify the extent of data variance, model fit, and predictive capacity of the relationship. The squared correlation coefficient served as a metric to evaluate the reproducibility of the experimental data (R2). The robustness of a model is assessed by statistical metrices. Specifically, a model demonstrates a strong predictive capability when its p-value, calculated from F-statistics, falls below 0.05 and its R-value meets or exceeds 0.6. In this context, N denotes the number of data points in the sample.
3.1 Linear regression
Statistical analysis showed all indicators were significant, with p-values below 0.05 and correlation coefficients over 0.78. The metrics confirmed the model’s significance and fit, with the best fit linear regression models and their metrics summarized in Table 4. Figure 3 shows the obtained results.
TABLE 4
| Property and TI | BP and RLI | Enthalpy of vaporization and ISLI | Molar refraction and ABC | Polarizability and ABC | Monoisotopic Mass and SLI |
|---|---|---|---|---|---|
| NEs Statistical Parameter | |||||
| N | 11 | 10 | 11 | 11 | 11 |
| R2 | 0.8381 | 0.9100 | 0.95382 | 0.95389 | 0.9748 |
| F- statistics | 46.61 | 80.96836 | 185.8923 | 186.1926 | 348.299 |
| p | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 11.778 | 2.4211 | 4.1407 | 1.641 | 6.7562 | |
| 239.32 | 37.426 | 11.964 | 4.7596 | 33.822 |
Statistical Metrics obtained from Linear Regression Analysis.
FIGURE 3

Best fit plots: Linear regression of neem phytochemicals.
3.2 Quadratic regression
The statistical parameters listed in Table 5 corroborate the optimal fit and the statistical significance of the quadratic regression model. Visual depictions of the result are in Figure 4.
TABLE 5
| Property and TI | BP and RLI | Enthalpy of vaporization and AUZ | Molar refraction and MDI | Polarizability and MDI | Monoisotopic Mass and RLI |
|---|---|---|---|---|---|
| NEs Statistical Parameter | |||||
| N | 11 | 10 | 11 | 11 | 11 |
| R2 | 0.83912 | 0.918,456 | 0.9779 | 0.97774 | 0.97952 |
| F- statistics | 20.8633 | 39.42159 | 177.3291 | 175.7358 | 191.3124 |
| p | 0.0006 | 0.000155 | 2.37E-07 | 2.45E-07 | 1.76E-07 |
| C | −0.0001 | -5E-10 | -8E-06 | -3E-06 | -9E-05 |
| 0.4394 | 0.0004 | 0.0626 | 0.0248 | 0.4608 | |
| 295.97 | 57.2 | 36.32 | 14.422 | 135.94 |
Statistical Metrics obtained from Quadratic Regression Analysis.
FIGURE 4

Best Fit Quadratic Regression Models: Azadirachta indica compounds.
3.3 VIKOR ranking of neem chemicals
TIs yielding superior regression model fits were used to rank compounds. The relevant data for the alternatives and criteria employed in this ranking procedure are presented in Table 6.
TABLE 6
| S. No | Compound | ABC | AUZ | RLI | SLI | MDI | ISLI |
|---|---|---|---|---|---|---|---|
| 1 | Azadirone | 25.983 | 301.78 | 27.3274 | 64.193 | 44 | 21.547 |
| 2 | Azadirachtin | 40.875 | 542.53 | 44.4979 | 99.675 | 68 | 35.887 |
| 3 | Stigmasterol | 23.67 | 271.2 | 25.6985 | 50.483 | 40 | 20.273 |
| 4 | Tiglic Acid | 4.53 | 37.516 | 1.6684 | 7.9502 | 8 | 4.8263 |
| 5 | Catechin | 16.647 | 174.44 | 20.7007 | 39.726 | 23 | 11.686 |
| 6 | Scopoletin | 10.744 | 97.656 | 9.7478 | 25.705 | 13 | 10.646 |
| 7 | Odoratone | 27.692 | 381.57 | 31.1197 | 58.012 | 54 | 24.777 |
| 8 | Tirucalol | 30.342 | 278.09 | 24.5695 | 59.16 | 42 | 17.359 |
| 9 | Nimbin | 29.388 | 350.3 | 32.2078 | 73.497 | 52 | 24.929 |
| 10 | Nimbolide | 27.275 | 270 | 31.4463 | 70.153 | 40 | 22.897 |
| 11 | Sugiol | 17.538 | 187.65 | 16.8485 | 41.18 | 30 | 15.01 |
Compounds (alternates) and indices (criterion).
The distribution of criterion weights is predicated on the effectiveness of the TIs in constructing the optimal linear and quadratic regression models. Table 7 delineates the allocated weights, which are collectively summed to unity.
TABLE 7
| Index | ABC | AUZ | RLI | SLI | MDI | ISLI |
|---|---|---|---|---|---|---|
| Weight | 0.2 | 0.1 | 0.3 | 0.1 | 0.2 | 0.1 |
Assigned criterion weights.
The positive and negative ideal solutions of this study are listed in Table 8.
TABLE 8
| Compound | ABC | AUZ | RLI | SLI | MDI | ISLI |
|---|---|---|---|---|---|---|
| Maximum | 40.875 | 542.53 | 44.4979 | 99.675 | 68 | 35.887 |
| Minimum | 4.53 | 37.516 | 1.6684 | 7.9502 | 8 | 4.8263 |
The positive and negative ideal solutions.
The normalized decision matrix used for the VIKOR is derived from the decision matrix is in Table 9.
TABLE 9
| S. No | Compound | ABC | AUZ | RLI | SLI | MDI | ISLI |
|---|---|---|---|---|---|---|---|
| 1 | Azadirone | 0.0819 | 0.0477 | 0.1203 | 0.0387 | 0.0800 | 0.0462 |
| 2 | Azadirachtin | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| 3 | Stigmasterol | 0.0947 | 0.0537 | 0.1317 | 0.0536 | 0.0933 | 0.0503 |
| 4 | Tiglic Acid | 0.2000 | 0.1000 | 0.3000 | 0.1000 | 0.2000 | 0.1000 |
| 5 | Catechin | 0.1333 | 0.0729 | 0.1667 | 0.0654 | 0.1500 | 0.0779 |
| 6 | Scopoletin | 0.1658 | 0.0881 | 0.2434 | 0.0806 | 0.1833 | 0.0813 |
| 7 | Odoratone | 0.0725 | 0.0319 | 0.0937 | 0.0454 | 0.0467 | 0.0358 |
| 8 | Tirucalol | 0.0580 | 0.0524 | 0.1396 | 0.0442 | 0.0867 | 0.0597 |
| 9 | Nimbin | 0.0632 | 0.0381 | 0.0861 | 0.0285 | 0.0533 | 0.0353 |
| 10 | Nimbolide | 0.0748 | 0.0540 | 0.0914 | 0.0322 | 0.0933 | 0.0418 |
| 11 | Sugiol | 0.1284 | 0.0703 | 0.1937 | 0.0638 | 0.1267 | 0.0672 |
Normalised decision matrix of bioactive Azadirachta indica compounds for VIKOR.
The utility and regret measures and the corresponding VIKOR rankings were calculated, as mentioned in the previous sections. The finalized decisions on ranking are listed in Table 10.
TABLE 10
| S. No | Compound | Rank | |||
|---|---|---|---|---|---|
| 1 | Azadirone | 0.4147 | 0.12027 | 0.40782 | 5 |
| 2 | Azadirachtin | 0 | 0 | 0 | 1 |
| 3 | Stigmasterol | 0.4773 | 0.1316 | 0.45812 | 7 |
| 4 | Tiglic Acid | 1 | 0.3 | 1 | 11 |
| 5 | Catechin | 0.6662 | 0.16668 | 0.61089 | 8 |
| 6 | Scopoletin | 0.8425 | 0.24340 | 0.82695 | 10 |
| 7 | Odoratone | 0.326 | 0.09370 | 0.31917 | 3 |
| 8 | Tirucalol | 0.4404 | 0.13958 | 0.45284 | 6 |
| 9 | Nimbin | 0.3045 | 0.08608 | 0.29573 | 2 |
| 10 | Nimbolide | 0.3876 | 0.09333 | 0.34933 | 4 |
| 11 | Sugiol | 0.65 | 0.19367 | 0.64779 | 9 |
VIKOR ranking of bioactive Azadrichta indica compounds.
3.4 SAW ranking of neem chemicals
The matrix calculated for the alternates and criteria was normalized to reduce the dominance of data with higher numerals. This process was achieved using the second phase of the SAW ranking. The resulting normalized matrix is listed in Table 11.
TABLE 11
| S. No | Compound | ABC | AUZ | RLI | SLI | MDI | ISLI |
|---|---|---|---|---|---|---|---|
| 1 | Azadirone | 0.6357 | 0.5562 | 0.6141 | 0.6440 | 0.6471 | 0.6004 |
| 2 | Azadirachtin | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 | 1.0000 |
| 3 | Stigmasterol | 0.5791 | 0.4999 | 0.5775 | 0.5065 | 0.5882 | 0.5649 |
| 4 | Tiglic Acid | 0.1108 | 0.0692 | 0.0375 | 0.0798 | 0.1176 | 0.1345 |
| 5 | Catechin | 0.4073 | 0.3215 | 0.4652 | 0.3986 | 0.3382 | 0.3256 |
| 6 | Scopoletin | 0.2629 | 0.1800 | 0.2191 | 0.2579 | 0.1912 | 0.2967 |
| 7 | Odoratone | 0.6775 | 0.7033 | 0.6994 | 0.5820 | 0.7941 | 0.6904 |
| 8 | Tirucalol | 0.7423 | 0.5126 | 0.5521 | 0.5935 | 0.6176 | 0.4837 |
| 9 | Nimbin | 0.7190 | 0.6457 | 0.7238 | 0.7374 | 0.7647 | 0.6947 |
| 10 | Nimbolide | 0.6673 | 0.4977 | 0.7067 | 0.7038 | 0.5882 | 0.6380 |
| 11 | Sugiol | 0.4291 | 0.3459 | 0.3786 | 0.4131 | 0.4412 | 0.4183 |
Normalised matrix of bioactive Azadirachta indica compounds for SAW ranking.
The data in the normalized matrix are aggregated as mentioned in the phase narration. From the obtained aggregation, the SAW ranking was assigned, as shown in Table 12.
TABLE 12
| S. No | Compound | ABC | AUZ | RLI | SLI | MDI | ISLI | Summation | Rank |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Azadirone | 0.1271 | 0.0556 | 0.1842 | 0.0644 | 0.1294 | 0.0600 | 0.6209 | 5 |
| 2 | Azadirachtin | 0.2000 | 0.1000 | 0.3000 | 0.1000 | 0.2000 | 0.1000 | 1.0000 | 1 |
| 3 | Stigmasterol | 0.1158 | 0.0500 | 0.1733 | 0.0506 | 0.1176 | 0.0565 | 0.5638 | 7 |
| 4 | Tiglic Acid | 0.0222 | 0.0069 | 0.0112 | 0.0080 | 0.0235 | 0.0134 | 0.0853 | 11 |
| 5 | Catechin | 0.0815 | 0.0322 | 0.1396 | 0.0399 | 0.0676 | 0.0326 | 0.3932 | 9 |
| 6 | Scopoletin | 0.0526 | 0.0180 | 0.0657 | 0.0258 | 0.0382 | 0.0297 | 0.2300 | 10 |
| 7 | Odoratone | 0.1355 | 0.0703 | 0.2098 | 0.0582 | 0.1588 | 0.0690 | 0.7017 | 3 |
| 8 | Tirucalol | 0.1485 | 0.0513 | 0.1656 | 0.0594 | 0.1235 | 0.0484 | 0.5966 | 6 |
| 9 | Nimbin | 0.1438 | 0.0646 | 0.2171 | 0.0737 | 0.1529 | 0.0695 | 0.7216 | 2 |
| 10 | Nimbolide | 0.1335 | 0.0498 | 0.2120 | 0.0704 | 0.1176 | 0.0638 | 0.6471 | 4 |
| 11 | Sugiol | 0.0858 | 0.0346 | 0.1136 | 0.0413 | 0.0882 | 0.0418 | 0.4054 | 8 |
SAW ranking of bioactive Azadirachta indica compounds.
A comparative graphical depiction of the hierarchical arrangement obtained from different techniques is shown in Figure 5.
FIGURE 5
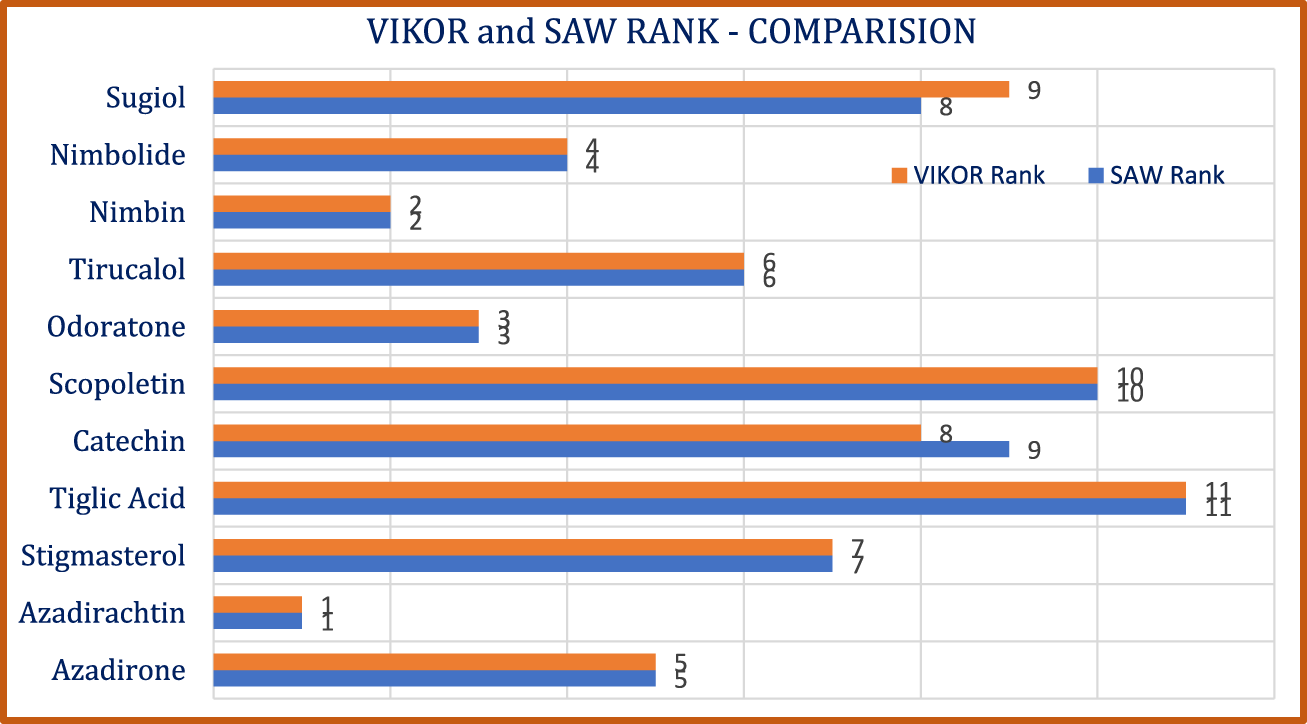
Ranking comparison of Bioactive Azadirachta indica Compounds -VIKOR and SAW.
4 Discussion
This research incorporated a range of parameters, such as correlation coefficients between indices and properties, to extract insights from both linear and quadratic regression analyses along with MCDM methodologies. These insights are instrumental in establishing mathematical models that enable the prediction of pharmaceutical compound behavior and hierarchical ranking.
The potential outcomes of the study were evaluated to underpin further investigations aimed at the development of effective neem-based drug formulations. From the statistical metrics of the linear and quadratic regression analyses and MCDM, it can be concluded that.
a) Predictive ability based on reproducibility of the TIs can be ordered as
b) Prediction accuracy of the properties considered in the study can be ordered as
c) A nuanced distinction in drug performance was evident, as exemplified by the contrasting rankings of catechin and sugiol. The association between VIKOR and SAW rankings, quantified using the Spearman rank correlation coefficient is 0.98, where represents the difference between compound rankings.
Regression analysis demonstrated predictive accuracies for various molecular properties, potentially accelerating the identification of promising neem-derived compounds for further development. The compound rankings derived from VIKOR and SAW techniques lend substantial credence and a notable degree of congruence, underscoring the reliability of the proposed ranking system.
To address biodiversity challenges, researchers can use these results to guide the creation of synthetic compounds that replicate the active components of neem. This approach enables drug development in regions in which neem are not readily available. Furthermore, these indices can serve as a bridge between scientists in neem-rich areas and those in countries with limited plant resources, thereby fostering worldwide collaborations in drug discovery. Given the limited global availability of neem, the use of regression models to make analogous predictions could aid pharmacists in creating synthetic compounds that replicate the properties of neem chemicals. The potential outcomes of this study could greatly enhance the clinical application of neem-derived compounds. Drug repurposing for new therapeutic applications, exploration of combination therapies for synergistic effects, and natural product-inspired drug design.
The QSPR model and compound ranking contribute to a) forecasting enzyme inhibition data that align with ligand physicochemical properties, b) elucidating supplementary information extracted from three-dimensional structures, c) minimizing the number of compounds required for synthesis, d) anticipating properties of structurally similar molecules, and e) ascertaining drug composition. Despite these valuable contributions, the development of QSPR models has limitations.
In QSPR, the formulation of optimal regression models is frequently challenged by issues such as overfitting and a lack of three-dimensional data. These challenges restrict the generalization ability of the models, particularly in scenarios involving multiple mechanisms. Overcoming these obstacles could facilitate the development of more robust QSPR models, which are vital for developing models that leverage extensive datasets. Neem is a plant containing over 300 phytochemicals, each exhibiting diverse biological activities, including insecticidal, antimicrobial, and medicinal properties. When ranking and QSPR models were integrated with machine learning techniques, it was possible to evaluate all neem phytochemicals. This integration allows pharmacists to explore their potential for disease treatment, optimize drug formulations, conduct virtual screenings, and perform ADMET profiling.
Chronic illnesses, including cardiovascular and neurological disorders, ischemic conditions, diabetes mellitus, renal impairment, skeletal muscle diseases, and certain cancers, continue to pose significant challenges in the medical field. However, despite these advancements, these conditions often result in drug resistance and adverse outcomes. The integration of experimental research on the bioactive compounds of neem with computational modeling can substantially advance the development of therapeutic strategies for chronic disorders.
5 Conclusion
In this study, the properties of neem phytochemicals were evaluated using regression analysis and multi-criteria decision-making methodologies. In regression models, RLI demonstrated the highest reproducibility with 0.83912 and 0.97952 for BP and MM, respectively, with significant p-values, indicating strong correlation with the desired metrics and proves effective in QSPR analysis. Hierarchical rankings demonstrated high concordance, with azadirachtin securing the first rank by providing positive ideal solutions of 40.875, 542.53, 44.4979, 99.675, 68, and 35.887, indicating its highest therapeutic potential. These results aid in rapid compound library assessment and guide the selectivity refinement of neem compounds. The results can be utilized to overcome geographical limitations, facilitate drug optimization, improve predictive accuracy, and enhance virtual screening, bridging drug development and clinical implementation while reducing effort, time, and resources.
Statements
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
DA: Conceptualization, Methodology, Visualization, Writing – original draft, Writing – review and editing. BJ: Supervision, Validation, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The article publication charges was covered by Vellore Institute of Technology, Chennai, India.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2025.1580267/full#supplementary-material
References
1
Abdullah A. Biswas P. Sahabuddin M. Mubasharah A. Khan D. A. Hossain A. et al (2023). Molecular dynamics simulation and pharmacoinformatic integrated analysis of bioactive phytochemicals from Azadirachta indica (neem) to treat diabetes mellitus. J. Chem.2023, 1–19. 10.1155/2023/4170703
2
Adnan Mr., Bokhary S. A. U. H. Abbas G. Iqbal T. (2022). Degree-based topological indices and QSPR analysis of Antituberculosis drugs. J. Chem.2022, 1–17. 10.1155/2022/5748626
3
Ali A. Furtula B. Gutman I. Vukičevi´c D. V. (2021). Augmented Zagreb index: extremal results and bounds article in match communications in mathematical and in computer chemistry. Available online at: https://www.researchgate.net/publication/341735696.
4
Antika L. D. Tasfiyati A. N. Hikmat H. Septama A. W. (2022). Scopoletin: a review of its source, biosynthesis, methods of extraction, and pharmacological activities. Z Naturforsch C J. Biosci.77, 303–316. 10.1515/znc-2021-0193
5
Anuradha D. S. Jaganthan B. (2023). Topological properties of boron triangular sheet for robotic finger flex motion through indices. AIP2946. 10.1063/5.0178070
6
Anuradha D. S. Julietraja K. Jaganathan B. Alsinai A. (2024). Curcumin-conjugated PAMAM dendrimers of two generations: comparative analysis of physiochemical properties using adriatic topological indices. ACS Omega9, 14558–14579. 10.1021/acsomega.4c00686
7
Anuradha S. Jaganathan B. (2023). Physiochemical properties of benzophenone and curcumin-conjugated PAMAM dendrimers using topological indices. Polycycl. Aromat. Compd.0, 1–23. 10.1080/10406638.2023.2234542
8
Arockiaraj M. Jeni Godlin J. J. Radha S. Aziz T. Al-Harbi M. (2025). Comparative study of degree, neighborhood and reverse degree based indices for drugs used in lung cancer treatment through QSPR analysis. Sci. Rep.15, 3639. 10.1038/s41598-025-88044-x
9
Asghar A. (2025). QSPR analysis of anti-hepatitis prescription drugs using degree based topological indices through M-polynomial and NM-polynomial. Chim. Techno Acta12. 10.15826/chimtech.2025.12.2.06
10
Ashraf T. Idrees N. (2024). Topological indices based VIKOR assisted multi-criteria decision technique for lung disorders. Front. Chem.12, 1407911. 10.3389/fchem.2024.1407911
11
Bajpai V. K. Sonwal S. Hwang S.-K. Shukla S. Khan I. Dey D. K. et al (2021). Sugiol, a diterpenoid: therapeutic actions and molecular pathways involved. Pharmacol. Res.163, 105313. 10.1016/j.phrs.2020.105313
12
Bakewell-Stone P. (2024). Azadirachta indica (neem tree). Available online at: https://cabidigitallibrary.org.
13
Bakrim S. Benkhaira N. Bourais I. Benali T. Lee L. H. El Omari N. et al (2022). Health benefits and pharmacological properties of stigmasterol. Antioxidants11, 1912–1932. 10.3390/antiox11101912
14
Balasubramaniyan D. Chidambaram N. (2023). On some neighbourhood degree-based topological indices with QSPR analysis of asthma drugs. Eur. Phys. J. Plus138, 823. 10.1140/epjp/s13360-023-04439-7
15
Braga T. M. Rocha L. Chung T. Y. Oliveira R. F. Pinho C. Oliveira A. I. et al (2021). Azadirachta indica a. Juss. in vivo toxicity—an updated review. Molecules26, 252. 10.3390/molecules26020252
16
Çolakoǧlu Ö. (2022). QSPR modeling with topological indices of some potential drug candidates against COVID-19. J. Math.2022. 10.1155/2022/3785932
17
Costa P. C. S. Evangelista J. S. Leal I. Miranda P. C. M. L. (2021). Chemical graph theory for property modeling in qsar and qspr—charming QSAR and QSPR. Mathematics9, 60–19. 10.3390/math9010060
18
Das S. Rai S. Kumar V. (2023). On topological indices of Molnupiravir and its QSPR modelling with some other antiviral drugs to treat COVID-19 patients. J. Math. Chem.62, 2581–2624. 10.1007/s10910-023-01518-z
19
Eze M. O. Ejike C. E. C. C. Ifeonu P. Udeinya I. J. Udenigwe C. C. Uzoegwu P. N. (2022). Anti-COVID-19 potential of Azadirachta indica (Neem) leaf extract. Sci. Afr.16, e01184. 10.1016/j.sciaf.2022.e01184
20
Farooq F. B. (2024). Implementation of multi-criteria decision making for the ranking of drugs used to treat bone-cancer. AIMS Math.9, 15119–15131. 10.3934/math.2024733
21
Farooq F. B. Idrees N. Noor E. Alqahtani N. A. Imran M. (2025). A computational approach to drug design for multiple sclerosis via QSPR modeling, chemical graph theory, and multi-criteria decision analysis. BMC Chem.19, 1. 10.1186/s13065-024-01374-1
22
Feng J. Yan Y. Huang M. Du Y. Lu Z. Li B. (2022). “A study on the multi-attribute decision theory and methods,” in Procedia computer science (Elsevier B.V.), 544–551. 10.1016/j.procs.2022.11.210
23
Fernandes S. R. Barreiros L. Oliveira R. F. Cruz A. Prudêncio C. Oliveira A. I. et al (2019). Chemistry, bioactivities, extraction and analysis of azadirachtin: state-of-the-art. Fitoterapia134, 141–150. 10.1016/j.fitote.2019.02.006
24
Gates L. E. Hamed A. A. (2020). The anatomy of the SARS-CoV-2 biomedical literature: introducing the CovidX network algorithm for drug repurposing recommendation. J. Med. Internet Res.22, e21169. 10.2196/21169
25
Gerdes H. Casado P. Dokal A. Hijazi M. Akhtar N. Osuntola R. et al (2021). Drug ranking using machine learning systematically predicts the efficacy of anti-cancer drugs. Nat. Commun.12, 1850. 10.1038/s41467-021-22170-8
26
Gnanaraj L. R. M. Ganesan D. Siddiqui M. K. (2023). Topological indices and QSPR analysis of NSAID drugs. Polycycl. Aromat. Compd.43, 9479–9495. 10.1080/10406638.2022.2164315
27
Hakami K. H. Khan A. R. Ali M. (2025). Mathematical modeling and QSPR analysis of hepatitis treatment drugs through connection indices an innovative approach. Heliyon11, e41234. 10.1016/j.heliyon.2024.e41234
28
Hakeem A. Nek M. K. Muhammad F. Ahmed N. (2023). On the molecular structure modelling of gamma graphyne and armchair graphyne nanoribbon via reverse degree-based topological indices. Mol. Phys.122. 10.1080/00268976.2023.2259510
29
Hakeem A. Ullah A. Zaman S. Jabeen S. (2023). QSPR analysis of some novel drugs used for cardiovascular diseases through degree-based topological indices and regression models. Res. Sq.10.21203/rs.3.rs-3576948/v1
30
Hasani M. Ghods M. Mondal S. Siddiqui M. K. Cheema I. Z. (2025). Modeling QSPR for pyelonephritis drugs: a topological indices approach using MATLAB. J. Supercomput81, 479. 10.1007/s11227-025-06967-8
31
Huang L. Wang Y. Pattabiraman K. Danesh P. Siddiqui M. K. Cancan M. (2023). Topological indices and QSPR modeling of new antiviral drugs for cancer treatment. Polycycl. Aromat. Compd.43, 8147–8170. 10.1080/10406638.2022.2145320
32
Huang R. Hanif M. F. Siddiqui M. K. Hanif M. F. Gegbe B. (2024). On analysis of silicon dioxide based on topological indices and entropy measure via regression model. Sci. Rep.14, 22478. 10.1038/s41598-024-73163-8
33
Husin M. N. Khan A. R. Awan N. U. H. Campena F. J. H. Tchier F. Hussain S. (2024). Multicriteria decision making attributes and estimation of physicochemical properties of kidney cancer drugs via topological descriptors. PLoS One19, e0302276. 10.1371/journal.pone.0302276
34
Idrees N. Noor E. Rashid S. Agama F. T. (2025). Role of topological indices in predictive modeling and ranking of drugs treating eye disorders. Sci. Rep.15, 1271. 10.1038/s41598-024-81482-z
35
Islas J. F. Acosta E. G-Buentello Z. Delgado-Gallegos J. L. Moreno-Treviño M. G. Escalante B. et al (2020). An overview of Neem (Azadirachta indica) and its potential impact on health. J. Funct. Foods74, 104171. 10.1016/j.jff.2020.104171
36
Johnson T. O. Adegboyega A. E. Adeyemi O. E. Okonkwo F. O. Ejembi S. A. Oche J.-R. I. et al (2021). Computer-aided identification of bioactive compounds of Azadirachta indica (neem) with potential activity against SARS-CoV-2 main protease. Res. Sq., 1–22. 10.21203/rs.3.rs-841180/v1
37
Karami T. K. Hailu S. Feng S. Graham R. Gukasyan H. J. (2022). Eyes on lipinski’s rule of five: a new “rule of thumb” for physicochemical design space of ophthalmic drugs. J. Ocular Pharmacol. Ther.38, 43–55. 10.1089/jop.2021.0069
38
Khalikova M. Jireš J. Horáček O. Douša M. Kučera R. Nováková L. (2023). What is the role of current mass spectrometry in pharmaceutical analysis?Mass Spectrom. Rev.43, 560–609. 10.1002/mas.21858
39
Khan A. Singh D. Waidha K. Sisodiya S. Gopinath P. Hussian S. et al (2024). Analysis of inhibition potential of nimbin and its analogs against NF-κB subunits p50 and p65: a molecular docking and molecular dynamics study. Anticancer Agents Med. Chem.24, 280–287. 10.2174/1871520623666230908101204
40
Khanpara P. Jadeja Y. (2022). A complete review on medicinal plant: Margosa Tree. J. Med. Plants Stud.10, 131–140. 10.22271/plants.2022.v10.i5b.1476
41
Kulli V. R. B C. T V. (2021). Computation of Adriatic (a, b)-KA index of some nanostructues KA index of some nanostrctures. Int. J. Math. Trends Technol.67, 79–87. 10.14445/22315373/ijmtt-v67i4p5110.14445/22315373/IJMTT-V67I4P511
42
Kumar Singh R. Nallaswamy D. Rajeshkumar S. Varghese S. S. (2025). Green synthesis of silver nanoparticles using neem and turmeric extract and its antimicrobial activity of plant mediated silver nanoparticles. J. Oral Biol. Craniofac Res.15, 395–401. 10.1016/j.jobcr.2025.02.005
43
Kuriachan G. Angamuthu P. (2025). Quantitative structure-property relationship analysis of physical and ADMET properties of anticancer drugs using domination topological indices. Int. J. Quantum Chem.125, e27525. 10.1002/qua.27525
44
Kuriachan G. Parthiban A. (2025). Computation of domination degree-based topological indices using python and QSPR analysis of physicochemical and ADMET properties for heart disease drugs. Front. Chem.13, 1536199. 10.3389/fchem.2025.1536199
45
Li Y. Aslam A. Saeed S. Zhang G. Kanwal S. (2022). Targeting highly resisted anticancer drugs through topological descriptors using VIKOR multi-criteria decision analysis. Eur. Phys. J. Plus137, 1245. 10.1140/epjp/s13360-022-03469-x
46
Magaji Yuguda Y. Unuebho I. E. kolli T. Dhanushya J. (2023). Emerging trends in computational approaches for drug discovery in molecular biology. GSC Biol. Pharm. Sci.24, 202–213. 10.30574/gscbps.2023.24.3.0340
47
Maji S. Modak S. (2021). Chemical science review and letters neem: treasure of natural phytochemicals. Chem. Sci. Rev. Lett.2021, 396–401. 10.37273/chesci.cs205205351
48
Modi G. Soni R. K. (2023). Azadirachta indica in focus: investigating neem ’ s diverse applications. J. Chem. Health Risks13 (3), 1500–1510. 10.52783/jchr.v13.i3.754
49
Mohan S. M. Singh N. Mishra D. Ehsan S. Chaturvedi V. Chaudhary A. et al (2023). Computational approaches to designing antiviral drugs against COVID-19: a comprehensive review. Curr. Pharm. Des.29, 2601–2617. 10.2174/0113816128259795231023193419
50
Monika P. Chandraprabha M. N. Murthy K. N. C. (2023). Catechin, epicatechin, curcumin, garlic, pomegranate peel and neem extracts of Indian origin showed enhanced anti-inflammatory potential in human primary acute and chronic wound derived fibroblasts by decreasing TGF-β and TNF-α expression. BMC Complement. Med. Ther.23, 181–216. 10.1186/s12906-023-03993-y
51
Nagar H. Ingle D. Kumar C. Kumar A. (2020). Thermodynamic properties of active pharmaceutical ingredients that are of interest in COVID-19. 10.1016/j.cdc.2021.100820
52
Nagarajan S. Priyadharsini G. Pattabiraman K. (2023). QSPR modeling of status-based topological indices with COVID-19 drugs. Polycycl. Aromat. Compd.43, 6868–6887. 10.1080/10406638.2022.2127803
53
Nagini S. Palrasu M. Bishayee A. (2024). Limonoids from neem (Azadirachta indica A. Juss.) are potential anticancer drug candidates. Med. Res. Rev.44, 457–496. 10.1002/med.21988
54
Nandi P. Ghosh S. N. (2023). Azadirachta indica – neem- A natural gift for prevention and treatment of chronic diseases- A review. Int. J. Minor Fruits, Med. Aromatic Plants9, 12–24. 10.53552/ijmfmap.9.2.2023.12-24
55
Nasir S. (2025). Topological descriptors of colorectal cancer drugs and characterizing physical properties via QSPR analysis. Int. J. Anal. Chem.2025, 5512172. 10.1155/ianc/5512172
56
Noviandy T. R. Maulana A. Irvanizam I. Idroes G. M. Maulydia N. B. Tallei T. E. et al (2025). Interpretable machine learning approach to predict Hepatitis C virus NS5B inhibitor activity using voting-based LightGBM and SHAP. Intelligent Syst. Appl.25, 200481. 10.1016/j.iswa.2025.200481
57
Nwanekezie M. N. Ndive J. N. Ogbonna I. L. Sebe G. O. (2023). Comprehensive physicochemical profiling and characterization of neem plant leaf extracts: insights for pharmaceutical and biomedical applications. Adv. Chem. Eng. Sci.13, 382–399. 10.4236/aces.2023.134026
58
Ogundipe O. J. Ojetola A. A. Akinpelu O. F. Sossou I. T. Ishola A. B. (2025). Aqueous leaf extract of Azadirachta indica protects against gentamicin-induced kidney injury via decreases in renal function, inflammation, and apoptosis markers. J. Med. Food28, 272–280. 10.1089/jmf.2023.0294
59
Ojha P. K. Vinay K. Joyita R. Roy K. (2021). Recent advances in quantitative structure–activity relationship models of antimalarial drugs. Expert Opin. Drug Discov.16, 659–695. 10.1080/17460441.2021.1866535
60
Onasanya A. K. Oderinde O. K. Akintunde T. Y. Shonekan O. O. Bankole I. S. Fadimu B. O. et al (2022). Bibliometric analysis of 100 top-cited articles on neem indexed in the web of science. Trop. J. Nat. Prod. Res.6, 95–108. 10.26538/tjnpr/v6i1.17
61
Ozge C. (2024). NM-Polynomials and neighborhood degree based topological indices of some benzenoid systems. J. Kadirli Fac. Appl. Sci.4, 166–178. Available online at: https://kadirliubfd.com/index.php/kubfd/article/view/106
62
Parmar G. Shah A. Chudasama J. M. Kashav P. James V. (2025). “Network-based methods in drug discovery,” in Computational methods for rational drug design, 255–284. 10.1002/9781394249190.ch12
63
Pattabiraman K. Cancan M. (2023). QSPR modeling with topological indices of some potential drugs against cancer. Polycycl. Aromat. Compd.0, 1–28. 10.1080/10406638.2023.2189270
64
Peng X. Gibbs E. Silverman J. M. Cashman N. R. Plotkin S. S. (2021). A method for systematically ranking therapeutic drug candidates using multiple uncertain screening criteria. Stat. Methods Med. Res.30, 1502–1522. 10.1177/09622802211002861
65
Preetha U. P. Suresh M. Tolasa F. T. Bonyah E. (2024). QSPR/QSAR study of antiviral drugs modeled as multigraphs by using TI’s and MLR method to treat COVID-19 disease. Sci. Rep.14, 13150. 10.1038/s41598-024-63007-w
66
Puttongsiri T. Manichart N. Teerarak M. Sikhao P. Somala N. Tongsri P. et al (2025). Siamese neem tree as a natural preservative: chemical profile, antioxidant properties, and antibacterial efficacy against foodborne pathogens and spoilage bacteria. J. Agric. Food Res.19, 101559. 10.1016/j.jafr.2024.101559
67
Qin H. Hussain M. Hanif M. F. Siddiqui M. K. Hussain Z. Fiidow M. A. (2025). On QSPR analysis of pulmonary cancer drugs using python-driven topological modeling. Sci. Rep.15, 3965. 10.1038/s41598-025-88419-0
68
Rahul M. P. Clement J. Singh Junias J. Arockiaraj M. Balasubramanian K. (2022). Degree-based entropies of graphene, graphyne and graphdiyne using Shannon’s approach. J. Mol. Struct.1260, 132797–132820. 10.1016/j.molstruc.2022.132797
69
Ramadhan D. Firdayani F. Karimah N. Permadi E. E. Muttaqien S.El Supriyono A. (2023). Identification of potential bioactive compounds from Azadirachta indica (Neem) as inhibitors of SARS-CoV-2 main protease: molecular docking and molecular dynamics simulation studies. J. Appl. Pharm. Sci.13, 40–49. 10.7324/JAPS.2023.139799
70
Saeed F. Idrees N. (2024). Integrating structural modeling and decision-making for anti-psychotic drugs through topological indices. Bionanoscience15, 59. 10.1007/s12668-024-01732-2
71
Saha O. Siddiquee N. H. Akter R. Sarker N. Bristi U. P. Sultana K. F. et al (2024). Antiviral activity, pharmacoinformatics, molecular docking, and dynamics studies of Azadirachta indica against nipah virus by targeting envelope glycoprotein: emerging strategies for developing antiviral treatment. Bioinform Biol. Insights18, 11779322241264145. 10.1177/11779322241264145
72
Sahu S. K. Ojha K. K. (2023). Applications of QSAR study in drug design of tubulin binding inhibitors. J. Biomol. Struct. Dyn.42, 12806–12821. 10.1080/07391102.2023.2273437
73
Sandhir R. Khurana M. Singhal N. K. (2021). Potential benefits of phytochemicals from Azadirachta indica against neurological disorders. Neurochem. Int.146, 105023. 10.1016/j.neuint.2021.105023
74
Sardar M. S. Ali M. A. Farahani M. R. Alaeiyan M. Cancan M. Campus Z. (2023). Topological Indices and QSPR/QSAR Analysis of some Drugs being investigated for the treatment of headaches. Eur. Chem. Bull.12, 69–83. 10.22541/au.166313352.29133526/v1
75
Sarkar S. Singh R. P. Bhattacharya G. (2021). Exploring the role of Azadirachta indica (neem) and its active compounds in the regulation of biological pathways: an update on molecular approach. 3 Biotech.11, 178–212. 10.1007/s13205-021-02745-4
76
Sorgun S. Birgin K. (2025). Vertex-edge-weighted molecular graphs: a study on topological indices and their relevance to physicochemical properties of drugs used in cancer treatment. J. Chem. Inf. Model65, 2093–2106. 10.1021/acs.jcim.4c02013
77
Stinguel P. Pratissoli D. de Oliveira R. C. Menini L. Piffer A. B. M. de Oliveira A. C. L. F. (2024). Characterization, formulation, and toxicity of plant oils on duponchelia fovealis caterpillars zeller 1847 (Lepidoptera: crambidae). Neotrop Entomol.54, 17. 10.1007/s13744-024-01229-3
78
Tembe-Fokunang E. A. Charles F. Kaba N. Donatien G. Michael A. Bonaventure N. (2019). The potential pharmacological and medicinal properties of neem (Azadirachta indica A. Juss) in the drug development of phytomedicine. J. Complementary Altern. Med. Res., 1–18. 10.9734/jocamr/2019/v7i130093
79
Tharmalingam G. Ponnusamy K. Govindhan M. Konsalraj J. (2023). On certain degree based and bond additive molecular descriptors of hexabenzocorenene. Biointerface Res. Appl. Chem.13, 1–14. 10.33263/BRIAC135.495
80
Tiwari N. (2023). Neem and its medicinal purpose: a review. Asian J. Pharmacogn.7, 51–60.
81
Tumanjong I. Cho Ngwa F. Mainsah E. N. F. Pascal M. T. (2024). In vitro anti-onchocercal activity, phytochemical analysis and toxicity studies of extracts of Azadirachta indica. J. Pharmacogn. Nat. Prod.10, 281.
82
Ullah A. Zaman S. Hamraz A. Muzammal M. (2023). On the construction of some bioconjugate networks and their structural modeling via irregularity topological indices. Eur. Phys. J. E46, 72. 10.1140/epje/s10189-023-00333-3
83
Ullah A. Zohra B. Zaman S. (2024). Computational aspects of two important biochemical networks with respect to some novel molecular descriptors. J. Biomol. Struct. Dyn.42, 791–805. 10.1080/07391102.2023.2195944
84
Wylie M. R. Merrell D. S. (2022). The antimicrobial potential of the neem tree Azadirachta indica. Front. Pharmacol.13, 891535–891616. 10.3389/fphar.2022.891535
85
Yang J. Konsalraj J. Raja S A. A. (2023). Neighbourhood sum degree-based indices and entropy measures for certain family of graphene molecules. Molecules28, 168. 10.3390/molecules28010168
86
Yu G. Siddiqui M. K. Hussain M. Hussain N. Saddique Z. Petros F. B. (2024). On topological indices and entropy measures of beryllonitrene network via logarithmic regression model. Sci. Rep.14, 7187. 10.1038/s41598-024-57601-1
87
Zaman S. Rasheed S. Alamer A. (2024). A quadratic regression model to quantify certain latest corona treatment drug molecules based on coindices of M-polynomial. J. Supercomput80, 26805–26830. 10.1007/s11227-024-06434-w
88
Zaman S. Yaqoob H. S. A. Ullah A. Sheikh M. (2023). QSPR analysis of some novel drugs used in blood cancer treatment via degree based topological indices and regression models. Polycycl. Aromat. Compd.44, 2458–2474. 10.1080/10406638.2023.2217990
89
Zuo X. Akhtar M. Aslam A. Tawfiq F. M. Kanwal S. (2023). Multiple-Criteria Decision-Making (MCDM) techniques to study the behavior of dendrimers using topological indices. PLoS One18, e0294515. 10.1371/journal.pone.0294515
Summary
Keywords
Azadirachta indica , neem, topological indices, regression models, QSPR, multi-criteria decision making, VIKOR, SAW
Citation
Anuradha DS and Jaganathan B (2025) Predictive modelling and ranking: Azadirachta indica compounds through indices and multi-criteria decision-making techniques. Front. Chem. 13:1580267. doi: 10.3389/fchem.2025.1580267
Received
20 February 2025
Accepted
08 April 2025
Published
29 April 2025
Volume
13 - 2025
Edited by
Micheal Arockiaraj, Loyola College, Chennai, India
Reviewed by
Fengwei Li, Ningbo University of Finance and Economics, China
Asad Ullah, Karakoram International University, Pakistan
Arul Jeya Shalini, Women’s Christian College, India
Updates
Copyright
© 2025 Anuradha and Jaganathan.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: B. Jaganathan, jaganathan.b@vit.ac.in
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.