- 1BioSensor Technologies, AIT Austrian Institute of Technology GmbH, Vienna, Austria
- 2Activartis Biotech GmbH, Vienna, Austria
The combination of microfabrication-based technologies with cell biology has laid the foundation for the development of advanced in vitro diagnostic systems capable of evaluating cell cultures under defined, reproducible, and standardizable measurement conditions. In the present review, we describe recent lab-on-a-chip developments for cell analysis and how these methodologies could improve standard quality control in the field of manufacturing cell-based vaccines for clinical purposes. We highlight in particular the regulatory requirements for advanced cell therapy applications using as an example dendritic cell-based cancer vaccines to describe the tangible advantages of microfluidic devices that overcome most of the challenges associated with automation, miniaturization, and integration of cell-based assays. As its main advantage, lab-on-a-chip (LoC) technology allows for precise regulation of culturing conditions, while simultaneously monitoring cell relevant parameters using embedded sensory systems. State-of-the-art lab-on-a-chip platforms for in vitro assessment of cell cultures and their potential future applications for cell-based strategies for cancer immunotherapy are discussed in the present review.
1. Cell-Based Strategies for Cancer Immuno-Therapy
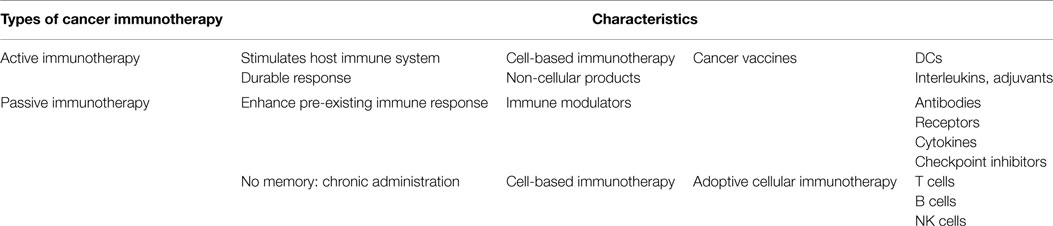
In recent years, cell therapies have been recognized as an important alternative to conventional medical care to alleviate human diseases. Cell therapy, defined as the treatment in which cells are injected into a patient, can be divided into mesenchymal stem cell therapy, hematopoietic stem cell transplantation, and allogeneic or autologous cell therapy (Gage, 1998). Over the past decades, cell therapy treatments have predominantly been applied in cancer immunotherapy (CIT) to control tumor growth and metastasis (Finn, 2003), following the specific activation of immune cells against tumors. In the case of adoptive, allogeneic cell therapy activation of the immune system is accomplished by passive immunization through transfusion of cytotoxic T cells into the patient. Over the years, various immune cell types have been employed for CIT, including natural killer T cells (NKT cells), lymphokine-activated killer cells (LAK cells), cytotoxic T lymphocytes (CTLs), and dendritic cells (DCs) (Zigler et al., 2013). Among these, cytotoxic T cells and DCs are at the forefront of cancer immunotherapies where immune cell activation can either be induced ex vivo or in vivo. Commonly used methods for ex vivo generation of cytotoxic T cells for anti-cancer treatment are based on the isolation of primary peripheral blood mononuclear cells (PBMC) followed by expansion and stimulation/activation of T cells. T cell stimulation can be achieved using anti-CD3/anti-CD28 monoclonal antibodies immobilized on planar substrates (Yamada-Ohnishi et al., 2004), artificial antigen-presenting cells, such as HLA-Ig-coated beads (Oelke et al., 2003) and paramagnetic anti-CD3/anti-CD28-coated beads (Trickett and Kwan, 2003). Alternatively, DC-based active cancer immunotherapy is based on ex vivo generation of “professional” antigen-presenting cells (APCs) that are able to offer specific tumor antigens to lymphocytes in vivo. One key features of matured antigen-presenting DCs for active cancer immunotherapy are their increased expression of MHC class II and co-stimulatory molecules as well as their ability to secrete cytokines, such as IL12, that enable recruitment and activation of cytotoxic antigen-specific CD8+ CTLs. A comparison between passive and active cancer immunotherapy is provided in Table 1.
In addition to the Nobel prize in Physiology and Medicine awarded in 2011 for discovery of DCs role in the immune system, latest advances in DC-based cancer immunotherapy were recently selected by Science as the breakthrough of the year 2013 (Couzin-Frankel, 2013). An advantage of DC-based therapeutic vaccines over passive immunotherapy strategies is the inherent ability of DC to prime naïve T cells and also expand memory T cells, which ultimately leads to the generation of a long-lived immune response capable of preventing tumor relapse (Kalinski et al., 2013). Although cancer immunotherapies are considered superior over conventional chemotherapy in terms of natural selectivity, specificity, and memory effect, a range of potential complications may still occur. For instance, immune-mediated therapies, which are priming the immune system to target tumors expressing certain tumor-associated antigens (TAA), may also induce a response to antigens that are expressed by normal tissues (Hung et al., 2008; Criscitiello, 2012). Potential safety concerns with DC-based cancer vaccines may further include local inflammatory reactions, systemic toxicity, and adverse effects on the host immune system, such as autoimmune responses caused by impurities, contaminants, or other components of the vaccine formulation (Kalantari et al., 2011). As a consequence, it is important to evaluate the safety, mechanism of action and efficacy of immune-mediated therapies. Accordingly, therapeutic vaccinations intended for treatment of cancer must be evaluated on its safety and clinical efficacy for both active and passive products. It is also important to note that clinical efficacy evaluations are compulsory for all pharmaceutical products based on living, functional cells that mediate therapeutic effects, including Alzheimers disease, Parkinsons disease, cardiovascular disorder, and autoimmune disease (Martínez-Morales et al., 2013). These include a large number of biotechnology and pharmaceutical industries, which are involved in several clinical trials using DC, NKT cells, and lymphocytes as well as stem cells derived from blood and bone marrow, liver, etc., to treat structural, metabolic, genetic, neurologic, orthopedic, and cardiovascular disorders. The status and outcome of current clinical trials involving safety of immunotherapeutic products, such as cancer vaccines, can be found at www.clinicaltrials.gov, where currently 797 studies in phase II and 787 studies in phase III are listed. Among these trials, several led to immune responses that are associated with overall survival (Kantoff and Higano, 2010; Walter et al., 2012) and disease-free survival (Schuster et al., 2011).
2. Regulatory Aspects and Quality Control Measures of Advance Therapy Medicinal Products
In all instances, the manufacturing process and biological material characteristics of the employed cell therapy must be rigorously controlled and demonstrated to be consistent with product specifications. This means that the manufacturing process of living, functional cells needs strict regulations based on validated methods and procedures. The three most important regulatory agencies responsible for the evaluation and supervision of medicines for human use are European Medicines Agency (EMA) in the European Union (EU), Food and Drug Administration (FDA) in the USA, and Ministry of Health, Labor and Welfare (MHLW) in Japan. Harmonization between these agencies is achieved through the International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH), which develops guidelines for quality, efficacy and safety of advanced therapy medicinal products (ATMP) (Dixon, 1999; ICH, 2015). In Europe, ATMP are further controlled by the Regulation (EC) No. 1394/2207 (Antunes and Pottering, 2007), which defines requirements of quality, efficacy, and safety for administration in humans. Consequently, good manufacturing practice (GMP) standards have to be applied for both production and quality control ensuring that medicinal products are consistently produced and controlled to the quality standards appropriate to their intended use and according to the requirements of the product specification (Alici and Blomberg, 2010). The quality criteria required by the European and American regulations for biological product characterization are based on tests for sterility, identity of the cellular and non-cellular components, purity, viability, potency, and reproducibility (Agency, 2007). A detailed description of product requirements is listed in Table 2. As an example, flow cytometry (FACS) and ELISA techniques have been extensively used to assess the expression density of functionally important cell membrane molecules and the amount of secreted molecules, such as interleukins, IFN-γ, IL-4, granzyme, perforin, and others. These tests need to provide information on an immune cells functional capacity, subset distribution, purity, viability, cytolytic immunity, and capacity to stimulate T-cells and attack target cancer cells.
3. Automation and Miniaturization of Cell-Based Assays for Quality Control
Automation is the most straightforward strategy for assuring a maximum of reproducibility, which is also a core request of regulators. It is also important to highlight that quality control (QC) of personalized cancer vaccines is by far the more labor intensive procedure in comparison to cell manufacturing, which has essentially become an engineering task (Hinz et al., 2006). Although robotic cell culture systems have been available since about 20 years (Sharma et al., 2011), to date no technological solution exists that allows for automation and miniaturization of QC measures to ensure product safety by simultaneously reducing manual labor steps, material costs, as well as sample and media requirement. Although recent advances in lab automation have brought several high throughput systems for automated cell culture analysis to the market, the applicability of these automated cell analysers for personalized medicine is still questionable. These commercially available automation standards are mainly based on the application of existing fluid handling systems for microtiter-plate technologies in combination with incubators, robotics, and external optical imaging systems. Table 3 lists a number of automated cell analysers developed for modern clinical laboratory environments. The main challenges with adapting these systems to quality control for immune cell therapy products are their inherent small sample volume requirements, expensive clinical grade reagents necessary to characterize cell phenotypes, and time-resolved analysis of cell population responses. Other drawbacks of existing automated cell analyser systems are their time consuming and expensive cell staining procedures that include endpoint detection methods to identify cell phenotypes (Kramer et al., 2013). End-point detection methods also often underestimate labeling artifacts and require complex handling steps and multiple reagents leading to low reproducibility and accuracy (Michelini et al., 2010). In this context, lab-on-a-chip (LoC) technology has the potential to provide the next generation of cell analysis tools capable of inexpensively testing large numbers of single cells or small numbers of cell populations under controlled and reproducible measurement conditions (Whitesides, 2006). Cell chips have initially been used to count and analyze cells in miniaturized flow cytometers which are commercially available today (e.g., Agilent 2100 bioanalyser) (McClain et al., 2001). More recently, microfluidic devices for cellular studies have been developed to investigate cell transport and cultivation in the absence and presence of concentration and temperature gradients or shear force conditions (Andersson and van den Berg, 2003). Overall, microfluidic systems have been used to perform cell sampling, cell trapping, sorting, patterning, treatment, and multi-parameter cell analysis under a defined set of conditions (Vilkner et al., 2004; Yi et al., 2006a,b; Charwat and Muellner, 2011; Charwat et al., 2013a,b; Ungerböck et al., 2013). The application of cell chip technology for automated quality control measures has therefore the potential to close the existing product-gap by providing fully automated and miniaturized analysis systems with improved reproducibility for (a) assuring compliance with specifications, (b) reduction of hand-on work and corresponding human errors, and (c) reduced usage of expensive clinical grade biological reagents.
The minimum requirements for cell chip technology to perform as a quality control measure for cell-based systems is the miniaturization, integration, and automation of crucial cell culture operations, such as liquid handling, proper cell culture, biosensing, as well as phenotype/secretome analysis within a single microdevice. Precise spatio-temporal control over the cellular microenvironment (e.g., physical and chemical stimulation) is mandatory for proper cell propagation (Oelke et al., 2003; Trickett and Kwan, 2003; Yamada-Ohnishi et al., 2004) as well as maintenance of cell phenotypes (Gallucci et al., 1999; Kodaira et al., 2000; Lewis et al., 2013). The integration of in-line biosensors enables dynamic and time-resolved monitoring of cell cultures during all stages of the production process of cell-based therapies in a non-destructive manner. Last but not least, automation and miniaturization of state-of-the-art quality control assays for monitoring of metabolites and cell phenotypes enable high-throughput analysis with reduction of manual labor.
The following three subsections focus on fundamental elements, which enable the microfabrication of chip-based quality control platforms, including fabrication methods and materials, liquid handling systems, and biosensing strategies for cell-based applications.
3.1. Fabrication Methods and Materials of Cell Chip Systems
Although studying in vitro cell cultures is an essential aspect of cell biology, its technological advancement has lagged behind compared with progress made in the fields of genomics, proteomics, and high-throughput testing of biochemicals. In this context, microfabrication technology has the potential to provide the next generation of cell analysis tools capable of inexpensively testing a large number of single cells or a small number of cell populations in environments of increased physiological relevance (Whitesides, 2003; Ionescu-Zanetti et al., 2005). Biochip technology is vital for cell analysis because it is capable of providing answers to dynamic and rapidly changing biological systems using label-free analytics and microfluidics (El-Ali et al., 2006). As seen in Figure 1 lab-on-a-chip and cell chip technology stems from semi-conductor fabrication technologies with the first microfluidic chips fabricated in the late 1980s. Later in 1993, Harrison and Manz achieved a large technological leap with on-chip capillary electrophoresis (Harrison et al., 1993). However, with the introduction of soft lithography by Xia and Whitesides 1998, biologists started to adapt these integrated microsystems for cell manipulation and analysis with the first dedicated journal lab-on-a-chip initiated in 2001 (Xia and Whitesides, 1998). In 2004, Michael Shuler and colleagues introduced micromachining techniques to fabricate multi-compartment cell culture systems in which microfluidic channels control fluid flow between the single tissue compartments (Sin et al., 2004). These microdevices, also known as micro cell culture analog (μCCA), animal-on-a-chip and body-on-a-chip, simulate the complex interplay between multiple organ and tissue types with a functional circulatory system. From that point on the development of highly complex cell-chip devices that incorporate micropumps and valves, mixers, actuators, degassers, biosensors, as well as multiple cell cultivation chambers for multiplexed cell analysis, adapting the concept of μTAS and labs-on-a-chip for cell-based applications has continuously increased to date (Reichen et al., 2013; Wagner et al., 2013; Frey et al., 2014).
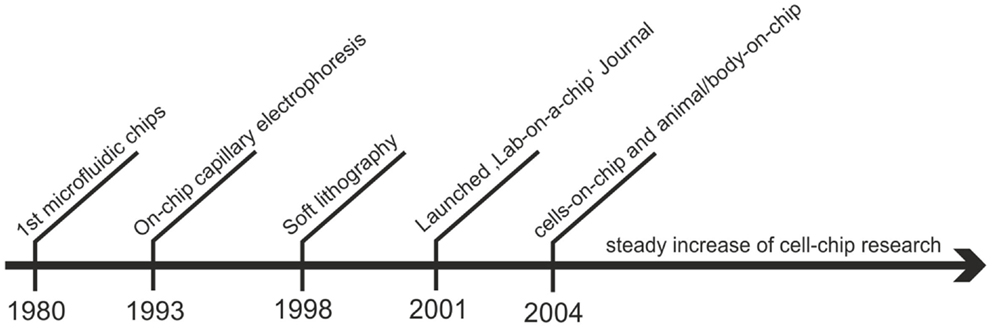
Figure 1. Overview over the historical development of lab-on-a-chip technology for cell-based assays.
The following section presents a short overview of methods and material commonly used to design and fabricate lab-on-a-chip systems for cell analysis. Fabrication methods used to build lab-on-a-chips systems are based on micro-electro-mechanical systems (MEMS) technology and include soft lithography, hot embossing, injection molding, laser micromachining, and photolithography as well as more recently introduced three-dimensional printing techniques (Fiorini and Chiu, 2005; Dragone et al., 2013; Ertl et al., 2014). The fabrication method of choice is guided by multiple factors, including available infrastructure (technology and equipment), fabrication speed, cost (multi-use or disposable devices), desired feature size, as well as the preferred fabrication material. While initially glass- and silicon-based materials were used to fabricate microfluidic devices (Harrison et al., 1992; Manz et al., 1992), replica molding has become a dominate trend in recent years due to its fast and inexpensive fabrication of microdevices using polydimethylsiloxane (PDMS) (McDonald and Whitesides, 2002; Zhang et al., 2009, 2010; Berthier et al., 2012), hydrogels, thermoset composites (Carlborg et al., 2011; Sollier et al., 2011), and thermoplastics (Duffy et al., 1998; Fiorini et al., 2003, 2004; Golden and Tien, 2007; Novak et al., 2013). The application of replica molding, also called “soft lithography” (Xia and Whitesides, 1998), eliminates the need for time- and cost-intensive clean room infrastructures. For high-throughput fabrication of microfluidics, a set of alternative replica molding procedures are available, including hot embossing and injection molding. Hot-embossing involves the use of a variety flat thermoplastic sheets, such as polymethylmethacrylate (PMMA), polycarbonate (PC), cyclic olefin copolymer (COC), polystyrene (PS), polyvinylchloride (PVC), and polyethyleneterephthalate (PETG) (Becker and Locascio, 2002; Novak et al., 2013; Ren et al., 2013), which are molded against a master using pressure and heat (Locascio et al., 2006). In turn, injection molding involves the high-pressure injection of pre-polymerized molten thermoplastic granules into a heated molding cavity (Mair et al., 2006; Attia et al., 2009), which allows the high-throughput industry-scale fabrication of thermoplastic devices. A detailed description of microfabrication methods of lab-on-a-chip devices can be found elsewhere (Fiorini and Chiu, 2005; Kim et al., 2008a; Coltro et al., 2010; Sollier et al., 2011; Wu and Gu, 2011).
Apart from the scientific aspects, these fabrication materials and processes require industrial and clinical compliance with respect to through-put, biostability, and feasibility. PDMS microdevices, for instance, cannot be fabricated in a high-throughput manner, because master molds are mechanically unstable and therefore the process is inadequate for industrial applications (van Kan et al., 2012). Furthermore, the disadvantages of PDMS (e.g., hydrophobicity, absorption of molecules toxicity of monomers, and vapor permeability) outweigh the advantages frequently reported in the literature (e.g., easy fabrication, gas permeability, and optical transparency). Even though some of the materials unpleasantries may be bypassed on a research level (Sasaki et al., 2010), strategies to date are not fit for industrial scale fabrication with respect to feasibility, as well as infrastructure and material costs. Therefore, state-of-the-art industrial fabrication processes, including hot embossing and injection molding, have been adopted for low-cost microdevice fabrication to meet the required production throughput as well as microdevice complexity (Roy et al., 2011, 2015). However, the major disadvantage of plastic chips is the integration of biosensors (especially metal electrodes) with only a few processes being fit for industrial application (Schrott et al., 2009; Huang et al., 2010).
3.2. Liquid Handling Systems for Microfluidic Cell Cultures
In this section, we review liquid handling systems and components for microfluidic cell cultures, including microvalves and micropumping systems. The main advantage of employing integrated liquid handling systems for cultivation of cell populations is the ability to continuously supply nutrients and soluble factors by simultaneously removing waste products, thus providing a stress free cellular microenvironment for optimum cell culture conditions. Recent technological advancements in lab-on-a-chip fabrication has enabled the creation of increasingly complex devices that include micropumps and valves, mixers, actuators, degassers, on-chip biosensors, as well as multiple cell cultivation chambers for multiplexed cell analysis (Reichen et al., 2013). The technology of handling nano- and picoliter volumes of fluids started in the 1980s (Whitesides, 2006) and over the years a variety of different methods and system have been reported for fluidic actuation and handling (Sackmann et al., 2014). The most commonly used fluid handling systems for cell cultures are based on either pressure driven or passive driven flow profiles using syringe- (Stevens et al., 2008; Sun et al., 2010; Chin et al., 2011; Song et al., 2014) and electrochemical pumps (Neagu, 1996; Neagu et al., 1997) as well as gravimetrically (Kim et al., 2008b) driven flow systems. Overall, the high response time (few seconds) and a very robust and long term reliability and consistency (together with relatively low power consumption) made syringe pump driven microfluidic systems one of the most commonly used for microfluidic cell cultures to date. Nevertheless the application of microvalves and micropumps to manipulate and transport solutions at the nanoliter scale (Unger et al., 2000; Grover et al., 2003; Kim et al., 2012) has opened new opportunities for improved cell culture handling (Figures 2A–E). For instance, the integration of computer-controlled pneumatically actuated (Figure 2A) micropumps may allow for automation of a number of crucial liquid handling steps, including cell seeding, flow direction control, the activation of cells, and their exposure to bioactive substances at specific solution concentrations. Additionally, passive fluid handling systems for cell cultures rely either on capillary (Figure 2C) (Zimmermann et al., 2007; Gervais and Delamarche, 2009) or gravimetrically (Figure 2D) (Morier et al., 2004) driven flow profiles exhibiting low (fixed) flow rates and very low response time. Although passive pumping eliminate the need for additional actuation systems and power consumption, these fluid handling systems are limited by their consistency and reusability.
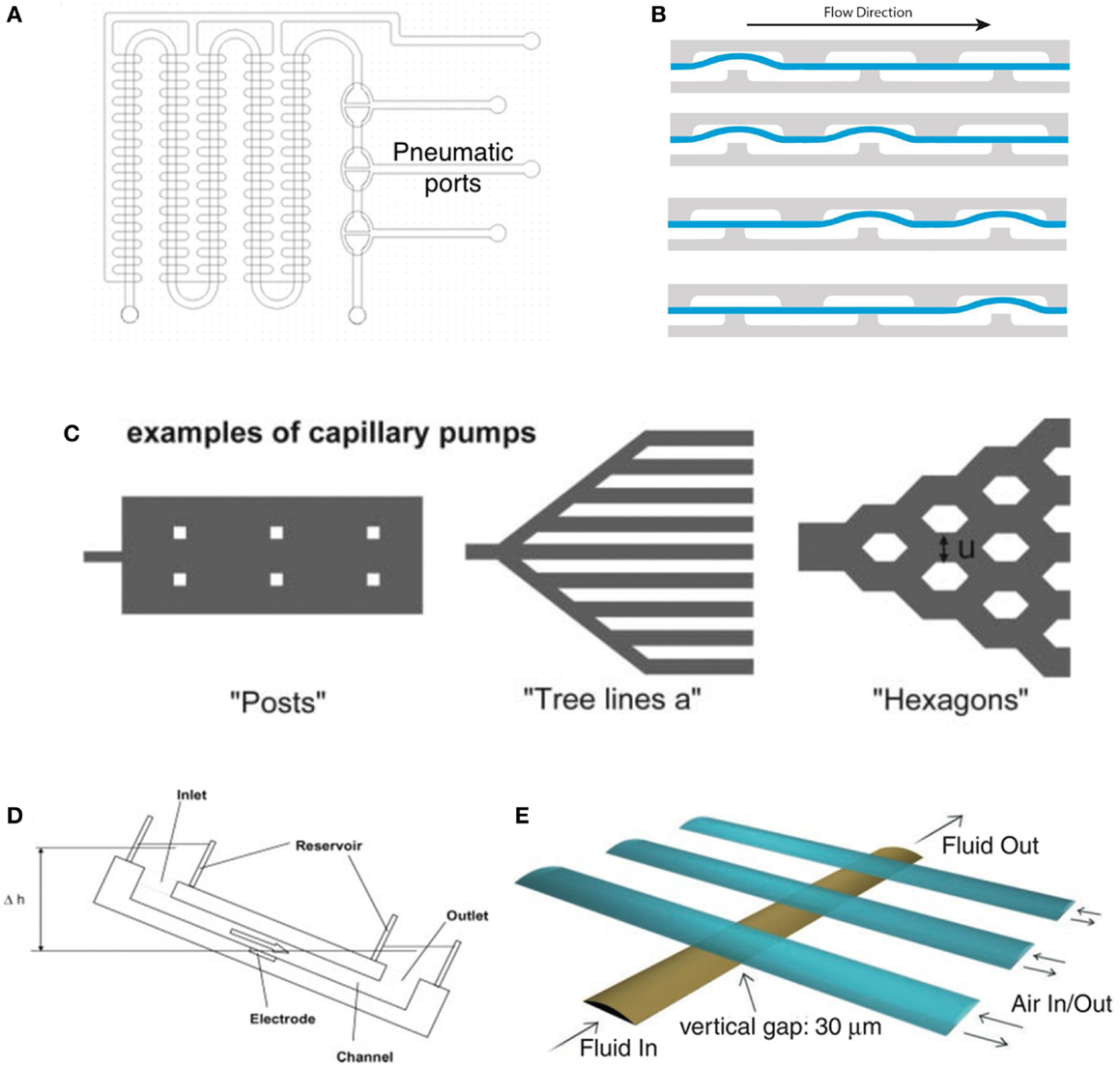
Figure 2. Various types of micropumps for automated and nanoliter liquid handling. (A) Micropumps based on the pneumatic actuation principle developed by Grover et al. (2003) allowing the pneumatic actuation of the individual pumping chambers via a programmable controller. Reproduced by permission of by Elsevier (B) shows the schematics of the fluid transport in the micropumps based on the design introduced by Grover et al. (2003). Kindly provided by Novak (2013), (C) capillary pumps can be actuated by the different geometry and density of post as well as the relative positioning of the channels thus creating different capillary pressures. For detailed information, see (Zimmermann et al., 2007). Reproduced by permission of The Royal Society of Chemistry. (D) Schematic drawing of a gravitational-driven pump using the height difference Δh between in- and outlet and thereby controlling the flowrate (Morier et al., 2004). Reprinted with permission of John Wiley & Sons, Inc. (E) Micropump design proposed by Unger et al. (2000) using a secondary gas-channel layer to control the flow-profile in the underlying liquid channel. Reprinted by permission of the American Association for the Advancement of Science.
The main advantage of employing on-chip microvalves and micropumps for quality control measures is that these integrated microsystems can easily be multiplexed during fabrication and enable high sample throughput (e.g., microfluidic microarray technology). Especially, the elimination of dead volumes stemming from tubing, macroscopic valves and fluidic connectors enables efficient sample and reagent consumption, thus reducing overall costs. Furthermore, spatio-temporal control over cells and fluids can be processed in a highly multi-plexed fashion using fully automated software scripts guaranteeing high reproducibility while reducing hands-on work, which is prone to user-to-user variations and failure.
3.3. Integrated Sensing Functions for Microfluidic Cell Analysis Systems
A major advantage of lab-on-a-chip technology is that it allows the facile integration of optical, electrical, magnetic, and acoustical sensors for cell analysis (Wu and Gu, 2011). Routinely used immunofluorescence end-point analysis can now be extended with complementary, continuous monitoring techniques (Charwat et al., 2013a,b, 2014; Picher et al., 2013; Novak et al., 2015). Several portable and miniaturized sensors have been developed over the past years to analyze rapidly changing biological systems. Among these biosensors optical-based systems, light scattering (Schafer and Jamieson, 1979; Wilson et al., 2005), absorption- and transmission- (Jindal and Cramer, 2004) or fluorescence spectroscopy (Reyes et al., 2002; Hata et al., 2003; Toma et al., 2011) are predominately used for cell analysis. A detailed review on integrated on-chip optical detection systems can be found at Kuswandi et al. (2007). Among cell analysis methods, fluorescence spectroscopy is still the most widely used optical technique for on chip cell detection, due to its superior selectivity and sensitivity and the availability of a broad range of optical labels (Reyes et al., 2002; Hata et al., 2003). Recently, a variety of integrated optical oxygen sensors (Schapper et al., 2009) for cell analysis have been developed for single point measurements (Vollmer et al., 2005; Lam et al., 2009) and 2D read out systems (Sud et al., 2006; Nock et al., 2008; Nock and Blaikie, 2010; Thomas et al., 2011; Ungerböck et al., 2013). For instance, Ungerboek et al. (Ungerböck et al., 2013) implemented a two-wavelength ratiometric imaging system (Stich et al., 2009; Wang and Meier, 2010; Larsen et al., 2011; Feng et al., 2012; Meier and Fischer, 2013) to combine low-cost and easily available imaging equipment with high-resolution imaging for oxygen sensing in microfluidic systems (Figures 3A,B). A CCD camera measuring the intensity at two different wavelengths of the sensors emitted light by using the different color channels of the camera one channel providing the oxygen sensitive intensity image, the other providing a so called reference image, from which ratiometric calculations were conducted to accurately detect respiratory activities. Although a variety of excitation sources are available for fluorescent detection laser-induced fluorescence (LIF) is most easily adapted to the dimensions of microchips (Auroux et al., 2002; Gao et al., 2004; Lee and Wen, 2005). Since laser systems are often expensive and wavelength specific (exhibiting very narrow bandwidth), a range of alternative light sources, including organic light-emitting diodes (OLEDs) have been successfully integrated into microfluidic devices (Choudhury et al., 2004; Cai et al., 2008, 2010; Shinar and Shinar, 2008; Liu et al., 2011; Sagmeister et al., 2013; Williams et al., 2014). Additionally, organic field-effect transistors (OFETs) (Figure 3C) as read-out devices for various physical (Sekitani et al., 2009), chemical, and biological (Lin and Yan, 2012) sensing applications can be used in the non-invasive, label free analysis system (Someya et al., 2010; Yun et al., 2014). Furthermore, the latest progress in OFET development points at the potential toward the fabrication of a low-cost, printable, flexible, highly sensitive, and selective detection platform for the integration into LoC analysis and sensor repertoire (Torsi et al., 2002; Someya et al., 2010). Another recent advancement includes the application of solution-processed fully spray-coated organic photodiodes (OPDs) (Figure 3E) (Tedde et al., 2009) for cell analysis, which allows the integration of disposable fluorescence and chemiluminescence sensors in cartridge-based analysis systems (Hofmann et al., 2005; Wang and Amatatongchai, 2009; Ryu et al., 2011). To study the electrical properties of mammalian cells, a variety of electroanalytical methods, such as voltammetry, potentiometry, and impedance spectroscopy, are available to provide information of cell viability, proliferation, and morphology changes. For instance, a current study by Yea et al. (2013) reported a newly developed electrochemical cyclic voltammetry (CV) system to determine the status of mouse embryonic stem (ES) cells. The proposed electrochemical analysis system can be applied to an electrical stem cell chip for diagnosis, drug detection, and on-site monitoring. A powerful non-invasive and label-free electro-analytical measurement technique to assess cell morphological changes, migration, stress responses, and differentiation is impedance spectroscopy, mainly known under the trademark ECIS ™(electrical cell-substrate impedance sensing). Microfluidic impedance cell cytometry (Sun and Morgan, 2010) and a great number of impedimetric cell culture analyzers have been published over the past decade including cytotoxicity (Xiao and Luong, 2003; Yeon et al., 2005), cell spreading (Wegener et al., 2000), endothelial cell stimulation and tight junctions (Wegener et al., 1999) IgE-mediated mast cell activation (Abassi et al., 2004), and stem cell differentiation (Cho et al., 2009; Hildebrandt et al., 2010). Another research group combined two well-established but usually independently used methods for the label-free analysis of living cells, impedance spectroscopy, and light scattering measurement (Charwat et al., 2013a) (Figure 3D). This novel dual-parameter cell-on-a-chip system detects light scattering from adherent cells providing information on cell number and intracellular granularity and simultaneously performs impedance spectroscopy to monitor cell adhesion and cell–cell interaction without disturbing normal cell behavior whereby mimicking physiological conditions, such as laminar flow and mechanical stress. Magnetic detection systems to analyze microfluidic cell cultures in real-time and label free are among others the magnetic lab-on-a-chip system (MAGLab) developed by Shoshi et al. (2012). This high-sensitive device allows the monitoring of phagocytosis in living cells (Figure 3F) in real time. This combination of magnetoresistive sensors, magnetic particles, and microfluidics has been developed for analyzing the uptake dynamics of the dorsal cell membrane at different physiological conditions. By tailoring the surface bio-chemistry and/or physical characteristics of the magnetic drug carriers, the developed methodology might be used to analyze the bioavailability of nanodrug carriers in cancerous cells/tissues, thus improving and expanding the biosensor repertoire for cell analysis (Shoshi et al., 2012). In the recent years, the development and fabrication of magnetic nanoparticles (MNPs) that can be effectively tailored molecule-specific, magnetism has become an attractive mechanism for bio-separation and bio-detection (Pankhurst et al., 2003; Cheon and Lee, 2008; Jun et al., 2008; Laurent et al., 2008; Gijs et al., 2010). MNPs can selectively bind to biological entities of interest, including nucleic acids, proteins, viruses, bacteria, and cells.
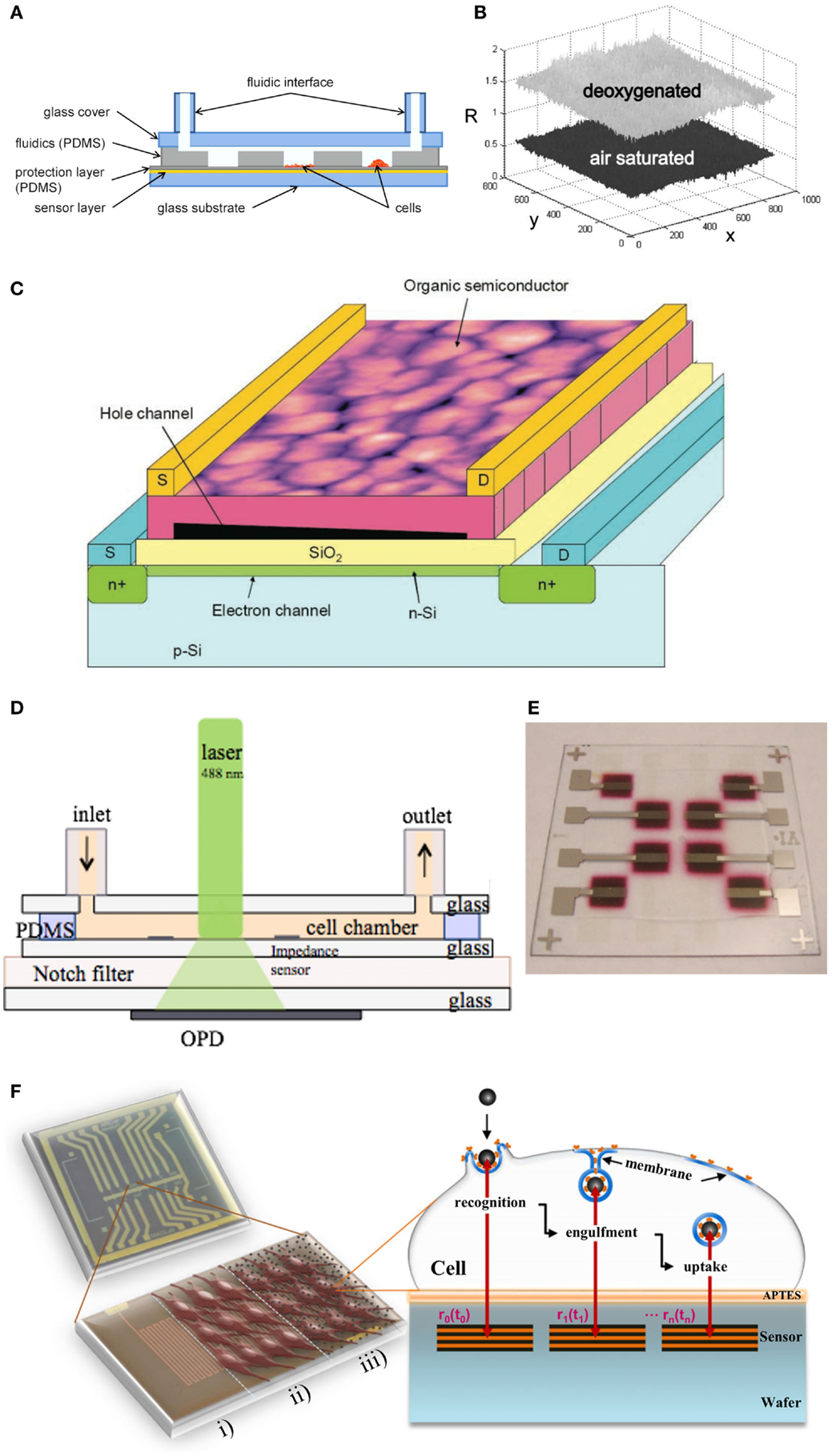
Figure 3. Different integrable biosensors for cell culture analysis. (A) Schematic cross section of the developed oxygen sensing chip assembly for high resolution oxygen imaging, which can be used to study 2D oxygen distribution inside microfluidic environments (Ungerböck et al., 2013). Reproduced by permission of The Royal Society of Chemistry. (B) Ratiometric (referenced) image obtained by division of the red by the green channel. (Ungerböck et al., 2013). Reproduced by permission of The Royal Society of Chemistry. (C) Schematic representation of organic field-effect transistors (OFETs) for label-free sensing of various chemical/biological species. A detailed description and application can be found elsewhere (Someya et al., 2010). Reprinted with permission of John Wiley & Sons, Inc. (D) Dual monitoring sensor system for continuous forward light scattering using OPDs and impedance measurement developed by (Charwat et al., 2013a). Reproduced by permission of The Royal Society of Chemistry. (E) Photograph of fully-spray coated organic photodetectors (OPDs) developed by Tedde et al. (2009). Reprinted with permission from American Chemical Society. (F) Sketch of the magnetoresistive-based phagocytosis monitoring methodology developed by Shoshi et al. (2012). Reproduced by permission of by Elsevier.
After almost half a century of biosensor research with promising application potential (e.g., agricultural, food and medical industries), a wide gap between research-based biosensing proof-of-principle and industrial application still persists. Even though a few industrial analysis systems have been commercialized so far, they are mainly confined to electrochemical methods (e.g., pH, conductivity, glucose, urea, lactate, etc). The main reason why only a few biosensors are commercialized to date is the inflexible and slow technology transfer toward commercialized products. Even though improvement of sensitivity, selectivity, and stability is driving the biosensor research field, the lack of in-line implementation within already existing production processes impede the application of microfabricated platforms for on-chip quality control. For future biosensing platforms to be able to act as enabling technology especially for the fields of medical/pharmaceutical diagnostics and quality control, existing challenges including multiplexed analysis (e.g., sensor arrays), improved signal-to-noise ratio, probe robustness (e.g., synthetic alternatives to natural antibodies), non-invasiveness, as well as sensor platform and assay validation need to be conquered prior successful application within the biomedical field. Within the area of cell-based cancer therapeutics, the concept of companion diagnostics and precision medicine can potentially be a main driving force for commercialization of new biosensor products within the pharmaceutical industry. Nonetheless, a highly interdisciplinary approach comprising biosensing, multiplexed assays, and miniaturized automated cell chip technology needs to be approached.
4. Flow Cytometry-on-Chip and Single Cell Manipulation and Isolation
Since fluorescence-activated cell sorting, commonly known as flow cytometry (FACS), is extensively used to assess viability, purity, and potency of cancer vaccines, alternative miniaturized flow cytometry systems are reviewed in the following sections. FACS is generally used to analyze sub-populations of a sample with single cell resolution to obtain information on cell size (Skierski, 2012) and structural complexity, such as the internal granularity of cells (Schafer and Jamieson, 1979; Wilson et al., 2005). Despite the many advantages of FACS techniques, there are also some drawbacks, such as size of commercially available flow cytometers, cost of equipment, maintenance, and throughput capability. The trend toward increased miniaturization has therefore led to the development of a range of modern cytometry techniques (Cho et al., 2010) (Figure 4A), the so called μFACS devices that exhibit improved portability and analysis time for various medical applications (Fiorini and Chiu, 2005). The main challenges of μFACS are concerned with miniaturization and integration of fluidic, optical, and electronic components. In particular, the reliable generation of disposable microchannel networks constitutes a fundamental requirement of μFACS applications for quality control measures for advanced cell therapies (Cvetković and Dittrich, 2013). Recent publications describe the development of advanced flow cytometry systems involving electroosmotic (Fu et al., 2004) (Figure 4B), dielectrophoretic (Lapizco-Encinas, 2004), magnetic (Pamme and Wilhelm, 2006) (Figure 4C), and hydrodynamic cell sorting application (Bang et al., 2006). While electroosmotic cell sorting allows precise flow switching (Fu, 2002; Chen et al., 2009), only low flow rates of about tens of particles per second can be applied. In turn, dielectrophoretic cell sorting allows manipulation and sorting at the level of single cells, but is characterized by a complex fabrication and low sensitivity relative to cell differentiation (Chen et al., 2009). Alternatively, magnetic sorting has become popular in recent years due to its high selectivity (Chen et al., 2009). Newly developed methods implement piezoelectric controlled actuators (Chen et al., 2009) and optical tweezers (Perroud et al., 2008) for high precision workflow in the μFACS environment. Here, hydrodynamically focused cells in the microchannels are selected and motion controlled by optical tweezers based on their fluorescence signal. The developed μFCS device by Perround et al. uses an infrared laser to laterally deflect and control cells into a collection channel (Perroud et al., 2008). Detection by forward-scattering and interrogating the signal against LIF allows for precise cell counting. The selected/labeled cells are captured by the trapping laser which then manipulates the flow direction into a different trajectory, thus sorting the cells into separate channels (Ligler and Kim, 2010). The on-chip integration of piezoelectric zirconate titanate films (PZT) actuators (Chen et al., 2009) in μFACS devices allows hydrodynamic focusing of single cells in the sub-nanoliter volume (Cho et al., 2010) (Figure 4A). When a targeted cell enters the sorting junction, the PZT actuator is activated using a voltage pulse to deflect the cell-containing fluid from the center position toward a side collection channel (Chen et al., 2011), thus enabling an operating limit of >1000 cells/s. Currently, commercially available is the miniaturized flow cytometer tabletop-box (NanoCellect, San Diego, CA, USA), which is based on integrated optical system, the microdevice is combined with PZT actuators (http://nanocellect.com/). The current developments in μFACS devices show good reliability and reasonable throughput to slowly replace the present conventional large and expensive cytometry machines.
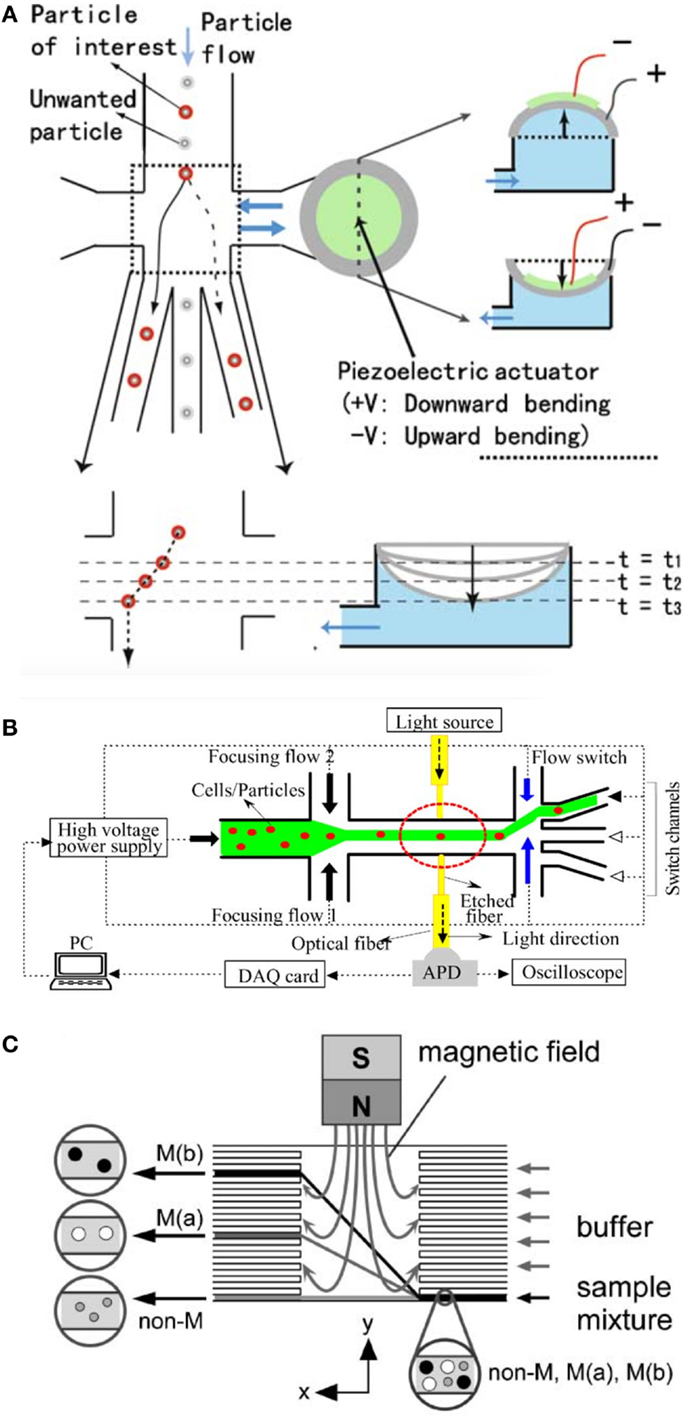
Figure 4. Different flow-cytometry cell sorting systems. (A) (Chen et al., 2009) and (Cho et al., 2010) developed a piezo actuated cell sorting system for cell separation. The flexible piezo actuator acts as a fast pump which pushes or pulls the previously detected cell into different sorting channels enabling the separation of up to 1000 cells/s. Reprinted from Chen et al. (2009) with permission of Springer Science + Business Media and Cho et al., 2010 reproduced by permission of The Royal Society of Chemistry. (B) The micro flow cytometer developed by (Fu et al., 2004) employs electrokinetic forces rather than the more conventional hydrodynamic forces technique for flow focusing and sample switching, and incorporates buried optical fibers for the on-line detection of cells or particles. Reproduced by permission of by Elsevier. (C) The group of Pamme and Wilhelm (2006) employed the approach of magnetic cell separation, which is one of the most efficient methods for bulk cell separation. The on-chip free-flow magnetophoresis magnetically labeled cells can be separated into subpopulations of different magnetization with high precision. Reproduced from Pamme et al. (2006a) by permission of The Royal Society of Chemistry.
5. Immunoassay-on-Chip for Quality Control Applications
Immunoassays are generally used to quantify molecules of biological interest based on the specificity and selectivity of antibodies against antigens (Cox, 2012). The identification of secreted biomolecules is also important in cell-based therapies, because they provide information on the activity of cancer vaccines. The most commonly employed immunoassays in use today is the enzyme-linked immunosorbent assay (ELISA) (Engvall and Perlmann, 1971; Van Weemen and Schuurs, 1971), which was developed in the 1960s. Since then ELISA has become a fundamental tool in biological research and the pharmaceutical industry (Fossceco et al., 1996; Lequin, 2005). ELISA is predominantly performed in standard 96-well plate formats which can easily be automated using standard liquid dispensing systems, robotics, and colorimetric detectors (Sun et al., 2010). The simplicity, relatively low cost, and large throughput still place the ELISA as the most commonly used assays today. Drawback of employing ELISA for routine quality control measurements for advanced cell therapy products is its large sample and reagent volumes, its susceptibility to contaminations and lack of providing information on dynamic changing biological systems. For instance, the time-dependent release of cytokines by DC can serve as an indicator of the ability to recruit and activate T-cells in vivo. Here, the application of microchip technology offers the opportunity to reduce the costs associated with clinical grade reagents, while ensuring faster, accurate, and time-resolved analysis (Sun et al., 2010; Chin et al., 2011; Eyer et al., 2013; Kim and Paczesny, 2013). A number of different ELISA-lab-on-a-chip (ELISA-LoC) systems with comparable sensitivities were recently designed, fabricated, and tested for immunological detection (Honda and Lindberg, 2005; Sista et al., 2008; Lee et al., 2009; Sun et al., 2010; Kim et al., 2011; Miller and Ng, 2011). A digital microfluidic (DMF) device by Miller and Ng (2011) was applied to a heterogeneous sandwich immunoassay (Figure 5A). The digital approach to microfluidics manipulates samples and reagents in the form of discrete droplets, as opposed to the streams of fluid used in microchannels. Sun et al. (2010) recently showed the combination of ELISA-LoC devices (Figure 5B) for immunological detection of staphylococcal enterotoxin B (SEB). This miniaturized 96-well ELISA plate, requiring 5 μl of sample combined with a CCD camera allows low-cost immunodetection without laboratory environment. A microfluidic device for solid-phase immunoassays based on microparticle labeling was developed by the group of Kim et al. (2011) for automated sample processing having programmable microvalve controls in a multilayer structure thus providing automated sample delivery, adjustable hydrodynamic washing, and compatibility with a wide range of substrates (Figure 5C). Additionally, Kim and Paczesny (2013) developed an ELISA-LoC device based on capillary fluid handling for passive immunoassay automation using multiple reagent and sample in sequential and parallel manner, thus reducing the total assay time to ~30 min with very low sample volume (Figure 5D). Another approach for single cell trapping and treatment (e.g., washing) prior immunoassays has been introduced by Eyer et al. (2013) and showed increased repeatability. The cells are controlled and directed using fluid control and hurdle-microstructures for individual cell trapping that also increase the overall sensitivity in detection. Overall, ELISA-LoC devices have increasingly improved in handling, selectivity, efficiency, and sensitivity, thus becoming an attractive tool for medicine and biotechnology as well as the pharmaceutical market. Besides the widely used ELISA immunoassay, there are also alternative assay formats capable of detecting biomarkers within a sample. For instance, Cesaro-Tadic et al. used microfluidic chips to detect the biologically important cytokine tumor-necrosis-factor-α (TNF-α) using a pre-coated PDMS surfaces and fluorescently tagged detection antibodies bound the captured analyte molecules (Cesaro-Tadic et al., 2004). Additionally, a recently developed method by Junkin and Tay (2014) enables the detection of cell signaling molecules with high resolution. Since the most direct way to observe cell activation is through the detection and quantification of transcription factors, the microdevices allows direct observation and quantification using fluorescence microscopy. Furthermore, activation of single CD4+ T-cells was studied by Zaretsky et al. (2012) using a microwell array in combination with fluorescent reporting of the regulatory T-cell transcription factor Foxp3 and surface staining of CD69. This LoC system was composed on a thin layer of a PDMS array of microwells, which can vary in size and depth, and is placed at the bottom of an optical 96-well plate. T cells activation and proliferation in the presence of antigen-coated microbeads was detected using standard optical microscopy. The microfluidic single-cell barcode chip (SCBC) (Figure 5E) developed by Ma et al. (2011) detects secreted proteins in a 1 nl volume microchambers, each loaded with single cells or small defined numbers of cells. Protein concentrations are measured with immunosandwich assays from a spatially encoded antibody barcode. A full barcode represents a complete panel of protein assays, and duplicate barcodes per microchamber enable measurement statistics at the single-cell level. The SCBC permits on-chip, highly multiplexed detection of less than 1000 copies of proteins and requires only ~1 × 104 cells for the assay (Ma et al., 2011). The group of Honda and Lindberg (2005) evaluated the commercially available multiple simultaneous immunoassay ELISA-LoC-CD for easy laboratory integration and ease of handling. Finally, García et al. (2013) recently developed a micromotor-based lab-on-chip immunoassays. This innovative LoC system uses a “on-the-fly” double-antibody sandwich assay (DASA) to selectively capture target protein, in the presence of excess of non-target proteins. This nanomotor-based microchip immunoassay offers many potential applications in clinical diagnostics, environmental and security monitoring fields, as well as further applications in the cell therapy and point-of-care diagnostics sector.
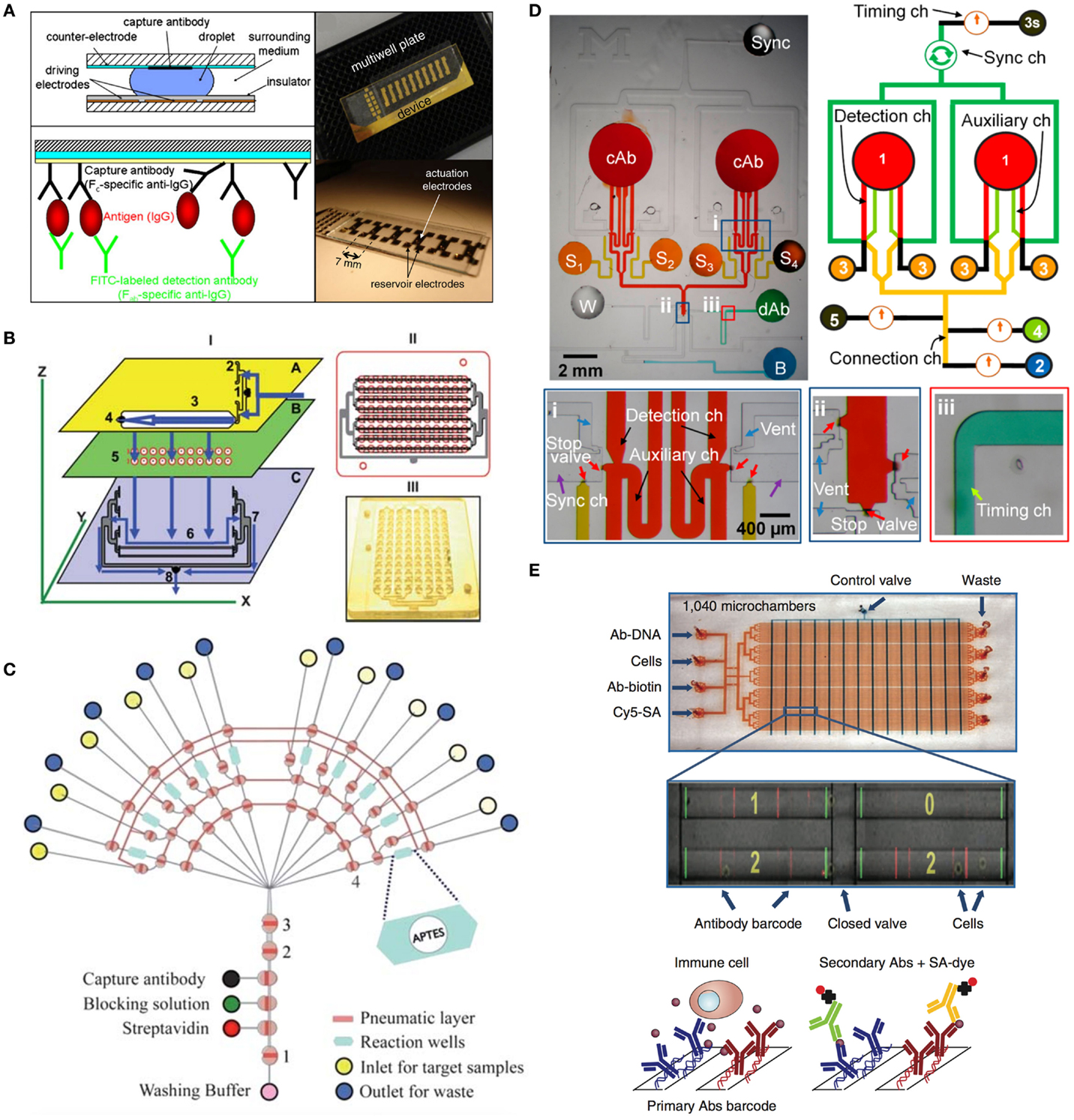
Figure 5. Facilitated automated immunoassay on chip. (A) A digital microfluidic (DMF) device by Miller and Ng (2011) was applied to a heterogeneous sandwich immunoassay. The digital approach to microfluidics manipulates samples and reagents in the form of discrete droplets, as opposed to the streams of fluid used in microchannels. DMF devices are straightforward to use, and are reconfigurable for any desired combination of droplet operations. This flexibility makes them suitable for a wide range of applications, especially those requiring long, multistep protocols such as immunoassays. Reprinted from Miller and Ng (2011) with permission of Springer Science + Business Media. (B) Schematic representation of the 96-well ELISA-LoC system developed by Sun et al. (2010) requires only 5 μl of sample and in the combination with a CCD camera allows low-cost immunodetection without laboratory environment. Reproduced from Sun et al. (2010) by permission of The Royal Society of Chemistry. (C) The microfluidic device by Kim et al. (2011) for solid-phase immunoassays based on microparticle labeling was developed using microvalve-control structures for automated sample processing. Programmable microvalve control in a multilayer structure provides automated sample delivery, adjustable hydrodynamic washing, and compatibility with a wide range of substrates. Reproduced from Kim et al., 2011 by permission of The Royal Society of Chemistry. (D) Capillarity-driven immunoassay device for sequential and parallel flow processing greatly saving time and labor with an inexpensive setup developed by Kim and Paczesny (2013). Reprinted with permission from American Chemical Society. (E) Design of the SCBC for single-cell protein secretome analysis developed by Ma et al. (2011). In their design flow, channels are shown in red and the control channels are shown in blue. An optical micrograph (zoomed picture) shows the loaded cells isolated within the microchambers, overlaid with the fluorescence micrograph of the developed assay barcode for those same microchambers. Reprinted by permission from Macmillan Publishers Ltd: Nature Medicine, Ma et al. (2011).
6. Concluding Remarks and Future Perspectives
In the advent of personalized cell therapies, the cell manufacturing industry will need to manipulate, cultivate, and expand patient-derived cells in vitro, thus requiring independent manufacturing and quality control measures for each patient. In this case, automation is the most effective strategy to assure reproducibility, which is a core prerequisite of regulating agencies. Although a number of robotic cell culture systems have been available for the past 20 years, no automation of quality control exists, which means that the cost of performing manual quality control becomes an influencing factor in cell manufacturing. In fact, regulatory approval may be inhibited by the lack of enabling technologies for automated quality control to (a) improve reproducibility for assuring compliance with specifications and (b) reduce hands-on work and corresponding human error. Consequently, translation of lab-on-a-chip technologies in the automation and miniaturization of quality control procedures is envisaged to improve cost effectiveness by reducing expensive clinical grade biological reagents and labor costs. Although in recent years a number of commercially available microfluidic systems have been introduced for cell analysis, their economic success and market acceptance has been slower than anticipated due to their unfamiliarity to the users. In summary, the reviewed lab-on-a-chip technologies containing integrated sensing functions and automated fluid handling systems have already been successfully employed for nucleic acid testing, immunoassays, and cell analysis.
Consequently, translation of lab-on-a-chip technology in the automation and miniaturization of quality control procedures is envisaged to improve cost-effectiveness by reducing expensive clinical grade reagents and hands-on labor costs. Figure 6 shows a summary of already available lab-on-a-chip elements for the integration within a single microfabricated platform capable of automated liquid handling, cell propagation, and stimulation, and of in-line biosensing and biomolecule analysis. For instance, the integration of fluid handling systems using micropumps and microvalves is likely to play a critical role in process automation because it will enable the programmable transportation and manipulation of microliter to nanoliter volumes in cell chips with high spatio-temporal precision, thus significantly improving assay robustness. Finally, merging of multiple biosensors and bioassays within one automated platform will be inevitable for lab-on-a-chip technology to become a competitive alternative to state-of-the-art industrial quality control measures of cell-based products and therapeutics.
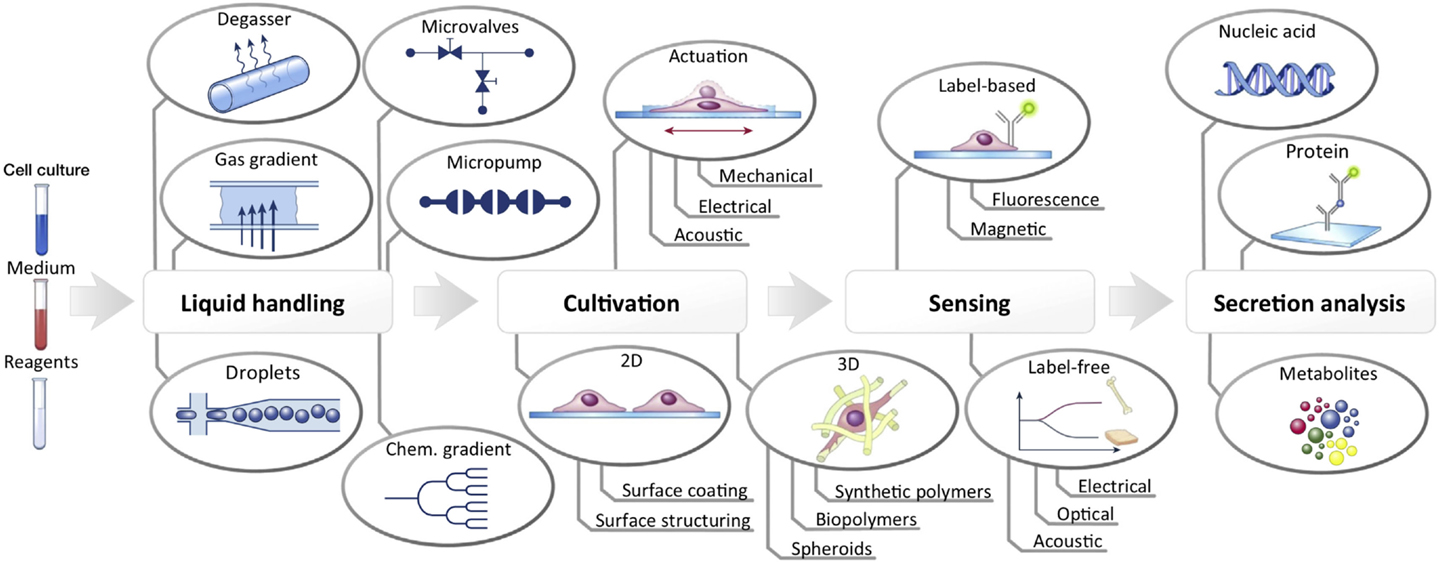
Figure 6. Schematic illustration of components and systems available to date that can be integrated into a fully automated lab-on-a-chip for cell culture analysis. Reproduced from Ertl et al. (2014) by permission of by Elsevier.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Funding of the Vienna Science and Technology Fund (WWTF) (LS13-092) is greatly acknowledged.
References
Abassi, Y. A., Jackson, J. A., Zhu, J. J., O’Connell, J., Wang, X., and Xu, X. (2004). Label-free, real-time monitoring of IgE-mediated mast cell activation on microelectronic cell sensor arrays. J. Immunol. Methods 292, 195–205. doi:10.1016/j.jim.2004.06.022
Agency, European Medicine. (2007). Guideline on Human Cell-Based Medicinal Products. Draft. Committee for Human Medicinal Product (CHMP). London: Agency, European Medicine. Available at: http://scholar.google.com/scholar?hl=en&btnG=Search&q=intitle:GUIDELINE+ON+HUMAN+CELLB ASED+MEDICINAL+PRODUCTS#0
Alici, E., and Blomberg, P. (2010). GMP facilities for manufacturing of advanced therapy medicinal products for clinical trials: an overview for clinical researchers. Curr. Gene Ther. 10, 508–515. doi:10.2174/156652310793797757
Andersson, H., and van den Berg, A. (2003). Microfluidic devices for cellomics: a review. Sens. Actuators B Chem. 92, 315–325. doi:10.1016/S0925-4005(03)00266-1
Antunes, M., and Pottering, H. (2007). Regulation (EC) No 1394/2007 of the European parliament and of the council of 13 November 2007 on advanced therapy medicinal products and amending directive 2001/83/EC and regulation (EC) No 726/2004. J. Eur. Union 324, 121–137.
Attia, U. M., Marson, S., and Alcock, J. R. (2009). Micro-injection moulding of polymer microfluidic devices. Microfluid. Nanofluidics 7, 1–28. doi:10.1007/s10404-009-0421-x
Auroux, P.-A. A., Iossifidis, D., Reyes, D. R., and Manz, A. (2002). Micro total analysis systems. 2. Analytical standard operations and applications. Anal. Chem. 74, 2637–2652. doi:10.1021/ac020239t
Bang, H., Chung, C., Kim, J., and Kim, S. (2006). Microfabricated fluorescence-activated cell sorter through hydrodynamic flow manipulation. Microsystem 12, 746–753. doi:10.1007/s00542-005-0076-9
Becker, H., and Locascio, L. E. (2002). Polymer microfluidic devices. Talanta 56, 267–287. doi:10.1016/S0039-9140(01)00594-X
Berthier, E., Young, E. W. K., and Beebe, D. (2012). Engineers are from PDMS-land, biologists are from polystyrenia. Lab. Chip 12, 1224–1237. doi:10.1039/C2lc20982a
Cai, Y., Shinar, R., Zhou, Z., and Shinar, J. (2008). Multianalyte sensor array based on an organic light emitting diode platform. Sens. Actuators B Chem. 134, 727–735. doi:10.1016/j.snb.2008.06.019
Cai, Y., Smith, A., Shinar, J., and Shinar, R. (2010). Data analysis and aging in phosphorescent oxygen-based sensors. Sens. Actuators B Chem. 146, 14–22. doi:10.1016/j.snb.2010.02.028
Carlborg, C. F., Haraldsson, T., Oberg, K., Malkoch, M., van der Wijngaart, W., and Öberg, K. (2011). Beyond PDMS: off-stoichiometry thiol-ene (OSTE) based soft lithography for rapid prototyping of microfluidic devices. Lab. Chip 11, 3136–3147. doi:10.1039/c1lc20388f
Cesaro-Tadic, S., Dernick, G., and Juncker, D. (2004). High-sensitivity miniaturized immunoassays for tumor necrosis factor a using microfluidic systems. Lab. Chip 4, 563–569. doi:10.1039/b408964b
Charwat, V., Joksch, M., Sticker, D., Purtscher, M., Rothbauer, M., and Ertl, P. (2014). Monitoring cellular stress responses using integrated high-frequency impedance spectroscopy and time-resolved ELISA. Analyst 139, 5271–5282. doi:10.1039/c4an00824c
Charwat, V., and Muellner, P. (2011). Monitoring light scattering characteristics of adherent cell cultures using a lab-on-a-chip. Photonics 2011, 1–2. doi:10.1109/ICO-IP.2011.5953790
Charwat, V., Purtscher, M., Tedde, S. F., Hayden, O., and Ertl, P. (2013a). Standardization of microfluidic cell cultures using integrated organic photodiodes and electrode arrays. Lab. Chip 13, 785–797. doi:10.1039/c2lc40965h
Charwat, V., Rothbauer, M., Tedde, S. F., Hayden, O., Bosch, J. J., Muellner, P., et al. (2013b). Monitoring dynamic interactions of tumor cells with tissue and immune cells in a lab-on-a-chip. Anal. Chem. 85, 11471–11478. doi:10.1021/ac4033406
Chen, C., Cho, S., Chiang, H., and Tsai, F. (2011). Specific sorting of single bacterial cells with microfabricated fluorescence-activated cell sorting and tyramide signal amplification fluorescence in situ hybridization. Anal. Chem. 83, 7269–7275. doi:10.1021/ac2013465
Chen, C., Cho, S., Tsai, F., Erten, A., and Lo, Y. (2009). Microfluidic cell sorter with integrated piezoelectric actuator. Biomed. Microdevices 11, 1223–1231. doi:10.1007/s10544-009-9341-5
Cheon, J., and Lee, J. H. (2008). Synergistically integrated nanoparticles as multimodal probes for nanobiotechnology. Acc. Chem. Res. 41, 1630–1640. doi:10.1021/ar800045c
Chin, C., Laksanasopin, T., and Cheung, Y. (2011). Microfluidics-based diagnostics of infectious diseases in the developing world. Nat. Med. 17, 1015–1019. doi:10.1038/nm.2408
Cho, S., Chen, C., Tsai, F., Godin, J., and Lo, Y. (2010). Human mammalian cell sorting using a highly integrated micro-fabricated fluorescence-activated cell sorter (μFACS). Lab. Chip 10, 1567–1573. doi:10.1039/c000136h
Cho, S., Gorjup, E. E. E., and Thielecke, H. H. (2009). Chip-based time-continuous monitoring of toxic effects on stem cell differentiation. Ann. Anat. 191, 145–152. doi:10.1016/j.aanat.2008.08.005
Choudhury, B., Shinar, R., and Shinar, J. (2004). Glucose biosensors based on organic light-emitting devices structurally integrated with a luminescent sensing element. J. Appl. Phys. 96, 2949. doi:10.1063/1.1778477
Coltro, W. K. T., de Jesus, D. P., da Silva, J. A. F., do Lago, C. L., and Carrilho, E. (2010). Toner and paper-based fabrication techniques for microfluidic applications. Electrophoresis 31, 2487–2498. doi:10.1002/elps.201000063
Couzin-Frankel, J. (2013). Cancer immunotherapy. Science 342, 1432–1433. doi:10.1126/science.342.6165.1432
Cox, K. L., Devanarayan, V., Kriauciunas, A., Manetta, J., Montrose, C., and Sittampalam, S. “Immunoassay methods,” in Assay Guidance Manual [Internet], eds G. S. Sittampalam, N. P. Coussens, H. Nelson, et al. (Bethesda, MD: Eli Lilly & Company and the National Center for Advancing Translational Sciences; 2004 [updated 2014 December 24]). Available at: http://www.ncbi.nlm.nih.gov/books/NBK92434/
Criscitiello, C. (2012). Tumor-associated antigens in breast cancer. Breast Care (Basel) 7, 262–266. doi:10.1159/000342164
Cvetković, B. Z., and Dittrich, P. S. (2013). A microfluidic device for open loop stripping of volatile organic compounds. Anal. Bioanal. Chem. 405, 2417–2423. doi:10.1007/s00216-012-6604-4
Dixon, J. R. (1999). The international conference on harmonization good clinical practice guideline. Qual. Assur. 6, 65–74. doi:10.1080/105294199277860
Dragone, V., Sans, V., Rosnes, M. H., Kitson, P. J., and Cronin, L. (2013). 3D-printed devices for continuous-flow organic chemistry. Beilstein J. Org. Chem. 9, 951–959. doi:10.3762/bjoc.9.109
Duffy, D. C., McDonald, J. C., Schueller, J. A., and Whitesides, G. M. (1998). Rapid prototyping of microfluidic system in poly(dimethylsiloxane). Anal. Chem. 70, 4974–4984. doi:10.1021/ac980656z
El-Ali, J. J., Sorger, P. K., and Jensen, K. F. (2006). Cells on chips. Nature 442, 403–411. doi:10.1038/nature05063
Engvall, E., and Perlmann, P. (1971). Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G. Immunochemistry 8, 871–874. doi:10.1016/0019-2791(71)90454-X
Ertl, P., Sticker, D., Charwat, V., Kasper, C., and Lepperdinger, G. (2014). Lab-on-a-chip technologies for stem cell analysis. Trends Biotechnol. 32, 245–253. doi:10.1016/j.tibtech.2014.03.004
Eyer, K., Stratz, S., Kuhn, P., Küster, S. K., and Dittrich, P. S. (2013). Implementing enzyme-linked immunosorbent assays on a microfluidic chip to quantify intracellular molecules in single cells. Anal. Chem. 85, 3280–3287. doi:10.1021/ac303628j
Feng, Y., Cheng, J., Zhou, L., Zhou, X., and Xiang, H. (2012). Ratiometric optical oxygen sensing: a review in respect of material design. Analyst 137, 4885–4901. doi:10.1039/c2an35907c
Finn, O. J. (2003). Cancer vaccines: between the idea and the reality. Nat. Rev. Immunol. 3, 630–641. doi:10.1038/nri1150
Fiorini, G. S., and Chiu, D. T. (2005). Disposable microfluidic devices: fabrication, function, and application. BioTechniques 38, 429–446. doi:10.2144/05383RV02
Fiorini, G. S., Jeffries, G. D., Lim, D. S., Kuyper, C. L., and Chiu, D. T. (2003). Fabrication of thermoset polyester microfluidic devices and embossing masters using rapid prototyped polydimethylsiloxane molds. Lab. Chip 3, 158–163. doi:10.1039/b305074m
Fiorini, G. S., Lorenz, R. M., Kuo, J. S., and Chiu, D. T. (2004). Rapid prototyping of thermoset polyester microfluidic devices. Anal. Chem. 76, 4697–4704. doi:10.1021/Ac0498922
Fossceco, S. L., Company, K. P., and Curtis, N. A. (1996). Exploring enzyme-linked immunosorbent assay (ELISA) data with the SAS® analyst application introduction. J. Pediatr. 146, 62–65.
Frey, O., Misun, P. M., Fluri, D. A., Hengstler, J. G., and Hierlemann, A. (2014). Reconfigurable microfluidic hanging drop network for multi-tissue interaction and analysis. Nat. Commun. 5, 4250. doi:10.1038/ncomms5250
Fu, A. Y.-C. (2002). Microfabricated Fluorescence-Activated Cell Sorters (μFACS) for Screening Bacterial Cells. Pasadena: Etd.Caltech.Edu. California Institute of Technology. Available at: http://thesis.library.caltech.edu/1760/
Fu, L., Yang, R., Lin, C.-H., Pan, Y., and Lee, G. (2004). Electrokinetically driven micro flow cytometers with integrated fiber optics for on-line cell/particle detection. Anal. Chim. Acta 507, 163–169. doi:10.1016/j.aca.2003.10.028
Gallucci, S., Lolkema, M., and Matzinger, P. (1999). Natural adjuvants: endogenous activators of dendritic cells. Nat. Med. 5, 1249–1255. doi:10.1038/15200
Gao, J., Yin, X.-F. F., and Fang, Z.-L. L. (2004). Integration of single cell injection, cell lysis, separation and detection of intracellular constituents on a microfluidic chip. Lab. Chip 4, 47–52. doi:10.1039/B310552k
García, M., Orozco, J., and Guix, M. (2013). Micromotor-based lab-on-chip immunoassays. Nanoscale 5, 1325–1331. doi:10.1039/c2nr32400h
Gervais, L., and Delamarche, E. (2009). Toward one-step point-of-care immunodiagnostics using capillary-driven microfluidics and PDMS substrates. Lab. Chip 9, 3330–3337. doi:10.1039/b906523g
Gijs, M. A. M., Lacharme, F., and Lehmann, U. (2010). Microfluidic applications of magnetic particles for biological analysis and catalysis. Chem. Rev. 110, 1518–1563. doi:10.1021/Cr9001929
Golden, A. P., and Tien, J. (2007). Fabrication of microfluidic hydrogels using molded gelatin as a sacrificial element. Lab. Chip 7, 720–725. doi:10.1039/b618409j
Grover, W. H., Skelley, A. M., Liu, C. N., Lagally, E. T., and Mathies, R. A. (2003). Monolithic membrane valves and diaphragm pumps for practical large-scale integration into glass microfluidic devices. Sens. Actuators B Chem. 89, 315–323. doi:10.1016/S0925-4005(02)00468-9
Harrison, D. J., Fluri, K., Seiler, K., Fan, Z., Effenhauser, C. S., and Manz, A. (1993). Micromachining a miniaturized capillary electrophoresis-based chemical analysis system on a chip. Science 261, 895–897. doi:10.1126/science.261.5123.895
Harrison, D. J., Manz, A., Fan, Z. H., Ludi, H., and Widmer, H. M. (1992). Capillary electrophoresis and sample injection systems integrated on a planar glass chip. Anal. Chem. 64, 1926–1932. doi:10.1021/Ac00041a030
Hata, K., Kichise, Y., Kaneta, T., and Imasaka, T. (2003). Hadamard transform microchip electrophoresis combined with diode laser fluorometry. Anal. Chem. 75, 1765–1768. doi:10.1021/Ac026330e
Hildebrandt, C. C., Büth, H. H., Cho, S. S., Impidjati, and Thielecke, H. H. (2010). Detection of the osteogenic differentiation of mesenchymal stem cells in 2D and 3D cultures by electrochemical impedance spectroscopy. J. Biotechnol. 148, 83–90. doi:10.1016/j.jbiotec.2010.01.007
Hinz, T., Buchholz, C. J., van der Stappen, T., Cichutek, K., and Kalinke, U. (2006). Manufacturing and quality control of cell-based tumor vaccines: a scientific and a regulatory perspective. J. Immunother. 29, 472–476. doi:10.1097/01.cji.0000211305.98244.56
Hofmann, O., Miller, P., Sullivan, P., Jones, T. S., DeMello, J. C., Bradley, D. D. C., et al. (2005). Thin-film organic photodiodes as integrated detectors for microscale chemiluminescence assays. Sens. Actuators B Chem. 106, 878–884. doi:10.1016/j.snb.2004.10.005
Honda, N., and Lindberg, U. (2005). Simultaneous multiple immunoassays in a compact discshaped microfluidic device based on centrifugal force. Clin. Chem. 1961, 1955–1961. doi:10.1373/clinchem.2005.053348
Huang, L.-B., Yung, K.-L., Xu, Y., and Xie, Y.-C. (2010). Injection Molded Plastic Microfluidic Biochips with Integrated Pumping Electrode. Xiamen: IEEE.
Hung, C. F., Wu, T. C., Monie, A., and Roden, R. (2008). Antigen-specific immunotherapy of cervical and ovarian cancer. Immunol. Rev. 222, 43–69. doi:10.1111/j.1600-065X.2008.00622.x
ICH. (2015). International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH) – Quality Guidelines.
Ionescu-Zanetti, C., Shaw, R. M., Seo, J. G., Jan, Y. N., Jan, L. Y., and Lee, L. P. (2005). Mammalian electrophysiology on a microfluidic platform. Proc. Natl. Acad. Sci. U.S.A. 102, 9112–9117. doi:10.1073/pnas.0503418102
Jindal, R., and Cramer, S. M. (2004). On-chip electrochromatography using sol-gel immobilized stationary phase with UV absorbance detection. J. Chromatogr. A 1044, 277–285. doi:10.1016/j.chroma.2004.05.065
Jun, Y. W., Seo, J. W., and Cheon, J. (2008). Nanoscaling laws of magnetic nanoparticles and their applicabilities in biomedical sciences. Acc. Chem. Res. 41, 179–189. doi:10.1021/ar700121f
Junkin, M., and Tay, S. (2014). Microfluidic single-cell analysis for systems immunology. Lab. Chip 14, 1246–1260. doi:10.1039/c3lc51182k
Kalantari, T., Kamali-Sarvestani, E., Ciric, B., Karimi, M. H., Kalantari, M., Faridar, A., et al. (2011). Generation of immunogenic and tolerogenic clinical-grade dendritic cells. Immunol. Res. 51, 153–160. doi:10.1007/s12026-011-8255-5
Kalinski, P., Muthuswamy, R., and Urban, J. (2013). Dendritic cells in cancer immunotherapy: vaccines and combination immunotherapies. Expert Rev. Vaccines 12, 285–295. doi:10.1586/erv.13.22
Kantoff, P., and Higano, C. (2010). Sipuleucel-T immunotherapy for castration-resistant prostate cancer philip. N. Engl. J. Med. 363, 411–422. doi:10.1056/NEJMoa1001294
Kim, J., Jensen, E. C., Megens, M., Boser, B., and Mathies, R. A. (2011). Integrated microfluidic bioprocessor for solid phase capture immunoassays. Lab. Chip 11, 3106–3112. doi:10.1039/c1lc20407f
Kim, J., Kang, M., Jensen, E., and Mathies, R. (2012). Lifting gate polydimethylsiloxane microvalves and pumps for microfluidic control. Anal. Chem. 84, 2067–2071. doi:10.1021/ac202934x.Lifting
Kim, P., Kwon, K. W., Park, M. C., Lee, S. H., Kim, S. M., and Suh, K. Y. (2008a). Soft lithography for microfluidics: a review. Korean BioChip Soc. 2, 1–11.
Kim, S., Lee, S., and Suh, K. (2008b). Cell research with physically modified microfluidic channels: a review. Lab. Chip 8, 1015–1023. doi:10.1039/b800835c
Kim, S., and Paczesny, S. (2013). Preprogrammed, parallel on-chip immunoassay using system-level capillarity control. Anal. Chem. 85, 6902–6907. doi:10.1021/ac401292d
Kodaira, Y., Nair, S. K., Wrenshall, L. E., Gilboa, E., and Platt, J. L. (2000). Phenotypic and functional maturation of dendritic cells mediated by heparan sulfate. J. Immunol. 165, 1599–1604. doi:10.4049/jimmunol.165.3.1599
Kramer, N., Walzl, A., Unger, C., Rosner, M., Krupitza, G., Hengstschläger, M., et al. (2013). In vitro cell migration and invasion assays. Mutat. Res. 752, 10–24. doi:10.1016/j.mrrev.2012.08.001
Kuswandi, B., Nuriman, Huskens, J., and Verboom, W. (2007). Optical sensing systems for microfluidic devices: a review. Anal. Chim. Acta 601, 141–155. doi:10.1016/j.aca.2007.08.046
Lam, R. H. W., Kim, M.-C., and Thorsen, T. (2009). Culturing aerobic and anaerobic bacteria and mammalian cells with a microfluidic differential oxygenator. Anal. Chem. 81, 5918–5924. doi:10.1021/ac9006864
Lapizco-Encinas, B. (2004). Dielectrophoretic concentration and separation of live and dead bacteria in an array of insulators. Anal. Chem. 76, 1571–1579. doi:10.1021/ac034804j
Larsen, M., Borisov, S., and Grunwald, B. (2011). A simple and inexpensive high resolution color ratiometric planar optode imaging approach: application to oxygen and pH sensing. Limnol. Oceanogr. Methods 2, 348–360. doi:10.4319/lom.2011.9.348
Laurent, S., Forge, D., Port, M., Roch, A., Robic, C., Elst, L. V., et al. (2008). Magnetic iron oxide nanoparticles: synthesis, stabilization, vectorization, physicochemical characterizations, and biological applications. Chem. Rev. 108, 2064–2110. doi:10.1021/Cr068445e
Lee, B. S., Lee, J.-N., Park, J.-M., Lee, J.-G., Kim, S., Cho, Y.-K., et al. (2009). A fully automated immunoassay from whole blood on a disc. Lab. Chip 9, 1548–1555. doi:10.1039/b820321k
Lee, S., and Wen, X. (2005). A dual gravity study of the (2+1) D compact U (1) gauge theory coupled with strongly interacting matter fields. Nucl. Phys. B 730, 164–178. doi:10.1016/j.nuclphysb.2005.09.033
Lequin, R. (2005). Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). Clin. Chem. 2418, 2415–2418. doi:10.1373/clinchem.2005.051532
Lewis, J. S., Dolgova, N., Chancellor, T. J., Acharya, A. P., Karpiak, J. V., Lele, T. P., et al. (2013). Dendritic cell activation is influenced by cyclic mechanical strain when cultured on adhesive substrates. Biomaterials 34, 9063–9070. doi:10.1016/j.biomaterials.2013.08.021
Ligler, F. S., and Kim, J. S. (2010). The Microflow Cytometer. Singapore: Pan Stanford Publishing Pte. Ltd. Available at: http://books.google.com/books?hl=en&lr=&id=Q9KX1LiLnhcC&oi=fnd&pg=PR5&dq=the+Microflow+Cytometer&ots=gT0JzUfZAr&sig=SUXsbA_NAuHONbs_qL7CnaQyAng
Lin, P., and Yan, F. (2012). Organic thin-film transistors for chemical and biological sensing. Adv. Mater. Weinheim 24, 34–51. doi:10.1002/adma.201103334
Liu, R., Cai, Y. K., Park, J.-M. M., Ho, K.-M. M., Shinar, J., and Shinar, R. (2011). Organic light-emitting diode sensing platform: challenges and solutions. Adv. Funct. Mater. 21, 4744–4753. doi:10.1002/adfm.201101536
Locascio, L. E., Ross, D. J., Howell, P. B., and Gaitan, M. (2006). Fabrication of polymer microfluidic systems by hot embossing and laser ablation. Methods Mol. Biol. 339, 37–46. doi:10.1385/1-59745-076-6:37
Ma, C., Fan, R., Ahmad, H., Shi, Q., Comin-Anduix, B., Chodon, T., et al. (2011). A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat. Med. 17, 738–743. doi:10.1038/nm.2375
Mair, D. A., Geiger, E., Pisano, A. P., Frechet, J. M. J., and Svec, F. (2006). Injection molded microfluidic chips featuring integrated interconnects. Lab. Chip 6, 1346–1354. doi:10.1039/B605911b
Manz, A., Harrison, D. J., Verpoorte, E. M. J., Fettinger, J. C., Paulus, A., Ludi, H., et al. (1992). Planar chips technology for miniaturization and integration of separation techniques into monitoring systems – capillary electrophoresis on a chip. J. Chromatogr. 593, 253–258. doi:10.1016/0021-9673(92)80293-4
Martínez-Morales, P. L., Revilla, A., Ocaña, I., González, C., Sainz, P., McGuire, D., et al. (2013). Progress in stem cell therapy for major human neurological disorders. Stem Cell. Rev. 9, 685–699. doi:10.1007/s12015-013-9443-6
McClain, M., Culbertson, C., Jacobson, S., and Ramsey, J. (2001). Flow cytometry of Escherichia coli on mirofluidic devices. Anal. Chem. 73, 5334–5338. doi:10.1021/ac010504v
McDonald, J. C., and Whitesides, G. M. (2002). Poly(dimethylsiloxane) as a material for fabricating microfluidic devices. Acc. Chem. Res. 35, 491–499. doi:10.1021/Ar010110q
Meier, R., and Fischer, L. (2013). Referenced luminescent sensing and imaging with digital color cameras: a comparative study. Sens. Actuators B Chem. 177, 500–506. doi:10.1016/j.snb.2012.11.041
Michelini, E. E., Cevenini, L. L., Mezzanotte, L. L., Coppa, A. A., and Roda, A. A. (2010). Cell-based assays: fuelling drug discovery. Anal. Bioanal. Chem. 398, 227–238. doi:10.1007/s00216-010-3933-z
Miller, E., and Ng, A. (2011). A digital microfluidic approach to heterogeneous immunoassays. Anal. Bioanal. Chem. 339, 337–345. doi:10.1007/s00216-010-4368-2
Morier, P., Vollet, C., and Michel, P. (2004). Gravityinduced convective flow in microfluidic systems: electrochemical characterization and application to enzymelinked immunosorbent assay tests. Electrophoresis 25, 3761–3768. doi:10.1002/elps.200406093
Neagu, C. (1996). An electrochemical microactuator: principle and first results. J. Microelectromech. Syst. 5, 2–9. doi:10.1109/84.485209
Neagu, C., Gardeniers, J., Elwenspoek, M., and Kelly, J. (1997). An electrochemical active valve. Electrochim. Acta 42, 3367–3373. doi:10.1016/S0013-4686(97)00189-8
Nock, V., and Blaikie, R. J. (2010). Spatially resolved measurement of dissolved oxygen in multistream microfluidic devices. IEEE Sens. J. 10, 1813–1819. doi:10.1109/JSEN.2010.2049016
Nock, V., Blaikie, R. J., and David, T. (2008). Patterning, integration and characterisation of polymer optical oxygen sensors for microfluidic devices. Lab. Chip 8, 1300–1307. doi:10.1039/B801879k
Novak, R. (2013). Microfluidic Technologies for Quantitative Single Cell Analysis. PhD thesis, University of California, Berkeley.
Novak, R., Ranu, N., and Mathies, R. A. (2013). Rapid fabrication of nickel molds for prototyping embossed plastic microfluidic devices. Lab. Chip 13, 1468–1471. doi:10.1039/c3lc41362d
Novak, R., Wartmann, D., Mathies, R. A., Dostálek, J., and Ertl, P. (2015). “Microfluidic platform for multiplexed cell sampling and time-resolved SPR-Based cytokine sensing,” in Proceedings of the 6th European Conference of the International Federation for Medical and Biological Engineering, eds I. Lacković and D. Vasic (Switzerland: Springer International Publishing), 45, 785–788. doi:10.1007/978-3-319-11128-5_195
Oelke, M. M., Maus, M. M. V., Didiano, D. D., June, C. H., Mackensen, A., Schneck, J. J. P., et al. (2003). Ex vivo induction and expansion of antigen-specific cytotoxic T cells by HLA-IGA-coated artificial antigen-presenting cells. Nat. Med. 9, 619–625. doi:10.1038/nm869
Pamme, N., and Wilhelm, C. (2006). Continuous sorting of magnetic cells via on-chip free-flow magnetophoresis. Lab. Chip 6, 974–980. doi:10.1039/b604542a
Pankhurst, Q. A., Connolly, J., Jones, S. K., and Dobson, J. (2003). Applications of magnetic nanoparticles in biomedicine. J. Phys. D Appl. Phys. 36, R167–R181. doi:10.1088/0022-3727/36/13/201
Perroud, T. D., Kaiser, J. N., Sy, J. C., Lane, T. W., Branda, C. S., Singh, A. K., et al. (2008). Microfluidic-based cell sorting of Francisella tularensis infected macrophages using optical forces microfluidic-based cell sorting of Francisella tularensis infected macrophages using optical forces. Anal. Chem. 80, 6365–6372. doi:10.1021/ac8007779
Picher, M. M. M., Küpcü, S. S., Huang, C.-J. C.-J., Dostalek, J. J., Pum, D. D., Sleytr, U. U. B., et al. (2013). Nanobiotechnology advanced antifouling surfaces for the continuous electrochemical monitoring of glucose in whole blood using a lab-on-a-chip. Lab. Chip 13, 1780–1789. doi:10.1039/c3lc41308j
Reichen, M., Veraitch, F. S., and Szita, N. (2013). Development of a multiplexed microfluidic platform for the automated cultivation of embryonic stem cells. J. Lab. Autom. 18, 519–529. doi:10.1177/2211068213499917
Ren, K. N., Zhou, J. H., and Wu, H. K. (2013). Materials for microfluidic chip fabrication. Acc. Chem. Res. 46, 2396–2406. doi:10.1021/Ar300314s
Reyes, D. R., Iossifidis, D. D., Auroux, P.-A. P.-A. A., and Manz, A. A. (2002). Micro total analysis systems. 1. Introduction, theory, and technology. Anal. Chem. 74, 2623–2636. doi:10.1021/ac0202435
Roy, E., Galas, J.-C., and Veres, T. (2011). Thermoplastic elastomers for microfluidics: towards a high-throughput fabrication method of multilayered microfluidic devices. Lab. Chip 11, 3193–3196. doi:10.1039/C1LC20251K
Roy, E., Stewart, G., Mounier, M., Malic, L., Peytavi, R., Clime, L., et al. (2015). From cellular lysis to microarray detection{,} an integrated thermoplastic elastomer (TPE) point of care lab on a disc. Lab. Chip 15, 406–416. doi:10.1039/C4LC00947A
Ryu, G., Huang, J., Hofmann, O., Walshe, C. A., Sze, J. Y. Y., McClean, G. D., et al. (2011). Highly sensitive fluorescence detection system for microfluidic lab-on-a-chip. Lab. Chip 11, 1664–1670. doi:10.1039/c0lc00586j
Sackmann, E. K., Fulton, A. A. L., and Beebe, D. J. D. (2014). The present and future role of microfluidics in biomedical research. Nature 507, 181–189. doi:10.1038/nature13118
Sagmeister, M., Tschepp, A., and Kraker, E. (2013). Enabling luminescence decay time-based sensing using integrated organic photodiodes. Anal. Bioanal. Chem. 405, 5975–5982. doi:10.1007/s00216-013-6998-7
Sasaki, H., Onoe, H., Osaki, T., Kawano, R., and Takeuchi, S. (2010). Parylene-coating in PDMS microfluidic channels prevents the absorption of fluorescent dyes. Sens. Actuators B Chem. 150, 478–482. doi:10.1016/j.snb.2010.07.021
Schafer, I., and Jamieson, A. (1979). Multiangle light scattering flow photometry of cultured human fibroblasts: comparison of normal cells with a mutant line containing cytoplasmic inclusions. J. Histochem. Cytochem. 58, 345–358. doi:10.1369/jhc.2009.954826
Schapper, D., Alam, M. N. H. Z., Szita, N., Eliasson Lantz, A., Gernaey, K. V., Schaepper, D., et al. (2009). Application of microbioreactors in fermentation process development: a review. Anal. Bioanal. Chem. 395, 679–695. doi:10.1007/s00216-009-2955-x
Schrott, W., Svoboda, M., Slouka, Z., and Šnita, D. (2009). Metal electrodes in plastic microfluidic systems. Microelectron. Eng. 86, 1340–1342. doi:10.1016/j.mee.2009.01.001
Schuster, S. J., Neelapu, S. S., Gause, B. L., Janik, J. E., Muggia, F. M., Gockerman, J. P., et al. (2011). Vaccination with patient-specific tumor-derived antigen in first remission improves disease-free survival in follicular lymphoma. J. Clin. Oncol. 29, 2787–2794. doi:10.1200/JCO.2010.33.3005
Sekitani, T., Yokota, T., and Zschieschang, U. (2009). Organic nonvolatile memory transistors for flexible sensor arrays. Science 326, 1516–1519. doi:10.1126/science.1179963
Sharma, S., Raju, R., Sui, S., and Hu, W.-S. (2011). Stem cell culture engineering – process scale up and beyond. Biotechnol. J. 6, 1317–1329. doi:10.1002/biot.201000435
Shinar, J., and Shinar, R. (2008). Organic light-emitting devices (OLEDs) and OLED-based chemical and biological sensors: an overview. J. Phys. D Appl. Phys. 41, 133001. doi:10.1088/0022-3727/41/13/133001
Shoshi, A., Schotter, J., Schroeder, P., Milnera, M., Ertl, P., Charwat, V., et al. (2012). Magnetoresistive-based real-time cell phagocytosis monitoring. Biosens. Bioelectron. 36, 116–122. doi:10.1016/j.bios.2012.04.002
Sin, A., Chin, K. C., Jamil, M. F., Kostov, Y., Rao, G., and Shuler, M. L. (2004). The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnol. Prog. 20, 338–345. doi:10.1021/bp034077d
Sista, R., Hua, Z., Thwar, P., Sudarsan, A., Srinivasan, V., Eckhardt, A., et al. (2008). Development of a digital microfluidic platform for point of care testing. Lab. Chip 8, 2091–2104. doi:10.1039/b814922d
Skierski, J. (2012). Cytometria wczoraj i dziś podróż przez dziesięciolecia. Cytometria Polska 8, 6–11.
Sollier, E., Murray, C., Maoddi, P., and Di Carlo, D. (2011). Rapid prototyping polymers for microfluidic devices and high pressure injections. Lab. Chip 11, 3752–3765. doi:10.1039/C1lc20514e
Someya, T., Dodabalapur, A., Huang, J., See, K. C., and Katz, H. E. (2010). Chemical and physical sensing by organic field-effect transistors and related devices. Adv. Mater. 22, 3799–3811. doi:10.1002/adma.200902760
Song, P., Hu, R., Tng, D., and Yong, K. (2014). Moving towards individualized medicine with microfluidics technology. RSC Adv. 4, 11499–11511. doi:10.1039/c3ra45629c
Stevens, D., Petri, C., and Osborn, J. (2008). Enabling a microfluidic immunoassay for the developing world by integration of on-card dry reagent storage. Lab. Chip 8, 2038–2045. doi:10.1039/b811158h
Stich, M. I. J., Borisov, S. M., Henne, U., and Schäferling, M. (2009). Read-out of multiple optical chemical sensors by means of digital color cameras. Sens. Actuators B Chem. 139, 204–207. doi:10.1016/j.snb.2008.12.036
Sud, D., Mehta, G., Mehta, K., Linderman, J., Takayama, S., and Mycek, M.-A. A. (2006). Optical imaging in microfluidic bioreactors enables oxygen monitoring for continuous cell culture. J. Biomed. Opt. 11, 50504. doi:10.1117/1.2355665
Sun, S., Yang, M., Kostov, Y., and Rasooly, A. (2010). ELISA-LOC: lab-on-a-chip for enzyme-linked immunodetection. Lab. Chip 10, 2093–2100. doi:10.1039/c003994b
Sun, T., and Morgan, H. (2010). Single-cell microfluidic impedance cytometry: a review. Microfluid. Nanofluidics 8, 423–443. doi:10.1007/s10404-010-0580-9
Tedde, S. F., Kern, J., Sterzl, T., Fürst, J., Lugli, P., Hayden, O., et al. (2009). Fully spray coated organic photodiodes. Nano Lett. 9, 980–983. doi:10.1021/nl803386y
Thomas, P. C., Raghavan, S. R., and Forry, S. P. (2011). Regulating oxygen levels in a microfluidic device. Anal. Chem. 83, 8821–8824. doi:10.1021/ac202300g
Toma, K., Dostalek, J., and Knoll, W. (2011). Long range surface plasmon-coupled fluorescence emission for biosensor applications. Opt. Express 19, 97–106. doi:10.1364/OE.19.011090
Torsi, L., Lovinger, A. J., Crone, B., Someya, T., Dodabalapur, A., Katz, H. E., et al. (2002). Correlation between oligothiophene thin film transistor morphology and vapor responses. J. Phys. Chem. B 106, 12563–12568. doi:10.1021/Jp021473q
Trickett, A. A., and Kwan, Y. L. Y. (2003). T cell stimulation and expansion using anti-CD3/CD28 beads. J. Immunol. Methods 275, 251–255. doi:10.1016/S0022-1759(03)00010-3
Unger, M. A., Chou, H. P., Thorsen, T., Scherer, A., and Quake, S. R. (2000). Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 288, 113–116. doi:10.1126/science.288.5463.113
Ungerböck, B., Charwat, V., Ertl, P., and Mayr, T. (2013). Microfluidic oxygen imaging using integrated optical sensor layers and a color camera. Lab. Chip 13, 1593–1601. doi:10.1039/c3lc41315b
van Kan, J. A., Zhang, C., Perumal Malar, P., and van der Maarel, J. R. C. (2012). High throughput fabrication of disposable nanofluidic lab-on-chip devices for single molecule studies. Biomicrofluidics 6, 36502. doi:10.1063/1.4740231
Van Weemen, B. K., and Schuurs, A. H. (1971). Immunoassay using antigen-enzyme conjugates. FEBS Lett. 15, 232–236. doi:10.1016/0014-5793(71)80319-8
Vilkner, T., Janasek, D., and Manz, A. (2004). Micro total analysis systems. Recent developments. Anal. Chem. 76, 3373–3385. doi:10.1021/ac040063q
Vollmer, A., Probstein, R., Gilbert, R., and Thorsen, T. (2005). Development of an integrated microfluidic platform for dynamic oxygen sensing and delivery in a flowing medium. Lab. Chip 5, 1059–1066. doi:10.1039/b508097e
Wagner, I., Materne, E.-M., Brincker, S., Süssbier, U., Frädrich, C., Busek, M., et al. (2013). A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab. Chip 13, 3538–3547. doi:10.1039/c3lc50234a
Walter, S., Weinschenk, T., Stenzl, A., Zdrojowy, R., Pluzanska, A., Szczylik, C., et al. (2012). Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat. Med. 18, 1254–1261. doi:10.1038/nm.2883
Wang, X., and Amatatongchai, M. (2009). Thin-film organic photodiodes for integrated on-chip chemiluminescence detection application to antioxidant capacity screening. Sens. Actuators B Chem. 140, 643–648. doi:10.1016/j.snb.2009.04.068
Wang, X.-D., and Meier, R. (2010). Photographing oxygen distribution. Angew. Chem. Int. Ed. Engl. 49, 4907–4909. doi:10.1002/anie.201001305
Wegener, J., Zink, S., Rösen, P., and Galla, H. H. (1999). Use of electrochemical impedance measurements to monitor beta-adrenergic stimulation of bovine aortic endothelial cells. Pflugers Arch. 437, 925–934. doi:10.1007/s004240050864
Wegener, J. J., Keese, C. R., and Giaever, I. I. (2000). Electric cell-substrate impedance sensing (ECIS) as a noninvasive means to monitor the kinetics of cell spreading to artificial surfaces. Exp. Cell Res. 259, 158–166. doi:10.1006/excr.2000.4919
Whitesides, G. M. (2003). The ‘right’ size in nanobiotechnology. Nat. Biotechnol. 21, 1161–1165. doi:10.1038/nbt872
Whitesides, G. M. (2006). The origins and the future of microfluidics. Nature 442, 368–373. doi:10.1038/nature05058
Williams, G., Backhouse, C., and Aziz, H. (2014). Integration of organic light emitting diodes and organic photodetectors for lab-on-a-chip bio-detection systems. Electronics 3, 43–75. doi:10.3390/electronics3010043
Wilson, J. D., Bigelow, C. E., Calkins, D. J., and Foster, T. H. (2005). Light scattering from intact cells reports oxidative-stress-induced mitochondrial swelling. Biophys. J. 88, 2929–2938. doi:10.1529/biophysj.104.054528
Wu, J., and Gu, M. (2011). Microfluidic sensing: state of the art fabrication and detection techniques. J. Biomed. Opt. 16, 080901. doi:10.1117/1.3607430
Xia, Y. N., and Whitesides, G. M. (1998). Soft lithography. Annu. Rev. Mater. Sci. 28, 153–184. doi:10.1146/annurev.matsci.28.1.153
Xiao, C. C., and Luong, J. H. (2003). On-line monitoring of cell growth and cytotoxicity using electric cell-substrate impedance sensing (ECIS). Biotechnol. Prog. 19, 1000–1005. doi:10.1021/bp025733x
Yamada-Ohnishi, Y., Azuma, H., Urushibara, N., Yamaguchi, M., Fujihara, M., Kobata, T., et al. (2004). Cytotoxic difference of T cells expanded with anti-CD3 monoclonal antibody in the presence and absence of anti-CD 28 monoclonal antibody. Stem Cells Dev. 13, 315–322. doi:10.1089/154732804323099244
Yea, C.-H., An, J. H., Kim, J., and Choi, J.-W. (2013). In situ electrochemical detection of embryonic stem cell differentiation. J. Biotechnol. 166, 1–5. doi:10.1016/j.jbiotec.2013.04.007
Yeon, J. H., Park, J. K., and Ju, H. Y. (2005). Cytotoxicity test based on electrochemical impedance measurement of HepG2 cultured in microfabricated cell chip. Anal. Biochem. 341, 308–315. doi:10.1016/j.ab.2005.03.047
Yi, C. Q., Li, C. W., Ji, S. L., and Yang, M. S. (2006a). Microfluidics technology for manipulation and analysis of biological cells. Anal. Chim. Acta 560, 1–23. doi:10.1016/j.aca.2005.12.037
Yi, C. Q., Zhang, Q., Li, C. W., Yang, J., Zhao, J. L., and Yang, M. S. (2006b). Optical and electrochemical detection techniques for cell-based microfluidic systems. Anal. Bioanal. Chem. 384, 1259–1268. doi:10.1007/s00216-005-0252-x
Yun, M., Sharma, A., Fuentes-hernandez, C., Hwang, D. K., Dindar, A., Singh, S., et al. (2014). Stable organic field-effect transistors for continuous and nondestructive sensing of chemical and biologically relevant molecules in aqueous environment. ACS Appl. Mater. Interfaces 6, 1616–1622. doi:10.1021/am404460j
Zaretsky, I., Polonsky, M., and Shifrut, E. (2012). Monitoring the dynamics of primary T cell activation and differentiation using long term live cell imaging in microwell arrays. Lab. Chip 12, 5007–5015. doi:10.1039/c2lc40808b
Zhang, M. Y., Wu, J. B., Wang, L. M., Xiao, K., and Wen, W. J. (2010). A simple method for fabricating multi-layer PDMS structures for 3D microfluidic chips. Lab. Chip 10, 1199–1203. doi:10.1039/B923101c
Zhang, W., Lin, S., Wang, C., Hu, J., Li, C., Zhuang, Z., et al. (2009). PMMA/PDMS valves and pumps for disposable microfluidics. Lab. Chip 9, 3088–3094. doi:10.1039/b907254c
Zigler, M., Shir, A., and Levitzki, A. (2013). Targeted cancer immunotherapy. Curr. Opin. Pharmacol. 13, 504–510. doi:10.1016/j.coph.2013.04.003
Keywords: microfluidics, lab-on-a-chip, immunoassay, automation, quality control, cell therapy, FACS, ATMP
Citation: Wartmann D, Rothbauer M, Kuten O, Barresi C, Visus C, Felzmann T and Ertl P (2015) Automated, miniaturized, and integrated quality control-on-chip (QC-on-a-chip) for cell-based cancer therapy applications. Front. Mater. 2:60. doi: 10.3389/fmats.2015.00060
Received: 02 February 2015; Accepted: 24 August 2015;
Published: 16 September 2015
Edited by:
Michael P. M. Jank, Fraunhofer Institute for Integrated Systems and Device Technology, GermanyReviewed by:
Llibertat Abad Muñoz, Institut de Microelectrònica de Barcelona (IMB-CNM-CSIC), SpainEmmanuel Roy, Centre National de la Recherche Scientifique, France
Eduardo Corton, Universidad de Buenos Aires and CONICET, Argentina
Copyright: © 2015 Wartmann, Rothbauer, Kuten, Barresi, Visus, Felzmann and Ertl. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peter Ertl, BioSensor Technologies, AIT Austrian Institute of Technology GmbH, Muthgasse 11, 1190 Vienna, Austria,cGV0ZXIuZXJ0bEBhaXQuYWMuYXQ=
 David Wartmann
David Wartmann Mario Rothbauer
Mario Rothbauer Olga Kuten
Olga Kuten Caterina Barresi
Caterina Barresi Carmen Visus
Carmen Visus Thomas Felzmann
Thomas Felzmann Peter Ertl
Peter Ertl