- Department of gastroenterology, The Third Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang, China
The accurate classification of gastrointestinal diseases from endoscopic images is essential for early detection and treatment. However, current methods face challenges in effectively integrating both global and local features, which limits their ability to capture both broad semantic information and subtle lesion details, ultimately affecting classification performance. To address this issue, this study introduces a novel deep learning framework, the Global and Local Interaction Network (GLI-Net). The GLI-Net consists of four main components: a Global Branch Module (GB) designed to extract global image features, a Local Branch Module (LB) focused on capturing detailed lesion features, an Information Exchange Module (LEM) that facilitates bidirectional information exchange and fusion between the global and local features, and an Adaptive Feature Fusion and Enhancement Module (AFE) aimed at optimizing the fused features. By integrating these modules, GLI-Net effectively captures and combines multi-level feature information, which improves both the accuracy and robustness of endoscopic image classification. Experiments conducted using the Kvasir and Hyper-Kvasir public datasets demonstrate that GLI-Net outperforms existing state-of-the-art models across several metrics, including accuracy, F1 score, precision, and recall. Additionally, ablation studies confirm the contribution of each module to the overall system performance. In summary, GLI-Net’s advanced feature extraction and fusion techniques significantly enhance medical endoscopic image classification, highlighting its potential for use in complex medical image analysis tasks.
1 Introduction
Gastrointestinal cancers are among the most common cancer types globally, affecting not only the United States but also many other countries. In 2023, it is estimated that there will be approximately 153,020 new cases of gastrointestinal cancer and 52,550 related deaths worldwide. Of these, colorectal cancer accounts for about 34.97% of gastrointestinal cancers. It is well-established that certain intestinal conditions, such as polyps and ulcers, play a significant role in the development of colorectal cancer. Early detection of cancer indicators is crucial for managing colorectal cancer, as it can notably improve patient outcomes and survival rates. Therefore, early diagnosis is a critical component in the fight against this cancer, offering hope for better prognoses and higher survival chances.
Endoscopy remains a key method for the initial identification and evaluation of colorectal cancer, demonstrating its effectiveness in reducing mortality rates. This diagnostic tool captures numerous visual frames during gastrointestinal examinations, which are typically reviewed manually. This manual process is not only labor-intensive and repetitive but also subject to human error, as the accuracy of diagnosis depends on the endoscopist’s expertise, experience, and mental acuity. Such variability can result in incorrect diagnoses or missed abnormalities. To address these challenges, there is an urgent need for a precise, advanced computer-assisted diagnostic system. This system would autonomously identify and flag suspicious images, reducing the significant manual workload for endoscopists and improving diagnostic accuracy. This technological innovation is poised to advance the early detection of colorectal cancer, potentially leading to better patient outcomes and increased survival rates.
For instance, Karargyris and Bourbakis [1] proposed a method using image processing techniques to detect polyps and ulcers in wireless capsule endoscopy videos, achieving improved detection rates. Mesejo et al. [2] developed a computer-aided system based on computer vision and machine learning for classifying gastrointestinal lesions in regular colonoscopy images, enhancing diagnostic accuracy. Charfi et al. [3] combined the local binary pattern variance and discrete wavelet transform to make texture extraction for wireless capsule endoscopy images. However, despite the fact that computer-aided diagnosis systems is beneficial for endoscopic image classification compared with human beings, it still encounters significant obstacles. Primarily, due to the high variability within the same class of samples, such as differences in size and shape of lesions, the extraction of consistent features from the same category is quite difficult. By contrast, the subtle differences between different classes also present a challenge in accurate classification, where the different samples from different classes may have the similar attributes. Furthermore, interference factors like bubbles, turbidity, and artifacts caused by the movement of the capsule camera during endoscopic procedures can also significantly reduce the detection rate of abnormal images. Obviously, these factors contribute to the overall difficulty in achieving high accuracy in endoscopic image classification, emphasizing the need for more advanced algorithms and techniques to address these challenges.
In recent years, deep learning, particularly convolutional neural networks (CNNs) [4–6], has made significant strides in the field of endoscopic image classification [7–9]. These technologies have automated medical image analysis, reducing the workload for physicians and enabling more efficient disease diagnosis through feature extraction and pattern recognition. Compared to traditional methods, deep learning models have demonstrated higher precision and recall. Deep learning’s ability to learn from data has made it superior in tasks such as polyp detection, lesion classification, and region recognition, outperforming traditional algorithms in terms of speed and accuracy [10]. However, despite these advancements, deep learning models in endoscopic image classification have yet to reach a level suitable for widespread clinical application. There are still many challenges such as the requirement for large annotated datasets and the difficulty in achieving higher diagnostic precision for rare or subtle pathologies. There is a need for more effective methods to enhance the classification accuracy of endoscopic images and address these limitations before deep learning can be fully integrated into clinical practice.
In this paper, we introduce a novel deep learning approach for classifying endoscopic images called GLI-Net (Global and Local Interaction Network). GLI-Net addresses the shortcomings of traditional methods in capturing both detailed features and global semantic information by effectively combining global and local features, leading to significant improvements in classification accuracy and robustness. The network is composed of four primary modules: the Global Branch Module (GB), which extracts global features and guides the Local Branch Module (LB); the Local Branch Module (LB), which focuses on extracting detailed features from lesion regions; the Information Interaction Module (LEM), which facilitates mutual information exchange and optimization between the global and local branches; and the Adaptive Feature Fusion and Enhancement Module (AFE), which adaptively fuses the global and local features, enhancing their representational power and boosting the model’s discriminative performance. The synergistic interaction of these modules enables GLI-Net to achieve superior results in medical image classification.
2 Related work
In this section, we will briefly describe the related works about classification on the endoscopic images. Due to different approaches used in this field, we divide the related works into two branches: human-crafted feature based methods and deep learning based methods.
2.1 Human-crafted feature based methods
For the human-crafted feature based methods, many machine learning methods with different images features designed by human beings were studied. For instance, Charfi and El Ansari demonstrated that their computer-aided diagnosis system can effectively detect colon abnormalities in wireless capsule endoscopy images [3]. The system employed image preprocessing to enhance quality, extracted key features such as color and texture, and used a support vector machine (SVM) classifier for abnormality detection. Their results verified that integrating color and texture features with SVM significantly can improve detection accuracy compared to manual analysis. This approach highlights the potential of feature-based machine learning methods for automating gastrointestinal disorder diagnosis in clinical practice. Furthermore, MeseJo et al. [2] made a study on how to apply the computer technology to diagnose gastrointestinal lesions from regular colonoscopic videos. Specifically, it exploited both computer vision and machine learning methods, conducting a virtual biopsy to differentiate hyperplastic lesions, serrated adenomas, and adenomas. Karargyris and Bourbakis [1] conducted a study on the detection of small bowel polyps and ulcers using wireless capsule endoscopy videos. Specifically, they developed an algorithm that leveraged image processing techniques to identify and analyze these gastrointestinal abnormalities, contributing to the advancement of non-invasive diagnostic methods.
Additionally, Li and Meng [11] developed an enhancement method based on adaptive contrast diffusion. This technique was designed to adjust the contrast in different regions of the image dynamically, which helped in highlighting the features of interest, particularly in the context of the gastrointestinal tract. By increasing the contrast, the method aimed to make it easier for medical professionals to identify and diagnose any abnormalities or pathologies within the small bowel. The enhancements are intended to facilitate a more accurate and reliable analysis of the endoscopic images, which is vital for effective clinical decision-making. The work by Souaidi and Ansari [12] delved into the detection of ulcer diseases from wireless capsule endoscopy images, employing a multi-scale analysis technique. Specifically, this approach involved examining images across various scales to identify ulcers of different sizes and shapes within the gastrointestinal tract, which enhanced the detection accuracy by capturing the nuances of ulcer appearances at multiple levels of detail.
2.2 Deep learning based methods
Different from human-crafted features based methods, deep learning based methods can automatically extract more semantic features for classification. For instance, Zhang et al. [13] focused on the automatic detection and classification of colorectal polyps by leveraging low-level CNN features from nonmedical domains. Specifically, the authors explored the transfer learning approach where pre-trained CNN models originally trained on nonmedical images were adapted for the task of polyp detection in endoscopic videos. The study aimed to demonstrate that features learned from large datasets in nonmedical domains could be effectively transferred to enhance the performance of medical image analysis tasks, particularly in the context of colorectal polyp identification. Shin and Balasingham [14] conducted a comparative study between a hand-crafted feature-based SVM and a CNN based deep learning framework for the automatic classification of polyps. They evaluated the performance of both methods in distinguishing polyps in endoscopic images, providing insights into the efficacy of deep learning versus traditional machine learning approaches for medical image classification. Zhao et al. [15] presented Adasan, an Adaptive Cosine Similarity Self-Attention Network for gastrointestinal endoscopy image classification, which integrated self-attention mechanisms with adaptive cosine similarity measures to enhance feature representation, improving classification accuracy of endoscopic images.
Furthermore, Zhu et al. [16] presented a method for lesion detection in endoscopy images leveraging features from CNNs. Also, a novel method for WCE video summarization was studied by using a Siamese neural network coupled with SVM, which condensed long WCE video sequences into shorter, representative summaries to facilitate faster and more efficient review by medical professionals. The Siamese network was employed to learn and compare image features, identifying similar frames within the video, while the SVM was utilized to classify these frames based on their medical relevance. Similarly [17], designed a network to identify and highlight potential lesions within the gastrointestinal tract by analyzing WCE video frames. By extending the Siamese network, Guo et al. [18] introduced the Triple ANet, an Adaptive Abnormal-Aware Attention Network designed for the classification of WCE images. It included three main components: an abnormal region detection module, an attention mechanism to highlight these regions, and a classification module, in which the attention mechanisms was introduced to focus on abnormal regions within the gastrointestinal tract, being crucial for accurate diagnosis. And The paper probably detailed the architecture of the network, how it was trained on WCE images, and its effectiveness in classifying normal versus abnormal images. This approach aimed to improve the accuracy and efficiency of WCE image analysis, providing a valuable tool for medical professionals to detect gastrointestinal abnormalities. Similarly, an Effectively Fused Attention Guided Convolutional Neural Network was proposed to integrated attention mechanisms to enhance feature extraction from endoscopic images, focusing on discriminative regions indicative of gastrointestinal conditions [19,20].
In recent years, many deep learning-based approaches have been applied to classify colorectal cancer and WCE images, yielding promising outcomes. However, due to the inherent characteristics of these images, such as considerable intra-class variations and subtle inter-class differences, there is still a need for more robust models to improve the accuracy and reliability of these algorithms. To overcome these challenges, future research should focus on developing models that are better equipped to handle the complexity and variability of endoscopic images. This could involve exploring advanced network architectures, integrating multi-modal data, or utilizing sophisticated feature extraction methods to capture subtle pathological changes more effectively.
3 Methods
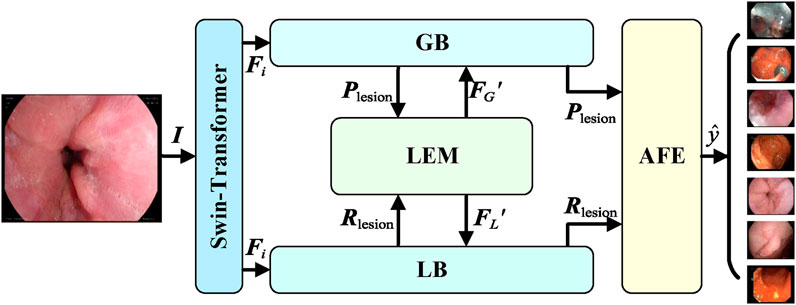
This section provides a detailed description of the overall architecture of GLI-Net (Global and Local Interaction Network). First, the main structure of the network and its global branch module (GB) and local branch module (LB) are introduced. Then, the structure and functionality of the Information Exchange Module (LEM) and the Adaptive Feature Fusion and Enhancement Module (AFE) are discussed in detail. The overall network architecture of GLI-Net is shown in Figure 1.
3.1 Overall network architecture
GLI-Net adopts a dual-branch global and local interaction network structure, as illustrated in Figure 1. The backbone of the network uses the Swin Transformer as a feature extractor, designed to extract both shallow and deep feature maps from the input endoscopic images and generate multi-scale feature representations. The feature sizes correspond to 1/4, 1/8, 1/16, and 1/32 of the input image size, as specified in Equation 1:
where
where
where
3.2 Global branch module (GB)
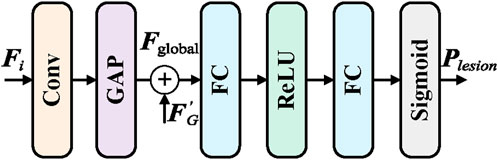
To effectively capture the overall lesion information in endoscopic images and guide the local branch module to focus on key regions, the Global Branch module (GB) is introduced. The goal of the GB module is to extract global lesion features from the deepest feature maps and generate lesion category prompts to guide the local branch, thereby enhancing the comprehensiveness of feature representations and improving classification accuracy. The GB module consists of convolutional layers, global adaptive pooling layers, and the Lesion Category Prompt Extractor (LCPE), with the specific structure shown in Figure 2.
The input global feature map
The module
3.3 Local branch module (LB)
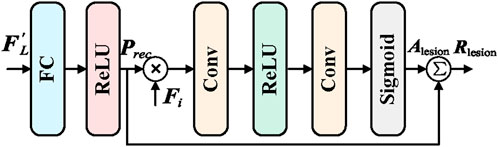
In order to further capture detailed lesion information in endoscopic images, and integrate guidance from the global features, the Local Branch Module (LB) is proposed. The main objective of the LB module is to receive enhanced lesion category prompts from the Information Exchange Module (LEM) through the Lesion Category Prompt Receiver (LCPR). These prompts are then used by the Lesion Region Detector (LRD) to identify detailed lesion features. The output detailed features are fed back into the Information Exchange Module and the subsequent Adaptive Feature Fusion and Enhancement (AFE) module, enabling the collaborative enhancement of both global and local features. The structure of the LB module is shown in Figure 3.
The Lesion Category Prompt Receiver (LCPR) module is responsible for receiving the enhanced lesion category prompt
where
3.4 Information exchange module (LEM)
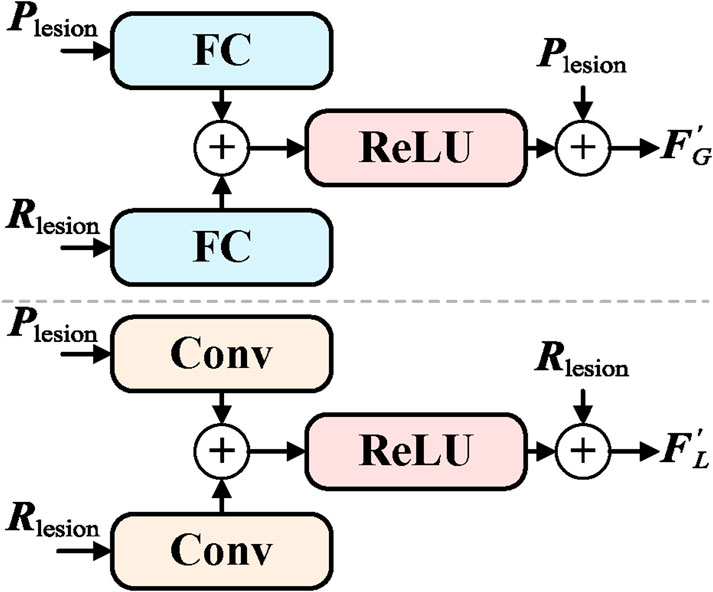
In order to enable efficient collaboration between the global and local networks and enhance the overall feature representation capability, an Information Exchange Module (LEM) has been proposed. The primary goal of the LEM module is to facilitate bidirectional information transfer and mutual supervision between the global branch (GB) and the local branch (LB), thereby improving the comprehensiveness of the features and the accuracy of classification. The detailed structure of the LEM module is shown in Figure 4.
The LEM module includes information transmission from global to local, feedback from local to global, and bidirectional information flow. The information transmission from global to local is responsible for passing the lesion category cue
where
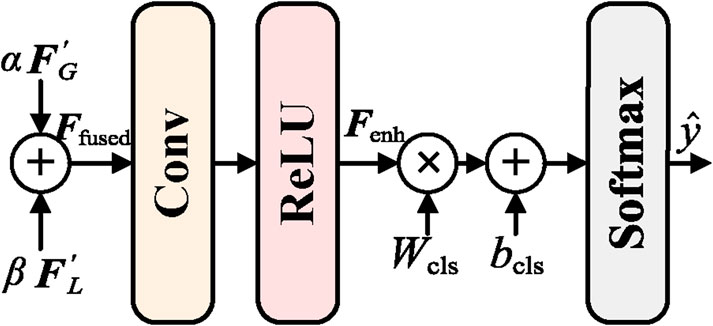
3.5 Adaptive feature fusion and enhancement (AFE) module
To fully integrate global and local features and further enhance the feature representation capability, an Adaptive Feature Fusion and Enhancement (AFE) module has been proposed. The primary objective of the AFE module is to effectively fuse the enhanced global features
First, the feature fusion component is responsible for adaptively fusing the enhanced global features
where
Finally, the enhanced features
where
3.6 Loss function
To effectively train GLI-Net, a comprehensive loss function has been designed, consisting of two main components: lesion region detection loss
where N is the total number of pixels or regions,
where M is the number of samples, C is the total number of classes,
4 Experiments
4.1 Experimental details
4.1.1 Dataset
In our experiments, the Kvasir dataset and the Hyper-Kvasir dataset were used. The Kvasir dataset contains 4,000 endoscopic images of gastrointestinal diseases, covering eight categories, with 500 images per category. The dataset includes both anatomical landmarks (such as the Z-line, pylorus, cecum, etc.) and pathological findings (such as esophagitis, polyps, ulcerative colitis, etc.). The image resolutions range from
4.1.2 Evaluation metrics
We use accuracy (ACC), F1 score, precision, and recall as classification evaluation metrics. These metrics are all derived from the confusion matrix, where the symbols are defined as True Positive (TP), True Negative (TN), False Positive (FP), and False Negative (FN). The specific calculation formulas are as follows:
4.1.3 Implementation details
The experiments in this study were conducted on a computer equipped with an NVIDIA RTX 4090 GPU with 24 GB of memory. During training, the Adam optimizer was used, with specific parameters set as
4.2 Experimental comparison
4.2.1 Kvasir public dataset
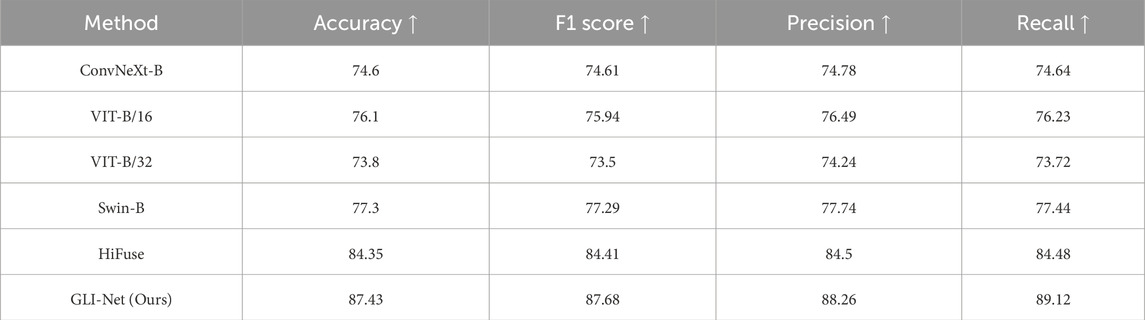
To validate the outstanding performance of our proposed GLI-Net on the Kvasir public dataset, we compared it with current state-of-the-art models. Specifically, as shown in Table 1, compared to ConvNeXt-B, ViT-B/16, ViT-B/32, and Swin-B models, our method achieved improvements of 13.31%, 10.25%, 14.81%, and 9.03% in Acc, respectively; 13.55%, 10.82%, 15.36%, and 9.39% in F1 score; 13.76%, 10.94%, 15.43%, and 9.51% in P; and 14.31%, 11.57%, 16.07%, and 10.10% in R. Moreover, compared to the HiFuse model, GLI-Net improved accuracy, F1 score, precision, and recall by 3.29%, 3.54%, 3.70%, and 4.28%, respectively. These results demonstrate that GLI-Net is more effective in capturing and integrating both global and local features, significantly enhancing the accuracy and robustness of medical endoscopic image classification, and showcasing its superior performance in complex medical image analysis tasks.
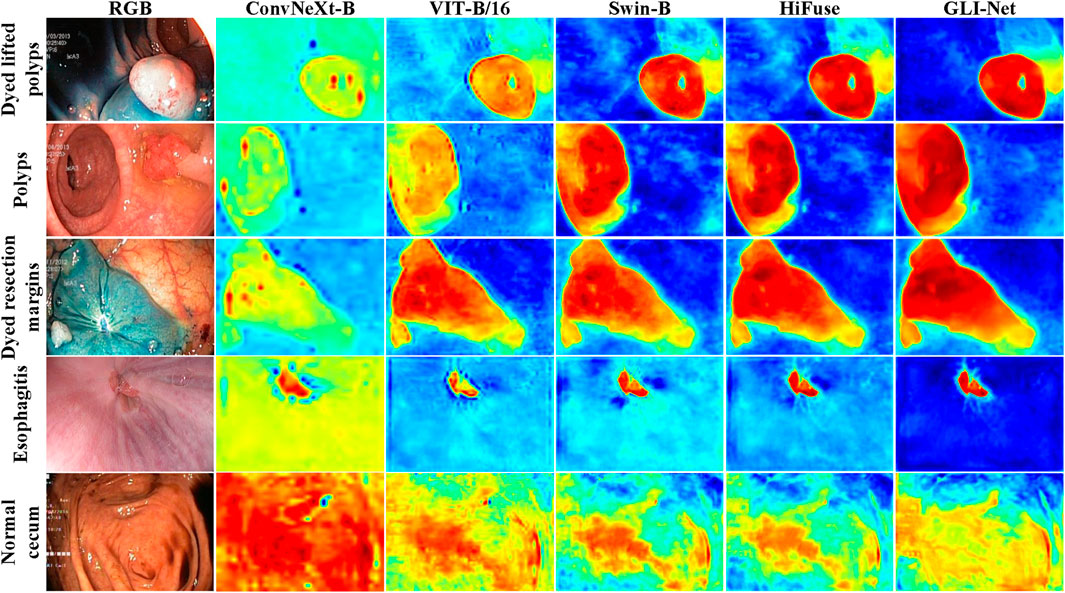
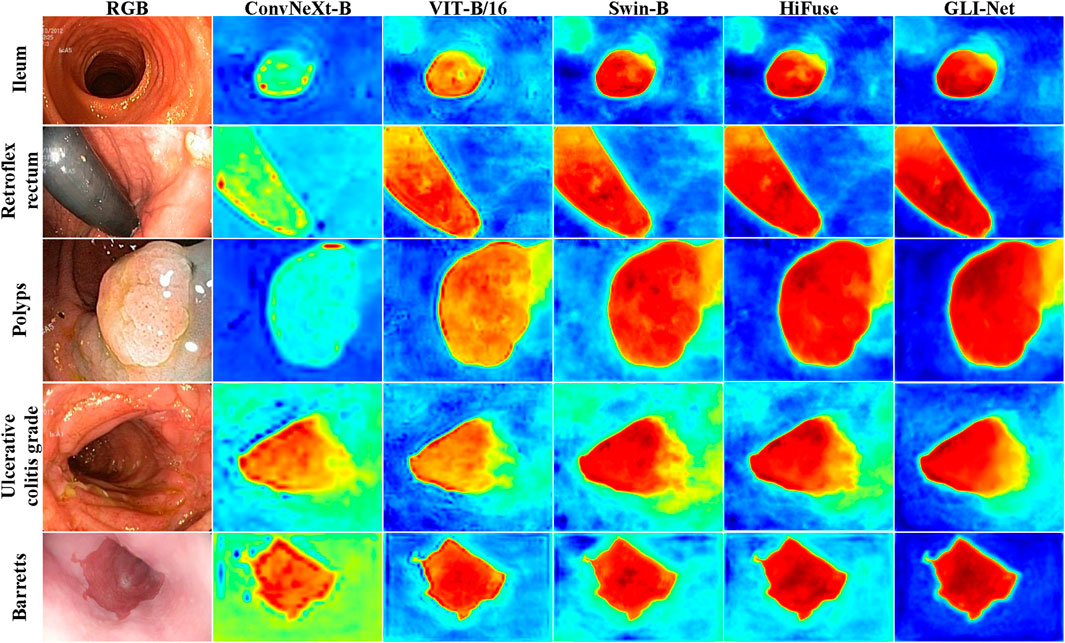
To further demonstrate the superior performance of GLI-Net on the Kvasir dataset, we applied the Grad-CAM method to visualize the model’s final layer, generating heatmaps that reflect the regions of the lesion the model focuses on. The specific details are shown in Figure 6. Compared to models such as ConvNeXt-B, ViT-B/16, ViT-B/32, Swin-B, and HiFuse, GLI-Net’s heatmaps show higher focus and coverage of the lesion regions, allowing for more accurate localization of lesions in endoscopic images. While other models can recognize some lesion areas, they exhibit discrepancies in precise localization and coverage. For example, ConvNeXt-B and ViT-B/32 show relatively blurred recognition, while Swin-B and HiFuse incorrectly label many non-lesion areas. GLI-Net, by effectively covering lesion regions and minimizing background interference, demonstrates significant advantages in feature extraction and region localization. These visualization results prove GLI-Net’s efficiency and reliability in medical image classification tasks.
4.2.2 Hyper-Kvasir public dataset
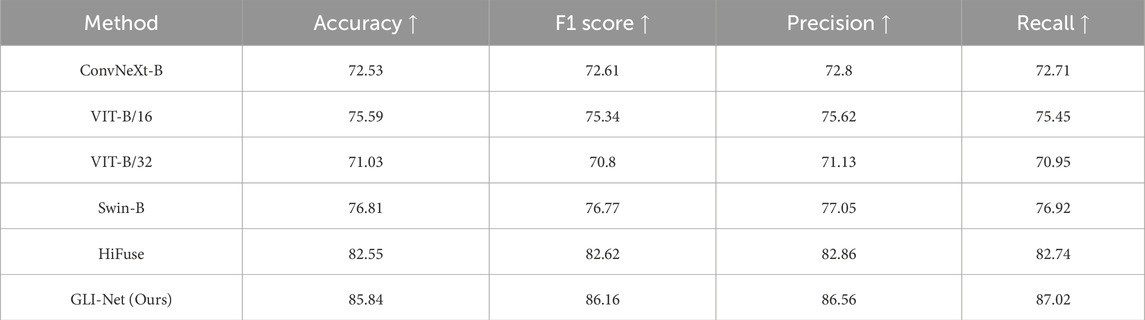
To validate the outstanding performance of our proposed GLI-Net on the Hyper-Kvasir public dataset, we compared it with current state-of-the-art models. The specific results are shown in Table 2. Compared to ConvNeXt-B, ViT-B/16, ViT-B/32, Swin-B, and HiFuse models, GLI-Net achieved improvements of 13.31%, 10.25%, 14.81%, 9.03%, and 3.29% in Acc, respectively; 13.55%, 10.82%, 15.36%, 9.39%, and 3.54% in F1 score; 13.76%, 10.94%, 15.43%, 9.51%, and 3.70% in P; and 14.31%, 11.57%, 16.07%, 10.10%, and 4.28% in R. These significant performance improvements indicate that GLI-Net is more effective in capturing and integrating both global and local features, significantly enhancing the accuracy and robustness of medical endoscopic image classification, and showcasing its superior performance in complex medical image analysis tasks.
To demonstrate the superior performance of GLI-Net on the Hyper-Kvasir dataset, we used the Grad-CAM method to generate heatmaps that visualize the lesion regions the model focuses on. The specific details are shown in Figure 7. Compared to models such as ConvNeXt-B, ViT-B/16, ViT-B/32, Swin-B, and HiFuse, GLI-Net’s heatmaps exhibit higher focus and coverage of the lesion areas, allowing for more accurate localization of lesions in endoscopic images. While other models can identify some lesion regions, they show discrepancies in localization and coverage. For example, ConvNeXt-B and ViT-B/32 exhibit relatively blurred recognition, while Swin-B and HiFuse incorrectly label non-lesion regions. GLI-Net, through more precise lesion region coverage and reduced background interference, demonstrates its advantages in feature extraction and region localization. These results show that GLI-Net can more effectively integrate global and local features, significantly improving the accuracy and robustness of medical endoscopic image classification, and proving its efficiency and reliability in real-world applications.
4.3 Ablation study
4.3.1 Ablation study of GLI-Net
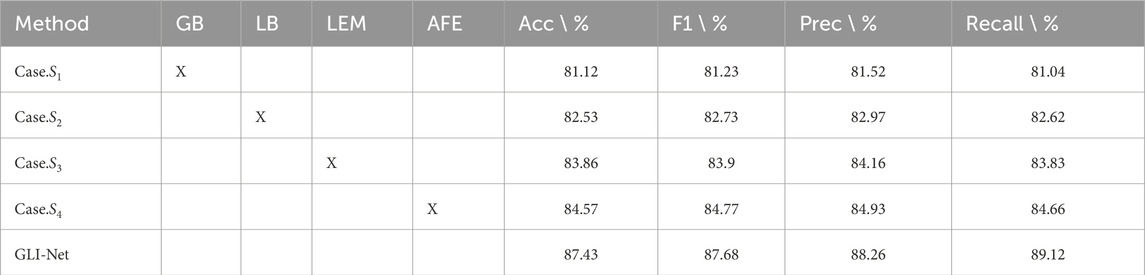
To evaluate the performance of GLI-Net on the Kvasir dataset, we conducted ablation experiments by sequentially removing the GB, LB, LEM, and AFE modules from the model. The results are shown in Table 3. The inclusion of each module significantly improved the model’s performance. The baseline model with the GB module removed achieved an accuracy of 81.12%. After removing the LB module, the accuracy increased to 82.53%, and further removal of the LEM module raised the accuracy to 83.86%. When the AFE module was removed, the accuracy reached 84.57%. Finally, the complete GLI-Net model achieved an accuracy of 87.43%, which is a 2.86% improvement over the model without the AFE module. In addition, GLI-Net also performed better in other evaluation metrics such as F1 score, precision, and recall, with improvements of 6.45%, 4.45%, 3.78%, 8.08%, 6.50%, and 5.29%, respectively. These experimental results demonstrate that the individual modules of GLI-Net play a critical role in enhancing feature extraction, feature fusion, and optimizing representation, significantly improving the accuracy and robustness of medical endoscopic image classification, and proving its superior performance in complex medical image analysis tasks.
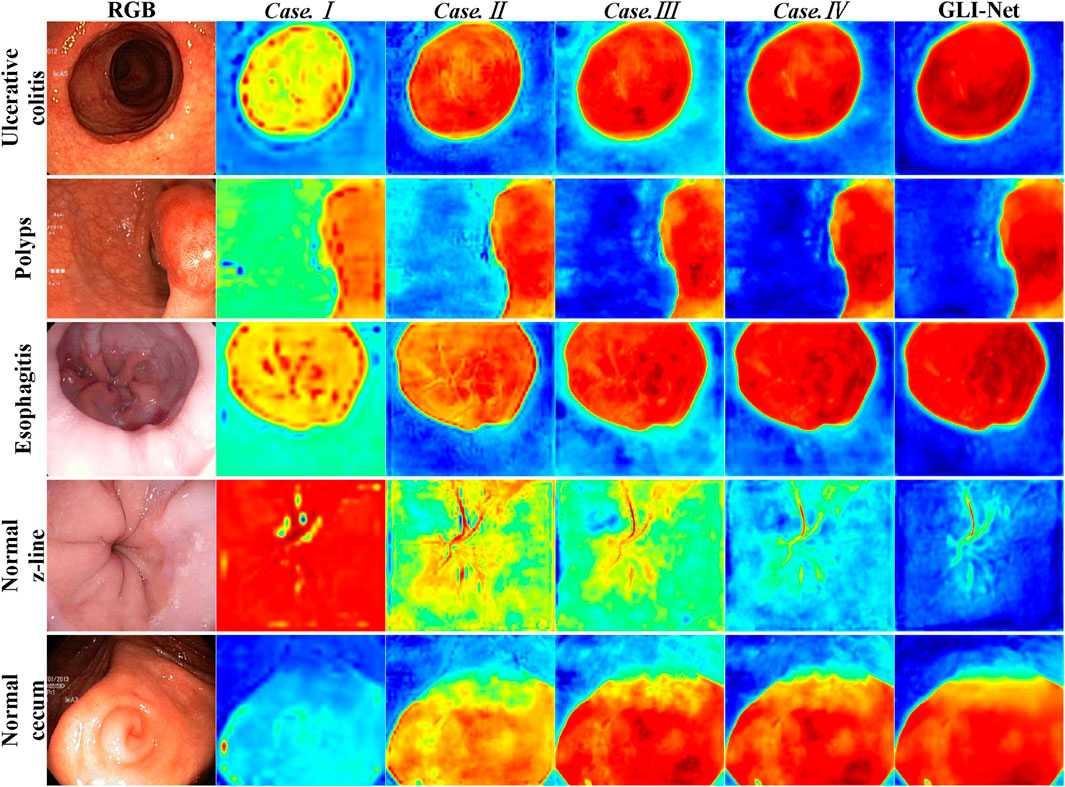
To verify the role of each module in GLI-Net, we conducted ablation experiments on the Kvasir dataset by sequentially removing the modules and used the Grad-CAM method to visualize the lesion regions the model focuses on under different configurations. The specific details are shown in Figure 8. The experimental results indicate that, after removing the Global Branch module (GB) (Case.
4.3.2 Ablation study of loss function
To evaluate the contribution of each component of the loss function in GLI-Net, we conducted ablation experiments on the Kvasir dataset by sequentially removing the lesion region detection loss, classification loss, and weight coefficient. The results are shown in Table 4. When only the lesion region detection loss was used (Case.
5 Conclusion
This paper presents GLI-Net, a novel network for medical endoscopic image classification, designed to enhance classification performance by effectively integrating both global and local features. GLI-Net utilizes a hierarchical multi-module architecture that includes a global branch module (GB), a local branch module (LB), an information exchange module (LEM), and an adaptive feature fusion and enhancement module (AFE) to facilitate comprehensive feature extraction and optimization. Evaluation on the Kvasir and Hyper-Kvasir public datasets showed that GLI-Net outperforms state-of-the-art models, including ConvNeXt-B, ViT-B/16, ViT-B/32, Swin-B, and HiFuse, across key metrics such as accuracy, F1 score, precision, and recall. Specifically, GLI-Net achieved accuracies of 87.43% and 85.84% on the Kvasir and Hyper-Kvasir datasets, respectively, surpassing the second-best models by 2.86% and 2.29%. Ablation studies confirmed the significant contribution of each module to the overall performance, as the removal of any module caused a notable performance decline, underscoring their synergistic interaction. Additionally, Grad-CAM visualization highlighted GLI-Net’s improved ability to accurately localize lesion areas, with better focus and coverage compared to other models, effectively reducing interference from background and non-lesion regions. These results demonstrate GLI-Net’s substantial advantages in feature extraction and region localization, leading to enhanced accuracy and robustness in medical endoscopic image classification.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
YZ: Writing – original draft, Writing–review and editing, Conceptualization, Data curation, Investigation, Methodology. MZ: Writing – original draft, Writing – review and editing, Conceptualization, Investigation. WC: Writing – original draft, Writing – review and editing, Data curation, Methodology. XW: Writing – original draft, Writing – review and editing, Formal Analysis. QS: Writing – original draft, Writing – review and editing, Data curation.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Karargyris A, Bourbakis N. Detection of small bowel polyps and ulcers in wireless capsule endoscopy videos. IEEE Trans Biomed Eng (2011) 58:2777–86. doi:10.1109/tbme.2011.2155064
2. Mesejo P, Pizarro D, Abergel A, Rouquette O, Beorchia S, Poincloux L, et al. Computer-aided classification of gastrointestinal lesions in regular colonoscopy. IEEE Trans Med Imaging (2016) 35:2051–63. doi:10.1109/tmi.2016.2547947
3. Charfi S, Ansari ME. Computer-aided diagnosis system for colon abnormalities detection in wireless capsule endoscopy images. Multimedia Tools Appl (2018) 77:4047–64. doi:10.1007/s11042-017-4555-7
5. Simonyan K. Very deep convolutional networks for large-scale image recognition (2014). arXiv preprint arXiv:1409.1556.
6. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition; 27-30 June 2016; Las Vegas, NV, USA (2016). p. 770–8.
7. Thambawita V, Strümke I, Hicks SA, Halvorsen P, Parasa S, Riegler MA. Impact of image resolution on deep learning performance in endoscopy image classification: an experimental study using a large dataset of endoscopic images. Diagnostics (2021) 11:2183. doi:10.3390/diagnostics11122183
8. Mukhtorov D, Rakhmonova M, Muksimova S, Cho Y-I. Endoscopic image classification based on explainable deep learning. Sensors (2023) 23:3176. doi:10.3390/s23063176
9. Yue G, Wei P, Liu Y, Luo Y, Du J, Wang T. Automated endoscopic image classification via deep neural network with class imbalance loss. IEEE Trans Instrumentation Meas (2023) 72:1–11. doi:10.1109/tim.2023.3264047
10. Bolhasani H, Jassbi SJ, Sharifi A. Dla-e: a deep learning accelerator for endoscopic images classification. J Big Data (2023) 10:76. doi:10.1186/s40537-023-00775-8
11. Li B, Meng MQ-H. Wireless capsule endoscopy images enhancement via adaptive contrast diffusion. J Vis Commun Image Representation (2012) 23:222–8. doi:10.1016/j.jvcir.2011.10.002
12. Souaidi M, Ansari ME. Multi-scale analysis of ulcer disease detection from wce images. IET Image Process (2019) 13:2233–44. doi:10.1049/iet-ipr.2019.0415
13. Zhang R, Zheng Y, Mak TWC, Yu R, Wong SH, Lau JY, et al. Automatic detection and classification of colorectal polyps by transferring low-level cnn features from nonmedical domain. IEEE J Biomed Health Inform (2016) 21:41–7. doi:10.1109/jbhi.2016.2635662
14. Shin Y, Balasingham I. Comparison of hand-craft feature based svm and cnn based deep learning framework for automatic polyp classification. In: 2017 39th annual international conference of the IEEE engineering in medicine and biology society (EMBC). IEEE (2017). p. 3277–80.
15. Zhao Q, Yang W, Liao Q. Adasan: adaptive cosine similarity self-attention network for gastrointestinal endoscopy image classification. In: 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI); 13-16 April 2021; Nice, France. IEEE (2021). p. 1855–9.
16. Zhu R, Zhang R, Xue D. Lesion detection of endoscopy images based on convolutional neural network features. In: 2015 8th International Congress on Image and Signal Processing (CISP); 14-16 October 2015; Shenyang, China. IEEE (2015). p. 372–6.
17. Chen J, Zou Y, Wang Y. Wireless capsule endoscopy video summarization: a learning approach based on siamese neural network and support vector machine. In: 2016 23rd International Conference on Pattern Recognition (ICPR); 04-08 December 2016; Cancun, Mexico. IEEE (2016). p. 1303–8.
18. Jeon Y, Cho E, Moon S, Chae S-H, Jo HY, Kim TO, et al. Deep convolutional neural network-based automated lesion detection in wireless capsule endoscopy. In: International forum on medical imaging in asia 2019, 11050. SPIE (2019). p. 64–296. doi:10.1117/12.2522159
19. Guo X, Yuan Y. Triple anet: adaptive abnormal-aware attention network for wce image classification. In: Medical image computing and computer assisted intervention–MICCAI 2019: 22nd international conference, shenzhen, China, october 13–17, 2019, proceedings, Part I 22. Springer (2019). p. 293–301.
Keywords: endoscopic image classification, deep learning, global and local feature fusion, global branch module, local branch module
Citation: Zhang Y, Zhuang M, Chen W, Wu X and Song Q (2025) GLI-Net: A global and local interaction network for accurate classification of gastrointestinal diseases in endoscopic images. Front. Phys. 13:1582245. doi: 10.3389/fphy.2025.1582245
Received: 24 February 2025; Accepted: 14 March 2025;
Published: 07 April 2025.
Edited by:
Huafeng Li, Kunming University of Science and Technology, ChinaReviewed by:
Bo Jiang, Northwest A&F University, ChinaChaoxun Guo, The Chinese University of Hong Kong, Shenzhen, China
Tianhao Peng, Moutai College, China
Copyright © 2025 Zhang, Zhuang, Chen, Wu and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuansen Zhang, emhhbmd5dWFuc2VuMDZAMTYzLmNvbQ==
 Yuansen Zhang
Yuansen Zhang Mengxiao Zhuang
Mengxiao Zhuang