- 1School of international education, Beijing University of Chemical Technology, Beijing, China
- 2School of Information Science and Technology, Beijing University of Chemical Technology, Beijing, China
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide, and early detection is crucial for improving cure rates. In recent years, object detection methods based on convolutional neural networks (CNNs) and transformers have made significant progress in medical image analysis. However, CNNs have limitations in capturing global contextual information, and while transformers can handle long-range dependencies, their high computational complexity limits their efficiency in practical applications. To address these issues, this paper proposes a novel object detection model—CoroYOLO. CoroYOLO builds upon the YOLOv10 architecture by incorporating the concept of State Space Model (SSM) and introduces the TSMamblock module, which dynamically models the input data, reduces redundant computations, and improves both computational efficiency and detection accuracy. Additionally, CoroYOLO integrates the Efficient Multi-Scale Attention (EMA) mechanism, which adaptively strengthens focus on critical regions, enhancing the model’s robustness in complex medical images. Experimental results show that after training on the SUN Polyp and PICCOLO datasets, CoroYOLO outperforms existing mainstream methods on the Etis-Larib dataset, achieving state-of-the-art performance and demonstrating the model’s effectiveness for early colorectal cancer detection.
1 Introduction
Colorectal Cancer (CRC) is one of the most common malignant tumors worldwide, with both its incidence and mortality rates showing an upward trend Jia et al. [1]; Davri et al. [2]. According to the World Health Organization (WHO), colorectal cancer causes millions of deaths annually, severely impacting global public health. Due to the lack of obvious early symptoms, many patients are diagnosed only when the tumor has progressed to advanced stages, which significantly reduces the effectiveness of treatment. Therefore, early screening and precise diagnosis are crucial for improving survival rates and reducing mortality Bousis et al. [3]; Foersch et al. [4].
Artificial Intelligence (AI), particularly deep learning technologies, has shown immense potential in medical image analysis, especially in the field of tumor detection Aruna Kumari et al. [5]; Daghrir et al. [6]; DeMatteo et al. [7]. Early detection is crucial in the diagnosis of colorectal cancer because timely identification of the disease can significantly improve treatment outcomes, enhance patient survival rates, and effectively reduce mortality. Deep learning techniques, such as Convolutional Neural Networks (CNN) and their improved algorithms, can precisely locate and identify cancerous regions from images such as CT, MRI, and endoscopy, helping doctors achieve early detection and timely treatment, thus significantly improving the prognosis of colorectal cancer Yu et al. [8]; Kiehl et al. [9]; Almayyan and AlGhannam [10]. Furthermore, common target detection methods, such as YOLO (You Only Look Once), Faster R-CNN, and DETR (Detection Transformer), have successfully been applied in tumor recognition in medical images, further demonstrating the broad application prospects of deep learning in cancer detection Echle et al. [11]; Tsai and Tao [12]; Sun et al. [13].
In different texture-based DICOM-related approaches for medical image processing, including Gray Level Co-occurrence Matrix (GLCM), Local Binary Pattern (LBP), Autocorrelation Function (ACF), and Histogram Patterns, these methods help doctors extract critical information by analyzing image texture features, thereby improving the accuracy of disease detection. Gray Level Co-occurrence Matrix (GLCM) is widely used for tasks such as tumor and tissue classification, as it extracts spatial relationships between pixels, making it especially suitable for colorectal cancer detection and classification Ramola et al. [14]. Local Binary Pattern (LBP) efficiently captures local texture features and is commonly used in facial recognition and lesion detection, with significant applications in early detection of colorectal cancer Shakya et al. [15]. Autocorrelation Function (ACF) helps analyze the periodicity and regularity of texture surfaces, supporting tumor texture analysis and cancer detection Shakya and Vidyarthi [16]. Histogram Patterns, by quantifying the distribution of pixel intensities, provide an intuitive way to reflect texture changes in an image, aiding in identifying various manifestations of colorectal cancer Shakya et al. [17]. These texture-based analysis methods not only enhance the precision of medical image processing but also offer crucial insights for colorectal cancer detection, particularly in early detection and prognosis prediction, which can effectively improve survival rates and treatment outcomes. Additionally, by combining emerging texture classification techniques and image compression algorithms, such as Discrete Cosine Transform (DCT) and Discrete Wavelet Transform (DWT), the storage and processing efficiency of images can be optimized while ensuring the preservation of key texture features, further improving the diagnostic accuracy and detection speed of colorectal cancer Shakya et al. [17].
However, traditional Convolutional Neural Networks (CNNs) and Transformer-based detection models still exhibit several limitations. First, while CNNs excel at local feature extraction, they face challenges in capturing global context due to the constraints of their localized receptive fields. This limitation is particularly critical in medical imaging, where the shape and location of tumors are often closely tied to surrounding tissues Liu et al. [18]; Wang et al. [19]; Xing et al. [20]; Zhang et al. [21]. In contrast, Transformers are capable of modeling long-range dependencies and effectively capturing global context in image processing. However, they come with high computational complexity, especially when processing high-resolution medical images, leading to substantial resource demands and efficiency bottlenecks during both training and inference Hatamizadeh and Kautz [22]; Qiao et al. [23]; Huang et al. [24]; Zhang et al. [25]. Additionally, both CNN and Transformer models are prone to overfitting, particularly in data-scarce scenarios, which is common in medical imaging where high-quality annotated datasets are often limited. Consequently, achieving a balance between precision and efficiency, while enhancing the model’s generalization ability, remains a key challenge in the application of deep learning to medical imaging.
In recent years, the Mamba network has effectively integrated a selection mechanism into its SSM, avoiding the secondary complexity of attention mechanisms in Transformers and significantly accelerating inference speed. By optimizing the state-space model, Mamba offers higher computational efficiency in dynamic modeling, enabling it to capture global context in medical images while reducing unnecessary computational overhead, thus improving the effectiveness of detection tasks Liu et al. [26]; Ma et al. [27]. However, the application of Mamba-based network frameworks in colorectal cancer detection is still relatively limited, and its full potential remains to be explored.
To address the limitations of existing methods, we propose a novel target detection approach—CoroYOLO. Based on the YOLOv10 architecture, this method introduces the concept of SSM and proposes a TSMamblock, which dynamically models the input data, effectively reducing redundant computations in YOLO and enhancing the model’s operational efficiency while maintaining stable detection accuracy. Furthermore, CoroYOLO incorporates the EMA attention mechanism, which adaptively strengthens focus on critical regions, thus improving the model’s robustness in complex medical images. We trained and evaluated the model on two widely used medical image datasets—the SUN Polyp Database and the PICCOLO Database. Experimental results show that CoroYOLO outperforms existing mainstream methods in both accuracy and efficiency, providing an effective solution for the early diagnosis of colorectal cancer. The contributions of this paper are as follows.
The structure of this paper is as follows: Section 2 presents the related work. Section 3 describes the methodology. Section 4 discusses the experiments. Finally, the conclusion is presented.
2 Related work
2.1 Applications and improvements of YOLO in colorectal cancer detection
YOLO, as an efficient object detection algorithm, has been widely applied in CRC detection and has achieved significant progress Haq et al. [28]. Firstly, a detection method based on YOLOv4 for endoscopic images improved the detection accuracy of colorectal cancer significantly, especially in identifying small polyps, through data augmentation and loss function improvements Lalinia and Sahafi [29]. However, this method showed an increased probability of false positives in noisy or low-quality images, which affected its practical application performance. Secondly, YOLOv5 was further optimized for colorectal cancer detection, particularly by introducing a spatial attention mechanism to enhance the model’s ability to detect small lesion areas, thereby improving the recognition accuracy of small lesions Zhang et al. [30]. However, due to the increased model complexity, the inference speed of this method became slower, limiting its real-time clinical application. Another method, based on an improved YOLOv3, adopted a transfer learning strategy and achieved high accuracy with limited annotated data, especially performing well on large-scale datasets Murugesan et al. [31]. However, due to insufficient training data, this method is prone to overfitting and requires a longer training time. To improve real-time performance, an optimization based on YOLOv4-tiny reduced the model parameters and optimized the network structure, thus improving inference speed and computational efficiency, making it particularly suitable for resource-constrained scenarios Reddy et al. [32]. However, this method experienced a drop in detection accuracy when processing high-resolution images, especially in complex backgrounds, where the model’s performance was somewhat limited. Lastly, some studies proposed a multi-scale detection method based on YOLOv8 to address the multi-scale issue in colorectal cancer imaging Zhang et al. [30]. By adaptively adjusting the input image scale, this method successfully improved the detection accuracy of lesions of various sizes Liu et al. [33]. However, this method still faces challenges in detecting complex backgrounds or low-contrast images, resulting in a decrease in accuracy.
In our work, we are the first to apply the improved YOLOv10 model to colorectal cancer detection. To ensure high accuracy while enhancing computational efficiency, we introduced the concept of state space modeling, which significantly optimizes the model’s computational process by dynamically modeling the input data.
2.2 Applications and improvements of Mamba in colorectal cancer detection
In recent years, significant progress has been made in integrating Mamba with YOLO in the field of object detection, particularly in improving computational efficiency and enhancing model robustness. One study, called ODMamba Wang et al. [34], proposed an innovative backbone network by introducing a state space model (SSM) to address the quadratic complexity of self-attention mechanisms. Unlike traditional Transformer-based or SSM-based methods, ODMamba can achieve efficient object detection through simple training without the need for pre-training. Additionally, to meet real-time requirements, ODMamba was designed with an optimized macro-structure, selecting the best scaling factors and level ratios, further improving inference speed and accuracy. The study also proposed an RG module based on a multi-branch structure to model the channel dimensions, overcoming the limitations of insufficient receptive fields and poor image localization in sequence modeling with SSM. Experimental results showed that ODMamba achieved state-of-the-art performance on the COCO dataset, demonstrating its potential in object detection. Another study introduced the FER-YOLO-Mamba model Ma et al. [35], which combines Mamba and YOLO principles for effective facial expression image recognition and localization. This model designed a dual-branch module (FER-YOLO-VSS), which combines the inherent advantages of convolutional layers in local feature extraction with the ability of SSM to capture long-range dependencies, thereby improving the accuracy and robustness of facial expression recognition. Furthermore, a YOLO-Mamba method Zhao and He [36] was proposed that integrates Mamba language with attention mechanisms, introducing a Mamba-based attention module. This module scans the feature and spatial dimensions of the image to comprehensively extract global contextual information, enhancing the model’s focus on key areas and reducing the impact of redundant information. Experiments showed that compared to traditional SE and CBAM attention mechanisms, this approach improved mAP50 by 0.8% and 1.3% on a public infrared aerial image dataset. Finally, another study introduced a YOLO-based Mamba-YOLO-World model Wang et al. [37], which employed the proposed MambaFusion Path Aggregation Network (MambaFusion-PAN) as its neck structure. This model introduced a novel feature fusion mechanism based on state space models, which consists of a parallel guided selective scanning algorithm and a serial guided selective scanning algorithm with linear complexity and a globally guided receptive field. This mechanism uses multimodal input sequences and Mamba hidden states to guide the feature fusion process. Experimental results demonstrated that this mechanism performed excellently in multiple object detection tasks, further enhancing the model’s ability for multimodal fusion and global context modeling. These studies show that models integrating Mamba and YOLO are becoming an effective solution in object detection tasks by optimizing computational processes, improving inference efficiency, and enhancing the model’s ability to perceive complex image features.
In this paper, we combine YOLOv10 with Mamba and propose the TSMamblock structure, which enhances the model’s ability to capture global contextual information by dynamically modeling the input data, while maintaining high-accuracy object detection. In addition to introducing TSMamblock, we also integrate the EMA attention mechanism, which adaptively strengthens the focus on key regions, thereby improving the model’s robustness in complex medical imaging, particularly in colorectal cancer detection tasks.
3 Methods
3.1 Preliminaries: SSM models
The State Space Model (SSM) Zhou et al. [38] is a modeling approach derived from continuous systems, used to map a one-dimensional signal
where the matrix
To meet the computational demands of discrete scenarios, S4 and Mamba introduce a time-scale parameter
where
After discretization, the continuous equations are reformulated as follows:
This formulation allows the state-space model to efficiently model sequences using discrete time steps, thus meeting the computational needs of practical applications. Furthermore, to compute the output more efficiently, the discretized state-space model often employs structured convolution kernels for global feature extraction. Specifically, the convolution kernel is defined as:
where
By combining discretization and convolution kernels, S4 and Mamba not only capture long-range dependencies but also significantly reduce computational complexity. This modeling approach effectively addresses the needs of large-scale data processing, demonstrating flexibility and efficiency in various task scenarios, and providing a solid technical foundation for global feature extraction and dynamic modeling of sequential data.
3.2 Overall network
As shown in Figure 1, we propose an innovative object detection network called CoroYOLO, which is composed of three main components: the Backbone, Neck, and Head. Through the collaborative design of multiple modules, CoroYOLO achieves enhanced feature extraction and detection capabilities.
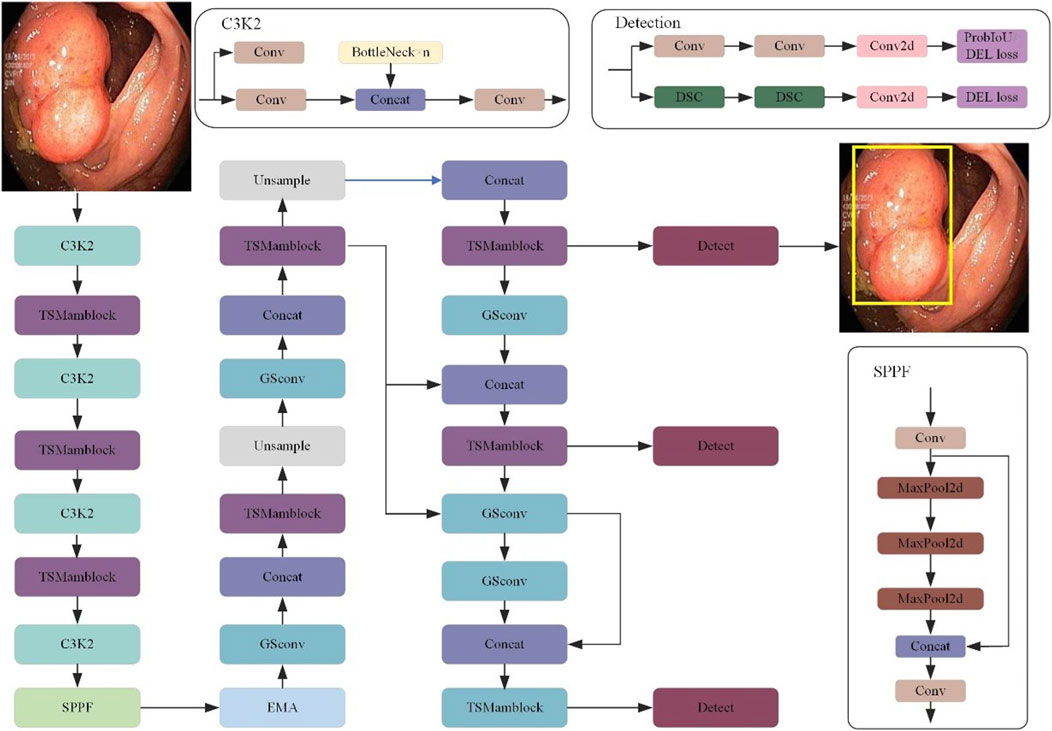
Figure 1. The overall architecture of the CoroYOLO network, illustrating the backbone, neck, and head components. Key modules include C3K2, TSMamblock, GSConv, SPPF, and EMA, designed for efficient feature extraction and detection.
In the Backbone section, we present an alternating stacked structure of TSMamblock modules designed to effectively extract multi-scale features from input images. The TSMamblock module integrates the SS2D module and the TS Block module. The SS2D module leverages the State Space Model (SSM) to perform global modeling of input features, capturing long-range dependencies. Meanwhile, the TS Block module employs temporal-spatial modeling mechanisms to decouple input features across temporal and spatial dimensions, further enhancing the model’s ability to represent features in complex scenarios. The combination of these two modules empowers the model with stronger global understanding and more robust local feature extraction capabilities, particularly when processing intricate medical images.
In the Neck section, we designed a multi-scale feature fusion mechanism to ensure accurate detection across feature maps of different resolutions. The SPPF module further enhances the model’s multi-scale feature aggregation capability by performing multiple MaxPooling operations to downsample and merge features, thereby enriching the contextual information available to the model. Additionally, we integrated the EMA module in this section. The EMA module employs a multi-branch and multi-scale structure to enhance feature interaction in both spatial and channel dimensions, significantly improving the model’s robustness in complex medical image detection tasks.
In the Head section, CoroYOLO adopts a multi-detection branch structure inspired by the YOLOv10 architecture. Specifically, by combining Convolution and DSC, the model effectively reduces computational overhead while maintaining high detection performance. Moreover, the detection head incorporates a combined optimization strategy of ProbIoU Loss and DEL Loss, which further strengthens the model’s ability in target localization and bounding box regression.
3.3 TSMamblock
This paper proposes the TSMamblock module (As shown in Figure 2), an efficient feature modeling module based on State Space Modeling (SSM). It is designed to enhance the model’s ability to capture global contextual information while improving the fine-grained representation of local features. The module is composed of two core sub-modules: the SS2D module and the TS Block module. Through a multi-level and multi-branch structural design, the TSMamblock module combines global and local feature modeling mechanisms, demonstrating outstanding detection performance in complex medical imaging tasks.
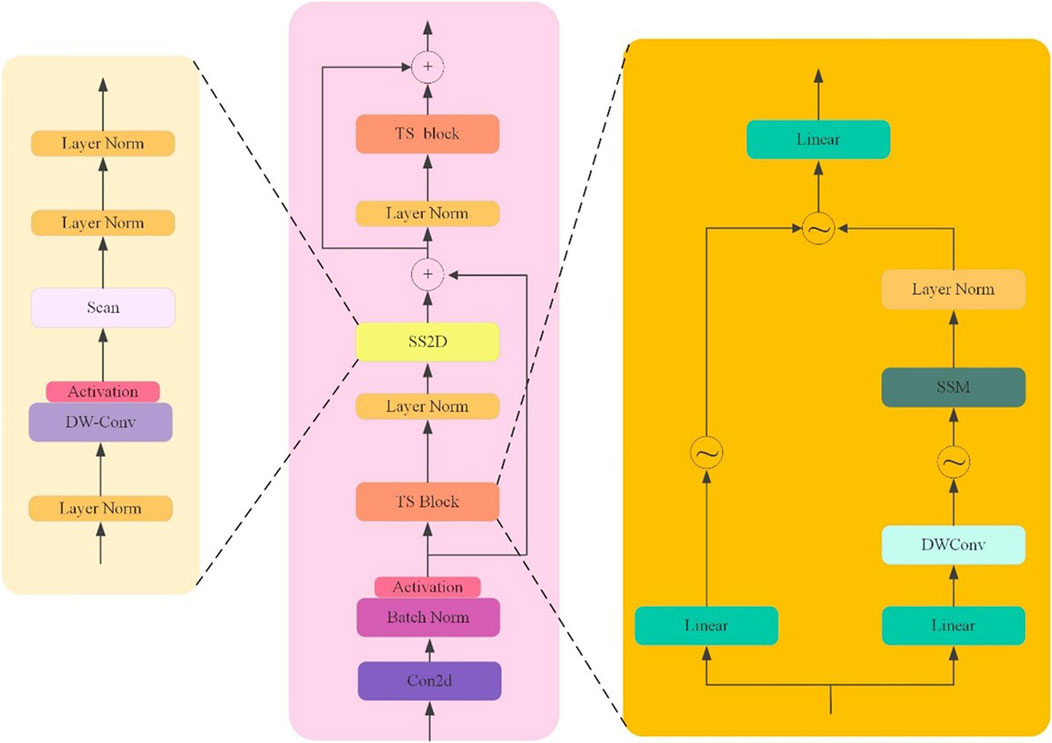
Figure 2. The architecture of the TSMamblock module, comprising the SS2D module for global modeling and the TS Block for temporal-spatial feature extraction.
The SS2D module is the core component of the TSMamblock module, focusing on state space modeling of input features while optimizing the feature extraction process by leveraging the characteristics of 2D convolution. The SS2D module employs a residual connection structure to map input features into a high-dimensional feature space, enabling multi-scale modeling to capture long-range dependencies. LayerNorm is applied to normalize the output of each layer, ensuring the stability of gradient propagation. To further enhance global feature modeling capabilities, the SS2D module integrates the dynamic properties of state space modeling with 2D convolution, effectively capturing global dependencies in the input data while avoiding high computational complexity. In its design, the SS2D module also incorporates activation functions and lightweight convolutional operations to improve the representation of input signals while suppressing background noise interference in object detection. Additionally, the multi-layer structure design allows the module to recursively aggregate contextual information, achieving efficient feature representation in complex scenarios.
The TS Block module is another key sub-module, designed for temporal and spatial modeling of input features. The TS Block employs a multi-branch structure to decompose input features along the temporal and spatial dimensions, separately extracting temporal and spatial dependencies, and ultimately unifying the multi-dimensional information through feature fusion. In this module, DW-Conv (Depthwise Convolution) is used for spatial feature extraction, reducing computational complexity while maintaining spatial feature resolution. Additionally, linear transformations are applied to enhance the representation of local features, enabling the model to achieve greater robustness in multi-scale object detection tasks. The output of the TS Block is combined with the output of the SS2D module through a residual connection, effectively integrating global and local features to ensure precise localization of target regions. Furthermore, the repeated use of LayerNorm and BatchNorm operations within the module enhances the stability of the model and improves training efficiency.
3.4 EMA attention mechanism
In this paper, we introduced the EMA attention mechanism Ouyang et al. [39] into the CoroYOLO framework, as shown in Figure 3. The EMA mechanism enhances feature interaction in spatial and channel dimensions through a multi-branch and multi-scale structure design, further optimizing the feature extraction process.
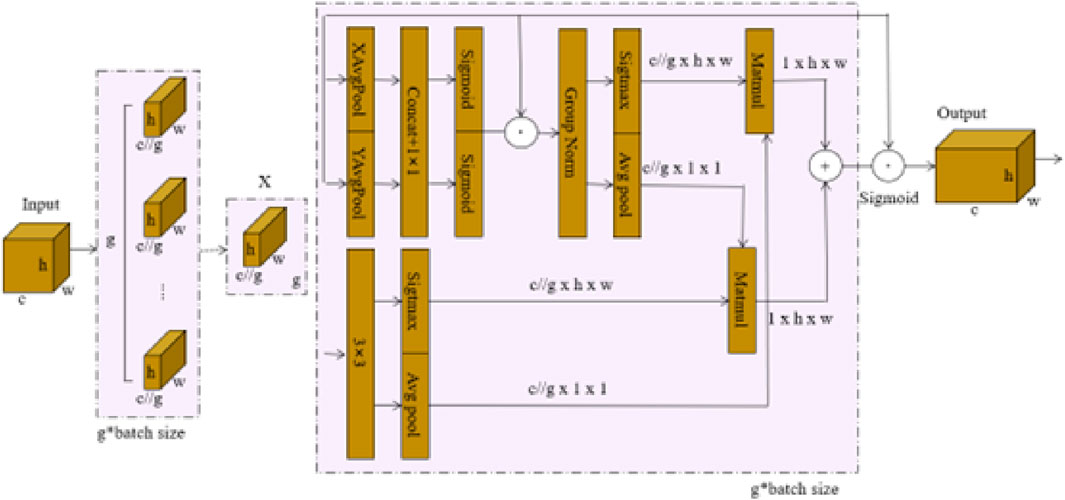
Figure 3. EMA attention mechanism network architecture Ouyang et al. [39].
As shown in the figure, the EMA mechanism first divides the input feature map
The attention weights generated by these branches are applied to the input features through pointwise multiplication, producing two spatial attention maps, which emphasize both local and global dependencies within the input features. Finally, the grouped feature maps are fused using the Sigmoid function to aggregate the outputs, with the resulting output feature map maintaining the same dimensions as the input
4 Experiment
4.1 Datasets
This In this study, we utilized three representative datasets: the SUN Colonoscopy Video Database Misawa et al. [40], the PICCOLO Widefield Dataset Peralta et al. [41], and the Etis-Larib Dataset Nogueira-Rodríguez et al. [42], which were used for training, evaluation, and testing of the proposed model, respectively. These datasets cover diverse clinical scenarios, enabling a comprehensive evaluation of the proposed method’s performance.
4.1.1 SUN colonoscopy video database
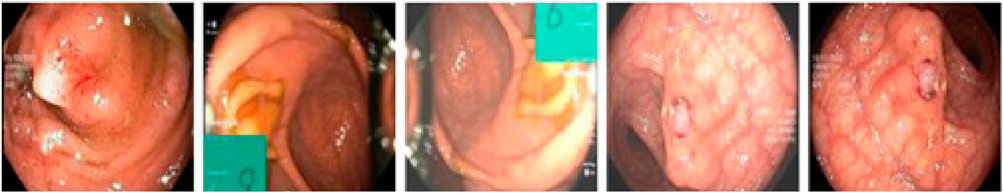
The SUN Colonoscopy Video Database is specifically designed for evaluating automated colorectal adenoma detection systems, with all images meticulously annotated by expert endoscopists. This dataset contains a total of 158,690 image frames, including 49,136 frames with annotated polyps (with precise bounding boxes) and 109,554 frames without polyps. These frames are derived from 100 fully annotated polyp samples. The dataset’s diversity and large-scale annotations make it an ideal choice for training and evaluating deep learning models, particularly in scenarios involving small target detection and challenging clinical conditions. Examples from the SUN Colonoscopy Video Database are shown in Figure 4.
4.1.2 PICCOLO widefield dataset
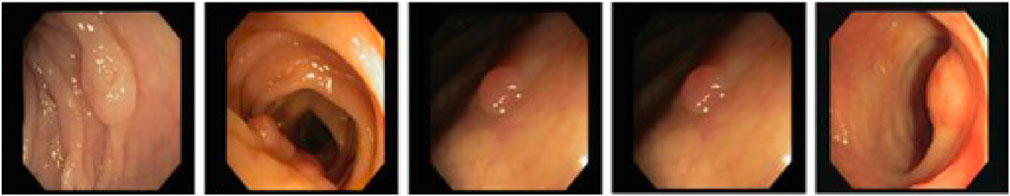
The PICCOLO Widefield Dataset is a publicly available dataset comprising 76 polyp lesions from 40 patients. Among these, 62 lesions include both White Light (WL) and Narrow Band Imaging (NBI) frames, while the remaining 14 lesions are represented by WL frames only. The dataset consists of a total of 3,433 images, including 2131 WL images and 1302 NBI images. The PICCOLO dataset is characterized by widefield colonoscopy images, which capture broader contextual information around polyps, aiding the model in learning both global and local features. The annotations, completed by expert endoscopists, are highly accurate and clinically reliable, making the dataset an essential tool for evaluating model performance in multimodal imaging scenarios. Examples from the PICCOLO Widefield Dataset are shown in Figure 5.
4.1.3 Etis-Larib dataset
The Etis-Larib Dataset is used exclusively in the testing phase and consists of images collected from real clinical environments. This dataset is highly challenging, as the polyps are often small, subtle, and located within complex backgrounds. It serves as a critical benchmark for evaluating the robustness of detection methods. In this study, the Etis-Larib dataset was used to compare the proposed CoroYOLO model with other detection algorithms, validating its performance in detecting small targets and handling complex medical imaging scenarios.
4.2 Implementation details
4.2.1 Experimental environment
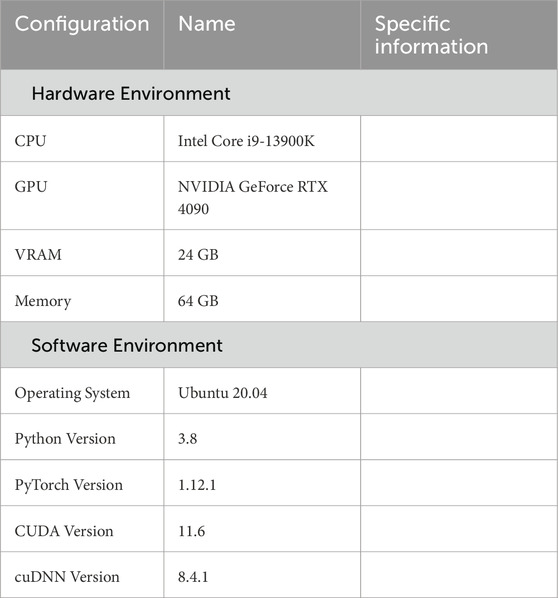
The experimental environment used in this study is shown in Table 1 including the hardware configuration and software platform. All experiments were conducted in a consistent environment to ensure reproducibility and fairness of the results.
4.2.2 Hyperparameter settings
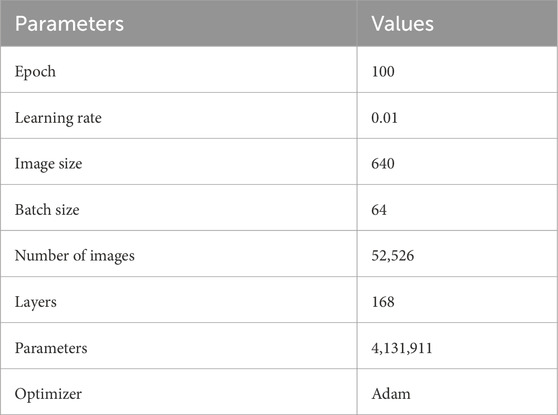
n this study, the hyperparameter settings for CoroYOLO were carefully tuned through multiple experiments to achieve an optimal balance between accuracy and efficiency. The learning rate and optimizer were selected to accelerate convergence while avoiding gradient oscillation, and the batch size and training epochs were configured considering hardware constraints and model complexity. Additionally, regularization parameters and the associated weights of the loss function were optimized to further enhance the robustness of the model. The specific hyperparameter settings are shown in Table 2.
4.2.3 Evaluation metrics
In this study, we adopted multiple evaluation metrics to comprehensively assess the performance of the model, including Precision (PR), Recall, F1 Score, and Mean Average Precision (mAP).
Precision (PR) measures the accuracy of the model’s predictions, indicating how many of the predicted positive samples are actual positives. A high precision score demonstrates the model’s ability to effectively control false positives.
Recall reflects the model’s ability to identify positive samples, indicating how many of the actual positive samples are correctly predicted as positives. A high recall score suggests the model is highly sensitive to detecting targets.
F1 Score is the weighted harmonic mean of precision and recall, balancing the trade-off between these two metrics. It is an essential metric when both prediction accuracy and detection capability are equally important.
Mean Average Precision (mAP) is a widely used evaluation standard in object detection tasks, measuring the overall detection performance of the model across different confidence thresholds. mAP takes into account the detection precision across multiple categories and serves as a core metric to evaluate the model’s overall performance.
4.2.4 Model training
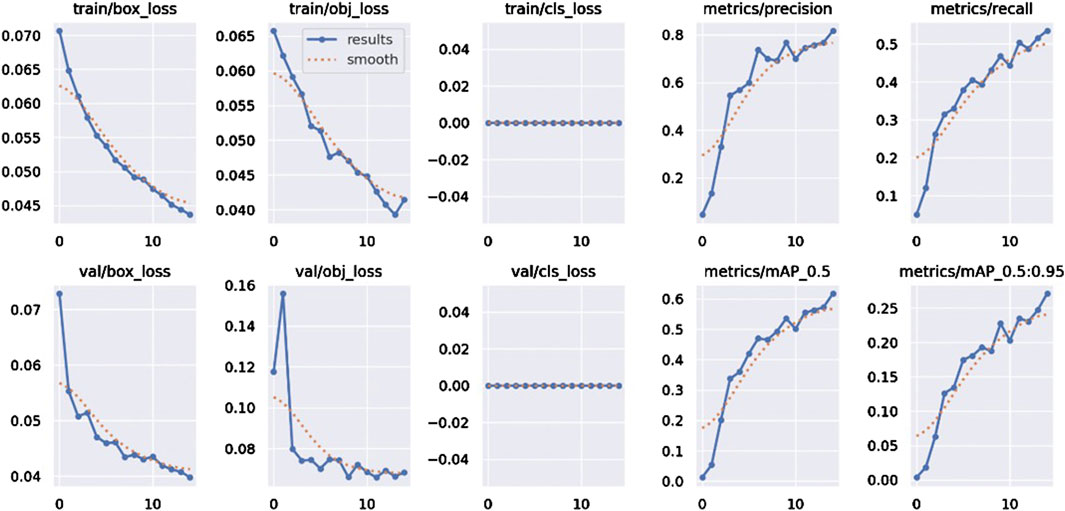
As shown in Figure 6, the training and validation processes demonstrate a good convergence trend, with the evolution of the loss functions and evaluation metrics effectively reflecting the optimization of the model’s performance.
During training, the box_loss (bounding box regression loss), obj_loss (object confidence loss), and cls_loss (classification loss) all show continuous decreases, indicating that the model is progressively reducing prediction errors in bounding box localization, object classification, and confidence estimation tasks. Notably, the rapid decrease in box_loss and obj_loss highlights the model’s ability to precisely locate spatial positions and effectively distinguish between foreground and background. Correspondingly, the validation losses val/box_loss and val/obj_loss also exhibit similar downward trends, aligning closely with the training losses, further demonstrating the model’s strong generalization ability and resistance to overfitting.
In terms of evaluation metrics, Precision and Recall gradually increase as the number of training epochs progresses, indicating ongoing improvements in the model’s accuracy and sensitivity in object detection. The significant increase in Precision reflects the model’s enhanced ability to control false positives, while the steady rise in Recall shows that the model’s ability to detect true positive targets is progressively improving.
The model’s detection performance at different Intersection over Union (IoU) thresholds is evaluated using mAP@0.5 and mAP@0.5:0.95. Both metrics exhibit a steady upward trend, with mAP@0.5 showing a faster growth rate, indicating that the model performs well under looser IoU thresholds. In contrast, the steady increase in mAP@0.5:0.95 suggests that the model maintains high accuracy even under stricter localization requirements. This demonstrates the model’s robustness and stability in multi-scale object detection tasks.
Algorithm 1 demonstrates the training and evaluation process of the CoroYOLO model. The algorithm outlines the steps for initializing the network, performing forward and backward propagation, computing the loss, and updating the model’s parameters.
4.3 Results
4.3.1 Comparison with state-of-the-art methods
As shown in Table 3, the CoroYOLO model clearly demonstrates superior performance compared to other advanced methods. In the evaluation on the Etis-Larib dataset, CoroYOLO achieved impressive results, with a precision of 96.31%, recall of 92.18%, F1 score of 93.15%, and mAP@0.5 of 98.89%. These results significantly outperform other models, such as YOLOv10, which achieved a precision of 89.65%, recall of 90.17%, F1 score of 91.32%, and mAP@0.5 of 90.28%. Compared to more advanced models like YOLOv8 and DETR, CoroYOLO shows clear advantages in both precision and recall, with a higher mAP@0.5 score. Moreover, CoroYOLO outperforms previous works based on CVC-ClinicDB and other datasets, such as Liu et al. [43] and Shen et al. [44]. It also surpasses traditional methods, including Wittenberg et al. [45], Wang et al. [46], and Qadir et al. [47], demonstrating its robustness and effectiveness in polyp detection in complex medical images. Figure 7 showcases the detection results of CoroYOLO on the Etis-Larib dataset, further validating the model’s outstanding performance in real-world applications.
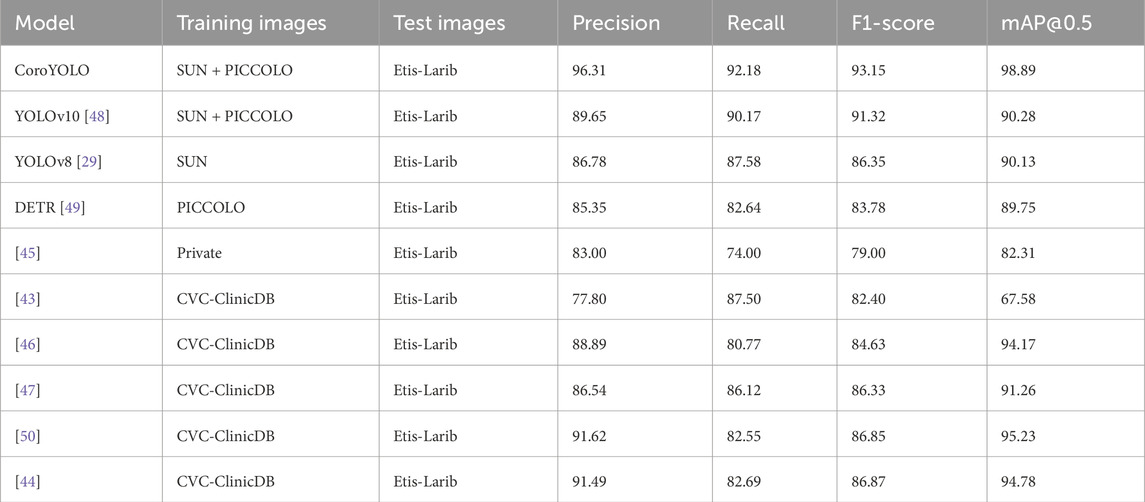
Table 3. Detection results of CoroYOLO on the Etis-Larib dataset, demonstrating its superior performance in identifying polyps in complex medical images.
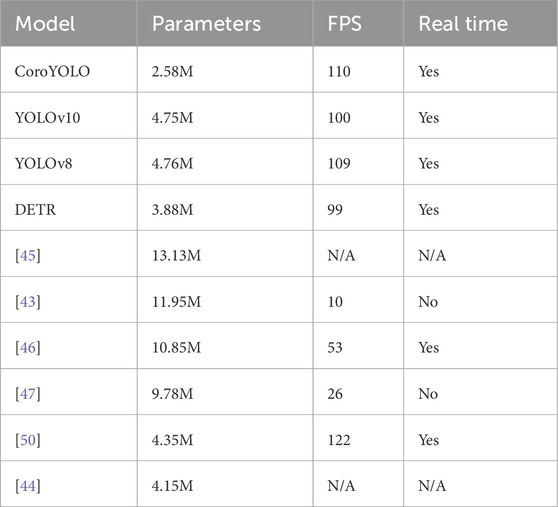
4.3.2 Computational complexity comparison
As shown in Table 4, CoroYOLO demonstrates excellent performance in terms of computational complexity and real-time capability. Compared to other models, CoroYOLO has fewer parameters while maintaining a high frame rate (FPS) and can efficiently run in real-time applications. Although other models such as YOLOv10 and YOLOv8 also perform well, CoroYOLO achieves a better balance between parameter count and processing speed. Additionally, some models like Liu et al. [43] and Qadir et al. [47] have larger computational costs and cannot achieve real-time inference, highlighting CoroYOLO’s advantage in efficiency.
4.4 Ablation study
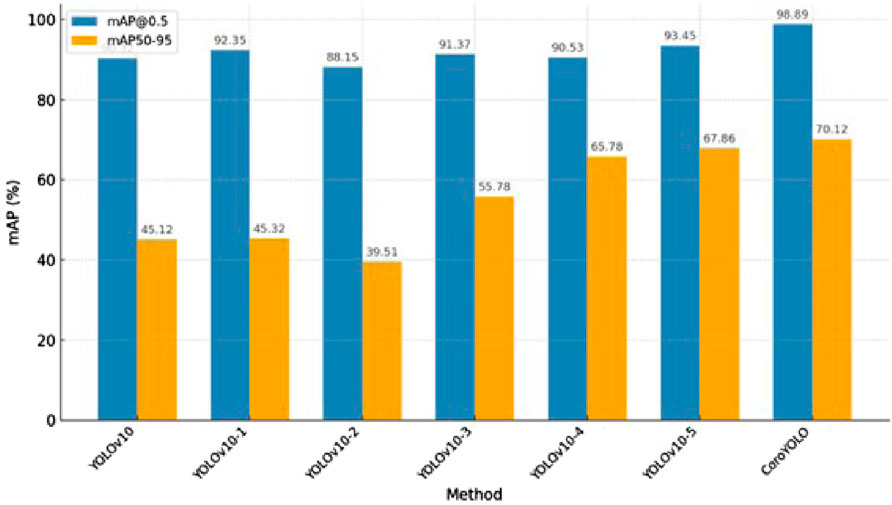
As shown in Table 5, the ablation study evaluates the contribution of different modules (TSMamblock, EMA, and GsConv) to the model’s performance. The baseline model YOLOv10, without any additional modules, achieves an mAP@0.5 of 90.32 and an mAP50-95 of 45.12, serving as the reference. When the EMA attention mechanism is added (YOLOv10-1), the mAP@0.5 improves to 92.35, although the increase in mAP50-95 is limited, indicating that EMA enhances the model’s focus on critical regions. Introducing the TSMamblock module (YOLOv10-2) results in an mAP@0.5 of 88.15 and an mAP50-95 of 39.51, demonstrating its effectiveness in capturing global contextual information, but its full potential is realized when combined with EMA.
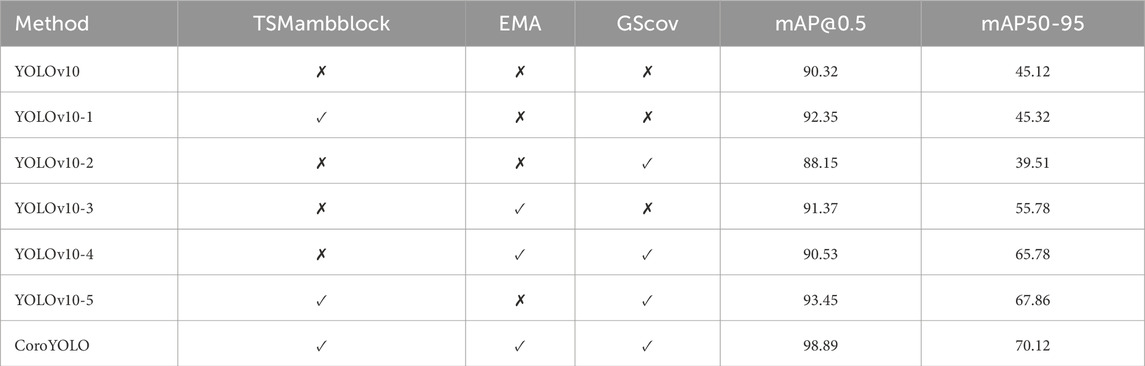
Table 5. Ablation study results showing the impact of different modules (TSMamblock, EMA, and GsConv) on the performance of CoroYOLO.
When TSMamblock and EMA are integrated (YOLOv10-3), the mAP@0.5 and mAP50-95 increase to 91.37 and 55.78, respectively, showing significant performance gains from their synergistic effect. Further inclusion of the GsConv module (YOLOv10-4) achieves an mAP@0.5 of 90.53 and an mAP50-95 of 65.78, validating the module’s efficiency in compressing feature channels and enhancing multi-scale feature modeling. The combination of all modules (YOLOv10-5) further improves the performance, reaching an mAP@0.5 of 93.45 and an mAP50-95 of 67.86, demonstrating the comprehensive optimization achieved by integrating these modules.
The final complete CoroYOLO model, integrating the TSMamblock, EMA, and GsConv modules, achieved the best performance with an mAP@0.5 of 98.89 and an mAP50-95 of 70.12. Figure 8 visualizes the table, providing a more intuitive representation of how CoroYOLO significantly improves the accuracy and robustness of object detection through the synergistic effects of its modules.
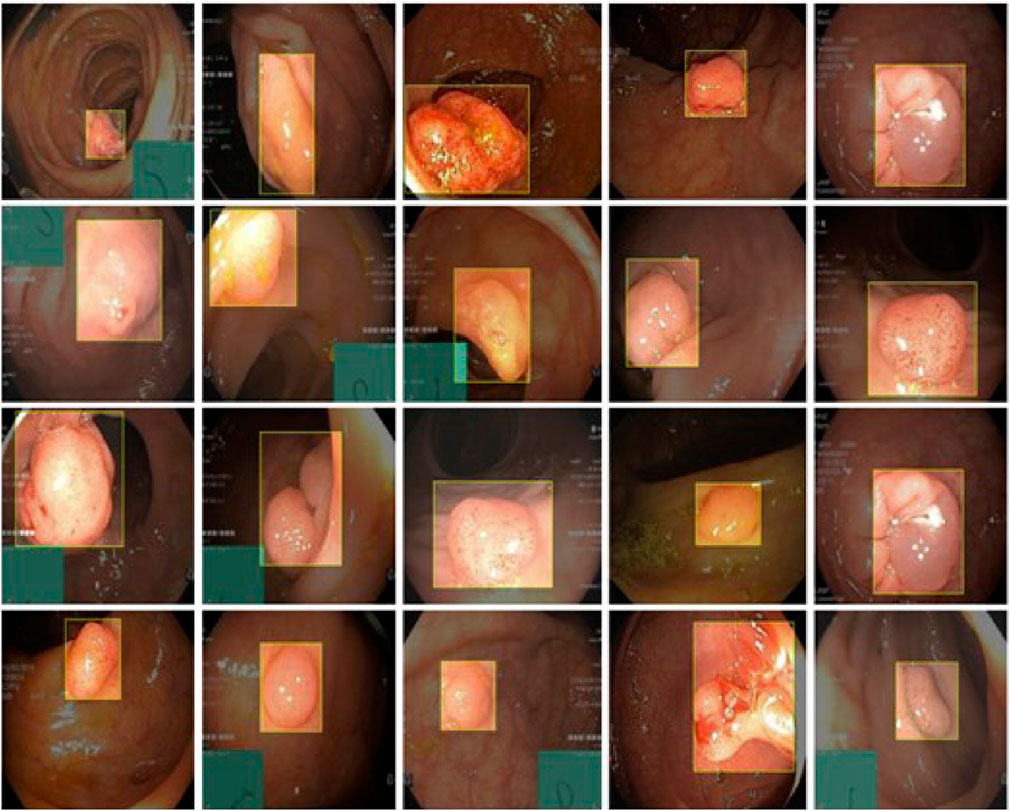
4.5 Small object detection
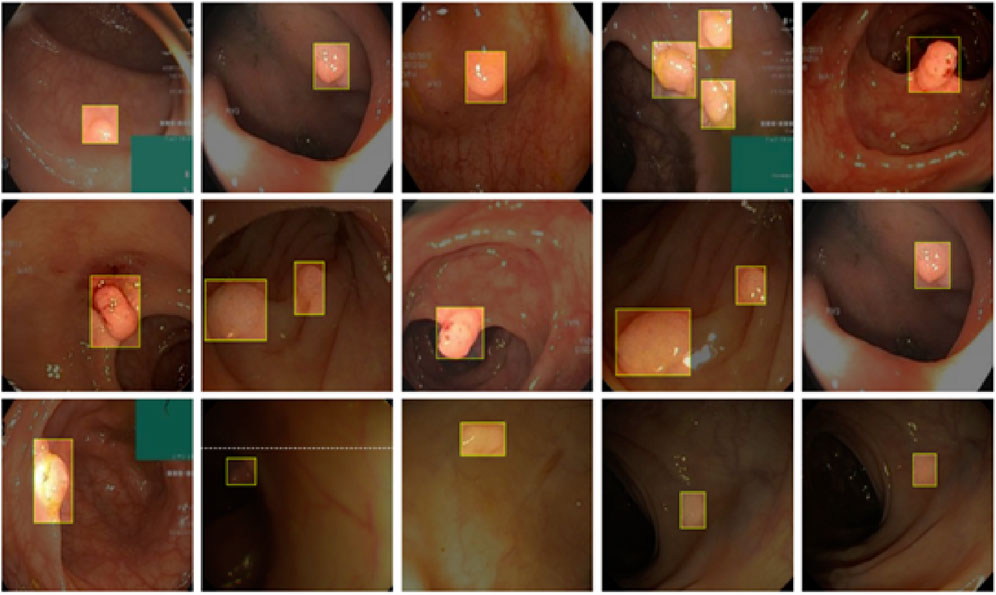
As shown in Figure 9, CoroYOLO demonstrates exceptional performance in small object detection tasks. By integrating the global modeling capability of the TSMamblock module and the EMA attention mechanism’s enhancement of key regions, the model is able to precisely locate small objects, reducing both missed detections and false positives. Additionally, the GsConv module further optimizes feature extraction and channel compression, enhancing adaptability to multi-scale targets. From the visualization results, it is evident that CoroYOLO exhibits high robustness and accuracy in small object detection tasks under complex backgrounds, significantly outperforming traditional methods and baseline models.
4.6 Limitations and future prospects
The proposed CoroYOLO model demonstrates excellent performance in object detection tasks within complex medical images; however, it still has certain limitations. First, the training and testing of the model primarily rely on the SUN, PICCOLO, and Etis-Larib datasets. While these datasets are representative to some extent, their diversity and scale are still limited, failing to comprehensively cover all clinical scenarios, such as more complex lighting conditions, images captured from different devices, and other types of lesion characteristics. Second, the current model has not undergone pruning optimization. Although it achieves high performance, its computational efficiency and hardware deployment can be further improved. This is particularly important for resource-constrained scenarios, where pruning could significantly reduce the model’s parameter size and inference time.
Future research can focus on two aspects to further enhance the model’s performance. On one hand, expanding the scale and diversity of the training and testing datasets by incorporating more real-world clinical data can improve the model’s generalization ability under various complex conditions. Specifically, the inclusion of multimodal data, such as Optical Coherence Tomography or ultrasound imaging, can further broaden the model’s applicability. On the other hand, pruning and quantization techniques can be applied to optimize the model’s computational efficiency by simplifying the network structure and reducing redundant computations. This would enable the model to run efficiently on embedded devices or mobile platforms, better meeting the demands of real-time clinical applications. These improvements would advance the practical use and development of CoroYOLO in the field of medical imaging.
5 Conclusion
This paper proposes a novel object detection model, CoroYOLO, specifically designed for colorectal cancer detection. By incorporating TSMamblock, EMA attention mechanism, and GsConv modules into the YOLOv10 framework, the model effectively enhances global modeling capabilities, attention to key regions, and multi-scale feature processing. CoroYOLO has undergone comprehensive training and evaluation on several medical imaging datasets (SUN, PICCOLO, and Etis-Larib), and experimental results demonstrate that it significantly outperforms existing mainstream methods in terms of precision, recall, F1-score, and mAP metrics. Furthermore, ablation studies validate the effectiveness and synergistic contribution of each module in improving model performance.
In future research, incorporating more real-world clinical data and optimizing the network structure can further enhance the model’s generalization ability and computational efficiency, providing stronger support for early CRC diagnosis and clinical applications. The introduction of CoroYOLO not only offers a new perspective for medical imaging detection tasks but also provides valuable insights for broader object detection applications.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
WC: Conceptualization, Formal Analysis, Data curation, Writing – original draft. FH: Investigation, Project administration, Writing – original draft, Methodology. YS: Visualization, Resources, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Jia Z, An J, Liu Z, Zhang F. Non-coding rnas in colorectal cancer: their functions and mechanisms. Front Oncol (2022) 12:783079. doi:10.3389/fonc.2022.783079
2. Davri A, Birbas E, Kanavos T, Ntritsos G, Giannakeas N, Tzallas AT, et al. Deep learning on histopathological images for colorectal cancer diagnosis: a systematic review. Diagnostics (2022) 12:837. doi:10.3390/diagnostics12040837
3. Bousis D, Verras G-I, Bouchagier K, Antzoulas A, Panagiotopoulos I, Katinioti A, et al. The role of deep learning in diagnosing colorectal cancer. Gastroenterol Review/Przegląd Gastroenterologiczny (2023) 18:266–73. doi:10.5114/pg.2023.129494
4. Foersch S, Glasner C, Woerl A-C, Eckstein M, Wagner D-C, Schulz S, et al. Multistain deep learning for prediction of prognosis and therapy response in colorectal cancer. Nat Med (2023) 29:430–9. doi:10.1038/s41591-022-02134-1
5. Aruna Kumari A, Bhagat A, Kumar Henge S. Classification of diabetic retinopathy severity using deep learning techniques on retinal images. Cybernetics Syst (2024) 1–25. doi:10.1080/01969722.2024.2375148
6. Daghrir J, Tlig L, Bouchouicha M, Litaiem N, Zeglaoui F, Sayadi M. Texture characterization fuzzy logic-based model for melanoma diagnosis. Cybernetics Syst (2023) 1–19. doi:10.1080/01969722.2023.2247262
7. DeMatteo C, Jakubowski J, Stazyk K, Randall S, Perrotta S, Zhang R. The headaches of developing a concussion app for youth: balancing clinical goals and technology. Int J E-Health Med Commun (Ijehmc) (2024) 15:1–20. doi:10.4018/ijehmc.352514
8. Yu G, Sun K, Xu C, Shi X-H, Wu C, Xie T, et al. Accurate recognition of colorectal cancer with semi-supervised deep learning on pathological images. Nat Commun (2021) 12:6311. doi:10.1038/s41467-021-26643-8
9. Kiehl L, Kuntz S, Höhn J, Jutzi T, Krieghoff-Henning E, Kather JN, et al. Deep learning can predict lymph node status directly from histology in colorectal cancer. Eur J Cancer (2021) 157:464–73. doi:10.1016/j.ejca.2021.08.039
10. Almayyan WI, AlGhannam BA. Detection of kidney diseases: importance of feature selection and classifiers. Int J E-Health Med Commun (Ijehmc) (2024) 15:1–21. doi:10.4018/ijehmc.354587
11. Echle A, Laleh NG, Schrammen PL, West NP, Trautwein C, Brinker TJ, et al. Deep learning for the detection of microsatellite instability from histology images in colorectal cancer: a systematic literature review. ImmunoInformatics (2021) 3:100008. doi:10.1016/j.immuno.2021.100008
12. Tsai M-J, Tao Y-H. Deep learning techniques for the classification of colorectal cancer tissue. Electronics (2021) 10:1662. doi:10.3390/electronics10141662
13. Sun C, Li B, Wei G, Qiu W, Li D, Li X, et al. Deep learning with whole slide images can improve the prognostic risk stratification with stage iii colorectal cancer. Computer Methods Programs Biomed (2022) 221:106914. doi:10.1016/j.cmpb.2022.106914
14. Ramola A, Shakya AK, Van Pham D. Study of statistical methods for texture analysis and their modern evolutions. Eng Rep (2020) 2:e12149. doi:10.1002/eng2.12149
15. Shakya AK, Ramola A, Vidyarthi A. Modeling of texture quantification and image classification for change prediction due to covid lockdown using skysat and planetscope imagery. Model Earth Syst Environ (2022) 8:2767–92. doi:10.1007/s40808-021-01258-6
16. Shakya AK, Vidyarthi A. Comprehensive study of compression and texture integration for digital imaging and communications in medicine data analysis. Technologies (2024) 12:17. doi:10.3390/technologies12020017
17. Shakya AK, Ramola A, Pandey DC. Polygonal region of interest based compression of dicom images. In: 2017 international conference on computing, communication and automation (ICCCA). IEEE (2017). p. 1035–40.
18. Liu X, Zhang C, Zhang L. Vision mamba: a comprehensive survey and taxonomy. (2024) arXiv preprint arXiv:2405.04404.
19. Wang F, Wang J, Ren S, Wei G, Mei J, Shao W, et al. Mamba-r: vision mamba also needs registers. (2024) arXiv preprint arXiv:2405.14858.
20. Xing X, Wang B, Ning X, Wang G, Tiwari P. Short-term od flow prediction for urban rail transit control: a multi-graph spatiotemporal fusion approach. Inf Fusion (2025) 118:102950. doi:10.1016/j.inffus.2025.102950
21. Zhang L, Liu J, Wei Y, An D, Ning X. Self-supervised learning-based multi-source spectral fusion for fruit quality evaluation: a case study in mango fruit ripeness prediction. Inf Fusion (2025) 117:102814. doi:10.1016/j.inffus.2024.102814
22. Hatamizadeh A, Kautz J Mambavision: a hybrid mamba-transformer vision backbone. (2024) arXiv preprint arXiv:2407.08083.
23. Qiao J, Liao J, Li W, Zhang Y, Guo Y, Wen Y, et al. Hi-mamba: hierarchical mamba for efficient image super-resolution. arXiv preprint arXiv:2410 (2024) :10140.
24. Huang J, Yu X, An D, Ning X, Liu J, Tiwari P. Uniformity and deformation: a benchmark for multi-fish real-time tracking in the farming. Expert Syst Appl (2025) 264:125653. doi:10.1016/j.eswa.2024.125653
25. Zhang H, Yu L, Wang G, Tian S, Yu Z, Li W, et al. Cross-modal knowledge transfer for 3d point clouds via graph offset prediction. Pattern Recognition (2025) 162:111351. doi:10.1016/j.patcog.2025.111351
26. Liu Y, Tian Y, Zhao Y, Yu H, Xie L, Wang Y, et al. Vmamba: visual state space model. Adv Neural Inf Process Syst (2024) 37:103031–63.
27. Ma J, Li F, Wang B. U-mamba: enhancing long-range dependency for biomedical image segmentation. (2024) arXiv preprint arXiv:2401.04722.
28. Haq I, Mazhar T, Asif RN, Ghadi YY, Ullah N, Khan MA, et al. Yolo and residual network for colorectal cancer cell detection and counting. Heliyon (2024) 10:e24403. doi:10.1016/j.heliyon.2024.e24403
29. Lalinia M, Sahafi A. Colorectal polyp detection in colonoscopy images using yolo-v8 network. Signal Image Video Process. (2024) 18:2047–58. doi:10.1007/s11760-023-02835-1
30. Zhang B, Wang Z, Zhang Y, Liu P. Ssm-yolo: lesion detection algorithm for colorectal polyps. In: 2024 IEEE 7th international conference on electronic information and communication Technology (ICEICT). IEEE (2024). p. 475–81.
31. Murugesan M, Arieth RM, Balraj S, Nirmala R. Colon cancer stage detection in colonoscopy images using yolov3 msf deep learning architecture. Biomed Signal Process Control (2023) 80:104283. doi:10.1016/j.bspc.2022.104283
32. Reddy JSC, Venkatesh C, Sinha S, Mazumdar S. Real time automatic polyp detection in white light endoscopy videos using a combination of yolo and deepsort. In: 2022 1st international conference on the paradigm shifts in communication, embedded systems, machine learning and signal processing (PCEMS). IEEE (2022). p. 104–6.
33. Liu Q, Liu Y, Lin D. Revolutionizing target detection in intelligent traffic systems: yolov8-snakevision. Electronics (2023) 12:4970. doi:10.3390/electronics12244970
34. Wang Z, Li C, Xu H, Zhu X. Mamba yolo: ssms-based yolo for object detection. (2024) arXiv preprint arXiv:2406.05835.
35. Ma H, Lei S, Celik T, Li H-C. Fer-yolo-mamba: facial expression detection and classification based on selective state space. (2024) arXiv preprint arXiv:2405.01828.
36. Zhao Z, He P. Yolo-mamba: object detection method for infrared aerial images. Signal Image Video Process. (2024) 18:8793–803. doi:10.1007/s11760-024-03507-4
37. Wang H, He Q, Peng J, Yang H, Chi M, Wang Y. Mamba-yolo-world: marrying yolo-world with mamba for open-vocabulary detection. In: ICASSP 2025-2025 IEEE international conference on acoustics, speech and signal processing (ICASSP). IEEE (2025). p. 1–5.
38. Zhou W, Kamata S-i., Wang H, Wong MS, Hou HC. Mamba-in-mamba: centralized mamba-cross-scan in tokenized mamba model for hyperspectral image classification. Neurocomputing (2025) 613:128751. doi:10.1016/j.neucom.2024.128751
39. Ouyang D, He S, Zhang G, Luo M, Guo H, Zhan J, et al. Efficient multi-scale attention module with cross-spatial learning. In: ICASSP 2023-2023 IEEE international conference on acoustics, speech and signal processing (ICASSP). IEEE (2023). p. 1–5.
40. Misawa M, Kudo S-e., Mori Y, Hotta K, Ohtsuka K, Matsuda T, et al. Development of a computer-aided detection system for colonoscopy and a publicly accessible large colonoscopy video database (with video). Gastrointest Endosc (2021) 93:960–7.e3. doi:10.1016/j.gie.2020.07.060
41. Peralta LFS, Pagador JB, Picón A, Calderón ÁJ, Polo F, Andraka N, et al. Piccolo white-light and narrow-band imaging colonoscopic dataset. PICCOLO White-Light and Narrow-Band Imaging Colonoscopic Dataset (2020).
42. Nogueira-Rodríguez A, Domínguez-Carbajales R, López-Fernández H, Iglesias Á, Cubiella J, Fdez-Riverola F, et al. Deep neural networks approaches for detecting and classifying colorectal polyps. Neurocomputing (2021) 423:721–34.
43. Liu X, Guo X, Liu Y, Yuan Y. Consolidated domain adaptive detection and localization framework for cross-device colonoscopic images. Med image Anal (2021) 71:102052. doi:10.1016/j.media.2021.102052
44. Shen Z, Fu R, Lin C, Zheng S. Cotr: convolution in transformer network for end to end polyp detection. In: 2021 7th international conference on computer and communications (ICCC). IEEE (2021). p. 1757–61.
45. Wittenberg T, Zobel P, Rathke M, Mühldorfer S. Computer aided detection of polyps in whitelight-colonoscopy images using deep neural networks. Curr Dir Biomed Eng (2019) 5:231–4. doi:10.1515/cdbme-2019-0059
46. Wang D, Zhang N, Sun X, Zhang P, Zhang C, Cao Y, et al. Afp-net: realtime anchor-free polyp detection in colonoscopy. In: 2019 IEEE 31st international conference on tools with artificial intelligence (ICTAI). IEEE (2019). p. 636–43.
47. Qadir HA, Shin Y, Solhusvik J, Bergsland J, Aabakken L, Balasingham I. Toward real-time polyp detection using fully cnns for 2d Gaussian shapes prediction. Med Image Anal (2021) 68:101897. doi:10.1016/j.media.2020.101897
48. Wang A, Chen H, Liu L, Chen K, Lin Z, Han J, et al. Yolov10: real-time end-to-end object detection. Adv Neural Inf Process Syst (2024) 37:107984–8011.
49. Mohammad F, Sharma V, Das PK. Polyp detection in colonoscopy images using improved deformable detr. In: TENCON 2022-2022 IEEE region 10 conference (TENCON). IEEE (2022). p. 1–6.
Keywords: colorectal cancer detection, state space model, deep learning, Mamba, YOLOv10
Citation: Chen W, Hou F and Shen Y (2025) CoroYOLO: a novel colorectal cancer detection method based on the Mamba framework. Front. Phys. 13:1597378. doi: 10.3389/fphy.2025.1597378
Received: 21 March 2025; Accepted: 30 April 2025;
Published: 22 May 2025.
Edited by:
Bhishma Karki, National Research Council Nepal, NepalReviewed by:
Yasuko Kawahata, Rikkyo University, JapanAmit Shakya, Sant Longowal Institute of Engineering and Technology, India
Copyright © 2025 Chen, Hou and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenfei Chen, MjAyMjA5MDA0NkBidWN0LmVkdS5jbg==
 Wenfei Chen
Wenfei Chen Fengrui Hou1
Fengrui Hou1