- 1Department of Plant Food Products and Biofortification, Institute of Food Biotechnology and Genomics NAS of Ukraine, Kyiv, Ukraine
- 2Department of Biology, University of York, York, United Kingdom
Salinity is a major threat to modern agriculture causing inhibition and impairment of crop growth and development. Here, we not only review recent advances in salinity stress research in plants but also revisit some basic perennial questions that still remain unanswered. In this review, we analyze the physiological, biochemical, and molecular aspects of Na+ and Cl− uptake, sequestration, and transport associated with salinity. We discuss the role and importance of symplastic versus apoplastic pathways for ion uptake and critically evaluate the role of different types of membrane transporters in Na+ and Cl− uptake and intercellular and intracellular ion distribution. Our incomplete knowledge regarding possible mechanisms of salinity sensing by plants is evaluated. Furthermore, a critical evaluation of the mechanisms of ion toxicity leads us to believe that, in contrast to currently held ideas, toxicity only plays a minor role in the cytosol and may be more prevalent in the vacuole. Lastly, the multiple roles of K+ in plant salinity stress are discussed.
General Aspects of Plant Salt Stress
Soil salinity is one of the most important global problems that negatively affects crop productivity. Salinity impairs plant growth and development via water stress, cytotoxicity due to excessive uptake of ions such as sodium (Na+) and chloride (Cl−), and nutritional imbalance. Additionally, salinity is typically accompanied by oxidative stress due to generation of reactive oxygen species (ROS) (Tsugane et al., 1999; Hernandez et al., 2001; Isayenkov, 2012).
Plant responses to salinity have been divided into two main phases. An ion-independent growth reduction, which takes place within minutes to days, causes stomatal closure and inhibition of cell expansion mainly in the shoot (Munns and Passioura, 1984; Munns and Termaat, 1986; Rajendran et al., 2009). A second phase takes place over days or even weeks and pertains to the build-up of cytotoxic ion levels, which slows down metabolic processes, causes premature senescence, and ultimately cell death (Munns and Tester, 2008; Roy et al., 2014). Tolerance to both types of stress is governed by a multitude of physiological and molecular mechanisms: osmotic tolerance, ionic tolerance, and tissue tolerance (Rajendran et al., 2009; Roy et al., 2014). Osmotic tolerance initiates relatively quickly and includes a rapid decrease in stomatal conductance to preserve water. It employs fast long-distance (root to shoot) signaling mechanisms (Ismail et al., 2007; Maischak et al., 2010; Roy et al., 2014), which largely do not discriminate between osmotic effects created by NaCl, KCl, mannitol, or polyethylene glycol (Yeo et al., 1991; Chazen et al., 1995).
The entering of salt into the root system triggers activation of several signal cascades that generate ionic tolerance by restricting (net) Na+ influx into the root and reduce (net) Na+ translocation. Lastly, tissue tolerance is enhanced by compartmentation of toxic ions into vacuoles to avoid detrimental effects on cytoplasmic processes. The above strategies have been observed in many types of plant, and differences in tolerance between glycophytes and halophytes are predominantly due to the greater robustness of the employed mechanisms in the latter, rather than a qualitative difference (Flowers and Colmer, 2008, 2015; Maathuis et al., 2014). Most of these aspects have been covered in previous reviews; here, we will focus particularly on the quantitative role of symplastic and the apoplastic pathways regarding salt influx, an evaluation of how mechanisms of chloride uptake, transport, and distribution, compare to that of sodium and a critical re-evaluation of ion toxicity.
How does Salt Enter the Plant?
Salinity creates a dilemma for plants; increased levels of inorganic minerals in the environment create osmotic and water stress but at the same time provide cheap osmoticum to lower the cell osmotic potential and hence prevent water loss. In spite of decades of research, one of the most enigmatic questions relating to plant salt stress remains the mechanism(s) by which Na+ and Cl− enter roots.
Ion uptake can occur via the symplastic and the apoplastic pathway (Figure 1; Gao et al., 2007; Negrão et al., 2011; Maathuis et al., 2014). The apoplastic pathway is a direct flow continuum between the outside and the xylem (Yeo et al., 1987; Anil et al., 2005; Krishnamurthy et al., 2009). In most conditions, the contribution of this “bypass” flux is less than 1% of the transpirational volume flow (Hanson et al., 1985; Moon et al., 1986; Yeo et al., 1987). Nevertheless, this can be much greater when transpirational demand is high (Pitman, 1982; Sanderson, 1983). In rice, it is particularly pronounced and could be responsible for up to 50% of total Na+ uptake (Yeo et al., 1987; Malagoli et al., 2008; Krishnamurthy et al., 2009; Kronzucker and Britto, 2011). Significant apoplastic Na+ flux has also been reported in other species (Peterson et al., 1981, 1986), and recently, it was shown that up to 50% of Cl− translocation to rice shoots is also apoplastic (Shi et al., 2013). Although particularly pronounced in rice, the combined data suggest that nonsymplastic Na+ and Cl− uptake may be very relevant in monocots. However, solute permeability coefficients in Arabidopsis roots are not that different from those in rice (Ranathunge and Schreiber, 2011), and detailed experimentation is needed to test the contribution of the apoplastic pathway in this model and other dicots (Figure 1).
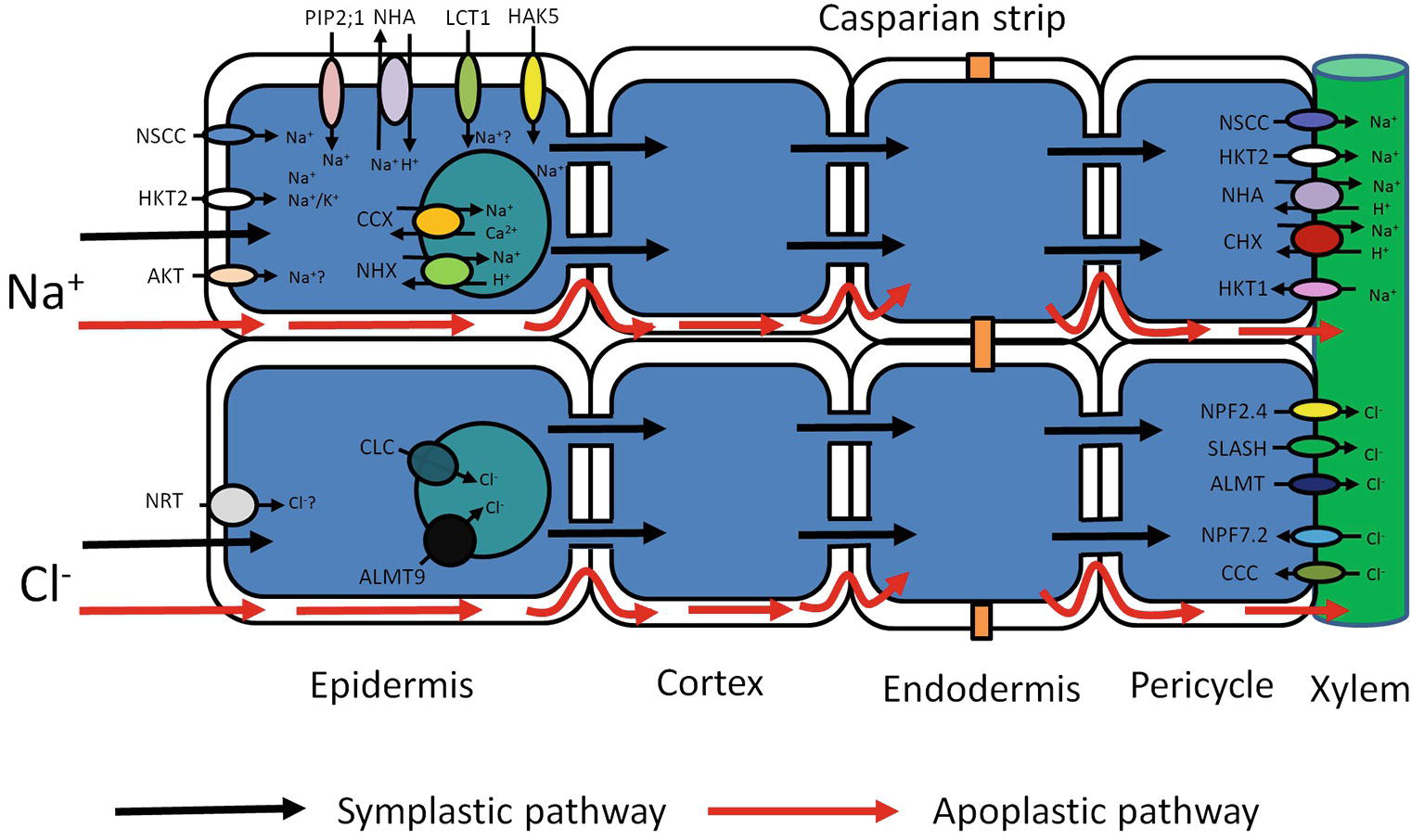
Figure 1. Schematic representation of possible transport pathways for Na+ and Cl− uptake and their cellular and long-distance distribution. Red arrows represent Na+ and Cl− entry sites and route through cell walls – apoplastic bypass flow. Black arrows represent Na+ and Cl− entry sites and cytoplasmic route through plasma membrane-symplastic pathway. Various transporters (AKT, HKT2, NSCC, PIP2;1, NHA, LCT1, HAK5) may be involved in Na+ uptake and movement through the plasma membrane. Compartmentalization of Na+ in vacuoles is mediated by tonoplast transporters (CCX, NHX). The further Na+ redistribution over long distances may rely on members of several membrane transporter families (NSCC, HKT, NHA, CHX). Cl− entry to the root cells through the plasma membrane may be mediated by Cl−/H+ co-transporter NRT. Vacuolar Cl− sequestration may possibly be performed by two anion tonoplast transporters (ALMT and CLC). Cl− membrane transport over long distances may be conducted by membrane transporters from different protein families (NPF, SLASH, ALMT, NPF, CCC). AKT, Arabidopsis K+ transporter; HKT, High-affinity K+ transporter Type; NSCC, Nonselective cation channels; PIP 2,1, Plasma membrane intrinsic protein (Aquaporin); NHA, Na+/H+ antiporter (SOS1); LCT1, Low-affinity cation transporter; HAK, High-affinity K+ uptake transporter; CHX, cation/H+ exchanger; NHX, Na+/H+ exchanger; NRT, Nitrate transporter; ALMT, Aluminum-activated malate transporter; CLC, Chloride channel; NPF, Nitrate transporter 1/peptide transporter; CCC, Cation/chloride cotransporter; SLASH, Anion channel associated homolog 1.
Net uptake via the symplastic pathway of Na+ (Cl−) into roots is assumed to be catalyzed by a specific complement of transporters (Figure 1). Evidence points to a large number of different systems, but their relative contribution, and therefore physiological relevance, is often unclear. Nonselective cation channels (NSCCs) are encoded by two gene families: glutamate receptor-like channels (GLRs) and cyclic nucleotide-gated channels (CNGCs) and blocked by Ca2+ (Leng et al., 2002; Demidchik et al., 2004, 2018; Demidchik and Maathuis, 2007). The apoplastic Ca2+ concentration in root cells is probably in the region of 0.2–0.4 mM (Legué et al., 1997), which is enough to reduce NSCC-mediated flux by 30–50% (Essah et al., 2003). The remaining flux can be further diminished not only by a number of channel blockers like Gd3+ and La3+ (Demidchik and Maathuis, 2007) but also by organic compounds like cyclic GMP. Thus, in plants like Arabidopsis, it appears that a large fraction of inward Na+ flux is carried by NSCCs but either the genetic identity of the contributing channels is obscure or their putative role has not been quantified. For example, though AtCNGC3 channels impacted on salt-related growth (Gobert et al., 2006), whether they directly affected Na+ uptake was not measured.
In monocotyledonous plants, the situation is likely to be different. In contrast to Arabidopsis, which contains only the subclass 1, Na+ selective, AtHKT1 isoform, monocots have multiple HKT isoforms. Arabidopsis HKT1 functions in long-distance transport of Na+ via xylem and phloem (Berthomieu et al., 2003; Sunarpi et al., 2005), but in several cereals HKTs can mediate Na+ uptake: In rice, OsHKT2;1 catalyzes Na+ uptake in low K+, low Na+ (<2 mM) conditions (Horie et al., 2007). Overexpression of HvHKT2;1 in barley causes increased Na+ uptake in salt stress conditions (Mian et al., 2011) with a Km for Na+ transport in the low-affinity region (3–6 mM). Similarly, altered expression of TaHKT2;1 in wheat affected Na+ accumulation in the low-affinity range (Laurie et al., 2002), but detailed flux studies are lacking (Figure 1).
Intriguingly, electrophysiological experiments in Xenopus oocytes showed considerable Na+ conductance when heterologously expressing the aquaporin AtPIP2;1 from Arabidopsis (Byrt et al., 2017). This interesting phenomenon was PIP isoform specific. A dual ion and water conducting capability, as suggested for AtPIP2;1, could couple ion and water flux, which would have obvious physiological relevance. However, in intact roots, unidirectional Na+ influx is typically in the range of 20–200 μmol gFW−1 h−1 (Kronzucker and Britto, 2011). Using a conversion factor of 5 × 10−4 m2 gFW−1 (Ahmad et al., 2015), this equates to 0.1–1 mol m−2 s−1. In comparison, PIP2;1 generated fluxes in oocytes were in the order of 0.2–0.5 μmol m−2 s−1 (Byrt et al., 2017), ~6 orders of magnitude smaller. Thus, the extent to which aquaporins contribute to Na+ uptake in intact plants appears to be negligible (Figure 1).
The low-affinity cation transporter LCT1 (Schachtman et al., 1997; Clemens et al., 1998; Kronzucker and Britto, 2011), when expressed in yeast, functions as a nonselective cation carrier and is capable of transporting K+, Rb+, Na+, and Ca2+ (Schachtman et al., 1997; Clemens et al., 1998). Moreover, its expression in yeast resulted in increased salt sensitivity (Amtmann et al., 2001), promoting the hypothesis that LCT1 could mediate Na+ uptake in planta. Nevertheless, more recent analyses suggest that LCT1 is not directly involved in Na+ transport (Figure 1; Plett and Moller, 2010).
A combination of high salinity and low K+ could cause Na/K ratios over 103 fold, a value that exceeds the K/Na selectivity of many K+ channels (Maathuis, 2014). Shaker type K+ channels, such as KAT1 and AKT1, are involved in K+ uptake and, on the basis of detailed electrophysiological studies, were deemed not to participate in Na+ transport (Schachtman et al., 1991; Amtmann and Sanders, 1998). However, later studies found that in intact tissue the Na-influx inhibitor profile mostly aligned with that of AKT1 type channels in the halophytic plant Suaeda maritima (Wang et al., 2007) and in rice (Kader and Lindbergh, 2005). Data from the halophyte study showed 30–40% reduction in unidirectional Na+ influx in the presence of “classical” K+ channel blockers such as Cs, Ba2+, and TEA, suggesting that up to 30–40% of Na+ influx could occur via AKT1 type channels. However, these types of studies are notoriously difficult to interpret because most blockers show only limited selectivity. Furthermore, flux assays did not show any lower Na+ influx in akt1 KO mutants compared to WT Arabidopsis (Figure 1; Essah et al., 2003).
In contrast to Na+, Cl− is an essential nutrient for plants. It has been postulated that Cl− is transported into the cell by a H+/Cl− symport, but its molecular nature is unknown. When plants are exposed to salinity, the external [Cl−] may be sufficiently high for a fraction of Cl− to enter passively through anion channels, but the relevant transport mechanism is this case too is unknown. Another class of potential Cl− transporters is the cation chloride cotransporters (CCCs), a notion that has so far received little attention but is supported by the observation that 100 μM bumetanide drastically reduces Na+ uptake in Suaeda maritima (Zhang et al., 2011). In Arabidopsis, AtCCC has been studied in some detail, showing that it is expressed in root and shoot tissues and most likely involved in coordinated K+, Na+, and Cl− symport (Colmenero-Flores et al., 2007; Zhang et al., 2010). However, loss of function in AtCCC led to an increase in Cl− uptake (Colmenero-Flores et al., 2007), arguing against a role of AtCCC in Cl− uptake (Figure 1). This study indicates that CCC transporters from grapevine and Arabidopsis are targeted to the Golgi and Trans-Golgi network and indirectly influence long-distance ion transport and plant salt tolerance (Henderson et al., 2015). According to these data, the AtCCC like VvCCC is involved in the transport of Na+, K+, Cl− and contributes to Na+ and Cl− homeostasis (Henderson et al., 2015).
In summary, for the majority of species and conditions, there is considerable evidence suggesting that NSCCs are a main pathway for Na+ influx into roots in glycophytic plants but these may subsume multiple channels from multiple channel families. This complexity impedes the construction of a detailed picture regarding “who does what” and by how much. In monocots, members of the HKT family could contribute to Na+ uptake. The usage of mutants should allow us to make progress regarding Cl− uptake. Existing data show that for aquatic plants like rice with its specific root anatomy, the apoplastic bypass of Na+ and Cl− can be considerable.
Early Components of Salinity Sensing in Plants
The plant response to salinity is complex but presumably includes some mechanism to report increasing levels of ions, either in the external medium or within the symplast. However, how Na+ or Cl− is sensed by plants remains unknown. In animal systems, primary Na+ sensors typically rely on functioning of specific Na+ selective ion channels with Na+ binding sites that modulate the gate and thus are capable of functioning as reporters of body fluid Na+ levels. In other cell types such as taste buds, Enac-type Na+ channels cause depolarizations proportionate to the amount of Na+ that is present (Maathuis, 2014) while sensory cells of nematode cilia have “transmembrane channel like” (TMC) channels activated by Na+ concentration higher than 140 mM causing an avoidance reaction (Chatzigeorgiou et al., 2013).
Thus, primary sensors in animals typically rely on Na+ specific binding sites that modulate transporter activity. As yet, no similar mechanisms have been identified in plant species but other, rapid responses such as salt-induced membrane depolarization and Ca2+ signals could form early components of salt sensing relays. However, membrane depolarizations do not confer any specificity so are unlikely to be physiologically relevant. Increases in extracellular NaCl cause rapid Ca2+ elevation in the cytosol (Knight et al., 1997), but these are often similar to signals induced by equiosmolar levels of osmotica such as mannitol. However, in some cases, the Ca2+ signals are salt specific (e.g. Choi et al., 2014), although tests for ionic specificity are usually lacking. Unfortunately, the upstream components of the Ca2+ signal are entirely unknown. New insights such as those from Choi et al. (2016) show that long-distance Ca2+ waves in response to salinization might form useful tools to devise mutant screens that could point to upstream components. Reactive oxygen species (ROS) may constitute another potential component upstream of the Ca2+ signal: Annexin1 (AtANN1) from Arabidopsis thaliana responds to high extracellular NaCl by mediating ROS-activated Ca2+ influx through the plasma membrane of plant cells (Laohavisit et al., 2013). Thus, annexin 1 could be an early key component of root cell adaptation to salinity (Laohavisit et al., 2013). Downstream, Ca2+-dependent signaling can be propagated by calcium-dependent protein kinases (CDPKs) and calcineurin B-like proteins (CBLs) (Weinl and Kudla, 2008) with CBL-interacting protein kinases (CIPKs) (Boudsocq and Sheen, 2013), which in turn modulate protein activity and gene transcription (Figure 2).
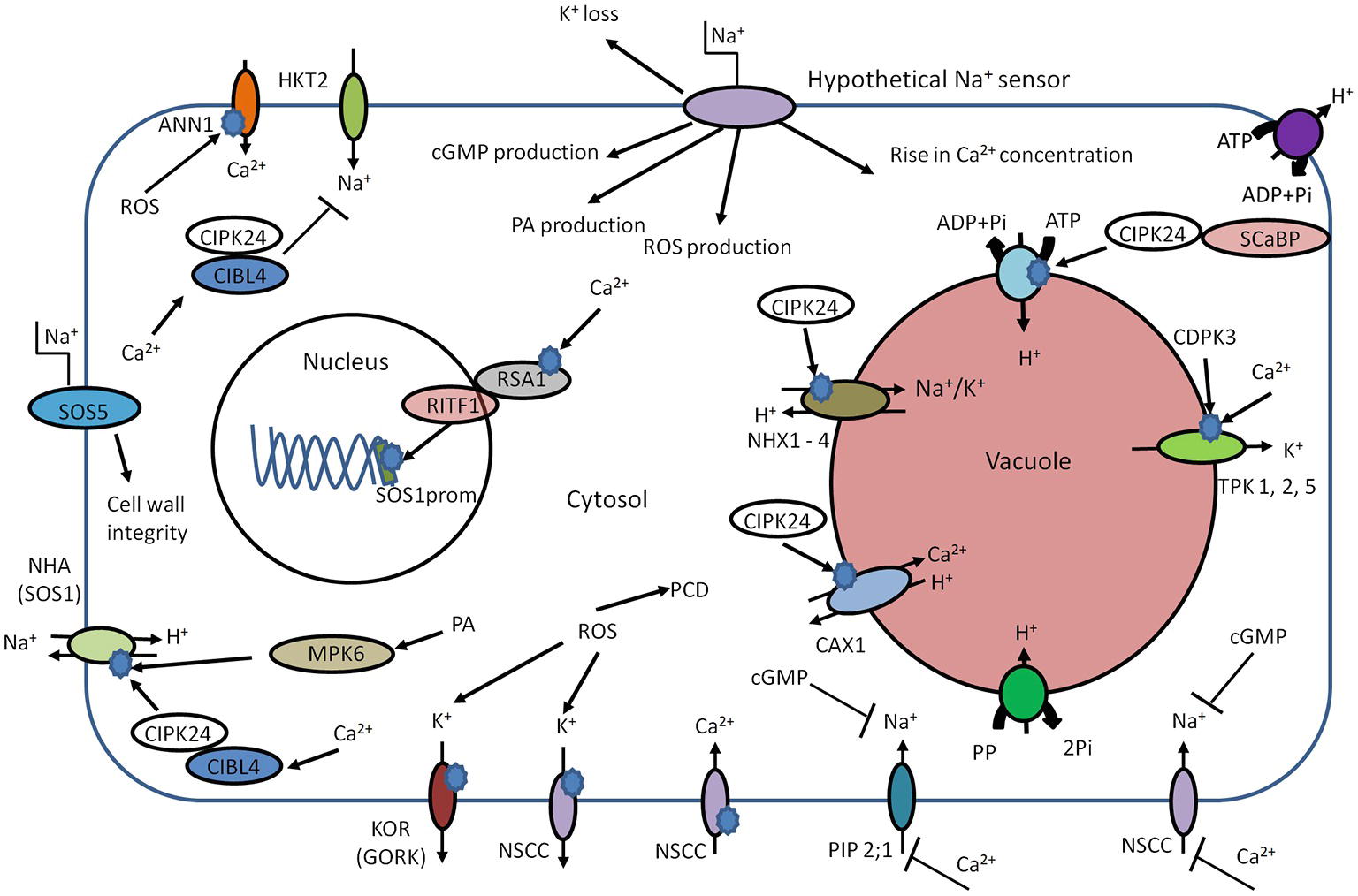
Figure 2. Schematic overview of early components involved in salt sensing. High external Na+ concentration leads to elevation of intracellular Ca2+, phosphatidic acid (PA), and cGMP. Cld PA can activate NHA (SOS1) in an independent manner. The main target of Ca2+ is CIBL4 (SOS3). The CIBL4 is capable to form the complex with CBL-interacting serine/threonine-protein kinase 24 (CIPK24, SOS2). The CIBL4-CIPK24 complex activates NHA (SOS1) and inhibits Na+ uptake by HKT2. CIPK24 together with SCaBP (SOS3 like protein) is involved in activation of the V-ATPase. CIPK24 participates in the activation of vacuolar transporters such as CAX and NHX. The rise in cytosolic Ca2+ concentration could trigger interaction of RSA1-RITF1. RSA1-RITF1 complex activates promoter of SOS1 gene. PA is involved in activation of mitogen-activated protein kinase 6 (MPK6). MPK6 can directly phosphorylate SOS1. The Ca2+-dependent kinase (CDPK3) and cytosolic Ca2+ lead to activation of vacuolar two-pore K+ channels (TPKs) and subsequent K+ release from vacuole. Due to the plasma membrane localization, SOS5 protein is considered to be potential candidate for extracellular Na+ sensing and helps maintain of cell wall integrity and architecture. Annexin1 (ANN1) is capable to sense the high concentrations of extracellular Na+ by mediating ROS-activated Ca2+ influx through the plasma membrane of plant cells. The rise of cGMP leads to inhibition of Na+ uptake, possibly via cyclic nucleotide–gated ion channels (CNGSs) and glutamate receptor (GLRs). PIP2;1, CNGCs, and GLRs could be blocked by exogenous Ca2+. The ROS production leads to K+ leak via activation of outward K+ channels – KOR (guard cells outward K+ channel, GORK) and NSCC. The intracellular accumulation of ROS at high levels can trigger programmed cell death (PCD).
One CBL (CBL4 or SOS3; salt over sensitive) is responsible for sensing calcium signals caused by salinity (Figure 2). The calcium binds to CBL4 causing dimerization of this protein and enhancing the activity of CIPK24 (SOS2) serine/threonine protein kinase. The resulting CBL4/CIPK24 (SOS3/SOS2) complex activates the Na+/H+ antiporter–SOS1 via phosphorylation (Zhu, 2002; Martínez-Atienza et al., 2007; Munns and Tester, 2008). Interestingly, the cytoplasmic C-terminal of SOS1 has been suggested to function as an intracellular Na+ sensor (Shi et al., 2002; Qiu et al., 2002; Shabala et al., 2005), but hard evidence for this is lacking. The SOS pathway has additional components – SOS4 and SOS5 (Shi et al., 2003; Mahajan and Tuteja, 2005), which may be involved in Na+ and K+ homeostasis (Mahajan et al., 2008). The sos4 mutants exhibit higher Na+/K+ ratio in comparison with wild-type plants (Mahajan et al., 2008). Due to the outer membrane localization, SOS5 is another potential candidate for (extracellular) Na+ sensing (Figure 2; Shi et al., 2003; Mahajan et al., 2008).
Thus, the SOS pathway is a key regulator of Na+ homeostasis, for example via SOS1. But, due to interaction with other regulatory proteins, it also participates in regulation of additional mechanisms of ion homeostasis: mutations in AtHKT1, which is responsible for Na+ translocation to the shoot (Halfter et al., 2000), suppress the sos3 mutation (Rus et al., 2001). Thus, the SOS2-SOS3 complex is involved in negative regulation of AtHKT1 during salinity stress. SOS2, in addition to modulating SOS1, can interact with vacuolar Na+/H+ exchanger (NHX) antiporters and significantly elevate their exchange activity (Zhu, 2002). SOS2 may also interact with the N-terminus of CAX1 (a H+/Ca2+ exchanger) (Qiu et al., 2002; Figure 3).
It seems likely that SOS1 activity induced by salinity not only relies on the SOS3–SOS2 complex but could be phosphorylated in a phospholipase D (PLD) signaling pathway-dependent manner (Yu et al., 2010): High Na+ concentrations cause an increase in enzyme activity of PLDα1 that lead to fast accumulation of phosphatidic acid (PA) as a lipid second messenger. PA in turn activates mitogen-activated protein kinase 6 (MPK6), which is capable of directly phosphorylating SOS1 (Yu et al., 2010). Loss-of-function mutants of PLDα1 and MPK6 exhibit sensitivity to salinity and accumulate more Na+ accumulation in the shoots.
A further SOS1 regulating mechanism originates in nuclear Ca2+ signaling in response to high salinity (Guan et al., 2013). Nuclear Ca2+ activates the Ca2+-binding protein RSA1, which complexes with RITF1 (RSA1 interacting transcription factor). Subsequently, the activated complex RSA1-RITF1 binds at the SOS1 promoter to augment its transcription (Figure 2; Guan et al., 2013).
Other early components in NaCl-induced sensing and signaling may be the previously mentioned ROS and cyclic nucleotides such as cGMP. Both cGMP and ROS show rapid transient increases in cytoplasmic levels after salinity stress onset (Kiegle et al., 2000; Donaldson et al., 2004). The rise of cellular cGMP can be detected within seconds after application of salinity and osmotic stress (Donaldson et al., 2004). Furthermore, cGMP inhibits Na+ influx in several plant species (Maathuis and Sanders, 2001; Essah et al., 2003; Rubio et al., 2003) while it can regulate transcription of various genes related to salinity stress and promote K+ uptake (Maathuis, 2006, 2014; Isner and Maathuis, 2016). Indeed, the work by Donaldson et al. (2004) strongly suggests cross talk between Ca2+ and cGMP signaling (Figure 2).
A rise in ROS is detected within minutes after the onset of salinity stress (Hong et al., 2008), and this phenomenon can activate downstream MAPK cascades (Miller et al., 2010; Maathuis, 2014). Recent studies demonstrate participation of ROS in transcriptional regulation. For example, ROS generated by plasma membrane-localized NADPH oxidase can help to stabilize AtSOS1 transcripts (Chung et al., 2008). The ROS-sensitive transcription factor ERF1 (ethylene response factor) in rice can bind to multiple promoters, including those of MAPKs, and improves general performance of plants under salinity stress (Schmidt et al., 2013). ROS are also thought to alter ion fluxes; for example, outward rectifying K+ channels are directly activated by ROS in Arabidopsis roots (Demidchik et al., 2010). The model further postulates that moderate K+ decrease in the cytosol will generate a low level of ROS designated for signaling, while high levels of salinity generate damaging ROS that activate K+ efflux channels and accelerate cellular K+ leak. Thus, fast and significant K+ loss could lead to acute ROS toxicity and development of programmed cell death (PCD). Indirectly, ROS such as the superoxide anion affect GORK channel transcription, providing yet another feedback loop (Tran et al., 2013).
Ion Toxicity: How Relevant is it and Where does it Occur?
Some measure of the two main components of salt damage, osmotic stress and ion toxicity, can be obtained by comparing plants exposed to salt and those treated with equiosmolar quantities of an inert osmoticum such as polyethylene glycol (PEG). Many of such studies are available, and the majority shows overwhelmingly that the osmotic stress component makes up a much larger fraction than the ion toxicity component (Castillo et al., 2007; Zhao et al., 2010).
Salt toxicity directly relates to ion concentrations and can manifest itself in all cell compartments though is usually assumed to be associated with the cytoplast. Thus, reliable measurements of cellular Na+ and Cl− concentrations are crucial for proper evaluation of toxicity but such measurements have only been conducted for relatively few plants. Values for [Na+]vac [Cl−]vac and [K+]vac using X-ray analyses (Chen et al., 2014), microelectrodes (Carden et al., 2003), dyes (Wissing and Smith, 2000; Wu et al., 2015a), and whole tissue extraction (Patishtan et al., 2018) show that both [Na+]vac and [Cl−]vac vary widely from tens of mM to 1 M in many halophytes (Zhao et al., 2005; Flowers and Colmer, 2008).
Obtaining truthful values of cytoplasmic ion concentrations is fraught with difficulty because of the small size of this compartment (see Kronzucker and Britto, 2011; Flowers et al., 2014) for a discussion of methodological aspects). Though all approaches generate some artifacts, techniques based on the use of ratiometric fluorescent dyes such as SBFI and ion selective electrodes (Carden et al., 2003) are preferable, since they record in real time and do not require any tissue preparation. Data for [Na+]cyt determined with SBFI (Halperin and Lynch, 2003; Kader and Lindbergh, 2005) vary between ~5 and 70 mM with external NaCl concentrations between 5 and 100 mM. Data obtained with electrodes (Carden et al., 2003) fall in the range 5–25 mM when barley was exposed to 200 mM NaCl. Data for cytoplasmic [K+] are less variable and approximately between 70 and 90 mM (Walker et al., 1996; Carden et al., 2003). To measure cytoplasmic Cl−, genetic GFP-based reporters such as Clomeleon (Markova et al., 2008) and fluorescent dyes have been exploited in animal cells showing values typically between 50 and 80 mM (Salomonsson et al., 1993). Unfortunately, these techniques have not (yet) been used to determine [Cl−]cyt in plant cells. Gerson and Poole (1972) used microelectrodes to measure [Cl−]cyt in nonvacuolate root tip cells and found ~30 mM in the presence of 60 mM KCl (comparable to 60 mM NaCl). Multiple studies from compartmental analysis and X-ray studies (White and Broadley, 2001; Flowers et al., 2014) show values ranging from ~45 to 140 mM in the presence of 50–100 mM NaCl. In all, these numbers suggest that during moderate salinity (~5–10 dS m−1), the maximum [Na+]cyt (60–70 mM) is similar to [K+]cyt (~70–90 mM) and cytoplasmic K:Na ratios are unlikely to drop far below unity. Data for [Cl−]cyt are more scarce but suggest a comparable or slightly higher range to that of Na+ and K+.
Data for the apoplast compartment vary greatly. Work with pea and spinach showed a substantial difference between these species (Speer and Kaiser, 1991). Pea apoplasts reached concentrations of around 90 and 200 mM for Na+ and Cl−, respectively, while corresponding levels in spinach did not exceed 10 and 15 mM. Comparative studies with canola and rice (Gao et al., 2016) determined apoplastic Na+ levels of ~130 (canola) and ~100 (rice) mM after 20 day treatment with 150 (canola) and 100 (rice) mM NaCl. Salinization of Vicia faba beans with 50, 75, or 100 mM NaCl caused apoplastic Na+ to rise to ~5, 30, and 100 mM (Shahzad et al., 2013).
The presence of high concentrations of Na+ and Cl− can disturb water structure via kosmo- and chaotropic effects, inhibit enzymes, and create nutritional imbalance. The higher charge density of Na+ compared to K+ means it behaves as a weak “kosmotrope” that organizes and immobilizes water structure around itself. Kosmotropy affects hydrogen bonding between water molecules and polar groups of proteins and nucleic acids, potentially interfering with their biochemical activity. K+ has a less tight hydration shell thus behaving as a weak “chaotrope.” A discernible kosmotropic effect of Na+ typically requires concentrations of >200 mM. Furthermore, molecular dynamic simulation shows that Na+ and K+ influence protein or DNA in a similar manner (Cheng et al., 2006). In fact, the tropic effects of anions are typically greater than those of cations but Cl− (like K+) is a very weak kosmotrope. Thus, in the presence of moderate salinity and the prevailing cytoplasmic concentrations, the potential impact of K+, Na+, and Cl− on solvent properties would be negligible.
Another potential ion toxicity mechanism that is often referred to in the context of salinity stress is the requirement of many enzymes to bind K+ which can be disrupted by Na+ displacing K+. What is striking is that in many cases the older literature reports almost identical effects of K+ and Na+ on enzyme and polysome activity (Johnson et al., 1968; Greenway and Osmond, 1972; Osmond and Greenway, 1972; Brady et al., 1984). Enzymes, such as starch synthase and glucose-6-phosphate dehydrogenase (Johnson et al., 1968), show maximum activity in the presence of 50–100 mM monovalents but do so irrespective of it being K+ or Na+. A recent report on the kinetic properties of rice pyrroline-5-carboxylate reductase agrees with this notion (Forlani et al., 2015). These findings indicate that Na+ can substitute K+ without significant problems for many biochemical activities. However, in other cases, the requirement for K+ is more specific; for example, the Kcat for Na+ activation of pyruvate kinase, a classical example of a K+ stimulated enzyme (Kachmar and Boyer, 1953), is only about 8% compared to that for K+. Nevertheless, the approximately 10-fold higher affinity of this enzyme for K+ (Kachmar and Boyer, 1953) compared to Na+ ensures that a Na:K ratio of more than 3 would be needed to significantly reduce enzyme activity. Similarly, studies on other enzymes (Shelp and Atkins, 1983; Gibrat et al., 1990) show Km values for K+ that are often very low (10–15 mM) while those for Na+ are much higher (>100 mM). In a few studies, the effect of anions was scrutinized and, as far as is known, in general Cl− has no detrimental effect on enzyme activity at [Cl−]cyt below ~80–100 mM (Greenway and Osmond, 1972; Brady et al., 1984), which is also implied by routine measurement of 60–90 mM [Cl−]cyt in animal cells.
In contrast to cytoplasmic concentrations, vacuolar levels of Na+ and Cl− readily reach several hundred mM. For example, a recent study on rice cultivars exposed to 50 mM NaCl showed tissue [Na+] of up to 600 mM (Patishtan et al., 2018). Consequently, vacuolar Na/K ratios can easily exceed values of 3 or 4. The central vacuole plays a role in ionic homeostasis, pH regulation, and osmotic adjustment. As lytic compartment, the vacuole contains hydrolases, phosphatases, and phosphoesterases (Boller and Kende, 1979) and thus is linked to protein turnover processes like ubiquitination. In spite of extensive information regarding the vacuolar proteome, to our knowledge no vacuole-specific enzymes have been tested for the functional implications of high Na+ and Cl− concentrations. The above data suggest that when glycophytes are exposed to moderate salinity stress (~50–150 mM) ion toxicity in the cytosol is unlikely to be problematic but, in contrast, whether vacuolar enzymes can maintain functionality when typically surrounded by much higher salt concentrations is a question that needs to be answered urgently.
In summary, in spite of considerable inward gradients for Na+, most data suggest that plants adequately prevent accumulation of Na+ and Cl− in the cytosol beyond 50–80 mM, even in the presence of 100–150 mM external Na+. Apoplastic levels are in the same order of magnitude. Consequently, if ion toxicity occurs in these compartments, it is probably of limited magnitude. The situation in the vacuole where ion concentrations can easily exceed 500 mM may be quite different. Alternatively, the ionic component of salinity stress could manifest itself via generic parameters such as the membrane potential (e.g. Gill et al., 2017) rather than specific ionic interactions. Since many transport processes are voltage dependent, depolarization could influence a plethora of processes including the uptake of essential nutrients.
What is the Role of Potassium in Salinity Stress?
K+ is the most abundant cation in plant cells and an essential nutrient that is important for many enzymatic reactions, ionic and pH homeostasis and maintaining adequate membrane potential (Maathuis 2009; Ahmad and Maathuis, 2014). Cytosolic K+ is also an important determinant of plant adaptive responses to a broad range of environmental stresses (e.g. Shabala and Pottosin, 2014). In hydrated form, Na+ and K+ are chemically and structurally very similar and some biophysical roles of K+, particularly generating turgor, can be fulfilled by Na+. Nevertheless, K+ is uniquely required for many physiological and biochemical processes, whereas Na+ is not. The transport systems involved in the uptake and distribution of K+ and Na+ in combination are key determinants of plant salinity tolerance due to their ability to determine tissue and cytosolic K+/Na+ ratios, parameters that are generally believed to impact greatly on salt tolerance (Maathuis and Amtmann, 1999; Shabala and Cuin, 2008). Influx of Na+ worsens the K/Na ratio, and this is further exacerbated by salt stress-induced K+ loss, a phenomenon that is often more pronounced in salt-sensitive species (Chen et al., 2007; Wu et al., 2018). GORK (guard cell outward rectifying K+ channel) type and ROS-activated NSCC-type channels are likely to mediate the main fraction of K+ efflux (Jayakannan et al., 2013; Wu et al., 2015b). In addition, salt stress-induced K+ leakage can cause PCD (Figure 2; Demidchik et al., 2014).
Interaction between K+ and Na+ transport has been described in many studies and many forms. For example, salinity affects K+ transporter transcription as exemplified by work on OsAKT1 (an inward rectifying K+ channel) where in situ hybridization showed that OsAKT1 transcription is downregulated after exposure to salinity in a cell type and cultivar-specific manner (Golldack et al., 2003). Other studies have shown upregulation of the phloem localized AKT2/3 and stelar root tissue located SKOR (an outward rectifying K+ channel) by salinity (Marten et al., 1999; Maathuis, 2006). These data in combination suggest that salinity may increase K+ circulation via the vascular bundles (Maathuis, 2006; Shabala and Cuin, 2008). Endomembrane channels such as the vacuolar TPKs (two pore K+ channels) are also likely to play an important role. The expression of tobacco TPK1a was increased around twofold by salt stress or osmotic shock (Hamamoto et al., 2008), whereas TPK overexpression in tobacco cells increased their resistance to salt stress (Wang et al., 2013). Post-translational modulation of TPK1 also impacts on salt tolerance as was shown for AtTPK1, which becomes phosphorylated by a Ca2+-dependent kinase (CDPK3; Latz et al., 2013) (Figure 2).
It is generally assumed that increased levels of K+ mitigate against salt stress, but this may be an oversimplification. Indeed, recent experimental data suggest that Na+ toxicity and water deficit are often not the key causes of plant growth inhibition by NaCl in Arabidopsis (Álvarez-Aragón et al., 2016). Rather, the overaccumulation of Na+ plus K+ might trigger growth reduction in NaCl-treated Arabidopsis plants, for example, via effects on stomatal regulation or systemic stress responses that lower growth (Álvarez-Aragón et al., 2016).
These salinity-induced effects on K+ transport are often variety, tissue and cell specific. For example, salt-tolerant barley varieties are capable to better retain K+ in the roots in comparison with sensitive genotypes (Chen et al., 2007), whereas different Arabidopsis ecotypes show widely varying K+/Na+ ratios (Álvarez-Aragón et al., 2016). Salt-induced K+ efflux in wheat and barley mesophyll cells of salt-sensitive varieties is significantly higher than that in salt-tolerant varieties (Wu et al., 2013, 2015c), and in contrast to cultivated barley, halophytic wild barley relatives exhibited much better tissue K+ retention (Garthwaite et al., 2005). At the tissue level, the higher salt sensitivity of barley root apex in comparison with the mature root zone was explained by a much greater NaCl-induced K+ efflux in root apex (Shabala et al., 2006). In turn such observations may be caused by the cell and tissue specificity of K+ transporters. In the root, mature zone efflux is mediated primarily by GORK-type channels (Chen et al., 2007; Shabala et al., 2007a; Wu et al., 2013; Chakraborty et al., 2016), whereas in the elongation zone NSCC channels fulfill this function (Bose et al., 2014). These findings suggest that the ability of plants to retain K+ in various tissues is an important feature of plant salt tolerance (Wu et al., 2018).
Multiple reports suggest that K+ could play important role in cell signaling during salinity (Shabala 2009, 2017; Anschütz et al., 2014; Shabala and Pottosin, 2014; Wu et al., 2018). In this context, K+ can cause cell- and tissue-specific metabolic changes and drive a “metabolic switch” to inhibit energy-dependent biosynthetic processes. The ensuing reduction or arrest of plant growth saves energy, which in turn augments the capacity for the synthesis of compounds that help in defence and repair of cellular systems (Demidchik et al., 2014). A K+-signaling function can also be envisaged in its capacity to generate PCD (Huh et al., 2002; Shabala, 2009). The physiological role of PCD during salinity is still discussed, but many experimental data directly link salinity stress, K+ leak, and PCD. For example, Arabidopsis mutants with loss of function in GORK channels exhibit slower development of salinity-related PCD (Demidchik et al., 2010), an observation that can be mimicked with channel blockers such as TEA (Osakabe et al., 2013). Interestingly, overexpression of the animal antiapoptotic CED-9 gene in tobacco mesophyll cells led to a reduction of stress-induced K+ efflux and improved plant salt tolerance (Shabala et al., 2007b). Thus, stress-induced K+ leakage, ROS, and PCD are likely to be tightly connected (Demidchik et al., 2010).
In all, K+ homeostasis is intricately linked to salt tolerance. At its most basic level, this involves substitution of K+ in its biochemical roles, but more complex relationships may exist such as K+-related PCD and the role of K+ as a signaling moiety that modulates metabolic pathways. Where the latter processes are concerned, more detailed studies at the molecular level are needed for example on the exact mechanism and cytosolic K+ concentrations that cause PCD.
Conclusions
The ever-increasing salinization of arable land will require multipronged solutions of which crops with increased tolerance is one. Exploiting genetic diversity will help achieving this objective but would be far more effective when combined with a comprehensive understanding of the molecular tolerance mechanisms. Great progress has been made in the last decades but yet many of the basic processes that contribute to tolerance are only partially understood. Further studies are urgently needed to unravel the details of Na+, and especially Cl−, uptake mechanisms. Mapping of toxicity at the cell and tissue level will aid in setting targets for tolerance improvement. Perception, sensing, and signaling chains lack important components in particular those at the beginning of pathways, whereas greater understanding of the role of other minerals such as K+ should enable us to mitigate salt stress by manipulating uptake and distribution of these nutrients.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahmad, I., and Maathuis, F. J. M. (2014). Cellular and tissue distribution of potassium: physiological relevance, mechanisms and regulation. J. Plant Physiol. 171, 708–714. doi: 10.1016/j.jplph.2013.10.016
Ahmad, I., Devonshire, J., Mohamed, R., Schultze, M., and Maathuis, F. J. M. (2015). Overexpression of the potassium channel TPKb in small vacuoles confers osmotic and drought tolerance to rice. New Phytol. 209, 1040–1048. doi: 10.1111/nph.13708
Álvarez-Aragón, R., Haro, R., Benito, B., and Rodríguez-Navarro, A. (2016). Salt intolerance in Arabidopsis: shoot and root sodium toxicity, and inhibition by sodium-plus-potassium overaccumulation. Planta 243, 97–114. doi: 10.1007/s00425-015-2400-7
Amtmann, A., and Sanders, D. (1998). Mechanisms of Na+ uptake by plant cells. Adv. Bot. Res. 29, 75–112. doi: 10.1016/S0065-2296(08)60310-9
Amtmann, A., Fischer, M., Marsh, E. L., Stefanovic, A., Sanders, D., and Schachtman, D. P. (2001). The wheat cDNA LCT1generates hypersensitivity to sodium in a salt-sensitive yeast strain. Plant Physiol. 126, 1061–1071. doi: 10.1104/pp.126.3.1061
Anil, V. S., Krishnamurthy, P., Kuruvilla, S., Sucharitha, K., Thomas, G., and Mathew, M. (2005). Regulation of the uptake and distribution of Na+ in shoots of rice (Oryza sativa) variety Pokkali: role of Ca2+ in salt tolerance response. Physiol. Plant 124, 451–464. doi: 10.1111/j.1399-3054.2005.00529.x
Anschütz, U., Becker, D., and Shabala, S. (2014). Going beyond nutrition: regulation of potassium homoeostasis as a common denominator of plant adaptive responses to environment. J. Plant Physiol. 171, 670–687. doi: 10.1016/j.jplph.2014.01.009
Berthomieu, P., Conéjéro, G., Nublat, A., Brackenbury, W. J., Lambert, C., Savio, C., et al. (2003). Functional analysis of AtHKT1 in Arabidopsis shows that Na(+) recirculation by the phloem is crucial for salt tolerance. EMBO J. 22, 2004–2014. doi: 10.1093/emboj/cdg207
Boller, T., and Kende, H. (1979). Hydrolytic enzymes in the central vacuole of plant cells. Plant Physiol. 63, 1123–1132. doi: 10.1104/pp.63.6.1123
Bose, J., Shabala, L., Pottosin, I., Velarde-Buendía, A. M., Massart, A., Poschenrieder, C., et al. (2014). Kinetics of xylem loading, membrane potential maintenance, and sensitivity of K+-permeable channels to reactive oxygen species: physiological traits that differentiate salinity tolerance between pea and barley. Plant Cell Environ. 37, 589–600. doi: 10.1111/pce.12180
Boudsocq, M., and Sheen, J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40. doi: 10.1016/j.tplants.2012.08.008
Brady, C. J., Gibson, T. S., Barlow, E. W. R., Speirs, J., and Wyn Jones, R. G. (1984). Salt tolerance in plants. I. Ions, compatible organic solutes and the stability of plant ribosomes. Plant Cell Environ. 7, 571–578. doi: 10.1111/1365-3040.ep11591840
Byrt, C. S., Zhao, M., Kourghi, M., Bose, J., Henderson, S. W., Qiu, J., et al. (2017). Non-selective cation channel activity of aquaporin AtPIP2;1 regulated by Ca2+ and pH. Plant Cell Environ. 40, 802–815. doi: 10.1111/pce.12832
Carden, D. E., Walker, D. J., Flowers, T. J., and Miller, A. J. (2003). Single-cell measurements of the contributions of cytosolic Na+ and K+ to salt tolerance. Plant Physiol. 131, 676–683. doi: 10.1104/pp.011445
Castillo, E. G., Tuong, T. Ph., Ismail, A. M., and Inubushi, K. (2007). Response to salinity in rice: comparative effects of osmotic and ionic stresses. Plant Prod. Sci. 10, 159–170. doi: 10.1626/pps.10.159
Chakraborty, K., Bose, J., Shabala, L., Eyles, A., and Shabala, S. (2016). Evaluating relative contribution of osmotolerance and tissue tolerancemechanisms toward salinity stress tolerance in three Brassica species. Physiol. Plant 158, 135–151. doi: 10.1111/ppl.12447
Chatzigeorgiou, M., Bang, S., Hwang, S. W., and Schafer, W. R. (2013). tmc-1 encodes a sodium-sensitive channel required for salt chemosensation in C. elegans. Nature 494, 95–99. doi: 10.1038/nature11845
Chazen, O., Hartung, W., and Neumann, P. M. (1995). The different effects of PEG 6000 and NaCl on leaf development are associated with differential inhibition of root water transport. Plant Cell Environ. 18, 727–735. doi: 10.1111/j.1365-3040.1995.tb00575.x
Chen, S., Diekmann, H., Janz, D., and Polle, A. (2014). Quantitative X-ray elemental imaging in plant materials at the subcellular level with a transmission electron microscope: applications and limitations. Materials 7, 3160–3175. doi: 10.3390/ma7043160
Chen, Z., Pottosin, I. I., Cuin, T. A., Fuglsang, A. T., Tester, M., Jha, D., et al. (2007). Root plasma membrane transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiol. 145, 1714–1725. doi: 10.1104/pp.107.110262
Cheng, Y., Korolev, N., and Nordenskiöld, L. (2006). Similarities and differences in interaction of K+ and Na+ with condensed ordered DNA. A molecular dynamics computer simulation study. Nucleic Acids Res. 34, 686–696. doi: 10.1093/nar/gkj434
Choi, W. G., Toyota, M., Kim, S. H., Hilleary, R., and Gilroy, S. (2014). Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. USA 111, 6497–6502. doi: 10.1073/pnas.1319955111
Choi, W. G., Hilleary, R., Swanson, S. J., Kim, S. H., and Gilroy, S. (2016). Rapid, long distance electrical and calcium signaling in plants. Annu. Rev. Plant Biol. 67, 287–307. doi: 10.1146/annurev-arplant-043015-112130
Chung, Y. M., Lee, S. B., Kim, H. J., Park, S. H., Kim, J. J., Chung, J. S., et al. (2008). Replicative senescence induced by Romo1-derived reactive oxygen species. J. Biol. Chem. 283, 33763–33771. doi: 10.1074/jbc.M805334200
Clemens, S., Antosiewicz, D. M., Ward, J. M., Schachtman, D. P., and Schroeder, J. I. (1998). The plant cDNA LCT1 mediates the uptake of calcium and cadmium in yeast. Proc. Natl. Acad. Sci. USA 95, 12043–12048. doi: 10.1073/pnas.95.20.12043
Colmenero-Flores, J. M., Martinez, G., Gamba, G., Vázquez, N., Iglesias, D. J., Brumós, J., et al. (2007). Identification and functional characterization of cation chloride cotransporters in plants. Plant J. 50, 278–292. doi: 10.1111/j.1365-313X.2007.03048.x
Demidchik, V., Essah, P. A., and Tester, M. (2004). Glutamate activates cation currents in the plasma membrane of Arabidopsis root cells. Planta 219, 167–175. doi: 10.1007/s00425-004-1207-8
Demidchik, V., and Maathuis, F. J. M. (2007). Physiological roles of nonselective cation channels in plants: from salt stress to signaling and development. New Phytol. 175, 387–404. doi: 10.1111/j.1469-8137.2007.02128.x
Demidchik, V., Cuin, T. A., and Svistunenko, D. (2010). Arabidopsis root K+ efflux conductance activated by hydroxyl radicals: single-channel properties, genetic basis and involvement in stress-induced cell death. J. Cell Sci. 123, 1468–1479. doi: 10.1242/jcs.064352
Demidchik, V., Straltsova, D., Medvedev, S. S., Pozhvanov, G. A., Sokolik, A., and Yurin, V. (2014). Stress-induced electrolyte leakage: the role of K+-permeable channels and involvement in programmed cell death and metabolic adjustment. J. Exp. Bot. 65, 1259–1270. doi: 10.1093/jxb/eru004
Demidchik, V., Shabala, S., Isayenkov, S., Cuin, T. A., and Pottosin, I. (2018). Calcium transport across plant membranes: mechanisms and functions. New Phytol. 220, 49–69. doi: 10.1111/nph.15266
Donaldson, L., Ludidi, N., Knight, M. R., Gehring, C., and Denby, K. (2004). Salt and osmotic stress cause rapid increases in Arabidopsis thaliana cGMP levels. FEBS Lett. 569, 317–320. doi: 10.1016/j.febslet.2004.06.016
Essah, P. A., Davenport, R., and Tester, M. (2003). Sodium influx and accumulation in Arabidopsis. Plant Physiol. 133, 307–318. doi: 10.1104/pp.103.022178
Flowers, T. J., and Colmer, T. D. (2008). Salinity tolerance in halophytes. New Phytol. 179, 945–963. doi: 10.1111/j.1469-8137.2008.02531
Flowers, T. J., and Colmer, T. D. (2015). Plant salt tolerance: adaptations in halophytes. Annu. Bot. 115, 327–331. doi: 10.1093/aob/mcu267
Flowers, T. J., Munns, R., and Colmer, T. D. (2014). Sodium chloride toxicity and the cellular basis of salt tolerance in halophytes. Annu. Bot. 115, 419–431. doi: 10.1093/aob/mcu217
Forlani, G., Bertazzini, M., Zarattini, M., Funck, D., Ruszkowski, M., and Nocek, B. (2015). Functional properties and structural characterization of rice δ1-pyrroline-5-carboxylate reductase. Front. Plant Sci. 6:565. doi: 10.3389/fpls.2015.00565
Gao, J. P., Chao, D. Y., and Lin, H. X. (2007). Understanding abiotic stress tolerance mechanisms: recent studies on stress response in rice. J. Integ. Plant Biol. 49, 742–750. doi: 10.1111/j.1744-7909.2007.00495.x
Gao, L., Liu, M., Wang, M., Shen, Q., and Guo, S. (2016). Enhanced salt tolerance under nitrate nutrition is associated with apoplast Na+ content in canola (Brassica. napus L.) and rice (Oryza sativa L.) plants. Plant Cell Physiol. 57, 2323–2333. doi: 10.1093/pcp/pcw141
Garthwaite, A. J., von Bothmer, R., and Colmer, T. D. (2005). Salt tolerance in wild Hordeum species is associated with restricted entry of Na+ and Cl− into the shoots. J. Exp. Bot. 56, 2365–2378. doi: 10.1093/jxb/eri229
Gerson, D. F., and Poole, R. J. (1972). Chloride accumulation by mung bean root tips. Plant Physiol. 50, 603–607. doi: 10.1104/pp.50.5.603
Gibrat, R., Grouzis, J. P., Rigaud, J., and Grignon, C. (1990). Potassium stimulation of corn root plasmalemma ATPase II. H+-pumping in native and reconstituted vesicles with purified ATPase. Plant Physiol. 93, 1183–1189. doi: 10.1104/pp.93.3.1183
Gill, M. B., Zeng, F., Shabala, L., Zhang, G., Fan, Y., Shabala, S., et al. (2017). Cell-based phenotyping reveals QTL for membrane potential maintenance associated with hypoxia and salinity stress tolerance in barley. Front. Plant Sci. 16:1941. doi: 10.3389/fpls.2017.01941
Gobert, A., Park, G., Amtmann, A., Sanders, D., and Maathuis, F. J. M. (2006). Arabidopsis thaliana cyclic nucleotide gated channel 3 forms a non-selective ion transporter involved in germination and cation transport. J. Exp. Bot. 57, 791–800. doi: 10.1093/jxb/erj064
Golldack, D., Quigley, F., Michalowski, C. B., Kamasani, U. R., and Bohnert, H. J. (2003). Salinity stress-tolerant and -sensitive rice (Oryza sativa L.) regulate AKT1-type potassium channel transcripts differently. Plant Mol. Biol. 51, 71–81. doi: 10.1023/A:1020763218045
Greenway, H., and Osmond, C. B. (1972). Salt responses of enzymes from species differing in salt tolerance. Plant Physiol. 49, 256–259. doi: 10.1104/pp.49.2.256
Guan, Q., Wu, J., Yue, X., Zhang, Y., and Zhu, J. (2013). A nuclear calcium sensing pathway is critical for gene regulation and salt stress tolerance in Arabidopsis. PLoS Genet. 9:e1003755. doi: 10.1371/journal.pgen.1003755
Halperin, S. J., and Lynch, J. P. (2003). Effects of salinity on cytosolic Na+ and K+ in root hairs of Arabidopsis thaliana: in vivo measurements using the fluorescent dyes SBFI and PBFI. J. Exp. Bot. 54, 2035–2043. doi: 10.1093/jxb/erg219
Halfter, U., Ishitani, M., and Zhu, J. K. (2000). The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl. Acad. Sci. USA 97, 3735–3740. doi: 10.1073/pnas.040577697
Hamamoto, S., Marui, J., Matsuoka, K., Higashi, K., Igarashi, K., Nakagawa, T., et al. (2008). Characterization of a tobacco TPK-type K+ channel as a novel tonoplast K+ channel using yeast tonoplasts. J. Biol. Chem. 283, 1911–1920. doi: 10.1074/jbc.M708213200
Hanson, P. J., Sucoff, E. I., and Markhart, A. H. (1985). Quantifying apoplastic flux through red pine root systems using trisodium, 3-hydroxy-5,8,10-pyrenetrisulfonate. Plant Physiol. 77, 21–24. doi: 10.1104/pp.77.1.21
Henderson, S. W., Wege, S., Qiu, J., Blackmore, D. H., Walker, A. R., Tyerman, S. D., et al. (2015). Grapevine and arabidopsis cation-chloride cotransporters localize to the golgi and trans-golgi network and indirectly influence long-distance ion transport and plant salt tolerance. Plant Physiol. 169, 2215–2229. doi: 10.1104/pp.15.00499
Hernandez, J. A., Ferrer, M. A., Jimenez, A., Barcelo, A. R., and Sevilla, F. (2001). Antioxidant systems and O2−/H2O2 production in the apoplast of pea leaves. Its relation with salt-induced necrotic lesions in minor veins. Plant Physiol. 127, 817–831. doi: 10.1104/pp.010188
Hong, J. K., Yun, B. W., Kang, J. G., Raja, M. U., Kwon, E., Sorhagen, K., et al. (2008). Nitric oxide function and signaling in plant disease resistance. J. Exp. Bot. 59, 147–154. doi: 10.1093/jxb/erm244
Horie, T., Costa, A., Kim, T. H., Han, M. J., Horie, R., Leung, H. -Y., et al. (2007). Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO J. 26, 3003–3014. doi: 10.1038/sj.emboj.7601732
Huh, G. H., Damsz, B., Matsumoto, T. K., Reddy, M. P., Rus, A. M., Ibeas, J. I., et al. (2002). Salt causes ion disequilibrium-induced programmed cell death in yeast and plants. Plant J. 29, 649–659. doi: 10.1046/j.0960-7412.2001.01247.x
Isayenkov, S. V. (2012). Physiological and molecular aspects of salt stress in plants. Cytol. Genet. 46, 302–318. doi: 10.3103/S0095452712050040
Ismail, A. M., Heuer, S., Thomson, M. J., and Wissuwa, M. (2007). Genetic and genomic approaches to develop rice germplasm for problem soils. Plant Mol. Biol. 65, 547–570. doi: 10.1007/s11103-007-9215-2
Isner, J. C., and Maathuis, F. J. M. (2016). cGMP signalling in plants: from enigma to main stream. Functional Plant Biol. 45, 93–101. doi: 10.1071/FP16337
Jayakannan, M., Bose, J., Babourina, O., Rengel, Z., and Shabala, S. (2013). Salicylic acid improves salinity tolerance in Arabidopsis by restoring membrane potential and preventing salt-induced K+ loss via a GORK channel. J Exp. Bot. 64, 2255–2268. doi: 10.1093/jxb/ert085
Johnson, M. K., Johnson, E. J., MacElroy, R. D., Speer, H. L., and Bruff, B. S. (1968). Effects of salts on the halophilic alga Dunaliella viridis. J. Bacteriol. 95, 1461–1468.
Kachmar, J. F., and Boyer, P. D. (1953). The potassium activation and calcium inhibition of pyruvic phosphoferase. J. Biol. Chem. 200, 669–682.
Kader, M. A., and Lindberg, S. (2005). Uptake of sodium in protoplasts of salt-sensitive and salt-tolerant cultivars of rice, Oryza sativa L. determined by the fluorescent dye SBFI. J. Exp. Bot. 56, 3149–3158. doi: 10.1093/jxb/eri312
Kiegle, E., Moore, C. A., Haseloff, J., Tester, M. A., and Knight, M. R. (2000). Cell-type-specific calcium responses to drought, salt and cold in the Arabidopsis root. Plant J. 23, 267–278. doi: 10.1046/j.1365-313x.2000.00786.x
Knight, H., Trewavas, A. J., and Knight, M. R. (1997). Calcium signaling in Arabidpsis thaliana responding to drought and salinity. Plant J. 12, 1067–1078. doi: 10.1046/j.1365-313X.1997.12051067.x
Krishnamurthy, P., Ranathunge, K., Franke, R., Prakash, H., Schreiber, L., and Mathew, M. (2009). The role of root apoplastic transport barriers in salt tolerance of rice (Oryza sativa L.). Planta 230, 119–134. doi: 10.1007/s00425-009-0930-6
Kronzucker, H. J., and Britto, D. T. (2011). Sodium transport in plants: a critical review. New Phytol. 189, 54–81. doi: 10.1111/j.1469-8137.2010.03540.x
Laohavisit, A., Richards, S. L., Shabala, L., Colaço, R. D. D. R., Swarbreck, S. M., et al. (2013). Salinity-induced calcium signaling and root adaptation in Arabidopsis require the calcium regulatory protein annexin1. Plant Physiol. 163, 253–262. doi: 10.1104/pp.113.217810
Latz, A., Mehlmer, N., Zapf, S., Mueller, T. D., Wurzinger, B., Pfister, B., et al. (2013). Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol. Plant 6, 1274–1289. doi: 10.1093/mp/sss158
Laurie, S., Feeney, K. A., Maathuis, F. J., Heard, P. J., Brown, S. J., and Leigh, R. A. (2002). A role for HKT1 in sodium uptake by wheat roots. Plant J. 32, 139–49. doi: 10.1046/j.1365-313X.2002.01410.x
Legué, V., Blancaflor, E., Wymer, C., Perbal, G., Fantin, D., and Gilroy, S. (1997). Cytoplasmic free Ca2+ in Arabidopsis roots changes in response to touch but not gravity. Plant Physiol. 114, 789–800. doi: 10.1104/pp.114.3.789
Leng, Q., Mercier, R. W., Hua, B. G., Fromm, H., and Berkowitz, G. A. (2002). Electrophysiological analysis of cloned cyclic nucleotide-gated ion channels. Plant Physiol. 128, 400–410. doi: 10.1104/pp.010832
Maathuis, F. J. M. (2006). The role of monovalent cation transporters in plant responses to salinity. J. Exp. Bot. 57, 1137–1147. doi: 10.1093/jxb/erj001
Maathuis, F. J. M. (2009). Physiological functions of mineral macronutrients. Curr. Opin. Plant. Biol. 12, 250–258. doi: 10.1016/j.pbi.2009.04.003
Maathuis, F. J. M. (2014). Sodium in plants: perception, signalling, and regulation of sodium fluxes. J. Exp. Bot. 65, 849–858. doi: 10.1093/jxb/ert326
Maathuis, F. J. M., and Amtmann, A. (1999). K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Ann. Bot. 84, 123–133. doi: 10.1006/anbo.1999.0912
Maathuis, F. J., and Sanders, D. (2001). Sodium uptake in Arabidopsis roots is regulated by cyclic nucleotides. Plant Physiol. 127, 1617–1625. doi: 10.1104/pp.010502
Maathuis, F. J. M., Ahmad, I., and Patishtan, J. (2014). Regulation of Na+ fluxes in plants. Front. Plant Sci. 5:467. doi: 10.3389/fpls.2014.00467
Mahajan, S., Pandey, G. K., and Tuteja, N. (2008). Calcium- and salt-stress signaling in plants: shedding light on SOS pathway. Arch. Biochem. Biophys. 471, 146–158. doi: 10.1016/j.abb.2008.01.010
Mahajan, S., and Tuteja, N. (2005). Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophys. 444, 139–158. doi: 10.1016/j.abb.2005.10.018
Maischak, H., Zimmermann, M. R., Felle, H. H., Boland, W., and Mithofer, A. (2010). Alamethicin-induced electrical long distance signaling in plants. Plant Signal. Behav. 5, 988–990. doi: 10.1104/pp.108.133884
Malagoli, P., Britto, D. T., Schulze, L. M., and Kronzucker, H. J. (2008). Futile Na+ cycling at the root plasma membrane in rice (Oryza sativa L.) –kinetics, energetics, and relation to salinity tolerance. J. Exp. Bot. 59, 4109–4117. doi: 10.1093/jxb/ern249
Markova, O., Mukhtarov, M., Real, E., Jacob, Y., and Bregestovski, P. (2008). Genetically encoded chloride indicator with improved sensitivity. J. Neurosci. Methods 170, 67–76. doi: 10.1016/j.jneumeth.2007.12.016
Marten, I., Hoth, S., Deeken, R., Ache, P., Ketchum, K. A., Hoshi, T., et al. (1999). AKT3, a phloem-localized K1 channel, is blocked by protons. Proc. Natl. Acad. Sci. USA 96, 7581–7586. doi: 10.1073/pnas.96.13.7581
Martínez-Atienza, J., Jiang, X., Garciadeblas, B., Mendoza, I., Zhu, J. K., Pardo, J. M., et al. (2007). Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 143, 1001–1012. doi: 10.1104/pp.106.092635
Mian, A., Oomen, R. J., Isayenkov, S., Sentenac, H., Maathuis, F. J. M., and Véry, A. A. (2011). Over-expression of an Na+-and K+-permeable HKT transporter in barley improves salt tolerance. Plant J. 68, 468–79. doi: 10.1111/j.1365-313X.2011.04701.x
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., and Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–67. doi: 10.1111/j.1365-3040.2009.02041.x
Moon, G. J., Clough, B. F., Peterson, C. A., and Allaway, W. G. (1986). Apoplastic and symplastic pathways in Avicennia marina (Forsk.) Vierh. roots revealed by fluorescent tracer dyes. Aust. J. Plant Physiol. 13, 637–648. doi: 10.1071/PP9860637
Munns, R., and Passioura, J. B. (1984). Hydraulic resistance of plants. Effects of NaCl in barley and lupin. Aust. J. Plant Physiol. 11, 351–359. doi: 10.1071/PP9840351
Munns, R., and Termaat, A. (1986). Whole-plant responses to salinity. Aust. J. Plant Physiol. 13, 143–160. doi: 10.1071/PP9860143
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Negrão, S., Courtois, B., Ahmadi, N., Abreu, I., Saibo, N., and Oliveira, M. (2011). Recent updates on salinity stress in rice: from physiological to molecular responses. Crit. Rev. Plant Sci. 30, 329–377. doi: 10.1080/07352689.2011.587725
Osakabe, Y., Arinaga, N., Umezawa, T., Katsura, S., Nagamachi, K., Tanaka, H., et al. (2013). Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25, 609–624. doi: 10.1105/tpc.112.105700
Osmond, C. B., and Greenway, H. (1972). Salt responses of carboxylation enzymes from species differing in salt tolerance. Plant Physiol. 49, 260–263. doi: 10.1104/pp.49.2.260
Patishtan, J., Hartley, T. N., Fonseca de Carvalho, R., and Maathuis, F. J. M. (2018). Genome wide association studies to identify rice salt-tolerance markers. Plant Cell Environ. 41, 970–982. doi: 10.1111/pce.12975
Peterson, C. A., Emanuel, M. E., and Humphreys, G. B. (1981). Pathway of movement of apoplastic fluorescent dye tracers through the endodermis at the site of secondary root formation in corn (Zea mays) and broad bean (Vicia faba). Canad. J. Bot. 59, 618–625. doi: 10.1139/b81-087
Peterson, T. A., Swanson, E. S., and Hull, R. J. (1986). Use of lanthanum to trace apoplastic solute transport in intact plants. J. Exp. Bot. 37, 807–822. doi: 10.1093/jxb/37.6.807
Pitman, M. G. (1982). Transport across plant roots. Q. Rev. Biophys. 15, 481–554. doi: 10.1017/S0033583500003437
Plett, D. C., and Moller, I. S. (2010). Na+ transport in glycophytic plants: what we know and would like to know. Plant Cell Environ. 33, 612–626. doi: 10.1111/j.1365-3040.2009.02086.x
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., and Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. USA 99, 8436–8441. doi: 10.1073/pnas.122224699
Rajendran, K., Tester, M., and Roy, S. J. (2009). Quantifying the three main components of salinity tolerance in cereals. Plant Cell Environ. 32, 237–249. doi: 10.1111/j.1365-3040.2008.01916.x
Ranathunge, K., and Schreiber, L. (2011). Water and solute permeabilities of Arabidopsis roots in relation to the amount and composition of aliphatic suberin. J. Exp. Bot. 62, 1961–74. doi: 10.1093/jxb/erq389
Roy, S. J., Negrão, S., and Tester, M. (2014). Salt resistant crop plants. Curr. Opin. Biotechnol. 26, 115–124. doi: 10.1016/j.copbio.2013.12.004
Rubio, F., Flores, P., Navarro, J. M., and Martı̀nez, V. (2003). Effects of Ca2+, K+ and cGMP on Na+ uptake in pepper plants. Plant Sci. 165, 1043–1049. doi: 10.1016/S0168-9452(03)00297-8
Rus, A., Yokoi, S., Sharkhuu, A., Reddy, M., Lee, B. H., Matsumoto, T. K., et al. (2001). AtHKT1 is a salt tolerance determinant that controls Na entry into plant roots. Proc. Natl. Acad. Sci. USA 98, 14150–14155. doi: 10.1073/pnas.241501798
Salomonsson, M., Gonzalez, R., Kornfeld, M., and Persson, A. E. G. (1993). The cytosolic chloride concentration in macula densa and cortical thick ascending limb cells. Acta Physiol. 147, 305–313. doi: 10.1111/j.1748-1716.1993.tb09503.x
Sanderson, J. (1983). Water uptake by different regions of the barley root. Pathways of radial flow in relation to development of the endodermis. J. Exp. Bot. 34, 240–253. doi: 10.1093/jxb/34.3.240
Schachtman, D. P., Tyerman, S. D., and Terry, B. R. (1991). The K+/Na+ selectivity of a cation channel in the plasma membrane of root cells does not differ in salt-tolerant and salt-sensitive wheat species. Plant Physiol. 97, 598–605. doi: 10.1104/pp.97.2.598
Schachtman, D. P., Kumar, R., Schroeder, J. I., and Marsh, E. L. (1997). Molecular and functional characterization of a novel low-affinity cation transporter (LCT1) in higher plants. Proc. Natl. Acad. Sci. USA 94, 11079–11084. doi: 10.1073/pnas.94.20.11079
Schmidt, R., Mieulet, D., Hubberten, H. M., Obata, T., Hoefgen, R., Fernie, A. R., et al. (2013). Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25, 2115–2131. doi: 10.1105/tpc.113.113068
Shabala, S. (2009). Salinity and programmed cell death: unravelling mechanisms for ion specific signalling. J. Exp. Bot. 60, 709–712. doi: 10.1093/jxb/erp013
Shabala, S. (2017). Signalling by potassium: another second messenger to add to the list? J. Exp. Bot. 68, 4003–4007. doi: 10.1093/jxb/erx238
Shabala, L., Cuin, T. A., Newman, I., and Shabala, S. (2005). Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants. Planta 222, 1041–1050. doi: 10.1007/s00425-005-0074-2
Shabala, S., Demidchik, V., Shabala, L., Cuin, T. A., Smith, S. J., Miller, A. J., et al. (2006). Extracellular Ca2+ ameliorates NaCl-induced K+ loss from Arabidopsis root and leaf cells by controlling plasma membrane K+-permeable channels. Plant Physiol. 141, 1653–1665. doi: 10.1104/pp.106.082388
Shabala, S., Cuin, T. A., and Pottosin, I. (2007a). Polyamines prevent NaCl induced K+ efflux from pea mesophyll by blocking nonselective cation channels. FEBS Lett. 581, 1993–1999. doi: 10.1016/j.febslet.2007.04.032
Shabala, S., Cuin, T. A., Prismall, L., and Nemchinov, L. G. (2007b). Expression of animal CED-9 anti-apoptotic gene in tobacco modifies plasma membrane ion fluxes in response to salinity and oxidative stress. Planta 227, 189–197. doi: 10.1007/s00425-007-0606-z
Shabala, S., and Cuin, T. A. (2008). Potassium transport and plant salt tolerance. Physiol. Plant. 133, 651–669. doi: 10.1111/j.1399-3054.2007.01008.x
Shabala, S., and Pottosin, I. (2014). Regulation of potassium transport in plants under hostile conditions: implications for abiotic and biotic stress tolerance. Physiol. Plant. 151, 257–279. doi: 10.1111/ppl.12165
Shahzad, M., Zörb, C., Geilfus, C. -M., and Mühling, K. H. (2013). Apoplastic Na+ in Vicia faba leaves rises after short-term salt stress and is remedied by silicon. J. Agron. Crop Sci. 199, 161–170. doi: 10.1111/jac.12003
Shelp, B. J., and Atkins, C. A. (1983). Role of inosine monophosphate oxidoreductase in the formation of ureides in nitrogen-fixing nodules of cowpea (Vigna unguiculata L. Walp). Plant Physiol. 72, 1029–1034. doi: 10.1104/pp.72.4.1029
Shi, H., Quintero, F. J., Pardo, J. M., and Zhu, J. K. (2002). The putative plasma membrane Na(+)/H(+) antiporter SOS1 controls long-distance Na(+) transport in plants. Plant Cell 14, 465–477. doi: 10.1105/tpc.010371
Shi, H., Lee, B. H., Wu, S. J., and Zhu, J. K. (2003). Overexpression of a plasmamembrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 21, 81–85. doi: 10.1038/nbt766
Shi, Y., Wang, Y., Flowers, T. J., and Gong, H. (2013). Silicon decreases chloride transport in rice (Oryza sativa L.) in saline conditions. J. Plant Physiol. 170, 847–853. doi: 10.1016/j.jplph.2013.01.018
Speer, M., and Kaiser, W. M. (1991). Ion relations of symplastic and apoplastic space in leaves from Spinacia oleracea L. and Pisum sativum L. under salinity. Plant Physiol. 97, 990–997. doi: 10.1104/pp.97.3.990
Sunarpi, , Horie, T., Motoda, J., Kubo, M., Yang, H., Yoda, K., et al. (2005). Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 44, 928–938. doi: 10.1111/j.1365-313X.2005.02595.x
Tran, D., El-Maarouf-Bouteau, H., Rossi, M., Biligui, B., Briand, J., Kawano, T., et al. (2013). Posttranscriptional regulation of GORK channels by superoxide anion contributes to increases in outward-rectifying K+ currents. New Phytol. 198, 1039–1048. doi: 10.1111/nph.12226
Tsugane, K., Kobayashi, K., Niwa, Y., Ohba, Y., Wada, K., and Kobayashi, H. (1999). A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell 11, 1195–1206. doi: 10.2307/3870742
Walker, D. J., Leigh, R. A., and Miller, A. J. (1996). Potassium homeostasis in vacuolate plant cells. Proc. Natl. Acad. Sci. USA 93, 10510–10514. doi: 10.1073/pnas.93.19.10510
Wang, F., Deng, S., Ding, M., Meijuan, J. S., Huipeng, W., Yansha, Z., et al. (2013). Overexpression of a poplar two-pore K+ channel enhances salinity tolerance in tobacco cells. Plant Cell Tissue Organ Cul. 112, 19–31. doi: 10.1007/s11240-012-0207-9
Wang, S. M., Zhang, J. L., and Flowers, T. J. (2007). Low-affinity Na+ uptake in the Halophyte Suaeda maritime. Plant Physiol. 145, 559–571. doi: 10.1104/pp.107.104315
Weinl, S., and Kudla, J. (2008). The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol. 179, 675–686. doi: 10.1111/j.1469-8137.2009.02938.x
White, P. J., and Broadley, M. R. (2001). Chloride in soils and its uptake and movement within the plant: a review. Annu. Bot. 88, 967–988. doi: 10.1006/anbo.2001
Wissing, F., and Smith, J. A. C. (2000). Vacuolar chloride transport in Mesembryanthemum crystallinum L. measured using the fluorescent dye lucigenin. J. Membrane Biol. 177, 199–208. doi: 10.1007/s002320010003
Wu, H., Shabala, L., Barry, K., Zhou, M., and Shabala, S. (2013). Ability of leaf mesophyll to retain potassium correlates with salinity tolerance in wheat and barley. Physiol. Plant 149, 515–527. doi: 10.1111/ppl.12056
Wu, H., Shabala, L., Liu, X., Azzarello, E., Zhou, M., Pandolfi, C., et al. (2015a). Linking salinity stress tolerance with tissue-specific Na+ sequestration in wheat roots. Front. Plant. Sci. 6:71. doi: 10.3389/fpls.2015.00071
Wu, H., Shabala, L., Zhou, M., and Shabala, S. (2015b). Chloroplast generated ROS dominate NaCl- induced K+ efflux in wheat leaf mesophyll. Plant Signal. Behav. 10:e1013793. doi: 10.1080/15592324.2015.1013793
Wu, H., Zhu, M., Shabala, L., Zhou, M., and Shabala, S. (2015c). K+ retention in leaf mesophyll, an overlooked component of salinity tolerance mechanism: a case study for barley. J. Integr. Plant Biol. 57, 171–185. doi: 10.1111/jipb.12238
Wu, H., Zhang, X., Giraldo, J. P., and Shabala, S. (2018). It is not all about sodium: revealing tissue specificity and signalling roles of potassium in plant responses to salt stress. Plant Soil 431, 1–17. doi: 10.1007/s11104-018-3770-y
Yeo, A., Yeo, M., and Flowers, T. (1987). The contribution of an apoplastic pathway to sodium uptake by rice roots in saline conditions. J. Exp. Bot. 38, 1141–1153. doi: 10.1093/jxb/38.7.1141
Yeo, A. R., Lee, K. -S., Izard, P., Boursier, P. J., and Flowers, T. J. (1991). Short and long-term effects of salinity on leaf growth in rice (Oryza sativa L.). J. Exp. Bot. 42, 881–889. doi: 10.1093/jxb/42.7.881
Yu, L., Nie, J., Cao, C., Jin, Y., Yan, M., Wang, F., et al. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188, 762–773. doi: 10.1111/j.1469-8137.2010.03422.x
Zhang, J. L., Flowers, T. J., and Wang, S. M. (2010). Mechanisms of sodium uptake by roots of higher plants. Plant Soil 326, 45–60. doi: 10.1007/s11104-009-0076-0
Zhang, J. L., Wetson, A. M., Wang, S. M., Gurmani, A. R., Bao, A. K., and Wang, C. M. (2011). Factors associated with determination of root 22Na (+) influx in the salt accumulation halophyte Suaeda maritima. Biol. Trace Elem. Res. 139, 108–117. doi: 10.1007/s12011-010-8644-y
Zhao, K. -F., Fan, H., Song, J., Sun, M. X., Wang, B. Z., Zhang, S. Q., et al. (2005). Two Na+ and Cl− hyperaccumulators of the Chenopodiaceae. J. Integr. Plant Biol. 47, 311–318. doi: 10.1111/j.1744-7909.2005.00057.x
Zhao, K. -F., Song, J., Fan, H., Zhou, S., and Zhao, M. (2010). Growth response to ionic and osmotic stress of NaCl in salt-tolerant and salt-sensitive maize. J. Integr. Plant Biol. 52, 468–475. doi: 10.1111/j.1744-7909.2010.00947.x
Keywords: salt stress, role of K+, transport of Na+ and Cl−, mechanisms of salt tolerance, membrane transporters, ion uptake, symplastic and apoplastic pathway
Citation: Isayenkov SV and Maathuis FJM (2019) Plant Salinity Stress: Many Unanswered Questions Remain. Front. Plant Sci. 10:80. doi: 10.3389/fpls.2019.00080
Edited by:
Meixue Zhou, University of Tasmania, AustraliaReviewed by:
Jayakumar Bose, University of Adelaide, AustraliaFrancisco Rubio, Centro de Edafología y Biología Aplicada del Segura (CEBAS), Spain
Copyright © 2019 Isayenkov and Maathuis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stanislav V. Isayenkov, stan.isayenkov@gmail.com
 Stanislav V. Isayenkov
Stanislav V. Isayenkov Frans J. M. Maathuis
Frans J. M. Maathuis