- 1National Key Laboratory of Crop Genetic Improvement, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
- 2Department of Biochemistry, University of Saskatchewan, Saskatoon, SK, Canada
The ICK/KRP cyclin-dependent kinase (CDK) inhibitors are important plant cell cycle regulators sharing only limited similarity with the metazoan CIP/KIP family of CDK inhibitors. Information is still limited regarding the specific functions of different ICK/KRP genes in planta. We have shown previously that down-regulation of multiple CDK inhibitor ICK/KRP genes up-regulates the E2F pathway and increases cell proliferation, and organ and seed sizes in Arabidopsis. In this study, we observed that the quintuple ick1/2/5/6/7 mutant had more cells in the cortical layer of the root apical meristem (RAM) than the wild type (Wt) while its RAM length was similar to that of the Wt, suggesting a faster cell cycle rate in the quintuple mutant. We further investigated the effects of down-regulating ICK genes on tissue culture responses. The cotyledon explants of ick1/2/5/6/7 could form callus efficiently in the absence of cytokinin and also required a lower concentration of 2,4-D for callus induction compared to the Wt plants, suggesting increased competence for callus induction in the mutant. In addition, the quintuple ick mutant showed enhanced abilities to regenerate shoots and roots, suggesting that increased competence to enter the cell cycle in the quintuple mutant might make it possible for more cells to become proliferative and be utilized to form shoots or roots. These findings indicate that CDK activity is a major factor underlying callus induction and increased cell proliferation can enhance in vitro organogenesis.
Introduction
Cell division is fundamental to plant growth, development, and reproduction. In eukaryotes, cyclin-dependent kinases (CDKs) control cell division cycle, and their activities are in turn modulated by different factors (Morgan, 1997). Among them, CDK inhibitors are crucial negative regulators which inhibit CDK activity through direct protein binding. CDK inhibitor genes were initially identified in mammalian and yeast (Sherr and Roberts, 1995). The first plant CDK inhibitor gene ICK1 (Inhibitor of CDK) was discovered in Arabidopsis (Wang et al., 1997) and there are seven ICK genes (also refered to as KRPs) in Arabidopsis (De Veylder et al., 2001). To date, ICK/KRP genes have been identified from different plant species such as tobacco (Jasinski et al., 2003), maize (Coelho et al., 2005), rice (Barrôco et al., 2006), tomato (Bisbis et al., 2006), apple (Malladi and Johnson, 2011), and avocado (Sabag et al., 2013).
Tissue culture and plant regeneration from explants have many different applications. When proper stimuli are given, somatic plant cells may form adventitious embryos, root, or shoots (De Klerk et al., 1997). Plant regeneration usually takes one of the two pathways: somatic embryogenesis and organogenesis (Davey and Anthony, 2010). The plant regeneration process can be divided conceptually into the following three phases: (1) dedifferentiation, during which the cells acquire competence to respond to the induction stimuli; (2) induction, during which the competent cells are induced to enter particular morphologic pathways; and (3) realization, during which the calli undergo morphological differentiation and development (see review by Duclercq et al., 2011). The organogenesis pathway is more often the route encountered in micropropagation, haploid production and plant transformation (Duclercq et al., 2011). During in vitro organogenesis, callus induction is followed by shoot and root regeneration. It has been demonstrated in a wide range of plant species that generally a high cytokinin (CK) to auxin ratio induces shoot organogenesis, whereas a low ratio results in root development (Sangwan and Sangwan-Norreel, 1990; De Klerk et al., 1997; Davey and Anthony, 2010). In addition to the exogenous plant hormones, other conditions such as nutrient concentrations, sugar sources, and induction time on culture media can also affect the frequency of plant regeneration (Christianson and Warnick, 1983).
In recent years, considerable progress has been made in understanding the developmental events during de novo organogenesis and its underlying molecular mechanisms. Certain genes involved in shoot and root organogenesis processes have been identified (see reviews: Duclercq et al., 2011; Motte et al., 2014). Most of these genes are involved in auxin and CK pathways or shoot meristem maintenance. In addition, ESR1 and ESR2 encoding transcriptional factors of the AP2/ERF family, are identified as enhancers of shoot regeneration in Arabidopsis (Banno et al., 2001; Ikeda et al., 2006), and CUP-SHAPED COTYLEDON1 (CUC1), CLAVATA3/EMBRYO SURROUNDING REGION-RELATED PEPTIDE (CLE2) and GCN5-related N-acetyltransferase 1 (GNAT1) were identified as ESR1 up-regulated genes (Matsuo et al., 2009), while CUC1, Cyclin D1;1 and ARABIDOPSIS PHOSPHOTRANSMITTER 6 (AHP6) were identified as ESR2 up-regulated genes (Ikeda et al., 2006), which might also be involved in shoot regeneration.
Cell division is a prerequisite to both callus induction and shoot/root regeneration during organogenesis. Although there is considerable amount of knowledge on the functions of cell cycle regulators in the cell cycle, relative little is known about their involvement in vitro plant regeneration. Several previous studies showed that cell cycle regulators can affect callus induction. Overexpression of a D-type cyclin has been shown to increase callus induction frequency and callus growth rate in Arabidopsis (Riou-Khamlichi et al., 1999; Cockcroft et al., 2000). In rice, inducible expression of a rice CDK-activating kinase also increases callus induction of tobacco leaf explants (Yamaguchi et al., 2003).
In a previous study, we have reported the effects of down-regulating ICK/KRP CDK inhibitors on plant growth and development using a series of ick mutants (Cheng et al., 2013a). The multiple ick mutants particularly the quintuple mutant had increased CDK activity, up-regulated E2F-RB pathway and enhanced cell proliferation. In this study, we investigated the effects of ICK down-regulation on root cell proliferation and further on callus induction and plant regeneration.
Materials and Methods
Plant Materials and Growth Conditions
The T-DNA insertion lines for the five Arabidopsis ICK/KRP genes used in this study have been described previously (Cheng et al., 2013a). For plants in soil, Arabidopsis plants were grown at 21°C under 16 h/8 h day/night photoperiod in a plant growth room. For seedlings in plates, seeds were sterilized as described (Valvekens et al., 1988) and sowed on ½-strength solid MS medium (1/2 MS, 1% sucrose, 0.7% agar, pH 5.8). The plates were placed vertically in the tissue culture room with the temperature of 22°C and photoperiod of 16 h/8 h day/night.
Root Length and Root Meristem Size Analysis
The seeds of wild type (Wt) and ick1/2/5/6/7 quintuple mutant were sterilized and sowed on square plates containing ½-strength solid MS medium. The plates were placed vertically in the plant tissue culture room. Everyday from 2 to 6 days after germination (DAG), 16–25 seedlings from each line were removed from the plates for root length and meristem size analyses. The root length was measured with a ruler. To measure root meristem size, the root was removed and immersed in Hoyer’s solution chloral hydrate/water/glycerol (3:0.8:0.4) on a glass slide. After 30 min treatment, the slide was covered with a coverslip. The root meristem was observed with DIC (differential interference contrast) under a microscope (Nikon ECLIPSE 80i). The root meristem size was represented by the number of meristematic cortex cells, which was counted as described (Casamitjana-Martíneznez et al., 2003; Ioio et al., 2007). The length of cortex cells in the mature zone was determined at 6 DAG. For each root, an image was taken and cell length measured for 6–10 cortex cells along the mature zone using Image J (http://rsb.info.nih.gov/ij).
Callus Induction and Growth Analyses
Sterilized Wt and mutant Arabidopsis seeds were sowed on square plates containing solid 1/2 MS medium. Seven days after sowing, cotyledons were cut into explants of approximately 4 mm × 4 mm in size. The explants were placed onto 1/2 MS medium containing 0.2 mg/ml 2,4-D (2,4-dichlorophenoxy-acetic acid) or containing both 0.2 mg/ml 2,4-D and 0.2 mg/ml 6-BA (6-benzylaminopurine) solidified with 0.7% agarose. Ten plates for each treatment (about 15 explants in each plate) were used for callus induction and growth analyses. Every the seventh day, the explants were examined to obtain the frequency of explants with callus. Then, the explants were transferred to a fresh plate containing the same medium. The weight of the plate before and after the transfer was measured and the average weight of calli was obtained [=(weight of plate after transfer – weight of plate before transfer)/number of the calli].
To determine the minimal 2,4-D concentration for callus induction, root segments, and excised cotyledons were incubated on ½-strength solid MS medium supplemented with different concentrations of 2,4-D. (for the first batch: 0 mg/L, 0.1mg/L, 0.15 mg/L, and 0.2 mg/L; for the second batch: 0.005 mg/L, 0.01 mg/L, 0.02mg/L, 0.03 mg/L, 0.04 mg/L, 0.05 mg/L). Callus induction frequency was obtained after 10 and 20 days of culture for root explants and after 20 days for cotyledon explants. Fresh weigh was obtained after 20 days of culture. For each treatment, 5–6 plates with 32 root or cotyledon explants in each plate were used.
Root and Shoot Regeneration Analysis
Sterilized Wt and mutant seeds were sowed on square plates containing ½-strength MS solid medium, with the plates placed vertically in the tissue culture room. Seven days after sowing, excised roots were cut into 3–5 mm segments and transferred onto callus induction medium (CIM) containing Gamborg’s B5 salt and vitamins (Gamborg et al., 1968), 2% sucrose, 0.5 g/L MES, 0.48 mg/L 2,4-D, 0.043 mg/L kinetin (KT), and 0.7% agarose. After 7 days of culture on CIM, the root explants with callus were transferred either onto shoot induction medium (SIM) containing Gamborg’s B5 and vitamins, 2% sucrose, 0.5 g/L MES, 1 mg/L isopentenyladenine (2-ip), 0.15 mg/L indole-3-acetic acid (IAA), and 0.7% agarose for regenerating shoots, or onto root induction medium (RIM) containing Gamborg’s B5 and vitamins, 2% sucrose, 0.5 g/L MES, 0.87 mg/L IAA and 0.7% agarose for regenerating roots (Yasutani et al., 1994). For shoot regeneration analysis, each line had 9–10 SIM plates with each plate having ∼50 explants. After 30 days of culture, the calli were surveyed to obtain the frequency of shoot regeneration for each plate.
To determine shoot and root regeneration in different ick mutant lines, calli were first induced from root explants as described above. The explants with callus were then transferred onto SIM for shoot induction or RIM for root induction, with 9–10 plates for each treatment. For shoot induction each plate had ∼40 cultured root explants (root explants with callus), while for root induction each plate about 50 cultured root explants transferred from the CIM. After 20 days of culture on SIM or RIM, the number of calli with regenerating shoots was counted and shoot regeneration frequency obtained for each plate. For root regeneration, we determined the frequency of explants with root induction (=number of calli with regenerating roots/total number of calli). Since the number of roots on each callus varied greatly, we grouped the explants into four categories: (1) no root, (2) 1–5 roots, (3) 6–10 roots and (4) more than 10 roots. The percentages of the four categories were calculated for each line.
RNA Extractions and Real-time PCR
Arabidopsis total RNA was isolated using TRIzol reagent (Invitrogen) according to manufacturer’s instructions. First-strand cDNA synthesis and real-time PCR analysis were performed as described previously (Zhang et al., 2015). The primers for real-time PCR are listed in Supplementary Table S1.
Statistical Analysis
The Mann–Whitney two-tailed U test was used for analyzing the differences in callus induction rates from cotyledon explants and root induction rates from root explants. Fisher’s least significant difference (LSD) method was used for multiple comparisons of shoot regeneration rates of the Wt and mutants. For the other differences between the mutant and Wt, The Student’s t-test (T-test) method was used and the analysis was performed.
Results
Cell Division was Accelerated in Root Cortex Meristematic Cells of ick1/2/5/6/7 Mutant
In our previous study, we established a series of T-DNA insertion lines in which one to five ICK genes were knocked out, and observed phenotypical changes in triple, quadruple and quintuple mutants. Notably the quintuple mutant ick1 ick2 ick5 ick6 ick7 (referred to as ick1/2/5/6/7 for shoot) has larger leaves, petals, and seeds than the Wt (Cheng et al., 2013a), suggesting that cell proliferation is promoted in the ick1/2/5/6/7 quintuple mutant, as a result of down-regulating ICK genes.
To determine more specifically how cell proliferation is affected in the mutant, we examined cell production in the root. First, we analyzed the root growth of the ick1/2/5/6/7 mutant and Wt plants from 2 to 6 days after seed plating. As shown in Figure 1A, the primary root length for ick1/2/5/6/7 quintuple mutant was very similar to that of the Wt. We then investigated the cortex cells in the mature zone of the roots at 6-day stage after germination (DAG). The length of cortex cells in different positions along each root was measured, and the total cortex cell number estimated based on the root length and average cortex cell length. The results showed that the average length of the cortex cells in the mature zone of ick1/2/5/6/7 quintuple mutant was reduced compared with that of Wt (137.7 ± 3.6 um compared to 168.9 ± 4.1 um in Wt). Since the mutant and the Wt lines had a similar root length (36.3 ± 3.9 mm and 36.3 ± 3.2 mm at 6 DAG, respectively), the total cell number in a cortex cell file along the mature zone of ick1/2/5/6/7 quintuple mutant (264.0 ± 8.8) was significantly higher than that of the Wt (229.1 ± 8.2; Figures 1B,C; Figure 1B shows the relative ratio of the mutant to Wt).
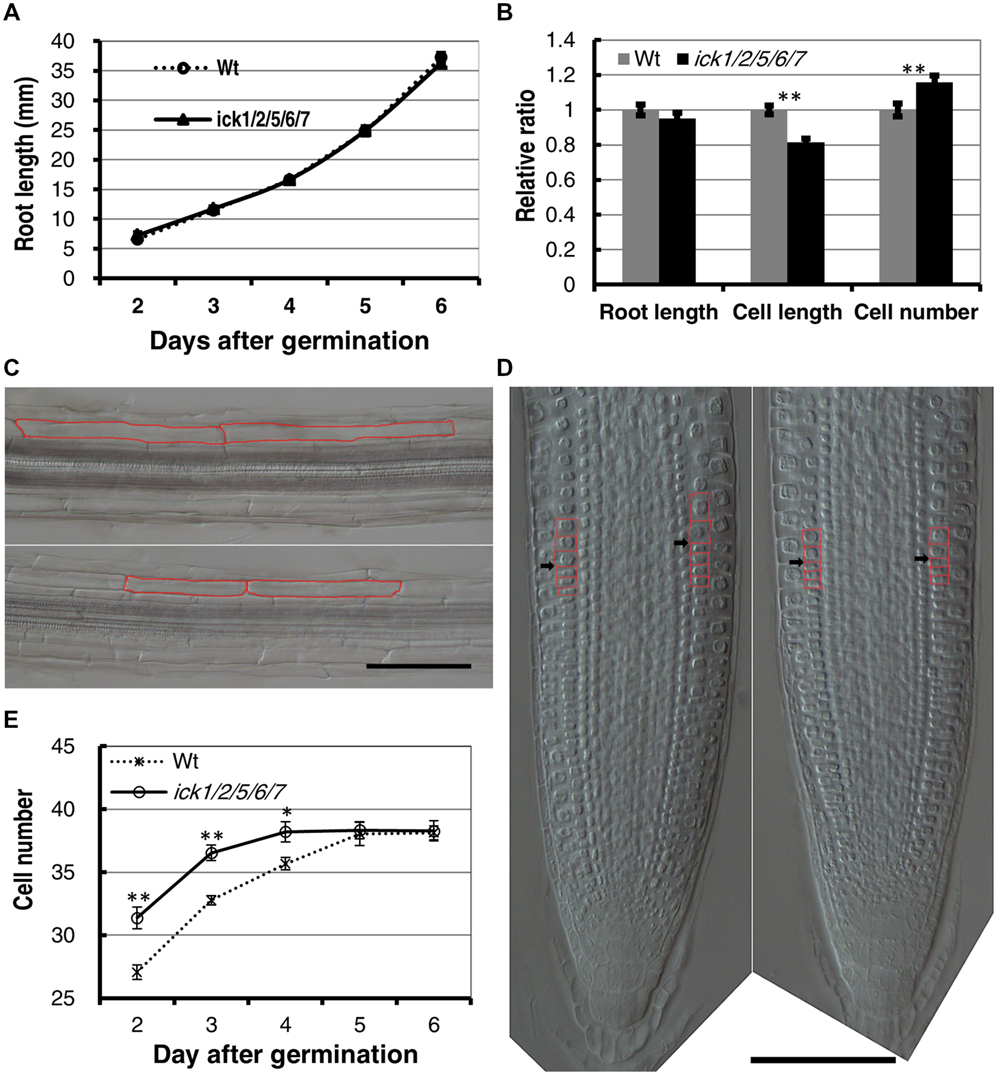
FIGURE 1. Cell length, cell number, and root apical meristem size of wild type (Wt) and ick1/2/5/6/7 mutant Arabidopsis plants. (A) Primary root length of the Wt and ick1/2/5/6/7 quintuple mutant at 2–6 days after germination (DAG). Each datum point represents the average of 16–25 seedlings. (B) Relative root length, cortex cell length and cortex cell number of the mature zone of the Wt and ick1/2/5/6/7 mutant plants at 6 DAG (relative ratio = value of mutant/value of Wt). For root length and cell length analyses, 20–22 seedlings were used. For cell length analysis, at least six cells in the mature zone were measured for each root tip. (C) Differential interference contrast (DIC) images showing the cortex cells of the Wt (upper) and ick1/2/5/6/7 quintuple mutant (lower) roots. Two cortex cells of each line are marked in red. (D) Longitudinal view of root tips of 6-day-old Wt and ick1/2/5/6/7 quintuple (right) plants. The root tips were placed on a glass slide, covered with a coverslip, and viewed under a DIC microscope. The cortex cells around the transition zone are marked in red, and black arrowheads indicate the boundary between meristem and elongation zone. (E) Meristem size of Wt and ick1/2/5/6/7 quintuple mutant plants at 2-6 DAG. The root meristem is expressed as the number of cortex cells in a file extending from the quiescent center (QC) to the first elongated cell. The Student’s t-test was used for analyzing the difference between the Wt and mutant [error bar = SE (standard error); ∗P < 0.05, ∗∗P < 0.01]. Each datum point represents the average of 16–25 seedlings. Bars = 100 um.
To determine whether the quintuple mutant has a larger root meristem, we performed a time-course analysis on root meristem size following Ioio’s method (Ioio et al., 2007). In this assay, the root-meristem size was expressed as the number of cortex cells in a file extending from the quiescent center (QC) to the first elongated cell (Figure 1D). We found that the roots of ick1/2/5/6//7 quintuple mutant and Wt had a similar final root meristem size. However, the ick1/2/5/6/7 quintuple mutant reached the final size 4 DAG, while the Wt reached this final size 5 DAG (Figure 1E), suggesting an accelerated rate of cell division and reduced cell elongation in the mutant. These results imply that more cells in the cortex of the quintuple mutant are likely due to a faster cell production rate, instead of a larger root meristem.
Down-regulation of Five ICKs Increased Callus Induction
To further understand the impact of ICK down-regulation, we examined tissue culture responses since cell proliferation is critical for callus and plant regeneration. Cotyledon explants of both Wt and ick1/2/5/6/7 mutant produced calli efficiently on 1/2 MS medium containing both 0.2 mg/ml 2,4-D and 0.2 mg/ml 6-BA. On 1/2 MS medium containing 0.2 mg/ml 2,4-D, however, there was a higher frequency of callus induction for the mutant explants (Figures 2A,B). For instance, 98.4% of the ick1/2/5/6/7 mutant explants produced calli, compared 69.1% for the Wt (Figure 2C). In addition, the calli of quintuple mutants were much larger with lightly greenish color, while those of the Wt yellower and smaller (Figure 2B). These results indicate that down-regulation of the five ICK genes enhances callus formation and reduces CK requirement for callus induction.
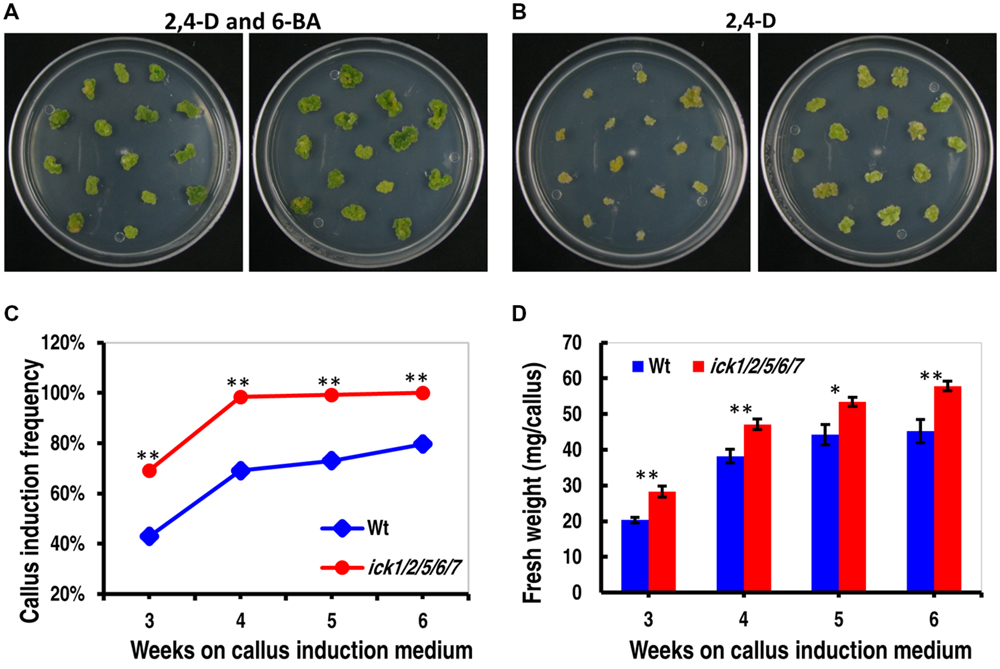
FIGURE 2. Callus induction from cotyledon explants and callus growth of Wt and ick1/2/5/6/7 plants. (A,B) Representative plates showing callus induction from cotyledon explants of Wt (left) and ick1/2/5/6/7 mutant (right) in the presence of 2,4-D and 6-BA (A) or 2,4-D alone (B). Similar results were obtained in three independent experiments. In each experiment, 10 plates were used for each treatment, with each plate having around 15 explants. The data from one experiment are shown here. (C) Callus induction frequency of the Wt and ick1/2/5/6/7 mutant explants on culture medium containing 2,4-D alone. The difference in the induction rate between the Wt and mutant at each time point was compared by Mann–Whitney U test. (D) Fresh weight of calli from the Wt and ick1/2/5/6/7 mutant cotyledon explants after 3–7 weeks of culture on culture medium containing both 2,4-D and 6-BA. The differences between the Wt and mutant were analyzed by Student’s t-test (error bar = SE; ∗P < 0.05, ∗∗P < 0.01).
To determine callus growth rate, the explants were transferred to fresh callus induction plates every week, and the callus growth was obtained by weighing the plate immediately after the transfer and on the seventh day of culture. As shown in Figure 2D and Supplementary Figure S1, the calli of ick1/2/5/6/7 grew faster than those of Wt in the presence of 6-BA and 2,4-D or 2,4-D only. Those results indicate that down-regulation of the five ICK genes also enhances callus growth.
Auxin Dependency for Callus Induction was Decreased in the ick1/2/5/6/7 Mutant
To further confirm that ICK down-regulation reduces auxin requirement for callus induction, we determined the minimal 2,4-D concentration for callus induction from root explants of both Wt and mutant plants. In this assay, the root segments (about 5 mm in length) were incubated on the 1/2 MS medium supplemented with different concentrations of 2,4-D. We first used 2,4-D concentrations of 0, 0.05, 0.1, 0.15, and 0.2 mg/L. Neither the Wt nor the quintuple mutant showed callus induction on 1/2 MS medium without 2,4-D after 20 days of culture; whereas, on the culture plates containing 0.05, 0.1, 0.15, or 0.2 mg/L 2,4-D, almost all segments of both lines generated calli (Supplementary Figures S2A,B). We then used a series of lower 2,4-D concentrations of 0.005, 0.01, 0.02, 0.03, 0.04, and 0.05 mg/L. The root explants of both lines produced no callus at 0 and 0.005 mg/L 2,4-D, and almost all root explants of both lines produced calli at 0.02, 0.03, 0.04, and 0.05 mg/L 2,4-D after 20 days of culture (Figure 3A). At 0.01 mg/ml 2,4-D, the callus induction frequency of ick1/2/5/6/7 mutant (61.1%) was significantly higher than that of the Wt (19.9%; Figure 3B). This observation suggests that the ick1/2/5/6/7 mutant needs a lower concentration of auxin for callus induction compared to Wt. Moreover, for the 2,4-D concentrations at which the callus induction frequencies of the two lines were comparable, the fresh weights of calli from the mutant root explants were significantly higher than those of Wt (Figure 3C and Supplementary Figure S2C), consistent with observation made with the cotyledon explants.
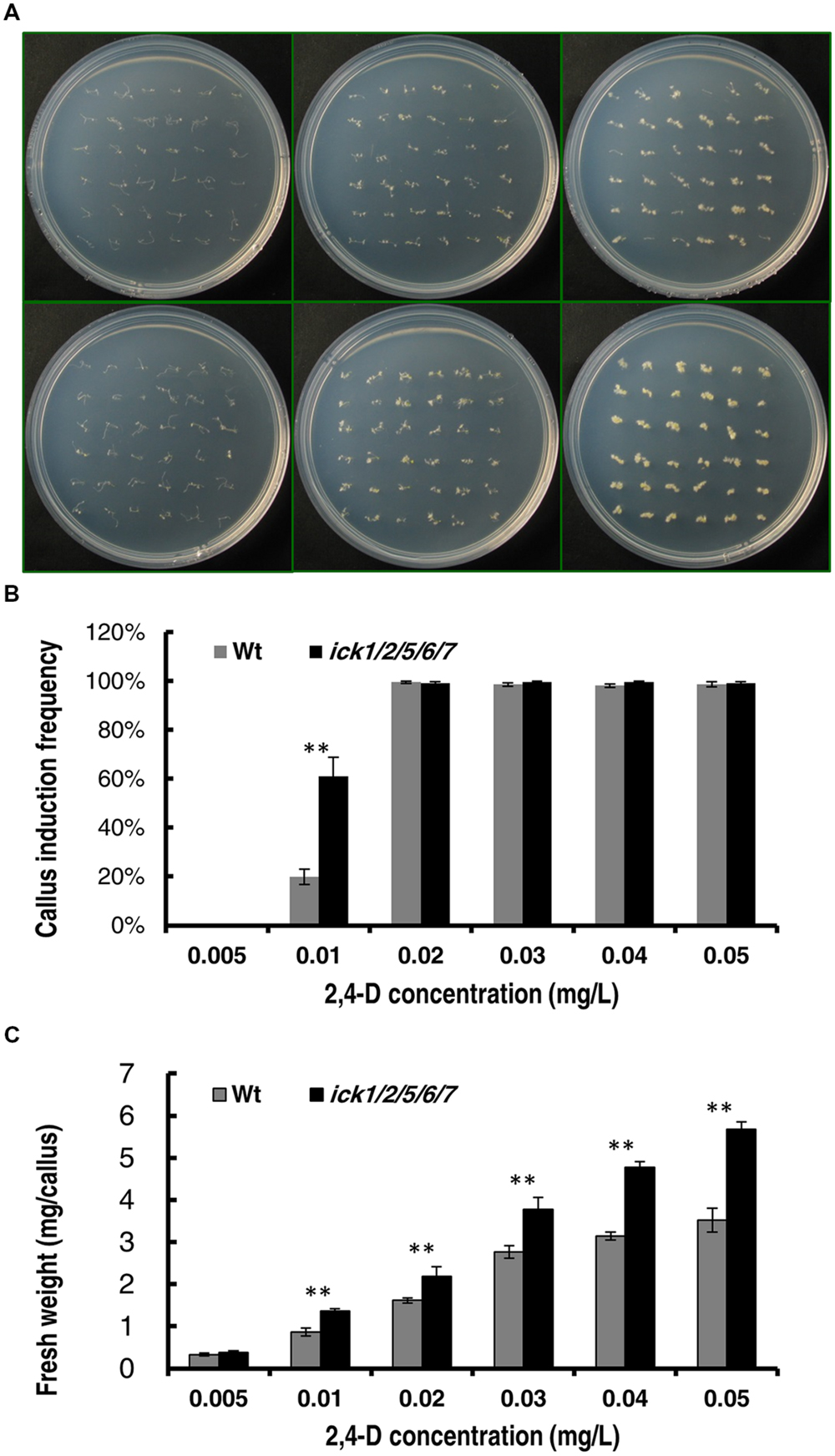
FIGURE 3. Callus induction from root explants of the Wt and ick1/2/5/6/7 mutant at different 2,4-D concentrations. Root segments were cultured on 1/2 MS medium containing the indicated concentrations of 2,4-D for 20 days. (A) Representative plates showing the root explants of Wt (the upper row) and ick1/2/5/6/7 mutant (the lower row) on 1/2 MS medium containing 0.05, 0.01, and 0.05 mg/L (the first–third column, respectively) 2,4-D. (B) Callus induction rate of root explants of Wt and ick1/2/5/6/7 mutant after 20 days of culture. (C) Fresh weight of root explants with callus of Wt and ick1/2/5/6/7 after 20 days of culture. The bars show the mean values of 4–5 plates. Student’s t-test was used for analyzing the differences in induction rate and fresh weight between the Wt and mutant (error bar = SE; ∗∗P < 0.01).
We also determined the minimal 2,4-D concentration required for callus induction from cotyledonary petiole of both lines. When the 2,4-D concentration was higher than 0.05 mg/ml, almost all of the cotyledonary petiole explants of ick1/2/5/6/7 produced calli (Supplementary Figures S3A,B). When the 2,4-D concentration was lower than 0.1 mg/ml, the callus induction frequency of the ick1/2/5/6/7 mutant was significantly higher than that of the Wt (Figures 4A,B and Supplementary Figures S3A,B). At 0.005 mg/ml 2,4-D, 33% of ick1/2/5/6/7 cotyledonary petioles showed callus induction, while no cotyledonary petioles of the Wt did (Figures 4A,B). Also, at various concentrations of 2,4-D the calli of the ick1/2/5/6/7 mutant were significantly larger than those of Wt (Figure 4C and Supplementary Figure S3C), further confirming that the callus of ick1/2/5/6/7 mutant was growing faster than the Wt callus.
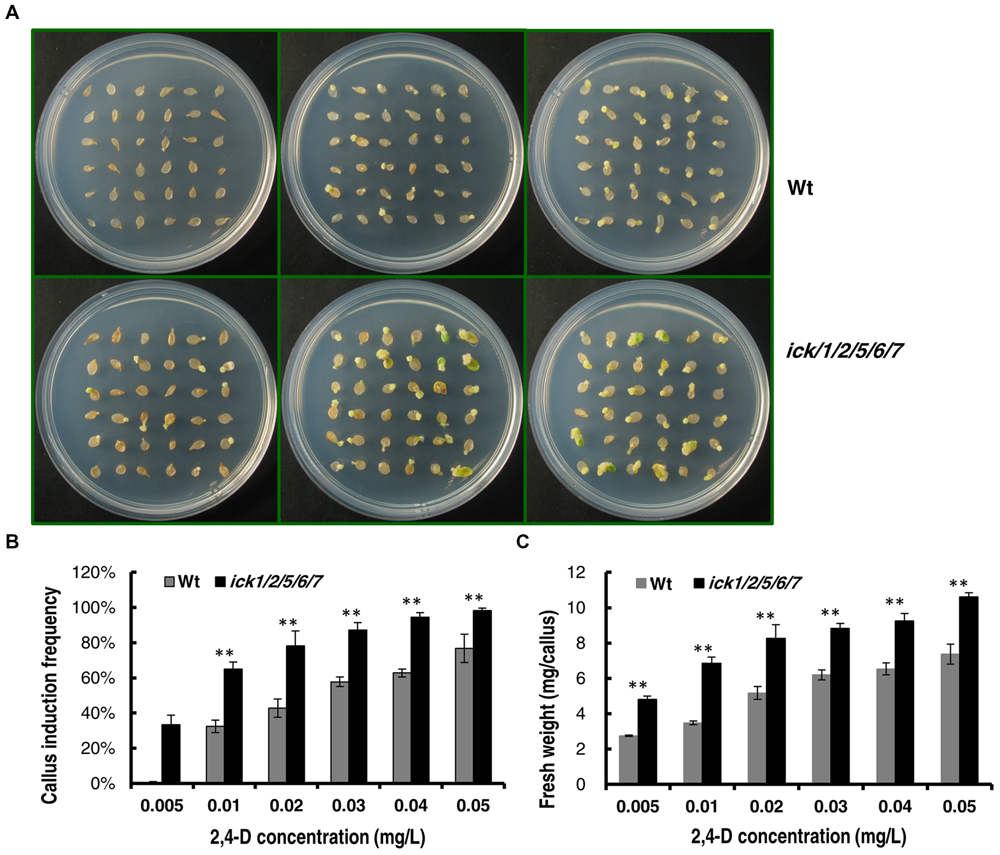
FIGURE 4. Callus induction from cotyledon explants of the Wt and ick1/2/5/6/7 mutant at different 2,4-D concentrations. The cotyledons were excised form 7-day-old seedlings and cultured on 1/2 MS medium for 20 days. (A) Representative plates showing the cotyledon explants of Wt (the upper row) and ick1/2/5/6/7 mutant (the lower row) on 1/2 MS medium containing 0.005 mg/L, 0.02g/L, 0.04 mg/L (the first–third column) 2,4-D. (B) Callus induction rate of the cotyledon explants of ick1/2/5/6/7 and Wt on the indicated media. (C) Fresh weight of the cotyledon explants with callus of ick1/2/5/6/7 and Wt on the indicated media. The bars show the mean induction rates of 4–5 plates. Student’s t-test was used for analyzing the differences in induction rate and fresh weight between the Wt and mutant (error bar = SE; ∗∗P < 0.01).
Shoot and Root Regeneration was Enhanced in ick1/2/5/6/7 Mutant
In addition to callus induction, shoot and root regeneration is another important aspects of plant tissue culture. Thus, we investigated the ability of the root explants to regenerate shoots. Root explants (about 5 mm segments) were first cultured on CIM. After 7 days, explants with callus were transferred to SIM. As shown in Supplementary Figure S4, the shoot regeneration frequency of ick1/2/5/6/7 quintuple mutant was significantly higher than that of Wt after 30 days of culture. Also, each of the explants of ick1/2/5/6/7 quintuple mutant on average regenerated more roots than that of Wt. Those results indicate that the ick1/2/5/6/7 quintuple mutant has a stronger ability to regenerate shoots and roots.
Disruption of ICK Genes Additively Promoted Shoot and Root Regeneration
Our previous results on a series T-DNA mutants showed that the effects from down-regulating ICK genes become more evident as more ICK genes are disrupted (Cheng et al., 2013a). Therefore, we selected a series of single, double, triple, quadruple, and quintuple mutants to determine whether such an additive effect of multiple loci also exists for shoot and root regeneration. For this analysis, root explants with callus were transferred onto SIM or RIM (RIM) for shoot or root regeneration. After culturing on SIM for 20 days, the single ick1 mutant had a similar frequency of calli with regenerating shoots to the Wt, and there was a trend of increasing regeneration frequency from the single to the quintuple mutant (Figure 5A). Although the differences among Wt, ick1/2, ick1/2/7, ick1/2/6/7 did not reach a significant level, the quintuple ick1/2/5/6/7 mutant with the highest frequency showed significant differences from the other lines (Figure 5B).
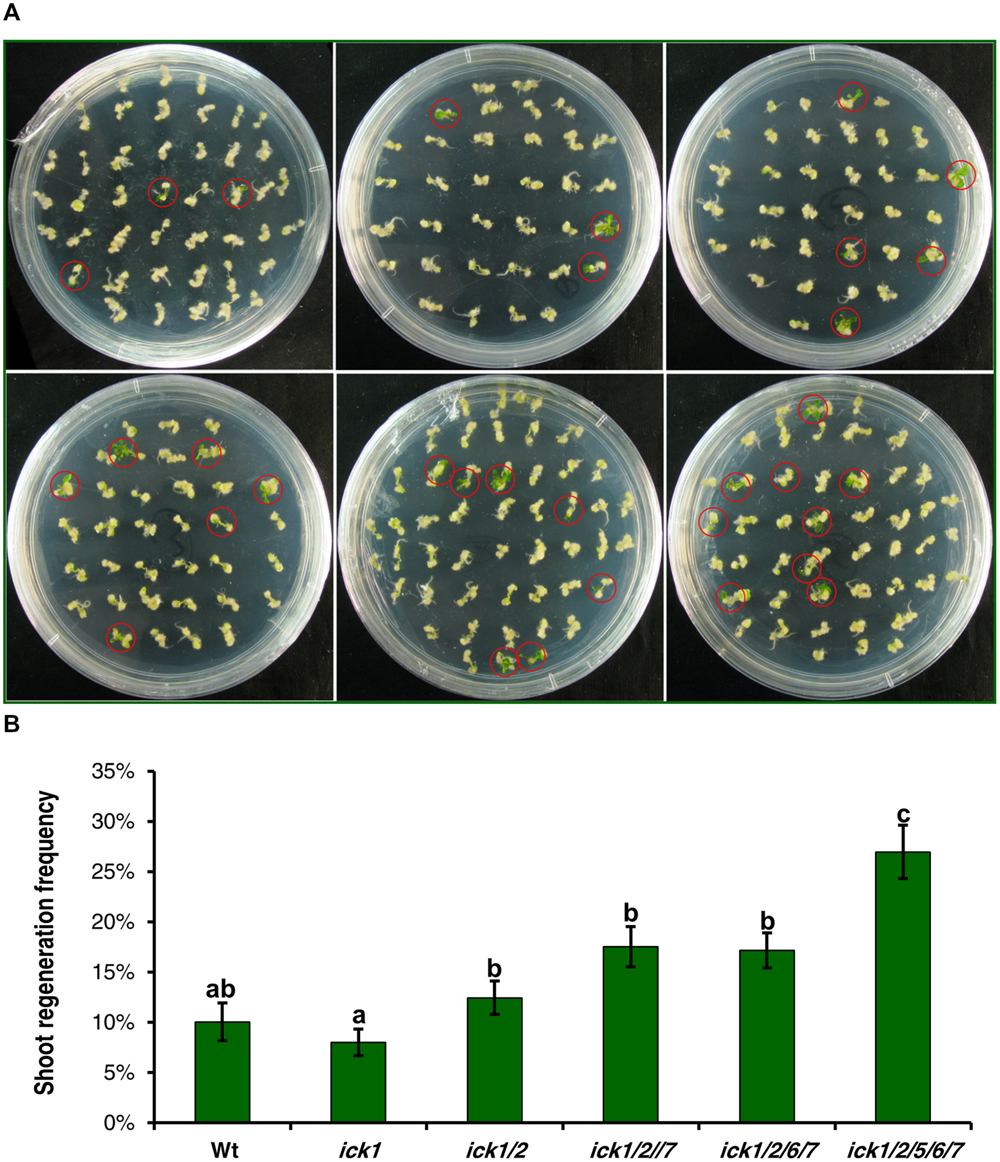
FIGURE 5. Shoot regeneration from root-derived calli of Wt and various ick mutant plants. Root explants were cultured on callus induction medium (CIM) first. After 7 days, root explants with callus were transferred onto shoot induction medium (SIM) and cultured for 30 days. For each line, 9–10 plates were used with each plate having about 40 explants. (A) Representative plates showing shoot regeneration on SIM. The plates in the first row are: Wt (left), ick1 (middle), ick1/2 (right), and in the second row, ick1/2/7 (left), ick1/2/6/7 (middle), and ick1/2/5/6/7 (right). The red circles mark the calli with regenerating shoots. (B) Frequency of shoot regeneration in Wt and ick mutants (Mean ± SE). The significant differences among different lines were analyzed by Fisher’s least significant difference (LSD) method, and are indicated by different lowercase letters (P < 0.05).
Calli were also cultured on RIM for 20 days, although most calli of all the lines produced roots, the calli of ick1/2/6/7 and ick1/2/5/6/7 mutants had visibly more roots than Wt and ick1, ick1/2 and ick1/2/7 mutants (Supplementary Figure S5A). We first compared the root regeneration rate of the six lines. The root regeneration frequencies of ick1/2/6/7 and ick1/2/5/6/7 were significantly higher than those of other lines (Supplementary Figure S5B). To better characterize the number of roots per callus, the calli were grouped into four categories, with 0, 1–5, 6–10, and more than 10 roots per callus, respectively. The calli of ick1/2/6/7 and ick1/2/5/6/7 mutants had significant more roots than the calli of other lines (Supplementary Figure S5C). Those results indicate that down-regulation of ICK genes additively enhances both the abilities for shoot and root regeneration.
E2F-dependent Genes and Shoot Regeneration Related Genes were Mostly Up-regulated During Shoot Regeneration of ick1/2/5/6/7
In our previous studies, we have demonstrated that the E2F-dependent genes are up-regulated in the ick1/2/5/6/7 seedlings (Cheng et al., 2013a). The expression levels of the same group of E2F-dependent genes that function in cell cycle, DNA synthesis, chromatin structure, metabolism, plant development, cell structure, and light signaling/photosynthesis (Ramirez-Parra et al., 2003; de Jager et al., 2009) were analyzed in the SIM incubated callus of ick1/2/5/6/7 mutant and Wt. Of the 24 genes analyzed, 20 had a higher level of expression in the mutant, with 17 of them having a relative fold expression higher than 1.19 (equal to the Log2 value of 0.25). The four down-regulated genes are MCM3, HTH/EDA17, KICP-02, and CCA1 (Figure 6A). This result suggests that E2F-pathway was also enhanced in the callus of ick1/2/5/6/7 mutant as observed in the seedlings (Cheng et al., 2013a).
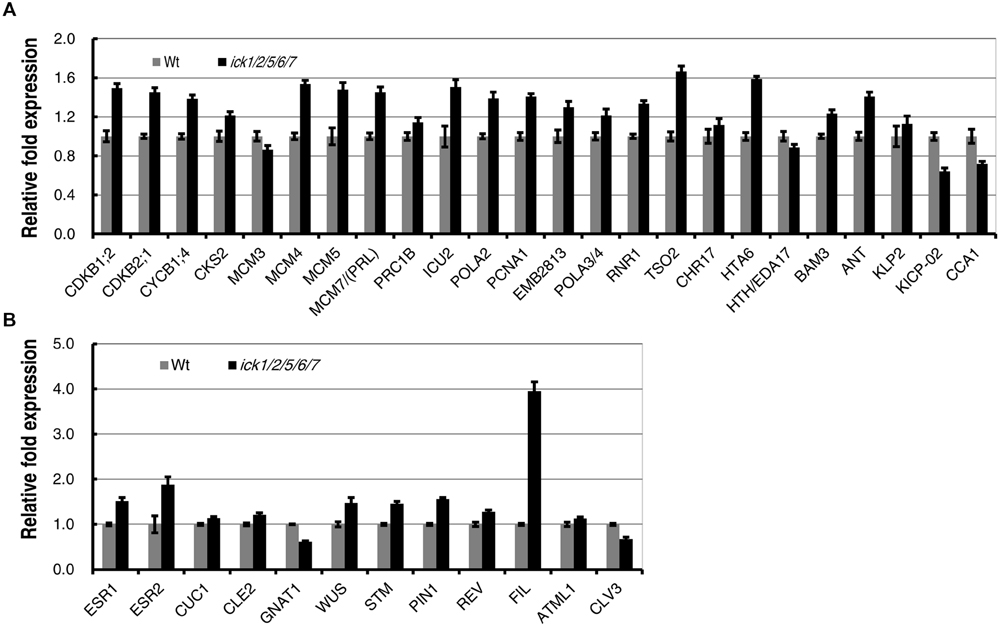
FIGURE 6. Expression of E2F-dependent and shoot regeneration related genes during in vitro shoot regeneration of the Wt and ick1/2/5/6/7 mutant. Root explants were cultured on CIM for 7 days and then transferred onto SIM. Total RNA was extracted after 7 days of culture on SIM. Each value on the vertical axes indicates a relative fold expression level calculated by scaling to the Ubq10c transcript level. Data present the average of four biological repeats and error bars indicated standard errors. Each biological repeat has four technical replicates. (A) E2F-dependent genes. (B) Shoot regeneration related genes.
It has been demonstrated that a number of genes were up-regulated during de novo shoot regeneration (Duclercq et al., 2011; Motte et al., 2014). To investigate whether the increased shoot regeneration ability of ick1/2/5/6/7 mutant is accompanied by the up-regulation of these de novo regeneration related genes, a group of 12 genes were selected and their expression levels analyzed in the calli cultured on SIM for shoot regeneration. Of the 12 genes, ESR1 and ESR2 are known as the enhancers of shoot regeneration (Banno et al., 2001; Matsuo et al., 2009), CUC1, CLE2, and GNAT1 are induced by ESR1 (Matsuo et al., 2009), and WUS, STM, PIN1, REV, FIL, ATML1, and CLV3 are up-regulated during de novo shoot formation (Gordon et al., 2007; Duclercq et al., 2011). Among these 12 genes, 10 were up-regulated in the calli of ick1/2/5/6/7 mutant, while only 2 (GNAT1 and CLV3) were down-regulated (Figure 6B). Interestingly, the FIL was highly up-regulated in the callus of ick1/2/5/6/7 with about fourfold of expression relative to that in the Wt calli.
Discussion
Down-regulation of Five CDK Inhibitor Genes Promotes Cell Proliferation in Roots
Root growth is determined by the balance between cell division and cell elongation (Beemster and Baskin, 1998). The defined cortical layer development in Arabidopsis provides a good tool to investigate the cell proliferation rate (Ivanov and Dubrovsky, 1997). Several studies using this approach have revealed that a reduced root apical meristem (RAM) size is responsible for the observed inhibition of primary root growth under different conditions (Westet al., 2004; Ubeda-Tomás et al., 2009; González-García et al., 2011). In this study, we observed that the same number of cortex meristematic cells in ick1/2/5/6/7 quintuple mutant generated more but smaller cells in the mature zone compared to the Wt, indicating an accelerated rate of cell division in the mutant. Previously, we have shown that CDK activity (most likely CDKA) is enhanced in the ick mutants (Cheng et al., 2013a), which is likely to be responsible for the increased rate of cell division in roots.
CDK Kinase Activity is a Major Factor Underlying Callus Induction
In various plant species, callus induction depends on exogenous application of both auxin and CK. The factors underlying callus induction/repression and the genes involved in these processes have been reviewed recently by Ikeuchi et al. (2013). It has been observed that the transgenic Arabidopsis overexpressing a putative CK receptor CKI1 could produce callus efficiently in the absence of CK (Kakimoto, 1996), indicating the significance of CK in callus induction. Also, leaf explants of transgenic Arabidopsis constitutively expressing a cell cycle regulator gene CYCD3 could also produce calli in the absence of exogenous CK (Riou-Khamlichi et al., 1999). It has been shown that CK promotes G1/S and G2/M transitions through regulating CDK activities (Sieberer et al., 2003; Del Pozo et al., 2005). Furthermore, overexpression of rice R2, a CDK-activating kinase, also results in CK-independent callus induction in tobacco (Yamaguchi et al., 2003). In this study, we showed that callus could be induced in the ick1/2/5/6/7 quintuple mutant in the absence of CK. The increased CDK activity in the ick1/2/5/6/7 mutant must have lowered the threshold requirement for CK as well as auxin for cells in the explants to enter and progress through the cell cycle. In addition, antisense expression of Nicto;CYCA3;2 in tobacco has been observed to impairs callus formation (Yu et al., 2003). Together, these results suggest that regulation of CDK activity is a key determinant of callus induction.
It is well known that auxin along with auxin signaling modules is required for callus formation (Fan et al., 2012; Perianez-Rodriguez et al., 2014). Part of the auxin requirement may be for up-regulating certain cell cycle genes. It has been shown that Arabidopsis CDKA is induced by auxin (Hemerly et al., 1993). Our results showing that knockdown of ICK genes also lowers the threshold requirement for auxin in terms of callus induction suggests that the effect of auxin on callus induction is at least partially through CDK.
It has been reported that transgenic Arabidopsis plants overexpressing two TFs genes, HB52 and CRF3, exhibit spontaneous callus formation without exogenous phytohormone in some organs (Xu et al., 2012). It is not known whether CDK activity is enhanced in those transgenic plants. However, genome-wide transcriptome profiling during callus initiation has revealed the up-regulation of many cell-cycle related genes (Xu et al., 2012). Thus, it is possible that HB52 and CRF3 promote callus formation in the absence of exogenous hormones through these cell cycle genes.
Increased Cell Proliferation Enhances In Vitro Organogenesis
In the classical scheme for plant regeneration, the explants undergo dedifferentiation to obtain pluripotency during callus induction. The calli are then transferred to SIM or RIM to induce shoots and roots, respectively. Sometimes, shoots and roots can be induced at the same time. During callus induction, founder meristem cells arise in the pericycle of root explants (Atta et al., 2009). Depending on the subsequent culture conditions, the cell fate of organ primordia is determined to be either shoot or root identity (Christianson and Warnick, 1983; Atta et al., 2009). During in vitro organogenesis, auxin is the main phytohormone for root organogenesis, while CK promotes shoot organogenesis (Skoog and Miller, 1957; Duclercq et al., 2011; da Rocha Correa et al., 2012). Thus, the auxin-CK crosstalk is important for organ formation and identity determination (Gordon et al., 2009; Besnard et al., 2011; Cheng et al., 2013b; Zhao et al., 2013). Motte et al. (2014) reviewed the mutants with altered regeneration phenotypes, and noticed that most of the genes are related to meristem maintenance, and auxin and CK signaling.
Our findings that the quintuple ick mutant with increased CDK activity showed increased shoot/root organogenesis from both the root and cotyledonary explants indicate enhanced competence for cell proliferation can also promote organogenesis. We speculate that under the auxin and CK conditions favoring organ formation, increased competence to enter the cell cycle in the quintuple mutant makes it possible for more cells to become proliferative and capable of forming shoots or roots, while under conditions favoring unorganized growth increased competence of cell proliferation makes it easier to induce callus formation. Consistent with this suggestion, application of cell cycle inhibitors during SIM incubation was shown to significantly impair organogenesis in Arabidopsis (Che et al., 2007). In addition, Arabidopsis ESR1 and ESR2, belonging to the AP2/EAR family transcription factors, both could enhance shoot regeneration (Banno et al., 2001; Ikeda et al., 2006). Overexpression of ESR2 has been shown to up-regulate cell cycle genes (Ikeda et al., 2006), suggesting that ESRs may promote shoot regeneration by up-regulating cell cycle machinery and cell proliferation.
We further observed that E2F-dependent genes and shoot regeneration related genes generally showed higher levels of expression in the ick1/2/5/6/7 during shoot regeneration compared to Wt plants (Figures 6A,B) suggesting that enhanced shoot regeneration is accompanied by the up-regulation of a consort of genes involved in the regeneration process. Interestingly, among them, FIL was highly up-regulated in ick1/2/5/6/7 (Figure 6B). Although the regulation of FIL expression in the quintuple mutant during shoot regeneration is unknown, it is interesting to note that FIL gene encodes a YABBY (YAB) family putative transcription factor that has been implicated in specifying abaxial cell identities and thus being involved in development of leaves and floral organs, and in meristem activity (Sawa et al., 1999; Lugassi et al., 2010).
Conclusion
In this study, we have demonstrated that down-regulation of CDK inhibitor genes results in enhanced shoot/root regeneration. To date, while efficient plant regeneration system has been established in a range of plant species, other plant species remain recalcitrant. Since regulation of the cell cycle by CDK is conserved through plants and all eukaryotes, it is tempting to speculate that callus induction can be enhanced through modulating CDK activity in other plants as well. In vitro plant regeneration has been optimized most empirically by testing a variety of hormonal and culture conditions. The realization that CDK regulation plays a key role provides molecular means to enhance plant regeneration and possibly plant transformation for applications in different plants, particularly crop species.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Arabidopsis Biological Resource Center and Nottingham Arabidopsis Stock Centre for the T-DNA insertion mutants. HW is grateful for financial support (Discovery grant) from the Natural Sciences and Engineering Research Council of Canada, YZ is grateful for financial support from the Ministry of Science and Technology of China (grants 2006CB101604 and 2011AA10A104) and the National Natural Science Foundation of China (30471097), and YC is grateful for financial support from China Postdoctoral Science Foundation (2014M562036).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00825
References
Atta, R., Laurens, L., Boucheron-Dubuisson, E., Guivarc’h, A., Carnero, E., Giraudat-Pautot, V., et al. (2009). Pluripotency of Arabidopsis xylem pericycle underlies shoot regeneration from root and hypocotyl explants grown in vitro. Plant J. 57, 626–644. doi: 10.1111/j.1365-313X.2008.03715.x
Banno, H., Ikeda, Y., Niu, Q. W., and Chua, N. H. (2001). Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 13, 2609–2618. doi: 10.1105/tpc.13.12.2609
Barrôco, R. M., Peres, A., Droual, A. M., De Veylder, L., De Wolf, J., Mironov, V., et al. (2006). The cyclin-dependent kinase inhibitor Orysa. KRP1 plays an important role in seed development of rice. Plant Physiol. 142, 1053–1064.
Beemster, G. T., and Baskin, T. I. (1998). Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 116, 1515–1526. doi: 10.1104/pp.116.4.1515
Besnard, F., Vernoux, T., and Hamant, O. (2011). Organogenesis from stem cells in planta: multiple feedback loops integrating molecular and mechanical signals. Cell. Mol. Life Sci. 68, 2885–2906. doi: 10.1007/s00018-011-0732-4
Bisbis, B., Delmas, F., Joubès, J., Sicard, A., Hernould, M., Inzé, D., et al. (2006). Cyclin-dependent kinase (CDK) inhibitors regulate the CDK-cyclin complex activities in endoreduplicating cells of developing tomato fruit. J. Biol. Chem. 281, 7374–7383. doi: 10.1074/jbc.M506587200
Casamitjana-Martínez, E., Hofhuis, H. F., Xu, J., Liu, C. M., Heidstra, R., and Scheres, B. (2003). Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13, 1435–1441. doi: 10.1016/S0960-9822(03)00533-5
Che, P., Lall, S., and Howell, S. H. (2007). Developmental steps in acquiring competence for shoot development in Arabidopsis tissue culture. Planta 226, 1183–1194. doi: 10.1007/s00425-007-0565-4
Cheng, Y., Cao, L., Wang, S., Li, Y., Shi, X., Liu, H., et al. (2013a). Downregulation of multiple CDK inhibitor ICK/KRP genes upregulates the E2F pathway and increases cell proliferation, and organ and seed sizes in Arabidopsis. Plant J. 75, 642–655. doi: 10.1111/tpj.12228
Cheng, Z. J., Wang, L., Sun, W., Zhang, Y., Zhou, C., Su, Y. H., et al. (2013b). Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol. 161, 240–251. doi: 10.1104/pp.112.203166
Christianson, M., and Warnick, D. (1983). Competence and determination in the process of in vitro shoot organogenesis. Dev. Biol. 95, 288–293. doi: 10.1016/0012-1606(83)90029-5
Cockcroft, C. E., den Boer, B. G., Healy, J. S., and Murray, J. A. (2000). Cyclin D control of growth rate in plants. Nature 405, 575–579. doi: 10.1038/35014621
Coelho, C. M., Dante, R. A., Sabelli, P. A., Sun, Y., Dilkes, B. P., Gordon-Kamm, W. J., et al. (2005). Cyclin-dependent kinase inhibitors in maize endosperm and their potential role in endoreduplication. Plant Physiol. 138, 2323–2336. doi: 10.1104/pp.105.063917
da Rocha Correa, L., Troleis, J., Mastroberti, A., Mariath, J., and Fett-Neto, A. (2012). Distinct modes of adventitious rooting in Arabidopsis thaliana. Plant Biol. 14, 100–109. doi: 10.1111/j.1438-8677.2011.00468.x
Davey, M. R., and Anthony, P. (2010). Plant Cell Culture: Essential Methods. Hoboken, NJ: John Wiley & Sons.
de Jager, S. M., Scofield, S., Huntley, R. P., Robinson, A. S., den Boer, B. G., and Murray, J. A. (2009). Dissecting regulatory pathways of G1/S control in Arabidopsis: common and distinct targets of CYCD3; 1. E2Fa and E2Fc. Plant Mol. Biol. 71, 345–365. doi: 10.1007/s11103-009-9527-5
De Klerk, G. J., Arnholdt-Schmitt, B., Lieberei, R., and Neumann, K. H. (1997). Regeneration of roots, shoots and embryos: physiological, biochemical and molecular aspects. Biol. Plant. 39, 53–66. doi: 10.1023/A:1000369309303
Del Pozo, J. C., Lopez-Matas, M., Ramirez-Parra, E., and Gutierrez, C. (2005). Hormonal control of the plant cell cycle. Physiol. Plant. 123, 173–183. doi: 10.1111/j.1399-3054.2004.00420.x
De Veylder, L., Beeckman, T., Beemster, G. T., Krols, L., Terras, F., Landrieu, I., et al. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653–1668. doi: 10.2307/3871392
Duclercq, J., Sangwan-Norreel, B., Catterou, M., and Sangwan, R. S. (2011). De novo shoot organogenesis: from art to science. Trends Plant Sci. 16, 597–606. doi: 10.1016/j.tplants.2011.08.004
Fan, M., Xu, C., Xu, K., and Hu, Y. (2012). Lateral organ boundaries domain transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 22, 1169–1180. doi: 10.1038/cr.2012.63
Gamborg, O. L. C., Miller, R. A., and Ojima, K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50, 151–158. doi: 10.1016/0014-4827(68)90403-5
González-García, M. P., Vilarrasa-Blasi, J., Zhiponova, M., Divol, F., Mora-García, S., Russinova, E., et al. (2011). Brassinosteroids control meristem size by promoting cell cycle progression in Arabidopsis roots. Development 138, 849–859. doi: 10.1242/dev.057331
Gordon, S. P., Chickarmane, V. S., Ohno, C., and Meyerowitz, E. M. (2009). Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. U.S.A. 106, 16529–16534. doi: 10.1073/pnas.0908122106
Gordon, S. P., Heisler, M. G., Reddy, G. V., Ohno, C., Das, P., and Meyerowitz, E. M. (2007). Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134, 3539–3548. doi: 10.1242/dev.010298
Hemerly, A. S., Ferreira, P., de Almeida Engler, J., Van Montagu, M., Engler, G., and Inzé, D. (1993). cdc2a expression in Arabidopsis is linked with competence for cell division. Plant Cell 5, 1711–1723. doi: 10.1105/tpc.5.12.1711
Ikeda, Y., Banno, H., Niu, Q. W., Howell, S. H., and Chua, N. H. (2006). The ENHANCER OF SHOOT REGENERATION 2 gene in Arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol. 47, 1443–1456. doi: 10.1093/pcp/pcl023
Ikeuchi, M., Sugimoto, K., and Iwase, A. (2013). Plant callus: mechanisms of induction and repression. Plant Cell 25, 3159–3173. doi: 10.1105/tpc.113.116053
Ioio, R. D., Linhares, F. S., Scacchi, E., Casamitjana-Martinez, E., Heidstra, R., Costantino, P., et al. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17, 678–682. doi: 10.1016/j.cub.2007.02.047
Ivanov, V. B., and Dubrovsky, J. G. (1997). Estimation of the cell-cycle duration in the root apical meristem: a model of linkage between cell-cycle duration, rate of cell production, and rate of root growth. Int. J. Plant Sci. 158, 757–763. doi: 10.1086/297487
Jasinski, S., Leite, C. S., Domenichini, S., Stevens, R., Raynaud, C., Perennes, C., et al. (2003). NtKIS2, a novel tobacco cyclin-dependent kinase inhibitor is differentially expressed during the cell cycle and plant development. Plant Physiol. Biochem. 41, 667–676. doi: 10.1016/S0981-9428(03)00082-2
Kakimoto, T. (1996). CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274, 982–985. doi: 10.1126/science.274.5289.982
Lugassi, N., Nakayama, N., Bochnik, R., and Zik, M. (2010). A novel allele of FILAMENTOUS FLOWER reveals new insights on the link between inflorescence and floral meristem organization and flower morphogenesis. BMC Plant Biol. 10:131. doi: 10.1186/1471-2229-10-131
Malladi, A., and Johnson, L. K. (2011). Expression profiling of cell cycle genes reveals key facilitators of cell production during carpel development, fruit set, and fruit growth in apple (Malus × domestica Borkh). J. Exp. Bot. 62, 205–219. doi: 10.1093/jxb/erq258
Matsuo, N., Mase, H., Makino, M., Takahashi, H., and Banno, H. (2009). Identification of ENHANCER OF SHOOT REGENERATION 1-upregulated genes during in vitro shoot regeneration. Plant Biotechnol. 26, 385–393. doi: 10.5511/plantbiotechnology.26.385
Morgan, D. O. (1997). Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13, 261–291. doi: 10.1146/annurev.cellbio.13.1.261
Motte, H., Vereecke, D., Geelen, D., and Werbrouck, S. (2014). The molecular path to in vitro shoot regeneration. Biotechnol. Adv. 32, 107–121. doi: 10.1016/j.biotechadv.2013.12.002
Perianez-Rodriguez, J., Manzano, C., and Moreno-Risueno, M. A. (2014). Post-embryonic organogenesis and plant regeneration from tissues: two sides of the same coin? Front. Plant Sci. 5:219. doi: 10.3389/fpls.2014.00219
Ramirez-Parra, E., Fründt, C., and Gutierrez, C. (2003). A genome-wide identification of E2F-regulated genes in Arabidopsis. Plant J. 33, 801–811. doi: 10.1046/j.1365-313X.2003.01662.x
Riou-Khamlichi, C., Huntley, R., Jacqmard, A., and Murray, J. A. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283, 1541–1544. doi: 10.1126/science.283.5407.1541
Sabag, M., Ari, G. B., Zviran, T., Biton, I., Goren, M., Dahan, Y., et al. (2013). PaKRP, a cyclin-dependent kinase inhibitor from avocado, may facilitate exit from the cell cycle during fruit growth. Plant Sci. 213, 18–29. doi: 10.1016/j.plantsci.2013.08.007
Sangwan, R., and Sangwan-Norreel, B. (1990). “Genetic transformation and plant improvement,” in The Impact of Biotechnology on Agriculture, eds A. Altman and P. M. Hasegawa (Berlin: Springer), 299–337.
Sawa, S., Watanabe, K., Goto, K., Kanaya, E., Morita, E. H., and Okada, K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13, 1079–1088. doi: 10.1101/gad.13.9.1079
Sherr, C. J., and Roberts, J. M. (1995). Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 9, 1149–1163. doi: 10.1101/gad.9.10.1149
Sieberer, T., Hauser, M. T., Seifert, G. J., and Luschnig, C. (2003). PROPORZ1, a putative Arabidopsis transcriptional adaptor protein, mediates auxin and cytokinin signals in the control of cell proliferation. Curr. Biol. 13, 837–842. doi: 10.1016/S0960-9822(03)00327-0
Skoog, F., and Miller, C. (1957). Chemical regularion of growth and organ formation in plant fissue cultured. In vitro. Symp. Soc. Exp. Biol. 11, 118–131. doi: 10.1038/jid.2014.273
Ubeda-Tomás, S., Federici, F., Casimiro, I., Beemster, G. T., Bhalerao, R., Swarup, R., et al. (2009). Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 19, 1194–1199. doi: 10.1016/j.cub.2009.06.023
Valvekens, D., Van Montagu, M., and Van Lijsebettens, M. (1988). Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. U.S.A. 85, 5536–5540. doi: 10.1073/pnas.85.15.5536
Wang, H., Zhou, Y., Gilmer, S., Whitwill, S., and Fowke, L. C. (1997). A plant cyclindependent kinase inhibitor gene. Nature 386, 451–452. doi: 10.1038/386451a0
West, G., Inzé, D., and Beemster, G. T. (2004). Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol. 135, 1050–1058. doi: 10.1104/pp.104.040022
Xu, K., Liu, J., Fan, M., Xin, W., Hu, Y., and Xu, C. (2012). A genome-wide transcriptome profiling reveals the early molecular events during callus initiation in Arabidopsis multiple organs. Genomics 100, 116–124. doi: 10.1016/j.ygeno.2012.05.013
Yamaguchi, M., Kato, H., Yoshida, S., Yamamura, S., Uchimiya, H., and Umeda, M. (2003). Control of in vitro organogenesis by cyclin-dependent kinase activities in plants. Proc. Natl. Acad. Sci. U.S.A. 100, 8019–8023. doi: 10.1073/pnas.1332637100
Yasutani, I., Ozawa, S., Nishida, T., Sugiyama, M., and Komamine, A. (1994). Isolation of temperature-sensitive mutants of Arabidopsis thaliana that are defective in the redifferentiation of shoots. Plant Physiol. 105, 815–822.
Yu, Y., Steinmetz, A., Meyer, D., Brown, S., and Shen, W. H. (2003). The tobacco A-type cyclin. Nicta; CYCA3; 2, at the nexus of cell division and differentiation. Plant Cell 15, 2763–2777.
Zhang, Y., Li, B., Huai, D., Zhou, Y., and Kliebenstein, D. J. (2015). The conserved transcription factors, MYB115 and MYB118, control expression of the newly evolved benzoyloxy glucosinolate pathway in Arabidopsis thaliana. Front. Plant Sci. 6:343. doi: 10.3389/fpls.2015.00343
Keywords: Arabidopsis thanliana, cell cycle, cyclin-dependent kinase, CDK inhibitor, ICK/KRP, cell proliferation, callus induction, plant regeneration
Citation: Cheng Y, Liu H, Cao L, Wang S, Li Y, Zhang Y, Jiang W, Zhou Y and Wang H (2015) Down-regulation of multiple CDK inhibitor ICK/KRP genes promotes cell proliferation, callus induction and plant regeneration in Arabidopsis. Front. Plant Sci. 6:825. doi: 10.3389/fpls.2015.00825
Received: 04 July 2015; Accepted: 22 September 2015;
Published: 13 October 2015.
Edited by:
Bo Liu, University of California, Davis, USAReviewed by:
Ming Yang, Oklahoma State University, USASherryl Bisgrove, Simon Fraser University, Canada
Copyright © 2015 Cheng, Liu, Cao, Wang, Li, Zhang, Jiang, Zhou and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Wang, Department of Biochemistry, University of Saskatchewan, Saskatoon, SK S7N 5E5, Canada, hong.wang@usask.ca; Yongming Zhou, National Key Laboratory of Crop Genetic Improvement, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, China, ymzhou@mail.hzau.edu.cn
 Yan Cheng
Yan Cheng Han Liu
Han Liu Ling Cao1,2
Ling Cao1,2 Yuanyuan Zhang
Yuanyuan Zhang Yongming Zhou
Yongming Zhou Hong Wang
Hong Wang