- 1Key Laboratory of Horticultural Plant Biology (MOE), Key Laboratory of Horticultural Crop Biology and Genetic Improvement (Central Region), MOA, Huazhong Agricultural University, Wuhan, China
- 2School of Biology and Food Engineering, Fuyang Teachers College, Fuyang, China
ZFPs play important roles in many biological processes, including plant development, stress response, and phytohormone response. RanBP2-type zinc finger transcription factors have been characterized in animals and humans. However, their functions remain largely unknown in plants. In this study, we identified a RanBP2-type zinc finger protein gene (SlRBZ) in tomato. SlRBZ was constitutively expressed in roots, stems, leaves, flowers, and fruits. The SlRBZ-GFP fused protein was localized in the nucleus. Overexpression of SlRBZ resulted in chlorosis and dwarf phenotypes in tomato. Determination of physiological index showed that chlorophyll, carotenoid, and GAs contents were evidently decreased in transgenic plants. Furthermore, the qRT-PCR and RNA-Seq analyses demonstrated that the transcription of the genes involved in these biosynthesis pathways obviously decreased in SlRBZ-OE plants. In addition, ultrastructural observation by transmission electron microscopy indicated that plastids could not develop into mature chloroplasts with normal chloroplast membrane and thylakoid membrane system in SlRBZ-OE plants. The results suggest that overexpression of SlRBZ may impair the biosynthesis of chlorophyll, carotenoid, and gibberellin through blocking chloroplast development, resulting in chlorosis and dwarfism in tomato.
Introduction
Chlorophyll (Chl) has important function in light harvesting and photosynthetic energy transduction, which is closely related to chloroplast-nuclear signaling and chloroplast development (Eckhardt et al., 2004). Chl biosynthesis is a complex process, including the formation of 5-aminolevulinic acid (ALA), protoporphyrin IX from eight molecules of ALA, and Chl in the magnesium branch (Beale, 1999; Eckhardt et al., 2004). A limiting factor of the whole pathway is glutamyl tRNA reductase (GluTR), which is the key enzyme for ALA formation. HEMA1, HEMA2, and HEMA 3 encode GluTR and are expressed in different tissues and environmental conditions (McCormac et al., 2001). Eight molecules of ALA are first converted into uroporphyrinogen III. After three steps, uroporphyrinogen III is oxidized into Proto under the function of protoporphyrinogen oxidase (Tanaka and Tanaka, 2007). Mg–chelatase consists of CHLI, CHLD, and CHLH (Papenbrock et al., 1997). Under the combined effect of these three enzymes, MgProtoMe cyclase, NADPH-Pchlide oxidoreductase, and Chl synthase, Proto is transferred into Chl (Eckhardt et al., 2004). Inhibition of each enzymatic step can affect the synthesis of Chl and result in leaf color change. The transcription of these enzymes required for Chl biosynthesis is individually and independently regulated. Meanwhile, light and developmental processes could induce the expression of the majority of these enzymes. Chloroplast formation influences leaf color in plants. Many chloroplastid proteins can partially block chloroplast development and result in a chlorotic phenotype (Carol et al., 1999; Wu et al., 2007). Chloroplasts are arrested and Chl is almost absent in the insertion mutant of the HST gene, which encodes an important enzyme participating in the biosynthesis of the PSII mobile electron transport co-factor PQ (Chao et al., 2014).
Gibberellins (GAs) are important for plant growth and development. Decreased GA content can cause plant dwarfism. Many different isoforms of GAs exist in nature, whereas only a few of them possess biological activity and regulate plant development, including GA1, GA3, GA4, and GA7 (Hedden and Phillips, 2000). GA12 and GA53 as precursors can be converted into many GA intermediates and bioactive GAs, wherein GA 20-oxidases (GA20ox) and GA 3-oxidases (GA3ox) have key roles (Ross et al., 1999). In addition, GA3ox catalyzes the final step in the synthesis of bioactive GAs. However, GA 2-oxidase (GA2ox) functions in converting active GAs and their precursors into inactive isoforms (Thomas et al., 1999). The mechanisms of GA biosynthesis and catabolism have been well-characterized in plants. As reported, GAs are synthesized from the common diterpene precursor geranylgeranyl diphosphate (GGPP) in plastids (Okada et al., 2000). During this process, GGPP is first separately converted into ent-kaurene by ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS; Duncan and West, 1981). Then, the ent-kaurene is converted into GA12 by ent-kaurene oxidase (KO) and ent-kaurenoic oxidase (KAO; Yamaguchi, 2008). Furthermore, GGPP is also the precursor necessary for carotenoids and Chl biosynthesis (Rodríguez-Concepción et al., 2001). Overexpression of PSY results in dwarfism and chlorosis in tomato, as it is the first gene for a carotenoid biosynthetic enzyme. Chl, GA, and carotenoid are synthesized in plastids, most of which are controlled by nuclear genes (Fray et al., 1995).
Chloroplasts are organelles that differentiate from plastids in plant cells (Gruissem, 1989). Different species, tissues, and environmental conditions can influence their ultimate shape and structure (Izawa and Good, 1966; Dodge, 1970). Notably, chloroplasts are surrounded by outer and inner envelope membranes. Chloroplast biogenesis is coordinately controlled by the proteins encoded in the nuclear and plastid genomes (Chen and Schnell, 1999). Moreover, ~80% of chloroplast proteins are encoded by nuclear genes (Ohyama et al., 1986). These proteins participate in protein transport, translation, and folding during chloroplast development (Klein and Mullet, 1986; Hendrick and Hartl, 1993; Cline and Henry, 1996; Fuks and Schnell, 1997). Many genes involved in the chloroplast development have been isolated by analyzing Arabidopsis mutants. A nuclear-encoded sigma factor, AtSIG6 controls early chloroplast development in Arabidopsis cotyledons, the mutation of which results in a cotyledon-specific pale green phenotype (Ishizaki et al., 2005). Co-suppression of two highly conserved heat shock protein genes, namely, cpHsc70-1 and cpHsc70-2, causes a white and stunted phenotype (Latijnhouwers et al., 2010). Huang et al. (2009) uncovered that the EMB1303 gene is required for chloroplast development, which encodes a chloroplast-localized protein in Arabidopsis (Huang et al., 2009). The Arabidopsis MDA1 gene affects chloroplast morphology and mda1 mutants exhibit reduced pigmentation of cotyledons, leaves, stems, and sepals (Robles et al., 2012). Moreover, a guanylate kinase encoded by a nuclear gene VIRESCENT 2 participates in chloroplast differentiation in rice (Sugimoto et al., 2007). Impairment of chloroplast development brings about chlorophyll (Chl) reduction.
Zinc finger protein (ZFP) was initially discovered in Xenopus oocytes (Miller et al., 1985). And many ZFPs have been identified in eukaryotes. Previous studies demonstrated that ZFPs participate in many biological processes, including plant growth and development, stress response, and phytohormone response (Laity et al., 2001). Ran-binding proteins (RanBPs), a novel type of zinc finger transcription factors, also broadly existed in higher eukaryotes. RanBP ZFP has three Ran-binding domains and two zinc finger motifs, which is a component of the nuclear pore complex and important in nuclear pore function (Chang et al., 2007). Further studies have demonstrated that RanBP2 complex is required for E3 activity (Werner et al., 2012). In humans, RanBP ZFPs have been implicated in the regulation of mRNA processing. However, little is known about the functions of RanBP ZFPs in plants. To date, only one gene of this type has been characterized in cotton, RanBP2 ZFP, which has been identified to participate in the different development stages of glands and may function in the development of the cotton gland (Chang et al., 2007). Therefore, the characterization of the functions of RanBP ZFPs in plants will shed new insights into the roles of these transcription factors in developmental processes.
In this study, the SlRBZ gene was characterized as RanBP-type transcription factor, which was localized in the nucleus. Overexpression of SlRBZ resulted in chlorosis and dwarfism phenotypes in tomato. The synthesis of Chl, carotenoid and GA, as well as the expression levels of genes related to these pathways, were evidently suppressed in SlRBZ-OE plants. Meanwhile, ultrastructural observation revealed that chloroplast formation was blocked. This conclusion was further supported by RNA-Seq analyses and qRT-PCR. These results demonstrate that overexpression of SlRBZ may inhibit the biosynthesis of Chl, carotenoid, and GA through affecting chloroplast formation, resulting in chlorosis and dwarfism.
Materials and Methods
Plant Materials and Treatments
Tomato (Solanum lycopersicum cv. Ailsa Craig) plants and transgenic lines in this background were grown in a growth chamber (16 h light/8 h dark photoperiod; 25°C). In order to investigate the response of SlRBZ gene to different growth regulator, the four-leaf-stage seedlings were separately sprayed with 100 μM GA3, 100 μM IAA, and 100 μM ABA. Tissues were collected from the roots, stems, leaves, flowers, and fruits of AC plants for analyzing the expression pattern of SlRBZ. Dwarf phenotypes of transgenic plants were restored by spraying with 100 μM GA3 at an interval of 3 d. Experiments were carried out with three independent biological replicates per treatment (each group with eight plants). The seedlings treated with distilled water were used as controls.
Isolation of SlRBZ Gene, Construction of Expression Vectors, and Transgenic Analysis
The full-length coding sequence of tomato SlRBZ gene was amplified from the AC cDNA using gene-specific primers (forward primer: 5′-GCTCTAGA ATGGGACGAGAAGGAGATTG-3′; reverse primer: 5′-GCGGTACC CTATGCAACAGTAACAGGTTGAGT-3′) based on the gene sequence (Solyc03g033560). The fragment of the SlRBZ gene was inserted into the pMV2 vector under control of the CaMV 35S promoter. The RNAi vector was constructed by amplifying a 116 bp fragment and ligating with pHellsgate 2 through BP reaction. Both constructs were verified by double digestion and transformed into AC mediated by Agrobacterium tumefaciens strain C58. The transgenic plants were further detected with PCR using genomic DNA as templates and CaMV35S forward and gene-specific reverse primers for SlRBZ. We cloned a 2.083-kb SlRBZ endogenous promoter fragment upstream of the start codon using the primers listed in Table S2. And the promoter SlRBZ:GUS fusion construct was generated by inserting the promoter fragment of SlRBZ in front of the GUS coding sequence. Transgenic plants carrying this construct were acquired as described earlier. The tissues of positive transformants were submerged in GUS staining solution overnight in the dark. Chlorophyll was then removed by incubation in 75% ethanol and the expression patterns were observed under a microscope.
Subcellular Localization of SlRBZ in Tomato
To analyze the subcellular localization of SlRBZ, the full-length open reading frame (ORF) without the stop codon of SlRBZ was PCR-amplified using the primers containing Kpn I and BamH I recognition sites (Table S2). The plasmids containing the correct sequence of SlRBZ were digested with Kpn I and BamH I and introduced into pU1391 to create a fusion construct (pU1391-SlRBZ). The fusion construct and the control (pU1391) were bombarded into onion epidermal cells using the PDS-1000 system (Bio-Rad). The onion cells were cultured on MS medium and then observed with a confocal laser microscope (Leica TCSST2) 24 h after bombardment.
RNA Isolation and qRT-PCR Analysis
Total RNA was extracted by TRIzol reagent (Invitrogen, USA). First-strand cDNA was synthesized using M-MLV reverse transcriptase (Toyobo, Japan) according to the supplier's protocols. The qRT-PCR was performed using the SYBR Green I Master Kit (Roche, Switzerland) on a LightCycler 480 Real-Time PCR Detection System, with β-actin transcripts as internal controls. The primers used for qRT-PCR analysis are listed in Table S1. The PCR amplification included a 30 s denaturation at 95°C, followed by 40 cycles of 95°C for 10 s, 58°C for 15 s, and 72°C for 20 s. Each cDNA sample was subjected to a qRT-PCR analysis in triplicate. The data were normalized using β-actin as the reference gene.
Measurement of Plant Growth, Chl Content, and Net Photosynthesis Rate
Six randomly selected 1-month-old seedlings were collected to measure morphological indices, including plant height, leaf length, and leaf width. Chl content was measured following the procedure described by Wellburn (1994). First, leaf tissues were ground using liquid nitrogen. Chl was then extracted with 80% (v/v) acetone for an hour under low light intensity. Extraction was carried out several times during the reverse centrifugal tube to accelerate the process. Second, the samples were centrifuged at 12000 g for 10 min, and then the clear liquid was collected to determine Chl content. Finally, Chl content was determined by spectrophotometry. Net photosynthesis rate was measured by a gas exchange system of TPS-1 (PP Systems Company, UK). The experiments were performed thrice using independent biological replicates. The analysis of variance (ANOVA) with Bonferroni's post-test was conducted using SPSS.17.
Carotenoid Assay by HPLC
Carotenoids were extracted following the method described by Liu et al. with slight modifications (Liu et al., 2007). Carotenoids were determined on a reverse phase Analytical YMC Carotenoid Column C30 (150 × 4.6 mm i.d., 3 μm, Wilmington, NC, USA) using a Waters HPLC system with a photodiode array detector (Waters, Milford, MA). Operation was conducted under subdued light to avoid carotenoid degradation. Identification of carotenoids was performed by comparison with standard spectra. Quantification was performed using the calibration curve generated with commercially available lycopene, β-carotene, β-cryptoxanthin, lutein, and violaxanthin standards (Sigma-Aldrich).
Quantification of Endogenous GAs
Tomato leaf tissues were homogenized in liquid nitrogen and then the GA was extracted in 4 mL 80% (v/v) ice-cold aqueous methanol containing butylated hydroxytoluene (1 mmol/L) and polyvinylpyrrolidone (60 mg/g fresh weight). The samples were incubated overnight at 4°C and centrifuged at 10,000 g for 10 min. The resulting supernatants were individually collected and filtered through C18 Sep-Pak cartridges (Waters, Millford, MA, USA). The efflux was collected, and then dried in N2. Concentrations of GA1+3 and GA4+7 were measured by ELISA following methods described in previous publications (Zhu et al., 2005).
Scanning Electron Microscope (SEM) Observation
For SEM, the samples from transgenic and control seedlings were cut into small pieces of ~0.1 cm3 size, placed in 2.5% glutaraldehyde fluid, vacuum fixed for ~24 h, and then washed three times with 0.1 M phosphate buffer for 10 min. The samples were dehydrated in a series of ethanol, dried in HCP-2 (Hitachi), and coated with palladium–gold in an ion injection apparatus (JFC-1600). Observations were carried out on a scanning electron microscope (JEOL JEM-6390 LV).
Transmission Electron Microscope (TEM) Observation
The samples (cotyledons of 2-week-stage seedlings and true leaves from top to bottom of 6-week-stage seedlings) were fixed in 2.5% glutaraldehyde, vacuum fixed overnight, washed three times with 0.1 M phosphate buffer for 30 min, and then post-fixed with 1% osmiophilic tetroxide for 2 h. The fixed samples were dehydrated with a series of alcohol solutions, and then infiltrated and embedded in Spurr resin (SPI-812) with an acetone mixture. The ultra-thin sections were prepared with a Reichert Ultracut-6 (Leica Microsystems, Bannockburn, IL, USA), stained with uranyl acetate and lead citrate before TEM (Hitachi H-7650, Tokyo, Japan), and then photographed with Gatan 832 digital imaging system.
RNA-Sequencing and Functional Analysis of DEGs
Transgenic tomato plants overexpressing SlRBZ (OE-5) and wild-type AC were used for transcriptomic analysis. Total RNA was isolated from these materials, and the poly-A containing mRNA was purified from the total RNA using poly-T oligo-attached magnetic beads. The mRNA was fragmented into small pieces (250–350 bp) and then reversely transcribed into first-strand cDNA using random hexamers, followed by second-strand cDNA synthesis using DNA Polymerase I and RNase H. End repair of the double-stranded cDNA was performed to convert the overhangs into blunt ends and then purified. The libraries were constructed and sequenced using Illumina HiSeq® 2000. Clean reads were gained by removing the 3′ adaptor, low-quality reads, and reads less than 20 nt or containing two N. Gene expression levels were calculated using FPKM method (fragments per kilobase of transcript per million mapped reads). An absolute value of the log2 ratio ≥ 1 and P ≤ 0.05 were applied as thresholds to characterize the significance of gene expression level. The DEGs were annotated by aligning against the NCBI non-redundant nucleotide database. For further identification of pathways related to the SlRBZ gene, DEGs were compared with the Kyoto Encyclopedia of Genes and Genomes database (KEGG).
Results
Isolation and Molecular Characterization of SlRBZ
We cloned and identified a Ran-binding protein zinc finger gene (named as SlRBZ) in tomato. Based on the nucleotide sequence of this gene (http://solgenomics.net/), the primers for the full length were designed. We isolated the full-length ORF from the tomato cultivar Ailsa Craig (AC) using RT-PCR. The predicted coding sequence of SlRBZ was 996 bp and encoded a protein of 331 amino acid residues. Through genomic and cDNA sequence alignment, we determined that SlRBZ contains seven exons and six introns. SlRBZ harbors three dispersed RanBP-type zinc fingers that conform to the RanBP2-type consensus sequence pattern (W-XC-X(2)-C-X(3)-N-X(6)-C-X(2)-C). Carrying out the pBLAST with SlRBZ amino acid sequence, the homologous proteins of other species were retrieved from the NCBI database. Alignments of SlRBZ with these homologous genes, including potato (AFX67024.1), ricinus (XP_0025307), soybean (XP_0035202), crowtoe (AFK38574.1), medicago (XP_0036293), cucumber (XP_0041348), Vitis vinifera (XP_0022686), strawberry (XP_0042878), Arabidopsis (NP_179388.), rice (EEE60388.1), and maize (AFW57699.1), indicated that all these genes contain three conserved domains (Figure 1). The amino acid sequence of SlRBZ has 90% sequence identity with the homolog in potato but only 51% sequence identity with the homolog in maize. The other homologs displayed ~51–90% sequence identity to SlRBZ.
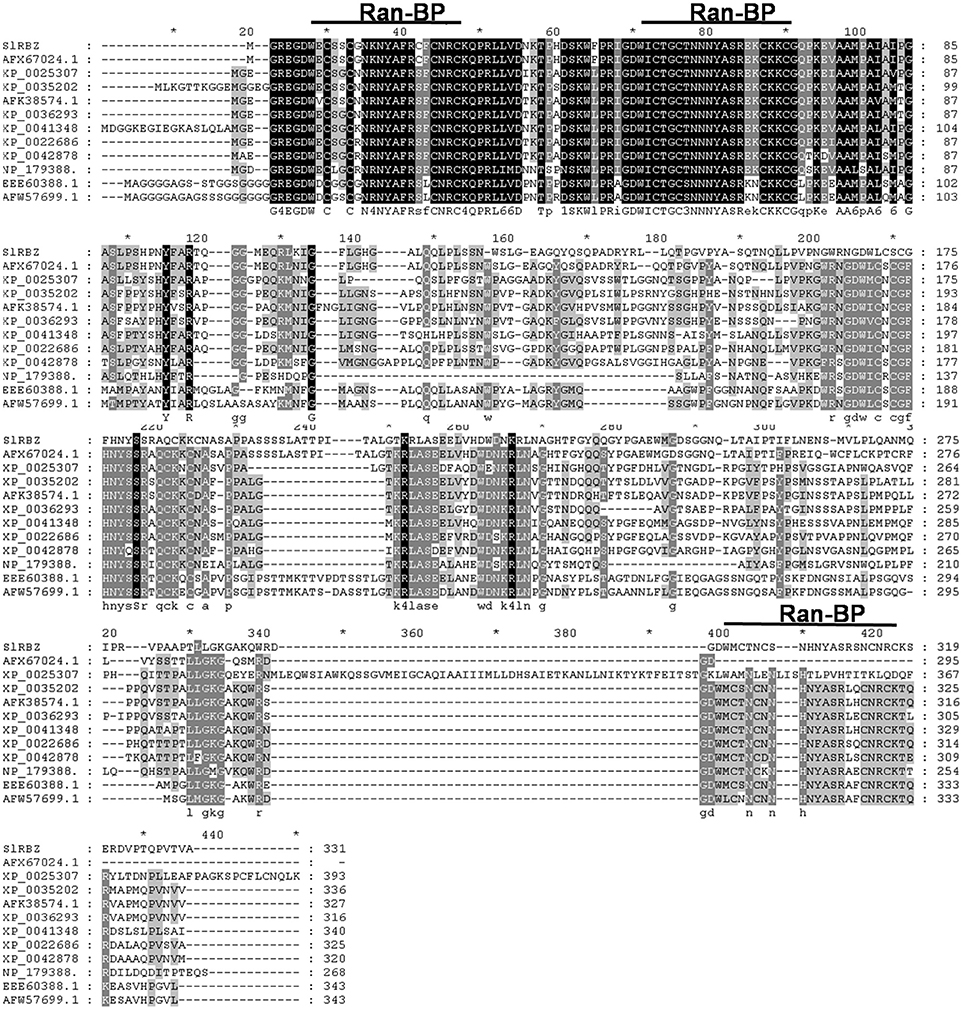
Figure 1. Amino acid alignments of the protein encoded by SlRBZ with 11 homologs from other species. Identical residues are highlighted in black, conserved residues in solid line.
Subcellular Localization and Expression Pattern of SlRBZ
To study the subcellular localization of SlRBZ, we examined the GFP fluorescence in onion epidermal cells transformed with a fusion construct (pU1391-SlRBZ-GFP) and a control construct (pU1391-GFP). The fusion protein SlRBZ-GFP was localized in the nucleus, whereas GFPs alone were observed in the membrane and cytoplasm, manifesting that SlRBZ is a nuclear protein (Figure S1). To examine the expression pattern of SlRBZ, we carried out qRT-PCR analysis of the total RNA extracted from several tissues of AC, including roots, young stems, functional leaves, flowers, and green fruits. SlRBZ was expressed in various tissues, and the expression was much higher in the young stems (Figure 2A). We further carried out the transformation of SlRBZ promoter::GUS construct and analyzed the spatial expression pattern of SlRBZ. GUS was mainly detected in stems and petioles (Figure 2B).
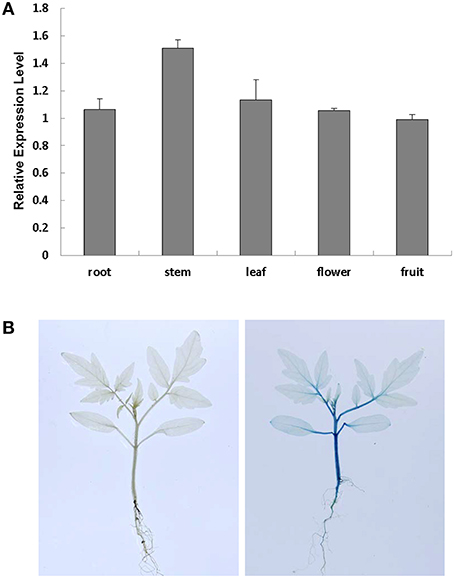
Figure 2. Analysis of SlRBZ expression in tomato. (A) Real-time RT-PCR analysis of SlRBZ expression in different tissues. (B) GUS expression in young seedlings, including roots, stems, leaves. Error bars indicate standard error (SE) of three replicates.
SlRBZ is Induced by GA and IAA but Suppressed by ABA
We evaluated the SlRBZ expression changes after treatment with different phytohormones. The investigation indicated that the transcript levels of SlRBZ substantially increased 1 h after GA treatment (Figure 3A). However, SlRBZ expression was evidently repressed from 1 to 24 h after ABA treatment (Figure 3B). In addition, we found that the transcription level of this gene reached a peak value 1 h after IAA treatment and then quickly returned to the lower levels 2 h later (Figure 3C). These results indicated that SlRBZ was positively regulated by GA and IAA, whereas negatively by ABA.
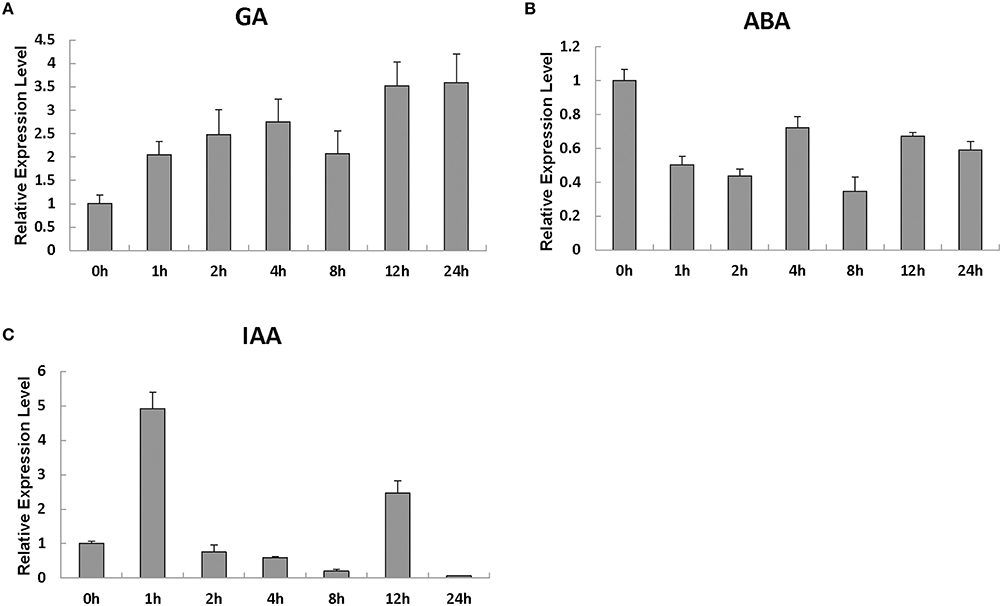
Figure 3. Real-time RT-PCR analysis of the expression of SlRBZ in response to the phytohormones GA3 (A), ABA (B), and IAA (C). All samples were prepared at the indicated time points from three biological replicates in each group. Error bars indicate the SE of three replicates.
Overexpression of SlRBZ Caused Chlorosis and Dwarfism in Tomato
We characterized the function of SlRBZ by generating transgenic tomato plants with overexpression or RNAi silencing of SlRBZ. 12 SlRBZ overexpression (OE) and 17 RNAi knock-down (Ri) tomato lines were obtained, respectively. The transgenic plants and AC plants grew under the same conditions. And the overexpression plants exhibited a severely etiolated and dwarf phenotype, whereas no obvious change of morphology was observed in SlRBZ-Ri lines (Figure 4A). The expression level of the SlRBZ gene in transgenic and AC plants was examined by qRT-PCR. The expression of SlRBZ was evidently upregulated in SlRBZ-OE lines and downregulated in SlRBZ-Ri lines (Figure 4B). Therefore, we selected the overexpression transgenic lines and AC for further studies. We measured plant height, leaf length and width of SlRBZ-OE plants. The average plant height of OE-5 and OE-10 plants was 1.91 and 1.67 cm, respectively, which was considerably lower than that of AC (6.04 cm; Figure 4C). The leaves of the SlRBZ-overexpression plants were considerably smaller than those of AC. The mean leaflet length of AC, OE-5, and OE-10 lines was 4.12, 1.28, and 1.06 cm, respectively, suggesting that OE-5 and OE-10 plants were 31.07 and 25.73% smaller than AC, respectively (Figure 4C). In addition, the mean leaflet width was considerably smaller in transgenic plants compared with controls (Figure 4C). To investigate whether the cell size was influenced in SlRBZ-OE plants, the cell number per unit area in the upper epidermis were observed by SEM. The average cell number was 10 in each field of view (1500 × : ≈0.0015 mm2) in AC, whereas that was 25 in SlRBZ-OE plants (Figure S2). We thus, concluded that the cell size was depressed in SlRBZ-OE plants.
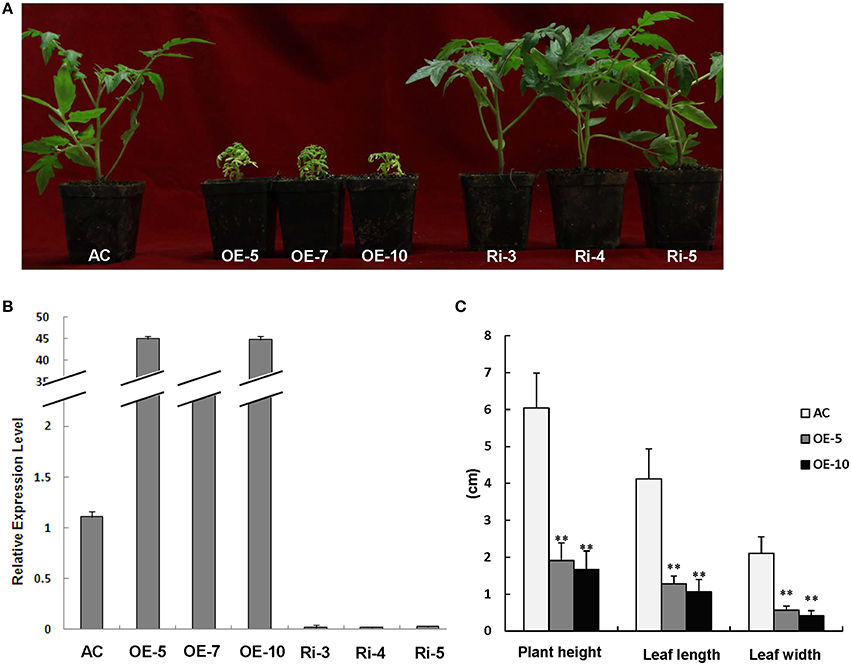
Figure 4. Overexpression of SlRBZ results in chlorosis and dwarfism in tomato. (A) Phenotypes of seedlings of SlRBZ-overexpressing tomato OE-5, OE-7, and OE10, and RNAi-inhibiting expression lines Ri-3, Ri-4, and Ri-5, and AC plants. (B) The expression level of SlRBZ in transgenic and AC plants. (C) Phenotype analysis (C) in wild type AC and SlRBZ-OE transgenic tomato plants. Error bars indicates SE (n = 3). Asterisks indicate significant differences compared with WT (**P < 0.01).
Chl Synthesis and Photosynthesis were Inhibited in SlRBZ Overexpressing Plants
To determine the reasons for leaf chlorosis, we measured the Chl content and found that Chl levels were apparently reduced in SlRBZ overexpressing plants compared with the wild-type AC (Figure 5A). In addition, expression of Chl biosynthetic genes, including HEMA, HEML1, HEMB1, HEMC, HEME1, HEMF1, HEMG1, CHLD, CHLM, CRD, DVR, PORA, and CAO, was compared between SlRBZ transgenic plants and AC seedlings, and all genes in the Chl biosynthetic tetrapyrrole pathway were downregulated in SlRBZ transgenic plants (Figure 5B). To establish whether or not SlRBZ impacts photosynthetic capacity, leaf net CO2 assimilation rate (Pn) was measured by a gas exchange system of TPS-1. Results showed that the Pn of overexpressed SlRBZ plants was inhibited in comparison with AC (Figure 5C).
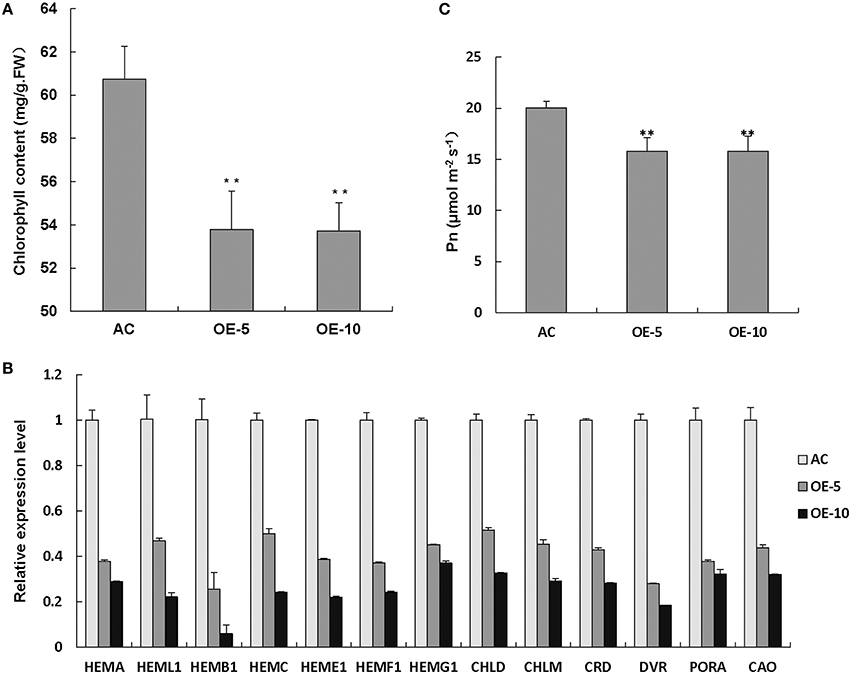
Figure 5. Chl synthesis and photosynthesis are inhibited in SlRBZ overexpressing plants. Chlorophyll content (A), expression levels of chlorophyll biosynthetic genes (B) and leaf net CO2 assimilation rate (C) in wild type AC and SlRBZ-OE transgenic tomato plants. Error bars indicates SE (n = 3). Asterisks indicate significant differences compared with WT (**P < 0.01).
Carotenoid Contents and Transcription Level of Genes Related to Carotenoid Biosynthesis were Decreased in SlRBZ-OE Plants
As we know, both Chl and carotenoids are synthesized in plant leaves and contribute to the leaf color. Therefore, the total carotenoid content was also measured by the spectrophotometer. The level of carotenoids was markedly decreased in SlRBZ transgenic plants in comparison with AC (Figure 6A). For details, several compounds of carotenoids, such as lutein, β-carotene, and β-crytoxanthin, was further detected by HPLC. Lutein, β-carotene, and β-crytoxanthin contents were 71.83, 289.46, and 22.60 μg/g·FW, respectively, in AC leaves (Figure 6B). By contrast, these compounds were 6.54, 28.94, and 3.66 μg/g·FW, respectively, in SlRBZ overexpressing plants (Figure 6B). In other words, the content of lutein, β-carotene, and β-crytoxanthin was 9.1, 10, and 16.2% lower than AC. In addition, violaxanthin, lycopene, and 9-cis-violaxanthin could be detected in AC, whereas these compounds were absent in SlRBZ overexpressing plants (Figure 6B). Furthermore, we examined the transcriptional changes of genes correlated with carotenoid biosynthesis, including PSY2, PDS, CRTR-B1, and LYC-B. Results showed that the expression levels of these genes evidently decreased in SlRBZ overexpressing plants (Figure 6C). Furthermore, several genes involved in the early steps of Chl, carotenoid, and GA biosynthesis, like DXS, DXR, and GGPS, were also considerably lower in the SlRBZ overexpressing plants than that in AC (Figure 6C).
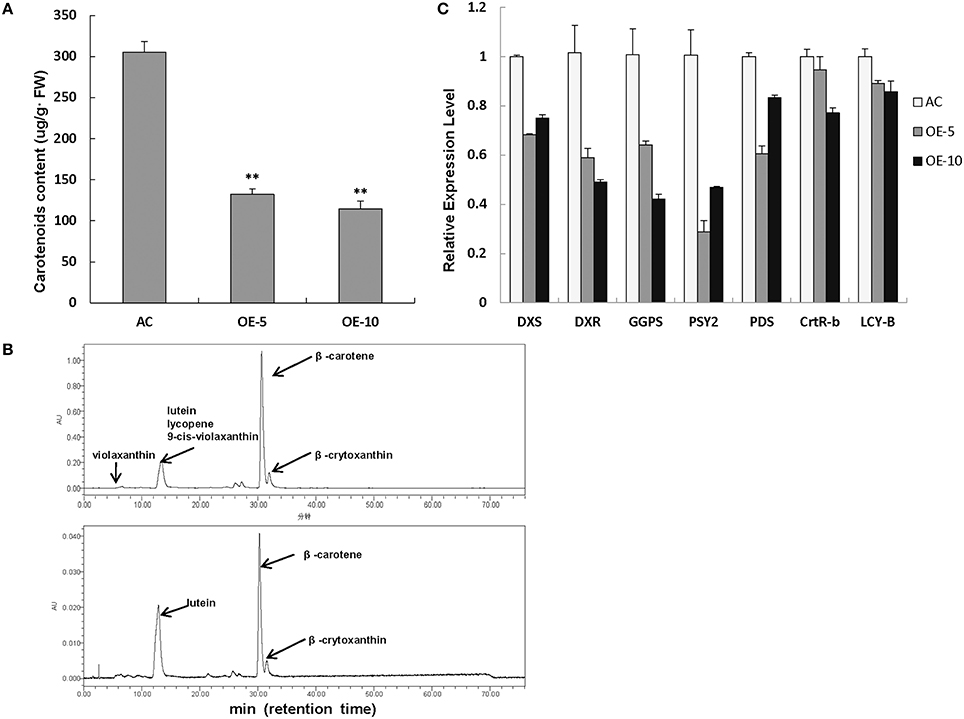
Figure 6. Carotenoid contents are decreased in SlRBZ-OE plants. (A) Carotenoids content were measured by spectrophotometer. (B) Carotenoids content were measured by HPLC (Upper: wild type AC plants; Lower: SlRBZ-OE transgenic tomato plants OE-5). (C) Expression levels of carotenoids biosynthetic genes and expression levels of genes involved in the early steps of carotenoid, chlorophyll, and GA biosynthesis pathways. Error bars indicates SE (n = 3). Asterisks indicate significant differences compared with WT (**P < 0.01).
SlRBZ Overexpressing Plants Exhibit a Classical Reduced GA Phenotype
The SlRBZ overexpressing transgenic plants showed dwarf phenotypes. We thus, measured the content of endogenous GAs. Results indicated that the two types of GAs (GA1+3 and GA4+7) were evidently decreased in transgenic plants compared with AC (Figure 7A). To further determine whether or not the synthesis of GA was influenced by SlRBZ, we compared the expression of GA biosynthetic genes between SlRBZ transgenic and AC plants. Previous studies indicated that KO, KAO, CPS, and KS have important functions in the early steps of GA biosynthesis. The qRT-PCR analysis showed that the expression of these genes was dramatically downregulated in SlRBZ transgenic plants (Figure 7B). In addition, the transcriptional levels of five genes in the GA biosynthetic pathway, namely, SlGA2ox1, SlGA2ox2, SlGA2ox5, SlGA20ox2, and SlGA20ox4, which determine GA concentration in many plants, were also repressed in transgenic plants (Figure 7B). Furthermore, spraying with 100 μM GA3 on SlRBZ overexpressing plants every 3 d can significantly rescue the plant height back to control levels, demonstrating that SlRBZ overexpressing plants exhibit a classical reduced GA phenotype (Figure S3).
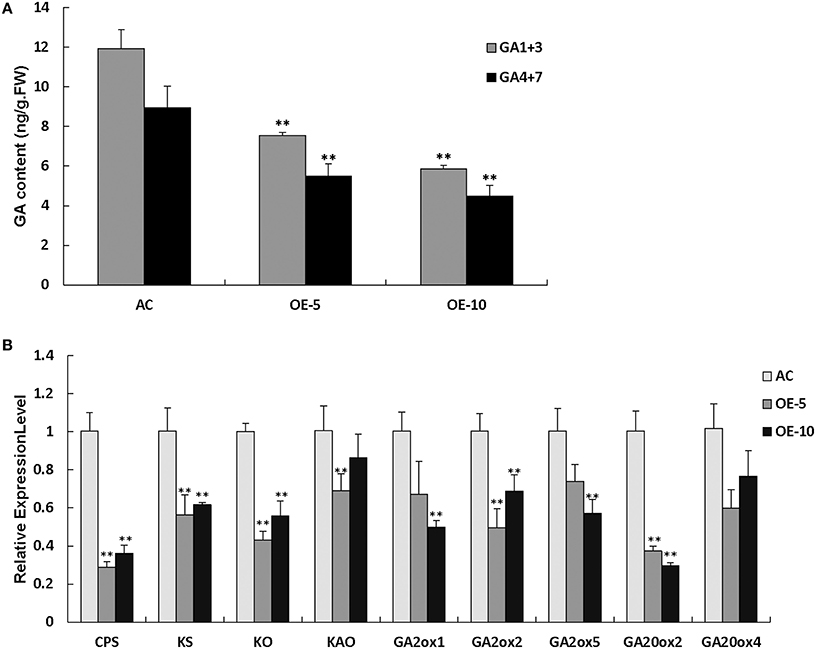
Figure 7. GA1+3 and GA4+7 are evidently decreased in transgenic plants compared with AC. (A) GA1+3 and GA4+7 content were measured by an indirect ELISA method with anti-GA antibodies. (B) qRT-PCR analysis of GAs biosynthetic genes. Error bars indicate the SE of three replicates. Asterisks indicate significant differences compared with WT (**P < 0.01).
Overexpressing SlRBZ Can Affect the Chloroplast Development in Tomato
Overexpression of SlRBZ resulted in leaf chlorosis and reduced total Chl content. Moreover, chloroplast development was closely related with Chl content. We thus, observed the chloroplast ultrastructure by TEM in the transgenic tomato leaves. Observations indicated that chloroplasts in AC leaves were normal, including the thylakoid membrane system, lamellar layer system of the thylakoid, and the inner and outer membrane system (Figure 8). However, the ultrastructure of chloroplasts in SlRBZ overexpressing plants was evidently blocked. For example, the membranes of chloroplast and grana and stroma thylakoid exhibited different degrees of rupture and collapse; grana lamellae were significantly less than AC; the stromata were disintegrated (Figure 8). In addition, the thylakoid was cracked, the number and volume of osmiophilic globules increased and the configurations of the thylakoid systems nearly disappeared (Figure 8). Furthermore, the number of granum-stroma thylakoid membranes in overexpressed SlRBZ plants significantly decreased (Figure 8). Taken together, these results demonstrated that SlRBZ has a critical function in chloroplast formation.
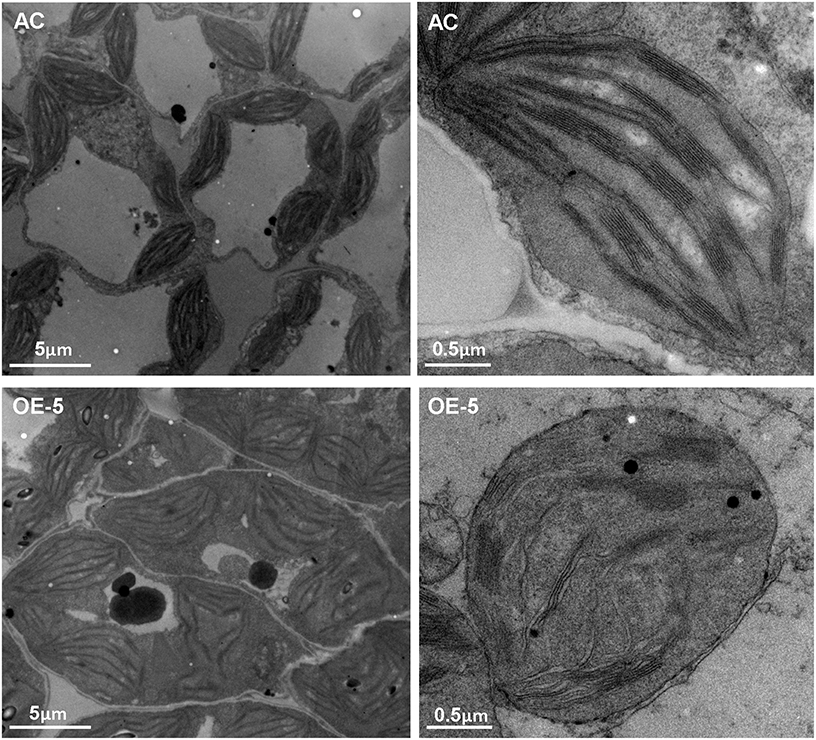
Figure 8. Transmission electron microscopy of chlopolasts in wild type AC and SlRBZ-OE transgenic tomato plants (OE-5).
SlRBZ Inhibits the Expression of Chl and Photosynthesis-Related Genes
To further characterize the molecular mechanism that triggers the phenotype of chlorosis and dwarfism mediated by SlRBZ, we further compared the gene expression profiles of SlRBZ transgenic and AC plants by RNA-Seq analysis (The transcriptome data about the transgenic tomato plants SlRBZ-OE and wild-type AC are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE77340). Clean reads were obtained by discarding low-quality reads, resulting in 22,824,878 clean reads for AC and 22,958,774 clean reads for SlRBZ-OE. Among them, 13,836,447 (60.62%) clean reads for AC, and 12836008 (55.91%) clean reads for SlRBZ-OE were mapped to the reference genome ITAG2.4, suggesting that the RNA-seq transcriptomes were sufficient for subsequent gene expression analyses. The putative differentially expressed genes (DEGs) between SlRBZ-OE and AC plants were identified by applying fold changes (FC ≥ 2 or FC ≤ 0.5), and (P ≤ 0.01) were applied as standards to determine the significance levels of DEGs. On the basis of these criteria, 515 genes were identified to be upregulated in the SlRBZ-OE plants compared with AC. Meanwhile, the transcription of 568 genes was decreased by more than twofold in the SlRBZ-OE plants. For biological explanation of these DEGs, we analyzed these genes using the KEGG pathway. Results indicated that overexpression of SlRBZ can repress the expression of genes involved in Chl synthesis pathway, including por, hemE, and chlM (Table S1). Furthermore, the expression levels of photosynthetic genes, such as Psa E, Psa F, Psa H (Photosystem I), Psb P, Psb Q, Psb W, Psb 27 (Photosystem II), and Pet F (photosynthetic electron transport), were evidently repressed in the SlRBZ-OE plants (Table S1). In addition, in accordance to our previous results, some GA biosynthesis-related genes were also detected and showed significantly lower expression levels in SlRBZ-OE plants than in Wt, such as GA20ox-2 (Solyc06g035530) and GA2ox-1 (Solyc05g053340; Figure 7B).
SlRBZ Inhibits the Expression of Many Light-Harvesting Chl a/b-Binding Protein Genes (Lhca/b) and Chloroplast Protein Genes
The Lhcb genes encoding membrane proteins, which consist of the antennae complexes and capture and transfer light energy to the reaction centers of photosystem I and photosystem II (Armond et al., 1977), are abundant in plants. The transcriptional levels of the Lhcb genes are induced by many developmental cues and environmental signals, such as chloroplast formation, light, and oxidative stress (McCormac and Terry, 2002; Staneloni et al., 2008). Therefore, their expression is severely repressed in tissues without mature chloroplasts. In accordance to this conclusion, we found that the expression of many Lhca/b genes were significantly downregulated in SlRBZ-OE plants, including Solyc03g005790, Solyc06g069730, Solyc04g082920, Solyc04g082930, Solyc12g011280, Solyc12g009200, Solyc08g067320, Solyc08g067330, CAB and Solyc08g007180 (Table S1). As Lhcb genes are reporters of chloroplast formation, we speculate that chloroplasts must be blocked in SlRBZ-OE plants. In addition, thylakoid lumenal proteins are necessary for the formation of photosystem II complexes (Hou et al., 2015). The expression of genes encoding thylakoid lumenal proteins must be repressed without normal photosystem II complexes in etiolated plants. Consistent with the foregoing, many genes encoding thylakoid lumenal proteins were also downregulated in transgenic plants, such as Solyc01g087040, Solyc03g082890, Solyc06g066620, and Solyc10g084040 (Table S1).
Discussion
ZFPs are important regulators and widespread in nature. They participate in multiple biological processes, including development, plant architecture, phytohormone response, and stress response (Laity et al., 2001; Li et al., 2013). Ran-binding proteins are also characterized as ZFPs that contain two conserved Ran-BP domains. This type of ZFPs was first discovered in the nuclear export protein RanBP2 (Gamsjaeger et al., 2007). Its functions have been studied in humans and animals (Higa et al., 2007). However, it remains largely uncharacterized in plants. The vast knowledge on ZFPs in animals benefits their discoveries in plants. Only a RanBP2 zinc finger protein gene was identified in upland cotton, which is expressed in the different development stages of glands (Chang et al., 2007). Similar to other ZFPs, SlRBZ is highly conserved in different species. In this study, we found that SlRBZ acts as a novel regulator controlling the biosynthesis of Chl, carotenoid, and GA in tomato. As previously reported, WRKY53 and OsDOS participate in the control of leaf senescence, which belongs to other subfamilies of ZFPs (Miao et al., 2004; Kong et al., 2006). Thus, we infer that SlRBZ may influence Chl synthesis in plants through the common mechanisms with other ZFPs. In addition, the expression level of SlRBZ is significantly higher in tomato plants treated with exogenous GA than in controls, suggesting that this gene is positively controlled by GA.
SlRBZ encodes a RanBP transcription factor that is localized in the nucleus. Overexpression of SlRBZ results in etiolated and dwarf phenotype in tomato. Notably, this phenotype appears at the early developmental stage. qRT-PCR analyses indicate that SlRBZ is expressed in almost all tissues. Chl, carotenoids, and GAs are synthesized in chloroplasts (Xing et al., 2010). We measured the contents of Chl, carotenoid and GAs in the SlRBZ-OE transgenic plants, and confirmed that these three chemical substances were evidently decreased. Moreover, the expression of the genes involved in their biosynthesis pathways were also inhibited in SlRBZ overexpressing plants, such as por, hemE, chlM, PSY2, PDS, ZDS, CRTR-B1, LYC-B, KO, KAO, CPS, and KS, suggesting that chloroplast development must be repaired.
Ultrastructure observation showed that chloroplasts are blocked in SlRBZ overexpressing plants, particularly the membranes of chloroplast and grana, and thylakoid stroma appeared to be ruptured and collapsed. Previous studies have demonstrated that mutations in pds3 and zds mutants block chloroplast development (Dong et al., 2007; Qin et al., 2007). HCF164, a thioredoxin-like protein, also participates in chloroplast development, and its mutation causes abnormal chloroplast morphology (Lennartz et al., 2001). The expression of its homologs (Solyc07g064940, Solyc08g006720, Solyc02g087850, Solyc01g087520, Solyc09g074570, etc.) was downregulated in SlRBZ-OE tomato plants. In addition, many thylakoid lumenal proteins were evidently affected in SlRBZ-OE tomato plants, which were encoded by the nuclear genes and localized in chloroplasts. Koussevitzky et al. (2007) showed that abnormal chloroplasts influenced the expression of plastid-localized genes (Koussevitzky et al., 2007). Furthermore, transcriptome analyses indicated that many genes correlated with photosynthesis and chloroplast differentiation are repressed in SlRBZ-OE plants. More importantly, the Lhcbs are used as the reporter genes for chloroplast development, and their expression are severely repressed in plants without mature chloroplasts (Larkin et al., 2003). In accordance with this result, numerous Lhcbs were significantly downregulated in SlRBZ-OE plants. From these results, we infer that the chlorosis and dwarfism phenotype in SlRBZ-OE tomato plants result from the impaired chloroplasts and then the reduction of Chl, carotenoid, and GAs synthesis.
Chloroplasts are essential for photosynthesis, and then plant vitality and growth (Pogson and Albrecht, 2011). The formation of the chloroplasts is codetermined by the nuclear- and plastid-encoded genes, such as SLP, APG2, and PAC (Taylor, 1989). However, to date, very few transcription factors have been identified to control chloroplast formation. Golden2-like genes, which belong to the GARP family of transcription factors, are required for chloroplast development (Waters et al., 2008). In addition, the GATA transcription factors (GNC and CGA1) also participate in chloroplast development in Arabidopsis (Chiang et al., 2012). In this study, we determined that the overexpression of SlRBZ can block chloroplast formation and the membranes of chloroplast and grana and stroma thylakoid in SlRBZ overexpressing plants ruptured and collapsed, demonstrating that SlRBZ regulates the formation of chloroplast in tomato. Similarly, CYO1, which encodes a protein disulfide isomerase, specifically regulates chloroplast biogenesis in Arabidopsis (Shimada et al., 2007). The number of grana thylakoids is closely related to Chl content (Anderson et al., 1973). Moreover, the genes relevant to chloroplastid proteins can reduce the stack of thylakoid and result in pale plants and etiolated phenotype (Jarvis et al., 1998; Huang et al., 2009; Barry et al., 2012). Consistent with this conclusion, our results showed that the Chl content was evidently decreased in SlRBZ-OE transgenic plants. In a previous report, the Lhc gene family was repressed in tissues lacking mature chloroplasts (Larkin et al., 2003). In accordance with this result, Lhcb genes as reporters of the formation of chloroplast, were repressed in SlRBZ-OE plants. In the present study, we found that net photosynthetic rate in SlRBZ-OE transgenic plants was just 78.91% of the Wt rate. Photosynthetic rate was repressed if grana lamella was reduced (Osborne and Raven, 1986). In general, Chl, carotenoid, and gibberellin biosyntheses were impaired in chloroplast, which is consistent with the reduced expression of most of genes involved in the tetrapyrrole biosynthetic pathway, as well as MEP and GA pathways in SlRBZ-OE transgenic plants (Figure S4). Therefore, we speculate that overexpression of SlRBZ may inhibit Chl, carotenoid, and GA biosynthesis by destroying chloroplast structure resulting in chlorosis and dwarfism in transgenic tomato plants. In accordance with this conclusion, depression of PDS, DXR, and HST participating in MEP, the PQ pathway, and carotenoid biosynthesis resulted in albino and dwarf phenotypes in Arabidopsis (Qin et al., 2007; Xing et al., 2010; Chao et al., 2014). These conclusions provide new insights the role of this novel RanBP2 zinc finger protein in tomato Chl, carotenoid, and GA biosynthesis. Future studies on this gene using ChIP-seq will provide more information underlying its regulatory mechanisms.
Author Contributions
ZY designed research; MF, SG, JR, and QY performed research; MF and CY analyzed data and wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National High-Tech Research Development Program in China (2012AA100104), the Natural Science Foundation of China (31572128, 31230064, 31372081), the Foundation for the Author of National Excellent Doctoral Dissertation of China (No. 201468).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00907
Figure S1. Subcellular localization of SlRBZ. Constructs of GFP and SlRBZ-GFP were transiently expressed in onion epidermal cells. GFP was localized in the entire onion cells (A-C), SlRBZ -GFP was localized in nucleus (D-F). Bright-field images (A,D), GFP fluorescent images (B,E), and merged images (C,F).
Figure S2. (A) Cell morphology of leaf epidermis in AC and OE-5 plants. (B) Cell number was counted in each field of view (1500 × : ≈0.0015mm2) on at least 20 microscopes from three plants. Asterisks indicate significant differences compared with WT (**P < 0.01).
Figure S3. The dwarf phenotype of SlRBZ-OE plants was rescued by spraying with GA3. A Growth response of treated with 100 μM GA3 on AC and SlRBZ-OE plants. b Plant height was measured before and after spraying GAs.
Figure S4. The expression of genes participating in carotenoid, chlorophyll and GA biosynthesis pathways were obviously decreased in SlRBZ-OE plants.
Table S1. Expression of genes involved in photosynthesis and chlorophyll biosynthesis in the SlRBZ-OE plants.
Table S2. Primers sequences used for functional and expression analysis in this study.
References
Anderson, J. M., Goodchild, D., and Boardman, N. (1973). Composition of the photosystems and chloroplast structure in extreme shade plants. Biochim. Biophys. Acta 325, 573–585. doi: 10.1016/0005-2728(73)90217-X
Armond, P., Staehelin, L., and Arntzen, C. (1977). Spatial relationship of photosystem I, photosystem II, and the light-harvesting complex in chloroplast membranes. J. Cell Biol. 73, 400–418. doi: 10.1083/jcb.73.2.400
Barry, C. S., Aldridge, G. M., Herzog, G., Ma, Q., McQuinn, R. P., Hirschberg, J., et al. (2012). Altered chloroplast development and delayed fruit ripening caused by mutations in a zinc metalloprotease at the lutescent2 locus of tomato. Plant Physiol. 159, 1086–1098. doi: 10.1104/pp.112.197483
Beale, S. I. (1999). Enzymes of chlorophyll biosynthesis. Photosyn. Res. 60, 43–73. doi: 10.1023/A:1006297731456
Carol, P., Stevenson, D., Bisanz, C., Breitenbach, J., Sandmann, G., Mache, R., et al. (1999). Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 11, 57–68. doi: 10.1105/tpc.11.1.57
Chang, P. A., Li, B., Ni, X. M., Xie, Y. F., and Cai, Y. F. (2007). Molecular cloning and expression analysis of a RanBP2 zinc finger protein gene in upland cotton (Gossypium hirsutum L.). Colloids Surf. B 55, 153–158. doi: 10.1016/j.colsurfb.2006.11.042
Chao, Y., Kang, J., Zhang, T., Yang, Q., Gruber, M. Y., and Sun, Y. (2014). Disruption of the homogentisate solanesyltransferase gene results in albino and dwarf phenotypes and root, trichome and stomata defects in Arabidopsis thaliana. PLoS ONE 9:e94031. doi: 10.1371/journal.pone.0094031
Chen, X., and Schnell, D. J. (1999). Protein import into chloroplasts. Trends Cell Biol. 9, 222–227. doi: 10.1016/S0962-8924(99)01554-8
Chiang, Y. H., Zubo, Y. O., Tapken, W., Kim, H. J., Lavanway, A. M., Howard, L., et al. (2012). Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 160, 332–348. doi: 10.1104/pp.112.198705
Cline, K., and Henry, R. (1996). Import and routing of nucleus-encoded chloroplast proteins. Annu. Rev. Cell Dev. Biol. 12, 1–26. doi: 10.1146/annurev.cellbio.12.1.1
Dodge, J. (1970). Changes in chloroplast fine structure during the autumnal senescence of Betula leaves. Ann. Bot. 34, 817–824.
Dong, H., Deng, Y., Mu, J., Lu, Q., Wang, Y., Xu, Y., et al. (2007). The Arabidopsis spontaneous cell death1 gene, encoding a ζ-carotene desaturase essential for carotenoid biosynthesis, is involved in chloroplast development, photoprotection and retrograde signalling. Cell Res. 17, 458–470. doi: 10.1038/cr.2007.51
Duncan, J. D., and West, C. A. (1981). Properties of kaurene synthetase from Marah macrocarpus endosperm: evidence for the participation of separate but interacting enzymes. Plant Physiol. 68, 1128–1134. doi: 10.1104/pp.68.5.1128
Eckhardt, U., Grimm, B., and Hörtensteiner, S. (2004). Recent advances in chlorophyll biosynthesis and breakdown in higher plants. Plant Mol. Biol. 56, 1–14. doi: 10.1007/s11103-004-2331-3
Fray, R. G., Wallace, A., Fraser, P. D., Valero, D., Hedden, P., Bramley, P. M., et al. (1995). Constitutive expression of a fruit phytoene synthase gene in transgenic tomatoes causes dwarfism by redirecting metabolites from the gibberellin pathway. Plant J. 8, 693–701. doi: 10.1046/j.1365-313X.1995.08050693.x
Fuks, B., and Schnell, D. J. (1997). Mechanism of protein transport across the chloroplast envelope. Plant Physiol. 114, 405.
Gamsjaeger, R., Liew, C. K., Loughlin, F. E., Crossley, M., and Mackay, J. P. (2007). Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 32, 63–70. doi: 10.1016/j.tibs.2006.12.007
Gruissem, W. (1989). Chloroplast gene expression: how plants turn their plastids on. Cell 56, 161–170. doi: 10.1016/0092-8674(89)90889-1
Hedden, P., and Phillips, A. L. (2000). Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 5, 523–530. doi: 10.1016/S1360-1385(00)01790-8
Hendrick, J. P., and Hartl, F. (1993). Molecular chaperone functions of heat-shock proteins. Annu. Rev. Biochem. 62, 349–384. doi: 10.1146/annurev.bi.62.070193.002025
Higa, M. M., Alam, S. L., Sundquist, W. I., and Ullman, K. S. (2007). Molecular characterization of the Ran-binding zinc finger domain of Nup153. J. Biol. Chem. 282, 17090–17100. doi: 10.1074/jbc.M702715200
Hou, X., Fu, A., Garcia, V. J., Buchanan, B. B., and Luan, S. (2015). PSB27: a thylakoid protein enabling Arabidopsis to adapt to changing light intensity. Proc. Nat. Acad. Sci. U.S.A. 112, 1613–1618. doi: 10.1073/pnas.1424040112
Huang, X., Zhang, X., and Yang, S. (2009). A novel chloroplast-localized protein EMB1303 is required for chloroplast development in Arabidopsis. Cell Res. 19, 1205–1216. doi: 10.1038/cr.2009.84
Ishizaki, Y., Tsunoyama, Y., Hatano, K., Ando, K., Kato, K., Shinmyo, A., et al. (2005). A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. Plant J. 42, 133–144. doi: 10.1111/j.1365-313X.2005.02362.x
Izawa, S., and Good, N. E. (1966). Effect of salts and electron transport on the conformation of isolated chloroplasts. II. electron microscopy. Plant Physiol. 41, 544–552. doi: 10.1104/pp.41.3.544
Jarvis, P., Chen, L. J., Li, H., Peto, C. A., Fankhauser, C., and Chory, J. (1998). An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282, 100–103. doi: 10.1126/science.282.5386.100
Klein, R. R., and Mullet, J. E. (1986). Regulation of chloroplast-encoded chlorophyll-binding protein translation during higher plant chloroplast biogenesis. J. Biol. Chem. 261, 11138–11145.
Kong, Z., Li, M., Yang, W., Xu, W., and Xue, Y. (2006). A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 141, 1376–1388. doi: 10.1104/pp.106.082941
Koussevitzky, S., Stanne, T. M., Peto, C. A., Giap, T., Sjögren, L. L., Zhao, Y., et al. (2007). An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol. Biol. 63, 85–96. doi: 10.1007/s11103-006-9074-2
Laity, J. H., Lee, B. M., and Wright, P. E. (2001). Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11, 39–46. doi: 10.1016/S0959-440X(00)00167-6
Larkin, R. M., Alonso, J. M., Ecker, J. R., and Chory, J. (2003). GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906. doi: 10.1126/science.1079978
Latijnhouwers, M., Xu, X.-M., and Møller, S. G. (2010). Arabidopsis stromal 70-kDa heat shock proteins are essential for chloroplast development. Planta 232, 567–578. doi: 10.1007/s00425-010-1192-z
Lennartz, K., Plücken, H., Seidler, A., Westhoff, P., Bechtold, N., and Meierhoff, K. (2001). HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b6f complex in Arabidopsis. Plant Cell 13, 2539–2551. doi: 10.1105/tpc.13.11.2539
Li, W.-T., He, M., Wang, J., and Wang, Y.-P. (2013). Zinc finger protein (ZFP) in plants. Plant Omics 6, 474–480.
Liu, Q., Xu, J., Liu, Y., Zhao, X., Deng, X., Guo, L., et al. (2007). A novel bud mutation that confers abnormal patterns of lycopene accumulation in sweet orange fruit (Citrus sinensis L. Osbeck). J. Exp. Bot. 58, 4161–4171. doi: 10.1093/jxb/erm273
McCormac, A. C., Fischer, A., Kumar, A. M., Söll, D., and Terry, M. J. (2001). Regulation of HEMA1 expression by phytochrome and a plastid signal during de-etiolation in Arabidopsis thaliana. Plant J. 25, 549–561. doi: 10.1046/j.1365-313x.2001.00986.x
McCormac, A. C., and Terry, M. J. (2002). Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. Plant J. 32, 549–559. doi: 10.1046/j.1365-313X.2002.01443.x
Miao, Y., Laun, T., Zimmermann, P., and Zentgraf, U. (2004). Targets of the WRKY53 transcription factor and its role during leaf senescence in Arabidopsis. Plant Mol. Biol. 55, 853–867. doi: 10.1007/s11103-005-2142-1
Miller, J., McLachlan, A., and Klug, A. (1985). Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 4, 1609.
Ohyama, K., Fukuzawa, H., Kohchi, T., Shirai, H., Sano, T., Sano, S., et al. (1986). Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 322, 572–574. doi: 10.1038/322572a0
Okada, K., Saito, T., Nakagawa, T., Kawamukai, M., and Kamiya, Y. (2000). Five geranylgeranyl diphosphate synthases expressed in different organs are localized into three subcellular compartments in Arabidopsis. Plant Physiol. 122, 1045–1056. doi: 10.1104/pp.122.4.1045
Osborne, B. A., and Raven, J. A. (1986). Light absorption by plants and its implications for photosynthesis. Biol. Rev. 61, 1–60. doi: 10.1111/j.1469-185X.1986.tb00425.x
Papenbrock, J., Gräfe, S., Kruse, E., Hänel, F., and Grimm, B. (1997). Mg-chelatase of tobacco: identification of a Chl D cDNA sequence encoding a third subunit, analysis of the interaction of the three subunits with the yeast two-hybrid system, and reconstitution of the enzyme activity by co-expression of recombinant CHL D, CHL H and CHL I. Plant J. 12, 981–990. doi: 10.1046/j.1365-313X.1997.12050981.x
Pogson, B. J., and Albrecht, V. (2011). Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol. 155, 1545–1551. doi: 10.1104/pp.110.170365
Qin, G., Gu, H., Ma, L., Peng, Y., Deng, X. W., Chen, Z., et al. (2007). Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 17, 471–482. doi: 10.1038/cr.2007.40
Robles, P., Micol, J. L., and Quesada, V. (2012). Arabidopsis MDA1, a nuclear-encoded protein, functions in chloroplast development and abiotic stress responses. PLoS ONE 7:e42924. doi: 10.1371/journal.pone.0042924
Rodríguez-Concepción, M., Ahumada, I., Diez-Juez, E., Sauret-Güeto, S., Lois, L. M., Gallego, F., et al. (2001). 1-Deoxy-d-xylulose 5-phosphate reductoisomerase and plastid isoprenoid biosynthesis during tomato fruit ripening. Plant J. 27, 213–222. doi: 10.1046/j.1365-313x.2001.01089.x
Ross, J. J., MacKenzie-Hose, A. K., Davies, P. J., Lester, D. R., Twitchin, B., and Reid, J. B. (1999). Further evidence for feedback regulation of gibberellin biosynthesis in pea. Physiol. Plant. 105, 532–538. doi: 10.1034/j.1399-3054.1999.105319.x
Shimada, H., Mochizuki, M., Ogura, K., Froehlich, J. E., Osteryoung, K. W., Shirano, Y., et al. (2007). Arabidopsis cotyledon-specific chloroplast biogenesis factor CYO1 is a protein disulfide isomerase. Plant Cell 19, 3157–3169. doi: 10.1105/tpc.107.051714
Staneloni, R. J., Rodriguez-Batiller, M. J., and Casal, J. J. (2008). Abscisic acid, high-light, and oxidative stress down-regulate a photosynthetic gene via a promoter motif not involved in phytochrome-mediated transcriptional regulation. Mol. Plant 1, 75–83. doi: 10.1093/mp/ssm007
Sugimoto, H., Kusumi, K., Noguchi, K., Yano, M., Yoshimura, A., and Iba, K. (2007). The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. Plant J. 52, 512–527. doi: 10.1111/j.1365-313X.2007.03251.x
Tanaka, R., and Tanaka, A. (2007). Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346. doi: 10.1146/annurev.arplant.57.032905.105448
Taylor, W. C. (1989). Regulatory interactions between nuclear and plastid genomes. Annu. Rev. Plant Biol. 40, 211–233. doi: 10.1146/annurev.pp.40.060189.001235
Thomas, S. G., Phillips, A. L., and Hedden, P. (1999). Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. U.S.A. 96, 4698–4703. doi: 10.1073/pnas.96.8.4698
Waters, M. T., Moylan, E. C., and Langdale, J. A. (2008). GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 56, 432–444. doi: 10.1111/j.1365-313X.2008.03616.x
Wellburn, A. R. (1994). The spectral determination of chlorophylls a andb, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 144, 307–313. doi: 10.1016/S0176-1617(11)81192-2
Werner, A., Flotho, A., and Melchior, F. (2012). The RanBP2/RanGAP1 SUMO1/Ubc9 Complex Is a Multisubunit SUMO E3 Ligase. Mol. Cell 46, 287–298. doi: 10.1016/j.molcel.2012.02.017
Wu, Z., Zhang, X., He, B., Diao, L., Sheng, S., Wang, J., et al. (2007). A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol. 145, 29–40. doi: 10.1104/pp.107.100321
Xing, S., Miao, J., Li, S., Qin, G., Tang, S., Li, H., et al. (2010). Disruption of the 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) gene results in albino, dwarf and defects in trichome initiation and stomata closure in Arabidopsis. Cell Res. 20, 688–700. doi: 10.1038/cr.2010.54
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Keywords: SlRBZ, chlorosis, dwarfism, chloroplast, tomato
Citation: Fan M, Gao S, Ren J, Yang Q, Li H, Yang C and Ye Z (2016) Overexpression of SlRBZ Results in Chlorosis and Dwarfism through Impairing Chlorophyll, Carotenoid, and Gibberellin Biosynthesis in Tomato. Front. Plant Sci. 7:907. doi: 10.3389/fpls.2016.00907
Received: 13 April 2016; Accepted: 08 June 2016;
Published: 22 June 2016.
Edited by:
Jianjun Chen, University of Florida, USAReviewed by:
Antonio Ferrante, Università degli Studi di Milano, ItalyYi Li, University of Connecticut, USA
Copyright © 2016 Fan, Gao, Ren, Yang, Li, Yang and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changxian Yang, yangcx0915@mail.hzau.edu.cn;
Zhibiao Ye, zbye@mail.hzau.edu.cn
 Mingqin Fan1,2
Mingqin Fan1,2 Changxian Yang
Changxian Yang