- 1State Key Laboratory of Crop Stress Biology in Arid Areas, College of Horticulture, Northwest A&F University, Yangling, China
- 2Key Laboratory of Horticultural Plant Biology and Germplasm Innovation in Northwest China, Ministry of Agriculture, Northwest A&F University, Yangling, China
WRKY transcription factors are known to play important roles in plant responses to biotic stresses. We previously showed that the expression of the WRKY gene, VqWRKY52, from Chinese wild Vitis quinquangularis was strongly induced 24 h post inoculation with powdery mildew. In this study, we analyzed the expression levels of VqWRKY52 following treatment with the defense related hormones salicylic acid (SA) and methyl jasmonate, revealing that VqWRKY52 was strongly induced by SA but not JA. We characterized the VqWRKY52 gene, which encodes a WRKY III gene family member, and found that ectopic expression in Arabidopsis thaliana enhanced resistance to powdery mildew and Pseudomonas syringae pv. tomato DC3000, but increased susceptibility to Botrytis cinerea, compared with wild type (WT) plants. The transgenic A. thaliana lines displayed strong cell death induced by the biotrophic powdery mildew pathogen, the hemibiotrophic P. syringe pathogen and the necrotrophic pathogen B. cinerea. In addition, the relative expression levels of various defense-related genes were compared between the transgenic A. thaliana lines and WT plants following the infection by different pathogens. Collectively, the results indicated that VqWRKY52 plays essential roles in the SA dependent signal transduction pathway and that it can enhance the hypersensitive response cell death triggered by microbial pathogens.
Introduction
Grapevine (Vitis vinifera L.) is an important fruit crop that is cultivated world-wide, however, lots of grapevine varieties are highly susceptible to infection by a large variety of pathogens. For example, powdery mildew, Botrytis cinerea and downy mildew all affect the growth of grapevine and reduce its fruit quality (Mzid et al., 2007). Chemicals are often used in vineyards to prevent, or limit, disease out breaks and to increase production, but this practice leads to an increase in production costs and increased risk of environmental pollution and pesticide residues, with consequent deleterious effects on human health (Zhu et al., 2012). There is therefore considerable interest in developing new cultivated grapevine varieties that are highly resistant to pathogens and that retain high quality fruit. To this end, classical crossbreeding is commonly used, but this is time consuming and the phenotypic traits of the filial generation are typically unstable, so this approach has limited potential. In contrast, the use of molecular breeding to obtain new cultivated grapevine varieties is, in many ways, easier than conventional methods. However, molecular breeding of disease-resistant grape varieties has, to date, been limited by the rudimentary understanding of the networks of defense related genes, and so identifying these in grape is an important objective.
Plants are both lack of mobile defender cells and the somatic adaptive immune system to fight against microbial pathogens that may impair plant growth and reproduction. Instead, they have developed their unique immunity mechanisms to protect themselves, which include two-branched innate immune system, namely the PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI) (Dangl, 2001; Jones and Dangl, 2006). These plant immunities may share some common signaling components such as hypersensitive response (HR), reactive oxygen species (ROS), activating the expression of PATHOGENESIS-RELATED (PR) genes (Tena et al., 2011). Pathogen induced HR and ROS play important roles in plant defense. In grape, HR could limit the supply of nutrients required by the biotrophic fungus for further growth and development (Qiu et al., 2015). Meanwhile, ROS, which are generated in response to pathogen attacks, play an important role in regulating HR cell death. Besides, they are also involved in local and systemic resistance to different plant pathogens (Torres et al., 2002; Mur et al., 2008; Miller et al., 2009; Mersmann et al., 2010; Adachi et al., 2015). HR and the induction of PR proteins can be triggered by SA regulated defense mechanisms at the infection site or in distal parts of the plant, leading to the development of systemic acquired resistance (SAR) (Thomma et al., 1998; Wu et al., 2015).
Hypersensitive response and ROS are often associated with hormone regulated defense signaling pathways such as salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) (Tena et al., 2011). SA signaling pathways play important roles in plant defense responses (Wu et al., 2015). In Arabidopsis, enhanced disease susceptibility 1 (EDS1) and phytoalexin deficient 4 (PAD4) are involved in SA signaling pathways. They play essential roles upstream of SA biosynthesis and HR (Rustérucci and Parker, 2001). Arabidopsis isochorismate synthase functional (ICS1) is involved in pathogen-induced accumulation of SA and plays essential roles in diverse stress responses (Strawn et al., 2007). In grape, EDS1 like and PAD4 of two grapevine species, Vitis vinifera cv. Cabernet Sauvignon and V. aestivalis cv. Norton, are associated with the SA pathway, also play important roles in grapevine defenses against powdery mildew (Gao et al., 2014).
In addition to phytohormones, diverse families of transcription factors are known to regulate plant defenses. As an example, the WRKY gene family, which is one of the largest families of transcription factors in plants (Ülker and Somssich, 2004), had been shown to modulate plant defense signaling pathways (Eulgem and Somssich, 2007; Phukan et al., 2016). Arabidopsis thaliana WRKY18, WRKY40, and WRKY60 are all involved in responses to Pseudomonas syringae and B. cinerea, with WRKY18 playing a more substantial part in the process (Xu et al., 2006). In addition, A. thaliana WRKY33 is required for defense against necrotrophic fungi, and the wrky33 mutant is highly susceptible to B. cinerea (Liu et al., 2015). However, loss of function of WRKY57 has been reported to enhance host resistance to this pathogen, since WRKY57 usually compromises B. cinerea resistance by competing with WRKY33 to regulate the expression levels of jasmonate ZIM-domain (JAZ) genes, JAZ1 and JAZ5, which in turn act as repressors of the JA signaling pathway (Yan and Yu, 2016).
Although little is known regarding disease resistance-related genes in grapevine, several WRKY genes have been identified that may have essential roles in defense responses. VvWRKY1 can increase the resistance to downy mildew through jasmonic acid signaling pathway (Marchive et al., 2013). Ectopic expression of VvWRKY2 in tobacco was reported to result in high resistance to B. cinerea (Mzid et al., 2007), while Chinese wild grapevine Vitis pseudoreticulata (Vp) WRKY3 was shown to be specifically induced by pathogen infection, SA and ET, and its over-expression in tobacco enhanced resistance to Ralstonia solanacearum (Zhu et al., 2012). In addition, the expression of VpWRKY1 and VpWRKY2 is strongly induced by Erysiphe necator infection, and VpWRKY1 or VpWRKY2 over-expressing transgenic A. thaliana lines had increased resistance to powdery mildew (Li et al., 2010). The V. pseudoreticulata EIRP1 E3 ligase has been shown to interact with VpWRKY11 and this interaction may affect disease resistance by mediating proteolysis of the protein (Yu et al., 2013).
WRKY proteins can be phosphorylated by mitogen activated protein kinases (MAPKs) at specific sites to regulate plant defense signals (Ishihama and Yoshioka, 2012). For example, when WRKY7, WRKY8, WRKY9, and WRKY11 are phosphorylated by a MAPK, they can regulate the expression of NADPH oxidase, which triggers a ROS burst and cell death in Nicotiana benthamiana (Adachi et al., 2015). Other protein modifications, such as acetylation, may also affect WRKY protein function. For example, two effectors, PopP2 and AvrRps4, which are delivered by plant pathogens to suppress host defense, have evolved to block the function of WRKY transcription factors, potentially, though acetylating lysine residues in the WRKY domain, thereby affecting binding activity (Leroux et al., 2015; Sarris et al., 2015). The WRKY transcription factors can be classified into groups I–III, based on their WRKY domains and zinc-finger motifs (Eulgem et al., 2000; Wang et al., 2014). Group III genes are thought to be the most evolutionarily advanced and exhibit a high degree of adaptability (Zhang and Wang, 2005). In grapevine, 59 VvWRKY genes have been identified and classified into the three main groups (I–III) (Guo et al., 2014).
A range of wild grape genotypes have been identified in China, some of which show far greater resistance than cultivated grapevine varieties to some microbial pathogens (Wang et al., 1995). For example, Chinese wild Vitis quinquangularis clone Shang-24 was shown to be resistant to a number of fungal pathogens, particularly to E. necator (Wang et al., 1995; Wan et al., 2007). This wild grape species therefore has considerable potential as a resource for identifying disease resistance genes. In the current study, we characterized the expression of WRKY52 from Shang-24 that had been treated with SA or methyl-jasmonate (MeJA). We also over-expressed VqWRKY52 in A. thaliana and analyzed the responses of the transgenic lines to inoculation with Golovinomyces cichoracearum, B. cinerea, and P. syringae pv. tomato DC3000 (PstDC3000). The results are presented and discussed in the context of a role for VqWRKY52 in an SA dependent signal transduction pathway and in HR cell death.
Materials and Methods
Plant Materials, Growth Condition, and Pathogen
Chinese wild V. quinquangularis clone Shang-24 seedlings were grown in the grape germplasm resources orchard at the Northwest A&F University, Yangling, Shaanxi, China. Wild type (WT) A. thaliana (ecotype type, Columbia- 0), the pad4 mutant and N. benthamiana were preserved in our lab. A. thaliana plants were grown under the following conditions: 21°C, 50% relative humidity and a long-day photoperiod (16 h- light/ 8 h- dark). N. benthamiana was grown in a growth chamber under the following conditions: 26°C, 50% relative humidity and a long-day photoperiod (16 h- light/ 8 h- dark). G. cichoracearum was cultured on A. thaliana pad4 mutant plants at 21°C and a photoperiod of 16 h light/8 h dark. PstDC3000 was preserved at -80°C. B. cinerea was maintained at 22°C on Potato Glucose Agar as described by Wan et al. (2015).
Grape Hormone Treatments
Young leaves of 2-year-old grapes were sprayed with 100 μM SA or 50 μM MeJA (Repka et al., 2004; Wang and Li, 2006). Sterile distilled water was used as a mock control. Samples were collected at 1, 12, 24, and 48 h post treatment (hpt) and frozen at -80°C.
Quantitative Real-Time PCR
Quantitative real-time PCR analysis was performed as previously described (Tu et al., 2016). The E.Z.N.A.® Plant RNA Kit (Omega Bio-tek, USA, R6827-01) was used to extract grapevine total RNA and the RNA prep plant kit (Tiangen Biotech., China) was used to extract A. thaliana RNA. Prime Script TMR Tase (TaKaRa Biotechnology, Dalian, China) was used to synthesize first-strand cDNA. We used SYBR green (TaKaRa Biotechnology) and an IQ5 real-time PCR instrument (Bio-Rad, Hercules, CA, USA) to conduct quantitative real-time PCR (qRT-PCR) analysis. All of the above procedures were carried out according to the manufacturers’ instructions. VvActin1 or AtActin2 were used as references genes. Primers used for the qRT-PCR analyses are listed in Supplementary Table S1. Three biological replicates were analyzed for each experiment and three technical replicates for each biological replicate. Relative expression levels were analyzed with the IQ5 software using the Normalized Expression Method.
Vector Construction
Total RNA extractions from leaves of V. quinquangularis clone Shang-24 and first-strand cDNA synthesis were performed as described above. The open reading frame (ORF) of VqWRKY52 was amplified by PCR using the specific primers F1 (5′-CGGGATCCATGGAGAACATGGGAAGTTGGGAAC-3′) and R1 (5′-GGGGTACCTTAAAAGAATCCCAGGTGGTCGAAGTTA-3′). The PCR product was cloned into the pGEM-T easy vector (Promega, Madison, WI, USA), to give the construct pGEM-Teasy-VqWRKY52, which was then sequenced. To obtain the over-expression vector, the VqWRKY52 ORF from pGEM-Teasy-VqWRKY52 was inserted immediately downstream of the CaMV35S promoter in the plant over-expression vector, pCambia 2300 (Cambia, Brisbane, QLD, Australia) using the BamHI and KpnI restriction endonucleases.
Grapevine DNA was extracted from leaves of V. quinquangularis clone Shang-24 as previously described (Tu et al., 2016). A 2107 bp VqWRKY52 promoter fragment was amplified by PCR from genomic DNA using the gene specific primers F2 (5′-CCCAAGCTTCGGAATTCGCGTGATCAAAGTAATTGAGG-3′) and R2 (5′-TCCCCCGGGTTTTAAACCACCCAAAGAAGAAGAA-3′), and inserted into the binary vector pBI121 (Clontech, Palo Alto, CA, USA), to replace the CaMV 35S promoter, upstream from the β-glucuronidase (GUS) reporter gene, using the HindIII and SmaI restriction endonucleases. The resulting vector was named proV qWRKY 52:GUS. pBI121 was used as a positive control and renamed pro35S:GUS.
Plant Transformation
Each of the above constructs was introduced into Agrobacterium tumefaciens strain GV3101, and these resulting A. tumefaciens were used to transform A. thaliana, using the floral dip method (Clough and Bent, 1998). Transient expression in N. benthamiana was performed by infiltration as previously described (Guan et al., 2014). The infiltrated plants were maintained for an additional 3 days under the same conditions and the hormone treatments were performed as described above. For the A. thaliana transformation, the three T3 homozygous lines with the strongest resistance to powdery mildew (#28, #30, and #33) were selected and used for all subsequent experiments. To assess GUS activity in the transgenic proV qWRKY 52:GUS plants, three independent transgenic T3 lines were analyzed.
GUS Assays
β-Glucuronidase activity assays were performed as previously described (Jefferson et al., 1987). A vacuum was applied to the samples for 30 min prior to incubation at 37°C for 24–48 h in the staining solution [1 mM 5-bromo-4-chloro-3-indolyl-β-D-glucuronide (X-gluc; Biosynth AG), 100 mM sodium phosphate (pH 7.0), 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.1% Triton X-100 and 0.1 mM EDTA]. Chlorophyll was then cleared from the samples with 70% ethanol, and the samples were viewed under a light microscope (BX53, Olympus, Japan).
Inoculation of A. thaliana with Pathogen
Four-week-old T3 transgenic and WT plants were inoculated with G. cichoracearum, and the number of conidiophores per colony was counted at 7 days post inoculation (dpi), as previously described (Wang et al., 2011). Samples for the expression profile analysis of defense related genes and the accumulation of O2- were collected at 0, 24, 48, and 72 h post inoculation (hpi), samples used for analysis of fungal structures and monitoring cell death were collected at 7 dpi.
Four-week-old plants were inoculated with PstDC3000 by dipping whole rosettes in PstDC3000 solutions (108 cfu/mL, in 10 mM Mg2SO4 supplemented with 0.025% Silwet77) as previously described (Wen et al., 2015; Guo et al., 2016). The inoculated plants were maintained under 90% relative humidity for 24 h before being moved to normal growth conditions. Samples used for morphological observation were taken at 5 dpi. Samples used for the expression analysis of defense related genes were collected at 0, 6, 12, and 24 hpi. Leaves inoculated with PstDC3000 by infiltration (Varet et al., 2003) were used to monitor the bacterial growth at 3 dpi, and the detection of cell death was performed at 0, 24, 48, and 72 hpi, while accumulation of O2- and H2O2 was measured at 0 and 72 hpi.
The B. cinerea conidial suspension (1.5 × 106 conidia/ml) used for inoculation was prepared as previously described (Wan et al., 2015). Detached leaves were used for morphological observation and the lesion diameter analysis, which were performed by droplet inoculating with 10 μL of the conidial suspension, as previously described (Guo et al., 2016). Samples used for morphological observation and lesion diameters analysis were photographed and measured at 3 dpi. Adult plants were inoculated by spraying, as previously described (Wan et al., 2015), and were then used for analyzing the expression of defense related genes at 0, 12, 24, and 48 hpi. Cell death was measured at 0, 24, 48, and 72 hpi and the accumulation of O2- and H2O2 was measured at 0 and 48 hpi.
ROS Levels and Cell Death Assay
Cell death and the fungal structures were visualized by staining with trypan blue as previously described (Vogel and Somerville, 2000). Diaminobenzidine (DAB) staining was used to detect the accumulation of H2O2 (Fryer, 2002), and nitro blue tetrazolium (NBT) staining was used to detect the accumulation of O2- (Kim et al., 2011), as previously described. All samples were imaged with a light microscope (Olympus, Japan). At least six leaves were used for each independent experiment and three biological replicates were analyzed.
Statistical Analysis
Data analysis and plotting were performed using Microsoft Excel (Microsoft Corporation, USA) and Sigma plot (v. 10.0, Systat, Inc., Point Richmond, CA, USA). Significant differences were assessed through paired t-test using the SPSS Statistics software (IBM China Company, Ltd, Beijing, China) as previously described (Tu et al., 2016). All experiments were performed using three biological replicates, with each biological replicate having three technical replicates.
Accession Numbers
The accession numbers of the genes used in this paper are found in The Arabidopsis Information Resource1 and the grape genome Sequence2: AtActin2 (AT3G18780), AtEDS1 (AT3G48090), AtPR1 (AT2G14610), AtPR2 (AT3G57260), AtPR5 (AT1G75040), AtPDF1.2 (At5g44420), AtICS1 (At1g74710), VvActin1 (AY680701), VqWRKY52 (KY411919).
Results
VqWRKY52 Expression is Induced by SA Treatment
In previous studies, a transcriptome analysis of Chinese wild V. quinquangularis clone Shang-24 at different time points after inoculation with E. necator indicated that VqWRKY52 was highly induced by this treatment (Jiao et al., 2015). To identify which hormones could affect the expression of VqWRKY52, we measured its expression levels in V. quinquangularis clone Shang-24 at 1, 12, 24, and 48 h post treatment with SA or MeJA. We observed that the expression of VqWRKY52 was strongly induced by SA treatment, but not by MeJA, at 12 h post treatment, compared to the mock treatment, followed by a decrease at 24 h compared with the expression levels at 1 and 12 h (Figure 1A).
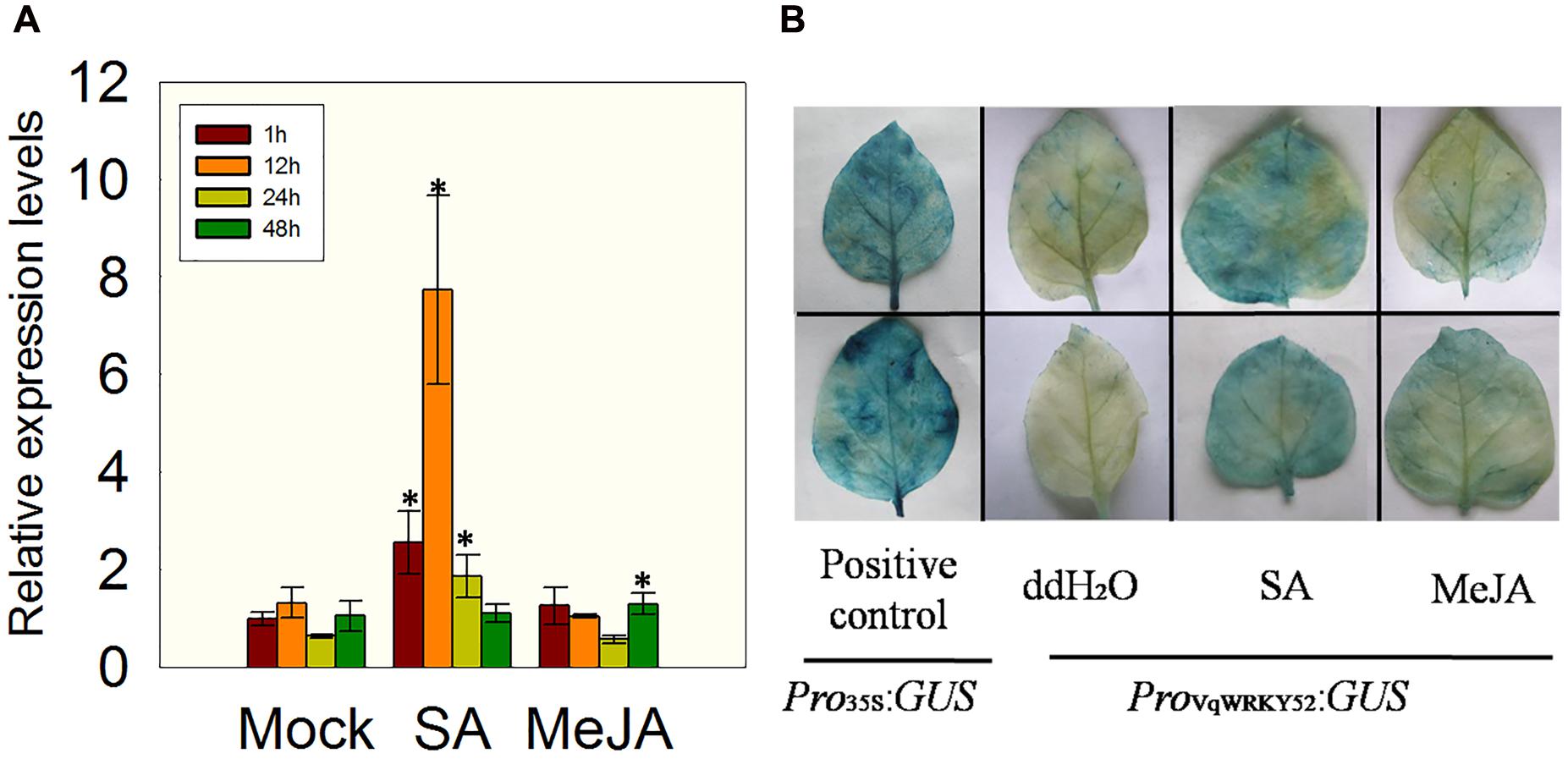
FIGURE 1. Expression of VqWRKY52 following salicylic acid (SA) and methyl jasmonate (MeJA) treatments. (A) Relative expression levels in Vitis pseudoreticulata by qRT-PCR. (B) Analysis of VqWRKY52 promoter activity in Nicotiana benthamiana. Bars represent the mean ± SD from three independent experiments. Asterisks indicate statistical significance between treatment and mock (∗0.01 < P < 0.05, Student’s t-test).
To validate this result, a 2107 bp VqWRKY52 promoter fragment was fused to the GUS reporter gene and transiently expressed in N. benthamiana, using a Pro35S:GUS construct as a positive control. The transiently expressing leaves were treated with SA, MeJA or a negative ddH2O control and then subjected to GUS staining. Leaves expressing the Pro35S:GUS construct showed strong GUS activity, while little activity was detected in the leaves transiently expressing the ProV qWRKY 52:GUS construct. However, when ProV qWRKY 52:GUS expressing leaves were treated with SA, high GUS activity levels were observed, while only low levels of activity were detected in MeJA treated leaves (Figure 1B). This was consistent with the expression results presented in Figure 1A.
Cloning and Sequence Analysis of VqWRKY52
Gene specific primers were designed according to the VvWRKY52 (GSVIVT01028718001) cDNA sequence from the Grape Genome Database (12×2) and used to amplify the VqWRKY52 ORF. The VqWRKY52 coding sequence (CDS) is 1092 bp, encoding a 364 amino acid protein, and the nucleotide sequence had 99.27% identity to the V. vinifera homolog, with only eight single nucleotide polymorphisms (SNPs) found between the CDS from the two grape genotypes (Supplementary Figure S1). The corresponding deduced amino acid sequences shared 99.18% identity (Supplementary Figure S2).
VqWRKY52 Expression Patterns
To further understand the temporal and spatial expression profile of VqWRKY52, transgenic A. thaliana lines constitutively expressing the ProV qWRKY 52:GUS construct were generated. Plants from the T3 generation were stained for GUS activity and while not activity was detected at the germination stage (Figure 2A), during early seedling growth, low GUS activity was observed in the cotyledon tips and roots (Figure 2B). Two-week-old plants grown on Murashige-Skoog (MS) basal medium showed strong GUS activity in all organs, but especially in leaves (Figure 2C). Aging leaves of 3-week-old plants in soil showed more GUS activity than the young leaves (Figure 2D), and strong GUS activity was observed in flowers and siliques, but not in the seeds and anthers (Figures 2E–H).

FIGURE 2. Expression of VqWRKY52 in Arabidopsis thaliana. T3 homozygous ProV qWRKY 52:GUS plants were stained with 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid at different growth stages. (A) Mature embryos were cultivated on Murashige-Skoog (MS) basal medium for 24 h. Scale bar = 200 μm. (B) 5-day-old seedling. Scale bar = 500 μm. (C) 2-week-old plant. Scale bar = 2 mm. (D) 3-week-old plant. (E) Stalk. Scale bar = 2 mm. (F) Inflorescence. Scale bar = 1 mm. (G) Flower. Scale bar = 200 μm. (H) Silique. Scale bar = 1 mm.
Over-Expressing VqWRKY52 in A. thaliana Enhances Resistance to Powdery Mildew
To further investigate the putative function of VqWRKY52 in defense process, three T3 generation transgenic A. thaliana lines expressing VqWRKY52 (Supplementary Figure S3), together with WT, and the A. thaliana pad4 mutant, which is susceptible to powdery mildew, were inoculated with G. cichoracearum. Over-expressing lines showed enhanced resistance to G. cichoracearum at 7 dpi (Figure 3A) and showed a large number of dead cells, while minimal cell death was observed in the WT plants and no obvious cell death occurred in the pad4 mutant (Figure 3B). The G. cichoracearum colonies growing on the pad4 mutant were the largest, followed by those on the WT plant, while the three over-expressing lines had the smallest colonies. This correlated with the extent of the cell death triggered by G. cichoracearum that was observed at the infection site, which is consistent with cell death restricting fungal growth (Figure 3B). We also counted the number of conidiophores per colony from the five different genotypes (Figure 3C) and determined that the three transgenic lines had a significantly fewer than the WT plants.
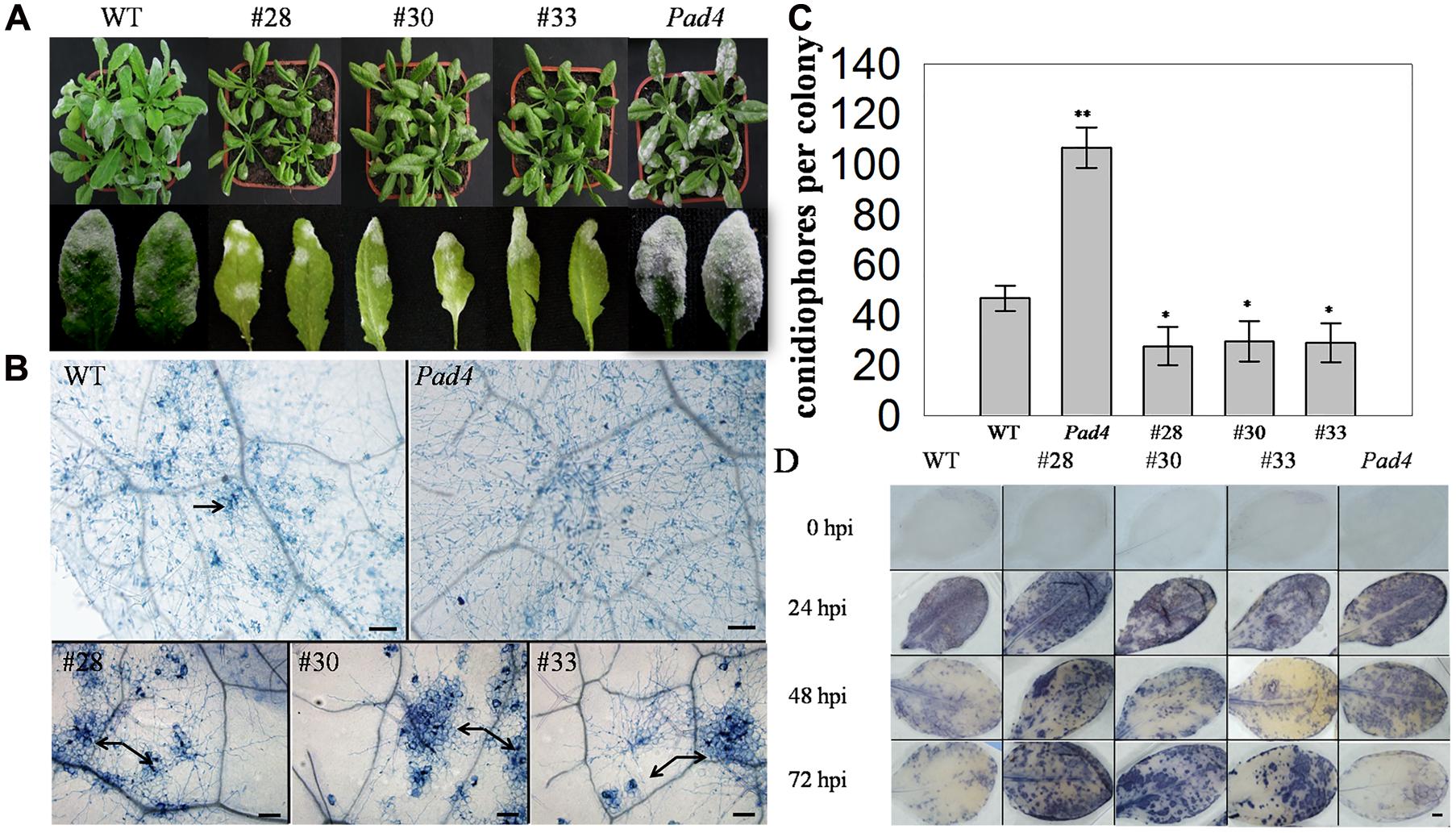
FIGURE 3. Phenotype of VqWRKY52 over-expressing lines inoculated with powdery mildew (G. cichoracearum). (A) Over-expressing lines (#28, #30, #33), pad4 mutant plants and wild type (WT) plants were infected with powdery mildew. Plants were photographed 7 days post-inoculation (dpi). (B) Fungal structures and plant cell death were stained with trypan blue at 7 dpi. The cell death induced by G. cichoracearum colonies is was highlighted by the arrow. Scale bar = 100 μm. (C) The accumulation of O2- in different plants at 0, 24, 48, and 72 h post-inoculation (hpi). O2- was visualized with nitro blue tetrazolium (NBT). Scale bar = 2 cm. (D) Quantitative analysis of conidiophore formation on different plants at 7 dpi. Bars represent the mean ± SD from three independent experiments. Asterisks indicate statistical significance between the over-expressing lines, the pad4 mutant plants and WT plants (∗0.01 < P < 0.05, ∗∗P < 0.01, Student’s t-test).
Since the accumulation of the superoxide anion (O2-) is associated with cell death (Yi et al., 2011), we measured O2- levels in the different genotypes at 0, 24, 48, and 72 hpi. A significant difference was observed between WT, pad4, and the three transgenic lines at 48, 72 hpi, with high levels in the latter. Interestingly, almost all of the leaves from the tested lines accumulated O2- at 24 hpi, while the mottled leaves with spots of O2- accumulation were appeared at 48 and 72 hpi (Figure 3D).
The Expression of Defense Related Genes Post Inoculation with G. cichoracearum
Since VqWRKY52 expression was induced by SA (Figures 1A,B), we hypothesized that it operates via SA mediated signaling pathways and the expression levels of marker genes involved in SA signaling pathways in Arabidopsis will be affected in three over-expressing lines. To test this, we measured the expression of four marker genes. The expression of AtICS1, which is involved in SA biosynthesis and affects SA accumulation (Strawn et al., 2007), was higher in the over-expressing lines than in the WT at all three time point post-inoculation, showing an initial increase at 24 hpi, peaking at 48 hpi and declining again at 72 hpi. AtEDS1 is involved in the SA related signaling pathway and plays essential roles upstream of SA biosynthesis (Rustérucci and Parker, 2001). The expression of this gene was similar to AtICS1 at 24 and 48 hpi; however, its expression levels were lower in the over-expressing lines than in the WT at 72 hpi. In addition, the expression of AtPR1 and AtPR5 increased at 24, 48, and 72 hpi compared to 0 hpi, with expression being higher in the three over-expression lines than in the WT (Figure 4).
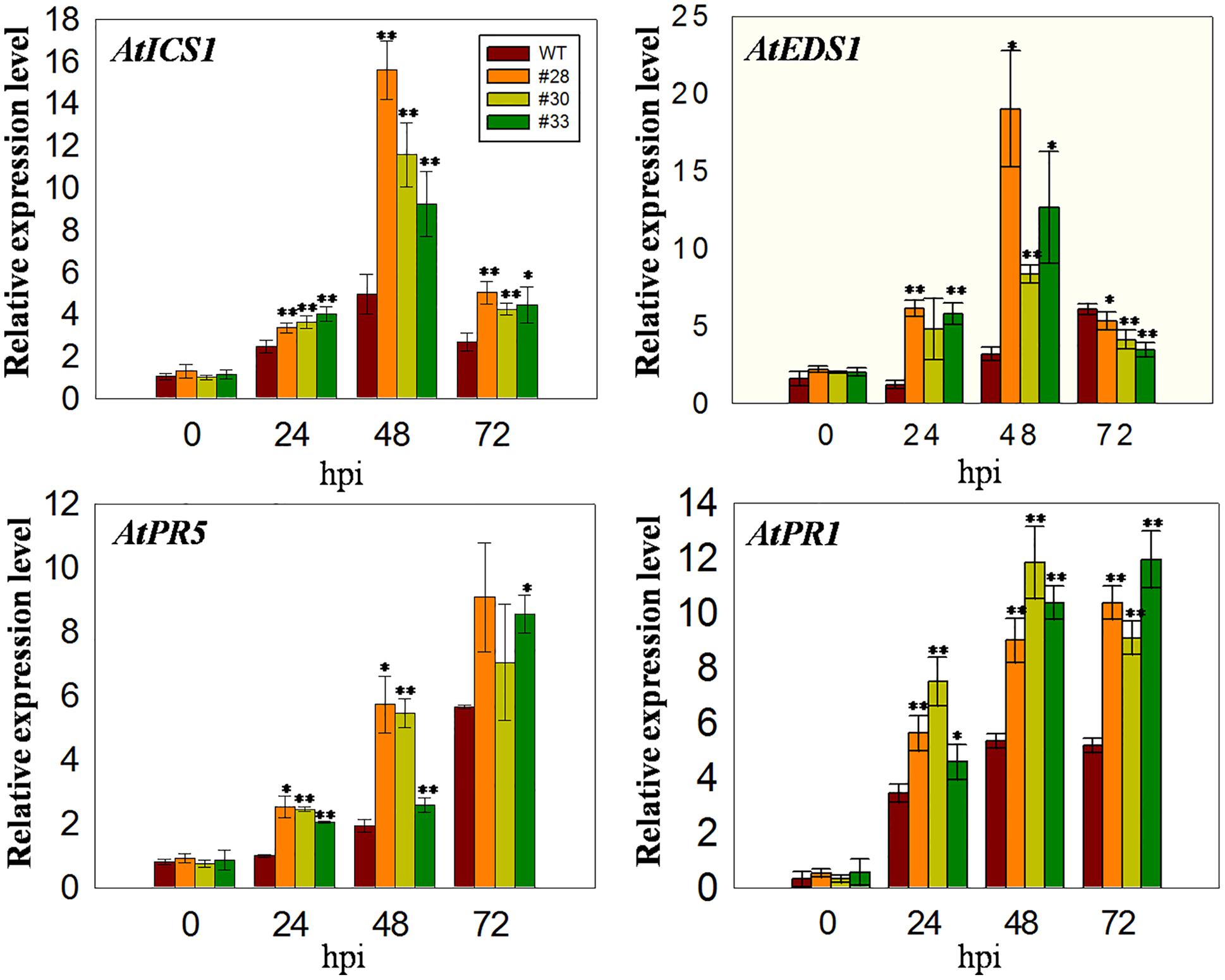
FIGURE 4. Quantitative analysis of the expression of defense-related genes in VqWRKY52 over-expressing lines and WT plants following G. cichoracearum infection. Relative expression levels of AtICS1, AtEDS1, AtPR1, and AtPR5 were analyzed using qRT-PCR. Bars represent the mean ± SD from three independent experiments. Asterisks indicate statistical significance between the over-expressing lines and WT plants (∗0.01 < P < 0.05, ∗∗P < 0.01, Student’s t-test).
Over-Expressing VqWRKY52 in A. thaliana Enhances Resistance to Pseudomonas
To examine the association between VqWRKY52 and responses to infection by PstDC3000, three T3 generation transgenic lines over-expressing VqWRKY52 and WT plants were inoculated with PstDC3000 and examined at 5 dpi. The three transgenic lines showed increased resistance to PstDC3000, based on less severe disease symptoms and fewer diseased leaves than the WT plants (Figure 5A). We also measured the growth of PstDC3000 by counting the bacterial numbers per unit leaf area at 3 dpi. As shown in Figure 5B, we observed less growth in the three over-expressing lines than in WT plants, which suggested VqWRKY52 mediated suppression of bacterial growth. The transgenic lines showed enhanced resistance to PstDC3000 and strongly induced cell death. Specifically, no cell death was detected in the three transgenic lines and WT plants at 0 hpi, and less cell death was observed in the transgenic lines than in the WT plants at 24 hpi. Strong cell death was apparent at 48 hpi and was enhanced in the overexpression lines at 72 hpi. At this time point less cell death was detected in the WT plants to an extent that was similar to that observed in the transgenic lines at 24 hpi (Figure 5C). Strong cell death may be associated with a burst of ROS production and so we examined the accumulation of O2- and H2O2 at 72 hpi, where more cell death was detected in the transgenic lines than in the WT plants. We found a larger accumulation of O2- and H2O2 in the transgenic lines than in the WT plants (Figure 5D).
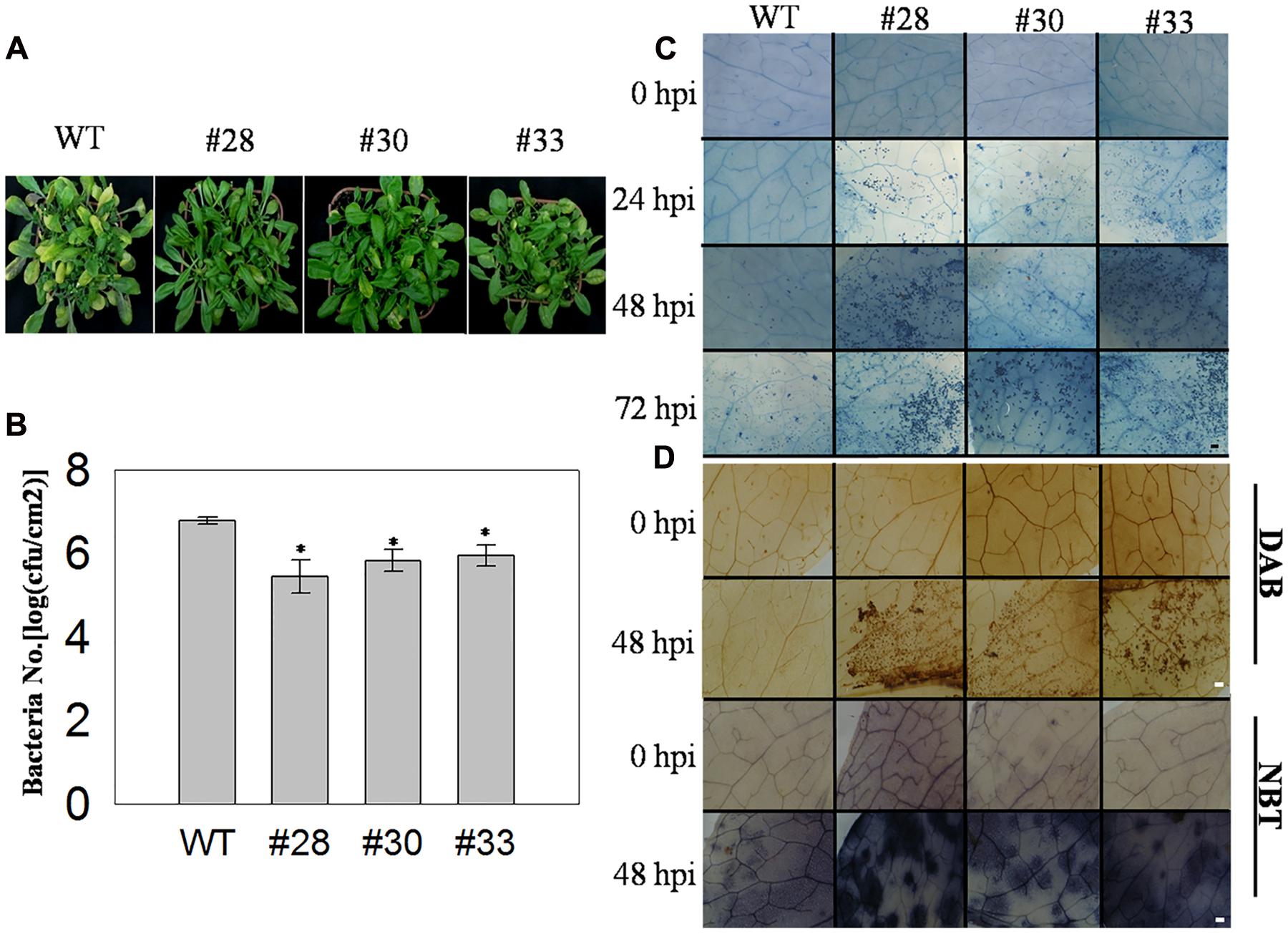
FIGURE 5. The response of VqWRKY52 over-expressing lines and WT to Pst DC3000 infection. (A) Infected leaves from over the overexpression lines and WT plants were photographed 5 dpi. (B) The number of bacterial cells in the leaves was determined at 3 dpi. (C) The plant cell death induced by PstDC3000 at 0, 24, 48, and 72 hpi was visualized by staining with trypan blue. Scale bar = 200 μm. (D) H2O2 and O2- accumulation in the overexpression lines and WT plants at 0 and 48 hpi. H2O2 were stained with DAB and O2- with NBT in WT and overexpression plants. The experiment was repeated three times, and at least six leaves were used in each independent experiment. Scale bar = 200 μm. The experiments have three independent biological replicates, and for each biological replicate three technical replicates were analyzed. Data represent mean values ± SD from three independent experiments. Asterisks indicate statistical significance between the overexpression lines and WT (∗0.01 < P < 0.05, Student’s t-test).
The Expression Levels of Defense Related Genes Post Inoculation with PstDC3000
Since cell death induced by PstDC3000 first appeared at 24 hpi in the three over-expressing lines and WT plants, we analyzed the expression levels of defense related genes at earlier times points, specifically 0, 6, 12, and 24 hpi. The expression of AtPR1, AtPR5, and AtEDS1 was induced in WT plants, but inhibited in the transgenic lines to varying degrees, which is the opposite of their expression pattern following the powdery mildew infection. This was also true for the AtPDF1.2 gene, which is associated with the MeJA signaling (Xu et al., 2006) (Figure 6).
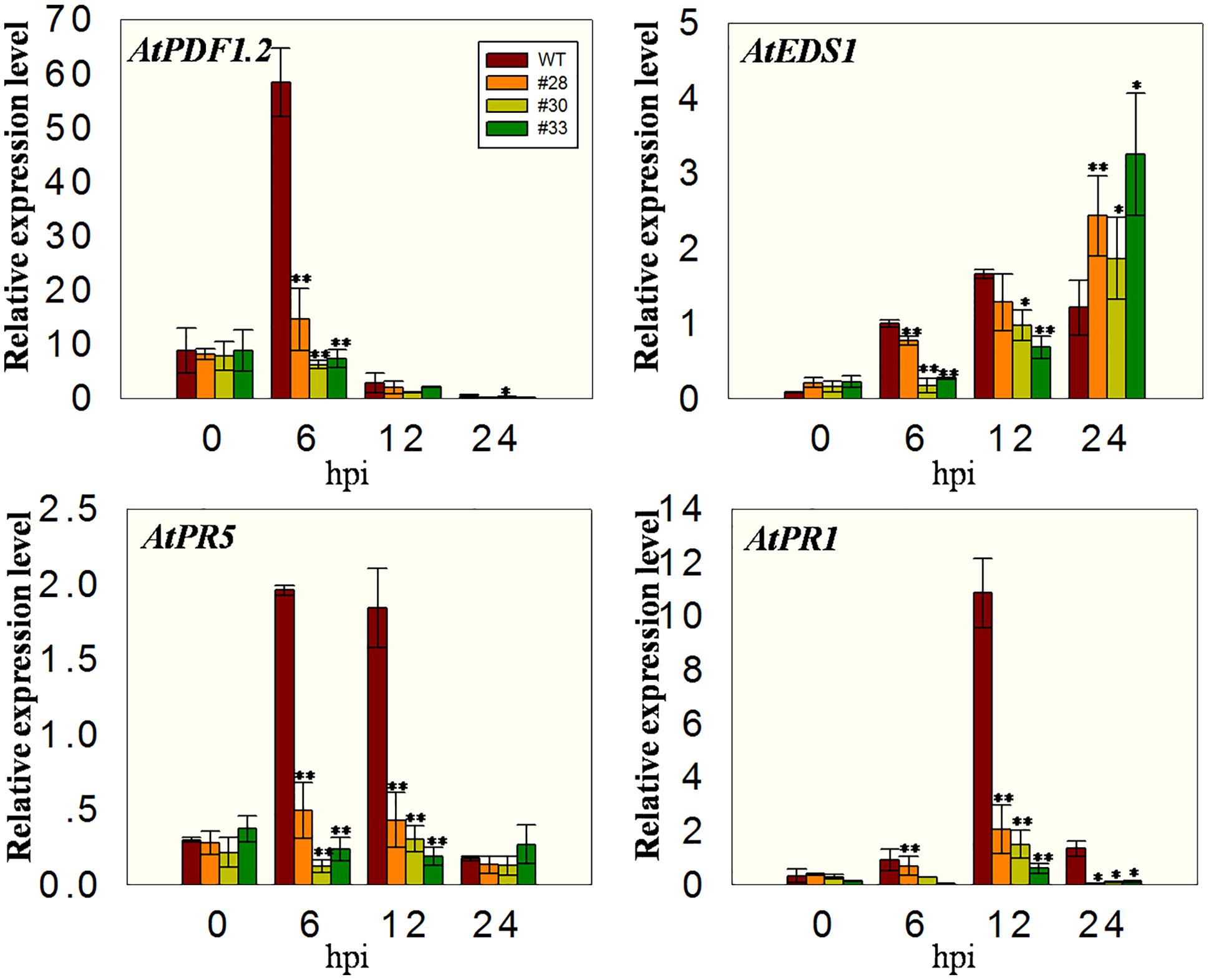
FIGURE 6. Quantitative analysis of defense-related genes in VqWRKY52 over-expressing lines and WT plants following PstDC3000 inoculation. Relative expression levels of AtEDS1, AtPR1, AtPR5, and AtPDF1.2 were analyzed using qRT-PCR. Bars represent the mean ± SD of three independent experiments. Asterisks indicate statistical significance between transgenic lines and WT (∗0.01 < P < 0.05, ∗∗P < 0.01, Student’s t-test).
Over-Expressing VqWRKY52 in A. thaliana Enhances Susceptibility to B. cinerea
Cell death induced by pathogens is important in the resistance to biotrophic pathogens, and we also wanted to determine whether this was also true for necrotrophic pathogens, such as B. cinerea. We observed the detached leaves of three overexpression lines and WT plants that had been inoculated with B. cinerea at 3 dpi and measured the lesion diameters of infected leaves, which were larger in the overexpression lines (Figures 7A,B). Subsequently, three plants from each genotype were incubated with B. cinerea by spraying, and cell death was detected at 0, 24, 48, and 72 hpi, with ROS staining at 0 and 48 hpi. Compared with the WT plants, cell death was more extensive in the transgenic lines at 24, 48, and 72 hpi, but was not observed at 0 hpi (Figure 7C). Minimal O2- and H2O2 accumulation was observed at 0 hpi in the VqWRKY52 over-expressing lines and WT plants, while higher levels were detected in the three over-expressing lines than in WT plants at 48 hpi (Figure 7D).
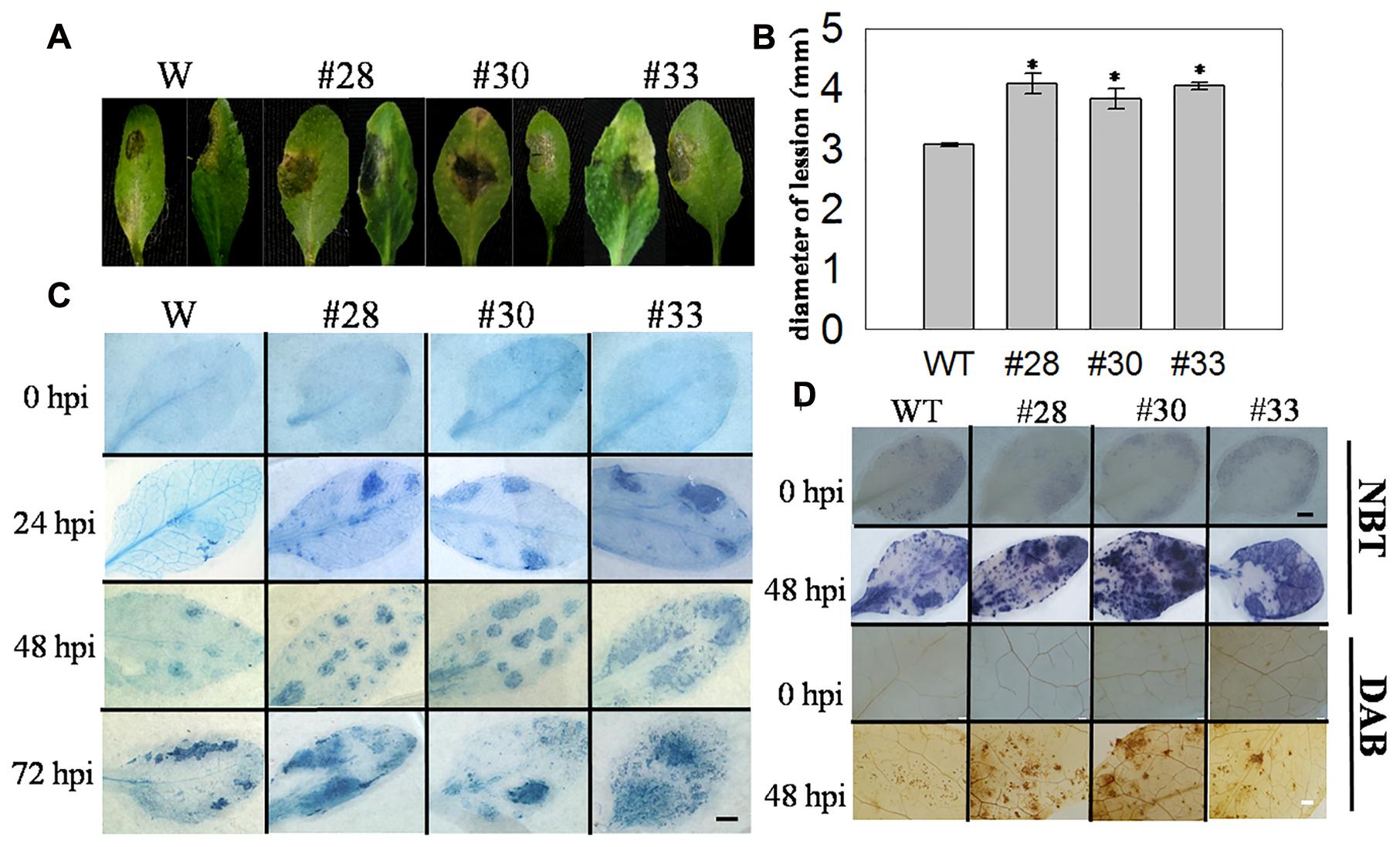
FIGURE 7. The responses of VqWRKY52 over-expressing lines and WT to Botrytis cinerea inoculation. (A) Infected leaves from transgenic lines and WT plants were photographed 3 dpi. (B) The average diameters of lesions of infected leaves at 3 dpi. (C) The plant cell death induced by B. cinerea at 0, 24, 48, and 72 hpi were stained with trypan blue. Scale bar = 2 cm. (D) H2O2 and O2- accumulation in transgenic lines and WT at 0 and 48 hpi. H2O2 were stained with DAB and O2- with NBT in WT and over-expressing plants. The experiment was repeated three times, at least six leaves were used in each independent experiment. Black bar = 2 cm; White bar = 200 μm. Data represent mean values ± SD from three independent experiments. Asterisks indicate statistical significance between transgenic lines and WT (∗0.01 < P < 0.05, Student’s t-test).
Relative Expression Levels of Defense Related Genes Post Inoculation with B. cinerea
To investigate whether the over-expressing lines had altered expression of defense-related genes, we measured the transcript levels of AtPR1, AtPR2, AtPR5, and AtPDF1.2 at different time points after B. cinerea inoculation. All plants showed low expression of AtPDF1.2 before inoculation and an induction of expression at 12 hpi, with the three transgenic lines having higher levels than the WT plants. At 24 hpi, the expression in WT plants was high, whereas it declined in the three transgenic lines such that the WT plants had the highest levels. Low AtPDF1.2 expression levels were detected at 48 hpi in all the plants, while the expression of AtPR2 and AtPR5 was low at 12, 24, and 48 hpi in the WT plants compared with 0 hpi, but were significantly higher in the overexpression lines than in the WT at all three time point, indicating less inhibition. Although the expression of AtPR1 was more highly induced in the overexpression lines than in the WT plants following the incubation with powdery mildew, no significant differences were observed in these plants following incubation with B. cinerea (Figure 8).
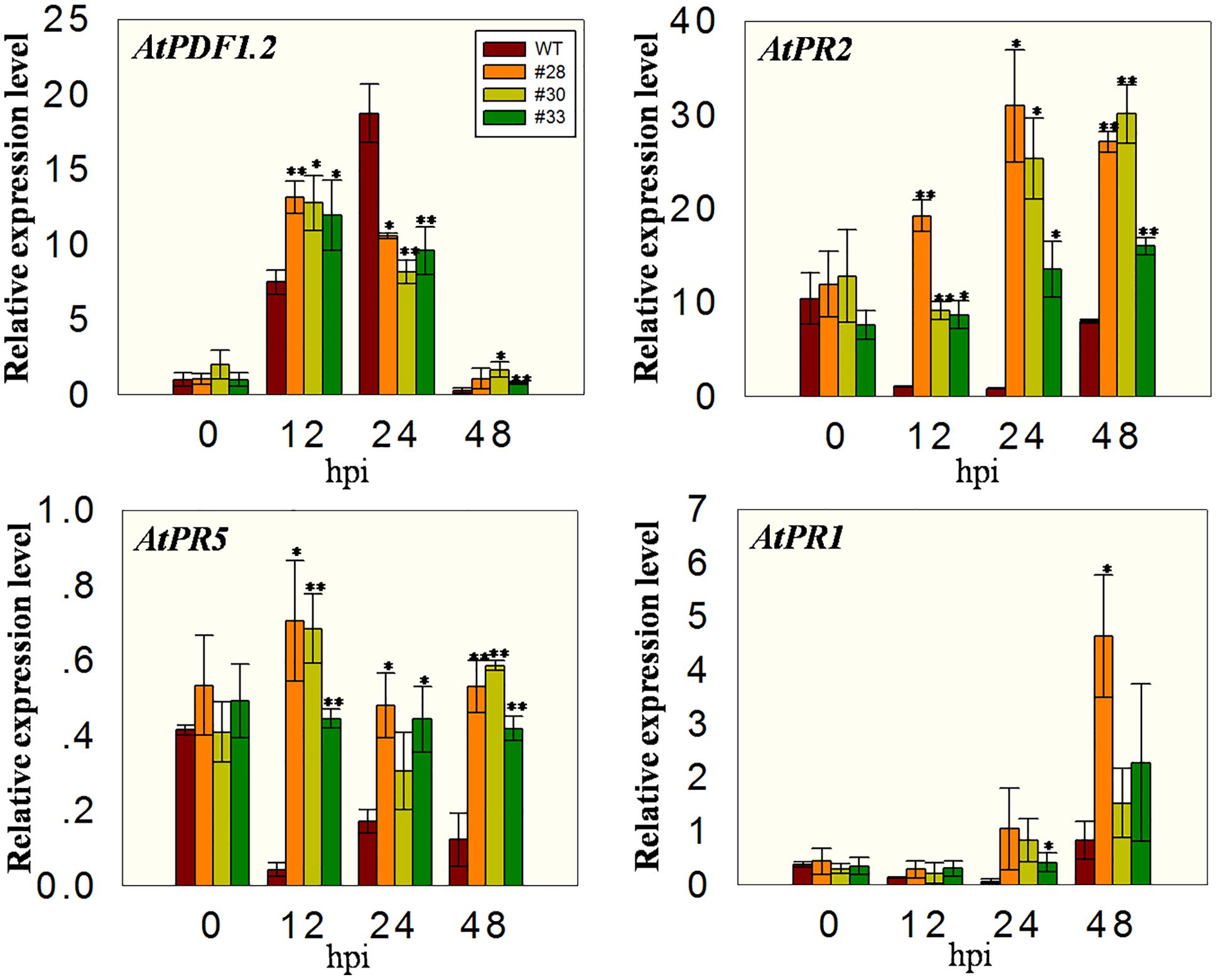
FIGURE 8. Quantitative analysis of defense-related genes in VqWRKY52 over-expressing lines and WT plants following B. cinerea inoculation. Relative expression levels of AtPR1, AtPR2, AtPR5, and AtPDF1.2 were analyzed using qRT-PCR. Bars represent the mean ± SD from three independent experiments. Asterisks indicate statistical significance between transgenic lines and WT (∗0.01 < P < 0.05, ∗∗P < 0.01, Student’s t-test).
Discussion
The WRKY transcription factor family is one of the largest in plants and members play important roles in signaling networks that regulate many plant processes, including defense signaling (Rushton et al., 2010). In grape, 59 grape WRKY genes (VvWRKY) have been identified and classified into three main groups (I–III) (Guo et al., 2014). Recent studies have shown that the WRKY domain of the NOD-like receptor RRS1-R, which is blocked by the pathogen effectors PopP2 and AvrRps4, belong to Group III (Leroux et al., 2015; Sarris et al., 2015), indicating the importance of the Group III genes in plant defense. The VqWRKY52 gene belongs to Group III (Guo et al., 2014). An earlier analysis revealed that VqWRKY52 is strongly induced post inoculation with E. necator (Guo et al., 2014). Here, we observed a strong increase in expression by SA treatment, suggesting a role for VqWRKY52 in disease resistance. So we tested the responses to powdery mildew, PstDC3000 and B. cinerea in A. thaliana VqWRKY52 over-expressing lines and WT plant.
It is known that SA signaling plays important roles in plant defense. Previous studies of WRKY70, a member of the group III in A. thaliana, indicated that its expression was strongly induced by SA (Li et al., 2004). Furthermore, WRKY70 over-expressing lines showed an increased resistance to the biotroph fungi Erysiphe cichoracearum (Eci), but decreased resistance to a necrotroph fungi (Li et al., 2006). In this current study, the expression of VqWRKY52 in V. quinquangular was induced by SA treatment but not by MeJA (Figure 1A), suggesting that it may be involved in the SA related defense signaling. The expression analysis of VqWRKY52 promoter in N. benthamiana together with the enhanced expression of AtEDS1 and AtICS1 post inoculation of G. cichoracearum were consistent with it being involved in the SA signaling pathway but not in JA mediated responses (Figure 1B), further indicating that it plays a role in resistance to biotrophs.
When WT plants, the pad4 mutant and the VqWRKY52 over-expressing lines were incubated with G. cichoracearum, the latter showed greater resistance than the others (Figure 3). Interestingly, G. cichoracearum inoculation induced strong cell death and, accordingly, fungal growth was inhibited to a greater degree, in the three over-expressing lines (Figures 3A,B). Pathogen-induced HR cell death is thought to be an important plant defense response (Wang et al., 2011), and many genes involved in cell death and plant defense have been identified. These genes can be grouped into two classes: genes involved in spontaneous cell death and genes involved in enhanced pathogen-induced cell death. The A. thaliana enhanced disease resistance 1 (EDR1) is a suppressor of plant defenses, and associated with pathogen-induced cell death, and the edr1 mutant shows enhanced disease resistance and powdery mildew-induced cell death (Serrano et al., 2014). This suggests an important role for VqWRKY52 in resistance of powdery mildew associated with pathogen-induced cell death.
PAD4 is also involved in defense responses, such as oxidative stress-related events and the HR (Serrano et al., 2014). In our studies, the pad4 mutant showed decreased resistance to powdery mildew, and had the largest number of fungi growth of the tested lines, while no obvious cell death was detected (Figure 3B). This indicated that the cell death induced by the pathogen was an active defense, rather than affecting by fungal growth. We generated pad4 mutant plants over-expressing VqWRKY52, but the pathogen-induced cell death was blocked compared with the overexpression lines in WT plants (Supplementary Figure S4), suggesting that the function of VqWRKY52 in pathogen-induced cell death relies on the AtPAD4 gene in A. thaliana.
There are two VqWRKY52 homologs in A. thaliana, AtWRKY41 and AtWRKY53. AtWRKY41 has been shown to be involved in seed development and defense response (Higashi et al., 2008; Zhong et al., 2014), while AtWRKY53 mainly plays a role in leaf development, senescence and defense response (Murray et al., 2007; Ay et al., 2009; Ying and Zentgraf, 2010; Xie et al., 2011). Over-expressing WRKY41 transgenic lines had increased resistance to PstDC3000 and enhanced susceptibility to Erwinia carotovora, but reduced expression of AtPDF1.2, which was induced by MeJA (Higashi et al., 2008). AtWRKY53 was found to promote basal resistance and wrky53 mutants had increased susceptibility to PstDC3000 inoculation (Murray et al., 2007). When we analyzed the response of the VqWRKY52 over-expressing lines to PstDC3000, similar results were obtained. The VqWRKY52 overexpression lines showed an increased resistance to PstDC3000 (Figure 5), and suppressed AtPDF1.2 expression (Figure 6). Strong cell death induced by PstDC3000 was also detected (Figure 5C). In contrast to AtEDS1, AtPR1 and AtPR5 expression following powdery mildew inoculation (Figure 4), expression of these genes was inhibited following PstDC3000 inoculation compared with WT plants at an early stage (Figure 6), consistent with previous studies (Huang et al., 2016). This indicated that the cell death induced by PstDC3000 and the expression of AtPR1 and AtPR5 in the overexpression lines are uncoupled. PopP2 and AvrRps4 are believed to block the functions of WRKY transcription factors, potentially, through acetylating lysine residues in the WRKY domain (Leroux et al., 2015; Sarris et al., 2015). Therefore, the inhibition of AtPR1 and AtPR5 post PstDC3000 inoculation in the three overexpression lines may be affected by PopP2 or AvrRps4. Alternatively, VqWRKY52 proteins without DNA binding activity may partly interfere with regulating the expression of AtPR1 and AtPR5.
We also tested the response of the VqWRKY52 over-expressing lines to B. cinerea, a necrotrophic pathogen. It is known that necrotrophic pathogens have virulence strategies to promote host cell death and acquire nutrients from dead cells (Kan, 2006; Mengiste, 2012). Here, a stronger cell death was also induced by B. cinerea in the overexpression lines than in WT plants, and it is possible that they had an increased susceptibility, since the strong cell death (Figure 7C) promoted the growth of B. cinerea. In addition, the expression of AtPR2 and AtPR5 was strongly inhibited in the WT plants post inoculation compared to the transgenic lines. This suggested that overexpression of VqWRKY52 can reduce the inhibition of AtPR2 and AtPR5 expression. The expression levels of AtPR1 showed significant difference with post inoculation of G. cichoracearum and PstDC3000 among WT and three overexpression lines (Figure 6). However, no significant difference was found on post B. cinerea inoculation (Figure 8). This indicated that over-expressed VqWRKY52 can’t affect the expression of AtPR1. The expression levels of AtPDF1.2 was enhanced at 24 hpi while suppressed at 48 hpi (Figure 8). Since AtPDF1.2 was induced by MeJA (Higashi et al., 2008), this suggested that MeJA signaling was enhanced in an early stage. However, strong cell death facilitated the infection progress, which finally resulted in the fact that even enhanced MeJA signaling did not increase the resistance. Our results also indicated that over-expressing VqWRKY52 increased the expression of SA signaling pathway related genes, which promote B. cinerea infection.
The ROS burst occurs hours after pathogen attacks, and is essential in regulating HR cell death (Mur et al., 2008). WRKY7, 8, 9, and 11 can regulate the ROS burst and cell death in N. benthamiana through controlling the expression of NADPH oxidase (Adachi et al., 2015). In our studies, the accumulation of O2- and H2O2 was detected following the inoculation with three different pathogens (Figures 3D, 5D, and 7D) in the transgenic lines and WT plants, and accumulation levels correlated positively with HR related cell death. This suggested that the ROS burst may control pathogen-induced cell death in VqWRKY52 overexpression lines.
Interestingly, leaves of 3-week-old plants in soil showed low promoter activity. This was consistent with our result (Figure 1). However, 2-week-old plants grown on Murashige-Skoog (MS) basal medium showed strong promoter activity. Although the age of the plants may impact the promoter activity, we suppose that the high humidity also plays important roles. High humidity often occurred together with pathogen infection and can promote pathogen growth and compromise plant disease responses (Zhou et al., 2004; Cai et al., 2015). This may suggest that VqWRKY52 plays essential roles in plant defense. In conclusion, our results suggest that VqWRKY52 may be involved in SA dependent signaling and pathogen-induced cell death. Future studies are needed to investigate the regulatory mechanisms of VqWRKY52 mediated HR related cell death.
Author Contributions
XipW and XiaW designed the study. XiaW, RG, and MT contributed to most of the experiments. XiaW and DW constructed the vectors, DW and CG performed data analysis. ZL and RW assisted with the analysis of the results. XiaW and XipW wrote the manuscript. All of the authors approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31572110 and 31501740), as well as the Program for Innovative Research Team of Grape Germplasm Resources and Breeding (2013KCT-25). We thank PlantScribe (www.plantscribe.com) for careful editing of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00097/full#supplementary-material
Footnotes
References
Adachi, H., Nakano, T., Miyagawa, N., Ishihama, N., Yoshioka, M., Katou, Y., et al. (2015). WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 27, 2645–2663. doi: 10.1105/tpc.15.00213
Ay, N., Irmler, K., Fischer, A., Uhlemann, R., Reuter, G., and Humbeck, K. (2009). Epigenetic programming via histone methylation at WRKY53 controls leaf senescence in Arabidopsis thaliana. Plant J. 58, 333–346. doi: 10.1111/j.1365-313X.2008.03782.x
Cai, H., Sheng, Y., Yan, Y., Xiao, Z., Cheng, J., Wu, J., et al. (2015). CaWRKY6 transcriptionally activates CaWRKY40, regulates ralstonia solanacearum resistance, and confers high-temperature and high-humidity tolerance in pepper. J. Exp. Bot. 66, 3163–3174. doi: 10.1093/jxb/erv125
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium –mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dangl, J. L. (2001). Plant pathogens and integrated defense responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Eulgem, T., and Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Fryer, M. J. (2002). Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 53, 1249–1254. doi: 10.1093/jexbot/53.372.1249
Gao, F., Dai, R., Pike, S. M., Qiu, W., and Gassmann, W. (2014). Functions of EDS1-like and PAD4 genes in grapevine defenses against powdery mildew. Plant Mol. Biol. 86, 381–393. doi: 10.1007/s11103-014-0235-4
Guan, Q., Yue, X., Zeng, H., and Zhu, J. (2014). The protein phosphatase RCF2 and its interacting partner NAC019 are critical for heat stress-responsive gene regulation and hermos tolerance in Arabidopsis. Plant Cell 26, 438–453. doi: 10.1105/tpc.113.118927
Guo, C., Guo, R., Xu, X., Gao, M., Li, X., Song, J., et al. (2014). Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 65, 1513–1528. doi: 10.1093/jxb/eru007
Guo, R., Tu, M., Wang, X., Jiao, Z., Ran, W., Li, Z., et al. (2016). Ectopic expression of a grape aspartic protease gene, AP13, in Arabidopsis thaliana, improves resistance to powdery mildew but increases susceptibility to Botrytis cinerea. Plant Sci. 248, 17–27. doi: 10.1016/j.plantsci.2016.04.006
Higashi, K., Ishiga, Y., Inagaki, Y., Toyoda, K., Shiraishi, T., and Ichinose, Y. (2008). Modulation of defense signal transduction by flagellin-induced WRKY41 transcription factor in Arabidopsis thaliana. Mol. Genet. Genomics. 279, 303–312. doi: 10.1007/s00438-007-0315-0
Huang, L., Zhang, S., Singer, S. D., Yin, X., Yang, J., Wang, Y., et al. (2016). Expression of the grape VqSTS21 gene in Arabidopsis confers resistance to osmotic stress and biotrophic pathogens but not Botrytis cinerea. Front. Plant Sci. 7:1379. doi: 10.3389/fpls.2016.01379
Ishihama, N., and Yoshioka, H. (2012). Post-translational regulation of WRKY transcription factors in plant immunity. Curr. Opin. Plant Biol. 15, 431–437. doi: 10.1016/j.pbi.2012.02.003
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907.
Jiao, C., Gao, M., Wang, X., and Fei, Z. (2015). Transcriptome characterization of three wild Chinese Vitis, uncovers a large number of distinct disease related genes. BMC Genomics 16:223. doi: 10.1186/s12864-015-1442-3
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kan, J. A. L. V. (2006). Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 11, 247–253. doi: 10.1016/j.tplants.2006.03.005
Kim, S. H., Woo, D. H., Kim, J. M., Lee, S. Y., Chung, W. S., and Moon, Y. H. (2011). Arabidopsis, MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem. Biophys. Res. Commun. 412, 150–154. doi: 10.1016/j.bbrc.2011.07.064
Leroux, C., Huet, G., Jauneau, A., Camborde, L., Trémousaygue, D., Kraut, A., et al. (2015). A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 161, 1074–1088. doi: 10.1016/j.cell.2015.04.025
Li, H., Xu, Y., Xiao, Y., Zhu, Z., Xie, X., Zhao, H., et al. (2010). Expression and functional analysis of two genes encoding transcription factors, VpWRKY1 and VpWRKY2, isolated from Chinese wild Vitis pseudoreticulata. Planta 232, 1325–1337. doi: 10.1007/s00425-010-1258-y
Li, J., Brader, G., Kariola, T., and Palva, E. T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46, 477–491. doi: 10.1111/j.1365-313X.2006.02712.x
Li, J., Brader, G., and Palva, E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331. doi: 10.1105/tpc.016980
Liu, S., Kracher, B., Ziegler, J., Birkenbihl, R. P., and Somssich, I. E. (2015). Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. Elife 4:e07295. doi: 10.7554/eLife.07295
Marchive, C., Léon, C., Kappel, C., Coutos-Thévenot, P., Corio-Costet, M. F., Delrot, S., et al. (2013). Over-expression of VvWRKY1 in grapevines induces expression of jasmonic acid pathway-related genes and confers higher tolerance to the downy mildew. PLoS ONE 8:e54185. doi: 10.1371/journal.pone.0054185
Mengiste, T. (2012). Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50, 267–294. doi: 10.1146/annurev-phyto-081211-172955
Mersmann, S., Bourdais, G., Rietz, S., and Robatzek, S. (2010). Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 154, 391–400. doi: 10.1104/pp.110.154567
Miller, G., Schlauch, K., Tam, R., Cortes, D., Torres, M. A., Shulaev, V., et al. (2009). The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2:ra45. doi: 10.1126/scisignal.2000448
Mur, L. A., Kenton, P., Lloyd, A. J., Ougham, H., and Prats, E. (2008). The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59, 501–520. doi: 10.1093/jxb/erm239
Murray, S. L., Ingle, R. A., Petersen, L. N., and Denby, K. J. (2007). Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol. Plant Microbe Interact. 20, 1431–1438. doi: 10.1094/MPMI-20-11-1431
Mzid, R., Marchive, C., Blancard, D., Deluc, L., Barrieu, F., Corio-Costet, M. F., et al. (2007). Overexpression of VvWRKY2 in tobacco enhances broad resistance to necrotrophic fungal pathogens. Physiol. Plant 131, 434–447. doi: 10.1111/j.1399-3054.2007.00975.x
Phukan, U. J., Jeena, G. S., and Shukla, R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7:760. doi: 10.3389/fpls.2016.00760
Qiu, W., Feechan, A., and Dry, I. (2015). Current understanding of grapevine defense mechanisms against the biotrophic fungus (Erysiphe necator), the causal agent of powdery mildew disease. Hortic. Res. 2, 41–46. doi: 10.1038/hortres.2015.20
Repka, V., Fischerová, I., and Šilhárová, K. (2004). Methyl jasmonate is a potent elicitor of multiple defense responses in grapevine leaves and cell-suspension cultures. Biol. Plant. 48, 273–283. doi: 10.1023/B:BIOP.0000033456.27521.e5
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Rustérucci, C., and Parker, J. E. (2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13, 2211–2224. doi: 10.1105/tpc.13.10.2211
Sarris, P., Duxbury, Z., Huh, S. U., Ma, Y., Segonzac, C., Sklenar, J., et al. (2015). A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 161, 1089–1100. doi: 10.1016/j.cell.2015.04.024
Serrano, I., Gu, Y., Qi, D., Dubiella, U., and Innes, R. W. (2014). The Arabidopsis EDR1 protein kinase negatively regulates the ATL1 E3 ubiquitin ligase to suppress cell death. Plant Cell 26, 4532–4546. doi: 10.1105/tpc.114.131540
Strawn, M. A., Marr, S. K., Inoue, K., Inada, N., Zubieta, C., and Wildermuth, M. C. (2007). Arabidopsis isochorismate synthase functional in pathogen-induced salicylate biosynthesis exhibits properties consistent with a role in diverse stress responses. J. Biol. Chem. 282, 5919–5933. doi: 10.1074/jbc.M605193200
Tena, G., Boudsocq, M., and Sheen, J. (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14, 519–529. doi: 10.1016/j.pbi.2011.05.006
Thomma, B. P., Eggermont, K., Penninckx, I. A., Mauch-Mani, B., Vogelsang, R., Cammue, B. P., et al. (1998). Separate jasmonate-dependent and salicylate-dependent defense response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. U.S.A. 95, 15107–15111. doi: 10.1073/pnas.95.25.15107
Torres, M. A., Dangl, J. L., and Jones, J. D. G. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. U.S.A. 99, 517–522. doi: 10.1073/pnas.012452499
Tu, M., Wang, X., Feng, T., Sun, X., Wang, Y., Huang, L., et al. (2016). Expression of a grape (Vitis vinifera) bZIP transcription factor, VlbZIP36, in Arabidopsis thaliana, confers tolerance of drought stress during seed germination and seedling establishment. Plant Sci. 252, 311–323. doi: 10.1016/j.plantsci.2016.08.011
Ülker, B., and Somssich, I. E. (2004). WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7, 491–498. doi: 10.1016/j.pbi.2004.07.012
Varet, A., Hause, B., Hause, G., Scheel, D., and Lee, J. (2003). The Arabidopsis NHL3 gene encodes a plasma membrane protein and its overexpression correlates with increased resistance to pseudomonas syringae pv. tomato DC3000. Plant Physiol. 132, 2023–2033. doi: 10.1104/pp.103.020438
Vogel, J., and Somerville, S. (2000). Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. U.S.A. 97, 1897–1902. doi: 10.1073/pnas.030531997
Wan, R., Hou, X., Wang, X., Qu, J., Singer, S. D., Wang, Y., et al. (2015). Resistance evaluation of Chinese wild Vitis genotypes against Botrytis cinerea and different responses of resistant and susceptible hosts to the infection. Front. Plant Sci. 6:854. doi: 10.3389/fpls.2015.00854
Wan, Y., Schwaninger, H., He, P., and Wang, Y. (2007). Comparison of resistance to powdery mildew and downy mildew in Chinese wild grapes. Vitis 46, 132–136.
Wang, L. J., and Li, S. H. (2006). Thermo tolerance and related antioxidant enzyme activities induced by heat acclimation and salicylic acid in grape (Vitis vinifera L.) leaves. Plant Growth Regul. 48, 137–144. doi: 10.1007/s10725-005-6146-2
Wang, M., Vannozzi, A., Wang, G., Liang, Y. H., Tornielli, G. B., Zenoni, S., et al. (2014). Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hortic. Res. 1, 221–240. doi: 10.1038/hortres.2014.16
Wang, Y., Liu, Y., He, P., Chen, J., Lamikanra, O., and Lu, J. (1995). Evaluation of foliar resistance to Uncinula necator in Chinese wild Vitis species. Vitis 34, 159–164.
Wang, Y., Nishimura, M. T., Zhao, T., and Tang, D. (2011). ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J. 68, 74–87. doi: 10.1111/j.1365-313X.2011.04669.x
Wen, Z., Yao, L., Wan, R., Li, Z., Liu, C., and Wang, X. (2015). Ectopic expression Aarabidopsis thaliana of an NB-ARC encoding putative disease resistance gene from wild Chinese Vitis pseudoreticulata enhances resistance to phytopathogenic fungi and bacteria. Front. Plant Sci. 6:1087. doi: 10.3389/fpls.2015.01087
Wu, G., Liu, S., Zhao, Y., Wang, W., Kong, Z., and Tang, D. (2015). Enhanced disease resistance 4 associates with clathrin heavy chain2 and modulates plant immunity by regulating relocation of EDR1 in Arabidopsis. Plant Cell 27, 857–873. doi: 10.1105/tpc.114.134668
Xie, Y., Huhn, K., Brandt, R., Potschin, M., Bieker, S., Bieker, S., et al. (2011). Revoluta and WRKY53 connect early and late leaf development in Arabidopsis. Development 141, 4772–4783. doi: 10.1242/dev.117689
Xu, X., Chen, C., Fan, B., and Chen, Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18, 1310–1326. doi: 10.1105/tpc.105.037523
Yan, J., and Yu, D. (2016). The WRKY57 transcription factor affects the expression of Jasmonate ZIM-Domain genes transcriptionally to compromise Botrytis cinerea resistance. Plant Physiol. 171, 2771–2782. doi: 10.1104/pp.16.00747
Yi, S. Y., Lee, D. J., Yeom, S. I., Yoon, J., Kim, Y. H., Kwon, S.-Y., et al. (2011). A novel pepper (Capsicum annuum) receptor-like kinase functions as a negative regulator of plant cell death via accumulation of superoxide anions. New Phytol. 185, 701–715. doi: 10.1111/j.1469-8137.2009.03095.x
Ying, M., and Zentgraf, U. (2010). A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J. 63, 179–188. doi: 10.1111/j.1365-313X.2010.04233.x
Yu, Y., Xu, W., Wang, J., Wang, L., Yao, W., Yang, Y., et al. (2013). The Chinese wild grapevine (Vitis pseudoreticulata) E3 ubiquitin ligase erysiphe necator-induced ring finger protein 1 (EIRP1) activates plant defense responses by inducing proteolysis of the VpWRKY11 transcription factor. New Phytol. 200, 834–846. doi: 10.1111/nph.12418
Zhang, Y., and Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5:1. doi: 10.1186/1471-2148-5-1
Zhong, J. D., Jing, Y. Y., Gui, X. L., Zhong, C. W., Shu, Q. Z., and Shao, J. Z. (2014). WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3, transcript levels not downstream of ABA. Plant J. 79, 810–823. doi: 10.1111/tpj.12597
Zhou, F., Menke, F. L., Yoshioka, K., Moder, W., Shirano, Y., and Klessig, D. F. (2004). High humidity suppresses ssi4-mediated cell death and disease resistance upstream of map kinase activation, H2O2 production and defense gene expression. Plant J. 39, 920–932. doi: 10.1111/j.1365-313X.2004.02180.x
Keywords: WRKY, salicylic acid, hypersensitive response, cell death, reactive oxygen species, Chinese wild Vitis
Citation: Wang X, Guo R, Tu M, Wang D, Guo C, Wan R, Li Z and Wang X (2017) Ectopic Expression of the Wild Grape WRKY Transcription Factor VqWRKY52 in Arabidopsis thaliana Enhances Resistance to the Biotrophic Pathogen Powdery Mildew But Not to the Necrotrophic Pathogen Botrytis cinerea. Front. Plant Sci. 8:97. doi: 10.3389/fpls.2017.00097
Received: 16 November 2016; Accepted: 17 January 2017;
Published: 31 January 2017.
Edited by:
Essaid Ait Barka, University of Reims Champagne-Ardenne, FranceReviewed by:
Roel Rabara, New Mexico Consortium, USAJérôme Crouzet, University of Reims Champagne-Ardenne, France
Copyright © 2017 Wang, Guo, Tu, Wang, Guo, Wan, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiping Wang, wangxiping@nwsuaf.edu.cn
 Xianhang Wang
Xianhang Wang Rongrong Guo1,2
Rongrong Guo1,2 Mingxing Tu
Mingxing Tu Ran Wan
Ran Wan Zhi Li
Zhi Li Xiping Wang
Xiping Wang