- 1Department of Drug Design and Pharmacology, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark
- 2Department of Medicinal Chemistry, Key Laboratory of Chemical Biology (Ministry of Education), School of Pharmaceutical Sciences, Shandong University, Jinan, Shandong, China
- 3Center for Structural and Functional Neuroscience, Center for Biomolecular Structure and Dynamics, Division of Biological Sciences, University of Montana, Missoula, MT, United States
N-Methyl-d-aspartate (NMDA) receptors play critical roles in central nervous system function and are involved in variety of brain disorders. We previously developed a series of (R)-3-(5-furanyl)carboxamido-2-aminopropanoic acid glycine site agonists with pronounced variation in activity among NMDA receptor GluN1/2A-D subtypes. Here, a series of (R)-2-amino-3-triazolpropanoic acid analogues with a novel chemical scaffold is designed and their pharmacological properties are evaluated at NMDA receptor subtypes. We found that the triazole can function as a bioisostere for amide to produce glycine site agonists with variation in activity among NMDA receptor subtypes. Compounds 13g and 13i are full and partial agonists, respectively, at GluN1/2C and GluN1/2D with 3- to 7-fold preference in agonist potency for GluN1/2C-D over GluN1/2A-B subtypes. The agonist binding mode of these triazole analogues and the mechanisms by which the triazole ring can serve as a bioisostere for amide were further explored using molecular dynamics simulations. Thus, the novel (R)-2-amino-3-triazolpropanoic acid derivatives reveal insights to agonist binding at the GluN1 subunit of NMDA receptors and provide new opportunities for the design of glycine site agonists.
1 Introduction
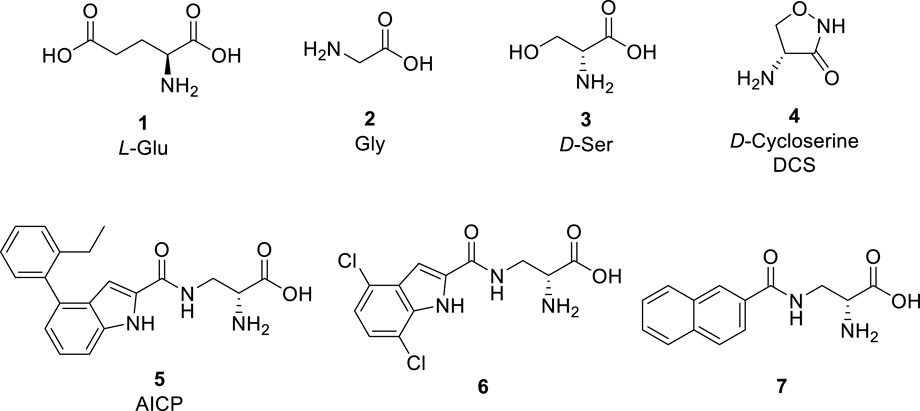
l-Glutamate (Glu, 1, Figure 1) is the principle excitatory neurotransmitter in the mammalian brain (Lemoine et al., 2012). When released within the synaptic cleft, Glu binds to both ionotropic and metabotropic Glu receptors, which results in activation and/or modulation of the postsynaptic neuron (Niciu et al., 2012). The ionotropic Glu receptors (iGluRs) are divided into three main functional classes, namely, N-methyl-d-aspartate (NMDA) receptors, (S)-amino-3-hydroxy-5-methyl-4-isoxazolepropionate (AMPA) receptors, and kainic acid (KA) receptors (Hansen et al., 2021). NMDA receptors play vital roles in learning, memory, and cognitive functions in the normal brain, but dysfunction of NMDA receptors has been associated with a variety of neurological and psychiatric disorders, including pain, stroke, epilepsy, schizophrenia, depression, and various neurodegenerative diseases, such as Parkinson’s and Alzheimer’s diseases (Paoletti et al., 2013; Sanz-Clemente et al., 2013; Yamamoto et al., 2015; Hansen et al., 2021). Therefore, NMDA receptors are desirable therapeutic targets for developing novel treatments in a number of central nervous system disorders.
NMDA receptors are tetrameric ion channels that comprise two GluN1 and two GluN2A−D or two GluN3A-B subunits (Karakas and Furukawa, 2014; Lee et al., 2014). The four GluN2 subunits have distinct developmental and regional expression in the central nervous system and endow NMDA receptor subtypes with different functional properties (Hansen et al., 2018; Hansen et al., 2021). Thus, the physiological roles of NMDA receptor subtypes and their involvement in brain disorders are primarily determined by the GluN2 subunits, and ligands that can discriminate between NMDA receptor subtypes based on the GluN2 subunit are attractive in the development of new therapeutic agents.
The activation of NMDA receptors composed of GluN1 and GluN2 subunits (GluN1/2A-D) requires simultaneous binding of two distinct agonists at the agonist binding domain (ABD), namely Glycine (Gly, 2, Figure 1) or d-Serine (d-Ser, 3) to GluN1 and Glu to GluN2 (Benveniste and Mayer, 1991; Clements and Westbrook, 1991; Hansen et al., 2018; Hansen et al., 2021). The endogenous agonists Gly and d-Ser show similar activity at GluN1/2A-D receptors (Figure 1) (Chen et al., 2008; Zhao et al., 2022). By contrast, d-Cycloserine (DCS, 4) is a full agonist at GluN1/2A (i.e. similar response compared to Gly, Rmax ≈ 100%), a partial agonist at GluN1/2B and GluN1/2D (i.e., lower response compared to Gly, Rmax < 100%), and a superagonist at GluN1/2C (i.e., higher response compared to Gly, Rmax > 100%) (Sheinin et al., 2001; Chen et al., 2008; Dravid et al., 2010; Jessen et al., 2017). Urwyler et al. developed amino-3-(4-(2-ethylphenyl)-1H-indole-2-carboxamido)propanoic acid (AICP, 5) as a highly potent NMDA receptor Gly site agonist (Urwyler et al., 2009), that was also identified as a full agonist at GluN1/2A (Rmax ≈ 100%), a partial agonist at GluN1/2B and GluN1/2D, and a highly efficacious superagonist at GluN1/2C subunit (Jessen et al., 2017; Zhao et al., 2020). Furthermore, we reported a small series of amido-, ether and thioether (R)-alanine analogues showing variation among GluN2A-D NMDA receptor subtypes (Maolanon et al., 2017).
Based on these studies, we developed a diverse set of Gly site agonists with structural variation in the aromatic part of the amido analogues 5–7 (Figure 1). Thus, (R)-3-(5-furanyl)carboxamido-2-aminopropanoic acid derivatives were designed as Gly site agonists that display pronounced variation in potency, efficacy, and GluN2 subunit-specific activity (Figure 2). Among them, 8p and 8r are functionally selective GluN1/2C agonists with nanomolar potencies. Besides, compound 8h and 8q showed high agonist potencies at GluN1/2A, GluN1/2C and GluN1/2D receptors. Compound 8d was identified as a partial agonist at GluN1/2A and GluN1/2C receptors (Zhao et al., 2022).
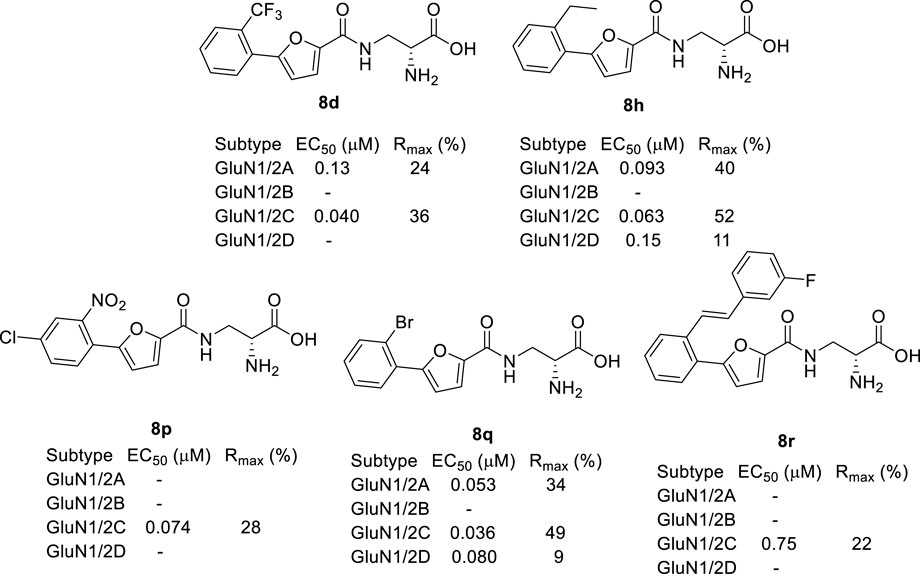
FIGURE 2. Recently developed NMDA receptor Gly site agonists. EC50 is the concentration that produces 50% response at the GluN1 subunit in the continuous presence of a saturating concentration of glutamate. Rmax is the maximal response compared to the maximal agonist response to Gly. Data are previously published and were measured using two-electrode voltage-clamp recordings of responses from recombinant NMDA receptor subtypes (Zhao et al., 2022).
Triazoles are well-known chemical scaffolds that have been employed widespread in many biologically active compounds (Sahu et al., 2013). Moreover, the structural features of triazoles enable these chemical scaffolds to mimic different functional groups, justifying the wide use of triazoles as bioisostere of amide bonds in the development of new active molecules (Valverde et al., 2013; Valverde et al., 2015). In this study, we designed a series of 1,4-disubstituted-1,2,3-triazole analogues as NMDA receptor Gly site agonists (Figure 3A). We maintained the essential α-amino acid part of AICP and furan analogues, and replaced the amide bond with the bioisostere 1,2,3-triazole. Different groups were introduced on the C-4 position of 1,2,3-triazole ring, including alkyl chains, (hetero)aromatic rings with or without substitutions, and thiophene rings. Despite some differences in the overall dipolar moment and distance between the substituents, their structural features aligned with the amide-binding moiety through overlaying of the lowest energy conformations for β-amide-d-alanine and β-triazole-d-alanine (Figure 3B). Moreover, the C-4 atom is electronegative, allowing the C-H bond to act as a hydrogen bond donor, and the lone pair of N-3 electrons can act as a hydrogen bond acceptor. We also replaced the amide bond with 1,2,4-triazole to fulfill the scaffold diversities.
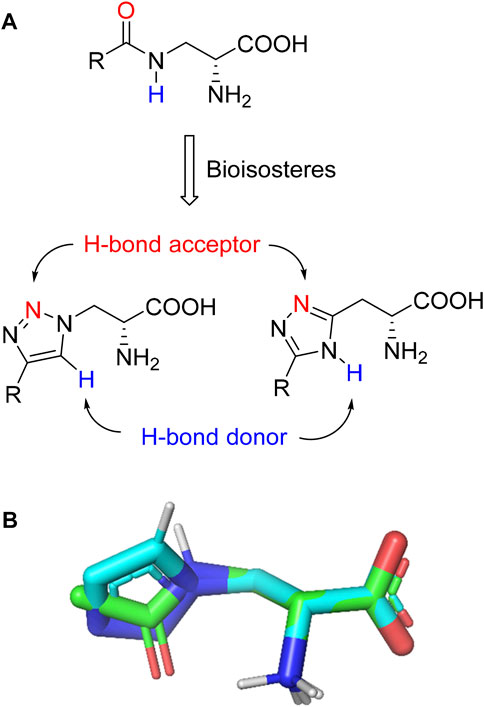
FIGURE 3. (A) Design strategy for novel triazole analogues as NMDA receptor Gly site agonists. (B) Overlay of lowest energy conformations for β-amide-d-alanine (green) and β-triazole-d-alanine (cyan) with amide carbonyl or 1,2,3-triazole. The C-4 atom is electronegative, allowing the C-H bond to act as a hydrogen bond donor, and the lone pair of N-3 electrons can act as a hydrogen bond acceptor.
Here, we demonstrate that these new Gly site agonists display variation in activity among GluN1/2A-D NMDA receptor subtypes. These findings expand the pharmacology of GluN1 Gly site agonists and provide a novel scaffold for the development of NMDA receptor ligands.
2 Chemistry
The synthesis of newly designed compounds began with an esterification between N-Boc-d-serine methyl ester 9 and methanesulfonyl chloride to give sulfonate 10, which was then substituted by NaN3 to yield the key azide intermediate 11 (Scheme 1). The azide 11 was then treated with various alkynes to afford 1,2,3-triazoles 12a-j. The Boc and methyl group of 12a-j were removed with 6M HCl and the final pure products 13a-j were obtained by purification using prep-HPLC.
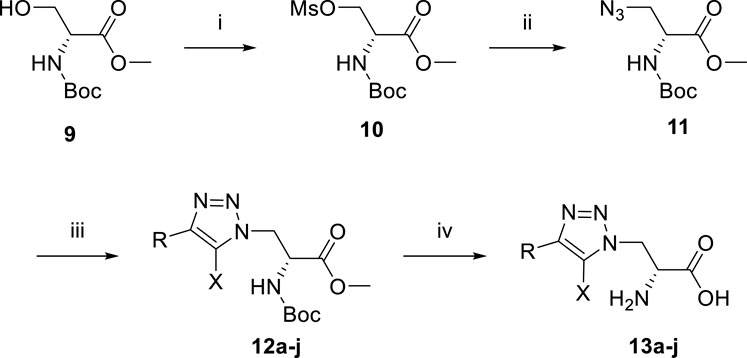
SCHEME 1. Reagents and conditions: (i) MsCl, NEt3, DCM, 0°C. Yield: 61%. (ii) NaN3, DMF, 70°C. Yield: 38%. (iii) various alkyne, sodium ascorbate, copper sulfate, i-PrOH, H2O. (iv) 6M HCl, reflux.
The synthesis of compound 16 started from the protection of N-Boc-d-asparagine 14 (Scheme 2). The protected intermediate 15 was then treated with N,N-dimethylformamide dimethyl acetal at 75°C for 2 h. After that, the reaction mixture was redissolved in AcOH and treated with hydrazine hydrate to give the 1,2,4-triazole compound. The protecting groups were cleaved by BBr3, and the final product was afforded after purification by prep-HPLC.
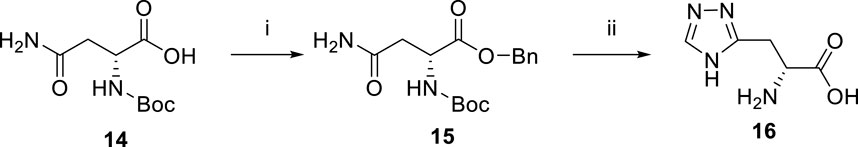
SCHEME 2. Reagents and conditions: (i) Cs2CO3, MeOH, rt; then benzyl bromide, DMF, rt. Yield: 60%. (ii) N,N-dimethylformamide dimethyl acetal, 75°C; then AcOH, hydrazine hydrate; then BBr3, DCM, −10°C. Yield: 90%.
For general compounds with a CH-CH2 motif, the 1H-NMR spectra should give t- and d-peaks for the CH and CH2 motif, respectively. Due to the chiral center and the neighboring groups, the α-CH and the β-CH2 of the final compounds form two groups of complicated peaks; generally, a broad single peak for the α-proton and multiplets for the β-protons.
3 Results and discussion
3.1 Pharmacological evaluation
The pharmacological properties of analogues 13a-j and 16 were determined with two-electrode voltage-clamp recordings of responses from recombinant NMDA receptor subtypes (Table 1; Figure 4). Compound 13f was identified as agonist with lower maximal response relative to glycine (Rmax) at GluN1/2A (47%) and GluN1/2B (23%), but Rmax similar to glycine at GluN1/2C (94%) and GluN1/2D (96%). Similarly, compound 13g was a partial agonist at GluN1/2A (66%) and GluN1/2B (56%) with a higher agonist efficacy at GluN1/2C (97%) and GluN1/2D (88%). The potency of 13g was improved compared to 13f, and 13g displayed 4- to 7-fold preference in agonist potency for GluN1/2C-D over GluN1/2A-B subtypes. Compound 13i was a partial agonist at all subtypes with the highest agonist potency in the series and 3- to 6-fold preference in agonist potency for GluN1/2C-D over GluN1/2A-B subtypes. Interestingly, compound 16 was a partial agonist GluN1/2A-C (57%–78%) and a superagonist at GluN1/2D (168%), albeit with low potency compared to Gly (Table 1; Figure 4).
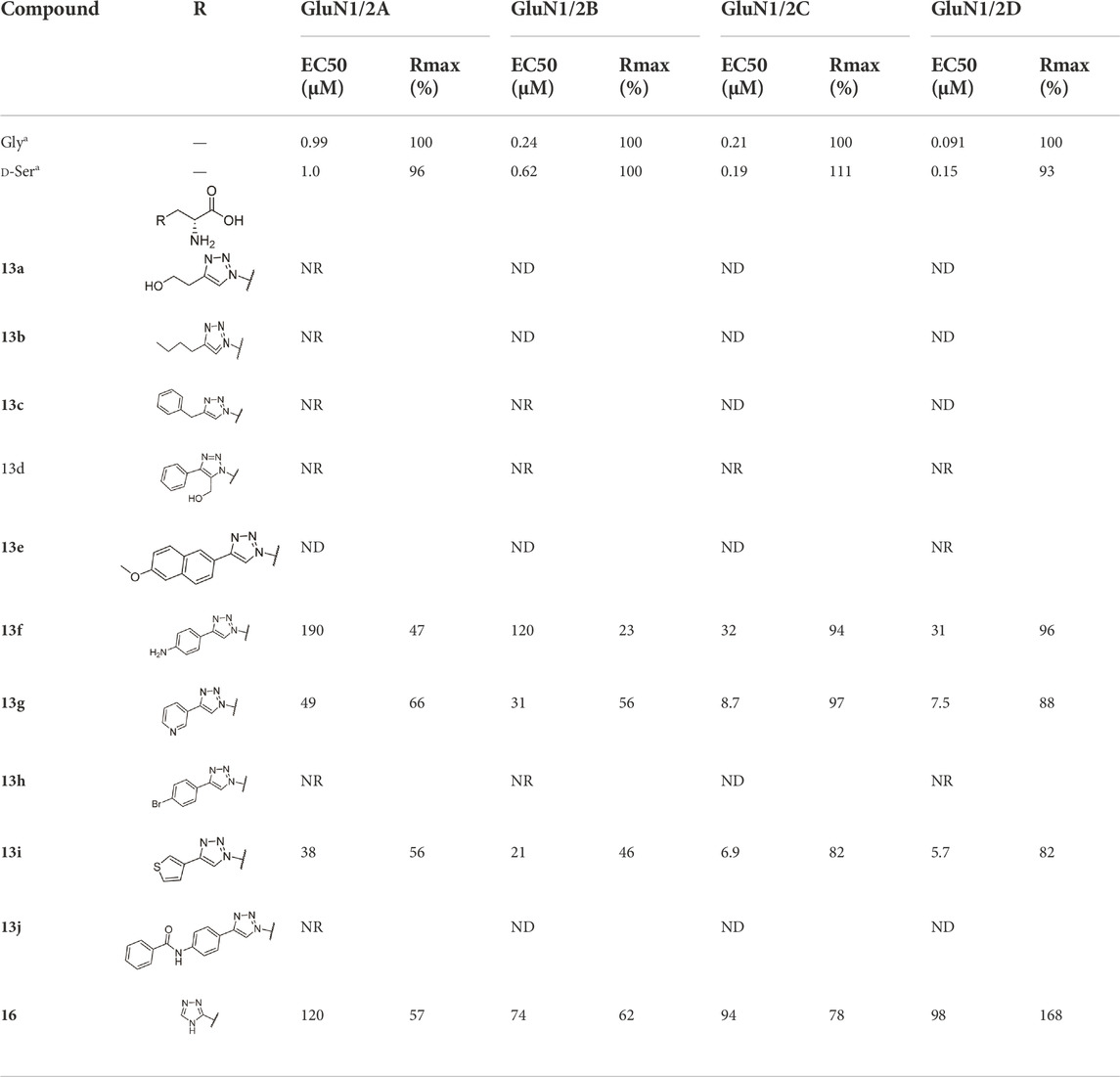
TABLE 1. EC50 values for activation of rat NMDA receptor subtypes measured using two-electrode voltage-clamp electrophysiology in the presence of 300 µM Glu. Relative efficacy is the fitted maximal response to the agonist compared to maximal response to 100 µM Gly in the same recording. ND means that the EC50 could not be determined, but some agonist activity was observed. NR indicates less than 5% response at 100 µM of the agonist. Data are from 3–13 oocytes.a Data for Gly and d-Ser are previously published (Zhao et al., 2022). See Supplementary Table S1 in the Supporting Information for sample sizes, Hill slopes, and standard error of mean.
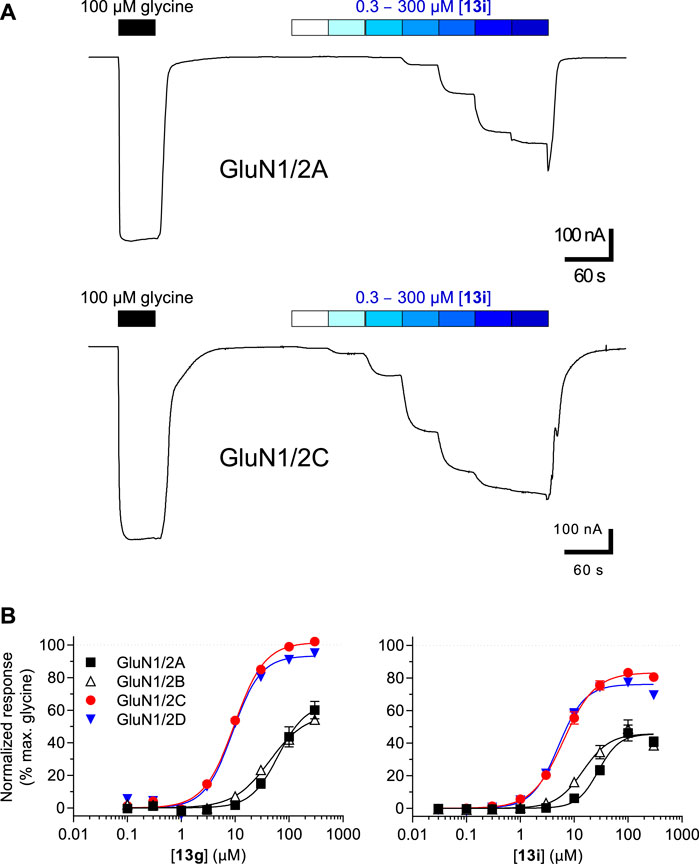
FIGURE 4. (A) Representative recordings of 13i concentration-response data in the presence of 300 µM Glu at GluN1/2A and GluN1/2C subtypes. Responses were recorded using two-electrode voltage-clamp electrophysiology. (B) Concentration-response relationships for 13g and 13i at GluN1/2A-D subtypes. The fitted maximal agonist response is normalized to the maximal Gly response. Data from 8–11 oocytes are shown as mean of ± SEM. See Table 1 for EC50 values.
The structure-activity relationship study shows that pyridine (13g) and thiophene rings (13i) on the C-4 position of the 1,2,3-triazole ring improve potencies. Alkyl chains (13a and 13b), bromobenzene (13h), benzyl ring (13c) and additional substitutions (13j) on the aniline result in inactive compounds or compounds with very low potency.
3.2 Molecular modeling
The pharmacological study identified compounds 13g and 13i among the new triazole analogues as the most potent Gly site agonists at NMDA receptor subtypes. We therefore performed computational modeling to estimate binding modes for these agonists.
Briefly, a homology model of the GluN1/2C agonist binding domain (ABD) heterodimer was constructed using the GluN1/2A ABD heterodimer crystal structure with bound Gly and Glu (PDB ID: 5I57; Yi et al., 2016) as template. Induced-fit docking of 13g and 13i into the Gly binding site in the resulting GluN1/2C model was then performed to obtain docking poses. Subsequently, molecular dynamics simulations (500 ns each) were performed using the best scoring docking pose for each ligand. Finally, the most abundant binding conformations were clustered from the simulation trajectories.
In the most abundant conformation for each ligand, the key interactions between receptor and the α-amino acid part were conserved for analogues 13g and 13i (Figures 5A,B), and these interactions were identical to interactions observed in GluN1 structures with bound Gly or d-Ser (Furukawa and Gouaux, 2003). GluN1 residues T518, S688 and D732 formed a narrow tunnel in the agonist binding pocket and interacted with the triazole spacers of these analogues via hydrogen bonds, similar to the amide spacer of 8h and 8p docked into the GluN1 ABD (Zhao et al., 2022). The pyridine or thiophene ring of analogues 13g and 13i interacted with residues Y692 and F754 (GluN1) and bound close to the GluN1 and GluN2 subunits interface (Figures 5A,B). These additional hydrophobic interactions with surrounding residues might improve binding affinity in the series.
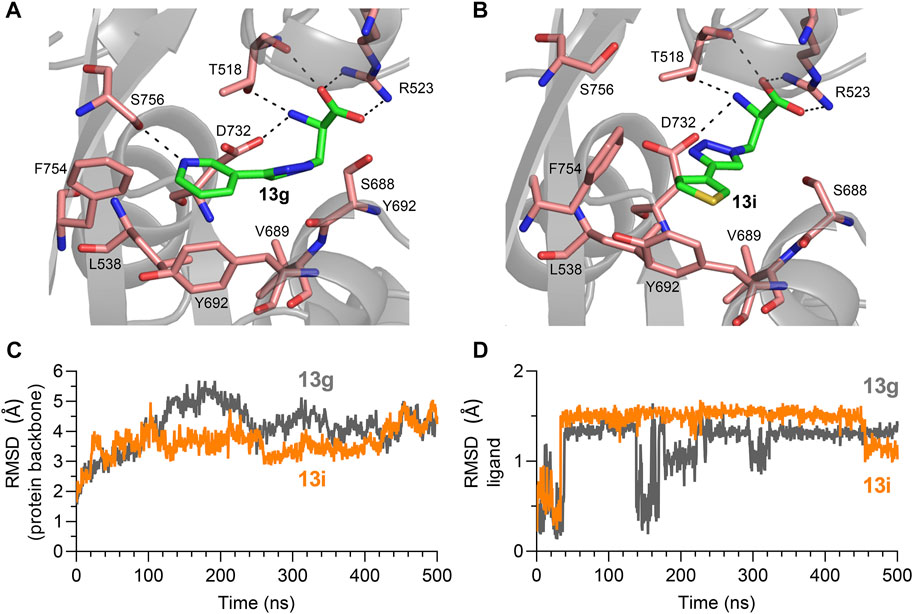
FIGURE 5. (A,B) Most abundant binding mode of 13g and 13i in the Gly binding site of GluN1 during molecular dynamics simulation from in initial docking model. (C,D) RMSD of protein backbone and ligand compared to the initial docking model for molecular dynamics simulations with 13g (grey) and 13i (orange).
Movement of GluN1 L538, V689, Y692, F753, and F754 was required to accommodate binding of the triazole analogues, while the GluN2C ABD remained stable compared to the Gly/Glu-bound GluN1/2C-model structure. Root mean square deviations (RMSD) from the initial docking model remained relatively stable for the protein backbone over the 500 ns simulations (Figure 5C). Similarly, RMSD from the initial docking poses became relatively stable after 30 ns simulation time for both 13g and 13i (Figure 5D). The binding conformation of 13i remained stable for 400 ns, while the binding pose of 13g was fluctuating according to the RMSD plot, indicating that binding of 13i is more stable than 13g at the NMDA receptor Gly binding site. Furthermore, molecular mechanics with generalized born and surface area (MM/GBSA) were calculated to estimate binding free energy for 13g and 13i to the Gly binding pocket. The average binding energy is -39.49 kcal/mol and -47.91 kcal/mol for 13g and 13i, respectively. These results indicate that binding of 13i is stronger compared to 13g, consistent with potencies measured using two-electrode voltage-clamp electrophysiology (Table 1).
4 Conclusion
In the present work, a series of triazole analogues (13a-j and 16) were designed, synthesized, and characterized at GluN1/2A-D NMDA receptor subtypes. Compounds 13g and 13i are full and partial agonists, respectively, at GluN1/2C and GluN1/2D with 3- to 7-fold preference and low micromolar agonist potencies for GluN1/2C-D over GluN1/2A-B subtypes. This profile is different from d-cycloserine (DCS, 4) in terms of agonist efficacies, but similar in terms of agonist potencies (Sheinin et al., 2001; Chen et al., 2008; Dravid et al., 2010; Jessen et al., 2017). DCS has intriguing neuroactive properties, and administration of DCS can enhance extinction of fear in both rodents and humans, which may be beneficial in the treatment of some psychiatric disorders (Davis et al., 2006; Hofmann et al., 2015; Singewald et al., 2015). However, it remains unresolved whether partial agonism at GluN2B- and GluN2D-containing receptors or superagonism at GluN2C-containing NMDA receptors mediates the behavioral effects of DCS (Sotres-Bayon et al., 2007; Sotres-Bayon et al., 2009; Hillman et al., 2011; Dalton et al., 2012; Gupta et al., 2013; Ogden et al., 2014). To this end, it would desirable to develop novel glycine site NMDA receptor agonists with nanomolar potencies, which would enable effective competition with endogenous glycine or d-serine, but with variation in GluN2 subunit-specific profiles (i.e., agonist efficacies). These novel agonists would enable new studies to determine the optimal GluN2 subunit-specific profiles for the treatment of a range of neurological and psychiatric disorders (Liu et al., 2019; Liu et al., 2021; Shelkar et al., 2021).
The pharmacological results and in silico study show that the triazole ring can serve as the bioisostere for the amide in Gly site agonists. Compared to previously reported amido Gly site agonists, such as AICP analogues and furan analogues (Maolanon et al., 2017; Zhao et al., 2022), the new triazole analogues were generally less potent, but more potent than ether and thioether derivatives. However, there are striking similarities between EC50 values for 13g (pyridyl; 7.5–49 µM) and 13i (thienyl; 5.7–38 µM) and previously published Ki values for amido Gly site agonists compounds 8 (pyridyl; 40 µM) and 11 (thienyl; 7.9 µM) in Urwyler et al. (Urwyler et al., 2009). Although the methods to evaluate their biological activity are very different, the fact that these compounds (with triazoles or amides) all display activity at micromolar concentrations illustrate that 1,4 triazoles are suitable replacements for the central amide of 2-aminopropionic acid-based NMDA receptor agonists. Thus, triazole derivatives are good starting points for designing new analogues in the development of NMDA receptor glycine site ligands with increased potencies and GluN2 subunit-specific activity.
5 Materials and methods
5.1 Chemistry
An inert atmosphere of N2 using standard Schlenk and syringe-septum technique was applied for reactions requiring inert conditions. All solvents used in reactions were pre-dried with a glass contour solvent system (SG Water, United States LCC). Silica gel 60 (Merck, 0.015–0.040 mm) was used for normal phase column chromatography. Thin layer chromatography (TLC) for monitoring of reactions and chromatography fractions used silica gel 60 F254 aluminum plates (Merck). Reagents and starting materials were purchased from commercial vendors. The 1H and 13C NMR spectra were generated with Bruker 400 and Bruker 600 spectrometers at 400 and 600 MHz for proton and at 100 and 150 MHz for carbon using signals of residual nondeuterated solvent as internal standards. LC/MS spectra were obtained on an Agilent 1200 system using a Zorbax Eclipse XBD-C18 column (4.6 × 50 mm) and a UV detector equipped with an API ion source (HP 1100 MSD). Analytical HPLC spectra were determined on an Ultimate 3000 system with a Gemini-NX C18 column (4.6 × 250 mm). Prep-HPLC purification was performed on an Ultimate 3000 system with a Gemini-NX C18 column (21 × 250 mm). All compounds were obtained as highly viscous oils, unless otherwise stated. For high-resolution mass spectroscopy (HRMS), compounds were analyzed by ultra-high-performance liquid chromatography (Agilent 1260 UPLC, Agilent Technologies, Santa Clara, CA, United States ) coupled to high-resolution mass spectrometry (6520 Q-TOF, Agilent, United States). Chromatographic separation was carried out using a C8 column (Avantor ACE C8, 3 μm, 3.0 × 50 mm, Radnor, PA, United States) and a mobile phase constituted of aqueous solution A (water, 0.1% formic acid) and organic solution B (acetonitrile, 0.1% formic acid). The column was held at ambient temperature and a gradient method (1 ml/min) was applied over 6 min, as follows: 0–4 min, 3%–95% B; 4–6 min, 95% B. The autosampler was maintained at ambient temperature and 10 µL of each sample was injected for HRMS analysis in positive mode. Samples were ionized by electrospray with the ionization parameters: capillary voltage, 4.0 kV (positive mode); gas temperature, 325°C; gas flow, 8.0 L/min, fragmentor, 180 V. Data was collected in high-resolution mode (4 GHz) over a mass range of 100–1,700 m/z. Mass correction was employed using reference compounds m/z 322.048121 and 1221.990637.
Procedure A: 1,3-Dipolar cycloaddition
3-Azido-N-Boc-d-alanine methyl ester 11 (244 mg, 1 mmol) and corresponding alkynes (1 mmol) were dissolved in a mixture of isopropanol and H2O (3 ml, v/v, 2:1). Sodium ascorbate (198 mg, 0.1 mmol) was added into the reaction mixture and a solution of CuSO4 (8 mg, 0.05 mmol) in H2O (0.5 ml) was added later with vigorous stirring. The reaction mixture was then stirred overnight. After concentration, the crude was diluted with H2O (15 ml) and extracted with EtOAc (3 × 10 ml). The organic layer was washed by brine twice and the combined organic layers were dried over MgSO4 and concentrated in vacuum. The pure product was obtained by dry column vacuum chromatography.
Procedure B: Deprotection of the amino acid
The corresponding protected compounds (1 mmol) were dissolved in 6M HCl (5 ml) and stirred overnight under reflux. The aqueous layer was washed with EtOAc (10 ml) and concentrated under vacuum. The crude product was further purified with prep-HPLC to afford the final products as highly viscous oils or amorphous powder without defined melting point.
N-Boc-O-(methylsulfonyl)-d-serine methyl ester (10)
To a solution of N-Boc-d-serine methyl ester 9 (2.96 g, 13.5 mmol) in DCM (35 ml) at 0°C, NEt3 (3.78 ml, 27 mmol) was added. Then, methanesulfonyl chloride (1.72 ml, 17.6 mmol) was added dropwise. The reaction mixture was stirred for additional 30 min. Saturated NaHCO3 (20 ml) was added to quench the reaction and the organic layer was washed with brine 3 times. The combined aqueous layers were extracted with DCM (2 × 25 ml). The combined organic layers were dried over MgSO4 and concentrated in vacuum. The crude product was purified by dry column vacuum chromatography to give the title product as yellow oil (2.62 g). 1H NMR (400 MHz, Chloroform-d) δ 5.37 (s, 1H), 4.62–4.59 (m, 1H), 4.58–4.48 (m, 2H), 3.82 (s, 3H), 3.02 (s, 3H), 1.46 (s, 9H). 13C NMR (101 MHz, Chloroform-d) δ 169.2, 155.2, 80.7, 68.9, 60.4, 53.1, 37.5, 28.3. TLC: (Hept-EtOAc 6:4) Rf: 0.31. Yield: 61%.
3-Azido-N-Boc-d-alanine methyl ester (11)
To a solution of mesylate serine 10 (100 mg, 0.343 mmol) in anhydrous DMF (5 ml), NaN3 (56 mg, 0.858 mmol) was added and heated to 70°C for 10 min. The mixture was transferred to a beaker with cold water (10 ml) and then extracted by EtOAc (3 × 10 ml). The combined organic layers were washed with brine (25 ml), dried over MgSO4 and concentrated in vacuum. The crude product was purified by dry column vacuum chromatography to give the title product (32 mg). 1H NMR (600 MHz, Chloroform-d) δ 5.36 (d, J = 7.5 Hz, 1H), 4.48 (dt, J = 8.1, 3.6 Hz, 1H), 3.80 (s, 3H), 3.72 (d, J = 3.8 Hz, 2H), 1.46 (s, 9H). 13C NMR (151 MHz, Chloroform-d) δ 170.4, 155.2, 80.7, 53.7, 53.0, 32.0, 28.4. TLC: (Hept-EtOAc 6:4) Rf: 0.66. Yield: 38%.
(R)-2-Amino-3-(4-(2-hydroxyethyl)-1H-1,2,3-triazol-1-yl)propanoic acid (13a)
Procedure A and B. 1H NMR (400 MHz, D2O) δ 7.86 (s, 1H), 4.98 (dd, J = 4.9, 1.9 Hz, 2H), 4.52 (dd, J = 5.4, 4.4 Hz, 1H), 3.81 (t, J = 6.4 Hz, 2H), 2.90 (t, J = 6.3 Hz, 2H). 13C NMR (101 M Hz, D2O) δ 169.2, 125.1, 125.0, 60.4, 53.2, 49.2, 27.5. LC-MS m/z [M + H]+ calculated for C7H13N4O3 201.10, found 201.3. HRMS (ESI) m/z [M + H]+ calculated for C7H13N4O3 201.0982, found 201.0987. Yield: 27%, 2 steps.
(R)-2-Amino-3-(4-butyl-1H-1,2,3-triazol-1-yl)propanoic acid (13b)
Procedure A and B. 1H NMR (600 MHz, D2O) δ 8.16 (s, 1H), 5.13 (qd, J = 15.3, 4.8 Hz, 2H), 4.72 (t, J = 5.2 Hz, 1H), 2.79 (t, J = 7.6 Hz, 2H), 1.63 (p, J = 7.5 Hz, 2H), 1.30 (h, J = 7.4 Hz, 2H), 0.86 (t, J = 7.4 Hz, 3H). 13C NMR (101 MHz, D2O) δ 168.6, 125.6, 107.2, 52.6, 49.8, 30.1, 23.3, 21.2, 12.8. LC-MS m/z [M + H]+ calculated for C9H17N4O2 213.13, found 213.5. HRMS (ESI) m/z [M + H]+ calculated for C9H17N4O2 213.1346, found 213.1343. Yield: 61%, 2 steps.
(R)-2-Amino-3-(4-benzyl-1H-1,2,3-triazol-1-yl)propanoic acid (13c)
Procedure A and B. 1H NMR (600 MHz, Methanol-d4) δ 8.07 (s, 1H), 7.30 (s, 4H), 7.23 (s, 1H), 5.06 (s, 2H), 4.70 (s, 1H), 4.15 (s, 2H). 13C NMR (151 MHz, Methanol-d4) δ 168.2, 147.8, 138.6, 129.8, 127.9, 127.4, 53.6, 51.5, 49.4, 49.3, 49.1, 49.0, 48.9, 48.7, 48.6, 31.9. LC-MS m/z [M + H]+ calculated for C12H15N4O2 247.12, found 247.5. HRMS (ESI) m/z [M + H]+ calculated for C12H15N4O2 247.1190, found 247.1187. Yield: 37%, 2 steps.
(R)-2-Amino-3-(5-(hydroxymethyl)-4-phenyl-1H-1,2,3-triazol-1-yl)propanoic acid (13d)
Procedure A and B. 1H NMR (600 MHz, D2O) δ 7.56–7.52 (m, 2H), 7.49–7.41 (m, 3H), 5.05 (dd, J = 5.4, 3.2 Hz, 2H), 4.78 (s, 2H), 4.76–4.74 (m, 1H). 13C NMR (151 MHz, D2O) δ 176.5, 168.6, 145.6, 133.8, 129.1, 128.6, 127.8, 52.3, 51.0, 47.3. LC-MS m/z [M + H]+ calculated for C12H15N4O3 263.11, found 263.5. HRMS (ESI) m/z C12H15N4O3 [M + H]+ calculated 263.1139, found 263.1137. Yield: 50%, 2 steps.
(R)-2-Amino-3-(4-(6-hydroxynaphthalen-2-yl)-1H-1,2,3-triazol-1-yl)propanoic acid (13e)
Procedure A and B. 1H NMR (400 MHz, DMSO-d6) δ 8.56 (s, 1H), 8.25 (d, J = 19.0 Hz, 1H), 7.90–7.80 (m, 2H), 7.76 (dd, J = 31.2, 8.5 Hz, 1H), 7.27–7.11 (m, 2H), 5.13 (d, J = 4.0 Hz, 2H), 4.77 (s, 1H), 3.31 (s, 3H). 13C NMR (101 MHz, DMSO-d6) δ 167.2, 156.0, 147.7, 135.2, 129.5, 128.3, 127.4, 124.4, 124.2, 123.7, 122. 7, 119.2, 108.6, 54.5, 52.4, 49.3. LC-MS m/z [M + H]+ calculated for C16H17N4O3 313.13, found 313.5. HRMS (ESI) m/z [M + H]+ calculated for C16H17N4O3 3313.1295, found 313.1294. Yield: 79%, 2 steps.
(R)-2-Amino-3-(4-(4-aminophenyl)-1H-1,2,3-triazol-1-yl)propanoic acid (13f)
Procedure A and B. 1H NMR (600 MHz, Methanol-d4) δ 8.55 (s, 1H), 8.04 (d, J = 8.5 Hz, 2H), 7.52 (d, J = 8.5 Hz, 2H), 5.10 (d, J = 5.0 Hz, 2H), 4.74 (t, J = 5.0 Hz, 1H). 13C NMR (151 MHz, Methanol-d4) δ 168.6, 147.6, 132.7, 131.8, 128.4, 124.9, 124.4, 53.7, 50.2. LC-MS m/z [M + H]+ calculated for C11H14N5O2 248.11, found 248.4. HRMS (ESI) m/z [M + H]+ calculated for C11H14N5O2 248.1142, found 248.1142. Yield: 72%, 2 steps.
(R)-2-Amino-3-(4-(pyridin-3-yl)-1H-1,2,3-triazol-1-yl)propanoic acid (13g)
Procedure A and B. 1H NMR (600 MHz, D2O) δ 9.27 (d, J = 2.0 Hz, 1H), 9.01 (dt, J = 8.3, 1.7 Hz, 1H), 8.82 (dt, J = 5.8, 1.1 Hz, 1H), 8.69 (d, J = 2.0 Hz, 1H), 8.21 (dd, J = 8.3, 5.8 Hz, 1H), 5.23–5.14 (m, 2H), 4.65–4.60 (m, 1H). 13C NMR (151 MHz, D2O) δ 169.4, 143.3, 141.7, 140.4, 138. 2, 130.1, 127.8, 125.4, 49.6. LC-MS m/z [M + H]+ calculated for C10H12N5O2 234.10, found 234.5. HRMS (ESI) m/z [M + H]+ calculated for C10H12N5O2 234.0986, found 234.0986. Yield: 65%, 2 steps.
(R)-2-Amino-3-(4-(4-bromophenyl)-1H-1,2,3-triazol-1-yl)propanoic acid (13h)
Procedure A and B. 1H NMR (600 MHz, Methanol-d4) δ 8.39 (s, 1H), 7.77 (d, J = 8.5 Hz, 2H), 7.62 (d, J = 8.5 Hz, 2H), 5.09–5.03 (m, 2H), 4.67 (t, J = 4.7 Hz, 1H). 13C NMR (151 MHz, Methanol-d4) δ 168.7, 148.3, 133.2, 130.6, 128.5, 123.7, 123.4, 53.8, 50.2, 49.4, 49.3, 49.1, 49.0, 48.9, 48.7, 48.6. LC-MS m/z [M + H]+ calculated for C11H12BrN4O2 311.01, found 311.4. HRMS (ESI) m/z [M + H]+ calculated for C11H12BrN4O2 311.0138, found 311.0133. Yield: 94%, 2 steps.
(R)-2-Amino-3-(4-(thiophen-3-yl)-1H-1,2,3-triazol-1-yl)propanoic acid (13i)
Procedure A and B. 1H NMR (600 MHz, Methanol-d4) δ 8.29 (s, 1H), 7.81–7.77 (m, 1H), 7.56–7.48 (m, 2H), 5.08–5.03 (m, 2H), 4.71 (t, J = 5.0 Hz, 1H). 13C NMR (151 MHz, Methanol-d4) δ 168.6, 145.6, 132.5, 127.8, 126.7, 123.3, 122.6, 53.8, 50.1, 49.4, 49.3, 49.1, 49.0, 48.9, 48.7, 48.6. LC-MS m/z [M + H]+ calculated for C9H11N4O2S 239.06, found 239.5. HRMS (ESI) m/z [M + H]+ calculated for C9H11N4O2S 239.0597, found 239.0592. Yield: 80%, 2 steps.
(R)-2-Amino-3-(4-(4-benzamidophenyl)-1H-1,2,3-triazol-1-yl)propanoic acid (13j)
Procedure A and B. 1H NMR (600 MHz, Methanol-d4) δ 8.37 (s, 1H), 7.94 (d, J = 7.3 Hz, 2H), 7.83 (q, J = 8.3, 7.8 Hz, 4H), 7.58 (t, J = 7.2 Hz, 1H), 7.51 (t, J = 7.3 Hz, 2H), 5.07 (s, 2H), 4.71 (s, 1H). 13C NMR (151 MHz, Methanol-d4) δ 168.9, 168.6, 148.9, 140.3, 136.1, 132.9, 129.6, 128.6, 127.3, 127.3, 123.4, 122.4, 53.9, 50.4, 49.4, 49.3, 49.1, 49.0, 48.9, 48.7, 48.6. LC-MS m/z [M + NH4]+ calculated C18H21N6O3 369.17, found 369.6. HRMS (ESI) m/z [M + H]+ calculated for C18H18N5O3 352.1404, found 352.1407. Yield: 72%, 2 steps.
Benzyl-N-Boc-d-asparaginate (15)
N-Boc-d-asparagine (1 g, 1 eq) was dissolved in MeOH (20 ml) and aqueous Cs2CO3 solution (10 ml, 20% w/v) was added. After stirring for 1 h, the solvent was removed and re-dissolved in DMF (20 ml). Benzyl bromide (770 μL, 1.5 eq) was added into the mixture and stirred overnight at rt. The solution was concentrated, and diluted in water (50 ml) to form a precipitate. The precipitate was then collected, dissolved in EtOAc (20 ml) and washed with water (10 ml) to afford the final product 16 as a white powder (830 mg, 60%). 1H NMR (600 MHz, Methanol-d4) δ 7.37–7.29 (m, 5H), 5.16 (s, 2H), 4.51 (d, J = 6.2 Hz, 1H), 2.73 (d, J = 5.9 Hz, 2H), 1.42 (s, 9H). 13C NMR (151 MHz, Methanol-d4) δ 174.9, 173.1, 157.8, 137.2, 129.5, 129.2, 129.1, 80.8, 68.1, 51.9, 49.6, 37.9, 28.7. LC-MS m/z [M + H-Boc]+ calculated for C11H15N2O3 223.11, found 223.5. Yield: 60%.
(R)-2-Amino-3-(4H-1,2,4-triazol-3-yl)propanoic acid (16)
A solution of benzyl-N-Boc-d-asparaginate 16 (100 mg, 1 eq) and DMF-DMA (4 ml) was stirred at 75°C for 2 h. The mixture was concentrated and redissolved in AcOH (5 ml). To the resulting solution, hydrazine hydrate (35.3 μL, 2 eq) was added and stirred overnight at rt. The solution was then concentrated to dryness. The crude was dissolved in anhydrous DCM (10 ml), and cooled to −10°C under N2 atmosphere. BBr3 (5 ml) was then added dropwise and stirred for another 2 h. The resulting mixture was dissolved in methanol (10 ml) and further purified by prep-HPLC. 1H NMR (600 MHz, Methanol-d4) δ 8.85 (s, 1H), 4.49 (dd, J = 7.4, 5.3 Hz, 1H), 3.55–3.42 (m, 2H). 13C NMR (151 MHz, Methanol-d4) δ 170.2, 156.6, 145.6, 52.2, 49.6, 28.4. LC-MS m/z [M + H]+ calculated for C5H9N4O2 157.07, found 157.4. HRMS (ESI) m/z [M + H]+ calculated for C5H9N4O2 157.0720, found 157.0719. Yield: 90%.
5.2 Pharmacological evaluation
For expression in Xenopus laevis oocytes purchased from Rob Weymouth (Xenopus 1, Dexter, MI), cDNAs encoding rat NMDA receptor subunits, GluN1-1a (Genbank accession number U11418 and U08261), GluN2A (D13211), GluN2B (U11419), GluN2C (M91563), and GluN2D (L31611), were used to produce cRNAs using the mMessage mMachine kit (Ambion, Life Technologies, Paisley, United Kingdom) according to the manufacturer’s protocol. The open reading frame of the GluN2B subunit was altered without changing the amino acid sequence to remove a termination site for the T7 RNA polymerase used to synthesize cRNA (Hansen et al., 2014).
Oocytes were injected with cRNAs for GluN1 and GluN2 subunits at a 1:2 ratio, and incubated as previously described (Hansen et al., 2013). The injected oocytes were used for two-electrode voltage-clamp recordings at room temperature (21–23°C) after 2–4 days of expression. as previously described (Hansen et al., 2013). The holding potential was −40 mV in all recordings. The recording solution was comprised of (in mM) 90 NaCl, 1 KCl, 10 HEPES, 0.5 BaCl2 and 0.01 EDTA (pH 7.4). For experiments with GluN1/2A or GluN1/2B receptors, the oocytes were injected at least 10 min before recordings with 50 mM BAPTA (30–50 nL) to minimize use-dependent increases in response amplitudes (Williams, 1993). Compounds were dissolved as 20–100 mM stock solutions in DMSO, and DMSO concentrations were constant in all recording solutions and never exceeded 1%.
GraphPad Prism (GraphPad Software, La Jolla, CA) was used to analyze agonist concentration-response data. Briefly, the data for each oocyte were fitted to the following Hill equation, I = Rmax/(1 + 10^((logEC50-log [A])*nH)). Rmax is the maximum agonist current response normalized to the maximal response to 100 µM Gly, nH is the Hillslope, [A] is the concentration of agonist, and EC50 is the concentration of agonist that activates a half-maximum response.
5.3 Molecular modeling
Computational modeling experiments were performed in the Schrödinger suite, Release 2019–1, Maestro version 11.9.011.
The full-length model of GluN2C (AlphaFold, AF-Q14957-F1) was downloaded from Uniport (ID: Q14957), which was then aligned with the GluN1/2A ABD crystal structure (PDB ID: 5I57) with PyMol. The GluN2C ABD domain (amino acids 401–535, 660–798) was extracted, and combined with GluN1 and ligands (Gly and Glu). This GluN1/2C ABD heterodimer model was then prepared using the Protein Preparation Wizard with default parameters and the resulting protein structure was used for induced-fit docking.
Compounds were prepared using the LigPrep module with default parameters. The highest scoring conformation from the LigPrep results was used for conformational search, which was performed with OPLS3e force field and H2O as solvent. Charges were obtained from force field and cutoff was set to extended. No constrains and substructures was used and the Mini method was set to PRCG (Polak-Ribier Conjugate Gradient; Polak and Ribiere, 1969) with 2500 maximum interactions. Convergence was set to Gradient with the threshold as 0.05. The conformational search method was set to mixed torsional/low-mode sampling. And the search was customized with intermediate torsion sampling option. Maximum number of steps were set to 1000 and 100 steps were set per rotatable bond. All other options and parameters were set as default.
Induced-fit docking of compound 13g and 13i were performed with default parameters. XP Glide Redocking Precision was applied as the Glide redocking precision and the other parameters was set as default. All docking results were treated with energy minimization, and the binding pose of each compound was selected based on alignment with Gly and the IFDScore.
Molecular dynamic simulations were performed with the Desmond module of Schrödinger Suites. The highest scoring docking pose of each compound was used as the initial input structure, and the docking complex was solvated into a TIP3P orthorhombic water box. The distance between box edge and the protein was set to ≥10 Å. The whole system was neutralized with corresponding number of Na+ or Cl−, and 150 mM NaCl was added into the system. The production simulation was performed with NPT (constant particle number, pressure and temperature) ensemble at 300 K and 1.0 bar. The whole system was relaxed before production simulation and all other parameters were set as default. The most abundant binding conformations were generated by clustering the production trajectory. The RMSD plots of ligands and protein backbone were generated via the simulation event analysis panel of Maestro with all hydrogens neglected, and the MM/GBSA of the protein-ligand complexes were calculated.
Amino acid residues are numbered based on the full-length polypeptide sequence, including the signal peptide (initiating methionine is 1).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
FZ and GM synthesized compounds for the study and analyzed compound purity. FZ, FY, JL, ML, and KH performed electrophysiology experiments and data analysis. ES. performed HRMS experiments. FZ performed molecular modeling experiments. FZ, FY, ES, LB, KH, and RC. designed and planned experiments. All authors contributed to writing the manuscript.
Funding
The authors acknowledge financial support from the National Institutes of Health [NS097536, GM140963] to KH and a Summer Fellowship to ML. from the Center for Structural and Functional Neuroscience at the University of Montana. FZ acknowledge financial support from the National Natural Science Foundation of China [82204200], the China Scholarship Council, the International Postdoctoral Exchange Fellowship Program (Talent-Introduction Program) [YJ20210,279], the China Postdoctoral Science Foundation [2022M711939], and the Natural Science Foundation of Shandong Province [ZR2022QH287 and ZR2022QH312].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2022.1008233/full#supplementary-material
References
Benveniste, M., and Mayer, M. L. (1991). Kinetic analysis of antagonist action at N-methyl-D-aspartic acid receptors. Two binding sites each for glutamate and glycine. Biophys. J. 59 (3), 560–573. doi:10.1016/s0006-3495(91)82272-x
Chen, P. E., Geballe, M. T., Katz, E., Erreger, K., Livesey, M. R., O'Toole, K. K., et al. (2008). Modulation of glycine potency in rat recombinant NMDA receptors containing chimeric NR2A/2D subunits expressed in Xenopus laevis oocytes. J. Physiology 586 (1), 227–245. doi:10.1113/jphysiol.2007.143172
Clements, J. D., and Westbrook, G. L. (1991). Activation kinetics reveal the number of glutamate and glycine binding sites on the N-methyl-D-aspartate receptor. Neuron 7 (4), 605–613. doi:10.1016/0896-6273(91)90373-8
Dalton, G. L., Wu, D. C., Wang, Y. T., Floresco, S. B., and Phillips, A. G. (2012). NMDA GluN2A and GluN2B receptors play separate roles in the induction of LTP and LTD in the amygdala and in the acquisition and extinction of conditioned fear. Neuropharmacology 62 (2), 797–806. doi:10.1016/j.neuropharm.2011.09.001
Davis, M., Ressler, K., Rothbaum, B. O., and Richardson, R. (2006). Effects of D-cycloserine on extinction: Translation from preclinical to clinical work. Biol. Psychiatry 60 (4), 369–375. doi:10.1016/j.biopsych.2006.03.084
Dravid, S. M., Burger, P. B., Prakash, A., Geballe, M. T., Yadav, R., Le, P., et al. (2010). Structural determinants of D-cycloserine efficacy at the NR1/NR2C NMDA receptors. J. Neurosci. 30 (7), 2741–2754. doi:10.1523/jneurosci.5390-09.2010
Furukawa, H., and Gouaux, E. (2003). Mechanisms of activation, inhibition and specificity: Crystal structures of the NMDA receptor NR1 ligand-binding core. Embo J. 22 (12), 2873–2885. doi:10.1093/emboj/cdg303
Gupta, S. C., Hillman, B. G., Prakash, A., Ugale, R. R., Stairs, D. J., and Dravid, S. M. (2013). Effect of D-cycloserine in conjunction with fear extinction training on extracellular signal-regulated kinase activation in the medial prefrontal cortex and amygdala in rat. Eur. J. Neurosci. 37 (11), 1811–1822. doi:10.1111/ejn.12197
Hansen, K. B., Ogden, K. K., Yuan, H., and Traynelis, S. F. (2014). Distinct functional and pharmacological properties of Triheteromeric GluN1/GluN2A/GluN2B NMDA receptors. Neuron 81 (5), 1084–1096. doi:10.1016/j.neuron.2014.01.035
Hansen, K. B., Tajima, N., Risgaard, R., Perszyk, R. E., Jorgensen, L., Vance, K. M., et al. (2013). Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-D-aspartate receptors. Mol. Pharmacol. 84 (1), 114–127. doi:10.1124/mol.113.085803
Hansen, K. B., Wollmuth, L. P., Bowie, D., Furukawa, H., Menniti, F. S., Sobolevsky, A. I., et al. (2021). Structure, function, and pharmacology of glutamate receptor ion channels. Pharmacol. Rev. 73 (4), 1469–1658. doi:10.1124/pharmrev.120.000131
Hansen, K. B., Yi, F., Perszyk, R. E., Furukawa, H., Wollmuth, L. P., Gibb, A. J., et al. (2018). Structure, function, and allosteric modulation of NMDA receptors. J. General Physiology 150 (8), 1081–1105. doi:10.1085/jgp.201812032
Hillman, B. G., Gupta, S. C., Stairs, D. J., Buonanno, A., and Dravid, S. M. (2011). Behavioral analysis of NR2C knockout mouse reveals deficit in acquisition of conditioned fear and working memory. Neurobiol. Learn. Mem. 95 (4), 404–414. doi:10.1016/j.nlm.2011.01.008
Hofmann, S. G., Otto, M. W., Pollack, M. H., and Smits, J. A. (2015). D-Cycloserine augmentation of cognitive behavioral therapy for anxiety disorders: An update. Curr. Psychiatry Rep. 17 (1), 532. doi:10.1007/s11920-014-0532-2
Jessen, M., Frederiksen, K., Yi, F., Clausen, R. P., Hansen, K. B., Brauner-Osborne, H., et al. (2017). Identification of AICP as a GluN2C-selective N-Methyl-d-Aspartate receptor superagonist at the GluN1 Glycine site. Mol. Pharmacol. 92 (2), 151–161. doi:10.1124/mol.117.108944
Karakas, E., and Furukawa, H. (2014). Crystal structure of a heterotetrameric NMDA receptor ion channel. Science 344 (6187), 992–997. doi:10.1126/science.1251915
Lee, C. H., Lü, W., Michel, J. C., Goehring, A., Du, J., Song, X., et al. (2014). NMDA receptor structures reveal subunit arrangement and pore architecture. Nature 511 (7508), 191–197. doi:10.1038/nature13548
Lemoine, D., Jiang, R., Taly, A., Chataigneau, T., Specht, A., and Grutter, T. (2012). Ligand-gated ion channels: New insights into neurological disorders and ligand recognition. Chem. Rev. 112 (12), 6285–6318. doi:10.1021/cr3000829
Liu, J., Shelkar, G. P., Sarode, L. P., Gawande, D. Y., Zhao, F., Clausen, R. P., et al. (2021). Facilitation of GluN2C-containing NMDA receptors in the external globus pallidus increases firing of fast spiking neurons and improves motor function in a hemiparkinsonian mouse model. Neurobiol. Dis. 150, 105254. doi:10.1016/j.nbd.2021.105254
Liu, J., Shelkar, G. P., Zhao, F., Clausen, R. P., and Dravid, S. M. (2019). Modulation of burst firing of neurons in nucleus reticularis of the thalamus by GluN2C-containing NMDA receptors. Mol. Pharmacol. 96 (2), 193–203. doi:10.1124/mol.119.116780
Maolanon, A. R., Risgaard, R., Wang, S.-Y., Snoep, Y., Papangelis, A., Yi, F., et al. (2017). Subtype-specific agonists for NMDA receptor Glycine binding sites. ACS Chem. Neurosci. 8 (8), 1681–1687. doi:10.1021/acschemneuro.7b00117
Niciu, M. J., Kelmendi, B., and Sanacora, G. (2012). Overview of glutamatergic neurotransmission in the nervous system. Pharmacol. Biochem. Behav. 100 (4), 656–664. doi:10.1016/j.pbb.2011.08.008
Ogden, K. K., Khatri, A., Traynelis, S. F., and Heldt, S. A. (2014). Potentiation of GluN2C/D NMDA receptor subtypes in the amygdala facilitates the retention of fear and extinction learning in mice. Neuropsychopharmacology 39 (3), 625–637. doi:10.1038/npp.2013.241
Paoletti, P., Bellone, C., and Zhou, Q. (2013). NMDA receptor subunit diversity: Impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 14 (6), 383–400. doi:10.1038/nrn3504
Polak, E., and Ribiere, G. (1969). Note sur la convergence de méthodes de directions conjuguées. R. I. R. O. 3 (16), 35–43. doi:10.1051/m2an/196903r100351
Sahu, J. K., Ganguly, S., and Kaushik, A. (2013). Triazoles: A valuable insight into recent developments and biological activities. Chin. J. Nat. Med. 11 (5), 456–465. doi:10.3724/sp.j.1009.2013.00456
Sanz-Clemente, A., Nicoll, R. A., and Roche, K. W. (2013). Diversity in NMDA receptor composition: Many regulators, many consequences. Neuroscientist 19 (1), 62–75. doi:10.1177/1073858411435129
Sheinin, A., Shavit, S., and Benveniste, M. (2001). Subunit specificity and mechanism of action of NMDA partial agonist D-cycloserine. Neuropharmacology 41 (2), 151–158. doi:10.1016/s0028-3908(01)00073-9
Shelkar, G. P., Liu, J., and Dravid, S. M. (2021). Astrocytic NMDA receptors in the basolateral amygdala contribute to facilitation of fear extinction. Int. J. Neuropsychopharmacol. 24 (11), 907–919. doi:10.1093/ijnp/pyab055
Singewald, N., Schmuckermair, C., Whittle, N., Holmes, A., and Ressler, K. J. (2015). Pharmacology of cognitive enhancers for exposure-based therapy of fear, anxiety and trauma-related disorders. Pharmacol. Ther. 149, 150–190. doi:10.1016/j.pharmthera.2014.12.004
Sotres-Bayon, F., Bush, D. E., and LeDoux, J. E. (2007). Acquisition of fear extinction requires activation of NR2B-containing NMDA receptors in the lateral amygdala. Neuropsychopharmacology 32 (9), 1929–1940. doi:10.1038/sj.npp.1301316
Sotres-Bayon, F., Diaz-Mataix, L., Bush, D. E., and LeDoux, J. E. (2009). Dissociable roles for the ventromedial prefrontal cortex and amygdala in fear extinction: NR2B contribution. Cereb. Cortex 19 (2), 474–482. doi:10.1093/cercor/bhn099
Urwyler, S., Floersheim, P., Roy, B. L., and Koller, M. (2009). Drug design, in vitro pharmacology, and structure-activity relationships of 3-acylamino-2-aminopropionic acid derivatives, a novel class of partial agonists at the glycine site on the N-methyl-D-aspartate (NMDA) receptor complex. J. Med. Chem. 52 (16), 5093–5107. doi:10.1021/jm900363q
Valverde, I. E., Bauman, A., Kluba, C. A., Vomstein, S., Walter, M. A., and Mindt, T. L. (2013). 1, 2, 3-triazoles as amide bond mimics: Triazole scan yields protease-resistant peptidomimetics for tumor targeting. Angew. Chem. Int. Ed. 52 (34), 8957–8960. doi:10.1002/anie.201303108
Valverde, I. E., Vomstein, S., Fischer, C. A., Mascarin, A., and Mindt, T. L. (2015). Probing the backbone function of tumor targeting peptides by an amide-to-triazole substitution strategy. J. Med. Chem. 58 (18), 7475–7484. doi:10.1021/acs.jmedchem.5b00994
Williams, K. (1993). Ifenprodil discriminates subtypes of the N-methyl-D-aspartate receptor: Selectivity and mechanisms at recombinant heteromeric receptors. Mol. Pharmacol. 44 (4), 851–859.
Yamamoto, H., Hagino, Y., Kasai, S., and Ikeda, K. (2015). Specific roles of NMDA receptor subunits in mental disorders. Curr. Mol. Med. 15 (3), 193–205. doi:10.2174/1566524015666150330142807
Yi, F., Mou, T. C., Dorsett, K. N., Volkmann, R. A., Menniti, F. S., Sprang, S. R., et al. (2016). Structural basis for negative allosteric modulation of GluN2A-containing NMDA receptors. Neuron 91 (6), 1316–1329. doi:10.1016/j.neuron.2016.08.014
Zhao, F., Atxabal, U., Mariottini, S., Yi, F., Lotti, J. S., Rouzbeh, N., et al. (2022). Derivatives of (R)-3-(5-Furanyl)carboxamido-2-aminopropanoic acid as potent NMDA receptor Glycine site agonists with GluN2 subunit-specific activity. J. Med. Chem. 65 (1), 734–746. doi:10.1021/acs.jmedchem.1c01810
Keywords: ionotropic glutamate receptors, ligand-gated ion channel, two-electrode voltage-clamp electrophysiology, co-agonist, subtype selectivity
Citation: Zhao F, Mazis G, Yi F, Lotti JS, Layeux MS, Schultz EP, Bunch L, Hansen KB and Clausen RP (2022) Discovery of (R)-2-amino-3-triazolpropanoic acid derivatives as NMDA receptor glycine site agonists with GluN2 subunit-specific activity. Front. Chem. 10:1008233. doi: 10.3389/fchem.2022.1008233
Received: 31 July 2022; Accepted: 19 October 2022;
Published: 17 November 2022.
Edited by:
Simone Brogi, University of Pisa, ItalyReviewed by:
Nela Durisic, The University of Queensland, AustraliaAaron Nilsen, Oregon Health and Science University, United States
Copyright © 2022 Zhao, Mazis, Yi, Lotti, Layeux, Schultz, Bunch, Hansen and Clausen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fabao Zhao, ZmFiYW8uemhhb0BzdW5kLmt1LmRr; Kasper B. Hansen, a2FzcGVyLmhhbnNlbkB1bW9udGFuYS5lZHU=; Rasmus P. Clausen, cmFjQHN1bmQua3UuZGs=
 Fabao Zhao
Fabao Zhao Georgios Mazis1
Georgios Mazis1 James S. Lotti
James S. Lotti Michael S. Layeux
Michael S. Layeux Kasper B. Hansen
Kasper B. Hansen Rasmus P. Clausen
Rasmus P. Clausen