- 1State Key Laboratory of North China Crop Improvement and Regulation, College of Plant Protection, Hebei Agricultural University, Baoding, China
- 2College of Horticulture, Hebei Agricultural University, Baoding, China
- 3College of Life Sciences, Hebei Agricultural University, Baoding, China
Plant apoplast serves as the frontier battlefield of plant defense in response to different types of pathogens. Many pathogenesis-related (PR) proteins are accumulated in apoplastic space during the onset of plant–pathogen interaction, where they act to suppress pathogen infection. In this study, we found the expression of Triticum aestivum lipid transfer protein 3 (TaLTP3) gene was unregulated during incompatible interaction mediated by leaf rust resistance genes Lr39/41 at the early infection stage. Stable transgenic wheat lines overexpressing TaLTP3 exhibited enhanced resistance to leaf rust pathogen Puccinia triticina. Transcriptome analysis revealed that overexpression of TaLTP3 specifically activated the transcription of pathogenesis-related protein 1a (TaPR1a) and multiple plant hormone pathways, including salicylic acid (SA), jasmonic acid (JA), and auxin, in response to the infection of the model bacterial pathogen Pseudomonas syringae pv. tomato DC3000. Further investigation indicated that TaLTP3 physically associated with wheat TaPR1a protein in the apoplast. Transgenic wheat lines overexpressing TaLTP3 and TaPR1a showed higher accumulations of reactive oxygen species (ROS) during plant defense responses. All these findings suggested that TaLTP3 is involved in wheat resistance against leaf rust pathogen infection and forming a TaLTP3-TaPR1a complex in apoplast against this pathogen, which provides new insights into the functional roles of PR proteins.
Introduction
Plants utilize complex defense mechanisms to fight against pathogen infections. Apoplastic space represents an essential site for pathogen infection and the frontier battlefield of plant defense response, where either plant-secreted proteins, such as proteases and pathogenesis-related (PR) proteins, directly interact with pathogens or cell surface-localized pattern recognition receptors (PRRs) perceive pathogen-associated molecular patterns (PAMPs) to initiate PAMP-triggered immunity (PTI) (Jashni et al., 2015; Qi et al., 2017). PR protein-encoding genes are highly induced by pathogen infections, leading to the enrichment of PR proteins in the apoplast that participate in various plant defense responses (Van Loon et al., 2006; Wang et al., 2018). It has been documented that induction of PR genes expression is considered as the indicator of the activation of phytohormone signaling pathways such as salicylic acid (SA) and jasmonic acid (JA) (Van Loon et al., 2006). PR proteins and peptides have also been utilized as promising tools for engineering plants with multiple stress tolerance (Ali et al., 2018).
Among all the designated PR proteins, PR1 can be induced by various stresses and considered as a hallmark of plant defense responses with multiple functions (Breen et al., 2017). A quantitative peptidomics study revealed that the CAP-derived peptide 1 (CAPE1) peptide cleavage from C-terminus of PR1 protein acts as a damage-associated molecular pattern (DAMP) triggering plant defense responses such as the burst of reactive oxygen species (ROS) and expression of other PR genes (Chen et al., 2014). More recently, a study showed that TaPR1-mediated host defense in wheat against septoria nodorum blotch (SNB) (caused by Parastagonospora nodorum) requires the cleavage of the C-terminal region, while toxin 3 (SnTox3) effector suppresses TaPR1-mediated plant defense by preventing the cleavage of the C-terminal region via direct interaction with overexpression of pathogenesis-related protein 1a (TaPR1a) (Sung et al., 2021). Interestingly, a recent investigation demonstrated that wheat TaPR1 associates with thaumatin-like protein TaTLP1 [also known as pathogenesis-related protein 5 (TaPR5)] in the apoplastic space and exerts enhanced antifungal activities (Wang et al., 2020). Overexpression of wheat homolog of PR1 gene (TaPR1a) in transgenic wheat line resulted in significantly enhanced resistance against both stripe rust and leaf rust fungi (Bi et al., 2020). Interestingly, they found that the model bacterial pathogen Pseudomonas syringae (P. syringae) pv. tomato DC3000 was sufficient to activate the transcriptional response of TaPR1a-mediated plant defense.
Lipid transfer proteins (LTP) belonging to PR14 family constitute a large protein family that exist in all the land plants and they are involved in various biological processes (Salminen et al., 2016). A growing body of evidence showed that LTPs are involved in plant defense against biotic stress. An Arabidopsis apoplastic LTP protein encoded by defective in induced resistance 1 (DIR1) is proposed to interact with a lipid-derived molecule to trigger long-distance signaling (Maldonado et al., 2002). The expression of Arabidopsis LTP3 is directly regulated by the transcription factor MYB96 (Guo et al., 2013), the latter which serves as a key transcriptional regulator bridging signal pathways of abscisic acid (ABA) and SA (Seo et al., 2009; Seo and Park, 2010). Barley lipid transfer protein 4 (HvLTP4), firstly designated as PR14, is highly induced by infection of powdery mildew (Molina and García-Olmedo, 1993). Several Pichia pastoris-expressed wheat LTPs showed direct antifungal activities against multiple pathogens (Sun et al., 2008). Wheat non-specific LTPs exhibit differential inhibition to wheat and non-wheat pathogens in vitro (Sun et al., 2008). Overexpression of wheat LTP gene TaLTP5 results in increased resisance against Cochliobous sativus and Fusarium graminearum (Zhu et al., 2012). Wang et al. showed that overexpression of Triticum aestivum lipid transfer protein (TaLTP3) in Arabidopsis confers heat stress tolerance (Wang et al., 2014). More recently, heterologous overexpression of Arabidopsis AtLTP4.4 in transgenic wheat results in enhanced resistance to Fusarium head blight and detoxification of mycotoxin deoxynivalenol (DON) (McLaughlin et al., 2020). Although wheat LTPs have been widely studied, the molecular mechanism underlying LTP-mediated defense response in apoplast remains unclear.
Our previous studies showed that TaLTP3 protein was targeted by a wheat rust effector PNPi (Puccinia NPR1 interactor) (Bi et al., 2020). Therefore, we hypothesized that TaLTP3 might play an important role in wheat resistance against rust infections. In this study, we generated transgenic wheat lines overexpressing TaLTP3 to explore its fuctions during wheat defense responses and found transgenic lines exhibited enhanced resistance to leaf rust. Resistance mechanism of TaLTP3 was further investigated by transcriptome sequencing. TaLTP3 localized to the apoplast and interacted with TaPR1a suggesting that a TaLTP3-TaPR1a protein complex is involved in wheat resistance against leaf rust pathogen.
Materials and Methods
Gene Cloning, Generation of Wheat Transgenic Lines, and Pathogen Inoculation
The complete coding region of TaLTP3 (GenBank accession AY226580.1) was amplified from complementary DNA (cDNA) synthesized from RNA extracted from the seedlings of wheat line “Chinese Spring.” The complete coding region of TaLTP3 was constructed to binary T-DNA vector pLGY-02 (Ubi:gene, T-DNA). Transgenic wheat line overexpressing TaLTP3 gene was generated in the genetic background of spring common wheat cultivar “JW1” (selected from a segregating population of the cross of spring common wheat cultivars “Fielder” and “NB1”) with technique support from Bangdi Ltd. Company, Shandong, China. Transgenic wheat lines overexpressing TaPR1a in the same genetic background were derived from a previous study (Bi et al., 2020).
Seedlings of the wheat isogenic line carrying leaf rust resistance gene Lr39/41 in background of common wheat cultivar “Thatcher” were inoculated with Puccinia triticina (Pt), a virulent pathotype THTT following previously described procedure (Li et al., 2018). A high-resistant phenotype with the hypersensitive response (HR) was observed in Lr39/41 isogenic line after inoculation with Pt pathotype THTT. RNA samples for quantitative PCR (qPCR) were collected from wheat leaves inoculated with leaf rust or water control at 0, 1, 2, 5, and 8 days postinoculation (dpi). Four independent biological replicates were included for each time point and treatment. Seedlings of transgenic wheat line TaLTP3-OE and wild-type plants were inoculated with Pt virulent pathotype FHJQ following a similar procedure. The wild-type plant “JW1” showed a moderate susceptible phenotype (rust sporulation with necrotic spots on wheat leaf) toward most of the collected Pt pathotypes including FHJQ and THTT. The disease symptoms were photographed at 14 dpi. An image analysis software ASSESS version 2.0 (American Phytopathological Society, St. Paul, Minisota, MN, United States) was employed to evaluate the percentage of Pt sporulation area for each inoculated leaf (Lamari, 2008; Bi et al., 2020). Two independent transgenic lines of TaLTP3-OE with 9–13 biological replicates were included for the Pt inoculation. The Dunnett’s test was conducted by using statistical analysis software SAS version 9.4.
Model bacterial pathogen P. syringae DC3000 was inoculated in second leaves of wheat seedlings according to the previously described method (Bi et al., 2020). P. syringae DC3000 was cultivated in low-salt Luria-Bertani (LS-LB) medium with 50 μg/mL rifampicin (Rif) antibiotics for 48 h (Jacob et al., 2017) and then diluted to optical density (OD) 600 = 0.6 in distilled water. Second leaves of transgenic wheat line TaLTP3-OE were infiltrated with bacterial suspensions by using a 1-ml needless syringe through the abaxial surface. Each transgenic line consisted of 6–8 independent biological replicates. Seedlings of wild-type plants and water-infiltration served as controls. The infiltration region was marked by using a marker pen. RNA samples for qPCR and RNA-sequencing (RNA-seq) assays were collected from the infiltration region at 6 h post-inoculation (hpi).
Ribonucleic Acid-Sequencing
Ribonucleic acid samples were sent to Novogene Corporation Ltd. for KAPA library construction and transcriptome sequencing following the default procedure. The Illumina HiSeq 1000 System was employed for the 12-Gb sequencing for each RNA sample. Transcriptome was assembled with reference genome of Triticum aestivum (common wheat “Chinese Spring” TGACv1 version) (Appels et al., 2018) by using the HISAT2 FDR and StringTie software (Kim et al., 2019). Assembled transcripts that could not be found in the gene models of the reference genome were annotated as “novel” transcripts. Gene expressions were profiled by using the HTSeq version 0.9.1 software (Trapnell et al., 2010). The expected number of fragments per kilobase of transcript sequence per million base pairs (FPKM) for each gene was used to evaluate the expression of assembled genes. FPKM-based heatmaps were generated by the MeV software. Differentially expressed genes (DEGs) were filtered the expression levels of genes between different groups for an false discover rate (FDR)-adjusted p-value < 0.05 and |log2-fold change| > 1 by using the DESeq2 software (Love et al., 2014). The Gene Ontology (GO) annotations were assigned to each DEGs with the GO sequencing (Young et al., 2010). The KEGG (Kyoto Encyclopedia of Genes and Genomes) annotations were employed to profile the regulatory pathways enriched with DEGs (Kanehisa and Goto, 2000).
Quantitative Real Time-PCR Assay
Ribonucleic acids were prepared by using the RNA Extraction Kit (QIAGEN, Hilden, Germany, United Kingdom) and the first-strand cDNA was synthesized by using the Reverse Transcription Kit (Takara, Dalian, China). Primers for qRT-PCR assay were designed (Supplementary Table 1) and a preliminary test was performed on the Roche LightCycler 96 qRT-PCR machine by using a series of 2-fold diluted cDNA templates (1:1, 1:2, 1:4, 1:8, and 1:32) to evaluate the amplification efficiency for each pair of primers. Melting curves were generated to ensure the specificity of the amplification. The wheat reference gene TaActin (GenBank accession AB181991) was used as the endogenous control (Paolacci et al., 2009). The transcript level was expressed relative to that of TaActin based on the 2–ΔCt method as described in a previous study (Wu et al., 2019). Mean and SE were calculated by using the Microsoft Excel software. The two-sample t-test and the generalized linear model (GLM) ANOVA were performed by using the SAS software version 9.4.
Subcellular Localization
Full-length open reading frame (ORF) of TaLTP3 was cloned into transient expression vectors of pGWB5 (35S:gene-GFP, T-DNA) and pGWB454 (35S:gene-RFP, T-DNA), respectively. The recombinant vectors were transformed into Agrobacterium strain GV3101 and then used for infiltration of Nicotiana benthamiana (N. benthamiana). Corresponding empty vectors expressing only green fluorescent protein (GFP) and red fluorescent protein (RFP) served as controls. Green or red fluorescence was checked 48 hpi by using a Nikon Ti-2 microscope with a GFP or RFP filter, respectively. Epidermal peels of tobacco leaves were prepared from the infiltration region and soaked in 800 mM mannitol for 6 min to induce the plasmolysis (Ma et al., 2012).
Yeast Two-Hybrid Assay
Primers that were used to amplify the full-length ORF of TaLTP3 and five other plant defense-related LTP genes from cDNA synthesized from RNA isolated from the common wheat line “Chinese Spring” are listed in Supplementary Table 1. A neighbor-joint tree including all the LTP proteins in this study was generated by using the MEGA software version 7.0. The signal peptides of wheat LTPs were predicted by SignalP 5.0 server.1 The amplified DNA fragment lacking the sequence encoding the signal peptide was cloned into a Gateway yeast two-hybrid (Y2H) vector pLAW10 bait domain (BD). Recombined Y2H activation domain (AD) vectors carrying different regions of the TaPR1a gene were derived from a previous study (Bi et al., 2020). The coding regions of TaPR1a(25–93aa) and TaPR1a(25–128aa) were cloned into a Gateway Y2H vector pLAW11 (AD). Yeast transformation was performed by using lithium acetate and polyethylene glycol 3350 as previously described (Gietz and Schiestl, 2007). Cotransformed yeast strains were spotted on SD-Leu-Trp-His-Ade selective medium.
Bimolecular Fluorescence Complementation Assay
Full-length ORF of TaLTP3 with signal peptide was constructed to pDEST-GWVYNE (35S:gene-YFPN, T-DNA). The recombinant construct of TaPR1a-pDEST-GWVYCE carrying complete coding region of TaPR1a was derived from our previous study (Bi et al., 2020). The fusion genes were coexpressed in tobacco leaves by Agrobacterium infiltration. Yellow fluorescence was checked 48 hpi by using a Nikon Ti-2 microscope with a yellow fluorescent protein (YFP) filter. Coexpression of AvrLm1-pDEST-GWVYNE and BnMPK9-pDEST-GWVYCE in the same vectors was used as a positive control (Ma et al., 2018). Signal peptide-fused empty vectors of SP(TaPR1a)-pDEST-GWVYNE and SP(TaPR1a)-pDEST-GWVYCE were derived from our previous study (Bi et al., 2020) and coexpressed with TaPR1a-pDEST-GWVYCE and TaLTP3-pDEST-GWVYNE, respectively, as negative controls.
Co-immunoprecipitation Assay
Deoxyribonucleic acid fragment encoding the signal peptide-truncated TaLTP3 protein was cloned into pGWB5 as 35S:TaLTP3(30–122)-GFP. A specific reverse primer of TaPR1a with stop codon and sequence encoding cMYC tag was designed and used to amplify the DNA fragment encoding the signal peptide-truncated TaPR1a protein. The resulting DNA segment was constructed into pGWB5 as 35S:TaPR1a(25–164)-cMYC. Recombinant pGWB5:TaLTP3(30–122)-GFP and pGWB5: TaPR1a(25–164)-cMYC were coexpressed in tobacco leaves by using Agrobacterium. Coexpression of GFP and TaPR1a(25–164)-cMYC served as a negative control. Total proteins were extracted from Agrobacterium-infiltrated tobacco leaves at 3 dpi and a portion of the total protein was reserved as an input sample. α-GFP IP samples were prepared by using GFP-trap beads (LABLEAD, Beijing, China). Input and IP samples were separated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto membranes made of polyvinylidene difluoride (PVDF). Immunoblots were performed by using α-GFP or α-cMYC antibody (Solarbio, Beijing, China).
Reactive Oxygen Species Staining
To visualize the accumulation of ROS in wheat leaves infiltrated with P. syringae DC3000, two chemical reagents, nitroblue tetrazolium (NBT) and 3,3-diaminobenzidine (DAB), were applied to stain the superoxide anion (O2–) and hydrogen peroxide (H2O2), respectively. The staining protocols were modified from a previous study (Wang et al., 2007). Leaf regions infiltrated with P. syringae DC3000 were collected at 2, 4, and 8 hpi. For O2– visualization, samples were soaked in 10 mM sodium azide (NaN3) and 10 mM potassium phosphate buffer (pH 7.8) with 0.1% NBT (w/v) for 24 h. For H2O2 staining, samples were incubated in hydrochloric acid (HCl)-acidified solution (pH 3.8) containing 1 mg/mL of DAB for 24 h. Thereafter, samples were decolored in boiling 95% ethanol for 10 min. The percentage of stained area in each leaf was determined by using the ASSESS software (Lamari, 2008). The whole experiment was repeated twice with two independent transgenic lines and each repeat consisted of 6–10 biological replicates. The Dunnett’s test was conducted by using the SAS software version 9.4.
Results
Stable Overexpression of TaLTP3 in Transgenic Wheat Line Results in Enhanced Resistance to Leaf Rust Pathogen Puccinia triticina
To determine the role of TaLTP3 in plant resistance, we first quantified the expression change of this gene in leaf rust resistance mediated by Lr39/41 gene. Interestingly, TaLTP3 was significantly upregulated at the wheat early-stage resistance toward leaf rust (Figure 1A). To further explore the function of TaLTP3, transgenic wheat lines overexpressing TaLTP3 (TaLTP3-OE) were generated in the genetic background of commonspring wheat line “JW1.” The expression levels of TaLTP3 in TaLTP3-OE reached 1.7- to 2.8-fold of those in the wild-type (Figure 1B). The third leaves of TaLTP3-OE and wild-type plants were inoculated with urediniospores of the Pt pathotype FHJQ and FHJQ exhibited high virulence on wild-type plants (Figure 1C). The sporulation of leaf rust pathogen was observed in both the TaLTP3-OE and wild-type plants. The percentage of Pt sporulation area for each inoculated leaf was evaluated by using the ASSESS software. Compared with wild-type plants, significantly enhanced resistance to Pt was observed in TaLTP3-OE as evidenced by decreased sporulation area (**p < 0.01, ***p < 0.0001, 9–30 biological replicates) (Figure 1C and Supplementary Table 2). We further investigated several agronomic traits of transgenic wheat line TaLTP3-OE including plant height, spike length, spikelet number per spike, and seed number per spike under greenhouse conditions. We did not observe any significant difference in these agronomic traits between TaLTP3-OE and wild-type plants (Figure 1D). However, when we checked the seed size and thousand grain weight, we found smaller and lighter seeds harvested from TaLTP3-OE than wild-type plants (Figure 1E), indicating overexpression of TaLTP3 might have influence on wheat production.
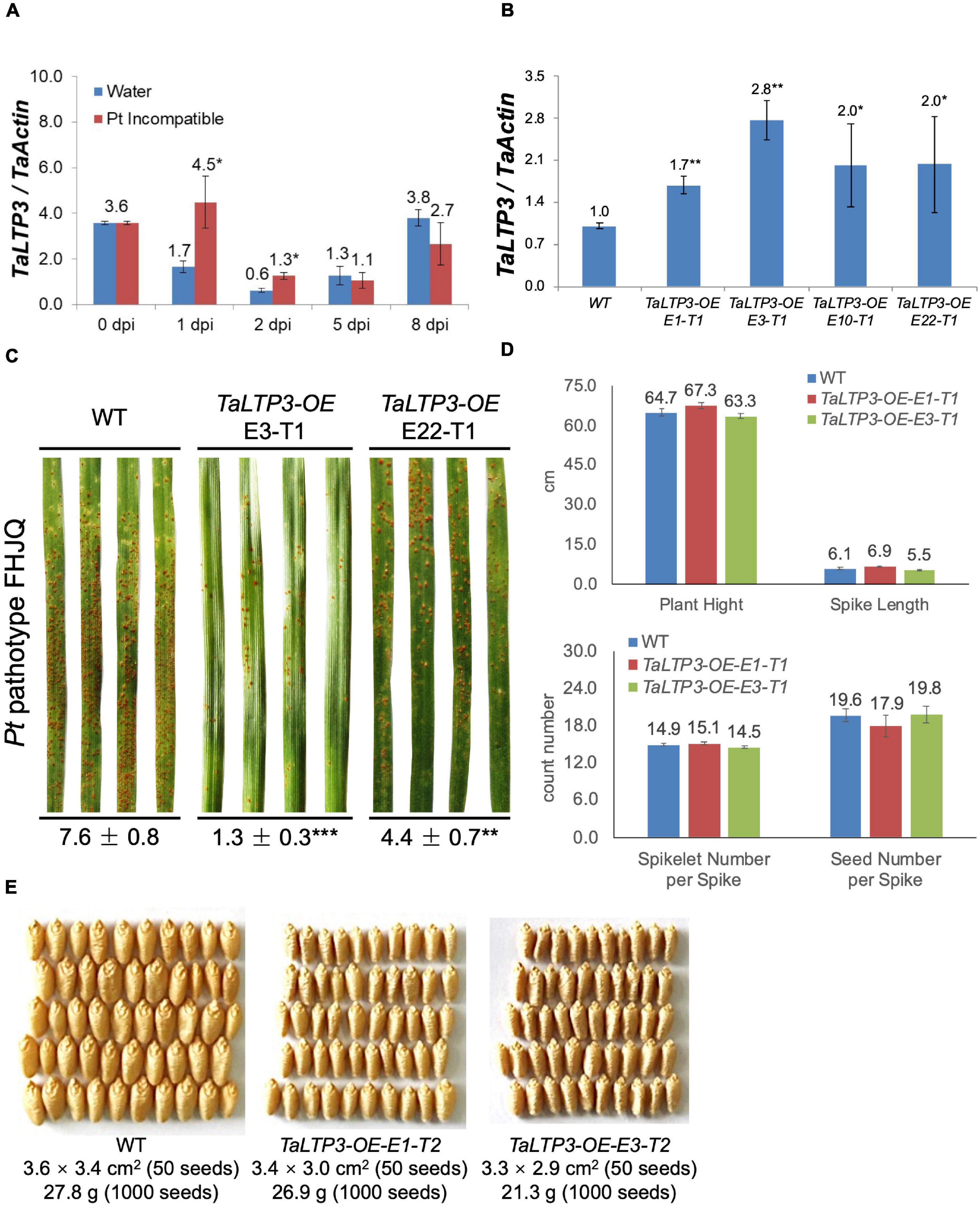
Figure 1. TaLTP3 was involved in wheat resistance to leaf rust. (A) Expression levels of TaLTP3 during Lr39/41-mediated wheat resistance toward leaf rust were evaluated by quantitative PCR (qPCR) assay. Leaf samples were collected at 0, 1, 2, 5, and 8 days postinoculation (dpi) with water (control) and avirulent Pt pathotype THTT. The transcript levels for all the genes were expressed as linearized fold-TaActin levels, which were calculated based on the formula 2(Actin Ct –Target Ct). Data were expressed as mean ± SE from four biological replicates. The asterisk indicated a significant difference (p < 0.05) between the water control and Pt-inoculated samples by using the t-test. (B) The transcript level of TaLTP3 in transgenic wheat line overexpressing TaLTP3 and wild-type plants were evaluated by qPCR assay. (C) The susceptibility phenotypes of TaLTP3-OE and wild-type plants to the Pt pathotype FHJQ. The numbers below the images of the Pt-infected wheat leaves represent the average percentages of the rust sporulation area in each leaf. The asterisks (**p < 0.01, ***p < 0.0001) were annotated based on the comparison between the transgenic line and wild-type plants by the Dunnett’s test. (D) Agronomic traits of transgenic wheat line TaLTP3-OE and wild-type plants were investigated in greenhouse condition. (E) Seeds harvested from transgenic wheat line TaLTP3-OE and wild-type plants were compared in size and thousand grain weight.
Overexpression of TaLTP3 Positively Regulates Multiple Signaling Pathways Mediated by Plant Hormones of Auxin, Jasmonic Acid, and Salicylic Acid
To reveal the mechanism of TaLTP3 during plant resistance, we have used model bacterial pathogen P. syringae pv. tomato DC3000 to trigger the immune response in wheat leaves. Although P. syringae DC3000 is not a wheat pathogen under a natural environment, a typical cell death was observed in wheat leaf infiltrated with P. syringae DC3000 at 48 hpi (Figure 2A), indicating a successful induction of non-host resistance. We applied RNA-seq on samples collected from P. syringae DC3000-infiltrated third leaves of wheat transgenic line TaLTP3-OE and wild-type plants at 6 hpi. Samples from the water-infiltrated leaves served as a control. Each of the treatments included three biological replicates and, in total, 12 samples were subjected to a 12-Gb Illumina sequencing (Supplementary Table 3). The reference genome of Triticum astivum from the Ensembl Genomes (TGACv1 version) was used to assemble the transcriptome. A total of 122,130 transcripts were mapped to the genome sequence (Supplementary Table 4). Significant (R2 > 0.92) correlations of the overall gene expressions among biological replicates were detected (Supplementary Table 5). We used the FPKM value to predict the expression levels of transcripts. Raw reads from the RNA-seq were deposited at the National Center for Biotechnology Information (NCBI) BioProject PRJNA746113.
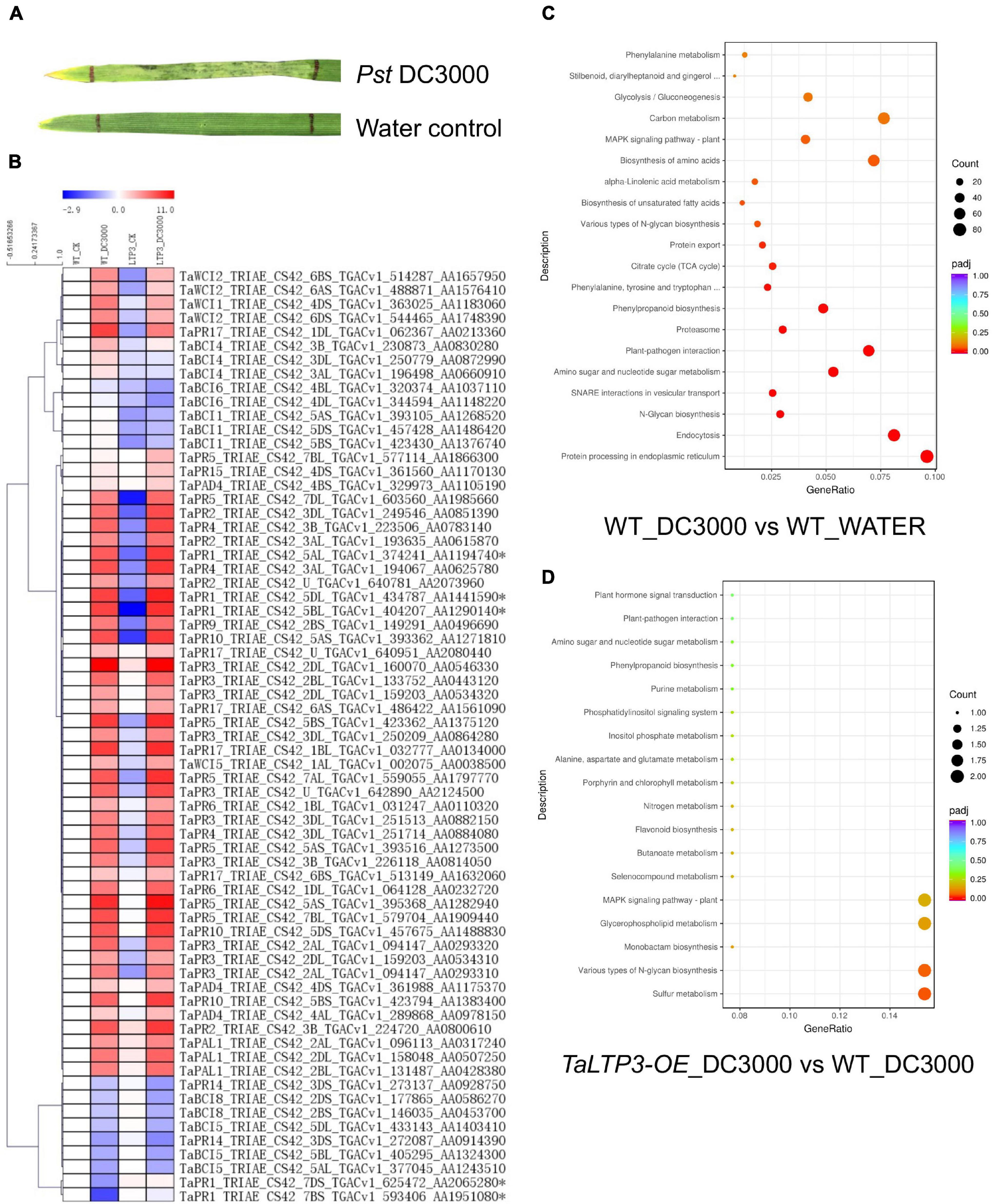
Figure 2. Transcriptome analysis revealed the regulatory network of TaLTP3 during plant defense. (A) A typical cell death was observed in wheat leaf infiltrated with P. syringae DC3000 at 48 h postinfiltration. (B) Expression profile of PR genes upon P. syringae DC3000 infection in TaLTP3-OE transgenic wheat line. FPKM values of WT_DC3000, LTP_CK, and LTP_DC3000 were relative to that of WT_CK. Data were calculated as log2-fold change format by using the Microsoft Excel software. Heatmap was generated by using the MeV software. Genes with similar expression patterns were clustered. (C,D) The Gene Ontology (GO) annotations for upregulated differentially expressed genes (DEGs) in comparisons of “WT_DC3000 vs. WT_WATER”and “TaLTP3-OE_DC3000 vs. WT_DC3000.” DEGs were classified based on their GO annotations into three main categories including biological process, cellular component, and molecular function.
The expression levels of all 18 PR gene families were displayed in the heat map based on their FPKM values derived from the transcriptome data (Figure 2B). Most of the PR genes were highly induced by the infection of P. syringae DC3000, indicating an activation of a broad-range plant defense response by this model pathogen. Compared with the wild-type plants, TaPR1a, TaPR5, and TaPR15 showed higher levels of expression in TaLTP3-OE. DEGs in comparison of “WT_DC3000 vs. WT_WATER” were identified (|log2-fold change| > 1 and q-value < 0.01) by using DESeq2. In the wild-type plants, we found 9,178 genes were significantly upregulated upon P. syringae DC3000 infection. The KEGG annotations of these P. syringae DC3000-sensitive genes indicated the activation of a broad-range plant defense responses including Ca2+-dependent ROS induction, mitogen-activated protein kinase (MAPK) signaling, and effector-triggered immunity (Supplementary Figure 1 and Supplementary Table 6). The GO annotations for these DEGs enriched in “plant–pathogen interaction” and “protein processing in endoplasmic reticulum” (Figure 2C).
From the comparison between “TaLTP3-OE_DC3000” and “WT_DC3000,” only 214 genes were significantly higher expressed (log2-fold change > 1 and q-value < 0.01) in TaLTP3-OE transgenic upon P. syringae DC3000 infection. The GO annotations of these upregulated DEGs enriched in “Various types of N-glycan biosynthesis” and “Glycerophospholipid metabolism” (Figure 2D). Among all these TaLTP3-responsive upregulated DEGs, TaPR1a was the only gene annotated into the KEGG pathway of “plant–pathogen interaction” (Table 1). Furthermore, we found 173 genes with significantly lower (log2-fold change < -1 and q-value < 0.01) expressions in TaLTP3-OE transgenic line upon P. syringae DC3000 infection. Several key regulators in the hormone pathway of auxin and JA, including auxin response factor 9 (ARF9), A-type Arabidopsis response regulator (A-ARR), and protein TIFY 6a [JAZ (jasmonate ZIM-domain)], were downregulated in the KEGG pathway of “plant hormone signal transduction” (Table 1 and Supplementary Figure 2).
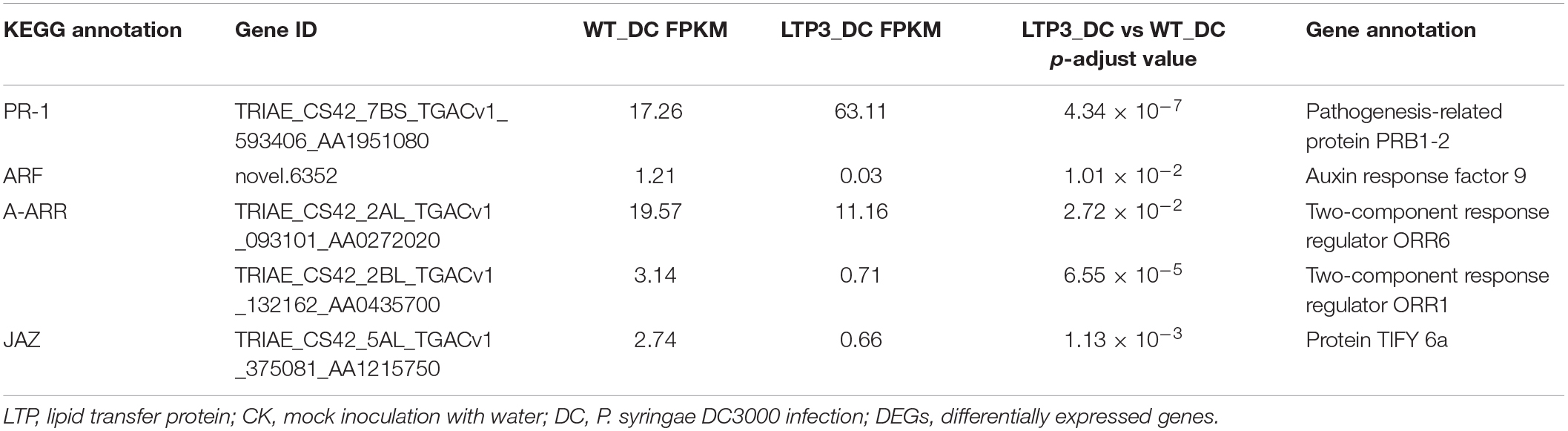
Table 1. DEGs of “LTP3_DC vs. WT_DC” annotated in the KEGG pathway of “plant–pathogen interaction” and “plant hormone signal transduction.”
We further evaluated the expression levels of two SA synthesis genes, TaPAL and TaPAD4, during P. syringae DC3000 infection in TaLTP3-OE transgenic line by using qRT-PCR assay (Figure 3). qPCR showed that the expression levels of both the TaPAL and TaPAD4 were elevated upon P. syringae DC3000 infection in TaLTP3-OE transgenic line, which was consistent with those in RNA-seq (Figure 2B), suggesting that our RNA-seq data are reliable.
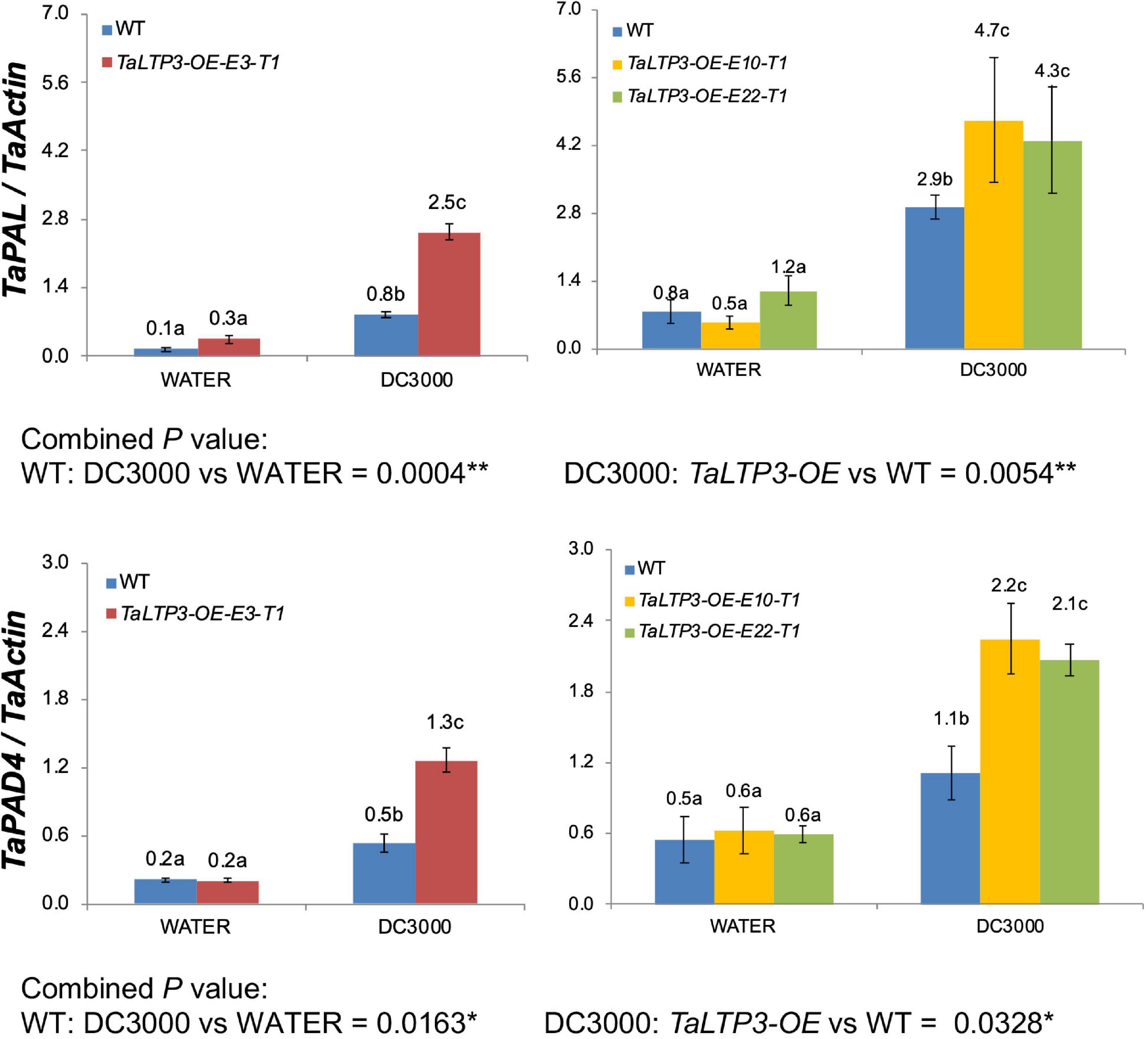
Figure 3. The transcriptional levels of TaPAL and TaPAD4 were determined by qPCR in transgenic line TaLTP3-OE upon P. syringae DC3000 infection. The transcript levels for all the genes were expressed as linearized fold-TaActin levels, which were calculated based on the formula 2(Actin Ct –Target Ct). Three independent transgenic lines of TaLTP3-OE were tested. Each experiment, consisting of 4–9 biological replicates, was considered as a block. Calculations of the mean and SE were performed by using the Microsoft Excel software. Generalized linear model (GLM) ANOVA (*p < 0.05, **p < 0.01) was conducted by using the SAS software version 9.4.
TaLTP3 Interacts With Pathogenesis-Related Protein 1a in the Apoplastic Space
As overexpression of TaLTP3 affected TaPR1a expression during plant defense response (Figure 2B, Supplementary Figure 2, and Table 1) and both TaLTP3 and TaPR1a are predicted as secreted proteins by signal P, we hypothesized that TaLTP3 might function through physical interaction with TaPR1a in the apoplastic space. The apoplast localization of TaPR1a has been validated in our previous study (Bi et al., 2020). GFP- and RFP-tagged TaLTP3 were generated, respectively. The recombinant constructs TaLTP3-GFP and TaLTP3-RFP were transientlyexpressed in N. benthamiana by agroinfiltration. Plasmolysis was induced by treatment with 800 mM mannitol. Pronounced green and red fluorescent signals for TaLTP3-GFP and TaLTP3-RFP were visualized in the apoplastic space, respectively (Figure 4), but not for the empty vector (EV-GFP and EV-RFP) controls (Figure 4), suggesting that TaLTP3 localizes in the apoplastic space.
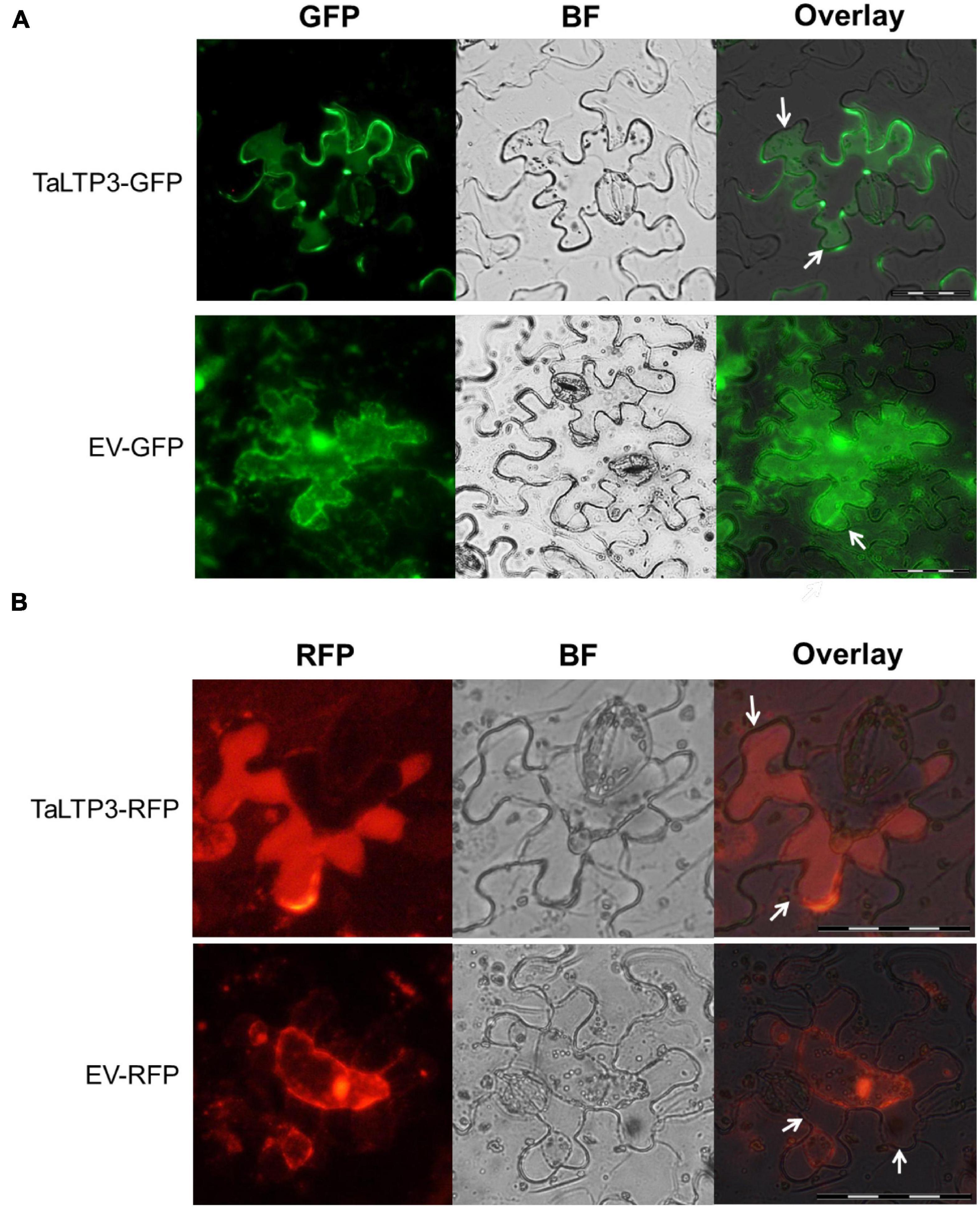
Figure 4. TaLTP3 protein was localized in the apoplastic space. The TaLTP3-GFP (A) and TaLTP3-RFP (B) Recombinant proteins were transiently expressed in N. benthamiana leaves by agroinfiltration. GFP or RFP alone was used as a control. Leaf epidermal peels were plasmolyzed by incubation in 800 mM mannitol for 6 min. White arrows indicating the separation between the plant cell wall and plasma membrane during plasmolysis. Scale bar = 100 μm. BF, bright field; EV, empty vector; GFP, green fluorescent protein; RFP, red fluorescent protein.
To examine the interaction between TaLTP3 and TaPR1a, a Y2H assay was performed by using a signal peptide-truncated version of these two proteins. Strong interaction was observed between TaPR1a(25–164) and TaLTP3(30–122) as yeast transformed with both the constructs was able to grow on the stringent synthetic dropout (SD) selective media lacking leucine, tryptophan, histidine, and adenine (SD-Leu-Trp-His-Ade) (Figure 5A). Because the C-terminal region of TaPR1a contains the conserved CAPE1 peptide in most PR1 proteins that might act as a DAMP in triggering plant defense response, we generated several truncated versions of TaPR1a protein, in which different C-terminal regions were deleted (Figure 5B). As shown in Figure 5A, an N-terminal region of TaPR1a ranging from 25 to 93 aa was sufficient to mediate the interaction with TaLTP3. However, we did not observe any interaction between TaPR1a (94–164 aa) and TaLTP3. As many wheat LTP genes, including TaLTP2, TaLTP3F1, TaLTP4.3, TaLTP4.1, and TaPR14, have been previously reported in wheat resistance to various pathogens, we also checked the interaction of TaPR1a with these LTP proteins and found that TaPR1a interacts with all these LTP proteins (Supplementary Figure 3). Collectively, these initial Y2H results indicated a conserved physical interaction between TaPR1a and LTP proteins.
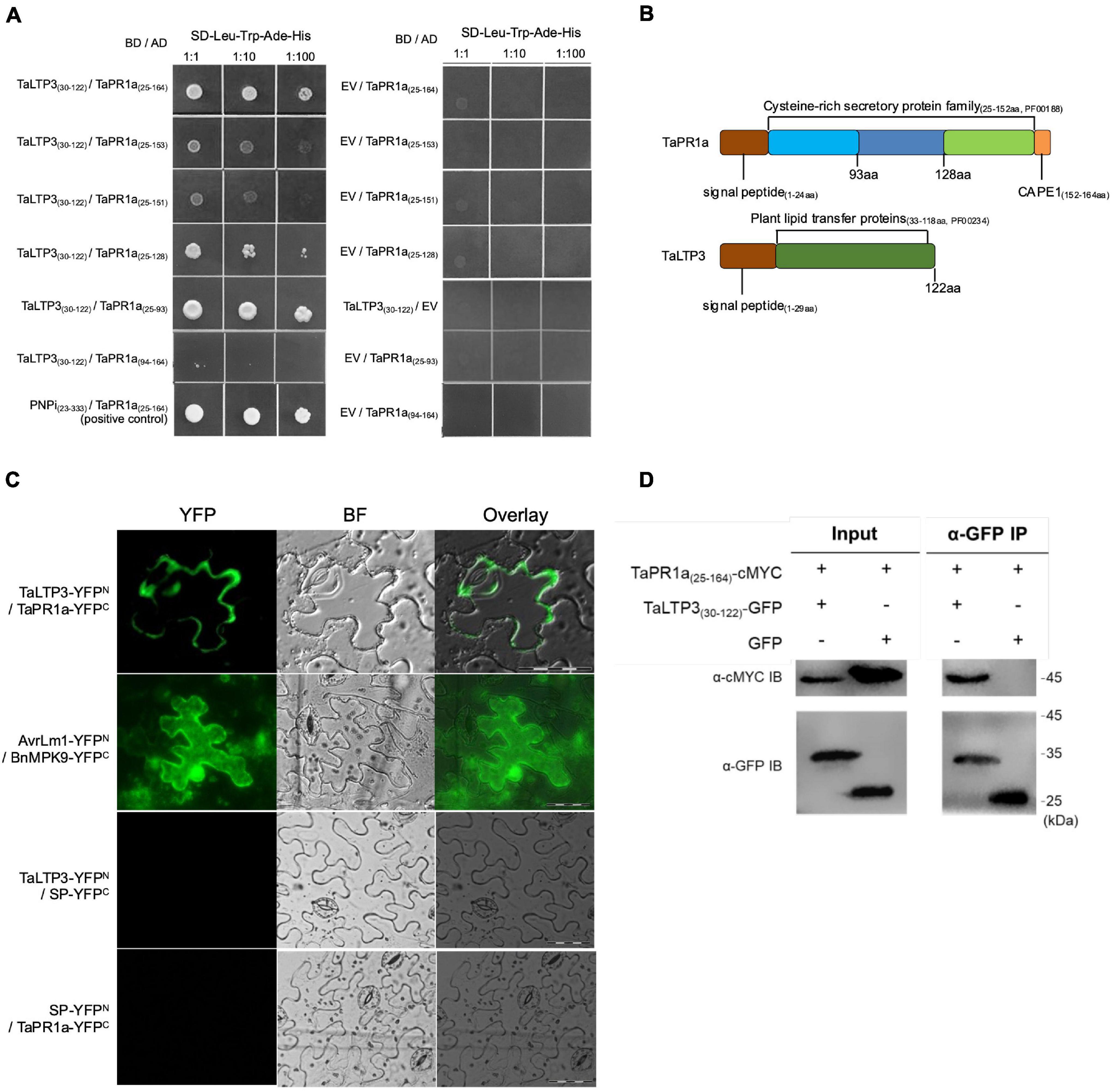
Figure 5. TaLTP3 physically associated with TaPR1a in vitro and in vivo. (A) Interaction of TaLTP3 and TaPR1a was detected by Y2H. Yeast transformants coexpressing different bait and prey recombinant constructs were tested on selective medium of SD-Leu-Trp-His-Ade, EV, and empty vector. (B) Signal peptide and conserved domain for TaPR1a and TaLTP3 proteins were schematically presented. (C) Bimolecular fluorescence complementation (BiFC) assay showed that the interaction between TaPR1a and TaLTP3 occurred in the apoplastic space. YFP signals were observed in the tobacco leaves coexpressing TaLTP3-YFPN and TaPR1a-YFPC. Recombinant constructs of AvrLm1-YFPN and BnMPK9-YFPC were coexpressed as a positive control. Segment encoding signal peptide of TaPR1a was cloned into BiFC vectors as SP-YFPN and SP-YFPC as negative controls. Scale bar = 100 μm. Due to the fixed microscopy settings for visualizing yellow and green fluorescent signals, the yellow fluorescent signals were presented in green color. (D) Validation of the interaction between TaPR1a and TaLTP3 by co-immunoprecipitation (Co-IP) assay. Total proteins from tobacco leaves transiently expressing TaPR1a(25–164)-cMYC and TaLTP3(30–122)-GFP were extracted and immunoprecipitated with an α-GFP antibody (α-GFP IP). Input and bound proteins were immunoblotted with α-GFP and α-cMYC antibodies, respectively. Original, uncropped blots for the Co-IP assay were displayed as Supplementary Figure 4. BF, bright field; EV, empty vector; YFP, yellow fluorescent protein; BiFC, bimolecular fluorescence complementation; Y2H, yeast-two hybrid.
To validate the in-planta interaction between TaLTP3 and TaPR1a, a bimolecular fluorescence complementation (BiFC) assay was performed in N. benthamiana. Strong yellow fluorescence signals in the apoplast were observed in N. benthamiana leaves coexpressing TaLTP3-YFPN and TaPR1a-YFPC (Figure 5C), whereas fluorescence in cytoplasm and nucleus was found in positive control coexpressing AvrLm1-YFPN and BnMPK9-YFPC (Ma et al., 2018). No YFP signals were observed in the N. benthamiana leaves coexpressing the negative controls: TaLTP3-YFPN and SP(TaPR1a)-YFPC nor SP(TaPR1a)-YFPN and TaPR1a-YFPC.
To further confirm the interaction in vivo, a co-immunoprecipitation (co-IP) assay was performed following transient expression of TaPR1a(25–164)-cMYC and TaLTP3(30–122)-GFP constructs in N. benthamiana. GFP was used as a negative control. cMYC-tagged TaPR1a co-immunoprecipitated with GFP-tagged TaLTP3, but not with GFP (Figure 5D and Supplementary Figure 4), indicating that TaLTP3 physically interacts with TaPR1a.
Overexpression of Pathogenesis-Related Protein 1a and TaLTP3 in Wheat Increases Accumulation of Reactive Oxygen Species Triggered by P. syringae DC3000
To explore the coeffects of TaLTP3 and TaPR1a during plant defense response, we evaluate the accumulation of H2O2 and O2– in the transgenic wheat lines TaLTP3-OE and TaPR1a-OE during P. syringae DC3000 infection. Leaf samples were collected at 2, 4, and 8 h after P. syringae DC3000 inoculation and stained with DAB and NBT to visualize H2O2 and O2–, respectively. Percentage of the stained area in each infiltrated leaf was calculated by using the ASSESS software. Compared with the wild-type plants, we observed significantly higher (*p < 0.05, **p < 0.01, 6–10 biological replicates) accumulation of H2O2 in the transgenic wheat lines TaPR1a-OE and TaLTP3-OE at 4 and 8 hpi (Figure 6A and Supplementary Table 7). For the NBT staining, the transgenic wheat line TaPR1a-OE showed a fast and higher accumulation of O2– at 2 and 4 hpi, whereas the transgenic wheat line TaLTP3-OE accumulated more O2– only at 4 hpi (Figure 6B and Supplementary Table 8). These results indicated that both the TaLTP3 and TaPR1a might function through a ROS-dependent plant defense response.
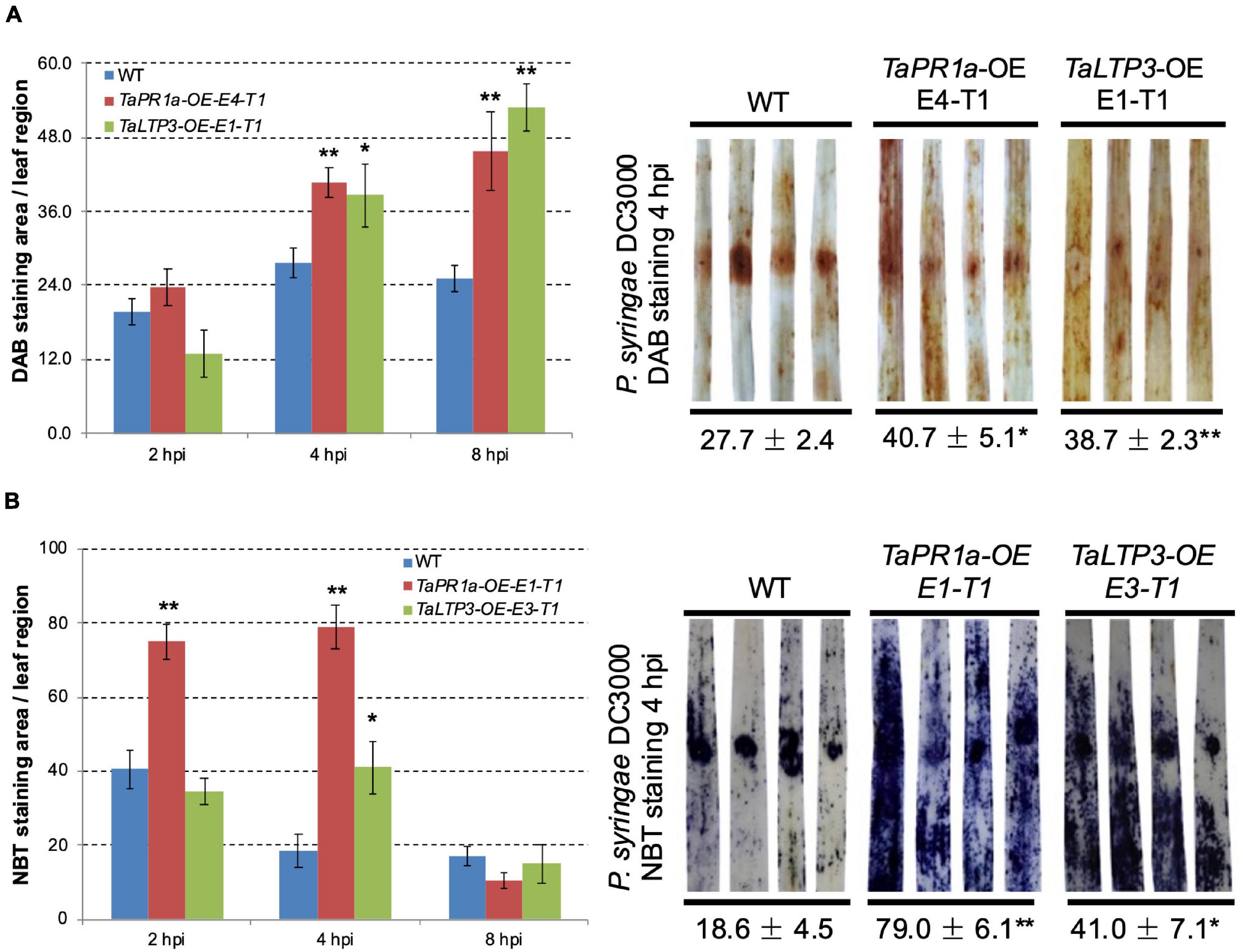
Figure 6. Transgenic wheat lines TaPR1a-OE and TaLTP3-OE showed higher accumulation of reactive oxygen species (ROS) upon infection of model bacterial pathogen P. syringae DC3000. (A) Samples from the infiltrated region were collected at 2, 4, and 8 hours postinoculation (hpi) and further stained with 3,3-diaminobenzidine (DAB). (B) Corresponding samples were stained with nitroblue tetrazolium (NBT) to visualize the accumulation of superoxide anion (O2–). The percentage of stained area in the infiltrated region was calculated By using the ASSESS software. Asterisk indicates the significance of the differences between the transgenic lines and wild-type plants established by using the Dunnett’s test (*p < 0.05, **p < 0.01, Supplementary Tables 7, 8).
Discussion
Many designated PR proteins are secreted into the apoplastic space to function. Therefore, apoplast represents the initial battlefield between plant and phytopathogens interaction and apoplastic immunity plays an important role in plant defense responses. TaLTP3, a member of the PR14 gene family, was upregulated during wheat resistance to leaf rust infections (Figure 1A). Our studies showed that both the TaLTP3 and TaPR1a are secreted into the apoplastic space (Figure 4; Bi et al., 2020). PR1 protein functions in multiple ways during plant defense response including direct antimicrobe activity, triggering DAMP-triggered immunity, and targeting pathogen effectors (Chen et al., 2014; Gamir et al., 2017; Sung et al., 2021). LTP also has antifungal and antioxidant activities (McLaughlin et al., 2020). In Arabidopsis, LTP was reported as upstream of SA-mediated resistance to biotic and abiotic stresses (Maldonado et al., 2002; Guo et al., 2013). Previous studies showed that PR1 interacts with pathogenesis-related proteins from plants and effectors from pathogens (Breen et al., 2017), suggesting that PR1 has a broad-spectrum interactors in plant–pathogen interaction. Therefore, we speculated that PR1 might form different complexes with various plant defense-related proteins targeting phytopathogens. In this study, we identified and validated a direct interaction between TaLTP3 and TaPR1a in the apoplastic space (Figure 5). Further study showed that this interaction was conserved among TaPR1a and other plant-defense-related LTP proteins (Supplementary Figure 3). The CAPE1 peptide in the C-terminal region of PR1 is conserved and acts as a DAMP-triggered plant defense-related transcriptional change (Chen et al., 2014). However, our studies showed the N-terminal region of TaPR1a, excluding the CAPE1 peptide, is required for the interaction between TaLTP3 and TaPR1a (Figures 5A,B). We also found that the interaction of TaLTP3 and TaPR1a occurs in the apoplast (Figure 5C). These observations implicated that TaLTP3 protein forms a complex with TaPR1a in the apoplast without occupying the CAPE1 peptide region of TaPR1a, suggesting that TaLTP3 might stabilize TaPR1a via physical interaction to potentiate the CAPE1-triggered plant immune response. Further studies to determine our hypothesis are needed.
Our findings showed that transgenic lines TaLTP3-OE and TaPR1a-OE exhibited improved resistance to leaf rust (Figure 1C; Bi et al., 2020) and increased accumulation of ROS upon P. syringae DC3000 infection (Figure 6 and Supplementary Tables 7, 8). ROS has been widely reported to have a direct antimicrobial effect and can be involved in cell wall stiffening. Moreover, ROS acts as local and systemic signal molecules triggering plant defense response (Waszczak et al., 2018). Our results indicated that both the TaPR1a and TaLTP3 may promote ROS accumulation during pathogen infection. A total of 330 LTP genes were identified from hexaploid genome of wheat and many of which were highly induced by abiotic stresses of drought and salinity (Fang et al., 2020). Close homologs in the LTP and PR1 gene families might also have functional redundancy. Future studies focusing on dissecting the redundant functions between LTP and PR1 genes are required.
In our previous study, we noticed that infection of model bacterial pathogen P. syringae DC3000 in wheat was sufficient to induce the expression of CAPE1-responsive genes TaAdi3 and TaPR7 (Bi et al., 2020). Therefore, we challenged the TaLTP3-OE line with model bacterial P. syringae for RNA-seq analysis. A specific upregulation of TaPR1a was detected in the transcriptome database by the KEGG annotation in TaLTP3-OE line compared to that in the WT plant (Table 1), suggesting that overexpression of TaLTP3 can affact the expression of TaPR1a that may explain the earlier induction of TaLTP3 than TaPR1a during wheat resistance to leaf rust mediated by the Lr39/41 gene (Figure 1A; Li et al., 2018). Furthermore, we found suppressions of key regulator genes in auxin and JA pathways (ARF9, A-ARR, and JAZ) in TaLTP3-OE transgenic line (Table 1). Both the ARF9 and A-ARR are negative regulators suppressing transcription of specific auxin-responsive genes (Leyser, 2006). JAZ proteins are repressors of JA signaling and inhibit the expression of JA-responsive genes by interaction with jasmonate-insensitive 1 (JIN1) transcription factor transcription factor (Chini et al., 2007). In addition, gene expressions of TaPAL and TaPAD4 in upstream of SA pathway were highlyinduced in TaLTP3-OE transgenic line (Figures 2B and 3). Taken together, we speculated that TaLTP3 acts as a positive regulator in the activation of crosstalk among SA, JA, and auxin. Although SA and JA are normally considered to control antagonistic pathways (Pieterse et al., 2009; Li et al., 2019), wheat plants seem to have a unique mechanism of hormone-mediated regulation of disease resistance. For instance, infection of spot blotch in resistance wheat genotype “Yangmai#6” elicited both the SA and JA signaling (Sahu et al., 2016). Spray treatments of both the SA and JA, but not ABA, on wheat leaves improved resistance to leaf rust (Kim et al., 2020).
Conclusion
We validated a positive role of TaLTP3 in plant defense response. Stable expression of TaLTP3 in transgenic wheat line improved resistance to leaf rust infection. Overexpression of TaLTP3 elevates the expression of TaPR1a and activates multiple plant defense-related hormone pathways including SA, JA, and auxin. A novel interaction between TaLTP3 and TaPR1a in the apoplast was identified and confirmed. TaLTP3 might play multiple roles in plant defense response, such as functioning with antimicrobe activities, forming a TaLTP3-TaPR1a complex in apoplast against pathogen and activating defense signaling. The presented findings have greatly expanded our knowledge on the molecular mechanism of PR genes during wheat resistance to pathogen infections. The generated transgenic wheat lines with enhanced defense responses may also serve as valuable resources for the genetic improvement of wheat resistance.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA746113.
Author Contributions
XW and LM designed the experiment. JZ and WB conducted most of the experiments. SZ, JS, and ML worked on the RT-qPCR validation and rust inoculation. XW drafted the manuscript with contributions from LM and XY. All the authors read and approved the final manuscript.
Funding
XW was supported by the Provincial Natural Science Foundation of Hebei (C2021204008), Independent Project of State Key Laboratory of North China Crop Improvement and Regulation (NCCIR2021ZZ-4), the Provincial Supporting Program of Hebei for the Returned Oversea Scholars (C20190180), and the Modern Agricultural Industry System Wheat Innovation Team in Hebei Province (HBCT2018010204). LM was supported by the “Hundred Talents Program” for the introduction of high-level overseas talents in Hebei Province (E2020100004). XY was supported by the Natural Science Foundation of Hebei Province (C2020204050) and the Scientific and Technological Research Projects of Higher Education of Hebei Province (ZD2019086).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Professor Genying Li (Shandong Academy of Agricultural Sciences, China) for the excellent support in generating transgenic wheat materials.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.771806/full#supplementary-material
Footnotes
References
Ali, S., Ganai, B. A., Kamili, A. N., Bhat, A. A., Mir, Z. A., Bhat, J. A., et al. (2018). Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 212, 29–37. doi: 10.1016/j.micres.2018.04.008
Appels, R., Eversole, K., Feuillet, C., Keller, B., Rogers, J., Stein, N., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. doi: 10.1126/science.aar7191
Bi, W., Zhao, S., Zhao, J., Su, J., Yu, X., Liu, D., et al. (2020). Rust effector PNPi interacting with wheat TaPR1a attenuates plant defense response. Phytopathol. Res. 2, 1–14.
Breen, S., Williams, S. J., Outram, M., Kobe, B., and Solomon, P. S. (2017). Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 22, 871–879. doi: 10.1016/j.tplants.2017.06.013
Chen, Y. L., Lee, C. Y., Cheng, K. T., Chang, W. H., Huang, R. N., Nam, H. G., et al. (2014). Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signaling in tomato. Plant Cell 26, 4135–4148. doi: 10.1105/tpc.114.131185
Chini, A., Fonseca, S., Fernández, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Fang, Z., He, Y., Liu, Y., Jiang, W., Song, J., Wang, S., et al. (2020). Bioinformatic identification and analyses of the non-specific lipid transfer proteins in wheat. J. Integr. Agr. 19, 1170–1185. doi: 10.1186/1471-2164-9-86
Gamir, J., Darwiche, R., Van’t Hof, P., Choudhary, V., Stumpe, M., Schneiter, R., et al. (2017). The sterol-binding activity of PATHOGENESIS-RELATED PROTEIN 1 reveals the mode of action of an antimicrobial protein. Plant J. 89, 502–509. doi: 10.1111/tpj.13398
Gietz, R. D., and Schiestl, R. H. (2007). High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34. doi: 10.1038/nprot.2007.13
Guo, L., Yang, H., Zhang, X., and Yang, S. (2013). Lipid transfer protein 3 as a target of MYB96 mediates freezing and drought stress in Arabidopsis. J. Exp. Bot. 64, 1755–1767. doi: 10.1093/jxb/ert040
Jacob, C., Panchal, S., and Melotto, M. (2017). Surface inoculation and quantification of Pseudomonas syringae population in the Arabidopsis leaf apoplast. Bio. Protoc. 7:e2167. doi: 10.21769/BioProtoc.2167
Jashni, M. K., Mehrabi, R., Collemare, J., Mesarich, C. H., and De Wit, P. J. (2015). The battle in the apoplast: further insights into the roles of proteases and their inhibitors in plant–pathogen interactions. Front. Plant. Sci. 6:584. doi: 10.3389/fpls.2015.00584
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30.
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Kim, M., Lee, A., Roh, Y. J., Lee, H. M., Jo, Y., Cho, H., et al. (2020). Gene Expression and Metabolomics Profiling of the Common Wheat Obtaining Leaf Rust Resistance by Salicylic or Jasmonic Acid through a Novel Detached Leaf Rust Assay. Agronomy 10:1668. doi: 10.3390/agronomy10111668
Lamari, L. (2008). Assess 2.0: image analysis software for plant disease quantification. St Paul: The American Phytopathological Society (APS).
Leyser, O. (2006). Dynamic Integration of Auxin Transport and Signalling. Curr. Biol. 16, R424–R433.
Li, N., Han, X., Feng, D., Yuan, D., and Huang, L.-J. (2019). Signaling Crosstalk between Salicylic Acid and Ethylene/Jasmonate in Plant Defense: Do We Understand What They Are Whispering? Int. J. Mol. Sci. 20:671. doi: 10.3390/ijms20030671
Li, X., Wang, X., Kang, Z., Ren, Z., Bi, W., Yang, W., et al. (2018). Suppression subtractive hybridization and microarray analysis reveal differentially expressed genes in the Lr39/41 -mediated wheat resistance to Puccinia triticina. Eur. J. Plant Pathol. 152, 479–492.
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21. doi: 10.1186/s13059-014-0550-8
Ma, L., Djavaheri, M., Wang, H., Larkan, N. J., Haddadi, P., Beynon, E., et al. (2018). Leptosphaeria maculans effector protein AvrLm1 modulates plant immunity by enhancing MAP kinase 9 phosphorylation. iScience 3, 177–191. doi: 10.1016/j.isci.2018.04.015
Ma, L., Lukasik, E., Gawehns, F., and Takken, F. L. (2012). The use of agroinfiltration for transient expression of plant resistance and fungal effector proteins in Nicotiana benthamiana leaves. Plant Fungal Pathogens 2012, 61–74.
Maldonado, A. M., Doerner, P., Dixon, R. A., Lamb, C. J., and Cameron, R. K. (2002). A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419, 399–403. doi: 10.1038/nature00962
McLaughlin, J. E., Al Darwish, N., Garcia-Sanchez, J., Tyagi, N., Trick, H. N., Mccormick, S., et al. (2020). A lipid transfer protein has antifungal and antioxidant activity and suppresses Fusarium head blight disease and DON accumulation in transgenic wheat. Phytopathology 111, 671–683. doi: 10.1094/PHYTO-04-20-0153-R
Molina, A., and García-Olmedo, F. (1993). Developmental and pathogen-induced expression of three barley genes encoding lipid transfer proteins. Plant J. 4:983. doi: 10.1046/j.1365-313x.1993.04060983.x
Paolacci, A. R., Tanzarella, O. A., Porceddu, E., and Ciaffi, M. (2009). Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 10:11. doi: 10.1186/1471-2199-10-11
Pieterse, C. M. J., Leon-Reyes, A., Van Der Ent, S., and Van Wees, S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. doi: 10.1038/nchembio.164
Qi, J., Wang, J., Gong, Z., and Zhou, J.-M. (2017). Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 38, 92–100. doi: 10.1016/j.pbi.2017.04.022
Sahu, R., Sharaff, M., Pradhan, M., Sethi, A., Bandyopadhyay, T., Mishra, V. K., et al. (2016). Elucidation of defense-related signaling responses to spot blotch infection in bread wheat (Triticum aestivum L.). Plant J. 86, 35–49. doi: 10.1111/tpj.13149
Salminen, T. A., Blomqvist, K., and Edqvist, J. (2016). Lipid transfer proteins: classification, nomenclature, structure, and function. Planta 244, 971–997.
Seo, P. J., and Park, C. M. (2010). MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 186, 471–483. doi: 10.1111/j.1469-8137.2010.03183.x
Seo, P. J., Xiang, F., Qiao, M., Park, J.-Y., Lee, Y. N., Kim, S.-G., et al. (2009). The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 151, 275–289. doi: 10.1104/pp.109.144220
Sun, J.-Y., Gaudet, D. A., Lu, Z.-X., Frick, M., Puchalski, B., and Laroche, A. (2008). Characterization and antifungal properties of wheat nonspecific lipid transfer proteins. Mol. Plant Microbe Interact. 21, 346–360. doi: 10.1094/MPMI-21-3-0346
Sung, Y.-C., Outram, M. A., Breen, S., Wang, C., Dagvadorj, B., Winterberg, B., et al. (2021). PR1-mediated defence via C-terminal peptide release is targeted by a fungal pathogen effector. New Phytol. 229, 3467–3480. doi: 10.1111/nph.17128
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., Van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Van Loon, L. C., Rep, M., and Pieterse, C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Wang, C., Huang, L., Buchenauer, H., Han, Q., Zhang, H., and Kang, Z. (2007). Histochemical studies on the accumulation of reactive oxygen species (O2– and H2O2) in the incompatible and compatible interaction of wheat-Puccinia striiformis f. sp. tritici. Physiol. Mol. Plant Pathol. 71, 230–239. doi: 10.1016/j.pmpp.2008.02.006
Wang, F., Yuan, S., Wu, W., Yang, Y., Cui, Z., Wang, H., et al. (2020). TaTLP1 interacts with TaPR1 to contribute to wheat defense responses to leaf rust fungus. PLoS Genet. 16:e1008713. doi: 10.1371/journal.pgen.1008713
Wang, F., Zang, X.-S., Kabir, M. R., Liu, K.-L., Liu, Z.-S., Ni, Z.-F., et al. (2014). A wheat lipid transfer protein 3 could enhance the basal thermotolerance and oxidative stress resistance of Arabidopsis. Gene 550, 18–26. doi: 10.1016/j.gene.2014.08.007
Wang, X., Bi, W., Gao, J., Yu, X., Wang, H., and Liu, D. (2018). Systemic acquired resistance, NPR1, and pathogenesis-related genes in wheat and barley. J. Integr. Agr. 17, 2468–2477. doi: 10.1016/s2095-3119(17)61852-5
Waszczak, C., Carmody, M., and Kangasjärvi, J. (2018). Reactive Oxygen Species in Plant Signaling. Annu. Rev. Plant Biol. 69, 209–236.
Wu, J., Gao, J., Bi, W., Zhao, J., Yu, X., Li, Z., et al. (2019). Genome-wide expression profiling of genes associated with the Lr47-mediated wheat resistance to leaf rust (Puccinia triticina). Int. J. Mol. Sci. 20:4498. doi: 10.3390/ijms20184498
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, 1–12.
Keywords: wheat, pathogenesis-related protein, lipid transfer protein, apoplastic, resistance
Citation: Zhao J, Bi W, Zhao S, Su J, Li M, Ma L, Yu X and Wang X (2021) Wheat Apoplast-Localized Lipid Transfer Protein TaLTP3 Enhances Defense Responses Against Puccinia triticina. Front. Plant Sci. 12:771806. doi: 10.3389/fpls.2021.771806
Received: 07 September 2021; Accepted: 19 October 2021;
Published: 25 November 2021.
Edited by:
Wen-Ming Wang, Sichuan Agricultural University, ChinaReviewed by:
Zhonglin Mou, University of Florida, United StatesPramod Prasad, Indian Institute of Wheat and Barley Research (ICAR), India
Copyright © 2021 Zhao, Bi, Zhao, Su, Li, Ma, Yu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaodong Wang, zhbwxd@hebau.edu.cn; Lisong Ma, lisong.ma@anu.edu.au
†These authors have contributed equally to this work
 Jiaojie Zhao1†
Jiaojie Zhao1† Lisong Ma
Lisong Ma Xiaodong Wang
Xiaodong Wang