- Key Laboratory of Integrated Management of Crop Diseases and Pests, Ministry of Education, Department of Plant Pathology, College of Plant Protection, Nanjing Agricultural University, Nanjing, China
Fatty acid metabolism is important for the maintenance of fatty acid homeostasis. Free fatty acids, which are toxic in excess, are activated by esterification with coenzyme A (CoA) and then subjected to β-oxidization. Fatty acid β-oxidation-related genes play critical roles in the development and virulence of several phytopathogens. In this study, we identified and characterized a peroxisomal-CoA synthetase in the rice blast fungus Magnaporthe oryzae, MoPCS60, which is a homolog of PCS60 in budding yeast. MoPCS60 was highly expressed during the conidial and early infectious stages and was induced under oleate treatment. Targeted deletion of MoPCS60 resulted in a significant reduction in growth rate when oleate and olive oil were used as the sole carbon sources. Compared with the wild-type strain Guy11, the ΔMopcs60 mutant exhibited fewer peroxisomes, more lipid droplets, and decreased pathogenicity. The distribution of MoPcs60 varied among developmental stages and was mainly localized to peroxisomes in the hyphae, conidia, and appressoria when treated with oleate. Our results suggest that MoPcs60 is a key peroxisomal-CoA synthetase involved in fatty acid β-oxidation and pathogenicity in rice blast fungi.
Introduction
Fatty acids have various biological functions in cell metabolism and signaling; they are also components of complex membrane lipids and act as precursors for the synthesis of bioactive lipids (Calder, 2015). Fatty acid metabolism is a multistep process critical for maintaining fatty acid homeostasis that involves the breakdown of fatty acids to produce energy. Fatty acid metabolism occurs via the well-characterized β-oxidation cycle to produce acetyl-coenzyme A (CoA), which is further metabolized to generate energy and precursors used in other metabolic pathways (Listenberger et al., 2003; van Roermund et al., 2003; Wysham et al., 2016). CoA is a pantothenic acid-containing coenzyme involved in the oxidation of fatty acids in the β-oxidation cycle, and also of pyruvate in the citric acid cycle. Free fatty acids are activated by esterification with CoA and then degraded by β-oxidation. Excess free fatty acids are toxic to cells (Rajvanshi et al., 2017).
Peroxisomes are involved in the degradation of fatty acids, especially long-chain fatty acids (Wang et al., 2007). Peroxisomes are single membrane-bound microbodies that present in all eukaryotic cells. Peroxisomes exert several effects according to environmental cues and metabolic requirements (Koch et al., 2010; Chen et al., 2016; Francisco et al., 2017). Peroxisomes play crucial roles in various catabolic processes including methanol and fatty acid metabolism, among other biosynthetic processes (Yan et al., 2005; Goh et al., 2011; Hagen et al., 2015; Chen et al., 2016; Li L. et al., 2017). Peroxisome biogenesis, matrix protein transport, and peroxisome-associated protein degradation are essential for cellular growth and differentiation (Lingard et al., 2009; Zhong et al., 2016). Peroxisome dysfunction in humans can be classified as deficiencies in a single peroxisomal enzyme, or peroxisomal biogenesis disorders such as Zellweger syndrome, neonatal adrenoleukodystrophy, and infantile Refsum disease (Subramani et al., 2000; Liu et al., 2012; Walter and Erdmann, 2019).
Magnaporthe oryzae is the causal agent of rice blast, a devastating disease affecting rice plants worldwide. M. oryzae is a filamentous fungus that exhibits high adaptability to external environments and can rapidly mobilize lipids via the MAP kinase and cAMP signaling pathways during appressorium development (Wang et al., 2005; Ramos-Pamplona and Naqvi, 2006; Patkar et al., 2012). Long-chain fatty acids are degraded by peroxisomal β-oxidation pathways to yield acetyl-CoA (which is released from triglycerides), glycerol (which is essential for appressorium turgor pressure), and energy (de Jong et al., 1997; Patkar et al., 2012). The organelles and biochemical pathways that break down fatty acids, such as peroxisomal β-oxidation and the glyoxylate cycle, are involved in the regulation of infection-related morphogenesis (Fernandez et al., 2014). Two conserved members of the Zn2-Cys6 family of transcriptional regulators, Far1 and Far2, are essential for lipid droplet mobilization during appressorium development and maturation (bin Yusof et al., 2014). The peroxisomal carnitine acetyl transferase activity protein Pth2, which is induced by acetate and lipids, maintains the acetyl-CoA pools necessary for appressorium function (Bhambra et al., 2006).
Peroxisomal-CoA synthetase 60 (Pcs60) belongs to the family of AMP-binding proteins (Fabian and Ralf, 1996). Pcs60 contains a C-terminal tripeptide peroxisomal targeting signal 1 (PTS1) motif, which is required for targeting peroxisomes via Pex5; its expression is induced under oleic acid growth conditions (Fabian and Ralf, 1996; Hagen et al., 2015). Furthermore, Pcs60 is an acyl-activating enzyme that converts oxalate to oxalyl-CoA and regulates responses to biotic and abiotic stresses in plants and yeast (Foster et al., 2012; Foster and Nakata, 2014; Frohlich et al., 2015; Mulleder et al., 2016; Yang et al., 2018). However, the function of Pcs60 proteins in filamentous fungi remains unknown. In this study, we characterized the function of M. oryzae Pcs60 (MoPcs60) in rice blast fungus and found that Pcs60 plays an important role in the utilization of long-chain fatty acids, peroxisome biosynthesis, lipid droplet degradation, and invasive hyphal (IH) growth in M. oryzae.
Materials and Methods
Fungal Strains and Culture Conditions
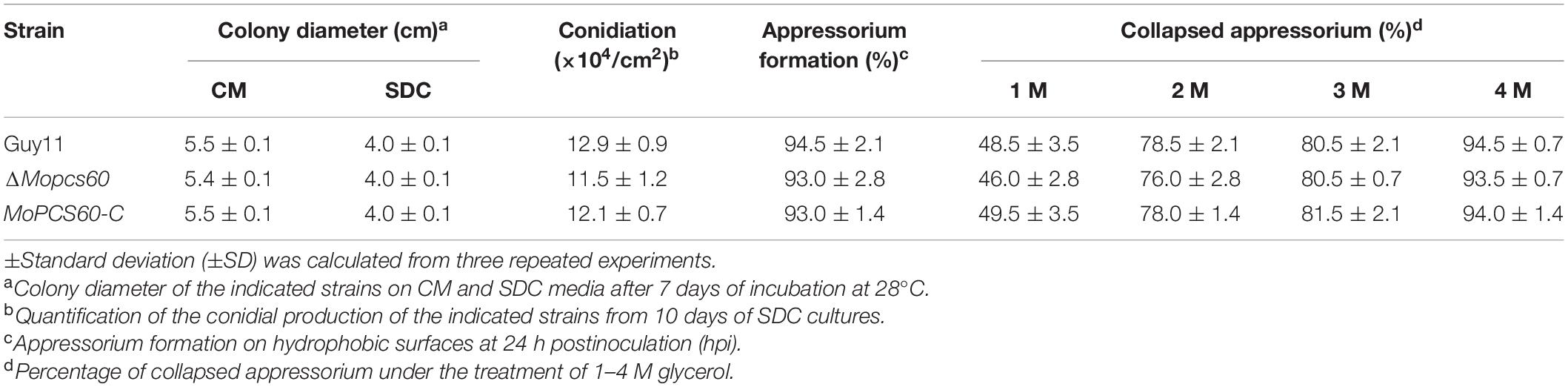
The M. oryzae Guy11 isolate was used as the wild-type strain throughout. The ΔMopcs60 mutant and the complemented strains MoPCS60-C were generated in this study. For vegetative growth assay, 3 mm × 3 mm agar blocks of the indicated strains were placed onto CM, SDC, and minimal media (MM) supplementing with 1% (w/v) glucose, 2% (w/v) olive, 1 mM sodium butyrate, sodium laurate, and sodium oleate as sole carbon source, respectively. All indicated strains were prior to incubation at 28°C for 7 days and photographed.
Complementation of the S. cerevisiae ΔScpcs60 Mutant
The cDNA of MoPCS60 was amplified using primers pYES2-F/pYES2-R (Supplementary Table 1) and then cloned into the yeast expression vector pYES2 plasmid (Invitrogen). Colonies were cultured on synthetic dropout (SD) medium without uracil. The empty pYES2 vector was transformed into the ΔScpcs60 mutant as a control. All strains were diluted to an OD600 = 0.1, after that 5 μl of 10-fold serial dilutions was cultured on YPG plates containing 6 μg/ml adriamycin (Lum et al., 2004) and 2% galactose at 30°C for 3 days.
MoPCS60 Gene Deletion and Complementation
To construct the MoPCS60 gene knockout vector, approximately 1-kb fragments franking the targeted gene were amplified with primers (Supplementary Table 1). Then, the PCR products were digested with SalI/EcoRI and BamHI/SacI and cloned into the pCX62 plasmid. The resulting gene knockout vector was transformed into protoplasts of Guy11. The hygromycin-resistant transformants were screened by PCR and verified by Southern blot (Supplementary Figure 1). To generate the complemented transformants, fragment containing the putative promoter and MoPCS60 encoding region was amplified and cloned into pYF11 (bleomycin resistance) using the yeast gap repair approach (Bruno et al., 2004). The resulting vector was sequenced and transformed into protoplasts of the ΔMoPcs60 mutant.
Quantification of Gene Expression by qRT-PCR
Total RNA samples were extracted from mycelia, conidia, and infected rice leaves using PureLink™ RNA mini kit (Invitrogen) according to the manufacturer’s protocol. Reverse transcriptase HiScript III RT SuperMix for qPCR (Vazyme Biotech Co., Ltd., Nanjing, China) was used to prepare cDNA. qRT-PCR was run on an Applied Biosystems (Foster City, CA, United States) 7500 Real-time PCR System with SYBR Premix Ex Taq (Vazyme Biotech Co., Ltd., Nanjing, China). The 2–ΔΔCT method was used to calculate the relative quantification of each transcript (Livak and Schmittgen, 2001) with the M. oryzae actin gene as the internal control. The experiment was repeated three times with three biological replicates. The primers used in this section are listed in Supplementary Table 1.
Conidiation, Appressorium Formation, and Turgor Analysis
For conidiation, 10-day-old SDC agar cultures were washed with 3 ml H2O and counted with a hemocytometer (Zhang et al., 2010). For appressorium formation assays, 25 μl of conidial suspensions (5 × 104 spores/ml) was placed onto a hydrophobic surface (Fisher brand, Germany) and cultured at 28°C. Appressorium formation rate was counted at 24 hpi under a microscope (Zhang et al., 2011; Liu et al., 2018). Appressorium turgor was measured as previously described (Zhong et al., 2016).
Observation of Lipid Droplet Degradation and Glycogen Translocation During Appressorial Development
Conidial suspensions (25 μl) were incubated on hydrophobic surface. The lipid droplets during appressoria development were stained with Nile red solution as previously described (Zhong et al., 2016). The glycogen during appressorial development was stained with KI solution as previously described (Zhong et al., 2016). The samples were observed under a Zeiss Axio Observer A1 inverted microscope after 3-min incubation.
Pathogenicity, Aniline Blue, and 3,3′-Diaminobenzidine Staining Assays
For spraying assay, 2-week-old rice seedlings (Oryza sativa cv. CO39) were sprayed with 4 ml of conidial suspensions (1 × 105 spores/ml, 0.2% gelatin, w/v) of each strain. The inoculated plants were placed into a moist chamber at 28°C for 24 h and then transferred to another moist chamber under a 12-h light/12-h dark photoperiod for 4 days and photographed (Zhang et al., 2014; Zhong et al., 2016). For detached barley assay, three droplets of conidial suspensions (20 μl, 1 × 105 spores/ml) were drop-inoculated onto 7-day-old detached barley leaves (Hordeum vulgare cv. Four-arris) for 5 days and photographed (Zhong et al., 2016). For infectious growth observation, detached rice sheaths were injected with 1 ml of conidial suspensions (1 × 105 spores/ml) and placed into a moist chamber at 28°C. Infectious hyphae (IH) types were examined under a confocal microscope (Zeiss LSM710, 63 × oil) (Liu et al., 2016). Papillary callose deposits were examined by staining with solution (0.067 M K2HPO4, pH = 9.0) containing 0.05% (w/v) aniline blue (Sigma) at 32 hpi. ROS accumulation was examined by staining with 3,3′-diaminobenzidine (DAB, Sigma-Aldrich, 1 mg/ml) solution (pH = 3.8) for 12 h and destained with a solution (ethanol/acetic acid = 98:2, v/v) for 2 h at 24 hpi.
Subcellular Localization, Peroxisome, and Lipid Droplet Observation
To investigate the cellular localization, MoPCS60-GFP and RFP-Pts1 (a fluorescent marker appended with a type I peroxisomal targeting signal) were constructed and cotransformed into the ΔMopcs60 mutants. The resulting transformants were observed under a confocal fluorescence microscope (Zeiss LSM710, 63 × oil) (Zhong et al., 2016). For peroxisome quantity and morphology observation, RFP-Pts1 was transformed into the wild-type Guy11, ΔMopcs60 mutant and the complemented strain MoPCS60-C, and observed under a confocal fluorescence microscope (Zhong et al., 2016). For lipid droplet staining, vegetative hyphae were cultured in liquid CM for 16 h and then stained with BODIPY™ 493/503 (1 μg/ml) in the dark at 25°C for 3 min (Rajvanshi et al., 2017). Fluorescent images of peroxisomes and lipid droplets were taken under a confocal fluorescence microscope (Zeiss LSM710, 63 × oil).
Results
MoPcs60 Is a Homolog of Pcs60 in Budding Yeast
First, we used the Pcs60 peptide sequence from budding yeast as the query in a BlastP search within the M. oryzae genome database in FungiDB1. We identified a Pcs60 homolog (MGG_06199) that shared 49% amino acid similarity with Pcs60, which we named MoPcs60. MoPCS60 encodes a peroxisomal-CoA synthetase comprising 522 amino acids, which contains two conserved AMP-binding domains (Fabian and Ralf, 1996) and a C-terminal PTS1 tripeptide motif required for peroxisomal targeting (Hagen et al., 2015). Our phylogenetic analysis revealed that Pcs60 is well-conserved across various organisms including yeast, filamentous fungi, plants, mammals, and homo sapiens (Figure 1A). Heterologous complementation assays revealed that MoPCS60 rescued the phenotype of a Saccharomyces cerevisiaeΔpcs60 mutant exposed to adriamycin (Figure 1B), indicating that MoPcs60 is a homolog of yeast Pcs60. Moreover, MoPCS60 expression was higher during the conidial (2.8-fold) and early infectious (2.9-fold) stages compared with in the mycelial stage (Figure 1C), indicating a potential role of MoPCS60 in conidiation and infection in rice blast fungus.
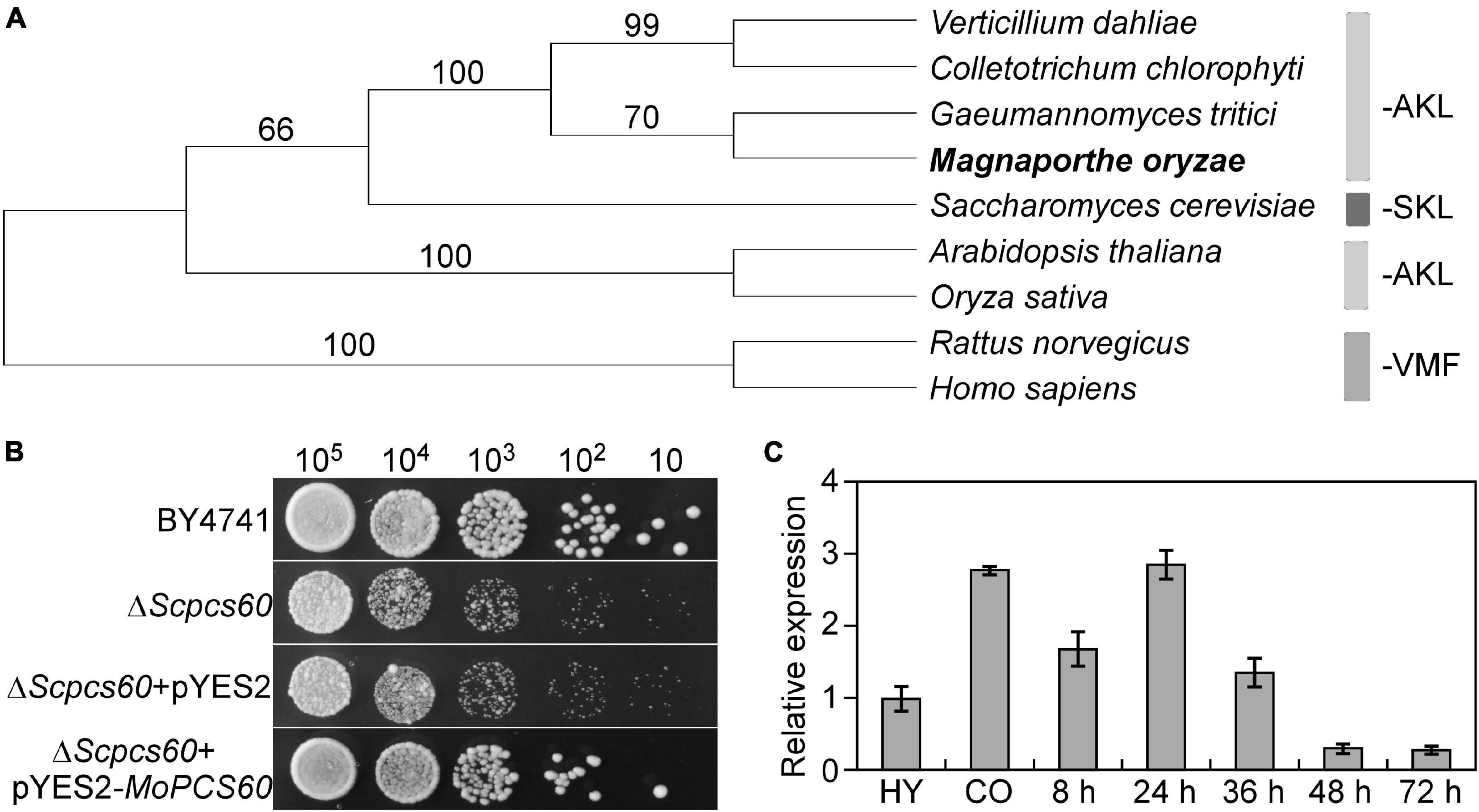
Figure 1. Phylogenetic analysis, yeast complementation, and expression profiles of MoPCS60. (A) Phylogenetic trees of MoPcs60 proteins from different organisms were constructed using the CLUSTAL_W and MEGA 5.1 programs by the neighbor-joining method with 1000 bootstrap replicates. Species names and GenBank accession numbers are as follows: XP_003712086.1 (M. oryzae); OLN86417.1 (C. chlorophyti); XP_009223944.1 (G. tritici R3-111a-1); XP_009649893.1 (V. dahliae); NP_001438.1 (H. sapiens); NP_075243.1 (R. norvegicus); NP_001054304.1 (O. sativa); NP_190468.1 (A. thaliana); NP_009781 (S. cerevisiae). AKL and SKL: major PTS1 tripeptides, VMF: minor PTS1 tripeptides. The rice blast fungus was showed in bold. (B) MoPcs60 partially rescued the growth defect of the yeast ΔScpcs60 mutant. Serial dilutions of BY4741, ΔScpcs60, and ΔScpcs60 transformed with pYES2 or pYES2-MoPCS60 were grown on YPG (galactose) with 6 μg/ml adriamycin plates at 30°C for 3 days and then photographed. (C) qRT-PCR analyses the expression levels of MoPCS60 at different fungal developmental stages. RNA was extracted from hyphae cultured for 36 h (HY), conidia (CO), and rice leaves inoculated with the wild-type conidia for 8, 24, 36, 48 and 72 h, respectively. Error bars represent the standard deviations of three replicates.
MoPcs60 Is Not Involved in Vegetative Growth, Conidiation, or Appressorium Formation
To investigate the biological function of MoPcs60 in M. oryzae, we generated a MoPCS60 deletion mutant by replacing the MoPCS60 coding region with the hygromycin phosphotransferase gene, through polyethylene glycol-mediated protoplast transformation. The MoPCS60 gene deletion in the ΔMopcs60 mutant was confirmed by Southern blot analysis (Supplementary Figure 1). The complemented transformant strain MoPCS60-C was generated by transforming a genetic construct containing MoPCS60 fused to green fluorescent protein (MoPCS60-GFP) into ΔMopcs60 protoplasts. The vegetative growth, conidial production, and appressorium formation phenotypes of ΔMopcs60 and the complemented transformant strain MoPCS60-C were similar to those of the wild-type strain Guy11 (Table 1), which suggests that MoPcs60 plays no role in vegetative growth, conidiation, or appressorium formation in M. oryzae.
MoPcs60 Is Important for Infectious Hyphal Growth
To examine the role of MoPcs60 in fungal pathogenicity, conidial suspensions of Guy11, ΔMopcs60, and MoPCS60-C were sprayed onto susceptible rice seedlings. After 5 days of incubation, numerous typical lesions had formed on rice leaves inoculated with Guy11 and MoPCS60-C. By contrast, many smaller, restricted lesions had formed on rice leaves inoculated with the ΔMopcs60 mutant (Figure 2A). The lesions on the rice leaves were graded based on a “lesion-type” assay (Liu et al., 2018). We found that the ΔMopcs60 mutant formed more type 1, and fewer type 2 and type 3 lesions compared with Guy11 and MoPCS60-C (Figure 2A); no type 4 or type 5 lesions were present on leaves inoculated with ΔMopcs60. We then performed a barley infection assay, which showed that the pathogenicity was reduced in ΔMopcs60 compared with Guy11 and MoPCS60-C, as shown by the smaller lesions (Figure 2B). Furthermore, we examined the penetration and IH growth of ΔMopcs60, Guy11, and MoPCS60-C in rice sheath cells. More than 60% of the rice cells showed type 4 IH growth when inoculated with Guy11 and MoPCS60-C conidia, whereas over 70% of rice cells showed type 1, type 2, or type 3 IH growth when inoculated with ΔMopcs60 conidia (Figure 2C). In addition, we examined endogenous reactive oxygen species (ROS) production in infected cells and found that there was no significant difference in ROS production following infection with Guy11 or ΔMopcs60 (Supplementary Figures 2A,B). These results suggest that MoPcs60 plays an important role in infectious hyphal growth.
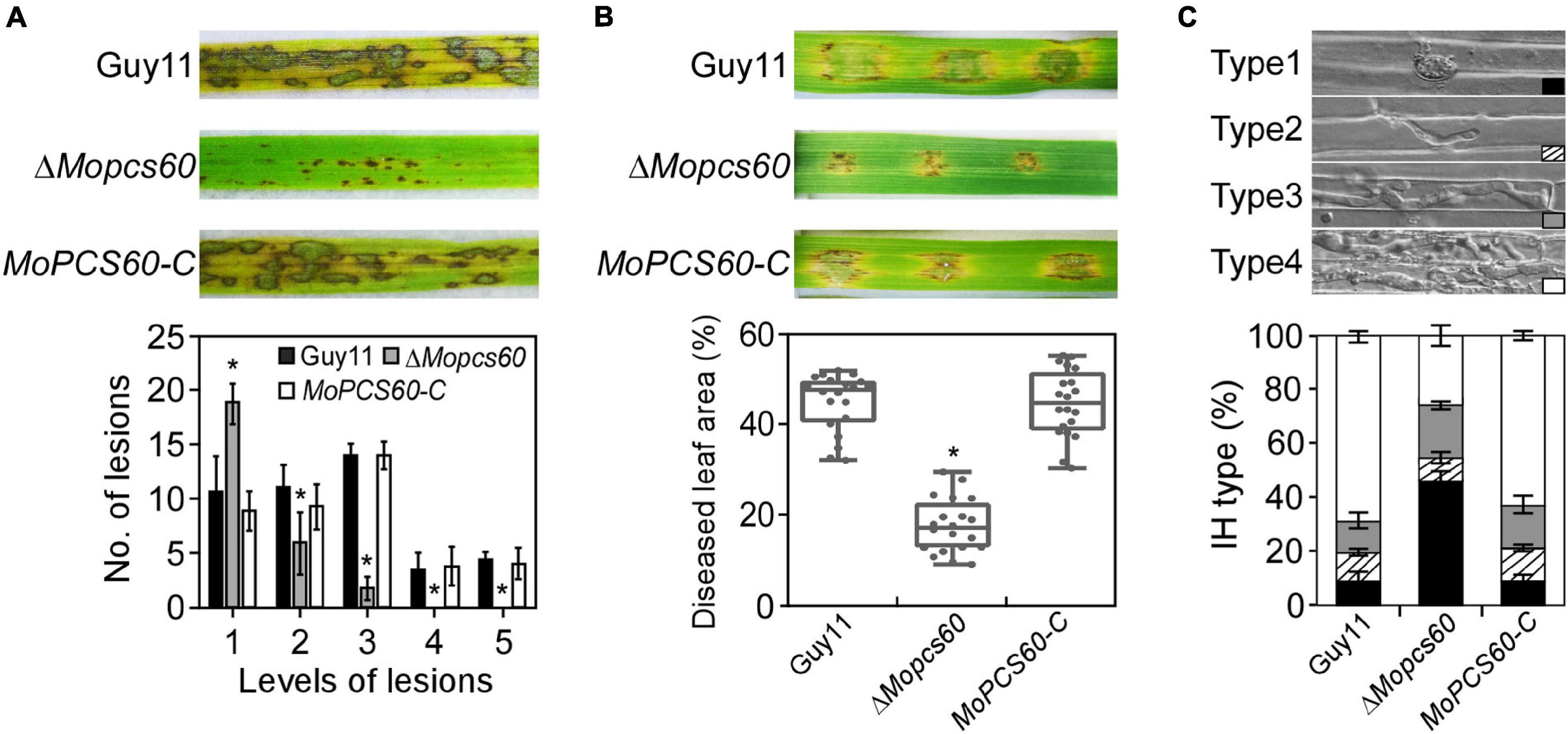
Figure 2. The ΔMopcs60 mutant is defective in pathogenicity. (A) Conidial suspensions were sprayed onto rice seedlings and examined at 5 dpi (days postinoculation) (Top). Lesion types were counted within an area of 4 cm2 leaf from each strain (Bottom). Error bars represent the standard deviations of three replicates; asterisk represents significant difference (*p < 0.01). (B) Conidial suspensions were inoculated onto detached barley leaves and examined at 5 dpi (Top). Diseased leaf area (Bottom) was analyzed by ImageJ (Media Cybernetics Inc., Shanghai, China). The percentage was represented by a box-dot-plot. Statistical analysis was performed using GraphPad Prism 8.0.1, and asterisk represents significant difference (*p < 0.01). (C) Detached rice sheaths were injected with conidial suspensions. IH types were counted and statistically analyzed at 32 hpi. Error bars represent the standard deviations of three replicates.
The Pathogenicity of ΔMopcs60 Is Not Due to Appressorium Turgor or Penetration
Appressorium-mediated penetration of M. oryzae is a turgor-driven process associated with substantial glycerol accumulation in the infected cell (Ramos-Pamplona and Naqvi, 2006). To investigate whether the reduced pathogenicity of ΔMopcs60 was associated with appressorium-mediated penetration, we first determined the rate of appressorium collapse in the presence of various hyperosmotic concentrations of glycerol by performing an incipient cytorrhysis assay. We found that there was no significant difference in appressorium turgor between ΔMopcs60 and Guy11 (Table 1), which suggests that MoPcs60 is not necessary for the generation or maintenance of appressorium turgor.
Glycogen translocation and lipid degradation are required for appressorial maturation and appressorium-mediated host penetration (Wang et al., 2005; Zhong et al., 2016; Li X. et al., 2017). Therefore, we examined the cellular degradation of lipid droplets during appressorium development by Nile red staining. Lipid droplets were gradually degraded during appressorial maturation, and there was no difference in intracellular lipid storage between ΔMopcs60 and Guy11 (Supplementary Figure 3A). Moreover, we examined glycogen storage by iodine solution staining and found that glycogen translocation was not significantly different between ΔMopcs60 mutant and Guy11 (Supplementary Figure 3B). These results indicated that MoPcs60 is not necessary for lipid droplet degradation or glycogen translocation in the appressorium.
Next, we evaluated the penetration pegs using aniline blue staining, which stains papillary callose deposits (Sun et al., 2006; Shi et al., 2021). The callose deposits formed by ΔMopcs60 were not significantly different to those of Guy11 at 32 h postinoculation (Figures 3A,B). These results indicated that the pathogenicity of ΔMopcs60 was not associated with appressorium turgor or penetration.
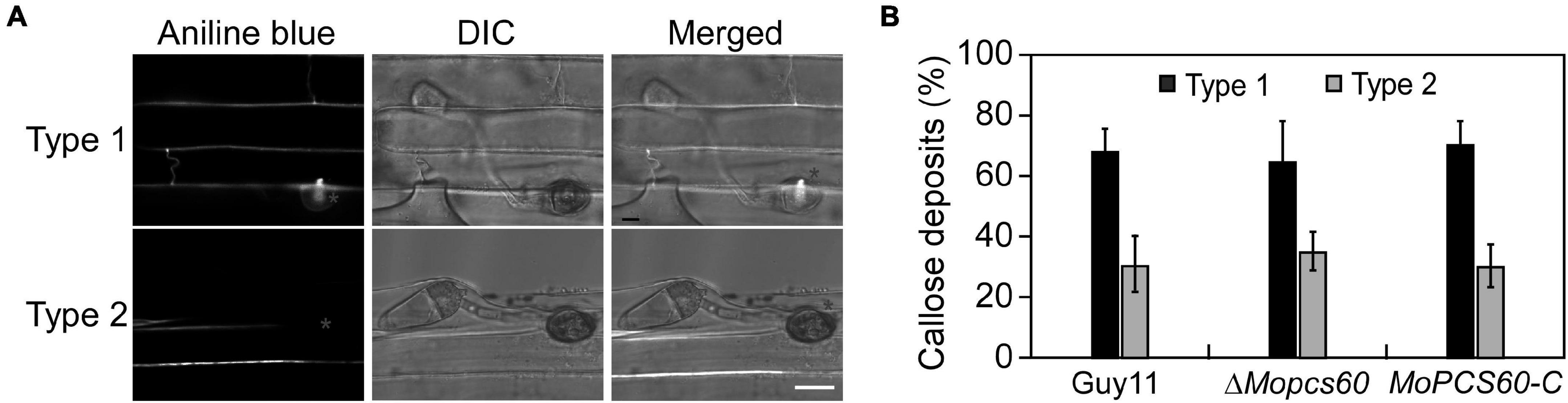
Figure 3. Papillary callose deposits are not changed in the ΔMopcs60 mutant. (A) Aniline blue staining to visualize appressorium of the wild-type Guy11, ΔMopcs60 mutant, and the complemented transformant MoPCS60-C at 32 hpi on rice leaves. Bar = 5 μm. (B) Statistically analyzed the number of papillary callose deposits formed underneath appressoria. Error bars represent the standard deviations of three replicates.
MoPcs60 Is Involved in the Utilization of Long-Chain Fatty Acids
To investigate the role of MoPcs60 in fatty acid metabolism, we inoculated minimal medium plates containing short-chain (butyrate), medium-chain (laurate), or long-chain (oleate and olive oil) fatty acids as the sole carbon source with Guy11, ΔMopcs60, or MoPCS60-C. After 7 days of incubation, the growth rate of the ΔMopcs60 mutant was significantly lower on oleate and olive oil plates compared with Guy11 and MoPCS60-C. There was no significant difference in growth rate among the three tested strains on butyrate or laurate plates (Figures 4A,B). Our results suggested that MoPcs60 is involved in long-chain fatty acid utilization.
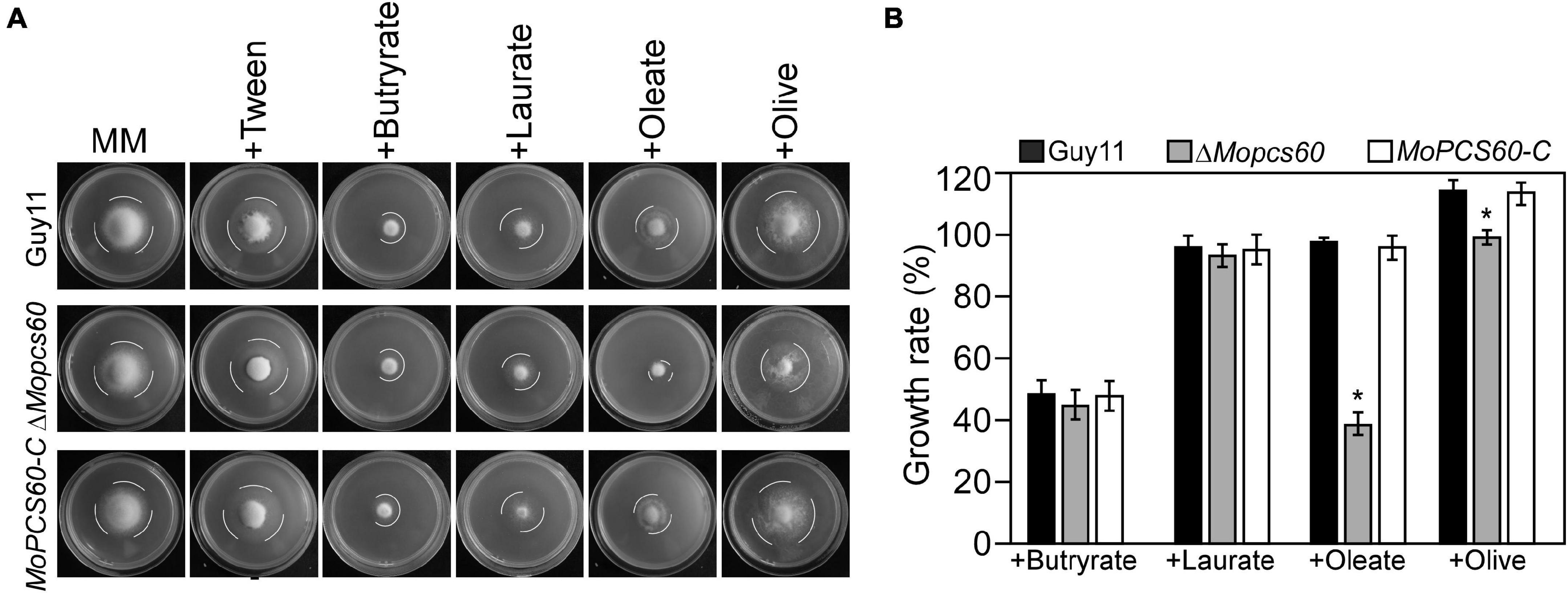
Figure 4. The ΔMopcs60 mutant shows defect in utilization of long-chain fatty acids. (A) The wild-type strain Guy11, ΔMopcs60 mutant, and the complemented transformant MoPCS60-C were grown on minimal medium and MM with butyrate (1 mM), laurate (1 mM), olive (1% w/v), and oleate (1 mM) as the sole carbon sources and incubated at 28°C for 7 days. (B) Statistical analysis of the growth rate of the indicated strains on different media. Error bars are standard deviations from three biological repeats, and asterisk represents significant differences at *p < 0.01.
MoPCS60 Deletion Reduces Peroxisome Quantity and Increases the Number of Lipid Droplets
Peroxisomes are ubiquitous organelles intimately involved in fatty acid metabolism (Yan et al., 2005; Wang et al., 2007). To investigate whether the dysfunctional fatty acid metabolism in ΔMopcs60 was related to peroxisomes, we examined the morphology and quantity of peroxisomes in Guy11, ΔMopcs60, and MoPCS60-C, all of which expressed red fluorescent protein (RFP) fused to a “PTS1 targeting peptide” (RFP-Pts1). We found that the number of peroxisomes was markedly lower in the ΔMopcs60 mutant compared with the other two strains (Figures 5A,B). Defective fatty acid metabolism causes an increase in non-polar lipid content (Rajvanshi et al., 2017). Therefore, we quantified the lipid droplets and examined their morphology in Guy11, ΔMopcs60, and MoPCS60-C by staining with BODIPY™ 493/503; the number of droplets was significantly higher in the ΔMopcs60 mutant than the other two strains (Figures 5C,D). However, the morphologies of the peroxisomes and lipid droplets were similar among all three strains examined (Figures 5A,C). These results suggest that MoPcs60 plays a critical role in peroxisome biogenesis and lipid droplet degradation in rice blast fungus.
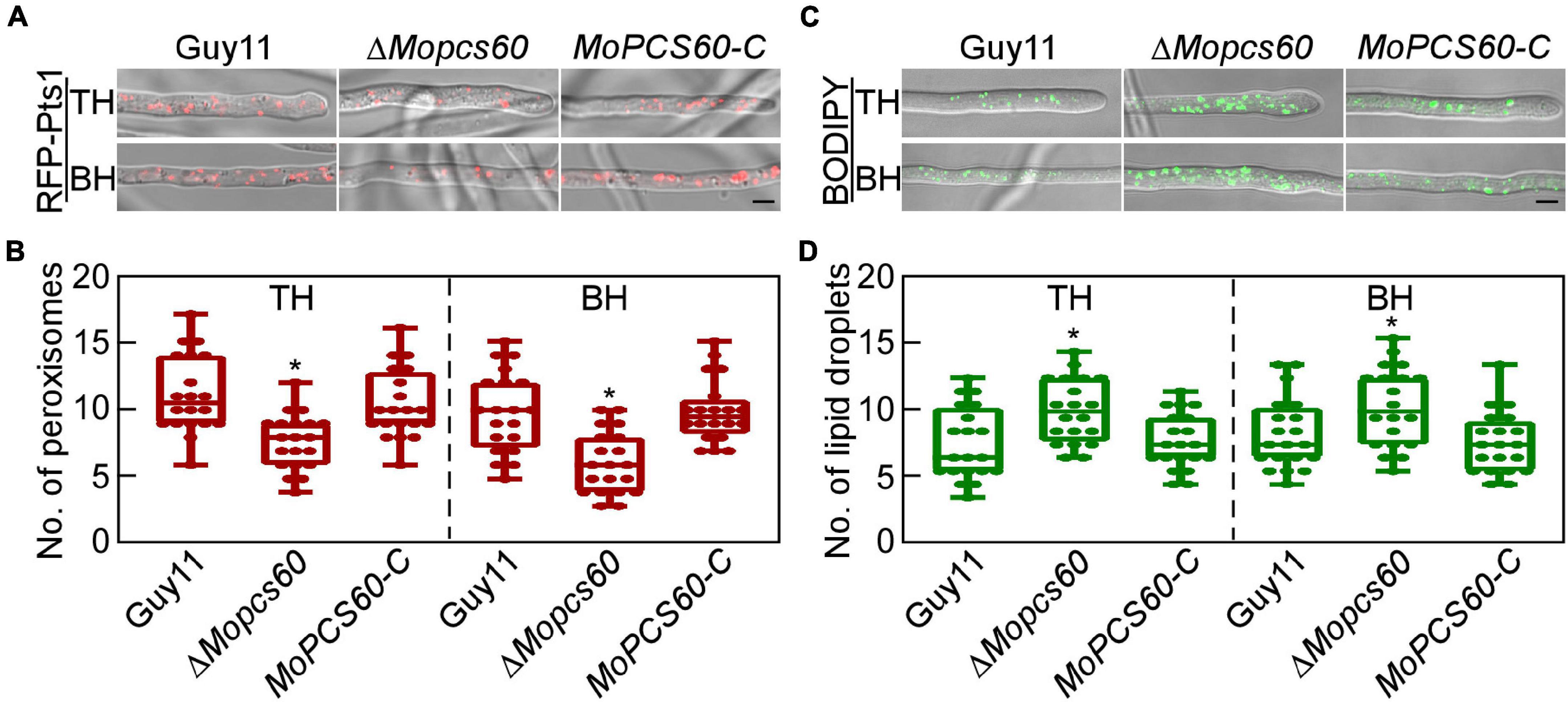
Figure 5. The ΔMoPcs60 mutant shows a decreased number of peroxisomes and an increased number of lipid droplets. (A) Peroxisomes were examined in tip (TH) and basal (BH) hyphae of Guy11, ΔMopcs60, and MoPCS60-C strains expressing RFP-Pts1 protein. Differential interference contrast (DIC) and fluorescent images were taken under a confocal microscope. Bar = 5 μm. (B) Statistically analyzed the number of peroxisomes in 1 × 103 μm from 20 fields of view in each strain and represented by a box-dot-plot. Statistical analysis was performed using GraphPad Prism 8.0.1, and asterisks represent significant differences at *p < 0.01. (C) Lipid droplets were examined in TH and BH of Guy11, ΔMopcs60, and MoPCS60-C strains stained by BODIPY 493/503. Bar = 5 μm. (D) Statistically analyzed the number of lipid droplets in 1 × 103 μm from 20 fields of view in each strain and represented by a box-dot-plot. Statistical analysis was performed using GraphPad Prism 8.0.1, and asterisks represent significant differences at *p < 0.01.
MoPcs60 Localizes to Peroxisomes Under Oleate Induction
To investigate the subcellular localization of MoPcs60, we generated a ΔMopcs60 mutant strain that coexpressed MoPcs60-GFP and RFP-Pts1, which we examined using confocal microscopy. MoPcs60-GFP was distributed throughout the cells, being present in the hyphae, conidium, appressorium, and IH (Figures 6A,B). Due to the involvement of MoPcs60 in oleate metabolism, we examined the localization of MoPcs60-GFP following oleate treatment. We found that MoPcs60-GFP was mainly distributed in the punctate structures, which colocalized with RFP-Pts1 in the hyphae, conidium, and appressorium under oleate treatment (Figures 6A,B). In addition, we analyzed the expression levels of MoPCS60 under oleate and olive oil treatment and found that MoPCS60 expression was greater than 2-fold higher under the long-chain fatty acid treatment (Figure 6C). These results indicated that the expression and localization of MoPcs60 were related to oleate metabolism in M. oryzae.
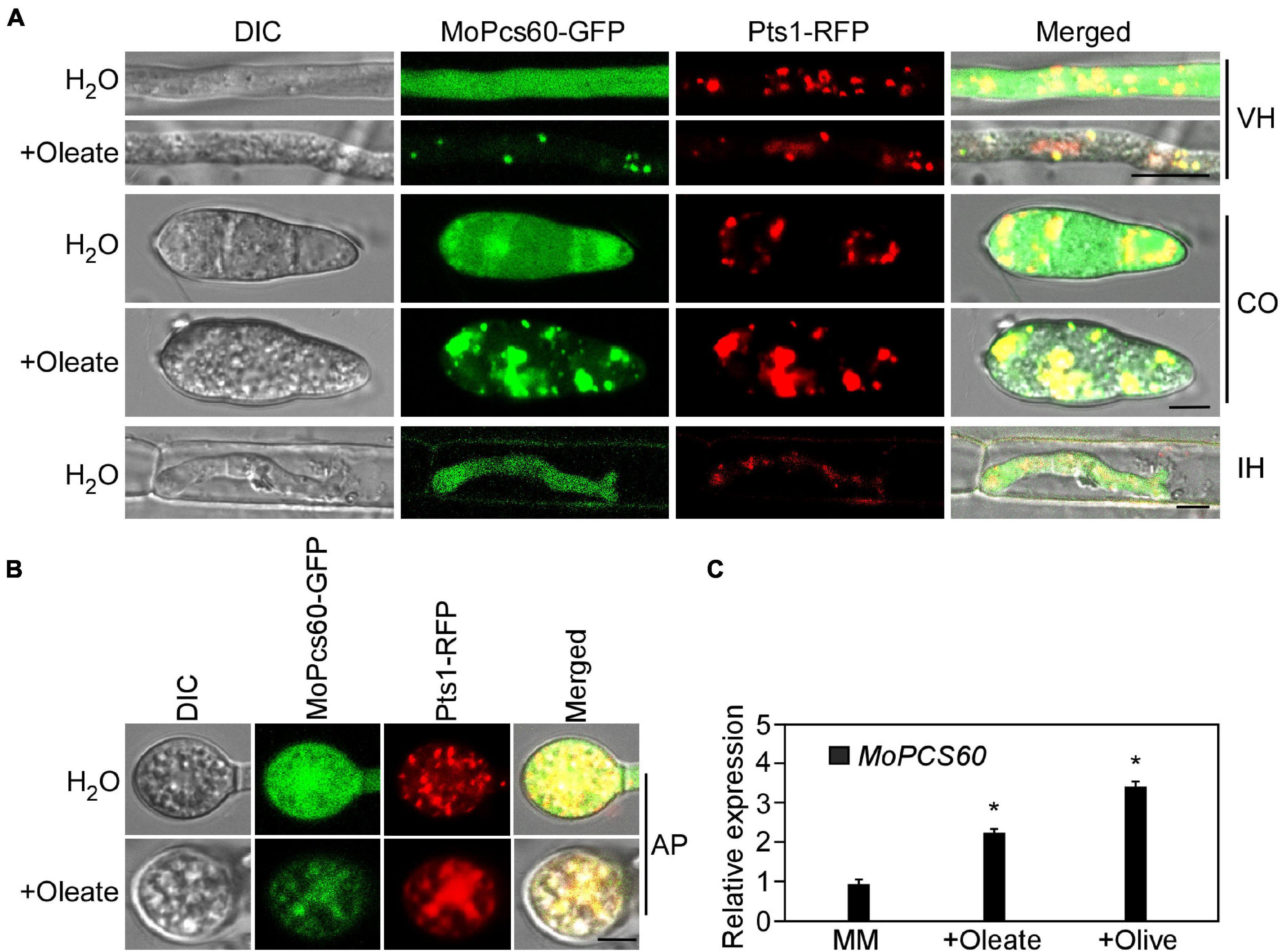
Figure 6. MoPcs60 localizes to peroxisomes under oleate induction. (A,B) Hypha, conidium, invasive hyphae, and appressorium of the MoPCS60-C strain coexpressing MoPCS60-GFP and RFP-Pts1 with or without oleate treatment were observed under a confocal microscope. VH, vegetative hyphae; CO, conidium; IH, invasive hyphae; AP, appressorium. Bar = 5 μm. (C) qRT-PCR analyses the expression levels of MoPCS60 in wild-type Guy11 with or without oleate and olive treatment. Error bars are standard deviations from three biological repeats, and asterisk represents significant difference (*p < 0.01).
Discussion
In this study, we identified and characterized the peroxisomal-CoA synthetase MoPcs60 in M. oryzae and found that it plays an important role in long-chain fatty acid utilization, peroxisome biosynthesis, lipid droplet degradation, and invasive growth. Our findings demonstrated that fatty acid metabolism mediated by MoPcs60 is critical for the development and pathogenicity of M. oryzae, and provide new insight into the fundamental molecular relationship between fatty acid metabolism and phytopathogen pathogenicity.
Free fatty acids are activated by esterification with CoA and then oxidized and metabolized (Daum et al., 2007). Fatty acid metabolism is critical for the maintenance of fatty acid and energy homeostasis, and excess free fatty acids are toxic (Carballeira, 2008; Rajvanshi et al., 2017). Indeed, fungal strains with dysfunctional fatty acid metabolism exhibit growth, peroxisome biogenesis, and mitochondrial defects when grown on fatty acid-supplemented media. In M. oryzae, an MoMFP1 (FOX2, a multifunctional enzyme) deletion prevented the utilization of fatty acids as the sole carbon source and attenuated M. oryzae virulence (Wang et al., 2007). Furthermore, the fatty acid synthase β-subunit dehydratase Fas1 is essential for peroxisome biogenesis, β-oxidation, and pathogenesis (Sangappillai and Nadarajah, 2020). Consistent with these findings, our data showed that deletion of MoPCS60 led to defects in the utilization of long-chain fatty acids, peroxisome biogenesis, and infectious growth in M. oryzae. Moreover, lipid catabolism genes are highly expressed during the early stages of infection in the phytopathogen Blumeria graminis (Both et al., 2005). Taken together, these findings suggest that fatty acid metabolism is important for fungal pathogenesis.
In M. oryzae, lipid droplets move to the appressorium following conidial germination. Lipid droplet degradation is associated with glycerol accumulation, which is required for generating turgor pressure during penetration (Wang et al., 2007; Sangappillai and Nadarajah, 2020). Although the ΔMopcs60 mutant exhibited defects in fatty acid metabolism and virulence, the mobilization and breakdown of lipid droplets during turgor generation were not impaired in the appressorium; this is in contrast to the mutants ΔMomfp1, Δfar1, Δfar2, and Δfar1Δfar2 (Wang et al., 2007; bin Yusof et al., 2014). This indicates that MoPcs60 is not involved in lipid droplet mobilization during appressorium development, and that MoPcs60 is not essential for appressorium-mediated infection in M. oryzae. In addition, there were no apparent differences in callose deposits or ROS accumulation between the wild-type and mutant strains, indicating that MoPcs60 was not involved in penetration peg formation or host immunity suppression. Therefore, MoPcs60 deletion likely disrupted CoA synthesis and peroxisome biogenesis, which in turn prevented the oxidation of free fatty acids, causing fatty acid toxicity in IH.
Peroxisomal-CoA synthetase 60 is a PTS1-containing protein that can be induced under oleic acid growth conditions in S. cerevisiae (Fabian and Ralf, 1996). In contrast to the yeast Pcs60, which is constitutively expressed and localized in peroxisomes, MoPcs60 should be treated with oleate and is distributed outside of peroxisomes. This disparity in expression and localization suggests that the function of Pcs60 differs between S. cerevisiae and M. oryzae. In addition, MoPCS60 expression levels were high under oleate treatment and were comparable to the MoPCS60 expression levels observed during the early stage of infection. Therefore, MoPcs60 might play a role in the response to host fatty acids during infectious growth. However, MoPcs60 is mainly distributed in the cytosol of IH, which suggests that there are unknown MoPcs60-related regulatory mechanisms at play during infection. MoPcs60 functions as an acyl-activating enzyme involved in oxalic acid metabolism in yeast and plants (Foster et al., 2012; Foster and Nakata, 2014; Xian et al., 2020); such potential roles for MoPcs60 should be investigated in M. oryzae.
Conclusion
We demonstrated that the peroxisomal-CoA synthetase MoPcs60 plays a critical role in fatty acid metabolism, peroxisome biosynthesis, lipid droplet degradation, and invasive growth in M. oryzae. MoPcs60 mainly localizes to peroxisomes in the hyphae, conidia, and appressorium in response to extracellular oleate. These results provide new insight into the pathogenic mechanisms of M. oryzae.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
TZ, ZZ, and HZ designed the research and wrote the manuscript. TZ, YL, XL, WG, EM, and RZ performed the experiments. TZ, HZ, and XZ analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (grant numbers 31871912 and 32061143045 to HZ) and Fundamental Research Funds for the Central Universities (grant number KYZ201816 to HZ).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.811041/full#supplementary-material
Supplementary Figure 1 | Targeted deletion of MoPCS60 in M. oryzae. (A) MoPCS60 gene replacement strategy in M. oryzae genome. (B) MoPCS60 gene knockout mutants were verified by Southern blot analysis with probe 1 (gene fragment) and probe 2 (HPH), respectively. The genomic DNA was digested with EcoRI.
Supplementary Figure 2 | MoPcs60 is not involved in scavenging host-derived ROS. (A) DAB staining to visualize infection hyphae of the wild-type Guy11, ΔMopcs60 mutant, and the complemented transformant MoPCS60-C at 24 hpi in infected cells. Bar = 100 μm. (B) Number of no reddish-brown precipitate in infected cells was counted and analyzed. Error bars represent the standard deviations of the three replicates.
Supplementary Figure 3 | MoPcs60 is not necessary for lipid droplets degradation and glycogen translocation during appressorium development. (A) Lipid droplets of conidia and appressoria were stained by Nile red at different time points and observed under a fluorescence microscope. Error bars represent the standard deviations of three replicates. Bar = 20 μm. (B) Glycogen deposits of conidia and appressoria were stained by iodine solution at different time points and observed under a fluorescence microscope. Error bars represent the standard deviations of three replicates. Bar = 20 μm.
Footnotes
References
Bhambra, G. K., Wang, Z. Y., Soanes, D. M., Wakley, G. E., and Talbot, N. J. (2006). Peroxisomal carnitine acetyl transferase is required for elaboration of penetration hyphae during plant infection by Magnaporthe grisea. Mol. Microbiol. 61, 46–60. doi: 10.1111/j.1365-2958.2006.05209.x
bin Yusof, M. T., Kershaw, M. J., Soanes, D. M., and Talbot, N. J. (2014). FAR1 and FAR2 regulate the expression of genes associated with lipid metabolism in the rice blast fungus Magnaporthe oryzae. PLoS One 9:e99760. doi: 10.1371/journal.pone.0099760
Both, M., Csukai, M., Stumpf, M. P. H., and Spanu, P. D. (2005). Gene expression profiles of Blumeria graminis indicate dynamic changes to primary metabolism during development of an obligate biotrophic pathogen. Plant Cell 17, 2107–2122. doi: 10.1105/tpc.105.032631
Bruno, K. S., Tenjo, F., Li, L., Hamer, J. E., and Xu, J. R. (2004). Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot. Cell 3, 1525–1532. doi: 10.1128/EC.3.6.1525-1532.2004
Calder, P. C. (2015). Functional roles of fatty acids and their effects on human health. JPEN J. Parenter. Enteral Nutr. 39, 18–32. doi: 10.1177/0148607115595980
Carballeira, N. M. (2008). New advances in fatty acids as antimalarial, antimycobacterial and antifungal agents. Prog. Lipid Res. 47, 50–61. doi: 10.1016/j.plipres.2007.10.002
Chen, X. L., Wang, Z., and Liu, C. (2016). Roles of peroxisomes in the rice blast fungus. Biomed Res. Int. 2016:9343417. doi: 10.1155/2016/9343417
Daum, G., Wagner, A., Czabany, T., and Athenstaedt, K. (2007). Dynamics of neutral lipid storage and mobilization in yeast. Biochimie 89, 243–248. doi: 10.1016/j.biochi.2006.06.018
de Jong, J. C., Mccormack, B. J., Smirnoff, N., and Talbot, N. J. (1997). Glycerol generates turgor in rice blast. Nature 389, 244–245. doi: 10.1038/38418
Fabian, B., and Ralf, E. (1996). Identification of a yeast peroxisomal member of the family of AMP-binding proteins. Eur. J. Biochem. 240, 468–476.
Fernandez, J., Marroquin-Guzman, M., and Wilson, R. A. (2014). Evidence for a transketolase-mediated metabolic checkpoint governing biotrophic growth in rice cells by the blast fungus Magnaporthe oryzae. PLoS Pathog. 10:e1004354. doi: 10.1371/journal.ppat.1004354
Foster, J., and Nakata, P. A. (2014). An oxalyl-CoA synthetase is important for oxalate metabolism in Saccharomyces cerevisiae. FEBS Lett. 588, 160–166. doi: 10.1016/j.febslet.2013.11.026
Foster, J., Kim, H. U., Nakata, P. A., and Browse, J. (2012). A previously unknown oxalyl-CoA synthetase is important for oxalate catabolism in Arabidopsis. Plant Cell 24, 1217–1229. doi: 10.1105/tpc.112.096032
Francisco, T., Rodrigues, T. A., Dias, A. F., Barros-Barbosa, A., Bicho, D., and Azevedo, J. E. (2017). Protein transport into peroxisomes: knowns and unknowns. Bioessays 39, 1–8. doi: 10.1002/bies.201700047
Frohlich, F., Petit, C., Kory, N., Christiano, R., Hannibal-Bach, H. K., Graham, M., et al. (2015). The GARP complex is required for cellular sphingolipid homeostasis. eLife 4:e08712. doi: 10.7554/eLife.08712
Goh, J., Jeon, J., Kim, K. S., Park, J., Park, S. Y., and Lee, Y. H. (2011). The PEX7-mediated peroxisomal import system is required for fungal development and pathogenicity in Magnaporthe oryzae. PLoS One 6:e28220. doi: 10.1371/journal.pone.0028220
Hagen, S., Drepper, F., Fischer, S., Fodor, K., Passon, D., Platta, H. W., et al. (2015). Structural insights into cargo recognition by the yeast PTS1 receptor. J. Biol. Chem. 290, 26610–26631. doi: 10.1074/jbc.M115.657973
Koch, J., Pranjic, K., Huber, A., Ellinger, A., Hartig, A., Kragler, F., et al. (2010). PEX11 family members are membrane elongation factors that coordinate peroxisome proliferation and maintenance. J. Cell Sci. 123, 3389–3400. doi: 10.1242/jcs.064907
Li, L., Wang, J., Chen, H., Chai, R., Zhang, Z., Mao, X., et al. (2017). Pex14/17, a filamentous fungus-specific peroxin, is required for the import of peroxisomal matrix proteins and full virulence of Magnaporthe oryzae. Mol. Plant Pathol. 18, 1238–1252. doi: 10.1111/mpp.12487
Li, X., Gao, C., Li, L., Liu, M., Yin, Z., Zhang, H., et al. (2017). MoEnd3 regulates appressorium formation and virulence through mediating endocytosis in rice blast fungus Magnaporthe oryzae. PLoS Pathog. 13:e1006449. doi: 10.1371/journal.ppat.1006449
Lingard, M. J., Monroe-Augustus, M., and Bartel, B. (2009). Peroxisome-associated matrix protein degradation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 4561–4566.
Listenberger, L. L., Han, X., Lewis, S. E., Cases, S., Farese, R. V., Ory, D. S., et al. (2003). Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. U.S.A. 100, 3077–3082.
Liu, X., Ma, C., and Subramani, S. (2012). Recent advances in peroxisomal matrix protein import. Curr. Opin. Cell Biol. 24, 484–489. doi: 10.1016/j.ceb.2012.05.003
Liu, X., Qian, B., Gao, C., Huang, S., Cai, Y., Zhang, H., et al. (2016). The putative protein phosphatase MoYvh1 functions upstream of MoPdeH to regulate the development and pathogenicity in Magnaporthe oryzae. Mol. Plant Microbe Interact. 29, 496–507. doi: 10.1094/MPMI-11-15-0259-R
Liu, X., Yang, J., Qian, B., Cai, Y., Zou, X., Zhang, H., et al. (2018). MoYvh1 subverts rice defense through functions of ribosomal protein MoMrt4 in Magnaporthe oryzae. PLoS Pathog. 14:e1007016. doi: 10.1371/journal.ppat.1007016
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lum, P. Y., Armour, C. D., Stepaniants, S. B., Cavet, G., Wolf, M. K., Butler, J. S., et al. (2004). Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell 116, 121–137.
Mulleder, M., Calvani, E., Alam, M. T., Wang, R. K., Eckerstorfer, F., Zelezniak, A., et al. (2016). Functional metabolomics describes the yeast biosynthetic regulome. Cell 167, 553–565. doi: 10.1016/j.cell.2016.09.007
Patkar, R. N., Ramos-Pamplona, M., Gupta, A. P., Fan, Y., and Naqvi, N. I. (2012). Mitochondrial beta-oxidation regulates organellar integrity and is necessary for conidial germination and invasive growth in Magnaporthe oryzae. Mol. Microbiol. 86, 1345–1363. doi: 10.1111/mmi.12060
Rajvanshi, P. K., Arya, M., and Rajasekharan, R. (2017). The stress-regulatory transcription factors Msn2 and Msn4 regulate fatty acid oxidation in budding yeast. J. Biol. Chem. 292, 18628–18643. doi: 10.1074/jbc.M117.801704
Ramos-Pamplona, M., and Naqvi, N. I. (2006). Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Mol. Microbiol. 61, 61–75. doi: 10.1111/j.1365-2958.2006.05194.x
Sangappillai, V., and Nadarajah, K. (2020). Fatty acid synthase beta dehydratase in the lipid biosynthesis pathway is required for conidiogenesis, pigmentation and appressorium formation in Magnaporthe oryzae S6. Int. J. Mol. Sci. 21, 1–25. doi: 10.3390/ijms21197224
Shi, H. B., Meng, S., Qiu, J. H., Wang, C. C., Shu, Y. Z., Luo, C. X., et al. (2021). MoWhi2 regulates appressorium formation and pathogenicity via the MoTor signalling pathway in Magnaporthe oryzae. Mol. Plant Pathol. 22, 969–983. doi: 10.1111/mpp.13074
Subramani, S., Koller, A., and Snyder, W. B. (2000). Import of peroxissomal matrix and membrane protein. Annu. Rev. Biochem. 69, 399–418.
Sun, C. B., Suresh, A., Deng, Y. Z., and Naqvi, N. I. (2006). A multidrug resistance transporter in Magnaporthe is required for host penetration and for survival during oxidative stress. Plant Cell 18, 3686–3705. doi: 10.1105/tpc.105.037861
van Roermund, C. W., Waterham, H. R., Ijlst, L., and Wanders, R. J. (2003). Fatty acid metabolism in Saccharomyces cerevisiae. Cell. Mol. Life Sci. 60, 1838–1851. doi: 10.1007/s00018-003-3076-x
Walter, T., and Erdmann, R. (2019). Current advances in protein import into peroxisomes. Protein J. 38, 351–362. doi: 10.1007/s10930-019-09835-6
Wang, Z. Y., Jenkinson, J. M., Holcombe, L. J., Soanes, D. M., Veneault-Fourrey, C., Bhambra, G. K., et al. (2005). The molecular biology of appressorium turgor generation by the rice blast fungus Magnaporthe grisea. Biochem. Soc. Trans. 33, 384–388. doi: 10.1042/BST0330384
Wang, Z. Y., Soanes, D. M., Kershaw, M. J., and Talbot, N. J. (2007). Functional analysis of lipid metabolism in Magnaporthe grisea reveals a requirement for peroxisomal fatty acid beta-oxidation during appressorium-mediated plant infection. Mol. Plant Microbe Interact. 20, 475–491. doi: 10.1094/MPMI-20-5-0475
Wysham, W. Z., Roque, D. R., Han, J., Zhang, L., Guo, H., Gehrig, P. A., et al. (2016). Effects of fatty acid synthase inhibition by orlistat on proliferation of endometrial cancer cell lines. Target. Oncol. 11, 763–769. doi: 10.1007/s11523-016-0442-9
Xian, P., Cai, Z., Cheng, Y., Lin, R., Lian, T., Ma, Q., et al. (2020). Wild soybean oxalyl-CoA synthetase degrades oxalate and affects the tolerance to Cadmium and Aluminum Stresses. Int. J. Mol. Sci. 21:8869. doi: 10.3390/ijms21228869
Yan, M., Rayapuram, N., and Subramani, S. (2005). The control of peroxisome number and size during division and proliferation. Curr. Opin. Cell Biol. 17, 376–383. doi: 10.1016/j.ceb.2005.06.003
Yang, J., Fu, M., Ji, C., Huang, Y., and Wu, Y. (2018). Maize oxalyl-CoA decarboxylase1 degrades oxalate and affects the seed metabolome and nutritional quality. Plant Cell 30, 2447–2462. doi: 10.1105/tpc.18.00266
Zhang, H., Liu, K., Zhang, X., Song, W., Zhao, Q., Dong, Y., et al. (2010). A two-component histidine kinase, MoSLN1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Curr. Genet. 56, 517–528. doi: 10.1007/s00294-010-0319-x
Zhang, H., Tang, W., Liu, K., Huang, Q., Zhang, X., Yan, X., et al. (2011). Eight RGS and RGS-like proteins orchestrate growth, differentiation, and pathogenicity of Magnaporthe oryzae. PLoS Pathog. 7:e1002450. doi: 10.1371/journal.ppat.1002450
Zhang, H., Zhao, Q., Guo, X., Guo, M., Qi, Z., Tang, W., et al. (2014). Pleiotropic function of the putative zinc-finger protein MoMsn2 in Magnaporthe oryzae. Mol. Plant Microbe Interact. 27, 446–460. doi: 10.1094/MPMI-09-13-0271-R
Zhong, K., Li, X., Le, X., Kong, X., Zhang, H., Zheng, X., et al. (2016). MoDnm1 dynamin mediating peroxisomal and mitochondrial fission in complex with MoFis1 and MoMdv1 is important for development of functional appressorium in Magnaporthe oryzae. PLoS Pathog. 12:e1005823. doi: 10.1371/journal.ppat.1005823
Keywords: MoPcs60, peroxisome, β-oxidation, fatty acid metabolism, pathogenicity
Citation: Zhang T, Li Y, Li X, Gu W, Moeketsi EK, Zhou R, Zheng X, Zhang Z and Zhang H (2022) The Peroxisomal-CoA Synthetase MoPcs60 Is Important for Fatty Acid Metabolism and Infectious Growth of the Rice Blast Fungus. Front. Plant Sci. 12:811041. doi: 10.3389/fpls.2021.811041
Received: 08 November 2021; Accepted: 01 December 2021;
Published: 26 January 2022.
Edited by:
Xiao-Ren Chen, Yangzhou University, ChinaReviewed by:
Lu Zheng, Huazhong Agricultural University, ChinaGuoqing Chen, China National Rice Research Institute, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2022 Zhang, Li, Li, Gu, Moeketsi, Zhou, Zheng, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haifeng Zhang, hfzhang@njau.edu.cn
 Ting Zhang
Ting Zhang Emily Kolojane Moeketsi
Emily Kolojane Moeketsi Zhengguang Zhang
Zhengguang Zhang Haifeng Zhang
Haifeng Zhang