- State Key Laboratory of Subtropical Silviculture, Zhejiang A&F University, Hangzhou, China
The kernel of Torreya grandis cv. ‘Merrillii’ (Cephalotaxaceae) is a rare nut with a variety of bioactive compounds and a high economic value. β-sitosterol is not only the most abundant plant sterol but also has various biological effects, such as antimicrobial, anticancer, anti-inflammatory, lipid-lowering, antioxidant, and antidiabetic activities. In this study, a squalene synthase gene from T. grandis, TgSQS, was identified and functionally characterized. TgSQS encodes a deduced protein of 410 amino acids. Prokaryotic expression of the TgSQS protein could catalyze farnesyl diphosphate to produce squalene. Transgenic Arabidopsis plants overexpressing TgSQS showed a significant increase in the content of both squalene and β-sitosterol; moreover, their drought tolerance was also stronger than that of the wild type. Transcriptome data from T. grandis seedlings showed that the expression levels of sterol biosynthesis pathway-related genes, such as HMGS, HMGR, MK, DXS, IPPI, FPPS, SQS, and DWF1, increased significantly after drought treatment. We also demonstrated that TgWRKY3 directly bound to the TgSQS promoter region and regulated its expression through a yeast one-hybrid experiment and a dual luciferase experiment. Together, these findings demonstrate that TgSQS has a positive role in β-sitosterol biosynthesis and in protecting against drought stress, emphasizing its importance as a metabolic engineering tool for the simultaneous improvement of β-sitosterol biosynthesis and drought tolerance.
Introduction
Sterols are isoprenoid-derived molecules found in bacteria, fungi, insects, mammals, and plants (Yuan et al, 2010). In plants, more than 250 kinds of sterols and sterol conjugates have been identified, including free sterols, sterol esters, sterol glucosides, and acylated sterol glucosides. The most abundant plant sterols are sitosterol, followed by stigmasterol and campesterol (Goldstein and Brown, 1990; Schluttenhofer and Yuan, 2015). Sitosterol is not only a component of the plant cell membrane, which plays an important role in seed germination and organ development; it can also participate in the plant’s response to low temperatures, wounds, salt, and drought stress (Elkeilsh et al, 2019). To initiate plant sterol biosynthesis, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) are generated from either the cytosolic mevalonate (MVA) pathway or the plastidial methylerythritol phosphate (MEP) pathway (Guan et al, 1998; Jiang et al, 2017). Then, the “head-to-tail” condensation of IPP and DMAPP forms C15 farnesyl diphosphate (FPP). Subsequently, two molecules of FPP are condensed head-to-head by squalene synthase (SQS) to form squalene (Nguyen et al, 2013). Finally, sterols, such as sitosterol, stigmasterol, and campesterol, are produced through a series of reactions (Singh et al., 2017). Because FPP is also the precursor of other non-sterol isoprenoids, such as ubiquinones and sesquiterpenoids, regulation of SQS has been considered important for sterol biosynthesis (Nie et al, 2019).
Some studies have shown that SQS expression significantly increases in roots, stems, and leaves when Panax ginseng and Portulaca oleracea are treated with methyl jasmonate (Kumar et al., 2015). In addition, several transcription factors have been reported to regulate SQS expression. For example, two bHLH family transcription factors, SREBP1a and SREBP2 (sterol regulatory element binding proteins), are involved in cholesterol synthesis by regulating the expression of SQS in human cells (He et al., 2016). TPO1 negatively regulates Saccharomyces cerevisiae SQS (ERG9) expression, while YER064C and SLK19 positively regulate its expression (Tai et al., 2023; Kennedy and Bard, 2001). WRKY1 increases the content of triterpenes, such as withaferin, by positively regulating SQS expression in Withania somnifera (Sun et al., 2012).
Torreya grandis cv. ‘Merrillii’ is an excellent variety, with a cultivation history of more than one thousand years. The kernel of T. grandis cv. ‘Merrillii’ is a precious nut that is rich in unsaturated fatty acids and β-sitosterol, with insecticidal, anti-inflammatory, and antioxidant effects (Huang et al., 2007; Divi and Krishna, 2009). Several studies have indicated that β-sitosterol can protect against breast cancer (Awad et al., 2007; Ju et al., 2004), prostate cancer (Awad et al., 2001), colon cancer (Awad et al., 1997; Clough and Bent, 1998), and gastric cancer (Zhao et al., 2017) by inhibiting cell proliferation and inducing apoptosis. In addition, β-sitosterol can inhibit the increase in blood lipids caused by a high-fat diet (Singh et al., 2015; Salehi et al., 2020). With the given benefits, improving the β-sitosterol content in T. grandis can increase the kernel’s nutritional values. In this study, we cloned the SQS of T. grandis for the first time and proved that it encodes an active squalene synthase. The β-sitosterol content increased significantly in mature seeds of TgSQS-overexpressing homozygous lines of Arabidopsis thaliana; moreover, their drought tolerance was also stronger than that of the wild type. We also demonstrated that TgWRKY3 directly bound to the TgSQS promoter region and regulated its expression through a yeast one-hybrid experiment and a dual luciferase experiment.
Materials and methods
Plant materials, growth conditions, and drought treatments
For transcriptome analyses, one-year-old T. grandis seedlings were transplanted to pots with 300 g of a soil mixture and grown in a shady plastic greenhouse. After two weeks, the surviving seedlings were irrigated for 12 h, and then irrigation was withdrawn to start drought treatment.
For drought resistance tests, plants were grown in a potting soil mixture in growth chambers at 22°C with 16 h/day illumination. The relative humidity was approximately 70% ( ± 5%). After growing for one week, the plants were irrigated with 1 L of water per tray for 6 h, and then drought treatment was imposed by withdrawing irrigation for half of the plants until most died. The other half were grown under a standard irrigation regime as a control. After 20 days of drought treatment, representative pots were placed to take photos.
For DAB and NBT staining, Arabidopsis seeds were surface sterilized and cold treated at 4°C for three days. Then, the seeds were plated on an MS medium containing 3% (w/v) sucrose and 0.8% (w/v) agar and grown at 22°C with a 16-h daily light period. The 10-day-old seedlings were transferred to the MS (Murashige and Skoog) medium with 10% PEG (with controls) for three days before staining. For DAB staining, the samples were immersed in a DAB solution (SL1805, Coolaber) for 4 h and then in 95% ethanol for decoloring. For NBT staining, the samples were first immersed in NBT staining solution (S19048, Yuanye) until a dark blue color appeared (approximately 1 h) and then in 95% ethanol for decoloring. Photographs were taken using a stereo microscope (SZX16, Olympus).
Phylogenetic analysis
Phylogenetic analysis generated a bootstrap neighbor-joining evolutionary tree using MEGA 7.0 with 1,000 bootstrap replicates. Amino acid sequence alignment was conducted using DNAMAN software.
Gene cloning, construction, and transformation in A. thaliana
To obtain TgSQS overexpression in transgenic plants, the coding sequence of TgSQS was PCR-amplified with the primers listed in Table S1 and introduced into the Super1300 vector driven by a Super promoter (multiple CaMV35S are connected in series). The construct was transformed into Agrobacterium strain GV3101 for transfection of Arabidopsis using the floral-dip method (Devarenne et al, 2002). Seeds were screened on an MS agar medium containing 50 mg/L of hygromycin. The selected T3-generation transgenic plants with 100% resistance to hygromycin were considered homozygous lines and harvested for further analysis.
Subcellular localization
The coding sequence of TgSQS was cloned into a modified pCAMBIA1300, which had GFP fused to the N-terminal, and introduced into Agrobacterium tumefaciens strain GV3101. The transient gene expression analysis of TgSQS and an endoplasmic reticulum marker in Nicotiana benthamiana was performed according to a previous study (Nakashima et al., 1995). Agrobacteria were grown overnight in LB (Luria-Bertani) media and brought to an OD600 of 0.8 in the injection solution. After three days of injection, green fluorescent protein (GFP) and red fluorescent protein (RFP) fluorescence was observed via confocal laser scanning microscopy (LSM510, Carl Zeiss).
Prokaryotic protein expression and enzyme activity analysis
The coding sequence of TgSQS was introduced into the pET-32a vector (provided by Zuying Zhang) and transformed into competent Rosetta (DE3) to produce a recombinant protein containing a Trx-His-tag at the N-terminus. The cloning sites were BamHI and SacI. The transformed bacteria were grown to an OD600 of 0.6 at 37°C in an LB medium, and then 0.4 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to induce TgSQS expression. After culturing for 20 h at 16°C, the cells were collected and lysed by sonication. The crude enzyme solution was subjected to Trx-His-Tag purification using a Ni Sepharose 6 Fast Flow gravity column to obtain purified protein. Imidazole was removed from the purified protein using centrifugal ultrafiltration (Amicon Ultra 30 kDa, Millipore, Burlington, MA, USA). After BCA protein quantification, 10-μl samples were used to run a 10% SDS-PAGE gel.
The enzyme reaction of the recombinant protein TgSQS was conducted as follows: 50 µg of purified protein was incubated at 30°C with 20 µg of FPP triammonium salt (Sigma–Aldrich, St. Louis, MO, USA), 50 mM Tris–HCl (pH7.5), 25 mM MgCl2, 1 mM DTT, 2% glycine, and 3 mM NADPH in a total reaction volume of 500 µl. After 8 h, the reaction product was extracted three times with 500 µl of n-hexane, concentrated to 100 µl, and analyzed using GC–MS. GC–MS detection was performed using Trace GC Ultra-ISQ (Thermo Scientific) with a DB-5MS column (30 m × 0.25 mm × 0.25 μm; Agilent). The GC temperature program was as follows: 80°C, raised to 300°C at a rate of 15°C/min, and held at this temperature for 18 min. The carrier gas was ultrahigh-purity helium at a flow rate of 1.0 ml/min. The GC interface temperature was 290°C, and the sample injection volume used was 1 μl.
Squalene analysis
Dry A. thaliana seeds were accurately weighed to 0.039 g and ground with liquid nitrogen. Ultrasonically extracted samples were extracted for 20 min three times, and the solvent was removed under low temperature and pressure and dissolved in 1 ml of n-hexane. A total of 1.5 ml of a 2 mol/L KOH-ethanol solution was added to 0.4 ml of the extraction solution. Samples were saponified ultrasonically for 10 min in a 60°C water bath and placed in a 60°C oven for 60 min after vortexing. They were shaken for 1 min, and 1 ml of water and 1 ml of n-hexane were added after cooling. Samples were extracted for 2 min, and then the supernatants were removed. We added 0.25 g of anhydrous sodium sulfate to purify the supernatants, dried the supernatants with liquid nitrogen, and then dissolved the precipitates in 0.1 ml of n-hexane to start GC–MS analysis. The GC–MS detection was performed using a Trace GC Ultra-ISQ (Thermo Scientific) with a DB-5MS column (30 m × 0.25 mm × 0.25 μm: Agilent). The GC temperature program was as follows: 80°C, raised to 300°C at a rate of 15°C/min, and held at this temperature for 18 min. The carrier gas was ultrahigh-purity helium at a flow rate of 1.0 ml/min. The GC interface temperature was 290° C, and the sample injection volume used was 1 μl.
RNA extraction and transcriptome analysis
Total RNA was isolated from the leaves of T. grandis seedlings treated with drought stress using the RNAprep Pure Plant Kit (DP441, Tiangen); mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. First-strand cDNA was synthesized using a random hexamer primer and M-MuLV reverse transcriptase (RNase H-). The library fragments were purified with the AMPure XP system (Beckman Coulter, Beverly, USA) to select cDNA fragments that were preferentially 370–420 bp in length. Library quality was assessed using a Qubit2.0 Fluorometer, Agilent Bioanalyzer 2100 system, and qRT-PCR. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using the TruSeq PE Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Novaseq platform, and 150-bp paired-end reads were generated.
Raw reads of the fastq format were first processed using in-house perl scripts. At the same time, the Q20, Q30, and GC content of the clean data were calculated. Differential expression analysis of two conditions/groups (two biological replicates per condition) was performed using the DESeq2 R package (1.20.0). The obtained P-values were adjusted using the Benjamini–Hochberg method, which is designed to control the false discovery rate. Genes with an adjusted P-value <0.05 found by DESeq2 were assigned as differentially expressed. Gene Ontology (GO) enrichment analysis and KEGG pathways of differentially expressed genes were implemented using the cluster Profiler R package, in which gene length bias was corrected.
Quantitative real-time PCR (qRT-PCR)
qRT-PCR was performed with a C1000 Touch™ Thermal Cycler system (Bio-Rad) and the ChamQ SYBR qPCR Master Mix kit (Vazyme). The relative expression level was calculated according to the 2−△△Ct method. The actin gene was used as a reference gene. Ct represents the PCR cycle number at which the amount of target reaches a fixed threshold. The corresponding primers are listed in Table S2.
Sterol extraction and analysis
We accurately weighed 0.039 g of dry A. thaliana seeds and 0.5 g of fresh leaves (from T. grandis seedlings treated with drought stress) and ground them in liquid nitrogen. Samples were extracted with ultrasound for 20 min three times, and the solvent was removed under low temperature and pressure and dissolved in 1 ml of n-hexane. A volume of 1.5 ml of a 2 mol/L KOH-ethanol solution was added to a 0.4 ml extraction solution. Samples were ultrasonically saponified for 10 min in a 60°C water bath and placed in a 60°C oven for 60 min after vortexing. They were shaken for 1 min, and 1 ml of water and 1 ml of n-hexane were added after cooling. Samples were extracted for 2 min, and the supernatants were removed. We added 0.25 g of anhydrous sodium sulfate to purify the supernatants, dried the supernatants with liquid nitrogen, and dissolved the precipitates with 0.1 ml of n-hexane for silylation to form trimethylsilyl ether derivatives of sterols, which were used for GC–MS.
Samples (1 µl) were injected into a GCMS-QP201PLUS apparatus (Shimadzu), which consisted of an automated sampler injection system, a split/splitless injector, and a DB-5MS column (30 m × 0.25 mm × 0.25 μm: Agilent). The GC temperature program was as follows: 80°C, raised to 290°C at a rate of 20°C/min, and held at this temperature for 18 min. The carrier gas was ultrahigh-purity helium at a flow rate of 1.0 ml/min and a 1:10 split injection ratio. Authentic campesterol, stigmasterol, β-sitosterol, and cycloartenol were purchased from Sigma-Aldrich. For quantification, a standard curve for individual authentic standards was generated.
Vector construction and dual luciferase assay
The coding sequence of TgWRKYs was obtained from RNA sequencing and cloned into the pCAMBIA 1300-GFP vector to express the effector. The 1870-bp promoter fragments of TgSQS were sub-cloned into the pGreenII 0800-LUC vector as a reporter. The corresponding primers are listed in Table S1. Individual effector vectors and recombinant reporter vectors were transferred to A. tumefaciens GV3101. The reporter was mixed with each kind of effector or the empty pCAMBIA 1300-GFP vector (control) at a 1:1 (v:v) ratio and then injected into tobacco leaves as described previously (Nakashima et al., 1995). A dual-luciferase assay was carried out using the Dual-Lumi™ Luciferase Reporter Gene Assay Kit (Beyotime, RG089). The firefly luciferase (FLUC) and Renilla luciferase (RLUC) values were measured using a Promega GloMax20/20 Luminescence Detector. The relative LUC activity was calculated as the ratio between FLUC and RLUC activities, and three biological replicates were applied.
Yeast one-hybrid assay
Yeast one-hybrid (Y1H) assays were carried out with the Matchmaker Gold Yeast One-Hybrid System Kit (Clontech) according to the manufacturer’s protocol (PT4087-1, Clontech, USA). The corresponding primers are listed in Table S1. Based on the distribution of predicted W-box binding sites in the TgSQS promoter region, three short fragments of the TgSQS promoter (the adenine residue of the translational start codon ATG) were assigned position +1, and the numbers flanking the sequences of the TgSQS promoter fragments were counted based on this number. The positions of three 200-bp sequences containing a W-box were as follows: W1, 1775 to 1575 bp; W2, 1275 to 1075 bp; and W3, 375–175 bp). They were subcloned into the pAbAi vector to obtain pAbAi-W1/W2/W3. Figure S2 provides the sequence of the TgSQS promoter. The coding sequence of TgWRKY3/6/7 was inserted into the pGADT7 vector to construct the prey-AD vector. Then, the linearized pAbAi-W1/W2/W3 vector was transformed into Y1HGold. After determining the minimal inhibitory concentration of Aureobasidin A (AbA) for positive transformants, the prey-AD vector was transformed into the bait yeast strain. Successfully transformed yeast strains were grown on the corresponding SD medium (SD/−Leu, SD/−Leu + AbA) for 3–5 days to take photos.
Results
Identification of TgSQS
Since the genome of T. grandis has not yet been sequenced, we searched the transcriptome data (NCBI accession Nos. SRX9417001–SRX9417009) and found a unigene annotated as squalene synthase. This sequence was used as a query in the blastx of NCBI, and the results showed that this unigene had high identity (>80%) with squalene synthase from Taxus cuspidate, Ginkgo biloba L., and Pinus massoniana, indicating that this unigene likely encodes squalene synthase in T. grandis; thus, it was named TgSQS. The 1230-bp coding length of TgSQS was determined using sequence alignment in blastx. Phylogenetic tree analysis of TgSQS with SQSs from angiosperms, gymnosperms, algae, and animals showed that TgSQS was most closely related to gymnosperms, such as T. cuspidate, and furthest from animals (Figure 1A). Multiple alignments of TgSQS with squalene synthase from A. thaliana, T. cuspidate, and P. massoniana showed that there were six highly conserved regions, I–VI, in its amino acid sequence, among which II and IV contained aspartic acid (DXXXD) active sites (Figure 1B), which were reported to bind to isopentene phosphate groups (Rushton et al., 2010), and VI was reported to perform the function of membrane targeting and anchoring (Ishihama et al., 2011). The DeepTMHMM-based predictions of transmembrane domains revealed that TgSQS had one C-terminal transmembrane domain (amino acid residues 389–407), which was highly like those predicted for A. thaliana (Figure S1B, C) and Taraxacum koksaghyz SQS (Valitova et al, 2016). We further investigated the subcellular localization of TgSQS through transient expression in N. benthamiana leaves. As can be seen in Figure S1A, the TgSQS protein fused with GFP at the N-terminal colocalized with the endoplasmic reticulum marker, which was consistent with the literature (Valitova et al., 2016). These results suggest that TgSQS encodes functional squalene synthase.
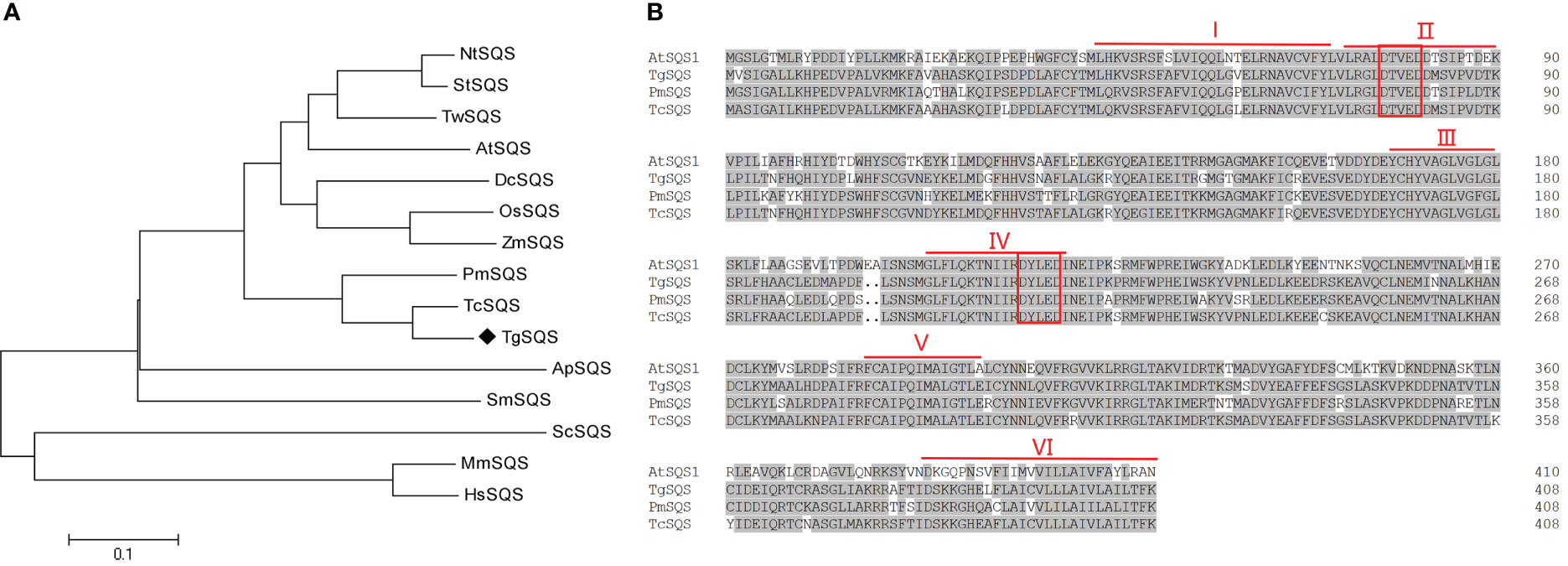
Figure 1 Analysis of the TgSQS amino acid sequence. (A) Construction of the phylogenetic tree of squalene synthase from different species using the neighbor-joining method. The accession numbers of the genes used were: NtSQS, Nicotiana tabacum, AAB08578.1; StSQS, Solanum tuberosum, BAA82093.1; AtSQS1, Arabidopsis thaliana, AEE86403.1; OsSQS, Oryzasativa Japonica Group, BAA22557.1; DcSQS, Dendrobium catenatum, AGI56082.1; ZmSQS, Zea mays, BAA22558.1; PmSQS, Pinus massoniana, AHI96421.1; TcSQS, Taxus cuspidate, ABI14439.1; ApSQS, Auxenochlorella protothecoides, KFM22694.1; HsSQS, Homo sapiens, AAB33404.1; ScSQS, Saccharomyces cerevisiae, AAA34597.1; TwSQS, Tripterygium wilfordii, AMR60779.1; SmSQS, Selaginella moellendorffii, XP024532546.1; MmSQS, Mus musculus, AP_034321.2. (B) Alignment of the full-length amino acid sequences of TgSQS and its homologs in Arabidopsis thaliana, Taxus cuspidate, and Pinus massoniana. The six conserved regions (I, II, III, IV, V, and VI) of squalene synthase are marked by red lines, and two aspartate rich domains (DXXXD) are marked by red squares.
Functional characterization of TgSQS in vitro and in vivo
To understand the catalytic function of TgSQS, the recombinant expression plasmid pET-32a-TgSQS was constructed and transformed into E. coli Rosetta (DE3) to produce a recombinant protein containing a Trx-His-tag at the N-terminus. The pET-32a empty vector served as a control. After induced expression, extraction, and purification, the purified recombinant proteins were analyzed using SDS-PAGE. TgSQS was successfully extracted from the supernatant of lysates with a molecular weight of around 70 kDa (Figure 2A). The enzyme activity of TgSQS was assayed using the purified protein with the cofactors NADPH and Mg2+, while Trx-His-tag was used as a control. The catalytic products were analyzed using GC–MS; the peak at 13.02 min in the profile of TgSQS reaction products (Figure 2B) corresponded to that observed in the standard squalene sample (Figure 2C), but no such peak was detected in the control (Figure 2D). Moreover, the characteristics of peaks in the mass spectrum of TgSQS (Figure 2E) were consistent with those of standard squalene (Figure 2F). These results proved that TgSQS has the catalytic activity of squalene synthase in vitro.
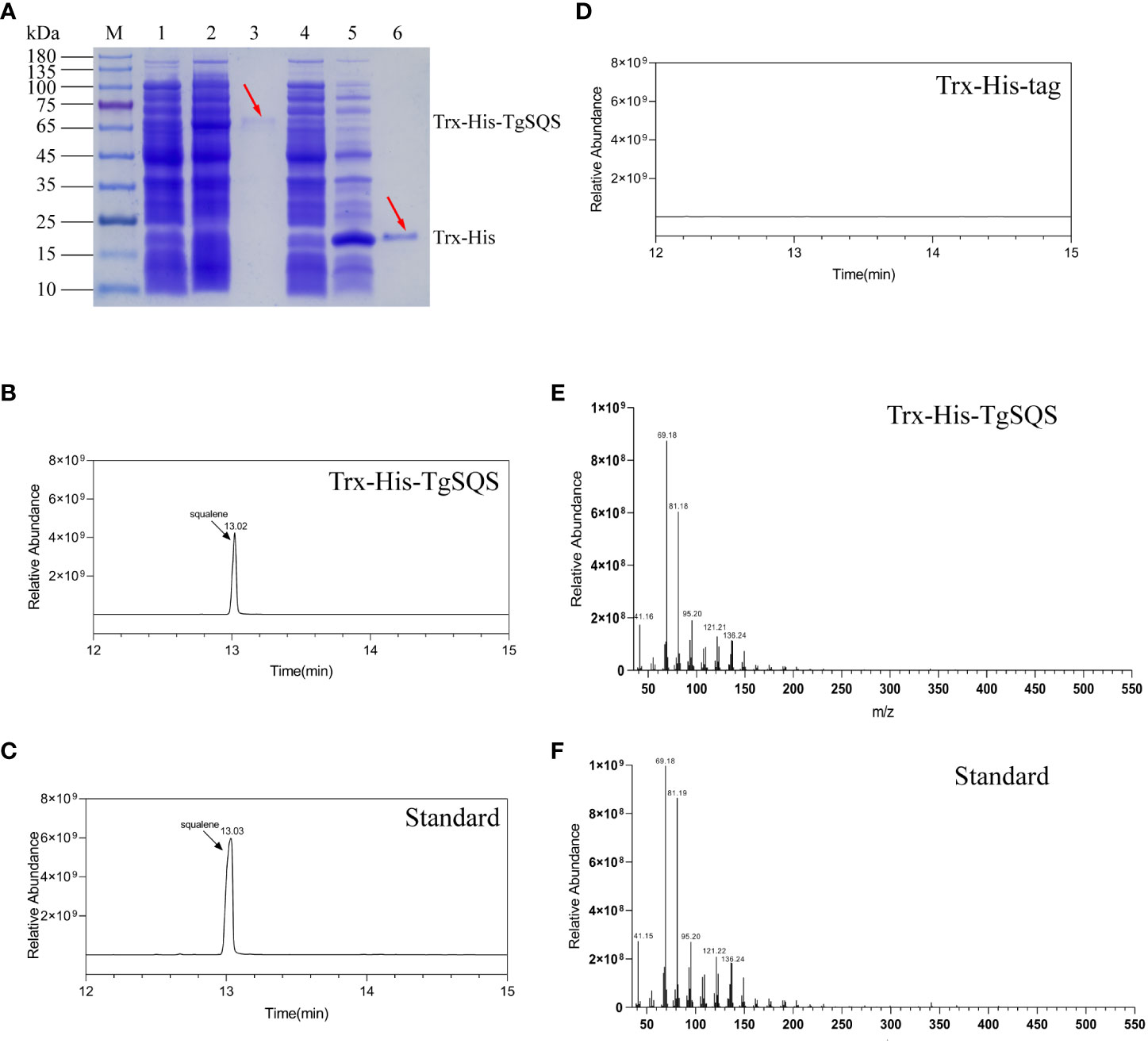
Figure 2 Identification of recombinant TgSQS protein and gas chromatography-mass spectroscopy (GC–MS) analysis of squalene in the catalytic products of TgSQS. (A) 10% SDS-PAGE detection of recombinant TgSQS protein expressed in (E) coli Rosetta (DE3). M, protein marker; Line 1, induced pET-32a-TgSQS bacteria; Line 2, supernatant of induced pET-32a-TgSQS bacteria after ultrasound; Line 3, purified pET-32a-TgSQS protein; Line 4, induced pET-32a bacteria; Line 5, supernatant of induced pET-32a bacteria after ultrasound; Line 6, purified Trx-His-Tag; (B) GC–MS detection of the reaction products of Trx-His-TgSQS; (C) GC–MS detection of the standard squalene; (D) GC–MS detection of the reaction products of Trx-His-Tag (control); (E) MS analysis of the catalytic products of Trx-His-TgSQS; (F) MS analysis of standard squalene.
To further verify the function of TgSQS, the overexpression plasmid super1300-TgSQS was transferred into A. thaliana, and three constitutive overexpression lines with significantly higher expression levels of TgSQS were obtained (Figure 3A). The content of squalene in mature seeds was detected, and the results showed that the content of squalene in the three overexpression lines was significantly higher than that of the wild type (Figure 3B), indicating that TgSQS also has the catalytic activity of squalene synthase in vivo.
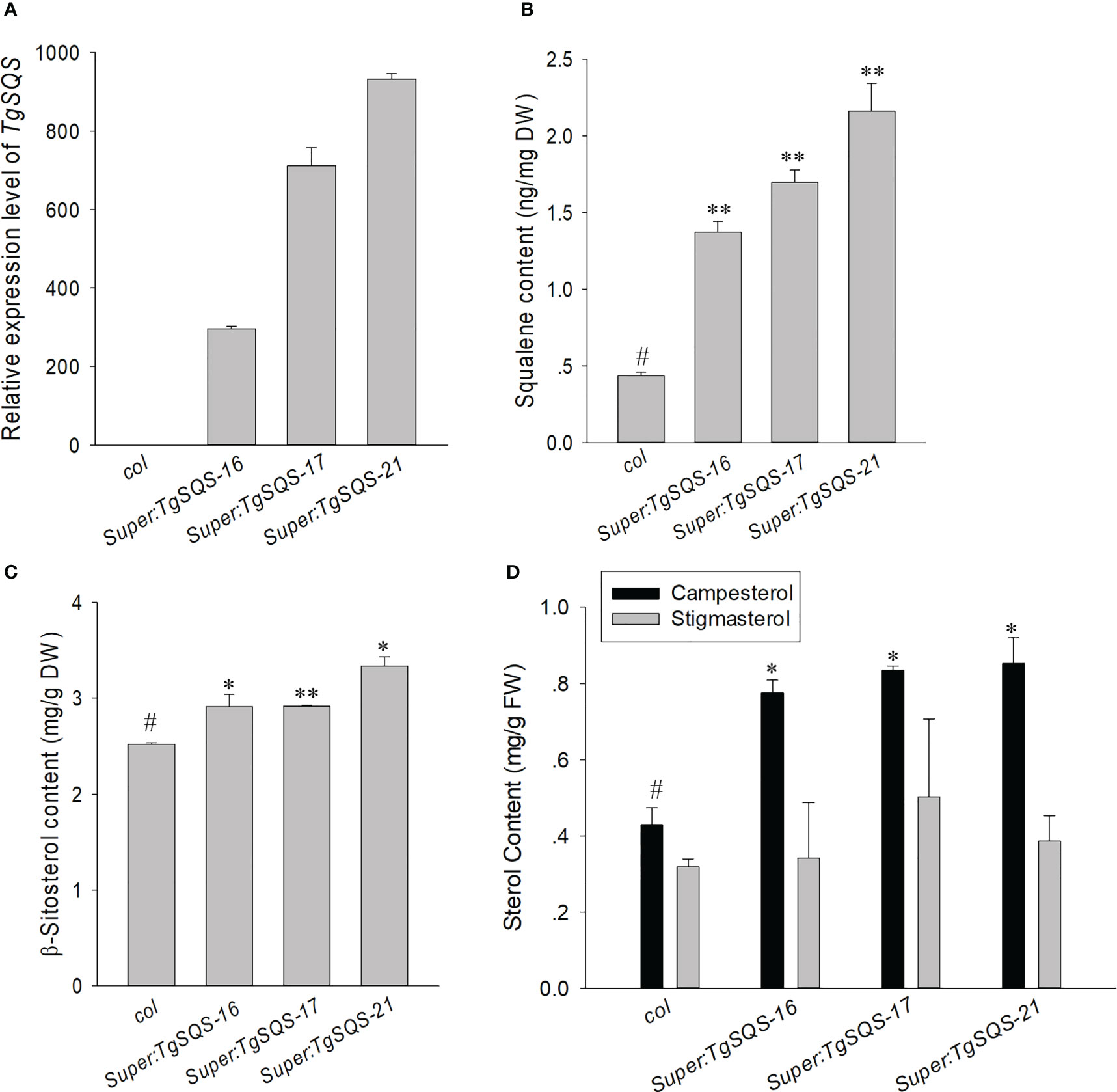
Figure 3 Increased squalene and β-sitosterol content in the seeds of TgSQS-overexpressing Arabidopsis thaliana lines. (A) Transcript level of TgSQS, (B) seed squalene content, (C) seed β-sitosterol content, and (D) stigmasterol and campesterol content in col and three TgSQS-overexpressing Arabidopsis thaliana lines. Data are mean ± SD (n = 3); asterisks indicate significant differences relative to the control by a two-tailed Student’s t-test. #, control*p <0.05; **p <0.001.
Overexpression of TgSQS increased β-sitosterol content in Arabidopsis seeds
In plants, the dominant sterols comprise β-sitosterol, campesterol, and stigmasterol, while β-sitosterol is the most abundant. It has been evidenced in many in vitro and in vivo studies that β-sitosterol possesses various biological actions, such as immunomodulatory, antimicrobial, anticancer, anti-inflammatory, lipid-lowering, antioxidant, and anti-diabetic activities (Babu and Jayaraman, 2020). Therefore, increasing the β-sitosterol content in T. grandis seeds is very important to improve their nutritional value. As it is difficult to achieve genetic transformation in T. grandis, to verify the role of TgSQS in the sterol biosynthetic pathway, we used the super promoter to express TgSQS in A. thaliana (Figure 3A) and tested the β-sitosterol content in mature seeds, as can be seen from Figure 3C, which indicates that the β-sitosterol content in the mature seeds of three TgSQS-overexpressing lines increased by 15%, 15%, and 30% compared with the wild type, indicating that constitutively increasing TgSQS expression in Arabidopsis significantly increased β-sitosterol content in mature seeds. At the same time, the campesterol and stigmasterol content were also detected in these materials (Figure 3D); the results showed that the campesterol content in three TgSQS-overexpressing lines were also significantly higher than that of the wild type. However, the stigmasterol content showed no difference between the three TgSQS-overexpressing lines and the wild type.
TgWRKY3 bound the W-box elements in the TgSQS promoter
Due to the important role of TgSQS in sterol biosynthesis, we studied its transcriptional regulation mechanism. Through cloning and analysis of its promoter (1,860 bp before ATG), three W-box cis-acting elements were found in the entire promoter region. Since the W-box element was the DNA binding site of WRKY family transcription factors, WRKY TFs were regarded as candidates to regulate TgSQS expression. In total, 23 TgWRKYs with full-length coding sequences were found by searching the transcriptome data of T. grandis (NCBI accession Nos. SRX9417001–SRX9417009). All of them were constructed in the overexpression vector pCAMBIA1300-GFP, and the dual luciferase assay was carried out with the empty vector as the control. TgWRKY3, TgWRKY6, and TgWRKY7 significantly increased the expression level of the reporter gene (Figures 4B, C), indicating that they may regulate the expression of TgSQS.
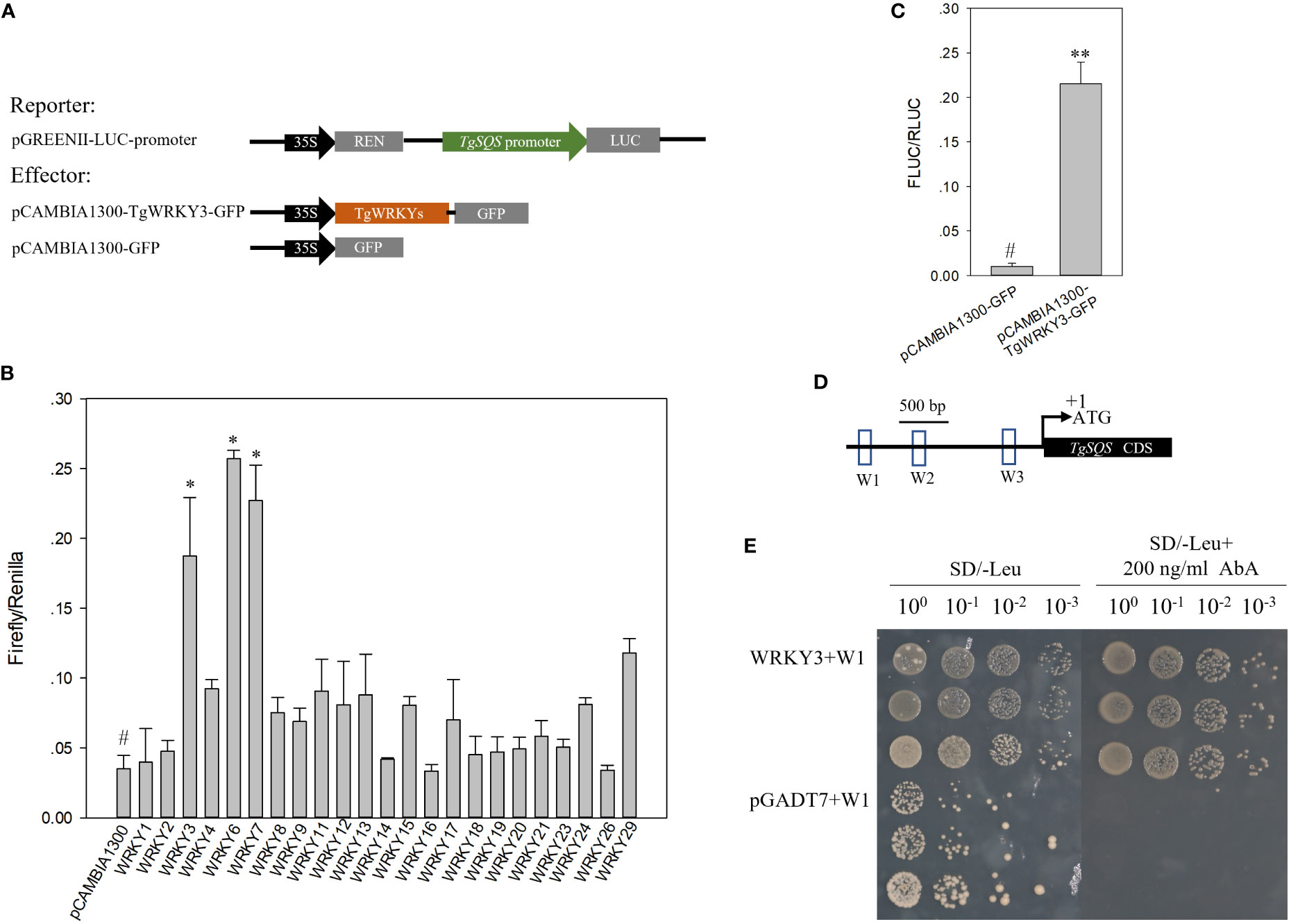
Figure 4 TgWRKY3 directly bound to the promoter of TgSQS and positively regulated TgSQS expression in tobacco leaves. (A) Schematic diagrams of vectors used for the dual luciferase assay; (B) Effects of TgWRKYs on the activities of the TgSQS promoter. Each column represents the mean ± SD of three biological replicates. Asterisks indicate significant differences relative to the control as determined by a two-tailed Student’s t-test. #, control; *p <0.05. (C) Effects of TgWRKY3 on the activities of the TgSQS promoter. Data are the mean ± SD (n = 4); asterisks indicate significant differences relative to the control by a two-tailed Student’s t-test. **p < 0.001. (D) Cis-element analysis in the sequence of the TgSQS promoter (the bait); W1, W2, and W3 represent 200-bp nucleotide sequences containing the W-box. The nucleotide sequences of W1, W2, and W3 are shown in Figure S2. (E) Y1H verification of the interaction between TgWRKY3 and the TgSQS promoter. The pAbAi vector carrying W1, W2, and W3 (200-bp sequences containing the W-box from the TgSQS promoter) and the pGADT7-TgWRKY3 recombinant vector were used to co-transfect Y1HGold receptive cells. Y1HGold cells co-transfected with pAbAi-W2/W3 and pGADT7-TgWRKY3 did not grow normally on the SD/−Leu medium supplied with 200 ng/ml AbA; thus, the pictures were not shown. From left to right, the dilutions of the bacterial solution are 1, 0.1, 0.01, and 0.001. Three monoclonal yeast strains are repeated.
To verify whether TgWRKY3, TgWRKY6, and TgWRKY7 could directly bind to the promoter of TgSQS, a yeast one-hybrid experiment was carried out. In the Y1H assay, the pAbAi vector carrying W1, W2, and W3 (200-bp sequences containing W-box from the TgSQS promoter, Figures 4D, S2) and the pGADT7-TgWRKY3/6/7 recombinant vector served as the reporter and the effector, respectively. Only Y1HGold yeast co-transformed with pGADT7-TgWRKY3 and pAbAi-W1 grew normally on the SD/−Leu medium supplied with 200 ng/ml AbA, whereas the yeast carrying the pGADT7-TgWRKY6/7 and pGADT7 empty vector did not (Figure 4E), indicating that TgWRKY3 directly bound the W-box-containing region in the TgSQS promoter.
Sterol biosynthesis-related genes in T. grandis can respond to drought stress
Preliminary experimental results showed that T. grandis seedlings had strong drought tolerance. Therefore, we carried out drought treatment on T. grandis seedlings and sampled the leaves at the D40 stage (drought treatment for 40 days when soil moisture was zero but seedlings stayed green) and D60 stage (drought treatment for 60 days when seedlings became wilted) to perform transcriptome sequencing with normally watered seedlings (CK, sampled at the D60 stage) as a control. PCA showed low variability among biological repeats, which indicated that there was a high correlation between repetitions (Figure 5A). The nine libraries produced over 6G of clean bases, with Q30 percentages (percentage of sequences with sequencing error rates <0.1%) ranging from 94% to 95% (Table S3). All bases were assembled into 66,798 unigenes with a mean length of 1,241 bp (Table S4). The GO, NT, NR, SwissProt, PFAM, KO, and KOG databases were used to annotate the predicted protein sequence, and 33,318 unigenes were annotated by at least one database (Table S5). These data showed that RNA-seq was of high quality and could be used for further analysis.
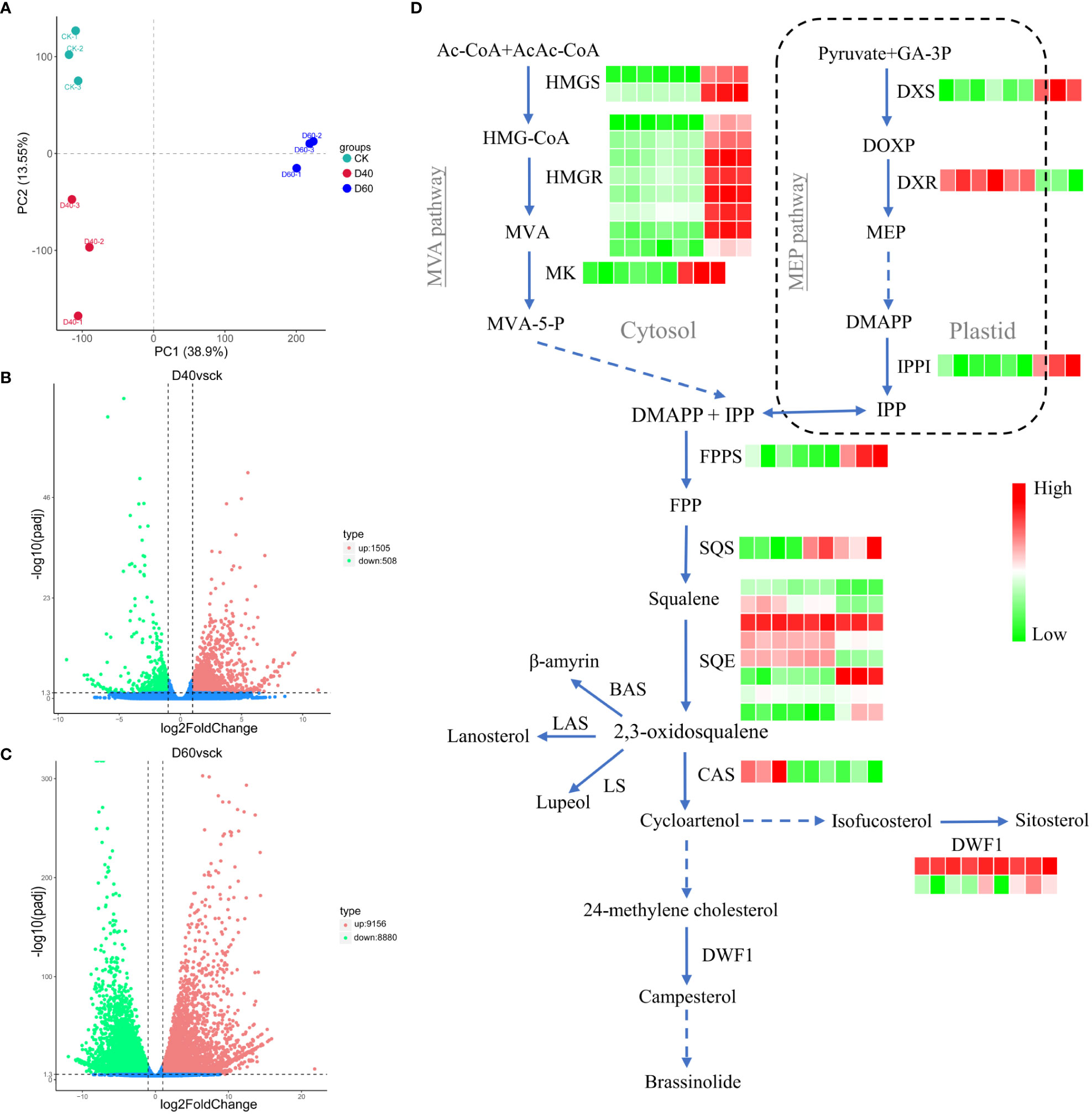
Figure 5 Sterol biosynthesis-related genes in Torreya grandis responded to drought stress. (A) Principal component analysis (PCA) of the genes. (B) Volcano plots for DEGs from CKvsD40; (C) Volcano plots for DEGs from CKvsD60. (D) Simplified view of isoprenoid biosynthesis in plants. Solid blue arrows indicate single-step reactions, dashed arrows denote several steps, and the double arrow between the cytosolic and plastid compartments indicates metabolic crosstalk between them. In the heat map, the first three squares from left to right represent CK samples, the three in the middle represent D40 samples, and the three on the right side represent the three D60 samples. Ac-CoA, acetyl CoA; AcAc-CoA, acetoacetyl CoA; BAS, b-amyrin synthase; BR6OX2, brassinosteroid-6-oxidase 2; CAS, cycloartenol synthase; DMAPP, dimethylallyl diphosphate; DWF1, D24 sterol reductase; DOXP, 1-deoxy-D-xylulose 5-phosphate; DXR, 1-deoxy-Dxylulose 5-phosphate reductoisomerase; DXS, 1-deoxy-D-xylulose-5-phosphate synthase; FPP, farnesyl diphosphate; FPPS, FPP synthase; GA-3P, glyceraldehyde 3-phosphate; HMG-CoA, 3-hydroxy-3-methylglutaryl-CoA; HMGR, HMG-CoA reductase; HMGS, HMG-CoA synthase; IPP, isopentenyl diphosphate; IPPI, isopentenyl diphosphate isomerase; LAS, lanosterol synthase; LS, lupeol synthase; MEP, 2-C-methyl-D-erythritol 4-phospahte; MVA, mevalonate; MVA-5-P, 5-phosphomevalonate; MK, mevalonate kinase; SQE, squalene monooxygenase/epoxidase; SQS, squalene synthase.
A total of 2,013 DEGs, including 1,505 up and 508 downregulated genes, were identified in the D40 stage compared with CK (Figure 5B). In the D60 stage, compared with CK, 9,156 genes were upregulated and 8,880 genes were downregulated (Figure 5C), indicating that there were many more DEGs in the latter. Moreover, the DEGs of D40vsCK and D60vsCK did not completely overlap. As shown in Figure S6, a total of 1,336 DEGs overlapped, of which 461 were jointly upregulated and 266 were jointly downregulated. Brassinosteroid biosynthesis and terpenoid backbone biosynthesis were found in the KEGG path enrichment scatter plot at the D60 stage compared with CK (Figures S4, S5), indicating that genes related to terpenoid and sterol biosynthesis were involved in the response of T. grandis to drought stress, considering that campesterol is a precursor of brassinosteroids. Consequently, we combined the sterol biosynthesis pathway with the T. grandis transcriptome data in this study to draw a simplified map and mark the expression changes of related genes in CK, D40, and D60 stages in the form of a heat map (Figure 5D). The expression levels of HMGS, HMGR, MK, DXS, IPPI, FPPS, SQS, and DWF1 increased significantly at the D60 stage (Table S6). At the same time, the sterol content in the leaves of the CK, D40, and D60 stages was detected, and the results showed that the content of sitosterol, campesterol, and stigmasterol in the D60 stage was significantly higher than in the CK, while there was no significant difference between the D40 stage and the CK (Figure 6). This result was consistent with the fact that the differential genes of the D40 stage in the transcriptome were far less than those in the D60 stage, which might be because the soil moisture at the D40 stage had just dropped to zero and the T. grandis seedlings were still in the early stages of drought stress. Together, these results indicate that the sterol biosynthesis pathway is involved in the response of T. grandis to drought stress.
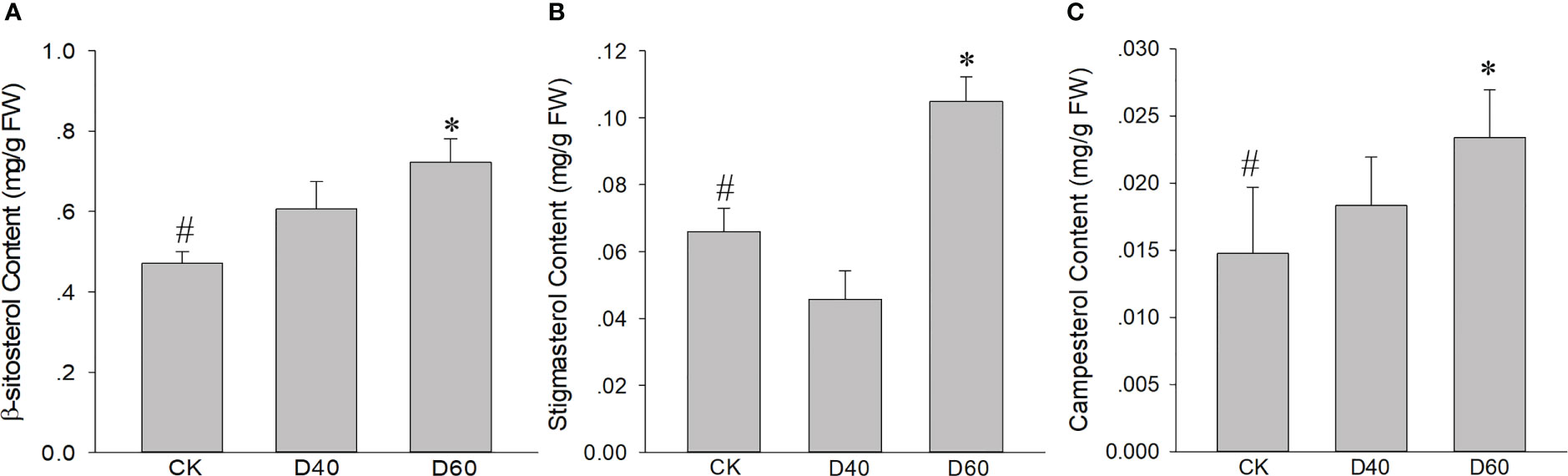
Figure 6 Increased (A) β-sitosterol content, (B) stigmasterol content, and (C) campesterol content in drought-treated Torreya grandis seedlings. Data are the mean ± SD (n = 3); asterisks indicate significant differences relative to the control by a two-tailed Student’s t-test. #, control; *p <0.05.
TgSQS responds to drought stress by affecting ROS accumulation in A. thaliana
To verify the function of TgSQS under drought stress, three homozygous overexpressing A. thaliana lines and the wild type were used for drought treatment. After 20 days of drought, the wild-type leaves began to wilt, while the leaves of TgSQS-overexpressing lines stretched normally, indicating that they were more drought-tolerant than the wild type (Figure 7C). To deeply understand the drought tolerance mechanism, the accumulation of H2O2 and superoxide in the overexpressing lines and the wild type after simulated drought stress was observed by DAB and NBT staining. In this study, 10-day-old seedlings were treated with 10% PEG for 72 h, followed by 3,3-diaminobenzidine (DAB) staining to determine the presence of H2O2 and nitro blue tetrazolium (NBT) staining to show the presence of the superoxide anion. As shown in Figures 7A, B, under control conditions, wild-type and transgenic plants showed similar basal levels of H2O2 and superoxide, but under 10% PEG treatment, TgSQS-OE leaves and roots showed less H2O2 and superoxide accumulation than wild-type leaves and roots. These results suggest that TgSQS improves drought tolerance by reducing ROS accumulation.
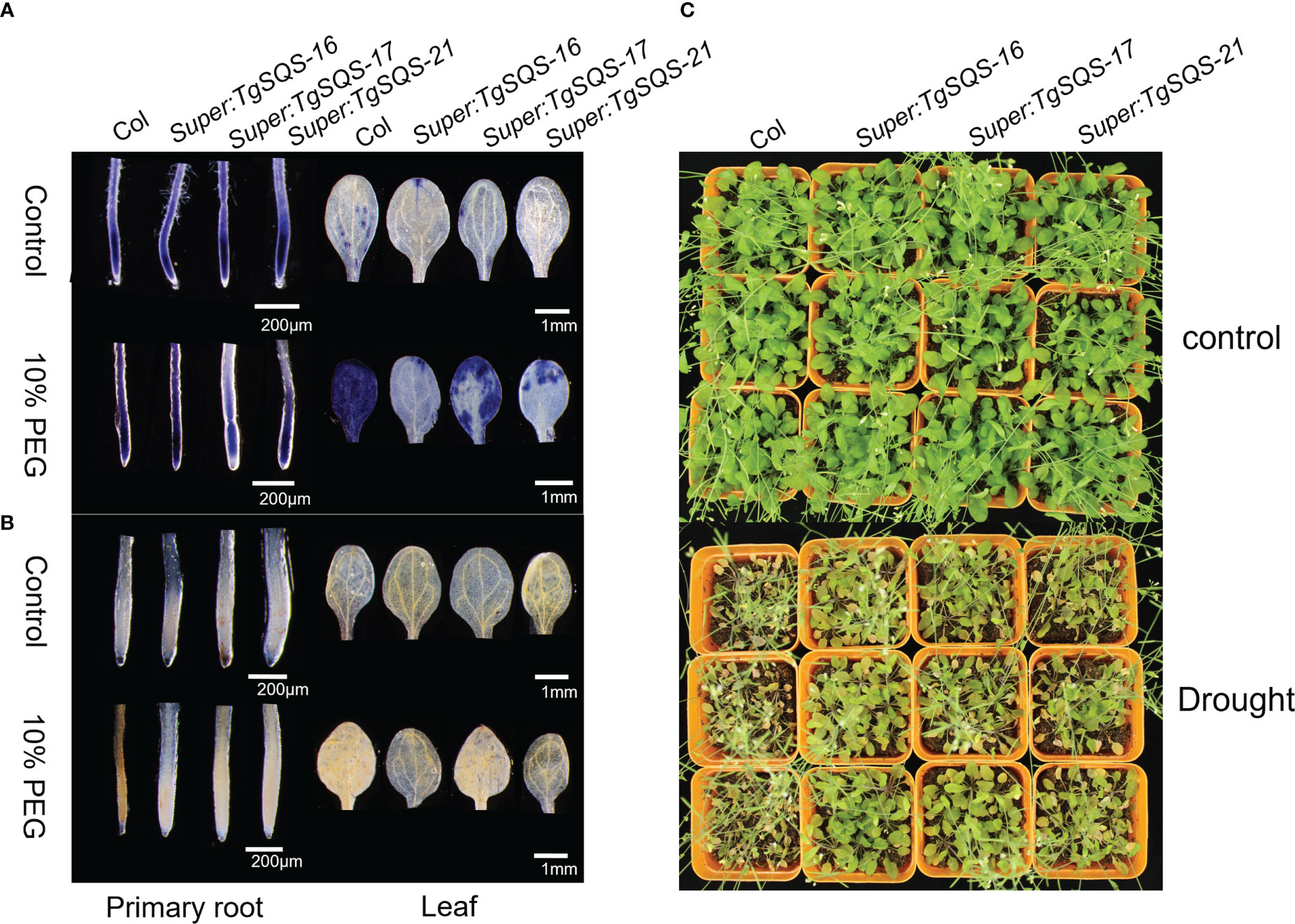
Figure 7 TgSQS responded to drought stress by affecting ROS accumulation in Arabidopsis thaliana. (A) Nitroblue tetrazolium (NBT) staining in the primary root tip and leaf of TgSQS-OE and col 13-day-old seedlings after treatment with 10% PEG for three days. The color strength shows the O2− concentration in the root tips or leaves. (B) 3,3-Diaminobenzidine (DAB) staining in the primary root tip and leaf of TgSQS-OE and col 13-day-old seedlings after treatment with 10% PEG for three days. The color strength shows the H2O2 concentration in the root tips or leaves. (C) The phenotypes of TgSQS-OE and Arabidopsis col seedlings under drought stress treatment. Three-week-old col and TgSQS-OE seedlings were subjected to drought stress without water for 20 days.
Discussion
In this study, we found that a unigene in the transcriptome data of T. grandis might be the gene encoding functional squalene synthase, and we verified its function in vitro and in vivo (Figures 1–3). Since AtSQS was cloned in 1995 (Nguyen et al., 2013), the SQS of many species, such as Tripterygium wilfordii (Zhang et al., 2016), soybean (Nguyen et al., 2013), P. ginseng (Kim et al., 2011), Solanum nigrum (Susan et al., 1991), Siraitia grosvenorii (Zhou et al., 2012), birch (Zhao et al., 2009), and persimmon (Zhou et al., 2012), have been cloned. Based on these reports, the number of SQS varies among different species, which may be a way for plants to meet changes in different developmental and environmental situations. Several literature reports have shown that SQS is a key enzyme for sitosterol synthesis, and changes in both SQS activity and gene expression levels can affect the phytosterol content. For example, treatment of tobacco suspension cells with SQS inhibitors and fungal elicitors could reduce their SQS activity, thereby significantly reducing their sterol content (Ding et al., 2020; Wollam and Antebi, 2011). The content of β-sitosterol significantly increased after PgSQS overexpression in P. ginseng and Acanthopanax senticosus (Li et al., 2019; Sikder et al., 2014). After reducing the expression of SQS by virus-induced gene silencing in W. somnifera leaves, a significant decrease in sterol content was detected (Singh et al., 2017). In this paper, we demonstrated that TgSQS overexpression in Arabidopsis significantly increased the content of β-sitosterol and campesterol in mature seeds (Figure 3), which was consistent with the literature (Nguyen et al., 2013), suggesting that we could increase the content of β-sitosterol and campesterol in T. grandis nuts to improve its nutritional value by increasing TgSQS expression.
Until now, only WsWRKY1 had been known to regulate WsSQS and WsSQE in W. somnifera, according to reports on transcriptional regulation of SQS (Sun et al., 2012). In the present study, we demonstrated that TgWRKY3 directly bound to the promoter region of TgSQS and upregulated its expression (Figure 4). WRKY domain-containing genes comprise one of the largest TF families in plants and are characterized by a highly conserved WRKYGQK motif at the N-terminal end, together with a novel zinc-finger-like motif (Ferrer et al., 2017). In A. thaliana, the WRKY family contains 72 members, which can be divided into three categories, and the second category can be further divided into five subclasses (a–e) (Rushtone et al., 2010). The phylogenetic tree analysis of TgWRKY3, WsWRKY1, and WRKY TFs in A. thaliana showed that TgWRKY3 has a close relationship with AtWRKY21, AtWRKY74, and AtWRKY39 in Group II-d (Figure S3) and that WsWRKY1 has a close relationship with AtWRKY41 and AtWRKY53 in Group III, indicating that we found another WRKY that regulates SQS expression. Multiple sequence alignment results showed that TgWRKY3 has a WRKYGQK conserved domain and the characteristic zinc finger domain C2H2, indicating that TgWRKY3 indeed belongs to the WRKY TFs.
As one of the largest transcription factor families in plants, WRKY TFs have been found to play a role in plant development and tolerance to a variety of abiotic stressors, including wounding, drought, salt, heat, and cold pressure (Choi et al., 2003; Ju et al., 2004; Wentzinger et al., 2002; Chen et al., 2012). Recent research has also shown that WRKY TFs can participate in regulating the biosynthesis of multiple metabolites, such as sesquiterpenes, alkaloids, and terpenes (Seo et al, 2005). For example, MrWRKY1 interacts with the promoter of MrFPS to regulate the biosynthesis of α-Bisabolol in Matricaria recutita L. (Unland et al., 2018), and NbWRKY8 can regulate capsidiol biosynthesis by binding the promoter of NbHMGR2 in N. benthamiana (Jadaun et al., 2017). In our study, TgWRKY3, TgWRKY6, and TgWRKY7 both significantly increased the expression level of the reporter gene, but only TgWRKY3 directly bound to the promoter of TgSQS (Figure 4), indicating that TgWRKY6 and TgWRKY7 regulate TgSQS in a manner that interacts with other transcription factors. However, the expression level of TgWRKY3 in T. torreya seedlings after drought treatment significantly increased, while TgWRKY6 and TgWRKY7 did not (Figures 8B–D), suggesting that TgWRKY3 is involved in both the synthesis of β-sitosterol and the plant’s response to drought stress, while TgWRKY6 and TgWRKY7 might also regulate TgSQS and be involved in the response to different stressors.
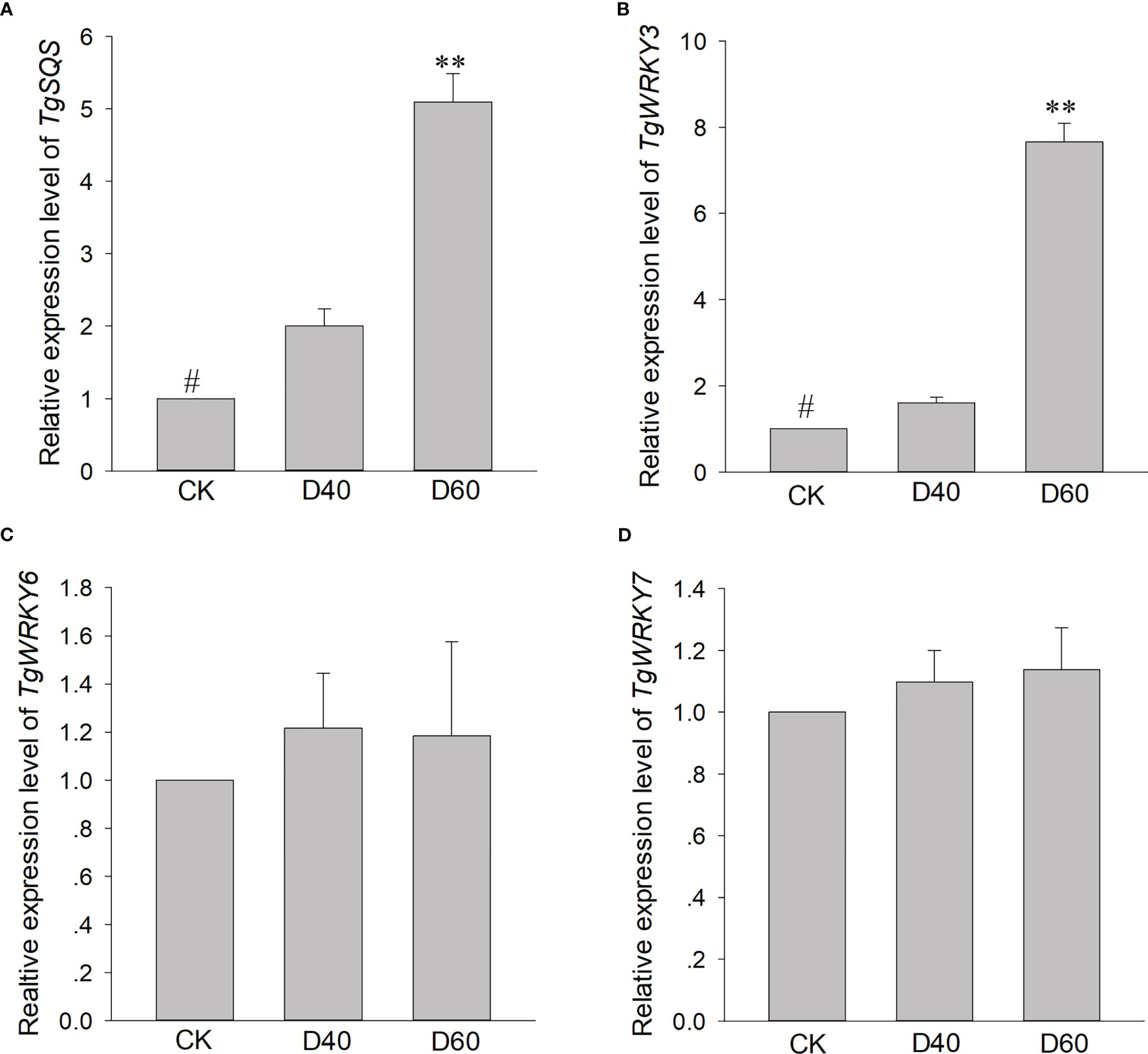
Figure 8 qRT-PCR analysis of gene expression patterns of TgSQS (A), TgWRKY3 (B), TgWRKY6 (C), and TgWRKY7 (D) among drought treatment stages. Data are the mean ± SD (n = 3); asterisks indicate significant differences relative to the control by a two-tailed Student’s t-test. #, control; **p <0.0001.
In this study, we found that TgSQS-overexpressing lines showed stronger drought tolerance and less ROS accumulation than the wild type (Figure 7), indicating that TgSQS probably responded to drought stress by reducing ROS accumulation. However, the specific mechanism is still unclear. Our results also showed that the content of phytosterols (β-sitosterol, campesterol, and stigmasterol) in T. grandis seedlings after drought treatment was significantly higher than that in the control; furthermore, the β-sitosterol content was at least one order of magnitude higher than that of campesterol and stigmasterol (Figure 6). Studies have shown that an increase in β-sitosterol content can be observed in rice under drought conditions (Lee et al., 2004; Kumar et al., 2015). Additionally, drought tolerance and total antioxidant capacity were significantly improved with 100 µM sitosterol treatment in T. aestivum and 120 µM sitosterol treatment in Trifolium repens (Eulgem et al., 2000; Lou et al., 2019). These results imply that β-sitosterol is involved in the response to drought stress. β-sitosterol has been reported as a component of the cell membrane (Vriet et al., 2012); thus, the response of β-sitosterol to drought is probably realized by changing the membrane fluidity and permeability. However, the effect of TgSQS on ROS accumulation is perhaps related to brassinosteroids (BRs). As a plant hormone, BRs control several traits of agronomic importance, such as seed germination, plant architecture, seed yield, and tolerance to various abiotic and biotic stressors (Du et al, 2022; Wani et al., 2021). Several studies have shown that plants treated with exogenous BRs are drought tolerant, and in tomato, it has been clearly demonstrated that a rise in the level of BR biosynthesis is the key to enhancing the tolerance capacity (Nolan et al., 2020; Pandit et al., 2000). In another study, BR treatment rescued the heavy aggregation of ROS in the case of drought stress (Zhang et al., 2018). Therefore, TgSQS might affect ROS accumulation by regulating BR biosynthesis.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/ , SAMN32775921 https://www.ncbi.nlm.nih.gov/, SAMN32775922 https://www.ncbi.nlm.nih.gov/, SAMN32775923 https://www.ncbi.nlm.nih.gov/, SAMN32775924 https://www.ncbi.nlm.nih.gov/, SAMN32775925 https://www.ncbi.nlm.nih.gov/, SAMN32775926 https://www.ncbi.nlm.nih.gov/, SAMN32775927 https://www.ncbi.nlm.nih.gov/, SAMN32775928 https://www.ncbi.nlm.nih.gov/, SAMN32775929.
Author contributions
FZ conceived and guided the experiments, analyzed the data, and wrote the draft. CK performed the experiment and analyzed the data. ZM performed the experiment. WC performed the experiment. YL performed the experiment. HL conceived and guided the experiments. JW is responsible for resources and supervision. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Natural Science Foundation of China (31901339).
Acknowledgments
We are grateful to Dr. Zuying Zhang for providing pET-32a vector. We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1136643/full#supplementary-material
References
Awad, A. B., Chinnam, M., Fink, C. S., Bradford, P. G. (2007). Beta-sitosterol activates fas signaling in human breast cancer cells. Phytomedicine 14, 747–754. doi: 10.1016/j.phymed.2007.01.003
Awad, A. B., Fink, C. S., Williams, H. (2001). In vitro and in vivo (SCID mice) effect of phytosterols on the growth and dissemination of human prostate cancer PC-3 cells. Eur. J. Cancer Prev. 10, 507–513. doi: 10.1097/00008469-200112000-00005
Awad, A. B., Hernandez, A. Y. T., Fink, C. S., Mendel, S. L. (1997). Effect of dietary phytosterols on cell proliferation and protein kinase cactivity in rat colonic mucosa. Nutr. Cancer 27, 210–215. doi: 10.1080/01635589709514527
Babu, S., Jayaraman, S. (2020). An update on β-sitosterol: a potential herbal nutraceutical for diabetic management. BioMed. Pharmacother. 131, 110702. doi: 10.1016/j.biopha.2020.110702
Chen, L., Song, Y., Li, S., Zhang, L., Zou, C., Yu, D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819, 120–128. doi: 10.1016/j.bbagrm.2011.09.002
Choi, Y. H., Kong, K. R., Kim, Y. A., Jung, K. O., Kil, J. H., Rhee, S. H., et al. (2003). Induction of bax and activation of caspases during beta-sitosterol-mediated apoptosis in human colon cancer cells. Int. J. Oncol. 23, 1657–1662. doi: 10.3892/ijo.23.6.1657
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Devarenne, T. P., Ghosh, A., Chappell, J. (2002). Regulation of squalene synthase, a key enzyme of sterol biosynthesis, in tobacco. Plant Physiol. 129, 1095–1106. doi: 10.1104/pp.001438
Ding, M., Lou, H., Chen, W., Zhou, Y., Zhang, Z., Xiao, M., et al. (2020). Comparative transcriptome analysis of the genes involved in lipid biosynthesis pathway and regulation of oil body formation in Torreya grandis kernels. Ind. Crop Prod. 145, 112051. doi: 10.1016/j.indcrop.2019.112051
Divi, U. K., Krishna, P. (2009). Brassinosteroid: a biotechnological target for enhancing crop yield and stress tolerance. New Biotechnol. 26, 131–136. doi: 10.1016/j.nbt.2009.07.006
Du, Y., Fu, X., Chu, Y., Wu, P., Liu, Y., Ma, L., et al. (2022). Biosynthesis and the roles of plant sterols in development and stress responses. Int. J. Mol. Sci. 23 (4), 2332. doi: 10.3390/ijms23042332
Elkeilsh, A., Awad, Y. M., Soliman, M. H., Abu-Elsaoud, A., Abdelhamid, M. T., El-Metwally, I. M. (2019). Exogenous application of β-sitosterol mediated growth and yield improvement in water-stressed wheat (Triticum aestivum) involves up-regulated antioxidant system. J. Plant Res. 132, 881–901. doi: 10.1007/s10265-019-01143-5
Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/s1360-1385(00)01600-9
Ferrer, A., Altabella, T., Arró, M., Boronat, A. (2017). Emerging roles for conjugated sterols in plants. Prog. Lipid Res. 67, 27–37. doi: 10.1016/j.plipres.2017.06.002
Goldstein, J. L., Brown, M. S. (1990). Regulation of the mevalonate pathway. Nature 343, 425–430. doi: 10.1038/343425a0
Guan, G. M., Dai, P. H., Shechter, I. (1998). Differential transcriptional regulation of the human squalene synthase gene by sterol regulatory element-binding proteins (SREBP) 1a and 2 and involvement of 5′DNA sequence elements in the regulation. J. Biol. Chem. 273, 12526–12535. doi: 10.1074/jbc.273.20.12526
He, Z. Y., Zhu, H. D., Li, W. L., Zeng, M. M., Wu, S. F., Chen, S. W., et al. (2016). Chemical components of cold pressed kernel oils from different Torreya grandis cultivars. Food Chem. 209, 196–202. doi: 10.1016/j.foodchem.2016.04.053
Huang, Z. S., Jiang, K. J., Pi, Y., Hou, R., Liao, Z. H., Cao, Y., et al. (2007). Molecular cloning and characterization of the yew gene encoding squalene synthase from Taxus cuspidata. J. Biochem. Mol. Biol. 40, 625–635. doi: 10.5483/bmbrep.2007.40.5.625
Ishihama, N., Yamada, R., Yoshioka, M., Katou, S., Yoshioka, H. (2011). Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell 23, 1153–1170. doi: 10.1105/tpc.110.081794
Jadaun, J. S., Sangwan, N. S., Narnoliya, L. K., Singh, N., Bansal, S., Mishra, B., et al. (2017). Over-expression of DXS gene enhances terpenoidal secondary metabolite accumulation in rose-scented geranium and Withania somnifera: active involvement of plastid isoprenogenic pathway in their biosynthesis. Physiol. Plant 159, 381–400. doi: 10.1111/ppl.12507
Jiang, J. J., Ma, S. H., Ye, N. H., Jiang, M., Cao, J. S., Zhang, J. H. (2017). WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 59 (2), 86–101. doi: 10.1111/jipb.12513
Ju, Y. H., Clausen, L. M., Allred, K. F., Almada, A. L., Helferich, W. G. (2004). β-sitosterol, β -sitosterol glucoside, and a mixture of β -sitosterol and β-sitosterol glucoside modulate the growth of estrogen-responsive breast cancer cells in vitro and in ovariectomized athymic mice. J. Nutr. 134, 1145–1151. doi: 10.1093/jn/134.5.1145
Kennedy, M. A., Bard, M. (2001). Positive and negative regulation of squalene synthase (erg9), an ergosterol biosynthetic gene, in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1517, 177–189. doi: 10.1016/s0167-4781(00)00246-3
Kim, T. D., Han, J. Y., Huh, G. H., Choi, Y. E. (2011). Expression and functional characterization of three squalene synthase genes associated with saponin biosynthesis in Panax ginseng. Plant Cell Physiol. 52, 125–137. doi: 10.1093/pcp/pcq179
Kumar, M. S., Ali, K., Dahuja, A., Tyagi, A. (2015). Role of phytosterols in drought stress tolerance in rice. Plant Physiol. Biochem. 96, 83–89. doi: 10.1016/j.plaphy.2015.07.014
Lee, M. H., Jeong, J. H., Seo, J. W., Shin, C. G., Kim, Y. S., In, J. G., et al. (2004). Enhanced triterpene and phytosterol biosynthesis in Panax ginseng overexpressing squalene synthase gene. Plant Cell Physiol. 45 (8), 976–984. doi: 10.1093/pcp/pch126
Li, Z., Cheng, B., Yong, B., Liu, T., Peng, Y., Zhang, X., et al. (2019). Metabolomics and physiological analyses reveal β-sitosterol as an important plant growth regulator inducing tolerance to water stress in white clover. Planta 250, 2033–2046. doi: 10.1007/s00425-019-03277-1
Lou, H., Ding, M., Wu, J., Zhang, F., Chen, W., Yang, Y., et al. (2019). Full-length transcriptome analysis of the genes involved in tocopherol biosynthesis in Torreya grandis. J. Agric. Food Chem. 67, 1877–1888. doi: 10.1021/acs.jafc.8b06138
Nakashima, T., Inoue, T., Oka, A., Nishino, T., Osumi, T., Hata, S. (1995). Cloning, expression, and characterization of cDNAs encoding Arabidopsis thaliana squalene synthase. Proc. Natl. Acad. Sci. U.S.A. 92 (6), 2328–2332. doi: 10.1073/pnas.92.6.2328
Nguyen, H. T., Neelakadan, A. K., Quach, T. N., Valliyodan, B., Kumar, R., Zhang, Z., et al. (2013). Molecular characterization of glycine max squalene synthase genes in seed phytosterol biosynthesis. Plant Physiol. Biochem. 73, 23–32. doi: 10.1016/j.plaphy.2013.07.018
Nie, S., Huang, S., Wang, S., Mao, Y., Liu, J., Ma, R., et al. (2019). Enhanced brassinosteroid signaling intensity via SlBRI1 overexpression negatively regulates drought resistance in a manner opposite of that via exogenous BR application in tomato. Plant Physiol. Biochem. 138, 36–47. doi: 10.1016/j.plaphy.2019.02.014
Nolan, T. M., Vukasinović, N., Liu, D., Russinova, E., Yin, Y. (2020). Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell 32, 298–318. doi: 10.1105/tpc.19.00335
Pandit, J., Danley, D. E., Schulte, G. K., Mazzalupo, S., Pauly, T. A., Hayward, C. M., et al. (2000). Crystal structure of human squalene synthase, a key enzyme in cholesterol biosynthesis. J. Biol. Chem. 275 (39), 30610–30617. doi: 10.1074/jbc.M004132200
Rushton, J. P., Somssich, E. I., Ringler, P., Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15 (5), 247–258. doi: 10.1016/j.tplants.2010.02.006
Salehi, B., Quispe, C., Sharifi-Rad, J., Cruz-Martins, N., Nigam, M., Mishra, A. P., et al. (2020). Phytosterols: from preclinical evidence to potential clinical applications. Front. Pharmacol. 11. doi: 10.3389/fphar.2020.599959
Schluttenhofer, C., Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167 (2), 295–306. doi: 10.1104/pp.114.251769
Seo, J. W., Jeong, J. H., Shin, C. G., Lo, S. C., Han, S. S., Yu, K. W., et al. (2005). Overexpression of squalene synthase in Eleutherococcus senticosus increases phytosterol and triterpene accumulation. Phytochemistry 66, 869–877. doi: 10.1016/j.phytochem.2005.02.016
Sikder, K., Das, N., Kesh, S. B., Dey, S. (2014). Quercetin and beta-sitosterol prevent high fat diet induced dyslipidemia and hepatotoxicity in Swiss albino mice. Indian J. Exp. Biol. 52, 60–66. doi: 10.3109/03630269.2013.855936
Singh, A. K., Dwivedi, V., Rai, A., Pal, S., Reddy, S. G. E., Rao, D. K. V., et al. (2015). Virus-induced gene silencing of Withania somnifera squalene synthase negatively regulates sterol and defence-related genes resulting in reduced withanolides and biotic stress tolerance. Plant Biotechnol. J. 13, 1287–1299. doi: 10.1111/pbi.12347
Singh, A. K., Kumar, S. R., Dwivedi, V., Rai, A., Pal, S., Shasany, A. K., et al. (2017). A WRKY transcription factor from Withania somnifera regulates triterpenoid withanolide accumulation and biotic stress tolerance through modulation of phytosterol and defense pathways. New Phytol. 215, 1115–1131. doi: 10.1111/nph.14663
Sun, Y., Zhao, Y., Wang, L., Lou, H. X., Cheng, A. X. (2012). Cloning and expression analysis of squalene synthase, a key enzyme involved in antifungal steroidal glycoalkaloids biosynthesis from Solanum nigrum. Drug Discovery Ther. 6 (5), 242–248. doi: 10.5582/ddt.2012.v6.5.242
Susan, M. J., Yim, H. T., Tobe, M. F., Gordon, W. R. (1991). Molecular cloning and characterization of the yeast gene for squalene synthetase. Proc. Natl. Acad. Sci. U.S.A. 88, 6038–6042. doi: 10.1073/pnas.88.14.6038
Tai, Y., Wang, H., Yao, P., Sun, J., Guo, C., Jin, Y., et al. (2023). Biosynthesis of α-bisabolol by farnesyl diphosphate synthase and α-bisabolol synthase and their related transcription factors in matricaria recutita l. Int. J. Mol. Sci. 24 (2), 1730. doi: 10.3390/ijms24021730
Unland, K., Pütter, K. M., Vorwerk, K., Deenen, N. V., Twyman, R. M., Prüfer, D., et al. (2018). Functional characterization of squalene synthase and squalene epoxidase in Taraxacum koksaghyz. Plant Direct 2 (6), e00063. doi: 10.1002/pld3.63
Valitova, J. N., Sulkarnayeva, A. G., Minibayeva, F. V. (2016). Plant sterols: diversity, biosynthesis, and physiological functions. Biochem. (Moscow) 81 (8), 819–834. doi: 10.1134/S0006297916080046
Vriet, C., Russinova, E., Reuzeau, C. (2012). Boosting crop yields with plant steroids. Plant Cell 24, 842–857. doi: 10.1105/tpc.111.094912
Wani, S. H., Anand, S., Singh, B., Bohra, A., Joshi, R. (2021). WRKY transcription factors and plant defense responses: latest discoveries and future prospects. Plant Cell Rep. 40 (7), 1071–1085. doi: 10.1007/s00299-021-02691-8
Wentzinger, L. F., Bach, T. J., Hartmann, M. A. (2002). Inhibition of squalene synthase and squalene epoxidase in tobacco cells triggers an up-regulation of 3-hydroxy-3-methylglutaryl coenzyme a reductase. Plant Physiol. 130, 334–346. doi: 10.1104/pp.004655
Wollam, J., Antebi, A. (2011). Sterol regulation of metabolism, homeostasis, and development. Annu. Rev. Biochem. 80, 885–916. doi: 10.1146/annurev-biochem-081308-165917
Yuan, G. F., Jia, C. G., Li, Z., Sun, B., Zhang, L. P., Liu, N., et al. (2010). Effect of brassinosteroids on drought resistance and abscisic acid concentration in tomato under water stress. Sci. Hortic. 126, 103–108. doi: 10.1016/j.scienta.2010.06.014
Zhang, B., Liu, Y., Chen, M., Feng, J., Ma, Z., Zhang, X., et al. (2018). Cloning, expression analysis and functional characterization of squalene synthase (SQS) from Tripterygium wilfordii. Molecules 23 (2), 269. doi: 10.3390/molecules23020269
Zhang, M., Wang, S., Yin, J., Li, C., Zhan, Y., Xiao, J., et al. (2016). Molecular cloning and promoter analysis of squalene synthase and squalene epoxidase genes from Betula platyphylla. Protoplasma 253 (5), 1347–1363. doi: 10.1007/s00709-015-0893-3
Zhao, Y., Chang, S. K., Qu, G., Li, T., Cui, H. (2009). Beta-sitosterol inhibits cell growth and induces apoptosis in SGC-7901 human stomach cancer cells. J. Agr. Food Chem. 57, 5211–5218. doi: 10.1021/jf803878n
Zhao, H., Tang, Q., Mo, C., Bai, L., Tu, D., Ma, X. (2017). Cloning and characterization of squalene synthase and cycloartenol synthase from Siraitia grosvenorii. Acta Pharm. Sin. B 7, 215–222. doi: 10.1016/j.apsb.2016.06.012
Keywords: Torreya grandis, transcriptional regulation, squalene synthase, sitosterol biosynthesis, drought
Citation: Zhang F, Kong C, Ma Z, Chen W, Li Y, Lou H and Wu J (2023) Molecular characterization and transcriptional regulation analysis of the Torreya grandis squalene synthase gene involved in sitosterol biosynthesis and drought response. Front. Plant Sci. 14:1136643. doi: 10.3389/fpls.2023.1136643
Received: 03 January 2023; Accepted: 04 May 2023;
Published: 20 June 2023.
Edited by:
Meng Kou, Xuzhou Institute of Agricultural Sciences in Jiangsu Xuhuai District, ChinaReviewed by:
Chen Junfeng, Shanghai University of Traditional Chinese Medicine, ChinaAlbert Ferrer, University of Barcelona, Spain
Copyright © 2023 Zhang, Kong, Ma, Chen, Li, Lou and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heqiang Lou, 20170030@zafu.edu.cn; Jiasheng Wu, wujs@zafu.edu.cn
†Present address: Feicui Zhang Institute of Maize and Featured Dryland Crops, Zhejiang Academy of Agricultural Sciences, Dongyang, China
‡These authors have contributed equally to this work
 Feicui Zhang
Feicui Zhang Congcong Kong†‡
Congcong Kong†‡ Zhenmin Ma
Zhenmin Ma Heqiang Lou
Heqiang Lou Jiasheng Wu
Jiasheng Wu