- 1College of Horticulture, Shenyang Agricultural University, Shenyang, China
- 2National & Local Joint Engineering Research Center of Northern Horticultural Facilities Design & Application Technology (Liaoning), Shenyang, China
- 3Key Laboratory of Protected Horticulture (Shenyang Agricultural University), Ministry of Education, Shenyang, China
- 4Key Laboratory of Horticultural Equipment, Ministry of Agriculture and Rural Affairs, Shenyang, China
- 5Collaborative Innovation Center of Protected Vegetable Provincial Co-construction Surrounds Bohai Gulf Region, Shenyang, China
Plant height is an important agronomic trait. Dwarf varieties present several advantages, such as lodging resistance, increased yield, and suitability for mechanized harvesting, which are crucial for crop improvement. However, limited research is available on dwarf tomato varieties suitable for production. In this study, we report a novel short internode mutant named “short internode and pedicel (sip)” in tomato, which exhibits marked internode and pedicel shortening due to suppressed cell elongation. This mutant plant has a compact plant structure and compact inflorescence, and has been demonstrated to produce more fruits, resulting in a higher harvest index. Genetic analysis revealed that this phenotype is controlled by a single recessive gene, SlSIP. BSA analysis and KASP genotyping indicated that ERECTA (ER) is the possible candidate gene for SlSIP, which encodes a leucine-rich receptor-like kinase. Additionally, we obtained an ER functional loss mutant using the CRISPR/Cas9 gene-editing technology. The 401st base A of ER is substituted with T in sip, resulting in a change in the 134th amino acid from asparagine (N) to isoleucine (I). Molecular dynamics(MD) simulations showed that this mutation site is located in the extracellular LRR domain and alters nearby ionic bonds, leading to a change in the spatial structure of this site. Transcriptome analysis indicated that the genes that were differentially expressed between sip and wild-type (WT) plants were enriched in the gibberellin metabolic pathway. We found that GA3 and GA4 decreased in the sip mutant, and exogenous GA3 restored the sip to the height of the WT plant. These findings reveal that SlSIP in tomatoes regulates stem elongation by regulating gibberellin metabolism. These results provide new insights into the mechanisms of tomato dwarfing and germplasm resources for breeding dwarfing tomatoes.
1 Introduction
Ideal plant architecture is a significant breeding objective for numerous crops (Reinhardt and Kuhlemeier, 2002). Dwarfism is a desirable characteristic in crop breeding because plant biomass is strongly correlated with its height, confers enhanced resistance to lodging damage from wind and rain, and is associated with stable and increased yields by improving the harvest index (Law et al., 1978; Miralles et al., 1998; Li et al., 2018). In particular, in the 1960s and 1970s, the Green Revolution, which introduced semi-dwarf wheat and rice varieties, substantially increased grain yield throughout Asia (Peng et al., 1999; Khush, 2001). Presently, a considerable number of dwarfing genes have been identified, characterized, and successfully cloned in diverse crop species. For instance, more than 60 dwarfing genes have been identified and cloned in rice, including sd-1, D-h, and OsKS2 (Liu et al., 2017). In wheat, 26 dwarfing genes have been identified, primarily belonging to the Rht family (Wang et al., 2014). In tomatoes, genes associated with gibberellin and brassinosteroid synthesis and signaling pathways, such as PROCERA, JMJ524, and DWARF, have been identified to regulate plant height (Bassel et al., 2008; Li et al., 2015; Livne et al., 2015; Li et al., 2016). Cavasin et al. (2021) evaluated a tomato line carrying the dwarf gene and reported a yield gain of 8.04%. Many dwarf and semi-dwarf cultivars, such as Micro-Tom and Micro-Gold, have been developed as ornamental cultivars or used in genetic transformation studies because of their greater spatial efficiency (Scott et al., 1995; Meissner et al., 1997; Gerszberg et al., 2014). Dwarf breeding is a beneficial option for adjusting different crops to a particular cropping system without compromising productivity or other traits. However, dwarf tomato varieties suitable for crop production have yet to be explored and developed.
Several dwarfing mutants are accompanied by shortening of other tissues and organs. Pedicels are specialized internodes that support and orient flowers and fruits at an upward angle on the inflorescence stem (Douglas et al., 2002). The length of the pedicel is related to traits, such as mechanized harvesting. Additionally, the length and orientation of pedicels directly determine the architecture of the inflorescence or spikes, which can significantly affect crop yield (Prenner et al., 2009). A single ERECTA (ER) mutation in Arabidopsis thaliana resulted in short stature and compact inflorescences (Torii et al., 1996). The ER family (ERf) is considered a crucial signal in plant development, among the key players during morphogenesis, and generates different responses in different tissues and organs (Shpak, 2013). ERECTA family of receptors is an ancient family of leucine-rich repeat RLKs that contain a signal peptide, leucine-rich repeat in the extracellular domain, transmembrane domain, and cytoplasmic serine/threonine protein kinase domain (Lease et al., 2001). Previous research on ER mutants in Arabidopsis thaliana has revealed that ER is one of the most important genes for promoting localized cell proliferation. ER controls multiple aspects of plant growth and development, such as stomatal development and differentiation (Shpak et al., 2005), plant morphogenesis, inflorescence morphology, leaf initiation (DeGennaro et al., 2022), multiple organ differentiation, and drought resistance (Shen et al., 2015; Li et al., 2021).
The function and mechanism of ERfs are best comprehended in Arabidopsis thaliana concerning stomatal development. ERf receptors activity are regulated by the secretion of peptides, including those of the EPF/EPFL family, which can function as agonists or antagonists during stomatal development (Shpak, 2013). Ligands that bind to the LRR domain can activate the ER protein and subsequently trigger the mitogen-activated protein kinase (MAPK) signal transduction cascade involving YDA, MKK4/MKK5, and MPK3/MPK6 (Meng et al., 2012). The ER kinase domain is relatively conserved across species, whereas the extracellular LRR domain is markedly different (Kosentka et al., 2017). This suggests that the ER-MAPK module exhibits a relatively stable behavior, and that mutations in the LRR domain are crucial for investigating ER function. The mechanism by which ER regulates stem cell development is similar to that of stomata. ERfs are expressed in the epidermis and xylem, and EPF/EPFL act as upstream regulators of ER activity (Uchida et al., 2012), whereas the MAPK cascade functions as a downstream regulator of ER signaling (Meng et al., 2012). ER expression under a series of promoters can rescue elongation defects of stems, pedicels, epidermis, and leaves when expressed in the phloem (Uchida et al., 2012). EPFL4 (also known as CHALLAH-LIKE2 [CLL2]) and EPFL6 (CHALLAH [CHAL]) are upstream of ERfs and redundantly regulate stem elongation and inflorescence growth. The epfl4 epfl6 double mutant does not have any apparent epidermal defects but shows dwarfing and compact inflorescences (Abrash et al., 2011). PACLOBUTRAZOL RESISTANCE (PRE) is downstream of the MAPK cascade, which promotes pedicel elongation (Cai et al., 2017). HOMOLOG OF BEE2 INTERACTING WITH IBH 1(HBI1) is downstream of PRE1. HBI1 can promote the expression of the BR biosynthesis genes CYP85A2 and AUXIN RESPONSE FACTOR 3 (ARF3) by directly binding to their promoters, which influences the expression of genes involved in auxin biosynthesis and signaling. Although the process of signal transmission from the extracellular to the intracellular space is complex, its ultimate outcome is determined by hormone signals. In melons, ER regulates stem elongation through auxin signaling by directly affecting polar auxin transport (Yang et al., 2020). Xu et al. (2023) found that an ER mutant in cucumber led to the formation of short internodes by reducing the expression of auxin genes and decreasing endogenous auxin content. However, the regulatory processes in tomatoes remain unclear.
In this study, we aimed to identify and functionally characterize a novel dwarf and short pedicels mutant named “short internode and pedicel (sip)” in tomato. Through BSA analysis, we found that the mutation in sip was due to the base substitution A134T in ERECTA, which encodes an LRR receptor-like serine/threonine protein kinase. This mutation induces conformational changes in the LRR domain, resulting in up-regulation of gibberellin degradation genes and changes in endogenous gibberellin levels. These findings will help to elucidate the mechanism of dwarfing in tomatoes and enable the development of potential dwarf-breeding genes.
2 Materials and methods
2.1 Plant materials and population development
Sequenced tomato ‘Heinz 1706’ as wild-type (WT) and ‘Alisa Craig’(AC) as parentage of another population were obtained from Tomato Genetics Resource Center (https://tgrc.ucdavis.edu/). sip is the mutant line of ‘Heinz 1706’ that has been self-crossed for six generations through ethyl methanesulfonate (EMS) mutagenization. For genetic analysis, sip was crossed with WT to obtain the F1 and F2 generations. The F1 was backcrossed with the parents to obtain BC1 generations. Similarly, sip was crossed with AC to obtain F1, F2, and BC1 progeny. All plants were grown in greenhouses at 15–28 °C at the Shenyang Agriculture University(Shenyang, China).
2.2 Microscopic observation of internode and pedicel
The central elongating sections of the stems of the WT and sip plants were sampled at the seedling stage with six true leaves. The stalks sampled at the flowering stage contained out-of-zone parts from the WT and sip plants for comparison of their microscopic structures. Tissues were fixed overnight in formalin-acetic alcohol (FAA). The samples were then dehydrated in a gradient alcoholic solution and soaked in different proportions of ethanol-xylene solution. These tissues were embedded in paraffin, and transverse and longitudinal sections were stained with toluidine blue and examined under a Zeiss microscope (MicroBeam IV, Germany).
2.3 Pool construction and bulked-segregant analysis
We used two F2 populations to identify SlSIP: cross sip × WT for MutMap and cross sip × AC for QTL-seq. For MutMap analysis, we used 28 F2 individuals (WT-sip hybrid population) with shortened internodes and pedicels to construct the DNA pools “F2dw-pool”. For QTL-seq, 45 individuals with short internodes and flower stalks were selected from the isolated population of AC and sip hybrid in the F2 generation to construct “SF-pool”. In comparison, 50 individuals with normal height and long pedicels were selected to construct “HL-pool”. Genomic DNA from the parents (WT, AC, and sip) and F2 was extracted from fresh leaves using a DNA Secure Plant Kit (Tiangen, Beijing, China).
Qualified total genomic DNA was used to construct paired-end libraries with an insert size of 300–400 bp using a Paired-End DNA Sample Prep kit (Illumina Inc., San Diego, CA, USA). These libraries were sequenced using a NovaSeq6000 (Illumina Inc., San Diego, CA, USA) at GeneDenovo (Guangzhou, China). To identify SNPs and InDels, the BWA software (Li and Durbin, 2009) was used to align the clean reads from each sample to the tomato reference genome SL4.0 (https://solgenomics.net/organism/Solanum_lycopersicum/genome). The analytical method was the same as that described by (Wang et al., 2017). The QTL-seq method was referenced and improved on the method proposed by Takagi et al. (2013). Metadata for the bioproject are available at NCBI under the accession number PRJNA1006521 and PRJNA1007367.
2.4 SNP genotyping by KASP and PARMS
According to the SNPs between the WT and sip identified via DNA sequencing (MutMap). Kompetitive allele-specific PCR (KASP) markers in the 20 and 40 Mb regions of chromosome 8 (chr.8) that are specific for different subgenomes were designed and named M20 and M40, respectively. Based on mutations in the candidate genes Solyc08g061560 and Solyc08g062600, we designed markers K1 and K2 for genotyping. For genotyping analysis, DNA was extracted from 3 WT, 3 sip, and 165 F2 individuals showing the sip phenotype.
The penta-primer amplification refractory mutation system (PARMS) is a KASP-like SNP genotyping technique that combines ARMS, also known as allele-specific PCR (Heim and Meyer, 1990), with universal energy transfer–labeled primers (Nuovo et al., 1999). Based on the QTL-seq results, we designed markers every 5 Mb within a range of 10–50 Mb on chromosome 8. These markers were named P1–P10. SNP calling and plotting were performed using the online software snpdecoder (http://www.snpway.com/snpdecoder/) with manual modifications. All primer sequences are listed in Supplementary Table S1.
2.5 CRISPR/Cas9Pubi-H-ER vector construction and tomato genetic transformation
The two SG sequences used were SG61560-1: 5’-GCTGATTACTGTGCCTGGAG-3’ and SG61560-2: 5’-GGGGAGTTGTCTCCTGCTAT-3’. Oligo dimer was prepared using a 20-µL reaction mixture containing 16 µL of annealing buffer, 1 µL of SG61560-UP1, 1 µL of SG61560-LW1, 1 µL of SG61560-UP2, and 1 µL of SG61560-LW2. The reaction mixture was centrifuged at 12000 rpm for 1 min and was diluted to 10 µM by adding deionized water. The reaction conditions were denaturation at 95 °C for 3 min and annealing. The oligo dimer was inserted into the target vector in a reaction mixture containing 1 µL of saCas9/g RNA Vector, 1 µL of oligo dimer, 1 µL of Solution1, 1 µL of Solution2, and 6 µL of H2O. It was incubated at 16 °C for 2 h. The sequencing primer used was PUV4-R: 5′-TCCCAGTCACGACGTTGTAA-3′. The transgenic tomato lines were selected based on their hygromycin resistance. Tomato genetic transformation method can be referred to Sun et al. (2020).
2.6 Molecular dynamics simulation of ER and SlSIP
The protein sequences of ER and SlSIP were obtained from NCBI (National Center for Biotechnology Information https://www.ncbi.nlm.nih.gov/). RosetaTTAFold (Baek et al., 2021) was used for modeling, and confidence scores were 0.81 and 0.78, respectively. A good-quality model was used for the subsequent analyses. Gromacs2019.6 (Van Der Spoel et al., 2005) was selected as the kinetic simulation software and amber14sb was selected as the protein force field. A TIP3P water model was applied to the system, a water box was established (with its edge at least 1.2 nm from the protein edge), and a sodium ion balance system was added. Particle-mesh Ewald (PME) handles electrostatic interactions using the steepest descent method for energy minimization over 50,000 steps. The cutoff distances of the Coulomb force and van der Waals radius were 1.4 nm. A canonical ensemble (NVT) and constant-pressure and constant-temperature (NPT) were used to balance the system. Subsequently, 100 ns MD simulations were performed at normal temperature and pressure. The V-rescale temperature coupling method and Berendsen (Berendsen et al., 1984) method were used to control the simulated temperature at 300 K and pressure at 1 bar. The results were analyzed using various built-in Gromacs package functions, such as root mean square deviation (RMSD), radius of gyration (RoG), root mean square fluctuation (RMSF), and solvent-accessible surface area (SASA). PyMOL 2.5.1 (Mooers, 2019) was used for the graphical display and LigPlot 2.1 (Laskowski and Swindells, 2011) was used for the visual display.
2.7 RNA extraction and transcriptome sequencing
Approximately 500 mg of the third internode of the WT and sip was collected at four true leaf stage for transcriptome analysis. Total RNA from six samples (three biological replicates per sample) was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). The RNA-seq analysis was performed as described previously (Sun et al., 2020). Metadata for the bioproject are available at NCBI under the accession number PRJNA1003223.
2.8 Real-time quantitative PCR
Stems from 50 WT and sip plants were collected from 3–4 true leaves for mRNA extraction using an RNA extraction kit (Kangwei, China) and cDNA synthesis using reverse transcriptase (TaKaRa, China). The tomato gene SlActin was used as a control (all primer sequences are listed in Supplementary Table S2). A 20-μL reaction mixture was prepared including 2 µL of cDNA (1:10 dilution), 10 µL of 2× TransStart Top Green qPCR SuperMix (TransGen, China), 0.5 µL of each primer (10 µmol/µL), and ddH2O. The reaction conditions were: 95°C for 7 min; 40 cycles at 95°C for 10 s, 58°C for 30 s, and 72°C for 20 s; and then 71 cycles at 95°C for 10 s, 60. 5°C for 10 s, and 95°C for 10 s. The relative gene expression levels were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
2.9 Plant hormone treatment and endogenous GA determination
Seedlings at the 3–4 leaf stage of sip and WT were treated with foliar sprays of GA1 (100 µM), GA3 (100 µM), GA4 (100 µM), GA7 (100 µM), IAA (50 µM), and H2O (as a control). Spraying was performed every three days for one week, and plant height was recorded. Sampling was performed, during the four-leaf stage, approximately two weeks after the seeds were planted. Approximately 1 g of young and tender stems (specifically the second internode) was collected from both sip mutants and WT plants, with three biological replicates for each. Phytohormones were quantified following Li et al. (2019) with slight modifications, using a liquid chromatography-mass spectrometry system (Agilent 1290, AB company Qtrap6500; California, CA, USA).
2.10 Subcellular localization
The coding sequence (CDS) of ER was cloned into the binary vector pCAMBIA-1300-35S-GFP. Primer sequences: LRR-1300-BAMI-F:5’-GCTCGGTACCCGGGGATCCATGGCATCATTTTTACTCCAAAGAT-3’ (Tm: 60.8 °C); and LRR-1300-SALI-R: 5’-GCCCTTGCTCACCATGTCGACGCCACTATTCTGGGATATGACCT-3’ (Tm: 59.3 °C). The recombinant constructs were transformed into Agrobacterium tumefaciens strain GV3101. Subsequently, transformed material was cultured in YEP liquid medium (containing 50 mg·mL-1 kanamycin and 100 mg·mL-1 rifampicin) at 28°C for 16 h. The bacteria were then centrifuged and slowly resuspended in a suspension medium (10 mM MES, 10 mM MgCl2, 1 M acetosyringone). After 48h of incubation in the dark, the fluorescence signals were observed using a Leica laser confocal microscope (TCS SP82400301, Germany).
2.11 In situ hybridization
The unique sequence of the ER transcript was amplified from WT cDNA using ExTaq polymerase (TaKaRa) and ligated into pSPT18 and pspt19 vectors. The constructs were linearized and subjected to RNA labeling reaction using T7 or SP6 RNA polymerase. Slice were prepared and in situ hybridization analysis was performed following the methods described by Cheng et al. (2022).
2.12 Prediction of promoter binding elements
The promoter sequences (2000 bp before the coding region) of genes related to gibberellin synthesis and degradation were obtained from NCBI. The promoter sequences were then predicted using the online promoter prediction software PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and PlantRegMap (http://plantregmap.gao-lab.org/binding_site_prediction_result.php).
3 Results
3.1 Phenotypic identification and genetic analysis of short internodes and pedicels mutants sip
In this study, Heinz ‘1706’ was used as WT. Additionally, a stably inheritable dwarf mutant line “sip,” exhibiting significantly shortened internodes and petioles, was obtained through EMS mutagenesis. From the seedling stage, sip showed significant dwarfism compared to WT (Figure 1B), which persisted throughout the growth period (Figures 1A, B, K). At the fifth true leaf stage, the plant height of the sip was only 58% that of the WT (Figures 1A, B). However, the number of internodes in sip and WT remained constant throughout the different periods (Figure 1C), and have the same number of leaves during the seedling stage (Supplementary Figure 1A), suggesting that dwarfing was due to internode shortening rather than a growth lag. Observation of paraffin sections of stems indicates that the elongation of stem cells are significantly inhibited in sip (Figures 1D, E), but the cross-section showed no significant difference in cell area (Supplementary Figures 1B, C, H). In addition to internode shortening, the most prominent phenotype of sip was a compact inflorescence structure with severely shortened flower stalks (Figure 1G). The shortening stalk was caused by both the shortening of the proximal axial end length and the abnormal development of the position of the abscission zone, which made the distal axial end almost disappear (Figures 1G–J). Considering the abnormal morphology of the abscission zone, we compared the abscission rates of WT and sip and found that sip had a lower abscission rate than that of WT. Paraffin sections of petioles revealed that the small cell layer of the abscission zone of sip was discontinuous between the medulla and cortex compared with that of the WT (Figures 1D, F), which might explain why sip was less likely to have abscission than the WT. sip exhibits a compact cluster of inflorescences, interestingly, the single inflorescence of sip can form more flowers (Figures 1G, M).This characteristic enables sip to produce more fruits, resulting in a significantly higher yield index (Figures 1K, M; Supplementary Figure 1L). We have noticed that compared to WT, the fruits of sip have a more flattened round shape, and the ratio of fruit diameter to length has decreased, as well as the weight of individual fruits (Supplementary Figures 1D-G, K). However, this does not affect the yield per plant of sip, as it has more fruits.
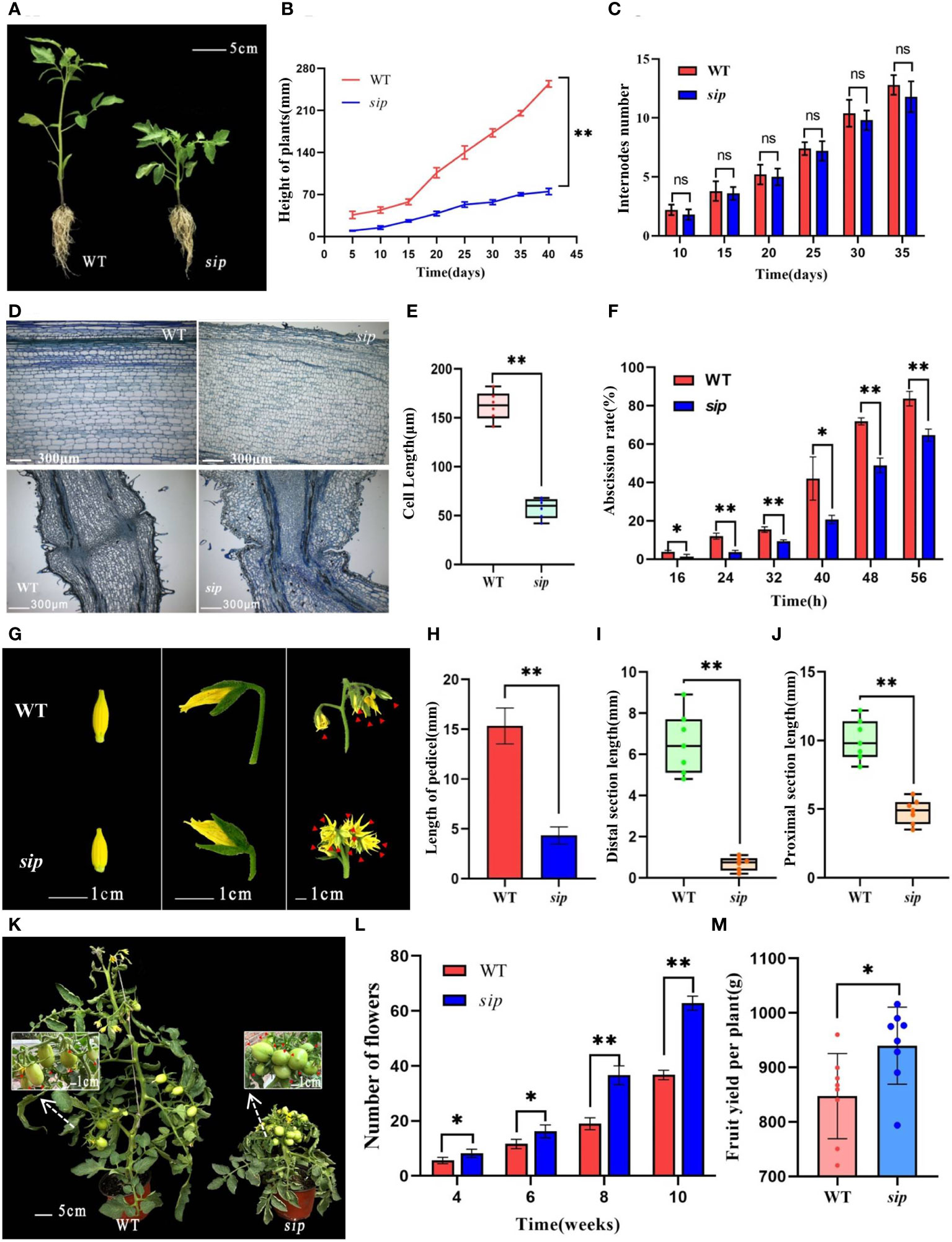
Figure 1 Phenotypic identification of sip mutants. (A) Plant height of WT and sip at five true leaves stage (n=3). (B, C) Plant height and stem internode number of two tomato lines at different stages (the beginning of sowing is regarded as 0 days, n=3). (D) Microscopic observation of longitudinal sections of stems and peduncles in WT and sip(the third internode of WT and sip at five true leaves stage and pedicels at the stage of full petal expansion). (E) Length of stem cells. (F) Abscission rate of WT and sip (three replicates, with 50 flower stalks per replicate). (G) Morphology of inflorescences and peduncles in WT and sip (flowers within the inflorescence marked with red arrows). (H–J) Length of the pedicels and lengths of the proximal section and distal section in WT and sip (n=7). (K) Plant morphology and fruit-setting capacity at the fruiting stage (fruits marked with red arrows). (L) Statistical analysis of the number of flowers in WT and sip plants. (M) Yield per plant. Student’s t-test was used to determine a significant difference. *p < 0.05 and **p < 0.01; ns, no significant difference. All data are presented as mean ± SD.
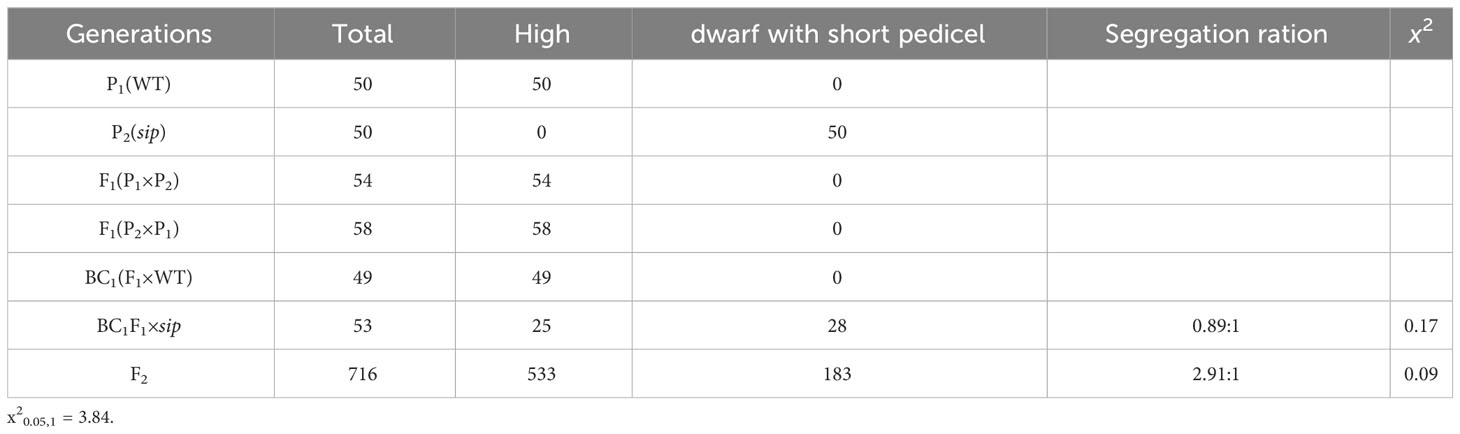
To genetically characterize the short internodes with the loss-abaxial pedicel phenotype of the sip mutant, we generated an F2 population of 716 individuals by crossing the WT with sip. All F1 plants showed normal plant height and pedicels. Specifically, 533 individuals out of 716 from the F2 population displayed a normal phenotype similar to the WT, whereas the remaining 183 individuals were similar to sip with a 3:1 segregation ratio, as revealed by the chi-square test (Table 1, χ2 = 0.09). This indicates that a single recessive gene is responsible for sip phenotype. This hypothesis is further supported by the 1:1 Mendelian ratio observed in the BC1F1 population (Table 1; χ2 = 0.17).
3.2 Mapping of the SlSIP by BSA-seq
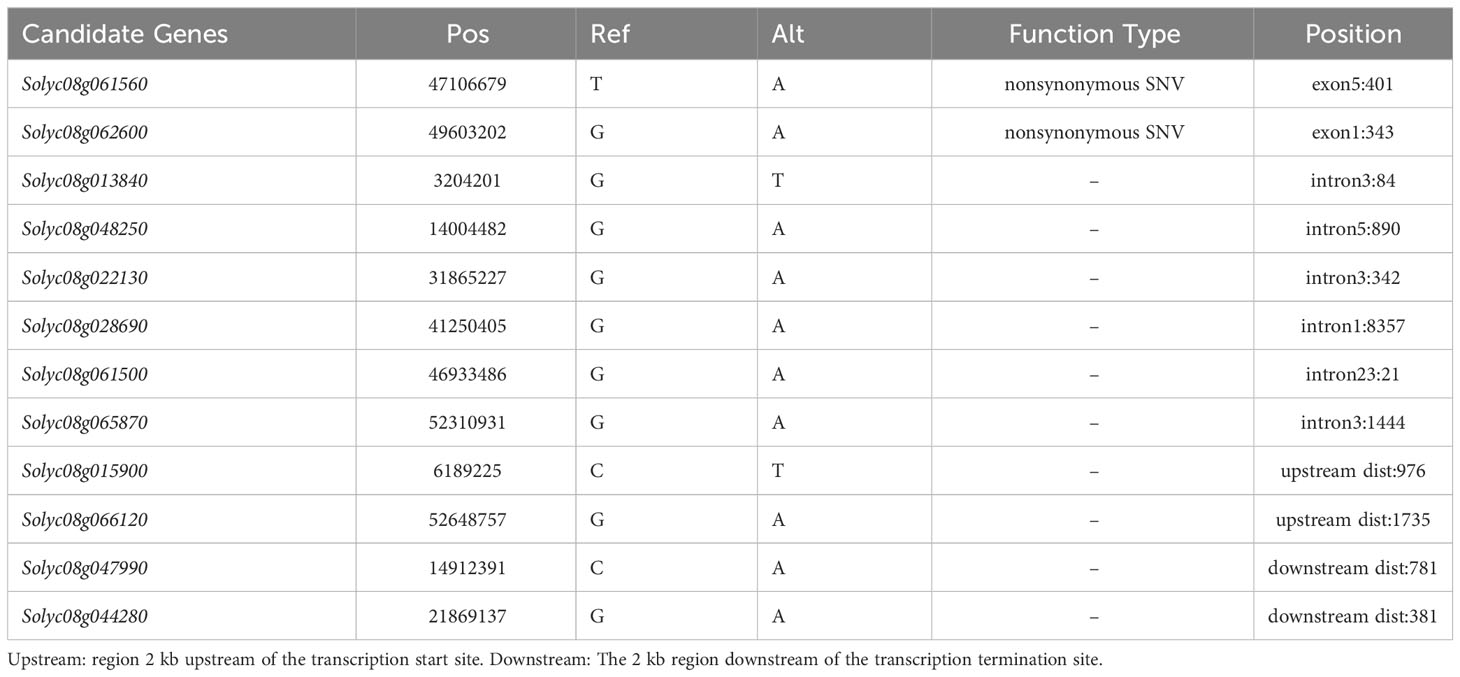
Candidate genes were identified using MutMap analysis. In total, 181746682 and 175293598 clean reads (150 + 150 bp) were acquired from the F2sip-pool (average read depth > 34.27×depth coverage or 99.84% coverage) and the WT (33.05×depth coverage or 99.83% coverage), respectively. To reduce background noise, we used the sliding window method to fit the SNP index and related calculation results. The allele frequency differences of 4,233 filtered SNPs from the two pools were calculated and mapped across the 12 tomato chromosomes to create a Manhattan plot (Figure 2A). Only the 3rd and 8th chromosomes had SNPs with confidence intervals exceeding 99%. Surprisingly, a large number of SNP-index peak values of 1 in chr.8 were observed. After excluding the F2-sip heterozygous sites and sites congruent with the reference genome, we obtained 226 SNPs on chr.08. Two SNPs were found in exons, six in introns (two upstream and two downstream), and the remaining 214 in intergenic regions. Candidate genes were defined based on 12 single nucleotide polymorphisms (SNPs) located outside the intergenic region (Table 2).
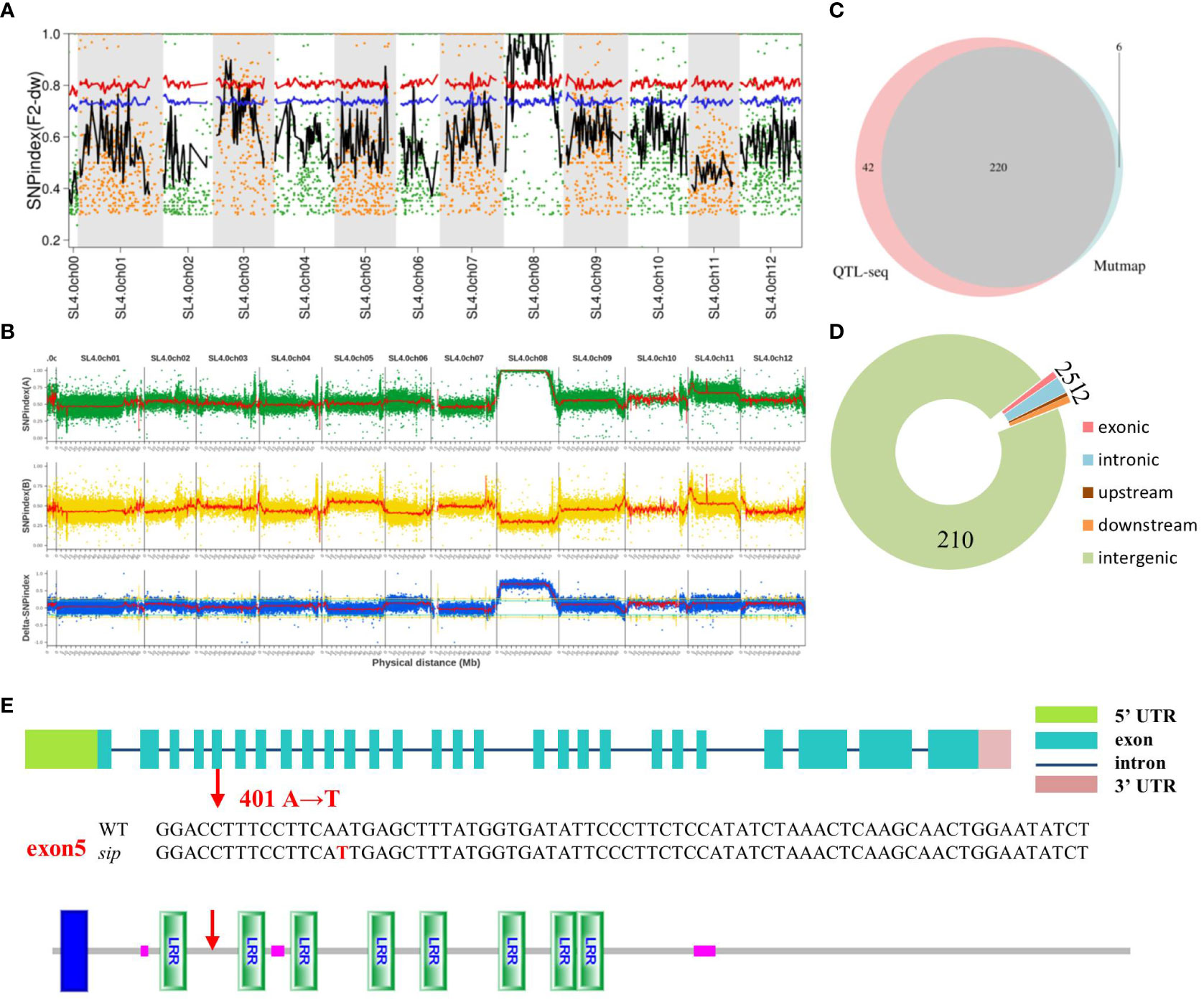
Figure 2 Genome-wide distribution of SNP-index. (A) Differences in allele frequencies among 28 lines with the sip phenotype in the F2 population and WT (the red and blue line represents the 99% and 95% confidence interval, respectively). (B) Differences in allele frequencies between 45 lines with the sip phenotype in the F2 population and AC. The X-axis represents the 12 chromosomes of tomato. The Y-axis represents the difference in allele frequencies (SNP values) between the two pools. (C) The Venn diagram of SNP-index of the two databases. (D) The distribution of SNPs in the intersection of the two databases. (E) Analysis of CDS sequence and protein sequence structure of mutation site in sip. (Red arrows represent mutation sites; protein structure prediction using online prediction tool: http://smart.embl-heidelberg.de/).
To obtain fewer candidate genes and shorter candidate regions, another F2 generation isolated population was constructed by crossing sip with the distantly derived variety (AC). The F2 generation had a 3:1 separation ratio, 304 tall plants with normal pedicels, and 121 short internodes with loss-abaxial plants (χ2 = 0.09). The shortened internodes at the seedling stage and the structure of the absent flower stalk made it easy to identify extreme phenotypes in the F2-isolated populations. QTL-seq was used and 282.43 Gb of clean data were obtained by Illumina NovaSeq, including 30.47 Gb, 29.25 Gb, 104.04 Gb, and 118.67 Gb from the AC, sip, F2SF-pool, and F2HL-pool mixed pools, respectively, all of which were of high quality (94.56% > Q30 > 92.84%) and had stable GC content (37.14% > GC > 36.45%). The average sequencing depths for the parent and F2 pools were 32.09× and 110.04×, respectively. A total of 293,701 SNPs were obtained from the parents and two mixed pools. The result positioned SlSIP to a large interval on chromosome 8 (Figure 2B). Although the candidate region remains extensive, the sequencing results from QTL-seq indicated that of the 17,713 SNPs on chr.8, 10,427 SNPs with differences were found to be in concordance with the reference genome. The 6,138 heterozygous loci in sip were also disregarded because it was previously determined that the mutated genes were recessive. Since there were no dwarfing or short pedicel plants in the parent AC under normal cultivation conditions, SNPs that exhibited the same genotype as sip in AC could also be excluded. After excluding heterozygous SNPs and those that were consistent with the reference genome in the F2SF-pool, and then removing SNPs in the F2HL-pool that had the same genotype as sip, 262 remaining SNPs were identified as potential candidates. The SNP distribution and annotation statistics are presented in Supplementary Table S3.
We developed PARMS markers every 5 Mb between 10 Mb and 50 Mb on chromosome 8 and validated them in F2 individuals with sip phenotype, yielding consistent results indicating significant linkage of large segments (Supplementary Figure 2). To narrow the range of candidate genes, we compared the data from the two sequencing results. Interestingly, we observed a substantial overlap (Figure 2C), which further confirmed the accuracy of both the sampling and sequencing processes. The overlapping portions of the two results included two SNPs located in the exonic region, five SNPs in the intronic region, one in the upstream region, two in the downstream region, and 210 in the intergenic region (Figure 2D).
3.3 Screening and identification of candidate genes as ERECTA
Candidate genes were present in the shared regions between the two sequencing results, and we temporarily ignored SNPs in the intergenic regions. Mutations in both exons were nonsynonymous. Solyc08g061560 encodes an LRR receptor-like serine/threonine-protein kinase (ERECTA), and Solyc08g062600 is an Exostosin family-like protein probable arabinosyltransferase ARAD1. We analyzed SNPs located in introns and none at intron boundaries (Table 2). Mutations in the upstream region may result in changes in the transcription recognition sites that alter gene expression at the transcriptional level. There may be some lncRNA recognition sites in downstream regions that regulate gene transcription. Additional quantitative fluorescence verification showed that only Solyc08g044280 expressed in the stems of seedlings among these three genes and the mutation did not affect their transcription at the seedling stage (Supplementary Figure 3).
These results suggest that the two nonsynonymous mutations in this exon may be candidate genes for the sip phenotype. Thus, we designed the KASP probe marking and classification to determine whether the two genes had a mutation phenotype separation. According to the WT and sip hybrid F2 populations, 92 individuals had short internodes and flower stalks for genotyping. Only SNP with ERECTA mutations were co-isolated from the mutant phenotype (Supplementary Table S4). These results indicated that ERECTA is a candidate gene responsible for the sip phenotype. Sequencing results showed a specific alteration was induced in exon 5 of ERECTA, resulting in the replacement of 401st base of the CDS from A to T (Figure 2E). This alteration resulted in the substitution of amino acid 134 from N to I. This amino acid change results in the conversion of the polar hydrophilic side chain into a nonpolar hydrophobic side chain. This suggests that the spatial conformation of the receptor was modified to some extent.
3.4 Protein structure analysis of the LRR domains of ER and SlSIP by MD simulations
The overall structure remained rigid after mutation, with an RMSD of 1.05 nm (Figures 3A–D, Supplementary Figure 4). Further examination of the mutated amino acids in the stick format revealed that ASN134 and ILE134 were located in the coil region of the overall structure. Specifically, they were positioned at the interface between the interior and exterior of the structure. Notably, Asn134 was more exposed than Ile134, indicating a higher degree of surface accessibility (Figures 3A, B). In particular, ASN134 in the extracellular region of a protein can interact with other amino acids. For the WT protein, hydrogen bonds were observed between Asn134 and Met107, Leu112, Leu131, and Ser132, all of which were located in parallel coil structures (Figure 3C). However, only contact with Asn158 has been observed at the mutant site Ile134 (Figure 3D). Calculations and simulations revealed differences in the secondary structures under stable conditions (Figure 3E). When comparing horizontally, we found that the mutated structures, β-sheets, turns, and 3-Helix, increased, whereas coils and bends decreased. In summary, this mutation significantly disrupts the original internal contacts of the protein, leading to increased flexibility and decreased rigidity. This alteration results in the emergence of new secondary protein structures.
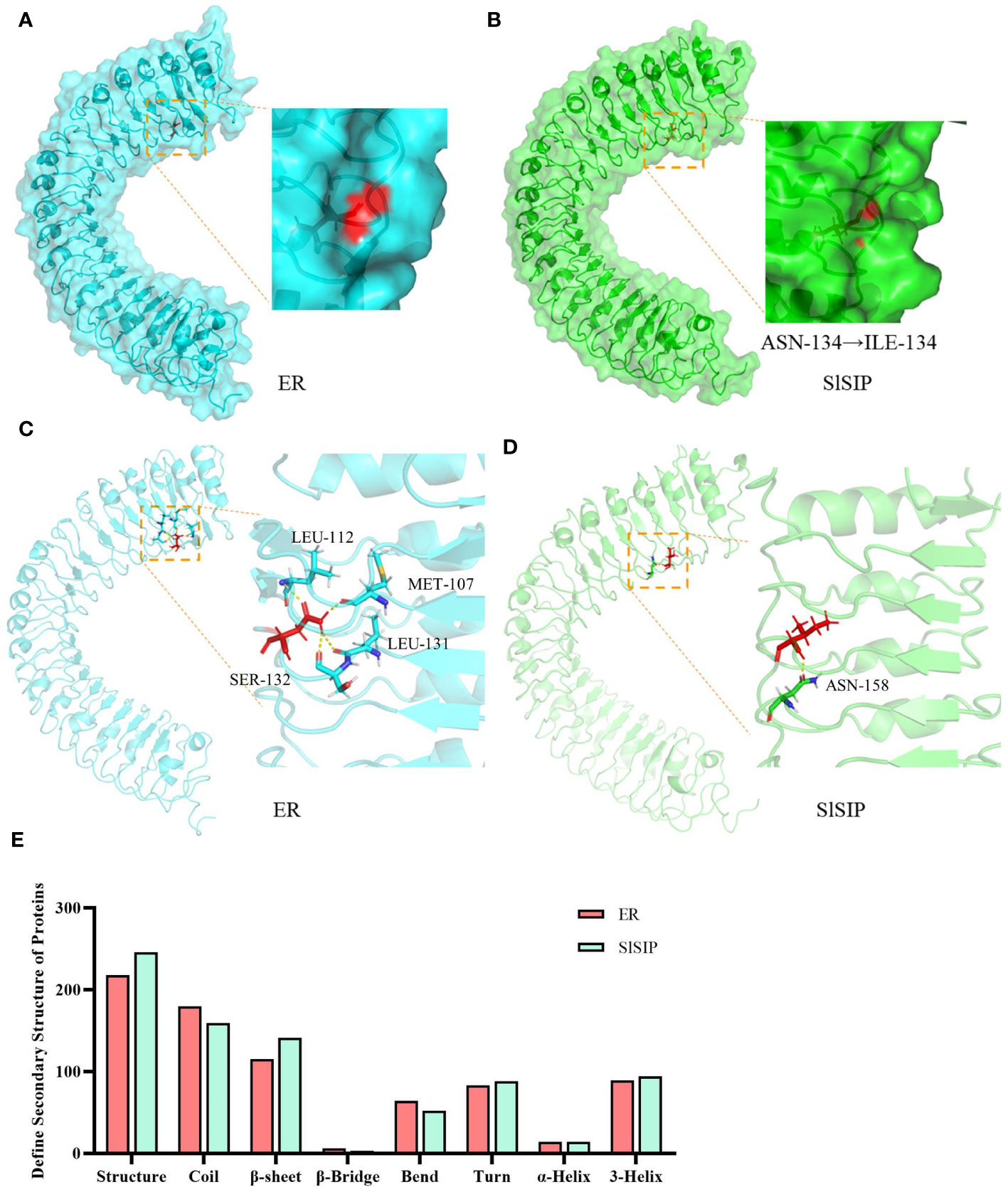
Figure 3 Protein structures of the LRR structural domains from ER and SlSIP by MD simulations. (A, B) The surface structure of the ER and SlSIP proteins, with 134 amino acid highlighted in red. (C, D) The rigid structure and internal binding of the ER and SlSIP proteins; the 134th amino acid marked in red, and the amino acids that form hydrogen bonds with the 134th residue are marked in different colors. (E) The differences in secondary structure between two LRR domains.
3.5 Function verification of SlSIP
To validate the function of SlSIP, we deleted the ER gene from the AC background using the CRISPR/Cas9 technology. The specific locations of the two target sites are shown in Figure 4A. After performing sequencing analyses on 30 plants in the T0 generation, we confirmed the presence of two homozygous er mutant plants, er#15 and er#16, and obtained the T1 generation. Non-edited negative plants were used as controls. Notably, plant er#15 and er#16 underwent precise editing at target site 1, resulting in a 1-bp deletion that caused the formation of a premature stop codon (Figure 4A). Consistent with the phenotype of sip, er#15 and er#16 both show obvious internode shortening, compact inflorescence, and shortened pedicels (both proximal section length and distal section length are significantly reduced), but the number of internodes has not decreased (Figure 4B; Supplementary Figures 5A–F). Compared to AC, the number of flowers in each inflorescence in er mutants have also increased, and correspondingly, the mutants produce more fruits per inflorescence (Figures 4C, D; Supplementary Figures S5G, S6). Similar to sip, er mutants also undergo changes in fruit morphology, with no significant difference in fruit diameter compared to AC, but a decrease in fruit length leads to a decrease in the fruit index and fruit weight (Supplementary Figures S5H–L). This consistent phenotype further supported the hypothesis that SlSIP functions as ER.
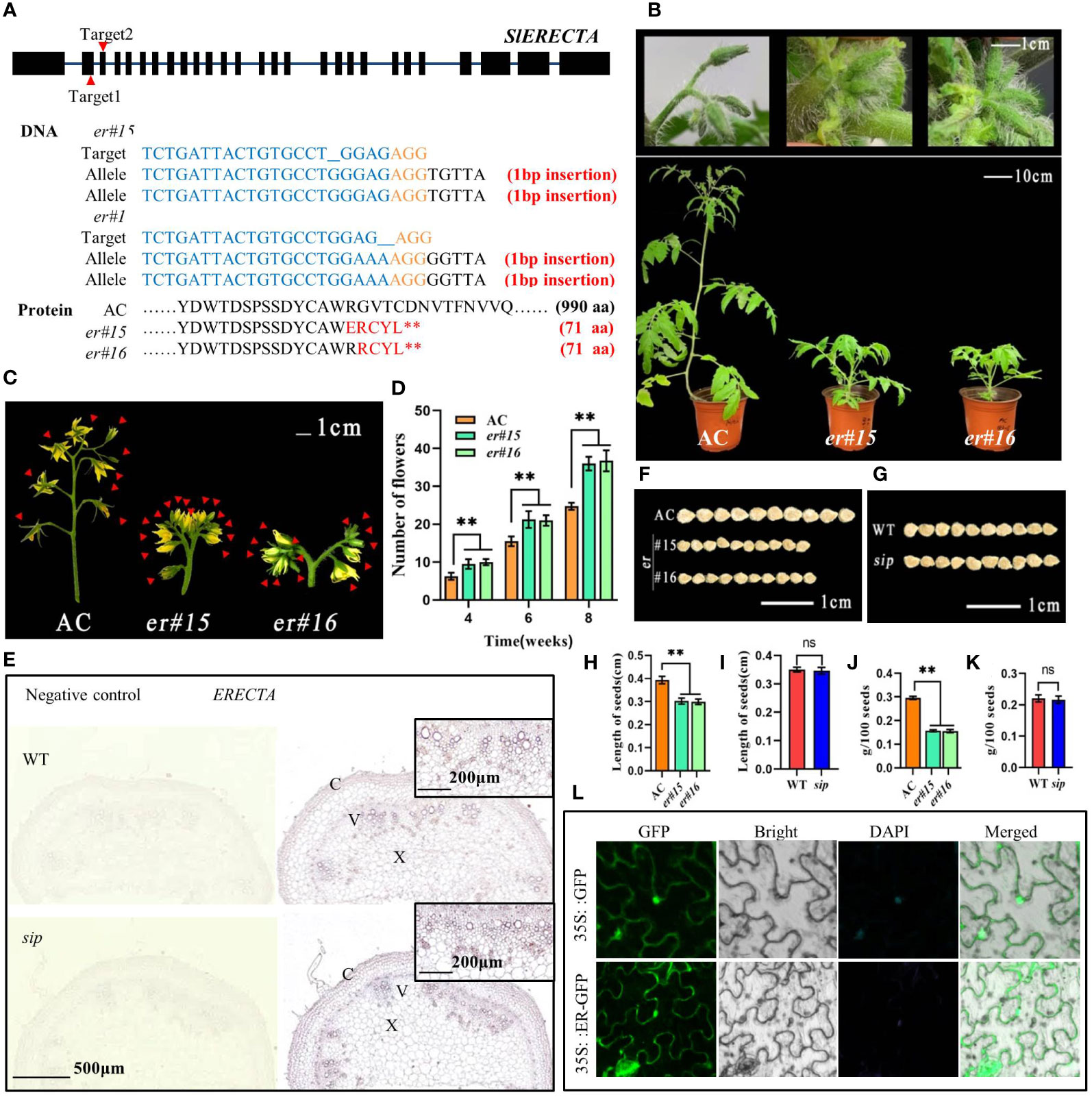
Figure 4 Functional verification of candidate gene SlSIP. (A) Targeting information of CRISPR/Cas9Pubi-H-ER vector and sequence alignment of ER mutant and background material. (B) Comparison of the phenotypes of ER loss-of-function mutants er#15 and er#16 with AC. (C) Comparison of inflorescences between AC and er mutant (flowers marked with red arrows). (D) Statistical analysis of the number of flowers in AC and er plants. (E) In situ hybridization of ER expression in the stems of WT and sip. (F, G) Comparison of seed size. (H–K) Length of seed (n=10) and hundred-seed weight (n=3). (L) Subcellular localization of ER. Student’s t-test was used to determine a significant difference. **P < 0.01; ns, no significant difference. All data are presented as mean ± SD.
Subcellular localization findings indicate that ER in tomato is positioned on the cell membrane (Figure 4L). In situ hybridization results showed that, similar to other species, ER was highly expressed in vascular bundles. However, there was no significant difference in the expression of ER in sip stems compared to that in the WT (Figure 4E). This indicated that the short internodes in sip stems were not caused by a decrease in SlSIP expression. Additionally, we have discovered an intriguing phenomenon. We compared the sip and ER knockout mutants and found that some ER functions were unaffected in sip. Specifically, we found that the seeds of the er mutant plants from both strains displayed a significant size reduction compared to that of AC (Figure 4F) and exhibited a significant decrease in seed length and 100-seed weight (Figures 4H, J). However, this phenomenon was not observed in the WT and sip (Figures 4G, I, K). Intriguingly, we also found that the disruption of ER function in tomatoes had a direct impact on seed size., suggesting that the mutated domain in sip plays a specific role in regulating stem elongation in tomato.
3.6 ER regulates internode elongation by modulating gibberellin levels in tomatoes
To gain insight into the mechanism by which ER regulates stem elongation, total mRNA was extracted from the young and delicate stems of WT and sip plants for transcriptome profiling. High-throughput sequencing was performed on 6 RNA-seq libraries, with three replicates of WT obtaining 52,976,758, 44,569,918, and 45,661,190 clean reads, and sip obtaining 45,345,936, 47,858,102, and 51,281,884 clean reads, respectively. A total of 18,504 genes were expressed in the young stems. Using a threshold of log2FC absolute value > 1 and p value < 0.05, 86 genes were down-regulated and 184 genes were up-regulated in expression (Supplementary Figure 7A, Supplementary Table S5). Cluster analysis demonstrated a high level of consistency between the three biological replicates of both WT and sip (Supplementary Figure 7B). To analyze the functional significance of these differentially expressed genes (DEGs), Gene Ontology (GO) term enrichment analyses and KEGG pathway enrichment analysis were conducted to investigate the potential biological functions of the DEGs. GO enrichment analysis showed that they were mainly enriched in three categories: biological process, cellular component, and molecular function (Supplementary Figure 8). KEGG pathway enrichment analysis reveals that up-regulated differentially expressed genes are mainly enriched in diterpenoid biosynthesis pathways, phenylalanine metabolism and RNA polymerase, etc. KEGG pathway map of The down-regulated genes are mainly enriched in various metabolic pathways such as linoleic acid metabolism and diterpenoid biosynthesis pathways (Figure 5A; Supplementary Figure 9, Supplementary Table S6). Several transcription factor expression levels are changed in the sip variant juvenile stems, with 11 up-regulated and 7 down-regulated, including 6 members of the MYB family, 2 members of the bHLH family, 2 members of the WARK family, 2 members of the AP2 family, 2 members of the ASR family, 1 member of the TGA family, 1 member of the HSF family, 1 member of the MADS family, and Nuclear Transcription Factor Y subunit B-5-like (Figure 5C).
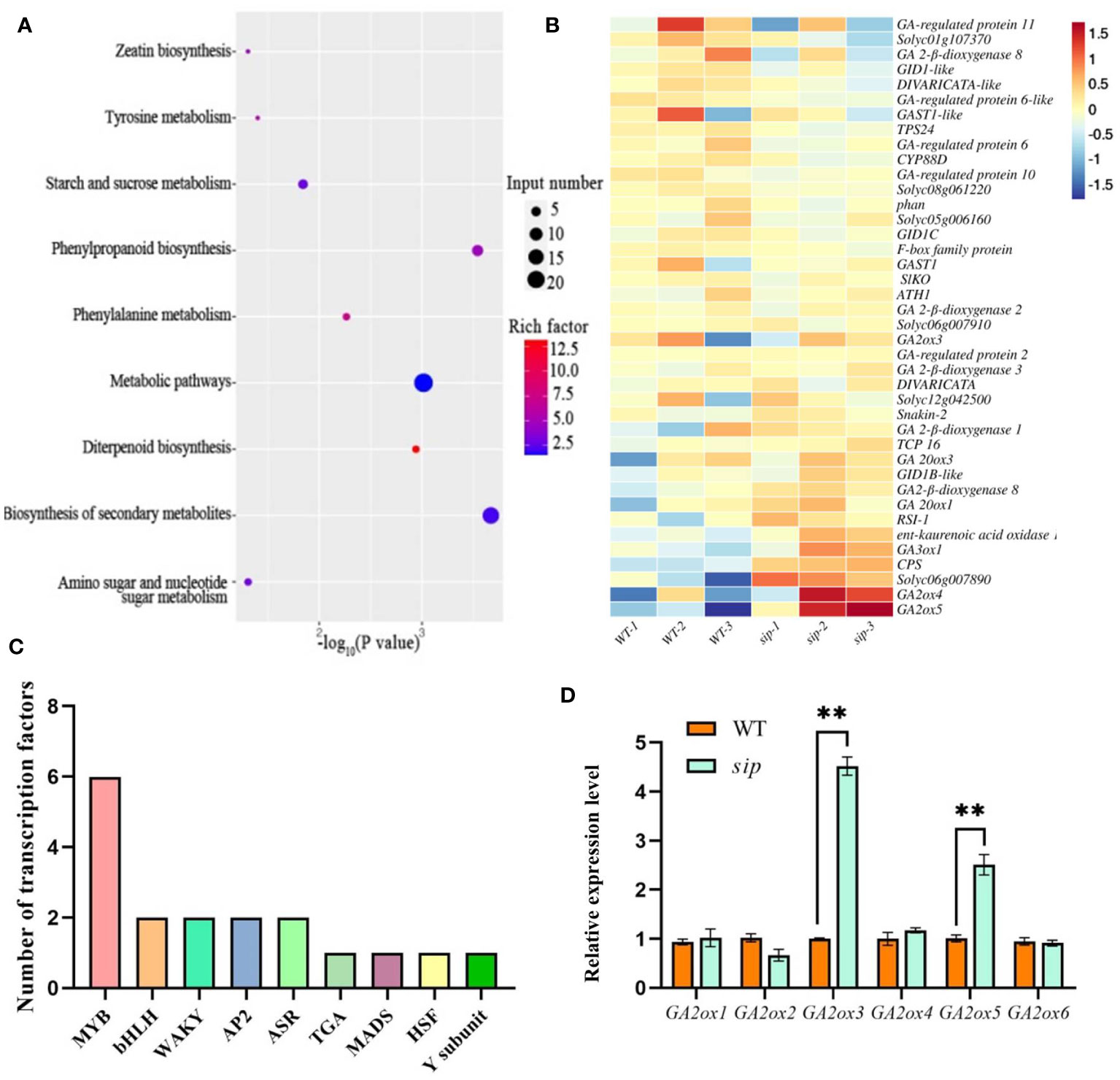
Figure 5 Differentially expressed genes between WT and sip. (A) KEGG pathway map of up-regulated genes. (B) Heat map of gibberellin-related genes in RNA-seq. (C) Number of differentially expressed transcription factors. (D) Real-time quantitative validation of differentially expressed genes (n=3). A Student’s t-test indicated a significant difference. **P < 0.01. All data are presented as mean ± SD.
Diterpenoids are precursors of gibberellin synthesis, and several DEGs have been implicated in the gibberellin signaling pathway, including gibberellin biosynthesis, degradation, and polar transport genes (Figure 5B). Quantitative real-time PCR analysis verified that the relative expression levels of gibberellin degradation genes were significantly higher in sip than in WT (Figure 5D). Is the short internode of sip caused by the up-regulated expression of gibberellin-degrading genes? We further measured the gibberellin content in the young stems of both WT and sip. Measurement of endogenous gibberellin levels revealed a significant reduction in the levels of the active gibberellins GA3 and GA4 in sip (Figure 6A). Exogenous application of GA3 restored the height of sip to that of WT plants, and ER knockout mutants er#15 and er#16 responded similarly to gibberellin (Figures 6B–E). Quantitative real-time PCR result reveals that the expression levels of GA2oxs are elevated in the stems of both er#15 and er#16 (Figure 6F). These lines of evidence suggest that ER may regulate internode elongation in tomatoes by modulating gibberellin metabolism.
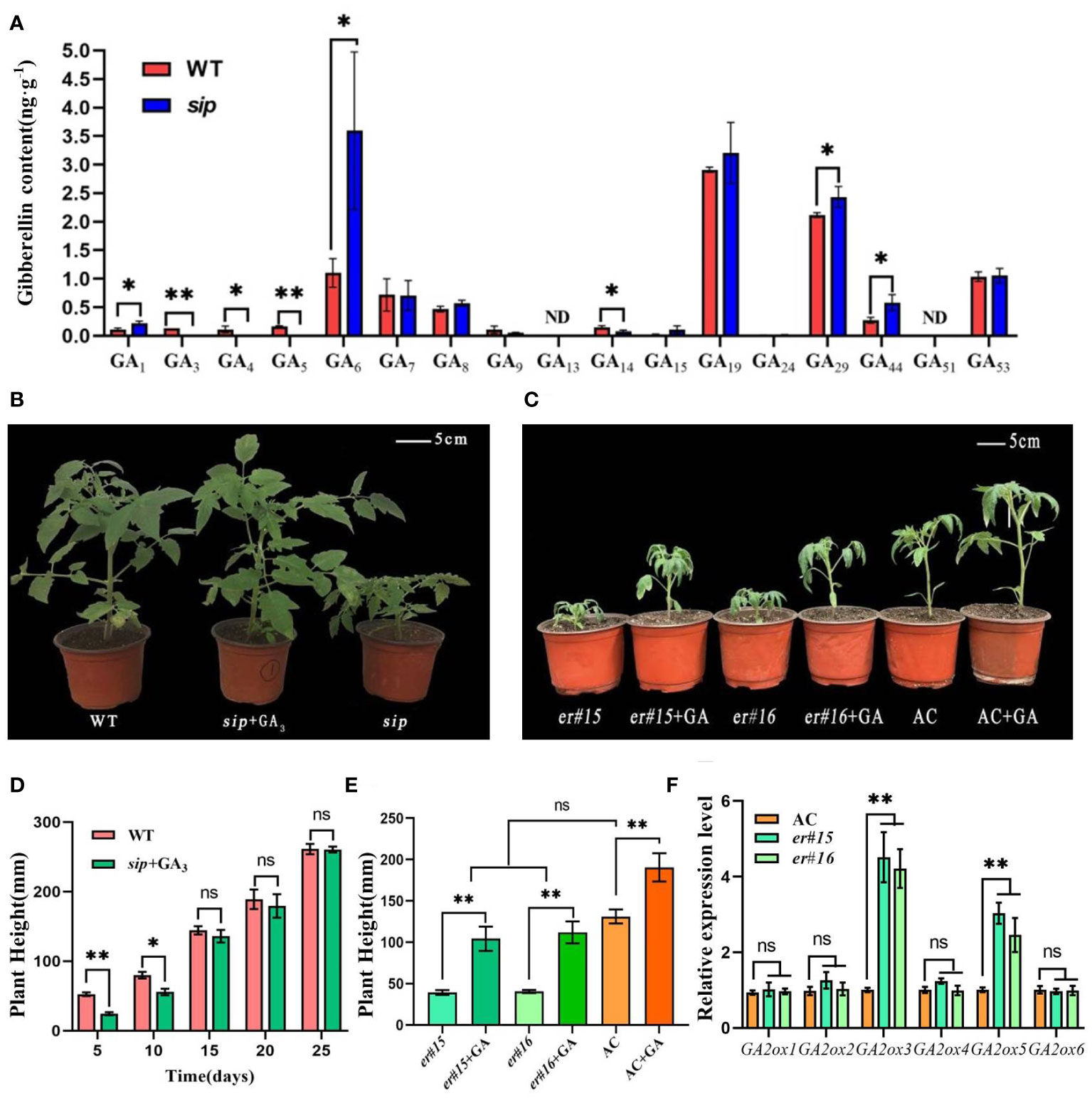
Figure 6 Gibberellin content and hormone recovery experiments. (A) Gibberellin content in stems of WT and sip (n=3). (B, D) Exogenous application of gibberellin restores the height of sip to that of WT. (C, E) Exogenous application of gibberellin restores the height of er to that of AC. (F) Expression of GA2oxs in the stems of er mutant. Student’s t-test was used to determine significance. *p < 0.05 and **p < 0.01; ns, no significant difference. All data are presented as mean ± SD.
4 Discussion
4.1 Identification of a novel dwarf mutant in tomato
Dwarfing varieties possess significant characteristics such as being space-saving, being resistant to lodging, and being suitable for mechanized harvesting. Most commercial tomato varieties have long stems and many lateral branches, necessitating a considerable amount of time and labor for pruning. The tomato mutant we identified, sip, exhibits a compact phenotype reminiscent of a bouquet, while effectively reducing flower pedicel shedding and producing more fruit. These unique traits not only make it suitable for mechanized harvesting but also hold potential for its utilization as an ornamental variety. We constructed two populations for mapping and analyzed the BSA sequencing results using bioinformatics. The findings from both populations exhibited a high degree of similarity. Both BSA results and the PRAMS markers suggested the presence of a significant linked region on chr.8, which makes the mapping of sip challenging. In this case, a comparison of the BSA results of the two populations proves to be highly valuable. We identified only two SNPs located within the exons that had the potential to contribute to the sip phenotype. To verify this, KASP genotyping was performed, and the results showed that only SlSIP co-segregated with the short internode and pedicel phenoty. Subsequent validation of SlSIP’s function was carried out with ER loss-of-function mutants created through CRISPR/Cas9 gene-editing technology, demonstrating that SlSIP was responsible for the sip phenotype.
SlSIP encodes a leucine-rich repeat receptor-like kinase ER, which is involved in regulating multiple aspects of plant growth and development. In Arabidopsis, the homologous gene AtERECTA exhibits similar phenotypic characteristics to sip and confers compact inflorescence, blunt fruits, and short petioles (Torii et al., 1996; Shpak et al., 2004; Meng et al., 2012; Uchida et al., 2012; Du et al., 2018). The reports on ER in tomato also support the aforementioned mapping results in terms of phenotype (Villagarcia et al., 2012; Kwon et al., 2020). Interestingly, when compared to ER signaling mutants in tomatoes that were created by expressing the dominant-negative, signal disrupter, truncated ERECTA (AtΔKinase) from Arabidopsis (Villagarcia et al., 2012), we did not observe any difference in stomatal density and flowering time in sip (Figure 1L; Supplementary Figure 11), nor did we notice a decrease in fruit set or the number of leaves due to reduction of leaf formation and growth. This could be attributed to the functional redundancy between tomato ER and its homologous gene ERL in regulating stomatal development. Compared to the study conducted by Kwon et al. (2020) we did observe additional phenotypes, such as an increase in the number of flowers and fruits in the mutant varieties. Considering the different phenotypes observed in er mutants caused by different mutation sites reported in Arabidopsis (Kosentka et al., 2017), we speculate that this difference may be attributed to different genetic backgrounds and mutation sites. These mutants are very important for exploring the function and mechanism of tomato.
4.2 ER is involved in the biosynthesis and signal transduction pathways of gibberellin
Numerous studies (Qu et al., 2017; Cai et al., 2020; Yang et al., 2020; Xu et al., 2023) have revealed a strong correlation between the inhibition of cell elongation observed in ER mutants and disturbances in auxin synthesis and transport processes. The er erl1 erl2 triple mutant exhibited lower levels of auxin, accompanied by a significant decrease in the expression levels of key auxin biosynthesis genes. However, we did not observe a decrease in indol-3-acetic acid (IAA) levels in sip mutants (Supplementary Figure 10B). Is this possibly caused by abnormalities in auxin transport processes? We examined the expression levels of the PIN gene family, which is involved in auxin transport, in the stems of the mutant plants. The expression levels of these genes showed no difference compared to the WT plants (Supplementary Figure 10C). Instead, we observed significant changes in the gibberellin content, including a slight increase in GA1 and trace amounts of GA3 and GA4 (Figure 5A). External application of IAA did not restore the height of sip mutants to that of WT plants (Supplementary Figure 10A), whereas exogenous GA application did (Figures 5E, H). According to reports on gibberellin biosynthesis pathway, the intermediate product GA12-aldehyde can be converted into GA12 and GA53 through non-hydroxylation and hydroxylation pathways (Helliwell et al., 2001). GA53 is further catalyzed by a series of GA2 oxidases (GA2oxs) to form GA20. GA20 can then be degraded into GA29 by the action of GA2oxs or converted into GA5 by the catalysis of GA3oxs. GA5 subsequently undergoes further modifications to form the active gibberellin, GA3 (Yamaguchi, 2008). We found an increase in GA29 and a decrease in GA5 in sip (Figure 5D), which is consistent with the up-regulation of GA2oxs expression (Figure 5C). The increased GA29 suggests that more GA20 is being formed as a substrate, while the decreased GA5 and GA3 indicate a reduction in their synthesis from GA20 catalyzed by GA3oxs. These findings suggest that ER may regulate the gibberellin pathway by modulating the expression of GA2 oxidases.
Because of the presence of an extracellular LRR domain in ER, it is expected that there are ligands that can bind to the LRR domain. The protein kinase domains of ER in different species are relatively conserved, whereas the transmembrane and extracellular domains show less conservation (Kosentka et al., 2017). This suggests that there may be different ligands and co-receptors in different species but that they all likely utilize MAPK cascades as downstream pathways. ERf receptor activity is regulated by a group of cysteine-rich peptides belonging to the EPF/EPFL family (Hara et al., 2007; Shpak, 2013). More than 11 ligands have been identified in Arabidopsis, including members of the EFL family and TOOMANY MOUTHS (TMM) (Berger and Altmann, 2000). However, there are few genes with high homology to these secreted peptides in tomatoes. We hypothesized that different structural domains of ER may contain distinct ligand binding sites to perform their respective functions. The mutation N134I may result in the preventing the binding of ligands or reducing their activity, thus inhibiting the activation of the MAPK-mediated pathway that regulates plant height. Then, we performed MD simulations to study the folding of SlSIP and found that the mutated site could form hydrogen bonds with other internal amino acids. The alteration of N134I leads to changes in the hydrogen bonds formed within this particular site (Figures 3C, D), which provides support for this hypothesis.
In Arabidopsis, it has been reported that the ER-MAPK module regulates plant height by controlling the activity of DELLA through KNAT1/BREVIPEDICELLUS(BP) (Milhinhos et al., 2019). However, there are no homologous genes of BP in tomato, we speculate that there may be similar transcription factors that directly or indirectly regulate the expression of GA2oxs downstream of the ER-MAPK module in tomato. Through transcriptomic analysis, we have identified several differentially expressed transcription factors, including members of the MYB, WAKY, and MADS families. Members of these families reportedly participate in regulating inflorescence architecture and plant growth in Arabidopsis. We predicted whether the transcription factors from the differential expression can bind to the promoters of gibberellin-related genes. We found that Solyc10g005080 (MYB family) and MADS3 have binding sites for ent-kaurenoic acid oxidase 1-like, GA2ox5, gibberellin-regulated protein 6, gibberellin-regulated protein 11, CPS. This suggests that they may act as downstream regulators of the ER-MAPK module. These speculations and the mechanism of ER regulation of gibberellin need to be further elucidated in future studies.
4.3 ER mutant varieties offer potential for breeding strategies
ER is a receptor-like protein kinase of leucine-rich repeat RLKs that regulates diverse biological processes during plant growth and development, including seed germination, stem elongation, stomatal development, and inflorescence architecture. ER is involved in various functions, and its regulatory mechanisms are highly complex. There is extensive research on ER in Arabidopsis, but the research currently on ER in tomato is very limited. In Arabidopsis, four phosphorylation sites Thr807, Thr812, Tyr815, and Tyr820 are critical for the functionality of the ER kinase domain. Thr807 and Thr812 have a positive effect on ER function, whereas phosphorylation of Tyr815 and Tyr820 at these sites has an inhibitory effect on ER function (Kosentka et al., 2017). This suggests that ER signal transduction varies across different regulatory pathways, which explains the significant differences in the phenotypes observed among the different ER mutants. Interestingly, we compared the sip mutant and the knockout mutant er, and found that they exhibit consistent phenotypes in terms of internode length, inflorescence structure, fruit morphology, and so on. However, the mutations in sip still retain some of the functions of ER.
In Arabidopsis, Wu et al. isolated three er mutants, er563, er795, and er1214, containing 124, 99, and 632 amino acids, respectively (Wu et al., 2022). The seeds of er563 and er795 mutants are significantly smaller than those of WT, while er1214 mutant shows normal seed size and weight. Only er1214 contains intact extracellular and transmembrane domains but lacks the kinase domain. Consistent with these findings, we observed the same phenomenon in sip mutants and er#15,16 lines. SlSIP did not affect seed size (Figure 4E), we speculate that this is because the kinase domain in SlSIP is not disrupted. Additionally, we found that if all inflorescences, including those generated from lateral branches, are retained, sip has more inflorescences compared to er (Supplementary Figures 1J, 5C). It seems that sip has a stronger ability to differentiate floral buds from lateral branches compared to er. Considering the previously reported role of ER in stress responses, we subjected sip to drought and cold stress, and it showed stronger tolerance compared to WT (not shown in this article). These findings suggest that sip carries a novel mutation in tomato that could separate this compact structure from its other regulatory functions.
We observe that both sip and er can produce more fruits, which is undoubtedly beneficial for production. Whether it is compact plant type or producing more fruits, it can be applied in tomato breeding. Concentrated flowering and fruiting are significant for processing tomatoes in production, as mechanical harvesting requires concentrated flowering and fruiting to adapt. As ER serves as an upstream central signal for plant growth and development (Shpak, 2013), and considering its role in response to both biotic (Godiard et al., 2003) and abiotic stresses (Du et al., 2018; Juneidi et al., 2020), would knocking out ER potentially have any adverse effects in production? The mutants like sip have more potential for application in breeding, it provides a possibility to separate the plant height and the ability to produce more fruits from other ER-controlled traits in tomato breeding.
In this study, we identified a shortened internodes and pedicel mutant sip, and conducted mapping through BSA analysis. The mutation that separates plant height from other traits controlled by ER, this separation allows for a combination of favorable traits, and providing a valuable foundation for a more thorough examination of ER function in tomato. These findings contribute to optimized breeding strategies for cultivating tomato varieties with desirable dwarfing traits and can serve as a reference for further investigations and applications in crop improvement and breeding programs targeting plant architecture and yield potential.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
XZ designed and carried out the experiments and wrote the manuscript. KZ, HZ, MB analyzed the results, YH, YC, TC provided scientific advice, and revised the manuscript. MQ and JM conceived the research area, provided scientific advice, and supervised the project. All authors read and approved the final manuscript.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant Nos. 31991184, 31972397, and 31861143045), and the Major Agricultural Project of Liaoning Province (2022020768-JH1/102-01).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1283489/full#supplementary-material
References
Abrash, E. B., Davies, K. A., Bergmann, D. C. (2011). Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand–receptor interactions. Plant Cell 23, 2864–2879. doi: 10.1105/tpc.111.086637
Baek, M., DiMaio, F., Anishchenko, I., Dauparas, J., Ovchinnikov, S., Lee, G. R., et al. (2021). Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876. doi: 10.1126/science.abj8754
Bassel, G. W., Mullen, R. T., Bewley, J. D. (2008). Procera is a putative DELLA mutant in tomato (Solanum lycopersicum): effects on the seed and vegetative plant. J. Exp. Bot. 59, 585–593. doi: 10.1093/jxb/erm354
Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., DiNola, A., Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690. doi: 10.1063/1.448118
Berger, D., Altmann, T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14, 1119–1131.
Cai, H., Chai, M., Chen, F., Huang, Y., Zhang, M., He, Q., et al. (2020). HBI1 acts downstream of ERECTA and SWR1 in regulating inflorescence architecture through the activation of the brassinosteroid and auxin signaling pathways. New Phytol. 229, 414–428. doi: 10.1111/nph.16840
Cai, H., Zhao, L., Wang, L., Zhang, M., Su, Z., Cheng, Y., et al. (2017). ERECTA signaling controls Arabidopsis inflorescence architecture through chromatin-mediated activation of PRE1 expression. New Phytol. 214, 1579–1596. doi: 10.1111/nph.14521
Cavasin, P. Y., Fumes, L. A. A., Fonseca, A. R., Silva, D. J. H. (2021). Evaluation of families derived from backcrosses of processed tomato with dwarfism gene. Crop Breed. Appl. Biotechnol. 21 (1), e362221113. doi: 10.1590/1984-70332021v21n1n13
Cheng, L., Li, R., Wang, X., Ge, S., Wang, S., Liu, X., et al. (2022). A SlCLV3-SlWUS module regulates auxin and ethylene homeostasis in low light-induced tomato flower abscission. Plant Cell 34, 4388–4408. doi: 10.1093/plcell/koac254
DeGennaro, D., Urquidi Camacho, R. A., Zhang, L., Shpak, E. D. (2022). Initiation of aboveground organ primordia depends on combined action of auxin, ERECTA family genes, and PINOID. Plant Physiol. 190, 794–812. doi: 10.1093/plphys/kiac288
Douglas, S. J., Chuck, G., Dengler, R. E., Pelecanda, L., Riggs, C. D. (2002). KNAT1 and ERECTA regulate inflorescence architecture in arabidopsis. Plant Cell 14, 547–558. doi: 10.1105/tpc.010391
Du, J., Jiang, H., Sun, X., Li, Y., Liu, Y., Sun, M., et al. (2018). Auxin and gibberellins are required for the receptor-like kinase ERECTA regulated hypocotyl elongation in shade avoidance in Arabidopsis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00124
Gerszberg, A., Hnatuszko-Konka, K., Kowalczyk, T., Kononowicz, A. K. (2014). Tomato (Solanum lycopersicum L.) in the service of biotechnology. Plant Cell Tissue Organ Culture (PCTOC) 120, 881–902. doi: 10.1007/s11240-014-0664-4
Godiard, L., Sauviac, L., Torii, K. U., Grenon, O., Mangin, B., Grimsley, N. H., et al. (2003). ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J. 36 (3), 353–365. doi: 10.1046/j.1365-313X.2003.01877.x
Hara, K., Kajita, R., Torii, K. U., Bergmann, D. C., Kakimoto, T. (2007). The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 21, 1720–1725. doi: 10.1101/gad.1550707
Heim, M., Meyer, U. A. (1990). Genotyping of poor metabolisers of debrisoquine by allele-specific PCR amplification. Lancet 336, 529–532. doi: 10.1016/0140-6736(90)92086-W
Helliwell, C., Chandler, P. M., Poole, A., Dennis, E. S., Peacock, W. J. (2001). The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. 98 (4), 2065–2070. doi: 10.1073/pnas.98.4.2065
Juneidi, S., Gao, Z., Yin, H., Makunga, N. P., Chen, W., Hu, S., et al. (2020). Breaking the summer dormancy of pinellia ternata by introducing a heat tolerance receptor-like kinase ERECTA gene. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00780
Khush, G. S. (2001). Green revolution: the way forward. Nat. Rev. Genet. 2, 815–822. doi: 10.1038/35093585
Kosentka, P. Z., Zhang, L., Simon, Y. A., Satpathy, B., Maradiaga, R., Mitoubsi, O., et al. (2017). Identification of critical functional residues of receptor-like kinase ERECTA. J. Exp. Bot. 68, 1507–1518. doi: 10.1093/jxb/erx022
Kwon, C.-T., Heo, J., Lemmon, Z. H., Capua, Y., Hutton, S. F., Van Eck, J., et al. (2020). Rapid customization of Solanaceae fruit crops for urban agriculture. Nat. Biotechnol. 38, 182–188. doi: 10.1038/s41587-019-0361-2
Laskowski, R. A., Swindells, M. B. (2011). LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Modeling 51, 2778–2786. doi: 10.1021/ci200227u
Law, C. N., Snape, J. W., Worland, A. J. (1978). The genetical relationship between height and yield in wheat. Heredity 40 (1), 133–151. doi: 10.1038/hdy.1978.13
Lease, K. A., Lau, N. L., Schuster, R. A., Torii, K. U., Walker, J. C. (2001). Receptor serine/Threonine protein kinases in signalling: analysis of the erecta receptorLike kinase of arabidopsis thaliana. New Phytol. 151 (1), 133–143. doi: 10.1046/j.1469-8137.2001.00150.x
Li, X. J., Chen, X. J., Guo, X., Yin, L. L., Ahammed, G. J., Xu, C. J., et al. (2016). DWARF overexpression induces alteration in phytohormone homeostasis, development, architecture and carotenoid accumulation in tomato. Plant Biotechnol. J. 14, 1021–1033. doi: 10.1111/pbi.12474
Li, H., Durbin, R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, Z.-F., Guo, Y., Ou, L., Hong, H., Wang, J., Liu, Z.-X., et al. (2018). Identification of the dwarf gene GmDW1 in soybean (Glycine max L.) by combining mapping-by-sequencing and linkage analysis. Theor. Appl. Genet. 131, 1001–1016. doi: 10.1007/s00122-017-3044-8
Li, Y., Sun, M., Xiang, H., Liu, Y., Li, H., Qi, M., et al. (2019). Low overnight temperature-induced gibberellin accumulation increases locule number in tomato. Int. J. Mol. Sci. 20 (12), 3042. doi: 10.3390/ijms20123042
Li, H., Yang, Y., Wang, H., Liu, S., Jia, F., Su, Y., et al. (2021). The receptor-like kinase ERECTA confers improved water use efficiency and drought tolerance to poplar via modulating stomatal density. Int. J. Mol. Sci. 22 (14), 7245. doi: 10.3390/ijms22147245
Li, J., Yu, C., Wu, H., Luo, Z., Ouyang, B., Cui, L., et al. (2015). Knockdown of a JmjC domain-containing gene JMJ524 confers altered gibberellin responses by transcriptional regulation of GRAS protein lacking the DELLA domain genes in tomato. J. Exp. Bot. 66, 1413–1426. doi: 10.1093/jxb/eru493
Liu, F., Wang, P., Zhang, X., Li, X., Yan, X., Fu, D., et al. (2017). The genetic and molecular basis of crop height based on a rice model. Planta 247, 1–26. doi: 10.1007/s00425-017-2798-1
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Livne, S., Lor, V. S., Nir, I., Eliaz, N., Aharoni, A., Olszewski, N. E., et al. (2015). Uncovering DELLA-independent gibberellin responses by characterizing new tomato procera mutants. Plant Cell 27, 1579–1594. doi: 10.1105/tpc.114.132795
Meissner, R., Jacobson, Y., Melamed, S., Levyatuv, S., Shalev, G., Ashri, A., et al. (1997). A new model system for tomato genetics. The Plant Journal 12 (6), 1465–1475. doi: 10.1046/j.1365-313x.1997.12061465.x
Milhinhos, A., Vera-Sirera, F., Blanco-Touriñán, N., Mari-Carmona, C., Carrió-Seguí, À., Forment, J., et al. (2019). SOBIR1/EVR prevents precocious initiation of fiber differentiation during wood development through a mechanism involving BP and ERECTA. Proc. Natl. Acad. Sci. 116 (37), 18710–18716. doi: 10.1073/pnas.1807863116
Meng, X., Wang, H., He, Y., Liu, Y., Walker, J. C., Torii, K. U., et al. (2012). A MAPK cascade downstream of ERECTA receptor-like protein kinase regulates Arabidopsis inflorescence architecture by promoting localized cell proliferation. Plant Cell 24, 4948–4960. doi: 10.1105/tpc.112.104695
Miralles, D. J., Katz, S. D., Colloca, A., Slafer, G. A. (1998). Floret development in near isogenic wheat lines differing in plant height. Field Crops Res. 59, 21–30. doi: 10.1016/S0378-4290(98)00103-8
Mooers, B. H. M. (2019). Shortcuts for faster image creation in PyMOL. Protein Sci. 29, 268–276. doi: 10.1002/pro.3781
Nuovo, G. J., Hohman, R. J., Nardone, G. A., Nazarenko, I. A. (1999). In situ amplification using universal energy transfer-labeled primers. J. Histochem. Cytochem. 47, 273–279. doi: 10.1177/002215549904700301
Peng, J., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., et al. (1999). 'Green revolution' genes encodemutant gibberellin responsemodulators. Nature 400, 256–261.
Prenner, G., Vergara-Silva, F., Rudall, P. J. (2009). The key role of morphology in modelling inflorescence architecture. Trends Plant Sci. 14 (6), 302–309. doi: 10.1016/j.tplants.2009.03.004
Qu, X., Zhao, Z., Tian, Z. (2017). ERECTA regulates cell elongation by activating auxin biosynthesis in arabidopsis thaliana. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01688
Reinhardt, D., Kuhlemeier, C. (2002). Plant architecture. EMBO Rep. 3 (9), 846–851. doi: 10.1093/embo-reports/kvf177
Scott, J. W., Harbaugh, B. K., Baldwin, E. A. (1995). ‘Micro-Gold’ miniature dwarf tomatoes. HortScience 30, 643–644. doi: 10.21273/HORTSCI.30.3.643
Shen, H., Zhong, X., Zhao, F., Wang, Y., Yan, B., Li, Q., et al. (2015). Overexpression of receptor-like kinase ERECTA improves thermotolerance in rice and tomato. Nat. Biotechnol. 33, 996–1003. doi: 10.1038/nbt.3321
Shpak, E. D. (2013). Diverse roles of ERECTA family genes in plant development. J. Integr. Plant Biol. 55, 1238–1250. doi: 10.1111/jipb.12108
Shpak, E. D., Berthiaume, C. T., Hill, E. J., Torii, K. U. (2004). Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131, 1491–1501. doi: 10.1242/dev.01028
Shpak, E. D., McAbee, J. M., Pillitteri, L. J., Torii, K. U. (2005). Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309 (5732), 290–293. doi: 10.1126/science.1109710
Sun, M., Li, H., Li, Y., Xiang, H., Liu, Y., He, Y., et al. (2020). Tomato YABBY2b controls plant height through regulating indole-3-acetic acid-amido synthetase (GH3.8) expression. Plant Sci. 297, 110530. doi: 10.1016/j.plantsci.2020.110530
Takagi, H., Abe, A., Yoshida, K., Kosugi, S., Natsume, S., Mitsuoka, C., et al. (2013). QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 74, 174–183. doi: 10.1111/tpj.12105
Torii, K. U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R. F., et al. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8 (4), 735–746. doi: 10.1105/tpc.8.4.735
Uchida, N., Lee, J. S., Horst, R. J., Lai, H. H., Kajita, R., Kakimoto, T., et al. (2012). Regulation of inflorescence architecture by intertissue layer ligand-receptor communication between endodermis and phloem. Proc. Natl. Acad. Sci. 109, 6337–6342. doi: 10.1073/pnas.1117537109
Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., Berendsen, H. J. C. (2005). GROMACS: Fast, flexible, and free. J. Comput. Chem. 26, 1701–1718. doi: 10.1002/jcc.20291
Villagarcia, H., Morin, A. C., Shpak, E. D., Khodakovskaya, M. V. (2012). Modification of tomato growth by expression of truncated ERECTA protein from Arabidopsis thaliana. J. Exp. Bot. 63, 6493–6504. doi: 10.1093/jxb/ers305
Wang, Y., Chen, L., Du, Y., Yang, Z., Condon, A. G., Hu, Y. (2014). Genetic effect of dwarfing gene Rht13 compared with Rht-D1b on plant height and some agronomic traits in common wheat (Triticum aestivum L.). Field Crops Res. 162, 39–47. doi: 10.1016/j.fcr.2014.03.014
Wang, N., Liu, Z., Zhang, Y., Li, C., Feng, H. (2017). Identification and fine mapping of a stay-green gene (Brnye1) in pakchoi (Brassica campestris L. ssp. chinensis). Theor. Appl. Genet. 131, 673–684. doi: 10.1007/s00122-017-3028-8
Wu, X., Cai, X., Zhang, B., Wu, S., Wang, R., Li, N., et al. (2022). ERECTA regulates seed size independently of its intracellular domain via MAPK-DA1-UBP15 signaling. Plant Cell 34 (10), 3773–3789. doi: 10.1093/plcell/koac194
Xu, X., Hu, Q., Wang, J., Wang, X., Lou, L., Xu, J., et al. (2023). A 2-bp deletion in the protein kinase domain region of the ERECTA-like receptor kinase gene in cucumber results in short internode phenotype. Plant Sci. 327, 111536. doi: 10.1016/j.plantsci.2022.111536
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Keywords: tomato, short internodes, short pedicels, ERECTA, gibberellin
Citation: Zhao X, Zhang K, Zhang H, Bi M, He Y, Cui Y, Tan C, Ma J and Qi M (2023) Tomato short internodes and pedicels encode an LRR receptor-like serine/threonine-protein kinase ERECTA regulating stem elongation through modulating gibberellin metabolism. Front. Plant Sci. 14:1283489. doi: 10.3389/fpls.2023.1283489
Received: 26 August 2023; Accepted: 27 October 2023;
Published: 17 November 2023.
Edited by:
Elisabetta Mazzucotelli, Council for Agricultural Research and Economics- Research Centre for Genomics and Bioinformatics, ItalyReviewed by:
Choon-Tak Kwon, Kyung Hee University Global Campus, Republic of KoreaMing Luo, Southwest University, China
Copyright © 2023 Zhao, Zhang, Zhang, Bi, He, Cui, Tan, Ma and Qi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingfang Qi, qimingfang@126.com; Jian Ma, mj76@163.com
 Xueya Zhao
Xueya Zhao Kunpeng Zhang1,2,3,4,5
Kunpeng Zhang1,2,3,4,5 Changhua Tan
Changhua Tan Mingfang Qi
Mingfang Qi