- 1Department of Plant Pathology, Faculty of Agriculture, Kasetsart University, Bangkok, Thailand
- 2Food Biotechnology Research Team, Functional Ingredients and Food Innovation Research Group, National Center for Genetic Engineering and Biotechnology, National Science and Technology Development Agency, Pathum Thani, Thailand
- 3Enzyme Technology Research Team, Biorefinery and Bioproducts Technology Research Group, National Center for Genetic Engineering and Biotechnology, National Science and Technology Development Agency, Pathum Thani, Thailand
- 4Microbiological and Molecular Biological Laboratory, Scientific Instruments Center, School of Science, King Mongkut’s Institute of Technology Ladkrabang, Bangkok, Thailand
- 5Biodiversity Research Centre, Research and Development Group for Bio-Industries, Thailand Institute of Scientific and Technological Research, Pathum Thani, Thailand
- 6Department of Biochemistry and Microbiology, Faculty of Pharmaceutical Sciences, Chulalongkorn University, Bangkok, Thailand
- 7Center of Excellence in Microbial Diversity and Sustainable Utilization, Chiang Mai University, Chiang Mai, Thailand
- 8Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 9Graduate School of Infection Control Sciences, Kitasato University, Tokyo, Japan
- 10Ōmura Satoshi Memorial Institute, Kitasato University, Tokyo, Japan
A novel endophytic actinomycete, strain MEP2-6T, was isolated from scab tissues of potato tubers collected from Mae Fag Mai Sub-district, San Sai District, Chiang Mai Province, Thailand. Strain MEP2-6T is a gram-positive filamentous bacteria characterized by meso-diaminopimelic acid in cell wall peptidoglycan and arabinose, galactose, glucose, and ribose in whole-cell hydrolysates. Diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, and hydroxy-phosphatidylethanolamine were the major phospholipids, of which MK-9(H6) was the predominant menaquinone, whereas iso-C16:0 and iso-C15:0 were the major cellular fatty acids. The genome of the strain was 10,277,369 bp in size with a G + C content of 71.7%. The 16S rRNA gene phylogenetic and core phylogenomic analyses revealed that strain MEP2-6T was closely related to Amycolatopsis lexingtonensis NRRL B-24131T (99.4%), A. pretoriensis DSM 44654T (99.3%), and A. eburnea GLM-1T (98.9%). Notably, strain MEP2-6T displayed 91.7%, 91.8%, and 87% ANIb and 49%, 48.8%, and 35.4% dDDH to A. lexingtonensis DSM 44653T (=NRRL B-24131T), A. eburnea GLM-1T, and A. pretoriensis DSM 44654T, respectively. Based on phenotypic, chemotaxonomic, and genomic data, strain MEP2-6T could be officially assigned to a novel species within the genus Amycolatopsis, for which the name Amycolatopsis solani sp. nov. has been proposed. The type of strain is MEP2-6T (=JCM 36309T = TBRC 17632T = NBRC 116395T). Amycolatopsis solani MEP2-6T was strongly proven to be a non-phytopathogen of potato scab disease because stunting of seedlings and necrotic lesions on potato tuber slices were not observed, and there were no core biosynthetic genes associated with the BGCs of phytotoxin-inducing scab lesions. Furthermore, comparative genomics can provide a better understanding of the genetic mechanisms that enable A. solani MEP2-6T to adapt to the plant endosphere. Importantly, the strain smBGCs accommodated 33 smBGCs encoded for several bioactive compounds, which could be beneficially applied in the fields of agriculture and medicine. Consequently, strain MEP2-6T is a promising candidate as a novel biocontrol agent and antibiotic producer.
Introduction
The genus Amycolatopsis belongs to the phylum Actinomycetota, class Actinomycetia (Oren and Garrity, 2021), order Pseudonocardiales, and the family Pseudonocardiaceae. This species was first proposed by Lechevalier et al. (1986) as Amycolatopsis orientalis. The taxonomic description of this genus was amended based on the 16S rDNA, chemotaxonomic characteristics, and genome sequences by Lee (2009); Tang et al. (2010), and Nouioui et al. (2018). Most species in the genus Amycolatopsis form long chains of substrate and aerial hyphae, which may differentiate into chains of squarish to oval fragments as spore-like structures. These characteristics constitute key morphological characteristics (Lechevalier et al., 1986). This genus is mycolate-less and contains meso-diaminopimelic acid in the peptidoglycan wall (Lechevalier and Lechevalier, 1970; Embley et al., 1988). Arabinose and galactose are diagnostic sugars found in whole-cell hydrolysates. The predominant isoprenoid quinone observed in Amycolatopsis is MK-9(H4) with phosphatidylethanolamine as a diagnostic phospholipid (Lechevalier et al., 1977). The G + C content in the genome of Amycolatopsis species is generally within the range of 66 to 75 mol% (Li et al., 2021). At the time of writing, the genus Amycolatopsis included 87 species with validly published names (Parte et al., 2020, https://lpsn.dsmz.de/genus/amycolatopsis).
Numerous species of the genus Amycolatopsis have adapted to occupy many diverse biological niches, such as soil (Camas et al., 2013), plants (Mingma et al., 2020), salt mines (Tatar et al., 2013), lakes (Li et al., 2021), marine environments (Bian et al., 2009), insects (Beemelmanns et al., 2017), animals (Labeda et al., 2003), and humans (Huang et al., 2004). The planta Amycolatopsis species have been discovered inside the tissues of different plant species: A. samaneae recovered from the roots of Samanea saman (Duangmal et al., 2011), and A. jiangsuensis was isolated from the stems of Dendranthema indicum (Xing et al., 2013), A. stemonae isolated from the stems of Stemona sp. (Klykleung et al., 2015), A. anabasis isolated from the roots of Anabasis elatior (Wang et al., 2020), and A. dendrobii isolated from the roots of Dendrobium heterocarpum Lindl (Tedsree et al., 2021). However, their ability to adapt to plants is not fully understood.
Members of the genus Amycolatopsis are closely connected with a history of antibiotic drug discovery as invaluable commercial producers of secondary metabolites with antibacterial, antifungal, or antiviral activities. They have continued to gain considerable attention in the search for new drugs (Kisil et al., 2021). Metabolites are synthesized by several diverse gene clusters in the genome, called biosynthetic gene clusters (BGCs). Fortunately, the reduced costs of whole-genome sequencing and public access to genome databases are now allowing the scientific community to sequence and share thousands of prokaryotic genomes worldwide. This can help to comprehensively detect and compare BGCs between organisms, as well as to identify their chemical structures using robust bioinformatics tools (Alam et al., 2022), such as antiSMASH 7.0 (Blin et al., 2023), BAGEL4 (van Heel et al., 2018), and PRISM 4 (Skinnider et al., 2020). This knowledge and these tools can guide us in predicting where to find promising new compounds and help us determine if the search for new producers should be based on phylogeny, geography, or specific ecological niches (Adamek et al., 2018).
In the present study, the novel endophytic Amycolatopsis strain MEP2-6T was isolated from scab lesions on potato tuber surfaces; however, there have been no reports of its association with potato scab disease. Thus, we aimed to comprehensively characterize Amycolatopsis strain MEP2-6T based on polyphasic and genome-based taxonomy and evaluate the pathogenicity of strain MEP2-6T in plants. In addition, we determine how this strain can adapt to reside in plants, study the diversity of the secondary metabolite biosynthetic gene clusters (smBGCs) of the strain MEP2-6T, and compare the distribution pattern of smBGCs with its closest type strains. This work represents the first of its kind to report on the Amycolatopsis species inhabiting potato scab lesions and can provide a better understanding of the genetic mechanisms of this species that occupy plants. We also reported the genetic potential of this species to produce various types of secondary metabolites. These outcomes can contribute to the identification of specialized biosynthetic pathways that are of particular interest and serve as a guide for antibiotic drug discovery.
Materials and methods
Bacterial isolation and preservation
Scabby tubers of potato (Solanum tuberosum L.) (Supplementary Figure 1) collected from Mae Fag Mai Sub-district, San Sai District, Chiang Mai Province, northern Thailand (18○58’56.3’’N, 98○58’45.8’’E), were washed with running tap water and then surface sterilized according to the method described by Kuncharoen et al. (2018) with slight modifications. Briefly, the scab-infected potato tubers were washed with running tap water to remove soil particles, soaked in 75 % (v/v) ethanol for 1 min, drenched in 1 % (v/v) NaOCl for 3 min, rinsed three times in sterile distilled water, and air-dried in a laminar flow. Single lesions appearing at the border between healthy and scab tissues of the surface-sterilized tubers were aseptically cut into small pieces, homogenized using a sterile mortar with 1 ml of 0.85% (w/v) NaCl, and incubated in a water bath at 60°C for 10 min to eliminate any competing rhizobacteria (Fyans et al., 2016). The homogenate was diluted 100-fold. Subsequently, 0.1 ml was spread onto 2.5% (w/v) water agar (Arai, 1975) supplemented with 25 µg nalidixic acid ml−1 and 50 µg cycloheximide ml−1 and then incubated at 30°C in the dark for 14 days. An interesting colony of strain MEP2-6T was isolated and purified on International Streptomyces Project medium 2 (ISP 2) agar (Shirling and Gottlieb, 1966), maintained on ISP 2 agar slant, stored at −20°C and −80°C in the ISP 2 broth supplemented with 20 % (v/v) glycerol, and lyophilized for long-term preservation.
Genomic DNA extraction
Genomic DNA of strain MEP2-6T was extracted using the FavorPrepTM Tissue Genomic DNA Extraction Mini Kit (Favorgen, Taiwan), according to the manufacturer’s instructions. DNA quality and quantity were determined using 1% (w/v) agarose gel electrophoresis and NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The purified genomic DNA was stored at −20°C until use.
Genome sequencing, assembly, and annotation
The genome sequence of strain MEP2-6T was successfully constructed by combining Illumina HiSeq 2,500 paired-end sequencing (Illumina Inc., San Diego, CA, USA) at Novogene (Biopolis Way, Singapore) and the GridION sequencer (Oxford Nanopore Technologies—ONT, UK) at Siriraj Long-read Lab (Siriraj Medical Research Center, Thailand). The genome assembly consists of four steps: first, raw Illumina reads were performed the quality control by removing low-quality sequences and trimming the adapter and primer sequences using fastp v0.20.0 (Chen et al., 2018), and then evaluated their sequence quality with FastQC v0.12.0 (https://github.com/s-andrews/FastQC) and MultiQC v1.17 (Ewels et al., 2016); second, ONT raw signals were base called and demultiplexed using Guppy v3.4.5 (ONT) with the use of a specific high-accuracy model (-c dna_r9.4.1_450bps_hac.cfg) to obtain raw ONT reads; third, the raw ONT long reads were filtered by their quality and sequence length using the Nanofilt program (De Coster et al., 2018), and then adapters were trimmed using Porechop v0.2.4 (https://github.com/rrwick/Porechop). Finally (step 4), the cleaned short and long sequences were assembled de novo using SPAdes v3.15.4 (Antipov et al., 2016; Prjibelski et al., 2020) with a minimum contig size of 500 bps.
The assembled genome of strain MEP2-6T and the publicly available genome assemblies of the closely related Amycolatopsis species, as well as an outgroup with validly published names downloaded from the NCBI database (3 June 2022) using the E-utilities Command (Kans, 2022), were estimated for genome completeness and contamination using CheckM v1.1.6 (Parks et al., 2015). Contiguity and the completeness of universal single-copy orthologs were assessed using QUAST v5.2.0 (Gurevich et al., 2013) and BUSCO v5.4.7 (Simão et al., 2015; Manni et al., 2021).
The genome of strain MEP2-6T was annotated using Rapid Annotation with the Subsystem Technology (RAST) server (Aziz et al., 2008) using the RASTtk algorithm (Brettin et al., 2015) and re-annotated according to the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (Tatusova et al., 2016). The genomes of the closest Amycolatopsis species were obtained from NCBI GenBank and annotated using the RAST server. The accession no. of the genomes are shown in Table 1.
Analysis of 16S rRNA gene sequence
PCR amplification of the 16S rRNA gene was performed as described by Suriyachadkun et al. (2009). The purified PCR product of the 16S rRNA gene was sequenced by Macrogen (Seoul, Republic of Korea) using universal primers, as previously described by Lane (1991). The sequence was trimmed manually using the BioEdit software (Hall, 1999) to obtain an almost complete 16S rRNA gene sequence (1,407 bp). The sequence was aligned with the sequences of available valid type strains in the genus Amycolatopsis, and any sequence similarities on the EzBioCloud server (https://www.ezbiocloud.net/) were determined (Yoon et al., 2017).
16S rRNA gene and genome phylogenies
Phylogenetic trees based on the 16S rRNA gene sequence were generated using the neighbor-joining (Saitou and Nei, 1987), maximum-likelihood (Felsenstein, 1981), and maximum-parsimony (Fitch, 1972) tree-making methods using MEGA X (Kumar et al., 2018). Evolutionary distance matrices were computed based on Kimura’s two-parameter model (Kimura, 1980). The confidence of the tree topologies was statistically evaluated using 1,000 bootstrap resampling replicates (Felsenstein, 1985).
OrthoFinder v2.5.4 (Emms and Kelly, 2017, 2018, 2019) was used to construct species phylogeny based on 1,104 orthologs found in genomes of strain MEP2-6T and its 19 closely related Amycolatopsis species within an outgroup, Streptomyces scabiei 87.22T. The genome phylogeny was then visualized using the interactive Tree of Life (iTOL) (Letunic and Bork, 2019) and further modified using Inkscape (https://inkscape.org/).
Phenotypic characterization
Cell morphology of strain MEP2-6T was observed using scanning electron microscopy (JEOL, JSM-IT500HR, Tokyo, Japan) after being grown on yeast malt extract agar (ISP 2) at 30°C for 21 days. The cultural characteristics of strain MEP2-6T and its closest type strains were determined after 14 days of incubation at 30°C on various agar media: ISP 2 agar, oatmeal agar (ISP 3), inorganic salt-starch agar (ISP 4), glycerol-asparagine agar (ISP 5), peptone-yeast extract iron agar (ISP 6), tyrosine agar (ISP 7), and nutrient agar (NA) (Shirling and Gottlieb, 1966). The color of the colonies and diffusible pigments were assigned based on the ISCC-NBS color system (Kelly, 1964). Growth at different temperatures (15°C, 25°C, 30°C, 37°C, 45°C, and 50°C) and tolerance levels to NaCl (1%–10%, w/v) were assessed using ISP 2 agar as the basal medium, and the effect of pH on growth ranging from 4 to 10 (at intervals of 1 pH unit) was examined in ISP 2 broth at 30°C for 14 days using the following buffer system: acetate buffer (pH 4–5), phosphate buffer (pH 6–8), and glycine–sodium hydroxide buffer (pH 9–10). Utilization of carbohydrates as the sole carbon source was observed using ISP 9 as the basal medium supplemented with a final concentration of 1% (w/v) of the carbon sources (Shirling and Gottlieb, 1966). Acid production from carbohydrates was determined using a basal inorganic nitrogen medium, according to the method described by Gordon et al. (1974). Starch hydrolysis, nitrate reduction, milk peptonization, milk coagulation, gelatin liquefaction, and H2S production were assessed on ISP 4 agar, ISP 8 broth (0.5% peptone, 0.3% beef extract, and 0.1% KNO3, pH 7.0), 10% (w/v) skimmed milk agar, 10% (w/v) skimmed milk broth, glucose-peptone-gelatin medium (2.0% glucose, 0.5% peptone, and 20% gelatin, pH 7.0), and ISP 6 agar, respectively. Enzymatic activity was assayed using the API ZYM (bioMérieux) commercial kit, according to the manufacturer’s instructions.
Chemotaxonomic characterization
Freeze-dried whole cells of strain MEP2-6T and its closest type strains for chemotaxonomic analyses were obtained after growth in yeast extract dextrose broth (1% yeast extract, 1% dextrose, pH 7.0) at 30°C (200 rpm) for 7 days. Isomers of cell wall diaminopimelic acid (A2pm) and reducing sugars of strain MEP2-6T whole-cell hydrolysates were determined using thin-layer chromatography (TLC) (Staneck and Roberts, 1974). The N-acyl group of muramic acid in the cell wall peptidoglycan of strain MEP2-6T was examined according to the method described by Uchida and Aida (1984). The cellular phospholipids of strain MEP2-6T were extracted and identified using 2-dimensional TLC, as previously described by Minnikin et al. (1984). Mycolic acid was extracted and monitored by TLC following the method described by Tomiyasu (1982). Cellular fatty acid methyl esters of all strains were prepared according to the method described by Sasser (1990) and analyzed using gas chromatography (MIDI, Sherlock Microbial Identification System, TSBA6 Sherlock Version 6.2B, USA). Isoprenoid quinones were extracted using the method previously employed by Collins et al. (1977) and were detected by LC-DAD-ESI-MS (AB Sciex, Framingham, MA, USA) equipped with a CAPCELL CORE C18 column (3.0 mm i.d. × 100 mm), OSAKA SODA Co., Ltd., using methanol–isopropanol (7:3, v/v). Finally, UV detection was performed at 270 nm wavelength.
Comparative genomic analyses
The genome of strain MEP2-6T was used to determine the taxonomic parameters between its closely related strains. This process involved average nucleotide identity based on BLAST (ANIb) and MUMmer (ANIm), and digital DNA–DNA hybridization (dDDH) values using JSpeciesWS (Richter et al., 2016) and the Genome-to-Genome Distance Calculator (GGDC) 3.0, with the recommended formula 2 (Meier-Kolthoff et al., 2013, 2022), respectively, to verify its taxonomic status. OrthoVenn3 (Sun et al., 2023, https://orthovenn3.bioinfotoolkits.net) was used to analyze shared and strain-specific orthologous clusters. A cluster of orthologous genes (COGs) in the unique orthologous cluster was functionally annotated and identified using eggNOG v5.0 (Huerta-Cepas et al., 2019), a database of orthology relationships, functional annotations, and gene evolutionary histories, as well as the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al., 2016), a database resource used to gain an in-depth understanding of the high-level functions and utilities of the biological system. AntiSMASH 7.0 (Blin et al., 2023) was used to identify secondary metabolite biosynthetic gene clusters (smBGCs) with default settings. Gene clusters with a BLAST identity of >80% were determined to belong to the same smBGCs. The results were collected in a presence/absence matrix to establish the number of genes designed for individual smBGCs in each bacterial strain. Hierarchical cluster analysis using the DICE coefficient with Unweighted Pair Group Method with Arithmetic (UPGMA) mean value was implemented with PAST (Hammer et al., 2001).
Plant-pathogenicity test
Strain MEP2-6T was examined for pathogenicity using the plant seedling method previously described by Flores-Gonzalez et al. (2008) and Dees et al. (2013), with slight modifications. Cherry tomato seeds (Solanum lycopersicum var. cerasiforme) were surface-disinfected with 6% (v/v) NaOCl for 10 min, rinsed three times with sterile distilled water, placed on sterile tissue paper, and air-dried under laminar flow. Subsequently, sterilized seeds were aseptically placed on an 8-day-old culture of strain MEP2-6T grown on ISP 2 agar medium and incubated at room temperature for 8 days. Streptomyces scabiei WSLK1-9 and ISP 2 agar plates without bacteria were used as controls. The appearance of seedlings after bacterial growth was recorded. The strain was considered phytopathogenic if the seeds displayed stunting or did not germinate. This experiment was performed in triplicate. A potato tuber slice technique was also used to determine the pathogenicity of strain MEP2-6T following the procedure previously described by Loria et al. (1995) and Henao et al. (2021), with slight modifications. The potato tubers cv. Spunta were surface-sterilized in 3% (v/v) NaOCl for 5 min, rinsed three times with sterile distilled water to eliminate the disinfectant, and air-dried under laminar flow. The surface-sterilized tubers were cut into slices (0.5 cm thick) and placed on moist sterile membrane filter paper in sterile Petri dishes. Strain MEP2-6T and S. scabiei WSLK1-9 were grown on ISP2 agar medium for 8 days at 30°C to produce high mycelia and spore masses. Agar plugs of the sporulated strain MEP2-6T were flipped and placed at the center (pith tissue) of potato slices. The agar plugs of Streptomyces scabiei WSLK1-9 and ISP 2 without bacteria were used as controls. The moist Petri dishes were incubated in the dark at 30°C for six days. Each isolate was tested in triplicate.
In planta colonization of Solanum tuberosum L. cv. Spunta and microscopic observation
The axillary buds of sprouted potato (S. tuberosum L. cv. Spunta) were excised and surface sterilized by soaking in 10% and 5% (v/v) NaClO solution for 15 min and 10 min, respectively, and rinsed twice thoroughly with sterile distilled water to eliminate the sterilizing agent (Barker, 1953). The sterilized buds were dried in a flow hood, aseptically cut into small pieces, placed on Murashige–Skoog agar media supplemented with 0.1% (w/v) activated charcoal (Murashige and Skoog, 1962; Feyissa et al., 2005), and incubated at 25°C under 1,000 lx illumination using 40-watt TL 33 Philips fluorescent lamps for 14 days (Roca et al., 1978). The prepared 8-day-old strain MEP2-6T agar plug was directly applied at the wounding (pin-prick) stem node site of 14-day-old S. tuberosum L. cv. Spunta cultures and incubated at 25°C under 1,000 lx illumination for five days. Colonization was monitored five days post-inoculation using a bright-field microscopic technique (Thomas and Reddy, 2013). A ZEISS Axiolab 5 optical microscope, together with the microscope camera Axiocam 208 color and ZEISS Labscope Imaging App v4.2.1 (Carl Zeiss Microscopy Deutschland GmbH, Oberkochen, Germany) was used for bright-field microscopy. Thin stem tissues were prepared using a free-hand-cut sectioning technique with a fine razor blade. The tissues were examined after mounting with Shear’s mounting medium (6.0 g potassium acetate, 120 ml glycerol, 180 ml 95% (v/v) ethanol, and 300 ml distilled water) on acetone-washed and autoclaved microscope slides under oil immersion (×100). Images were captured with the ZEISS Labscope Imaging App and processed using Adobe Photoshop 2021 v22.5.9.1101 (Adobe Systems Inc., San Jose, CA, USA) software.
Determination of extracellular carbohydrate-degrading enzyme production
Strain MEP2-6T and its closest strains were investigated for their production of extracellular carbohydrate-degrading enzymes, including endoglucanase, chitinase, and pectinase, according to the method described by Kumla et al. (2020). An agar mycelial plug of each strain was inoculated into the tested agar media. Endoglucanase, chitinase, and pectinase production were investigated on carboxymethyl cellulose (CMC) agar, colloidal chitin agar, and pectin agar, respectively. The plates were incubated at 25°C in the dark for 5 days. Colonies of all strains were immersed in 1% (w/v) Congo red for 30 min and then rinsed with 1 M NaCl for 15 min for chitinase and endoglucanase tests. For the pectinase test, colonies of all strains were immersed in 1% (w/v) hexadecyltrimethylammonium bromide for 15 min and rinsed with sterile deionized (DI) water. A positive result for the production of each enzyme was indicated by the presence of a clear transparent zone around the colony. Four replicates were performed for each enzyme.
Results
Sequence analysis and 16S rRNA gene and genome phylogenies
16S rRNA gene sequence analysis revealed that strain MEP2-6T (1,407 bp, accession no. OR762507) is a member of the genus Amycolatopsis, the order Pseudonocardiales, and the family Pseudonocardiaceae. Sequence analysis revealed that strain MEP2-6T shared a close relationship with Amycolatopsis lexingtonensis NRRL B-24131T, A. pretoriensis DSM 44654T, A. kentuckyensis NRRL B-24129T, A. rifamycinica DSM 46095T, A. tolypomycina DSM 44544T, and A. eburnea GLM-1T, with sequence similarities of 99.4% (9 nt difference at 1,400), 99.3% (10 nt difference at 1,401), 99.0% (14 nt difference at 1,400), 99.1% (12 nt difference at 1,400), 99.1% (12 nt difference at 1,400), and 98.9% (15 nt difference at 1,400), respectively. A maximum-likelihood phylogenetic tree based on the 16S rRNA gene sequence (Figure 1) indicated that strain MEP2-6T formed a tightly independent cluster with the closest species, A. lexingtonensis NRRL B-24131T and A. pretoriensis DSM 44654T. The clusters that could be recovered in neighbor-joining and maximum parsimony trees are presented in Supplementary Figures 2, 3, respectively. Genome phylogeny (Figure 2) suggested that strain MEP2-6T formed a robustly liberated clade with A. lexingtonensis DSM 44653T (=NRRL B-24131T) and A. eburnea GLM-1T. Therefore, based on a combination of sequence analysis, 16S rRNA gene phylogenetic tree, and phylogenomic tree, A. lexingtonensis NRRL B-24131T (=JCM 12672T =DSM 44653T), A. pretoriensis DSM 44654T (=JCM 12673T), and A. eburnea GLM-1T (=TBRC 9315T) were used to further clarify the phenotypic characteristics, chemotaxonomic properties, and genome comparisons.
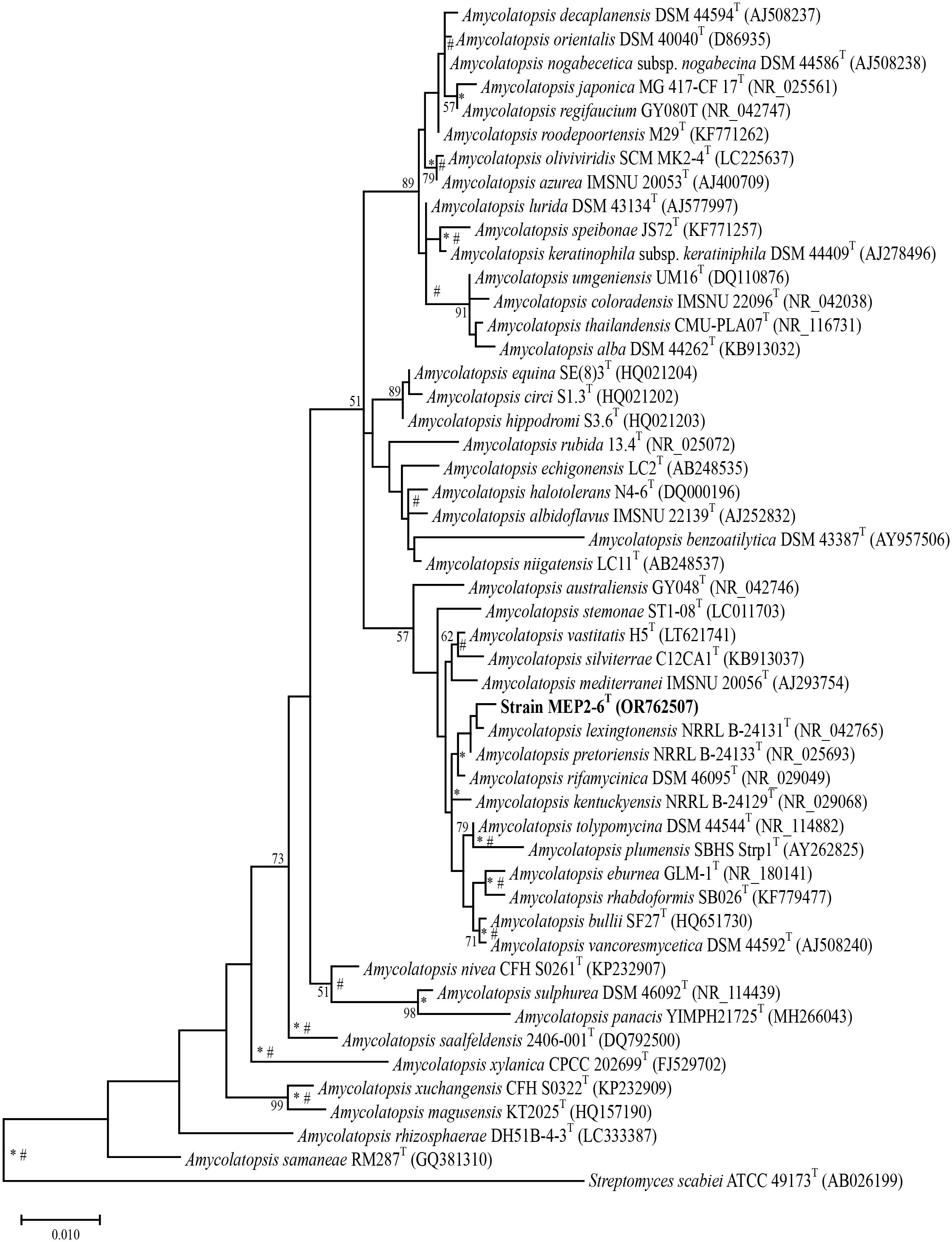
Figure 1 Maximum-likelihood phylogenetic tree based on the 16S rRNA gene sequences of strain MEP2-6T and its closely related type strains with validly published names. Streptomyces scabiei 87.22T was used as the outgroup. Asterisks and sharps (*, #) indicate that the corresponding nodes were also recovered in the neighbor-joining and maximum-parsimony trees, respectively. Bootstrap values of ≥50% (percentages of 1,000 replications) are shown at branch nodes Bar, 0.01 substitutions per nucleotide position.
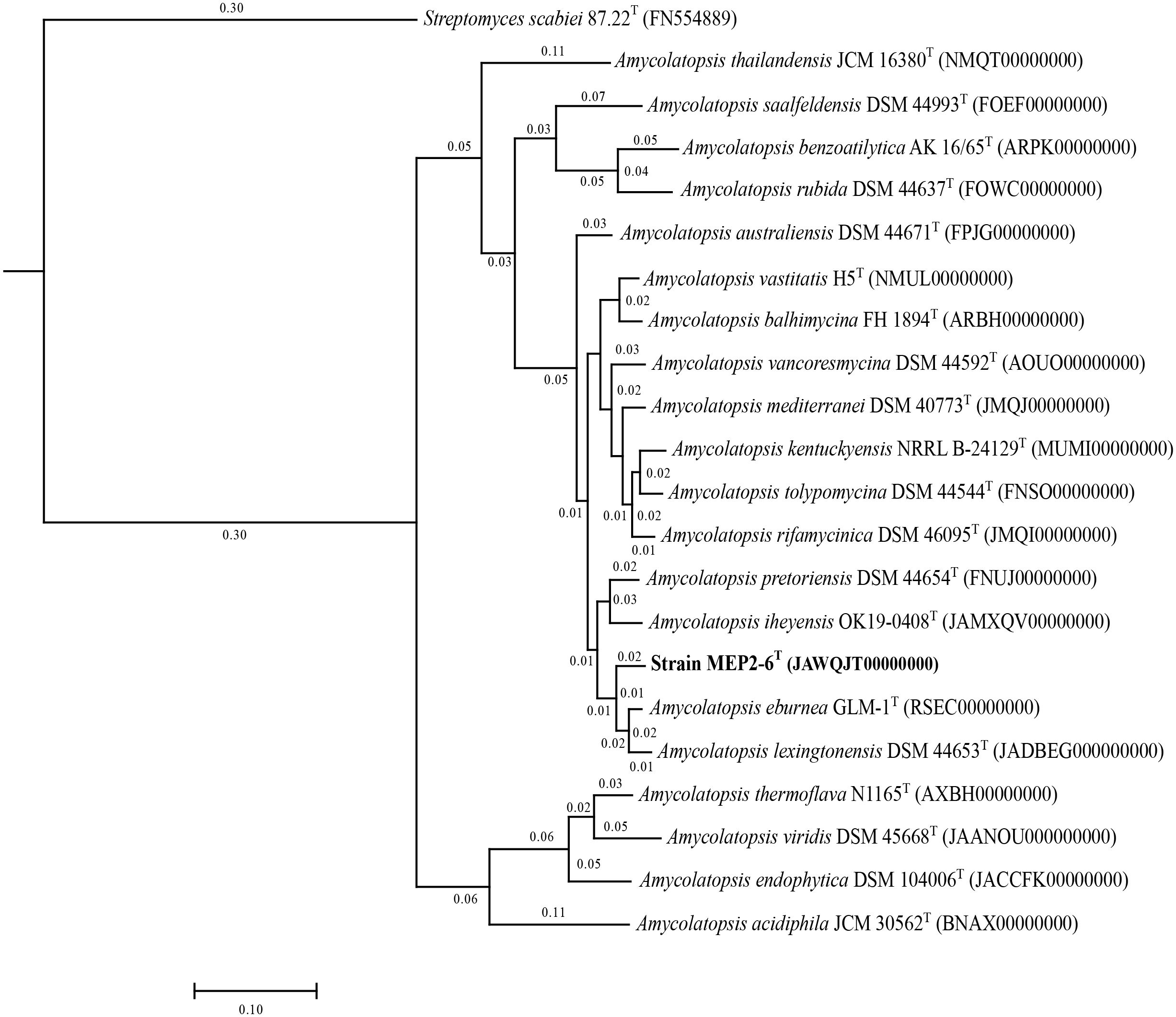
Figure 2 A core phylogenomic tree based on 1,104 orthologous proteins illustrates the evolutionary relationship between strain MEP2-6T and its closely related Amycolatopsis species. Streptomyces scabiei 87.22T was used as the outgroup. The distance matrices between species are shown at the node.
Phenotypic characteristics
Strain MEP2-6T produced septal substrate and aerial hyphae (0.4–0.5 µm × 1.1–1.4 µm in size) that fragmented into rod-like elements (Figure 3). The strain adequately developed moderate orange-colored substrate mycelia and pale orange yellow aerial mycelia on ISP 2 agar medium but did not form aerial hyphae on ISP 5 and nutrient agar media. Soluble pigments were not observed in any of the media tested. Strain MEP2-6T also grew well on ISP 2, ISP 3, ISP 4, and ISP 7, whereas it grew moderately and/or poorly grew on ISP 5, ISP 6, and nutrient agar media. The cultural characteristics of strain MEP2-6T and the phylogenetically related type strains on ISP 2 agar medium are shown in Figure 4, and other media are presented in Supplementary Table 1, Supplementary Figure 4. Growth occurred at 15°C–37°C (optimum, 30°C), pH 5–9 (optimum, 7), and 1%–4% (w/v) NaCl. Strain MEP2-6T utilized amygdalin, L-arabinose, D-fructose, D-galactose, D-glucose, D-melezitose, myo-inositol, L-rhamnose, D-sucrose, and D-xylose as the sole carbon sources. The strain had the ability to reduce nitrate, coagulate, and peptonize milk, and to liquefy gelatin, but did not hydrolyze starch and produce H2S. In the API ZYM test, it was positive for leucine arylamidase, valine arylamidase, cystine arylamidase, α-chymotrypsin, and Naphthol-AS-BI-phosphohydrolase, while acid phosphatase, N-acetyl-β-glucosaminidase, and α–fucosidase were weakly positive. The phenotypic properties that distinguished strain MEP2-6T from other closely related strains are listed in Table 2.

Figure 3 Scanning electron micrograph of strain MEP2-6T grown on ISP 2 agar medium at 30°C for 14 days showing the morphology of the substrate and aerial mycelia. Bar, 2 µm.
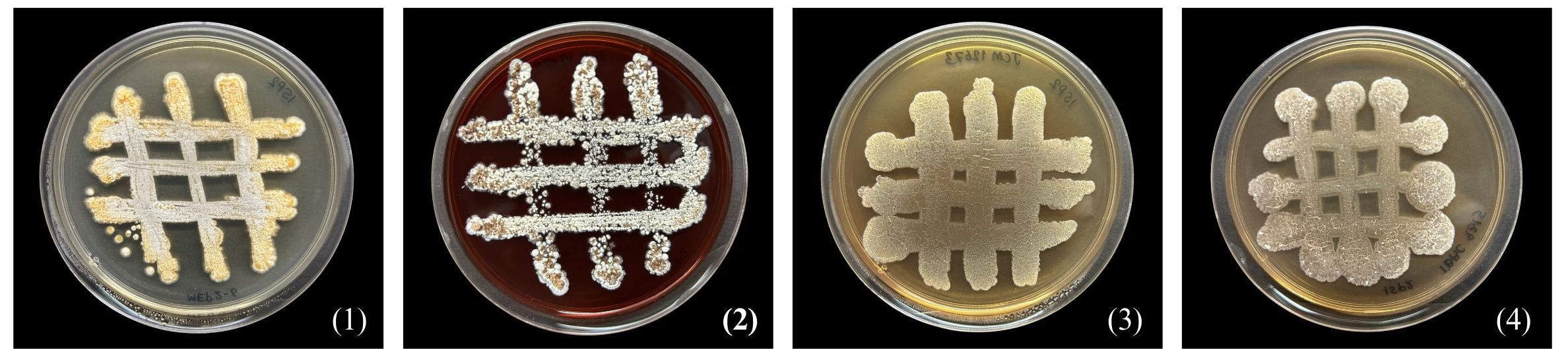
Figure 4 Differential colonial appearances of strain MEP2-6T and its closest type strains grown on ISP 2 agar medium at 30°C for 14 days. Strain: 1, MEP2-6T; 2, Amycolatopsis lexingtonensis JCM 12672T; 3, A. pretoriensis JCM 12673T; 4, A. eburnea TBRC 9315T.
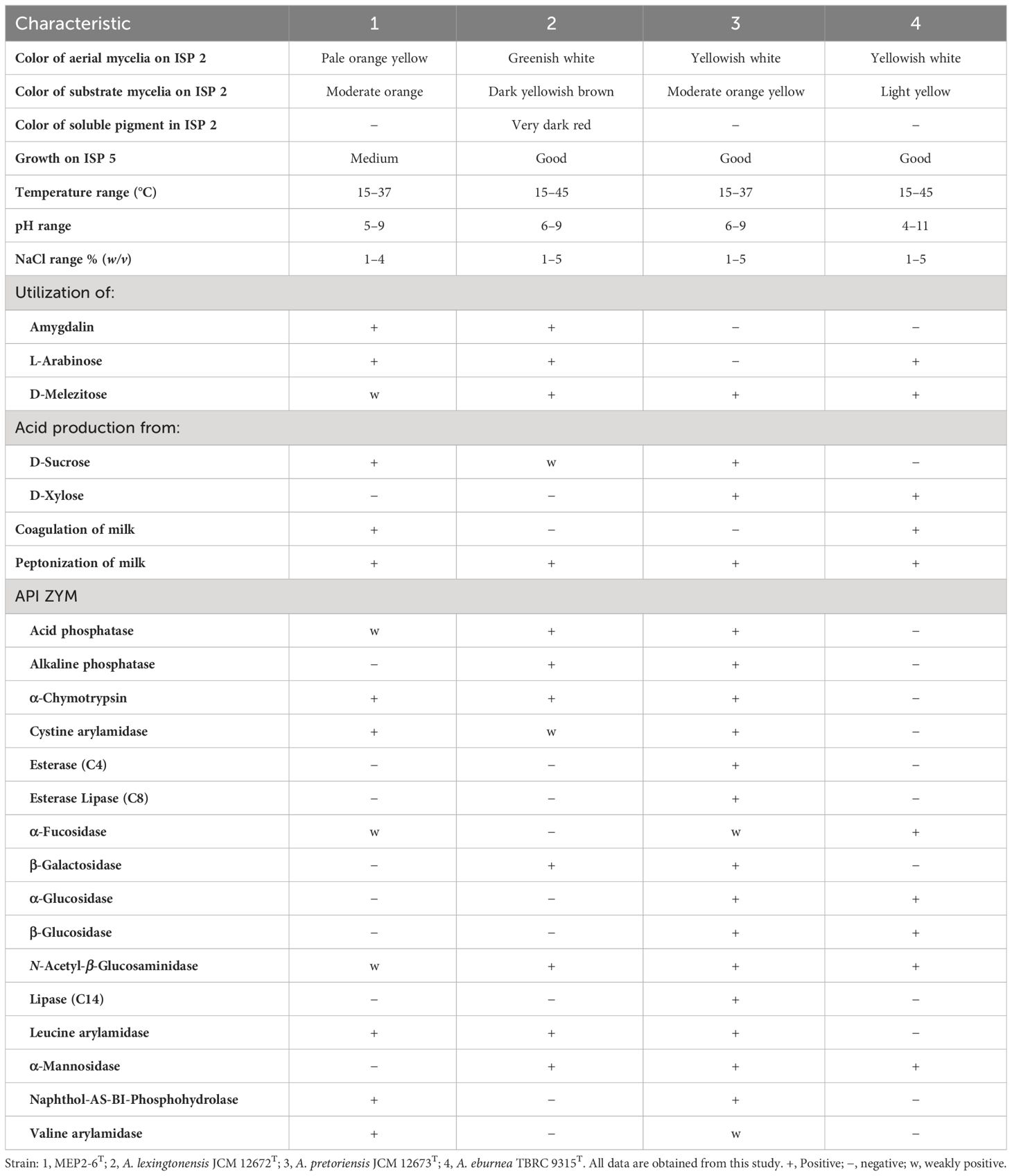
Table 2 Differential phenotypic characteristics between strain MEP2-6T and its closely related strains.
Chemotaxonomy
Strain MEP2-6T comprises meso-diaminopimelic acid as the diagnostic diamino acid in the cell wall peptidoglycan. Arabinose, galactose, glucose, and ribose were identified as the diagnostic sugars in the whole-cell hydrolysate. The N-acyl group of muramic acid in peptidoglycan is acetyl, whereas mycolic acids are absent. The polar lipid profile consisted of diphosphatidylglycerol (DPG), phosphatidylglycerol (PG), phosphatidylethanolamine (PE), hydroxy-phosphatidylethanolamine (OH−PE), an unidentified aminophospholipid (APL), six unidentified phospholipids (PL1−PL6), an unidentified glycolipid (GL), and five unidentified lipids (L1−L5) (Supplementary Figure 5). The major menaquinone in strain MEP2-6T was MK-9(H6) (90.1%), while MK-9(H4) (9.9%) was a minor component. The iso-C16:0 (37.9%) and iso-C15:0 (13.7%), which accounted for >10% of the total fatty acids, were the predominant cellular fatty acids in the strain profile. Differences in the types and quantities of cellular fatty acids of strain MEP2-6T and its closely related species are presented in Table 3.
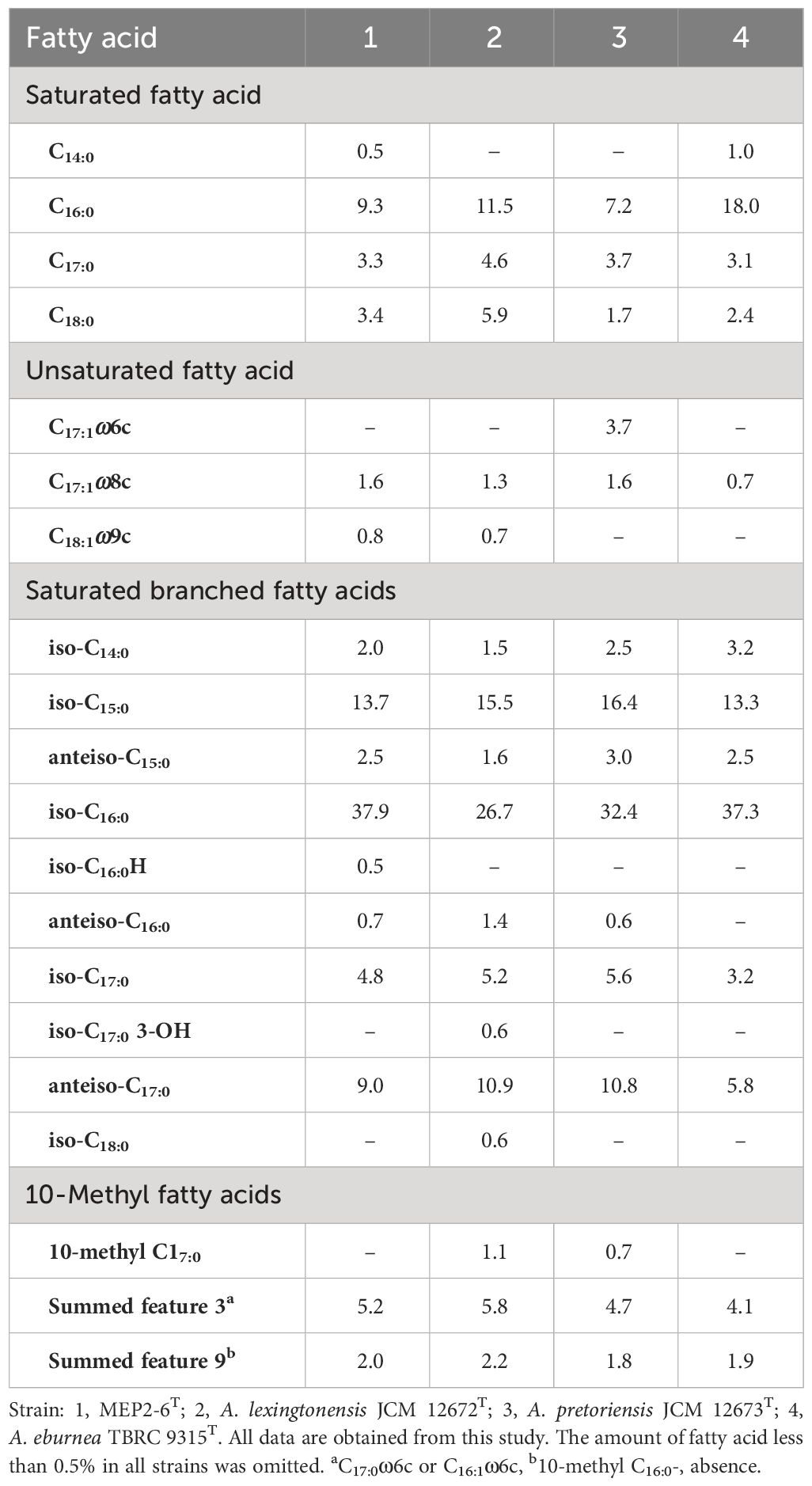
Table 3 Different cellular fatty acid profiles (%) of strain MEP2-6T and its closely related type strains.
Genomic feature and comparison
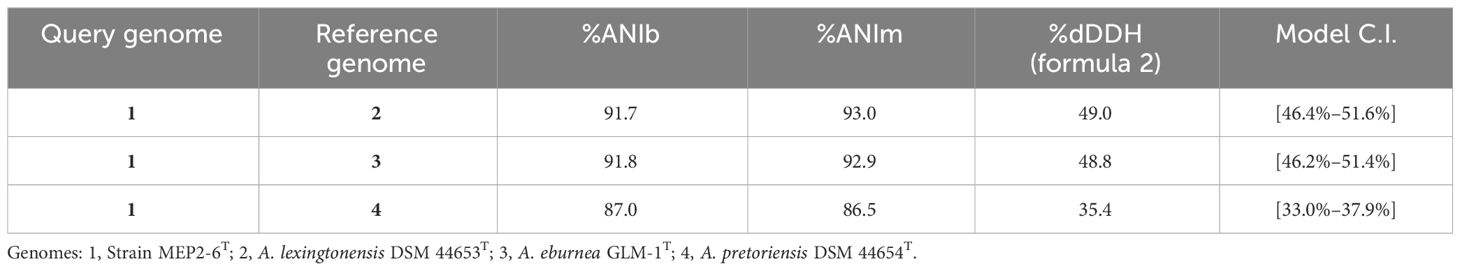
The genome sequence of strain MEP2-6T was 10,277,369 bp in size with a GC content of 71.74 mol% (accession no. JAWQJT000000000). The genomic features of the genome and the closest species of Amycolatopsis are summarized in Table 1. Pairwise genome-level comparisons between strain MEP2-6T and its phylogenetically closest relatives, including ANIb, ANIm, and dDDH values, were calculated to accurately delineate the species (Table 4). The strains exhibited 91.7%, 91.8%, and 87% ANIb and 93%, 92.9%, and 86% ANIm to A. lexingtonensis DSM 44653T (=JCM 12672T), A. eburnea GLM-1T (=TBRC 9315T), and A. pretoriensis DSM 44654T (=JCM 12673T), respectively. The dDDH values for the comparison of strain MEP2-6T to A. lexingtonensis DSM 44653T, A. eburnea GLM-1T, and A. pretoriensis DSM 44654T were 49%, 48.8%, and 35.4%, respectively. Both values were significantly lower than the threshold values of 95%–96% ANI (Richter and Rosselló-Móra, 2009) and 70% dDDH (Wayne et al., 1987; Goris et al., 2007), which is recommended for use in species discrimination. Consequently, strain MEP2-6T can be officially recognized as a novel species within the genus Amycolatopsis.
To characterize core and strain-specific genes, orthologous groups were determined using the translated proteomes of strain MEP2-6T compared to those of the three closest Amycolatopsis species: A. lexingtonensis DSM 44653T, A. eburnea GLM-1T, and A. pretoriensis DSM 44654T (Figure 5). In total, 8,824 orthologous clusters and 39,478 proteins were identified. The core genome shared by the four strains was depicted by 6,336 orthologous clusters. In a pairwise comparison, the largest number of orthologous clusters was found for A. eburnea GLM-1T/A. lexingtonensis DSM 44653T (308), followed by strain MEP2-6T/A. lexingtonensis DSM 44653T (266), A. eburnea GLM-1T/A. pretoriensis DSM 44654T (212), MEP2-6T/A. pretoriensis DSM 44654T (183), and strain MEP2-6T/A. eburnea GLM-1T (158). These findings agreed well with the taxonomic position of strain MEP2-6T in the core phylogenomic tree (Figure 2).
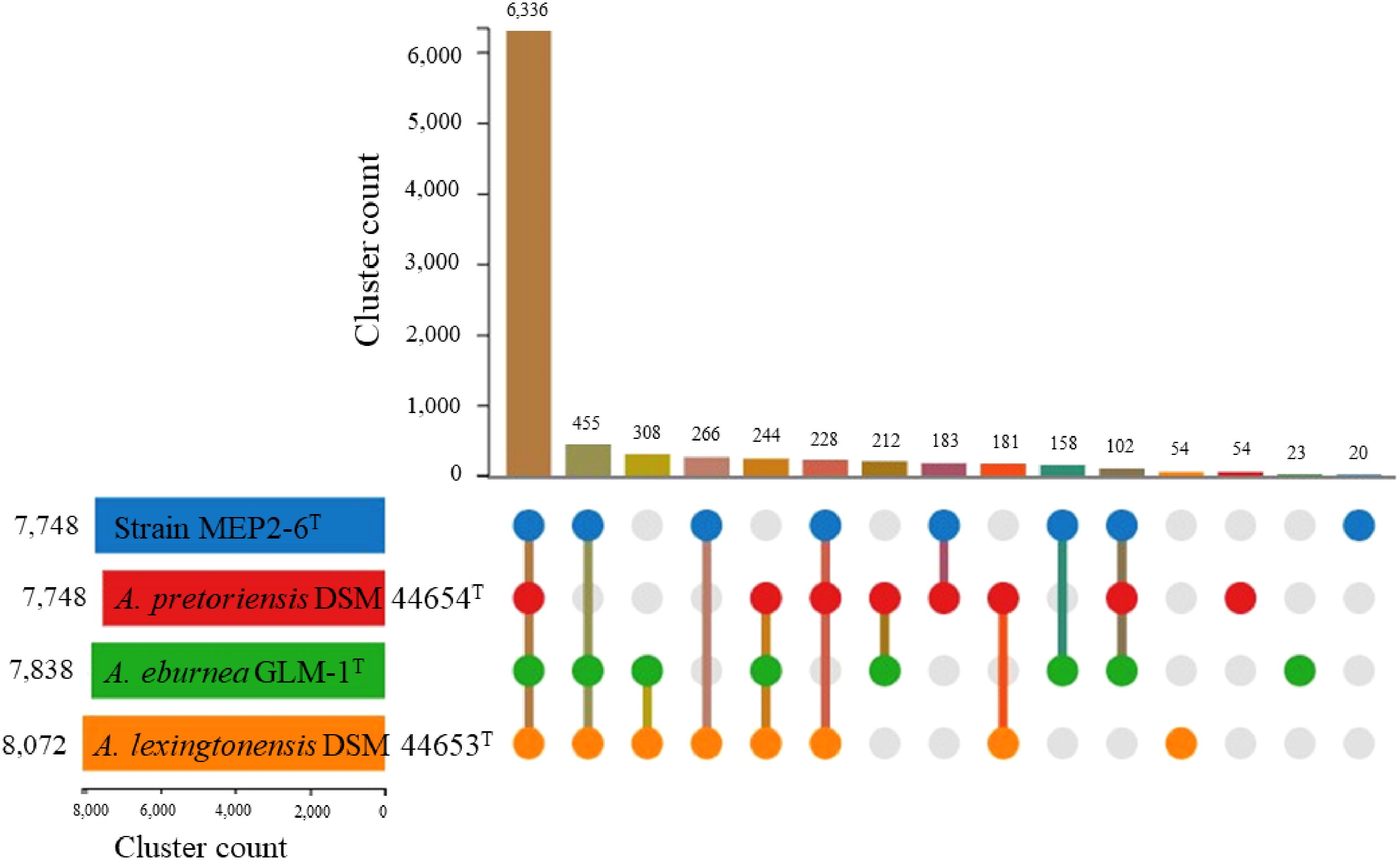
Figure 5 Proteome comparison of strain MEP2-6T and closely related type strains of the genus Amycolatopsis based on OrthoVenn3. An UpSet plot showed unique and shared orthologous clusters among species. The left horizontal bar chart depicts the number of orthologous clusters per species, whereas the right vertical bar chart illustrates the number of orthologous clusters shared among the species.
As shown in Figure 5, the number of strain-specific clusters for each strain was 20 for MEP2-6T, 23 for A. eburnea GLM-1T, 54 for A. lexingtonensis DSM 44653T, and 54 for A. pretoriensis DSM 44654T. Strain MEP2-6T uniquely contained an orthologous cluster of the mycothiol biosynthesis process that was not found in the closest relatives. Functional annotation based on the eggNOG and KEGG databases revealed that the strain completely contained all genes encoded for the key enzymes in the biosynthetic pathway of mycothiol: ino1 (myo-inositol-1-phosphate synthase, EC 5.5.1.4), mshA (D-inositol-3-phosphate glycosyltransferase, EC 2.4.1.250), mshB (N-acetyl-1-D-myo-inositol-2-amino-2-deoxy-alpha-D-glucopyranoside deacetylase, EC 3.5.1.103), mshC (L-cysteine:1D-myo-inositol-2-amino-2-deoxy-alpha-D-glucopyranoside ligase, EC 6.3.1.13), and mshD (mycothiol synthase, EC 2.3.1.189). The protein sequences of the genes encoding mycothiol biosynthetic enzymes showed sequence identities ranging from 94.0% to 98.9% to the reference protein based on UniProt BLAST (Coudert et al., 2023). Proteins with over 90% sequence identity typically share the same biological processes (Joshi and Xu, 2007). The organization of the gene cluster and biosynthetic pathways is illustrated in Figure 6.
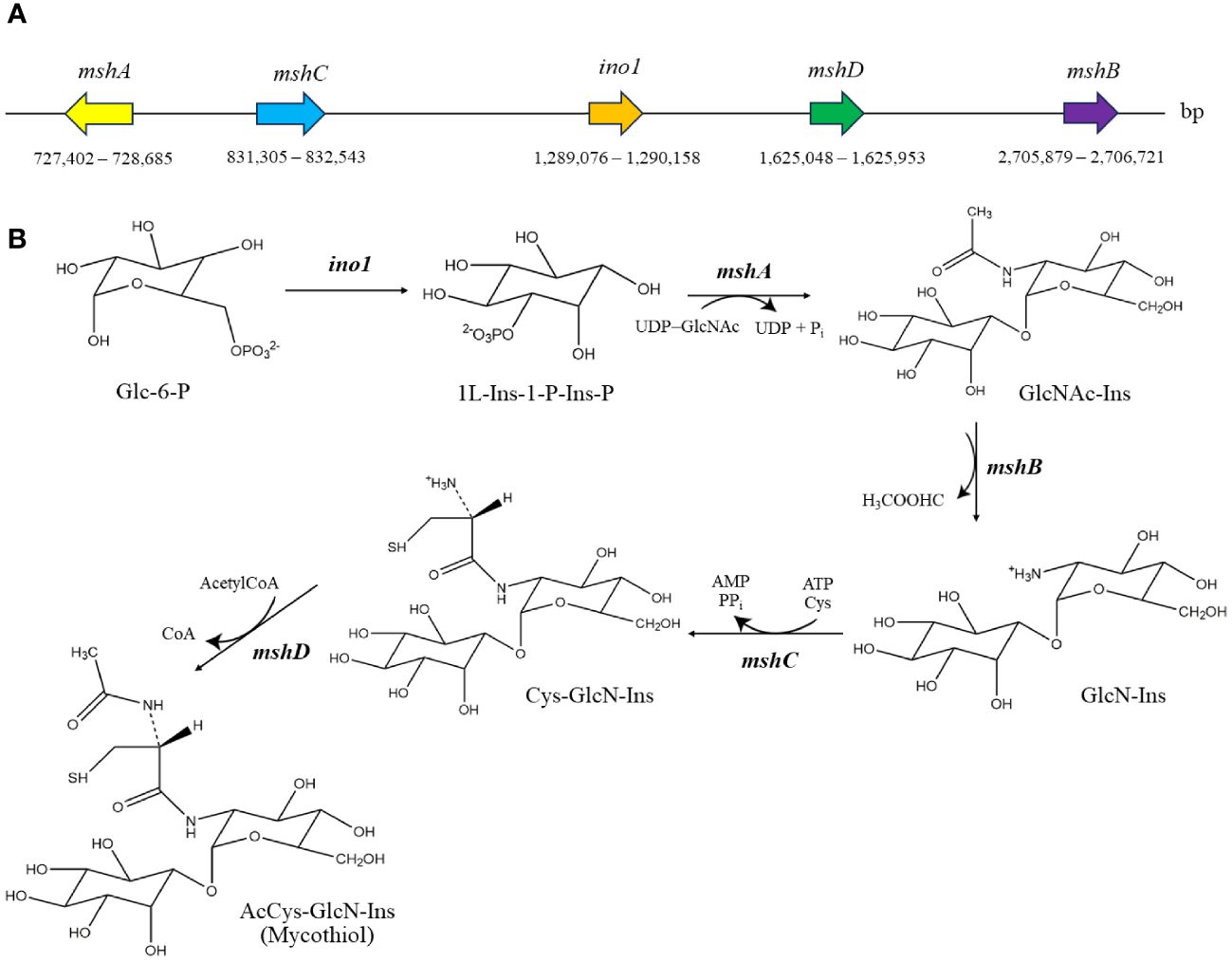
Figure 6 Genomic organization of mycothiol biosynthetic gene cluster (A) and biosynthetic pathway of mycothiol (B) in strain MEP2-6T. The structures were drawn using the ChemDraw program. Glc-6-P, glucose-6-phosphate; 1L-Ins-1-P-Ins-P, 1L-myo-inositol-1-phosphate; GlcNAc-Ins, 1-O-(2-acetamido-2-deoxy-α-D-glucopyranosyl)-D-myo-inositol; GlcN-Ins, 1-O-(2-amino-2-deoxy-α-D-glucopyranosyl)-D-myo-inositol; Cys-GlcN-Ins, 1-O-[2-[[(2R)-2-amino-3-mercapto-1-oxopropyl]amino]-2-deoxy-α-D-glucopyranosyl]-D-myo-inositol.
Predictive functional signatures to live inside plant
Based on RAST annotation and enzyme prediction by KEGG, a plant-derived strain MEP2-6T illustrated an important enrichment of genes related to encoding enzymes for plant-synthesized sugar interconversions, which were absent in A. eburnea GLM-1T isolated from arbuscular mycorrhizal fungi and A. lexingtonensis DSM 44653T and A. pretoriensis DSM 44654T isolated from equine placentas. These genes included galA, xynD, cel74a, cbhA, and bglB-bglX, which encode α-galactosidase (EC 3.2.1.22), arabinoxylan arabinofuranohydrolase (EC 3.2.1.55), xyloglucan-specific exo-β-1,4-glucanase (EC 3.2.1.155), cellulose-1,4-beta-cellobiosidase (exoglucanases, EC 3.2.1.91), and β-D-glucosidases (EC 3.2.1.21), respectively. The genes malZ, chiA, and one encoded maltodextrin glucosidase (EC 3.2.1.20), chitinase (E 3.2.1.14), and endoglucanase (EC 3.2.1.4), respectively, were found in all strains. Nonetheless, the gene coding for endoglucanase was over-represented in strain MEP2-6T with seven copies.
Several transporters of oligo- and monosaccharides were overexpressed in strain MEP2-6T. ABC transporter genes for various sugars, such as chitobiose (dasA, dasB, and dasC), raffinose/stachyose/melibiose (msmE, msmF, msmG, msmX, and msmK), fructose (frcC, frcB, and frcA), trehalose/maltose (thuE, thuF, and thuG), and multiple sugars (e.g., malK, sugC, and msiK), were found to be more than or equal to a two-fold copy among the closest non-plant-associated Amycolatopsis species. The ABC transporter genes cebE, cebF, and cebG, which encode the cellobiose transport system, were only present in strain MEP2-6T. Interestingly, strain MEP2-6T takes up fructose into the cell via the fructose ABC transport system, but the three closest species, A. lexingtonensis DSM 44653T, A. eburnea GLM-1T, and A. pretoriensis DSM 44654T, ingest fructose into the cells through the phosphotransferase system (PTS) encoded by four genes: fruA, fruB, fruK, and ptsI. These genes located in the genome of strain MEP2-6T prove that the strain can import and utilize various sugars as sole carbon sources.
The amino acids excreted by the host plant can serve as nitrogen sources for plant-derived actinobacteria. Genes associated with isoleucine, leucine, and valine were distributed in both plant-associated and non-plant-associated actinobacteria. However, it is fascinating that genes encoding branched-chain amino acid transporters (livF, livG, livH, and livM) were found to be overexpressed in strain MEP2-6T, with over two copies per genome. As an insight into the stress response, all strains employed a similar system, for instance, glutathione peroxidase (gpx, EC 1.11.1.9) and superoxide dismutase Fe–Mn family (sod2, EC 1.15.1.1). Interestingly, only strain MEP2-6T (plant-associated) contained the superoxide dismutase Cu–Zn family (sod1, EC:1.15.1.1) and rubredoxin, which are utilized for its ability to respond to oxidative stress.
Protein secretion plays a crucial role in modulating bacteria–niche interactions, particularly in the symbiotic (parasitic, mutualistic, or commensal) colonization of bacteria. In Gram-positive bacteria, secretion proteins are exported out of the cytoplasm by the conserved Sec translocase system, twin-arginine translocation (TAT) system, or, alternately, by the type VII system (Tseng et al., 2009). The genome of strain MEP2-6T encodes 1,050 (11.3%) secreted proteins involved in several protein secretion systems (Table 5). The strain comprises all genes responsible for coding the protein-conducting channel SecYEG, the ATP-dependent motor protein SecA, and the ancillary membrane protein complex SecDF, which delivers secretory proteins across the plasma membrane through the translocase (Lycklama et al., 2012). In addition, strain MEP2-6T contained all genes, tatA, tatB, and tatC, which encode twin-arginine translocation proteins that transport folded proteins across the plasma membrane (Palmer and Berks, 2012) and included four genes as part of the type VII secretion system: eccB, eccC, eccD, and mycP to promote host colonization.
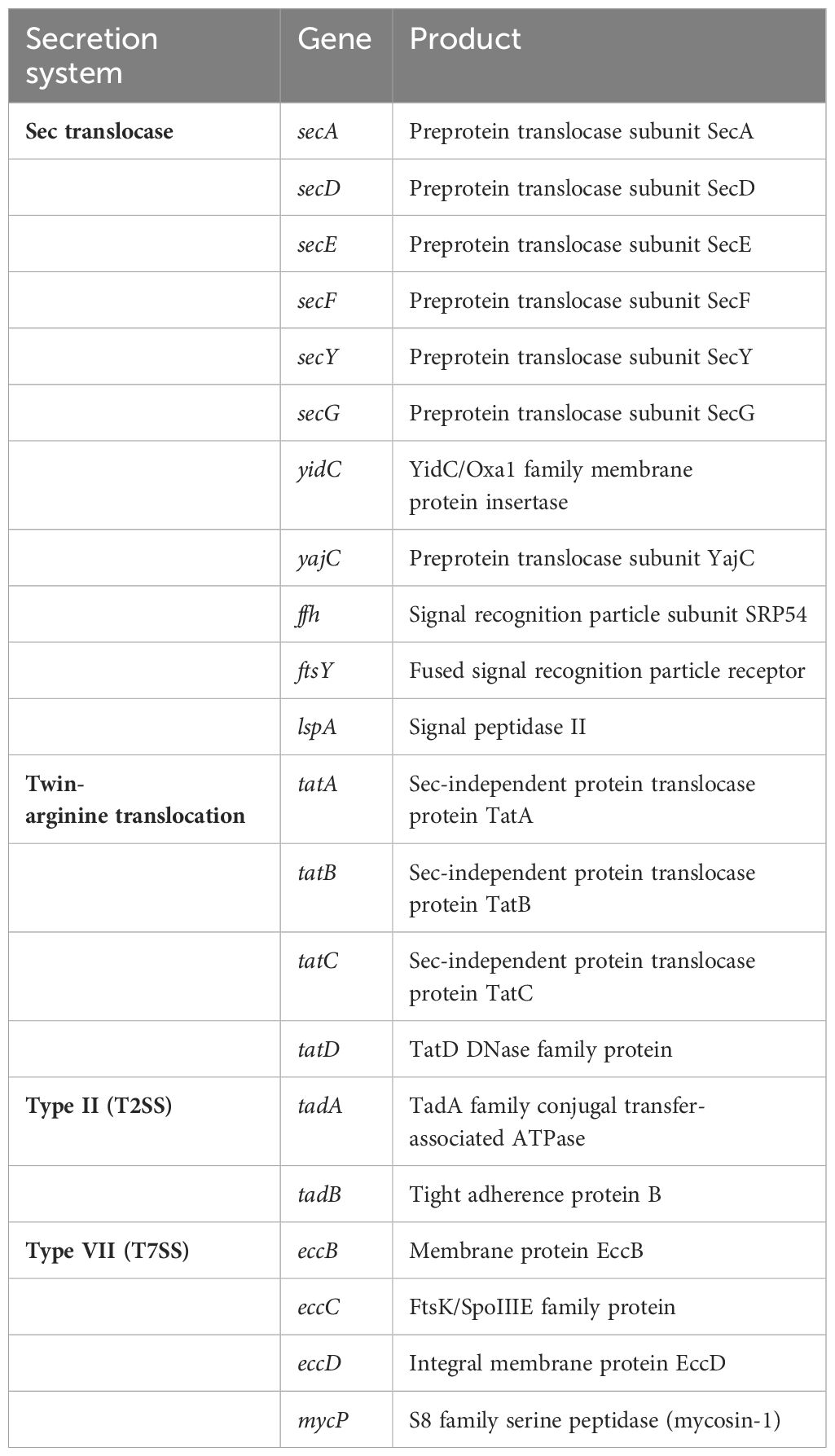
Table 5 Genes responsible for encoding protein secretion systems present in the genome of strain MEP2-6T.
Diversity of secondary metabolite biosynthetic gene clusters
The genomes of the strain MEP2-6T and its closest Amycolatopsis species were evaluated for candidate secondary metabolite biosynthetic gene clusters (BGCs) using antiSMASH 7.0, a pipeline for secondary metabolite identification. The genome of S. scabiei 87.22T (accession no. FN554889) was also compared to prove that strain MEP2-6T had no BGCs association in causing scab disease in potatoes. The number of identified BGCs per species, based on antiSMASH, ranged from 27 to 33. Strain MEP2-6T comprised 33 BGCs exhibiting different similarities to gene clusters, with known functions ranging from 4% to 100%. The BGCs that exhibited ≥50% homology to known functional gene clusters are shown in Table 6. Based on the antiSMASH version 7.0 annotation, S. scabiei 87.22T, which is a well-known causative agent of potato scab disease, contained BGCs encoded for the phytotoxins associated with the occurrence of scab lesions on potato tubers, including thaxtomin, bottromycin, and concanamycin A (Supplementary Table 2) (Li et al., 2019a). To prove that strain MEP2-6T is not a potato scab-causing pathogen, the protein sequences of the gene clusters responsible for synthesizing thaxtomin, bottromycin, and concanamycin A from S. scabiei 87.22T were subjected to a BLASTP search against all proteins of strain MEP2-6T. S. scabiei 87.22T comprised 52, 17, and 54 protein-coding genes in the BGCs of thaxtomin, bottromycin, and concanamycin A, respectively. Strain MEP2-6T contained 7, 2, and 25 protein-coding genes for the thaxtomins, bottromycin, and concanamycin A BGCs, respectively. Although protein-coding genes related to the BGCs of thaxtomin, bottromycin, and concanamycin A were present, the strain could not produce the compounds because those of the protein-coding genes were not the core biosynthetic gene clusters. For instance, two genes clustered in bottromycin BGC and found in strain MEP2-6T were identified as btmA. This gene encodes the phosphotransferase system (PTS) transporter subunits EIIC and IIE (Franz et al., 2021), which are responsible for selecting and transporting sugar molecules across the bacterial cytoplasmic membrane (McCoy et al., 2015). According to the seven protein-coding genes of strain MEP2-6T that are associated with thaxtomin BGC, two significant genes were identified as txtD and txtH, which encode nitric oxide synthase and MbtH family non-ribosomal peptide synthase (NRPS) accessory proteins, respectively. The txtH gene functions as a chaperone by promoting proper folding and stimulation of the two crucial NRPS enzymes encoded by txtA and txtB (Li et al., 2019b). However, strain MEP2-6T was unable to produce thaxtomin because it had no txtA, txtB, or txtC, which are the core biosynthetic genes responsible for catalyzing the conversion of L-tryptophan to thaxtomin (Jiang et al., 2018). Based on the 25 protein-coding genes of strain MEP2-6T related to the BGC of concanamycin A, four significantly encoded type I polyketide synthases and two encoded acyl carrier proteins (ACP). Nonetheless, it could not produce concanamycin A because it lacks the core synthesis domains of ketosynthase (KS) and acyltransferase (AT) (Haydock et al., 2005). Consequently, based on the analysis of the BGCs, it can be concluded that strain MEP2-6T is not a phytopathogen, even though it inhabits the scab lesions of potato.
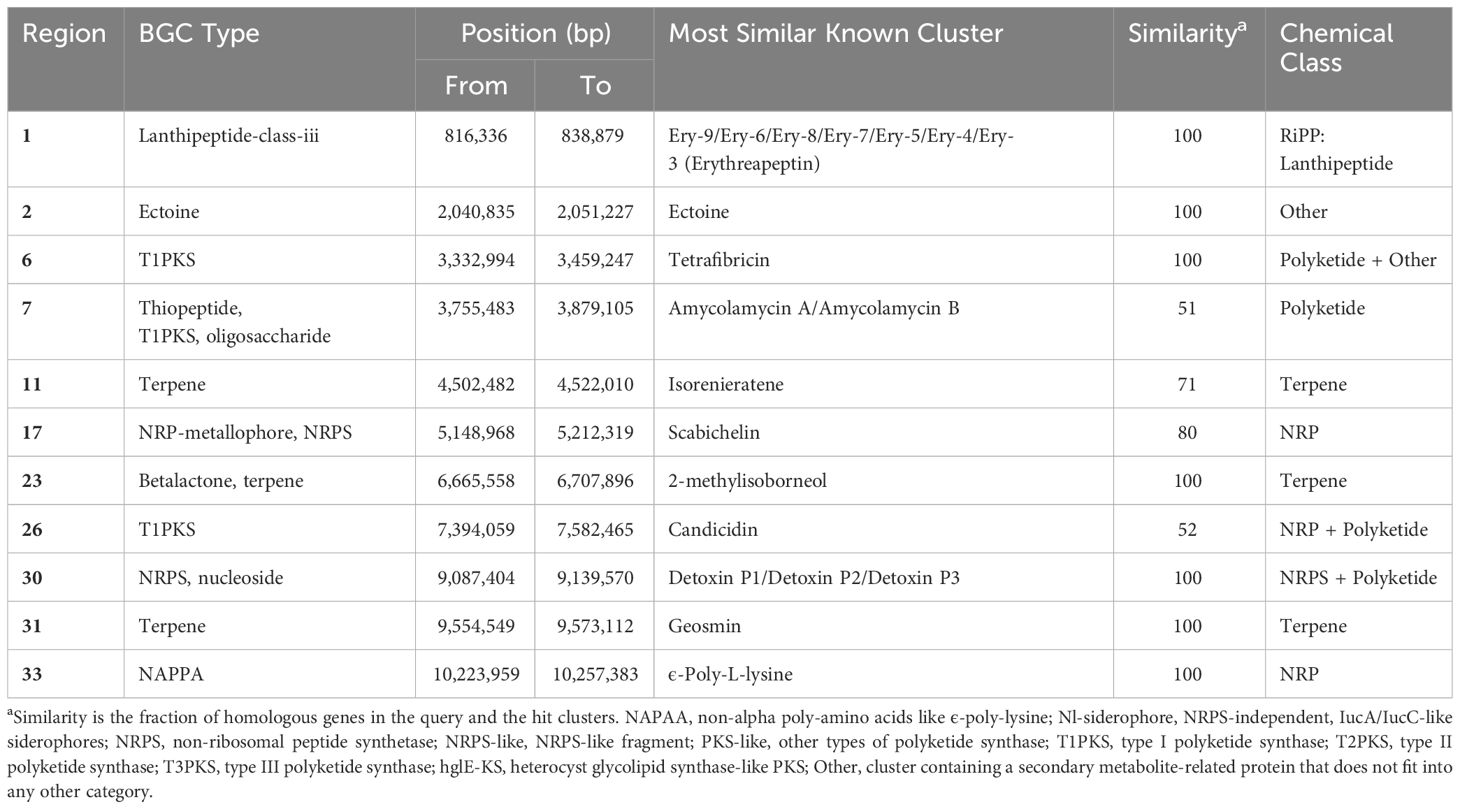
Table 6 Distribution of identified biosynthetic gene clusters (≥50% homology with known BGCs) encoding for secondary metabolites in strain MEP2-6T.
The distribution of secondary metabolite biosynthetic gene clusters (BGCs) among the strains in this study is presented in Figure 7 as hierarchical clusters. Among the four Amycolatopsis species, the three most frequently presented classes of BGCs encode genes for the production of type I polyketide synthases (T1PKS), non-ribosomal peptide synthases (NRPS), and terpenes. It can be determined that the pattern of BGCs is correlated with species phylogeny. Strain MEP2-6T, isolated from scab lesions on potato tubers, shared a monophyletic clade with A. eburnea GLM-1T, isolated from spores of Funneliformis mosseae RYA08, an arbuscular mycorrhizal fungus that inhabits Aquilaria crassna Pierre ex Lec (Chaiya et al., 2019) and a polyphyletic clade with A. lexingtonensis DSM 44653T isolated from lesions on horse placentas (Labeda et al., 2003). This phylogenetic cluster was consistent with the genomic similarity and the core phylogenomic tree. Although the BGCs seemed to be correlated based on species phylogeny, some BGCs and their products were different among the four Amycolatopsis strains; for example, ladderane and thioamitide BGCs were not found in strain MEP2-6T and A. eburnea GLM-1T but were present in A. lexingtonensis DSM 44653T and A. pretoriensis DSM 44654T. Compounds encoded by BGCs (≥50% homology with known functions) were found in all and/or were unique in the four Amycolatopsis strains, as shown in Table 7.
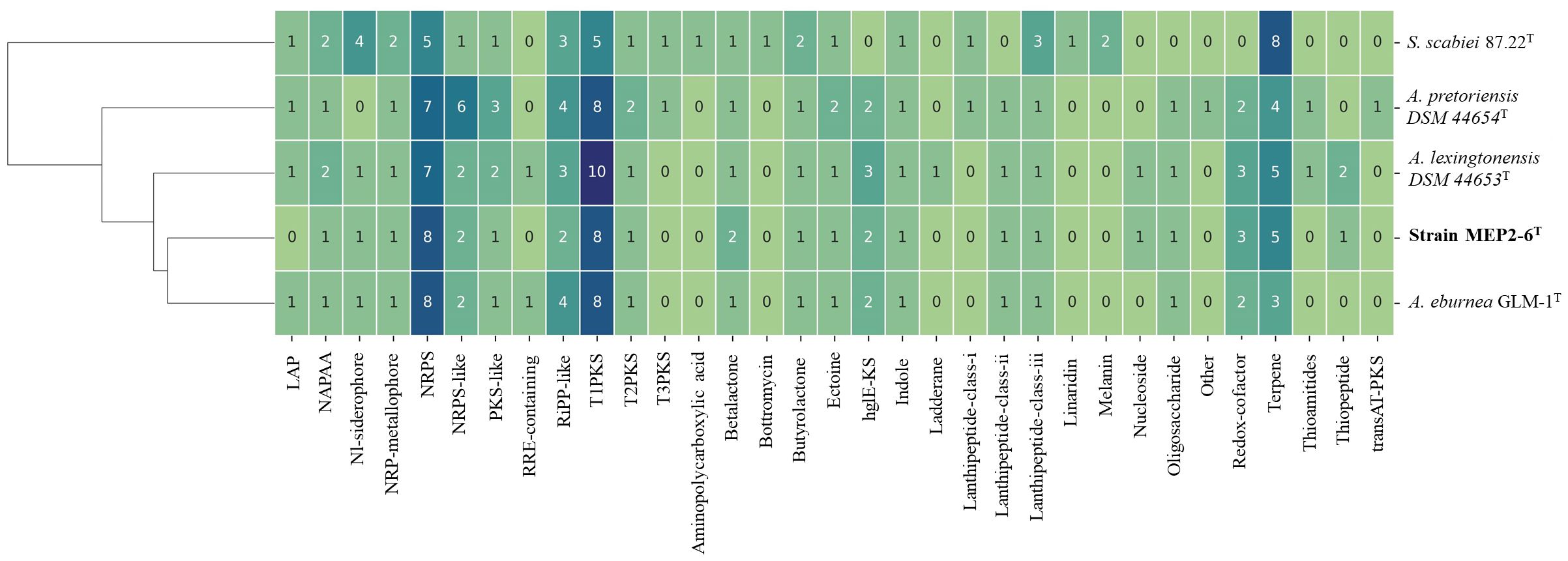
Figure 7 Distribution of BGCs across the genome of strain MEP2-6T and its closest relatives. The hierarchical heatmap depicts the number of genes assigned to the individual smBGCs. Rows were clustered using Euclidean distances. LAP, linear azol(in)e-containing peptides; NAPAA, non-alpha poly-amino acids such as ϵ-poly-lysine; Nl-siderophore, NRPS-independent, IucA/IucC-like siderophores; NRPS, non-ribosomal peptide synthetase; NRPS-like, NRPS-like fragment; PKS-like, other types of polyketide synthase; T1PKS, type I polyketide synthase; T2PKS, type II polyketide synthase; T3PKS, type III polyketide synthase; hglE-KS, heterocyst glycolipid synthase-like PKS; Other, cluster containing a secondary metabolite-related protein that does not fit into any other category.
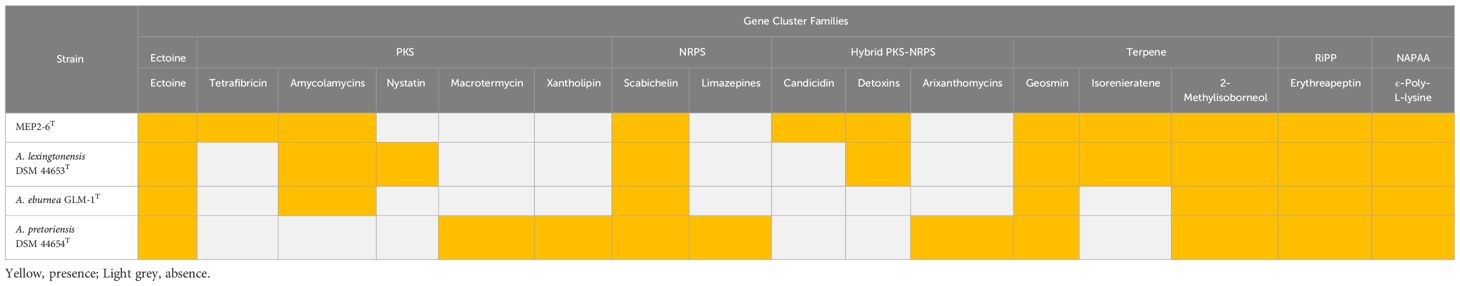
Table 7 Predicted compounds encoded by biosynthetic gene cluster families (GCFs) identified in the genomes of strain MEP2-6T and its closest Amycolatopsis species with ≥50% homology with known functions.
Pathogenicity on plant
A tomato seedling test was conducted to confirm the pathogenicity of strain MEP2-6T, and the findings are shown in Supplementary Figure 6. Tomato seeds cultured with strain MEP2-6T germinated as expected, whereas those cultured with S. scabiei WSLK1-9 did not germinate. Moreover, the potato tuber slice test revealed that strain MEP2-6T could not necrotize tissues on potato tuber slices compared to the control S. scabiei WSLK1-9. Thus, it can be concluded that strain MEP2-6T is non-phytopathogenic and unrelated to the cause of potato scab disease.
Endophytic colonization of Solanum tuberosum L. cv. Spunta by strain MEP2-6T
As strain MEP2-6T encompassed genes related to niche colonization in its genome, we examined where it enters the endosphere of S. tuberosum L. cv. Spunta to gain insights into the endophytic biology of the genus Amycolatopsis. Consequently, we inoculated mycelia with spore masses of Amycolatopsis strain MEP2-6T into the wounding stem node site of 14-day-old S. tuberosum L. cv. Spunta cultures (Figures 8A, B). At 5 days post-inoculation, the potato culture grew regularly (Figure 8C), and strain MEP2-6T attached to the stem node in dense white mycelia (Figure 8D) without damage. Colonized S. tuberosum L. cv. Spunta stems were then excised using a free-hand-cut technique with a fine razor blade for sectioning and visualization with high-resolution bright-field microscopy. The findings showed that strain MEP2-6T colonized not only the stem surface but also the internal stem tissue and, notably, the intracellular space (Figures 8E, F). Incredibly, no plant cellular membrane separates Amycolatopsis from its intracellular space.
Figure 8 Endophytic colonization of Solanum tuberosum L. cv. Spunta by the Amycolatopsis strain MEP2-6T. The potato culture before inoculation of strain MEP2-6T (A), hyphae with spore masses of strain MEP2-6T inoculated into the stem node wounding site (B), the normal potato culture after inoculation of strain MEP2-6T for five days (C), attachment of white mycelia of strain MEP2-6T on the stem node (D), sections of S. tuberosum cv. Spunta stem epidermal cells invaded by Amycolatopsis MEP2-6T (E, F). Scale bar 20 μm. The boxed part of the image is shown as a magnification on the right side of (E). Red arrows indicate white mycelia in (B, D) and indicate vegetative cells in (E, F).
Production of extracellular carbohydrate-degrading enzymes
The genomes of strain MEP2-6T and its closest type strains exhibit many genes encoding enzymes potentially involved in carbohydrate-degrading enzymes, particularly cellulose-binding related genes. Genes encoding endoglucanases and chitinases were observed in strain MEP2-6T and its closest relatives, whereas cbhA and bglB-bglX encoded exoglucanases and β-D-glucosidases, respectively, were uniquely detected in strain MEP2-6T. These results clearly indicate that all strains could hydrolyze cellulose. Nevertheless, the symbiotic actinobacterium Frankia sp. has reduced a set of carbohydrate-degrading enzyme genes in its genome, especially pectinase (Pujic et al., 2012). Strain MEP2-6T and its closest neighbors were examined for extracellular production of endoglucanases, pectinases, and chitinases. All strains produced endoglucanases by observing the clear transparent zone on carboxymethyl cellulose (CMC) agar (Figure 9) but did not produce pectinase and chitinase (Supplementary Table 3).
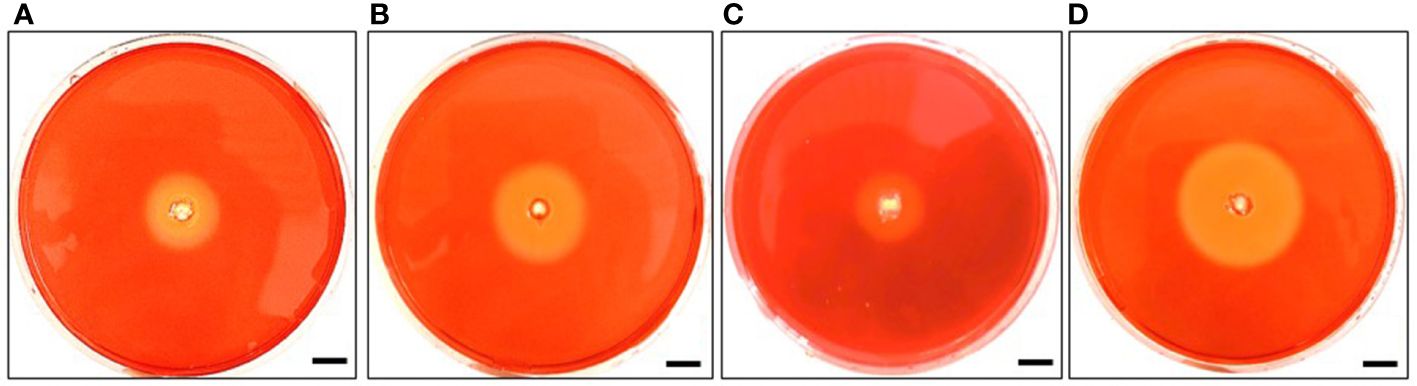
Figure 9 Diameter of the clear transparent zone illustrating endoglucanase activity of strain MEP2-6T and its closest type strains. Strain MEP2-6T (A), A. pretoriensis JCM 12673T (B), A. lexingtonensis JCM 12672T (C), and A. eburnea TBRC 9315T (D). Scale bar 10 mm.
Discussion
Recently, the integration of polyphasic taxonomy and genome sequence-based taxonomy has provided precision, reliability, and reproducibility for bacterial classification (Nouioui et al., 2018). In the present study, we unambiguously identified a novel endophytic actinomycete species, strain MEP2-6T, isolated from scab lesions on potato tubers. Sequence analysis of the 16S rRNA gene revealed that strain MEP2-6T belonged to the family Pseudonocardiaceae, order Pseudonocardiales, class Actinomycetia, and phylum Actinomycetota. Results from chemotaxonomic characteristics, including a type IV cell wall, a type A whole-cell sugar pattern (Lechevalier et al., 1971), and a type PII phospholipid type II (Lechevalier et al., 1977), also indicated that the strain was a member of the genus Amycolatopsis. Based on a combination of 16S rRNA gene phylogenetic and core phylogenomic analyses (Figures 1, 2), strain MEP2-6T distinctly shared the closest relationship with A. lexingtonensis NRRL B-24131T (=JCM 12672T =DSM 44653T), A. pretoriensis DSM 44654T (=JCM 12673T), and A. eburnea GLM-1T (=TBRC 9315T). ANIb, ANIm, and dDDH values were used to confirm the novelty of strain MEP2-6T. The values of the two types of ANI and dDDH between the strain and its closest strains (Table 4) were significantly below the cut-off values recommended for species delineation: <95% for ANIb, <96% for ANIm (Richter and Rosselló-Móra, 2009), and <70% for dDDH (Wayne et al., 1987; Goris et al., 2007). Thus, it should be noted that strain MEP2-6T represents a new species within the genus Amycolatopsis, for which the name Amycolatopsis solani sp. nov. was proposed.
In this study, no significant correlation was observed between genome size and environment. The genome sizes of plant-derived Amycolatopsis strain MEP2-6T and its closest Amycolatopsis species were very similar (10.2 Mb ± 0.5 Mb). This finding is in contrast to that of Kitasatospora sp. SUK42, which occupies the stems of Antidesma neurocarpum Miq, adapted to an endophytic lifestyle via genome reduction (Zin et al., 2021). On one hand, the genomic reduction did not occur in all endophytic actinobacterial genera. Micromonospora and Streptomyces appeared to have developed so as to adapt to multiple ecological niches, which could be altered to larger genomes to shelter different lifestyles (Trujillo et al., 2014; Quach et al., 2022; Zhou et al., 2023).
Identifying orthologous clusters is crucial for comparative genomic studies because it allows comparison of evolutionary relationships between genes across different species (Sun et al., 2023). This study illustrates the novel strain MEP2-6T and its closest Amycolatopsis species, which shared 6,336 orthologous clusters associated with biological, molecular, and cellular functions (Figure 5). Although most of the functions relied on orthologous clusters that were similar among the strains, the function of the mycothiol biosynthesis process was unique in strain MEP2-6T. It effectively comprises all the essential genes in the biosynthetic pathway of mycothiol (Figure 6). Living within plants of microbial endophytes often induces stress-responsive genes to generate reactive oxygen species (ROS) scavengers (Bosamic et al., 2020). Mycothiol plays a vital role in the detoxification of alkylating agents, reactive oxygen and nitrogen species, and antibiotics and also acts as a thiol buffer, which is crucial for maintaining a highly reducing environment within the cell (Newton et al., 2008). Consequently, strain MEP2-6T was able to live inside the plant tissues.
Living as a plant endophyte, bacteria must have a genetic system to utilize plant-synthesized carbohydrates as a nutritional source (Fabryová et al., 2018; Jiménez-Gómez et al., 2019). Our study revealed that the plant-associated actinobacteria strain MEP2-6T was rich in genes related to encoding plant-synthesized polysaccharide interconversion enzymes: galA, xynD, cel74a, cbhA, and bglB-bglX, which are depleted in A. eburnea GLM-1T residing in arbuscular mycorrhizal fungi, A. lexingtonensis DSM 44653T, and A. pretoriensis DSM 44654T, occupying the equine placenta. The xynD gene encodes arabinoxylan arabinofuranohydrolase (EC 3.2.1.55), which plays a crucial role in the conversion of arabinoxylan to L-arabinose (Lee et al., 2001). Altogether, cel74a plays a vital role in encoding xyloglucan-specific exo-β-1,4-glucanase (EC 3.2.1.155), which is responsible for converting polymeric xyloglucan to heptasaccharides (Wong et al., 2010). The genome of strain MEP2-6T uniquely contained the cbhA and bglB-bglX genes encoding exoglucanases and β-D-glucosidases, respectively. These genes are typically found in endophytic actinomycetes such as Fodinicola acacia, Frankia sp. (Phạm et al., 2020), and Streptomyces sp. (El-Gendy et al., 2022). Moreover, the gene encoding endoglucanase (EC 3.2.1.4), which is responsible for randomly cleaving the cellulose polymer into more petite sugar and oligomeric polysaccharides (Rahman et al., 2018), was found to be higher in copies of the strain MEP2-6T than in the closest non-plant-associated Amycolatopsis species. These findings are in accordance with those of previous studies, which indicated that Micromonospora lupini Lupac 08, M. noduli GUI 43, and M. saelicesensis Lupac 09 isolated from the root nodules of Leguminosae plants as endophytic actinobacterial models comprised a significant number of functional genes related to plant polysaccharide-degrading enzymes (Trujillo et al., 2014; Trujillo et al., 2007; Riesco et al., 2022). Similar reports have revealed that although plant-derived and non-plant-derived bacterial genomes differ in the presence and absence of functional genes associated with carbohydrate degradation, they are phylogenetically related (Levy et al., 2018; Riesco et al., 2022).
Membrane transport systems are remarkably related to behavior and are intrinsic for a microbe to survive in a given environment (Ren and Paulsen, 2007). Numerous oligo- and monosaccharide transporter systems, including chitobiose, raffinose/stachyose/melibiose, fructose, trehalose/maltose, and multiple sugar transporters, were found to be overexpressed in plant-derived Amycolatopsis MEP2-6T. These results correlate well with the carbohydrate metabolism of several endophytic bacteria that requires the introduction of sugars released by plants in the form of root exudates into cells to serve as carbon sources (Badri and Vivanco, 2009). Moreover, a recent study showed that several endophytic and rhizosphere species of the genus Pseudomonas responded to root exudates by inducing several transport systems that encode a Major Facilitator Superfamily (MFS) transporter (Mavrodi et al., 2021). In the present study, the cellobiose transporter was individually expressed in the plant-derived Amycolatopsis strain MEP2-6T. This transporter is typically found in Sinorhizobium and Rhizobium, root nodule endosymbionts (Iyer et al., 2016). Strain MEP2-6T actively ingested fructose into cells via the fructose ABC transporter, while the three closest species used the phosphotransferase system (PTS). This finding is in good agreement with those of previous studies conducted by Lambert et al. (2001) and Pinedo and Gage (2009), who reported that the root nodule, an endosymbiotic Sinorhizobium meliloti, lacked transport-related PTS proteins necessary for sugar transport and fructose uptake by the fructose ABC transporter.
Plant-associated and rhizosphere bacteria typically use amino acids released by plants as carbon and nitrogen sources (Riesco et al., 2022). In our study, liv genes, encoded as transporters for branched-chain amino acids were overexpressed in the potato tuber-associated strain MEP2-6T. Branched-chain amino acids were identified as key factors in the relationship between bacteria and Leguminosae plants by serving as nitrogen sources for the bacteria. Their transporters are necessary to facilitate their movement across the symbiosomal membrane to make nitrogen available to the bacteria (Prell et al., 2009). The transporters of branched-chain amino acids were also abundant in the bacterial community of root colonizers in maize and sugarcane, indicating that the metabolism and transport of amino acids play a critical role in plant–microbe interactions and are not limited to the symbiosis between Rhizobium and legumes (de Souza et al., 2019).
As indicated above, in addition to mycothiol, the plant-associated Amycolatopsis strain MEP2-6T possesses two major genes, sod1 and sod2, encoded the superoxide dismutase Cu-Zn family and superoxide dismutase Fe-Mn family, respectively, to protect itself from reactive oxygen species produced by the plant’s immune system. This result is in line with that of a previous study, which revealed that the mutualist endophyte Paraburkholderia phytofirmans PsJN triggered a weak and temporal defense reaction with an oxidative eruption, and the bacterium protected itself by producing superoxide dismutase (Brader et al., 2017). Our research also revealed that genes encoding rubredoxin were found only in strain MEP2-6T. This compound is a non-heme iron protein found in some actinobacterial species of the genera Mycobacterium, Dietzia, and Saccharomonospora, and it plays a critical role in the reduction of superoxide and in the adaptation of plants to changing environmental conditions (Nie et al., 2014; Sushko et al., 2021).
In Gram-positive bacteria, the cell wall peptidoglycan acts as a surface structure for transporting and assembling secretory proteins that interact with the environment, especially the infected host tissues (Schneewind and Missiakas, 2012). Secretory proteins associated with host colonization are exported from the cytoplasmic membrane and interact with the host at the cell wall via the Sec-dependent pathway, Sec-independent twin-arginine translocation (TAT) system, or type VII secretion system (Sutcliffe, 2011). To the best of our knowledge, this is the first analysis of genes related to host plant colonization of the genus Amycolatopsis. The Amycolatopsis strain MEP2-6T contains four pathways to colonize plant tissues: Sec-dependent, Sec-independent (TAT), T2SS, and T7SS. All genes responsible for encoding membrane protein channel (secY, secE, and secG), ancillary proteins (secD, yidC, and yajC), and the ATPase (secA) in the Sec-dependent pathway were found in the strain MEP2-6T genome but lacks secB gene for coding the chaperone that targets proteins to the Sec translocon for passage via the plasma membrane (Scott and Barnett, 2006). Although the secB gene was absent in the genome of the strain, the fused signal recognition particle receptor encoded by the ftsY gene can guide proteins to the translocon for passage through the cytoplasmic membrane (Crowther et al., 2012). Significant genes related to the Sec-independent TAT pathway, including tatA, tatB, and tatC, were also located in the genome of Amycolatopsis strain MEP2-6T. Like other actinobacteria (i.e., Frankia sp. strain CcI3) and other bacteria (e.g., Vibrio fischeri), the tatABC operon encodes translocase proteins, which play an essential role in the excretion of fully folded proteins across the cytoplasmic membrane via the transmembrane proton gradient as the main driving force for translocation as well as function for host symbiotic colonization (Normand et al., 2007; Dunn and Stabb, 2008). Four genes, eccB, eccC, eccD, and mycP, defined as part of the T7SS, were located in strain MEP2-6T genome (Table 5). These are organized in the same cluster and involve the crucial proteins for secreting conserved membrane component proteins EccB, EccC, EccD, and S8 family serine peptidase (MycP), which were similar to the T7SS gene cluster of other Gram-positive bacteria: Mycobacterium, Streptomyces, Micromonospora, Bifidobacterium, Bacillus, and Streptococcus (Fyans et al., 2013; Houben et al., 2014; Trujillo et al., 2014; Rivera-Calzada et al., 2021). This secretion system plays an essential role in promoting the colonization of niches and host–microbe interactions between members in Actinobacteria and Firmicutes (Abdallah et al., 2007; Liu et al., 2023). Two genes, tadA and tadB, in Amycolatopsis strain MEP2-6T were identified as components of the Type II secretion system (T2SS), which encodes the TadA family conjugal transfer-associated ATPase and tight adherence protein B, respectively. Similar to other members of the phylum Actinobacteria, Mycobacterium smegmatis, Streptomyces coelicolor, Thermobifida fusca, and Bifidobacterium breve, these genes are essential for successful colonization of various environmental niches (Kachlany et al., 2001; Tomich et al., 2007; O’Connell Motherway et al., 2011).
The genome sequences of actinomycetes have a much higher potential for the production of secondary metabolites (Bentley et al., 2002). Based on our insight into the genome of strain MEP2-6T, we found fascinating niches of secondary metabolite BGCs, which had the potential to encode metabolites with five major chemical classes: PKS, NRPS, hybrid PKS-NRPS, terpene, and saccharide (Table 6). Moreover, the diversity of compounds encoded by BGCs in each strain was different (Table 7). Ectoine and scabichelin were ubiquitously detected in all Amycolatopsis strains in this study. They play crucial roles in stress protection and iron acquisition (Jones et al., 2019; Richter et al., 2019). The BGC encoding ϵ-poly-L-lysine (ϵ-PL) was also present in the genomes of all the strains. This compound, a homopoly(amino acid) comprised of 25–35 L-lysine residues with amide linkages formed between the ϵ-amino and α-carboxy groups, is edible, bacteriostatic and non-toxic to humans and the environment. Consequently, they have been extensively used in the food, feed, and pharmaceutical industries as both food and feed preservatives, dietary agents, and gene/drug/vaccine carriers (Wang et al., 2021). BGCs coding for tetrafibricin, a fibrinogen receptor antagonist (Kamiyama et al., 1993), and candicidin, a compound that has the ability to control cucumber Rhizoctonia rot (Yao et al., 2021) and inhibit some species of Rhizopus, Mucor, Pythium, Phytophthora, Penicillium, and Candida (Muller, 1958; Jørgensen et al., 2009), were only found in strain MEP2-6T. Notably, strain MEP2-6T is a promising biocontrol agent and candidate with strong potential as a novel antibiotic producer. Further studies on this strain are recommended for high-value drug discovery and development.
Secondary metabolite biosynthesis pathways and their associated gene clusters have been determined based on predictions drawn from bioinformatic algorithms and can thereby guide the discovery of interesting compounds (Medema and Fischbach, 2015). However, little is known about the evolution of BGCs, as they are correlated with a species source or phylogeny (Jensen, 2016). According to our work (Figure 6), it can be evidently observed that the Amycolatopsis strain MEP2-6T and its closest species exhibited a high similarity in their BGC patterns in the hierarchical cluster. The distribution patterns of BGCs were evolutionarily correlated with the species phylogeny. This result is in line with those of studies conducted by Adamek et al. (2018) and Chase et al. (2023), who reported that the BGCs distribution patterns of bacteria were mainly driven by species phylogeny.
Amycolatopsis strain MEP2-6T was isolated from the scab tissues on the surface of potato tubers, potato tuber slices, and tomato seedling tests were used to verify its pathogenicity. The strain did not necrotize potato tissue or inhibit tomato seed germination (Supplementary Figure 6). This finding is in accordance with the study conducted by Croce et al. (2021), who reported that non-pathogenic actinomycetes had no ability to induce stunting of plant seedlings. At present, evidence suggests that thaxtomins and other secreted phytotoxins, such as bottromycin and concanamycin A, play an important role in the development or severity of potato scab disease rather than other mechanisms (Li et al., 2019a). The genomes of strains MEP2-6T and S. scabiei 87.22T were annotated with the antiSMASH 7.0. Protein-coding sequences of strain MEP2-6T were subjected to a BLASTP search against the protein sequences of thaxtomin, bottromycin, and concanamycin A BGCs of S. scabiei 87.22T. Strain MEP2-6T comprised several protein-coding genes associated with the BGCs of these compounds, yet they are not the core biosynthetic genes; therefore, it was unable to produce three significant phytotoxins: thaxtomin, bottromycin, and concanamycin A (Covington et al., 2021). Based on a combination of the pathogenicity test on plants and the analysis of phytotoxin BGCs, strain MEP2-6T can be regarded as a non-phytopathogenic actinomycete.
Previous studies have revealed colonization of the root surfaces of Arabidopsis by actinobacteria (Bulgarelli et al., 2012) and chickpea and sorghum by Amycolatopsis strain BCA-696 (Alekhy and Gopalakrishnan, 2016). Moreover, it has been reported that Streptomyces strains LUP30 and LUP47B, isolated from lucerne plants, can colonize germinating seeds of wheat (Franco et al., 2017). To the best of our knowledge, the present study is the first to show the presence of a non-streptomycete, Amycolatopsis, in S. tuberosum L. cv. Spunta stem epidermis cell. Mycelia appeared denser on the plant surface than in the endosphere, which may reflect different physiological characteristics between life outside and inside the plant. Amycolatopsis strain MEP2-6T invaded potato stem epidermis cells through minor wounds and lived in vegetative mycelial forms without spore formation. Additionally, potato culture remains asymptomatic. This event agrees with Hallmann et al. (1997) and Rosenblueth and Martínez-Romero (2006), who reported that endophytic bacteria can occupy the plant endosphere during all or part of their life cycle and do not harm the host plant. Endophytic bacteria can be classified as obligate or facultative based on their lifestyle. Obligate endophytes have a complete life cycle in the host plant, and transmission to other plants occurs either vertically or by vectors. In contrast, facultative endophytes have a biphasic life cycle that alternates between plants and soils (Hardoim et al., 2008). Therefore, based on life strategies, Amycolatopsis strain MEP2-6T can be assumed to be a facultative endophyte.
Most bacterial endophytes can produce and secrete carbohydrate-degrading enzymes, especially those that are active against cellulose and pectin, to locally disrupt the plant cell wall, facilitate colonization, and spread to other plant parts (Pinski et al., 2019). Based on these results, the strain MEP2-6T and its closest relatives can produce endoglucanase. This enzyme randomly cleaves cellulose polymer into more petite sugars and oligomeric polysaccharides (Menéndez et al., 2015; Rahman et al., 2018). However, no pectinolytic activity was observed. Similar to the findings of the genome analysis, the genes encoding endoglucanase enzymes were present in all strains, while genes related to pectinase production were absent. Although endophytic bacteria colonize host plants via wounds and natural openings such as the stomata and lenticels, endoglucanase activity helps them to colonize successfully; for example, the endophytic Azoarcus sp. The BH72 mutant, lacking endoglucanase activity, had a decreased capability to colonize rice roots and could not spread to the plant’s aboveground compartments (Reinhold-Hurek et al., 2006). Endoglucanase activity was also found in plant-symbiotic actinomycetes, Frankia AcN14a, Frankia Ar112.2 (Igual et al., 2001), and other facultative endophytic actinomycetes, Micromonospora lupini Lupac 08 (Trujillo et al., 2014), and Streptomyces endus OsiSh-2 (Xu et al., 2017). Strain MEP2-6T and its closest neighbors had no chitinase activity, even though their genomes included the chitinase gene. This phenotype was similar to that of Bacillus licheniformis N1, DSM13, and ATCC 14580, in which the chitinase gene in their genomes was silent. This event may be caused by an inactive promoter of the chitinase gene in organisms (Lee et al., 2009).
Here, we report that the potato tuber-derived actinomycete strain MEP2-6T is a novel species of the genus Amycolatopsis, whose name was proposed to be Amycolatopsis solani sp. nov., and the type of strain is MEP2-6T. Comparative genomics reliably provides a better understanding of the underlying genetic mechanisms of the adaptation of Amycolatopsis solani MEP2-6T to endophytic lifestyles. Comparative smBGCs exhibited a fascinating genetic potential to synthesize different chemical classes of bioactive secondary metabolites and undoubtedly indicated that the distribution patterns of smBGCs are mainly related to species phylogeny. Moreover, strain MEP2-6T can produce endoglucanase, which is an important enzyme in plant-based biofuels and food-feed industries (Behera et al., 2017), and is also a critical enzyme that inhibits Phytophthora infestans, a causative agent of potato late blight disease (Zhang et al., 2023). Thus, we suggest that A. solani MEP2-6T could be further investigated as a promising candidate for the discovery of novel bioactive compounds, biotechnological applications, and potato probiotics.
Description of Amycolatopsis solani sp. nov.
Amycolatopsis solani (so.la′ni. L. gen. n. solani of Solanum, the generic name of potato).
A gram-positive, aerobic, mesophilic endophytic actinomycete produced septal substrate and aerial mycelia that fragmented into rod-like elements (0.4–0.5 µm × 1.1–1.4 µm in size). Moderate orange-colored substrate mycelia and pale orange yellow aerial mycelia were well-developed on ISP 2 agar medium. Diffusible pigments were not produced in any of the agar media. Good growth on ISP 2, ISP 3, ISP 4, and ISP 7; moderate growth on ISP 5 and ISP 6; poor growth on nutrient agar medium. Growth occurred at 15°C–37°C (optimal at 30° C), pH 5–9 (optimal at 7) and was tolerated up to 4% (w/v) NaCl. Amygdalin, L-arabinose, D-fructose, D-galactose, D-glucose, D-melezitose, myo-inositol, L-rhamnose, D-sucrose, and D-xylose were used as sole carbon sources. The acids were produced only from D-sucrose. Coagulation and peptonization of milk, nitrate reduction, and gelatin liquefaction were all positive. Starch hydrolysis and H2S production tests were negative. Importantly, α-chymotrypsin, cystine arylamidase, leucine arylamidase, naphthol-AS-BI-phosphohydrolase, and valine arylamidase were positive, whereas acid phosphatase, α-fucosidase, and N-acetyl-β-glucosaminidase were weakly positive. Tests for alkaline phosphatase, esterase (C4), esterase lipase (C8), α-glucosidase, α-mannosidase, trypsin, α-galactosidase, β-galactosidase, β-glucosidase, β-glucuronidase, and lipase (C14) were negative. Cell wall peptidoglycan is composed of meso-diaminopimelic. Whole-cell sugars include arabinose, galactose, glucose, and ribose. The N-acyl group of muramic acid in peptidoglycan is an acetyl group. No mycolic acid was detected. The polar lipid profile consists of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, hydroxyphosphatidylethanolamine, an unidentified aminophospholipid, six unidentified phospholipids, an unidentified glycolipid, and five unidentified lipids. MK-9(H6) and MK-9(H4) are the major and minor menaquinones, respectively. The predominant fatty acids are iso-C16:0 and iso-C15:0.
The type of strain, MEP2-6T (=JCM 36309T =TBRC 17632T =NBRC 116395T), was isolated from single lesions at the borders between healthy and scab tissues of surface-sterilized potato tubers collected from Chiang Mai Province, Thailand. The DNA G + C content of the type strain calculated from the genome sequence was 71.7 mol%.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
TW: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review & editing. WM: Investigation, Methodology, Software, Validation, Writing – review & editing. PM: Data curation, Formal analysis, Investigation, Writing – review & editing. NN: Formal analysis, Writing – review & editing. JS: Formal analysis, Writing – review & editing. NC: Formal analysis, Writing – review & editing. SU: Formal analysis, Writing – review & editing. ST: Resources, Software, Validation, Writing – review & editing. NS: Conceptualization, Resources, Supervision, Writing – review & editing. YA: Formal analysis, Writing – review & editing. NK: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Software, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by Kasetsart University through the Graduate School Fellowship Program and National Research Council of Thailand (NRCT), Grant number: N42A650203 and supported by Chiang Mai University.
Acknowledgments
We sincerely thank Professor Aharon Oren for his advice regarding epithet derivation. We also gratefully acknowledge the Department of Plant Pathology, Faculty of Agriculture, Kasetsart University, and the Center of Excellence in Microbial Diversity and Sustainable Utilization, Chiang Mai University, for providing the research facilities. We also express our gratitude to Noppadol Komolmis for his kind advice during sample collection. The LC-MS study was partially supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS), Japan Agency for Medical Research and Development (AMED) under Grant Number JP22ama121035 (Phase II).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1346574/full#supplementary-material
References
Abdallah, A. M., Gey van Pittius, N. C., DiGiuseppe Champion, P. A., Cox, J., Luirink, J., Vandenbroucke-Graul, C. M. J. E., et al. (2007). Type VII secretion — mycobacteria show the way. Nat. Rev. Microbiol. 5, 883–889. doi: 10.1038/nrmicro1773
Adamek, M., Alanjary, M., Sales-Ortells, H., Goodfellow, M., Bull, A. T., Winkler, A., et al. (2018). Comparative genomics reveals phylogenetic distribution patterns of secondary metabolites in Amycolatopsis species. BMC Genom. 19, 426. doi: 10.1186/s12864-018-4809-4
Alam, K., Mazumder, A., Sikdar, S., Zhao, Y. M., Hao, J., Song, C., et al. (2022). Streptomyces: The biofactory of secondary metabolites. Front. Microbiol. 13, e968053. doi: 10.3389/fmicb.2022.968053
Alekhy, G., Gopalakrishnan, S. (2016). Exploiting plant growth-promoting Amycolatopsis sp. in chickpea and sorghum for improving growth and yield. J. Food Legumes. 29, 225–231.
Antipov, D., Korobeynikov, A., McLean, J. S., Pevzner, P. A. (2016). hybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics 32, 1009–1015. doi: 10.1093/bioinformatics/btv688
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genom 9, 75. doi: 10.1186/1471-2164-9-75
Badri, D. V., Vivanco, J. M. (2009). Regulation and function of root exudates. Plant Cell Environ. 32, 666–681. doi: 10.1111/j.1365-3040.2009.01926.x
Barker, W. G. (1953). A method for the in vitro culturing of potato tubers. Science 118, 384–385. doi: 10.1126/science.118.3066.384
Beemelmanns, C., Ramadhar, T. R., Kim, K. H., Klassen, J. L., Cao, S., Wyche, T. P., et al. (2017). Macrotermycins A-D, glycosylated macrolactams from a termite-associated Amycolatopsis sp. M39. Org Lett., 19, 1000–1003. doi: 10.1021/acs.orglett.6b03831
Behera, B. C., Sethi, B. K., Mishra, R. R., Dutta, S. K., Thatoi, H. N. (2017). Microbial cellulases - diversity & biotechnology with reference to mangrove environment: A review. J. Genet. Eng. Biotechnol. 15, 197–210. doi: 10.1016/j.jgeb.2016.12.001
Bentley, S. D., Chater, K. F., Cerdeño-Tárraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147. doi: 10.1038/417141a
Bian, J., Li, Y., Wang, J., Song, F. H., Liu, M., Dai, H. Q., et al. (2009). Amycolatopsis marina sp. nov., an actinomycete isolated from an ocean sediment. Int. J. Syst. Evol. Microbiol. 59, 477–481. doi: 10.1099/ijs.0.000026-0
Blin, K., Shaw, S., Augustijn, H. E., Reitz, Z. L., Biermann, F., Alanjary, M., et al. (2023). antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 51, W46–W50. doi: 10.1093/nar/gkad344
Bosamic, C. T., Barbadikar, K. M., Modi, A. (2020). “Genomic insights of plant endophyte interaction: prospective and impact on plant fitness,” in Microbial Endophytes Functional Biology and Applications. Eds. Kumar, A., Radhakrishnan, E. K. (Cambridge: Woodhead Publishing), 227–249.
Brader, G., Compant, S., Vescio, K., Mitter, B., Trognitz, F., Ma, L. J., et al. (2017). Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Annu. Rev. Phytopathol. 55, 61–83. doi: 10.1146/annurev-phyto-080516-035641
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365. doi: 10.1038/srep08365
Bulgarelli, D., Rott, M., Schlaeppi, K., Ver Loren van Themaat, E., Ahmadinejad, N., Assenza, F., et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488, 91–95. doi: 10.1038/nature11336
Camas, M., Sahin, N., Sazak, A., Spröer, C., Klenk, H. P. (2013). Amycolatopsis magusensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 63, 1254–1260. doi: 10.1099/ijs.0.042770-0
Chaiya, L., Matsumoto, A., Wink, J., Inahashi, Y., Risdian, C., Pathom-Aree, W., et al. (2019). Amycolatopsis eburnea sp. nov., an actinomycete associated with arbuscular mycorrhizal fungal spores. Int. J. Syst. Evol. Microbiol. 69, 3603–3608. doi: 10.1099/ijsem.0.003669
Chase, A. B., Bogdanov, A., Demko, A. M., Jensen, P. R. (2023). Biogeographic patterns of biosynthetic potential and specialized metabolites in marine sediments. ISME J. 17, 976–983. doi: 10.1038/s41396-023-01410-3
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Collins, M. D., Pirouz, T., Goodfellow, M., Minnikin, D. E. (1977). Distribution of menaquinones in actinomycetes and corynebacteria. J. Gen. Microbiol. 100, 221–230. doi: 10.1099/00221287-100-2-221
Coudert, E., Gehant, S., de Castro, E., Pozzato, M., Baratin, D., Neto, T., et al. (2023). UniProt Consortium. annotation of biologically relevant ligands in UniProtKB using ChEBI. Bioinformatics 39, btac793. doi: 10.1093/bioinformatics/btac793
Covington, B. C., Xu, F., Seyedsayamdost, M. R. (2021). A natural product chemist’s guide to Unlocking silent biosynthetic gene clusters. Annu. Rev. Biochem. 90, 763–788. doi: 10.1146/annurev-biochem-081420-102432
Croce, V., López-Radcenco, A., Lapaz, M. I., Pianzzola, M. J., Moyna, G., Siri, M. I. (2021). An integrative approach for the characterization of plant-pathogenic Streptomyces spp. Strains Based on metabolomic, bioactivity, and phylogenetic analysis. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.643792
Crowther, G. J., Quadri, S. A., Shannon-Alferes, B. J., Van Voorhis, W. C., Rosen, H. (2012). A mechanism-based whole-cell screening assay to identify inhibitors of protein export in Escherichia coli by the Sec pathway. J. Biomol Screen. 17, 535–541. doi: 10.1177/1087057111431606
De Coster, W., D’Hert, S., Schultz, D. T., Cruts, M., Van Broeckhoven, C. (2018). NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669. doi: 10.1093/bioinformatics/bty149
Dees, M. W., Sletten, A., Hermansen, A. (2013). Isolation and characterization of Streptomyces species from potato common scab lesions in Norway. Plant Pathol. 62, 217–225. doi: 10.1111/j.1365-3059.2012.02619.x
de Souza, R. S. C., Armanhi, J. S. L., Damasceno, N., de, B., Imperial, J., Arruda, P. (2019). Genome sequences of a plant beneficial synthetic bacterial community reveal genetic features for successful plant colonization. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01779
Duangmal, K., Mingma, R., Pathom-Aree, W., Thamchaipenet, A., Inahashi, Y., Matsumoto, A., et al. (2011). Amycolatopsis samaneae sp. nov., isolated from roots of Samanea saman (Jacq.) Merr. Int. J. Syst. Evol. Microbiol. 61, 951–955. doi: 10.1099/ijs.0.022699-0
Dunn, A. K., Stabb, E. V. (2008). The twin arginine translocation system contributes to symbiotic colonization of Euprymna scolopes by Vibrio fischeri. FEMS Microbiol. Lett. 279, 251–258. doi: 10.1111/fml.2008.279.issue-2
El-Gendy, M. M. A. A., Yahya, S. M. M., Hamed, A. R., Mohamed, A., El-Bondkly, A. (2022). Assessment of the phylogenetic analysis and antimicrobial, antiviral, and anticancer activities of marine endophytic Streptomyces species of the soft coral Sarcophyton convolutum. Int. Microbiol. 25, 133–152. doi: 10.1007/s10123-021-00204-x
Embley, M. T., Smida, J., Stackebrandt, E. (1988). The phylogeny of mycolate-less wall chemotype IV actinomycetes and description of Pseudonocardiaceae fam. nov. Syst. Appl. Microbiol. 11, 44–52. doi: 10.1016/S0723-2020(88)80047-X
Emms, D. M., Kelly, S. (2017). STRIDE: species tree root inference from gene duplication events. Mol. Biol. Evol. 34, 3267–3278. doi: 10.1093/molbev/msx259
Emms, D. M., Kelly, S. (2018). STAG: species tree inference from all genes. bioRxiv, 267914. doi: 10.1101/267914
Emms, D. M., Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238. doi: 10.1186/s13059-019-1832-y
Ewels, P., Magnusson, M., Lundin, S., Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinform. 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Fabryová, A., Kostovčík, M., Díez-Méndez, A., Jiménez-Gómez, A., Celador-Lera, L., Saati-Santamaría, Z., et al. (2018). On the bright side of a forest pest-the metabolic potential of bark beetles’ bacterial associates. Sci. Total Environ. 619–620, 9–17. doi: 10.1016/j.scitotenv.2017.11.074
Felsenstein, J. (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376. doi: 10.1007/BF01734359
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Feyissa, T., Welander, M., Negash, L. (2005). Micropropagation of Hagenia abyssinica: a multipurpose tree. Plant Cell Tiss Organ Cult. 80, 119–127. doi: 10.1007/s11240-004-9157-1
Fitch, W. M. (1972). Toward defining the course of evolution: minimum change for a species tree topology. Syst. Zool. 20, 406–416. doi: 10.1093/sysbio/20.4.406
Flores-Gonzalez, R., Velasco, I., Montes, F. (2008). Detection and characterization of Streptomyces causing potato common scab in Western Europe. Plant Pathol. 57, 162–169. doi: 10.1111/j.1365-3059.2007.01734.x
Franco, C. M. M., Adetutu, E. M., Le, H. X., Ballard, R. A., Araujo, R., Tobe, S. S., et al. (2017). Complete genome sequences of the endophytic Streptomyces sp. strains LUP30 and LUP47B, isolated from lucerne plants. Genome Announc. 5, e00556–17. doi: 10.1128/genomeA.00556-17
Franz, L., Kazmaier, U., Truman, A. W., Koehnke, J. (2021). Bottromycins - biosynthesis, synthesis and activity. Nat. Prod Rep. 38, 1659–1683. doi: 10.1039/D0NP00097C
Fyans, J. K., Bignell, D., Loria, R., Toth, I., Palmer, T. (2013). The ESX/type VII secretion system modulates development, but not virulence, of the plant pathogen Streptomyces scabies. Mol. Plant Pathol. 14, 119–130. doi: 10.1111/j.1364-3703.2012.00835.x
Fyans, J. K., Bown, L., Bignell, D. R. D. (2016). Isolation and characterization of plant-pathogenic Streptomyces species associated with common scab-infected potato tubers in Newfoundland. Phytopathol. 106, 123–131. doi: 10.1094/PHYTO-05-15-0125-R
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., Tiedje, J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Gordon, R. E., Barnett, D. A., Handerhan, J. E., Pang, C.-N. (1974). Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int. J. Syst. Bacteriol. 24, 54–63. doi: 10.1099/00207713-24-1-54
Gurevich, A., Saveliev, V., Vyahhi, N., Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hall, T. A. (1999). BioEdit: a user- friendly biological sequence alignment editor and analysis program for Windows 95/98 NT. Nucleic Acids Symp Ser. 41, 95–98.
Hallmann, J., Quadt-Hallmann, A., Mahaffee, W. F., Kloepper, J. W. (1997). Bacterial endophytes in agricultural crops. Can. J. Microbiol. 43, 895–914. doi: 10.1139/m97-131
Hammer, Ø., Harper, D. A. T., Ryan, P. D. (2001). PAST: paleontological statistics software package for education. Palaeontol Electron. 4, 9.
Hardoim, P. R., van Overbeek, L. S., Elsas, J. D. (2008). Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 16, 463–771. doi: 10.1016/j.tim.2008.07.008
Haydock, S. F., Appleyard, A. N., Mironenko, T., Lester, J., Scott, N., Leadlay, P. F. (2005). Organization of the biosynthetic gene cluster for the macrolide concanamycin A in Streptomyces neyagawaensis ATCC 27449. Microbiology 151, 3161–3169. doi: 10.1099/mic.0.28194-0
Henao, L., Guevara, M., Restrepo, S., Husserl, J. (2021). Genotypic and phenotypic characterization of Streptomyces species associated with potato crops in the central part of Colombia. Plant Pathol. 71, 750–761. doi: 10.1111/ppa.13485
Houben, E. N., Korotkov, K. V., Bitter, W. (2014). Take five - Type VII secretion systems of mycobacteria. Biochim. Biophys. Acta 1843, 1707–1716. doi: 10.1016/j.bbamcr.2013.11.003
Huang, Y., Paściak, M., Liu, Z., Xie, Q., Gamian, A. (2004). Amycolatopsis palatopharyngis sp. nov., a potentially pathogenic actinomycete isolated from a human clinical source. Int. J. Syst. Evol. Microbiol. 54, 359–363. doi: 10.1099/ijs.0.02685-0
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Igual, J. M., Velázquez, E., Mateos, P. F., Rodríguez-Barrueco, C., Cervantes, E., Martínez-Molina, E. (2001). Cellulase isoenzyme profiles in Frankia strains belonging to different cross-inoculation groups. Plant Soil 229, 35–39. doi: 10.1023/A:1004835313723
Iyer, B., Rajput, M. S., Jog, R., Joshi, E., Bharwad, K., Rajkumar, S. (2016). Organic acid mediated repression of sugar utilization in rhizobia. Microbiol. Res. 192, 211–220. doi: 10.1016/j.micres.2016.07.006
Jensen, P. R. (2016). Natural products and the gene cluster revolution. Trends Microbiol. 24, 968–977. doi: 10.1016/j.tim.2016.07.006
Jiang, G., Zhang, Y., Powell, M. M., Zhang, P., Zuo, R., Zhang, Y., et al. (2018). High-yield production of herbicidal thaxtomins and thaxtomin analogs in a nonpathogenic Streptomyces strain. Appl. Environ. Microbiol. 84, e00164–e00118. doi: 10.1128/AEM.00164-18
Jiménez-Gómez, A., Saati-Santamaría, Z., Igual, J. M., Rivas, R., Mateos, P. F., García-Fraile, P. (2019). Genome insights into the novel species Microvirga brassicacearum, a rapeseed endophyte with biotechnological potential. Microorganisms 7, 354. doi: 10.3390/microorganisms7090354
Jones, S. E., Pham, C. A., Zambri, M. P., McKillip, J., Carlson, E. E., Elliot, M. A. (2019). Streptomyces volatile compounds influence exploration and microbial community dynamics by altering iron availability. mBio 10, e00171–e00219. doi: 10.1128/mBio.00171-19
Jørgensen, H., Fjaervik, E., Hakvåg, S., Bruheim, P., Bredholt, H., Klinkenberg, G., et al. (2009). Candicidin biosynthesis gene cluster is widely distributed among Streptomyces spp. isolated from the sediments and the neuston layer of the Trondheim fjord, Norway. Appl. Environ. Microbiol. 75, 3296–3303. doi: 10.1128/AEM.02730-08
Joshi, T., Xu, D. (2007). Quantitative assessment of relationship between sequence similarity and function similarity. BMC Genom. 8, e222. doi: 10.1186/1471-2164-8-222
Kachlany, S. C., Planet, P. J., DeSalle, R., Fine, D. H., Figurski, D. H. (2001). Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 9, 429–437. doi: 10.1016/S0966-842X(01)02161-8
Kamiyama, T., Umino, T., Fujisaki, N., Fujimori, K., Satoh, T., Yamashita, Y., et al. (1993). Tetrafibricin, a novel fibrinogen receptor antagonist. I. Taxonomy, fermentation, isolation, characterization and biological activities. J. Antibiot. 46, 1039–1046. doi: 10.7164/antibiotics.46.1039
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., Tanabe, M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462. doi: 10.1093/nar/gkv1070
Kans, J. (2022). Entrez Direct: E-utilities on the Unix Command line (National Center for Biotechnology Information (US) Maryland: Bethesda.
Kelly, K. L. (1964). Inter-Society Color Council - National Bureau of Standards Color Name Charts Illustrated with Centroid Colors (Washington, DC: US Government Printing Office).
Kimura, M. A. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kisil, O. V., Efimenko, T. A., Efremenkova, O. V. (2021). Looking back to Amycolatopsis: history of the antibiotic discovery and future prospects. Antibiotics 10, 1254. doi: 10.3390/antibiotics10101254
Klykleung, N., Tanasupawat, S., Pittayakhajonwut, P., Ohkuma, M., Kudo, T. (2015). Amycolatopsis stemonae sp. nov., isolated from a Thai medicinal plant. Int. J. Syst. Evol. Microbiol. 65, 3894–3899. doi: 10.1099/ijsem.0.000509
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kumla, J., Nundaeng, S., Suwannarach, N., Lumyong, S. (2020). Evaluation of multifarious plant growth promoting trials of yeast isolated from the soil of Assam tea (Camellia sinensis var. assamica) plantations in northern Thailand. Microorganisms 8, e1168. doi: 10.3390/microorganisms8081168
Kuncharoen, N., Pittayakhajonwut, P., Tanasupawat, S. (2018). Micromonospora globbae sp. nov., an endophytic actinomycete isolated from roots of Globba winitii C. H. Wright. Int. J. Syst. Evol. Microbiol. 68, 1073–1077. doi: 10.1099/ijsem.0.002625
Labeda, D. P., Donahue, J. M., Williams, N. M., Sells, S. F., Henton, M. M. (2003). Amycolatopsis kentuckyensis sp. nov., Amycolatopsis lexingtonensis sp. nov. and Amycolatopsis pretoriensis sp. nov., isolated from equine placentas. Int. J. Syst. Evol. Microbiol., 53, 1601–1605. doi: 10.1099/ijs.0.02691-0
Lambert, A., Østerås, M., Mandon, K., Poggi, M. C., Le Rudulier, D. (2001). Fructose uptake in Sinorhizobium meliloti is mediated by a high-affinity ATP-binding cassette transport system. J. Bacteriol. 183, 4709–4717. doi: 10.1128/JB.183.16.4709-4717.2001
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in The Nucleic Acid Techniques in Bacterial Systematics. Eds. E. Stackebrandt, E., Goodfellow, M. (John Wiley & Sons, Chichester), 115–148.
Lechevalier, M. P., De Bievre, C., Lechevalier, H. A. (1977). Chemotaxonomy of aerobic actinomycetes: phospholipid composition. Biochem. Syst. Ecol. 5, 249–260. doi: 10.1016/0305-1978(77)90021-7
Lechevalier, M. P., Lechevalier, H. (1970). Chemical composition as a criterion in the classification of aerobic actinomycetes. Int. J. Syst. Bacteriol 20, 435–443. doi: 10.1099/00207713-20-4-435
Lechevalier, H. A., Lechevalier, M. P., Gerber, N. N. (1971). Chemical composition as a criterion in the classification of actinomycetes. Adv. Appl. Microbiol. 14, 47–72. doi: 10.1016/S0065-2164(08)70539-2
Lechevalier, M. P., Prauser, H., Labeda, D. P., Ruan, J.-S. (1986). Two new genera of nocardioform actinomycetes: Amycolata gen. nov. and Amycotlatopsis gen. nov. Int. J. Syst. Bacteriol. 36, 29–37. doi: 10.1099/00207713-36-1-29
Lee, S. D. (2009). Amycolatopsis ultiminotia sp. nov., isolated from rhizosphere soil, and emended description of the genus Amycolatopsis. Int. J. Syst. Evol. Microbiol. 59, 1401–1404. doi: 10.1099/ijs.0.006577-0
Lee, R. C., Burton, R. A., Hrmova, M., Fincher, G. B. (2001). Barley arabinoxylan arabinofuranohydrolases: purification, characterization and determination of primary structures from cDNA clones. Biochem. J. 356, 181–189. doi: 10.1042/bj3560181
Lee, K. Y., Heo, K. R., Choi, K. H., Kong, H. G., Nam, J. S., Yi, Y. B., et al. (2009). Characterization of a chitinase gene exhibiting antifungal activity from a biocontrol bacterium Bacillus licheniformis N1. Plant Pathol. J. 25, 344–351. doi: 10.5423/PPJ.2009.25.4.344
Letunic, I., Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Levy, A., Salas Gonzalez, I., Mittelviefhaus, M., Clingenpeel, S., Herrera Paredes, S., Miao, J., et al. (2018). Genomic features of bacterial adaptation to plants. Nat. Genet. 50, 138–150. doi: 10.1038/s41588-017-0012-9
Li, Y., Liu, J., Adekunle, D., Bown, L., Tahlan, K., Bignell, D. R. D. (2019b). TxtH is a key component of the thaxtomin biosynthetic machinery in the potato common scab pathogen Streptomyces scabies. Mol. Plant Pathol. 20, 1379–1393. doi: 10.1111/mpp.12843
Li, Y., Liu, J., Díaz-Cruz, G., Cheng, Z., Bignell, D. R. D. (2019a). Virulence mechanisms of plant-pathogenic Streptomyces species: an updated review. Microbiology 165, 1025–1040. doi: 10.1099/mic.0.000818
Li, R., Wang, M., Ren, Z., Ji, Y., Yin, M., Zhou, H., et al. (2021). Amycolatopsis aidingensis sp. nov., a halotolerant actinobacterium, produces new secondary metabolites. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.743116
Liu, Y., Shu, X., Chen, L., Zhang, H., Feng, H., Sun, X., et al. (2023). Plant commensal type VII secretion system causes iron leakage from roots to promote colonization. Nat. Microbiol. 8, 1434–1449. doi: 10.1038/s41564-023-01402-1
Loria, R., Bukhalid, R. A., Creath, R. A., Leiner, R. H., Olivier, M., Steffens, J. C. (1995). Differential production of thaxtomins by pathogenic Streptomyces species in vitro. Phytopathol. 85, 537–541. doi: 10.1094/Phyto-85-537
Lycklama, A., Nijeholt, J. A., Driessen, A. J. (2012). The bacterial Sec-translocase: structure and mechanism. Philos. Trans. R Soc. Lond B Biol. Sci. 367, 1016–1028. doi: 10.1098/rstb.2011.0201
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A., Zdobnov, E. M. (2021). BUSCO Update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38, 4647–4654. doi: 10.1093/molbev/msab199
Mavrodi, O. V., McWilliams, J. R., Peter, J. O., Berim, A., Hassan, K. A., Elbourne, L. D. H., et al. (2021). Root exudates alter the expression of diverse metabolic, transport, regulatory, and stress response genes in rhizosphere Pseudomonas. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.651282
McCoy, J. G., Levin, E. J., Zhou, M. (2015). Structural insight into the PTS sugar transporter EIIC. Biochim. Biophys. Acta 1850, 577–585. doi: 10.1016/j.bbagen.2014.03.013
Medema, M. H., Fischbach, M. A. (2015). Computational approaches to natural product discovery. Nat. Chem. Biol. 11, 639–648. doi: 10.1038/nchembio.1884
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 14, 60. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., Carbasse, J. S., Peinado-Olarte, R. L., Göker, M. (2022). TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 7, 50, D801–D807. doi: 10.1093/nar/gkab902
Menéndez, E., García-Fraile, P., Rivas, R. (2015). Biotechnological applications of bacterial cellulases. AIMS Bioeng. 2, 163–182. doi: 10.3934/bioeng.2015.3.163
Mingma, R., Inahashi, Y., Matsumoto, A., Takahashi, Y., Duangmal, K. (2020). Amycolatopsis pithecelloba sp. nov., a novel actinomycete isolated from roots of Pithecellobium dulce in Thailand. J. Antibiot. 73, 230–235. doi: 10.1038/s41429-019-0271-z
Minnikin, D. E., O’Donnell, A. G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., et al. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241. doi: 10.1016/0167-7012(84)90018-6
Muller, W. H. (1958). The influence of antibiotics on microorganisms causing fruit and vegetable rots. Am. J. Bot. 45, 183–190. doi: 10.2307/2446448
Murashige, T., Skoog, F. (1962). A Revised medium for rapid growth and bio assays with tobacco tissue cultures. Plant Physiol. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Newton, G. L., Buchmeier, N., Fahey, R. C. (2008). Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol. Mol. Biol. Rev. 72, 471–494. doi: 10.1128/MMBR.00008-08
Nie, Y., Chi, C. Q., Fang, H., Liang, J. L., Lu, S. L., Lai, G. L., et al. (2014). Diverse alkane hydroxylase genes in microorganisms and environments. Sci. Rep. 4, 4968. doi: 10.1038/srep04968
Normand, P., Lapierre, P., Tisa, L. S., Gogarten, J. P., Alloisio, N., Bagnarol, E., et al. (2007). Genome characteristics of facultatively symbiotic Frankia sp. strains reflect host range and host plant biogeography. Genome Res. 17, 7–15. doi: 10.1101/gr.5798407
Nouioui, I., Carro, L., Garcia-Lopez, M., Meier-Kolthoff, J. P., Woyke, T., Kyrpides, N. C., et al. (2018). Genome-based taxonomic classification of the phylum Actinobacteria. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02007
O’Connell Motherway, M., Zomer, A., Leahy, S. C., Reunanen, J., Bottacini, F., Claesson, M. J., et al. (2011). Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U.S.A. 108, 11217–112122. doi: 10.1073/pnas.1105380108
Oren, A., Garrity, G. M. (2021). Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 71, 5056. doi: 10.1099/ijsem.0.005056
Palmer, T., Berks, B. (2012). The twin-arginine translocation (Tat) protein export pathway. Nat. Rev. Microbiol. 10, 483–496. doi: 10.1038/nrmicro2814
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Parte, A. C., Sardà Carbasse, J., Meier-Kolthoff, J. P., Reimer, L. C., Göker, M. (2020). List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612. doi: 10.1099/ijsem.0.004332
Phạm, H. T. T., Suwannapan, W., Koomsiri, W., Inahashi, Y., Také, A., Matsumoto, A., et al. (2020). Fodinicola acaciae sp. nov., an endophytic actinomycete isolated from the roots of Acacia mangium Willd. and its genome analysis. Microorganisms 8, 467. doi: 10.3390/microorganisms8040467
Pinedo, C. A., Gage, D. J. (2009). HPrK regulates succinate-mediated catabolite repression in the gram-negative symbiont Sinorhizobium meliloti. J. Bacteriol. 191, 298–309. doi: 10.1128/JB.01115-08
Pinski, A., Betekhtin, A., Hupert-Kocurek, K., Mur, L. A. J., Hasterok, R. (2019). Defining the genetic basis of plant endophytic bacteria interactions. Int. J. Mol. Sci. 20, 1947. doi: 10.3390/ijms20081947
Prell, J., White, J. P., Bourdes, A., Bunnewell, S., Bongaerts, R. J., Poole, P. S. (2009). Legumes regulate Rhizobium bacteroid development and persistence by the supply of branched-chain amino acids. Proc. Natl. Acad. Sci. U.S.A. 106, 12477–12482. doi: 10.1073/pnas.0903653106
Prjibelski, A., Antipov, D., Meleshko, D., Lapidus, A., Korobeynikov, A. (2020). Using SPAdes de novo assembler. Curr. Protoc. Bioinf. 70, e102. doi: 10.1002/cpbi.102
Pujic, P., Fournier, P., Alloisio, N., Hay, A. E., Maréchal, J., Anchisi, S., et al. (2012). Lectin genes in the Frankia alni genome. Arch. Microbiol. 194, 47–56. doi: 10.1007/s00203-011-0770-1
Quach, N. T., Vu, T. H. N., Bui, T. L., Le, T. T. X., Nguyen, T. T. A., Cuong, C., et al. (2022). Genomic and physiological traits provide insights into ecological niche adaptations of mangrove endophytic Streptomyces parvulus VCCM 22513. Ann. Microbiol. 72, 27. doi: 10.1186/s13213-022-01684-6
Rahman, M. S., Fernando, S., Ross, B., Wu, J., Qin, W. (2018). Endoglucanase (EG) activity assays. Methods Mol. Biol. 1796, 169–183. doi: 10.1007/978-1-4939-7877-9_13
Reinhold-Hurek, B., Maes, T., Gemmer, S., Van Montagu, M., Hurek, T. (2006). An endoglucanase is involved in infection of rice roots by the not-cellulose-metabolizing endophyte Azoarcus sp. strain BH72. Mol. Plant Microbe Interact. 19, 181–188. doi: 10.1094/MPMI-19-0181
Ren, Q., Paulsen, I. T. (2007). Large-scale comparative genomic analyses of cytoplasmic membrane transport systems in prokaryotes. J. Mol. Microbiol. Biotechnol. 12, 165–179. doi: 10.1159/000099639
Richter, A. A., Mais, C.-N., Czech, L., Geyer, K., Hoeppner, A., Smits, S. H. J., et al. (2019). Biosynthesis of the stress-protectant and chemical chaperon ectoine: biochemistry of the transaminase EctB. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02811
Richter, M., Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U S A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Richter, M., Rosselló-Móra, R., Oliver Glöckner, F., Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinform. 32, 929–931. doi: 10.1093/bioinformatics/btv681
Riesco, R., Ortúzar, M., Fernández-Ábalos, J. M., Trujillo, M. E. (2022). Deciphering genomes: genetic signatures of plant-associated Micromonospora. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.872356
Rivera-Calzada, A., Famelis, N., Llorca, O., Geibel, S. (2021). Type VII secretion systems: structure, functions and transport models. Nat. Rev. Microbiol. 19, 567–584. doi: 10.1038/s41579-021-00560-5
Roca, W. M., Espinoza, N. O., Roca, M. R., Bryan, J. E. (1978). A tissue culture method for the rapid propagation of potatoes. Am. Potato J. 55, 691–701. doi: 10.1007/BF02852143
Rosenblueth, M., Martínez-Romero, E. (2006). Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 19, 827–837. doi: 10.1094/MPMI-19-0827
Saitou, N., Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sasser, M. (1990). Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101 (Newark: DE: MIDI Inc).
Schneewind, O., Missiakas, D. M. (2012). Protein secretion and surface display in Gram-positive bacteria. Philos. Trans. R. Soc Lond B Biol. Sci. 367, 1123–1139. doi: 10.1098/rstb.2011.0210
Scott, J. R., Barnett, T. C. (2006). Surface proteins of gram-positive bacteria and how they get there. Annu. Rev. Microbiol. 60, 397–423. doi: 10.1146/annurev.micro.60.080805.142256
Shirling, E. B., Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinform. 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Skinnider, M. A., Johnston, C. W., Gunabalasingam, M., Merwin, N. J., Kieliszek, A. M., MacLellan, R. J., et al. (2020). Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Commun. 11, 6058. doi: 10.1038/s41467-020-19986-1
Staneck, J. L., Roberts, G. D. (1974). Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl. Microbiol. 28, 226–231. doi: 10.1128/am.28.2.226-231.1974
Sun, J., Lu, F., Luo, Y., Bie, L., Xu, L., Wang, Y. (2023). OrthoVenn3: an integrated platform for exploring and visualizing orthologous data across genomes. Nucleic Acids Res. 51, W397–W403. doi: 10.1093/nar/gkad313
Suriyachadkun, C., Chunhametha, S., Thawai, C., Tamura, T., Potacharoen, W., Kirtikara, K., et al. (2009). Planotetraspora Thailandica sp. nov., isolated from soil in Thailand. Int. J. Syst. Evol. Microbiol. 59, 992–997. doi: 10.1099/ijs.0.003228-0
Sushko, T., Kavaleuski, A., Grabovec, I., Kavaleuskaya, A., Vakhrameev, D., Bukhdruker, S., et al. (2021). A new twist of rubredoxin function in M. tuberculosis. Bioorg Chem. 109, 104721. doi: 10.1016/j.bioorg.2021.104721
Sutcliffe, I. C. (2011). New insights into the distribution of WXG100 protein secretion systems. Antonie Van Leeuwenhoek. 99, 127–131. doi: 10.1007/s10482-010-9507-4
Tang, S.-K., Wang, Y., Guan, T.-W., Lee, J.-C., Kim, C.-J., Li, W.-J. (2010). Amycolatopsis halophila sp. nov., a halophilic actinomycete isolated from a salt lake. Int. J. Syst. Evol. Microbiol. 60, 1073–1078. doi: 10.1099/ijs.0.012427-0
Tatar, D., Sazak, A., Guven, K., Cetin, D., Sahin, N. (2013). Amycolatopsis cihanbeyliensis sp. nov., a halotolerant actinomycete isolated from a salt mine. Int. J. Syst. Evol. Microbiol. 63, 3739–3743. doi: 10.1099/ijs.0.050963-0
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Tedsree, N., Tanasupawat, S., Sritularak, B., Kuncharoen, N., Likhitwitayawuid, K. (2021). Amycolatopsis dendrobii sp. nov., an endophytic actinomycete isolated from Dendrobium heterocarpum Lindl. Int. J. Syst. Evol. Microbiol. 71, 4902. doi: 10.1099/ijsem.0.004902
Thomas, P., Reddy, K. M. (2013). Microscopic elucidation of abundant endophytic bacteria colonizing the cell wall–plasma membrane peri-space in the shoot-tip tissue of banana. AoB Plants. 5, plt011. doi: 10.1093/aobpla/plt011
Tomich, M., Planet, P., Figurski, D. (2007). The tad locus: postcards from the widespread colonization island. Nat. Rev. Microbiol. 5, 363–375. doi: 10.1038/nrmicro1636
Tomiyasu, I. (1982). Mycolic acid composition and thermally adaptative changes in Nocardia asteroides. J. Bacteriol. 151, 828–837. doi: 10.1128/jb.151.2.828-837.1982
Trujillo, M. E., Bacigalupe, R., Pujic, P., Igarashi, Y., Benito, P., Riesco, R., et al. (2014). Genome features of the endophytic actinobacterium Micromonospora lupini strain Lupac 08: on the process of adaptation to an endophytic life style? PloS One 9, e108522. doi: 10.1371/journal.pone.0108522
Trujillo, M. E., Kroppenstedt, R. M., Fernández-Molinero, C., Schumann, P., Martínez-Molina, E. (2007). Micromonospora lupini sp. nov. and Micromonospora saelicesensis sp. nov., isolated from root nodules of Lupinus angustifolius. Int. J. Syst. Evol. Microbiol. 57, 2799–2804. doi: 10.1099/ijs.0.65192-0
Tseng, T. T., Tyler, B. M., Setubal, J. C. (2009). Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 9, S2. doi: 10.1186/1471-2180-9-S1-S2
Uchida, K., Aida, K. O. (1984). An improved method for the glycolate test for simple identification of the acyl type of bacterial cell walls. J. Gen. Appl. Microbiol. 30, 131–134. doi: 10.2323/jgam.30.131
van Heel, A. J., de Jong, A., Song, C., Viel, J. H., Kok, J., Kuipers, O. P. (2018). BAGEL4: a user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 46, W278–W281. doi: 10.1093/nar/gky383
Wang, H. F., Li, X. Y., Gao, R., Xie, Y. G., Xiao, M., Li, Q. L., et al. (2021). Amycolatopsis anabasis sp. nov., a novel endophytic actinobacterium isolated from roots of Anabasis elatior. Int. J. Syst. Evol. Microbiol. 70, 3391–3398. doi: 10.1099/ijsem.0.004184
Wang, L., Zhang, C., Zhang, J., Rao, Z., Xu, X., Mao, Z., et al. (2021). Epsilon-poly-L-lysine: recent advances in biomanufacturing and applications. Front. Bioeng Biotechnol. 9. doi: 10.3389/fbioe.2021.748976
Wayne, L. G., Brenner, D. J., Colwell, R. R., Grimont, P. A. D., Kandler, O., Krichevsky, M. I., et al. (1987). Report of the Ad Hoc Committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 468–464. doi: 10.1099/00207713-37-4-463
Wong, D. D., Chan, V. J., McCormack, A. A., Batt, S. B. (2010). A novel xyloglucan-specific endo-beta-1,4-glucanase: biochemical properties and inhibition studies. Appl. Microbiol. Biotechnol. 86, 1463–1471. doi: 10.1007/s00253-009-2364-2
Xing, K., Liu, W., Zhang, Y. J., Bian, G. K., Zhang, W. D., Tamura, T., et al. (2013). Amycolatopsis Jiangsuensis sp. nov., a novel endophytic actinomycete isolated from a coastal plant in Jiangsu, China. Antonie Van Leeuwenhoek. 103, 433–439. doi: 10.1007/s10482-012-9823-y
Xu, T., Li, Y., Zeng, X., Yang, X., Yang, Y., Yuan, S., et al. (2017). Isolation and evaluation of endophytic Streptomyces endus OsiSh-2 with potential application for biocontrol of rice blast disease. J. Sci. Food Agric. 97, 1149–1157. doi: 10.1002/jsfa.7841
Yao, X., Zhang, Z., Huang, J., Wei, S., Sun, X., Chen, Y., et al. (2021). Candicidin isomer production is essential for biocontrol of Cucumber Rhizoctonia Rot by Streptomyces albidoflavus W68. Appl. Environ. Microbiol. 87, e03078–e03020. doi: 10.1128/AEM.03078-20
Yoon, S. H., Ha, S. M., Kwon, S., Lim, J., Kim, Y., Seo, H., et al. (2017). Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617. doi: 10.1099/ijsem.0.001755
Zhang, J., Huang, X., Hou, Y., Xia, X., Zhu, Z., Huang, A., et al. (2023). Isolation and screening of antagonistic endophytes against Phytophthora infestans and preliminary exploration on anti-oomycete mechanism of Bacillus velezensis 6-5. Plants (Basel). 12, e909. doi: 10.3390/plants12040909
Zhou, S., Zhou, Y., Li, C., Wu, W., Xu, Y., Xia, W., et al. (2023). Identification and genomic analyses of a novel endophytic actinobacterium Streptomyces endophytica sp. nov. with potential for biocontrol of yam anthracnose. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1139456
Keywords: Amycolatopsis, biosynthetic gene cluster, comparative genomics, endophytic actinomycetes, potato scabby tuber, pathogenicity
Citation: Wannawong T, Mhuantong W, Macharoen P, Niemhom N, Sitdhipol J, Chaiyawan N, Umrung S, Tanasupawat S, Suwannarach N, Asami Y and Kuncharoen N (2024) Comparative genomics reveals insight into the phylogeny and habitat adaptation of novel Amycolatopsis species, an endophytic actinomycete associated with scab lesions on potato tubers. Front. Plant Sci. 15:1346574. doi: 10.3389/fpls.2024.1346574
Received: 29 November 2023; Accepted: 07 March 2024;
Published: 27 March 2024.
Edited by:
Katsuharu Saito, Shinshu University, JapanReviewed by:
Ken-ichi Kucho, Kagoshima University, JapanAdeel Malik, Sangmyung University, Republic of Korea
Copyright © 2024 Wannawong, Mhuantong, Macharoen, Niemhom, Sitdhipol, Chaiyawan, Umrung, Tanasupawat, Suwannarach, Asami and Kuncharoen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nattakorn Kuncharoen, fagrnoku@ku.ac.th; Nakarin Suwannarach, suwan.462@gmail.com
 Thippawan Wannawong1
Thippawan Wannawong1 Somboon Tanasupawat
Somboon Tanasupawat Nakarin Suwannarach
Nakarin Suwannarach Yukihiro Asami
Yukihiro Asami Nattakorn Kuncharoen
Nattakorn Kuncharoen