- 1College of Life and Environmental Science, Minzu University of China, Beijing, China
- 2The National Engineering Laboratory of Crop Molecular Breeding, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing, China
- 3Jiushenghe Seed Industry Co. Ltd., Changji, China
- 4Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China
- 5State Key Laboratory of Crop Stress Adaptation and Improvement, School of Life Sciences, Henan University, Kaifeng, China
- 6Wheat Research Institute, Gansu Academy of Agricultural Sciences, Lanzhou, China
- 7Qinghai Academy of Agricultural and Forestry Sciences, Qinghai University/Key Laboratory of Agricultural Integrated Pest Management, Xining, China
- 8Datong Hui and Tu Autonomous County Agricultural Technology Extension Center, Xining, China
- 9Institute of Biotechnology, Xianghu Laboratory, Hangzhou, China
A recombinant inbred line (RIL) population derived from wheat landrace Qingxinmai and breeding line 041133 exhibited segregation in resistance to powdery mildew and stripe rust in five and three field tests, respectively. A 16K genotyping by target sequencing (GBTS) single-nucleotide polymorphism (SNP) array-based genetic linkage map was used to dissect the quantitative trait loci (QTLs) for disease resistance. Four and seven QTLs were identified for adult-plant resistance (APR) against powdery mildew and stripe rust. QPm.caas-1B and QPm.caas-5A on chromosomes 1B and 5A were responsible for the APR against powdery mildew in line 041133. QYr.caas-1B, QYr.caas-3B, QYr.caas-4B, QYr.caas-6B.1, QYr.caas-6B.2, and QYr.caas-7B detected on the five B-genome chromosomes of line 041133 conferred its APR to stripe rust. QPm.caas-1B and QYr.caas.1B were co-localized with the pleiotropic locus Lr46/Yr29/Sr58/Pm39/Ltn2. A Kompetitive Allele Specific Polymorphic (KASP) marker KASP_1B_668028290 was developed to trace QPm/Yr.caas.1B. Four lines pyramiding six major disease resistance loci, PmQ, Yr041133, QPm/Yr.caas-1B, QPm.caas-2B.1, QYr.caas-3B, and QPm.caas-6B, were developed. They displayed effective resistance against both powdery mildew and stripe rust at the seedling and adult-plant stages.
Introduction
Obligate biotrophic fungi are serious pathogens that constantly endanger the production of wheat (Triticum aestivum L.) in China, for example, Blumeria graminis (DC.) Speer f. sp. tritici (Bgt) and Puccinia striiformis Westend. f. sp. tritici Erikss. (Pst), inciting wheat powdery mildew and stripe rust, respectively. Powdery mildew is primarily found in wheat-growing areas with maritime climates. It became one of the epidemic diseases since the 1970s, spreading from the southwestern to eastern and northern regions of the country within a few decades. Outbreaks of powdery mildew have affected an average of 7 million hectares of wheat annually from 2002 to 2020 (Xiao et al., 2022). Historically, stripe rust has long been a serious biotic constraint in wheat production and continues to be a yield-limiting disease at present. In 2023, stripe rust occurred on an area of 2.67 million hectares (https://www.natesc.org.cn/).
Assessment of resistance to powdery mildew and stripe rust is a mandated task in national and provincial wheat yield trials before commercializing any cultivars given the importance of these diseases in limiting wheat production in China. The improvement of resistance to these diseases is one of the most important objectives in many wheat breeding programs. Two types of genetic mechanisms conferring disease resistance are available to breeders. One is all-stage resistance (ASR), showing race-specific effectiveness and qualitative inheritance. This type of gene may not be effective when virulent pathotypes of pathogens occur. Genes associated with ASR are characterized by seedling tests with artificial inoculation of a single or a few isolates of pathogens in condition-controlled greenhouse or growth chambers. The other is adult-plant resistance (APR), showing effectiveness at the later growth stage of crops, non-race specific mode, and quantitative inheritance. The loci governing APR are usually characterized in field disease nurseries either artificially inoculated or naturally infected at the adult-plant stage.
Most officially designated powdery mildew (Pm) and stripe rust (Yr) resistance genes are characterized as ASR genes. Among the designated Pm genes, Pm38 (Spielmeyer et al., 2005), Pm39 (Lillemo et al., 2008), Pm46 (Herrera-Foessel et al., 2014), Pm55 (Zhang et al., 2016), and Pm62 (Zhang et al., 2018) are identified as the APR genes. Twenty out of 56 Yr genes are reported to perform APR to stripe rust. Yr18 (Singh and Rajaram, 1993), Yr36 (Uauy et al., 2005), Yr39 (Lin and Chen, 2007), Yr52 (Ren et al., 2012), Yr59 (Zhou et al., 2014), Yr62 (Lu et al., 2014), and Yr79 (Feng et al., 2018) are the APR genes against stripe rust. Additionally, 222 and 505 quantitative trait loci (QTLs) for resistance to powdery mildew and stripe rust have been characterized using genetic linkage maps constructed with biparental recombinant inbred line (RIL) populations or genome-wide association study (GWAS) with natural populations (Singh et al., 2022; Kumar et al., 2023).
Wheat diseases caused by different pathogens, such as Bgt and Pst, often occur concurrently, which complicates the genetic improvement of disease resistance in wheat breeding (Liu et al., 2015). Stacking genes or QTL conferring resistance to different diseases in single genotypes offers a feasible option. For example, winter wheat cultivar Jimai 22 carries YrJ22 (Chen et al., 2016) and Pm52 (Qu et al., 2020) for resistance to stripe rust and powdery mildew, respectively. YrZH84 (Li et al., 2006), LrZH84 (Zhao et al., 2008), and QPm.caas-3BS (Jia et al., 2018) are responsible for resistance to stripe rust, leaf rust, and powdery mildew in winter wheat breeding line Zhou 8425B. Pm64 for powdery mildew resistance and Yr5 for stripe rust resistance are tightly linked in wheat-wild emmer (Triticum turgidum var. dicoccoides) introgression line WE35 (Zhang et al., 2019a). Wheat cultivars or lines resistant to multiple diseases are preferably developed by breeders for broader adaption of cultivars.
We have identified the ASR gene PmQ against powdery mildew in wheat landrace Qingxinmai (Li et al., 2020) and the ASR gene Yr041133 against stripe rust in breeding line 041133 (Li et al., 2022). To stack the resistance to both diseases, we developed a RIL population from the cross of Qingxinmai × 041133. The objectives of this study were to 1) dissect APR loci and 2) develop wheat lines pyramiding loci conferring resistance to both powdery mildew and stripe rust.
Materials and methods
Plant materials and field planting
The mapping population of cross Qingxinmai × 041133 comprised 228 F2:9 RILs. Wheat landrace Qingxinmai from Xinjiang carries gene PmQ on the chromosome arm 2BL for its resistance to powdery mildew at the seedling stage (Li et al., 2020). Wheat breeding line 041133 (pedigree: Jining 13 × Tongmai 2) from Qinghai Province carries gene Yr041133 on the chromosome arm 7BL for its seedling resistance tests to stripe rust (Li et al., 2022). Field disease nurseries were established at Beijing (BJ; 116.33°E 39.96°N) and Changping, Beijing (CP; 116.26°E 40.17°N) for assessing resistance to powdery mildew and Qingshui, Gansu Province (QS; 105.80°E 34.60°N) for assessing resistance to stripe rust at the adult-plant stage (Hu et al., 2023). All the field trials were carried out using a randomized complete block design with two replicates. Each plot consisted of a single row 1 m in length in sites BJ and CP and 1.5 m in length in QS. Approximately 40–50 seeds were sown in each row.
Assessments of APR and ASR to powdery mildew
Field trials were carried out to assess powdery mildew resistance at the adult-plant stage at the CP site in 2018–2019 (2019CP), 2019–2020 (2020CP), and 2020–2021 (2021CP) and the BJ site in 2021–2022 (2022BJ) and 2022–2023 (2023BJ). Plants of susceptible wheat spreader, Zhongzuo 9504, were grown every 20 rows and around the experimental plots. They were inoculated with a mixture of isolates designated Bgt27 at the jointing stage [growth stage (GS) 26] (Zadoks et al., 1974). Bgt27 was produced by mixing prevalent isolates E09, E15, E21, E23-(2), and E31, and it was virulent on 20 known Pm genes (Wang et al., 2022; Hu et al., 2023). Maximum disease severity (MDS) on penultimate leaves was rated as the percentage of leaf area covered by Bgt colonies at the late grain-filling stage (GS 77) (Lan et al., 2009). The mean MDS value from five plants randomly selected in each plot was calculated to represent the phenotype of each entry.
The seedling test for assessing powdery mildew resistance was carried out twice following a previously described method (Li et al., 2020). Ten seedlings per line at two-leaf-stage were inoculated with isolates Bgt27 and Bgt1 collected from Yuncheng, Shandong Province. After incubation in a dew plastic bag for 24 h, inoculated seedlings were grown in a greenhouse set at 15°C–18°C to allow symptom development for 2 weeks. Infection types (ITs) on primary leaves were rated on a 0–4 scale. Plants with ITs 0 (immune), 0; (hypersensitive reaction), 1 (highly resistant), and 2 (moderately) were categorized into the resistant group, and those with ITs 3 (moderately susceptible) and 4 (highly susceptible) were classified into the susceptible group.
Assessments of APR and ASR to stripe rust
The adult-plant resistance to stripe rust was assessed in the field disease nursery set at the QS site in 2018–2019 (2019QS), 2019–2020 (2020QS), and 2020–2021 (2021QS). Spreader plants of Huixianhong were planted every 30 rows and around the plots and inoculated by spraying with urediniospores of a mixture of Pst races CYR32, CYR33, and CYR34 at GS 26 (Bai et al., 2022). At GS 77 when the susceptible control Huixianhong was fully infected, disease severity (DS) for the RILs and their parents were scored on the modified Cobb’s scale as described by Peterson et al. (1948).
The Pst race CYR34 was used to perform the seedling resistance test to stripe rust as described previously (Li et al., 2022). Briefly, wheat seedlings at the two-leaf stage were inoculated with Pst urediniospores suspended in light mineral oil (Novec 7200) at 4 mg/mL. After incubation at 9°C–13°C for 24 h in a dew chamber, inoculated plants were grown in a greenhouse for symptom development. A previously established 0–9 scale was used to rate the ITs on primary leaves (McNeal et al., 1971).
Genetic map construction and QTL detection
A genetic linkage map was previously constructed using the same RIL population as described in another report (Li et al., 2024). The RIL population and the parents were genotyped using the wheat 16K genotyping by target sequencing (GBTS) single-nucleotide polymorphism (SNP) array (MolBreeding Biotechnology Co. Ltd., Shijiazhuang, China, http://www.molbreeding.com). A genetic linkage map (3,113.1 cM in length) was constructed using 2,398 bin SNP markers polymorphic between parents Qingxinmai and line 041133. Analysis of QTLs for resistance to powdery mildew and stripe rust was performed using IcIMapping 4.2 software (Meng et al., 2015). The QTLs repeatedly detected in at least two environments and the best linear unbiased estimate (BLUE) datasets were considered the stable QTLs. The physical positions of the QTLs identified were determined by projecting the closely linked markers on the Chinese Spring reference genome sequence RefSeq v1.0 (IWGSC, 2018). MapChart v2.3 software was used to draw the genetic maps for the regions where stable QTLs reside (Voorrips, 2002).
Development of KASP markers
The SNP marker closely linked to the QTLs on chromosome 1BL was converted to a Kompetitive Allele Specific Polymorphic (KASP) marker. Primer was designed using the web-based tool Polymarker (http://www.polymarker.info/). The reaction mixture (10 µL) was generated by mixing 4 µL genomic DNA (50 ng/µL), 5 µL 2× KASP master mix, 0.7 µL primer mix (12 mM of each allele-specific primer and 30 mM of the common primer), and 0.3 µL ddH2O. The following profile for DNA amplification was run in the ABI 7500 device (Applied Biosystems, Foster City, CA, USA): initial denaturation at 94°C for 15 min, 35 cycles of 94°C for 20 s, and 60°C for 1 min. Blue (521 nm) and red (556 nm) fluorescent signals were read at 25°C for 2 min in a FLUOstar Omega microplate reader (BMG Labtech, Durham, NC, USA). They were transformed into FAM homozygote, HEX homozygote, and FAM/HEX heterozygote genotypes with Klustering Caller software (http://www.lgcgroup.com/).
Prediction of candidate genes
Genes within the mapping interval of the QTLs on chromosome 1BL were extracted from the Chinese Spring reference genome sequence RefSeq v1.0 annotations (https://wheat-urgi.versailles.inra.fr/). Spatiotemporal expression patterns of candidate genes were analyzed in the Wheat Expression Browser (http://www.wheat-expression.com/).
Statistical analysis
The BLUE and the broad-sense heritability (H2) were calculated using the Aov (ANOVA of multi-environmental trials) function in the QTL IciMapping software (Meng et al., 2015). Phenotypic correlations and frequency distributions were computed from different environments and the BLUE value of each line in SPSS v. 20.0 for Windows (IBM SPSS, Armonk, NY, USA).
Results
Phenotypic performances of APR against powdery mildew and stripe rust
Reactions of the Qingxinmai × 041133 RIL population and the parents to isolate Bgt27 at the adult-plant stage were assessed in five field trials conducted in two farms at Changping and Beijing during the wheat growing seasons of 2019–2023. The mean MDS of line 041133 (4.62% ± 4.04%) across sites and years was significantly smaller than that of Qingxinmai (40% ± 4.47%) (p < 0.05) (Figure 1A). Values of MDS for the RILs displayed a continuous distribution in a range of 0% to 100% with the coefficients of variation (CVs) from 0.84 to 1.11 in different trials (Table 1, Figure 1C). Frequency distributions of MDS for the RILs were nearly normal in all environments, except for 2022BJ. This was demonstrated by the absolute values of the Skewness and Kurtosis coefficients (Table 1). A high value of broad-sense heritability (H2 = 0.94) across environments was observed for the APR against powdery mildew.
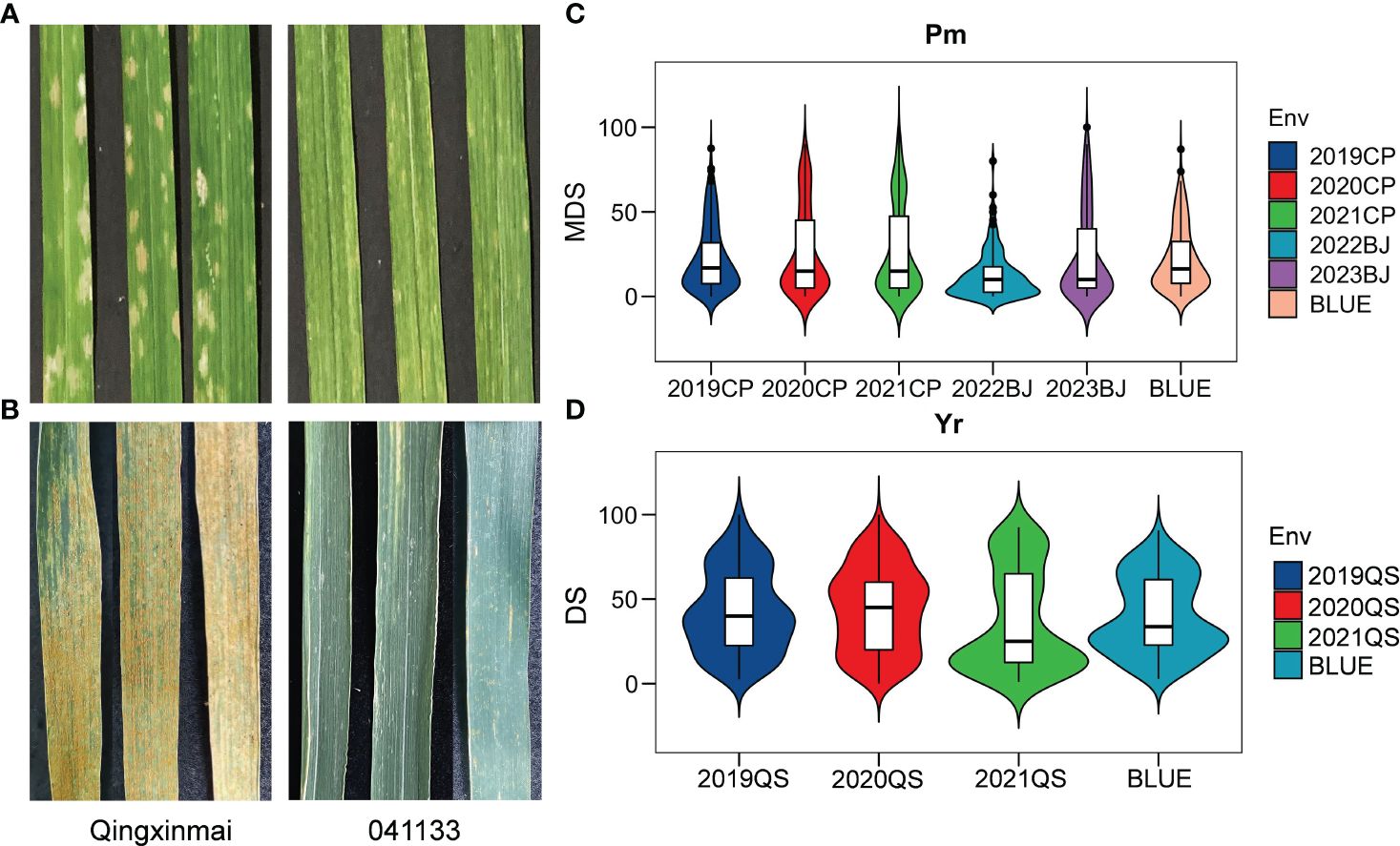
Figure 1 Phenotypes of parents Qingxinmai and line 041133 and their RIL population to Blumeria graminis f. sp. tritici isolate mixture Bgt27 and Puccinia striiformis f. sp. tritici race mixture (CYR32, CYR33, and CYR34) at the adult-plant stage. (A, B) The powdery mildew-infected (A) and stripe rust-infected (B) penultimate leaves of Qingxinmai and line 041133 sampled at the grain-filling stage when disease severity was scored. (C, D) Disease scores of powdery mildew (C) and stripe rust (D) of the RIL population obtained in the field tests at the disease nurseries set at sites Changping (CP), Beijing (BJ), and Qingshui (QS). BLUE, best linear unbiased estimate; RIL, recombinant inbred line.
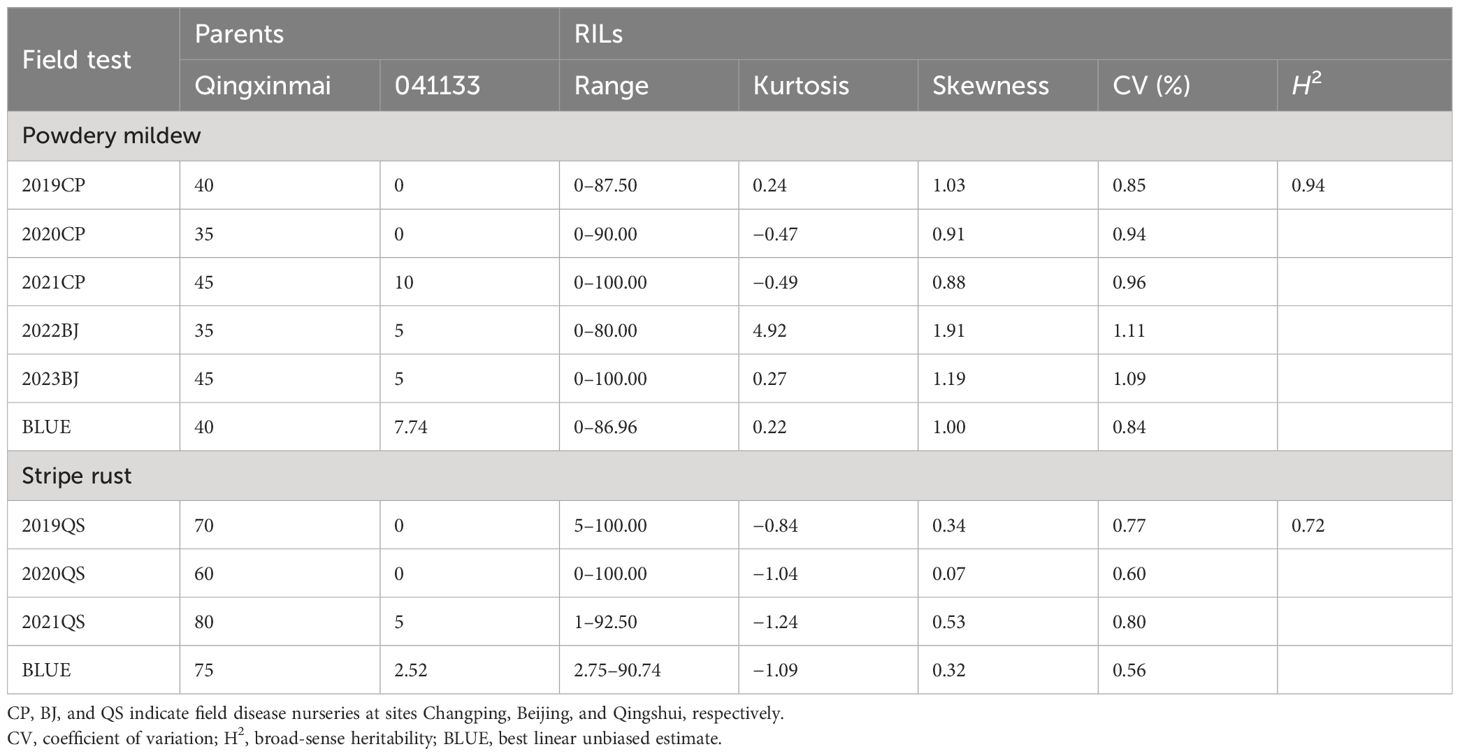
Table 1 Phenotypic variation of powdery mildew and stripe rust of the parents and the RIL population in different environments.
The RIL population of cross Qingxinmai × 041133 was grown in the field disease nursery established at site QS to assess their reactions to stripe rust during the three wheat growing seasons of 2019–2021. Line 041133 produced a mean DS value of 1.88% ± 2.40%, which was significantly smaller than that of Qingxinmai (71.25% ± 8.54%) (p < 0.05) (Figure 1B). The DS values of the RILs across different years ranged from 0% to 100% with the CVs of 0.56–0.80 (Figure 1D). Frequency distributions of DS values for the RILs were nearly normal across years and the BLUE dataset demonstrated by the absolute values of the Skewness and Kurtosis coefficients (Table 1). The H2 of stripe rust resistance was 0.72.
A significant correlation of MDS for powdery mildew was observed among the five field trials and the BLUE dataset with a range of Pearson’s correlation coefficients from 0.41 to 0.99 (p < 0.01) (Figure 2A). Pearson’s correlation coefficients for the stripe rust DS values across different years ranged from 0.27 to 0.94 (p < 0.01) (Figure 2B). Significant correlations were also observed among most of the disease rating scores for powdery mildew and stripe rust (Supplementary Table 1).
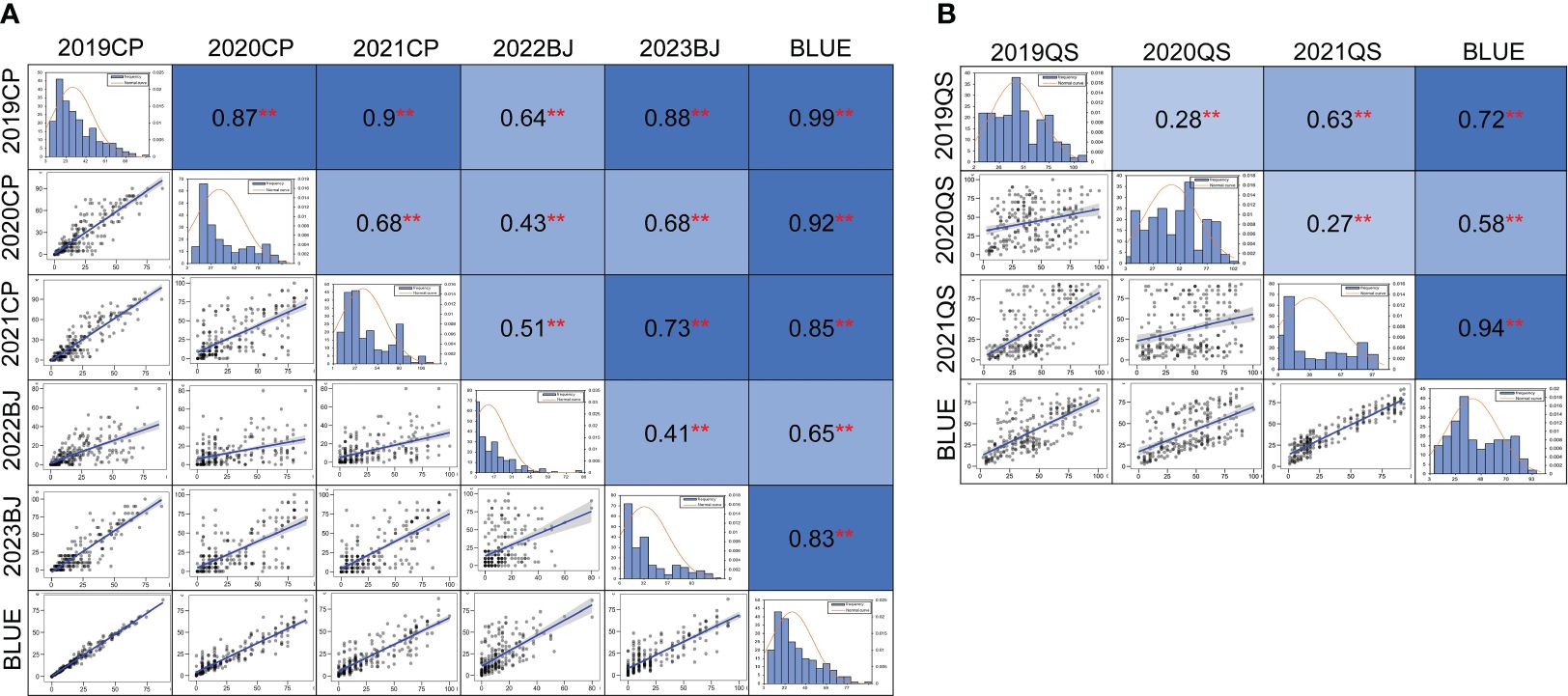
Figure 2 Phenotypic distribution and correlation coefficients of powdery mildew (A) and stripe rust (B) in the Qingxinmai × 041133 RIL population based on the best linear unbiased estimate (BLUE) datasets. **Significant at p < 0.01. The field tests at the disease nurseries set at Changping (CP), Beijing (BJ), and Qingshui (QS) sites. BLUE, best linear unbiased estimate; RIL, recombinant inbred line.
QTL detection
Four and seven QTLs for APR against powdery mildew and stripe rust were identified, respectively (Table 2). Five of them, located on chromosomes 1B, 2B, 3B, and 6B, were stably detected in at least two field trials and the BLUE datasets, explaining 3.73%–20.48% of the phenotypic variations.
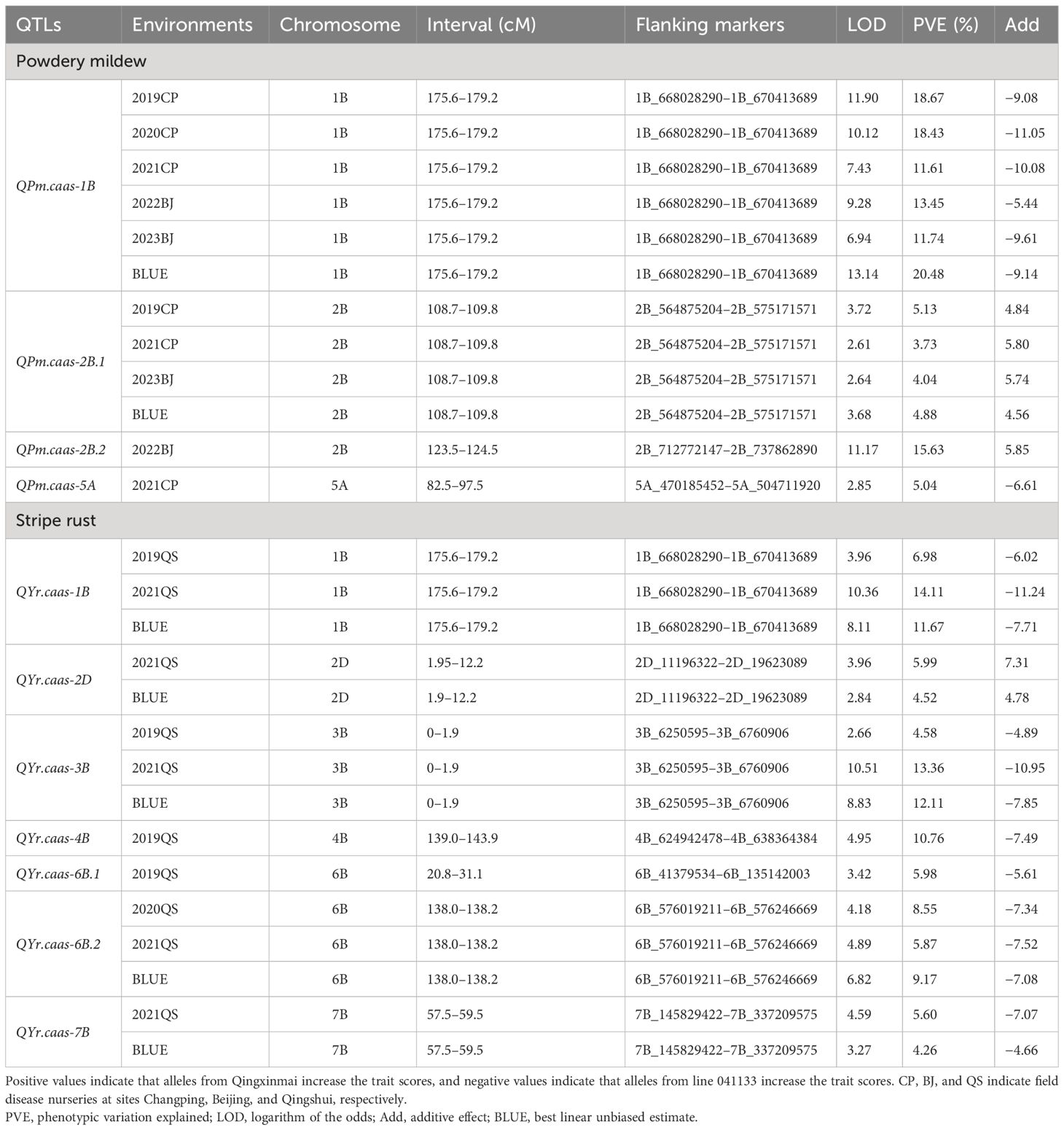
Table 2 Quantitative trait loci (QTLs) for powdery mildew and stripe rust identified from different environments in the Qingxinmai × 041133 RIL population.
QTLs for powdery mildew resistance
Two stable QTLs for powdery mildew resistance were identified on chromosomes 1B and 2B. QPm.caas-1B was detected in all five field trials and the BLUE dataset. It explained 11.61%–20.48% of the phenotypic variation with a range of logarithm of the odds (LOD) values from 6.94 to 13.14 (Table 2, Figure 3A). The resistance allele of these QTLs was derived from line 041133. QPm.caas-2B.1 was detected in three field trials, 2019CP, 2021CP, and 2023BJ, and the BLUE datasets, showing minor effects by explaining 3.73%–5.13% of the phenotypic variations (LOD = 2.61–3.72) (Table 2; Supplementary Figure 1A). The resistance allele of this locus was contributed by Qingxinmai. QPm.caas-2B.2 and QPm.caas-5A were detected in only one environment, explaining 15.63% (LOD = 11.17) and 5.04% (LOD = 2.85) of the phenotypic variations, respectively. The positive alleles were contributed by Qingxinmai and line 041133, respectively (Table 2).
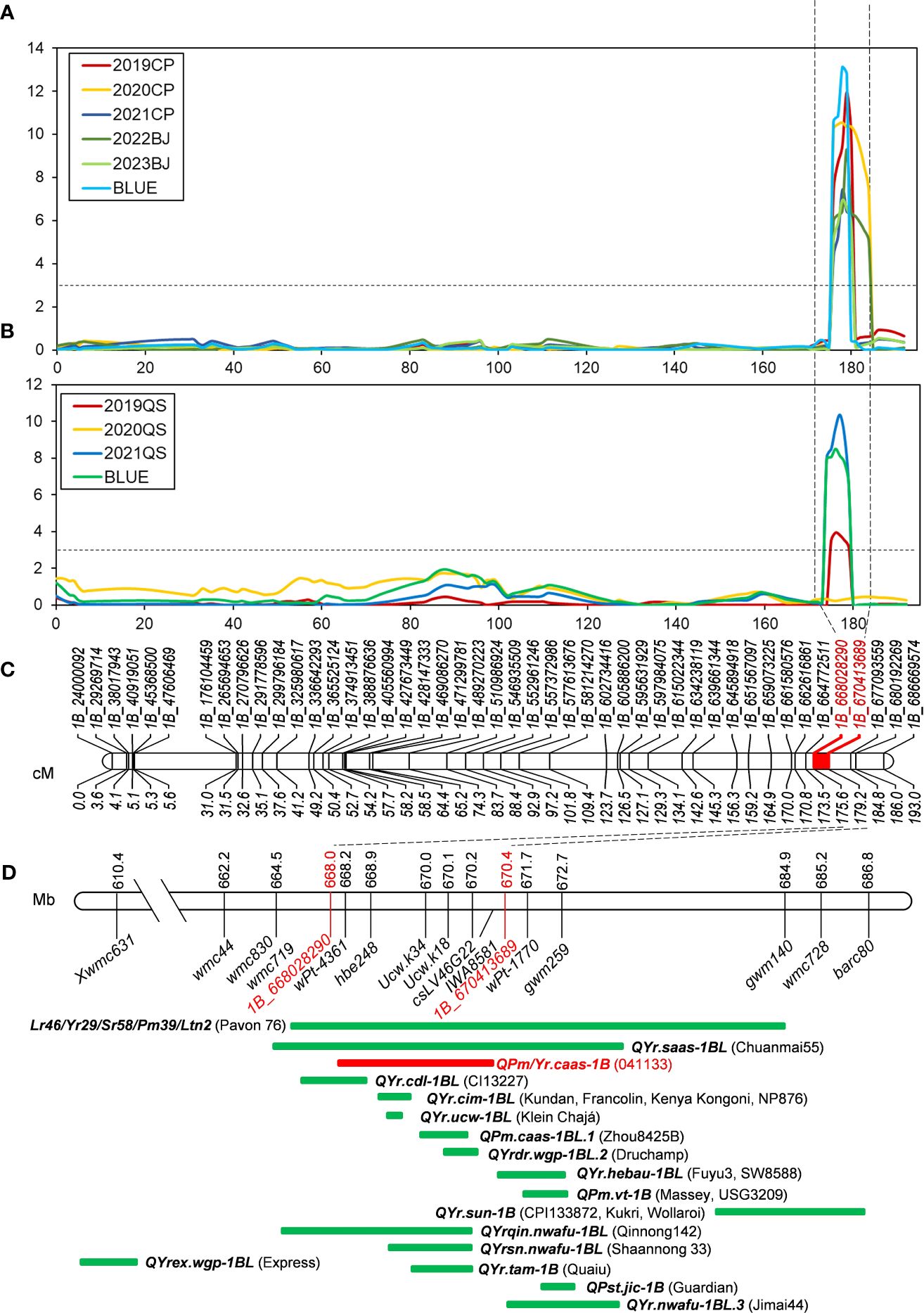
Figure 3 QTL mapping of QPm/Yr.caas-1B. QPm/Yr.caas-1B for resistance to powdery mildew (A) and stripe rust (B) in the Qingxinmai × 041133 RIL population at the logarithm of the odds (LOD) of 3.0. (C) Linkage map of chromosome 1BL constructed using SNP markers generated by the 16K GBTS SNP array. (D) Comparison of QYr.caas-3B (red bar) identified in this study and genes/QTLs (green bars) previously mapped on chromosome 1BL for resistance to powdery mildew and stripe rust based on the physical positions of linked molecular markers projected in the Chinese Spring reference genome RefSeq v1.0. QTL, quantitative trait locus; RIL, recombinant inbred line; SNP, single-nucleotide polymorphism; GBTS, genotyping by target sequencing.
QTLs for stripe rust resistance
Five stable QTLs for APR against stripe rust were detected on chromosomes 1B, 2D, 3B, 6B, and 7B. Three of them, QYr.caas-1B, QYr.caas-3B, and QYr.caas-6B.2, were detected in two environments and the BLUE datasets, explaining 6.98%–14.11%, 4.58%–13.36%, and 5.87%–9.17% of the phenotypic variations, respectively (Table 2). The LOD values for these loci ranged from 2.66 to 10.51 (Table 2, Figure 3B; Supplementary Figures 2A, 3A). Another two QTLs, QYr.caas-2D and QYr.caas-7B explaining 4.52%–5.99% and 4.26%–5.60% of the phenotypic variations, respectively, were detected in single environments and the BLUE dataset. The resistance alleles of all the stable QTLs were contributed by line 041133, except for QYr.caas-2D, which was contributed by Qingxinmai. The remaining two loci, QYr.caas-4B and QYr.caas-6B.1, were detected in single environments, accounting for 5.98%–10.76% of the phenotypic variations. The resistance alleles of both loci were contributed by line 041133 (Table 2).
Develop KASP marker tightly linked to QPm/Yr.caas-1B
The DNA sequences flanking the SNP marker 1B_668028290 obtained from the CS reference genome RefSeq v1.0 were used to design the KASP marker, KASP_1B_668028290, linked to QPm/Yr.caas-1B (Supplementary Table 2). This KASP marker was verified by genotyping the complete RIL population (Supplementary Figure 4). As expected, lines with the positive allele derived from 041133 produced significantly smaller disease severity scores of powdery mildew and stripe rust than those with the negative allele in all environments and the BLUE datasets, except for 2020QS (P < 0.01) (Supplementary Figure 5).
Effects of major QTLs on disease resistance
We analyzed the effects of major QTLs, QPm.caas-1B, QPm.caas-2B.1, QYr.caas-1B, QYr.caas-3B, and QYr.caas-6B.2, on their resistance to powdery mildew and stripe rust by comparing the BLUE datasets of disease scores observed in the RIL population. Lines with QPm.caas-1B and QPm.caas-2B.1 alleles significantly reduced MDS values of powdery mildew (p < 0.01) (Figure 4A). The stripe rust resistance appeared to be associated with the number of QTLs in the RILs. Compared to the lines that were free of any of QTLs QYr.caas-1B, QYr.caas-3B, and QYr.caas-6B.2, lines with two and three alleles significantly reduced the DS values of stripe rust (p < 0.01) (Figure 4B).
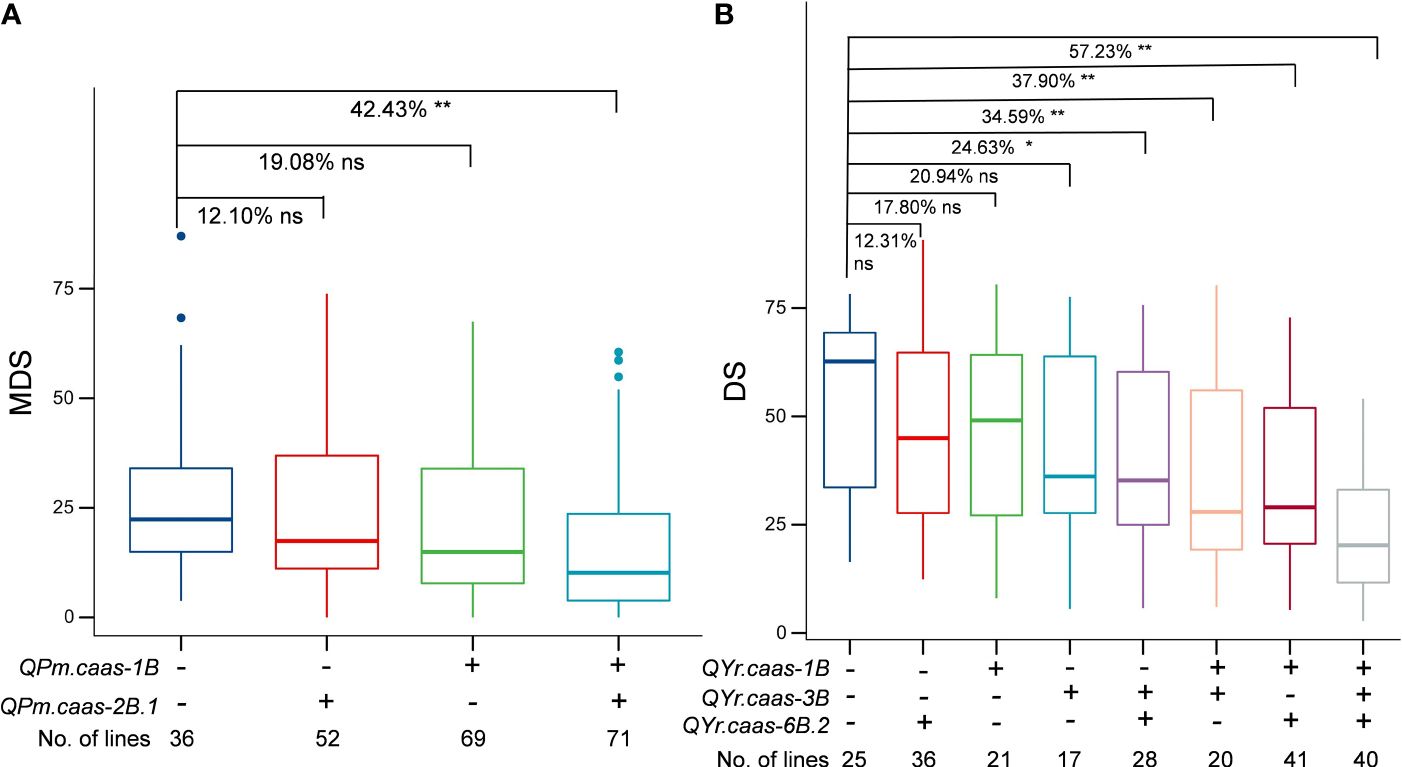
Figure 4 Additive effects of powdery mildew-related QTLs (A) and stripe rust-related QTLs (B) in the Qingxinmai × 041133 RIL population. + and − indicate lines with and without the positive alleles of the target QTLs based on the flanking markers of the corresponding QTLs, respectively. QTLs, quantitative trait loci; RIL, recombinant inbred line. ns: not significant, *: significant at P < 0.05, **: significant at P < 0.01.
Pyramiding of the loci conferring powdery mildew and stripe rust resistance
Based on the genotypes of the molecular markers linked to the two ASR genes, PmQ and Yr041133, and four APR loci, QPm/Yr.caas-1B, QPm.caas-2B.1, QPm.caas-6B, and QYr.caas-3B, four lines pyramiding all loci and one line without any loci were selected. The four pyramided lines QH18, QH19, QH82, and QH202 were resistant to the mixtures of Bgt isolates (Bgt27) and Pst races (CYR32, CYR33, and CYR34) at the adult-plant plant stage with the mean disease scores of 1.7%–8.7% and 2.8%–11.0%, respectively (Supplementary Table 3). These lines were also resistant to Bgt1 and CYR34 pathogens at the seedling stage showing ITs 0, 0; or 1. However, line QH102, which was free of any resistance locus, was susceptible to any of these pathogens at the adult-plant stage (mean disease scores of 68.3% and 69.9%) and the seedling stage (ITs 4 and 3) in the powdery mildew and stripe rust tests. Qingxinmai was susceptible to isolate Bgt27 at both the adult-plant and seedling stages. It was also susceptible to the Pst race CYR34 at both growth stages. Line 041133 was susceptible at the seedling stage but resistant at the adult-plant stage to Bgt27 (Supplementary Table 3).
Potential candidate genes of QPm/Yr.caas-1B
Sequence alignment of the QPm/Yr.caas-1B-flanking markers 1B_668028290 and 1B_670413689 resulted in a corresponding physical interval of 2.38 Mb (668.03–670.41 Mb) in the chromosome arm 1BL of Chinese Spring reference genome sequence RefSeq v1.0 (Table 2, Figure 3). Seven genes for disease resistance were annotated in this genomic interval, including a CC-NBS-LRR (CNL) and six protein kinases (Supplementary Table 4). The expression patterns of these disease resistance genes were analyzed using the publicly available database at Wheat Expression Browser (http://www.wheat-expression.com/) (Figure 5). The expression of TraesCS1B01G451600 was induced by Bgt isolate E09, but it was not expressed until 7 d post inoculation (dpi) of the Pst race CYR31. A varying degree of expression was observed for gene TraesCS1B01G451700 after inoculation with both Bgt and Pst pathogens. The expression levels of genes TraesCS1B01G454000, TraesCS1B01G454100, and TraesCS1B01G454400 peaked at 24 h after inoculation with Bgt isolate E09. Neither Bgt nor Pst was able to induce the expression of TraesCS1B01G452600 (CNL) and TraesCS1B01G454900.
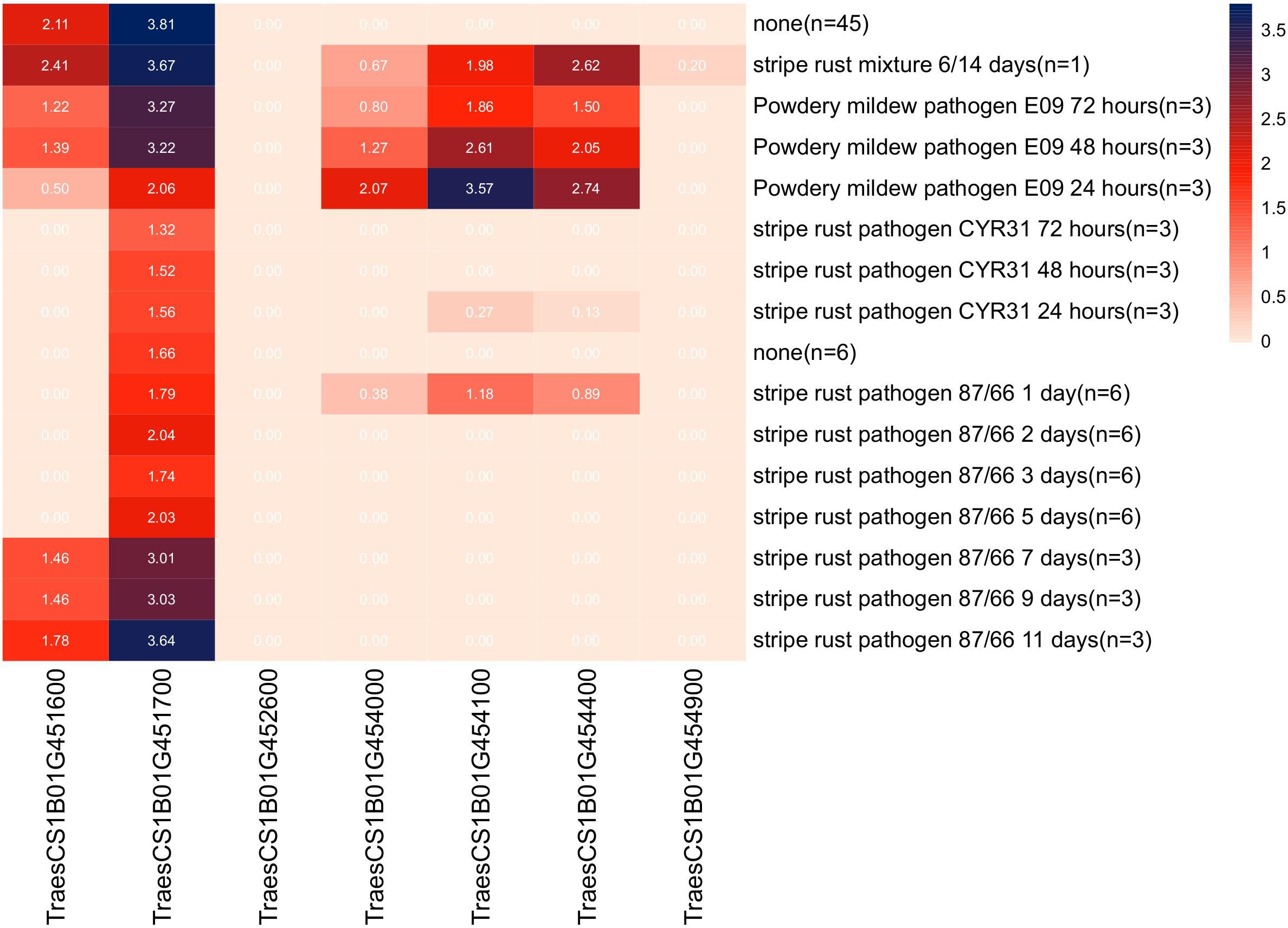
Figure 5 Expression patterns of seven candidate disease resistance genes annotated in the genomic interval of locus QPm/Yr.caas-1B in the Wheat Expression Browser (http://www.wheat-expression.com/).
Previously, gene TraesCS1B01G454100 encoding a receptor-like protein kinase (RLK) was regarded as one of the candidate genes of QYr.ucw-1BL (Cobo et al., 2019), which shares a similar genetic interval as QPm/Yr.caas.1B (Figure 3D). However, the flanking markers of these QTLs, ucw.k34 and ucw.k18, exhibited monomorphism between Qingxinmai and line 041133 (data not shown). The predicted open reading frame (ORF) sequence of TraesCS1B01G454100 was also identical between Qingxinmai and line 041133 (Supplementary Figure 6). This indicates that it is most likely not the candidate gene for QPm/Yr.caas-1B. Based on these findings, TraesCS1B01G451600, TraesCS1B01G451700, and TraesCS1B01G454000 are possible candidate genes for QPm/Yr.caas.1B, which warrants further functional verification.
Discussion
Genetic control of powdery mildew and stripe rust resistance in line 041133
The seedling resistance of line 041133 to Pst race CYR34 was controlled by the ASR gene Yr041133 on chromosome 7BL (Li et al., 2022). The current study observed the adult-plant resistance to a mixture of Pst races CYR32, CYR33, and CYR34 in this breeding line. The stripe rust resistance in line 041133 was governed by seven QTLs, QYr.caas-1B, QYr.caas-3B, QYr.caas.6B.2, QYr.caas-2D, QYr.caas-4B, QYr.caas.6B.1, and QYr.caas-7B, with the first three loci repeatedly detected in multiple environments. Although QYr.caas-7B was observed on chromosome 7B (145.83–337.20 Mb), it appeared not to overlap with Yr041133 (608.90–609.70 Mb). None of the QTLs were located on the same chromosome region as the ASR gene Yr041133.
Line 041133 was susceptible at the seedling stage to Bgt27 and another 23 Bgt isolates tested in a previous study (Li et al., 2020). However, it was resistant to powdery mildew at the adult-plant stage when tested with isolate Bgt27. Results of QTL mapping detected QPm.caas-1B on chromosome 1B and QPm.caas-5A on chromosome 5A of line 041133, which conferred its adult-plant resistance against powdery mildew.
Genetic control of powdery mildew in wheat landrace Qingxinmai
Qingxinmai was previously characterized to be resistant to isolate Bgt1 due to the presence of the ASR gene PmQ on the chromosome arm 2BL (Li et al., 2020). The Bgt isolate mixture Bgt27 was not suitable to determine the effect of PmQ on its adult-plant resistance to powdery mildew, as Qingxinmai was not as effective against powdery mildew at the adult-plant stage as that at the seedling stage. However, we detected two minor QTLs, QPm.caas-2B.1 and QPm.caas-2B.2, on the chromosome arm 2BL of Qingxinmai. QPm.caas-2B.1 contributes 3.73% to 5.13% of phenotypic variations for reducing MDS in three environments. Locus QPm.caas-2B in Japanese cultivar Fukuhokomugi (Liang et al., 2006), locus QPm.inra-2B in French breeding line RE9001 (Bougot et al., 2006), locus QPm.vt-2B in US cultivars Massey (Liu et al., 2001), and locus Pm.vt-2BL in USG3209 (Tucker et al., 2007) did not overlap with the mapping interval of QPm.caas-2B.1. The physical positions of QPm.caas-2BL (546.94–685.76 Mb) in Lumai 21 (Lan et al., 2010b) and QPmtj.caas-2BL (568.44–706.97 Mb) in breeding line Tianmin 668 (Hu et al., 2023) coincided with those of QPm.caas-2B.1. However, no pedigree connection was recorded between Lumai 21, Tianmin 668, and Qingxinmai. Another QTL, QPm.caas-2B.2, is likely identical to PmQ, as they share the same genomic interval on the chromosome arm 2BL of Qingxinmai. Since Qingxinmai was not effective against isolate Bgt27 at both the seedling and adult-plant stages, we were not able to accurately determine the effectiveness of PmQ on the adult-plant resistance using isolate Bgt27 and the Qingxinmai × 041133 RIL population in the current study.
Pleiotropic locus for resistance to stripe rust and powdery mildew on chromosome 1BL of line 041133
QPm.caas-1B and QYr.caas-1B for the adult-plant resistance of line 041133 to powdery mildew and stripe rust were co-localized in the same genetic interval on the chromosome arm 1BL. Correlations between the disease severity scores of powdery mildew and stripe rust measured in the Qingxinmai × 041133 RIL population demonstrated that there may exist a common genetic mechanism conferring the resistance to the two diseases (Lillemo et al., 2008).
The chromosomal region where QPm/Yr.caas-1B was detected appears to be rich in genetic loci for resistance to wheat powdery mildew and stripe rust. A well-characterized locus Lr46/Yr29/Sr58/Pm39/Ltn2, conferring multiple fungal pathogens and leaf tip necrosis, was identified in this genomic region in Mexican spring wheat cultivar Pavon 76 and several other wheat cultivars from different countries (Li et al., 2014). For example, the first designated gene in this region is Lr46 for leaf rust resistance (Singh et al., 1998). A gene that is closely linked to Lr46 was characterized as Yr29 for resistance to stripe rust (William et al., 2003). Gene Pm39 was detected at the locus Lr46/Yr29 in a CIMMYT breeding line Saar (Lillemo et al., 2008). The leaf tip necrosis gene Ltn2 was co-segregated with these rust and powdery mildew resistance genes (Rosewarne et al., 2006), which can serve as a phenotypic marker trait for these disease resistance genes.
There are also some other sources of locus Lr46/Yr29 from different countries. For example, the Uruguayan wheat cultivar Americano 25e proved to carry Lr46, which was derived from a landrace (Kohli, 1986; Kolmer, 2015). Locus Yr29/Lr46 was detected in Indian cultivars New Pusa 876 and Sujata (Lan et al., 2015; Ponce-Molina et al., 2018a). Chuanmai 55 selected from the cross SW3243 × SW8688 carries QYr.saas-1BL co-localized with Yr29/Lr46 (Yang et al., 2019). Additionally, the QTLs for stripe rust resistance detected in Fuyu 3 (Gebrewahid et al., 2020), SW8588 (Zhang et al., 2019b), Qinnong 142 (Zeng et al., 2019), Shaannong 33 (Huang et al., 2021), and Jimai 44 (Liu et al., 2023) overlapped with Yr29/Lr46. However, it is still not determined with certainty whether this multi-pathogen resistance locus is caused by the pleiotropic effects of a single gene or multiple-linked genes (Cobo et al., 2019).
In addition to Lr46/Yr29/Sr58/Pm39/Ltn2, there are another 15 identified QTLs near this locus, including QYr.ucw-1BL (Cobo et al., 2018, 2019), QYr.saas-1BL (Yang et al., 2019), QYr.cdl-1BL (Kolmer et al., 2012), QYr.tam-1B (Basnet et al., 2014), QYr.cim-1BL (Lan et al., 2014; Calvo-Salazar et al., 2015; Ren et al., 2017; Ponce-Molina et al., 2018b), QPm.caas-1BL.1 (Jia et al., 2018), QYrdr.wgp-1BL.2 (Hou et al., 2015), QYr.hebau-1BL (Zhang et al., 2019b; Gebrewahid et al., 2020), QPm.vt-1B (Liu et al., 2001; Tucker et al., 2007), QPst.jic-1B (Melichar et al., 2008), QYrex.wgp-1BL (Lin and Chen, 2009), QYr.sun-1B (Bariana et al., 2010; Zwart et al., 2010; Bansal et al., 2014), QYrsn.nwafu-1BL (Huang et al., 2021), QYrqin.nwafu-1BL (Zeng et al., 2019), and QYr.nwafu-1BL.3 (Liu et al., 2023). Ten of them were co-localized with QPm/Yr.caas-1B detected in this study.
Cobo et al. (2019) mapped locus QYr.ucw-1BL within a genetic interval of 0.06 cM flanking by molecular markers ucw.k34 and ucw.k18. The physical location of QYr.ucw-1BL in the Chinese Spring reference genome was 115 kb (670.03–670.14 Mb). The RLK-encoding gene TraesCS1B01G454100 is the high-confidence gene annotated in this genomic interval only. However, KASP markers ucw.k34 and ucw.k18 detected no polymorphism between Qingxinmai and line 041133, and no variation in DNA sequence was observed in the ORF coding region at TraesCS1B01G454100 between the two parental lines. These results indicate that TraesCS1B01G454100 is most likely not the candidate gene of QPm/Yr.caas-1B.
Comparison of QYr.caas-3B and QYr.caas-6B.1 with known QTLs
QYr.caas-3B and QYr.caas-6B.2 for stripe rust resistance were stably detected on chromosomes 3BS and 6BL of line 041133. There exist 12 reported QTLs conferring APR and four genes for ASR against stripe rust on 3BS. Eight of them, including QYr.ucw-3BS in UC1110 (Lan et al., 2017), QYr.tam-3B in Quaiu (Basnet et al., 2014), QYr.cim-3BS.2 in Frankolin (Lan et al., 2014), QYr.inra-3BS in Renan (Dedryver et al., 2009), QYr.ar-3BS in VA96W-270 (Subramanian et al., 2016), QYr.nwafu-3BS in FDC12 (Liu et al., 2022), QYr‐3B.1 in Pavon76 (William et al., 2006), and QYr.ucw‐3B.2 in IWA5202 (Maccaferri et al., 2015), overlapped with QYr.caas-3B detected in the current study. The interval of QYr.caas-3B in the Chinese Spring reference genome sequence is 510 kb (6.25–6.76 Mb), which is similar to locus Sr2/Lr27/Yr30 (Spielmeyer et al., 2003). Therefore, QYr.caas-3B is likely Yr30.
Nine loci conferring stripe rust resistance have been detected on the chromosome arm 6BL. They included QYrpav.cim-6BL in Pavon 76 (William et al., 2006), QYr.inra-6B in Renan (Dedryver et al., 2009), QYr.caas-6BL in Pingyuan 50 (Lan et al., 2010a), QYr.cim-6BL in Pastor (Rosewarne et al., 2012), QYrdr.wgp-6BL.2 in Druchamp (Hou et al., 2015), QYr.wsu-6BL in IWA7257 (Bulli et al., 2016), QYr.nwafu-6BL.1 in Fried (Wu et al., 2018), QYr.nwafu-6BL.2 in P10078 (Zeng et al., 2019), and QYr.nwafu-6BL.3 in Xinong1376 (Mu et al., 2019). The physical location of QYr.caas-6BL.2 in the Chinese Spring reference genome is 230 kb (576.02–576.25 Mb). It coincides with QYr.inra-6B and QYr.wsu-6BL but differs from the other loci previously reported on this chromosome arm.
Pyramiding QTLs for resistance to powdery and stripe rust
Genes or QTLs performing APR usually last a long period of time. For example, locus Lr46/Yr29 has been used in breeding and remained effective against stripe rust and leaf rust for several decades in different countries, such as Mexico (Li et al., 2014; Lan et al., 2015), India (Ponce-Molina et al., 2018a), Uruguay (Kohli, 1986), and Argentina (Kolmer, 2015; Cobo et al., 2018). Wheat breeding line 041133 has maintained its stripe rust for approximately two decades since it was developed in Qinghai Province (L. Ma, unpublished data). We confirmed that it was resistant to stripe rust, as well as powdery mildew, at the adult-plant stage throughout the course of this study from 2019 to 2023 in all the field tests. As the shift in virulence patterns of Pst races, virulent pathogen pathotypes may accumulate, resulting in the defeat of the disease resistance loci. Pyramiding different loci for resistance to the same disease or even different diseases can mitigate the risk of ineffectiveness for resistance loci. We have developed a KASP marker KASP_1B_668028290 for tracing QPm/Yr.caas-1B in molecular marker-assisted selection. We stacked six loci for resistance to powdery mildew and stripe rust in four breeding lines. These lines demonstrate excellent disease resistance and will be useful as a new source of disease resistance in wheat breeding.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
YL: Data curation, Methodology, Software, Writing – original draft, Writing – review & editing. JH: Data curation, Methodology, Software, Writing – review & editing. HLL: Data curation, Writing – review & editing. DQ: Data curation, Methodology, Writing – review & editing. YQ: Data curation, Writing – review & editing. JD: Data curation, Writing – review & editing. LH: Data curation, Writing – review & editing. LM: Data curation, Writing – review & editing. QW: Methodology, Writing – review & editing, Funding acquisition, Software, Supervision. ZL: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. YZ: Funding acquisition, Supervision, Writing – review & editing. HJL: Funding acquisition, Supervision, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The financial support from the National Natural Science Foundation (32172001 and 32301834) and the Key Research and Development Program of Zhejiang (2024SSYS0099) is gratefully acknowledged.
Conflict of interest
Author HLL was employed by Jiushenghe Seed Industry Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1397274/full#supplementary-material
Supplementary Figure 1 | Molecular mapping of QPm.caas-2B.1 for powdery mildew resistance at the adult-plant stage. (A) QPm.caas-2B.1 for powdery mildew in the RIL population of Qingxinmai × 041133 at the logarithm of the odd (LOD) of 2.5. (B) Linkage map of chromosome 2B constructed using SNP markers generated by the 16K GBTS SNP array. (C) Comparison of the QTL (red bar) identified in this study and the genes/QTL (green bars) for resistance to powdery mildew previously mapped on chromosome 2BL based on the physical positions of linked molecular markers projected in the Chinese Spring reference genome RefSeq v1.0.
Supplementary Figure 2 | Molecular mapping of QYr.caas-3B for stripe rust resistance at the adult-plant stage. (A) QYr.caas-3B for stripe rust in the RIL populations at the logarithm of the odd (LOD) of 3.0. (B) Linkage map of chromosome 3B constructed using SNP markers generated by the 16K GBTS SNP array. (C) Comparison of QYr.caas-3B (red bar) identified in this study and genes/QTL (green bars) previously mapped on chromosome 3BS for resistance to stripe rust based on the physical positions of linked molecular markers projected in the Chinese Spring reference genome RefSeq v1.0.
Supplementary Figure 3 | Molecular mapping of QYr.caas-6B.2 for stripe rust resistance at the adult-plant stage. (A) QYr.caas-6B.2 for stripe rust in the RIL populations at the logarithm of the odd (LOD) of 3.0. (B) Linkage map of chromosome 6B constructed using SNP markers generated by the 16K GBTS SNP array. (C) Comparison of QYr.caas-6B.2 (red bar) identified in this study and genes/QTL (green bars) previously mapped on chromosome 6BL for resistance to stripe rust based on the physical positions of linked molecular markers projected in the Chinese Spring reference genome RefSeq v1.0.
Supplementary Figure 4 | Scatter plots of the RILs from cross Qingxinmai × 041133 for the Kompetitive allele-specific PCR (KASP) assay using the QPm/Yr.caas-1B linked marker KASP_1B_668028290. The blue and red dots represent the RILs with the CC (Qingxinmai) and AA (041133) genotypes, respectively. The non-template control is indicated by the black dots.
Supplementary Figure 5 | Box plot of MDS values about QPm.caas-1B and DS values about QYr.caas-1B with genotypes of line 041133 and Qingxinmai in different environments. ns: not significant. **P < 0.01.
Supplementary Figure 6 | Alignment of the predicted open reading form region sequences of gene TraesCS1B01G454100. The comparative genomes include 10+ genomes (Walkowiak et al., 2020), Zang1817 (Guo et al., 2020), Fielder (Sato et al., 2021), KN9204 (Shi et al., 2022), AK58 (Jia et al., 2023), Kariega (Athiyannan et al., 2022), and Renan (Aury et al., 2022).
References
Athiyannan, N., Abrouk, M., Boshoff, W. H. P., Cauet, S., Rodde, N., Kudrna, D., et al. (2022). Long-read genome sequencing of bread wheat facilitates disease resistance gene cloning. Nat. Genet. 54, 227–231. doi: 10.1038/s41588-022-01022-1
Aury, J. M., Engelen, S., Istace, B., Monat, C., Lasserre-Zuber, P., Belser, C., et al. (2022). Long-read and chromosome-scale assembly of the hexaploid wheat genome achieves high resolution for research and breeding. GigaScience 11, 1–18. doi: 10.1093/gigascience/giac034
Bai, B., Li, Z. M., Wang, H. M., Du, X. L., Wu, L., Du, J. Y., et al. (2022). Genetic analysis of adult plant resistance to stripe rust in common wheat cultivar “Pascal”. Front. Plant Sci. 13, 918437. doi: 10.3389/fpls.2022.918437
Bansal, U. K., Kazi, A. G., Singh, B., Hare, R. A., Bariana, H. S. (2014). Mapping of durable stripe rust resistance in a durum wheat cultivar Wollaroi. Mol. Breed. 33, 51–59. doi: 10.1007/s11032-013-9933-x
Bariana, H. S., Bansal, U. K., Schmidt, A., Lehmensiek, A., Kaur, J., Miah, H., et al. (2010). Molecular mapping of adult plant stripe rust resistance in wheat and identification of pyramided QTL genotypes. Euphytica 176, 251–260. doi: 10.1007/s10681-010-0240-x
Basnet, B. R., Singh, R. P., Ibrahim, A., Herrera-Foessel, S. A., Huerta-Espino, J., Lan, C. X., et al. (2014). Characterization of Yr54 and other genes associated with adult plant resistance to yellow rust and leaf rust in common wheat Quaiu 3. Mol. Breed. 33, 385–399. doi: 10.1007/s11032-013-9957-2
Bougot, Y., Lemoine, J., Pavoine, M. T., Guyomarch, H., Gautier, V., Muranty, H., et al. (2006). A major QTL effect controlling resistance to powdery mildew in winter wheat at the adult plant stage. Plant Breed. 125, 550–556. doi: 10.1111/j.1439-0523.2006.01308.x
Bulli, P., Zhang, J. L., Chao, S. M., Chen, X. M., Pumphrey, M. (2016). Genetic architecture of resistance to stripe rust in a global winter wheat germplasm collection. G3-Genes Genomes Genet. 6, 2237–2253. doi: 10.1534/g3.116.028407
Calvo-Salazar, V., Singh, R. P., Huerta-Espino, J., Cruz-Izquierdo, S., Lobato-Ortiz, R., Sandoval-Islas, S., et al. (2015). Genetic analysis of resistance to leaf rust and yellow rust in spring wheat cultivar Kenya Kongoni. Plant Dis. 99, 1153–1160. doi: 10.1094/PDIS-07-14-0718-RE
Chen, C., He, Z. H., Lu, J. L., Li, J., Ren, Y., Ma, C. X., et al. (2016). Molecular mapping of stripe rust resistance gene YrJ22 in Chinese wheat cultivar Jimai 22. Mol. Breed. 36, 118. doi: 10.1007/s11032-016-0540-5
Cobo, N., Pflüger, L., Chen, X. M., Dubcovsky, J. (2018). Mapping QTL for resistance to new virulent races of wheat stripe rust from two Argentinean wheat cultivars. Crop Sci. 58, 2470–2483. doi: 10.2135/cropsci2018.04.0286
Cobo, N., Wanjugi, H., Lagudah, E., Dubcovsky, J. (2019). A high-resolution map of wheat QYrucw-1BL, an adult plant stripe rust resistance locus in the same chromosomal region as Yr29. Plant Genome 12, 180055. doi: 10.3835/plantgenome2018.08.0055
Dedryver, F., Paillard, S., Mallard, S., Robert, O., Trottet, M., Negre, S., et al. (2009). Characterization of genetic components involved in durable resistance to stripe rust in the bread wheat “Renan”. Phytopathology 99, 968–973. doi: 10.1094/PHYTO-99-8-0968
Feng, Y. J., Wang, M. N., See, D. R., Chao, S., Zheng, Y. L., Chen, X. M. (2018). Characterization of novel gene Yr79 and four additional quantitative trait loci for all-stage and high-temperature adult-plant resistance to stripe rust in spring wheat PI 182103. Phytopathology 108, 737–747. doi: 10.1094/PHYTO-11-17-0375-R
Gebrewahid, T. W., Zhang, P. P., Zhou, Y., Yan, X. C., Xia, X. C., He, Z. H., et al. (2020). QTL mapping of adult plant resistance to stripe rust and leaf rust in a Fuyu 3/Zhengzhou 5389 wheat population. Crop J. 8, 655–665. doi: 10.1016/j.cj.2019.09.013
Guo, W. L., Xin, M. M., Wang, Z. H., Yao, Y. Y., Hu, Z. R., Song, W. J., et al. (2020). Origin and adaptation to high altitude of Tibetan semi-wild wheat. Nat. Commun. 11, 5085. doi: 10.1038/s41467-020-18738-5
Herrera-Foessel, S. A., Singh, R. P., Lillemo, M., Huerta-Espino, J., Bhavani, S., Singh, S., et al. (2014). Lr67/Yr46 confers adult plant resistance to stem rust and powdery mildew in wheat. Theor. Appl. Genet. 127, 781–789. doi: 10.1007/s00122-013-2256-9
Hou, L., Chen, X. M., Wang, M. N., See, D. R., Chao, S., Bulli, P., et al. (2015). Mapping a large number of QTL for durable resistance to stripe rust in winter wheat Druchamp using SSR and SNP markers. PloS One 10, e0126794. doi: 10.1371/journal.pone.0126794
Hu, J. H., Gebremariam, T. G., Zhang, P., Qu, Y. F., Qiu, D., Shi, X. H., et al. (2023). Resistance to powdery mildew is conferred by different genetic loci at the adult-plant and seedling stages in winter wheat line Tianmin 668. Plant Dis. 107, 2133–2143. doi: 10.1094/PDIS-11-22-2633-RE
Huang, S., Liu, S. J., Zhang, Y. B., Xie, Y. Z., Wang, X. T., Jiao, H. X., et al. (2021). Genome-wide wheat 55K SNP-based mapping of stripe rust resistance loci in wheat cultivar Shaannong 33 and their alleles frequencies in current Chinese wheat cultivars and breeding lines. Plant Dis. 105, 1048–1056. doi: 10.1094/PDIS-07-20-1516-RE
International Wheat Genome Sequencing Consortium (IWGSC) (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, 7191. doi: 10.1126/science.aar7191
Jia, A. L., Ren, Y., Gao, F. M., Yin, G. H., Liu, J. D., Guo, L., et al. (2018). Mapping and validation of a new QTL for adult-plant resistance to powdery mildew in Chinese elite bread wheat line Zhou8425B. Theor. Appl. Genet. 131, 1063–1071. doi: 10.1007/s00122-018-3058-x
Jia, J. Z., Zhao, G. Y., Li, D. P., Wang, K., Kong, C. Z., Deng, P. C., et al. (2023). Genome resources for the elite bread wheat cultivar Aikang 58 and mining of elite homeologous haplotypes for accelerating wheat improvement. Mol. Plant 16, 1893–1910. doi: 10.1016/j.molp.2023.10.015
Kohli, M. M. (1986). Wheat varieties of the Southern Cone Region of South America: names, parentage, pedigrees and origins. J. Phys. D Appl. Phys. 44, 5625–5634. doi: 10.1088/0022-3727/43/44/442002
Kolmer, J. A. (2015). A QTL on chromosome 5BL in wheat enhances leaf rust resistance of Lr46. Mol. Breed. 35, 74. doi: 10.1007/s11032-015-0274-9
Kolmer, J. A., Lin, M., Bai, G. (2012). Genetics of leaf rust resistance in the winter wheat line CI13227. Crop Sci. 52, 2166–2172. doi: 10.2135/cropsci2012.02.0136
Kumar, S., Saini, D. K., Jan, F., Jan, S., Tahir, M., Djalovic, I., et al. (2023). Comprehensive meta-QTL analysis for dissecting the genetic architecture of stripe rust resistance in bread wheat. BMC Genomics 24, 259. doi: 10.1186/s12864-023-09336-y
Lan, C. X., Hale, I. L., Herrera-Foessel, S. A., Basnet, B. R., Randhawa, M. S., Huerta-Espino, J., et al. (2017). Characterization and mapping of leaf rust and stripe rust resistance loci in hexaploid wheat lines UC1110 and PI610750 under Mexican environments. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01450
Lan, C. X., Liang, S. S., Wang, Z. L., Yan, J., Zhang, Y., Xia, X. C., et al. (2009). Quantitative trait loci mapping for adult-plant resistance to powdery mildew in Chinese wheat cultivar Bainong 64. Phytopathology 99, 1121–1126. doi: 10.1094/PHYTO-99-10-1121
Lan, C. X., Liang, S. S., Zhou, X. C., Zhou, G., Lu, Q. L., Xia, X. C., et al. (2010a). Identification of genomic regions controlling adult-plant stripe rust resistance in Chinese landrace Pingyuan 50 through bulked segregant analysis. Phytopathology 100, 313–318. doi: 10.1094/PHYTO-100-4-0313
Lan, C. X., Ni, X. W., Yan, J., Zhang, Y., Xia, X. C., Chen, X. M., et al. (2010b). Quantitative trait loci mapping of adult-plant resistance to powdery mildew in Chinese wheat cultivar Lumai 21. Mol. Breed. 25, 615–622. doi: 10.1007/s11032-009-9358-8
Lan, C. X., Rosewarne, G. M., Singh, R. P., Herrera-Foessel, S. A., Huerta-Espino, J., Basnet, B. R., et al. (2014). QTL characterization of resistance to leaf rust and stripe rust in the spring wheat line Francolin1. Mol. Breed. 34, 789–803. doi: 10.1007/s11032-014-0075-6
Lan, C. X., Zhang, Y. L., Herrera-Foessel, S. A., Basnet, B. R., Huerta-Espino, J., Lagudah, E. S., et al. (2015). Identification and characterization of pleiotropic and co-located resistance loci to leaf rust and stripe rust in bread wheat cultivar Sujata. Theor. Appl. Genet. 128, 549–561. doi: 10.1007/s00122-015-2454-8
Li, Y. H., Hu, J. H., Qu, Y. F., Qiu, D., Lin, H. L., Du, J. Y., et al. (2024). Alleles on locus chromosome 4B from different parents confer tiller number and the yield-associated traits in wheat. BMC Plant Biol. in press. doi: 10.1186/s12870-024-05079-4
Li, Z. F., Lan, C. X., He, Z. H., Singh, R. P., Rosewarne, G. M., Chen, X. M., et al. (2014). Overview and application of QTL for adult plant resistance to leaf rust and powdery mildew in wheat. Crop Sci. 54, 1907–1925. doi: 10.2135/cropsci2014.02.0162
Li, Y. H., Lin, R. M., Hu, J. H., Shi, X. H., Qiu, D., Wu, P. P., et al. (2022). Mapping of wheat stripe rust resistance gene Yr041133 by BSR-Seq analysis. Crop J. 10, 447–455. doi: 10.1016/j.cj.2021.06.009
Li, Y. H., Shi, X. H., Hu, J. H., Wu, P. P., Qiu, D., Qu, Y. F., et al. (2020). Identification of a recessive gene PmQ conferring resistance to powdery mildew in wheat landrace Qingxinmai using BSR-Seq analysis. Plant Dis. 104, 743–751. doi: 10.1094/PDIS-08-19-1745-RE
Li, Z. F., Zheng, T. C., He, Z. H., Li, G. Q., Xu, S. C., Li, X. P., et al. (2006). Molecular tagging of stripe rust resistance gene YrZH84 in Chinese wheat line Zhou 8425B. Theor. Appl. Genet. 112, 1098–1103. doi: 10.1007/s00122-006-0211-8
Liang, S. S., Suenaga, K., He, Z. H., Wang, Z. L., Liu, H. Y., Wang, D. S., et al. (2006). Quantitative trait loci mapping for adult-plant resistance to powdery mildew in bread wheat. Phytopathology 96, 784–789. doi: 10.1094/PHYTO-96-0784
Lillemo, M., Asalf, B., Singh, R. P., Huerta-Espino, J., Chen, X. M., He, Z. H., et al. (2008). The adult plant rust resistance loci Lr34/Yr18 and Lr46/Yr29 are important determinants of partial resistance to powdery mildew in bread wheat line Saar. Theor. Appl. Genet. 116, 1155–1166. doi: 10.1007/s00122-008-0743-1
Lin, F., Chen, X. M. (2007). Genetics and molecular mapping of genes for race-specific all-stage resistance and non-race-specific high-temperature adult-plant resistance to stripe rust in spring wheat cultivar Alpowa. Theor. Appl. Genet. 114, 1277–1287. doi: 10.1007/s00122-007-0518-0
Lin, F., Chen, X. M. (2009). Quantitative trait loci for non-race-specific, high-temperature adult-plant resistance to stripe rust in wheat cultivar Express. Theor. Appl. Genet. 118, 631–642. doi: 10.1007/s00122-008-0894-0
Liu, S. X., Griffey, C. A., Maroof, M. A. S. (2001). Identification of molecular markers associated with adult plant resistance to powdery mildew in common wheat cultivar Massey. Crop Sci. 41, 1268–1275. doi: 10.2135/cropsci2001.4141268x
Liu, S. J., Liu, D., Zhang, C. L., Zhang, W. J., Wang, X. T., Mi, Z. W., et al. (2023). Slow stripe rusting in Chinese wheat Jimai 44 conferred by Yr29 in combination with a major QTL on chromosome arm 6AL. Theor. Appl. Genet. 136, 175. doi: 10.1007/s00122-023-04420-z
Liu, T. G., Qiu, J., Zhou, Y. L., Xu, S. C., Chen, H. G., Liu, Y., et al. (2015). Multi-disease resistance evaluation of Chinese advanced winter wheat lines for the national regional test. Sci. Agric. Sin. 48, 2967–2975. doi: 10.3864/j.issn.0578-1752.2015.15.006
Liu, S. J., Wang, X. T., Zhang, Y. Y., Jin, Y. G., Xia, Z. H., Xiang, M. J., et al. (2022). Enhanced stripe rust resistance obtained by combining Yr30 with a widely dispersed, consistent QTL on chromosome arm 4BL. Theor. Appl. Genet. 135, 351–365. doi: 10.1007/s00122-021-03970-4
Lu, Y., Wang, M. N., Chen, X. M., See, D., Chao, S., Jing, J. X. (2014). Mapping of Yr62 and a small-effect QTL for high-temperature adult-plant resistance to stripe rust in spring wheat PI 192252. Theor. Appl. Genet. 127, 1449–1459. doi: 10.1007/s00122-014-2312-0
Maccaferri, M., Ricci, A., Salvi, S., Milner, S. G., Noli, E., Martelli, P. L., et al. (2015). A high-density, SNP-based consensus map of tetraploid wheat as a bridge to integrate durum and bread wheat genomics and breeding. Plant Biotechnol. J. 13, 648–663. doi: 10.1111/pbi.12288
McNeal, F. H., Konzak, C. F., Smith, E. P., Tate, W. S., Russel, T. S. (1971). A uniform system for recording and processing. Cereal Res. Data USDA ARS Bull. 42, 34–121.
Melichar, J. P., Berry, S., Newell, C., MacCormack, R., Boyd, L. A. (2008). QTL identification and microphenotype characterisation of the developmentally regulated yellow rust resistance in the UK wheat cultivar Guardian. Theor. Appl. Genet. 117, 391–399. doi: 10.1007/s00122-008-0783-6
Meng, L., Li, H. H., Zhang, L. Y., Wang, J. K. (2015). QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283. doi: 10.1016/j.cj.2015.01.001
Mu, J. M., Wu, J. H., Liu, S. J., Dai, M. F., Sun, D. J., Huang, S., et al. (2019). Genome-wide linkage mapping reveals stripe rust resistance in common wheat (Triticum aestivum) Xinong1376. Plant Dis. 103, 2742–2750. doi: 10.1094/PDIS-12-18-2264-RE
Peterson, R. F., Campbell, A. B., Hannah, A. E. (1948). A diagrammatic scale for estimating rust intensity on leaves and stems of cereals. Can. J. Res. 26, 496–500. doi: 10.1139/cjr48c-033
Ponce-Molina, L. J., Huerta-Espino, J., Singh, R. P., Basnet, B. R., Alvarado, G., Randhawa, M. S., et al. (2018b). Characterization of leaf rust and stripe rust resistance in spring wheat 'Chilero'. Plant Dis. 102, 421–427. doi: 10.1094/PDIS-11-16-1545-RE
Ponce-Molina, L. J., Huerta-Espino, J., Singh, R. P., Basnet, B. R., Lagudah, E., Aguilar-Rincón, V. H., et al. (2018a). Characterization of adult plant resistance to leaf rust and stripe rust in Indian wheat cultivar 'New Pusa 876'. Crop Sci. 58, 630–638. doi: 10.2135/cropsci2017.06.0396
Qu, Y. F., Wu, P. P., Hu, J. H., Chen, Y. X., Shi, X. H., Qiu, D., et al. (2020). Molecular detection of the powdery mildew resistance genes in winter wheats DH51302 and Shimai 26. J. Integr. Agric. 19, 931–940. doi: 10.1016/S2095-3119(19)62644-4
Ren, Y., Singh, R. P., Basnet, B. R., Lan, C. X., Huerta-Espino, J., Lagudah, E. S., et al. (2017). Identification and mapping of adult plant resistance loci to leaf rust and stripe rust in common wheat cultivar Kundan. Plant Dis. 101, 456–463. doi: 10.1094/PDIS-06-16-0890-RE
Ren, R. S., Wang, M. N., Chen, X. M., Zhang, Z. J. (2012). Characterization and molecular mapping of Yr52 for high-temperature adult-plant resistance to stripe rust in spring wheat germplasm PI 183527. Theor. Appl. Genet. 125, 847–857. doi: 10.1007/s00122-012-1877-8
Rosewarne, G. M., Singh, R. P., Huerta-Espino, J., Herrera-Foessel, S. A., Forrest, K. L., Hayden, M. J., et al. (2012). Analysis of leaf and stripe rust severities reveals pathotype changes and multiple minor QTLs associated with resistance in an Avocet × Pastor wheat population. Theor. Appl. Genet. 124, 1283–1294. doi: 10.1007/s00122-012-1786-x
Rosewarne, G. M., Singh, R. P., Huerta-Espino, J., William, H. M., Bouchet, S., Cloutier, S., et al. (2006). Leaf tip necrosis, molecular markers and beta1-proteasome subunits associated with the slow rusting resistance genes Lr46/Yr29. Theor. Appl. Genet. 112, 500–508. doi: 10.1007/s00122-005-0153-6
Sato, K., Abe, F., Mascher, M., Haberer, G., Gundlach, H., Spannagl, M., et al. (2021). Chromosome-scale genome assembly of the transformation-amenable common wheat cultivar ‘Fielder’. DNA Res. 28, 1–7. doi: 10.1093/dnares/dsab008
Shi, X. L., Cui, F., Han, X. Y., He, Y. L., Zhao, L., Zhang, N., et al. (2022). Comparative genomic and transcriptomic analyses uncover the molecular basis of high nitrogen use efficiency in the wheat cultivar Kenong 9204. Mol. Plant 15, 1440–1456. doi: 10.1016/j.molp.2022.07.008
Singh, R. P., Mujeeb-Kazi, A., Huerta-Espino, J. (1998). Lr46: a gene conferring slow rusting resistance to leaf rust in wheat. Phytopathology 88, 890–894. doi: 10.1094/PHYTO.1998.88.9.890
Singh, R. P., Rajaram, S. (1993). Genetics of adult plant resistance to stripe rust in ten spring bread wheats. Euphytica 72, 1–7. doi: 10.1007/BF00023766
Singh, K., Saini, D. K., Saripalli, G., Batra, R., Gautam, T., Singh, R., et al. (2022). WheatQTLdb V2.0: a supplement to the database for wheat QTL. Mol. Breed. 42, 56. doi: 10.1007/s11032-022-01329-1
Spielmeyer, W., Mcintosh, R. A., Kolmer, J., Lagudah, E. S. (2005). Powdery mildew resistance and Lr34/Yr18 genes for durable resistance to leaf and stripe rust cosegregate at a locus on the short arm of chromosome 7D of wheat. Theor. Appl. Genet. 111, 731–735. doi: 10.1007/s00122-005-2058-9
Spielmeyer, W., Sharp, P. J., Lagudah, E. S. (2003). Identification and validation of markers linked to broad-spectrum stem rust resistance gene Sr2 in wheat (Triticum aestivum L.). Crop Sci. 43, 333–336. doi: 10.2135/cropsci2003.3330
Subramanian, N. K., Mason, R. E., Milus, E. A., Moon, D. E., Brown-Guedira, G. (2016). Characterization of two adult-plant stripe rust resistance genes on chromosomes 3BS and 4BL in soft red winter wheat. Crop Sci. 56, 143–153. doi: 10.2135/cropsci2015.01.0043
Tucker, D. M., Griffey, C. A., Liu, S., Brown-Guedira, G., Marshall, D. S., Maroof, M. A. S. (2007). Confirmation of three quantitative trait loci conferring adult plant resistance to powdery mildew in two winter wheat populations. Euphytica 155, 1–13. doi: 10.1007/s10681-006-9295-0
Uauy, C., Brevis, J. C., Chen, X. M., Khan, I., Jackson, L., Chicaiza, O., et al. (2005). High-temperature adult-plant (HTAP) stripe rust resistance gene Yr36 from Triticum turgidum ssp. dicoccoides is closely linked to the grain protein content locus Gpc-B1. Theor. Appl. Genet. 112, 97–105. doi: 10.1007/s00122-005-0109-x
Voorrips, R. E. (2002). Mapchart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Walkowiak, S., Gao, L. L., Monat, C., Haberer, G., Kassa, M. T., Brinton, J., et al. (2020). Multiple wheat genomes reveal global variation in modern breeding. Nature 588, 277–283. doi: 10.1038/s41586-020-2961-x
Wang, A. L., Zhao, Y. N., Zhang, M. H., Yuan, J. H., Liu, W., Fan, J. R., et al. (2022). The quantitative analyses for the effects of two wheat varieties with different resistance levels on the fungicide control efficacies to powdery mildew. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.864192
William, M., Singh, R. P., Huerta-Espino, J., Islas, S. O., Hoisington, D. (2003). Molecular marker mapping of leaf rust resistance gene Lr46 and its association with stripe rust resistance gene Yr29 in wheat. Phytopathology 93, 153–159. doi: 10.1094/PHYTO.2003.93.2.153
William, H. M., Singh, R. P., Huerta-Espino, J., Palacios, G., Suenaga, K. (2006). Characterization of genetic loci conferring adult plant resistance to leaf rust and stripe rust in spring wheat. Genome 49, 977–990. doi: 10.1139/g06-052
Wu, J. H., Liu, S. J., Wang, Q. L., Zeng, Q. D., Mu, J. M., Huang, S., et al. (2018). Rapid identification of an adult plant stripe rust resistance gene in hexaploid wheat by high-throughput SNP array genotyping of pooled extremes. Theor. Appl. Genet. 131, 43–58. doi: 10.1007/s00122-017-2984-3
Xiao, J., Liu, B., Yao, Y. Y., Guo, Z. F., Jia, H. Y., Kong, L. R., et al. (2022). Wheat genomic study for genetic improvement of traits in China. Sci. China Life Sci. 65, 1718–1775. doi: 10.1007/s11427-022-2178-7
Yang, M. Y., Li, G. R., Wan, H. S., Li, L. P., Li, J., Yang, W. Y., et al. (2019). Identification of QTLs for stripe rust resistance in a recombinant inbred line population. Int. J. Mol. Sci. 20, 3410. doi: 10.3390/ijms20143410
Zadoks, J. C., Chang, T. T., Konzak, C. F. (1974). A decimal code for the growth stages of cereals. Weed Res. 14, 415–421. doi: 10.1111/j.1365-3180.1974.tb01084.x
Zeng, Q. D., Wu, J. H., Liu, S. J., Chen, X. M., Yuan, F. P., Su, P. P., et al. (2019). Genome-wide mapping for stripe rust resistance loci in common wheat cultivar Qinnong 142. Plant Dis. 103, 439–447. doi: 10.1094/PDIS-05-18-0846-RE
Zhang, R. Q., Fan, Y. L., Kong, L. N., Wang, Z. J., Wu, J. Z., Xing, L. P., et al. (2018). Pm62, an adult-plant powdery mildew resistance gene introgressed from Dasypyrum villosum chromosome arm 2VL into wheat. Theor. Appl. Genet. 131, 2613–2620. doi: 10.1007/s00122-018-3176-5
Zhang, P. P., Li, X., Gebrewahid, T. W., Liu, H. X., Xia, X. C., He, Z. H., et al. (2019b). QTL Mapping of adult-plant resistance to leaf and stripe rust in wheat cross SW 8588/Thatcher using the wheat 55K SNP array. Plant Dis. 103, 3041–3049. doi: 10.1094/PDIS-02-19-0380-RE
Zhang, R. Q., Sun, B. X., Chen, J., Cao, A. Z., Xing, L. P., Feng, Y. G., et al. (2016). Pm55, a developmental-stage and tissue-specific powdery mildew resistance gene introgressed from Dasypyrum villosum into common wheat. Theor. Appl. Genet. 129, 1975–1984. doi: 10.1007/s00122-016-2753-8
Zhang, D. Y., Zhu, K. Y., Dong, L. L., Liang, Y., Li, G. Q., Fang, T. L., et al. (2019a). Wheat powdery mildew resistance gene Pm64 derived from wild emmer (Triticum turgidum var. dicoccoides) is tightly linked in repulsion with stripe rust resistance gene Yr5. Crop J. 7, 761–770. doi: 10.1016/j.cj.2019.03.003
Zhao, X. L., Zheng, T. C., Xia, X. C., He, Z. H., Liu, D. Q., Yang, W. X., et al. (2008). Molecular mapping of leaf rust resistance gene LrZH84 in Chinese wheat line Zhou 8425B. Theor. Appl. Genet. 117, 1069–1075. doi: 10.1007/s00122-008-0845-9
Zhou, X. L., Wang, M. N., Chen, X. M., Lu, Y., Kang, Z. S., Jing, J. X. (2014). Identification of Yr59 conferring high-temperature adult-plant resistance to stripe rust in wheat germplasm PI 178759. Theor. Appl. Genet. 127, 935–945. doi: 10.1007/s00122-014-2269-z
Keywords: wheat (Triticum aestivum L.), powdery mildew, stripe rust, adult-plant resistance, QTL mapping
Citation: Li Y, Hu J, Lin H, Qiu D, Qu Y, Du J, Hou L, Ma L, Wu Q, Liu Z, Zhou Y and Li H (2024) Mapping QTLs for adult-plant resistance to powdery mildew and stripe rust using a recombinant inbred line population derived from cross Qingxinmai × 041133. Front. Plant Sci. 15:1397274. doi: 10.3389/fpls.2024.1397274
Received: 07 March 2024; Accepted: 16 April 2024;
Published: 08 May 2024.
Edited by:
Diego Rubiales, Spanish National Research Council (CSIC), SpainReviewed by:
Yunfeng Xu, Kansas State University, United StatesDinesh Kumar Saini, Texas Tech University, United States
Yinguang Bao, Shandong Agricultural University, China
Copyright © 2024 Li, Hu, Lin, Qiu, Qu, Du, Hou, Ma, Wu, Liu, Zhou and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongjie Li, lihongjie@xhlab.ac.cn; Yijun Zhou, zhouyijun@muc.edu.cn; Zhiyong Liu, zyliu@genetics.ac.cn
†These authors have contributed equally to this work
 Yahui Li
Yahui Li Jinghuang Hu2†
Jinghuang Hu2† Dan Qiu
Dan Qiu Lu Hou
Lu Hou Yijun Zhou
Yijun Zhou Hongjie Li
Hongjie Li