- 1Laboratoire de Recherche en Sciences Veígeítales - Génomique et Biotechnologie des Fruits, Universiteí de Toulouse, Centre national de la recherche scientifique (CNRS), Université Toulouse III - Paul Sabatier (UPS), Toulouse-Institut National Polytechnique (INP), Toulouse, France
- 2Key Laboratory of Plant Hormones and Development Regulation of Chongqing, School of Life Sciences, Chongqing University, Chongqing, China
- 3Center of Plant Functional Genomics, Institute of Advanced Interdisciplinary Studies, Chongqing University, Chongqing, China
Fruit development can be viewed as the succession of three main steps consisting of the fruit initiation, growth and ripening. These processes are orchestrated by different factors, notably the successful fertilization of flowers, the environmental conditions and the hormones whose action is coordinated by a large variety of transcription factors. Among the different transcription factor families, TEOSINTE BRANCHED 1, CYCLOIDEA, PROLIFERATING CELL FACTOR (TCP) family has received little attention in the frame of fruit biology despite its large effects on several developmental processes and its action as modulator of different hormonal pathways. In this respect, the comprehension of TCP functions in fruit development remains an incomplete puzzle that needs to be assembled. Building on the abundance of genomic and transcriptomic data, this review aims at collecting available TCP expression data to allow their integration in the light of the different functional genetic studies reported so far. This reveals that several Class I TCP genes, already known for their involvement in the cell proliferation and growth, display significant expression levels in developing fruit, although clear evidence supporting their functional significance in this process remains scarce. The extensive expression data compiled in our study provide convincing elements that shed light on the specific involvement of Class I TCP genes in fruit ripening, once these reproductive organs acquire their mature size. They also emphasize their putative role in the control of specific biological processes such as fruit metabolism and hormonal dialogue.
1 Introduction
In addition to their major importance in human nutrition, fruits have become a vast area of investigation in recent decades given their contribution to the understanding of the evolutionary history of angiosperms (Seymour et al., 2013) through their role in seed development and dispersal. A deeper understanding of the mechanisms behind the development and ripening of fleshy fruits is of major interest considering the wide range of applications it opens up for plant breeding, improvement of quality attributes and post-harvest handling as well as technological value. Transcription factors (TFs) are instrumental in mediating the transcriptomic reprogramming driving the developmental processes underpinning organ and tissue differentiation. Among the many TF families involved in fruit development, MADS-box TFs have been documented as the core contributors, as evidenced by the role of the master regulators, RIN, TAGL1, and MBP3 in the control of fruit ripening and locular gel differentiation in tomato (Vrebalov et al., 2002, 2009; Zhang et al., 2019b; Huang et al., 2021). The involvement of other TF families such as NAC, ERF, ARF or DOF has also been well documented (Hao et al., 2015; Liu et al., 2020; Forlani et al., 2021; Chirinos et al., 2023; Zou and Sun, 2023) and likewise, MYB TFs have been reported to actively contribute to fruit quality through the control of texture, flavor and phenolic and carotenoid metabolism (Allan and Espley, 2018). Overall, it is now well established that fleshy fruit development is made of a series of developmental transitions involving almost all types of TF families even though little is known about the specific role of many of these transcriptional regulators. In this view, the TCP transcription family exemplifies this lack of knowledge. Members of this transcription family genes have largely been described and reviewed for their effect on several developmental processes such as cell growth and proliferation, germination, embryogenesis, leaf and flower morphogenesis, biotic and abiotic stress responses (Dhaka et al., 2017; Lan and Qin, 2020; Viola and Gonzalez, 2023; Viola et al., 2023). In the previous decade, the active expression of TCP genes has been reported in a large number of species cultivated for its fruits such as tomato (Parapunova et al., 2014), Prunus mume (Zhou et al., 2016), strawberry (Wei et al., 2016), grapevine (Jiu et al., 2019; Leng et al., 2019), banana (Wang et al., 2022a), apple (Tabarelli et al., 2022), eggplant (Li et al., 2022), citrus (Liu et al., 2022a), passion fruit (Rizwan et al., 2022), cucumber (Ren et al., 2023), kiwifruit (Li et al., 2024) and pepper (Dong et al., 2024). Despite these numerous inventories, the impacts of TCP on fruit development remain enigmatic.
The TCP gene family was first described as a small group of plant-specific transcription factors sharing a conserved domain, named TCP after the three first characterized family members, TEOSINTE BRANCHED (TB) 1 from maize, CYCLOIDEA (CYC) from Antirrhinum majus, and PROLIFERATING CELL FACTORS (PCFs) from rice (Cubas et al., 1999). The TCP domain consists of a non-canonical basic-helix-loop-helix (bHLH) structure that allows DNA binding and protein-protein interactions (Cubas et al., 1999; Nicolas and Cubas, 2016). The protein crystal structure of rice TCP OsPCF6 revealed that the TCP domain functions as a dimer, with two TCP domains closely intertwined into a stable conformation (Sun et al., 2020). In addition, the DNA-TCP-domain complex structures were determined on AtTCP15 and AtTCP10: the homodimeric TCP domains adopt a three-site recognition mode for double-stranded DNA through a short pair of β-strands formed in the dimer interface completed with two basic flexible loops from each monomer (Zhang et al., 2023). Based on phylogenetic analysis, TCPs can be classified into two homology classes: Class I and Class II with the latter being further divided into two subclasses: the CIN-TCP subclass and the CYC/TB1 subclass (Cubas et al., 1999). Members of Class I TCP display a shorter TCP domain with a 4 amino acid deletion in the basic region and also exhibit a conserved region close to the TCP domain C-terminal extremity (Zhou et al., 2022). Both CIN and CYC/TB1 proteins are highly conserved and have been shown to be involved in controlling leaf morphology and size, petal development, trichome formation, and plant flowering (Bresso et al., 2018; Sarvepalli and Nath, 2018; Lan and Qin, 2020; Rath et al., 2022). In addition, the expression of CIN genes is controlled by the well-conserved microRNA319 (Bresso et al., 2018). To our knowledge, few studies reported the involvement of Class II TCPs in fruit development or ripening; only Class II AtTCP4 was recently found to contribute to the determination of ovule identity (Lan et al., 2023) and to the specification of apical gynoecium (Wang et al., 2024). Therefore, the current review will focus on Class I TCP functions in the frame of fruit development and ripening following flower fertilization. It aims at integrating recent literature and available expression data on Class I TCPs in order to draw a first picture of the potential role of this class of transcription factors in fruit biology.
1.1 Expression patterns of Class I TCP genes suggesting multiple actions in fruit development and ripening
Over the past decade, NGS technologies have enabled the generation of ever-increasing amounts of genomic and transcriptomic data from a large panel of plant species. Taking advantage of these resources, we performed a comprehensive survey to gain insight into the expression patterns in fruit organs of TCP-encoding genes. Interestingly, monitoring the expression of Class I TCP TF genes either by qRT-PCR or by RNAseq in plant species representative of a large diversity of botanical families (Table 1), revealed that TCPs are expressed in several fleshy fruits such as the dicots apple, pear, strawberry, blueberry, cucumber, tomato and grape and the monocots banana.
Table 1 shows that several Class I TCP genes are preferentially expressed at early stages of fruit development while others exhibit higher expression levels at late developmental stages, from ripening initiation to full-ripe fruit (Table 1). To further refine the classification of the TCP genes inventoried in Table 1, we built a dedicated phylogenetic tree based on the genome annotations of five species representative of Angiosperm families: Arabidopsis, tomato, grapes, rice and maize (Arabidopsis thaliana, Solanum lycopersicum, Vitis vinifera, Oryza sativa and Zea mays respectively). The tree divides Class I TCP family into 4 sub-groups encompassing the majority of the annotated Class I TCP (Figure 1). Interestingly, these sub-groups can be associated to the presence or absence of conserved motifs predicted with the MEME software (Bailey and Elkan, 1994), thus consolidating the proposed phylogeny (Supplementary Table S1). When available, all the TCP genes listed in Table 1 were classified using the groups deduced from the phylogeny (Supplementary Table S2). TCP genes from each group are found to be expressed during fruit development or ripening, thus suggesting a coordinated action of TCP factors as reported in other physiological processes in vegetative organs (Danisman, 2016; Viola et al., 2023).
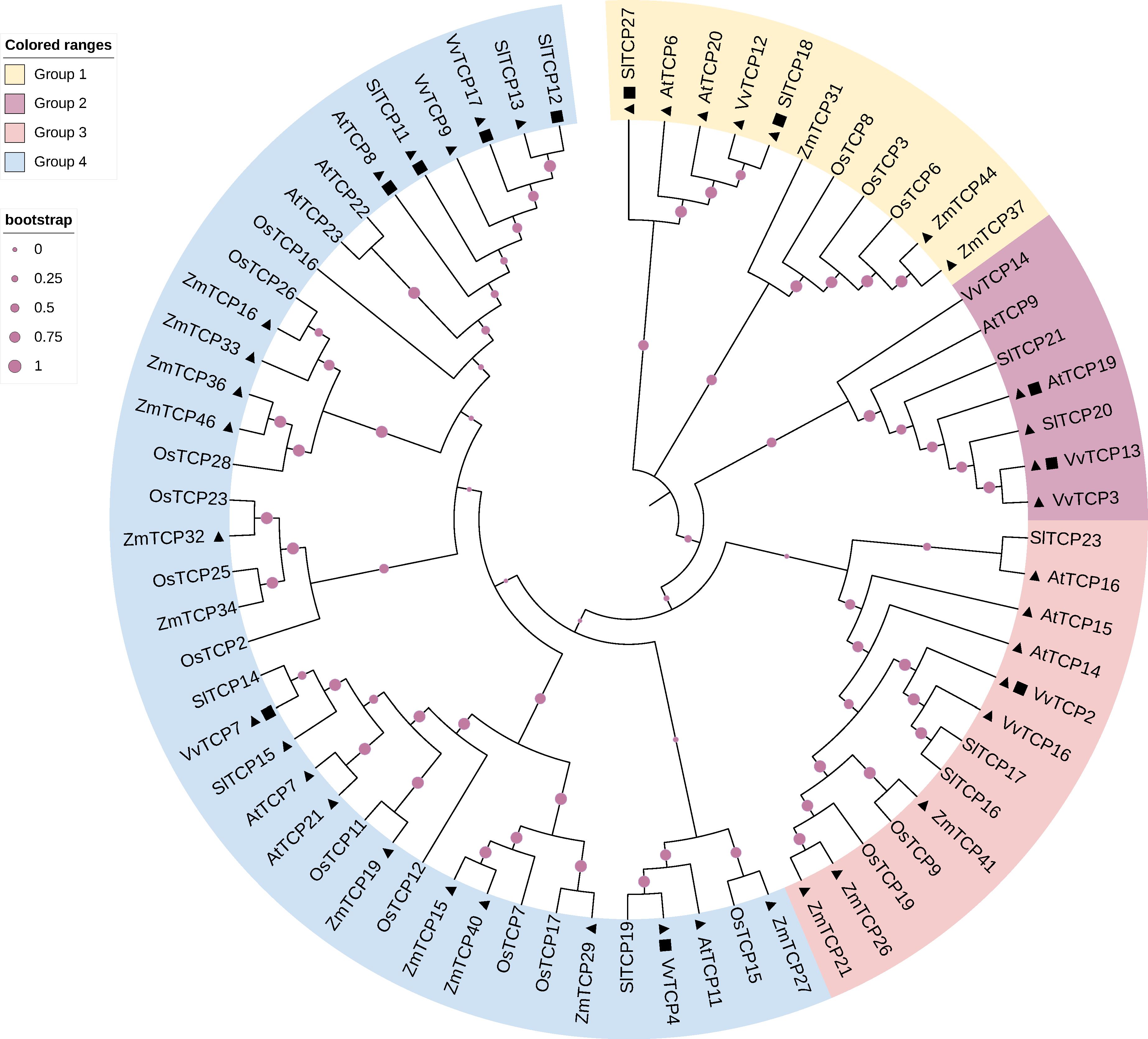
Figure 1 Phylogenetic tree of Class I TCP proteins. The phylogenetic analysis of TCP gene family among grape (Vitis vinifera), Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), maize (Zea mays), and tomato (Solanum lycopersicum). The circular phylogenetic tree was constructed using the full-length protein sequences of TCPs from five species by MEGAX using the MUSCLE alignment and Neighbor-Joining (NJ) method. Bootstrap analysis was performed using 1000 replicates. Triangles represent genes specifically expressed in the early stages of fruit development, and squares represent genes specifically expressed in the late stages of fruit development. Gene information shown in the Supplementary Table S1.
Massive amounts of transcriptomics data generated either by RNAseq or microarrays, can be easily accessed via different datamining and visualization platforms such as BAR-ePlant (The Bio-Analytic Resource for Plant Biology) (Waese et al., 2017), GEO DataSets (Barrett et al., 2009, 2011), TomExpress (Zouine et al., 2017) or Tomato Expression Atlas (TEA) (Fernandez-Pozo et al., 2017; Shinozaki et al., 2018). Notably, these platforms allow the normalization of the signals from large datasets, enabling the comparative assessment of expression levels in different conditions (including different development stages). We collected the expression patterns of Class I TCP genes from four plant species: tomato (Solanum lycopersicum) and grapes (Vitis vinifera) fleshy berries, maize (Zea mays) kernel and Arabidopsis thaliana dry silique. The expression data were extracted from BAR-ePlant for maize and Arabidopsis, from TomExpress for tomato and from GEO DataSets for grape and used to build heatmaps on representative stages for each fruit or imbedded seeds (Figure 2). For tomato, the interpretation of these heatmaps was completed by the analysis of TEA and TomExpress data to highlight the tissues and fruit developmental stages exhibiting preferential expression (Supplementary Table S1). These expression data suggest the involvement of several Class I TCP genes in fruit development, either at early (growth, immature fruit) or late stages (ripening, from its initiation to senescence). Among the TCP isoforms highly expressed in fruits, some display high expression levels in both vegetative and reproductive tissues (stems, shoot, leaves, seeds and fruits) while others exhibit preferential expression during fruit development or ripening (Supplementary Table S1). Notably, the TCPs expressed at the early stages of fruit development are also expressed in vegetative tissues. For instance, the Arabidopsis AtTCP14 and AtTCP15 are highly expressed in the carpel and the silique, but also in germinating seeds, shoot apex and leaves (Figure 2A; Supplementary Table S1). The expression of SlTCP12, SlTCP15, SlTCP18 have been reported in tomato fruit ripening, with SlTCP12 and SlTCP18 expression being related to the actions of the master regulators of ripening RIN, Cnr and AP2a (Parapunova et al., 2014). Supporting these data, SlTCP12 and SlTCP18 display a preferential expression during fruit ripening (Figure 2B). By contrast, SlTCP11 and SlTCP15 genes are expressed in a wide variety of stages with a maximum in young fruits (3cm fruits in Figure 2B). Noteworthy, the TCP genes highly expressed in early fruit development exhibit a high expression in root, leaf, meristems and flower buds. More recently it was reported that SlTCP15 is a direct target of the kinase CDK8 and that the phosphorylated form is active in inducing pollen development (Xu et al., 2024). Altogether, these data suggest that SlTCP15 is involved in several developmental processes. In grape, Class I TCP genes are highly expressed in green berry (VvTCP7 and VvTCP12) and exhibit also high expression in leaves and vegetative buds, while VvTCP13 expression peaks only in ripe berries (Figure 2C). As for maize, the structure of the kernel with the fused pericarp and seed tegument did not allow to discriminate between early and late fruit development, although it appears that all the Class I TCPs expressed in endosperm growth display also expression in the pericarp (Figure 2D).
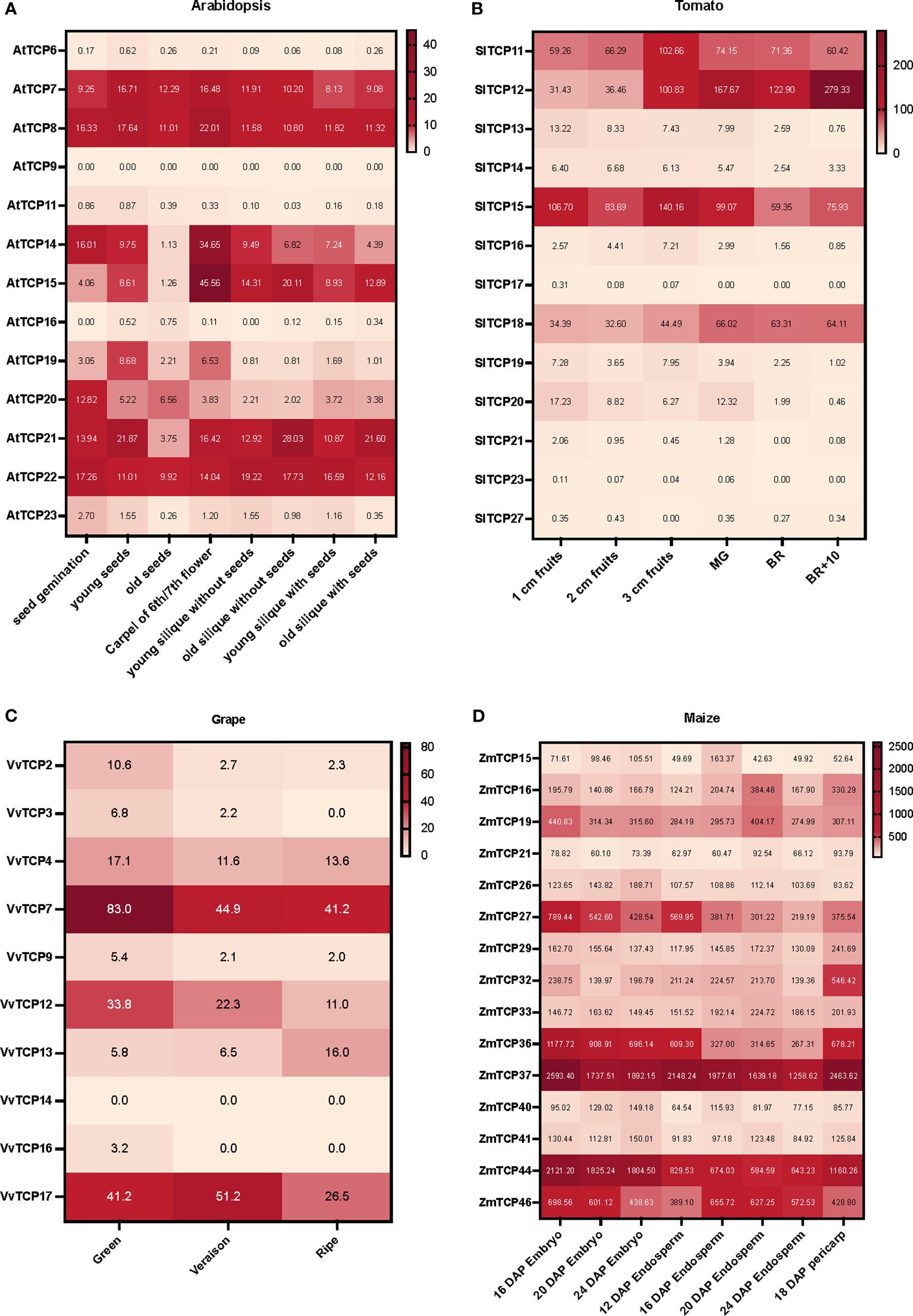
Figure 2 Heatmaps of transcript accumulation profiles of Class I TCP genes in (A) Arabidopsis (Arabidopsis thaliana), (B) tomato (Solanum lycopersicum), (C) grape (Vitis vinifera cv ‘Corvina’), and (D) maize (Zea mays). The transcript accumulation data were obtained from BAR-ePlant for maize and Arabidopsis, from TomExpress for tomato and from GEO DataSets for grapes. It was visualized as heat maps using Prism 9. The color scale represents the transcript levels with increased (red) or decreased (white) transcript abundance.
Highlighting the isoforms highly expressed in fruit development on the phylogenetic tree (Figure 1) failed to reveal any obvious correlation between the territories of expression and the clustering of the TCP isoforms within the different groups. Likewise, no obvious correlation was found when considering the expression studies reported in the literature (Supplementary Table S2). It is important to mention that the majority of Class I subclades are found in a wide range of land plant species, including the bryophyte Sphagnum fallax (Zhou et al., 2022), suggesting that unlike MADS-box transcription factors, the expression territory in reproductive organs is not a main driver of primary sequence evolution within Class I TCPs.
2 What is known about the functions of Class I TCP in fruit development?
The expression data presented in the first section enable to discriminate between the TCPs expressed in early fruit development encompassing the pre-mature stages and those expressed during the ripening phase. Remarkably, TCP genes expressed at early stages of fruit development display high expression level in vegetative tissues. In contrast, TCPs expressed at later stages of fruit development show greater tissue specificity and reach their expression peak during the ripening process.
Class I TCPs have been reported to act as modulators of cell proliferation, expansion and endoreplication, as component of hormone’s signaling cascades, often in response to environmental conditions, and also as regulators of leaf senescence or pigments accumulation (Viola et al., 2023). Very often, a single isoform can be involved in multiple mechanisms. For instance, AtTCP15 was shown to operate in seed germination (Resentini et al., 2015), in auxin-mediated cell elongation (Ferrero et al., 2021), in cotyledon opening (Alem et al., 2022), in stamen development (Gastaldi et al., 2023), in the regulation of potato tuber sprouting via the modulation of the abscisic acid/gibberelic acid balance or in the modulation of anthocyanin biosynthesis (Viola et al., 2016; Ding et al., 2022; Wang et al., 2022b). Considering that all these mechanisms are also operating during fleshy fruit growth, tissues’ differentiation and ripening, Class I TCPs can be viewed as potential regulators of different aspects of fruit development.
2.1 Class I TCP factors in early fruit development
Early fruit development can be seen as a series of processes starting with the flower fertilization and leading to an immature fruit. These sequential developmental processes encompass the fruit set, fruit growth, tissue differentiation and modification of cell metabolism and coincides with seed formation and maturation. In fleshy fruits, the growth and accumulation of metabolites often occur in carpels, but can also be in inner tissues such as locular tissue, placenta and columella in tomato, or in seed outgrowth (like the Litchi chinensis aril) or in flower receptacle (like strawberry). In tomato, early fruit development includes three physiological processes: fruit set, cell division and cell expansion (Gillaspy et al., 1993). Interestingly, the fruit growth relies on a combination of cell divisions and cell expansion, the latter being boosted by endoreplication leading to an increase of cell ploidy mainly in pericarp and locular tissue cells (Azzi et al., 2015; Tourdot et al., 2023).
Given that Class I TCPs act as modulators of cell division and elongation in a wide range of biological processes, it is tempting to speculate on the potential conservation of this regulatory roles for TCP expressed during fruit development. In Arabidopsis, AtTCP14, AtTCP15 and AtTCP8 are known to activate several cell-cycle and cell-proliferation genes while inhibiting endoreplication, since modifications of ploidy number has been reported in various mutant backgrounds (Kieffer et al., 2011; Li et al., 2012; Peng et al., 2015; Zhang et al., 2019a). Interestingly, these three genes correspond to three Class I TCP genes highly expressed in the carpel and the silique (Figure 2). Although altered silique architecture has been reported in AtTCP15 SRDX dominant negative lines, such as replum enlargement in plants (Uberti-Manassero et al., 2012), clear data demonstrating the impact of AtTCPs regulation on Arabidopsis silique development are still missing.
Evidence documenting the function of Class I TCPs in early fruit development is also limited in other species. In wheat (Triticum aestivum L.), the phenotype displayed by the Ta-TCP9-A mutants suggests the existence of an action of this gene on seed and fruit growth (Zhao et al., 2018). Indeed, two independent loss-of-function mutants carrying distinct mutations in TaTCP9-A gene display an increased grain length, width, and weight associated with an increase in the spike length. It is noteworthy that Ta-TCP9 belongs to the same group than AtTCP14 and AtTCP15 (Ta-TCP9 being close to OsTCP9, Figure 1), but its contribution to the regulation of the elongation processes occurring in the grain and the floral pieces still requires solid validation given that the mutant lines harbor several mutations in their genome in addition to the one affecting the TCP9 gene.
TCPs have also received attention regarding their role in cotton fiber development from the ovule surface. GhTCP14, a Class I TCP also belonging to the AtTCP14/AtTCP15 clade, was proposed to regulate fiber development, particularly during the initiation and elongation stages which coincides with early fruit ripening (Wang et al., 2013). GhTCP14 action seems to interfere with auxin signaling and may involve other Class I TCP genes co-expressed with GhTCP14, including GhTCP7a, 9b, 15a/b/c, 21, and 22 (Li et al., 2017). This action can also be paralleled with the presumed function of AtTCP14 and AtTCP15 in trichomes’ formation (Camoirano et al., 2020).
2.2 Class I TCP factors in fruit ripening
Mining the transcriptomic data highlighted the expression of different TCP genes at different stages of fruit ripening (Figure 2), showing that some isoforms are preferentially expressed during the ripening process of tomato and grape fruit (see section 1). While similarities between fruit ripening and leaf senescence mechanisms have been suggested by several authors (Picton et al., 2011; Gapper et al., 2013; Wang et al., 2017; Ma et al., 2018, 2019), it would, however, be appropriate to distinguish between the ripening process sensu stricto and the over-ripening phase (Gapper et al., 2013). Obviously, both ripening and senescence involve large gene expression and metabolism reprogramming. A striking similarity between the two processes is the important role of the plant hormone ethylene, known to be a key regulator of the two processes (Picton et al., 2011; Sharma et al., 2021). Moreover, ABA, known as a plant senescence hormone, has been documented to impact fruit ripening (Ye et al., 2017; Kou et al., 2021; Gupta et al., 2022). At the cellular level, ripening and senescence share several common processes like chlorophyll breakdown and cell wall modifications (Forlani et al., 2019; Ling et al., 2021). In addition, several senescence-associated proteins are also associated with fruit ripening, including type 2C phosphatase SlPP2C (Jiang et al., 2023), isopentenyl transferase SlIPT4 (Zhang et al., 2018) and the recently characterized VviNAC60 in grape (D'Inca et al., 2023).
In the last decade, several studies have shed light on the regulatory effect exerted by TCP on leaf senescence, indicating that different TCP isoforms can act either as negative or positive regulators. For instance, overexpression of chrysanthemum gene CmTCP14 in Arabidopsis results in delayed leaf senescence (Zhang et al., 2017), and AtTCP20 inhibits the expression of the lipoxygenase AtLOX2, encoding a key enzyme in jasmonic acid biosynthesis, thus leading to impaired leaf senescence (Danisman et al., 2012). Similarly, cabbage BrTCP21 delays leaf senescence by influencing the GA biosynthetic pathway (Xiao et al., 2019). By contrast, BrTCP7 binds to the promoter regions of BrOPR3 and BrRCCR, involved in jasmonic acid (JA) biosynthesis and chlorophyll breakdown respectively, thus suggesting that this TCP acts as a positive regulator of leaf senescence (Xu et al., 2019).
The study of TCP expression in fruit peach (Prunus persicum) development and ripening (Guo et al., 2018) highlighted the potential contribution of PpTCP.A2. Indeed, PpTCP.A2 is expressed in fruit pre-ripening and its VIGS-induced silencing in peach fruit reveals an increase in ethylene production coinciding with an upregulation of PpACS1, encoding the enzyme that catalyzes the synthesis of ACC, the direct precursor of ethylene. The study suggested that the action of PpTCP.A2 is reminiscent of AtTCP20, CmTCP14 or BrTCP21 action as negative regulator of leaf senescence (see above).
In banana (Musa acuminata), several TCP factors (MaTCP15, MaTCP19, MaTCP20, and MaTCP22) were found to be ethylene-inducible (Song et al., 2018). Interestingly, these authors suggested that MaTCP20 could activate the promoters of two xyloglucan endotransglucosylase/hydrolases (MaXTH10 and MaXTH11) since they contain the TCP-conserved (T/C) GGNCCCA regulatory elements and they can effectively bind MaTCP20 protein as shown by EMSA and dual-luciferase assays. The data also suggest that TCP may impact fruit quality, since XTHs are involved in cell wall modification processes that accompany fruit softening.
Fruit color, an important quality trait, is controlled by TCP as suggested by different studies on anthocyanin accumulation. The overexpression of FaTCP11 in strawberries (Fragaria × ananassa), a gene first identified by expression correlation network analysis on the transcriptomes of red ripe fruits from two parental lines and their progeny (Pillet et al., 2015), leads to enhanced expression of Leucoanthocyanidin Reductase (LAR) and Flavonone 3’-Hydroxylase (F3’H). This finding suggests that FaTCP11 is critical in regulating the flavan-3-ols accumulation, thereby exerting a significant impact on strawberry quality. More recently the woodland strawberry FvTCP9 gene was also shown to regulate positively anthocyanin accumulation using VIGS-induced RNAi silencing or gene overexpression (Xie et al., 2020). FvTCP9 expression level is correlated with anthocyanin production, with the expression of phenolic-metabolism related genes and with that of other ripening-related genes involved in fruit softening or sugar accumulation.
Further sustaining the involvement of TCPs in the regulation of pigment accumulation in fleshy fruit species, the MdTCP46 gene in apple (Malus domestica), has been reported to promote anthocyanin biosynthesis in response to high light exposure (An et al., 2020; You et al., 2023). Accordingly, overexpression of MdTCP46 in fruit peel via infiltration effectively leads to a dramatic increase of anthocyanin production in fruit under high light. The same study demonstrated that MdTCP46 interacts with MdMYB1 and MdBT2 to form a regulatory module controlling anthocyanin accumulation under different light intensities. Interestingly, this regulatory module is also connected to hormone signaling cascades since MdTCP46 was shown to interact with the ABA signaling effectors MdABI5 or MdRGL2a DELLA protein while conversely, its expression is inhibited by GA treatment (Liu et al., 2022b; You et al., 2023).
2.3 TCP regulation at the crossroad of hormonal signaling
The complex process of fruit development, from the fertilized flower to the final ripening phase, is regulated by an intricate interplay between multiple plant hormones. At early stages of fruit growth, gibberellic acid (GA) and auxin have synergistic action in promoting cell division and expansion, thereby regulating fruit development and enlargement after fertilization (Serrani et al., 2008; He and Yamamuro, 2022). Later, the transition from immature stage to ripening stage is characterized by shifts in phytohormone profiles such as auxin, ethylene and abscisic acid (ABA) accompanied by transcriptional reprogramming (Forlani et al., 2019). Several studies suggest that the hormonal regulation of the expression of TCP genes is a frequent and widespread phenomenon in plants despite the fact that the physiological significance of these TCPs remains obscure to date.
In tomato, SlTCP12, SlTCP15 and SlTCP18 genes that are expressed in fruit ripening display Cis-elements in their promoter regions shown to be typical binding sites of the RIN master regulator of fruit ripening as evidenced by yeast one-hybrid experiments (Parapunova et al., 2014).There are also reports supporting that ethylene acts as a direct or indirect regulator of TCPs expressed during fruit ripening. Indeed, co-expression studies demonstrated recently the existence of a strong correlation between the expression of SlTCP15 and the ethylene-responsive transcription factor 4 SlERF4 while a second correlation was observed between SlTCP18 and Auxin-responsive Factor 5 (SlARF5), suggesting an interaction with multiple hormones signaling effectors (Edris et al., 2023).
In grape and blueberry, the expression patterns of TCP genes in fruits treated with different hormones also suggest a strong link between Class I TCP genes and hormone signaling. For instance, ABA treatment of grape berry at the veraison stage enhances VvTCP14 and VvTCP16 transcript levels while reducing that of VvTCPs 6/7/15/17 (Jiu et al., 2019). On the other hand, in blueberry, expression of VcTCP19/38/43/49/61 is sensitive to methyl-jasmonate treatment (Li et al., 2021).
3 Concluding remarks
The TCP family received less attention than many other transcriptions factor families like MADS, MYB, AP2-ERF, NAC with regard to their role in fruit biology. Yet, the increasing number of studies on different fruit species and the massive release of transcriptomic data produced by NGS technologies provided clues pointing to the putative involvement of Class I TCP transcription factors as active regulators of fruit development and ripening. While the puzzle is far from complete, the expression data reviewed in this manuscript support the idea that TCP genes expressed at early stages of fruit development, including growth and differentiation, correspond to those already shown to be involved in several growth or differentiation processes in vegetative organs. The exact nature of the processes involving TCP in fruit growth remains to be elucidated. Nevertheless, based on the current model related to germination or internode elongation, it is tempting to speculate on a potential role of TCPs from different phylogenetic groups on cell division, expansion or endoreplication. Among the TCP genes expressed during fleshy fruit development several display preferential expression at the ripening phase. While these genes still await functional characterization, studies in strawberry and banana suggest that TCPs can impact various determinants of fruit quality such as pigment accumulation or modification of texture associated with ripening. In addition, various studies suggest a link between TCPs and hormone signaling, notably ethylene, ABA and auxin, three phytohormones known as key regulators in the frame of fruit biology. Addressing the functional characterization of TCP genes in fruit will be a key challenge in the future, as it may open new avenues for improving fruit quality, and provide novel targets for breeding strategies or biotechnical engineering.
Author contributions
YG: Conceptualization, Writing – original draft, Writing – review & editing. FR: Conceptualization, Formal Analysis, Writing – review & editing. LZ: Conceptualization, Formal Analysis, Validation, Writing – review & editing. JP: Formal Analysis, Validation, Writing – review & editing. MB: Conceptualization, Formal Analysis, Validation, Writing – review & editing, Writing – original draft. BV: Conceptualization, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. YG was supported by the Chinese Scholarship Council. The research was supported by the European Union grants H2020 HARNESSTOM 101000716, the Labex TULIP ANR-10-LABX-41 and by OxyFruit ANR. Project also supported by graduate scientific research and innovation foundation of Chongqing, China (Grant No. CYS18064) and support from the Institut National Polytechnique de Tououse.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1411341/full#supplementary-material
Supplementary Table 1 | Inventory, classification and preferential expression of Class I TCP genes in Arabidopsis, tomato, grape and maize. Class I TCP proteins were classified into four groups based on protein sequences from Arabidopsis thaliana, Solanum lycopersicum, Vitis vinifera, and Zea mays. Expression data for each protein group were extracted from the following databases: BAR-ePlant, TEA, and TomExpress for Solanum lycopersicum; BAR-ePlant for Arabidopsis thaliana and Zea may; GEO DataSets for Vitis vinifera. Protein domains were predicted using the MEME software.
Supplementary Table 2 | Classification of TCP isoforms described in the literature (listed in ). The categorization was deduced from a phylogenetic analysis performed on the MEGAX software on amino acids sequence using the MUSCLE alignment and the Neighbor Joining algorithm.
References
Alem, A. L., Ariel, F. D., Cho, Y., Hong, J. C., Gonzalez, D. H., Viola, I. L. (2022). TCP15 interacts with GOLDEN2-LIKE 1 to control cotyledon opening in Arabidopsis. Plant J. 110, 748–763. doi: 10.1111/tpj.15701
Allan, A. C., Espley, R. V. (2018). MYBs drive novel consumer traits in fruits and vegetables. Trends Plant Sci. 23, 693–705. doi: 10.1016/j.tplants.2018.06.001
An, J. P., Liu, Y. J., Zhang, X. W., Bi, S. Q., Wang, X. F., You, C. X., et al. (2020). Dynamic regulation of anthocyanin biosynthesis at different light intensities by the BT2-TCP46-MYB1 module in apple. J. Exp. Bot. 71, 3094–3109. doi: 10.1093/jxb/eraa056
Azzi, L., Deluche, C., Gevaudant, F., Frangne, N., Delmas, F., Hernould, M., et al. (2015). Fruit growth-related genes in tomato. J. Exp. Bot. 66, 1075–1086. doi: 10.1093/jxb/eru527
Bailey, T. L., Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in bipolymers. UCSD Tech. Rep. CS94–351.
Barrett, T., Troup, D. B., Wilhite, S. E., Ledoux, P., Evangelista, C., Kim, I. F., et al. (2011). NCBI GEO: archive for functional genomics data sets-10 years on. Nucleic Acids Res. 39, D1005–D1010. doi: 10.1093/nar/gkq1184
Barrett, T., Troup, D. B., Wilhite, S. E., Ledoux, P., Rudnev, D., Evangelista, C., et al. (2009). NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 37, D885–D890. doi: 10.1093/nar/gkn764
Bresso, E. G., Chorostecki, U., Rodriguez, R. E., Palatnik, J. F., Schommer, C. (2018). Spatial control of gene expression by miR319-regulated TCP transcription factors in leaf development. Plant Physiol. 176, 1694–1708. doi: 10.1104/pp.17.00823
Camoirano, A., Arce, A. L., Ariel, F. D., Alem, A. L., Gonzalez, D. H., Viola, I. L. (2020). Class I TCP transcription factors regulate trichome branching and cuticle development in Arabidopsis. J. Exp. Bot. 71, 5438–5453. doi: 10.1093/jxb/eraa257
Chirinos, X., Ying, S., Rodrigues, M. A., Maza, E., Djari, A., Hu, G., et al. (2023). Transition to ripening in tomato requires hormone-controlled genetic reprogramming initiated in gel tissue. Plant Physiol. 191, 610–625. doi: 10.1093/plphys/kiac464
Cubas, P., Lauter, N., Doebley, J., Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313X.1999.00444.x
D'Inca, E., Foresti, C., Orduna, L., Amato, A., Vandelle, E., Santiago, A., et al. (2023). The transcription factor VviNAC60 regulates senescence- and ripening-related processes in grapevine. Plant Physiol. 192, 1928–1946. doi: 10.1093/plphys/kiad050
Danisman, S. (2016). TCP transcription factors at the interface between environmental challenges and the plant's growth responses. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01930
Danisman, S., van der Wal, F., Dhondt, S., Waites, R., de Folter, S., Bimbo, A., et al. (2012). Arabidopsis Class I and Class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. doi: 10.1104/pp.112.200303
Dhaka, N., Bhardwaj, V., Sharma, M. K., Sharma, R. (2017). Evolving tale of TCPs: new paradigms and old lacunae. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00479
Ding, A. M., Xu, C. T., Xie, Q., Zhang, M. J., Yan, N., Dai, C. B., et al. (2022). ERF4 interacts with and antagonizes TCP15 in regulating endoreduplication and cell growth in Arabidopsis. J. Integr. Plant Biol. 64, 1673–1689. doi: 10.1111/jipb.13323
Dong, Z., Hao, Y., Zhao, Y., Tang, W., Wang, X., Li, J., et al. (2024). Genome-wide analysis of the TCP transcription factor gene family in pepper (Capsicum annuum L.). Plants-Basel 13, 641. doi: 10.3390/plants13050641
Edris, S., Abulfaraj, A. A., Makki, R. M., Abo-Aba, S., Algandaby, M. M., Sabir, J., et al. (2023). Early fruit development regulation-related genes concordantly expressed with TCP transcription factors in tomato (Solanum lycopersicum). Curr. Issues Mol. Biol. 45, 2372–2380. doi: 10.3390/cimb45030153
Fernandez-Pozo, N., Zheng, Y., Snyder, S. I., Nicolas, P., Shinozaki, Y., Fei, Z., et al. (2017). The tomato expression atlas. Bioinformatics 33, 2397–2398. doi: 10.1093/bioinformatics/btx190
Ferrero, L. V., Gastaldi, V., Ariel, F. D., Viola, I. L., Gonzalez, D. H. (2021). Class I TCP proteins TCP14 and TCP15 are required for elongation and gene expression responses to auxin. Plant Mol. Biol. 105, 147–159. doi: 10.1007/s11103-020-01075-y
Forlani, S., Masiero, S., Mizzotti, C. (2019). Fruit ripening: the role of hormones, cell wall modifications, and their relationship with pathogens. J. Exp. Bot. 70, 2993–3006. doi: 10.1093/jxb/erz112
Forlani, S., Mizzotti, C., Masiero, S. (2021). The NAC side of the fruit: tuning of fruit development and maturation. BMC Plant Biol. 21, 238. doi: 10.1186/s12870-021-03029-y
Gapper, N. E., McQuinn, R. P., Giovannoni, J. J. (2013). Molecular and genetic regulation of fruit ripening. Plant Mol. Biol. 82, 575–591. doi: 10.1007/s11103-013-0050-3
Gastaldi, V., Alem, A. L., Mansilla, N., Ariel, F. D., Viola, I. L., Lucero, L. E., et al. (2023). BREVIPEDICELLUS/KNAT1 targets TCP15 to modulate filament elongation during Arabidopsis late stamen development. Plant Physiol. 191, 29–34. doi: 10.1093/plphys/kiac502
Gillaspy, G., Ben-David, H., Gruissem, W. (1993). Fruits: a developmental perspective. Plant Cell 5, 1439–1451. doi: 10.2307/3869794
Guo, Z.-H., Shu, W.-S., Cheng, H.-Y., Wang, G.-M., Qi, K.-J., Zhang, S.-L., et al. (2018). Expression analysis of TCP genes in peach reveals an involvement of PpTCP.A2 in ethylene biosynthesis during fruit ripening. Plant Mol. Biol. Rep. 36, 588–595. doi: 10.1007/s11105-018-1105-z
Gupta, K., Wani, S. H., Razzaq, A., Skalicky, M., Samantara, K., Gupta, S., et al. (2022). Abscisic acid: role in fruit development and ripening. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.817500
Hao, Y., Hu, G., Breitel, D., Liu, M., Mila, I., Frasse, P., et al. (2015). Auxin response factor SlARF2 is an essential component of the regulatory mechanism controlling fruit ripening in tomato. PloS Genet. 11, e1005649. doi: 10.1371/journal.pgen.1005649
He, H., Yamamuro, C. (2022). Interplays between auxin and GA signaling coordinate early fruit development. Hortic. Res. 9, uhab078. doi: 10.1093/hr/uhab078
Huang, B., Hu, G., Wang, K., Frasse, P., Maza, E., Djari, A., et al. (2021). Interaction of two MADS-box genes leads to growth phenotype divergence of all-flesh type of tomatoes. Nat. Commun. 12, 6892. doi: 10.1038/s41467-021-27117-7
Jiang, Y., Shi, Z., Liu, W., Qi, M., Xu, T., Wu, Y., et al. (2023). Silencing SlPP2C expression delayed plant senescence and fruit ripening in tomato. Physiol. Plant 175, e13925. doi: 10.1111/ppl.13925
Jiu, S., Xu, Y., Wang, J., Wang, L., Wang, S., Ma, C., et al. (2019). Genome-wide identification, characterization, and transcript analysis of the TCP transcription factors in Vitis vinifera. Front. Genet. 10. doi: 10.3389/fgene.2019.01276
Kieffer, M., Master, V., Waites, R., Davies, B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68, 147–158. doi: 10.1111/j.1365-313X.2011.04674.x
Kou, X., Zhou, J., Wu, C. E., Yang, S., Liu, Y., Chai, L., et al. (2021). The interplay between ABA/ethylene and NAC TFs in tomato fruit ripening: a review. Plant Mol. Biol. 106, 223–238. doi: 10.1007/s11103-021-01128-w
Lan, J., Qin, G. (2020). The regulation of CIN-like TCP transcription factors. Int. J. Mol. Sci. 21, 4498. doi: 10.3390/ijms21124498
Lan, J., Wang, N., Wang, Y., Jiang, Y., Yu, H., Cao, X., et al. (2023). Arabidopsis TCP4 transcription factor inhibits high temperature-induced homeotic conversion of ovules. Nat. Commun. 14, 5673. doi: 10.1038/s41467-023-41416-1
Leng, X., Wei, H., Xu, X., Ghuge, S. A., Jia, D., Liu, G., et al. (2019). Genome-wide identification and transcript analysis of TCP transcription factors in grapevine. BMC Genomics 20, 786. doi: 10.1186/s12864-019-6159-2
Li, Y., An, S., Cheng, Q., Zong, Y., Chen, W., Guo, W., et al. (2021). Analysis of evolution, expression and genetic transformation of TCP transcription factors in blueberry reveal that VcTCP18 negatively regulates the release of flower bud dormancy. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.697609
Li, Z. Y., Li, B., Dong, A. W. (2012). The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol. Plant 5, 270–280. doi: 10.1093/mp/ssr086
Li, D., Li, H., Feng, H., Qi, P., Wu, Z. (2024). Unveiling kiwifruit TCP genes: evolution, functions, and expression insights. Plant Signal Behav. 19, 2338985. doi: 10.1080/15592324.2024.2338985
Li, W., Li, D. D., Han, L. H., Tao, M., Hu, Q. Q., Wu, W. Y., et al. (2017). Genome-wide identification and characterization of TCP transcription factor genes in upland cotton (Gossypium hirsutum). Sci. Rep. 7, 10118. doi: 10.1038/s41598–017-10609–2
Li, D., Tang, X., Dong, Y., Wang, Y., Shi, S., Li, S., et al. (2022). Comparative genomic investigation of TCP gene family in eggplant (Solanum melongena L.) and expression analysis under divergent treatments. Plant Cell Rep. 41, 2213–2228. doi: 10.1007/s00299-022-02918-2
Ling, Q., Sadali, N. M., Soufi, Z., Zhou, Y., Huang, B., Zeng, Y., et al. (2021). The chloroplast-associated protein degradation pathway controls chromoplast development and fruit ripening in tomato. Nat. Plants 7, 655–666. doi: 10.1038/s41477-021-00916-y
Liu, Y. J., An, J. P., Gao, N., Wang, X., Chen, X. X., Wang, X. F., et al. (2022b). MdTCP46 interacts with MdABI5 to negatively regulate ABA signalling and drought response in apple. Plant Cell Environ. 45, 3233–3248. doi: 10.1111/pce.14429
Liu, M., Li, Z., Zhang, Y., Gao, J. (2020). Role of ethylene response factors (ERFs) in fruit ripening. Food Qual Saf-Oxford 4, 15–20. doi: 10.1093/fqsafe/fyz042
Liu, D.-H., Luo, Y., Han, H., Liu, Y.-Z., Alam, S. M., Zhao, H.-X., et al. (2022a). Genome-wide analysis of citrus TCP transcription factors and their responses to abiotic stresses. BMC Plant Biol. 22, 325. doi: 10.1186/s12870-022-03709-3
Ma, X., Balazadeh, S., Mueller-Roeber, B. (2019). Tomato fruit ripening factor NOR controls leaf senescence. J. Exp. Bot. 70, 2727–2740. doi: 10.1093/jxb/erz098
Ma, X., Zhang, Y., Tureckova, V., Xue, G. P., Fernie, A. R., Mueller-Roeber, B., et al. (2018). The NAC transcription factor SlNAP2 regulates leaf senescence and fruit yield in tomato. Plant Physiol. 177, 1286–1302. doi: 10.1104/pp.18.00292
Nicolas, M., Cubas, P. (2016). TCP factors: new kids on the signaling block. Curr. Opin. Plant Biol. 33, 33–41. doi: 10.1016/j.pbi.2016.05.006
Parapunova, V., Busscher, M., Busscher-Lange, J., Lammers, M., Karlova, R., Bovy, A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 14, 157. doi: 10.1186/1471-2229-14-157
Peng, Y., Chen, L., Lu, Y., Wu, Y., Dumenil, J., Zhu, Z., et al. (2015). The ubiquitin receptors DA1, DAR1, and DAR2 redundantly regulate endoreduplication by modulating the stability of TCP14/15 in Arabidopsis. Plant Cell 27, 649–662. doi: 10.1105/tpc.114.132274
Picton, S., Barton, S. L., Bouzayen, M., Hamilton, A. J., Grierson, D. (2011). Altered fruit ripening and leaf senescence in tomatoes expressing an antisense ethylene-forming enzyme transgene. Plant J. 3, 469–481. doi: 10.1111/j.1365–313X.1993.tb00167.x
Pillet, J., Yu, H. W., Chambers, A. H., Whitaker, V. M., Folta, K. M. (2015). Identification of candidate flavonoid pathway genes using transcriptome correlation network analysis in ripe strawberry (Fragaria x ananassa) fruits. J. Exp. Bot. 66, 4455–4467. doi: 10.1093/jxb/erv205
Rath, M., Challa, K. R., Sarvepalli, K., Nath, U. (2022). CINCINNATA-like TCP transcription factors in cell growth -an expanding portfolio. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.825341
Ren, J., Yang, L., Cao, R., Wang, Y., Zhang, C., Yu, X., et al. (2023). Integrated metabolome and transcriptome analysis provides new insights into the glossy graft cucumber fruit (Cucumis sativus L.). Int. J. Mol. Sci. 24, 12147. doi: 10.3390/ijms241512147
Resentini, F., Felipo-Benavent, A., Colombo, L., Blazquez, M. A., Alabadi, D., Masiero, S. (2015). TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant 8, 482–485. doi: 10.1016/j.molp.2014.11.018
Rizwan, H. M., Shaozhong, F., Li, X., Bilal Arshad, M., Yousef, A. F., Chenglong, Y., et al. (2022). Genome-wide identification and expression profiling of KCS gene family in passion fruit (Passiflora edulis) under fusarium kyushuense and drought stress conditions. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.872263
Sarvepalli, K., Nath, U. (2018). CIN-TCP transcription factors: transiting cell proliferation in plants. IUBMB Life 70, 718–731. doi: 10.1002/iub.1874
Serrani, J. C., Ruiz-Rivero, O., Fos, M., Garcia-Martinez, J. L. (2008). Auxin-induced fruit-set in tomato is mediated in part by gibberellins. Plant J. 56, 922–934. doi: 10.1111/j.1365-313X.2008.03654.x
Seymour, G. B., Ostergaard, L., Chapman, N. H., Knapp, S., Martin, C. (2013). Fruit development and ripening. Annu. Rev. Plant Biol. 64, 219–241. doi: 10.1146/annurev-arplant-050312-120057
Sharma, K., Gupta, S., Sarma, S., Rai, M., Sreelakshmi, Y., Sharma, R. (2021). Mutations in tomato 1-aminocyclopropane carboxylic acid synthase2 uncover its role in development beside fruit ripening. Plant J. 106, 95–112. doi: 10.1111/tpj.15148
Shinozaki, Y., Nicolas, P., Fernandez-Pozo, N., Ma, Q., Evanich, D. J., Shi, Y., et al. (2018). High-resolution spatiotemporal transcriptome mapping of tomato fruit development and ripening. Nat. Commun. 9, 364. doi: 10.1038/s41467-017-02782-9
Song, C. B., Shan, W., Yang, Y. Y., Tan, X. L., Fan, Z. Q., Chen, J. Y., et al. (2018). Heterodimerization of MaTCP proteins modulates the transcription of MaXTH10/11 genes during banana fruit ripening. Biochim. Biophys. Acta Gene Regul. Mech. 1861, 613–622. doi: 10.1016/j.bbagrm.2018.06.005
Sun, L., Zou, X., Jiang, M., Wu, X., Chen, Y., Wang, Q., et al. (2020). The crystal structure of the TCP domain of PCF6 in Oryza sativa L. reveals an RHH-like fold. FEBS Lett. 594, 1296–1306. doi: 10.1002/1873-3468.13727
Tabarelli, M., Malnoy, M., Janik, K. (2022). Chasing consistency: an update of the TCP gene family of Malus x Domestica. Genes-Basel 13, 1696. doi: 10.3390/genes13101696
Tourdot, E., Mauxion, J. P., Gonzalez, N., Chevalier, C. (2023). Endoreduplication in plant organogenesis: a means to boost fruit growth. J. Exp. Bot. 74, 6269–6284. doi: 10.1093/jxb/erad235
Uberti-Manassero, N. G., Lucero, L. E., Viola, I. L., Vegetti, A. C., Gonzalez, D. H. (2012). The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J. Exp. Bot. 63, 809–823. doi: 10.1093/jxb/err305
Viola, I. L., Alem, A. L., Jure, R. M., Gonzalez, D. H. (2023). Physiological roles and mechanisms of action of Class I TCP transcription factors. Int. J. Mol. Sci. 24, 5437. doi: 10.3390/ijms24065437
Viola, I. L., Camoirano, A., Gonzalez, D. H. (2016). Redox-dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in Arabidopsis. Plant Physiol. 170, 74–85. doi: 10.1104/pp.15.01016
Viola, I. L., Gonzalez, D. H. (2023). TCP transcription factors in plant reproductive development: juggling multiple roles. Biomolecules 13, 750. doi: 10.3390/biom13050750
Vrebalov, J., Pan, I. L., Arroyo, A. J., McQuinn, R., Chung, M., Poole, M., et al. (2009). Fleshy fruit expansion and ripening are regulated by the Tomato SHATTERPROOF gene TAGL1. Plant Cell 21, 3041–3062. doi: 10.1105/tpc.109.066936
Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., et al. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346. doi: 10.1126/science.1068181
Waese, J., Fan, J., Pasha, A., Yu, H., Fucile, G., Shi, R., et al. (2017). ePlant: visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell 29, 1806–1821. doi: 10.1105/tpc.17.00073
Wang, N., Guo, T., Wang, P., Sun, X., Shao, Y., Liang, B., et al. (2017). Functional analysis of apple MhYTP1 and MhYTP2 genes in leaf senescence and fruit ripening. Sci. Hortic-amsterdam 221, 23–32. doi: 10.1016/j.scienta.2017.04.018
Wang, J., Wang, Z., Jia, C., Miao, H., Zhang, J., Liu, J., et al. (2022a). Genome-wide identification and transcript analysis of TCP gene family in banana (Musa acuminata L.). Biochem. Genet. 60, 204–222. doi: 10.1007/s10528-021-10100-8
Wang, Y., Wang, N., Lan, J., Pan, Y., Jiang, Y., Wu, Y., et al. (2024). Arabidopsis transcription factor TCP4 controls the identity of the apical gynoecium. Plant Cell, koae107. doi: 10.1093/plcell/koae107
Wang, K., Zhang, N., Fu, X., Zhang, H., Liu, S., Pu, X., et al. (2022b). StTCP15 regulates potato tuber sprouting by modulating the dynamic balance between abscisic acid and gibberellic acid. Front. Plant Sci. 13, 1009552. doi: 10.3389/fpls.2022.1009552
Wang, M. Y., Zhao, P. M., Cheng, H. Q., Han, L. B., Wu, X. M., Gao, P., et al. (2013). The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol. 162, 1669–1680. doi: 10.1104/pp.113.215673
Wei, W., Hu, Y., Cui, M. Y., Han, Y. T., Gao, K., Feng, J. Y. (2016). Identification and transcript analysis of the TCP transcription factors in the diploid woodland strawberry Fragaria vesca. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01937
Wen, H., Chen, Y., Du, H., Zhang, L., Zhang, K., He, H., et al. (2020). Genome-wide identification and characterization of the TCP gene family in cucumber (Cucumis sativus L.) and their transcriptional responses to different treatments. Genes-Basel 11, 1379. doi: 10.3390/genes11111379
Xiao, X. M., Xu, Y. M., Zeng, Z. X., Tan, X. L., Liu, Z. L., Chen, J. W., et al. (2019). Activation of the transcription of BrGA20ox3 by a BrTCP21 transcription factor is associated with gibberellin-delayed leaf senescence in Chinese flowering cabbage during storage. Int. J. Mol. Sci. 20, 3860. doi: 10.3390/ijms20163860
Xie, Y. G., Ma, Y. Y., Bi, P. P., Wei, W., Liu, J., Hu, Y., et al. (2020). Transcription factor FvTCP9 promotes strawberry fruit ripening by regulating the biosynthesis of abscisic acid and anthocyanins. Plant Physiol. Biochem. 146, 374–383. doi: 10.1016/j.plaphy.2019.11.004
Xu, R., Chong, L., Zhu, Y. (2024). Mediator kinase subunit CDK8 phosphorylates transcription factor TCP15 during tomato pollen development. Plant Physiol. 195, 865–878. doi: 10.1093/plphys/kiae079
Xu, R., Sun, P., Jia, F., Lu, L., Li, Y., Zhang, S., et al. (2014). Genomewide analysis of TCP transcription factor gene family in Malus domestica. J. Genet. 93, 733–746. doi: 10.1007/s12041-014-0446-0
Xu, Y. M., Xiao, X. M., Zeng, Z. X., Tan, X. L., Liu, Z. L., Chen, J. W., et al. (2019). BrTCP7 transcription factor is associated with MeJA-promoted leaf senescence by activating the expression of BrOPR3 and BrRCCR. Int. J. Mol. Sci. 20, 3963. doi: 10.3390/ijms20163963
Ye, Y., Zhou, L., Liu, X., Liu, H., Li, D., Cao, M., et al. (2017). A novel chemical inhibitor of ABA signaling targets all ABA receptors. Plant Physiol. 173, 2356–2369. doi: 10.1104/pp.16.01862
You, C., Han, Y., An, J. (2023). The RGL2a-TCP46-MYB1 module regulates GA-mediated anthocyanin biosynthesis in apple. Fruit Res. 3, 21. doi: 10.48130/FruRes-2023-0021
Zhang, Y., Li, Z., Tu, Y., Cheng, W., Yang, Y. (2018). Tomato (Solanum lycopersicum) SlIPT4, encoding an isopentenyltransferase, is involved in leaf senescence and lycopene biosynthesis during fruit ripening. BMC Plant Biol. 18, 107. doi: 10.1186/s12870-018-1327-0
Zhang, T., Qu, Y., Wang, H., Wang, J., Song, A., Hu, Y., et al. (2017). The heterologous expression of a chrysanthemum TCP-P transcription factor CmTCP14 suppresses organ size and delays senescence in Arabidopsis thaliana. Plant Physiol. Biochem. 115, 239–248. doi: 10.1016/j.plaphy.2017.03.026
Zhang, J., Wang, Y., Naeem, M., Zhu, M., Li, J., Yu, X., et al. (2019b). An AGAMOUS MADS-box protein, SlMBP3, regulates the speed of placenta liquefaction and controls seed formation in tomato. J. Exp. Bot. 70, 909–924. doi: 10.1093/jxb/ery418
Zhang, Y., Xu, Y. P., Nie, J. K., Chen, H., Qin, G., Wang, B., et al. (2023). DNA-TCP complex structures reveal a unique recognition mechanism for TCP transcription factor families. Nucleic Acids Res. 51, 434–448. doi: 10.1093/nar/gkac1171
Zhang, G., Zhao, H., Zhang, C., Li, X., Lyu, Y., Qi, D., et al. (2019a). TCP7 functions redundantly with several Class I TCPs and regulates endoreplication in Arabidopsis. J. Integr. Plant Biol. 61, 1151–1170. doi: 10.1111/jipb.12749
Zhao, Y., Su, X., Wang, X., Wang, M., Chi, X., Aamir Manzoor, M., et al. (2021). Comparative genomic analysis of TCP genes in six rosaceae species and expression pattern analysis in Pyrus bretschneideri. Front. Genet. 12. doi: 10.3389/fgene.2021.669959
Zhao, J., Zhai, Z., Li, Y., Geng, S., Song, G., Guan, J., et al. (2018). Genome-wide identification and expression profiling of the TCP family genes in spike and grain development of wheat (Triticum aestivum L.). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01282
Zhou, H., Hwarari, D., Ma, H., Xu, H., Yang, L., Luo, Y. (2022). Genomic survey of TCP transcription factors in plants: phylogenomics, evolution and their biology. Front. Genet. 13. doi: 10.3389/fgene.2022.1060546
Zhou, Y., Xu, Z., Zhao, K., Yang, W., Cheng, T., Wang, J., et al. (2016). Genome-wide identification, characterization and expression analysis of the TCP gene family in Prunus mume. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01301
Zou, X., Sun, H. (2023). DOF transcription factors: specific regulators of plant biological processes. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1044918
Keywords: teosinte branched, cycloidea, proliferating cell factor (TCP), class I TCP, transcription factor, fruit development, fruit ripening
Citation: Gao Y, Regad F, Li Z, Pirrello J, Bouzayen M and Van Der Rest B (2024) Class I TCP in fruit development: much more than growth. Front. Plant Sci. 15:1411341. doi: 10.3389/fpls.2024.1411341
Received: 02 April 2024; Accepted: 13 May 2024;
Published: 28 May 2024.
Edited by:
Claudio Bonghi, University of Padua, ItalyReviewed by:
Livio Trainotti, University of Padua, ItalyNeftali Ochoa-Alejo, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, Mexico
Copyright © 2024 Gao, Regad, Li, Pirrello, Bouzayen and Van Der Rest. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Benoît Van Der Rest, benoit.van-der-rest@toulouse-inp.fr
 Yushuo Gao
Yushuo Gao Farid Regad
Farid Regad Zhengguo Li
Zhengguo Li Julien Pirrello1
Julien Pirrello1 Benoît Van Der Rest
Benoît Van Der Rest