- 1The Key Laboratory of Medicinal Plant Biology of Yunnan Province, National & Local Joint Engineering Research Center on Germplasms Innovation & Utilization of Chinese Medicinal Materials in Southwest China, Yunnan Agricultural University, Kunming, China
- 2Yunnan Characteristic Plant Extraction Laboratory, Kunming, Yunnan, China
The transcription factors of WRKY genes play essential roles in plant growth, stress responses, and metabolite biosynthesis. Erigeron breviscapus, a traditional Chinese herb, is abundant in flavonoids and has been used for centuries to treat cardiovascular and cerebrovascular diseases. However, the WRKY transcription factors that regulate flavonoid biosynthesis in E. breviscapus remain unknown. In this study, a total of 75 EbWRKY transcription factors were predicted through comprehensive genome-wide characterization of E. breviscapus and the chromosomal localization of each EbWRKY gene was investigated. RNA sequencing revealed transient responses of 74 predicted EbWRKY genes to exogenous abscisic acid (ABA), salicylic acid (SA), and gibberellin 3 (GA3) after 4 h of treatment. In contrast, the expression of key structural genes involved in flavonoid biosynthesis increased after 4 h in GA3 treatment. However, the content of flavonoid metabolites in leaves significantly increased at 12 h. The qRT-PCR results showed that the expression patterns of EbWRKY11, EbWRKY30, EbWRKY31, EbWRKY36, and EbWRKY44 transcription factors exhibited a high degree of similarity to the 11 structural genes involved in flavonoid biosynthesis. Protein-DNA interactions were performed between the key genes involved in scutellarin biosynthesis and candidate WRKYs. The result showed that F7GAT interacts with EbWRKY11, EbWRKY36, and EbWRKY44, while EbF6H has a self-activation function. This study provides comprehensive information on the regulatory control network of flavonoid accumulation mechanisms, offering valuable insights for breeding E. breviscapus varieties with enhanced scutellarin content.
1 Introduction
Erigeron brevisapus is a traditional medicinal plant in the Asteraceae family and is mainly distributed in Southwest China. The sales revenue of traditional Chinese medicine preparations derived from E. breviscapus as a primary ingredient in China reached 3 billion RMB in 2020 (http://yn.chinadaily.com.cn/a/202007/27/WS5f1e9bb2a310a859d09da571.html). The main active flavonoid in E. brevisapus, scutellarin, is extracted from the leaves and has been extensively utilized in prescription injections for the treatment of cardiovascular diseases (Chen et al., 2021; Ju et al., 2021; Yang et al., 2022b; Zhang et al., 2022). We successfully elucidated the complete biosynthesis pathway of scutellarin and constructed the high-level production yeast factory (Liu et al., 2018; Wang et al., 2022). Recently, we reported the transcription factors that regulate scutellarin (R2R3-MYB) and anthocyanin (bHLH) biosynthesis in E. brevisapus as well (Gao et al., 2022; Zhao et al., 2022). However, the regulatory mechanism of the flavonoid pathway governed by the WRKY transcription factor family in E. breviscapus remains elusive.
WRKY transcription factor is the seventh largest TF family in higher plants and is named for its characteristic WRKY domain (Rushton et al., 2010). The typical structure of WRKY is the N-terminal, which contains conserved amino acid sequence WRKYGQK, whereas the C-terminal contains a zinc finger motif (C2H2 or C2HC) (Eulgem et al., 2000). According to the number of WRKY domains and the type of zinc finger motif, WRKY can be divided into three categories: Group I contains two WRKY domains, whereas Groups II and III have a single WRKY domain, WRKY domains of Group II and III family members are more similar in sequence to the C-terminal than to the N-terminal WRKY domain of Group I proteins (Eulgem et al., 2000; Dong et al., 2003). The members of Group II WRKY were further divided into five subgroups: IIa, including IIa, IIc, IId, and IIe, based on additional conserved structural motifs (Eulgem et al., 2000; Zhang and Wang, 2005). WRKY transcription factors play important roles in plant growth and development, defense regulation, stress, and synthesis of secondary metabolites (Eulgem et al., 1999; Johnson et al., 2002; Yu et al., 2012, Yu et al, 2013).
Since the first WRKY gene (SPL1) was cloned from sweet potatoes (Ishiguro and Nakamura, 1994), the identification and functional analysis of WRKY genes has developed rapidly in plants, especially in crops, fruits, and medicinal plants. Several WRKY transcription factors have been identified in Arabidopsis thaliana, Glycine max, Vitis vinifera, Panax ginseng, and Salvia miltiorrhiza (Wang et al., 2011; Guo et al., 2014; Li et al., 2015; Yang et al., 2017; Di et al., 2021). A total of 14549 WRKY genes were recorded in the Plant Transcription Factor Database (PlantTFDB) (Jin et al., 2017). Numerous studies have substantiated the close association between WRKY transcription factors and the biosynthesis of flavonoid metabolites (Amato et al., 2017; Duan et al., 2018; Wang et al., 2023). The overexpression of AeWRKY32 (Okra) induced anthocyanin accumulation, with higher expression levels of AtCHS1, AtCHI4, AtF3H1, and AtDFR2 in transgenic Arabidopsis (Zhu et al., 2023). AtWRKY23 transcription factors regulate flavonol accumulation, auxin transport, root growth, and development (Grunewald et al., 2012). VvWRKY70 and NtWRKY11b have been identified as regulators involved in flavonol biosynthesis, the content of flavonol significant decreased in VvWRKY70-overexpressing grape calli lines by inhibiting the promoter VvCHS2, VvCHS3, and VvFLS4. Conversely, overexpression of NtWRKY11b led to a substantial increase in flavonol content ranging from 37.8% to 80.7%. (Wang et al., 2021; Wei et al., 2023). BcWRKY1 significantly increased the transcript of CHS to regulate flavonoid biosynthesis (Zeng et al., 2022). However, the current literature rarely reports on the regulatory mechanisms by which WRKY transcription factors regulate flavone and flavonol biosynthesis in medicinal plants.
Plant hormones have a prominent function in the modulation of the growth, development, reproduction, and secondary metabolism of plants, such as SA, ABA, GA3, and MeJA, shown to be involved in the regulation of flavonoid biosynthesis (Khan et al., 2015; Lucho-Constantino et al., 2017; Li and Ahammed, 2023). Exogenous ABA could promote the synthesis of ABA in Artemisia argyi leaves and up-regulated the content of chlorogenic acid, nevertheless significantly down-regulated other flavonoid metabolites after ABA treatment (Yang et al., 2022a). Exogenous GA3 evidently decreased the contents of naringin and naringenin in P. chinense Schneid seedlings (Yang et al., 2023). Recently, an increasing interest has focused on WRKY transcription factors response to plant hormones and involved in flavonoid metabolism (Schluttenhofer and Yuan, 2015; Vives-Peris et al., 2018; Xu et al., 2020; Yamamoto et al., 2020). LrWRKY3 transcription factor response to MeJA may specifically interact with the ANR and LAR gene and might be involved in anthocyanins synthesis in L. radiate, regulated the content of pelargonidin-3-O-glucoside-5-O-arabinoside in L. radiate (Wang et al., 2023). VqWRKY31 also activated SA defense signaling and changed the accumulation of stilbenes, flavonoids, and proanthocyanidins (Yin et al., 2022). Flavonoid compounds are involved in the defence of plants against biotic and abiotic stresses, WRKY transcription factors can respond to hormone signal transduction pathways, improve the accumulation of flavonoids, and play key roles in the regulation of various stressful stresses (drought, low temperature, wounds, disease-resistant, etc.) in plants (Pourcel et al., 2007; Agati et al., 2012; Han et al., 2018a, Han et al., 2018b). The main medicinal active ingredients of E. breviscapu are flavonoids, the study of the response mechanisms of WRKY transcription factors and flavonoids under hormonal stress, and can effectively analyze and identify WRKY transcription factors involved in the synthesis of scutellarin, and elucidate the molecular mechanisms by which WRKY transcription factors regulate the synthesis of scutellarin.
In this study, the conserved motifs, gene structure, chromosome location, phylogenetic trees, gene expression profile, and function of WRKY genes were identified based on the whole genome of E. breviscapus. Additionally, integrated metabolomic and transcriptomic analyses were performed to study the expression patterns of WRKY genes and flavonoid metabolites in response to exogenous hormone treatments. Our study revealed the expression of EbWRKYs and the accumulation of flavonoids differed under the treatment of three exogenous hormones in E. breviscapus leaves. We also identified three candidate WRKY genes potentially involved in the regulation of scutellarin biosynthesis. This study provided valuable guiding information for growth and development research and functional identification of WRKY transcription factors involved in scutellarin biosynthesis in E. breviscapus.
2 Methods
2.1 Plant treatment
The two-month-old E. breviscapus seedlings were germinated and cultivated in a growth chamber under controlled conditions at 22 °C with a photoperiod of 16 hours light and 8 hours dark. Furthermore, the leaves of E. breviscapus were treated with 200 mL of ABA, SA, and gibberellin 3 (GA3) solution at a concentration of 200 μmol/L. Leaf samples were collected at time points of 0 h, 4 h, 12 h, and 24 h, immediately frozen in liquid nitrogen, and stored at -80 °C. Each experimental sample has three biological repetitions.
2.2 Identification and physicochemical properties of WRKY proteins
The PfamScan v1.6 tool was employed to annotate the protein domains of the entire genome sequence of E. breviscapus, utilizing the Pfam 35.0 database. Sequences exhibiting E-values lower than Le-5 and encompassing the PF03106 domain were screened, while manually excluding any atypical characteristics observed in WRKY genes. The ProtParam tool (https://web.expasy.org/protparam/) was utilized for predicting various attributes of EbWRKY proteins, including molecular weights (MWs), isoelectric points (pIs), amino acid counts, open reading frame (ORF) lengths. Protein subcellular localization was predicted by PSORT (https://psort.hgc.jp/).
2.3 Protein domain and phylogenetic evolution analysis
Multiple sequence alignments were conducted using MAFFT v7.490 to elucidate the evolutionary relationship between E. breviscapus and A. thaliana. To investigate the interrelationship among E. breviscapus WRKY proteins, a phylogenetic tree encompassing both E. breviscapus and A. thaliana WRKY proteins was constructed through Phylipv3.698 software employing the neighbor-joining method with 1000 repetitions. Subsequently, EvolView (https://evolgenius.info/evolview/#/) was employed as an evolutionary tree visualization tool for further analysis. The WRKY protein sequences of A. thaliana were downloaded from the TAIR database (https://www.arabidopsis.org/).
2.4 Comprehensive analysis of WRKY genes
The intron-exon structures of the EbWRKY genes were determined using the gene structure display server provided by Peking University (http://gsds.cbi.pku.edu.cn/). The conserved motifs of WRKY proteins were predicted using MutipleEm for Motif Elicitation, a tool available at http://alternate.meme-suite.org/tools/meme. For motif prediction, we employed optimized parameters including any number of repetitions (20), minimum width (10), and maximum width (80). Gene structure and chromosome mapping analysis of WRKY family members were conducted using TBtools v1.098691, while collinearity between the WRKY gene family in E. breviscapus and A. thaliana, Daucus carota, Helianthus annuus, Lycopersicon esculentum, and Solanum tuberosum was analyzed with the one-step MCScanX tool. Collinearity within E. breviscapus was visualized using Advanced Circos software.
2.5 RNA-sequencing data analysis
The leaves of E. breviscapus were subjected to hormone treatment, followed by flash freezing in liquid nitrogen for RNA extraction and subsequent cDNA library construction. Transcriptome sequencing was performed using Illumina Hiseq 4000 platform. SkrTools (version 1.0) was employed to calculate the raw data generated from sequencing, which underwent filtration using Trimmomatic v0.39 and RiboDetector v0.2.4 (Bolger et al., 2014; Deng et al., 2022). Subsequently, rRNA sequences were eliminated from the raw data to obtain high-quality clean reads that were utilized for gene differential expression analysis.
2.6 Expression profiling analysis of EbWRKY genes in various tissues
The differential expression of EbWRKYs in roots, stems, leaves, and flowers was calculated using the Salmon software based on the previously assembled genomic data of E. breviscapus. Additionally, the expression levels in the transcriptome data treated with three exogenous hormones at different time points were analyzed. The TPM value (Transcripts Per Million) was used to calculate gene expression values. Clustering results and heat maps were generated using TBtools v1.098691 (Chen et al., 2020).
2.7 Metabolites analysis
The frozen and fresh leaves of E. breviscapus (100 mg) were ground in liquid nitrogen, and the homogenate was resuspended in pre-chilled 80% methanol and 0.1% formic acid by vortexing. The samples were incubated on ice for 5 min and then centrifuged at 15,000 g at 4°C for 20 min. The supernatant was diluted to a final methanol concentration of 53% for the LC-MS/MS analysis (Dunn et al., 2011). Samples were injected onto an Xselect HSS T3 column (2.1×150 mm, 2.5 μm) with a 20-min linear gradient at a 0.4 mL/min flow rate for the positive/negative polarity mode. The eluents used were eluent A (0.1% formic acid water) and eluent B (0.1% formic acid-acetonitrile). The solvent gradient was set as follows: 2% B, 2 min; 2-100% B, 15.0 min; 100% B, 17.0 min; 100-2% B, 17.1 min; 2% B, 20 min (Wang et al., 2014). The data files generated by HPLC-MS/MS were processed using the SCIEX OS Version.
The dried leaves of E. breviscapus powder sample (0.3 g) were dissolved in 50 mL of methanol, and the supernatant was extracted for 30 minutes by ultrasonic use for HPLC analysis. Samples were injected onto an Agilent EC-C18 column (4.6 x 100 mm, 2.7 μm), with a 50-min linear gradient at a 1 mL/min flow rate. The eluents used were eluent A (acetonitrile) and eluent B (0.1% phosphoric acid water). The solvent gradient was set as follows: 0-10 min, 12%- 15% A; 10- 32 min, 15% A; 32- 33 min, 15%- 20% A; 33- 50 min, 20%- 22% A. Scutellarin (SE), Chlorogenic acid (CGA), 3,5-dicaffeoylquinolinic acid (3,5-diCQA), and Erigoster B (EB) were quantified using the external standard method with standards purchased from Sigma-Aldrich (Shanghai, China). Variance significance analysis was conducted employing SPSS 20.0.
2.8 Co-expression network analysis
The expression of candidate EbWRKYs and key genes involved in flavonoid biosynthesis was extracted from the transcriptome data obtained from the roots, stems, leaves, and flowers of E. breviscapus. Initially, statistically significant correlations between differential metabolites were calculated using R (version 4.1.1). A significance level of p<0.05 was applied for statistical analysis. Subsequently, gene expression levels and relative metabolite contents were collected to identify correlation pairs with a Pearson product-moment correlation coefficient (PCC)≥0.6 and a p-value ≤ 0.05. The filtered genes were then utilized to construct a co-expression network which was visualized using Cytoscape version 3.3.0 software (https://www.cytoscape.org).
2.9 Protein-DNA interactions assays
The proteins-DNA interaction between EbWRKY11, EbWRKY36, EbWRKY44, EbF6H, and F7GAT was investigated using the bait construct pAbAi and prey construct PGADT7, which were generated through a BP reaction. The prey plasmid was transformed into the bait strain yeast Y1H and selected with supplemented medium containing SD/-Leu, SD/-Leu/Aba (Clontech). The binding domain was predicted using JASPAR (https://jaspar.elixir.no/).
2.10 Quantitative real-time PCR analysis
Leaves were collected, and total RNA was extracted from hormone-treated samples using a HiPure HP Plant RNA Mini Kit (R4165-02). Subsequently, cDNA synthesis was performed utilizing a PrimeScript RT Reagent Kit with gDNA Eraser (Takara, Japan). Gene-specific primers for qRT-PCR reactions were designed employing Primer3 web version 4.1.0 (https://primer3.ut.ee/) (Supplementary Table S6). A Quantstudio 5 Flex Real-Time PCR System (Thermo Fisher Scientific, USA) was employed to analyze three technical replicates. The expression levels of genes from different treatments were normalized to EbACTIN2. Finally, the relative expression levels were calculated using the 2–ΔΔCt method and visualized using GraphPad Prism 8.0.2.
3 Results
3.1 Identification and physicochemical properties of WRKY genes in E. breviscapus
A total of 75 putative EbWRKYs were identified from E. breviscapus genomic data, which were designated as EbWRKY1 to EbWRKY75. The number of amino acids ranged from 144 (EbWRKY51) to 756 (EbWRKY19), and the isoelectric points ranged from 4.97 (EbWRKY29/47) to 10.12 (EbWRKY73), including 45 acidic and 30 basic amino acids. In addition, the relative molecular weights ranged from 19.99 (EbWRKY68) to 80.89 kDa (EbWRKY25). Based on sequence analysis conducted using PSORT software, it was determined that the 75 EbWRKY proteins are localized within the nucleus, suggesting their potential regulatory roles as transcription factors in this cellular compartment. In addition, EbWRKY19 is located on the cell membrane and may be involved in the expression and regulation of genes related to membrane transport (Supplementary Table S1).
3.2 Evolution and sequence analysis of WRKY transcription factors
To gain a comprehensive understanding of plant biodiversity mechanisms and the regulatory role of WRKY genes in the network, we conducted an evolutionary analysis of WRKY transcription factors. Subsequently, phylogenetic analyses were performed on 75 E. breviscapus and 72 Arabidopsis WRKY transcription factors (Figure 1A). A total of 147 WRKYs were divided into seven branches. E. breviscapus and Arabidopsis WRKY proteins with the same classifications were classified into I, II, and III. Group I has 17 EbWRKYs, Group II can be divided into five subtypes according to the different zinc-finger structural sites: IIa, IIb, IIc, IId, and IIe. There were five EbWRKYs in Group IIa, eight EbWRKYs in Group IIb, eleven EbWRKYs in Group IIc, ten EbWRKYs in Group IId, nine EbWRKYs in Group IIe, and fifteen EbWRKYs in Group III. The three major classes and five subclasses in the phylogenetic tree contained both the WRKY genes from E. breviscapus and Arabidopsis, indicating that the WRKY families of Arabidopsis and E. breviscapus are highly similar at the evolutionary level. Additionally, the WRKY transcription factors of E.breviscapus exhibit a high degree of similarity within their respective branches of the phylogeny, suggesting an increased homogeneity in the WRKY gene family during evolutionary processes.
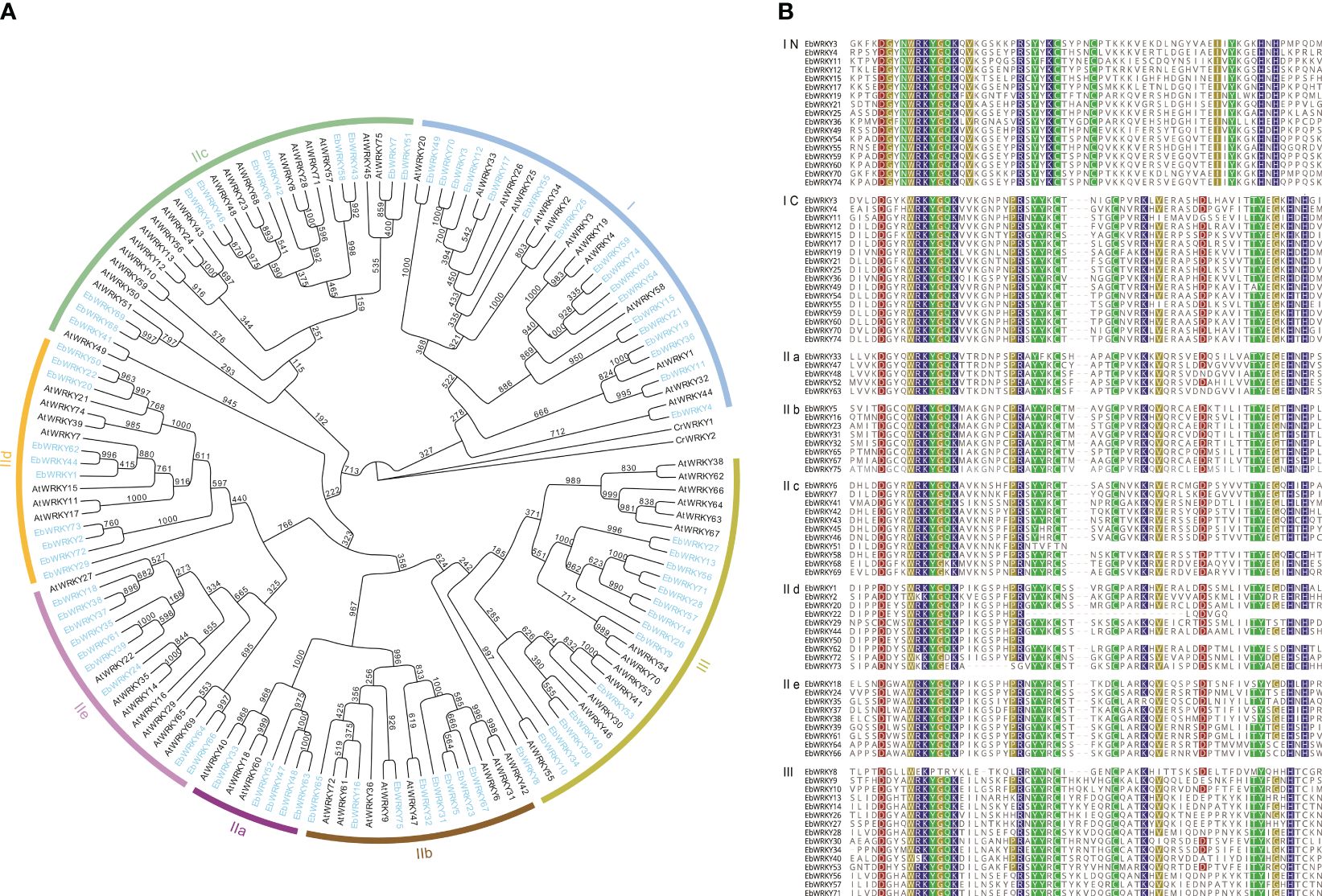
Figure 1 Phylogenetic and WRKY protein domain sequence analysis of E. breviscapus: (A) Phylogenetic analysis of E. breviscapus and Arabidopsis WRKY transcription factor; (B) WRKY protein domain sequence analysis in E. breviscapus. IN and IC represent the C-terminal and N-terminal WRKY domain of Group I, respectively.
In addition to WRKYGQK, the core motif of E. breviscapus WRKY heptapeptide contained five variants: WRKYGKK, WKKYGQK, WKKYGDK, WKKYGEK, and WSKYGQK (Figure 1B). Sequence comparison results showed that each WRKY protein, except EbWRKY8, contained a typical WRKY conserved domain at the N-terminal and a complete zinc finger structure at the C-terminal (CX4-5CX22-23HXH/C), which is an important feature for identifying WRKY transcription factors. All WRKY sequences of E. breviscapus showed a high similarity and conservation of the WRKY domain. Group I EbWRKYs contained the same heptapeptide core motif WRKYGQK at the N-and C-terminal and the zinc finger structure C2H2 behind the WRKY structure at the C-terminal. Group II and Group III had a WRKY domain at the N-terminal, but the zinc finger structure at the C-terminal differed (C2HX). EbWRKY8 was domain sequence was lost, and the C-terminal retained a zinc-finger structure. However, the sequences of EbWRKY8 are highly similar to the other EbWRKYs, and evolutionary analysis clustered them into Group III.
3.3 Gene structure and conserved motif analysis of WRKY proteins
75 WRKY protein motifs were analyzed using MEME and TBtools. The conserved domain of the WRKY motif was identified in motifs 1 and 3, while motif 2 exhibited a zinc-finger structure. Motifs 1 and 2 were found in almost all EbWRKY proteins, indicating their widespread presence. Notably, distinct EbWRKYs displayed diverse motif structures, with similar motifs observed within each branch clustering by the same type of EbWRKY. Motif 15 was exclusively present in the transcription factor genes of EbWRKY49, EbWRKY54, EbWRKY59, EbWRKY60, and EbWRKY74 in Group I. Motif 16 was identified as a characteristic motif of Group IIb EbWRKYs while motif 4 was found to be a common feature of Groups I and II (b,c). Only eight members of Group III contained motif 11 whereas motif 12 was detected both in the sequences of Groups III and IId EbWRKY (Figure 2A; Supplementary Table S2). Phylogenetic trees of WRKY proteins were established, and the three groups were clustered according to their sequence similarity. WRKY sequences with similar structures in the evolutionary tree clustered into a single branch, indicating that these WRKY proteins may have similar functions. TBtools were used to analyze the number and distribution of exons of CDS sequences of the 75 WRKYs (Supplementary Table S3). The results showed a significant difference in the number of introns and exons in the WRKY gene family of E. breviscapus. The numbers of introns and exons were 1-6 and 2-7. (Figure 2B).
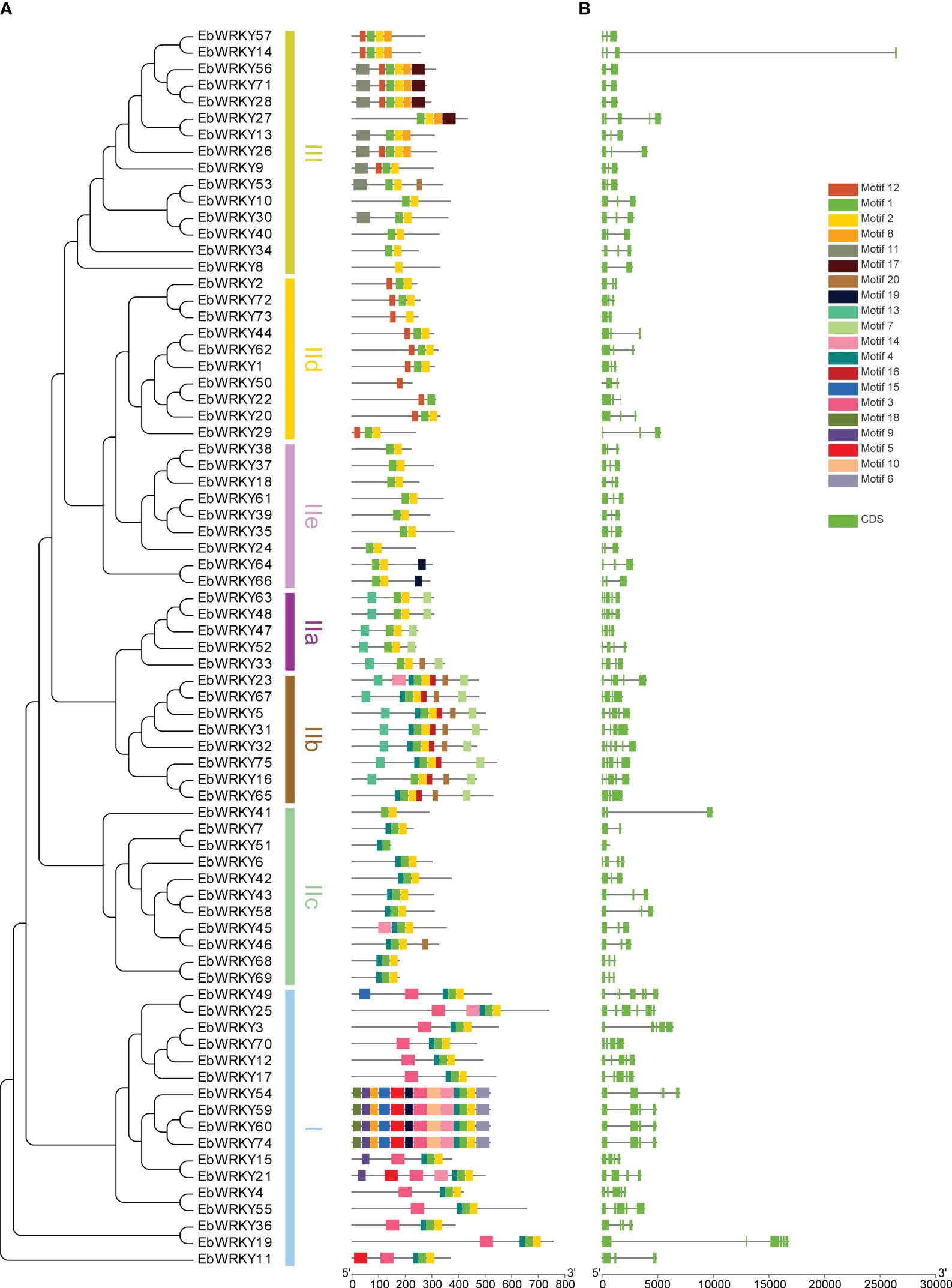
Figure 2 The evolutionary relationship, genetic structures, and motifs of WRKY genes in E. breviscapus: (A) Phylogenetic evolution and motif structure of E. breviscapus WRKY genes; (B) Gene structure of E. breviscapus WRKY genes (green box represents the exon).
3.4 Chromosomal mapping and collinearity analysis of WRKY genes
The distribution of the 75 EbWRKY genes across all nine E. breviscapus chromosomes exhibited irregular patterns (Figure 3A). A total of 21 EbWRKY genes were localized to chromosome 1, accounting for 28% of the EbWRKY gene family. Ten tandem duplications occurred on the six chromosomes. Eight WRKY members were collinear on chromosomes 1, 3, 4, and 6, respectively (Figure 3B). To further predict the potential evolutionary patterns of the EbWRKY gene family, we constructed comparative syntenic maps of E. breviscapus in associated with five representative species, including A. thaliana, D. carota, H. annuus, L. esculentum, and S.tuberosum (Figure 3C). The number of orthologous gene pairs between E. breviscapus and A. thaliana, D. carota, H.annuus, L.esculentum, and S. tuberosum were 21, 37, 33, 34, and 36, respectively (Supplementary Table S4). These results revealed that the identified orthologous events of EbWRKY-HaWRKY were considerably higher than those of other WRKY species based on their close evolutionary relationship. An extensive level of synteny conservation and an increased number of orthologous events in EbWRKY-HaWRKY indicated that EbWRKY genes in E. breviscapus shared a similar structure and function with HaWRKY genes.
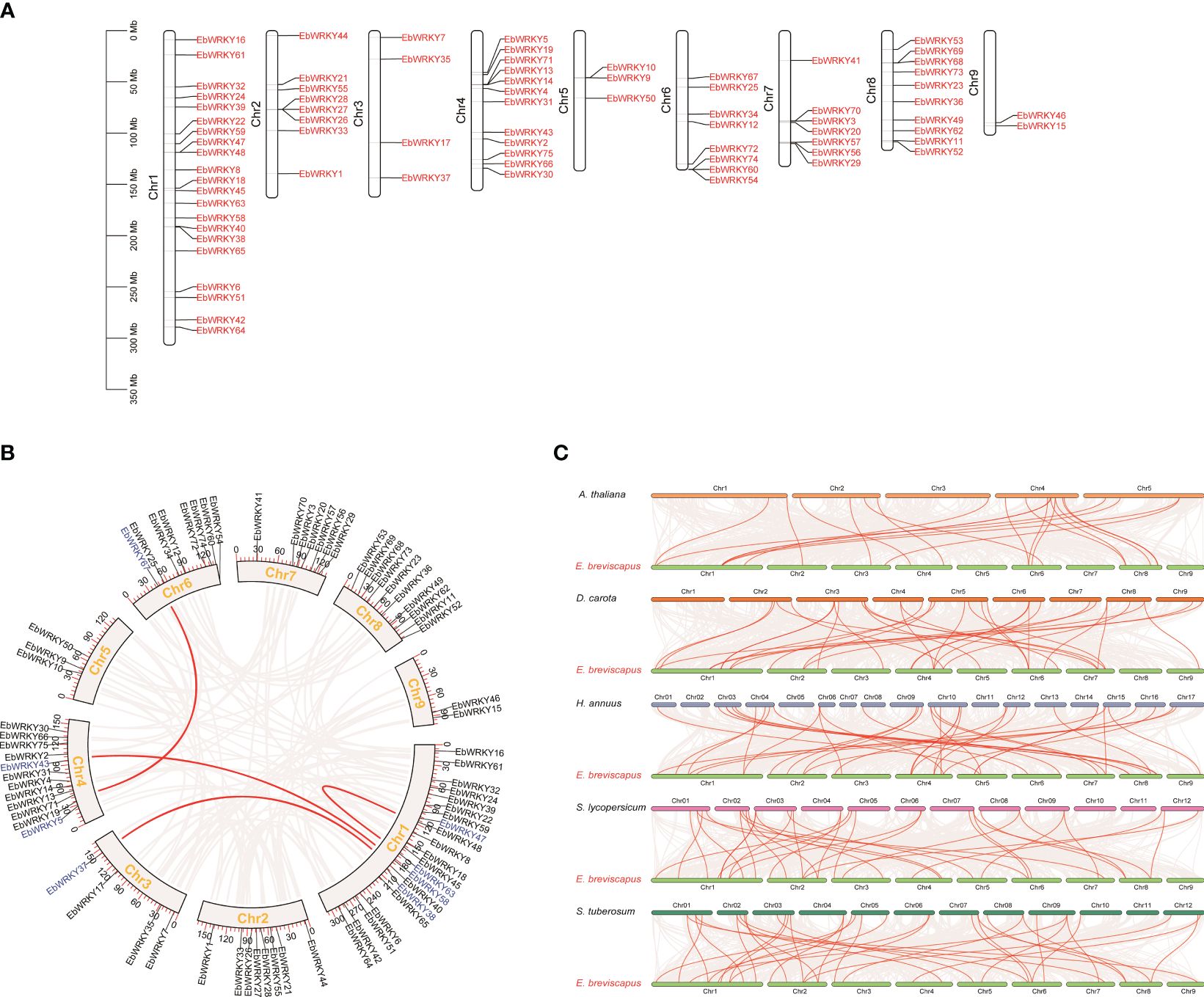
Figure 3 The Synteny analysis and chromosome location of WRKY genes in E. breviscapus: (A) Chromosomal location of WRKY genes; (B) The internal collinearity circle diagram of the E. breviscapus genome (the black line represents the position of genes on chromosomes; the arc represents the collinearity relationship between genes; the WRKY gene pair is highlighted with a red line; the gene name color represents different subgroups); (C) WRKY gene pairs are highlighted with red lines according to the analysis of collinearity between E. breviscapus and different plant genomes.
3.5 Expression pattern of WRKY genes in the different tissues and hormone treatment
The TPM values of the roots, stems, leaves, and flowers were extracted from the genomic database to clarify the expression of EbWRKY family genes. Except for seven EbWRKY genes that exhibited no expression in any tissue, the remaining EbWRKY genes demonstrated specific expression patterns in leaves, roots, stems, and flowers, respectively. (Supplementary Table S5; Figure 4A). In our previous study, exogenous application of SA, GA3, and ABA onto the leaves of E. breviscapus was found to enhance their scutellarin (SE) content, with ABA treatment showing the most significant effect (Supplementary Table S6). Therefore, transcriptomic analysis was employed to investigate the underlying expression mechanism of EbWRKYs in response to hormone induction. The results showed that the expression levels of EbWRKYs significantly altered after three hormone treatments. In the ABA treatment, the expression levels of EbWRKY69 and EbWRKY30 were significantly up-regulated after 4 h. In contrast, in the SA treatment assays, the gene expression levels of EbWRKY8, EbWRKY17, EbWRKY51, EbWRKY67, EbWRKY18, EbWRKY66, and EbWRKY64 were significantly up-regulated after 12 h of treatment, and EbWRKY52 and EbWRKY57 were significantly up-regulated after 24 h. The expression levels of EbWRKY3, EbWRKY41, EbWRKY47, EbWRKY2, and EbWRKY39 genes were significantly upregulated after 4 hours of GA treatment. However, a gradual decline in their expression levels was observed in the leaves over time. (Figure 4B).
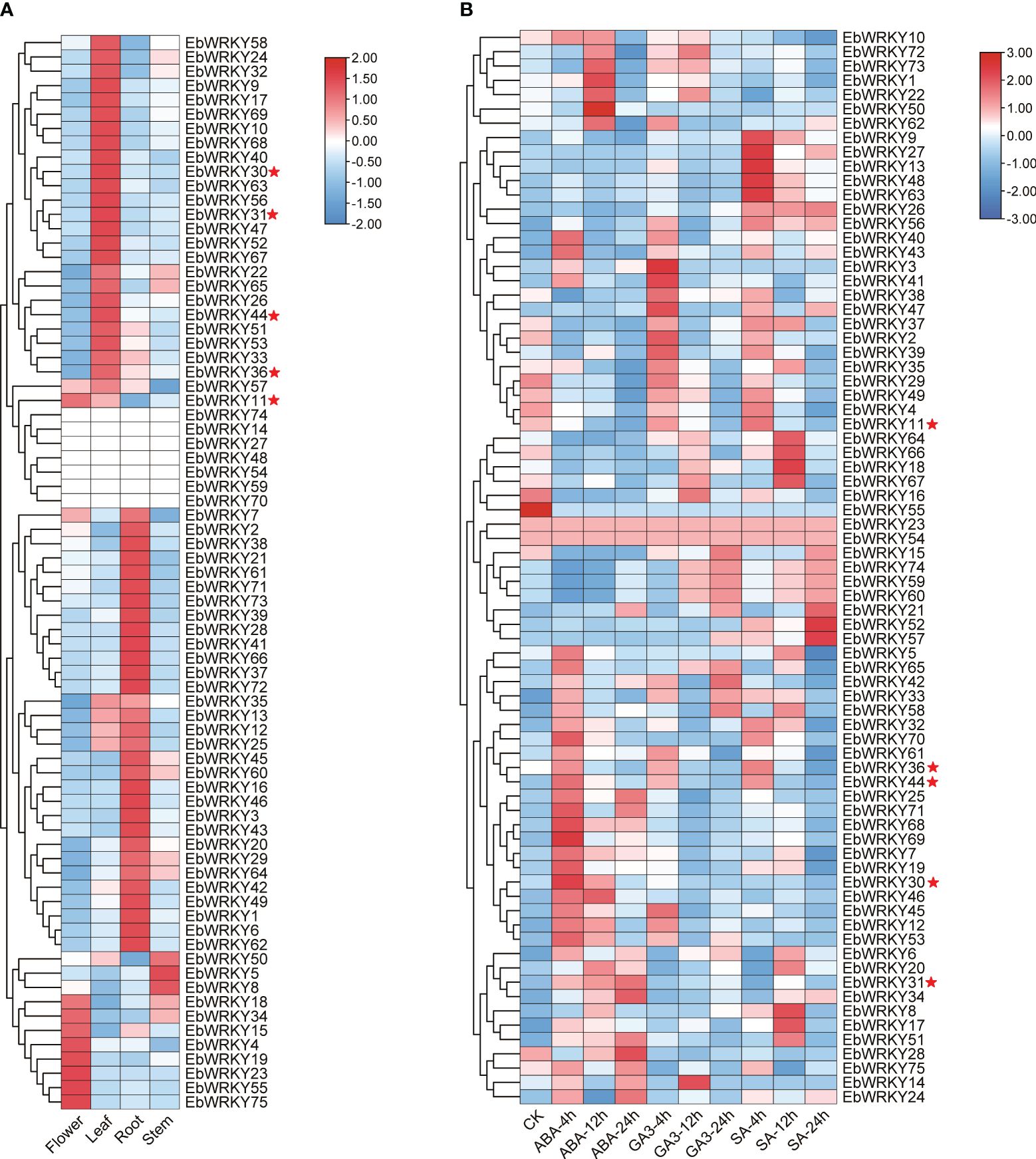
Figure 4 The expression profiles of WRKY genes in E. breviscapus: (A) Expression profiles of WRKY genes in different tissues of E. breviscapus; (B) Expression profiles of E. breviscapus WRKY under different hormone treatments at different times. The color scale on the right of each diagram represents TPM expression values: red indicates higher levels and blue indicates lower levels. CK represents the untreated sample. The red star represents five candidate EbWRKYs that may involved in the flavonoid metabolic pathway in E. breviscapus.
3.6 Hormone-induced expression analysis of structural gene and flavonoid metabolites in the leaves E. breviscapus
Ultra-high-performance liquid chromatography (UPLC) and tandem mass spectrometry (MS/MS) were used to determine dynamic changes in flavonoid metabolites in nine E. breviscapus treated with the three hormones. Pearson correlation coefficients of the QC samples were calculated based on the relative quantitative values of the metabolites. The R2 values of all the samples were close to 1, indicating better stability of the entire detection process and higher data quality (Supplementary Table S7; Figure 5A). Scutellarin biosynthesis commences with phenylalanine, followed by the enzymatic catalyzation of phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), coumaric acid coenzyme A ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavone synthase II (FSII), flavonoid 7-O-glucuronosyltransferase (F7GAT), and flavone-6 hydroxylase (F6H) to yield scutellarin. Furthermore, other flavonoids including kaempferol, quercetin hesperidin, and luteolin are biosynthesized via the flavanone 3-hydroxylase (F3H), flavonoid 3’-hydroxylase (F3’H), and flavonol synthase (FLS).
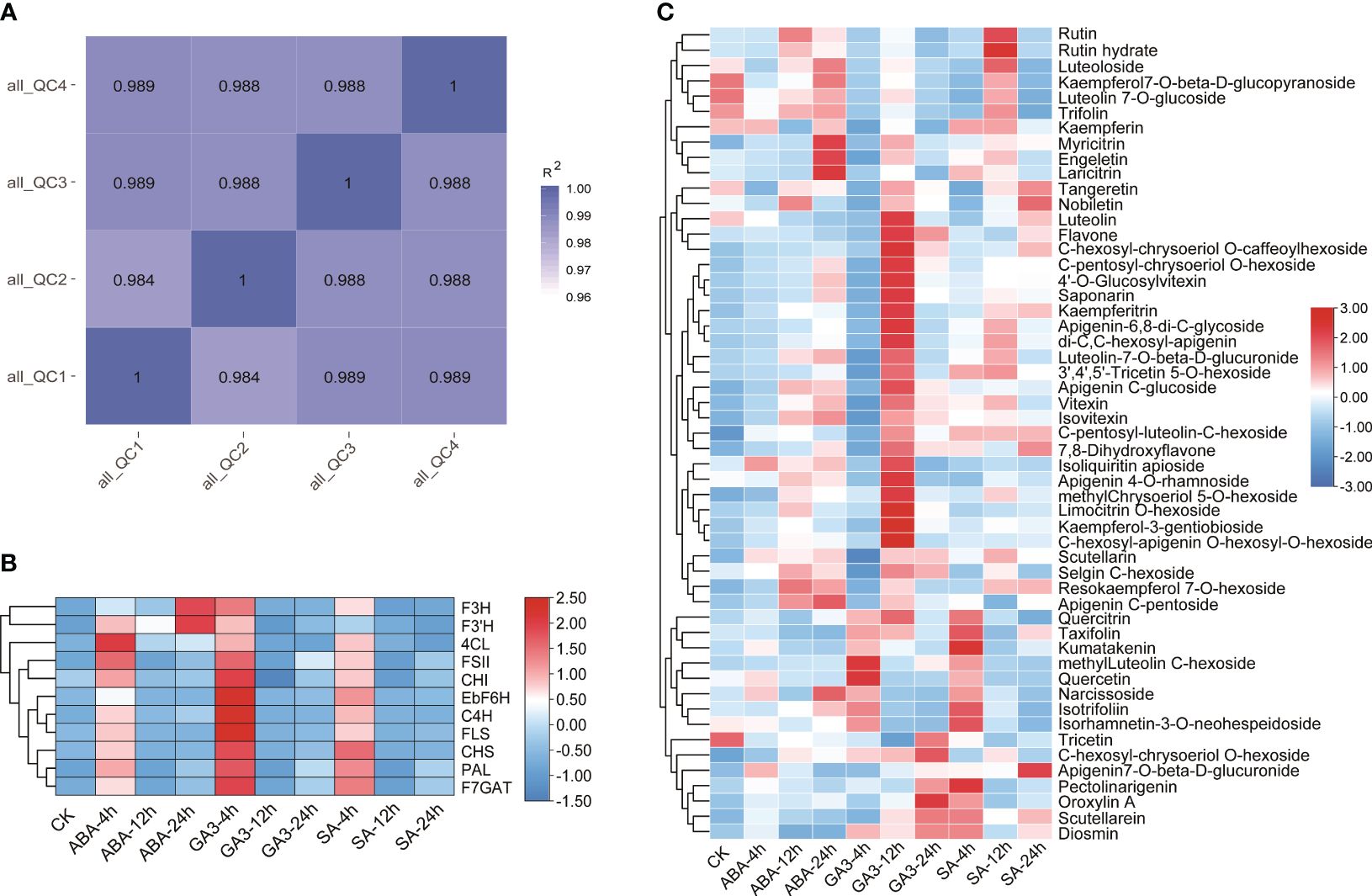
Figure 5 Quantitative and structural gene expression analysis of flavonoid pathway metabolites in the leaves of E. breviscapus: (A) Pearson correlation coefficient analysis of the QC of samples; (B) Structural gene expression analysis; (C) Contents of 54 flavone and flavonol metabolites in E. breviscapus under abscisic acid (ABA), salicylic acid (SA), and gibberellin 3 (GA3) hormone treatments at different times. CK represents the unprocessed sample.
The expression patterns of 11 key enzyme genes involved in flavonoid biosynthesis were analyzed following treatments with ABA, SA, and GA3. The results showed that the expression of 11 genes treated with the three hormones was up-regulated at 4 h. The downstream genes regulating flavonol biosynthesis, F3H and F3’H, were significantly up-regulated in ABA treatment at 24 h but down-regulated in GA3 treatment. The gene expression of GA3 exhibited its peak at 4 hours, while the response to SA did not manifest prominently. The expression levels of FLS, C4H, and F6H were significantly up-regulated after 4 h of GA3 treatment (Supplementary Table S8; Figure 5B).
A total of 159 flavonoids were identified, with three biological replicates set for each sample (Supplementary Table S7). Flavones and flavonols accounted for the majority (57.8%), followed by flavanones (15.7%), isoflavones (10.6%), and anthocyanins (8.1%). Chalcones, dihydrochalcones (4.4%), and other flavonoids (3.1%) were found in lower abundance. The analysis revealed the identification of 92 flavone and flavonol metabolites, with scutellarin exhibiting the highest content, followed by Apigenin7-O-β-D-glucuronide (Supplementary Table S7; Figure 5C). After treatment with ABA, SA, and GA3, the metabolism patterns of 54 flavone and flavonol metabolites exhibited differential changes, and the responses of flavonol and flavone compounds significantly increased after 12 h of GA3 treatment, with 14 compounds significantly increased compared to other levels. In addition, the content of pectolinarigenin significantly increased after 24 hours of SA treatment, while the content of oroxylin A showed a significant increase after 24 hours of GA3 treatment compared to other levels.
3.7 Integrated analysis of WRKYs involved in flavonoid metabolism
Transcriptome and metabolome data were integrated and analyzed to construct a co-expression network of key genes involved in flavonoid metabolism pathways. A co-expression network was constructed by screening relevant pairs through Pearson analysis of gene expression levels and compound content (PCC ≥ 0.6, p ≤ 0.05). A total of 231 related pairs were identified and visualized using Cytoscape software (version 3.3.0). The network revealed a total of 102 interconnected nodes connected by 231 edges, encompassing 10 key enzyme genes and 45 EbWRKYs, alongside the presence of 47 flavonoid metabolites comprising 26 flavone and flavonol derivatives. In addition, a positive correlation was observed in 143 pairs, while 88 pairs exhibited a negative correlation (Figure 6). Within the flavonoid metabolic pathway, five potential EbWRKYs were identified as candidates with positive associations, namely EbWRKY11, EbWRKY30, EbWRKY31, EbWRKY36, and EbWRKY44. Notably, among these candidates, EbWRKY11 demonstrated connections to four key genes (EbF6H, F7GAT, FLS, and CHI). The expression of EbWRKY30 showed a positive correlation with 4CL, EbWRKY31, and F3’H. Additionally, the presence of EbWRKY36 and EbWRKY44 was found to be associated with PAL, EbF6H, C4H, 4CL, F7GAT, CHI, CHS, and FLS. Notably, EbWRKY44 also exhibited a connection with FSII. (Figure 6).
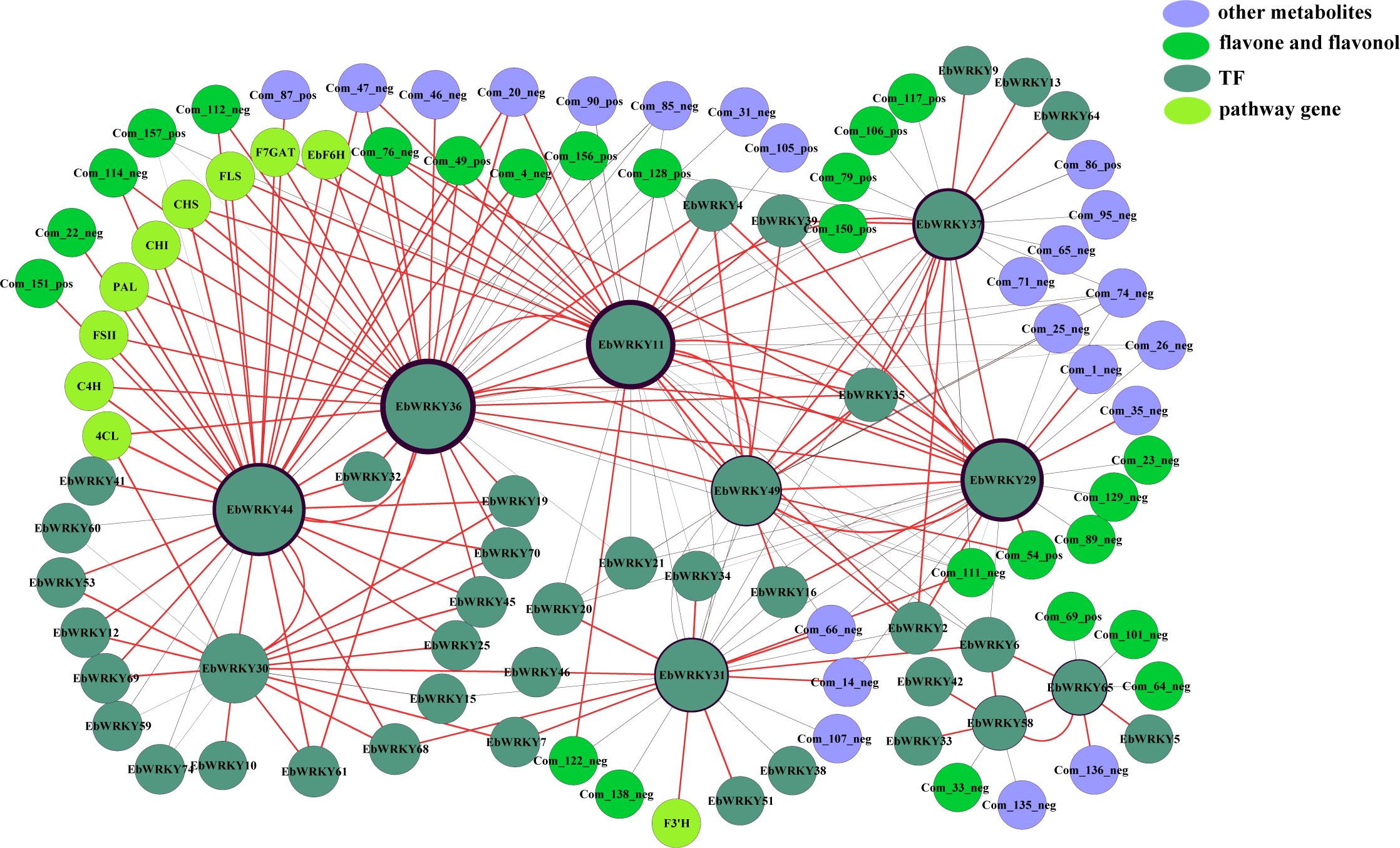
Figure 6 Co-expression analysis of structural genes involved in the flavonoid biosynthesis pathway and EbWRKYs in the leaves of E. breviscapus. Yellowish-green nodes represent genes; Blackish-green nodes represent WRKY TFs; Lavender nodes represent other flavonoid metabolites; Green nodes represent flavone and flavonol metabolites. The size of the circle is associated with the number of EbWRKY genes. Black circles outside the genes are associated with the number of metabolites.
Additionally, EbWRKY11 exhibited association with a total of 17 flavonoid metabolites, encompassing nine flavone and flavonol metabolites. Among the 13 EbWRKYs, eight pairs displayed positive correlation while five pairs showed negative correlation. EbWRKY31 was associated with six metabolites, including three flavones and flavonols. Seven pairs were positively correlated, and eight pairs were negatively correlated with 15 EbWRKYs. However, EbWRKY30 was not directly connected with metabolites but was related to 17 EbWRKYs. The EbWRKY36 gene was found to be associated with 18 flavonoid metabolites and 12 other EbWRKY genes, exhibiting positive correlations in 11 pairs and a negative correlation in one pair. The correlation analysis revealed that EbWRKY44 was associated with 12 flavonoid metabolites and sixteen EbWRKYs, among which thirteen pairs exhibited positive correlations while three pairs showed negative correlations. Overall, the co-expression analysis of the selected EbWRKY genes revealed that these genes might play an essential role in flavonoid synthesis.
3.8 Quantitative real-time PCR profiling characterization of genes under exogenous hormone treatment
To investigate the expression pattern responses of genes under ABA, SA, and GA3 exogenous hormones in E. breviscapus, eleven structural genes of the flavonoid biosynthesis pathway and five WRKY genes (EbWRKY11, EbWRKY30, EbWRKY31, EbWRKY36, and EbWRKY44) were selected for qRT-PCR analysis after exogenous hormone treatment. Eleven genes involved in the flavonoid synthesis pathway exhibited significant up-regulation, implying their potential functional role in this biological process. (Figure 7; Supplementary Table S9). The relative expression of the selected key genes exhibited distinct temporal patterns in response to different treatments. Notably, FLS displayed the highest expression level, with a 20-fold increase observed after 4 hours of GA3 treatment and a 10-fold increase after 4 hours of ABA and SA treatment, followed by a subsequent decrease at 12 hours. C4H, F6H, and F3H showed similar expression patterns after 4 h of treatment, indicating that these genes are sensitive to GA3, SA, and ABA. CHI and PAL showed relatively high expression levels after 4 h of ABA treatment (> 3.5-fold). CHS exhibited the highest expression level, with an 8-fold expression at 4 h of ABA treatment and a 6.8-fold expression with GA3 treatment. FSII showed a 2.9-fold higher expression after 4 h of SA treatment.
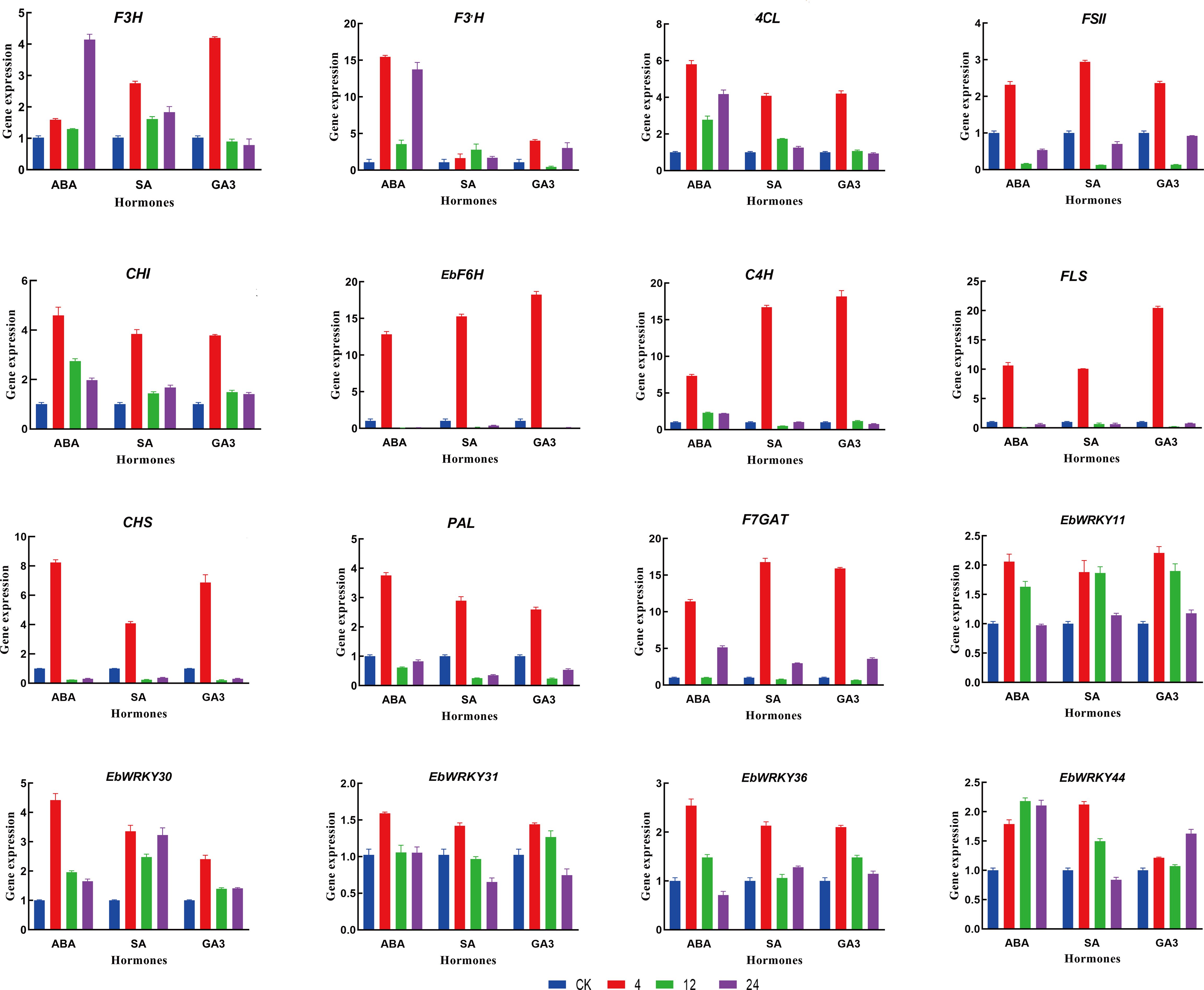
Figure 7 Relative expression of selected Ebgenes in response to exogenous hormone treatment. Genes expression was analyzed by RT-qPCR. Blue was used as the untreated control (expression = 1); Red, green, and purple represent 4h, 12h, and 24h. Error bars represent standard errors. Data were calculated using the 2–ΔΔCt method.
The expression patterns of EbWRKY11, EbWRKY30, EbWRKY31, EbWRKY36, and EbWRKY44 transcription factors related to the structural genes involved in flavonoid biosynthesis varied under different hormone treatments. EbWRKY11 showed the highest expression level after 4 h of hormone treatment and gradually decreased after 12 h, indicating that EbWRKY11 was sensitive to ABA, SA, and GA3. EbWRKY30 was sensitive to ABA and had the highest expression level at 4 h with a 4.3-fold increase. In the SA treatment, an increase was followed by a decrease in the volatility of EbWRKY30. EbWRKY31 showed a significant response to ABA treatment, and the expression level gradually decreased after SA and GA3 treatment at 4h. EbWRKY36 exhibited a > 2-fold change in expression after 4 h of hormone treatments. EbWRKY44 was more sensitive to SA than to ABA and GA3, with a 2.2-fold expression at 4 h. However, the gene expression level was highest at 12 h and 24 h of ABA treatment.
3.9 Protein-DNA interactions between EbWRKY11, EbWRKY36, EbWRKY44, and F7GAT and EbF6H
The yeast one-hybrid assay was conducted to validate the interaction between EbWRKY36, EbWRKY44, EbWRKY11 and two key structural genes, F7GAT and EbF6H, which encode key enzymes involved in the conversion of apigenin to scutellarin. The bait vector was constructed by utilizing the high GC% content of the F7GAT and EbF6H promoter domains. The results demonstrated that EbWRKY36, EbWRKY44, and EbWRKY11 exhibit binding affinity towards the promoter region F7GAT (1801-2500bp) (Figure 8A). Unfortunately, EbF6H was proven to have a self-activation function (Figure 8B). The two predicted regions with high scores containing WRKY-binding sites (ATAGTCAACT and TTCAAAGTCAAA) were truncated for verification. Notably, self-activation was observed in the 1501-2100bp region, while the 501-800bp region exhibited no self-activation but lacked interaction with the transcription factors EbWRKY36 and EbWRKY44, as well as EbWRKY11 (Figures 8A, B; Supplementary Table S10). These findings confirm that these three WRKY transcription factors of E. breviscapus play a role in the transcriptional regulation of key structural genes and regulated biosynthesis of scutellarin, a crucial active ingredient.
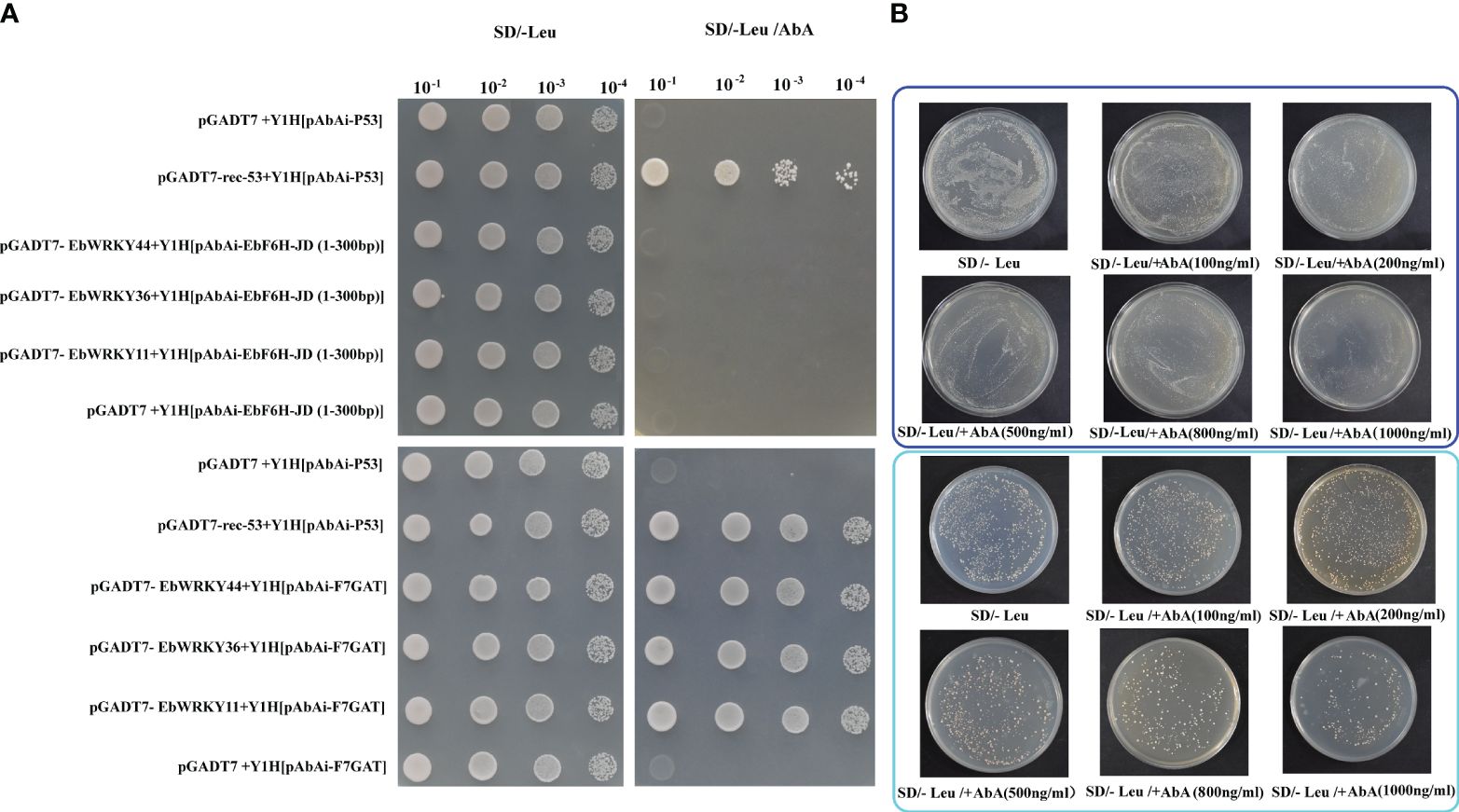
Figure 8 Protein-DNA interactions between EbWRKY11, EbWRKY36, EbWRKY44 with EbF6H and F7GAT. (A) Yeast One-Hybrid experiments. The pGADT7 was an AD empty vector; pGADT7-rec-53+Y1H[pAbAi-P53] was a positive control and pGADT7 +Y1H[pAbAi-P53] was a negative control. The EbF6H-JD (1-300bp) represents the promoter area of EbF6H 501-800bp. (B) EbF6H self-activation experiments. The upper blue box shows the full-length EbF6H promoter and the cyan box shows the area of 1501~2100bp promoter after the truncated EbF6H.
4 Discussion
The origins of WRKY transcription factors can be traced back to prokaryotes, with their presence limited to certain diplomonads, social amoebae, other amoebozoa species, and members of the fungal class incertae sedis (Rinerson et al., 2015; Chen et al., 2017). The 75 EbWRKYs were classified into two major branches: Group I was separated into a single branch, while the remaining EbWRKYs formed two complex branches consisting of individual sub-branches including Group IIc, Group IIa + IIb, Group IId + IIe, and Group III. Group II has the most numbers of EbWRKYs. Based on previous studies, WRKYs might evolve from the common ancestors, Group I WRKYs representing a more primitive form that subsequently evolved into Group II, encompassing subgroups IIa + IIb, IIc, and IId + IIe, Group III was closely related to Group IId and Group IIe (Zhang and Wang, 2005). In contrast, the present study revealed a closer genetic relationship between Group IIa + IIb and Group III compared to that between Group IId + IIe, suggesting a stronger evolutionary connection of EbWRKYs in Group IIa + IIb with those in Group III. These findings imply a shared ancestral origin among these genes.
The conserved domains of the EbWRKY protein were further evaluated according to the motif characteristics of three categories and five subtypes. All WRKY protein sequences exhibited a completely conserved domain (WRKYGQK) and zinc finger structure associated with the WRKY motif except for EbWRKY8. The gene family phylogenetic analyses were consistent with the results of the motif structure and sequence alignment. The findings further substantiate that the evolutionary pattern of Group I WRKYs exhibits a higher degree of conservatism compared to other types, which was reported in previous study (Eulgem et al., 2000). Multiple sequence alignments indicated that, regardless of the common WRKYGQK heptapeptide sequence, there were other variations, mainly distributed in Group IIc (EbWRKY68/69) and Group IId (EbWRKY2/72/73). In addition, the WRKY domain variations of EbWRKYs occurred in Group III (EbWRKY40). The differences in the conserved domain of the WRKY protein may be caused by variations in the WRKYGQK heptapeptide sequence and zinc finger structure during evolution or the deletion mutation of amino acid residues (Zhang and Wang, 2005; Wang et al., 2011; Llorca et al., 2014). Mutations in the conserved domain reflect the diversity of the evolution of the plant WRKY gene family, similar minor variations have been observed in Citrus and rice (Xie et al., 2005; Ayadi et al., 2016). Variations in WRKYGQKs affect its affinity for the W-box and further influence its function (Eulgem et al., 2000; Maeo et al., 2001). Therefore, the interactions between EbWRKY proteins with these variations and downstream target genes, and their binding preferences with cis-acting W-box elements, should be further investigated.
The bioactive flavonoids, particularly scutellarin, are predominantly distributed in the leaves of E. breviscapus, which serves as the primary raw material for pharmaceutical extraction. In this study, 54 flavone and flavonol compounds showed spatiotemporal accumulation in the leaves of E. breviscapus after hormone treatments, notably, GA3 significantly improved the accumulation of flavones and flavonols in the leaves. Meanwhile, EbWRKY30, EbWRKY31, EbWRKY36, and EbWRKY44 exhibited specific high expression levels in the leaves of E. breviscapus, while the expression level of EbWRKY11 was lower than that in the roots but higher than in other tissues after hormone treatment. EbWRKY44 and EbWRKY36 exhibit significant increases in response to three hormones and treatment for 4 hours, while EbWRKY30 and EbWRKY31 specifically respond to ABA treatment only, displaying an inverse expression pattern of transcripts. These tissue-specific expression patterns suggest the potential involvement of these five EbWRKY transcription factors in flavonoid metabolism within leaves, while also indicating a possible role for EbWRKY11 in root metabolism and development.
The WRKY transcription factor can bind to the promoter regions of functional genes or be induced by external stimuli, thereby regulating gene transcription levels involved in secondary metabolite accumulation (Liu et al., 2015; Bray, 1997; Shinozaki and Yamaguchi, 2000). Our previous work showed that PAL, EbF6H, C4H, 4CL, F7GAT, CHI, CHS, FLS, FSII, F3H, and F3’H were key structural genes regulating flavonoid biosynthesis in the leaves of E. breviscapus (Gao et al., 2022; Zhao et al., 2022). MdWRKY11 regulates anthocyanin synthesis through directly binding to the flavonoid 3-O-glycosyl transferase promoter in apple (Liu et al., 2019). McWRKY71 controls McANR and proanthocyanidin synthesis in Malus crabapple (Zhang et al., 2022). Other reports demonstrated that FaWRKY71 stimulates anthocyanin accumulation in strawberry (Fragaria×ananassa) by up-regulating FaF3’H, FaLAR, and FaANR (Yue et al., 2022). In this study, the structural genes of flavonoid biosynthesis of E. breviscapus significantly increased after 4 h of exogenous hormone treatment. The 11 structural gene expression patterns are basically consistent with the experimental results of qRT-PCR analysis in hormone treatment. EbWRKY11, EbWRKY30, EbWRKY31, EbWRKY36, and EbWRKY44 are closely related to the genes of flavonoid biosynthesis. qRT-PCR analysis further verified the expression patterns of the structural genes involved in the flavonoid biosynthesis pathway are consistent with EbWRKYs. Therefore, these five EbWRKY transcription factors may participate in the transcription of key structural genes that regulate flavonoid metabolite accumulation.
The phylogenetic analysis revealed that EbWRKY11, EbWRKY30, EbWRKY31, EbWRKY36, and EbWRKY44 were distributed across all three WRKY groups. The homologous genes of EbWRKY44 in Group IId were AtWRKY7/11/15/17 in Arabidopsis. In Group IIb, AtWRKY6, AtWRKY31, and AtWRKY42 were co-orthologous to EbWRKY31. EbWRKY30 in Group III was homologous to AtWRKY30/46/41/53. EbWRKY36 and EbWRKY11 were orthologs of AtWRKY1 and AtWRKY32, which belonged to different nodes in the same branch of Group I. Previous studies have shown that WRKY transcription factors in Arabidopsis are widely involved in the regulation of biotic and abiotic stress, plant growth, and development. AtWRKY15 regulates Arabidopsis growth and the salt stress response, whereas AtWRKY7/11/17 are negative regulators of the PAMP immune system, which could enhance plant resistance to pathogens (Vanderauwera et al., 2012; Arraño-Salinas et al., 2018). In addition, AtWRKY7 contains a CaM-binding domain, a new CaM-binding transcription factor that regulates plant growth and development and plays an important role in Ca2+signal transduction (Park et al., 2005). Moreover, MxWRKY55 and VvWRKY28 from Malus xiaojinensis and grape respectively, belong to Group II with WRKY TFs playing a role in plant resistance and contributing to higher salt tolerance (Han et al., 2020; Liu et al., 2022). EbWRKY31 and EbWRKY44 both belong to Group II and may be involved in salt stress response. Overexpression of AtWRKY30 enhances abiotic stress tolerance in A. thaliana at the early growth stage (Scarpeci et al., 2013). AtWRKY1 and AtWRKY41 can resist Pseudomonas syringae (Mukhi et al., 2021). AtWRKY42 regulates Pi homeostasis to adapt to environmental changes, and AtWRKY6 participates in Pi transportation (Robatzek and Somssich, 2001; Su et al., 2015). AtWRKY53 regulates stomatal movement and negatively regulates drought resistance, whereas AtWRKY46 is involved in the sensitivity of Arabidopsis to drought and salt stress (Ma et al., 2017; Freeborough et al., 2021). Based on the evolutionary analysis results of EbWRKYs and AtWRKYs, it was speculated that EbWRKYs homologous to various branches of Arabidopsis might participate in plant growth, development, and responses to biological and abiotic stresses with other members in the branch.
Transcription factors can act alone or in conjunction with other proteins to form complex binding complexes at gene promoters, thereby exerting control over gene expression through physical interactions that span long distances and coordinate the transcriptional activation of specific genes (Banerji et al., 1981; Karin, 1990; Latchman, 1997; Yao et al., 2015);. In apple, MdWRKY1 increases anthocyanin accumulation by activating MdLNC499 and MDERF109 expression (Ma et al., 2021). The interaction between PyWRKY26 and PybHLH3 targeting the promoter of PyMYB114 may potentially modulate the accumulation of anthocyanins in red-skinned pears (Li et al., 2020). In this study, protein-DNA interactions between EbWRKY11, EbWRKY36, EbWRKY44, and F7GAT and EbF6H, which were typical glycosylase and hydroxylase involved in scutellarin biosynthesis in E. breviscapus. The results revealed that EbWRKY36, EbWRKY44, and EbWRKY11 demonstrate direct binding to the promoter of F7GAT, whereas these three EbWRKYs did not exhibit interaction with the promoter region of EbF6H(501-800bp). EbF6H cannot be directly regulated by EbWRKY36, EbWRKY44, and EbWRKY11, however, it has been demonstrated in grape that the indirect regulation of structure genes on flavonoid biosynthesis hydroxylation occurs through the regulation of other regulatory elements. VvWRKY26 is preferentially recruited by a VvMYB5a-driven MBW complex to regulate flavonoid hydroxylation (Amato et al., 2019).
5 Conclusion
In this study, a total of 75 EbWRKY transcription factors were predicted from the genome of E. breviscapus. The amino acid number, molecular weight, predicted isoelectric point (PI) value, chromosome position, domain pattern, and conservative motif of EbWRKYs were revealed by bioinformatics-based analyses. The specificity of EbWRKYs gene expression in different tissues and their expression pattern under hormone treatment were determined based on RNA sequencing. Combining metabolome and transcriptome results revealed the regulatory mechanism between the WRKY transcription factor and key genes involved in flavonoid biosynthesis. The expression patterns of EbWRKY11, EbWRKY30, EbWRKY31, EbWRKY36, and EbWRKY44 transcription factors were similar to those of the 11 key structural genes involved in flavonoid biosynthesis. EbWRKY36, EbWRKY44, and EbWRKY11 can interact with the promoter of F7GAT, which was the key glycosyltransferase involved in scutellarin biosynthesis. We provided comprehensive information about the WRKY gene family of E. breviscapus and the mechanism of EbWRKY genes involved in flavonoid metabolism regulation.
Data availability statement
The raw data of E. breviscapus transcriptome in the current study are available in the National Center for Biotechnology Information (NCBI) database under project number PRJNA971382 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA971382). The genomic data of E. breviscapus is downloaded from the Medicinal Plants multi-Omics Database (http://medicinalplants.ynau.edu.cn/genome/detail/68).
Ethics statement
Experimental research and field studies on plants comply with relevant institutional, national, and international guidelines and legislation, and all methods were performed according to the relevant guidelines and regulations. The cultivated E. breviscapus were collected with official permissions of Yunnan Hongling Biological Technology Co., LTD. Honghe, China.
Author contributions
WS: Formal analysis, Writing – original draft. SZ: Data curation, Writing – original draft. QL: Software, Writing – review & editing. GX: Software, Writing – review & editing. YZ: Formal analysis, Project administration, Writing – review & editing. FW: Methodology, Writing – review & editing. GZ: Methodology, Writing – review & editing. SY: Funding acquisition, Investigation, Writing – review & editing. BH: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Fundamental Research Project of Yunnan (202101AS070037). The Science and Technique Programs in Yunnan Province (202102AE090042). Yunnan Characteristic Plant Extraction Laboratory (2022YKZY001), the First Projects of Science and Technology Plan in the Biomedical field in 2021 (202102AA310048).
Acknowledgments
The authors thank the lab members for their assistance.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1412574/full#supplementary-material
Abbreviations
ABA, abscisic acid; SA, salicylic acid; GA3, gibberellin 3; AtWRKY, Arabidopsis thaliana WRKY Transcription factor; EbWRKY, Erigeron breviscapus (Vant.) Hand.-Mazz WRKY Transcription factor; HaWRKY, Helianthus annuus WRKY Transcription factor; qRT-PCR, Quantitative RT-PCR; PAL: Phenylalanine ammonia-lyase; C4H: Cinnamate 4-hydroxylase; 4CL, 4-coumarate, CoA ligase; CHS, Chalcone synthase; CHI, Chalcone isomerase; FSII, Flavone synthase II; EbF6H, Flavone 6-hydroase; F7GAT: Flavonoid 7-O-glucuronosyltransferase; F3H: Flavanone 3-hydroxylase; F3’H, Flavonoid 3’-hydroxylase; FLS, Flavonol synthase; UPLC, Ultra-high-performance liquid chromatography; SE, Scutellarin.
References
Agati, G., Azzarello, E., Pollastri, S., Tattini, M. (2012). Flavonoids as antioxidants in plants: location and functional significance. Plant science. 196, 67–76. doi: 10.1016/j.plantsci.2012.07.014
Amato, A., Cavallini, E., Walker, A. R., Pezzotti, M., Bliek, M., Quattrocchio, F., et al. (2019). The MYB5-driven MBW complex recruits a WRKY factor to enhance the expression of targets involved in vacuolar hyper-acidification and trafficking in grapevine. Plant J. 99, 1220–1241. doi: 10.1111/tpj.14419
Amato, A., Cavallini, E., Zenoni, S., Finezzo, L., Begheldo, M., Ruperti, B., et al. (2017). A Grapevine TTG2-Like WRKY transcription factor is involved in regulating vacuolar transport and flavonoid biosynthesis. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01979
Arraño-Salinas, P., Domínguez-Figueroa, J., Herrera-Vásquez, A., Zavala, D., Medina, J., Vicente-Carbajosa, J., et al. (2018). WRKY7,-11 and-17 transcription factors are modulators of the bZIP28 branch of the unfolded protein response during PAMP-triggered immunity in Arabidopsis thaliana. Plant Sci. 277, 242–250. doi: 10.1016/j.plantsci.2018.09.019
Ayadi, M., Hanana, M., Kharrat, N., Merchaoui, H., Marzoug, R. B., Lauvergeat, V., et al. (2016). The WRKY transcription factor family in Citrus: valuable and useful candidate genes for Citrus breeding. Appl. Biochem. Biotechnol. 180, 516–543. doi: 10.1007/s12010-016-2114-8
Banerji, J., Rusconi, S., Schaffner, W. (1981). Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 27, 299–308. doi: 10.1016/0092-8674(81)90413-X
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bray, E. A. (1997). Plant responses to water deficit. Trends Plant Sci. 2, 48–54. doi: 10.1016/S1360-1385(97)82562-9
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, F., Hu, Y., Vannozzi, A., Wu, K., Cai, H., Qin, Y., et al. (2017). The WRKY transcription factor family in model plants and crops. Crit. Rev. Plant Sci. 36, 311–335. doi: 10.1080/07352689.2018.1441103
Chen, Y. J., Chen, C., Li, M. Y., Li, Q. Q., Zhang, X. J., Huang, R., et al. (2021). Scutellarin reduces cerebral ischemia reperfusion injury involving in vascular endothelium protection and PKG signal. Nat. Prod Bioprospect. 11, 659–670. doi: 10.1007/s13659-021-00322-z
Deng, Z. L., Münch, P. C., Mreches, R., McHardy, A. C. (2022). Rapid and accurate identification of ribosomal RNA sequences via deep learning. Nucleic Acids Res. 50, e60. doi: 10.1093/nar/gkac112
Di, P., Wang, P., Yan, M., Han, P., Huang, X., Yin, L., et al. (2021). Genome-wide characterization and analysis of WRKY transcription factors in Panax ginseng. BMC Genomics 22, 1–15. doi: 10.1186/s12864-021-08145-5
Dong, J., Chen, C., Chen, Z. (2003). Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Mol. Biol. 51, 21–37. doi: 10.1023/A:1020780022549
Duan, S., Wang, J., Gao, C., Jin, C., Li, D., Peng, D., et al. (2018). Functional characterization of a heterologously expressed Brassica napus WRKY41-1 transcription factor in regulating anthocyanin biosynthesis in Arabidopsis thaliana. Plant Sci. 268, 47–53. doi: 10.1016/j.plantsci.2017.12.010
Dunn, W. B., Broadhurst, D., Begley, P., Zelena, E., Francis-McIntyre, S., Anderson, N., et al. (2011). Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 6, 1060–1083. doi: 10.1038/nprot.2011.335
Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Eulgem, T., Rushton, P. J., Schmelzer, E., Hahlbrock, K., Somssich, I. E. (1999). Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J. 18, 4689–4699. doi: 10.1093/emboj/18.17.4689
Freeborough, W., Gentle, N., Rey, M. E. C. (2021). WRKY transcription factors in cassava contribute to regulation of tolerance and susceptibility to cassava mosaic disease through stress responses. Viruses. 13 (9), 1820. doi: 10.3390/v13091820
Gao, Q., Song, W., Li, X., Xiang, C., Chen, G., Xiang, G., et al. (2022). Genome-wide identification of bHLH transcription factors: Discovery of a candidate regulator related to flavonoid biosynthesis in Erigeron breviscapus. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.977649
Grunewald, W., Smet, I., Lewis, D. R., Löfke, C., Jansen, L., Goeminne, G., et al. (2012). Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc. Natl. Acad. Sci. U S A. 109, 1554–1559. doi: 10.1073/pnas.1121134109
Guo, C., Guo, R., Xu, X., Gao, M., Li, X., Song, J., et al. (2014). Evolution and expression analysis of the grape (Vitis vinifera L.) WRKY gene family. J. Exp. Bot. 65 (6), 1513–1528. doi: 10.1093/jxb/eru007
Han, D., Ding, H., Chai, L., Liu, W., Zhang, Z., Hou, Y., et al. (2018a). Isolation and characterization of MbWRKY1, a WRKY transcription factor gene from Malus baccata (L.) Borkh involved in drought tolerance. Can. J. Plant Sci. 98, 1023–1034. doi: 10.1139/cjps-2017-0355
Han, D., Hou, Y., Ding, H., Zhou, Z., Li, H., Yang, G. (2018b). Isolation and preliminary functional analysis of MbWRKY4 gene involved in salt tolerance in transgenic tobacco. Int. J. Agric. Biol. 20, 433–441. doi: 10.1080/17429145.2018.1499145
Han, D., Zhou, Z., Du, M., Li, T., Wu, X., Yu, J., et al. (2020). Overexpression of a Malus xiao**ensis WRKY transcription factor gene (MxWRKY55) increased iron and high salinity stress tolerance in Arabidopsis thaliana. In Vitro Cell. Dev. Biology-Plant 56, 600–609. doi: 10.1007/s11627-020-10129-1
Ishiguro, S., Nakamura, K. (1994). Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ Upstream regions of genes coding for sporamin and β-amylase from sweet potato. Mol. Gen. Genet. 244, 563–571. doi: 10.1007/BF00282746
Jin, J., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045. doi: 10.1093/nar/gkw982
Johnson, C. S., Kolevski, B., Smyth, D. R. (2002). TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 14, 1359–1375. doi: 10.1105/tpc.001404
Ju, S. H., Tan, L. R., Liu, P. W., Tan, Y. L., Zhang, Y. T., Li, X. H., et al. (2021). Scutellarin regulates osteoarthritis in vitro by inhibiting the PI3K/AKT/mTOR signaling pathway. Mol. Med. Rep. 23, 83. doi: 10.3892/mmr.2020.11722
Karin, M. (1990). Too many transcription factors: Positive and negative interactions. New Biol. 2, 126–131.
Khan, M. I., Fatma, M., Per, T. S., Anjum, N. A., Khan, N. A. (2015). Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.00462
Latchman, D. S. (1997). Transcription factors: An overview. Int. J. Biochem. Cell Biol. 29, 1305–1312. doi: 10.1016/S1357-2725(97)00085-X
Li, Z., Ahammed, G. J. (2023). Hormonal regulation of anthocyanin biosynthesis for improved stress tolerance in plants. Plant Physiol. Biochem. 201, 107835. doi: 10.1016/j.plaphy.2023.107835
Li, C., Li, D., Shao, F., Lu, S. (2015). Molecular cloning and expression analysis of WRKY transcription factor genes in Salvia miltiorrhiza. BMC Genomics 16, 200. doi: 10.1186/s12864-015-1411-x
Li, C., Wu, J., Hu, K. D., Wei, S. W., Sun, H. Y., Hu, L. Y., et al. (2020). PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Hortic. Res. 7, 37. doi: 10.1038/s41438-020-0254-z
Liu, X., Cheng, J., Zhang, G., Ding, W., Duan, L., Yang, J. (2018). Engineering yeast for the production of breviscapine by genomic analysis and synthetic biology approaches. Nat. Commun. 9, 448. doi: 10.1038/s41467-018-02883-z
Liu, W., Liang, X., Cai, W., Wang, H., Liu, X., Cheng, L., et al. (2022). Isolation and functional analysis of VvWRKY28, a Vitis vinifera WRKY transcription factor gene, with functions in tolerance to cold and salt stress in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 23, 13418. doi: 10.3390/ijms232113418
Liu, J., Osbourn, A., Ma, P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708. doi: 10.1016/j.molp.2015.03.012
Liu, W. J., Wang, Y. C., Yu, L., Jiang, H. Y., Guo, Z. W., Xu, H. F., et al. (2019). MdWRKY11 participates in anthocyanin accumulation in red-fleshed apples by affecting MYB transcription factors and the photoresponse factor MdHY5. J. Agric. Food Chem. 67, 8783–8793. doi: 10.1021/acs.jafc.9b02920
Llorca, C. M., Potschin, M., Zentgraf, U. (2014). bZIPs and WRKYs: two large transcription factor families executing two different functional strategies. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00169
Lucho-Constantino, G. G., , Zaragoza-Martínez., F., Ponce-Noyola, T., Carlos, M., Cerda-García-, R., Ramos-Valdivia, A. C. (2017). Antioxidant responses under jasmonic acid elicitation comprise enhanced production of flavonoids and anthocyanins in Jatropha curcas leaves. Acta Physiol. Plant 39, 165. doi: 10.1007/s11738-017-2461-2
Ma, J., Gao, X., Liu, Q., Shao, Y., Zhang, D., Jiang, L., et al. (2017). Overexpression of TaWRKY146 increases drought tolerance through inducing stomatal closure in Arabidopsis thaliana. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02036
Ma, H., Yang, T., Li, Y., Zhang, J., Wu, T., Song, T., et al. (2021). The long noncoding RNA MdLNC499 bridges MdWRKY1 and MdERF109 function to regulate early-stage light-induced anthocyanin accumulation in apple fruit. Plant Cell. 33, 3309–3330. doi: 10.1093/plcell/koab188
Maeo, K., Hayashi, S., Kojima-Suzuki, H., Morikami, A., Nakamura, K. (2001). Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins. Biosci. Biotechnol. Biochem. 65, 2428–2436. doi: 10.1271/bbb.65.2428
Mukhi, N., Brown, H., Gorenkin, D., Ding, P., Bentham, A. R., Stevenson, C. E. M., et al. (2021). Perception of structurally distinct effectors by the integrated WRKY domain of a plant immune receptor. Proc. Natl. Acad. Sci. U S A. 118 (50), e2113996118. doi: 10.1073/pnas.2113996118
Park, C. Y., Lee, J. H., Yoo, J. H., Moon, B. C., Choi, M. S., Kang, Y. H., et al. (2005). WRKY group IId transcription factors interact with calmodulin. FEBS Lett. 579, 1545–1550. doi: 10.1016/j.febslet.2005.01.057
Pourcel, L., Routaboul, J. M., Cheynier, V., Lepiniec, L., Debeaujon, I. (2007). Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci. 12, 29–36. doi: 10.1016/j.tplants.2006.11.006
Rinerson, C. I., Rabara, R. C., Tripathi, P., Shen, Q. J., Rushton, P. J. (2015). The evolution of WRKY transcription factors. BMC Plant Biol. 15, 66. doi: 10.1186/s12870-015-0456-y
Robatzek, S., Somssich, I. E. (2001). A new member of the Arabidopsis WRKY transcription factor family, AtWRKY6, is associated with both senescence- and defence-related processes. Plant J. 28, 123–133. doi: 10.1046/j.1365-313X.2001.01131.x
Rushton, P. J., Somssich, I. E., Ringler, P., Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Scarpeci, T. E., Zanor, M. I., Mueller-Roeber, B., Valle, E. M. (2013). Overexpression of AtWRKY30 enhances abiotic stress tolerance during early growth stages in Arabidopsis thaliana. Plant Mol. Biol. 83, 265–277. doi: 10.1007/s11103-013-0090-8
Schluttenhofer, C., Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 167, 295–306. doi: 10.1104/pp.114.251769
Shinozaki, K., Yamaguchi, K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. doi: 10.1016/S1369-5266(00)80068-0
Su, T., Xu, Q., Zhang, F. C., Chen, Y., Li, L. Q., Wu, W. H., et al. (2015). WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in Arabidopsis. Plant Physiol. 167, 1579–1591. doi: 10.1104/pp.114.253799
Vanderauwera, S., Vandenbroucke, K., Inzé, A., Cotte, B., Mühlenbock, P., Rycke, R., et al. (2012). AtWRKY15 perturbation abolishes the mitochondrial stress response that steers osmotic stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U S A. 109 (49), 20113–20118. doi: 10.1073/pnas.1217516109
Vives-Peris, V., Marmaneu, D., Gómez-Cadenas, A., Pérez-Clemente, R. M. (2018). Characterization of Citrus WRKY transcription factors and their responses to phytohormones and abiotic stresses. Springer Netherlands. 62, 33–44. doi: 10.1007/s10535-017-0737-4
Wang, Y., Liu, X., Chen, B., Liu, W., Guo, Z., Liu, X., et al. (2022). Metabolic engineering of Yarrowia lipolytica for scutellarin production. Synth Syst. Biotechnol. 7, 958–964. doi: 10.1016/j.synbio.2022.05.009
Wang, Z., Luo, Z., Liu, Y., Li, Z., Liu, P., Bai, G., et al. (2021). Molecular cloning and functional characterization of NtWRKY11b in promoting the biosynthesis of flavonols in Nicotiana tabacum. Plant Sci. 304, 110799. doi: 10.1016/j.plantsci.2020.110799
Wang, J. B., Pu, S. B., Sun, Y., Li, Z. F., Niu, M., Yan, X. Z., et al. (2014). Metabolomic profiling of autoimmune hepatitis: the diagnostic utility of nuclear magnetic resonance spectroscopy. J. Proteome Res. 13, 3792–3801. doi: 10.1021/pr500462f
Wang, N., Song, G., Zhang, F., Shu, X., Cheng, G., Zhuang, W., et al. (2023). Characterization of the WRKY gene family related to anthocyanin biosynthesis and the regulation mechanism under drought stress and methyl jasmonate treatment in Lycoris radiata. Int. J. Mol. Sci. 24, 2423. doi: 10.3390/ijms24032423
Wang, Q., Wang, M., Zhang, X., Hao, B., Kaushik, S. K., Pan, Y. (2011). WRKY gene family evolution in Arabidopsis thaliana. Genetica. 139, 973–983. doi: 10.1007/s10709-011-9599-4
Wei, Y., Meng, N., Wang, Y., Cheng, J., Duan, C., Pan, Q. (2023). Transcription factor VvWRKY70 inhibits both norisoprenoid and flavonol biosynthesis in grape. Plant Physiol. 193, 2055–2070. doi: 10.1093/plphys/kiad423
Xie, Z., Zhang, Z. L., Zou, X., Huang, J., Ruas, P., Thompson, D., et al. (2005). Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol. 137, 176–189. doi: 10.1104/pp.104.054312
Xu, N., Liu, S., Lu, Z., Pang, S., Wang, L., Wang, L., et al. (2020). Gene expression profiles and flavonoid accumulation during salt stress in Ginkgo biloba seedlings. Plants. 9 (9), 1162. doi: 10.3390/plants9091162
Yamamoto, R., Ma, G., Zhang, L., Hirai, M., Yahata, M., Yamawaki, K., et al. (2020). Effects of salicylic acid and methyl jasmonate treatments on flavonoid and carotenoid accumulation in the juice sacs of satsuma mandarin in vitro. Appl. Sci. 10 (24), 8916. doi: 10.3390/app10248916
Yang, L., Luo, S., Jiao, J., Yan, W., Zeng, B., He, H., et al. (2023). Integrated transcriptomic and metabolomic analysis reveals the mechanism of gibberellic acid regulates the growth and flavonoid synthesis in phellodendron chinense schneid seedlings. Int. J. Mol. Sci. 24, 16045. doi: 10.3390/ijms242216045
Yang, L., Tao, Y., Luo, L., Zhang, Y., Wang, X., Meng, X. (2022a). Dengzhan Xixin injection derived from a traditional Chinese herb Erigeron breviscapus ameliorates cerebral ischemia/reperfusion injury in rats via modulation of mitophagy and mitochondrial apoptosis. J. Ethnopharmacol. 288, 114988. doi: 10.1016/j.jep.2022.114988
Yang, L., Yan, Y., Zhao, B., Xu, H., Su, X., Dong, C. (2022b). Study on the regulation of exogenous hormones on the absorption of elements and the accumulation of secondary metabolites in the medicinal plant artemisia argyi leaves. Metabolites. 12, 984. doi: 10.3390/metabo12100984
Yang, Y., Zhou, Y., Chi, Y., Fan, B., Chen, Z. (2017). Characterization of soybean wrky gene family and identification of soybean WRKY genes that promote resistance to soybean cyst nematode. Sci. Rep. 7, 17804. doi: 10.1038/s41598-017-18235-8
Yao, L., Shen, H., Laird, P. W., Farnham, P. J., Berman, B. P. (2015). Inferring regulatory element landscapes and transcription factor networks from cancer methylomes. Genome Biol. 16, 105. doi: 10.1186/s13059-015-0668-3
Yin, W., Wang, X., Liu, H., Wang, Y., Nocker, S., Tu, M., et al. (2022). Overexpression of VqWRKY31 enhances powdery mildew resistance in grapevine by promoting salicylic acid signaling and specific metabolite synthesis. Hortic. Res. 9, uhab064. doi: 10.1093/hr/uhab064
Yu, Y., Hu, R., Wang, H., Cao, Y., He, G., Fu, C., et al. (2013). MlWRKY12, a novel Miscanthus transcription factor, participates in pith secondary cell wall formation and promotes flowering. Plantsci 212, 1–9. doi: 10.1016/j.plantsci.2013.07.010
Yu, F., Huaxia, Y., Lu, W., Wu, C., Cao, X., Guo, X., et al. (2012). GhWRKY15, a member of the WRKY transcription factor family identified from cotton (Gossypium hirsutum L.), is involved in disease resistance and plant development. BMC Plant Biol. 12, 144. doi: 10.1186/1471-2229-12-144
Yue, M., Jiang, L., Zhang, N., Zhang, L., Liu, Y., Wang, Y., et al. (2022). Importance of FaWRKY71 in strawberry (Fragaria x ananassa) fruit ripening. Int. J. Mol. Sci. 23, 12483. doi: 10.3390/ijms232012483
Zeng, M., Zhong, Y., Guo, Z., Yang, H., Zhu, H., Zheng, L., et al. (2022). Expression and functional study of bcWRKY1 in baphicacanthus cusia (Nees) bremek. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.919071
Zhang, Y., Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5, 1. doi: 10.1186/1471-2148-5-1
Zhang, J., Wang, Y., Mao, Z., Liu, W., Ding,., L., Zhang, X., et al. (2022). Transcription factor McWRKY71 induced by ozone stress regulates anthocyanin and proanthocyanidin biosynthesis in Malus crabapple. Ecotoxicol Environ. Saf. 232, 113274. doi: 10.1016/j.ecoenv.2022.113274
Zhao, Y., Zhang, G., Tang, Q., Song, W., Gao, Q., Xiang, G., et al. (2022). EbMYBP1, a R2R3-MYB transcription factor, promotes flavonoid biosynthesis in Erigeron breviscapus. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.946827
Keywords: flavonoid, Erigeron breviscapus, WRKY, hormones, structure gene
Citation: Song W, Zhang S, Li Q, Xiang G, Zhao Y, Wei F, Zhang G, Yang S and Hao B (2024) Genome-wide profiling of WRKY genes involved in flavonoid biosynthesis in Erigeron breviscapus. Front. Plant Sci. 15:1412574. doi: 10.3389/fpls.2024.1412574
Received: 05 April 2024; Accepted: 20 May 2024;
Published: 03 June 2024.
Edited by:
Huihui Li, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Wenjun Huang, Chinese Academy of Sciences (CAS), ChinaDeguo Han, Northeast Agricultural University, China
Copyright © 2024 Song, Zhang, Li, Xiang, Zhao, Wei, Zhang, Yang and Hao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Hao, Bing.Hao@hotmail.com; Shengchao Yang, 13099437499@163.com
†These authors have contributed equally to this work
 Wanling Song1,2†
Wanling Song1,2† Qi Li
Qi Li Guisheng Xiang
Guisheng Xiang Yan Zhao
Yan Zhao Fan Wei
Fan Wei Guanghui Zhang
Guanghui Zhang Shengchao Yang
Shengchao Yang Bing Hao
Bing Hao