- 1State Key Laboratory of Cotton Biology, Key Laboratory of Cotton Genetic Improvement, Ministry of Agriculture, Institute, Cotton Research of Chinese Academy of Agricultural Sciences, Anyang, China
- 2Zhengzhou Research Base, State Key Laboratory of Cotton Biology, School of Agricultural Sciences, Zhengzhou University, Zhengzhou, China
- 3Cotton Incorporated, Cary, NC, United States
- 4Department of Plant and Environmental Sciences, New Mexico State University, Las Cruces, NM, United States
Upland cotton (Gossypium hirsutum) is the world’s leading fiber crop and one of the most important oilseed crops. Genetic improvement of cotton has primarily focused on fiber yield and quality. However, there is an increased interest and demand for enhanced cottonseed traits, including protein, oil, fatty acids, and amino acids for broad food, feed and biofuel applications. As a byproduct of cotton production, cottonseed is an important source of edible oil in many countries and could also be a vital source of protein for human consumption. The focus of cotton breeding on high yield and better fiber quality has substantially reduced the natural genetic variation available for effective cottonseed quality improvement within Upland cotton. However, genetic variation in cottonseed oil and protein content exists within the genus of Gossypium and cultivated cotton. A plethora of genes and quantitative trait loci (QTLs) (associated with cottonseed oil, fatty acids, protein and amino acids) have been identified, providing important information for genetic improvement of cottonseed quality. Genetic engineering in cotton through RNA interference and insertions of additional genes of other genetic sources, in addition to the more recent development of genome editing technology has achieved considerable progress in altering the relative levels of protein, oil, fatty acid profile, and amino acids composition in cottonseed for enhanced nutritional value and expanded industrial applications. The objective of this review is to summarize and discuss the cottonseed oil biosynthetic pathway and major genes involved, genetic basis of cottonseed oil and protein content, genetic engineering, genome editing through CRISPR/Cas9, and QTLs associated with quantity and quality enhancement of cottonseed oil and protein.
Introduction
Upland cotton (Gossypium hirsutum L.) is the world’s leading fiber crop, as well as one of the most important oilseed crops along with soybean, rapeseed, sunflower and peanut.1 The production of the cotton fiber and cottonseed is normally at the ratio of 1:1.65, and cottonseed oil accounts for about 20% of the whole seed weight, and the oil is the second most valuable component of the cotton crop behind fiber, on a price per unit weight basis (O’Brien et al., 2005). Because of its rather neutral flavor, cottonseed oil is commonly desired by the food industry as it does not mask the natural flavor of the food used to cook or process. Cottonseed is also rich in high quality protein containing amino acids that are important for both human consumption (if the toxic gossypol is removed) and animal feeds, especially farm raised fish. The global production of cottonseed protein is estimated to be about 11 million metric tons annually. In fact, cottonseed is the second most important potential source of plant proteins after soybean (Spadaro and Gardner, 1979). However, cottonseed and its derivative products are traditionally regarded as a by-product of the more valuable cotton fiber production, providing only about 14–19% of farm-gate value in cotton production.2 The fact that cottonseed is a by-product of cotton production greatly improves its sustainability metrics compared to other oilseeds. Cotton research has thus far been understandably focused primarily on the yield and quality of cotton fiber, while the seed traits, except for seed germination and seed size, are relatively neglected. Consequently, the research and development focus on cottonseeds has been lagging behind other oilseed crops in spite of its abundance in availability and excellent potentials for improvement.
There is a long history of cottonseed oil utilization going back more than 100 years. This arose along with the cotton plantation in the new world and cottonseed oil dominated the vegetable oil market until the rise of soybean oil and canola oil in the 1950s. As Upland cotton production was expanded from the United States to other countries, the use of cottonseed oil for food and protein for animal feed became common in all the cotton growing areas in the world. Cottonseed oil is generally favored due to not only its ready availability and specifically developed extraction technology, but also its bland flavor, does not mask the true flavor of the food that it cooks. Its high smoke point makes it ideal and somewhat superior to other vegetable oils and animal fats for frying applications. As it contains a relatively high level of saturated fatty acids that confers high oxidative stability and high melting point, cottonseed oil has also commonly be used in food industry as “an invisible oil” in the processed snack foods, margarine making and various confectionery applications (Liu et al., 2008; Liu, 2011). More recently, the use of cottonseed oil for renewal fuels (mostly biodiesel) has also attracted considerable attention, as it has a negative carbon profile and could significantly reduce CO2 emission in comparison to fossil fuels (Karaosmanoglu et al., 1999; Meneghetti et al., 2007). The whole cottonseed or the meal following oil extraction is rich in proteins and used as popular source of animal feed. Globally, approximately 10 million metric tons of protein is produced by cottonseed (Kumar et al., 2021). Cottonseed protein is endowed with a high level of arginine relative to most plant-based proteins, which has been shown to slow down cancer progression, to act a principal regulator of blood pressure, and to cause a relaxation of cardiovascular smooth muscle cells following conversion to nitric oxide (Lowell et al., 1990; Moncada and Higgs, 1993). Lysine is an important amino acid for humans and animals; and cottonseed kernels contain on average 2.3% lysine (dry weight of kernel powder basis), higher than rice (2.15%) and lower than wheat (2.7%) (Chen et al., 1986). There is an increasing trend of using whole intact cottonseed for feeding lactating dairy cows by leveraging the rumen bypass effects offered by the thick seed coat and remaining fuzz (i.e., linters) following ginning. In addition, cotton is also rich in antioxidants such tocopherols with vitamin E as its main form (Smith and Creelman, 2001).
Despite the continued research focus on cotton fiber, the prospects of increased utilization of cottonseed oil as food, feed and biofuels, have encouraged researchers to develop ways to genetically improve cottonseed products and maximize the outcome for enhanced fiber production and quality, improved nutritional value and expanded industrial applications. Furthermore, there is an environmental impetus to develop such a sustainable byproduct of a valuable fiber crop because of its abundant availability without the need for additional land use and detrimental greenhouse gas emission (Zucker and Zucker, 1943; Ory and Flick, 1994; Alford et al., 1996).
Cotton has a complex genetic base as an allotetraploid species and complicated genetic mechanisms underpinning the accumulation of various valuable metabolites in cottonseed and the development of fibers which cover the seeds. Nevertheless, considerable progress has been made to elucidate the molecular and biochemical mechanism, which has also been used in numerous attempts in genetically enhancing the accumulation or alteration of the relative levels of protein, amino acids, oil, and fatty acid composition in cottonseed. The objective of this review is to summarize and discuss the cottonseed oil biosynthetic pathways and major genes involved, genetic basis of cottonseed oil and protein content, genetic engineering, genome editing through CRISPR/Cas9, and QTLs associated with quantity and quality enhancement of cottonseed oil and protein.
Cottonseed Oil and Storage Proteins Biosynthesis and Accumulation
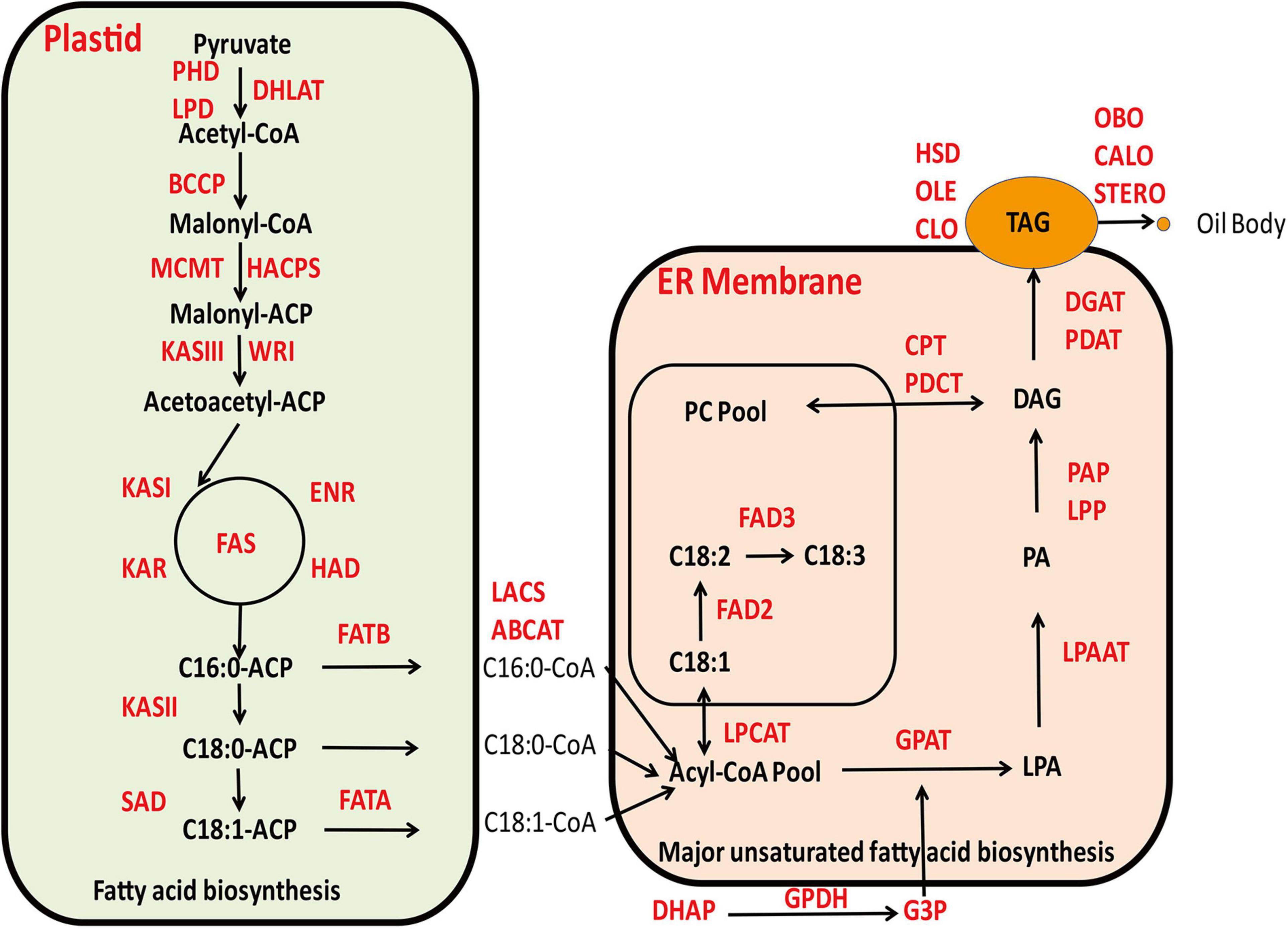
The biochemical processes involved in the biosynthesis of seed oil are relatively well known (Browse and Ohlrogge, 1995; Ohlrogge and Jaworski, 1997; Voelker and Kinney, 2001). Currently, there is a profound understanding on the biochemical and molecular functions of most steps in the lipid biosynthetic pathway, as well as the inheritance of phenotypic performance of various mutants corresponding to these metabolic steps in model plants (Byrne et al., 1996; McMullen et al., 1998). With the advent of numerous high quality genome sequence databases derived from cotton and associated Gossypium species, attempts have been made to identify key genes and their interactive gene networks that are involved in oil biosynthesis in cotton (Liu et al., 2009; Jiao et al., 2013; Hovav et al., 2015; Hu et al., 2016; Xu et al., 2016; Zhao et al., 2018c; Ma et al., 2021; Zhu et al., 2021). The current understanding for the general biosynthetic pathway of cottonseed oil is shown in Figure 1.
The major constituent of cottonseed oil is triacylglycerols (TAG) that is comprised of three fatty acids esterified on a glycerol backbone. Cottonseed oil accumulates during the maturation phase of the embryo, which is a highly compartmentalized process including de novo biosynthesis of fatty acids mainly occurring in plastids, production of glycerol 3-phoshpate (G-3-P) in cytoplasm and TAG assembly by dehydration condensation of acyl-CoA and G-3-P in endoplasmic reticulum (ER) (Nikolau et al., 2003). The TAG molecules contain the same acyl groups that are also found in membrane lipids, which are predominantly linoleate (18:2), followed by palmitate (16:0), oleate (18:1), stearate (18:0), and linolenate (18:3), in addition to a number of minor fatty acids (Cherry, 1983). The final step of TAG assembly, the acylation of the sn-3 position of 1,2 diacylglycerol catalyzed by diacylglycerol acyltransferase (DGAT) to form TAG is commonly regarded as a rate-limiting step (Guo et al., 2017) and plays a substantial role in determining oil content in cottonseed. The resulting TAG molecules in ER will accumulate within a sphere structure known as lipid droplets or oil bodies covered by a monolayer of phospholipids membrane that is decorated with numerous lipid droplets associated proteins, such as oleosin, caleosin, stereoleosin and others. When a lipid droplet reaches a certain size, it will bud off and be released into the cytoplasm (Voelker and Kinney, 2001). It has been generally recognized that DGAT1 plays a crucial role in determining TAG production as it catalyzes the rate-limiting step to convert DAG into TAG. However, there are reports that DGAT3 may play a more active role in promoting TAG biosynthesis in light of transcriptome analysis of developing cottonseeds, although empirical evidence is required (Hovav et al., 2015; Zhao et al., 2018c). A considerable body of literature is now available to imply that a number of transcription factors, such as WRI1, are playing an imperative role in fostering the carbon reallocation toward to de novo fatty acids biosynthesis in plastids and rendering increased availability for TAG biosynthesis (Kong et al., 2019). In congruence, WRI1 and NF-YB6 were highly expressed and displayed coordinated temporal patterns with oil accumulation in cottonseeds (Zhao et al., 2018b). That the key genes involved in de novo fatty acid biosynthesis, such as SAD6 and FATA, showed clear differential expression concomitant with oil accumulation, which were substantially highly expressed in G. barbadense than in G. hirsutum, highlighting the divergence between these two closely related allotetraploid cottons (Zhu et al., 2021). Some other enzymes in plastids, such as GhPEPC1, are not only involved in photosynthesis but also key to the inflowing of carbon turnover to fatty acid biosynthesis attributable to the accumulation of cottonseed oil (Xu et al., 2016). In higher plants, seed storage proteins are synthesized on the rough ER, using amino acids directly taken up by the embryo, or obtained after transamination reactions. Subsequently, they are transported into protein storage vacuoles by a vesicle-mediated pathway (Jolliffe et al., 2005). In cottonseed, two major classes of storage proteins are globulins and albumins, which differ in their solubility properties. Both globulins and albumins are synthesized and compartmentalized in storage protein vacuoles during cottonseed maturation (Dure and Chlan, 1981).
Factors Affecting Cottonseed Oil and Protein Contents
The contents of oil and protein in cottonseeds are quantitative traits that are simultaneously affected by genetic and environmental factors and their interactions. Cottonseed oil and protein contents often vary among different growing seasons, growing locations and years (Singh et al., 1985; Dani and Kohel, 1989; Ye et al., 2003). The interactive effects of the genotype × environment depend on not only environmental factors such as water, fertilizer, soil, light and temperature, but also on the relative contribution of the parent genotypes to the trait (Wang, 1992; Hom et al., 2015). However, the fatty acid composition of seed oil is mainly determined by the genotype of the developing embryo (embryogenic control) (Downey and Harvey, 1963; Ecker and Yaniv, 1993; Velasco et al., 2004). Kohel (1980) estimated a moderate heritability based on a 20 × 5 NCII design and a low heritability based on F2/F3 regression for cottonseed oil content. Heritability estimates for oil content varied from low (Meredith et al., 2012) to moderate (Zeng et al., 2015; Campbell et al., 2016; Kothari et al., 2016) or high (Yu et al., 2012; Zhao et al., 2019), depending on different genetic backgrounds of cultivars and testing environmental conditions in these studies. Singh et al. (1985) showed that the contents of protein and oil exhibited non-additive genetic effects with a substantial environmental influence based on a set of diallel crosses involving ten parents. However, Yuan et al. (2001) showed that oil content was controlled mainly by maternal additive effect, while protein content by direct additive effect in F3 seeds harvested from F2 hybrids between four Upland lines with the double recessive (gl2gl3) glandless trait and five Upland lines with the dominant glandless (Gl2e) trait. Based on F3 hybrids between 13 cotton chromosome substitution lines (CSLs) each carrying a pair of chromosomes or arms from G. barbadense and five elite Upland cultivars, Wu et al. (2010) confirmed that seed oil content had significant cytoplasmic effects and also dominance effects, while protein content had significant embryo additive effects based on the additive and dominance (AD) genetic model with cytoplasmic effects. In 316 Upland cotton accessions genotyped by 390 K SNPs, Du et al. (2018b) indicated that cottonseed protein, oil, palmitic, linoleic, oleic, myristic and stearic acid contents exhibited significant additive and dominance effects; however, the epistatic effects and genotype-environment interactions were largely diverse across traits. Therefore, cottonseed oil and protein contents are heritable traits that are controlled by multiple genes with additive and dominant effects with variable heritability estimates.
The content of cottonseed oil is strongly and negatively correlated with protein content (Hanny et al., 1978; Kohel and Cherry, 1983; Wu et al., 2009; Yu et al., 2012; Hinze et al., 2015; Campbell et al., 2016), but positively correlated with fiber length, fiber uniformity and fiber strength (Kothari et al., 2016). A recent study by Yuan et al. (2019) confirmed the significant positive correlation of the total fatty acid content in cottonseeds with fiber length and strength and its significant negative correlation with fiber uniformity. Furthermore, palmitic acid content was significantly and positively correlated with fiber elongation. However, the study further showed that the reverse was true for the correlations of cottonseed protein content with fiber length, strength, and uniformity. The correlation analysis further suggested that the above well-documented negative association between seed protein and oil contents may be to some extent attributed to the negative correlation between oleic acid and protein content. Research efforts to increase the oil content of cottonseed, which will most likely also decrease the level of protein, will actually have a positive impact on cottonseed value and sustainability metrics due to being able to reduce the level of fertilizer (mainly supplemental nitrogen) applied to the plant to support protein production in the seed. Increasing seed oil and reducing seed protein will therefore positively impact the carbon footprint for cotton production and utilization.
Genetic Variation Within Gossypium and Classical Genetic Studies of Cottonseed Oil, Fatty Acid, and Protein Contents
One of the biggest challenges in improving the cottonseed quality traits is the limited amendable genetic variability within cotton germplasm, despite the existence of great genetic variation in cottonseed oil content (17–27%) and protein content (16–36%) among cotton species and cultivars (Kohel, 1980; Wu et al., 2009; Dowd et al., 2010; Kothari et al., 2016). Sharif et al. (2019) showed that oil content was the highest in G. lobatum (24.82%) and G. harknessii (24.22%), whereas the Old World wild species had lower oil contents including G. stocksii and G. somalense with the lowest oil content (11.22%). Among the four cultivated species, G. barbadense had the highest oil content, followed by G. hirsutum; and the two A-genome diploid species (G. herbaceum and G. arboreum) showed the lowest level. After comparing 33 Gossypium species, Hinze et al. (2015) confirmed that diploid species except for the A- and K- genome species possessed the lowest oil and protein contents. Tetraploids (21.7%) and the K-genome species (21.4%) had the highest oil content (21.7%). In addition, large ranges in oil contents within the D genome and each cultivated species were observed. Agarwal et al. (2003) and Khan et al. (2015) also found significant variability for oil content in the cotton germplasm collections in India and Pakistan, respectively. Therefore, sufficient genetic variability in cottonseed oil exists within the Gossypium genus which could be utilized for making genetic gains (Kohel, 1978; Horn et al., 2011). However, the genetic improvement in oil accumulation of cottonseed is constrained by the rather limited variation among the elite Upland cotton (G. hirsutum) cultivars and lines as the result of extensive selection within the species toward improving lint yield and quality. Significant variation in cottonseed oil and protein contents may exist between cultivars and race stocks within Upland cotton. Hinze et al. (2015) showed that cultivated tetraploid accessions had higher oil content (22.7%) than wild tetraploid accessions (20.9%). Among four Upland cotton breeding lines and four semi-wild G. hirsutum accessions tested in multi-environments, Kothari et al. (2016) showed that the oil content ranged from 13 to 27% and the protein values ranged from 16 to 36%. The variability for oil content in elite cultivars and lines of G. hirsutum ranged from 14.5 to 22.0% with mean of 19.2% (Pandey, 1977). A classic breeding approach through crosses between selected germplasm led to a moderate increase in oil content (21.20–26.30%) as compared to their parents (21.48–24.16%) (Dani, 1988). To overcome the bottleneck of low genetic variation in existing Upland cotton, the cultivated allotetraploid cotton can be crossed with diploid cottons, followed by selection to improve oil accumulation (Thiagarajan and Ramaswamy, 1982). In addition to oil content, natural variations in fatty acid components such as oleic acid, myristic acid, linoleic acid and linolenic acid in cottonseed oil, have also been reported (Lukonge et al., 2007). However, fatty acid profiles in large cotton germplasm collections and breeding populations remain to be analyzed.
As for cottonseed protein, based on results from a large cotton germplasm collection (1,335 and 1,234 accessions in the Mississippi and Texas location, respectively), Kohel et al. (1985) suggested that there was sufficient variability for genetic improvement. Hinze et al. (2015) detected a wide range of protein content (10–36%) and oil content (8–27%) in 2,256 accessions representing five tetraploid and 28 diploid Gossypium species, and the results showed that wild diploid species generally had extremely low cottonseed protein contents. The Old World A-genome species had the highest protein content (23.8%), followed by the tetraploid species (21.6%). Cultivated tetraploid accessions had a wider range (14.9–35.9%) protein content than wild tetraploid accessions (15.4–30.7%) although both groups had similar mean protein content. Variation in protein components and relative content of the protein subunits were also investigated among cultivars (Song and Zhang, 2007).
In general, considerable variation in oil content and proteins content have been identified in cottonseed; however, inconsistency and relatively small magnitude of the variations poses significant challenge for their utilization in cotton breeding programs. Nonetheless, the existence of the variation offers promise for the discovery of greater variation if a greater segment of cotton germplasm is explored, especially the wild Gossypium species and those beyond the mainstream germplasm collections such as exotic cotton germplasm.
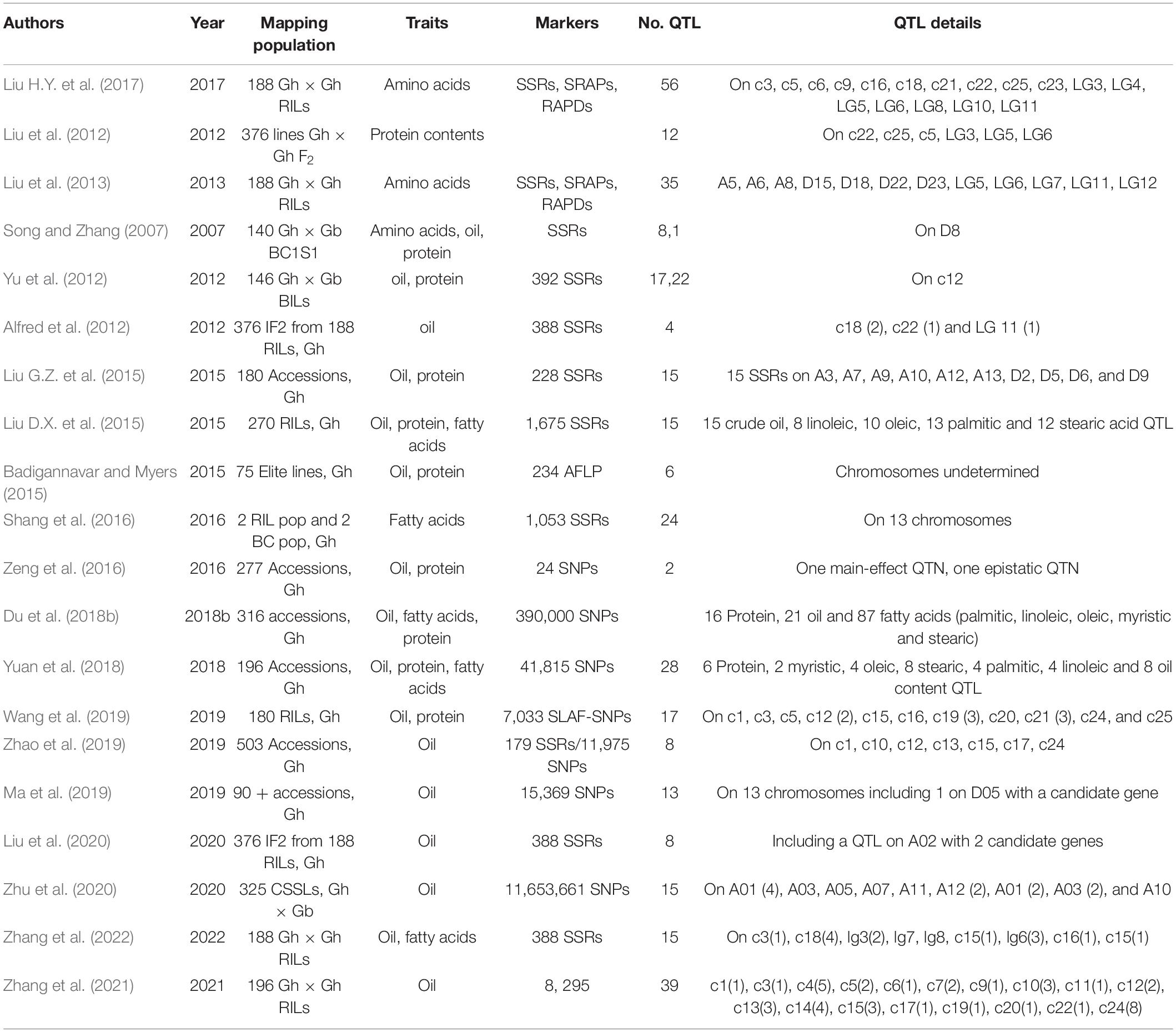
Quantitative Trait Loci Mapping and Genomewide Association Studies of Cottonseed Oil, Fatty Acid, and Protein Contents
Using linkage mapping and genomewide association studies (GWAS), QTLs associated with contents of cottonseed oil, fatty acids, protein, and amino acids have been detected in specifically designed genetic populations Zhang et al., 2022. Song and Zhang (2007) were among the first to report a single QTL for kernel oil percentage in an interspecific BC1S1 population derived from an interspecific G. hirsutum (Gh) TM-1 × G. barbadense (Gb) Hai 7124 cross based on simple sequence repeat (SSR) markers. This was followed by Yu et al. (2012) who mapped 12 QTLs in relevance to cottonseed oil, protein and gossypol contents using a different interspecific population comprised of backcross inbred lines (BILs). Up to date, a total of more than 160 QTLs for cottonseed oil content and more than 130 QTLs for different fatty acids were identified from at least 14 published studies (Table 1). These mapping populations included three Gh × Gb populations and five recombinant inbred lines (RIL) populations for linkage mapping, and six accession panels for GWAS. A meta-analysis was previously performed using the cotton QTL database with only a few reports on cottonseed oil and protein contents (Said et al., 2013, 2015a,b).3 More studies have been published since then, which requires a meta-analysis of QTLs to identify consistent QTLs and QTL hotspots or clusters across environments and genetic populations. It appears that some of the QTLs were in common or on similar chromosomal locations between/among studies. As a result, candidate genes for oil QTLs were identified or validated (Ma et al., 2019; Liu et al., 2020; Zhang et al., 2021). Furthermore, QTLs corresponding to various fatty acids were identified in other studies (Liu G.Z. et al., 2015; Du et al., 2018b; Yuan et al., 2018). However, only a small number of these QTLs were identified in multi-environments or multiple genetic backgrounds. For example, Ma et al. (2019) identified 19 QTLs for cottonseed oil content in multiple environments based on GWAS in Upland cotton, and a peroxidase (PRXR1) gene was confirmed to be the candidate gene within one of the QTL regions via virus induced gene silencing (VIGS). More recently, Zhang et al. (2021) reported that only five of 39 QTLs for cottonseed oil content were stable across different environments in a RIL population of 196 lines, and several genes including one coding for a transcription factor within the stable QTL regions were differently expressed during ovule development. Till now, none of the QTLs reported have been tracked using associated markers in genetic populations for cottonseed oil improvement. Hence, their direct application in marker-assisted selection (MAS) for oil content and quality is still unknown.
Chromosomes or genes introduced to G. hirsutum from G. barbadense and other tetraploid Gossypium species were found to affect cottonseed oil content substantially, based on CSLs (Wu et al., 2009, 2010; Bellaloui et al., 2020; Saha et al., 2020). Multiple QTL alleles from G. barbadense were demonstrated as highly promising for enhancing seed oil content in introgressed G. hirsutum lines (Zhu et al., 2020). Therefore, introgression breeding between G. hirsutum and G. barbadense may greatly improve the content of seed oil content and possibly fatty acid composition. These QTLs detected for seed quality traits in cotton are expected to be useful in cotton breeding to develop cotton with improved cottonseed nutrient quality. However, to date, all the genetic populations developed were small in size (100–200 progeny), which limited genetic recombination between parents. The extent to which cottonseed oil content can be increased and oil quality can be enhanced as the result of alteration in fatty acid composition through extensive introgression breeding remains unknown. The goal of research and breeding effects is to transfer the identified desirable QTLs into elite cotton cultivars for the improvements of both oil accumulation and oil quality without trade-offs in fiber yield or quality.
Gene Expression Studies During Cottonseed Oil and Protein Accumulation
Lipids and fatty acids are a large class of compounds existing in plants, and most edible vegetable oil consists of a few common fatty acids, including saturated, monounsaturated, and polyunsaturated fatty acids. Fatty acids are stored in seeds in the form of triacylglycerol (TAG). Therefore, the TAG biosynthetic pathway involving many enzymes has become one of the hallmarks of lipid biochemistry. Cottonseed oil contains 71% unsaturated fatty acids and 28% saturated fatty acids. Unsaturated fatty acids include 58% linoleic acid (18:2) and 13% oleic acid (18:1), and saturated fatty acids include 26% palmitic acid (16:0) and 2% stearic acid (18:0) (Cherry, 1983). In addition, there are many other minor fatty acids including dihydrosterculic acid (DHSA) (Dowd et al., 2010; Dowd, 2012).
Since the lipid and protein biosynthetic pathways compete for the same substrate using phosphoenolpyruvate through acetyl-CoA carboxylase (ACCase) and phosphoenolpyruvate carboxylase (PEPC), respectively, it is not surprising that cottonseed oil and protein contents are negatively correlated as reported previously (Hanny et al., 1978; Kohel and Cherry, 1983; Wu et al., 2009; Yu et al., 2012; Hinze et al., 2015; Campbell et al., 2016). Cui et al. (2017) showed that overexpression of GhACCase subunits resulted in increased cottonseed oil content by 17–22%. A large number of oil-related genes, such as fatty acyl-ACP thioesterase B (FATB), acyl carrier protein 5 (ACP5) (Yuan et al., 2018) and KASIII (Du et al., 2018a), have been identified by various approaches including GWAS (e.g., Du et al., 2018a,b), gene expression studies (e.g., Ma et al., 2021; Zhu et al., 2021), cloning and sequence-based in silico analysis (Zhang et al., 2009; Yu et al., 2011; Yurchenko et al., 2014; Shang et al., 2016; Cui et al., 2020). Among GhSAD genes coding for stearoyl-acyl carrier protein desaturase, GhSAD4 was found to stand out as the most relevant to determine the relative ratio of oleic acid and linoleic acid (Shang et al., 2017). Of 17 SAD gene family members identified in Upland cotton, GhA-SAD6 and GhD-SAD8 have strong substrate specificity for 16:0-ACP, and GhA-SAD5 and GhA-SAD7 exhibited a high specific activity on 18:0-ACP (Liu et al., 2019). Tetraploid cotton genomes contain 13 LPAAT genes, including five on Dt subgenome and eight on the At subgenome (Wang et al., 2017). Based on a further sequence variation and gene expression analysis, genetic modification to overexpress single genes like At-Gh13LPAAT5 was found to be effective in improving the production of total TAG and oil content (Wang et al., 2017). In addition to these genes that have been well known for their role in fatty acid biosynthesis, other genes that encode less studied proteins, such as a calcium-dependent lipid-binding (CaLB) protein (Zhao et al., 2019) and a peroxidase (PRXR1) (Ma et al., 2019) were also implicated for their roles in determining cottonseed oil content based on GWAS followed by confirmation using VIGS.
Genetic Engineering of Cotton for Improving Cottonseed Oil, Fatty Acid, and Protein Contents
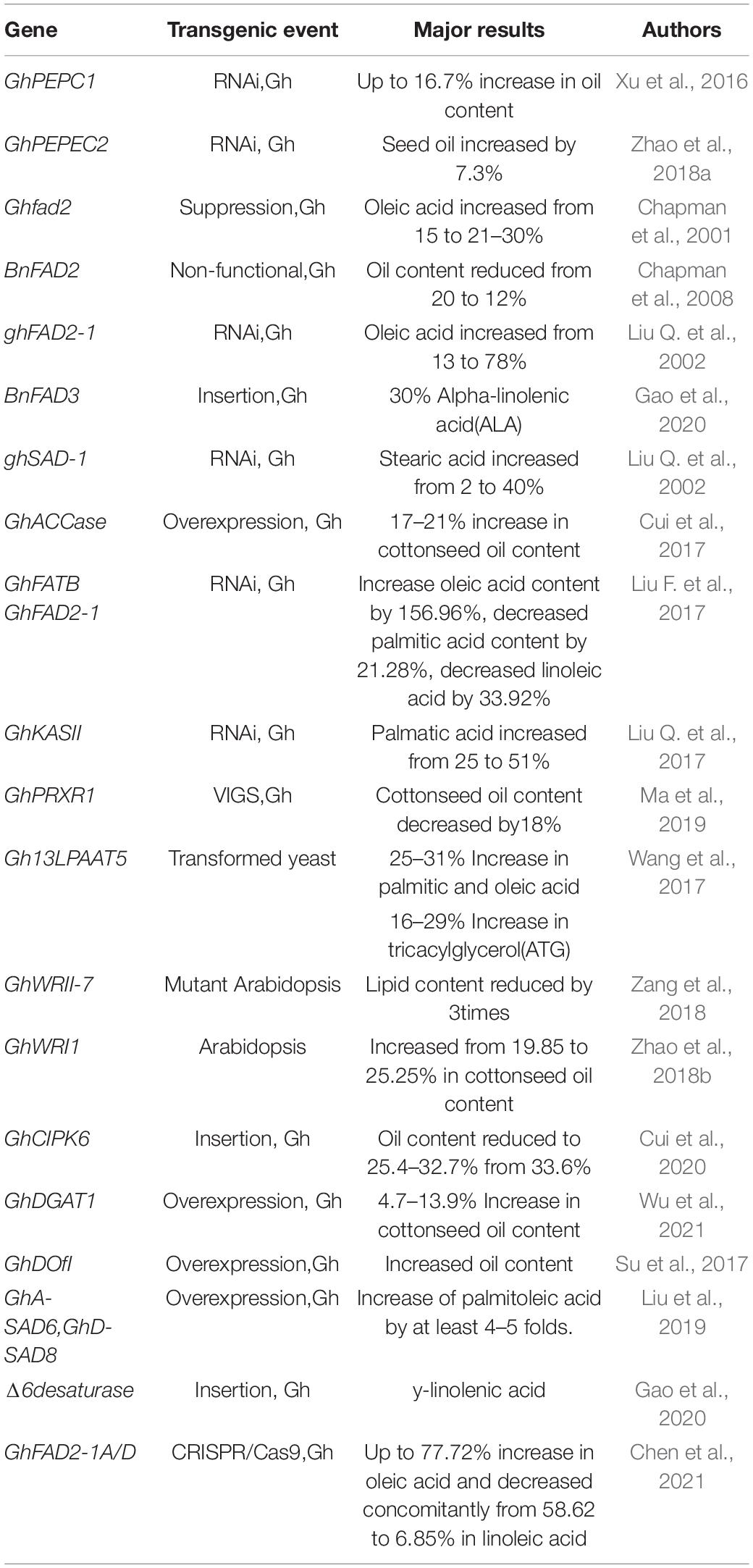
While the focus of cotton breeding on improving fiber quality will not change, there is an increased interest in enhancing the value of cottonseed by enhancing seed oil production and improving the nutritional and functional properties of the cottonseed oil (Liu et al., 2009). In the earliest attempts to genetically improve cottonseed oil, modest changes in oil content and fatty acid composition were achieved in Acala cotton through traditional breeding (Cherry et al., 1981; Cherry, 1983), reflecting the meager genetic variation available in natural germplasm and elite breeding lines. However, the improvements in molecular mechanisms underpinning the genetic variation and biochemical pathways, as well as the advent of genetic engineering approaches provide an alternative to rapidly alter carbon metabolism and manipulate lipid composition in cottonseed. Genetic modification of cottonseed oil has also been made more efficient through a series of methodological advancements in transgene expression systems, plant regeneration from tissue culture and gene transformation via Agrobacterium tumefaciens or particle bombardment (Zhang, 2015).
RNA interference (RNAi) attenuations in the expressions of genes coding for fatty acid desaturase (FAD2) on chromosome D12, stearoyl-ACP desaturase 1 (SAD1) on D9 and β-ketoacyl-acyl carrier protein synthase (KASII) in cotton resulted in substantially altered fatty acid composition, with particularly enhanced levels in oleic acid (from 13 to 78%), stearic acid (from 2 to 40%) and palmitic acid (from 26 to 15%), respectively, in cottonseed (Liu Q. et al., 2002, 2017). In addition, RNAi-directed down-regulation of PEPC2 up-regulated most lipid synthesis-related genes, resulting in 7.3% increase in cottonseed oil content (Zhao et al., 2018a). Shockey et al. (2017) and Sturtevant et al. (2017) identified a natural mutant allele of FAD2-1D in G. barbadense with high oleic acid in cottonseed oil, and its incorporation into G. hirsutum doubled oleic acid content (Dowd et al., 2020). Most recently, Chen et al. (2021) confirmed that knockout mutants of the GhFAD2 genes in Upland cotton by CRISPR/Cas9 editing increased the oleic acid level to 77.7% with a concomitant decrease in linoleic acid (from 58.6 to 6.9%) and palmitic acid (from 23.95 to 13.18%). Transforming Upland cotton with an FAD3 gene from Brassica napus and a D6D gene from Echium plantagineum resulted in approximately 30% α-linolenic acid (ALA) and 20% γ-linolenic acid (GLA), respectively, with no change in total oil content (Gao et al., 2020). Table 2 presents a summary of the genes that have been genetically engineered to improve cottonseed oil content and fatty acid composition.
Cottonseed oil is featured with a small amount of rare cyclic fatty acids, including DHSA and its downstream products sterculic acid and malvalic acid, all of which have been found to suppress mammalian Stearoyl-CoA desaturase activity and improve liver metabolomic profiles in high fat fed mice (Paton et al., 2017). The key genes encoding for cyclopropane fatty acid synthases converting oleic acid to DHSA have been identified in Arabidopsis (Bao et al., 2002) and cotton (Yu et al., 2011). Although the proof of concept has been made in producing DHSA in transgenic model plants (Yu et al., 2018; Okada et al., 2020), associated genes have yet to be modified in the cotton genome to raise DHSA production in cottonseeds.
It should be recognized that a radical modification of fatty acid composition may have deleterious effects on membrane integrity and impede seed germination under conventional farming practice, even though the modification was transcriptionally controlled by seed-specific promoters. For example, in the case that a leaky “seed-specific” promoter was used, severe compromises in plant growth and development, especially under environmental stresses, and penalty in yield may occur as a result (Lindgren et al., 2003). Further, the commercial planting of genetically modified crops generated by transgenic approaches especially those by agrobacterium mediated transformation assisted by selectable markers such as kanamycin resistance, has met substantial public skepticism and resistance in addition to lengthy and heavy regulatory burdens (Shockey et al., 2017). However, the recent availability of versatile genome editing techniques, such as transcription activator-like effector nucleases (TALEN) and clustered regulatory interspaced short palindromic repeats (CRISPR)/Cas9 systems has allowed scientists to precisely edit the expression of target genes without T-DNA insertion. Most importantly, the use of genome editing techniques may circumvent the lengthy regulatory processes and renders its products for rapid commercialization (Wang et al., 2020; Zhang et al., 2020).
Although cottonseed storage proteins are generally deficient in essential amino acids, especially lysine, which can be inadequate from a nutrition point of view, synthetic forms of lysine and other essential amino acids can be added to the diet to correct the deficiency. Cottonseed protein also tends to be deficient in isoleucine and the sulfur-rich amino acids such as methionine and cysteine (Capdevila and Dure, 1977). The sulfur-rich proteins, such as albumin, constitute a low fraction of the total cottonseed proteins (Galau et al., 1992; Hu et al., 2011). Genetic improvement of cottonseed storage protein and amino acid profiles is clearly long overdue, which could be developed in concert with the development of gossypol-free trait to meet the nutritional requirement for use as a source of high quality plant protein for non-ruminant animals or humans. The broad application of cottonseeds for human consumption and as animal feed is considerably constrained by the presence of gossypol which is sequestered in the pigment glands of cottonseed and other plant tissues. Gossypol is in a class of polyphenol compounds (terpenoids) that can be toxic and nutritionally undesirable, if safe levels in the diet are exceeded. Natural glandless (devoid of gossypol) cotton mutants exist in cotton and have been extensively studied and used in breeding (Zhang and Wedegaertner, 2021). Genetic modified glandless cottonseed has also been developed by RNAi down-regulation of cadinene synthase (Sunilkumar et al., 2006), which is currently being incorporated into elite Upland cotton cultivars to enable broad applications of cotton proteins for human consumption and monogastric animals. Commercialization of this technology has been slow due to international regulatory hurdles for genetically modified crops. Most recent reviews on genetics, breeding and genetic engineering to develop glandless cotton can be found in Rathore et al. (2020) and Zhang and Wedegaertner (2021).
Natural genetic variation in vitamin E also exists within cotton (Smith and Creelman, 2001). For example, several long-staple Acala 1517 cultivars were higher in α-tocopherol than medium-staple Upland cultivars. However, the genetic and genomic basis of the variation is currently not understood. Radcliffe and Czajka-Narins (2006) showed that cottonseed oil had a lowering effect on total cholesterol content for both male and female rats, but on high-density lipoprotein cholesterol for male rats only, and the replacement of corn oil with cottonseed oil resulted in changes in tocopherol status. A follow-up study in human by Radcliffe et al. (2009) further showed that cottonseed oil used in muffins and potato chips even increased vitamin E intake. Recently, Salimath et al. (2021) reported that genetic modification by converting tocopherols into more potent form of tocotrienols via introducing homogentisate geranylgeranyl transgenic coding sequence under the control of the Brassica napus seed-specific promoter from barley through genetic engineering. Transgenic cottonseeds had a 2–3-fold increase in the accumulation of total vitamin E (tocopherols + tocotrienols), with more than 60% γ-tocotrienol.
Prospective
As a byproduct in cotton production, cottonseed has excellent potential for use as a source of sustainable, high quality vegetable oil, biofuel and proteins because of its abundance that is expected to grow as the demand for cotton fiber continues to increase. Genetic improvements in nutritional value and functional properties of cottonseeds are being leveraged by the rapid advancements in biotechnology and genomics-based molecular breeding. In this review, we have summarized the most recent advances in genetic improvement of cottonseeds in relevance to the content of oil or protein, fatty acid composition that have been demonstrated to be amendable. Genetic improvements of cottonseed traits have proven to be particularly challenging as cottonseed is relatively low value product compared to cotton fiber that commends more than 85% of the farm-gate value of cotton production. This necessitates the employment of high precision genome editing technology and molecular breeding strategies to enable achieving genetic improvements in seed traits without trade-offs in fiber production and quality, as well as regulatory hurdles. Although cotton is among the earliest crops being grown commercially, the path leading to a successful commercialization and public acceptance of genetic modified cottonseed oils or whole seeds as a novel source of food grade proteins with improved nutritional value may not be an easier task in comparison to other genetic modified non-food crops. Nevertheless, more and more proof of concept studies have been conducted in model plants and recently in cotton that renders cotton industry standing on a new threshold of research and development, equipped with ever increasing knowledge in the intricate relationships and carbon reallocation between seed and fiber, and new sets of tools with high precision for modifying the cotton genome. It could be envisioned that the development of nutritionally improved and functionally versatile cottonseed, perhaps led by the development of high oleic cottonseed oil that could emulate the success of high oleic soybean oils, such as Plenish, Vistive Gold, and Calyxt, will come to fore, along with the continuous and synchronized development in cotton fiber.
Author Contributions
MW finalized the summary of all the publications used in this review and wrote the manuscript. JY directed the study and contributed to the writing of the manuscript. JZ provided Tables 1, 2 and an early incomplete draft of the manuscript, and finalized the manuscript. TW edited the manuscript. WP contributed to the writing of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported in part by grants from the National Natural Science Foundation of China (grant nos. 31621005, 31301368, and 31301367), the Natural Science Foundation of Xinjiang Uygur Autonomous Region of China (grant nos. 2021D01B113 and 2020D01A135), the Scientific Research Project of Henan Province, and Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences. The authors also acknowledged the support from Cotton Incorporated and New Mexico Agricultural Experiment Station, United States.
Conflict of Interest
TW was employed by Cotton Incorporated.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
- ^ www.statista.com/statistics/267271/worldwide-oilseed-production-since-2008
- ^ www.cotton.org/econ/cropinfo/costsreturns/usa.cfm
- ^ www.cottonqtldb.org
References
Agarwal, D., Singh, P., Chakrabarty, M., Shaikh, A., and Gayal, S. (2003). Cotton seed oil quality, utilization and processing. Cicr Tech. Bull. 25:16.
Alford, B. B., Liepa, G. U., and Vanbeber, A. N. (1996). Cotton seed protein: What does the future hold? Plant Foods for Human. Nutr. 49, 1–11. doi: 10.1007/BF01092517
Alfred, Q., Liu, H. Y., Xu, H. M., Li, J. R., Wu, J. G., Zhu, S. J., et al. (2012). Mapping of quantitative trait loci for oil content in cottonseed kernel. J. Genet. 91, 289–295. doi: 10.1007/s12041-012-0184-0
Bao, X. M., Katz, S., Pollard, M., and Ohlrogge, J. (2002). Carbocyclic fatty acids in plants: biochemical and molecular genetic characterization of cyclopropane fatty acid synthesis of Sterculiafoetida. Proc. Natl. Acad. Sci. U. S. A. 99, 7172–7177. doi: 10.1073/pnas.092152999
Badigannavar, A., and Myers, G. O. (2015). Genetic diversity, population structure and marker trait associations for seed quality traits in cotton (Gossypium hirsutum). J. Genet. 94, 87–94.
Bellaloui, N., Saha, S., Tonos, J. L., Scheffler, J. A., Jenkins, J. N., McCarty, J. C., et al. (2020). Effects of interspecific chromosome substitution in Upland cotton on cottonseed micronutrients. Plants 9:1081. doi: 10.3390/plants9091081
Byrne, P. F., McMullen, M. D., Snook, M. E., Musket, T. A., Theuri, J. M., Widstrom, N. W., et al. (1996). Quantitative trait loci and metabolic pathways: genetic control of the concentration of maysin, a corn earworm resistant factor, in maize silks. Proc. Natl. Acad. Sci. U. S. A. 93, 8820–8825. doi: 10.1073/pnas.93.17.8820
Campbell, B. T., Chapman, K. D., Sturtevant, D., Kennedy, C., Horn, P., Chee, P., et al. (2016). Genetic analysis of cottonseed protein and oil in a diverse cotton germplasm. Crop Sci. 55, 2457–2464. doi: 10.2135/cropsci2015.12.0742
Capdevila, A. M., and Dure, L. (1977). Developmental biochemistry of cottonseed embryogenesis and germination. Plant Physiol. 59, 268–273.
Chapman, K. D., Austin, B. S., Sparace, S. A., Kinney, A. J., Ripp, K. G., Pirtle, I. L., et al. (2001). Transgenic cotton plants with increased seed oleic acid content. J. Am. Oil Chem. Soc. 78, 941–947.
Chapman, K. D., Neogi, P. B., Hake, K. D., Stawska, A. A., Speed, T. R., Cotter, M. Q., et al. (2008). Reduced oil accumulation in cottonseeds transformed with a Brassica nonfunctional allele of a delta-12 fatty acid desaturase (FAD2). Crop Sci. 48, 1470–1481.
Chen, Y. Z., Fu, M. C., Li, H., Wang, L. G., Liu, R. Z., Liu, Z. J., et al. (2021). High-oleic acid content, nontransgenic allotetraploid cotton (Gossypium hirsutum L.) generated by knockout of GhFAD2 genes with CRISPR/Cas9 system. Plant Biotechnol. J. 19, 424–426. doi: 10.1111/pbi.13507
Chen, Z. F., Zhang, Z. W., and Cheng, H. L. (1986). The analysis of upland cotton quality. Acta Agron. Sin. 12, 195–200.
Cherry, J. P., Kohel, R. J., Jones, L. A., and Powell, W. H. (1981). “Cottonseed quality: factors affecting feed and food uses” in Proceedings of Beltwide Cotton Production Research Conference. ed. J. M. Brown (New Orleans: National Cotton Council of Amercia). 266–282.
Cui, Y. P., Liu, Z. J., Zhao, Y. P., Wang, Y. M., Huang, Y., Li, L., et al. (2017). Overexpression of heteromeric GhAccase subunits enhanced oil accumulation in upland cotton. Plant Mol. Biol. Rep. 35, 287–297. doi: 10.1007/s11105-016-1022-y
Cui, Y. P., Su, Y., Wang, J. J., Jia, B., Wu, M., Pei, W. F., et al. (2020). Genome-Wide characterization and analysis of CIPK gene family in two cultivated allopolyploid cotton species: sequence variation, association with seed oil content, and the role of GhCIPK6. Int. J. Mol. Sci. 21:863. doi: 10.3390/ijms21030863
Dani, R. G., and Kohel, R. J. (1989). Maternal effects and generation mean analysis of seed-oil content in cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 77, 569–575. doi: 10.1007/BF00274282
Dowd, M. K. (2012). Identification of the unsaturated heptadecyl fatty acids in the seed oils of Thespesiapopulnea and Gossypium hirsutum. J. Am. Oil. Chem. Soc. 89, 1599–1609. doi: 10.1007/s11746-012-2071-5
Dowd, M. K., Boykin, D. L., Meredith, W. R. Jr., Campbell, B. T., Bourland, F. M., Gannaway, J. R., et al. (2010). Fatty acid profiles of cottonseed genotypes from the national cotton variety trials. J. Cotton Sci. 14, 64–73.
Dowd, M. K., McCarty, J. C. Jr., Shockey, J., and Jenkins, J. N. (2020). Registration of four upland cotton germplasm lines with elevated levels of seed oil oleic acid. J. Plant Reg. 14, 64–71. doi: 10.1002/plr2.20017
Downey, R. K., and Harvey, B. L. (1963). Methods of breeding for oil quality in rape. Can. J. Plant Sci. 43, 271–275. doi: 10.4141/cjps63-054
Du, X. M., Huang, G., He, S. P., Yang, Z. E., Sun, G. F., Ma, X. F., et al. (2018a). Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 50, 796–802. doi: 10.1038/s41588-018-0116-x
Du, X. M., Liu, S. Y., Sun, J. L., Zhang, G. Y., Jia, Y. H., Pan, Z. E., et al. (2018b). Dissection of complicate genetic architecture and breeding perspective of cottonseed traits by genome-wide association study. BMC Genomics 19:451. doi: 10.1186/s12864-018-4837-0
Dure, L., and Chlan, C. (1981). Developmental biochemistry of cottonseed embryogenesis and germination. XII. Purification and properties of principal storage proteins. Plant Physiol. 68, 180–186. doi: 10.1104/pp.68.1.180
Ecker, R., and Yaniv, Z. (1993). Genetic control of fatty acid composition in seed oil of Sinapis alba L. Euphytica 69, 45–49. doi: 10.1007/bf00021724
Galau, G. A., Wang, H. Y. C., and Hughes, D. W. (1992). Cotton Mat5-A (C164) gene and Mat5-D cDNAs encoding methionine-rich 2S albumin storage proteins. Plant Physiol. 99, 779–782. doi: 10.1104/pp.99.2.779
Gao, L. H., Chen, W., Xu, X. Y., Zhang, J., Singh, T. K., Liu, S. M., et al. (2020). Engineering trienoic fatty acids into cottonseed oil improves low-temperature seed germination, plant photosynthesis and cotton fiber quality. Plant Cell Physiol. 61, 1335–1347. doi: 10.1093/pcp/pcaa062
Guo, X., Fan, C., Chen, Y., Wang, J., Yin, W., Wang, R. C., et al. (2017). Identification and characterization of an efficient acyl-CoA: diacylglycerol acyltransferase 1 (DGAT1) gene from the microalga Chlorella ellipsoidea. BMC Plant Biol. 17:48. doi: 10.1186/s12870-017-0995-5
Hanny, B. W., Meredith, W. R., Bailey, J. C., and Harvey, A. J. (1978). Genetic relationships among chemical constituents in seeds, flower buds, terminals, and mature leaves of cotton. Crop Sci. 18, 1071–1074.
Hinze, L. L., Horn, P. J., Kothari, N., Dever, J. K., Frelichowski, J., Chapman, K. D., et al. (2015). Nondestructive measurements of cottonseed nutritional trait diversity in the US National Cotton Germplasm Collection. Crop Sci. 55, 770–782. doi: 10.2135/cropsci2014.04.0318
Hom, N. H., Schierholt, A., Mollers, C., and Becker, H. C. (2015). Pollen genotype effects on seed quality traits in winter oil seed rape. Crop Sci. 55, 493–500.
Horn, P. J., Neogi, P., Tombokan, X., Ghosh, S., Campbell, B. T., and Chapman, K. D. (2011). Simultaneous quantification of oil and protein in cottonseed by low-field time-domain nuclear magnetic resonance. J. Am. Oil Chem. Soc. 88, 1521–1529.
Hovav, R., Faigenboim, D. A., Kadmon, N., Hu, G. J., Zhang, X., Gallagher, J. P., et al. (2015). A transcriptome profile for developing seed of polyploid cotton. Plant Genom. 8, 1–15. doi: 10.3835/plantgenome2014.08.0041
Hu, G. J., Houston, N. L., Pathak, D., Schmidt, L., Thelen, J. J., and Wendel, J. F. (2011). Genomically biased accumulation of seed storage proteins in allopolyploid cotton. Genetics 189, 1103–1115. doi: 10.1534/genetics.111.132407
Hu, G. J., Hovav, R., Grover, C. E., Faigenboim-Doron, A., Kadmon, N., Page, J. T., et al. (2016). Evolutionary conservation and divergence of gene coexpression networks in Gossypium (cotton) seeds. Genome Biol. Evol. 8, 3765–3783. doi: 10.1093/gbe/evw280
Jiao, X. M., Zhao, X. C., Zhou, X. R., Green, A. G., Fan, Y. L., Wang, L., et al. (2013). Comparative transcriptomic analysis of developing cotton cotyledons and embryo axis. PLoS One 8:e71756. doi: 10.1371/journal.pone.0071756
Jolliffe, N. A., Craddock, C. P., and Fridgerio, L. (2005). Pathways for protein transport to seed storage vacuoles. Biochem. Soc. Trans. 33, 1016–1018. doi: 10.1042/BST20051016
Karaosmanoglu, F., Tuter, M., Gollu, E., Yanmaz, S., and Altintig, E. (1999). Fuel properties of cottonseed oil. Energy Sources 21, 821–828. doi: 10.1080/00908319950014371
Khan, K., Agarwal, P., Shanware, A., and Sane, V. A. (2015). Heterologous expression of two Jatropha Aquaporins imparts drought and salt tolerance and improves seed viability in transgenic Arabidopsis thaliana. PLoS One 10:e0128866. doi: 10.1371/journal.pone.0128866
Kohel, R. J. (1978). Survey of Gossypium Hirsutum L. Germplasm Collections for Seed-oil Percentage and Seed Characteristics. US: US Department of Agriculture. 187.
Kohel, R. J., and Cherry, J. P. (1983). Variation of cottonseed quality with stratified harvests. Crop Sci. 23, 1119–1124. doi: 10.2527/2005.8371715x
Kohel, R. J., Glueck, J., and Rooney, L. W. (1985). Comparison of cotton germplasm collections for seed protein content. Crop Sci. 25, 961–963. doi: 10.1186/s12870-017-0981-y
Kong, Q., Yuan, L., and Ma, W. (2019). WRINKLED1, a “Master Regulator” in transcriptional control of plant oil biosynthesis. Plants 8:238. doi: 10.3390/plants8070238
Kothari, N., Campbell, B. T., Dever, J. K., and Hinze, L. L. (2016). Combining ability and performance of cotton germplasm with diverse seed oil content. Crop Sci. 56, 19–29.
Kumar, M., Tomar, M., Punia, S., Grasso, S., Arrutia, F., Choudhary, J., et al. (2021). Cottonseed: a sustainable contributor to global protein requirements. Trend Food Sci Tech 111, 100–113. doi: 10.1016/j.tifs.2021.02.058
Lindgren, L. O., Stalberg, K. G., and Hoglund, A. S. (2003). Seed-specific overexpression of an endogenous Arabidopsis phytoene synthase gene results in delayed germination and increased levels of carotenoids, chlorophyll, and abscisic acid. Plant Physiol. 132, 779–785. doi: 10.1104/pp.102.017053
Liu, B. L., Sun, Y., Xue, J. A., Mao, X., Jia, X. Y., and Li, R. Z. (2019). Stearoyl-ACP Δ9 desaturase 6 and 8 (GhA-SAD6 and GhD-SAD8) are responsible for biosynthesis of palmitoleic acid specifically in developing endosperm of upland cotton seeds. Front. Plant Sci. 10:703. doi: 10.3389/fpls.2019.00703
Liu, D. X., Liu, F., Shan, X. R., Zhang, J., Tang, S. Y., Fang, X. M., et al. (2015). Construction of a high-density genetic map and lint percentage and cottonseed nutrient trait QTL identification in upland cotton(Gossypium hirsutum L.). Mol. Genet. Genom. 290, 1683–1700. doi: 10.1007/s00438-015-1027-5
Liu, F., Zhao, Y. P., Zhu, H. G., Zhu, Q. H., and Sun, J. (2017). Simultaneous silencing of GhFAD2-1 and GhFATB enhances the quality of cottonseed oil with high oleic acid. J. Plant Physiol. 215, 132–139.
Liu, G. Z., Mei, H. X., Wang, S., Li, X. H., Zhu, X. F., and Zhang, T. Z. (2015). Association mapping of seed oil and protein contents in upland cotton. Euphytica 205, 637–645. doi: 10.1007/s10681-015-1450-z
Liu, H. Y., Quampah, A., Chen, J. H., Li, J. R., Huang, Z. G., He, Q. L., et al. (2013). QTL mapping based on different genetic systems for essential amino acid contents in cottonseeds in fifferent environments. PLoS One 8:e57531. doi: 10.1371/journal.pone.0057531
Liu, H. Y., Quampah, A., Chen, J. H., Li, J. R., Huang, Z. R., He, Q. L., et al. (2012). QTL analysis for gossypol and protein contents in upland cottonseeds with two different genetic systems across environments. Euphytica 188, 453–463. doi: 10.1007/s10681-012-0733-x
Liu, H. Y., Quampah, A., Chen, J. H., Li, J. R., Huang, Z. R., He, Q. L., et al. (2017). QTL mapping with different genetic systems for nine nonessential amino acids of cottonseeds. Mol. Genet. Genom. 292, 671–684. doi: 10.1007/s00438-017-1303-7
Liu, H. Y., Zhang, L., Mei, L., Quampah, A., He, Q. L., Zhang, B. S., et al. (2020). qOil-3, a major QTL identification for oil content in cottonseed across genomes and its candidate gene analysis. Ind. Crops Prod. 145:112070. doi: 10.1016/j.indcrop.2019.112070
Liu, Q. (2011). Functional and Topological Analysis of Acyl-CoA:Diacylglycerol Acyltransferase 2 from Sacchromyces Cerevisiae. Edmonton: Department of Agricultural, Food and Nutritional Science.
Liu, Q., Singh, S. P., and Green, A. (2008). “Genetic manipulation of fatty acid composition in cottonseed oil,” in Proceeding of the 18th International Symposium on Plant Lipids, Bordeaux, 20–25.
Liu, Q., Singh, S., and Green, A. (2002). High-oleic and high-stearic cottonseed oils: nutritionally improved cooking oils developed using gene silencing. J. Am. Coll. Nutr. 21, 205–211. doi: 10.1080/07315724.2002.10719267
Liu, Q., Singh, S., Chapman, K., and Green, A. (2009). “Bridging Traditional and Molecular Genetics in Modifying Cottonseed oil” in Genetics and Genomics of Cotton. Ed. A.H., Paterson (New York: Springer). 353–384. doi: 10.1007/978-0-387-70810-2_15
Liu, Q., Wu, M., Zhang, B. L., Shrestha, P., Petrie, J., Green, G. A., et al. (2017). Genetic enhancement of palmitic acid accumulation in cotton seed oil through RNAi down-regulation of ghKAS2 encoding b-ketoacyl-ACP synthase II (KASII). Plant Biotechnol. J. 15, 132–143. doi: 10.1111/pbi.12598
Lowell, J. A., Parnes, H. L., and Blackburn, G. L. (1990). Dietary immunomodulation: Beneficial effects on oncogenesis and tumor growth. Crit. Care Med. 18, 5145–5148.
Lukonge, E., Labuschagne, M. T., and Hugo, A. (2007). The evaluation of oil and fatty acid composition in seed of cotton accessions from various countries. J.Sci. Food and Agric. 87, 340–347. doi: 10.1002/jsfa.2731
Ma, J. J., Liu, J., Pei, W. F., Ma, Q. F., Wang, N. H., Zhang, X., et al. (2019). Genome-wide association study of the oil content in upland cotton (Gossypium hirsutum L.) and identification of GhPRXR1, a candidate gene for a stable QTL qOC-Dt5-1. Plant Sci. 286, 89–97. doi: 10.1016/j.plantsci.2019.05.019
Ma, L. H., Cheng, X. Q., Wang, C., Zhang, X. Y., Xue, F., Li, Y. J., et al. (2021). Explore the gene network regulating the composition of fatty acids in cottonseed. BMC Plant Biol. 21:177. doi: 10.1186/s12870-021-02952-4
McMullen, M. D., Byrne, P. F., Snook, M. E., Wiseman, B. R., Lee, E. A., Widstrom, N. W., et al. (1998). Quantitative trait loci and metabolic pathways. Proc. Natl. Acad. Sci. U. S. A. 95, 1996–2000.
Meneghetti, S. M. P., Meneghetti, M. R., Serra, T. M., Barbosa, D. C., and Wolf, C. R. (2007). Biodiesel production from vegetable oil mixtures: cottonseed, soybean, and castor oils. Energy Fuels. 21, 3746–3747.
Meredith, W. R. Jr., Boykin, D. R., Bourland, F. M., Caldwell, D. L., Campbell, B. T., Gannaway, J. R., et al. (2012). Genotype x environment interactions over seven years for yield, yield components, fiber quality and gossypol traits in the regional high quality tests. J. Cotton Sci. 16, 160–169.
Moncada, S., and Higgs, A. (1993). The L-Arginine-nitric oxide pathway. N. Engl. J. Med. 329, 2002–2012.
Nikolau, B. J., Ohlrogge, J. B., and Wurtele, E. S. (2003). Plant biotin-containing carboxylases. Arch Biochem. Biophys. 414, 211–222. doi: 10.1016/s0003-9861(03)00156-5
O’Brien, R. D., Jones, L. A., King, C. C., Wakelyn, P. J., and Wan, P. J. (2005). “Cottonseed oil” in Bailey’s Industrial Oil and Fat Products. 6th Edn. ed. F. Shahidi (New Jersey: Wiley).
Ohlrogge, J. B., and Jaworski, J. G. (1997). Regulation of fatty acid synthesis. Annu. Rev. Plant Biol. 48, 109–136.
Okada, S., Taylor, M., Zhou, X. R., Naim, F., Marshall, D., Blanksby, S. J., et al. (2020). Producing cyclopropane fatty acid in plant leafy biomass via expression of bacterial and plant cyclopropane fatty acid synthases. Front Plant Sci. 11:30. doi: 10.3389/fpls.2020.00030
Ory, R. L., and Flick, G. J. (1994). “Peanut and cotton seed protein for food uses” in New and Developing Sources of Food Proteins. ed. B. J. F. Hudson (London: Chapman and Hall). 195–240. doi: 10.1002/food.19850290312
Paton, C. M., Vaughan, R. A., Alpergin, E. S. S., Assadi-Porter, F., and Dowd, M. K. (2017). Dihydrosterculic acid from cottonseed oil suppresses desaturase activity and improves liver metabolomic profiles of high fat fed mice. Nutr. Res. 45, 52–62. doi: 10.1016/j.nutres.2017.06.008
Radcliffe, J. D., and Czajka-Narins, D. M. (2006). Lipids and tocopherols in serum and liver of female rats fed diets containing corn oil or cottonseed oil. Plant Foods Hum. Nutr. 61, 33–36. doi: 10.1007/s11130-006-0011-y
Radcliffe, J. D., Martin, S., and Imrhan, V. (2009). Consumption of muffins and potato chips containing cottonseed oil (CSO) increases the intake of vitamin E (VE) without affecting fat intake. FASEB J. 23, 729–729.
Rathore, K. S., Pandeya, D., Campbell, L. M., Wedegaertner, T. C., Puckhaber, L., Stipanovic, R. D., et al. (2020). Ultra-low gossypol cottonseed: selective genesilencing opens up a vast resource of plant-based protein to improve human nutrition. Crit. Rev. Plant Sci. 39, 1–29. doi: 10.1080/07352689.2020.1724433
Saha, S., Bellaloui, N., Jenkins, J. N., McCarty, J. C., and Stelly, D. M. (2020). Effect of chromosome substitutions from Gossypium barbadense L., G. tomentosum Nutt. Ex seem and G. mustelinum Watt into G. hirsutum L. on cottonseed protein and oil content. Euphytica 216:118.
Said, J. I., Lin, Z. X., Zhang, X. L., Song, M. Z., and Zhang, J. F. (2013). A comprehensive meta QTL analysis for fiber quality, yield, yield related and morphological traits, drought tolerance, and disease resistance in tetraploid cotton. BMC Genomics 14:776–798. doi: 10.1186/1471-2164-14-776
Said, J. I., Knapka, J. A., Song, M. Z., and Zhang, J. F. (2015a). Cotton QTLdb: a cotton QTL database for QTL analysis, visualization, and comparison between Gossypium hirsutum and G. hirsutum × G. barbadense populations. Mol. Genet. Genom. 290, 1615–1625. doi: 10.1007/s00438-015-1021-y
Said, J. I., Song, M. Z., Wang, H. T., Lin, Z. X., Zhang, X. L., Fang, D. D., et al. (2015b). A comparative meta-analysis of QTL between intraspecific Gossypium hirsutum and interspecific G. hirsutum × G. barbadense populations. Mol. Genet. Genom. 290, 1003–1025. doi: 10.1007/s00438-014-0963-9
Salimath, S. S., Romsdahl, T. B., Konda, A. R., Zhang, W., Cahoon, E. B., Dowd, M. K., et al. (2021). Production of tocotrienols in seeds of cotton (Gossypium hirsutum L.) enhances oxidative stability and offers nutraceutical potentia. Plant Biotechnol. J. 19, 1268–1282. doi: 10.1111/pbi.13557
Shang, L. G., Abduweli, A., Wang, Y. M., and Hua, J. P. (2016). Genetic analysis and QTL mapping of oil content and seed index using two recombinant inbred lines and two backcross populations in Upland cotton. Plant Breed. 135, 224–223. doi: 10.1111/pbr.12352
Shang, X. G., Cheng, C. Z., Ding, J., and Guo, W. Z. (2017). Identification of candidate genes from the SAD gene family in cotton for determination of cottonseed oil composition. Mol. Genet. Genom. 292, 173–186. doi: 10.1007/s00438-016-1265-1
Sharif, I., Farooq, J., Chohan, S. M., Saleem, S., Kainth, R. A., Mahmood, A., et al. (2019). Strategies to enhance cottonseed oil contents and reshape fatty acid profile employing different breeding and genetic engineering approaches. J. Integ. Agric. 18, 2205–2218. doi: 10.1016/s2095-3119(18)62139-2
Shockey, J., Dowd, M., Mack, B., Gilbert, M., Scheffler, B., Ballard, L., et al. (2017). Naturally occurring high oleic acid cottonseed oil: identification and functional analysis of a mutant allele of Gossypium barbadense fatty acid desaturase-2. Planta 245, 611–622. doi: 10.1007/s00425-016-2633-0
Singh, M., Singh, T. H., and Chahal, G. S. (1985). Genetic analysis of some seed quality characters in upland cotton (Gossypium hirsutum L.). Theor. Appl. Genet. 71, 126–128. doi: 10.1007/BF00278264
Smith, C. W., and Creelman, R. A. (2001). Vitamin E concentration in upland cotton seeds. Crop Sci. 41, 577–579. doi: 10.2135/cropsci2001.412577x
Song, X. L., and Zhang, T. Z. (2007). Identification of quantitative trait loci controlling seed physical and nutrient traits in cotton. Seed Sci. Res. 17, 243–251.
Spadaro, J. J., and Gardner, H. K. Jr. (1979). Food Uses for Cottonseed Protein. New Orlean: Southern Regional Research Center. 422–424.
Sturtevant, D., Horn, P., Kennedy, C., Hinze, L., Percy, R., and Chapman, K. (2017). Lipid metabolites in seeds of diverse Gossypium accessions: molecular identification of a high oleic mutant allele. Planta 245, 595–610. doi: 10.1007/s00425-016-2630-3
Su, Y., Liang, W., Liu, Z. J., Wang, Y. M., Zhao, Y. P., Ijaz, B., et al. (2017). Overexpression of GhDof1 improved salt and cold tolerance and seed oil content in Gossypium hirsutum. J. Plant Physiol. 218, 222–234. doi: 10.1016/j.jplph.2017.07.017
Sunilkumar, G., Campbell, L. M., Puckhaber, L., Stipanovic, R. D., and Rathore, K. S. (2006). Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc. Natl. Acad. Sci. 103, 18054–18059. doi: 10.1073/pnas.0605389103
Thiagarajan, C. P., and Ramaswamy, K. R. (1982). Varietal variation and seasonal influence on the pattern of accumulation of oil and protein in the developing cotton seed. Madras Agric. J. 69, 223–228.
Velasco, L., Vich, B. P., and Fernandez, M. J. M. (2004). Novel variation for tocopherol profile in sunflower created by mutagenesis and recombination. Plant Breed 123, 490–492. doi: 10.1111/j.1439-0523.2004.01012.x
Voelker, T., and Kinney, A. J. (2001). Variation in the biosynthesis of seed-storage lipids. Ann. Rev. Plant Physiol. 52, 335–361. doi: 10.1146/annurev.arplant.52.1.335
Wang, N. H., Ma, J. J., Pei, W. F., Wu, M., Li, H. J., Li, X. L., et al. (2017). A genome-wide analysis of the lysophosphatidate acyltransferase (LPAAT) gene family in cotton: organization, expression, sequence variation, and association with seed oil content and fiber quality. BMC Genom. 18:218. doi: 10.1186/s12864-017-3594-9
Wang, Q. Q., Alariqi, M., Wang, F. Q., Li, B., Ding, X., Rui, H. P., et al. (2020). The application of a heat-inducible CRISPR/Cas12b (C2c1) genome editing system in tetraploid cotton (G. hirsutum) plants. Plant Biotechnol. J. 18, 2436–2443. doi: 10.1111/PBJ.13417
Wang, T. Q. (1992). The heredity of oil content and its heterosis in rapeseed. J. Guizhou Agric. Sci. 6, 37–40. doi: 10.1038/hdy.1958.3
Wang, W. W., Sun, Y., Yang, P., Cai, X. Y., Yang, L., Ma, J. R., et al. (2019). A high density SLAF-seq SNP genetic map and QTL for seed size, oil and protein content in upland cotton. BMC Genom. 20:599. doi: 10.1186/s12864-019-5819-6
Wu, J. X., Jenkins, J. N., McCarty, J. C., and Thaxton, P. (2009). Seed trait evaluation of Gossypium barbadense L. chromosomes/arms in a G. hirsutum L. background. Euphytica 167, 371–380. doi: 10.1007/s10681-009-9896-5
Wu, J. X., McCarty, J. C., and Jenkins, J. N. (2010). Cotton chromosome substitution lines crossed with cultivars: Genetic model evaluation and seed trait analyses. Theor. Appl. Genet. 120, 1473–1483. doi: 10.1007/s00122-010-1269-x
Wu, P., Xu, X. L., Li, J. W., Zhang, J., Chang, S. Y., Yang, X. Y., et al. (2021). Seed-specific overexpression of cotton GhDGAT1 gene leads to increased oil accumulation in cottonseed. Crop J. 9, 487–490.
Xu, Z. P., Li, J. W., Guo, X. P., Jin, S. X., and Zhang, X. L. (2016). Metabolic engineering of cottonseed oil biosynthesis pathway via RNA interference. Sci. Rep. 6:33342. doi: 10.1038/srep33342
Ye, Z. H., Lu, Z. Z., and Zhu, J. (2003). Genetic analysis for developmental behavior of some seed quality traits in upland cotton (Gossypium hirsutum L.). Euphytica 129, 183–191.
Yu, J. W., Yu, S. X., Fan, S. L., Song, M. Z., Zhai, H. H., Li, X. L., et al. (2012). Mapping quantitative trait loci for cottonseed oil, protein and gossypol content in a Gossypium hirsutum × Gossypium barbadense backcross inbred line population. Euphytica 187, 191–201.
Yu, X. H., Cahoon, R. E., Horn, P. J., Shi, H., Prakash, R. R., Cai, Y. H., et al. (2018). Identification of bottlenecks in the accumulation of cyclic fatty acids in camelina seed oil. Plant Biotechnol. J. 16, 926–938. doi: 10.1111/pbi.12839
Yu, X. H., Rawat, R., and Shanklin, J. (2011). Characterization and analysis of the cotton cyclopropane fatty acid synthase family and their contribution to cyclopropane fatty acid synthesis. BMC Plant Biol. 11:97. doi: 10.1186/1471-2229-11-97
Yuan, Y. C., Zhang, H. J., Wang, L. Y., Xing, H. X., Mao, L. L., Tao, J. C., et al. (2019). Candidate quantitative trait loci and genes for fiber quality in Gossypium hirsutum L. detected using single- and multi-locus association mapping. Ind. Crops Prod. 134, 356–369.
Yuan, Y. L., Zhang, T. Z., Jing, S. R., Pan, J. J., Xing, C. Z., Guo, L. R., et al. (2001). Studies of the inheritance of seed qualities and the exploitation of F2 heterosis in low hossypol strains in Upland cotton. Acta Genet. Sin. 28, 471–481.
Yuan, Y., Wang, X., Wang, L., Xing, H., Wang, Q., Saeed, M., et al. (2018). Genome-wide association study identifies candidate genes related to seed oil composition and protein content in Gossypium hirsutum L. Front. Plant Sci. 9:1359.
Yurchenko, O. P., Park, S., Ilut, D. C., Inmon, J. J., Millhollon, J. C., Liechty, Z., et al. (2014). Genome-wide analysis of the omega-3 fatty acid desaturase gene family in Gossypium. BMC Plant Biol. 14:312. doi: 10.1186/s12870-014-0312-5
Zang, X. S., Pei, W. F., Wu, M., Geng, Y. H., Wang, N. H., Liu, G. Y., et al. (2018). Genome-scale analysis of the WRI-like family in Gossypium and functional characterization of GhWRI1a controlling triacylglycerol content. Front. Plant Sci. 9:1516. doi: 10.3389/fpls.2018.01516
Zeng, L. H., Campbell, B. T., Bechere, E., Dever, J. K., Zhang, J. F., Jones, A. S., et al. (2015). Genotypic and environmental effects on cottonseed oil, nitrogen, and gossypol contents in eighteen years Regional High Quality tests. Euphytica. 206, 815–824. doi: 10.1007/s10681-015-1523-z
Zeng, Y. D., Sun, J. L., Bu, S. H., Deng, K. S., Tao, T., Zhang, Y. M., et al. (2016). EcoTILLING revealed SNPs in GhSus genes that are associated with fiber- and seed-related traits in upland cotton. Sci. Rep. 6:29250. doi: 10.1038/srep29250
Zhang, D. B., Hussain, A., Manghwar, H., Xie, K. B., Xie, S. S., Zhao, S. H., et al. (2020). Genome editing with the CRISPR-Cas system: an art, ethics and global regulatory perspective. Plant Biotechnol. J. 18, 1651–1669. doi: 10.1111/pbi.13383
Zhang, J. F. (2015). “Transgenic cotton breeding” in Cotton. eds D. Fang and R. G. Percy (USA: Agronomy Monographs). 229–253.
Zhang, J. F., and Wedegaertner, T. (2021). Genetics and breeding for glandless Upland cotton with improved yield potential and disease resistance: a review. Front. Plant Sci. 12:753426. doi: 10.3389/fpls.2021.753426
Zhang, M., Fan, J. L., Taylor, D. C., and Ohlrogge, J. B. (2009). DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell. 21, 3885–3901. doi: 10.1105/tpc.109.071795
Zhang, Y. B., Wang, Y., Feng, G. Y., Duan, H. R., and Liu, H. Y. (2022). QTLs analysis of oil and three main fatty acid contents in cottonseeds. Acta Agron. Sin. 48, 380–395. doi: 10.3389/fpls.2018.01359
Zhang, Z. B., Gong, J. W., Zhang, Z., Gong, W. K., Li, J. W., Shi, Y. Z., et al. (2021). Identification and analysis of oil candidate genes reveals the molecular basis of cottonseed oil accumulation in Gossypium hirsutum L. Theor. Appl. Genet. Epub online ahead of print. doi: 10.1007/s00122-021-03975-z
Zhao, W. X., Kong, X. H., Yang, Y., Nie, X. H., and Lin, Z. X. (2019). Association mapping seed kernel oil content in upland cotton using genome-wide SSRs and SNPs. Mol. Breed. 39:105.
Zhao, Y. P., Huang, Y., Wang, Y. M., Cui, Y. P., Liu, Z. J., and Hua, J. P. (2018a). RNA interference of GhPEPC2 enhanced seed oil accumulation and salt tolerance in Upland cotton. Plant Sci. 271, 52–61. doi: 10.1016/j.plantsci.2018.03.015
Zhao, Y. P., Liu, Z. J., Wang, X. W., Wang, Y. M., and Hua, J. P. (2018b). Molecular characterization and expression analysis of GhWRI1 in upland cotton. J. Plant Biol. 61, 186–197.
Zhao, Y. P., Wang, Y. M., Huang, Y., Cui, Y. P., and Hua, J. P. (2018c). Gene network of oil accumulation reveals expression profiles in developing embryos and fatty acid composition in Upland cotton. J. Plant Physiol. 228, 101–112. doi: 10.1016/j.jplph.2018.06.002
Zhu, D., Le, Y., Zhang, R., Li, X. J., and Lin, Z. X. (2021). A global survey of the gene network and key genes for oil accumulation in cultivated tetraploid cottons. Plant Biotechnol. J. 19, 1170–1182. doi: 10.1111/pbi.13538
Zhu, D., Li, X. M., Wang, Z. W., You, C. Y., Nie, X. H., Sun, J., et al. (2020). Genetic dissection of an allotetraploid interspecific CSSLs guides interspecific genetics and breeding in cotton. BMC Genom. 21:431. doi: 10.1186/s12864-020-06800-x
Keywords: seed oil content (SOC), fatty acid, seed protein content (SPC), amino acids, genome editing, quantitative trait loci (QTLs)
Citation: Wu M, Pei W, Wedegaertner T, Zhang J and Yu J (2022) Genetics, Breeding and Genetic Engineering to Improve Cottonseed Oil and Protein: A Review. Front. Plant Sci. 13:864850. doi: 10.3389/fpls.2022.864850
Received: 28 January 2022; Accepted: 15 February 2022;
Published: 10 March 2022.
Edited by:
Linghe Zeng, United States Department of Agriculture (USDA), United StatesReviewed by:
Sandra E. Branham, Clemson University, United StatesFred Bourland, University of Arkansas, United States
Copyright © 2022 Wu, Pei, Wedegaertner, Zhang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinfa Zhang, jinfazhang@nmsu.edu; Jiwen Yu, yujw666@hotmail.com
 Man Wu
Man Wu Wenfeng Pei
Wenfeng Pei Tom Wedegaertner3
Tom Wedegaertner3 Jinfa Zhang
Jinfa Zhang Jiwen Yu
Jiwen Yu