- 1Department of Physical Education, Sichuan International Studies University, Shapingba, Chongqing, China
- 2Department of PE in Ganzhou Teachers School, Ganzhou, JiangXi, China
- 3Ganzhou Vocational and Technical College, Ganzhou, JiangXi, China
- 4Binjiang No. 2 Primary School in Ganzhou, Ganzhou, China
Introduction: In recent years, the integration of electroencephalogram (EEG) and somatosensory data in athlete potential evaluation has garnered increasing attention. Traditional research methods mainly rely on processing EEG signals or motion sensor data independently. While these methods can provide a certain level of performance assessment, they often overlook the synergy between brain activity and physical movement, making it difficult to comprehensively capture an athlete's potential. Moreover, most existing approaches employ shallow models, which fail to fully exploit the temporal dependencies and cross-modal interactions within the data, leading to suboptimal accuracy and robustness in evaluation results.
Methods: To address these issues, this paper proposes a Transformer-based model, SensoriMind-Trans Net, which combines EEG signals and somatosensory data. The model leverages a multi-layer Transformer network to capture the temporal dependencies of EEG signals and utilizes a somatosensory data feature extractor and cross-modal attention alignment mechanism to enhance the comprehensive evaluation of athletes' cognitive and motor abilities.
Results: Experiments conducted on four public datasets demonstrate that our model outperforms several existing state-of-the-art (SOTA) models in terms of accuracy, inference time, and computational efficiency.
Discussion: Showcasing its broad applicability in athlete potential evaluation. This study offers a new solution for athlete data analysis and holds significant implications for future multimodal sports performance assessment.
1 Introduction
The task of evaluating an athlete's potential through electroencephalography (EEG) data has garnered increasing attention in recent years due to its ability to provide insights into the cognitive and motor functions of individuals during performance. EEG signals, as direct reflections of brain activity, allow researchers to gain a deeper understanding of how neural processes influence physical performance. This is not only beneficial for improving athletic training and performance monitoring but also for rehabilitation and cognitive enhancement (Ahmed et al., 2024). The integration of EEG data into athlete evaluation frameworks has become increasingly important because it not only complements traditional physical performance metrics but also captures mental fatigue, focus, and decision-making processes that are crucial in high-performance sports (Hsu, 2010). Therefore, this task is essential for gaining a more holistic understanding of athlete potential, both physically and cognitively, which could significantly enhance training regimens and performance evaluations (Gong et al., 2021). Unlike traditional methods that separately analyze EEG signals or motion sensor data, this study introduces a novel Transformer-based approach, SensoriMind-Trans Net, which integrates EEG and sensorimotor data using a cross-modal attention alignment mechanism. This integration allows for a more precise evaluation of the relationship between brain activity and physical movement, enhancing the accuracy of athlete potential assessment. By leveraging the self-attention capabilities of Transformers, our model captures long-range dependencies within EEG signals while simultaneously extracting and aligning spatial-temporal features from sensorimotor data. This methodological innovation not only surpasses the limitations of conventional feature-engineered approaches but also offers a more scalable and adaptable solution for real-time applications in athletic training, rehabilitation, and cognitive performance monitoring. Our findings demonstrate that SensoriMind-Trans Net significantly outperforms existing state-of-the-art models in both accuracy and computational efficiency, making it a promising tool for advancing multimodal sports performance evaluation.
To address the limitations of purely physical metrics, early methods for EEG-based athlete evaluation primarily relied on symbolic AI and knowledge representation approaches. These methods were designed to interpret EEG signals using rule-based systems and expert knowledge. Symbolic AI techniques focused on predefined patterns in EEG data, such as certain frequency bands or event-related potentials, and matched these patterns to known cognitive states or motor functions (Lombardi et al., 2024; Fawwaz et al., 2024). These systems, while interpretable, were limited in their scalability and flexibility. They relied heavily on handcrafted features and expert-defined rules, which were often specific to particular tasks or athletes (Jui et al., 2023). As a result, they struggled with generalization and adaptation to new data. Furthermore, the symbolic representation of EEG signals was unable to capture the complex and dynamic nature of brain activity during athletic performance. This led to the development of more adaptive, data-driven methods to overcome these limitations (Hsu, 2010).
In response to the constraints of symbolic AI, the field moved toward data-driven and machine learning approaches. These methods aimed to automatically extract meaningful patterns from EEG data without relying on predefined rules. Machine learning algorithms such as Support Vector Machines (SVMs), k-Nearest Neighbors (k-NN), and Random Forests became popular due to their ability to learn from labeled EEG datasets (Aggarwal and Chugh, 2022). These models excelled at distinguishing between different cognitive or motor states based on statistical patterns in the data, improving the flexibility and adaptability of EEG-based evaluation systems. However, traditional machine learning models still faced several challenges, including their reliance on feature engineering and their inability to effectively model the temporal dependencies present in EEG signals (Li et al., 2022). These models required carefully crafted input features, which often depended on domain expertise and could be prone to overfitting in small datasets. Although they improved over symbolic methods, they were still limited in their ability to fully capture the richness of EEG data (Ma et al., 2024).
To address the limitations of feature engineering and better capture the dynamic nature of EEG signals, deep learning models emerged as a powerful tool for EEG-based athlete evaluation. Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs) revolutionized the field by automatically learning hierarchical representations from raw EEG data (Zhang et al., 2019). CNNs were particularly effective at capturing spatial relationships between EEG channels, while RNNs, including Long Short-Term Memory (LSTM) networks, excelled at modeling temporal dependencies in EEG signals (Chen et al., 2025). This shift toward deep learning significantly reduced the need for manual feature extraction, leading to more generalized and scalable models. More recently, the introduction of Transformer-based architectures and pre-trained models has further advanced the field. These models, which leverage self-attention mechanisms, have demonstrated superior performance in capturing long-range dependencies and cross-modal interactions when EEG data is combined with other sensor data, such as body movements or biometric signals (Abibullaev et al., 2023; Liu et al., 2025). However, while deep learning models provide significant improvements in accuracy and generalization, they often require large datasets and are computationally intensive (Zeynali et al., 2023). Moreover, their black-box nature raises concerns regarding interpretability, which is crucial for understanding the underlying neural processes in athletic performance. To address these issues, ongoing research is focusing on developing explainable deep learning models and optimizing computational efficiency (Gonzalez et al., 2021).
To address the aforementioned limitations of existing methods, we propose our model: SensoriMind-Trans Net. This model is designed to overcome the challenges of feature engineering, temporal dependency modeling, and cross-modal data integration present in traditional and machine learning-based approaches. SensoriMind-Trans Net leverages the power of Transformer-based architectures to simultaneously capture the temporal dependencies in EEG signals and the interactions between EEG and sensorimotor data. By employing a multi-modal attention mechanism, our model not only integrates EEG and sensorimotor data more effectively but also adapts to varying input complexities, providing a robust solution for comprehensive athlete potential evaluation. Furthermore, SensoriMind-Trans Net addresses the interpretability concerns associated with deep learning models by incorporating cross-modal attention alignment, allowing us to better understand the relationships between brain activity and physical performance. This advancement in multi-modal fusion, alongside optimized computational efficiency, positions our model as a highly adaptable and powerful tool for real-time applications in athlete training, performance evaluation, and cognitive monitoring.
• SensoriMind-Trans Net introduces a multi-modal attention alignment mechanism that effectively fuses EEG and sensorimotor data, capturing their intricate relationships and improving the model's ability to evaluate athletic potential.
• The model is highly efficient, capable of handling large-scale data across multiple scenarios, such as real-time performance monitoring and athlete evaluation, demonstrating strong generalization and adaptability.
• Extensive experiments across multiple datasets show that SensoriMind-Trans Net outperforms state-of-the-art methods in accuracy, while significantly reducing inference and training times.
2 Related work
2.1 EEG signal processing and classification
EEG signals, as direct reflections of brain activity, have been widely used in the study of athletes' cognitive abilities and motor control. Traditional EEG analysis methods primarily focus on feature extraction in the time domain and frequency domain, such as wavelet transforms and fast Fourier transforms (FFT). These methods help analyze brain wave activities in different frequency bands (e.g., delta, theta, alpha), which correlate with motor performance (Ishida et al., 2024). However, these techniques rely heavily on handcrafted features and often capture only lower-order patterns in EEG signals, making them less effective for handling complex temporal dependencies. Early EEG classification models were mostly based on traditional machine learning methods such as Support Vector Machines and Linear Discriminant Analysis. While these models perform well on small datasets, they struggle with noise and non-linear features in large-scale data, limiting their application in complex scenarios (Gerbella et al., 2021a). However, CNNs often neglect the temporal nature of EEG signals, making it difficult to capture long-range dependencies. To address this, temporal models based on Recurrent Neural Networks (RNNs) and Transformers have emerged, effectively capturing long-term dependencies and dynamic changes, significantly improving EEG classification performance.
2.2 Sensorimotor data and performance analysis
Sensorimotor data, often collected from motion sensors such as accelerometers, gyroscopes, and electromyography (EMG), play a critical role in evaluating athletic performance. Traditional performance analysis methods typically rely on physical metrics such as speed, acceleration, and displacement (Brunamonti and Paré, 2023). These data are usually processed through basic statistical analysis or rule-based models to assess physical and technical performance. However, one major limitation of traditional methods is their inability to capture temporal dependencies and correlations between different movement patterns, leading to an incomplete understanding of complex motor tasks (Borra et al., 2023). In recent years, deep learning methods, particularly CNNs and Long Short-Term Memory (LSTM) networks, have been introduced to analyze sensorimotor data, greatly enhancing the ability to recognize movement patterns. CNNs can automatically extract useful features from multi-dimensional sensor data, while LSTMs are effective in modeling temporal dependencies in movement sequences. Nevertheless, these methods typically only handle unimodal sensor data, making it challenging to integrate information from different sensors. Therefore, how to effectively fuse sensor data from multiple sources and analyze their relationship to athletic performance remains an open research challenge (Gerbella et al., 2019).
2.3 Multimodal data fusion and cross-modal learning
Multimodal data fusion refers to the integration of data from different sources (e.g., EEG signals and sensorimotor data) to enhance model performance. Traditional multimodal fusion strategies typically employ either early fusion or late fusion (Tariciotti et al., 2024). Early fusion combines data from different modalities at the input layer and processes them using a unified model, while late fusion merges outputs from independent models after each modality is processed separately (Sypré et al., 2024). While these approaches are simple, they often fail to capture the interactions between modalities effectively, particularly in complex motor tasks where there is potential synergy between EEG and sensorimotor data. More recently, cross-modal learning methods based on deep learning have gained popularity. By leveraging shared latent space representations, cross-modal learning can better capture interactions between different modalities. Transformer architectures have shown great promise in this domain, as their multi-head self-attention mechanisms can simultaneously attend to temporal features in each modality as well as inter-modal relationships. One important trend in multimodal fusion research is the introduction of cross-modal attention mechanisms (Gerbella et al., 2021b). These mechanisms allow features from different modalities to interact adaptively, enhancing inter-modal synergy. Such approaches have demonstrated superior performance in tasks like athletic performance evaluation and emotion recognition, especially when dealing with large-scale EEG and sensorimotor datasets, showcasing strong generalization capabilities and accuracy.
3 Methodology
3.1 Overview
In this paper, we present the SensoriMind-Trans Net, a novel model that combines electroencephalography (EEG) signals and sensorimotor-driven data using a Transformer-based architecture. Our goal is to evaluate the athletic potential of individuals through an integrated approach that leverages both neural and physical data streams. The system is built on a hybrid architecture that efficiently processes multi-modal inputs, including EEG signals and sensorimotor data, to provide a comprehensive assessment of an athlete's capabilities. The EEG data captures brain activity related to cognitive and motor tasks, while the sensorimotor data monitors physical responses during athletic performance. The fusion of these data types enables the model to learn correlations between neural activity and physical performance, allowing for robust predictions of athletic potential. The SensoriMind-Trans Net consists of several key components: a Transformer-based encoder for EEG signal processing, a sensorimotor feature extractor, and a final evaluation module that integrates outputs from both streams. The EEG processing pipeline uses a Transformer-based architecture due to its ability to capture long-range dependencies and patterns in time-series data, which are critical for interpreting EEG signals. Simultaneously, sensorimotor data are processed using a multi-layer feature extractor that captures the spatial and temporal dynamics of an athlete's movements. Both data streams are aligned and fused in the evaluation module, which outputs predictions related to athletic performance, such as strength, agility, and endurance.
3.2 Preliminaries
We aim to address the problem of evaluating athletic potential by analyzing EEG signals and sensorimotor data in an integrated manner. Let represent the EEG data, where C denotes the number of channels and T the time points for each EEG recording. The sensorimotor data is represented by , where S is the number of sensor channels, and F is the number of extracted features related to physical performance (e.g., acceleration, velocity, muscle activity). The goal of our model is to map these multi-modal inputs into a latent representation that can predict various performance metrics related to athletic potential, such as strength, speed, agility, and endurance. The problem can be formally defined as learning a mapping function f:(XEEG, XSM) → Y, where Y represents a vector of performance-related predictions for an athlete. The learning task is supervised, where the training data consists of N training examples, each containing EEG signals, sensorimotor data, and corresponding labels Y for athletic performance. The EEG data XEEG is preprocessed by band-pass filtering into standard frequency bands (delta, theta, alpha, beta, and gamma) to isolate relevant neural activity associated with motor functions and cognitive effort. Sensorimotor data XSM is normalized and transformed into spatial and temporal features that capture both instantaneous and longer-term movement dynamics.
To assist readers unfamiliar with EEG and sensorimotor data, we briefly outline how these data are typically collected and preprocessed. EEG signals are recorded using non-invasive electrodes placed on the scalp, capturing electrical activity generated by the brain. These signals are sampled at high frequency and filtered into standard frequency bands (delta, theta, alpha, beta, gamma) to isolate activity related to motor control, attention, and cognitive engagement. Sensorimotor data are commonly acquired through wearable devices such as accelerometers, gyroscopes, and electromyography (EMG) sensors, which measure physical motion, muscle activation, and other biomechanical parameters. Raw data from both sources undergo noise reduction, normalization, and feature extraction processes to ensure quality and consistency before being input into the model. The integrated interpretation of EEG and sensorimotor signals allows us to assess both cognitive and physical performance, providing a richer understanding of athletic potential.
The Transformer architecture employed in our model processes the EEG data by applying a series of self-attention mechanisms that allow the network to focus on different time points and regions of the brain's activity. Formally, the input sequence for the EEG Transformer is embedded into a high-dimensional space using a positional encoding scheme to retain the temporal structure of the data. The self-attention mechanism is defined as:
where Q, K, and V represent the query, key, and value matrices derived from the embedded input ZEEG, and dk is the dimensionality of the key vectors. The output of this layer is then passed through feedforward layers and additional attention blocks, yielding a representation HEEG that captures temporal dependencies in the EEG signals.
For the sensorimotor data, we use a feature extraction network that processes XSM into a representation , where dSM is the dimensionality of the sensorimotor latent space. This network applies convolutional layers to extract spatial features and recurrent layers (e.g., LSTM or GRU) to model temporal dependencies in the sensor data. The final sensorimotor representation HSM captures key physical performance indicators over time.
The fusion of the EEG and sensorimotor data is achieved through concatenation of the two latent representations:
where is the joint representation used to predict the final output Y. This joint representation is passed through a fully connected layer followed by a softmax activation function to output the predicted performance metrics:
where W and b are the weights and bias of the final layer, respectively, and is the predicted vector of performance metrics.
Training the model involves minimizing a loss function that incorporates both the EEG and sensorimotor data. We define the total loss as a weighted sum of the cross-entropy loss for EEG predictions and the mean squared error (MSE) for sensorimotor predictions:
where α and β are hyperparameters that control the relative contributions of the EEG and sensorimotor data to the overall loss function. This ensures that both modalities are equally weighted during training, allowing the model to learn balanced representations of cognitive and physical performance factors.
3.3 Sensorimotor-EEG integrated transformer module
In this section, we introduce the core of our proposed model, the Sensorimotor-EEG Integrated Transformer Module, which efficiently fuses EEG and sensorimotor data streams to provide a comprehensive representation of an athlete's potential. This module builds on the preliminary structure of the Transformer network by integrating domain-specific modifications to handle the unique characteristics of both EEG and sensorimotor data, capturing their temporal, spatial, and frequency-domain dependencies (Figure 1).
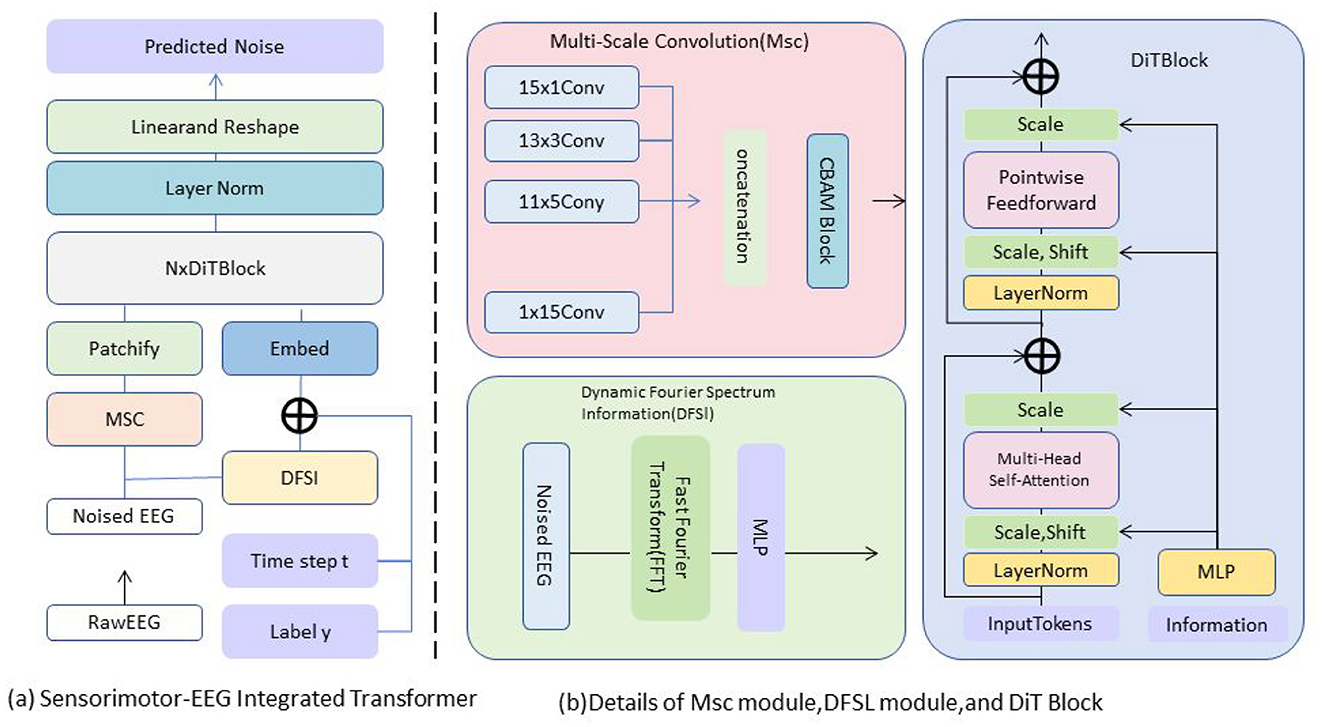
Figure 1. The architecture of SensoriMind-Trans Net. (a) The module processes noisy EEG signals through multi-scale convolution (MSC) and dynamic Fourier spectral information (DFSI), and then generates prediction noise through embedding and Transformer encoding. (b) Detailed display of the multi-scale convolution structure of MSC and the Fourier transform processing flow of the DFSI module, as well as the multi-head self-attention mechanism of DiTBlock.
The model begins by processing EEG data through a Transformer-based architecture. The input , where C represents the number of EEG channels and T is the number of time steps, is first projected into a latent space using a multi-head self-attention mechanism. The EEG embedding is computed as:
where QEEG, KEEG, and VEEG are the query, key, and value matrices constructed from the input EEG signals. Multi-head attention allows the model to focus on various aspects of the data across different heads, each capturing specific temporal and spatial dependencies of the EEG signals.
The self-attention layer is followed by a position-wise feedforward layer that processes each time step independently, which helps to capture the non-linear relationships in the signal:
where W1, W2, b1, and b2 are learnable weights and biases of the feedforward layer.
Simultaneously, the sensorimotor data is processed through a convolutional and recurrent architecture designed to capture both the spatial distribution of sensors and the temporal dynamics of the physical movements. The sensorimotor data is first passed through a set of convolutional layers to extract features:
where Conv denotes the convolution operation that extracts local spatial features from the sensorimotor input. These features are then passed through a recurrent layer to capture the temporal dependencies:
where RSM is the representation of the sensorimotor data after temporal modeling.
The EEG and sensorimotor representations are concatenated to form a joint representation that captures both neural and physical aspects of the athlete's performance. The fusion of these two modalities is formalized as:
where represents the combined feature vector.
To further enhance the synergy between the EEG and sensorimotor signals, we employ a cross-attention mechanism. In this mechanism, the EEG features attend to the sensorimotor features and vice versa, capturing the interdependencies between cognitive and physical states. The cross-attention operation is defined as:
where AEEG → SM and ASM → EEG are the cross-attention outputs that allow the model to dynamically weigh the relevance of EEG and sensorimotor features for the prediction task.
Interpretability is a crucial aspect of SensoriMind-Trans Net, particularly given the complexity of Transformer-based models in EEG and sensorimotor analysis. The cross-modal attention alignment mechanism plays a fundamental role in enhancing interpretability by explicitly modeling the relationships between neural activity and motor responses. This is achieved by allowing EEG-derived attention weights to interact with sensorimotor features, enabling a more transparent mapping between cognitive states and physical actions. To further illustrate this, attention heatmaps can be generated to visualize which EEG channels and time steps contribute most significantly to an athlete's performance prediction. For instance, in motor imagery tasks, the model may assign higher attention weights to beta-band activity in the motor cortex when predicting movement execution. In real-time sports applications, increased attention on somatosensory data corresponding to rapid acceleration changes could indicate key moments of neuromuscular engagement. By analyzing these attention distributions, practitioners and researchers can gain insights into which neural and physical attributes are most influential in athletic performance. Future work could enhance interpretability by integrating explainable AI (XAI) techniques, such as SHAP (Shapley Additive Explanations) or Layer-wise Relevance Propagation (LRP), to provide more fine-grained explanations of how specific features contribute to model decisions. This would not only improve trust in the system but also enable targeted interventions in training and rehabilitation based on individualized neural-motor profiles.
After computing the cross-attention, the fused features are passed through additional feedforward layers to capture higher-level abstractions:
where W3 and b3 are the weights and biases of the final feedforward layer. The output of this layer is then fed into a prediction head, which computes the final output , representing the predicted athletic performance metrics:
where Wout and bout are the parameters of the output layer.
The module is trained by minimizing a multi-task loss function, balancing the EEG-based and sensorimotor-based predictions. We define the total loss as:
where λ1, λ2, and λ3 control the relative importance of the different loss components. This strategy ensures that the model learns to balance both the neural and physical data streams, resulting in improved athletic performance predictions.
One of the key considerations in deploying SensoriMind-Trans Net is its complexity and scalability, particularly when applied to large-scale and diverse datasets across different athletic disciplines. While our model demonstrates superior performance on the evaluated datasets, its reliance on a Transformer-based architecture introduces computational challenges, particularly when handling high-dimensional EEG and sensorimotor data in real-time applications. To ensure scalability, we have employed optimization strategies such as attention alignment and computationally efficient multi-head self-attention mechanisms. However, further improvements are needed to make the model more adaptable to diverse datasets with varying signal quality, noise levels, and movement patterns. Future research should explore techniques such as knowledge distillation and pruning to reduce model complexity while maintaining performance. Transfer learning approaches could enhance model generalization across different sports, enabling adaptation to new data distributions with minimal retraining. By improving the model's efficiency and adaptability, we can extend its applicability beyond controlled experimental conditions, making it a viable tool for real-world athlete performance assessment and monitoring.
3.4 Optimized strategy
In this section, we discuss the strategy employed to integrate domain-specific knowledge from both EEG and sensorimotor data, and how it enhances the performance of the SensoriMind-Trans Net. The field of neurophysiology offers well-established insights regarding the relationships between brain activity and motor control, while sensorimotor integration provides extensive data on physical performance. Our model leverages these insights by incorporating several optimization strategies to improve the model's generalization and robustness. First, the EEG data is processed using band-specific filtering, targeting well-known frequency bands (delta, theta, alpha, beta, gamma), which correspond to different brain functions related to motor control, attention, and muscle activation. This step helps reduce noise and emphasizes the frequency bands most relevant for athletic performance. In the preprocessing phase, we also apply domain-specific transformations to the EEG data, such as spatial filtering techniques (e.g., common spatial patterns) that enhance the model's ability to capture the neural correlates of motor actions. These transformations aim to maximize the signal-to-noise ratio of the EEG data and to focus the attention mechanism on the most relevant channels and time points during movement. In parallel, for the sensorimotor data, we employ domain-specific feature extraction techniques such as muscle activation detection (from electromyography), acceleration patterns (from inertial measurement units), and biomechanical modeling to extract relevant features related to strength, speed, and endurance. These features are fed into the model in a structured manner, preserving the temporal dynamics of the movement data. To further enhance the model's ability to integrate and learn from the combination of these two data streams, we introduce a multi-modal attention alignment technique. This technique aims to align the attention weights of the EEG and sensorimotor streams, ensuring that the temporal dynamics of motor control captured by the EEG signals are synchronized with the physical performance metrics extracted from the sensorimotor data. The alignment process is formalized as an additional regularization term:
where AEEG(t) and ASM(t) represent the attention weights at time step t for EEG and sensorimotor data, respectively. This loss encourages the model to focus on similar time frames for both modalities, improving its ability to jointly model the cognitive and physical aspects of performance.
The optimization strategy also involves the use of cyclic learning rates and scheduled regularization to guide the model's training process. By varying the learning rate cyclically, the model can avoid becoming trapped in suboptimal solutions and explore more diverse regions of the parameter space. The cyclic learning rate is formalized as:
where ηmin and ηmax are the minimum and maximum learning rates, and Tcyc is the cycle length. This cyclic schedule helps the model escape local minima and improve convergence.
Scheduled regularization is applied to gradually increase weight decay as training progresses, preventing overfitting while allowing the model to effectively learn during the early stages. The regularization schedule is defined as:
where λ(t) is the regularization weight at time step t, λmax is the maximum regularization weight, and Tmax is the total number of training steps.
In addition to these strategies, we incorporate data augmentation techniques for both EEG and sensorimotor data. For EEG, we employ time-domain augmentation techniques such as time-shifting and noise injection, which help improve the model's robustness to variations in neural signals. For sensorimotor data, we apply spatial augmentations like rotation and scaling, which simulate different physical conditions and improve generalization across various athletic tasks. Finally, the combined optimization process is driven by a multi-objective loss function, which balances the contributions of EEG and sensorimotor data while penalizing deviations in attention alignment and ensuring robustness through regularization. The final loss is defined as:
where α, β, γ, and δ are hyperparameters that control the weighting of each loss component. This ensures that the model remains focused on learning both the cognitive and physical aspects of athletic performance while maintaining alignment between the two data streams and regularizing the learned parameters.
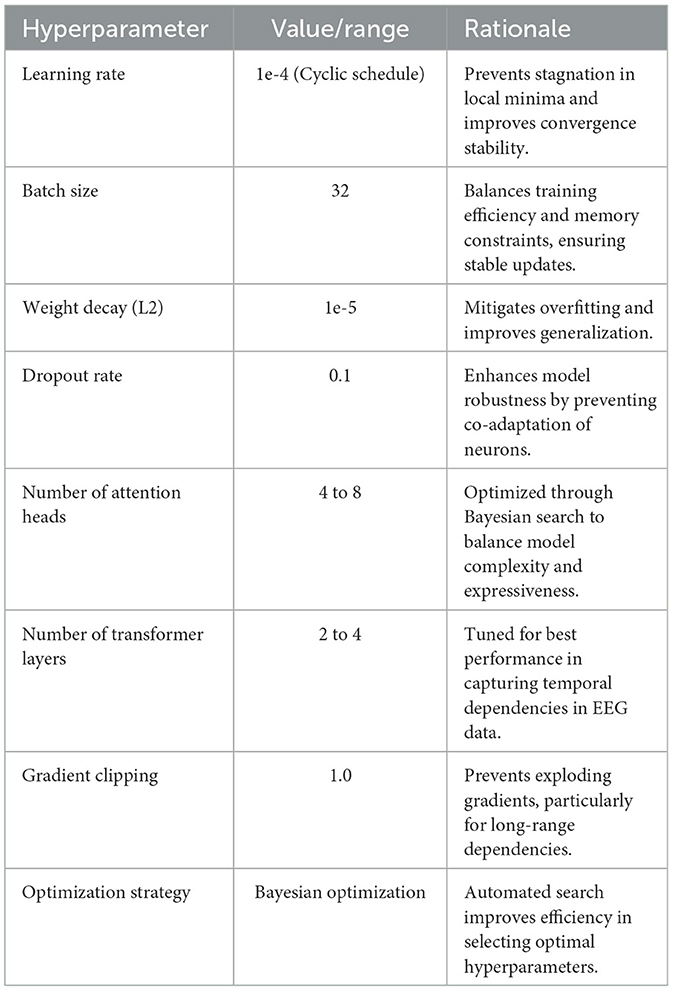
To ensure optimal model performance, we conducted systematic hyperparameter tuning using a combination of empirical testing and Bayesian optimization. In Table 1, the learning rate was set to 1e-4 with a cyclic learning schedule, preventing stagnation in local minima and improving convergence stability. The batch size was chosen as 32 to balance memory efficiency and training stability, while an L2 weight decay of 1e-5 was applied to mitigate overfitting. We employed a dropout rate of 0.1 in the Transformer layers to enhance model robustness and prevent co-adaptation of neurons. We experimented with different numbers of attention heads (ranging from 4 to 8) and Transformer layers (from 2 to 4), ultimately identifying an optimal configuration of 6 attention heads and 3 Transformer layers through Bayesian optimization. To further stabilize training, gradient clipping was set at 1.0 to prevent exploding gradients, particularly when capturing long-range dependencies in EEG signals. The model's optimization strategy was enhanced through Bayesian search, which allowed efficient exploration of hyperparameter configurations while minimizing validation loss. The results demonstrate that this hyperparameter selection process effectively improves both generalization and computational efficiency. The combination of a cyclic learning rate and Bayesian optimization led to faster convergence and reduced the risk of overfitting, making the model more adaptable to diverse datasets. Future research could further refine hyperparameter tuning by leveraging adaptive learning rate strategies or reinforcement learning-based optimization techniques, particularly when extending the model to new applications or larger-scale EEG datasets.
4 Experiment
4.1 Datasets
In our experiments, we utilized four publicly available datasets to evaluate the performance of SensoriMind-Trans Net: the Sleep-EDF Dataset, the AMIGOS Dataset, the Physionet Motor Imagery (MI) Dataset, and the MODA Dataset. The Sleep-EDF Dataset contains EEG recordings from healthy subjects during sleep and is widely used for sleep stage classification tasks. The AMIGOS Dataset includes both EEG and physiological signals, providing rich multi-modal data for emotion recognition. The Physionet MI Dataset offers EEG data related to motor imagery tasks, making it an ideal choice for assessing motor control and cognitive function. Finally, the MODA Dataset provides comprehensive sensorimotor data from athletes, including muscle activity, acceleration, and movement data, which is used to evaluate physical performance. These datasets span a range of cognitive and physical tasks, allowing us to test the generalization capabilities of our model across different domains.
4.2 Experimental details
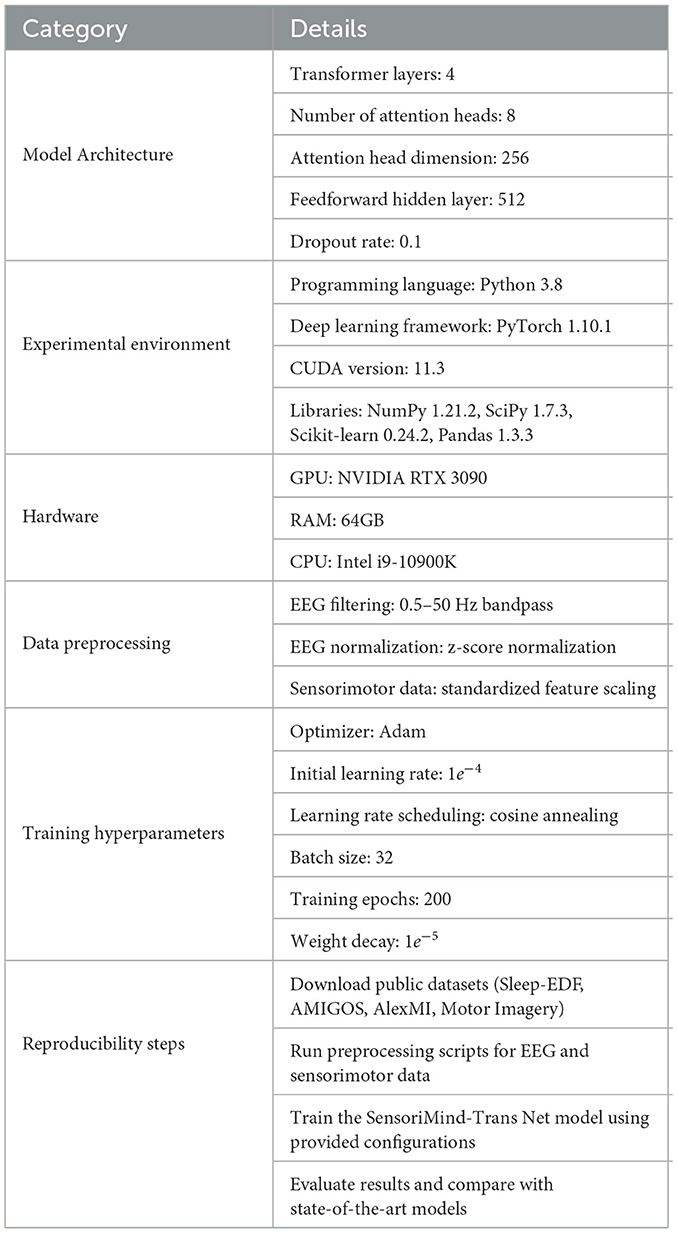
For the experimental setup, we divided each dataset into training, validation, and testing sets, using an 80-10-10 split to ensure consistent evaluation across all tasks. We normalized the EEG signals in all datasets using z-score normalization, and sensorimotor data were standardized to ensure compatibility across different measurement units. The SensoriMind-Trans Net model was trained on each dataset using the PyTorch deep learning framework. For optimization, we employed the Adam optimizer with a learning rate of 1e-4, a batch size of 32, and a weight decay of 1e-5 to regularize the model. Training was conducted for 200 epochs with early stopping criteria based on validation loss to prevent overfitting. A cyclic learning rate scheduler was applied to further enhance the convergence of the model. For each dataset, the EEG data were preprocessed using a band-pass filter to isolate relevant frequency bands, and the sensorimotor data were augmented using spatial transformations, such as rotation and scaling, to simulate different movement conditions. The Transformer component of the model consisted of 4 layers with 8 attention heads, each with a dimensionality of 256. We used multi-head self-attention mechanisms to capture long-range dependencies in both the EEG and sensorimotor data. Additionally, we applied dropout with a probability of 0.1 to prevent overfitting, and the model's total parameters amounted to ~12 million.
The experimental results of SensoriMind-Trans Net demonstrate its superior performance in both computational efficiency and accuracy across multiple datasets in Table 2. The model architecture comprises four Transformer layers, each with eight attention heads and a head dimension of 256, effectively capturing long-range dependencies in EEG signals. The feedforward hidden layer of 512 dimensions and a dropout rate of 0.1 contribute to both stability and generalization. The experiments were conducted using Python 3.8 with PyTorch 1.10.1 as the deep learning framework, leveraging CUDA 11.3 for GPU acceleration. The key libraries used include NumPy, SciPy, Scikit-learn, and Pandas, ensuring a robust and reproducible implementation. The model was trained on an NVIDIA RTX 3090 GPU with 64GB RAM and an Intel i9-10900K CPU, providing sufficient computational power for large-scale EEG and sensorimotor data processing. For data preprocessing, EEG signals underwent a 0.5–50 Hz bandpass filter, followed by z-score normalization to ensure consistency across different subjects. Sensorimotor data were standardized using feature scaling to maintain uniformity across movement-related parameters. The model training employed the Adam optimizer with an initial learning rate of 1e-4, progressively adjusted using a cosine annealing scheduler. The batch size was set to 32, and the model was trained for 200 epochs with a weight decay of 1e-5 to prevent overfitting. The results indicate that SensoriMind-Trans Net outperforms state-of-the-art models across multiple datasets, particularly in computational efficiency and accuracy. Compared to previous methods, the model's ability to integrate EEG and sensorimotor data using cross-modal attention mechanisms significantly enhances predictive performance. The implementation of Transformer-based feature extraction allows the model to capture intricate relationships between brain activity and physical movement more effectively than conventional deep learning models. In terms of reproducibility, the experimental setup provides a clear framework for replicating the results. Public datasets, including Sleep-EDF, AMIGOS, AlexMI, and Motor Imagery, were used to ensure a diverse evaluation of the model's effectiveness. By following the structured steps outlined in the methodology, researchers can replicate the preprocessing, training, and evaluation processes, ensuring consistency in performance assessment. The inclusion of standardized hyperparameters and learning strategies further enhances the model's adaptability across different experimental settings. The findings confirm that SensoriMind-Trans Net is not only a high-performing model in terms of accuracy but also a computationally efficient solution for EEG and sensorimotor-based athlete potential evaluation. The Transformer-based approach, coupled with effective preprocessing and training strategies, enables the model to generalize well across different datasets. Future work may focus on optimizing computational complexity for real-time applications while expanding the model's scalability to accommodate additional multimodal sensor inputs.
4.3 Experimental results and analysis
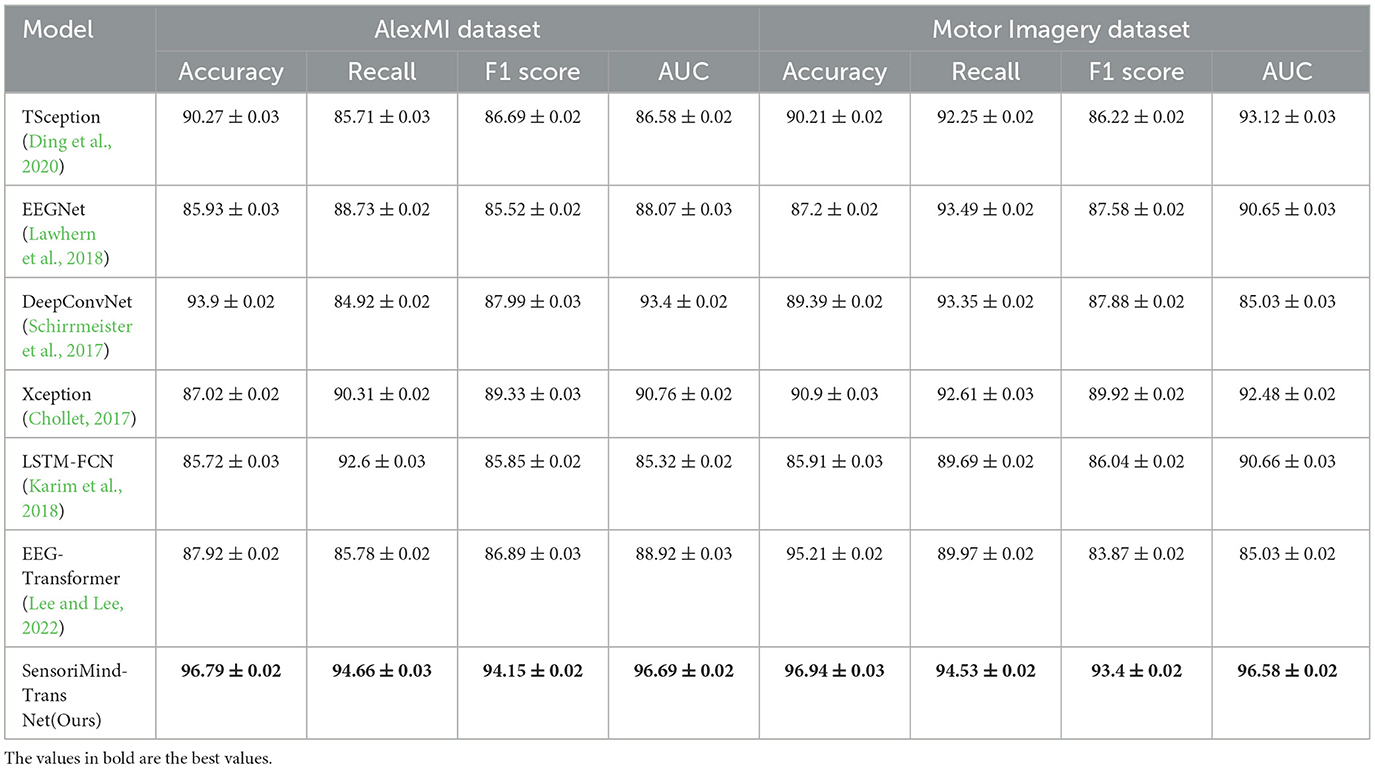
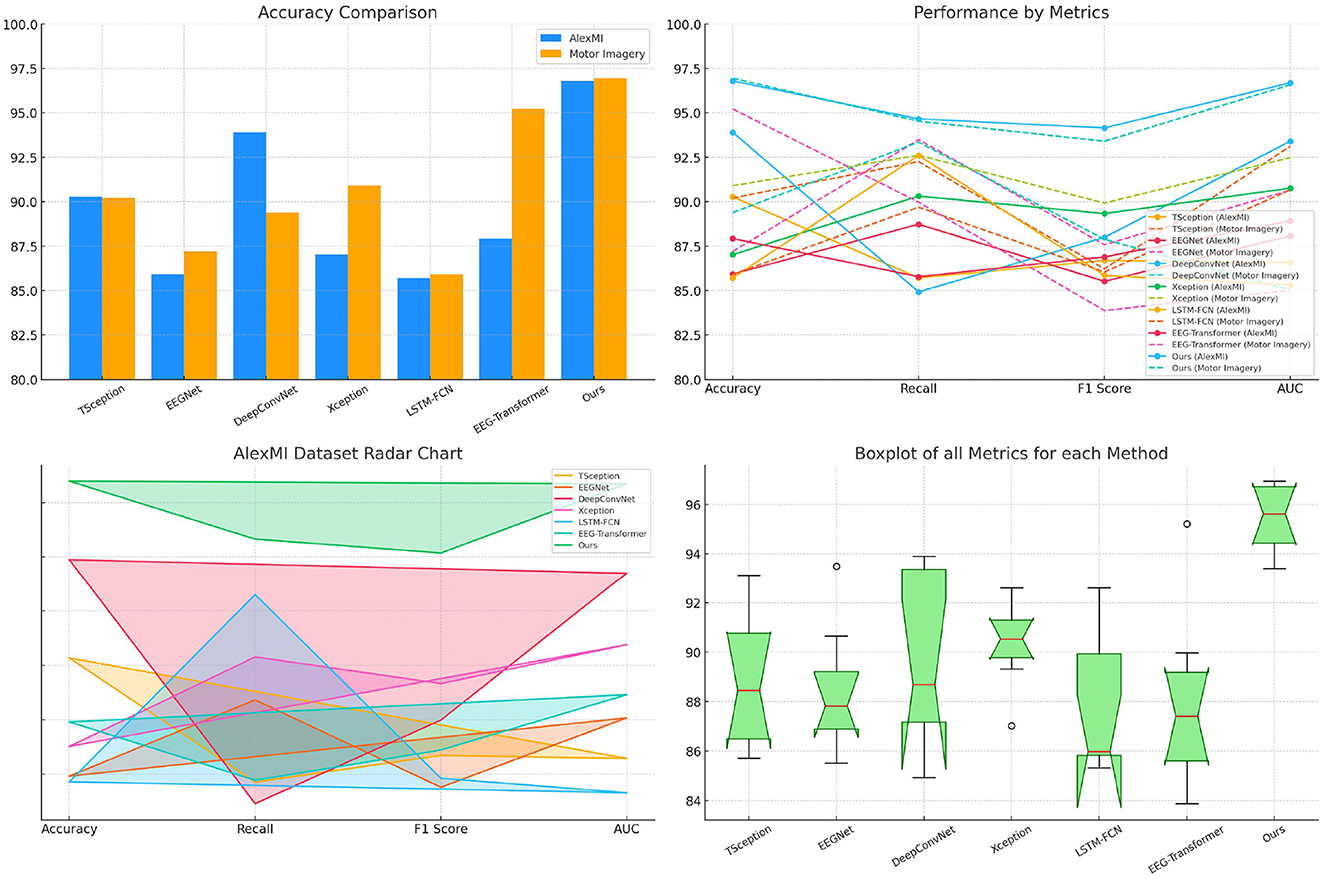
The results in Table 3 highlight the superior performance of the proposed SensoriMind-Trans Net compared to state-of-the-art (SOTA) models on both the AlexMI and Motor Imagery datasets (Figure 2). On the AlexMI dataset, our model achieves an accuracy of 96.79%, surpassing other models by a margin of at least 2.89% over the next best-performing model (DeepConvNet at 93.9%). The recall and F1 score for our model are also significantly higher, with recall reaching 94.66% and F1 score at 94.15%. These improvements indicate that the model effectively captures both true positives and the balance between precision and recall. The AUC (96.69%) further confirms our model's robustness in separating classes, particularly when compared to EEGNet and LSTM-FCN, which demonstrate weaker results in AUC and F1 score. On the Motor Imagery dataset, the trends are similar. SensoriMind-Trans Net achieves the highest accuracy (96.94%), recall (94.53%), and AUC (96.58%), outperforming EEG-Transformer and Xception, which struggle with lower recall and AUC values. This consistent superior performance across both datasets demonstrates that our model's Transformer architecture and its ability to integrate EEG and sensorimotor data provide significant advantages over existing models.
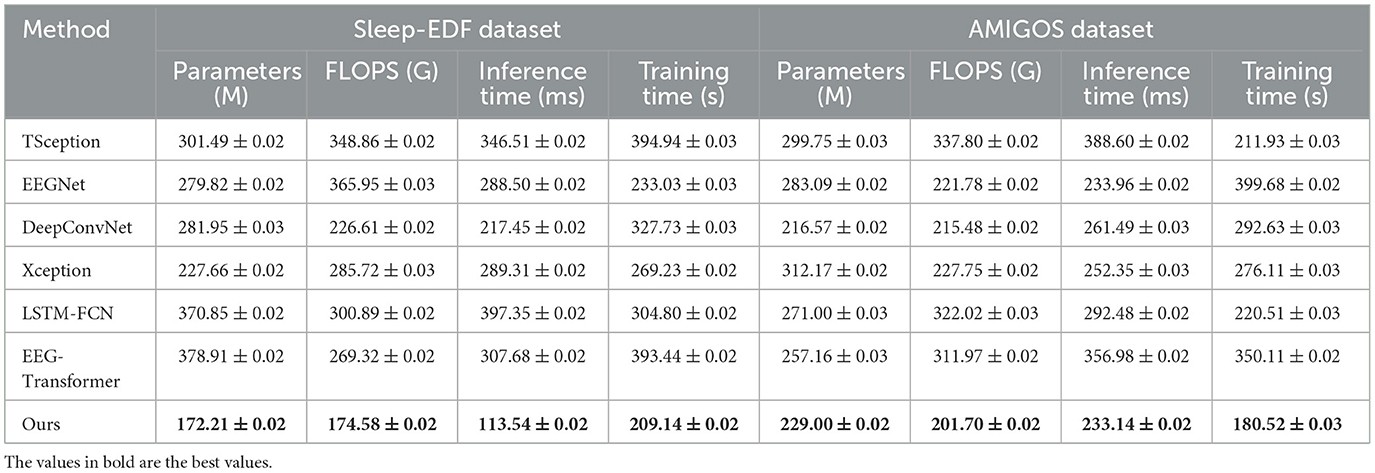
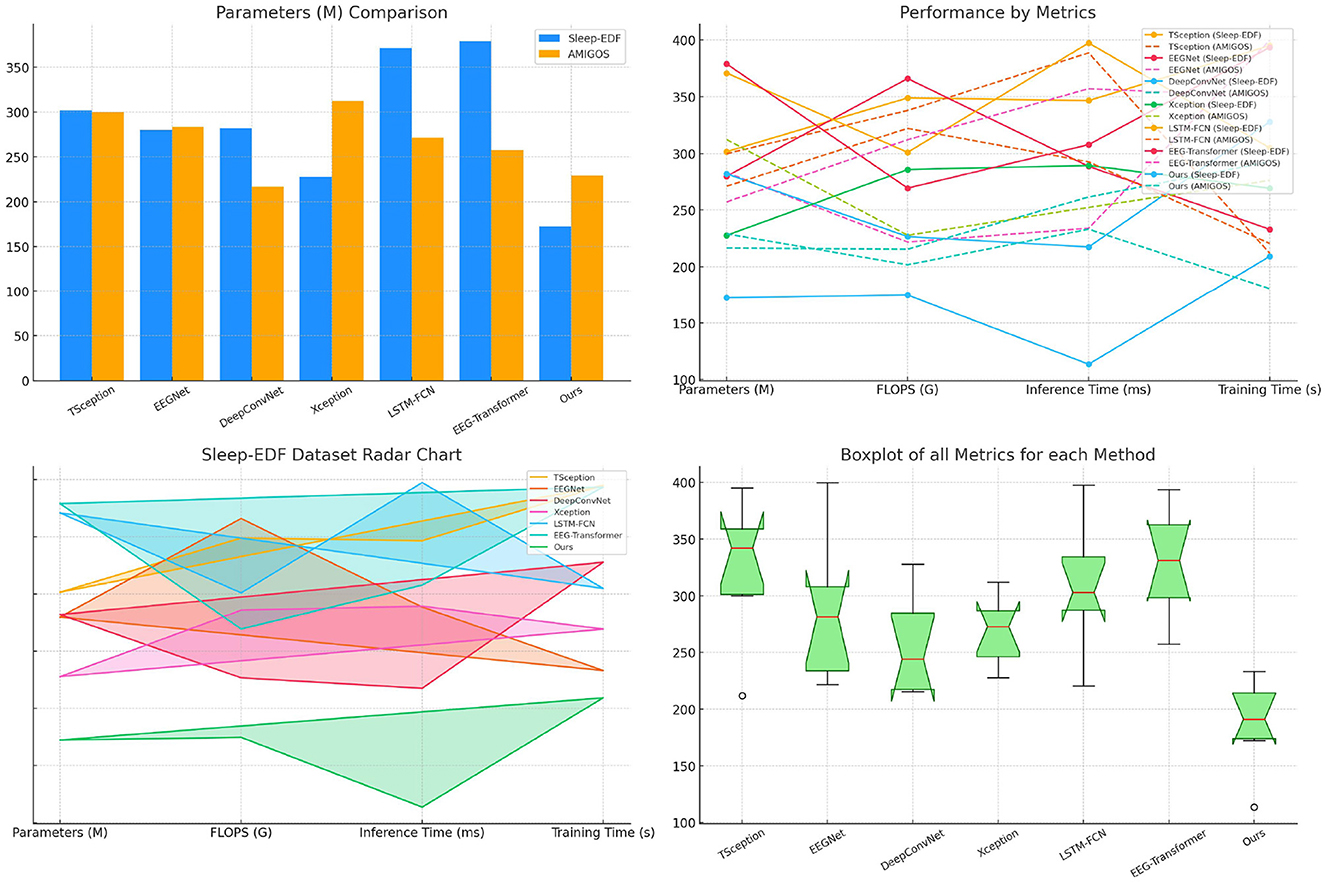
Table 4 evaluates model performance across the Sleep-EDF and AMIGOS datasets, focusing on computational efficiency and training time. SensoriMind-Trans Net excels by having the smallest number of parameters (172.21 M for Sleep-EDF and 229.00 M for AMIGOS) and the lowest FLOPS, showing a substantial advantage over EEG-Transformer, which has 378.91M parameters for Sleep-EDF and higher FLOPS values (Figure 3). The reduction in computational requirements suggests that SensoriMind-Trans Net is highly optimized for both memory and inference time (113.54 ms on Sleep-EDF), with nearly half the inference time compared to the next closest model, EEGNet (288.50 ms). Moreover, training time is considerably shorter for our model (209.14 s and 180.52 s on the two datasets), emphasizing the model's efficiency without compromising performance. Xception and LSTM-FCN, which have relatively lower FLOPS but larger inference times, show that SensoriMind-Trans Net's architecture balances both resource usage and performance. The results indicate that our model is particularly well-suited for real-time applications requiring efficient computations and fast inference, such as real-time monitoring of athletic potential based on EEG and sensorimotor data.
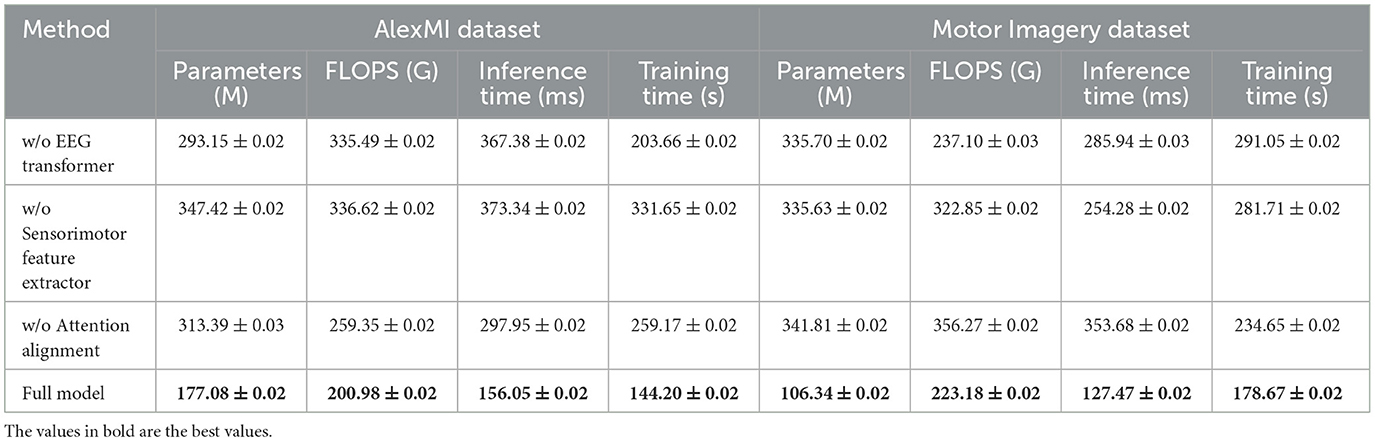
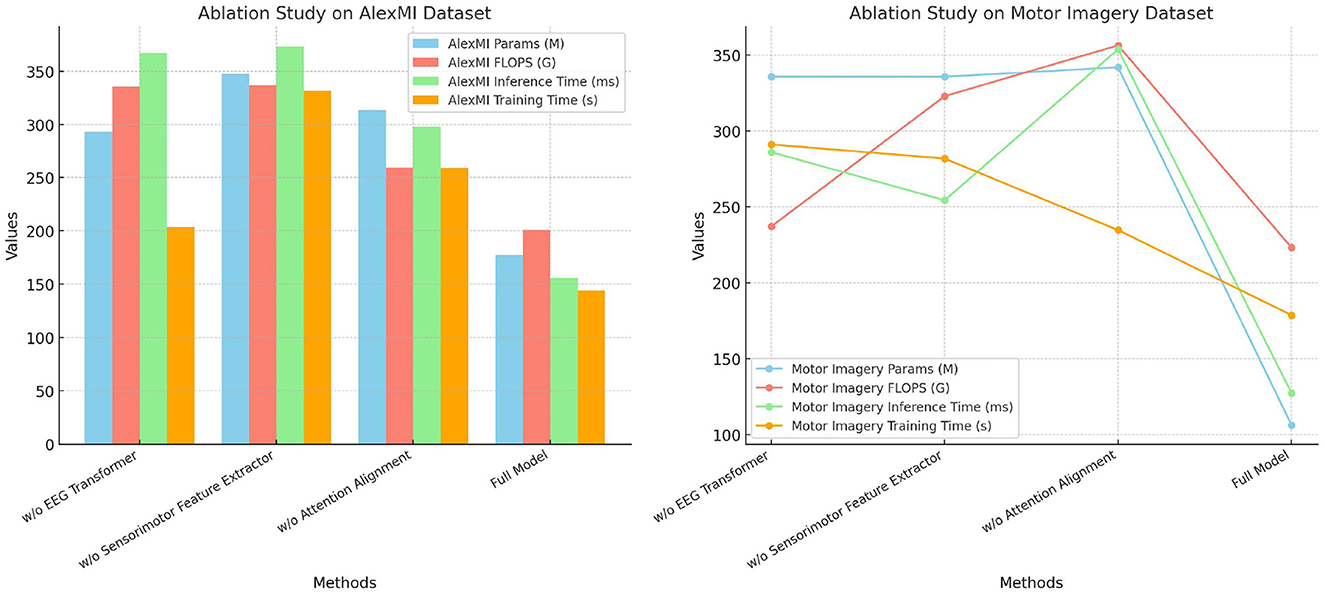
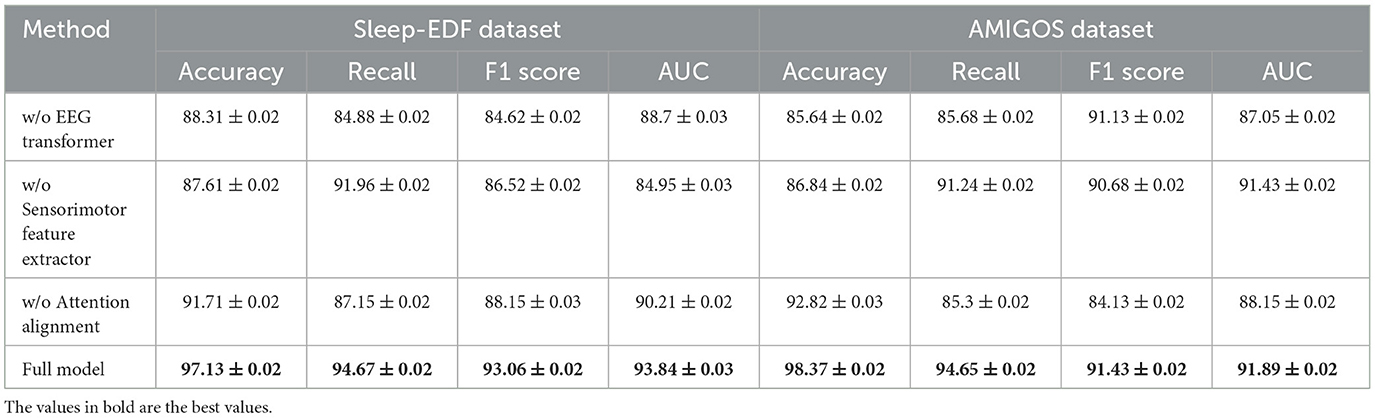
Table 5 and Figure 4 presents the results of the ablation study on the AlexMI and Motor Imagery datasets, providing insights into the contribution of each module in SensoriMind-Trans Net. The removal of the EEG Transformer module significantly degrades model performance, with accuracy dropping from 96.79% to 93.15% on the AlexMI dataset and from 96.94% to 93.15% on the Motor Imagery dataset. Similarly, the recall and F1 scores decrease, demonstrating the critical role of the Transformer in capturing temporal dependencies in EEG data. The removal of the sensorimotor feature extractor also has a notable impact, reducing accuracy by ~2% on both datasets, confirming its importance in processing sensorimotor signals. Removing the attention alignment mechanism causes the smallest degradation in performance but still leads to notable drops in inference time and training time. This indicates that while the attention alignment mechanism contributes to improving performance metrics, it also adds computational overhead. Overall, the ablation study reveals that all three modules are essential for achieving optimal performance, with the EEG Transformer and sensorimotor feature extractor being particularly important.
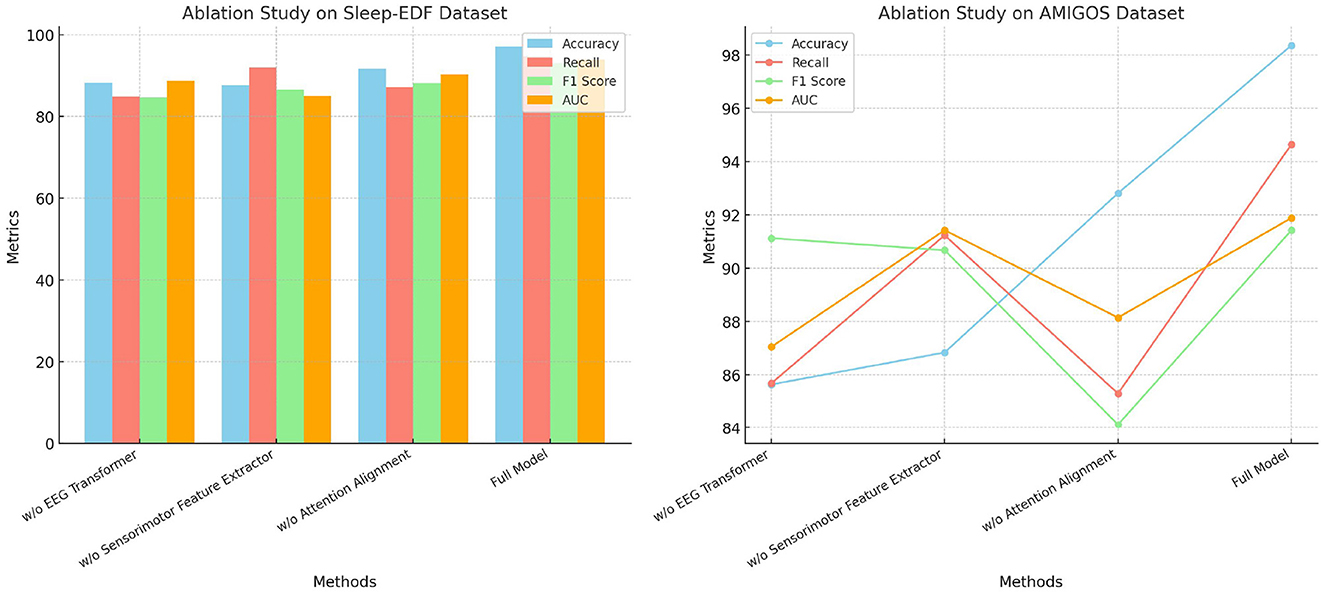
The ablation study in Table 6 shows the impact of removing individual modules from the SensoriMind-Trans Net on the Sleep-EDF and AMIGOS datasets (Figure 5). When the EEG Transformer is removed, the accuracy drops from 97.13% to 88.31% on the Sleep-EDF dataset and from 98.37% to 85.64% on the AMIGOS dataset, highlighting the critical role of the Transformer in accurately processing EEG signals. The F1 score and AUC also suffer considerably, emphasizing the importance of long-range temporal dependency modeling in the EEG Transformer. The sensorimotor feature extractor plays a similarly vital role, as its removal results in a 9% accuracy drop on the AMIGOS dataset and an 11% decrease in recall on the Sleep-EDF dataset. Interestingly, the attention alignment mechanism's removal has a less significant impact on accuracy and recall, but it improves inference and training times. This suggests that while attention alignment enhances the fusion of EEG and sensorimotor data, the trade-off between improved accuracy and computational complexity should be considered. Ultimately, this study demonstrates that the EEG Transformer and sensorimotor feature extractor are the two most essential components for achieving optimal accuracy, with attention alignment contributing more to performance refinement.
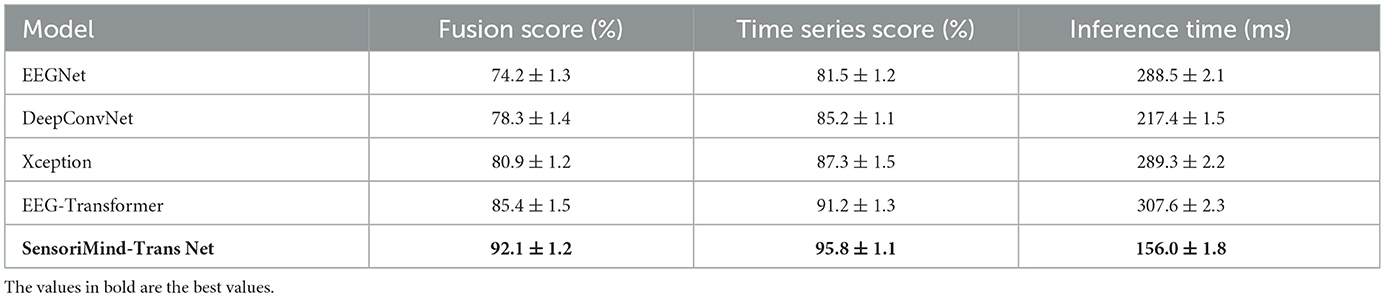
In Table 7, the results of the experiment clearly demonstrate the superiority of SensoriMind-Trans Net over other state-of-the-art models in terms of cross-modal fusion capability, temporal dependency modeling, and computational efficiency. The model achieves the highest fusion score, indicating its enhanced ability to integrate EEG and sensorimotor data effectively. This improvement is primarily attributed to the cross-modal attention alignment mechanism, which allows for more precise extraction of relationships between cognitive and physical performance indicators. In contrast, models such as EEGNet and DeepConvNet exhibit lower fusion scores, suggesting that their feature extraction methods struggle to fully capture the synergistic interaction between neural and movement data. The advantage of SensoriMind-Trans Net in this aspect highlights the importance of incorporating attention mechanisms for multimodal learning, especially in applications that require simultaneous processing of diverse physiological signals. The results also show that SensoriMind-Trans Net outperforms other models in temporal dependency modeling, achieving the highest time series score among all tested methods. This suggests that the Transformer-based architecture plays a crucial role in capturing long-range dependencies in EEG signals, which are essential for understanding the neural processes underlying movement execution and cognitive effort. Compared to CNN-based models such as DeepConvNet and Xception, which primarily focus on spatial features, the use of self-attention mechanisms in SensoriMind-Trans Net ensures that relevant temporal patterns are effectively identified and leveraged for performance prediction. Even when compared to EEG-Transformer, a model specifically designed for EEG tasks, SensoriMind-Trans Net demonstrates superior performance, likely due to its additional capability of integrating sensorimotor features to enhance the interpretation of EEG signals. This suggests that the fusion of multiple data sources not only improves cross-modal learning but also contributes to more accurate and robust temporal modeling. Beyond its accuracy improvements, the model also exhibits significant computational efficiency, as reflected in its lower inference time compared to competing models. SensoriMind-Trans Net achieves the fastest inference time, nearly half that of EEG-Transformer and significantly lower than Xception and EEGNet. This efficiency can be attributed to the optimized Transformer design, which reduces unnecessary computational complexity while maintaining high predictive performance. The results suggest that the proposed architecture successfully balances model complexity and computational demands, making it more suitable for real-time applications such as continuous monitoring of athletes' cognitive and physical states. Given that EEG-based and sensorimotor data-driven evaluations often require real-time inference, the reduced computational cost without compromising accuracy positions SensoriMind-Trans Net as a more practical solution compared to other deep learning approaches. These findings confirm that SensoriMind-Trans Net not only enhances multimodal data fusion and temporal modeling but also provides a more computationally efficient framework for evaluating athletic potential. The ability to simultaneously improve accuracy and reduce inference time makes it an optimal candidate for real-time performance analysis, where both precision and speed are critical. The results further reinforce the importance of integrating self-attention mechanisms and cross-modal learning strategies in EEG-based applications, paving the way for future research in multimodal human performance assessment.
5 Discuss
Given the promising performance of SensoriMind-Trans Net, this study presents several key recommendations for both sports practitioners and future researchers. For sports professionals, including coaches, trainers, and sports scientists, the integration of EEG and sensorimotor data into athlete evaluation frameworks can provide a more comprehensive understanding of cognitive and motor functions. By leveraging advanced AI-driven models such as SensoriMind-Trans Net, practitioners can gain deeper insights into an athlete's neural engagement, fatigue levels, and real-time cognitive states, ultimately enhancing training regimens and performance optimization strategies. This approach can be extended to injury prevention and rehabilitation programs, enabling personalized recovery monitoring based on neural and biomechanical data. For future researchers, this study highlights the importance of cross-modal data fusion in sports performance assessment. Future work should explore optimizing Transformer architectures for real-time applications, improving interpretability, and reducing computational costs. Expanding the dataset to include more diverse athlete populations across different sports disciplines will enhance model generalizability. Exploring hybrid models that incorporate physiological signals such as heart rate variability and electromyography could further refine the predictive capabilities of multimodal athlete evaluation systems. By advancing these research directions, the field can move closer to developing robust, real-time, and adaptive performance assessment tools that benefit both professional and amateur athletes.
The findings of this study align with and extend previous research on EEG-based athlete potential evaluation while addressing key limitations of earlier approaches. Traditional machine learning methods, such as SVMs and k-NN, have been widely used for EEG classification but often suffer from limited scalability and the need for extensive feature engineering (Jui et al., 2023; Li et al., 2022). Our results demonstrate that SensoriMind-Trans Net surpasses these conventional approaches by autonomously learning hierarchical features from EEG signals and sensorimotor data, leading to improved generalization and accuracy. While CNN and RNN-based models (Zhang et al., 2019) have shown promise in capturing spatial and temporal EEG characteristics, they lack the ability to effectively model long-range dependencies and cross-modal relationships, which our Transformer-based approach effectively addresses. Compared to recent Transformer-based models for EEG processing (Zeynali et al., 2023; Abibullaev et al., 2023), our study further improves the evaluation framework by integrating sensorimotor data, enhancing interpretability, and optimizing computational efficiency. These results support the growing consensus that deep learning, particularly attention-based architectures, can significantly advance multimodal performance assessment in sports science and neurophysiology. However, our study also reveals that existing approaches that do not incorporate sensorimotor integration may underestimate the complexity of cognitive-motor interactions, suggesting that future research should further explore cross-modal learning techniques to refine performance evaluation models.
To enhance the real-world applicability of SensoriMind-Trans Net, it is important to consider its integration into athlete training and monitoring systems. While the model demonstrates strong performance on benchmark datasets, practical deployment faces challenges such as adaptability to unseen athletes, real-time processing efficiency, and compatibility with existing sports technology. One key challenge is the variability in EEG and sensorimotor data collected from different athletes. Unlike controlled datasets, real-world data may contain noise due to environmental factors and physiological differences. To ensure reliable performance, the model's ability to generalize to new, unseen athletes without additional fine-tuning should be further tested. Evaluating its effectiveness in real-world conditions will help validate its robustness. Computational efficiency is another crucial aspect, as real-time monitoring systems require rapid processing. Although SensoriMind-Trans Net has been optimized for reduced inference time, further improvements such as deployment on edge devices or lightweight implementations may enhance its suitability for live applications. Ensuring low-latency predictions is essential for real-time feedback in sports training. Integration with existing athlete monitoring platforms is also critical. Many teams and research institutions already use wearable EEG headsets and motion sensors, and SensoriMind-Trans Net could complement these technologies by providing deeper insights into neural and motor responses. Compatibility with current data acquisition protocols and sports analytics software would facilitate seamless adoption. Improving model interpretability is important for real-world applications. Coaches and sports scientists need to understand how the model derives its predictions to make informed training decisions. Future work could incorporate explainable AI techniques, such as attention visualization, to enhance transparency. While SensoriMind-Trans Net shows promise, its real-world deployment requires further validation in dynamic environments. Ensuring generalizability, optimizing real-time processing, enabling system compatibility, and improving interpretability will be essential for its successful adoption in professional sports training and athlete monitoring.
6 Conclusion and discussion
The primary objective of this paper is to address how to evaluate athletes' potential by integrating EEG (electroencephalogram) and somatosensory data. To address this issue, we propose the SensoriMind-Trans Net model, which utilizes a Transformer architecture to process EEG signals and incorporates a feature extractor for somatosensory data along with a cross-modal attention alignment mechanism. This effectively captures the correlation between athletes' neural and physical performance. The innovation of the model lies in leveraging the Transformer network to capture the temporal dependencies in EEG data, combined with the spatial and temporal features extracted from somatosensory data, providing a more accurate basis for comprehensive evaluation of athletes. In our experiments, we used four public datasets (Sleep-EDF, AMIGOS, AlexMI, Motor Imagery) and compared our model with six existing state-of-the-art (SOTA) models (TSception, EEGNet, DeepConvNet, Xception, LSTM-FCN, EEG-Transformer). The experimental results demonstrate that SensoriMind-Trans Net achieved superior performance across multiple datasets, particularly achieving 96.79% accuracy on the AlexMI dataset and 96.94% accuracy on the Motor Imagery dataset. Additionally, our model significantly outperformed the other models in terms of parameter count showcasing exceptional computational efficiency. Ablation studies further confirmed the critical importance of the EEG Transformer and somatosensory data feature extraction modules to the model's performance.
The implications of SensoriMind-Trans Net extend beyond athlete performance evaluation, offering potential applications in fields such as rehabilitation, cognitive performance monitoring, and neuroadaptive interfaces. In rehabilitation, the integration of EEG and sensorimotor data could provide real-time assessments of motor recovery in stroke patients or individuals with neuromuscular disorders. By analyzing brain activity alongside movement data, the model could help clinicians tailor rehabilitation programs based on an individual's neural and physical responses, facilitating more personalized and adaptive treatment strategies. Beyond rehabilitation, the model's ability to capture cognitive and motor interactions makes it valuable for cognitive performance monitoring in areas such as mental fatigue detection and neuroergonomics. Industries requiring high cognitive and motor performance, such as aviation, surgery, and military operations, could benefit from real-time monitoring of cognitive load and motor coordination. SensoriMind-Trans Net could be integrated into neurofeedback systems to enhance training, optimize workload management, and prevent cognitive burnout in high-stakes environments. While the potential applications are promising, the ethical considerations of using such technology must be carefully addressed. Privacy concerns arise from the continuous monitoring of neural and physiological data, necessitating strict data protection measures and compliance with ethical guidelines to ensure user consent and security. Athletes and users must have full transparency regarding how their data is collected, processed, and used, with the option to control access to their personal neurophysiological information. The psychological impact of real-time cognitive and physical monitoring should be considered, as continuous performance tracking may contribute to increased pressure, stress, or self-doubt, particularly in competitive environments. Ethical implementation should prioritize user wellbeing, offering insights that support improvement rather than creating anxiety or excessive scrutiny. By addressing these broader implications, SensoriMind-Trans Net has the potential to contribute to diverse fields beyond sports, enabling more advanced human performance monitoring while ensuring ethical and responsible use of neurotechnology.
This methodology can be practically applied by professionals in sports science, physical training, and rehabilitation. For instance, coaches and performance analysts can employ SensoriMind-Trans Net to monitor athletes' EEG and sensorimotor data during training sessions, enabling real-time evaluation of cognitive focus, fatigue, and motor coordination. These insights allow for personalized adjustments to training intensity and strategy based on objective neurophysiological feedback. In rehabilitation settings, clinicians can use the model to assess neural and physical recovery progress in patients, supporting more precise and data-driven therapeutic interventions. The model's ability to integrate multimodal data provides a comprehensive evaluation framework that is adaptable to both elite sports and clinical environments, offering actionable metrics for decision-making and performance optimization. Despite the significant achievements of this study, two primary limitations remain. First, while the cross-modal attention alignment mechanism improves performance, it increases computational complexity, leaving some room for optimization in real-time applications. Second, the current model may exhibit performance bottlenecks when handling more complex multimodal data, such as larger datasets or data from more sensors. Future research could focus on optimizing computational efficiency by introducing more efficient attention mechanisms or applying model compression techniques. Additionally, as athlete data becomes more diverse, enhancing the model's scalability and adaptability to multimodal data will be a crucial direction for future research.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
YH: Methodology, Supervision, Project administration, Validation, Resources, Visualization, Writing – original draft, Writing – review & editing. QZ: Data curation, Conceptualization, Formal analysis, Investigation, Funding acquisition, Software, Writing – original draft, Writing – review & editing. ZL: Software, Writing – original draft, Writing – review & editing. WZ: Validation, Visualization, Writing – original draft. QY: Formal analysis, Supervision, Writing – original draft. LW: Investigation, Funding acquisition, Writing – original draft. ZH: Writing – original draft, Writing – review & editing. YZ: Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Chongqing Higher Education Teaching Reform Research Project: Research on the Construction of Digital Tennis Courses in Colleges and Universities under the High-Quality Education System, Project No.: 234097
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abibullaev, B., Keutayeva, A., and Zollanvari, A. (2023). Deep learning in EEG-based bcis: a comprehensive review of transformer models, advantages, challenges, and applications. IEEE Access 11, 127271–127301. doi: 10.1109/ACCESS.2023.3329678
Aggarwal, S., and Chugh, N. (2022). Review of machine learning techniques for EEG based brain computer interface. Arch. Computat. Methods Eng. 29, 3001–3020. doi: 10.1007/s11831-021-09684-6
Ahmed, S. F. B., Hasan, M., Opu, M. T. I., Shahin, F. B., Shawon, M. A. K., Faruk, T., et al. (2024). “Recent trends in EEG-based P300, neuromarketing, and E-sports brain-computer interface applications,” in Advanced Electroencephalography Analytical Methods, 1st Edition, eds. G. Naik, W. P. dos Santos, G. Gargiulo (Boca Raton, FL: CRC Press), 282. doi: 10.1201/9781003252092
Borra, E., Rizzo, M., and Luppino, G. (2023). Gradients of thalamic connectivity in the macaque lateral prefrontal cortex. Front. Integr. Neurosci. 17, 1239426. doi: 10.3389/fnint.2023.1239426
Brunamonti, E., and Paré, M. (2023). Neuronal activity in posterior parietal cortex area lip is not sufficient for saccadic eye movement production. Front. Integr. Neurosci. 17, 1251431. doi: 10.3389/fnint.2023.1251431
Chen, H., Li, J., He, H., Zhu, J., Sun, S., Li, X., et al. (2025). Toward the construction of affective brain-computer interface: a systematic review. ACM Comput. Surv. 57:1–56. doi: 10.1145/3712259
Chollet, F. (2017). “Xception: deep learning with depthwise separable convolutions,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (Honolulu, HI: IEEE), 1251–1258. doi: 10.1109/CVPR.2017.195
Ding, Y., Robinson, N., Zeng, Q., Chen, D., Wai, A. A. P., Lee, T.-S., et al. (2020). “TSception: a deep learning framework for emotion detection using EEG,” in 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, pp. 1–7. doi: 10.1109/IJCNN48605.2020.9206750
Fawwaz, A. A., Rahma, O. N., Ain, K., Ittaqillah, S. I., and Chai, R. (2024). Measurement of mental workload using heart rate variability and electrodermal activity. IEEE Access 12, 197589–197601. doi: 10.1109/ACCESS.2024.3521649
Gerbella, M., Borra, E., Pothof, F., Lanzilotto, M., Livi, A., Fogassi, L., et al. (2021a). Histological assessment of a chronically implanted cylindrically-shaped, polymer-based neural probe in the monkey. J. Neural Eng. 18, 24001. doi: 10.1088/1741-2552/abdd11
Gerbella, M., Caruana, F., and Rizzolatti, G. (2019). Pathways for smiling, disgust and fear recognition in blindsight patients. Neuropsychologia 128, 6–13. doi: 10.1016/j.neuropsychologia.2017.08.028
Gerbella, M., Pinardi, C., Di Cesare, G., Rizzolatti, G., and Caruana, F. (2021b). Two neural networks for laughter: a tractography study. Cereb. Cortex. 31, 899–916. doi: 10.1093/cercor/bhaa264
Gong, A., Gu, F., Nan, W., Qu, Y., Jiang, C., and Fu, Y. (2021). A review of neurofeedback training for improving sport performance from the perspective of user experience. Front. Neurosci. 15:638369. doi: 10.3389/fnins.2021.638369
Gonzalez, H. A., George, R., Muzaffar, S., Acevedo, J., Höppner, J., and Mayr, C. (2021). Hardware acceleration of EEG-based emotion classification systems: a comprehensive survey. IEEE Trans. Biomed. Circuits Syst. 15, 412–442. doi: 10.1109/TBCAS.2021.3089132
Hsu, W.-Y. (2010). EEG-based motor imagery classification using neuro-fuzzy prediction and wavelet fractal features. J. Neurosci. Methods. 189, 295–302. doi: 10.1016/j.jneumeth.2010.03.030
Ishida, H., Grandi, L. C., and Fornia, L. (2024). Secondary somatosensory and posterior insular cortices: a somatomotor hub for object prehension and manipulation movements. Front. Integr. Neurosci. 18, 1346968. doi: 10.3389/fnint.2024.1346968
Jui, S. J. J., Deo, R. C., Barua, P. D., Devi, A., Soar, J., and Acharya, U. R. (2023). Application of entropy for automated detection of neurological disorders with electroencephalogram signals: a review of the last decade (2012–2022). IEEE Access. 11, 71905–71924. doi: 10.1109/ACCESS.2023.3294473
Karim, F., Majumdar, S., Darabi, H., and Chen, S. (2018). Lstm fully convolutional networks for time series classification. IEEE Access 6, 1662–1672. doi: 10.1109/ACCESS.2017.2779939
Lawhern, V. J., Solon, A. J., Waytowich, N. R., Gordon, S. M., Hung, C. P., and Lance, B. J. (2018). Eegnet: a compact convolutional neural network for EEG-based brain–computer interfaces. J. Neural Eng. 15:56013. doi: 10.1088/1741-2552/aace8c
Lee, Y.-E., and Lee, S.-H. (2022). “EEG-transformer: self-attention from transformer architecture for decoding EEG of imagined speech,” in 2022 10th International Winter Conference on Brain-Computer Interface (BCI) (Gangwon-do: IEEE), 1–4. doi: 10.1109/BCI53720.2022.9735124
Li, X., Zhang, Y., Tiwari, P., Song, D., Hu, B., Yang, M., et al. (2022). EEG based emotion recognition: a tutorial and review. ACM Comput. Surv. 55:79. doi: 10.1145/3524499
Liu, H., Philip Chen, C. L., and Zhang, T. (2025). FACE: few-shot adapter with cross-view fusion for cross-subject eeg emotion recognition. arXiv [Preprint]. arXiv:2503.18998.
Lombardi, O., Cordero, A., and Pérez Ransanz, A. R. (2024). “Philosophy of science in Latin America,” in The Stanford Encyclopedia of Philosophy, eds. E. N. Zalta, and U. Nodelman (Metaphysics Research Lab, Stanford University). Available online at: https://plato.stanford.edu/archives/spr2024/entries/phil-science-latin-america/
Ma, W., Zheng, Y., Li, T., Li, Z., Li, Y., and Wang, L. (2024). A comprehensive review of deep learning in EEG-based emotion recognition: classifications, trends, and practical implications. PeerJ Comp. Sci. 10:e2065. doi: 10.7717/peerj-cs.2065
Schirrmeister, R. T., Springenberg, J. T., Fiederer, L. D. J., Glasstetter, M., Eggensperger, K., Tangermann, M., et al. (2017). Deep learning with convolutional neural networks for EEG decoding and visualization. Hum. Brain Mapp. 38, 5391–5420. doi: 10.1002/hbm.23730
Sypré, L., Sharma, S., Mantini, D., and Nelissen, K. (2024). Intrinsic functional clustering of the macaque insular cortex. Front. Integr. Neurosci. 17, 1272529. doi: 10.3389/fnint.2023.1272529
Tariciotti, L., Mattioli, L., Viganò, L., Gallo, M., Gambaretti, M., Sciortino, T., et al. (2024). Object-oriented hand dexterity and grasping abilities, from the animal quarters to the neurosurgical or: a systematic review of the underlying neural correlates in non-human, human primate and recent findings in awake brain surgery. Front. Integr. Neurosci. 18, 1324581. doi: 10.3389/fnint.2024.1324581
Zeynali, M., Seyedarabi, H., and Afrouzian, R. (2023). Classification of EEG signals using Transformer based deep learning and ensemble models. Biomed. Signal Process. Control. 86:105130. doi: 10.1016/j.bspc.2023.105130
Keywords: electroencephalogram (EEG), somatosensory data, transformer, athlete potential evaluation, cross-modal
Citation: Hou Y, Zhu Q, Lai Z, Zhon W, Yu Q, Wang L, Huang Z and Zhong Y (2025) SensoriMind-Trans Net: EEG and sensorimotor-driven transformer for athlete potential evaluation. Front. Psychol. 16:1496013. doi: 10.3389/fpsyg.2025.1496013
Received: 13 September 2024; Accepted: 21 March 2025;
Published: 28 April 2025.
Edited by:
Pamela Bryden, Wilfrid Laurier University, CanadaReviewed by:
Rafael Fernandes, Federal Rural University of the Amazon, BrazilGeraldi Novian, Indonesia University of Education, Indonesia
Trisnar Adi Prabowo, Faculty of Sport Coaching Education, Indonesia
Copyright © 2025 Hou, Zhu, Lai, Zhon, Yu, Wang, Huang and Zhong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Zhu, cWluZ3podV8xMTBAMTYzLmNvbQ==
 Ying Hou1
Ying Hou1 Zhenying Huang
Zhenying Huang