- 1Department of Biotechnology, Khalsa College for Women, Amritsar, Punjab, India
- 2Center of Excellence in Genomics and Synstem Biology (CEGSB) and Center for Pre-Breeding Research (CPBR), International Crops Research Institute for the Semi-Arid Tropics, Hydrabad, India
- 3Department of Bioengineering and Biosciences, School of Agriculture, Lovely Professional University, Phagwara, Punjab, India
In the global food sector, extending shelf life and ensuring food safety continue to be major concerns that call for innovative approaches that go beyond traditional preservation methods. RNA interference (RNAi) and CRISPR have become cutting-edge biotechnological techniques with enormous potential for food preservation. CRISPR-mediated gene editing allows for precise modifications in food crops, livestock, and microbial systems to delay ripening, increase resistance to oxidative stress, and suppress enzymes responsible for rancidity. We have tried addressed this by introducing a comparative analysis of CRISPR and RNAi efficiency across climacteric and non-climacteric fruits, highlighted emerging targets (e.g., ethylene-independent ripening regulators and cell wall-modifying enzymes), and identified critical gaps in regulatory frameworks and delivery methods in less-explored crops like guava and papaya. This integration aims to present a more forward-looking perspective beyond existing literature. Similarly, to improve food stability and manage post-harvest degradation, RNAi-based techniques help to silence genes. By reducing mycotoxin contamination, improving disease resistance in livestock and aquaculture, and focussing on foodborne pathogens, these technologies provide revolutionary solutions for food safety that go beyond preservation. Despite its potential, the commercialisation and adoption of gene-edited food items are heavily influenced by legal frameworks, ethical issues, and public opinion. The mechanics and uses of CRISPR and RNA interference in food safety and preservation are examined in this review, along with ethical and legal issues and potential future developments for these technologies to ensure sustainable food security.
1 Introduction
Food spoilage and waste are significant global issues that contribute to food insecurity, economic losses, and environmental degradation. According to the Food and Agriculture Organisation (FAO), roughly 1.3 billion tonnes of food—almost one-third of all food produced globally—are lost or wasted each year, resulting in nearly $940 billion in economic losses (FAO, 2019). Food spoiling is caused primarily by microbial contamination, enzymatic activity, and oxidative degradation, which result in changes in texture, colour, flavour, and nutritional content. Perishable foods, such as fruits, vegetables, dairy, and meat, are especially vulnerable, with post-harvest losses accounting for 40%–50% of total food waste in developing nations due to inadequate storage and preservation measures. Traditional food preservation techniques such as refrigeration, chemical preservatives, and irradiation serve to reduce spoiling, but they frequently raise issues about food safety, environmental effect, and consumer health (Vermeulen et al., 2012).
In recent years, biotechnology breakthroughs have provided fresh techniques to combat food spoiling, with CRISPR and RNA interference (RNAi) emerging as game-changing genome editing tools. CRISPR-Cas technology allows for precise alterations in food crops, livestock, and microbial ecosystems to delay ripening, increase oxidative stress resistance, and reduce lipid oxidation (Jinek et al., 2012). Similarly, RNAi-mediated gene silencing provides a tailored strategy to inhibit spoilage-related pathways and microbial contamination, hence improving food stability and safety (Baulcombe, 2004). Beyond preservation, these technologies provide unique food safety options, such as CRISPR-based bacteriophage engineering to target foodborne bacteria and RNAi-mediated inhibition of antibiotic resistance genes (Bikard et al., 2014; Kim et al., 2021). Furthermore, CRISPR and RNAi applications in minimising mycotoxin contamination and avoiding viral infections in cattle and aquaculture offer promising approaches to safer food production (Shin et al., 2018).
Despite the growing interest in CRISPR and RNAi in food biotechnology, most available reviews concentrate on their fundamental mechanisms or applications in agriculture and health (Barrangou and Doudna, 2016; Miao et al., 2019). However, there has been little extensive consideration of how these technologies are related to food preservation and safety. Our review fills this gap by thoroughly investigating CRISPR and RNAi applications for delaying ripening, increasing oxidative stability, reducing lipid oxidation, and preventing microbial contamination. We also present a critical assessment of the regulatory frameworks, ethical considerations, and consumer viewpoints that influence the commercialisation of gene-edited food products. Unlike earlier studies, which focused on either CRISPR or RNAi, we provide a comparative review of their processes and efficacy in food preservation. This review seeks to present a comprehensive view of how cutting-edge technology can revolutionise food sustainability while maintaining consumer safety by tackling both technological breakthroughs and socio-regulatory issues.
2 Fundamentals of CRISPR and RNA interference
Recent progress in gene-editing technologies has introduced a new generation of CRISPR and RNAi platforms tailored for food biotechnology applications. Tools like Cas12a, which cleaves DNA at staggered positions and enables multiplex editing in AT-rich genomes, and Cas13, which targets RNA for reversible gene silencing, have significantly broadened the scope of post-harvest food regulation (Tang et al., 2019; Li J et al., 2020). The discovery of CasΦ, a hypercompact enzyme from bacteriophages, enhances delivery possibilities in constrained systems like viral vectors (Batra et al., 2020). Moreover, base editors (e.g., cytidine and adenine deaminases) and prime editors enable precise nucleotide substitutions and insertions without double-strand DNA breaks, thus reducing genomic disruption while optimizing traits such as shelf life and flavor (Gaudelli et al., 2017; Anzalone et al., 2019). In microbial control and post-processing safety, CRISPR interference (CRISPRi) and activation (CRISPRa) offer reversible control of gene expression by targeting transcriptional regulators, allowing fine-tuning of microbial pathways or food quality traits such as oxidative stability and aroma (Qi et al., 2013). These have been coupled with Cas12a- and Cas13-based biosensors integrated into food packaging or cold-chain diagnostics for rapid detection of foodborne pathogens and spoilage biomarkers (Gootenberg et al., 2018; Li et al., 2022). Advances in RNAi delivery systems, notably topical sprays containing synthetic siRNAs or double-stranded RNAs supported by nanoparticles, enable gene silencing in crops without transgenic modification, thus aligning with non-GMO regulatory frameworks (Dalakouras et al., 2020). Likewise, virus-induced gene silencing (VIGS) facilitates temporary suppression of ripening and spoilage-related genes in produce such as tomatoes and melons (Senthil-Kumar and Mysore, 2011).
Both technologies are also driving biosensor innovation and microbial safety efforts. CRISPRi and base editors are being used to suppress virulence factors in spoilage microbes without eliminating beneficial flora (Tonutti et al., 2023), and RNAi-based sprays are in development for controlling mycotoxin-producing genes in fungi like Fusarium and Aspergillus, enhancing food safety from source to shelf (Stakheev et al., 2024). Moreover, CRISPR-mediated editing of lipoxygenase (LOX) genes and RNAi-mediated silencing of polyphenol oxidase (PPO) are now applied to reduce lipid oxidation and browning, respectively, extending the freshness and appeal of legumes, fruits, and dairy (Bhowmik et al., 2023).
2.1 CRISPR-cas system
The CRISPR-Cas system is an advanced genome-editing technology derived from bacterial adaptive immunity. It involves two primary components: CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats), which store genetic information from past viral infections, and Cas (CRISPR-associated) proteins, such as Cas9, which act as molecular scissors to precisely cut DNA at designated locations. The process begins with target recognition, where a guide RNA (gRNA) directs the Cas enzyme to a specific DNA sequence. Once the enzyme binds to the target sequence, it induces a double-strand break (DSB) in the DNA. The cell then repairs this break via non-homologous end joining (NHEJ) or homology-directed repair (HDR), leading to genetic modifications.
CRISPR technology has numerous applications in food biotechnology. It enhances crop improvement by increasing drought resistance, yield, and nutritional value (Touzdjian Pinheiro Kohlrausch Távora et al, 2022). It also contributes to disease resistance, allowing engineered plants to withstand pathogens and pests, thereby reducing reliance on pesticides (Okita et al., 2023). In terms of food safety, CRISPR is used to eliminate foodborne pathogens, enhancing food security and reducing the risk of contamination (Jinek et al., 2012). In livestock, genetic modifications improve disease resistance, growth rates, and nutritional composition, contributing to a more sustainable food supply (Velez et al., 2024). Additionally, CRISPR optimizes bioprocessing by engineering microbes to enhance fermentation efficiency in food production. Another significant application is allergen reduction, where CRISPR can silence allergenic genes in food products, decreasing allergic reactions. Furthermore, the technology extends shelf-life by delaying ripening and degradation, preserving food freshness for longer periods.
The potential of CRISPR technology in food biotechnology is immense, offering solutions to global food production challenges by promoting sustainability, reducing chemical dependency, and improving food quality. However, despite its benefits, CRISPR faces regulatory concerns, ethical considerations, and public acceptance challenges (Fire et al., 1998). The ongoing research and development in this field will determine the extent to which CRISPR can be effectively integrated into food biotechnology to address food security and agricultural sustainability.
2.2 RNA interference (RNAi)
RNA interference (RNAi) is a gene-silencing mechanism that regulates gene expression at the post-transcriptional level. It is a natural cellular process used by organisms to control gene expression and protect against viral infections. The RNAi mechanism involves two key types of small RNA molecules: small interfering RNA (siRNA) and microRNA (miRNA). These molecules guide the RNA-induced silencing complex (RISC) to target and degrade specific messenger RNA (mRNA), thereby preventing the production of particular proteins (Fire et al., 1998).
The applications of RNAi in food biotechnology are extensive. In crop improvement, RNAi has been used to develop plants with enhanced resistance to viruses, fungi, and pests, reducing the need for chemical pesticides (Zhang et al., 2020). It also plays a role in improving food quality by regulating genes involved in nutrient composition, such as reducing allergenicity in peanuts and tomatoes (Gelaye and Luo, 2025). In food safety, RNAi technology is employed to prevent the expression of genes responsible for toxin production in fungi and bacteria, mitigating food contamination risks (Velez et al., 2024). Moreover, RNAi can be used to extend the shelf-life of perishable food items by controlling genes responsible for ripening and spoilage.
RNAi technology also has significant regulatory implications. Due to its precision and ability to silence specific genes without introducing foreign DNA, RNAi-based modifications face fewer regulatory challenges compared to traditional genetic engineering methods. However, public perception and regulatory policies vary across countries, influencing the commercialization of RNAi-based food products (Velez et al., 2024). As research progresses, RNAi continues to be a promising tool for ensuring food security, sustainability, and enhanced agricultural practices.
2.3 Comparisons between CRISPR and RNAi for food safety and preservation
CRISPR and RNAi are both powerful gene-editing tools, but they function through distinct mechanisms. CRISPR directly edits DNA by introducing targeted cuts, while RNAi regulates gene expression at the mRNA level without altering the genetic sequence (Jinek et al., 2012; Fire et al., 1998). In food safety, CRISPR is more efficient for eliminating foodborne pathogens by permanently disabling genes responsible for virulence and toxin production, whereas RNAi provides a reversible gene silencing approach that can be used for temporary suppression of harmful traits (Touzdjian Pinheiro Kohlrausch Távora et al., 2022).
For food preservation, both technologies play essential roles. CRISPR can be used to extend shelf-life by modifying genes responsible for ripening and spoilage, whereas RNAi achieves similar results by silencing the expression of these genes without genetic modifications (Okita et al., 2023). Additionally, RNAi has been widely adopted in post-harvest preservation strategies, such as controlling enzymatic browning in fruits like apples and potatoes (Velez et al., 2024). In contrast, CRISPR offers a more precise and long-term solution for enhancing food longevity by permanently altering metabolic pathways involved in degradation processes. For instance, CRISPR/Cas9 has emerged as a more efficient and precise tool than RNAi, especially in climacteric fruits like tomato and banana, where targeted editing of key ethylene pathway genes such as ACS2 and ACO1 has led to significant improvements in shelf life and fruit quality (Martín-Pizarro et al., 2018; Hu et al., 2021). In contrast, RNAi continues to be widely applied in non-climacteric fruits like strawberry and grape, where ripening is often regulated by polygenic networks and partial gene silencing remains effective (Xue et al., 2020). Recent studies have also identified emerging gene targets beyond the classical ethylene pathway, including ethylene-independent regulators such as NOR-like1, SlAP2a, and F-box proteins, which offer broader applicability across fruit types (Tonutti et al., 2023). In addition, both CRISPR and RNAi have been employed to modify the expression of cell wall-modifying enzymes like polygalacturonase, pectin methylesterase, and expansin, which are crucial for fruit softening. For example, CRISPR-mediated knockout of PG2a in tomato resulted in firmer fruits and extended shelf life, while RNAi-based suppression of similar genes in papaya showed moderate improvements (Wang et al., 2019). Furthermore, CRISPR enables multiplex editing—for instance, simultaneous modification of SlAP2a and RIN in tomato has successfully enhanced both shelf life and flavor (Yuan et al., 2024). The development of transgene-free CRISPR delivery systems, such as ribonucleoprotein (RNP) complexes and virus-based vectors, offers a regulatory advantage over RNAi, positioning CRISPR as a superior platform for post-harvest trait improvement in fruits (Gao et al., 2023) (Table 3). Despite their advantages, both CRISPR and RNAi face regulatory and public acceptance challenges. CRISPR-modified foods often undergo stricter regulatory scrutiny due to permanent genetic modifications, while RNAi-based foods are generally perceived as more natural and face fewer regulatory hurdles (Touzdjian Pinheiro Kohlrausch Távora et al., 2022). However, ethical concerns and biosafety regulations remain critical aspects that need to be addressed before widespread adoption of both technologies in food biotechnology.
In conclusion, CRISPR and RNAi each offer unique advantages for food safety and preservation. CRISPR is best suited for permanent genetic modifications to enhance food security, while RNAi provides a flexible and reversible approach for temporary gene suppression. The choice between these technologies depends on the specific requirements of food biotechnology applications, regulatory considerations, and consumer acceptance.
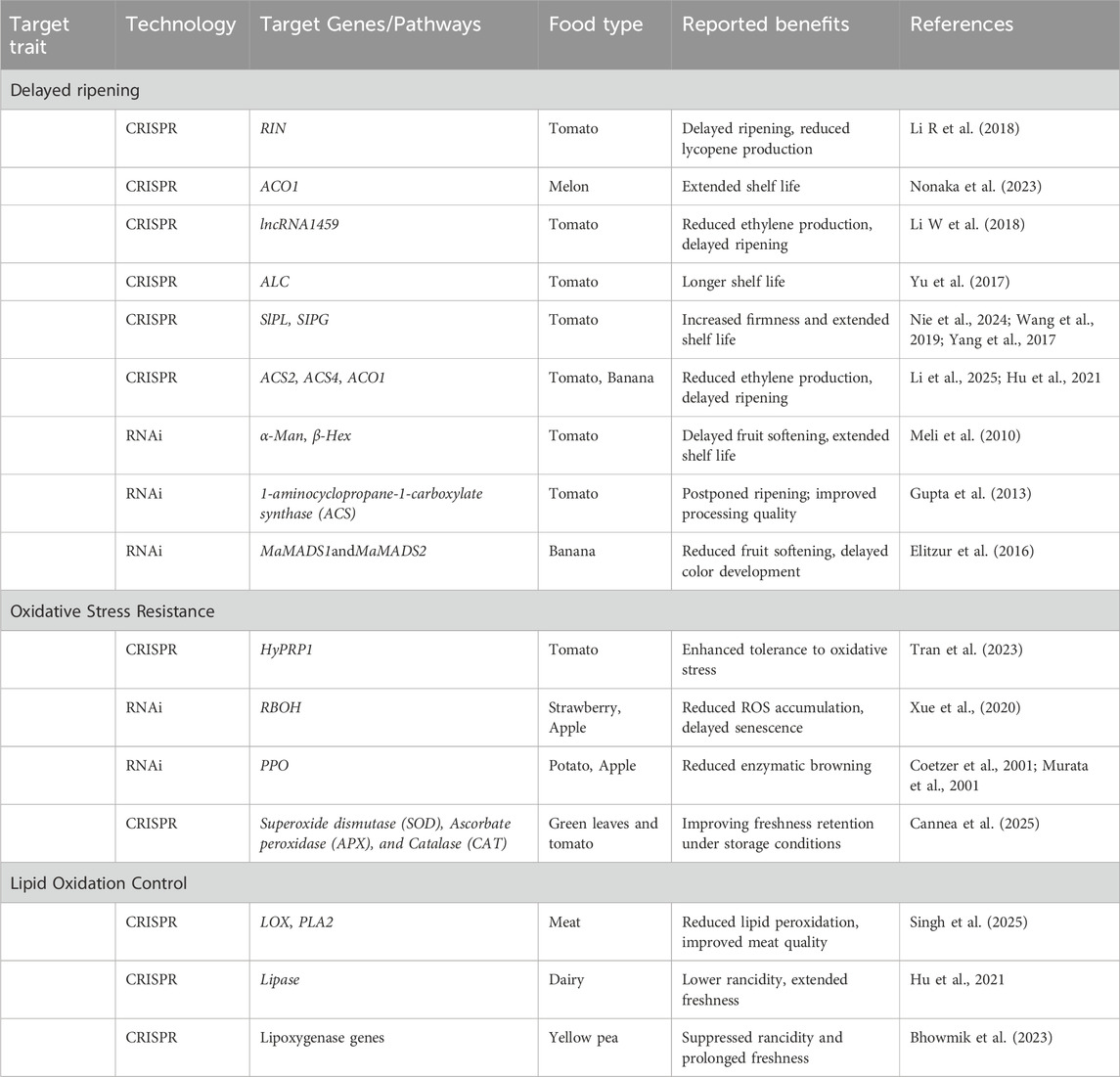
3 CRISPR and RNAi for Enhancing Food Shelf Life
The extension of shelf life in perishable food products such as fruits, vegetables, meat, and dairy is a critical goal in post-harvest biotechnology. Among various molecular tools, CRISPR-Cas-based gene editing and RNA interference (RNAi) have emerged as promising technologies for modulating physiological processes related to ripening, senescence, oxidative stress, and lipid peroxidation. These approaches offer targeted, efficient, and heritable modifications for enhancing the quality and longevity of food products (Figure 1).
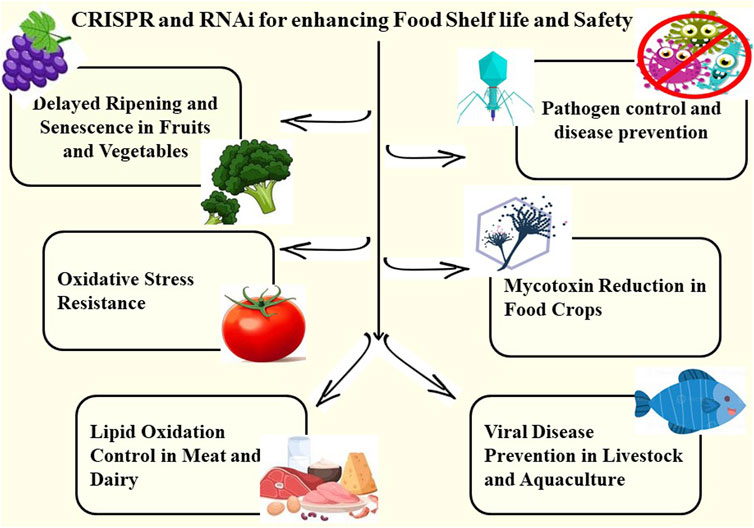
Figure 1. CRISPR and RNAi Technologies for Enhancing Food Shelf Life and EnsuringSafety. This image depicts the dual applications of CRISPR and RNA interference (RNAi) in the food industry, which aim to extend shelf life while also improving food safety. On the left, it shows how gene editing and gene silencing techniques are used to delay ripening and senescence in fruits and vegetables, improve oxidative stress resistance, and control lipid oxidation in meat and dairy products, resulting in longer freshness and less spoilage. On the right, the graphic depicts food safety applications such as the use of CRISPR and RNAi for pathogen control and disease prevention, RNA interference to reduce antimicrobial resistance, gene editing to prevent mycotoxin biosynthesis in crops, and interventions against viral infections in livestock and aquaculture.
3.1 Delayed ripening and senescence in fruits and vegetables
3.1.1 CRISPR-mediated gene editing
CRISPR-Cas9 technology has enabled precise genome modifications that can suppress or activate specific genes involved in ripening pathways. A significant area of focus has been the regulation of fruit ripening and senescence, processes tightly controlled by hormonal and genetic factors. CRISPR/Cas9-mediated genome editing has shown promising results in modifying ripening-related genes to delay post-harvest deterioration. For instance, the targeted mutagenesis of the SlPL (Solanum lycopersicum Pectate lyase), SlPG gene in tomato (S. lycopersicum) has led to enhanced fruit firmness and extended shelf life without compromising essential ripening characteristics (Nie et al., 2022; Wang et al., 2019; Yang et al., 2017). Moreover, knockout of the RIN (Ripening Inhibitor) transcription factor in tomato delayed the onset of ripening, resulting in firmer fruits with prolonged shelf life without compromising taste or nutritional value (Li S et al., 2020). Similarly, CRISPR-based editing of ethylene biosynthesis genes, such as ACS2 (1-Aminocyclopropane-1-carboxylic acid Synthase 2), ACS4 (1-Aminocyclopropane-1-carboxylic acid Synthase 4), and ACO1 (1-Aminocyclopropane-1-carboxylic acid Oxidase 1), has been shown to significantly delay the climacteric phase in tomatoes and bananas (Li et al., 2025; Sharma et al., 2024; Hu et al., 2021).
3.1.2 RNAi-based approaches
RNAi has been extensively utilized to suppress ripening-associated genes post-transcriptionally. Silencing of ACC (1-aminocyclopropane-1-carboxylate) oxidase and ACC synthase,a key enzymes in the ethylene biosynthetic pathwayhas led to delayed ethylene production and extended shelf life in tomato (Gao et al., 2007). Additionally, the downregulation of ripening-specific N-glycoprotein processing genes, such as α-mannosidase (α-Man) and β-D-N-acetylhexosaminidase (β-Hex), has resulted in reduced cell wall degradation and softer fruits with longer post-harvest storage stability (Meli et al., 2010). The enzyme 1-aminocyclopropane-1-carboxylate (ACC) oxidase catalyzes the final step in ethylene biosynthesis from its precursor ACC. RNA interference (RNAi)-mediated suppression of ACC oxidase in tomato has successfully produced transgenic lines with significantly extended shelf life (Batra et al., 2010). Similarly, suppression of three homologs of ACC synthase (ACS) during ripening led to reduced ethylene production and delayed fruit ripening in tomato (Gupta et al., 2013). Similarly, RNAi technology has been effectively employed to silence ethylene biosynthesis genes, such as those encoding 1-aminocyclopropane-1-carboxylate synthase (ACS). Silencing of ACS gene homologs in tomato has resulted in delayed ripening and improved suitability for industrial processing (Gupta et al., 2013). These interventions enable better synchronization of ripening during transportation and retail display, thereby reducing post-harvest losses. Additionally, the SISGR1 gene, which encodes the STAY GREEN protein involved in ethylene signalling and fruit color development, was suppressed in transgenic tomato lines, leading to delayed ripening and prolonged shelf life (Luo et al., 2013). In banana (Musa acuminata), RNAi-mediated silencing of two SEPALLATA3-like MADS-box genes, MaMADS1 and MaMADS2, resulted in reduced fruit softening, delayed color development, and overall enhancement of shelf life (Elitzur et al., 2016) (Table 3).
3.2 Oxidative stress resistance
Post-harvest oxidative stress, resulting from the accumulation of reactive oxygen species (ROS), significantly impacts the shelf life of fruits and vegetables. CRISPR-based modifications have targeted genes involved in antioxidant biosynthetic pathways, leading to enhanced detoxification capacity and improved oxidative stress tolerance in crops (Cannea et al., 2025). Additionally, RNAi strategies have been deployed to downregulate genes implicated in ROS production or insufficient scavenging, thereby preserving cellular integrity during storage. Such modifications extend the functional shelf life of produce and improve their visual and nutritional quality by mitigating oxidative browning, tissue softening, and metabolic degradation.
3.2.1 CRISPR-modified antioxidant pathways
Oxidative stress, resulting from reactive oxygen species (ROS) accumulation, accelerates senescence and quality degradation. CRISPR-Cas9 has been used to upregulate antioxidant enzyme activity, such as superoxide dismutase (SOD), ascorbate peroxidase (APX), and catalase (CAT), by disrupting negative regulatory elements or editing transcriptional repressors. This strategy has been applied in leafy greens and tomatoes, improving freshness retention under storage conditions (Cannea et al., 2025).
3.2.2 RNAi strategies for regulating oxidative stress-related genes
RNAi-mediated silencing of genes involved in ROS overproduction, such as respiratory burst oxidase homologs (RBOHs), has led to reduced oxidative damage and delayed senescence in fruits like strawberry and apple (Zhang et al., 2020). Moreover, targeting polyphenol oxidase (PPO) via RNAi has been effective in reducing browning in produce such as potatoes and apples, contributing to extended visual and nutritional shelf life (Coetzer et al., 2001).
3.3 Lipid oxidation control in meat and dairy
Lipid oxidation is a major determinant of quality deterioration and off-flavor development in meat and dairy products. Although the application of CRISPR and RNAi in this sector is relatively nascent, initial studies indicate promising avenues. Targeted editing or silencing of genes encoding lipoxygenases and other rancidity-related enzymes can suppress lipid peroxidation, thereby maintaining flavor integrity and extending product shelf life. This molecular approach provides an alternative to synthetic antioxidants, aligning with consumer demand for natural and minimally processed foods.
3.3.1 Targeting rancidity-related enzymes through CRISPR and RNAi
Lipid peroxidation is a primary cause of quality deterioration in meat and dairy products. Both CRISPR and RNAi have shown potential in regulating genes involved in lipid degradation, such as lipoxygenases (LOX) and phospholipase A2 (PLA2). In dairy, the application of CRISPR has targeted the lipase genes responsible for free fatty acid accumulation and off-flavor development during storage (Hu et al., 2021). Similarly, RNAi-mediated suppression of rancidity-associated enzymes in meat products has resulted in reduced oxidative spoilage and improved shelf stability under refrigeration (Singh et al., 2025). These biotechnological innovations underscore the potential of CRISPR and RNAi not only in improving shelf life but also in reducing food waste, enhancing food security, and supporting sustainable supply chains. Table 1 summarizes recent advances and their functional outcomes.
4 Application of CRISPR and RNAi in food safety
Globally, one of the most significant public concerns is food safety. Throughout the supply chain, food safety problems can arise from physical, chemical, and microbiological risks. One of the essential steps for ensuring food safety is the use of quick, precise, targeted, and field-deployable detection techniques that satisfy a variety of requirements. This will help address food safety concerns and protect consumer health. The CRISPR-Cas system and RNA interference (RNAi), two recently developed technologies, have been effectively used to biosensing and have shown great promise for developing theoretically new detection techniques with high sensitivity and specificity (Figure 2). With their unique combination of target recognition specificity, signal transduction, and effective signal amplification capabilities, the newly developed CRISPR-Cas systems and RNAi exhibit exceptional specificity and sensitivity, indicating great promise for resolving the aforementioned issues and creating next-generation methods for food safety detection (Table 3).
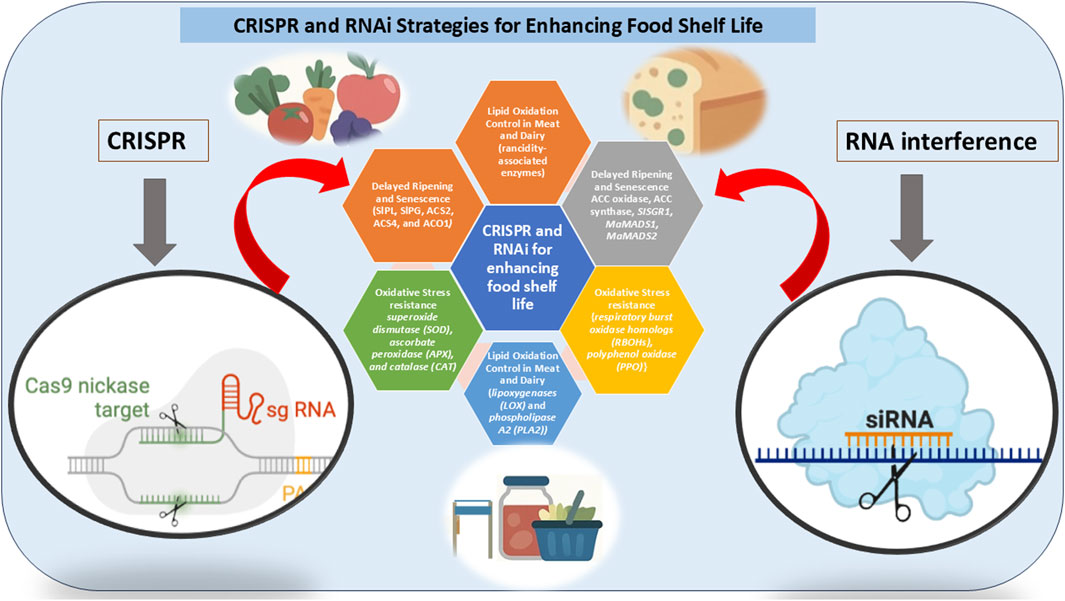
Figure 2. The diagram illustrates the comparative mechanisms of CRISPR/Cas9 and RNAi in regulating fruit ripening genes, highlighting their roles in gene editing and gene silencing, respectively. It further presents a farm-to-table pipeline, starting from target gene identification and laboratory transformation, progressing through greenhouse validation and field application, and culminating in improved post-harvest traits. The final stages depict enhanced storage, distribution, and delivery of shelf-stable fruits to consumers. This integrated view emphasizes the translational potential of both technologies in reducing post-harvest losses and improving food safety.
4.1 Pathogen control and foodborne disease prevention
Food safety events are mostly caused by foodborne microorganisms. It is crucial to detect foodborne pathogens quickly and accurately in order to protect human health and lives. There are many potentials uses for CRISPR-based technologies in the identification of foodborne pathogens. The characteristics of CRISPR-based nucleic acid detection and RNAi are high sensitivity, high specificity, modularization, programmability, ease, low cost, and fast time consumption that have made it a popular study topic in the field of food rapid detection (Alizadeh et al., 2021; Li R et al., 2018). For instance, in tomato (Solamnumlycopersicum), by using microRNA (miRNA)/artificial miRNA, the regulation of the expression of genes involved in biotic stress have been identified (Zuo et al., 2011; Jatan and Lata, 2019). Moreover, in Achyranthes bidentata the counter defence mechanism was to promote root growth and development and enhances transport activity in various stresses by integrated miRNA-mRNA (Yang et al., 2021). The enhancement of resistance against Fusarium ear rot (FER) in maize (Zea mays) has been achieved by miRNA (Zhou et al., 2020). In barley, wheat (Triticum astivum L.), and Arabidopsis enhancement of the resistance to Fusarium head blight (FHB) and Fusarium seedling blight (FSB) by host-induced gene silencing of the fungal chitin gene has been achieved with the help of RNAi technology (Cheng et al., 2015).
Additionally, using CRISPR-Cas9, the S gene and mildew resistance locus O (MLO)-resistant gene were developed along with rice ethylene response factor 922. (Das et al., 2019). Using CRISPR-Cas9, Tomelo is a nontransgenic tomato cultivar that is resistant to Oidium neolycopersici-caused powdery mildew disease (Nekrasov et al., 2017). Another example is rice resistance to the blast pathogen Magnaporthe has been created by employing CRISPR-Cas9 technology to target the OsERF922 gene (Das et al., 2019). The approach with the highest potential for controlling citrus pathogens was thought to be CRISPR-Cas combined with RNA interference (Goulin et al., 2019). Pto DC3000, a tomato variety resistant to bacterial speck disease, was developed using the CRISPR-Cas gene editing method (Ortigosa et al., 2019).
Moreover, the application of the CRISPR-Cas genome editing system has been documented in various industrially and technologically significant bacterial species, including Clostridium beijerinckii (a metabolic host for acetone and alcohol production), Lactobacillus reuteri (employed as a probiotic), and Streptomyces species (recognized as antimicrobial producers (Luo et al., 2016; Liu Z et al., 2020). Conversely, the food industry, particularly agricultural crops, has significantly profited from recent advancements in genetic engineering, which encompass enhancements in grain yield, herbicide tolerance, plant biomass, insect resistance, and even sensory and nutritional characteristics of the crops (Dong et al., 2021).
Furthermore, lactic acid-producing microbes, which are common in probiotics and starter cultures, are particularly common CRISPR-Cas systems; loci show up in 77% of bifidobacteria genomes and 62.9% of lactobacilli genomes that were examined (Briner et al., 2015; Sun et al., 2015). These bacteria’ diversity and distribution of CRISPR-Cas systems offer a historical perspective on the phage-microbial ecosystems of large-scale fermentations. Furthermore, CRISPR-Cas can be an effective technique for managing fermentation processes, with uses in genome editing, phage resistance, plasmid vaccination, strain-typing, and antibacterial activity. Numerous studies have demonstrated the potential of CRISPR-based sensors to target all food contaminants. For instance, Li J et al. (2020) developed a CRISPR-based biosensor to detect Pb2+ in the presence of other interfering cations including Ca2+, K+, Zn2+, Mn2+, Fe3+, Cd2+, Ni2+, Co2+, and Cu2+. Moreover, the liposome amplification technique in conjunction with CRISPR-Cas12a was used to distinguish between meat adulteration.
4.2 Mycotoxin reduction in food crops
Fungal diseases rank among the leading causes of agricultural losses and challenges to global food security (Almeida et al., 2019; Fones et al., 2020). Both the formation of new harmful species and the spread of existing ones are caused by climate change, transportation, and trade. Fungi belonging to the genera Fusarium, Aspergillus, Penicillium, and Alternaria can produce a variety of harmful secondary metabolites called Mycotoxins in addition to having direct effects on the amount and quality of yields. Mycotoxin contamination poses serious health risks to both humans and animals. The global issue of mycotoxin contamination of food and feed requires the use of highly effective and biologically safe methods. RNA interference (RNAi) is a naturally occurring mechanism that is crucial to many eukaryotic functions, such as controlling gene expression, preserving genomic stability, and defending against viruses, among others. The treatment of plant diseases, notably those brought on by fungi that produce mycotoxin, and food safety have recently seen a significant increase in the application of RNAi-based approaches (Stakheeve et al., 2024). For instance, although Monascus Red, derived from Monascus purpureus, is widely used as a natural food colorant, its application is restricted due to the co-production of citrinina nephrotoxic mycotoxin also synthesized by the fungus (Liu W et al., 2020). In recent years, the emergence of toxic fungi and the mycotoxins they create has been a huge agricultural disaster to provide a safe, wholesome food supply and a serious health concern (Thipe et al., 2021).
For instance, it has been established that certain fungi, such as Botrytis cinerea, create siRNAs during fungal infections. These siRNAs are then transmitted to the plant host, where they occupy its RNAi machinery, downregulate defence genes, and suppress an innate immune response. According to Rampersad (2020), host-derived sRNAs have been found to target the virulence genes of the pathogen Verticillium dahliae in order to prevent fungal invasion. Furthermore, research has demonstrated that RNA silencing signals in plants can travel both through short-range transport, which covers roughly 10–15 cells, and long-range transport, which covers the entire tissue, to neighbouring cells. Moreover, the exact process by which siRNA is transferred from pathogen to host is still unknown. Exosomes or extracellular vesicles, which start in intraluminal plant vesicles, are thought to mediate the exchange (Rutter and Innes, 2018). The finding that plant cell exosomes proliferate during an infection process provides evidence in support of this theory. The clathrin-mediated endocytosis pathway in the necrotrophic fungus Sclerotinia sclerotiorum has been identified during RNAi-based therapies (Šečić and Kogel, 2021; Wytinck et al., 2020), suggesting that endocytosis may then facilitate fungal uptake.
4.3 Viral disease prevention in livestock and aquaculture
Virus-resistant crops that express sense/antisense constructs are examples of gene-silenced GM crops that use RNA interference (RNAi) for pest management. For example, sequences of virus coat proteins from invasive viruses have been used to modify a variety of crops. For instance, cucurbits have been made resistant to the cucumber mosaic virus or zucchini yellowing mosaic virus, potatoes to the potato leaf roll virus or potato virus Y, and papaya to the papaya ringspot virus (Khalid et al., 2017). The range of marketed GM crops that impose RNA interference in pests has been expanded to include GM maize that targets Western maize rootworm larvae and expresses double-stranded RNA (dsRNA; with a hairpin loop). This insect belongs to the Coleoptera order, which is known to be more sensitive to the effects of dsRNA administered orally, in contrast to other insects, such as Lepidoptera, which may be less sensitive (Baum et al., 2007).
Aquaculture is the industry that produces food at the quickest rate in the world, and as production has increased, viral infections have surfaced, posing a problem for sustainable growth. Therefore, it is essential to control and prevent sickness. The habitat, reservoir hosts, susceptibility of farmed species, transmission dynamics, viral pathogenicity, and viral characteristics will all play a role in this. For instance, the characteristics of some viruses that have been found in recent years, including piscine orthoreovirus (PRV) (Palacios et al., 2010) and piscine myocarditis virus (PMCV) (Haugland et al., 2011), have not yet been thoroughly described. Similar to this, a new virus that has been known to cause a significant mortality rate in Israeli farmed tilapia has recently been discovered (Eyngor et al., 2014) and described as an orthomyxo-like virus that most likely belongs to a new genus within the Orthomyxoviridae family (Bacharach et al., 2016).
5 Regulatory, ethical, and consumer perspectives
The commercialisation of CRISPR and RNA interference (RNAi)-modified foods is influenced by differing global regulatory frameworks, which affect their acceptance and market availability. Countries such as the United States and Brazil have comparatively lenient rules, with gene-edited crops that do not contain foreign DNA not categorised as genetically modified organisms (GMOs) and so exempt from severe regulatory scrutiny (Waltz, 2018). In contrast, the European Union (EU) has taken a strict stance, finding that all genome-edited foods are subject to existing GMO legislation and must undergo comprehensive safety evaluations and traceability procedures before approval (Court of Justice of the European Union, 2018). Similarly, countries like China and India are building regulatory frameworks that strike a balance between biotechnological developments and biosafety concerns, with a focus on comprehensive risk assessments prior to commercialisation (Table 2).
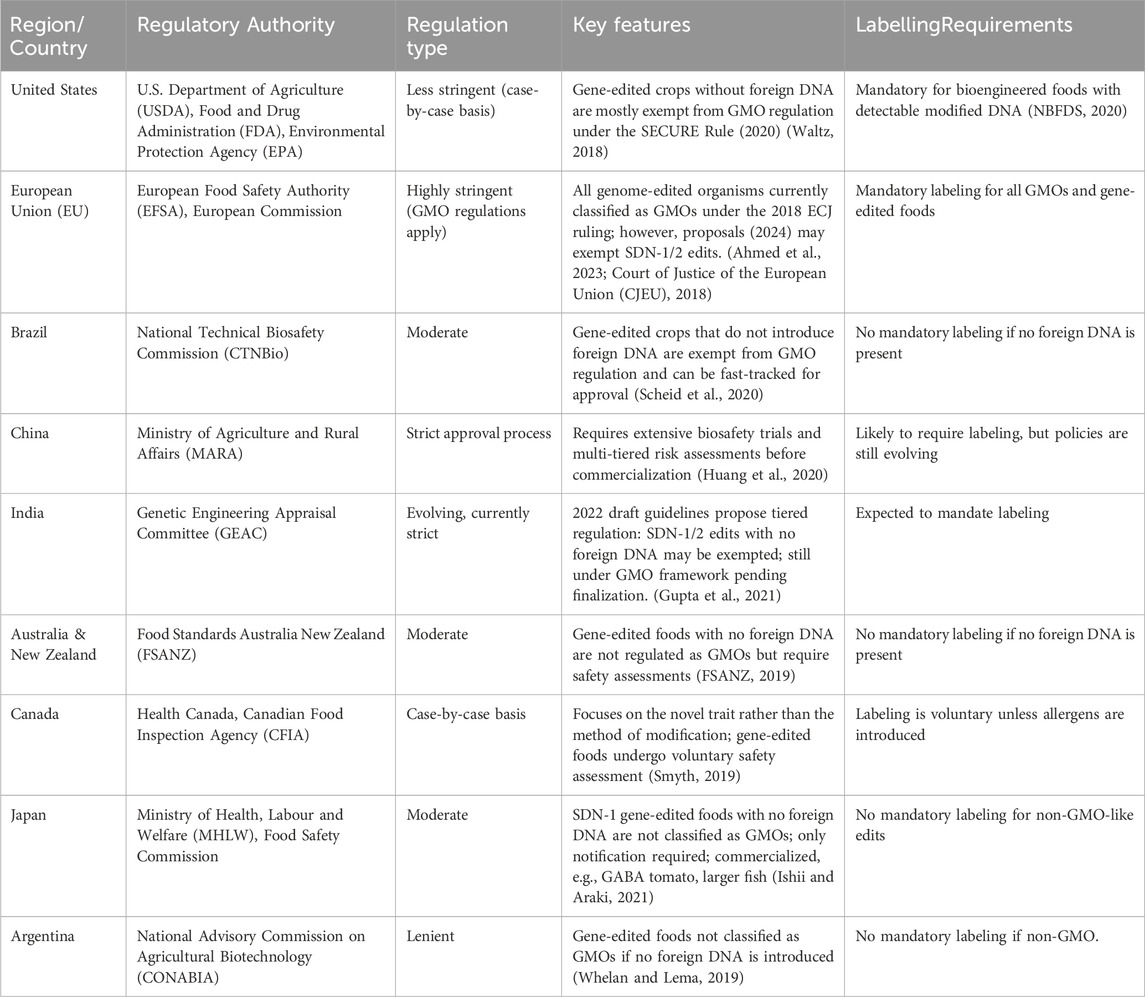
Table 2. International regulatory frameworks for foods modified by CRISPR and RNAi: Regulations, authorisations, and labelling needs.
CRISPR and RNA interference (RNAi) have slightly different regulatory approaches due to their distinct mechanisms and outcomes. CRISPR introduces permanent, heritable changes at the DNA level through targeted genome editing, making the resulting organisms subject to GMO regulations in many countries, particularly when foreign DNA is involved (Wolt et al., 2016; Podevin et al., 2013). In contrast, RNAi works by temporarily silencing gene expression at the mRNA level, resulting in non-heritable effects that are frequently transient and limited to the treated organism or generation (Jagtap et al., 2011). Regulatory agencies are more stringent with CRISPR-edited crops, particularly in the EU, where all genome-edited organisms are currently regulated as GMOs under the 2018 ECJ ruling. In the United States, however, CRISPR-edited crops that do not contain foreign DNA and mimic natural mutations may be exempt from GMO regulation under the USDA’s SECURE rule (USDA, 2020). In contrast, RNAi-based productsparticularly when used externally as biopesticidesare generally subject to less stringent regulation. For example, in the United States, RNAi-based sprays are regulated as biopesticides by the EPA (EPA, 2014), whereas non-transgenic RNAi approaches may be exempt from GMO labelling or oversight in many regions. Thus, the primary regulatory difference arises from the permanence and heritability of CRISPR modifications versus the transient, reversible nature of RNAi-based gene silencing.
Beyond regulatory concerns, ethical considerations and public perception are critical in deciding consumer acceptability of gene-edited food products. While CRISPR and RNAi provide more precise and targeted alterations than traditional genetic engineering, public worries remain about unintended genetic effects, long-term health consequences, and environmental dangers. Ethical conflicts also centre on corporate control over biotechnology, intellectual property rights, and access to genome-editing technologies, generating concerns about food sovereignty and equitable rewards for farmers and consumers (Feeney et al., 2021). Public view varies by area, with consumers in North America and parts of Asia being more accepting of gene-edited foods, whilst European consumers are sceptical, frequently demanding clear labelling and comprehensive safety evaluations.
Before CRISPR- and RNAi-modified foods reach the market, safety studies and risk management techniques must be implemented to ensure their reliability. To address potential safety concerns, regulatory agencies such as the United States Food and Drug Administration (FDA), the European Food Safety Authority (EFSA), and the Codex Alimentarius Commission emphasise the importance of comprehensive molecular characterisation, allergenicity testing, and environmental risk assessments. Furthermore, developments in bioinformatics and off-target effect studies are being used to improve risk evaluation processes, ensuring that gene-edited foods meet stringent safety requirements (Tang et al., 2019). Another key problem is the labelling and commercialisation of CRISPR and RNAi-modified food products. While clear labelling may increase customer trust and allow for more informed decisions, there is no global agreement on labelling regulations for gene-edited goods. The EU requires strict labelling for all genetically modified foods, whereas the US National Bioengineered Food Disclosure Standard (NBFDS) only requires labelling for items with detectable changed DNA (National Academies of Science, Engineering, and Medicine, 2016). The lack of standardised labelling policies impedes international trade and raises worries about gene-edited foods’ market accessibility. To successfully integrate CRISPR and RNAi technologies into the food sector, global cooperation, clear communication, and comprehensive safety assessments will be required (Table 3).
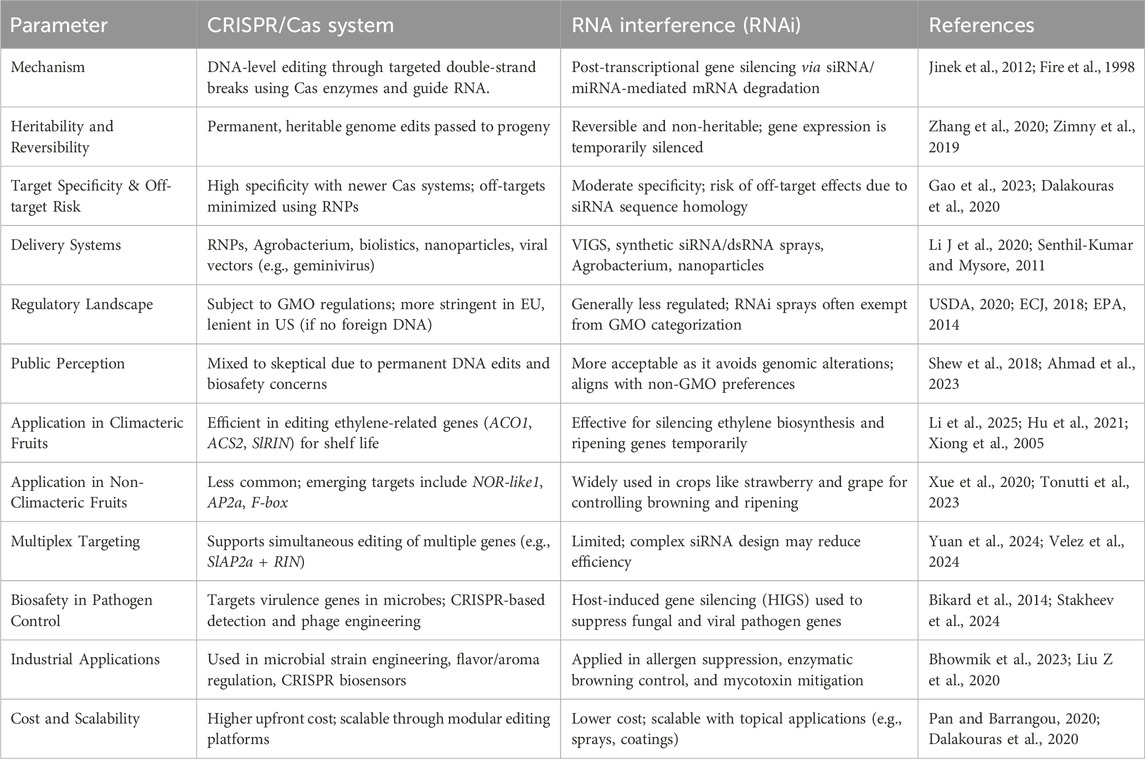
Table 3. Comparative analysis of CRISPR and RNAi technologies in food shelf-life and safety applications.
6 Future prospects and challenges
In the imminent future, it will be feasible to produce novel foods with enhanced characteristics on an industrial scale (Pan and Barrangou, 2020). The safe management of food-associated microbes, a subject of extensive research, is the primary concern in the food industry, focusing on the regulation of spoilage bacteria, pathogens, and beneficial microbes such as probiotics and starter cultures. Due to its origins in food microbiology, CRISPR-Cas research has exploded in order to examine its ability to execute specific DNA sequences and its potential uses in genome editing (Ahmad A et al., 2021). The advantages of genome-edited crops for human health and agriculture, such as preventing the spread of diseases, eradicating invasive species that harm the environment and agriculture, and combating herbicide and pesticide-resistant plants, must be weighed against the ethical and ecological issues surrounding their use (Chapman et al., 2017). Researchers can ascertain the DNA sequence of foodborne pathogens, investigate their genes and activities, and perform a worldwide analysis of their gene expression thanks to the ongoing improvement of detection techniques and intervention strategies (Taniguchi et al., 2021). Foodborne pathogen gene editing and extremely specific detection are made possible by CRISPR-based technology. The detection and identification of food safety issues has undergone a fresh revolution thanks to omics-based and CRISPR-based technology. Despite their immense potential, the widespread application of CRISPR and RNAi in food biotechnology remains constrained by regulatory discrepancies, ethical concerns, and varied consumer perceptions across regions. Moving forward, harmonized global regulations, robust safety evaluations, transparent communication, and inclusive public engagement will be essential to foster the responsible adoption of these technologies. As research advances and public awareness grows, CRISPR and RNAi are poised to play a pivotal role in revolutionizing food sustainability, reducing post-harvest losses, and ensuring global food security. The CRISPR-Cas system for food and agricultural engineering will grow even more in the near future, as evidenced by the simultaneous description of newly found CRISPR-Cas techniques and the creation of innovative gene editing tools.
7 Conclusion
CRISPR and RNA interference (RNAi) represent transformative biotechnological tools in addressing critical challenges related to food preservation and safety. Their precise gene-editing and gene-silencing capabilities have been successfully leveraged to delay ripening, enhance oxidative stress resistance, and control lipid oxidation in perishable food products, thereby significantly extending shelf life. Additionally, these technologies offer novel approaches to mitigate foodborne pathogens, reduce mycotoxin contamination, and improve disease resistance in both crops and livestock, promoting a safer and more resilient food system. The development of functional foods that satisfy consumer expectations may be made possible by the CRISPR-Cas technology, which can already predict how DNA can be engineered to address issues related to the agriculture and food industries at all levels of food manufacturing, from farm to fork (Stout et al., 2017). Given the relatively unexplored reservoir of novel and distinct CRISPR-Cas arrays found in the human and food microbiomes, CRISPR-Cas systems may be a good tool for use in food-grade systems. Furthermore, because diverse bacterial species are present in all aspects of food production and consumption, the CRISPR-Cas system may be able to regulate every type of bacterium found in food, including fermentative, probiotic, pathogenic, and spoilage bacteria. By using these technologies, the food sector can create next-generation food cultures and modify various microbial populations, among other benefits. Numerous CRISPR-Cas applications have been enhanced for use in food science, including genome editing of food-grade bacterial strains, identification of closely related bacterial strains, protection of starter cultures from phages, modulation of specific strains, and vaccination of starter cultures against plasmid uptake. The flexible programmability of CRISPR in relation to targeted killing enables food scientists to combat foodborne bacteria. Even while CRISPR-Cas technology offers remarkable benefits for real-time detection, there are still technological obstacles in the way of its transition from cutting-edge to useful technology. Employing CRISPR technology in food packaging applications requires answers to two important questions: economic viability and societal acceptability. In light of these concerns, it is imperative that the next-generation of food packaging cut waste in food and packaging materials and its negative environmental effects (such as pollution, greenhouse gas emissions, and resource consumption) by 2050.
Author contributions
AS: Conceptualization, Supervision, Writing – review and editing, Writing – original draft, Visualization. MS: Writing – original draft, Conceptualization, Supervision. PS: Writing – original draft, Data curation. SS: Writing - review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
MS would like to acknowledge the award of DST-WISE fellowship to pursue post-doctoral studies.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmad A, A., Ghouri, M. Z., Munawar, N., Ismail, M., Ashraf, S., and Aftab, S. O. (2021). “Regulatory, ethical, and social aspects of CRISPR crops,” in CRISPR crops: the future of food security, 261–287.
Ahmad, A., Jamil, A., and Munawar, N. (2023). GMOs or non-GMOs? The CRISPR conundrum. Front. Plant Sci. 9, 1232938. doi:10.3389/fpls.2023.1232938
Ahmad S, S., Tang, L., Shahzad, R., Mawia, A. M., Rao, G. S., Jamil, S., et al. (2021). CRISPR-Based crop improvements: a way forward to achieve zero hunger. J. Agric. Food Chem. 69 (30), 8307–8323. doi:10.1021/acs.jafc.1c02653
Alizadeh, A. M., Hashempour-Baltork, F., Khaneghah, A. M., and Hosseini, H. (2021). New perspective approaches in controlling fungi and mycotoxins in food using emerging and green technologies. Curr. Opin. food Sci. 39, 7–15. doi:10.1016/j.cofs.2020.12.006
Almeida, F., Rodrigues, M. L., and Coelho, C. (2019). The still underestimated problem of fungal diseases worldwide. Front. Microbiol. 10, 214. doi:10.3389/fmicb.2019.00214
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576 (7785), 149–157. doi:10.1038/s41586-019-1711-4
Bacharach, E., Mishra, N., Briese, T., Zody, M. C., KembouTsofack, J. E., Zamostiano, R., et al. (2016). Characterization of a novel orthomyxo-like virus causing mass die-offs of tilapia. MBio 7 (2), e00431–e00416. doi:10.1128/mBio.00431-16
Barrangou, R., and Doudna, J. A. (2016). Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 34 (9), 933–941. doi:10.1038/nbt.3659
Batra, A., Sane, V. A., Trivedi, P. K., Sane, A. P., and Nath, P. (2010). Suppression of ACC oxidase expression in tomato using heterologous gene from banana prolongs shelf-life both on vine and post-harvest. Curr. Sci. 99, 1243–1250.
Batra, K., Singh, T. P., Sharma, M., Batra, R., and Schvaneveldt, N. (2020). Investigating the psychological impact of COVID-19 among healthcare workers: a meta-analysis. Int. J. Environ. Res. public health 17 (23), 9096. doi:10.3390/ijerph17239096
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25 (11), 1322–1326. doi:10.1038/nbt1359
Bhowmik, P., Yan, W., Hodgins, C., Polley, B., Warkentin, T., Nickerson, M., et al. (2023). CRISPR/Cas9-mediated lipoxygenase gene-editing in yellow pea leads to major changes in fatty acid and flavor profiles. Front. Plant Sci. 14, 1246905. doi:10.3389/fpls.2023.1246905
Bikard, D., Euler, C. W., Jiang, W., Nussenzweig, P. M., Goldberg, G. W., Duportet, X., et al. (2014). Exploiting CRISPR-cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 32 (11), 1146–1150. doi:10.1038/nbt.3043
Briner, A. E., Lugli, G. A., Milani, C., Duranti, S., Turroni, F., Gueimonde, M., et al. (2015). Occurrence and diversity of CRISPR-cas systems in the genus Bifidobacterium. PloS one 10 (7), e0133661. doi:10.1371/journal.pone.0133661
Cannea, F. B., and Padiglia, A. (2025). Antioxidant defense systems in plants: mechanisms, regulation, and biotechnological strategies for enhanced oxidative stress tolerance. Life 15 (8), 1293. doi:10.3390/life15081293
Chapman, J. E., Gillum, D., and Kiani, S. (2017). Approaches to reduce CRISPR off-target effects for safer genome editing. Appl. Biosaf. 22 (1), 7–13. doi:10.1177/1535676017694148
Cheng, W., Song, X. S., Li, H. P., Cao, L. H., Sun, K., Qiu, X. L., et al. (2015). Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to F usarium head blight and seedling blight in wheat. Plant Biotechnol. J. 13 (9), 1335–1345. doi:10.1111/pbi.12352
Court of Justice of the European Union (CJEU) (2018). Judgment in case C-528/16. Luxembourg: Court of Justice of the European Union.
Coetzer, C., Corsini, D., Love, S., Pavek, J., and N, T. (2001). Control of enzymatic browning in potato (Solanum tuberosum L.) by sense and antisense RNA from tomato polyphenol oxidase. J. Agric. Food Chem. 49 (2), 652–657. doi:10.1021/jf001217f
Dalakouras, A., Wassenegger, M., Dadami, E., Ganopoulos, I., Pappas, M. L., and Papadopoulou, K. (2020). Genetically modified organism-free RNA interference: exogenous application of RNA molecules in plants. Plant Physiol. 182, 38–50. doi:10.1104/pp.19.00570
Das, A., Sharma, N., and Prasad, M. (2019). CRISPR/Cas9: a novel weapon in the arsenal to combat plant diseases. Front. Plant Sci. 9, 2008. doi:10.3389/fpls.2018.02008
Dong, H., Huang, Y., and Wang, K. (2021). The development of herbicide resistance crop plants using CRISPR/Cas9-mediated gene editing. Genes 12 (6), 912. doi:10.3390/genes12060912
Elitzur, T., Vrebalov, J., Giovannoni, J. J., Goldschmidt, E. E., Friedman, H., Khayat, E., et al. (2016). Banana MaMADS transcription factors are necessary for fruit ripening and molecular tools to promote shelf-life and food security. Plant Physiol. 171 (1), 380–391. doi:10.1104/pp.15.01866
EPA (2014). White paper on RNAi technology as a pesticide: problem formulation for human health and ecological risk assessment. Washington, DC, United States: U.S. Environmental Protection Agency.
European Union (2018). Organisms obtained by mutagenesis are GMOs and are, in principle, subject to the obligations laid down by the GMO Directive. Available online at: https://curia.europa.eu/jcms/upload/docs/application/pdf/2018-07/cp180111en.pdf.
Eyngor, M., Zamostiano, R., KembouTsofack, J. E., Berkowitz, A., Bercovier, H., Tinman, S., et al. (2014). Identification of a novel RNA virus lethal to tilapia. J. Clin. Microbiol. 52 (12), 4137–4146. doi:10.1128/JCM.00827-14
FAO (2019). The State of food and agriculture 2019: moving forward on food loss and waste reduction. Food Agric. Organ. U. N. doi:10.4060/CA6030EN
Feeney, O., Cockbain, J., and Sterckx, S. (2021). Ethics, patents and genome editing: a critical assessment of three options of technology governance. Front. Political Sci. 3, 731505. doi:10.3389/fpos.2021.731505
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditiselegans. Nature 391 (6669), 806–811. doi:10.1038/35888
Fones, H. N., Bebber, D. P., Chaloner, T. M., Kay, W. T., Steinberg, G., and Gurr, S. J. (2020). Threats to global food security from emerging fungal and oomycete crop pathogens. Nat. Food 1 (6), 332–342. doi:10.1038/s43016-020-0075-0
Food Standards Australia New Zealand (FSANZ) (2019). Regulation of genetically modified and genome-edited foods. FSANZ Rep. doi:10.1007/s11248-019-00159-w
Gao, H. Y., Zhu, B. Z., Zhu, H. L., Zhang, Y. L., Xie, Y. H., Li, Y. C., et al. (2007). Effect of suppression of ethylene biosynthesis on flavor products in tomato fruits. Russ. J. Plant Physiol. 54, 80–88. doi:10.1134/s1021443707010128
Gao, C., et al. (2023). CRISPR–Cas delivery strategies for sustainable agriculture. Nat. Rev. Mol. Cell Biol. 24, 131–146.
Gelaye, Y., and Luo, H. (2025). Application of epigenetics for allergen-free peanut production: a comprehensive review. Epigenetics Insights 18 (1), 0. doi:10.48130/epi-0025-0006
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature 551 (7681), 464–471. doi:10.1038/nature24644
Goulin, E. H., Galdeano, D. M., Granato, L. M., Matsumura, E. E., Dalio, R. J. D., and Machado, M. A. (2019). RNA interference and CRISPR: promising approaches to better understand and control citrus pathogens. Microbiol. Res. 226, 1–9. doi:10.1016/j.micres.2019.03.006
Gootenberg, J. S., Abudayyeh, O. O., Kellner, M. J., Joung, J., Collins, J. J., and Zhang, F. (2018). Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360 (6387), 439–444. doi:10.1126/science.aaq0179
Gupta, A., Pal, R. K., and Rajam, M. V. (2013). Delayed ripening and improved fruit processing quality in tomato by RNAi-mediated silencing of three homologs of 1-aminopropane-1-carboxylate synthase gene. J. Plant Physiol. 170, 987–995. doi:10.1016/j.jplph.2013.02.003
Gupta, M., Dey, A., and Verma, P. (2021). Genome editing regulations in India: current status and future perspectives. Front. Plant Sci. 12, 643981. doi:10.3389/fpls.2021.643981
Gustavsson, J., Cederberg, C., Sonesson, U., Van Otterdijk, R., and Meybeck, A. (2011). Global food losses and food waste: extent, causes, and prevention. Rome, Italy: FAO Report.
Haugland, Ø., Mikalsen, A. B., Nilsen, P., Lindmo, K., Thu, B. J., Eliassen, T. M., et al. (2011). Cardiomyopathy syndrome of Atlantic salmon (Salmo salar L.) is caused by a double-stranded RNA virus of the Totiviridae family. J. virology 85 (11), 5275–5286. doi:10.1128/JVI.02154-10
Hu, C., Sheng, O., Deng, G., He, W., Dong, T., Yang, Q., et al. (2021). CRISPR/Cas9-mediated genome editing of MaACO1 (aminocyclopropane-1-carboxylate oxidase 1) promotes the shelf life of banana fruit. Plant Biotechnol. J. 19 (4), 654–656. doi:10.1111/pbi.13534
Huang, S., Weigel, D., Beachy, R. N., and Li, J. (2020). A proposed regulatory framework for genome-edited crops. Nat. Genet. 52 (8), 791–797. doi:10.1038/s41588-020-0694-1
Ishii, T., Araki, M., and Sharma, A. K. (2021). Toxicity of boric acid, borax and other boron containing compounds: a review. Regul. Toxicol. Pharmacol. 121, 104873. doi:10.1016/j.yrtph.2021.104873
Jagtap, U. B., Gurav, R. G., and Bapat, V. A. (2011). Role of RNA interference in plant improvement. Naturwissenschaften 98, 473–492. doi:10.1007/s00114-011-0798-8
Jatan, R., and Lata, C. H. A. R. U. (2019). Role of microRNAs in abiotic and biotic stress resistance in plants. Proc. Indian Natl. Sci. Acad. 85, 553–567. doi:10.16943/PTINSA/2019/49586
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337 (6096), 816–821. doi:10.1126/science.1225829
Khalid, A., Zhang, Q., Yasir, M., and Li, F. (2017). Small RNA based genetic engineering for plant viral resistance: application in crop protection. Front. Microbiol. 8, 43. doi:10.3389/fmicb.2017.00043
Kim, S. M., Ryu, M. Y., and Kim, J. S. (2021). CRISPR as a promising tool to combat antimicrobial resistance. Front. Cell. Infect. Microbiol. 11, 685348. doi:10.3389/fcimb.2021.685348
Li R, R., Fu, D., Zhu, B., Luo, Y., and Zhu, H. (2018). CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. 94 (3), 513–524. doi:10.1111/tpj.13872
Li W, W., Wu, S., Fu, P., Liu, J., Han, H., Bai, L., et al. (2018). National molecular tracing network for foodborne disease surveillance in China. Food control. 88, 28–32. doi:10.1016/j.foodcont.2017.12.032
Li, Y., Man, S., Ye, S., Liu, G., and Ma, L. (2022). CRISPR-Cas-based detection for food safety problems: current status, challenges, and opportunities. Compr. Rev. Food Sci. Food Saf. 21 (4), 3770–3798. doi:10.1111/1541-4337.13000
Li, J., Lu, Y., Cheng, K., Zhu, G., Wang, X., Lin, T., et al. (2025). ACS4 exerts a pivotal role in ethylene biosynthesis during the ripening of tomato fruits in comparison to ACS2. Plant J. 121 (5), e70043. doi:10.1111/tpj.70043
Li J, J., Yang, S., Zuo, C., Dai, L., Guo, Y., and Xie, G. (2020). Applying CRISPR-Cas12a as a signal amplifier to construct biosensors for non-DNA targets in ultralow concentrations. Acs Sensors 5 (4), 970–977. doi:10.1021/acssensors.9b02305
Li S, S., Zhu, B., Pirrello, J., Xu, C., Zhang, B., Bouzayen, M., et al. (2020). Roles of RIN and ethylene in tomato fruit ripening and ripening-associated traits. New Phytol. 226 (2), 460–475. doi:10.1111/nph.16362
Liu Z, Z., Dong, H., Cui, Y., Cong, L., and Zhang, D. (2020). Application of different types of CRISPR/Cas-based systems in bacteria. Microb. cell factories 19, 172–14. doi:10.1186/s12934-020-01431-z
Liu W, W., An, C., Shu, X., Meng, X., Yao, Y., Zhang, J., et al. (2020). A dual-plasmid CRISPR/Cas system for mycotoxin elimination in polykaryotic industrial fungi. ACS Synth. Biol. 9 (8), 2087–2095. doi:10.1021/acssynbio.0c00178
Luo, Y., Ma, B., Zeng, Q., and Zhang, Y. (2013). Suppression of SlSGR1 gene delays fruit ripening and affects ethylene signaling in tomato. Plant Cell Rep. 32 (6), 913–923. doi:10.1007/s00299-013-1417-5
Luo, M. L., Leenay, R. T., and Beisel, C. L. (2016). Current and future prospects for CRISPR-based tools in bacteria. Biotechnol. Bioeng. 113 (5), 930–943. doi:10.1002/bit.25851
Martín-Pizarro, C., and Posé, D. (2018). Genome editing as a tool for fruit ripening manipulation. Front. Plant Sci. 9, 1415. doi:10.3389/fpls.2018.01415
Meli, V. S., Ghosh, S., Prabha, T. N., Chakraborty, N., Chakraborty, S., and Datta, A. (2010). Enhancement of fruit shelf life by suppressing N-glycan processing enzymes. Proc. Natl. Acad. Sci. 107 (6), 2413–2418. doi:10.1073/pnas.0909329107
Miao, C., Xiao, L., Hua, K., Zou, C., Zhao, Y., Bressan, R. A., et al. (2019). Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. 116 (23), 11246–11255. doi:10.1073/pnas.1901769116
Murata, M., Nishimura, M., Murai, N., Haruta, M., Homma, S., and Itoh, Y. (2001). A transgenic apple callus showing reduced polyphenol oxidase activity and lower browning potential. BiosciBiotechnolBiochem 65 (2), 383–388. doi:10.1271/bbb.65.383
National Academies of Sciences, Engineering, and Medicine (2016). Genetically engineered crops: experiences and prospects. Washington, DC, United States: National Academies Press. doi:10.17226/23395
National Bioengineered Food Disclosure Standard (NBFDS) (2020). Mandatory disclosure of bioengineered foods. U.S. Department of Agriculture (USDA) Agricultural Marketing Service. Available online at: https://www.ams.usda.gov/rules-regulations/national-bioengineered-food-disclosure-standard.
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., and Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7 (1), 482. doi:10.1038/s41598-017-00578-x
Nie, H., Shi, Y., Geng, X., and Xing, G. (2022). CRISRP/Cas9-MediatedTargeted mutagenesis of TomatoPolygalacturonase gene (SlPG)Delays fruit softening. Front. Plant Sci. 13, 729128. doi:10.3389/fpls.2022.729128
Nie, H., Yang, X., Zheng, S., and Hou, L. (2024). Gene-Based developments in improving quality of tomato: focus on firmness, Shelf life, and pre-and post-harvest stress adaptations. Horticulturae 10 (6), 641. doi:10.3390/horticulturae10060641
Nonaka, S., Ito, M., and Ezura, H. (2023). Targeted modification of CmACO1 by CRISPR/Cas9 extends the shelf-life of Cucumis melo var. reticulatus melon. Front. Genome Ed. 5, 1176125. doi:10.3389/fgeed.2023.1176125
Ortigosa, A., Gimenez-Ibanez, S., Leonhardt, N., and Solano, R. (2019). Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of Sl JAZ 2. Plant Biotechnol. J. 17 (3), 665–673. doi:10.1111/pbi.13006
Okita, T. W., and Delseny, M. (2023). Genome editing in plants: new advances and applications in plant biology and agriculture. Plant Sci. 328, 111577. doi:10.1016/j.plantsci.2022.111577
Palacios, G., Lovoll, M., Tengs, T., Hornig, M., Hutchison, S., Hui, J., et al. (2010). Heart and skeletal muscle inflammation of farmed salmon is associated with infection with a novel reovirus. PLoS one 5 (7), e11487. doi:10.1371/journal.pone.0011487
Pan, M., and Barrangou, R. (2020). Combining omics technologies with CRISPR-based genome editing to study food microbes. Curr. Opin. Biotechnol. 61, 198–208. doi:10.1016/j.copbio.2019.12.027
Piergentili, R., Del Rio, A., Signore, F., Umani Ronchi, F., Marinelli, E., and Zaami, S. (2021). CRISPR-Cas and its wide-ranging applications: from human genome editing to environmental implications, technical limitations, hazards and bioethical issues. Cells 10 (5), 969. doi:10.3390/cells10050969
Podevin, N., Davies, H. V., Hartung, F., Nogue, F., and Casacuberta, J. M. (2013). Site-directed nucleases: a paradigm shift in predictable, knowledge-based plant breeding. Trends Biotechnol. 31 (6), 375–383. doi:10.1016/j.tibtech.2013.03.004
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152 (5), 1173–1183. doi:10.1016/j.cell.2013.02.022
Rutter, B. D., and Innes, R. W. (2018). Extracellular vesicles as key mediators of plant–microbe interactions. Curr. Opin. plant Biol. 44, 16–22. doi:10.1016/j.pbi.2018.01.008
Sharma, M., Sidhu, A. K., Samota, M. K., Shah, P., Pandey, M. K., and Gangurde, S. S. (2024). Technological advancements in the CRISPR toolbox for improving plant salt tolerance. Discov. Agric. 2 (1), 102. doi:10.1007/s44279-024-00105-3
Scheid, J. F., de Figueiredo, M. C., and de Barros, I. C. (2020). The regulatory landscape for genome-edited crops in Brazil. Plant Cell Rep. 39 (4), 379–388. doi:10.1007/s00299-020-02535-5
Šečić, E., and Kogel, K. H. (2021). Requirements for fungal uptake of dsRNA and gene silencing in RNAi-based crop protection strategies. Curr. Opin. Biotechnol. 70, 136–142. doi:10.1016/j.copbio.2021.04.001
Singh, P. K., Agrawal, N., and Yadav, S. (2025). “Meat products with modified fatty acid profile,” in Healthier meat products (Cham: Springer Nature Switzerland), 227–246.
Senthil-Kumar, M., and Mysore, K. S. (2011). Virus-induced gene silencing can persist for more than 2 years and also be transmitted to progeny seedlings in Nicotiana benthamiana and tomato. Plant Biotechnol. J. 9 (7), 797–806. doi:10.1111/j.1467-7652.2011.00589.x
Shew, A. M., Nalley, L. L., Snell, H. A., Nayga, R. M., and Dixon, B. L. (2018). CRISPR versus GMOs: public acceptance and valuation. Glob. Food Secur. 19, 71–80. doi:10.1016/j.gfs.2018.10.005
Shin, J., Jiang, W., and Liu, J. (2018). Genome editing for food safety. Trends Biotechnol. 36 (5), 442–456. doi:10.1016/j.tibtech.2018.01.008
Smyth, S. J. (2019). Canadian regulatory perspectives on genome edited crops. New Biotechnol. 49, 45–50. doi:10.1016/j.nbt.2018.12.002
Stakheev, A. A., Taliansky, M., Kalinina, N. O., and Zavriev, S. K. (2024). RNAi-Based approaches to control mycotoxin producers: challenges and perspectives. J. Fungi 10 (10), 682. doi:10.3390/jof10100682
Stout, E., Klaenhammer, T., and &Barrangou, R. (2017). CRISPR-Cas technologies and applications in food bacteria. Annu. Rev. food Sci. Technol. 8 (1), 413–437. doi:10.1146/annurev-food-072816-024723
Sun, Z., Harris, H. M., McCann, A., Guo, C., Argimón, S., Zhang, W., et al. (2015). Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 6 (1), 8322. doi:10.1038/ncomms9322
Tang, X., Lowder, L. G., Zhang, T., Malzahn, A. A., Zheng, X., Voytas, D. F., et al. (2019). A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 5 (7), 778–788. doi:10.1038/s41477-019-0430-1
Taniguchi, T., Ohki, M., Urata, A., Ohshiro, S., Tarigan, E., Kiatsomphob, S., et al. (2021). Detection and identification of adhesins involved in adhesion of Campylobacter jejuni to chicken skin. Int. J. Food Microbiol. 337, 108929. doi:10.1016/j.ijfoodmicro.2020.108929
Thipe, V. C., Maloney, V., Klein, A., Gokul, A., Keyster, M., and Katti, K. V. (2021). “RNA interference and CRISPR/Cas9 techniques for controlling mycotoxins,” in CRISPR and RNAi systems (Elsevier), 463–490.
Tonutti, P., Brizzolara, S., and Beckles, D. M. (2023). Reducing crop losses by gene-editing control of organ developmental physiology. Curr. Opin. Biotechnol. 81, 102925. doi:10.1016/j.copbio.2023.102925
Touzdjian Pinheiro Kohlrausch Távora, F., de Assis dos Santos Diniz, F., de Moraes Rêgo-Machado, C., Chagas Freitas, N., Barbosa Monteiro Arraes, F., Chumbinho de Andrade, E., et al. (2022). CRISPR/Cas-and topical RNAi-based technologies for crop management and improvement: reviewing the risk assessment and challenges towards a more sustainable agriculture. Front. Bioeng. Biotechnol. 10, 913728. doi:10.3389/fbioe.2022.913728
Tran, M. T., Son, G. H., Song, Y. J., Nguyen, N. T., Park, S., Thach, T. V., et al. (2023). CRISPR-Cas9-based precise engineering of SlHyPRP1 protein towards multi-stress tolerance in tomato. Front. Plant Sci. 14, 1186932. doi:10.3389/fpls.2023.1186932
USDA (2020). SECURE rule: United States Department of Agriculture. Washington, DC, United States: U.S. Department of Agriculture.
USDA (2020). “SECURE rule: sustainable, ecological, consistent, uniform, responsible,” in Efficient. Washington, DC, United States: United States Department of Agriculture – APHIS.
Vermeulen, S., Campbell, B. M., and Ingram, J. S. (2012). Climate change and food systems. Annu. Rev. Environ. Resour. 37, 195–222. doi:10.1146/annurev-environ-020411-130608
Velez, A., Darlington, M., Jurat-Fuentes, J., Kogel, K. H., Smagghe, G., and Whyard, S. (2024). RNA interference in agriculture: methods, applications, and governance. Counc. Agric. Sci. Technol. Available online at: https://doi.org/10.62300/IRNE9191.
Waltz, E. (2018). With a free pass, CRISPR-edited plants reach market in record time. Nat. Biotechnol. 36 (1), 6–7. doi:10.1038/nbt0118-6b
Wang, D., Samsulrizal, N. H., Yan, C., Allcock, N. S., Craigon, J., Blanco-Ulate, B., et al. (2019). Characterization of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol. 179 (2), 544–557. doi:10.1104/pp.18.01187
Whelan, A. I., and Lema, M. A. (2019). Regulatory framework for genome-edited crops in Argentina. Transgenic Res. 28 (2), 129–135. doi:10.1007/s11248-019-00133-3
Wolt, J. D., Wang, K., and Yang, B. (2016). The regulatory status of genome-edited crops. Plant Biotechnol. J. 14 (2), 510–518. doi:10.1111/pbi.12444
Wytinck, N., Sullivan, D. S., Biggar, K. T., Crisostomo, L., Pelka, P., Belmonte, M. F., et al. (2020). Clathrin mediated endocytosis is involved in the uptake of exogenous double-stranded RNA in the white mold phytopathogen Sclerotinia sclerotiorum. Sci. Rep. 10 (1), 12773. doi:10.1038/s41598-020-69771-9
Xie, X., Yin, X., and Liu, X. (2006). RNAi-mediated silencing of ACC synthase gene results in altered ripening in tomato. J. Plant Physiology Mol. Biol. 32 (1), 46–52.
Xin, Y., Guo, T., and Qiao, M. (2025). Current application and future prospects of CRISPR-Cas in lactic acid Bacteria: a review. Food Res. Int. 209, 116315. doi:10.1016/j.foodres.2025.116315
Xiong, C., Zhu, H., Bai, Y., Liu, Y., and Liu, Y. (2005). Suppression of ACC oxidase gene expression extends shelf life of tomato fruit. J. Agric. Biotechnol. 13 (5), 625–628.
Xue, C., Guan, S. C., Chen, J. Q., Wen, C. J., Cai, J. F., and Chen, X. (2020). Genome-wide identification and functional characterization of strawberry pectin methylesterases related to fruit softening. BMC Plant Biol. 20, 13. doi:10.1186/s12870-019-2225-9
Yang, L., Huang, W., Xiong, F., Xian, Z., Su, D., Ren, M., et al. (2017). Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol. J. 15 (12), 1544–1555. doi:10.1111/pbi.12737
Yang, Y. H., Li, M. J., Yi, Y. J., Li, R. F., Li, C. X., Yang, H., et al. (2021). Integrated miRNA-mRNA analysis reveals the roles of miRNAs in the replanting benefit of Achyranthes bidentata roots. Sci. Rep. 11 (1), 1628. doi:10.1038/s41598-021-81277-6
Yu, Q. H., Wang, B., Li, N., Tang, Y., Yang, S., Yang, T., et al. (2017). CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci. Rep. 7 (1), 11874. doi:10.1038/s41598-017-12262-1
Yuan, L., Gai, W., Xuan, X., Ahiakpa, J. K., Li, F., Ge, P., et al. (2024). Advances in improving tomato fruit quality by gene editing. Hortic. Plant J. doi:10.1016/j.hpj.2024.04.008
Zhang, D., Hussain, M., and Yan, L. (2020). Advances in CRISPR and RNAi technologies for improving food quality and safety. Trends Biotechnol. 38 (10), 1234–1245.
Zhou, Z., Cao, Y., Li, T., Wang, X., Chen, J., He, H., et al. (2020). MicroRNAs are involved in maize immunity against Fusarium verticillioides ear rot. Genomics, Proteomics and Bioinforma. 18 (3), 241–255. doi:10.1016/j.gpb.2019.11.006
Zimny, T., Sowa, S., and Racovita, M. (2019). Genome editing applications in plant breeding and agriculture. Biotechnol. Adv. 37 (1), 107–121.
Keywords: CRISPR, RNA interference, food preservation, food safety, gene editing, biotechnology
Citation: Sidhu AK, Sharma M, Shah P and Sandhu SK (2025) CRISPR and RNA interference: revolutionary tools for extending food shelf life and ensuring safety. Front. Food Sci. Technol. 5:1609948. doi: 10.3389/frfst.2025.1609948
Received: 11 April 2025; Accepted: 25 August 2025;
Published: 24 October 2025.
Edited by:
Miguel Rebollo-Hernanz, Universidad Autónoma de Madrid, SpainReviewed by:
Guoliang Yuan, Pacific Northwest National Laboratory (DOE), United StatesWadzani Palnam Dauda, Federal University Gashua Yobe, Nigeria
Copyright © 2025 Sidhu, Sharma, Shah and Sandhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Madhvi Sharma, bWFkaHZpc2hhcm1hNDEzQGdtYWlsLmNvbQ==
 Amanpreet K. Sidhu
Amanpreet K. Sidhu Madhvi Sharma
Madhvi Sharma Priya Shah
Priya Shah Simrandeep Kaur Sandhu3
Simrandeep Kaur Sandhu3