- College of Life Science, Capital Normal University, Beijing, China
NUCLEAR FACTOR Y transcription factors belong to a multimember family and consist of NF-YA/B/C subunits. Members of the NF-Y family have been reported to regulate physiological processes in plant. In this study, we identified and annotated two NF-YA, nine NF-B, and twelve NF-YC proteins in the genome of Physcomitrella patens. Analyses of conserved domains demonstrated that PpNF-YA/B/C shared the same conserved domains with their orthologous proteins in Arabidopsis, O. sativa and mouse. Expression profiles indicated that PpNF-Ys were widely expressed in different tissues and developmental stages of P. patens throughout protonema and gametophores. The majority of PpNF-Y genes were responsive to abiotic stress via either ABA-independent or -dependent pathways. Some of ABA-regulated PpNF-Y expression were mediated by ABI3. To our knowledge, this study was the first to evaluate NF-Y families in Physcomitrella patens, and provides a foundation to dissect the function of PpNF-Ys.
Introduction
NUCLEAR FACTOR Y (NF-Y) transcription factors (TFs), also known as Heme-associated proteins (HAPs) and CCAAT box binding factors (CBFs), are important regulators of numerous plant developmental processes and stress-induced responses. The NF-Y transcription factor is consists of three subunits: NF-YA (also known as CBF-B/HAP2), NF-YB (CBF-A/HAP3), and NF-YC (CBF-C/HAP5) (Mantovani, 1999). Each subunit of the NF-Y complex is encoded by a gene family in angiosperms (Stephenson et al., 2007; Siefers et al., 2009). For example, the A. thaliana genome encodes 36 NF-Ys paralogs (Siefers et al., 2009) and O. sativa harbors at least 30 NF-Y genes (Thirumurugan et al., 2008). At present, the NF-Ys families have been studied in animals and flowering plants (Matuoka and Chen, 2002; Dolfini et al., 2012; Rípodas et al., 2014), they were identified to play important roles in controlling florescence (Kumimoto et al., 2010; Li et al., 2011), nodule formation (Combier et al., 2006), ABA (abscisic acid)/blue light responses (Chen et al., 2007; Nelson et al., 2007; Li et al., 2008; Liu and Howell, 2010; Liang et al., 2014), chloroplast biosynthesis (Miyoshi et al., 2003) and stress tolerance (Warpeha et al., 2007; Yamamoto et al., 2009).
Increasing evidence shows that NF-Y subunits are involved in abiotic stress responses in monocots and dicots. Microarray data showed that accumulation of TaNF-YA1 reduced statistically in response to drought stress in wheat leaves (Stephenson et al., 2007). The nf-ya5 mutants are failed to bear drought stress, whereas, overexpression of this gene enhances drought tolerance (Li et al., 2008). In soybean (G. max), overexpression of GmNFYA3 was found to reduce leaf water loss and enhanced drought tolerance (Ni et al., 2013). AtNFYA1-RNAi lines were insensitive to salt stress (Li et al., 2013).
There is also research on the NF-YB family. For example, AtNF-YB2 was found to be up-regulated by osmotic stress (Chen et al., 2007). In barley (H. vulgar), HvNF-YB3 transcript levels were induced by drought and HvNF-YB1 and HvNF-YB4 showed a significant decrease in transcription level after application of ABA. HvNF-YB5, however, showed a great increase in transcription level in response to high salt stress (Liang et al., 2012). LEC1 (LEAFY COTYLEDON1, also NF-YB9) and L1L (LEAFY COTYLEDON1-LIKE, also NF-YB6) are involved in A. thaliana embryo development and response to ABA (Warpeha et al., 2007). Similar, to AtNF-YA5 (Li et al., 2008), overexpression of AtNF-YB1, a non-LEC1 member of the NF-YB family, increased plant tolerance to drought stress (Nelson et al., 2007). Overexpression of ZmNF-YB2 not only improved the drought resistance, but also increased production in withholding water conditions (Nelson et al., 2007).
NF-YCs were responsive to abiotic stress. AtNF-YC2 could be statistically increased in response to many environmental stimuli, such as light, heat, and drought stress (Hackenberg et al., 2012). Another study found that NF-YC1, -YC5, -YC2, and -YC3 greatly increased under dry, salt and ABA treatments in B. napus (Liang et al., 2014). Even though there are not any functional studies of these genes so far, it is likely that this gene family should also be associated with drought and/or ABA treatments in angiosperms.
The moss Physcomitrella patens is an emerging land plant model with a fully sequenced genome (Rensing et al., 2008). Its unique phylogenetic position, between green algae and angiosperms, enables the study of the evolution of gene function in plants (Rensing et al., 2008) and it has been widely used to study defense mechanisms against abiotic stress (Frank et al., 2005; Cuming et al., 2007; Qudeimat et al., 2008; Komatsu et al., 2013). It has been reported that phytohormone ABA is required in both vascular plants and non-vascular bryophytes to protect vegetative tissue during water stress by activating downstream gene transcription through transcription factors, such as ABSCISICACID INSENSITIVE 3 (ABI3) (Khandelwal et al., 2010). So far, six NF-YB genes had were cloned in P. patens (Xie et al., 2008) and the expression of PpNF-YAs, -YB1-5, and -YC1-5 in response to ABA in protonemal tissues was examined (Yotsui et al., 2013). Also, PpNF-YC1 and -YB4 act synergistically with ABI3A to promote transcriptional levels of PpLEA1 with/without ABA treatment (Yotsui et al., 2013). However, the functions of other NF-Y genes have not been systematically investigated in P. patens, especially in response to abiotic stress, such as salt, osmotic and desiccation stress. Unlike protonema, mature gametophyte has more complicated structure, including rhizoid, buds, stems, and leaves, and is more similar to the structure of higher land plants. Therefore, we selected mature gametophyte tissues as materials to exploit the functions of those PpNF-Y genes in response to abiotic stress.
In this study, we identified two NF-YAs, nine NF-YBs and twelve NF-YCs in P. patens using sequence information from A. thaliana, and analyzed the evolutionary relationship of these genes with their ortholog in O. sativa and A. thaliana. Subsequently, we systematically determined the expression profile of each NF-Y gene in different developmental stages and under abiotic stress, including NaCl, mannitol, and desiccation treatments. Also, our results suggest that most PpNF-Y members are regulated by abiotic stress in an ABI3-mediated ABA-dependent or ABA-independent manner.
Materials and Methods
Identification PpNF-Y Family Members in P. patens
Thirty A. thaliana NF-Y CDS sequences were retrieved from the A. thaliana TAIR database (http://www.arabidopsis.org). The BLASTN program was used with an E-value cut-off of 3.8e−25 to identify predicted PpNF-Y sequences using P. patens database (http://www.phytozome.net/search.php?show=blast&method=Org_Ppatens_v1.6). The primers of each PpNF-Y cDNA sequence were designed with Primer Premier 5.0 (Table S1). And the PCR products obtained from cDNA amplification using PrimeSTAR HS DNA polymerase (TaKaRa, R010A, Japan) and followed by sequencing.
Phylogenetic Trees and Conservation Domains Analyses
The phylogenetic trees of KfNF-Y, PpNF-Y, AtNF-Y, and OsHAP were constructed by the Neighbor-Joining method using conserved and full-length amino acid sequences, respectively (MEGA; version 5.05), and the parameters were p-distance model and 1000 bootstrap replicates. Multiple sequence alignments were implemented using the conserved and full-length amino acid sequences, respectively by Clustal_X software. The sequences of KfNF-Ys, AtNF-Ys and OsNF-Ys were gained from NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi), TAIR database (http://www.arabidopsis.org/) and Orygenes_DB (http://orygenesdb.cirad.fr/), respectively.
Plant Material and Growth Conditions
The Gransder Wood strains of P. patens was used as the wild type (Rensing et al., 2008). The triple deletion mutants of the PpABI3 (abi3a/b/c) was described previously (Khandelwal et al., 2010). Cultures were grown in 9 cm Petri dish on BCDAT medium with 0.7% Agar (Sigma-Aldrich, A7002, USA), which corresponds to the minimal medium described by Sakakibara et al. (2003). The culture conditions were as follows: 16 h/8 h light/dark cycles, 80 μmol s−1 m−2 photosynthetically active radiation, 23°C constant temperature. Tissues from distinct stages were harvested, including protonemata tissue which is compose of chloronema and caulonema cells. Initial cells divided to generate buds, making the transition from protonema to juvenile gametophores (1-week old). Juvenile gametophores is compose of young stem, leafy and rhizoid (2-week old). Mature and late stage gametophores were represented by 3-, 4-, and 8-week old plant, respectively. Sporophytes that produce haploid spores (s) (Arazi, 2012).
Stress Treatment
Gametophores tissues grown for 4 weeks on cellophane-layered BCD medium without AT, then treated with 10 μM ABA (Sigma-Aldrich, A4906, USA), 400 mM NaCl (Sigma-Aldrich, S7653, USA), 600 mM mannitol (Sigma-Aldrich, M9546, USA), and were transferred to an empty Petri dish and desiccated inside a laminar flow hood. Then these tissues were collected in 0/2/4/8/12/24 h(s).
Total RNA Isolation and cDNA Synthesis
Total RNA was extracted from various tissues using the RNeasy plant mini Kit (QIAGEN, 74106, USA). RNA samples were treated by RNase-Free DNase I (Thermo, EN0525, USA). 1 μg of total RNA was used for the synthesis of first-strand cDNA using oligo (dT) as a primer and SuperScript III Reverse Transcriptase (Invitrogen, 18080-044, USA) according to the manufacturers' protocols.
Quantitative PCR
qRT-PCR was performed to examine mRNA levels of PpNF-Y genes in different developmental stages and various tissues using the 2−ΔΔCT formula described in Applied Biosystems User Bulletin No. 2. Relative mRNA levels of target genes were normalized to Tublin and Actin5. Real-time PCR was performed on an Applied CFX96 Touch™ realtime PCR system using the SYBR®Premix ExTaqTM Kit (TaKaRa, RR820L, Japan). Each 20.0 μl qRT-PCR reaction included 10.0 μl SYBR® Premix Ex Taq TM (2 ×), 0.5 μl Forward Primer (10 μM), 0.5 μl Reverse Primer (10 μM), and 0.2 μl cDNA. The reaction was initial denaturation at 95°C for 30 s, followed by 42 cycles of 95°C for 5 s and 60°C for 30 s with a final dissociation stage. The data were the mean value from three biological replicates, and every biological replicate was the average value from three technical repeats. Primers for qRT-PCR reactions are reported in Table S1. As a statistical analysis, Student's t-test were used with two-tailed distribution.
Results
Identification of P. patens NF-Y Genes
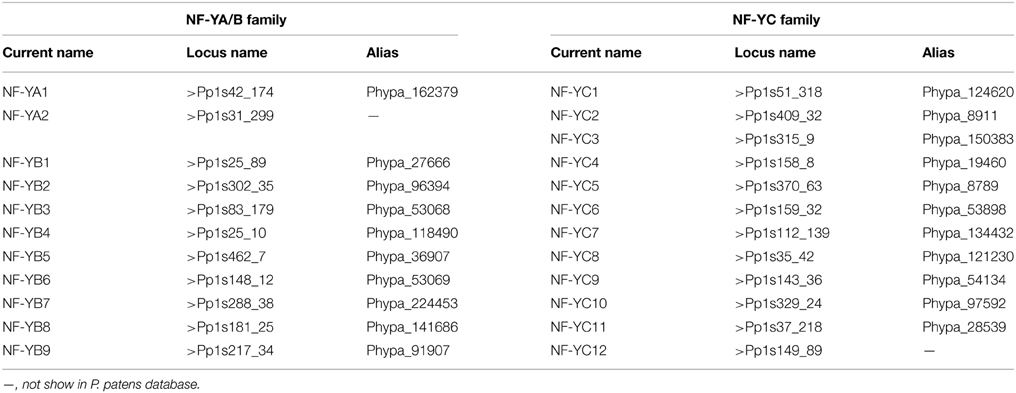
In order to identify P. patens analogs of NF-Y proteins, amino acid sequences of A. thaliana NF-Ys (Siefers et al., 2009; Petroni et al., 2012) were used to search against the P. patens database (http://www.phytozome.net/search.php?show=blast&method=Org_Ppatens_v1.6). The sequences with high similarity were acquired as homologs. Two tentative consensus (TC) sequences similar to AtNF-YAs, nine to AtNF-YBs, and twelve to AtNF-YCs were obtained and their full length cDNA were cloned and confirmed by sequencing. According to nomenclature of NF-Ys in A. thaliana, T. aestivum and B. distachyon, the genes were named as follows (Table 1).
Phylogenetic Classification of PpNF-Ys
The comprehensive phylogenetic analysis was performed to understand the evolutionary status of PpNF-Ys. As showed in the phylogenetic tree (Figure 1A), Pp/Os/AtNF-YAs could be divided into three discrete groups (I–III) comprised of 16, 5, and 1 proteins, respectively. Both two PpNF-YAs subunits were clustered into major group I, and were counterparts with OsNF-YAs. The Pp/Os/AtNF-YB subunits were clustered into four distinct clades (I–IV) consisted of 13, 4, 1, and 15 proteins, respectively (Figure 1B). All PpNF-YBs were clustered into group I or IV, while group II includes AtNF-YB9 (LEAFY COTYLEDON1, AtLEC1), AtNF-YB6 (LEC1-LIKE, L1L), OsNF-YB7, and OsNF-YB9, which agrees with previous work (Siefers et al., 2009; Petroni et al., 2012). Compared to the Pp/Os/AtNF-YB subunits, the Pp/Os/AtNF-YC subunits could be grouped into three clusters (cluster I–III), comprised of 21, 7, and 4 proteins, respectively (Figure 1C). Just as in the PpNF-YA subunits, -YB and -YC also clustered as a closer homolog with A. thaliana than OsNF-Y counterparts. The results only minimally change using full-length amino acid sequences (Figures S1A–C).
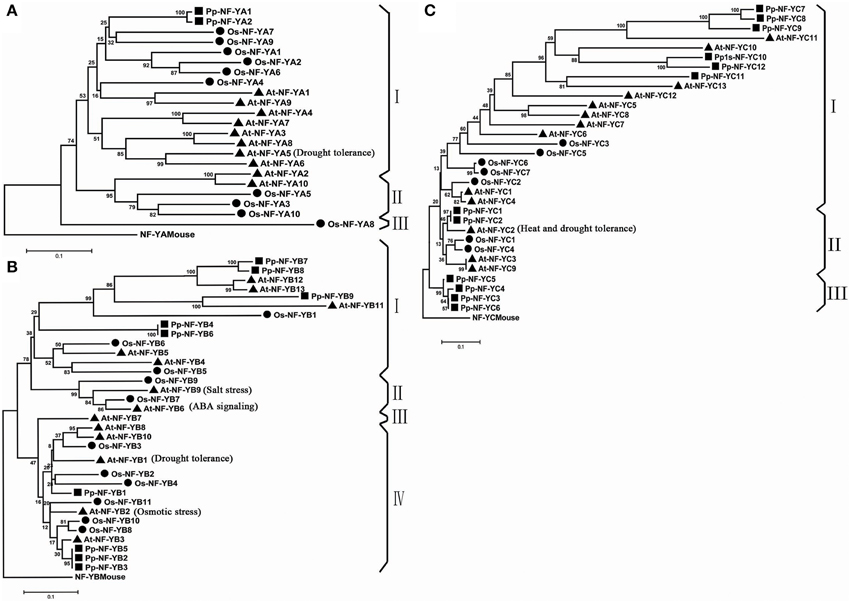
Figure 1. Phylogenetic tree of NF-YA/B/C subunits. (A) Phylogenetic tree of NF-YA families. (B) Phylogenetic tree of NF-YB families. (C) Phylogenetic tree of NF-YC families.
Identification of Domain Conservation
The length of three PpNF-Y subunit families was from 153 aa to 377 aa. Each member of the NF-Y family contains heterodimerization domain and DNA binding domain at the CCAAT site. In order to explore the evolution of the conserved domains in PpNF-Ys, multiple sequence alignment for NF-Ys was applied using full-length protein sequences. The conserved core region of PpNF-YA, -YB, and -YC were ~53, 90, and 77 amino acids in length (Figures 2A–C), respectively, which was similar to Maity and de Crombrugghe (Maity and de Crombrugghe, 1992). The PpNF-YA domain consisted of two subdomains, an A1 domain for the NF-YB and -YC interaction and an A2 domain for DNA binding (Xing et al., 1993, 1994; Mantovani et al., 1994). Three histidine (H) (red box) and an arginine (R) residues (green box) of NF-YA proteins are certainly conserved, and are requisite for DNA binding in mammalian and S. cerevisiae (Xing et al., 1993). These residues are also conserved in PpNF-YAs (Figure 2A). PpNF-YB and -YC contained both DNA binding domain and the protein interaction domain (Sinha et al., 1996; Romier et al., 2003). A gray box indicates that the lysine (K) is conserved in non-LEC1-type NF-YB, whereas it changed into an aspartate (D) in a LEC1-type NF-YB. The HFM (histone-fold motif) of the core histone H2B (Figure 2B light blue box) (Mantovani, 1999), which is made up of three alpha-helices and two beta strand-loop domains (solid blue rectangles and black lines were represented on the top of the alignment, respectively) (Arents and Moudrianakis, 1993), was observed (light blue box) in PpNF-YBs. However, it bore slight differences, such as the 66–75 residues in the conserved core in PpNF-YB7, -YB8, and -YB9 (Figure 2B). PpNF-YC were also characterized by a core histone, but were more similar to the HFM of the core histone H2A (light blue box) than H2B (Mantovani, 1999) (Figure 2C).
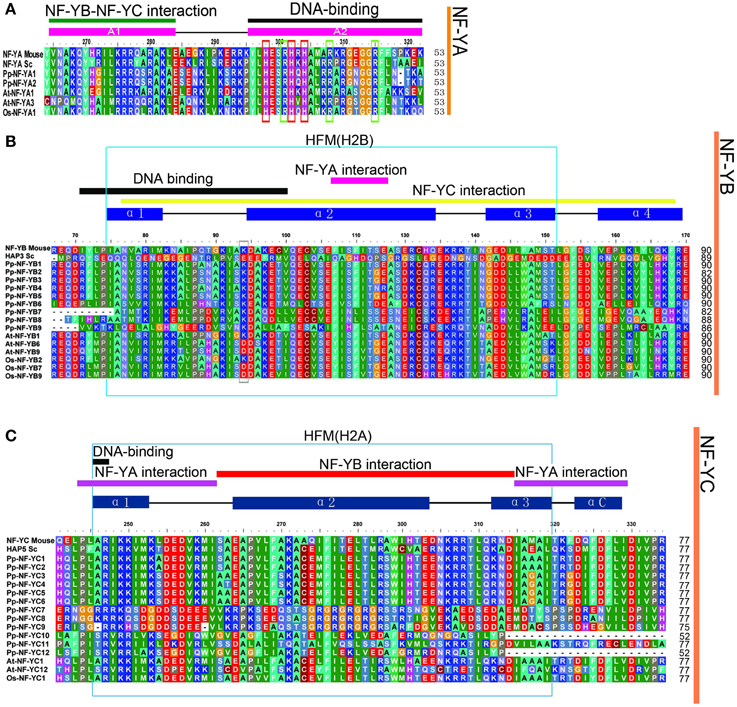
Figure 2. Multiple sequences alignment of the conversed domains of NF-Ys from P. patens, S. cerevisiae, O. sativa, and A. thaliana. The functions of referred genes were written in parentheses. (A) PpNF-YA family alignment. (B) BnNF-YB family alignment. (C) PpNF-YC family alignment.
Temporal and Spatial Expression Patterns of PpNF-Y Genes
NF-Y subunit family members are reported to be involved in developmental processes. We examined the developmental- and tissue-specific expression patterns of PpNF-Y genes using qRT-PCR. Plants of different developmental stages were harvested: protonema stage, transition stage from protonema to juvenile gametophores (1-week old), juvenile gametophores (2-week old), mature gametophores (3-, 4-, and 8-week old) and sporophytes (s) (Arazi, 2012). The PpNF-YAs, -YB4, -YB8, -YC3, -YC4, -YC6, -YC7, -YC9, and -YC11 were constitutively expressed at all developmental stages (Figure 3A, Figure S6A). PpNF-YB2, -YB5, -YB7, -YB9, -YC1, and -YC10 genes were expressed prominently in protonema (Figures 3B,C, Figures S6B,C). Accumulation of PpNF-YC5 and -YC12 genes were increased significantly at the transition stage, and was further induced in 2-week old gametophores (Figure 3C, Figure S6C). mRNA level of PpNF-YB1, -YB6, -YC5, and -YC8 expression was highest in adult gametophores (Figures 3B,C, Figures S6B,C) and started to drop in 8-week old gametophore. The expression of -YC2 was higher in protonema and young gametophores, and lower in adult gametophores. However, its transcriptional level increased dramatically in mature gametophores (Figures 3B,C, Figures S6B,C). Also, the expressions of PpNF-YB6 and -YC5 were increased strongly in sporophytes (Figures 3B,C, Figures S6B,C).
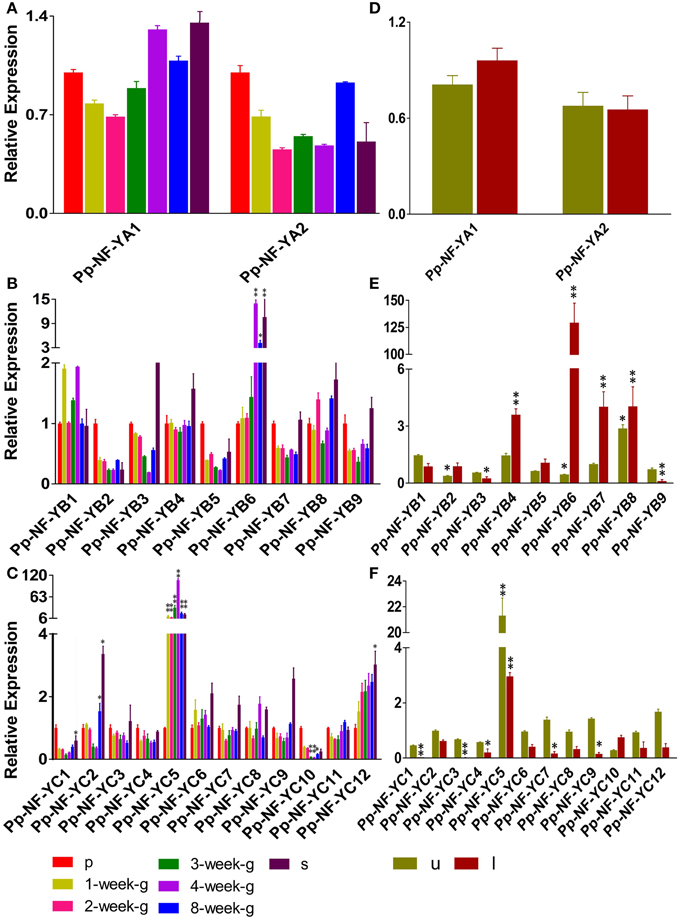
Figure 3. Temporal and spatial expression patterns of PpNF-Ys genes. Total RNA was extracted from protonema (p), from 1-week-/2-week-/3-week-/4-week-/8-week-old gametophores (g), sporophytes (s), and from the upper-part (u), including shoot tip, and lower-part (l), including rhizoids, of 1-week-old gametophores. (A,D) Relative transcription abundance of PpNF-YA. (B,E) PpNF-YB. (C,F) PpNF-YC. The expression level of each gene was normalized against that of the protonema. Tublin was the housekeeping gene. Error bars indicate SD from three biological replicates. Significant differences were analyzed by t-test (*P ≤ 0.05 and **P ≤ 0.01).
The promoter::GUS assay of AtNF-Ys showed that each member was detected in a variety of tissues, such as roots, rosette leaves and flowers (Siefers et al., 2009). We wanted to determine whether PpNF-Ys exhibited tissue-specific expression patterns as well. We obtained specific tissues of 1-month-old gametophores including the upper-part (u) with the shoot tip, and the lower-part (l) with rhizoids as described previously (Xu et al., 2014a). Accumulation of two PpNF-YA genes was not different between the upper and lower part of the gametophore (Figure 3D, Figure S6D). PpNF-YB2, -YB4, -YB5, -YB6, -YB7, and -YB8 were expressed at higher levels in lower-parts of gametophores compared to upper parts, and -YB6 mRNA level in the lower-part was 125 times higher than that in the upper part. However, PpNF-YB1, -YB3, and -YB9 expressions exhibited the opposite pattern (Figure 3E, Figure S6E). Most of the PpNF-YCs, except for -YC10, were expressed at lower levels in the lower-part of gametophores compare to that in the upper-part, and -YC1 and -YC3 transcriptional levels were too low to be examined in the lower-part (Figure 3F, Figure S6F).
Expression of P. patens NF-Ys Transcription in Response to Different Stress
Increasing evidence have indicated that At/TaNF-Y genes play important roles in response to abiotic stress (Nelson et al., 2007; Stephenson et al., 2007; Li et al., 2008). Accordingly, in order to understand whether PpNF-Ys could respond to abiotic stress, we determined the expression of PpNF-Y genes under different stress treatments, including salt, desiccation, osmotic stress, and applied exogenous ABA. Only genes showed high level (8 fold and up) of transcriptional alteration before and after treatments were presented in Figure 4, otherwise in the Supplemental Data.
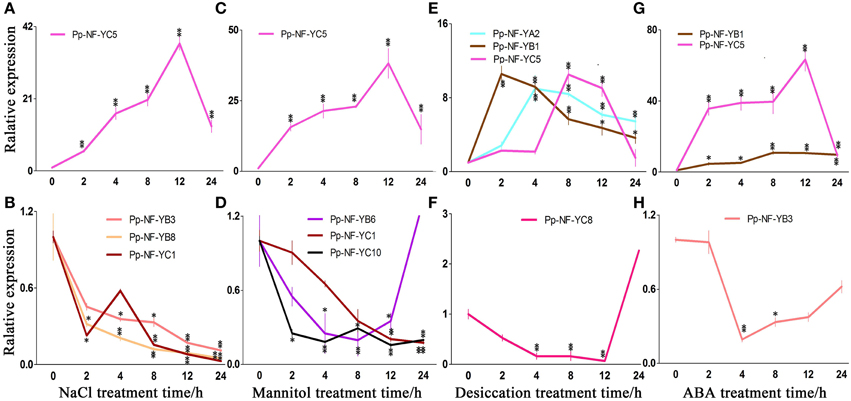
Figure 4. Typical expression patterns of PpNF-Ys in wild type plants in response to abiotic stress. (A,B) PpNF-Ys by salt stress. (C,D) PpNF-Ys by mannitol treatment. (E,F) PpNF-Ys by desiccation stress. (G,H) PpNF-Ys by ABA treatment. The expression level of each gene was normalized against that of the untreated control. Tublin was the housekeeping gene. Error bars indicate SD from three biological replicates. Significant differences were analyzed by t-test (*P ≤ 0.05 and **P ≤ 0.01).
The relative accumulation of PpNF-YB1, -YB4, -YB6, and -YC5 up-regulated after 2 h of NaCl treatment and remained induced up to 12 h of treatment, -YC5 exhibited a 40-fold of increase after 12 h of treatment (Figure 4A, Figures S2B,C, S7B,C). By contrast, PpNF-YB3, -YB8, and -YC10 reduced significantly with NaCl treatment (Figure 4B, Figures S2B,C, S7B,C). Transcriptional levels of PpNF-YB2, -YB5, -YC3, -YC4, -YC6, and -YC7 were initially inhibited during 2 or 4 h under salt stress, whereas -YAs, -YC8, -YC9, -YC11, and -YC12 up-regulated slightly (Figures S2A–C, S7A–C).
In response to osmotic stress, the transcriptional level of PpNF-YB1 and -YB4 was induced to more than 6-fold of the level observed in the untreated control at 12 and 24 h of treatment (Figure 4C, Figures S3B,C, S8B,C). PpNF-YC5 transcription up-regulated significantly when treated for 12 h and its mRNA level was 48-fold higher compared to the control (Figure 4C, Figures S3C, S8C). Accumulation of PpNF-YB8, -YC1, -YC2, and -YC10 continuously declined, while -YB6 reduced until 24 h (Figure 4D, Figures S3B,C, S8B,C). Osmotic stress-responsive PpNF-YB2, -YB3, -YB7, -YB9 -YC6, and -YC11 showed a modest decrease after 8 h of mannitol treatment, while -YC4, -YC7, -YC8, -YC9, and -YC12 increased (Figures S3B,C, S8B,C). The expression of PpNF-YC3 and -YAs remained constant under osmotic stress (Figures S3A,C, S8A,C).
Part of the physiological effects caused by desiccation is due to osmotic stress, but responses to these two kinds of stress are not quite the same. We also identified many PpNF-Ys that responded to desiccation. The expression of PpNF-YB1 and -YC10 was significantly induced compared to the control after 2 h, while the accumulation of PpNF-YA2, -YB5, -YC4, and -YC6 was induced after 4 h, of -YB6 and -YC5 after 8 h and of -YC7 after 12 h (Figure 4E, Figures S4A–C, S9A–C). The expression of PpNF-YC8 decreased until 12 h by desiccation (Figure 4F, Figures S4C, S9C). Other desiccation-responsive PpNF-YA1, -YB3, -YB4, -YC1,-YC2, and -YC3 only increased slightly, whereas -YC11 decreased upon desiccation (Figures S4A–C, S9A–C). PpNF-YB2, -YB7, -YB8, -YB9, and -YC9 transcripts were mostly unaffected by desiccation stress (Figures S4B,C, S9B,C).
Regulation of NF-Y Transcription by ABA
Plants respond to abiotic stress through ABA-dependent and ABA-independent manner. To identify whether PpNF-Ys function via the ABA pathway, the expression profiles of all PpNF-Ys under ABA treatments were further examined. The relative accumulation of PpNF-YB1 and -YC5 was dramatically induced after 2 h of ABA treatment, especially -YC5, which exhibited a 60-fold of increase (Figure 4G, Figures S5B,C, S10B,C). In contrast, the accumulation of PpNF-YB3, -YC1, and -YC2 decreased after 2 h and then recovered to untreated levels at 24 h of ABA treatment (Figure 4H, Figures S5B,C, S10B,C). The transcriptional level of PpNF-YB5, -YB6, -YB8, -YB9, -YC3, -YC4, -YC6, -YC7, and -YC8 was induced slightly, but others did not respond to ABA treatment (Figures S5A–C, S10A–C).
P. patens NF-Y Members Function Via the ABI3-Dependent Pathway
ABI3 is an essential component in the ABA-dependent regulatory network of abiotic stress responses of mosses and angiosperms (Khandelwal et al., 2010; Mönke et al., 2012). AtNF-YA5 was involved in ABI3-dependent pathways in drought resistance (Li et al., 2008), and PpNF-YC1 was reported to activate the PpLEA1 promoter in an ABI3-dependent manner (Yotsui et al., 2013). In order to assess whether there are any other PpNF-Ys that act via ABI3 pathway, we compared the expression of PpNF-Y genes in wild type and abi3 loss-of-function mutants (Khandelwal et al., 2010) under abiotic stress.
In NaCl treatments, the accumulation of PpNF-YB4 and PpNF-YC5 mRNA was lower in abi3 mutants than that of wild type, while PpNF-YB1, -YC8, -YC9, and -YC12 was higher in abi3 mutants (Figure 5A, Figures S2E,F, S7E,F). The expression of PpNF-YB6 and -YC2 exhibited an opposite pattern (Figure 5A, Figures S2E,F, S7E,F). In response to osmotic stress, transcriptional levels of most PpNF-Y genes decreased in abi3 mutants, with only -YC9 enhanced after a long time (12 h) of treatment (Figure 5B, Figures S3E,F, S8E,F). The expression of PpNF-YB1, -YB7, and -YC12 had an opposite trend for abi3 mutants and wild type plants during mannitol treatment (Figure 5B, Figures S3E,F, S8E,F). Many PpNF-Ys were induced during the desiccation process (Figure 5C, Figures S4A–C, S9A–C). By contrast, in abi3 mutants, most PpNF-Ys exhibited less alteration of transcriptional levels (Figures S4D–F, S9D–F). For example, PpNF-YA2, -YB1, -YB5, -YC5, -YC6, and -YC10 decreased dramatically in abi3 mutants under desiccation stress, while PpNF-YB7, -YB8, and -YB9 remained constant in wild type plants (Figure 5C, Figures S4E,F, S9E,F).
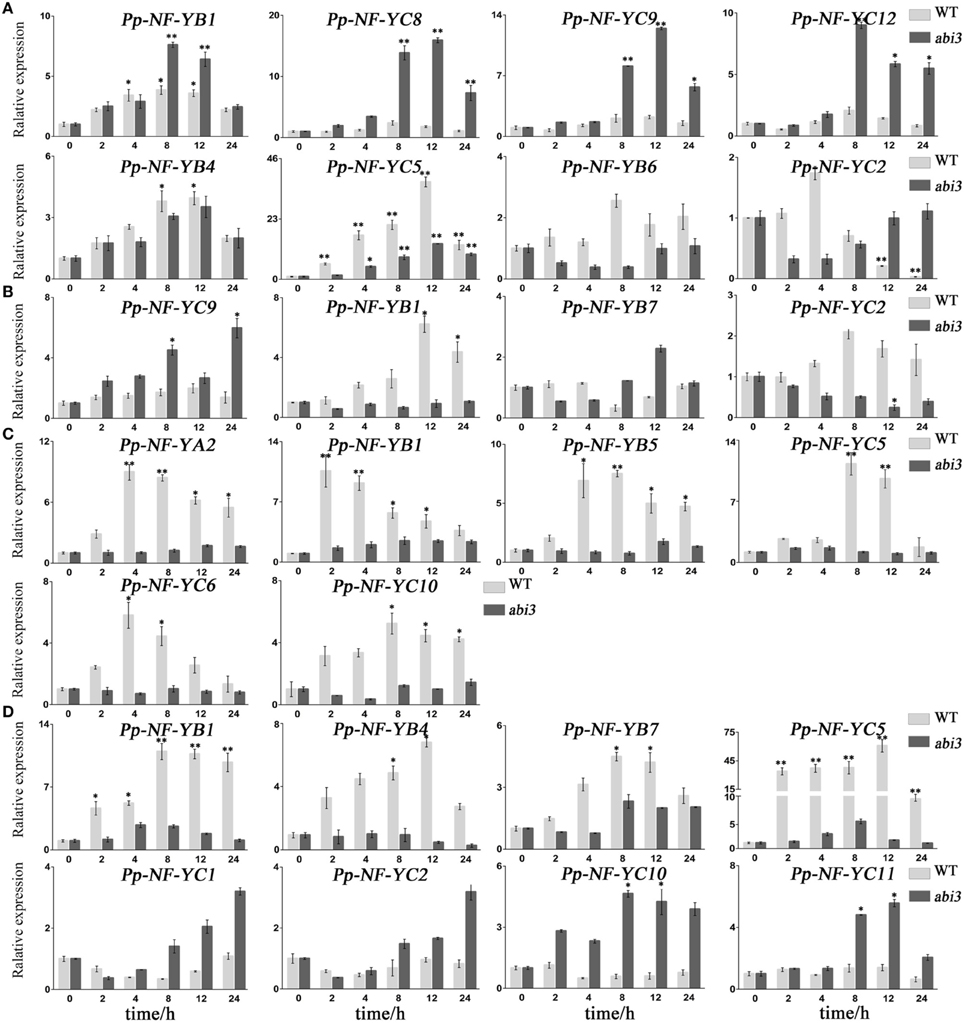
Figure 5. Typical expression patterns between abi3 mutants and wild type plants in response to abiotic stress. (A) PpNF-Ys by salt stress. (B) PpNF-Ys by mannitol treatment. (C) PpNF-Ys by desiccation stress. (D) PpNF-Ys by ABA treatment. The expression level of each gene was normalized against that of the untreated control. Tublin was the housekeeping gene. Error bars indicate SD from three biological replicates. Significant differences were analyzed by t-test (*P ≤ 0.05 and **P ≤ 0.01).
To determine whether ABA regulation of PpNF-Ys expression is ABI3-dependent, we treated abi3 mutants with exogenous ABA and compared the mRNA levels of PpNF-Ys with that of wild type. Transcriptional levels of PpNF-YB1, -YB4, -YB7, and -YC5 were lower in abi3 mutants than wild type under ABA treatment (Figure 5D, Figures S5E,F, S10E,F). PpNF-YC1, -YC2, -YC10, and -YC11 had opposite patterns (Figure 5D, Figures S5F, S10F).
Discussion
Duplication Diversification of NF-Ys Genes in P. patens
Multiple alignments showed that the conserved domains of PpNF-Y family members, with their protein interaction and DNA binding domains (Xing et al., 1994; Sinha et al., 1996; Romier et al., 2003), are highly similar from mammals (such as mouse) to plants (such as angiosperm). Whereas they vary greatly in N- and C-terminal domains of the same NF-Y family (Lee et al., 2003).
The phylogenetic tree indicated that PpNF-Y genes formed several monophyletic groups. This implied that NF-Y genes expanded in the moss and angiosperm lineages in parallel. AtLEC1 (PpNF-YB9) is a seed-specific gene, is involved in the late stages of embryogenesis (Xie et al., 2008). The fact that the LEC1-type NF-YB gene is absent in the moss genome suggests that functional divergence of LEC1 emerged in seed plants.
Plant Transcriptional factor Database V3.0 (http://planttfdb.cbi.pku.edu.cn/index.php?sp=Ppa) collected 69,450 transcriptional factors from 87 species, demonstrating that there are 2 NF-YAs, 16 NF-YBs, and 19 NF-YCs in P. patens, which are different with 23 NF-Y members in this study (Table S2). In order to explain the reasons, we collected all PpNF-Y gene sequences predicted in PlantTFBD (version 3.0), and found that our cloned NF-Y genes were predicted as transcriptional factors in PlantTFBD, The fact that the different member of PpNF-Y genes due to alternative splicing. For example: Pp1s25_89V6.1 and Pp1s25_89V6.2 (PpNF-YB1); Pp1s302_35V6.1, Pp1s302_35V6.2, Pp1s302_35V6.3, Pp1s302_35V6.4, Pp1s302_35V6.5 and Pp1s302_35V6.6 (PpNF-YB2); Pp1s83_179V6.1 and Pp1s83_179V6.2 (PpNF-YB3); Pp1s159_32V6.1, Pp1s159_32V6.2; Pp1s315_9V6.1, Pp1s315_9V6.2 and Pp1s315_9V6.3 (PpNF-YC3); Pp1s370_63V6.1, Pp1s370_63V6.2 and Pp1s370_63V6.3 (PpNF-YC5); Pp1s159_32V6.2, Pp1s159_32V6.3, and Pp1s159_32V6.4 (PpNF-YC6). Alternative splicing occurs as a normal phenomenon in eukaryotes and lead to the rapid expansion of genes (Yuan et al., 2009). Also, this view is agreed with the point of new gene functions, which consider that replication of the gene lead to new gene function, subsequently evolution of the genes number in response to environmental stimuli also result in the new gene function (Francino, 2005).
PpNF-Y Genes Were Expressed in Various Developmental Stage and Tissues
Previous studies have shown that BnNF-YA3,-YA13,-YA14 (Liang et al., 2014) and AtNF-YA2 play roles in root development and in abiotic stress tolerance (Leyva-González et al., 2012). However, we found that accumulation of PpNF-YAs was barely changed in different developmental stages and tissues (Figures 3A,D). Based on the phylogenetic trees presents in Figure 1A, PpNF-YAs have less sequence similarity with B. napus and A. thaliana members, implied that PpNF-YAs may have distinct functions that are still unknown. Or due to still being at the primary stage of evolution, the function of PpNF-YAs might not be quite established yet. It is reported that NF-YB family members in rice, such as OsNF-YB2, -YB3, and -YB4 were ubiquitously expressed (Miyoshi et al., 2003), but their homologs in Physcomitrella, PpNF-YB2, -YB5, -YB7, and -YB9, accumulated high mRNA levels in protonema (Figure 3B), suggesting the functional divergence of NF-YB between mosses and angiosperms. AtNF-YC3, -YC4, and -YC9 express strongly in vascular tissue and regulate flowering time (Siefers et al., 2009; Laloum et al., 2013). BnNF-YC2 has a relatively high level of expression in the silique stage (Liang et al., 2014). Our data indicate that PpNF-YB6 plays functions in mature gametophore and sporophytes stage (Figure 3B), PpNF-YC5 plays roles in the P. patens-specific transition stages from protonema to juvenile gametophores, mature gametophore and sporophytes stage (Figure 3C).
Tissue-specific expression patterns indicated that PpNF-YB2, -YB4, -YB5, -YB6, -YB7, and PpNF-YC10 accumulate more than in the lower-parts of mature gametophores (Figures 3E,F). We assumed that these PpNF-Ys might play roles in rhizoid, especially for PpNF-YB6, which was induced significantly in the lower-part of gametophores. Whereas, PpNF-YB1, -YB3, and -YB9 expressed more in the upper-part of gametophores. Most PpNF-YCs, except for PpNF-YC1 and -YC3, showed higher mRNA level in the upper-part of gametophores. Based on the expression patterns, we assumed that PpNF-YB1, -YB3, -YB9, and -YCs might perform the functions in shoot tips.
Regulatory Network of Stress-Responsive NF-Ys in P. patens
Transcription regulation is critical in plant responses to abiotic stress, and transcription factors are important components in these regulatory networks (Saibo et al., 2009). The fact that the transcriptional factor NF-Ys could be induced by different abiotic stress suggests a general role in stress responses. There are reported that support this assumption (Leyva-González et al., 2012). According to their expressing patterns during stress treatments, we classified the PpNF-Ys into several groups. PpNF-YB1 and -YC5 up-regulated rapidly within 2 h of stress or ABA treatments (Figure 4). PpNF-Ys reached transcriptional peaks after 4 or 8 h of stress or ABA treatments, such as PpNF-YB4, PpNF-YA2, -YB5, -YB6, -YC4, -YC5, -YC6 -YB1, and -YB7. PpNF-YC5 and PpNF-YC7 were induced after 12 h of treatment. These genes might play roles at different stages of stress and/or ABA responses. By contrast, some PpNF-Ys genes decreased under abiotic stress, including PpNF-YB8 in mannitol, PpNF-YC8 in desiccation and NF-YB3, -YB8, -YC1, YC10 in NaCl. We assumed that these genes act as negative regulators in responses to abiotic stress.
PpNF-Ys Function in Abiotic Stress Responses Via ABA-dependent and ABA-independent Pathways
Two kinds of signaling pathways are reported to respond to plant abiotic stress: ABA-dependent and ABA-independent pathway. Previous studies showed that some NF-Y genes could be activated in an ABA-dependent way, including AtNF-YA5, ZmNF-YB2, six BnNF-YAs, seven Bn-NF-YBs, and two BnNF-YCs (Nelson et al., 2007; Li et al., 2008; Xu et al., 2014b).
In Physcomitrella and flowering plants, stress-related genes were induced in an ABA-dependent pathway, in which, ABI3 is a key upstream transcriptional factor that induces a series of transcriptional events (Khandelwal et al., 2010; Hauser et al., 2011). It has been reported that transgenic plants overexpressing AtNF-YA1 greatly up-regulated expression of ABI3, implying that NF-YA1 is located upstream of ABI3 (Li et al., 2013). And overexpression lines of AtNF-YA2, -YA7, and -YA10 enhance abiotic stress tolerance via altered ABA perception (Leyva-González et al., 2012). The expression of PpNF-YB4 and -YB7 increased in NaCl, mannitol, and ABA treatments and it was higher in wild type plants compared with abi3 mutants. Accumulation of PpNF-YC5 was dramatically induced by abiotic stress and ABA treatment and it was lower in abi3 mutants compared with wild type plants, suggesting that these genes are involved in an ABA-dependent pathway and regulated by ABI3 (Figures S2–S5). The transcriptional level of PpNF-YA2, -YB5, and -YB6 was high under desiccation stress and ABA application (Figures S2, S5); we speculated that these genes responded to desiccation in an ABA-dependent manner. BnNF-YB11 and -YB14 are strongly induced by abiotic stress via MYB (myeloblastosis oncogene) and MYC (myelocytomatosis oncogene) factors (Xu et al., 2014b). PpNF-YB1 was induced by ABA treatment, while its expressional level was higher in abi3 mutants than wild type plants under salt stress (Figures S2, S5); thus we speculated that PpNF-YB1 might be regulated by other ABA-related pathways, rather than the ABI3-dependent ones. When ABI3 is compromised, the regulatory effects of these pathways are revealed. By contrast, PpNF-YB1 was responsive to osmotic and desiccation stress through an ABI3-dependent manner. Environmental stress causes plants to over- and under-express several genes to cope with stress (Khraiwesh et al., 2012). PpNF-YB3 was a negative regulatory factor because it reduced in response to NaCl, mannitol and ABA treatments (Figures S2, S5).
It has been suggested that soybean NF-YA3 involved in ABA-independent pathways (Ni et al., 2013), and NaCl- and mannitol-responsive BnNF-YB1 and -YB9 are not affected by ABA treatment either (Xu et al., 2014b). The existence of ABA-independent pathways in mosses has been intensively proved (Cuming et al., 2007). In our study, NaCl-, mannitol-responsive PpNF-YC8, -YC9 and desiccation-responsive PpNF-YC1, -YC2, -YC6, -YC10, -YC11 were not response to ABA treatment (Figures S2, S5), implying that these genes respond to abiotic stress via an ABA-independent pathway. Also, previous report has shown that expression of PpNF-YAs, -YB1-5, and -YC1-4 in response to ABA were not significantly changed, however, PpNF-YC5 was induced strongly between the wild type and abi3 mutants by northern blotting (Yotsui et al., 2013). These results have some inconsistency with our study, especially, compared with wild type, the expression of PpNF-YC5 was a dramatically decreased in abi3 mutants (Figure 5D). The reason might lie in the materials selected, the protonamal tissues were used in Yotsui's paper, but the gametophores was selected in our study. Based on temporal and spatial expression pattern of PpNF-Ys presented in Figure 3 and Figure S6, expressions of those PpNF-Y genes were different in various developmental process, implied that they exhibited different degree expression pattern in response to same treatment in various developmental stages.
In conclusion, our work confirmed the existence of PpNF-Y genes in the Physcomitrella genome, and analyzed the transcription pattern of PpNF-Y family members. The present study offers a useful foundation for further studies of PpNF-Y proteins.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Quatrano RS (Washington University, St. Louis, USA) for providing abi3(a/b/c) mutants. This study was supported by funds from NSFC (31470357), Chinese Ministry of Agriculture (2014ZX08009-23B, 2009ZX08009-058B), Chinese Ministry of Science and Technology (2007AA021405) and PHR (IHLB) to YH.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00642
References
Arazi, T. (2012). MicroRNAs in the moss Physcomitrella patens. Plant Mol. Biol. 80, 55–65. doi: 10.1007/s11103-011-9761-5
Arents, G., and Moudrianakis, E. N. (1993). Topography of the histone octamer surface: repeating structural motifs utilized in the docking of nucleosomal DNA. Proc. Natl. Acad. Sci. U.S.A. 90, 10489–10493. doi: 10.1073/pnas.90.22.10489
Chen, N. Z., Zhang, X. Q., Wei, P. C., Chen, Q. J., Ren, F., Chen, J., et al. (2007). AtHAP3b plays a crucial role in the regulation of flowering time in Arabidopsis during osmotic stress. J. Biochem. Mol. Biol. 40, 1083–1089. doi: 10.5483/BMBRep.2007.40.6.1083
Combier, J. P., Frugier, F., de Billy, F., Boualem, A., El-Yahyaoui, F., Moreau, S., et al. (2006). MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 20, 3084–3088. doi: 10.1101/gad.402806
Cuming, A. C., Cho, S. H., Kamisugi, Y., Graham, H., and Quatrano, R. S. (2007). Microarray analysis of transcriptional responses to abscisic acid and osmotic, salt, and drought stress in the moss, Physcomitrella patens. New Phytol. 176, 275–287. doi: 10.1111/j.1469-8137.2007.02187.x
Dolfini, D., Gatta, R., and Mantovani, R. (2012). NF-Y and the transcriptional activation of CCAAT promoters. Crit. Rev. Biochem. Mol. Biol. 47, 29–49. doi: 10.3109/10409238.2011.628970
Francino, M. P. (2005). An adaptive radiation model for the origin of new gene functions. Nat. Genet. 37, 573–577. doi: 10.1038/ng1579
Frank, W., Ratnadewi, D., and Reski, R. (2005). Physcomitrella patens is highly tolerant against drought, salt and osmotic stress. Planta 220, 384–394. doi: 10.1007/s00425-004-1351-1
Hackenberg, D., Keetman, U., and Grimm, B. (2012). Homologous NF-YC2 subunit from arabidopsis and tobacco is activated by photooxidative stress and induces flowering. Int. J. Mol. Sci. 13, 3458–3477. doi: 10.3390/ijms13033458
Hauser, F., Waadt, R., and Schroeder, J. I. (2011). Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21, R346–R355. doi: 10.1016/j.cub.2011.03.015
Khandelwal, A., Cho, S. H., Marella, H., Sakata, Y., Perroud, P. F., Pan, A., et al. (2010). Role of ABA and ABI3 in desiccation tolerance. Science 327, 546. doi: 10.1126/science.1183672
Khraiwesh, B., Zhu, J. K., and Zhu, J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819, 137–148. doi: 10.1016/j.bbagrm.2011.05.001
Komatsu, K., Suzuki, N., Kuwamura, M., Nishikawa, Y., Nakatani, M., Ohtawa, H., et al. (2013). Group A PP2Cs evolved in land plants as key regulators of intrinsic desiccation tolerance. Nat. Commun. 4, 2219. doi: 10.1038/ncomms3219
Kumimoto, R. W., Zhang, Y., Siefers, N., and Holt, B. F. III. (2010). NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. 63, 379–391. doi: 10.1111/j.1365-313X.2010.04247.x
Laloum, T., De Mita, S., Gamas, P., Baudin, M., and Niebel, A. (2013). CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 18, 157–166. doi: 10.1016/j.tplants.2012.07.004
Lee, H., Fischer, R. L., Goldberg, R. B., and Harada, J. J. (2003). Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 100, 2152–2156. doi: 10.1073/pnas.0437909100
Leyva-González, M. A., Ibarra-Laclette, E., Cruz-Ramirez, A., and Herrera-Estrella, L. (2012). Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS ONE 7:e48138. doi: 10.1371/journal.pone.0048138
Li, C., Distelfeld, A., Comis, A., and Dubcovsky, J. (2011). Wheat flowering repressor VRN2 and promoter CO2 compete for interactions with NUCLEAR FACTOR-Y complexes. Plant J. 67, 763–773. doi: 10.1111/j.1365-313X.2011.04630.x
Li, W. X., Oono, Y., Zhu, J., He, X. J., Wu, J. M., Iida, K., et al. (2008). The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20, 2238–2251. doi: 10.1105/tpc.108.059444
Li, Y. J., Fang, Y., Fu, Y. R., Huang, J. G., Wu, C. A., and Zheng, C. C. (2013). NFYA1 is involved in regulation of postgermination growth arrest under salt stress in Arabidopsis. PLoS ONE 8:e61289. doi: 10.1371/journal.pone.0061289
Liang, M., Hole, D., Wu, J., Blake, T., and Wu, Y. (2012). Expression and functional analysis of NUCLEAR FACTOR-Y, subunit B genes in barley. Planta 235, 779–791. doi: 10.1007/s00425-011-1539-0
Liang, M., Yin, X., Lin, Z., Zheng, Q., Liu, G., and Zhao, G. (2014). Identification and characterization of NF-Y transcription factor families in Canola (Brassica napus L.). Planta 239, 107–126. doi: 10.1007/s00425-013-1964-3
Liu, J. X., and Howell, S. H. (2010). bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22, 782–796. doi: 10.1105/tpc.109.072173
Maity, S. N., and de Crombrugghe, B. (1992). Biochemical analysis of the B subunit of the heteromeric CCAAT-binding factor. A DNA-binding domain and a subunit interaction domain are specified by two separate segments. J. Biol. Chem. 267, 8286–8292.
Mantovani, R. (1999). The molecular biology of the CCAAT-binding factor NF-Y. Gene 239, 15–27. doi: 10.1016/S0378-1119(99)00368-6
Mantovani, R., Li, X. Y., Pessara, U., Hooft van Huisjduijnen, R., Benoist, C., and Mathis, D. (1994). Dominant negative analogs of NF-YA. J. Biol. Chem. 269, 20340–20346.
Matuoka, K., and Chen, K. Y. (2002). Transcriptional regulation of cellular ageing by the CCAAT box-binding factor CBF/NF-Y. Ageing Res. Rev. 1, 639–651. doi: 10.1016/S1568-1637(02)00026-0
Miyoshi, K., Ito, Y., Serizawa, A., and Kurata, N. (2003). OsHAP3 genes regulate chloroplast biogenesis in rice. Plant J. 36, 532–540. doi: 10.1046/j.1365-313X.2003.01897.x
Mönke, G., Seifert, M., Keilwagen, J., Mohr, M., Grosse, I., Hähnel, U., et al. (2012). Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res. 40, 8240–8254. doi: 10.1093/nar/gks594
Nelson, D. E., Repetti, P. P., Adams, T. R., Creelman, R. A., Wu, J., Warner, D. C., et al. (2007). Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. U.S.A. 104, 16450–16455. doi: 10.1073/pnas.0707193104
Ni, Z., Hu, Z., Jiang, Q., and Zhang, H. (2013). GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 82, 113–129. doi: 10.1007/s11103-013-0040-5
Petroni, K., Kumimoto, R. W., Gnesutta, N., Calvenzani, V., Fornari, M., Tonelli, C., et al. (2012). The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 24, 4777–4792. doi: 10.1105/tpc.112.105734
Qudeimat, E., Faltusz, A. M., Wheeler, G., Lang, D., Holtorf, H., Brownlee, C., et al. (2008). A PIIB-type Ca2+-ATPase is essential for stress adaptation in Physcomitrella patens. Proc. Natl. Acad. Sci. U.S.A. 105, 19555–19560. doi: 10.1073/pnas.0800864105
Rensing, S. A., Lang, D., Zimmer, A. D., Terry, A., Salamov, A., Shapiro, H., et al. (2008). The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. doi: 10.1126/science.1150646
Rípodas, C., Castaingts, M., Clúa, J., Blanco, F., and Zanetti, M. E. (2014). Annotation, phylogeny and expression analysis of the nuclear factor Y gene families in common bean (Phaseolus vulgaris). Front. Plant Sci. 5:761. doi: 10.3389/fpls.2014.00761
Romier, C., Cocchiarella, F., Mantovani, R., and Moras, D. (2003). The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 278, 1336–1345. doi: 10.1074/jbc.M209635200
Saibo, N. J., Lourenço, T., and Oliveira, M. M. (2009). Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann. Bot. 103, 609–623. doi: 10.1093/aob/mcn227
Sakakibara, K., Nishiyama, T., Sumikawa, N., Kofuji, R., Murata, T., and Hasebe, M. (2003). Involvement of auxin and a homeodomain-leucine zipper I gene in rhizoid development of the moss Physcomitrella patens. Development 130, 4835–4846. doi: 10.1242/dev.00644
Siefers, N., Dang, K. K., Kumimoto, R. W., Bynum, W. E. IV. Tayrose, G., and Holt, B. F. III. (2009). Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 149, 625–641. doi: 10.1104/pp.108.130591
Sinha, S., Kim, I. S., Sohn, K. Y., de Crombrugghe, B., and Maity, S. N. (1996). Three classes of mutations in the A subunit of the CCAAT-binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol. Cell. Biol. 16, 328–337.
Stephenson, T. J., McIntyre, C. L., Collet, C., and Xue, G. P. (2007). Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol. Biol. 65, 77–92. doi: 10.1007/s11103-007-9200-9
Thirumurugan, T., Ito, Y., Kubo, T., Serizawa, A., and Kurata, N. (2008). Identification, characterization and interaction of HAP family genes in rice. Mol. Genet. Genomics 279, 279–289. doi: 10.1007/s00438-007-0312-3
Warpeha, K. M., Upadhyay, S., Yeh, J., Adamiak, J., Hawkins, S. I., Lapik, Y. R., et al. (2007). The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 143, 1590–1600. doi: 10.1104/pp.106.089904
Xie, Z., Li, X., Glover, B. J., Bai, S., Rao, G. Y., Luo, J., et al. (2008). Duplication and functional diversification of HAP3 genes leading to the origin of the seed-developmental regulatory gene, LEAFY COTYLEDON1 (LEC1), in nonseed plant genomes. Mol. Biol. Evol. 25, 1581–1592. doi: 10.1093/molbev/msn105
Xing, Y., Fikes, J. D., and Guarente, L. (1993). Mutations in yeast HAP2/HAP3 define a hybrid CCAAT box binding domain. EMBO J. 12, 4647–4655.
Xing, Y., Zhang, S., Olesen, J. T., Rich, A., and Guarente, L. (1994). Subunit interaction in the CCAAT-binding heteromeric complex is mediated by a very short alpha-helix in HAP2. Proc. Natl. Acad. Sci. U.S.A. 91, 3009–3013. doi: 10.1073/pnas.91.8.3009
Xu, B., Ohtani, M., Yamaguchi, M., Toyooka, K., Wakazaki, M., Sato, M., et al. (2014a). Contribution of NAC transcription factors to plant adaptation to land. Science 343, 1505–1508. doi: 10.1126/science.1248417
Xu, L., Lin, Z., Tao, Q., Liang, M., Zhao, G., Yin, X., et al. (2014b). Multiple NUCLEAR FACTOR Y transcription factors respond to abiotic stress in Brassica napus L. PLoS ONE 9:e111354. doi: 10.1371/journal.pone.0111354
Yamamoto, A., Kagaya, Y., Toyoshima, R., Kagaya, M., Takeda, S., and Hattori, T. (2009). Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 58, 843–856. doi: 10.1111/j.1365-313X.2009.03817.x
Yotsui, I., Saruhashi, M., Kawato, T., Taji, T., Hayashi, T., Quatrano, R. S., et al. (2013). ABSCISIC ACID INSENSITIVE3 regulates abscisic acid-responsive gene expression with the nuclear factor Y complex through the ACTT-core element in Physcomitrella patens. New Phytol. 199, 101–109. doi: 10.1111/nph.12251
Keywords: transcription factors, temporal and spatial expression, abiotic stress, gene regulation, abscisic acid
Citation: Zhang F, Han M, Lv Q, Bao F and He Y (2015) Identification and expression profile analysis of NUCLEAR FACTOR-Y families in Physcomitrella patens. Front. Plant Sci. 6:642. doi: 10.3389/fpls.2015.00642
Received: 09 April 2015; Accepted: 02 August 2015;
Published: 19 August 2015.
Edited by:
Keqiang Wu, National Taiwan University, TaiwanReviewed by:
Adriana Garay, Universidad Nacional Autónoma de México, MexicoYoichi Sakata, Tokyo University of Agriculture, Japan
Xingliang Hou, South China Botanical Garden (CAS), China
Copyright © 2015 Zhang, Han, Lv, Bao and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yikun He, College of Life Science, Capital Normal University, #105 XiSanHuan N. Rd., Beijing 100048, China,eWhlQGNudS5lZHUuY24=
 Fang Zhang
Fang Zhang Min Han
Min Han Qiang Lv
Qiang Lv Fang Bao
Fang Bao Yikun He
Yikun He