- Plant Molecular Biology Group, International Centre for Genetic Engineering and Biotechnology, New Delhi, India
SRO1 is an important regulator of stress and hormonal response in plants and functions by interacting with transcription factors and several other proteins involved in abiotic stress response. In the present study, we report OsRBD1, an RNA binding domain 1- containing protein as a novel interacting partner of OsSRO1a from rice. The interaction of OsSRO1a with OsRBD1 was shown in yeast as well as in planta. Domain–domain interaction study revealed that C-terminal RST domain of OsSRO1a interacts with the N-terminal RRM1 domain of OsRBD1 protein. Both the proteins were found to co-localize in nucleus. Transcript profiling under different stress conditions revealed co-regulation of OsSRO1a and OsRBD1 expression under some abiotic stress conditions. Further, co-transformation of both OsSRO1a and OsRBD1 in yeast conferred enhanced tolerance toward salinity, osmotic, and methylglyoxal treatments. Our study suggests that the interaction of OsSRO1a with OsRBD1 confers enhanced stress tolerance in yeast and may play an important role under abiotic stress responses in plants.
Introduction
RCD1 is a nuclear protein that interacts with several transcription factors and other stress responsive proteins and has been shown to protect plants against oxidative damage and other stresses (Belles-Boix et al., 2000; Katiyar-Agarwal et al., 2006; Jaspers et al., 2009, 2010; Vainonen et al., 2012). RCD1 belongs to a plant-specific SIMILAR to RCD1 (SRO) gene family, and was first isolated as a CEO (CLONE EIGHTY-ONE) protein, based on its ability to complement ROS sensitivity in yeast cells (Belles-Boix et al., 2000). The Arabidopsis genome encodes one RCD1 and five SROs proteins and the loss-of-function mutation in RCD1 results in highly pleiotropic phenotypes (Overmyer et al., 2000; Jaspers et al., 2009; Teotia and Lamb, 2009). In Arabidopsis, rcd1 mutant is known to be defective in plant development, but mutant of its closest Arabidopsis homolog, sro1 displays normal development (Jaspers et al., 2009). Notably, rcd1-sro1 double mutant exhibits severe growth defects, indicating unequal genetic redundancy in RCD1 and SRO1 functions. In plants, a new unified nomenclature system has been proposed for SRO genes since naming conventions based on Arabidopsis thaliana were not found to be suitable for most other plant species (Jaspers et al., 2010). However, the previous annotations of Arabidopsis SRO genes have been retained in the new nomenclature.
SRO has been classified as a novel sub-family of proteins involved in ubiquitin and ADP-ribose conjugation systems. The members of SRO family contain a conserved globular domain, WWE (named after three of its conserved residues, W and E; PF02825) which is predicted to mediate specific protein–protein interactions (Aravind, 2001) and a region similar to the catalytic domain of poly (ADP-ribose) polymerase proteins (PARP signature; PF00644; Ahlfors et al., 2004) that mediates attachment of ADP-ribose units from NAD+ to target proteins and have implications in a number of processes, such as DNA repair, apoptosis, transcription, and chromatin remodeling (Hassa and Hottiger, 2008). The domain composition of SRO proteins is unique within plants as SRO proteins apart from WWE and PARP domains, possess a C-terminal RCD1-SRO-TAF4 domain (RST domain; PF12174) that is believed to be critical for interaction with several, mostly plant-specific, transcription factors (Jaspers et al., 2010). In plants, SRO genes have been classified into two structural types based on their domain composition (Jaspers et al., 2010). The genome of the monocot plants including rice, have been found to contain only group I SRO genes (hence, named as SRO1) which possess all the three domains, viz. WWE, PARP, and RST whereas eudicots in addition to group I SRO genes, contain group II SRO genes (named as SRO2) as well, characterized by the absence of N-terminal WWE domain but having PARP and RST domains. In rice genome annotation database RGAP7, SRO1 is named as Radical induced cell death (RCD1) gene, but as per the nomenclature proposed by Jaspers et al. (2010), we will now refer rice RCD1 genes as SRO1.
The first reported interacting partner of RCD1 protein was STO (salt tolerance; Belles-Boix et al., 2000), a protein that confers salt tolerance to yeast and contains two putative zinc fingers (Lippuner et al., 1996) similar to those found in the transcription regulators, GATA-1 (Putterill et al., 1995) and CONSTANS (Orkin, 1996). The other RCD1 interacting partner includes a putative protein, similar to the members from the Ethylene Responsive Element Binding Protein (EREBP) sub-family of AP2/EREBP plant transcription factors. Members of this protein sub-family are involved in the induction of defense genes in response to biotic and abiotic stresses (Zhou et al., 1997; Fujimoto et al., 2000). Further, an interaction between the predicted cytoplasmic tail of SOS1, a sodium transporter, and RCD1 has been proposed in A. thaliana (Katiyar-Agarwal et al., 2006) and its interaction with DREB2A is also reported (Belles-Boix et al., 2000; Vainonen et al., 2012), which reveals a function for RCD1 in stress tolerance. In Arabidopsis, it has been shown that loss of rcd1 expression leads to malfunctioned control of cell death in response to apoplastic ROS and that WRKY70 and SGT1b work as cell death regulators downstream of RCD1 (Brosché et al., 2014). Further another study reports that RCD1 protects plant cells from activating ROS-triggered programs, such as cell death and induction of pathogen-responsive genes (PR genes) and extra-plastidic antioxidant enzymes, by supporting the induction of chloroplast antioxidant system via interactions with the transcription factor Rap2.4a (Hiltscher et al., 2014). In rice, a member of SRO family, OsSRO1c, is reported to play an important role in drought and oxidative stress tolerance of rice by promoting stomatal closure and H2O2 accumulation through a novel pathway involving SNAC1 (stress-responsive NAC 1) and a zinc-finger gene, DST (You et al., 2013).
The rice SRO gene family consists of five members named as OsSRO1a, OsSRO1b, OsSRO1c, OsSRO1d, and OsSRO1e. In the present study, we have identified interacting partners of rice OsSRO1a. Our results show that OsSRO1a interacts with several proteins. Of these, we have characterized a novel interacting partner, the RNA binding domain (RBD) containing protein, OsRBD1. Detailed studies suggest that both the proteins are localized in nucleus and their interaction confers stress tolerance in yeast. Altogether, our results suggest that this interaction may have relevance in stress physiology.
Materials and Methods
Genomic Distribution of OsSRO Genes on Rice Chromosomes
Position of each of the OsSRO genes on rice chromosome available at RGAP version 7 was determined1 and multiple alignment of all the isoforms of rice SRO1 proteins was performed using ClustalW2 (Larkin et al., 2007).
Expression Analysis Using Rice Genome Database
To analyze the expression of OsSRO1 genes in silico, we used Gene Expression Omnibus platform2 accession number GSE6893 and GSE6901 for reproductive development and stress response, respectively. Expression data was further depicted by heatmap generated with the help of MeV software package.
Plant Material, Growth Conditions, and Sample Collection
Seedlings of IR64 rice cultivar were grown under controlled conditions in growth chamber at 28 ± 2°C and 16 h light/8 h dark photoperiod. After sterilization with 1% Bavistin for 20 min, seeds were germinated hydroponically in modified Yoshida et al. (1972) medium. Various treatments, including low and high temperature (4 and 42°C, respectively), desiccation (air dry), salinity (200 mM NaCl), MG (5 mM), oxidative (5 mM H2O2) and wounding (pricking the leaf with a needle) were given to the 12 days-old seedlings and shoot tissue was harvested after 6 h and 24 h of stress treatment. The shoots of untreated seedlings served as control.
Cloning of OsSRO1a and OsRBD1 from Rice
OsSRO1a and OsRBD1 were amplified from rice cDNA using gene specific primers (Supplementary Table S1) and cloned into TOPO-TA vector (Invitrogen, USA). For localization assay, each of the OsSRO1a and OsRBD1 cDNA were cloned in pMBPII vector as done previously (Kumar et al., 2012) at BamH1 and XbaI/BamH1 sites, respectively. For in planta interaction studies, OsSRO1a and OsRBD1 cDNA were cloned in BiFC1 and BiFC2 vectors at Not1 and Nco1/Not1 sites, respectively. OsSRO1a and OsRBD1 full length cDNA were also cloned in pYES2 yeast expression vector (Invitrogen, USA) at BamH1 and HindIII/BamH1 for yeast expression studies.
Yeast Two Hybrid Assay (YTH) for Identification of Interacting Partners in Rice Library
A Gal4-based two-hybrid system (Clontech, USA) was used for YTH assay as described by Kumar et al. (2012). The pGAD plasmid which contains the DNA activation domain of Gal4 (AD) and/or pGBD which contains the DNA binding domain of Gal4 (BD) were used to express the AD and BD fusion proteins, respectively. In brief, OsSRO1a encoding the bait protein was ligated into the pGBD vector using EcoRl/Sal1 sites. AH109 strain (MATa trp1-901 leu2-3, 112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS -MEL1TATA -lacZ) was used for transformation. Competent cells were made from glycerol stocks of the rice cDNA library (already cloned in pGAD vector), followed by its transformation with the bait plasmid. The transformants were grown on the two drop out (-Leu-Trp-) and three drop-out (-Leu-Trp-His) medium, followed by four drop-out (-Leu-Trp-His-Ade) medium supplied with 20 mM 3-AT (3-amino-1,2,4-Triazole) to find out the interacting partners.
Yeast monohybrid assay was performed by co-transforming OsSRO1a-pGAD and pGBD vector or co-transforming OsSRO1a-pGBD and pGAD vector followed by assessing the growth of transformants on two drop out (-Leu-Trp-) medium to confirm that if is any self interaction.
Construction of Deletion Mutants of OsSRO1a and OsRBD1
Deletion mutants for OsSRO1a and OsRBD1 were made to investigate domain to domain interactions. For this purpose, a set of six primers were designed to differentially amplify different domains (Supplementary Table S1). For OsSRO1a, three sets of primers were designed which were used for amplification of partial fragments of OsSRO1a, such that first fragment contained N-terminal (OsSRO1a_N), second contained PARP domain (OsSRO1a_PARP), while the third fragment contained RST domain (OsSRO1a_C). Primers for OsRBD1 deletion studies were also designed in a similar way so as to amplify different regions of the gene in three fragments (Supplementary Table S1). First fragment contained N-terminal region (OsRBD1_N), second containing RRM1 domain [OsRBD1_RRM1; RNA recognition motif (RRM) domain] and third containing C-terminus (OsRBD1_C). For checking one to one interaction between OsSRO1a and OsRBD1, both the genes were cloned in frame into both pGAD and pGBD vectors, resulting in various combinations of constructs.
Protoplast Isolation and Subcellular Localization
Protoplast isolation and transfection was performed following the method of Sheen (2001). In brief, soft and young leaves from 25 days-old tobacco plants were chopped finely and lysed with enzyme solution containing 1% cellulase Y-C, 0.1% macerozyme, (Kyowa Chemical Products, Osaka, Japan), 0.4 M mannitol, 5 mM MES, pH 5.7, filter sterilized. The mixture was incubated in a small petridish at 25°C for 3 h in dark and centrifuged at low speed (145 g, Eppendorf 5810R) followed by washing with wash buffer (0.4 M mannitol, 2.5 mM CaCl2, 1 mM MES, pH 5.7) and finally, the protoplasts were resuspended in 20 ml solution containing 0.4 m mannitol, 15 mM MgCl2, 5 mM MES, pH 5.7. The protoplasts were then diluted in a solution containing 154 mM NaCl, 125 mM CaCl2 5.0 mM KCl, 2.0 mM MES (pH 5.0) and 2.0 μg of OsSRO1a-pMBPII and OsRBD1-pMBPII plasmids were added along with 40% PEG and mixed gently. The mixture was then incubated for 20 h at 23°C with gentle agitation. The incubation buffer was subsequently removed and protoplasts were viewed via fluorescence microscopy (Zeiss observer Z1).
Particle Bombardment and Fluorescence Microscopy
Onion peel bombardment assay was performed as described previously (Kaur et al., 2014). About 3 μg of OsSRO1a-pMBPII and OsRBD1-pMBPII plasmids were coated separately on 1 μm gold particles and introduced into onion epidermal cells by microprojectile bombardment, using a Bio-Rad PDS/1000 helium-driven particle accelerator, as per the manufacturer’s instructions. Plates were incubated at 28°C for 18 h in dark before microscopic analysis. Transformed epidermal onion peels were observed under fluorescence microscope (Zeiss observer Z1). For nucleus staining, onion peels were briefly incubated with 100 nM DAPI (4, 6-diamidino-2-phenylindole, dihydrochloride) stain (Invitrogen, USA), prior to microscopic analysis.
Bimolecular Fluorescence Complementation Analysis
To confirm interactions in planta, OsSRO1a and OsRBD1 were cloned in BiFC1 and BiFC2 vectors, respectively and transformed into Agrobacterium (LBA4404). Empty vectors were also transformed as controls. All the constructs were then inoculated in YPD media (containing yeast extract, peptone, and dextrose) till absorbance reached 1.2 and the cells were collected by centrifugation at 5000 g. Cultures were then resuspended in 10 mM of MgCl2 and 10 mM MES containing buffer and incubated for 4 h at 28°C. Cell cultures containing OsSRO1a and OsRBD1 constructs were mixed and agro-infiltrated into 25 days-old tobacco leaves. Both the empty vectors were also pooled together and infiltrated as control.
Real Time PCR
Real Time PCR was performed as described previously (Mustafiz et al., 2011). Total RNA was isolated from the shoot tissues of control and stressed seedlings using RaFlexTM solution I and solution II (GeNei, India), and cDNA synthesis was performed with RevertAidTM RNAse H minus cDNA synthesis kit (Fermentas Life Sciences, USA). Manufacturer’s protocol was strictly followed in both the above procedures. Real time PCR primers for OsSRO1a and OsRBD1 were designed from 3′ UTR regions using Primer3 software (see Supplementary Table S2 for primer sequences). Three replicates of each sample (control and stress treated) were used. Mean Ct values were calculated for each sample and further normalized against the maximum expression value obtained for the control sample (Livak and Schmittgen, 2001). eEF-1α gene was used as internal control. The statistical significance of stress-induced change in OsSRO1a and OsRBD1 transcript levels at each time point compared to control was tested by paired t-test (one-sided) using Q-Gene (Simon, 2003).
Stress Tolerance Assay for the Significance of Interaction Between OsSRO1 and OsRBD1
OsSRO1a and OsRBD1 cloned in pYES2 vectors were transformed, individually as well as together in yeast BY4741 (MATa his31Δ leu2Δ1 met 15Δ0 ura3Δ0) strain. Transformed strains were grown overnight in YPD medium at 30°C and diluted to 0.5 OD600. Serial dilutions (1:10, 1:100, 1:1000, and 1:10,000) of 0.5 OD600 were then spotted on solid YPD medium supplemented with various stress-inducers to study the significance of OsSRO1a and OsRBD1 interaction in response to salinity (1.2 M NaCl), oxidative (10 mM H2O2), osmotic (1 M mannitol), and methylglyoxal (8 mM) treatments. Growth pattern for each of the yeast transformants was also observed by streaking assay on stress and control media Empty vector was also transformed and used as control.
Results
Identification and Chromosomal Localization of OsSRO Genes
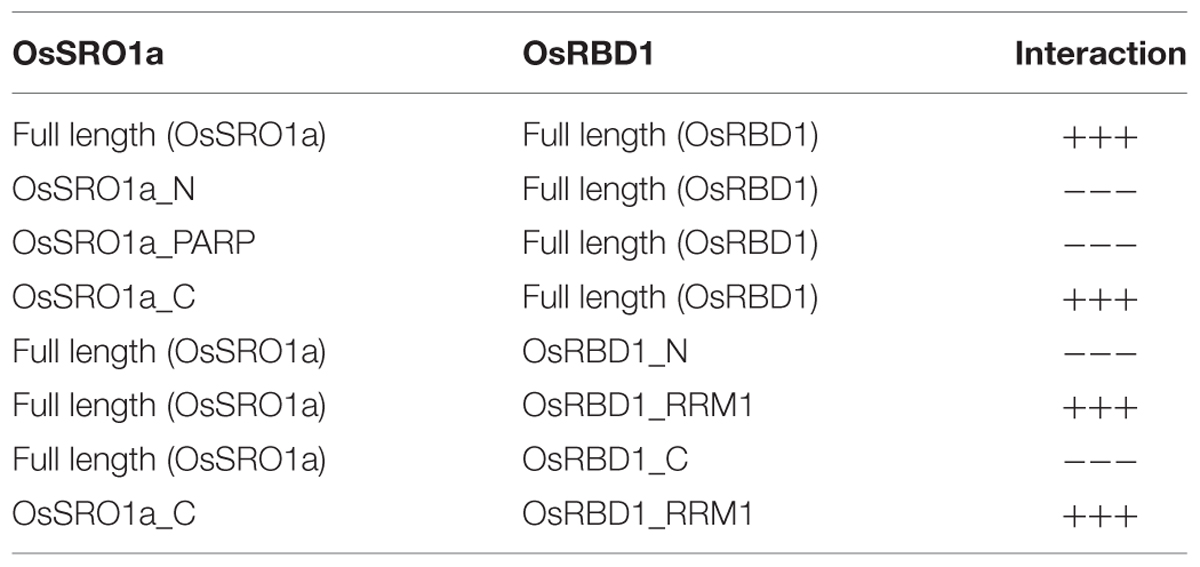
Five OsSRO genes were identified from the RGAP 7 database, distributed on chromosome X, III, VI and IV, and named as OsSRO1a, OsSRO1b, OsSRO1c, OsSRO1d, and OsSRO1e, respectively (Figure 1A) with both OsSRO1b and OsSRO1c located on the chromosome III. Multiple alignments of the amino acid sequences of all the five SRO1 proteins revealed about 40–61% similarity among them. OsSRO1a was most similar to OsSRO1b sharing 61% homology in amino acid sequence, followed by OsSRO1e possessing 49% sequence similarity. Domain search analysis3 showed that all the OsSRO1 proteins contain an N-terminal WWE domain, a catalytic core, comprising poly (ADP-ribose) polymerase (PARP) domain and a C-terminal RST (RCD1-SRO-TAF4) domain.
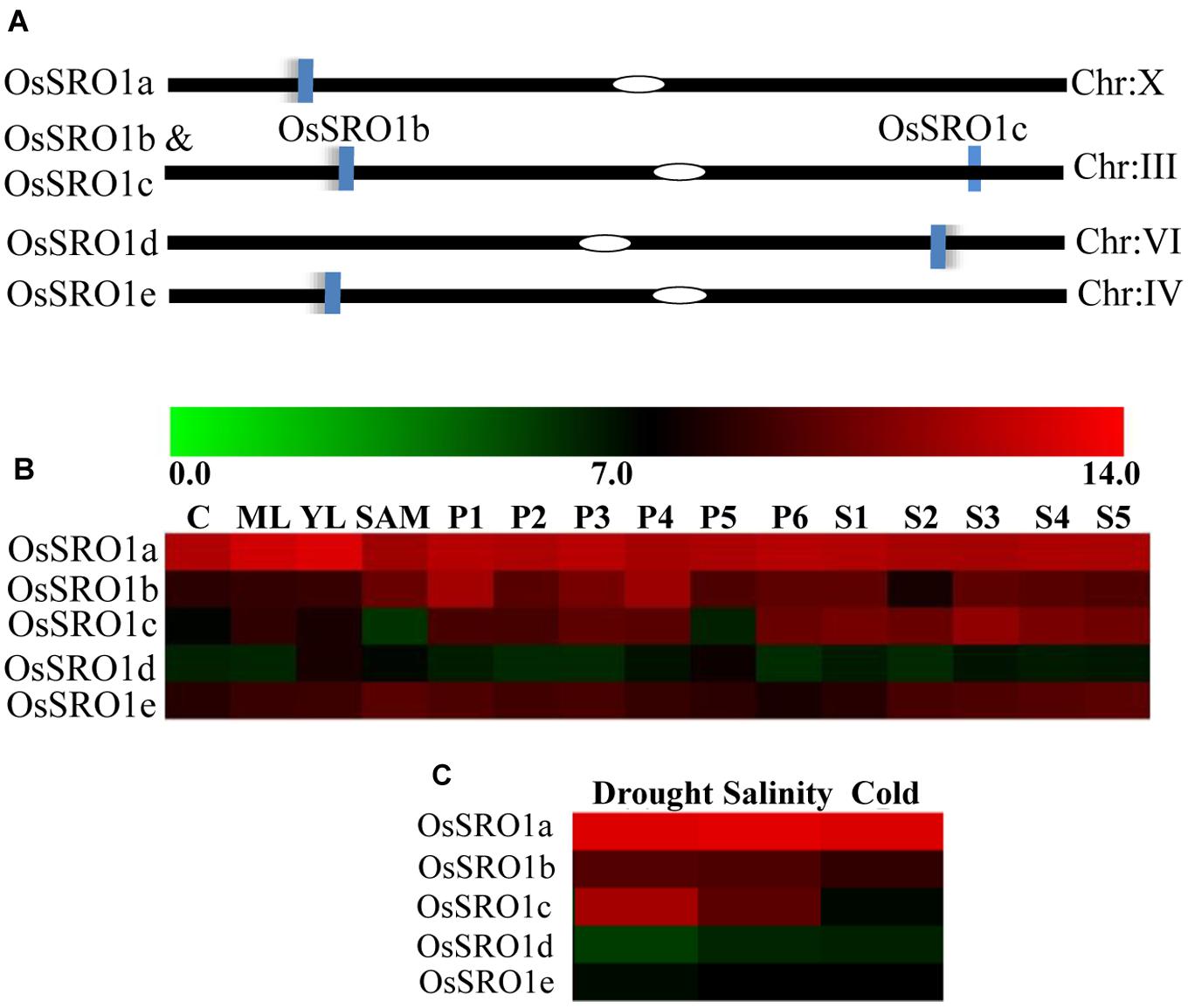
FIGURE 1. Rice SRO1 genes and their genomic distribution on chromosomes. (A) Genomic distribution of OsSRO1 genes on rice chromosomes. Chromosome number is indicated at the right and blue bars represent position of the OsSRO1 genes. (B) Expression profile of OsSRO1 genes at different developmental stages as mentioned on top of each column viz. 7 days seedlings taken as control (C), mature leaf (ML), young leaf (YL), shoot apical meristem (SAM), young inflorescence (PI, up to 3 cm), inflorescence (P2, 3–5 cm; P3, 5–10 cm; P4, 10–15 cm; P5, 15–22 cm; P6, 22–30 cm) and seed (S1, 0–2 dap; S2, 3–4 dap; S3, 5–10 dap; S4, 11–20 dap; S5, 21–29 dap), and (C) stress conditions such as drought, salinity, and cold using microarray data. Color bar at the top represents expression values, thereby green color representing lowest expression levels and red signifying highest expression level.
Developmental and Stress-Specific Regulation of Rice SRO1 Genes
To examine the expression profile of rice SRO1 genes at various stages of development and under various abiotic stress conditions, publicly available microarray data4 was analyzed. Analysis of microarray dataset indicated that OsSRO1a, OsSRO1b, and OsSRO1e genes expressed constitutively across various developmental stages, as compared to OsSRO1c and OsSRO1d (Figure 1B). In response to drought, cold and salt stress, OsSRO1a was significantly up-regulated in comparison to other OsSRO1 genes (Figure 1C). Thus, being highly stress-inducible, OsSRO1a was selected for detailed studies to identify its role in abiotic stress response.
Identification of Interacting Partners of OsSRO1a
Domain analysis revealed the presence of a typical RST domain in OsSRO1a at its C-terminus, which suggests that OsSRO1a may associate with other proteins to elicit its regulatory roles. In an attempt to identify the potential interacting partners, full-length ORF of OsSRO1a (LOC_Os10g42710) was amplified and cloned. Yeast two hybrid assay (YTH) was performed using OsSRO1a-pGBD construct as bait protein, and yeast library of rice as prey. A number of clones grew on four drop-out medium (Trp- His- Ade- Leu-) and further confirmed by transferring the colonies to medium containing increasing concentration (10, 15, and 20 mM, respectively) of 3-amino-1,2,4-triazole (3AT). Even in the presence of 20 mM 3AT, a large number of colonies appeared. In order to reconfirm if this was not due to trans-activation, monohybrid assays were performed which suggested that OsSRO1a does not act as a trans-activator (data not shown). Most of the colonies that grew were found to be positive, with varying length of inserts. Plasmid from about 100 colonies was isolated, sequenced and analyzed using NCBI-BLAST search, which revealed that OsSRO1a may interact with several proteins such as TBC domain-containing protein, RRM containing protein, CAX-interacting protein, universal stress protein domain containing protein and many more (Supplementary Table S2). Among all the interacting partners, OsRBD1 was found to be predominantly present and hence selected for further studies. The OsRBD1 gene (LOC_Os12g01010) is located on chromosome number 12 and domain analysis using Pfam showed the presence of RRM1 at its N-terminus. Sequence analysis of OsRBD1 using BindN tool5 predicted strong binding sites for RNA where about 84 out of 227 residues were predicted to be RNA binding amino acids.
RST Domain of OsSRO1a Interacts with the RRM1 Domain of OsRBD1
To study specific domain–domain interactions between OsSRO1a and OsRBD1, different deletion constructs were designed. Based on the domain architecture of OsSRO1a and OsRBD1 proteins, different deletion fragments were prepared, selectively amplifying different domains (Figure 2A). Firstly, a protein to protein interaction study was carried out using constructs containing full-length OsSRO1a and OsRBD1 proteins as bait and prey, respectively. Colonies co-transformed with OsSRO1a and OsRBD1 constructs could grow well on four drop-out + 3-AT medium (Figure 2B). Next, domain wise interactions were studied using constructs containing different deletion fragments of OsSRO1a and OsRBD1 in different combinations. Out of all combinations tested, OsRBD1_RRM1 and OsSRO1a_C could successfully grow on four drop-out + 3-AT medium (Figure 2C). The growth of other co-transformants was severely hampered even in the three drop-out medium (data not shown). For further confirmation, full-length and deletion constructs which could grow well on four drop-out medium were then spotted on four drop-out + 3-AT (20 mM) medium (Figure 2D). The combination of OsSRO1a-pGAD + OsSOS1-pGBD constructs was used as a positive control. N, N1, and N2 indicate negative controls which represented co-transformed empty vectors, OsSRO1a_BD + pGAD and OsRBD1-BD + pGAD constructs, respectively. The data clearly indicated that the RST domain at the C-terminus of OsSRO1a (OsSRO1a_C) interacts with the RRM1 domain at N-terminus of OsRBD1 (OsRBD1_RRM1) protein. The summary of interactions between OsSRO1a and OsRBD1 proteins is shown in Table 1.
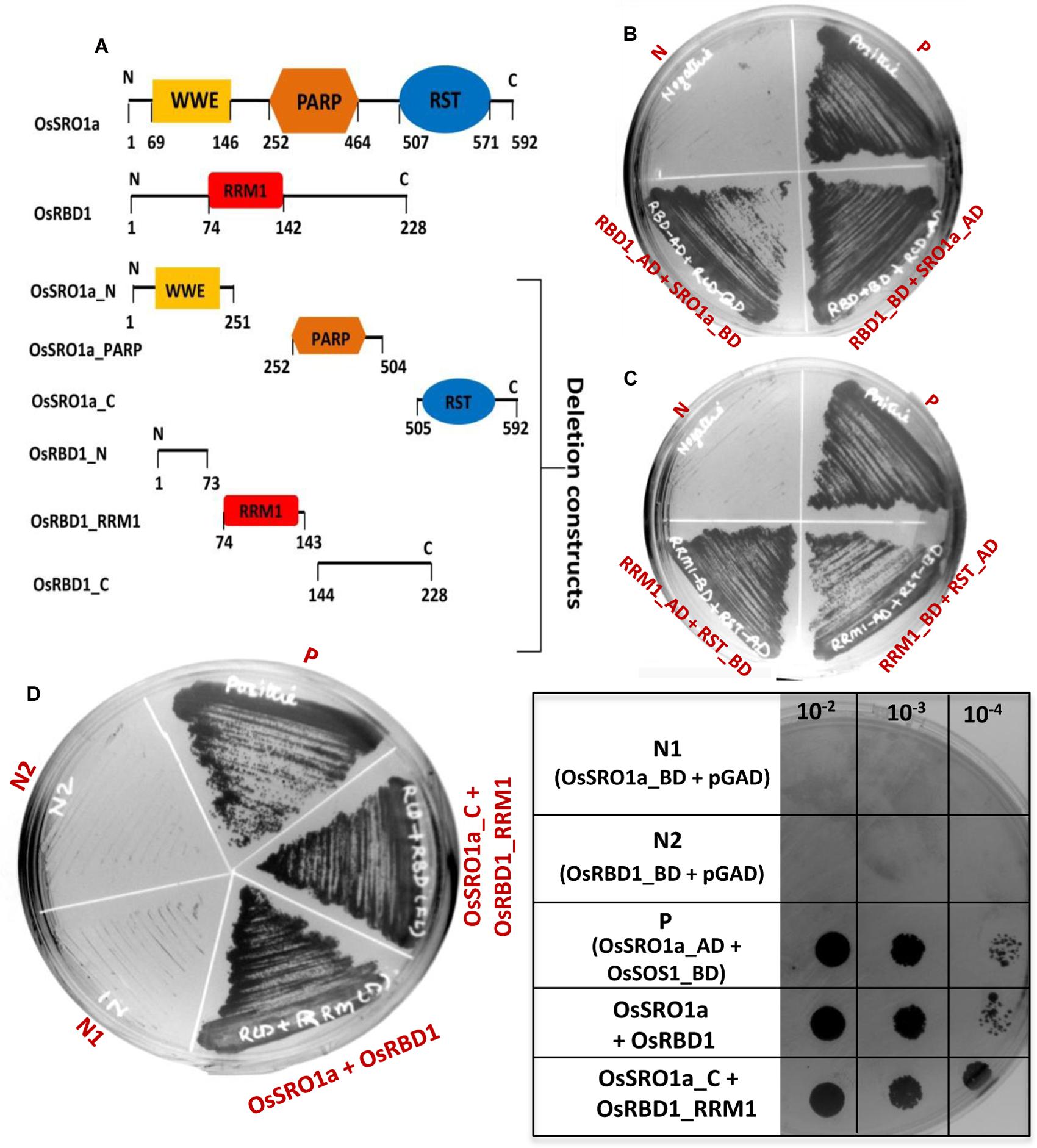
FIGURE 2. RST domain of OsSRO1a interacts with RRM1 domain of OsRBD1. (A) Schematic representation of the domains present in OsSRO1a and OsRBD1 proteins and various deletions constructs prepared for domain–domain interaction studies. (B) Growth observed after co-transforming full-length OsRBD1 (in pGAD vector) and OsSRO1a (in pGBD vector), and OsRBD1 (in pGBD vector) and OsSRO1a (in pGAD vector), and (C) after co-transforming OsRBD1_RRM1 (in pGAD vector) and OsSRO1a_RST (in pGBD vector), and OsRBD1_RRM1 (in pGBD vector) and OsSRO1a_RST (in pGAD vector) on four drop-out + 3-AT (20 mM) medium. (D) Growth observed after streaking co-transformed full-length OsRBD1 and OsSRO1a (OsSRO1a + OsRBD1) and OsSRO1a_C and OsRBD1_RRM1 (SRO1_C + RRM1) on four drop-out + 20 mM 3-AT medium (left panel) and spotting serial dilutions of the same (right panel). P is the positive control taken as OsSRO1a-pGAD + OsSOS1-pGBD, and N, N1, and N2 are the negative controls, representing co-transformed empty vectors, OsSRO1a_BD + pGAD and OsRBD1-BD + pGAD, respectively. The data shown is representative of three clones used for each construct and experiment was repeated thrice.
Subcellular Localization of OsSRO1a and OsRBD1
For localization studies, OsSRO1a and OsRBD1 cDNAs were amplified and cloned into pMBPII vector in translational fusion with GFP at the C-terminus. The resulting pMBPII-OsSRO1a and pMBPII-OsRBD1 constructs were used for particle bombardment in onion peel epidermal cells. Both OsSRO1a and OsRBD1 were found to be localized in nucleus, as confirmed by superposition of fluorescence image of the nucleus staining dye DAPI with that of GFP (Figures 3A,B)
FIGURE 3. In vivo localization of OsSRO1a and OsRBD1. Onion peel epidermal cells showing expression of (A) OsSRO1a-GFP and (B) OsRBD1-GFP driven by CaMV 35S promoter in the nucleus, as indicated by the superposition of GFP and DAPI dye fluorescence. (C) Tobacco leaf protoplasts transfected with pMBPII vector alone (for free GFP visualization; left panel) and, OsSRO1a-pMBPII (middle panel) and OsRBD1-pMBPII (right panel) to confirm the nuclear localization of OsSRO1a and OsRBD1, respectively.
Localization of OsSRO1a and OsRBD1 was also analyzed in tobacco protoplasts. Transiently transfected protoplasts with vector alone (pMBPII) showed green fluorescence of free GFP throughout the protoplasts (Figure 3C). However, transfection with either OsSRO1a-GFP or OsRBD1-GFP showed green fluorescence restricted to nucleus only thereby, indicating nuclear localization of OsSRO1a and OsRBD1 (Figure 3C).
In planta Interaction of OsSRO1a and OsRBD1
The bimolecular fluorescence complementation (BiFC) assay is based on the observation that the association of fluorescent protein fragments can be facilitated by an interaction between proteins fused to the fragments in a split YFP system. In principle, the BiFC assay can be used to visualize interactions between any proteins that can be fused to fluorescent protein fragments. To confirm the interaction in planta, both the interacting partners were cloned in BiFC vectors resulting in OsSRO1a-BiFC1 and OsRBD1-BiFC2 constructs. Recombinant OsSRO1a-BiFC1 and OsRBD1-BiFC2 constructs were transformed in Agrobacterium strain and positive clones were selected for further studies. Before infiltration, culture of Agrobacterium cells containing OsSRO1a-BiFC1 and OsRBD1-BiFC2 plasmids were mixed and then infiltrated into tobacco leaf. Different constructs, including recombinant plasmids and control, were infiltrated in the same leaf to avoid any artifact due to the age or position of the leaf. Suspensions of A. tumefaciens carrying both the constructs were infiltrated into the left half of the leaf and agro-suspension containing empty vectors were also mixed and infiltrated on the right side of the leaf. Highly fluorescent left half of the leaf was observed under UV after 24 h of infiltration while there was no fluorescence on the right side where empty vectors were infiltrated even when checked at 36 and 48 h. This showed that a strong interaction occurs between OsSRO1a and OsRBD1 proteins in planta (Figure 4A).
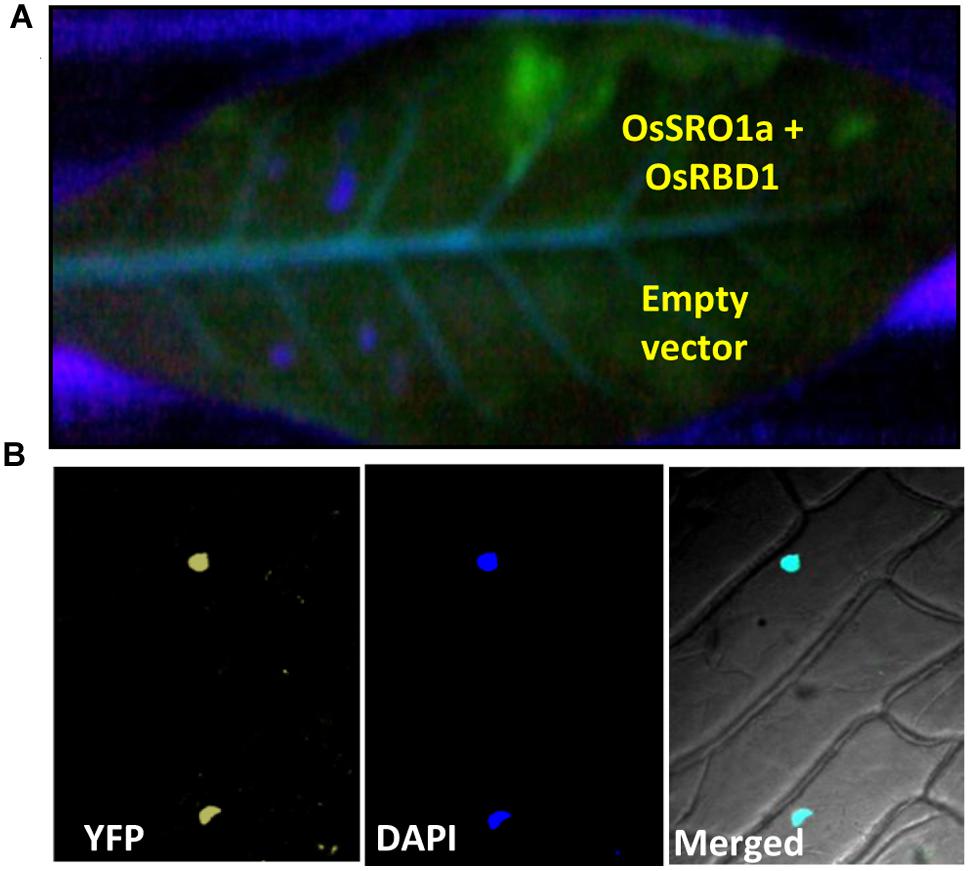
FIGURE 4. Confirmation of OsSRO1a and OsRBD1 interaction in planta. (A) Agroinfiltration of OsSRO1a-BiFC1 + OsRBD1-BiFC2 constructs in Nicotiana tabacum leaves. Upper side of the midrib shows co-infiltration of OsSRO1a-BiFC1 and OsRBD1-BiFC2 constructs and the lower side shows co-infiltration of BiFC1 and BiFC2 empty vectors. (B) Onion-peel bombardment assay to confirm in vivo interaction between OsRBD1-BiFC1 and OsSRO1a-BiFC2.
To reconfirm in vivo OsSRO1a-OsRBD1 interactions, OsSRO1a-BiFC1 and OsRBD1-BiFC2 constructs were also transformed in onion peel epidermal cells. When viewed under fluorescence microscope, bombarded peels containing both the proteins showed yellow fluorescence exclusively in nucleus while no fluorescence was detected where empty vectors were bombarded (Figure 4B).
Expression Analysis of OsSRO1a and OsRBD1 in Response to Different Abiotic Stresses
To study the relative expression levels of OsSRO1a in response to various stresses, quantitative real-time (qRT) PCR analysis was performed in shoots of rice seedlings (Cultivar IR64). OsSRO1a was found to be differentially regulated under stress (Figure 5A). OsSRO1a expression increased threefold in response to methylglyoxal (MG) treatment (a cytotoxic metabolite whose levels rise in response to abiotic stress) throughout the time course of stress treatments and about twofold up-regulation was observed in response to oxidative and salinity stress. Further, wounding, cold and desiccation stress led to ∼1.5-fold increase in OsSRO1a expression. However, OsSRO1a was down-regulated throughout following heat stress treatment (Figure 5A).
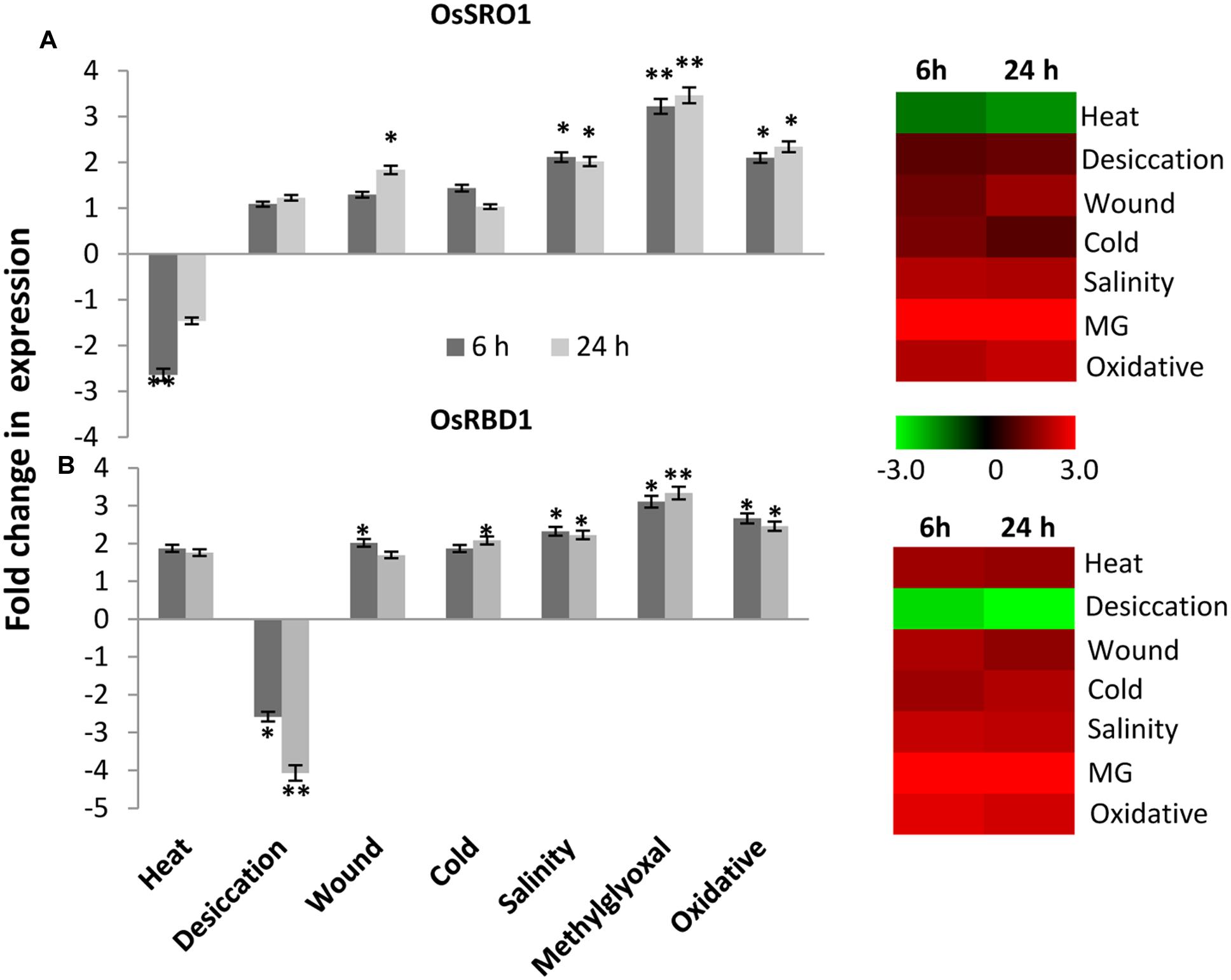
FIGURE 5. Expression analysis of (A) OsSRO1a and (B) OsRBD1 in response to various abiotic stresses. Analysis was done using cDNA prepared from shoot tissues of 12 days-old seedlings at control conditions or seedlings subjected to heat (42°C), desiccation (air dry), wound (mechanical injury), cold (4°C), salinity (200 mM NaCl), methylglyoxal (5 mM MG) and oxidative stress (5 mM H2O2) for 6 h and 24 h. Corresponding heatmaps were generated using TIGR MeV software package. Color bar represents relative expression values; thereby, green color represents lower expression levels, black represents no change, and red signifies higher expression level. Three replicates of each sample were used. Statistically significant change in transcript levels is indicated (∗P < 0.05; ∗∗P < 0.001).
Stress-induced expression profile of OsRBD1 was also examined in order to explore the significance of OsSRO1a-OsRBD1 interactions in stress. For this, qRT-PCR was performed using same set of cDNA as used for OsSRO1a expression profiling. Upon comparison, a co-regulation in OsSRO1a and OsRBD1 expression was observed in response to various stress treatments. Like OsSRO1a, OsRBD1 was also found to be up-regulated under most of the stress treatments (Figure 5B). About threefold increase in OsRBD1 levels was observed in response to MG treatment while oxidative stress led to ∼2 fold up-regulation in its expression. In addition, a positive co-regulation in OsSRO1a and OsRBD1 expression was also observed in response to wounding and salinity stress. However, unlike OsSRO1a which showed reduced expression on exposure to high temperature, OsRBD1 expression increased under such condition. In response to desiccation stress, OsRBD1 transcript levels declined at both 6 h and 24 h (Figure 5B).
OsSRO1a-OsRBD1 Interaction Imparts Stress Tolerance in Yeast
To explore the significance of OsSRO1a-OsRBD1 interactions in stress response, OsSRO1a and OsRBD1 cDNA were cloned in pYES2 vector and transformed, either individually or together, in yeast BY4741 strain. Empty vector (pYES2) was used as control. The positive colonies were confirmed by colony PCR and spotted following serial dilution on solid YPD-agar medium or YPD-agar medium supplemented with stress inducing agents (Figures 6A–F). Growth pattern was also compared following streaking the constructs on control and stress media. It was observed that co-transformed constructs containing both OsSRO1a and OsRBD1 genes could grow better in response to salinity and MG treatment as compared to the empty vector or separately transformed OsSRO1a and OsRBD1 constructs (Figures 6C,F). However, growth of interacting clones was not significantly different from the individually transformed OsSRO1a and OsRBD1 constructs in case of osmotic stress (Figure 6E) but severally hampered in case of oxidative stress (Figure 6D), though single clones or empty vector could survive well under similar conditions.
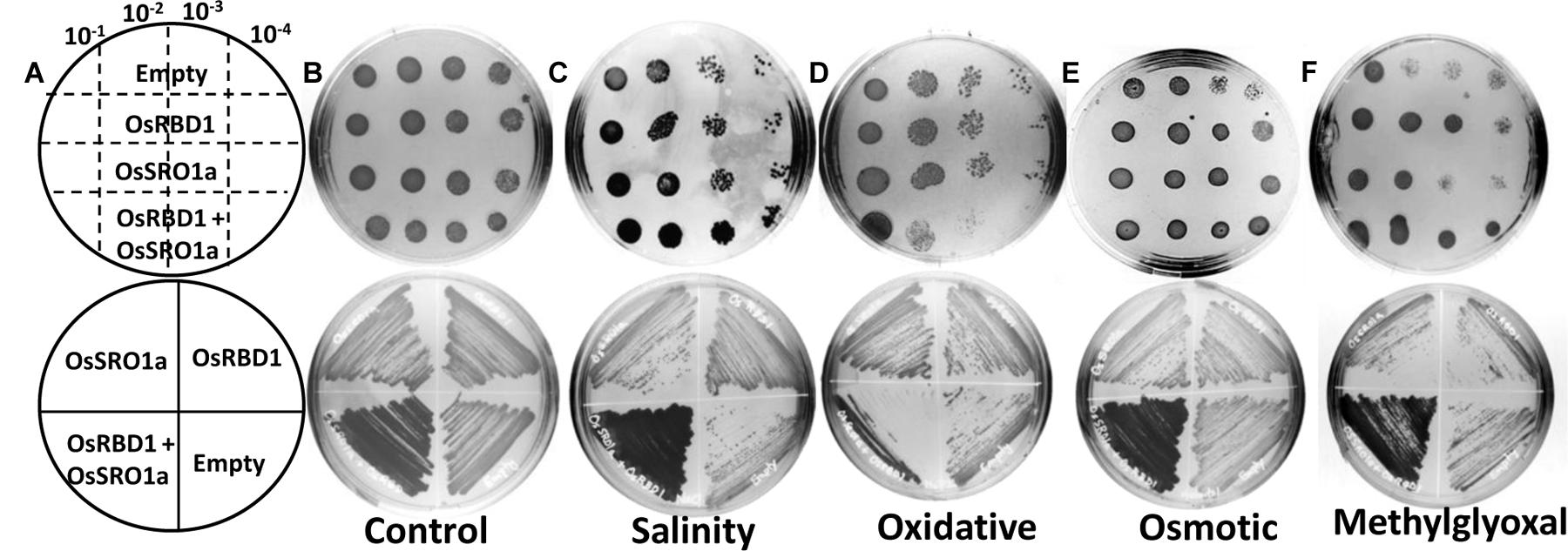
FIGURE 6. Interaction between OsSRO1a and OsRBD1 provides stress tolerance to Saccharomyces cerevisiae. (A) Schematic depiction of various constructs used for spotting serial dilutions (top panel) and streaking (bottom panel) assays. Comparison of growth and stress tolerance of individually transformed OsSRO1a and OsRBD1 constructs and co-transformed cells containing both the constructs under (B) non-stress (control) conditions, (C) salinity (1.2 M NaCl), (D) oxidative (10 mM H2O2), (E) osmotic (1 M mannitol), and (F) methylglyoxal (8 mM) treatments. Empty vector was also transformed and used as control. Top panel in each case represents spotting of serially diluted cultures of the constructs used in the study and bottom panel shows growth after streaking the same constructs. Three clones of each construct were used and the experiment was repeated thrice.
Discussion
SRO proteins regulate abiotic stress-related and developmental responses by interacting with various transcription factors. In the present study, we report a novel interacting partner of OsSRO1a protein, i.e., an RNA binding domain-containing protein OsRBD1 from rice, which interacts with OsSRO1a through its RRM. OsSRO1a, in addition to poly (ADP-ribose) polymerase domain (PARP), possesses a conserved C-terminal RST domain through which all interactions are mediated; and a WWE domain, present in several RCD1 and SRO proteins. The combination of PARP-RST domain is specific to plants and RST domain-bearing C-terminus sequence of SRO proteins is suggested to be critical for interaction with several, mostly plant specific transcription factors (Jaspers et al., 2009). As reported by various groups, our results also suggest several interacting partners of OsSRO1a in rice, most of which are either transcription factors or related to stress response such as, universal stress domain containing protein (USPs), which is known to play a key role in ethylene mediated stress response in rice (Sauter et al., 2002); a cold acclimation protein WCOR413 involved in low temperature stress (Allard et al., 1998); ubiquitin conjugating enzymes involved in ubiquitination, and many more.
Among several interacting partners of OsSRO1a, we studied detailed interaction with an RNA binding domain-containing protein, OsRBD1, being predominantly present among all the colonies screened. We found that both OsSRO1a and OsRBD1 were nucleus-localized and showed interaction in nucleus. OsRBD1 shows 86% amino acid sequence similarity to ALY2 (AT5G02530) from Arabidopsis (Uhrig et al., 2004) and about 46% similarity with YRA1 from yeast (Sträßer and Hurt, 2000), which are export factors playing a key role in splicing-coupled RNA transport from the nucleus and also in transcriptional activation. The RBD proteins, in general, comprise a large family in rice and are recognized as key regulatory factors in the post-transcriptional regulation of gene expression in eukaryotes (Li et al., 2014), and are also found to be actively involved in stress response as well as in development and growth of plants (Teotia and Lamb, 2009). In fact, regulatory mechanisms such as RNA synthesis, processing, transport, translation, storage, stability, and degradation are emerging as important processes involved in the manipulation of cellular responses to stress (Ambrosone et al., 2012).
Our results suggest that OsSRO1a may interact through its RST domain with the RRM1 domain of OsRBD1. Previously, it has been shown that RRM domain of ALY2, the closest Arabidopsis homolog of OsRBD1 is central for its interaction with P19 protein of tomato bushy stunt virus in yeast (Uhrig et al., 2004). RRM1 domains possess both RNA and protein binding abilities, and thus, binding of OsSRO1a at the RRM1 domain may alter its RNA binding capacity or its interaction with other proteins, thereby contributing to a mode of regulation of OsRBD1 function. Although function of RNA binding proteins (RBPs) is not characterized in detail, it has been suggested that some members may play a role in stress response, as their mRNA levels are reported to increase following exposure to various stresses (Ambrosone et al., 2012). For instance, OsDEG10 predicted to encode a small RBP, is strongly induced under several abiotic stress treatments such as, high light, anoxia, salinity, ABA, methyl viologen, and cold; and OsDEG10 RNAi transgenic plants are more sensitive to high light and cold stresses as compared to wild type plants (Park et al., 2009). Further, an RBP AtRGGA from Arabidopsis has also been shown to regulate tolerance to salt and drought stress (Ambrosone et al., 2015). The plants over-expressing AtRGGA gene were found to exhibit greater tolerance to ABA and salt stress on plates and in soil and accumulated lower levels of proline when exposed to drought stress. However, molecular mechanisms of how RBPs contribute to plant responses under different stresses are largely unknown.
In order to investigate the significance of OsSRO1a and OsRBD1 interaction in stress, we analyzed the transcriptional profile of both these genes in response to various abiotic stress factors. A positive correlation between OsSRO1a and OsRBD1 expression was seen in response to various stresses such as, low temperature, salinity, wounding, and oxidative stress along with methylglyoxal treatment. However, in case of high temperature and desiccation conditions, OsSRO1a and OsRBD1 expression was negatively correlated. SRO1 is known to function in various stress conditions either in a positive or negative fashion, thereby acting as a regulator of gene expression (Ahlfors et al., 2004; Fujibe et al., 2004; Katiyar-Agarwal et al., 2006; Vainonen et al., 2012). In Arabidopsis, RCD1 protein levels are reported to reduce significantly after 30 min of heat shock, whereas its interacting partner DREB2A which is known to be involved in transient and rapid response to heat stress but gradual response to salt and drought stress, was found to be stabilized after 1 h of heat shock. This opposite nature of protein accumulation suggests a negative impact of RCD1–DREB2A interaction, implying that RCD1 may be involved in targeting DREB2A for degradation (Vainonen et al., 2012). Hence, a similar regulation is possible even in case of OsSRO1a and OsRBD1 interaction as suggested by a negative correlation in their expression levels under heat stress.
Though role of OsRBD1 and its counterparts in other organisms has not been earlier investigated in stress response, yet it has been clearly demonstrated that loss of YRA1, an yeast homolog of OsRBD1 gene results in impaired nuclear poly(A)+ RNA export at restrictive growth conditions, with YRA1 null mutants being non-viable (Sträßer and Hurt, 2000). Yra1p, an intranuclear protein contains an RRM domain and shows in vitro RNA–RNA annealing activity. In addition, it is able to directly bind Mex67p, a nuclear mRNA export factor and is considered to be essential for nuclear export of mRNA. Further, YRA1 is known to interact with several other proteins involved in RNA metabolism, some of which are mutually exclusive interactions (Kelly and Corbett, 2009). In view of its role in other organisms, we can speculate that increased OsRBD1 expression under stress and its interaction with multi-stress inducible OsSRO1a at the central RRM1 domain may selectively regulate OsRBD1 protein’s RNA binding ability and hence, nuclear RNA stability and export, or may even modify its transcriptional co-activation ability, thereby regulating protein synthesis in the long run. But this hypothesis needs to be experimentally validated.
In order to investigate the significance of such interaction and its effect on stress response, we checked the growth pattern of yeast strains transformed with OsSRO1a and OsRBD1 under various stresses. The results suggested enhanced tolerance of yeast cells co-transformed with OsSRO1a and OsRBD1 to various stresses such as, salinity and methylglyoxal stress. It has been earlier shown that OsSRO1 and RBDs play a crucial role in salt tolerance (Katiyar-Agarwal et al., 2006; Ambrosone et al., 2015). Further, in response to osmotic stress, the growth of yeast cells co-transformed with OsSRO1a and OsRBD1 was not found to be significantly better than the individually transformed OsSRO1 or OsRBD1 or empty vector. Few reports on RBDs however, suggest that these proteins are not actively involved in response to osmotic stress conditions (Kim et al., 2007a,b). For instance, an RNA binding domain-containing protein, AtGRP2 from Arabidopsis, has been shown not to affect seed germination under osmotic stress but accelerates seed germination and seedling growth under cold stress (Kim et al., 2007a). In fact, loss of both rcd1 and sro1 in Arabidopsis results in increased resistance to osmotic stress (Teotia and Lamb, 2009). Here, we could see that though the growth of OsSRO1a over-expressing yeast cells was not hampered but even its interaction with OsRBD1 did not result in any growth difference. Furthermore, oxidative stress resulted in a negative response that is, decreased stress tolerance of the co-transformed yeast strain despite a positive correlation in expression between the two interacting partners in oxidative stress, thereby depicting differential regulatory effect of OsSRO1a binding at the RRM1 domain of OsRBD1. The reduced oxidative stress tolerance of OsSRO1a upon interaction with OsRBD1 finds a support from the existing literature. For instance, a co-dominant rcd1 mutant has been shown to accumulate superoxide ions and transient spreading lesions in response to ozone and superoxides but not H2O2 (Overmyer et al., 2000). Hydrogen peroxide at concentration as high as 10 mM was found to be ineffective in inducing cell death in the rcd1 mutant (Overmyer et al., 2000). On similar lines, it has been also shown that rcd1 mutant of Arabidopsis is more resistant to methyl viologen but very sensitive to ozone as compared to wild type (Fujibe et al., 2006), suggesting that RCD1 shows a differential response to different elicitors of oxidative stress response. Further, OsSRO1c-overexpressing rice has been found to be highly sensitive to oxidative stress but shows enhanced tolerance toward drought conditions since it promotes H2O2-mediated stomatal closure (You et al., 2013). Though role of SRO proteins has been described in different stresses, such as salinity (Katiyar-Agarwal et al., 2006), dehydration (Vainonen et al., 2012), and oxidative stress (Fujibe et al., 2004; Jaspers et al., 2009), this study might demonstrate another aspect of SRO-mediated regulation by affecting mRNA stability through its interaction with OsRBD1, a close homolog of RNA export factors (Uhrig et al., 2004).
Taken together, this study describes a novel interacting partner of SRO proteins, i.e., OsRBD1, which interacts through its RRM1 motif with the RST domain of OsSRO1a. Further, we have shown that this interaction has implications in stress response in yeast. However, detailed studies are needed to functionally validate such role in plants.
Author Contributions
SS and CK performed the experiments and wrote the manuscript. SLS-P and SKS conceived the idea, designed the experiments and edited the manuscript. All the authors approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by internal grants of International Centre for Genetic Engineering and Biotechnology (ICGEB), India and Indo-Finnish project from the Department of Biotechnology (DBT), Government of India. Authors thank Dr. Subhasis Samanta for providing yeast library of rice.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00062
Footnotes
- ^http://rice.plantbiology.msu.edu/index.shtml
- ^http://www.ncbi.nlm.nih.gov/geo/
- ^http://pfam.xfam.org/
- ^http://www.ricearray.org/
- ^http://bioinfo.ggc.org/bindn/
References
Ahlfors, R., Lång, S., Overmyer, K., Jaspers, P., Brosché, M., Tauriainen, A., et al. (2004). Arabidopsis radical-induced Cell Death1 belongs to the WWE protein-protein interaction domain protein family and modulates abscisic acid, ethylene, and methyl jasmonate responses. Plant Cell 16, 1925–1937. doi: 10.1105/tpc.021832
Allard, F., Houde, M., Kr, M., Ivanov, A., Huner, N. P. A., and Sarhan, F. (1998). Betaine improves freezing tolerance in wheat. Plant Cell Physiol. 39, 1194–1202. doi: 10.1093/oxfordjournals.pcp.a029320
Ambrosone, A., Batelli, G., Nurcato, R., Aurilia, V., Punzo, P., Bangarusamy, D. K., et al. (2015). The Arabidopsis RNA-binding protein AtRGGA regulates tolerance to salt and drought stress. Plant Physiol. 168, 292–306. doi: 10.1104/pp.114.255802
Ambrosone, A., Costa, A., Leone, A., and Grillo, S. (2012). Beyond transcription: RNA-binding proteins as emerging regulators of plant response to environmental constraints. Plant Sci. 182, 12–18. doi: 10.1016/j.plantsci.2011.02.004
Aravind, L. (2001). The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem. Sci. 26, 273–275. doi: 10.1016/S0968-0004(01)01787-X
Belles-Boix, E., Babiychuk, E., Van Montagu, M., Inzé, D., and Kushnir, S. (2000). CEO1, a new protein from Arabidopsis thaliana, protects yeast against oxidative damage. FEBS Lett. 482, 19–24. doi: 10.1016/S0014-5793(00)02016-0
Brosché, M., Blomster, T., Salojärvi, J., Cui, F., Sipari, N., Leppälä, J., et al. (2014). Transcriptomics and functional genomics of ROS-Induced cell death regulation by RADICAL-INDUCED CELL DEATH1. PLoS Genet. 10:e1004112. doi: 10.1371/journal.pgen.1004112
Fujibe, T., Saji, H., Arakawa, K., Yabe, N., Takeuchi, Y., and Yamamoto, K. T. (2004). A methyl viologen-resistant mutant of Arabidopsis, which is allelic to ozone-sensitive SRO1a, is tolerant to supplemental ultraviolet-b irradiation. Plant Physiol. 13, 275–285. doi: 10.1104/pp.103.033480
Fujibe, T., Saji, H., Watahiki, M. K., and Yamamoto, K. T. (2006). Overexpression of the RADICAL-INDUCED CELL DEATH1 (RCD1) gene of Arabidopsis causes weak rcd1 phenotype with compromised oxidative-stress responses. Biosci. Biotechnol. Biochem. 70, 1827–1831. doi: 10.1271/bbb.50673
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404. doi: 10.1105/tpc.12.3.393
Hassa, P. O., and Hottiger, M. O. (2008). The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front. Biosci 13:3046–3082. doi: 10.2741/2909
Hiltscher, H., Rudnik, R., Shaikhali, J., Heiber, I., Mellenthin, M., Meirelles Duarte, I., et al. (2014). The radical induced cell death protein 1 (RCD1) supports transcriptional activation of genes for chloroplast antioxidant enzymes. Front. Plant Sci. 5:475. doi: 10.3389/fpls.2014.00475
Jaspers, P., Blomster, T., Brosche, M., Saloja, J., Ahlfors, R., Vainonen, J. P., et al. (2009). Radical-induced cell death1 (SRO1a) is an important regulator of stress and hormonal and developmental responses in Arabidopsis thaliana. Plant J. 60, 268–279. doi: 10.1111/j.1365-313X.2009.03951.x
Jaspers, P., Overmyer, K., Wrzaczek, M., Vainonen, J. P., Blomster, T., Salojarvi, J., et al. (2010). The RST and PARP-like domain containing SRO protein family: analysis of protein structure, function and conservation in land plants. BMC Genomics 11:170. doi: 10.1186/1471-2164-11-170
Katiyar-Agarwal, S., Zhu, J., Kim, K., Agarwal, M., Fu, X., Huang, A., et al. (2006). The plasma membrane Na+/H+ antiporter SOS1 interacts with SRO1A and functions in oxidative stress tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 18816–18821. doi: 10.1073/pnas.0604711103
Kaur, C., Mustafiz, A., Sarkar, A., Ariyadasa, T. U., Singla-Pareek, S. L., and Sopory, S. K. (2014). Expression of abiotic stress inducible ETHE1-like protein from rice is higher in roots and is regulated by calcium. Physiol. Plant. 152, 1–16. doi: 10.1111/ppl.12147
Kelly, S. M., and Corbett, A. H. (2009). Messenger RNA export from the nucleus: a series of molecular wardrobe changes. Traffic 10, 1199–1208. doi: 10.1111/j.1600-0854.2009.00944.x
Kim, Y., Park, S. J., Jang, B., Jung, C. H., Ahn, S. J., Goh, C. H., et al. (2007a). Functional characterization of a glycine-rich RNA-binding protein 2 in Arabidopsis thaliana under abiotic stress conditions. Plant J. 50, 439–451. doi: 10.1111/j.1365-313X.2007.03057.x
Kim, Y. O., Pan, S. O., Jung, C. H., and Kang, H. (2007b). A zinc finger-containing glycine-rich RNA-binding protein, atRZ-1a, has a negative impact on seed germination and seedling growth of Arabidopsis thaliana under salt or drought stress conditions. Plant Cell Physiol. 48, 1170–1181. doi: 10.1093/pcp/pcm087
Kumar, R., Mustafiz, A., Sahoo, K. K., Sharma, V., Samanta, S., Sopory, S. K., et al. (2012). Functional screening of cDNA library from a salt tolerant rice genotype Pokkali identifies mannose-1-phosphate guanyl transferase gene (OsMPG1) as a key member of salinity stress response. Plant Mol. Biol. 79, 555–568. doi: 10.1007/s11103-012-9928-8
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Li, D., Zhang, H., Hong, Y., Huang, L., Li, X., Zhang, Y., et al. (2014). Biochemical characterization and expression expression analyses of the YTH domain-containing RNA-binding protein family in Arabidopsis and rice. Plant Mol. Biol. Rep. 32, 1169–1186. doi: 10.1007/s11105-014-0724-2
Lippuner, V., Cyert, M. S., and Gasser, C. S. (1996). Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J. Biol. Chem. 271, 12859–12866. doi: 10.1074/jbc.271.22.12859
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mustafiz, A., Singh, A. K., Pareek, A., Sopory, S. K., and Singla-Pareek, S. L. (2011). Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genomics 11, 293–305. doi: 10.1007/s10142-010-0203-2
Orkin, S. H. (1996). Development of the hematopoietic system. Curr. Opin. Genet. Dev. 6, 597–602. doi: 10.1016/S0959-437X(96)80089-X
Overmyer, K., Tuominen, H., Kettunen, R., Betz, C., Langebartels, C., Sandermann, H. Jr., et al. (2000). The ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12, 1849–1862. doi: 10.1105/tpc.12.10.1849
Park, H. Y., Kang, I. S., Han, J. S., Lee, C. H., An, G., and Moon, Y. H. (2009). OsDEG10 encoding a small RNA-binding protein is involved in abiotic stress signaling. Biochem. Biophys. Res. Commun. 380, 597–602. doi: 10.1016/j.bbrc.2009.01.131
Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. doi: 10.1016/0092-8674(95)90288-0
Sauter, M., Rzewuski, G., Marwedel, T., and Lorbiecke, R. (2002). The novel ethylene-regulated gene OsUsp1from rice encodes a member of a plant protein family related to prokaryotic universal stress proteins. J. Exp. Bot. 53, 2325–2331. doi: 10.1093/jxb/erf096
Sheen, J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127, 1466–1475. doi: 10.1104/pp.010820
Simon, P. (2003). Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 19, 1439–1440. doi: 10.1093/bioinformatics/btg157
Sträßer, K., and Hurt, E. (2000). Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 19, 410–420. doi: 10.1093/emboj/19.3.410
Teotia, S., and Lamb, R. S. (2009). The paralogous genes radical-induced cell death1 and similar to SRO1a have partially redundant functions during Arabidopsis development. Plant Physiol. 151, 180–198. doi: 10.1104/pp.109.142786
Uhrig, J. F., Canto, T., Marshall, D., and MacFarlane, S. A. (2004). Relocalization of nuclear ALY proteins to the cytoplasm by the tomato bushy stunt virus P19 pathogenicity protein. Plant Physiol. 135, 2411–2423. doi: 10.1104/pp.104.046086
Vainonen, J. P., Jaspers, P., Wrzaczek, M., Lamminmaki, A., Reddy, R. A., Vaahtera, L., et al. (2012). SRO1A-DREB2A interaction in leaf senescence and stress responses in Arabidopsis thaliana. Biochem. J. 442, 573–581. doi: 10.1042/BJ20111739
Yoshida, S., Forno, D. A., Cock, J. H., and Gomez, K. A. (1972). Laboratory Manual for Physiological Studies of Rice. Manila: International Rice Research Institute.
You, J., Zong, W., Li, X., Ning, J., Hu, H., Li, X., et al. (2013). The SNAC1-targeted gene OsSRO1c modulates stomatal closure and oxidative stress tolerance by regulating hydrogen peroxide in rice. J. Exp. Bot. 64, 569–583. doi: 10.1093/jxb/ers349
Keywords: abiotic stress, radical induced cell death, rice, RNA binding domain-containing protein, similar to RCD1
Citation: Sharma S, Kaur C, Singla-Pareek SL and Sopory SK (2016) OsSRO1a Interacts with RNA Binding Domain-Containing Protein (OsRBD1) and Functions in Abiotic Stress Tolerance in Yeast. Front. Plant Sci. 7:62. doi: 10.3389/fpls.2016.00062
Received: 21 October 2015; Accepted: 14 January 2016;
Published: 03 February 2016.
Edited by:
Oscar Vicente, Universitat Politécnica de Valéncia, SpainReviewed by:
Avinash Mishra, CSIR-Central Salt & Marine Chemicals Research Institute, IndiaHao Peng, Washington State University, USA
Juan Francisco Jimenez Bremont, Instituto Potosino de Investigacion Cientifica y Tecnologica, Mexico
Copyright © 2016 Sharma, Kaur, Singla-Pareek and Sopory. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sneh L. Singla-Pareek, c25laEBpY2dlYi5yZXMuaW4=; Sudhir K. Sopory, c29wb3J5QGljZ2ViLnJlcy5pbg==
 Shweta Sharma
Shweta Sharma Charanpreet Kaur
Charanpreet Kaur Sneh L. Singla-Pareek
Sneh L. Singla-Pareek Sudhir K. Sopory*
Sudhir K. Sopory*